Abstract
Recent technological advancements and genome-wide studies provide compelling evidence that dynamic chromatin interaction and three-dimensional genome organization in nuclei play an important role in regulating gene expression. Mammalian genomes consist of many small functional domains termed topologically associated domains (TADs), many of them organized by CCCTC-binding factor (CTCF) and the cohesion complex. Changes in genome TADs might result in inappropriate promoter/enhancer communications leading to activation of oncogenes or suppression of tumor suppressors. During normal hematopoiesis and leukemogenesis, genome structure alters considerably to facilitate normal and malignant hematopoiesis, respectively. Delineating theses normal and abnormal processes will evolve our understanding of disease pathogenesis and development of potential treatment strategies. This review highlights the role of CTCF and its associated protein complexes in three dimensional genome organization in development and leukemogenesis, as well as the roles of CTCF boundary defined TAD in transcription regulation. We further explore the function of chromatin modulators, such as CTCF, cohesin, and long non-coding RNAs (lncRNAs) in chromosomal interactions and hematopoietic genome organization. Finally, we focus on the implication of 3D genome alteration in the pathogenesis of leukemia and provide a scientific basis for targeted intervention.
Keywords: chromatin, chromatin loops, genome organization, transcription, CTCF, cohesion, transcription factors, epigenetic regulators, lncRNAs, lineage development, oncogenesis
Introduction
Organized in three dimensions inside the nucleus, chromatin orchestrates mammalian gene transcription into distinct expression signatures/patterns. Hierarchically organized genomic information needs to be readily available for duplication, transmission and expression, all of which allow dynamic transition between the relatively uncompact state and highly compacted state during nuclear processes while assuring successful interpretation of genetic and epigenetic information. However, accessing and interpreting hierarchical genome organization in the nucleus has been obscured by previous limitations in light microscopy techniques. The development of 3C (chromosome conformation capture) and its derivatives have made it possible to study the relationship between nuclear organization and gene expression. Since then various studies have highlighted that topologically associated domains (TADs) are both structural and functional chromosomal units that constrain and regulate enhancer/ promoter communication for specific gene expression programs1. Changes in TADs might result in inappropriate promoter/enhancer communications, leading to altered gene transcription programs that include activation of oncogenes or suppression of tumor suppressors2, 3. Although TADs are functionally important in gene expression, it is difficult to assess how they are dynamically organized and whether the conformation effects are a cause or consequence of gene regulation. However, supportive epigenetic evidence together with loss-of-function studies of necessary factors has provided a functional role for chromatin interactions and organization. Furthermore, chromatin insulators, CTCF binding sites in many cases, play a critical role in defining TADs and chromatin signatures within the defined domains4, 5. Understanding of the topological organization of chromatin and genome may provide valuable insight into how the genome and epigenome act collaboratively to orchestrate lineage-specific transcription and fate decision during development and disease states.
Recent advances in genome biology have shed light on 3D genome organization in lineage specific gene regulation in normal and malignant hematopoiesis. In this review we will discuss recent findings and principles of genome organization and its role in transcriptional regulation, focusing on the role of chromatin insulator proteins including CTCF and cohesin as well as lncRNAs and their involvement in pathogenesis of hematopoietic malignancies.
CTCF/cohesin mediated chromatin boundary and principle of genome organization
The theory of chromosomal territory was proposed as early as 1982, using traditional light microscopic techniques6‘7. Later, these findings were confirmed by DNA fluorescence in-situ hybridization (FISH) analysis, notably at the mouse beta-globin gene cluster8. The power of FISH and microscopic techniques lies in their ability to do single-cell analysis but is limited by poor resolution and lack of overall whole cell population overview as well. These limitations were overcome by the introduction of chromosome conformation capture (3C) and its variants. With the advancement of 3C-based techniques, it is now possible to not only map long-range ‘cis’ and ‘trans’ chromosomal interactions at specific genomic regions, but also provide comprehensive views of genome-wide chromatin organization. This information allows us to generate three-dimensional maps of the genome inside the nucleus and to understand the relationship between genome organization and function.
Recent studies revealed that CCCTC-binding factor (CTCF) and its associated cohesin complex have emerged as a master regulator of mammalian genome organization9–12. CTCF was first demonstrated to act as enhancer, blocker, and barrier insulator in the chicken β-globin locus where it binds to chicken HS4 (cHS4) insulator located in the 5’ upstream boundary of the β-globin locus control region (LCR) (Figure 1)13, 14. CTCF is a highly conserved zinc-finger protein across species involved in transcription activation/repression, insulation, imprinting, and X chromosome inactivation9, 15–17, all of which are attributed to two properties of CTCF insulator: enhancer blocking when placed between enhancer and promoter and chromatin barrier when located in the boundary between euchromatin and heterochromatin. However, genome wide CTCF binding data revealed that although CTCF mostly interacts with the same DNA-sites in different cell types, often it functions as a chromatin barrier in one cell type but not in the other18. Whether and how these boundary elements (CTCF-binding sites) are directly linked to its biological function remains largely unknown. This diversity in its role is in part due to CTCF’s ability to homodimerize with itself and to heterodimerize with other proteins19, 20. Certain CTCF genome functions required cohesin complex that catalyzes the folding of the genome into loops that are anchored by CTCF21, 22. Indeed, cohesin is essential for CTCF anchored TAD boundary formation, though it may not be required for CTCF mediated enhancer/promoter interactions.
Figure 1. The schematic presentation of chicken β-globin locus, in which the first vertebrate chromatin insulator, cHS4, was identified.
