Abstract
Long non-coding RNAs (lncRNAs) are a largely uncharacterized group of non-coding RNAs with diverse regulatory roles in various biological processes. Recent observations have elucidated the functional roles of lncRNAs in cutaneous biology, e.g. in proliferation and differentiation of epidermal keratinocytes and in cutaneous wound repair. Furthermore, the role of lncRNAs in keratinocyte-derived skin cancers is emerging, especially in cutaneous squamous cell carcinoma (cSCC), which presents a significant burden to health care services worldwide and causes high mortality as metastatic disease. Elucidation of the functions of keratinocyte-specific lncRNAs will improve understanding of the molecular pathogenesis of epidermal disorders and skin cancers and can be exploited in development of new diagnostic and therapeutic applications for keratinocyte carcinomas. In this review, we summarize the current evidence of functionally important lncRNAs in cutaneous biology and in keratinocyte carcinomas.
Keywords: Skin cancer, Basal cell carcinoma, Cutaneous squamous cell carcinoma, Epidermis, Wound repair, Ultraviolet radiation
Introduction
Skin cancers are the most common cancer types globally with increasing incidence [1, 2]. Melanoma, basal cell carcinoma (BCC), and cutaneous squamous cell carcinoma (cSCC) are the three major types of skin cancer. Cumulative exposure to ultraviolet radiation (UVR) is a common risk factor for skin cancers, but they differ with respect to mutational profiles and alterations in cellular signaling pathways [3]. Melanoma originates from melanocytes, whereas BCC and cSCC originate from epidermal keratinocytes and are, therefore, called keratinocyte carcinomas (KC). The best preventive measure against skin cancer is avoiding excessive and cumulative exposure to sunlight and other sources of UVR. In addition, early detection and treatment is pivotal for the prognosis of the disease. The mortality rates for skin cancers vary between populations. However, taking into account the considerably higher incidence of KC over melanoma, it is estimated that the global mortality rate for all non-melanoma skin cancers (NMSCs) including BCC and cSCC, is even higher than for melanoma [4].
A significant proportion of human genome encodes non-coding RNAs (ncRNAs), including ribosomal RNA (rRNA) and transfer RNA (tRNA), and other functionally relevant ncRNAs, roughly categorized to small (sncRNAs) and long non-coding RNAs (lncRNAs) [5]. MicroRNAs (miRNAs) present an evolutionary conserved subgroup of sncRNAs deregulated in different cancers, including BCC and cSCC [6, 7]. LncRNAs are single-stranded RNA molecules larger than 200 nucleotides in size, lacking protein-coding capacity and sequence conservation [8]. It has become increasingly evident that they regulate a variety of cellular functions, and that aberrant expression of lncRNAs plays a role in various pathological conditions including cancer [9].
The mutational background for cSCC and BCC is well documented, and several driver mutations in protein coding genes have been identified [10–17]. These same driver gene mutations are also found frequently in epidermal keratinocytes in normal sun-exposed skin [18], indicating, that also other factors, e.g. changes in non-coding genes and the microenvironment, are also necessary for development of cSCC [19]. Mutations in the non-coding regions of genome can affect chromatin structure, transcription factor binding, and gene expression [20]. Moreover, these mutations may alter expression or secondary structure of lncRNAs or interfere with lncRNA interaction with other regulatory factors [21]. The consequence of non-coding mutations in lncRNA expression and function in cutaneous carcinogenesis and skin cancer development is largely unknown. However, recent evidence suggests that lncRNAs participate in the complex cancer signaling network in skin malignancies. Elucidation of their role in cutaneous biology is likely to reveal new molecular targets for diagnostics and therapeutic intervention. In this review, we summarize the current findings of the function of lncRNAs in cutaneous biology and in keratinocyte carcinomas.
Keratinocyte carcinomas
Keratinocyte carcinomas BCC and cSCC are the most common forms of skin cancer with increasing incidence globally [22, 23]. The primary cause for KCs is chronic exposure to UVR, and other important risk factors include immunosuppression, human papillomavirus infection, and chronic cutaneous ulceration [22–24]. While BCC is the most common human malignancy, cSCC accounts for the majority of deaths among KCs [25, 26]. In addition, a personal history of KCs is associated with a risk for other cancers [27]. In contrast to BCC, which rarely metastasizes, the risk of metastasis for cSCC is estimated as 1–4% and the prognosis of metastatic cSCC is poor [26]. Overall, the high prevalence of KCs poses a marked burden on health care worldwide and has a major impact on the patients’ quality of life [28].
