Abstract
Alcohol drinking is a leading risk factor for the development of esophageal squamous cell carcinoma (ESCC). However, the molecular mechanisms of alcohol-associated ESCC remain poorly understood. One of the most commonly mutated genes in ESCC is nuclear factor erythroid 2 like 2 (NFE2L2 or NRF2), which is a critical transcription factor regulating oxidative stress response and drug detoxification. When NRF2 is hyperactive in cancer cells, however, it leads to metabolic reprogramming, cell proliferation, chemoradioresistance, and poor prognosis. In this study, hyperactive NRF2 was found to upregulate acetyl-CoA synthetase short-chain family members 2 (ACSS2), an enzyme that converts acetate to acetyl-CoA, in ESCC cells and mouse esophagus. We also showed that knockdown of NRF2 or ACSS2 led to decreased ACSS2 expression, which in turn reduced the levels of acetyl-CoA and ATP with or without ethanol exposure. In addition, ethanol exposure enhanced lipid synthesis in ESCC cells. Moreover, we observed a change in the metabolic profile of ESCC cells exposed to ethanol as a result of their NRF2 or ACSS2 status. We further showed that ACSS2 contributed to the invasive capability of NRF2high ESCC cells exposed to ethanol. In conclusion, the NRF2/ACSS2 axis mediates the metabolic effect of alcohol drinking on ESCC.
Keywords: ACSS2, NRF2, Ethanol, Acetate, Esophageal squamous cell carcinoma
Introduction
Esophageal squamous cell carcinoma (ESCC) is the 6th most common cancer worldwide [1]. Globally, the highest incidences of ESCC are found in the African esophageal cancer corridor (Eastern and Southern Africa) and the Asian esophageal cancer belt (East and Central Asia) [1, 2]. Alcohol drinking is an established risk factor for ESCC [3–6]. The risk of ESCC development is strongly associated with an individual’s daily average alcohol consumption, thus, individuals who drink ≥ 70g/day have the highest risk of developing ESCC [7–10]. Current molecular mechanisms of alcohol-associated ESCC focus primarily on the effects of acetaldehyde. Ethanol is metabolized to acetaldehyde by alcohol dehydrogenase, cytochrome P450 2E1, and, to a much lesser extent by catalase, and is further oxidized from acetaldehyde to acetate by acetaldehyde dehydrogenase 2 [11]. Acetaldehyde is a highly reactive compound that causes various damages to DNA [12–16] and proteins [17, 18].
Recently, studies on human ESCC samples from China, Japan and Malawi with next-generation sequencing have identified driver mutations of ESCC [19–22]. One of the most common mutated genes is nuclear factor (erythroid-derived 2)-like 2 (NFE2L2 or NRF2), which is a transcription factor regulating oxidative stress response and drug detoxification genes [23–28]. Under normal conditions, NRF2 is bound to Kelch-like ECH-associated protein 1 (KEAP1) dimers, for ubiquitinylation by the CUL3 E3 ubiquitin-protein ligase, and subsequent proteasomal degradation. In normal esophagus, NRF2 interacts with the Wnt pathway, Pparβ/δ, and PI3K/Akt pathway, all of which are involved in the development of esophageal epithelium [29]. Moreover, NRF2 is important in normal squamous differentiation of basal progenitor cells [30], and maintaining esophageal barrier function through its regulation of energy-dependent tight junction [31]. As a result, NRF2 is involved in the pathogenesis of eosinophilic esophagitis and gastroesophageal reflux disease [30, 31].
Somatic mutations of NRF2, KEAP1, or CUL3 result in the constitutive transcription of NRF2 cytoprotective genes in cancer cells, which leads to increased metabolism, proliferation, and chemoradioresistance in these cells [32, 33]. NRF2 is mutated in ~10% of ESCC cases, while genes encoding its regulators CUL3 and KEAP1 are mutated in ~3% and 4% of ESCC cases, respectively [34]. KEAP1 and CUL3 mutations may be found all over the length of the protein. NRF2 mutations are mostly located in the KEAP1 binding domain at the N-terminus of the NRF2 protein, and therefore decrease the binding affinity of KEAP1 and subsequent degradation of NRF2 [35–37]. More recently, it has been reported that ESCC patients with high nuclear NRF2 expression have significantly poorer prognosis [38]. Through NRF2 ChIP-seq of mouse esophageal samples, we previously showed that hyperactive NRF2 bound to the promoter regions of many metabolic genes, one of which was acetyl-CoA synthetase short-chain family member 2 (Acss2). Moreover, microarray analysis of differential gene expression in Keap1−/− esophagus in comparison with Nrf2−/−;Keap1−/− esophagus identified Acss2 as one of the genes upregulated due to NRF2 hyperactivation [39].
ACSS2 belongs to a family of acetyl-CoA synthetase short-chain enzymes involved in metabolizing acetate to acetyl-CoA [40–42]. ACSS1 and ACSS3 are located in the mitochondria, while ACSS2 is cytosolic and nuclear [42–46]. ACSS2 is critical for tumor metabolism in hypoxic and glucose-limited environments as cancer cells utilize acetate as a carbon source, leading to a metabolic switch from aerobic glycolysis to oxidative phosphorylation (OXPHOS) [40, 41, 45, 47]. ACSS2 controls acetate’s contribution to fatty acid synthesis and supports the biosynthesis of membrane phospholipids in breast cancer [47]. It helps cancer cells survive in a hypoxic environment through lipogenesis (45). It also promotes the transcription of lipid synthesis and cell proliferation genes in breast cancer and hepatocellular carcinoma cells [40, 48, 49].
In this study, we showed that NRF2 regulated ACSS2 expression in esophageal squamous epithelial cells in vitro and in vivo. The NRF2/ACSS2 axis and ethanol exposure promoted OXPHOS and lipid synthesis, led to metabolic reprogramming, and enhanced invasion, in ESCC cells.
Materials and Methods
Cell culture
ESCC cell lines were selected based on NRF2 expression. KYSE410 cells carry wild-type NRF2 and express a low level of NRF2, thus are defined as NRF2low. KYSE70 cells carry a homozygous point mutation (NRF2W24C) and express a high level of NRF2, and thus are defined as NRF2high [19, 50]. When NRF2 was knockdown by siRNA in KYSE70 cells, these cells were defined as NRF2low-KYSE70 cells. When KEAP1 was knockdown by siRNA in KYSE410 cells, these cells were defined as NRF2high-KYSE410 cells.
RPMI 1640 Glutamax media (Gibco, Gaithersburg, MD) supplemented with 10% FBS and 0.1% penicillin/streptomycin was used to culture cells under normal conditions. For cell-based assays where starvation media was used, cells were either cultured in nutrient-free DMEM media (Gibco) for 4 h or RPMI 1640 without glucose (Gibco) supplemented with 10% dFBS, 5mM glucose and 300 μM acetate for assays that run for 24 or 72 h. In these long-term ethanol exposure studies, 5mM glucose rather than 10 mM glucose was used as heavy alcohol drinkers have been shown to consume less dietary glucose, and absorb less glucose from dietary sources [51–54]. After a dose-response experiment with ethanol, 50 mM ethanol was chosen for subsequent experiments that required ethanol exposure.
siRNA transfection
siRNA transfection was done using Lipofectamine RNAiMax (Invitrogen, Waltham, MA), Optimem limited serum media (Gibco), ACSS2 siRNA (AM16708, ID177990, Invitrogen), NFE2L2 siRNA (4392421, IDs9491, Invitrogen), or KEAP1 siRNA (4392420, IDs18982, Invitrogen). Transfection was conducted according to the manufacturer’s protocol. Gene knockdown was achieved 48 to 72 h after transfection.
CRISPR Cas9 knockdown
CRISPR Cas9 knockdown was done by Synthego (Redwood City, CA). The sequence targeted was 482 bp from the UTR on exon 2 of NRF2. The guide RNA sequence used was UAUUUGACUUCAGUCAGCGA. The guide target was found near a CGG PAM sequence. Results were analyzed through an ICE analysis tool. CRISPR generated a mixed population of cells (Fig. S3). Western blotting confirmed the knockdown result (Fig. 1C, D).
