Summary
The RAV (related to ABI3/viviparous 1) group of transcription factors (TFs) play multifaceted roles in plant development and stress responses. Here, we show that strawberry (Fragaria × ananassa) FaRAV1 positively regulates anthocyanin accumulation during fruit ripening via a hierarchy of activation processes. Dual‐luciferase assay screening of all fruit‐expressed AP2/ERFs showed FaRAV1 had the highest transcriptional activation of the promoter of FaMYB10, a key activator of anthocyanin biosynthesis. Yeast one‐hybrid and electrophoretic mobility shift assays indicated that FaRAV1 could directly bind to the promoter of FaMYB10. Transient overexpression of FaRAV1 in strawberry fruit increased FaMYB10 expression and anthocyanin production significantly. Correspondingly, transient RNA interference‐induced silencing of FaRAV1 led to decreases in FaMYB10 expression and anthocyanin content. Transcriptome analysis of FaRAV1‐overexpressing strawberry fruit revealed that transcripts of phenylpropanoid and flavonoid biosynthesis pathway genes were up‐regulated. Luciferase assays showed that FaRAV1 could also activate the promoters of strawberry anthocyanin biosynthetic genes directly, revealing a second level of FaRAV1 action in promoting anthocyanin accumulation. These results show that FaRAV1 stimulates anthocyanin accumulation in strawberry both by direct activation of anthocyanin pathway gene promoters and by up‐regulation of FaMYB10, which also positively regulates these genes.
Keywords: RAV, anthocyanin, activator, strawberry, MYB
Introduction
The octoploid cultivated strawberry (Fragaria × ananassa) is a typical non‐climacteric fruit and an economically important horticultural crop worldwide. Its popularity is principally due to its sweet taste, unique fragrance, nutritional value and bright colour, all of which are pivotal factors in determining fruit quality.
Strawberry fruit when ripe are distinguished by a high content of anthocyanins, which are water‐soluble flavonoid compounds that generate the characteristic reddish, purple and bluish hues of many fruits, leaves, flowers and seeds. Extensive studies have revealed diverse biological functions for anthocyanins, including an association with reduced incidence of chronic diseases, an ability to confer stress resistance, and attraction of pollinators and seed dispersers (Schaefer et al ., 2004). Anthocyanins are synthesized from phenylalanine by a series of enzymes: phenylalanine ammonia‐lyase (PAL), cinnamate 4‐hydroxylase (C4H), 4‐coumarate: CoA ligase (4CL). The specific flavonoid pathway is initiated by the condensation of one molecule of 4‐coumaroyl‐coenzyme A (CoA) and three molecules of malonyl‐CoA, catalysed by chalcone synthase (CHS) to produce naringenin chalcone. Other early anthocyanin biosynthetic genes (EBG) include chalcone isomerase (CHI), flavonoid 3‐hydroxylase (F3H/FHT) and flavonoid 3′‐hydroxylase (F3′H), which produce naringenin, dihydrokaempferol and dihydroquercetin, respectively. The late anthocyanin biosynthetic pathway genes (LBG) include dihydroflavonol‐4‐reductase (DFR), anthocyanidin synthase (ANS) and 3‐glycosyltransferase (GT1) to produce leucoanthocyanidins, anthocyanidins and anthocyanins, respectively (Almeida et al ., 2007; Griesser et al., 2008; Lin et al ., 2013). The activity of the MYB‐bHLH‐WD40 (MBW) ternary transcriptional complex is central to the regulation of the pathway. This consists of three classes of regulatory proteins, R2R3‐MYBs, bHLHs and TTG1 (also termed WD40), which can act independently or in cooperation with each other as a complex (Baudry et al ., 2004; Lloyd et al ., 2017; Xu et al ., 2015). MYB transcription factors are the most important regulators of anthocyanin biosynthesis (Allan et al ., 2008). In maize, the R2R3 MYB C1 protein interacts with a bHLH TF to activate the promoter of DFR (Sainz et al ., 1997). In Arabidopsis, anthocyanin production is transcriptionally regulated by an R2R3‐MYB protein, for example PAP1, PAP2, MYB113 or MYB114; one bHLH protein, for example TT8, GL3 or EGL3; and one TTG1 protein (Gonzalez et al ., 2008; Li, 2014). In apple, MdMYB1/MYBA TF transcripts are correlated with apple fruit skin colour and MdMYB10 is responsible for production of anthocyanin in red‐fleshed fruit (Ban et al., 2007; Espley et al., 2007; Takos et al., 2006). In grapevine, VvMYB1 and VvMYB2 act mainly on the expression of VvUFGT to regulate anthocyanin biosynthesis (Kobayashi et al., 2002, 2004; Walker et al., 2007). In pear, PpMYB10 enhances anthocyanin accumulation via regulation of genes encoding enzymes of the anthocyanin pathway (Feng et al., 2010). In tomato, LeANT1 (Sapir et al., 2008) and LeAN2 (Boches et al., 2009; Mes et al., 2008) can also regulate anthocyanin biosynthesis. Many other anthocyanin‐regulating MYBs have been isolated from other species, for example Petunia hybrida AN2 (Quattrocchio et al., 1999), Gerbera hybrida GhMYB10 (Elomaa et al., 2003), Oryza sativa C1 (Saitoh et al., 2004), Antirrhinum majus ROSEA1, ROSEA2 and VENOSA (Schwinn et al., 2006) and Garcinia mangostana GmMYB10 (Palapol et al., 2009).
In strawberry, several transcription factors have been identified and functionally confirmed as regulators of anthocyanin biosynthesis. FaMYB1 and FcMYB1 (from Fragaria chiloensis) both act as repressors of genes that catalyse the few steps of anthocyanin biosynthesis in strawberry (Aharoni et al., 2001; Salvatierra et al., 2013). Diploid (Fragaria vesca) transgenic strawberry plants in which FvMYB10 was inhibited had undetectable levels of anthocyanin, while overexpression of FvMYB10 significantly elevated anthocyanin levels, confirming the crucial role of MYB10 in regulating anthocyanin accumulation (Lin‐Wang et al., 2014). High‐throughput transcriptome analysis showed that FaMYB10 is a general regulator of EBG and LBG in the flavonoid/phenylpropanoid pathway during the ripening of strawberry (Medina‐Puche et al., 2014). Moreover, FvMYB10 interacts with FvbHLH33 to activate the FvDFR and FvUFGT promoter (Lin‐Wang et al., 2014). Through systematic analysis of SNP variants, a candidate SNP in FvMYB10 was confirmed to be the cause of the loss of red colour in yellow strawberry fruits (Hawkins et al., 2016). An ACTTATAC insertion introduces a predicted premature termination codon in FaMYB10, which suggested the loss of FaMYB10 intact protein accounts for the loss of red colour in white octoploid strawberry (Wang et al., 2019a). Thus, all the evidence indicates that MYB10 plays a key role in strawberry anthocyanin biosynthesis.
AP2/ERFs are plant‐specific transcription factors with diverse functions in plant growth, development and responses to environmental stresses. They are divided into four categories: the APETALA2 (AP2), ERF, related to ABI3/VP1 (RAV), and Soloist families (Licausi et al., 2013; Mizoi et al., 2012; Zhu et al., 2010). AP2/ERFs have been implicated in determining different aspects of fruit quality, including fruit aroma and flavour (Xie et al., 2016). Our previous research characterized citrus CitAP2.10 as a regulator of (+)‐valencene synthesis (Shen et al., 2016) while citrus CitERF13 interacts with CitVHA‐c4 to regulate citric acid accumulation (Li et al., 2016). Several AP2/ERFs have been shown to be involved in anthocyanin biosynthesis in different species. In pear, PpERF3 interacts with PpMYB114 and PpbHLH3 to co‐regulate the coloration of red pear fruit (Yao et al., 2017) and Pp4ERF24 and Pp12ERF96 regulate blue light‐induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruit by interacting with PpMYB114, thus enhancing the expression of PpUFGT (Ni et al ., 2019). In apple, the regulator MdERF1B not only interacts with MdMYB9 and MdMYB11 but also binds to the promoters of MdMYB9 and MdMYB11 to promote anthocyanin and proanthocyanin accumulation (Zhang et al., 2018a). Another ERF transcription factor MdERF38 promotes drought stress‐induced anthocyanin biosynthesis via interaction with MdMYB1 and can be degraded by MdBT2 at the post‐translational level (An et al., 2019). Several AP2/ERFs have been identified as having roles in determining strawberry fruit quality. FaABI4 is an AP2‐type protein and a positive regulator of strawberry ripening (Chai and Shen, 2016), and an ERF–MYB complex including FaERF9 and FaMYB98 activates the FaQR promoter and increases furaneol content in cultivated strawberry (Zhang et al., 2018b). However, it is still unknown whether FaAP2/ERFs participate in anthocyanin biosynthesis in strawberry.
In this study, we screened all AP2/ERF genes expressed in strawberry fruit by dual‐luciferase assay to identify those capable of activating the promoter of FaMYB10 and found FaRAV1 had the highest activation effect. FaRAV1 was found to directly bind to the FaMYB10 promoter. Transient overexpression and RNA interference (RNAi) of FaRAV1 in strawberry fruit validated its significant role in anthocyanin biosynthesis. In addition, FaRAV1 also directly transactivates the promoters of anthocyanin biosynthesis‐related genes, which illustrates the importance of FaRAV1 in strawberry anthocyanin biosynthesis.
