Abstract
Neural circuit analysis relies on having molecular markers for specific cell types. However, for a cell type identified only by its circuit function, the process of identifying markers remains laborious. We developed physiological optical tagging sequencing (PhOTseq), a technique for tagging and expression-profiling cells based on their functional properties. PhOTseq was capable of selecting rare cell types and enriching them by nearly one hundred-fold. We applied PhOTseq to the challenge of mapping receptor-ligand pairings among pheromone-sensing neurons in mice. Together with in vivo ectopic expression of vomeronasal chemoreceptors, PhOTseq identified the complete combinatorial receptor code for a specific set of ligands.
One Sentence Summary:
Physiology-driven phototagging allows expression profiling of rare cell types and reveals structure in olfactory coding
Molecular markers are a powerful tool for labeling and analyzing neuronal cell types. However, in many cases a single marker is insufficient to define a unique cell type, and may label a few, or a few hundred, physiologically-distinguishable cell types (1–3). In such cases, one wishes to select specific physiological populations and discover their molecular identities. However, tools for proceeding from function to molecular markers are not fully mature. Markers such as c-fos label active neurons promiscuously across many cell types (4). Techniques like patch-seq enable expression-profiling of single recorded neurons (5–8); however, this approach faces obstacles for rare cell types or when one needs many cells of the same type to profile low-abundance transcripts.
We reasoned that the key bottlenecks of this pipeline could be replaced by an all-optical approach. One prototype is CaMPARI (9), which uses the simultaneous presence of calcium and excitation by short-wavelength light under the control of the experimenter. While CaMPARI allows one to control labeling of excited neurons temporally, as with c-fos any active neuron will be labeled, so that this may comprise dozens or more cell types. There is considerable need for tools that provide the specificity of patch-seq with the convenience and high-throughput of optical methods.
Here we report a method called physiological optical tagging sequencing (PhOTseq). PhOTseq allows specific cell types to be defined intersectionally via their physiological activity under diverse conditions. We exploited two coexpressed fluorescent proteins, GCaMP and photoactivatable mCherry (PAmCherry) (10–12): GCaMP permits recording neuronal activity by large-scale calcium imaging, and PAmCherry enables long-term labeling of selected neurons by photoactivation (PA). Using these reporters, we performed calcium imaging using a diverse set of stimuli or experiences, and then selected a specific collection of cells for tagging by photoactivation. Finally, the tagged neurons were harvested and profiled for their mRNA expression (Fig. 1A).
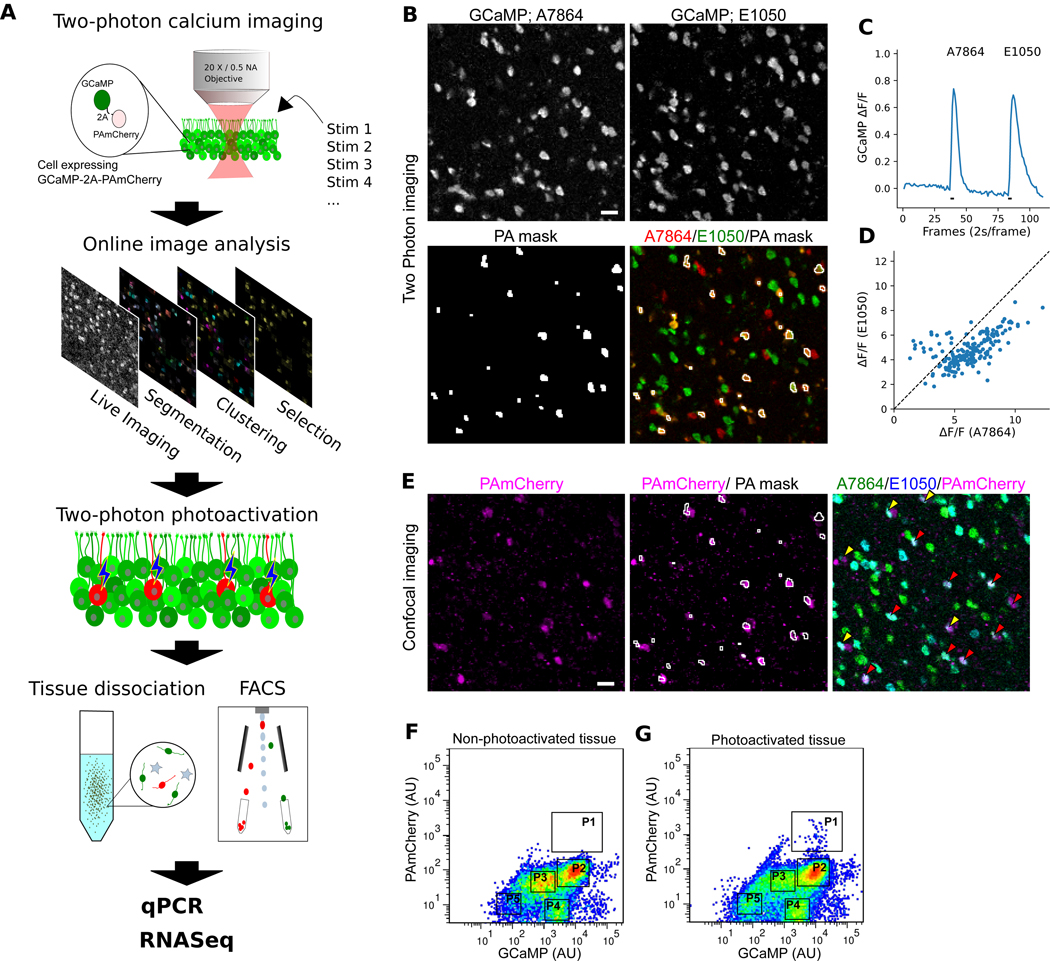
Fig. 1. Two-photon photoactivation can tag neurons chosen by activity pattern.
