Abstract
In their role as molecular chaperones, heat shock proteins (Hsps) mediate protein folding thereby mitigating cellular damage caused by physiological and environmental stress. Nauplii of the crustacean Artemia franciscana respond to heat shock by producing Hsps; however, the effects of cold shock on Hsp levels in A. franciscana have not been investigated previously. The effect of cold shock at 1 °C followed by recovery at 27 °C on the amounts of ArHsp90, Hsp70, ArHsp40, and ArHsp40-2 mRNA and their respective proteins in A. franciscana nauplii was examined by quantitative PCR (qPCR) and immunoprobing of western blots. The same Hsp mRNAs and proteins were also quantified during incubation of nauplii at their optimal growth temperature of 27 °C. qPCR analyses indicated that the abundance of ArHsp90, Hsp70, and ArHsp40 mRNA remained relatively constant during both cold shock and recovery and was not significantly different compared with levels at optimal temperature. Western blotting revealed that ArHsp90, ArHsp40, and ArHsp40-2 were generally below baseline, but at detectable levels during the 6 h of cold shock, and persisted in early recovery stages before declining. Hsp70 was the only protein that remained constant in quantity throughout cold shock and recovery. By contrast, all Hsps declined rapidly during 6 h when nauplii were incubated continuously at 27 °C optimal temperature. Generally, the amounts of ArHsp90, ArHsp40, and ArHsp40-2 were higher during cold shock/recovery than those during continuous incubation at 27 °C. Our data support the conclusion that low temperature preserves Hsp levels, making them available to assist in protein repair and recovery after cold shock.
Electronic supplementary material
The online version of this article (10.1007/s12192-020-01147-4) contains supplementary material, which is available to authorized users.
Keywords: Cold shock, Molecular chaperones, qPCR, Western blotting, Nauplii, Brine shrimp
Introduction
An organism’s ability to tolerate extreme temperatures determines its abundance and geographical distribution by affecting growth, motility, and reproduction (Ramløv 2000; Huang et al. 2007; Bale and Hayward 2010). Changes in temperature beyond physiologically normal levels adversely affect metabolism, gene expression, and protein synthesis, ultimately decreasing the overall performance of an organism within its environment (Ramløv 2000; Teets and Denlinger 2013; Overgaard et al. 2014). At high and low temperatures, such effects may damage lipids, proteins, and DNA and disrupt cellular stability (Lesser 2006; Lalouette et al. 2011). An important response to cellular stress is the expression of heat shock proteins (Hsps), along with other stress response factors that help preserve proteostasis (Lindquist and Craig 1988; Morimoto 1998; Ostankovitch and Buchner 2015).
Heat shock proteins (Hsp) are molecular chaperones synthesized during exposure to and recovery from physiological or environmental stress such as temperature, anoxia, and UV exposure, as well as during diapause, a state of dormancy typical of arthropods (Lindquist and Craig 1988; Denlinger 2002; Colinet and Hoffmann 2012; Kim et al. 2013). In relation to temperature and cold stress, Hsps have been shown as the first line of defense in insects, with only a few studies in crustaceans (Colinet et al. 2007, 2010; Rinehart et al. 2007; Koštál and Tollarová-Borovanská 2009; Li et al. 2009a; Zhang et al. 2011, 2018; Teets et al. 2012; Kim et al. 2013; Loc et al. 2013; King and MacRae 2015; Chen et al. 2018b; Wu et al. 2018). Hsps are divided into families based on molecular mass and amino acid sequence (King and MacRae 2015; Ostankovitch and Buchner 2015; Nillegoda et al. 2018); they generally range in molecular mass from ~ 15 to 110 kDa.
The Hsp90 (Schopf et al. 2017; Genest et al. 2019), Hsp70 (Fernández-Fernández et al. 2017; Genest et al. 2019; Mayer and Gierasch 2019), Hsp60 (Lopez et al. 2015; Skjærven et al. 2015), and cochaperone Hsp40 (Qiu et al. 2006; Li et al. 2009b) families are each composed of subfamilies depending on cellular localization either in the cytoplasm, mitochondria, or endoplasmic reticulum. Hsp90, Hsp70, and Hsp60 require ATP to participate in protein folding and degradation (Clare and Saibil 2013; Kim et al. 2013), whereas Hsp40 acts as a cochaperone in the ATPase activity of Hsp70 (Alderson et al. 2016; Nillegoda et al. 2015, 2017, 2018; Qiu et al. 2006). Conversely, the family of small heat shock proteins (sHsps), characterized by an α-crystallin domain, function independently of ATP and interact with cellular proteins to prevent their irreversible denaturation (Strauch and Haslbeck 2016; Haslbeck et al. 2019). Ultimately, the activation of Hsps protects organisms from death caused by cellular injury and apoptosis (Angelidis et al. 1991; Morimoto 1998; Beere 2004; Powers et al. 2009; King and MacRae 2015; Mahat et al. 2016; Sottile and Nadin 2018).
Artemia franciscana, an extremophile crustacean, is distributed globally excluding Antarctic regions, and it dominates hypersaline waters (Ruebhart et al. 2008; Muñoz and Pacios 2010; Eimanifar et al. 2014). A. franciscana embryos undergo either oviparous or ovoviviparous development, leading to diapause-destined cysts or swimming nauplii (larvae), respectively (Liang and MacRae 1999; MacRae 2003, 2016). Thermotolerance, the survival of an otherwise lethal heat shock following exposure to a sublethal temperature, occurs in Artemia nauplii (Clegg et al. 2000; Frankenberg et al. 2000; Sung et al. 2008), as does cross-tolerance, the increased resistance to stress after exposure to a different stress (Sung et al. 2008, 2011; Norouzitallab et al. 2015). Artemia adults, but not cysts and nauplii, upregulate the sHsp ArHsp22 upon heat shock (Qiu and MacRae 2008b). By contrast, Hsp90 and Hsp70 exhibit different induction patterns and cell localizations when heat shocked preemergence cysts, larvae, and adults are compared (Frankenberg et al. 2000; Willsie and Clegg 2001). Very little is known, however, about the influence of cold shock on Hsps in A. franciscana and, for that matter, in any crustacean. With this in mind, the response of ArHsp90, Hsp70, ArHsp40, and ArHsp40-2 to short-term cold shock and recovery was investigated in second instar nauplii of A. franciscana.
Materials and methods
Culturing of A. franciscana nauplii
Oviparously produced, quiescent, and desiccated A. franciscana cysts from the Great Salt Lake (INVE Aquaculture, Inc., Ogden, UT, USA) were hydrated overnight in distilled water at 4 °C with constant aeration. Hydrated cysts were collected by suction filtration, and 5 g of cysts was incubated in 500 ml 33.0 ppt, filtered, and UV-treated sea water (Halifax Harbour, NS, Canada) for 20 h at 27 °C with shaking at 150 rpm. Second instar nauplii that hatched from cysts were used immediately in the experimental procedures described below. Second instar nauplii have begun mRNA transcription and the synthesis of essential proteins such as heat shock proteins (McClean and Warner 1971; Conte et al. 1973; Miller and McLennan 1988).
This research was performed in accordance with the ethical guidelines provided by the Canadian Council on Animal Care (CCAC). The University Committee on Laboratory Animals (UCLA) of Dalhousie University approved the research under assigned Protocol Number 117-36.
Cold shock and recovery of A. franciscana nauplii
A. franciscana nauplii were incubated for 6 h in 500 ml seawater with aeration, held in a water bath maintained at 1 °C (the lowest temperature at which all animals survived), and then immediately allowed to recover at 27 °C for 6 h. The duration of experiments was 12 h with nauplii harvested before cold shock (0 h), as well as during cold shock and recovery, as noted in Fig. 1. In parallel experiments, second instar nauplii, incubated continuously at 27 °C, were harvested at the same time points. The cold shock and recovery experiment was conducted for three biological replicates.
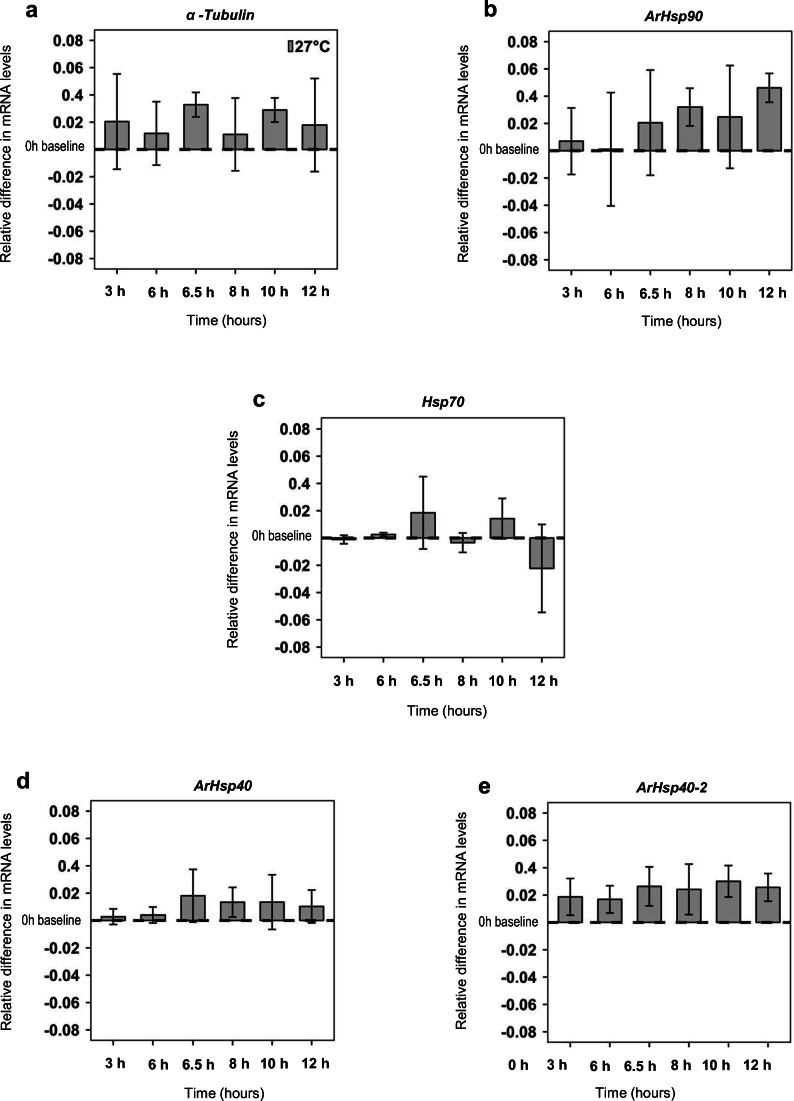
Fig. 1.
