Infecting 90% of the global population, HSV-1 and HSV-2 represent some of the most prevalent viruses in the world. Much of their success can be attributed to their ability to establish lifelong latent infections in the dorsal root ganglia (DRG). It is still largely unknown, however, how host transcription factors are involved in establishing this latency. Here, we report that RUNX1, expressed highly in DRG, binds HSV-1 genome, represses transcription of numerous viral genes, and suppresses productive in vitro infection. Our computational work further suggests this strategy may be used by other herpesviruses to reinforce latency in a cell-specific manner.
KEYWORDS: herpesviruses, transcriptional repression, virus-host interactions
ABSTRACT
Herpes simplex virus 1 (HSV-1) and HSV-2 can efficiently establish lifelong, transcriptionally silent latency states in sensory neurons to escape host detection. While host factors have previously been associated with long-range insulators in the viral genome, it is still unknown whether host transcription factors can repress viral genes more proximately to promote latency in dorsal root ganglion (DRG) neurons. Here, we assessed whether RUNX (runt-related transcription factor) transcription factors, which are critical in the development of sensory neurons, could be binding HSV-1 genome directly to suppress viral gene expression and lytic infection. Using previously published transcriptome sequencing data, we confirmed that mouse DRG neurons highly express Runx1 mRNA. Through computational analysis of HSV-1 and HSV-2 genomes, we observed that putative RUNX consensus binding sites (CBSs) were more enriched and more closely located to viral gene transcription start sites than would be expected by chance. We further found that RUNX CBSs were significantly more enriched among genomes of herpesviruses compared to those of nonherpesviruses. Utilizing an in vitro model of HSV-1 infection, we found that overexpressed RUNX1 could bind putative binding sites in the HSV-1 genome, repress numerous viral genes spanning all three kinetic classes, and suppress productive infection. In contrast, knockdown of RUNX1 in neuroblastoma cells induced viral gene expression and increased HSV-1 infection in vitro. In sum, these data support a novel role for RUNX1 in directly binding herpesvirus genome, silencing the transcription of numerous viral genes, and ultimately limiting overall infection.
IMPORTANCE Infecting 90% of the global population, HSV-1 and HSV-2 represent some of the most prevalent viruses in the world. Much of their success can be attributed to their ability to establish lifelong latent infections in the dorsal root ganglia (DRG). It is still largely unknown, however, how host transcription factors are involved in establishing this latency. Here, we report that RUNX1, expressed highly in DRG, binds HSV-1 genome, represses transcription of numerous viral genes, and suppresses productive in vitro infection. Our computational work further suggests this strategy may be used by other herpesviruses to reinforce latency in a cell-specific manner.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) and HSV-2 infect about 90% of the world’s population, classically resulting in oral and genital lesions (1). Much of their success as viruses can be attributed to their relatively unique ability in establishing transcriptionally silent, lifelong infections in the sensory neurons of dorsal root ganglia (DRG). While current treatments inhibit active DNA replication during reactivation (2), there are currently no approved treatments targeting HSV-1 or HSV-2 in their latent infection, reflecting the still-incomplete understanding of the mechanisms of latency.
After cell entry, HSV-1 and HSV-2 inject their double-stranded DNA genomes into the host nucleus, where they can freely associate with histones and host factors (3). One of the major regulators of latency is thought to be host-derived CTCF, commonly associated with chromatin insulators. Clusters of CTCF binding sites have been identified flanking immediate early (IE) genes required for the initiation of lytic infection (4), and a subset of these sites has been shown to produce an enhancer blocking effect by recruiting PRC2 and SUZ12 (5). While these interactions help to explain the repression of IE genes that occurs during latency, it is unclear whether there are more proximally based mechanisms for the repression of other viral genes. Moreover, it is not obvious how this mechanism might mediate latency in a cell-specific manner in DRG neurons, especially given the ubiquitous expression of CTCF (6). Thus, we hypothesized that cell-lineage-specific transcription factors (TFs), particularly repressive ones, could bind more proximally to promoter regions of viral genes and help promote latency.
In light of this hypothesis, we decided to investigate the RUNX (runt-related transcription factor) family of transcription factors, which are critical in the development of DRG sensory neurons, as potential mediators of HSV latency. While RUNX3 controls the differentiation of proprioceptive, position-sensing DRG neurons, RUNX1 plays an analogous role in the development of nociceptive, pain-sensing DRG neurons (7). RUNX1 remains highly expressed in mature TRPV1+ nociceptive neurons (8), which are required for HSV-1 infection and subsequent lethality (9). (RUNX2, on the other hand, is a major regulator of bone development [10] and is not classically associated with sensory neuron development.) At a molecular level, RUNX1 and RUNX3 can each heterodimerize with CBFβ and act as repressors at target genes by recruiting histone deacetylases and methyltransferases (11). Moreover, RUNX1 has already been shown to bind HIV-1 long terminal repeat to promote latency, and clinically, its expression in T cells inversely correlates with HIV-1 viral load in patients (12). Considering these findings, then, we thought that RUNX TFs could be mediating latent infections in HSV-1 and HSV-2 in an analogous fashion.
To assess this hypothesis, we first utilized previously published RNA-Seq data to verify that Runx1 and Runx3 mRNA are highly expressed in DRG neurons compared to enteric neurons, which are readily infected by HSV-1 and are killed as a result of lytic infection (9). Through computational analysis, we found that the genomes of HSV-1 and HSV-2 contain more putative RUNX consensus binding sites (CBSs) than would be expected by chance and that these sites are significantly closer to transcription start sites (TSSs) of viral genes compared to random simulations. We repeated similar analyses with other viruses and found that RUNX CBSs are broadly enriched among herpesviruses but not among nonherpesviruses. To validate these in silico findings, we infected HEK293T cells overexpressing RUNX1 and RUNX3 with HSV-1 and found that overexpression of RUNX1, but not RUNX3, repressed numerous viral genes—spanning immediate early (alpha) genes, early (beta) genes, and late (gamma) genes—and that this repression was significantly reduced with the DNA-binding mutant D198G RUNX1. Consistent with these findings, we found by using chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) that overexpressed RUNX1, but not RUNX3 or mutant RUNX1, bound to multiple putative CBSs located upstream of viral gene TSSs. Lastly, we confirmed that stable overexpression of RUNX1, but not RUNX3 or mutant RUNX1, significantly decreased cellular HSV-1 infection, as measured by flow cytometry and plaque assays. Taken together, these data suggest that RUNX1 binds the genome of HSV-1 and suppresses transcription of viral genes from all three kinetic classes, which may ultimately confer a survival advantage to the virus by reinforcing cell-type-specific latent infection and promoting the accumulation of CBSs in its genome.
RESULTS
Putative RUNX binding sites are more enriched and more closely located to viral gene TSSs in HSV-1 and HSV-2 genomes than would be expected by chance.
