Abstract
The essential role of reverse transcription in the HIV life cycle is illustrated by the fact that half of the ~30 FDA-approved drugs for HIV treatment target HIV-1 reverse transcriptase (RT). Even though more than 160 structures of RT deposited in the Protein Data Bank (PDB) have revealed the molecular architecture of RT in great detail, some key states of RT function and inhibition remain still unknown. Recent structures of RT initiation complexes, RT poised for RNA hydrolysis, and RT with approved drugs and investigational compounds have provided a deeper understanding of RT function and inhibition, suggesting novel avenues for targeting this central enzyme of HIV.
Introduction
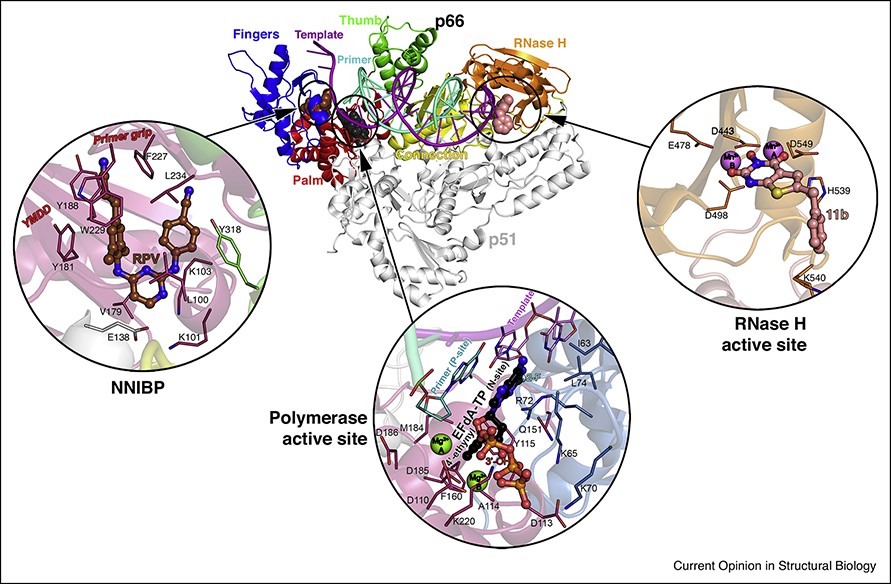
Human immunodeficiency virus infection (HIV/AIDS) came into the spotlight as a silent, deadly, and mysterious killer during the 1980s. Nearly 40 years later, AIDS has been transformed from a lethal infection to a manageable chronic disease, largely because of highly active antiretroviral therapy (HAART). However, HIV/AIDS remains a major threat to global health (~40 million people infected in the world). HIV-1 reverse transcriptase (RT) is central to HAART and in pre-exposure prophylaxis. Standard therapies include at least two RT inhibitors, and often three [1]. RT is a characteristic enzyme in all retroviruses, with polymerase and RNase H activities, and is essential for HIV replication. Figure 1 shows the overall arrangement of the HIV-1 RT p66/p51 heterodimer. p66 has an N-terminal RNA-dependent and DNA-dependent DNA polymerase domain with a hand-like arrangement, with fingers, palm, thumb and connection subdomains; and an RNase H domain, that digests the RNA. Both activities require Mg2+ for catalysis. The smaller subunit, p51, a proteolytic product of p66, has a very different arrangement (despite the same amino acid sequence) and plays a structural role. HIV-1 RT is the best understood polymerase in terms of structure, mechanisms of catalysis, inhibition, and resistance [2–4]. Nevertheless, the recent years have unveiled the detailed molecular architecture of some key uncharted functional states of HIV-1 RT, as well as advances in inhibitor and drug development. Here, we will discuss those in which structural biology has played a prominent role.
Figure 1. Overview of HIV-1 reverse transcriptase (RT) structure and main binding sites for substrates and inhibitors.
Structure of RT in complex with dsDNA and incoming EFdA-TP (PDB ID 5J2M), with template-primer, with p66 polymerase fingers, palm, thumb, connection subdomains and RNase H domain, and with p51 indicated, with the RT/rilpivirine (brown spheres, PDB ID 4G1Q) and RT/11b (light pink spheres, PDB ID 6AOC) structures superposed. Zoom-ins of: i) the NNIBP (NNRTI binding pocket) of the RT/rilpivirine (RPV, circled) complex (PDB ID 4G1Q) superposed to the former structure, with residues, catalytic motif YMDD and primer grip domain location highlighted; ii) the polymerase active site in the presence of incoming EFdA-TP (circled, 2-fluorine (2-F), 4’-ethynyl and 3’-OH moieties indicated), residues, template-primer, catalytic metals, N-site, and P-site indicated; iii) the RNase H active site with 11b bound, catalytic metals and residues indicated.
Structure and function of HIV-1 reverse transcriptase
Initiation of reverse transcription
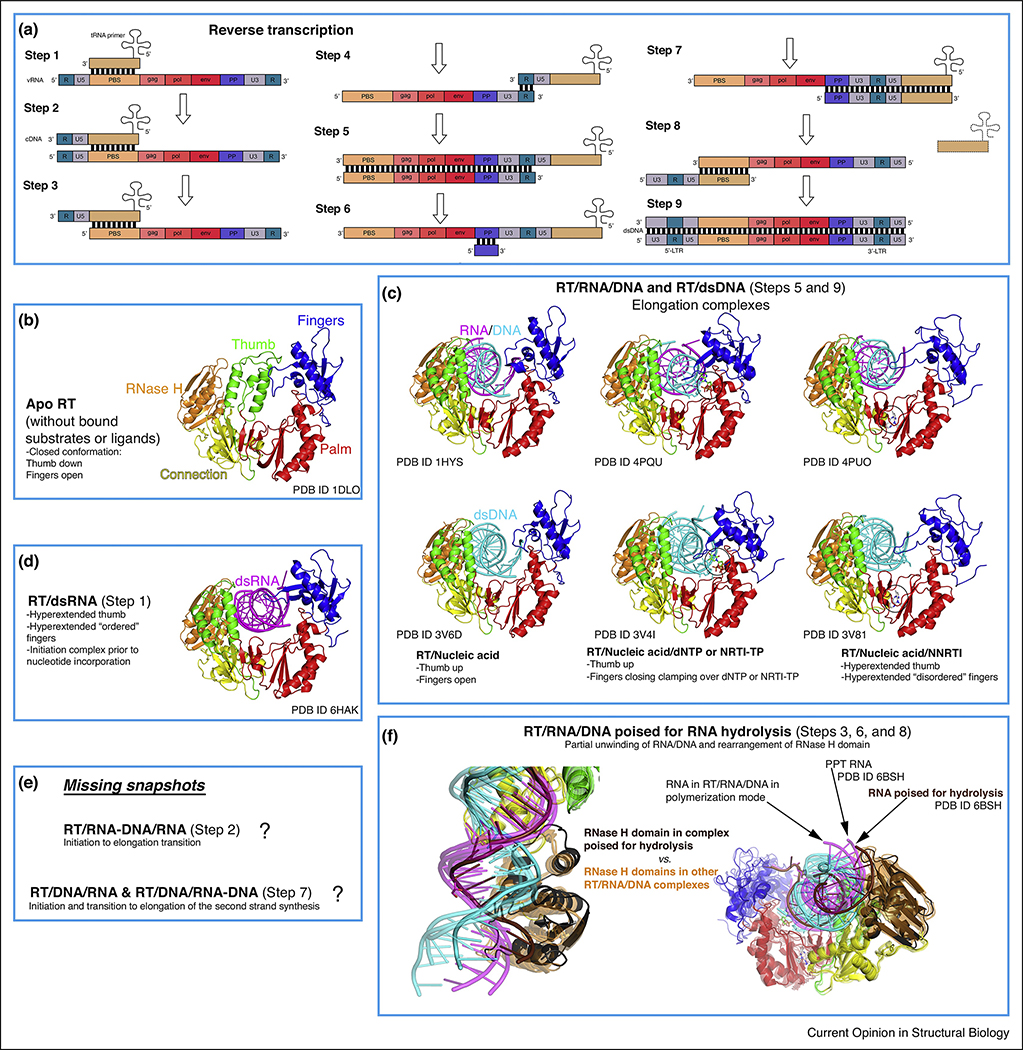
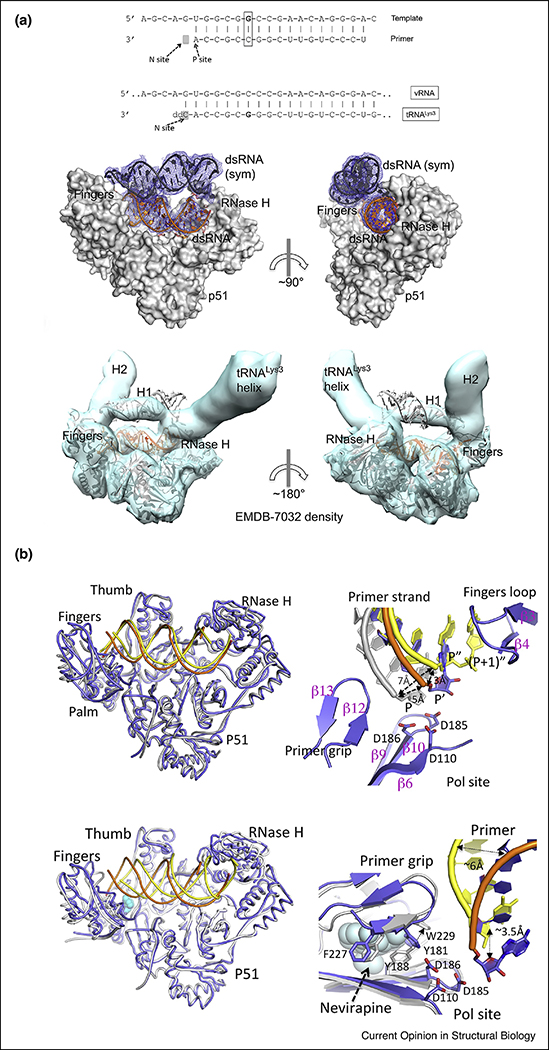
