Abstract
COVID-19 is a respiratory disease caused by SARS-CoV-2, an enveloped positive sense RNA virus. The SARS-CoV-2 spike glycoprotein, human angiotensin-converting enzyme 2 (ACE2) and human transmembrane protease serine 2 (TMPRSS2) are essential for the host cell-mediated viral entry. Targeting these proteins represent viable options to stop the first stage of infection and transmission. Hence, 97 alkaloids from African medicinal plants with reported antiviral activity were evaluated for this purpose via in silico studies. These alkaloids were docked for their interactions with SARS-CoV-2 spike glycoprotein, ACE2, and TMPRSS2. Top 20 alkaloids with highest binding affinities were further screened for their interactions with spike glycoprotein of SARS-CoV and MERS-CoV, and with ACE2-SARS-CoV-2 receptor-binding domain complex (ACE2-RBD). The energy profiling, molecular dynamics simulation (MDS), binding free energy base on Molecular Mechanics/Generalized Born Surface Area (MMGBSA), clustering of MDS trajectories, and virtual physicochemical and pharmacokinetic screening of the best docked alkaloids were performed. Results revealed that more than 15 alkaloids interacted better than the reference compounds. 10–Hydroxyusambarensine and Cryptospirolepine were docked in a similar binding pattern to the S1-specificy pocket of TMPRSS2 as camostat (reference inhibitor). The strong binding affinities, stability of the alkaloid-protein complexes and amino acid interactions displayed by cryptospirolepine, 10-hydroxyusambarensine, and cryptoquindoline with important binding hotspots of the proteins suggest these alkaloids have the potential of altering the capacity of SARS-CoV-2 membrane mediated host cell entry. Further in vitro and in vivo evaluation of these “drug-like” alkaloids as potential inhibitors of coronavirus cell entry is proposed.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, ACE2, TMPRSS2, spike glycoprotein, alkaloid
1. Introduction
Coronaviruses refer to a family of viruses that cause diverse forms of diseases in multiple animal hosts. Four main genera of coronaviruses – alpha, beta, gamma, and delta – infect animals but two of these genera, alpha and beta, transmit to humans (Paules et al., 2020). Human coronavirus (HCoV) causes respiratory tract complications which vary in severity. Prevalence of HCoVs and their association with upper and lower respiratory tract diseases ranges from 3.3% to 16% (Cabeça et al., 2013; Gaunt et al., 2010; Lu et al., 2012; Walsh et al., 2013). Alpha coronaviruses (HCoV-NL63 and HCoV-229E) and beta coronaviruses (HCoV-OC43 and HCoV-HKU1) are prevalent in human population and responsible for mild respiratory illnesses such as sore throat, cough, and common cold (Lu et al., 2012; Walsh et al., 2013), while beta coronaviruses like MERS-CoV (Middle East respiratory syndrome coronavirus), SARS-CoV (severe acute respiratory syndrome coronavirus), and SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) are rarer in human but more deadly (Letko et al., 2020). MERS-CoV, SARS-CoV, and SARS-CoV-2 are believed to have initially transmitted from a natural host, purported to be bat, via intermediate mammalian hosts to humans (Al-Tawfiq and Memish, 2014; Bolles et al., 2011; Zhou et al., 2020). The novel human coronavirus, SARS-CoV-2, was first identified in Wuhan, China in the year 2019 (WHO, 2020) and was reported as the causative agent of COVID-19 (coronavirus disease 2019).
SARS-CoV-2, an enveloped positive-sense RNA virus (Gorbalenya et al., 2020), utilizes its spike glycoprotein for receptor recognition and membrane fusion to initiate infection (Gallagher and Buchmeier, 2001; Simmons et al., 2013). The S1 subunit of coronavirus spike glycoprotein contains the receptor binding domain (RBD), which binds to the peptidase domain (PD) of host cell angiotensin-converting enzyme 2 (ACE2) (Li et al., 2005), while S2 subunit of the spike glycoprotein ensures membrane fusion with host cell (Tong, 2009). The human ACE2 is a type I integral membrane glycoprotein whose biological function is to process angiotensin, a peptide hormone that modulates vasoconstriction and blood pressure, into its matured form. But ACE2 is hijacked by some coronaviruses, like SARS-CoV and SARS-CoV-2, as entry point to host cell (Hoffmann et al., 2020). Relative to SARS-CoV, SARS-CoV-2 is believed to bind human ACE2 more efficiently, thereby increasing its chance of human to human transmission (Wan et al., 2020). In addition, SARS-CoV-2 just like SARS-CoV, utilizes host cell transmembrane protease serine 2 (TMPRSS2) to cleave its spike glycoprotein (Glowacka et al., 2011; Heurich et al., 2014; Hoffmann et al., 2020; Shulla et al., 2011) to ensure the fusion of host cell and virus membranes (Hoffmann et al., 2020). These interactions of SARS-CoV-2 spike glycoprotein with ACE2 and TMPRSS2 ensure the entry of the viral RNA genome into the host cell, where viral replication takes place. Both ACE2 and TMPRSS2 are well expressed in nasal and bronchial epithelium (Bertram et al., 2012).
Potential therapeutic approaches to curtail COVID-19 include development of SARS-CoV-2 spike glycoprotein-based vaccine; usage of transmembrane protease serine 2 (TMPRSS2) inhibitor to block the cleavage of the spike glycoprotein; usage of anti-ACE2 peptides or antibody to block ACE2 receptor surface; and introduction of soluble form of ACE2 to competitively bind with SARS-CoV-2, slow down viral entry into cells, and protect the lung from injury through its unique enzymatic function (Zhang et al., 2020). Computational techniques have been applied for virtual screening of several FDA approved drugs (Elmezayen et al., 2020; Lobo-Galo et al., 2020), natural agents (Aanouz et al., 2020), and repurposing of clinically approved antiviral drugs (Adeoye et al., 2020; Boopathi et al., 2020; Hendaus, 2020; Khan et al., 2020; Muralidharan et al., 2020).
Exploration of natural products presents the option of identifying compounds with minimal side effects. Alkaloids are secondary metabolites of plants used to manage diverse diseases. Given the antiviral potentials of alkaloids from Africa medicinal plants (Dhama et al., 2018; Kudi and Myint, 1999; Meyer et al., 1997; Ogbole et al., 2018), this research seeks to investigate the potentials of alkaloids to interact and alter binding function of coronavirus spike glycoprotein, block the receptor function of ACE2 and/or inhibit cleavage function of TMPRSS2. These disruptions could serve to prevent SARS-CoV-2 binding and fusion to cell, and consequently prevent infection of cells and viral replication.
2. Materials and methods
2.1. Protein preparation
The crystal structures of proteins for the docking studies were retrieved from Protein Databank (http://www.rcsb.org) with their various identification codes: ACE2 (1R42) (Towler et al., 2004); TMPRSS2 (2OQ5) (Kyrieleis et al., 2007); SARS-CoV-2 chimeric receptor-binding domain complexed with ACE2 tagged as ACE2-RBD (6VW1) (Shang et al., 2020); SARS-CoV-2 spike glycoprotein (6VSB) (Wrapp et al., 2020); SARS-CoV spike glycoprotein (5X5B) (Yuan et al., 2017); and MERS-CoV spike glycoprotein (5X5C) (Yuan et al., 2017). All the crystal structures were prepared by removing existing ligands and water molecules while missing hydrogen atoms were added using Autodock version 4.2 program (Scripps Research Institute, La Jolla, CA). Thereafter, nonpolar hydrogens were merged while polar hydrogens were added to each protein. The process was repeated for each protein and subsequently saved into dock-able PDBQT format.
2.2. Ligand preparation
Structure Data Format (SDF) structures of reference inhibitors (MLN-4760, N-Acetyl-D-Glucosamine, Camostat, and Nafamostat) and 97 alkaloids reportedly present in African plants were retrieved from the PubChem database (www.pubchem.ncbi.nlm.nih.gov), while compounds that were not available on the database were drawn with Chemdraw version 19, and converted to mol2 chemical format using Open babel (O’Boyle et al., 2011). Polar hydrogen charges of the Gasteiger-type were assigned while the nonpolar hydrogen molecules were merged with the carbons, and the internal degrees of freedom and torsions were set to zero. The ligand molecules were then converted to dock-able format using Autodock tools.
2.3. Molecular docking
The initial virtual screening of the alkaloids for active regions of the human host cell receptor, protease and the coronaviruses spike glycoprotein, were carried out using AutoDock vina (Trott and Olson, 2010), and validated with BINDSURF (https://bio-hpc.ucam.edu/achilles/) (Sánchez-Linares et al., 2012). AutoDock vina provides a one step and faster docking analysis which relies on both empirical and knowledge-based scoring functions, for this reason it was selected for the initial virtual screening. The estimated inhibition constant for the top-ranked alkaloids and reference compounds was calculated with AutoDock 4.2 (Morris et al., 2009). Twenty-five million energy evaluations were performed for each compound with a total of ten runs using AutoDock 4.2. Lamarckian Genetic Algorithm was used and Gasteiger partial charges were added to the proteins. PDBQT form of each protein and ligand were used for exhaustive docking calculations to find the spots with best binding affinities. The pose with the best affinity was taken as the representation of the cluster. The top 20 alkaloids with higher binding affinity for ACE2 were further docked with the complex formed between ACE2 and SARS-CoV-2 spike glycoprotein receptor-binding domain (ACE2-RBD). Also, these top 20 alkaloids were docked with the spike glycoproteins of SARS-CoV and MERS-CoV. The molecular interactions between these alkaloids and proteins were viewed with Discovery Studio Visualizer version 16.
2.4. Energy profile calculations
The energy profiles of the ligand-protein complex in the selected clusters with the best docked poses were calculated by BINDSURF. The number of poses and best poses in the selected clusters from the population cluster for each docked ligand and the binding coordinates in the cluster were calculated by the clustering tool.
2.5. Molecular dynamics simulation
Molecular Dynamics Simulation (MDS) using NAMD software (Phillips et al., 2005) was done on the resulted complexes from AutoDockVina docking step. The files required for the MDS were prepared using CHARMM-GUI web server (Brooks et al., 2009; Lee et al., 2016). Each complex was minimized for 10,000 steps then a production run for 100 ns was performed. The simulation temperature was set at 310 K, and salt concentration was set at physiological concentration of 0.154 M NaCl.
The results from the MDS were analyzed using VMD TK console scripts for calculation of Backbone-Root Mean Square Deviation (RMSD), Per residue Root Mean Square Fluctuations (RMSF), Radius of Gyration (RoG), and Surface Accessible Surface Area (SASA) (Humphrey et al., 1996). In addition, Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) (Miller et al., 2012) calculation was performed using AmberTools 20 (Case et al., 2020). MDS Trajectories were clustered automatically using TTClust Version 4.7.2 by utilizing the elbow method to calculate the number of clusters and a representative structure for each cluster was produced (Tubiana et al., 2018). Following that was the analysis of the representative structures using Protein Ligand Interaction Profiler (PLIP) webserver to know the types and number of bonds that participated in the interaction (Salentin et al., 2015).
2.6. Physicochemical and pharmacokinetic study
The alkaloids that demonstrated highest binding affinities to the proteins were subjected to Lipinski filter to predict their membrane permeability and ease of intestinal absorption via passive diffusion. To pass this test, the alkaloids were expected not to violate more than one criteria for drug-likeness of Lipinski’s rule of five: octanol-water partition coefficient (log P) ≤5; molecular mass ≤500; hydrogen bond donors (OH + NH count) ≤5; and hydrogen bond acceptors (O + N atom count) ≤10 (Lipinski et al., 1997). The pharmacokinetic studies were performed with the SDF file and canonical SMILES of the selected alkaloids. The predicted absorption, distribution, metabolism, excretion, and toxicity (ADMET) studies were analyzed using the admetSAR webserver (Cheng et al., 2012).
3. Results
3.1. Molecular docking
The screened 97 alkaloids from African medicinal flora demonstrated varying degrees of binding with SARS-CoV-2 spike glycoprotein, human ACE2, and protease TMPRSS2 (Table S1, Supporting material). From the docking scores obtained from the analysis, ranking based on negative low value of binding free energy, and binding orientation in the respective proteins, a hit list of top 20 alkaloids was defined. The binding energies of alkaloids with highest binding affinities for complex formed between ACE2 and SARS-CoV-2 spike glycoprotein receptor-binding domain (ACE2-RBD) are presented in Table 1. The binding energies of alkaloids with highest binding affinities for spike glycoproteins of SARS-CoV-2, SARS-CoV, and MERS-CoV are presented in Table 2, while the estimated inhibition constant of the three top-ranked alkaloids with highest binding affinities for ACE2, TMPRSS2, and SARS-CoV-2 spike glycoprotein is presented in Table 3.
Table 1.
