Abstract
Retinol-binding protein 2 (RBP2; originally cellular retinol-binding protein, type II (CRBPII)) is a 16 kDa cytosolic protein that in the adult is localized predominantly to absorptive cells of the proximal small intestine. It is well established that RBP2 plays a central role in facilitating uptake of dietary retinoid, retinoid metabolism in enterocytes, and retinoid actions locally within the intestine. Studies of mice lacking Rbp2 establish that Rbp2 is not required in times of dietary retinoid-sufficiency. However, in times of dietary retinoid-insufficiency, the complete lack of Rbp2 gives rise to perinatal lethality owing to RBP2 absence in both placental (maternal) and neonatal tissues. Moreover, when maintained on a high-fat diet, Rbp2-knockout mice develop obesity, glucose intolerance and a fatty liver. Unexpectedly, recent investigations have demonstrated that RBP2 binds long-chain 2-monoacylglycerols (2-MAGs), including the canonical endocannabinoid 2-arachidonoylglycerol, with very high affinity, equivalent to that of retinol binding. Crystallographic studies establish that 2-MAGs bind to a site within RBP2 that fully overlaps with the retinol binding site. When challenged orally with fat, mucosal levels of 2-MAGs in Rbp2 null mice are significantly greater than those of matched controls establishing that RBP2 is a physiologically relevant MAG-binding protein. The rise in MAG levels is accompanied by elevations in circulating levels of the hormone glucose-dependent insulinotropic polypeptide (GIP). It is not understood how retinoid and/or MAG binding to RBP2 affects the functions of this protein, nor is it presently understood how these contribute to the metabolic and hormonal phenotypes observed for Rbp2-deficient mice.
Keywords: Vitamin A, retinoid, monoacylglycerol, obesity, intestine, enteroendocrine signaling, endocannabinoid, glucose-dependent insulinotropic polypeptide (GIP) and dietary fat
Introduction
Retinol-binding protein 2 (RBP2) was identified by Ong in the mid-1980s as a low molecular weight (~16 kDa) cytosolic protein present in the small intestine that binds tightly all-trans-retinol (Ong 1984). Ong originally named this protein cellular retinol-binding protein, type 2, (CRBPII) to distinguish it from another cytosolic retinol-binding protein that had been identified several years earlier and named simply cellular retinol-binding protein (CRBP) (Bashor et al. 1973; Ong and Chytil 1978; Ross et al. 1979; Takase et al. 1979). With the advent of newer genetic nomenclature, CRBPII became RBP2, and CRBP became RBP1. Although the earlier literature uses CRBPII and CRBP to identify these proteins, throughout this review we will employ solely the now accepted genetic nomenclature of RBP2 and RBP1. We note that at least eight other proteins are well established in the literature to bind specifically retinoids (retinol, retinaldehyde (retinal) or retinoic acid) facilitating their transport, metabolism and/or actions. The identities of these others are summarized in Table 1. Our intent is to provide readers with some insight into these other retinoid-binding proteins, allowing them to be distinguished from RBP2.
Table 1.
| Original nomenclature | Genetic nomenclature | Ligands | Tissue distribution |
|---|---|---|---|
| Retinol-binding protein (RBP) | RBP4 | Retinol | Serum/plasma |
| Cellular retinol-binding protein I (CRBPI) | RBP1 | Retinol and retinaldehyde | Most tissues |
| Cellular retinol-binding protein II (CRBPII) | RBP2 | Retinol and retinaldehyde | Small intestine |
| Cellular retinol-binding protein IV (CRBPIV) | RBP5 | Retinol and retinaldehyde | Liver, kidney, spleen |
| Cellular retinol-binding protein III (CRBPIII) | RBP7 | Retinol& retinaldehyde | Adipose and muscle |
| Interphotoreceptor retinol-binding protein (IRBP) | RBP3 | Retinol and retinaldehyde and other lipids | Eye, retina retinal pigmented epithelium (RPE) |
| Cellular retinaldehyde-Binding protein (CRALBP) | RLBP1 | 11-cis-retinol and 11-cis-retinaldehyde | Retina, RPE |
| Cellular retinoic acid-binding protein I (CRABPI) | CRABP1 | Retinoic acid | Many tissues |
| Cellular retinoic acid-binding protein II (CRABPII) | CRABP2 | Retinoic acid | Skin, adipose, others |
| Fatty acid-binding protein 5 (FABP5) | FABP5 | Fatty acids and retinoic acid | Adipose, liver, others |
A listing of retinoid-binding proteins that are canonically accepted to be importantly involved in facilitating retinoid metabolism and/or actions.
RBP1, RBP2, RBP5, RBP7, CRABP1, CRABP2, and FABP5 are all fatty acid-binding proteins. These along with RBP4 are member of the lipocalin superfamily.
Until very recently, the sole known action of RBP2 in the adult was to bind retinol or retinaldehyde facilitating retinoid (vitamin A and its metabolites) uptake into the gut, retinoid metabolism within the intestinal epithelium, and retinoid actions locally within the intestine (Blaner and Li 2015). How RBP2 acts in these processes is depicted in Figure 1. The preponderance of the published literature is focused on these aspects of RBP2 biology. However, recently published findings establish that RBP2 also binds tightly with selected monoacylglycerols (MAGs) (Lee et al. 2020), affecting intestinal levels and actions of these neutral lipids. These new data establish that RBP2 is also a physiologically relevant intestinal binding protein for MAGs and acts more broadly in dietary fat metabolism.
Figure 1.
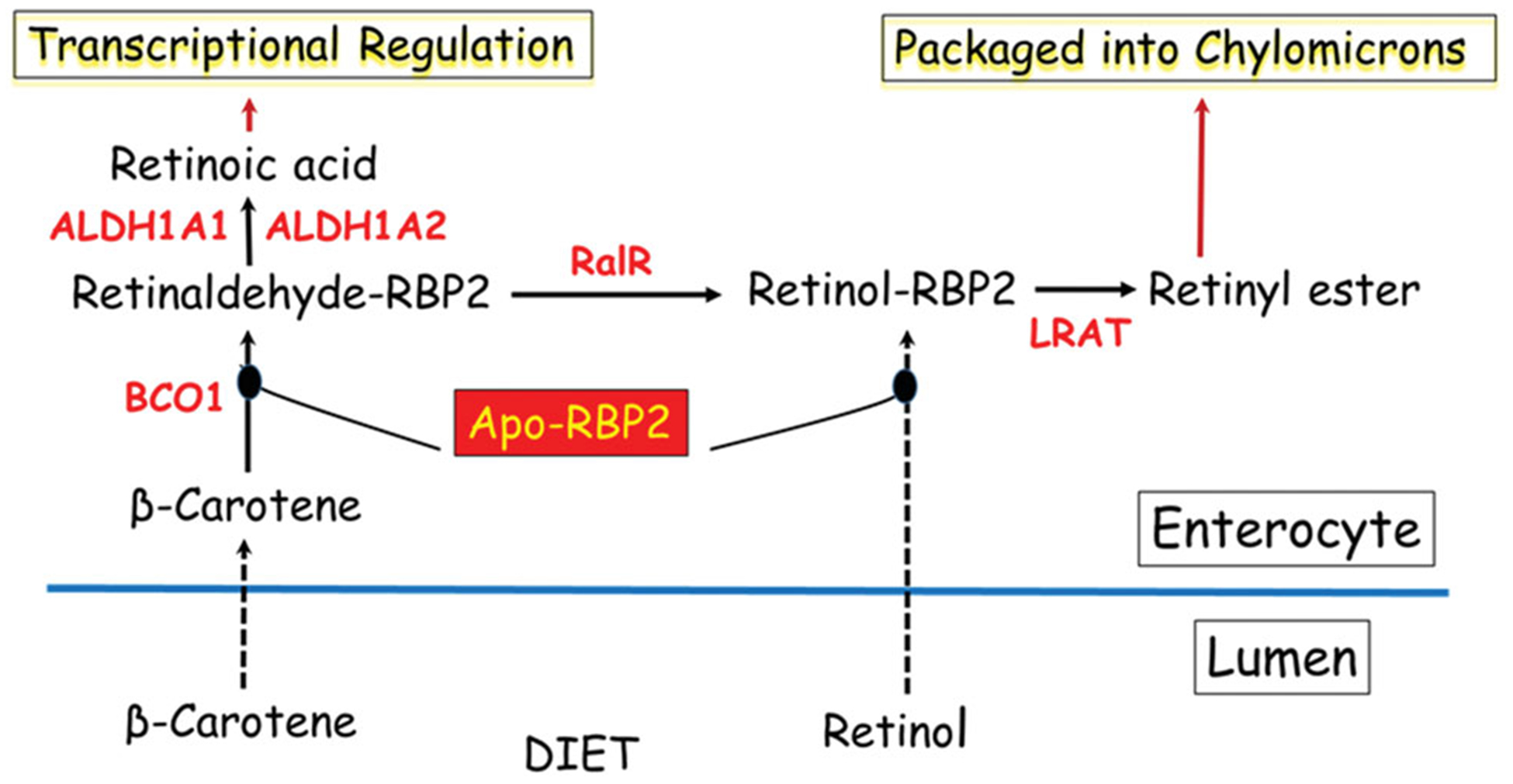
Longstanding understanding of the role of RBP2 in retinoid uptake, metabolism and actions within the absorptive cells (enterocytes) of the small intestine. These arise due to the ability of RBP2 to bind retinol or retinaldehyde with high affinity. This allows apo-RBP2 to bind retinol as it is newly absorbed by enterocytes from the intestinal lumen after arriving there as a component of the diet. Alternatively, apo-RBP2 can bind retinaldehyde formed from newly absorbed β-carotene by β-carotene 15,15′-dioxygenase (BCO1) within the enterocyte. Retinaldehyde bound to RBP2 is a substrate for the intestinal retinaldehyde reductase (RalR) allowing for retinaldehyde conversion to retinol, while bound to RBP2. Retinol bound to RBP2 is a substrate for lecithin:retinol acyltransferase (LRAT) producing retinyl esters that are packaged into nascent chylomicrons for uptake into the body. Thus, retinol or retinaldehyde obtained from the diet is metabolically channeled by RBP2 to the chylomicrons for uptake along with other dietary fat into the circulation. Some retinaldehyde bound to RBP2 is converted to retinoic acid by the actions of retinaldehyde dehydrogenases 1 and 2 (ALDH1A1 and ALDH1A2). The retinoic acid can be used locally to regulate retinoic acid-dependent transcription.
Identification of RBP2
RBP2 was identified by Ong (1984) in a survey for previously unidentified cytosolic proteins able to bind retinol, retinaldehyde and/or retinoic acid. At that time, a number of intra- and extracellular proteins that bound one or more of these retinoids had been identified. Consequently, it was reasonable to hypothesize that other important retinoid-binding proteins remained to be discovered. To this end, Ong (1984) prepared cytosols from 1-day-old rat pups, added a moderate amount of retinol (1 μmol) to the cytosol preparations, and using classical column chromatography protocols following the fluorescence of bound retinol, was able to purify RBP2 to homogeneity. The properties of RBP2 were reported to be similar to, but yet distinct from those of the other known cytosolic retinol-binding protein, RBP1. RBP2 has a similar molecular weight of about 16,000 Da and like RBP1 binds all-trans-retinol in a one-to-one molar ratio. Retinol bound to the newly purified protein exhibited considerably altered absorbance and fluorescence excitation spectra compared to free retinol in organic solvents suggesting that the retinol was bound within a hydrophobic internal pocket in RBP2.
Rabbit antiserum was raised against the homogeneously purified rat RBP2 (Ong 1984). This allowed for the development of a radioimmunoassay able to measure quantitatively RBP2 levels and for the immunohistochemical localization of RBP2 within tissues (Ong 1984). RBP2 was found by radioimmunoassay to be expressed in a number of 1-day-old neonatal tissues, but liver and intestine had levels that were 100-fold higher than any other neonatal tissues examined. The small intestine of the adult rat possessed levels of RBP2 that were 500-fold greater than any other tissue examined, with decreasing RBP2 concentrations observed from jejunum to colon (Ong 1984). The amount of RBP2 present in the rat intestine was reported to be 0.04% of the intestinal wet weight (Ong 1984) and to account for approximately 1% of cytosolic protein mass in the adult jejunum (Ong 1985). Immunohistochemical localization studies established that RBP2 is expressed in absorptive cells, and more highly near the tips of the villi than at the base of the villi (Crow and Ong 1985). Proliferating cells in the crypts of Liberkühn stained only lightly for RBP2 and goblet cells showed no RBP2 staining. The cellular localization of RBP1 in the small intestine was also reported (Crow and Ong 1985). Unlike for RBP2, the intestinal epithelial cells showed no staining for RBP1. RBP1 staining was restricted to connective tissue cells in the lamina propria and to cells within gut-associated lymphoid tissue. Based on these localization studies, it was proposed that RBP2, but not RBP1, plays a central role in the absorption of dietary retinoid (Crow and Ong 1985).
Interestingly, during the purification of RBP2, chromatography on an anion exchange column allowed for the resolution of two “essentially identical” forms of RBP2 (Ong 1984). It was noted at the time that the only difference observed between the two forms was their degree of saturation with retinol. A subsequent detailed structural study aimed at identifying possible differences between the two forms of RBP2, employing RBP2 purified from the adult rat proximal small intestine, established that the primary sequences for the two RBP2 forms were identical, but one was found to be acetylated at its NH2 terminus (Schaefer et al. 1989). The binding affinities of all-trans-retinol for the two RBP2 species were not reported. It is tempting to speculate now that these observations presage the findings discussed below that RBP2 is also able to bind with high affinity MAGs (Lee et al. 2020).
Human RBP2 was purified from postmortem whole small intestine and partially characterized (Inagami and Ong 1992). The reported characteristics of human RBP2 were very similar to those of purified rat RBP2. Like rat RBP2, two forms of human RBP2 were purified and these differed only in that one form had a blocked N-terminus. Sequence analysis of the first 36 residues in the human RBP2 N-terminal found that only a single residue differed with that of rat RBP2 and this difference did not affect the charge of the protein. Purified human RBP2 bound all-trans-retinol and all-trans-retinaldehyde but not all-trans-retinoic acid. Purified human RBP1 only bound all-trans-retinol. Thus, the ligand binding properties of purified rat RBP2 and human RBP2 were identical. Immunologic analyses identified human RBP2 as being most highly expressed in the proximal jejunum (Inagami and Ong 1992). Like rat RBP2, human RBP2 also was found by radioimmunoassay to be present at high concentrations in individual human intestinal biopsies, ranging up to 1% of total mucosal protein (Inagami and Ong 1992).
The primary structure of rat RBP2 was deduced from a cloned cDNA (Li et al. 1986). RBP2 was found to be a 15,580 Da protein containing 134 amino acids. Seventy-five of 134 residues (56%) are identical to those present in rat RBP1. Later, in the mouse (Vogel et al. 2001) and in the human (Folli et al. 2002), another cytosolic retinol-binding protein, RBP7 or cellular retinol-binding protein, type III, was identified. It was determined that mouse RBP2 shares 56% sequence identity with mouse RBP7 (Vogel et al. 2001). A similar degree of sequence identity was also reported for a fourth cellular retinol-binding protein, cellular retinol-binding protein, type IV (RBP5) upon its identification in the human genome (Folli et al. 2001). Interestingly, unlike RBP1, RBP2 and RBP7, RBP5 is not expressed in the mouse. The sequence alignments for the four intracellular retinol-binding proteins (RBPs) are provided in Figure 2(C).