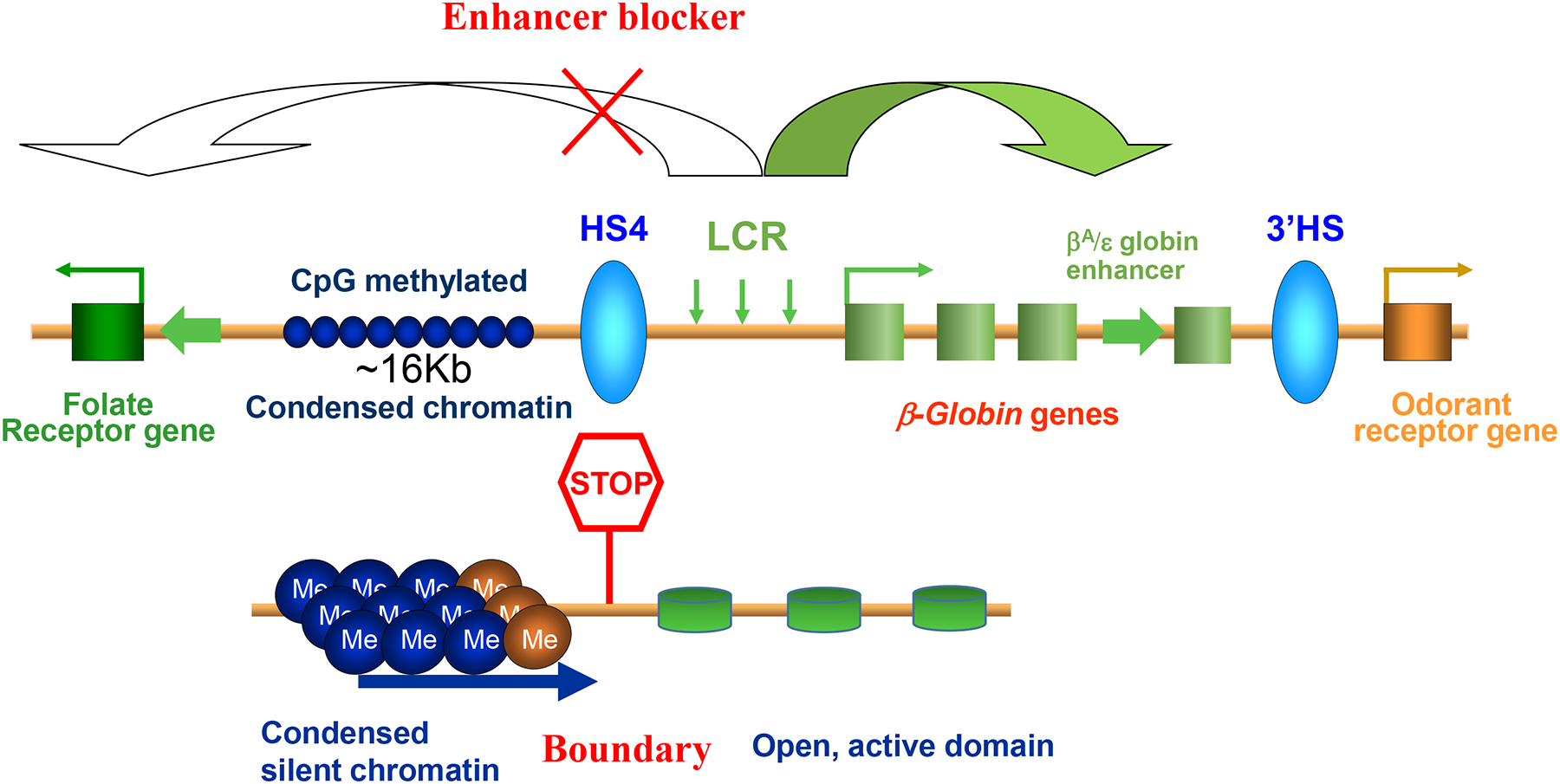
The cHS4 located at the 5’ boundary of chicken β-globin genes possesses both enhancer blocking and chromatin barrier activities in endogenous and ectopic chromatin sites.
Cohesin functions to tether chromosomal DNA during mitosis and meiosis to allow proper DNA segregation into daughter cells. The cohesin complex comprises of two parts: bridging its open is protein RAD21 complexing with the open-ended heterodimer, Smc1 and Smc3, which is made up of the two long coiled-coil molecules; the second part comprises the stromal antigen 1 (STAG1 or SA1) and stromal antigen 2 (STAG2 or SA2) proteins that also interact with Rad21 and outside of the Smc1-Smc3-Rad21 trimer. Genome-wide ChIP-seq analysis for CTCF and cohesin revealed that these two proteins mostly overlap in their binding patterns and CTCF is required for cohesin to bind chromatin, perhaps through its interaction with the SCC3/SA1 subunit of the cohesin complex23–25. This suggests that cohesin may also play a non-canonical role in enhancer/promoter interactions and gene expression. A recent crystal structural study revealed that a segment within the N-terminus of the CTCF protein also directly interacts with the SA2-SCC1 subunits of human cohesin complex, a structural basis required for the formation of CTCF anchored loops22. This complex is believed to cause chromatin cohesion by trapping progressive and dynamic movement DNA loop into its ring-like structure and become stalled in the TAD boundary bound by CTCF, a so called loop extrusion, thereby influencing chromatin structure dynamics including forming or stabilizing chromatin loops26, 27. Depletion of cohesin resulted in reduced intra-domain interactions in TADs, while CTCF loss led to increased inter-domain interactions28. However, not all sites occupied by cohesin are CTCF dependent. In CTCF-depleted cells, most of the cohesin bound sites are unperturbed, indicating overlapping as well as distinct functions of CTCF and cohesin in genome organization24, 29, 30.
