Links between trimethylamine-N-oxide (TMAO) and cardiovascular disease (CVD) have focused attention on mechanisms by which animal-based diets have negative health consequences. In a meta-analysis of data from foundational gut microbiome studies, we found evidence that specialized bacteria have and express a metabolic pathway that circumvents TMAO production and is often misannotated because it relies on genetic code expansion. This naturally occurring mechanism for TMAO attenuation is negatively correlated with CVD. Ultimately, these findings point to new avenues of research that could increase microbiome-informed understanding of human health and hint at potential biomedical applications in which specialized bacteria are used to curtail CVD development.
KEYWORDS: cardiovascular disease, microbiome, molecular genetics
ABSTRACT
Cardiovascular disease (CVD) has been linked to animal-based diets, which are a major source of trimethylamine (TMA), a precursor of the proatherogenic compound trimethylamine-N-oxide (TMAO). Human gut bacteria in the genus Bilophila have genomic signatures for genetic code expansion that could enable them to metabolize both TMA and its precursors without production of TMAO. We uncovered evidence that the Bilophila demethylation pathway is actively transcribed in gut microbiomes and that animal-based diets cause Bilophila to rapidly increase in abundance. CVD occurrence and Bilophila abundance in humans were significantly negatively correlated. These data lead us to propose that Bilophila, which is commonly regarded as a pathobiont, may play a role in mitigating cardiovascular disease. Human gut microbiomes have been shown to affect the development of a myriad of disease states, but mechanistic connections between diet, health, and microbiota have been challenging to establish. The hypothesis that Bilophila reduces cardiovascular disease by circumventing TMAO production offers a clearly defined mechanism with a potential human health impact, but investigations of Bilophila cell biology and ecology will be needed to fully evaluate these ideas.
IMPORTANCE Links between trimethylamine-N-oxide (TMAO) and cardiovascular disease (CVD) have focused attention on mechanisms by which animal-based diets have negative health consequences. In a meta-analysis of data from foundational gut microbiome studies, we found evidence that specialized bacteria have and express a metabolic pathway that circumvents TMAO production and is often misannotated because it relies on genetic code expansion. This naturally occurring mechanism for TMAO attenuation is negatively correlated with CVD. Ultimately, these findings point to new avenues of research that could increase microbiome-informed understanding of human health and hint at potential biomedical applications in which specialized bacteria are used to curtail CVD development.
INTRODUCTION
The human gut microbiome is increasingly recognized for its influential role in health. The gut microbiome has been implicated in cardiovascular disease (CVD), the leading cause of death in developed countries (1). Metabolic end products associated with the consumption of animal products have been shown to promote CVD; dietary sources of proatherogenic compounds include choline, phosphatidylcholine, and l-carnitine (2–11). Gut microorganisms convert these compounds to trimethylamine (TMA), which in turn is converted to trimethylamine-N-oxide (TMAO) through the action of host hepatic flavin monooxygenase 3 (12, 13). TMAO is a causative agent of CVD pathogenesis; therefore, elucidating pathways relevant to this compound is central to understanding human health (10, 14).
The canonical view of CVD and TMAO involves the action of the gut microbiota in transforming precursor compounds from a range of animal-based dietary sources to TMA, which can then be converted to TMAO in the liver (Fig. 1A). While TMAO is frequently referred to as the sole breakdown product of TMA, a subset of methanogenic archaea in the gut have the ability to utilize TMA by an alternative pathway (15, 16). This pathway is enabled by genetic code expansion (GCE), via insertion of the 22nd amino acid, pyrrolysine (Pyl), in place of a TAG amber codon at conserved sites of the tri-, di-, and monomethylamine methyltransferase genes, with methane as the end product (17–19). TMA metabolism that relies on proteins that use an expanded code has also been described in some bacteria from environmental settings, including symbionts of gutless marine worms (20), as well as the Firmicutes bacterium Acetohalobium arabaticum isolated from a Crimean lagoon (21). Despite its potential importance, bacterial metabolism of TMA remains largely unexplored in the gut microbiome.