Reverse transcription is the process by which HIV converts its single-stranded (ss)RNA genome to double-stranded (ds)DNA (Figure 2a). As RT cannot initiate synthesis de novo, HIV-1 recruits the host tRNALys3 as a primer, which anneals to the primer-binding site (PBS) near the 5’ end of the HIV genome. Initiation is slow and non-processive, contrary to a much faster elongation phase that has been extensively characterized by biochemical and structural studies. Initiation has been studied in some depth, but its structural characterization remained elusive because of the much lower affinity of dsRNA to RT compared to dsDNA and RNA/DNA (template-primer, T-P) [5]. The first structure of an RT initiation complex (RTIC, 8 Å global resolution, 4.5 Å duplex region) was reported in 2018 by the Puglisi laboratory [6••], using cryo-EM, followed by the 3.95 Å resolution crystal structure this year by our laboratory [7••]. These remarkable structures provide highly complementary information (Figure 3a). The crystal structure describes the HIV-1 RT/dsRNA initiation complex poised for nucleotide incorporation (P-site, primer position 0), while the cryo-EM structure captures the RTIC right after incorporation of one nucleotide and before translocation (N-site, primer position +1). The cryo-EM structure positions the RNA elements protruding outside of the RT nucleic acid-binding cleft, and the crystal structure provides a detailed model for RT and the dsRNA. The cryo-EM structure consists of a 101 bp portion of HIV-1 genomic RNA (vRNA) and a full-length tRNALys3, whereas the crystal structure represents the core of the RTIC formed by a 23–17 T-P dsRNA to mimic the PBS region of the duplex. Both structures were enabled by disulfide cross-linking between a thioalkyl-modified G of the RNA and mutated thumb residue Q258C.
Figure 2. Conformational states of RT during reverse transcription.
(a) Schematic representation of retroviral reverse transcription, displaying the genomic and host nucleic acids (modified from https://en.wikipedia.org/wiki/Reversetranscriptase#/media/File:Reversetranscription.svg, Creative Commons Attribution License CC BY 3.0 license). In brief, the different steps correspond to: 1- Initiation of first strand synthesis; 2- Initiation to elongation transition (RNA/DNA hybrid primer); 3- Hydrolysis of the 5’ region of the viral RNA (vRNA); 4- tRNA jump to the 3’ region of the vRNA; 5- Elongation (dsDNA); 6- vRNA degradation (except polypurine tract, PPT); 7- Initiation of the second strand synthesis; 8- PPT hydrolysis and second jump (tRNA exit); 9- Second strand elongation and addition of the long terminal repeats (LTRs). (b-f) The rest of the boxes (except from the one indicating the challenges remaining, that is, uncharacterized conformational states) contain all the described conformations of RT complexed with nucleic acids (and apo form) during reverse transcription, with color coding and annotations included for clarity. The notion of ‘ordered’ versus ‘disordered’ hyperextended fingers subdomain (boxes C and D) is based on B-factors (lower versus higher) and number of interactions with the nucleic acid strand (more versus less).
Figure 3. Comparison of the cryo-EM and X-ray crystallographic structures of the RT initiation complex (RTIC).
(a) On top, nucleotide sequences of i) dsRNA 23-mer-17-mer template-primer used in X-ray crystallography structure (a template G was cross-linked to the Q258C position of RT without primer extension and with the G and C positions of the sixth base pair switched compared with the naturally annealed PBS-tRNALys3 sequence), and ii) the annealed PBS region of vRNA-tRNALys3 in the RTIC for which the cryo-EM structure was reported (the vRNA-tRNALys3 was cross-linked to RT at ‘G’ and primer extended with catalytic incorporation of a ddCMP). Below, comparison of the crystal lattice dsRNA/dsRNA interaction reminiscent of vRNA-tRNALys3 RNA interactions in the RTIC complex and fitting of the above complex in the 8 Å cryo-EM density (EMBD-7032) by aligning the RT/dsRNA complex and the dsRNA (sym) positioned exactly on the density for the H1 helix and connecting loop of vRNA.