Results
Regulatory effect of FaAP2/ERFs on the promoter of FaMYB10
In a previous study, 120 individual FaAP2/ERF genes from ‘Yuexin’ strawberry were isolated and identified, consisting of 95 ERFs, 18 AP2s, 6 RAVs and 1 soloist member (Zhang et al., 2018b). It was found that 86 of these AP2/ ERFs were expressed in strawberry fruit. Using this information, we screened all the fruit‐expressed AP2/ERFs for their ability to transactivate the promoter of FaMYB10. The results (threshold was set as 2) showed that 5 members could activate the promoter of FaMYB10, including FaRAV1, FaRAV6, FaERF61, FaERF85 and FaERF86 (Figure 1), with FaRAV1 displaying the strongest activation effect of approximately 4.0‐fold. According to a phylogenetic tree containing strawberry AP2/ERFs and other ERFs which had been characterized in anthocyanin biosynthesis, FaERF85 and FaERF86 were similar to MdERF1B (Figure S1).
Figure 1.

Regulatory effects of FaAP2/ERFs on the promoter of FaMYB10. SK refers to empty vector and is set as 1. Error bars represent SE based on three biological replicates.
The strawberry RAV family has six members. A phylogenetic tree of strawberry RAV genes was constructed by aligning the full‐length amino acid sequences with other plant RAVs, including Arabidopsis, Populus, rice, apple and tomato (Figure S2). FaRAV1 clustered with FaRAV2 and FaRAV3 and was similar to SlRAV3, although none of these genes have been characterized to date. Transient expression in tobacco leaves of 35S‐FaRAV1‐GFP showed strong fluorescence in the nucleus, and the red nucleus signal of the mCherry marker merged with the green fluorescence (Figure S3).
The interaction between FaRAV1 and the FaMYB10 promoter
According to the results of the dual‐luciferase assay, FaRAV1, which showed the strongest activation effect on the FaMYB10 promoter, was chosen for further study. The promoter sequence of FaMYB10 was inserted into the pAbAi vector (Clontech, Japan) and transformed into the Y1H strain. The FaMYB10 promoter was not activated without protein binding (Figure 2a). The interaction between FaRAV1 and the FaMYB10 promoter was analysed; yeast containing FaRAV1 exhibited normal growth under 125 ng/mL AbA, while growth of the negative control cells, containing the empty vector, was inhibited, which indicated that FaRAV1 could directly bind to the promoter of FaMYB10 (Figure 2b). The specificity of FaRAV1 binding to the FaMYB10 promoter was confirmed by electrophoretic mobility shift assay (EMSA). It has been reported that RAV genes contain an AP2 domain and a B3 domain, which can bind specifically to DNA sequences with the consensus motif, 5′‐CAACA‐3′ and 5′‐CACCTG‐3′ respectively (Kagaya et al., 1999). After searching the promoter of FaMYB10, we found one CAACA motif within a 402‐bp region upstream of the start codon (Figure 2c). EMSA results indicated that FaRAV1 can bind to this specific CAACA motif and mutating the putative binding sites eliminated FaRAV1 protein binding, while increasing the concentration of cold probe significantly reduced the binding affinity of the biotinylated probe (Figure 2d).
Figure 2.
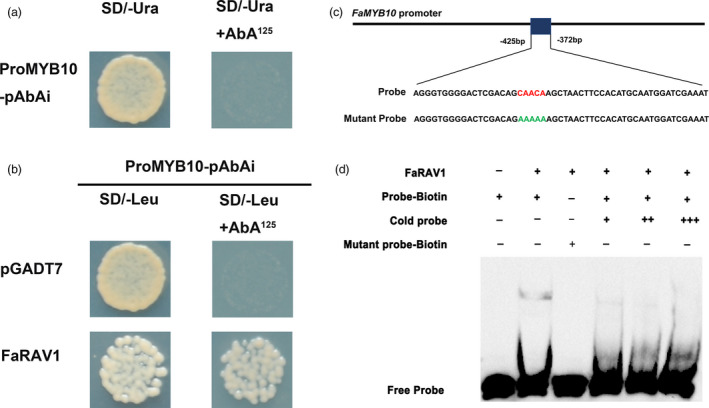
The interaction between FaRAV1 and the FaMYB10 promoter. a, Autoactivation was tested on SD/‐Ura in the presence of 125 ng/mL aureobasidin A (AbA). b, Physical interaction was determined on SD medium lacking Leu in the presence of 125 ng/mL AbA. The empty pGADT7 vector was applied as a negative control. c, The probe used for EMSA with the RAV core sequence is in red, and the mutated nucleotides indicated in green. D, EMSA of 3′‐biotin‐labelled dsDNA probes with the purified FaRAV1 protein. The presence (+) or absence (–) of specific probes is marked. The concentration of the cold probe was 16 nm (+), 32 nm (++) or 64 nm (+++) while that of the biotinylated probe was 8 nm. Water was added in place of FaRAV1 protein as a control.
Relative expression of FaRAV1, FaMYB10 and total anthocyanin content, during strawberry fruit development and ripening
The process of strawberry fruit development and ripening were divided into four major stages: G (green), T (turning), IR (intermediate red) and R (full red). Fruit from each stage were cut into three parts (apical, middle and basal) for further analysis (Figure 3a). Anthocyanin accumulation was initiated in the apical sections. At the turning stage, anthocyanin accumulated mainly in the apical region (21.3 µg/g), less than 10% (1.76 µg/g) in the middle section and no detectable anthocyanin in the basal region (Figure 3b). At the ripening stage, the anthocyanin content was 130.8, 131.1 and 69.3 µg/g in the apical, middle and basal sections, respectively. The transcript of FaRAV1 increased during colour change in the apical, middle and basal sections and increased steadily during fruit ripening. The transcript of FaMYB10, in contrast, showed an expression pattern which preceded the accumulation of anthocyanin (Figure 3c).
Figure 3.
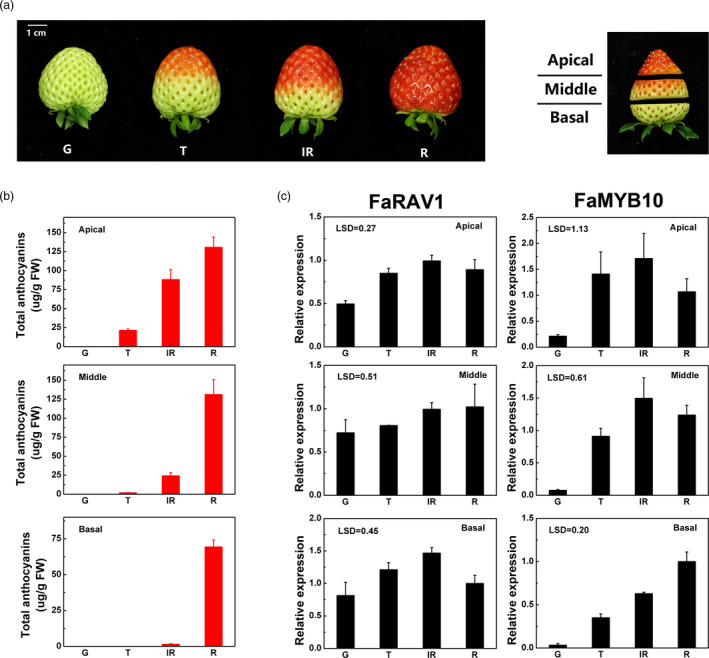
Relative expression of FaRAV1, FaMYB10 and total anthocyanins during strawberry (cv. Yuexin) fruit development and ripening. a, Fruits were collected at four stages: G, green; T, turning; IR, intermediate red; and R, full red. Three sections were sampled: apical; middle; and basal. b, Changes in the contents of total anthocyanins in each of the three sections at four developmental stages. FW, fresh weight. C, Relative expression levels of FaRAV1 and FaMYB10. The expression levels were calculated relative to corresponding values in the basal section at the R stage. Error bars represent SE based on three biological replicates. LSD values represent LSD at P = 0.05.