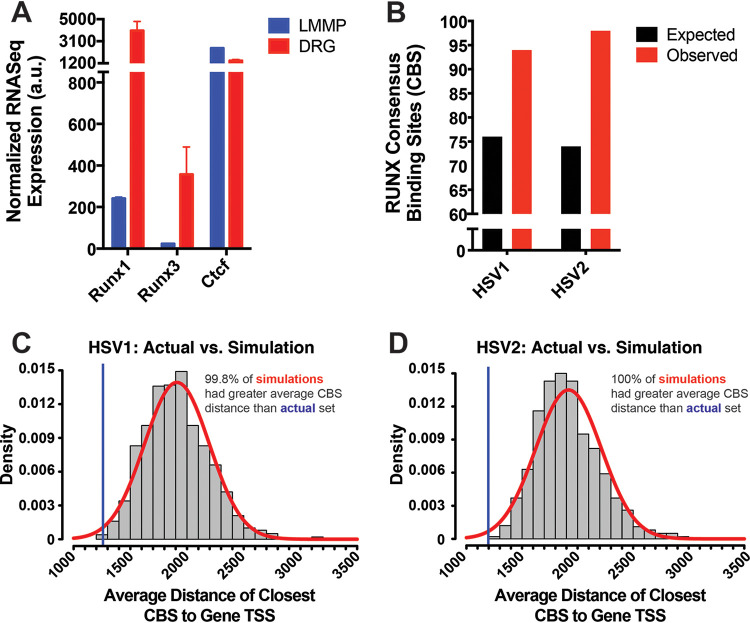
In order to first confirm that RUNX transcription factors are highly expressed by DRG neurons, we utilized previously published transcriptome sequencing (RNA-Seq) data that included analysis of host genes in mouse DRG neurons and longitudinal muscle myenteric plexus (LMMP) neurons (9). Here, we found that HSV-1 undergoes lytic infection in LMMP neurons, eventually leading to their destruction through a partially neutrophil-dependent process. Because of this stark contrast with the more quiescent infection in DRG neurons, we used LMMP neurons as a comparator in our analysis and subsequently found that both Runx1 and Runx3 mRNA expressions were significantly higher in DRG neurons (Fig. 1A). Consistent with this finding in mice, fully differentiated human DRG neurons also have also been reported to highly express RUNX1 (13–15). Importantly, Ctcf mRNA expression in mouse DRG neurons was comparable, if not lower, compared to mouse LMMP neurons (Fig. 1A), calling into question whether CTCF is sufficient in explaining DRG-specific latency.
FIG 1.
Putative RUNX CBSs are more enriched and more closely localized to the TSSs of viral genes in HSV-1 and HSV-2 genomes than would be expected by chance. (A) Normalized RNA-Seq expression of Runx1, Runx3, and Ctcf mRNA in longitudinal muscle myenteric plexus (LMMP) and dorsal root ganglion (DRG) neurons of B6 mice from the data set previously published by Khoury-Hanold et al. (9). The data are means ± the standard errors (SE; n = 2). (B) Quantification of observed and expected putative RUNX consensus binding sites (CBS) in HSV-1 and HSV-2 genomes. The sequences utilized for CBS analysis are listed in Table 1. The expected numbers were calculated accounting for each genome’s GC content. (C) Average distance between TSS of viral genes and closest CBS from actual set of binding sites (blue line) and 1,000 simulated random distributions of CBSs (red curve).
Having confirmed the high expression of host RUNX TFs in DRG, we assessed whether RUNX consensus binding sites (CBSs), which can bind both RUNX1 and RUNX3, are present in the viral genomes of HSV-1 and HSV-2. Utilizing computational methods in R, we found that both genomes contain about 25% more binding sites than would be expected by chance, even when considering the GC content of each genome (Fig. 1B). Furthermore, when we analyzed the localization of RUNX CBSs, we observed that the average distance between viral TSSs and the closest upstream CBS was ∼1,200 bp (Fig. 1C and D), with more than 75% of viral genes containing a CBS within 2,000 bp upstream of their TSS (data not shown), which is thought to be a common location where silencer elements are found (16, 17). For comparison, we simulated random distributions of RUNX binding sites and found that the actual distribution of CBSs was closer to the TSS than the vast majority (>99%) of simulations in either genome, raising the possibility that these putative sites may be functional (18). In sum, these results suggest that there may be some selective pressure on HSV-1 and HSV-2 toward the enrichment of RUNX binding sites, particularly in regions upstream of viral genes.
RUNX CBSs are more highly enriched in herpesviruses than in nonherpesviruses.
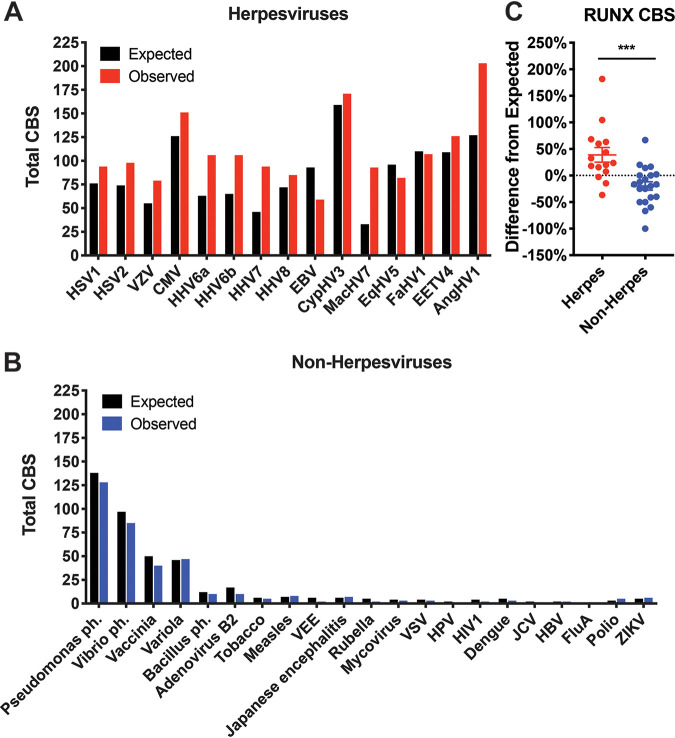
Considering that other herpesviruses also feature latent infections in specific cell types such as sensory neurons (e.g., VZV) or leukocytes (e.g., CMV), which also highly express RUNX1 (19), we were interested in seeing whether trends observed with HSV-1 and HSV-2 might extend to other herpesviruses. To this end, we repeated our quantification analysis of RUNX CBSs in genomes of major herpesviruses available through the National Center for Biotechnology Information (NCBI). As in HSV-1 and HSV-2, herpesviruses (including those infecting nonhuman hosts) harbor significantly more RUNX CBSs than would be expected by chance (Fig. 2A and C). In contrast, nonherpesviruses—including RNA viruses which should not bind transcription factors and thus should represent a more random distribution of binding sites, free from TF-mediated selective pressures—have fewer putative binding sites than predicted by chance (Fig. 2B and C). These data indicate that whatever selective pressure may exist for RUNX CBS enrichment is unique to herpesviruses.
FIG 2.
RUNX CBSs are found more frequently than would be expected by chance across herpesviruses but less frequently in nonherpesviruses. (A) Expected and observed numbers of RUNX CBSs across human and nonhuman herpesviruses. (B) Expected and observed numbers of RUNX CBSs across major nonherpesviruses. Expected numbers were calculated accounting for each genome’s GC content. (C) Percent difference of observed from expected numbers of RUNX CBS calculated for each evaluated virus. The data are means ± the SE. ***, P < 0.001 (versus nonherpesviruses assessed by the Student t test). In addition to being significantly different compared to each other, the herpesvirus difference is significantly greater than zero (P = 0.01), while the nonherpesvirus difference is significantly less than zero (P = 0.02).
