In the early 2000s, a binary toxin (CDT)-producing strain of Clostridium difficile, ribotype 027 (RT027), caused extensive outbreaks of diarrheal disease in North America and Europe. This strain has not become established in Australia, and there is a markedly different repertoire of circulating strains there compared to other regions of the world. The C. difficile Antimicrobial Resistance Surveillance (CDARS) study is a nationwide longitudinal surveillance study of C. difficile infection (CDI) in Australia.
KEYWORDS: Clostridium difficile, molecular epidemiology, ribotyping, surveillance
ABSTRACT
In the early 2000s, a binary toxin (CDT)-producing strain of Clostridium difficile, ribotype 027 (RT027), caused extensive outbreaks of diarrheal disease in North America and Europe. This strain has not become established in Australia, and there is a markedly different repertoire of circulating strains there compared to other regions of the world. The C. difficile Antimicrobial Resistance Surveillance (CDARS) study is a nationwide longitudinal surveillance study of C. difficile infection (CDI) in Australia. Here, we describe the molecular epidemiology of CDI in Australian health care and community settings over the first 5 years of the study, 2013 to 2018. Between 2013 and 2018, 10 diagnostic microbiology laboratories from five states in Australia participated in the CDARS study. From each of five states, one private (representing community) and one public (representing hospitals) laboratory submitted isolates of C. difficile or PCR-positive stool samples during two collection periods per year, February-March (summer/autumn) and August-September (winter/spring). C. difficile was characterized by toxin gene profiling and ribotyping. A total of 1,523 isolates of C. difficile were studied. PCR ribotyping yielded 203 different RTs, the most prevalent being RT014/020 (n = 449; 29.5%). The epidemic CDT+ RT027 (n = 2) and RT078 (n = 6), and the recently described RT251 (n = 10) and RT244 (n = 6) were not common, while RT126 (n = 17) was the most prevalent CDT+ type. A heterogeneous C. difficile population was identified. C. difficile RT014/020 was the most prevalent type found in humans with CDI. Continued surveillance of CDI in Australia remains critical for the detection of emerging strain lineages.
INTRODUCTION
Clostridium (Clostridioides) difficile is an important cause of infectious diarrhea in health care settings in high-income countries. Prior to the 2000s, C. difficile infection (CDI) was underappreciated because few hospitalized patients progressed to fulminant disease (1). In the last 2 decades, an increase in the frequency and severity of CDI was noted, initially in Canada, then the United States and Europe, owing to the emergence of an epidemic strain of C. difficile, ribotype 027 (RT027) (1). The incidence and severity of CDI continue to impose a significant burden on global health care systems due to substantial costs associated with extended hospital stays and treatment. In their report, “Antibiotic Resistance Threats in the United States,” the Centers for Disease Control and Prevention indicated that C. difficile remained an urgent threat to public health that required aggressive action (2). In the United States, the national burden of CDI was estimated at 462,199 cases (3), costing the government $1 billion in health care expenditure in 2017 (2).
CDI is not a notifiable disease in Australia; however, monitoring of CDI rates in hospitals has been mandated by the Australian Commission on Safety and Quality in Healthcare for hospital accreditation since 2010 (4). All states and territories in Australia have reported a significant increase in rates of hospital-identified (HI) CDI since mid-2011 (5). HI CDI was defined as CDI diagnosed in a patient attending any area of an acute-care public hospital (i.e., patients admitted to inpatient wards or units, including psychiatry, rehabilitation, and aged care, and those attending emergency and outpatient departments). Increased rates of CDI may reflect more testing or use of more sensitive diagnostic algorithms; however, enhanced surveillance suggested that much of the increase (24%) was attributable to more community-associated (CA) CDI (5).
Despite this increase in the incidence of CDI in Australia (5) and the emergence of new virulent RTs (6, 7), most clinical microbiology laboratories in Australia do not culture or further characterize strains of C. difficile causing disease. The C. difficile Antimicrobial Resistance Surveillance (CDARS) study was initiated to address this problem with nationwide longitudinal surveillance of CDI in Australia. Here, we describe the molecular epidemiology of CDI in Australian health care and community settings over the first 5 years of the study, 2013 to 2018. We previously reported antimicrobial susceptibility data for 2013 to 2014, including typing of selected isolates (∼30%) (8), and we now present complete molecular typing results for all isolates from 2013 to 2018.
MATERIALS AND METHODS
Sample and data collection.
