Abstract
Introduction:
The chromosome 19 miRNA cluster (C19MC) encodes a large family of microRNAs (miRNAs) that are abundantly expressed in the placenta of higher primates and also in certain cancers. In the placenta, miRNAs from this cluster account for nearly 40% of all miRNAs present in trophoblasts. However, the function of these miRNAs in the placenta remains poorly understood. Recent observations reveal a role for these miRNAs in cell migration, and suggest that they are involved in the development and function of the human placenta. Here, we examine the placenta in transgenic mice expressing the human C19MC miRNAs.
Methods:
We produced transgenic mice using pronuclear microinjection of a bacterial artificial chromosome plasmid carrying the entire human C19MC locus and derived a homozygous line using crossbreeding. We performed morphological characterization and profiled gene expression changes in the placentas of the transgenic mice.
Results:
C19MC transgenic mice delivered on time with no gross malformations. The placentas of transgenic mice expressed C19MC miRNAs and were larger than wild type placentas. Histologically, we found that the transgenic placenta exhibited projections of spongiotrophoblasts that penetrated deep into the labyrinth. Gene expression analysis revealed alterations in the expression of several genes involved in cell migration, with evidence of enhanced cell proliferation.
Discussion:
Mice that were humanized for transgenically overexpressed C19MC miRNAs exhibit enlarged placentas with aberrant delineation of cell layers. The observed phenotype and the related gene expression changes suggest disrupted migration of placental cell subpopulations.
Keywords: Placenta, C19MC, microRNA, spongiotrophoblast, migration
Introduction
Human placental trophoblasts express a broad repertoire of microRNAs (miRNAs) [1,2]. MiRNAs constitute a large family of small regulatory RNA molecules, typically 19–24 nucleotides long, which are derived from stem-loop structures embedded in longer transcripts. Numerous miRNA species have been described in most eukaryotes and viruses. Whereas many miRNAs are conserved across species, some families show a more restricted evolutionary distribution. One such miRNA family is expressed from the chromosome 19 miRNA cluster (C19MC) locus, which is only found in primates [3,4]. This locus harbors one of the largest miRNA clusters, containing 46 genes that encode 58 mature miRNAs. These C19MC miRNAs are predominantly expressed in placental trophoblasts [3,5], where they account for approximately 40% of the miRNA molecules produced in these cells [6]. Outside the placenta, expression of these miRNAs has been restricted to human embryonic stem cells [7–9] and also discrete types of cancers, including hepatocellular carcinoma [10,11], brain tumors [12,13], testicular and breast cancer [14,15].
The function of C19MC miRNAs remains poorly understood. We previously found that villous primary human trophoblasts (PHT), which release a high amount of C19MC miRNA to the culture medium, are relatively resistant to replication of diverse types of DNA and RNA viruses, and that this resistance can be conferred to other, non-placental cell types by transferring the medium from cultured PHT cells or by transfecting these non-trophoblastic cells with the C19MC-expressing bacterial artificial chromosome (BAC). We also found that this effect is mediated by stimulation of cellular autophagy [16,17]. Other studies suggest that C19MC miRNAs regulate the differentiation and migration properties of trophoblasts [18,19]. For example, C19MC miRNAs appear to be expressed mostly in villous trophoblasts and at a much lower level in invasive extravillous trophoblasts (EVTs), and overexpression of C19MC miRNAs in the EVT-like HTR8/SVneo cell line reduce their migration and invasion properties [18]. Overexpression of members of the C19MC family has been implicated in the development of preeclampsia, a common hypertensive disorder of pregnancy [19]. It was recently found that miR-518b, a member of the C19MC family, targets EGR1/VEGF and regulates trophoblast migration in a cellular model of early embryonic arrest [20].
Outside the placenta, high expression of C19MC miRNAs has been associated with enhanced cell proliferation [11,13,21–24]. These observations suggest a role for C19MC miRNAs in the molecular regulation of cell physiology. Because these miRNAs are primate-specific, there is no easily accessible animal model that can enable the study of C19MC miRNA function at the organismal level. We recently reported the generation of transgenic mice carrying a BAC plasmid comprising 160 kb of the human C19MC genomic DNA locus. Importantly, transgenic mice expressed the C19MC miRNAs mainly in placentas in a way that is highly reminiscent of the C19MC expression pattern in humans, and released these miRNAs to the maternal and fetal compartments [25]. To assess the effect of C19MC overexpression in the placenta, we generated a transgenic mouse line, made homozygous for the human C19MC transgene. These mice appear normal and have a normal lifespan, but exhibit placental morphological alterations. We found that the placentas of transgenic animals are commonly enlarged compared to wild-type animals, and exhibit invaginations of spongiotrophoblasts in the labyrinth layer. Gene expression profiling experiments revealed altered expression of several genes that may regulate placental development.
Methods
Generation and analysis of transgenic mice
The generation of the C19MC transgenic lines has been previously described [26]. Briefly a 160-kb BAC clone (BAC #RP11–1055O17) containing the entire C19MC locus was microinjected into 1-cell-stage B6SJLF1 embryos, using standard procedures. The transgenic founders were backcrossed to C57BL/6 mice and were PCR-genotyped. To obtain transgenic mice that are homozygous for the C19MC cluster, transgene hemizygous males and females were crossbred, and the progeny were tested by mating with wild-type (WT) mice, confirming homozygosity by producing only homozygous pups. Timed pregnancies were established using homozygous C19MC transgenic and C57BL/6 WT mice, as controls. Analysis of the mouse placentas was usually performed at E17.5, unless otherwise indicated.