Hsp mRNA in A. franciscana nauplii during continuous incubation at 27 °C. Differences in the mean relative amounts of a α-tubulin, b ArHsp90, c Hsp70, d ArHsp40, and e ArHsp40-2 mRNA relative to the baseline at 0 h. The x-axis depicts continuous time of experiments for 12 h. Error bars represent the s.d. for n = 3. Letters above and below bars indicate Games-Howell pairwise groupings between time points (Tukey’s post hoc tests, P < 0.05) following one-way ANOVA tests
Quantification of mRNA in A. franciscana nauplii
At each sampling time, approximately 150 mg of A. franciscana nauplii was recovered from seawater, washed with distilled water on 5-μm nylon mesh filters (Spectrum Labs Inc), flash frozen, and homogenized in 600 μl of TRIzol® (Invitrogen, Burlington, ON, Canada). RNA was extracted using the Ribopure™ RNA Purification Kit (Invitrogen), and cDNA was reverse-transcribed from 3 μg total RNA using the SuperScript® IV First-Strand Synthesis Kit for RT-PCR (Invitrogen).
qPCRs were performed using 15 ng of cDNA as template, primers specific to ArHsp90, Hsp70, ArHsp40, ArHsp40-2, and α-tubulin at 10 μM (Table 1) and SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad). α-tubulin was the reference gene against which other mRNAs were compared. The qPCR protocol was as follows 3 min at 95 °C, 40 cycles of 10 s at 95 °C, 10 s at 51 °C, and 30 s at 72 °C, with a final 10 s at 95 °C, in a CFX 96 Touch™ Real-Time Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). cDNA copy numbers were determined from a standard curve of Cq values (R2 > 0.98) generated using cDNA diluted in a 4-fold series, and the standard curve was fitted by linear regression for all Hsps and α-tubulin. Melt curve analyses were performed at the end of qPCR. Cq and melt curve values were analyzed with CFX Maestro™ Software. The qPCR experiments were performed using three biological replicates.
Table 1.
Primers used in qPCR
| Gene | Forward sequence (5′ to 3′) | Reverse sequence (5′ to 3′) | Length of amplicon (bpa) |
|---|---|---|---|
| Hsp90 | GGTGTGGGTTTCTATTCTGC | GCAGCAGATTCCCACACA | 95 |
| Hsp70 | AGACAAGCCACAAAGGATGC | GCAGCAGTTGGTTCGTTGA | 71 |
| ArHsp40-1 | GTGCATCAGTTGAGCGTCAC | TGCTGAACCATTCCAGGAGC | 194 |
| ArHsp40-2 | TGACCCATTCGGTGGGTTTG | TCGTGTTCAATGGGTGGGTC | 144 |
| α-tubulinb | CGACCATAAAAGCGCAGTCA | CTACCCAGCACCACAGGTCTCT | 276 |
Primers were produced by Integrated DNA Technologies, Coralville, IA, USA
abp, base-pairs
bKing et al. (2013)
Quantification of Hsps in A. franciscana nauplii
Approximately 150 mg of A. franciscana nauplii from each experimental treatment was washed with distilled water on 5-μm nylon mesh filters (Spectrum Labs Inc) and suspended (1:1 w/v) in cold Pipes Buffer (100 mM Pipes, 1 mM MgCl2, 1 mM EGTA, pH 7.4) containing 1:100 w/v Halt protease inhibitor cocktail (Thermo Scientific, Rockford, IL, USA), before flash-freezing in liquid nitrogen. Frozen tissues were homogenized on ice with a micropestle (Kimble Chase, Vineland, NJ, USA) for 1 min, followed by centrifugation at 4 °C for 15 min at 16,000g. Protein concentrations of the resulting supernatants were determined by Bradford assay (Bradford 1976). Thirty micrograms of each protein extract was then resolved in 8–16% Mini-Protean TGX™ gels (Bio-Rad) for 30 min at 250 V. These gels were either stained with Colloidal Coomassie Brilliant Blue (10% (w/v) ammonium sulfate, 0.1% (w/v) Coomassie G-250, 3% (v/v) phosphoric acid, 20% (v/v) ethanol) or transferred to nitrocellulose membranes (Bio-Rad). The GeneDireX BLUelf Prestained Protein Ladder (Froggabio, Toronto, ON, Canada) was used as a molecular mass size marker. Gels were stained with Colloidal Coomassie Brilliant Blue to verify that proteins were present in equal quantities in each lane, and successful transfer of proteins to nitrocellulose was verified with Ponceau S stain (0.1% (w/v) Ponceau, 1.5% (w/v) trichloroacetic acid; Online Resource 1).
To detect molecular chaperones, blots were incubated in 5% Carnation nonfat milk in TBS (10 mM Tris, 1 M NaCl, pH 7.4) for 1 h at room temperature followed by incubation for 20 min at room temperature with antibodies to either ArHsp90 diluted 1:2000, Hsp70 diluted 1:5000 (SMC-164; StressMarq, Victoria, BC, Canada), ArHsp40 (Jiang et al. 2016) diluted 1:2000, or ArHsp40-2 (Rowarth and MacRae 2018a) diluted 1:2000. All dilutions were in TBS. Blots were then incubated for 16 h with antibodies in 4% Carnation nonfat milk in TBS. The antibody to ArHsp90 was raised in a rabbit (ABBIOTEC, San Diego, CA, USA) using the peptide 77-CLELFEEIAEDKENYKKFYE-96, which was based on the sequence of an ArHsp90 cDNA cloned from A. franciscana (manuscript in preparation). Membranes were washed with four changes of TBST (10 mM Tris, 140 mM NaCl, 0.1% Tween 20, pH 7.4) once for 1 min, then 3 times for 5 min each. Subsequently, membranes were washed in four changes of HST (10 mM Tris, 1 M NaCl, 0.5% Tween 20, pH 7.4) following the same procedure as with TBST. Washed membranes were incubated for 20 min in HRP-conjugated goat antirabbit IgG antibody (Life Technologies, Burlington ON, Canada) diluted 1:5000 in TBS for ArHsp90, ArHsp40, and ArHsp40-2 or HRP-conjugated goat antimouse IgG antibody (Life Technologies) diluted 1:5000 in TBS for Hsp70. The membranes were washed in TBST, followed by HST as before and antibody-reactive proteins were visualized with Clarity™ Western ECL Substrate (Bio-Rad) using a DNR Bio-Imaging Systems MF-ChemiBIS 3.2 gel documentation system (Montreal Biotech, Montreal, QC, Canada). After visualization, immunoreactive protein bands were analyzed using the Image Studio Lite version 5.2 software (Li-Core Biosciences, Lincoln, NE, USA). Signal values were obtained by drawing a box around each protein band appearing as dark rectangular areas against a white background. The smallest box on the blot was chosen as a standard size for measuring other protein bands on the same blot. The western blotting experiments were performed using three biological replicates.
Full blots revealing antibody-reactive protein bands for each Hsp of interest, including reactive bands smaller than the Hsp being examined and thus likely to represent degradation products, are also shown in Online Resource 2. Other than for the cold shock/recovery samples of ArHsp90, where it appeared that the upper band was susceptible to breakdown (Online Resource 2) and for ArHsp40-2 where nonspecific probes were detected below the original protein bands (Online Resource 2), degradation of Hsps was minimal and thus unlikely to influence the results.
Statistical analysis
The optimal temperature and cold shock/recovery treatments were analyzed for changes in Hsp and tubulin mRNA and Hsp levels. For Hsp and tubulin mRNA, a measure of “relative difference in mRNA” was calculated. In both cold shock/recovery and optimal temperature groups, Cq values at each time point were averaged for three technical replicates. Average Cq values at each time point were then divided by the average Cq value at 0 h (the baseline) to obtain a relative value. This relative Cq value was subtracted from 1 to obtain the relative difference in mRNA levels. Similarly, for Hsps, signal values for protein bands were quantified at each time point in both cold shock/recovery and optimal temperature groups. For each time point, signal values were divided by the signal value at 0 h to obtain relative values. These relative values were subtracted from 1 to obtain a “relative difference in Hsp level”.