Development of KC involves accumulation of several molecular and cellular changes. Both BCC and cSCC harbor a substantial mutational burden, mainly due to cumulative UV exposure typically observed as C → T transitions in the DNA [12–19]. Several studies using BCC and cSCC murine models suggest that these cancers arise from multiple cellular origins, e.g. from different stem cell populations in the basal layer of the epidermis, hair follicle bulge or sebaceous gland [29].
Despite a high frequency of UV-induced mutations, BCCs and cSCCs do not harbor many common genetic alterations, except inactivation of tumor suppressor p53 [12–19]. Several driver gene mutations have been identified for cSCC, resulting in constitutive activation of HRAS and inactivation of tumor suppressors p53 and NOTCH1 [12–16]. Conversely, BCC is strongly associated with aberrant activation of the Hedgehog signaling pathway due to loss of PTCH1 receptor function and activation of the G protein-coupled receptor SMO [17–19]. Like many other cancers, cSCCs and BCCs are associated with epigenetic deregulation and aberrant DNA methylation, which also contribute to cancer progression [30–34].
Actinic keratoses (AKs) are early precursors of cSCC and Bowen’s disease is in situ cSCC (cSCCIS), where atypical keratinocytes extend throughout the epidermis [35]. If left untreated, these lesions develop to invasive cSCCs. In general, patients with BCCs or resectable primary cSCCs have a good prognosis, whereas metastatic cSCC is associated with poor outcome [26]. Radiation and chemotherapy can be used for advanced and recurrent high-risk tumors that cannot be excised, especially those located in the facial area [36]. Recently, targeted therapies have been approved for therapy of advanced BCC and cSCC. Vismodegib, an inhibitor of Hedgehog pathway, is available for treatment of locally advanced BCC [37]. Immune checkpoint inhibitor, programmed cell death protein-1 (PD-1) blocking monoclonal antibody cemiplimab, has been approved for treatment of patients with locally advanced or metastatic cSCC [38]. Nevertheless, there is an urgent need for additional targeted therapies for advanced cSCCs and for prognostic biomarkers for predicting the risk of recurrence and metastatic potential of cSCC.
Long non-coding RNAs
Long non-coding RNAs (lncRNAs) are single-stranded RNAs mainly transcribed by RNA polymerase II, which undergo post-transcriptional processing, such as 5′-capping, splicing and polyadenylation [8]. This way lncRNAs closely resemble messenger RNAs (mRNA), but they are not translated to proteins. Some lncRNAs are rapidly degraded after transcription, whereas others are extremely stable [39, 40]. A rapid turnover of lncRNAs enables a dynamic cellular response via specifically induced lncRNAs, for instance in DNA damage, immune response, and cellular differentiation [41–43].
LncRNAs are poorly conserved between species [44–46]. In general, lncRNAs are considered larger than 200 nucleotides in size. This division, however, is not strict, as some lncRNAs are less than 200 nucleotides in size, and some lncRNAs can function both as regulatory lncRNAs and can be processed to sncRNAs [47]. Classification of lncRNAs into distinct subgroups is commonly based on their genomic location.
Long intergenic or intervening non-coding RNAs (lincRNAs) are transcribed from distinct loci, often from their own promoters, whereas intronic lncRNAs are transcribed from intronic regions within protein-coding gene [45, 47]. Sense lncRNAs are transcribed from the sense strand also containing exons of protein-coding genes [47]. Natural antisense transcripts (NATs) are transcribed from the antisense strand of a protein-coding gene, overlapping either exonic or intronic regions [47]. Bidirectional lncRNAs are produced divergently from the same promoter of a protein-coding gene. Circular RNAs (circRNA) are a recently discovered group of lncRNAs structurally different from most lncRNAs. They are produced by back splicing of precursor mRNAs or lncRNAs, resulting in covalently closed circular RNAs without polyadenylation, and they can originate from intronic or exonic transcripts [48].
