Abstract
The Acinetobacter calcoaceticus-baumannii (Acb) complex is regarded as a group of phenotypically indistinguishable opportunistic pathogens responsible for mainly causing hospital-acquired pneumonia and bacteremia. The aim of this study was to determine the frequency of isolation of the species that constitute the Acb complex, as well as their susceptibility to antibiotics, and their distribution at the Hospital Infantil de Mexico Federico Gomez (HIMFG). A total of 88 strains previously identified by Vitek 2®, 40 as Acinetobacter baumannii and 48 as Acb complex were isolated from 52 children from 07, January 2015 to 28, September 2017. A. baumannii accounted for 89.77% (79/88) of the strains; Acinetobacter pittii, 6.82% (6/88); and Acinetobacter nosocomialis, 3.40% (3/88). Most strains were recovered mainly from patients in the intensive care unit (ICU) and emergency wards. Blood cultures (BC) provided 44.32% (39/88) of strains. The 13.63% (12/88) of strains were associated with primary bacteremia, 3.4% (3/88) with secondary bacteremia, and 2.3% (2/88) with pneumonia. In addition, 44.32% (39/88) were multidrug-resistant (MDR) strains and, 11.36% (10/88) were extensively drug-resistant (XDR). All strains amplified the blaOXA-51 gene; 51.13% (45/88), the blaOXA-23 gene; 4.54% (4/88), the blaOXA-24 gene; and 2.27% (2/88), the blaOXA-58 gene. Plasmid profiles showed that the strains had 1–6 plasmids. The strains were distributed in 52 pulsotypes, and 24 showed identical restriction patterns, with a correlation coefficient of 1.0. Notably, some strains with the same pulsotype were isolated from different patients, wards, or years, suggesting the persistence of more than one clone. Twenty-seven sequence types (STs) were determined for the strains based on a Pasteur multilocus sequence typing (MLST) scheme using massive sequencing; the most prevalent was ST 156 (27.27%, 24/88). The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas I-Fb system provided amplification in A. baumannii and A. pittii strains (22.73%, 20/88). This study identified an increased number of MDR strains and the relationship among strains through molecular typing. The data suggest that more than one strain could be causing an infection in some patient. The implementation of molecular epidemiology allowed the characterization of a set of strains and identification of different attributes associated with its distribution in a specific environment.
Keywords: Acinetobacter baumannii, Acinetobacter calcoaceticus-Acinetobacter baumannii complex, intensive care unit, resistance, molecular typing
Introduction
The Acinetobacter genus includes species widely disseminated in nature, mostly in water and soil; some of these species are considered opportunistic pathogens that are relevant for their association with health-care associated infections (HAIs). The clinically important species in this genus are Acinetobacter pittii, Acinetobacter nosocomialis, and Acinetobacter baumannii, with the last being the most important epidemiologically and frequently isolated from the intensive care unit (ICU), causing infections such as ventilator-associated pneumonia, bacteremia, urinary tract infection, meningitis, and wound infection (Dexter et al., 2015; Wong et al., 2017).
The Acinetobacter calcoaceticus-baumannii complex (Acb) harbored six species: A. calcoaceticus, A. baumannii, A. pittii, A. nosocomialis, Acinetobacter seifertii, and Acinetobacter dijkshoorniae (Gerner-Smidt et al., 1991; Nemec et al., 2011, 2015). The Acb complex cause hospital-acquired pneumonia and bacteremia in critically ill or immunocompromised patients (Wong et al., 2017; Vazquez and Kollef, 2018). The species of the complex are phenotypically indistinguishable and molecular methods are required for its correct identification (Gerner-Smidt et al., 1991). The participation of A. calcoaceticus in clinical infections remains unclear compared with other species, in which clinical importance has been demonstrated (Peleg et al., 2008; Nemec et al., 2015; Cosgaya et al., 2016). A. baumannii is one of the most difficult bacteria to contain in a hospital environment. Additionally, it is included in the list of priority pathogens resistant to antibiotics, owing to the resistance it has acquired to different antibiotics such as carbapenems and cephalosporins limiting the therapeutic options for the treatment of infections caused by this pathogen (Higgins et al., 2010; Shrivastava et al., 2018).
The pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and amplified fragment length polymorphism (AFLP) methods have been used to type clinical strains of A. baumanni (Limansky et al., 2004; Rafei et al., 2014). In hospitals in Spain and Germany, a total of 20 allelic profiles or sequence types (STs) were identified through MLST, and these results agreed with those generated by PFGE, suggesting MLST as a tool for the molecular epidemiological study of clinical strains of A. baumannii (Bartual et al., 2005). However, there are other studies where according to the tool used, the typing of the strains differs completely; the relationships between clinical isolates that were determined through PFGE revealed that isolates were not closely related (Evans et al., 2008). Through nucleotide analysis of the blaOXA-51 gene sequence, two closely related groups were identified. The sequencing of the blaOXA-51 gene, being only one genetic element, yielded a less-defined clonal relationship than PFGE analyses, where the whole genome is digested and analyzed to establish whether there is a clonal relationship between the strains under study (Evans et al., 2008).
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas system has been proposed for typing bacterial strains. Two CRISPR-Cas systems have been identified in strains of A. baumannii from military and children’s hospitals. An analysis of the nucleotide sequences of the CRISPR-Cas systems (CRISPR-AYE and Acinetobacter baylyi ADP systems) grouped the isolates into two clonal complexes and provided information about the evolution of these complexes (Hauck et al., 2012). At the same time, the CRISPR-Cas I-Fb system has been proposed for the subtyping of strains (Karah et al., 2015).
Genome sequencing is used to characterize and establish genetic relationships among isolates. Clinical strains identified as Acb complex were typed using PFGE, MLST, and single nucleotide polymorphism (SNP) analyses. SNP analysis was more discriminatory than those obtained by PFGE and MLST for the identification of clones and their association with outbreaks (Fitzpatrick et al., 2016).
A. baumannii mortality is approximately 14.5% and it has been associated with HAIs in tertiary level hospitals in Mexico (Bocanegra-Ibarias et al., 2015). Furthermore, the Mexican strains that have been characterized are distinguished by their resistance to imipenem and meropenem, as well as being associated with the amplification of carbapenemases such as OXA-23, OXA-239, and OXA-58. An epidemiological aspect of relevance is the different STs distributed among Mexican hospitals, belonging to clonal complexes 636 and 92, presenting ST208, ST369, and ST758 (Alcántar-Curiel et al., 2014; Tamayo-Legorreta et al., 2014; Gonzalez-Villoria et al., 2016). However, information about other species related to the Acb complex has not been available. The Acb complex poses a major challenge to this genus, suggesting a very recent diversification of those species, and events of homologous recombination can probably be contributed to a homogenous gene composition (Mateo-Estrada et al., 2019).
The aim of this study was to determine the frequency of isolation of the species that constitute the Acb complex, as well as the susceptibility to antibiotics, and their distribution at the Hospital Infantil de Mexico Federico Gomez (HIMFG). To achieve this aim, the Acinetobacter species were distinguished when the strains were identified as the Acb complex using a collection of 88 strains from 52 children from 07, January 2015 to 28, September 2017, previously identified as the A. baumannii and Acb complex. Then, after the susceptibility profile was determined, four of the most frequent blaOXA genes were detected, the plasmid profile, pulsotype, and sequence type were established and the CRISPR-Cas system was carried out for all strains.
Materials and Methods
Identification by MALDI-TOF Biotyper
In this study, all strains among 07, January 2015 to 28, September 2017 were considered and identified as the A. baumannii or Acb complex, from patients with or without HAIs. If one patient had more than one strain, all strains were included in this study. The strains were previously identified at the Central Clinical Laboratory at the HIMFG using the Vitek® 2 automated system (BioMérieux, Marcy l’Étoile France), and they were subsequently reidentified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) Biotyper (mass spectrometer, Bremen, Germany). Each strain was spread onto Brucella blood agar (BD Difco, Madrid, Spain), and one colony was placed on a metallic card for analysis (Bruker Daltonics Ultraflextreme, Bremen, Germany). The sample allowed to dry at room temperature; 1 μl of formic acid (70%) was placed on each well to dry at room temperature, and 1 μl of matrix [saturated solution of α-cyano-4-hydroxycinnamic acid (HCCA; Bruker Daltonics Ultraflextreme, Bremen, Germany) in 50% acetonitrile (Sigma, California, United States) and 2.5% trifluoroacetic acid were subsequently added (Sigma, California, United States)]. The spectra were analyzed using the MALDI Biotyper software Bruker Daltonics Ultraflextreme 3.1 (Bremen, Germany) and were compared with a database using identification criteria at the species level with a score between 1.7 and 1.9.
Antibiotic Susceptibility Test
Antibiotic susceptibility testing was performed using a Vitek® 2 automated system (BioMériux, Marcy l’Étoile, France). The antibiotics considered included penicillins (piperacillin); β-lactam combination agents (ampicillin-sulbactam and piperacillin-tazobactam); cephems (cefepime and ceftriaxone); carbapenems (imipenem); a lipopeptide (colistin); aminoglycosides (gentamicin); fluoroquinolones (ciprofloxacin); and folate pathway antagonists (trimethoprim-sulfamethoxazole), according to the Clinical and Laboratory Standards Institute (CLSI, 2018). Colistin susceptibility by the broth microdilution method according to CLSI was determined. Escherichia coli ATCC®25922 and Pseudomonas aeruginosa ATCC®27853 were used as quality controls; including A. baumannii ATCC®19606 as an internal control. Susceptibility to tigecycline was interpreted according to the United States Food and Drug Administration (FDA) breakpoints for Enterobacteriaceae. The multidrug-resistant (MDR) profile was defined as the strains resistant to three or more antimicrobial classes, and the extensively drug-resistant (XDR) profile was defined as the strains nonsusceptible to ≥1 agent in all but ≤2 categories (Magiorakos et al., 2012).