Although both SA1 and SA2 containing cohesin complexes were found to associate with CTCF biding sites in the human genome, the SA1 containing cohesin complexes are preferentially involved in stabilization of CTCF-defined TAD boundaries, whereas the SA2 containing cohesin complexes facilitate cell-type specific enhancer/promoter communications in a CTCF independent manner31. Furthermore, it was shown that cohesin associates with a mediator to bridge the enhancer-promoter contacts within and between larger TADs and is required for cell lineage specific genome organization and transcription events in lineage commitment32, underscoring the importance of cohesin-mediated genome organization in cell type specific transcription. In addition to genome organization, cohesin also co-localizes with transcription activator complexes and facilitates re-establishment of transcription factor clusters after S-phase and chromatin condensation33. Thus, cohesin, believed to be important during M-phase, has also emerged to play essential roles in chromatin and genome organization during G1 and S phases.
Long-noncoding RNAs (lncRNAs) regulate CTCF boundary activity
The notion that lncRNAs are involved in CTCF mediated genome organization stems from a seminal paper by Dr. Gary Felsenfeld’s group showing that DEAD-box RNA helicase p68 and steroid receptor RNA activator (SRA) interact with both CTCF and cohesin. Depletion of p68 or SRA results in loss of cohesin binding to CTCF34, suggesting that p68/SRA stabilizes the CTCF and cohesin interaction, and RNA may be required for proper chromatin boundary function. It has also been shown that lncRNAs Tsix and Xite target CTCF to specific genome loci to mediate long range chromosomal interactions in a locus specific manner during X chromosome inactivation35. Given that many imprinted lncRNAs are cis acting36 and associated with CTCF35, it is reasonable to speculate that tethering of CTCF to the imprinting control regions may be attributed to RNA-mediated mechanisms. Two recent back-to-back publications demonstrated that the RNA binding region (RBR) of CTCF is required for CTCF dimerization/clustering and CTCF-mediated long-range chromatin interactions37, 38, suggesting that RNA molecules are essential components of CTCF directed genome organization. The RBR of CTCF is located in the C-terminal that includes Zinc finger 10 (ZF10). ZF10 is dispensable for the DNA recognition, and binding and deletion of this domain disrupt half of all CTCF loops38. Thus, CTCF contributes to chromatin loops and genome organization via RNA-dependent and RNA-independent mechanisms. This makes all the more interesting the question of what type(s) of RNA contribute to CTCF genome action and, more importantly, what are the underlying molecular mechanisms and biological consequences.
In a recent interesting studies, it has been shown that a HOXA gene associated RNA, HOTTIP, coordinates with CTCF to promote aberrant posterior HOXA TAD and chromatin signature in acute myeloid leukemia (AML)39. HOTTIP is a long noncoding RNA (lncRNA) transcribed from the 5’ tip of the HOXA locus and coordinate posterior end of the HOXA genes40. Posterior HOXA genes are aberrantly activated in specific types of AML, especially in those patients and cells carrying mixed lineage leukemia (MLL) gene rearrangement or the C-terminal of Nucleophosmin 1 (NPM1C+) mutation41, 42. HOXA and HOXB genes are critical for maintaining the balance between self-renewal and differentiation of hematopoietic stem cells (HSCs)43–45. Dysregulation of HOXA and/or HOXB genes is a dominant mechanism of leukemic transformation46. Epigenetic analysis of the HOX gene loci revealed that distinct chromatin/epigenetic signatures defined by CTCF boundary in the posterior HOXA domain contribute to the aberrant HOXA gene expression4, 47. Attenuation of CTCF boundary located in the beginning of the posterior HOXA domain impairs HOXA TAD and HOXA gene transcription, leading to blockage of AML leukemogenesis4. Overexpression of HOTTIP lncRNA restores CTCF mediated HOXA TAD and leukemogenesis in the boundary attenuated AML cells,39 supporting that collaboration of HOTTIP and CTCF stratifies CTCF boundary and its mediated oncogenic TAD to promote leukemogenesis. However, it remains unknown through which mechanism lncRNAs such as HOTTIP are targeted to specific CBSs to modulate CTCF boundary activity.
Spatial organization of genome for enhancer/promoter interactions and gene regulation
Recently, it has become possible to determine chromatin interactions in a truly unbiased and genome-wide manner by the Hi-C related technology, which can reveal the overall folding of the genome48. Recent studies implicated that CTCF regulates intra- and inter-chromosomal contacts within the nucleus at several developmentally regulated genomic loci49, 50 and suggested a primary function for CTCF in global organization of chromatin architecture9, 51. Currently, for large genomes such as those of humans and mice, Hi-C analysis will produce an interaction map with a general range of resolution from ~ 10 Kb to 1 Mb depending on the depth of sequencing reads. Such a long-range interaction landscape occupies individual territories/chromatin neighborhood and is compartmentalized in mammalian genomes in distinct levels called topologically associated domains (TADs) or sub-TADs, which were first observed with 5C data48, 52, 53. Although TADs are mostly conserved across cell types and species, TADs are indeed structural and functional chromosomal units that constrain enhancer/promoter communication for specific transcription programs1, 48. With transcription programs controlled by enhancers and super-enhancers that are key DNA regulatory elements engaged in physical long-range communications with their target promoters, organizing the genome into TADs clearly has an advantage in that regions within TADS are in close proximity for required nuclear processes such as transcriptional co-regulation, while the regions between TADs rarely interact. The transition regions between TADs are frequently bound by CTCF and cohesin, forming chromatin boundaries. In addition to CTCF-mediated TAD boundary, CTCF binding sites located within TADs facilitate and stabilize enhancer-promoter interactions to control cell-type specific transcription and cell identity54. The distinct functions of CTCF binding sites in TAD boundary establishment and enhancer/promoter interactions are dependent on the ability of CTCF to recruit distinct cohesin components to the genome chromatin sites31. Thus, CTCF recruits distinct cohesin complex in its function in eukaryotic genome organization and gene expression. Interestingly, it was recently demonstrated that apart from the role of cohesin in CTCF-mediated TAD boundaries, Stag2 (SA2) is involved in intra-TAD interactions and hematopoietic lineage specific transcription programs to control HSC self-renewal and differentiation in hematopoiesis55. It remains to be seen whether CTCF boundaries are differentially dependent on the chesin-SA1 complex for TAD formation while intra-TAD CTCF binding sites only specifically recruit cohesin-SA2 for cell type specific gene regulation, how CTCF differentially recruits different cohesin complexes for its distinct functions, and whether an RNA mechanism is involved in these processes. These outstanding questions warrant further investigation.
