Abstract
Epigenetic reprogramming during germ cell formation is essential to gain pluripotency and thus embryogenic potential. The histone modification H3K27me3, which is catalysed by the Polycomb repressive complex 2 (PRC2), regulates important developmental processes in both plants and animals, and defects in PRC2 components cause pleiotropic developmental abnormalities. Nevertheless, the role of H3K27me3 in determining embryogenic potential in gymnosperms is still elusive. To address this, we generated H3K27me3 profiles of Norway spruce (Picea abies) embryonic callus and non-embryogenic callus using CUT&RUN, which is a powerful method for chromatin profiling. Here, we show that H3K27me3 mainly accumulated in genic regions in the Norway spruce genome, similarly to what is observed in other plant species. Interestingly, H3K27me3 levels in embryonic callus were much lower than those in the other examined tissues, but markedly increased upon embryo induction. These results show that H3K27me3 levels are associated with the embryogenic potential of a given tissue, and that the early phase of somatic embryogenesis is accompanied by changes in H3K27me3 levels. Thus, our study provides novel insights into the role of this epigenetic mark in spruce embryogenesis and reinforces the importance of PRC2 as a key regulator of cell fate determination across different plant species.
Keywords: Embryogenic callus, gymnosperm, H3K27me3, histone modification, Norway spruce, somatic embryos
PRC2-mediated H3K27me3 is associated with embryogenic potential in Norway spruce, and the early phase of somatic embryogenesis is accompanied by changes in H3K27me3 levels.
Introduction
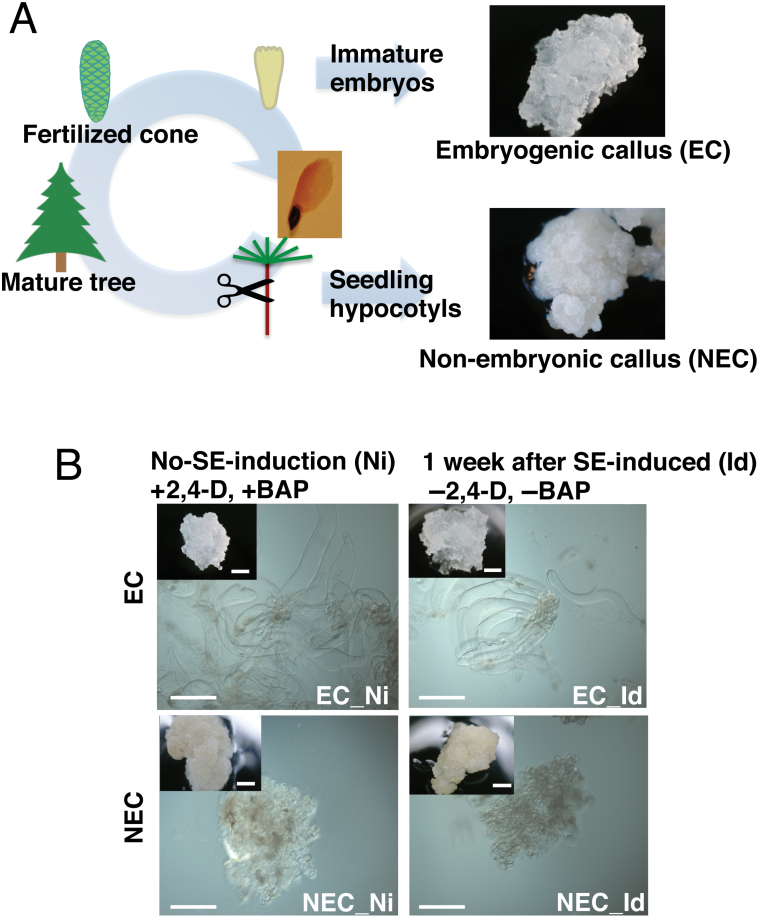
Micropropagation through somatic embryogenesis in spruce species was established nearly three decades ago, and nowadays this method is commercially applied to propagate several coniferous species (reviewed in Klimaszewska et al., 2015). This technology enables an efficient proliferation of spruce individuals, bypassing the long reproductive cycle of this species. Norway spruce (Picea abies) is a common conifer species that is ecologically and economically important in Europe. In Norway spruce, two different types of callus can be generated: embryogenic callus (EC), usually induced from immature embryos, and non-embryogenic callus (NEC), induced from vegetative seedling tissues (Nagmani et al., 1987; Becwar et al., 1988; Verhagen & Wann, 1989). Interestingly, the success rate of EC formation is largely dependent on the tissue of the original embryos and their developmental stages (Becwar et al., 1988; Ramarosandratana & Van Staden, 2003). Immature embryos have a higher potential for EC formation compared with mature tissues (Jain et al., 1988; von Arnold et al., 1995). After callus formation, EC and NEC are different in their appearance and biochemical properties (Becwar et al., 1988) (Fig. 1A), but both can be maintained in a growth medium containing 2,4-dichlorophenoxyacetic acid (2,4-D) and 6-benzylaminopurine (BAP). Transferring these calli to a medium without these phytohormones induces somatic embryogenesis exclusively in EC, showing that EC and NEC have distinct embryogenic potentials (Nagmani et al., 1987; Becwar et al., 1988). Nevertheless, little is known about the molecular mechanisms underlying the distinct embryonic competencies between EC and NEC.
Fig. 1.
Embryogenic callus (EC) and Non-embryogenic callus (NEC) in Norway spruce. (A) Explants from immature embryo form EC and explants from seedling hypocotyl form NEC. (B) Morphology of EC and NEC before and 1 week after somatic embryogenesis induction. Both types of calli can be induced and maintained in the presence of 9 µM 2,4-dichlorophenoxyacetic acid (2,4-D) and 4.4 µM 6-benzylaminopurine (BAP). Transferring to medium without 2,4-D or BAP triggers somatic embryogenesis in EC. Scale bar: 200 μm; inset scale bar: 2 mm.
Polycomb repressive complex 2 (PRC2) contains histone methyltransferase activity, and is responsible for the tri-methylation of lysine 27 of histone H3 (H3K27me3). This histone modification is a repressive epigenetic mark observed in developmental regulatory genes and environmental responsive genes (reviewed in (Schwartz & Pirrotta, 2007; Hennig & Derkacheva, 2009; Margueron & Reinberg, 2011). The H3K27me3 mark plays a crucial role in cell differentiation and tissue morphogenesis. In Arabidopsis, H3K27me3 profiles change when cells convert from a differentiated to a multipotency state, and vice versa (Lafos et al., 2011; He et al., 2012). Depletion of PRC2 function causes disruption of post-embryonic development, resulting in the formation of callus- or embryo-like cell masses after germination (Chanvivattana et al., 2004; Bouyer et al., 2011). Strikingly, in PRC2 mutants, leaf tissue explants have no capacity of callus formation, while callus formation from root is unaffected (He et al., 2012). Because the process of callus formation from leaves transits through a root-like developmental program (Sugimoto et al., 2010), callus formation has been recently considered as a transdifferentiation process rather than a dedifferentiation process (Sugimoto et al., 2011). Thus, the callus formation process is different depending on the original tissue. Interestingly, PRC2 mutants show a higher competency of somatic embryo induction from vegetative tissues when compared with wild-type plants (Mozgová et al., 2017). This suggests that H3K27me3 plays a crucial role in repression of pluripotency in Arabidopsis. In contrast, to date, knowledge about the influence of chromatin status, in particular of H3K27me3 deposition, in gymnosperm embryogenesis is very limited.
Here, we surveyed the differences of H3K27me3 deposition in EC and NEC of Norway spruce. Most genome-wide analyses of histone modifications in plants have been accomplished in species with relatively small genomes, with the exception of maize, barley, and wheat (Zhang et al., 2007; Makarevitch et al., 2013; Widiez et al., 2014; Baker et al., 2015; Hussey et al., 2017; Latrasse et al., 2017; Chica et al., 2017; Lü et al., 2018; Qi et al., 2018; Li et al., 2019; Montgomery et al., 2020). In contrast to many angiosperm studies, genome-wide chromatin profiling in other plant species has been limited to moss, liverwort, and unicellular algae (Widiez et al., 2014; Mikulski et al., 2017; Montgomery et al., 2020). Gymnosperm species have generally very large (4–30 Gbp) and poorly assembled genomes, making this type of profiling laborious and challenging. To explore the roles of H3K27me3 in embryogenic competency of gymnosperms and chromatin dynamics during early phases of somatic embryogenesis, we identified genes with H3K27me3 marks in EC and NEC using CUT&RUN, which is an antibody-based technology for profiling targeted DNA–protein interactions (Skene & Henikoff, 2017). This method reduces background signals compared with the conventional chromatin immunoprecipitation (ChIP) method (Skene et al., 2017) and is thus more suitable for whole genome analyses of histone modifications in organisms with large genome, such as gymnosperms.
