Abstract
The role of the selenoproteome, which is the collection of all proteins containing selenium in an organism, in cancer development, growth and progression requires further investigation, due to the importance of selenium in both cancer and immune system function. Data about the selenoproteome, including its differential expression, single nucleotide variations, copy number variations, methylation, pathways and overall survival (OS) in five leading types of cancer are available from the GSCALite website. Subsequent to the analysis of these datasets, it was revealed that there was increased expression of GPX3 in stomach adenocarcinoma and lung squamous cell carcinoma, SELENOV in oesophageal carcinoma, GPX8 and GPX4 in colon adenocarcinoma, TXNRD1 and SEPHS1 in hepatocellular carcinoma and GPX8 in lung adenocarcinoma were associated with poor survival. Decreased gene expression of SELENOP was indicated in liver hepatocellular carcinoma and GPX3, and SELENOW, SELENOK, SELENBP1 and SECISBP2 in lung adenocarcinoma were associated with a poor prognosis. OS data suggested that hypermethylation of GPX4 in colon adenocarcinoma, GPX8 in lung squamous cell carcinoma, GPX1 in stomach adenocarcinoma and GPX3 in lung adenocarcinoma was associated with low survival, as is hypomethylation of GPX5 in lung adenocarcinoma. The selenoproteome is heterogeneous, especially in its effect on the OS of patients with cancer. The present study demonstrated that the roles of GPX4 in colon adenocarcinoma, SCLY and SELENOV in oesophageal carcinoma, SEPHS1 in liver hepatocellular carcinoma, SELENOK in lung cancer, as well as SELENOM and SELENOW in stomach adenocarcinoma requires further research. The present study may lead to the identification of novel biomarkers or potential therapeutic targets for use in the treatment of cancers, such as colon adenocarcinoma, oesophageal carcinoma, liver hepatocellular carcinoma, lung cancer and stomach adenocarcinoma.
Keywords: selenoproteome, selenoprotein, methylation, overall survival, cancer
Introduction
Cancer has become one of the world's major disease burdens. Its incidence, and the mortality due to the disease, are increasing rapidly. In 2018 the cancer with the highest death rate globally was lung cancer for both sexes, followed by colorectal cancer, stomach cancer, liver cancer, female breast cancer and oesophageal cancer (1,2). Cancer incidence and mortality in China are about 50% of those in Asia. The five leading types of cancer death of China in 2013, 2015 and 2018 were lung cancer, liver cancer, stomach cancer, oesophageal cancer, and colorectal cancer (3-5). The main treatments used for cancer are surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy. The genes and proteins which are altered in cancer cells, immune cells and other cells in the tumour microenvironment can be targeted for treatment (6), and the selenoproteome may be targeted in both cancer cells and immune cells for therapy, due to the important role of selenium in cancer and immunity (7-10).
It has been demonstrated that selenoproteins, including selenoprotein P (SELP, SELENOP), glutathione peroxidases (GPX), thioredoxin reductases (TXNRD) and selenoprotein F (SEP15, SELENOF), can regulate tumourigenesis and progression through their effects on cancer-related signalling pathways (11). The relationship between single nucleotide polymorphisms in selenoprotein genes and cancer risk has been studied for SELENOP and GPX (12), as well as TXNRD, selenoprotein N (SEPN1, SELENON), selenoprotein S (VIMP, SELENOS), and selenoprotein W (SEPW1, SELENOW) (13). It has been suggested that SELENOP is decreased in various cancers, except for metastatic melanoma, in which it is elevated (11), and low concentrations of SELENOP are related to poor survival in renal cancer (14). TXNRD has been shown to be overexpressed in aggressive tumours, including breast cancer and melanomas (15), but whether there is a relationship between TXNRD levels and prognosis is unknown. Methylation of GPX1, GPX3, selenium binding protein 1 (SELENBP1) and methionine sulfoxide reductase B1 (MSRB1, SEPX1, SELENOR) have been observed in human cancers and human cancer cell lines, and GPX3 methylation can be a tumour biomarker in prostate cancer (16).
Previous studies have focused primarily on the link between a single selenoprotein and a specific cancer risk, including changes in gene expression, single nucleotide polymorphisms, and methylation. However, the selenoproteome contain 25 selenoproteins and seven proteins related to selenoprotein synthesis, and changes in selenoproteome expression levels and methylation, may be valuable for tumour identification and prognosis. The Gene Set Cancer Analysis website GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/) (17) is a useful resource for the analysis of the roles of selenoproteomes in various cancers.
In the present study, GSCALite was used to evaluate differential expression and survival analysis of the selenoproteome in the five leading types of cancer. Single Nucleotide Variations (SNVs) and overall survival (OS) affected by SNVs were analysed. Copy Number Variation (CNV), methylation and the relationship between CNV, methylation and gene expression, as well as the effects of methylation on OS were also studied. Finally, the pathways involved in cancer development, growth, and progression were evaluated.
Materials and methods
Selenoproteome gene set collection
The HUGO Gene Nomenclature Committee (HGNC) symbol gene set for transcription and translation of selenoprotein was accessed. The selenoprotein gene set included iodothyronine deiodinase 1 (DIO1), DIO2, DIO3, GPX1, GPX2, GPX3, GPX4, GPX6, SELENOR, SELENOF, selenoprotein H (C11orf31, SELENOH), selenoprotein I (EPT1, SELENOI), selenoprotein K (SELK, SELENOK), selenoprotein M (SELM, SELENOM), SELENON, selenoprotein O (SELO, SELENOO), SELENOP, SELENOS, selenoprotein T (SELT, SELENOT), selenoprotein V (SELV, SELENOV), SELENOW, TXNRD1, TXNRD2, TXNRD3 and selenophosphate synthetase 2 (SEPHS2). The selenoprotein expression-related gene set included selenocysteine lyase (SCLY), SEPHS1, SELENBP1, selenocysteine insertion sequence-binding protein 2 (SECISBP2), tRNA selenocysteine associated protein (SECp43), Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (SEPSECS) and tRNA selenocysteine 1 associated protein 1 (TRNAU1AP). In addition, homologues of selenium-containing GPX, including GPX5, GPX7, GPX8, and homologues of SELENOR, such as MSRB2 and MSRB3, were also contained in the gene set.