Between 2013 and 2018, 10 diagnostic microbiology laboratories participated in the CDARS study. The laboratories comprised one private (representing community) and one public (representing hospitals) site from each of five states of Australia (Western Australia [WA], New South Wales [NSW], Victoria [VIC], South Australia [SA], and Queensland [QLD]). There were up to two collection phases per calendar year, in February-March and August-September, representing the summer-autumn and winter-spring seasons, respectively. During each phase, sites were asked to save up to 15 nonduplicate isolates of C. difficile or stool samples based on the criteria previously described (8). All samples and isolates were stored at −70°C at participating sites, shipped to the reference laboratory (PathWest Laboratory Medicine, Nedlands, WA) on transport swabs under ambient conditions, and processed within 24 h of arrival.
Specimens from private laboratories largely represented CA CDIs, as these facilities served patients from general practitioners (40 to 50%), aged-care facilities (1 to 3%), and private (community) hospitals (50 to 60%), some of which are large tertiary facilities with intensive care units. Conversely, specimens from public laboratories that were based in large tertiary-care medical centers (public hospital sites) represented HI CDIs that, by definition (5), may have included some CA CDI cases also. Basic demographic data (gender and age) were collected, and children <1 year of age were excluded from the analysis. Chi-square tests of significance between data sets were performed in IBM SPSS Statistics version 26.
C. difficile culture and epidemiological typing.
Stool samples and isolates of C. difficile that were PCR positive for a toxin gene(s) were cultured, and DNA was extracted using methods previously described (8). All recovered C. difficile strains underwent toxin gene profiling and PCR ribotyping. The toxin genes tcdA (toxin A), tcdB (toxin B), cdtA, and cdtB (CDT) were amplified by PCR as previously described (8). C. difficile RTs were determined by amplification of the 16S-23S rRNA intergenic spacer region, and PCR products were separated on the QIAxcel capillary electrophoresis platform (Qiagen GmbH, Hilden, Germany). The BioNumerics software package v.6.5 (Applied Maths, Saint-Martens-Latem, Belgium) was used for dendrogram and cluster analysis of PCR ribotyping band patterns. Isolates that could not be identified with the available reference library were designated with internal nomenclature, prefixed with QX.
RESULTS
Isolate collection.
A total of 1,675 eligible samples (stool or isolates) were received during the 10 collection phases, of which 46% (n = 778) were submitted by private laboratories. From these samples, 1,523 isolates of C. difficile (90.9%) were recovered (Table 1). For the entire study, 60.6% of cases were female, the median age was 69 years (interquartile range, 50 to 81 years), and the majority (62%) of patients were ≥65 years old.
TABLE 1.
Summary of sample collection and C. difficile recovery
| Phase | Site type | No. of specimens or strains receiveda
|
C. difficile recovery |
||||||
|---|---|---|---|---|---|---|---|---|---|
| NSW | QLD | SA | VIC | WA | Total | n | % | ||
| 1 | Private | 19 | 21 | 13 | 15 | 8 | 76 | 66 | 86.8 |
| Public | 31 | 5 | 21 | 22 | 20 | 99 | 87 | 87.9 | |
| Total | 50 | 26 | 34 | 37 | 28 | 175 | 153 | 87.4 | |