In both EC and NEC, the H3K27me3 mark was enriched in genic regions, rather than in intergenic regions or transposable elements. Nearly half of H3K27me3-enriched genes were common between EC and NEC; however, many genes showed lower H3K27me3 levels in EC before somatic embryo induction when compared with other samples. For example, some homologues of AINTEGUMENTA-LIKE (AIL) and ABSCISIC ACID INSENSITIVE 3 (ABI3) transcription factor genes display low H3K27me3 in EC compared with NEC. Interestingly, a 1-week period in embryo-inducing medium was sufficient to provoke alterations in H3K27me3 profiles of both EC and NEC. Notwithstanding, these two types of callus displayed large differences in H3K27me3 profiles in response to somatic embryo induction. Thus, we revealed that two different types of calli in Picea abies have distinct chromatin profiles, suggesting that the genome-wide H3K27me3 status in EC underlies its high pluripotency of cell differentiation.
Materials and methods
Plant materials and growth conditions
The Norway spruce (Picea abies L. Karst) embryogenic culture cell line (EC) was established from immature embryos by a previously described method (von Arnold & Clapham, 2008). EC was established and proliferated on half-strength LP medium at 22 °C in the dark. For somatic embryo induction, calli were transferred into solidified BMI-SI medium (von Arnold & Clapham, 2008) and were harvested 1 week later. Two-month-old seedlings were grown in quarter-strength SH medium (von Arnold & Clapham, 2008). For NEC formation, explants from hypocotyls of 2-month-old seedlings were cultured on the half-strength LP medium at 22 °C in the dark. NEC were generated in April 2017 and harvested in February 2018.
Imaging of culture cell morphology
Subcultured calli from EC and NEC were crushed with forceps in 0.3 M sorbitol solution. Images were captured through a differential interference contrast microscope (Axioscope A1, Zeiss, Germany) and a Leica camera DFC490.
CUT&RUN and sequencing
Nuclei were isolated as described previously with the exception that the MEB buffer contained no EGTA (Shu et al., 2014). CUT&RUN (Skene & Henikoff, 2017) was carried out using isolated nuclei (from EC and NEC before and after induction and from seedlings) as described previously with modifications as follows. Triton X-100 was added to the washing buffer at a final concentration of 0.1%. After the extraction of digested DNA, larger fragments were removed from extracted DNA by ×0.6 volume Agencourt AMPure XP (Beckman Coulter). Subsequently, the DNA was recovered by ×3.0 volume AMpure XP. Major modifications from the original CUT&RUN protocol are as follows. Extracted nuclei were immobilized onto concanavalin A (ConA) beads. Chromatin binding to beads was confirmed by comparing recovered amounts of genomic DNA between flow-through and pull-down fractions. To avoid chelation of calcium ions, which are required for ConA bead binding, MEB buffer without EGTA was used for nuclear extraction. Triton X-100 was added to the washing buffer to ensure permeability of pA-MNase. After pA-MNase digestion, DNA size distribution corresponding to mono-, di-, and tri-nucleosomes was confirmed using the Bioanalyzer system (Agilent Technologies). Anti-H3K27me3 antibody was used (Cell Signaling Technology cat. no. 9733S). The libraries were prepared using MicroPlex Library Preparation Kit v2 (Diagenode) following the manufacturer’s instructions. Paired-end reads of 125 bp were sequenced with an Illumina HiSeq2500 on the National Genomics Infrastructure—SciLifeLab. The same amount of spike-in DNA (Saccharomyces cerevisiae genomic DNA) was added to each sample after extracting MNase-digested DNA. The read counts of spike-in DNA, which mapped to the S. cerevisiae reference genome, were well correlated with total read counts in each sample.
Data analysis
H3K27me3 signal peaks were called as follows. (i) The paired-end reads were separately mapped by BWA (v0.7.17) (Li & Durbin, 2009) to the P. abies whole-genome v1.0 (Congenie.org) and merged if they were mapped in the same contigs. (ii) The merged fragments with a size longer than 700 bp were discarded. (iii) The contigs of genome sequences were split by 500 bp bins. (iv) The read numbers overlapping each bin were counted and P-values were calculated using a binominal test by each bin. (v) Obtained P-values were adjusted by p.adjust packages in R using the Benjamini–Hochberg method. (vi) The bins with false discovery rate ≤10−25 were selected. Peaks were merged if their distances were shorted than 100 bp. The genes that overlapped with H3K27me3 peaks were defined as H3K27me3-enriched genes. The H3K27me3 signal intensity per gene was defined as reads per kilobase per billion mapped reads over gene regions, including 500 bp extensions in both upstream and downstream orientations. For genomic features, transposable element sequences were annotated as described before (Nakamura et al., 2019). For drawing signal distribution over genes, deepTools computeMatrix and plotHeatmap were used (Ramírez et al., 2014). Shannon entropy was calculated by the R package ‘entropy’ using transcriptome data retrieved from the ConGenIE resource (http://congenie.org/). To calculate the Shannon entropy the following transcriptome datasets were used for organ specificity: ENA ID: ERS235801 (male flower), ERS235809 (female flower), ERS235803 (needles from 2009), ERS235807 (pineapple galls), ERS235808 (buds, early season), ERS235811 (stem from vegetative shoots), ERS235802 (vegetative shoots), and ERS235821 (wood, June), and for condition specificity: ENA ID: ERS235803 (needles from 2009), ERS235804 (needles from 2008), ERS235805 (infected needles), ERS235814 (needles from dried twig), ERS235815 (needles girdled twig), ERS235817 (early morning needles), ERS235818 (mid-day needles), ERS235819 (late afternoon needles), and ERS235820 (night needles) (Nystedt et al., 2013). Homologues of Arabidopsis proteins were identified as previously described (Nakamura et al., 2019).
Phylogenetic analysis of H3K27 methyltransferase homologues
CLF, SWN, and MEA amino acid sequences were obtained from TAIR10 and used as query sequences to identify E(z) homologues in the plant kingdom, using tblastx (Camacho et al., 2009). The dataset of coding sequences (CDS) in PLAZA gymnosperm (v1.0), Plaza Monocots (v4.5), ConGenIE (v1.0), Coge (https://genomevolution.org/coge/), Phytozome (v1), and NCBI, were searched for homologues in Amborella trichopoda, Persea americana, Populus trichocarpa, Oryza sativa ssp. japonica, Zea mays, Picea abies, Picea glauca, Picea sitchensis, Pinus pinaster, Pinus sylvestris, Pseudotsuga menziesii, Taxus baccata, Cycas micholitzii, Ginkgo biloba, Gnetum montanum, Selaginella moellendorffii, Sphagnum fallax, Marchantia polymorpha, Physcomitrella patens, and Chlamydomonas reinhardtii (Goodstein et al., 2012; Van Bel et al., 2018). Amino acid sequences of the SET domain were aligned using ClustalW and the phylogenetic tree was constructed by PHYML and drawn in MEGA7.0 (Thompson et al., 1994; Guindon et al., 2010; Kumar et al., 2016). The E(z) of Drosophila melanogaster and MA_9310110g0010, which is a homologue of Arabidopsis SDG7, were used as an outgroup.
Gene ontology analysis
A list of Arabidopsis genes homologous to the P. abies genes of interest was used for gene ontology (GO) analysis. The Arabidopsis GO annotation was downloaded from TAIR (https://www.arabidopsis.org). The hypergeometric P-values were calculated based on the frequency of each GO term in P. abies genes of interest with Arabidopsis homologues over the whole set of P. abies genes with Arabidopsis homologues. Enriched GO terms were summarized using REVIGO (Supek et al., 2011).
DNA motif identification
After mapping, the read counts per base pair were normalized by total reads and then converted into Bedgraph format by Bedtools (Quinlan & Hall, 2010). Subsequently, the regions with more than 8.0×10−7 read/total reads were considered signal peaks. If the distance between adjacent peaks was less than 101 bp, the peaks were merged. To calculate the motif enrichment, background DNA sequences were randomly picked from the P. abies genome to have the same length distribution as the peak regions. Selected DNA sequences were used as a query to run MEME-ChIP (Machanick & Bailey, 2011). The JASPAR core (2018) plant dataset was used as known motifs.