(A) The work flow of physiological optical tagging sequencing (PhOTseq). (B–D) Two-photon calcium imaging of the VNE explanted from a GCaMP5g-p2a-PAmCherry transgenic mouse. (B) Representative images of calcium chemosensory response evoked by either 10 μM A7864 (top left) or 10 μM E1050 (top right). Photoactivation mask (bottom) selects cells responsive to both stimuli. Scale bar: 20 μm. (C) The average GCaMP intensity obtained from masked cells. Black bars: delivery time-course of stimuli. (D) Individual responses (ΔF/F) among masked cells to 10 μM A7864 and 10 μM E1050. (E) Confocal imaging after online two-photon photoactivation. PAmCherry signals (left), PAmCherry signals with two-photon PA mask (middle), and PAmCherry signals with calcium responses to either 10 μM A7864 (cyan) or 10 μM E1050 (green) (right). Red and yellow arrow heads: the examples of PA mask regions associated with single and double PAmCherry-positive neurons, respectively. Scale bar: 20 μm. (F and G) FACS analysis of cells from non-photoactivated tissue (F) or from photoactivated tissue (G). Clusters were marked as P1, P2, P3, P4, and P5. P1 and P2 represent a photoactivated and an experimental control group, respectively. Of the total population, P1 and P2 respectively accounted for 0.013% and 33.9% in (F) or 0.218% and 37.3% in (G).
We created several plasmids and viruses all expressing variants of GCaMP-2A-PAmCherry. The self-cleaving 2A peptide (13) allowed both proteins to be expressed from a single mRNA; the tight coupling facilitated ratiometric analysis to compensate for variability in expression. Initial in vitro and in vivo experiments demonstrated the feasibility of performing both calcium imaging and photo-tagging using this reporter (Fig. S1 and Fig. S2). To test the performance of the complete pipeline (Fig. 1A), we turned to mouse vomeronasal sensory neurons (VSNs), whose primary function is pheromone sensing and social communication. VSNs exhibit dense cell packing (14) (thus challenging photo-tagging accuracy), extreme functional heterogeneity, and an unambiguous relationship between molecular identity and physiological function (15–18). We generated a transgenic tetO-GCaMP5g-2A-PAmCherry mouse and drove expression in all VSNs via OMP-IRES-tTA (Fig. S3A). We observed calcium responses from the VSNs, and two-photon photoactivation resulted in photo-tagging at single-cell resolution (Fig. S3 and Fig. S4).
We evoked combinatorial VSN responses using two ligands, 5-androsten-3β, 17β-diol disulphate (A7864) and 1, 3, 5(10)-estratrien-3, 17β-diol disulfate (E1050). A custom online algorithm automatically segmented responsive neurons; out of 430 neurons responding to at least one ligand, 192 neurons responded to both ligands. Voxels corresponding to these dually-responsive neurons were chosen to create a mask so that only these voxels were illuminated during photoactivation (Fig. 1, B–D). Photoactivated neurons were readily distinguished from the background, and PA tagged an average of 1.32 cells per region (Fig. 1E and Fig. S5). The photoactivated vomeronasal epithelia (VNEs) were subsequently subjected to dissociation and fluorescence-activated cell sorting (FACS; Fig. 1, F and G). In FACS analysis, 4–5 abundant clusters were observed, of which only one (P1) was almost entirely dependent upon photoactivation. This population represents the one selected later for sequencing.
