Summary
NPR1 has been found to be a key transcriptional regulator in some plant defence responses. There are nine NPR1 homologues (TaNPR1) in wheat, but little research has been done to understand the function of those NPR1‐like genes in the wheat defence response against stem rust (Puccinia graminis f. sp. tritici) pathogens.
We used bioinformatics and reverse genetics approaches to study the expression and function of each TaNPR1.
We found six members of TaNPR1 located on homoeologous group 3 chromosomes (designated as TaG3NPR1) and three on homoeologous group 7 chromosomes (designated as TaG7NPR1). The group 3 NPR1 proteins regulate transcription of SA‐responsive PR genes. Downregulation of all the TaNPR1 homologues via virus‐induced gene co‐silencing resulted in enhanced resistance to stem rust. More specifically downregulating TaG7NPR1 homeologues or Ta7ANPR1 expression resulted in stem rust resistance phenotype. By contrast, knocking down TaG3NPR1 alone did not show visible phenotypic changes in response to the rust pathogen. Knocking out Ta7ANPR1 enhanced resistance to stem rust. The Ta7ANPR1 locus is alternatively spliced under pathogen inoculated conditions.
We discovered a new mode of NPR1 action in wheat at the Ta7ANPR1 locus through an NB‐ARC–NPR1 fusion protein negatively regulating the defence to stem rust infection.
Keywords: mapping, mutant, NPR1, RT‐qPCR, Rust, Triticum aestivum, virus‐induced gene silencing (VIGS)
Introduction
Plants constantly battle with a variety of pathogens in the environment via their complex and effective innate immune systems (Spoel & Dong, 2012). A hypersensitive response (HR) is one of the strategies used to defend against biotrophic pathogens by which rapid programmed cell death occurs immediately surrounding the infection sites (Morel & Dangl, 1997) to restrict the pathogens from further spreading and replication. HR also activates a series of signals that are transduced to remote regions of the plants and generates systemic acquired resistance (SAR) with a broad‐spectrum resistance to subsequent pathogen attacks (Pajerowska‐Mukhtar et al., 2013).
A central positive regulator of SAR signalling in Arabidopsis is NPR1 (Non‐expresser of Pathogenesis‐Related genes 1), also known as Non‐Immunity 1 (NIM1)). The gene is essential for transducing the salicylic acid (SA) signal to activate Pathogenesis‐Related (PR) gene expression (Cao et al., 1994; Dong, 2004). In addition, NPR1 is required by diverse immune signalling pathways, including basal defence, effector‐triggered immunity (ETI) and induced systemic resistance (Rate & Greenberg, 2001; Shirano et al., 2002; Spoel et al., 2003; Pajerowska‐Mukhtar et al., 2013). NPR1 also mediates crosstalk between SA‐mediated and Jasmonic acid (JA)‐mediated signalling pathways (Spoel et al., 2003). The plant‐specific transcription factor WRKY70 is identified as a common component downstream of NPR1 in both SA‐mediated and JA‐mediated signalling pathways (Li et al., 2004). WRKY70 expression is activated by SA and repressed by JA.
NPR1 protein contains an ankyrin repeat domain and a broad complex, tramtrack and bric‐à‐brac/poxvirus and zinc‐finger (BTB/POZ) domain (Cao et al., 1997; Aravind & Koonin, 1999). Since the first cloning of Arabidopsis NPR1 in 1997 (Cao et al., 1997), a significant amount of work has been done to understand the mode of NPR1 action. In the absence of infection, or at low concentration of SA, NPR1 predominantly exists as oligomers through intermolecular disulphide bonds and is retained in the cytoplasm. After pathogen challenge, with elevated SA level, NPR1 converts to a monomeric state by reduction of the redox‐sensitive disulphide bonds. It is then translocated to the nucleus, where NPR1 physically interacts with TGA‐bZIP transcriptional factors and activates the expression of defence response genes (Mou et al., 2003). Nuclear accumulation of NPR1 is needed for basal defence gene expression and resistance, whereas its subsequent turnover is required for establishing SAR (Spoel et al., 2009). Two NPR1 paralogues, NPR3 and NPR4, are required to be the SA receptors (Fu et al., 2012). Both NPR3 and NPR4 contain the BTB domain and ankyrin repeats, which are typical adaptors for CUL3 substrate. Either NPR3 or NPR4 can directly bind to SA and modulate their interactions with NPR1 that results in NPR1 degradation through CUL3‐mediated ubiquitination (Fu et al., 2012; Moreau et al., 2012).
The monomeric state of NPR1 and the subsequent turnover and ubiquitination of the protein in Arabidopsis require a high level of SA often triggered by ETI. In plants, a superfamily of nucleotide‐binding domain and leucine‐rich repeat‐containing proteins (NLRs) can recognise pathogen effectors and initiate ETI. Decades of studies on plant NLRs have revealed two modes of actions, recognising and signalling by itself, or forming a heterogenous protein complex of NLR pairs often arranged in a head‐to‐head orientation, in which one acts as a sensor and the other as a transducer (Césari et al., 2014a; Williams et al., 2014; Saucet et al., 2015; Grund et al., 2019). Several of the sensor NLRs have been found to contain unconventional domains other than their conserved NLR multidomains (Kanzaki et al., 2012; Césari et al., 2014b). These integrated domain (ID)‐containing NLRs are termed NLR‐IDs of which the unusual integrated domains serve as decoys of effector targets in facilitating pathogen detection by the sensor NLR.
Given the pivotal role of NPR1 in defence signalling, studies have been conducted to investigate the role of NPR1‐like genes in other plants. For example, GhNPR1 was shown to play a key role in the SA‐dependent SAR in Gladiolus (Zhong et al., 2015). In wheat (Triticum aestivum L.), elevated PR1 gene induction in transgenic lines carrying the Arabidopsis (AtNPR1) gene suggested that a similar SA‐dependent NPR1‐mediated defence pathway exists in monocots. However, diversified functions of NPR1‐like genes have also been revealed, suggesting that the regulation of defence gene induction between dicots and monocots is quite different (Silverman et al., 1995). In dicots, overexpression of NPR1 orthologues in apple or grapevine has been shown to provide broad‐spectrum resistance (Malnoy et al., 2007; Le Henanff et al., 2011). Overexpression of a rice NPR1 homologue led to constitutive activation of defence response and hypersensitivity to light (Chern et al., 2005). Overexpression of AtNPR1 in wheat enhanced resistance to Fusarium graminearum, a necrotrophic pathogen causing wheat head blight (Makandar et al., 2006). A study on wheat‐stripe rust pathogen Puccinia striiformis f. sp. tritici (Pst) interaction indicated that a Pst effector interacted with wheat NPR1 during infection, implicating a role of wheat NPR1 during the defence response to rust pathogens (Wang et al., 2016).
There are three fungal pathogens from the genus Puccinia that cause wheat‐rust diseases, namely leaf rust (P. triticina), stem rust (P. graminis f. sp. tritici), and stripe rust (Pst). These three diseases combined can cause estimated annual losses of US$2–5 billion to wheat production worldwide (http://www.usda.gov/nass) depending on the varieties grown and developmental stage at which infection occurred. Wheat‐rust pathogens are biotrophs that only survive in living cells and sequester nutrients from their host via haustoria (Dodds et al., 2004). HR is the most common phenotype observed among resistant wheat lines. In this study, we aimed to identify all the wheat homologues of NPR1 (designated as TaNPR1) and explore their roles in the host defence response to rust pathogens. Our research revealed that homoeologous group 3 TaNPR1 proteins regulated transcription of SA‐responsive PR genes. We also discovered a new mode of NPR1 action in wheat at the Ta7ANPR1 locus through an NB‐ARC–NPR1 fusion protein that negatively regulated the defence response to stem rust infection.
Materials and Methods
Plant materials
Alpowa (PI 566596), a soft, white, spring wheat cultivar, was obtained from the USDA National Plant Germplasm System (NPGS). Chinese Spring (CS) and CS + Sr33 were from Evans Lagudah at Commonwealth Scientific and Industrial Research Organization (CSIRO, Australia). CS nulli‐tetra lines and deletion lines were provided by Bikram S. Gill at Kansas State University and Adam Lukazewski at UC Riverside (USA). The ethyl methane sulfonate (EMS) mutagenised Alpowa population was generated by the Giroux laboratory at Montana State University (Feiz et al., 2009).
Mapping populations consisted of 400 F2 lines derived from a cross between the mutant AlR805Q and Alpowa.
Sequence analysis
All Blasts and the sequences downloads were conducted at either the National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov (Altschul et al., 1990) or the International Wheat Genome Sequencing Consortium (IWGSC, 2014, 2018) (https://urgi.versailles.inra.fr/blast_iwgsc/blast.php). Softberry database Fgenesh software (Solovyev et al., 2006) was used for gene prediction. Dnastar software (www.dnastar.com) was used to analyse DNA sequences. InterProScan (http://www.ebi.ac.uk/InterProScan/), Pfam (http://pfam.xfam.org/) (El‐Gebali et al., 2019), and Prosite Scan (http://npsa‐pbil.ibcp.fr) (Hulo et al., 2006) were used to predict the conserved domains and motifs. Multiple sequence alignments were created using clustalw software (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Plant growth conditions
For the race specificity screens, seeds were directly planted in 4‐inch pots (five seedlings per pot) containing SunGro Horticulture Sunshine mix (HeavyGardens Co., Denver, CO, USA). Before inoculation, all the seedlings were grown in a growth room of the Plant Growth Center at Montana State University (PGC‐MSU) under the following conditions: 22°C : 14°C, day : night temperatures and a 16‐h photoperiod. Plants were watered every day and fertilised with Peters General Purpose Plant Food (Scotts‐Miracle‐Gro Co., Marysville, OH, USA), at the concentration of 150 ppm every other day.
