Abstract
Chloroplast genomes (cpDNA) in angiosperms are usually highly conserved. Although rearrangements have been observed in some lineages, such as Passiflora, the mechanisms that lead to rearrangements are still poorly elucidated. In the present study, we obtained 20 new chloroplast genomes (18 species from the genus Passiflora, and Dilkea retusa and Mitostemma brevifilis from the family Passifloraceae) in order to investigate cpDNA evolutionary history in this group. Passiflora cpDNAs vary in size considerably, with ∼50 kb between shortest and longest. Large inverted repeat (IR) expansions were identified, and at the extreme opposite, the loss of an IR was detected for the first time in Passiflora, a rare event in angiosperms. The loss of an IR region was detected in Passiflora capsularis and Passiflora costaricensis, a species in which occasional biparental chloroplast inheritance has previously been reported. A repertory of rearrangements such as inversions and gene losses were detected, making Passiflora one of the few groups with complex chloroplast genome evolution. We also performed a phylogenomic study based on all the available cp genomes and our analysis implies that there is a need to reconsider the taxonomic classifications of some species in the group.
Keywords: chloroplast genome (plastome) rearrangements, Passiflora, inverted repeat (IR) loss, Passiflora classification
Significance
Chloroplast genomes (plastomes) have been extensively investigated, and wide plastome diversity has been reported to occur in Passiflora. This genus has long attracted attention due to its broad geographic distribution and remarkable species diversity, particularly with regard to flower morphology. There are around 500 species of Passiflora, most of which are distributed pantropically. In this study, we sequenced, assembled, and annotated new plastomes. Our analysis revealed some interesting findings. First, extensive sequence rearrangements were confirmed. Most noteworthy of all, the two smallest chloroplast genomes in Passiflora were found to exhibit the loss of a region, an inverted repeat region, that is a rare event among angiosperms. Finally, we provided one of the first pictures into the evolutionary history of plastome structure in this genus.
Introduction
In higher plants, most chloroplast (cp) genomes have a quadripartite structure consisting of two copies of inverted repeats (IRs) separating two single copy regions, a large (LSC) and a small (SSC) region (Sugiura 1992; Yang et al. 2010). Although highly conserved in their structural organization, cp genomes may exhibit deletions, including gene loss, and rearrangements, such as IR inversions and expansions or retractions. These rearrangements are well documented, for instance, in Hevea brasiliensis (Euphorbiaceae) (Tangphatsornruang et al. 2011) and in species of riverweeds (Podostemaceae) (Bedoya et al. 2019), both families belonging to the Malpighiales. Some other plant families, such as Fabaceae (Cai et al. 2008), Campanulaceae (Haberle et al. 2008), Pinaceae and other conifers (Wu, Lin, et al. 2011; Wu, Wang, et al. 2011), and Geraniaceae (Guisinger et al. 2011; Weng et al. 2014), also exhibit a high number of rearrangements in their cp genomes.
However, the mechanisms which lead to rearrangements in plastid genomes (plastomes) are still poorly elucidated. Among these mechanisms, intramolecular homologous recombination mediated by the presence of repeat structures at the boundaries of the rearranged region plays a role in bringing about structural changes (Ogihara et al. 1988; Wu, Lin, et al. 2011), as well as recombination between tRNA genes (Haberle et al. 2008). In addition, foreign DNA insertions (large open reading frames, for instance) have led to extensive cp genome rearrangements in Campanulaceae sensu stricto (Knox 2014). Interestingly, for some angiosperms in which highly rearranged cpDNAs occur, occasional biparental chloroplast inheritance has been reported, as in Campanulaceae (Cosner et al. 2004; Haberle et al. 2008; Barnard-Kubow et al. 2017) and Geraniaceae (Metzlaff et al. 1981; Chumley et al. 2006; Weng et al. 2014).
In the genus Passiflora (Passifloraceae, Malpighiales), the results of artificial intraspecific crosses revealed the prevalence of maternal and the potential for biparental chloroplast inheritance, whereas interspecific crosses in turn showed the occurrence of paternal chloroplast inheritance (Muschner et al. 2006; Hansen et al. 2007). Passiflora could be regarded as an excellent model for studying the evolution of chloroplast genomes. Apart from the potential for different patterns of chloroplast inheritance, the genus has been described as having a syndrome of features related to chloroplast genome changes (Shrestha et al. 2019). Rearrangements in the cpDNA structure, such as inversions and gene losses, have been reported for the first time in Passiflora edulis (Cauz-Santos et al. 2017), but in later studies, rearrangements were also identified in different Passiflora species (Rabah et al. 2019; Shrestha et al. 2019).
To place this in context, Passiflora is the richest genus in the Passifloraceae family, consisting of some 520 species, popularly known as passionflowers, with great diversity in the size and shape of flowers pollinated by insects, hummingbirds, or bats. The geographical distribution of passionflowers is mainly Neotropical, and they are found mainly in the South and Central Americas. However, occurrences have been documented in Southeast Asia, Oceania, and Australia (Ulmer and MacDougal 2004). The Brazilian biomes (Amazon, Caatinga, Cerrado, Atlantic forest, Pampa, and Pantanal) harbor 147 species, including 85 that are endemic to the country. Some species are cultivated for medicinal (e.g., Passiflora incarnata) and ornamental purposes due to their exotic flowers, but most are for fresh fruit consumption and industrialized juice production (e.g., P. edulis). Morphological characters have been used to subdivide the classical intrageneric division of the genus into 22 subgenera (Killip 1938). Nowadays, the most well-accepted classification has reduced the number of subgenera to four: Astrophea (57 species), Decaloba (214), Deidamioides (13), and Passiflora (236) (Ulmer and MacDougal 2004). Phylogenetic studies have also confirmed the subdivision of Passiflora into four subgenera (Muschner et al. 2003, 2012; Hansen et al. 2006). However, the position of the subgenus Deidamioides is poorly resolved and it has been recognized as a paraphyletic group (Muschner et al. 2003, 2012), suggesting that further analysis is needed using phylogenomic approaches and larger volumes of data (e.g., a set of chloroplast genes).
In our study, we sequenced, assembled, and annotated 18 chloroplast genomes representing the four subgenera in the genus Passiflora, along with the Passifloraceae Dilkea retusa and Mitostemma brevifilis, and extensive sequence rearrangements were found. We were able to address the following questions: Are the frequency and type of cpDNA sequence rearrangements particular to each subgenus? And what is the significance of these rearrangements for Passiflora intrageneric classification?
Materials and Methods
Plant Material
In total, 18 species of the genus Passiflora were analyzed, belonging to the four subgenera: Astrophea (3), Decaloba (5), Deidamioides (2), and Passiflora (8). Of these species, 17 were field collected, and Passiflora costaricensis (Decaloba), a species native to Central America, was obtained from the Italian National Collection of Passiflora. In addition, two species of Passifloraceae (D. retusa and M. brevifilis) were also field collected and comparative studies conducted (supplementary table S1, Supplementary Material online). All plant accessions are registered in the Sistema Nacional de Gestão do Patrimônio Genético e do Conhecimento Tradicional Associado (SISGEN, Brazil).
Intact Chloroplast Isolation by Sucrose Gradient Centrifugation and cpDNA Extraction
Intact chloroplast organelles were isolated using the sucrose gradient method (Takamatsu et al. 2018). Some 20 g of fresh leaves from each plant were frozen in liquid nitrogen and macerated. The material was resuspended in 200 ml of isolation buffer (50 mM Tris–HCl [pH 8.0], 0.35 M sucrose, 7 mM ethylenediaminetetraacetic acid, 5 mM 2-mercaptoethanol, and 0.1% bovine serum albumin) and incubated for 10 min in the dark. The suspension was filtered through two layers of gauze and then two layers of Miracloth (Merck), and the filtrate centrifuged at 1,000 × g for 10 min. Finally, the pellet was washed in 50 ml of isolation buffer and centrifuged at 1,000 × g for 10 min.
The pellet was resuspended in 5 ml of isolation buffer and the suspension slowly laid out in a 20/45% sucrose density batch gradient in 50 mM Tris–HCl (pH 8.0), 0.3 M sorbitol, and ethylenediaminetetraacetic acid 7 mM. The gradient was centrifuged at 2,000 × g for 30 min. The green band formed at the interface containing the intact chloroplasts was collected, diluted with three volumes of isolation buffer, and centrifuged at 3,000 × g for 10 min to obtain the purified chloroplasts in the pellet.