Histology and in situ hybridization
Timed breeding pairs were established, and pregnant mice were sacrificed at the indicated times. Placentas were harvested and fixed overnight in 4% paraformaldehyde in phosphate buffered saline (PBS). For standard histology, fixed placentas were embedded in paraffin, and 5 μm sections were cut using a microtome (Leica RM2255, Wetzlar, Germany) and stained with SelecTech Hematoxylin and Eosin Staining System (Surgipath, Richmond, IL). Stained sections were examined using a Nikon 90i microscope (Nikon, Tokyo, Japan). For in situ hybridization, placentas were fixed overnight in 4% paraformaldehyde in PBS and cryoprotected through sucrose gradients (15% in PBS and 30% in PBS) before embedding in Tissue-Tek O.C.T Compound (Sakura Finetek, Torrance, CA). Cryosections (10 μM) were cut at −20ºC and collected on Superfrost Plus slides (Fisher Scientific, Waltham, MA). Cryosections were rehydrated in PBS, digested with proteinase K (10 μg/ml, Sigma-Aldrich, St. Louis, MO), for 5 min at 37ºC, and processed with 0.2 N HCL for 10 min at room temperature (RT), acetylated (0.25% acetic anhydride [Sigma-Aldrich] in triethanolamine-hydrochloride, TEA) for 10 min at RT, and then hybridized with cRNA probes overnight at 60ºC. Slides were washed four times with 4×SSC, digested with RNaseA (5 μg/ml, Thermo-Fisher) for 15 min at 37ºC, and washed twice with 0.5×SSC for 15 min at 60ºC. Slides were blocked using 1% blocking reagent (Sigma-Aldrich) in maleic acid buffer, followed by incubation with anti-DIG-AP antibody (0.5 U/ml) (Sigma-Aldrich) for 2 h at RT, washed with maleic acid buffer with 0.2% Tween20, and then reacted with BM purple (Sigma-Aldrich) with 1 mM levamisole (Sigma-Aldrich) overnight. The sections were examined using a Nikon 90i microscope.
Immunohistochemistry (IHC)
Immunohistochemical staining was performed on 5 μm paraffin-embedded sections of 4% paraformaldehyde-fixed mouse placentas. Ki67 staining was performed using a rabbit polyclonal antibody (1:1500, NB110–89717, Novus Biologicals, Centennial, CO). Antibody binding was detected using the Vectastain ABC Elite kit (Vector Laboratories, Burlingham, CA). Slides were counterstained with hematoxylin and mounted with VectaMount AQ Aqueous Mounting Medium (Vector).
RNA extraction
For cellular mRNA and miRNA analysis, total RNA was extracted from mouse placenta using TRI Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s instructions and purified using EconoSpin spin columns (Epoch Life Science, Missouri City, TX). RNA samples were exposed to RNase-free DNase (Qiagen, Valencia, CA). The quantity and quality of total RNA was determined by a NanoDrop 1000 spectrometer (Thermo-Fisher) and by an Agilent bioanalyzer (Agilent, Santa Clara, CA).
Microarray and data analysis
Total RNA samples from WT and transgenic placentas were analyzed using Agilent SurePrint G3 mouse gene expression microarray. RNA samples were obtained from two individual WT placentas and from six individual transgenic placentas. Raw microarray data were acquired using an Agilent microarray scanner and processed with Agilent Extraction Image Analysis software. After normalizing the samples by loess normalization and log2 -transforming intensities, we applied the empirical Bayesian algorithm, implemented in the R package “limma” [27], to identify genes that were differentially expressed between normal and transgenic mouse placentas. Benjamini and Hochberg’s method [28] was used to calculate the adjusted p-values to control for false discovery rate. Microarray data have been submitted to the GEO database for public access (GSE141131).
Quantitative real-time PCR analysis
Reverse transcription and quantitative PCR (RT-qPCR) was performed in duplicate, using the ViiA 7 Sequence Detection System (Thermo-Fisher) as previously described [29,30]. For mRNA analysis, total RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription kit (Thermo-Fisher) according to the manufacturer’s protocol. Quantitative PCR was performed by means of SYBER Select (Thermo-Fisher). For miRNA, cDNA synthesis and qPCR were performed with the miRScript PCR system (Qiagen) according to the manufacturer’s protocols. PCR primers are given in Supplementary Table 1. Dissociation curves were run on all reactions, and samples were normalized to the expression of the ribosomal protein L32 for mRNAs and to RNU6B small nuclear RNA for miRNAs, respectively. The fold increase relative to control samples was determined by the 2-ΔΔCt method [31].
Statistics
All statistical analysis was carried out using the R computing environment. Mouse embryo weight, placenta weight, and litter size were analyzed using a paired t-test. For RT-qPCR data, ANOVA or ANOVA with repeated measures was applied to the delta-delta Ct values to determine whether a gene was differentially expressed between the wild type and C19MC transgenic placentas, and whether sex has a significant effect on the expression of the selected genes. The statistical analysis of the microarray data is provided above. All p-values were adjusted either by Tukey’s method to control for family-wise error rate, or Benjamini-Hochberg’s method to control for false discovery rate, where applicable.
