Abstract
Plasmid-mediated quinolone resistance (PMQR) genes located on conjugative plasmids can be transferred to other bacteria in the absence of antimicrobial selective pressure. To elucidate the prevalence of resistance, including PMQR in an egg-producing commercial layer farm in western Japan where no antimicrobials were used, minimum inhibitory concentrations (MIC) for a total of 375 Escherichia coli isolates obtained from chicken houses in the farm between 2012 and 2017 were determined using the agar dilution methods. Eighty-seven isolates resistant to oxytetracycline (OTC) accounted for 23.0% of the tested isolates, followed by isolates resistant to dihydrostreptomycin (DSM) (18.4%), sulfisoxazole (18.1%), ampicillin (AMP) (14.4%), trimethoprim (TMP) (14.4%), and nalidixic acid (10.1%). The prevalence rate of multidrug-resistant (MDR) isolates—which are resistant to 3 or more antimicrobial classes, including β-lactams, aminoglycosides, quinolones, folate pathway inhibitors, tetracyclines, and phenicols—was inversely related to the age of chickens at the time of bacterial examination. Probably, the prevalence of MDR isolates in layer chickens may have decreased with age owing to the absence of selective pressure. Furthermore, 45 isolates exhibiting enrofloxacin MICs of more than 0.25 μg/mL were examined for PMQR genes. The transfer of PMQR genes was tested by conjugation analysis. Southern blot analysis of genomic DNA revealed that the qnrS1 (5 isolates), qnrS2 (1 isolate), and qnrS13 genes (1 isolate) were located on plasmids with sizes ranging from approximately 60 to 120 kpb. In 1 of the 5 qnrS1-positive isolates and in an isolate with qnrS13, the qnrS genes were transferred to recipient strains. The plasmid harboring the qnrS1 gene was typed as IncF by PCR-based replicon typing. On this plasmid, the blaTEM, aadA, tetA, and dfrA1 genes responsible for resistance to AMP, DSM, OTC, and TMP, respectively, were detected. The tetA gene was detected in the plasmid harboring the qnrS13 gene, which was typed as IncI1. These results suggest that despite the low prevalence of quinolone resistance in this farm, various PMQR genes, located on diverse plasmids, exist.
Key words: Escherichia coli, layer chicken, multidrug resistance, plasmid, quinolone
Introduction
Plasmid-mediated quinolone resistance (PMQR) is conferred by the genes located on plasmids and usually causes low-level resistance to quinolones. The qnrS gene was first discovered in a transferable plasmid in Shigella flexneri 2b isolated from a human in Japan in 2003 (Hata et al., 2005). Since then, PMQR genes have been detected in Escherichia coli and Klebsiella oxytoca isolates from human clinical specimens in Japan (Ode et al., 2009). They have also been found in enterobacteria from various species of animals, including dairy cows (Asai et al., 2010), companion animals (Harada et al., 2017), and zoo animals (Ahmed et al., 2007). Although PMQR genes in bacterial isolates from broiler chickens have been occasionally reported (Kawanishi et al., 2013, Ozaki et al., 2017, Nishikawa et al., 2019), studies on the prevalence and characterization of isolates with PMQR genes from layer chickens around the world are very few (Niero et al., 2018, Seo and Lee, 2019). The PMQR genes can be located on conjugative plasmids along with other resistance genes conferring extended-spectrum and plasmid-mediated AmpC β-lactamases targeting third-generation cephalosporins (Ozaki et al., 2017, Nishikawa et al., 2019), which are considered to be clinically important. Therefore, these resistance genes can potentially spread and persist in the absence of selective pressure. This is especially important as usually, antimicrobial agents are not used for growth promotion and therapeutic purposes in layer farms during egg production cycles. Thus, monitoring of such farms in which rearing practices do not use antimicrobials may be essential to investigate the prevalence of antimicrobial resistance in commensal E. coli. Moreover, E. coli is suitable for such long-term prevalence analyses because it is considered to be a member of the healthy intestinal flora (Van den Bogaard et al., 2000). In the present study, antimicrobial susceptibilities of E. coli isolates obtained from chicken houses in a commercial layer farm during the 5-year study were determined, and the PMQR genes were characterized.
Materials and methods
Bacterial Isolates
Environmentally controlled windowless houses in a commercial egg-producing chicken farm in western Japan were monitored for antimicrobial resistance in E. coli from July 2012 to June 2017. Approximately 26,000 layer hens were reared in each of the houses that were connected with egg belts. Environmental samples were collected once a month from the 3 houses (houses A, B, and C) except for the periods when chickens were between production cycles. Approximately 120-day-old flocks were brought from young chicken-rearing farms. The flocks were molted when they were older than 450 D. After molting, the flocks were reared for additional 200–250 D. No antimicrobials were administered in this farm. Houses A and B were adjacently situated, and house C was 30 m apart from house A and B. In each of the houses, 2 swabs each were obtained from spots on the wall, floor, egg belts, and feed trough. A total of 1,328 swab samples were subjected to bacterial examinations. Each of the swabs was inoculated into 2 mL of nutrient broth (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) and incubated for 6 h. The cultures were spread onto deoxycholate hydrogen sulfide lactose agar plates (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) and incubated at 37°C for 20 h. Two suspect colonies from each sample were picked and identified as E. coli using API20 E (bioMérieux, Marcy-l’Étoile, France). One isolate from each of the sampling point was arbitrarily selected, and a total of 375 isolates were used.