The data were analyzed using one-way ANOVA to test the effect of sampling time on Hsp mRNA and protein. Data for cold shock/recovery and 27 °C experiments were analyzed separately because of potential confounding issues caused by differences among developmental stages, and thus, the patterns in the data are described independently. For all analyses, each sampling point was used as a categorical factor to facilitate Tukey’s post hoc tests to determine differences between specific pairs of time points. Relative differences in mRNA and protein levels were plotted as mean ± SD. Representative plots and statistical analysis were performed using RStudio, an adjacent platform of the statistical software R, version 3.6.0 (R Core Team 2018).
Results
Hsp mRNA quantities remained constant during continuous incubation of A. franciscana at 27 °C
At 27 °C, mRNA remained constant relative to 0 h for α-tubulin (one-way ANOVA, F6,13 = 0.38; P = 0.86; Fig. 1a), ArHsp90 (one-way ANOVA, F6,13 = 0.88; P = 0.52; Fig. 1b), Hsp70 (one-way ANOVA, F6,13 = 1.87; P = 0.17; Fig. 1c), ArHsp40 (one-way ANOVA, F6,13 = 0.58; P = 0.72; Fig. 1d), and ArHsp40-2 (one-way ANOVA, F6,13 = 0.42; P = 0.83; Fig. 1e). These patterns are similar to those observed during the cold shock/recovery experiments.
Hsps were reduced in amount during continuous incubation of A. franciscana at 27 °C
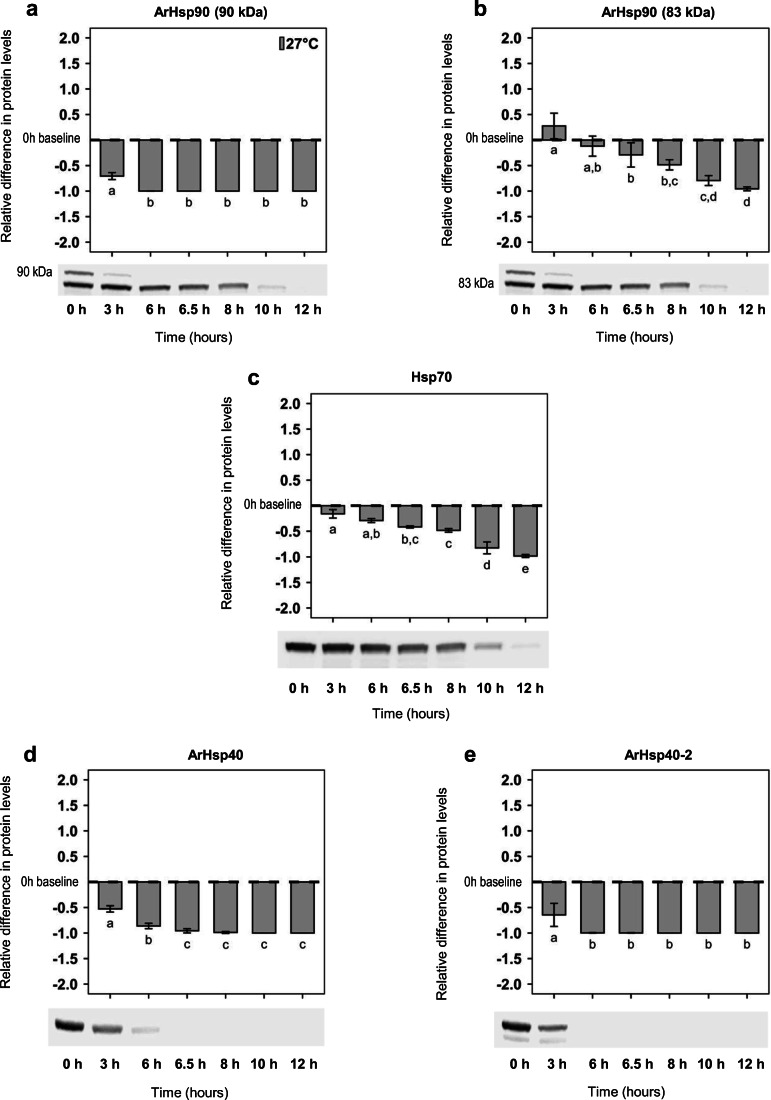
Generally, when compared with their loss during cold shock and recovery, Hsps declined faster during continuous incubation at 27 °C. ArHsp90 (90 kDa) decreased significantly after 3-h incubation at 27 °C and then weakened further such that it was difficult to detect on blots (one-way ANOVA, F6,13 = 56.00; P < 0.001; Fig. 2a). By contrast, the smaller 83 kDa protein did not change significantly relative to the amount present at 0 h until 6.5 h of incubation at 27 °C and then it continued to decrease, reaching low levels by the end of the incubation (one-way ANOVA, F6,13 = 20.44; P < 0.001; Fig. 2b). Hsp70 was reduced after 3 h of incubation and continued significantly downward until the incubation ended (one-way ANOVA, F6,13 = 73.02; P < 0.001; Fig. 2c). ArHsp40 (one-way ANOVA, F6,13 = 71.38; P < 0.001; Fig. 2d) and ArHsp40-2 (one-way ANOVA, F6,13 = 7.33; P = 0.0023; Fig. 2e) underwent substantial reductions after 3 h of incubation and subsequently declined until they were difficult to detect on blots.
Fig. 2.
Hsps in A. franciscana nauplii during continuous incubation at 27 °C. Differences in the mean amounts of a ArHsp90 (90 kDa), b ArHsp90 (83 kDa), c Hsp70, d ArHsp40, and e ArHsp40-2 relative to baseline at 0 h with representative immunoblots. The x-axis depicts continuous time of experiments for 12 h. Error bars represent the s.d. for n = 3. Letters above and below bars indicate Games-Howell pairwise groupings between time points (Tukey’s post hoc tests, P < 0.05) following one-way ANOVA tests
Hsp mRNA levels remained constant during cold shock and recovery of A. franciscana
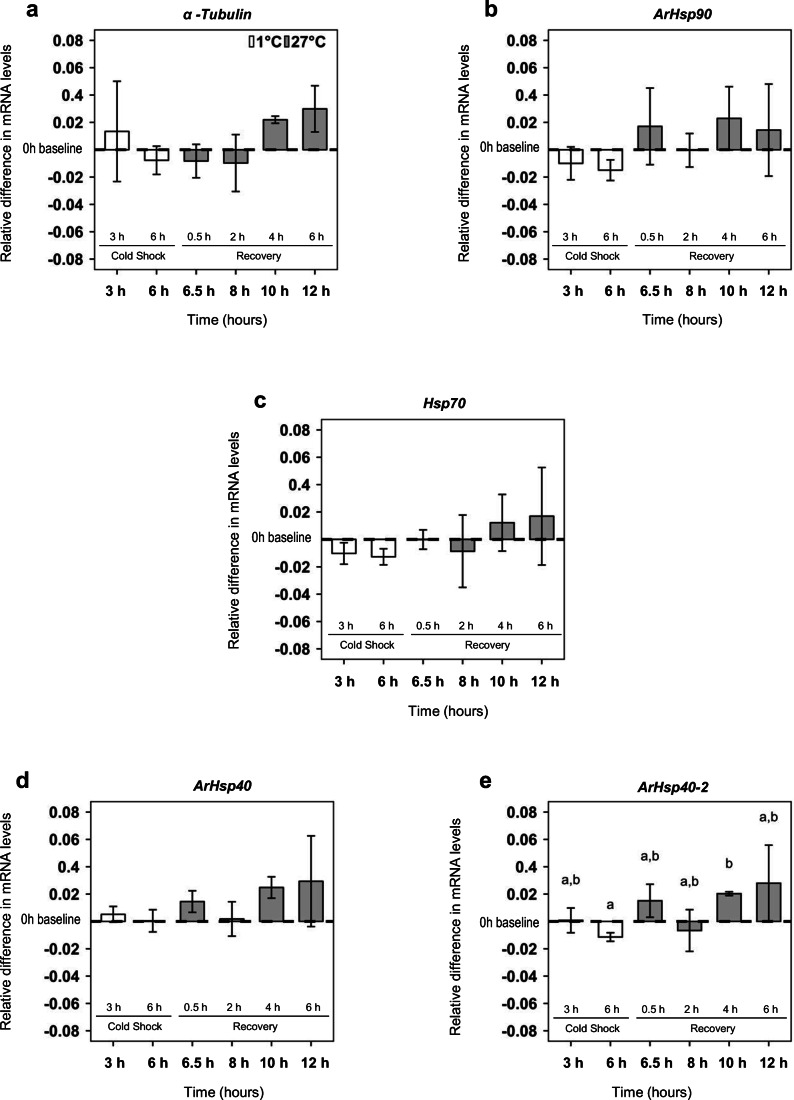
Quantification of mRNA by qPCR revealed that during cold shock and recovery, the amounts of α-tubulin mRNA (one-way ANOVA, F6,13 = 2.34; P = 0.11, Fig. 3a), ArHsp90 mRNA (one-way ANOVA, F6,13 = 1.55; P = 0.25, Fig. 3b), Hsp70 mRNA (one-way ANOVA, F6,13 = 1.10; P = 0.41, Fig. 3c), and ArHsp40 mRNA (one-way ANOVA, F6,13 = 1.83; P = 0.18, Fig. 3d) in A. franciscana nauplii were generally constant. ArHsp40-2 mRNA was the only mRNA that differed statistically between time points during cold shock and recovery (one-way ANOVA, F6,13 = 3.58; P = 0.03; Fig. 3e). A Tukey post hoc test revealed that ArHsp40-2 mRNA at 6 h of cold shock was significantly lower than at 4 h of recovery (Fig. 3e).