Binding energies of reference inhibitors and top 20 bioactive alkanoids with human Angiotensin-Converting Enzyme 2 (ACE2), Transmembrane Protease Serine 2 (TMPRSS2), and ACE2-Spike Receptor Binding Domain complex (ACE2-RBD).
| S/No | Plant species (Family) | Class of compound | Compounds | Binding energies (kcal/mol) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| ACE2 |
TMPRSS2 |
ACE2-RBD |
|||||||
| Vina | Bindsurf | Vina | Bindsurf | Vina | Bindsurf | ||||
| R1 | MLN-4760 | –7.0 | 7.2 | ND | ND | ND | ND | ||
| R2 | N-Acetyl-d-glucosamine | –5.6 | –5.7 | ND | ND | ND | ND | ||
| R3 | Camostat | ND | ND | –7.6 | –7.8 | ND | ND | ||
| 1 | Cryptolepis sanguinolenta (Periplocaceae) | Cryptolepines | Cryptospirolepine | –10.7 | –10.8 | –9.9 | –10.2 | –10.7 | –10.9 |
| 2 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | 10-Hydroxyusambarensine | –10.4 | –10.3 | –10.4 | –10.7 | –10.5 | –9.9 |
| 3 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | Strychnopentamine | –9.9 | –10.3 | –8.8 | –8.9 | –8.4 | –9.8 |
| 4 | Cryptolepis sanguinolenta (Periplocaceae) | Cryptolepines | Biscryptolepine | –9.8 | –9.9 | –8.9 | –8.9 | –10.1 | –9.9 |
| 5 | Cryptolepis sanguinolenta (Periplocaceae) | Cryptolepines | Cryptoquindoline | –9.7 | –9.9 | –9.7 | –9.9 | –9.9 | –9.8 |
| 6 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | Isostrychnopentamine | –9.5 | –9.8 | –8.7 | –8.8 | –9.2 | –9.8 |
| 7 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | Chrysopentamine | –9.4 | –9.7 | –8.6 | –8.9 | –10.5 | –8.8 |
| 8 | Triphyophyllum peltatum, | Naphthoisoquinolines | Jozipeltine A | –9.3 | –9.7 | –8.4 | –8.7 | –10.4 | –8.9 |
| 9 | Monodora angolensis (Annonaceae) | Indole alkaloids | Annonidine F | –9.0 | –9.3 | –7.8 | –7.9 | –9.2 | –8.8 |
| 10 | Atalantia monophylla Corrêa | Acridone alkaloid | Atalaphylline | –8.8 | –8.9 | –7.9 | –7.6 | –8.2 | –8.6 |
| 11 | Corydalis saxicola Bunting | Protoberberine-type | Coptisine | –8.7 | –8.9 | –8.2 | –8.6 | –8.7 | –8.5 |
| 12 | Triphyophyllum peltatum (Dioncophyllaceae) | Naphthoisoquinolines | Dioncophylline B | –8.6 | –8.9 | –7.7 | –7.8 | –8.6 | –8.6 |
| 13 | Corydalis saxicola Bunting | Protoberberine-type | dehydroapocavidine | –8.6 | –8.7 | –7.6 | –7.9 | –7.8 | –8.4 |
| 14 | Fagara zanthoxyloides (Rutaceae) | Indole alkaloids | Alstonine | –8.5 | –8.8 | –7.9 | –7.8 | –8.9 | –8.6 |
| 15 | Triphyophyllum peltatum (Dioncophyllaceae) | Naphthoisoquinolines | 5′-O-Demethyl-dioncophylline A | –8.5 | –8.5 | –7.3 | –7.6 | –8.6 | –7.9 |
| 16 | Triphyophyllum peltatum (Dioncophyllaceae) | Naphthoisoquinolines | Dioncophylline A | –8.5 | –8.9 | –7.0 | –7.4 | –8.3 | –8.3 |
| 17 | Glossocalyx brevipes (Siparunaceae) | Indole alkaloids | Liriodenine | –8.4 | –8.7 | –7.3 | –7.8 | –8.6 | –8.1 |
| 18 | Camptotheca acuminate Decaisne | Quinoline | Camptothecin | –8.4 | –8.9 | –8.1 | –8.8 | –7.8 | –8 |
| 19 | Magnolia gradiflora | Aporphine | Lanuginosine. | –8.4 | –8.8 | –7.5 | –7.9 | –8.5 | –8.5 |
| 20 | Rhigiocarya racemifera | Oxoaporphine | Ancistrocladidine | –8.4 | –8.9 | –7.5 | –7.5 | –8.6 | –8 |
R1, R2, and R3 are reference inhibitors. Values in bold are for alkaloids with highest binding affinities for the corresponding proteins.
ND = not determined.
Table 2.
Binding energies of reference inhibitor and top 20 bioactive alkaloids from African plants with the spike glycoprotein of coronaviruses.
| Binding energies (kcal/mol) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 |
SARS-CoV |
MERS-CoV |
|||||||
| S/No | Plant species (Family) | Class of compound | Compounds | Vina | Bindsurf | Vina | Bindsurf | vina | Bindsurf |
| R1 | Nafamostat | –7.0 | –7.2 | –8.5 | –8.7 | 9.7 | 9.6 | ||
| 1 | Cryptolepis sanguinolenta (Periplocaceae) | Cryptolepines | Cryptospirolepine | –10.6 | –10.9 | –12.0 | –12.1 | –11.3 | –11.4 |
| 2 | Cryptolepis sanguinolenta (Periplocaceae) | Cryptolepines | Isocryptolepine | –9.7 | –9.9 | –8.6 | –8.8 | –9.1 | –9.2 |
| 3 | Cryptolepis sanguinolenta (Periplocaceae) | Cryptolepines | Cryptoquindoline | –9.5 | –9.8 | –9.7 | –9.5 | –10.1 | –10.4 |
| 4 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | 10 –Hydroxyusambarensine | –9.4 | –9.9 | –10.0 | –10.1 | –11.4 | –11.1 |
| 5 | Fagara zanthoxyloides (Rutaceae) | Indole alkaloids | Fagaronine | –9.3 | –9.8 | –7.4 | –7.6 | –8.2 | –8.6 |
| 6 | Triphyophyllum peltatum, | Naphthoisoquinolines | Jozipeltine A | –9.3 | –9.8 | –8.8 | –8.8 | –9.4 | –9.3 |
| 7 | Triphyophyllum peltatum (Dioncophyllaceae) | Naphthoisoquinolines | 5′–O–Demethyl–dioncophylline A | –8.7 | –8.8 | –7.5 | –7.2 | –8.3 | –8.6 |
| 8 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | Chrysopentamine | –8.6 | –8.9 | –9.0 | –9.1 | –10.3 | –10.1 |
| 9 | Monodora angolensis (Annonaceae) | Indole alkaloids | Annonidine F | –8.4 | –8.8 | –8.3 | –8.5 | –10.0 | –10.2 |
| 10 | Triphyophyllum peltatum (Dioncophyllaceae) | Naphthoisoquinolines | Dioncopeltine A | –8.3 | –8.6 | –7.5 | –7.6 | –9.2 | –9.9 |
| 11 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | Isostrychnopentamine | –8.2 | –8.5 | –8.6 | –8.7 | –9.7 | –9.8 |
| 12 | Sida acuta (Malvaceae) | Cryptolepines | Cryptolepine | –8.2 | –8.6 | –7.2 | –7.4 | –9.7 | –9.3 |
| 13 | Ancistrocladus tanzaniensis (Acistrocladaceae) | Naphthoisoquinolines | Ancistrotanzanine C | –8.1 | –8.4 | –7.9 | –7.0 | 9.3 | 9.2 |
| 14 | Cryptolepis sanguinolenta (Periplocaceae) | Cryptolepines | Quindoline | –8.1 | –8.6 | –7.2 | –7.8 | –9.2 | –9.7 |
| 15 | Strychnos usambarensis (Loganiaceae) | Indole alkaloids | Strychnopentamine | –7.9 | –7.9 | –8.7 | –8.3 | –9.4 | –9.4 |
| 16 | Ancistrocladus robertsoniorum (Acistrocladaceae) | Naphthoisoquinolines | Ancistrobertsonine A | –7.9 | –8.3 | –8.0 | –8.5 | –7.5 | –7.7 |
| 17 | Ancistrocladus robertsoniorum (Acistrocladaceae) | Naphthoisoquinolines | Ancistrobertsonine B | –7.9 | –8.1 | –8.1 | –8.0 | –8.0 | –8.8 |
| 18 | Ancistrocladus tanzaniensis (Acistrocladaceae) | Naphthoisoquinolines | Ancistrocladidine | –7.9 | –8 | –7.5 | –7.3 | –8.5 | –8.9 |
| 19 | Ancistrocladus tanzaniensis (Acistrocladaceae) | Naphthoisoquinolines | Ancistrotectorine | –7.9 | –8.5 | –7.2 | –7.2 | –8.1 | –8.3 |
| 20 | Triphyophyllum peltatum (Dioncophyllaceae) | Naphthoisoquinolines | Dioncophylline A | –7.9 | –8 | –7.4 | –7.7 | –9.1 | –9.8 |
R1 is reference inhibitor. Values in bold are for alkaloids with highest binding affinities for the corresponding proteins.
Table 3.
Inhibition constant (Ki) of 3 top-ranked alkaloids with highest affinities for ACE2, TMPRSS2 and SARS-COV-2 spike glycoprotein.
| S/No | Compounds | Inhibition constant (µM) |
||
|---|---|---|---|---|
| ACE2 | TMPRSS2 | SARS-COV-2 S glycoprotein | ||
| R1 | MLN-4760 | 25.62 | ND | ND |
| R2 | N-Acetyl-d-glucosamine | 65.02 | ND | ND |
| R3 | Camostat | ND | 1.56 | ND |
| R4 | Nafamostat | ND | ND | 63.03 |
| 1 | Cryptospirolepine | 2.67 | 6.55 | 23.21 |
| 2 | 10 -Hydroxyusambarensine | 5.81 | 3.20 | ND |
| 3 | Strychnopentamine | 4.45 | ND | ND |
| 4 | Isocryptolepine | ND | ND | 18.11 |
| 5 | Cryptoquindoline | ND | 3.40 | 15.80 |
R1, R2, R3, and R4 are reference inhibitors. ND = not determined.
The results from this study revealed that MLN-4760 and N-Acetyl-d-glucosamine (reference inhibitors) interacted with human ACE2 with binding energy of −7.0 and −5.6 Kcal/mol and estimated inhibition constant of 25.62 and 65.02 µM, respectively (Tables 1 and 3). It was observed that all the top 20 alkaloids interacted with ACE2 with binding affinity higher than the two reference inhibitors used in this study. Cryptospirolepine, 10–hydroxyusambarensine, and strychnopentamine showed the best binding to ACE2 with energies of −10.7, −10.4, and −9.9 Kcal/mol and estimated inhibition constant of 2.67, 5.81, and 4.45 µM, respectively (Tables 1 and 3).
Base on the estimated inhibition constant, the 3 top-ranked alkaloids especially cryptospirolepine had lower values than the reference compounds (MLN-4760 and N-Acetyl-d-glucosamine). Furthermore, cryptospirolepine, 10–hydroxyusambarensine, and chrysopentamine showed the best binding affinities to the ACE2-RBD with energies of −10.7, −10.5, and −10.5 Kcal/mol, respectively (Table 1).
Fifteen of the top-ranked alkaloids interacted with TMPRSS2 with binding affinities better than that camostat (–7.6 Kcal/mol), a reference inhibitor of TMPRSS2. The interaction of 10–hydroxyusambarensine, cryptospirolepine, and cryptoquindoline to TMPRSS2 demonstrated highest binding affinities with energies of −10.4, −9.9, and −9.7 Kcal/mol (Table 1). Though the estimated inhibition constant of camostat for TMPRSS2 (1.56 µM) was lower compared to these 3 top-ranked alkaloids (Table 3).
The result also revealed that nafamostat, the reference inhibitor for spike glycoprotein, had better binding tendency to the spike glycoprotein of SARS-CoV and MERS-CoV than SARS-CoV-2. Also, more than ten of the top-ranked alkaloids have higher binding affinities to the spike glycoproteins than nafamostat (Table 2). The alkaloids, cryptospirolepine, isocryptolepine, and cryptoquindoline interacted with the spike glycoprotein of SARS-CoV-2 with better binding affinities and lower estimated inhibition constant than the reference compound (Tables 2 and 3). Cryptospirolepine and 10–hydroxyusambarensine demonstrated highest affinities to the spike glycoprotein of SARS-CoV and MERS-CoV (Table 2).
3.2. Interaction of alkaloids with amino acid residues of target proteins
The alkaloids with the highest binding affinities interacted with various the amino acid residues of human ACE2 and TMPRSS2, and these are represented in Table 4. Likewise, the amino acid residues of spike glycoprotein of SARS-CoV-2, SARS-CoV, and MERS-CoV interacted with top-binding alkaloids as presented in Table 5.
Table 4.
Interacting amino acid residues of human ACE2 and TMPRSS2 with the top-binding alkaloids from African plants.
|
Compound |
Protein target | Residues involved in hydrophobic interactions (bond distance, Å) | Residues involved in hydrogen bonding (bond distance, Å) |
|---|---|---|---|
| MLN-4760 | ACE2 | ASP382 (4.33) HIS401 (2.17) PHE40 (4.7) PHE390 (5.04,4.20) ARG393(5.45) | ASN394 (2.17) ALA348 (2.37) ASP382 (2.61) ASP350 (2.90) TRY385 (3.17) |
| N-acetyl-D-glucosamine | ND | LYS470 (2.13) ILE468 (2.87) TRP473(2.23) PHE473 (3.18) ASN188 (2.97) | |
| Cryptospirolepine | HIS378 (4.43) HIS401 (5.15) PHE40 (4.84) ARG393(5.68) | ALA348 (2.83) ASN394 (3.18) | |
| 10-Hydroxyusambarensine | ALA348 (5.21) HIS378 (4.54) HIS401 (4.72) PHE40 (5.33) PHE390 (3.88) | PRO346 (2.32) GLU375 (2.77) ASN394 (1.98) | |
| Camostat | TMPRSS2 | ND | ARG41 (3.23, 3.57) SER195 (2.88) ALA190 (2.61) ASP189 TRP215 (2.90) GLN192 (2.76, 2.49) |
| Cryptospirolepine | SER151 (2.55) TYR149 (5.54, 5.26, 5.22) ILE75 (4.78) | SER151 (2.02, 4.03) HIS40 SER39 (4.10) | |
| 10-Hydroxyusambarensine | ALA190 (5.09) HIS96 (4.68) HIS57 (4.65) ARG41 (4.371) TRP215 (3.63) GLN192 (4.12) CYS191 (4.53) | SER195 (2.32) SER214 (2.72) ASP189 (2.62) ALA190(2.27) |
ACE2 = Angiotensin-Converting Enzyme 2; TMPRSS2 = Transmembrane protease serine 2.
ND = Not detected.
Table 5.