Herpesviruses are selectively enriched with RUNX CBSs, but not binding sites of other transcription factors.
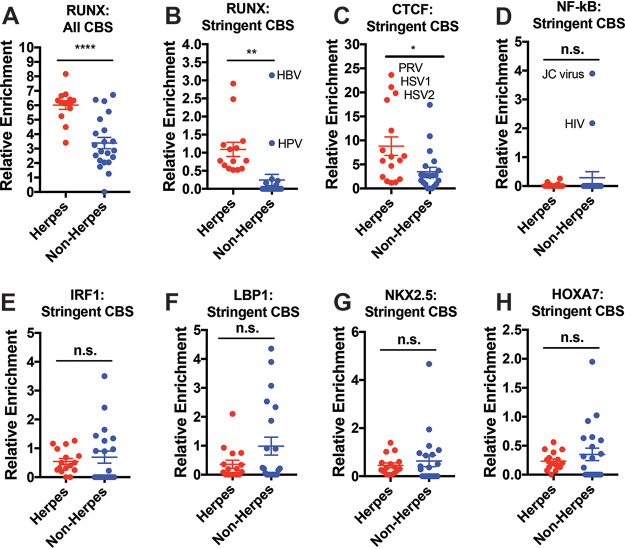
To see how many of the putative sites are more likely to be functional, we tried applying more stringent conditions in analyzing CBSs. By quantifying only CBSs that are within 2,000 bp upstream of viral gene TSSs and not part of a gene body, we found that herpesviruses continue to be more highly enriched with RUNX CBSs than nonherpesviruses (Fig. 3A and B). Interestingly, the two evaluated nonherpesviruses that have comparable relative CBS enrichment to herpesviruses (hepatitis B virus [HBV] and human papillomavirus [HPV]) are also DNA viruses that have chronic, indolent courses of infection, often persisting for years. It is also important to note that this stringent metric, while potentially useful in predicting some novel functional relationships, is not sensitive in detecting all binding interactions. HIV-1, for instance, does not present with a particularly high enrichment score in these calculations despite reports of being bound by RUNX1 (12).
FIG 3.
Herpesviruses are more highly enriched with RUNX CBS, but not other comparable transcription factors. (A) Relative genomic enrichment of all RUNX CBS in herpesviruses and nonherpesviruses. Enrichment calculated as the total CBS/genome size × 10,000. (B) Relative genomic enrichment of RUNX CBS that are located within 2,000 bp of a gene TSS and not present inside a gene body. (C) CBS analysis for CTCF, previously implicated in the latency of HSV-1. (D) CBS analysis for NF-κB, a TF that has already been described to bind to sites within HIV-1 and JC viruses. (E to H) CBS analyses for IRF1, LBP1, NKX2.5, and HOXA7, which have CBS of same length as RUNX (and thus the same expected enrichment). The data are means ± the SE. *, P < 0.05; ***, P < 0.01; *****, P < 0.0001 (versus nonherpesviruses assessed by the Student t test).
We next assessed whether the CBS enrichment seen with herpesviruses is truly specific to RUNX and repeated binding site analysis for other TFs. We first examined CTCF and found that its CBS (which is more frequently detected overall given its shorter 5-mer length) was significantly more enriched in genomes of herpesviruses than in nonherpesviruses (Fig. 3C). This effect, however, was primarily driven by HSV-1, HSV-2, and pseudorabies virus (PRV), which were previously identified to contain clusters of CTCF binding sites (4). The exclusion of these viruses rendered the difference non-significant (data not shown), suggesting that a CTCF-driven mechanism may apply only to a subset of herpesviruses.
Next, we evaluated NF-κB, which has previously been reported to bind directly to HIV-1 and JC virus (20, 21). As expected, HIV-1 and JC virus showed particularly high enrichment when stringent criteria were applied (Fig. 3D), providing some validation for this computational methodology. Aside from these two viruses, however, there were no significant differences in NF-κB CBS enrichment between herpesviruses and nonherpesviruses (Fig. 3D). We also did not observe any significant differences in enrichment of other transcription factors with 7-mer CBSs of comparable expected frequencies (Fig. 3E to H).
RUNX1 overexpression represses transcription of HSV-1 viral genes and binds HSV-1 directly.
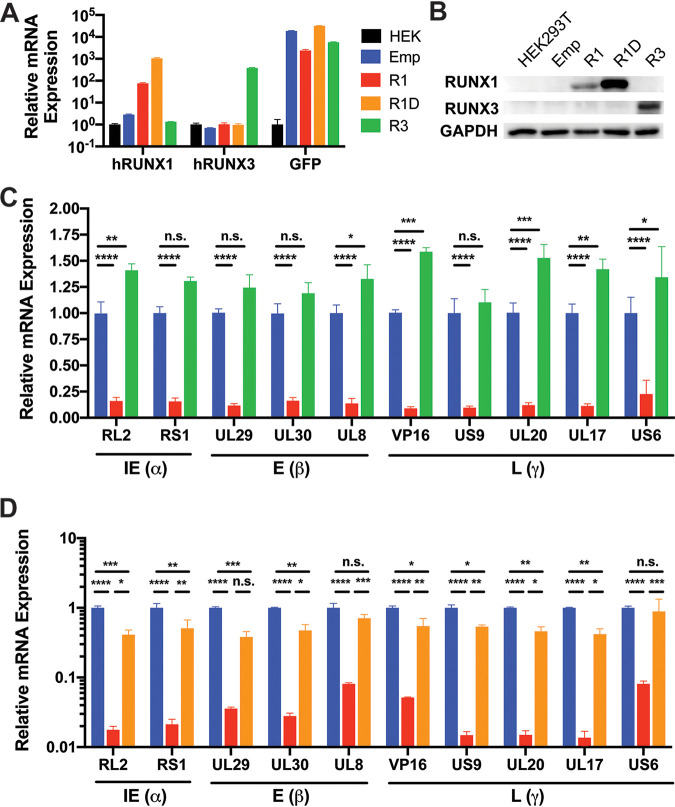
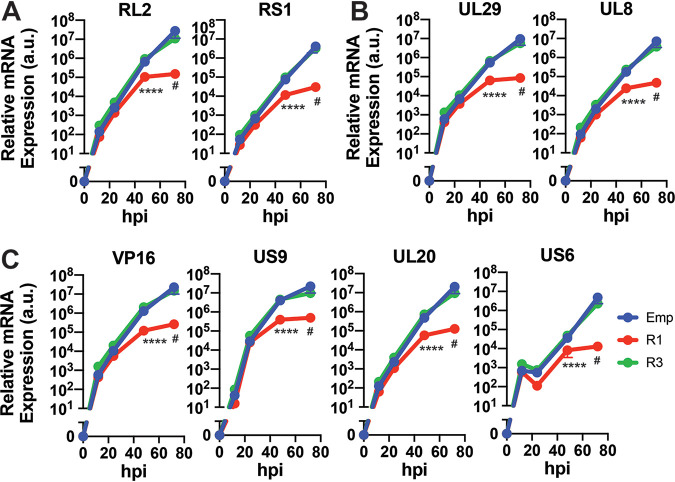
In order to functionally validate these in silico findings, we generated cell lines stably overexpressing WT RUNX1, DNA-binding mutant D198G RUNX1 (22), or RUNX3 (Fig. 4A and B). After infecting these cells with HSV-1 in vitro for 48 h, we measured mRNA expression of two immediate early genes, three early genes, and five late genes by RT-qPCR and found that overexpression of RUNX1, but not RUNX3, resulted in repression of viral genes spanning all kinetic classes (Fig. 4C; see also Fig. 5). We further found that overexpression of mutant D198G RUNX1 significantly impaired viral gene silencing (Fig. 4D), indicating that the DNA-binding activity of RUNX1 is required for this effect.