Mapping data
Mapping data of each tissue are listed in Supplementary Table S1 at JXB online.
Reverse transcription–quantitative PCR
Total RNA was extracted as previously described (Nakamura et al., 2019). The cDNA was synthesized by RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) with both oligo(dT) and oligo hexamer primers. Quantitative real-time PCR was performed as described (Nakamura et al., 2019). Primers used are listed in Supplementary Table S2.
Results
Characterization of H3K27me3-rich regions in Norway spruce
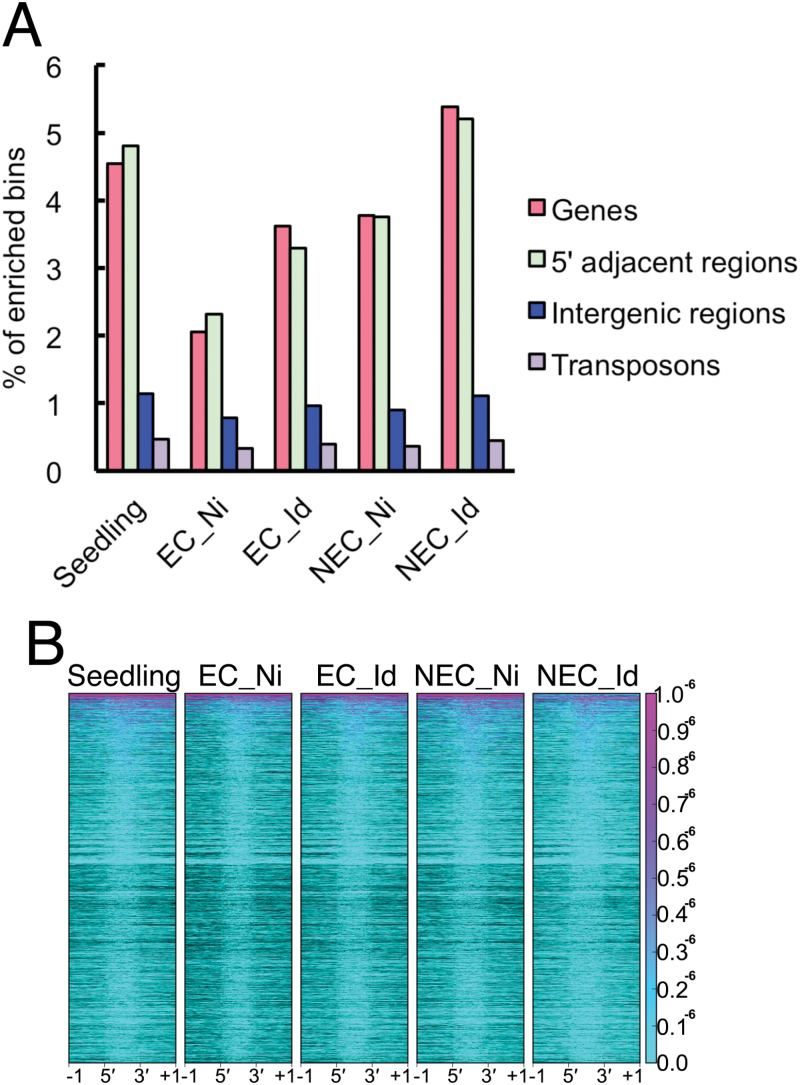
To profile the H3K27me3 histone modification in the Norway spruce genome (19.6 Gbp), we adapted the previously developed CUT&RUN method (Skene & Henikoff, 2017) to this species. Given our interest in unravelling the role of this histone modification on the embryogenic potential of Norway spruce, we sampled EC and NEC, before and after somatic embryo induction (Fig. 1B). Our main focus was on early changes, before cell morphological changes were visible. We therefore sampled calli 1 week after somatic embryo induction prior to morphological changes (Filonova et al., 2000). EC consisted of two different cell types: elongated cells and small and compacted cells (Fig. 1B) (Filonova et al., 2000). On the other hand, NEC consisted of only round small cells. After 2 weeks of SE induction, compacted cells of EC proliferated and aggregated to generate proembryonic cell masses (Filonova et al., 2000), whereas cells of NEC turned brown (see Supplementary Fig. S1). As a control, we also sampled seedlings. Then, we investigated the genomic features that overlapped with H3K27me3 peaks. We found that H3K27me3 was highly enriched in coding regions and presumable promoter regions, across all sample types (Fig. 2A–B). On the other hand, repetitive sequences and intergenic regions were under-represented among H3K27me3-enriched regions (Fig. 2A). These observations are consistent with previous studies in angiosperms, moss, and unicellular algae (Zhang et al., 2007; Makarevitch et al., 2013; Widiez et al., 2014; Baker et al., 2015; Hussey et al., 2017; Latrasse et al., 2017; Mikulski et al., 2017; Chica et al., 2017; Lü et al., 2018; Qi et al., 2018; Li et al., 2019). This shows that (i) the CUT&RUN method can be successfully applied to large genomes, such as that of Norway spruce, and (ii) despite the significant differences in genome size between angiosperms and gymnosperms, deposition of H3K27me3 is detected in similar genomic features across different plant species.
Fig. 2.
H3K27me3 accumulates in genic regions. (A) H3K27me3-enriched genomic features. (B) H3K27me3 accumulation over target genes ±1 kb adjacent regions.
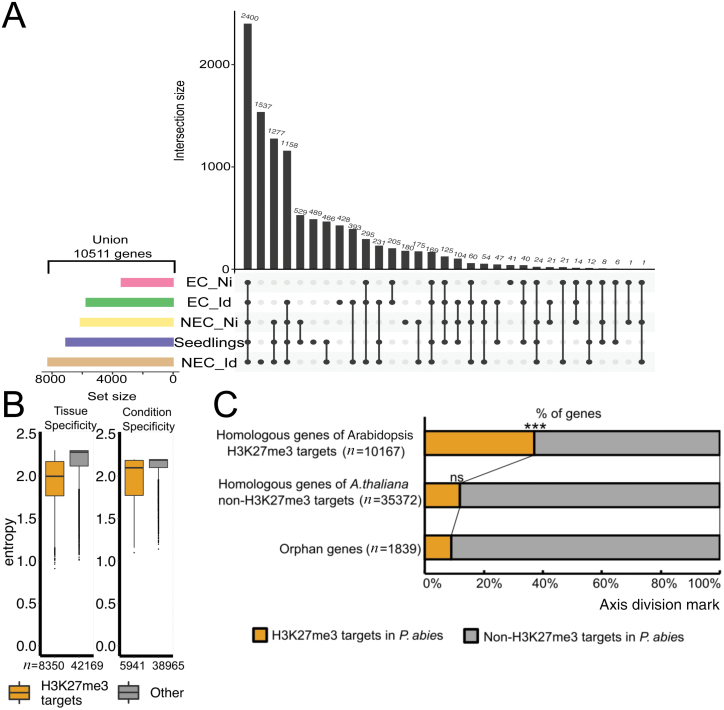
Among the five tissue samples, a total of 10 511 genes had H3K27me3 peaks within the gene body or in adjacent regions (Fig. 3A). Each tissue had more than 3000–8000 genes marked with H3K27me3 (Fig. 3A). NEC tissue subject to somatic embryo induction (NEC_Id) had the highest number of H3K27me3-marked genes. On the other hand, non-induced EC (EC_Ni) had only 3422 H3K27me3-marked genes. Among all five samples there were 2400 genes that overlapped. The H3K27me3 modification is frequently present in developmental regulatory genes, in both animals and plants (Hennig & Derkacheva, 2009; Margueron & Reinberg, 2011). To test the developmental roles of H3K27me3 in Norway spruce, we investigated the expression specificity of H3K27me3-enriched genes across different tissues or environmental conditions. We calculated the Shannon entropy between H3K27me3-enriched genes and non-enriched genes. This measure can be used to estimate the specificity of expression of a given gene (Schug et al., 2005). We used eight different tissue/organ transcriptome datasets (male cone, female cone, wood, stems, among others), and nine transcriptome datasets of different conditions of needles (daytime, infection, wounding, among others) (see ‘Materials and methods’ for details). In both cases, the subset of H3K27me3-enriched genes showed a lower Shannon entropy compared with the subset that was not marked by H3K27me3. These results indicate that genes targeted by H3K27me3 are expressed in specific organs or conditions. Thus, H3K27me3-enriched genes in gymnosperms show responsiveness to developmental and environmental regulation, similarly to what is observed in angiosperms (Zhang et al., 2007; Makarevitch et al., 2013; Liu et al., 2015; Akter et al., 2019).