The pellet was resuspended in 2% CTAB buffer to promote lysis. The suspension was then incubated at 65 °C for 1 h with stirring. The supernatant was extracted 2× with an equal volume of chloroform:isoamyl alcohol (24:1) and centrifuged at 10,000 × g for 20 min. An equal volume of isopropanol was added to the upper layer and incubated at 20 °C for 1 h. Finally, the aqueous phase was centrifuged at 10,000 × g for 20 min, and the cpDNA pellet washed with ethanol (70%), dried, and resuspended in 40 μl of Tris–ethylenediaminetetraacetic acid buffer.
Chloroplast Genome Sequencing and Assembly
Illumina sequencing libraries were constructed using a total of 100 ng of input cpDNA and the Nextera DNA Flex library Kit, following the manufacturer’s instructions. The sequencing was performed on the Illumina NextSeq platform (Fundação Hemocentro de Ribeirão Preto, Brazil) in two distinct runs, the first containing six species using paired-end sequencing (2× 76 bp), and the second 13 species using paired-end sequencing (2× 150 bp) (supplementary table S2, Supplementary Material online). The paired-end reads were initially trimmed and filtered in Trimmomatic v. 0.39 (Bolger et al. 2014). Adapter trimming was performed based on ILLUMINACLIP:NexteraPE-PE.fa:2:30:10:2, and using the following quality filtering parameters: sliding window of 10:20, leading of 20, trailing of 20, and minimum length of 36 bp.
The filtered reads were de novo assembled in NOVOPlasty v. 3.8.3 (Dierckxsens et al. 2017), using the protein-coding genes psbA, rbcL, and rpoB from the P. edulis cp genome (NC_034285.1) as seeds. The k-mer sizes used in the assembly varied from 23 to 39 (supplementary table S2, Supplementary Material online). In order to obtain the final assembly, and taking into account that there were overlapping regions, the cp sequences were merged in Geneious v. 2019.1.2 (https://www.geneious.com, last accessed April 2020) using the “de novo Assembly” function. Finally, the correctness of the assembly was checked, and coverage estimated using the “Map to reference” function to map the paired-end raw reads onto the final assembled cp genome.
The plastid DNA sequence of P. costaricensis was obtained by long-read sequencing. Barcoded, large-insert (10 kb) libraries were constructed using 150 ng of pure high-molecular-weight DNA. Sequencing was performed in an SMRT cell using P6 polymerase with C4 chemistry on the PacBio RSII instrument at the Uppsala NGI Platform (Uppsala University, Sweden). The data were filtered to obtain high-quality reads (reads with quality < 0.75 and length < 500 bp were removed). The sequences were assembled using the CANU assembler with default parameters (Koren et al. 2017). Subsequently, the chloroplast contigs were extracted in Geneious by mapping the resulting contigs to the Passiflora complete cp genomes obtained in this study. A final assembled cp genome of P. costaricensis was obtained in Geneious by joining the contigs using the “de novo Assembly” function (supplementary table S2, Supplementary Material online). Finally, the raw reads were mapped onto the final contig using the “Map to reference” function.
Primer design and polymerase chain reaction (PCR) were applied to confirm the positioning of some distinct cpDNA arrangements (supplementary table S3, Supplementary Material online). Amplification reactions were performed using 20 ng template DNA, 1× buffer, 1 mM MgCl2, 0.2 mM of each dNTP, 0.3 μM of the forward and reverse primers, 1.2 U Go Taq Flex DNA polymerase (Promega, Madison, WI), and ultra-pure water added to bring the final volume up to 20 μl. The amplification program was as follows: 95 °C for 5 min, 35 cycles at 95 °C for 40 s, 55 °C for 40 s, and 72 °C for 1 min, followed by a final 8 min of incubation at 72 °C. The amplified fragments were checked on 1% (w/v) agarose gel with a 1,000-bp molecular standard Invitrogen ladder (Carlsbad, CA).
Chloroplast Genome Annotation, Identification of Repeated Elements, and Comparative Analysis
The annotation of cp genomes was carried out in the GeSeq (Organellar Genome Annotation) online program (Tillich et al. 2017) with default settings to identify protein-coding gene sequences, rRNAs, and tRNAs based on the chloroplast reference sequences and BLAST homology searches, followed by manual corrections for start and stop codons and intron positions in GenomeView software. All tRNA genes were further confirmed using the tRNAscan-SE and ARAGORN online search server (Laslett and Canback 2004; Lowe and Chan 2016). Pseudogenes were classified based on nucleotide losses in sequences or the presence of internal stop codons. OGDRAW software was used for constructing the circular cp genome map (Greiner et al. 2019).
REPuter software (Kurtz et al. 2001) was used to identify direct and palindromic repeat elements based on the following criteria: minimum repeat size ≥30 bp and sequence identities ≥90% (Hamming distance equal to 3).
Two different multiple sequence alignments were run in Progressive Mauve v.2.4.0 (Darling et al. 2010) to identify possible rearrangements in the cpDNA molecules. The first contained 11 cp genomes to represent the diversity in size and structure in each Passiflora subgenus, in addition to the cp genomes of D. retusa and M. brevifilis (Passifloraceae), with Populus trichocarpa (Salicaceae) as a reference. The second alignment contained all 20 cp genomes studied herein. Subsequently, to detect possible expansions or contractions in the IR regions, the IR boundaries of both LSC and SSC regions with full annotations for the adjacent genes were analyzed across the cp molecules used in the first multiple sequence alignment. In addition to reconstructing the evolutionary history of Passiflora chloroplast genomes, we analyzed the plastome structures run in the first multiple sequence alignment against the available GenBank cp genomes of Passiflora foetida, Passiflora tetrandra, Passiflora obovata, Passiflora auriculate, and Passiflora biflora, studied by Shrestha et al. (2019) (supplementary table S4, Supplementary Material online).
Phylogenomic Studies
We performed a phylogenomic study based on all the available cp genomes of passionflowers (49 species in total, 20 generated in this study and 29 species whose cp DNA sequences were imported from the GenBank database, supplementary table S4, Supplementary Material online).
A set of 68 passionflower chloroplast protein-coding genes was used in a phylogenetic analysis, and the species Po. trichocarpa (Salicaceae) used as the outgroup, with the aim of obtaining a rooted tree (supplementary table S5, Supplementary Material online).
A local python pipeline was used to extract each gene in the data set (consisting of 50 taxa), align them individually at nucleotide level in MUSCLE and make an interleaved Nexus matrix consisting of all individual alignments. The resulting matrix was then analyzed in ModelFinder (Kalyaanamoorthy et al. 2017) to determine the best partition scheme and evolutionary models in accordance with the Akaike information criterion (AIC) , and treating each gene alignment as a charset. Because the partition scheme indicated by ModelFinder is an a priori test based on point estimates, and a single model was selected with GTR + G + I for the whole data set, we decided to include an alternative model with partition based on codon position with separate parameters in GTR + G + I for each codon position, and then subsequently test the two-partition scheme using Bayes Factors. In both partitioned and nonpartitioned analyses, the Markov chain Monte Carlo algorithm was run in MrBayes (Ronquist et al. 2012) for 10,000,000 generations, sampling one tree every 1,000 generations and discarding the first 25% of trees as burn-in, to estimate the values of posterior probabilities (PPs). The convergence of the runs was monitored based on an average standard deviation of split frequencies below 0.01, potential scale reduction factor values close to 1.0, and ESS values above 1,000. Finally, the phylogenetic trees were visualized using FigTree version 1.4.4.
Results
Chloroplast Genome Features in Passifloraceae
In Passiflora, a large variation in cpDNA sequence length was observed (∼55 kb), ranging from 113,114 bp in Passiflora capsularis (subgenus Decaloba) to 167,953 bp in Passiflora deidamioides (subgenus Deidamioides) (supplementary fig. S1, Supplementary Material online, and table 1). The majority had the typical quadripartite structure; the length of the LSC region ranged from 57,244 bp in Passiflora suberosa (subgenus Decaloba) to 90,064 bp in Passiflora rhamnifolia (subgenus Astrophea), whereas the SSC region varied from 12,854 in Passiflora cerradensis (subgenus Astrophea) to 13,744 bp in P. deidamioides. IRs ranged from 21,928 in Passiflora edmundoi (subgenus Passiflora) to 43,626 bp in P. suberosa (subgenus Decaloba). The cpDNA sequence lengths of the Passifloraceae D. retusa and M. brevifilis were similar to those of the Astrophea species, which had the shortest SSC region sequenced in this study. The GC content of all 20 cp genomes was very similar (average 36.7%).
Table 1.