We used PhOTseq to begin unraveling the logic of chemosensation. Fewer than a dozen vomeronasal receptors (VRs) have ligands of known structure (5, 19, 20), and these are scattered widely across the gene family. We reasoned that a saturation analysis of vomeronasal receptors responsible for encoding a focused region of “chemical space” will provide new insights into the molecular logic of chemosensation. First, we examined functional types of VSNs broadly by light sheet calcium imaging while presenting 15 different sulfated steroids, of which six were also delivered over a range of concentrations, for a total of 27 different chemosensory stimuli. Approximately 15,000 neurons, ~10% of the total VSN population per hemisphere, were visualized in each imaging volume (3). From three imaging volumes, more than 5000 neurons responded to at least one of the ligands, and they fell naturally into 20 different response types (Fig. 2, A and B). From this reference clustering, we chose to photo-tag the most abundant cell type, cluster1, and three others showing similar chemoreceptive fields (cluster2, 3, and 6) (Fig. S6), among which cluster3 comprised fewer than 2% of the responsive neurons and presumably ~0.2% of the total VSN population (Fig. 2B). As a control group, we selected one of the most dissimilar cell types, cluster19, which strongly responded to sulfated pregnanolones.
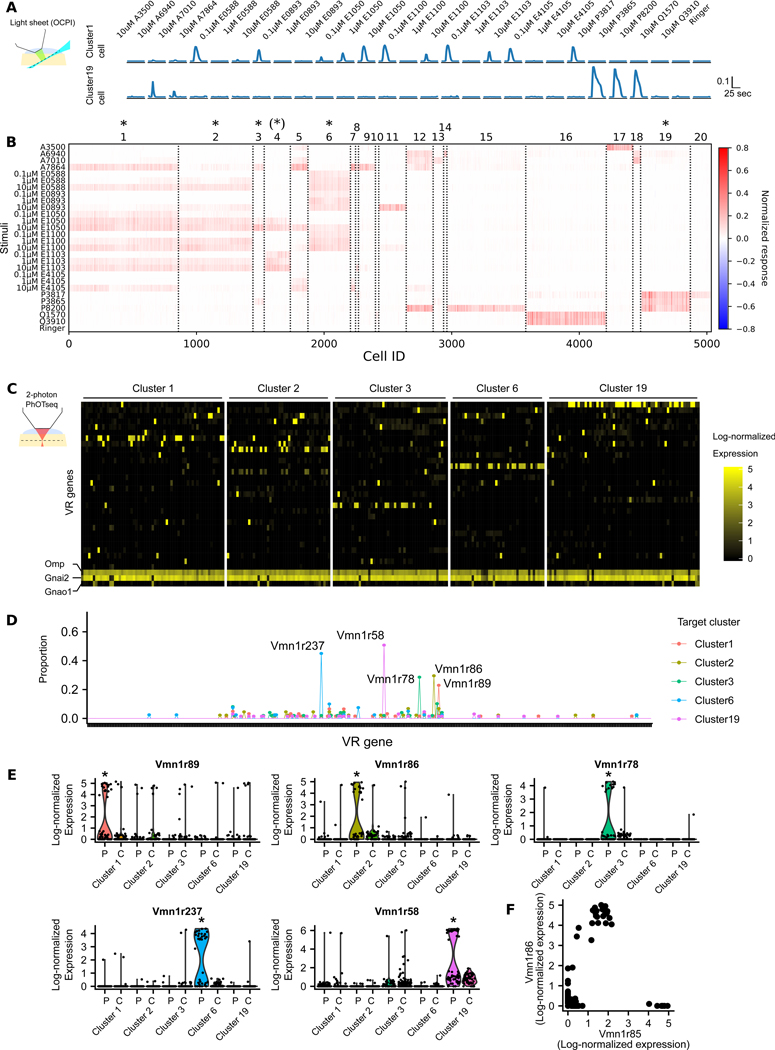
Fig. 2. PhOTseq identified of VR genes of overlapping cell types.
(A) Example calcium traces (normalized ΔF) from two cells. Amplitude was normalized by the maximum amplitude of all cells recorded. Ligands are listed at the top. (B) Neuronal responses to sulfated steroids. Cells are on columns, stimuli on rows. If not indicated, the ligand concentration is 10 μM. The color bar indicates normalized response. Cluster identities are reported at the top. “*” marks PhOTseq target cell types. “(*)” marks the functional type whose receptor identity was discovered during analysis by ectopic expression in Figure 3. (C) Expression of the 30 most highly expressed VR genes when averaged across all sequenced cells; also shown are three marker genes (Omp, Gnai2, and Gnao1). Cells are on columns, genes on rows. PhOTseq-targeted functional types are shown at the top; cells belonging to these functional clusters were specifically photoactivated and sequenced. The colorbar indicates log-normalized expression level. (D) The proportion of a VSN type in each group shown in (C). Each tick on the horizontal axis represents a different VR gene. Each functional type exhibited only one common VSN type. (E) The expression of five VR genes across different experiment groups. “P” indicates photoactivated cells and “C” indicates non-photoactivated control cells. Asterisk (*) indicates padj < 0.01 (Wilcoxon rank-sum test; padj< 0.01 (Wilcoxon rank-sum test; padj: 2.7×10−11, 2.4×10−10, 1.1×10−20, 1.3×10−40, 2.7×10−30; average fold difference: 6.2, 23.8, 80.7, 75.7, 45.9 for Vmn1r89, Vmn1r86, Vmn1r78, Vmn1r237, Vmn1r58, respectively). (F) Single-cell expression of Vmn1r85 and Vmn1r86 is non-exclusive.