Interacting amino acid residues of spike glycoprotein of coronaviruses with the top-binding alkaloids from African plants.
|
Compound |
Coronavirus | Residues involved in hydrophobic interactions (bond distance, Å) | Residues involved in hydrogen bonding (bond distance, Å) |
|---|---|---|---|
| Nafamostat | SARS-Cov-2 | THR286 (3.02) GLN218 (3.15) PHE220 (5.87) | LYS300 (3.36) SER297 (2.33) |
| Cryptospirolepine | VAL860 (5.12) LEU828 (4.12) THR732 (4.14) HIS1058 (5.01) PRO863 (4.11) PRO862 (3.80) ASP867 (2.90) | ND | |
| Isocryptolepine | LYS417 (4.33) ARG403 (2.33) ILE418 (4.79) TYR453 (6.13) | (2.33) | |
| Nafamostat | SARS-CoV | PRO575 (4.00, 5.39) SER574 (5.00) PHE558 (2.00) ASP560 (2.02) ASP554 (3.72) |
ASP554 (2.51) |
| Cryptospirolepine | TYR300 (2.02) LEU948 (4.88, 5.78) PHE837 (5.79, 4.52, 3.75) LYS836 (5.38) ARG982 (4.64) | ND | |
| 10-Hydroxyusambarensine | THR535 (5.15, 5.38) PRO575 SER574 (3.95, 3.69, 4.76) PHE578 (4.31) CYS576 VAL308 (5.33) | THR535 (2.24) CYS576 (3.72) | |
| Nafamostat | MERS-CoV | SER51 (3.48) HIS348 (5.43) HIS670 (4.12) ILE337 (5.37) PHE354 (3.89) LEU344 (5.12) ARG335 (5.07) | SER51 (1.95) ASN342 (2.67) HIS670 (2.80) HIS348 (3.55) |
| 10-Hydroxyusambarensine | ASN342 (2.72) HIS348 (5.11, 4.88) HIS670 (5.19, 5.38) ILE337(5.03, 5,08, 5.15) LEU344 (5.36, 5,35) LYS668 (4.25) LYS668 (4.42, 5.23) PHE341(4.97) | SER51 (3.36, 4.13) PHE354 (2.72) | |
| Cryptospirolepine | ILE337 (5.50) PHE354 (5.03) ASP49 (3.79) | GLN78 (4.05) SER51 (3.14) |
ND = Not detected.
The interacted residues of ACE2 and TMPRSS2 with respective ligand groups were majorly through hydrophobic and hydrogen bond interactions (Table 4). Few hydrogen bonds below bond distance of 3.40 Å were observed with coronaviruses spike glycoproteins (Table 5).
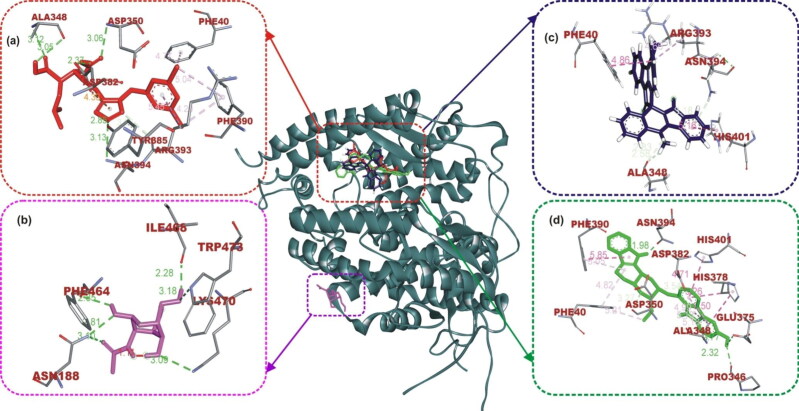
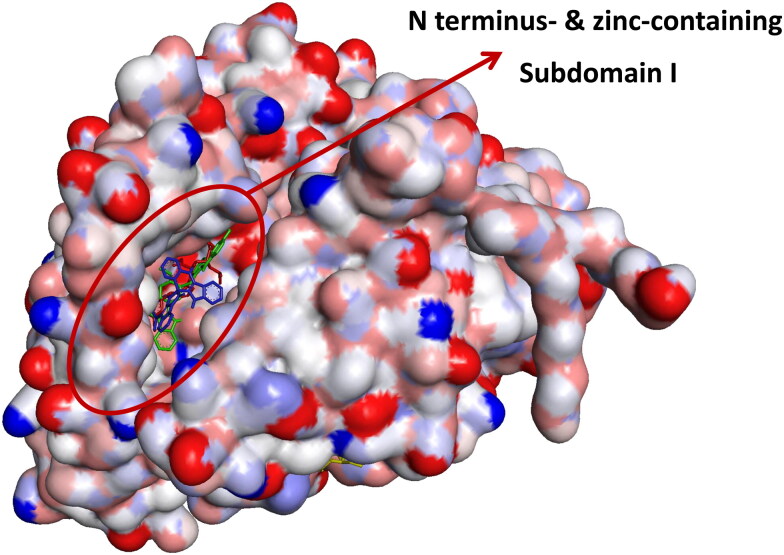
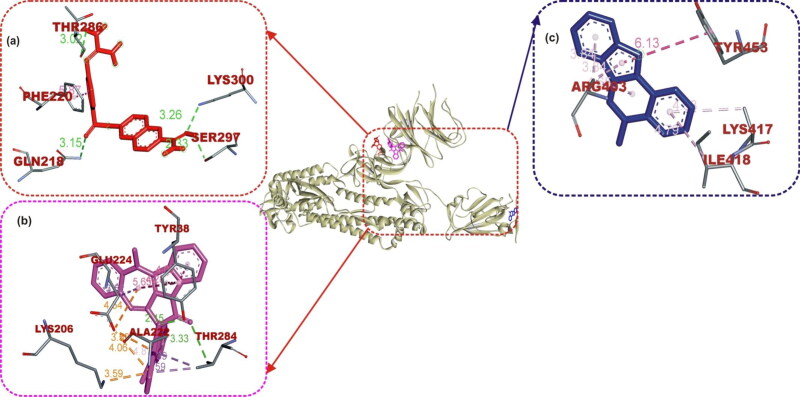
The ligand-protein binding interaction showed that MLN-4760 was docked into the N terminus- and zinc-containing subdomain I of ACE2 (Figure 1(a) and 2), while N-acetyl-D-glucosamine interacted with the amino residues in the C terminus-containing subdomain II (Figure 1(b)). MLN-4760 had Pi-Alkyl interaction with Phe40, Phe390, and His401, a carbon hydrogen interaction to Arg393, a Pi-Anion interaction to Asp382, while the remaining residue interacted via hydrogen bond (Figure 1(a)). N-Acetyl-D-glucosamine interacted solely via conventional hydrogen bond to the entire amino acid residue (Figure 1(b)). Cryptospirolepine the topmost docked alkaloid and 10-hydroxyusambarensine was docked into the subdomain I of ACE2 in a similar manner as MLN-4760 (Figures 1(c,d) and 2). Cryptospirolepine interacted via carbon and Pi-donor hydrogen bond to Ala348 and Arg393, respectively. It further interacted via Pi-Pi stacking to Phe40 and His401 and via Pi-Alkyl interaction to Arg393 (Figure 1(c)). 10–hydroxyusambarensine interacted to Pro346, Glu375, and Asn394 via hydrogen bond. Ala348, Asp382, and Asp350 were seen in a carbon hydrogen interaction, while a Pi-Pi T-shaped interaction was observed between His401, Phe40, Phe390, and 10–hydroxyusambarensine (Figure 1(d)).
Figure 1.
The interactive view of ligands in binding cavity of human ACE2. Ligands in sticks representation are presented by colors: (a) Red: MLN-4760, (b) Yellow: N-Acetyl-D-Glucosamine, (c) Blue: Cryptospirolepine, (d) Green: 10–Hydroxyusambarensine. Types of interactions are represented by Green-dotted lines: hydrogen bond interactions, light purple-dotted line: hydrophobic interactions (Pi-Alkyl, Alkyl, and pi-stacking) purple-dotted line: Pi-Pi T Shaped, yellow-dotted lines: Pi-sulfur interactions, pi-stacking interactions. Three-letter amino acids are in red color.
Figure 2.
Surface view of ligands in binding cavity of human ACE2. (a) Green: 10–Hydroxyusambarensine; (b) Blue: Cryptospirolepine; and (c) Red: MLN-4760.
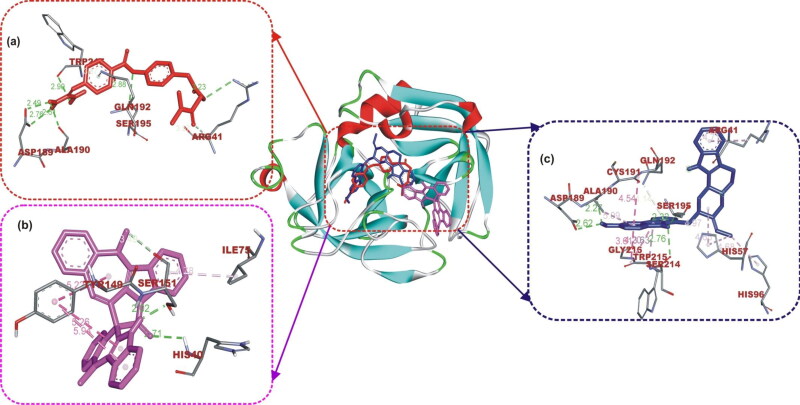
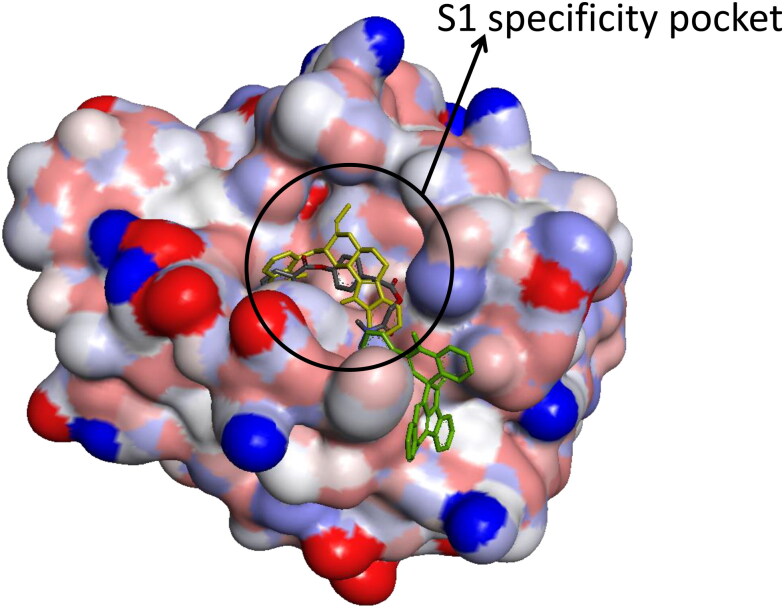
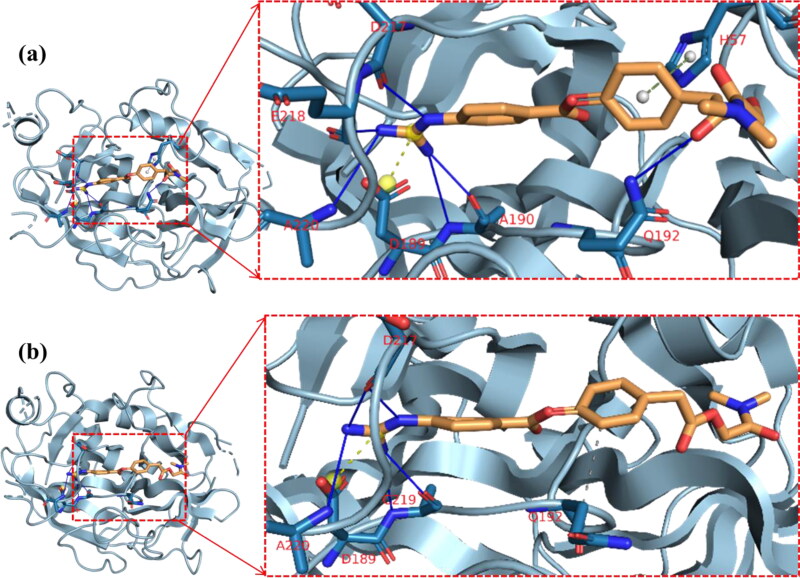
Camostat was docked into the S1-specificity pocket of TMPRSS2 (Figures 3(a) and 4). It interacted via carbon hydrogen interaction to Gln192 and via conventional hydrogen bond to five amino residues (Arg41, Ser195, Trp215, Ala190, and Asp189) to TMPRSS2. The conventional hydrogen bond was formed in the direction of the guanidine group in this order: first ester bond, second ester bond, while the last three residues interacted with amidinonitrogen of guanidine group, respectively. The phenyl ring was responsible for the carbon hydrogen interaction with Gln192 (Figure 3(a)). While cryptospirolepine was docked close to the binding pocket, 10–hydroxyusambarensine (the highest docked alkaloid to TMPRSS2) was observed in the S1-specificity pocked of TMPRSS2 (Figures 3(b) and 4), in a similar manner as camostat (Figure 3(a)). The difference observed was a Pi-Sigma interaction to Trp215, an Amide-Pi Stacking to Cys191, and an additional Pi-Alkyl interaction to His96 and His57 (Figure 3(c)).
Figure 3.
The interactive view of ligands in binding cavity of human TMPRSS2. Ligands in sticks representation are presented by colors: (a) Yellow: Camostat, (b) Black: Cryptospirolepine, (c) Green: 10-Hydroxyusambarensine. Types of interactions are represented by Green-dotted lines: hydrogen bond interactions, light purple-dotted line: hydrophobic interactions (Pi-Alkyl, Alkyl, and pi-stacking) purple-dotted line: Pi-Pi T Shaped, yellow-dotted lines: Pi-sulfur interactions, pi-stacking interactions. Three-letter amino acids are in red color.
Figure 4.
Surface view of ligands in binding cavity of human TMPRSS2. (a) Yellow: 10–Hydroxyusambarensine; (b) Black: Cryptospirolepine; (c) Green: Camostat.
Nafamostat, a reference inhibitor of the spike glycoprotein of SARS-CoV and MERS-CoV interacted with the spike glycoprotein of SARS-CoV-2 in a different manner. Nafamostat was docked into the subdomain SD1 and SD2 region of the S1 subunit of SARS-CoV spike glycoprotein (Figure 5(a)). The same inhibitor was docked into to the N-terminal domain (NTD) region of the S1 subunit of MERS-CoV spike glycoprotein (Figure 6(a)).