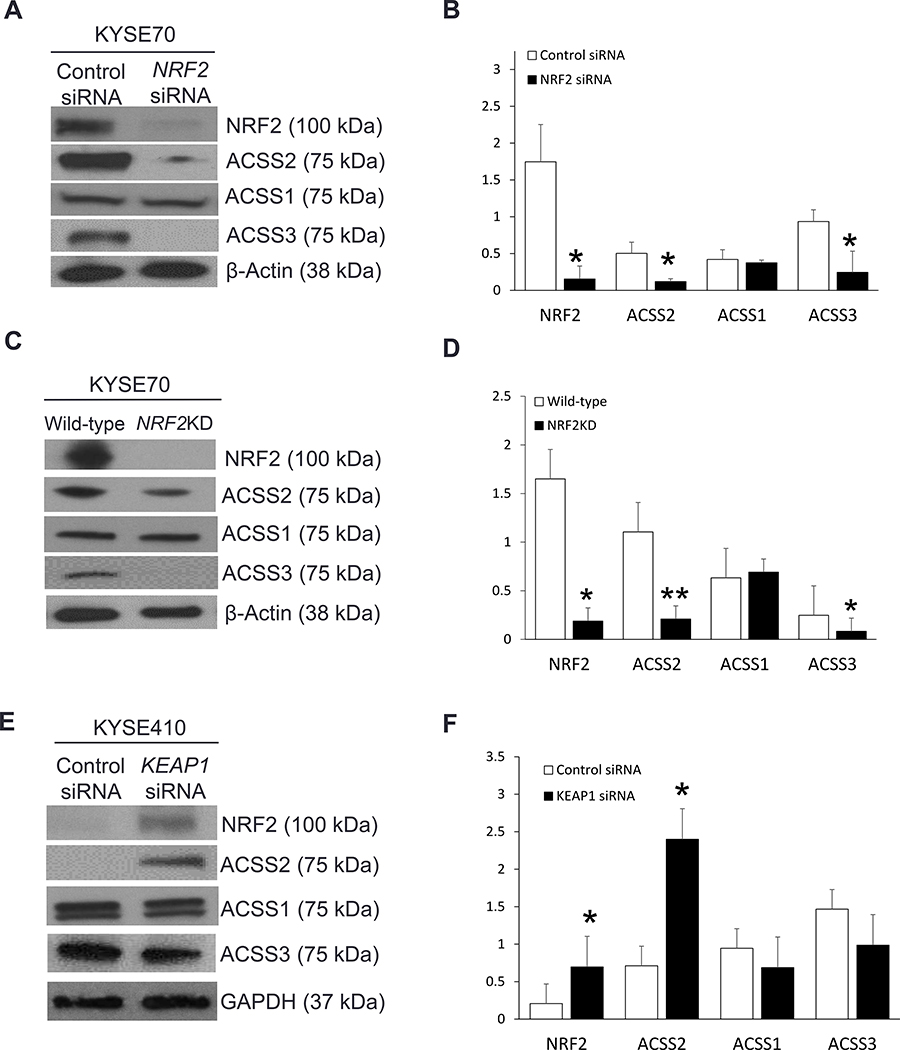
Figure 1. NRF2 regulates the expression of ACSS2 in ESCC cells.
(A, B) NRF2KD in KYSE70 cells through NRF2 siRNA transfection led to a significant decrease in ACSS2 and ACSS3. (C, D) NRF2KD in KYSE70 cells through CRISPR-Cas9 also led to a significant decrease in ACSS2 and ACSS3. (E, F) A significant increase in NRF2 and ACSS2 expression was observed in NRF2high KYSE410 cells due to KEAP1 siRNA transfection as compared to control.
Western blotting
Total protein was isolated from human ESCC cells and mouse tissues using a standard method. Antibodies to ACSS1 and ACSS3 were purchased from Proteintech (Rosemont, IL) and were used at a concentration of 1:3,000 and 1:600, respectively. ACSS2 antibody (Cell Signaling, Danvers, MA) was used at a concentration of 1:3,000. NRF2 antibody (Abcam, Cambridge, MA) was used at a 1:2,000 concentration, while GAPDH (Genetex, Irvine, CA) and β-actin (Cell Signaling) were used as loading controls at 1:10,000 and 1:8,000, respectively. All antibodies were diluted using SuperBlock T20 blocking buffer (Thermofisher Scientific, Waltham, MA). Data was based on biologic triplicates.
Cell-based assays
Acetate was measured in KYSE70 and KYSE410 cell lysates using acetate colorimetric assay kit (ab204719, Abcam). Cells were seeded at a 2.0 × 106 density in three 75 cm2 flasks per group, after which siRNA transfection was conducted 24 h after seeding. Cells were cultured in a normal medium.
An acetyl-CoA fluorometric assay kit (ab87546, Abcam) and an ATP assay kit (MAK190, Sigma-Aldrich, St. Louis, MO) were used for acetyl-CoA and ATP measurements. Cells were either cultured in nutrient-free DMEM media (Gibco) for 4 h (Fig. 4C–H) or with RPMI 1640 with no glucose (Gibco) supplemented with 10% dFBS, 5 mM glucose, and 300 μM acetate with or without 50 mM ethanol for 72 h, with media and ethanol replaced every 24 h (Fig. 4I, L). 1-(2,3-di(Thiophen-2-yl)quinoxalin-6-yl)-3-(2-methoxyethyl)urea, an ACSS2 inhibitor (ACSS2i), was purchased from Millipore-Sigma (Burlington, MA).
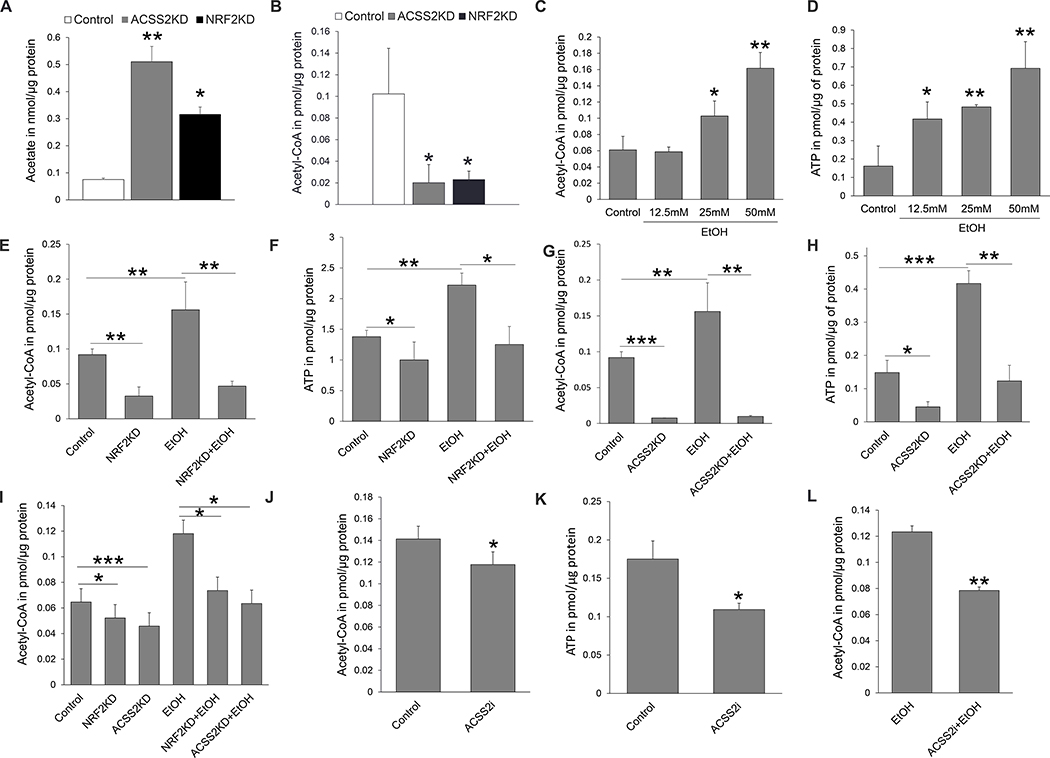
Figure 4. NRF2/ACSS2 axis regulates acetate metabolism in NRF2high KYSE70 ESCC cells.
. (A) Acetate metabolism was significantly inhibited when ACCS2 or NRF2 was knocked down. (B) Acetyl-CoA was significantly decreased when ACSS2 or NRF2 was knocked down. (C, D) Ethanol exposure significantly increased acetyl-CoA and ATP in a dose-dependent manner. Based on these results, 50mM ethanol was used to treat cells in the subsequent experiments. (E, F) 50mM ethanol exposure significantly increased acetyl-CoA and ATP, whereas NRF2 knockdown counteracted such effects. (G, H) ACSS2 knockdown inhibited acetyl-CoA and ATP. (I) Knockdown of NRF2 or ACSS2 led to a significant decrease in acetyl-CoA in ESCC cells in the presence or absence of ethanol exposure. (J, K) ESCC cells treated with an ACSS2 inhibitor had significantly decreased acetyl-CoA and ATP as compared to control. (L) ACSS2 inhibitor significantly decreased cellular acetyl-CoA when cells were exposed to ethanol over 72 h. *P <0.05, **P <0.01, ***P <0.001.
To determine the effects of ACSS2 and ethanol on fatty acid synthesis in KYSE70 cells, cells were first transfected with siRNA in 75 cm2 flasks under normal cell culture conditions for 72 h. Afterwards, these transfected cells were seeded in 100 mm plates at a concentration of 6.0×106 cells per plate. Cells were cultured in RPMI 1640 media with no glucose (Gibco) supplemented with 10% dFBS, 5 mM glucose and 300 μM acetate with or without 50 mM ethanol for 24 h. At the end of the 24 h period, cells were collected using trypsin and washed with cold 1X PBS before running the assay. Fatty acid synthesis was measured using a free fatty acid fluorometric assay kit (ab65341, Abcam). Data was based on biologic triplicates to ensure reproducibility.
Immunohistochemical staining (IHC)
Deparaffinized sections were pre-treated to retrieve antigens before detection with a rabbit polyclonal anti-NRF2 (Invitrogen) at a 1:200 dilution, and a rabbit polyclonal anti-ACSS2 (Sigma-Aldrich) at a 1:25 dilution. Cytokeratin 5 immunohistochemistry with anti-CK5 (Invitrogen) was used to validate the antigenicity of human tissue sections.
Human ESCC formalin-fixed paraffin-embedded tissue sections were decoded without patient information. IHC staining was scored based on a positive staining area and staining intensity. Numeric scores were provided for staining area and staining intensity: 3 for high intensity or a high percentage of positive staining (>75% cancer cells); 2 for moderate staining or moderate percentage (50–75%); 1 for low staining intensity or low percentage of positive staining (25–50%); and 0 for no staining. A combined numerical score for each antigen was generated by adding the numerical score of the percentage of positive staining to that of the staining intensity.