Selenoproteome gene set analysis in cancer
Selenoproteome gene set analysis in cancer was performed using the GSCALite website. Cancer genomic and normal tissue data were downloaded from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) database in the GSCALite website. To access these data, we entered the gene set into the search box at the top of the web page, then selected the databases of colon adenocarcinoma (COAD), oesophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and stomach adenocarcinoma (STAD) from TCGA and the normal tissues of colon, oesophagus, liver, lung and stomach from GTEx in the left search box. Finally, mRNA Expression, SNV, CNV, Methylation, Pathway Activity and GTEx Expression were selected in the right search box followed by clicking the button Start Gene Set Analysis. A detailed description of the method can be seen on the website (http://bioinfo.life.hust.edu.cn/web/GSCALite/). For survival analysis, the OS in lung and liver were further verified using Kaplan-Meier plots (http://kmplot.com/analysis/) (18,19).
Statistical analysis
The differential expression of the selenoproteome was analysed using GSCALite. For mRNA Expression, the significant conditions used were fold change (FC) >2 and FDR <0.05. For survival analysis, genes with a Kaplan-Meier log-rank test P-value <0.05 were used. For Methylation and Pathway activity, FDR ≤0.05 was considered significant.
Results
Selenoproteome mRNA and survival
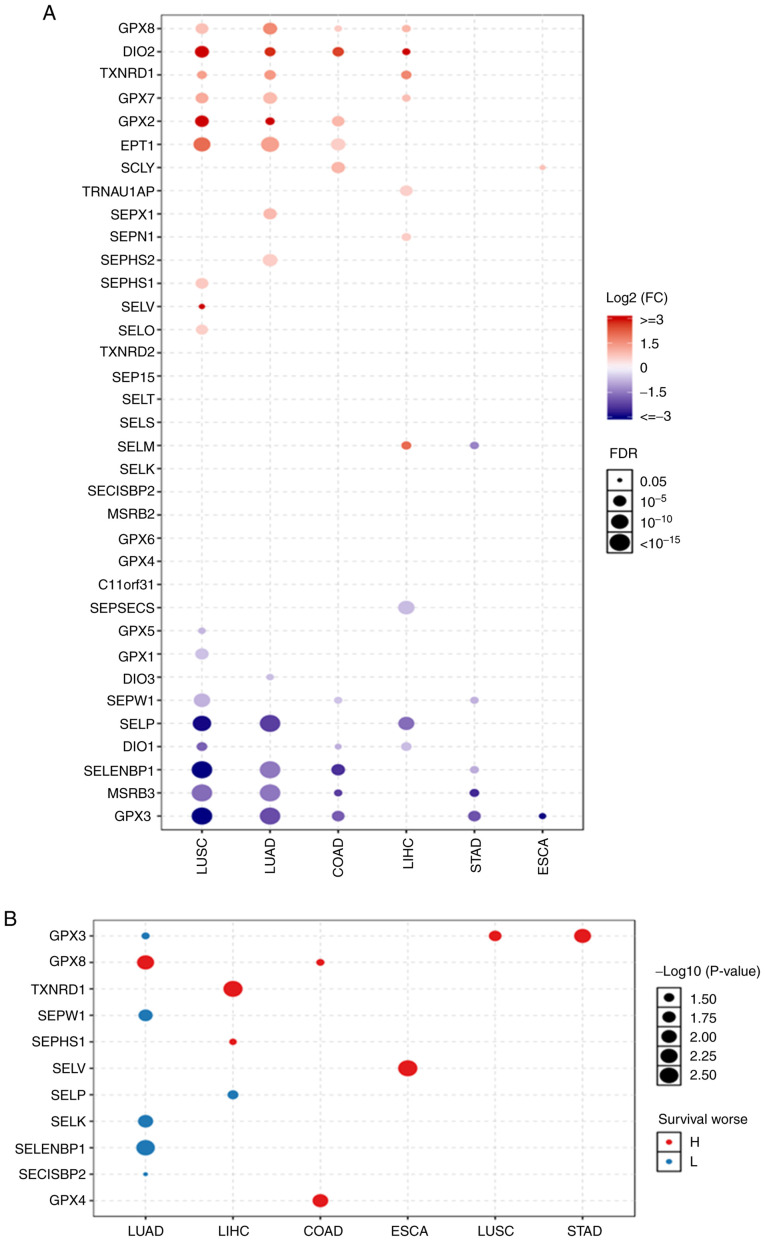
Selenoproteome expression levels in the normal tissues were analysed. The results shown in Fig. S1 suggested that the expression of the selenoproteome was tissue specific. The three most highly expressed genes in the colon were GPX3, SELENBP1 and SELENOW; in the oesophagus they were GPX3, SELENOM and SELENOW; in the liver were GPX3, GPX2 and GPX1; in the lung were GPX3, GPX1 and GPX4; and in the stomach were GPX3, GPX2 and SELENOM. The differing expression levels of selenoproteins between normal tissues and tumours were analysed (Fig. 1A), and it was found that the expression levels of GPX8, DIO2, GPX2, SELENOI and SCLY were significantly increased, while those of SELENOW, DIO1, SELENBP1, MSRB3 and GPX3 were significantly decreased in colon adenocarcinoma. In oesophageal carcinoma, the only gene with increased expression was SCLY, and the only gene with decreased expression was GPX3. In liver hepatocellular carcinoma, the genes with increased expression were GPX8, DIO2, TXNRD1, GPX7, TRNAU1AP, SELENON and SELENOM, while the genes with decreased expression were SEPSECS, SELENOP and DIO1. In stomach adenocarcinoma, no genes had increased expression levels, and the expression of SELENOM, SELENOW, SELENBP1, MSRB3 and GPX3 was significantly decreased. In lung adenocarcinoma and lung squamous cell carcinoma, the genes with increased expression were GPX8, DIO2, TXNRD1, GPX7, GPX2 and SELENOI; the genes with decreased expression were SELENOP, SELENBP1, MSRB3 and GPX3. SEPX1 and SEPHS2 had increased expression in lung adenocarcinoma, SEPHS1, SELENOV and SELENOO were increased in lung squamous cell carcinoma, and DIO3 was decreased in lung adenocarcinoma, Levels of GPX5, GPX1 and DIO1 were decreased in lung squamous cell carcinoma. Expression survival analysis (Fig. 1B) showed that the GPX3, which had an increased level of expression in stomach adenocarcinoma and lung squamous cell carcinoma, SELENOV in oesophageal carcinoma, GPX8 and GPX4 in colon adenocarcinoma, TXNRD1 and SEPHS1 in liver hepatocellular carcinoma and GPX8 in lung adenocarcinoma were associated with poor survival. The genes with decreased expression: SELENOP in liver hepatocellular carcinoma and GPX3, and SELENOW, SELENOK, SELENBP1 and SECISBP2 in lung adenocarcinoma had a poor prognosis. OS in lung and liver cancer supported these findings, and are shown in Figs. S2 and S3. The expression of SELENOM was significantly increased in liver hepatocellular carcinoma, while it was significantly decreased in stomach adenocarcinoma. GPX3 expression was significantly decreased in lung adenocarcinoma, lung squamous cell carcinoma and stomach adenocarcinoma. However, lower expression of GPX3 in lung adenocarcinoma and the high expression of GPX3 in lung squamous cell carcinoma and stomach adenocarcinoma had a poor prognosis.