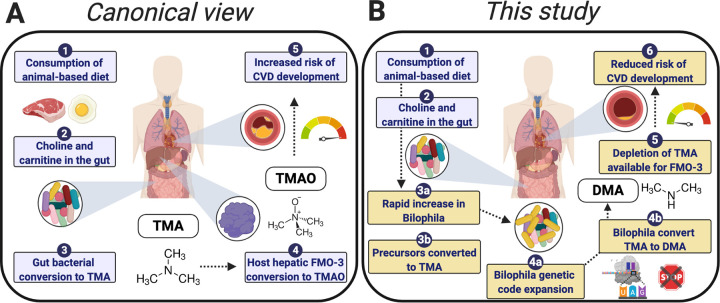
FIG 1.
(A) Canonical view of TMA: this compound is converted to TMAO in the liver via a host hepatic flavin monooxygenase 3, leading to increased risk of cardiovascular disease. (B) A new view of TMA metabolism proposed in this study: the specialized gut bacteria, Bilophila, increase in abundance and use genetic code expansion to augment metabolism, thereby reducing the amount of TMA available for conversion to TMAO. TMA, trimethylamine; FMO-3, flavin monooxygenase 3; TMAO, trimethylamine-N-oxide; CVD, cardiovascular disease; DMA, dimethylamine.
A Deltaproteobacterium commonly found in the human gut, Bilophila wadsworthia, also has genes necessary for encoding pyrrolysine (22). First identified in an appendicitis infection in 1989, this “bile-loving,” taurine-degrading bacterium is commonly referred to as a pathobiont associated with abscesses (23, 24). Additionally, Bilophila is able to produce hydrogen sulfide, and these bacteria may be linked to inflammatory bowel disease, colorectal cancer, and systematic inflammation (24–28). Despite its classification as a pathobiont, Bilophila is also commonly present in healthy human microbiomes (29, 30). There is uncertainty about GCE and TMA metabolic pathway characteristics in these organisms; notably, whether the pyrrolysine pathway is functional in trans and whether the pathway is expressed. The function of this pathway is questioned (21) because of organizational differences in the pyrrolysyl-tRNA synthetase gene (required for encoding Pyl): in archaea, this synthetase is encoded by a single gene, while in Bilophila, the N- and C-terminal domains are encoded by two distinct genes. Second, in previous reports of Pyl pathways, the trimethylamine methyltransferase and Pyl genes are commonly adjacent to one another (21, 22), while this is not the case in Bilophila. Third, the TMA methyltransferase protein is prematurely truncated at the TAG codon in public Bilophila data sets available on the NCBI and JGI-IMG webservers. Despite the ubiquity of this bacterium, and its potential importance in modulating TMAO levels (and thus CVD risk), these ambiguities remain unresolved, and studies of the human gut microbiome make no mention of the possibility of a bacterial pathway for TMA utilization. In this study, we investigate this alternative pathway of trimethylamine metabolism in Bilophila, the change in abundance of this taxon in response to an animal-based diet, and the correlation between Bilophila abundance and CVD pathology (Fig. 1B).
RESULTS
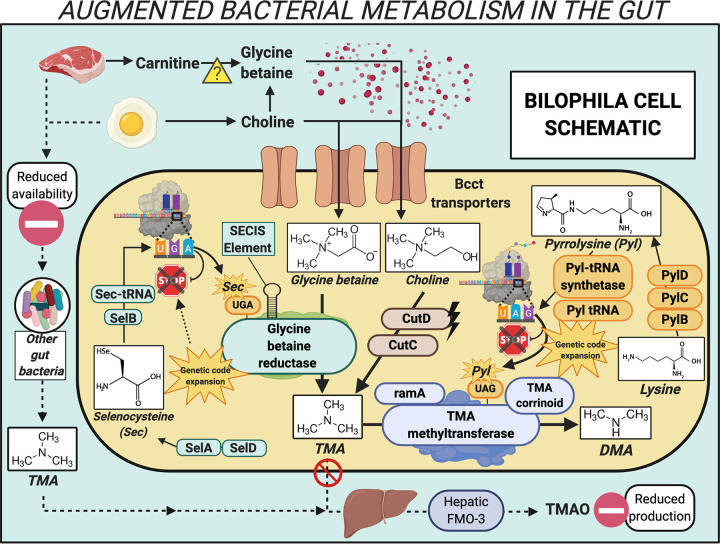
To explore the potential for Bilophila TMA metabolism, we retranslated the protein-coding sequences from publicly available Bilophila genomes (Table S1) using a custom translation table with the TAG codon as a readthrough rather than a stop codon (see Materials and Methods). We found an in-frame TAG codon at a conserved position in the TMA methyltransferase, corresponding to the pyrrolysine residue of this protein in archaeal methanogens (17) and bacteria (20, 21) (Fig. 2 and Table S2). All genes known to have functions specific to pyrrolysine production were also identified in the Bilophila genomes: PylB, PylC, and PylD and the pyrrolysyl-tRNA synthetase N and C termini, as well as the Pyl-specific tRNA, with the corresponding CUA anticodon (Fig. 2; Fig. S1). One exception was that Bilophila wadsworthia ATCC 49260 is missing one component: a Pyl tRNA was not located. In addition to the Pyl-containing TMA methyltransferase, accessory genes for the TMA methyltransferase pathway, including ramA (the activating gene) and the TMA methyltransferase corrinoid protein, are encoded in all of the Bilophila genomes (Fig. 2 and Table S3). The dimethylamine methyltransferase gene is not encoded in any of the genomes, and a gene similar to monomethylamine methyltransferase, while present, does not have the conserved in-frame TAG codon that is a hallmark of this gene. In summary, metabolic inference from genomic data indicates that Bilophila have the potential to convert TMA to dimethylamine (DMA) in the human gut.