Pathogens
The Pgt race QFCSC was used for the stem rust assay and was provided by Yue Jin from the Cereal Disease Laboratory (USDA‐ARS, St Paul, MN, USA). Leaf rust race Pt PBJJG was provided by Robert Bowden (USDA‐ARS, Manhattan, KS, USA) and is maintained at Montana State University. Stripe rust isolates were from the Plant Pathology Research Center at Yangling, Shaanxi, in China.
Pathogen inoculation
For leaf and stem rust tests, seedlings were inoculated with either Pt or Pgt at the 2‐leaf stage. A video protocol detailing the process can be accessed at https://vimeo.com/48605764. Uredospores were mixed in Soltrol 170 Isoparaffin (Chempoint, Bellevue, WA, USA) at a concentration of 0.5 mg ml−1. The suspension was sprayed onto the leaves using a Badger 350 Air‐brush gun and Propel propellant (Badger Air‐Brush Co., Franklin Park, IL, USA). The inoculated seedlings were then placed in a Percival I‐60D dew chamber (Percival Scientific Inc., Perry, IA, USA) at an ambient air temperature of 15–17°C for leaf rust and 19–22°C for stem rust, respectively. After 24 h of incubation, an additional 3 h high humidity and light intensity conditions were added for stem rust inoculated plants. All inoculated plants were then placed back in the growth chamber or glasshouse. Disease responses were assessed when rust symptoms were fully expressed on Alpowa 8–22 dpfi using the seedling 0–4 infection type (IT) scale (Stakman et al., 1962; McIntosh et al., 1995). In detail, IT0: no visible uredia; IT; hypersensitive flecks; IT1: small uredia with necrosis; IT2: small‐sized to medium‐sized uredia with green islands surrounded by necrosis; IT3: medium‐sized uredia without necrosis; IT4: large‐sized uredia without necrosis. The variations within each class are indicated by the use of ‘−’ (less than average for the class) and ‘+’ (more than average for the class). When variable reactions were observed, IT ranges are listed from lowest to highest.
Stripe rust inoculation and assessment
Stripe rust inoculations were conducted at the Institute of Plant Pathology of Northwest A&F University, China. Freshly collected uredospores were applied with a paintbrush to the surface of primary leaves of 7‐d‐old wheat seedlings. After inoculation, plants were incubated for 24 h in the dark in a 100% humidity dew chamber and were subsequently transferred to a growth chamber with a 16‐h photoperiod. Stripe rust infection types were assessed based on a 0 (immune) to 9 (highest susceptible) scale (Line & Qayoum, 1992).
Quantification of pustule density on leaves
Infection types of wheat lines were quantified using imagej software (http://rsb.info.nih.gov/ij) and presented by the average of the percentage of pustule area/leaf area from three leaves per treatment.
Virus‐induced gene silencing
The BSMV vectors utilised in these experiments were obtained from Andrew O. Jackson at UC Berkeley. The fragments used to silence TaG7NPR1, TaG3NPR1 and Ta7ANPR1 were generated by PCR amplification from two synthesised oligonucleotides primers containing 10 overlapping base pairs at the 3′ terminus (Supporting Information Table S1). Overlap Extension PCR amplification of dsDNA fragment was performed using the program as follows: 8 min at 95°C, followed by 34 cycles of 30 s at 95°C, 30 s at 32°C and 40 s at 72°C, and finally 2 min at 72°C. The target fragments were inserted into the modified γ vector ready for direct PCR cloning as described by Campbell & Huang (2010). Infectious RNA transcripts were synthesised in vitro using T7 RNA polymerase (New England Biolabs, Ipswich, MA, USA) from linearised α, β and γ plasmids. The BSMV inoculum was prepared with 1 µl of each of the in vitro transcription reactions and 22.5 µl inoculation FES buffer. The inoculum was then used to rub‐inoculate the first leaf of the two‐leaf stage plants. For simplicity, the BSMV‐derived construct with no insert was named as γ00, and each BSMV silencing construct was named as γtarget. For example, a BSMV silencing construct carried a 185‐bp fragment of the wheat phytoene desaturase (PDS) gene was named as γPDS. The concurrent silencing BSMV inoculum was made by combining the α : β : (γtarget1 : γtarget2) transcripts in a 2 : 2 : 2 (1 + 1) ratio with excess FES. For example, for silencing multiple genes both PDS and G7ANPR1, the BSMV inoculum was made by combining an equimolar ratio of α, β, and (γPDS : γG7ANPR1) at a 2 : 2 : 2(1 + 1) ratio with excess inoculation buffer (named as FES) containing a wounding agent.
Gene expression analysis by RT‐qPCR and RNA‐seq
Expression of the genes targeted for silencing was quantified by comparative RT‐qPCR. Sampled tissues for the time‐course study were immediately frozen in liquid nitrogen and stored at −80°C before extraction of total RNA. Three independent biological replications were performed for each experiment. The time course of G7ANPR1 gene expression was assessed by RT‐qPCR. Wheat cultivar CS and CS + Sr33, were inoculated with the Pgt race QFCSC suspended in inoculation buffer Soltrol 170 Isoparaffin (Chempoint) and, furthermore, inoculation with Soltrol 170 Isoparaffin alone was used as a mock control. Total RNA was isolated and treated with DNase I on a column using the Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) and following the manufacturer's instruction. The quality and concentration of total RNA were assessed via agarose gels and 260/280ABS measurements on a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). Threshold values (Ct) were generated from the CFX96 real‐time PCR detection system (Bio‐Rad, Hercules, CA, USA) using the iScript One‐Step RT‐PCR Kit with SYBR Green (Bio‐Rad) and following the manufacturers recommended protocol. We used gene‐specific primers (Livak & Schmittgen, 2001) and the relative gene expression 2–ΔΔCT method for gene quantification. Amounts of RNA in each reaction were calculated using the average ΔCt normalised to three reference genes 18S rRNA, actin and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). Primers specific for each gene are listed in Table S2. Each reaction was conducted in triplicate for three biological replicates. Relative expression of the G7ANPR1 gene is presented as expression level of this gene in the leaf rust inoculated plants relative to that in the control (buffer Soltrol170 Isoparaffin inoculated plants). Standard deviations were calculated among different biological replicates. Mean relative expression levels were calculated using the ∆Ct method between biological replicates ± SD.
Wheat cultivar CS and CS + Sr33, were inoculated with BSMV inoculum combining an equimolar ratio of α, β and γ transcripts suspended in the inoculation buffer containing a wounding agent (FES). Inoculation with FES buffer alone was used as a mock control. All the leaf tissues snap‐frozen in liquid nitrogen and stored at −80°C until RNA isolation. Real‐time PCR was conducted similarly for the time‐course stem rust inoculation study.
Expression abundance of TaNPR1 homeologues was based on RNA sequence data already available in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) via kallisto software (Bray et al., 2016). Transcripts of Ta7ANPR1,Ta4ANPR1 and Ta7DNPR1 in the RNA‐seq data generated from Columbus (susceptible) and Columbus‐NS765 (Resistant) inoculated with Pgt (Pujol et al., 2016), Avocet+Yr5 and Vuka (susceptible) inoculated with Pst, and Lr48 near‐isogenic lines in Lal Bahadur (susceptible) background inoculated with Pt inoculated wheat (Saini et al., 2002).Sequence quality control checks were conducted via FastQC tool in interactive mode. Paired‐end sequences were split using fastq‐dump before transcript quantification. Wheat transcriptome data were downloaded from the IWGSC, indexed and reads were pseudoaligned. Transcript abundance was then quantified and recorded as transcript per million (tpm).
Mutant screen
The mutagenised population was generated by EMS (Feiz et al., 2009). The population was selfed and advanced to M8 generation. The primers used in mutant identification are listed in Table S2.
For the A genome mutation screening, the A genome‐specific primers G7A‐MF + G7A‐MR were designed to locate in the deletion variations among the A, B and D genomes to ensure A genome specificity (Fig. S1). To detect the A genome mutation, G7A‐MF + G7A‐MR were used to screen the EMS induced population first. PCR products were then purified using the Qiagen gel purification kit, sequenced and compared. First, sequence from wild‐type Alpowa was compared with the G7ANPR1 gene sequences of CS from IWGSC, and then sequences from individual mutagenised lines were compared with the sequence of the wild‐type Alpowa for mutation identification.
SA/JA level analysis with the LC‐MS
Extraction of SA/JA was performed according to Wang's method (Wang et al., 2017). Frozen samples were then ground in liquid N2 with mortar and pestle. An amount of c. 200 mg fresh leaves was extracted with 750 µl mixture of MeOH : H2O : HOAc (90 : 9 : 1, v/v/v) and centrifuged for 1 min at 9600 g. The supernatant was collected and the extraction was repeated twice. Pooled supernatants were dried in N2, re‐suspended in 1000 μl of pure chromatographic grade MeOH, and finally filtered with a Millex‐HV 0.22 μm filter from Millipore (Bedford, MA, USA). Quantitation was carried out using the standard addition method by spiking control plant samples with SA and JA solutions (ranging from 50 to 1000 ng ml−1), and extracting as described above. Analyses were carried out using an LC‐30A + TripleTOF5600+ (AB Sciex, Singapore) machine in Life Science Instrument Shared Platform of Northwest A&F University, China. Three biological replicates were carried out for each assay.