Amplification of blaOXA-LIKE Genes
Genomic DNA of strains was obtained with the Quick-DNA Universal kit (Zymo, Irvine, California, United States). blaOXA-LIKE genes were amplified by PCR using the specific primers listed in Table 1. PCR assays were performed using the following thermocycling conditions: 94°C for 5 min; 30 cycles at 94°C for 25 s, 52°C for 40 s, and 72°C for 50 s; and a final step at 72°C for 6 min. A. baumannii ATCC®19606 was used as a positive control for blaOXA-51.
Table 1.
Primers used to amplify blaOXA-LIKE genes.
| Gene | Sequence 5' – 3' | Amplified size (bp) | Reference |
|---|---|---|---|
| blaOXA-23 | F: GATCGGATTGGAGAA CAGA R: ATTTCTGACCGCATTTCCAT |
501 | Hujer et al. (2006) |
| blaOXA-24 | F: GGTTAGTTGGCCCCCTTAAA R: AGTTGACGCAAAAGGGGATT |
246 | Hujer et al. (2006) |
| blaOXA-51 | F: TAATGCTTTGATCGGCCTTG R: TGCATTGCACTTCATCTTGG |
353 | Hujer et al. (2006) |
| blaOXA-58 | F: AAGTATTGGGGCTTGTGCTG R: CCCCTCTGCGCTCTACATAC |
599 | Hujer et al. (2006) |
OXA-23 (blaOXA-23), OXA-24 (blaOXA-24), OXA-51 (blaOXA-51), and OXA-58 (blaOXA-58).
Plasmid DNA Profiles
The extraction of plasmid DNA was performed using the technique of Eckhardt (1978). A colony of each strain grown first on Brucella blood agar was cultured in 3 ml of Luria-Bertani (LB) broth (BD Difco, Madrid, Spain) with constant stirring (200 rpm) at 37°C for 15 h. Subsequently, 100 μl of this bacterial culture was incubated in 5 ml of LB broth under agitation at 37°C for 2.5 h. Finally, 1 ml bacterial culture was taken and centrifuged for 8 min at 14,000 rpm. The pellet was dissolved in 500 μl of cold sterile water, mixed with 1 ml of 0.3% sarcosyl solution, and then centrifuged at 14,000 rpm for 6 min. The pellet was incubated with 40 μl of 20% Ficoll in 10:1 TE buffer, kept on ice for 15 min and mixed with 20 μl of lysis solution [0.4 mg/ml RNase, 1 mg/ml bromophenol blue, 80 μl lysozyme (20 mg/ml in water)]. After 30 μl of SDS (10%) was added to each well, the samples were run on a 0.75% agarose gel (Promega, Wisconsin, United States) under the following conditions: 100 V for 15 min in 1X TBE buffer (AMRESCO, United States) without completely covering the gel and with the negative polarity inverted. After this time, 1X TBE was added to the chamber in a cold room until the gel was covered, and the samples were placed. The electrophoretic shift was performed with the poles in the standard orientation at 40 V for 90 min and thereafter at 100 V for 21 h. The gel was stained with ethidium bromide for visualization.
PFGE Assay
The 88 strains were seeded and incubated at 37°C for 18 h. PFGE assay was performed for all strains as described by Mancilla-Rojano et al. (2019). Briefly, colonies cultured on Brucella blood agar were selected and suspended in 1 ml of negative Gram suspension buffer (100 mM Tris-HCl and 100 mM EDTA 100 pH 8). The bacterial suspension was embedded into 1% agarose plugs (SeaKem, Cambrex, Rockland, United States) and lysed with 5 ml of lysis buffer at pH 8.0 [0.5 M Tris-HCl, 0.5 M EDTA, 1% N-lauryl sarcosine sodium salt, and 25 μl of proteinase K (20 mg/ml)]. Afterward, the samples were digested with the ApaI enzyme (Promega, Wisconsin, United States), and the chromosomal DNA obtained was subjected to electrophoresis on 1% agarose gels (Bio-Rad, Hercules, California, United States) using in the CHEF MAPPER system (Bio-Rad, Hercules, California, United States) using 0.5X TBE (AMRESCO, United States) under the following conditions: initial time 5.0 s, final time 30.0 s, 6 V/cm, inclination angle 120, and running time 24 h. The lambda marker (Biolabs, Hertfordshire, England, United Kingdom) was used as a molecular weight marker. The electrophoresis gels were stained with 0.5 mg/ml ethidium bromide for 40 min and visualized under UV light. The DNA fragment patterns generated by PFGE were analyzed and compared using NTSYS software version 2.2 (Applied Biostatistics, Setauket, New York, United States) with the unweighted pair group method using the arithmetic average (UPGMA) algorithm and the DICE correlation coefficient. The relatedness degree was assessed according to the criteria established by Tenover et al. (1995).
MLST Assay
Amplification of the fragments was carried out according to the MLST protocol with some modifications (Diancourt et al., 2010). The primers to amplify the fusA (elongation factor EF-G) gene were designed using the genome of A. baumannii AYE (GCA_000069245.1). To perform massive sequencing, the following adaptors were incorporated in each primer: F: 5'- TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG-3' and R: 5'- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA-3' (Table 2).
Table 2.
Primers used to amplify genes to determine STs by multilocus sequence typing (MLST).
| Gene | Sequence 5'-3' | Amplified size (bp) | |
|---|---|---|---|
| cpn60 | F: TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG ACT GTA CTT GCT CAA GC R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GTT CAG CGA TGA TAA GAA GTG G |
480 | Diancourt et al. (2010) |
| fusA | F: TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG ACA ATT ACC TCT GCT GCA ACA R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA TCA TCG CAG GTT TAC GTG CT |
633 | This study |
| gltA | F: TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG AGA TGT ATT GGC CTC AGG TCA CTT R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACG GGT TTA CTT TGT AGT CAC GGT CTG |
545 | Diancourt et al. (2010) |
| pyrG | F: TCG TCG GCA GCG TCA GTA GTG TAT AAG AGA CAG GGT GTT GTT TCA TCA CTA GGW AAA GG R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GAT AAA TGG TAA AGA YTC GAT RTC ACC |
434 | Diancourt et al. (2010) |
| recA | F: TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT GAA TCT TCY GGT AAA AC R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGT TTC TGG GCT GCC AAA CAT TAC |
425 | Diancourt et al. (2010) |
| rplB | F: TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GTA GAG CGT ATT GAA TAC GAT CCT AAC C R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GCA CCA CCA CCR TGY GGG TGA TC |
472 | Diancourt et al. (2010) |
| rpoB | F: TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GGC TTC TGA AGT ACG TGA CGT R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACG TCA CGT GCA ACG TTC GCT T |
502 | Diancourt et al. (2010) |
Citrate synthase (gltA); DNA gyrase subunit B (gyrB); glucose dehydrogenase B (gdhB); homologous recombination factor (recA); 60-kDa chaperonin (cpn60); glucose-6 phosphate isomerase (gpi); and RNA polymerase sigma factor (rpoD).
The genes were amplified from genomic DNA (1 μg), and PCR was performed under the following conditions: 94°C for 2 min, 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s, and a final step of 72°C for 5 min. To verify the amplified products via electrophoresis, 1.8% agarose gel electrophoresis was run using 1X TAE buffer at 120 V, and then the gel was stained with ethidium bromide to observe the amplified products with a transilluminator (Bio-Rad, CA, United States).
Constitutive gene sequencing was carried out using the Illumina Nextseq500 platform on 1 μg of genomic DNA. The readings obtained for each of the strains were analyzed through the bioinformatics tool FASTQC (Andrews, 2010) and were filtered with AfterQC (Chen et al., 2017). The sequences that had a depth of less than 20X and those that did not have the necessary length for the analysis were eliminated. Assembly of the readings was carried out from the sequences deposited in PubMLST using the aTRAM 2.0 program (Allen et al., 2018) with default parameters for paired readings and was verified by mapping the raw readings using BWA software (Li and Durbin, 2009). The mapped readings were filtered with SAMtools (Li et al., 2009) with the -m3 option to preserve only those readings that properly mapped onto the sequences, and the mappings were edited using SeaView (Gouy et al., 2010). The sequences obtained from each gene and the concatemer were analyzed using the database for A. baumannii.1 Each strain was characterized by a pattern of numbers that define its ST.
CRISPR-Cas System Identification
The CRISPR-Cas I-Fb system (95°C for 5 min, 30 cycles at 95°C for 1 min, 58°C for 1 min, 72°C for 7 min, and a final step of 72°C for 10 min), the CRISPR AYE system (95°C for 5 min, 30 cycles at 95°C for 1 min, 45°C for 1 min, 72°C for 7 min, and a final step of 72°C for 10 min), and the gene cas1 (95°C for 5 min, 35 cycles at 95°C for 30 s, 48°C for 30 s, 72°C for 30 s, and a final step of 72°C for 7 min) were identified by PCR (Table 3). To verify the presence of the amplified DNA, electrophoresis was performed on a 1% agarose gel using 1X TBE buffer at 120 V, and then the gel was stained with ethidium bromide to observe the amplification products with a transilluminator. The PCR products were purified and subsequently sequenced by capillary electrophoresis following the Sanger method. The sequences obtained were compared with the sequence of the CRISPR loci in strains of A. baumannii, which is available on the CRISPR web server.2 This server has 12 genomes of A. baumannii with structures confirmed by the CRISPR systems. The repeated sequences were analyzed by multiple alignments using the MultAlin interface page3 to determine the similarity between the strains.
Table 3.