NRF2 ChIP-PCR assay
These assays were performed using the EZ-ChIP PCR kit (17–371, Millipore-Sigma) as previously described [31]. Immunoprecipitations were performed using control mouse IgG or 2 μg/ml rabbit anti-NRF2 polyclonal antibody (Santa-Cruz Biotech, Santa Cruz, CA). The NRF2 binding region - antioxidant response element (ARE) in mouse Acss2 gene was identified from our previous ChIP-seq data [39]. The PCR control used was a Gapdh primer provided with the kit. The Nqo1 primer sequences were based on a previous publication and served as a positive control [55]. The PCR primer sequences used were 5’-TACACCCTCACCAGCACATT-3’, and 5’-TTTCTGCTGGATGTGGTGG-3’. The predicted sizes of PCR products of Gapdh, Nqo1, and Acss2 were 166 bp, 186 bp, and 595 bp, respectively.
Seahorse cell mito-stress assay
The Seahorse cell mito-stress assay was used to characterize the effect of NRF2, ACSS2, and ethanol on cell energy phenotype and oxidative phosphorylation (OXPHOS) in ESCC. KYSE70 cells were seeded at a density of 2 × 104 cells/well, and KYSE410 cells were seeded at a density of 2.5 × 104 cells/well in XFp plates and allowed to stabilize overnight. XF media was supplemented with 5mM D-glucose and 2 mM L-glutamine as described in the manufacturer’s instructions for the XF Cell Mito-Stress assay with or without 20mM ethanol. Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were next measured using a Seahorse XFp analyzer (Agilent Inc., Santa Clara, CA). Cells were then treated with or without ethanol for 1.5 hours. For the cells treated with ethanol, ethanol was added not only to the media but also to the surrounding moat. Time 0 in the Seahorse graphs (Fig. 5) indicated the beginning of Seahorse measurement (i.e., once the plates were placed in the instrument). For KYSE70 cells, 1 μM oligomycin, 0.5 μM FCCP, and 0.5 μM rotenone were simultaneously applied to measure the glycolysis and OXPHOS in the cells. Injections of 1.5 μM oligomycin, 3.0 μM FCCP, and 0.5 μM rotenone were simultaneously applied to measure the same energy-producing pathways in KYSE410 cells. Injections were determined by cell characterization methods stated in the manufacturer’s instructions for the XF Cell Mito-Stress assay. Incucyte® (Sartorius, Göttingen, Germany) was used to determine cell number in each well. The results were normalized to cell number and analyzed using Wave software (Agilent Inc.). Data was based on biological triplicates to ensure reproducibility.
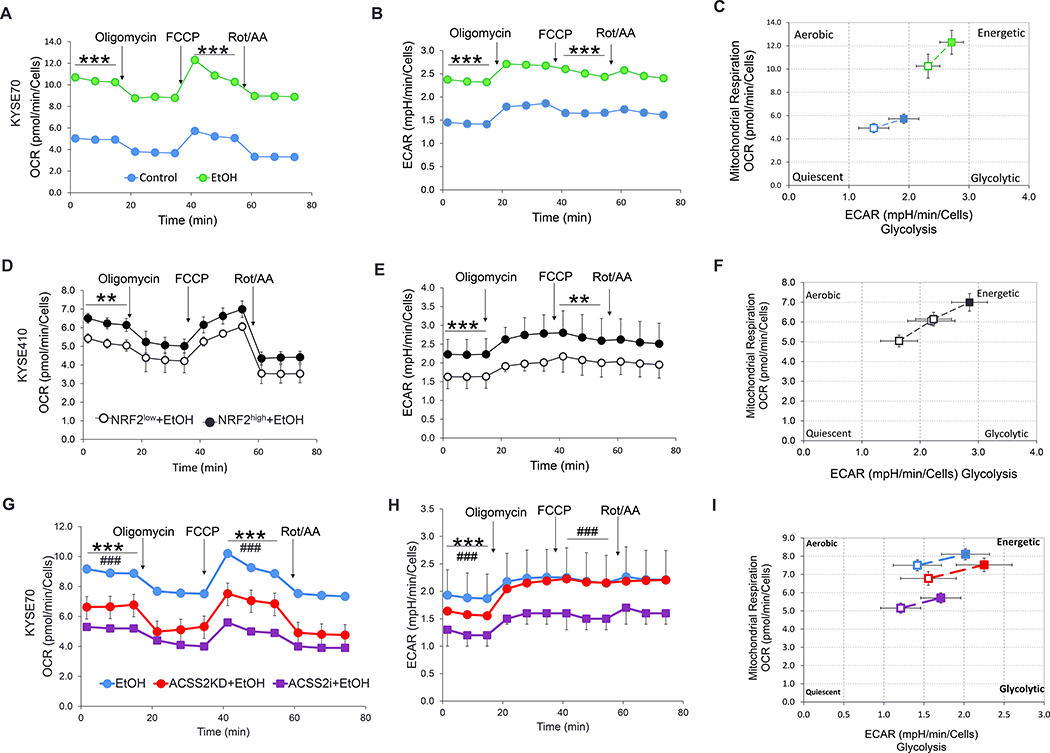
Figure 5. ACSS2 and ethanol induce a metabolic shift in NRF2high ESCC cells.
(A, B) A significant increase in OCR and ECAR were observed when KYSE70 cells were exposed to ethanol. (C) KYSE70 cells became more energetic when exposed to ethanol. (D, E) Increased OCR and ECAR in NRF2high- KYSE410 cells were observed after ethanol exposure as compared to control NRF2low-KYSE410 cells. (F) Hyperactive NRF2 led to a more energetic phenotype in KYSE410 cells. (G, H) Downregulation or inhibition of ACSS2 decreased OCR and ECAR in KYSE70 cells. The statistical differences between wild-type and ACSS2KD were indicated by asterisks (*), while those between control and ACSS2i treatment was indicated by hashtags (#). (I) ACSS2 inhibition led to a less energetic phenotype in KYSE70 cells. *P <0.05, **P <0.01, ***P <0.001. ###P <0.001.
Cell invasion assay
The assay was done according to the Corning cell invasion assay protocol. Control, ACSS2KD, NRF2KD, and ACSS2i treated KYSE70 cells were loaded at a density of 2 × 104 cells/insert in growth factor reduced Matrigel invasion chamber cell culture inserts (8 μm pore size; Corning; Corning, NY). Cells were then incubated for 36 h with either control or 50 mM ethanol containing serum‐free medium in the upper chamber. The bottom chamber was filled with 750 μl of 10% FBS‐containing RPMI medium. Cells at the bottom of the plate were stained with 1 μg/ml Hoechst, and analysis was done using Pico ImageXpress imaging (Molecular Devices, San Jose, CA). Data was based on biological triplicates to ensure reproducibility.
Animal experiments
Animal experiments were approved by the Institutional Animal Care and Use Committees at the North Carolina Central University (protocol number XC-12–03-2008). Animals in these experiments were bred and maintained in our animal facility. Wild-type, Nrf2−/− and Keap1−/− esophageal tissues were obtained as previously reported [39]. Sox2CreER mice were obtained from the Jackson Laboratory (Bar Harbor, ME), and LSL-Nrf2E79Q/+ mice were obtained from Dr. Bernard Weissman’s lab at the University of North Carolina at Chapel Hill, Chapel Hill, NC [56]. Cre recombinase was activated by intraperitoneal injection of tamoxifen (75 mg/kg in corn oil, once per day, 5 days). Mice were then sacrificed by CO2 asphyxiation at 5 weeks after tamoxifen induction. No anesthetics were used for mice.
Statistical analyses
Student’s t-test was used to analyze Western blot, cell-based assay, Seahorse, and cell-invasion results from biologic triplicates. A Pearson correlation test was used to analyze the relationship between NRF2 and ACSS2 expression in human samples.
Results
NRF2 regulates expression of ACSS2 in vitro
To determine whether NRF2 regulates ACSS2 in vitro, we selected two human ESCC cell lines, NRF2high-KYSE70 and NRF2low-KYSE410, based on the level of NRF2 expression [22, 50, 57]. When KYSE70 cells were transfected with NRF2 siRNA, there was a significant decrease in the expression of NRF2, ACSS2, and ACSS3 (Fig. 1A). A similar result was observed when NRF2 was knocked down using CRISPR-Cas9 in KYSE70 cells (Fig. 1B). On the contrary, KEAP1 siRNA led to an increase in NRF2 and ACSS2 expression in KYSE410 cells (Fig. 1C). These results indicate that NRF2 regulates ACSS2 protein expression in vitro.
NRF2 regulates ACSS2 expression and binds to the promoter of Acss2 in vivo
We next determined whether NRF2 regulated ACSS2 expression in vivo. Using immunohistochemistry (IHC), we examined the expression of NRF2 and ACSS2 in esophageal squamous epithelial cells of Nrf2−/−, wild-type, and Keap1−/− mice. As expected, when NRF2 was constitutively hyperactive in Keap1−/− esophagus, ACSS2 expression was upregulated as compared to wild-type and Nrf2−/− esophagi (Fig. 2A). Similar results were also observed in the lung, liver, and kidney tissues (Fig. S1). We also used Sox2CreER;LSL-Nrf2E79Q/+ mice to determine whether induced hyperactivation of NRF2 in the adult esophagus may upregulate ACSS2 expression. Sox2CreER;LSL-Nrf2E79Q/+mouse expressed a high level of NRF2 in the esophagus due to the expression of a mutant allele (E79Q) after tamoxifen induction. Indeed, ACSS2 was overexpressed in the Sox2CreER;LSL-Nrf2E79Q/+ esophagus (Fig. 2B).