Antimicrobial Susceptibility Testing
Susceptibilities to ampicillin (AMP), cefazolin (CEZ), ceftiofur (CTF), dihydrostreptomycin (DSM), gentamicin (GEN), kanamycin (KAN), oxytetracycline (OTC), chloramphenicol (CHL), nalidixic acid (NAL), enrofloxacin (ERFX), sulfisoxazole (SUL), and trimethoprim (TMP) were determined by the agar dilution method (Clinical Laboratory Standards Institute, 2009). E. coli ATCC 25922 was used as the quality control strain. Minimum inhibitory concentrations (MIC) were interpreted using the resistance breakpoints defined in the previous study (Nishikawa et al., 2019).
Classification of Isolates
The antimicrobials used represented 6 antimicrobial classes (classification was done according to the Canadian Integrated Program for Antimicrobial Resistance Surveillance) (Government of Canada, 2015). Because CTF-resistant isolates were not obtained in this study (see Results), it was not included in these classes. The antimicrobial classes and the antimicrobial(s) they comprise (in parentheses) are as follows: β-lactams (AMP and CEZ), aminoglycosides (DSM, KAN, and GEN), quinolones (NAL and ERFX), folate pathway inhibitors (SUL and TMP), tetracyclines (OTC), and phenicols (CHL). An isolate was considered to be multidrug-resistant (MDR) if it was resistant to 3 or more antimicrobial classes described previously (MacKinnon et al., 2018).
Statistical Analysis
Isolates obtained during each of 4 months were grouped according to the age of chickens (4–23 months) at the time of bacterial examination to include more than 50 isolates in each of the group (Table 1). Differences in the prevalence rate of MDR isolates in E. coli obtained between each of the periods of 4 months were evaluated by application of a chi-square test. Differences were considered significant at P < 0.05. Fifteen isolates obtained when the chickens were 24-month old were excluded from this analysis because it is a general practice to remove chickens older than 700 D from the farm.
Table 1.
The prevalence of multidrug-resistant (MDR) isolates and distribution for E. coli isolates from layer chicken houses.
| Age of chickens at the time of isolation | Total number of isolates | Number of antimicrobial classes1 |
Prevalence of MDR isolates | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |||
| 4–7 months | 76 | 39 | 10 | 3 | 4 | 14 | 6 | 0 | 31.6%a |
| 8–11 months | 81 | 46 | 15 | 5 | 8 | 2 | 4 | 1 | 18.5%a |
| 12–15 months | 81 | 49 | 16 | 5 | 5 | 4 | 2 | 0 | 13.6%b |
| 16–19 months | 54 | 36 | 9 | 4 | 1 | 0 | 4 | 0 | 9.3%b |
| 20–23 months | 68 | 47 | 10 | 3 | 5 | 1 | 2 | 0 | 11.8%b |
a,bValues that share no common letters differ significantly (P < 0.05).
“0” signifies that isolates within this column had no resistance against any of 6 antimicrobial classes (β-lactams, aminoglycosides, quinolones, folate pathway inhibitors, tetracyclines, and phenicols) (see Materials and Methods). “39” indicates the number of isolates that were not resistant to the aforementioned antimicrobials. Italicized numbers below the lanes with 3 through 6 represent MDR isolates.
PCR Detection of Antimicrobial Resistance Genes and Pulsed-Field Gel Electrophoresis
Forty-five isolates exhibiting more than 0.25 μg/mL of ERFX MIC were screened for 8 PMQR genes (qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, qepA, and oqxAB) by PCR (Park et al., 2006, Robicsek et al., 2006, Chmelnitsky et al., 2009, Ciesielczuk et al., 2013) (Supplementary Table 1). Because the qnrS-positive isolates were resistant to AMP, DSM, OTC, or TMP (see Results and discussion section), genes responsible for this resistance along with integrons were screened using PCR with primer pairs described by Carlson et al., 1999, Lévesque et al., 1995, Madsen et al., 2000, Ng et al., 2001, and White et al. (2001) (Supplementary Table 1). Sequencing of the whole qnrS gene and class 1 integrons was performed with primers described by Dolejska et al. (2011) and Lévesque et al. (1995), respectively (Supplementary Table 1). Isolates with PMQR genes were subjected to XbaI-digested pulsed-field gel electrophoresis (PFGE), as previously described (Ozaki et al., 2011).
