Abstract
An automated on-line isolation and fractionation system including controlling software was developed for selected nanosized biomacromolecules from human plasma by on-line coupled immunoaffinity chromatography-asymmetric flow field-flow fractionation (IAC-AsFlFFF). The on-line system was versatile, only different monoclonal antibodies, anti-apolipoprotein B-100, anti-CD9, or anti-CD61, were immobilized on monolithic disk columns for isolation of lipoproteins and extracellular vesicles (EVs). The platelet-derived CD61-positive EVs and CD9-positive EVs, isolated by IAC, were further fractionated by AsFlFFF to their size-based subpopulations (e.g., exomeres and exosomes) for further analysis. Field-emission scanning electron microscopy elucidated the morphology of the subpopulations, and 20 free amino acids and glucose in EV subpopulations were identified and quantified in the ng/mL range using hydrophilic interaction liquid chromatography-tandem mass spectrometry (HILIC-MS/MS). The study revealed that there were significant differences between EV origin and size-based subpopulations. The on-line coupled IAC-AsFlFFF system was successfully programmed for reliable execution of 10 sequential isolation and fractionation cycles (37–80 min per cycle) with minimal operator involvement, minimal sample losses, and contamination. The relative standard deviations (RSD) between the cycles for human plasma samples were 0.84–6.6%.
Human biomacromolecules are complex and structurally diverse. They are excellent biomarkers, needed for the recognition and detection of the early stages of diseases, to understand the pathogenesis of the disease, and to find the treatment solutions. Their fast and reliable isolation and purification are often challenging or even a bottleneck for their diagnostic and therapeutic applications.1 The challenges are caused by their stability requirements and unique nature, especially when isolated from human biofluids. New systems are needed to eliminate the problems, such as operator dependent errors,2 aggregation,3,4 oxidation, shear and mechanical stress, and contamination,5 present in the current methods and techniques, which are often also tedious and time consuming.
The study of subpopulations of lipoproteins and extracellular vesicles (EVs) has proven to be important for detection of different diseases6,7 (e.g., small dense low-density lipoprotein (LDL) particles are considered unfavorable for health8,9), or for finding a new extracellular vesicle subpopulation called exomere,10 whose effects on health are still unclear. EV is a general term that includes both ectosomal and endosomal EVs (exosomes). Their size and other biophysical characteristics are similar, and thus EVs and exosomes cannot be separated based on size or density alone.11 Therefore specific immunobased separation techniques are needed for future clinical diagnostic and therapeutic applications of exosomes,6,11,12 and especially the study of subpopulations of EVs has lately attracted great attention.6,10,13
Selective and high yield isolation of nanosized biomacromolecules (e.g., lipoproteins and EVs) from human biofluids, such as plasma, is a challenging task due to the complex nature of sample matrices. Immunoaffinity chromatography (IAC) with polymer-based monolithic columns has proven to be a valuable tool for selective isolation of these biomacromolecules.13,14 However, the need for the clarification of characteristics of heterogeneous subpopulations, and for the deep understanding of their effects on human health, calls for further fractionation and analysis. Asymmetrical flow field-flow fractionation (AsFlFFF) combined with light-scattering detectors has already demonstrated its importance in off-line fractionation of biomacromolecules,15,16 lipoproteins,3 and EVs by helping to shed light on the nature of their subpopulations.10,13 Furthermore, the on-line coupling of different techniques has offered extra advantages, such as ease of automation and high reproducibility, usually resulting in a short analysis time.2
In this work, a novel automated on-line isolation and fractionation IAC-AsFlFFF system was constructed for the isolation and fractionation of EVs, exosomes, exomeres, and apolipoprotein B-100 (apoB-100) containing lipoproteins from challenging human plasma. The system was equipped with multiple detectors, such as ultraviolet, multiangle light-scattering, dynamic light-scattering, and diode array detectors (UV, MALS, DLS, and DAD) to provide more information on the chemical and physical characteristics of biomacromolecules. Field-emission scanning electron microscopy (FESEM) gave morphology information of the fractionated EV subpopulations. Their surface charge was revealed by ζ potential measurements. The free amino acid and glucose composition of the isolated EV subpopulations was elucidated by hydrophilic interaction liquid chromatography–tandem mass spectrometry (HILIC-MS/MS), and these results were subjected to statistical analysis for obtaining extra information on EV subpopulations.
Experimental Section
Chemicals and Reagents
The details are given in the Supporting Information.
Instrumentation
The details are given in the Supporting Information.
Preparation of Solutions
Phosphate buffered saline (PBS) solution was prepared by dissolving a PBS tablet in 200 mL of MilliQ (MQ) water and filtered through a 0.2 μm membrane filter (Supor-200). Carbonate-bicarbonate solution (0.1 M, pH 11.3) was prepared by mixing Na2CO3 solution (90 mL of 0.1 M) with NaHCO3 solution (10 mL, 0.1 M). The pH of the solution was adjusted to 11.3 with 1 M NaOH. NH4OH (0.15 M, pH 11.5) was prepared by diluting with 1.13 mL of 25% ammonia solution to a final volume of 50 mL with MQ water. The HILIC-MS/MS mobile phase A was prepared by adding 1 mL of formic acid to acetonitrile (999 mL) and the mobile phase B was prepared by adding 1 mL of formic acid to MQ water (999 mL). All standards and internal standards were prepared in mobile phase B.
