Abstract
TP53 mutations (TP53mut) in AML patients associate with poor prognosis that may affect therapy and outcome. In addition to TP53 mut patients, TCGA AML patient sequencing data shows that there are around 3% of patients have detectable low frequency TP53mut reads. Importantly, these patients showed worse outcome as compared to the TP53 wild type (TP53wt) patients. We have studied the effect of low frequency TP53mut in two AML cell lines, OCI-AML2 and MV4–11. Both cells have low frequency single hotspot TP53mut. Interestingly, the resistant cells derived from both lines have homogeneous TP53mut. TP53mut clones isolated from the parental cells also show increased chemoresistance potential and have higher population of leukemia stem cell (LSC) maker positive cells, a characteristic of chemoresistant cells. When mixed with TP53wt cells, the TP53mut cells show survival advantage suggesting its potential to develop chemoresistance. We previously showed that histone deacetylase inhibitor Romidepsin can re-sensitize chemoresistant cells by eradicating LSC marker positive cells. Here we further show that Romidepsin can reactivate p53 targeted genes which are dysregulated in TP53mut cells and preferentially targets TP53mut subpopulation. Therefore, our study shows that low frequency TP53mut is linked to chemoresistance and shed lights on therapeutic strategies for treatments on chemoresistance.
Keywords: TP53 mutation, chemoresistance, AML
Introduction
Chemoresistance is a major burden for the treatment of many cancers, including acute myeloid leukemia (AML). Overall survival of AML patients, despite intensive treatment with high-dose chemotherapy, stem cell transplantation, and radiation therapy, remains poor with the five-year overall survival less than 30%. The major burden for therapy is chemoresistance. AML is a clonal disease and chemoresistant cells may either evolved from the expansion of a subclone of the primary cells or from the clone gained additional mutations during therapy1. The mechanism underlying resistance remains to be elucidate.
Tumor suppressor p53 is the first in line guardian of the genome in cells. In response to cellular stress, such as DNA damage, UV irradiation or serum deprivation, p53 is stabilized and promotes cell-cycle arrest, apoptosis, or other anti-proliferative programs2, 3. p53 is a transcription factor and numerous p53-responsive target genes, such as CDKN1A, GADD45, MDM2, and BAX, contain the p53 consensus sequence in their respective promoters4–8. TP53 is the most frequently mutated gene in human cancers. Most TP53mut occur in the DNA binding domain and disrupt its transcriptional activity for its wild-type target genes, therefore preventing stress responses and enabling aberrant proliferation and survival of TP53 mutated (TP53mut) cells. The mutant p53 protein is often stabilized due to reduced interaction with negative regulators and degradation9, 10. In addition, these mutant proteins display gain-of-function (GOF) activities beyond TP53 loss11, 12. Growing evidences show p53 GOF can promote stemness of the cells13, 14, enhancing cell self-renewal15, promoting cancer stem cell related gene expression in variety of cancers13, 16, which leading to dramatic promotion of invasion, metastasis and chemoresistance through activation of these genes for survival and multidrug resistance10, 12, 17
TP53 mutation occurs in about 30% of therapy related AML/MDS (t-AML/MDS) and less than 10% of de novo AML patients and are strongly associates with the resistance to chemotherapy and shorten survival 1, 18–21. In this study, we found that patients with low frequency TP53mut also show poor prognoses. We therefore studied the effect of low frequency TP53mut in the development of chemoresistance. Two cell lines from AML patients were analyzed and found with low frequency TP53mut. We show TP53mut subpopulation of cells exhibits increased drug resistance and the population possessing LSC properties expands while cells gaining chemoresistance. Our results suggest that low frequency mutation is an important factor promoting the development of chemoresistance.
Material and method
Cell lines
OCI-AML2, OCI-AML3, KG-1, MV4–11, THP-1 and Molm-13 were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen and American type Culture Collection repositories and cultured under their instructions. Chemo-resistant cell lines were generated as described previously22. All cell lines were verified by short tandem repeat (STR) analysis and tested for mycoplasma contamination by ICBR sequencing core at University of Florida.
Chemicals and antibodies
Chemicals and antibodies used for this study are listed in Supplementary Table 1
Cell viability
Cell viabilities were tested as previously described22. Briefly, 1×104 cells/well were seeded into either vehicle or drug containing culture medium for 72h. The viable cells were tested with MTS assay kit (G5430, Promega). The absorbance was measured at 490nm wavelength. Cell viability was calculated by the comparison of the absorbance reading obtained from treated versus control cells after subtraction of the background.
RNA-sequencing analysis
RNA libraries were prepared using the TruSeq RNA sample prep kit (Illumina, San Diego) and sequenced using Illumina HiSeq 2000 Sequencer (Illumina). RNA-seq data were processed using Tophat-Cufflinks pipeline23. Gene set enrichment analysis was performed with GSEA software 24. Variants detection and TP53 mutation sequencing data visualization was carried out by Partek flow.
Interested genes were confirmed by Realtime RT-PCR. Primers are listed in Supplementary Table 2.
Raw data and normalized gene expression data are deposited in the Gene Expression Omnibus database under accession number GSE108142 and GSE114649 as previously described 22.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed as described previously 25, 26. Briefly, 5x106 cells/sample were cross-linked by 1% formaldehyde and quenched with 0.125M glycine. Cells were sonicated to obtain approximately 300bp-500bp chromatin fragments. And subsequently immunoprecipitated with indicated antibodies or lgG as control. The purified DNA from precipitated chromatin was subjected to qPCR amplification. The primers for ChIP are listed in Supplementary Table 2.