Transient overexpression of FaRAV1 promotes anthocyanin accumulation
To test the relationship between FaRAV1 and anthocyanin accumulation, we transiently expressed FaRAV1 by injection of Agrobacterium tumefaciens containing the FaRAV1 OE construct driven by the 35S promoter into attached green strawberry fruit. Overexpression of FaRAV1 significantly promoted anthocyanin accumulation (Figure 4a); 5 days after infiltration FaRAV1 OE fruit started to turn red at the apical end and became almost fully red after 7 days, whereas the control fruit was just beginning to turn red. On the ninth day, FaRAV1 OE fruit became totally red, while the control fruit reached approximately intermediate red stage. The fruit were harvested 9 days after infiltration (Figure S4) and the relative expression of FaRAV1 was increased in OE fruit, up to 60‐fold compared to the control, indicating the transient overexpression was very effective (Figure 4b). FaMYB10, which has been shown to be a direct target of FaRAV1 (Figures 1 and 2) was induced 8.1‐fold (Figure 4b). The total anthocyanin content of FaRAV1 OE fruit was up to 3.6‐fold higher compared with the control fruit (Figure 4b), supporting a role for FaRAV1 in promoting anthocyanin accumulation. KEGG pathway analysis of the differentially expressed genes (DEGs) between FaRAV1 OE and control fruit revealed enrichment of genes involved in phenylpropanoid biosynthesis and flavonoid biosynthesis using octoploid cultivated strawberry as reference genome (Edger et al., 2019) (Figure S5), which is consistent to results using diploid strawberry as reference genome (data not shown). Thus, we checked the expression levels of genes encoding enzymes of the anthocyanin pathway by RT‐qPCR and found CHS, CHI, F3H, DFR, ANS and GT1 showed 6.7‐, 5.0‐, 4.8‐, 2.4‐, 4.5‐ and 3.3‐fold higher levels of expression respectively (Figure 4c), which was consistent with the RNA‐seq data.
Figure 4.
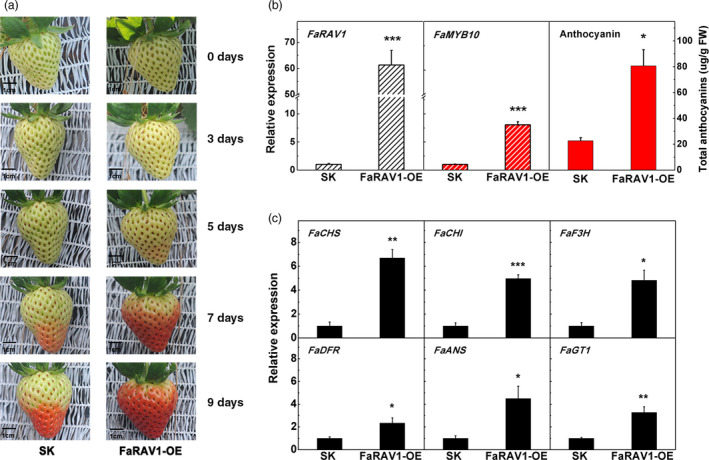
Transient overexpression of FaRAV1 promotes anthocyanin accumulation in strawberry fruit. a, Transient overexpression of FaRAV1 (right) and control (left) fruit at different times after injection, SK refers to empty vector. b, Transcript analysis of FaRAV1, FaMYB10 and anthocyanin quantification of FaRAV1 OE fruit and control fruit. C, RT‐qPCR verification of transcript levels of the main anthocyanin‐related genes. Error bars represent SE based on three biological replicates. Asterisks denote significant differences using Student’s t‐test, *P < 0.05, **P < 0.01, ***P < 0.001.
Transient RNAi of FaRAV1 down‐regulates anthocyanin biosynthesis
For a better understanding of the function of FaRAV1, we performed transient RNAi (Wang et al., 2019b) . Agrobacterium tumefaciens strain GV3101 harbouring FaRAV1 RNAi construct and empty vector pHB were separately injected into attached green strawberry fruit. RNAi fruit showed less coloration, and the content of anthocyanin was reduced to 65% compared to the control (Figure 5a,b). The expression level of FaRAV1 was reduced to 68% (Figure 5b). Transcripts of FaMYB10, the direct target gene of FaRAV1, were decreased significantly (Figure 5b). RT‐qPCR analysis showed that transcripts of all selected anthocyanin biosynthesis‐related genes were also decreased in FaRAV1 RNAi fruit (Figure 5c). Consistent with the transient overexpression of FaRAV1, RNAi results demonstrated that FaRAV1 plays an important role in the regulation of strawberry anthocyanin biosynthesis.
Figure 5.
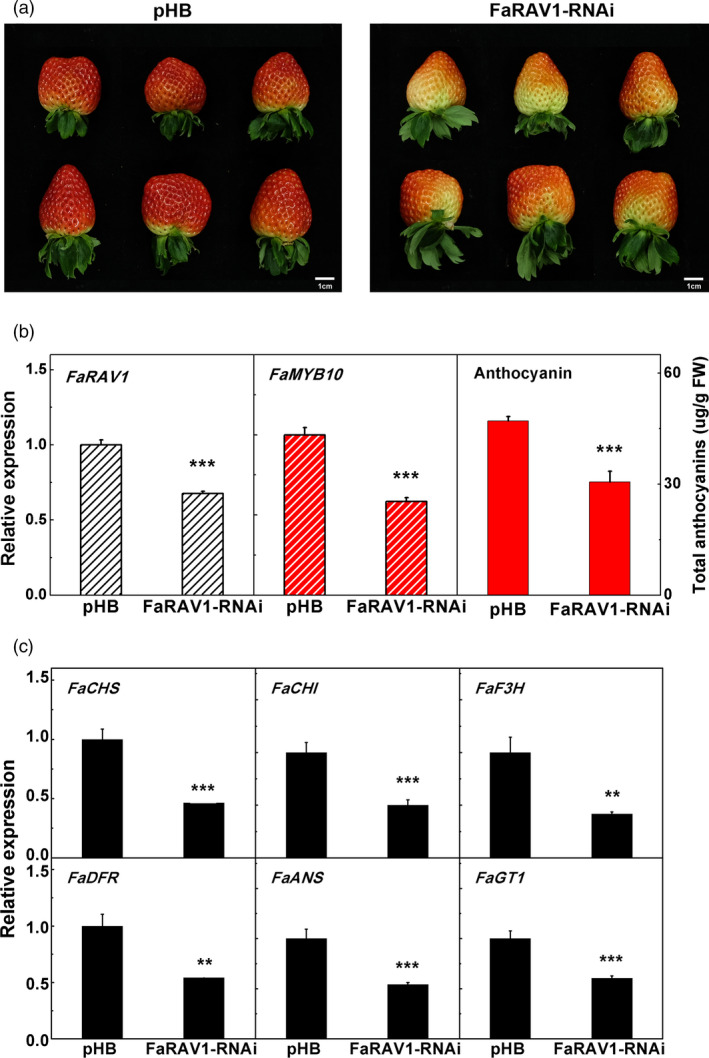
Transient RNAi of FaRAV1 reduced anthocyanin contents in strawberry fruit. a, The phonotype of transient RNAi of FaRAV1 (right) and control (left) fruit. b, Transcript analysis of FaRAV1, FaMYB10 and anthocyanin quantification of FaRAV1 RNAi fruit and control fruit. c, RT‐qPCR verification of transcript levels of the main anthocyanin‐related genes. Error bars represent SE based on three biological replicates. Asterisks denote significant differences using Student’s t‐test, **P < 0.01, ***P < 0.001.
Regulatory role of FaRAV1 in activating anthocyanin biosynthetic gene promoters
To further explore the trans‐regulatory role of FaRAV1 in anthocyanin biosynthesis, we assayed the transactivation effect of FaRAV1 on the promoters of anthocyanin biosynthetic genes. Dual‐luciferase assays revealed that FaRAV1 could significantly activate the promoters of CHS (1.53‐fold), CHI (2.3‐fold), F3H (1.95‐fold), DFR (3.6‐fold), ANS (1.31‐fold) and GT1 (2.3‐fold), respectively and that FaRAV1 showed a higher activation effect towards the DFR promoter compared with the other promoters (Figure 6a). Yeast one‐hybrid assay showed that FaRAV1 directly bind to the CHS, F3H, DFR and GT1 promoters but not promoters of CHI and ANS (Figure 6b), indicating that FaRAV1 would promote anthocyanin accumulation by direct regulation of CHS, F3H, DFR, GT1 and indirect regulation of CHI, ANS, in addition to regulating FaMYB10 transcript accumulation.
Figure 6.