LncRNAs are specifically expressed during normal physiological processes including cell differentiation and tissue development, whereas untimely and aberrant expression of lncRNAs in various pathological conditions is becoming evident [9–11]. Thus, alternative classification has been suggested, for instance by grouping them into lncRNAs regulating gene expression locally (cis) or in distance (trans) [49], or by other criteria such as subcellular localization, association with DNA-elements, or functional mechanism [50].
Molecular functions of lncRNAs
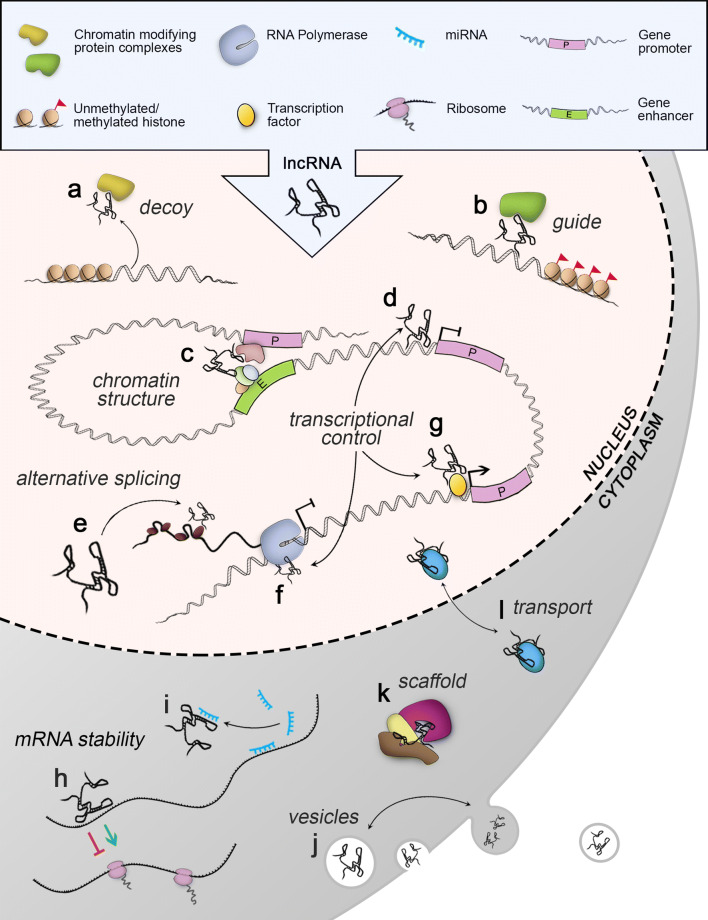
In general, the regulatory role of lncRNAs is based on binding to specific effector molecules by sequence complementarity or structural recognition to mediate gene expression. The single-stranded structure of lncRNAs and folding into unique secondary and tertiary structures gives them the ability to bind to RNA, DNA or proteins and this way control diverse cellular functions [8, 51, 52] (Fig. 1). LncRNAs typically exhibit a strict cell and tissue-specific expression and subcellular localization, indicating strictly controlled regulatory role for distinct lncRNAs [44, 53]. Specific localization of distinct lncRNAs to cytoplasm, nucleus, or other cellular compartments is likely to reflect their function (Fig. 1). In addition, some lncRNAs are secreted in extracellular vesicles and exosomes, and can exert their effect in adjacent cells and in cells in other tissues [54, 55]. In general, lncRNA mechanism of action can be divided into four main types: signals, guides, decoys, and scaffolds [51]. Simply, they can also be classified as nuclear lncRNAs in mediating gene transcription [56] or cytoplasmic lncRNAs controlling post-transcriptional events and mRNA stability [57] (Fig. 1).
Fig. 1.
