Abstract
The unfolded protein response (UPR) is an adaptive response that is implicated in multiple metabolic pathologies, including hepatic steatosis. In the present study, we analyzed publicly available RNAseq data to explore how the execution of the UPR is orchestrated in specimens that exhibit hepatocyte ballooning, a landmark feature of steatosis. By focusing on a panel of well-established UPR genes, we assessed how the UPR is coordinated with the whole transcriptome in specimens with or without hepatocyte ballooning. Our analyses showed that neither average levels nor correlation in expression between major UPR genes such as HSPA5 (BiP/GRP78), HSP90b1 (GRP94), or DDIT3 (CHOP) is altered in different groups. However, a panel of transcripts depending on the stringency of the analysis ranged from 16 to 372 lost its coordination with HSPA5, the major UPR chaperone, when hepatocyte ballooning occurred. In 13 genes, the majority of which is associated with metabolic processes, and the coordination with the HSPA5 was reversed from positive to negative in livers with ballooning hepatocytes. In order to examine if during ballooning, UPR genes abolish established and acquire novel functionalities, we performed gene ontology analyses. These studies showed that among the various UPR genes interrogated, only DDIT3 was not associated with conventional functions linked to endoplasmic reticulum stress during ballooning, while HSPA90b1 exhibited the highest function retention between the specimens with or without ballooning. Our results challenge conventional notions on the impact of specific genes in disease and suggest that besides abundance, the mode of coordination of UPR may be more important for disease development.
Electronic supplementary material
The online version of this article (10.1007/s12192-020-01132-x) contains supplementary material, which is available to authorized users.
Keywords: Liver, ER stress, Correlation network, Pathogenesis
Introduction
During an individual’s lifetime and in response to certain environmental and other challenges, the cells continuously adapt their transcriptional program in order to attain homeostasis, adequate physiological performance, and normal function (van Dam et al. 2018). In disease, homeostasis is abolished, and this is associated with organ dysfunction and ultimately with the onset of pathology. A classic example of a pathology linked to loss of homeostasis is hepatic steatosis or fatty liver disease. Hepatic steatosis refers to the accumulation of lipids in the liver, typically exceeding 5% of liver weight or when 5% of hepatocytes contain lipid vacuoles without this being associated with excess alcohol intake, viral infection, or drug treatments (Nassir et al. 2015); can be the result of excessive lipid uptake from the liver, enhanced lipogenesis, or reduced clearance; and affects about 33% of adult population in the USA (Mehta et al. 2008).
Hepatic steatosis can progress to nonalcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and hepatocellular carcinoma. Ballooning degeneration of hepatocytes represents a landmark for the transition of steatosis to NASH and describes a special form of hepatocyte degeneration at which the cells exhibit characteristic swelling and enlargement (Caldwell et al. 2010; Lackner et al. 2008).
A major adaptive signaling pathway that is responsible for tissue homeostasis is the unfolded protein response (UPR) that follows stress of endoplasmic reticulum (ER) (Frakes and Dillin 2017). At conditions of excessive accumulation of misfolded and unfolded proteins, the UPR is initiated to resolve ER stress by triggering a precisely orchestrated response involving overexpression of chaperones inhibition of protein translation and eradication of severely misfolded proteins (Frakes and Dillin 2017; Zhu and Lee 2015). This response is guided by the activation of 3 intracellular receptors, IRE1, ATF6, and PERK, each of which defines the 3 major UPR branches that are associated by unique and redundant cellular activities (Frakes and Dillin 2017).
UPR is involved in the onset of various pathologies such as cancer, diabetes, and neurodegeneration (Back and Kaufman 2012; Hetz and Saxena 2017; Cheretis et al. 2006; Hetz et al. 2013). Several lines of evidence also indicate that deregulation of the UPR is implicated in the development of hepatic steatosis (Han and Kaufman 2016; Song and Malhi 2019). In the majority of the studies, the phenotype of animals subjected to genetic ablation of UPR targets was investigated and was consistent with the protective role of various UPR-associated genes against the development of fatty liver disease (Yamamoto et al. 2010; Ji et al. 2011; Chen et al. 2014a; Chen et al. 2014b; Kammoun et al. 2009; Rutkowski et al. 2008). These studies offer valuable mechanistic insights on how specific UPR genes are associated with the disease. However, how the UPR is collectively regulated beyond the loss of function of predetermined gene targets, in hepatic steatosis, is poorly understood. Furthermore, which UPR branch or specific UPR-associated gene is primarily involved in hepatic steatosis remains unclear.