| 2 | Private | 15 | 10 | 14 | 15 | 14 | 68 | 67 | 98.5 |
| Public | 15 | 6 | 15 | 15 | 15 | 66 | 62 | 93.9 | |
| Total | 30 | 16 | 29 | 30 | 29 | 134 | 129 | 96.3 | |
| 3 | Private | 20 | 24 | 8 | 17 | 13 | 82 | 71 | 86.6 |
| Public | 23 | 4 | 17 | 22 | 17 | 83 | 78 | 94.0 | |
| Total | 43 | 28 | 25 | 39 | 30 | 165 | 149 | 90.3 | |
| 4 | Private | 15 | 15 | 8 | 12 | 15 | 65 | 61 | 93.8 |
| Public | 15 | 21 | 15 | 15 | 15 | 81 | 76 | 93.8 | |
| Total | 30 | 36 | 23 | 27 | 30 | 146 | 137 | 93.8 | |
| 5 | Private | 19 | 15 | 15 | 17 | 10 | 76 | 66 | 86.8 |
| Public | 15 | 15 | 16 | 19 | 20 | 85 | 79 | 92.9 | |
| Total | 34 | 30 | 31 | 36 | 30 | 161 | 145 | 90.1 | |
| 6 | Private | 19 | 26 | 10 | 20 | 5 | 80 | 75 | 93.8 |
| Public | 20 | 23 | 12 | 19 | 25 | 99 | 90 | 90.9 | |
| Total | 39 | 49 | 22 | 39 | 30 | 179 | 165 | 92.2 | |
| 7 | Private | 20 | 23 | 15 | 20 | 1 | 79 | 76 | 96.2 |
| Public | 20 | 22 | 15 | 19 | 20 | 96 | 94 | 97.9 | |
| Total | 40 | 45 | 30 | 39 | 21 | 175 | 170 | 97.1 | |
| 8 | Private | 19 | 26 | 12 | 23 | 4 | 84 | 74 | 88.1 |
| Public | 18 | 24 | 23 | 16 | 22 | 103 | 97 | 94.2 | |
| Total | 37 | 50 | 35 | 39 | 26 | 187 | 171 | 91.4 | |
| 9 | Private | 16 | 20 | 15 | 27 | 4 | 82 | 71 | 86.6 |
| Public | 20 | 16 | 19 | 15 | 70 | 59 | 84.3 | ||
| Total | 36 | 36 | 15 | 46 | 19 | 152 | 130 | 85.5 | |
| 10 | Private | 25 | 26 | 13 | 15 | 7 | 86 | 74 | 86.0 |
| Public | 22 | 23 | 27 | 19 | 24 | 115 | 100 | 87.0 | |
| Total | 47 | 49 | 40 | 34 | 31 | 201 | 174 | 86.6 | |
| Total | 386 | 365 | 284 | 366 | 274 | 1,675 | 1,523 | 90.9 | |
NSW, New South Wales; QLD, Queensland; SA, South Australia; VIC, Victoria; WA, Western Australia.
Epidemiological typing and toxin profiling.
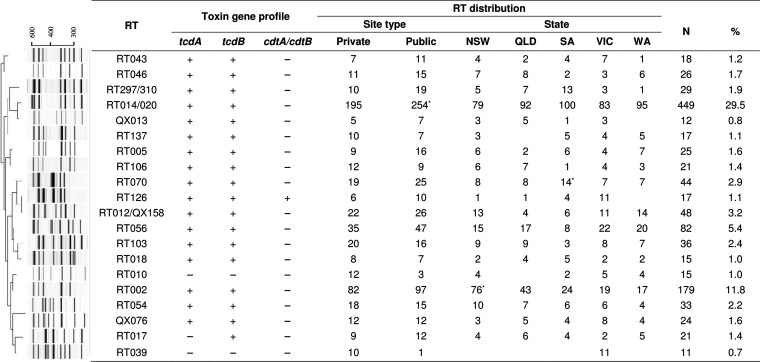
There were 1,523 C. difficile isolates recovered, and PCR ribotyping yielded 203 unique RTs. Of these isolates, 1,197 (78.6%) were assigned to one of 51 internationally recognized RTs or were given an internal QX number (n = 275; 18.1%). A small number of isolates were singleton strains unique to our laboratory and could not be identified with the available reference library (n = 51; 3.3%). C. difficile RT014 and RT020 and C. difficile RT297 and RT310 were grouped as RT014/020 and RT297/310, respectively, due to similarities in their banding patterns. The 20 most prevalent RTs of C. difficile, which comprised 76.1% of all isolates (n = 1,159), along with their distribution between states and laboratory type, are shown in Fig. 1. Overall, C. difficile RT014/020 (n = 449; 29.5%) was the most prevalent, followed by RT002 (n = 179; 11.8%), a majority of which was found in a public laboratory in NSW (n = 48; 26.8%; P < 0.05), and RT056 (n = 82; 5.4%). The distribution of the 12 most common RTs was consistent throughout the 5-year period with only slight variations between laboratory types (Fig. 2). The epidemic C. difficile RT027 (n = 2) and RT078 (n = 6) and the recently described RT251 (n = 10) and RT244 (n = 6) were found in low numbers.
FIG 1.
Dendrogram summary, toxin profiles, and distributions of the 20 most prevalent C. difficile PCR RTs. PCR ribotyping pattern cluster analysis using both the Dice coefficient and the neighbor-joining method was used. Proportions were compared by χ2 test. *, P < 0.05. NSW, New South Wales; QLD, Queensland; SA, South Australia; VIC, Victoria; WA, Western Australia.