(b) On top, superposition of the RT/dsRNA structure (blue protein, orange RNA) on the structure of the RTIC core (PDB ID 6B19; yellow protein and RNA) showing high structural resemblance. Detail of the relative locations of the primer 3′ ends: P′ of RT/dsRNA (blue/orange), (P+ 1)′′ of cryo-EM RTIC N complex (yellow), and P of RT/dsDNA (gray) structures. The incorporated ddCMP is at (P + 1)′′ and the nucleotide corresponding to P′ of RT/dsRNA is located at P′′ in the RTIC structure. The primer 3′ end has to reach the P-site for nucleotide incorporation by RT. Detail of the RT/dsRNA complex (blue/orange), closely resembling RT in the RT/dsDNA/nevirapine complex (white/yellow/cyan spheres, PDB ID 3V81). Close-up view of the polymerase site and NNRTI pocket shows that the primer 3′ ends in both structures are displaced away from the polymerase site, and also about 3.5 Å apart. Only minimal structural arrangements of the NNRTI pocket region of RT/dsRNA structure are needed to accommodate nevirapine; the major adjustment required is the repositioning of W229 as indicated by an arrow. The side chains of Y181 and Y188 have disordered electron density, and therefore are not included in the RT/dsRNA structure. Figure adapted from Ref. [7••].
A key finding in both structures is the large displacement of the primer terminus (~7 Å) from the polymerase active site (Figure 3b), which might explain the very slow rate of addition of the first DNA nucleotides. Both thumb and fingers are hyperextended, owing to the wider dsRNA versus RNA/DNA and dsDNA. Unexpectedly, the RTIC active site region is most similar to that in RT/dsDNA/NNRTI, albeit differing widely in their nucleic acid tracks and contacts with RT [7••]. The RTIC crystal structure shows the NNRTI-binding pocket in an open configuration (Figures 2b,c and 3b), never visualized before in an RT structure not containing an NNRTI. The cryo-EM structure reveals the vRNA forms two helical stems protruding above the RT active site, with a connecting loop extending toward the RNase H region, while the 5′ end of the tRNA refolds and stacks on the PBS. Surprisingly, in the crystal structure, weak dsRNA: dsRNA lattice interactions mimic these protruding RNA elements observed in the cryo-EM structure (Figure 3a). The RNA topology thus forces RT to adopt an unusual conformation. Reciprocally, RT limits the basal conformational heterogeneity of the vRNA/tRNA, as observed by single-molecule Forster resonance energy transfer [8•]. This publication reports that extending the duplex PBS region from 18 to 22 bp decreases the RTIC koff significantly. Overall, the RTIC is a very dynamic complex, and significant conformational rearrangements of both RT and the vRNA/tRNA constituents are required for the start of retroviral reverse transcription.
Relationship between polymerase and ribonuclease activities
The RNase H domain hydrolyzes the vRNA in different stages of reverse transcription (Figure 2a) [9]. Several RT/RNA/DNA structures were available, with the substrate interacting either with the polymerase or the RNase H active site. However, all had the scissile phosphate distance >4 Å, that is, not poised for efficient catalysis. In 2018, the Yang laboratory reported the first structure of RT in complex with a RNA/DNA substrate poised for catalysis (Figure 2f) [10••]. They found that previous failures were due to unfavorable sequences for RNase H cleavage: 2 residues located 4 bp upstream of the cleavage site, and especially when position −4 has a rA-dT pair, block the unwinding needed for proper positioning of the substrate for catalysis.
The spatial-temporal coordination of polymerase and ribonuclease activities within RT, essential for reverse transcription, is not fully understood. An important question is whether an RNA/DNA substrate can simultaneously engage both active sites. The Johnson laboratory (by pre-steady-state kinetics [11•]) and Nowotny laboratory (via detection of RNase H cleavage of RNA/DNA cross-linked to the polymerase active site [12•]) have found compelling evidences supporting existence of this simultaneous state. The first work also suggests the basis of the coordination between both active sites: polymerization is fast and processive, whereas RNA hydrolysis is slow and periodic (i.e. an RNase H cut takes 6 times longer than 1 nucleotide incorporation, but every cut hydrolyzes ~6 nucleotides).
Maturation of RT
RT initially exists as homodimer and undergoes maturation, leading to a heterodimer with a sole RNase H domain (reviewed in Ref. [13]). Maturation starts with the transition of monomeric RT from a compact (p51-like) to an extended (p66-like) conformation. Next, dimerization yields a p66/p66’ homodimer in which just p66’ (but not p66) will unfold and become exposed, allowing cleavage by protease, finishing RT maturation. The unfolding step remains controversial. London and colleagues have solved the structure of an isolated RNase H domain-swapped dimer [14•] (it captures a partially unfolded monomer stabilized by the second monomer), that points to instability of the Y427 binding pocket as the trigger of RNase H unfolding. Electron paramagnetic resonance and modeling experiments by the Clore laboratory also support the unfolding of a single RNase H domain and the existence of an asymmetric homodimer [15•]. The Ishima laboratory (in collaboration with Sarafianos, Parniak and Sluis-Cremer laboratories) has also shown that tRNA (packaged in HIV-1 virions) may enhance maturation by interacting with the homodimer and promoting conformational asymmetry [16•]. According to their NMR data, though, the unfolding may not occur just with p66/p66’ alone, requiring presence of either nucleic acid or protease [17•]. Our laboratory in collaboration with that of Dmitry Lyumkis (Salk) has recently solved a structure of the HIV-1 Pol polyprotein precursor by cryo-EM, revealing a dimeric organization in which the RT dimer is very similar in structure to the mature p66/p51 heterodimer (Harrison et al., unpublished), with many fascinating implications about RT maturation and assembly, as well as protease activation during virion morphogenesis.
Structure and inhibition of HIV-1 reverse transcriptase
Research efforts for HIV inhibitor/drug discovery and development, remain very numerous and vigorous [18]. Given the large number of papers on the topic in the review period, we will address those works comprising or based on structures.
Nucleoside RT inhibitors (NRTIs)
Classic NRTIs are chain terminators, lacking the 3’–OH group necessary for catalysis, thus halting reverse transcription. Medicinal chemistry, enzymology and crystallography enabled thorough understanding of most drug resistance mechanisms, that is, exclusion and excision. Nevertheless, NRTIs with alternative mechanisms and with higher genetic barrier toward drug resistance are needed [2,3].
