SUMMARY
The glycan ligands recognized by Siglecs, influenza viruses, and galectins, as well as many plant lectins, are not well defined. To explore their binding to Asparagine (Asn)-linked N-glycans we synthesized a library of isomeric multiantennary N-glycans that vary in terminal non-reducing sialic acid, galactose, and N-acetylglucosamine residues, as well as core fucose. We identified specific recognition of N-glycans by several plant lectins, human galectins, influenza viruses, and Siglecs and explored the influence of sialic acid linkages and branching of the N-glycans. These results show the unique recognition of complex-type N-glycans by a wide variety of glycan-binding proteins and their abilities to distinguish isomeric structures, which provides new insights into the biological roles of these proteins and the uses of lectins in biological applications to identify glycans.
Keywords: Chemoenzymatic synthesis, N-glycan isomer, microarray, N-glycan binding specificity, glycosyltransferases, glycan binding proteins
INTRODUCTION
N-glycosylation is a complex yet exquisitely controlled post-translational modification of glycoproteins. Complex N-glycans with core fucose and terminal sialic acids residues are implicated in many biological and pathological events. Altered branching and core fucosylation of N-glycans are associated with cancer progression(Taniguchi and Kizuka, 2015). Additionally, sialylated N-glycans directly serve as ligands for influenza virus recognition(Byrd-Leotis, et al., 2014), and are suggested to function as signaling receptors for endogenous cell communication molecules, such as Siglecs(Peng and Paulson, 2017). However, due to the structural complexity, it has been difficult to define the molecular mechanisms by which these N-glycans exert their functions. A library of naturally occurring complex N-glycans, therefore, would be valuable for investigating the structural requirements for recognition and probing molecular mechanisms associated with key biological processes.
Preparation of N-glycans typically relies on either isolation from natural sources or complete chemical synthesis, each with its own limitations. Complete resolving of N-glycan pools obtained from natural sources to harvest each N-glycan isomer remains challenging. De novo chemical synthesis of complex N-glycans requires significant expertise to overcome the difficulties in generating regio- and stereoselective glycosidic linkages(Steffen, et al., 2007; Sun, et al., 2008; Walczak and Danishefsky, 2012).
To address such difficulties and inefficiencies, chemoenzymatic approaches have been developed in order to synthesize symmetrical(M., et al., 2012; Peng, et al., 2017; Shivatare, et al., 2013; Unverzagt, 1996; Wu, et al., 2016) and asymmetrical(Calderon, et al., 2016; Hanashima, et al., 2005; Li, et al., 2015; Shivatare, et al., 2016; Wang, et al., 2013) N-glycans. However, most of these approaches rely on chemical synthesis to produce a number of initial core structures in the early steps, which is often time-consuming and difficult to scale up. In addition, many of the product N-glycans either contain aglycone such as azidoethyl and aminopentyl, or need derivatization with bifunctional “linkers” such as AEAB(Song, et al., 2009) and 2-amino-methyl N,O-hydroxyethyl(Bohorov, et al., 2006) to facilitate functional assays. However, there is evidence that the linker used to immobilize the glycan may affect the ability of a glycan to be recognized by its binding partner(Grant, et al., 2014; Padler-Karavani, et al., 2012; Tessier, et al., 2013). As the natural linker, asparagine (Asn) retains, at least to some extent, the presentation of the glycans within the context of the peptide backbone and the amide nitrogen in correct anomeric linkage. It has also been proven to be one of the preferable linkers in terms of its favorable glycan presentation properties(Grant, et al., 2014). Preserving the Asn-linkage in N-glycan synthesis, therefore, becomes the preferred choice. Thus far, N-glycans with full range of topological branches, sialic acid linkages, and core fucosylation have not been readily available, and in particular, Asn-linked N-glycans are scarce. Only derivatives of biantennary N-glycans have been successfully synthesized with Asn linkage(Calderon, et al., 2016; M., et al., 2012). Although the comprehensive array from Consortium for Functional Glycomics (CFG) contains Asn-linked N-glycans, this array lacks multiantennary complex type N-glycans. Thus, a systematic approach that can efficiently produce those highly complex Asn-linked N-glycans in a reasonable amount is highly desirable.
We herein report an integrated chemoenzymatic approach to efficiently generate a library of complex multiantennary Asn-linked N-glycan isomers in sub-milligram quantities. Our strategy takes advantage of a sialylated glycopeptide (SGP)(Seko, et al., 1997), which can be purified from chicken egg yolk powder in large quantities, and a set of recombinant human glycosyltransferases. This approach yields a library of 32 N-glycosylasparagine isomers, all of which occur naturally in human and other mammals (See the database of UniCarbKB(Campbell, et al., 2014) http://www.unicarbkb.org/ and CFG http://www.functionalglycomics.org for information). These compounds can be conveniently converted to free reducing-glycans. These sequence-defined N-glycans also bring insights into the unique binding specificities of N-glycan binding partners including plant lectins, human galectins, influenza viruses, and Siglecs.
RESULTS
We designed an N-glycan library composed of 32 naturally occurring bi-, tri- and tetraantennary, complex type structures for synthesis (Fig. 1). Sixteen (1-16) of them are precursors with no sialic acid. The other sixteen (17-32) are fully capped with either α2,3- or α2,6-sialic acid. Each non-core fucosylated glycan has a core fucosylated counterpart in the library. The branching patterns of the triantennary N-glycans are isomers of either the so-called 2,2,4- (with β1,4-linked GlcNAc branch on the α3-linked Man) or 2,2,6-form (with β1,6-linked GlcNAc branch on the α6-linked Man).
Figure 1.
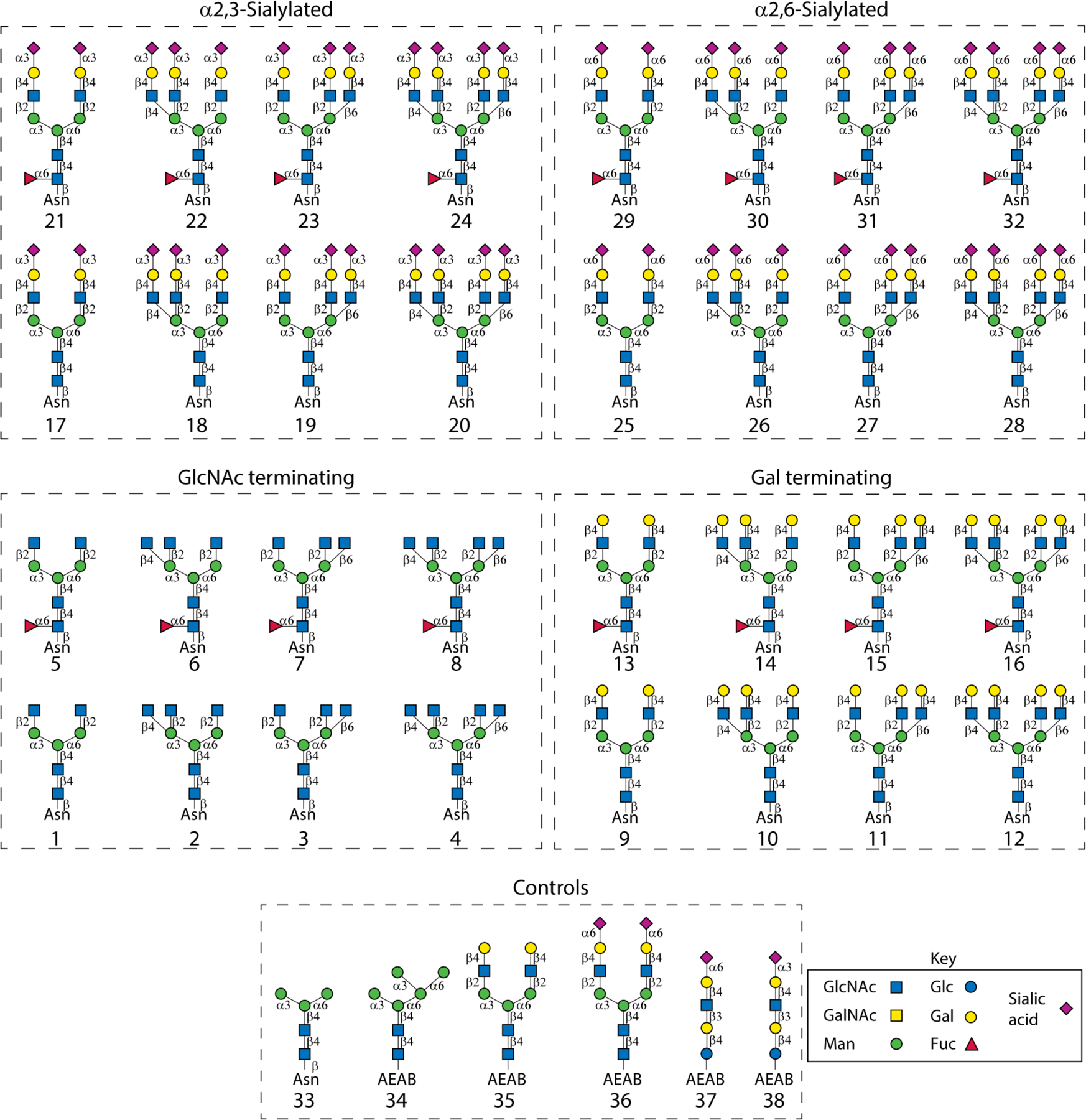
Composition of the 32 complex-type naturally occurring N-glycans and controls used in this study. Sixteen (1-16) of the N-glycans are precursors with no sialic acids. Half of them (1-8) are GlcNAc-terminating and the other half (9-16) are Gal-terminating structures. The other sixteen (17-32) are fully capped with either α2,3- (17-24) or α2,6-sialic acid (25-32). Both non-fucosylated and fucosylated species are present in each subgroup. Six glycans from the archival collections of our lab were printed as controls on the array including two mannose-terminating compounds Man3-Asn (33) and Man5-AEAB (34), two complex-type N-glycans NA2-AEAB (35) and A2-AEAB (36), and α2,6- and α2,3-sialic acid containing glycans LSTc-AEAB (37) and LSTd-AEAB (38). The symbolic nomenclature for glycans is shown.
Generation of a sialylated biantennary glycopeptide SGP and the Fmoc-labelled N-glycosylasparagine core structure
To generate the library, we reasoned that the GlcNAc-terminating biantennary structure 1 would be a suitable starting compound. This compound can be conveniently generated by a few steps of chemoenzymatic reactions from a sialylated biantennary glycopeptide designated SGP (39), which is abundantly available in chicken egg yolk(Seko, et al., 1997; Sun, et al., 2014; Zou, et al., 2012). Around 1.5 g SGP was harvested from 1.36 kg egg yolk powder (Fig. 2a), which is comparable to the previous report(Sun, et al., 2014). The compound was analyzed by MALDI-MS and ESI-MS (Fig. 2b) and the results are in good accordance with the predicted value.
Figure 2.