The fact that the UPR involves the concomitant activation of specific signaling branches that regulate overlapping yet distinct cellular functions and biochemical cascades provides a unique model system to explore how transcriptional reprogramming operates during disease pathogenesis (Gardner et al. 2013). By using outbred deer mice as a model, we recently showed that the UPR is highly variable among individuals and this variability is associated with the onset of hepatic steatosis (Havighorst et al. 2019). Noteworthy, despite this variability, a high degree of coordination is maintained in the expression of different chaperones in different individuals (Havighorst et al. 2019). This coordination in gene expression was retained beyond specific chaperones, to the whole transcriptome, in a manner that specific functions of the UPR attributed to particular UPR branches could be predicted (Zhang et al. 2019). These notions led us to hypothesize that analysis of the profile of coordination of gene expression, as opposed to the levels of expression, between the UPR and the whole transcriptome could unveil important clues regarding the pathogenesis of hepatic steatosis. To that end, we analyzed a previously published comprehensive RNAseq data set of human liver specimens that had been characterized for a set of features associated with fatty liver disease (Hoang et al. 2019). We focused particularly on the comparison between the specimens that developed hepatocyte ballooning as opposed to those that did not and explored the coordination in the expression of a roster or eight UPR genes that have been recognized as UPR targets. Those genes can be used to monitor the profile of the UPR and included HSPA5 (BiP/GRP78) (Ji et al. 2011; Chen et al. 2014a; Luo et al. 2006), HSP90B1 (GRP94) (Chen et al. 2014b), DDIT3 (CHOP) (Fornace et al. 1989), ATF4 (Fusakio et al. 2016), calnexin (CANX) (Guérin et al. 2008), EDEM2 (Mast et al. 2005), DNAJB9 (Lee et al. 2003), and PDIA4 (Joshi-Tope et al. n.d.). Our results suggest that while expression of these UPR markers remained unaltered between groups, the transcriptome correlated with them was distinct. Furthermore, our analyses unveiled a roster of transcripts for which the mode of coordination showed unique profiles in the hepatocytes exhibiting ballooning degeneration and identified DDIT3 as the major regulator of this process.
Methods
Data retrieval and classification
RNAseq data were downloaded from GSE130970 (Hoang et al. 2019). Transcripts per million (TPM) data were used for the analyses. Specimens were divided into two groups according to the original authors’ (Hoang et al. 2019) classification, those exhibiting hepatocyte ballooning (grade 1 and 2) and those that did not (grade 0).
Analyses
Average values were analyzed and graphed by using the Prism 8 software (GraphPad, San Diego, CA). Association analyses were performed by using the Pearson’s test for the criteria described.
Results
UPR expression levels
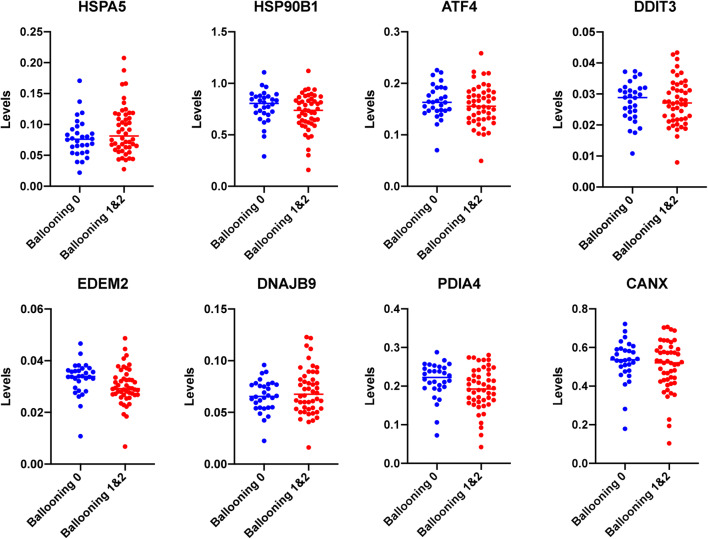
Seventy-eight specimens were included in this analysis that had been characterized before in terms of hepatocyte ballooning (Hoang et al. 2019). Among them, 30 were characterized as ballooning grade 0 and 48 specimens as ballooning grade 1 or 2 (Hoang et al. 2019). Following normalization with GAPDH levels, the expression of each of HSPA5, HSP90B1, DDIT3, ATF4, CANX, EDEM2, DNAJB9, and PDIA4 was evaluated and compared in the groups exhibiting ballooning degeneration and those that did not. As shown in Fig. 1, no significant difference between the groups for any of the genes tested was revealed. A trend for lower expression in EDEM2 and PDIA4 in the specimens exhibiting ballooning degeneration was observed but remained insignificant (P = 0.06 and P = 0.07, respectively, unpaired t test).
Fig. 1.