Results
Identification of wheat NPR1 homologues
To identify NPR1‐like genes in bread wheat (T. aestivum L.), a cDNA of NPR1‐like sequence, named as wheat chromosome 3 short arm NPR1 (W3SNPR1), was used as a probe to search its homologues via genomic blot Southern hybridisation. The W3SNPR1 shares a 99% sequence identity to another mRNA sequence deposited in the NCBI database (accession XM_020328292.1) predicted as a BTB/POZ and ankyrin repeat‐containing NPR1‐like protein amplified from Aegilops tauschii (D genome donor of bread wheat). Nine hybridisation fragments were detected and the chromosome location of each fragment was determined using the wheat CS nulli‐tetrasomic (NT) lines (Fig. 1a). In each NT line, a pair of chromosomes was missing and compensated for by a pair of its homoeologous chromosomes. For example, in N3AT3B, the two 3A chromosomes are missing and compensated for by four 3B chromosomes. When a fragment is missing in N3AT3B compared with the rest of the NT lines, it suggests the missing fragment is located on 3A chromosomes. By such an analysis, two fragments were assigned to each of the chromosomes 3B, 3D and 7A and one was assigned to each of the chromosomes 3A, 4A (7B translocated region) and 7D (Fig. 1a). The results revealed that all the wheat NPR1 (referred as TaNPR1 thereafter) homologues were located on six chromosomes of the two homoeologous groups: 3A/3B/3D of group 3 (designated as TaG3NPR1) and 7A/4A/7D of group 7 (designated as TaG7NPR1). One TaNPR1 homologue was found on chromosome 4A instead of 7B due to an ancient translocation between 4AL and 7BS in the tetraploid progenitor of hexaploid wheat (Liu et al., 1992; Devos et al., 1995).
Fig. 1.
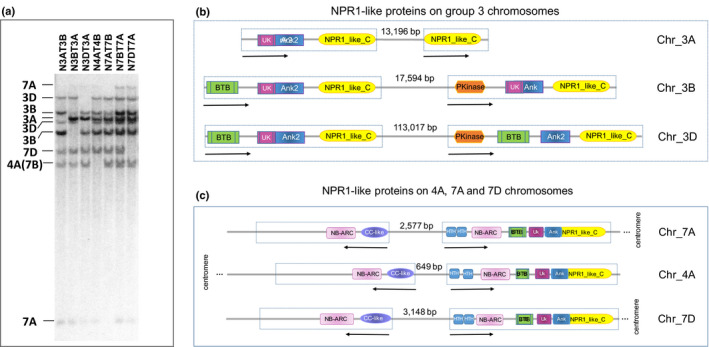
Wheat NPR1‐like genes and organisation and structures of the proteins. (a) Genomic blot (Southern) hybridisation result from genomic DNA of seven wheat NT lines and W3SNPR1 as a probe. Lines indicate the chromosome location of each fragment. (b) Arrangement and structures of six NPR1 homologues in wheat group 3 chromosomes. One dashed box represents one predicted gene and the arrow under each box indicates the orientation of the gene. The distance between the two NPR1 homologues in the same chromosome is indicated in base pairs, but not related to the scale in the figure. Each coloured box represents a region of a known functional domain. BTB: Broad complex, Tramtrack and Bric‐à‐brac; UK: DUF3420 unknown domain; Ank: Ankyrin repeats; NPR1_like_C: A region conserved at the NPR1 C‐terminal; PKinase: Protein Kinase. The sizes of the domains are not according to scale. (c) Arrangement and structures of the three NPR1 homologues and the three NB‐CC‐like genes in wheat chromosomes 7A, 7D and 4A(7B).
The W3SNPR1 sequence was then used to Blast search the International Wheat Genomic Sequence Consortium database (IWGSC‐CS RefSeq v1.0). Nine NPR1‐like proteins were identified (Notes S1). On chromosomes 3A, 3B and 3D (based on the prediction of Fgenesh + genome annotation pipeline at Softberry.com), they each carried a gene that resembled a classical type of NPR1 protein with domains of BTB, ankyrin repeat and a highly conserved NPR1‐like C‐terminal (Fig. 1b). In addition, a gene encoding an integrated protein of a kinase fused with NPR1 was found at c. 17–113 kb proximal to the classical TaG3NPR1 in the same orientation on each of chromosomes 3B and 3D. At the same homoeologous locus of 3A there was only the C‐terminal part of the NPR1‐like gene left, the sequence for protein kinase and most of the NPR1 domains were missing (Fig. 1b), explaining why only one fragment was detected on chromosome 3A in the Southern blotting result (Fig. 1a). Additional search of the entire 3A sequences revealed no hits for the DNA sequence corresponding to the integrated kinase domain of the TaG3NPR1.
On 7A, 4A (7B) and 7D, each chromosome carried an NPR1 with additional domains. The N‐terminal regions of the TaG7NPR1 proteins have domains of two consecutive DNA binding sites for MYC4 transcription factor and an NB‐ARC (Fig. 1c). In close proximity (650–3000 bp) of each of the integrated TaG7NPR1, a gene encoding for a CC‐like + and NB‐ARC domain was found in the opposite orientation (Fig. 1c). Similarly, TaG7NPR1 genes in the same arrangement with a CC+NB‐ARC were found in the A genome donor of Triticum urartu (http://plants.ensembl.org/Triticum_urartu/Info/Index) and A. tauschii (http://plants.ensembl.org/Aegilops_tauschii/Info/Index).
Silencing of the TaNPR1 genes
After cloning Sr33 (Periyannan et al., 2013), we sought to test the involvement of the TaNPR1 genes in the Sr33‐mediated defence response in wheat; we knocked down the endogenous TaNPR1 genes in two wheat lines, CS and CS + Sr33, using a barley stripe mosaic virus‐induced gene silencing (BSMV‐VIGS) assay. Several silencing constructs were made for the assay. A construct containing a 170‐bp fragment conserved among the TaG3NPR1 was used to silence all six NPR1 homologues on group 3 chromosomes and labelled as BSMV:G3. Another distinct construct containing a 170‐bp fragment conserved among the TaG7NPR1 was used to silence the NPR1 homologues on chromosomes 7A, 4A(7B) and 7D, and labelled as BSMV:G7. The sequences (Table S2) and locations (Notes S2, S3) of the primers are provided as Supporting Information. A construct carrying only the BSMV genome was used as a nontarget control and labelled as BSMV:00, and a construct carrying a 183‐bp PDS gene was used as a nontarget control for the assay and labelled as BSMV:PDS. Three rounds of silencing assays were conducted: Round 1, silence all the homologues simultaneously via co‐inoculation of two silencing constructs of BSMV:G3 and BSMV:G7 (labelled as BSMV:G3 + G7); Round 2, silence only one group of TaNPR1 at a time with inoculation of either BSMV:G3 or BSMV:G7; Round 3, silence a specific gene when needed. In each assay, CS and CS + Sr33 seedlings inoculated with BSMV:00, BSMV:PDS or only the inoculation buffer (mock) were included as controls. At 6 d post BSMV (dpb) inoculation, viral symptoms were visualised on the newly emerged leaves of plants inoculated with BSMV. At 9 dpb, plants inoculated with BSMV:PDS started to show photo‐bleaching phenotype, and plants inoculated with other BSMV constructs showed viral‐symptom‐free leaf segments, indicating BSMV‐induced gene silencing had been initiated. Three viral‐symptom‐free leaf segments were randomly sampled from plants inoculated with each targeting construct to check the expression levels of the target genes through quantitative real‐time PCR analysis with the corresponding primers (Table S2). The results confirmed a c. 30% reduction in the relative expression of the TaG3NPR1 and TaG7NPR1 genes (Table S3). Stem rust pathogen Pgt race QFCSC was inoculated at 10 dpb. CS controls showed susceptible infection type scored as IT3 at 14 d post fungus inoculation (dpfi) (Fig. 2a). Resistant control CS + Sr33 plants displayed a resistant phenotype score as IT1. A similar level of resistance was observed in CS + Sr33 when both TaG3NPR1 and TaG7NPR1 were knocked down. Surprisingly, CS inoculated with either BSMV:G3 + G7 or BSMV:G7 showed an enhanced level of resistance to the pathogen on knocking down TaG7NPR1 genes that enhanced CS resistance to the pathogen. However, when the six copies of TaG3NPR1 were silenced in CS or CS + Sr33, ITs of silenced CS were as susceptible as their corresponding nonsilenced or mock controls, and silenced CS + Sr33 were as resistant as their controls, suggesting that Sr33‐mediated resistance is TaG3NPR1 independent (Fig. 2a,b).
Fig. 2.