Figure 2.
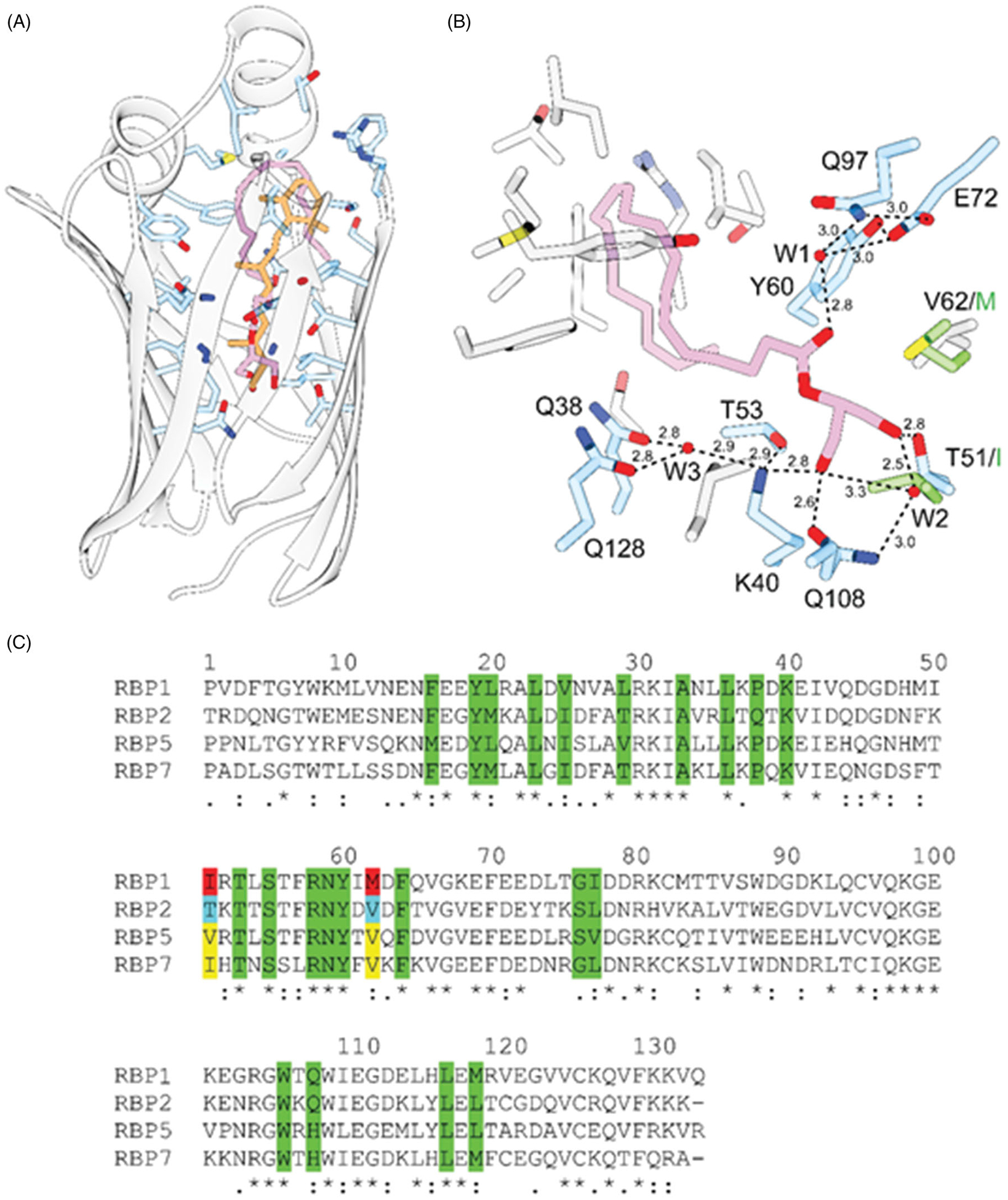
Interaction of 2-AG with RBP2. A, the overall orientation of 2-AG (purple) within the binding-pocket of human RBP2 (PDB #6BTH) in comparison to all-trans-retinol (orange) (PDB #4QZT). B, hydrogen bond network that contribute to the high affinity of 2-AG. Hydrogen bonds are indicated by dashed lines. Distances are shown in Å. Ordered water molecules (W) are depicted as red sphered. C, sequence alignment of human RBPs. Amino acid that line the inside of the binding cavities are marked in green. Residues in position 51 and 62 that prevent binding of monoacylglycerols to RBP1 are labeled in red. Yellow background highlights residues found in the sequences of RBP5 and RBP7. They may indicate an alternative ligand specificity to RBP1 and RBP2.
Based on sequence homologies RBP2 was identified as being a member of the fatty acid-binding protein (FABP) family (Li et al. 1986). The FABP family comprises 9 known 14–15 kDa intracellular proteins that bind unesterified fatty acids with high affinity (Storch and Corsico 2008; Gajda and Storch 2015; Hotamisligil and Bernlohr 2015). In addition, the FABP family now includes the four intracellular RBPs (RBP1, RBP2, RBP5, and RBP7) and two intracellular retinoic acid-binding proteins (CRABP1 and CRABP2) (Ong et al. 1994; Storch and Corsico 2008; Gajda and Storch 2015; Hotamisligil and Bernlohr 2015). Until recently, none of the retinoid-binding proteins had been shown to bind with high affinity ligands other than retinol, retinaldehyde, or retinoic acid (see Ligand binding section). However, a number of FABPs are reported to bind strongly to 2-MAGs or N-acylethanolamides (NEAs) (Newberry et al. 2012; Lagakos et al. 2013; Thumser et al. 2014; Gajda and Storch 2015), and this in turn influences the levels of these compounds in cells and tissues.
The early work on RBP2 has been extensively summarized in several excellent prior reviewers. The reader is referred to these for more insight into the early work on RBP2 (Ong et al. 1994; Li and Norris 1996; Li and Tso 2003).
RBP2 expression
In the adult
There is now consensus that in the adult RBP2 is expressed at high levels only in the small intestine (Ong 1984; Li et al. 1986). There is some early evidence that Rbp2 mRNA may be expressed at relatively low levels in a few other adult tissues, including adrenals, testes and brain (Li et al. 1986), but this has not been confirmed by subsequent studies.
Ong’s first description of RBP2 reported that the protein localizes in the adult rat to the small intestine with only trace immunoreactivity for the protein being detected in the adult colon, liver and whole eye (Ong 1984). No immunoreactive RBP2 could be detected in kidney, muscle, lung, heart, brain, spleen, adrenal, uterus, testis or skin (Ong 1984). RBP1 was reported to be expressed at low levels in the rat small intestine and at greater levels in the liver, kidney, lung, brain, spleen, eye, uterus, and testis (Ong 1984). When Li et al. first cloned the cDNA for rat Rbp2, Rbp2 mRNA was found predominantly in the adult rat small intestine (Li et al. 1986). It was reported to be ~2-fold more abundant in the proximal compared to the distal half of the small intestine. Rbp2 mRNA was first detectable in intestinal total RNA during the 19th day of gestation, a time that corresponds to the appearance of an absorptive columnar epithelium. No Rbp2 mRNA was detected in total RNA obtained from the adult colon, liver, kidney, spleen, heart, or lung. Dot blot analyses of adrenal, testes, and brain RNA gave signals that were 2.3%, 0.8%, and 1.5% of the signal produced by small intestine total RNA (Li et al. 1986). Unlike Rbp2 expression, Rbp1 mRNA was found to be abundant in adult liver, kidney, testes, lung, adrenal, spleen, colon, brain and heart (Levin et al. 1987).
Cryostat sectioning of jejunal segments from 6-week-old rats revealed that Rbp2 mRNA is expressed maximally in the lower-villus, and RBP2 protein maximally in the mid-villus. Feeding of a high-fat diet (70% calories from fat) for 48 h after previous feeding for 7 days of a low-fat diet (7% of calories from fat) resulted in a pronounced increase within 12 h for Rbp2 mRNA from the lower- to mid-villus (Ogura et al. 2008). This was taken to indicate that Rbp2 mRNA is maximally expressed in the lower-villus, and that dietary fat causes an enhancement of Rbp2 gene expression in villus cells. RBP2 protein had earlier been localized to absorptive cells located in portions of the villus closer to its tip (Crow and Ong 1985). Presumably, Rbp2 mRNA is expressed first in lower, newly formed enterocytes closer to the crypt, giving rise to cells that express RBP2 protein that then migrate up the villus to its tip.
Recent evidence suggests that rat Rbp2 mRNA may be expressed in the liver during pregnancy and that diet-induced iron-deficiency during pregnancy further increases Rbp2 expression (Cottin et al. 2016). However, neither the presence of RBP2 protein in the livers of pregnant dams nor the time course of Rbp2 mRNA expression were reported. The increase in hepatic Rbp2 expression upon pregnancy was associated with a decrease in mRNA expression of other retinoid metabolic genes and decreased retinoid levels. This was taken to suggest that Rbp2 expression is unlikely to favor retinoid storage in the liver during pregnancy (Cottin et al. 2016). This interesting finding requires confirmation and further investigation.
Following the identification of mammalian RBP2, Finlay et al. (1990) reported the purification and characterization of an 18 kDa chick duodenal protein that displays a high degree of sequence homology with rat RBP2. This protein bound retinol and displayed a similar anatomic distribution within the chick intestine to that of rat RBP2. Thus, RBP2 appears to be expressed across vertebrate species. Interestingly, 1,25-dihydroxyvitamin D3, provided at concentrations that stimulate calcium uptake into embryonic chick duodenal organ cultures, was found to decrease the rate of synthesis of chick RBP2. A similar effect for 1,25-dihydroxyvitamin D3 on rodent or human RBP2 expression has not been reported.
Expression during the perinatal period
Rbp2 mRNA and protein are more widely expressed in tissues during the perinatal period (Ong 1984; Levin et al. 1987). Ong originally identified that the highest levels of immunoreactive RBP2 protein in the 1-day-old rat were present in the small intestine and liver (Ong 1984). RBP2 protein was also detected, albeit at concentrations that were one to two orders of magnitude lower, in kidney, muscle, lung, heart, brain, and spleen of 1-day-old rats (Ong 1984).
Rbp2 mRNA is reported to be expressed highly in the mouse embryonic liver and intestine and at lower levels in lung, uterus, vagina and placental tissue (Ong 1984; Sapin et al. 1997; Hind et al. 2002; Matsuda et al. 2004). Expression in many of these tissues continues into the postnatal period, gradually declining to undetectable by about 3 weeks of age. Rbp2 is expressed in both the embryonic and the early postnatal lung (Ong 1984; Hind et al. 2002). Immunohistochemical studies have demonstrated that RBP2 protein expression in the mouse lung peaks at postnatal day 4 and progressively declines through postnatal day 15 (Hind et al. 2002). Investigations exploring the expression pattern of Rbp2 in mouse placental tissue ranging from 6.5 to 19.5 days postcoitum (dpc) established that Rbp2 mRNA is expressed in the visceral yolk sac, appearing at 12.5 dpc and gradually increasing to a steady level by 15.5 dpc (Sapin et al. 1997).
Rbp2 is also expressed in the developing mouse uterus and vagina during the neonatal period (Matsuda et al. 2004). Treatment of neonatal female mice with estrogens (estradiol-17β or diethylstilbestrol), for 5 successive days starting on the first day after birth, markedly increased expression of Rbp2 mRNA in the vagina and uterus (Matsuda et al. 2004). Exposure to estrogenic substances during this perinatal period is known to cause irregular development of the female reproductive tract that leads to ovary-independent proliferation and cornification in the vaginal epithelium in mice (Matsuda et al. 2004). Since these authors had earlier shown that simultaneous administration of retinol with estrogens diminished the development of the irregular vaginal epithelium, they proposed that increased expression of Rbp2 in response to perinatal estrogen exposure could be a compensation aimed at assuring retinol availability in the developing tissues and consequently normal development of the uterine and vaginal epithelium (Matsuda et al. 2004).
The patterns of expression of RBP2 and RBP1 protein and activities for two microsomal retinol-metabolizing enzymes lecithin:retinol acyltransferase (LRAT) and retinaldehyde reductase (RalR) were evaluated in the rat small intestine during the perinatal period (Ong et al. 1991). RBP2 protein appears initially immediately prior to birth, rising to its highest level by day 3 after birth declining by 50% during the late suckling period and remaining at this level throughout adult life. Immunostaining for RBP2 indicated that it was expressed evenly in all villus-associated enterocytes. RBP1 was present in the villus layer during gestation but absent in the villus at birth. Intestinal LRAT activity increased rapidly 2 days prior to birth, reaching a peak 6 to 12 days after birth, and slowly declining through postnatal day 28. Intestinal RalR activity was detectable at birth, but declined in the early suckling period only to reappear at postnatal day 21. The overlapping temporal expression patterns for RBP2, RBP1, LRAT, and RalR in the perinatal rat small intestine led Ong et al. (1991) to suggest that the expression of these proteins prepares the small intestine to process retinoid that will be obtained from the diet after birth.
Rbp2 mRNA levels significantly increase in the rat small intestine starting at birth and during the postnatal period (Ogura et al. 2005). The temporal pattern for the increased Rbp2 mRNA expression is paralleled by mRNA levels for hepatocyte nuclear factor-4 (HNF-4), one of the transcription factors proposed to regulate Rbp2 expression (see Gene structure of RBP2 and its regulation section) (Ogura et al. 2005). Expression levels of two retinaldehyde dehydrogenases (Aldh1a1 and Aldh1a2), enzymes needed for the synthesis of retinoic acid are very high at birth (Bhat 1998; Ogura et al. 2005). Similarly, the human gene for HNF-4α possesses a retinoic acid response element that is activated by 9-cis-retinoic acid via the action of retinoid X receptors (RXRs) (Qian et al. 2000). This led to the proposal that perinatal retinoic acid signaling, through its effects on HNF-4α levels, may be a pivotal factor leading to enhanced Rbp2 expression in the perinatal period (Ogura et al. 2005).
Ligand binding
Retinoid binding
In Ong’s original identification of rat RBP2, he found that retinol remained bound to RBP2 throughout its purification to homogeneity (Ong 1984). This was taken to indicate that retinol is the endogenous ligand for RBP2. Four subsequent published reports examined the binding properties of different retinoids toward purified apo-RBP2 and apo-RBP1 (MacDonald and Ong 1987; Levin et al. 1988; Li et al. 1991; Kane et al. 2011). The findings of these independent investigations are in agreement that all-trans-retinol binds with high affinity to both RBP2 and RBP1 and that all-trans-retinoic acid is unable to bind either protein. RBP2 purified from 1-day-old rat pups and RBP1 purified from rat liver were both found to bind all-trans-retinol with high affinity (Kd < 50 nM) (MacDonald and Ong 1987). 13-cis-Retinol, 3-dehydroretinol and all-trans-retinaldehyde were reported also to bind apo-RBP2 with high affinity, based on their ability to displace all-trans-retinol from holo-RBP2 (MacDonald and Ong 1987). Apo-RBP1 bound both 13-cis-retinol and 3-dehydroretinol, but all-trans-retinaldehyde did not displace all-trans-retinol from RBP1 (MacDonald and Ong 1987). It was reported further that neither protein bound all-trans-retinoic acid nor 9-cis- or 11-cis-retinol (MacDonald and Ong 1987). Similar studies, but employing recombinant rat RBP2 and RBP1 expressed in Escherichia coli and assessing ligand binding by monitoring changes in endogenous tryptophan fluorescence (both RBP2 and RBP1 contain 4 tryptophan residues), found that all-trans-retinol binds each protein with a Kd value in the 50 to 100 nM range (Levin et al. 1988). All-trans-retinaldehyde was reported to bind recombinant RBP2, but not RBP1. Neither recombinant protein was able to bind all-trans-retinoic acid.