Figure 1.
The differential selenoproteome between (A) normal tissues and tumors, and (B) the expression survival analysis in COAD, ESCA, LIHC, LUAD, LUSC and STAD. The selenoproteome expression levels and survival analysis in the five leading types of cancer death were performed using GSCALite, and the results with significant differences were presented in the figures. Blue and red represent lower expression and higher expression, respectively, in (A) blue and red indicates the worse of the low or high expression in the cancer types. The size dot indicates the significance. COAD, colon adenocarcinoma; ESCA, oesophageal carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma.
Single nucleotide variations in the selenoproteome and survival
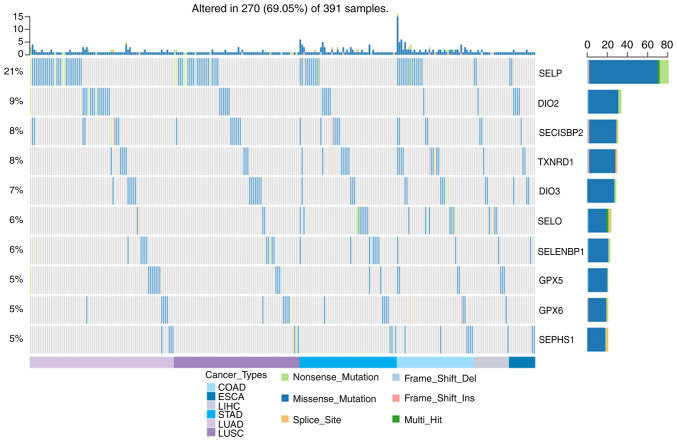
Fig. 2 show that the SNV frequency of the selenoproteome was 69.05% (270 of 391 tumours). The top 10 mutated genes were SELENOP, DIO2, SECISBP2, TXNRD1, DIO3, SELENOO, SELENBP1, GPX5, GPX6 and SEPHS1, and the most frequent type of SNV was a missense mutation. The SNV frequency of the selenoproteome was increased in oesophageal carcinoma, liver hepatocellular carcinoma, colon adenocarcinoma, stomach adenocarcinoma, lung squamous cell carcinoma and lung adenocarcinoma. SNV survival analysis found no significant difference between mutated and non-mutated genes.
Figure 2.
The SNV of selenoproteome in COAD, ESCA, LIHC, LUAD, LUSC and STAD. The SNV of selenoproteome in the five leading types of cancer associated mortality were performed using GSCALite, and the top ten genes and the SNV type were presented in the figure. COAD, colon adenocarcinoma; ESCA, oesophageal carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma.
Copy number variation of the selenoproteome
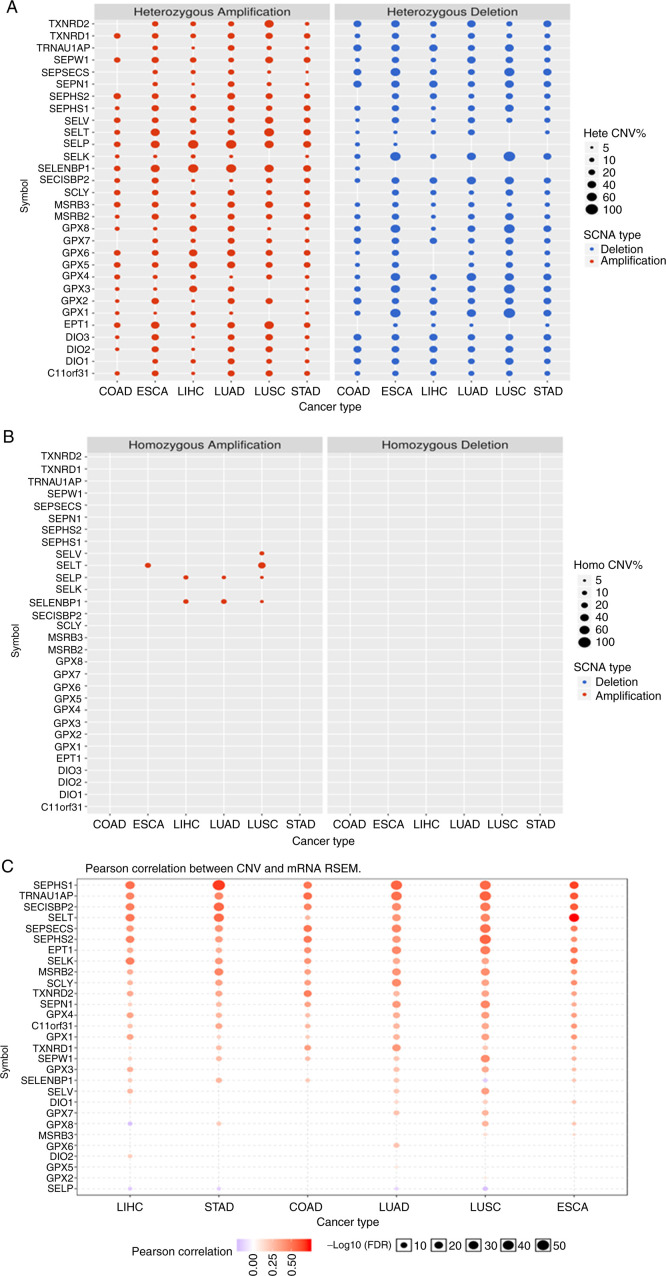
The results shown in Fig. 3A suggested that the CNV was very different for each gene in each type of cancer. The main type of CNV was heterozygous amplification and deletion, and only a few genes, such as SELENOP and SELENBP1, had a low frequency of heterozygous deletion. The genes with homozygous amplification were SELENOT in oesophageal carcinoma and lung squamous cell carcinoma; SELENOP and SELENBP1 in liver hepatocellular carcinoma, lung squamous cell carcinoma and lung adenocarcinoma; and SELENOV in lung squamous cell carcinoma. There were no homozygous deletions (Fig. 3B). Pearson correlation showed a strong correlation between CNV and SELENOT, SELENOV mRNA RSEM in oesophageal carcinoma and lung squamous cell carcinoma, and a poor correlation for SELENOP and SELENBP1 (Fig. 3C).
Figure 3.