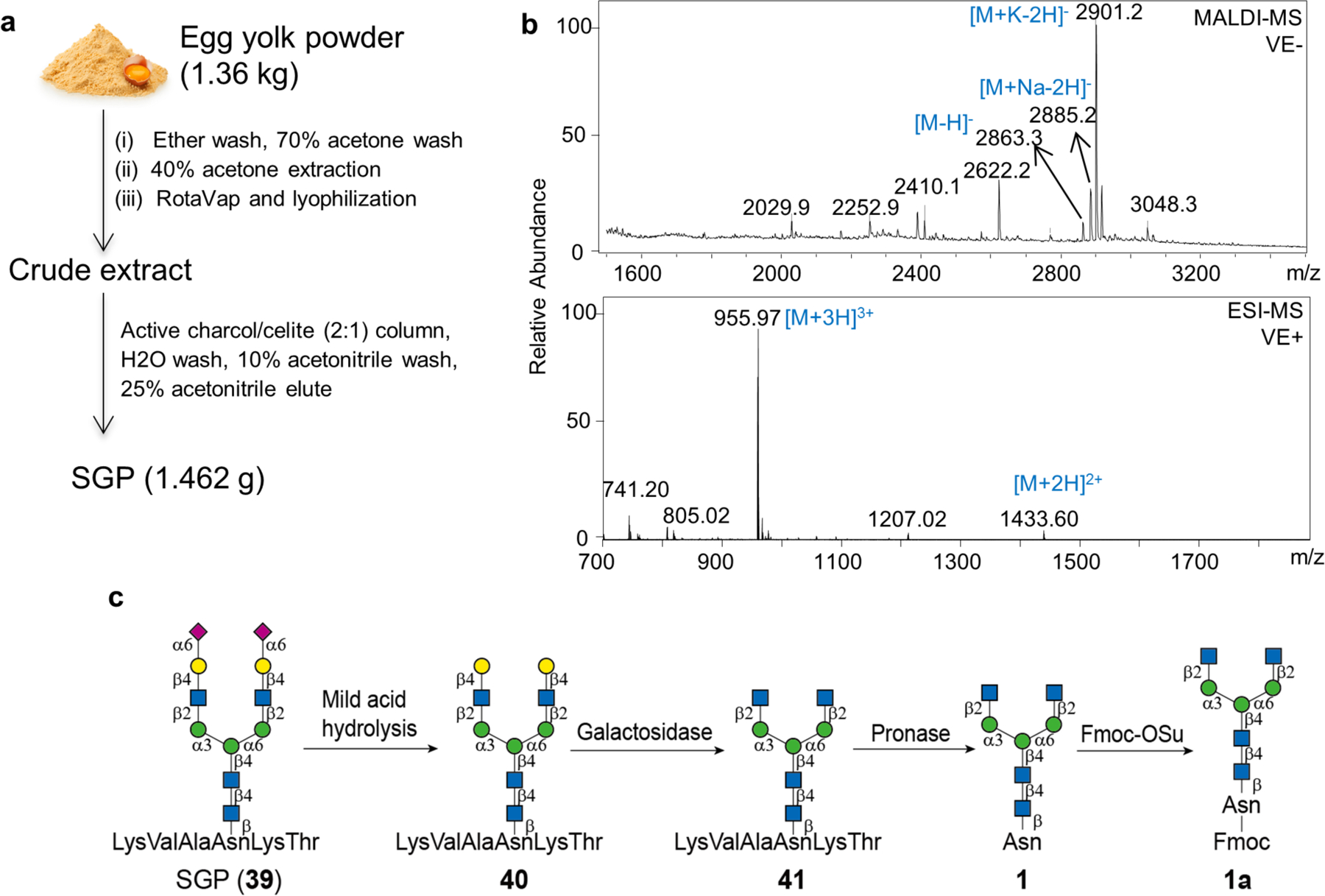
Generation of the SGP and the Fmoc protected biantennary glycosylasparagine compound 1a. (a) A sialylated biantennary glycopeptide designated SGP (39) is abundantly available in chicken egg yolk and can be extracted by 40% acetone and purified by active charcoal/celite (2:1) column. Around 1.5 g SGP was harvested from 1.36 kg egg yolk powder. (b) Negative ion-mode MALDI-MS and positive ion-mode ESI-MS analyses of the purified SGP. (c) The SGP was converted to the target, Fmoc protected biantennary compound 1a, by mild acid hydrolysis, galactosidase and pronase digestion and Fmoc derivatization.
The SGP was then converted to the Asn-linked GlcNAc-terminating biantennary compound 1 by sequential removal of terminal sialic acid, Gal, and trimming the peptide to one amino acid (Fig 2c). This was achieved by mild acid hydrolysis, galactosidase and pronase digestion, respectively. This reaction order minimizes the requirement for purification with >90% conversion. Fmoc was thereafter selectively installed at the amine of the Asn residue to facilitate subsequent purification and MS analysis. The product (1a) gave a characteristic fluorescence signal at excitation (Ex) and emission (Em) at 254 and 340 nm, respectively.
N-glycan chemo-enzymatic synthesis
Starting from the biantennary compound 1a, a set of six human-derived recombinant glycosyltransferases were used to expand the library (Supplemental Scheme 1). The compound 1a was branched by either MGAT4a or MGAT5 to produce the 2,2,4- (2a) and 2,2,6-form (3a) tri-antennary N-glycan precursors, respectively. The compound 1a was also sequentially treated with MGAT4a and MGAT5 to harvest a tetra-antennary precursor 4a. Compounds 1a to 4a were then galactosylated by B4GALT1 to harvest 9a through 12a, and subsequently sialylated by either ST3GAL4 or ST6GAL1 to produce the Neu5Acα2,3- and Neu5Acα2,6-terminating structures 17a to 20a, and 25a to 28a, respectively. The precursor 1a was also successfully fucosylated by FUT8 to generate the precursor 5a. This compound was similarly branched, elongated and capped to produce the other 16 core-fucosylated compounds. We also treated the precursor 1a with hexosaminidase to harvest the Man-terminating compound 33a. In all reactions, the substrates were quantitatively converted and the yields were >90%. All of the Fmoc-labelled Asn-linked N-glycans generated were analyzed by HPLC (Supplemental Fig. S1) and MALDI-MS. The detected m/z were in accord with the predicted values (Supplemental Table S1). The Asn-linked oligosaccharides 1 to 33 can be easily generated by incubation with 5% piperidine (Fig. 3a and b). All of the purified Asn-linked N-glycans were analyzed by NMR and the protons were successfully assigned (Supporting Information). Those Asn-linked N-glycan isomers were used to generate a new sequence-defined N-glycan array in order to study their biological functions, which will be discussed below.
Figure 3.
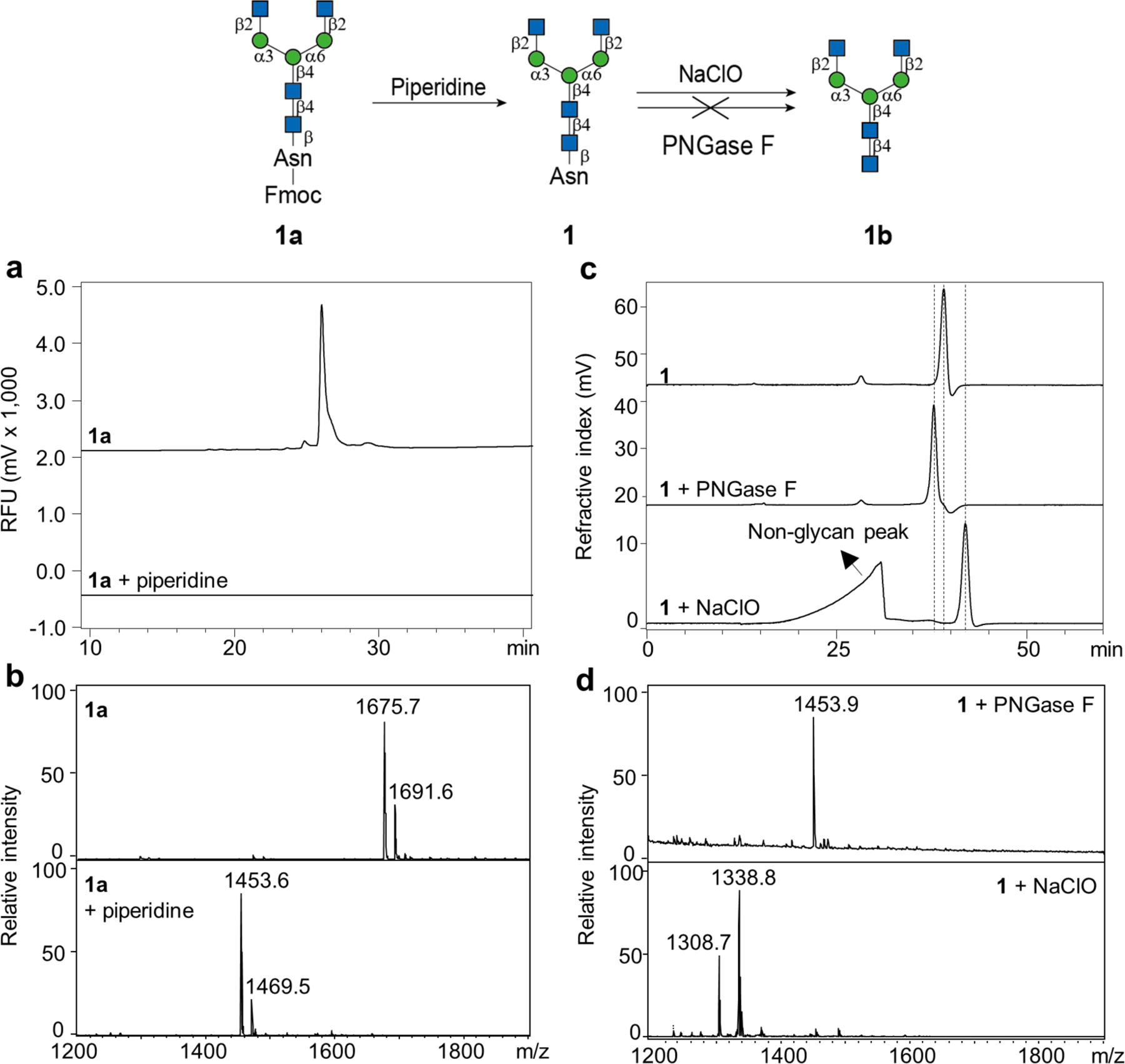
Generation of the asparagine-linked N-glycans and reducing N-glycans. (a) and (b) Normal phase HPLC and MALDI-MS analyses of the substrate 1a and the product after 5% piperidine treatment. (c) and (d) Size exclusion chromatography and MALDI-MS analyses of the substrate 1 and the products after PNGase F or NaClO treatment. Dashes indicated the shift of the elution time of the product peak.
Unexpectedly and importantly, we identified a method to convert the Asn-linked glycans to free reducing oligosaccharides (Fig. 3c and d). MALDI-MS showed no change in the size of compound 1 after PNGase F incubation (Fig. 3d), suggesting the enzyme is unable to release glycans from a single asparagine, as would be predicted(Fan and Lee, 1997). By comparison, brief treatment with sodium hypochlorite (NaClO - bleach) shifted the peak to the right on size exclusion chromatography (Fig. 3c). MALDI-MS (Fig. 3d) indicates the successful removal of the Asn residue and the generation of the free reducing glycan 1b. The remaining thirty-one Asn-linked compounds were easily converted to reducing glycans in the same manner using bleach.
Microarray construction and interrogation using sequence-specific N-glycan binding partners
Sequence-defined N-glycans are ideal standard compounds for unambiguous detection of N-glycans in biochemical and clinical studies. We thus sought to define the binding specificities of various N-glycan binding partners. To perform the task, we constructed an N-glycan microarray with the 33 Asn-linked N-glycans, together with five glycan conjugates (34-38) from the collection in our laboratory (Fig. 1).
Lectins
Plant lectins have been widely used for decades to detect N-glycans with certain structural features(Cummings and Kornfeld, 1982). However, the specificity of many lectins was originally characterized using hapten inhibition assays or affinity chromatography with limited number of N-glycan variants. Thus, re-evaluation of the lectin specificities in direct binding assays with a larger number of sequence-defined N-glycan isomers is needed. We assayed fifteen plant lectins in this array at two concentrations and the binding of each individual lectin was strikingly different (Fig. 4, Supplemental Fig. S2, Supplemental Table S2). Each lectin has individual structural preferences, some of which have not been adequately demonstrated previously. As the binding patterns of each lectin tested at higher concentrations (Supplemental Fig. S2) were very similar to those at lower concentration (Fig. 4), we discuss below only the data at lower concentrations unless otherwise specified.
Figure 4.