Fig. 3.
Hsp mRNA in A. franciscana nauplii during cold shock and recovery. Differences in the mean amounts of a α-tubulin, b ArHsp90, c Hsp70, d ArHsp40, and e ArHsp40-2 mRNA relative to baseline at 0 h. The x-axis depicts specific cold shock and recovery treatment times of experiments. Error bars represent the s.d. for n = 3. Letters above and below bars indicate Games-Howell pairwise groupings between time points (Tukey’s post hoc tests, P < 0.05) following one-way ANOVA tests
The amounts of Hsps varied during cold shock and recovery of A. franciscana
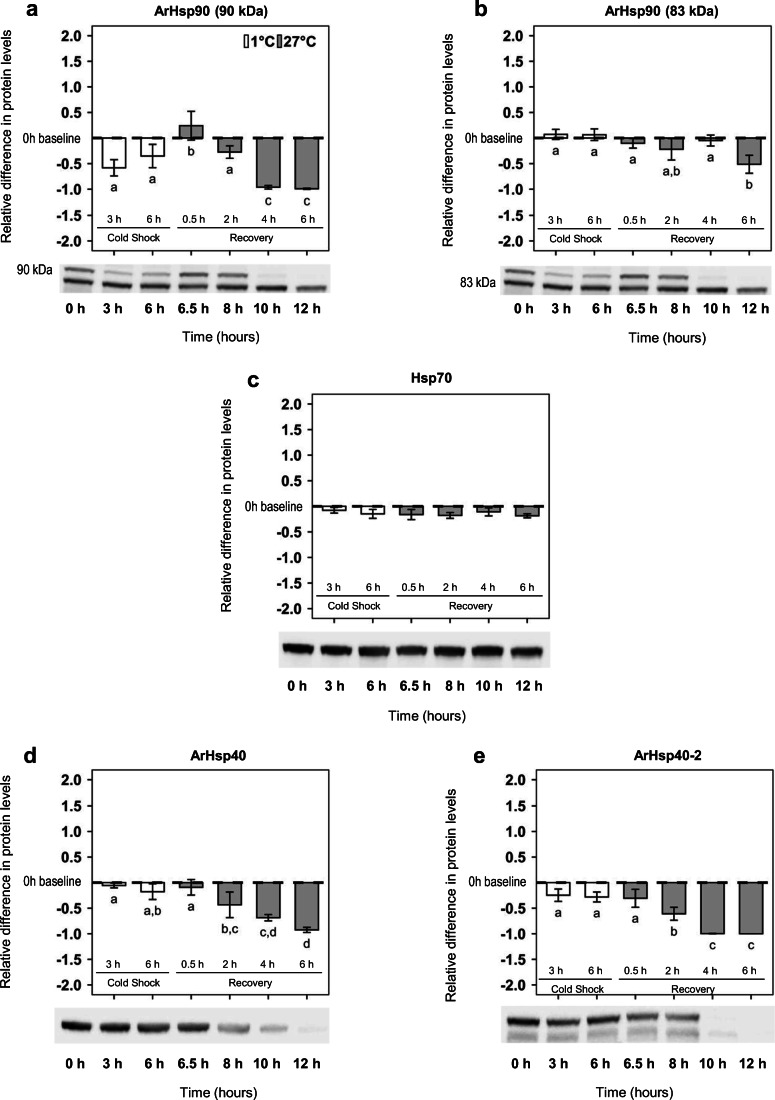
Two immunoreactive protein bands of 90 kDa and 83 kDa were detected in all samples probed with the antibody to ArHsp90. The larger 90 kDa protein was reduced upon cold shock and then increased abruptly at 0.5 h of recovery, before declining again at 4 h recovery (one-way ANOVA, F6,13 = 22.58; P < 0.001; Fig. 4a). The smaller 83 kDa protein was unaffected by cold shock and did not decrease significantly until 6 h of recovery (one-way ANOVA, F6,13 = 7.40; P = 0.0022; Fig. 4b). Hsp70 remained constant throughout cold shock and recovery (one-way ANOVA, F6,13 = 1.03; P = 0.44; Fig. 4c), whereas ArHsp40 (one-way ANOVA, F6,13 = 19.02; P < 0.001; Fig. 4d) and ArHsp40-2 (one-way ANOVA, F6,13 = 31.73; P < 0.001; Fig. 4e) were maintained during cold shock, but waned upon recovery.
Fig. 4.
Hsps in A. franciscana nauplii during cold shock and recovery. Differences in the mean amounts of a ArHsp90 (90 kDa), b ArHsp90 (83 kDa), c Hsp70, d ArHsp40, and e ArHsp40-2 relative to baseline at 0 h with representative immunoblots. The x-axis depicts specific cold shock and recovery treatment times of experiments. Error bars represent the s.d. for n = 3. Letters above and below bars indicate Games-Howell pairwise groupings between time points (Tukey’s post hoc tests, P < 0.05) following one-way ANOVA tests
Discussion
The tolerance of organisms to changes in temperature varies and can ultimately affect their growth, reproduction, and survival (Ramløv 2000; Huang et al. 2007; Bale and Hayward 2010). Production of molecular chaperones upon heat stress has been examined in many animals, but considerably less is known about their synthesis in response to cold; this is especially true for crustaceans (Stillman and Tagmount 2009; Zhang et al. 2009; Ronges et al. 2012). To address this issue, we examined the effect of cold shock and recovery on Hsps in A. franciscana nauplii. Our results indicate that, with the exception of ArHsp40-2 mRNA, all other Hsp mRNAs in our study were unaffected by cold shock. The levels of the corresponding Hsps, ArHsp90, Hsp70, ArHsp40, and ArHsp40-2, were all preserved and sustained throughout cold shock and recovery beyond observed optimal levels in nauplii.
ArHsp90, Hsp70, and ArHsp40 mRNA are not cold-inducible in A. franciscana nauplii
Physiological adaptations to thermal stress in organisms may result in chronic expression of Hsp genes (Chen et al. 2018a). In our study, ArHsp90, Hsp70, or ArHsp40 mRNA remained in continuously steady and unchanged levels during cold shock and recovery, and at an optimal temperature of 27 °C. These results are in contrast with findings that indicate that either Hsp90, Hsp70, or Hsp40 mRNA have been upregulated in response to heat or cold shock in other crustaceans, such as the Pacific white shrimp (Litopenaeus vannamei), the white leg shrimp (Penaeus vannamei), the mud crab (Scylla serrata), and in many insect species (Fu et al. 2013; King and MacRae 2015; Chen et al. 2018b; Sung et al. 2018). However, in larvae of the Antarctic midge (Belgica antarctica), Hsp90 and Hsp70 were constitutively expressed at both optimal and suboptimal temperatures, such that heat or cold shock and recovery did not influence the expression of Hsps in this species (Rinehart et al. 2006). It appears that as for B. antarctica, ArHsp90, Hsp70, or ArHsp40 mRNA may have preexisted in high amounts at optimal temperature and prior to cold shock. Pretranscribed, rather than new transcription of Hsps mRNA, might help A. franciscana to quickly synthesize Hsps in the event of cold stress. This may be a favorable evolutionary adaptation for A. franciscana nauplii to thrive in thermally variable environments such as the Great Salt Lake, where annual temperatures fluctuate between 0.5 °C in January (winter) and 22.5 °C in August (summer) (Crosman and Horel 2009). Additionally, Artemia are extremophiles dominating hypersaline environments otherwise unsuitable for other species, and as a result, Artemia may possess an array of genes already being constitutively expressed to help them cope with recurring environmental threats including cold stress (Clegg et al. 2001; Gajardo and Beardmore 2012).
A. franciscana nauplii used in our experiments were hatched from cysts in quiescence, a state of dormancy preceded by diapause (Clegg and Jackson 1998; Clegg and Trotman 2002). During diapause, which is in itself a stressful event, protein synthesis and development are halted, while Hsp expression is increased to protect A. franciscana from cold harsh winters (MacRae 2003, 2010; Qiu et al. 2007). ArHsp40, ArHsp40-2, ArHsp21, ArHsp22, Hsp70, and their heat shock factor, HSF1, have all been associated with stress tolerance in diapausing A. franciscana cysts (Qiu and MacRae 2008a, b; Rowarth and MacRae 2018a; Tan and MacRae 2018), and ArHsp21, ArHsp22, and Hsp70 are upregulated in A. franciscana reactivated postdiapause cysts (Wang et al. 2007). We suspect that the consistent amounts of Hsp mRNA observed in nauplii at optimal and cold/recovery stages could also be an extension of prior elevation of Hsp expression in diapause and quiescence (Clegg and Trotman 2002; MacRae 2003, 2016). Perhaps Hsp mRNA are maintained at constantly high amounts until favorable conditions return to break out of dormancy and nauplii have survived through early development (MacRae 2003, 2010; Qiu et al. 2007). As with ArHsp40-2 in our cold shock/recovery experiment, Hsp90 mRNA also increased in larval onion maggot (Delia antiqua) and pupal flesh fly (Sarcophaga crassipalpis) during winter diapause to promote cold hardiness while preserving the development and physiology of the insects (Sonoda et al. 2006; Zhang and Denlinger 2010).