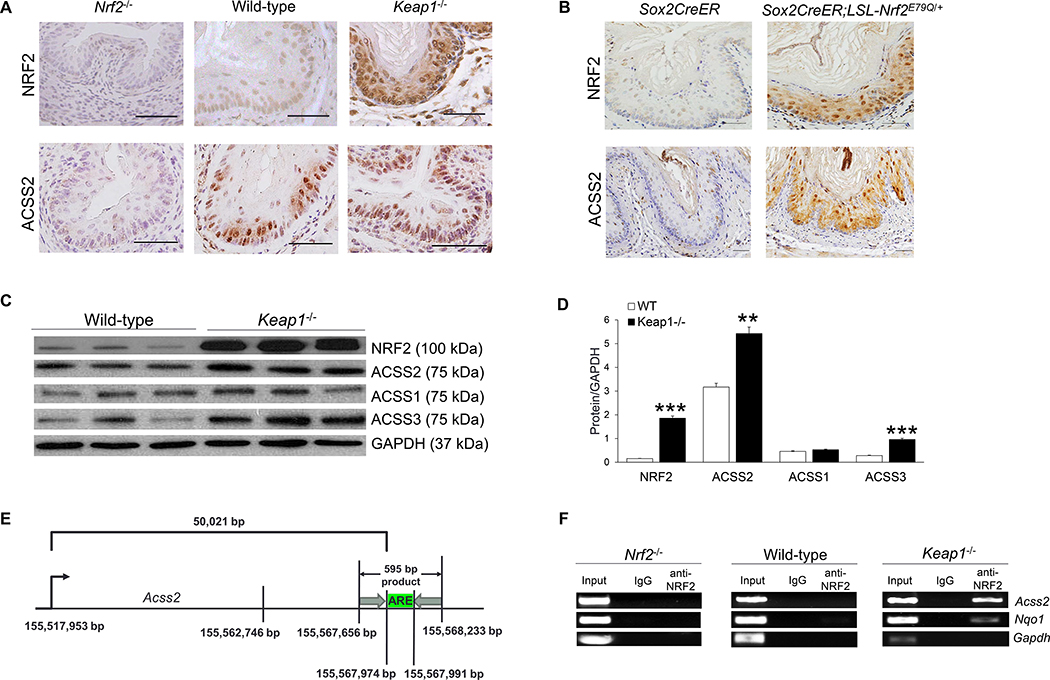
Figure 2. NRF2 regulates ACSS2 expression and binds to the promoter of Acss2 in vivo.
(A) Immunohistochemistry of mouse esophagus showed that NRF2 and ACSS2 expression increased when NRF2 expression was activated (Keap1−/−) and decreased when NRF2 was deficient (Nrf2−/−); scale bar = 50 μm. (B) Immunohistochemistry also showed ACSS2 upregulation due to NRF2 hyperactivation in the esophagus that expressed a constitutively active mutant Nrf2 (Sox2CreER;LSL-Nrf2E79Q+) as compared to control (Sox2CreER); scale bar = 50 μm. (C) Western blot of wild-type and Keap1−/− mouse esophagi showed increased NRF2 expression and overexpression of ACSS2 and ACSS3. (D) Statistical analysis of the Western blot revealed significant increases in ACSS2 and ACSS3 expression in Keap1−/− mouse esophagi. (E) Schematic diagram showing ARE region in mouse Acss2 gene, and the ChIP-PCR primer target (595 bp). (F) NRF2 ChIP-PCR of wild-type, Keap1−/− and Nrf2−/− mouse esophagi identified Acss2 as a target gene of hyperactive NRF2. *P <0.05, **P <0.01, ***P <0.001.
Using Western blotting, we confirmed ACSS2 and ACSS3 upregulation in Keap1−/− esophagi as compared to wild-type esophagi (Fig. 2C, D). Our previous NRF2 ChIP-seq data using mouse esophagus indicated that hyperactive NRF2 bound to mouse Acss2 gene [39]. To further validate this observation, we performed NRF2 ChIP-PCR using mouse esophageal tissues. Consistent with our ChIP-seq data, NRF2 ChIP-PCR revealed NRF2 binding to mouse Acss2 gene in Keap1−/− esophagus, but not in Nrf2−/− and wild-type esophagi (Fig. 2E, F). The regulation of ACSS2 expression by NRF2 was further supported through IHC of human ESCC samples. A significantly positive correlation between NRF2 and ACSS2 expression was observed in our human samples (Fig. 3). These results using the mouse and human tissue samples indicate that NRF2 regulates ACSS2 expression in vivo.
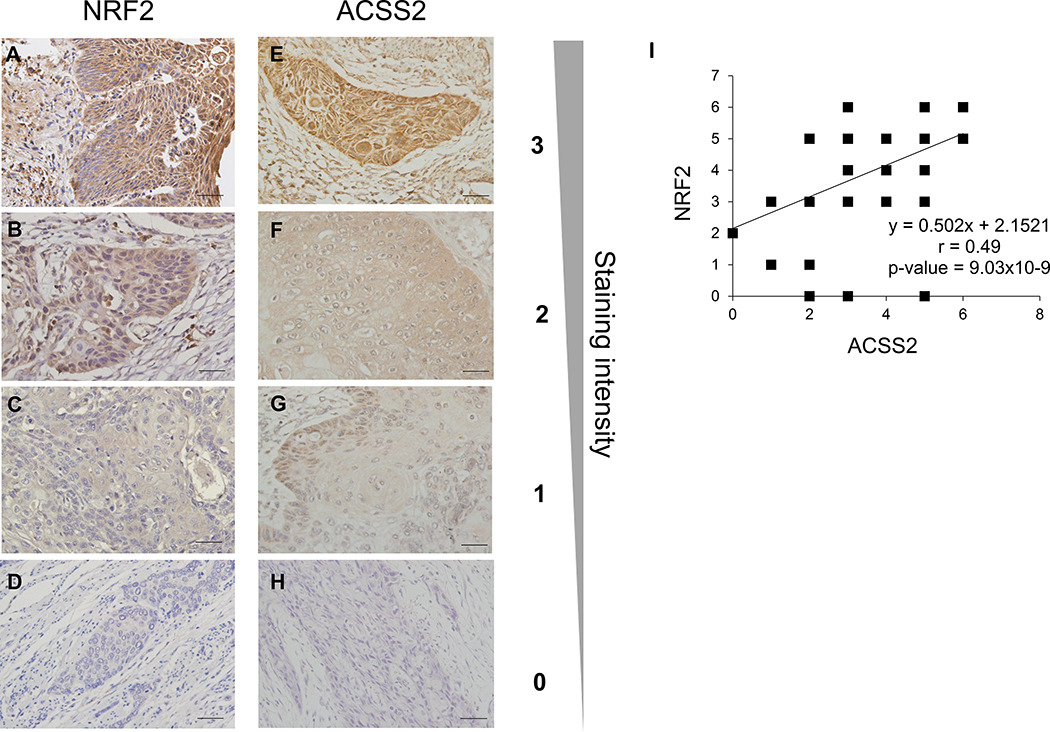
Figure 3. NRF2 and ACSS2 expression are positively correlated in human ESCC tissue samples.
. ESCC samples were categorized based on the staining intensity (Scale bar = 50 μm). Images in panel A-H came from different patients. (A) high NRF2 staining intensity; (B) moderate NRF2 staining intensity; (C) low NRF2 staining intensity; (D) no NRF2 staining; (E) high ACSS2 staining intensity; (F) moderate ACSS22 staining intensity; (G) low ACSS2 staining intensity; (H) no ACSS2 staining. (I) IHC of human samples revealed a significantly positive correlation between NRF2 and ACSS2 expression in these samples.
NRF2/ACSS2 axis mediates ethanol metabolism in ESCC cells
To gain insight into the relationship between NRF2, ACSS2, and ethanol metabolism in ESCC, we first investigated the roles of NRF2 and ACSS2 in acetate metabolism. We measured cellular acetate and acetyl-CoA in KYSE70 cells transfected with ACSS2 or NRF2 siRNA, and KYSE410 cells transfected with KEAP1 siRNA (Fig. S2A, B). We did not further investigate ACSS3 because it has a low affinity for acetate but a higher affinity for propionate. Hence, ACSS3 is not a major enzyme in the acetate metabolism pathway [58]. As observed in Fig. 4, ACSS2KD led to a significant increase in acetate and a significant decrease in acetyl-CoA. Similar findings were also observed in NRF2KD KYSE70 cells (Fig. 4A, B). When NRF2 was upregulated in KYSE410 cells, cellular acetate decreased, while acetyl-CoA production increased (Fig. S2C, D). These results indicate that NRF2 and ACSS2 regulate acetate metabolism in ESCC cells.