Results
Generation of homozygous mice for the C19MC transgene
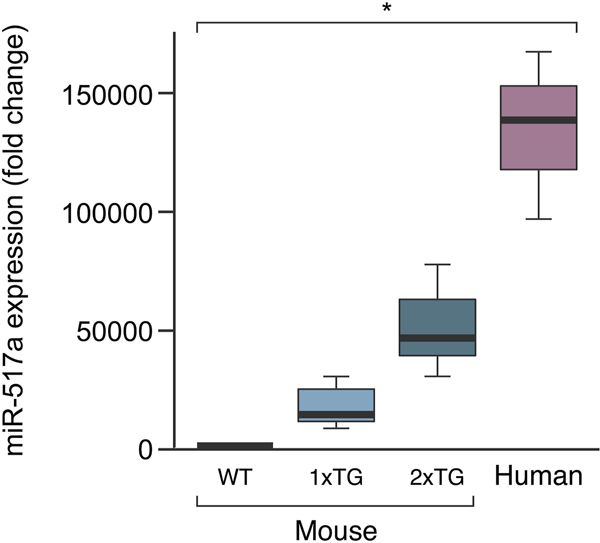
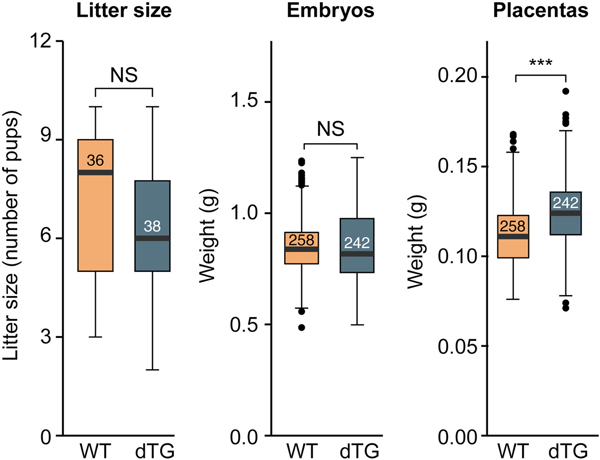
We recently reported the generation of transgenic lines carrying a BAC that harbors the entire human C19MC locus [29]. These mice express human C19MC miRNAs primarily in the placenta, thus recapitulating the expression pattern in humans [26]. Our C19MC hemizygous mice did not exhibit any obvious morphologic anomalies. To obtain mice that express higher placental levels of C19MC we crossbred the hemizygous females and males and established two independent lines. RT-qPCR assessment of miR-517a, one of the most abundant C19MC miRNAs in these placentas, showed that homozygous mice placentas express this transgenic C19MC miRNA at a level that is roughly four times higher than in the hemizygous mice (Fig. 1), but lower than its levels in the human placenta. Expression profiles of another member of this family, miR-518b revealed a similar increase in homozygous mice (not shown). Homozygous animals were viable and exhibited no gross abnormalities, and the litter size was insignificantly smaller among the homozygous transgenics (Fig. 2), with no evidence of a higher rate of fetal death or resorption. Fetuses and placentas from WT and homozygous transgenic animals (hereinafter referred to as dTG) were collected at E17.5. Whereas the mean dTG fetal weight was insignificantly different from WT controls, we observed that mean placental weight in dTG fetuses was higher (by a mean of 14%, Fig. 2). Notably, we observed a wide range of weight in each genotypic category, with some dTG placentas nearly twice as big as a typical WT placenta and others in the range of normal variation. This variability was observed in placentas from different litters and in placentas within a single litter, indicating that the phenotype is not fully penetrant.
Fig. 1: RT-qPCR evaluation of miR-517a expression in mouse placentas.
Shown are wild-type (WT), transgenic lines (TG), and a human placenta. The box-plots show arbitrary values, representing C19MC miRNA expression levels. Mouse WT, n=3, Mouse 1xTg, n=6, Mouse 2xTg, n=13, Human, n=3. P<0.02 for all pairwise comparisons, determined by one-way ANOVA analysis, with Tukey’s post hoc test.
Fig. 2: The effect of C19MC transgene overexpression on litter size and feto-placental weight.
Measurements, shown as box-plots, were performed at E17.5, and include litter size (WT, n=36; dTG, n=38), embryo weight, and placenta weight (WT, n=258; dTG, n=242). The bottom and top edges indicate the 25th and 75th percentiles, respectively. The numbers inside the boxes represent the number of observations. Statistical significance was determined using a t-test. NS, non-significant, * denotes p<0.05, ** denotes p<0.01, *** denotes p<0.001.