Mouse xenograft studies
Xenograft studies were performed following the protocol approved by the Institutional Animal Care and Use Committee of the University of Florida (IACUC #201909309). MV4–11 single cell clone with TP53mut were first infected by lentivirus produced from pMSCV-GFP plasmid, then mixed with MV4–11 parental cells. Then, 1×106 mixture cells were injected intravenously into tail vein of 8 to 10-week-old NSG mice (The Jackson Laboratory, 005557). Mice were treated by either 1.2 mg/kg Ara-C or vehicle from day 0 to day 9 and euthanized on day 42.
Statistical analysis
Student’s t-test was used to analyze data from gene expression, cell counting, cell viability and in vivo xenograft experiments. Values of P< 0.05 were considered statistically significant. Pearson correlation coefficient analysis was carried out by GraphPad Prism 6.
Results
p21 gene expression and protein level were decreased in chemo-resistant cells
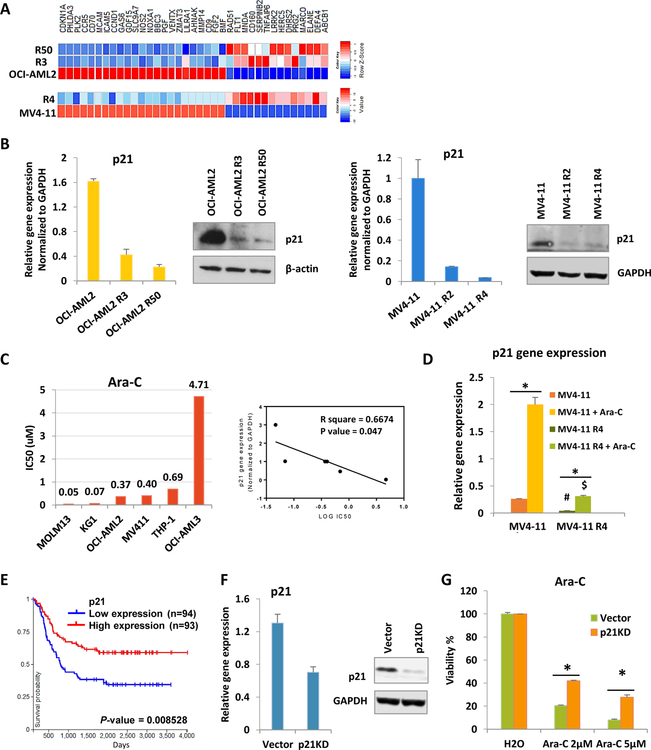
In order to study chemoresistance mechanism, Cytarabine (Ara-C) resistant cell lines were generated from parental AML cells, OCI-AML2 and MV4–11(Fig. S1A, B). RNA-seq analysis was performed to examine global changes in gene expressions in chemoresistant cells. Compared to parental cells, one of highly changed genes in resistant cells is CDKN1A (Fig. 1a). RT-PCR and western blot further confirmed that both mRNA and protein level of p21 were reduced in resistant cells (Fig. 1b). Then we investigated whether the decrease of p21 directly links to drug sensitivity. First, we examined the Ara-C IC50 and their p21 expression across 6 human AML cell lines. p21 expression is negatively correlated with drug sensitivity (Fig. 1c). Interestingly, Ara-C treatment failed to induce p21 expression in resistant cells to the level as wild type cells (Fig. 1d). Cell cycle analysis shows cell cycle arrests at G0/G1 stage after Ara-C treatment in sensitive cells but not in resistant cells (Fig. S1C). The patient survival data from TCGA also shows that a lower level of p21 is strongly correlated with poor survival (Fig. 1e). Further, the knocked down of p21 gene expression in OCI-AML2 cells increases drug resistance (Fig. 1f, 1g). Consistently, p21 knock down cells fail to trigger cell cycle arrest upon Ara-C treatment (Fig. S1C). Together, resistant cells have lower p21 expression and Ara-C fails to activate p21 in these cells to trigger cell cycle arrest eventually leading to cell death. Therefore decreased p21 expression in AML chemoresistant cells is linked to Ara-C drug resistance.
Figure 1.
Lower p21 expressions were correlated with drug resistance.