Summary of the 20 Passifloraceae Chloroplast Genomes Sequenced
| Species | Taxonomic Classification Subgenus/Supersections, Sections, or Series | cp Genome Size (bp) | LSC (bp) | SSC (bp) | IR (bp) | Total GC % | No. Unique Genes | Protein-Coding Genes | tRNAs | rRNAs | NCBI Accession No. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Passiflora cerradensis | Subgenus Astrophea/section Capreolata | 164,515 | 84,143 | 12,854 | 33,759 | 36.6 | 107 | 73 | 30 | 4 | MT525871 |
| Passiflora haematostigma | Subgenus Astrophea/section Pseudoastrophea | 163,775 | 89,717 | 12,876 | 30,591 | 36.5 | 106 | 72 | 30 | 4 | MT525875 |
| Passiflora rhamnifolia | Subgenus Astrophea/section Pseudoastrophea | 162,217 | 90,064 | 12,921 | 29,616 | 36.4 | 106 | 72 | 30 | 4 | MT525882 |
| Passiflora candollei | Subgenus Decaloba/section Decaloba | 138,081 | 72,565 | 13,506 | 26,005 | 37.2 | 104 | 70 | 30 | 4 | MT525870 |
| Passiflora capsularis | Subgenus Decaloba/section Xerogona | 113,114 | a | a | a | 36.1 | 102 | 68 | 30 | 4 | MT525883 |
| Passiflora costaricensis | Subgenus Decaloba/section Xerogona | 114,230 | a | a | a | 36.1 | 102 | 68 | 30 | 4 | MT473979 |
| Passiflora suberosa | Subgenus Decaloba/section Cieca | 158,313 | 57,244 | 13,817 | 43,626 | 37.2 | 103 | 69 | 30 | 4 | MT525868 |
| Passiflora vespertilio | Subgenus Decaloba/section Decaloba | 138,456 | 72,902 | 13,158 | 26,196 | 37.1 | 104 | 70 | 30 | 4 | MT525880 |
| Passiflora contracta | Subgenus Deidamioides/section Tetrastylis | 166,558 | 87,313 | 13,513 | 32,866 | 36.7 | 107 | 73 | 30 | 4 | MT533196 |
| Passiflora deidamioides | Subgenus Deidamioides/section Deidamioides | 167,953 | 82,571 | 13,744 | 35,819 | 36.8 | 108 | 74 | 30 | 4 | MT525873 |
| Passiflora alata | Subgenus Passiflora/supersection Laurifolia/section Quadrangularis | 147,773 | 85,535 | 13,494 | 24,372 | 36.9 | 105 | 71 | 30 | 4 | MT525869 |
| Passiflora cristalina | Subgenus Passiflora/supersection Distephana | 145,054 | 85,662 | 13,530 | 22,931 | 36.9 | 104 | 70 | 30 | 4 | MT525872 |
| Passiflora edmundoi | Subgenus Passiflora/supersection Stipulata/section Kermesinae | 142,737 | 85,567 | 13,314 | 21,928 | 37.2 | 105 | 71 | 30 | 4 | MT525874 |
| Passiflora loefgrenii | Subgenus Passiflora/supersection Stipulata/section Kermesinae | 146,537 | 86,370 | 13,267 | 23,450 | 37.1 | 104 | 70 | 30 | 4 | MT525876 |
| Passiflora miniata | Subgenus Passiflora/supersection Distephana | 151,920 | 85,863 | 13,477 | 26,290 | 37.0 | 105 | 71 | 30 | 4 | MT525877 |
| Passiflora mucronata | Subgenus Passiflora/supersection Stipulata/section Granadillastrum | 150,839 | 84,839 | 13,552 | 26,224 | 36.9 | 104 | 70 | 30 | 4 | MT525878 |
| Passiflora recurva | Subgenus Passiflora/supersection Passiflora/series Passiflora | 151,837 | 85,863 | 13,504 | 26,235 | 37.0 | 105 | 71 | 30 | 4 | MT525879 |
| Passiflora watsoniana | Subgenus Passiflora/supersection Stipulata/section Kermesinae | 146,520 | 86,139 | 13,355 | 23,513 | 37.0 | 105 | 71 | 30 | 4 | MT525881 |
| Dilkea retusa | Dilkea genus | 161,923 | 88,575 | 12,686 | 30,331 | 36.2 | 109 | 75 | 30 | 4 | MT525866 |
| Mitostemma brevifilis | Mitostemma genus | 163,032 | 88,837 | 12,695 | 30,750 | 36.1 | 109 | 75 | 30 | 4 | MT525867 |
Loss of an IR region.
Comparing the four subgenera, Decaloba was the subgenus with the highest variation, ranging from 113,114 bp in P. capsularis and 114,230 in P. costaricensis, the smallest cp molecules sequenced herein due to the loss of an IR region (fig. 1), up to 158,313 bp in P. suberosa. This loss of the IR region in P. capsularis and P. costaricensis, particularly the IRa, was confirmed by mapping the raw reads onto the assembled genomes that resulted in continuity of coverage along the cp genome molecule. In addition, the IR loss was confirmed by PCR (supplementary table S3, Supplementary Material online). In contrast, the largest cp genomes were observed for the two species of the Deidamioides subgenus. The Astrophea subgenus also had large cpDNA molecules, whereas in the eight species of the Passiflora subgenus (which contains the vast majority of the species described in the genus), the cpDNA ranged from 142,737 bp (P. edmundoi) to 151,920 bp (Passiflora miniata).
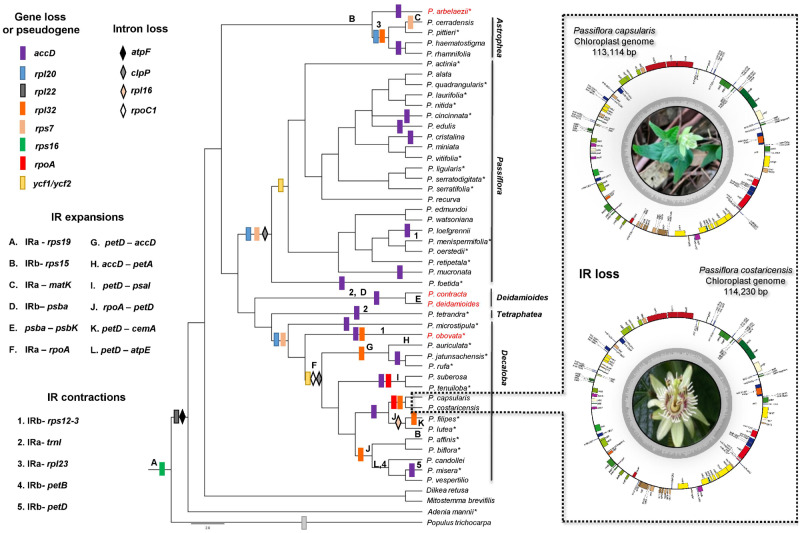
Fig. 1.
Gene losses, IR expansions/contractions mapped onto the cladogram of the Bayesian inference with plastid genes. *Species analyzed by Shrestha et al. (2019). The species in red belong to the subgenus Deidamioides. The chloroplast genomes of Passiflora capsularis and Passiflora costaricensis, the smallest in Passiflora due to the loss of an IR region, are shown on the right. Genes are represented as boxes inside or outside the large circle to indicate clockwise (inside) or counter clockwise (outside) transcription. The flower image of P. costaricensis was kindly provided by Maurizio Vecchia, 2005.
The cp genomes contained between 102 and 109 unique genes, and this variation is related to the protein-coding genes identified when species were compared (68–75 protein-coding genes), because all species were found to have the same tRNA (30) and rRNA (4) gene content. Chloroplast genes are involved in different biological processes and were annotated accordingly with functional categories. Due to duplication and the emergence of IR regions during the evolutionary history of the genus, 18–38 genes were found to have two copies, including protein-coding genes, tRNAs, and all four rRNAs. The total number of genes ranged from 102 (P. capsularis and P. costaricensis) to 141 (P. suberosa), as a result of expansions and retractions of the IR regions, and also the gene losses described below.
Introns were identified in 10–15 sequences of protein-coding and tRNA genes, mainly in the Astrophea and Deidamioides subgenera (15 introns). The intron within the atpF gene was not found in the Passifloraceae species analyzed in this study, but it was found in Po. trichocarpa, the species used in the comparative analysis (fig. 1). Additionally, the Astrophea and Deidamioides subgenera harbored an intron in the clpP gene, which was not found in the Decaloba and Passiflora subgenera, revealing the loss of this region. In the Decaloba subgenus, we found the lowest number of introns, because of losses in the rpoC1 and rpl16 genes.
Repetitive sequence analysis detected between 115 (Passiflora alata) and 445 (Passiflora contracta) repeats (table 2). The majority were in forward orientation (ranging from 65 repeats in P. cerradensis to 286 in Passiflora haematostigma), followed by palindromic repeats (ranging from 22 repeats in P. edmundoi and Passiflora loefgrenii to 183 in P. contracta). The length of the repeats varied between 116 and 1,070 bp, respectively, in Passiflora miniata and P. costaricensis. In most of the species, the highest number of repeat structures was found in the LSC region, with some detected in the IR and SSC regions (supplementary table S6, Supplementary Material online). Comparing the subgenera, high numbers of repeats were found in the Deidamioides subgenus (a respective 405 and 445 total repeats in P. deidamioides and P. contracta) and Decaloba subgenus (a respective 199 and 280 total repeats in Passiflora candollei and P. costaricensis) in which the largest repeat (1,070 bp identified in P. costaricensis) possibly corresponds to a piece of the IR region that was lost in this species.