We then examined the role of the NRF2/ACSS2 axis in ethanol metabolism. We first determined the dose-dependent effects of ethanol on acetyl-CoA and ATP in KYSE70 cells (Fig. 4C, D). We observed that ethanol increased acetyl-CoA and ATP production in a dose-dependent manner (Fig. 4C, D). When KYSE70 cells were transfected with either NRF2 or ACSS2 siRNA, significant decreases in acetyl-CoA and ATP were observed in the presence or absence of ethanol exposure (Fig. 4E–H). A significant decrease in acetyl-CoA could still be observed after 72 h (Fig. 4I). Similarly, when ACSS2 was inhibited with a chemical inhibitor in KYSE70 cells, significant decreases in acetyl-CoA and ATP were also observed (Fig. 4J–L). On the contrary, when KYSE410 cells were transfected with KEAP1 siRNA and exposed to ethanol over 72 h, there was a significant increase in acetyl-CoA compared to control KYSE410 cells (Fig. S2E). These results demonstrate that ethanol is used as an energy source in NRF2high ESCC cells and that the NRF2/ACSS2 axis contributes to ethanol metabolism in ESCC cells.
Ethanol exposure causes a metabolic shift in NRF2high ESCC cells
Our previous report showed that hyperactive NRF2 caused metabolic reprogramming in the esophagus [39]. Together with our current findings, we hypothesized that ethanol exposure promoted OXPHOS in NRF2high ESCC cells. To test this hypothesis, we ran a Seahorse Mito-stress assay on KYSE70 cells exposed to ethanol. As shown in Fig. 5A, KYSE70 cells exposed to ethanol upregulated OXPHOS at both the baseline and stressed levels as compared to control. Extracellular acidification rate (ECAR), a measure of glycolysis, was also upregulated during ethanol exposure (Fig. 5B). Additionally, ethanol exposure of these NRF2high cells made the cells adopt a more energetic phenotype (Fig. 5C).
We then determined whether the NRF2/ACSS2 axis played a role in this metabolic shift. We first tested the impact of NRF2 on OXPHOS in NRF2high and NRF2low ESCC cells exposed to ethanol. As shown in Fig. 5D, NRF2 hyperactivation led to a significant increase in baseline OXPHOS in KYSE410 cells. A much greater change in OXPHOS was observed in KYSE70 cells when NRF2 was knocked down using a CRISPR-Cas9 approach (Fig. S4A). NRF2KD in KYSE70 cells led to a significant decrease in OXPHOS at both the baseline and stressed levels. NRF2 hyperactivation led to increased ECAR, while its downregulation led to decreased ECAR (Fig. 5E, Fig. S4B). These results were in agreement with our previous work, in which hyperactive NRF2 upregulated glycolysis in the mouse esophagus [39].
NRF2 hyperactivation also produced a more energetic phenotype in KYSE410 cells (Fig. 5F). Conversely, downregulation of NRF2 in KYSE70 cells decreased their mitochondrial and glycolytic activity (Fig. S4C). We also investigated the role of ACSS2 on OXPHOS and ECAR in ESCC cells exposed to ethanol. ACSS2KD or ACCS2 inhibition led to a significant decrease in mitochondrial respiration in KYSE70 cells (Fig. 5G). Baseline ECAR was significantly decreased when ACSS2 was either knocked down or when the ACSS2 enzyme was inhibited (Fig. 5G). Finally, metabolic phenotype analysis showed that ACSS2KD or ACSS2 inhibition led to a decrease of mitochondrial respiration (Fig. 5I). These results show that ACSS2 and ethanol upregulate OXPHOS in NRF2high ESCC
Ethanol exposure promotes lipid synthesis in NRF2high ESCC cells
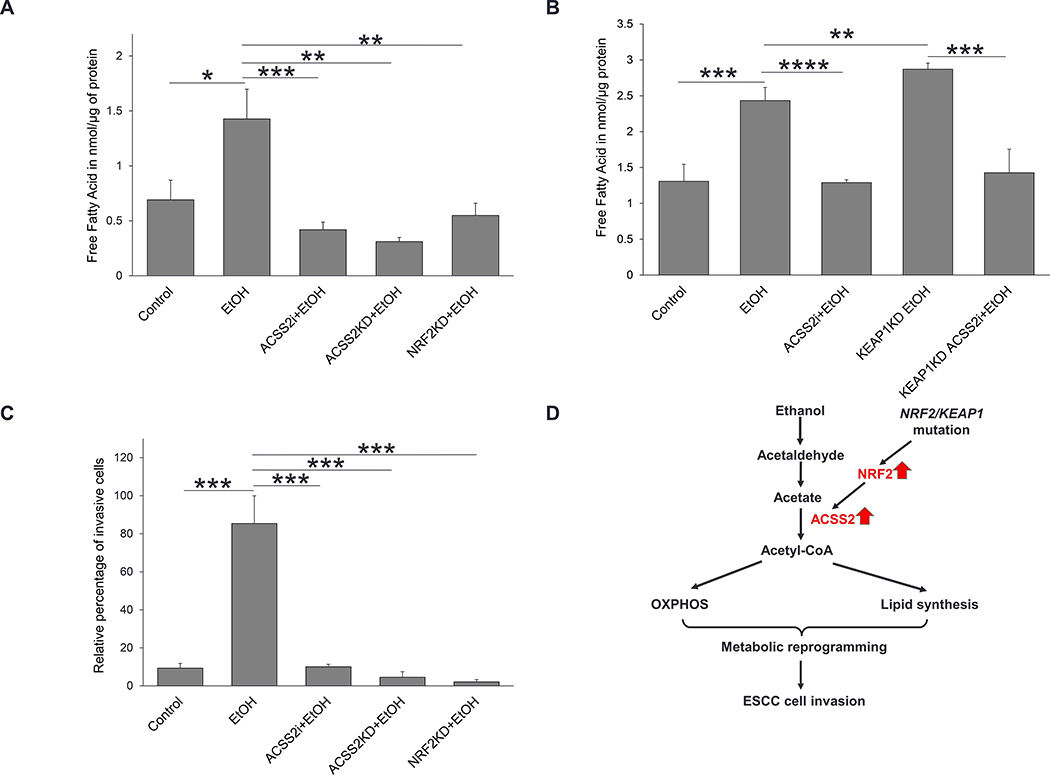
Acetate metabolism via ACSS2 has been shown to promote lipid synthesis in different cancers [40, 47]. We hypothesized that ESCC cells would increase lipid synthesis when exposed to ethanol and that this process would be dependent on the NRF2/ACSS2 axis. We exposed KYSE70 and KYSE410 cells to 50 mM ethanol over 24 h, and then measured the amount of cellular free fatty acid. As shown in Fig. 6A and B, there was an increase in free fatty acids when ESCC cells were exposed to ethanol. The increase of free fatty acid was significantly decreased when either ACSS2 or NRF2 was knocked down (Fig. 6A), or when ACSS2 was chemically inhibited (Fig. 6A, B). However, NRF2 hyperactivation led to significantly increased lipid synthesis after ethanol exposure (Fig. 6B). This increase in lipid synthesis, however, was significantly suppressed when ACSS2 was chemically inhibited in KEAP1KD KYSE410 cells (Fig. 6B). These data suggest that ethanol can be used for lipid synthesis via the NRF2/ACSS2 axis in ESCC cells.
Figure 6. NRF2/ACSS2 axis facilitates lipid biosynthesis and cell invasion in ESCC cells exposed to ethanol.
(A) Ethanol exposure increased free fatty acid in KYSE70 cells. Lipid synthesis decreased in KYSE70 cells when NRF2 or ACCS2 was knocked down or when ACSS2 was inhibited. (B Ethanol exposure increased lipid synthesis in KYSE410 cells, ACSS2 inhibition during ethanol exposure led to a significant decrease in lipid synthesis. Upregulation of NRF2 during ethanol exposure led to a significant increase in lipid synthesis compared to control KYSE410 cells exposed to ethanol. Inhibition of ACSS2, however, after NRF2 upregulation led to a significant decrease in lipid synthesis. (C) Ethanol exposure led to an increase in the KYSE70 cell invasion. NRF2KD, ACSS2KD, and ACSS2i led to a significant decrease in cell invasion. (D) A schematic showing that the NRF2/ACSS2 axis mediates metabolic reprogramming and promotes invasion of ESCC cells when exposed to ethanol. NRF2/KEAP1 mutations result in NRF2 hyperactivation, which further upregulates ACSS2. ACSS2 overexpression enhances acetyl-CoA turnover, which is used for lipid synthesis and energy production through OXPHOS. These metabolic changes support ESCC cell invasion. *P <0.05, **P <0.01, ***P <0.001.
Ethanol exposure facilitates invasion of NRF2high ESCC cells
Previous reports have indicated that ethanol exposure increased cancer cell invasion [59–62]. Acetate metabolism by ACSS2 has been implicated in vascular invasion and metastasis of glioblastoma [61]. We hypothesized that ethanol-induced metabolic reprogramming mediated by the NRF2/ACSS2 axis in ESCC cells would impact cell invasion. As shown in Fig. 6C, NRF2high cells exposed to ethanol had the highest percentage of invasive cells compared to control. However, cell invasion significantly decreased in ethanol-exposed cells when ACSS2 was chemically inhibited, or when NRF2 or ACSS2 was knocked down by siRNA. These results suggest that ethanol exposure facilitates invasion of NRF2high ESCC cells through the NRF2/ACSS2 axis.
Discussion
In this study, we show a novel mechanism of alcohol-associated ESCC. Previous studies on alcohol-associated cancer have mainly focused on acetaldehyde due to its reactive nature and its ability to induce genetic mutations as well as protein aberrations [17, 18, 63–65]. Our data indicate that the NRF2/ACSS2 axis mediates the metabolic effects of ethanol exposure, i.e., increased OXPHOS and lipid synthesis, on NRF2high ESCC. This discovery is consistent with previous studies showing that acetate supports cell proliferation, invasion, and metastases in other cancers [40, 41, 47, 66].