Another methodology used to investigate retinoid binding affinities to apo- and holo-RBP2 and to apo- and holo-RBP1, involved the use of recombinant rat RBP2 and RBP1 expressed in E. coli but labeled with 6-flurotrypthophan to allow for the use in 19F NMR spectroscopy to assess ligand binding (Li et al. 1991). These investigations led to the conclusion that all-trans-retinol binds with a slightly lesser affinity to RBP2 than to RBP1, but that all-trans-retinaldehyde bound with approximately equal affinities to RBP2 and RBP1. This later finding does not agree with earlier data (MacDonald and Ong 1987; Levin et al. 1987) and contradicts the earlier proposals that the ability of RBP2 but not RBP1 to bind all-trans-retinaldehyde is physiologically important, since dietary carotenoids like β-carotene are metabolized primarily within the small intestine by the enzyme β-carotene 15,15′-dioxygenase (BCO1) to all-trans-retinaldehyde that can then be converted locally into other retinoid species (Figure 1).
Later studies explored the binding affinities of recombinant RBP2 and RBP1 for 9-cis-retinoids (Kane et al. 2011). RBP2 and RBP1 both were found to bind 9-cis-retinol with high affinities (Kd values of 68 nM and 11 nM, respectively). This finding disagrees with the earlier report regarding 9-cis-retinol binding to purified 1-day-old rat pup RBP2 where no binding to RBP2 was observed (MacDonald and Ong 1987). 9-cis-Retinaldehyde also was reported to bind both RBP2 and RBP1 with similar high affinities, but 9-cis-retinoic acid was unable to bind either purified protein (Kane et al. 2011).
As can be surmised above, there are discrepancies in the Kd values reported by the different research groups for the retinoids that were studied. Some of this lack of concurrence is undoubtedly due to the different sources of the RBP2 (purified from tissues versus expressed recombinantly) that were studied, as well as differences in the protocols (tryptophan fluorescence vs. NMR spectroscopy) used for assessing retinoid binding. Never-the-less, it is clear for all of the reported values that retinol and retinaldehyde bind RBP2 very tightly.
Binding of nonretinoid ligands
None of the published studies exploring retinoid binding to RBP2 investigated whether purified RBP2 is able to bind nonretinoid ligands (MacDonald and Ong 1987; Levin et al. 1988; Li et al. 1991; Kane et al. 2011). To address this possibility, Lee et al. investigated whether RBP2 can bind with high affinity other physiologically relevant ligands (Lee et al. 2020). To avoid potential experimental bias associated with studying the binding of hydrophobic ligands to a protein specialized for interaction with small hydrophobic molecules, we developed a retinol-competition assay that measured the ability of a tested compound to outcompete retinol from the binding site (Silvaroli et al. 2019). The principle of this assay is based on the fact that protein fluorescence of RBP2 in complex with retinol is quenched due to the fluorescence resonance energy transfer between the tryptophan residues and the retinoid moiety. The assay was adopted for a high-throughput screening format to search for any RBP2 binding partners by screening commercially available compound libraries comprising 986 bio-lipids. After elimination of compounds with spectral properties interfering with the fluorescence assay, the screen revealed several unique hits that clustered on three groups of lipids: 2-monoacylglycerols (2-MAGs), 1-monoacylglycerols (1-MAGs), and N-acylethanolamides (NAEs).
To precisely determine the affinity of RBP2 toward MAGs and other neutral lipids like the NAEs, we performed a dose-response assay. Changes in the fluorescence intensity of the protein or the retinoid moiety plotted as a function of lipid concentration were best fitted with one-site saturation binding model. The values of the apparent dissociation constant (Kd) revealed affinity at low nanomolar ranges for 2-arachidonoylglycerol (2-AG) and 1-arachidonoylglycerol (1-AG) (27.1 ± 2.5 and 31.0 ± 2.7 nM, respectively). Somewhat higher Kd values were determined for 2-lineoylglycerol (2-LG) and 2-oleoylglycerol (2-OG) (40.0 ± 4.9 and 65.4 ± 4.4 nM, respectively), whereas affinity for N-acrachidonoylethanolamine (AEA) was over 20-fold lower as compared to 2-MAGs (Lee et al. 2020). Intestinal MAG concentrations following a conventional meal will be several orders of magnitude greater than that of retinol or of retinaldehyde. Since RBP2 binds 2-AG, 2-OG, 2-LG, and 1-AG with high affinity, with approximately the same order of magnitude as that of retinol binding and, moreover the absence of RBP2 in the intestine increases enterocyte 2-MAG concentrations following a fat challenge (see Physiological actions of RBP2 in health and disease section), RBP2 must act physiologically within the intestinal mucosa as a MAG-binding protein.
The finding that RBP2 binds tightly MAGs in addition to retinol and retinaldehyde is both surprising and puzzling. This is because another member of the FABP family that binds 2-MAGs, liver-fatty acid-binding protein (FABP1), co-localizes with RBP2 in the absorptive cells of the jejunum (Newberry et al. 2012; Lagakos et al. 2013; Thumser et al. 2014; Gajda and Storch 2015). Moreover, still another FABP member, intestinal-fatty acid-binding protein (FABP2) also colocalizes to the jejunum and like FABP1 binds tightly unesterified fatty acids (Newberry et al. 2012; Lagakos et al. 2013; Thumser et al. 2014; Gajda and Storch 2015). Both FABP1 and FABP2 are expressed abundantly in jejunum, as is RBP2. This raises important questions regarding to what extent the actions of RBP2, FABP1, and FABP2 in the intestine are redundant and/or complementary.
Protein structure
RBP2 and the three other closely related intracellular RBPs (RBP1, RBP5, and RBP7) as well as the other FABP family members belong to a ubiquitous and functionally diverse superfamily of proteins collectively called lipocalins. In vertebrates, this protein superfamily is comprised of several retinoid-binding proteins that, in addition to the intercellular RBPs, include serum retinoid-binding protein (RBP4) (Lakshmi et al. 2015; Schiefner and Skerra 2015). These relatively small (130–180 amino acids) monomeric proteins share a common compact architecture composed of 8 to 10 antiparallel β-strands, which wined around to form a well-defined β-barrel that dominates the structure (Figure 3(A,B)) (Flower et al. 2000; Newcomer and Ong 2000; Skerra 2000). This shields the central hydrophobic cavity that constitutes a versatile lipid binding site. At the N-terminus, this binding packet is enclosed and protected from the solvent by short loops and densely packed hydrophobic side chains. The opposite side of the β-barrel is composed of four flexible loops, each connecting one pair of β-strands that together surround the entry way for the ligand and form a so-called ‘portal site’ (Schiefner and Skerra 2015). This basic motif constitutes a versatile structural platform that can be easily modified for the purpose of binding specific lipid molecules. Thus, the ligand binding specificity of individual lipocalins solely depends on the number of β-strands (size of the β-barrel), amino acid composition lining the inside of the binding cavity, and length of the flexible loops at the entry site.
Figure 3.
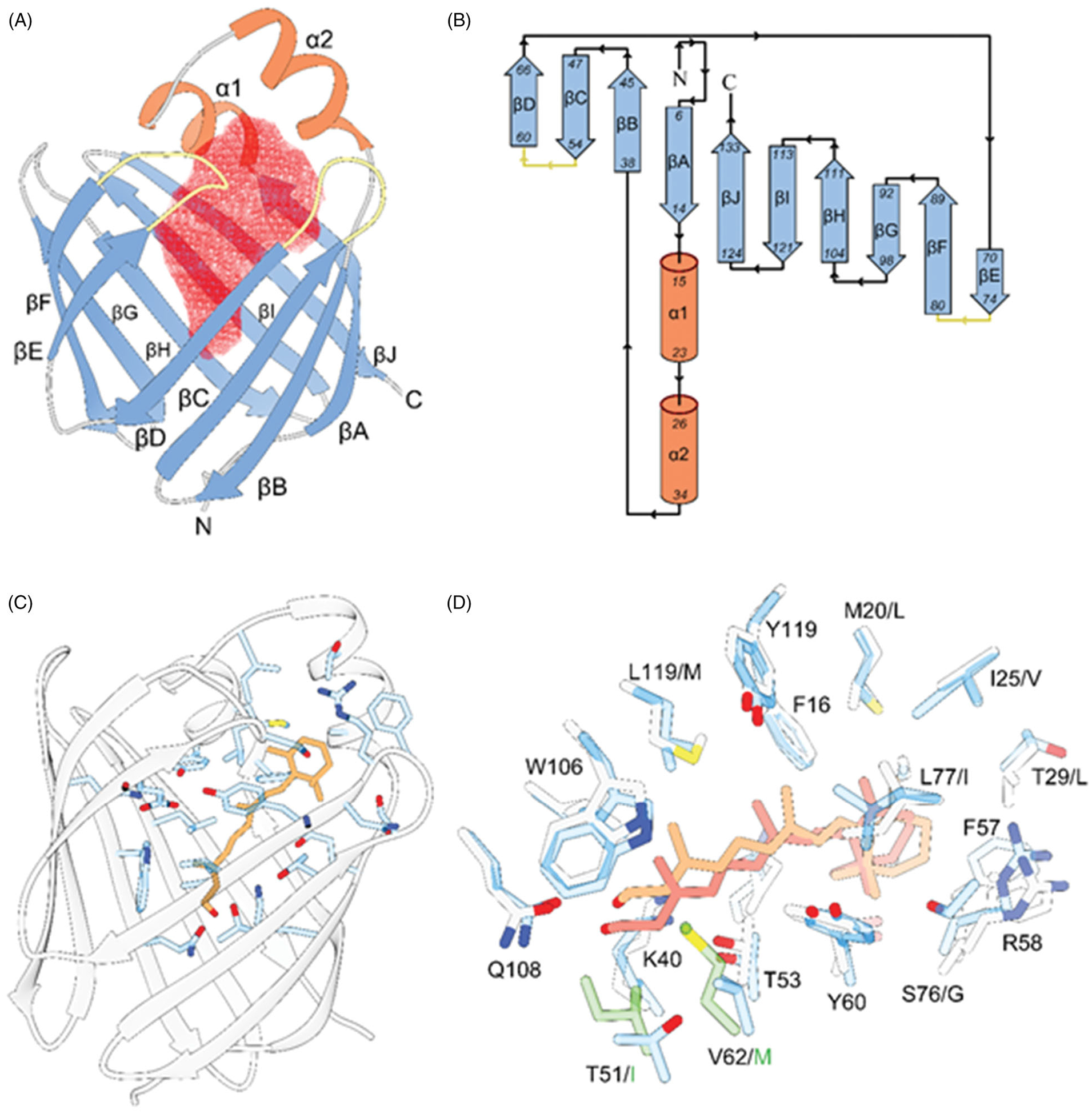
The overall structural motive and the mode of all-trans-retinol interaction with RBPs. A, ribbon diagram of human RBP2 (PDB #6BTH). The red mesh represents the ligand-binding cavity inside the β-barrel composed of ten antiparallel β-strands. The entry portal region is defined by two α-helices (shown in orange) and loops connecting and β-strands C-D and E-F (colored yellow). B, topology diagrams for the RBPs. The color scheme is identical as in panel A. C, position of the retinoid moiety within the binding pocket of RBP2 (PDB #4QZT). The side chains of amino acids in the vicinity of all-trans-retinol (orange) are highlighted in blue. D, overlay of the binding sites structures and ligand orientation found in human RBP1 (PDB #5HBS) and human RBP2 (PDB #4QZT). The side chains of RBP1 and RBP2 are colored gray and blue, respectively. The position of all-trans-retinol seen in RBP1 is shown in light orange, whereas the retinoid moiety found in RBP2 is colored in pink. The side chains shown in green correspond to the substitutions of T51/I and V62/M in RBP1 that contribute to the different in the position of the ligand.
Structural determinants of the interaction of RBP2 with retinol
Specifically, the structural fold of the RBPs consists of ten β-strands organized in five-stranded β-sheets. Two short α-helices that are part of extended loop connecting the βA and βB strands together with hairpin turns between βC-βD and βE-βF define the portal region (Figure 3(B,C)). Although the structures of all four cytosolic RBPs present in the human genome have been solved, only RBP2 and RBP1 were crystalized in complex with retinoids (Zanotti et al. 1993; Tarter et al. 2008; Wang et al. 2012; Nossoni et al. 2014; Silvaroli et al. 2016; Menozzi et al. 2017, 2018). This echoes significant differences in dissociation constants (Kd) for all-trans-retinol observed between these closely related proteins (MacDonald and Ong 1987; Levin et al. 1988; Cheng et al. 1991; Folli et al. 2001, 2002). Upon binding, the hydroxyl group of all-trans-retinol is positioned at the bottom of the binding pocket where it forms hydrogen bonds with the polar side chains of conserved Lys40 and Glu108. In this orientation, the β-ionone ring of the retinoid moiety is located at the entrance to the binding cavity making extensive hydrophobic and van der Waals interaction with the residues of the portal region (Figure 3(C)). Importantly, the contacts of the β-ionone ring with the protein scaffold greatly reduces the conformational flexibility of the entry portal, prompting changes in the orientation of several side chains and locking retinoid inside the binding pocket (Silvaroli et al. 2016). A comparison of the holo structures of RBP2 and RBP1 reveals subtle difference in the position of the retinoid moiety. In RBP2, the retinol molecule is moved toward the bottom of the binding site by about 1.2 Å in comparison to the location of the ligand in RBP1 (Figure 3(D)). This shift in position results from two amino acid substitutions: T51/I and V62/M in RBP1. The much larger and hydrophobic side chains of isoleucine and methionine alter the environment of the polar patch that accommodates the hydroxyl group on the retinol, shifting the position of the molecule upwards. Consequently, retinol is located deeper within the binding pocket of RBP2 as compared to RBP1. Nevertheless, this shift in the relative position of the ligands and variation in the residues that line the interior of the β-barrel does not explain the 3.3- to 100-fold difference in the Kd value for all-trans-retinol reported for these two proteins (Li et al. 1991; Kane et al. 2011). Interestingly, the recent comparison of the conformational flexibilities of the apo- and holo-forms of RBP2 and RBP1 suggest that the difference in binding affinity is mainly determined by the energy required for the transition between the open and closed conformation of the portal region (Estarellas et al. 2019). Stronger interactions between the retinoid moiety and the residues of the portal site in RBP1 disfavor the release of the ligand. Thus, increased residence time in the binding pocket translates into a measurably higher affinity. Surprisingly, seemingly minor changes within the amino acid composition of the portal site can contribute to functionally meaningful changes in the conformational flexibility and the overall binding affinity, as exemplified by the conservative mutation of I78 in RBP1 to Leu78 in RBP2 (Estarellas et al. 2019). Discovery of this fine tuning of RBP properties via changes in specific residues away from the canonical binding site sheds new light on the functional diversity of RBPs as well as the molecular mechanisms responsible for acquisition and release of retinoids by this class of carrier proteins in biological systems.