FIG 4.
Overexpression of RUNX1, but not RUNX3, represses HSV-1 viral gene expression. Validation of RUNX (A) mRNA and (B) protein in stable cell lines overexpressing empty vector (Emp), RUNX1 (R1), DNA-binding mutant D198G RUNX1 (R1D), or RUNX3 (R3). (C) After Emp, R1, and R3 cell lines were infected with HSV-1 at an MOI of 1 for 48 h, the mRNA expression of viral genes was measured by RT-qPCR and normalized to HPRT mRNA. (D) The same experiment as in panel C was repeated for Emp, R1, and R1D cell lines. The data are means ± the SE (n = 3). *, P < 0.05; ***, P < 0.01; ****, P < 0.001; *****, P < 0.0001 (as assessed by two-way ANOVA).
FIG 5.
RUNX1 overexpression blunts HSV-1 viral gene transcription over time. After Emp, R1, and R3 cell lines were infected with HSV-1 at an MOI of 1, the mRNA expression of immediate early genes (A), early genes (B), and late genes (C) was measured by RT-qPCR and normalized to HPRT mRNA at 12, 24, 48, and 72 hpi. The data are means ± the SE (n = 3). ##, P < 0.1; *****, P < 0.0001 (as assessed by two-way ANOVA tests).
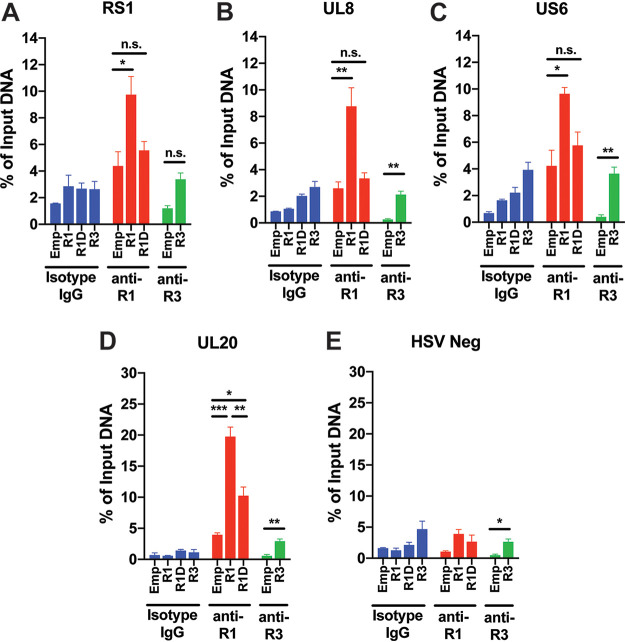
To assess whether RUNX1 directly binds the HSV-1 genome, we infected HEK293T cells transiently overexpressing WT RUNX1, D198G RUNX1, or RUNX3 and performed ChIP-qPCR using primers flanking CBSs present upstream of RS1 (at bp 1162 relative to TSS), UL8 (at bp −240), US6 (at bp −70), and UL20 (at bp −30). While overexpressed WT RUNX1 was significantly enriched at these putative sites, neither mutant RUNX1 nor RUNX3 yielded a comparable increase in enrichment compared to cells transfected with empty vector (Fig. 6A to D). In contrast, WT RUNX1 was not significantly enriched at a “RUNX desert region” of the HSV-1 genome that was more than 1,000 bp from the nearest putative binding sites (Fig. 6E). These data indicate that, much like host target genes, putative RUNX sites in the viral genome are indeed bound by RUNX1.
FIG 6.
RUNX1 binds to putative binding sites upstream of HSV-1 viral genes. HEK293T cells transiently overexpressing empty vector (Emp), RUNX1 (R1), DNA-binding mutant D198G RUNX1 (R1D), or RUNX3 (R3) were infected with HSV-1 at an MOI of 10. After 5 h infection, nuclear DNA was immunoprecipitated with isotype IgG, anti-RUNX1, or anti-RUNX3 antibodies. After elution and purification of bound single-stranded DNA, RUNX enrichment at putative binding sites in the promoter regions of RS1 (A), UL8 (B), US6 (C), UL20 (D), and “RUNX desert region” (HSV Neg) (E) was assessed by ChIP-qPCR and normalized to input DNA. The data are means ± the SE (n = 3). *, P < 0.05; ***, P < 0.01; ****, P < 0.001 (as assessed by two-way ANOVA).
RUNX1 overexpression decreases HSV-1 infection in vitro.
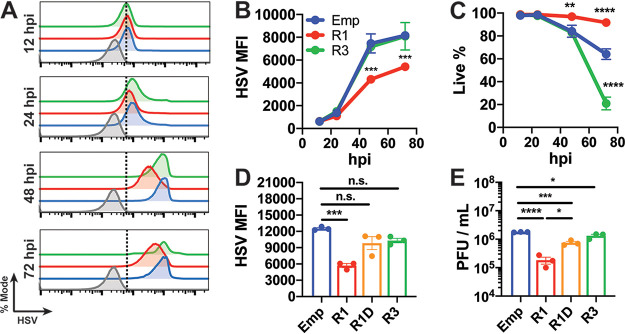
Next, we evaluated whether RUNX1 overexpression could suppress active HSV-1 infection in vitro. After infecting cell lines stably overexpressing RUNX1 or RUNX3, we measured the presence of cellular HSV-1 by flow cytometry as a marker of successful infection over time. We found that cells overexpressing RUNX1, but not RUNX3, presented with significantly lower HSV-1 and higher viability at both 48 and 72 h postinfection (hpi) (Fig. 7A to C). Repeating this experiment, we also found that overexpression of the RUNX1 DNA-binding mutant failed to repress HSV-1 replication compared to cells overexpressing functional RUNX1 (Fig. 7D). Finally, plaque assays revealed that RUNX1 overexpression significantly decreased infectious virions in supernatants by ∼10-fold and that this effect was significantly reduced with mutant RUNX1 or RUNX3 overexpression (Fig. 7E).
FIG 7.
Overexpression of RUNX1, but not RUNX3, decreases HSV-1 infection in vitro. HEK293T cells overexpressing empty vector, RUNX1, and RUNX3 were infected with HSV-1 at an MOI of 1. (A) Cells were stained with anti-HSV-1 polyclonal antibody, and fluorescence was measured by flow cytometry at 12, 24, 48, and 72 hpi. Histograms are shown normalized to mode. Mock-infected cells are displayed in gray. Dotted line indicates cutoff for HSV-1+ cells. (B) Quantification of HSV-1 MFI at each time point. (C) Live proportion of singlets as measured by live-dead staining at each time point. (D and E) The same experiment as in panels A to C was performed with stable cell lines overexpressing R1D. (D) HSV-1 MFI was measured by flow cytometry at 48 hpi. (E) Viral titers of supernatant 48 hpi were measured by plaque assay. The data are means ± the SE (n = 3). *, P < 0.05; ***, P < 0.01; ****, P < 0.001; *****, P < 0.0001 (as assessed by two-way ANOVA and one-way ANOVA).