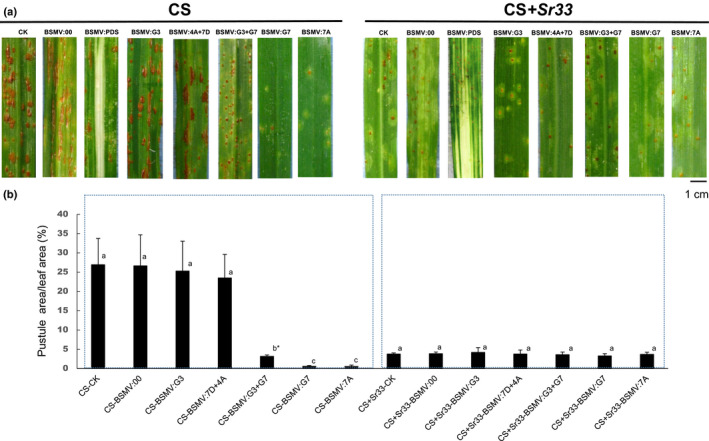
Infection types of TaNPR1 knockeddown plants. (a) Infection types of Sr33 near‐isogenic lines on a Chinese Spring (CS) background when challenged with stem rust race QFCSC 14 d post inoculation. Leaves labelled with BSMV:G3, BSMV:4A + 7D, BSMV:G3 + G7; BSMV:G7 and BSMV:7A were from the TaNPR1 plants; groups 3, 4A + 7D, groups 3 and 7, only group 7 or only chromosome 7A were silenced, respectively. BSMV:00 plants were inoculated with BSMV without the target gene. BSMV:PDS plants were inoculated with BSMV plus PDS gene. CK plants that were not inoculated with BSMV are the mock control. (b) Infection types were quantified by percentage of pustule area/leaf area. Each number is the average of three leaves. The same letter indicates that differences are not significant, and different letters indicate that differences are significant. *, P < 0.05. Each error bar shows the standard deviations among the three leaves.
TaG7NPR1 includes three homologues, Ta7ANPR1, Ta4ANPR1 and Ta7DNPR1. To test which homeologue was vital for the enhanced resistance in CS, we aimed to silence each homeologue one at a time. However, the cDNAs of Ta4ANPR1 and Ta7DNPR1 are highly similar (Notes S3), we could not find a region to silence only one individual of the two, so the two genes were simultaneously silenced and the construct was labelled as BSMV:4A + 7D. To silence Ta7ANPR1 specifically, a region‐specific to this gene was synthesised using two overlapped oligos named G7AoligoF and G7AoligoR and listed in Table S1 and was labelled as BSMV:7A. Only the Ta7ANPR1 silenced plants showed enhanced resistance (Fig. 2a,b), whereas plants silenced both Ta4ANPR1 + Ta7DNPR1 had no changes in ITs in response to Pgt QFCSC (Fig. 2a). Real‐time PCR assays of three biological replicates revealed that the transcript abundance of each target gene in silenced CS plants was reduced c. 20–41% (Table S3). The experiments suggested that the Ta7ANPR1 gene is the one negatively involved in the defence response to Pgt QFCSC. However, resistance of CS in either TaG7NPR1 or Ta7ANPR1 silenced plants was better compared with when both group 3 and 7 TaNPR1 genes were silenced (Fig. 2a,b), suggesting a positive role of TaG3NPR1 in defence against Pgt QFCSC.
Expression of TaG7NPR1 post rust inoculations
Time‐course expression levels of the three TaG7NPR1 genes were analysed using RNA‐seq datasets downloaded from NCBI. Notably, Ta7ANPR1 was the only one among the three genes that showed differential expression during stem rust infection in wheat plants (Fig. S2). Transcript abundances of Ta4ANPR1 and Ta7DNPR1 were low and unchanged during the time course post three rust inoculations in both compatible and incompatible interactions (Fig. S2).
To confirm the expression of Ta7ANPR1 via real‐time PCR, the transcript of the gene in wheat line CS was analysed with three pairs of overlapped primers. In the absence of pathogen infection conditions, only a 5785‐bp cDNA (labelled as Ta7ANPR1 cDNA‐1) was amplified (Fig. 3). The protein encoded by this mRNA had only 881 amino acids with a stop codon appearing after exon 3, resulting in a short protein without any signature domains for NPR1 (Fig. 3). To understand why the NPR1 domain was absent from the transcript, we investigated possible alternative splicing at the locus using the primers flanking the stop codon and two RNA samples of CS grown under biotic‐stressed conditions such as rust/virus infection. An additional fragment was discovered from CS RNA extracted from leaf tissues 24 h after inoculation with either stripe rust or barley stripe mosaic virus (Fig. 4a) or Cadenza RNA 24 h after stem rust inoculation (Fig. 4b). This alternative spliced transcript of Ta7ANPR1 had the intron (including the stop codon) that was retained between exons 3 and 4 in the cDNA‐1 spliced out and translated to a protein of 1437 amino acids with fused domains of NB‐ARC and NPR1 (Fig. 3). The same alternative spliced transcript was also identified from RNA‐seq data generated from a pair of the wheat Lr47‐cultivar ‘Scholar’ near isogeneic lines (NILs) post leaf rust pathogen inoculation (Fig. 4c). Interestingly, the transcript encoding levels for the protein without NPR1 domains were upregulated about eight‐fold in the susceptible NIL (Scholar/‐Lr47), whereas the levels of the two types of Ta7ANPR1 transcripts were about the same as in the resistant NIL (Scholar/+Lr47) and double the expression level at 2 d post Pt‐inoculation (Fig. 4c).
Fig. 3.
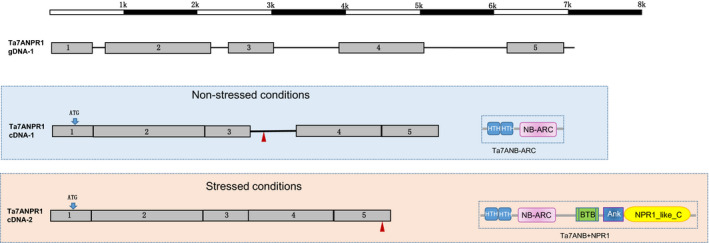
Genomic DNA and two transcripts of the Ta7ANPR1 gene. Ta7ANPR1 has five exons. Under nonstressed growth conditions, the transcript retains the intron between exons 3 and 4, including a stop codon. The encoded peptide has 881 amino acids containing only the NB‐ARC domain. Under stressed conditions, an additional transcript of Ta7ANPR1 was detected. The alternative spliced isoform has the retained intron, and the stop codon removed, so the encoded peptide is 1431 amino acids and contains NB‐ARC and NPR1. The red arrowheads indicate the locations of the stop codons.
Fig. 4.
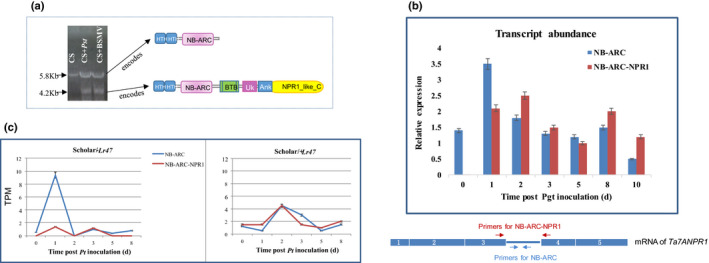
Alternative splicing of the Ta7ANPR1 locus. (a) Reverse transcription amplified cDNAs from total RNAs extracted from CS under the nonstressed condition, stripe rust inoculated or BSMV inoculated condition. An additional fragment was amplified from CS under stressed conditions. (b) Transcripts of two isoforms of Ta7ANPR1 in Cadenza during the time course of Pgt TPMKC infection. (c) Detection of two transcripts of Ta7ANPR1 from RNA‐seq data generated from Lr47 near‐isogenic lines post‐Pt PBJJG inoculation at six time points. TPM, transcripts per million. Each error bar shows the standard deviation of three biological replicates.
Transcript abundances of the two mRNA isoforms encoding NB‐ARC (the long isoform) and NB‐ARC–NPR1 (the short isoform) were measured quantitatively by real‐time qRT‐PCR in Cadenza during the time course for Pgt TPMKC (Cadenza was susceptible) inoculation. Two sets of primers were designed, as shown in Fig. 4(b). One pair flanked the borders of the spliced‐out region (925 bp), the other pair mapped inside the spliced fragment. For qRT‐PCR, we set up a program with a 20 s extension time to only allow a c. 200‐bp fragment to be amplified. Under the given conditions, the primers flanking the 925‐bp alternatively spliced region could only be amplified from the short isoform when the 925‐bp fragment was spliced out.
The primers located inside the spliced fragment could only be amplified from the long intron‐retention mRNA. The transcript abundance of the two isoforms could be measured in the same RNA sample when two rounds of PCR were set up. The short NB‐ARC–NPR1 isoform was undetectable in the absence of the rust infection (Fig. 4b). However, when NB‐ARC–NPR1 mRNA was detectable at 1 d post inoculation, the two isoforms had similar level expressions over the course of the Pgt TPMKC infection (Fig. 4b).