(A and B) The CNV of selenoproteome and (C) the correlation between CNV and mRNA RSEM. The CNV of selenoproteome and the correlation between CNV and mRNA RSEM in the five leading types of cancer associated mortality were performed using GSCALite, the size dot indicates the frequency of CNV in (A) and (B), and the brightness of red indicates the strength of the correlation between CNV and mRNA RSEM. COAD, colon adenocarcinoma; ESCA, oesophageal carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma.
Methylation of the selenoproteome and survival
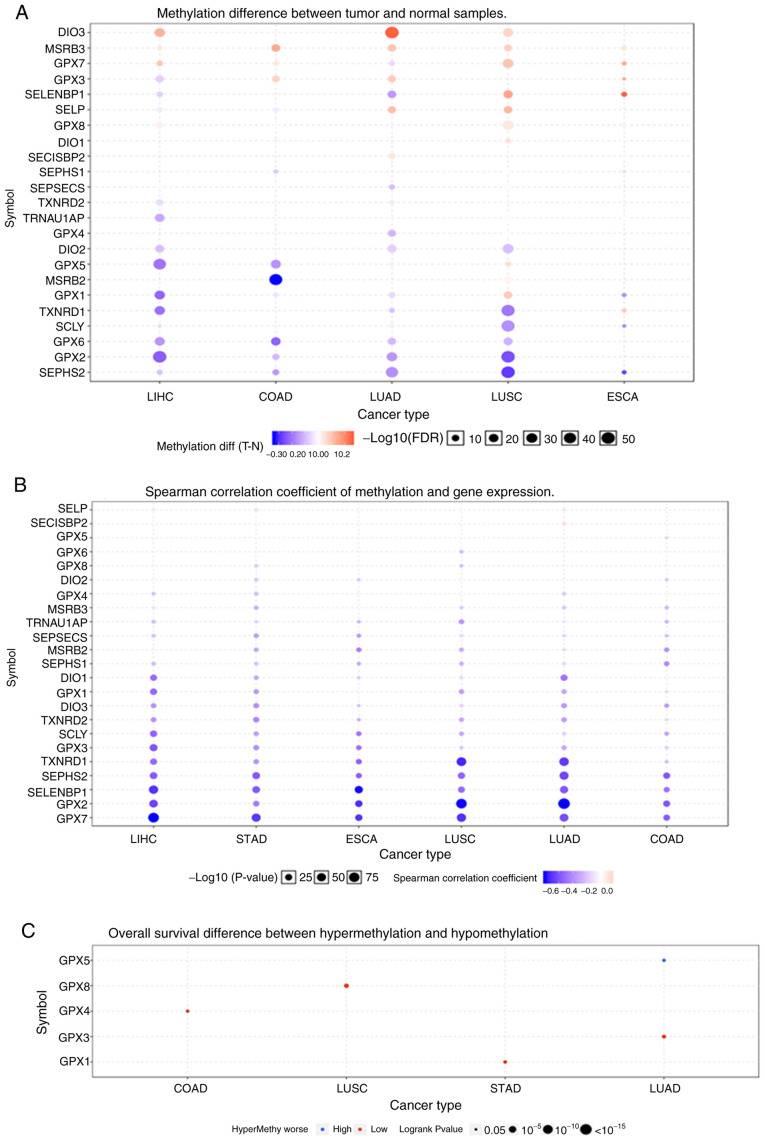
Fig. 4A shows that the methylation of the selenoproteome in different tumours was highly heterogeneous. There were more hypermethylated than hypomethylated genes in lung squamous cell carcinoma and oesophageal carcinoma, and there were more hypomethylated than hypermethylated genes in colon adenocarcinoma, liver hepatocellular carcinoma and lung adenocarcinoma. There was no differential methylation in stomach adenocarcinoma. The Spearman correlation coefficient indicated that most of the genes were negatively correlated, and only SELENOP in liver hepatocellular carcinoma and stomach adenocarcinoma, and SECISBP2 in lung adenocarcinoma showed a positive correlation between methylation and gene expression (Fig. 4B). OS analysis suggested that hypermethylation of GPX4 in colon adenocarcinoma, GPX8 in lung squamous cell carcinoma, GPX1 in stomach adenocarcinoma and GPX3 in lung adenocarcinoma had poor prognosis, and hypomethylation of GPX5 in lung adenocarcinoma had poor prognosis (Fig. 4C). GPX5 and GPX8, which were associated with OS are not selenoproteins. The selenoproteins, GPX1 and GPX3 had lower expression levels in cancer tissues, and GPX4 showed no change.
Figure 4.
The differential methylation of selenoproteome between (A) normal tissues and tumors and (B) the correlation between methylation and gene expression, as well as (C) the overall survival between hypermethylation and hypomethylation. The methylation of selenoproteome and survival, as well as the correlation between methylation and gene expression, in the five leading types of cancer associated mortality were performed using GSCALite. The blue indicates the hypomethylation and the red indicates the hypermethylation in (A). Blue and red represent negative correlation and positive correlation respectively in (B). Blue indicates a higher hypermethylated survival rate and red indicates a lower hypermethylated survival rate in (C). COAD, colon adenocarcinoma; ESCA, oesophageal carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma.
Pathway activity
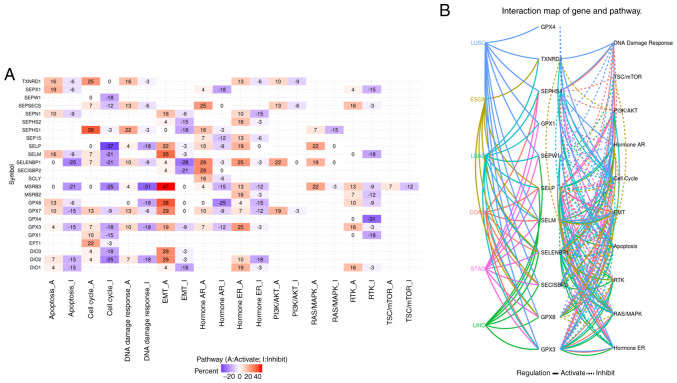
The pathway activity shown in Fig. 5A suggested that there were 24 genes involved in tumour-related signalling pathways. For GPX3, the expression level of which was decreased in cancer, the main pathways were apoptosis inhibition, cell cycle inhibition, DNA damage response inhibition, EMT activation, and RTK activation. For TXNRD1, which was increased in some cancers, the main pathways were apoptosis activation, cell cycle activation, and DNA damage response activation. The relationship network between the 13 survival-related genes, SELENOM, which is increased in liver hepatocellular carcinoma and decreased in stomach adenocarcinoma, and pathways in the five leading types of cancer death, was evaluated. The results shown in Fig. 5B revealed that GPX3 was not involved in the tumour-related signalling pathways of oesophageal carcinoma, and TXNRD was not involved in the tumour-related signalling pathways of colon adenocarcinoma, stomach adenocarcinoma or liver hepatocellular carcinoma. In colon adenocarcinoma, GPX4 was not involved in any of the tumour-related signalling pathways, and GPX8 was involved in the activation of EMT; in oesophageal carcinoma, neither reduced GPX3 or the survival-related SELENOV genes were involved in tumour-related signalling pathways; in liver hepatocellular carcinoma, only one survival-related gene, SELENOP, was involved in HormoneAR and RTK activation; for lung squamous cell carcinoma, both GPX3 and GPX8 were involved in EMT activation; for lung adenocarcinoma, survival-related genes were involved in RAS/MAPK, RTK, EMT and PI3K/AKT activation, as well as apoptosis inhibition; and in stomach adenocarcinoma, survival-related genes were involved in PI3K/AKT activation and HormoneAR inhibition.