Abnormal placental morphology in dTG mice
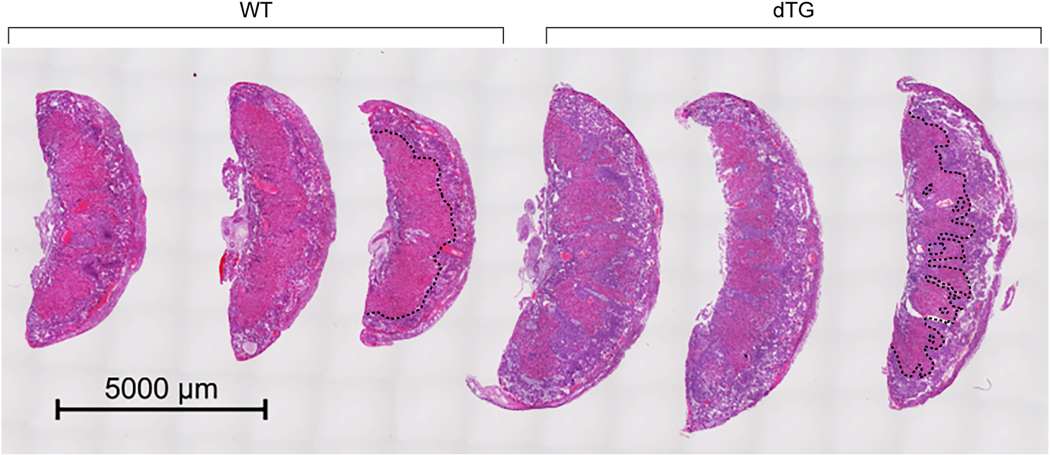
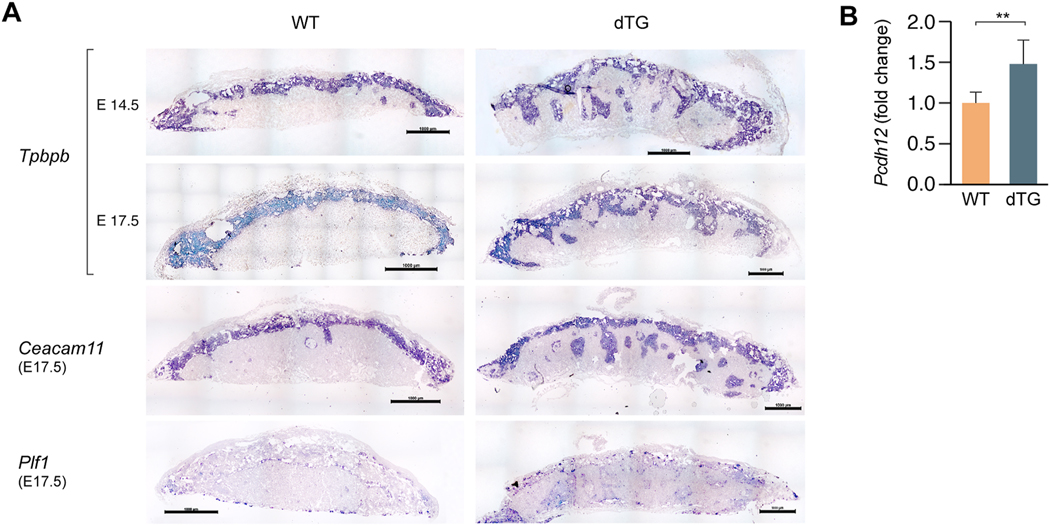
Histological inspection of hematoxylin-eosin stained sections revealed abnormal morphology in dTG placentas. The boundary between the labyrinthine layer and the junctional zone was irregular, with deep spongiotrophoblast invaginations into the labyrinth (Fig. 3). To validate the phenotype and determine whether some trophoblast subpopulations are more affected than others, we conducted an in situ hybridization assessment of dTG placentas. Using probes for Tpbpb and Ceacam11, which are mostly expressed in spongiotrophoblast cells in the junctional zone [32,33], we confirmed the presence of Tpbpb- and Caecam11-positive invaginations in the E17.5 labyrinth layer (Fig. 4A). We observed ectopic clusters of spongiotrophoblasts in the labyrinth at E14.5 (Fig. 4A, top panel), suggesting abnormal cell proliferation and/or migration in tissues overexpressing C19MC. The junctional zone comprises two main cell types, including spongiotrophoblasts and glycogen cells. Using RT-qPCR, we determined that the mRNA for Protocadherin 12 (pcdh12), a marker of junctional zone glycogen cells, exhibits a higher expression in dTG placentas (Fig. 4B). We also assessed the expression of Plf1, a marker of trophoblast giant cells (TGCs) that is expressed in different TGC subpopulations [34], including parietal TGCs, canal TGCs, and spiral artery TGCs (Fig. 4), and identified discrete cell clusters at the expected locations for these TGC subtypes, which were similar in WT and dTG placentas.
Fig. 3: Placenta histology from WT and dTG mice,
The images show 3 placentas from a WT litter juxtaposed to 3 placentas from a dTG litter, obtained at E17.5 and stained with hematoxylin and eosin. The border between the spongiotrophoblast and the labyrinth is outlined by a dashed black line in one WT placenta and one dTG placenta.
Fig. 4: In situ hybridization for delineation of the placental junctional zone.