For cell type selection under two-photon microscopy, based on Figure 2B we used a subset of stimuli that distinguish cells among these five selected clusters (Fig. S7, Fig. S8, and Movie S1). Following online two-photon photoactivation, we collected photoactivated or non-photoactivated experimental control cells by FACS and performed single cell RNA sequencing. We obtained a total of 622 qualified cells (Fig. S9), among which 61, 44, 49, 40, and 65 photoactivated cells were from experiments aiming at functionally-defined cluster1, cluster2, cluster3, cluster6, and cluster19 cells, respectively (Fig. 2C).
Physiological responses of chemosensory neurons are thought to be largely determined by the VR genes they express (21). Therefore, our investigation was focused on the expression of VR genes. Among photoactivated cells, any given cluster exhibited substantial enrichment of at least one VR gene, with different VR genes enriched in different clusters (Fig. 2C). In contrast, among control cells VR gene expression was sporadic (Fig. S10). Because our targeting accuracy was less than 100% (Fig. S5C), we also expected some sporadic expression among cluster-selected photoactivated neurons (Fig. 2C). When the type of a VSN was defined by its maximally-expressed VR gene, only one abundant type was found among cluster-selected photoactivated neurons (Fig. 2D; cluster1: Vmn1r89 type (25%); cluster2: Vmn1r86 type (30%); cluster3: Vmn1r78 type (27%); cluster6: Vmn1r237 type (50%); cluster19: Vmn1r58 type (58%)). These five VR genes were the only significantly enriched receptor gene in each photoactivated group compared to all the other cells (Fig. 2E). During these analyses, we updated the Vmn1r237 gene model as the read coverage of Vmn1r237 and cloning revealed 3’UTRs missing from existing gene models (Fig. S11 and Data S1). We also unexpectedly observed coexpression of Vmn1r85 in Vmn1r86 neurons (Figs. 2F and S12A–C). Including data from our control and sporadic cells, coexpression was also observed for several other VR gene pairs, in each case consisting of a genomically-adjacent pair (Fig. S12, D–F). Taken together, we identified six putative VR genes mediating a focused set of responses.
To test whether PhOTseq identified true receptor-ligand pairs, we performed a gain-of-function study. Due to abnormal localization of VR proteins (22), in vitro heterologous expression systems have had little success, particularly for the V1R family (23). We reasoned that a VSN cell’s endogenous machinery would allow functional expression of a VR gene; seeking a more efficient route than making a transgenic mouse (5), we explored a virus-mediated approach. Among viral vectors we tested, intravenous injection of rAAV2/8-CAG-GFP was able to induce expression of GFP in a subset of VSNs (Fig. S13). We delivered rAAV2/8-CAG-GCaMP-2A-VR, where VR was one of the genes identified by PhOTseq, and for each obtained multiple neurons showing statistically significant responses to at least one ligand (Fig. 3, A and B, and Movie S2). In each case, the dominant response pattern matched the expected PhOTseq response pattern (Fig. 3, C and D). We also tested Vmn1r85, because this gene was found to be coexpressed with Vmn1r86 (Fig. 2C). Unexpectedly, the response pattern matched that of cluster4, which was another group similar to cluster1 but not chosen for analysis by PhOTseq (Fig. 3, C and D). Because sequencing revealed control neurons that expressed only Vmn1r85 (Fig. 2F), functional cluster4 cells likely expressed Vmn1r85.