Molecular functions of lncRNAs. Nuclear lncRNAs can regulate epigenetic changes by a decoying or b guiding chromatin-modifying complexes to specific genomic loci. c lncRNAs can induce chromosomal looping to control gene expression by simultaneously binding to protein complexes or specific DNA elements. LncRNAs can inhibit gene transcription d by blocking a transcription factor binding site or f by binding to RNA polymerase. g LncRNAs may contribute to transcriptional activation by guiding transcription factors or other co-factors to gene promoters. e LncRNAs can regulate alternative splicing that can occur by lncRNA binding to mRNA and blocking the splice-site. LncRNAs can also recruit and guide splicing factors to the sites of transcription. Cytoplasmic lncRNAs can regulate mRNA stability h directly by binding to mRNAs or i indirectly by sequestering miRNAs by complementary base pairing. j Some lncRNAs can be secreted to extracellular vesicles or exosomes allowing them to mediate intercellular signaling. k LncRNAs can serve as scaffolds to promote the assembly of active ribonucleoprotein complexes in the cytoplasm or nucleus. l LncRNAs can aid intracellular translocation of proteins
LncRNAs in cutaneous biology
Regulation of epidermal differentiation by lncRNAs
The skin serves as a protective barrier against several environmental threats, including microbes, chemicals, and physical insults, and it also controls water loss and thermoregulation. Skin consists of several different cell types and stem cell populations, which co-operate to maintain and regenerate its structure and function [58]. The epidermal layer of skin is under continuous turnover, as the cells generated from the basal keratinocytes lose their proliferative capacity, commit to terminal differentiation, and move towards skin surface [59]. During this process, keratinocytes undergo major morphological and mechanical changes due to spatiotemporal alteration in their transcriptional program [60]. Several markers for keratinocyte differentiation have been identified and the chromatin dynamics play a crucial role during this process [61–63].
Transcriptional changes during differentiation of epidermal keratinocytes also involve alterations in the expression of non-coding RNAs and accordingly specific lncRNAs have been implicated in keratinocyte differentiation [64, 65] (Fig. 2). Differentiation antagonizing non-protein coding RNA (DANCR) is downregulated during terminal differentiation of keratinocytes and it is required for maintaining the undifferentiated phenotype of epidermal progenitor cells [66]. DANCR is a negative regulator of MAF and MAFB transcription factors, which are important regulators of differentiation in various cell types [67]. DANCR represses the expression of MAF and MAFB epigenetically by guiding a chromatin-modifying protein complex to the promoters of their genes [66]. In contrast to DANCR, terminal differentiation-induced ncRNA (TINCR) is highly expressed in differentiating keratinocytes specifically in the suprabasal layers of human epidermis [68]. TINCR promotes keratinocyte differentiation by stabilizing mRNAs coding for proteins involved in keratinocyte differentiation e.g. transcription factors MAF and MAFB, together with an RNA-binding protein STAU1 [68]. Together, DANCR and TINCR are able to regulate the expression of a broad range of genes in keratinocytes and this way function as pivotal regulators of epidermal differentiation.
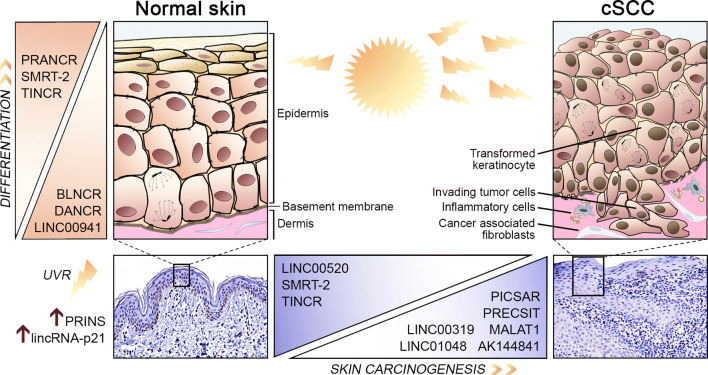
Fig. 2.