RUNX1 knockdown increases HSV-1 infection in vitro.
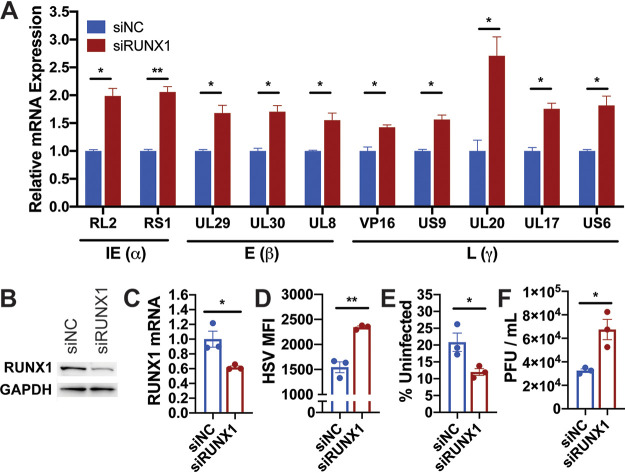
To validate our findings in a more relevant model, we repeated our experiments in the human neuroblastoma cell line IMR-32. Like DRG neurons, IMR-32 cells are derived from neural crest cells and express high levels of RUNX1 at baseline (23). IMR-32 cells have also been used to establish a model of HSV-2 latency in vitro (24). Once confirming successful knockdown of RUNX1 by small interfering RNA (siRNA) (Fig. 8B and C), we infected IMR-32 cells with HSV-1 at a multiplicity of infection (MOI) of 1 and found that RUNX1 suppression led to the transcriptional upregulation of all tested HSV-1 genes (Fig. 8A). Moreover, RUNX1 knockdown enhanced HSV-1 infection in vitro by increasing HSV-1 MFI (Fig. 8D), decreasing the proportion of uninfected cells (Fig. 8E), and increasing viral titers in the supernatant (Fig. 8F). Collectively, these data demonstrate that high cellular expression of RUNX1 by neuronal cells may represent a viable strategy to limit productive HSV-1 infection.
FIG 8.
RUNX1 knockdown in IMR-32 induces HSV-1 viral gene expression and promotes HSV-1 infection. After 24 h transfection with negative-control siRNA (siNC) or siRNA specific to RUNX1 (siRUNX1), IMR-32 neuroblastoma cells were infected with HSV-1 at an MOI of 1 for 48 h. (A) The mRNA expression of viral genes was measured by RT-qPCR and normalized to HPRT. (B and C) Validation of RUNX1 knockdown at (B) protein and (C) mRNA levels 24 h after transfection with siRNA. (D to E) Quantification of HSV MFI (D) and uninfected proportion of singlets (E). (F) Viral titers of HSV-1 present in supernatant 48 hpi was measured by plaque assay. The data are means ± the SE (n = 3). *, P < 0.05; ***, P < 0.01 (as assessed by two-way ANOVA and Student t test).
DISCUSSION
In the present study, we assessed the hypothesis that RUNX transcription factors, expressed in DRG neurons, might bind HSV-1 directly to repress transcription and, thereby, promote a more latent infection. We found that RUNX putative binding sites are selectively enriched among herpesviruses, compared to nonherpesviruses. Furthermore, we observed that these binding sites tend to be distributed tightly around TSSs of viral genes. In our in vitro validation studies, we found that overexpressed RUNX1 binds putative binding sites in HSV-1, represses transcription of viral genes spanning all three kinetic classes, and attenuates overall HSV-1 infection. Finally, we observed that siRNA knockdown of RUNX1 enhanced HSV-1 infection in IMR-32 neuroblastoma cells by inducing viral gene expression and increasing viral titers. In light of such findings, we find it plausible that RUNX1 binding might be used more generally by intracellular herpesviruses to escape host detection and survive in an otherwise hostile host environment.
Using a DNA binding mutant of RUNX1, we found that DNA binding activity was required for the suppression of HSV-1 gene expression and overall infection. However, it is still not clear which RUNX1 binding sites in the HSV-1 genome, specifically, are required for this effect. Therefore, future studies could mutate putative RUNX binding sites in the viral genome and assess whether these mutant viruses nullify the suppressive effects of RUNX1. Moreover, given that RUNX1 knockout mice exhibit embryonic lethality (25), such a mutant virus would be particularly helpful in determining whether RUNX1 binding is required for establishing HSV-1 latency in vivo. Still, even without such studies, the statistically anomalous enrichment and distribution of RUNX CBSs (Fig. 1 and 2), combined with prior reports of RUNX1 mediating HIV-1 latency (12), strongly argue for the parsimonious explanation that RUNX1 directly binds HSV-1 to promote its survival inside host neurons.
While our in vitro studies were limited to HSV-1, our computational findings suggest that herpesviruses, more generally, may be utilizing RUNX as a means to promote a more latent infection. Indeed, although the repression of viral genes by RUNX could be seen as an example of cell-autonomous immunity, the fact that RUNX CBSs were found more frequently than expected by chance argues for a more virally driven evolution, one in which the virus dynamically adapts to a cellular environment saturated with RUNX (as opposed to the host creating a RUNX-rich environment to counteract the virus). Moreover, when we survey the latent sites of infection for each herpesvirus, we find that they segregate either into sensory neurons (as with VZV, HSV-1, HSV-2, and PRV) or leukocytes (as with CMV, EBV, HHV-6, HHV-7, and HHV-8) (26). As in neurons, RUNX1 has been shown to be a master regulator of hematopoietic development, in both myeloid and lymphoid lineages (19), so much so that RUNX1 mutations feature heavily in various hematologic malignancies (27). Hence, the same rationale that led us to consider RUNX1 as a mediator of latency in the context of HSV-1 and DRG should naturally extend to other members of the same family, or at least the neurotropic subset. To underscore the specificity of this strategy for herpesviruses, future work could also evaluate how successfully, if at all, RUNX1 suppresses the infection of nonherpesviruses that harbor very few putative binding sites.
In contrast to RUNX1, we found that CTCF may not be a relevant mechanism across all herpesviruses. In addition to Ctcf mRNA not being significantly higher in DRG neurons (Fig. 1A), CTCF CBSs were found particularly enriched only in the previously identified subset of herpesviruses (HSV-1, HSV-2, and PRV) but not in others (Fig. 3C). In light of these data, it seems likely that CTCF plays a complementary role with RUNX, at least within a subset of herpesviruses: while CTCF acts more distally to block enhancers through insulator sites, RUNX operates more proximally by binding to promoter regions of specific genes. Future studies could directly compare the relative effects of these two host-factor-mediated strategies and how they may potentially interact with one another during infection.
In conclusion, our data demonstrate a novel role for host RUNX1 in directly binding and suppressing HSV-1, one that might help inform the DRG-specific latent infection that is observed. These experiments provide a basis for further exploring RUNX1-mediated latency not only in the context of HSV-1 but also across other herpesviruses more generally.
MATERIALS AND METHODS
Viruses.