Inside epithelial cells, ethanol is metabolized to acetaldehyde by alcohol dehydrogenase (ADH), cytochrome P450 2E1 and, to a much lesser extent by catalase, and is further oxidized to acetate by acetaldehyde dehydrogenase. ADH-mediated ethanol metabolism results in the generation of reducing equivalents in the form of reduced nicotinamide adenine dinucleotide and acetaldehyde, whereas ethanol oxidation by CYP2E1 leads to the production of acetaldehyde, but also the generation of reactive oxygen species [67]. Systemic metabolism of ethanol causes the reduction of retinoid, zinc and methyl groups, and the accumulation of iron. Through the bloodstream, ethanol is circulated to the esophageal epithelium and salivary glands. In the saliva, ethanol is oxidized by microbes to acetaldehyde. When ethanol and acetaldehyde contact esophageal epithelial cells, ethanol perturbs the lipid bilayer of the cell membrane and interferes with the function of intrinsic membrane proteins, such as TLR4, Notch, Shh, and Wnt pathway receptors, and thus activates or inhibits downstream signaling. Inside epithelial cells, ethanol metabolism causes oxidative damages to DNA, proteins, and lipids, and modulates fatty acid metabolism as well. These systemic and local effects of ethanol and acetaldehyde stimulate cell proliferation, inflammation, and angiogenesis, and suppress squamous cell differentiation, and therefore promote ESCC [11]. Our previous study demonstrated PAX9 inhibition as a mechanism of alcohol-associated ESCC [68].
Our current study is the first to directly link alcohol drinking, acetate metabolism, and ESCC. Ethanol metabolism has also been shown to generate oxidative stress and thus activates NRF2. This relationship between ethanol exposure and NRF2, however, remains complex, as ethanol has been shown to suppress NRF2 expression in alveolar epithelial cells and an NRF2 activator (sulforaphane) blocks the effects of ethanol [69]. Activation of NRF2 prevents ethanol-induced oxidative stress, lipid accumulation, and accelerated acetaldehyde metabolism [70, 71]. Overall, the effect of ethanol on NRF2 expression depends on the concentration of ethanol, duration of ethanol exposure, cellular context, and potentially some other factors. When we exposed human ESCC KYSE510 cells (NRF2 wild-type) to 100mM ethanol for 24 hours, we observed significant activation of NRF2 transcriptional activity using a luciferase reporter assay (unpublished data). However, it remains to be determined whether ethanol exposure can act in the same way in normal esophageal squamous epithelial cells in which NRF2 activity can be activated, but not to the level in NRF2-mutant ESCC cells.
Our previous microarray analysis comparing Keap1−/− to Nrf2−/−;Keap1−/− mouse esophagi identified Acss2 as one of the genes differentially expressed due to NRF2 hyperactivation. In this study, we further established ACSS2 as a transcriptional target of NRF2 in the esophagus in vitro and in vivo. Additionally, our prior ChIP-seq data and our current ChIP PCR data, indicate that NRF2 is a transcriptional regulator of ACSS2 [39].
The NRF2/ACSS2 axis allows ESCC cells to employ ethanol-derived acetate as an energy source and may thus facilitate alcohol-associated ESCC. Acetate metabolism is most prevalent in cancers when there is a lack of glucose or oxygen required for aerobic glycolysis [40, 47]. It has been proposed that cancer cells consume acetate from carbohydrate fermentation in the human gut. In carbohydrate fermentation studies, the highest acetate concentration recorded was 181.3 ± 23.9 μM [41, 72]. More recently, it has been established that acetate may be synthesized from glucose metabolism and mitochondrial coupling. This process involves the conversion of pyruvate to acetate by coupling either reactive oxygen species to pyruvate decarboxylation or by neomorphic enzyme activity from keto acid dehydrogenases that enable them to function as pyruvate decarboxylases [73]. Such production of acetate can only occur if there is a sufficient amount of glucose. The normal concentration of acetate in the blood is within 50–200 μM [72, 74–76]. Ethanol intake from alcohol drinking would produce a high level of acetate in the blood. Ethanol consumption has been reported to increase plasma acetate to 750 μM. Moreover, this elevated concentration of plasma acetate may be persistent over 24 h, unlike endogenous acetate produced from carbohydrate fermentation which remains elevated at 200 μM for <1 h [72, 77]. Furthermore, alcoholics and heavy drinkers oxidize ethanol at much faster rates than occasional drinkers. The fast oxidation of ethanol led to significantly elevated blood acetate levels of up to 1 mM [74, 75].
Our schematic model (Fig. 6D) explains how ESCC cells utilize acetate as an energy source originally derived from ethanol. According to our data, the concentration of acetyl-CoA and ATP in ESCC cells significantly increased after ethanol exposure. Acetyl-CoA and ATP production from endogenous acetate and exogenous acetate (via ethanol) was shown to be dependent on the NRF2/ACSS2 axis (Fig. S2 and Fig. 6). Elevated acetyl-CoA production from ethanol exposure was used for OXPHOS leading to a significant metabolic shift (Fig. 4A–C). NRF2/ACSS2 axis was involved in the upregulation of OXPHOS after ethanol exposure (Fig. S4 and Fig. 5D–I). Increased acetate metabolism from ethanol exposure also led to increased lipid synthesis in both NRF2high and NRF2low ESCC cells (Fig. 6A, B). This upregulated lipid synthesis in KYSE70 cells was dependent on the NRF2/ACSS2 axis as shown by the significant decrease in lipid synthesis due to NRF2KD, ACSS2KD or ACSS2 inhibition (Fig. 6A). Additionally, the significant increase of lipid synthesis when NRF2 was upregulated in KYSE410 cells, as well as the significant decrease when ACSS2 was chemically inhibited during ethanol exposure, provide further support for the important role of the NRF2/ACSS2 axis in ethanol-induced lipid synthesis in ESCC cells (Fig. 6B). These findings are in agreement with our prior metabolomic analysis of Keap1−/− versus wild-type mouse esophagi [39]. Taken together, NRF2 hyperactivity cooperates with alcohol drinking to facilitate metabolic changes in ESCC cells.
Metabolic reprogramming of cancer cells is expected to impact cancer cell behaviors [78]. An increase in OXPHOS has been reported to be important for cancer cell invasion and migration by providing necessary energy for microtubule motility [79, 80]. In this study, NRF2KD, ACSS2KD and ACSS2 inhibition in KYSE70 cells exposed to ethanol, not only led to a significant decrease in OXPHOS but also a significant decrease in cell invasion (Fig. S4, Fig. 5G and Fig 6C). Lipid synthesis has also been shown to serve important roles in cancer cell invasion and metastasis including structural support, and synthesis of chemical signals in the tumor microenvironment (70–74). Our data shows that ethanol exposure fuels lipid synthesis. This is in agreement with other reports that increased acetate metabolism resulted in significant increases in cholesterol and phospholipid in breast and hepatocellular carcinoma leading to more severe disease [40, 47]. In our data, both ethanol-induced lipid synthesis and cancer cell invasion were facilitated by the NRF2/ACSS2 axis. Nevertheless, our data using the fatty acid fluorometric assay does not provide direct evidence to support acetate as a carbon source of fatty acid synthesis. A carbon tracing experiment is warranted in the future to further validate ethanol and acetate as carbon sources for ESCC cells.
In summary, the NRF2/ACSS2 axis regulates downstream ethanol metabolism and metabolic reprogramming in ESCC, which may promote alcohol-associated ESCC (Fig. 6D).
Supplementary Material
Acknowledgments:
This work was supported by research grants from the National Institutes of Health (grant numbers U54 AA019765, U54 CA156735, U54 MD012392, and R21 AA028047).