Transferability and Plasmid DNA Analysis
Isolates carrying the qnrS gene were resistant to the aforementioned drugs, namely, AMP, DSM, OTC, or TMP. These isolates were tested for their ability to transfer resistance to recipient strains, spontaneous rifampin-resistant mutants of E. coli strains DH5α and ML1410. Overnight cultures of the donor and recipient strains were cocultured in a freshly prepared broth and incubated at 37°C for 6 h. Transconjugants were selected on Mueller–Hinton agar (MHA) plates containing 0.25 μg/mL of ERFX, 50 μg/mL of AMP, 10 μg/mL of DSM, or 50 μg/mL of OTC along with 25 μg/mL of rifampin. Identity of transconjugants was confirmed by looking for XbaI-digested PFGE profiles identical to those of the recipient strains. Plasmids of the transconjugants were typed using PCR-based replicon typing (Carattoli et al., 2005). To determine the chromosomal or plasmid location of the qnrS, blaTEM, and tetA genes and class 1 integron, Southern blot analysis of the transconjugants was performed using S1 nuclease–digested genomic DNA separated by PFGE in accordance with previously described methods (Shahada et al., 2011). DNA from the PFGE gel was transferred onto a Hybond-N+ membrane (Amersham Biosciences UK Ltd., Little Chalfont, UK). The PCR-amplified DNA fragments from isolates carrying the qnrS, blaTEM, and tetA genes and class 1 integron were labeled with digoxigenin using a DIG High Prime Labeling and Detection Starter Kit (Roche Diagnostics Corp., Indianapolis, IN) and used as a specific probe for these determinants. To determine molecular weights of the IncI1 and IncF plasmids, PCR-amplified fragments from each of the isolates positive for the IncI1 and IncF plasmids were labeled with digoxigenin as described previously, and Southern blot analysis of the transconjugants was performed using S1 nuclease–digested genomic DNA separated by PFGE.
Results and discussion
Prevalence of Antimicrobial Resistance in E. coli
Eighty-seven OTC-resistant isolates accounted for 23.0% of the total of 375 E. coli isolates, followed by DSM- (69; 18.4%), SUL- (68; 18.1%), AMP- (54; 14.4%), TMP- (54; 14.4%), NAL- (38; 10.1%), ERFX- (26; 6.9%), GEN- (9; 2.4%), KAN- (7; 1.9%), CHL- (7; 1.9%), and CEZ- (2; 0.5%) resistant strains; 226 isolates (60.3%) were susceptible to all the drugs tested. No isolates were resistant to CTF. The prevalence of E. coli resistant to antimicrobial drugs tested in this study was comparable with results obtained in the Japanese Veterinary Antimicrobial Resistance Monitoring System (National Veterinary Assay Laboratory, 2016a, National Veterinary Assay Laboratory, 2018), in accordance with which resistance to tetracycline was most frequently detected in 37.9 and 24.6% of the E. coli isolates obtained from the feces of layer chickens, between the years, 2012–2013 and 2014–2015, respectively. Rates of resistance to AMP, streptomycin, NAL, and TMP were 9.5–18.4% during the same period. The incidence of resistant strains in layer chickens is due to the selection of resistant bacteria within the intestinal flora of chickens during the growth phase before being picked out for laying. Substantial amounts of tetracyclines, penicillins, and sulfamonomethoxine were sold for the use in layer chickens (National Veterinary Assay Laboratory, 2016b) which may have been administered to growing chickens. However, information about the actual amounts of these drugs administered to young chickens is unavailable. The results in the present study obtained from monitoring environmental swab samples could in actual be reflecting the fecal carriage of antimicrobial-resistant E. coli. This consideration is also supported by a previous study that showed that the incidence of Salmonella in the environmental samples in layer chicken houses increased after induced molting (Murase et al., 2001).
The most frequently encountered phenotype among MDR isolates was resistance to β-lactams, aminoglycosides, folate pathway inhibitors, and tetracyclines (20 isolates). The second most frequent phenotype was the resistance to β-lactams, aminoglycosides, folate pathway inhibitors, tetracyclines, and quinolones, which was detected in 18 isolates. Twelve isolates were resistant to aminoglycosides, folate pathway inhibitors, and tetracyclines. Of the 76 isolates, which were obtained when the chickens were 4- to 7-month-old, 24 isolates were MDR, accounting for 31.6% (Table 1), which was significantly (P < 0.05) higher than the rates of MDR isolates at the ages of 12–15 months (13.6%), 16–19 months (9.3%), and 20–23 months (11.8%). The observation that the prevalence rate of MDR isolates decreased along with the age of the layers in this study may be partly due to the rearing practice at this farm because antimicrobial agents were not used during the study period. The birds may have carried these MDR strains at the time of introduction to this farm, and the prevalence may have decreased in the absence of the selection pressure.