Fig. 3.
Characterization of H3K27me3 target genes regarding their expression specificity and conservation. (A) UpSet plot of H3K27me3-enriched genes of five different tissues. (B) Shannon entropy distribution between subsets of H3K27me3-target and non-target genes. (C) Conservation of H3K27me3 target genes between Arabidopsis and Norway spruce. Statistical significance was determined using the hypergeometric test: ***P<0.0005; ns: P>0.05.
Next, we wondered whether the sets of genes marked with H3K27me3 are similar between angiosperms and gymnosperms. To test this, we assigned an Arabidopsis homologue to each P. abies gene. Approximately 65% of the total P. abies gene models had homology to at least one Arabidopsis gene based on a BLAST search. We then grouped the Arabidopsis homologues based on the presence of H3K27me3. Nearly 40% of P. abies genes showing homology to an Arabidopsis H3K27me3-marked gene were also H3K27me3-marked (Fig. 3C). This ratio is significantly higher when compared with that in P. abies homologous genes of non-H3K27me3 targets in Arabidopsis (less than 15%). Approximately 10% of orphan genes, which are specific for the gymnosperm lineage, had H3K27me3 marks (Fig. 3C), suggesting that newly evolved genes become integrated into the transcriptional network regulated by PRC2.
Proliferation of EC and NEC requires phytohormones, such as 2,4-D and BAP. A previous study in Arabidopsis showed that gene families related to auxin pathways, such as YUCCAs, IAAs, and PIN-FORMEDs, are targeted by H3K27me3, but ARFs are not (Lafos et al 2011). To investigate whether homologues of these gene families in Norway spruce have a similar tendency, we scored the percentage of genes with H3K27me3 in auxin-related gene families. Interestingly, we found that Norway spruce ARF homologues also lacked H3K27me3, while YUCCA homologues tended to be marked by H3K27me3, similarly to Arabidopsis. In contrast, PIN-FORMED homologues were depleted of H3K27me3 in all tissues examined (see Supplementary Fig. S2).
PRC2 core components are conserved throughout the plant kingdom (Hennig & Derkacheva, 2009). Angiosperm genomes have three to four H3K27 methyltransferase genes, which are called E(z) homologues (Hennig & Derkacheva, 2009). In Arabidopsis, the E(z) homologues are SWINGER (SWN), CURLY LEAF (CLF), and MEDEA (MEA). The latter is expressed only during the reproductive phase and is responsible for parent-of-origin-specific gene expression in the endosperm (reviewed in Batista & Köhler, 2020). Because previous phylogenetic analyses of E(z) homologues lack information on gymnosperm species, we performed a phylogenetic analysis of H3K27 methyltransferase genes of gymnosperms. All the species examined here had at least two putative H3K27 methyltransferase genes (see Supplementary Fig. S3), which are in the same clade as the Arabidopsis CLF gene. In Arabidopsis, CLF and SWN have partially redundant functions (Chanvivattana et al., 2004). Interestingly, we could only detect basal angiosperm, eudicot, and monocot homologues within the SWN orthologue clade (Supplementary Fig. S3). Thus, these data suggest that E(z) homologues underwent radiation and subfunctionalization in angiosperms after the divergence of angiosperms and gymnosperms.
In Arabidopsis, some H3K27me3 targets are known to be associated with Polycomb response elements (PRE), cis-regulatory elements responsible for the targeting of PRC2 (Xiao et al., 2017; Zhou et al., 2018). To test whether specific DNA sequences are associated with H3K27me3 deposition in gymnosperms, we performed a de novo motif identification using the MEME-ChIP algorithm (Machanick & Bailey, 2011). In both EC and NEC, most identified motifs were short AT-rich features, including a sequence similar to the target site of the DOF-domain-binding transcription factor (AT5G66940) (see Supplementary Fig. S4). Interestingly, we could also identify a motif partially similar to the one used by the Arabidopsis histone demethylase REF6 to bind its targets. Other DNA motifs were not shared between EC and NEC. Collectively, these data indicate that H3K27me3 in P. abies tends to localize at AT-rich sequences; nevertheless, additional studies will be required to further characterize and validate cis-regulatory elements responsible for PRC2 targeting and subsequent H3K27me3 deposition.
H3K27me3 profiles and response to somatic embryo induction in EC are largely different from those in NEC
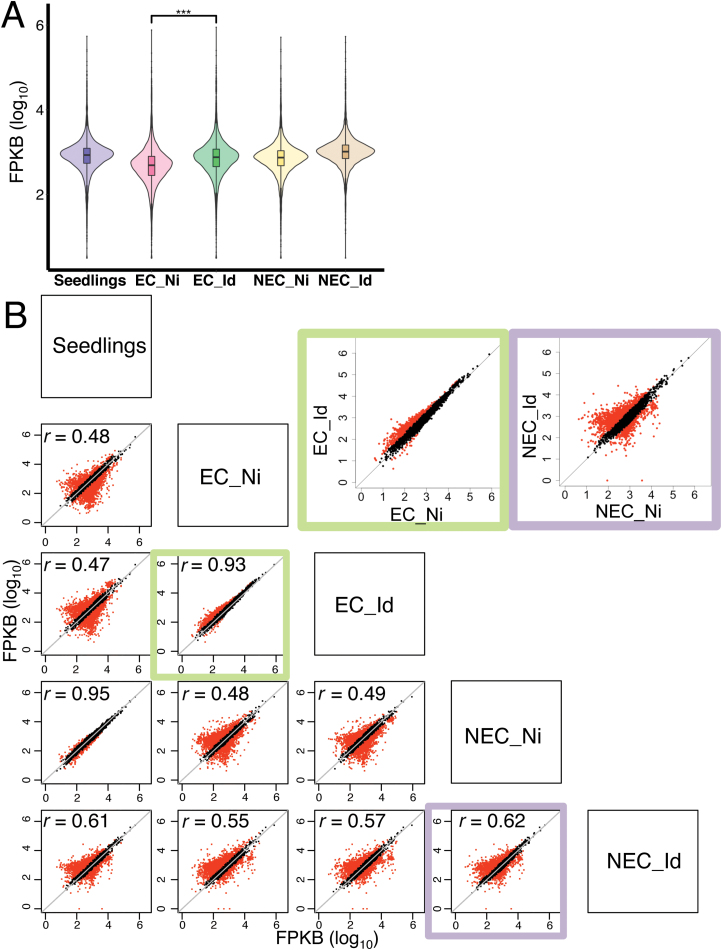
Pervious work revealed that H3K27me3 is present in callus tissue of Arabidopsis, and there is even de novo H3K27me3 present in callus (He et al., 2012). Consistent with this, both spruce EC and spruce NEC had detectable H3K27me3 (Figs 1–3). Nevertheless, the non-induced EC sample (EC_Ni) had the lowest levels of H3K27me3, which increased after embryogenesis induction (Fig. 4A). To directly compare the H3K27me3 signal per gene, and between different tissues, we plotted H3K27me3 reads in fragments per kilobase billion (FPKB) of each H3K27me-enriched gene in a pair-wise manner (Fig. 4B). We identified a considerable subset of genes showing increased H3K27me3 levels upon induction in EC, while only a few genes showed reduced H3K27me3 after induction (Fig. 4B, EC_Id versus EC_Ni). Unexpectedly, H3K27me3 profiles were largely similar between non-induced NEC and seedlings (Fig. 4B, seedling versus NEC_Ni), indicating that callus formation in NEC does not have a major impact on H3K27me3 accumulation. Nevertheless, somatic embryo induction of NEC also caused a dramatic alteration of H3K27me3 trends (Fig. 4B, NEC_Id versus NEC_Ni). Thus, these data indicate that the genes marked with H3K27me3 are largely different between EC and NEC. Moreover, H3K27me3 changes in response to somatic embryo induction are also distinct between EC and NEC.
Fig. 4.
Non-induced EC shows reduced H3K27me3 signals. (A) Distributions of H3K27me3 signal intensity. ***P<0.0005, statistical significance using a paired Student’s t-test. (B) Pair-wise comparisons of H3K27me3 signals in target genes. Red dots indicate genes with a difference in H3K27me3 accumulation of twice or more.