Primers used to amplify CRISPR-Cas systems.
| Gene | Sequence 5'-3' | Reference |
|---|---|---|
| aye-cris | F: CCGTAGTTGAATCAACACGTA R: TTTGATTGGGTAAAATGCCAAA |
Hauck et al. (2012) |
| aye-cas1 | F: TCAAGCTGCGATGCGAATGT R: ATCCGGGCAAATTGAAACGC |
Hauck et al. (2012) |
| ab-cris | F: AGTCCCAGAGTTTTGACCCA R: TTGGATTGGGTCATCATTGGT |
Karah et al. (2015) |
aye-cris: CRISPR-AYE system, aye-cas1: cas1 gene, ab-cris: CRISPR-Cas I-Fb system.
Statistical Analysis
The data were analyzed using the chi square test to evaluate the relationship between variables, with p < 0.05 considered significant. The descriptive statistics included percentages and frequencies.
Results
The MALDI-TOF-MS Biotyper Allowed the Differentiation of Acb Complex
The eighty-eight strains firstly identified as A. baumannii and 48 as Acb complex were reidentified with the MALDI-TOF-MS Biotyper, allowing us to differentiate between the species with the Acb complex. Additionally, 89.77% (79/88) of the strains were identified as A. baumannii, 6.82% (6/88) as A. pittii, and 3.40% (3/88) as A. nosocomialis.
Patients in the Intensive Care Unit and Blood Culture Samples Were the Most Frequent Sources of A. baumannii Isolation
In this study was included 52 patients (children between 1 and 15 years old) retained in 14 wards at the HIMFG from January 2015 to September 2017. Patients located in the ICU and emergency wards [26.92% (14/52)] made up the largest number of patients carrying the Acinetobacter strain (Figure 1).
Figure 1.
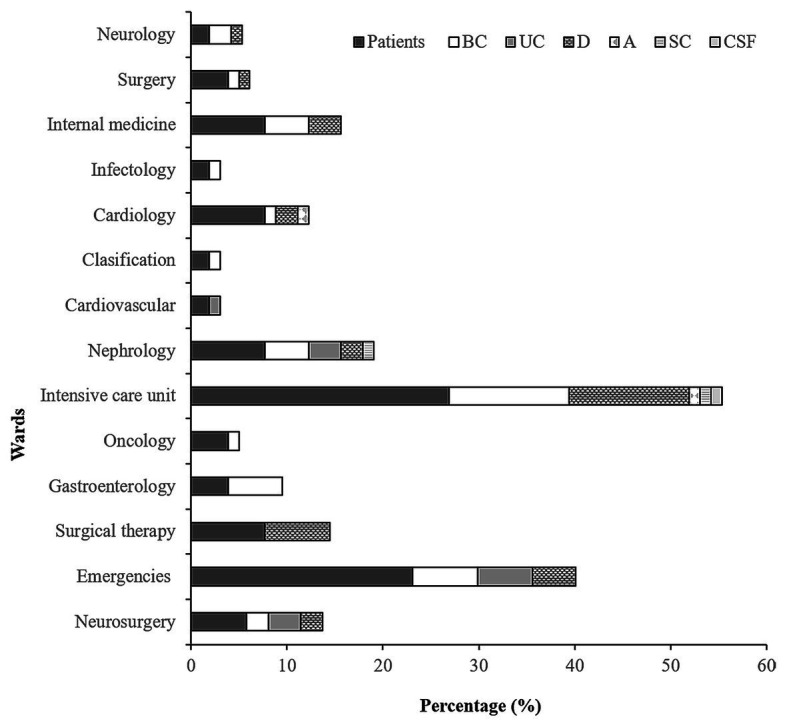
Frequency of isolation of the Acinetobacter baumannii, Acinetobacter pittii, and Acinetobacter nosocomialis strains. Percentage of patients from wards in which at least one of the strains was isolated in the period of study. BC, blood cultures; D, different samples; UC, urine cultures; SC, stool culture; A, autopsy; and CSF, cerebrospinal fluid.
From the 52 patients were recovered 88 strains with 37.5% (33/88), 25% (22/88), and 37.5% (33/88) obtained from the 2015, 2016, and 2017 samples, respectively. A. baumannii strains were identified in patients from several services wards, but mainly in emergency (13.63%, 12/88) and ICU (27.27%, 24/88). A. pittii strains were recovered from samples from patients in emergency, nephrology, and infectology. A. nosocomialis strains were obtained from samples from patients in surgery, gastroenterology, and internal medicine (Supplementary Table 1).
The isolation frequency by sample origin was as follows: 44.32% (39/88) from BC, 36.36% (32/88) from different (D) samples (including bronchial aspirate, catheter, mediastinal tissue, cholesteatoma culture, and peritoneal fluid), 13.63% (12/88) from urine cultures (UC), 2.27% (2/88) from a stool culture (SC) and an autopsy (A), and 1.14% (1/88) from cerebrospinal fluid (CSF, Figure 1).
According to the epidemiology analysis from 2015, nine strains (568BC, 173BC, 180BC, 181BC, 49BC, 50BC, 470BC, 471BC, and 800D) isolated from five patients were related to HAIs. Eight strains (219BC, 182BC, 183BC, 144D, 600BC, 928BC, 940BC, and 136BC) isolated from five patients were related to HAIs in 2017. The 13.63% (12/88) of strains were associated with primary bacteremia, 3.4% (3/88) with secondary bacteremia, and 2.3% (2/88) with pneumonia. The aforementioned infections were caused by identified strains such as A. baumannii, with the exception of one strain of A. nosocomialis (182BC) that was associated with primary bacteremia. Unlike these years in 2016, no HAIs were associated with Acinetobacter species.
The A. baumannii and A. pittii Strains Were Multidrug Resistant
The resistance profile was as follows: 97.73% (86/88) to a penicillin (piperacillin), 50% (44/88) to β-lactam combination agents (ampicillin-sulbactam), 35.23% (31/88) to β-lactam combination agents (piperacillin-tazobactam), 87.5% (77/88) to a third-generation cephem (ceftriaxone), 42.05% (37/88) to a fourth-generation cephem (cefepime), 38.64% (34/88) to a carbapenem (imipenem), 14.77% (13/88) to aminoglycosides (gentamicin), 40.91% (36/88) to fluoroquinolone (ciprofloxacin), 26.14% (23/88) to folate pathway antagonists (SXT), and 31.82% (28/88) to a glycylcycline (tigecycline; Figure 2). All strains were susceptible to colistin.
Figure 2.
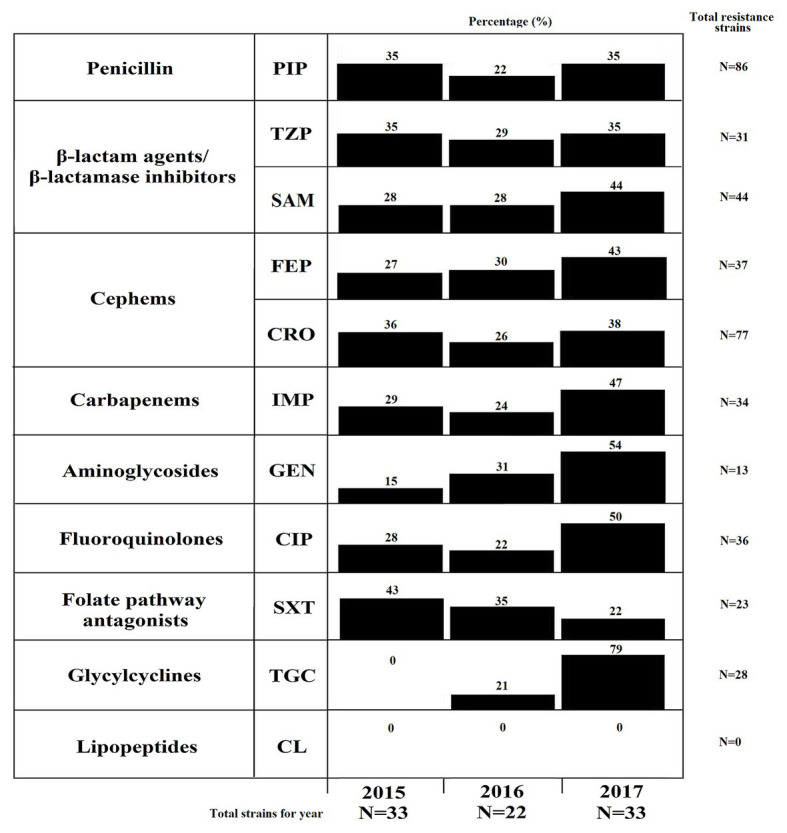
Resistance profiles of the A. baumannii, A. pittii, and A. nosocomialis strains. Percentages of strains resistant to at least one antibiotic in the following groups: penicillins (piperacillin), β-lactam agents/beta-lactamase inhibitors (ampicillin-sulbactam and piperacillin-tazobactam), third- and fourth-generation cephalosporins (cefepime and ceftriaxone), carbapenems (imipenem), aminoglycosides (gentamicin), fluoroquinolones (ciprofloxacin), folates (trimethoprim/sulfamethoxazole), glycylcyclines (tigecycline), and lipopeptides (colistin). PIP, piperacillin; TZP, piperacillin/tazobactam; SAM, ampicillin-sulbactam; FEP, cefepime; CRO, ceftriaxone; IMP, imipenem; GEN, gentamicin; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; TGC, tigecycline; and CL, colistin.
Additionally, 44.32% (39/88) of the strains showed a MDR resistance profile. The MDR profile distribution according to species was as follows: 94.87% (37/39) for A. baumannii and 5.13% (2/39) for A. pittii strains (Figure 3; Supplementary Table 1). Interestingly, the number of resistant strains for imipenem and tigecycline increased (p < 0.05) during 2017 (Supplementary Table 2). This is in contrast to folate pathway antagonists, for decreases in the number of resistant strains were observed (p < 0.05). On the other hand, 11.36% (10/88) of strains showed an XDR profile. Interestingly, all strains with this profile were identified as A. baumannii (Figure 3; Supplementary Table 1).
Figure 3.