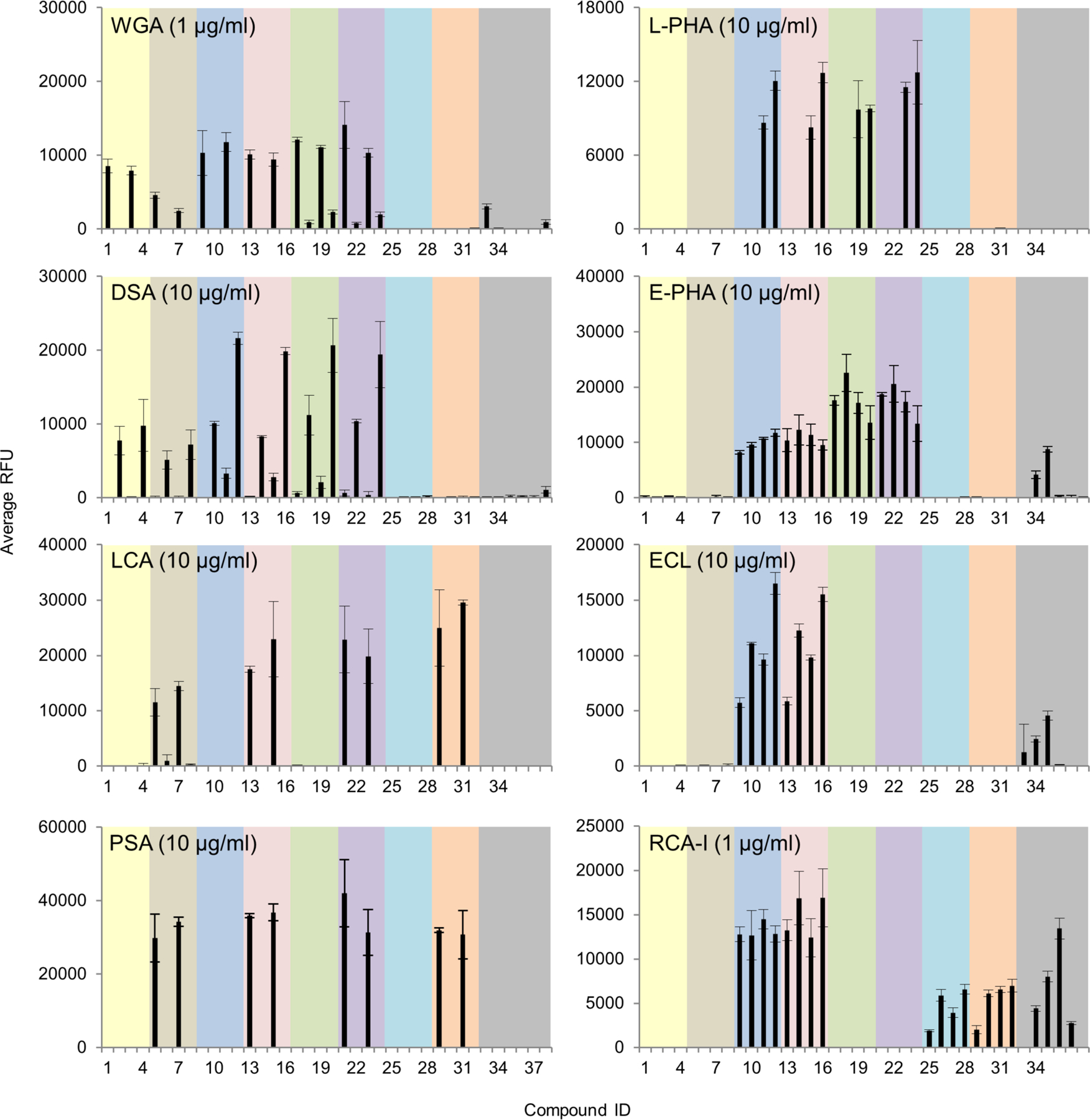
Microarray analyses of sequence-specific lectins using the newly prepared N-glycan array. Results are shown as relative fluorescence units (RFUs) by averaging the background-subtracted fluorescence signals of 4 replicate spots, error bars represent the standard deviation among the 4 values. The RFUs and the standard deviation were listed in Supplemental Table S2. These lectins shown here were also tested at a higher concentration and the data were shown in Supplemental Fig. S2. The probes are numbered as shown in Fig. 1 and grouped as indicated in the colored panels.
Interestingly, although known to bind to terminal sialic acid and GlcNAc(Gallagher, et al., 1985), Wheat Germ agglutinin (WGA) showed binding to N-glycans terminated with GlcNAc-, Galβ1–4GlcNAc- and Neu5Acα2,3- in a similar manner, which is consistent with a recent report(Wu, et al., 2016). Furthermore, binding of WGA was only detected to the biantennary and the 2,2,6-form triantennary, but not to the 2,2,4-counterparts nor the tetraantennary N-glycans, suggesting a branching preference of this lectin.
By contrast, Datura stramonium agglutinin (DSA) exhibited preference for the 2,2,4-form tri- and tetraantennary, both of which contain the C4 GlcNAc on the α1,3-linked mannose. This is consistent with the previous report that DSA bound certain types of branched N-glycans(Crowley, et al., 1984; Cummings and Kornfeld, 1984; Endo, et al., 1988; Green and Baenziger, 1989).
Our results of Lens culinaris agglutinin (LCA) concurred with the prior finding that its binding is restricted to N-glycans with α1,6-linked core fucose(Kornfeld, et al., 1981). In addition, we found LCA only bound the biantennary and the 2,2,6-form triantennary, which resembled the branching preference of WGA. Our data provides direct evidence for the tolerance of LCA to the C6 GlcNAc on the α1,6-linked mannose which confirmed the previous observation through cell transfection assays using MGAT5(Nakagawa, et al., 2008). As a comparison, Pisum sativum agglutinin (PSA)(van Wauwe, et al., 1975) was also tested on the array and it showed an almost identical binding pattern with LCA. This is consistent with the previous report that the two lectins had similar binding preference for core fucosylated N-glycans(Tateno, et al., 2009).
Phaseolus vulgaris leukoagglutinin (L-PHA) and its isolectin, Phaseolus vulgaris erythroagglutinin (E-PHA) have distinct binding specificities. L-PHA is known to bind to the β1,6-branch of the N-glycan backbone(Cummings and Kornfeld, 1982; Hammarström, et al., 1982) and indeed, only the 2,2,6-form triantennary and the tetraantennary were bound. E-PHA has a broader tolerance to branching patterns. In addition to those that were bound by L-PHA, it also bound biantennary and the 2,2,4-form triantennary N-glycans. It has been reported that E-PHA binds bisected complex-type N-glycans(Cummings and Kornfeld, 1982), but these are not yet available for testing on our array.
Concanavalin A (ConA) is known to bind biantennary and high mannose N-glycans(Baenziger and Fiete, 1979; Kornfeld and Ferris, 1975; OGATA, et al., 1975). Interestingly, in addition to those, weak binding was also observed to GlcNAc-terminating triantennary species (compounds 2, 3, and 6, 7). Elongation with Gal (compounds 10, 11 and 14, 15) seemed to block this binding.
Of note, among all the lectins above, LCA and ConA were the only two that could accommodate the non-reducing terminal α2,6-sialic acid; all could tolerate α2,3-sialic acid. This is important to take into consideration when these lectins are used in biochemical or clinical applications.
Erythrina cristagalli lectin (ECL) and Ricinus communis agglutinin (RCA-I), which recognize terminal β-1,4-linked Gal(Debray, et al., 1986; Iglesias, et al., 1982) and terminal Gal(Baenziger and Fiete, 1979), respectively, bound strongly to all Gal-terminating glycans. RCA-I, but not ECL, tolerated the presence of α2,6-sialic acid especially used at higher concentration, whereas neither could accommodate the α2,3- counterparts.
Sambucus nigra agglutinin (SNA) and Maackia amurensis lectin I (MAL-I) bound to α2,6- and α2,3-linked sialylated species, respectively, consistent with previous observations(Shibuya, et al., 1987; Shibuya, et al., 1987; Wang and Cummings, 1988). Interestingly, MAL-I had a slight preference towards the β1,4-linked branch which resembles DSA. In addition, non-sialylated Gal-terminating glycans were also bound by MAL-I, but the intensity was much lower than those with α2,3-sialic acid. The tolerance of the Gal-terminating glycans was prominent when using at a higher concentration (Supplemental Fig. S2). This was also confirmed by binding assays followed by Arthrobacter ureafaciens neuraminidase treatment of the array (Supplemental Fig. S3), which will be discussed in the section on influenza viruses.
Aleuria aurantia lectin (AAL), which is reported to recognize terminal fucose residues(Kochibe and Furukawa, 1980; Yamashita, et al., 1985), showed strong binding to all core-fucosylated glycans. Compared to LCA and PSA, AAL did not show preference towards particular branching patterns. Another fucose-binding lectin, Ulex europaeus agglutinin I (UEA-I), which is reported to recognize fucose in the H-antigen Fucα1–2Gal-(Matsumoto and Osawa, 1969), did not show any binding as that determinant is not present on this array.
Griffonia simplicifolia lectin (GSL-II) only bound strongly to those GlcNAc-terminating sequences as previously reported(Iyer, et al., 1976). Galanthus nivalis agglutinin (GNA), which is reported to recognize specific alpha-mannose-containing N-glycans(Shibuya, et al., 1988), did not show binding signals to any complex-type N-glycans but only to the two mannose-terminating N-glycans.
Altogether, these studies have identified unique recognition of complex N-glycans and the nuanced specificities of a group of important lectins that are widely used in glycan analysis.