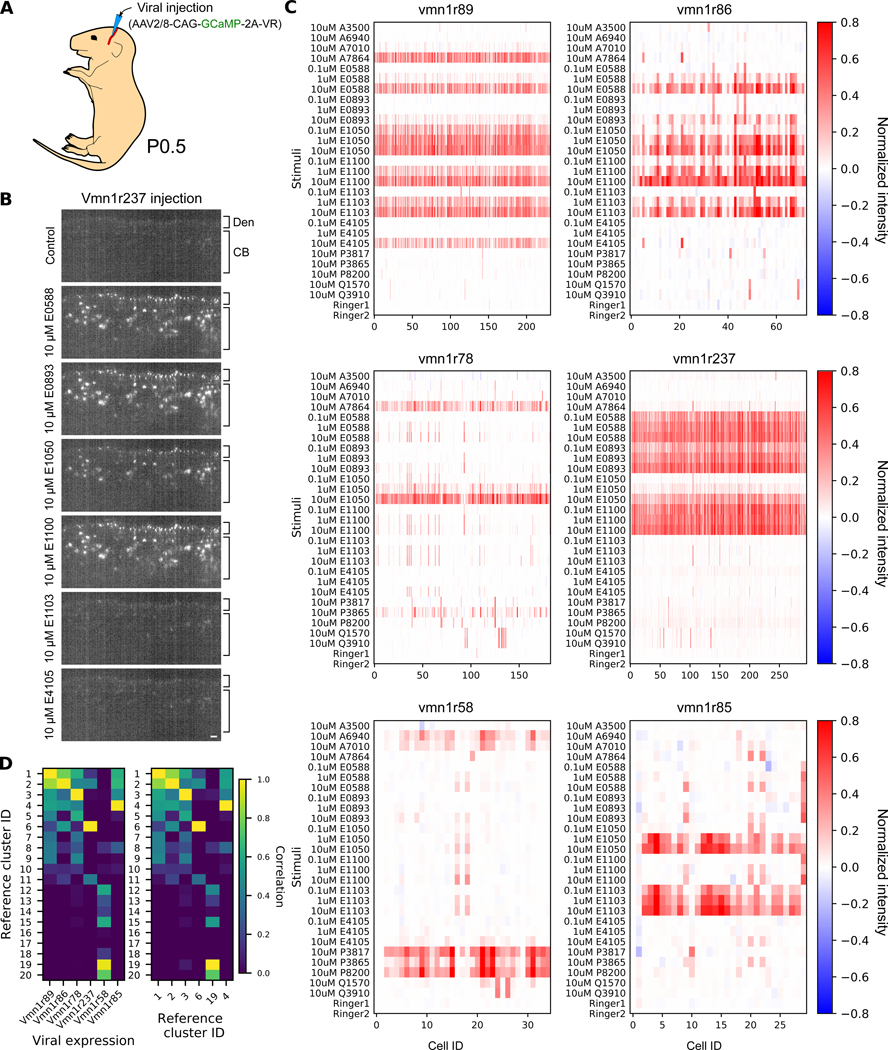
Fig. 3. Ectopic expression enabled functional analysis of vomeronasal receptors.
(A) Expression via AAV injection in the temporal vein of newborn pups. (B) Optical section from light sheet calcium imaging after the ectopic expression of GCaMP-2A-Vmn1r237 in response to different ligands. The apical layer is occupied by dendritic tips (Den) with cell bodies (CB) below. Scale bar: 20 μm (C) Calcium response after ectopic expression of GCaMP-2A-Vmn1r89, -Vmn1r86, -Vmn1r78, -Vmn1r237, -Vmn1r58, or -Vmn1r85. Ligands are identical to those in Figure 2B. (D) Pairwise correlation between the reference clustering and the ectopic responses shown in (C) (left) or autocorrelogram of the reference clustering (right).
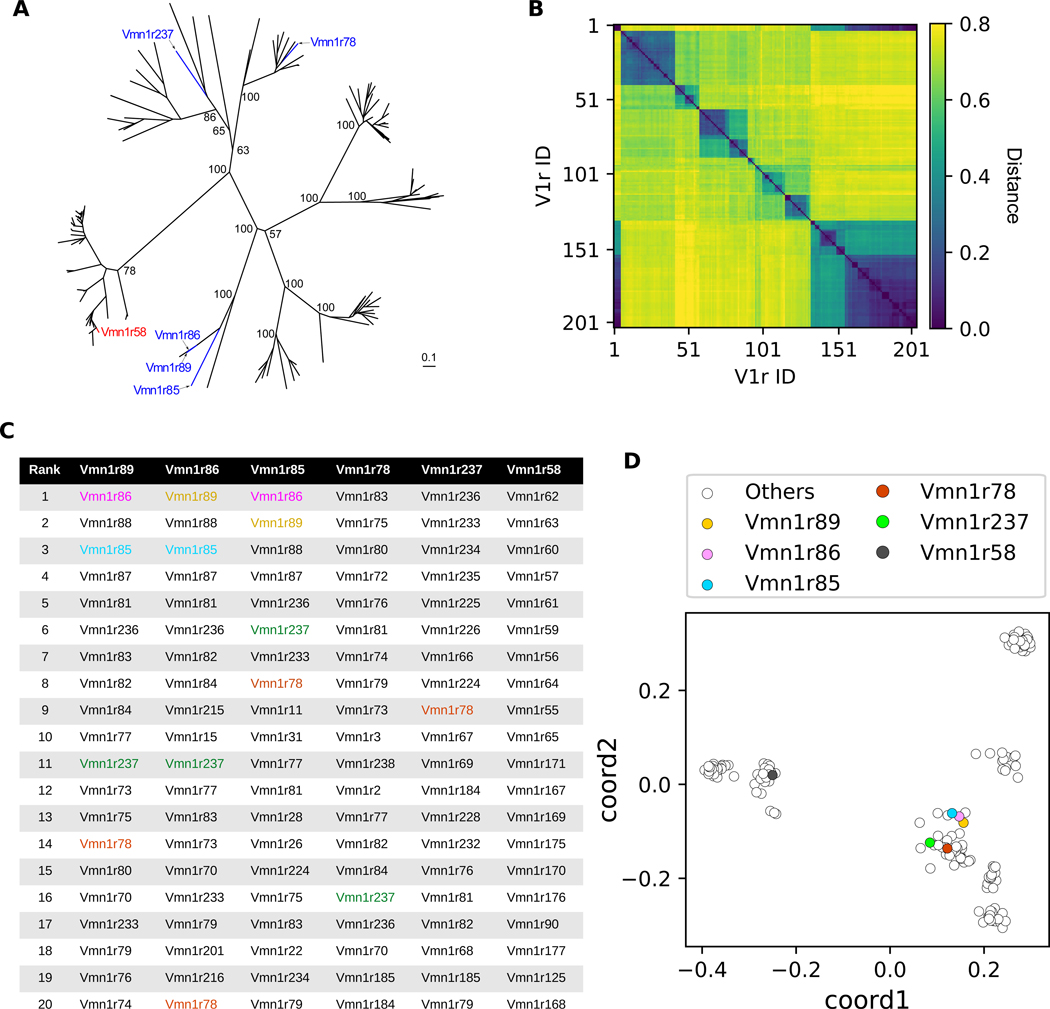
Only a few studies have examined the relationship between VR sequence and chemosensory function (5, 19) and never from the vantage point of complete knowledge of the genes that underlie a set of nearest neighbors in terms of function. To investigate this relationship, we performed multiple sequence alignment (MSA) of V1R protein sequences followed by phylogenetic tree analysis (Fig. 4A). Among V1R genes expressed in functionally similar types, Vmn1r89, Vmn1r86, and Vmn1r85 were close to each other in the sense of putative evolutionary distance. On the other hand, two of the functionally similar cell types, Vmn1r78 and Vmn1r237, belonged to different branches. In this representation Vmn1r58, which was the chosen outlier in terms of function, was not notably more divergent from Vmn1r89, Vmn1r86, and Vmn1r85 than the more functionally-similar Vmn1r78 and Vmn1r237. Consequently this tree structure representation did not suggest a particularly close correspondence between function and primary sequence.
Fig. 4. Sequence- and chemoreceptive-similarity are strongly correlated.