In order to further understand the relationship between TAD and gene regulation, several studies have employed CRISPR-CAS9 derived genome editing technology to manipulate TAD boundaries for understanding its role in genome organization and gene regulation. Deletion of these boundary regions disturbs neighboring TADs and causes ectopic long-range chromosomal interactions and transcriptional misregulation52. The HOX genes, especially HOXA and HOXB, regulate ordinary hematopoietic stem and progenitor cell (HS/PC) function by controlling the balance between proliferation and differentiation43, 44, 56. CTCF establishes a discrete functional chromatin domain in the HOX cluster during development5, 57. Disruption of CTCF boundaries in the central region of the HOXA locus alters functional chromatin domain and gene expression in mouse ES cell differentiation. This suggests that CTCF mediated TADs are not only structural components, but also regulatory units required for proper enhancer action5, 58.
According to the loop extrusion model, the progressive and dynamic movement of loops driven by cohesin are stalled at chromatin boundaries that require convergent orientation to form TADs by interacting with boundary proteins including CTCF26. It is conceivable that alteration of orientation of CTCF binding sites in the TAD boundary may disrupt the TAD formation as well as chromatin signature/gene transcription within the TAD protective chromatin neighborhood. The mouse Pcdh cluster are organized into two CTCF/cohesion dependent TAD-like chromatin domains. Guo et al. utilized CRISPR-Cas9 based genome editing to demonstrate that orientation of CTCF binding sites within the Pcdh enhancer determine chromatin loop topology and enhancer directionality59, implicating that the orientation of CTCF binding sites help to shape 3 dimensional genome architecture and regulate gene transcription. In a similar study, de wit et al. showed that CTCF binding site orientation is important for chromatin looping, although inversion of CTCF binding sites does not impair CTCF and cohesion recruitment60. Thus, CTCF is a master regulator of genome topology and gene regulation. CTCF is also extensively involved in modulating long-range chromatin interactions in pluripotent stem cells in which CTCF dependent loops demarcate chromatin-nuclear membrane attachments and transcriptional active gene cluster through extensive cross-talk between promoters and genomic regulatory elements. Thus, CTCF mediated genome-wide chromatin topology plays an important role in gene expression and cell fate commitment61, 62. The most direct evidence that long-range interactions between promoters and distal regulatory elements are involved in transcription comes from the recent genome-wide ChIA-PET analysis of the RNA polymerase II bound chromatin interaction networks63. In this study authors demonstrated that promoter directed interactions are mostly active and the actively transcribed genes with specific function are physically clustered in close proximity63, 64.
CTCF has a multi-faceted role in genome organization, in which it creates local chromatin hubs allowing coordinated expression within gene cluster, mediates interactions between distal enhancers and promoters, and specifies boundaries between active and repressive chromatin domains61. By analyzing CTCF associated chromatin loops using ChIA-PET in pluripotent mouse embryonic stem cells (mESCs), the authors defined 1,816 high-confidence interactions mediated by CTCF, of which 1,480 are intra-chromosomal and 336 are inter-chromosomal interactions. These interactions were further confirmed by 4C analysis for selected anchor points and DNA FISH techniques. Furthermore, CTCF-associated chromatin interactome were defined into five distinct chromatin domains based on distribution of seven distinct histone modifications. The interaction map was then overlapped with genome-wide RNA polymerase II (PolII) and p300 binding, which revealed that 28% of the genes whose promoters located far away from enhancers and were bound by PolII are brought into close proximity to the p300-bound enhancers through CTCF-mediated looping. They are also upregulated in mES cells compared with more differentiated neuronal stem cells,61 suggesting that CTCF mediated enhancer/promoter long-range interactions coordinate transcriptional activation. Furthermore, recent studies on genome organization during the mitosis to G1 phase cell cycle transition of erythroid cells revealed that prior to the formation of CTCF/cohesin defined large domain TADs, cohesin independent erythroid-specific sub-TADs have formed immediately after mitosis65. Important questions remain as to whether and how CTCF carries out distinct actions in TAD boundaries and enhancer/promoter contacts within TADs. It warrant determining whether different CTCF functions rely on its ability to interact with different protein complexes, for example cohesin versus Mediator complexes or Stag1 versus Stag2.
Organization of 3D genome in hematopoiesis and leukemia
Nuclear organization of the genome is a complex and dynamic process that compartmentalizes molecular machines to regulate nuclear processes including transcription, replication, and DNA repair in the nucleus66. Proper nuclear organization is particularly important for expression of many developmentally regulated complex gene loci in which long-range chromatin topologies coordinate the transcription at specific developmental stages or in specific tissue (Figure 2). Disregulation of genome topology may perturb normal cellular differentiation programs leading to cancer including leukemia (Figure 2). Genome structural alteration, including mutations, deletions, copy-number variations, and translocations are very common in leukemia67, 68. Many of these alterations are involved in perturbation of the chromatin/epigenetic landscape that becomes a driver of leukemogenesis68, 69. Hematopoiesis is a dynamic cellular process in which pluripotent hematopoietic stem cells (HSCs) give rise to diverse blood cell types. Throughout hierarchical hematopoietic development, genome organization and histone/DNA modifications play critical roles in translating cell fate decisions into epigenetic information that determines dynamic gene expression patterns in specific hematopoietic cell lineages70. In the transition from fetal to adult hematopoiesis, TAD boundaries and chromatin compartmentalization are gradually strengthened, which correlates with formation of intra-TAD enhancer/promoter loops involved in thousands of genes across the genome in adult HSCs71. It is notable that STAG1 and STAG2 have distinct functions: stabilizing CTCF-defined TAD boundaries and facilitating cell type specific enhancer/promoter communications within CTCF-defined TADs, respectively31. Although concurrent depletion of Stag1 and Stag2 revoked hematopoiesis, loss of Stag2 alone reduced chromatin accessibility and transcription of genes required for hematopoietic lineage specification55.