(A) Heatmap shows gene expressions of overlapped genes with similar fold change pattern from both resistant cells compared with their parental cells. R3, R50 and R4 represent for cells that can resistant to 3µM, 50 µM and 4 µM Ara-C. (B) p21 gene expressions and protein levels were tested in OCI-AML2, MV4–11 cells and their resistant lines. (C) Ara-C IC50 were tested across AML cell lines (Left). Pearson correlation coefficient of p21 gene expressions and Ara-C IC50. (D) MV4–11 and resistant cells were treated with either vehicle or 1µM Ara-C for 24h. #P<0.05 compared with MV4–11 group. $P<0.05 compared with MV4–11 treated with Ara-C group. (E) Kaplan plot for p21 gene expression in AML patients. (F) p21 knockdown efficiency in OCI-AML2 cells. (G) MTS assay after 72h drug treatment with indicated concentrations. Bars indicate the standard error of mean. *P < 0.05
TP53mut detected in both resistant cell lines was originated from a small subpopulation of parental cells
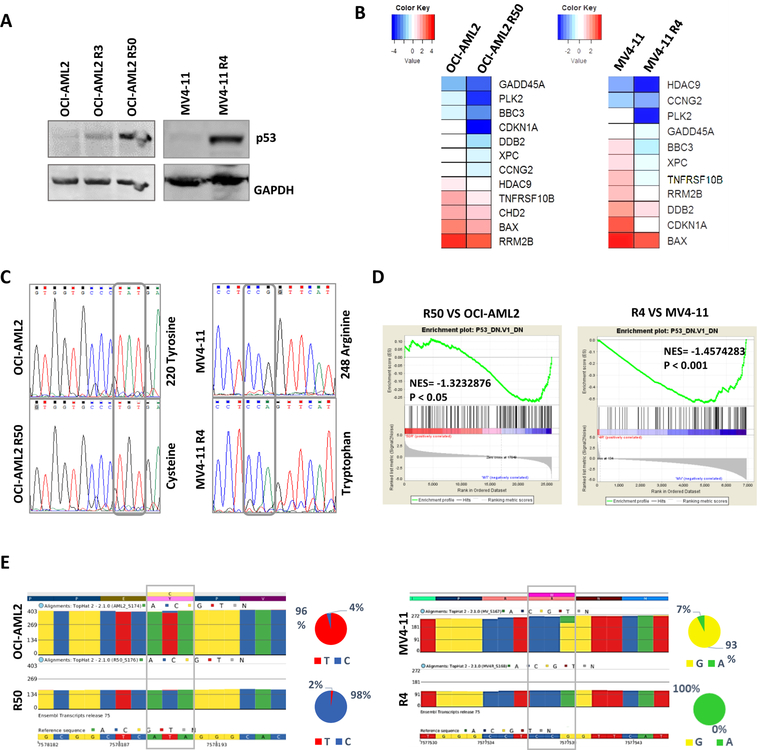
p53 is one of the major regulator of CDKN1A gene27, 28. Both AML cells, OCI-AML2 and MV4–11, are characterized as TP53 wild type (TP53wt) cells29. We therefore tested if p53 protein level is altered in chemoresistant cells. The result shows there is in fact, an increase of protein level of p53 in drug resistant cells, instead of decrease (Fig. 2a). In addition, RNA-seq results show gene expressions of a panel of p53 target genes were decreased in resistant cells (Fig. 2b). Giving the fact that p53 protein is expressed but p53 target gene is not activated, we suspect that TP53 gene is mutated in the resistant cells. Sanger sequencing results confirmed that both resistant cell lines have TP53mut, OCI-AML2 R50 has Y220C mutation and MV4–11 R4 has R248W mutation (Fig. 2c). Both mutations are hotspot mutations for TP53 10, 30. We then performed gene set enrichment assay on gene expression profiles of both chemo-resistant cell lines compared to their parental cells. The result shows that in both resistant cells, changed genes enriched in genes down-regulated in the NCI-60 with mutated TP53 gene set (Fig. 2d). Furthermore, patient data from TCGA also shows patients with TP53mut have lower p21 levels (Fig. S2A). Therefore, lower expression of p53 target genes is linked to TP53mut in resistant cells. There are two possible scenarios for TP53mut in resistant cells. One is that mutations are induced by Ara-C treatment, and second is that the mutation is originated from a small population of parental cells which was not detectable by conventional Sanger sequencing. Through analysis of our RNA sequencing data, we found that p53 mutant transcripts exist before chemo-drug treatment at 4% in OCI-AML2 cells and 7% in MV4–11 cells, which expended to nearly 100% after chemotherapy (Fig. 2e). We then turn into clinical data to investigate whether there are population of patients who considered TP53wt are in fact have low frequency TP53mut and whether these patients have poor prognosis compare to wild type TP53 patients. The dataset obtained from TCGA20, 31, 32 has 8.5% of AML patients with TP53 alteration, and the rest of 91.5% of patients have been classified as TP53wt (Figure S2B, Left). However, through the analysis of the sequence data on TP53wt patients, we found that there are over 3% (6 out of 174 patients) of patients has low frequency TP53mut reads under current sequencing depth (Fig. S2B, right panel). The mutation frequency range from 10 to 23 percent, indicating these patients have subclonal cells with TP53mut (Table. S3). We therefore divided all AML patients into three groups, TP53wt, TP53mut and patients with subclonal TP53mut. The Kaplan–Meier curve shows patients with TP53mut have significantly lower survival rate compared with TP53wt group. Similarly, despite lack of survival data on 2 patients, 3 out of 4 patients with subclonal TP53mut also have worse survival outcomes (Fig. S2C).
Figure 2.
AML cells with TP53mut were significantly enriched after chemo drug selection.
(A) p53 protein level in AML and chemoresistant cells. (B) p53 target genes in both resistant cells compared to their parental cells. (C) Sanger sequencing were performed on p53 DNA binding domain. Point mutations were found in resistant cells. (D) GSEA shows genes changed in resistant cells are enriched in p53 depletion regulated gene set. (E) Snapshot showing TP53 sequence reads in indicated cell lines. Top line is protein sequence, bottom is the DNA sequence, bar graph in between is visualized sequencing reads. Percentages of each base at the mutation site in total reads were shown in pie chart.