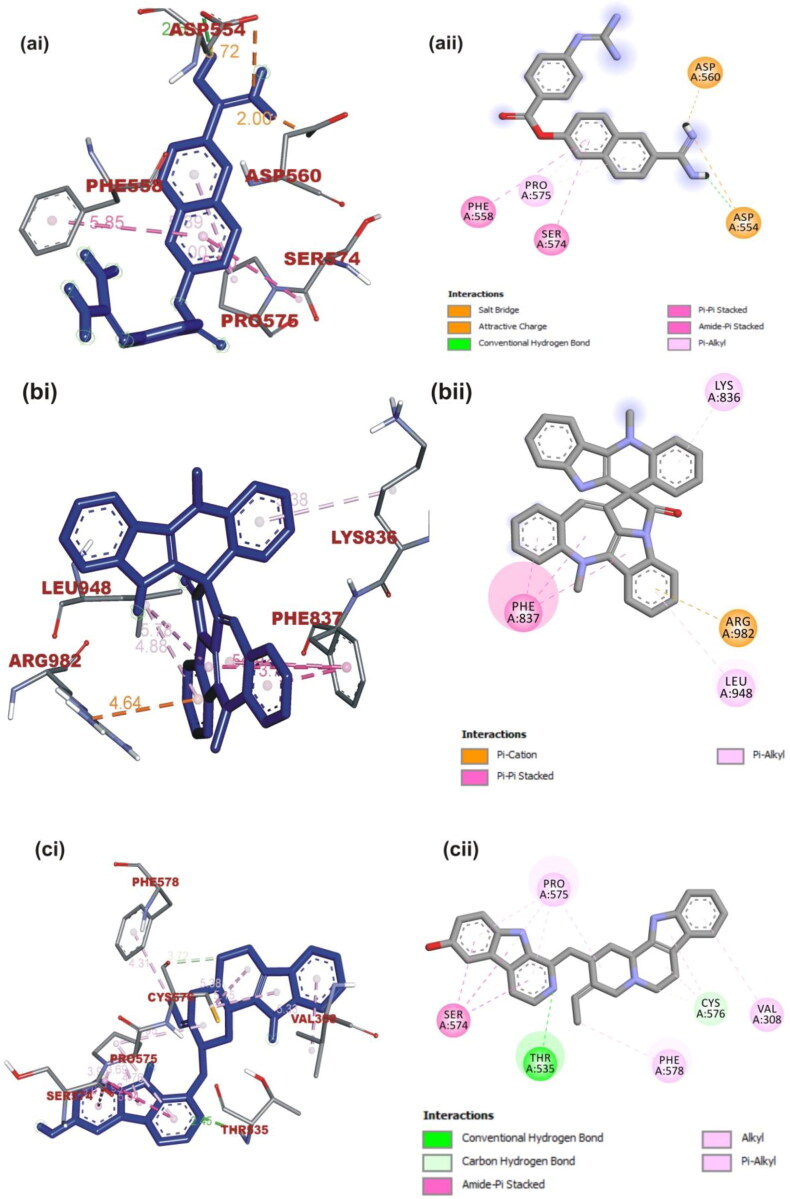
Figure 5.
(i) 3D and (ii) 2D visualization of interacting amino acid residues of SARS-CoV spike glycoprotein with ligands: (a) Nafamostat, (b) Cryptospirolepine, (c) 10–Hydroxyusambarensine. The ligands in stick representation are presented in (3D) as blue color while the (2D) as gray color.
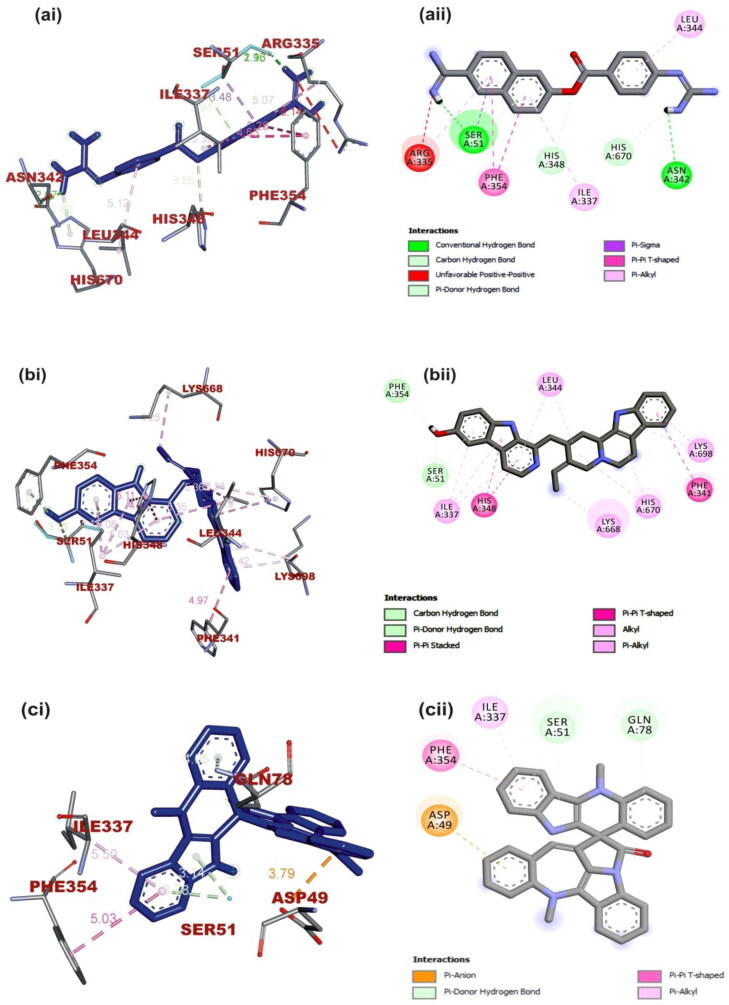
Figure 6.
(i) 3D and (ii) 2D visualization of interacting amino acid residues of MERS-CoV spike glycoprotein with ligands: (a) Nafamostat, (b) 10–Hydroxyusambarensine, (c) Cryptospirolepine. The ligands in stick representation are presented in (3D) as blue color while the (2D) as gray color.
Cryptospirolepine with the highest binding affinity for SARS-CoV-2 spike glycoprotein also interacted with SARS-CoV and MERS-CoV spike glycoprotein in a different binding pattern. It formed mainly hydrophobic interaction with SARS-CoV-2 spike glycoprotein (Figure 7(b)) A Pi-Sigma interaction with Val860, Leu828, and Thr732 was observed between cryptospirolepine and SARS-CoV-2 spike glycoprotein, others include Pi-cation, Pi-alkyl, and Pi-Pi T-Shaped interactions with His1058, Pro863, and Pro862, respectively (Figure 7(b)). Cryptospirolepine was docked into the S2 subunit region of SARS-CoV spike glycoprotein. Most of the interaction was with amino acid residue in the heptad repeat (HR1) domain of the S2 subunit (Figure 5(b)). Ala748 and Gln936 interacted via a carbon and Pi-donor hydrogen bond respectively; Arg996 and Gln939 and Asp932 via Pi-cation and Pi-anion, respectively (Figure 7(b) and Table 5). Cryptospirolepine was docked into the receptor-binding domain (RBD) of the S1 Subunit of MERS-CoV spike glycoprotein through hydrophobic interaction. A P-Pi T-shaped, Pi-alkyl, Pi-anion interaction, and Pi-donor hydrogen bond was observed with Phe354, Ile337, Asp49, and Ser51, Gln78, respectively (Figure 6(c)). 10-Hydroxyusambarensine interacted with the amino acid residue of the SD1 and SD2 region of the spike glycoprotein of SARS-CoV interacting via conventional carbon-hydrogen interaction to Thr535 and Cys576 respectively; Amide-Pi stacking to SER574 and Pi-alkyl interaction to Pro575, Phe578, and Val308 (Figure 5(c)). The same ligand interacted to the spike glycoprotein of MERS-CoV via hydrogen bond to Ser51 and Phe354; Pi-Pi T and Pi-Pi stacking Phe341 and His348, respectively; Pi-alkyl and alkyl to His670, Lys698, and Leu344 Lys668, respectively. The binding pattern exhibited by 10-hydroxyusambarensine was similar to that of Nafamostat to MERS-CoV.
Figure 7.
The interactive view of ligands in binding cavity of SARS-Cov-2 spike glycoprotein. Ligands in sticks representation are presented by colors: (a) Blue: Nafamostat, (b) Yellow: Cryptospirolepine, and (c) Red: Isocryptolepine. Types of interactions are represented by Green-dotted lines: hydrogen bond interactions, light purple-dotted line: hydrophobic interactions (Pi-Alkyl, Alkyl, and pi-stacking) purple-dotted line: Pi-Pi T Shaped, yellow-dotted lines: Pi-sulfur interactions, pi-stacking interactions. Three-letter amino acids are in red color.
3.3. Energy profile of best docked alkaloids to respective proteins
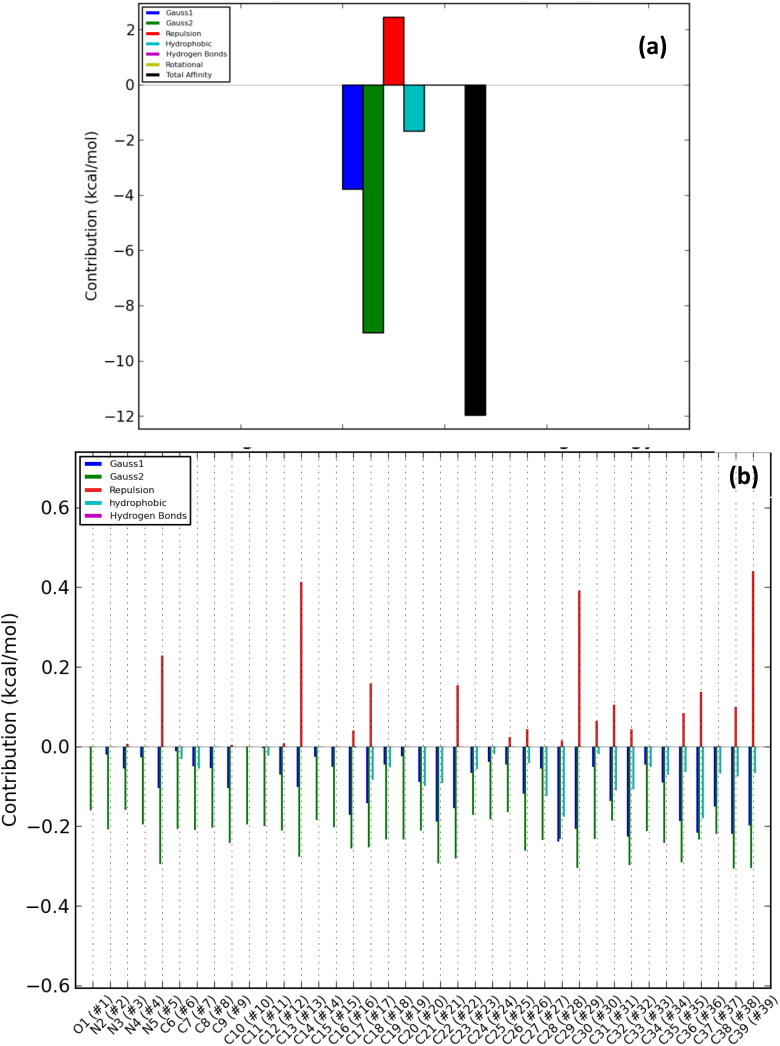
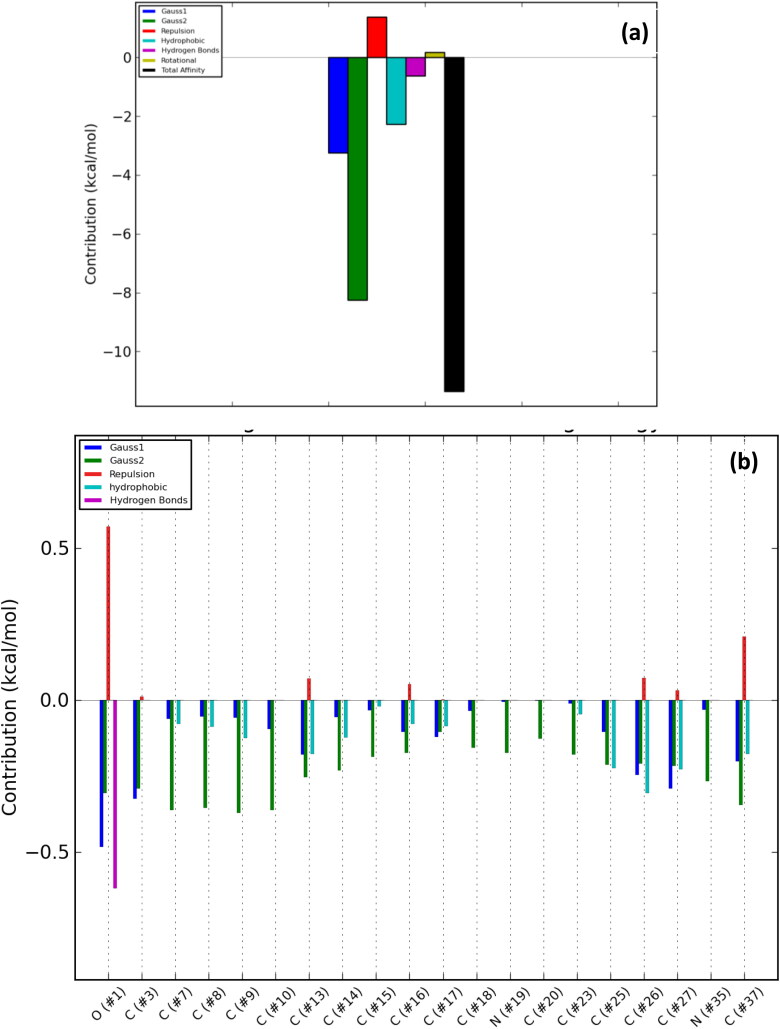
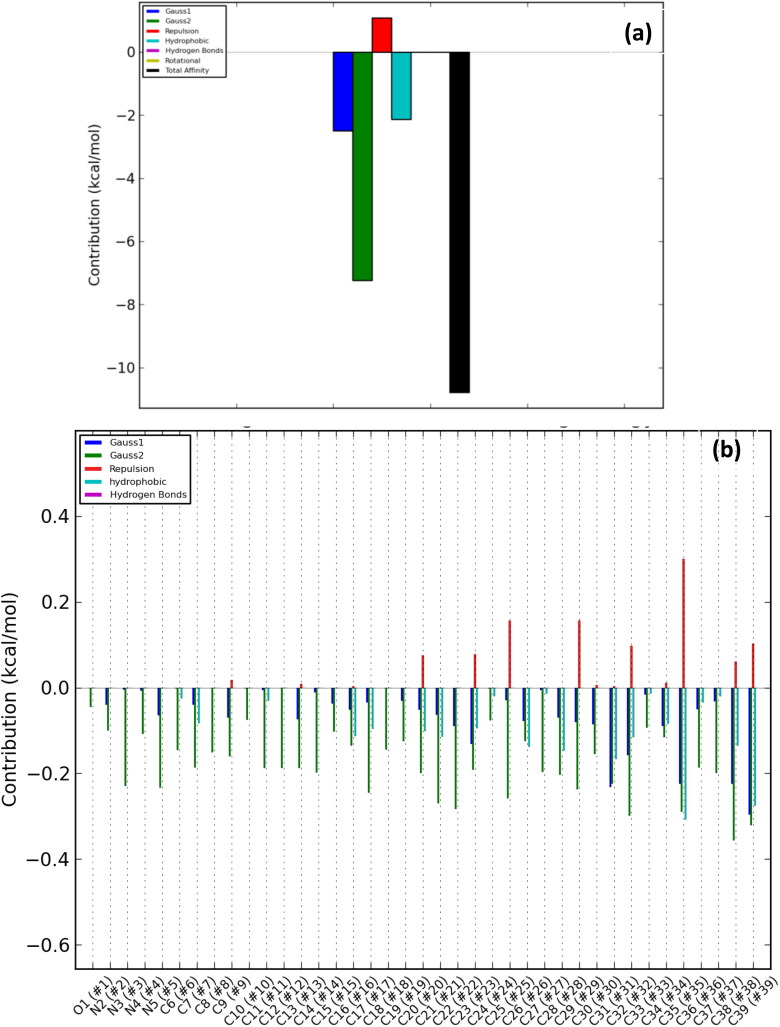
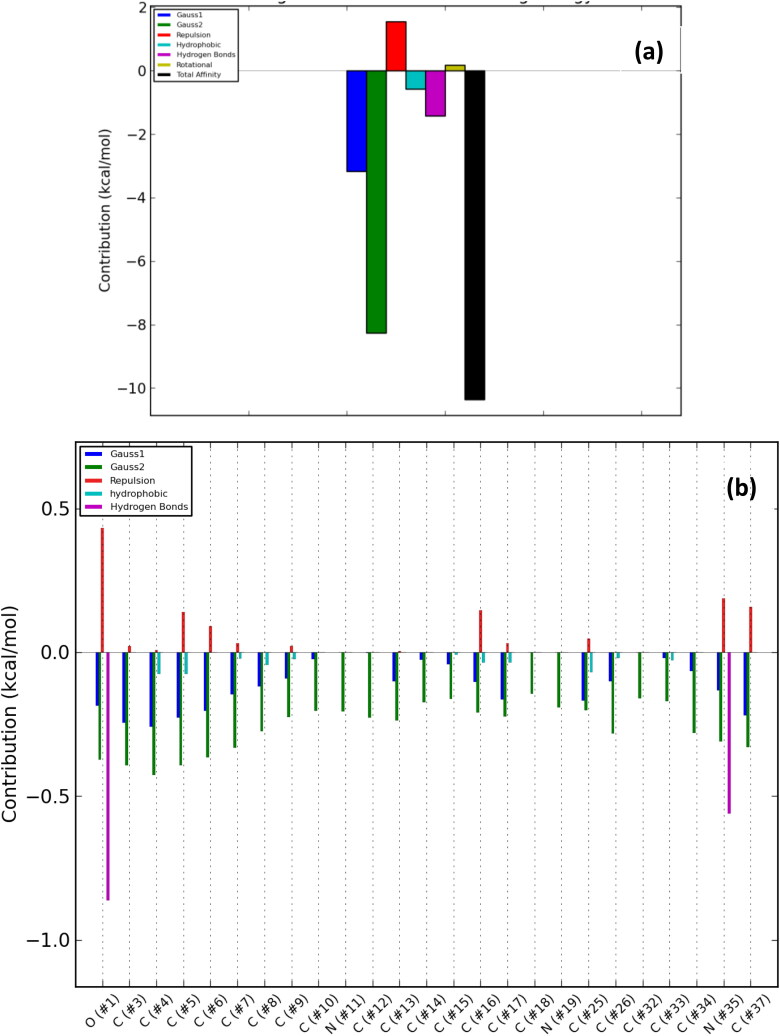
The number of poses and best poses in the selected clusters from the population cluster for each docked ligand and the binding coordinates in the cluster are shown in Table 6. The energy profiles of alkaloid-protein complex in the selected clusters with the best docked poses are shown in Figures 8–12. Gauss 1 (blue) and 2 (leaf green) bars represent the nonbonding interactions, red bar represents repulsion, light blue bar represents hydrophobic interaction, purple bar represents hydrogen bonds, light green bar represents rotational forces, while the black bar represents the total binding affinity which is a representative contribution of all bonding and nonbonding interactions between the alkaloids and the protein residues (Figures 8(a)–12(a)).