PMQR-Positive Isolates
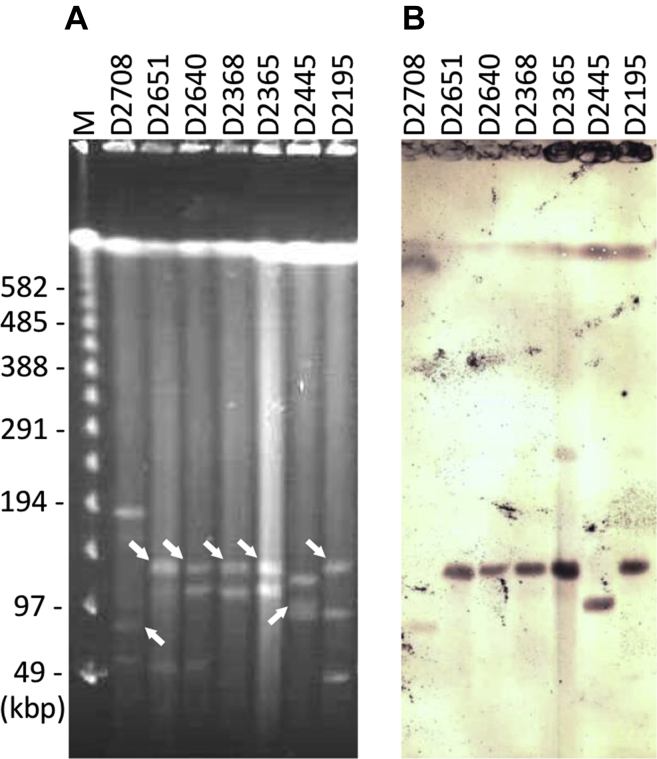
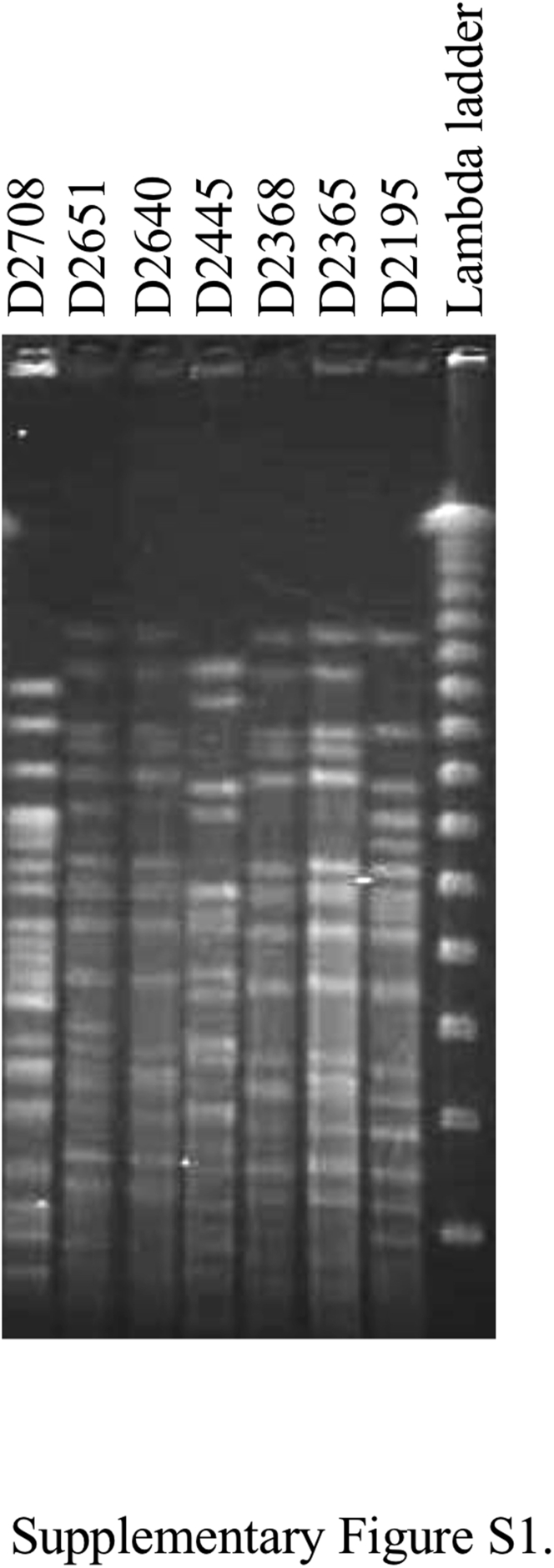
The qnrS1, qnrS2, and qnrS13 genes were detected in 5, 1, and 1 isolate(s), respectively (Table 2). Southern blot analysis revealed that the qnrS genes were located on plasmids with sizes ranging from approximately 60 to 120 kilobase pairs (Figure 1). Five isolates harboring the qnrS1 gene were obtained from 3 houses and had an identical resistance phenotype. The PFGE analysis (Supplementary Figure 1) revealed that 2 qnrS1-positive isolates from house B in 2013 and additional 2 isolates from house A in 2015 were genetically related to each other according to the established criteria for bacterial isolate typing by PFGE (Tenover et al., 1995). For 2 years, the qnrS1-positive E. coli isolates were repeatedly obtained from chicken houses of proximity. Similarly, samples of Salmonella with identical PFGE patterns were isolated for more than 1 year in layer chicken houses have been reported (Murase et al., 2001, Murase et al., 2004).