Cells with TP53mut have survival advantage under chemo-drug selection
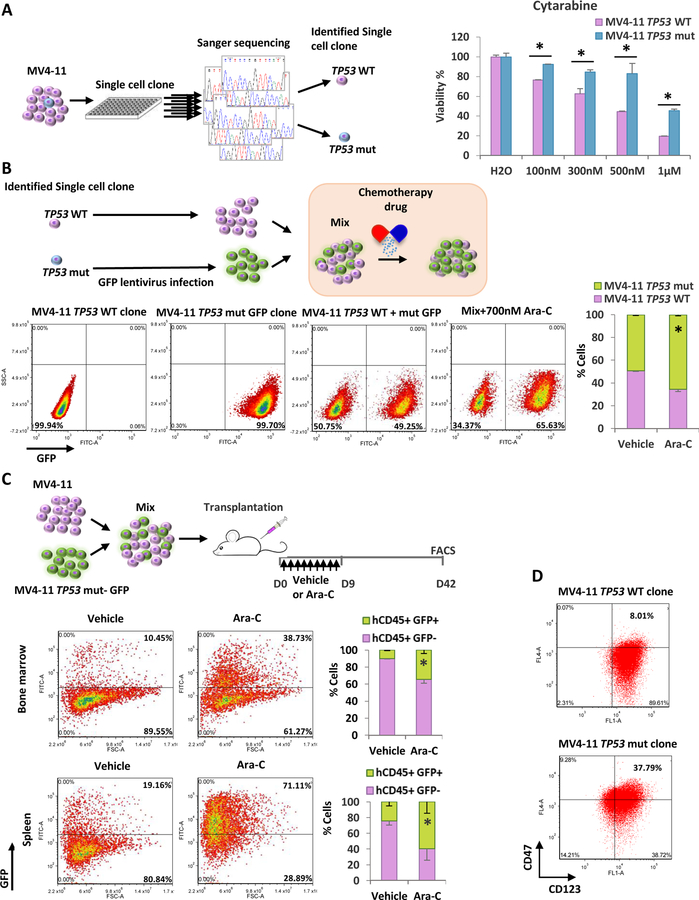
Based on results from our chemoresistant AML cells and patient clinic data, we hypothesize that TP53mut subclone in AML cells has survival advantage and therefore expend after chemotherapy. To test this, we first selected single cell clones which have wild type TP53 or mutation from parental MV4–11 cells (Fig. 3a). The TP53 mutation single clones grow in a similar rate as the parental cells (Fig. S3A), but less sensitive upon drug treatment (Fig. 3a, Fig. S3B). Further, we labeled TP53 mutant cells with GFP, and then mixed with TP53wt cells or MV4–11 parental cells to test whether mutated cells have growth advantage under low dose Ara-C treatment. The results show GFP positive TP53 mutant clone increases cell population dramatically after drug treatment (Fig. 3b, Fig. S3C). Consistent with the in vitro data, when the mixture of GFP TP53 mutant cells and MV4–11 parental cells were transplanted into irradiated NSG mice and subsequently treated with vehicle or Ara-C, the hCD45/GFP positive TP53mut leukemic cell population was significantly expanded in BM and Spleen of recipients upon Ara-C treatment, while percentage of TP53 WT cells significantly decreased over six weeks period (Fig. 3c). Thus, the data suggest that TP53 mutant population undergoes clonal expansion in response to the chemotherapy.
Figure 3.
Cells with TP53mut have survival advantage under drug pressure and have higher leukemia stem cells percentage.
(A) Left: Flow diagram showing the experimental procedures. Right: MTS assay in TP53wt and mutant cells after 72h Ara-C treatment. (B) Top: Flow diagram showing the experimental procedures. Bottom: Single cell clones of MV4–11 TP53wt and TP53mut labeled with GFP were first tested by FACS separately. Then cells were mixed and treated with either vehicle or Ara-C and tested by FACS on day 5. (C) Top: Flow diagram showing the experimental design. Middle and bottom: cells from bone marrow or spleen were gated for hCD45 positive and tested for GFP signal. Results were summarized in the right panel. Bars indicate the standard error of mean. *P < 0.05 (D) Cells were analyzed by FACS for LSC markers.
Our previous study shows that resistant cells have higher percentage of stem-cell surface marker positive cells 22. Since p53 GOF mutant regulates stem cell-associated genes and transcriptional programs5, 33, 34, we further tested whether stem-cell surface marker expression is correlated with TP53mut. The result shows that TP53mut clone has higher stem cell marker positive cells than TP53wt cells (Fig. 3d). Interestingly, the percentage of stem cell maker positive cells in TP53mut clone is similar to the resistant cells which have homogeneous TP53mut (Fig. S3D). Therefore, these results suggest that TP53mut contribute to stem-like phenotype and chemoresistance.