Table 6.
The binding profile of the ligand in the selected cluster.
| Receptor-ligand complex | Poses in Cluster | Best Pose | Binding site coordinate |
|---|---|---|---|
| Cryptospirolepine-6VSB | 62 | 1547 | 214.25, 205.16, 213.99 |
| Cryptospirolepine-5X5B | 89 | 1746 | 13.00, −7.77, −9.61 |
| 10 –Hydroxyusambarensine-5X5C | 87 | 338 | 34.87, −14.23, 0.60 |
| Cryptospirolepine-IR42 | 102 | 28 | 65.46, 70.54, 30.12 |
| Cryptospirolepine-2OQ5 | 92 | 132 | (–2.85, 27.80, 22.38) |
Figure 8.
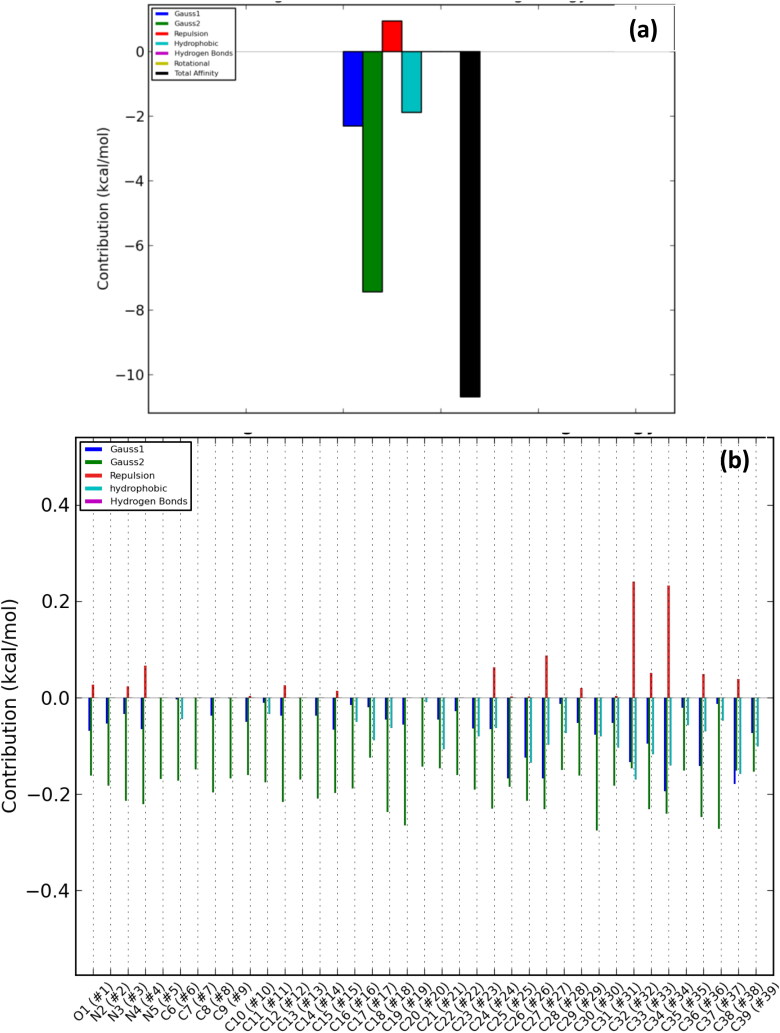
Overall energy profile of cryptospirolepine binding groups in SARS-Cov-2 spike glycoprotein: (a) Energetic contribution to the binding and (b) Energetic contributions for each atom in the ligand.
Figure 9.
Overall energy profile of cryptospirolepine binding groups in SARS-CoV spike glycoprotein: (a) Energetic contribution to the binding energy and (b) Energetic contributions for each atom in the ligand.
Figure 10.
Overall energy profile of 10 –Hydroxyusambarensine binding groups in MERS-CoV spike glycoprotein: (a) Energetic contribution to the binding energy and (b) Energetic contributions for each atom in the ligand.
Figure 11.
Overall energy profile of Cryptospirolepine binding groups in human ACE2: (a) Energetic contribution to the binding energy and (b) Energetic contributions for each atom in the ligand.
Figure 12.
Overall energy profile of Cryptospirolepine binding groups in human TMPRSS2: (a) Energetic contribution to the binding energy and (b) Energetic contributions for each atom in the ligand.
The contributions of the various type of interaction shows that of the total binding energy of −10.6 Kcal/mol exhibited by the binding of cryptospirolepine to the spike glycoprotein of SARS-CoV-2, −2.2 Kcal/mol was contributed by hydrophobic interaction, while the rest of the binding energy was contributed by nonbonding interaction mainly van der Waals, repulsive and rotational forces. A hydrophobic interaction and hydrogen bond energy of −0.5 and −1.5 Kcal/mol, respectively were contributed to the total binding energy between cryptospirolepine and spike glycoprotein of SARS-CoV. A hydrophobic interaction of −1.8, −1.6, and −2.3 Kcal/mol was contributed to the total binding energy of the spike glycoprotein of MERS-CoV, ACE2, and TMPRSS2 and respective alkaloids, while the rest of the energy was contributed by nonbonding interactions. The overall energy profile of the ligand-receptor complex of selected cluster with display of individual energetic contributions for each atom in the ligand are shown in Figures 8(b)–12(b). These reveal that the hydrogen bond energy that was contributed to the overall binding energy of MERS-CoV spike glycoprotein and respective ligand was mainly contributed by the interaction of oxygen atom with SER51 and PHE354, while oxygen and nitrogen atom contributed the total hydrogen bond energy of −1.4 Kcal/mol.
3.4. Results from molecular dynamics simulation
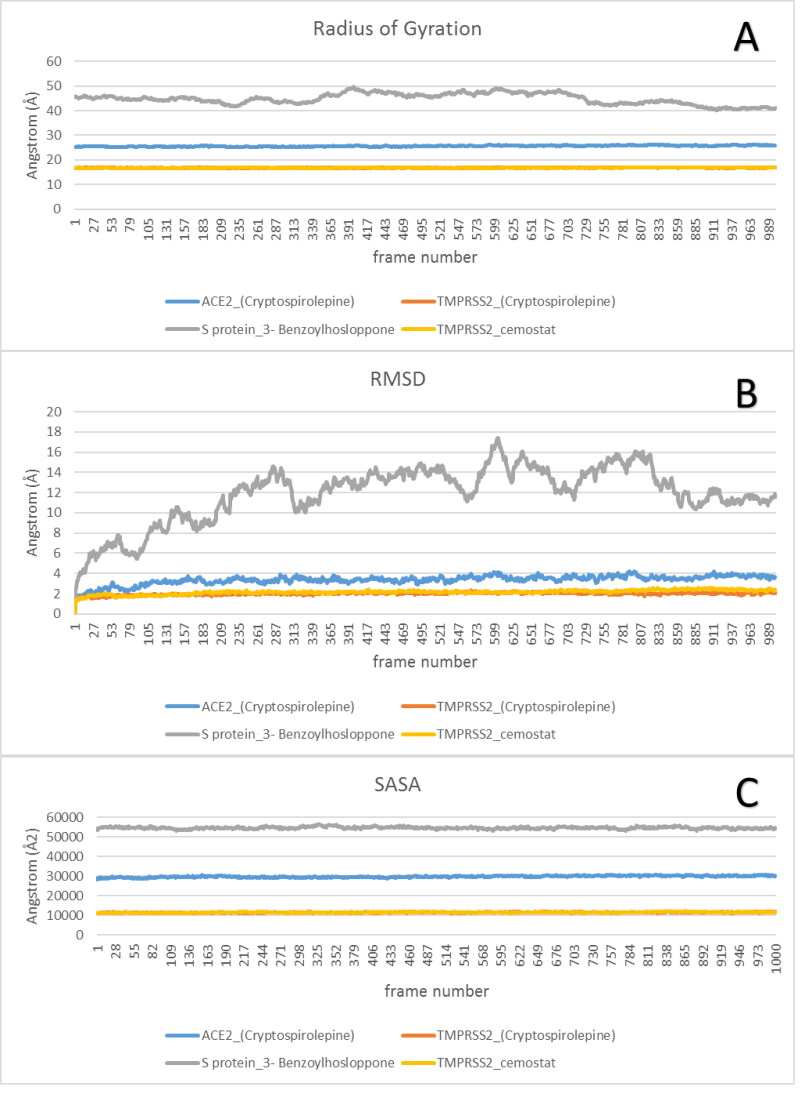
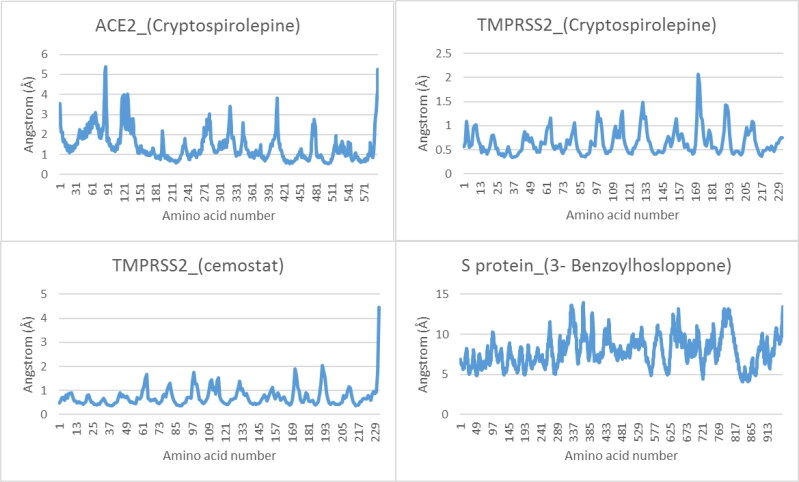
The complexes of best docked alkaloids to the target proteins were analyzed for MDS at 100 ns run. The results were analyzed using VMD Tk console scripts to calculate RMSD, SASA, RoG (Figure 13), and RMSF (Figure 14). TMPRSS2_Cryptospirolepine, and TMPRSS2_camostat complexes expressed similar RoG. The values of RoG of the S protein_3-Benzoylhosloppone complex, fluctuated about certain values towards the end of the run. The RMSD of the four protein-drug complexes has mean values of 3.36 Å, 1.99 Å, 11.9 Å, and 2.14 Å, and RMSF with mean values of 1.43 Å, 0.66 Å, 7.92 Å, and 0.73 Å for ACE2_(Cryptospirolepine), TMPRSS2_(Cryptospirolepine), S protein_3-Benzoylhosloppone, and TMPRSS2_camostat, respectively. The RMSF results for each protein-drug complex shows spikes at the end, which corresponds to the motion of the terminals (Figure 14).