Table 2.
Characteristics of the qnrS-positive E. coli isoates.
| Isolate ID1 | Isolation date | House | ERFX MIC | qnrS allele | Resistance phenotype2 |
|---|---|---|---|---|---|
| D2195 | Jul, 2012 | C | 2 μg/ml | qnrS1 | AMP, DSM, OTC, ERFX, TMP |
| D2365* | Jul, 2013 | B | 2 μg/ml | qnrS1 | AMP, DSM, OTC, ERFX, TMP |
| D2368* | Jul, 2013 | B | 2 μg/ml | qnrS1 | AMP, DSM, OTC, ERFX, TMP |
| D2445 | Apr, 2014 | B | 2 μg/ml | qnrS13 | OTC, NAL, ERFX |
| D2640* | May, 2015 | A | 2 μg/ml | qnrS1 | AMP, DSM, OTC, ERFX, TMP |
Pulsed-field gel electrophoresis (PFGE) patterns (Supplementary Figure 1) of isolates with an asterisk reveals that these were genetically related to each other according to the established criteria for bacterial isolate typing by PFGE (Tenover et al., 1995).
AMP, ampicillin; DSM, dihydrostreptomycin; OTC, oxytetracycline; NAL, nalidixic acid; ERFX, enrofloxacin; TMP, trimethoprim.
Figure 1.
(A) Pulsed-field gel electrophoresis patterns of S1 nuclease–digested genomic DNA of the qnrS-positive E. coli isolates, and (B) Southern blot hybridization with a probe prepared from the PCR amplicon using primer pairs specific for qnrS genes; the isolate number is indicated atop each of the lanes (see Table 2); Lane M-lambda ladder; the sizes of the markers are indicated on the left side of the panel. The bands in (B) hybridized with probes specific for the qnrS genes are indicated with arrows.
Transferability and Plasmid DNA Analysis
Transconjugants were obtained when they were selected using not only ERFX but also DSM and OTC. Transconjugant named TC2195-DH-OTC—which was selected with OTC—was derived from a recipient strain DH5α and received multidrug resistance from isolate D2195 (Table 3). The other transconjugants were named accordingly based on the abbreviation of the recipient and donor strains and the antimicrobial used for selection. The MICs of AMP, DSM, OTC, ERFX, and TMP in transconjugants TC2195-DH-OTC and TC2195-ML-DSM were comparable with those of the donor strain D2195 and interpreted as resistant, except for MICs of DSM and ERFX in TC2195-DH-OTC. Similarly, TC2445-ML-ERFX exhibited increased MICs of OTC and ERFX compared with that in the recipient strain ML1410. The blaTEM, aadA, and tetA genes, which are responsible for resistance to AMP, DSM, and OTC, respectively, were detected in isolates D2195 or D2445 by PCR. In isolate D2195, class 1 integron was detected, and sequencing analysis revealed the presence of the dfrA1 gene, which confers resistance to TMP, in the gene cassette of the class 1 integron. In addition, PCR using the forward primer specific to the aadA gene and the reverse primer specific to class 1 integron yielded an amplicon, indicating that the aadA gene was also integrated into the gene cassette of the integron. Southern blot analysis revealed that the blaTEM and tetA genes and class 1 integron were located on the same plasmids carrying the qnrS genes in each of the transconjugants (Supplementary Figure 2). Plasmids in TC2195-DH-OTC and TC2195-ML-DSM were typed as IncF, and that in TC2445-ML-ERFX was typed as IncI1. Probes specific to these plasmids hybridized with the plasmids carrying the qnrS and the other resistance genes in Southern blot analysis (Supplementary Figure 2).
Table 3.
Characteristics of transconjugants and the donor and recipient strains.
| Type of strain and name | MIC (μg/ml) of antimicrobial2 and resistance gene3 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CEZ | CTF | DSM | GEN | KAN | OTC | CHL | NAL | ERFX | SUL | TMP | |
| Transconjugant1 | ||||||||||||
| TC2195-DH-OTC | 256 (blaTEM) | 1 | ≤0.13 | 4 (GC4) | ≤0.13 | 1 | 256 (tetA) | 8 | 64 | 0.5 (qnrS) | ≤32 | >512 (GC) |
| TC2195-ML-DSM | 512 (blaTEM) | 4 | 0.5 | 32 (GC) | 0.5 | 4 | 256 (tetA) | 4 | 512 | 8 (qnrS) | ≤32 | >512 (GC) |
| TC2445-ML-ERFX | 2 | 2 | 0.25 | 4 | 0.5 | 4 | 128 (tetA) | 4 | 512 | 8 (qnrS) | 64 | 1 |
| Donor | ||||||||||||
| D2195 | >512 (blaTEM) | 2 | 0.5 | 32 (aadA) | 0.5 | 8 | 512 (tetA) | 8 | 16 | 2 (qnrS) | ≤32 | >512 (dfrA1) |
| D2445 | 2 | 1 | 0.5 | 8 | 0.5 | 8 | 256 (tetA) | 8 | 16 | 2 (qnrS) | 64 | 1 |
| Recipient | ||||||||||||
| DH5α | 2 | 1 | ≤0.13 | 2 | ≤0.13 | 1 | 2 | 8 | 64 | ≤0.13 | ≤32 | 1 |
| ML1410 | 2 | 1 | 0.25 | 4 | 0.5 | 4 | 2 | 4 | 512 | 0.5 | ≤32 | 2 |
Transconjugants were named accordingly based on the abbreviation of the recipient and donor strains, and the antimicrobial used for selection.