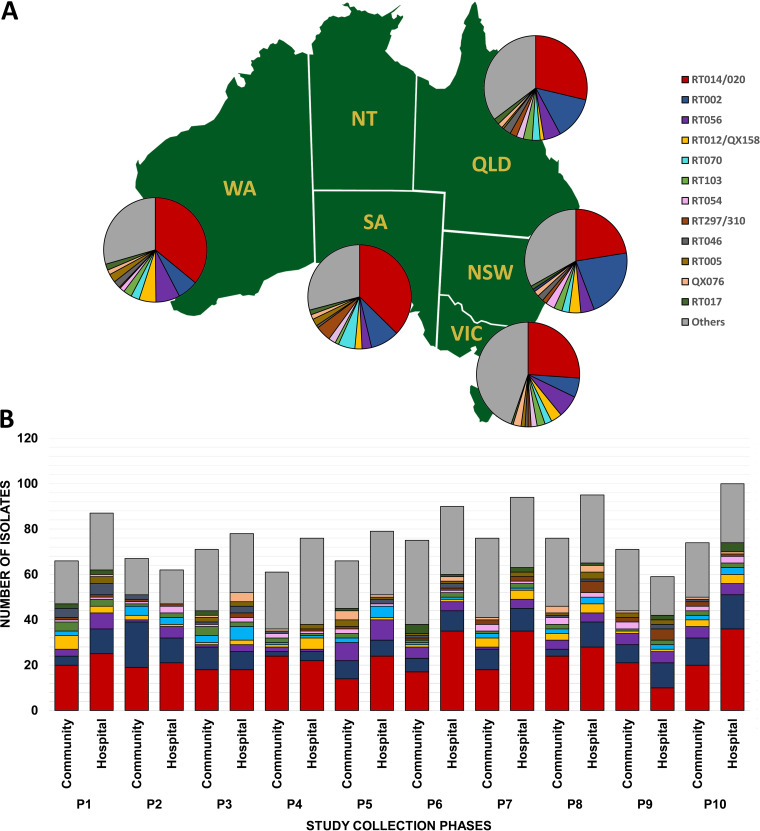
FIG 2.
Molecular epidemiology of CDI in Australia. Distribution of C. difficile RTs by Australian state (A) and by private or public collection site (B) over 10 collection phases, 2013 to 2018 (n = 1,523). NSW, New South Wales; QLD, Queensland; SA, South Australia; VIC, Victoria; WA, Western Australia. (The map was created with mapchart.net and is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License [https://creativecommons.org/licenses/by-sa/4.0/].)
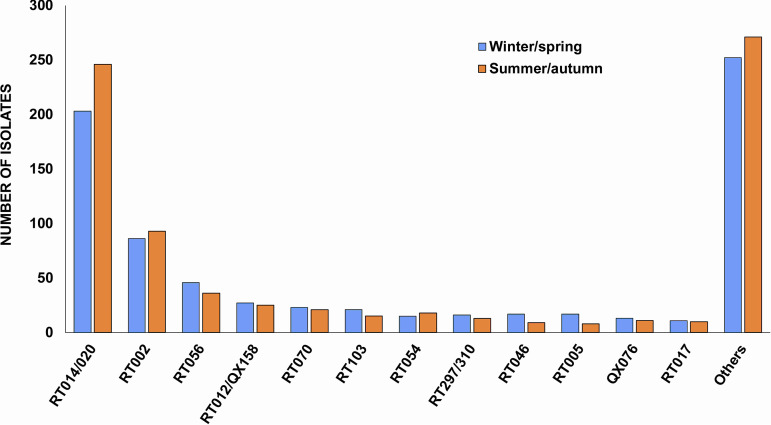
The vast majority of C. difficile strains (93.4%; n = 1,423) were positive for the major toxin genes tcdA and tcdB (A+B+), and 4.1% (n = 63) also contained cdtA and cdtB (CDT+) genes. Twenty-two strains had a variant toxin profile, A−B+CDT− (RT017, n = 21; QX134, n = 1), while five were positive for tcdB and cdtA and cdtB, resulting in the rare toxin profile A−B+CDT+. The overall prevalence of CDT+ C. difficile strains was 4.5% (n = 69). One strain of C. difficile RT033 was positive for cdtA and cdtB genes only. Interestingly, C. difficile RT126 was the most prevalent CDT+ type in Australia (n = 17), with 64.7% (n = 11) of isolates originating from Victoria during winter-spring of 2016 and the majority (72.7%; n = 8; P < 0.05) coming from the public laboratory. The six C. difficile RT078 strains were mainly from NSW (n = 4), with no obvious temporal clustering. Notably, the majority of CDT+ C. difficile strains were isolated from public laboratories. In contrast, 71 nontoxigenic C. difficile strains were submitted, and 77.5% (n = 55) were from private laboratories. Two nontoxigenic strains, RT010 and RT039 (n = 12 and n = 10, respectively), ranked in the top 20 most prevalent C. difficile strains in Australia (Fig. 1). There was no evidence of seasonality in RT distribution (Fig. 3).