Figure 2. CTCF mediated 3D genome organization regulates developmental specific transcription program.

Specific nuclear process such as transcription is occurred in the compartmentalized nucleus where chromatin is formed TADs to allow coregulation of developmental/lineage specific gene expression. Dysregulation of TADs often perturbs normal gene regulatory networks leading to alteration of normal cellular function and malignant transformation.
In T-cell acute lymphoblastic leukemia (T-ALL), aberrant activation of TAL1 is found in up to 60% of T-ALL patients, making the TAL1 gene the most frequent gain-of-function mutation observed in T-ALL. Interestingly, in the TAL1 locus there are four CTCF sites that separate TAL1 from neighboring genes and regulatory elements (Figure 3). How these regulatory elements differentially regulate TAL1 expression in normal versus malignant hematopoiesis, and whether they play a role in malignant transformation is an open question. Recently several studies revealed that the −31Kb CTCF binding site (−31CBS) located between the STIL gene and the TAL1 gene plays a critical role in controlling differential enhancer/promoter contacts and TAL1 gene activation in normal and malignant hematopoiesis72, 73. Thus, questions remain as to whether and how aberrant TAL1 activation is dependent on the CTCF defined chromatin neighborhood. A recent interesting experiment using CRIPR-Cas9 genome editing demonstrated that inversion of the −31CBS orientation alters TAL1 three-dimensional genome organization through changing CTCF defined TADs and chromatin signature in the TAL1 locus, resulting in inhibition of the TAL1-driven oncogenic transcription program and T-cell leukemogenesis74. In B-ALL cells, acute loss of CTCF disrupts TAD integrity and intra-TAD enhancer/promoter loops that resulted in specific blockage of oncogene MYC and MYC target gene expression75. Thus, targeting the CTCF-mediated chromatin neighborhood provides an opportunity to correct the aberrant oncogene transcription program and to develop new molecular therapy for acute leukemia. By mapping CTCF insulated chromatin neighborhoods in T-ALL, it is quite common to find that loss of the CTCF boundary and its’ protecting chromatin neighborhoods of proto-oncogenes underly ectopic activation of T-ALL leukemic oncogenes76. Consistent with the role of CTCF chromatin boundary in regulation of oncogenic transcription programs, combined analysis of whole genome sequencing and ChIP-exo data revealed CTCF/cohesion colocalized CBSs are a major mutational hotspot in the noncoding cancer genome77.
Figure 3. The model depicts that cell type specific formation of TADs/sub-TADs at the TAL1 locus modulates expression of TAL1 oncogene.
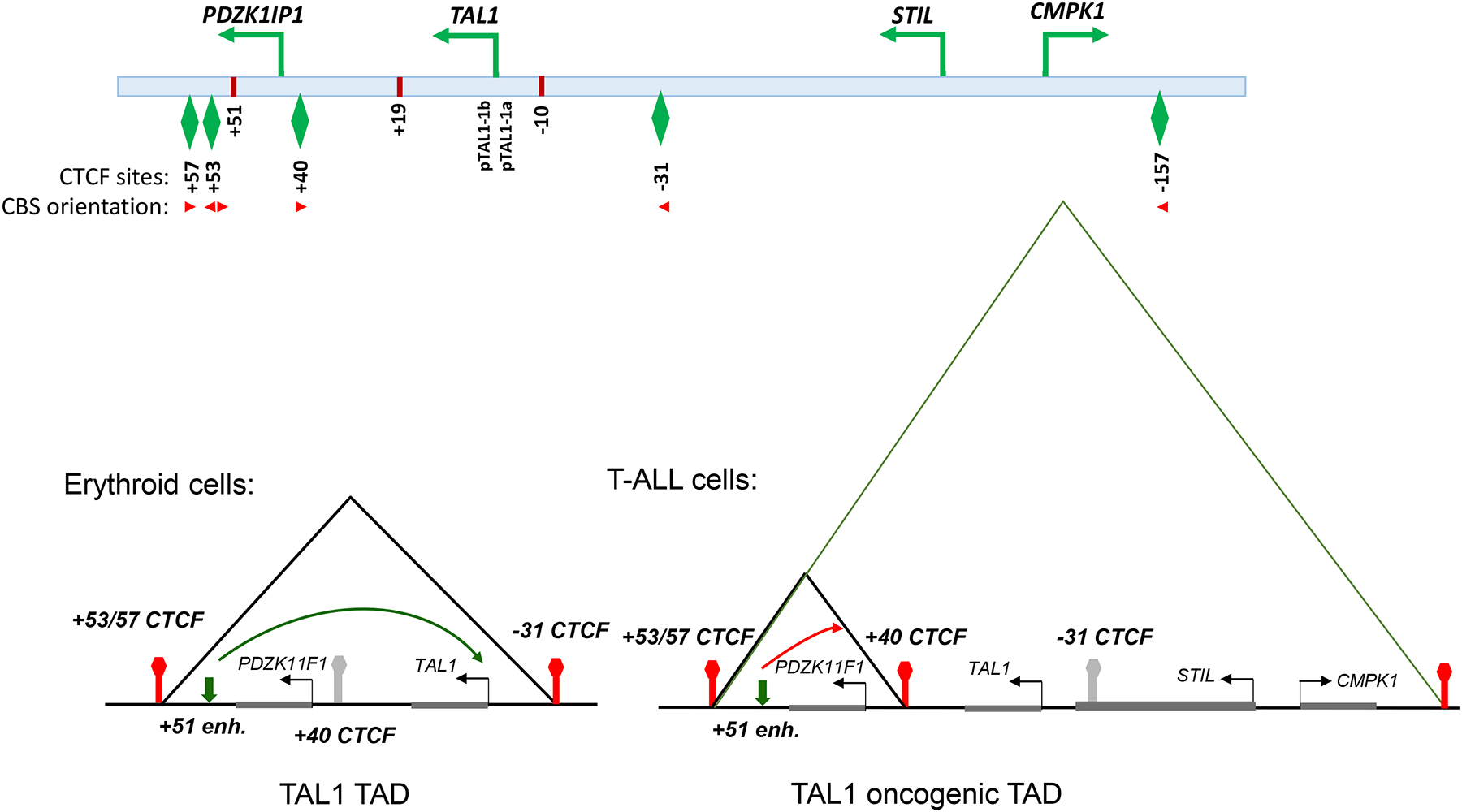
In erythroid cells, −31CBS interacts with +53CBS to form TAD/sub-TAD that allow proper activation of TAL1 gene by +51 erythroid enhancer. In contrast, +53CBS predominantly interacts with +40CBS and −157CBS to form larger TAD and sub-TAD that block the +51 enhancer action and allow T- and/or stem cell specific enhancers to aberrantly activate TAL1 gene.