AMP, ampicillin; CEZ, cephazolin; CTF, ceftiofur; DSM, dihydrostreptomycin; GEN, gentamicin; KAN, kanamycin; OTC, oxytetracycline; CHL, chloramphenicol; NAL, nalidixic acid; ERFX, enrofloxacin; SUL, sulfisoxazole; TMP, trimethoprim.
Resistance genes in the donor strains were detected by PCR and sequencing analysis. These genes and the gene cassette (GC) in the transconjugants were detected by Southern blot analysis (Supplementary Figure 2) using probes specific to the blaTEM, tetA, and qnrS genes, and the GC described below.
GC represents the gene cassette of the class 1 integron. PCR and sequencing analysis of isolate D2195 reveals that the aadA and dfrA1 genes were integrated in the gene cassette.
Transfer of resistance, caused by conjugation, occurred in two of the seven PMQR-positive isolates. As in our previous reports (Ozaki et al., 2017, Nishikawa et al., 2019), infrequent transfer of plasmids with PMQR genes in E. coli from broiler chickens was observed in the present study. These results may partly explain the low prevalence of PMQR-positive isolates among chickens in Japan (Kawanishi et al., 2013, Ozaki et al., 2017, Nishikawa et al., 2019). However, resistance genes detected in 2 PMQR-positive isolates were transferred to the recipient strains, and all the genes were located on the plasmids harboring the PMQR genes. Therefore, the plasmids found in the present study can be selected under the usage of drugs other than quinolones.
Conclusion
The prevalence of resistance in E. coli isolates from environmental samples obtained from a commercial layer farm in this study was comparable to the results obtained in the Japanese Veterinary Antimicrobial Resistance Monitoring System (National Veterinary Assay Laboratory, 2016a, National Veterinary Assay Laboratory, 2018). The prevalence rate of MDR isolates decreased along with the age of the layers, possibly due to nonusage of antimicrobial agents during the study period. Three distinct alleles of the qnrS gene were detected on the conjugative plasmids, with 2 replicon types in seven of the 378 isolates tested. We have isolated E. coli having other PMQR genes including oqxAB and aac(6′)-Ib-cr from broiler chickens, although the replicon types were unavailable (Ozaki et al., 2017, Nishikawa et al., 2019). The present study, together with the previous ones, suggests that in Japan, despite the low prevalence of PMQR-positive E. coli in poultry, various PMQR genes have been located on diverse plasmids.
Acknowledgments
This work was supported in part by a grant from MEXT for the Joint Research Program of the Research Center for Zoonosis Control, Hokkaido University. The authors would like to thank Editage (www.editage.jp) for English language editing.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2019.09.005.
Supplementary data
List of primers used in this study.
Supplementary Figure S1.
PFGE patterns of XbaI-digested genomic DNA isolated from the qnrS-positive Escherichia coli isolates.
Supplementary Figure S2.
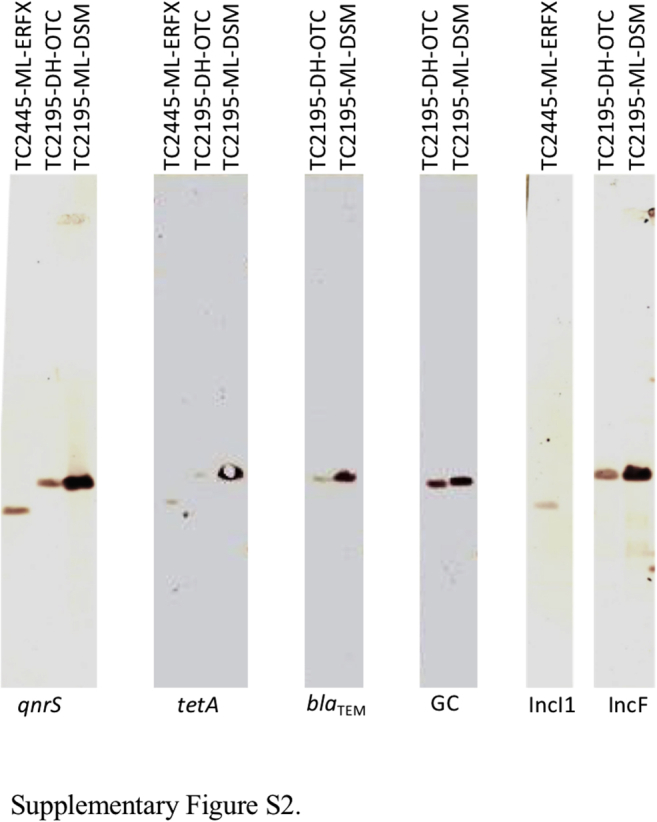
Southern blot hybridization with a probe prepared from the PCR amplicon using primer pairs specific to the qnrS, tetA, and blaTEM genes, the gene cassette, and the IncI1 and IncF plasmids.