To address the functional attributes of differentially marked genes (DMGs) between EC and NEC, we identified orthologues of DMGs in Arabidopsis and then analysed whether specific GO terms were enriched among these orthologous genes. Among the 789 genes that were H3K27me3-marked in non-induced EC, but not marked in non-induced NEC, GO terms related to response to metabolism, and in particular flavonoid metabolism, were most highly enriched (Table 1). On the other hand, somatic embryogenesis, positive regulation of cell differentiation, and negative regulation of cell proliferation were among the GO terms enriched in the 3498 genes that were H3K27me3 marked in non-induced NEC, but not marked in non-induced EC (Table 2). These data indicate that genes required to confer somatic embryogenesis competence are repressed by H3K27me3 in non-induced NEC tissues in Norway spruce (Table 2). After somatic embryo induction, 1741 genes more than doubled the signal of H3K27me3 in EC. In this gene subset, GO terms belonging to cell wall metabolism were enriched (Table 3).
Table 1.
The GO enrichment of genes marked by H3K27me3 specifically in EC_Ni but not in NEC_Ni
| GO term | Annotation | Frequency in sample | Enrichment | P-value |
|---|---|---|---|---|
| Biological process | ||||
| GO:0009892 | Negative regulation of metabolic process | 6 | 16.3 | 9.35×10−8 |
| GO:0009800 | Cinnamic acid biosynthetic process | 6 | 11.4 | 1.13×10−6 |
| GO:1901601 | Strigolactone biosynthetic process | 5 | 8.0 | 4.28×10−5 |
| GO:0009411 | Response to UV | 11 | 6.8 | 1.33×10−7 |
| GO:0010089 | Xylem development | 6 | 5.3 | 1.58×10−4 |
| GO:0009813 | Flavonoid biosynthetic process | 17 | 5.1 | 1.55×10−8 |
| GO:0042744 | Hydrogen peroxide catabolic process | 18 | 4.7 | 1.98×10−8 |
| GO:0002239 | Response to oomycetes | 6 | 4.6 | 3.74×10−4 |
| GO:0009739 | Response to gibberellin stimulus | 12 | 4.6 | 3.13×10−6 |
| Molecular function | ||||
| GO:0016711 | Flavonoid 3′-monooxygenase activity | 11 | 11.7 | 2.94×10−10 |
| GO:0042973 | Glucan endo-1,3-β-D-glucosidase activity | 5 | 8.0 | 4.28×10−5 |
| GO:0008134 | Transcription factor binding | 5 | 5.4 | 3.81×10−4 |
| GO:0004620 | Phospholipase activity | 5 | 4.9 | 6.06×10−4 |
| GO:0008757 | S-Adenosylmethionine-dependent methyltransferase activity | 7 | 4.4 | 2.47×10−4 |
| GO:0016747 | Transferase activity, transferring acyl groups other than amino-acyl groups | 9 | 4.2 | 7.15×10−5 |
| GO:0020037 | Heme binding | 48 | 4.1 | 3.27×10−16 |
| GO:0016762 | Xyloglucan:xyloglucosyl transferase activity | 6 | 4.1 | 7.77×10−4 |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 7 | 3.7 | 7.15×10−4 |
| GO:0019825 | Oxygen binding | 29 | 4.0 | 1.91×10−10 |
| GO:0004601 | Peroxidase activity | 18 | 4.0 | 2.14×10−7 |
| GO:0005506 | Iron ion binding | 30 | 3.6 | 1.39×10−9 |
Table 2.
The GO enrichment of genes marked by H3K27me3 specifically in NEC_Ni but not in EC_Ni
| GO term | Annotation | Frequency in sample | Enrichment | P-value |
|---|---|---|---|---|
| Biological process | ||||
| GO:0033231 | Carbohydrate export | 7 | 7.8 | 1.93×10−6 |
| GO:0045597 | Positive regulation of cell differentiation | 8 | 7.8 | 5.07×10−7 |
| GO:0035336 | Long-chain fatty-acyl-CoA metabolic process | 7 | 6.1 | 1.49×10−5 |
| GO:0010345 | Suberin biosynthetic process | 36 | 5.4 | 3.11×10−17 |
| GO:0009963 | Positive regulation of flavonoid biosynthetic process | 6 | 5.2 | 1.27×10−4 |
| GO:0016102 | Diterpenoid biosynthetic process | 15 | 4.6 | 1.68×10−7 |
| GO:0010262 | Somatic embryogenesis | 9 | 3.7 | 1.56×10−4 |
| GO:0052325 | Cell wall pectin biosynthetic process | 8 | 3.6 | 3.93×10−4 |
| GO:0008285 | Negative regulation of cell proliferation | 7 | 3.4 | 9.88×10−4 |
| GO:0048544 | Recognition of pollen | 29 | 3.2 | 1.82×10−8 |
| GO:0010268 | Brassinosteroid homeostasis | 16 | 3.0 | 2.37×10−5 |
| Molecular function | ||||
| GO:0009674 | Potassium:sodium symporter activity | 5 | 7.1 | 5.16×10−5 |
| GO:0103075 | Indole-3-pyruvate monooxygenase activity | 6 | 6.7 | 2.25×10−5 |
| GO:0080019 | Fatty-acyl-CoA reductase (alcohol-forming) activity | 15 | 6.5 | 9.07×10−10 |
| GO:0102406 | Omega-hydroxypalmitate O-sinapoyl transferase activity | 8 | 5.9 | 6.03×10−6 |
| GO:0047560 | 3-Dehydrosphinganine reductase activity | 9 | 4.8 | 1.44×10−5 |
| GO:0034768 | (E)-β-Ocimene synthase activity | 25 | 4.8 | 1.63×10−11 |
| GO:0009905 | ent-Copalyl diphosphate synthase activity | 15 | 4.7 | 1.27×10−7 |
| GO:0051119 | Sugar transmembrane transporter activity | 20 | 4.5 | 3.74×10−9 |
| GO:0008429 | Phosphatidylethanolamine binding | 10 | 4.3 | 1.80×10−5 |
| GO:0008395 | Steroid hydroxylase activity | 10 | 4.3 | 1.80×10−5 |
| GO:0047372 | Acylglycerol lipase activity | 15 | 4.3 | 4.86×10−7 |
| GO:0016628 | Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | 8 | 4.2 | 1.19×10−4 |
| GO:0050105 | L-Gulonolactone oxidase activity | 14 | 4.0 | 2.52×10−6 |
| GO:0047890 | Flavanone 4-reductase activity | 8 | 3.8 | 2.51×10−4 |
| GO:0045552 | Dihydrokaempferol 4-reductase activity | 8 | 3.8 | 2.51×10−4 |
| GO:0102483 | Scopolin β-glucosidase activity | 16 | 3.5 | 3.47×10−6 |
| GO:0052716 | Hydroquinone:oxygen oxidoreductase activity | 23 | 3.2 | 3.64×10−7 |
| GO:0016747 | Transferase activity, transferring acyl groups other than amino-acyl groups | 35 | 3.1 | 1.07×10−9 |
| Cellular component | ||||
| GO:0000325 | Plant-type vacuole | 9 | 3.9 | 9.87×10−5 |
| GO:0031012 | Extracellular matrix | 11 | 3.8 | 3.12×10−5 |
Table 3.