It is well recognized that chromosomal rearrangements, including the rearrangements without gene fusion, are frequently implicated in leukemogenesis. Chromosomal rearrangement often disrupts or merges genome TADs and results in enhancer hijacking or redirection of new regulatory circuits underlying pathogenesis of hematopoietic malignancies. AML with inv(3)/t(3.3) is aggressively associated with ectopic activation of stem cell oncogene EVI1. In this case, chromosomal inversion alters genome TAD structure and brings a distal GATA-2 enhancer into proximity with the EVI1 oncogene leading to aberrant activation of EVI1 and reducing its own target GATA2 tumor suppressor gene2. It has been reported that there is a marked association between transcription start sites and translocation targeting regions by comparison of translocation junctions with genome-wide nuclear run-on data,78 suggesting that active transcription events organized by TAD arrangement may be correlated with translocation.
CTCF/Cohesin mediated organization of AML genome in pathogenesis of myeloid neoplasms
The clinical importance and well-defined genetic feature of MLL-rearranged (MLLr+) leukemia accounts for up to 50% of infant and 10% of adult acute leukemia that are associated with very poor prognosis and chemo-resistance79, 80. The specific chromosomal translocation involved in MLL gene has been linked to aberrant activation of posterior HOXA genes and leukemic transformation in AML81, 82. In order to identify the important CBSs involved in AML leukemogenesis, a targeted pooled CRISPR-Cas9 knockout lentivirus screening library that specifically targeted non-coding CBSs in four HOX gene loci was generated to screen MLL-rearranged AML cells for critical CBSs regulating oncogenic HOXA gene activation4. A critical CTCF chromatin boundary (CBS7/9) located at the edge of a TAD encompassing the posterior HOXA genes was identified. The CBS7/9 boundary maintains oncogenic TAD and expression of posterior HOXA genes. Impaired CBS7/9 leads to expansion of repressive chromatin structure into the posterior HOXA domain, and blocks posterior HOXA TAD and enhancer/promoter contacts, leading to decreases in posterior HOXA-associated oncogenic transcription and prolonged survival in transplanted AML mouse models4, 39. Thus, CTCF boundary not only constrains the normal gene transcription, but is hijacked to activate oncogenic genome topology and transcription programs for leukemic transformation.
As described above, the multimeric cohesin complex plays an important role in transcriptional insulation along with CTCF. Several genes of the cohesin complex such as STAG2, RAD21, SMC1A and SMC3 were recurrently mutated or deleted in myeloid neoplasms, with 12.1% in AML; 8.0% in Myelodysplastic syndromes (MDS); 10.2% in chronic myelomonocytic leukemia; and 6.3% in chronic myelogenous leukemia83. These mutations or deletions reduce the binding of cohesin complex to chromatin83, presumably altering chromatin boundary activity and leukemic genome organization. It is especially interesting that cohesin gene mutations in AML are recurrently associated with Nucleophosmin 1 (NPM1) mutation, which characterizes 20%−30% of AML cases with normal cytogenetics84. However, the prognostic impact of cohesin mutations on patients remains unclear. NPM1 belongs to a histone chaperones family, the Nucleophosmin/nucleoplasmin (NPM) family, of proteins that are involved in ribosome biogenesis, mRNA processing, and chromatin remodeling. It is particularly interesting to note that NPM1 interacts with CTCF and acts as an anchor for CTCF mediated chromatin loops85. A mutation harboring a TCTG insertion in exon 12 of NPM1 gene results in cytoplasmic translocation of the NPM1C+ protein and becomes an AML driver mutation. Blockage of NPM1C+ cytoplasmic translocation reduced HOX gene expression and promoted AML differentiation, which may lead to a potential therapeutic approach for NPM1C+ AML86. Although it was proposed that cytoplasmic mislocalization of CTCF by NPM1C+ mutation attribute to the pathogenesis of AML87, it remains to be determined whether recurrent mutations of genes of the cohesin complex and NPM1 impede CTCF function and AML genome TAD formation.