Hepatic expression of various UPR genes in human liver specimens exhibiting hepatocyte ballooning grade 0 or grades 1 and 2. Levels indicate arbitrary units normalized to GAPDH expression. Average expression for each group is indicated. For all genes, no difference between the groups was observed except for EDEM2 and PDIA4 at which moderately lower expression in specimens with ballooning degeneration was observed but remained insignificant (P = 0.06 and P = 0.07, respectively, unpaired t test)
Coordination between HSPA5 and different UPR targets is maintained during ballooning degeneration
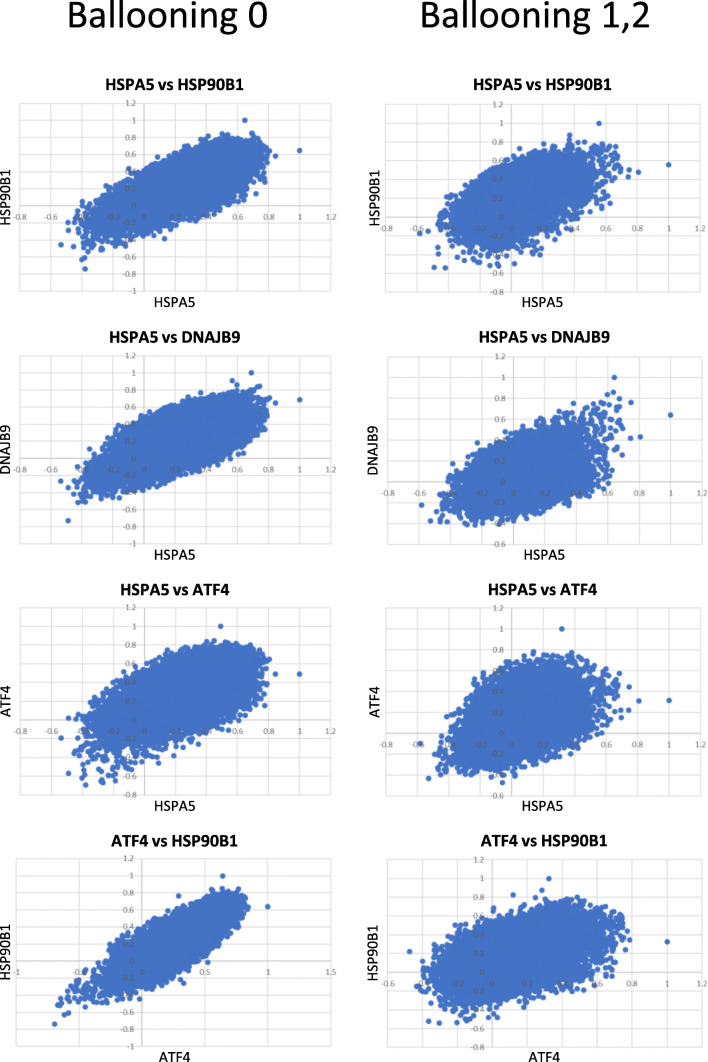
HSPA5 encodes for the chaperone BiP/GRP78 that is the master regulator of the response to ER stress since its dissociation from PERK, IRE1, and ATF6 triggers the execution of the UPR (Frakes and Dillin 2017; Gardner et al. 2013). Our earlier studies showed that HSPA5 expression is highly coordinated with the expression of different UPR-associated genes in fibroblasts from genetically diverse deer mice, irrespectively of the induction of the UPR (Havighorst et al. 2019). Furthermore, this coordination transcends individual genes and applies to the whole transcriptome (Zhang et al. 2019). In order to explore if this coordination is maintained in livers and whether it is preserved in samples that exhibit hepatocyte ballooning, we developed pairwise scatterplots depicting the correlation between HSPA5 and each of ATF4, DNAJB9, and HSP90b1 and the whole transcriptome. As shown in Fig. 2, in all pairwise comparisons, the correlation was maintained indicating appropriate execution of the UPR irrespectively of the ballooning degeneration. Similar trends were revealed with all UPR targets analyzed (data not shown) as well as between pairwise comparisons not involving HSPA5, such as between ATF4 and HSP90B1 (Fig. 2). Thus, we conclude that the development of ballooning does not compromise, at the transcriptome level, the overall coordination of the UPR.
Fig. 2.
Pairwise scatter plots indicating the P value (Pearson’s) in expression levels between each of HSPA5, ATF4, HSP90B1, and DNAJB9 and whole transcriptome at various combinations. A positive correlation was retained in both the groups without or with ballooning degeneration of hepatocytes
Some genes abolish coordination with HSPA5 during ballooning degeneration
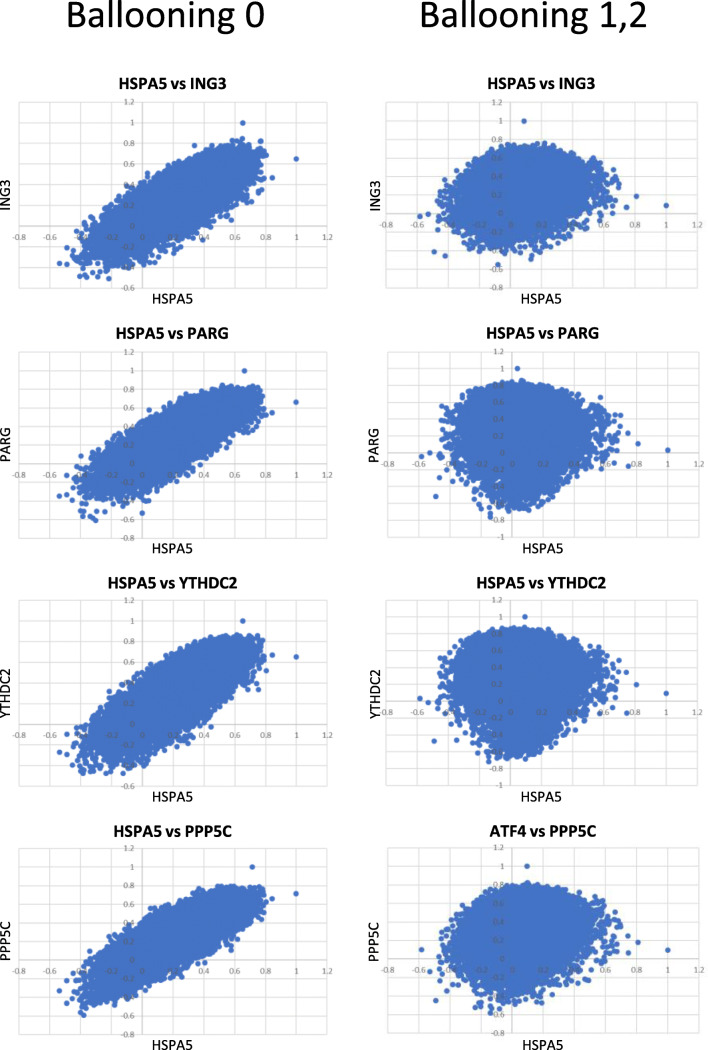