FIG 2.
Mechanistic overview of genetic code expansion augmented metabolism performed by Bilophila in the human gut. Bcct transporters, betaine/carnitine/choline transporter family proteins; CutD, choline trimethylamine lyase activating enzyme; CutC, choline trimethylamine lyase; TMA, trimethylamine; DMA, dimethylamine; PylB, 3-methylornithine synthase; PylC, 3-methylornithine-l-lysine ligase; PylD, 3-methylornithyl-N6-l-lysine dehydrogenase; Pyl-tRNA synthetase, pyrrolysyl-tRNA synthetase; Pyl tRNA, pyrrolysine tRNA; ramA, methylamine methyltransferase corrinoid protein reductive activase; TMA corrinoid, methyltransferase cognate corrinoid protein; SelD, selenide, water dikinase; SelA, l-seryl-tRNA(Sec) selenium transferase; SelB, selenocysteine-specific translation elongation factor; Sec-tRNA, selenocysteine tRNA; SECIS Element, selenocysteine insertion sequence element; TMAO, trimethylamine-N-oxide; FMO-3, flavin monooxygenase 3.
The secondary structures of two tRNAs, for selenocysteine and pyrrolysine, from two Bilophila genomes. Structure of the pyrrolysine tRNA with CTA (CUA) anticodon (left) and selenocysteine tRNA with TCA (UCA) anticodon and extra arm (right) for (A) Bilophila sp. 4_1_30 and (B) Bilophila wadsworthia 3_1_6. Download FIG S1, PDF file, 0.2 MB (163KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers for Bioprojects, corresponding publications, and reference numbers for Bilophila genomes used in this study. Download Table S1, PDF file, 0.04 MB (44.8KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Amino acid sequences of TMA methyltransferase proteins from Bilophila genomes, with the pyrrolysine residue (O) highlighted in green. Download Table S2, PDF file, 0.04 MB (41.8KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TIGRFAM identity and gene names of proteins relevant for Bilophila genetic code expansion and TMA metabolism. Download Table S3, PDF file, 0.04 MB (36KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The TMA-utilizing pathway we observed may benefit Bilophila by enabling them to use TMA directly and also TMA produced metabolically within the cell from two precursor compounds, choline and glycine betaine (Fig. 2). Briefly, Bilophila genomes encode the glycyl radical choline-TMA lyase and its associated activating protein (CutC and CutD, respectively), which convert choline to TMA (31). Glycine betaine (sources of which includes choline [32] and possibly carnitine [33]) can be converted to TMA via the selenocysteine (Sec)-containing glycine betaine reductase (GRD) pathway (34). The GRD pathway and the full set of machinery required for the noncanonical amino acid selenocysteine, encoded with a repurposed UGA stop codon, are also present in the Bilophila genomes (Table S3). Multiple copies of proteins belonging to the betaine/carnitine/choline family of transporters are also encoded in the Bilophila genomes. We conclude that the genomic data indicate that Bilophila has the ability to use choline and glycine betaine, converting these compounds to TMA, and subsequently to DMA. In doing so, these bacteria may deplete precursor compounds and TMA that would otherwise be available for host hepatic processes, thereby reducing or circumventing production of TMAO (Fig. 2).