On-Line IAC-AsFlFFF System
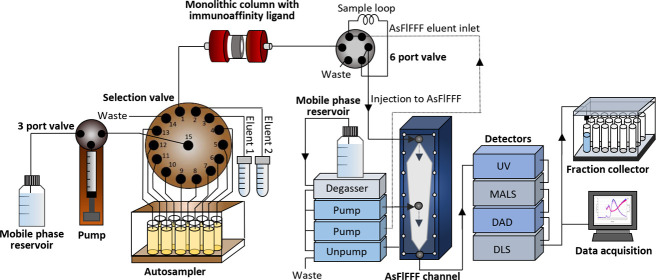
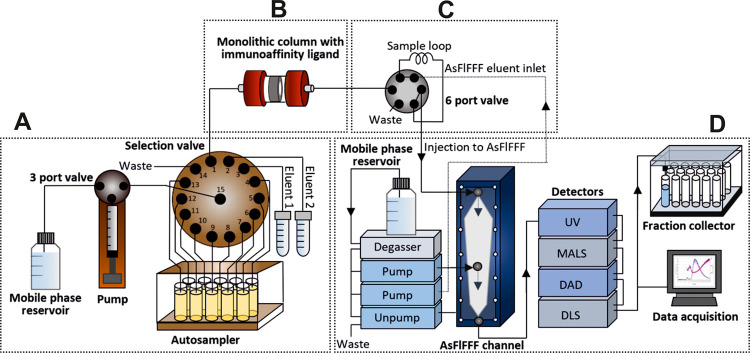
An on-line IAC-AsFlFFF system is depicted in Figure 1. In addition to a monolithic disk column and AsFlFFF, the system was composed of: (i) an in-house built pump with a 5 mL glass syringe driven by a stepping motor and a three-port valve with an electrical actuator for dispensing the solutions, (ii) a Cheminert C25Z-31814D (Vici AG, Schenkon, Switzerland) 14 position stream selection valve connected to an EMHMA-CE microelectric valve actuator (Vici AG, Schenkon, Switzerland) for selection of samples and eluents, and (iii) a six-port medium pressure injection valve V-451 (IDEX Upchurch Scientific, Oak Harbor, WA) connected to a Model E60 actuator (Vici AG, Schenkon, Switzerland) for the introduction of the sample to AsFlFFF. The setup was controlled by a Raspberry Pi (Model B Rev. 2.0) single board computer running on in-house written Python scripts on Raspbian (ver.joo ot 9 Stretch) operating system.
Figure 1.
Automated on-line system for the isolation of nanosized biomacromolecules. The system consisted of a selection valve for controlling the isolation process (A), a monolithic column for IAC (B), an automated six port valve for injection to AsFlFFF (C), and AsFlFFF with UV, MALS, DAD, and DLS detectors, and a fraction collector (D).
The selection valve was used for controlling the isolation process (Figure 1A), which included a 5 mL syringe pump and an autosampler for 10 samples. Process automation and control of the isolation process were performed using customized software. A three-port valve was used to connect the mobile phase reservoir to the syringe pump. The IAC based isolation included a monolithic column with immobilized immunoaffinity ligand (Figure 1B). The elution conditions to release the isolates from the column, immobilized with antibodies, were optimized in our previous studies.13,14 With an automated six port valve we connected monolithic column on-line to AsFlFFF (Figure 1C). The injection steps were controlled by a timer in our software that turned the valve and started the AsFlFFF run after the biomolecules entered the sample loop. This minimized the time under high pH conditions. AsFlFFF was utilized for characterization and provided valuable information on the size distribution and concentration of different subpopulations of the biomacromolecules with MALS, DLS, UV, and DAD detectors, and gentle fractionation of the isolates (Figure 1D). The fractions were collected with a fraction collector in a physiological buffer (PBS) for further analysis and characterization.
Scanning Electron Microscopy (SEM)
Preconcentrated combined fractions of EV subpopulations were dried on clean polished silicon wafer surfaces. The samples were then coated with a 3 nm Au–Pd alloy using a Cressington 208HR high resolution sputter coater and imaged at 3 kV with secondary electrons.
Hydrophilic Interaction Liquid Chromatography–Tandem Mass Spectrometry (HILIC-MS/MS)
Amino Acids and Glucose Extraction
CD9- and CD61-positive (CD9+ and CD61+) EV fractions were subjected to preconcentration and salt removal with disposable Nanosep centrifugal devices with 10 K molecular weight cutoff membrane filters at 14 000g for 2 min for each 500 μL fraction at room temperature. The filtrate was discarded. Thereafter, cold acetonitrile (50 μL) was added to the membrane for EV lysis and removal of precipitated proteins. Cold acetonitrile precipitated proteins effectively from human plasma17 and lysed the lipid bilayer membranes.18 After vortexing, the filter unit was centrifuged for another 2 min at 14 000g. The filtrate was then collected, and the ISTD mixture which yielded the final concentration of 1 ppm of amino acids (glycine-2,2-d2, l-phenylalanine-3,3-d2, and l-lysine-4,4,5,5-d4) and 5 ppm of d-fructose-13C6 in MQ water containing 0.1% formic acid was added to the filtrate for HILIC-MS/MS analysis.
Determination of Amino Acids and Glucose by HILIC-MS/MS
The method used for the determination of amino acids and glucose was based on our previously developed method19 with some modifications. The column temperature was set to 50 °C. Mobile phase A was acetonitrile with 0.1% formic acid, and mobile phase B was MQ water with 0.1% formic acid. The separation of the target analytes was performed using the following gradient program 20% B for 15 min (0.4 mL/min), 20–80% B for 5 min (0.3 mL/min), followed by 80–20% B for 3 min (0.3 mL/min). The injection volume was 3 μL for all samples. The effluent was electrosprayed, ionized (positive and negative mode for amino acids and sugars, respectively), and monitored by MS2 detection in the multiple reaction monitoring mode (MRM), with the exception of glucose, which was analyzed in the single ion monitoring mode. Ionization conditions and MRM parameters for different compounds are given in Table S1.
Total Protein and Total Cholesterol Analysis
Total cholesterol (free and esterified) concentrations in samples were measured using the Roche Cholesterol CHOD-PAP reagent (Kit no. 1489232; Roche, Germany) according to the manufacturer’s protocol. The absorbance was measured with an EnSpire multimode plate reader (PerkinElmer Inc.) at 510 nm. Total protein concentrations (concentration range: 0.1–1.0 mg/mL) were measured using a Bio-Rad DC Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA) based on the Lowry method20 at 750 nm and PierceTM BCA Protein Assay Kit (Item no. 23225; Thermofisher Scientific) (for concentrations lower than 0.1 mg/mL) based on bicinchoninic assay (BCA)21 at 550 nm according to the manufacturer’s protocols. Calibration curves and sample concentrations were calculated using EnSpire multilabel analyzer version 4.13.3005.1482.