Table 2.
Summary of the Short Direct-Repeats Identified in the cp Genomes of 18 Passiflora Species and the Passifloraceae, Dilkea retusa and Mitostemma brevifilis
| Species | Taxonomic Classification Subgenus/Supersections, Sections, or Series | cp Genome (bp) | REPuter Total Number | Palindromic | Forward | Reverse | Complement | Largest Repeat (bp) |
|---|---|---|---|---|---|---|---|---|
| Passiflora cerradensis | Subgenus Astrophea/section Capreolata | 164,515 | 118 | 38 | 65 | 9 | 6 | 193 |
| Passiflora haematostigma | Subgenus Astrophea/section Pseudoastrophea | 163,775 | 399 | 67 | 286 | 28 | 18 | 476 |
| Passiflora rhamnifolia | Subgenus Astrophea/section Pseudoastrophea | 162,217 | 398 | 67 | 222 | 56 | 53 | 486 |
| Passiflora candollei | Subgenus Decaloba/section Decaloba | 138,081 | 199 | 87 | 112 | 201 | ||
| Passiflora capsularis | Subgenus Decaloba/section Xerogona | 113,114 | 207 | 55 | 155 | 1 | 791 | |
| Passiflora costaricensis | Subgenus Decaloba/section Xerogona | 114,230 | 280 | 38 | 240 | 2 | 1,070 | |
| Passiflora suberosa | Subgenus Decaloba/section Cieca | 158,313 | 279 | 108 | 154 | 9 | 8 | 760 |
| Passiflora vespertilio | Subgenus Decaloba/section Decaloba | 138,456 | 238 | 73 | 164 | 1 | 151 | |
| Passiflora contracta | Subgenus Deidamioides/section Tetrastylis | 166,558 | 445 | 183 | 262 | 427 | ||
| Passiflora deidamioides | Subgenus Deidamioides/section Deidamioides | 167,953 | 405 | 129 | 261 | 15 | 352 | |
| Passiflora alata | Subgenus Passiflora/supersection Laurifolia/section Quadrangulares | 147,773 | 115 | 33 | 78 | 4 | 123 | |
| Passiflora cristalina | Subgenus Passiflora/supersection Distephana | 145,054 | 128 | 37 | 87 | 4 | 123 | |
| Passiflora edmundoi | Subgenus Passiflora/supersection Stipulata/section Kermesinae | 142,737 | 185 | 22 | 145 | 18 | 118 | |
| Passiflora loefgrenii | Subgenus Passiflora/supersection Stipulata/section Kermesinae | 146,537 | 133 | 22 | 104 | 5 | 2 | 137 |
| Passiflora miniata | Subgenus Passiflora/supersection Distephana | 151,920 | 152 | 35 | 114 | 3 | 116 | |
| Passiflora mucronata | Subgenus Passiflora/supersection Stipulata/section Granadillastrum | 150,839 | 172 | 49 | 97 | 22 | 4 | 372 |
| Passiflora recurva | Subgenus Passiflora/supersection Passiflora/series Passiflora | 151,837 | 158 | 41 | 116 | 1 | 143 | |
| Passiflora watsoniana | Subgenus Passiflora/supersection Stipulata/section Kermesinae | 146,520 | 158 | 25 | 128 | 5 | 147 | |
| Dilkea retusa | Dilkea genus | 161,923 | 214 | 91 | 110 | 7 | 6 | 106 |
| Mitostemma brevifilis | Mitostemma genus | 163,032 | 398 | 189 | 209 | 299 |
Most of the repeats were found in the intergenic sequences of the LSC or IR regions, but some repeats were located in gene sequences, including accD, clpP, psaA, psaB, rps18, ycf1, ycf2, and ycf3 (supplementary table S6, Supplementary Material online). Furthermore, in all species analyzed, a repeat was identified in the ndhA gene located in the respective SSC region. Note in particular that a high number of repeats in the accD gene and its promoter or intergenic regions was identified in all 20 cp genomes of Passifloraceae, with up to 151 repeats in P. costaricensis (supplementary table S6, Supplementary Material online).
A Repertory of Rearrangements: Inversions, Expansions/Losses of IR Regions, and Gene Losses
The results of the genomic comparison revealed a highly rearranged structure of cpDNAs in Passiflora. To detect all rearrangements, Po. trichocarpa (Salicaceae) was used as a reference because it is a phylogenetically close species with the same cpDNA genome pattern as most angiosperms. The chloroplast genome comparison performed showed different inversions (fig. 2 and supplementary fig. S2, Supplementary Material online). In total, 22 synteny blocks were identified among the cp genomes aligned.
Fig. 2.
Synteny and structural rearrangements detected in the chloroplast genomes of 11 Passiflora species in addition to the Passifloraceae, Dilkea retusa and Mitostemma brevifilis, and the Salicaceae, Populus trichocarpa. Colored bars indicate syntenic blocks and connecting lines indicate the correspondence between blocks. *Species of the Passiflora subgenus obtained in this study share the same structure with Passiflora alata.
Inversions were identified in the Astrophea subgenus compared with Dilkea and Mitostemma. They differed only with regard to one inversion (∼15 kb) in the LSC region of P. haematostigma and P. rhamnifolia. In fact, a high level of synteny was found between Passifloraceae Dilkea and Mitostemma, and Salicaceae Po. trichocarpa, revealing that fewer rearrangements have occurred, a feature common to most angiosperms. A large inversion in the LSC region flanking the clpP gene was found only in P. cerradensis, differing from the earlier rearrangement of the clpP and accD genes in P. haematostigma and P. rhamnifolia.
Rearrangements were also found in the Decaloba subgenus, with five species exhibiting a large inversion in the LSC region (fig. 2). Small sequence structures were also found inverted in the LSC region of P. suberosa. Interestingly, these small inversions are different from the very distinct rearrangement located at the beginning of the LSC region of P. candollei and Passiflora vespertilio, including the petB and clpP genes. Passiflora vespertilio also exhibited a different arrangement in the IR region, with an inversion between the rpl2 and rrn5 genes, and this arrangement was confirmed by PCR (supplementary table S3, Supplementary Material online).
The two species of the subgenus Deidamioides (P. deidamoides and P. contracta) differed due to the presence of a small inverted block in the LSC region (fig. 2). Comparing all 20 cp genomes, the species in the subgenus Passiflora differed by the presence of a large inversion in the LSC region.
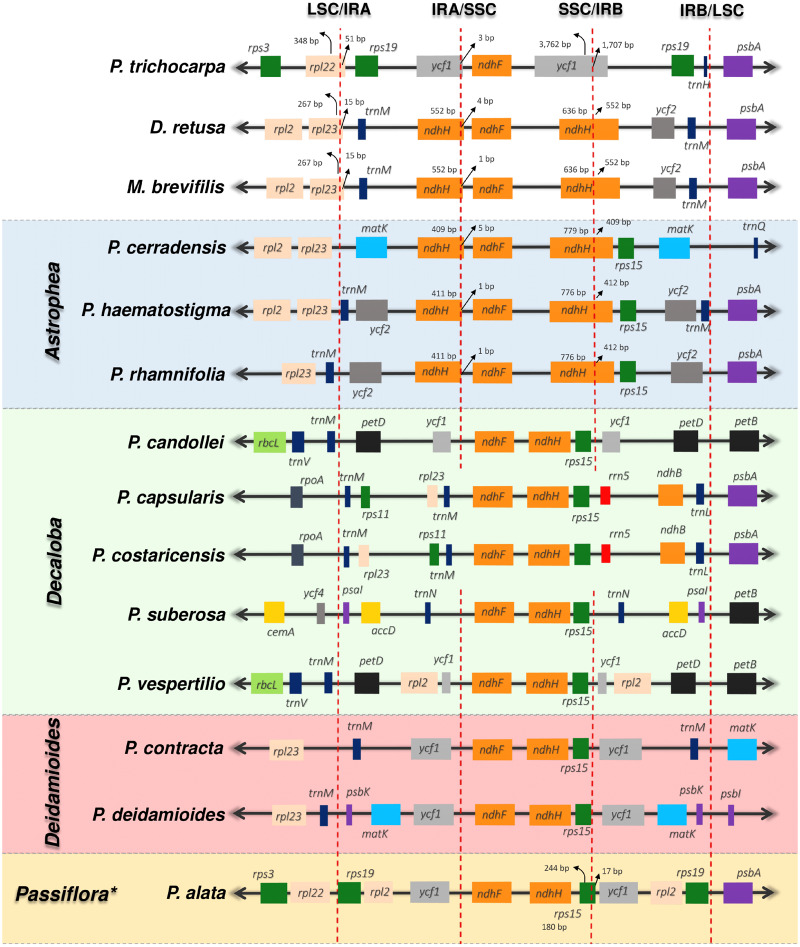
We also examined IR boundaries and detected different expansions and contractions (fig. 3). By comparing them with Po. trichocarpa, used as a reference, it was possible to detect expansions in the Passifloraceae D. retusa and M. brevifilis, in which the ndhH gene had expanded from the boundary of the SSC region to the IRA region (∼550 bp) creating a duplicated small fragment copy of the ndhH gene in the IRB region. Furthermore, the LSC region contained an expansion including a portion (15 bp) of the rpl23 sequence up to the IR region, and interestingly this arrangement seems to be particular to D. retusa and M. brevifilis and does not occur in the Passiflora genus, not even in the reference, Po. trichocarpa.