TP53mut results in a reduction of GADD45A and other p53 target genes, and their decrease contribute to drug resistant
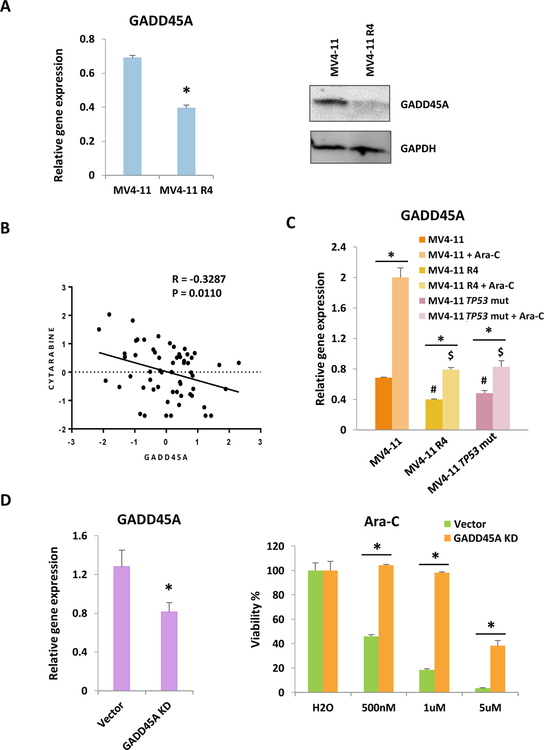
GADD45A (Growth Arrest And DNA Damage Inducible Alpha) is a p53 target gene and many evidence implicated that GADD45A functions as a stress sensor that results in cell cycle arrest, DNA repair or apoptosis 35, 36. However, its role for chemoresistance is not studied. We first examine the gene expression and protein level in chemoresistant cells. GADD45A was decreased in both gene expression and protein level in resistant cells, consistent with the notion that p53 activity is required for GADD45A gene activation (Fig. 4a). The NCI-60 human tumor cell Ara-C drug response data37 shows negative correlation of GADD45A expression level and drug resistance (Fig. 4b). In addition, we tested the correlation of drug sensitivity and GADD45A expression level across six AML cell lines and seen consistent results (Fig. S4A). These results implicate that lower GADD45A level is linked to chemoresistance. Importantly, Ara-C failed to activate GADD45A expression in resistant R4 cells compare to the level elevated in sensitive cells (Fig. 4c). Interestingly, TP53mut clone isolated from parental cells, which showed higher resistance to Ara-C, also has lower level of GADD45A expression and fails to response to Ara-C to levels compare to parental cells (Fig. 4c). Further, the knocked down of GADD45A in OCI-AML2 cells increases drug resistance (Fig. 4d). These data shows that reduction of GADD45A in TP53mut cells contributes to chemoresistance.
Figure 4.
Decrease of p53 target gene GADD45A also contributes to drug resistance.
(A) Gene expression and protein level were tested in chemoresistant and their parental cells. (B) Correlation of GADD45A gene transcript z-scores and Ara-C compound activities (-log values) z-scores in NCI60 cells. (C) MV4–11, MV4–11 R4 and MV4–11 single cell clone with TP53mut were treated with either vehicle or 1µM Ara-C for 24h. #P<0.05 compared with MV4–11 group. $P<0.05 compared with MV4–11 treated with Ara-C group. (D) shRNA knocking down efficiency of GADD45A in OCI-AML2 cells was tested (left). MTS assay was performed to test drug response in control and knockdown cells after 72h drug treatment (right). Bars indicate the standard error of mean. *P < 0.05
We also examined whether Ara-C also failed to activate other p53 target genes in resistant cells and TP53mut parental cell clones. polo-like kinase 2 (PLK2) and BAX protein has been shown are direct targets of p53 and are linked to chemoresistance 38–41. Our study shows that the expression of both genes are reduced in resistant cells as well as TP53mut cells. In addition, the expression of both genes fail to be fully activated by Ara-C in these cells (Fig. S4B). Therefore, mutated p53 failed to activate its target genes which are important for cell death and chemosensitivity.
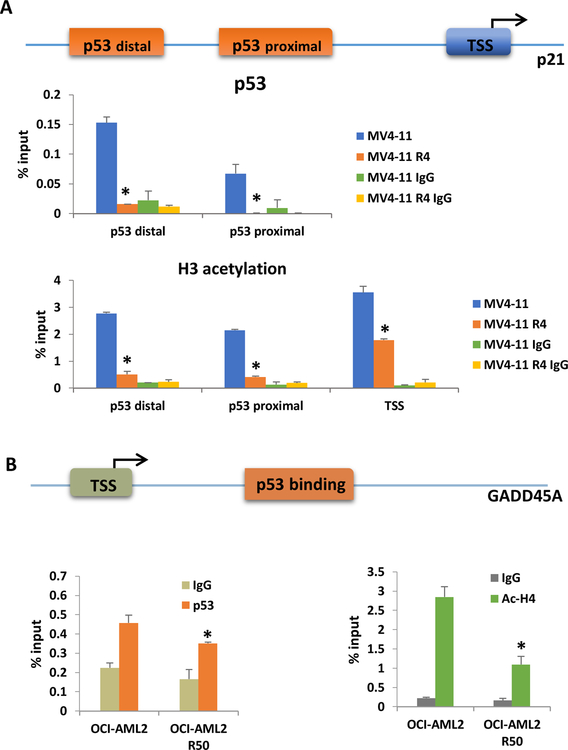
TP53mut causes reduction of p53 recruitment at both p21 and GADD45A promoters leading to a decrease of histone acetylation in chemoresistant cells
As both TP53mut in resistant cell lines are at DNA binding domain, we next want to test if dysregulation of downstream p53 target genes result from the loss of its chromatin recruitment. Chromatin Immunoprecipitation was performed in both chemoresistant cell lines and their parental cell lines. There is a significant reduction of p53 recruitment at p21 promoter regulatory regions (Fig. 5a, Fig. S5) and p53 binding site near GADD45A promoter (Fig. 5b). Consistent with low p53 recruitment, histone acetylation at same regions is also reduced, leading to repression of gene expression (Fig. 5a, b and Fig. S5).