Statistical Analysis
Different R 3.6.3 statistical analysis tools were used in this research. Skewness and Kurtosis tests for data distribution evaluation, principal component analysis (PCA) for visualization of differences between CD9+ and CD61+ EV subpopulations, and linear discriminant analysis (LDA) for statistical confirmation of these differences, including clarification of the variables involved in the process. Additional studies were also made to evaluate the potential differences between EVs of different sizes (PCA and LDA).22
In all of the cases, the concentrations of the free amino acids present in the EVs normalized by the total amount of protein were exploited as input variables for the development of the statistical models. Additional root square transformation was needed to provide normal data distribution of the input variables.
Results and Discussion
In this section, we describe the automated on-line IAC-AsFlFFF system and its application for the isolation and fractionation of nanosized biomacromolecules. Because in our previous studies13,14 the IAC method was developed for the isolation of LDL particles in plasma, we tested the applicability of the on-line IAC-AsFlFFF system first to lipoproteins. Then the study was focused on the isolation and fractionation of the subpopulations of CD9+ and CD61+ EVs, further characterized by FESEM and surface charge measurements. In addition, more in-depth vesicular free amino acid and glucose composition was clarified by HILIC-MS/MS.
Isolation and Fractionation of Human apoB-100 Containing Lipoproteins, and CD9+, CD61+ EVs by the On-line IAC-AsFlFFF System
Three different monolithic disk columns for the IAC were immobilized with anti-apoB-100, anti-CD9, and anti-CD61 according to our previous protocols.13,14 IAC process cycles (Figure S1) for the isolation of apoB-100 containing lipoproteins, CD9+, and CD61+ EVs can be found in Table S2. The repetition of the experiments is shown in Figure S1, where the regeneration and waiting periods were taken into account for the AsFlFFF to be ready for the next fractionation. The total time for apoB-100 lipoprotein isolation and regeneration of the disk column was 16.5 min (1 mL sample injection, 3 mL PBS wash, elution with 2 mL of NH4OH, and 3 mL PBS wash). Isolation and regeneration of the EV disks took 51 min. The major difference between the isolations was that EV isolations had a larger sample volume (5 mL) and an additional NH4OH regeneration step (Eluent 1, Figure 1A), and carbonate-bicarbonate solution (Eluent 2, Figure 1A) was used to elute the EVs. In our previous studies,14 we found that the carbonate-bicarbonate solution (pH 11.3) was able to elute LDL particles more specifically compared to NH4OH (pH 11.3) with minimal nonspecifically bound particles, and other possible protein contaminants. Therefore, the carbonate-bicarbonate solution was also used for selective elution of EVs in this study, while NH4OH was used for the elution of all apoB-100-containing lipoproteins from the anti-apoB-100 disk, and not only LDL. The flow rate for the apoB-100 containing lipoprotein isolation and elution was set to 0.5 mL/min, whereas it was 0.25 mL/min for the EV isolation. The regeneration step for EV disks before the analysis of the next sample was performed with 2 mL of NH4OH and 3 mL of PBS at a flow rate of 1 mL/min. No additional regeneration step was needed for the apoB-100 disk, since NH4OH was already used for elution. AsFlFFF run was automatically started after the eluent of IAC filled the sample loop (500 μL) in the six port valve. The injection flow of 0.1 mL/min was applied over 5 min during the focus mode at a cross-flow rate of 3 mL/min. PBS was used as an eluent in the AsFlFFF. The detector flow rate was set to 0.5 mL/min. A transition time of 1 min followed the focusing step. Biomolecules were subjected to high pH for only a few minutes before the buffer of the eluate was replaced with PBS.
Separation and fractionation with AsFlFFF of apoB-100 containing lipoproteins was achieved with 2 min linear decrease in cross-flow to 0.5 mL/min, followed by a linear decrease over 1–0 mL/min. After the cross-flow reached 0 mL/min, only the detector flow was applied for 15 min. The total run time was 24 min and the fraction collection needed extra 2 min.
Separation and fractionation with AsFlFFF of CD9+ and CD61+ EVs (including their subpopulations of the size range of exosomes and exomeres) were carried out with 5 min linear decrease in cross-flow to 1.0 mL/min after the focusing step, followed by a linear decrease over 15–0 mL/min. After the cross-flow reached 0 mL/min, only the detector flow was applied for 14 min. The total run time was 40 min and extra 2 min was added for the fraction collector. The optimal AsFlFFF conditions are given in Table S3.
Optimization of the Six Port Valve Connection from IAC to AsFlFFF
The six port valve timer of the system for transferring the sample to AsFlFFF from the IAC (anti-apoB-100 monolithic disk column) was optimized with 1 mL of 100 μg/mL LDL samples (n = 25, isolated by ultracentrifugation). The biomolecules were eluted into the loop (500 μL) of the six port valve, and after the transfer, the AsFlFFF run was started with the signal of the timer. The time which assured the highest sample concentrations (highest UV peak areas at 280 nm) was selected (Figure S2) for all of the following experiments. The UV peak areas showed repeatability of 0.3–6.6% for each selected valve time.