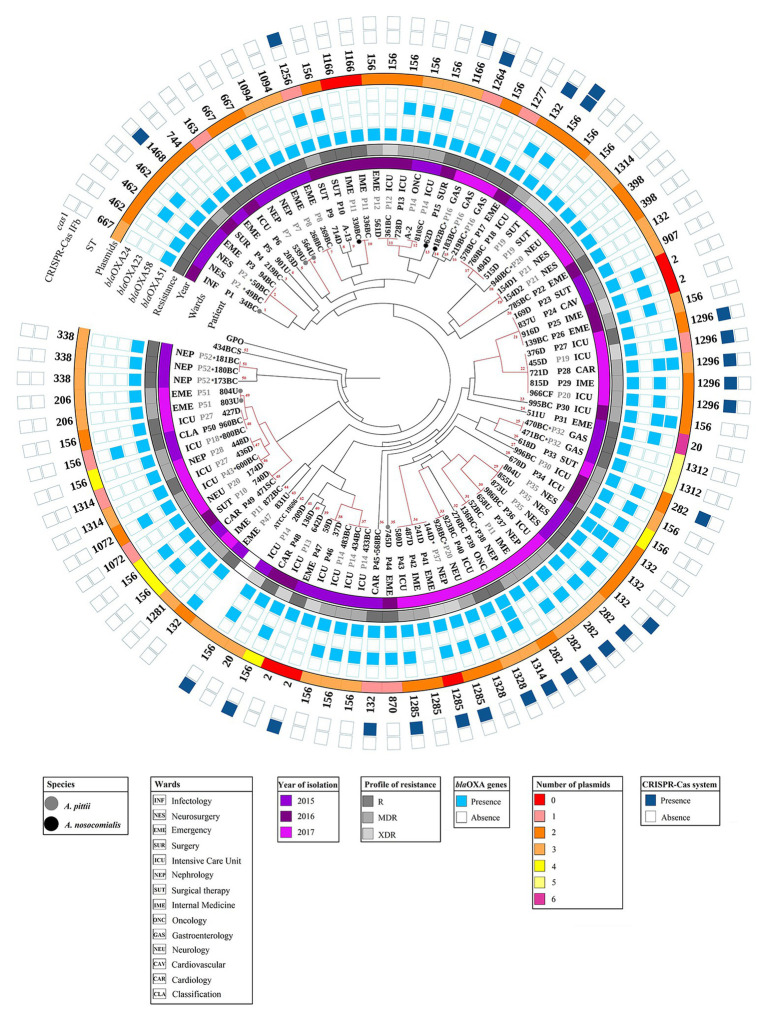
Dendrogram profile of the A. baumannii, A. pittii, and A. nosocomialis complex strains. Dendrogram profile displaying the genetic relatedness among the 88 strains based on the pulsed-field gel electrophoresis (PFGE) pattern. The red line represents the strains with the same pulsotype. The species other than A. baumannii are shown with circles in gray (A. pittii) and black (A. nosocomialis). Internal data represent strain with the origin of isolation, followed by the patient and the ward of origin for each strain. The asterisk symbol shows the strains associated with HAIs (bacteremia and pneumonia). The first band represents the year of isolation, and the second represents the profile of resistance: Resistant (R), multidrug-resistant (MDR), and extensively drug-resistant (XDR). The strains with a R profile showed resistance to PIP, SAM, or TZP. The detection of blaoxa genes is represented by clear blue clear squares and the amplification of the CRISPR-Cas system with dark blue squares. The number of plasmids associated with each strain is shown in the third band. The type sequences (ST) for each strain were added. The color codes for each analyzed characteristic are shown in the legends below the figure. The visualization was performed with the iTol program (Letunic and Bork, 2007).
The blaOXA-23 Gene was Amplified in the A. baumannii and A. pittii Strains
The presence of blaOXA-LIKE genes related to carbapenem resistance was determined by PCR. All strains amplified the blaOXA-51 gene, 51.13% (45/88) the blaOXA-23 gene, 4.54% (4/88) the blaOXA-24 gene, and 2.27% (2/88) the blaOXA-58 gene (Figure 3). Interestingly, the blaOXA-23 gene was amplified in two species (A. pittii and A. baumannii). The blaOXA-24 and blaOXA-58 genes were amplified only A. baumannii strains (Figure 3; Supplementary Table 1).
One to Six Plasmids Were Identified in A. baumannii, A. pittii, and A. nosocomialis Strains
The plasmid profiles obtained through the Eckardt method showed that the strains had from one to six plasmids with sizes between 2.4 and 121 kb. The results were the following: 37.5% (33/88) of the strains presented two plasmids, 34.09% (30/88) presented three plasmids, and only one of the strains presented six plasmids; however, in 7.95% (7/88) of the strains, these mobile elements were not detected (Figure 3; Supplementary Table 1). Briefly, A. baumannii strains carried one to six plasmids, A. pittii strains carried two or three plasmids, and A. nosocomialis carried one or three plasmids. No correlation was found between the resistance profile and the plasmid number; however, the largest number of MDR strains harbored between two or three plasmids.
The A. baumannii Strains Were Genetically Diverse in Comparison to A. pittii, A. nosocomialis
Macrorestriction patterns obtained by PFGE showed between 16 and 23 fragments with sizes of 48–339 kb. For construction of a dendrogram, a sensitive strain (434S), and the ATCC®19606 strain were included. In agreement with the macrorestriction pattern, the strains were distributed in 52 pulsotypes, and 24 showed were closely related with a correlation coefficient of 1.0 (Figure 3). Most of the strains maintained a correlation between the number of plasmids obtained by the Eckard technique and its pulsotype.
Strains recovered from the same patient were grouped into pulsotypes 6, 7, 10, 12, 19, 25, 37, 49, and 51 (Figure 3). The isolation period of these strains in the same patient was a maximum of 5 days according to PFGE and other determinants analyzed (resistance profile, blaoxa-like genes, plasmid number, ST, and CRISPR-Cas system) in this study, suggesting the presence of the same strain.
Interestingly, 12 pulsotypes (2, 11, 17, 21, 22, 31, 33, 34, 38, 46, 47, and 48) harbored strains with an identical macrorestriction pattern, which were isolated from at least two different patients with a minimum difference of 1 month between each strain. However, some changes were observed between these strains, mainly in profile resistance and blaoxa-like gene amplification.
Notably, some of the strains grouped in the same pulsotypes were isolated from different patients and/or wards and in some cases from samples from different years, such as, pulsotypes 17, 21, 31, and 45, suggesting the persistence of more than one clone in the hospital environment (Figure 3).
A. pittii strains were clustered into pulsotypes 1, 6, 35, and 49; while A. nosocomialis strains were clustered into pulsotypes 10, 13, and 14. Interestingly, all A. nosocomialis strains were closely related to A. baumannii strains. In addition, pulsotypes 2, 14, 15, 16, 18, 25, 31, 33, 34, 36, 46, 48, 50, and 51 harbored the strains related with HAIs (Figure 3; Supplementary Table 1).
ST 156 was Widely Distributed in the A. baumannii Strains
MLST results from Acinetobacter strains showed 27 STs, of which the most prevalent was ST 156, at 27.27% (24/88), followed by ST 132, 7.95% (7/88); STs 1296 and 282, 5.68% (5/88); STs 2, 1285, and 1314, 4.54% (4/88); STs 338, 462, 667, and 1166, 3.41% (3/88); STs 20, 206, 398, 1072, 1094, 1312, and 1328, 2.27% (2/88); and STs 163, 744, 870, 907, 1256, 1264, 1277, 1281, and 1468, 1.14% (1/88).
The relationships between the STs obtained in this study were analyzed through eBURST, which found that they belong to 12 clonal complexes (CC20, CC64, CC79, CC132, CC163, CC214, CC462, CC629, CC782, CC1094, CC1264, and CC2), and the last clonal complex (CC2) harbored most of the STs (Figure 3; Supplementary Table 1). The results obtained with MLST remained correlated with the data generated data through the other typing tools used in this study. The strains that presented the same plasmid, pulsotype, and resistance profiles also showed the same STs. Two inconsistencies were found between the PFGE and MLST results; more than one ST in the strains grouped into 17 and 21 pulsotypes, suggesting the presence of different strains in the same pulsotype.
In relation to the A. pittii strains, three STs (206, 667, and 870) were identified and regarding the A. nosocomialis strains, two STs (1166 and 1264) were identified (Figure 3; Supplementary Table 1). Interestingly, ST 1166 identified in A. nosocomialis (strains 330BC and 62D) has been described for A. baumannii, and according to the pulsotypes, these strains showed a macrorestriction pattern related to A. baumannii strains (pulsotypes 10 and 13). A. nosocomialis ST 1264 was associated with CC1264, while in A. pittii, only ST 667 was found in CC21; the other ST has not been associated with a CC.
Strains associated with HAIs showed STs 132, 462, 1312, 156, 338, 282, 1264, 1277, 1285, 1072, 1314, and 1328 (Figure 3; Supplementary Table 1)4.
CRISPR-Cas I-Fb Systems Were Identified in the A. baumannii, and A. nosocomialis Strains
The CRISPR-Cas I-Fb system was amplified in 22.73% (20/88) of the A. baumannii and A. nosocomialis strains; however, the other CRISPR-AYE system was not amplified (Figure 3; Supplementary Table 1). The cas1 gene was identified in 7.95% (7/88) of the strains.
The CRISPR-Cas I-Fb system was identified based on the sizes of the amplified PCR products, which was determined through sequencing spacers and repeated sequences. The sizes of the amplified sequences were approximately 700 bp (data not shown). Thereafter, they were sequenced, and when the bioinformatics search was carried out on the CRISPR server, it was found that the CRISPR-Cas I-Fb system sequences had 10 spacers with identical sequences to the nucleotide sequences of the strains in which they were performed5.
Discussion
A. baumannii is an opportunistic pathogen that is associated with severe infections worldwide and is related to the different attributes that allow it to emerge as a nosocomial microorganism, i.e., mainly resistance to antibiotics and its ability to persist in hospital environments due to its ability to resist drying (Antunes et al., 2014; Harding et al., 2017).