Despite the retention of the coordination between the UPR and the transcriptome, it is conceivable that the UPR-associated genes that are implicated in ballooning will exhibit positive correlation in the absence of ballooning but as soon as this pathology emerges, correlation will be abolished. To identify such genes, we calculated the correlation between HSPA5 and the transcriptome in the different groups and identified those that lost their coordination with HSPA5 in the ballooning group. Depending on the stringency of the filtering criteria applied, we identified a set of genes that ranged from 16 to 372 and for which correlation in expression with HSPA5 was positive in the samples without ballooning but was abolished during hepatocyte ballooning (Supplementary Table 1). This loss of correlation with HSPA5 and the corresponding genes extended to the whole transcriptome as depicted in Fig. 3, in the pairwise scatterplots at which the linear correlations in specimens without ballooning, were transformed to cyclical diagrams, typical when no association exists. Thus, for a subset of genes, the transcriptome associated with them is not associated with the UPR anymore when ballooning emerges.
Fig. 3.
Pairwise scatter plots depicting the P value (Pearson’s) in expression levels between HSPA5 and each of ING3, PARG, YTHDC2, and PPP5C. In these genes, the expression profile with HSPA5 showed positive correlation in the specimens without ballooning but no correlation in specimens with ballooning degeneration of hepatocytes
Genes for which coordination with HSPA5 exhibits opposite profiles during ballooning degeneration
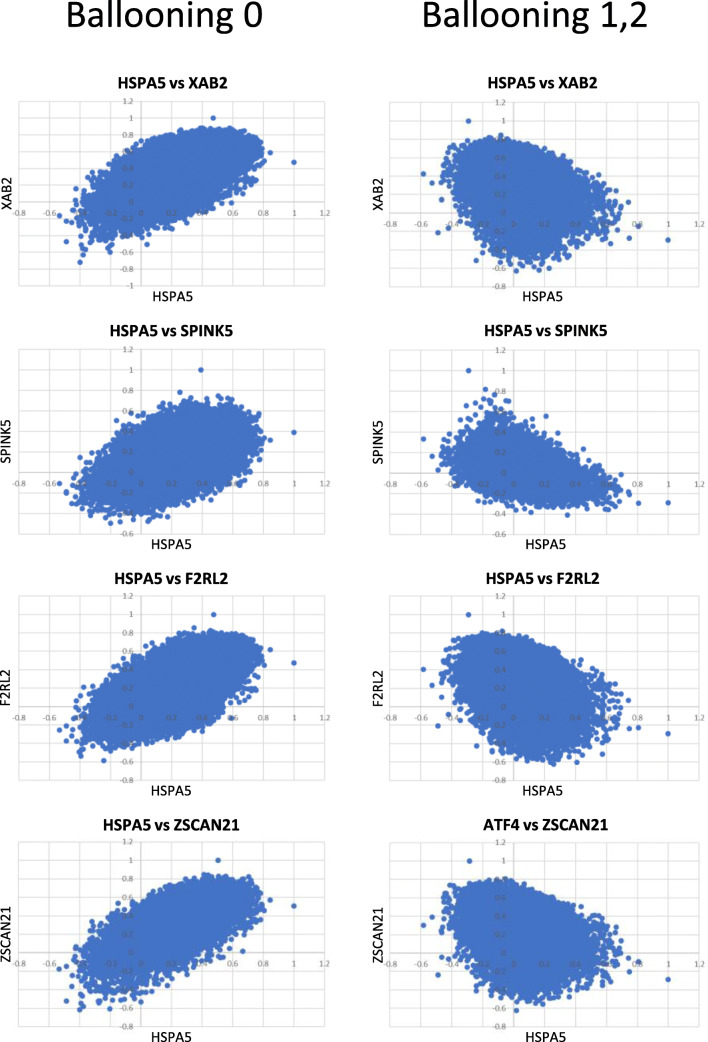
Although for most of the genes that did not retain the positive coordination recorded in the non-ballooning group, coordination was abolished in the specimens with ballooning degeneration; in 13 genes, the profile of coordination was reversed: These genes exhibited positive correlation with HSPA5 in the non-ballooning group but negative with the group having ballooning degeneration (Supplementary Table 2, Fig. 4). No genes exhibited the opposite trend, being negative in the ballooning and positive in the non-ballooning group. Interestingly, most of the genes for which correlation with HSPA5 reversed from positive to negative were associated with the regulation of metabolic processes according to REACTOME database (Joshi-Tope et al. n.d.).