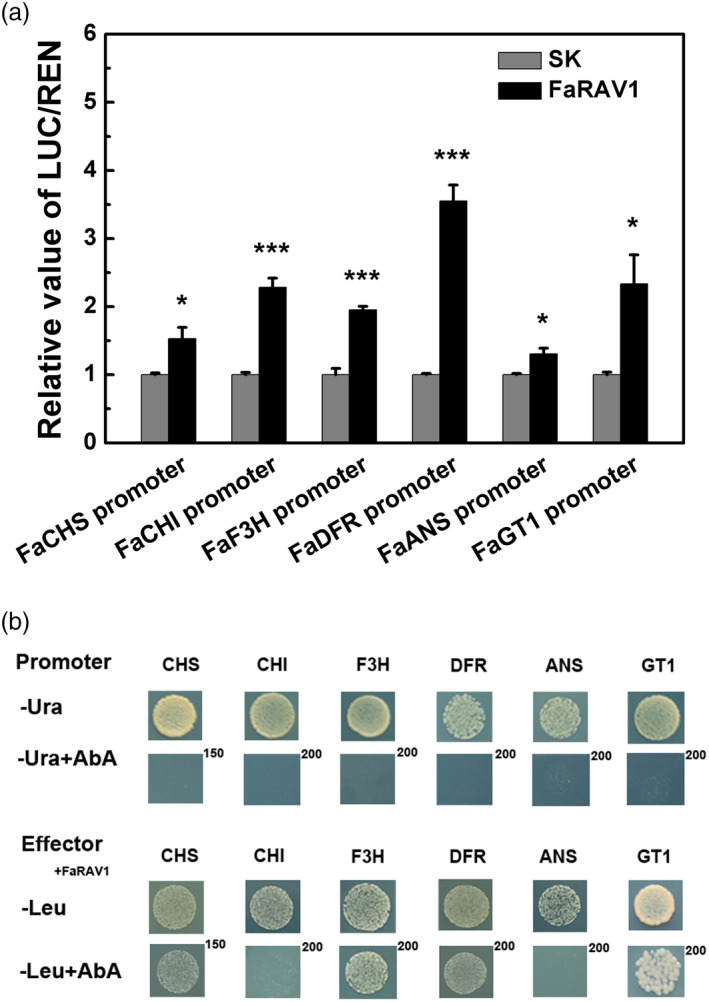
Effect of FaRAV1 on the promoters of anthocyanin biosynthetic genes. a, Dual‐luciferase assay of FaRAV1 on the promoters of anthocyanin biosynthetic genes. Error bars represent SE based on three biological replications. Asterisks denote significant differences using Student’s t‐test, *P < 0.05, ***P < 0.001. b, Yeast one‐hybrid analysis of the interaction of FaRAV1 and anthocyanin biosynthetic gene promoters. All promoters were used for autoactivation tests in the presence of different concentrations of aureobasidin A (AbA) on SD/‐Ura medium, and physical interaction was determined on SD/‐Leu medium in the presence of corresponding AbA concentrations.
Discussion
FaRAV1 positively regulates the biosynthesis of anthocyanin by direct activation of FaMYB10 transcription
Previous studies have shown that accumulation of RAV transcripts can be induced by darkness, wounding, low temperature, drought stress and pathogen attack (Fowler et al., 2005; Lee et al., 2005; Li et al., 2011; Sohn et al., 2006). Moreover, RAVs have been implicated in different aspects of plant physiological and developmental responses. SlRAV2, for example, increases bacterial wilt tolerance by inducing the expression of PR genes in tomato (Li et al., 2011). RAV1 can also regulate hypocotyl elongation (Ma et al., 2005) and has been suggested to inhibit plant growth (Hu et al., 2004), regulate leaf senescence (Woo et al., 2010) and control bud outgrowth in poplar (Moreno‐Cortés et al., 2012). TEMPRANILLO genes (TEM1 and TEM2) repress floral induction in the photoperiod pathway in Arabidopsis (Castillejo and Pelaz, 2008).
RAVs have not been implicated in fruit anthocyanin production or other aspects of fruit quality. In apple fruit, MdRAV1 is regulated by MdWRKY31 and is involved in mediating abscisic acid (ABA) sensitivity (Zhao et al., 2019). Anthocyanin can be induced by some abiotic stresses, such as drought (Hughes et al., 2010), ABA (Jia et al., 2011), light (Azuma et al., 2012), cold temperature (Steyn et al., 2009) and wounding stress (Gan et al., 2014; Saltveit, 2000). Cis‐elements related to abiotic stress responses are present in the promoter of FaRAV1, such as those implicated in response to ABA, methyl jasmonic acid (MeJA) and light response, with cis‐elements involved in light responsiveness appearing most frequently, providing clues for investigating the role of FaRAV1 (Table S3). Previous studies have reported that ABA is an internal signal for strawberry fruit ripening (Jia et al., 2011), and RAV transcription factors also play an important role in stress responses. Thus, we examined the expression of FaRAV1 in response to ABA in fruits. As is shown in Figure S6, FaRAV1 expression was sensitive to ABA, and transcripts were induced by 50 μm (6 h treatment) and 100 μm ABA (1 or 6 h treatment), indicting FaRAV1 might be involved in ABA‐mediated coloration and the mechanism of how FaRAV1 regulates needs further study. Here, we showed that FaRAV1 participates in anthocyanin accumulation via direct binding to the promoter of FaMYB10 and had the strongest activation effect on the FaMYB10 promoter compared with other fruit‐expressed FaAP2/ERFs (Figures 1 and 2). In addition, expression of FaMYB10 was induced in FaRAV1 OE fruit and reduced in FaRAV1 RNAi fruit correspondingly, indicating that FaRAV1 positively regulates anthocyanin biosynthesis via FaMYB10.
Other potential FaAP2/ERFs regulators of anthocyanin production in strawberry
By dual‐luciferase assay (Figure 1), we found FaRAV6, FaERF61, FaERF85, FaERF86 could also activate the FaMYB10 promoter (with a cut‐off of two‐fold). There were also four FaAP2/ERFs that appear to repress transcriptional activity of the FaMYB10 promoter: FaERF15, FaERF17, FaERF20 and FaERF29 (threshold set as 0.4). Based on transcript analysis of fruit‐expressed FaAP2/ERFs in a previous paper (Zhang et al ., 2018b), it could be suggested that FaERF85 might be a positive anthocyanin regulator, as its transcript greatly increased at the turning stage of strawberry fruit ripening. Based on phylogenetic analysis, FaERF85 clustered with MdERF1B, a positive anthocyanin regulator in apple (Zhang et al., 2018a), further supporting the suggestion of the involvement of FaERF85 in anthocyanin accumulation. Transcripts of FaERF29 gradually decreased during fruit development, which is negatively related to anthocyanin content, implying it might be a repressor of anthocyanin biosynthetic genes.
Hierarchical regulation of the MBW complex regulating anthocyanin biosynthesis
Several transcription factors have been reported to regulate anthocyanin biosynthesis via interaction with an activating MYB or MBW complex. For example, AtLBD37, AtLBD38 and AtLBD39 are negative regulators of anthocyanin biosynthesis via regulating PAP1 and PAP2 in Arabidopsis thaliana (Rubin et al., 2009). VmTDR4, a MADS box transcription factor, promotes anthocyanin accumulation through direct or indirect control of the R2R3 MYB family in bilberry (Jaakola et al., 2010). In apple, MdHY5 promotes anthocyanin accumulation by directly binding to the MdMYB10 promoter (An et al., 2017). In blood‐fleshed peach, BL, a NAC transcription factor, forms a heterodimer with PpNAC1 and activate the transcription of PpMYB10.1 (Zhou et al., 2015). In addition, some TFs play roles in anthocyanin accumulation via competing with the MYB or bHLH to interrupt or stabilize the MBW complex, such as AtMYBL2 and AtSPL9 (Dubos et al., 2008; Gou et al., 2011; Matsui et al., 2008). Another anthocyanin activator AtTCP3, which interacts separately with PAP1, PAP2 and TT2, promotes anthocyanin accumulation via stabilizing the formation of the MBW complex and thus stimulating the expression of LBGs (Li and Zachgo, 2013). Further research has characterized TFs in the MYB, LBD, SPL, TCP, MADS, bZIP and NAC families affecting anthocyanin levels. Here, we characterized FaRAV1 involvement in anthocyanin biosynthesis via upstream activation of FaMYB10. Unlike MdERF1B, FaRAV1 activates FaMYB10 transcription, but does not form a protein complex with FaMYB10 (Figure S7). FaERF85, the homologue of MdERF1B, might regulate anthocyanin via protein–DNA and protein–protein interaction, which needs further validation.
FaRAV1 positively regulates anthocyanin accumulation at multiple levels
As mentioned above, FaRAV1 promotes anthocyanin accumulation by activating the FaMYB10 promoter (Figure 1). Moreover, further investigation by luciferase transactivation assays, showed that FaRAV1 could also directly activate the promoters of anthocyanin biosynthetic structural genes. FaRAV1 showed the highest activation effect towards the DFR promoter compared with the other promoters (Figure 6a). Transcripts of FaMYB10 and anthocyanin biosynthesis‐related genes were altered in transient OE and RNAi fruit accordingly (Figures 4 and 5). We also demonstrated that FaRAV1 can directly bind to the CHS, F3H, DFR and GT1 promoters by yeast one‐hybrid assays (Figure 6b). However, FaRAV1 would not directly bind to the promoters of CHI and ANS (Figure 6b), indicating the activation of CHI and ANS is not directly driven by FaRAV1 and other TFs may be involved (e.g. FaMYB10). We also carried out cis‐element analysis of these anthocyanin biosynthesis promoters (Figure S8). CAACA motif existed in all promoters of anthocyanin biosynthetic genes, which would not explain why FaRAV1 could not bind to the promoters of CHI and ANS, implying other AP2/ERFs might also be involved. Interestingly, another differentially expressed anthocyanin gene RAP (reduced anthocyanin in petioles) is significantly up‐regulated 8.1‐fold in OE fruit and down‐regulated 0.57‐fold in RNAi fruit (Figure S9). RAP encodes a glutathione S‐transferase (GST) gene, which is the principal transporter of anthocyanins and whose transcript is up‐regulated in FvMYB10 OE fruit (Lin‐Wang et al., 2014) and down‐regulated in Yellow Wonder fruit, a natural mutant of the MYB10 gene (Gao et al., 2019; Hawkins et al., 2016; Luo et al., 2018), suggesting RAP operates downstream of FaMYB10. Dual‐luciferase experiments showed that the promoter activity of RAP cannot be triggered by FaRAV1 (Figure S10), suggesting FaRAV1 regulates its transactivation via FaMYB10. CRISPR/Cas9 has become a tool for studying the gene function in plant and is now available in cultivated strawberry (Gao et al., 2019). The use of CRISPR to generate new mutants will be one important feature of future work.