Figure 5.
(A) The pathways involved in cancer development, growth and progression of selenoproteome, and (B) cancer pathway analysis of overall survival related selenoproteome in COAD, ESCA, LIHC, LUAD, LUSC and STAD. The cancer pathway of selenoproteome in the five leading types of cancer associated mortality were performed using GSCALite. The luminance of red and blue represents the degree of activation and inhibition of related pathways, respectively, in (A). The solid line indicates that the pathway is activated, the dotted line indicates that the pathway is inhibited, and different colors represent different cancers in (B). COAD, colon adenocarcinoma; ESCA, oesophageal carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma.
Discussion
In the present study a global analysis of differential expression, SNV, CNV, methylation and pathways in the selenoproteomes of the five leading types of cancer death was performed using GSCALite. The selenoproteomes were highly heterogeneous, especially with respect to the effect on the OS of cancer patients, although some genes performed the same role in different tumours. To better understand the role of the selenoproteome in the development, growth, and progression of different tumours, we discuss the potential relationships between the selenoproteome and cancer, one by one.
In this study, it was found that the expression levels of GPX8, DIO2, GPX2, SELENOI and SCLY were significantly increased, and those of SELENOW, DIO1, SELENBP1, MSRB3 and GPX3 were significantly decreased in colon adenocarcinoma. Previously, TXNRD1, GPX1 and GPX4 have been found to be significantly increased compared with corresponding normal tissues in 32 colon adenocarcinoma patients from Germany (20), and the mRNA and protein levels of TXNRD were increased several fold in 50% (5/10) of colon adenocarcinoma patients (21). A study in Japan showed that GPX1, GPX3 and SELENOP were decreased and GPX2 was increased in colorectal cancer (CRC) (22). A study reviewed by Peters et al found that the redox homeostasis related selenoproteins GPX1-4, TXNRD1, SELENOF and SELENOP, and the WNT/β-catenin signalling pathway associated selenoproteins DIO3, GPX2, TXNRD3 and SELENOP may effect CRC risk and development (23). To the best of our knowledge, there are many selenoproteins whose roles in CRC are unknown (23). These observations indicate that the selenoproteomes which exhibited significant changes in this study are a promising research target, which should have high priority. Of particular interest are genes associated with survival, such as GPX4 and GPX8. Although there was no observed relationship between SNV and colorectal cancer/colon adenocarcinoma risk based on big genomic data (24), the SNV of GPX1, GPX2, GPX4, TXNRD1, TXNRD2, TXNRD3, SELENOF, SELENOP, SELENOS and SELENOW have been well studied in CRC, and the SNV of SELENOP and SELENOS had a strong effect on CRC risk (23-25). For colon adenocarcinoma, the top eight potential risk selenoprotein-related genes for SNV analysis were SELENOP, TXNRD1, DIO3, SELENOO, SEPHS1, SCLY, TXNRD2 and SELENOI (Fig. S4). The results of methylation analysis showed no difference in GPX4 methylation between tumour and normal samples, but the hypermethylation of GPX4 in colon adenocarcinoma was associated with poor survival. There was a negative correlation between GPX4 methylation and expression. Ferroptosis, which can be regulated by GPX4, has great prospects for application in tumour immunotherapy (26) and tumour death (27), and might play important roles in colon adenocarcinoma development, growth and progression due to the potential correlation between higher GPX4 and lower survival. Although GPX4 was not involved in the tumour-related signalling pathways, we hypothesized that higher GPX4 expression inhibits tumour cell ferroptosis, while GPX8 activates EMT, so patients with higher GPX4 and GPX8 have poor survival.
In oesophageal carcinoma, levels of SCLY were increased and those of GPX3 were decreased. The survival related gene was SELENOV, which is associated with a low survival rate at high levels, and there were no genes identified as being involved in any relationship between methylation and OS. In a study of oesophageal squamous cell carcinoma (ESCC) in China, it was found that the mRNA and protein levels of GPX2 were significantly increased compared with normal tissue, and lack of GPX2 expression was associated with poor prognosis (28). The expression of SELENBP1 was significantly decreased in oesophageal adenocarcinoma (EA) in the USA (29). It was suggested that there was a positive correlation between plasma SELENOP and EA risk, and there were no genetic variants related to EA risk (30). There was no relationship observed between selenium and Barrett's oesophagus in a study by Steevens et al (31). In this study, GPX3 and SELENOV did not appear to be involved in tumour-related signalling pathways. The potential risk of SCLY, which is involved in energy metabolism and therefore essential for tumour development (32), needs more research.
In liver hepatocellular carcinoma, the present study found that the levels of GPX8, DIO2, TXNRD1, GPX7, TRNAU1AP, SELENON and SELENOM were increased; and those of SEPSECS, SELENOP and DIO1 were decreased. SELENOP mRNA was consistently significantly decreased in hepatocellular carcinoma (HCC) in a study conducted in Xi'an, China (33), and TXNRD1 expression level was also increased in data from the GEO and TCGA databases (34). TXNRD1 mRNA was significantly increased in HCC patients from Hong Kong (35), Zhengzhou (34), Chongqing (36,37), and Guangzhou (38), China. High levels of TXNRD1 were associated with poor prognosis (34-38), and Auranofin, a TXNRD1 inhibitor, was shown to inhibit tumour growth in a mouse model, and in HCC cells (35-37). SELENOP also plays an important role in HCC development (39), and low levels of SELENOP were associated with poor prognosis (39-41). In this study, it was also demonstrated that higher levels of TXNRD1 and SEPHS1, as well as lower levels of SELENOP, were associated with poor prognosis. It was shown that SELENOP and SELENBP1 had homozygous mutations, and there were no genes involved in a relationship between methylation and OS. The expression levels of TXNRD1, SELENOP and SELENBP1 were associated with hypomethylation. Previously, it was suggested that SELENBP1 is downregulated in HCC (42) and is correlated with tumour prognosis (43). SELENOM, which is upregulated in HCC cells, may also be involved in HCC development (44), through apoptosis, the cell cycle, and EMT pathways. These results indicate that changes in the selenoproteome in liver hepatocellular carcinoma should be the subject of more attention. In particular, high expression of SEPHS1 can reduce redox damage and promote cell proliferation (45), leading to a poor prognosis for the patients.