(A) An unrooted phylogenetic tree of V1R genes. The five functionally similar (blue) and one distant (red) receptors studied here are marked. The scale bar indicates the number of amino acid substitutions per site. Numbers indicate bootstrap values. (B) Pairwise distances among 204 V1R protein sequences (see Methods). (C) For these deorphanized VRs, the top 20 nearest VRs, based on (B), are rank-ordered. The deorphanized VRs are colored if shown in the table. (D) A 2-dimensional representation of the distance matrix shown in (B). Each dot represents a single VR protein.
However, when the pairwise distances acquired from the MSA analysis were examined carefully, we noticed that despite their apparent evolutionary distance, Vmn1r78 and Vmn1r237 were consistently among the most closely-related VR genes to Vmn1r89, Vmn1r86, and Vmn1r85 (Fig. 4B and C). Among 204 V1R genes examined, the median neighbor rank among these VRs was 11, implying that these genes are typically among the top few-percent closest matches to one another. We therefore considered the possibility that phylogeny, in attempting to construct plausible ancestry, inaccurately models relatedness between sequences that are not nearest neighbors. To more comprehensively evaluate and visualize all pairwise distances, we performed a classical multidimensional scaling analysis to project the high-dimensional sequence-based distance relationships into two dimensions (Fig. 4D). This resulted in a different picture of the gene family, whose dominant feature was the presence of seven to nine apparent clusters. The functionally similar receptors, including Vmn1r78 and Vmn1r237, were near neighbors, whereas the functionally dissimilar receptor Vmn1r58 was positioned in a distant cluster. We conclude that relationships among the primary sequences of these VR receptors are strongly correlated with their degree of functional similarity.