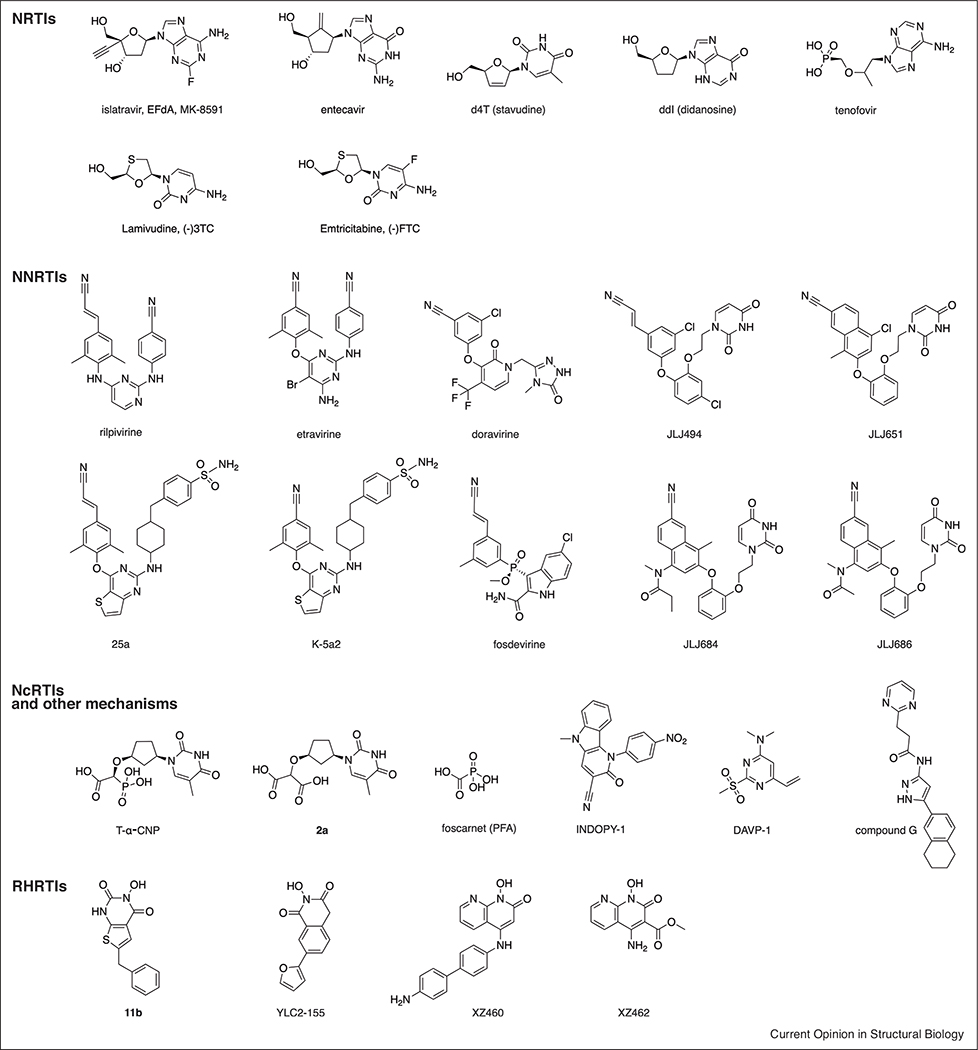
4’-ethynyl-2-fluoro-2’-deoxyadenosine (EFdA, MK-8591, islatravir, Chart 1) is a novel NRTI in Phase 2b trials for HIV treatment. It has a very long half-life, with great potential as long-acting drug [19]. The Sarafianos laboratory has been instrumental in characterizing its unusual mechanisms of action biochemically [20] and structurally. In 2016, they presented a set of structures of RT/DNA with EFdA that unraveled these mechanisms in atomic detail [21••]. EFdA bears a 3’-OH group; however, it acts as immediate or delayed chain terminator (ICT or DCT). The RT/dsDNA/EFdA-triphosphate (-TP) structure reveals the 4’-ethynyl group binds to a preformed hydrophobic pocket defined by conserved residues A114, Y115, F160, and M184 (Figure 1). No conformational change is needed for its opening, with a remarkable shape complementarity, also illustrated by structure-activity relationship (SAR) studies with 4’-modified derivatives [22]. The 3’–OH also forms polar interactions with RT. The tight binding thus explains its ICT mechanism, leading to a deficiency in translocation. Additionally, the 4’-ethynyl group causes primer distortion by steric hindrance when it is at the P-site or upstream of it. Finally, facile EFdA misincorporation stems from its ability to form Hoogsteen base pairing. All these features explain EFdA’s high genetic barrier for resistance [20].
Chart 1.
Molecular formulas of the compounds cited in the manuscript.
Entecavir (ETV) is an FDA-approved drug for treating hepatitis B virus (HBV) infections. It has also a 3’–OH group (Chart 1), and acts as a DCT [23•]. Because of intractability of producing recombinant HBV RT, the Yasutake and Mitsuya groups have solved HIV-1 RT/DNA/ETV structures as surrogates, where key substitutions present in HBV are introduced [23•,24•]. The structures reveal that the ETV-TP ethenyl appendage to the cyclopentyl ring, instead of binding to the same pocket as EFdA, is accompanied by a displacement of the M184 side chain (M204V/I in HBV RT confers resistance to ETV). They also provide evidence to explain the different binding specificity of 4’-substituted derivatives between HIV-1 and HBV: the preformed pocket observed in HIV-1 RT (Figure 1) may be shallower in HBV RT, explaining its preference for 4’-cyano over the bulkier 4’-ethynyl groups.
The Anderson laboratory in a collaboration with Schinazi recently solved RT/dsDNA structures bound to the TP forms of the very widely used NRTI drugs lamivudine [(−) 3TC] and emtricitabine [(−)FTC] (Chart 1), hinting that the distorted conformation of the TP in these L stereochemistry nucleotides may be responsible for their slower incorporation rate [25•]. These structures provide a framework for designing novel L stereochemistry NRTIs. Our laboratory has also contributed two recent publications describing RT/dsDNA/NRTI structures and resistance mechanisms. First, we have unveiled the Q151M and Q151M complex RT resistance mechanism that arise after treatment with dideoxynucleosides (ddNTPs) [26]. In essence, Q151M allows discrimination between the flexible ring of ddNTPs versus the canonical 3’-endo conformation in dNTPs. The disruption of the key Q151-R72 interaction (Figure 1), though, may explain the reduced viral fitness of the Q151M variant and presumably to existence of alternative conformations of the dNTP-binding site. The accompanying mutations seem to compensate the Q151M mutation and improve fitness by stabilizing a single dNTP-binding site conformer. Next, we have solved ternary complexes with the TP version of discontinued drug d4T (Chart 1). The stronger Kd of d4TTP relative to dTTP can be rationalized by more efficient stacking displayed by the d4TTP planar ring. In this work we also report an efficient and cost-effective method for nucleic acid cross-linking to RT, through the fingers subdomain I63C mutation and a thioalkylated adenine base in the upstream template strand [27•].
Non-nucleoside RT inhibitors (NNRTIs)
There are 6 FDA-approved NNRTIs, including recently approved doravirine (DOR, Chart 1), with > 50 classes of structurally diverse NNRTIs being investigated. Yet, developing new NNRTIs remains desirable, as resistance mutations and related adverse effects continue to emerge in patients [28]. Diarylpyrimidines (DAPYs, led by etravirine (ETV) and rilpivirine (RPV), Chart 1), structurally characterized by the Arnold laboratory, established the paradigm for the features that an NNRTI must have [29]: conformational flexibility and positional adaptability (‘wiggling and jiggling’) to impose a high genetic barrier to mutation, which first generation NNRTIs lacked. The NNRTI-binding pocket (NNIBP, Figure 1), contains three channels: entrance, tunnel and groove (where central ring, left and right wings of DAPYs lay, respectively). Entrance and groove are solvent-accessible, providing chemical space for NNRTI modification. Jorgensen recently reported a computational simulation of NNRTI entrance into and exit from the NNIBP [30], extending a related analysis from Jiang [31]. A crystal fragment screening campaign by our laboratory has revealed two additional inhibitory binding sites in the polymerase active site region: Knuckles and NNRTI Adjacent [32].