For lung cancer, the question of whether mutations in the GPX1 gene are associated with lung cancer risk is difficult to answer, due to differences in factors such as age, population, and lifestyle (46). In the present study, GPX1 was significantly decreased in lung squamous cell carcinoma, but was unchanged in lung adenocarcinoma. Lower levels of SELENOP were associated with higher lung cancer risk among Black patients in the south-eastern USA (47), and SELENOP was also decreased in Polish patients (48). SELENOP was consistently decreased in both lung squamous cell carcinoma and lung adenocarcinoma in this study. Lower levels of GPX3, SELENOW, SELENOK, SELENBP1 and SECISBP2 in lung adenocarcinoma were associated with poor survival, while high levels of GPX3 in lung squamous cell carcinoma were associated with poor survival, possibly related to EMT activation. Recently, it was shown that GPX3 can inhibit the proliferation of lung cancer cells (49), and GPX3 levels, which can be regulated by methylation, were lower in lung cancer patients (50,51). In this study, hypermethylation of GPX3 can inhibit GPX3 expression in lung adenocarcinoma, while there was no difference in methylation between lung squamous cell carcinoma and normal tissue. OS analysis showed that GPX3 hypermethylation and GPX5 hypomethylation in lung adenocarcinoma, and GPX8 hypermethylation in lung squamous cell carcinoma were associated with poor survival. More studies are needed to uncover the role of GPX3, including its expression levels and methylation status, in the development of lung squamous cell carcinoma and lung adenocarcinoma. It has also been demonstrated that low levels of SELENBP1 were associated with poor survival in lung adenocarcinoma (52), SELENOK may regulate the growth and migration of lung cancer through calcium ions (53), SELENOW may have antioxidant activity in lung cancer (54), and SECISBP2 appears to be associated with the prognosis of the whole cancer population (55). We think the role of SELENOK in lung cancer is worth further study, due to its important role in the immune system and cancer (56).
For stomach adenocarcinoma, unlike the other four types of cancer, there were no significantly upregulated selenoprotein genes. The downregulated genes were SELENOM, SELENOW, SELENBP1, MSRB3 and GPX3. High levels of GPX3 and GPX1 hypermethylation were associated with poor survival. It has been suggested that hypermethylation of GPX1 and GPX3 resulted in decreased mRNA expression of GPX1 and GPX3 in gastric cancer cells, and downregulation of GPX1 expression due to hypermethylation was related to poor survival in cancer patients from Korea. Downregulation of GPX3 has been associated with advanced gastric cancer and lymph node metastasis, but no correlation with survival was found (57). Another study demonstrated that downregulation of GPX3 due to hypermethylation was related to lymph node metastasis, while the overexpression of GPX3 in gastric cancer cells did not inhibit cell growth but did inhibit cell migration (58). High levels of GPX3 expression associated with poor survival in stomach adenocarcinoma may act through PI3K/AKT activation. Recently, it was shown that SELENOM and SELENOW were significantly decreased in gastric cancer in Harbin, China (59). Downregulation of SELENBP1 and associated poor survival were also observed in stomach adenocarcinoma in Wuhan, China (60). In this study, decreased levels of SELENBP1 and GPX3 were correlated with hypermethylation of the two genes, but the effects upon the levels of SELENOM and SELENOW were unclear. Although levels of SELENOP were lower in stomach adenocarcinoma, and were associated with the degree of stomach adenocarcinoma differentiation (61), the roles of SELENOM and SELENOW in stomach adenocarcinoma development needs further research.
In conclusion, we conducted a preliminary analysis of the differences in expression of the selenoproteome in five cancers, together with a comprehensive analysis of the effects of SNV, CNV, methylation and cancer pathways on gene expression. The effects of gene expression and methylation of the selenoproteome on survival were investigated. In the context of previous research, we concluded that the roles of GPX4 in colon adenocarcinoma, SCLY and SELENOV in oesophageal carcinoma, SEPHS1 in liver hepatocellular carcinoma, SELENOK in lung cancer, and SELENOM and SELENOW in stomach adenocarcinoma need further research. This study provides a valuable basis for the comprehensive understanding of the role of the selenoproteome in the development of cancer, and may lead to the identification of new biomarkers or potential therapeutic targets for cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 21561006 and 21867007); and the Science and Technology Foundation of Guizhou Province [grant nos. (2019)1258, LH-(2016)7372 and J-(2014)2028].