Identification of Ta7ANPR1 mutants
Six independent mutants of Ta7ANPR1 were identified from two spring wheat backgrounds, three from ‘Alpowa’ and three from ‘Cadenza’. The Alpowa mutants were identified by a set of primers, G7A‐MF1 + G7A‐MR1 (Table S2), specifically designed for Ta7ANPR1 after screening an EMS mutagenised Alpowa population (Feiz et al., 2009). The specificity of the primers was verified on three group 7 NT lines. A 508‐bp fragment was amplified from N7BT7D and N7DT7B but not from N7AT7D (Fig. S1), confirming that the primers specifically amplified Ta7ANPR1. After screening 576 individuals from the mutagenised population, three mutations between the primers were identified. One mutant was a G to A transition resulting in a codon of GGG to GAG, but was synonymous at the amino acid level. For the other two mutants each one had a missense mutation on Ta7ANPR1, resulting in an S to L change at location 736 (named as AlS736L) and an R to Q change at location 805 (AlR805Q). Both mutations were located within the NB‐ARC domain (aa 525–814). The mutants were then tested with Pgt TPMKC, along with CS and Alpowa as susceptible controls, and CS + Sr33 as a resistance control. As expected, the mutant with a silent mutation had a similar IT as CS and Alpowa and so did AlS736L (data not shown). AlR805Q was resistant to the pathogen (Fig. 5a).
Fig. 5.
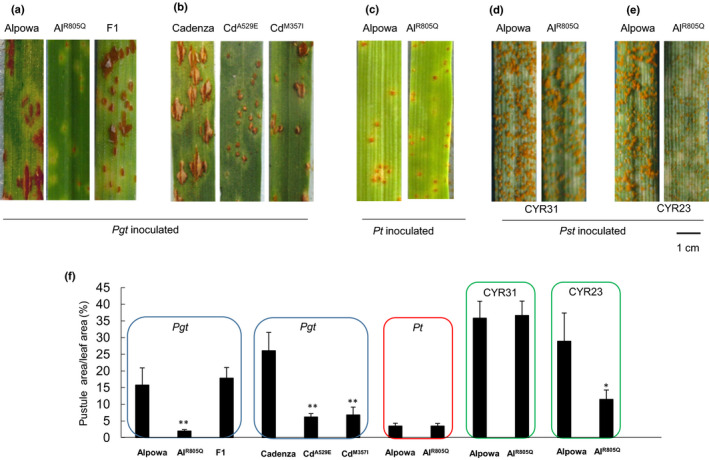
Infection types of Ta7ANPR1 mutants. (a) Infection types of wild‐type Alpowa, mutant AlR805Q and an F1 of Alpowa/ AlR805Q when challenged with stem rust race TPMKC 14 d post‐inoculation. (b) Infection types of wild‐type Cadenza and mutants of CdA529E and CdM357I at 14 d post‐Pgt TPMKC inoculation. (c) Infection types of wild‐type Alpowa and mutant AlR805Q at 9 d post‐Pt PBJJG inoculation. (d) Infection types of wild‐type Alpowa and mutant AlR805Q at 17 d post‐Pst CYR31 inoculation. (e) Infection types of wild‐type Alpowa and mutant AlR805Q at 17 d post‐Pst CYR23 inoculation. (f) Infection types were quantified by percentage pustule area/leaf area. Each number is an average of three leaves. * , P < 0.05. ** , P < 0.01. Each error bar shows the standard deviations of three leaves.
To confirm that the new resistance of AlR805Q was due to the mutation, we made a cross between the mutant (as a male parent) and the wild‐type Alpowa. Here, 10 F1 individuals were tested with Pgt TPMKC and showed a susceptible infection phenotype at 14 d postinoculation, whereas AlR805Q was highly resistant and wild‐type Alpowa was susceptible (Fig. 5a). The 10 F1 individuals were bagged and selfed to produce F2 seeds. In total, 400 F2 plants were screened with Pgt TPMKC at the seedling stage, the segregation of resistant to susceptible fitted a 1 : 3 ratio. Here, 75 susceptible and 21 resistant individuals were sampled to amplify the region of mutation at Ta7ANPR1 via PCR using the gene‐specific primers. Sequence data confirmed that the new resistance completely co‐segregated with a single nucleotide polymorphism (SNP) between AlR805Q and Alpowa.
The three mutants from wheat cultivar ‘Cadenza’ were identified from the wheat‐TILLING database (Krasileva et al., 2017) (https://www.seedstor.ac.uk/shopping‐cart‐tilling.php) using the Ta7ANPR1 sequence as a query. The database included more than 50 lines carrying a mutation on the Ta7ANPR1 gene in a heterozygous state. We requested three lines CdA529E, CdM357I and CdL1224R and selected homozygous mutations of each line after genotyping. The homozygous CdA529E and CdM357I mutants showed resistance to Pgt TPMKC (Fig. 5b), whereas CdL1224R (mutation located in the DUF3420 unknown domain of the NPR1 portion) was as susceptible as the wild‐type (data not shown).
Homozygous mutant AlR805Q was also tested with Pt PBJJG and two Pst races. The mutant was as susceptible as the wild‐type to Pt PBJJG (Fig. 5c) and Pst race CYR31 (Fig. 5d), but was less susceptible to Pst CYR23 than the wild‐type (Fig. 5e). The infection type of each genotype was also quantified as the percentage of pustule area/leaf area (Fig. 5f). These results suggested that the resistance of the mutant was rust pathogen‐species specific and race specific.
Levels of SA/JA and PR genes when the TaG3NPR1 genes were downregulated
A known function of NPR1 is associated with SA‐mediated signalling pathway, and no visible infection type changes were observed with Pgt inoculation when TaG3NPR1 were downregulated, so we decided to measure the levels of SA/JA and five PR genes in CS leaf segments in which the six copies of TaG3NPR1 were silenced, and post‐Pst inoculation. Leaf segments of the same stage as the silenced ones from CS seedlings without any treatments (mock) or treated with BSMV alone (BSMV:00) were used as controls. At 10 d post BSMV inoculation, six viral‐symptom‐free leaf segments were sampled from each treatment. Each ground leaf sample was divided into three portions; one of each was used for RNA extraction, SA and JA measurements. After confirming that the TaG3NPR1 levels were reduced at least 30% compared with the BSMV:00 control, the top three best TaG3NPR1‐silenced samples (Fig. S3a) were selected for further tests of the SA, JA and the PR genes. We observed significant high levels of SA in the leaves with BSMV‐inoculations (Fig. S3b) compared with the mock control. However, the levels of three SA‐mediated PR genes were similar among the high‐SA and low‐SA samples (Fig. S3d). When we measured the SA/JA and PR genes at 24 h post‐Pst inoculation (hpi), the level of SA was slightly reduced compared with the levels without Pst, but levels of the three PR genes were significantly increased (Fig. S3d). We learned from these observations that high SA level does not equal high PR1 level, but that PR1 expression did associate with certain SA concentrations, suggesting that SA was essential for PR1 expression. SA alone was not sufficient to achieve high levels of PR1, PR2 and PR5. Interestingly, at 24 h post inoculation, the SA levels detected in TaG3NPR1 silenced leaves (Fig. S3b) were significantly higher than that in the nonsilenced leaves, but the levels of PR1, PR2 and PR5 genes were significantly lower than the levels in the nonsilenced leaves (Fig. S3d), suggesting some of the TaG3NPR1 genes were required for SA‐mediated PR gene expression.
By contrast, the level of JA in the BSMV:00 control was significantly lower than those from the mock and TaG3NPR1 silenced leaves without Pst inoculation (Fig. S3c); no differences in the PR3 and PR10 gene levels were detected among the treatments (Fig. S3e). Post‐Pst inoculation, the JA levels were slightly decreased at 24 h post inoculation (Fig. S3c), but PR3 expression level increased, and the level was significantly higher in the TaG3NPR1 silenced leaves than that in nonsilenced controls (Fig. S3e).
Expression of TaG3NPR1 post‐Pst‐inoculation
It was impossible to silence each TaG3NPR1 gene individually because of their high levels of cDNA similarity (Notes S2). We, therefore, explored their expression levels post‐Pst‐inoculation using the RNA‐seq datasets generated from a Yr5 line on an Avocet background and a highly stripe rust susceptible wheat variety ‘Vuka’ (Dobon et al., 2016) downloaded from the NCBI database. Interestingly, four of the six TaG3NPR1 genes had detectable transcripts (Fig. S4). The Ta3ANPR1 and Ta3BNPR1 genes have similar expression profiles, upregulated at 1 d post inoculation in the Yr5 resistant line, and no change in the susceptible line over the time course. The two kinase‐TaG3NPR1 fusion genes in 3B and 3D also had similar profiles with an increased expression in the resistant line starting at 3 d post inoculation (Fig. S4). Expression profiles of the four TaG3NPR1 genes led us to infer that Ta3ANPR1 and Ta3BNPR1 were more likely to be responsible for regulating SA‐responsive genes based on the timing of their upregulation.
Transcript abundance of PR genes when the Ta7ANPR1 gene was knocked down or knocked out
The same five PR genes were measured when the Ta7ANPR1 gene was knocked down under two conditions, 48 h post‐inoculation (h post inoculation) with Pst CYR32 or without Pst inoculation. All five genes had relatively low expression levels without rust, but PR2 and PR3 genes had even lower expression levels in silenced leaves (Fig. S5a) than that in the control (Fig. S5b,c). Three of the five PR genes (PR1, PR2, and PR3) showed a significant upregulation at 48 h post inoculation (Fig. S5b,c). All except PR3 had a similar level of increase between silenced and nonsilenced leaves. Upregulation of PR3 was affected when the Ta7ANPR1 gene was downregulated, suggesting a decisive role for Ta7ANPR1 in this JA‐responsive PR gene.