FIG 3.
Seasonal distribution for the top 12 most prevalent RTs in Australia, 2013 to 2018.
DISCUSSION
Here, we report the results of nationwide longitudinal laboratory-based surveillance of C. difficile in health care and community settings in Australia. The study was performed on a collection of 1,675 specimens and isolates submitted by 10 laboratories in five states of Australia from 2013 to 2018, yielding 1,523 strains of C. difficile. Between 2011 and 2016, the average rate of CDI across Australia was 4 cases/10,000 patient bed-days, corresponding to approximately 10,000 cases of CDI per year (9). Thus, for every year of our study, approximately 3% of strains of C. difficile recovered from these cases were analyzed. A heterogeneous C. difficile population was identified with an RT distribution similar to those reported in 2013-2014 (8) and our earlier studies in 2010 (10) and 2012 (11). Our findings in the current study suggest that while temporal shifts occurred in some circulating strains of C. difficile, the most common RTs in Australia did not change. However, understanding these temporal shifts is important, as is indicated by RT244 emerging as a cause of severe community-associated infection and subsequently becoming the third most common RT detected in Australia in 2012 (6, 12).
C. difficile RT014/020 has remained the most common type causing CDI in humans in Australia since it was first reported as such in 2010, with a prevalence of 29.5%, similar to previous reports (8, 10, 11). Moreover, the proportions of RT014/020 isolated from private and public laboratories were comparable (43.4% and 56.6%, respectively), although significantly more strains were isolated from public laboratories based in tertiary hospitals (Fig. 1). C. difficile RT014 was reported as the most prevalent RT in pig herds in Australia (13), and more recently, genome analysis of the RT014 lineage in piglets and humans showed close genetic relatedness by core genome single-nucleotide polymorphism (cgSNP) analysis, suggesting long-range interspecies transmission and implying that CDI was a zoonoses (14).
C. difficile RT297/310 emerged as a group that may have been underestimated or overlooked in previous studies in Australia. The banding patterns of RT297/310 and RT014/020 differ only in the size of their largest amplicon (∼550 to 575 bp, RT014/020; ∼650 to 675 bp, RT297/310) (see Fig. S1 in the supplemental material). These small differences demonstrate the difficulty in distinguishing between potentially closely related C. difficile strains using conventional band-based typing methods and highlight the need to use sequenced-based methods to reliably determine strain relatedness.
In Europe, RT014/020 was the most common type isolated in 2008 (15); however, epidemic C. difficile RT027 emerged to become the most prevalent RT from mid-2011 to 2014 (16). Only two isolates of C. difficile RT027 were recovered from specimens submitted from public laboratories in this study over a 5-year period, one each from NSW and QLD. C. difficile RT027 has been reported rarely in Australia (17), and it has been postulated that the restricted use of fluoroquinolones in Australia has not favored the establishment of RT027 (18). Also, the geographic isolation of Australia may have contributed to the delayed appearance of this type. However, other CDT+ RTs of C. difficile, RT244 and RT251, were detected in the present study at low prevalences of 0.5% (n = 7) and 0.7% (n = 10), respectively. C. difficile RT244 emerged in Australia and New Zealand in 2011 and was first identified as a “presumptive RT027 strain” (19, 20). CDI caused by RT244 was associated with severe disease and a high mortality rate and predominantly occurred in the community (6). Similarly, C. difficile RT251 strains were detected around the same time as RT244 in Australia, and severe disease and death in younger patients have been described (7). Genomic analyses revealed that C. difficile RT244, RT251, and RT027 were genetically distinct but all belonged in the same phylogenetic multilocus sequence type (MLST) clade 2, suggesting a recent shared evolutionary ancestry (6, 7, 19).
C. difficile RT126 was the most prevalent CDT+ type recovered in this study, and the majority of isolates were isolated in VIC in 2016. This RT has been isolated from humans with CDI as well as from pigs and cattle, a trait shared with other RTs belonging to the same ST11 lineage (21). ST11 strains infect and colonize both humans and animals, with strong evidence of long-range inter- and intraspecies transmission (13). Although the ST11 type RT078 is commonly isolated from humans and animals in Europe and the United States (22, 23), only six isolates were recovered in this study. These cases were sporadic and mostly from NSW. The finding of low numbers of RT078 isolates circulating in NSW was consistent with our previous reports (8, 10, 11). Furthermore, RT078 and RT126 give very similar banding patterns (21) and are sometimes reported together as RT078/126 or not differentiated.