Isolation and Fractionation of apoB-100 Containing Lipoproteins by IAC-AsFlFFF
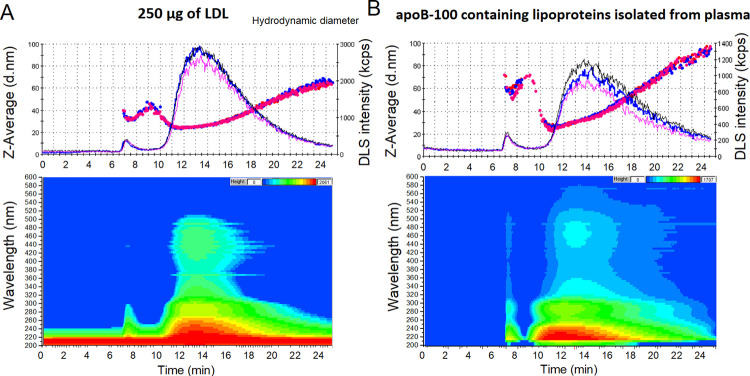
To study the recovery of the system, three LDL samples of 1 mL with concentration of 250 μg/mL (isolated from human plasma by ultracentrifugation, diluted with PBS to the required concentration) were captured and fractionated by IAC-AsFlFFF. The recovery was 99.6% based on DC protein assay and the samples contained 0.43 ± 0.01 mg of cholesterol. In addition, raw flow DLS data showed good repeatability of the isolation and fractionation (Figure 2A), while the RSD of the UV peak areas was 6.6%. The hydrodynamic diameter (24–28 nm) of LDL particles at a retention time of 10–15 min agreed well the size range reported in the literature.8 Isoabsorbance plots of the DAD detector also allowed the detection of carotenoids (11–18 min) in the LDL core (430–500 nm).4 Most of the carotenoids were found at 12–15 min, where most of the LDLs were retained. The tail of the DLS peak (Figure 2A) with a bigger size corresponds probably to fused LDL particles and LDL aggregates found in the sample.
Figure 2.
IAC-AsFlFFF analysis profiles after fractionating the anti-apoB-100 monolithic disk isolates. Technical replicates (n = 3) of raw flow DLS data, hydrodynamic diameter (dots as Z-Average) on the top, and isoabsorbance plot of selected run in the bottom, of preisolated LDLs (A) and apoB-100 containing lipoproteins isolated from human plasma (B).
The system was also used to study human plasma samples (1 mL, dilution factor 1:10 in PBS) to verify the applicability of the system, giving 0.84% (n = 3) for the RSD of UV peak areas. Not only the DLS and DAD analysis profiles resembled those of pure LDL, but also bigger sized particles were detected (Figure 2B), since the isolation with anti-apoB-100 monolithic disk was based on the recognition of the epitope of the apoB-100. Chylomicrons, very-low-density lipoprotein (VLDL) particles, and their remnants (intermediate-density lipoprotein (IDL) particles) were also captured by the anti-apoB-100 disk, as confirmed by the hydrodynamic diameter of 95 nm obtained from the DLS data. The results agreed with our previous experiments.14 The IAC also trapped small LDL particles of under 24 nm (retention time 11–12 min, Figure 2B), which were not present in our ultracentrifugation method for LDL isolation described in Ref 23. The on-line system could successfully capture all major lipoproteins,8 except HDL, that went through the monolithic column. The carotenoids were also found in the plasma isolates (at retention time 11–22 min), with the highest concentration at a retention time of 13 min.
Isolation and Fractionation of CD9+ and CD61+ EVs by IAC-AsFlFFF
The system was utilized to study subpopulations of EVs. The CD61 antibody was used for platelet-derived EVs in the exosomal size range13 and the CD9 antibody was used for both EVs originating from multivesicular bodies (MVBs),23,24 as well as those CD9+ EVs not originating from MVBs, but having the size range of exosomes.6,25 Subpopulations were divided based on their sizes: <50 nm for the size range of exomeres, and 50–80 and 80–120 nm for the size range of exosomes.10,13,26 Due to the pore size of monolithic disk columns (1.3 μm in diameter), bigger sized EVs were excluded from being captured by the IAC (Figure 3A). Isoabsorbance plots indicated no contamination of the isolates from lipoproteins, since no carotenoids were detected by the DAD. The concentration of platelet-derived EVs in the exosomal size range was significantly lower compared to CD9+ EVs (Figure 3B). The system could successfully and reproducibly isolate and fractionate the EVs, and FESEM of combined fractions of the subpopulations (Figure 3C) agreed with hydrodynamic diameters obtained by DLS. The RSD of UV peak areas of CD9+ EVs was 2.9%, while it was 4.2% for CD61+ EVs. The EVs in the size range of exomeres (<50 nm) were collected by combining fractions at retention times of 16–26 min, 50–80 nm sized exosomes at 27–34 min, and 80–120 nm sized exosomes at 35–40 min. Surprisingly, the exomere sized EVs were also detected at relatively high concentrations, while they are reported10 to contain low levels of tetraspanin CD9 and integrins like CD61. The previously reported results on exomeres were, however, obtained from samples originating from cell lines and cell culture, whereas our IAC disks enriched exomere sized CD9+ and CD61+ EVs from all possible cells that secrete exomeres to plasma, thus collecting together all possible CD9+ and CD61+ exomeres. Even smaller sized particles than exomeres were also found at the retention time of 10–15 min in both cases (Figure 3B). The mean surface charges (ζ potential) of the isolated exomere and exosome sized EVs were negatively charged (Figure S3), also agreeing with previously reported results.10 The CD9+ < 50 nm EVs had a mean charge of −14.1 mV (n = 9 independent measurements for each technical replicate n = 3), −16.2 mV for 50–80 nm, and −16.9 mV for 80–120 nm subpopulations. While the CD61+ < 50 nm subpopulation had a mean charge of −15.1 and −14.2 mV for 50–80 nm, and −16.6 mV for 80–120 nm subpopulations.
Figure 3.