To explore why the mutations on Ta7ANPR1 enhanced resistance to stem rust, we also analysed the five PR genes in the two mutants CdA529E and AlR805Q in the absence of pathogens. All five PR genes had low expression levels between the mutant AlR805Q and the wild‐type Alpowa (Fig. S6). The mutant CdA529E also had low expression levels in four of the five PR genes (Fig. S6) except that the PR10 level in CdA529E was significantly higher than that in the wild‐type Cadenza (Fig. S6).
Because the PR1 gene was undetectable in Cadenza and the mutant, and PR10 expression was different between the two lines without the pathogen, we measured the transcript abundances of the two PR genes post‐Pgt TPMKC at five time points (Fig. S7). PR1 became detectable at 1 d post inoculation but at very low levels until 5 d post inoculation in both CdA529E and Cadenza (Fig. S7a). A different pattern was observed with PR10 with a relatively high base‐level expression; mutant CdA529E had an upregulated PR10 expression at 1–2 d post inoculation and then gradually returned to the base level from 3 d post inoculation (Fig. S7b). By contrast, Cadenza had a low PR10 level during the early time points (1–2 d post inoculation) and slowly increased the level at later time points (3–5 d post inoculation) (Fig. S7b).
Discussion
A requirement of NPR1 in wheat defence response to Puccinia is selective
Bread wheat has nine homologues of the NPR1‐like gene in the genome. Three of them, named as wNPR1 by Cantu et al. (2013), are the three classical types of NPR1‐like genes in homoeologous group 3 chromosomes (Fig. 1b) with similar functional domains as the Arabidopsis AtNPR1. wNPR1 has a similar mode of action as AtNPR1, interacting with a TGA transcription factor for transduction of the SA signal (Cantu et al., 2013). During wheat–P. striiformis interaction, wNPR1 was targeted by a stripe rust effector protein PNPi (for Puccinia NPR1 interactor) (Wang et al., 2016). The PNPi competes with wNPR1 for interaction with TGA2.2 and reduced pathogenesis‐related gene expression (Wang et al., 2016). Similarly, in our study, when the TaG3NPR1 gene was knocked down with a BSMV:G3NPR1 construct, we found that three SA‐mediated PR genes (PR1, PR2 and PR5) had significantly lower expression levels compared with the control (Fig. S3f). These results suggested that TaG3NPR1 was involved in the wheat defence response against stripe rust, as the classical NPR1 gene for transducing the SA signal to activate PR gene expression (Cao et al., 1994; Dong, 2004). However, downregulating TaG3NPR1 did not alter the IT phenotype of the Sr33‐mediated stem rust resistance (Fig. 2), suggesting that the defence response to stem rust conferred by Sr33 did not require TaG3NPR1. Similarly, NPR1 is not always required in resistance in Arabidopsis. An NPR1‐independent resistance to a bacterial pathogen Pseudomonas syringae pv. maculicola and an oomycete Peronospora parasitica were identified in Arabidopsis (Li et al., 2001). This pathway bypasses NPR1 but requires SA and is an EDS1‐mediated pathway (Li et al., 2001). Our study suggested that Sr33‐mediated resistance is NPR1 independent, but this should not infer the implication that NPR1 is not required for immunity to stem rust based on only one gene. Clearly, more studies on other Sr genes are necessary to draw such a conclusion. The function of AtNPR1 in other plant species may not be the same as observed in Arabidopsis. AtNPR1 in rice has a similar function for its NPR1 homologue as it could enhance resistance against the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo) (Chern et al., 2005), but was more susceptible to herbivore attack (Yuan et al., 2007). AtNPR1‐expressing wheat was resistant to Fusarium head blight (Makandar et al., 2006), but was more susceptible to Fusarium seedling blight (Gao et al., 2013). Given the importance of NPR1 in host defence, the protein also becomes a target of pathogens. In rice, the NPR1 interacting protein NRR negatively regulates the defence response to Xoo. In brief, NPR1 is an important component of plant defence, but its involvement in SA defence signalling is not a universal phenomenon.
NPR1 proteins with IDs in the wheat genome
Furthermore for the classical type of NPR1, five homologues of NPR1‐like gene in the bread wheat genome occur as gene fusions with exogenous domains such as a protein kinase (Fig. 1b) or an NB‐ARC domain of the most common plant immune receptors (Fig. 1c). IWGSC and the EnsemblPlants databases have the sequences at all loci but the search tools failed to reveal them as fused NPR1 proteins. It was only in a recent study by Bailey et al. (2018) in which three NB‐ARC–NPR1 fusion proteins in hexaploid wheat based on gene predictions were reported (Bailey et al., 2018). Our findings back up this prediction. A possible reason why the NPR1 fused proteins were overlooked may be attributed to the findings in our study in which the mRNA of the Ta7ANPR1 gene in the absence of pathogen infection has a stop codon before the sequences coding the NPR1‐like domains (Fig. 4). Consequently, the gene was predicted only as an NB‐ARC containing protein. The NPR1 proteins with IDs are produced only when the gene is alternatively spliced under certain stresses, for example, biotic stresses (Fig. 4a). Because the sequences of TaG7NPR1 in bread wheat can be traced to its diploid ancestors (alternatively spliced variants annotated for the diploid D genome as AETGV20038900.1, AETGV20038900.2 and AETGV20038900.3), this indicated that the NB‐ARC–NPR1 fused protein already existed in the diploid donor species of bread wheat. Evolutionary events to create this NB‐ARC–NPR1 fusion and selection sweeps have maintained these NPR1 fused proteins, implying the benefits of these proteins for wheat. Several findings have suggested that NPR1 might contribute to defence against pathogens. Classic TaG3NPR1 proteins have been shown to be involved in stripe rust resistance and a target of the pathogen (Wang et al., 2016). This raises the possibility that the NPR1 domain of Ta7ANB‐ARC–NPR1 is used as a decoy to monitor the wNPR1. Organisation of the group 7 NB‐ARC–NPR1 genes in a head‐to‐head orientation with another NB‐ARC‐like gene (Fig. 1c) is indicative of some disease resistance gene pairs that require sensing and signalling partners to confer a resistance function, for example, the RRS1/RPS4 pair (Narusaka et al., 2009) and the Pi5‐1/Pi5‐2 pair (Lee et al., 2009).
The negative regulation of Ta7ANBS‐NPR1 is specific
The function of the Ta7ANB‐ARC–NPR1 fusion protein does not appear to enhance a general defence to all pathogens because the mutants showed resistance to stem rust (Fig. 5a,b), stripe rust CYR23 (Fig. 5e) but not to leaf rust PBJJG (Fig. 5c) and stripe rust CYR31 (Fig. 5d). It appeared that the mutations lifted the suppression on specific rust resistance genes. Among the six mutants identified, we only identified one mutant that had a mutation in the NPR1 portion of the fusion protein, the resistant mutants all had a mutation located in the NB‐ARC region of the gene. We considered that resistance against Pgt TPMKC in the mutants was due to the sequence changes in the NB region of the protein that altered the formation of the R protein complex. The resulting activated defence was hypothesised as mutation based rather than recognition based (Fig. 6a). The biotic stress caused by the rust infection triggered alternative splicing at the Ta7ANPR1 locus. It is likely the NLR–NPR1 fusion protein may not recognise Pgt TPMKC in the wheat cultivars CS and Cadenza because these two wheat lines were susceptible to the pathogen. The mutation‐based preformed defence was only sufficient against some rusts but not others, suggesting other components of the defence pathway might be the subjects of pathogen attack. Our previous study at the MNR220 locus also revealed that different rust pathogens recognised by the same R gene locus activated different PR genes, suggesting that different signalling pathways were used (Zhang et al., 2018). A wide range of NLR fusion IDs suggested the complexity of the exquisite nature of the plant pathogen surveillance systems and different defence signalling to cope with the highly sophisticated pathogen invasion strategies to stay healthy.
Fig. 6.
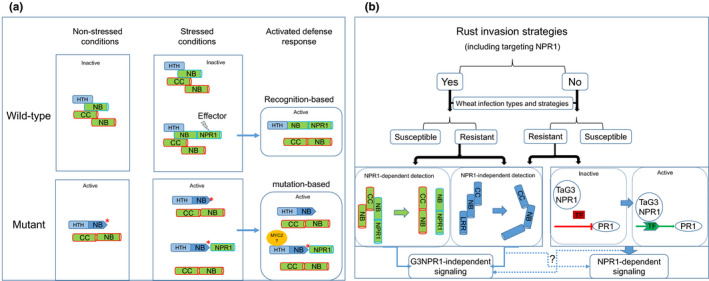
Models of TaNPR1‐induced resistance and the potential role of TaNPR1 proteins during wheat–rust interactions. (a) Model of resistance conferred by NPR1‐dependent detection in an R protein complex by a pair of head‐to‐head NB‐ARC genes. In wild‐type during the absence of rust, the two NB‐ARC genes fold together in an inactive formation. In the presence of a rust pathogen, one of the NB‐ARC loci produces an isoform protein containing NPR1 as a decoy. Two types of R protein complexes will be formed. If rust secretes an effector to attack TaNPR1, the decoy NPR1 could be mistakenly attacked, and then the NPR1‐decoy R protein complex will change to an active formation to induce a defence response. More NB‐NPR1 isoform will be produced to distract the NPR1‐attacking effector. In a mutant in which a mutation results in a change in the R protein complex to an active formation, the defence response is activated by the mutation. The mutant will be resistant to all rusts, except ones that have effectors to suppress other components of the defence signalling pathway. (b) During wheat–rust interactions, rust pathogens may apply invasion strategies that target or not target TaG3NPR1 (shown as Yes or No, respectively, on the figure). Without an ineffective detection system or compromised components for inducing defence signals, wheat will be susceptible to the pathogens. Resistant wheat in the ‘Yes’ group of rusts may use TaNPR1 as a decoy (e.g. Ta7ANPR1) in detection and have an alternative pathway to bypass TaNPR1 once the integrity of the NPR1 is compromised by the pathogens. Resistant wheat in the ‘No’ group of rusts may use either NPR1‐independent (e.g. Sr33) or NPR1‐dependent signalling through TaG3NPR1. Whether defence signalling could be transduced by both NPR1‐dependent or NPRI‐independent pathways is still a question to be resolved.