Conclusions and Future Prospective
Hematopoietic lineage commitment and differentiation require the coordination of external stimuli, signal transduction pathways, and lineage-restricted transcription factors for lineage fate determination. All information and knowledge that we gather suggest that many, if not all, transcription related events within the nucleus take place at clusters of genome enriched with chromatin regulators that facilitate long-range chromatin interactions between genes and their regulatory sites. Such a transcription event requires dynamic genome organization to form large and sub-TADs. It becomes clear that in this way, CTCF acts as chromatin boundary to demarcate independent chromatin domains and facilitate nucleus compartmentalization to regulate gene expression. However, many important questions remain to be answered. It is still largely unknown how CTCF boundary activity is perturbed in leukemia. Recent studies shed light on the potential role of non-coding RNAs and CTCF interactions in CTCF chromatin insulation37, 38. Whether and how non-coding RNAs regulate CTCF functions such as chromatin binding, cohesin interaction, TAD boundary formation, or enhancer/promoter communications remains to be determined and characterized. Although dynamic long-range interactions in the nucleus are implicated in regulated gene expression, a central question remains as to what the underlying mechanism is by which CTCF differentially modulates TAD boundaries versus enhancer/promoter contacts within CTCF defined TAD. Do distinct CTCF functions rely on recruitment of different cohesion components, transcription factors, or non-coding RNAs? Furthermore, understanding how dysregulated CTCF function leads to perturbation of the hematopoietic genome and gene transcription networks resulting leukemic pathogenesis has become particularly interesting. Given that manipulation of CTCF protein or its binding sites shows strong effects on the leukemic oncogene expression program4, 74, 75, we must ask whether new genome editing technologies can be utilized to target CTCF mediated genome organization or enhancer/promoter interactions for correction of oncogenic transcription programs and for targeted therapeutic applications.
Acknowledgements
We thank our colleague in the Huang and Qiu Labs for their comments on this manuscript. The authors also thank Rachael Mills for editing the manuscript. This work was supported by the grants from the National Institute of Health (R01DK110108, R01CA204044, R01HL141950 to S.H.; R01HL144712 to Y.Q.) and the Four Diamonds Fund (S.H.).
Footnotes
Conflict of interest disclosure:
The authors declare no competing financial interests.
Reference:
- 1.Valton AL, Dekker J. TAD disruption as oncogenic driver. Curr Opin Genet Dev 2016. February; 36: 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014. April 10; 157(2): 369–381. [DOI] [PubMed] [Google Scholar]
- 3.Taberlay PC, Achinger-Kawecka J, Lun AT, Buske FA, Sabir K, Gould CM, et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res 2016. June; 26(6): 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo H, Wang F, Zha J, Li H, Yan B, Du Q, et al. CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood 2018. August 23; 132(8): 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 2015. February 27; 347(6225): 1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum Genet 1982; 62(3): 201–209. [DOI] [PubMed] [Google Scholar]
- 7.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol 2006. June; 18(3): 307–316. [DOI] [PubMed] [Google Scholar]
- 8.Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, et al. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet 2008. March; 4(3): e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell 2009. June 26; 137(7): 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet 2018. December; 19(12): 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 2014. October 9; 159(2): 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 2015. December 17; 163(7): 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999. August 6; 98(3): 387–396. [DOI] [PubMed] [Google Scholar]
- 14.Chung JH, Whiteley M, Felsenfeld G. A 5’ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 1993. August 13; 74(3): 505–514. [DOI] [PubMed] [Google Scholar]
- 15.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 2008. October 10; 32(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 2006. September; 7(9): 703–713. [DOI] [PubMed] [Google Scholar]
- 17.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev 2002. February 1; 16(3): 271–288. [DOI] [PubMed] [Google Scholar]
- 18.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome research 2009. January; 19(1): 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci 2009. May 1; 122(Pt 9): 1275–1284. [DOI] [PubMed] [Google Scholar]
- 20.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Molecular cell 2004. January 30; 13(2): 291–298. [DOI] [PubMed] [Google Scholar]
- 21.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell 2016. March 10; 164(6): 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Haarhuis JHI, Sedeno Cacciatore A, Oldenkamp R, van Ruiten MS, Willems L, et al. The structural basis for cohesin-CTCF-anchored loops. Nature 2020. February; 578(7795): 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 2008. February 8; 132(3): 422–433. [DOI] [PubMed] [Google Scholar]
- 24.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008. February 14; 451(7180): 796–801. [DOI] [PubMed] [Google Scholar]
- 25.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A 2008. June 17; 105(24): 8309–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 2016. May 31; 15(9): 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A 2018. July 17; 115(29): E6697–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 2014. January 21; 111(3): 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 2009. July 16; 460(7253): 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 2009. November; 5(11): e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojic A, Cuadrado A, De Koninck M, Gimenez-Llorente D, Rodriguez-Corsino M, Gomez-Lopez G, et al. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol 2018. June; 25(6): 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013. June 6; 153(6): 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 2013. August 15; 154(4): 801–813. [DOI] [PubMed] [Google Scholar]
- 34.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 2010. November 15; 24(22): 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kung JT, Kesner B, An JY, Ahn JY, Cifuentes-Rojas C, Colognori D, et al. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell 2015. January 22; 57(2): 361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013. March 14; 152(6): 1308–1323. [DOI] [PubMed] [Google Scholar]
- 37.Saldana-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jacome-Lopez K, Nora EP, et al. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Mol Cell 2019. November 7; 76(3): 412–422 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldana-Meyer R, Reinberg D, et al. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol Cell 2019. November 7; 76(3): 395–411 e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y, et al. HOTTIP lncRNA Promotes Hematopoietic Stem Cell Self-Renewal Leading to AML-like Disease in Mice. Cancer Cell 2019. December 9; 36(6): 645–659 e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011. April 7; 472(7341): 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia 2002. February; 16(2): 186–195. [DOI] [PubMed] [Google Scholar]