Fig. 3.
IR boundary comparison of 11 Passiflora species in addition to the Passifloraceae, Dilkea retusa and Mitostemma brevifilis, and the reference species Populus trichocarpa. The red dotted lines indicate the border of the cp genome regions, and the colored boxes indicate the gene structures. *Species of the Passiflora subgenus obtained in this study share the same structure with Passiflora alata.
In the Astrophea subgenus, as in the Dilkea and Mitostemma genera, an expansion was observed in the SSC/IRA extending into part of the ndhD gene, whereas P. cerradensis showed an additional expansion (∼3.5 kb) of IRA/LSC that includes the matK gene, which was confirmed by PCR (supplementary table S3, Supplementary Material online). Comparing all the species sequenced, Decaloba is the subgenus with the highest number of rearrangements related to IR boundaries. For instance, in P. candollei and P. vespertilio, the expansion of IRA/LSC extends up to the petD gene, and in P. suberosa this expansion is even larger, extending up to the psaI gene and comprising important genes such as rbcL. As a result of this significant expansion, P. suberosa has an IR of ∼43 kb that includes 38 genes, a difference of 17 kb compared with the IRs of other Decaloba subgenus species analyzed in this study.
A different expansion of IRB/LSC was observed in P. deidamioides (subgenus Deidamioides), including the protein-coding genes matK, psbA, and psbK, and this expansion was confirmed by amplification by PCR (supplementary table S3, Supplementary Material online). This latter expansion in P. deidamioides has an additional fragment length of 3,126 bp compared with the IRB/LSC of P. contracta (subgenus Deidamioides). Finally, the species that belong to the subgenus Passiflora exhibited the smallest rearrangements related to IR boundaries, with part of the rps15 gene expanding from the SSC to the IRA region (64 bp in P. edmundoi).
Comparatively, there were no large variations in the length of the SSC region, and the difference in IR sizes between species was ∼21.5 kb, whereas in the LSC region it reached 33 kb. These differences were due to not only IR expansions but also gene and intron losses. All the cp genomes showed gene losses compared with the reference, Po. trichocarpa (fig. 1). The rpl22 gene was not found to be complete in any of the species analyzed. Additionally, accD was found as a pseudogene, containing just a piece of the sequence or up to 18 stop codons in some species of the different subgenera. Some events may have occurred independently alongside chloroplast genome evolution in the Passiflora genus, such as rpl32 gene loss in some species of the Decaloba and Astrophea subgenera (fig. 1). In addition, rpl20 was absent in the cp genomes of the Astrophea, Decaloba, and Passiflora subgenera. In the subgenus Passiflora, the rps7, ycf1, and ycf2 genes were found as pseudogenes. Finally, Decaloba subgenus exhibited most of the gene losses, over and above the loss of rps7, ycf1, and ycf2 (the last two were sometimes detected as small pseudogenes), and the loss of rpoA was detected in P. capsularis, P. costaricensis, and P. suberosa.
Phylogenomic Studies
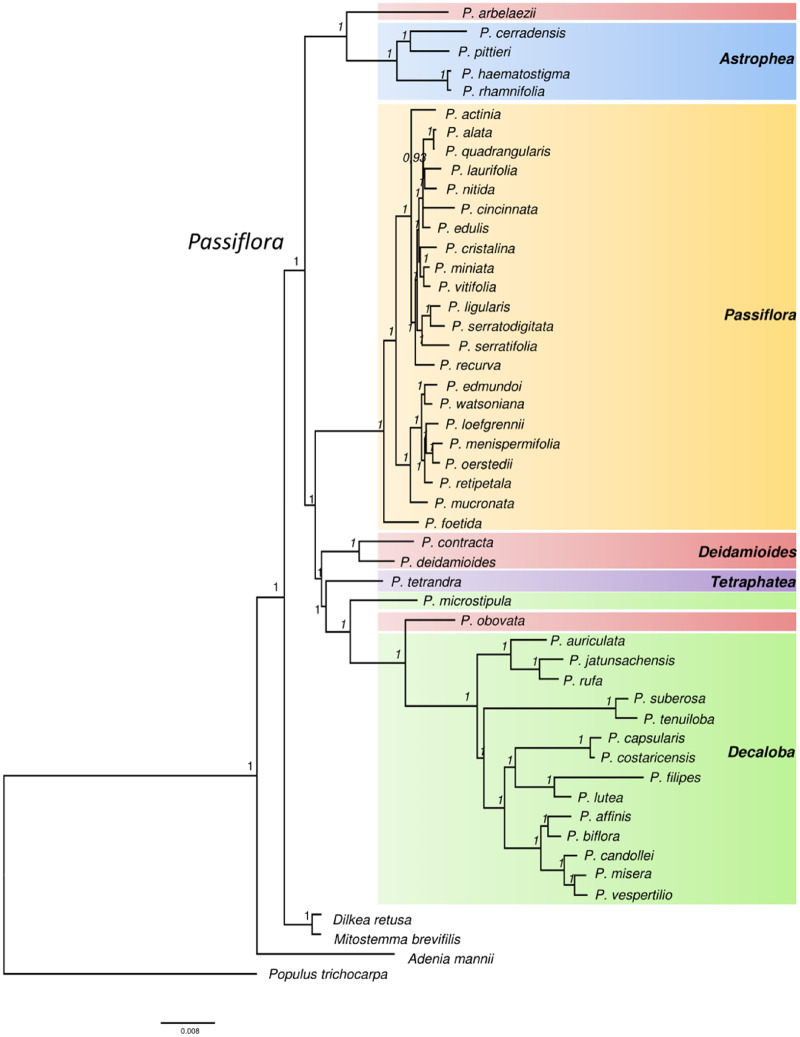
A phylogenomic study was performed based on 68 protein-coding genes of 49 Passifloraceae species with available chloroplast genomes. These species were used as the ingroup and Po. trichocarpa (Salicaceae) was the outgroup to obtain a rooted phylogenetic tree.
The two different partition schemes for Bayesian analysis had no substantial effect on the topology of the resulting trees. The analysis partitioned by codon position was highly favored in relation to the single model based on Bayes Factors (=5,448.8) and also showed slightly higher support values in nodes with PPs < 1.0. The accuracy of the inferred species phylogeny was strongly supported by the stability of the main clades generated and high PP values, with 97% (42 of 43) reaching 1 (fig. 4).
Fig. 4.
Bayesian phylogenetic reconstruction of Passiflora evolutionary history based on 68 chloroplast protein-coding genes.
Dilkea and Mitostemma formed a clade at the position (Adenia, ((Dilkea, Mitostemma), Passiflora)), all with high support in the phylogenetic tree. The Astrophea clade was supported placed on the tree as a monophyletic with high PPs = 1 and was split into two distinct subclades, one containing P. haematostigma and P. rhamnifolia (species from section Pseudastrophea) and the other grouping P. cerradensis and Passiflora pittieri. However, Passiflora arbelaezii (subgenus Deidamioides) was grouped as a sister taxon to the Astrophea species, dismembering the polyphyletic group of Deidamioides.
Deidamioides, as currently defined, is a polyphyletic subgenus, and despite the clade formed by P. contracta and P. deidamioides, other species assigned to Deidamioides were placed in different positions on the tree. Passiflora arbelaezii was placed as sister to the Astrophea subgenus whereas P. obovata (subgenus Deidamioides) was embedded in the Decaloba subgenus. Most species from the Decaloba subgenus clustered into a monophyletic clade with high support, having as successive sister groups Tetrapathea and two species of Deidamioides (P. contracta and P. deidamioides). Subgenus Passiflora was also recovered as a monophyletic group, but its sections display paraphyletic patterns. This subgenus contains 236 morphologically diverse species, which are classified in supersections, sections, and series (Feuillet and MacDougal 2003).