Figure 5.
p53 mutation causes a reduction of p53 recruitment at both p21 and GADD45A promoters and leading to a decrease of histone acetylation at these regions.
(A) Top: Diagram representation of p21 promoter. p53 distal and p53 proximal are two p53 binding site on p21 promoter, TSS represents for transcription start site. Middle and bottom: ChIP for p53 and ac-H3 in MV4–11 and MV4–11 R4 cells at p21 promoter region. (B) Top: Diagram representation of GADD45A promoter. Bottom: ChIP for p53 and ac-H4 at GADD45A promoter region. Bars indicate the standard error of mean. *P<0.05 compared to parental cell group.
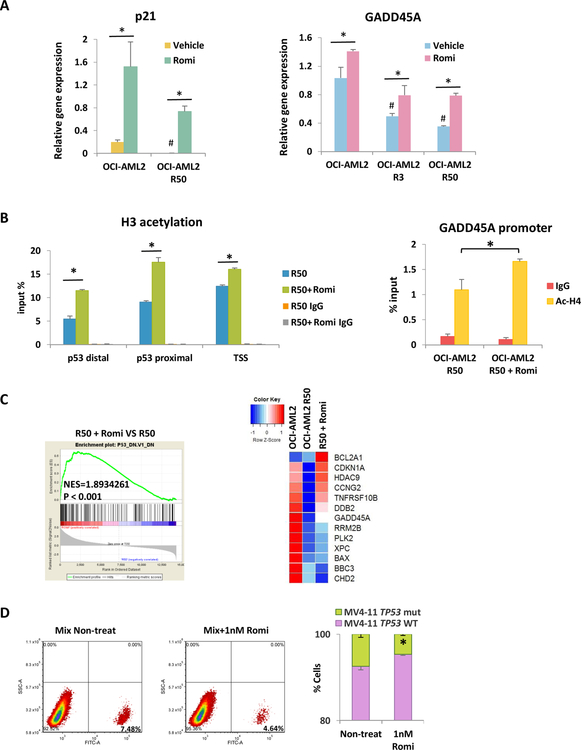
Romidepsin can restore genes dysregulated by TP53mut by increase histone acetylation and preferentially targets TP53mut cells
It has been shown HDAC inhibitors (HDACi) can elevate p21 expression 42, 43. Therefore we are interested to examine whether HDACi can restore p21 gene expression in TP53mut cells. The HDAC1/2 inhibitor Romidepsin treatment activates p21 expression in both parental OCI-AML2 cells and chemoresistant cells (Fig. 6a). Similarly, GADD45A gene expression increased upon Romidepsin treatment (Fig. 6a). We further demonstrate that Romidepsin restores gene expression by increasing histone acetylation at gene promoter regions (Fig. 6b, Fig. S6A). Interestingly, p53 recruitment was not restored by Romidepsin treatment (Fig. S6B, S6C). We then further examined whether Romidepsin can rescue p53 targeted genes in general. Gene set enrichment assay was performed with RNA seq data from resistant cells with or without Romidepsin. Interestingly, Romidepsin activated genes are enriched in genes downregulated by TP53mut in NCI-60 cancer cell data set (Fig. 6c). Heatmap shows majority of representative p53 target genes that are decreased in chemoresistant cells were reactivated by the Romidepsin treatment (Fig. 6c).
Figure 6.
Romidepsin can restore genes dysregulated by p53 mutation by increase histone acetylation and preferentially targets TP53mut cells
(A) p21 and GADD45A gene expressions upon 24h Romidepsin treatment. #P<0.05 compare to parental cells. (B) ChIP for ac-H3 in OCI-AML2 R50 cells with or without Romidepsin treatment at p21 and GADD45A promoter region. (C) GSEA was performed based on RNA-seq data for Romidepsin treatment in OCI-AML2 R50 cells. Heatmap shows expression of p53 target genes in OCI-AML2, OCI-AML2 50R and OCI-AML2 50R Romidepsin treated cells. (D) Single cell clone of MV4–11 TP53wt cells and TP53mut labeled with GFP were mixed and treated with either vehicle or 1nM Romidepsin. Cells were tested by FACS analysis on day 4. Bars indicate the standard error of mean. *P < 0.05
Since Romidepsin can resensitize resistant AML cells in response to chemotherapy 22, and restore the expression of p53 target genes in TP53mut cells, we next wanted to investigate how Romidepsin influences TP53 mutant subpopulation in AML cells. GFP positive TP53mut cells isolated from MV4–11 parental cells were mixed with MV4–11 TP53 WT cells and then treated with low dose Romidepsin for 4 days. The percent of GFP positive cells were significantly reduced (Fig. 6d), indicating Romidepsin preferentially targets TP53mut cells. In contrast, hypomethylating agent decitabine can not target TP53 mut cells as efficient as Romidepsin under similar cell killing rate (Fig. S7). Therefore the Romidepsin treatment potentially can suppress the clonal expansion of TP53mut cells and prevent or slowdown the development of chemoresistance during chemotherapy.