Galectins
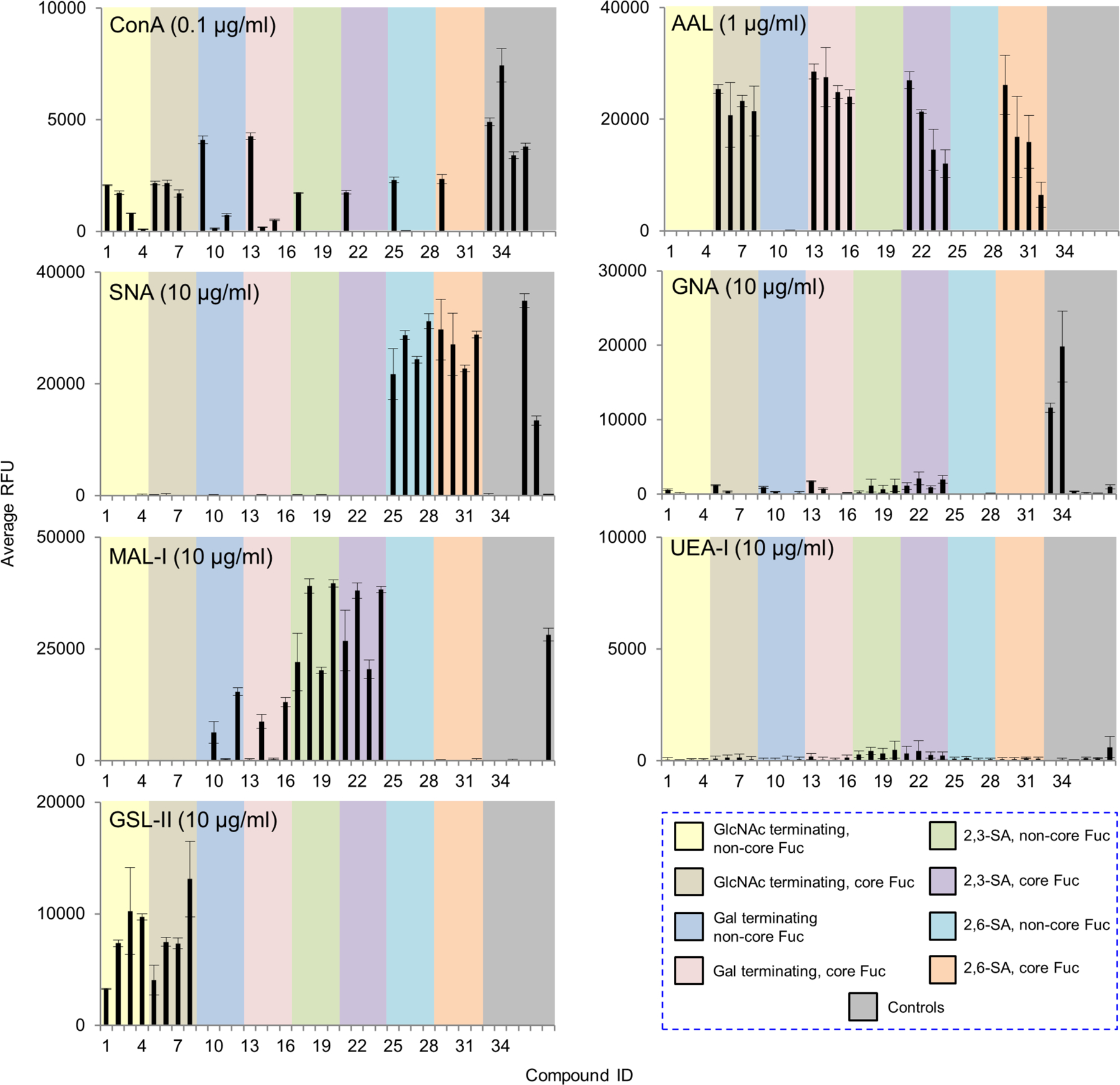
Galectin-1 and -3 are arguably the two most well-studied human galectins and potentially interact with endogenous and exogenous glycans. Human galectin-1 plays an important role in apoptosis, and tumor progression and metastasis. It was suggested that complex N-glycans are ligands for human galectin-1(Patnaik, et al., 2006; Stowell, et al., 2008). Indeed, binding by the recombinant human galectin-1 was observed at 50 μg/ml with the LacNAc (Galβ1–4GlcNAc)-terminating glycans, with or without core fucose (Fig. 5, Supplemental Table S2). The tetraantennary N-glycan with four LacNAc sequences was most strongly bound. Importantly, our result clearly shows the tolerance of galectin-1 to α2,3- but not to α2,6-linked sialic acid, which is consistent with the previous observations by microarray using small glycan epitopes(Stowell, et al., 2008) and in cell-based assays(Earl, et al., 2010; Patnaik, et al., 2006).
Figure 5.
Microarray analyses of recombinant human galectin-1 and -3. (a) and (b), The recombinant galectin-1 and -3 were tested at 5 and 50 μg/mL. Galectin-1 was detected by mouse anti-human galecin-1 followed by Alexa Fluor 488 labelled anti-mouse IgG (H+L). Galectin-3 was directly labelled with biotin and detected by cyanine 5-streptavidin. Results are shown as relative fluorescence units (RFUs) by averaging the background-subtracted fluorescence signals of 4 replicate spots, error bars represent the standard deviation among the 4 values. The RFUs and the standard deviation of 50 μg/mL were listed in Supplemental Table S2. The probes are grouped as indicated in the colored panels.
Human galectin-3 is involved in early development, tissue regeneration, cancer, immune and inflammatory responses, as well as recognition/effector functions against potential pathogens. As the only chimera-type in the galectin family, human galectin-3 is known to bind poly-N-acetyllactosamine containing glycans(Noll, et al., 2016; Song, et al., 2009; Stowell, et al., 2008) and the core 1 O-glycan (Galβ1–3GalNAcα-)(Yu, et al., 2007). However, although studies suggest that galectin-3 binding is associated with the expression of highly branched complex N-glycans(Guo, et al., 2008; Hirabayashi, et al., 2002), its N-glycan binding preference has not been clear. We observed strong binding to the LacNAc containing N-glycans with the recombinant human galectin-3 at 50 μg/ml concentration (Fig. 5, Supplemental Table S2). Weak or no binding was detected with the biantennary and the 2,2,6-form isomeric triantennary structure, which resembles the lectin DSA, although it differs in the lack of binding to the GlcNAc terminating sequences. Similar to galectin-1, galectin-3 can tolerant the presence of α2,3- but not α2,6-linked sialic acid. Our result suggests that galectin-3 can preferentially bind the N-glycan backbone sequences in the absence of poly-LacNAc extension.
Influenza viruses
Influenza viruses bind to terminal sialic acid in a linkage specific manner, as mammalian strains exhibit binding to α2,6-linked sialic acid receptors while avian strains show a preference for α2,3-linked receptors(Air, 2014; Byrd-Leotis, et al., 2017; Xiong, et al., 2014). Existing glycan microarrays have been utilized for the receptor binding characterization of various strains of influenza(Byrd-Leotis, et al., 2017; Byrd-Leotis, et al., 2014; Gulati, et al., 2014; McBride, et al., 2016; Nycholat, et al., 2012; Smith and Cummings, 2014), however such arrays as the CFG array do not have pairwise combinations of α2,3- vs. α2,6-sialic acid terminating structures or fucosylated vs. non-fucosylated structures.
Our sequence-defined N-glycan array enables quick diagnostic appraisal of influenza A virus receptor specificity. Two well-characterized strains of recombinant viruses, X-31 and X-31 HAM, were chosen for investigation. X-31 expresses the HA and NA genes from A/Aichi/2/68 in combination with the internal genes of the A/PR/8/34 strain(Kilbourne, 1969), while X-31 HAM contains an HA L226Q mutation which alters the specificity to recognize 2,3-linked sialic acid receptors(Rogers, et al., 1983). On the sequence-defined N-glycan microarray, X-31 bound to glycans 26, 28, and 31, which all terminate in α2,6-linked sialic acid (Fig. 6, Supplemental Table S3). The presence of a core fucose, on glycan 31, had no impact on the glycan recognition. By contrast, X-31 HAM bound most strongly to the α2,3-sialylated glycans 19, 20, 23, and 24, which is consistent with the reported specificity. Treatment of the array with Arthrobacter ureafaciens neuraminidase completely removes X-31 binding (Fig. 6), indicating the requirement for terminal sialic acid for recognition. This neuraminidase treatment was completed under conditions that result in reduction of SNA binding to background levels (Supplemental Fig. S3). Interestingly, the same treatment revealed that MAL-I binds to selected de-sialylated structures that were originally α2,6-sialylated (Compounds 17-32), in agreement with our observation that this lectin recognizes non-sialylated glycan structures (Fig. 4 and Supplemental Fig. S2).
Figure 6.
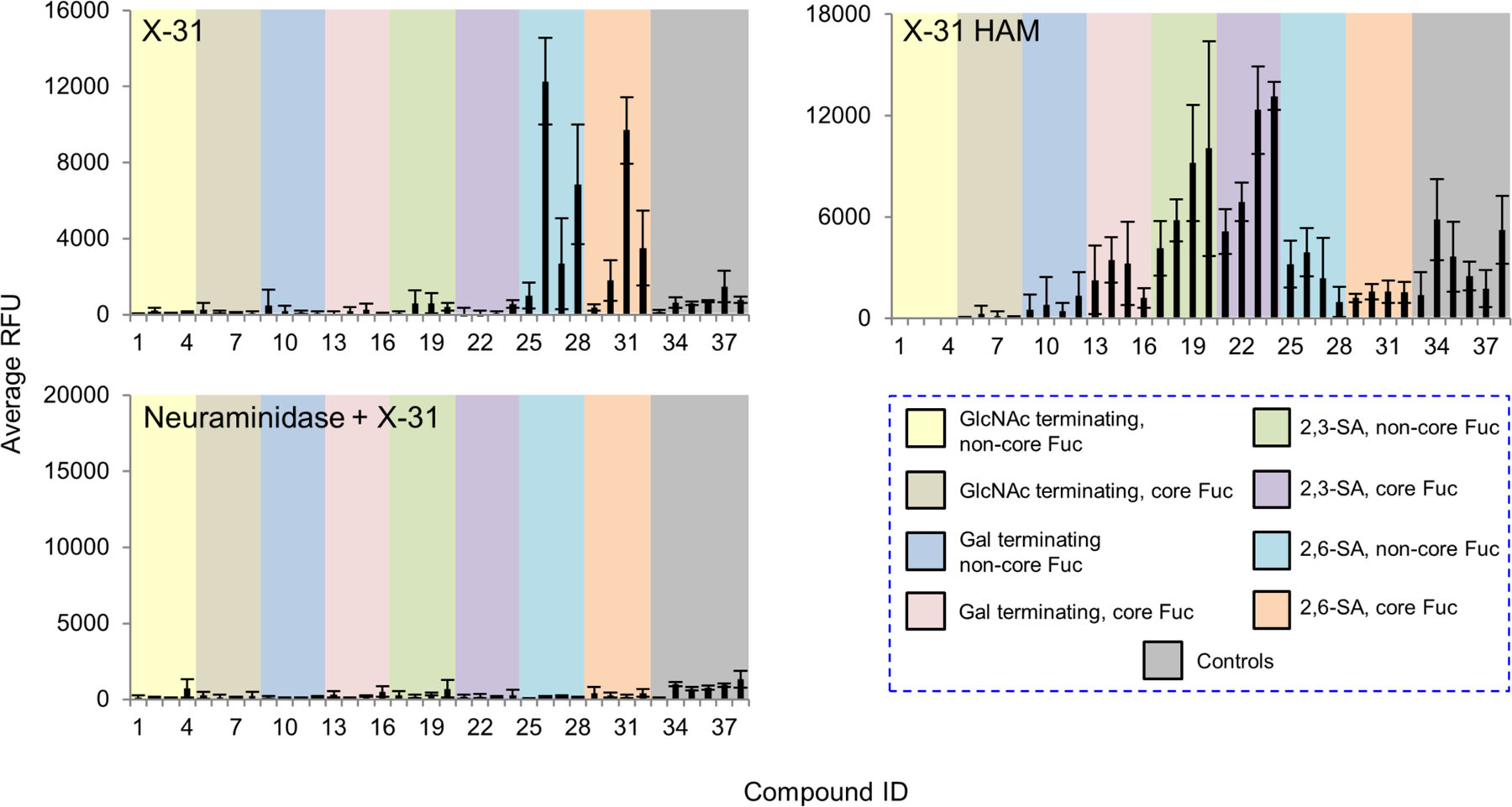
Microarray analyses of two strains of recombinant influenza viruses. Two strains of influenza viruses X-31 and X-31 HAM with HA titers of 128 HAU/50 μL, were labeled with Alexa Fluor-488 NHS ester and probed with the array. The slides were also pretreated with Arthrobacter ureafaciens neuraminidase or buffer alone 18 hours prior to incubation with X-31 to confirm the sialic acid-dependence. The RFUs and the standard deviation were listed in Supplemental Table S3. The probes are grouped as indicated in the colored panels.
Siglecs
Siglecs are endogenous sialic acid binding lectins that participate in cell signaling, adhesion, and are particularly important in immune cell regulation(Bornhofft, et al., 2018; Varki, et al., 2017). We investigated 11 recombinant human Siglecs, Siglec-1 to -11 and rat-derived Siglec-4 to explore their N-glycan binding. As the affinity of Siglec-ligand interaction is generally thought to be low, we also performed the binding assay with Siglecs precomplexed with the secondary antibodies to increase the binding avidity.