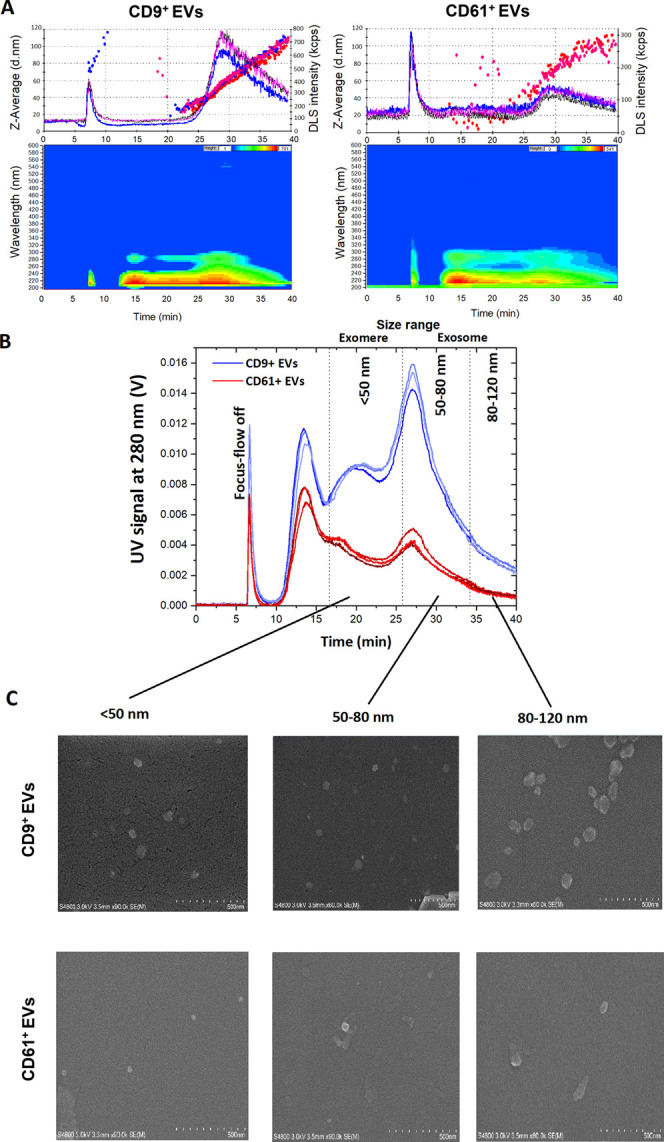
IAC-AsFlFFF analysis profiles after fractionating the CD9+ and CD61+ EV isolates. Technical replicates (n = 3) of raw flow DLS data, the hydrodynamic diameter (dots as Z-Average) on top, and isoabsorbance plot of selected run at the bottom (A). Overlaid UV spectra (280 nm) from the technical replicates (B), and FESEM morphology of fractionated subpopulations (C) for CD9+ and CD61+ EVs.
The IAC-AsFlFFF system resulted in faster and more selective isolation and fractionation for lipoproteins (38 samples/24 h) and EVs (18 samples/24 h) when compared to conventional standard methods, such as ultracentrifugation,8,27 with high repeatability, and minor sample loss and contamination. The IAC utilized high pH13,14 for the elution of biomacromolecules. However with on-line AsFlFFF the elution solution was immediately exchanged to PBS, avoiding long exposure to high pH and nonphysiological environment. The system was cost-effective and produced well-controlled final products needed especially in the EV-field.1 Additionally, it added significantly to the productivity of personnel and instruments, and quality of the data compared to our previous approaches for the isolation of same biomacromolecules.13,14,28
Amino Acid and Glucose Analysis of EV Subpopulations Isolated by On-line IAC-AsFlFFF
In plasma, amino acids are fundamentally involved in physiological activities, and their levels are used clinically for diagnostic purposes. EVs have also been found to carry amino acids along with other small metabolites.29 Therefore, the study of free amino acids found in EVs may elucidate their distinct biological roles and properties. In this study, we also developed an HILIC-MS/MS method for the analysis of free amino acids and sugars found in CD9+ and CD61+ EV subpopulations. Chromatograms with linearity and limit of detection of the method, and effect of the sample matrix on the results are found in the Supporting Information (Figures S4–S6, Tables S4 and S5).
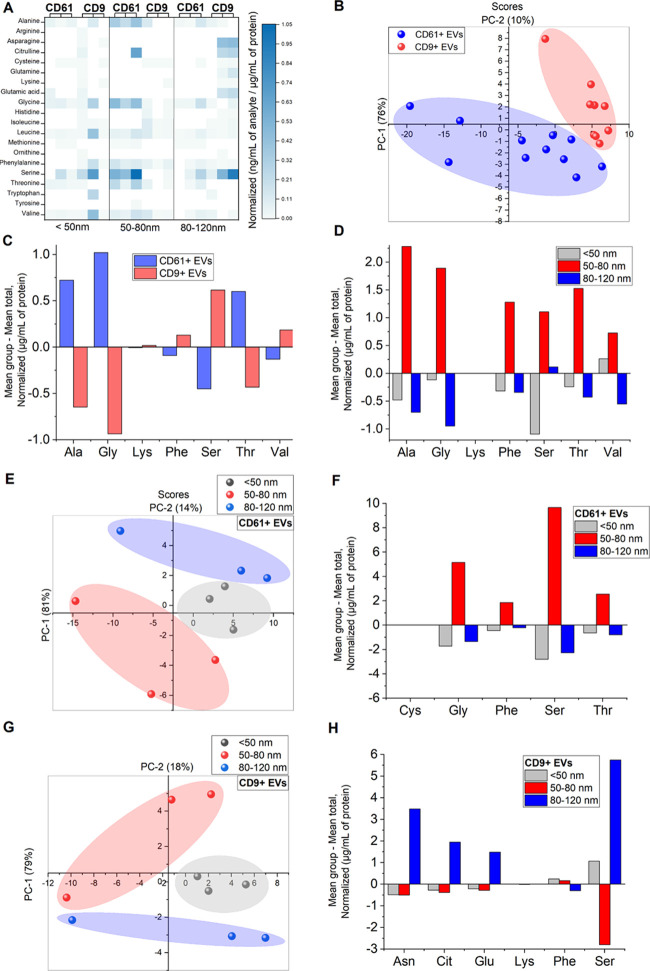
We found that both CD9+ and CD61+ EVs shared not only similarities but also differences in terms of free amino acid concentrations as displayed in Figure 4A. Among all free amino acids, Ser was found to be the most abundant. It is worth mentioning that Ser is a nonessential amino acid, and is an important component in the synthesis of membrane lipids, including sphingolipids and phosphatidylserine. Both lipids are enriched in exosomal membranes.30,31 It is thus possible that free Ser was incorporated inside the exosomes during exosome formation processes. In addition, CD61+ EV subpopulations of exosomal size range (50–80 nm) contained the highest concentrations of free amino acids, while for CD9+ EVs, the levels were the highest for the exosomal size range of 80–120 nm. The EVs < 50 nm (size range of exomeres) were least abundant in Ser, which indicates their nonmembranous nature as described by Zhang et al.10
Figure 4.