Next, we asked whether the pyrrolysine pathway is functional and whether GCE-enabled TMA metabolism is active in Bilophila from the gut environment. To do so, we reexamined data sets from a recent human gut metatranscriptomic study (35) and a mouse model system study (36) (Table S1). Applying a minimum threshold of 98% amino acid sequence identity with annotated Bilophila proteins, we identified expression of the TMA methyltransferase and pyrrolysine machinery proteins in both human fecal and mouse cecum samples (Fig. 2 and Tables S4 and S5). The fraction of TMA consumed via this bacterial metabolic process in the human gut microbiome remains uncertain, but expression data support the conclusion that this metabolic process is active.
The metatranscriptomic data sets from mouse and human studies, the SRA accession numbers, and the number of reads for each data set. Download Table S4, PDF file, 0.06 MB (62.6KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The TMA and Pyl genes from metatranscriptomic data, their respective amino acid sequences, and the percent identity with corresponding proteins in the Bilophila genomes. Download Table S5, PDF file, 0.05 MB (53.5KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
This survey of published microbiome sequence data uncovered evidence that bacteria in the genus Bilophila use genetic code expansion in the human gut to produce a TMA methyltransferase. We hypothesized that this mechanism could be used to compete with other TMA-utilizing processes, potentially decreasing the production of TMAO from the proatherogenic precursor trimethylamine. To explore this hypothesis, we reexamined additional publicly available data (Table S1) to determine whether this naturally occurring mechanism for TMAO attenuation is correlated with CVD. In a recent study describing the gut microbiome in atherosclerotic cardiovascular disease, Jie et al. (29) report that Bilophila is one of the 20 most abundant genera in the samples examined for this project. Their data also show that the abundance of Bilophila is significantly enriched in the microbiomes of individuals in the healthy/control group (n = 187) compared to the CVD group (n = 218). Second, in a study describing a rapid diet-induced change in the human gut microbiome, David et al. (37) reveal that Bilophila significantly increase in abundance in response to an animal-based diet compared to a plant-based diet. Finally, in a study detailing the transmission of atherosclerosis susceptibility via gut microbial transplanation, Gregory et al. (38) show that mice with certain taxa have increased TMAO levels and develop atherosclerotic lesions and postulate that this is a microbiome-dependent, transmissible trait. Bilophila is one of only six taxa that are significantly enriched in both the cecal and fecal microbiome of the healthy group compared to the mice that developed atherosclerotic lesions. The observations we report may challenge the widely held idea that members of this taxon act exclusively as pathobionts; their role in the microbiome and human health may be context dependent, and their potential to mitigate the impacts of animal products on CVD warrants further study.
MATERIALS AND METHODS
Microbiome data selection and accession numbers.
Genomic, metagenomic, and metatranscriptomic data sets used for this study were accessed on 15 January 2020 from publicly available sequencing projects (see Table S1 in the supplemental material). The Bilophila genomes analyzed are reference genomes from the Human Microbiome Project (39) as well as the type strain for this genus, from the Refseq database (40). All of the available genomes from the genus Bilophila were examined. BioProject accession no. PRJEB33885 was excluded because it includes a duplicate of a previously published genome (included in accession no. PRJNA41963). Paired metagenomic-metatranscriptomic data were accessed from a large cohort study of adult men (35), and additional metatranscriptomic data were used from a mouse model system (36). Metatranscriptomic and metagenomic read data were obtained using the SRA-toolkit (https://github.com/ncbi/sra-tools) v2.9.1 data via the fastq-dump option.
Genomic analysis and implementation of a custom protein translation table.
Predicted proteins in each genome were initially identified using Prodigal (42) v.2.6.3 using single genome mode, and functional annotation was determined using Hmmer (43) v.3.1, with the hmmscan option (1E−10 cutoff), with top hits only, against the Pfam (44) database v.31 and Tigrfam (45) database v.15.0.