Of the 12 tested, only four Siglecs showed binding to the N-glycan array (Fig. 7, Supplemental Table S4). Siglec-1 preferentially bound to NeuAcα2–3Galβ1–4GlcNAc- containing N-glycans, irrespective of the presence of core fucosylation (Fig. 7a). The binding was independent of the branching pattern. Structures with more α2,3-linked NeuAc elicited stronger signals. Siglec-9 showed almost identical binding preference to Siglec-1 (Fig. 7b). Binding signals of the two Siglecs without precomplex were higher than those of precomplexed, suggesting that the affinities of Siglec-1 and -9 are higher. Human Siglec-9 has been reported to selectively bind 6-sulfo LeX (Neu5Acα2–3Galβ1–4(Fucα2–3)(6S)GlcNAc-)(Macauley, et al., 2014), a structure which is not on this N-glycan array. We thus corroborated this finding using the CFG glycan microarray v5.4 (Supplemental Fig. S4, Supplemental Table S5). In addition to 6-sulfo LeX, Siglec-9 also exhibited strong binding to α2,3-sialic acid-containing glycans that lack either sulfate or fucose, suggesting that sulfation and fucosylation are not essential for Siglec-9 binding. Although no binding was detected to the α2,3-sialylated tri- and tetraantennary N-glycan structures on the CFG array, these glycans also contain a bisecting GlcNAc, which may prevent the binding. It is also possible that the binding determinant is inappropriately presented by the synthetic 2-amino-methyl N,O-hydroxyethyl linker used for those N-glycan structures on the CFG array.
Figure 7.
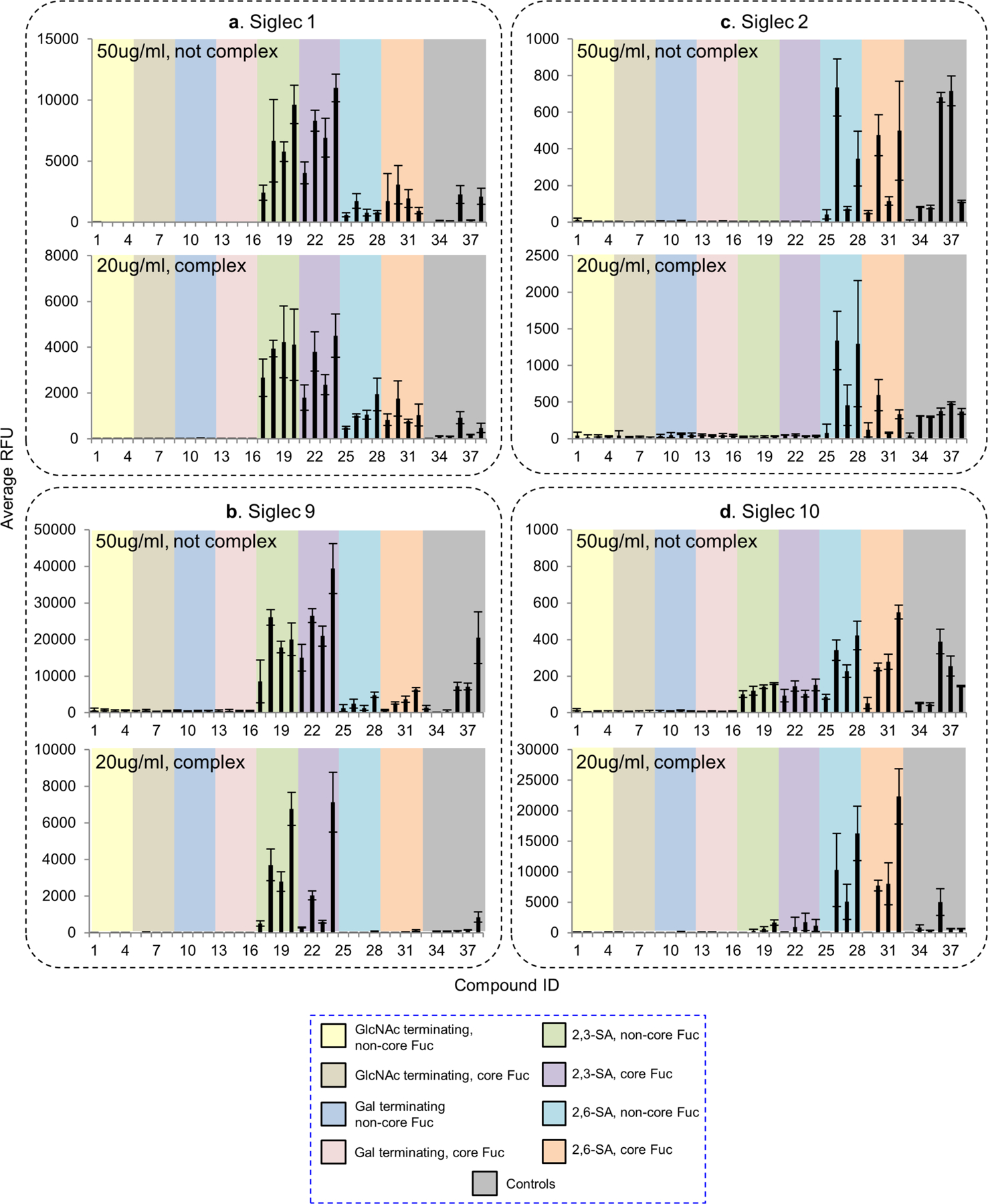
Microarray analyses of recombinant human Siglec-1, -2, -9 and -10 with N-glycans in the N-glycan array. (a-d) Human Siglec-1, -2, -9 and -10 were tested at 50 μg/mL without precomplex or at 20 μg /mL with preincubation with either anti-His (for Siglec-1) or anti-human IgG (for the other Siglecs). The RFUs and the standard deviation were listed in Supplemental Table S4. The probes are grouped as indicated in the colored panels.
In comparison to Siglec-1 and -9, previous data indicated that Siglec-2 and -10 preferentially bound to Neu5Acα2–6-containing glycans, although Siglec-2 may require the presence of C6-sulfate on the GlcNAc to enhance the binding(Macauley, et al., 2014). Our data showed that Siglec-2 and -10 both preferentially bound α2,6-sialylated N-glycans although some weak binding was detected to the α2,3-sialylated N-glycans with Siglec-10 (Fig. 7c and d). Preincubation with the secondary antibody significantly increased the binding intensity, suggesting multivalency may be important to achieve the avidity required for the recognition of these two Siglecs. This was also reflected by the observation that glycans containing higher numbers of branches (and thus more binding epitopes) elicited higher binding signals.
No binding was detected with the other Siglecs on the N-glycan array. However, Siglec-8 showed strong and exclusive binding towards one compound, Neu5Acα2–3(6S)Galβ1–4GlcNAc- among 600 probes on the CFG array (Supplemental Fig. S4, Supplemental Table S5). The N-glycan containing this sulfated epitope was not present on our N-glycan array.
Collectively, our data indicates that sialylated multiantennary N-glycans are potential endogenous receptors for Siglecs-1, -2, -9 and -10, and more binding partners may be discovered as we grow the diversity of this N-glycan array.
DISCUSSION
The results of our study provide new information about the glycan-binding specificities of a number of important molecules including human and plant proteins and influenza viruses. Insights into their recognition of N-glycans was greatly facilitated by the successful development of an integrated chemoenzymatic strategy for synthesis of a library of highly complex multiantennary N-glycan isomers, which encompasses variations in branching pattern, core fucosylation and terminal sialic acid linkage, and coupling of the N-glycan library to a microarray platform.
A major advantage of our synthetic strategy to generate useful N-glycans for this study is the preservation of the reducing terminal natural spacer asparagine, which increases the efficiency of glycosyltransferase reactions. In two direct comparisons, the reaction by ST3GAL4 and ST6GAL1 with an AEAB conjugate of the biantennary N-glycan was slow and the yield was not satisfactory (Supplemental Fig. S5). It is therefore difficult to achieve quantitative sialylation with the AEAB-linked tri- and tetraantennary N-glycans. In addition, it has been shown that FUT8 requires the presence of an appropriate peptide or other structural elements for its reactivity(Yang, et al., 2017). Thus Asn-linked N-glycans are ideal substrates for core-fucosylation reactions. Moreover, beta-linked Asn would more closely resemble the natural configuration of the N-glycans on glycoproteins, and thus present a more biologically relevant conformation for binding interrogation.
The sequence-defined N-glycan microarray platform sheds light on the fine specificities of the widely-used N-glycan binding plant lectins (Table 1, see Supplemental Discussion for detailed comparison) and provides guidance for their applications. Although the backbone-binding lectins, WGA, LCA, PSA, ConA, E-PHA, L-PHA and DSA can accommodate the α2,3-sialic acid, only LCA, PSA and ConA can tolerate the existence of α2,6-sialic acid. While lectins such as E-PHA, L-PHA and DSA are widely used in many histology studies to evaluate the expression of N-glycans with particular branching patterns(Croci, et al., 2014; Dennis, et al., 1987), desialylation is essential prior to staining with these lectins, as the binding epitope may be masked by the α2,6-sialic acid, and lead to false-negative results. Enhanced lectin staining, on the other hand, may result from either increased expression of the corresponding branches or simply loss of α2,6-linked sialic acid capping.
Table 1.
Summary of lectin specificities observed in this study. Shaded in grey indicates the binding epitope for each lectin. The red x shows the additional sequence in certain positions significantly reduces lectin binding. The symbol +/− means the substitution can be tolerated by the lectin. The table also shows whether α2,3-, α2,6-sialic acid and core fucose are tolerated by each lectin. The symbolic nomenclature for glycans are shown in Fig. 1.
| Lectin | Sequence | Tolerance of other modifications | ||
|---|---|---|---|---|
| α2,3-Neu5Ac | α2,6-Neu5Ac | core Fuc | ||
| WGA | 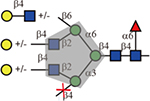 |
Yes | No | Yes |
| DSA | 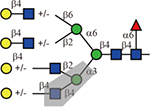 |
Yes | No | Yes |
| LCA or PSA | 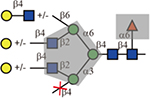 |
Yes | Yes | Must |
| L-PHA | 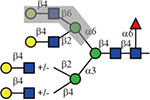 |
Yes | No | Yes |
| E-PHA | 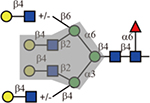 |
Yes | No | Yes |
| ConA | 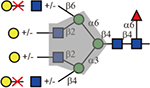 |
Yes | Yes | Yes |
| GNA | high mannose N-glycans | unknown | unknown | unknown |
| GSL-II | No | No | Yes | |
| ECL | 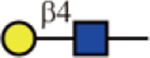 |
No | No | Yes |
| RCA-I | 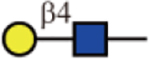 |
No | Yes | Yes |
| SNA | 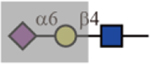 |
No | N/A | Yes |
| MAL-I | 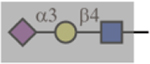 |
N/A | No | Yes |
| AAL | 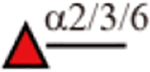 |
Yes | Yes | Yes |
| UEA-I | Not bound on this array | |||
Similarly, LCA stained α-fetoprotein (designated AFP-L3) has been suggested a good biomarker for hepatocellular carcinoma in clinical settings(Pinho and Reis, 2015; Taniguchi and Kizuka, 2015). Patients with high AFP-L3 have been associated with elevated expression of core fucosylation and the glycosyltransferase gene FUT8(Li, et al., 2001; Taketa, et al., 1993). Our study suggested that only core fucosylated N-glycans with biantennary and the 2,2,6-isoform triantennary but not the 2,2,4-isoform or the tetraantennary were positively bound by LCA. Thus, positive expression of AFP-L3 in HCC patients may be due to the combination of a relatively lower level of MGAT4 and a higher level of FUT8. Thus, our results improve the understanding in tumor-specific glycan expression and cancer biology.
Our newly synthesized N-glycan library also provides valuable insights into specificities of other glycan binding partners. It has been known for years that the linkage conformation of the terminal sialic acid to the penultimate galactose is important for species specificity of influenza viruses. However, there have been no N-glycan libraries that contain naturally occurring bi-, tri- and tetraantennary structures with pairwise combinations of α2,3- vs. α2,6-sialic acid terminating structures, or core-fucosylated vs. non-fucosylated structures. Our data with X-31 and X-31 HAM are good examples to show that, the sequence-defined N-glycan array enables quick interrogation of the receptor binding characteristics of newly isolated influenza viruses, which helps to identify the adaptation markers that are predictors of cross-species transmission and potential pandemic threats. Screening of a larger cohort of influenza viruses is now underway.