The prevalence of RT056 increased in the current study (8). C. difficile RT056 is well established in both humans and livestock in Australia (10, 24). It is also one of the most common RTs in Australian food (organic potatoes, and carrots) and the environment (compost and roll-out lawns) (25, 26). More recently, cgSNP analysis of C. difficile RT056 strains from humans, food, and compost demonstrated a clonal relationship between these strains consistent with recent transmission events (S. C. Lim, unpublished data).
The overall prevalence of C. difficile RT002 in this study was lower than previously reported in Australia (8). Almost half the RT002 strains reported in this study were isolated in NSW (n = 76), the majority (63.2%) from the laboratories based in a tertiary hospital; however, there was no temporal clustering over the 5-year period (P > 0.05). This RT was reported as the predominant clone recovered from patients with CDI in Hong Kong and was associated with high morbidity and mortality (27). C. difficile RT002 appeared to have a higher sporulation rate and higher rates of fluoroquinolone resistance, similar to epidemic RT027 (27). This type is among the most common C. difficile RTs in Europe and the United States (16, 28); however, its significance in Australia is unknown.
A small group of C. difficile RT017 strains was identified. This type has a variant toxin profile (A−B+CDT−) and is a major cause of CDI across Asia (29). Notably, two of the top 20 most prevalent C. difficile strains in Australia, RT010 and RT039, were nontoxigenic. These RTs were predominately from private laboratories and from patients with symptoms of CDI, suggesting that they may have been simultaneously colonized with toxigenic strains. Coinfection with and carriage of multiple C. difficile strains are not common (30, 31). A possible source of nontoxigenic C. difficile is the environment, and RT010 and RT039 have been found in lawns and compost in WA (26).
This study has some limitations. As reported in our earlier publication, there was a lack of information about the clinical significance of isolates (8). Diagnostic testing remains controversial in Australia and, in recent years, many laboratories that moved to nucleic acid amplification tests (NAATs) for the detection of C. difficile toxin genes have changed to a 2-step algorithm due to lack of specificity of NAATs (32). Thus, while many CDI isolates in this study were derived from PCR-positive stool specimens, the Australian Commission on Safety and Quality in Health Care describes NAATs as suitable for surveillance purposes (4). Last, as mentioned earlier, there may be some classification inconsistencies for HI and CA isolates due to the organizational structure of the health care systems in Australia. However, this is an ongoing surveillance study for C. difficile with the aim of monitoring emerging strains (8), and consistency in methodology remains critical for the purpose of conducting surveillance (33).
To summarize, a heterogeneous C. difficile strain population was identified in Australia between 2013 and 2018. C. difficile RT014/020 remained the most prevalent type found in humans with CDI. The detection of identical RTs commonly isolated from humans, animals, and the environment supports a zoonotic paradigm for CDI and the need for a One Health approach to CDI management. Future work using high-resolution whole-genome-based typing will determine the true extent of genetic relatedness of identical C. difficile RTs as well as possible bidirectional transmission of C. difficile between health care and community settings. Continued surveillance of CDI in Australia remains important for the detection of emerging strain lineages and for developing improved diagnostic tools and therapeutic options.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the laboratory scientists at each study site who contributed to the processing and/or shipment of patient specimens or C. difficile isolates to PathWest Laboratory Medicine, Nedlands, Australia.
Performed experiments: S.H., D.R.K. Analyzed data: S.H., D.R.K., P.P., T.V.R. Contributed patient samples, isolates and metadata: N.G., C.H., P.G.H., T.M.K., D.K., M.L., R.M., C.M., G.R.N., L.P., J.R., L.W., M.C.W., G.F.W., R.M.W. Drafted manuscript: S.H. Revised manuscript: D.R.K., P.P., T.V.R. All authors edited and approved the final version of the manuscript.
T.V.R. reports grants from Merck, grants from Otsuka, grants from Summit, and grants from Roche, outside the present work.