Fig. 4.
Pairwise scatter plots depicting the P value (Pearson’s) in expression levels between HSPA5 and each of XAB2, SPINK5, F2RL2, and ZSCAN21. In these genes, correlation with the HSPA5-associated transcriptome was positive in the specimens without ballooning but reversed to negative in the specimens exhibiting ballooning degeneration of hepatocytes
The functions of DDIT3-associated transcriptome shift to regulation of cell cycle phase transition in ballooning hepatocytes
We have recently showed that by analyzing the transcriptome correlated with specific UPR genes using a gene ontology platform such as GOrilla (Eden et al. 2009; Eden et al. 2007), we can unveil functions that for a given context are specifically associated with these genes, such as endoplasmic reticulum-associated degradation or apoptosis (Zhang et al. 2019). Based on this notion, we explored if and at which extent the UPR genes selected for analysis in this study retained their functionality as UPR-associated genes in the specimens without and the specimens with ballooning degeneration. To that end, we calculated the function retention index (FRI) and the function acquisition index (FAI). FRI reflects the ratio of the different functions in the non-ballooning group that were retained in the ballooning group. FAI reflects the ratio of the novel functions in the ballooning group that were absent from the non-ballooning group. We postulated that as pathology emerges, only the genes that are mechanistically linked to this pathology will exhibit plasticity in their transcriptional program, will abolish physiological functions, and will acquire novel functions. As expected, for all genes tested in the group without ballooning degeneration, the predicted functions frequently involved those linked to some aspects of the UPR. Many of these were retained in the ballooning group (expressed by FRI), while novel functions were also occasionally acquired (expressed by FAI). For most of the genes tested, the functions that characterized the associated transcriptome during conditions without ballooning were retained during ballooning degeneration, implying that their functionality remained intact and associated with their operation as UPR regulators (Fig. 5). For DDIT3 though, while its associated transcriptome predicted protein localization and transport in the specimens without ballooning, in the specimens with ballooning degeneration, this was abolished, and novel functions were introduced that in their majority were associated with cell cycle phase transition (Supplementary Tables 3 and 4). Since, for the genes tested including DDIT3 (CHOP), no considerable differences in expression levels were identified between groups, we hypothesize that differences in transcriptome represent truly transcriptional reprogramming. To that end, at different conditions, the same transcription factor, namely, DDIT3, is correlated with transcripts exhibiting distinct activities and functionality, despite overall expression levels are retained.
Fig. 5.
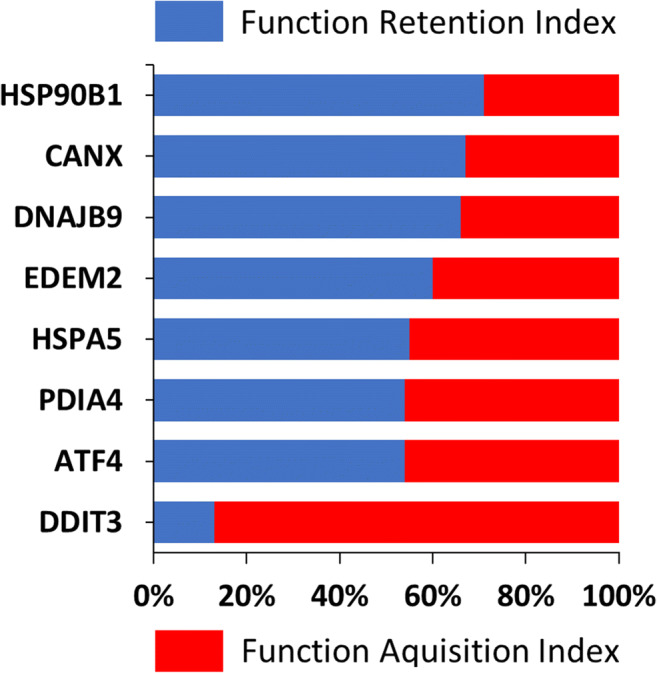
Function retention index (FRI) and function acquisition index (FAI) for each of HSP90B1, CANX, DNAJB9, EDEM2, HSPA5, PDIA4, ATF4, and DDIT3. FRI reflects the ratio of the different functions in the non-ballooning that were retained in the ballooning group. FAI reflects the ratio of the novel functions in the ballooning group that were absent from the non-ballooning group. To identify the corresponding functions, the transcriptome that was correlated significantly (P < 0.05, Pearson’s) with each of these genes was analyzed with the ShinyGO v0.61 platform (100 of most significant terms were analyzed)
Discussion
The UPR constitutes a highly versatile adaptive response that attains tissue homeostasis when unfolded and misfolded proteins accumulate. Deregulation of the UPR is involved in multiple pathologies including metabolic disease, cancer, aging, and neurodegeneration. Experimental evidence obtained by laboratory animals that had been subjected to loss of expression in specific genes has provided insights regarding how the particular targets are linked to specific phenotypes and metabolic pathologies. Yet, despite their power in addressing mechanistic aspects of disease pathogenesis, the relative contribution of various UPR-related transcripts in metabolic disease remains elusive. To address this, we have analyzed publicly available RNA sequencing data from human liver samples in terms of UPR coordination, at the whole transcriptome level. The phenotype we focused on was the ballooning degeneration of hepatocytes, a landmark for the transition of steatosis to NASH. For our analyses, we considered not the relative levels of expression of specific genes in different groups but rather the mode of coordination of the whole transcriptome with respect to different UPR-associated transcripts. The premise of this approach was that the potential implication of specific genes in disease would be reflected on how tightly the transcriptome associated with these genes would maintain its coordination in different experimental groups. Such approach not only would reflect the robustness of UPR coordination but will also unveil genes that deviate and presumably are causatively linked to the phenotype of interest.