For genomes in which enzymes were identified for the synthesis of pyrrolysine, we applied an alternate protein prediction procedure that reassigned TAG codons from stop to readthrough. To accomplish this task, we modified the source code for Prodigal by adding a custom translation table that has TAG readthrough and retains all three canonical bacterial start codons. Proteins with in-frame stop codons in the relevant genes were then manually inspected to determine whether the region containing and following the stop codon was conserved in comparisons to homologues containing pyrrolysine at a similar position (see Fig. S2 in the supplemental material). The modified code and documentation for the modified translation table are freely available at https://github.com/VeronikaKivenson/Prodigal. Protein searches for the full-length TMA methyltransferase amino acid sequence from Bilophila were performed on the NCBI and JGI-IMG webservers on 1 July 2020. For searching and identifying putative selenocysteine-containing proteins, the bSECIS (46) webserver was used and results were inspected and compared with previously identified selenoproteins. Protein sequence alignment was performed using Muscle (47), with Geneious 2020.1 used for visualization of the alignments. The Aragorn (48) web server was used to locate the tRNA sequences from each genome and to determine the secondary structure of Sec- and Pyl-specific tRNAs, as well as their corresponding anticodon sequences. tRNAscan-SE (49) v.1.23 was also used to search for the tRNAs for Sec. Chemdraw and Biorender software were used to create figures.
Alignment of amino acid sequences from trimethylamine methyltransferase following the repurposed stop for Bilophila and an archaeon (Methanosarcina barkeri) that have pyrrolysine biosynthesis proteins; the sequence following the repurposed codon (pyrrolysine residue O) is conserved. Download FIG S2, PDF file, 0.07 MB (67.3KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metatranscriptomic data analysis.
Preliminary mapping of metatranscriptomic sequence data to Bilophila genomes was done using Bowtie2 (50) v2.3.4.1. This approach identified samples in which these Bilophila bacteria were present and active at detectable levels. Next, metatranscriptomic reads were coassembled using SRA data from select samples belonging to each BioProject (Table S4) using Megahit (51) v.1.1.1, with default parameters. Functional annotation and identification of stop codon readthrough were performed as described earlier. In addition to Prodigal, FragGeneScan was used to identify partial genes (52). The computational biology data processing and analysis workflow were completed using the Extreme Science and Engineering Discovery Environment (XSEDE) Bridges resource at the Pittsburgh Supercomputing Center (53, 54).
ACKNOWLEDGMENTS
This work was supported by a grant from the Simons Foundation (SFARI 649176 to V.K.). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges system, which is supported by NSF award number ACI-1445606, at the Pittsburgh Supercomputing Center (PSC), through allocation TG-DEB170007.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
We thank Grace Deitzler and Maude David for helpful discussion of the gut microbiome data sets.
We declare that we have no competing interests.
The project was conceived by V.K. and S.J.G. V.K. performed the bioinformatic data analysis, and interpretation was performed by V.K. and S.J.G. V.K. and S.J.G. wrote the manuscript.
REFERENCES
- 1.Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Das SR, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Jordan LC, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, O’Flaherty M, Pandey A, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Spartano NL, Stokes A, Tirschwell DL, Tsao CW, Turakhia MP, VanWagner LB, Wilkins JT, Wong SS, Virani SS, American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Committee. 2019. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation 139:e56–e528. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 2.Ufnal M, Zadlo A, Ostaszewski R. 2015. TMAO: a small molecule of great expectations. Nutrition 31:1317–1323. doi: 10.1016/j.nut.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Falony G, Vieira-Silva S, Raes J. 2015. Microbiology meets big data: the case of gut microbiota-derived trimethylamine. Annu Rev Microbiol 69:305–321. doi: 10.1146/annurev-micro-091014-104422. [DOI] [PubMed] [Google Scholar]