Figure 13.
The Radius of Gyration (RoG), Root Mean Square Deviation (RMSD), and Surface Accessible Surface Area (SASA) for each of protein-ligand complex.
Figure 14.
The Root Mean Square Fluctuation for ACE2_(Cryptospirolepine), TMPRSS2_(Cryptospirolepine), TMPRSS2_(Camostat), and S glycoprotein_(3-Benzoylhosloppone), respectively. The spikes at the end of TMPRSS2_camostat, and ACE2_(Cryptospirolepine) corresponds to the motion of the terminals.
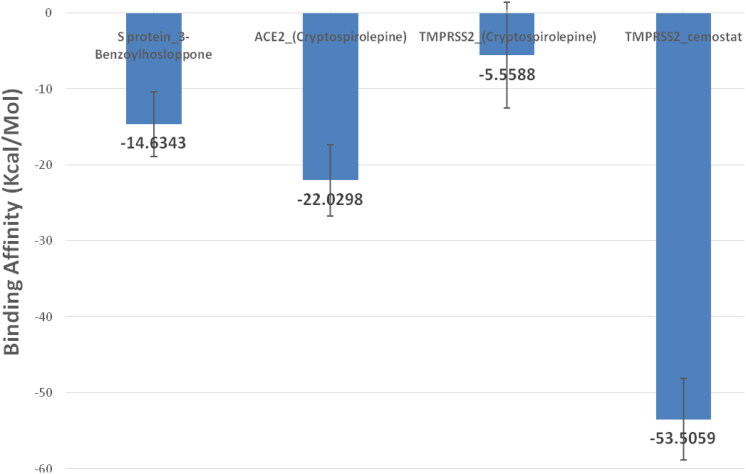
Molecular Mechanics/Generalized Born Surface Area (MMGBSA) algorithm implemented in AmberTools 20 was utilized to find the binding affinity between protein and drug in the four complexes. All frames (∼1000 frame) were used in this calculation for each protein-ligand complex. The binding energy (Kcal/mol) produced from the MM-GBSA analysis with the standard deviation as error bars revealed TMPRSS2_camostat complex (–53.5059 Kcal/mol) had the best binding affinity (Figure 15).
Figure 15.
The binding energy and standard deviation in (Kcal/mol) produced from the MM-GBSA analysis.
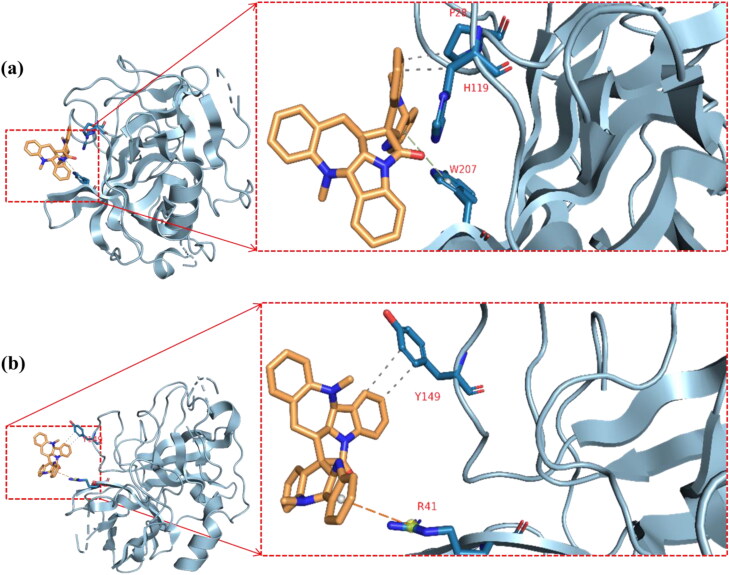
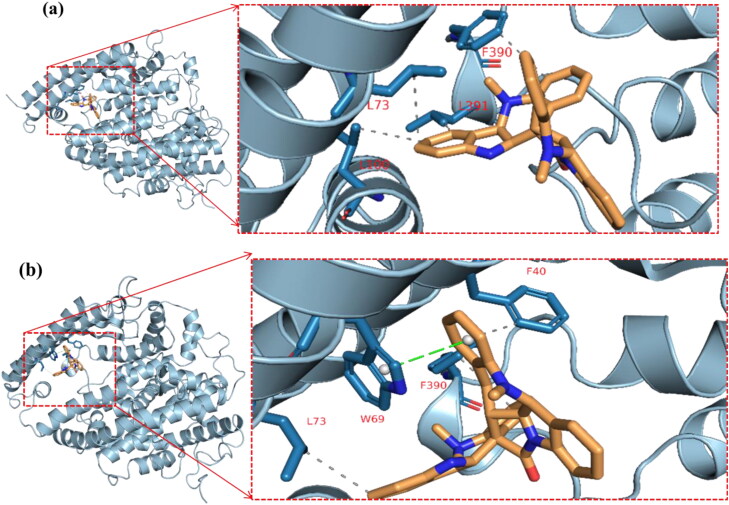
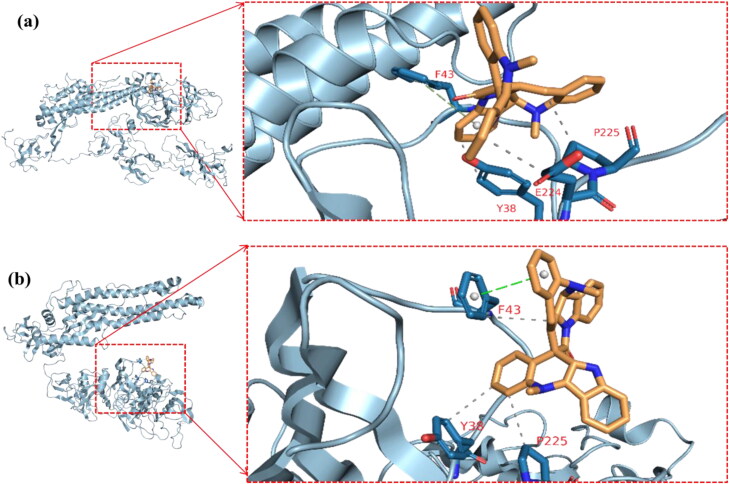
The cluster representatives for each trajectory with the number of bonds between the drug and the protein are shown in Table 7. Compared to camostat, cryptospirolepine in the representative cluster displayer week interaction with TMPRSS2, and this coincides with the MM-GBSA results (Figures 15–17). The most common interactions are hydrophobic interactions in case of ACE2_cryptospirolepine complex, while the predominant interaction was hydrogen bond in TMPRSS2_Camostat complex. Figures 16–19 show the first and last representative frame (whenever possible) of the complexes.
Table 7.
Clusters, its representative frame and the protein-ligand complexes interactions.
| TMPRSS2_ camostat complex | ||||
|---|---|---|---|---|
| Cluster number (representative frame) |
Hydrophobic |
Hydrogen bond |
Salt-bridges |
Pi-cation |
| Cluster 1 (frame 92) | None | A190 (2) Q192 D217 E218 A220 | D189 | H57 |
| Cluster 2 (frame 618) | Q192–V213 | R41 A190 S195 D217 E218 | H57–D189 | None |
| Cluster 3 (frame 284) | Q192 | A190 (2) S195 D217 (2) | D189 | None |
| Cluster 4 (frame 728) | None | |||
| Cluster 5 (frame 915) |
Q192 |
|
|
|
| TMPRSS2_(Cryptospirolepine) complex | ||||
| Cluster number (representative frame) |
Hydrophobic |
Pi-stacking |
Pi-cation |
|
| Cluster 1 (frame 747) | No Interactions | |||
| Cluster 2 (frame 121) | Y149(2) | None | R41 | |
| Cluster 3 (frame 515) | No Interactions | |||
| Cluster 4 (frame 966) | P28, H119 | W207 | None | |
| Cluster 5 (frame 699) |
No Interactions |
|
||
| ACE2_(Cryptospirolepine) complex | ||||
| Cluster number (representative frame) |
Hydrophobic |
Hydrogen bond |
Pi-stacking |
|
| Cluster 1 (Frame 39) | L73, L100, F390, L391 | None | None | |
| Cluster 2 (Frame157) | L39, F40 (2), W69, L73, F390 (2), L391 | N394 | F40, W69 | |
| Cluster 3 (Frame433) | L39, F40 (2), W69 (3), L73, A99, F390 (2) | None | F40, W69 | |
| Cluster 4 (Frame736) | F40, W69 (2), L73, L100, F390 (2) | None | F40, W69 | |
| Cluster 5 (Frame843) |
F40, L73, F390 |
None |
W69 |
|
| S protein_(3-Benzoylhosloppone) complex | ||||
| Cluster number (representative frame) | Hydrophobic | Hydrogen bond | Pi-stacking | |
| Cluster 1 (Frame316) | Y38, E224, P225 | Y38 | F43 | |
| Cluster 2 (Frame577) | F43 | None | F43 | |
The amino acids in bold are the most common in each complex.
Figure 16.
The representative structure for each cluster for TMPRSS2_Camostat and the types of interactions. Gray-dotted line is hydrophobic interactions, blue lines are hydrogen bond interactions, yellow-dotted lines represent salt-bridges, and green-dotted lines represent pi-stacking interactions. Single-letter amino acids are in red color. (a) Cluster 1 representative frame: 92 (b) Cluster 5 representative frame: 915.
Figure 17.
The second and fourth clusters in TMPRSS2_(Cryptospirolepine) complex. Gray-dotted line represents hydrophobic interactions, orange-dotted lines represent pi-cation interactions, and green-dotted lines represent pi-stacking interactions. Single-letter amino acids are in red color. (a) Cluster 2 Representative frame: 121 and (b) Cluster 4 representative frame: 966.
Figure 18.
The first and last clusters in ACE2_(Cryptospirolepine) complex. Gray-dotted line represents hydrophobic interactions, and green-dotted lines represent pi-stacking interactions. Single-letter amino acids are in red color. (a) Cluster 1 representative frame: 39 and (b) Cluster 5 Representative frame: 843.
Figure 19.
The first and last clusters in spike glycoprotein_(3-Benzoylhosloppone) complex. Gray-dotted line represent hydrophobic interactions, blue lines represents hydrogen bond interactions, and green-dotted lines represent pi-stacking interactions. (a) Cluster 1 representative frame: 316 and (b) Cluster 3 Representative frame: 772.
3.5. Pharmacokinetic properties of selected alkaloids
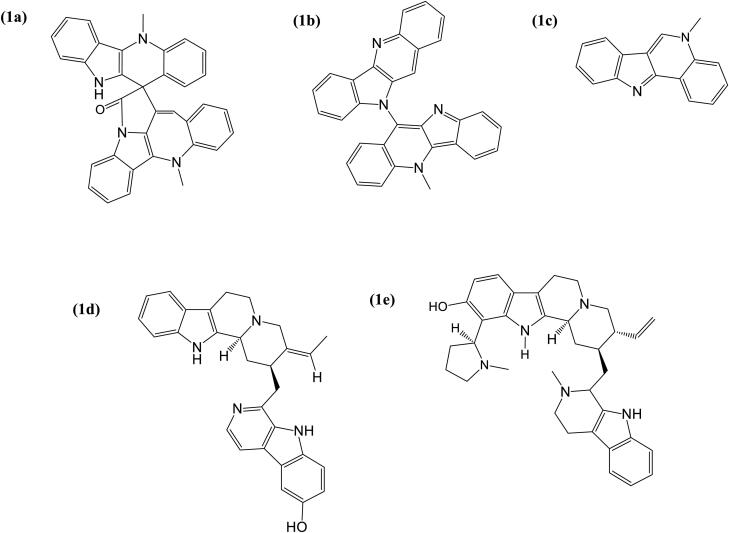
Five of the best docked alkaloids: cryptospirolepine, 10-hydroxyusambarensine, cryptoquindoline, isocryptolepine, and strychnopentamine (Figure 20) fulfilled the requirement for Lipinski analysis of the rule of-five with corresponding favorable predicted ADME/tox parameters (Table 8). The physicochemical and pharmacokinetics analyses suggested these five alkaloids (Figure 20) have a high probability of absorption, subcellular distribution, except for AMES toxicity parameter which indicated cryptoquindoline to be toxic at a probability of 0.89. All the top docked alkaloids were indicated to be non-carcinogenic, with very low acute toxicity and aqueous solubility of <0. The gastrointestinal absorption index was indicated to be high for 10–hydroxyusambarensine, isocryptolepine and 6-oxoisoiguesterin but low for cryptospirolepine and cryptoquindoline (Table 8).
Figure 20.
Structure of alkaloids with highest binding affinity to ACE2, TMPRSS2 and SARS-CoV-2 spike glycoprotein (a) Cryptospirolepine, (b) Cryptoquindoline, (c) Isocryptolepine, (d) 10–Hydroxyusambarensine, and (e) Strychnopentamin.