Multivariate analysis of amino acids found in CD61+ and CD9+ EV subpopulations. Heat map visualization of amino acids corresponding to their normalized concentrations (ng/mL per μg/mL of total protein) (A). Scoring plot of PCA analysis showing differences between CD61+ and CD9+ EVs (B). Discriminant analysis of differences between amino acids found in CD61+ and CD9+ EVs (C). Discriminant analysis of differences between amino acids found between different sizes of combined EV subpopulations (D). Scoring plot of PCA analysis revealing differences between subpopulations of different sizes of CD61+ EVs (E). Discriminant analysis of differences between amino acids found between CD61+ EV subpopulations (F). Scoring plot of PCA analysis revealing differences between subpopulations of different sizes of CD9+ EVs (G). Discriminant analysis of differences between amino acids found between CD9+ EV subpopulations (H).
To further visualize and clarify the differences between the two EV subpopulations, PCA and LDA analyses were used. The scoring plot obtained from PCA analysis (Figure 4B) revealed clear differences between CD9+ and CD61+ EVs. Based on the LDA results (Figure 4C), in which over 93% of samples were correctly classified, the differences between these two groups were majorly contributed by the following amino acids: Ala, Gly, Lys, Phe, Ser, Thr, and Val. The levels of Ala, Gly, and Thr, were significantly higher in CD61+ EVs compared to CD9+ EVs. Ala, Cys, Gly, Ser, and Thr are potentially involved in the biochemical pathway of gluconeogenesis from amino acids and can be directly converted to pyruvate.32 In addition, Ala and Gly play major roles in immune responses by inhibiting apoptosis,33 as well as serving as an anti-inflammatory and immunomodulatory agent.34 CD61+ EVs have originated from platelets, and these findings support the role of platelets in defense mechanism and inflammation.35 Interestingly, based on the Western blot analysis previously reported,13 part of the CD61+ EVs also contained CD9. Thus, the differences from statistical analyses of CD61+ and CD9+ EVs are most likely due to subpopulation sets (containing only CD61 or only CD9) that do not overlap. Moreover, based on subpopulations, regardless of the EV origin, 50–80 nm subpopulation (in the size range of small exosomes) contained the highest levels of Ala, Gly, Ser, and Thr (Figure 4D), indicating that EVs in the size range of small exosomes are more likely to take part in gluconeogenesis compared to other subpopulations.
The differences among EVs of < 50 nm for the size range of exomeres and 50–80 and 80–120 nm for the size range of exosomes for both CD9+ and CD61+ EVs were further investigated (Figure 4E–H). The PCA analysis suggested a clear distinction among the three subpopulations of different sizes (Figure 4E,G). The good classification of the samples by the LDA models (90 and 100% of the samples correctly classified for CD9+ and CD61+ subpopulations, respectively) confirmed these results. In CD61+ subpopulations, Cys, Gly, Phe, Ser, and Thr, contributed to the differences in the size. These free amino acids were dominant in the 50–80 nm subpopulation, indicating that among CD61+ EVs in the size range of small exosomes take part in gluconeogenesis. On the other hand, CD9+ EV subpopulations were different due to other free amino acids. Asn, Cit, Glu, Lys, Phe, and Ser contributed significantly in CD9+ EVs (Figure 4H). The levels of Asn, Cit, Glu, and Ser, were the highest in the 80–120 nm subpopulation of the large exosomal size range. Interestingly the levels of Ser were significantly higher in the 80–120 nm subpopulation compared to 50–80 nm subpopulation among CD9+ EVs. The differences also confirm distinct cellular origins of CD9+ and CD61+ EVs in the exosomal size range.
In addition to amino acids, glucose levels of EVs were measured. It was found that all subpopulations of CD61+ EVs contained glucose in the concentration range of pmol/mL. The presence of glucose can possibly be ascribed to the CD61+ EV fractions being rich in amino acids essential for gluconeogenesis. However, in CD9+ subpopulations only 50–80 nm subpopulation contained glucose. Importantly, these findings revealed that different free amino acids and monosaccharide concentrations were found in EV subpopulations isolated and fractionated by on-line IAC-AsFlFFF. These differences can be further utilized as key factors to differentiate the subpopulations of EVs (exomeres, and small and large exosomes) from different cellular origin, as well as to confirm distinct properties and functions of these particles in the human physiology. The functional activity of isolated and fractionated EVs still needs to be assessed. The results of this study demonstrated that on-line IAC-AsFlFFF can be utilized for biomarker and composition studies.
Conclusions
On-line coupled IAC-AsFlFFF allowed the reliable and fast isolation and fractionation of challenging biomacromolecules from human plasma, resulting in high purity subpopulations with high yields. The fully automated on-line system could process 18–38 samples in 24 h with only minor operator involvement. Due to a gentle fractionation step, apoB-100 containing lipoproteins, used in the testing step, and intact EV subpopulations under 120 nm were successfully obtained as confirmed by DLS and FESEM. The EV subpopulations were further exposed to particle surface charge as well as free amino acid and sugar composition studies. The results revealed that there were significant differences between cellular origin of EVs and even within subpopulations <120 nm. In addition to three different ligands employed in this study, the flexible system is applicable to any other ligand that can be immobilized on the monolithic disk column.
Acknowledgments
This study was supported by the Research Council for Natural Sciences and Engineering, Academy of Finland (grant number 1311369). The authors would like to thank Dr. Matti Jauhiainen, Jari Metso, and other personnel of Minerva Foundation Institute for Medical Research for providing plasma samples and for their support during the studies. Dr. Katariina Öörni from Wihuri Research Institute is thanked for providing lipoprotein samples. Shu Yi Fu is thanked for assisting with part of the experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c01986.