The Liu and De Clercq groups have jointly performed a remarkable SAR screening of the NNIBP [33–35]. Most notably, they have developed thiophene[3,2-d]pyrimidine NNRTIs, for example, K-5a2 and 25a (Chart 1). Basically, they kept a ‘privileged scaffold’ in the left wing (from ETV and RPV, respectively), and evolved the central ring and right wing with a thiophene moiety and a piperidine-linked benzenesulfonamide group. In collaboration with the Steitz lab [36••], they determined crystal structures of wild-type and mutant RTs with these lead compounds, pinpointing their binding modes and how they evade the main NNRTI-resistance mutations through conformational flexibility and positional adaptability. The thiophene group occupies a larger space in the entrance and makes water-mediated interactions with main-chain atoms of E138 and K101. Binding of the larger right wing moiety is accompanied by readjustment of the flanking residues, filling the groove channel. These leads keep the virtues of ETV and RPV and add significantly more hydrogen bonds with main-chain atoms, making them less susceptible to resistance mutations, as also observed for DOR [37]. Additionally, Zhan, Liu et al. have further developed these leads and explored substitutions in the three channels, with a focus on improving bioavailability [38,39]. Worth noting is their development of DAPYs targeting simultaneously the NNIBP and the aforementioned NNRTI Adjacent pocket [40•].
Another outstanding effort for NNRTI development has come from the collaboration between the Jorgensen and Anderson groups. They previously developed the catechol diether NNRTI class [41]. Lead JLJ494 (Chart 1) has a cyanovinyl group, susceptible to generation of off-target covalent adducts. For RPV this was not an issue, but fosdevirine, which also contains this group, was abandoned in Phase 2 clinical trials because of adverse effects [42]. Jorgensen then generated an alternative containing a cyano-naphthyl substituent [43]. Interestingly, the C–Cl bond in JLJ651 (Chart 1) points towards Y181, which is often mutated to C181 as one of the most prevalent NNRTI-resistance mutations, suggested potential for covalent bonding to the Cys residue upon replacement by an appropriate electrophile. They synthesized 5 compounds based on this premise and found 2 covalent Y181C NNRTIs (JLJ684 and JLJ686, Chart 1) [44••]. The crystal structures show continuous electron density between the C181 sulfur and the compounds, allowing visualization of the covalent bond. These compounds completely abrogated activity of C181-bearing RTs, but had a significant potency drop against wild-type and other mutant RTs.
Nucleotide-competing RT inhibitors (NcRTIs) and other inhibition mechanisms
NcRTIs are a class of different chemotypes that inhibit RT by competing with dNTP incorporation.
The first group is formed by substrate mimics, such as the α-carboxynucleoside phosphonates (α-CNPs, Chart 1), and product mimics (pyrophosphate, PPi) such as foscarnet (PFA, used to treat herpes infections, Chart 1). Collaboration between the Balzarini, Maguire, and Arnold groups has yielded development of α-CNPs as viral DNA polymerase inhibitors without requiring metabolic conversion. They have three moieties: nucleobase, carboxy-phosphonate, and linker [45]. Our laboratory has solved structures of T-α-CNP and a T-α-malonate derivative (2a, Chart 1) complexed to RT/dsDNA [46–48]. The structures show that α-CNPs perfectly mimic dNTPs, while malonates and bisphosphonates present weaker divalent metal ion chelation, explaining their lower inhibition. Replacement of the cyclic group by an acyclic linker shifts the selectivity from RT to herpetic DNA polymerases [49]. Our group has also determined the RT/DNA/PFA structure [47], expected to bind to a transient N-site RT/DNA complex. Key for solving it was using a 38-mer DNA T-P aptamer (developed by the DeStefano laboratory [50]): it mimics a dsDNA with pM binding to RT, and is catalytically active but translocation-incompetent in crystals (due to hairpin interaction with the RNase H domain). This structure shows PFA binding requires chelation with the active site, but its pattern is distinct for the expected for PPi poised for nucleotide excision. More structural information with further PPi analogs could facilitate their improvement.
Indolopyridones (INDOPYs) and dimethylamino-6-vinylpyrimidines (DAVPs) are chelation-independent NcRTIs (Chart 1). INDOPYs were discovered through cell-based high-throughput screening [51]. The Gotte laboratory also contributed to characterizing their mechanism, including the role of ATP in enhancing their binding to RT/dsDNA P-site complexes [52]. The lack of a structure with INDOPY-1 bound to RT/DNA hindered further progress. We have very recently solved this structure (PDB 6O9E, [53••]), which reveals how INDOPY-1 blocks the dNTP-binding site through a unique mode of binding, not been seen before in any polymerase: it stacks with the terminal T-P, wedges underneath the first template overhang base, and binds to conserved RT residues. DAVPs, discovered by the Botta laboratory, do not bind in the catalytic center but in a hinge region between the thumb and palm subdomains. They can bind to apo RT (structure solved by the Ennifar laboratory [54]), RT/dsDNA, or RT/dsDNA/dNTP complexes. Their mode of action is not well understood, being even affected by NNRTI-resistance mutations. Finally, Merck has identified and solved a structure with a novel ‘bifunctional’ compound (Chart 1) [55•]. Previous bifunctional compounds [56] were formed by NRTI and NNRTI moieties connected by a linker. This new scaffold is positioned between the NNRTI and NRTI pockets, on one side entering in the upper part of the NNIBP tunnel channel, and on the other, close to D186 and the DAVP-1 binding site. Interestingly, this compound may enter through the top of the tunnel channel without requiring opening of the NNIBP.
RNase H RT inhibitors (RHRTIs)
RHRTIs remain an attractive target for HIV inhibition. Most of them are active site binders with a metal chelator and a hydrophobic moiety, similarly to HIV-1 integrase strand transfer inhibitors. Despite several chemotypes displaying biochemical inhibition, this did not translate into antiviral activity, explaining the absence of RHRI approved drugs. The competition of the much larger RNA/DNA with the small molecule is presumed to be one of the most important hurdles for RHRTIs. A collaboration between the Sarafianos and Wang groups has made recent noteworthy progress on this front. They have found a novel crystal form for RT/RHRTIs [57••,58], bearing two copies related by non-crystallographic symmetry in its asymmetric unit. Remarkably, crystal structures of RT with 11b and YLC2–155 (Chart 1 and Figure 1) show different binding modes in each active site in the same crystal lattice. While the metal chelator module remains in a very similar position, the external hydrophobic moiety can be either oriented towards key residue H539 or in the opposite direction. Superposition of RT/YLC2–155 with a RT/RNA/DNA complex suggests that the binding mode with the peripheral moiety pointing to H539 would be favorable for interaction with RNA/DNA (Figure 1), while the other conformation would clash. They have already designed compounds to favor this second conformation [59]. Independently, the Burke, Hughes and Arnold groups have described another RHRTI chemotype, exemplified by compound XZ460 (Chart 1), with antiviral activity related to an optimized peripheral moiety [60•], similarly to those developed by Sarafianos and Wang. The crystal structure with XZ462 (Chart 1), suggest that this chemotype may also bind in the presence of the RNA/DNA substrate.