(A) A Tpbpb riboprobe shows invaginations of the junctional zone in the dTG labyrinthine layer at E14.5 and E17.5; spongiotrophoblast-specific riboprobe Ceacam11 exhibits a similar pattern. A riboprobe against Plf1 delineates the distribution of E17.5 trophoblast giant cells in WT and dTG placentas. Scale bar=1000 μm. Sense probes were used as negative controls showed no signal (not shown). (B) RT-qPCR of Pcdh12 in WT and dTG placentas. The relative expression level is presented as fold change in dTG vs WT placenta. The bars represent the mean relative expression in WT (n=3) vs dTG (n=14) placentas. P<0.01, determined by t-test.
Altered gene expression in TG placentas
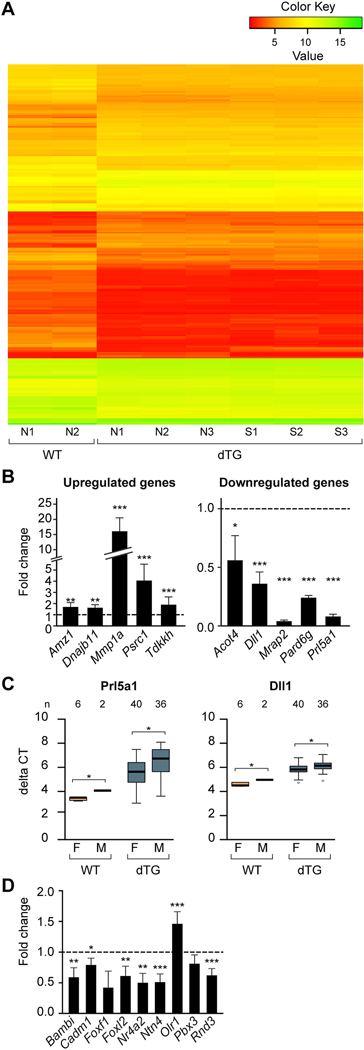
To define the transcriptional profiles of the dTG placentas, we used microarrays to profile mRNA expression in dTG and WT placentas. We identified 108 probes, corresponding to 84 transcripts that are downregulated in dTG mice compared to WT mice, and 101 probes, corresponding to 73 transcripts that are upregulated (Fig. 5A). We then selected 10 genes that were the most significantly upregulated (n=5) or downregulated (n=5), and validated their expression change using RT-qPCR (Fig. 5B). Among the upregulated genes in dTG placentas, Mmp1a, a placental metalloprotease involved in cell migration and invasion, exhibited a marked increase in dTG compared to WT placentas. In contrast, Prl5a1, a member of the prolactin family expressed in a subset of spongiotrophoblasts [34], showed a striking reduction of its expression levels in TG placentas. Melanocortin receptor accessory protein-2 (Mrap2), an important regulator of brain energy homeostasis, was expressed at less than 10% of the signal in dTG vs WT placentas. We used Ingenuity Pathway Analysis to identify pathways that might be affected by C19MC miRNA expression. The five top cellular and molecular functions from our analysis are shown in Supplementary Table 2, supporting our data interpretation. In addition, analysis of transcripts that exhibited the most marked expression difference (Fig. 5B) revealed that the expression of Dll1 and Prl5a1, but not other transcripts, was different between male and female dTG placentas, with a slightly higher level in male placentas for both genes (Fig. 5C). We noted the same difference in WT placentas, suggesting that the effect of fetal sex was transgene-independent.
Fig. 5: The expression profiles of mRNA in the C19MC dTG placentas.
(A) A heat map of expression profiles in 2 WT placentas and 6 dTG placentas. Each column represents a separate placenta. Gene expression was analyzed in 3 placentas of dTG conceptuses in which the embryo was of normal size (N1, N2, N3) and in 3 placentas of dTG conceptuses in which the embryo was smaller than the average (S1, S2, S3). Placentas and embryos from the WT conceptuses (N1 and N2) were of normal size. (B). RT-qPCR validation of expression changes in selected transcripts from the microarray results, including 5 upregulated (upper panel) and 5 downregulated genes (lower panel). RT-qPCR was performed using WT (n=6) and dTG placentas (n=8). Significance was determined by one-way ANOVA with repeated measures and p values adjusted by the Benjamini and Hochberg’s method to control the false discovery rate. (C) Analysis of sex-dependent expression of Prl5a1 and Dll1, showing a significant sex-dependent expression of Dll1 and Prl5a1 (p <0.05 two-way ANOVA, with Holm’s method to adjust for repeated measures). (D) The expression of C19MC miRNA targets in WT vs dTG mouse placentas. Our analysis focused on human-mouse orthologs, as described in the text. The relative expression level, analyzed by RT-qPCR, is presented as fold change in dTG compared to WT placenta. The bars represent the mean of fold change of dTG (n=7) relative to WT (n=3) placentas. Statistical significance was determined by one-way ANOVA with repeated measures then p values adjusted by the Benjamini and Hochberg’s method to control the false discovery rate. * denotes p<0.05, ** denotes p<0.01, *** denotes p<0.001.