Chemicals and reagents, instrumentation, optimized multiple reaction monitoring parameters for amino acids and sugar, IAC-AsFlFFF cycles, process cycles used in the IAC, optimal AsFlFFF conditions, optimal time for six port valve timer, ζ potential of CD9+ and CD61+ EV subpopulations, extracted-ion chromatograms based on MRM of a standard mixture containing amino acids, glucose, internal standards, and 50–80 nm CD61+ EV subpopulation, total ion chromatograms of blank, calibration curves for amino acids and glucose, recovery percentages of target amino acids and glucose based on standard addition analyses (PDF)
Author Contributions
The manuscript was written through contributions of all authors.
The authors declare the following competing financial interest(s): Patent application.
Supplementary Material
References
- Colao I. L.; Corteling R.; Bracewell D.; Wall I. Manufacturing Exosomes: A Promising Therapeutic Platform. Trends Mol. Med. 2018, 24, 242–256. 10.1016/j.molmed.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Hyötyläinen T.; Riekkola M.-L. Approaches for On-Line Coupling of Extraction and Chromatography. Anal. Bioanal. Chem. 2004, 378, 1962–1981. 10.1007/s00216-004-2545-x. [DOI] [PubMed] [Google Scholar]
- Yohannes G.; Sneck M.; Varjo S. J. O.; Jussila M.; Wiedmer S. K.; Kovanen P. T.; Öörni K.; Riekkola M. L. Miniaturization of Asymmetrical Flow Field-Flow Fractionation and Application to Studies on Lipoprotein Aggregation and Fusion. Anal. Biochem. 2006, 354, 255–265. 10.1016/j.ab.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Lu M.; Gursky O. Aggregation and Fusion of Low-Density Lipoproteins in Vivo and in Vitro. Biomol. Concepts 2013, 4, 501–518. 10.1515/bmc-2013-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sódar B. W.; Kittel Á.; Pálóczi K.; Vukman K. V.; Osteikoetxea X.; Szabó-Taylor K.; Németh A.; Sperlágh B.; Baranyai T.; Giricz Z.; Wiener Z.; Turiák L.; Drahos L.; Pállinger É.; Vékey K.; Ferdinandy P.; Falus A.; Buzás E. I. Low-Density Lipoprotein Mimics Blood Plasma-Derived Exosomes and Microvesicles during Isolation and Detection. Sci. Rep. 2016, 6, 24316 10.1038/srep24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R.; LeBleu V. S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth P. Triglyceride-Rich Lipoproteins as a Causal Factor for Cardiovascular Disease. Vasc. Health Risk Manage. 2016, 171–183. 10.2147/VHRM.S104369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki M.; Usui S.; Hosaki S.. Handbook. Lipoprotein Test, 2000.
- Toft-Petersen A. P.; Tilsted H. H.; Aarøe J.; Rasmussen K.; Christensen T.; Griffin B. A.; Aardestrup I. V.; Andreasen A.; Schmidt E. B. Small Dense LDL Particles - A Predictor of Coronary Artery Disease Evaluated by Invasive and CT-Based Techniques: A Case-Control Study. Lipids Health Dis. 2011, 10, 21. 10.1186/1476-511X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Freitas D.; Kim H. S.; Fabijanic K.; Li Z.; Chen H.; Mark M. T.; Molina H.; Martin A. B.; Bojmar L.; Fang J.; Rampersaud S.; Hoshino A.; Matei I.; Kenific C. M.; Nakajima M.; Mutvei A. P.; Sansone P.; Buehring W.; Wang H.; Jimenez J. P.; Cohen-Gould L.; Paknejad N.; Brendel M.; Manova-Todorova K.; Magalhães A.; Ferreira J. A.; Osório H.; Silva A. M.; Massey A.; Cubillos-Ruiz J. R.; Galletti G.; Giannakakou P.; Cuervo A. M.; Blenis J.; Schwartz R.; Brady M. S.; Peinado H.; Bromberg J.; Matsui H.; Reis C. A.; Lyden D. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cell Biol. 2018, 20, 332–343. 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M.; Martin-Jaular L.; Lavieu G.; Théry C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- de Jong O. G.; Kooijmans S. A. A.; Murphy D. E.; Jiang L.; Evers M. J. W.; Sluijter J. P. G.; Vader P.; Schiffelers R. M. Drug Delivery with Extracellular Vesicles: From Imagination to Innovation. Acc. Chem. Res. 2019, 52, 1761–1770. 10.1021/acs.accounts.9b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multia E.; Tear C. J. Y.; Palviainen M.; Siljander P.; Riekkola M. L. Fast Isolation of Highly Specific Population of Platelet-Derived Extracellular Vesicles from Blood Plasma by Affinity Monolithic Column, Immobilized with Anti-Human CD61 Antibody. Anal. Chim. Acta 2019, 1091, 160–168. 10.1016/j.aca.2019.09.022. [DOI] [PubMed] [Google Scholar]
- Liangsupree T.; Multia E.; Metso J.; Jauhiainen M.; Forssén P.; Fornstedt T.; Öörni K.; Podgornik A.; Riekkola M. L. Rapid Affinity Chromatographic Isolation Method for LDL in Human Plasma by Immobilized Chondroitin-6-Sulfate and Anti-ApoB-100 Antibody Monolithic Disks in Tandem. Sci. Rep. 2019, 9, 11235 10.1038/s41598-019-47750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohannes G.; Jussila M.; Hartonen K.; Riekkola M.-L. Asymmetrical Flow Field-Flow Fractionation Technique for Separation and Characterization of Biopolymers and Bioparticles. J. Chromatogr. A 2011, 1218, 4104–4116. 10.1016/j.chroma.2010.12.110. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Li Y.; Shen S.; Lee S.; Dou H. Field-Flow Fractionation: A Gentle Separation and Characterization Technique in Biomedicine. TrAC, Trends Anal. Chem. 2018, 108, 231–238. 10.1016/j.trac.2018.09.005. [DOI] [Google Scholar]