Discussion
A Repertory of cpDNA Rearrangements Concomitant with Passiflora Evolution
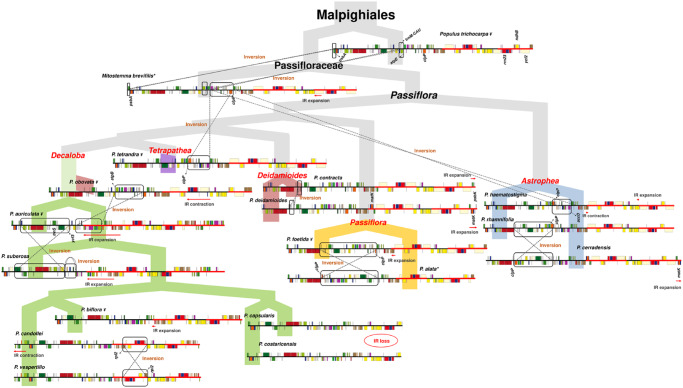
The evolutionary history of the Passiflora chloroplast genome revealed a high number of structural rearrangements (fig. 5). Inversions were detected in the subgenus Astrophea (P. haematostigma), but surprisingly not in the Dilkea, Mitostemma, and even Adenia genera in the Passifloraceae family (Shrestha et al. 2019). This suggests that the inversions in the Astrophea subgenus occurred after the separation of the Passiflora genus from its ancestors. The other three genera have fewer species (1–102) and exhibit cp genomes more closely resembling that of Po. trichocarpa. It is possible that the abundance of inversions occurred only in Passiflora and not in other Passifloraceae. Interestingly, compared with Po. trichocarpa, the passionflowers show two small inversions in the LSC region that could be used to characterize the species in this group.
Fig. 5.
Evolutionary history of chloroplast genome structure in the genus Passiflora. The structure of the chloroplast genomes was plotted on a tree representing the evolution of the Passiflora genus. In the tree, each subgenus was differentiated by colors: blue for Astrophea, green for Decaloba, red for Deidamioides, orange for Passiflora, and violet for Tetrapathea. In the comparison of the cp genomes, the boxes and dotted lines indicate the direction of the structural inversions, and the red arrows indicate the direction of IR expansions/contractions. *Mitostemma brevifilis shares the same structure with Dilkea retusa, and the species of the Passiflora subgenus obtained in this study share the same structure with Passiflora alata. ¥cp genomes obtained from the GenBank database.
Some inversions in the Passiflora genus could possibly be the result of intramolecular recombination of repeats, a mechanism that has been reported to impact the generation of rearrangements (Ogihara et al. 1988; Milligan et al. 1989; Gray et al. 2009; Ruhlman et al. 2017). In our study, short direct-repeat structures in the flanking regions between the accD and clpP genes were identified in both P. haematostigma and P. rhamnifolia (Astrophea). These kinds of repeats are also present in wheat (Ogihara et al. 1988), Asteraceae (Kim et al. 2005), and Geraniaceae (Guisinger et al. 2011), the latter showing highly rearranged cp genomes similar to the trends observed in Passiflora.
In addition, a distinct inversion was found in P. cerradensis (subgenus Astrophea), but because this inversion was flanked by clpP and caused repositioning of this gene, in evolutionary terms our results indicate that the rearrangement of accD and clpP genes in P. haematostigma and P. rhamnifolia occurred before the unique inversion found in P. cerradensis. Interestingly, all the inversions in Astrophea are different from those found in the other subgenera. Therefore, it is clear that Astrophea underwent further cpDNA changes after its separation from the clade ((Decaloba, Deidamioides), Passiflora) (∼40 Ma) (Muschner et al. 2012).
The Decaloba subgenus exhibits many different inversions, possibly because of the high number of repeat structures, as well as large IR expansions that are typical of this subgenus. On the other hand, the Deidamioides species have a similar cpDNA structure to Dilkea and Mitostemma, with just one small inversion in the LSC region. However, the Deidamioides subgenus differs from the other two Passifloraceae in that it has large IR expansions. Finally, species from subgenus Passiflora exhibit conserved structures and the same rearrangements in the LSC region as those previously described for P. edulis, the main cultivated species (Cauz-Santos et al. 2017).
Chloroplast genome IR expansions have been found in different plant groups, including Geraniaceae (Guisinger et al. 2011), Euphorbiaceae (Li et al. 2017), Solanaceae (Amiryousefi et al. 2018), and Bignoniaceae (Thode and Lohmann 2019). Decaloba species harbor larger variations, but P. capsularis lacks one of the IRs; P. suberosa exhibits a large expansion of the IRa/LSC up to the psaI gene. The latter IR expansions have already been described in previous studies on the Decaloba subgenus (Rabah et al. 2019; Shrestha et al. 2019), except for the lack of IR region in P. capsularis and P. costaricencis. In Astrophea species, IRb/SSC has expanded to part of the ndhH gene, and remarkably an IRa/LSC expansion was found to include the matK gene in P. deidamioides and P. cerradensis. However, these expansions do not have a common ancestor and supposedly occurred independently in the Astrophea and Deidamioides subgenera.
All species of the Passiflora subgenus exhibited an expansion to the rps19 gene, like that of Po. trichocarpa, but this expansion was not found in its sister group, suggesting that independent events occurred in the distinct subgenera. Expansions/contractions and loss of IRs are the main causes of variations in cpDNA sequence length, as in the Decaloba subgenus and other plants, such as Annona (Blazier, Ruhlman, et al. 2016) and Lamprocapnos (Park et al. 2018), as well as in monocots (Wang et al. 2008). Different mechanisms have been proposed to explain IR expansions, such as gene conversion or double-strand DNA breaks (Goulding et al. 1996; Wang et al. 2008).
Distinct genes have been lost during the evolution of cp genomes in the Passiflora genus, such as rpl20 and rpl22, the latter absent in all the cp genomes studied herein. Because the chloroplast organelle is responsible for vital processes like photosynthesis, gene loss can impair the efficiency of some metabolic pathways, plant growth, and cell survival (Neuhaus and Emes 2000; Kode et al. 2005; Rogalski et al. 2006; Romani et al. 2012). Furthermore, rpl20 and rpl22 encode proteins necessary for chloroplast translational apparatus and have been proven to be essential for cell viability in tobacco knockout mutants (Rogalski et al. 2008; Fleischmann et al. 2011). Similarly, the absence of rpl22 in legumes is offset by the existence of a functional copy transferred from the chloroplast to the nuclear genome (Gantt et al. 1991).
Gene transfer between chloroplast and nuclear genomes has been described in some species (Millen et al. 2001; Rousseau-Gueutin et al. 2013; Hong et al. 2017) and observed experimentally (Stegemann et al. 2003; Stegemann and Bock 2006; Lloyd and Timmis 2011). Despite the fact that the organelle gene needs a eukaryotic promoter to maintain its functionality in the nuclear genome, studies have revealed that some cp promoters are weakly active in the nucleus, which would render gene transfer viable without a nuclear promoter (Cornelissen and Vandewiele 1989; Wang et al. 2014). The functional transfer of rpl22 from cp to the nuclear genome has been observed in Fagaceae and it has been suggested that this happens in Passiflora (Jansen et al. 2011).
The Loss of an IR Region in Subgenus Decaloba
The cp genomes analyzed herein have the quadripartite structure common to almost all angiosperms. However, a complete loss of one IR was identified in P. capsularis and P. costaricensis, both species in the Xerogona section within the Decaloba subgenus (Espinoza et al. 2018). It is worthy of note that this kind of arrangement has not previously been reported in Passiflora cpDNAs (Cauz-Santos et al. 2017; Rabah et al. 2019; Shrestha et al. 2019). Bearing in mind that the loss of a complete IR region is rare in angiosperms and it was identified in species belonging to the same section (i.e., Xerogona), this event may have occurred in the ancestral chloroplast genome of the Xerogona section (subgenus Decaloba).
The loss of a complete IR has been described in few species of Geraniaceae and Cactaceae (Guisinger et al. 2011; Sanderson et al. 2015; Blazier, Jansen, et al. 2016), and in the large group of Fabaceae (legumes) in which the loss of an IR occurred once, leading to a monophyletic group within the subfamily Papilionideae, designated the IR-lacking clade, or IRLC (Lavin et al. 1990). It was suggested that this IR deletion could provide a means of testing the traditional evidence used to reconstruct Papilionideae phylogeny (Lavin et al. 1990). In addition, the rates of nucleotide substitution in genes of the IRLC-papilionoids are generally higher than those found in the IR-containing papilionoids (Schwarz et al. 2017).