The GO enrichment of genes that gain H3K27me3 after somatic embryo induction in EC
| GO term | Annotation | Frequency in sample | Enrichment | P-value |
|---|---|---|---|---|
| Biological process | ||||
| GO:0016102 | Diterpenoid biosynthetic process | 12 | 7.8 | 5.23×10−9 |
| GO:0002679 | Respiratory burst involved in defense response | 6 | 7.4 | 1.63×10−5 |
| GO:0080163 | Regulation of protein serine/threonine phosphatase activity | 7 | 6.7 | 1.06×10−5 |
| GO:0006666 | 3-Keto-sphinganine metabolic process | 5 | 5.7 | 2.36×10−4 |
| GO:0048544 | Recognition of pollen | 19 | 4.4 | 2.04×10−8 |
| GO:0006949 | Syncytium formation | 10 | 4.4 | 2.15×10−5 |
| GO:0009607 | Response to biotic stimulus | 6 | 4.0 | 8.11×10−4 |
| GO:0009625 | Response to insect | 12 | 3.9 | 1.74×10−5 |
| GO:0042744 | Hydrogen peroxide catabolic process | 35 | 3.8 | 1.43×10−11 |
| GO:0042545 | Cell wall modification | 14 | 3.6 | 1.08×10−5 |
| GO:0010089 | Xylem development | 9 | 3.3 | 5.19×10−4 |
| GO:0009740 | Gibberellic acid mediated signaling pathway | 17 | 3.1 | 1.29×10−5 |
| Molecular function | ||||
| GO:0009905 | ent-Copalyl diphosphate synthase activity | 12 | 8.0 | 4.09×10−9 |
| GO:0004864 | Protein phosphatase inhibitor activity | 8 | 6.1 | 7.55×10−6 |
| GO:0010333 | Terpene synthase activity | 16 | 6.1 | 1.93×10−9 |
| GO:0047560 | 3-Dehydrosphinganine reductase activity | 5 | 5.7 | 2.36×10−4 |
| GO:0010427 | Abscisic acid binding | 7 | 5.4 | 4.81×10−5 |
| GO:0050502 | cis-Zeatin O-β-D-glucosyltransferase activity | 6 | 5.1 | 1.78×10−4 |
| GO:0052716 | Hydroquinone:oxygen oxidoreductase activity | 13 | 3.8 | 1.03×10−5 |
| GO:0045330 | Aspartyl esterase activity | 14 | 3.8 | 5.56×10−6 |
| GO:0030599 | Pectinesterase activity | 14 | 3.7 | 8.19×10−6 |
| GO:0004601 | Peroxidase activity | 39 | 3.6 | 5.01×10−12 |
| GO:0016614 | Oxidoreductase activity, acting on CH-OH group of donors | 10 | 3.4 | 2.31×10−4 |
| GO:0030246 | Carbohydrate binding | 54 | 3.2 | 5.41×10−14 |
| GO:0016709 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD or NADPH as one donor, and incorporation of one atom of oxygen | 17 | 3.0 | 1.99×10−5 |
Homologues of somatic embryogenesis-related genes are differentially methylated between NEC and EC
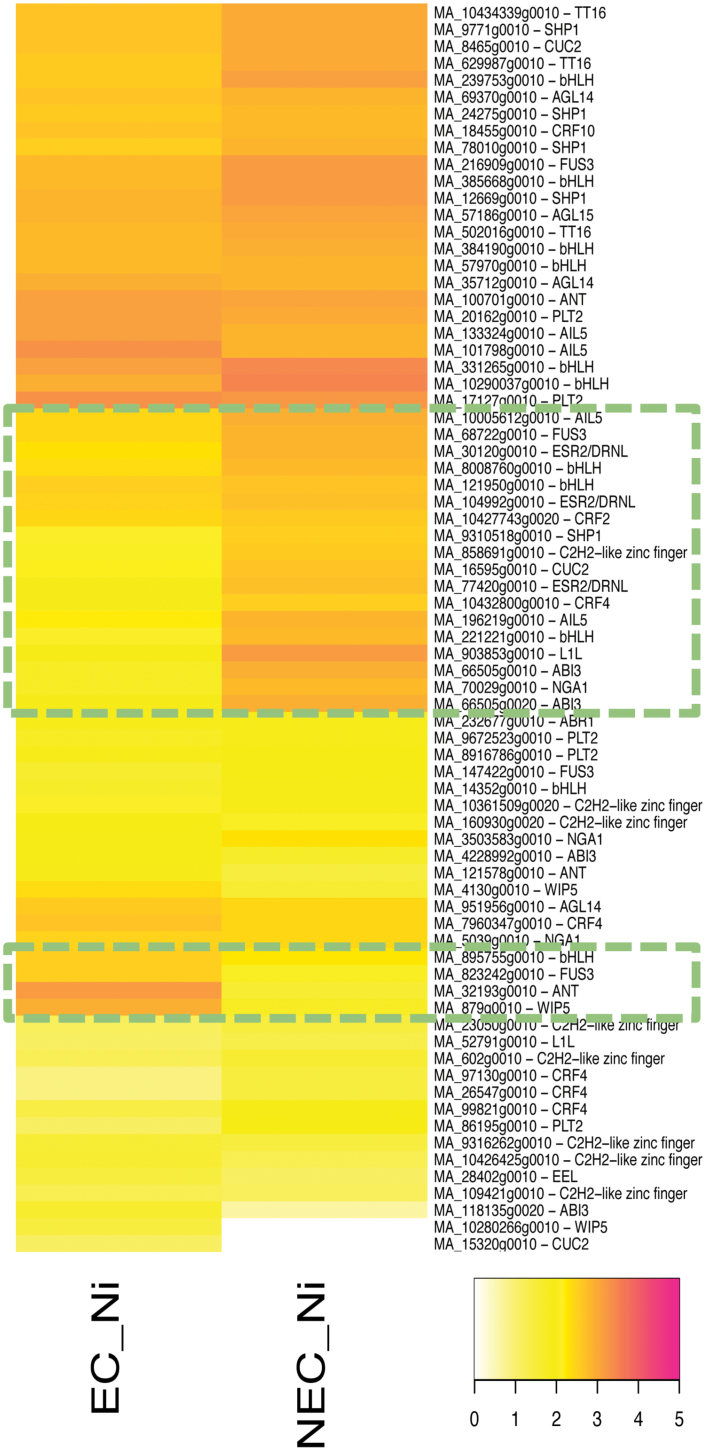
Studies in angiosperm species have identified several key regulators of somatic embryogenesis, among which are PLETHORA 4/BABY BOOM (PLT4/BBM) and PLETHORA 5/EMBRYO MAKER (PLT5/EMK), which are AP2-type transcription factors (Boutilier et al., 2002; Tsuwamoto et al., 2010). In addition, AGAMOUS-LIKE 15 (AGL15) and the LEAFY COTYLEDON genes LEC1 and LEC2 are also known embryonic markers, and overexpression of these genes increases the embryogenic potential of Arabidopsis vegetative tissues (Lotan et al., 1998; Stone et al., 2001; Harding et al., 2003; Thakare et al., 2008). Beside these genes, we also investigated homologues of other somatic embryogenesis-related transcription factors that were identified in a transcriptome analysis of Arabidopsis somatic embryogenesis (Wickramasuriya & Dunwell, 2015; Mozgová et al., 2017). To explore key regulators of somatic embryogenesis competency in gymnosperms, we compared the levels of H3K27me3 in somatic embryogenesis-related gene homologues, between non-induced NEC and EC samples (Fig. 5). Nearly half of the genes showed high H3K27me3 levels in both non-induced NEC and EC, whereas, gene subsets containing homologues of ABI3 (MA_66505g0020), DRNL (MA_30120g0010, MA_104992g0010, MA_77420g0010), CRF4 (MA_10432800g0010), CUC2 (MA_16595g0010), and FUS3 (MA_68722g0010) accumulated higher levels of this histone mark in NEC. Interestingly, a second homologue of FUS3 (MA_823242g0010) and WIP5 (MA_849g0010), as well as one of the ANT homologues (MA_32193g0010) had higher H3K27me3 levels in EC (Fig. 5). H3K27me3 modification is generally associated with gene repression. Consistently, expression of genes with high H3K27me3 in NEC was strongly repressed in NEC (see Supplementary Fig. S5). Likewise, genes with high H3K27me3 in EC were transcriptionally repressed in EC (Supplementary Fig. S5). Thus, many of the somatic embryogenesis-related gene homologues showed differential H3K27me3 enrichment between NEC and EC, possibly explaining the difference of somatic embryogenesis potential among these tissues.
Fig. 5.
Differential accumulation of H3K27me3 on homologues of genes related to somatic embryogenesis. Heatmap shows the H3K27me3 level of homologous genes of transcription factors highly expressed in Arabidopsis somatic embryogenesis.
Discussion
CUT&RUN as a suitable tool to profile large genomes
So far, most studies in chromatin profiling in plants have been limited to relatively small genomes (e.g. different angiosperms, a moss, and a liverwort) (Zhang et al., 2007; Widiez et al., 2014; Hussey et al., 2017; Latrasse et al., 2017; Chica et al., 2017; Lü et al., 2018; Montgomery et al., 2020). However, genome-wide analysis in gymnosperms has remained a challenge due to the large genome sizes of these species (Pellicer et al., 2018). Here, we successfully applied the CUT&RUN method to perform a genome-wide detection of H3K27me3 in Norway spruce. The lack of a complete genome assembly prevents us from providing a chromosomal map of H3K27me3 distribution; nevertheless, we identified genes marked by this modification and can show that the H3K27me3 modification is localized in genic regions of the Norway spruce genome, similar to what is observed in other plant species (Fig. 2). Our validation of the CUT&RUN method will facilitate an in-depth analysis of the distribution of H3K27me3 once a complete spruce genome assembly is available and open the door for epigenetic profiling studies in other gymnosperm species.