The genus Acinetobacter have six species with very similar phenotypes they and have been grouped into the Acb complex. Of this complex, A. pittii, A. nosocomialis and A. baumannii are the species associated with a greater number of infections and mortality; in addition, the isolation frequency of A. pittii and A. nosocomialis strains as etiological agents of HAIs is high (Wisplinghoff et al., 2012). The mortality rates of A. pittii are greater than those of A. nosocomialis but lower than those of A. baumannii, suggesting that a future A. pittii strain could also emerge as an important nosocomial pathogen (Yang et al., 2012). For this reason, the molecular typing of the Acb complex using different techniques is essential to generate information about the epidemiology of this complex and to learn more about the distribution of these species in hospital environments. The differentiation of Acb complex strains was carried out through MALDI-TOF MS in this study. This method allowed for the identification of the A. baumannii strains and differentiation between the species that belong to the Acb complex (Lin et al., 2008); three species were identified in this study: A. baumannii, A. pittii, and A. nosocomialis.
The ward with the highest recurrence of A. baumannii was the ICU, which corresponded to the data from other regions such as Europe, Asia, the United States, Latin America, and Morocco (Martins et al., 2009; Vincent et al., 2009; Uwingabiye et al., 2017). The high recurrence of isolating A. baumannii in the ICU has been attributed to risk factors for the development of infections, including invasive procedures, catheter placement, and intubation endotracheal, which may cause urinary tract and respiratory tract infections (Cisneros et al., 1996; Lynch et al., 2017). However, the development of A. baumannii infections is also related to the immune systems of patients since it has been observed in individuals with serious diseases such as hematological malignancies and diabetes mellitus, and even those subjected to prolonged antimicrobial therapy with broad-spectrum antibiotics (Vincent et al., 2009; Kempf and Rolain, 2012; Lin et al., 2016). A. pittii and A. nosocomialis strains were identified in patients at the HIMFG, but none of these strains were isolated from the ICU ward. The resistance percentage found in this study was low compared with those in other studies (Alcántar-Curiel et al., 2014, 2019; Xie et al., 2018).
The low resistance percentages could be associated with the source of strains from pediatric patients; however, an increase in the number of imipenem-resistant strains was found in 2017. The frequency of MDR strains in this study was high (44.32%), and this phenotype was identified in A. baumannii, and A. pittii strains. This MDR profile contrasted with the XDR profile, as only A. baumannii strains had this phenotype, and resistance has been associated with high epidemic potential and high mortality (Song et al., 2011; Lee et al., 2014). The main mechanism of A. baumannii resistance to β-lactams is enzymatic degradation by β-lactamases; oxacillinases (OXAs) give it the ability to hydrolyze oxacillin (Evans and Amyes, 2014). In this study, the blaOXA-51 gene, in addition to the blaOXA-23, blaOXA-24 and blaOXA-58 genes (identified more frequently in A. baumannii), were identified by PCR. All of the strains amplified the blaOXA-51 gene, which has been reported to be intrinsic in A. baumannii, with blaOXA-69 (Brown et al., 2005). The chromosomal location and the detection of these genes allow for identification at the species level (Turton et al., 2006).
Strains with an amplified blaOXA-23 gene formed the first group identified in A. baumannii, and the production of this enzyme is sufficient to confer resistance to carbapenems. This group is mostly distributed and has been detected in clinical strains in Brazil, Belgium, Singapore, and France, where have been related to outbreaks. In addition, the presence of the blaOXA-23 gene is limited to not only clinical strains but also environmental isolates, suggesting that the propagation of these genes occurs in different environments and under different selective pressures (Girlich et al., 2010). Moreover, 27% of the strains (A. pittii and A. baumannii) that amplified the blaOXA-23 gene showed resistance to imipenem, which have been reported in Latin America and Mexico. Other determinants of resistance have been associated with these species, such as the blaOXA-58 gene and metallo-β-lactamases, which are predominantly responsible for carbapenems resistance in A. nosocomialis and A. pittii; however, blaOXA-23 and blaOXA-24 have recently become more common in carbapenems resistance for both species (Yang et al., 2012; Cayô et al., 2014; Silva et al., 2018).
Most of the elements that encode for resistance can be found in mobile elements, such as plasmids. The determination of plasmid profiles has been used for comparisons of strains, since these can be found in identical numbers, sizes, and molecular weights in bacteria with different isolation origins. Our results agreed with other studies in which the authors included the characterization of 132 clinical strains of A. baumannii harboring of 1 to 7 plasmids (Singh et al., 2006). Furthermore, the strains that presented the same pulsotype, and ST had identical numbers of plasmids and molecular weights, with the exception of pulsotypes 17, 22, and 34, which showed the same type sequence and pulsotype but different in resistance profiles, blaOXA-LIKE genes, and plasmid profiles. The plasmid profile has been used as a complementary typing method to others such as PFGE; in this study, we observed congruence between the results obtained through each one; however, one of the limitations of this method is the use of mobile elements to carry out the typing of strains. Nevertheless, it provides a general overview of whether the strains can be carriers of mobile elements, from which their roles can be determined as mechanisms underlying the transfer of virulence and resistance determinants based on their sequencing.
Due to its high reproducibility, one of the most commonly used tools in the typing of clinical strains is PFGE. According to Tenover et al. (1995), when isolates do not differ in band pattern, they are considered indistinguishable, and therefore, the strains can be related. We found that 24 pulsotypes presented identical macrorestriction patterns; some strains were recovered from the same patient, but the others were isolated from different patients. The results of PFGE were consistent with the plasmid profiles, but when the advantages and disadvantages of these methods were compared, PFGE is clearly a technique that can be time consuming, laborious and expensive with respect to the profiling of plasmids; however, the results by PFGE were more accurate in terms of the typing of the strains, its reproducibility is high, and we can also obtain information regarding genetic diversity and clonal relationships.
Another one of the most common tools for typing A. baumannii strains is MLST, which is based on sequencing the variable regions of seven genes. For A. baumannii, two protocols based on different constitutive genes have been proposed, with some discrepancies (Tomaschek et al., 2016; Gaiarsa et al., 2019). Among the disadvantages that have been reported when using this method is the consideration of only seven constitutive genes of the total chromosomal information of A. baumannii strains, a bacterium with high genetic variation and would be disadvantageous to consider only a portion of the total information (Wang et al., 2013; Feng et al., 2016; Castillo-Ramírez and Graña-Miraglia, 2019). In this study, the strains were analyzed using the modified Pasteur protocol because in the case of the Oxford scheme, complications have been reported in the amplification of genes such as gdhB and gpi (Hamouda et al., 2010; Hamidian et al., 2017; Gaiarsa et al., 2019). The nucleotide sequences for the determination of STs were obtained through massive sequencing using next-generation platforms. However, under the Pasteur protocol, it was difficult to amplify and sequence the fusA gene; therefore, we designed other primers that allowed us to sequence this gene.
The most prevalent STs were STs 156, 132, 1296, 2, 282, 1285, and 1314. Interestingly, ST 156 belongs to the CC79 complex, which has been associated with outbreaks in several countries (Mexico, Canada, Honduras, Colombia, and even in Europe) and is epidemiologically important in Mexico (Loewen et al., 2014; Kubo et al., 2015; López-Leal et al., 2019). ST 156 strains are producers of carbapenemases (blaOXA-23) and recently included was the new variant OXA-239 (Tamayo-Legorreta et al., 2014; Graña-Miraglia et al., 2017; Mancilla-Rojano et al., 2019). According to our results, there is an association between ST 156 and blaOXA-23 gene amplification and between MDR and XDR profiles. Interestingly, ST 156 was identified during all 3 years of this study, which indicates that this lineage has been maintained and distributed within the hospital. However, other STs were associated with strains of a specific year. These data suggest that there is more than one strain distributed simultaneously within the HIMFG. STs 132 and particularly ST 2 the most common clone globally, are distributed in Europe, Asia, and Latin America, associated with outbreaks and resistance to carbapenems due to the presence of the blaOXA-66 and blaOXA-120 genes (Bakour et al., 2014; Dahdouh et al., 2017; Nowak et al., 2017; Levy-Blitchtein et al., 2018; Villalón et al., 2019). Conversely, we report a strain associated with ST 1468 that has recently been entered into the MLST A. baumannii database.
This technique has been used to distinguish the species of the Acb complex and designed to study the population of various bacteria, showing the presence of different clonal lineages, which could, for example, be associated with different species of the Acb complex (Bartual et al., 2005; Yamada et al., 2016). This finding agrees with our results, since the STs obtained for A. pittii were described in the MLST database for this species. Interestingly, the STs identified for A. pittii and A. nosocomialis are associated with different clonal complexes from those associated with A. baumannii. The MLST protocol is expensive and time consuming, and the analysis of the data can be slow; however, it should be noted that it allows the typing of strains and the differentiation of some species of the Acb complex (such as A. pittii). Similarly, the comparison with strains in other parts of the world enabled us to recognize the STs and clonal complexes associated with this hospital, determining that ST 156 is a clone that persists in the HIMFG.
The PFGE results were consistent with the MLST results, the strains with the same pulsotype presented the same ST, and except for the strains with pulsotypes 17 and 21, which were related despite being associated with different patients and years. The pulsotypes 17 and 21were grouped with the same restriction patterns; nevertheless, we could observe that they differed with respect to their resistance profile, plasmid profile, and amplification of blaOXA genes. However, the determination of different STs allows us to suggest that these pulsotypes group different strains; the opposite occurred in some of the strains that were isolated from different patients, since there were cases in which, according to the agreement of the data obtained by PFGE, the same patient could harbor different pulsotypes; nevertheless, they could present the same ST (P14, and P30).