Our results demonstrate the utility of PhOTseq, an all-optical solution to the problem of cell type selection and labeling. It provides an opportunity to study the relationships between genes and physiological function even in extremely rare cell types that can be defined only through extensive functional characterization. PhOTseq will be applicable when individual neurons respond combinatorially to various conditions, including sensory stimulation, emotional status, and behavior. We further anticipate that in vivo ectopic expression will be widely applicable for characterizing VR-ligand pairings. In addition, our data show unexpected receptor coexpression, a specific and systematic departure from the “one neuron–one receptor rule” (21) whose potential roles will need to be evaluated in many aspects of olfactory function. Lastly, our study provides biological insight on chemosensation by comprehensively mapping receptor-ligand pairings for chosen subsets of the vomeronasal sensory population, and suggests that saturation analyses can reveal sequence-function coupling unanticipated by single-ligand studies (24, 25).
Supplementary Material
Acknowledgments:
We thank Ron Yu for sharing the TetO vector and OMP-IRES-tTA mouse, Paul H. Taghert for the HEK cell line, Heide Schoknecht for mouse care, and Dae Woo Kim for developing imaging software. We thank Terra Barnes, Juntao Chen, Joseph Corbo, Xiaoyan Fu, Cody Greer, Alessandro Livi, Richard Roberts, and Manning Zhang for suggestions and comments. This work was supported by the Hope Center Viral Vectors Core and the Mouse Genetics Core at Washington University School of Medicine.
Funding: This work was funded by NIH/NIDCD R01 DC005964 and DC010381.
Footnotes
Competing interests: The authors have no competing interests.
Data and materials availability: Accession numbers are available in Supplementary Materials.
References and Notes:
- 1.Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C, Interneurons of the neocortical inhibitory system. Nat. Rev. Neurosci 5, 793–807 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Nassar M, Simonnet J, Lofredi R, Cohen I, Savary E, Yanagawa Y, Miles R, Fricker D, Diversity and overlap of parvalbumin and somatostatin expressing interneurons in mouse presubiculum. Front. Neural Circuits 9, 1–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu PS, Lee D, Holy TE, Experience-Dependent Plasticity Drives Individual Differences in Pheromone-Sensing Neurons. Neuron. 91, 878–892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okuno H, Regulation and function of immediate-early genes in the brain: Beyond neuronal activity markers. Neurosci. Res 69, 175–186 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Haga-Yamanaka S, Ma L, He J, Qiu Q, Lavis LD, Looger LL, Yu CR, Integrated action of pheromone signals in promoting courtship behavior in male mice. Elife. 2014, 1–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuzik J, Zeisel A, Máté Z, Calvigioni D, Yanagawa Y, Szabó G, Linnarsson S, Harkany T, Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat. Biotechnol 34, 175–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu S, Luo S, Evgrafov O, Li R, Schroth GP, Levitt P, Knowles JA, Wang K, Single-neuron RNA-Seq: Technical feasibility and reproducibility. Front. Genet 3, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, Sandberg R, Tolias AS, Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol 34, 199–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fosque BF, Sun Y, Dana H, Yang CT, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, Jayaraman V, Looger LL, Schreiter ER, Labeling of active neural circuits in vivo with designed calcium integrators. Science (80-. ). 347, 755–760 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Akerboom J, Chen T-W, Wardill TJ, Tian L, Marvin JS, Mutlu S, Carreras Caldéron N, Esposti F, Borghuis BG, Sun XR, Gordus A, Orger MB, Portugues R, Engert F, Macklin JJ, Filosa A, Aggarwal A, Kerr RA, Takagi R, Kracun S, Shigetomi E, Khakh BS, Baier H, Lagnado L, Wang SS-H, Bargmann CI, Kimmel BE, Jayaraman V, Svoboda K, Kim DS, Schreiter ER, Looger LL, Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci 32, 13819–13840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS, Ultrasensitive fluorescent proteins for imaging neuronal activity. 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.V Subach F, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, V Verkhusha V, Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan MD, King AMQ, Thomas GP, Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol 72, 2727–2732 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Wilson KCP, Raisman G, Age-related changes in the neurosensory epithelium of the mouse vomeronasal organ: extended period of post-natal growth in size and evidence for rapid cell turnover in the adult. Brain Res. 185, 103–113 (1980). [DOI] [PubMed] [Google Scholar]