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YJ, JD and ZZ performed the experiments; YJ and JD contributed to the data analysis and presentation. YJ and ZZ designed the experiment and wrote the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: Good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39(22) doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zheng R, Zhang S, Zeng H, Xia C, Zuo T, Yang Z, Zou X, He J. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Gatzka MV. Targeted tumor therapy remixed-an update on the use of small-molecule drugs in combination therapies. Cancers (Basel) 2018;10(155) doi: 10.3390/cancers10060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avery JC, Hoffmann PR. Selenium, selenoproteins, and immunity. Nutrients. 2018;10(1203) doi: 10.3390/nu10091203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Z, Rose AH, Hoffmann PR. The role of selenium in inflammation and immunity: From molecular mechanisms to therapeutic opportunities. Antioxid Redox Sign. 2012;16:705–743. doi: 10.1089/ars.2011.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairweather-Tait SJ, Bao Y, Broadley MR, Collings R, Ford D, Hesketh JE, Hurst R. Selenium in human health and disease. Antioxid Redox Sign. 2011;14:1337–1383. doi: 10.1089/ars.2010.3275. [DOI] [PubMed] [Google Scholar]
- 10.Rayman MP. Selenium and human health. Lancet. 2012;379:1256–1268. doi: 10.1016/S0140-6736(11)61452-9. [DOI] [PubMed] [Google Scholar]
- 11.Short SP, Williams CS. Selenoproteins in tumorigenesis and cancer progression. Adv Cancer Res. 2017;136:49–83. doi: 10.1016/bs.acr.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Méplan C. Association of single nucleotide polymorphisms in selenoprotein genes with cancer risk. Methods Mol Biol. 2018;1661:313–324. doi: 10.1007/978-1-4939-7258-6_22. [DOI] [PubMed] [Google Scholar]
- 13.Hatfield DL, Carlson BA, Tsuji PA, Tobe R, Gladyshev VN. Selenium and cancer. In: Molecular, Genetic, and Nutritional Aspects of Major and Trace Minerals. Collins J (ed). Academic Press, New York, NY, pp463-473, 2017. [Google Scholar]
- 14.Meyer HA, Endermann T, Stephan C, Stoedter M, Behrends T, Wolff I, Jung K, Schomburg L. Selenoprotein P status correlates to cancer-specific mortality in renal cancer patients. PLoS One. 2012;7(e46644) doi: 10.1371/journal.pone.0046644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lincoln DT, Ali EME, Tonissen KF, Clarke FM. The thioredoxin-thioredoxin reductase system: Over-expression in human cancer. Anticancer Res. 2003;23:2425–2433. [PubMed] [Google Scholar]
- 16.Jabłońska E, Reszka E. Selenium and epigenetics in cancer: Focus on DNA methylation. Adv Cancer Res. 2017;136:193–234. doi: 10.1016/bs.acr.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: A web server for gene set cancer analysis. Bioinformatics. 2018;34:3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 18.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(e82241) doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5(181006) doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagublu V, Arthur JR, Babayeva SN, Nicol F, Post S, Keese M. Expression of selenium-containing proteins in human colon carcinoma tissue. Anticancer Res. 2011;31:2693–2698. [PubMed] [Google Scholar]
- 21.Berggren M, Gallegos A, Gasdaska JR, Gasdaska PY, Warneke J, Powis G. Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 1996;16:3459–3466. [PubMed] [Google Scholar]
- 22.Murawaki Y, Tsuchiya H, Kanbe T, Harada K, Yashima K, Nozaka K, Tanida O, Kohno M, Mukoyama T, Nishimuki E, et al. Aberrant expression of selenoproteins in the progression of colorectal cancer. Cancer Lett. 2008;259:218–230. doi: 10.1016/j.canlet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Peters KM, Carlson BA, Gladyshev VN, Tsuji PA. Selenoproteins in colon cancer. Free Radic Biol Med. 2018;127:14–25. doi: 10.1016/j.freeradbiomed.2018.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Méplan C, Hesketh J. The influence of selenium and selenoprotein gene variants on colorectal cancer risk. Mutagenesis. 2012;27:177–186. doi: 10.1093/mutage/ger058. [DOI] [PubMed] [Google Scholar]
- 25.Méplan C, Hughes DJ, Pardini B, Naccarati A, Soucek P, Vodickova L, Hlavatá I, Vrána D, Vodicka P, Hesketh JE. Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis. 2010;31:1074–1079. doi: 10.1093/carcin/bgq076. [DOI] [PubMed] [Google Scholar]
- 26.Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassannia B, Vandenabeele P, Berghe TV. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Lei Z, Tian D, Zhang C, Zhao S, Su M. Clinicopathological and prognostic significance of GPX2 protein expression in esophageal squamous cell carcinoma. BMC Cancer. 2016;16(410) doi: 10.1186/s12885-016-2462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silvers AL, Lin L, Bass AJ, Chen G, Wang Z, Thomas DG, Lin J, Giordano TJ, Orringer MB, Beer DG, Chang AC. Decreased selenium-binding protein 1 in esophageal adenocarcinoma results from posttranscriptional and epigenetic regulation and affects chemosensitivity. Clin Cancer Res. 2010;16:2009–2021. doi: 10.1158/1078-0432.CCR-09-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takata Y, Kristal AR, Santella RM, King IB, Duggan DJ, Lampe JW, Rayman MP, Blount PL, Reid BJ, Vaughan TL, Peters U. Selenium, selenoenzymes, oxidative stress and risk of neoplastic progression from Barrett's esophagus: Results from biomarkers and genetic variants. PLoS One. 2012;7(e38612) doi: 10.1371/journal.pone.0038612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steevens J, Schouten LJ, Driessen ALC, Huysentruyt CJ, Keulemans YC, Goldbohm RA, van den Brandt PA. Toenail selenium status and the risk of Barrett's esophagus: The Netherlands Cohort Study. Cancer Causes Control. 2010;21:2259–2268. doi: 10.1007/s10552-010-9651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seale LA. Selenocysteine β-Lyase: Biochemistry, regulation and physiological role of the selenocysteine decomposition enzyme. Antioxidants (Basel) 2019;8(357) doi: 10.3390/antiox8090357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li CL, Nan KJ, Tian T, Sui CG, Liu YF. Selenoprotein P mRNA expression in human hepatic tissues. World J Gastroenterol. 2007;13:2363–2368. doi: 10.3748/wjg.v13.i16.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y, Liu Y, Zhao S, Zheng Z, Shen C, An L, Yuan Y. Large-scale analysis reveals a novel risk score to predict overall survival in hepatocellular carcinoma. Cancer Manag Res. 2018;10:6079–6096. doi: 10.2147/CMAR.S181396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D, Xu IMJ, Chiu DKC, Leibold J, Tse AP, Bao MH, Yuen VW, Chan CY, Lai RK, Chin DW, et al. Induction of oxidative stress through inhibition of thioredoxin reductase 1 is an effective therapeutic approach for hepatocellular carcinoma. Hepatology. 