- 42.Andreeff M, Ruvolo V, Gadgil S, Zeng C, Coombes K, Chen W, et al. HOX expression patterns identify a common signature for favorable AML. Leukemia 2008. November; 22(11): 2041–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dou DR, Calvanese V, Sierra MI, Nguyen AT, Minasian A, Saarikoski P, et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat Cell Biol 2016. June; 18(6): 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 2005. December 01; 106(12): 3988–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng C, Li Y, Liang S, Cui K, Salz T, Yang H, et al. USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet 2013. June; 9(6): e1003524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013. April; 27(5): 1000–1008. [DOI] [PubMed] [Google Scholar]
- 47.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O’Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia 2015. June; 29(6): 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012. April 11; 485(7398): 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nature genetics 2011. July; 43(7): 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 2006. November; 38(11): 1341–1347. [DOI] [PubMed] [Google Scholar]
- 51.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Current opinion in genetics & development 2007. October; 17(5): 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012. May 17; 485(7398): 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sexton T, Kurukuti S, Mitchell JA, Umlauf D, Nagano T, Fraser P. Sensitive detection of chromatin coassociations using enhanced chromosome conformation capture on chip. Nat Protoc 2012. July; 7(7): 1335–1350. [DOI] [PubMed] [Google Scholar]
- 54.Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, et al. CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Mol Cell 2017. September 21; 67(6): 1049–1058 e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viny AD, Bowman RL, Liu Y, Lavallee VP, Eisman SE, Xiao W, et al. Cohesin Members Stag1 and Stag2 Display Distinct Roles in Chromatin Accessibility and Topological Control of HSC Self-Renewal and Differentiation. Cell Stem Cell 2019. November 7; 25(5): 682–696 e688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng C, Li Y, Liang S, Cui K, Salz T, Yang H, et al. USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS genetics 2013. June; 9(6): e1003524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez-Carballo E, Lopez-Delisle L, Zhan Y, Fabre PJ, Beccari L, El-Idrissi I, et al. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev 2017. November 15; 31(22): 2264–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narendra V, Bulajic M, Dekker J, Mazzoni EO, Reinberg D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev 2016. December 15; 30(24): 2657–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 2015. August 13; 162(4): 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell 2015. November 19; 60(4): 676–684. [DOI] [PubMed] [Google Scholar]
- 61.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 2011. July; 43(7): 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 2019. August; 20(8): 437–455. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Wong CH, Birnbaum RY, Li G, Favaro R, Ngan CY, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 2013. December 12; 504(7479): 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature 2012. September 6; 489(7414): 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H, Emerson DJ, Gilgenast TG, Titus KR, Lan Y, Huang P, et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 2019. December; 576(7785): 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Misteli T Beyond the sequence: cellular organization of genome function. Cell 2007. February 23; 128(4): 787–800. [DOI] [PubMed] [Google Scholar]
- 67.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 2016. January 7; 127(1): 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012. March 22; 366(12): 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013. May 30; 368(22): 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haas S, Trumpp A, Milsom MD. Causes and Consequences of Hematopoietic Stem Cell Heterogeneity. Cell Stem Cell 2018. May 3; 22(5): 627–638. [DOI] [PubMed] [Google Scholar]
- 71.Chen C, Yu W, Tober J, Gao P, He B, Lee K, et al. Spatial Genome Re-organization between Fetal and Adult Hematopoietic Stem Cells. Cell Rep 2019. December 17; 29(12): 4200–4211 e4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patel B, Kang Y, Cui K, Litt M, Riberio MS, Deng C, et al. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia 2014. February; 28(2): 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, Kurukuti S, Saffrey P, Vukovic M, Michie AM, Strogantsev R, et al. Chromatin looping defines expression of TAL1, its flanking genes, and regulation in T-ALL. Blood 2013. December 19; 122(26): 4199–4209. [DOI] [PubMed] [Google Scholar]
- 74.Li Y, Liao Z, Luo H, Benyoucef A, Kang Y, Lai Q, et al. Alteration of CTCF-associated chromatin neighborhood inhibits TAL1-driven oncogenic transcription program and leukemogenesis. Nucleic Acids Res 2020. February 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyle J, Zhang Y, Wright S, Xu B, Shao Y, Easton J, et al. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer-promoter looping. Nucleic Acids Res 2019. July 26; 47(13): 6699–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 2016. March 25; 351(6280): 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 2015. July; 47(7): 818–821. [DOI] [PubMed] [Google Scholar]
- 78.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 2011. September 30; 147(1): 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeisig BB, Kulasekararaj AG, Mufti GJ, So CW. SnapShot: Acute myeloid leukemia. Cancer Cell 2012. November 13; 22(5): 698–698 e691. [DOI] [PubMed] [Google Scholar]
- 80.Winters AC, Bernt KM. MLL-Rearranged Leukemias-An Update on Science and Clinical Approaches. Front Pediatr 2017; 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rice KL, Licht JD. HOX deregulation in acute myeloid leukemia. J Clin Invest 2007. April; 117(4): 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia 2009. August; 23(8): 1490–1499. [DOI] [PubMed] [Google Scholar]
- 83.Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 2013. October; 45(10): 1232–1237. [DOI] [PubMed] [Google Scholar]
- 84.Heath EM, Chan SM, Minden MD, Murphy T, Shlush LI, Schimmer AD. Biological and clinical consequences of NPM1 mutations in AML. Leukemia 2017. April; 31(4): 798–807. [DOI] [PubMed] [Google Scholar]
- 85.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 2004. January 30; 13(2): 291–298. [DOI] [PubMed] [Google Scholar]
- 86.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell 2018. September 10; 34(3): 499–512 e499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang AJ, Han Y, Jia N, Chen P, Minden MD. NPM1c impedes CTCF functions through cytoplasmic mislocalization in acute myeloid leukemia. Leukemia 2019. December 12. [DOI] [PubMed] [Google Scholar]