WT HSV-1 (strain 17syn+ D68HR) was kindly provided by David Leib (Geisel School of Medicine at Dartmouth) (28). Virus was maintained and propagated using Vero cells.
Plasmids, antibodies, and reagents.
Human RUNX1 (SC123977), RUNX3 (SC302527), and CBFβ (SC110848) cDNA clones were purchased from OriGene. pIRES2-AcGFP1 plasmid (632435) was purchased from Clontech. RUNX1 siRNAs (s2460, s229352) and negative-control siRNA (4390844) were purchased from Thermo Fisher. Biotinylated anti-HSV-1/2 (0107) antibody was purchased from Virostat. SA-conjugated APC (405243) was purchased from BioLegend. Anti-RUNX1 (ab23980), anti-RUNX3 (ab11905), and normal rabbit IgG (ab172730) antibodies was purchased from Abcam. Anti-GAPDH antibody (GTX627408-01) was purchased from GeneTex.
Genomes.
GenBank and FASTA files of the following genomes were downloaded from NCBI: HSV-1 strain 17 (NC_001806), HSV-2 strain HG52 (NC_001798), VZV/HHV-3 (NC_001348), CMV/HHV-5 (NC_006273), HHV-6A (NC_001664), HHV-6B (NC_000898), HHV-7 (NC_001716), KSHV/HHV-8 (NC_009333), EBV/HHV-4 (NC_009334), Cyprinid herpesvirus 3 (NC_009127), Macaca namestrina herpesvirus 7 (NC_030200), Equid herpesvirus 5 strain 2-141/67 (NC_026421), Falconid herpesvirus 1 strain S-18 (NC_024450), Elephant endotheliotropic herpesvirus 4 (NC_028379), Anguillid herpesvirus 1 (NC_013668), Suid herpesvirus 1/pseudorabies virus (NC_006151), Macacine herpesvirus 5 (NC_003401), Pseudomonas phage 201phi2-1 (NC_010821), Vibrio phage KVP40 (NC_005083), Vaccinia virus (NC_006998), Variola virus (NC_001611), Bacillus phage phi4J1 (NC_029008), Human adenovirus B2 (NC_011202), Tobacco virus 1 (NC_027712), Measles virus (NC_001498), Venezuelan equine encephalitis virus (NC_001449), Japanese encephalitis virus (NC_001437), Rubella virus (NC_001545), Trichoderma atroviridae mycovirus (NC_033415), VSV (NC_001560), HPV type 16 (NC_001526), HIV-1 (NC_001802), Dengue virus 2 (NC_001474), JC polyomavirus (NC_001699), HBV strain ayw (NC_003977), Influenza A virus A/New York/392/2004 (NC_007373), poliovirus (NC_002058), and Zika virus (NC_012532).
CBS enrichment analysis.
Using R, FASTA files were loaded and searched for CBSs (and their reverse complement) listed in Table 1. For stringent searches (CBSs within 2,000 bp upstream of TSS and not within gene body), GenBank files were used to locate TSSs, calculate distance to closest TSS for each CBS, and exclude CBSs that did not meet the criteria. The expected number of CBS was calculated as the genome size × total combined probabilities of CBSs, taking into account each genome’s GC content. Relative enrichment was calculated as the number of CBSs/genome size × 10,000.
TABLE 1.
CBSs utilized for computational analyses
| TF | CBSs | Reference |
|---|---|---|
| RUNX | AACCACA, AACCGCA, GACCACA, GACCGCA | 31 |
| CTCF | CCCTC, CTCCC | 4 |
| NF-κB | GGGAATTTCC, GGGAATTCCC, GGAATTTCCC, GGGATTTCCC, GGAAAGTCCC, GGGAAGTCCC, GGAATGTCCC, GGGATGTCCC, GGAAATCCCC, GGGAATCCCC, GGAATTCCCC, GGGATTCCCC, GGAAAGCCCC, GGGAAGCCCC, GGAATGCCCC, GGGATGCCCC | 32 |
| IRF1 | TTCACTT, TTCAGTT, TTCTCTT, TTCTGTT | 32 |
| LBP1 | CAGCTGC, CAGCTGG, CAGCTTC, CAGCTTG | 32 |
| NKX2.5 | TCAAGTG, TCAAGTA, TTAAGTG, TTAAGTA | 32 |
| HOXA7 | CCAATCT, CCAATCG, TCAATCT, TCAATCG | 32 |
CBS TSS localization analysis.
For each evaluated genome, TSSs of each gene was located using the GenBank file and distance to the closest CBS was calculated for each gene TSS and averaged. Next, for each genome, 1,000 simulations were run as follows: (i) [number of CBSs] random numbers were chosen from 1 to [genome size/CBS size] without replacement; (ii) set of random numbers were multiplied by [CBS size]; and (iii) Distance to closest randomized CBS location was calculated for each gene TSS. The average distance of each simulation was plotted on a histogram and fitted with a normal distribution.
Generation of stable cell lines.
Coding regions of RUNX1 and RUNX3 cDNA clones were subcloned into IRES2-AcGFP1 plasmids using iProof high-fidelity DNA polymerase (Bio-Rad) and the primers listed in Table 2. After confirming sequences of plasmids, HEK293T cells were transfected with Empty-IRES2-AcGFP, RUNX1-IRES2-AcGFP, RUNX1-D198G-IRES2-AcGFP1, and RUNX3-IRES2-AcGFP1 using Lipofectamine 2000 (Invitrogen). After 24 h, cells were sorted for GFP+ cells using flow cytometry on BD FACSAria (Yale Flow Cytometry Core) and plated again for expansion. After five passages, GFP+ selection was performed again and expanded. Once overexpression was confirmed by RT-qPCR and Western blotting, polyclonal cells were frozen.
TABLE 2.
Cloning and mutagenesis primers
| Name | Primer |
|---|---|
| RUNX1-XhoI-F | GCACTCGAGATGGCTTCAGACAGCATATTTG |
| RUNX1-EcoRI-R | GCAGAATTCTCAGTAGGGCCTCCACAC |
| RUNX3-XhoI-F | GCACTCGAGATGGCATCGAACAGCATC |
| RUNX3-EcoRI-R | GCAGAATTCTCAGTAGGGCCGCCAC |
| RUNX1-D198G-F | GCCATCAAAATCACAGTGGGTGGGCCCCGAGAACCTCG |
| RUNX1-D198G-R | CGAGGTTCTCGGGGCCCACCCACTGTGATTTTGATGGC |
Point mutation of RUNX1.
Mutagenesis reaction was performed using TagMaster site-directed mutagenesis kit and the primers listed in Table 2. The coding region was subcloned into IRES2-AcGFP1 as described above, and the sequence was verified.
In vitro HSV-1 infection.
HEK293T cells were plated into 6-well plates at a density of 500,000 cells/well in Dulbecco modified Eagle medium (DMEM) CM (DMEM supplemented with 10% fetal bovine serum [FBS] and 1% penicillin-streptomycin). For transient-transfection experiments, cells were transfected 24 h after plating with described plasmids or siRNAs with Lipofectamine 2000 (Invitrogen). After 24 h, the plates were chilled for 30 min, and the cells were infected with HSV-1 at the described MOI in ABC buffer (0.5 mM MgCl2⋅6H2O, 0.9 mM CaCl2⋅2H2O, 1% glucose, 5% FBS, 1% penicillin-streptomycin) on ice for 1 h and at 37°C for 1 h. The cells were washed with acidic glycine buffer (0.14 M NaCl, 5 mM KCl, 1 mM MgCl2⋅6H2O, 0.7 mM CaCl2⋅2H2O, 0.1 M glycine [pH 3] in autoclaved water), and the supernatant was replaced with prewarmed DMEM CM. The cells were harvested at the described time for downstream analyses.