References
- Ahmed A.M., Motoi Y., Sato M., Maruyama A., Watanabe H., Fukumoto Y., Shimamoto T. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl. Environ. Microbiol. 2007;73:6686–6690. doi: 10.1128/AEM.01054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Sato C., Masani K., Usui M., Ozawa M., Ogino T., Aoki H., Sawada T., Izumiya H., Watanabe H. Epidemiology of plasmid-mediated quinolone resistance in salmonella enterica serovar typhimurium isolates from food-producing animals in Japan. Gut Pathog. 2010;2:17. doi: 10.1186/1757-4749-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Carlson S.A., Bolton L.F., Briggs C.E., Hurd H.S., Sharma V.K., Fedorka-Cray P.J., Jones B.D. Detection of multiresistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol. Cell. Probes. 1999;13:213–222. doi: 10.1006/mcpr.1999.0240. [DOI] [PubMed] [Google Scholar]
- Chmelnitsky I., Hermesh O., Navon-Venezia S., Strahilevitz J., Carmeli Y. Detection of aac(6΄)-Ib-cr in KPC-producing Klebsiella pneumoniae isolates from Tel Aviv, Israel. J. Antimicrob. Chemother. 2009;64:718–722. doi: 10.1093/jac/dkp272. [DOI] [PubMed] [Google Scholar]
- Ciesielczuk H., Hornsey M., Choi V., Woodford N., Wareham D.W. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol. 2013;62:1823–1827. doi: 10.1099/jmm.0.064428-0. [DOI] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute . Clinical Laboratory Standards Institute; Wayne: 2009. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard M7-A8. [Google Scholar]
- Dolejska M., Duskova E., Rybarikova J., Janoszowska D., Roubalova E., Dibdakova K., Maceckova G., Kohoutova L., Literak I., Smola J., Cizek A. Plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli isolates from an equine clinic and a horseback riding centre. J. Antimicrob. Chemother. 2011;66:757–764. doi: 10.1093/jac/dkq500. [DOI] [PubMed] [Google Scholar]
- Government of Canada . Public Health Agency of Canada; Guelph, Ontario: 2015. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2013 Annual Report – Chapter 2. Antimicrobial Resistance.http://publications.gc.ca/collections/collection_2015/aspc-phac/HP2-4- 2013-2-eng.pdf [Google Scholar]
- Harada K., Shimizu T., Mukai Y., Kuwajima K., Sato T., Kajino A., Usui M., Tamura Y., Kimura Y., Miyamoto T., Tsuyuki Y., Ohki A., Kataoka Y. Phenotypic and molecular characterization of antimicrobial resistance in Enterobacter spp. isolates from companion animals in Japan. PLoS One. 2017;12:e0174178. doi: 10.1371/journal.pone.0174178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata M., Suzuki M., Matsumoto M., Takahashi M., Sato K., Ibe S., Sakae K. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 2005;49:801–803. doi: 10.1128/AAC.49.2.801-803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanishi M., Ozawa M., Hiki M., Abo H., Kojima A., Asai T. Detection of aac(6')-Ib-cr in avian pathogenic Escherichia coli isolates in Japan. J. Vet. Med. Sci. 2013;75:1539–1542. doi: 10.1292/jvms.13-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque C., Piché L., Larose C., Roy P.H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon M.C., Pearl D.L., Carson C.A., Parmley E.J., McEwen S.A. Comparison of annual and regional variation in multidrug resistance using various classification metrics for generic Escherichia coli isolated from chicken abattoir surveillance samples in Canada. Prev. Vet. Med. 2018;154:9–17. doi: 10.1016/j.prevetmed.2018.03.010. [DOI] [PubMed] [Google Scholar]
- Madsen L., Aarestrup F.M., Olsen J.E. Characterisation of streptomycin resistance determinants in Danish isolates of Salmonella Typhimurium. Vet. Microbiol. 2000;75:73–82. doi: 10.1016/s0378-1135(00)00207-8. [DOI] [PubMed] [Google Scholar]
- Murase T., Nagato M., Shirota K., Katoh H., Otsuki K. Pulsed-field gel electrophoresis-based subtyping of DNA degradation-sensitive Salmonella enterica subsp. enterica serovar Livingstone and serovar Cerro isolates obtained from a chicken layer farm. Vet. Microbiol. 2004;99:139–143. doi: 10.1016/j.vetmic.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Murase T., Senjyu K., Maeda T., Tanaka M., Sakae H., Matsumoto Y., Kaneda Y., Ito T., Otsuki K. Monitoring of chicken houses and an attached egg-processing facility in a laying farm for Salmonella contamination between 1994 and 1998. J. Food Prot. 2001;64:1912–1916. doi: 10.4315/0362-028x-64.12.1912. [DOI] [PubMed] [Google Scholar]