Recently, the polymorphism observed between CRISPR-Cas systems has been used to genotype strains and establish phylogenetic relationships in different bacterial species; although, in some cases the spacers can be too diverse, hindering their use as a method for subtyping. On the other hand, the CRISPR-Cas system has been implemented as a successful typing method in species such as Salmonella enterica and Yersinia pestis (Cui et al., 2008; Shariat et al., 2015). The use of this system has also separated other bacteria such as Erwinia amylovora into different groups depending on their geographical origin (Rezzonico et al., 2011). Two systems have been described in A. baumannii: CRISPR-AYE and CRISPR-Cas subtype I-Fb (Hauck et al., 2012; Karah et al., 2015). In this study, the CRISPR-Cas subtype I-Fb system was identified in only 20 strains. Despite the amplification of the CRISPR-AYE system with primers reported previously by Karah et al. (2015), when the PCR products were sequenced, the nucleotide analyses did not show any homology with the CRISPR-Cas systems.
Despite the successful implementation of the CRISPR-Cas system as a typing method in other studies, we were unable to use it because not all of the strains had this system; however, two phenomena were observed. The first was that some strains that amplified the CRISPR-Cas I-Fb system had been grouped in the same pulsotypes and possessed the same STs. The second was that some strains had the same pulsotype but differed in STs or CRISPR-Cas system; interestingly, they provide us with information on the diversity within each of the subgroups obtained by PFGE and MLST. There was no ST that was specifically associated with some type of these systems, but interestingly, the CRISPR-Cas I-Fb system was identified with more frequency in strains recovered during 2017.
On the other hand, approximately 70% (14/20) of the strains with a CRISPR-Cas I-Fb system presented MDR and XDR profiles, which has not been observed in other studies since there is no correlation between the previous profiles (Hauck et al., 2012). Amazingly, we found that the strains that amplify the CRISPR-Cas I-Fb system presented from 1 to 4 plasmids. According to Mangas et al. (2019), there is an association between the presence of the CRISPR-Cas system and the absence of plasmids in A. baumannii genomes; as we could see in our results; these systems would limit the transfer of mobile elements such as plasmids.
In addition, the gene encoding the Cas1 protein of the CRISPR-Cas system was also identified only in seven strains. The gene encoding the Cas1 protein is one of the most conserved and is involved in the acquisition of spacers and it has been proposed that the gene evolves slower than other cas genes (Takeuchi et al., 2012; Makarova and Koonin, 2015). The strains that amplified this gene did not amplify any CRISPR-Cas system, so we could suggest the presence of another system different from those sought in this study. In contrast, 26 strains amplified at least one system but not the cas gene. For Enterococcus faecium strains, the cas1 gene has not been identified with CRISPR-Cas IIA systems, suggesting that these systems have lost their ability to acquire different spacers (van Schaik and Willems, 2010; Lyons et al., 2015), which could also have occurred in our strains. However, it is necessary to carry out other studies to demonstrate this hypothesis.
The amplified spacers were larger in the CRISPR-Cas I-Fb system. Strains that had more spacers in their CRISPR systems have been related to evolution in the environment (Hauck et al., 2012). The detection of these systems through PCR and amplification sequencing seems to be simple; however, something that must be highlighted is the probable implication of these systems in other processes. CRISPR-Cas systems in E. coli are related to the repair of DNA damage (Babu et al., 2011); this would be relevant in A. baumannii, since microorganisms are subjected to a series of environmental agents or possess resistance to desiccation. An important contribution of our work is the identification of the CRISPR-Cas system in clinical strains.
In the case of patients, there were two interesting conditions. First, in patients (14, 20, 27, 28, 37, 43, and 47), more than one strain was isolated, some with differences of days or 1 month. These strains were all different, locating themselves into different pulsotypes, STs, and with differences in the rest of the characteristics evaluated, suggesting the presence of different strains in the same patient over a short temporality. In the second case, was observed that patients (11, 16, and 19) had more than one strain; however, they were divided into two groups in the same patient that were identical, and that were not same in the characteristics evaluated, suggesting once again the presence of more than one strain over a longer period. In 11 patients, the strains were obtained on the same date but corresponded to different samples, presenting cases, in which the strains showed the same characteristics with respect to the pulsotype, blaOXA-LIKE genes, plasmid profile, and type sequence. Nevertheless, five patients (P16, P18, P27, P30, and P47) provided samples on the same date but with phenotypic and genotypic characteristics that were completely different. Also, there were cases presented in which the samples obtained more than 1 day apart exhibited the same characteristics.
Only 17 strains were associated with HAIs, such as bacteremia and pneumonia. According to the results, the patients: 2 (49BC and 50BC), 32 (470BC and 471BC), and 52 (173BC, 180BC, and 181BC) related with bacteremia, had more than one strain. The strains were distributed along the tree and only clustered when they were from the same patient such as pulsotypes 2 and 25, for which all of the phenotypic and genotypic characteristics were identical, with the exception of strain 173BC recovered from the patient 52, the only difference that had with the other two strains (189BC and 181BC) isolated of this patient was his macrorestriction pattern. Patients 18 (800BC) and 38 (136BC) developed bacteremia; while, patients 37 (144D), 43 (600BC), and 45 (568BC) developed pneumonia, the strains were randomly distributed in different pulsotypes, but apparently there was no related between them. Finally, patients 16 (182BC, 183BC, and 219BC) and 20 (928BC and 940BC) with bacteremia were all different from each other. Interestingly, these features were observed for strains identified as A. baumannii, with the exception of one strain of A. nosocomialis that was associated with a primary bacteremia. The latter was isolated together with two strains of A. baumannii from the same patient (P16) on the same date; however, they presented different genetic characteristics, which suggest that more than one different strain could be causing an infection in the patient. These strains were collected from a total of 10 patients and are genetically diverse since they presented different plasmid profiles, type sequences, and pulsotypes.
According to the results obtained in this study, an increase was identified an increase in the number of resistant carbapenems strains. The molecular typing methods allowed us to determine the relationships between clinical strains. PFGE data demonstrated that different patients could be infected by the same strain (identical pulsotypes were harbored by more than one patient). MLST showed that the strains in one group, i.e., pulsotypes 17 and 21, showed different STs and thus were not the same strain. ST 156 is a persistent clone at HIMFG. Finally, the CRISPR-Cas system identified in a low percentage of these strains could be associated with the nosocomial environment. The severity of the disease and the difference between strains that are closely related to those associated with HAIs shown in this study, were related to the immunocompromise patient and virulence factors of the strain, which were not determined in this study and will be examined in a future study. The clinical importance of A. baumannii in ICUs isolated from BCs has been well documented. A. baumannii infections (i.e., nosocomial pneumonia) tend to be more serious than those caused by A. nosocomialis and A. pittii (Lee et al., 2013). Therefore, it is necessary to identify and differentiate the species in the Acb complex to determine their epidemiological and clinical importance. The implementation of molecular epidemiology allows the characterization of a set of strains and identification of different attributes associated with their distribution in a specific environment. This study will allow future interventions for the recognition of risk factors related to opportunistic pathogens such as A. baumannii.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The Research Committee (Dr. Juan Garduño Espinosa), Ethics Committee (Dr. Luis Jasso Gutiérrez), and Biosecurity Committee (Dr. Marcela Salazar García) of the HIMFG granted approval for the development of the protocol HIM/2017/003 SSA.1299, HIM/2018/038 SSA.1513. The strains were provided by the Central Laboratory of the HIMFG, with prior informed consent of the patients to obtain the samples. Written informed consent was not required for this study according to the institutional ethical, biosecurity and investigation committees because the Central Laboratory from the HIMFG provided the A. baumannii clinical strain isolates from the child included in this study.
Author Contributions
AC-C had the initial idea and developed it into a project together with JX-C. JM-R done the experiments. AC-C, JX-C, SO, VF, and JA-G analyzed the data. OM-C performed identification by MALDI-TOF Biotyper. DR-Z reviewed the clinical data. AC-C, JX-C, SO, KE-M, MC, JR-G, JA-G, RH-C, and OM-C contributed reagents and materials. IP-O supplied the A. baumannii strains. MC-C carried out susceptibility. AC-C and JX-C wrote the manuscript, read and approved the final version. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge Gerardo Escalona Venegas and Isabel Franco Hernández for their technical assistance. Miriam Bobadilla del Valle and Francisco Leal Vega for sequencing support. Consejo Nacional de Ciencia y Tecnología, México for SNI EXP. AYTE17734.
Funding. JM-R received support from the Consejo Nacional de Ciencia y Tecnología, México, PDCPN 605309. This work was supported by Federal Founds HIM/2017/003 SSA.1299, HIM/2018/038 SSA.1513 at the HIMFG.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.576673/full#supplementary-material
References
- Alcántar-Curiel M. D., García-Torres L. F., González-Chávez M. I., Morfín-Otero R., Gayosso-Vázquez C., Jarillo-Quijada M. D., et al. (2014). Molecular mechanisms associated with nosocomial carbapenem-resistant Acinetobacter baumannii in Mexico. Arch. Med. Res. 45, 553–560. 10.1016/j.arcmed.2014.10.006, PMID: [DOI] [PubMed] [Google Scholar]
- Alcántar-Curiel M. D., Rosales-Reyes R., Jarillo-Quijada M. D., Gayosso-Vázquez C., Fernández-Vázquez J. L., Toledano-Tableros J. E., et al. (2019). Carbapenem-resistant Acinetobacter baumannii in three tertiary care hospitals in Mexico: virulence profiles, innate immune response and clonal dissemination. Front. Microbiol. 10:2116. 10.3389/fmicb.2019.02116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. M., LaFrance R., Folk R. A., Johnson K. P., Guralnick R. P. (2018). aTRAM 2.0: an improved, flexible locus assembler for NGS data. Evol. Bioinforma. 14:117693431877454. 10.1177/1176934318774546, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: una herramienta de control de calidad para datos de secuencia de alto rendimiento. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed April 2, 2020).