Flow cytometry.
Cells were trypsinized and washed with phosphate-buffered saline (PBS). After staining by using a LIVE/DEAD Fixable Aqua Dead Cell kit (Invitrogen) for 15 min at 4°C, the cells were washed with fluorescence-activated cell sorting (FACS) buffer (5% FBS in PBS) and incubated with anti-HSV-1 antibody (1:100) for 15 min at 4°C. The cells were washed again with FACS buffer and stained with SA-conjugated APC (1:1,000) for 15 min at 4°C. The cells were washed once more in FACS buffer and fixed with 1% paraformaldehyde. Samples were run on BD LSR Green (Yale Flow Cytometry Core).
ChIP-qPCR.
ChIP-qPCR was performed as described previously, with some modifications (29). Briefly, cells were transfected with RUNX1-IRES2-AcGFP1, RUNX1-D198G-IRES2-AcGFP1, or RUNX3-IRES2-AcGFP1 and infected with HSV-1 for 5 h at an MOI of 10. After infection, 1% formaldehyde was added to media for 5 min, and then the mixture was neutralized with 0.125 M glycine. The cells were lysed in cell lysis buffer (50 mM HEPES [pH 7.4], 1 mM EDTA, 85 mM KCl, 10% glycerol, 0.5% Nonidet P-40, protease inhibitor) and pelleted at 1,200 × g at 4°C for 5 min. The pellets were suspended in nucleus lysis buffer (50 mM Tris-HCl [pH 8.0], 2 mM EDTA, 150 mM NaCl, 5% glycerol, 1% Triton X-100, 1% SDS, protease inhibitor) and sonicated using a Bioruptor for 45 cycles (30 s on, 30 s off). Samples were incubated with 5 μg of anti-RUNX1 antibody, anti-RUNX3 antibody, or normal rabbit IgG and rotated overnight at 4°C. After incubation with Dynabeads-protein G (Thermo Fisher) for 1 h at room temperature, the beads were washed twice with ChIP wash buffer 1 (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 2 mM EDTA), twice with ChIP wash buffer 2 (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1% Triton X-100, 0.1% SDS, 2 mM EDTA), once with ChIP wash buffer 3 (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 500 mM LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mM EDTA), and once in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Bound DNA was eluted from beads using a ChIP elute kit (TaKaRa) and analyzed by RT-qPCR with primers flanking RUNX CBSs listed in Table 3 and normalized to a 5% input.
TABLE 3.
RT-qPCR primers
| Target | Fwd primer | Rev primer |
|---|---|---|
| HSV-1 RL2 | GTCGCCTTACGTGAACAAGAC | GTCGCCATGTTTCCCGTCTG |
| HSV-1 RS1 | CGGTGATGAAGGAGCTGCTGTTGC | CTGATCACGCGGCTGCTGTACA |
| HSV-1 UL29 | CATCAGCTGCTCCACCTCGCG | GCAGTACGTGGACCAGGCGG |
| HSV-1 UL30 | CATCACCGACCCGGAGAGGGAC | GGGCCAGGCGCTTGTTGGTG |
| HSV-1 UL8 | GGTGATGAGCGCAGTCC | GTCGTGCGTGTCTGTCC |
| HSV-1 US6 | TGTCGTCATAGTGGGCCTCCAT | AGACTTGTTGTAGGAGCATTCG |
| HSV-1 US9 | ACTCGGAAAGCGAAGACGAG | CGTCGACGCCTTAATACCGA |
| HSV-1 UL20 | ACCATCTCCAACGGCTTCAG | CCATACCCAGCCGGTCTTTT |
| HSV-1 VP16 | TCGGCGTGGAAGAAACGAGAGA | CGAACGCACCCAAATCGACA |
| HSV-1 UL17 | ACTGACGTCACCATCCATCG | ACTACAGCACAAGCGGAGAC |
| Human HPRT | CCTGGCGTCGTGATTAGTGAT | AGACGTTCAGTCCTGTCCATAA |
| RS1 CBS ChIP | GTGGACCGCTTCCTG | ATGTCGGCCATCCAG |
| UL8 CBS ChIP | GTGCGTTCCGGCAAC | TGGCCCATGATGCAG |
| US6 CBS ChIP | CCCCAATAAAGATCGCGGTAG | TACCCCTCCTCCTCGTAAAATG |
| UL20 CBS ChIP | GAGGAAGGTCATCCCGCATG | CAAGAACCCGGGTGTCTTTGATC |
| HSV Neg ChIP | CAAACACTTGGGGACTGTAGGTTTCTG | GGTCTCGTAACGCCAATCAAGATCG |
RNA extraction and RT-qPCR.
After the samples were lysed in RLT buffer and precipitated with ethanol, lysate was transferred to purification columns, and RNA was subsequently extracted by using an RNeasy minikit (Qiagen). After elution with RNase-free water, cDNA was synthesized using iScript (Bio-Rad) and analyzed with RT-qPCR as previously described (30) using the primers listed in Table 3.
Western blotting.
Cells were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% DOC, 0.1% SDS, 25 mM Tris [pH 7.4], protease inhibitor, phosphatase inhibitor) and incubated on ice for 45 min. After centrifugation at 15,000 rpm for 15 min at 4°C, protein samples were boiled at 95°C for 5 min and separated by SDS-PAGE on a 10% polyacrylamide gel. Samples were then transferred to a polyvinylidene difluoride membrane at 100 V for 60 min. Membrane was blocked for 1 h in blocking buffer (5% milk in TBS-Tween [TBST]) on an orbital shaker and incubated overnight with primary antibody diluted in 5% bovine serum albumin at 4°C. The blot was washed three times in TBST and incubated on orbital shaker for 1 h in secondary antibody diluted in blocking buffer. After three more washes in TBST, each blot was visualized with SuperSignal West Femto chemiluminescent substrate (Thermo Fisher, 34096) and ChemiDoc (Bio-Rad).
Plaque assays.
Once Vero cells were grown to confluence in 6-well plates, the cells were washed in PBS and infected with serial dilutions of supernatant samples (ranging from 10−1 to 10−6), diluted in ABC buffer in a total volume of 400 μl. After 1 h of incubation at 37°C with regular shaking, virus was aspirated and replaced with prewarmed DMEM CM supplemented with human IgG. After 48 h, the cells were fixed with crystal violet solution for 5 h, and the plates were dried overnight. Plaques were manually counted in each well, and dilutions yielding 5 to 100 plaques/well were used to subsequently calculate the PFU/ml.
Statistical analyses.
For statistical analysis, the data were analyzed by using the Student t test, one-way analysis of variance (ANOVA), or two-way ANOVA, as indicated (GraphPad Prism). A P value of <0.05 was considered statistically significant. All sample sizes (n) indicate the number of biological replicates used in the experiment. Conservative sample size calculations for a 2-fold effect were performed (μ1 = 100, μ2 = 50, σ = 20, α = 0.05, and β = 0.2) to confirm that three biological replicates would be sufficient in appropriately detecting an effect.