Discussion
In this study, we investigated genes that are important for AML chemoresistance. We found the expression of two p53 target genes, such as CDKN1A, GADD45A, are decreased in both resistant AML cell lines which are created from two parental sensitive cell lines, OCI-AML2 and MV4–11. p21 and GADD45A expression level directly affects drug sensitivity of AML cells. Further, RNA-seq result reveals that in fact, most of p53 targeted genes are repressed in resistant cells. These results prompted us to investigate the role of p53 in drug resistance in these cells. Since these two cell lines were characterized as TP53wt 29, we first considered whether p53 protein level is reduced in resistant cells. To our surprise, p53 protein level is not reduced, instead, is increased. Since TP53mut often cause increase of p53 protein level due to reduction of interaction with negative regulators 10, 11, we look into whether p53 is mutated in resistant cells. The Sanger sequencing results show that although parental cell lines are TP53wt, both resistant cells derived from the parental lines have point hotspot mutation at p53 DNA binding domain. We then investigated whether the mutation is originated from a subclone of parental cells. The RNA-seq result shows that both parental cells have subclonal TP53mut. Therefore the result indicates that the chemotherapy does not directly induce TP53 mutations, instead, subclonal TP53mut AML cells have better fitness under chemotherapy and preferentially expended after chemotherapy.
TP53mut occurs in less than 10% of de novo AML but patients with TP53mut exhibit lower complete remission rate, higher relapse rate and lower survival rate1, 44. We found six patients with low frequency TP53mut from TCGA database and the variant allele frequency ranging from 23% to 10%. Despite lack of survival data on 2 patients, 3 out of 4 patients with subclonal TP53mut have worse prognosis compared to wild type p53 patients. Similar results also seen in other studies. It is shown that there’s no significant difference of relapse rate and survival rate between high frequency mutation and subclonal mutations 44. Therefore, subclonal TP53mut will also be a predictor of therapy response and patient prognosis. Current targeted sequencing depth may not be sensitive enough for low frequency mutation detection. Future improved ultra-deep sequencing may be needed for subclonal TP53mut detection.
Since the collection of patient sample pairs at diagnose and relapse with low frequency TP53 mutation is not available to us, we isolated subclonal cells from patient derived cell lines and studied the chemoresistance of the mutant clones and also we showed that mutation clone has better fitness to sustain chemotherapy and become populated after chemotherapy. Therefore, these isolated p53 mutant clones from parental cells are good study model for studying how p53 mutation results in clonal expansion which lead to relapse and drug resistant disease.
Our previous study shows that histone deacetylase inhibitor Romidepsin can target stem cell marker positive cells and resensitize cells to chemotherapy. In this study, we show that TP53mut cells have higher stem cell marker positive cells, therefore Romidepsin may preferentially targets TP53mut cells. We further demonstrate that Romidepsin reactivated p53 targeted genes. The activation is not through the rescue of p53 binding, instead, it is through the increase of promoter acetylation on histone H3 and H4. These results provide further insight that reactivating p53 targeted genes may have therapeutic potential for TP53mut AML patients.
Supplementary Material
Acknowledgements
We thank Xutao Guo for excellent technical support and thoughtful discussions. We thank Suming Huang for the critical review of the manuscript. This research was supported by NIH/NHLBI grant HL144712 to YQ.
Footnotes
Conflict of Interest
All authors declare no conflict of interest.
References
- 1.Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015. February 26; 518(7540): 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soussi T The p53 tumor suppressor gene: from molecular biology to clinical investigation. Annals of the New York Academy of Sciences 2000. June; 910: 121–137; discussion 137–129. [DOI] [PubMed] [Google Scholar]
- 3.May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 1999. December 13; 18(53): 7621–7636. [DOI] [PubMed] [Google Scholar]
- 4.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 1994. June 16; 369(6481): 574–578. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen KT, Liu B, Ueda K, Gottesman MM, Pastan I, Chin KV. Transactivation of the human multidrug resistance (MDR1) gene promoter by p53 mutants. Oncol Res 1994; 6(2): 71–77. [PubMed] [Google Scholar]
- 6.Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, et al. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 1994. June; 9(6): 1799–1805. [PubMed] [Google Scholar]
- 7.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 1992. November 13; 71(4): 587–597. [DOI] [PubMed] [Google Scholar]
- 8.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet 1992. April; 1(1): 45–49. [DOI] [PubMed] [Google Scholar]
- 9.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nature reviews Cancer 2009. October; 9(10): 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue X, Zhao Y, Xu Y, Zheng M, Feng Z, Hu W. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J Mol Biol 2017. June 2; 429(11): 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014. March 17; 25(3): 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ 2019. January; 26(2): 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shetzer Y, Molchadsky A, Rotter V. Oncogenic Mutant p53 Gain of Function Nourishes the Vicious Cycle of Tumor Development and Cancer Stem-Cell Formation. Cold Spring Harb Perspect Med 2016. October; 6(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koifman G, Shetzer Y, Eizenberger S, Solomon H, Rotkopf R, Molchadsky A, et al. A Mutant p53-Dependent Embryonic Stem Cell Gene Signature Is Associated with Augmented Tumorigenesis of Stem Cells. Cancer Res 2018. October; 78(20): 5833–5847. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Yu H, Kobayashi M, Gao R, Boswell H, Liu Y. Gain-of-Function Mutant p53 Enhances Hematopoietic Stem Cell Self-Renewal. Blood 2014 December 6 2014; 124(21). [Google Scholar]