Alternative splicing has been reported to be associated with stresses, including abiotic and biotic stresses (Jordan et al., 2002; Zhang & Gassmann, 2007; Filichkin et al., 2015). Numerous TIR‐NB‐LRR and CC‐NB‐LRR plant R proteins have alternative isoforms (Ayliffe et al., 1999; Gassmann et al., 1999; Dinesh‐Kumar & Baker, 2000; Dodds et al., 2001; Jordan et al., 2002). The location of alternative splicing, so far, has only been seen between the sequence coding for NB and LRR domains (Jordan et al., 2002). In some cases, not only the presence of the alternative isoforms of the gene but also their ratios is crucial for active resistance against pathogen attack (Dinesh‐Kumar & Baker, 2000). In other incidences, for example L6, although an alternative form of L6 was detected, no functional relevance could be assigned (Dodds et al., 2001). Our studies revealed that alternative splicing from a locus of 7A is regulated by both the host and pathogen (Fig. 4b,c). The isoform of the Ta7ANB‐NPR1 fusion protein was promoted by the host and suppressed by the pathogen during the interaction, suggesting that alternative splicing is one of the strategies used by both host and pathogen during an interaction.
Expression profiles of the five PR genes in Ta7ANPR1 knocked down and knocked out lines seemed to reveal the same thing, that Ta7ANPR1 affected PR3 expression (Figs S5c, S6, S7a,b). These observations might suggest that the Ta7ANPR1 protein has both negative and positive functions. The negative regulatory role of the fusion protein is more likely to be located in the NB‐ARC region (Fig. 6a), and the NPR1 domains may be involved in regulating some PR gene expression. There are two Helix‐Turn‐Helix (HTH) motifs in both NB‐ARC and NB‐ARC–NPR1 proteins (Fig. 1c). As the HTH motif has G‐box DNA binding properties, we speculated that the function of these HTH motifs might be associated with MYC2 with negative regulation of PR3 and PR10. Upregulated expression of two JA‐responsive PR genes (PR3 and PR10) in the Ta7ANPR1 mutant (Fig. S6) suggested repression of JA‐mediated transcription in the wild‐type. However, downregulating Ta7ANPR1 proteins also affected PR3 expression (Fig. S5c), suggesting the requirement of Ta7ANPR1. Thus, we hypothesised a scenario in which the Ta7ANB‐ARC–NPR1 protein competes with the Ta7ANB‐ARC protein to bind the G‐box DNA of the regulated genes. Degradation of the Ta7ANB‐ARC–NPR1 protein by NPR1 directed‐ubiquitination releases the negative regulation. Further investigations on the association between Ta7ANB‐ARC–NPR1 and MYC2 and their association with the G‐box DNA of the two PR genes are required to substantiate this claim.
Potential roles of TaNPR1 proteins during wheat–rust interactions
We believe that the history of wheat–rust co‐evolution and the complexity of their interactions are partially revealed by the results of ours and others. Pathogens to be successful have to be able to sabotage host defence systems without being detected. Any components in the host defence pathways could be a pathogen target. We hypothesise the potential roles of TaNPR1‐like genes during wheat–rust interactions, as illustrated in Fig. 6(b). From the angle of the rusts, the pathogens could include targeting TaNPR1 as one of the strategies or not, namely the ‘Yes’ or the ‘No’ group. Wheat infection types will reflect the effectiveness of the host defence in terms of pathogen detection and defence signalling. Wheat plants that have neither an effective detection system nor components for inducing defence signals will be susceptible to the pathogens. Resistant wheat to the ‘Yes’ group of rusts are postulated to have an alternative defence signalling pathway that is independent of TaNPR1 to overcome the rust invasion strategy of attacking TaNPR1. A more advanced host may include using TaNPR1 as a decoy in detection and have an alternative pathway to bypass TaNPR1 once the integrity of the NPR1 component is compromised by the pathogens.
Author contributions
XW, ZK, EL and LH planned and designed the research; XW, HZ, YL, XM, BN and FW performed the experiments and analysed data; XW, EL and LH wrote the manuscript; XW, HZ and BN contributed equally to this work.
Supporting information
Fig. S1 Specificity of primers for screening mutations on Ta7ANPR1.
Fig. S2 Transcripts of Ta7ANPR1, Ta4ANPR1 and Ta7DNPR1 during a time course of rust infections.
Fig. S3 SA/JA concentrations and PR gene expression following downregulation of TaG3NPR1 genes.
Fig. S4 Transcripts of TaG3NPR1 and TaG3Kinase‐NPR1 during a time course of Pst infection.
Fig. S5 PR gene expression following downregulation of Ta7ANPR1 gene.
Fig. S6 PR gene expression in the wild‐types and mutants without pathogen inoculation.
Fig. S7 PR1 and PR10 expression during a time‐course post‐Pgt inoculation.
Notes S1 Amino acid sequences and the location of each predicted functional domain of the TaNPR1 proteins in Fig. 1(b, c).
Notes S2 Alignment of cDNAs of the TaG3NPR1 genes and location of the target region for silencing.
Notes S3 Alignment of cDNAs of the TaG7NPR1 genes and the location of the target region for silencing.
Table S1 Sequences of the oligos used to design silencing constructs.
Table S2 Sequences of the primers used for quantitative real‐time PCR and mutant screening.
Table S3 Real‐time qPCR analysis of TaNPR1‐like gene expression levels in silenced plants.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
XW wishes to thank the National Key Research and Development Program of China (no. 2018YFD0200405) and the Shaanxi Provincial Postdoctoral fund (2017). HZ and BN wish to thank the NSF BREAD program (grant no. IOS‐0965429) for financial support. The authors thank Hikmet Budak for help with analysis of the Ta7ANPR1 alternative isoform, and Michael Giroux for the use of his Alpowa mutagenesis population. We also would like to thank Northwest A&F University Life Science Research Core Services for letting us use the equipment for SA/JA tests.
Contributor Information
Xiaojing Wang, Email: wangxiaojing@nwsuaf.edu.cn.
Evans Lagudah, Email: evans.lagudah@csiro.au.