We compared these data to our recent findings regarding overexpression of C19MC miRNAs in HTR8/SVneo cells, which do not naturally express C19MC miRNAs [18]. In these human cells, transgenic overexpression of C19MC miRNAs attenuated their migration, and several C19MC mRNAs that were functionally related to cellular movement, migration, or invasion were downregulated [18]. We therefore analyzed the expression of some of the same genes in the C19MC transgenic placentas, and found that with the exception of Olr1, most genes were also downregulated in dTG placentas (Fig. 5D). A search for C19MC miRNA seed match in the sequence of these mouse transcripts revealed that most of them harbored several potential target sites (Supplementary Table 3). Foxf1, a transcript that did not change in the presence of transgenic miRNAs, lacked canonical C19MC miRNA sites. Together, these results suggest that C19MC miRNAs impact the expression of orthologous human and mouse genes that regulate cell migration.
Increased Ki67 staining in TG placentas
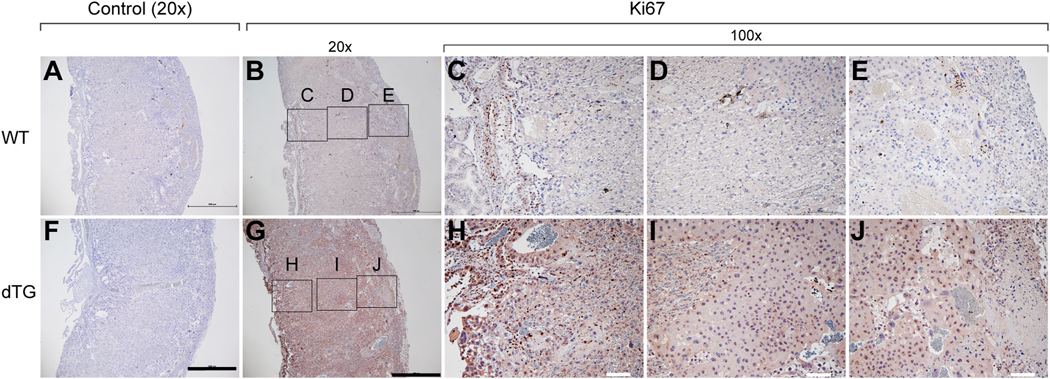
The size of the dTG placentas prompted us to assess if increased cell proliferation in these placentas might have contributed to their larger dimensions. Using immunohistochemical staining for the Ki67 antigen in sections of E17.5 WT and dTG placentas, we found a stronger Ki67 nuclear staining in the dTG placentas, localized mainly to the labyrinth (Fig. 6), suggesting an enhanced proliferation in the dTG labyrinth. Notably, we detected cytoplasmic Ki67 immunoreactivity in dTG sections that was absent in WT placentas. This staining pattern has been previously reported [35,36], and its relevance to cell proliferation is uncertain.
Fig. 6: The expression of Ki67 in the WT and dTG mouse placenta.
Immunohistochemistry was used to detect Ki67, counterstained with hematoxylin. Upper panel micrographs are from WT placentas and lower panel micrographs from dTG placentas. Panels A and F are no primary antibody negative control. Panels B and G are 20X magnification images (scale bar = 1mm) and panels C-E and H-J are 100X magnification images (scale bar = 100 μm). Images are representative of 3 independent experiments. Note that WT and dTG sections were on the same slide and were therefore exposed to identical experimental conditions.
Discussion
To study the effect of C19MC miRNAs on placental morphology and function, we produced a transgenic mouse model expressing the human C19MC miRNAs, using a BAC plasmid that harbors the entire locus. In these transgenic mice, the C19MC miRNAs are mainly expressed in placental tissue and recapitulate the expression patterns found in humans [26]. Our C19MC dTG mice have a C19MC expression level that resembles that in the human placenta. Importantly, placentas from dTG mice are larger than their WT counterparts and exhibit projections of the spongiotrophoblast into the labyrinthine zone. These invaginations of junctional zone cells were infrequently observed in the WT placentas, and usually did not penetrate the labyrinth as deeply as in dTG placentas.
Placental enlargement with protrusions of the spongiotrophoblast layer into the labyrinth have also been described in mice obtained after inter-subspecies hybridization [37] and also in cloned mice after somatic cell nuclear transfer (SCNT) [38]. In addition, knockout of Esx1, a paired-like homeobox gene located on the X chromosome and expressed in extraembryonic tissues during embryogenesis, also led to enlarged placentas with clusters of spongiotrophoblasts localized into the labyrinth [39]. PCDH12-deficient mice also exhibit placentas in which islets of spongiotrophoblasts are detected in the labyrinth, albeit the placentas were smaller [40].
Our mRNA expression profiles in dTG and WT placentas revealed several genes that were differentially expressed. Interestingly, one of the most upregulated genes is Mmp-1a, a member of the matrix metalloproteinase family that is mostly expressed in the placenta (Mouse ENCODE transcriptome data [41]. Despite high expression of Mmp-1a in the mouse placenta, there are no data on its function in the developing placenta. An Mmp-1 knockout mouse exhibits reduced incidence of lung cancer, but no obvious pregnancy-related phenotype [42]. It is thus possible that overexpression of Mmp-1a in the context of other alterations disrupts normal placental development and contribute to the observed phenotype. In contrast, Prl5a1, one of the 23 members of the placental prolactin gene family that are expressed in the mouse placenta [34], is markedly downregulated in dTG placentas. While the function of this gene is currently unknown, and a genetic knockout model has not been reported, it is expressed mostly in junctional zone cells during late gestation [34], suggesting that its lower expression in dTG mice represents gene downregulation rather than loss of Prl5a1-expressing trophoblasts. Notably, all 23 prolactin-related genes are expressed from a large cluster located on chromosome13 [43], with variable placental expression patterns among family members [34].