- 4.Janeiro MH, Ramírez MJ, Milagro FI, Martínez JA, Solas M. 2018. Implication of trimethylamine N-oxide (TMAO) in disease: potential biomarker or new therapeutic target. Nutrients 10:1398. doi: 10.3390/nu10101398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velasquez MT, Ramezani A, Manal A, Raj DS. 2016. Trimethylamine N-oxide: the good, the bad and the unknown. Toxins (Basel) 8:326. doi: 10.3390/toxins8110326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang WHW, Hazen SL. 2014. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest 124:4204–4211. doi: 10.1172/JCI72331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL. 2013. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JM, Hazen SL. 2018. Microbial modulation of cardiovascular disease. Nat Rev Microbiol 16:171–181. doi: 10.1038/nrmicro.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chittim CL, Martínez del Campo A, Balskus EP. 2019. Gut bacterial phospholipase Ds support disease-associated metabolism by generating choline. Nat Microbiol 4:155–163. doi: 10.1038/s41564-018-0294-4. [DOI] [PubMed] [Google Scholar]
- 12.Lang D, Yeung C, Peter R, Ibarra C, Gasser R, Itagaki K, Philpot R, Rettie A. 1998. Isoform specificity of trimethylamine N-oxygenation by human flavin-containing monooxygenase (FMO) and P450 enzymes: selective catalysis by FMO3. Biochem Pharmacol 56:1005–1012. doi: 10.1016/s0006-2952(98)00218-4. [DOI] [PubMed] [Google Scholar]
- 13.Zhang AQ, Mitchell SC, Smith RL. 1999. Dietary precursors of trimethylamine in man: a pilot study. Food Chem Toxicol 37:515–520. doi: 10.1016/s0278-6915(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. 2016. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 15.Borrel G, McCann A, Deane J, Neto MC, Lynch DB, Brugère J-F, O’Toole PW. 2017. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J 11:2059–2074. doi: 10.1038/ismej.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brugère J-F, Borrel G, Gaci N, Tottey W, O’Toole PW, Malpuech-Brugère C. 2014. Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes 5:5–10. doi: 10.4161/gmic.26749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul L, Ferguson DJ, Krzycki JA. 2000. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J Bacteriol 182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivasan G, James CM, Krzycki JA. 2002. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 19.Borrel G, Gaci N, Peyret P, O’Toole PW, Gribaldo S, Brugère J-F. 2014. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: implications for the evolution of a genetic code expansion cassette. Archaea 2014:374146. doi: 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Gladyshev VN. 2007. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res 35:4952–4963. doi: 10.1093/nar/gkm514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prat L, Heinemann IU, Aerni HR, Rinehart J, O’Donoghue P, Söll D. 2012. Carbon source-dependent expansion of the genetic code in bacteria. Proc Natl Acad Sci U S A 109:21070–21075. doi: 10.1073/pnas.1218613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaston MA, Jiang R, Krzycki JA. 2011. Functional context, biosynthesis, and genetic encoding of pyrrolysine. Curr Opin Microbiol 14:342–349. doi: 10.1016/j.mib.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laue H, Denger K, Cook AM. 1997. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl Environ Microbiol 63:2016–2021. doi: 10.1128/AEM.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baron EJ, Summanen P, Downes J, Roberts MC, Wexler H, Finegold SM. 1989. Bilophila wadsworthia, gen. nov. and sp. nov., a unique gram-negative anaerobic rod recovered from appendicitis specimens and human faeces. J Gen Microbiol 135:3405–3411. doi: 10.1099/00221287-135-12-3405. [DOI] [PubMed] [Google Scholar]
- 25.Peck SC, Denger K, Burrichter A, Irwin SM, Balskus EP, Schleheck D. 2019. A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc Natl Acad Sci U S A 116:3171–3176. doi: 10.1073/pnas.1815661116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. 2019. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med 25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 27.Feng Z, Long W, Hao B, Ding D, Ma X, Zhao L, Pang X. 2017. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog 9:59. [CrossRef] doi: 10.1186/s13099-017-0208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jie Z, Xia H, Zhong S-L, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen J-Y, Geng Q-S, Zhang Z-W, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K, Kristiansen K. 2017. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. 2012. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J 6:57–70. doi: 10.1038/ismej.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craciun S, Balskus EP. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A 109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andresen PA, Kaasen I, Styrvold OB, Boulnois G, Strøm AR. 1988. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J Gen Microbiol 134:1737–1746. doi: 10.1099/00221287-134-6-1737. [DOI] [PubMed] [Google Scholar]