ArHsp40-2 mRNA is cold-inducible in A. franciscana nauplii
The upregulation of ArHsp40-2 in A. franciscana nauplii, but not the other Hsp mRNAs, indicates that different Hsps vary in their response to cold stress. Several types of stress factors and genes are activated or deactivated depending on the type and intensity of stress and the functional need of the specific gene product (Fulda et al. 2010; King and MacRae 2015; Chen et al. 2018a). ArHsp40 and ArHsp40-2 are both heat-inducible J-domain homologs; however, only ArHsp40 influences diapause entry and in our experiments only ArHsp40-2 mRNA was cold-inducible (Jiang et al. 2016; Rowarth and MacRae 2018b). Similarly, Hsp60, Hsp67Ba, and Hsc70-1 transcript levels in D. melanogaster are unaffected by cold shock, whereas Hsp22, Hsp23, Hsp27, Hsp40, Hsp68, Hsp70Aa, and Hsp83 were significantly induced by cold (Colinet et al. 2010). We infer from this that while certain Hsps may be either nonessential for cold stress or their constitutive expression may be sufficient to mitigate the negative impacts of low temperatures, the upregulation of other Hsps may be required for defense, and to reduce the accumulation and extent of cold injury. Particular stress genes may only be induced when primary defense from constitutively expressed genes fail to provide protection in unpredictable and sudden changes in the environment (Sørensen 2010).
Hsps decline over time at optimal temperature in A. franciscana nauplii
All four Hsps in A. franciscana nauplii decreased rather rapidly when continuously incubated at an optimal temperature of 27 °C. Moreover, ArHsp90 (90 kDa), Hsp70, ArHsp40, and ArHsp40-2 were undetectable on blots by 6.5 h during recovery. In thermally optimal and nonstress conditions, Hsps are involved in multiple biological processes such as signalling, protein trafficking, cell cycle control, and cell division (Lindquist and Craig 1988; Hendrick and Hartl 1993). Hsps interact with client proteins and enzymes to drive metabolism, homeostasis, growth, and development (Lindquist and Craig 1988; Hendrick and Hartl 1993). Hsps are regulated by heat shock factors (HSFs), heat shock elements (HSEs), and other factors that control their levels in the cell (Morimoto 1993, 1998). Perhaps Hsps in A. franciscana nauplii are synthesized and regulated to differing amounts depending on their function during nauplii development. It appears that as A. franciscana nauplii developed through the instar II stage at optimal temperature, ArHsp90 (90 kDa), ArHsp40, Hsp70, and ArHsp40-2 may not have been relevant to their development, resulting in a quick decline in Hsp levels over time. Sørenson et al. (2010) propose that some Hsps may hinder normal growth in organisms and as a result are suppressed in stable nonstress instances. An extensive review by King and MacRae (2015) indicated that basal levels of Hsps exist in several insect species; however, Hsp levels can be altered by the influence of external factors such as cold, heat, diapause, desiccation, and anoxia. As well, synthesis of specific proteins may likely be kept to minimal levels to maximize cellular resources for normal cell function, but increase under stress conditions (Lackner et al. 2012).
ArHsp90, ArHsp40, and ArHsp40-2 levels were maintained during cold shock but decreased during recovery, while ArHsp70 remained constant
The key role played by Hsps in maintaining proteostasis becomes crucial under stress conditions (Feder and Hofmann 1999; Sørensen 2010). As temperatures drop, the rate of protein breakdown and aggregation increases, and without recycling or elimination of aggregates, the cell environment becomes toxic (Bukau et al. 2006; Liberek et al. 2008; Doyle et al. 2013). Hsps function to minimize this process. Zhang et al. (2011, 2018) suggest that this helps to enhance cold hardiness in insects by extending the half-lives of cellular proteins. All four Hsps in A. franciscana may have been preserved throughout cold shock and recovery to assist in folding and repairing cold-damaged proteins (Sørenson et al. 2010; Michaud and Denlinger 2004). It is worth noting the slower rate of decline of ArHsp90 (90 kDa), ArHsp40, and ArHsp40-2 for cold shock and recovery treatments compared with optimal temperature treatments, highlighting possible involvement of these Hsps in cold tolerance. In our study, ArHsp90 (90 kDa), the most ubiquitously expressed chaperone, increased briefly then declined when A. franciscana were in recovery at optimal temperature. The short increase in ArHsp90 immediately after nauplii were transferred from cold shock to recovery may have helped to initiate signalling responses for the repair of cold-damaged proteins. Hsp90 is known to associate with receptor kinases and steroid hormones in signalling cascades to facilitate several responses to environmental cues (Picard et al. 1990; Citri et al. 2006; Shapiro and Cowen 2010). With the exception of Hsp70, all Hsps declined in late recovery, at which point nauplii may have fully recovered from cold stress and had no need for their continued synthesis (Lackner et al. 2012).
The levels of Hsp70 in A. franciscana nauplii were static throughout cold shock and recovery. Similar studies by Zhang et al. (2011, 2018) found that larvae of the freeze-tolerant goldenrod gall moth (Epiblema scudderiana) and the gall fly (Eurosta solidaginis) maintain steady amounts of Hsp70 under lab-staged cold exposure at 4 °C for 24 h, upregulating Hsp70 only after further chilling to − 4 °C (Zhang et al. 2011, 2018). These results suggest that as for A. franciscana nauplii, E. scudderiana, and E. solidaginis conserve enough Hsp70 to protect and stabilize cold-denaturing macromolecules (Zhang et al. 2011, 2018). Increased translation of Hsp70 occurred in E. scudderiana and E. solidaginis for further protection at subzero temperatures (Zhang et al. 2011, 2018). The half-lives of Hsp70 and its mRNA transcripts were extended for 8 h following recovery from heat shock at 49 °C in Drosophila (Balakrishnan and De Maio 2006). Under optimal conditions, Hsp70 is present in low amounts as it forms part of a multiprotein complex that suppresses HSF1 and the downstream activation of heat shock genes by HSF1 (Shi et al. 1998; Gomez-Pastor et al. 2018). However, upon heat stress, and possibly cold stress, HSF1 is liberated by Hsp70 and associated proteins, which activates transcription of Hsp genes and increased translation Hsps such as Hsp70 in the cell for stress-protective roles (Shi et al. 1998). In addition to protein folding, Hsp70 helps transport proteins across membranes of organelles, preventing the aggregation of abnormal proteins that may have resulted from cold stress (Deshaies et al. 1988; Flynn et al. 1991; Bukau and Horwich 1998).
Hsp mRNA levels versus Hsp levels in A. franciscana
The abundance of Hsp mRNA was not correlated with that of Hsps in either optimal treatment or cold shock and recovery treatment. There are a few reasons to explain this discrepancy. First, Hsps may be produced on a “translation by demand” basis, where constitutive mRNAs are translated into protein according to their need for specific cellular processes or the onset of specific stress signalling (Beyer et al. 2004). For example, mRNA of the transcription factor Gcn4 is constitutively expressed in yeast under normal growth conditions; however, starvation activates translation of the protein Gcn4p that activates “survival genes” downstream (Hinnebusch and Natarajan 2002). Secondly, steady-state and stress-state expression of mRNA and protein may differ in the bid to maximize the cell’s energy resources (Cheng et al. 2016). Thirdly, posttranscriptional modifications which are crucial for acute stress adaptations could affect mRNA versus protein abundance (Lawless et al. 2009; Zhang et al. 2015). mRNA maturation and export take time and may cause delays in protein synthesis especially during stress (Lackner et al. 2012; Liu et al. 2016). Another likely reason for different Hsp mRNA versus protein amounts could be attributed to varying half-lives of these molecules under both optimal and cold shock states (Balakrishnan and De Maio 2006; Mauger et al. 2019). All or some of these phenomena may explain the discrepancy between Hsp mRNA abundance and Hsp levels.
Immunoprobing with ArHsp90 detects two separate bands of weights, 90 kDa and 83 kDa
Interestingly, in both optimal and cold shock/recovery treatments, two bands were detected at 90 kDa and 83 kDa for ArHsp90 after immunoprobing with the ArHsp90 antibody. We postulate that the 90 kDa protein band is the true ArHsp90 protein, as it corresponded with the expected molecular weight, while the bottom 83 kDa band may be a modified or degraded product of the upper band. ArHsp90 could be modified posttranslationally from its larger 90 kDa form to a smaller 83 kDa variant, or the larger 90 kDa protein is cleaved or degraded by proteases and regulatory proteins that modulate its function (Scroggins and Neckers 2007; Mollapour and Neckers 2012; Tanaka et al. 2012; Hanssum et al. 2014). The amounts of the 83 kDa protein were unchanged during cold shock and early recovery but decreased late in recovery. This is a slightly different trend than the 90 kDa product, which decreased more rapidly; these differences indicate that more research should focus on identifying the 83 kDa protein and its characteristics.