The phenotype observed in the dTG placenta suggests that overexpressed C19MC miRNAs impact the specification and development of placental cell layers or their migration. A similar conclusion was reported in the case of PCDH12-deficient mice [40]. Moreover, overexpression of the C19MC miRNAs in a human trophoblast cell line caused downregulation of a number of genes involved in cell migration [18]. Interestingly, orthologous mouse and human genes were downregulated in our dTG mice and in our extravillous trophoblast line that overexpresses C19MC, suggesting that despite the restricted expression of C19MC miRNAs to haplorhines (higher primates) with no homologous region in the mouse genome [4,5], corresponding transcripts are functionally conserved between humans and mice.
The ectopic expression of the C19MC miRNAs has been associated with certain cancers, and C19MC miRNAs have been described as oncomiRs in certain pediatric brain tumors [12,13,44,45]. Thus far, after prolonged observation (up to two years), none of our mice have developed any noticeable tumors, which might not be surprising because the transgenic expression of the C19MC miRNAs is mostly restricted to the placenta. Interestingly, we detected a higher nuclear ki67 staining in the transgenic labyrinth, suggesting an enhanced rate of cell proliferation in this layer. Thus, transgenic C19MC miRNAs in the mouse placenta appears to recapitulate some of the effects mediated by their ectopic expression in humans with increases in cell proliferation and migration.
Supplementary Material
Highlights.
C19MC miRNAs are highly expressed in the primate placenta
The function of the C19MC miRNAs is inadequately understood
We generated transgenic mice expressing the human C19MC miRNAs
Placentas of C19MC homozygous transgenic mice exhibit morphological abnormalities
Acknowledgements
We thank Tiffany Coon for technical assistance, Lori Rideout for assistance in manuscript preparation, and Bruce Campbell for editing. This project was supported by the Pennsylvania Department of Health Research Formula Funds (grants to J.-F.M and T.C.), the National Institutes of Health, (grant R21 HD089732 to J.-F.M. and Y.S. and R37 HD086916 to Y.S), and the Margaret Ritchie R. Battle Family Charitable Fund (to Y.S.). The generation of the C19MC BAC, as described, was made possible by National Institutes of Health (NIH) Grants UL1RR024153 and UL1TR000005.
Abbreviations
- C19MC
Chromosome 19 microRNA cluster
- miRNA
microRNA
- PHT
primary human trophoblasts
- BAC
bacterial artificial chromosome
- dTG
double (homozygous) transgenic animals
Footnotes
Declaration
Yoel Sadovsky is a consultant to Illumina, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mouillet JF, Chu T, Hubel CA, et al. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta, 31 (2010) 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sadovsky Y, Mouillet JF, Ouyang Y, et al. The function of trophomiRs and other microRNAs in the human placenta. Cold Spring Harb Perspect Med, 5 (2015) a023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet, 37 (2005) 766–770. [DOI] [PubMed] [Google Scholar]
- [4].Zhang R, Wang YQ, Su B. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol, 25 (2008) 1493–1502. [DOI] [PubMed] [Google Scholar]
- [5].Bortolin-Cavaille ML, Dance M, Weber M, et al. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res, 37 (2009) 3464–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Donker RB, Mouillet JF, Chu T, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod, 18 (2012) 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bar M, Wyman SK, Fritz BR, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells, 26 (2008) 2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Laurent LC. MicroRNAs in embryonic stem cells and early embryonic development. J Cell Mol Med, 12 (2008) 2181–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ren J, Jin P, Wang E, et al. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med, 7 (2009) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fornari F, Milazzo M, Chieco P, et al. In hepatocellular carcinoma miR-519d is upregulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol, 227 (2012) 275–285. [DOI] [PubMed] [Google Scholar]
- [11].Augello C, Colombo F, Terrasi A, et al. Expression of C19MC miRNAs in HCC associates with stem-cell features and the cancer-testis genes signature. Dig Liver Dis, 50 (2018) 583–593. [DOI] [PubMed] [Google Scholar]
- [12].Li M, Lee KF, Lu Y, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell, 16 (2009) 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kleinman CL, Gerges N, Papillon-Cavanagh S, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet, 46 (2014) 39–44. [DOI] [PubMed] [Google Scholar]
- [14].Flor I, Spiekermann M, Loning T, et al. Expression of microRNAs of C19MC in Different histological types of testicular germ cell tumour. Cancer Genom Proteom, 13 (2016) 281–289. [PubMed] [Google Scholar]