Perspectives
The RT/dsRNA initiation complex structures have revealed an unusual configuration of the polymerase active site (Figures 2d and 3). When and how this inefficient conformation switches to the more canonical in elongation complexes (Figures 2c and e)? What does the catalytically active conformation of an RTIC looks like? Further snapshots including RT/dsRNA/dNTP or NRTI-TP complexes and of RTIC complexes after incorporation of 2–8 dNTPs might shed light on that. For the time being, the reported RTIC structures have revealed a partially open NNIBP, which may explain RTIC unusual sensitivity to NNRTIs. In this sense, structural characterization of the initiation of the second strand synthesis (Figures 2a and e), also very sensitive to NNRTIs, may show whether the presence of the RNA primer along the DNA template distorts RT similarly to what is seen in the RTICs.
Next, the structure of RT poised for RNA hydrolysis (Figure 2f) provides a framework for designing improved RHRTIs that can bind in its presence. Moreover, as revealed by crystallographic fragment screening and other structural work, novel allosteric inhibitors can be envisioned that may bind in the channel between NRTIs and NNRTIs, and surrounding ones. The elucidation of a variety of novel RT/DNA/NcRTI complexes will also enable improved design of this underexploited inhibitor class.
Finally, an unexplored ‘wild west’ of RT structural biology continues to be its maturation. While the latest stage is reasonably understood, several lines of evidence suggest that polyprotein dimerization facilitates RT maturation and may be related to protease activation [61,62]. Additionally, a cryo-EM structure of the Pol polyprotein dimer shows RT in a conformation surprisingly similar to that of mature heterodimeric p66/p51 (Harrison, Lyumkis, Arnold et al., unpublished). Greater understanding of^ RT maturation could open the door to unprecedented opportunities for therapeutic intervention.
Acknowledgements
The authors thank colleagues of the Arnold group for helpful discussion, and former colleagues of the team, for their invaluable participation in the structural work cited in this review. We also thank the many investigators in the field for their continuing contributions to understanding the magnificent RT enzyme, in terms of structure, function, inhibition, and resistance, as well as making it one of the most important drug targets in medicine. We also want to apologize to those investigators whose papers have not been cited due to space limitation, given the amazing wealth of work on HIV RT. This work was supported by NIH MERIT Award R37 AI027690 and the HIVE Center Grant U54 GMl03368 (both to E.A.). The authors declare no conflict of interest.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.UNAIDS Fact Sheet July 2018. http://www.unaids.org/en/resources/fact-sheeton World Wide Web URL: http://www.unaids.org/en/resources/fact-sheet.
- 2.Das K, Arnold E: HIV-1 reverse transcriptase and antiviral drug resistance. Part 1. Curr Opin Virol 2013, 3:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das K, Arnold E: HIV-1 reverse transcriptase and antiviral drug resistance. Part 2. Curr Opin Virol 2013, 3:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E: Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol 2009, 385:693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isel C, Ehresmann C, Marquet R: Initiation of HIV reverse transcription. Viruses 2010, 2:213–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen KP, Mathiharan YK, Kappel K, Coey AT, Chen DH, Barrero D, Madigan L, Puglisi JD, Skiniotis G, Puglisi EV: Architecture of an HIV-1 reverse transcriptase initiation complex. Nature 2018, 557:118–122•• A remarkable cryo-EM structure of an RT initiation complex (RTIC, 8 Å global resolution, 4.5 Å duplex region). It describes the RT/dsRNA structure after incorporation of a dNTP and visualizes the organization of the protruding vRNA and tRNA elements involved in the RTIC. A significant displacement of the primer terminus from its usual catalytically efficient position is observed.
- 7.Das K, Martinez SE, DeStefano JJ, Arnold E: Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation. Proc Natl Acad Sci USA 2019, 116:7308–7313•• Crystal structure of RT with a 23-mer/17-mer T/P dsRNA that mimic the PBS region of the duplex at 3.95 Å resolution. It provides further detail of the duplex RT/dsRNA interface, significantly the displacement of the primer terminus (~7 Å) at the polymerase active site and the presence of an open NNBIP, likely accounting for the unusual sensitivity of NNRTIs by RTIC.
- 8.Coey AT, Larsen KP, Choi J, Barrero DJ, Puglisi JD, Puglisi EV: Dynamic interplay of RNA and protein in the human immunodeficiency virus-1 reverse transcription initiation complex. J Mol Biol 2018, 430:5137–5150• Single molecule FRET experiments of RTICs that show reduced RNA dynamics upon RT binding and the importance of an extended duplex in the primer-binding site (PBS) region, with 22 bp instead of 18 bp.
- 9.Hu WS, Hughes SH: HIV-1 reverse transcription. Cold Spring Harb Perspect Med 2012, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian L, Kim MS, Li H, Wang J, Yang W: Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid. Proc Natl Acad Sci USA 2018, 115:507–512•• First snapshot of an RT/RNA/DNA complex poised for RNA hydrolysis, as well as structural requirements for RNase H activity.
- 11.Li A, Li J, Johnson KA: HIV-1 reverse transcriptase polymerase and RNase H (ribonuclease H) active sites work simultaneously and independently. J Biol Chem 2016, 291:26566–26585• Pre-steady state kinetics work that provides evidence that nucleotide incorporation and RNase H cuts can occur simultaneously.
- 12.Figiel M, Krepl M, Poznanski J, Golab A, Sponer J, Nowotny M: Coordination between the polymerase and RNase H activity of HIV-1 reverse transcriptase. Nucleic Acids Res 2017, 45:3341–3352• This paper uses molecular dynamics simulations and detection of RNase H activity assays of RNA/DNA cross-linked to RT in the polymerase domain to prove the simultaneous binding of the substrate to RT.
- 13.London RE: HIV-1 reverse transcriptase: a metamorphic protein with three stable states. Structure 2019, 27:420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Pedersen LC, Gabel SA, Mueller GA, DeRose EF, London RE: Unfolding the HIV-1 reverse transcriptase RNase H domain-how to lose a molecular tug-of-war. Nucleic Acids Res 2016, 44:1776–1788• This paper reports the structure of the isolated RNase H domain swapped dimer that captures a partially unfolded RNase H domain.
- 15.Schmidt T, Schwieters CD, Clore GM: Spatial domain organization in the HIV-1 reverse transcriptase p66 homodimer precursor probed by double electron-electron resonance EPR. Proc Natl Acad Sci USA 2019, 116:17809–17816Using double electron-electron resonance EPR spectroscopy, Schmidt and coworkers find that in the p66/p66’ homodimer just one of the RNase H domains is extensively solvent-accessible (unfolded), consistent with the maturation model proposed by London and colleagues (see Ref. [14]).
- 16.Ilina TV, Slack RL, Elder JH, Sarafianos SG, Parniak MA, Ishima R: Effect of tRNA on the maturation of HIV-1 reverse transcriptase. J Mol Biol 2018, 430:1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slack RL, Ilina TV, Xi Z, Giacobbi NS, Kawai G, Parniak MA, Sarafianos SG, Sluis Cremer N, Ishima R: Conformational changes in HIV-1 reverse transcriptase that facilitate its maturation. Structure 2019. 10.1016/j.str.2019.08.004 Refs. [16] and [17] show the tRNA has a role in RT maturation, mainly by introducing conformational asymmetry that may facilitate the digestion of just one RNase H domain in the p66/p66’ homodimer.