ACKNOWLEDGMENTS
D.J.K. was a Paul and Daisy Soros Fellow and was supported by a grant from the National Cancer Institute (NCI) of the National Institutes of Health (NIH; F30CA236466). D.J.K. and J.K. were supported in part by an MSTP training grant from the NIH (T32GM007205, T32GM136651). W.K.-H. was supported by a virology training grant from the NIH (T32 AI 55403-10). This study was supported by funding from the NIH (AI054359, R01EB000487, R01AI127429, and R21AI131284 [to A.I.]). A.I. is an Investigator of the Howard Hughes Medical Institute.
W.K.-H., D.J.K., R.M., and A.I. designed research. D.J.K., W.K.-H., P.C.J., Y.K., J.K., S.D.P., and W.G. performed research. D.J.K., W.K.-H., and A.I. analyzed data. D.J.K., W.K.-H., and A.I. wrote the paper.
REFERENCES
- 1.Wald A, Corey L. 2007. Persistence in the population: epidemiology, transmission. Chapter 36 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Oxford University Press, Cambridge, United Kingdom: https://www.ncbi.nlm.nih.gov/books/NBK47447/#c36vdw-q6h-db5-de8. [Google Scholar]
- 2.Sauerbrei A. 2016. Herpes genitalis: diagnosis, treatment, and prevention. Geburtshilfe Frauenheilkd 76:1310–1317. doi: 10.1055/s-0042-116494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicoll MP, Proenca JT, Efstathiou S. 2012. The molecular basis of herpes simplex virus latency. FEMS Microbiol Rev 36:684–705. doi: 10.1111/j.1574-6976.2011.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amelio AL, McAnany PK, Bloom DC. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol 80:2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Washington SD, Musarrat F, Ertel MK, Backes GL, Neumann DM. 2018. CTCF binding sites in the herpes simplex virus 1 genome display site-specific CTCF occupation, protein recruitment, and insulator function. J Virol 92:e00156-18. doi: 10.1128/JVI.00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohlsson R, Renkawitz R, Lobanenkov V. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 7.Inoue K, Shiga T, Ito Y. 2008. Runx transcription factors in neuronal development. Neural Dev 3:20. doi: 10.1186/1749-8104-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, Ma Q. 2006. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron 49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 9.Khoury-Hanold W, Yordy B, Kong P, Kong Y, Ge W, Szigeti-Buck K, Ralevski A, Horvath TL, Iwasaki A. 2016. Viral spread to enteric neurons links genital HSV-1 infection to toxic megacolon and lethality. Cell Host Microbe 19:788–799. doi: 10.1016/j.chom.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komori T. 2018. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem Cell Biol 149:313–323. doi: 10.1007/s00418-018-1640-6. [DOI] [PubMed] [Google Scholar]
- 11.Durst KL, Hiebert SW. 2004. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- 12.Klase Z, Yedavalli VS, Houzet L, Perkins M, Maldarelli F, Brenchley J, Strebel K, Liu P, Jeang KT. 2014. Activation of HIV-1 from latent infection via synergy of RUNX1 inhibitor Ro5-3335 and SAHA. PLoS Pathog 10:e1003997. doi: 10.1371/journal.ppat.1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young GT, Gutteridge A, Fox H, Wilbrey AL, Cao L, Cho LT, Brown AR, Benn CL, Kammonen LR, Friedman JH, Bictash M, Whiting P, Bilsland JG, Stevens EB. 2014. Characterizing human stem cell-derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol Ther 22:1530–1543. doi: 10.1038/mt.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson R, Ahmmed AA, Poll A, Sakaue M, Laude A, Sieber-Blum M. 2018. Human peptidergic nociceptive sensory neurons generated from human epidermal neural crest stem cells (hEPI-NCSC). PLoS One 13:e0199996. doi: 10.1371/journal.pone.0199996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ. 2018. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 159:1325–1345. doi: 10.1097/j.pain.0000000000001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogbourne S, Antalis TM. 1998. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J 331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doni Jayavelu N, Jajodia A, Mishra A, Hawkins RD. 2020. Candidate silencer elements for the human and mouse genomes. Nat Commun 11:1061. doi: 10.1038/s41467-020-14853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. 2006. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res 16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mevel R, Draper JE, Lie ALM, Kouskoff V, Lacaud G. 2019. RUNX transcription factors: orchestrators of development. Development 146:dev148296. doi: 10.1242/dev.148296. [DOI] [PubMed] [Google Scholar]
- 20.Roulston A, Lin R, Beauparlant P, Wainberg MA, Hiscott J. 1995. Regulation of human immunodeficiency virus type 1 and cytokine gene expression in myeloid cells by NF-κB/Rel transcription factors. Microbiol Rev 59:481–505. doi: 10.1128/MMBR.59.3.481-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranganathan PN, Khalili K. 1993. The transcriptional enhancer element, κB, regulates promoter activity of the human neurotropic virus, JCV, in cells derived from the CNS. Nucleic Acids Res 21:1959–1964. doi: 10.1093/nar/21.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao LJ, Wang YY, Li G, Ma LY, Xiong SM, Weng XQ, Zhang WN, Wu B, Chen Z, Chen SJ. 2012. Functional features of RUNX1 mutants in acute transformation of chronic myeloid leukemia and their contribution to inducing murine full-blown leukemia. Blood 119:2873–2882. doi: 10.1182/blood-2011-08-370981. [DOI] [PubMed] [Google Scholar]
- 23.Inoue K, Ito Y. 2011. Neuroblastoma cell proliferation is sensitive to changes in levels of RUNX1 and RUNX3 protein. Gene 487:151–155. doi: 10.1016/j.gene.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Yura Y, Terashima K, Iga H, Yanagawa T, Yoshida H, Hayashi Y, Sato M. 1986. A latent infection of herpes simplex virus type 2 in a human neuroblastoma cell line IMR-32. Arch Virol 90:249–260. doi: 10.1007/BF01317374. [DOI] [PubMed] [Google Scholar]
- 25.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 26.Grinde B. 2013. Herpesviruses: latency and reactivation—viral strategies and host response. J Oral Microbiol 5. doi: 10.3402/jom.v5i0.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sood R, Kamikubo Y, Liu P. 2017. Role of RUNX1 in hematological malignancies. Blood 129:2070–2082. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. 2009. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol 83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taura M, Song E, Ho YC, Iwasaki A. 2019. Apobec3A maintains HIV-1 latency through recruitment of epigenetic silencing machinery to the long terminal repeat. Proc Natl Acad Sci U S A 116:2282–2289. doi: 10.1073/pnas.1819386116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi K, Sasai M, Iwasaki A. 2015. Toll-like receptor 9 trafficking and signaling for type I interferons requires PIKfyve activity. Int Immunol 27:435–445. doi: 10.1093/intimm/dxv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito Y, Bae SC, Chuang LS. 2015. The RUNX family: developmental regulators in cancer. Nat Rev Cancer 15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 32.Daily K, Patel VR, Rigor P, Xie X, Baldi P. 2011. MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics 12:495. doi: 10.1186/1471-2105-12-495. [DOI] [PMC free article] [PubMed] [Google Scholar]