Future experiments on Hsps expression and freeze-tolerance in A. franciscana
In our study, A. franciscana nauplii were cooled to 1 °C but not to freezing. Freezing temperatures can invoke upregulation of Hsps beyond what is observed at low temperatures (i.e., above 0 °C). For example, Hsp70 and Hsp40 expression in E. solidaginis were upregulated in larvae by 1.43- and 1.50-fold, respectively, at − 16 °C, but they were not upregulated at 3 °C (Zhang et al. 2018). These differences may manifest because below 0 °C metabolism and body mass content decrease quickly, ice crystals form, the cellular environment can become anoxic, and organism mortality occurs (Block et al. 2006; Tattersall et al. 2012). It is not surprising that Hsps would further increase, as observed in E. solidaginis and C. pipiens, to help rescue salvageable proteins. We predict that further freezing of A. franciscana nauplii could have elicited an upregulatory Hsp response, although further experimentation is required to test this prediction.
Conclusions
Hsps are integral to cellular processes under conditions of normal growth and during stress. In nauplii of A. franciscana, the levels of the Hsps examined in this study were generally maintained or increased in response to short-term cold shock followed by recovery at 27 °C, making them readily available for the repair of cold-damaged proteins in the cell. This suggests that Hsps may be essential for adaptation of A. franciscana to cold. Furthermore, the evidence presented highlights the importance of Hsps to cold tolerance and reveals at least one strategy that crustaceans utilize to tolerate cold. From an applied perspective, nauplii of A. franciscana are used as feed for commercially important aquatic animals and knowing how Artemia responds to cold may enhance its utilization in aquaculture, especially as nauplii are often stored cold prior to use.
Electronic supplementary material
Stained gels and blots containing proteins from A. franciscana nauplii. Gels were stained with Colloidal Coomassie Brilliant Blue to demonstrate equal protein loading in each lane (a, b) and blots were stained with Ponceau to confirm successful protein transfer (c, d). Faint staining at the upper right, especially in the cold shock/recovery gel and blot, indicates minor degradation of proteins in late instar II nauplii (PDF 2568 kb)
Full western blots of Hsps in A. franciscana nauplii. ArHsp90, Hsp70, ArHsp40, and ArHsp40-2 were detected after immunoprobing with antibodies during a, continuous incubation at 27 °C and b, cold shock/recovery at 1 °C followed by incubation at 27 °C. Protein bands detected at the bottom of some blots represent possible minimal degradation (PDF 370 kb)
Acknowledgments
We thank Jiabo Tan, Andrew Schofield, Sheethal Panchakshari, and two anonymous reviewers for their advice and contributions to this work.
Code availability
All codes are available for data analyzed with RStudio, an adjacent platform of the statistical software, R, version 3.6.0 (R Core Team 2018).
Authors’ contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by YAG. Data analyses were performed by YAG and LKW. The first draft of the manuscript was written by YAG, THM, and LKW and all authors commented on previous versions and reviews of the manuscript. All authors have read and approved the final manuscript.
Funding information
This project was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (Number RGPIN/04882-2016) to THM and a scholarship from Imhotep’s Legacy Academy at Dalhousie University to YAG.
Data availability
All data for Hsp mRNA and protein are available in Microsoft Excel files, version 16.35 (Microsoft 2020).
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Ethics approval
This research was performed in accordance with the ethical guidelines provided by the Canadian Council on Animal Care (CCAC). The University Committee on Laboratory Animals (UCLA) of Dalhousie University approved the research under assigned Protocol Number 117-36.
Consent for publication
All authors give their consent for the publication of this journal.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alderson TR, Kim JH, Markley JL. Dynamical structures of Hsp70 and Hsp70-Hsp40 complexes. Structure. 2016;24:1014–1030. doi: 10.1016/j.str.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidis CE, Lzaridis I, Pagoulatos GN. Constitutive expression of heat-shock protein 70 in mammalian cells confers thermoresistance. Eur J Biochem. 1991;199:35–39. doi: 10.1111/j.1432-1033.1991.tb16088.x. [DOI] [PubMed] [Google Scholar]
- Balakrishnan K, De Maio A. Heat shock protein 70 binds its own messenger ribonucleic acid as part of a gene expression self-limiting mechanism. Cell Stress Chaperones. 2006;11:44–50. doi: 10.1379/CSC-136R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale JS, Hayward SAL. Insect overwintering in a changing climate. J Exp Biol. 2010;213:980–994. doi: 10.1242/jeb.037911. [DOI] [PubMed] [Google Scholar]
- Beere HM. “The stress of dying”: the role of heat shock proteins in the regulation of apoptosis. J Cell Sci. 2004;117:2641–2651. doi: 10.1242/jcs.01284. [DOI] [PubMed] [Google Scholar]
- Beyer A, Hollunder J, Nasheuer HP, Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol Cell Proteomics. 2004;3:1083–1092. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- Block W, Baust JG, Franks F, et al. Cold tolerance of insects and other arthropods [and discussion] Philos Trans R Soc B Biol Sci. 2006;326:613–633. doi: 10.1098/rstb.1990.0035. [DOI] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chen B, Feder ME, Kang L. Evolution of heat-shock protein expression underlying adaptive responses to environmental stress. Mol Ecol. 2018;27:3040–3054. doi: 10.1111/mec.14769. [DOI] [PubMed] [Google Scholar]
- Chen T, Lin T, Li H, Lu T, Li J, Huang W, Sun H, Jiang X, Zhang J, Yan A, Hu C, Luo P, Ren C (2018b) Heat shock protein 40 (HSP40) in Pacific white shrimp (Litopenaeus vannamei): molecular cloning, tissue distribution and ontogeny, response to temperature, acidity/alkalinity and salinity stresses, and potential role in ovarian development. Front Physiol 9:1–13. 10.3389/fphys.2018.01784 [DOI] [PMC free article] [PubMed]
- Cheng Z, Teo G, Krueger S, et al. Differential dynamics of the mammalian mRNA and protein expression response to misfolding stress. Mol Syst Biol. 2016;12:855. doi: 10.15252/msb.20156423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, Lavi S, Eisenstein M, Kimchi A, Wallach D, Pietrokovski S, Yarden Y. Hsp90 recognizes a common surface on client kinases. J Biol Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- Clare DK, Saibil HR. ATP-driven molecular chaperone machines. Biopolymers. 2013;99:846–859. doi: 10.1002/bip.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg JS, Jackson SA. The metabolic status of quiescent and diapause embryos of Artemia franciscana. Arch Hydrobiol. 1998;52:425–439. [Google Scholar]
- Clegg JS, Trotman CNA. Physiological and biochemical aspects of Artemia biology. In: Abatzopoulos TJ, Beardmore JA, Clegg JS, Sorgeloos P, editors. Artemia, basic and applied biology. 1. Dordrecht: Kluwer Academic Publishers; 2002. [Google Scholar]
- Clegg JS, Jackson SA, Van Hoa N, Sorgeloos P. Thermal resistance, developmental rate and heat shock proteins in Artemia franciscana, from San Francisco Bay and southern Vietnam. J Exp Mar Biol Ecol. 2000;252:85–96. doi: 10.1016/S0022-0981(00)00239-2. [DOI] [PubMed] [Google Scholar]
- Clegg JS, Van Hoa N, Sorgeloos P. Thermal tolerance and heat shock proteins in encysted embryos of Artemia from widely different thermal habitats. Hydrobiologia. 2001;466:221–229. doi: 10.1023/A:1014580612237. [DOI] [Google Scholar]
- Colinet H, Hoffmann AA. Comparing phenotypic effects and molecular correlates of developmental, gradual and rapid cold acclimation responses in Drosophila melanogaster. Funct Ecol. 2012;26:84–93. doi: 10.1111/j.1365-2435.2011.01898.x. [DOI] [Google Scholar]
- Colinet H, Nguyen TTA, Cloutier C, Michaud D, Hance T. Proteomic profiling of a parasitic wasp exposed to constant and fluctuating cold exposure. Insect Biochem Mol Biol. 2007;37:1177–1188. doi: 10.1016/j.ibmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Colinet H, Lee SF, Hoffmann A. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. FEBS J. 2010;277:174–185. doi: 10.1111/j.1742-4658.2009.07470.x. [DOI] [PubMed] [Google Scholar]
- Conte FP, Peterson GL, Ewing RD. Larval salt gland of Artemia salina nauplii - regulation of protein synthesis by environmental salinity. J Comp Physiol. 1973;82:277–289. doi: 10.1007/BF00694240. [DOI] [Google Scholar]
- Crosman ET, Horel JD. MODIS-derived surface temperature of the Great Salt Lake. Remote Sens Environ. 2009;113:73–81. doi: 10.1016/j.rse.2008.08.013. [DOI] [Google Scholar]