The abbreviations used are:
- ARE
antioxidant response element
- ECAR
extracellular acidification rate
- ESCC
esophageal squamous cell carcinoma
- H&E
hematoxylin and eosin
- IHC
immunohistochemical staining
- KD
knockdown
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
Footnote: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68, 394–424 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 136, E359–386 [DOI] [PubMed] [Google Scholar]
- 3.Huang Q, Luo K, Yang H, Wen J, Zhang S, Li J, Ela Bella A, Liu Q, Yang F, Zheng Y, Hu R, Chen J and Fu J (2014) Impact of alcohol consumption on survival in patients with esophageal carcinoma: a large cohort with long-term follow-up. Cancer Sci. 105, 1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamangar F, Chow WH, Abnet CC and Dawsey SM (2009) Environmental causes of esophageal cancer. Gastroenterol Clin North Am. 38, 27–57, vii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Q, Liu W, Jia R, Long H, Zhang L, Lin P, Zhao H and Ma G (2016) Alcohol and survival in ESCC: prediagnosis alcohol consumption and postoperative survival in lymph node-negative esophageal carcinoma patients. Oncotarget. 7, 38857–38863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y and Muto M (2015) Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology. 149, 1700–1715 [DOI] [PubMed] [Google Scholar]
- 7.Castellsague X, Munoz N, De Stefani E, Victora CG, Castelletto R, Rolon PA and Quintana MJ (1999) Independent and joint effects of tobacco smoking and alcohol drinking on the risk of esophageal cancer in men and women. Int J Cancer. 82, 657–664 [DOI] [PubMed] [Google Scholar]
- 8.Launoy G, Milan CH, Faivre J, Pienkowski P, Milan CI and Gignoux M (1997) Alcohol, tobacco and oesophageal cancer: effects of the duration of consumption, mean intake and current and former consumption. Br J Cancer. 75, 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita M, Kumashiro R, Kubo N, Nakashima Y, Yoshida R, Yoshinaga K, Saeki H, Emi Y, Kakeji Y, Sakaguchi Y, Toh Y and Maehara Y (2010) Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: epidemiology, clinical findings, and prevention. Int J Clin Oncol. 15, 126–134 [DOI] [PubMed] [Google Scholar]
- 10.Sakata K, Hoshiyama Y, Morioka S, Hashimoto T, Takeshita T, Tamakoshi A and Group JS (2005) Smoking, alcohol drinking and esophageal cancer: findings from the JACC Study. J Epidemiol. 15 Suppl 2, S212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Chen H, Sun Z and Chen X (2015) Molecular mechanisms of ethanol-associated oro-esophageal squamous cell carcinoma. Cancer Lett. 361, 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang JL and Vaca CE (1997) Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis. 18, 627–632 [DOI] [PubMed] [Google Scholar]
- 13.Hecht SS, McIntee EJ and Wang M (2001) New DNA adducts of crotonaldehyde and acetaldehyde. Toxicology. 166, 31–36 [DOI] [PubMed] [Google Scholar]
- 14.Helander A and Lindahl-Kiessling K (1991) Increased frequency of acetaldehyde-induced sister-chromatid exchanges in human lymphocytes treated with an aldehyde dehydrogenase inhibitor. Mutat Res. 264, 103–107 [DOI] [PubMed] [Google Scholar]
- 15.Lambert B, Chen Y, He SM and Sten M (1985) DNA cross-links in human leucocytes treated with vinyl acetate and acetaldehyde in vitro. Mutat Res. 146, 301–303 [DOI] [PubMed] [Google Scholar]
- 16.Matsuda T, Kawanishi M, Yagi T, Matsui S and Takebe H (1998) Specific tandem GG to TT base substitutions induced by acetaldehyde are due to intra-strand crosslinks between adjacent guanine bases. Nucleic Acids Res. 26, 1769–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garaycoechea JI, Crossan GP, Langevin F, Mulderrig L, Louzada S, Yang F, Guilbaud G, Park N, Roerink S, Nik-Zainal S, Stratton MR and Patel KJ (2018) Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature. 553, 171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, Ahmed K, Constantinou S, Renaudin X, Lee M, Aebersold R and Venkitaraman AR (2017) A Class of Environmental and Endogenous Toxins Induces BRCA2 Haploinsufficiency and Genome Instability. Cell. 169, 1105–1118 e1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, Garg M, Liu LZ, Yang H, Yin D, Shi ZZ, Jiang YY, Gu WY, Gong T, Zhang Y, Xu X, Kalid O, Shacham S, Ogawa S, Wang MR and Koeffler HP (2014) Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 46, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Snell JM, Jeck WR, Hoadley KA, Wilkerson MD, Parker JS, Patel N, Mlombe YB, Mulima G, Liomba NG, Wolf LL, Shores CG, Gopal S and Sharpless NE (2016) Subtyping sub-Saharan esophageal squamous cell carcinoma by comprehensive molecular analysis. JCI Insight. 1, e88755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y, Iwaya T, Sudo T, Hayashi T, Takai H, Kawasaki Y, Matsukawa T, Eguchi H, Sugimachi K, Tanaka F, Suzuki H, Yamamoto K, Ishii H, Shimizu M, Yamazaki H, Yamazaki M, Tachimori Y, Kajiyama Y, Natsugoe S, Fujita H, Mafune K, Tanaka Y, Kelsell DP, Scott CA, Tsuji S, Yachida S, Shibata T, Sugano S, Doki Y, Akiyama T, Aburatani H, Ogawa S, Miyano S, Mori M and Mimori K (2016) Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology. 150, 1171–1182 [DOI] [PubMed] [Google Scholar]
- 22.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, Ma X, Liu L, Zhao Z, Huang X, Fan J, Dong L, Chen G, Ma L, Yang J, Chen L, He M, Li M, Zhuang X, Huang K, Qiu K, Yin G, Guo G, Feng Q, Chen P, Wu Z, Wu J, Ma L, Zhao J, Luo L, Fu M, Xu B, Chen B, Li Y, Tong T, Wang M, Liu Z, Lin D, Zhang X, Yang H, Wang J and Zhan Q (2014) Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 509, 91–95 [DOI] [PubMed] [Google Scholar]
- 23.Cullinan SB, Gordan JD, Jin JO, Harper JW and Diehl JA (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 24, 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furukawa M and Xiong Y (2005) BTB protein keap1 targets antioxidant transcription factor nrf2 for ubiquitination by the cullin 3-Roc1 ligase. Mol Cell Biol. 25, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K and Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate for proteasomal degradation of Nrf2. Mol Cell Biol. 24, 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tonelli C, Chio IIC and Tuveson DA (2018) Transcriptional Regulation by Nrf2. Antioxid Redox Signal. 29, 1727–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T and Yamamoto M (2006) Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol Cell Biol. 26, 2887–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang DD, Lo SC, Cross JV, Templeton DJ and Hannink M (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 24, 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Li J, Li H, Hu Y, Tevebaugh W, Yamamoto M, Que J and Chen X (2012) Transcript profiling identifies dynamic gene expression patterns and an important role for Nrf2/Keap1 pathway in the developing mouse esophagus. PLoS One. 7, e36504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang M, Ku WY, Zhou Z, Dellon ES, Falk GW, Nakagawa H, Wang ML, Liu K, Wang J, Katzka DA, Peters JH, Lan X and Que J (2015) BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J Clin Invest. 125, 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Hu Y, Fang Y, Djukic Z, Yamamoto M, Shaheen NJ, Orlando RC and Chen X (2014) Nrf2 deficiency impairs the barrier function of mouse oesophageal epithelium. Gut. 63, 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kansanen E, Kuosmanen SM, Leinonen H and Levonen AL (2013) The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 1, 45–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon EJ and Giaccia A (2015) Dual roles of NRF2 in tumor prevention and progression: possible implications in cancer treatment. Free Radic Biol Med. 79, 292–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du P, Huang P, Huang X, Li X, Feng Z, Li F, Liang S, Song Y, Stenvang J, Brunner N, Yang H, Ou Y, Gao Q and Li L (2017) Comprehensive genomic analysis of Oesophageal Squamous Cell Carcinoma reveals clinical relevance. Sci Rep. 7, 15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hast BE, Goldfarb D, Mulvaney KM, Hast MA, Siesser PF, Yan F, Hayes DN and Major MB (2013) Proteomic Analysis of Ubiquitin Ligase KEAP1 Reveals Associated Proteins That Inhibit NRF2 Ubiquitination. Cancer Res. 73, 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Namani A, Rahaman MM, Chen M and Tang XW (2018) Gene-expression signature regulated by the KEAP1-NRF2-CUL3 axis is associated with a poor prognosis in head and neck squamous cell cancer. Bmc Cancer. 18, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto S, Inoue J, Kawano T, Kozaki KI, Omura K and Inazawa J (2014) The Impact of miRNA-Based Molecular Diagnostics and Treatment of NRF2-Stabilized Tumors. Mol Cancer Res. 12, 58–68 [DOI] [PubMed] [Google Scholar]
- 38.Cui Y, Chen H, Xi R, Cui H, Zhao Y, Xu E, Yan T, Lu X, Huang F, Kong P, Li Y, Zhu X, Wang J, Zhu W, Wang J, Ma Y, Zhou Y, Guo S, Zhang L, Liu Y, Wang B, Xi Y, Sun R, Yu X, Zhai Y, Wang F, Yang J, Yang B, Cheng C, Liu J, Song B, Li H, Wang Y, Zhang Y, Cheng X, Zhan Q, Li Y and Liu Z (2020) Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu J, Xiong Z, Huang C, Li J, Yang W, Han Y, Paiboonrungruan C, Major MB, Chen KN, Kang X and Chen X (2019) Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J Biol Chem. 294, 327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, Horton JD, Hammer RE, McKnight SL and Tu BP (2014) Acetate Dependence of Tumors. Cell. 159, 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lakhter AJ, Hamilton J, Konger RL, Brustovetsky N, Broxmeyer HE and Naidu SR (2016) Glucose-independent Acetate Metabolism Promotes Melanoma Cell Survival and Tumor Growth. J Biol Chem. 291, 21869–21879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watkins PA, Maiguel D, Jia Z and Pevsner J (2007) Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res. 48, 2736–2750 [DOI] [PubMed] [Google Scholar]