This work was supported, in part, by funding from The Raine Medical Research Foundation (RPG002-19) and a fellowship from the National Health and Medical Research Council (APP1138257) awarded to D.R.K.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 3.Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, Wilson LE, Holzbauer SM, Phipps EC, Dumyati GK, Beldavs ZG, Kainer MA, Karlsson M, Gerding DN, McDonald LC, Emerging Infections Program Clostridioides difficile Infection Working Group. 2020. Trends in U.S. burden of Clostridioides difficile infection and outcomes. N Engl J Med 382:1320–1330. doi: 10.1056/NEJMoa1910215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Australian Commission on Safety and Quality in Health Care. 2011. Implementation guide for surveillance of Clostridium difficile infection. Commonwealth of Australia, Canberra, Australia. [Google Scholar]
- 5.Slimings C, Armstrong P, Beckingham WD, Bull AL, Hall L, Kennedy KJ, Marquess J, McCann R, Menzies A, Mitchell BG, Richards MJ, Smollen PC, Tracey L, Wilkinson IJ, Wilson FL, Worth LJ, Riley TV. 2014. Increasing incidence of Clostridium difficile infection, Australia, 2011–2012. Med J Aust 200:272–276. doi: 10.5694/mja13.11153. [DOI] [PubMed] [Google Scholar]
- 6.Eyre DW, Tracey L, Elliott B, Slimings C, Huntington PG, Stuart RL, Korman TM, Kotsiou G, McCann R, Griffiths D, Fawley WN, Armstrong P, Dingle KE, Walker AS, Peto TE, Crook DW, Wilcox MH, Riley TV. 2015. Emergence and spread of predominantly community-onset Clostridium difficile PCR ribotype 244 infection in Australia, 2010 to 2012. Euro Surveill 20:21059. doi: 10.2807/1560-7917.es2015.20.10.21059. [DOI] [PubMed] [Google Scholar]
- 7.Wehrhahn MC, Keighley C, Kurtovic J, Knight DR, Hong S, Hutton ML, Lyras D, Wang Q, Leong R, Borody T, Edye M, Riley TV. 2019. A series of three cases of severe Clostridium difficile infection in Australia associated with a binary toxin producing clade 2 ribotype 251 strain. Anaerobe 55:117–123. doi: 10.1016/j.anaerobe.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Knight DR, Giglio S, Huntington PG, Korman TM, Kotsanas D, Moore CV, Paterson DL, Prendergast L, Huber CA, Robson J, Waring L, Wehrhahn MC, Weldhagen GF, Wilson RM, Riley TV. 2015. Surveillance for antimicrobial resistance in Australian isolates of Clostridium difficile, 2013–14. J Antimicrob Chemother 70:2992–2999. doi: 10.1093/jac/dkv220. [DOI] [PubMed] [Google Scholar]
- 9.Australian Comission on Safety and Quality in Health Care. 2018. Clostridium difficile infection. Monitoring the national burden of Clostridium difficile. ACSQHC, Sydney, Australia. [Google Scholar]
- 10.Cheng AC, Collins DA, Elliott B, Ferguson JK, Paterson DL, Thean S, Riley TV. 2016. Laboratory-based surveillance of Clostridium difficile circulating in Australia, September–November 2010. Pathology 48:257–260. doi: 10.1016/j.pathol.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Collins DA, Putsathit P, Elliott B, Riley TV. 2017. Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012. Pathology 49:309–313. doi: 10.1016/j.pathol.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Huber CA, Hall L, Foster NF, Gray M, Allen M, Richardson LJ, Robson J, Vohra R, Schlebusch S, George N, Nimmo GR, Riley TV, Paterson DL. 2014. Surveillance snapshot of Clostridium difficile infection in hospitals across Queensland detects binary toxin producing ribotype UK 244. Commun Dis Intell Q Rep 38:E279–E284. [PubMed] [Google Scholar]
- 13.Knight DR, Squire MM, Riley TV. 2015. Nationwide surveillance study of Clostridium difficile in Australian neonatal pigs shows high prevalence and heterogeneity of PCR ribotypes. Appl Environ Microbiol 81:119–123. doi: 10.1128/AEM.03032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight DR, Squire MM, Collins DA, Riley TV. 2016. Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front Microbiol 7:2138. doi: 10.3389/fmicb.2016.02138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer MP, Notermans DW, Van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, Van Dissel JT, Kuijper EJ, Group ES. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 16.Freeman J, Vernon J, Pilling S, Morris K, Nicholson S, Shearman S, Longshaw C, Wilcox MH, Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes Study Group. 2018. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011–2014. Clin Microbiol Infect 24:724–731. doi: 10.1016/j.cmi.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Richards M, Knox J, Elliott B, Mackin K, Lyras D, Waring LJ, Riley TV. 2011. Severe infection with Clostridium difficile PCR ribotype 027 acquired in Melbourne, Australia. Med J Aust 194:369–371. doi: 10.5694/j.1326-5377.2011.tb03012.x. [DOI] [PubMed] [Google Scholar]