Conservation of H3K27me3 targets in Norway spruce and angiosperms
We found that some H3K27me3 target genes are similar between Arabidopsis and Norway spruce, supporting the conservation of the epigenetic mechanisms regulating these genes. At the same time, gymnosperm-specific genes also show H3K27me3, suggesting that H3K27me3 is an important epigenetic player in the regulation of species-specific pathways (Fig. 3C). Although the cis-elements associated with H3K27me3 deposition in Norway spruce are not similar to those described in Arabidopsis, AT-rich motifs and REF6 DNA-binding motifs were detected in our dataset. This co-localization of REF6 binding motif-like sequences and H3K27me3 suggests that, similarly to Arabidopsis, the H3K27me3 catalysis mechanism could be based on cis-regulatory sequences. Nevertheless, this remains to be formally tested: considering that the H3K27me3 distribution is usually broader than the PRC2 binding site itself (Xiao et al., 2017), the H3K27me3 signal peak might not be a good predictor for cis-regulatory elements for H3K27me3 target genes. Therefore, a high-quality genome assembly and direct binding assays using PRC2 components will provide a better resolution for the identification of cis-regulatory elements such as the PREs identified in Arabidopsis. A previous study of chromatin conformation in a relatively large genome revealed that H3K27me3 is associated with chromatin domain borders (Dong et al., 2017). Further analysis by Hi-C could provide a more complete view of H3K27me3 function in this large genome.
H3K27me3 target genes underlie differences in somatic embryogenesis potential
Previous empirical evidence showed that non-induced EC and NEC have different embryogenic potentials. This study validates these observations at the molecular level and adds a possible functional explanation. Here we detected that non-induced EC and NEC had largely different genes being marked by H3K27me3 (Fig. 4), and that non-induced NEC has a H3K27me3 profile that more closely resembles a seedling hypocotyl, indicating that the H3K27me3 profiles might be inherited from original explants to some extent. Although we could not determine the H3K27me3 profile of immature zygotic embryos due to limited source material, we speculate that, as in NEC, the low H3K27me3 observed in EC could be due to an already low accumulation of H3K27me3 in immature zygotic embryos. The megaspore mother cell of Arabidopsis has lower H3K27me3 than the surrounding sporophytic cells, suggesting that there is a reduction of H3K27me3 during female gametogenesis (She et al., 2013). Additionally, in Arabidopsis, it has been shown that PRC2 depletion followed by 2,4-D exposure is required for efficient somatic embryogenesis (Mozgová et al., 2017). Our observations that EC contains reduced levels of H3K27me3 and that a preculturing in 2,4-D-containing medium is essential for somatic embryogenesis in Norway spruce show that this developmental process might be similarly controlled in angiosperms and gymnosperms. Furthermore, it points to 2,4-D as an important component that has the ability to promote chromatin reprogramming in an already predisposed tissue, such as EC, allowing for the somatic embryogenesis program to be completed.
In some angiosperm species, it has been reported that heterochromatic epigenetic marks are associated with SE initiation rate. For example, perturbations of DNA methylation or H3K9me2 by chemicals facilitate embryogenesis from microspores in Brassica napus and Hordeum vulgare (Solís et al., 2015; Berenguer et al., 2017). A recent study reported that DNA methylation on the target sequences of REF6, which is a H3K27me3 demethylase, decreases REF6 binding (Qiu et al., 2019). If the global reduction of H3K27me3 in EC is associated with histone demethylase activity, it would be interesting to investigate whether loss of DNA methylation might be required preceding H3K27me3 reduction.
Associations between peroxidase activity and embryogenic potential have been observed in both angiosperms and gymnosperms (Kochba et al., 1977; Hrubcová et al., 1994; Kormutak & Vookova, 2001; Neves et al., 2003; Gallego et al., 2014). It is therefore interesting that GO terms related to (per)oxidase activities were enriched in response to somatic embryo induction (Table 3). Also genes involved in cell wall modification were significantly enriched among genes responding to somatic embryo induction (Table 3). Cell wall modification and the resulting cell wall dynamics are likely preceding organ initiation (Peaucelle et al., 2011; Francoz et al., 2015). Thus, the observed changes in H3K27me3 foreshadow the developmental processes of somatic embryo induction.
Generally, somatic embryogenesis takes 6–8 weeks after somatic embryo induction. We showed that both EC and NEC altered their H3K27me3 profiles only 1 week after somatic embryo induction, and that NEC and EC showed a different H3K27me3 accumulation profile in response to somatic embryo induction. While the H3K27me3 profile of induced EC resembled that of non-induced EC, NEC showed drastic changes in H3K27me3 target genes after induction. Considering that NEC reduces its proliferation and turns brown 2–3 weeks after SE induction, these large changes in the H3K27me3 profile might reflect a senescence program of the cells (Fig. 1). Indeed, in Arabidopsis, H3K27me3 is also associated with senescence-regulated genes (Ay et al., 2009; Brusslan et al., 2012).
In this study, we demonstrated that the genome-wide level of H3K27me3 in EC is extensively lower than in NEC and seedlings. Low H3K27me3 levels, together with the action of phytohormones such as 2,4-D might be the essential qualifications ensuring somatic embryogenesis potential in gymnosperms as well as in angiosperms (Mozgová et al., 2017). Validation of this hypothesis, coupled with further studies of the genes affected by H3K27me3 during somatic embryogenesis, could open the possibility of manipulating this developmental process, increasing the efficiency of somatic embryogenesis protocols, or even improving the competency of recalcitrant tissues.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Morphology of EC and NEC 2 weeks after somatic embryo induction.
Fig. S2. Percentage of genes with H3K27me3 modification in each auxin-related gene family.
Fig. S3. Phylogenetic analysis of putative H3K27me3 methyltransferases in gymnosperms.
Fig. S4. Identification of cis-element sequences associated with H3K27me3 peaks.
Fig. S5. Expression of representative SE-related genes is anti-correlated with their H3K27me3 level.
Table S1. Sequencing data list.
Table S2. List of primers used in this study.
Acknowledgements
The embryogenic culture cell line (EC) 11:18:1:1 (Picea abies L. Karst) was kindly provided by Dr Sara von Arnold. We thank the Henikoff lab for sharing pA-MNase and help to establish the CUT&RUN technique. Part of the computational work was performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at Uppmax (projects b2012015 and snic2017-7-223). Thanks to Dr M. R. Hatorangan, Dr J. Santos-González, and Dr M. S. Trejo-Arellano for technical advice on bioinformatic analyses. This work was supported by Svenska Forskningsrådet Formas (grant no. 2012-1519, 2016-00453 to LH), by the Knut och Alice Wallenbergs Stiftelse (grant no. 2012.0087 to LH), and by the Scandinavia-Japan Sasakawa Foundation (grant 16-13 to MN).
Author contributions
MN and LH designed the experiments and developed the H3K27me3 signal calling framework; MN performed the experiments and analysed data; RB and CK assisted in analysis and interpretation of data; MN, RB, and CK wrote the manuscript.
Data availability
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE145331. Mapping data of each tissue are listed in Supplementary Table S1.