The mode of RBP2 interactions with endocannabinoids and monoacylglycerols
The recent discovery of the interaction of RBP2 with non-retinoid ligands including the canonical endocannabinoids, 2-AG and AEA, or other selective monoacylglycerols rises an interesting question about the role of this protein in controlling intestinal lipid uptake and signaling (Lee et al. 2020). As evidenced by the crystal structures of human RBP2 in complex with 2AG and AEA (PDB # 6BTH and 6BTI, respectively), these ligands are accommodated in the same binding pocket as retinoids (Lee et al. 2020). Importantly, the affinity of the binding of 2-AG and related monoacylglycerols is comparable to that of all-trans-retinol. Thus, they can effectively outcompete the retinoid ligand from the binding site in biochemical assays (Lee et al. 2020). An analysis of the binding modes of all-trans-retinol and 2-AG revealed surprising similarities despite distinct structural differences between these compounds. Like all-trans-retinol, the hydroxyl terminus of the endocannabinoid interacts with the polar patch at the bottom of the binding site of RBP2 (Figure 2(A)). Noticeably, one of the free hydroxyl groups is placed at the canonical site between Lys40 and Glu108 and forms hydrogen bonds with the ζ-nitrogen and ε-oxygen atoms of these residues, whereas the other hydroxyl interacts with the γ-oxygen of T51. An additional contribution to the binding energy is provided by extended hydrogen bonding between the carbonyl oxygen of the ester group of the ligand, an ordered water molecule, and the side chains of Gln97, Glu72, and Tyr60. The significance of hydrogen bond interactions for overall binding affinity is underscored by the example of AEA. The presence of only one hydroxyl group in this molecule lowers the overall affinity of AEA by ~20-fold as compared to 2-AG. At this moment, it is unclear whether the presence of four double bonds in the Z-configuration characteristic of the arachidonoyl chain plays an important role for the affinity and/or selectivity of the ligand. In the complex with RBP2, the kinked configuration of the acyl chain of 2-AG or AEA mimics to some degree the β-ionone moiety, occupying the same position within the binding site and interacting with the side chains of the portal region (Figure 2(A)). Therefore, one may predict that there is a correlation between conformational flexibility (defined by the number of double bonds), the length of the acyl chain, and overall ligand affinity. However, systematic studies on the structure/function relationship are needed to precisely define the chemical boundaries that determine ligand specificity for RBP2. Such knowledge is essential for decoding the physiological role of this protein in the intestinal transport and metabolism of endogenous and dietary lipids.
Although structurally similar, the RBPs reveal surprising selectivity for their lipid binding partners. This phenomenon is not only reflected by the several fold differences in the Kd values for all-trans-retinol, but also in their ability to interact with nonretinoid compounds (Lee et al. 2020). For example, extensive screening for alternative binding partners for RBP1 revealed no interaction of this protein with monoacylglycerols or any other class of endogenous lipids, although these interactions are seen with RBP2 (Silvaroli et al. 2019). Conversely, RBP1 tightly binds abnormal cannabidiol (2-[(1R,6R)-6-Isopropenyl-3-methylcyclohex-2-en-1-yl]-5-pentylbenzene-1,3-diol), a xenobiotic compound that does not interact with RBP2 (Silvaroli et al. 2019). This ligand selectivity arises for very specific structural reasons. The overlapping of the spatial orientation of 2-AG bound to RBP2 with the structure of RBP1 reveals that the same amino acid substitutions that are responsible for the shift in the position in retinol in these proteins prevent the binding of monoacylglycerols by RBP1 (Figure 2(B)). Placing 2-AG within the binding pocket of RBP1 would imply the thermodynamically unfavorable positioning of one of the polar hydroxyl groups of the glycerol backbone right in-between the hydrophobic side chains of I51 and M62. Additionally, the absence of the threonine residue in position 51 of RBP1 eliminates one of the hydrogen bond interaction sites for the ligand, potentially lowering its binding affinity. Remarkably, the presence of T51 is characteristic only for RBP2. Similar to RBP1, this residue is substituted by hydrophobic valine or isoleucine in RBP5 and RBP7, respectively (Figure 2(C)). Thus, the binding of 2-MAGs seems to be a unique characteristic that distinguishes RBP2 from the other members of the RBP family. This finding supports the hypothesis that each member of the intracellular RBPs may selectively bind a different subset of lipid molecules. Therefore, the determination of physiologically relevant lipid binding partners for RBP2, RBP5, and RBP7 becomes essential for establishing the biological function of these proteins.
Gene structure of RBP2 and its regulation
The RBP2 gene
The first Rbp2 gene structure reported was for the rat (Demmer et al. 1987). Rat Rbp2 was found to consist of four exons spanning 0.65 kilobases that are interrupted by three introns with an aggregate length of 19.5 kilobases (Demmer et al. 1987). The gene encodes a 134-residue RBP2 (Demmer et al. 1987). Southern blot analysis indicated that the Rbp2 gene is highly conserved in rats, mice and humans. The mouse Rbp2 gene mapped to chromosome 9 and is located within 3.0 centimorgans of Rbp1. Based on syntenic homology, it was proposed that the human genes for RBP2 and RBP1 are located on chromosome 3 (Demmer et al. 1987).
Subsequent FISH analysis localized the human RBP2 gene to human chromosome band 3q23, in close proximity to the gene for RBP1 (De Baere et al. 1998). Interestingly, RBP2 maps distally to a chromosomal breakpoint that is associated with blepharophimosis-ptosis-epicanthus inversus (BPES) syndrome, a rare genetic eyelid malformation (De Baere et al. 1999). For three BPES patients harboring the 3q23 breakage, this was associated with congenital diaphragmatic hernia (CDH). Given the known association between retinoic acid signaling and congenital diaphragmatic hernia (Dalmer and Clugston 2019), a later literature review summarizing possible genetic origins of CDH, speculated that deletion of the RBP2 and/or RBP1 gene may provide a molecular basis for explaining the origins of this form of CDH (Goumy et al. 2010). However, a definitive role for RBP2 in either BPES or the associated CDH remains to be established.
Early investigations aimed at identifying elements present in the Rbp2 promoter that are important for regulating Rbp2 expression found that retinoic acid in the presence of retinoid X receptors (RXRs), but not retinoic acid receptors (RARs), markedly up-regulated Rbp2 expression (Mangelsdorf et al. 1991). These studies employed a rat Rbp2 promoter construct, from position −987 to +50, transiently transfected into mouse embryonal teratocarcinoma F9 cells. The promoter region needed to confer retinoic acid and RXR responsiveness was identified as a RXR-specific response element (RXRE) (Mangelsdorf et al. 1991). However, subsequent studies by others found that the RXRE present in the rat Rbp2 gene was not conserved in the mouse gene (Nakshatri and Chambon 1994). Rather, these later investigations identified two other conserved cis-acting elements (termed RE2 and RE3), within the promoters of both rat and mouse Rbp2, that mediate transactivation by RARs and RXRs in transfected Cos-1, CV-1 and HeLa cells. RE3 was determined to be the major promoter element conferring retinoic acid responsiveness. This element was reported also to bind the transcription factors hepatocyte nuclear receptor 4 (HNF-4) and apoA1 regulatory protein A1 (ARP-1) (Nakshatri and Chambon 1994). HNF-4 was found to constitutively activate the mouse Rbp2 promoter; whereas, ARP-1 was found to repress Rbp2 activation mediated by RARs, RXRs and HNF-4. When the rat Rbp2 promoter was transfected into human Caco-2 enterocytes that constitutively express RARα, RARγ, RXRα, and HNF-4, retinoic acid treatment of the cells had no effect on Rbp2 promoter activity. Based on these findings, it was proposed that the Rbp2 promoter is not induced by retinoic acid in tissues that express HNF-4 and ARP-1. Rather, it was suggested that retinoic acid inducibility observed in transfection experiments reflects the promiscuous binding of RARs/RXRs to HNF-4 and ARP-1 response elements present in the Rbp2 promoter (Nakshatri and Chambon 1994).
A number of investigations aimed at identifying regulatory elements present in the human RBP2 gene have been carried out using the human Caco-2 intestinal cell line. Caco-2 cells form polarized monolayers, possess many features of the mature small intestinal mucosa, endogenously express RBP2, and are able to absorb and metabolize retinol (Harrison and Hussain 2001; Harrison 2012). Treatment of Caco-2 cell monolayers with either all-trans- or 9-cis-retinoic acid increases RBP2 mRNA levels 2- to 3-fold, suggesting a role for retinoids in regulating RBP2 expression (Levin and Davis 1997). Retinoic acid induction of RBP2 was found to occur concomitantly with significant upregulations in both RARb and ARP-1 expression levels in treated cells. This led to the suggestion that both RARb and ARP-1 may have roles in regulating RBP2 expression (Levin and Davis 1997).
Later studies established that RBP2 expression is associated with differentiation of Caco-2 cells, as they reach confluence (Zhang et al. 2002). These investigations confirmed that retinoic acid treatment of cultured Caco-2 cells both increases RBP2 expression and retinol uptake compared to in the absence of exogenously added retinoids. Moreover, this effect of retinoic acid was reported to occur only for post-confluent differentiated Caco-2 cells and not in proliferating undifferentiated Caco-2 cells. Stable transfection experiments showed that the proximal 2.8 kb region of the human RBP2 gene is sufficient to confer retinoic acid inducibility in differentiated Caco-2 cells (Zhang et al. 2002). However, direct sequence analysis and transient transection experiments indicated that, like the rat Rbp2 promoter (Mangelsdorf et al. 1991; Nakshatri and Chambon 1994), the human RBP2 promoter is not a direct RXR target. Upregulation of the RBP2 promoter in Caco-2 monolayers was observed even when the promoter was truncated to within 103 base pairs upstream of the transcriptional start site. Based on these and other data, it was concluded that the retinoic acid-responsiveness of the human RBP2 promoter is mediated by an indirect mechanism that is associated with enterocyte differentiation (Zhang et al. 2002).
More detailed investigations of the human RBP2 gene promoter studied in Caco-2 cells found that the promoter contains a direct repeat 1 (DR-1) in the proximal promoter, at position −80 to −68, that binds endogenous HNF-4α as a homodimer (Yamaguchi et al. 2009). Transfection of HNF-4α into Caco-2 cells increased RBP2 promoter activity and mutation of the DR-1 abolished promoter activity. HNF-4α increased both endogenous RBP2 expression and retinol uptake and retinyl ester formation in transfected Caco-2 cells. Since the literature indicates that various nuclear receptors, including HNF-4α, RXRα, and peroxisome proliferator-activated receptor-α (PPARα) bind to DR-1 elements as homo- or heterodimers, these authors further explored whether this might also be true for the DR-1 in human RBP2. They found that the RXRα homodimer bound the element only very weakly in the presence of 9-cis-retinoic acid and that the DR-1 element failed completely to bind the PPARα:RXRα heterodimer (Yamaguchi et al. 2009). Collectively, these data led Yamaguchi et al. (2009) to conclude that HNF-4α regulates human RBP2 gene expression and that this contributes importantly toward regulating retinoid absorption and metabolism in human Caco-2 cells.
When post-confluent differentiated Caco-2 cells maintained in serum-free medium were treated with either arachidonic acid or its analogue, 5,8,11,14-eicosatetraynoic acid (ETYA), endogenous RBP2 mRNA was found to significantly rise and this was further enhanced when 9-cis-retinoic acid was given in a combined treatment (Suruga et al. 1999b). In Caco-2 cells, the transcripts of PPARa and RXRa, which were activated by their ligands ETYA and 9-cis-retinoic acid, were coexpressed. An electrophoretic mobility shift study using the same rat Rbp2 promoter construct first reported by Demmer et al. (1987) revealed that several nuclear proteins present in Caco-2 cells specifically bound to promoter elements present in this construct. Some of these protein/DNA complexes reacted to both anti-RXRα and anti-PPAR antibodies. Addition of in vitro synthesized RXRα and PPARα cooperatively bound to these elements as a heterodimer and the binding activities were enhanced by addition of ETYA or arachidonic acid, but not by addition of 9-cis-retinoic acid. These studies were taken to suggest that dietary fatty acids likely regulate RBP2 expression through PPAR:RXR heterodimers bound to response elements in the RBP2 gene (Suruga et al. 1999b).
Acetylation of histones H3 and H4 is reported to be involved in regulating expression of Rbp2 in the rat small intestine during the perinatal period (Ogura et al. 2007). This was determined using a ChIP assay which showed a rapid induction of acetylation of the histones H3 and H4 that interacted with the promoter/enhancer region of the Rbp2 gene. The binding of CBP and p300, which have histone acetyltransferse activity, as well as binding of RXRα increased on the Rbp2 promoter/ enhancer during the perinatal period. This was taken to suggest that Rbp2 gene expression during the perinatal period is associated with abrupt acetylation of histones H3 and H4 followed by the binding of CBP/p300 and RXRα (Ogura et al. 2007).
A number of single nucleotide polymorphisms (SNPs) in RBP2 have been identified. Investigations carried out in healthy human subjects exploring the possible roles of SNPs in genes involved in folate, retinoid, and vitamin E transport and metabolism found an association between RBP2 and plasma homocysteine levels (Clifford et al. 2012). One RBP2 SNP (rs2118981) was found to be associated with significantly diminished circulating homocysteine levels in males but not females. The authors did not provide commentary regarding their views on the molecular origins of this association. Possibly other RBP2 SNPs will be found to be associated with specific physiological responses and/or disease states.
Dietary modulation of RBP2 expression
A number of studies exploring the effects of dietary interventions, primarily ones involving high-fat diet feeding, on Rbp2 gene expression have been carried out in rats (Goda et al. 1994; Suruga et al. 1995, 1999a, 2005; Takase et al. 2000; Mochizuki et al. 2001, 2007). Some of the key findings from the rodent studies have been extended to humans through study of human Caco-2 cells manipulating the contents of the culture medium (Suzuki et al. 1998; Mochizuki et al. 2008).
Early investigations identified that jejunal Rbp2 levels are increased in rats fed a diet rich in long-chain triglycerides (Goda et al. 1994; Suruga et al. 1995) suggesting that interactions between RXR and PPARs may be one mechanism important for modulating Rbp2 gene expression. These investigations employed three diets that differed in their fat contents. Rats were fed either a high-fat diet rich in long-chain triacylglycerols, a high-fat diet rich in medium-chain triglycerides or a low-fat diet. Both Rbp2 mRNA and protein levels in the jejunum were more than twofold greater for rats fed the diet rich in long-chain triacylglycerols than in rats fed the low-fat diet or the diet rich in medium-chain triacylglycerols (Goda et al. 1994). The abundance of Rxra mRNA was elevated in the jejunums of rats fed the high-fat long-chain triglyceride diet but also in the group fed the high-fat diet rich in medium-chain triacylglycerols compared to the low-fat diet group. This suggests that expression levels of Rbp2 are modulated by dietary long chain fatty acids but not in a manner directly related to Rxra gene expression (Goda et al. 1994).