- [15].Jinesh GG, Flores ER, Brohl AS. Chromosome 19 miRNA cluster and CEBPB expression specifically mark and potentially drive triple negative breast cancers. PLoS One, 13 (2018) e0206008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Delorme-Axford E, Donker RB, Mouillet JF, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci USA, 110 (2013) 12048–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bayer A, Delorme-Axford E, Sleigher C, et al. Human trophoblasts confer resistance to viruses implicated in perinatal infection. Am J Obstet Gynecol, 212 (2015) 71 e71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xie L, Mouillet JF, Chu T, et al. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology, 155 (2014) 4975–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang M, Muralimanoharan S, Wortman AC, et al. Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc Natl Acad Sci USA, 113 (2016) E7069-E7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang W, Lu Z, Zhi Z, et al. Increased miRNA-518b inhibits trophoblast migration and angiogenesis by targeting EGR1 in early embryonic arrest. Biol Reprod, (2019). [DOI] [PubMed] [Google Scholar]
- [21].Ma W, Yu Q, Jiang J, et al. miR-517a is an independent prognostic marker and contributes to cell migration and invasion in human colorectal cancer. Oncol Lett, 11 (2016) 2583–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nguyen PN, Huang CJ, Sugii S, et al. Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J Biomed Sci, 24 (2017) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ren GB, Wang L, Zhang FH, et al. Study on the relationship between miR-520g and the development of breast cancer. Eur Rev Med Pharmacol Sci, 20 (2016) 657–663. [PubMed] [Google Scholar]
- [24].Zheng J, Sadot E, Vigidal JA, et al. Characterization of hepatocellular adenoma and carcinoma using microRNA profiling and targeted gene sequencing. PLoS One, 13 (2018) e0200776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chandler KJ, Chandler RL, Broeckelmann EM, et al. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome, 18 (2007) 693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chang G, Mouillet JF, Mishima T, et al. Expression and trafficking of placental microRNAs at the feto-maternal interface. FASEB J, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43 (2015) e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Benjamini Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol, 57 (1995) 289–300. [Google Scholar]
- [29].Mishima T, Miner JH, Morizane M, et al. The expression and function of fatty acid transport protein-2 and −4 in the murine placenta. PLoS One, 6 (2011) e25865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schaiff WT, Bildirici I, Cheong M, et al. Peroxisome proliferator-activated receptor-gamma and retinoid X receptor signaling regulate fatty acid uptake by primary human placental trophoblasts. J Clin Endocrinol Metab, 90 (2005) 4267–4275. [DOI] [PubMed] [Google Scholar]
- [31].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25 (2001) 402–408. [DOI] [PubMed] [Google Scholar]
- [32].Lescisin KR, Varmuza S, Rossant J. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev, 2 (1988) 1639–1646. [DOI] [PubMed] [Google Scholar]
- [33].Kataoka K, Takata Y, Nakajima A, et al. A carcinoembryonic antigen family cDNA from mouse placenta encoding a protein with a rare domain composition. Placenta, 21 (2000) 610–614. [DOI] [PubMed] [Google Scholar]
- [34].Simmons DG, Rawn S, Davies A, et al. Spatial and temporal expression of the 23 murine Prolactin/Placental Lactogen-related genes is not associated with their position in the locus. BMC Genomics, 9 (2008) 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kim YJ, Lee SH, Lee J, et al. Non-nuclear localization of Ki-67 in human colorectal cancer cells grown as multicellular layers. Arch Pharm Res, 36 (2013) 634–640. [DOI] [PubMed] [Google Scholar]
- [36].Faratian D, Munro A, Twelves C, et al. Membranous and cytoplasmic staining of Ki67 is associated with HER2 and ER status in invasive breast carcinoma. Histopathology, 54 (2009) 254–257. [DOI] [PubMed] [Google Scholar]
- [37].Zechner U, Reule M, Orth A, et al. An X-chromosome linked locus contributes to abnormal placental development in mouse interspecific hybrid. Nat Genet, 12 (1996) 398–403. [DOI] [PubMed] [Google Scholar]
- [38].Tanaka S, Oda M, Toyoshima Y, et al. Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod, 65 (2001) 1813–1821. [DOI] [PubMed] [Google Scholar]
- [39].Li Y, Behringer RR. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat Genet, 20 (1998) 309–311. [DOI] [PubMed] [Google Scholar]
- [40].Rampon C, Bouillot S, Climescu-Haulica A, et al. Protocadherin 12 deficiency alters morphogenesis and transcriptional profile of the placenta. Physiol Genomics, 34 (2008) 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mouse EC, Stamatoyannopoulos JA, Snyder M, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol, 13 (2012) 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fanjul-Fernandez M, Folgueras AR, Fueyo A, et al. Matrix metalloproteinase Mmp-1a is dispensable for normal growth and fertility in mice and promotes lung cancer progression by modulating inflammatory responses. J Biol Chem, 288 (2013) 14647–14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wiemers DO, Shao LJ, Ain R, et al. The mouse prolactin gene family locus. Endocrinology, 144 (2003) 313–325. [DOI] [PubMed] [Google Scholar]
- [44].Pfister S, Remke M, Castoldi M, et al. Novel genomic amplification targeting the microRNA cluster at 19q13.42 in a pediatric embryonal tumor with abundant neuropil and true rosettes. Acta Neuropathol, 117 (2009) 457–464. [DOI] [PubMed] [Google Scholar]
- [45].Sin-Chan P, Mumal I, Suwal T, et al. A C19MC-LIN28A-MYCN oncogenic circuit driven by hijacked super-enhancers is a distinct therapeutic vulnerability in ETMRs: A lethal brain tumor. Cancer Cell, 36 (2019) 51–67 e57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.