In land plants, IR regions were found to present slower rates of nucleotide substitution compared with those of the SC regions. However, the expansion of IRs did not necessarily decrease the substitution rates in some lineages, as in Pelargonium (Zhu et al. 2016; Weng et al. 2017).
Palmer and Thompson hypothesized that IR regions could play a part in the stabilization of cp genome structure (Palmer and Thompson 1982). However, later studies reported that the deletion of a complete IR would not necessarily lead to cp genome instability or the formation of new rearrangements (Palmer et al. 1987). Thus, an IR loss was considered to be a different pattern of rearrangement that occurs together with other rearrangements (e.g., inversion and gene loss) in the IRLC (Sabir et al. 2014). Interestingly, this pattern was also found in our study. The loss of a complete IR sequence in Xerogona section species is unique, alongside other different rearrangements that occur in the Decaloba subgenus, such as inversions, IR expansions, and gene and intron losses.
In Passiflora, previous studies revealed the occurrence of paternal inheritance in interspecific crosses (Muschner et al. 2006; Hansen et al. 2007). However, the potential for biparental inheritance in intraspecific crosses involving P. costaricensis (subgenus Decaloba) has also been reported (Hansen et al. 2007). Most surprisingly, when faced with the cp genome structure of P. costaricensis, we noted that our results unveiled the loss of an entire IR region. In other groups of seed plants that exhibit potential for biparental inheritance, a highly rearranged cpDNA structure also occurs, such as in Campanulaceae (Cosner et al. 2004; Haberle et al. 2008; Barnard-Kubow et al. 2017) and Geraniaceae (Metzlaff et al. 1981; Chumley et al. 2006; Weng et al. 2014).
Recent studies have shown that biparental inheritance could promote chloroplast competition mediated by accelerated rates of evolution in repeats located in the accD gene regulatory region (Sobanski et al. 2019). In our study, a high number of repeats within the accD gene sequence were observed in all species. Additionally, chloroplast biparental inheritance has the potential to restore cytonuclear incompatibility (Barnard-Kubow et al. 2017; Shrestha et al. 2019), and this type of incompatibility has also been reported in the genus Passiflora (Mracek 2006). Further studies are required to evaluate whether changes in the inheritance of chloroplast DNA could lead rearrangements in the nucleotide sequence.
Phylogenetic Relationships in Passiflora
Taxonomically speaking, Passiflora is the largest genus of Passifloraceae with ∼520 species, exhibiting high morphological diversity, especially in flower shape and color, and also in genome size. Although a wide variety of morphological traits has been applied in its traditional taxonomy, the species classification still has unresolved positions, particularly regarding species belonging to the Deidamioides subgenus. Molecular phylogenies did not clarify the position of this subgenus, but they did generally recover Astrophea, Decaloba, and Passiflora as well-supported monophyletic clades based on chloroplast, mitochondrial, or nuclear nucleotide sequences (Muschner et al. 2003, 2012; Krosnick et al. 2006, 2013). In addition to the four subgenera described, Tetrapathea (Passifloraceae) has been put forward as a new subgenus of Passiflora, with three species native to the Old World (Krosnick et al. 2009).
Our findings confirmed the Astrophea subgenus as a monophyletic clade, and also grouped together P. haematostigma and P. rhamnifolia, two species that belong to section Pseudoastrophea. Furthermore, the Astrophea subgenus was grouped as a sister clade to P. arbelaezii, a species of section Tryphostemmatoides in the subgenus Deidamioides, confirming previous phylogenetic studies (Krosnick et al. 2013). Based on the divergence times in the Passiflora genus, the separations of the clade Astrophea plus the Tryphostemmatoides section from the other Passiflora clades are very ancient, at around 40 Ma (Muschner et al. 2012).
In the past, due to its morphology, Tryphostemmatoides was considered a subgenus (Killip 1938), but later it was reduced to a section of the Deidamioides subgenus (Feuillet and MacDougal 2003). Previous studies using molecular phylogenetic inferences not only recovered section Tryphostemmatoides as a monophyletic clade but also revealed the paraphyletic position of this section in relation to other species of the Deidamioides subgenus (Muschner et al. 2012; Krosnick et al. 2013). Our findings also suggest the need for revisiting the taxonomic classification of Tryphostemmatoides.
The Deidamioides subgenus has been recovered as polyphyletic. Only the Brazilian endemic species P. contracta and P. deidamioides were grouped as a clade. Passiflora arbelaezii was sister to the Astrophea subgenus, and P. obovata placed in the Decaloba subgenus, confirming a previously phylogenomic inference (Shrestha et al. 2019). The classical taxonomic studies (Master 1871; Killip 1938) and the recent revision (Feuillet and MacDougal 2003) initially positioned some of these species in the Deidamioides subgenus and subsequently the Decaloba subgenus. Conversely, P deidamioides was initially considered as Decaloba (Harms 1923), before the creation of the Deidamioides section. Passiflora obovata was considered to belong to the subgenus Plectostemma, now known as the Decaloba subgenus (Harms 1923; Killip 1938), and was assigned to the Deidamioides subgenus in the classification of Feuillet and MacDougal (2003). However, molecular phylogenies still group this species in the Decaloba subgenus (Krosnick et al. 2006, 2013), corroborating our findings. This controversy suggests that the classification of P. obovata needs to be revised.
Passiflora tetrandra (subgenus Tetrapathea) was grouped as a sister to Decaloba, forming the group (Deidamioides, (Tetrapathea, Decaloba)), confirming previous findings (Krosnick et al. 2013; Shrestha et al. 2019).
On the other hand, heteroplasmy, the existence of divergent chloroplasts types, could be problematic and lead to the analysis of paralogous copies in phylogenetic studies (Wolfe and Randle 2004). For this reason, Hansen et al. (2007), discussing the implications of heteroplasmy in Passiflora, suggest that caution should be exercised when considering the interpretation of the chloroplast phylogenies. We should also point out that phylogenomics based on cpDNA sequences allow only one view of Passiflora evolution, and further reconstructions of phylogenetic trees based on nuclear genes will also be necessary.
Finally, regarding the Passiflora subgenus, incongruences were found in the positioning of some species compared with former phylogenetic studies. However, Passiflora is the largest subgenus (236 species), and due to the relevance of taxa sampling for phylogeny reconstruction accuracy (see Heath et al. 2008; Nabhan and Sarkar 2012), new analyses of an increased number of species may help elucidate the taxonomy within this subgenus. Interesting results were published by Sader et al. (2019) providing a robust and well-resolved time-calibrated phylogeny including some 100 taxa, and by means of phylogenetic comparative methods, they tested the relative importance of polyploidy in Passiflora evolution and diversification. According to these authors, changes in chromosome number and genome sizes may have contributed to morphological and ecological traits that explain the pattern of diversification observed in the genus.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Teonildes Sacramento Nunes for his assistance in Passiflora taxonomic identification; Maurizio Vecchia for giving us the Passiflora costaricensis specimen; and Dr Lucas C. Marinho for the Dilkea sample and Mr Steve Simmons for proofreading the manuscript. We also thank the biologist, Carlos A. de Oliveira, for helping with the field collections and technical assistance. This work was supported by the following Brazilian Institutions: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Pessoal de Ensino Superior (CAPES) [Finance Code 001], and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [grant numbers 2017/11815-6 to M.L.C.V.; 2017/04216-9 and 2019/09210-4 to L.A.C.-S.; 2019/07838-6 to Z.P.d.C.].