Investigations of 6-week-old male rats fed for 1 or 2 weeks diets that differed in the content of fat have provided evidence that dietary fat can induce Rbp2 transcription in the jejunum through increases in peroxisome proliferator-activated receptor-α (PPARα) mediated transcription (Suruga et al. 1999a, 1999b). Nuclear run-on assays revealed that increased Rbp2 mRNA levels upon high-fat diet feeding was, at least in part, triggered transcriptionally. Ppara mRNA levels were also increased in the jejunums of high-fat diet fed animals. Electrophoretic mobility shift assays showed that the binding activity of rat jejunal nuclear proteins to response elements in rat Rbp2 was greater in animals fed the high-fat diet and could be further enhanced by bacterially produced PPARα (Suruga et al. 1999a, 1999b). Subsequent studies employing electrophoretic mobility shift assays established that the PPARα:RXRα heterodimer binds to promoter elements within Rbp2 (Mochizuki et al. 2001). Based on these data, the authors suggested that the stimulation of Rbp2 expression elicited upon high-fat diet feeding was through a transcriptional mechanism mediated by the PPARα:RXRα heterodimer bound to elements present in the Rbp2 promoter (Suruga et al. 1999a, 1999b; Mochizuki et al. 2001). Other studies involving feeding of a high-fat diet rich in unsaturated fatty acids induced Rbp2 gene expression in the rat jejunum. This was associated with elevated Ppara expression (Takase et al. 2000). Nuclear run-on assays indicated that the increased Rbp2 mRNA levels resulted from increased transcription and that this was associated with PPARα:RXRα binding to a Rbp2 promoter element. Moreover, nuclear extracts from the jejunum of rats fed this high-fat diet gave greater densities of retarded bands than those of rats fed a fat-free diet. Expression was not affected by the diurnal rhythm of feeding. Collectively, these data were taken to suggest that Rbp2 gene expression is regulated predominantly by dietary fatty acids and PPARα mediated transcription (Takase et al. 2000).
The effect of dietary fat on jejunal RBP2 protein and mRNA levels was further studied in male rats (Suruga et al. 2005) fed for 2 weeks either a low-fat diet (containing 2.4% (w/w) corn oil) or a high-fat diet (containing 24.7% (w/w) corn oil). Dietary fat feeding rapidly increased both Rbp2 mRNA and protein levels, including a significant increase in Rbp2 mRNA levels within several hours after the start of the feeding period. Induction was found to be independent of whether the feeding period was restricted to the hours of darkness or of light, suggesting that Rbp2 expression is not subject to diurnal rhythms. Through transient transfection experiments, the authors further showed that unsaturated fatty acids increased rat Rbp2 promoter activity in a PPARα:RXRα heterodimer-dependent manner in transfected CV-1 cells (Suruga et al. 2005). Based on their data, these investigators agreed that the transcriptional induction of Rbp2 gene expression is mainly triggered by dietary fatty acids and PPARα actions (Suruga et al. 2005).
Follow-up studies by this same research group established that treatment of differentiated human Caco-2 cells with arachidonic acid, as well as other polyunsaturated fatty acids, increases PPARα:RXRα binding to elements present within human RBP2 promoter (Mochizuki et al. 2008). This effect was proposed to involve the actions of the coactivator p300 and it is binding to PPARα.
Fatty acids present in milk have been proposed to play an important role in intestinal maturation and gene expression in the postnatal small intestine. To explore the importance of fatty acids on Rbp2 expression in the postnatal period, rat pups at 17 days of age were weaned onto either a high- or low-fat diet (Mochizuki et al. 2007). Within 4 days after the start of feeding, Rbp2 mRNA levels were found to be significantly greater for the high-fat fed group. This was associated with elevations in both the expression of Fabp1 and Ppara gene expression. Administration of the synthetic PPARα agonist WY14.643 during 4 consecutive days of the weaning period caused a parallel increase in the mRNA levels of Rbp2, Fabp1, and Ppara (Mochizuki et al. 2007). These findings provide convincing evidence that Rbp2 expression in rodents during the postnatal period is also modulated by dietary fat affecting transcription mediated by PPARα (Mochizuki et al. 2007).
Taken in its entirety, the literature on the regulation of the human and murine genes encoding RBP2 does not allow for definitive conclusions to be drawn regarding the precise transcriptional mechanisms that modulate gene expression. Although the original reports proposing that this gene contains a retinoid-response element allowing for its direct regulation by retinoids, this now seems to be incorrect. Rather, other transcription factors, including PPARα and HNF-4α, along with RXR species are likely involved in controlling expression levels, definitive information of this process is still needed.
Physiological actions of rbp2 in health and disease
Role of RBP2 in ensuring normal perinatal development
Li and colleagues undertook the global knockout of Rbp2 in mice (Rbp2−/− mice) and concluded from studies of these mice that RBP2 plays a critical role in ensuring adequate transport of retinoid from the mother to the developing fetus, particularly when maternal dietary retinoid availability is limited (E et al. 2002). It was reported that when pregnant Rbp2−/− mice were switched at mid-gestation from a diet that contained a sufficient retinoid level to one with only a marginal level, this resulted in 100% mortality/liter within 24 h after birth in the Rbp2−/− line. But no mortality was observed for the wild-type line. Necropsy analysis suggested that neonatal mortality may have resulted from lesions in lung development but this was not conclusively established. It was further reported that neonatal mortality in heterozygote offspring of Rbp2−/− dams (79 ± 21%) was greater compared with the neonatal mortality in heterozygote offspring of wild-type dams (29 ± 25%). Li and colleagues took this to indicate that fetal as well as maternal RBP2 are required to ensure adequate delivery of retinoid to the developing organism during the neonatal period in times when dietary retinoid is limited (E et al. 2002).
Role of RBP2 in intestinal retinoid uptake and metabolism in the adult
With the initial identification of RBP2, Ong proposed that RBP2 plays a central role in facilitating the uptake and metabolism of dietary retinoids by enterocytes (Ong 1984). The very high level of RBP2 protein present in the small intestine and its immunolocalization to the absorptive cells of the villi fully supported this notion (Crow and Ong 1985). There has been much research aimed at confirming and extending this proposal (Napoli 2017).
A number of biochemical studies explored the possibility that retinol bound to RBP2 was available to an unidentified microsomal enzyme(s) that catalyzes retinyl ester formation (Ong et al. 1987). When microsomes prepared from 3-day-old rat liver (a postnatal time when RBP2 is still expressed in the liver) were incubated with purified retinol-RBP2, retinyl esters were found to accumulate in a time- and protein-dependent manner. Moreover, retinyl ester formation from retinol-RBP2 by the microsomal preparations was found to occur in an acyl-CoA-independent manner (Ong et al. 1987). More in-depth investigation of this observation showed that when retinol is bound to a nonspecific lipid-binding protein, like bovine serum albumin, it is rapidly esterified by rat liver microsomes in a reaction that utilized exogenous acyl-CoA (Ong et al. 1987). This led to the suggestion that retinol bound to RBP2 is not available for esterification by acyl-CoA-utilizing enzymes (Ong et al. 1987). With the identification of LRAT as an enzyme in the rat small intestine that catalyzes retinyl ester formation, it was shown that retinol bound to RBP2 is a substrate for LRAT catalyzed retinyl ester formation (MacDonald and Ong 1988; Ong et al. 1988). Later studies involving the use of Rbp2−/− mice, confirmed that RBP2 channels retinol to LRAT for retinyl ester synthesis (Wongsiriroj et al. 2008). Thus, it is now accepted that RBP2 acts within the small intestine to channel retinol to LRAT for retinyl ester formation, allowing for dietary retinoid uptake as retinyl ester in chylomicrons (Blaner and Li 2015; Mezaki et al. 2016).
As noted above, RBP2 binds all-trans-retinaldehyde with high affinity, comparable to that of all-trans-retinol (MacDonald and Ong 1987; Levin et al. 1988; Li et al. 1991; Kane et al. 2011). This observation had led to the proposal that retinaldehyde bound to RBP2 might, in a manner analogous to retinol-RBP2 and LRAT, serve as a substrate for intestinal enzymes that reduce retinaldehyde to retinol (MacDonald and Ong 1987; Levin et al. 1988). Indeed, when isolated rat intestinal microsomes were incubated with purified retinaldehyde-RBP2 complex, in the presence of either NADH or NADPH, this resulted in the enzymatic reduction of retinaldehyde to retinol (Kakkad and Ong 1988). This led to the conclusion that a RalR present in the small intestine specifically utilizes retinaldehyde bound to RBP2 to generate retinol from retinaldehyde that is derived from dietary provitamin A carotenoids.
Additional support for the notion that RBP2 plays a central role in facilitating intestinal retinoid uptake and metabolism, comes from studies that have established that identical gradients of RBP2 protein and RalR and LRAT activities exist anatomically across the adult small intestine (Herr et al. 1993). Similarly, dietary polyunsaturated triacylglycerol feeding was found to enhance both BCO1 activity and RBP2 protein levels in the rat intestine (During et al. 1998). Collectively, these observations are consistent with the notion that the expression of enzymes involved in retinoid metabolism in the intestine and RBP2 are coordinately regulated.
Study of global Rbp2−/− mice established that RBP2 is not absolutely required by the adult for ensuring the uptake of dietary retinoid (E et al. 2002). When these mice were maintained on a chow diet providing slightly elevated retinoid levels (25 IU retinol/g diet compared to 15 IU retinol/g diet present in a conventional chow diet), hepatic retinoid levels were found to be 40% of those of matched wild-type mice fed the same diet. Circulating retinol levels were not different for the two groups. Moreover, the Rbp2−/− mice fed the elevated retinoid diet displayed normal growth and reproduction (E et al. 2002). Subsequent studies of Rbp2−/− mice have established that even when these mice are fed a conventional chow diet they display normal blood retinol levels, and grow and reproduce normally (Wongsiriroj et al. 2008). These findings convincingly establish that the Rbp2−/− mice are not experiencing retinoid-deficiency when they consume sufficient levels of dietary retinoid. Thus, RBP2 acts to optimize retinoid uptake from the diet by channeling uptake and metabolism within absorptive cells (see Figure 1), but it is not essential for these processes in times of dietary retinoid-sufficiency (E et al. 2002).
In an effort to extend observations obtained from animal studies of RBP2 to humans, a number of studies of retinol uptake and metabolism have been carried out in human Caco-2 cells (Levin 1993; Lissoos et al. 1995; Levin and Davis 1997). Stably transfected cloned Caco-2 cells lines that overexpress RBP2 were found to take up two times more retinol from the culture medium than sham transfected control cells (Levin 1993; Lissoos et al. 1995). Overexpression of RBP2 also led to enhanced retinyl ester formation by the Caco-2 cultures. In other studies, when Caco-2 cells were treated with retinoic acid, this led to a 50% increase in retinol uptake (Levin and Davis 1997). These data suggest that RBP2 acts within the human intestine to channel retinoid uptake and metabolism in a manner similar to the rodent intestine.
Role of RBP2 in modulating intestinal monoacylglycerol levels in the adult
RBP2 binds to MAGs with very high affinity, similar to its affinity for retinol or retinaldehyde (Lee et al. 2020). Moreover, the absence of Rbp2, leads to elevations in mucosal levels of 2-AG, 2-LG, 2-OG and 2-palmitoylglycerol (2-PG) two hours after administration of a corn oil challenge to Rbp2−/− mice compared to wild-type mice. This is seen in Figure 4 for approximately 6- to 7-month-old male Rbp2−/− and wild-type mice that had been maintained on a chow diet throughout life. Thus, RBP2 has a role in modulating monoacylglycerol metabolism within the intestine. This role could involve the actions of RBP2 in facilitating bulk lipid uptake from the diet or 2-MAG signaling. It seems likely that RBP2 acts in the trafficking and metabolism of intestinal 2-MAGs given that RBP2 accounts for approximately 1% of enterocyte cytosolic protein (Ong 1985). MAGs generated in the intestinal lumen from dietary triglycerides are taken up by enterocytes where they are used primarily to resynthesize triglycerides that are then incorporated into nascent chylomicrons (Chon et al. 2012; Yen et al. 2015). Some MAGs will be degraded by monoacylglycerol lipase to nonesterified fatty acids that may be used for triglyceride synthesis and incorporation into chylomicrons (Chon et al. 2012; Douglass et al. 2015). It is possible that RBP2 plays a role in metabolically trafficking MAGs toward triglyceride synthesis and chylomicron formation. This putative action of RBP2 could affect both the quantity of triglyceride incorporated into chylomicrons and the acyl composition of chylomicron lipids.
Figure 4.
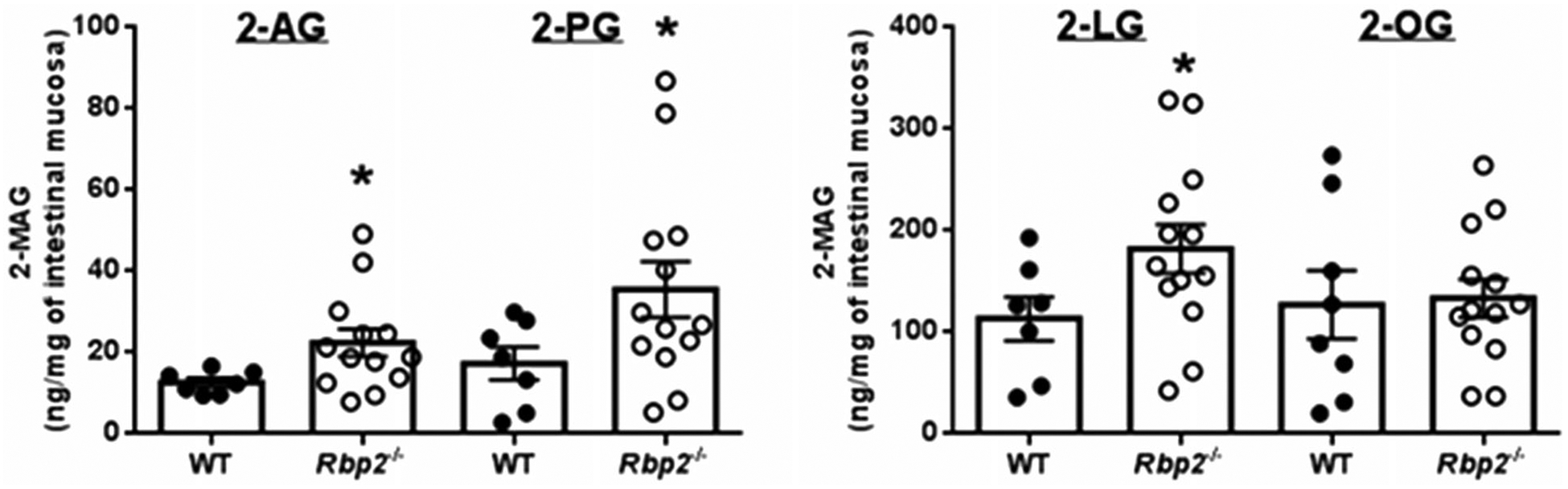
An oral fat challenge administered to Rbp2−/− mice results in elevated 2-MAG levels in intestinal absorptive cells. Two hours after administration of an oral challenge with corn oral, levels of 2-arachidonoylglycerol (2-AG) 2-palmitoylglycerol (2-PG), and 2-lineoylglycerol (2-LG) are significantly elevated in enterocyte scrapings for Rbp2−/− mice compared to matched wild type (WT) mice. Enterocyte levels of 2-oleoylglycerol (2-OG) were not significantly different for the two groups. *p < 0.05. (Taken from Lee et al. 2020).