- 18.Foster NF, Collins DA, Ditchburn SL, Duncan CN, van Schalkwyk JW, Golledge CL, Keed AB, Riley TV. 2014. Epidemiology of Clostridium difficile infection in two tertiary-care hospitals in Perth, Western Australia: a cross-sectional study. New Microbes New Infect 2:64–71. doi: 10.1002/nmi2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim SK, Stuart RL, Mackin KE, Carter GP, Kotsanas D, Francis MJ, Easton M, Dimovski K, Elliott B, Riley TV, Hogg G, Paul E, Korman TM, Seemann T, Stinear TP, Lyras D, Jenkin GA. 2014. Emergence of a ribotype 244 strain of Clostridium difficile associated with severe disease and related to the epidemic ribotype 027 strain. Clin Infect Dis 58:1723–1730. doi: 10.1093/cid/ciu203. [DOI] [PubMed] [Google Scholar]
- 20.De Almeida MN, Heffernan H, Dervan A, Bakker S, Freeman JT, Bhally H, Taylor SL, Riley TV, Roberts SA. 2013. Severe Clostridium difficile infection in New Zealand associated with an emerging strain, PCR-ribotype 244. N Z Med J 126:9–14. [PubMed] [Google Scholar]
- 21.Knight DR, Kullin B, Androga GO, Barbut F, Eckert C, Johnson S, Spigaglia P, Tateda K, Tsai PJ, Riley TV. 2019. Evolutionary and genomic insights into Clostridioides difficile sequence type 11: a diverse zoonotic and antimicrobial-resistant lineage of global one health importance. mBio 10:e00446-19. doi: 10.1128/mBio.00446-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, Bergwerff AA, Dekker FW, Kuijper EJ. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 23.Jhung MA, Thompson AD, Killgore GE, Zukowski WE, Songer G, Warny M, Johnson S, Gerding DN, McDonald LC, Limbago BM. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg Infect Dis 14:1039–1045. doi: 10.3201/eid1407.071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight DR, Riley TV. 2013. Prevalence of gastrointestinal Clostridium difficile carriage in Australian sheep and lambs. Appl Environ Microbiol 79:5689–5692. doi: 10.1128/AEM.01888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim SC, Foster NF, Elliott B, Riley TV. 2018. High prevalence of Clostridium difficile on retail root vegetables, Western Australia. J Appl Microbiol 124:585–590. doi: 10.1111/jam.13653. [DOI] [PubMed] [Google Scholar]
- 26.Moono P, Lim SC, Riley TV. 2017. High prevalence of toxigenic Clostridium difficile in public space lawns in Western Australia. Sci Rep 7:41196. doi: 10.1038/srep41196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong SH, Ip M, Hawkey PM, Lo N, Hardy K, Manzoor S, Hui WW, Choi KW, Wong RY, Yung IM, Cheung CS, Lam KL, Kwong T, Wu WK, Ng SC, Wu JC, Sung JJ, Lee N. 2016. High morbidity and mortality of Clostridium difficile infection and its associations with ribotype 002 in Hong Kong. J Infect 73:115–122. doi: 10.1016/j.jinf.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Tickler IA, Obradovich AE, Goering RV, Fang FC, Tenover FC, Consortium HAI. 2019. Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe 60:102050. doi: 10.1016/j.anaerobe.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Imwattana K, Wangroongsarb P, Riley TV. 2019. High prevalence and diversity of tcdA-negative and tcdB-positive, and non-toxigenic, Clostridium difficile in Thailand. Anaerobe 57:4–10. doi: 10.1016/j.anaerobe.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Behroozian AA, Chludzinski JP, Lo ES, Ewing SA, Waslawski S, Newton DW, Young VB, Aronoff DM, Walk ST. 2013. Detection of mixed populations of Clostridium difficile from symptomatic patients using capillary-based polymerase chain reaction ribotyping. Infect Control Hosp Epidemiol 34:961–966. doi: 10.1086/671728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neill GL, Beaman MH, Riley TV. 1991. Relapse versus reinfection with Clostridium difficile. Epidemiol Infect 107:627–635. doi: 10.1017/s0950268800049323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trubiano JA, Cheng AC, Korman TM, Roder C, Campbell A, May ML, Blyth CC, Ferguson JK, Blackmore TK, Riley TV, Athan E. 2016. Australasian Society of Infectious Diseases updated guidelines for the management of Clostridium difficile infection in adults and children in Australia and New Zealand. Intern Med J 46:479–493. doi: 10.1111/imj.13027. [DOI] [PubMed] [Google Scholar]
- 33.Hebden JN. 2012. Rationale for accuracy and consistency in applying standardized definitions for surveillance of health care-associated infections. Am J Infect Control 40:S29–S31. doi: 10.1016/j.ajic.2012.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.