- 15.Bozza TC, Feinstein P, Zheng C, Mombaerts P, Odorant receptor expression defines functional units in the mouse olfactory system. J. Neurosci (2002), doi:20026321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P, Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell (2004), doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Hallem EA, Ho MG, Carlson JR, The molecular basis of odor coding in the Drosophila antenna. Cell (2004), doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Grosmaitre X, Vassalli A, Mombaerts P, Shepherd GM, Ma M, Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: a patch clamp analysis in gene-targeted mice. Proc. Natl. Acad. Sci 103, 1970–1975 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, Dulac C, Molecular organization of vomeronasal chemoreception. Nature. 478, 241–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong WM, Cao J, Zhang X, Doyle WI, Mercado LL, Gautron L, Meeks JP, Physiology-forward identification of bile acid sensitive vomeronasal receptors. bioRxiv, 766592 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mombaerts P, Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci 5, 263–278 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Loconto J, Papes F, Chang E, Stowers L, Jones EP, Takada T, Kumánovics A, Fischer Lindahl K, Dulac C, Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 112, 607–18 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Stein B, Alonso MT, Zufall F, Leinders-Zufall T, Chamero P, Functional overexpression of vomeronasal receptors using a herpes simplex virus type 1 (HSV-1)-derived amplicon. PLoS One. 11, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, Matsunami H, Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat. Neurosci 18, 1446–1454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von der Weid B, Rossier D, Lindup M, Tuberosa J, Widmer A, Col JD, Kan C, Carleton A, Rodriguez I, Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat. Neurosci 18, 1455–1463 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Nagy A, Gertsenstein M, Vintersten K, Behringer R, Manipulating the mouse embryo: a laboratory manual (Firefly Books, 2003). [Google Scholar]
- 27.Zolotukhin S, Potter M, Zolotukhin I, Sakai Y, Loiler S, Fraites TJ Jr, Chiodo VA, Phillipsberg T, Muzyczka N, Hauswirth WW, others, Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. 28, 158–167 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Vadakkan TJ, Culver JC, Gao L, Anhut T, Dickinson ME, Peak multiphoton excitation of mCherry using an optical parametric oscillator (OPO). J. Fluoresc 19, 1103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu PS, Holy TE, in Pheromone Signaling: Methods and Protocols, Touhara K, Ed. (Humana Press, Totowa, NJ, 2013; 10.1007/978-1-62703-619-1_14), pp. 201–210. [DOI] [Google Scholar]
- 30.Comer ML, Delp EJ, The EM/MPM algorithm for segmentation of textured images: analysis and further experimental results. IEEE Trans. image Process 9, 1731–1744 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Holekamp TF, Turaga D, Holy TE, Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron. 57, 661–72 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Holy TE, BlockRegistration package (2019). [Google Scholar]
- 33.Turaga D, Holy TE, Organization of vomeronasal sensory coding revealed by fast volumetric calcium imaging. J. Neurosci 32, 1612–1621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holy TE, NeighborhoodClustering package (2019). [Google Scholar]
- 35.Kaur A, Dey S, Stowers L, in Pheromone Signaling: Methods and Protocols, Touhara K, Ed. (Humana Press, Totowa, NJ, 2013; 10.1007/978-1-62703-619-1_13), pp. 189–200. [DOI] [Google Scholar]
- 36.Arnson HA, Fu X, Holy TE, Multielectrode array recordings of the vomeronasal epithelium. J. Vis. Exp. JoVE (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leinders-Zufall T, Ishii T, Mombaerts P, Zufall F, Boehm T, Structural requirements for the activation of vomeronasal sensory neurons by MHC peptides. Nat. Neurosci 12, 1551–8 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, Regev A, Yanai I, CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Pyl PT, Huber W, HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahne F, Ivanek R, in Statistical Genomics: Methods and Protocols, Mathé E, Davis S, Eds. (Springer; New York, New York, NY, 2016; 10.1007/978-1-4939-3578-9_16), pp. 335–351. [DOI] [Google Scholar]
- 43.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, Integrative genomics viewer. Nat. Biotechnol 29, 24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibarra-Soria X, Levitin MO, Saraiva LR, Logan DW, The olfactory transcriptomes of mice. PLoS Genet. 10, e1004593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lampe SEG, Kaspar BK, Foust KD, Intravenous injections in neonatal mice. J. Vis. Exp. JoVE (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang K, Lee D, Hong S, Park S-G, Song M-R, The E3 ligase Mind bomb-1 (Mib1) modulates Delta-Notch signaling to control neurogenesis and gliogenesis in the developing spinal cord. J. Biol. Chem 288, 2580–2592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray PA, Hayes JA, Ling GY, Llona I, Tupal S, Picardo MCD, Ross SE, Hirata T, Corbin JG, Eugen J \’\in, others, Developmental origin of preBötzinger complex respiratory neurons. J. Neurosci 30, 14883–14895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawley J, Handbook of biological confocal microscopy (Springer Science & Business Media, 2010). [Google Scholar]
- 49.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, others, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A, RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y, ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol 8, 28–36 (2017). [Google Scholar]
- 52.Silvotti L, Cavalca E, Gatti R, Percudani R, Tirindelli R, A recent class of chemosensory neurons developed in mouse and rat. PLoS One. 6, e24462 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.