Conclusions
Despite the RAV gene family being characterized in many different physiological pathways in plants, the role of RAVs in anthocyanin biosynthesis has not been previously studied. Here, we found that FaRAV1 directly bound to and activated the promoter of FaMYB10. In addition, FaRAV1 can also directly bind to and activate CHS, F3H, DFR and GT1 promoters, showing a second effect of FaRAV1 on anthocyanin biosynthesis (Figure 7). Transient overexpression of FaRAV1 in strawberry fruit increased anthocyanin‐related gene expression and promoted anthocyanin production. Correspondingly, transient RNAi of FaRAV1 fruit contained less anthocyanin and lower level of anthocyanin‐related gene expression compared to control fruit. These results demonstrate that FaRAV1 functions positively in strawberry anthocyanin accumulation.
Figure 7.
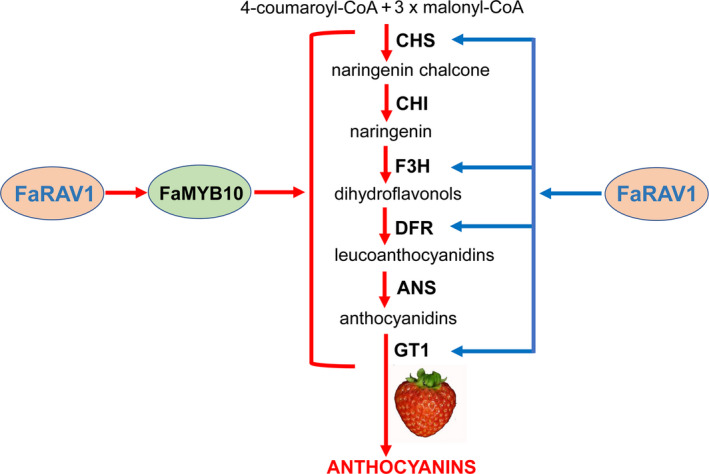
A working model of the role of FaRAV1 in promoting anthocyanin accumulation in strawberry fruit. FaRAV1 directly binds to the promoter of FaMYB10 and activates its transcription (approximately 4.0‐fold), marked by red arrows. Blue arrows indicate FaRAV1 can directly bind to and activate CHS (1.53‐fold), F3H (1.95‐fold), DFR (3.6‐fold) and GT1 (2.3‐fold) promoters.
Experimental procedures
Plant material and ABA treatment
Octoploid strawberry (Fragaria × ananassa ‘Yuexin’) plants were grown at the Zhejiang Academy of Agricultural Sciences in Haining (Zhejiang, China). Four stages of fruit (G, green; T, turning; IR, intermediate red; R, full red) were harvested at 25, 28, 33 and 37 days after anthesis and then transported to the laboratory within 2 h. Fruit of uniform size, absence of disease and mechanical wounding were selected. After removing the calyces, fruit were then separated into apical, middle and basal parts and frozen in liquid nitrogen rapidly and then stored at −80 °C for further use. Gene expression analyses are conducted with three biological replicates (representing ten fruit) for each time point. Green fruit were harvested for ABA treatment. Discs (10 mm in diameter and 2 mm in thickness) of fruit were prepared and infiltrated for 30 min in equilibration buffer (Archbold, 1999; Han et al., 2015), after which different concentrations of ABA (Sigma‐Aldrich; A1049, Germany) were added and then shaken for 1 or 6 h at 25 °C. Equilibration buffer consisted of 50 mm MES‐Tris (pH = 5.5), 10 mm MgCl2, 10 mm EDTA, 5 mm CaCl2, 200 mm mannitol and 5 mm vitamin C. After incubation, the residues were dried with tissue and frozen rapidly in liquid nitrogen and kept at −80 °C until further use.
RNA isolation and RT‐qPCR
Total RNA from strawberry fruit was extracted using the CTAB method (Chang et al., 1993). After elimination of genome DNA by gDNA eraser, 1 µg RNA was used for first‐strand cDNA synthesis by the PrimeScript™ RT reagent Kit (Takara, Dalian, China) and then diluted with water (1: 20). Real‐time PCR was carried out using a CFX96 instrument with SsoFast EvaGreen Supermix Kit (Bio‐Rad, America). The specificity of primers was assured by both melting curves and product sequencing before use. The PCR reactions and mixture were as described in our previous report (Yin et al., 2012). Data were analysed and relative expression level of the genes was calculated using the 2(‐△△ C t) method and using expression of the strawberry FaRIB413 (Zorrilla‐Fontanesi et al., 2012) as the internal control. The primers for RT‐qPCR analysis are listed in Table S1.
Gene isolation, promoter cloning and analysis
The SK vectors of FaAP2/ERFs were described in our previous report (Zhang et al., 2018b). Promoters of anthocyanin‐related genes were isolated according to genome databases (https://bioinformatics.psb.ugent.be/plaza/ or http://strawberry‐garden.kazusa.or.jp/index.html). The primers are listed in Table S2. These promoters were isolated and sequenced, and we performed an in silico analysis for FaMYB10 (1061 bp) (Delgado et al., 2018), CHS (1614 bp), CHI (2211 bp), F3H (1019 bp), DFR (1994 bp), ANS (830 bp), GT1 (1526 bp) and RAP (1427 bp) from upstream of the start codon. The analysis of cis‐elements within FaRAV1 promoter regions was conducted using the website http://bioinformatics.psb.ugent.be/webtools/plantcare/html/.
Dual‐luciferase assay
Dual‐luciferase assay was applied to investigate the transactivation activities of different TFs on target promoters. The full‐length sequences of FaAP2/ERFs transcription factors were amplified and inserted into pGreen II 0029 62‐SK vector and the promoters of eight anthocyanin‐related genes were constructed in the pGreen II 0800‐LUC vector. The primers used for vector construction are listed in Table S2. All constructs were electroporated into Agrobacterium tumefaciens GV3101, and the cultures were adjusted to an OD600 of 0.75 with infiltration buffer (10 mm MES, 10 mm MgCl2, 150 mm acetosyringone, pH 5.6). To research the activity of a specific transcription factor towards the target promoter, a mixture of A. tumefaciens containing TFs (1 mL) and promoters (100 μL) was infiltrated into tobacco (Nicotiana benthamiana) leaves by needleless syringe. Tobacco plants were grown in a greenhouse with a light/dark cycle of 16: 8 h at 24 °C. Three days after infiltration, discs from the tobacco leaves were collected and enzyme activity of firefly and renilla luciferases was measured using dual‐luciferase reagents (Promega, America). For every TF–promoter interaction, three biological replicates were performed for individual experiment.
Yeast one‐hybrid assay
The Matchmaker™ Gold Yeast One‐Hybrid Library Screening System (Clontech, Japan) was used to test the interaction of FaRAV1 with the FaMYB10 promoter. The promoter sequence was amplified and inserted into pAbAi vector, and the full‐length FaRAV1 was cloned into pGADT7 vector. The recombinant FaMYB10 promoter–pAbAi vector was linearized and transformed into the Y1HGold yeast strain to test the promoter autoactivation according to the system user manual. The Y1HGold strain carrying FaMYB10 promoter was transfected with the FaRAV1‐pGADT7 plasmid and the empty vector pGADT7 as a negative control separately.
Recombinant protein and EMSA analysis
The full‐length FaRAV1 was inserted into pET‐32a (Clontech, Japan) to generate the recombinant N‐terminal FaRAV1‐His fusion protein. The construct was purified and transformed into Escherichia coli strain Rosetta 2(DE3)pLysS (Novagen, Germany). The transformed cells were induced by 0.5 mm isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) followed by incubation at 16 °C for 20 h. Then, the cells were collected by centrifugation and resuspended in buffer (20 mm Tris‐HCl, pH = 8.0, 0.5 m NaCl), after which they were subjected to sonication on ice with 2‐s/4‐s on/off cycle for 20 min, centrifuged at 10000 rpm for 30 min at 4 °C and the supernatant was purified using a HisTALON™ Gravity Column (Clontech, Japan), following the steps described in the official user manual.
EMSA was performed by using the LightShift Chemiluminescent EMSA kit (Thermo, America ) according to the manufacturer’s instructions. Single‐strand oligonucleotides were synthesized and 3′‐biotin‐end‐labelled by HuaGene. The details of the EMSA experiment can be found in Ge et al. (2017). The EMSA probes are listed in Table S2.
Anthocyanin measurement
Strawberry fruit were ground to powder under liquid nitrogen. Approximately 1 g powder was added to 5 mL methanol‐0.05% HCl and then extracted at 4 °C in the dark for 12 h. The supernatant was collected by centrifugation for further analysis and the extraction procedure was repeated once (Wrolstad et al., 1982). The collected supernatants were pooled, filtered through 0.22 µm Millipore membranes and then evaporated at 30 °C in an evaporator machine. The residual material was resuspended in 1 mL methanol and filtered through a 0.22‐µm Millipore membrane for HPLC analysis using an HPLC (Agilent 1269, America) analytical column SB‐C18 (4.6 × 250 mm, 5 µm, Agilent Technologies, America).