- National Veterinary Assay Laboratory Report on the Japanese Veterinary antimicrobial resistance monitoring System: 2012 to 2013. 2016. http://www.maff.go.jp/nval/yakuzai/pdf/jvarm_report_2012_2013.pdf
- National Veterinary Assay Laboratory Annual report of Sales amount and Sales Volume of Veterinary drugs, Quasi-drugs and Medical Devices. Appendix: Sales amounts and Sales Volumes (active Substance) of Antibiotics, Synthetic Antibacterials, Anthelmintics and Antiprotozoals. 2016. http://www.maff.go.jp/nval/iyakutou/hanbaidaka/pdf/h29hanbaikoukin.pdf
- National Veterinary Assay Laboratory Report on the Japanese Veterinary antimicrobial resistance monitoring System: 2014 to 2015. 2018. http://www.maff.go.jp/nval/yakuzai/pdf/JVARM_Report_2014-2015.pdf
- Ng L.K., Martin I., Alfa M., Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes. 2001;15:209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- Niero G., Bortolaia V., Vanni M., Intorre L., Guardabassi L., Piccirillo A. High diversity of genes and plasmids encoding resistance to third-generation cephalosporins and quinolones in clinical Escherichia coli from commercial poultry flocks in Italy. Vet. Microbiol. 2018;216:93–98. doi: 10.1016/j.vetmic.2018.02.012. [DOI] [PubMed] [Google Scholar]
- Nishikawa R., Murase T., Ozaki H. Plasmid-mediated quinolone resistance in Escherichia coli isolates from commercial broiler chickens and selection of fluoroquinolone-resistant mutants. Poult. Sci. 2019 doi: 10.3382/ps/pez337. in press. [DOI] [PubMed] [Google Scholar]
- Ode T., Saito R., Kumita W., Sato K., Okugawa S., Moriya K., Koike K., Okamura N. Analysis of plasmid-mediated multidrug resistance in Escherichia coli and Klebsiella oxytoca isolates from clinical specimens in Japan. Int. J. Antimicrob. Agents. 2009;34:347–350. doi: 10.1016/j.ijantimicag.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Ozaki H., Esaki H., Takemoto K., Ikeda A., Nakatani Y., Someya A., Hirayama N., Murase T. Antimicrobial resistance in fecal Escherichia coli isolated from growing chickens on commercial broiler farms. Vet. Microbiol. 2011;150:132–139. doi: 10.1016/j.vetmic.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Ozaki H., Matsuoka Y., Nakagawa E., Murase T. Characteristics of Escherichia coli isolated from broiler chickens with colibacillosis in commercial farms from a common hatchery. Poult. Sci. 2017;96:3717–3724. doi: 10.3382/ps/pex167. [DOI] [PubMed] [Google Scholar]
- Park C.H., Robicsek A., Jacoby G.A., Sahm D., Hooper D.C. Prevalence in the United States of aac(6΄)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 2006;50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A., Strahilevitz J., Sahm D.F., Jacoby G.A., Hooper D.C. Qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 2006;50:2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo K.W., Lee Y.J. Detection of plasmid-mediated quinolone resistance genes in β-lactamase-producing Escherichia coli isolates from layer hens. Poult. Sci. 2019;98:1480–1487. doi: 10.3382/ps/pey545. [DOI] [PubMed] [Google Scholar]
- Shahada F., Sekizuka T., Kuroda M., Kusumoto M., Ohishi D., Matsumoto A., Okazaki H., Tanaka K., Uchida I., Izumiya H., Watanabe H., Tamamura Y., Iwata T., Akiba M. Characterization of Salmonella enterica serovar Typhimurium isolates harboring a chromosomally encoded CMY-2 β-lactamase gene located on a multidrug resistance genomic island. Antimicrob. Agents Chemother. 2011;55:4114–4121. doi: 10.1128/AAC.00560-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bogaard A.E.J.M., London N., Stobberingh E.E. Antimicrobial resistance in pig faecal samples from The Netherlands (five abattoirs) and Sweden. J. Antimicrob. Chemother. 2000;45:663–671. doi: 10.1093/jac/45.5.663. [DOI] [PubMed] [Google Scholar]
- White P.A., McIver C.J., Rawlinson W.D. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 2001;45:2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used in this study.