- 18.Waheed AA, Tachedjian G: Why do we need new drug classes for HIV treatment and prevention? Curr Top Med Chem 2016, 16:1343–1349. [DOI] [PubMed] [Google Scholar]
- 19.Singh K, Sarafianos SG, Sonnerborg A: Long-acting anti-HIV drugs targeting HIV-1 reverse transcriptase and integrase. Pharmaceuticals 2019, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markowitz M, Sarafianos SG: 4’-Ethynyl-2-fluoro-2’-deoxyadenosine, MK-8591: a novel HIV-1 reverse transcriptase translocation inhibitor. Curr Opin HIV AIDS 2018, 13:294–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salie ZL, Kirby KA, Michailidis E, Marchand B, Singh K, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG: Structural basis of HIV inhibition by translocation-defective RT inhibitor 4’-ethynyl-2-fluoro-2’-deoxyadenosine (EFdA). Proc Natl Acad Sci USA 2016, 113:9274–9279• This paper, through a set of RT/dsDNA structures with EFdA in different locations (N-site, P-site, misincorporated, upstream of the P-site), pinpoints the atomic details of the multiple EFdA mechanisms for RT inhibition.
- 22.Takamatsu Y, Das D, Kohgo S, Hayashi H, Delino NS, Sarafianos SG, Mitsuya H, Maeda K: The high genetic barrier of EFdA/MK-8591 stems from strong interactions with the active site of drug-resistant HIV-1 reverse transcriptase. Cell Chem Biol 2018, 25:1268–1278.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasutake Y, Hattori SI, Hayashi H, Matsuda K, Tamura N, Kohgo S, Maeda K, Mitsuya H: HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir. Sci Rep 2018, 8:1624.• Ref. [23], using HIV-1 RT as surrogate, allowed detailed characterization of the mechanism of action of anti-hepatitis B virus drug entecavir, which as well as EFdA is an NRTI that retains the 3’-OH group necessary for catalysis.
- 24.Yasutake Y, Hattori SI, Tamura N, Matsuda K, Kohgo S, Maeda K, Mitsuya H: Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4’-modified nucleoside RT inhibitors. Biochem Biophys Res Commun 2019, 509:943–948• Ref. [24], using HIV-1 RT as surrogate, allowed detailed characterization of the mechanism of action of anti-hepatitis B virus drug entecavir, which as well as EFdA is an NRTI that retains the 3’-OH group necessary for catalysis.
- 25.Bertoletti N, Chan AH, Schinazi RF, Yin YW, Anderson KS: Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: first crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine. Protein Sci 2019, 28:1664–1675• Structures of HIV-1 RT with the TP forms of the widely used drugs 3TC and FTC show an unusual conformation of the triphosphate groups, explaining the relatively slow incorporation of the active forms of these L-nucleoside drugs.
- 26.Das K, Martinez SE, Arnold E: Structural insights into HIV reverse transcriptase mutations Q151M and Q151M complex that confer multinucleoside drug resistance. Antimicrob Agents Chemother 2017, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez SE, Bauman JD, Das K, Arnold E: Structure of HIV-1 reverse transcriptase/d4TTP complex: novel DNA cross-linking site and pH-dependent conformational changes. Protein Sci 2019, 28:587–597•This paper describes the mechanism of inhibition of the discontinued drug d4T and also reports a novel and cost-effective method for cross-linking nucleic acids to RT through a mutation in the fingers residue I63 to C.
- 28.Namasivayam V, Vanangamudi M, Kramer VG, Kurup S, Zhan P, Liu X, Kongsted J, Byrareddy SN: The journey of HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) from lab to clinic. J Med Chem 2019, 62:4851–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das K, Bauman JD, Clark AD, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E: High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA 2008, 105:1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodda LS, Tirado-Rives J, Jorgensen WL: Unbinding dynamics of non-nucleoside inhibitors from HIV-1 reverse transcriptase. J Phys Chem B 2019, 123:1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen L, Shen J, Luo X, Cheng F, Xu Y, Chen K, Arnold E, Ding J, Jiang H: Steered molecular dynamics simulation on the binding of NNRTI to HIV-1 RT. Biophys J 2003, 84:3547–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauman JD, Patel D, Dharia C, Fromer MW, Ahmed S, Frenkel Y, Vijayan RSK, Eck JT, Ho WC, Das K et al. : Detecting allosteric sites of HIV-1 reverse transcriptase by X-ray crystallographic fragment screening. J Med Chem 2013, 56:2738–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang B, Chen W, Zhao T, Li Z, Jiang X, Ginex T, Vilchez D, Luque FJ, Kang D, Gao P et al. : Exploiting the tolerant region I of the non-nucleoside reverse transcriptase inhibitor (NNRTI) binding pocket: discovery of potent diarylpyrimidine-typed HIV-1 NNRTIs against wild-type and E138K mutant virus with significantly improved water solubility and favorable safety profiles. J Med Chem 2019, 62:2083–2098. [DOI] [PubMed] [Google Scholar]
- 34.Kang D, Wang Z, Zhang H, Wu G, Zhao T, Zhou Z, Huo Z, Huang B, Feng D, Ding X et al. : Further exploring solvent-exposed tolerant regions of allosteric binding pocket for novel HIV-1 NNRTIs discovery. ACS Med Chem Lett 2018, 9:370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Zhang J, Gao P, Sun L, Song Y, Kang D, Liu X, Zhan P: Efficient drug discovery by rational lead hybridization based on crystallographic overlay. Drug Discov Today 2018. 10.1016/j.drudis.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Kang D, Nguyen LA, Smithline ZB, Pannecouque C, Zhan P, Liu X, Steitz TA: Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors. eLife 2018, 7• This work shows in structural detail how an evolved diarylpyrimidine scaffold retains the potency of the parent compounds but presents higher resilience to resistance mutations, mainly the E138K mutation affecting second-generation NNRTIs.
- 37.Cote B, Burch JD, Asante-Appiah E, Bayly C, Bedard L, Blouin M, Campeau LC, Cauchon E, Chan M, Chefson A et al. : Discovery of MK-1439, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses. Bioorg Med Chem Lett 2014, 24:917–922. [DOI] [PubMed] [Google Scholar]
- 38.Kang D, Zhang H, Wang Z, Zhao T, Ginex T, Luque FJ, Yang Y, Wu G, Feng D, Wei F et al. : Identification of dihydrofuro[3,4-d] pyrimidine derivatives as novel HIV-1 non-nucleoside reverse transcriptase inhibitors with promising antiviral activities and desirable physicochemical properties. J Med Chem 2019, 62:1484–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang D, Zhao T, Wang Z, Feng D, Zhang H, Huang B, Wu G, Wei F, Zhou Z, Jing L et al. : Discovery of piperidine-substituted thiazolo[5,4-d]pyrimidine derivatives as potent and orally bioavailable HIV-1 non-nucleoside reverse transcriptase inhibitors. Commun Chem 2019, 2:74. [Google Scholar]
- 40.Huo Z, Zhang H, Kang D, Zhou Z, Wu G, Desta S, Zuo X, Wang Z, Jing L, Ding X et al. : Discovery of novel diarylpyrimidine derivatives as potent HIV-1 NNRTIs targeting the “NNRTI Adjacent” binding site. ACS Med Chem Lett 2018, 9:334–338• Taking advantage of previous structural work, these investigators achieved the design of an extended NNRTI reaching the nearby NNRTI Adjacent site.