Literature Cited
- Amiryousefi A, Hyvönen J, Poczai P. 2018. The chloroplast genome sequence of bittersweet (Solanum dulcamara): plastid genome structure evolution in Solanaceae. PLoS One 13(4):e0196069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard-Kubow KB, McCoy MA, Galloway LF. 2017. Biparental chloroplast inheritance leads to rescue from cytonuclear incompatibility. New Phytol. 213(3):1466–1476. [DOI] [PubMed] [Google Scholar]
- Bedoya AM, et al. 2019. Plastid genomes of five species of riverweeds (Podostemaceae): structural organization and comparative analysis in Malpighiales. Front Plant Sci. 10: 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier JC, Jansen RK, et al. 2016. Variable presence of the inverted repeat and plastome stability in Erodium. Ann Bot. 117(7):1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier JC, Ruhlman TA, et al. 2016. Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci Rep. 6:24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, et al. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 67(6):696–704. [DOI] [PubMed] [Google Scholar]
- Cauz-Santos LA, et al. 2017. The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: structural organization and phylogenomic studies in Malpighiales. Front Plant Sci. 8:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley TW, et al. 2006. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 23(11):2175–2190. [DOI] [PubMed] [Google Scholar]
- Cornelissen M, Vandewiele M. 1989. Nuclear transcriptiońal activity of the tobacco plastid psbA promoter. Nucleic Acids Res. 17(1):19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosner ME, Raubeson LA, Jansen RK. 2004. Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes. BMC Evol Biol. 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza TEB, Jørgensen PM, MacDougal JM. 2018. A taxonomic revision of Passiflora sect. Xerogona (Passifloraceae) using principal component analysis. Ann Mo Bot Gard.103(2):258–313. [Google Scholar]
- Feuillet C, MacDougal JM. 2003. A new infrageneric classification of Passiflora L. (Passifloraceae). J Newsl Passiflora Soc Int. 13:34–35. [Google Scholar]
- Fleischmann TT, et al. 2011. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23(9):3137–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD. 1991. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10(10):3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding SE, Olmstead RG, Morden CW, Wolfe KH. 1996. Ebb and flow of the chloroplast inverted repeat. Mol Gen Genet. 252:195–206. [DOI] [PubMed] [Google Scholar]
- Gray BN, Ahner BA, Hanson MR. 2009. Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res. 18(4):559–572. [DOI] [PubMed] [Google Scholar]
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28(1):583–600. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 66(4):350–361. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Escobar LK, Gilbert LE, Jansen RK. 2007. Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogenetic studies. Am J Bot. 94(1):42–46. [DOI] [PubMed] [Google Scholar]
- Hansen AK, et al. 2006. Phylogenetic relationships and chromosome number evolution in Passiflora. Syst Bot. 31(1):138–150. [Google Scholar]
- Harms H. 1923. Beiträge zur Kenntnis der amerikanischen Passifloraceen. I. Repert. Nov. specierum regni Veg.
- Heath TA, Hedtke SM, Hillis DM. 2008. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 46(3):239–257. [Google Scholar]
- Hong CP, et al. 2017. accD nuclear transfer of Platycodon grandiflorum and the plastid of early Campanulaceae. BMC Genomics. 18: 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H. 2011. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28(1):835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killip EP. 1938. The American species of Passifloraceae. Chicago (IL): Chicago Field Museum. [Google Scholar]
- Kim KJ, Choi KS, Jansen RK. 2005. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol Biol Evol. 22(9):1783–1792. [DOI] [PubMed] [Google Scholar]
- Knox EB. 2014. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc Natl Acad Sci U S A. 111(30):11097–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. 2005. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44(2):237–244. [DOI] [PubMed] [Google Scholar]
- Koren S, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosnick SE, Ford AJ, Freudenstein J V. 2009. Taxonomic revision of Passiflora subgenus Tetrapathea including the monotypic genera Hollrungia and Tetrapathea (Passifloraceae), and a new species of Passiflora. Syst Bot. 34(2):375–385. [Google Scholar]
- Krosnick SE, Harris EM, Freudenstein J V. 2006. Patterns of anomalous floral development in the Asian Passiflora (subgenus Decaloba: supersection Disemma). Am J Bot. 93(4):620–636. [DOI] [PubMed] [Google Scholar]
- Krosnick SE, Porter-Utley KE, MacDougal JM, Jørgensen PM, McDade LA. 2013. New insights into the evolution of Passiflora subgenus Decaloba (Passifloraceae): phylogenetic relationships and morphological synapomorphies. Syst Bot. 38(3):692–713. [Google Scholar]
- Kurtz S, et al. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Doyle JJ, Palmer JD. 1990. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution (N Y). 44(2):390–402. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. 2017. The complete chloroplast genome sequence of tung tree (Vernicia fordii): organization and phylogenetic relationships with other angiosperms. Sci Rep. 7:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AH, Timmis JN. 2011. The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol Biol Evol. 28(7):2019–2028. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master MT. 1871. Contributions to the natural history of the Passifloraceea. Trans Linn Soc Lond. 27:626–629. [Google Scholar]
- Metzlaff M, Börner T, Hagemann R. 1981. Variations of chloroplast DNAs in the genus Pelargonium and their biparental inheritance. Theor Appl Genet. 60:37–41. [DOI] [PubMed] [Google Scholar]
- Millen RS, et al. 2001. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13(3):645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan BG, Hampton JN, Palmer JD. 1989. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol Biol Evol. 6(4):355–368. [DOI] [PubMed] [Google Scholar]
- Mracek J. 2006. Investigation of interspecific genome-plastome incompatibility in Oenothera and Passiflora [Dissertation]. LMU Munich.
- Muschner VC, Zamberlan PM, Bonatto SL, Freitas LB. 2012. Phylogeny, biogeography and divergence times in Passiflora (Passifloraceae). Genet Mol Biol. 35(4 Suppl 1):1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschner VC, et al. 2003. A first molecular phylogenetic analysis of Passiflora (Passifloraceae). Am J Bot. 90(8):1229–1238. [DOI] [PubMed] [Google Scholar]
- Muschner VC, et al. 2006. Differential organellar inheritance in Passiflora’s (Passifloraceae) subgenera. Genetica 128(1–3):449–453. [DOI] [PubMed] [Google Scholar]
- Nabhan AR, Sarkar IN. 2012. The impact of taxon sampling on phylogenetic inference: a review of two decades of controversy. Briefings Bioinf. 13(1):122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ. 2000. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 51(1):111–140. [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Terachi T, Sasakuma T. 1988. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc Natl Acad Sci U S A. 85(22):8573–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Osorio B, Aldrich J, Thompson WF. 1987. Chloroplast DNA evolution among legumes: loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr Genet. 11:275–286. [Google Scholar]
- Palmer JD, Thompson WF. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29(2):537–550. [DOI] [PubMed] [Google Scholar]
- Park S, An B, Park SJ. 2018. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci Rep. 8(1):13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabah SO, et al. 2019. Passiflora plastome sequencing reveals widespread genomic rearrangements. J Syst Evol. 57(1):1–14. [Google Scholar]
- Rogalski M, Ruf S, Bock R. 2006. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34(16):4537–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R. 2008. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20(8):2221–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, et al. 2012. Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J. 72(6):922–934. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, et al. 2013. Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 161(4):1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman TA, Zhang J, Blazier JC, Sabir JSM, Jansen RK. 2017. Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am J Bot. 104(4):559–572. [DOI] [PubMed] [Google Scholar]
- Sabir J, et al. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 12(6):743–754. [DOI] [PubMed] [Google Scholar]
- Sader MA, et al. 2019. The role of chromosome changes in the diversification of Passiflora L. (Passifloraceae). Syst Biodivers Syst Biodivers. 17(1):7–21. [Google Scholar]
- Sanderson MJ, et al. 2015. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat . Am J Bot. 102(7):1115–1127. [DOI] [PubMed] [Google Scholar]
- Schwarz EN, et al. 2017. Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across legume subfamilies. J Mol Evol. 84(4):187–203. [DOI] [PubMed] [Google Scholar]
- Shrestha B, et al. 2019. Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol Phylogenet Evol. 138:53–64. [DOI] [PubMed] [Google Scholar]
- Sobanski J, et al. 2019. Chloroplast competition is controlled by lipid biosynthesis in evening primroses. Proc Natl Acad Sci U S A. 116(12):5665–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S, Bock R. 2006. Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell 18(11):2869–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S, Hartmann S, Ruf S, Bock R. 2003. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci U S A. 100(15):8828–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. 1992. The chloroplast genome. Plant Mol Biol. 19(1):149–168. [DOI] [PubMed] [Google Scholar]
- Takamatsu T, et al. 2018. Optimized method of extracting rice chloroplast DNA for high-quality plastome resequencing and de novo assembly. Front Plant Sci. 9:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangphatsornruang S, et al. 2011. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene 475(2):104–112. [DOI] [PubMed] [Google Scholar]
- Thode VA, Lohmann LG. 2019. Comparative chloroplast genomics at low taxonomic levels: a case study using Amphilophium (Bignonieae, Bignoniaceae). Front Plant Sci. 10: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, et al. 2017. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer T, MacDougal JM. 2004. Passiflora: passionflowers of the world. Portland (OR: ): Timber Press. [Google Scholar]
- Wang D, Qu Z, Adelson DL, Zhu JK, Timmis JN. 2014. Transcription of nuclear organellar DNA in a model plant system. Genome Biol Evol. 6(6):1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, et al. 2008. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng ML, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31(3):645–659. [DOI] [PubMed] [Google Scholar]
- Weng ML, Ruhlman TA, Jansen RK. 2017. Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214(2):842–851. [DOI] [PubMed] [Google Scholar]
- Wolfe AD, Randle CP. 2004. Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: implications for plant molecular systematics. Syst Bot. 29(4):1011–1020. [Google Scholar]
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 3:1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, et al. 2010. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS One 5(9):e12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209(4):1747–1756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.