- Denlinger DL. Regulation of diapause. Annu Rev Entomol. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol. 2013;14:617–629. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- Eimanifar A, Van Stappen G, Marden B, Wink M. Artemia biodiversity in Asia with the focus on the phylogeography of the introduced American species Artemia franciscana Kellogg, 1906. Mol Phylogenet Evol. 2014;79:392–403. doi: 10.1016/j.ympev.2014.06.027. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat shock proteins, molecular chaperones and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández MR, Gragera M, Ochoa-Ibarrola L, Quintana-Gallardo L, Valpuesta JM. Hsp70 – a master regulator in protein degradation. FEBS Lett. 2017;591:2648–2660. doi: 10.1002/1873-3468.12751. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Floccot MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Frankenberg MM, Jackson SA, Clegg JS. The heat shock response of adult Artemia franciscana. J Therm Biol. 2000;25:481–490. doi: 10.1016/S0306-4565(00)00013-9. [DOI] [PubMed] [Google Scholar]
- Fu W, Zhang F, Liao M, Liu M, Zheng B, Yang H, Zhong M. Molecular cloning and expression analysis of a cytosolic heat shock protein 70 gene from mud crab Scylla serrata. Fish Shellfish Immunol. 2013;34:1306–1314. doi: 10.1016/j.fsi.2013.02.027. [DOI] [PubMed] [Google Scholar]
- Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol. 2010;2010:1–23. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajardo GM, Beardmore JA (2012) The brine shrimp Artemia: adapted to critical life conditions. Front Physiol. 3. 10.3389/fphys.2012.00185 [DOI] [PMC free article] [PubMed]
- Genest O, Wickner S, Doyle SM. Hsp90 and Hsp70 chaperones: collaborators in protein remodeling. J Biol Chem. 2019;294:2109–2120. doi: 10.1074/jbc.REV118.002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssum A, Zhong Z, Rousseau A, Krzyzosiak A, Sigurdardottir A, Bertolotti A. An inducible chaperone adapts proteasome assembly to stress. Mol Cell. 2014;55:566–577. doi: 10.1016/j.molcel.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Weinkauf S, Buchner J. Small heat shock proteins: simplicity meets complexity. J Biol Chem. 2019;294:2121–2132. doi: 10.1074/jbc.REV118.002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Hartl F. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LH, Chen B, Kang L. Impact of mild temperature hardening on thermotolerance, fecundity, and Hsp gene expression in Liriomyza huidobrensis. J Insect Physiol. 2007;53:1199–1205. doi: 10.1016/j.jinsphys.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Jiang G, Rowarth NM, Panchakshari S, MacRae TH. ArHsp40, a type 1 J-domain protein, is developmentally regulated and stress inducible in post-diapause Artemia franciscana. Cell Stress Chaperones. 2016;21:1077–1088. doi: 10.1007/s12192-016-0732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- King AM, MacRae TH. Insect heat shock proteins during stress and diapause. Annu Rev Entomol. 2015;60:59–75. doi: 10.1146/annurev-ento-011613-162107. [DOI] [PubMed] [Google Scholar]
- King AM, Toxopeus J, Macrae TH. Functional differentiation of small heat shock proteins in diapause-destined Artemia embryos. FEBS J. 2013;280:4761–4772. doi: 10.1111/febs.12442. [DOI] [PubMed] [Google Scholar]
- Koštál V, Tollarová-Borovanská M. The 70 kDa heat shock protein assists during the repair of chilling injury in the insect, Pyrrhocoris apterus. PLoS One. 2009;4:e4546. doi: 10.1371/journal.pone.0004546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Schmidt MW, Wu S, Wolf DA, Bahler J. Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biol. 2012;13:R25. doi: 10.1186/gb-2012-13-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalouette L, Williams CM, Hervant F, Sinclair BJ, Renault D. Metabolic rate and oxidative stress in insects exposed to low temperature thermal fluctuations. Comp Biochem Physiol - A Mol Integr Physiol. 2011;158:229–234. doi: 10.1016/j.cbpa.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Lawless C, Pearson RD, Selley JN, et al. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. 2009 doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser MP. Oxidative stress iin marine environments: biochemistry and physiological ecology. Annu Rev Physiol. 2006;68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- Li F, Luan W, Zhang C, Zhang J, Wang B, Xie Y, Li S, Xiang J. Cloning of cytoplasmic heat shock protein 90 (FcHSP90) from Fenneropenaeus chinensis and its expression response to heat shock and hypoxia. Cell Stress Chaperones. 2009;14:161–172. doi: 10.1007/s12192-008-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Qian X, Sha B. Heat shock protein 40: structural studies and their functional implications. Protein Pept Lett. 2009;16:606–612. doi: 10.2174/092986609788490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, MacRae TH. The synthesis of a small heat shock/α-crystallin protein in Artemia and its relationship to stress tolerance during development. Dev Biol. 1999;207:445–456. doi: 10.1006/dbio.1998.9138. [DOI] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Ziȩtkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Loc NH, MacRae TH, Musa N, et al. Non-lethal heat shock increased Hsp70 and immune protein transcripts but not Vibrio tolerance in the white-leg shrimp. PLoS One. 2013;8:73199–73206. doi: 10.1371/journal.pone.0073199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez T, Dalton K, Frydman J. The mechanism and function of group II chaperonins. J Mol Biol. 2015;427:2919–2930. doi: 10.1016/j.jmb.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae TH. Molecular chaperones, stress resistance and development in Artemia franciscana. Semin Cell Dev Biol. 2003;14:251–258. doi: 10.1016/j.semcdb.2003.09.019. [DOI] [PubMed] [Google Scholar]
- MacRae TH. Gene expression, metabolic regulation and stress tolerance during diapause. Cell Mol Life Sci. 2010;67:2405–2424. doi: 10.1007/s00018-010-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae TH. Stress tolerance during diapause and quiescence of the brine shrimp, Artemia. Cell Stress Chaperones. 2016;21:9–18. doi: 10.1007/s12192-015-0635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger DM, Joseph Cabral B, Presnyak V, et al. mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci U S A. 2019;116:24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Gierasch LM. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J Biol Chem. 2019;294:2085–2097. doi: 10.1074/jbc.REV118.002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean DK, Warner AH. Aspects of nucleic acid metabolism during development of brine shrimp Artemia salina. Dev Biol. 1971;24:88–105. doi: 10.1016/0012-1606(71)90048-0. [DOI] [PubMed] [Google Scholar]
- Michaud MR, Denlinger DL. Molecular modalities of insect cold survival: current understanding and future trends. Int Congr Ser. 2004;1275:32–46. doi: 10.1016/j.ics.2004.08.059. [DOI] [Google Scholar]
- Miller D, McLennan AG. The heat shock response of the cryptobiotic brine shrimp Artemia – II. Heat shock proteins. J Therm Biol. 1988;13:125–134. doi: 10.1016/0306-4565(88)90023-X. [DOI] [Google Scholar]
- Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim Biophys Acta, Mol Cell Res. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Cells in stress : transcriptional activation of heat shock genes. Sci New Ser. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Muñoz J, Pacios F. Global biodiversity and geographical distribution of diapausing aquatic invertebrates: the case of the cosmopolitan brine shrimp, Artemia (Branchiopoda, Anostraca) Crustaceana. 2010;83:465–480. doi: 10.1163/001121610X489449. [DOI] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, Guilbride DL, Wade RC, Morimoto RI, Mayer MP, Bukau B. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524:247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Stank A, Malinverni D, Alberts N, Szlachcic A, Barducci A, de Los Rios P, Wade RC, Bukau B. Evolution of an intricate J-protein network driving protein disaggregation in eukaryotes. Elife. 2017;6:1–28. doi: 10.7554/elife.24560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Wentink AS, Bukau B. Protein disaggregation in multicellular organisms. Trends Biochem Sci. 2018;43:285–300. doi: 10.1016/j.tibs.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Norouzitallab P, Baruah K, Muthappa DM, Bossier P. Non-lethal heat shock induces HSP70 and HMGB1 protein production sequentially to protect Artemia franciscana against Vibrio campbelli. Fish Shellfish Immunol. 2015;42:395–399. doi: 10.1016/j.fsi.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Ostankovitch M, Buchner J. The network of molecular chaperones: insights in the cellular proteostasis machinery. J Mol Biol. 2015;427:2899–2903. doi: 10.1016/j.jmb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Overgaard J, Sørensen JG, Com E, Colinet H. The rapid cold hardening response of Drosophila melanogaster: complex regulation across different levels of biological organization. J Insect Physiol. 2014;62:46–53. doi: 10.1016/j.jinsphys.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- Powers MV, Clarke PA, Workman P. Death by chaperone: HSP90, HSP70 or both? Cell Cycle. 2009;8:518–526. doi: 10.4161/cc.8.4.7583. [DOI] [PubMed] [Google Scholar]