- 33.Lindstedt G, Lindstedt S. 1961. On the biosynthesis and degradation of carnitine. Biochem Biophys Res Commun 6:319–323. doi: 10.1016/0006-291x(61)90137-1. [DOI] [PubMed] [Google Scholar]
- 34.Andreesen JR. 1994. Glycine metabolism in anaerobes. Antonie Van Leeuwenhoek 66:223–237. doi: 10.1007/BF00871641. [DOI] [PubMed] [Google Scholar]
- 35.Abu-Ali GS, Mehta RS, Lloyd-Price J, Mallick H, Branck T, Ivey KL, Drew DA, DuLong C, Rimm E, Izard J, Chan AT, Huttenhower C. 2018. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol 3:356–366. doi: 10.1038/s41564-017-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natividad JM, Lamas B, Pham HP, Michel M-L, Rainteau D, Bridonneau C, da Costa G, van Hylckama Vlieg J, Sovran B, Chamignon C, Planchais J, Richard ML, Langella P, Veiga P, Sokol H. 2018. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun 9:2802. doi: 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA, Lusis AJ, Hazen SL. 2015. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem 290:5647–5660. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Human Microbiome Jumpstart Reference Strains Consortium, Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, Feldgarden M, Gevers D, Haas BJ, Madupu R, Ward DV, Birren BW, Gibbs RA, Methe B, Petrosino JF, Strausberg RL, Sutton GG, White OR, Wilson RK, Durkin S, Giglio MG, Gujja S, Howarth C, Kodira CD, Kyrpides N, Mehta T, Muzny DM, Pearson M, Pepin K, Pati A, Qin X, Yandava C, Zeng Q, Zhang L, Berlin AM, Chen L, Hepburn TA, Johnson J, McCorrison J, Miller J, Minx P, Nusbaum C, Russ C, Sykes SM, Tomlinson CM, Young S, et al. 2010. A catalog of reference genomes from the human microbiome. Science 328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. 2014. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res 42:D553–D559. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths‐Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. 2004. The Pfam protein families database. Nucleic Acids Res 32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res 31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Gladyshev VN. 2005. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics 21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
- 47.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 52.Rho M, Tang H, Ye Y. 2010. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, Roskies R, Scott JR, Wilkins-Diehr N. 2014. XSEDE: accelerating scientific discovery. Comput Sci Eng 16:62–74. doi: 10.1109/MCSE.2014.80. [DOI] [Google Scholar]
- 54.Nystrom NA, Levine MJ, Roskies RZ, Scott JR. 2015. Bridges: a uniquely flexible HPC resource for new communities and data analytics, p 1–8. In Proceedings of the 2015 XSEDE Conference: Scientific Advancements Enabled by Enhanced Cyberinfrastructure. Association for Computing Machinery, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The secondary structures of two tRNAs, for selenocysteine and pyrrolysine, from two Bilophila genomes. Structure of the pyrrolysine tRNA with CTA (CUA) anticodon (left) and selenocysteine tRNA with TCA (UCA) anticodon and extra arm (right) for (A) Bilophila sp. 4_1_30 and (B) Bilophila wadsworthia 3_1_6. Download FIG S1, PDF file, 0.2 MB (163KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers for Bioprojects, corresponding publications, and reference numbers for Bilophila genomes used in this study. Download Table S1, PDF file, 0.04 MB (44.8KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Amino acid sequences of TMA methyltransferase proteins from Bilophila genomes, with the pyrrolysine residue (O) highlighted in green. Download Table S2, PDF file, 0.04 MB (41.8KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TIGRFAM identity and gene names of proteins relevant for Bilophila genetic code expansion and TMA metabolism. Download Table S3, PDF file, 0.04 MB (36KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The metatranscriptomic data sets from mouse and human studies, the SRA accession numbers, and the number of reads for each data set. Download Table S4, PDF file, 0.06 MB (62.6KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The TMA and Pyl genes from metatranscriptomic data, their respective amino acid sequences, and the percent identity with corresponding proteins in the Bilophila genomes. Download Table S5, PDF file, 0.05 MB (53.5KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alignment of amino acid sequences from trimethylamine methyltransferase following the repurposed stop for Bilophila and an archaeon (Methanosarcina barkeri) that have pyrrolysine biosynthesis proteins; the sequence following the repurposed codon (pyrrolysine residue O) is conserved. Download FIG S2, PDF file, 0.07 MB (67.3KB, pdf) .
Copyright © 2020 Kivenson and Giovannoni.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.