Alternatively, RBP2 may play a role in facilitating 2-MAG signaling. 2-AG is a canonical endocannabinoid acting through the two cannabinoid receptors CB1 and CB2 (Chanda et al. 2019). The other long-chain 2-MAGs act through different cell surface G-protein coupled receptors that also regulate metabolism and energy expenditure (McIntosh et al. 2009; Hansen et al. 2011; Ekberg et al. 2016).
Role of RBP2 in the development of obesity and related metabolic disease
Recent investigations involving the use of Rbp2−/− mice have identified a previously unexpected linkage between RBP2 and the development of obesity, glucose intolerance and fatty liver (Lee et al. 2020). These studies establish that 6- to 7-month-old chow-fed male Rbp2−/− mice acquire more body fat, show an impaired response to a glucose challenge, and manifest elevated hepatic triglyceride levels compared to diet-, age-, gender-, and genetic background-matched wild-type mice. Similarly, 2-month-old Rbp2−/− fed a high-fat diet for 6–8 weeks gain significantly more body weight, display impaired glucose tolerance, and show higher fasting hepatic triglyceride levels than age-matched wild-type mice (Lee et al. 2020).
What accounts for the age- and diet-dependent increase in body fat, glucose insensitivity, and elevated fasting hepatic triglyceride levels observed for Rbp2−/− mice has not been fully established. As discussed above in this section, Rbp2−/− mice are not retinoid-deficient when maintained on a chow diet (E et al. 2002). Lee et al. reported that no differences in tissue retinoid-related parameters could be identified that would account for this Rbp2−/− phenotype (Lee et al. 2020). This implicates differences in mucosal 2-MAG levels in the causation of these metabolic phenotypes. Supporting this contention, the elevations in 2-MAG levels are accompanied by elevated blood levels of the intestinal hormone glucose-dependent insulinotropic polypeptide (GIP). As seen in Figure 5, 30 min after administration of an oral fat challenge, blood GIP levels are significantly elevated in chow fed 6- to 7-month-old Rbp2−/− mice compared to matched wild-type mice. 2-MAGs are potent agonists for the cell surface receptor GPR119 that is present on the enteroendocrine K-cells that are responsible for GIP synthesis and secretion (McIntosh et al. 2009; Hansen et al. 2011; Ekberg et al. 2016). GPR119 is known to signal enteroendocrine-cell release of GIP in both rodents and humans, affecting metabolism and energy expenditure (McIntosh et al. 2009; Hansen et al. 2011; Ekberg et al. 2016). As mentioned above, 2-AG is a canonical endocannabinoid and is able to bind and activate cannabinoid receptors 1 and 2 (CB1 and CB2) affecting energy expenditure and body weight (Hansen and Vana 2018; Chanda et al. 2019). A recent study in humans concluded that elevated GIP levels in obesity are likely a consequence of increased gut endocannabinoid levels (Chia et al. 2017). Several studies have reported elevated blood GIP levels in obese humans (Theodorakis et al. 2004; Calanna et al. 2013). Elevated GIP levels have been reported in diet induced obese mice (Sun et al. 2019). Based on this literature, it may not be too unreasonable to speculate that the impaired metabolic phenotypes observed for Rbp2−/− mice arise through RBP2’s 2-MAG ligands and their effects on blood GIP levels following fat intake.
Figure 5.
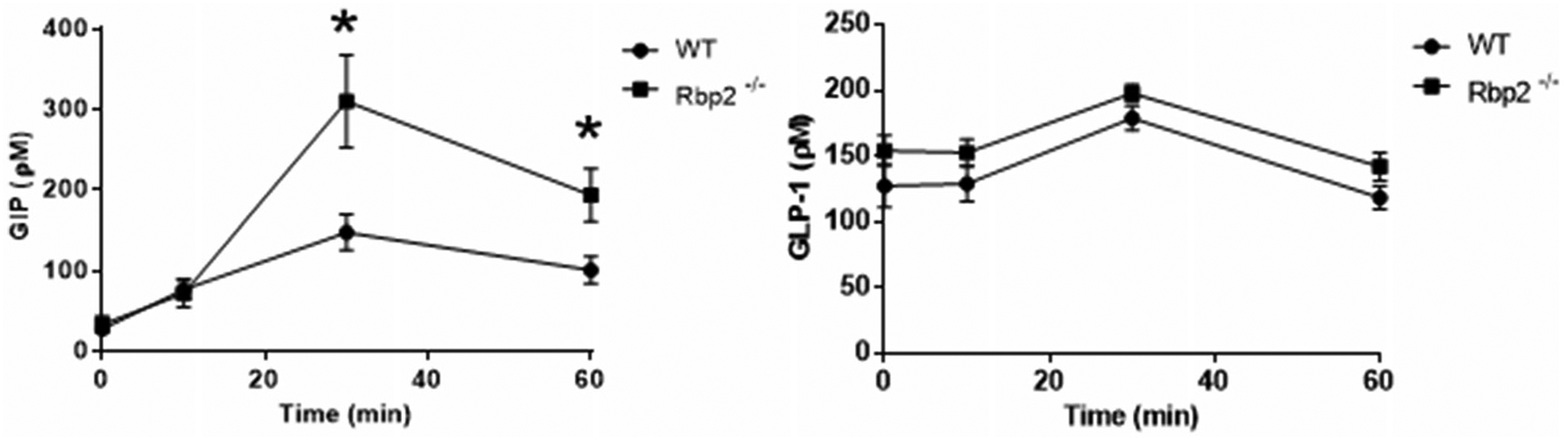
Time course showing plasma levels of glucose-dependent insulinotropic polypeptide (GIP) (left) and glucagon-like protein-1 (GLP-1) (right) following an oral challenge with corn oil. Plasma GIP levels are significantly elevated 30 and 60 min after the challenge for chow fed 6- to 7-month-old Rbp2−/− mice compared with matched wild-type mice. No statistically significant differences in plasma GLP-1 levels were observed for the same mice. *p < 0.05.
Role of RBP2 in maintaining intestinal immunity
There is evidence in the literature that RBP2 plays a central role in maintaining intestinal innate immunity (McDonald et al. 2012). Within the small intestine, dendritic cells (DCs) are key initiators and regulators of the adaptive immune response, affecting the balance between tolerance and immunity within the mucosa (Agace and Persson 2012). Over the last decade, it has become clear that DCs use all-trans-retinoic acid to promote intestine-specific immune responses, including Foxp3+ Treg conversion, lymphocyte gut homing molecule expression, and IgA production (Iliev et al. 2009; Feng et al. 2010; Molenaar et al. 2011). The literature indicates that both retinoids per se and RBP2 are required for imprinting intestinal lamina propria CD103+ DCs with the ability to generate gut tropic T cells (Iliev et al. 2009; Feng et al. 2010; Molenaar et al. 2011; Jaensson-Gyllenbäck et al. 2011; McDonald et al. 2012). Studies of isolated DCs have identified that granulocyte-macrophage colony-stimulating factor, IL-4 and all-trans-retinoic acid are needed to induce the enzyme retinaldehyde dehydrogenase 1 (ALDH1A1), which catalyzes the final enzymatic step needed for all-trans-retinoic acid formation (Iliev et al. 2009). DC expression of Aldh1a1 was found to be reduced in mice fed a retinoid-deficient diet and expression could be rescued upon feeding of a retinoid-sufficient diet (Molenaar et al. 2011). Interestingly, relatively high concentrations of retinoids present within bile, and consequently ones derived from hepatic retinoid pools, also have been proposed to be the source of retinoids used by the intestine to imprint intestinal CD103+ DCs (Jaensson-Gyllenbäck et al. 2011). Collectively, this published work establishes that luminal retinoids act as an environmental cue shaping the phenotype of intestinal DCs and attendant DC responses (Agace and Persson 2012).
Subsequent studies extending these findings regarding retinoid actions in intestinal immunity have shown that RBP2 plays a key role in maintaining normal DC responses. Investigations involving the use of Rbp2−/− mice have shown that Rbp2 expression is required to confer the capacity of CD103+ DCs present in the lamina propria of the small intestine to convert retinol to all-trans-retinoic acid (McDonald et al. 2012). Through study of Rbp2−/− mice, it was shown that imprinting of Aldh1a1 expression in CD103+ DCs requires epithelial cell expression of Rbp2. Thus, intestinal epithelial cell-specific expression of Rbp2 must play an integral role in transforming luminal retinoids into a local cue to imprint gut DCs with an intestinal phenotype. This presumably occurs by channeling either retinol and/or retinaldehyde from the intestinal epithelium to CD103+ DCs within the lamina propria (McDonald et al. 2012). The possibility that RBP2 channels retinaldehyde to CD103+ DCs is in keeping with published finding establishing that the transcription factor ISX, which is a key regulator of BCO1 expression in the gut, also affects gut homing and differentiation of lymphocytes within the small intestine (Widjaja-Adhi et al. 2017).
RBP2 and intestinal adaptive responses
A number of different studies have been carried out to assess whether intestinal RBP2 exhibits an adaptive response to removal of a portion of the bowel. All these studies employed rat models but each used different resection protocols and different time intervals of study. Never-the-less, each of these studies reached the same conclusion that Rbp2 expression is adaptively elevated following surgical resection of the small intestine and that this response benefits retinoid absorption by the intestine.
One study examined RBP2 expression in a shortened jejunum, where the distal 5 cm portion of the proximal jejunum of a 2-month-old rat was joined to the proximal end of the ileum (Takase et al. 1993). After allowing 3 weeks for recovery, the amount of RBP2 protein was assessed immunologically (Takase et al. 1993). A twofold increase in the total amount of RBP2 present in the proximal ileum was reported to occur, but in the setting of hyperplasia. However, for the proximal jejunum, preceding the bypassed segment, the RBP2 level per unit DNA increased by 80%. No differences in either circulating or hepatic total retinol content were observed for the bypass group. The authors took their data to suggest that the increase in RBP2 content following jejunum-bypass might be an adaptive response to enhance retinol absorption. Similar studies, involving a 70% resection of the proximal small intestine of an adult rat, explored adaptive responses 2 days after resection (Dodson et al. 1996). Rbp2 mRNA expression was reported to be elevated in the remnant ileum. This early adaptive response was accompanied by elevations in expression of other intestinal genes involved in nutrient uptake and metabolism including Fabp1 and apolipoprotein A-IV (Apoa4), a gene involved in chylomicron formation. This was taken to suggest that Rbp2 expression is regulated in a manner that reflects complex multifaceted regulatory responses to bowel resection that enhance nutrient uptake (Dodson et al. 1996).
Later investigations aimed at gaining insight into the expression of genes involved in retinoid absorption, transport and metabolism during bowel resection were carried out in a rat model for short bowel syndrome where 75% of the entire small intestine was removed and adaptive changes assessed 7 days later (Hebiguchi et al. 2015). Expression of both Rbp2 mRNA as well as mRNA for Apoa4 was found to be higher than those observed in sham-operated rats. However, the retinol content of the residual ilium and the retinyl ester content in the jejunum were lower than those observed in sham-operated rats. The authors took this to indicate that Rbp2 and Apoa4 in rats with a shortened bowel contribute to more effective esterification of dietary retinol and more effective uptake into the body in nascent chylomicrons. Thus, they proposed that the esterification and secretion of retinoids in nascent chylomicrons are enhanced in the absorptive epithelial cells of intestines of rats with experimentally-induced short bowel syndrome through the upregulation of Rbp2 and Apoa4 mRNA.
Future research directions and conclusions
It is now well established that RBP2 plays a central role in retinoid uptake from the diet, retinoid processing within the absorptive epithelium allowing for retinyl ester formation and its packaging in nascent chylomicrons and helping mediate retinoid actions within the intestine. It is also clear that RBP2 plays an important role in facilitating retinoid actions in maintaining normal cell and tissue differentiation early in life. Both the RBP2 protein and the RBP2 gene are well studied, although some details still need to be filled in for knowledge of the molecular properties of RBP2 to be complete. The recent identification that both human and mouse RBP2 are physiologically relevant intestinal 2-MAG-binding proteins and the possibly related observation that Rbp2 absence in mice affects release into the circulation of the hormone GIP by the enteroendrocrine K-cells raise a number of new and fundamentally important questions regarding RBP2 and its physiologic actions.
After a fat-rich meal, intestinal concentrations of 2-MAGs will normally be several orders of magnitude greater than those of retinol/retinaldehyde, yet the Kd values for retinol and retinaldehyde binding are the same order of magnitude as those reported for 2-MAG binding. Since MAGs can displace retinol from its binding site in RBP2 (Lee et al. 2020), this raises an important question regarding how RBP2 facilitates both retinol and MAG absorption and/or metabolism. Does the quality and/or quantity of dietary fat influence retinol-RBP2 interactions in a manner that affects retinoid absorption mediated by RBP2? Or, is the concentration of RBP2 within enterocytes sufficient to accommodate both retinoid and MAG absorption and metabolism? Understanding of these issues will be important for understanding RBP2 actions in intestinal neutral lipid metabolism and nutrient sensing.
The Rbp2−/− mice are more prone to developing obesity and related metabolic disease than matched wild-type mice (Lee et al. 2020). At 2 months of age, these mice when fed a high-fat diet gain significantly more body weight than matched wild-type mice and they accrue significantly greater body weights at 6 to 7 months of age when fed a control chow diet compared to matched wild-type mice. This raises a question regarding how dietary fat and retinoids may interact within the gut to influence the absorption of each other. Do differences in dietary retinoid contents influence fat absorption, and consequently calorie intake? And, if so, how might this involve the actions of RBP2 in binding MAGs and/or retinoids?
Many reports have established FABPs as important modulators of metabolism and inflammatory processes and consequently, contributors to metabolic disease development and prevention. Mice deficient in Fabp1, when fed a high-fat diet rich in long-chain saturated fatty acids, display significantly greater body weights and fat masses compared to matched WT mice. When mice lacking Fabp2 are fed this same diet, they remain lean, with lower body weights and fat masses than WT mice. Consistent with these experimental studies, polymorphisms in both the human FABP1 and FABP2 genes have been associated with increased incidence of obesity and glucose intolerance. Both FABPI and FABP2 are expressed abundantly in jejunum, as is RBP2. Unlike FABP1, RBP2 does not bind FFAs. However, both RBP2 and FABP1 are MAG-binding proteins that co-localize to the same anatomical region of the small intestine. Are their functions in MAG trafficking redundant or does each protein have a unique metabolic action within the enterocyte? Moreover, Rbp2−/−, Fabp1−/−, and Fabp2−/− mice each display clear adiposity phenotypes when challenged with a high-fat diet. Do the observed weight gain phenotypes all arise due to effects of these proteins on the same pathway or through multiple different pathways?
Are the biochemical and metabolic phenotypes observed for Rbp2−/− mice associated with enteroendocrine signaling? We do not yet understand the molecular basis for altered enterocyte levels of 2-MAGs in response to an oral fat challenge or the associated elevations in GIP levels in blood. It is unclear whether this involves a direct mechanism that affects enteroendocrine cells through only local actions within the intestine. Or, if these effects involve the gut-brain axis and central nervous system input into the regulatory process. These possibilities need to be resolved. In addition, we will need to gain understanding of the role of RBP2 in human intestinal physiology and enteroendocrine signaling, extending work in the mouse.
It is clear that much research focused on the actions of RBP2 in intestinal 2-MAG metabolism, in intestinal hormone production, and in the development of obesity and related metabolic phenotypes still needs to be undertaken before this intriguing protein can be understood.