References
- Akter A, Takahashi S, Deng W, et al. 2019. The histone modification H3 lysine 27 tri-methylation has conserved gene regulatory roles in the triplicated genome of Brassica rapa L. DNA Research 26, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. 2009. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. The Plant Journal 58, 333–346. [DOI] [PubMed] [Google Scholar]
- Baker K, Dhillon T, Colas I, Cook N, Milne I, Milne L, Bayer M, Flavell AJ. 2015. Chromatin state analysis of the barley epigenome reveals a higher-order structure defined by H3K27me1 and H3K27me3 abundance. The Plant Journal 84, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista RA, Köhler C. 2020. Genomic imprinting in plants—revisiting existing models. Genes & Development 34, 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becwar MR, Wann SR, Johnson MA, Verhagen SA, Feirer RP, Nagmani R. 1988. Development and characterization of in vitro embryogenic systems in conifers. In: Ahuja MR, ed. Somatic cell genetics of woody plants. Forestry Sciences, Vol 30, Dordrecht: Springer Netherlands, 1–18. [Google Scholar]
- Berenguer E, Bárány I, Solís MT, Pérez-Pérez Y, Risueño MC, Testillano PS. 2017. Inhibition of histone H3K9 methylation by BIX-01294 promotes stress-induced microspore totipotency and enhances embryogenesis initiation. Frontiers in Plant Science 8, 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, et al. 2002. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. The Plant Cell 14, 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Roudier F, Heese M, et al. 2011. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genetics 7, e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusslan JA, Rus Alvarez-Canterbury AM, Nair NU, Rice JC, Hitchler MJ, Pellegrini M. 2012. Genome-wide evaluation of histone methylation changes associated with leaf senescence in Arabidopsis. PLoS One 7, e33151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. 2004. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276. [DOI] [PubMed] [Google Scholar]
- Chica C, Louis A, Roest Crollius H, Colot V, Roudier F. 2017. Comparative epigenomics in the Brassicaceae reveals two evolutionarily conserved modes of PRC2-mediated gene regulation. Genome Biology 18, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Tu X, Chu PY, Lü P, Zhu N, Grierson D, Du B, Li P, Zhong S. 2017. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Molecular Plant 10, 1497–1509. [DOI] [PubMed] [Google Scholar]
- Filonova LH, Bozhkov PV, von Arnold S. 2000. Developmental pathway of somatic embryogenesis in Picea abies as revealed by time-lapse tracking. Journal of Experimental Botany 51, 249–264. [DOI] [PubMed] [Google Scholar]
- Francoz E, Ranocha P, Nguyen-Kim H, Jamet E, Burlat V, Dunand C. 2015. Roles of cell wall peroxidases in plant development. Phytochemistry 112, 15–21. [DOI] [PubMed] [Google Scholar]
- Gallego P, Martin L, Blazquez A, Guerra H, Villalobos N. 2014. Involvement of peroxidase activity in developing somatic embryos of Medicago arborea L. Identification of an isozyme peroxidase as biochemical marker of somatic embryogenesis. Journal of Plant Physiology 171, 78–84. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE. 2003. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiology 133, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L. 2012. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genetics 8, e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M. 2009. Diversity of Polycomb group complexes in plants: same rules, different players? Trends in Genetics 25, 414–423. [DOI] [PubMed] [Google Scholar]
- Hrubcová M, Cvikrová M, Eder J. 1994. Peroxidase activities and contents of phenolic acids in embryogenic and nonembryogenic alfalfa cell suspension cultures. Biologia Plantarum 36, 175. [Google Scholar]
- Hussey SG, Loots MT, van der Merwe K, Mizrachi E, Myburg AA. 2017. Integrated analysis and transcript abundance modelling of H3K4me3 and H3K27me3 in developing secondary xylem. Scientific Reports 7, 3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain SM, Newton RJ, Soltes EJ. 1988. Enhancement of somatic embryogenesis in Norway spruce (Picea abies L.). Theoretical and Applied Genetics 76, 501–506. [DOI] [PubMed] [Google Scholar]
- Klimaszewska K, Hargreaves C, Lelu-Walter MA, Trontin JF. 2015. Advances in conifer somatic embryogenesis since year 2000. In: Germana M, Lambardi M, eds. In vitro embryogenesis in higher plants. Methods in Molecular Biology, Vol 1359. New York: Humana Press, 131–162. [Google Scholar]
- Kochba J, Lavee S, Spiegel-Roy P. 1977. Differences in peroxidase activity and isoenzymes in embryogenic and non-embryogenic ‘Shamouti’ orange ovular callus lines. Plant and Cell Physiology 18, 463–467. [Google Scholar]
- Kormutak A, Vookova B. 2001. Peroxidase activity in non-embryogenic and embryogenic calli and in developing somatic embryos of white fir (Abies concolor Gord. et Glend). Plant Biosystems 135, 101–105. [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. 2011. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genetics 7, e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Rodriguez-Granados NY, Veluchamy A, et al. 2017. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics & Chromatin 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang M, Lin K, et al. 2019. The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biology 20, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhou S, Wang W, Ye Y, Zhao Y, Xu Q, Zhou C, Tan F, Cheng S, Zhou DX. 2015. Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem. The Plant Cell 27, 1428–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. 1998. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lü P, Yu S, Zhu N, et al. 2018. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nature Plants 4, 784–791. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Eichten SR, Briskine R, Waters AJ, Danilevskaya ON, Meeley RB, Myers CL, Vaughn MW, Springer NM. 2013. Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. The Plant Cell 25, 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulski P, Komarynets O, Fachinelli F, Weber APM, Schubert D. 2017. Characterization of the Polycomb-group mark H3K27me3 in unicellular algae. Frontiers in Plant Science 8, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SA, Tanizawa Y, Galik B, et al. 2020. Chromatin organization in early land plants reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Current Biology 30, 573–588.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgová I, Muñoz-Viana R, Hennig L. 2017. PRC2 represses hormone-induced somatic embryogenesis in vegetative tissue of Arabidopsis thaliana. PLoS Genetics 13, e1006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagmani R, Becwar MR, Wann SR. 1987. Single-cell origin and development of somatic embryos in Picea abies (L.) Karst. (Norway spruce) and P. glauca (Moench) Voss (white spruce). Plant Cell Reports 6, 157–159. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Köhler C, Hennig L. 2019. Tissue-specific transposon-associated small RNAs in the gymnosperm tree, Norway spruce. BMC Genomics 20, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves N, Segura-Nieto M, Blanco MA, Sánchez M, González A, González JL, Castillo R. 2003. Biochemical characterization of embryogenic and non-embryogenic calluses of sugarcane. In Vitro Cellular & Developmental Biology – Plant 39, 343–345. [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H. 2011. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Current Biology 21, 1720–1726. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Hidalgo O, Dodsworth S, Leitch IJ. 2018. Genome size diversity and its impact on the evolution of land plants. Genes 9, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Li Z, Liu C, et al. 2018. CGT-seq: epigenome-guided de novo assembly of the core genome for divergent populations with large genome. Nucleic Acids Research 46, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Mei H, Deng X, He K, Wu B, Yao Q, Zhang J, Lu F, Ma J, Cao X. 2019. DNA methylation repels targeting of Arabidopsis REF6. Nature Communications 10, 2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramarosandratana AV, Van Staden J. 2003. Tissue position, explant orientation and naphthaleneacetic acid (NAA) affect initiation of somatic embryos and callus proliferation in Norway spruce (Picea abies). Plant Cell, Tissue and Organ Culture 74, 249–255. [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Research 42, W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ Jr. 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biology 6, R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nature Reviews. Genetics 8, 9–22. [DOI] [PubMed] [Google Scholar]
- She W, Grimanelli D, Rutowicz K, Whitehead MW, Puzio M, Kotlinski M, Jerzmanowski A, Baroux C. 2013. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development 140, 4008–4019. [DOI] [PubMed] [Google Scholar]
- Shu H, Nakamura M, Siretskiy A, Borghi L, Moraes I, Wildhaber T, Gruissem W, Hennig L. 2014. Arabidopsis replacement histone variant H3.3 occupies promoters of regulated genes. Genome Biology 15, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís MT, El-Tantawy AA, Cano V, Risueño MC, Testillano PS. 2015. 5-Azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Frontiers in Plant Science 6, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. 2001. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences, USA 98, 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Gordon SP, Meyerowitz EM. 2011. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends in Cell Biology 21, 212–218. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM. 2010. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Developmental Cell 18, 463–471. [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakare D, Tang W, Hill K, Perry SE. 2008. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiology 146, 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Yokoi S, Takahata Y. 2010. Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Molecular Biology 73, 481–492. [DOI] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K. 2018. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research 46, D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen SA, Wann SR. 1989. Norway spruce somatic embryogenesis: high-frequency initiation from light-cultured mature embryos. Plant Cell, Tissue and Organ Culture 16, 103–111. [Google Scholar]
- von Arnold S, Clapham D. 2008. Spruce embryogenesis. Methods in Molecular Biology 427, 31–47. [DOI] [PubMed] [Google Scholar]
- von Arnold S, Egertsdotter U, Ekberg I, Gupta P, Mo H, Nörgaard J. 1995. Somatic embryogenesis in Norway spruce (Picea Abies). In: Jain SM, Gupta PK, Newton RJ, eds. Forestry sciences. Somatic embryogenesis in woody plants. Vol 3, Gymnosperms. Dordrecht: Springer Netherlands, 17–36. [Google Scholar]
- Wickramasuriya AM, Dunwell JM. 2015. Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genomics 16, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widiez T, Symeonidi A, Luo C, Lam E, Lawton M, Rensing SA. 2014. The chromatin landscape of the moss Physcomitrella patens and its dynamics during development and drought stress. The Plant Journal 79, 67–81. [DOI] [PubMed] [Google Scholar]
- Xiao J, Jin R, Yu X, et al. 2017. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nature Genetics 49, 1546–1552. [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. 2007. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biology 5, e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang Y, Krause K, Yang T, Dongus JA, Zhang Y, Turck F. 2018. Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nature Genetics 50, 638–644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE145331. Mapping data of each tissue are listed in Supplementary Table S1.