Our data also identified potential natural ligands for human Siglecs-1, -2, -9 and -10. We demonstrated that these Siglecs, although generally thought to require clustered display, can directly bind to multiantennary N-glycans with correct sialic acid conformation. Considering that the local concentration of sialylated epitopes on cell surface can be higher due to the presence of multiple copies of multiantennary N-glycans, engagement of Siglecs with ligands may be stronger than on the array, which could be enough to elicit the downstream cell signaling.
In summary, our strategy enables efficient and convenient production of highly complex asparagine-linked multiantennary N-glycans. The synthesis takes advantage of regio- and stereoselective glycosyltransferases to efficiently generate N-glycan isomers, which would otherwise be extremely difficult for chemical synthesis and separation. As additional glycosyltransferases become available, we anticipate that N-glycans with more modifications such as poly-N-acetyllactosamine, Lex, sialyl-Lex, and blood group antigens can be synthesized to quickly expand the N-glycan collection. The success in their in vitro biosynthesis allows for the generation of an N-glycan microarray, which offers an excellent high-throughput platform to elucidate N-glycan recognition specificities of various glycan binding partners. Our arrays, as a unique resource, will be available for other researchers to explore their own biologically relevant glycan binding partners in the future.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Richard Cummings (rcummin1@bidmc.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The glycosyltransferases were generated in the HEK293 cells (Freestyle 293-F cells, male, Thermo Fisher). Cells were maintained in serum-free medium (Freestyle-293 Expression Medium, Thermo Fisher) at 37°C at 5% CO2 on an orbital shaker at 150 rpm.
METHOD DETAILS
Materials
All chemicals were purchased from Sigma-Aldrich and used without further purification. HPLC solvents were purchased from Fisher Scientific. Details of the commercial substances are listed in the Key Resources Table. Plasmids of six glycosyltransferases, FUT8, MGAT4a, MGAT5, B4GalT1, ST3Gal4 and ST6Gal1(Moremen, et al., 2017) were kindly provided by Professor Kelley Moremen at the Complex Carbohydrate Research Center, University of Georgia, Athens, GA. In addition, Professor Moremen also kindly provided two purified glycosyltransferases, MGAT4a and MGAT5 that were directly used in the synthesis. The human galectin-3 was recombinantly expressed and purified as described previously(Noll, et al., 2016). Two strains of influenza virus, X-31 and X-31 HAM were reported previously(Benton, et al., 2015) and prepared in house.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-human galectin-1 (clone # 933133) | R&D Systems | Cat#MAB1152 |
| Goat polyclonal anti-Human IgG (H+L), Alexa Fluor 488 | Thermo Fisher | Cat#A-11013 |
| Rabbit polyclonal anti-Mouse IgG (H+L), Alexa Fluor 488 | Thermo Fisher | Cat#A-11059 |
| Mouse monoclonal anti penta-His, Alexa Fluor 488 | Qiagen | Cat#35310 |
| Bacterial and Virus Strains | ||
| X-31 | John W. McCauley (Benton, D.J. et. al. 2015) | N/A |
| X-31 HAM | John W. McCauley (Benton, D.J. et. al. 2015) | N/A |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Egg yolk powder | Magic Flavors | N/A |
| Pronase | EMD Millipore | Cat#53702-250KU |
| Phosphatase | Sigma | Cat#P5931-200UN |
| β-galactosidase | Sigma | Cat#G5160 |
| Neuraminidase (Arthrobacter ureafaciens) | New England Biolabs | Cat#P0722L |
| UDP-Gal | Sigma; Chemily | Cat#U4500; Cat#SN02006 |
| UDP-GlcNAc | Chemily | Cat#SN02009 |
| GDP-Fucose | Chemily | Cat#SN02002 |
| CMP-sialic acid | Sigma; Chemily | Cat#233264; Cat#SN02001 |
| Fmoc N-hydroxysuccinimide ester | Sigma | Cat#46920-5G-F |
| 293 Freestyle expression medium | Thermo Fisher | Cat#12338018 |
| Polyethylenimene | Polysciences, Inc | Cat#23966 |
| Valproic acid | Sigma | Cat#P4543-100G |
| His-Pur Ni-NTA resin | Thermo Fisher | Cat#88221 |
| WGA | Vector Labs | Cat#B-1025 |
| DSA | Vector Labs | Cat#B-1185 |
| LCA | Vector Labs | Cat#B-1045 |
| PSA | Vector Labs | Cat#B-1055 |
| L-PHA | Vector Labs | Cat#B-1115 |
| E-PHA | Vector Labs | Cat#B-1125 |
| ECL | Vector Labs | Cat#B-1145 |
| RCA-I | Vector Labs | Cat#B-1085 |
| Con A | Vector Labs | Cat#B-1005 |
| SNA | Vector Labs | Cat#B-1305 |
| MAL-I | Vector Labs | Cat#B-1315 |
| GSL-II | Vector Labs | Cat#B-1215 |
| AAL | Vector Labs | Cat#B-1395 |
| GNA | Vector Labs | Cat#B-1245 |
| UEA-I | Vector Labs | Cat#B-1065 |
| Recombinant human Siglec-1/CD169 | R&D Systems | Cat#5197-SL |
| Recombinant human Siglec-2/CD22 Fc Chimera | R&D Systems | Cat#1968-SL |
| Recombinant human Siglec-3/CD33 Fc Chimera | R&D Systems | Cat#1137-SL |
| Recombinant human MAG/Siglec-4a Fc Chimera | R&D Systems | Cat#8940-MG |
| Recombinant human Siglec-5/CD105 Fc Chimera | R&D Systems | Cat#1072-SL |
| Recombinant human Siglec-6/CD327 Fc Chimera | R&D Systems | Cat#2859-SL |
| Recombinant human Siglec-7/Fc Chimera | R&D Systems | Cat#1138-SL |
| Recombinant human Siglec-8/Fc Chimera | R&D Systems | Cat#9045-SL |
| Recombinant human Siglec-9/Fc Chimera | R&D Systems | Cat#1139-SL |
| Recombinant human Siglec-10/Fc Chimera | R&D Systems | Cat#2130-SL |
| Recombinant human Siglec-11/Fc Chimera | R&D Systems | Cat#3258-SL |
| Recombinant Rat MAG/Siglec-4a Fc Chimera | R&D Systems | Cat#538-MG |
| Recombinant Human Galectin-1 | R&D Systems | Cat#1152-GA |
| Recombinant Human Galectin-3 | Laboratory stock (Noll, et al., 2016) | N/A |
| Streptavidin - Cyanine 5 | Invitrogen | Cat#434316 |
| Sulfo-NHS-LC-LC-Biotin | Thermo Scientific | Cat#21338 |
| Alexa Fluor-488 NHS | Thermo Scientific | Cat#A20000 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human: Freestyle 293-F | Thermo Fisher | Cat#R79007 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Plasmid: B4GALT1-pGEn2-DEST | Kelley Moremen (Moremen et al, 2017) | HsCD00522296 |
| Plasmid: ST3GAL4-pGEn2-DEST | Kelley Moremen (Moremen et al, 2017) | HsCD00413174 |
| Plasmid: ST6GAL1-pGEn2-DEST | Kelley Moremen (Moremen et al, 2017) | HsCD00413052 |
| Plasmid: MGAT4A-pGEn2-DEST | Kelley Moremen (Moremen et al, 2017) | HsCD00413158 |
| Plasmid: MGAT5-pGEn2-DEST | Kelley Moremen (Moremen et al, 2017) | N/A |
| Plasmid: FUT8-pGEn2-DEST | Kelley Moremen (Moremen et al, 2017) | HsCD00413179 |
| Software and Algorithms | ||
| Other | ||
| Nexterion H NHS functionalized slides | Schott AG | Cat#1070936 |
Preparation of the N-glycan library
Detailed protocol regarding the extraction of SGP, synthesis of the Fmoc-labelled asialo-, agalacto-biantennary N-glycan core structure, production of the glycosyltransferases, and the enzymatic reactions for generating the N-glycan library are in Supporting Information. The products generated in this study were analyzed by HPLC, MS and NMR.
HPLC analysis and purification
HPLC analysis and semi-preparative purification of the Fmoc-labelled Asn-linked glycans were performed on a Shimadzu HPLC CBM-20A system. It contained a UV detector SPD-20AV and a fluorescence detector RF-20A. UV absorption at 254 nm and fluorescence at 254 nm excitation (Ex) and 340 nm emission (Em) were used to detect and quantify the Fmoc-tagged glyco-amino acid.
Analytical and semi-preparative HPLC were performed using 250 ˣ 4.6 mm and 250 ˣ 10 mm Zorbax NH2 columns, respectively. The mobile phases were acetonitrile, water, and 250 mM ammonium acetate. The concentration of water increased from 16% to 40% and the ratio of ammonium acetate from 4% to 50% over 60 min.
Mass spectrometry
An UltrafleXtreme MALDI-TOF/TOF system from Bruker Daltonics (Billerica, MA) was used for MALDI-TOF MS analysis of the glycans and their derivatives. MS was performed in reflective positive or reflective negative mode. 2,5-Dihydroxybenzoic acid (10 mg/ml in MeOH:H2O=1:1) was used as the matrix.
ESI-MS analysis was performed on an Orbitrap Fusion Lumos from Thermo Fisher Scientific (San Jose, CA). The sample after column clean-up was injected directly into the mass spectrometer using a syringe. Full MS data acquisition was set at 120K resolution. Electrospray settings for the direct infusion was set at 4.0KV in positive ion mode with the S-lens set at 30 volts; capillary temperature at 250°C for efficient desolvation and ionization of the positively charged ions. The mass range used for the study was m/z 150 to 2000. The AGC target setting for the Full MS was at 1E6. The Orbitrap Fill times were set at 120 ms. HCD energy values were set at 35 eV used as normalized collision energy for the glycan analyzed in this paper.
NMR
1H and 13C NMR spectra were recorded on a Bruker Avance II 600MHz and an Agilent 700MHz NMR Magnet System at the NMR core facility at Harvard Medical School. The compounds were deuterium oxide exchanged three times before reconstitution in deuterium oxide for analysis. The NMR data is presented in part VI of the Supplemental Information. Characterization of the generated glycans/glycan conjugates are as follows: Chemical shift (in parts per million (ppm) relative to water as the internal standard), multiplicity (s = singlet, d = doublet, t = triplet, dd = doublet of doublet, m = multiplet and/or multiple resonances), coupling constant in Hertz (Hz), integration. All NMR signals were assigned on the basis of 1H NMR, 1H-1H COSY, 1H-1H TCOSY, and 1H-13C HSQC experiments.
Microarray printing
Glycan conjugates were reconstituted in phosphate buffer (100 mM sodium phosphate, pH 8.5) at final concentration of 50 μM. Control glycan conjugates were reconstituted in phosphate buffer to a final concentration of 100 μM. All conjugates were arrayed onto Nexterion H NHS functionalized slides (Schott AG, Louisville, KY) using a sciFLEXARRAYER S11 from Scienion AG (Berlin, Germany). The average drop volume was within 5% variation of the target volume of 330pL. After printing, slides were incubated at room temperature in a humidity cabinet set at 80% relative humidity overnight. The slides were then blocked with 50 mM ethanolamine in 100 mM sodium tetraborate buffer (pH 8.5) for 1 hour and washed briefly with 1x phosphate buffered saline with 0.05% polysorbate 20 (Tween-20). This was followed by brief rinsing with ultrapure water before drying by centrifugation. The slides were stored desiccated at -20°C until use.