- Antunes L. C. S., Visca P., Towner K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. 10.1111/2049-632X.12125, PMID: [DOI] [PubMed] [Google Scholar]
- Babu M., Beloglazova N., Flick R., Graham C., Skarina T., Nocek B., et al. (2011). A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 79, 484–502. 10.1111/j.1365-2958.2010.07465.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakour S., Alsharapy S. A., Touati A., Rolain J. M. (2014). Characterization of Acinetobacter baumannii clinical isolates carrying blaOXA-23 carbapenemase and 16S rRNA methylase armA genes in Yemen. Microb. Drug Resist. 20, 604–609. 10.1089/mdr.2014.0018, PMID: [DOI] [PubMed] [Google Scholar]
- Bartual S. G., Seifert H., Hippler C., Luzon M. A. D., Wisplinghoff H., Rodriguez-Valera F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. 10.1128/JCM.43.9.4382-4390.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocanegra-Ibarias P., Peña-López C., Camacho-Ortiz A., Llaca-Díaz J., Silva-Sánchez J., Barrios H., et al. (2015). Genetic characterisation of drug resistance and clonal dynamics of Acinetobacter baumannii in a hospital setting in Mexico. Int. J. Antimicrob. Agents 45, 309–313. 10.1016/j.ijantimicag.2014.10.022, PMID: [DOI] [PubMed] [Google Scholar]
- Brown S., Young H. K., Amyes S. G. B. (2005). Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11, 15–23. 10.1111/j.1469-0691.2004.01016.x, PMID: [DOI] [PubMed] [Google Scholar]
- Castillo-Ramírez S., Graña-Miraglia L. (2019). Inaccurate multilocus sequence typing of Acinetobacter baumannii. Emerg. Infect. Dis. 25, 186–187. 10.3201/eid2501.180374, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayô R., Merino M., Ruiz del Castillo B., Cano M. E., Calvo J., Bou G., et al. (2014). OXA-207, a novel OXA-24 variant with reduced catalytic efficiency against carbapenems in Acinetobacter pittii from Spain. Antimicrob. Agents Chemother. 58, 4944–4948. 10.1128/AAC.02633-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Huang T., Zhou Y., Han Y., Xu M., Gu J. (2017). AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 18:80. 10.1186/s12859-017-1469-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros J. M., Reyes M. J., Pachon J., Becerril B., Caballero F. J., Garcia-Garmendia J. L., et al. (1996). Bacteremia due to Acinetobacter baumannii: epidemiology, clinical findings, and prognostic features. Clin. Infect. Dis. 22, 1026–1032. 10.1093/clinids/22.6.1026, PMID: [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2018). Performance standards for antimicrobial susceptibility testing. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cosgaya C., Marí-Almirall M., Van Assche A., Fernández-Orth D., Mosqueda N., Telli M., et al. (2016). Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int. J. Syst. Evol. Microbiol. 66, 4105–4111. 10.1099/ijsem.0.001318, PMID: [DOI] [PubMed] [Google Scholar]
- Cui Y., Li Y., Gorgé O., Platonov M. E., Yan Y., Guo Z., et al. (2008). Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One 3:e2652. 10.1371/journal.pone.0002652, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdouh E., Gómez-Gil R., Pacho S., Mingorance J., Daoud Z., Suárez M. (2017). Clonality, virulence determinants, and profiles of resistance of clinical Acinetobacter baumannii isolates obtained from a Spanish hospital. PLoS One 12:e0176824. 10.1371/journal.pone.0176824, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter C., Murray G. L., Paulsen I. T., Peleg A. Y. (2015). Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev. Anti-Infect. Ther. 13, 567–573. 10.1586/14787210.2015.1025055, PMID: [DOI] [PubMed] [Google Scholar]
- Diancourt L., Passet V., Nemec A., Dijkshoorn L., Brisse S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. (1978). A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid 1, 584–588. 10.1016/0147-619x(78)90016-1, PMID: [DOI] [PubMed] [Google Scholar]
- Evans B. A., Amyes S. G. B. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. 10.1128/CMR.00117-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. A., Hamouda A., Towner K. J., Amyes S. G. B. (2008). OXA-51-like β-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin. Microbiol. Infect. 14, 268–275. 10.1111/j.1469-0691.2007.01919.x, PMID: [DOI] [PubMed] [Google Scholar]
- Feng Y., Ruan Z., Shu J., Chen C. L., Chiu C. H. (2016). A glimpse into evolution and dissemination of multidrug-resistant Acinetobacter baumannii isolates in East Asia: a comparative genomics study. Sci. Rep. 6:24342. 10.1038/srep24342, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick M. A., Ozer E. A., Hauser A. R. (2016). Utility of whole-genome sequencing in characterizing Acinetobacter epidemiology and analyzing hospital outbreaks. J. Clin. Microbiol. 54, 593–612. 10.1128/JCM.01818-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiarsa S., Batisti Biffignandi G., Esposito E. P., Castelli M., Jolley K. A., Brisse S., et al. (2019). Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front. Microbiol. 10:930. 10.3389/fmicb.2019.00930, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner-Smidt P., Tjiernberg I., Ursing J. (1991). Reliability of phenotypic tests for identification of Acinetobacter species. J. Clin. Microbiol. 29, 277–282. 10.1128/JCM.29.2.277-282.1991, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. (2010). First isolation of the blaOXA-23 carbapenemase gene from an environmental Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 54, 578–579. 10.1128/AAC.00861-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Villoria A. M., Tamayo-Legorreta E., Garza-Ramos U., Barrios H., Sanchez-Pérez A., Rodríguez-Medina N., et al. (2016). A multicenter study in Mexico finds Acinetobacter baumannii clinical isolates belonging to clonal complexes 636B (113B) and 92B harboring OXA-72, OXA-239, and OXA-469. Antimicrob. Agents Chemother. 60, 2587–2588. 10.1128/AAC.02042-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. 10.1093/molbev/msp259, PMID: [DOI] [PubMed] [Google Scholar]
- Graña-Miraglia L., Lozano L. F., Velázquez C., Volkow-Fernández P., Pérez-Oseguera Á., Cevallos M. A., et al. (2017). Rapid gene turnover as a significant source of genetic variation in a recently seeded population of a healthcare-associated pathogen. Front. Microbiol. 8:1817. 10.3389/fmicb.2017.01817, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Nigro S. J., Hall R. M. (2017). Problems with the oxford multilocus sequence typing scheme for Acinetobacter baumannii: do sequence type 92 (ST92) and ST109 exist? J. Clin. Microbiol. 55, 2287–2289. 10.1128/JCM.00533-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda A., Evans B. A., Towner K. J., Amyes S. G. B. (2010). Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of blaOXA-51-like genes. J. Clin. Microbiol. 48, 2476–2483. 10.1128/JCM.02431-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. M., Hennon S. W., Feldman M. F. (2017). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. 10.1038/nrmicro.2017.148, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck Y., Soler C., Jault P., Mérens A., Gérome P., Nab C. M., et al. (2012). Diversity of Acinetobacter baumannii in four French military hospitals, as assessed by multiple locus variable number of tandem repeats analysis. PLoS One 7:e44597. 10.1371/journal.pone.0044597, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins P. G., Dammhayn C., Hackel M., Seifert H. (2010). Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 233–238. 10.1093/jac/dkp428, PMID: [DOI] [PubMed] [Google Scholar]
- Hujer K. M., Hujer A. M., Hulten E. A., Bajaksouzian S., Adams J. M., Donskey C. J., et al. (2006). Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter reed Army medical Center. Antimicrob. Agents Chemother. 50, 4114–4123. 10.1128/AAC.00778-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karah N., Samuelsen Ø., Zarrilli R., Sahl J. W., Wai S. N., Uhlin B. E. (2015). CRISPR-cas subtype I-Fb in Acinetobacter baumannii: evolution and utilization for strain subtyping. PLoS One 10:e0118205. 10.1371/journal.pone.0118205, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf M., Rolain J. M. (2012). Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int. J. Antimicrob. Agents 39, 105–114. 10.1016/j.ijantimicag.2011.10.004, PMID: [DOI] [PubMed] [Google Scholar]
- Kubo Y., Komatsu M., Tanimoto E., Sugimoto K., Tanaka S., Migita S., et al. (2015). Spread of OXA-23-producing Acinetobacter baumannii ST2 and ST246 in a hospital in Japan. J. Med. Microbiol. 64, 739–744. 10.1099/jmm.0.000077, PMID: [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Chen C. L., Wu S. R., Huang C. W., Chiu C. H. (2014). Risk factors and outcome analysis of Acinetobacter baumannii complex bacteremia in critical patients. Crit. Care Med. 42, 1081–1088. 10.1097/CCM.0000000000000125, PMID: [DOI] [PubMed] [Google Scholar]
- Lee Y. T., Kuo S. C., Yang S. P., Lin Y. T., Chiang D. H., Tseng F. C., et al. (2013). Bacteremic nosocomial pneumonia caused by Acinetobacter baumannii and Acinetobacter nosocomialis: a single or two distinct clinical entities? Clin. Microbiol. Infect. 19, 640–645. 10.1111/j.1469-0691.2012.03988.x, PMID: [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2007). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. 10.1093/bioinformatics/btl529, PMID: [DOI] [PubMed] [Google Scholar]
- Levy-Blitchtein S., Roca I., Plasencia-Rebata S., Vicente-Taboada W., Velásquez-Pomar J., Muñoz L., et al. (2018). Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg. Microbes Infect. 7:119. 10.1038/s41426-018-0127-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. 10.1093/bioinformatics/btp324, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limansky A. S., Zamboni M. I., Guardati M. C., Rossignol G., Campos E., Viale A. M. (2004). Evaluation of phenotypic and genotypic markers for clinical strains of Acinetobacter baumannii. Medicina 64, 306–312. PMID: [PubMed] [Google Scholar]