An overview of lncRNAs implicated in epidermal homeostasis in normal skin and in cSCC progression. Solar ultraviolet radiation (UVR) induces a stress response and altered expression of specific lncRNAs, such as PRINS and lincRNA-p21 in normal keratinocytes. Cumulative exposure to UVR predisposes epidermal keratinocytes to DNA damage and malignant transformation, which eventually lead to development of invasive cSCC. Several lncRNAs have been shown to be involved in cutaneous homeostasis. TINCR, PRANCR, and SMRT-2 promote and DANCR, BLNCR, and LINC00941 inhibit keratinocyte differentiation. Deregulation of lncRNAs is becoming evident in cSCC progression. The expression of TINCR, SMRT-2, and LINC00520 is downregulated and the expression of PICSAR, PRECSIT, MALAT1, AK144841, LINC00319, and LINC01048 is upregulated in cSCC. These lncRNAs could be used as new prognostic markers and as novel therapeutic targets in cSCC
The expression of LINC00941 is downregulated upon keratinocyte differentiation, and it antagonizes the function of small proline rich protein 5 (SPRR5), which promotes differentiation of keratinocytes [69]. The expression of beta1-adjacent long non-coding RNA (BLNCR) is also downregulated during keratinocyte differentiation, preceding downregulation of ITGB1, which codes for integrin β1, an epidermal stem cell marker adjacent to BLNCR gene [70, 71]. BLNCR and ITGB1 are both transcriptionally regulated by transcription factors p63 and AP-1, and loss of BLNCR and ITGB1 expression may be an early event resulting in loss of proliferative capacity of keratinocytes and in subsequent terminal differentiation [71].
Progenitor renewal associated non-coding RNA, (PRANCR), is one of the most recently characterized lncRNAs involved in epidermal homeostasis [72]. Depletion of PRANCR leads to reduced proliferative capacity and differentiation of keratinocytes. PRANCR regulates the expression of several genes coding for cell cycle regulators, including E2F transcription factor target genes [72]. In addition, H19 imprinted maternally expressed transcript (H19) and SCC misregulated transcript-2 (SMRT-2) are recently identified lncRNAs induced during differentiation of epidermal keratinocytes [73, 74]. Depletion of SMRT-2 results in repression of several genes associated with epidermal differentiation and development [74]. These genes are also regulated by zinc finger protein 750 (ZNF750) and Kruppel like factor 4 (KLF4), suggesting that SMRT-2 functions upstream of the ZNF750-KLF4-axis [74]. ZNF750 functions downstream of p63 in driving epidermal differentiation by upregulating KLF4 [75]. Moreover, ZNF750 upregulates expression of lncRNA TINCR [76]. Taken together, these observations provide a regulatory link between SMRT-2, ZNF750 and TINCR in regulation of epidermal keratinocyte differentiation.
As aberrant keratinocyte differentiation and stem-cell characteristics are involved in KC tumor development [77, 78], it is not surprising that the expression of keratinocyte differentiation inducing lncRNAs, SMRT-2 and TINCR, are strongly downregulated in cSCC [66, 74, 79]. Many of the lncRNAs associated with epidermal differentiation listed here are not implicated in cSCC and it will be important to investigate their mechanistic role in progression of cutaneous cancers.
LncRNAs in cutaneous wound repair
Cutaneous wound repair is a complex and strictly controlled process, which involves co-operation of several different cell types, including keratinocytes, fibroblasts, endothelial cells, and inflammatory cells [80]. Delayed wound healing resulting in chronic ulcers is usually associated with an underlying condition, such as insufficient arterial or venous circulation, diabetes or prolonged inflammation [80]. Chronic wounds also carry a risk of developing to cSCC [81].
The role of lncRNAs in normal wound repair or in pathogenesis of chronic ulcers is largely unknown. LncRNA expression profile in Marjolin ulcer, a rare, aggressive type of cSCC that evolves in scars or chronic wounds has been reported, but functional characterization of the lncRNAs in this condition is lacking [82]. Growth arrest-specific 5 lncRNA (GAS5) is a repressor of glucocorticoid receptor expression, which serves as a tumor suppressor in many cancers [83]. GAS5 has been shown to promote wound healing by inducing epithelialization and angiogenesis via c-myc inhibition [84]. Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), a tumor-promoting lncRNA in many cancers [85] has been shown to stimulate repair of ischemic wounds by promoting migration of human dermal fibroblasts through hypoxia-inducible factor-1α (HIF-1α) signaling [86, 87]. In addition, lncRNA H19 has been shown to promote wound healing via HIF-1α pathway [88, 89].