While conventional analyses failed to reveal major differences in UPR genes between the specimens that exhibited and those that did not, ballooning degeneration, several trends were revealed when coordination was evaluated. First, we noted that coordination of major UPR targets such as HSPA5, HSP90B1, and ATF4 is maintained in the different groups which in turn implies that UPR is responsive and functional in hepatocyte ballooning and, therefore, as such cannot constitute a therapeutic target.
By focusing on the transcriptome that abolishes its coordination with HSPA5/BiP/GRP78, the master regulator of the UPR, we were able to identify genes that were associated with BiP/GRP78 only in the specimens without ballooning but not in those exhibiting the ballooning degeneration. Depending on how stringent the criteria for establishing abolishment of coordination were, these genes ranged from 9 to 216. When highest stringency conditions were used, these genes included PPP5C, CEP83, ZNF714, ENOX2, SMTN, TUBB3, AKAP13, KIAA0754, and CRCT1 which behave as UPR targets (positive correlation with HSPA5) in the absence of ballooning but abolish this responsiveness when ballooning develops. In addition to those, we have identified a set of 17 genes that behaved as UPR targets in both specimens without and those exhibiting ballooning degeneration; however, this association was positive in the former and negative in the latter. These genes according to REACTOME (Joshi-Tope et al. n.d.) included 5 with yet undefined function and 6, namely, OPLAH, ADPRM, MIGA2, ZNF181, C1GALT1, and GOLIM4, that were associated with metabolic processes. The fact that for these genes the positive regulation becomes negative implies that they are activated by UPR under physiological conditions but inhibited by UPR when ballooning occurs.
Probably, the most intriguing finding of the present analysis is related to the assessment of specific UPR-associated transcriptomes for function. In most of the cases, the transcriptomes associated with the UPR genes assessed predicted specific functions relevant to protein processing and were retained during ballooning degeneration. Thus, even when the context changes, these genes continued operating as components of the ER stress response. For DDIT3, however, while functions related to protein localization and transport were predicted in the absence of ballooning degeneration, none of these were retained in ballooning hepatocytes and novel functions were acquired. These functions were now associated with the regulation of cell cycle phase transition. DDIT3 (DNA damage induced transcript 3), also known as GADD153 (growth arrest and DNA damage 153) or CHOP (C/EBP homologous protein), encodes a transcription factor belonging to the CCAAT/enhancer binding protein (C/EBP) family (Ramji and Foka 2002). It is activated by cellular stress conditions and induces cell cycle arrest and apoptosis (Barone et al. 1994; Han et al. 2013). CHOP regulates the cell cycle regulator p21/waf1 during ER stress facilitating the commitment of cells into a proapoptotic program (Mihailidou et al. 2010; Tang et al. 2019). CHOP deletion in pancreatic β cells of mouse diabetes models protected from apoptosis and improved β cell survival (Oyadomari et al. 2002; Song et al. 2008). Also, hepatocyte cell lines deficient in CHOP expression were protected from palmitate-induced apoptosis (Cazanave et al. 2010; Pfaffenbach et al. 2010). Interestingly, the loss of CHOP in mice led to the development of significant steatohepatitis and decreased macrophage apoptosis under conditions of FFA-induced ER stress (Malhi et al. 2013).