- Qiu Z, MacRae TH. ArHsp22, a developmentally regulated small heat shock protein produced in diapause-destined Artemia embryos, is stress inducible in adults. FEBS J. 2008;275:3556–3566. doi: 10.1111/j.1742-4658.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- Qiu Z, MacRae TH. ArHsp21, a developmentally regulated small heat-shock protein synthesized in diapausing embryos of Artemia franciscana. Biochem J. 2008;411:605–611. doi: 10.1042/bj20071472. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Tsoi SCM, MacRae TH. Gene expression in diapause-destined embryos of the crustacean, Artemia franciscana. Mech Dev. 2007;124:856–867. doi: 10.1016/j.mod.2007.09.001. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- Ramløv H. Aspects of natural cold tolerance in ectothermic animals. Hum Reprod. 2000;15:26–46. doi: 10.1093/humrep/15.suppl_5.26. [DOI] [PubMed] [Google Scholar]
- Rinehart JP, Hayward SAL, Elnitsky MA, Sandro LH, Lee RE, Denlinger DL. Continuous up-regulation of heat shock proteins in larvae, but not adults, of a polar insect. Proc Natl Acad Sci. 2006;103:14223–14227. doi: 10.1073/pnas.0606840103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart JP, Li A, Yocum GD, Robich RM, Hayward SAL, Denlinger DL. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc Natl Acad Sci. 2007;104:11130–11137. doi: 10.1073/pnas.0703538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronges D, Walsh JP, Sinclair BJ, Stillman JH. Changes in extreme cold tolerance, membrane composition and cardiac transcriptome during the first day of thermal acclimation in the porcelain crab Petrolisthes cinctipes. J Exp Biol. 2012;215:1824–1836. doi: 10.1242/jeb.069658. [DOI] [PubMed] [Google Scholar]
- Rowarth NM, MacRae TH. ArHsp40 and ArHsp40-2 contribute to stress tolerance and longevity in Artemia franciscana, but only ArHsp40 influences diapause entry. J Exp Biol. 2018;221:jeb189001. doi: 10.1242/jeb.189001. [DOI] [PubMed] [Google Scholar]
- Rowarth NM, MacRae TH. Post-diapause synthesis of ArHsp40-2, a type 2 J-domain protein from Artemia franciscana, is developmentally regulated and induced by stress. PLoS One. 2018;13:1–17. doi: 10.1371/journal.pone.0201477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruebhart DR, Cock IE, Shaw GR. Invasive character of the brine shrimp Artemia franciscana Kellogg 1906 (Branchiopoda: Anostraca) and its potential impact on Australian inland hypersaline waters. Mar Freshw Res. 2008;59:587. doi: 10.1071/mf07221. [DOI] [Google Scholar]
- Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- Scroggins BT, Neckers L. Post-translational modification of heat-shock protein 90: impact on chaperone function. Expert Opin Drug Discovery. 2007;2:1403–1414. doi: 10.1517/17460441.2.10.1403. [DOI] [PubMed] [Google Scholar]
- Shapiro RS, Cowen LE. Coupling temperature sensing and development: Hsp90 regulates morphogenetic signaling in Candida albicans. Virulence. 2010;1:45–48. doi: 10.4161/viru.1.1.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjærven L, Cuellar J, Martinez A, Valpuesta JM. Dynamics, flexibility, and allostery in molecular chaperonins. FEBS Lett. 2015;589:2522–2532. doi: 10.1016/j.febslet.2015.06.019. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Fukumoto K, Izumi Y, Yoshida H, Tsumuki H. Cloning of heat shock protein genes (hsp90 and hsc70) and their expression during larval diapause and cold tolerance acquisition in the rice stem borer, Chilo suppressalis walker. Arch Insect Biochem Physiol. 2006;63:36–47. doi: 10.1002/arch. [DOI] [PubMed] [Google Scholar]
- Sørensen JG. Application of heat shock protein expression for detecting natural adaptation and exposure to stress in natural populations. Curr Zool. 2010;56:703–713. doi: 10.1093/czoolo/56.6.703. [DOI] [Google Scholar]
- Sottile ML, Nadin SB. Heat shock proteins and DNA repair mechanisms: an updated overview. Cell Stress Chaperones. 2018;23:303–315. doi: 10.1007/s12192-017-0843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman JH, Tagmount A. Seasonal and latitudinal acclimatization of cardiac transcriptome responses to thermal stress in porcelain crabs, Petrolisthes cinctipes. Mol Ecol. 2009;18:4206–4226. doi: 10.1111/j.1365-294X.2009.04354.x. [DOI] [PubMed] [Google Scholar]
- Strauch A, Haslbeck M. The function of small heat-shock proteins and their implication in proteostasis. Essays Biochem. 2016;60:163–172. doi: 10.1042/ebc20160010. [DOI] [PubMed] [Google Scholar]
- Sung YY, Pineda C, MacRae TH, et al. Exposure of gnotobiotic Artemia franciscana larvae to abiotic stress promotes heat shock protein 70 synthesis and enhances resistance to pathogenic Vibrio campbellii. Cell Stress Chaperones. 2008;13:59–66. doi: 10.1007/s12192-008-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YY, MacRae TH, Sorgeloos P, Bossier P. Stress response for disease control in aquaculture. Rev Aquac. 2011;3:120–137. doi: 10.1111/j.1753-5131.2011.01049.x. [DOI] [Google Scholar]
- Sung YY, Rahman NA, Shazili NAM, Chen S, Lv A, Sun J, Shi H, MacRae TH. Non-lethal heat shock induces Hsp70 synthesis and promotes tolerance against heat, ammonia and metals in post-larvae of the white leg shrimp Penaeus vannamei (Boone, s1931) Aquaculture. 2018;483:21–26. doi: 10.1016/j.aquaculture.2017.09.034. [DOI] [Google Scholar]
- Tan J, MacRae TH. Stress tolerance in diapausing embryos of Artemia franciscana is dependent on heat shock factor 1 (Hsf1) PLoS One. 2018;13:1–18. doi: 10.1371/journal.pone.0200153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mizushima T, Saeki Y. The proteasome: molecular machinery and pathophysiological roles. Biol Chem. 2012;393:217–234. doi: 10.1515/hsz-2011-0285. [DOI] [PubMed] [Google Scholar]
- Tattersall GJ, Sinclair BJ, Withers PC, et al. Coping with thermal challenges: physiological adaptations to environmental temperatures. Compr Physiol. 2012;2:2151–2202. doi: 10.1002/cphy.c110055. [DOI] [PubMed] [Google Scholar]
- Teets NM, Denlinger DL. Physiological mechanisms of seasonal and rapid cold-hardening in insects. Physiol Entomol. 2013;38:105–116. doi: 10.1111/phen.12019. [DOI] [Google Scholar]
- Teets NM, Peyton JT, Ragland GJ, Colinet H, Renault D, Hahn DA, Denlinger DL. Combined transcriptomic and metabolomic approach uncovers molecular mechanisms of cold tolerance in a temperate flesh fly. Physiol Genomics. 2012;44:764–777. doi: 10.1152/physiolgenomics.00042.2012. [DOI] [PubMed] [Google Scholar]
- Wang W, Meng B, Chen W, Ge X, Liu S, Yu J. A proteomic study on postdiapaused embryonic development of brine shrimp (Artemia franciscana) Proteomics. 2007;7:3580–3591. doi: 10.1002/pmic.200700259. [DOI] [PubMed] [Google Scholar]
- Willsie JK, Clegg JS. Nuclear p26, a small heat shock/alpha-crystallin protein, and its relationship to stress resistance in Artemia franciscana embryos. J Exp Biol. 2001;204:2339–2350. doi: 10.1242/jeb.204.13.2339. [DOI] [PubMed] [Google Scholar]
- Wu YK, Zou C, Fu DM, Zhang WN, Xiao HJ. Molecular characterization of three Hsp90 from Pieres and expression patterns in response to cold and thermal stress in summer and winter diapause of Pieris melete. Insect Sci. 2018;25:273–283. doi: 10.1111/1744-7917.12414. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Denlinger DL. Molecular characterization of heat shock protein 90, 70 and 70 cognate cDNAs and their expression patterns during thermal stress and pupal diapause in the corn earworm. J Insect Physiol. 2010;56:138–150. doi: 10.1016/j.jinsphys.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Zhang MZ, Zheng CJ, Liu J, Hu HJ. Identification of two hsp90 genes from the marine crab, Portunus trituberculatus and their specific expression profiles under different environmental conditions. Comp Biochem Physiol - C Toxicol Pharmacol. 2009;150:465–473. doi: 10.1016/j.cbpc.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang G, Storey JM, Storey KB. Chaperone proteins and winter survival by a freeze tolerant insect. J Insect Physiol. 2011;57:1115–1122. doi: 10.1016/j.jinsphys.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Bhattacharya S, Pi J, Clewell RA, Carmichael PL, Andersen ME. Adaptive posttranslational control in cellular stress response pathways and its relationship to toxicity testing and safety assessment. Toxicol Sci. 2015;147:302–316. doi: 10.1093/toxsci/kfv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Storey JM, Storey KB. Elevated chaperone proteins are a feature of winter freeze avoidance by larvae of the goldenrod gall moth, Epiblema scudderiana. J Insect Physiol. 2018;106:106–113. doi: 10.1016/j.jinsphys.2017.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stained gels and blots containing proteins from A. franciscana nauplii. Gels were stained with Colloidal Coomassie Brilliant Blue to demonstrate equal protein loading in each lane (a, b) and blots were stained with Ponceau to confirm successful protein transfer (c, d). Faint staining at the upper right, especially in the cold shock/recovery gel and blot, indicates minor degradation of proteins in late instar II nauplii (PDF 2568 kb)
Full western blots of Hsps in A. franciscana nauplii. ArHsp90, Hsp70, ArHsp40, and ArHsp40-2 were detected after immunoprobing with antibodies during a, continuous incubation at 27 °C and b, cold shock/recovery at 1 °C followed by incubation at 27 °C. Protein bands detected at the bottom of some blots represent possible minimal degradation (PDF 370 kb)
Data Availability Statement
All data for Hsp mRNA and protein are available in Microsoft Excel files, version 16.35 (Microsoft 2020).