Li Huang, Email: li.huang@montana.edu.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. 1999. Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. Journal of Molecular Biology 285: 1353–1361. [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Frost DV, Finnegan EJ, Lawrence GJ, Anderson PA, Ellis JG. 1999. Analysis of alternative transcripts of the flax L6 rust resistance gene. The Plant Journal 17: 287–292. [DOI] [PubMed] [Google Scholar]
- Bailey PC, Schudoma C, Jackson W, Baggs E, Dagdas G, Haerty W, Moscou M, Krasileva KV. 2018. Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biology 19: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near‐optimal probabilistic RNA‐seq quantification. Nature Biotechnology 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Campbell J, Huang L. 2010. Silencing of multiple genes in wheat using Barley stripe mosaic virus. Journal of Biotech Research 2: 12–20. [Google Scholar]
- Cantu D, Yang BJ, Ruan R, Li K, Menzo V, Fu DL, Chern M, Ronald PC, Dubcovsky J. 2013. Comparative analysis of protein–protein interactions in the defense response of rice and wheat. BMC Genomics 14: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong XN. 1994. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired‐resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong XN. 1997. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63. [DOI] [PubMed] [Google Scholar]
- Césari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN. 2014b. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Frontiers in Plant Science 5: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T. 2014a. The NB‐LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO Journal 33: 1941–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC. 2005. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Molecular Plant–Microbe Interactions 18: 511–520. [DOI] [PubMed] [Google Scholar]
- Devos KM, Dubcovsky J, Dvorak J, Chinoy CN, Gale MD. 1995. Structural evolution of wheat chromosomes 4a, 5a, and 7b and its impact on recombination. Theoretical and Applied Genetics 91: 282–288. [DOI] [PubMed] [Google Scholar]
- Dinesh‐Kumar SP, Baker BJ. 2000. Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proceedings of the National Academy of Sciences, USA 97: 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobon A, Bunting DCE, Cabrera‐Quio LE, Uauy C, Saunders DGO. 2016. The host‐pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics 17: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti A‐M, Ayliffe MA, Ellis JGJTPC. 2004. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16: 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. 2001. Six amino acid changes confined to the leucine‐rich repeat beta‐strand/beta‐turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XN. 2004. NPR1, all things considered. Current Opinion in Plant Biology 7: 547–552. [DOI] [PubMed] [Google Scholar]
- El‐Gebali J, Mistry A, Bateman SR, Eddy A, Luciani SC, Potter M, Qureshi LJ, Richardson GA, Salazar A, Smart ELL et al 2019. The Pfam protein families database in 2019. Nucleic Acids Research 47(D1): D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiz L, Beecher BS, Martin JM, Giroux MJ. 2009. In planta mutagenesis determines the functional regions of the wheat puroindoline proteins. Genetics 183: 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S, Priest HD, Megraw M, Mockler TC. 2015. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Current Opinion in Plant Biology 24: 125–135. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan SP, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N et al 2012. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao CS, Kou XJ, Li HP, Zhang JB, Saad ASI, Liao YC. 2013. Inverse effects of Arabidopsis NPR1 gene on fusarium seedling blight and fusarium head blight in transgenic wheat. Plant Pathology 62: 383–392. [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. 1999. The Arabidopsis RPS4 bacterial‐resistance gene is a member of the TIR‐NBS‐LRR family of disease‐resistance genes. The Plant Journal 20: 265–277. [DOI] [PubMed] [Google Scholar]
- Grund E, Tremousaygue T, Deslandes L. 2019. Plant NLRs with integrated domains: unity makes strength. Plant Physiology 179: 1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk‐Genevaux PS, Pagni M, Sigrist CJA. 2006. The PROSITE database. Nucleic Acids Research. 34: D227–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium . 2014. A chromosome‐based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 286. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium . 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: 661. [DOI] [PubMed] [Google Scholar]
- Jordan T, Schornack S, Lahaye T. 2002. Alternative splicing of transcripts encoding Toll‐like plant resistance proteins‐what's the functional relevance to innate immunity? Trends Plant Science 7: 392–398. [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, Fournier E, Tharreau D, Terauchi R. 2012. Arms race co‐evolution of Magnaporthe oryzae AVR‐Pik and rice Pik genes driven by their physical interactions. The Plant Journal 72: 894–907. [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez‐Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, Simmonds J, Ramirez‐Gonzalez RH, Wang X et al. 2017. Uncovering hidden variation in polyploid wheat. Proceedings of the National Academy of Sciences, USA 114: E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Henanff G, Farine S, Kieffer‐Mazet F, Miclot AS, Heitz T, Mestre P, Bertsch C, Chong J. 2011. Vitis vinifera VvNPR1.1 is the functional ortholog of AtNPR1 and its overexpression in grapevine triggers constitutive activation of PR genes and enhanced resistance to powdery mildew. Planta 234: 405–417. [DOI] [PubMed] [Google Scholar]
- Lee S‐K, Song M‐Y, Seo Y‐S, Kim H‐K, Ko S, Cao P‐J, Suh J‐P, Yi G, Roh J‐H, Lee SJG. 2009. Rice Pi5‐mediated resistance to Magnaporthe oryzae requires the presence of two coiled‐coil–nucleotide‐binding–leucine‐rich repeat genes. Genetics 181: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. 2004. The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. Plant Cell 16: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang YL, Dong XN. 2001. Activation of an EDS1‐mediated R‐gene pathway in the snc1 mutant leads to constitutive, NPR1‐independent pathogen resistance. Molecular Plant–Microbe Interactions 14: 1131–1139. [DOI] [PubMed] [Google Scholar]
- Line RF, Qayoum A. 1992. Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America, 1968–87. Technical bulletin–United States Department of Agriculture 1788: 44. [Google Scholar]
- Liu C, Atkinson M, Chinoy C, Devos K, Gale M. 1992. Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theoretical and Applied Genetics 83: 305–312. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah JJMP‐MI. 2006. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Molecular Plant–Microbe Interactions 19: 123–129. [DOI] [PubMed] [Google Scholar]
- Malnoy M, Jin Q, Borejsza‐Wysocka E, He S, Aldwinckle H. 2007. Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus × domestica . Molecular Plant–Microbe Interactions 20: 1568–1580. [DOI] [PubMed] [Google Scholar]
- McIntosh RA, Wellings CR, Park RF. 1995. Wheat rusts: an atlas of resistance genes. Clayton, Vic, Australia: CSIRO Publishing. [Google Scholar]
- Moreau M, Tian M, Klessig DF. 2012. Salicylic acid binds NPR3 and NPR4 to regulate NPR1‐dependent defense responses. Cell Research 22: 1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Dangl JL. 1997. The hypersensitive response and the induction of cell death in plants. Cell Death and Differentiation 4: 671–683. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. 2003. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. 2009. RRS1 and RPS4 provide a dual Resistance‐gene system against fungal and bacterial pathogens. The Plant Journal 60: 218–226. [DOI] [PubMed] [Google Scholar]
- Pajerowska‐Mukhtar KM, Emerine DK, Mukhtar MS. 2013. Tell me more: roles of NPRs in plant immunity. Trends Plant Science 18: 402–411. [DOI] [PubMed] [Google Scholar]
- Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, Deal K, Luo M, Kong X, Bariana H. 2013. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341: 786–788. [DOI] [PubMed] [Google Scholar]
- Pujol V, Robles J, Wang P, Taylor J, Zhang P, Huamh L, Table L, Lagudah E. 2016. Cellular and molecular characterization of a stem rust resistance locus on wheat chromosome 7AL. BMC Research Notes 9: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Greenberg JT. 2001. The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. The Plant Journal 27: 203–211. [DOI] [PubMed] [Google Scholar]
- Saini R, Kaur M, Singh B, Sharma S, Nanda GS. 2002. Genes Lr48 and Lr49 for hypersensitive adult plant leaf rust resistance in wheat (Triticum aestivum L.). Euphytica 124: 365–370. [Google Scholar]
- Saucet SB, Ma Y, Sarris PF, Furzer OJ, Sohn KH, Jones JD. 2015. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nature Communications 6: 6338. [DOI] [PubMed] [Google Scholar]
- Shirano Y, Kachroo P, Shah J, Klessig DF. 2002. A gain‐of‐function mutation in an Arabidopsis Toll interleukin1 receptor–nucleotide binding site–leucine‐rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman P, Seskar M, Kanter D, Schweizer P, Metraux J‐P, Raskin I. 1995. Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiology 108: 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V, Kosarev P, Seledsov I, Vorobyev D. 2006. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biology 7 (Suppl. 1): 10.1–10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. 2012. How do plants achieve immunity? Defence without specialized immune cells. Journal of Nature Reviews Immunology 12: 89. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux J‐P, Brown R, Kazan K. 2003. NPR1 modulates cross‐talk between salicylate‐and jasmonate‐dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. 2009. Proteasome‐mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stakman EC, Stewart DM, Loegering WQ. 1962. Identification of physiologic races of Puccinia graminis var. tritici. U.S. Department of Agriculture . ARS E617: 5–53. [Google Scholar]
- Wang X, Wang Y, Liu P, Mu X, Liu X, Wang X, Zhao M, Huai B, Huang L, Kang Z. 2017. TaRar1 is inovolved in wheat defense against stripe rust pathogen mediated by YrSu . Frontiers in Plant Science 8: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang B, Li K, Kang Z, Cantu D, Dubcovsky J. 2016. A conserved Puccinia striiformis protein interacts with wheat NPR1 and reduces induction of pathogenesis‐related genes in response to pathogens. Molecular Plant–Microbe Interactions 29: 977–989. [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ. 2014. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhong S, Li Q, Zhu Z, Lou Y, Wang L, Wang J, Wang M, Li Q, Yang D. 2007. Functional analysis of rice NPR1‐like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnology 5: 313–324. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qiu Y, Yuan C, Chen X, Huang L. 2018. Fine‐tuning of PR genes in wheat responding to different Puccinia rust species. Journal of Plant Physiology and Pathology 6: 2. [Google Scholar]
- Zhang X‐C, Gassmann W. 2007. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiology 145: 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Xi L, Lian Q, Luo X, Wu Z, Seng S, Yuan X, Yi M. 2015. The NPR1 homolog GhNPR1 plays an important role in the defense response of Gladiolus hybridus . Plant Cell Reports 34: 1063–1074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Specificity of primers for screening mutations on Ta7ANPR1.
Fig. S2 Transcripts of Ta7ANPR1, Ta4ANPR1 and Ta7DNPR1 during a time course of rust infections.
Fig. S3 SA/JA concentrations and PR gene expression following downregulation of TaG3NPR1 genes.
Fig. S4 Transcripts of TaG3NPR1 and TaG3Kinase‐NPR1 during a time course of Pst infection.
Fig. S5 PR gene expression following downregulation of Ta7ANPR1 gene.
Fig. S6 PR gene expression in the wild‐types and mutants without pathogen inoculation.
Fig. S7 PR1 and PR10 expression during a time‐course post‐Pgt inoculation.
Notes S1 Amino acid sequences and the location of each predicted functional domain of the TaNPR1 proteins in Fig. 1(b, c).
Notes S2 Alignment of cDNAs of the TaG3NPR1 genes and location of the target region for silencing.
Notes S3 Alignment of cDNAs of the TaG7NPR1 genes and the location of the target region for silencing.
Table S1 Sequences of the oligos used to design silencing constructs.
Table S2 Sequences of the primers used for quantitative real‐time PCR and mutant screening.
Table S3 Real‐time qPCR analysis of TaNPR1‐like gene expression levels in silenced plants.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