How relevant to human disease is complete ablation of CHOP expression is debatable considering that in human specimens, CHOP expression remains unaltered (Fig. 1). Notwithstanding this notion, according to the results of the present analysis, CHOP activity appears to be highly relevant, considering that the functions of CHOP-associated transcriptome totally shift in specimens with different ballooning status (Supplementary Tables 3 and 4) despite the similar levels of DDIT3 expression (Fig. 1). Several questions are generated from these observations. Does the shift of the functions of DDIT3-associated transcriptome occur before the onset of hepatocellular ballooning, and in fact, is hepatocellular ballooning a consequence of this shift? Is CHOP a protective or destructive factor against the development of hepatocellular ballooning in human? What factors cause the functional shift of DDIT3-associated transcriptome? In view of the fact that CHOP is a transcription factor, it is plausible that the shift of CHOP-associated transcriptome during ballooning degeneration is not a direct consequence of differential CHOP expression but rather of its engagement in transcription complexes with different co-factors that alter CHOP’s specificity. The fact that its adaptive UPR-related activity is substituted by its cell cycle regulatory activity appears highly relevant in the context of degeneration.
Collectively, the present study identifies UPR as a major regulator of hepatic steatosis, not in terms of abundance of specific transcripts but rather with regard to alterations of the UPR-associated transcriptome during disease development. Furthermore, our strategy provides an example of how analysis of coordination in gene expression may illuminate unforeseen aspects of disease pathogenesis.
Electronic supplementary material
(DOCX 19 kb)
Authors’ contributions
YZ performed the analyses, interpreted results, and edited and approved MS; IC interpreted the results and edited and approved MS; and HK analyzed the data, interpreted the results, and drafted and approved MS.
Funding information
This study was supported by NSF (Award Number: 1736150).
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interest.
Ethics approval and consent to participate
N/A
Consent for publication
N/A
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Back SH, Kaufman RJ. Endoplasmic reticulum stress and type 2 diabetes. Annu Rev Biochem. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone MV, Crozat A, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G(1)/S arrest. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- Caldwell S, Ikura Y, Dias D, Isomoto K, Yabu A, Moskaluk C, Pramoonjago P, Simmons W, Scruggs H, Rosenbaum N, Wilkinson T, Toms P, Argo CK, al-Osaimi AMS, Redick JA. Hepatocellular ballooning in NASH. J Hepatol. 2010;53(4):719–723. doi: 10.1016/j.jhep.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazanave SC, Elmi NA, Akazawa Y, Bronk SF, Mott JL, Gores GJ. CHOP and AP-1 cooperatively mediate PUMA expression during lipoapoptosis. Am J Physiol Gastrointest Liver Physiol. 2010;299:G236–G243. doi: 10.1152/ajpgi.00091.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WT, Zhu G, Pfaffenbach K, Kanel G, Stiles B, Lee AS. GRP78 as a regulator of liver steatosis and cancer progression mediated by loss of the tumor suppressor PTEN. Oncogene. 2014;33(42):4997–5005. doi: 10.1038/onc.2013.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WT, Tseng CC, Pfaffenbach K, Kanel G, Luo B, Stiles BL, Lee AS. Liver-specific knockout of GRP94 in mice disrupts cell adhesion, activates liver progenitor cells, and accelerates liver tumorigenesis. Hepatology. 2014;59(3):947–957. doi: 10.1002/hep.26711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheretis C, Dietrich F, Chatzistamou I, Politi K, Angelidou E, Kiaris H, Mkrtchian S, Koutselini H. Expression of ERp29, an endoplasmic reticulum secretion factor in basal-cell carcinoma. Am J Dermatopathol. 2006;28(5):410–412. doi: 10.1097/01.dad.0000211521.49810.ac. [DOI] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007;3(3):e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace AJ, Nebert DW, Hollander MC, Luethy JD, Papathanasiou M, Fargnoli J, Holbrook NJ. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol. 1989;9:4196–4203. doi: 10.1128/MCB.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes AE, Dillin A. The UPRER: sensor and coordinator of organismal homeostasis. Mol Cell. 2017;66(6):761–771. doi: 10.1016/j.molcel.2017.05.031. [DOI] [PubMed] [Google Scholar]
- Fusakio ME, Willy JA, Wang Y, Mirek ET, al Baghdadi RJ, Adams CM, Anthony TG, Wek RC. Transcription factor ATF4 directs basal and Joshi-Tope G, Vastrik I, Gopinath GR, Matthews L, Schmidt E, Gillespie M, D’Eustachio P, Jassal B, Lewis S, Wu G, Birney E, Stein L. The genome knowledgebase: a resource for biologists and bioinformaticists. Cold Spring Harb Symp Quant Biol.68:237-43metabolism in the liver. Mol Biol Cell. 2016;27(9):1536–1551. doi: 10.1091/mbc.E16-01-0039. [DOI] [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM. Walter P Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5(3):a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin R, Arseneault G, Dumont S, Rokeach LA. Calnexin is involved in apoptosis induced by endoplasmic reticulum stress in the fission yeast. Mol Biol Cell. 2008;19(10):4404–4420. doi: 10.1091/mbc.E08-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Kaufman RJ. The role of ER stress in lipid metabolism and lipotoxicity. J Lipid Res. 2016;57(8):1329–1338. doi: 10.1194/jlr.R067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, Kilberg MS, Sartor MA, Kaufman RJ. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havighorst A, Zhang Y, Farmaki E, Kaza V, Chatzistamou I, Kiaris H. Differential regulation of the unfolded protein response in outbred deer mice and susceptibility to metabolic disease. Dis Model Mech. 2019;12(2):dmm037242. doi: 10.1242/dmm.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Saxena S. ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol. 2017;13(8):477–491. doi: 10.1038/nrneurol.2017.99. [DOI] [PubMed] [Google Scholar]
- Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov. 2013;12(9):703–719. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- Hoang SA, Oseini A, Feaver RE, Cole B, Asgharpour A, Vincent R, Siddiqui M, Lawson MJ, Day NC, Taylor JM, Wamhoff BR, Mirshahi F, Contos MJ, Idowu M, Sanyal AJ. Distinct molecular profiles of nonalcoholic fatty liver disease. Sci Rep. 2019;9(1):12541. doi: 10.1038/s41598-019-48746-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Kaplowitz N, Lau MY, Kao E, Petrovic LM, Lee AS. Liver-specific loss of glucose-regulated protein 78 perturbs the unfolded protein response and exacerbates a spectrum of liver diseases in mice. Hepatology. 2011;54(1):229–239. doi: 10.1002/hep.24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Tope G, Vastrik I, Gopinath GR, Matthews L, Schmidt E, Gillespie M, D’Eustachio P, Jassal B, Lewis S, Wu G, Birney E, Stein L The genome knowledgebase: a resource for biologists and bioinformaticists. Cold Spring Harb Symp Quant Biol 68:237–243 [DOI] [PubMed]
- Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferré P, Foufelle F. J Clin Invest. 2009 May;119(5):1201-15. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119(5):1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner C, Gogg-Kamerer M, Zatloukal K, Stumptner C, Brunt EM, Denk H (2008) Ballooned hepatocytes in steatohepatitis: the value of keratin immunohistochemistry for diagnosis. J Hepatol 48(5):821–828. https://doi.org/10.1016/j.jhep.2008.01.026.Epub2008Feb 22. [DOI] [PubMed]
- Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23(21):7448–7459. https://doi.org/10.1128/mcb.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed]
- Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26(15):5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Kropp EM, Clavo VF, Kobrossi CR, Han JS, Mauer AS, Yong J, Kaufman RJ. C/EBP homologous protein-induced macrophage apoptosis protects mice from steatohepatitis. J Biol Chem. 2013;288:18624–18642. doi: 10.1074/jbc.M112.442954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast SW, Diekman K, Karaveg K, Davis A, Sifers RN, Moremen KW. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 2005;15:421–436. doi: 10.1093/glycob/cwi014. [DOI] [PubMed] [Google Scholar]
- Mehta SR, Thomas EL, Bell JD, Johnston DG, Taylor-Robinson SD. Non-invasive means of measuring hepatic fat content. World J Gastroenterol. 2008;14(22):3476–3483. doi: 10.3748/wjg.14.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailidou C, Papazian I, Papavassiliou AG, Kiaris H. CHOP-dependent regulation of p21/waf1 during ER stress. Cell Physiol Biochem. 2010;25:761–766. doi: 10.1159/000315096. [DOI] [PubMed] [Google Scholar]
- Nassir F, Rector RS, Hammoud GM, Ibdah JA. Pathogenesis and prevention of hepatic steatosis. Gastroenterol Hepatol (N Y) 2015;11(3):167–175. [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109:525–532. doi: 10.1172/JCI0214550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenbach KT, Gentile CL, Nivala AM, Wang D, Wei Y, Pagliassotti MJ. Linking endoplasmic reticulum stress to cell death in hepatocytes: roles of C/EBP homologous protein and chemical chaperones in palmitate-mediated cell death. Am J Physiol Endocrinol Metab. 2010;298:E1027–E1035. doi: 10.1152/ajpendo.00642.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramji D, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/bj20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15(6):829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MJ, Malhi H. The unfolded protein response and hepatic lipid metabolism in non alcoholic fatty liver disease. Pharmacol Ther. 2019;13:107401. doi: 10.1016/j.pharmthera.2019.107401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BB, Scheuner D, Ron D, Pennathur S, Kaufman RJ. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J Clin Investig. 2008;118:3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q et al (2019) Cell Prolif e12706 [DOI] [PMC free article] [PubMed]
- van Dam S, Võsa U, van der Graaf A, Franke L, de Magalhães JP. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 2018;19(4):575–592. doi: 10.1093/bib/bbw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21(17):2975–2986. doi: 10.1091/mbc.e09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lucius MD, Altomare D, Havighorst A, Farmaki E, Chatzistamou I, Shtutman M, Kiaris H. Coordination analysis of gene expression points to the relative impact of different regulators during endoplasmic reticulum stress. DNA Cell Biol. 2019;38(9):969–981. doi: 10.1089/dna.2019.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Lee AS. Role of the unfolded protein response, GRP78 and GRP94 in organ homeostasis. J Cell Physiol. 2015;230(7):1413–1420. doi: 10.1002/jcp.24923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 19 kb)