- Lin C. Y., Chen Y. M., Lin M. C., Chang Y. P., Chao T. Y., Wang C. C., et al. (2016). Risk factors of multidrug-resistant Acinetobacter baumannii recurrence after successful eradication in ventilated patients. Biom. J. 39, 130–138. 10.1016/j.bj.2015.07.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C., Sheng W. H., Chang S. C., Wang J. T., Chen Y. C., Wu R. J., et al. (2008). Application of a microsphere-based array for rapid identification of Acinetobacter spp. with distinct antimicrobial susceptibilities. J. Clin. Microbiol. 46, 612–617. 10.1128/JCM.01798-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C., Alsaadi Y., Fernando D., Kumar A. (2014). Genome sequence of an extremely drug-resistant clinical isolate of Acinetobacter baumannii strain AB030. Genome Announc. 2, e01035–e01114. 10.1128/genomeA.01035-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Leal G., Zuniga-Moya J. C., Castro-Jaimes S., Graña-Miraglia L., Pérez-Oseguera Á., Reyes-García H. S., et al. (2019). Unexplored genetic diversity of multidrug- and extremely drug-resistant Acinetobacter baumannii isolates from tertiary hospitals in Honduras. Microb. Drug Resist. 25, 690–695. 10.1089/mdr.2018.0311, PMID: [DOI] [PubMed] [Google Scholar]
- Lynch J., Zhanel G., Clark N. (2017). Infections due to Acinetobacter baumannii in the ICU: treatment options. Semin. Respir. Crit. Care Med. 38, 311–325. 10.1055/s-0037-1599225, PMID: [DOI] [PubMed] [Google Scholar]
- Lyons C., Raustad N., Bustos M. A., Shiaris M. (2015). Incidence of type II CRISPR1-Cas systems in enterococcus is species-dependent. PLoS One 10:e0143544. 10.1371/journal.pone.0143544, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. 10.1111/j.1469-0691.2011.03570.x, PMID: [DOI] [PubMed] [Google Scholar]
- Makarova K., Koonin E. (2015). Annotation and classification of CRISPR-Cas systems. Methods Mol. Biol. 1311, 47–75. 10.1007/978-1-4939-2687-9_4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla-Rojano J., Castro-Jaimes S., Ochoa S. A., Bobadilla del Valle M., Luna-Pineda V. M., Bustos P., et al. (2019). Whole-genome sequences of five Acinetobacter baumannii strains from a child with leukemia M2. Front. Microbiol. 10:132. 10.3389/fmicb.2019.00132, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangas E. L., Rubio A., Álvarez-Marín R., Labrador-Herrera G., Pachón J., Pachón-Ibáñez M. E., et al. (2019). Pangenome of Acinetobacter baumannii uncovers two groups of genomes, one of them with genes involved in CRISPR/Cas defence systems associated with the absence of plasmids and exclusive genes for biofilm formation. Microb. Genom. 5:e000309. 10.1099/mgen.0.000309, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins A. F., Kuchenbecker R., Sukiennik T., Boff R., Reiter K. C., Lutz L., et al. (2009). Carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme: dissemination in Southern Brazil. Infection 37, 474–476. 10.1007/s15010-009-9003-9, PMID: [DOI] [PubMed] [Google Scholar]
- Mateo-Estrada V., Graña-Miraglia L., López-Leal G., Castillo-Ramírez S. (2019). Phylogenomics reveals clear cases of misclassification and genus-wide phylogenetic markers for Acinetobacter. Genome Biol. Evol. 11, 2531–2541. 10.1093/gbe/evz178, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec A., Krizova L., Maixnerova M., Sedo O., Brisse S., Higgins P. (2015). Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus – Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 65, 934–942. 10.1099/ijs.0.000043, PMID: [DOI] [PubMed] [Google Scholar]
- Nemec A., Krizova L., Maixnerova M., van der Reijden T. J. K., Deschaght P., Passet V., et al. (2011). Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittiii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162, 393–404. 10.1016/j.resmic.2011.02.006, PMID: [DOI] [PubMed] [Google Scholar]
- Nowak J., Zander E., Stefanik D., Higgins P. G., Roca I., Vila J., et al. (2017). High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 72, 3277–3282. 10.1093/jac/dkx322, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. 10.1128/CMR.00058-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafei R., Kempf M., Eveillard M., Dabboussi F., Hamze M., Joly-Guillou M. L. (2014). Current molecular methods in epidemiological typing of Acinetobacter baumannii. Future Microbiol. 9, 1179–1194. 10.2217/fmb.14.63, PMID: [DOI] [PubMed] [Google Scholar]
- Rezzonico F., Smits T. H. M., Duffy B. (2011). Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora. Appl. Environ. Microbiol. 77, 3819–3829. 10.1128/AEM.00177-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat N., Timme R. E., Pettengill J. B., Barrangou R., Dudley E. G. (2015). Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology 161, 374–386. 10.1099/mic.0.000005, PMID: [DOI] [PubMed] [Google Scholar]
- Shrivastava S., Shrivastava P., Ramasamy J. (2018). World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 32, 76–77. 10.4103/jms.jms_25_17 [DOI] [Google Scholar]
- Silva L., Mourão J., Grosso F., Peixe L. (2018). Uncommon carbapenemase-encoding plasmids in the clinically emergent Acinetobacter pittii. J. Antimicrob. Chemother. 73, 52–56. 10.1093/jac/dkx364, PMID: [DOI] [PubMed] [Google Scholar]
- Singh A., Goering R. V., Simjee S., Foley S. L., Zervos M. J. (2006). Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 19, 512–530. 10.1128/CMR.00025-05, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y., Cheong H. J., Choi W. S., Heo J. Y., Noh J. Y., Kim W. J. (2011). Clinical and microbiological characterization of carbapenem-resistant Acinetobacter baumannii bloodstream infections. J. Med. Microbiol. 60, 605–611. 10.1099/jmm.0.029439-0, PMID: [DOI] [PubMed] [Google Scholar]
- Takeuchi N., Wolf Y. I., Makarova K. S., Koonin E. V. (2012). Nature and intensity of selection pressure on CRISPR-associated genes. J. Bacteriol. 194, 1216–1225. 10.1128/JB.06521-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo-Legorreta E. M., Garza-Ramos U., Barrios-Camacho H., Sanchez-Perez A., Galicia-Paredes A., Meza-Chavez A., et al. (2014). Identification of OXA-23 carbapenemases: novel variant OXA-239 in Acinetobacter baumannii ST758 clinical isolates in Mexico. New Microbes New Infect. 2, 173–174. 10.1002/nmi2.60, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. 10.1128/JCM.33.9.2233-2239.1995, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaschek F., Higgins P. G., Stefanik D., Wisplinghoff H., Seifert H. (2016). Head-to-head comparison of two multi-locus sequence typing (MLST) schemes for characterization of Acinetobacter baumannii outbreak and sporadic isolates. PLoS One 11:e0153014. 10.1371/journal.pone.0153014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton J. F., Kaufmann M. E., Gill M. J., Pike R., Scott P. T., Fishbain J., et al. (2006). Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J. Clin. Microbiol. 44, 2630–2634. 10.1128/JCM.00547-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwingabiye J., Lemnouer A., Baidoo S., Frikh M., Kasouati J., Maleb A., et al. (2017). Intensive care unit-acquired Acinetobacter baumannii infections in a Moroccan teaching hospital: epidemiology, risk factors and outcome. Germs 7, 193–205. 10.18683/germs.2017.1126, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W., Willems R. J. (2010). Genome-based insights into the evolution of enterococci. Clin. Microbiol. Infect. 16, 527–532. 10.1111/j.1469-0691.2010.03201.x, PMID: [DOI] [PubMed] [Google Scholar]
- Vazquez C., Kollef M. H. (2018). Acinetobacter pneumonia: improving outcomes with early identification and appropriate therapy. Clin. Infect. Dis. 67, 1455–1462. 10.1093/cid/ciy375, PMID: [DOI] [PubMed] [Google Scholar]
- Villalón P., Ortega M., Sáez-Nieto J. A., Carrasco G., Medina-Pascual M. J., Garrido N., et al. (2019). Dynamics of a sporadic nosocomial Acinetobacter calcoaceticus – Acinetobacter baumannii complex population. Front. Microbiol. 10:593. 10.3389/fmicb.2019.00593, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. L., Rello J., Marshall J., Silva E., Anzueto A. (2009). International study of the prevalence and outcomes of infection in intensive care units. JAMA 302, 2323–2329. 10.1001/jama.2009.1754, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X., Qiao F., Yu R., Gao Y., Zong Z. (2013). Clonal diversity of Acinetobacter baumannii clinical isolates revealed by a snapshot study. BMC Microbiol. 13:234. 10.1186/1471-2180-13-234, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Paulus T., Lugenheim M., Stefanik D., Higgins P. G., Edmond M. B., et al. (2012). Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Inf. Secur. 64, 282–290. 10.1016/j.jinf.2011.12.008, PMID: [DOI] [PubMed] [Google Scholar]
- Wong D., Nielsen T. B., Bonomo R. A., Pantapalangkoor P., Luna B., Spellberg B. (2017). Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30, 409–447. 10.1128/CMR.00058-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R., Zhang X. D., Zhao Q., Peng B., Zheng J. (2018). Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg. Microbes Infect. 7:31. 10.1038/s41426-018-0038-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Endo K., Sawase K., Anetai M., Narita K., Hatakeyama Y., et al. (2016). Rapid species identification and epidemiological analysis of carbapenem-resistant Acinetobacter spp. by a PCR-based open reading frame typing method. J. Med. Microbiol. 65, 923–927. 10.1099/jmm.0.000314, PMID: [DOI] [PubMed] [Google Scholar]
- Yang J., Chen Y., Jia X., Luo Y., Song Q., Zhao W., et al. (2012). Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 18, E506–E513. 10.1111/1469-0691.12035, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.