Table 8.
In silico prediction of physicochemical and pharmacokinetics properties of top binding alkaloids.
| (a) Lipinski filter analysis | |||||
|---|---|---|---|---|---|
| Lipinski filters |
Cryptospirolepine |
10-Hydroxyusambarensine |
Cryptoquindoline |
Isocryptolepine |
Strychnopentamine |
| Molecular weight (g/mol) | 504.58 | 448.56 | 448.52 | 232.28 | 459.75 |
| Num. heavy atoms | 39 | 34 | 35 | 18 | 41 |
| Num. rotatable bonds | 2 | 2 | 1 | 0 | 4 |
| Num. H-bond acceptors | 1 | 3 | 2 | 1 | 4 |
| Hydrogen bond donor | 1 | 3 | 0 | 0 | 3 |
| cLogP | 3.75 | 3.31 | 4.02 | 4.02 | 3.51 |
| Molar refractivity | 161.96 | 142.46 | 145.65 | 76.01 | 180.14 |
| Lipinski violation |
1 |
0 |
0 |
0 |
1 |
| (b) Admet SAR | |||||
| Absorption (Probability) | |||||
| Blood-Brain Barrier | BBB+ (0.99) | BBB+ (0.83) | BBB+ (0.95) | BBB+ (0.97) | BBB+ (0.71) |
| Human intestinal absorption | HIA+ (0.97) | HIA+ (0.98) | HIA+ (0.99) | HIA+ (0.99) | HIA+ (0.98) |
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| Caco-2 permeability | Caco2+ (0.74) | Caco2+ (0.53) | Caco2+ (0.76) | Caco2+ (0.67) | Caco2+ (0.53) |
| P-glycoprotein substrate | Nonsubstrate (0.50) | Substrate (0.91) | Noninhibitor (0.69) | Noninhibitor (0.74) | Noninhibitor (0.87) |
| P-glycoprotein inhibitor | Noninhibitor (0.84) | Noninhibitor (0.60) | Noninhibitor (0.72) | Noninhibitor (0.74) | Noninhibitor (0.54) |
| Renal organic cation transporter | Noninhibitor (0.70) | Inhibitor (0.80) | Noninhibitor (0.67) | Noninhibitor (0.54) | Noninhibitor (0.70) |
| Distribution (Probability) | |||||
| Subcellular localization | Mitochondria (0.70) | Mitochondria (0.65) | Mitochondria (0.55) | Mitochondria (0.76) | Mitochondria (0.57) |
| Metabolism | |||||
| CYP450 Substrate | Substrate (0.69) | Substrate (0.53) | Substrate (0.86) | Substrate (0.65) | Nonsubstrate (0.79) |
| Noninhibitor (0.76) | Noninhibitor (0.83) | Noninhibitor (0.86) | Noninhibitor (0.81) | Inhibitor (0.54) | |
| Toxicity | |||||
| AMES toxicity | Non AMES toxic (0.61) | Non AMES toxic (0.75) | AMES toxic (0.89) | AMES toxic (0.80) | Non AMES toxic (0.68) |
| Carcinogens | Noncarcinogens (0.94) | Noncarcinogens (0.97) | Noncarcinogens (0.92) | Noncarcinogens (0.96) | Noncarcinogens (0.95) |
| Acute oral toxicity | III (0.50) | III (0.51) | III (0.67) | III (0.72) | III (0.56) |
| Rat acute toxicity LD50 (mol/kg) | 2.4992 | 2.7896 | 2.4420 | 2.3770 | 2.7631 |
| Aqueous solubility (LogS) | –3.5692 | –2.7626 | –3.1120 | –3.21 | –2.6192 |
| Pharmacokinetics | |||||
| GI absorption | Low | High | Low | High | High |
| Log Kp (skin permeation) (cm/s) | –4.82 | –5.70 | –3.97 | –5.38 | –5.82 |
4. Discussion
Recognition of receptor on the host cell by coronavirus is obligatory for infection to occur. Both SARS-CoV and SARS-CoV-2 have been reported to utilize sites on ACE2 of the human cell as receptor (Hoffmann et al., 2020). The massive surface area of the lung, and the vast distribution of ACE2 in human alveolar epithelial cells makes the lungs a vulnerable target organ for the virus (Zhao et al., 2020). Thus, compounds that hinder viral entry into cell, or directly disrupt viral replication processes will halt transmissibility and pathogenicity. Blocking the cell entry effectively ensures the virus is denied access to host cell mechanism for replication. We hypothesized that alkaloids from African medicinal plants with reported bioactivity including antiviral potentials may interrupt coronavirus cell entry and consequently halt viral replication, pathogenicity, and transmissibility. To understand this, we evaluated the virtual binding of the alkaloids to the coronavirus spike glycoprotein, human ACE2, TMPRSS2, and the complex formed between the spike glycoprotein RBD and ACE2 (ACE2-RBD).
Computer modeling reveals the spike glycoproteins of SARS-CoV and SARS-CoV-2 have about 80% amino acid similarity (Li et al., 2005; Xia et al., 2020; Xu et al., 2020). The receptor binding domain (RBD) in the spike glycoproteins of SARSCoV and SARSCoV-2 are similar, with a root mean square deviation (RMSD) of about 0.68 Å over 139 pairs of Cα atoms (Yan et al., 2020). The spike glycoprotein is a type I fusion protein. In addition to binding to the receptor with S1 subunit, it uses S2 subunit to facilitate fusion of viral envelope with host cell membrane (Tong, 2009; Zhou et al., 2020). These two domains could be therapeutic targets. The top 20 alkaloids demonstrated good binding affinities to the spike glycoprotein. Cryptospirolepine, isocryptolepine, cryptoquindoline, and 10-hydroxyusambarensine had better binding to the spike glycoprotein of SARS-CoV-2, SARS-CoV, and MERS-CoV compared to the reference compound nafamostat. The binding strength of cryptospirolepine to the coronaviruses was consistently better than that of other alkaloids except for 10-hydroxyusambarensine that slightly demonstrated better binding to the spike glycoprotein of MERS-CoV. The interactions of these alkaloids with the spike glycoprotein could alter the stabilizing effects that Leu455, Gln493, and Asn501 in the RBD offers to the hotspots of ACE2. Also, the insertion of Phe486 from SARS-CoV-2 RBD into the hydrophobic pocket (Shang et al., 2020) may be hindered, thereby weakening the possible interaction of SARS-CoV-2 RBD with ACE2. The strong binding affinities of these alkaloids for spike glycoproteins suggest their capacity to alter the conformation of the spike glycoproteins of these coronaviruses and halt their binding to host receptor.
Compounds that bind ACE2 have potential to block the interactions of SARS-CoV-2 and SARS-CoV. ACE2 is a receptor essential for viral cell entry. SARS-CoV-2 and SARS-CoV have been reported to utilize ACE2 as functional cell receptor (Hoffmann et al., 2020; Li et al., 2003; Wan et al., 2020; Zhou et al., 2020) but the spike glycoprotein of SARS-CoV-2 was reported to have more than 10 times higher binding affinity compared with SARS-CoV (Wrapp et al., 2020). This high affinity of SARS-CoV-2 for ACE2 appears to contribute to the efficient human to human transmission of the virus, and the rate at which it infects cells. The multiple binding of SARS-CoV to the receptor results in down regulation of ACE2 expression, thereby causing reduced protective mechanism of ACE2 against lung injury (Imai et al., 2005; Kuba et al., 2005), and enhanced renin-angiotensin pathway which favors tissue injury through ACE: these make the virus lethal. SARS-CoV spike protein binds strongly with human ACE2 (Li et al., 2005); Glutamine (residue 479) in the receptor-binding domain (RBD) of SARS-CoV which corresponds to residue 394 in SARS-CoV-2 (Wan et al., 2020) binds to essential lysine 31 on the human ACE2 receptor (Wu et al., 2012). The interactions of SARS-coronaviruses with ACE2 are similar. Each peptidase domain of ACE2 accommodates one receptor binding domain in S1 subunit of spike protein. This interaction is mediated mainly through polar interactions (Song et al., 2018; Yan et al., 2020). However, the receptor binding domain in S1 subunit of SARS-CoV and SARS-CoV-2 are reported to show sequence and conformational variations in their interfaces with ACE2: variations at the N terminus of α1 where Arg426, Tyr484, and Thr487 in SARS-CoV are respectively replaced by Asn439, Gln498, and Asn501 in SARS-CoV-2; variation at the C terminus of α1 where Leu472 in SARS-CoV is replaced by Phe486 in SARS-CoV-2 (Yan et al., 2020); and the replacement of Tyr442 and Asn479 in SARS-CoV by Leu455 and Gln493 respectively in SARS-CoV-2 (Walls et al., 2020). These variations appear to be responsible for the difference in affinity of these coronaviruses for ACE2. The affinity of the viral spike glycoprotein for the host cellular receptor dictates binding. Alteration of this affinity limits viral nucleocapsid access to the host cytosol. This alteration can be achieved by changing the conformation of the host cell receptor, or direct binding of the spike glycoprotein by chemical agents. The binding affinities of reference compounds (MLN-4760 and N-Acetyl-d-glucosamine) for ACE2 was lesser relative to the alkaloids cryptospirolepine, 10-Hydroxyusambarensine, and strychnopentamine which have binding energy of −10.7, −10.4, and −9.9 Kcal/mol, respectively. The stronger binding of these alkaloids to ACE2 could alter the receptor. Lys31 and Lys353 are reported as the hotspots of ACE2, essential for the binding of coronavirus (Li et al., 2005; Shang et al., 2020). Interactions of compounds with this region is proposed to affect the binding of coronavirus (Gaunt et al., 2010). The interaction of these alkaloids with adjacent residues rather than the hotspots of ACE2 suggest the ability of the alkaloids to alter the suitable hydrophobic environment that is needed for coronavirus binding. This possible alteration may halt the interaction of RBD of the spike glycoprotein with ACE2.
In addition to the ability of alkaloids to block access to ACE2, we tested the possibility of them binding to the complex formed between the spike glycoprotein RBD and ACE2 (ACE2-RBD). From the list of alkaloids, cryptospirolepine, 10–hydroxyusambarensine, and chrysopentamine demonstrated best binding with ACE2-RBD, with binding energy of −10.7, −10.5, and −10.5 Kcal/mol. These alkaloids affinities for ACE2-RBD indicate that cryptospirolepine, 10–hydroxyusambarensine, and chrysopentamine may disrupt cell entry of SARS-CoV-2 even at the point when the S1 subunit of spike glycoprotein binds to ACE2. Stabilization of the interface between RBD of SARS-CoV-2 and human ACE2 is essential, and achieved by coronavirus by its residues at the RBD. The hotspots (Lys31 and Lys353) on ACE2 need to be suitably accommodated in hydrophobic setting for effective binding of coronavirus. To achieve this, SARS-CoV-2 neutralizes the charges of lysine residues by stabilizing Lys31 of ACE2 with Leu455 and Gln493, and further stabilizing Lys353 of ACE2 by Asn501 (Shang et al., 2020). These contribute to favorable recognition of ACE2 receptor. Both cryptospirolepine and 10–hydroxyusambarensine, by their interactions, may be good candidates to block access of spike glycoprotein to ACE2, and destabilize the ACE2-RBD complex.
TMPRSS2 is a member of the human type II transmembrane protease serine which is defined by N-terminal transmembrane domain, and a C-terminal extracellular serine protease domain. The spike glycoprotein of SARS-CoV and SARS-CoV-2 are reported to be primed by host protease TMPRSS2 (Glowacka et al., 2011; Hoffmann et al., 2020); an essential step for fusion of viral and host cell membranes. The S2 subunit in the spike glycoprotein of SARS-CoV-2 appears to facilitate fusion of viral envelope with host cell membrane better than that of SARS-CoV (Xia et al., 2020). This promotes entry of viral content into host cell cytoplasm. Inhibiting this fusion will also limit the viral spread. In vitro study reveals that blocking the activity of TMPRSS2 inhibits cell entry of SARS-CoV (Kawase et al., 2012). The SARS-CoV-2 spike protein has several multi-basic arginine residues at the S1/S2 cleavage site. This indicates a high tendency of cleavage at this point (Hoffmann et al., 2020). Report suggests that annulment of the S1/S2 cleavage site in the spike glycoprotein of SARS-CoV-2 affects its mediated cell entry (Walls et al., 2020). This indicates the importance of the cleavage performed by the host cell protease TMPRSS2, as the cleavage activates fusion of viral and host cell membranes, to guarantee viral infectiveness. The alkaloids 10–hydroxyusambarensine, cryptospirolepine, and cryptoquindoline demonstrated binding energy of −10.4, −9.9, and −9.7 Kcal/mol, respectively to the host protease TMPRSS2. These affinities were better than the binding provided by the reference compound (camostat). Camostat mesylate, a serine protease inhibitor, was reported to block the activity of TMPRSS2 (Zhou et al., 2015) and compounds with similar antiviral activity could be considered as anti-SARS-CoV-2 (Yamamoto et al., 2016). While 10–hydroxyusambarensine had the best binding affinity, it interacted in similar manner as camostat: both were docked into the S1-specificity pocket of TMPRSS2. Both compounds interacted with residue Ala190, Asp189 and Gln192 which are amino acid located at the basement of the pocket. This essential interaction with Asp189 determines the specificity of the S1 pocket for basic residues Arg and Lys of the substrate (Kyrieleis et al., 2007). The amidino nitrogen and hydroxyl group of 9H-pyrido[3,4-b]indol-6-ol moiety of 10–hydroxyusambarensine were responsible for the hydrogen bond with the protein. Similar to the phenylquanidine of camostat, the 9H-pyrido[3,4-b]indol-6-ol moiety of 10–hydroxyusambarensine, with its hydroxyl group directed towards the carboxylate group of Asp189, formed strong hydrogen bond with Asp189 and other residue in the pocket. The phenyl group of the 9H-pyrido[3,4-b]indol-6-ol further had hydrophobic interactions with CYS119 and TRP215, just as the peptide planes of the bonds between Trp215–Gly216 and Cys191–Gln192 sandwiched the phenyl ring of benzamidine in the native ligand to TMPRSS2 (Kyrieleis et al., 2007). Apart from the 9H-pyrido[3,4-b]indol-6-ol moiety, other groups of 10–hydroxyusambarensine interacted with the imidazol ring of His57 of the S2 pocket that is found next to the S1 pocket and ARG41 which are outside the hydrophobic cleft. A similar interaction was observed with camostat. The additional hydrophobic interaction by 10–hydroxyusambarensine may be responsible for its higher binding affinity relative to camostat. In a similar docking study with SARS-CoV-2 3CLpro, 10–hydroxyusambarensine, cryptospirolepine, and cryptoquindoline were observed to be docked in strikingly similar pattern as ritonavir with even higher binding affinities (Gyebi et al., 2020). The interaction of these alkaloids with TMPRSS2 may limit its protease function, thereby preventing the fusion of viral and human cell membranes. The potential exhibited by 10–hydroxyusambarensine, cryptospirolepine, and cryptoquindoline to inhibit the cleavage of spike glycoprotein by interacting with TMPRSS2, suggest they may function as inhibitors of SARS-CoV-2 cell entry. The result from the MDS analysis of the top docked alkaloids with their complexed proteins showed that the complexes were stable and could be therefore subjected to experimental processes in further studies. The Lipinski filtering analysis showed that these five alkaloids are drug-like, with no more than one violation of Lipinski’s rule (Lipinski et al., 1997). Likewise, the predicted ADMET filtering evaluation shows that these alkaloids may be well absorbed, distributed, metabolized, and not toxic. Though 10–hydroxyusambarensine may be more absorbed in the gastrointestinal tract than cryptospirolepine. However, these alkaloids may be optimized more to improve their physicochemical and pharmacokinetics properties.