Microarray analysis
The glycans were printed on NHS-coated glass slides and assayed as previously described(Song, et al., 2016). The detailed procedures were in Supporting Information. After rehydration using TSM buffer (20 mM Tris–HCl, 150 mM sodium chloride, 0.2 mM calcium chloride, and 0.2 mM magnesium chloride), the microarray slides were probed with biotinylated lectins, human galectin-1, galectin-3, influenza viruses, or Siglecs. The bound lectins were detected with cyanine 5-streptavidin (Invitrogen) at 0.5 μg/mL. Human galectin-1 was detected with mouse anti-human galecin-1 followed with Alexa Fluor 488 labelled anti-mouse IgG (H+L). The recombinant human galectin-3 was labelled with biotin using EZ-Link™ Sulfo-NHS-LC-LC-biotin (Thermo Scientific) prior to the binding assay according to the previous report(Noll, et al., 2016). The bound galectin-3 was thereafter detected with cyanine 5-streptavidin at 0.5 μg/mL. Two strains of influenza viruses, X-31 and X-31 HAM, were labeled with Alexa Fluor-488 NHS ester 24 hours prior to the assay. Excess label was dialyzed away and the HA titers of each virus were obtained using chicken erythrocytes. The viruses were assayed at 128 HAU/50 μL on the microarray. The microarray slides were also pretreated with Neuraminidase A at 3000 U/mL or buffer alone for 18 hours followed by virus incubation. Binding was directly quantified after thorough washing. Twelve recombinant Siglecs were tested at two concentrations (5 and 50 μg/mL) without precomplexation. Binding was detected by overlaying with Alexa Fluor-488 labelled anti-human IgG or Penta-His. In addition, Siglecs were precomplexed with the secondary antibody at 1:3 (w/w) ratio to anti-Human IgG or Penta-His 15 minutes prior to addition of binding buffer towards the final concentration (20 μg/mL). After incubation, the binding signals were directly quantified.
Slides were scanned with a Genepix 4300A, microarray scanner from Molecular Devices (Sunnyvale, CA). Scanned images were quantified using Genepix Pro 7 software. For cyanine5 and Alexa488 fluorescence, a setting of 649 nm (Ex) and 670 nm (Em) and 495 nm (Ex) and 519 nm (Em) were used, respectively. All images obtained from the scanner were in grayscale and colored for easy discrimination. The results are quantified as shown in the data quantitation section.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data quantitation for the array analysis
The signal intensities were quantified by the GenePix Pro 7 that is associated with the microarray scanner and processed by Excel (Microsoft). The results are shown as relative fluorescence units (RFUs) by averaging the background-subtracted fluorescence signals of four replicate spots with error bars representing the standard deviation among the four values.
DATA AND SOFTWARE AVAILABILITY
All of the N-glycan microarray datasets related to this work are shown in the Supplemental Information and are publicly available at the National Center for Functional Glycomics (NCFG) website: https://ncfg.hms.harvard.edu/ncfg-data
ADDITIONAL RESOURCES
Not applicable.
Supplementary Material
SIGNIFICANCE.
N-glycosylation is an important post-translational modification that is involved in many biological and pathological events. The N-glycan ligands recognized by a group of important proteins such as Siglecs, galectins, influenza viruses, as well as many plant lectins, are elusive due to the lack of a library of well-characterized standards and a high-throughput platform. Taking a highly efficient chemoenzymatic approach, we produced a library of naturally occurring isomeric asparagine-linked glycans. They vary in non-reducing terminal sialic acid, galactose, and N-acetylglucosamine residues, as well as core fucose. The success of their synthesis enables a careful investigation of a number of N-glycan binding partners on microarrays. Our results elucidate the unique glycan-binding specificities of several plant lectins, human galectin-1 and -3, influenza viruses, and Siglecs, providing important guidance in their biological and clinical applications in identifying glycans with certain structural features. Our data also shed light on the natural ligands for those molecules, which may lead to the rational design of efficient inhibitors. Our N-glycan library is now poised as a discovery platform for other glycan binding partners that are associated with key biological processes.
Acknowledgements
We thank Richard H. Barnes for his technical support and Dr. Jamie Heimburg-Molinaro for manuscript editing and review. We thank Dr. Kelley Moremen (glycoenzymes.ccrc.uga.edu) and the Repository for providing the glycosyltransferases and the expression plasmid, and acknowledge funding through NIH grants P41GM103390 and P01GM107012.
Funding Sources
This work was supported by CEIRS grant contract HHSN272201400 to DS and RDC and NIH Grant P41GM103694 to RDC.
Footnotes
Supporting Information. Supplemental Methods; Supplemental Discussion; Supplemental Scheme S1; Supplemental Figures S1–S5; Supplemental Tables S1–S5; Supplemental Data- complete HPLC, MALDI-MS, and NMR profiles. Files: Supplemental Information main file (PDF); Supplemental Table S5 (Excel).
Declaration of interests: The authors declare no competing interests.
References
- Air GM (2014). Influenza virus-glycan interactions. Curr Opin Virol 7, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baenziger JU, and Fiete D (1979). Structural determinants of concanavalin A specificity for oligosaccharides. Journal of Biological Chemistry 254, 2400–2407. [PubMed] [Google Scholar]
- Baenziger JU, and Fiete D (1979). Structural determinants of Ricinus communis agglutinin and toxin specificity for oligosaccharides. Journal of Biological Chemistry 254, 9795–9799. [PubMed] [Google Scholar]
- Benton DJ, Martin SR, Wharton SA, and McCauley JW (2015). Biophysical Measurement of the Balance of Influenza A Hemagglutinin and Neuraminidase Activities. The Journal of Biological Chemistry 290, 6516–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohorov O, Andersson-Sand H, Hoffmann J, and Blixt O (2006). Arraying glycomics: a novel bi-functional spacer for one-step microscale derivatization of free reducing glycans. Glycobiology 16, 21C–27C. [DOI] [PubMed] [Google Scholar]
- Bornhofft KF, Goldammer T, Rebl A, and Galuska SP (2018). Siglecs: A journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev Comp Immunol 86, 219–231. [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L, Cummings RD, and Steinhauer DA (2017). The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, et al. (2014). Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci U S A 111, E2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon AD, Liu Y, Li X, Wang X, Chen X, Li L, and Wang PG (2016). Substrate specificity of FUT8 and chemoenzymatic synthesis of core-fucosylated asymmetric N-glycans. Organic & Biomolecular Chemistry 14, 4027–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, and Packer NH (2014). UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Research 42, D215–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci Diego O., Cerliani Juan P., Dalotto-Moreno T, Méndez-Huergo Santiago P., Mascanfroni Ivan D., Dergan-Dylon S, Toscano Marta A., Caramelo Julio J., García-Vallejo Juan J., Ouyang J, et al. (2014). Glycosylation-Dependent Lectin-Receptor Interactions Preserve Angiogenesis in Anti-VEGF Refractory Tumors. Cell 156, 744–758. [DOI] [PubMed] [Google Scholar]
- Crowley JF, Goldstein IJ, Arnarp J, and Lonngren J (1984). Carbohydrate binding studies on the lectin from Datura stramonium seeds. Arch Biochem Biophys 231, 524–533. [DOI] [PubMed] [Google Scholar]
- Cummings RD, and Kornfeld S (1982). Characterization of the structural determinants required for the high affinity interaction of asparagine-linked oligosaccharides with immobilized Phaseolus vulgaris leukoagglutinating and erythroagglutinating lectins. Journal of Biological Chemistry 257, 11230–11234. [PubMed] [Google Scholar]
- Cummings RD, and Kornfeld S (1984). The distribution of repeating [Gal beta 1,4GlcNAc beta 1,3] sequences in asparagine-linked oligosaccharides of the mouse lymphoma cell lines BW5147 and PHAR 2.1. Journal of Biological Chemistry 259, 6253–6260. [PubMed] [Google Scholar]
- Debray H, Montreuil J, Lis H, and Sharon N (1986). Affinity of four immobilized Erythrina lectins toward various N-linked glycopeptides and related oligosaccharides. Carbohydr Res 151, 359–370. [DOI] [PubMed] [Google Scholar]
- Dennis J, Laferte S, Waghorne C, Breitman M, and Kerbel R (1987). Beta 1–6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science 236, 582–585. [DOI] [PubMed] [Google Scholar]
- Earl LA, Bi S, and Baum LG (2010). N- and O-Glycans Modulate Galectin-1 Binding, CD45 Signaling, and T Cell Death. Journal of Biological Chemistry 285, 2232–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Iino K, Nozawa S, Iizuka R, and Kobata A (1988). Immobilized Datura stramonium agglutinin column chromatography, a novel method to discriminate the urinary hCGs of patients with invasive mole and choriocarcinoma from those of normal pregnant women and patients with hydatidiform mole. Jpn J Cancer Res 79, 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J-Q, and Lee YC (1997). Detailed Studies on Substrate Structure Requirements of Glycoamidases A and F. Journal of Biological Chemistry 272, 27058–27064. [DOI] [PubMed] [Google Scholar]
- Gallagher JT, Morris A, and Dexter TM (1985). Identification of two binding sites for wheat-germ agglutinin on polylactosamine-type oligosaccharides. Biochem. J 231, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant OC, Smith HM, Firsova D, Fadda E, and Woods RJ (2014). Presentation, presentation, presentation! Molecular-level insight into linker effects on glycan array screening data. Glycobiology 24, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, and Baenziger JU (1989). Characterization of oligosaccharides by lectin affinity high-performance liquid chromatography. Trends Biochem Sci 14, 168–172. [DOI] [PubMed] [Google Scholar]
- Gulati S, Lasanajak Y, Smith DF, Cummings RD, and Air GM (2014). Glycan array analysis of influenza H1N1 binding and release. Cancer Biomark 14, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H-B, Nairn A, Harris K, Randolph M, Alvarez-Manilla G, Moremen K, and Pierce M (2008). Loss of expression of N-acetylglucosaminyltransferase Va results in altered gene expression of glycosyltransferases and galectins. FEBS Letters 582, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström S, Hammarström ML, Sundblad G, Arnarp J, and Lönngren J (1982). Mitogenic leukoagglutinin from Phaseolus vulgaris binds to a pentasaccharide unit in N-acetyllactosamine-type glycoprotein glycans. Proceedings of the National Academy of Sciences 79, 1611–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima S, Manabe S, and Ito Y (2005). Divergent Synthesis of Sialylated Glycan Chains: Combined Use of Polymer Support, Resin Capture–Release, and Chemoenzymatic Strategies. Angewandte Chemie International Edition 44, 4218–4224. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J, Hashidate T, Arata Y, Nishi N, Nakamura T, Hirashima M, Urashima T, Oka T, Futai M, Muller WEG, et al. (2002). Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochimica et Biophysica Acta (BBA) - General Subjects 1572, 232–254. [DOI] [PubMed] [Google Scholar]
- Iglesias JL, Lis H, and Sharon N (1982). Purification and properties of a D-galactose/N-acetyl-D-galactosamine-specific lectin from Erythrina cristagalli. European journal of biochemistry / FEBS 123, 247–252. [DOI] [PubMed] [Google Scholar]
- Iyer PN, Wilkinson KD, and Goldstein LJ (1976). An -N-acetyl-D-glycosamine binding lectin from Bandeiraea simplicifolia seeds. Arch Biochem Biophys 177, 330–333. [DOI] [PubMed] [Google Scholar]
- Kilbourne ED (1969). Future influenza vaccines and the use of genetic recombinants. Bulletin of the World Health Organization 41, 643–645. [PMC free article] [PubMed] [Google Scholar]