The detection procedure was set as solvent A (formic acid: water, 1: 1000, v/ v) and solvent B (formic acid: acetonitrile, 1: 1000, v/ v) with the following gradient: 0–2 min, 5%; 2–7 min, 5–15%; 7–20 min, 15–20%; 20–25 min, 20–27%; 25–32 min, 27%; 31–41 min, 27%–35%; 41.01–43 min, 5%. The flow rate was 0.8 mL/min at 30 °C. The post‐run‐time was set at 5 min and the detection wavelength was 520 nm (Cheng et al., 2014). Pelargonidin‐3‐glucoside (P3G) and cyanidin‐3‐glucoside (C3G) were used as standards.
Phylogenetic tree construction
The phylogenetic tree was constructed with the FigTree v1.4.2 program, aligning the full‐length amino acid sequences of RAVs using the neighbour‐joining method for the ClustalX v2.0 program. The sequences using for the phylogenetic tree included Fragaria × ananassa FaRAV1, FaRAV2, FaRAV3, FaRAV4, FaRAV5, FaRAV6, Arabidopsis thaliana AtRAV1 (At1g13260), AtRAV1‐like (At3g25730), AtTEM1 (At1g25560), AtTEM2 (At1g68840), AtRAV3 (At1g50680), AtRAV3L (At1g51120), Solanum lycopersicum SlRAV1, SlRAV2, SlRAV3, Malus × domestica MdRAV1 (MDP0000939633), MdRAV2 (MDP0000128924), MDP0000945267, MDP0000321569, MDP0000223137, MDP0000153589, MDP0000165802, MDP0000534780, MDP0000526584, MDP0000485280, MDP0000207722, Oryza sativa OsRAV1 (Os01g04800.1), OsRAV2 (Os01g04750.1), OsRAV3 (Os05g47650.1), OsRAV4 (Os01g49830.1), Populus trichocarpa PtRAV1, PtRAV2 (GenBank_Number), PtRAV3, PtRAV4 and PtRAV5 .
Subcellular localization analysis
The FaRAV1 full‐length coding sequence without the stop codon was fused to the pCAMBIA1300‐sGFP vector (KpnI/SalI) at the C‐terminal and then expressed transiently in transgenic N. benthamiana (with nucleus‐located mCherry) leaves by A. tumefaciens infiltration (GV3101) using the same method as described above for the dual‐luciferase assay. Tobacco leaves were measured 2 days after infiltration and the fluorescence was imaged with a Nikon A1‐SHS confocal laser scanning microscope. The excitation wavelength for GFP fluorescence was 488 nm, and fluorescence was detected at 490 to 520 nm. The primers for GFP construction are listed in Table S2.
Transient overexpression and RNAi in strawberry fruit
The construct of pGreenII 0029 62‐SK containing FaRAV1 was used for transient overexpression. Forward and reverse PCR‐amplified cDNA fragments of FaRAV1 were inserted into the 2× CaMV35S‐driven vector pHB to produce the FaRAV1‐RNAi construct. All the constructs were independently transformed into A. tumefaciens strain GV3101. Attached fruit of similar size at the green (G) stage were selected and injected with A. tumefaciens, containing construct FaRAV1‐SK and empty vector SK, FaRAV1‐RNAi and empty vector pHB under the same infiltration conditions, which were performed in 2018 and 2019 respectively. The cultures were adjusted to an OD600 of 1.0 with infiltration buffer (10 mm MES, 10 mm MgCl2, 150 mm acetosyringone, pH 5.6). A. tumefaciens suspension was evenly injected into the basal part of fruit at two or three sites until the whole fruit became hydrophanous. The fruit were collected 9 days after transfection and each fruit was collected as an individual sample. Three biological replicates were sampled for analysis. The primers for RNAi construction are listed in Table S2.
RNA‐seq
The FaRAV1 transient overexpressing fruit and relevant control fruit were processed for Illumina RNA‐seq analysis. Cultivated strawberry genome‐based reads used a reference for transcriptome analysis. TPM (Transcripts per million reads) were used to estimate gene expression levels and a threshold of twofold change was applied to select differentially expressed genes.
Yeast two‐hybrid assay
Yeast two‐hybrid assay was performed to test the interaction between FaRAV1 and FaMYB10 using the Matchmaker™ Gold Yeast Two‐Hybrid System (Clontech, Japan). Full‐length coding sequences of FaRAV1 and FaMYB10 were separately cloned into pGADT7 and pGBKT7 vectors. The concentrations of aureobasidin A (AbA) to inhibit self‐transactivation were tested on SD/‐Trp medium. pGBKT7‐p53 and pGBKT7‐T were used as positive control, while pGBKT7‐Lam and pGBKT7‐T were used as negative control. pGBKT7 and pGADT7 vectors containing target genes were co‐transformed into the Y2H strain and the interactions were detected on QDO (SD/‐Ade/‐His/‐Leu/‐Trp) in the presence of AbA and X‐α‐Gal. The primers for vector construction are listed in Table S2.
Statistics
Student’s two‐tailed t‐test (*, P < 0.05; **, P < 0.01; ***, P < 0.001) was used to evaluate significant differences between two groups in this study. Figures were treated with Origin 8.0 (Microcal Software, America). Least significant differences (LSD) at the 5% level were conducted by DPS7.05 (Zhejiang University).
Accession numbers
GenBank accession number for the genes identified are FaRAV1, XM_011466945.1; FaMYB10, EU155162; CHS, AY997297; CHI, AB201755; F3H, AY691919; DFR, AY695812; ANS, AY695817; and GT1, AY575056.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
K.C. and G.J. conceived the research plans; Y.S., X.r.Y. and K.C. supervised the experiments; Z.Z., Y.M., Y.Z., Y.X., W.L. and Y.L. performed the experiments and analysis; S.L., X.L. and X.f.Y. provided technical assistance to Z.Z.; Z.Z. and Y.S. wrote the article. D.G. and A.C.A. were involved in the design, discussion and revision of the manuscript; all authors read and approved the final article.
Supporting information
Figure S1 Phylogenetic tree analysis of strawberry AP2/ERF proteins and related AP2/ERF TFs from apple and pear.
Figure S2 Phylogenetic analysis of FaRAV1 and 34 other RAV proteins.
Figure S3 Subcellular localization of FaRAV1. FaRAV1 was inserted into the pCAMBIA1300‐sGFP vector and transiently expressed in tobacco leaves.
Figure S4 The appearance of FaRAV1 overexpression fruit (right) 9 d after injection, compared to the control (left).
Figure S5 PEGG analysis of DEGs between FaRAV1 OE fruit and control fruit.
Figure S6 RT‐qPCR analysis of FaRAV1 expression in response to ABA treatment in strawberry fruit.
Figure S7 Yeast two‐hybrid analysis of the interactions between FaRAV1 and FaMYB10.
Figure S8 Cis regulatory elements in promoters of anthocyanin biosynthetic genes.
Figure S9 Relative expression of RAP in FaRAV1 OE and RNAi fruit compared with the control fruit.
Figure S10 Regulatory effect of FaRAV1 on the promoter of RAP.
Table S1 Primers used for reverse transcription quantitative PCR.
Table S2 Primers used for vector construction.
Table S3 Motifs in the FaRAV1 promoter identified in silico by PlantCARE.
Acknowledgement
We thank Dr. Liu YS (Anhui Agricultural University) for providing the RNAi vector and technical assistance. This research was supported by Zhejiang Provincial Science and Technology Project (2016C04001) and the 111 Project (B17039) and by the Fundamental Research Funds for the Central Universities (2019XZZX005‐1‐06).
Zhang, Z. , Shi, Y. , Ma, Y. , Yang, X. , Yin, X. , Zhang, Y. , Xiao, Y. , Liu, W. , Li, Y. , Li, S. , Liu, X. , Grierson, D. , Allan, A. C. , Jiang, G. and Chen, K. (2020) The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J., 10.1111/pbi.13382
Contributor Information
Guihua Jiang, Email: jgh2004267@sina.com.
Kunsong Chen, Email: akun@zju.edu.cn.