- Rico E.; González O.; Blanco M. E.; Alonso R. M. Evaluation of Human Plasma Sample Preparation Protocols for Untargeted Metabolic Profiles Analyzed by UHPLC-ESI-TOF-MS. Anal. Bioanal. Chem. 2014, 406, 7641–7652. 10.1007/s00216-014-8212-y. [DOI] [PubMed] [Google Scholar]
- Yoshida K.; Horii K.; Fujii Y.; Nishio I. Real-Time Observation of Liposome Bursting Induced by Acetonitrile. ChemPhysChem 2014, 15, 2909–2912. 10.1002/cphc.201402333. [DOI] [PubMed] [Google Scholar]
- Helin A.; Sietiö O.-M.; Heinonsalo J.; Bäck J.; Riekkola M.-L.; Parshintsev J. Characterization of Free Amino Acids, Bacteria and Fungi in Size-Segregated Atmospheric Aerosols in Boreal Forest: Seasonal Patterns, Abundances and Size Distributions. Atmos. Chem. Phys. 2017, 17, 13089–13101. 10.5194/acp-17-13089-2017. [DOI] [Google Scholar]
- Lowry O. H.; Rosebrough N. J.; Farr A. L.; Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [PubMed] [Google Scholar]
- Smith P. K.; Krohn R. I.; Hermanson G. T.; Mallia A. K.; Gartner F. H.; Provenzano M. D.; Fujimoto E. K.; Goeke N. M.; Olson B. J.; Klenk D. C. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 1985, 150, 76–85. 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Multia E.; Sirén H.; Andersson K.; Samuelsson J.; Forssén P.; Fornstedt T.; Öörni K.; Jauhiainen M.; Riekkola M.-L. Thermodynamic and Kinetic Approaches for Evaluation of Monoclonal Antibody - Lipoprotein Interactions. Anal. Biochem. 2017, 518, 25–34. 10.1016/j.ab.2016.10.024. [DOI] [PubMed] [Google Scholar]
- Witwer K. W.; Théry C. Extracellular Vesicles or Exosomes? On Primacy, Precision, and Popularity Influencing a Choice of Nomenclature. J. Extracell. Vesicles 2019, 8, 1648167 10.1080/20013078.2019.1648167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J.; Arras G.; Colombo M.; Jouve M.; Morath J. P.; Primdal-Bengtson B.; Dingli F.; Loew D.; Tkach M.; Théry C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E968–77. 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morani M.; Mai T. D.; Krupova Z.; Defrenaix P.; Multia E.; Riekkola M.-L.; Taverna M. Electrokinetic Characterization of Extracellular Vesicles with Capillary Electrophoresis: A New Tool for Their Identification and Quantification. Anal. Chim. Acta 2020, 1128, 42–51. 10.1016/j.aca.2020.06.073. [DOI] [PubMed] [Google Scholar]
- Coumans F. A. W.; Brisson A. R.; Buzas E. I.; Dignat-George F.; Drees E. E. E.; El-Andaloussi S.; Emanueli C.; Gasecka A.; Hendrix A.; Hill A. F.; Lacroix R.; Lee Y.; van Leeuwen T. G.; Mackman N.; Mäger I.; Nolan J. P.; van der Pol E.; Pegtel D. M.; Sahoo S.; Siljander P. R. M.; Sturk G.; de Wever O.; Nieuwland R. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017, 120, 1632–1648. 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- Gan N.; Multia E.; Sirén H.; Ruuth M.; Öörni K.; Maier N. M.; Jauhiainen M.; Kemell M.; Riekkola M. L. Tailor-Made Approach for Selective Isolation and Elution of Low-Density Lipoproteins by Immunoaffinity Sorbent on Silica. Anal. Biochem. 2016, 514, 12–23. 10.1016/j.ab.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Ramirez M. I.; Amorim M. G.; Gadelha C.; Milic I.; Welsh J. A.; Freitas V. M.; Nawaz M.; Akbar N.; Couch Y.; Makin L.; Cooke F.; Vettore A. L.; Batista P. X.; Freezor R.; Pezuk J. A.; Rosa-Fernandes L.; Carreira A. C. O.; Devitt A.; Jacobs L.; Silva I. T.; Coakley G.; Nunes D. N.; Carter D.; Palmisano G.; Dias-Neto E. Technical Challenges of Working with Extracellular Vesicles. Nanoscale 2018, 10, 881–906. 10.1039/C7NR08360B. [DOI] [PubMed] [Google Scholar]
- Inuzuka M.; Hayakawa M.; Ingi T. Serine, an Activity-Regulated Protein Family, Incorporates Serine into Membrane Lipid Synthesis. J. Biol. Chem. 2005, 280, 35776–35783. 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- Skotland T.; Hessvik N. P.; Sandvig K.; Llorente A. Exosomal Lipid Composition and the Role of Ether Lipids and Phosphoinositides in Exosome Biology. J. Lipid Res. 2019, 60, 9–18. 10.1194/jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Molecular Nutrition of Amino Acids and Proteins. In A Volume in the Molecular Nutrition Series, Dardevet D., Ed.; Academic Press, 2016. [Google Scholar]
- Li P.; Yin Y.-L.; Li D.; Kim S. W.; Wu G. Amino Acids and Immune Function. Br. J. Nutr. 2007, 98, 237–252. 10.1017/S000711450769936X. [DOI] [PubMed] [Google Scholar]
- Zhong Z.; Wheeler M.; Li X.; M F.; et al. L-Glycine: A Novel Antiinflammatory, Immunomodulatory, and Cytoprotective Agent. Curr. Opin. Clin. Nutr. Metab. Care 2003, 6, 229–240. 10.1097/00075197-200303000-00013. [DOI] [PubMed] [Google Scholar]
- Stokes K. Y.; Granger D. N. Platelets: A Critical Link between Inflammation and Microvascular Dysfunction. J. Physiol. 2012, 590, 1023–1034. 10.1113/jphysiol.2011.225417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.