- Kochibe N, and Furukawa K (1980). Purification and properties of a novel fucose-specific hemagglutinin of Aleuria aurantia. Biochemistry 19, 2841–2846. [DOI] [PubMed] [Google Scholar]
- Kornfeld K, Reitman ML, and Kornfeld R (1981). The carbohydrate-binding specificity of pea and lentil lectins. Fucose is an important determinant. J Biol Chem 256, 6633–6640. [PubMed] [Google Scholar]
- Kornfeld R, and Ferris C (1975). Interaction of immunoglobulin glycopeptides with concanavalin A. Journal of Biological Chemistry 250, 2614–2619. [PubMed] [Google Scholar]
- Li D, Mallory T, and Satomura S (2001). AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clinica Chimica Acta 313, 15–19. [DOI] [PubMed] [Google Scholar]
- Li L, Liu Y, Ma C, Qu J, Calderon AD, Wu B, Wei N, Wang X, Guo Y, Xiao Z, et al. (2015). Efficient chemoenzymatic synthesis of an N-glycan isomer library. Chemical Science 6, 5652–5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N.C. M, Ryan M, E.D. C, Rui X, Janani R, Wenjie P, Nahid R, Michel G, Warren W, W.I. A, et al. (2012). Recognition of Sialylated Poly‐N‐acetyllactosamine Chains on N‐ and O‐Linked Glycans by Human and Avian Influenza A Virus Hemagglutinins. Angewandte Chemie International Edition 51, 4860–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley MS, Crocker PR, and Paulson JC (2014). Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol 14, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, and Osawa T (1969). Purification and characterization of an anti-H(O) phytohemagglutinin of Ulex europeus. Biochim Biophys Acta 194, 180–189. [DOI] [PubMed] [Google Scholar]
- McBride R, Paulson JC, and de Vries RP (2016). A Miniaturized Glycan Microarray Assay for Assessing Avidity and Specificity of Influenza A Virus Hemagglutinins. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D, et al. (2017). Expression system for structural and functional studies of human glycosylation enzymes. Nature Chemical Biology 14, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Miyoshi E, Yakushijin T, Hiramatsu N, Igura T, Hayashi N, Taniguchi N, and Kondo A (2008). Glycomic Analysis of Alpha-Fetoprotein L3 in Hepatoma Cell Lines and Hepatocellular Carcinoma Patients. Journal of Proteome Research 7, 2222–2233. [DOI] [PubMed] [Google Scholar]
- Noll AJ, Gourdine JP, Yu Y, Lasanajak Y, Smith DF, and Cummings RD (2016). Galectins are Human Milk Glycan Receptors. Glycobiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk W, Wilson IA, et al. (2012). Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem Int Ed Engl 51, 4860–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OGATA S. i., MURAMATSU T, and KOBATA A (1975). Fractionation of Glycopeptides by Affinity Column Chromatography on Concanavalin A-Sepharose. Journal of Biochemistry 78, 687–696. [DOI] [PubMed] [Google Scholar]
- Padler-Karavani V, Song X, Yu H, Hurtado-Ziola N, Huang S, Muthana S, Chokhawala HA, Cheng J, Verhagen A, Langereis MA, et al. (2012). Cross-comparison of Protein Recognition of Sialic Acid Diversity on Two Novel Sialoglycan Microarrays. Journal of Biological Chemistry 287, 22593–22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik SK, Potvin B, Carlsson S, Sturm D, Leffler H, and Stanley P (2006). Complex N-glycans are the major ligands for galectin-1, -3, and -8 on Chinese hamster ovary cells. Glycobiology 16, 305–317. [DOI] [PubMed] [Google Scholar]
- Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ, et al. (2017). Recent H3N2 Viruses Have Evolved Specificity for Extended, Branched Human-type Receptors, Conferring Potential for Increased Avidity. Cell Host Microbe 21, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, and Paulson JC (2017). CD22 Ligands on a Natural N-Glycan Scaffold Efficiently Deliver Toxins to B-Lymphoma Cells. Journal of the American Chemical Society 139, 12450–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho SS, and Reis CA (2015). Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 15, 540–555. [DOI] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, and Wiley DC (1983). Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304, 76–78. [DOI] [PubMed] [Google Scholar]
- Seko A, Koketsu M, Nishizono M, Enoki Y, Ibrahim HR, Juneja LR, Kim M, and Yamamoto T (1997). Occurrence of a sialylglycopeptide and free sialylglycans in hen’s egg yolk. Biochimica et Biophysica Acta (BBA) - General Subjects 1335, 23–32. [DOI] [PubMed] [Google Scholar]
- Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, and Peumans WJ (1987). The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2–6)Gal/GalNAc sequence. Journal of Biological Chemistry 262, 1596–1601. [PubMed] [Google Scholar]
- Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, and Peumans WJ (1987). Fractionation of sialylated oligosaccharides, glycopeptides, and glycoproteins on immobilized elderberry (Sambucus nigra L.) bark lectin. Archives of Biochemistry and Biophysics 254, 1–8. [DOI] [PubMed] [Google Scholar]
- Shibuya N, Goldstein IJ, Van Damme EJ, and Peumans WJ (1988). Binding properties of a mannose-specific lectin from the snowdrop (Galanthus nivalis) bulb. J Biol Chem 263, 728–734. [PubMed] [Google Scholar]
- Shivatare SS, Chang S-H, Tsai T-I, Ren C-T, Chuang H-Y, Hsu L, Lin C-W, Li S-T, Wu C-Y, and Wong C-H (2013). Efficient Convergent Synthesis of Bi-, Tri-, and Tetra-antennary Complex Type N-Glycans and Their HIV-1 Antigenicity. Journal of the American Chemical Society 135, 15382–15391. [DOI] [PubMed] [Google Scholar]
- Shivatare SS, Chang S-H, Tsai T-I, Tseng SY, Shivatare VS, Lin Y-S, Cheng Y-Y, Ren C-T, Lee C-CD, Pawar S, et al. (2016). Modular synthesis of N-glycans and arrays for the hetero-ligand binding analysis of HIV antibodies. Nature Chemistry 8, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, and Cummings RD (2014). Investigating virus-glycan interactions using glycan microarrays. Curr Opin Virol 7, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Ju H, Lasanajak Y, Kudelka MR, Smith DF, and Cummings RD (2016). Oxidative release of natural glycans for functional glycomics. Nat Meth 13, 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lasanajak Y, Xia B, Smith DF, and Cummings RD (2009). Fluorescent Glycosylamides Produced by Microscale Derivatization of Free Glycans for Natural Glycan Microarrays. ACS Chemical Biology 4, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, and Cummings RD (2009). Novel Fluorescent Glycan Microarray Strategy Reveals Ligands for Galectins. Chemistry & Biology 16, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen E, Ralf S, Gislinde G, Joachim S, and Carlo U (2007). Synthesis of Pentaantennary N‐Glycans with Bisecting GlcNAc and Core Fucose. Angewandte Chemie International Edition 46, 4173–4175. [DOI] [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, and Cummings RD (2008). Galectin-1, -2, and -3 Exhibit Differential Recognition of Sialylated Glycans and Blood Group Antigens. Journal of Biological Chemistry 283, 10109–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Bao W, Tian X, Li M, Liu H, Dong J, and Huang W (2014). A simplified procedure for gram-scale production of sialylglycopeptide (SGP) from egg yolks and subsequent semi-synthesis of Man3GlcNAc oxazoline. Carbohydrate Research 396, 62–69. [DOI] [PubMed] [Google Scholar]
- Sun B, Srinivasan B, and Huang X (2008). Pre-Activation-Based One-Pot Synthesis of an α-(2,3)-Sialylated Core-Fucosylated Complex Type Bi-Antennary N-Glycan Dodecasaccharide. Chemistry – A European Journal 14, 7072–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketa K, Endo Y, Sekiya C, Tanikawa K, Koji T, Taga H, Satomura S, Matsuura S, Kawai T, and Hirai H (1993). A Collaborative Study for the Evaluation of Lectin-Reactive α-Fetoproteins in Early Detection of Hepatocellular Carcinoma. Cancer research 53, 5419–5423. [PubMed] [Google Scholar]
- Taniguchi N, and Kizuka Y (2015). Chapter Two - Glycans and Cancer: Role of N-Glycans in Cancer Biomarker, Progression and Metastasis, and Therapeutics In Advances in cancer research, Drake RR, and Ball LE, eds. (Academic Press; ), pp. 11–51. [DOI] [PubMed] [Google Scholar]
- Tateno H, Nakamura-Tsuruta S, and Hirabayashi J (2009). Comparative analysis of core-fucose-binding lectins from Lens culinaris and Pisum sativum using frontal affinity chromatography. Glycobiology 19, 527–536. [DOI] [PubMed] [Google Scholar]
- Tessier MB, Grant OC, Heimburg-Molinaro J, Smith D, Jadey S, Gulick AM, Glushka J, Deutscher SL, Rittenhouse-Olson K, and Woods RJ (2013). Computational Screening of the Human TF-Glycome Provides a Structural Definition for the Specificity of Anti-Tumor Antibody JAA-F11. PLOS ONE 8, e54874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unverzagt C (1996). Chemoenzymatic Synthesis of a Sialylated Undecasaccharide–Asparagine Conjugate. Angewandte Chemie International Edition in English 35, 2350–2353. [Google Scholar]
- van Wauwe JP, Loontiens FG, and de Bruyne CK (1975). Carbohydrate binding specificity of the lectin from the PEA (Pisum sativum). Biochimica et Biophysica Acta (BBA) - Protein Structure 379, 456–461. [DOI] [PubMed] [Google Scholar]
- Varki A, Schnaar RL, and Crocker PR (2017). I-Type Lectins In Essentials of Glycobiology, rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, et al. , eds. (Cold Spring Harbor (NY)), pp. 453–467. [Google Scholar]
- Walczak MA, and Danishefsky SJ (2012). Solving the Convergence Problem in the Synthesis of Triantennary N-Glycan Relevant to Prostate-Specific Membrane Antigen (PSMA). Journal of the American Chemical Society 134, 16430–16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WC, and Cummings RD (1988). The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked alpha-2,3 to penultimate galactose residues. Journal of Biological Chemistry 263, 4576–4585. [PubMed] [Google Scholar]
- Wang Z, Chinoy ZS, Ambre SG, Peng W, McBride R, de Vries RP, Glushka J, Paulson JC, and Boons G-J (2013). A General Strategy for the Chemoenzymatic Synthesis of Asymmetrically Branched N-Glycans. Science 341, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liu Y, Ma C, Li L, Bai J, Byrd-Leotis L, Lasanajak Y, Guo Y, Wen L, Zhu H, et al. (2016). Identification of the binding roles of terminal and internal glycan epitopes using enzymatically synthesized N-glycans containing tandem epitopes. Organic & Biomolecular Chemistry 14, 11106–11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, McCauley JW, and Steinhauer DA (2014). Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr Top Microbiol Immunol 385, 63–91. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Kochibe N, Ohkura T, Ueda I, and Kobata A (1985). Fractionation of L-fucose-containing oligosaccharides on immobilized Aleuria aurantia lectin. J Biol Chem 260, 4688–4693. [PubMed] [Google Scholar]
- Yang Q, Zhang R, Cai H, and Wang L-X (2017). Revisiting the substrate specificity of mammalian α1,6-fucosyltransferase reveals that it catalyzes core fucosylation of N-glycans lacking α1,3-arm GlcNAc. Journal of Biological Chemistry 292, 14796–14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L-G, Andrews N, Zhao Q, McKean D, Williams JF, Connor LJ, Gerasimenko OV, Hilkens J, Hirabayashi J, Kasai K, et al. (2007). Galectin-3 Interaction with Thomsen-Friedenreich Disaccharide on Cancer-associated MUC1 Causes Increased Cancer Cell Endothelial Adhesion. Journal of Biological Chemistry 282, 773–781. [DOI] [PubMed] [Google Scholar]
- Zou Y, Wu Z, Chen L, Liu X, Gu G, Xue M, Wang PG, and Chen M (2012). An Efficient Approach for Large-Scale Production of Sialyglycopeptides from Egg Yolks. Journal of Carbohydrate Chemistry 31, 436–446. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the N-glycan microarray datasets related to this work are shown in the Supplemental Information and are publicly available at the National Center for Functional Glycomics (NCFG) website: https://ncfg.hms.harvard.edu/ncfg-data