References
- Aharoni, A. , De Vos, C.H. , Wein, M. , Sun, Z.K. , Greco, R. , Kroon, A. , Mol, J.N.M. et al (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. [DOI] [PubMed] [Google Scholar]
- Allan, A.C. , Hellens, R.P. and Laing, W.A. (2008) MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. [DOI] [PubMed] [Google Scholar]
- Almeida, J.R.M. , D’Amico, E. , Preuss, A. , Carbone, F. , De Vos, C.H. , Deiml, B. , Mourgues, F. et al (2007) Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch. Biochem. Biophys. 465, 61–71. [DOI] [PubMed] [Google Scholar]
- An, J.P. , Qu, F.J. , Yao, J.F. , Wang, X.N. , You, C.X. , Wang, X.F. and Hao, Y.J. (2017) The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4, 17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J.P. , Zhang, X.W. , Bi, S.Q. , You, C.X. , Wang, X.F. and Hao, Y.J. (2019) The ERF transcription factor MdERF38 promotes drought stress‐induced anthocyanin biosynthesis in apple. Plant J. 101, 573–589. [DOI] [PubMed] [Google Scholar]
- Archbold, D.D. (1999) Carbohydrate availability modifies sorbitol dehydrogenase activity of apple fruit. Plant Physiol. 105, 391–395. [Google Scholar]
- Azuma, A. , Yakushiji, H. , Koshita, Y. and Kobayashi, S. (2012) Flavonoid biosynthesis‐related genes in grape skin are differentially regulated by temperature and light conditions. Planta, 236, 1067–1080. [DOI] [PubMed] [Google Scholar]
- Ban, Y. , Honda, C. , Hatsuyama, Y. , Igarashi, M. , Bessho, H. and Moriguchi, T. (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. [DOI] [PubMed] [Google Scholar]
- Baudry, A. , Heim, M.A. , Dubreucq, B. , Caboche, M. , Weisshaar, B. and Lepiniec, L. (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . Plant J. 39, 366–380. [DOI] [PubMed] [Google Scholar]
- Boches, P.S. , Peterschmidt, B.C. and Myers, J.R. (2009) Breeding tomato for increased fruit phenolics. J. Am. Soc. Hort. Sci. 44, 1055–1056. [Google Scholar]
- Castillejo, C. and Pelaz, S. (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. [DOI] [PubMed] [Google Scholar]
- Chai, L. and Shen, Y.Y. (2016) FaABI4 is involved in strawberry fruit ripening. Sci. Hortic‐Amsterdam. 210, 34–40. [Google Scholar]
- Chang, S.J. , Puryear, J. and Cairney, J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. [Google Scholar]
- Cheng, J. , Wei, G. , Zhou, H. , Gu, C. , Vimolmangkang, S. , Liao, L. and Han, Y. (2014) Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 166, 1044–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado, L.D. , Zúñiga, P.E. , Figueroa, N.E. , Pastene, E. , Escobar‐Sepúlveda, H.F. , Figueroa, P.M. , Garrido‐Bigotes, A. et al (2018) Application of a JA‐Ile biosynthesis inhibitor to methyl jasmonate‐treated strawberry fruit induces upregulation of specific MBW complex‐related genes and accumulation of proanthocyanidins. Molecules, 23, 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos, C. , Le Gourrierec, J. , Baudry, A. , Huep, G. , Lanet, E. , Debeaujon, I. , Routaboul, J.M. et al (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana . Plant J. 55, 940–953. [DOI] [PubMed] [Google Scholar]
- Edger, P.P. , Poorten, T.J. , VanBuren, R. , Hardigan, M.A. , Colle, M. , McKain, M.R. , Smith, R.D. et al (2019) Origin and evolution of the octoploid strawberry genome. Nat. Genet. 51, 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomaa, P. , Uimari, A. , Mehto, M. , Albert, V.A. , Laitinen, R.A. and Teeri, T.H. (2003) Activation of anthocyanin biosynthesis in Gerbera hybrida Asteraceae suggests conserved protein‐protein and protein‐promoter interactions between the anciently diverged monocots and Eudicots. Plant Physiol. 133, 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley, R.V. , Hellens, R.P. , Putterill, J. , Stevenson, D.E. , Kutty‐Amma, S. and Allan, A.C. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S.Q. , Wang, Y.L. , Yang, S. , Xu, Y.T. and Chen, X.S. (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3‐MYB transcription factor PyMYB10. Planta, 232, 245–255. [DOI] [PubMed] [Google Scholar]
- Fowler, S.G. , Cook, D. and Thomashow, M.F. (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 Is gated by the circadian clock. Plant Physiol. 137, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Y. , Li, H. , Xie, Y. , Wu, W. , Li, M. , Wang, X. and Huang, J. (2014) THF1 mutations lead to increased basal and wound‐induced levels of oxylipins that stimulate anthocyanin biosynthesis via COI1 signaling in Arabidopsis . J. Integr. Plant Biol. 56, 916–927. [DOI] [PubMed] [Google Scholar]
- Gao, Q. , Luo, H.F. , Li, Y.P. , Liu, Z.C. and Kang, C.Y. (2019) Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant biotechnology J. 10.1111/PBI.13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, H. , Zhang, J. , Zhang, Y.J. , Li, X. , Yin, X.R. , Grierson, D. and Chen, K.S. (2017) EjNAC3 transcriptionally regulates chilling‐induced lignification of loquat fruit via physical interaction with an atypical CAD‐like gene. J. Exp. Bot. 68, 5129–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, A. , Zhao, M.Z. , Leavitt, J.M. and Lloyd, A.M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Gou, J.Y. , Felippes, F. , Liu, C.J. , Weigel, D. and Wang, J.W. (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156‐targeted SPL transcription factor. Plant Cell, 23, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesser, M. , Hoffmann, T. , Bellido, M.L. , Rosati, C. , Fink, B. , Kurtzer, R. , Aharoni, A. et al (2008) Redirection of flavonoid biosynthesis through the down‐regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 146, 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. , Dang, R.H. , Li, J.X. , Jiang, J.Z. , Zhang, N. , Jia, M.R. , Wei, L.Z. et al (2015) SUCROSE NONFERMENTING1‐RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol. 167, 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, C. , Caruana, J. , Schiksnis, E. and Liu, Z.C. (2016) Genome‐scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep. 6, 29017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y.X. , Wang, Y.H. , Liu, X.F. and Li, J.Y. (2004) Arabidopsis RAV1 is down‐regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 14, 8–15. [DOI] [PubMed] [Google Scholar]
- Hughes, N.M. , Reinhardt, K. , Field, T.S. , Gerardi, A.R. and Smith, W.K. (2010) Association between winter anthocyanin production and drought stress in angiosperm evergreen species. J. Exp. Bot. 61, 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola, L. , Poole, M. , Jones, M.O. , Karppinen, T. , Koskimäki, J.J. , Hohtola, A. , Haggman, H. et al (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153, 1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, H.F. , Chai, Y.M. , Li, C.L. , Lu, D. , Luo, J.J. , Qin, L. and Shen, Y.Y. (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya, Y. , Ohmiya, K. and Hattori, T. (1999) RAV1, a novel DNA‐binding protein, binds to bipartite recognition sequence through two distinct DNA‐binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, S. , Ishimaru, M. , Hiraoka, K. and Honda, C. (2002) Myb‐related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta, 215, 924–933. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S. , Goto‐Yamamoto, N. and Hirochika, H. (2004) Retrotransposon‐induced mutations in grape skin color. Science,, 304, 982–982. [DOI] [PubMed] [Google Scholar]
- Lee, J.H. , Kim, S.H. , Jung, Y.H. , Kim, J.A. , Lee, M.O. , Choi, P.G. , Choi, W. et al (2005) Molecule cloning and functional analysis of rice (Oryza sativa L.) OsNDR1 on defense signaling pathway. Plant Pathology J. 21, 149–157. [Google Scholar]
- Li, S.T. (2014) Transcriptional control of flavonoid biosynthesis. Plant Signaling Behav. 9, e27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.T. and Zachgo, S. (2013) TCP3 interacts with R2R3‐MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana . Plant J. 76, 901–913. [DOI] [PubMed] [Google Scholar]
- Li, C.W. , Su, R.C. , Cheng, C.P. , Sanjaya, You, S.‐J. , Hsieh, T.‐H. , Chao, T.‐C. and Chan, M.‐T. (2011) Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP‐mediated defense pathway. Plant Physiol. 156, 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.J. , Yin, X.R. , Xie, X.L. , Allan, A.C. , Ge, H. , Shen, S.L. and Chen, K.S. (2016) The Citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein‐protein interaction with the vacuolar proton pump, CitVHA‐c4. Sci. Rep. 6, 20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi, F. , Ohme‐Takagi, M. and Perata, P. (2013) APETALA2/ethylene responsive factor AP2/ERF transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Lin, X. , Xiao, M. , Luo, Y. , Wang, J.Y. and Wang, H.Q. (2013) The effect of RNAi‐induced silencing of FaDFR on anthocyanin metabolism in strawberry (Fragaria × ananassa) fruit. Sci Hortic (Amsterdam). 160, 123–128. [Google Scholar]