2019;69:1768–1786. doi: 10.1002/hep.30467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuo L, Xiang J, Pan X, Gao Q, Zhang G, Yang Y, Liang L, Xia J, Wang K, Tang N. PCK1 downregulation promotes TXNRD1 expression and hepatoma cell growth via the Nrf2/Keap1 pathway. Front Oncol. 2018;8(611) doi: 10.3389/fonc.2018.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Q, Zhang G, Zheng Y, Yang Y, Chen C, Xia J, Liang L, Lei C, Hu Y, Cai X, et al. SLC27A5 deficiency activates NRF2/TXNRD1 pathway by increased lipid peroxidation in HCC. Cell Death Differ. 2020;27:1086–1104. doi: 10.1038/s41418-019-0399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu B, Meng W, Zeng X, Zhao H, Liu W, Zhang T. TXNRD1 is an unfavorable prognostic factor for patients with hepatocellular carcinoma. Biomed Res Int. 2017;2017(4698167) doi: 10.1155/2017/4698167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shang N, Wang X, Shu Q, Wang H, Zhao L. The functions of selenium and selenoproteins relating to the liver diseases. J Nanosci Nanotechnol. 2019;19:1875–1888. doi: 10.1166/jnn.2019.16287. [DOI] [PubMed] [Google Scholar]
- 40.Tarek M, Louka ML, Khairy E, Ali-Labib R, Zakaria Zaky D, Montasser IF. Role of microRNA-7 and selenoprotein P in hepatocellular carcinoma. Tumor Biol. 2017;39(1010428317698372) doi: 10.1177/1010428317698372. [DOI] [PubMed] [Google Scholar]
- 41.Hughes DJ, Duarte-Salles T, Hybsier S, Trichopoulou A, Stepien M, Aleksandrova K, Overvad K, Tjønneland A, Olsen A, Affret A, et al. Prediagnostic selenium status and hepatobiliary cancer risk in the European Prospective Investigation into Cancer and Nutrition cohort. Am J Clin Nutr. 2016;104:406–414. doi: 10.3945/ajcn.116.131672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DI Stasio M, Volpe MG, Colonna G, Nazzaro M, Polimeno M, Scala S, Castello G, Costantini S. A possible predictive marker of progression for hepatocellular carcinoma. Oncol Lett. 2011;2:1247–1251. doi: 10.3892/ol.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia Y, Dai J, Zhang LL, Xia H. Biological functions of selenium-binding protein 1 and its relationship with diseases. Prog Biochem Biophys. 2019;46:128–137. [Google Scholar]
- 44.Guariniello S, Colonna G, Raucci R, Costantini M, Di Bernardo G, Bergantino F, Castello G, Costantini S. Structure-function relationship and evolutionary history of the human selenoprotein M (SelM) found over-expressed in hepatocellular carcinoma. Biochim Biophys Acta. 2014;1844:447–456. doi: 10.1016/j.bbapap.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Na J, Jung J, Bang J, Lu Q, Carlson BA, Guo X, Gladyshev VN, Kim J, Hatfield DL, Lee BJ. Selenophosphate synthetase 1 and its role in redox homeostasis, defense and proliferation. Free Radic Biol Med. 2018;127:190–197. doi: 10.1016/j.freeradbiomed.2018.04.577. [DOI] [PubMed] [Google Scholar]
- 46.Jaworska K, Gupta S, Durda K, Muszyńska M, Sukiennicki G, Jaworowska E, Grodzki T, Sulikowski M, Waloszczyk P, Wójcik J, et al. A low selenium level is associated with lung and laryngeal cancers. PLoS One. 2013;8(e59051) doi: 10.1371/journal.pone.0059051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epplein M, Burk RF, Cai Q, Hargreaves MK, Blot WJ. A prospective study of plasma Selenoprotein P and lung cancer risk among low-income adults. Cancer Epidemiol Biomarkers Prev. 2014;23:1238–1244. doi: 10.1158/1055-9965.EPI-13-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gresner P, Gromadzinska J, Jablonska E, Kaczmarski J, Wasowicz W. Expression of selenoprotein-coding genes SEPP1, SEP15 and hGPX1 in non-small cell lung cancer. Lung Cancer. 2009;65:34–40. doi: 10.1016/j.lungcan.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 49.An BC, Choi YD, Oh IJ, Kim JH, Park JI, Lee SW. GPx3-mediated redox signaling arrests the cell cycle and acts as a tumor suppressor in lung cancer cell lines. PLoS One. 2018;13(e0204170) doi: 10.1371/journal.pone.0204170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh IJ, Kim HE, Song SY, Na KJ, Kim KS, Kim YC, Lee SW. Diagnostic value of serum glutathione peroxidase 3 levels in patients with lung cancer. Thorac Cancer. 2014;5:425–430. doi: 10.1111/1759-7714.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi JY, An BC, Jung IJ, Kim JH, Lee SW. MiR-921 directly downregulates GPx3 in A549 lung cancer cells. Gene. 2019;700:163–167. doi: 10.1016/j.gene.2019.02.086. [DOI] [PubMed] [Google Scholar]
- 52.Chen G, Wang H, Miller CT, Thomas DG, Gharib TG, Misek DE, Giordano TJ, Orringer MB, Hanash SM, Beer DG. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. J Pathol. 2004;202:321–329. doi: 10.1002/path.1524. [DOI] [PubMed] [Google Scholar]
- 53.Marciel MP, Khadka VS, Deng Y, Kilicaslan P, Pham A, Bertino P, Lee K, Chen S, Glibetic N, Hoffmann FW, et al. Selenoprotein K deficiency inhibits melanoma by reducing calcium flux required for tumor growth and metastasis. Oncotarget. 2018;9:13407–3422. doi: 10.18632/oncotarget.24388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong D, Kim TS, Chung YW, Lee BJ, Kim IY. Selenoprotein W is a glutathione-dependent antioxidant in vivo. FEBS Lett. 2002;517:225–228. doi: 10.1016/s0014-5793(02)02628-5. [DOI] [PubMed] [Google Scholar]
- 55.Xu X, Huang L, Chan CH, Yu T, Miao R, Liu C. Assessing the clinical utility of genomic expression data across human cancers. Oncotarget. 2016;7:45926–45936. doi: 10.18632/oncotarget.10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marciel MP, Hoffmann PR. Molecular mechanisms by which Selenoprotein K regulates immunity and cancer. Biol Trace Elem Res. 2019;192:60–68. doi: 10.1007/s12011-019-01774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min SY, Kim HS, Jung EJ, Do Jee C, Kim WH. Prognostic significance of glutathione peroxidase 1 (GPX1) down-regulation and correlation with aberrant promoter methylation in human gastric cancer. Anticancer Res. 2012;32:3169–3175. [PubMed] [Google Scholar]
- 58.Peng DF, Hu TL, Schneider BG, Chen Z, Xu ZK, El-Rifai W. Silencing of glutathione peroxidase 3 through DNA hypermethylation is associated with lymph node metastasis in gastric carcinomas. PLoS One. 2012;7(e46214) doi: 10.1371/journal.pone.0046214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lan X, Xing J, Gao H, Li S, Quan L, Jiang Y, Ding S, Xue Y. Decreased expression of selenoproteins as a poor prognosticator of gastric cancer in humans. Biol Trace Elem Res. 2017;178:22–28. doi: 10.1007/s12011-016-0908-8. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Dong W, Lin J. Reduced selenium-binding protein 1 is associated with poor survival rate in gastric carcinoma. Med Oncol. 2011;28:481–487. doi: 10.1007/s12032-010-9482-7. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q, Gong L, Dong R, Qiao Q, He XL, Chu YK, Du XL, Yang Y, Zang L, Nan J, et al. Tissue microarray assessment of selenoprotein P expression in gastric adenocarcinoma. J Int Med Res. 2009;37:169–174. doi: 10.1177/147323000903700120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.