Funding
This work was supported by grants R01DK068437 (WSB), R01DK101251 (WSB), R01EY023948 (MG), R01DK122071 (WSB and MG), and R21AA028110 (WSB) from the US Public Health Services, National Institutes of Health.
Abbreviations:
- AEA
N-arachidonoylethanolamide
- ALDH1A1
retinaldehyde dehydrogenase 1
- ALDH1A2
retinaldehyde dehydrogenase 2
- APOA4
apolipoprotein A-IV
- ARP-1
apoA1 regulatory protein A1
- BCO1
β-carotene 15,15’-dioxygenase
- BPES
blepharophimosis-ptosis-epicanthus inversus syndrome
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- CDH
congenital diaphragmatic hernia
- CRABP1
cellular retinoic acid-binding protein, Type 1
- CRABP2
cellular retinoic acid-binding protein, Type 2
- DC
dendritic cell
- DR-1
direct repeat 1
- ETYA
5,8,11,14-eicosatetraynoic acid
- FABP
fatty acid-binding protein
- FABP1
liver fatty acid-binding protein
- FABP2
intestinal fatty acid-binding protein
- GIP
glucose-dependent insulinotropic polypeptide
- HNF-4
hepatocyte nuclear factor-4
- LRAT
lecithin:retinol acyltransferase
- MAG
monoacylglycerol
- NEA
N-acylethanolamide
- PPAR
peroxisome proliferator-activated receptor
- RalR
retinaldehyde reductase
- RAR
retinoic acid receptor
- RBP
intracellular retinol-binding proteins
- RBP1
retinol-binding protein 1 (cellular retinol-binding protein)
- RBP2
retinol-binding protein 2 (cellular retinol-binding protein, Type 2)
- RBP5
retinol-binding protein 4 (cellular retinol-binding protein, Type 4)
- RBP7
retinol-binding protein 3 (cellular retinol-binding protein, Type 3)
- RXR
retinoid X receptor
- RXRE
retinoid X receptor response element
- 1-AG
1-arachidonoylglycerol
- 2-AG
2-arachidonoylglycerol
- 2-LG
2-lineoylglycerol
- 2-OG
2-oleoylglycerol
- 2-PG
2-palmitoylglycerol
Footnotes
Disclosure statement
The authors have nothing to disclose.
References
- Agace WW, Persson EK. 2012. How vitamin A metabolizing dendritic cells are generated in the gut mucosa. Trends Immunol. 33(1):42–48. [DOI] [PubMed] [Google Scholar]
- Bashor MM, Toft DO, Chytil F. 1973. In vitro binding of retinol to rat-tissue components. Proc Natl Acad Sci USA. 70(12):3483–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat PV. 1998. Retinal dehydrogenase gene expression in stomach and small intestine of rats during postnatal development and in vitamin A deficiency. FEBS Lett. 426(2):260–262. [DOI] [PubMed] [Google Scholar]
- Blaner WS, Li Y. 2015. Vitamin A metabolism, storage and tissue delivery mechanisms In: Doll e P, Niederreither K, editors. The retinoids; biology, biochemistry, and disease. Hoboken (NJ): Wiley Blackwell; p. 3–34. [Google Scholar]
- Calanna S, Christensen M, Holst JJ, Laferrère B, Gluud LL, Vilsbøll T, Knop FK. 2013. Secretion of glucose-dependent insulinotropic polypeptide in patients with type 2 diabetes: systematic review and meta-analysis of clinical studies. Diabetes Care 36(10):3346–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda D, Neumann D, Glatz J. 2019. The endocannabinoid system: overview of an emerging multi-faceted therapeutic target. Prostaglandins Leukot Essent Fatty Acids 140:51–56. [DOI] [PubMed] [Google Scholar]
- Cheng L, Qian SJ, Rothschild C, d’Avignon A, Lefkowith JB, Gordon JI, Li E. 1991. Alteration of the binding specificity of cellular retinol-binding protein II by site-directed mutagenesis. J Biol Chem. 266(36):24404–24412. [PubMed] [Google Scholar]
- Chia CW, Carlson OD, Liu DD, Gonzalez-Mariscal I, Calvo SS-S, Egan JM. 2017. Incretin secretion in humans is under the influence of cannabinoid receptors. Am J Physiol Endocrinol Metab. 313(3):E359–E366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon S-H, Douglass JD, Zhou YX, Malik N, Dixon JL, Brinker A, Quadro L, Storch J. 2012. Over-expression of monoacylglycerol lipase (MGL) in small intestine alters endocannabinoid levels and whole body energy balance, resulting in obesity. PLoS One 7(8):e43962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford AJ, Chen K, McWade L, Rincon G, Kim S-H, Holstege DM, Owens JE, Liu B, Müller H-G, Medrano JF, et al. 2012. Gender and single nucleotide polymorphisms in MTHFB, BHMT, SPLC1, CRBP2, CETP and SCARB1 are significant predictors of plasma homocysteine normalized by RBC folate in healthy adults. J Nutr. 142(9):1764–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottin SC, Gambling L, Hayes HE, Stevens VJ, McArdle HJ. 2016. Pregnancy and maternal iron deficiency stimulate hepatic CRBPII expression in rats. J Nutr Biochem. 32: 55–63. [DOI] [PubMed] [Google Scholar]
- Crow JA, Ong DE. 1985. Cell-specific immunohistochemical localization of a cellular retinol-binding protein (type two) in the small intestine of rat. Proc Natl Acad Sci USA. 82(14):4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmer TRA, Clugston RD. 2019. Gene ontology enrichment analysis of congenital diaphragmatic hernia-associated genes. Pediatr Res. 85(1):13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baere E, Speleman F, van Roy N, Mortier K, De Paepe A, Messiaen L. 1998. Assignment of the cellular retinol-binding protein 2 gene (RBP2) to human chromosome band 3q23 by in situ hybridization. Cytogenet Cell Genet. 83(3–4):240–241. [DOI] [PubMed] [Google Scholar]
- De Baere E, van Roy N, Speleman F, Fukushima Y, de Paepe A, Messiaen L. 1999. Closing in on the BPES gene on 3q23: mapping of a de novo reciprocal translocation t(3; 4)(q23;p15.2) breakpoint within a 45-kb cosmid and mapping of three candidate genes, RBP1, RBP2, and β’-COP, distal to the breakpoint. Genomics 57(1):70–78. [DOI] [PubMed] [Google Scholar]
- Demmer LA, Birkenmeier EH, Sweetser DA, Levin MS, Zollman S, Sparkes RS, Mohandas T, Lusis AJ, Gordon JI. 1987. The cellular retinol binding protein II gene. Sequence analysis of the rat gene, chromosomal localization in mice and humans, and documentation of its close linkage to the cellular retinol binding protein gene. J Biol Chem. 262(6):2458–2467. [PubMed] [Google Scholar]
- Dodson BD, Wang JL, Swietlicki EA, Rubin DC, Levin MS. 1996. Analysis of cloned cDNAs differentially expressed in adapting remnant small intestine after partial resection. Am J Physiol. 271(2 Pt 1):G347–G356. [DOI] [PubMed] [Google Scholar]
- Douglass JD, Zhou YX, Wu A, Zadroga JA, Gajda AM, Lackey AI, Lang W, Chevalier KM, Sutton SW, Zhang S-P, et al. 2015. Global deletion of MGL in mice delays lipid absorption and alters energy homeostasis and diet-induced obesity. J Lipid Res. 56(6):1153–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- During A, Nagao A, Terao J. 1998. β-Carotene 15,15’-dioxygenase activity and cellular retinol-binding protein type II level are enhanced by dietary unsaturated triacylglycerols in rat intestine. J Nutr. 128(10):1614–1619. [DOI] [PubMed] [Google Scholar]
- E X, Zhang L, Lu J, Tso P, Blaner WS, Levin MS, Li E. 2002. Increased neonatal mortality in mice lacking cellular retinol-binding protein II. J Biol Chem. 277(39):36617–36623. [DOI] [PubMed] [Google Scholar]
- Ekberg JH, Hauge M, Kristensen LV, Madsen AN, Engelstoft MS, Husted A-S, Sichlau R, Egerod KL, Timshel P, Kowalski TJ, et al. 2016. GPR119, a major enteroendocrine sensor of dietary triglyceride metabolites coacting in synergy with FFA1 (GPR40). Endocrinology 157(12):4561–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estarellas C, Scaffidi S, Saladino G, Spyrakis F, Franzoni L, Galdeano C, Bidon-Chanal A, Gervasio FL, Luque FJ. 2019. Modulating ligand dissociation through methyl isomerism in accessory sites: binding of retinol to cellular carriers. J Phys Chem Lett. 10(23):7333–7339. [DOI] [PubMed] [Google Scholar]
- Feng T, Cong Y, Qin H, Benveniste EN, Elson CO. 2010. Generation of mucosal dendritic cells from bone marrow reveals a critical role of retinoic acid. J Immunol. 185(10): 5915–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay JA, Strom M, Ong DE, DeLuca HF. 1990. Regulation of cellular retinol binding protein type II by 1,25-dihydroxyvitamin D3. Biochemistry 29(20):4914–4921. [DOI] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE. 2000. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta 1482(1–2):9–24. [DOI] [PubMed] [Google Scholar]
- Folli C, Calderone V, Ottonello S, Bolchi A, Zanotti G, Stoppini M, Berni R. 2001. Identification, retinoid binding, and x-ray analysis of a human retinol-binding protein. Proc Natl Acad Sci USA. 98(7):3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folli C, Calderone V, Ramazzina I, Zanotti G, Berni R. 2002. Ligand binding and structural analysis of a human putative cellular retinol-binding protein. J Biol Chem. 277(44): 41970–41977. [DOI] [PubMed] [Google Scholar]
- Gajda AM, Storch J. 2015. Enterocyte fatty acid-binding proteins (FABPs): different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot Essent Fatty Acids 93:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda T, Yasutake H, Takase S. 1994. Dietary fat regulates cellular retinol-binding protein II gene expression in rat jejunum. Biochim Biophys Acta 1200(1):34–40. [DOI] [PubMed] [Google Scholar]
- Goumy C, Gouas L, Marceau G, Coste K, Veronese L, Gallot D, Sapin V, Vago P, Tchirkov A. 2010. Retinoid pathway and congenital diaphragmatic hernia: hypothesis from the analysis of chromosomal abnormalities. Fetal Diagn Ther. 28(3):129–139. [DOI] [PubMed] [Google Scholar]
- Hansen KB, Rosenkilde MM, Knop FK, Wellner N, Diep TA, Rehfeld JF, Andersen UB, Holst JJ, Hansen HS. 2011. 2-Oleoyl glycerol is a GPR119 agonist and signals GLP-1 release in humans. J Clin Endocrinol Metab. 96(9): E1409–E1417. [DOI] [PubMed] [Google Scholar]
- Hansen HS, Vana V. 2018. Non-endocannabinoid N-acylethanolamines and 2-monoglycerols in the intestine. Br J Pharmacol. 176(10):1443–1454. DOI: 10.1111/bph.14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison EH. 2012. Mechanisms involved in the intestinal absorption of dietary vitamin A and provitamin A carotenoids. Biochim Biophys Acta 1821(1):70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison EH, Hussain MM. 2001. Mechanisms involved in the intestinal digestion and absorption of dietary vitamin A. J Nutr. 131(5):1405–1408. [DOI] [PubMed] [Google Scholar]
- Hebiguchi T, Mezaki Y, Morii M, Watanabe R, Yoshikawa K, Miura M, Imai K, Senoo H, Yoshino H. 2015. Massive bowel resection upregulates the intestinal mRNA expression levels of cellular retinol-binding protein II and apolipoprotein A-IV and alters the intestinal vitamin A status in rats. Int J Mol Med. 35(3):724–730. [DOI] [PubMed] [Google Scholar]
- Herr FM, Wardlaw SA, Kakkad B, Albrecht A, Quick TC, Ong DE. 1993. Intestinal vitamin A metabolism: coordinate distribution of enzymes and CRBP(II). J Lipid Res. 34(9): 1545–1554. [PubMed] [Google Scholar]
- Hind M, Corcoran J, Maden M. 2002. Pre- and Postnatal lung development, maturation, and plasticity. Temporal/spatial expression of retinoid binding proteins and RAR isoforms in the postnatal lung. Am J Physiol Lung Cell Mol Physiol. 282(3):L468–L476. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Bernlohr DA. 2015. Metabolic functions of FABPs-mechanisms and therapeutic implications. Nat Rev Endocrinol. 11(10):592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Mileti E, Matteoli G, Chieppa M, Rescigno M. 2009. Intestinal epithelial cells promote colitis-protective regulatory T-cell differentiation through dendritic cell conditioning. Mucosal Immunol. 2(4):340–350. [DOI] [PubMed] [Google Scholar]
- Inagami S, Ong DE. 1992. Purification and partial characterization of cellular retinol-binding protein, type two, from human small intestine. J Nutr. 122(3):450–456. [DOI] [PubMed] [Google Scholar]
- Jaensson-Gyllenbäck E, Kotarsky K, Zapata F, Persson EK, Gundersen TE, Blomhoff R, Agace WW. 2011. Bile retinoids imprint intestinal CD103+ dendritic cells with the ability to generate gut-tropic T cells. Mucosal Immunol. 4(4): 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkad BP, Ong DE. 1988. Reduction of retinaldehyde bound to cellular retinol-binding protein (type II) by microsomes from rat small intestine. J Biol Chem. 263(26): 12916–12119. [PubMed] [Google Scholar]
- Kane MA, Bright FV, Napoli JL. 2011. Binding affinities of CRBPI and CRBPII for 9-cis-retinoids. Biochim Biophys Acta 1810(5):514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagakos WS, Guan X, Ho S-Y, Sawicki LR, Corsico B, Kodukula S, Murota K, Stark RE, Storch J. 2013. Liver fatty acid-binding protein binds monoacylglycerol in vitro and in mouse liver cytosol. J Biol Chem. 288(27):19805–19815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi B, Mishra M, Srinivasan N, Archunan G. 2015. Structure-based phylogenetic analysis of the lipocalin superfamily. PLoS One 10(8):e0135507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-A, Yang KJZ, Brun P-J, Silvaroli JA, Yuen JJ, Shmarakov I, Jiang H, Feranil JB, Li X, Lackey AI, et al. 2020. Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight. Sci Adv. 6(11):eaay8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin MS, Davis AE. 1997. Retinoic acid increases cellular retinol binding protein II mRNA and retinol uptake in the human intestinal Caco-2 cell line. J Nutr. 127(1):13–17. [DOI] [PubMed] [Google Scholar]
- Levin MS, Li E, Ong DE, Gordon JI. 1987. Comparison of tissue-specific expression and developmental regulation of two closely linked rodent genes encoding cytosolic retinol-binding proteins. J Biol Chem. 262(15):7118–7124. [PubMed] [Google Scholar]
- Levin MS, Locke B, Yang NC, Li E, Gordon JI. 1988. Comparison of the ligand binding properties of two homologous rat apocellular retinol-binding proteins expressed in Escherichia coli. J Biol Chem. 263(33):17715–17723. [PubMed] [Google Scholar]
- Levin MS. 1993. Cellular retinol-binding proteins are determinants of retinol uptake and metabolism in stably transfected Caco-2 cells. J Biol Chem. 268(11):8267–8276. [PubMed] [Google Scholar]
- Li E, Norris AW. 1996. Structure/function of cytoplasmic vitamin A-binding proteins. Annu Rev Nutr. 16:205–234. [DOI] [PubMed] [Google Scholar]