- 16.Solomon H, Dinowitz N, Pateras IS, Cooks T, Shetzer Y, Molchadsky A, et al. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene 2018. 03; 37(12): 1669–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush JA, Li G. Cancer chemoresistance: the relationship between p53 and multidrug transporters. International journal of cancer 2002. March 20; 98(3): 323–330. [DOI] [PubMed] [Google Scholar]
- 18.Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood 1994. November 1; 84(9): 3148–3157. [PubMed] [Google Scholar]
- 19.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood 2017. August 10; 130(6): 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013. May 30; 368(22): 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015. February 26; 125(9): 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan B, Chen Q, Shimada K, Tang M, Li H, Gurumurthy A, et al. Histone deacetylase inhibitor targets CD123/CD47-positive cells and reverse chemoresistance phenotype in acute myeloid leukemia. Leukemia 2019. April; 33(4): 931–944. [DOI] [PubMed] [Google Scholar]
- 23.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012. March 1; 7(3): 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005. October 25; 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jian W, Yan B, Huang S, Qiu Y. Histone deacetylase 1 activates PU.1 gene transcription through regulating TAF9 deacetylation and transcription factor IID assembly. Faseb J 2017. September; 31(9): 4104–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Yan B, Liao D, Huang S, Qiu Y. Acetylation of HDAC1 and degradation of SIRT1 form a positive feedback loop to regulate p53 acetylation during heat-shock stress. Cell Death Dis 2015; 6: e1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorospe M, Wang X, Holbrook NJ. Functional role of p21 during the cellular response to stress. Gene Expr 1999; 7(4–6): 377–385. [PMC free article] [PubMed] [Google Scholar]
- 28.el-Deiry WS, Harper JW, O’Connor PM, Velculescu VE, Canman CE, Jackman J, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res 1994. March 1; 54(5): 1169–1174. [PubMed] [Google Scholar]
- 29.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic acids research 2019. January 8; 47(D1): D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer research 2000. December 15; 60(24): 6788–6793. [PubMed] [Google Scholar]
- 31.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012. May; 2(5): 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013. April 2; 6(269): pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfister NT, Prives C. Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of p53. Cold Spring Harb Perspect Med 2017. February 1; 7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loizou E, Banito A, Livshits G, Ho YJ, Koche RP, Sanchez-Rivera FJ, et al. A Gain-of-Function p53-Mutant Oncogene Promotes Cell Fate Plasticity and Myeloid Leukemia through the Pluripotency Factor FOXH1. Cancer Discov 2019. May 8. [DOI] [PMC free article] [PubMed]
- 35.Salvador JM, Brown-Clay JD, Fornace AJ Jr. Gadd45 in stress signaling, cell cycle control, and apoptosis. Advances in experimental medicine and biology 2013; 793: 1–19. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman B, Liebermann DA. Gadd45 in modulation of solid tumors and leukemia. Advances in experimental medicine and biology 2013; 793: 21–33. [DOI] [PubMed] [Google Scholar]
- 37.Wang S, Gribskov M, Hazbun TR, Pascuzzi PE. CellMiner Companion: an interactive web application to explore CellMiner NCI-60 data. Bioinformatics 2016. August 1; 32(15): 2399–2401. [DOI] [PubMed] [Google Scholar]
- 38.Valenti F, Fausti F, Biagioni F, Shay T, Fontemaggi G, Domany E, et al. Mutant p53 oncogenic functions are sustained by Plk2 kinase through an autoregulatory feedback loop. Cell cycle 2011. December 15; 10(24): 4330–4340. [DOI] [PubMed] [Google Scholar]
- 39.te Raa GD, Malcikova J, Pospisilova S, Trbusek M, Mraz M, Garff-Tavernier ML, et al. Overview of available p53 function tests in relation to TP53 and ATM gene alterations and chemoresistance in chronic lymphocytic leukemia. Leukemia & lymphoma 2013. August; 54(8): 1849–1853. [DOI] [PubMed] [Google Scholar]
- 40.Weller M Predicting response to cancer chemotherapy: the role of p53. Cell Tissue Res 1998. June; 292(3): 435–445. [DOI] [PubMed] [Google Scholar]
- 41.de Viron E, Knoops L, Connerotte T, Smal C, Michaux L, Saussoy P, et al. Impaired up-regulation of polo-like kinase 2 in B-cell chronic lymphocytic leukaemia lymphocytes resistant to fludarabine and 2-chlorodeoxyadenosine: a potential marker of defective damage response. Br J Haematol 2009. December; 147(5): 641–652. [DOI] [PubMed] [Google Scholar]
- 42.Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J, et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol 2010. March; 30(5): 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A 2000. August 29; 97(18): 10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prochazka KT, Pregartner G, Rucker FG, Heitzer E, Pabst G, Wolfler A, et al. Clinical implications of subclonal TP53 mutations in acute myeloid leukemia. Haematologica 2019. March; 104(3): 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.