- 43.Ariyannur PS, Moffett JR, Madhavarao CN, Arun P, Vishnu N, Jacobowitz DM, Hallows WC, Denu JM and Namboodiri AM (2010) Nuclear-cytoplasmic localization of acetyl coenzyme a synthetase-1 in the rat brain. J Comp Neurol. 518, 2952–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujino T, Kondo J, Ishikawa M, Morikawa K and Yamamoto TT (2001) Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 276, 11420–11426 [DOI] [PubMed] [Google Scholar]
- 45.Jaworski DM, Namboodiri AM and Moffett JR (2016) Acetate as a Metabolic and Epigenetic Modifier of Cancer Therapy. J Cell Biochem. 117, 574–588 [DOI] [PubMed] [Google Scholar]
- 46.Perez-Chacon G, Astudillo AM, Balgoma D, Balboa MA and Balsinde J (2009) Control of free arachidonic acid levels by phospholipases A2 and lysophospholipid acyltransferases. Biochim Biophys Acta. 1791, 1103–1113 [DOI] [PubMed] [Google Scholar]
- 47.Schug ZT, Peck B, Jones DT, Zhang QF, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, McGarry L, James D, Shanks E, Kalna G, Saunders RE, Jiang M, Howell M, Lassailly F, Thin MZ, Spencer-Dene B, Stamp G, van den Broek NJF, Mackay G, Bulusu V, Kamphorst JJ, Tardito S, Strachan D, Harris AL, Aboagye EO, Critchlow SE, Wakelam MJO, Schulze A and Gottlieb E (2015) Acetyl-CoA Synthetase 2 Promotes Acetate Utilization and Maintains Cancer Cell Growth under Metabolic Stress. Cancer Cell. 27, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulusu V, Tumanov S, Michalopoulou E, van den Broek NJ, MacKay G, Nixon C, Dhayade S, Schug ZT, Vande Voorde J, Blyth K, Gottlieb E, Vazquez A and Kamphorst JJ (2017) Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 18, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X, Lin SH, Ren F, Li JT, Chen JJ, Yao CB, Yang HB, Jiang SX, Yan GQ, Wang D, Wang Y, Liu Y, Cai ZW, Xu YY, Chen J, Yu WQ, Yang PY and Lei QY (2016) Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat Commun. 7, 11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shibata T, Kokubu A, Saito S, Narisawa-Saito M, Sasaki H, Aoyagi K, Yoshimatsu Y, Tachimori Y, Kushima R, Kiyono T and Yamamoto M (2011) NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia. 13, 864–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cha BS, Ahn CW, Song YD, Lim SK, Kim KR, Huh KB and Lee HC (2000) Chronic alcohol intake differently influences glucose metabolism according to nutritional status. J Clin Endocrinol Metab. 85, 3646–3652 [DOI] [PubMed] [Google Scholar]
- 52.Jain H, Beriwal S and Singh S (2002) Alcohol induced ketoacidosis, severe hypoglycemia and irreversible encephalopathy. Med Sci Monit. 8, CS77–79 [PubMed] [Google Scholar]
- 53.Turner BC, Jenkins E, Kerr D, Sherwin RS and Cavan DA (2001) The effect of evening alcohol consumption on next-morning glucose control in type 1 diabetes. Diabetes Care. 24, 1888–1893 [DOI] [PubMed] [Google Scholar]
- 54.Volkow ND, Kim SW, Wang GJ, Alexoff D, Logan J, Muench L, Shea C, Telang F, Fowler JS, Wong C, Benveniste H and Tomasi D (2013) Acute alcohol intoxication decreases glucose metabolism but increases acetate uptake in the human brain. Neuroimage. 64, 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nioi P, McMahon M, Itoh K, Yamamoto M and Hayes JD (2003) Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H: quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 374, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowman BMMSAS, T.P.; Simon J; Ptacek TS; Muvlaney KM; Weir S; Nguyen T; Murphy RM; Hayes DN; Chen XL; Randell SH; Weissman BE; Major MB (2020) A Conditional Mouse Expressing a Tumor-derived Mutation in the NRF2 Transcription Factor Displays Hyperplasia of the Upper Gastrointestinal Tract and Decreased White Adipose Fat. J Pathol. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, Zhao YD, Sun J, Zhou CC, Yao R, Wang SY, Wang P, Sun N, Zhang BH, Dong JS, Yu Y, Luo M, Feng XL, Shi SS, Zhoul F, Tan FW, Qiu B, Li N, Shao K, Zhang LJ, Zhang LJ, Xue Q, Gao SG and He J (2014) Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 46, 1097–1102 [DOI] [PubMed] [Google Scholar]
- 58.Yoshimura Y, Araki A, Maruta H, Takahashi Y and Yamashita H (2017) Molecular cloning of rat acss3 and characterization of mammalian propionyl-CoA synthetase in the liver mitochondrial matrix. J Biochem. 161, 279–289 [DOI] [PubMed] [Google Scholar]
- 59.Aye MM, Ma CL, Lin H, Bower KA, Wiggins RC and Luo J (2004) Ethanol-induced in vitro invasion of breast cancer cells: The contribution of MMP-2 by fibroblasts. Int J Cancer. 112, 738–746 [DOI] [PubMed] [Google Scholar]
- 60.Gelfand R, Vernet D, Bruhn KW, Sarkissyan S, Heber D, Vadgama JV and Gonzalez-Cadavid NF (2017) Long-term exposure of MCF-7 breast cancer cells to ethanol stimulates oncogenic features. Int J Oncol. 50, 49–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu M, Chen G, Fu W, Liao MJ, Frank JA, Bower KA, Fang SY, Zhang Z, Shi XL and Luo J (2012) Ethanol Disrupts Vascular Endothelial Barrier: Implication in Cancer Metastasis. Toxicol Sci. 127, 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao M, Howard EW, Parris AB, Guo Z, Zhao Q and Yang X (2017) Alcohol promotes migration and invasion of triple-negative breast cancer cells through activation of p38 MAPK and JNK. Mol Carcinog. 56, 849–862 [DOI] [PubMed] [Google Scholar]
- 63.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ and Patel KJ (2012) Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 489, 571–575 [DOI] [PubMed] [Google Scholar]
- 64.Mizumoto A, Ohashi S, Hirohashi K, Amanuma Y, Matsuda T and Muto M (2017) Molecular Mechanisms of Acetaldehyde-Mediated Carcinogenesis in Squamous Epithelium. Int J Mol Sci. 18, 1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seitz HK and Homann N (2007) The role of acetaldehyde in alcohol-associated cancer of the gastrointestinal tract. Novartis Found Symp. 285, 110–119; discussion 119–114, 198–119 [DOI] [PubMed] [Google Scholar]
- 66.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, Huang ZG, Barnett S, Mickey BE, DeBerardinis RJ, Tu BP, Maher EA and Bachoo RM (2014) Acetate Is a Bioenergetic Substrate for Human Glioblastoma and Brain Metastases. Cell. 159, 1603–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cederbaum AI (2012) Alcohol metabolism. Clin Liver Dis. 16, 667–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong Z, Ren S, Chen H, Liu Y, Huang C, Zhang YL, Odera JO, Chen T, Kist R, Peters H, Garman K, Sun Z and Chen X (2018) PAX9 regulates squamous cell differentiation and carcinogenesis in the oro-oesophageal epithelium. J Pathol. 244, 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ushida Y and Talalay P (2013) Sulforaphane accelerates acetaldehyde metabolism by inducing aldehyde dehydrogenases: relevance to ethanol intolerance. Alcohol Alcohol. 48, 526–534 [DOI] [PubMed] [Google Scholar]
- 70.Jensen JS, Fan X and Guidot DM (2013) Alcohol causes alveolar epithelial oxidative stress by inhibiting the nuclear factor (erythroid-derived 2)-like 2-antioxidant response element signaling pathway. Am J Respir Cell Mol Biol. 48, 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu KC, Liu J and Klaassen CD (2012) Role of Nrf2 in preventing ethanol-induced oxidative stress and lipid accumulation. Toxicol Appl Pharmacol. 262, 321–329 [DOI] [PubMed] [Google Scholar]
- 72.Pomare EW, Branch WJ and Cummings JH (1985) Carbohydrate Fermentation in the Human-Colon and Its Relation to Acetate Concentrations in Venous-Blood. J Clin Invest. 75, 1448–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu XJ, Cooper DE, Cluntun AA, Warmoes MO, Zhao S, Reid MA, Liu J, Lund PJ, Lopes M, Garcia BA, Wellen KE, Kirsch DG and Locasale JW (2018) Acetate Production from Glucose and Coupling to Mitochondrial Metabolism in Mammals. Cell. 175, 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Korri UM, Nuutinen H and Salaspuro M (1985) Increased Blood Acetate - a New Laboratory Marker of Alcoholism and Heavy Drinking. Alcohol Clin Exp Res. 9, 468–471 [DOI] [PubMed] [Google Scholar]
- 75.Nuutinen H, Lindros K, Hekali P and Salaspuro M (1985) Elevated Blood Acetate as Indicator of Fast Ethanol Elimination in Chronic-Alcoholics. Alcohol. 2, 623–626 [DOI] [PubMed] [Google Scholar]
- 76.Pomare EW, Scheppach W, Branch WJ and Cummings JH (1985) Contribution of Fermentation in the Human Large-Intestine to Metabolism. Gastroenterology. 88, 1541–1541 [Google Scholar]
- 77.Sarkola T, Iles MR, Kohlenberg-Mueller K and Eriksson CJ (2002) Ethanol, acetaldehyde, acetate, and lactate levels after alcohol intake in white men and women: effect of 4-methylpyrazole. Alcohol Clin Exp Res. 26, 239–245 [PubMed] [Google Scholar]
- 78.Chaffer CL and Weinberg RA (2011) A Perspective on Cancer Cell Metastasis. Science. 331, 1559–1564 [DOI] [PubMed] [Google Scholar]
- 79.Denisenko TV, Gorbunova AS and Zhivotovsky B (2019) Mitochondrial Involvement in Migration, Invasion and Metastasis. Front Cell Dev Biol. 7, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Senft D and Ronai ZA (2016) Regulators of mitochondrial dynamics in cancer. Curr Opin Cell Biol. 39, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.