5. Conclusion
In summary, this study provides evidence that alkaloids from Africa medicinal plants may disrupt cell entry of SARSCoV-2 by binding to the spike glycoprotein, by blocking the human cell receptor ACE2, and inhibiting the host cell serine protease TMPRSS2 utilized by SARS-CoV-2 for spike glycoprotein priming. Given the capacity to interact with the proteins essential for receptor-binding and membrane fusion, cryptospirolepine, 10-hydroxyusambarensine, and cryptoquindoline could serve as pan-SARS-coronavirus cell entry inhibitors. These alkaloids could serve as essential inhibitors to prevent and/or treat COVID-19, and other coronavirus diseases that may emerge in future.
Funding Statement
This study was not funded.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aanouz, I., Belhassan, A., & El-Khatabi, K. (2020). Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeoye, A. O., Oso, B. J., & Olaoye, I. F. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq, J. A., & Memish, Z. A. (2014). Middle East respiratory syndrome coronavirus: Transmission and phylogenetic evolution. Trends in Microbiology, 22(10), 573–579. 10.1016/j.tim.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, S., Heurich, A., Lavender, H., Gierer, S., Danisch, S., Perin, P., Lucas, J. M., Nelson, P. S., Pöhlmann, S., & Soilleux, E. J. (2012). Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PloS One, 7(4), e35876. 10.1371/journal.pone.0035876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles, M., Donaldson, E., & Baric, R. (2011). SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission. Current Opinion in Virology, 1(6), 624–634. 10.1016/j.coviro.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 Coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, B. R., Brooks, C. L., Mackerell, A. D., Nilsson, L., Petrella, R. J., Roux, B., Won, Y., Archontis, G., Bartels, C., Boresch, S., Caflisch, A., Caves, L., Cui, Q., Dinner, A. R., Feig, M., Fischer, S., Gao, J., Hodoscek, M., Im, W., … Karplus, M. (2009). CHARMM: The biomolecular simulation program. Journal of Computational Chemistry, 30(10), 1545–1614. 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeça, T. K., Granato, C., & Bellei, N. (2013). Epidemiological and clinical features of human coronavirus infections among different subsets of patients. Influenza and Other Respiratory Viruses, 7(6), 1040–1047. 10.1111/irv.12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D., Belfon, K., & Ben-Shalom, I. (2020). AMBER 2020.
- Cheng, F., Li, W., Zhou, Y., Shen, J., Wu, Z., Liu, G., Lee, P. W., & Tang, Y. (2012). AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. Journal of Chemical Information and Modeling, 52(11), 3099–3105. 10.1021/ci300367a [DOI] [PubMed] [Google Scholar]
- Dhama, K., Karthik, K., Khandia, R., Munjal, A., Tiwari, R., Rana, R., Khurana, S. K., Ullah, S., Khan, R. U., Alagawany, M., & Farag, M. R. (2018). Medicinal and therapeutic potential of herbs and plant metabolites/extracts countering viral pathogens-current knowledge and future prospects. Current Drug Metabolism, 19(3), 236–263. 10.2174/1389200219666180129145252 [DOI] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., & Şahin, A. T. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, T. M., & Buchmeier, M. J. (2001). Coronavirus spike proteins in viral entry and pathogenesis. Virology, 279(2), 371–374. 10.1006/viro.2000.0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, E. R. R., Hardie, A., Claas, E. C. C. J., Simmonds, P., & Templeton, K. E. E. (2010). Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. Journal of Clinical Microbiology, 48(8), 2940–2947. 10.1128/JCM.00636-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka, I., Bertram, S., Müller, M. A., Allen, P., Soilleux, E., Pfefferle, S., Steffen, I., Tsegaye, T. S., He, Y., Gnirss, K., Niemeyer, D., Schneider, H., Drosten, C., & Pöhlmann, S. (2011). Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. Journal of Virology, 85(9), 4122–4134. 10.1128/JVI.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Baker, S. C., & Baric, R. S. (2020). The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 5(4), 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi, A. G., Ogunro, B. O., Adegunloye, P. A., Ogunyemi, M. O., & Afolabi, O. S. (2020). Potential inhibitors of Coronavirus 3-Chymotrypsin-Like Protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, (in press) 10.1080/7391102.2021764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich, A., Hofmann-Winkler, H., Gierer, S., Liepold, T., Jahn, O., & Pöhlmann, S. (2014). TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. Journal of Virology, 88(2), 1293–1307. 10.1128/JVI.02202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus, M. A. (2020). Remdesivir in the treatment of Coronavirus Disease 2019 (COVID-19): A simplified summary. Journal of Biomolecular Structure and Dynamics, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N.-H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Imai, Y., Kuba, K., Rao, S., Huan, Y., Guo, F., Guan, B., Yang, P., Sarao, R., Wada, T., Leong-Poi, H., Crackower, M. A., Fukamizu, A., Hui, C.-C., Hein, L., Uhlig, S., Slutsky, A. S., Jiang, C., & Penninger, J. M. (2005). Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature, 436(7047), 112–116. 10.1038/nature03712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase, M., Shirato, K., van der Hoek, L., Taguchi, F., & Matsuyama, S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. Journal of Virology, 86(12), 6537–6545. 10.1128/JVI.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., & Amera, G. (2020). Targeting SARS-Cov-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2'-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba, K., Imai, Y., Rao, S., Gao, H., Guo, F., Guan, B., Huan, Y., Yang, P., Zhang, Y., Deng, W., Bao, L., Zhang, B., Liu, G., Wang, Z., Chappell, M., Liu, Y., Zheng, D., Leibbrandt, A., Wada, T., … Penninger, J. M. (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nature Medicine, 11(8), 875–879. 10.1038/nm1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudi, A. C., & Myint, S. H. (1999). Antiviral activity of some Nigerian medicinal plant extracts. Journal of Ethnopharmacology, 68(1–3), 289–294. 10.1016/S0378-8741(99)00049-5 [DOI] [PubMed] [Google Scholar]
- Kyrieleis, O. J. P., Huber, R., Ong, E., Oehler, R., Hunter, M., Madison, E. L., & Jacob, U. (2007). Crystal structure of the catalytic domain of DESC1, a new member of the type II transmembrane serine proteinase family. The FEBS Journal, 274(8), 2148–2160. 10.1111/j.1742-4658.2007.05756.x [DOI] [PubMed] [Google Scholar]
- Lee, J., Cheng, X., Swails, J. M., Yeom, M. S., Eastman, P. K., Lemkul, J. A., Wei, S., Buckner, J., Jeong, J. C., Qi, Y., Jo, S., Pande, V. S., Case, D. A., Brooks, C. L., MacKerell, A. D., Klauda, J. B., & Im, W. (2016). CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. Journal of Chemical Theory and Computation, 12(1), 405–413. 10.1021/acs.jctc.5b00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko, M., Marzi, A., & Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology, 5(4), 562–568. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Li, W., Farzan, M., & Harrison, S. C. (2005). Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (New York, N.Y.), 309(5742), 1864–1868. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., Choe, H., & Farzan, M. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J. (1997). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 23(1–3), 3–25. 10.1016/S0169-409X(96)00423-1 [DOI] [PubMed] [Google Scholar]
- Lobo-Galo, N., Terrazas-López, M., & Martínez-Martínez, A. (2020). FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. Journal of Biomolecular Structure and Dynamics, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Yu, X., Wang, W., Duan, X., Zhang, L., Zhou, W., Xu, J., Xu, L., Hu, Q., Lu, J., Ruan, L., Wang, Z., & Tan, W. (2012). Characterization of human coronavirus etiology in Chinese adults with acute upper respiratory tract infection by real-time RT-PCR assays. PloS One., 7(6), e38638. 10.1371/journal.pone.0038638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, J. J. M., Afolayan, A. J., Taylor, M. B., & Erasmus, D. (1997). Antiviral activity of galangin isolated from the aerial parts of Helichrysum aureonitens. Journal of Ethnopharmacology, 56(2), 165–169. 10.1016/S0378-8741(97)01514-6 [DOI] [PubMed] [Google Scholar]
- Miller, B. R., McGee, T. D., Swails, J. M., Homeyer, N., Gohlke, H., & Roitberg, A. E. (2012). MMPBSA.py: An efficient program for end-state free energy calculations. Journal of Chemical Theory and Computation, 8(9), 3314–3321. 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., & Velmurugan, D. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–7. [DOI] [PubMed] [Google Scholar]
- O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3(1), 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbole, O. O., Akinleye, T. E., Segun, P. A., Faleye, T. C., & Adeniji, A. J. (2018). In vitro antiviral activity of twenty-seven medicinal plant extracts from Southwest Nigeria against three serotypes of echoviruses. Virology Journal, 15(1), 110. 10.1186/s12985-018-1022-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules, C. I., Marston, H. D., & Fauci, A. S. (2020). Coronavirus infections-more than just the common cold. JAMA, 323(8), 707–708. 10.1001/jama.2020.0757 [DOI] [PubMed] [Google Scholar]
- Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, R. D., Kalé, L., & Schulten, K. (2005). Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26(16), 1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentin, S., Schreiber, S., Haupt, V. J., Adasme, M. F., & Schroeder, M. (2015). PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Research, 43(W1), W443–W447. 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Linares, I., Pérez-Sánchez, H., Cecilia, J. M., & García, J. M. (2012). High-Throughput parallel blind virtual screening using BINDSURF. BMC Bioinformatics, 13(Suppl 14), S13. 10.1186/1471-2105-13-S14-S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581(7807), 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla, A., Heald-Sargent, T., Subramanya, G., Zhao, J., Perlman, S., & Gallagher, T. (2011). A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. Journal of Virology, 85(2), 873–882. 10.1128/JVI.02062-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, G., Zmora, P., Gierer, S., Heurich, A., & Pöhlmann, S. (2013). Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antiviral Research, 100(3), 605–614. 10.1016/j.antiviral.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W., Gui, M., Wang, X., & Xiang, Y. (2018). Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathogens, 14(8), e1007236. 10.1371/journal.ppat.1007236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, T. R. (2009). Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections. Infectious Disorder Drug Targets, 9(2), 223–245. 10.2174/187152609787847659 [DOI] [PubMed] [Google Scholar]
- Towler, P., Staker, B., Prasad, S., Menon, S., Tang, J., Parsons, T., Ryan, D., Fisher, M., Williams, D., Dales, N., Patane, M., & Pantoliano, M. (2004). ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. The Journal of Biological Chemistry, 279(17), 17996–18007. 10.1074/jbc.M311191200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubiana, T., Carvaillo, J.-C., Boulard, Y., & Bressanelli, S. (2018). TTClust: A versatile molecular simulation trajectory clustering program with graphical summaries. Journal of Chemical Information and Modeling, 58(11), 2178–2182. 10.1021/acs.jcim.8b00512 [DOI] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–212. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, E. E., Shin, J. H., & Falsey, A. R. (2013). Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. The Journal of Infectious Diseases, 208(10), 1634–1642. 10.1093/infdis/jit393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y., Shang, J., Graham, R., Baric, R. S., Ralph, S., & Li, F. (2020). Receptor recognition by the Novel Coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology, 94(7), e00127. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020). Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19). https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.aax0902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Peng, G., Wilken, M., Geraghty, R. J., & Li, F. (2012). Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. The Journal of Biological Chemistry, 287(12), 8904–8911. 10.1074/jbc.M111.325803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., Qi, F., Bao, L., Du, L., Liu, S., Qin, C., Sun, F., Shi, Z., Zhu, Y., Jiang, S., & Lu, L. (2020). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion . Cell Research, 30(4), 343–355. 10.1038/s41422-020-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., Zhong, W., & Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China: Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., Matsuyama, S., Li, X., Takeda, M., Kawaguchi, Y., Inoue, J. I., & Matsuda, Z. (2016). Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome Coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrobial Agents and Chemotherapy, 60(11), 6532–6539. 10.1128/AAC.01043-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y., Cao, D., Zhang, Y., Ma, J., Qi, J., Wang, Q., Lu, G., Wu, Y., Yan, J., Shi, Y., Zhang, X., & Gao, G. F. (2017). Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature Communications, 8(1), 1–9. 10.1038/ncomms15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Penninger, J. M., Li, Y., Zhong, N., & Slutsky, A. S. (2020). Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Medicine, 46(4), 586–585. 10.1007/s00134-020-05985-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Zhao, Z., Wang, Y., Zhou, Y., Ma, Y., & Zuo, W. (2020). Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv, 10.1101/2020.01.26.919985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–274. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Vedantham, P., Lu, K., Agudelo, J., Carrion, R., Nunneley, J. W., Barnard, D., Pöhlmann, S., McKerrow, J. H., Renslo, A. R., & Simmons, G. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Research, 116, 76–84. 10.1016/j.antiviral.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]