- 41.Frey KM, Bollini M, Mislak AC, Cisneros JA, Gallardo-Macias R, Jorgensen WL, Anderson KS: Crystal structures of HIV-1 reverse transcriptase with picomolar inhibitors reveal key interactions for drug design. J Am Chem Soc 2012, 134:19501–19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dousson C, Alexandre FR, Amador A, Bonaric S, Bot S, Caillet C, Convard T, da Costa D, Lioure MP, Roland A et al. : Discovery of the aryl-phospho-indole IDX899, a highly potent anti-HIV non-nucleoside reverse transcriptase inhibitor. J Med Chem 2016, 59:1891–1898. [DOI] [PubMed] [Google Scholar]
- 43.Lee WG, Chan AH, Spasov KA, Anderson KS, Jorgensen WL: Design, conformation, and crystallography of 2-naphthyl phenyl ethers as potent anti-HIV agents. ACS Med Chem Lett 2016, 7:1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan AH, Lee WG, Spasov KA, Cisneros JA, Kudalkar SN, Petrova ZO, Buckingham AB, Anderson KS, Jorgensen WL: Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: from design to protein crystallography. Proc Natl Acad Sci U S A 2017, 114:9725-9730 •• A catechol ether NNRTI scaffold is converted into covalent Y181C NNRTI through structure-based design, a structures with the covalent compound bound to Y181C RT.
- 45.Balzarini J, Ford A, Maguire NM, John J, Das K, Arnold E, Dehaen W, Maguire A: Alpha-carboxynucleoside phosphonates: direct-acting inhibitors of viral DNA polymerases. Future Med Chem 2019, 11:137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balzarini J, Das K, Bernatchez JA, Martinez SE, Ngure M, Keane S, Ford A, Maguire N, Mullins N, John J et al. : Alpha-carboxy nucleoside phosphonates as universal nucleosidetriphosphate mimics. Proc Natl Acad Sci USA 2015, 112:3475–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das K, Balzarini J, Miller MT, Maguire AR, DeStefano JJ, Arnold E: Conformational states of HIV-1 reverse transcriptase for nucleotide incorporation vs pyrophosphorolysis-binding of foscarnet. ACS Chem Biol 2016, 11:2158–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullins ND, Maguire NM, Ford A, Das K, Arnold E, Balzarini J, Maguire AR: Exploring the role of the alpha-carboxyphosphonate moiety in the HIV-RT activity of alpha-carboxy nucleoside phosphonates. Org Biomol Chem 2016, 14:2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.John J, Kim Y, Bennett N, Das K, Liekens S, Naesens L, Arnold E, Maguire AR, Gotte M, Dehaen W et al. : Pronounced inhibition shift from HIV reverse transcriptase to herpetic DNA polymerases by increasing the flexibility of alpha-carboxy nucleoside phosphonates. J Med Chem 2015, 58:8110–8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller MT, Tuske S, Das K, DeStefano JJ, Arnold E: Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer. Protein Sci 2016, 25:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jochmans D, Deval J, Kesteleyn B, Van Marck H, Bettens E, De Baere I, Dehertogh P, Ivens T, Van Ginderen M, Van Schoubroeck B et al. : Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J Virol 2006, 80:12283–12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehteshami M, Nijhuis M, Bernatchez JA, Ablenas CJ, McCormick S, de Jong D, Jochmans D, Gotte M: Formation of a quaternary complex of HIV-1 reverse transcriptase with a nucleotide-competing inhibitor and its ATP enhancer. J Biol Chem 2013, 288:17336–17346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruiz FX, Hoang A, Das K, Arnold E: Structural basis of HIV-1 inhibition by the nucleotide-competing reverse transcriptase inhibitor INDOPY-1. J Med Chem 2019. 10.1021/acs.jmedchem.9b01289•• RT/DNA structure complexed with NcRTI INDOPY-1, which shows how the compound binds in an unprecedented way, both stacking and intercalating with the nucleic acid, and interacting with RT. It provides a path towards developing improved NcRTIs and to target other polymerases of pathogens.
- 54.Freisz S, Bec G, Radi M, Wolff P, Crespan E, Angeli L, Dumas P, Maga G, Botta M, Ennifar E: Crystal structure of HIV-1 reverse transcriptase bound to a non-nucleoside inhibitor with a novel mechanism of action. Angew Chem Int Ed Engl 2010, 49:1805–1808. [DOI] [PubMed] [Google Scholar]
- 55.Lai MT, Tawa P, Auger A, Wang D, Su HP, Yan Y, Hazuda DJ, Miller MD, Asante-Appiah E, Melnyk RA: Identification of novel bifunctional HIV-1 reverse transcriptase inhibitors. J Antimicrob Chemother 2018, 73:109–117• Structure with a novel chemotype binding partly to the upper part of the tunnel channel in the NNIBP and to the connecting pocket with the polymerase active site.
- 56.Bailey CM, Sullivan TJ, Iyidogan P, Tirado-Rives J, Chung R, Ruiz-Caro J, Mohamed E, Jorgensen WL, Hunter R, Anderson KS: Bifunctional inhibition of human immunodeficiency virus type 1 reverse transcriptase: mechanism and proof-of-concept as a novel therapeutic design strategy. J Med Chem 2013, 56:3959–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kankanala J, Kirby KA, Huber AD, Casey MC, Wilson DJ, Sarafianos SG, Wang Z: Design, synthesis and biological evaluations of N-hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H. Eur J Med Chem 2017, 141:149–161RT-RNHI structure at 1.8 Å resolution in a crystal form that has two complexes in the asymmetric unit and where the RNHI shows two different conformations, with one being compatible with RNA/DNA binding that could facilitate achieving inhibition in vivo.
- 58.Kirby KA, Myshakina NA, Christen MT, Chen YL, Schmidt HA, Huber AD, Xi Z, Kim S, Rao RK, Kramer ST et al. : A 2-hydroxyisoquinoline-1,3-dione active-site RNase H inhibitor binds in multiple modes to HIV-1 reverse transcriptase. Antimicrob Agents Chemother 2017, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Tang J, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Parniak MA, Sarafianos SG, Wang Z: 6-Biphenylmethyl-3-hydroxypyrimidine-2,4-diones potently and selectively inhibited HIV reverse transcriptase-associated RNase H. Eur J Med Chem 2018, 156:680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyer PL, Smith SJ, Zhao XZ, Das K, Gruber K, Arnold E, Burke TR Jr, Hughes SH: Developing and evaluating inhibitors against the RNase H active site of HIV-1 reverse transcriptase. J Virol 2018, 92• A novel RHRTI chemotype with antiviral activity is developed. A high resolution crystal structure proofs that it binds to the RNase H domain. As in Ref. 55, the binding seems compatible with with RNA/DNA binding.
- 61.London RE: Structural maturation of HIV-1 reverse transcriptase - a metamorphic solution to genomic instability. Viruses 2016, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wapling J, Srivastava S, Shehu-Xhilaga M, Tachedjian G: Targeting human immunodeficiency virus type 1 assembly, maturation and budding. Drug Target Insights 2007, 2:159–182. [PMC free article] [PubMed] [Google Scholar]