- Li E, Qian SJ, Winter NS, d’Avignon A, Levin MS, Gordon JI. 1991. Fluorine nuclear magnetic resonance analysis of the ligand binding properties of two homologous rat cellular-retinol-binding proteins expressed in Escherichia coli. J Biol Chem. 266(6):3622–3629. [PubMed] [Google Scholar]
- Li E, Tso P. 2003. Vitamin A uptake from foods. Curr Opin Lipidol. 14(3):241–247. [DOI] [PubMed] [Google Scholar]
- Li E, Demmer LA, Sweetser DA, Ong DE, Gordon JI. 1986. Rat cellular retinol-binding protein II: use of a cloned cDNA to define its primary structure, tissue-specific expression, and developmental regulation. Proc Natl Acad Sci USA. 83(16): 5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissoos TW, Davis AE, Levin MS. 1995. Vitamin A trafficking in Caco-2 cells stably transfected with cellular retinol binding proteins. Am J Physiol. 268(2 Pt 1):G224–G231. [DOI] [PubMed] [Google Scholar]
- MacDonald PN, Ong DE. 1987. Binding specificities of cellular retinol-binding protein and cellular retinol-binding protein, type II. J Biol Chem. 262(22):10550–10556. [PubMed] [Google Scholar]
- MacDonald PN, Ong DE. 1988. Evidence for a lecithin-retinol acyltransferase activity in rat small intestine. J Biol Chem. 263(25):12478–12482. [PubMed] [Google Scholar]
- Mangelsdorf DJ, Umesono K, Kliewer SA, Borgmeyer U, Ong ES, Evans RM. 1991. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell 66(3):555–561. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Masui F, Mori T. 2004. Neonatal estrogenization leads to increased expression of cellular retinol binding protein 2 in the mouse reproductive tract. Cell Tissue Res. 316(1):131–139. [DOI] [PubMed] [Google Scholar]
- McDonald KG, Leach MR, Brooke KWM, Wang C, Wheeler LW, Hanly EK, Rowley CW, Levin MS, Wagner M, Li E, Newberry RD. 2012. Epithelial expression of the cytosolic retinoid chaperone cellular retinol binding protein II is essential for in vivo imprinting of local gut dendritic cells by lumenal retinoids. Am J Pathol. 180(3):984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh CHS, Widenmaier S, Kim S-J. 2009. Glucose-dependent insulinotropic polypeptide (gastric inhibitory polypeptide; GIP). Vitam Horm. 80:409–471. [DOI] [PubMed] [Google Scholar]
- Menozzi I, Polverini E, Berni R. 2018. Deciphering protein dynamics changes along the pathway of retinol uptake by cellular retinol-binding proteins 1 and 2. Arch Biochem Biophys. 645:107–116. [DOI] [PubMed] [Google Scholar]
- Menozzi I, Vallese F, Polverini E, Folli C, Berni R, Zanotti G. 2017. Structural and molecular determinants affecting the interaction of retinol with human CRBP1. J Struct Biol. 197(3):330–339. [DOI] [PubMed] [Google Scholar]
- Mezaki Y, Fujimi TJ, Senoo H, Matsuura T. 2016. The coordinated action of lecithin:retinol acyltransferase and cellular retinol-binding proteins for regulation of vitamin A esterification. Med Hypotheses 88:60–61. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Mochizuki H, Kawai H, Ogura Y, Shimada M, Takase S, Goda T. 2007. Possible role of fatty acids in milk as the regulator of the expression of cytosolic binding proteins for fatty acids and vitamin A through PPARalpha in developing rats. J Nutr Sci Vitaminol. 53(6):515–521. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Suruga K, Kitagawa M, Takase S, Goda T. 2001. Modulation of the expression of peroxisome proliferator-activated receptor-dependent genes through disproportional expression of two subtypes in the small intestine. Arch Biochem Biophys. 389(1):41–48. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Suzuki T, Goda T. 2008. PPAR alpha and PPAR delta transactivity and p300 binding activity induced by arachidonic acid in colorectal cancer cell line Caco-2. J Nutr Sci Vitaminol. 54(4):298–302. [DOI] [PubMed] [Google Scholar]
- Molenaar R, Knippenberg M, Goverse G, Olivier BJ, de Vos AF, O’Toole T, Mebius RE. 2011. Expression of retinaldehyde dehydrogenase enzymes in mucosal dendritic cells and gut-draining lymph node stromal cells is controlled by dietary vitamin A. J Immunol. 186(4):1934–1942. [DOI] [PubMed] [Google Scholar]
- Nakshatri H, Chambon P. 1994. The directly repeated RB (G/T)TCA motifs of the rat and mouse cellular retinol-binding protein II genes are promiscuous binding sites for RAR, RXR, HNF-4 and ARP-1 homo-and heterodimers. J Biol Chem. 269:890–902. [PubMed] [Google Scholar]
- Napoli JL. 2017. Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: effects on retinoid metabolism, function and related diseases. Pharmacol Ther. 173:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry EP, Kennedy SM, Xie Y, Luo J, Crooke RM, Graham MJ, Fu J, Piomelli D, Davidson NO. 2012. Decreased body weight and hepatic steatosis with altered fatty acid ethanolamide metabolism in aged L-Fabp−/− mice . J Lipid Res. 53(4):744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer ME, Ong DE. 2000. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochim Biophys Acta 1482(1–2):57–64. [DOI] [PubMed] [Google Scholar]
- Nossoni Z, Assar Z, Yapici I, Nosrati M, Wang W, Berbasova T, Vasileiou C, Borhan B, Geiger J. 2014. Structures of holo wild-type human cellular retinol-binding protein II (hCRBPII) bound to retinol and retinal . Acta Crystallogr D Biol Crystallogr. 70(Pt 12):3226–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Mochizuki K, Goda T. 2007. Induction of histone acetylation on the CRBPII gene in perinatal rat small intestine. Biochim Biophys Acta 1770(9):1289–1296. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Suruga K, Takase S, Goda T. 2005. Developmental changes of the expression of the genes regulated by retinoic acid in the small intestine of rats. Life Sci. 77(22): 2804–2813. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Yasutake H, Mochizuki K, Yoshikawa S, Suruga K, Sugiyama H, Takase S, Goda T. 2008. Distribution and dietary induction of cellular retinol-binding protein type II along the villus-crypt axis of the rat jejunum. J Nutr Sci Vitaminol. 54(2):130–135. [DOI] [PubMed] [Google Scholar]
- Ong DE, Chytil F. 1978. Cellular retinol-binding protein from rat liver. Purification and characterization. J Biol Chem. 253(3):828–832. [PubMed] [Google Scholar]
- Ong DE, Kakkad B, MacDonald PN. 1987. Acyl-coA-independent esterification of retinol bound to cellular retinol-binding protein (type II) by microsomes from rat small intestine. J Biol Chem. 262(6):2729–2736. [PubMed] [Google Scholar]
- Ong DE, Lucas PC, Kakkad B, Quick TC. 1991. Ontogeny of two vitamin A-metabolizing enzymes and two retinol-binding proteins present in the small intestine of the rat. J Lipid Res. 32(9):1521–1527. [PubMed] [Google Scholar]
- Ong DE, MacDonald PN, Gubitosi AM. 1988. Esterification of retinol in rat liver. Possible participation by cellular retinol-binding protein and cellular retinol-binding protein II. J Biol Chem. 263(12):5789–5796. [PubMed] [Google Scholar]
- Ong DE, Newcomer ME, Chytil F. 1994. Cellular retinoid-binding proteins In: Sporn MB, Roberts AB, Goodman DS, editors. The retinoids: biology, chemistry and medicine. 2nd ed New York: Raven Press; p. 283–318. [Google Scholar]
- Ong DE. 1984. A novel retinol-binding protein from rat-purification and partial characterization. J Biol Chem. 259(3): 1476–1482. [PubMed] [Google Scholar]
- Ong DE. 1985. Vitamin A-binding proteins. Nutr Rev. 43(8): 225–232. [DOI] [PubMed] [Google Scholar]
- Qian A, Cai Y, Magee TR, Wan Y-JY. 2000. Identification of retinoic acid-responsive elements on the HNF1alpha and HNF4alpha genes. Biochem Biophys Res Commun. 276(3): 837–842. [DOI] [PubMed] [Google Scholar]
- Ross AC, Takahashi YI, Goodman DS. 1979. The binding protein for retinol from rat testis cytosol. Isolation and partial characterization. J Biol Chem. 253:6591–6598. [PubMed] [Google Scholar]
- Sapin V, Ward SJ, Bronner S, Chambon P, Doll e P. 1997. Differential expression of transcripts encoding retinoid binding proteins and retinoic acid receptors during placentation of the mouse. Dev Dyn. 208(2):199–210. [DOI] [PubMed] [Google Scholar]
- Schaefer WH, Kakkad B, Crow JA, Blair IA, Ong DE. 1989. Purification, primary structure characterization, and cellular distribution of two forms of cellular retinol-binding protein, type II from adult rat small intestine. J Biol Chem. 264(7):4212–4221. [PubMed] [Google Scholar]
- Schiefner A, Skerra A. 2015. The menagerie of human lipocalins: a natural protein scaffold for molecular recognition of physiological compounds. Acc Chem Res. 48(4):976–985. [DOI] [PubMed] [Google Scholar]
- Silvaroli JA, Arne JM, Chelstowska S, Kiser PD, Banerjee S, Golczak M. 2016. Ligand binding induces conformational changes in human cellular retinol-binding protein 1 (CRBP1) revealed by atomic resolution crystal structures . J Biol Chem. 291(16):8528–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvaroli JA, Widjaja-Adhi MAK, Trischman T, Chelstowska S, Horwitz S, Banerjee S, Kiser PD, Blaner WS, Golczak M. 2019. Abnormal cannabidiol modulates vitamin A metabolism by acting as a competitive inhibitor of CRBP1. ACS Chem Biol. 14(3):434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A 2000. Lipocalins as a scaffold. Biochim Biophys Acta 1482(1–2):337–350. [DOI] [PubMed] [Google Scholar]
- Storch J, Corsico B. 2008. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 28:73–95. [DOI] [PubMed] [Google Scholar]
- Sun EWL, Martin AM, Young RL, Keating DJ. 2019. The regulation of peripheral metabolism by gut-derived hormones. Frontiers Endocrinol. 9:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suruga K, Kitagawa M, Yasutake H, Takase S, Goda H. 2005. Diet-related variation in cellular retinol-binding protein type II gene expression in rat jejunum. Br J Nutr. 94(6): 890–895. [DOI] [PubMed] [Google Scholar]
- Suruga K, Mochizuki K, Kitagawa M, Goda T, Horie N, Takeishi K, Takase S. 1999a. Transcriptional regulation of cellular retinol-binding protein, type II gene expression in small intestine by dietary fat. Arch Biochem Biophys. 362(1):159–166. [DOI] [PubMed] [Google Scholar]
- Suruga K, Mochizuki K, Suzuki R, Goda T, Takase S. 1999b. Regulation of cellular retinol-binding protein type II gene expression by arachidonic acid analogue and 9-cis retinoic acid in Caco-2 cells. Eur J Biochem. 262(1):70–78. [DOI] [PubMed] [Google Scholar]
- Suruga K, Suzuki R, Goda T, Takase S. 1995. Unsaturated fatty acids regulate gene expression of cellular retinol-binding protein, type II in rat jejunum. J Nutr. 125(8):2039–2044. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Suruga K, Goda T, Takase S. 1998. Peroxisome proliferator enhances gene expression of cellular retinol-binding protein, type II in CACO-2 cells. Life Sci. 62(10): 861–871. [DOI] [PubMed] [Google Scholar]
- Takase S, Goda T, Shinohara H. 1993. Adaptive changes of intestinal cellular retinol-binding protein, type II following jejunum-bypass operation in the rat. Biochim Biophys Acta 1156(2):223–231. [DOI] [PubMed] [Google Scholar]
- Takase S, Ong DE, Chytil F. 1979. Cellular retinol-binding protein allows specific interaction of retinol with the nucleus in vitro. Proc Natl Acad Sci USA. 76(5):2204–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase S, Suruga K, Goda T. 2000. Regulation of vitamin A metabolism-related gene expression. Br J Nutr. 84(Suppl 2):S217–S221. [DOI] [PubMed] [Google Scholar]
- Tarter M, Capaldi S, Carrizo ME, Ambrosi E, Perduca M, Monaco HL. 2008. Crystal structure of human cellular retinol-binding protein II to 1.2 A resolution. Proteins 70(4): 1626–1630. [DOI] [PubMed] [Google Scholar]
- Theodorakis MJ, Carlson O, Muller DC, Egan JM. 2004. Elevated plasma glucose-dependent insulinotropic polypeptide associates with hyperinsulinemia in impaired glucose tolerance. Diabetes Care 27(7):1692–1698. [DOI] [PubMed] [Google Scholar]
- Thumser AE, Moore JB, Plant NJ. 2014. Fatty acid binding proteins: tissue-specific functions in health and disease. Curr Opin Clin Nutr Metab Care 17(2):124–129. [DOI] [PubMed] [Google Scholar]
- Vogel S, Mendelsohn CL, Mertz JR, Piantedosi R, Waldburger C, Gottesman ME, Blaner WS. 2001. Characterization of a new member of the fatty acid-binding protein family that binds all-trans-retinol. J Biol Chem. 276(2):1353–1360. [DOI] [PubMed] [Google Scholar]
- Wang W, Nossoni Z, Berbasova T, Watson CT, Yapici I, Lee KSS, Vasileiou C, Geiger JH, Borhan B. 2012. Tuning the electronic absorption of protein-embedded all-trans-retinal. Science 338(6112):1340–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja-Adhi MA, Palczewski G, Dale K, Knauss EA, Kelly ME, Golczak M, Levine AD, von Lintig J. 2017. Transcription factor ISX mediates the cross talk between diet and immunity. Proc Natl Acad Sci USA. 114(43):11530–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsiriroj N, Piantedosi R, Palczewski K, Goldberg IJ, Johnston TP, Li E, Blaner WS. 2008. The molecular basis of retinoid absorption: a genetic dissection. J Biol Chem. 283(20):13510–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Miyamoto S, Ogura Y, Goda T, Suruga K. 2009. Hepatocyte nuclear factor-4alpha regulates human cellular retinol-binding protein type II gene expression in intestinal cells. Am J Physiol Gastrointest Liver Physiol. 296(3): G524–G533. [DOI] [PubMed] [Google Scholar]
- Yen C-L, Nelson W, Yen M-I. 2015. Intestinal triacylglycerol synthesis in fat absorption and systemic energy metabolism. J Lipid Res. 56(3):489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti G, Berni R, Monaco HL. 1993. Crystal structure of liganded and unliganded forms of bovine plasma retinol-binding protein. J Biol Chem. 268(15):10728–10738. [PubMed] [Google Scholar]
- Zhang L, E X, Luker KE, Shao J-S, Levin MS, Suh E, Li E. 2002. Analysis of human cellular retinol-binding protein II promoter during enterocyte differentiation. Am J Physiol Gastrointest Liver Physiol. 282(6):G1079–G1087. [DOI] [PubMed] [Google Scholar]