- Lin‐Wang, K. , McGhie, T.K. , Wang, M. , Liu, Y.H. , Warren, B. , Storey, R. , Espley, R.V. et al (2014) Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Frontiers Plant Sci. 5, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A. , Brockman, A. , Aguirre, L. , Campbell, A. , Bean, A. , Cantero, A. and Gonzalez, A. (2017) Advances in the MYB–bHLH–WD repeat MBW pigment regulatory model: addition of a WRKY factor and co‐option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H.F. , Dai, C. , Li, Y.P. , Feng, J. , Liu, Z.C. and Kang, C.Y. (2018) Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 69, 2595–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.G. , Sun, N. , Liu, X.G. , Jiao, Y.L. , Zhao, H.Y. and Deng, X.W. (2005) Organ‐specific expression of Arabidopsis genome during development. Plant Physiol. 138, 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, K. , Umemura, Y. and Ohme‐Takagi, M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. [DOI] [PubMed] [Google Scholar]
- Medina‐Puche, L. , Cumplido‐Laso, G. , Amil‐Ruiz, F. , Hoffmann, T. , Ring, L. , Rodríguez‐Franco, A. , Caballero, J.L. et al (2014) MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J. Exp. Bot. 65, 401–417. [DOI] [PubMed] [Google Scholar]
- Mes, P.J. , Boches, P. , Myers, J.R. and Durst, R. (2008) Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hort. Sci. 133, 262–269. [Google Scholar]
- Mizoi, J.Y. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2012) AP2/ERF family transcription factors in plant abiotic stress responses. BBA‐Gene Regul. Mech. 1819, 86–96. [DOI] [PubMed] [Google Scholar]
- Moreno‐Cortés, A. , Hernández‐Verdeja, T. , Sánchez‐Jiménez, P. , González‐Melendi, P. , Aragoncillo, C. and Allona, I. (2012) CsRAV1 induces sylleptic branching in hybrid poplar. New Phytol. 194, 98–90. [DOI] [PubMed] [Google Scholar]
- Ni, J.B. , Bai, S.L. , Zhao, Y. , Qian, M.J. , Tao, R.Y. , Yin, L. , Gao, L. et al (2019) Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light‐induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 99, 67–78. [DOI] [PubMed] [Google Scholar]
- Palapol, Y. , Ketsa, S. , Lin‐Wang, K. , Ferguson, I.B. and Allan, A.C. (2009) A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen (Garcinia mangostana L.) fruit during ripening. Planta, 229, 1323–1334. [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F. , Wing, J. , van der Woude, K. , Souer, E. , de Vetten, N. , Mol, J. and Koes, R. (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell, 11, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. , Tohge, T. , Matsuda, F. , Saito, K. and Scheible, W.R. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis . Plant Cell, 21, 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz, M.B. , Grotewold, E. and Chandler, V.L. (1997) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell, 9, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, K. , Onishi, K. , Mikami, I. , Thidar, K. and Sano, Y. (2004) Allelic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics, 168, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltveit, M.E. (2000) Wound induced changes in phenolic metabolism and tissue browning are altered by heat shock. Postharvest Biol. Tec. 21, 61–69. [Google Scholar]
- Salvatierra, A. , Pimentel, P. , Moya‐León, M.A. and Herrera, R. (2013) Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry, 90, 25–36. [DOI] [PubMed] [Google Scholar]
- Sapir, M. , Oren‐Shamir, M. , Ovadia, R. , Reuveni, M. , Evenor, D. , Tadmor, Y. , Nahon, S. et al (2008) Molecular aspects of Anthocyanin fruit tomato in relation to high pigment‐1. J. Hered. 90, 292–303. [DOI] [PubMed] [Google Scholar]
- Schaefer, H.M. , Schaefer, V. and Levey, D.J. (2004) How plant‐animal interactions signal new insights in communication. Trends Ecol. Evol. 19, 577–584. [Google Scholar]
- Schwinn, K. , Venail, J. , Shang, Y.J. , Mackay, S. , Alm, V. , Butelli, E. , Oyama, R. et al (2006) A small family of MYB‐regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum . Plant Cell, 18, 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, S.L. , Yin, X.R. , Zhang, B. , Xie, X.L. , Jiang, Q. , Grierson, D. and Chen, K.S. (2016) CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)‐valencene in ‘Newhall’ orange. J. Exp. Bot. 67, 4105–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, K.H. , Lee, S.C. , Jung, H.W. , Hong, J.K. and Hwang, B.K. (2006) Expression and functional roles of the pepper pathogen‐induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 61, 897–915. [DOI] [PubMed] [Google Scholar]
- Steyn, W.J. , Wand, S.J. , Jacobs, G. , Rosecrance, R.C. and Roberts, S.C. (2009) Evidence for a photoprotective function of low‐temperature‐induced anthocyanin accumulation in apple and pear peel. Physiol. Plantarum, 136, 461–472. [DOI] [PubMed] [Google Scholar]
- Takos, A.M. , Jaffé, F. , Jacob, S.R. , Bogs, J. , Robinson, S.P. and Walker, A.R. (2006) Light‐induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, A.R. , Lee, E. , Bogs, J. , McDavid, D.A.J. , Thomas, M.R. and Robinson, S.P. (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zhang, H. , Yang, Y. , Li, M.F. , Zhang, Y.T. , Liu, J.S. , Dong, J. et al (2019a) The control of red color by a family of MYB transcription factors in octoploid strawberry (Fragaria × ananassa) fruits. Plant Biotechnology J. 10.1111/pbi.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.H. , Tang, W. , Hu, W. , Zhang, Y.B. , Sun, J.Q. , Guo, X.H. , Lu, H. et al (2019b) A MYB/bHLH complex regulates tissue‐specific anthocyanin biosynthesis in the inner pericarp of red‐centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 99, 359–378. [DOI] [PubMed] [Google Scholar]
- Woo, H.R. , Kim, J.H. , Kim, J.Y. , Kim, J. , Lee, U. , Song, I.J. , Kim, J.H. et al (2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis . J. Exp. Bot. 61, 3947–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrolstad, R.E. , Culbertson, J.D. , Cornwell, C.J. and Mattick, L.R. (1982) Detection of adulteration of in blackberry juice concentrations and wines. J. Assoc. Offic. Anal. Chem. 65, 1417–1423. [PubMed] [Google Scholar]
- Xie, X.L. , Yin, X.R. and Chen, K.S. (2016) Roles of APETALA2/ethylene‐response factors in regulation of fruit quality. Crit. Rev. Plant Sci. 35, 120–130. [Google Scholar]
- Xu, W.J. , Dubos, C. and Lepiniec, L. (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Yao, G.F. , Ming, M.L. , Allan, A.C. , Gu, C. , Li, L.T. , Wu, X. , Wang, R. et al (2017) Map‐based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–451. [DOI] [PubMed] [Google Scholar]
- Yin, X.R. , Shi, Y.N. , Min, T. , Luo, Z.R. , Yao, Y.C. , Xu, Q. , Ferguson, I.B. et al (2012) Expression of ethylene response genes during persimmon fruit astringency removal. Planta, 235, 895–906. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Xu, H.F. , Wang, N. , Jiang, S.H. , Fang, H.C. , Zhang, Z.Y. , Yang, G.X. et al (2018a) The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol. Biol. 98, 205–218. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.Y. , Yin, X.R. , Xiao, Y.W. , Zhang, Z.Y. , Li, S.J. , Liu, X.F. , Zhang, B. et al (2018b) An ETHYLENE RESPONSE FACTOR‐MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol. 178, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X.Y. , Qi, C.H. , Jiang, H. , You, C.X. , Guan, Q.M. , Ma, F.W. , Li, Y.Y. et al (2019) The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic. Res. 6, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Lin‐Wang, K. , Wang, H.L. , Gu, C. , Dare, A.P. , Espley, R.V. , He, H.P. et al (2015) Molecular genetics of blood‐fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. [DOI] [PubMed] [Google Scholar]
- Zhu, Q. , Zhang, J.T. , Gao, X.S. , Tong, J.H. , Xiao, L.T. , Li, W.B. and Zhang, H.X. (2010) The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene, 457, 1–12. [DOI] [PubMed] [Google Scholar]
- Zorrilla‐Fontanesi, Y. , Rambla, J.L. , Cabeza, A. , Medina, J.J. , Sánchez‐Sevilla, J.F. , Valpuesta, V. , Botella, M.A. et al (2012) Genetic analysis of strawberry fruit aroma and identification of O‐methyltransferase FaOMT as the locus controlling natural variation in mesifurane content. Plant Physiol. 159, 851–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic tree analysis of strawberry AP2/ERF proteins and related AP2/ERF TFs from apple and pear.
Figure S2 Phylogenetic analysis of FaRAV1 and 34 other RAV proteins.
Figure S3 Subcellular localization of FaRAV1. FaRAV1 was inserted into the pCAMBIA1300‐sGFP vector and transiently expressed in tobacco leaves.
Figure S4 The appearance of FaRAV1 overexpression fruit (right) 9 d after injection, compared to the control (left).
Figure S5 PEGG analysis of DEGs between FaRAV1 OE fruit and control fruit.
Figure S6 RT‐qPCR analysis of FaRAV1 expression in response to ABA treatment in strawberry fruit.
Figure S7 Yeast two‐hybrid analysis of the interactions between FaRAV1 and FaMYB10.
Figure S8 Cis regulatory elements in promoters of anthocyanin biosynthetic genes.
Figure S9 Relative expression of RAP in FaRAV1 OE and RNAi fruit compared with the control fruit.
Figure S10 Regulatory effect of FaRAV1 on the promoter of RAP.
Table S1 Primers used for reverse transcription quantitative PCR.
Table S2 Primers used for vector construction.
Table S3 Motifs in the FaRAV1 promoter identified in silico by PlantCARE.