Wound and keratinocyte migration-associated lncRNA 1 and 2 (WAKMAR1 and WAKMAR2) are two recently identified lncRNAs, which play an important role in cutaneous wound repair [90, 91]. Expression of WAKMAR1 is highly upregulated in keratinocytes during wound repair and its expression stimulates keratinocyte migration and wound re-epithelization [90]. Expression of both WAKMAR1 and WAKMAR2 is induced by TGF-β and downregulation of their expression inhibits migration of keratinocytes, resulting in delayed wound re-epithelization [90, 91]. Accordingly, the expression of both WAKMAR1 and WAKMAR2 is reduced in keratinocytes in the edge of chronic wounds in vivo [90, 91]. Moreover, upregulation of WAKMAR2 expression inhibits production of inflammatory chemokines by keratinocytes, and this way promotes wound healing [91]. WAKMAR1 exerts its function by sequestering DNA methyltransferases, resulting in upregulation of the expression of E2F1 transcription factor and subsequent regulation of the expression of its target genes [90].
Regulation of lncRNAs by UVR
Exposure of skin to UVR induces several cellular responses. Activation of the inflammatory response manifesting as erythema in skin is an acute response after UV exposure [92]. UVR induces DNA damage in epidermal keratinocytes, which triggers a stress response, activation of p53 and DNA repair [93]. UV-induced DNA damage leads to systemic immunosuppression [94–96] which is exploited in treatment of inflammatory skin diseases, such as psoriasis and atopic dermatitis [97].
UVR leads to altered lncRNA expression in epidermal keratinocytes [98], melanocytes, [99] and dermal fibroblasts in culture [100–103]. The biological response of skin to UVA and UVB is distinct due to their different penetration to the skin and they trigger distinct expression pattern of lncRNAs in HDFs [101]. It is possible, that some of these lncRNAs play a role in the early cellular stress response or acute inflammation following exposure to UV. Also, several UV-regulated lncRNAs in keratinocytes show a similar expression trend in cSCC and BCC, suggesting a role for them in epidermal carcinogenesis [98].
A subset of UV-induced lncRNAs has been functionally characterized [102–105]. In keratinocytes, the expression of lincRNA-p21 is markedly induced by UVB through a p53-dependent mechanism and it exerts a tumor suppressive role by triggering UVB-induced apoptosis and cell cycle arrest [105]. Accordingly, a tumor suppressive function for lincRNA-p21 has been reported in head and neck SCC [106]. Psoriasis susceptibility-related RNA gene induced by stress (PRINS) is a lncRNA induced by UVB and other stress signals, such as serum starvation or translational inhibition in HaCaT cells, an epidermal keratinocyte derived cell line, which lacks functional p53 [104]. Elevated expression of PRINS in psoriatic epidermis has also been reported, suggesting a role for PRINS in pathogenesis of psoriasis [104].
Vitamin D is photochemically synthesized in the skin by UVB and recent findings support a cancer protecting role for vitamin D [107]. Interestingly, keratinocytes lacking vitamin D receptor show a distinct lncRNA expression pattern with increased expression of oncogenic lncRNAs and decreased expression of tumor-suppressive lncRNAs, including lincRNA-p21 [108]. It appears, that UVR plays a dual role in skin by inducing the innate immune response, but predisposing to systemic immunosuppression and genomic mutations [1, 92, 97]. It is not known, what is the feasible level of UV exposure and to what extent lncRNAs can mediate the balance between skin homeostasis and carcinogenesis.
LncRNAs in keratinocyte carcinomas
The UV-induced alteration of lncRNA expression in epidermal cells suggests that some of these lncRNAs exert a protective role against carcinogenesis by triggering UV-induced early stress response [98–105] (Table 1). On the other hand, some of them may play a role at the early stage of epidermal carcinogenesis and loss of some differentiation-associated lncRNAs may serve as markers for tumor initiation. In keratinocyte carcinomas, particularly in cSCCs, several lncRNAs are differentially expressed as compared to normal skin or keratinocytes, suggesting a role for them in cSCC progression [109, 110]. Some of the deregulated lncRNAs may function in signaling pathways, which are already mutationally activated or suppressed in cSCC. On the other hand, it is likely that some of these lncRNAs are targeted by UV-induced mutations or by genomic alterations within the lncRNA gene itself, as has been observed in several cancer cell lines [111, 112]. As none of the BCC-associated lncRNAs have been functionally characterized so far, we will focus on lncRNAs implicated in cSCC (Table 1).
Table 1.
Long non-coding RNAs with a potential role in cSCC or BCC development
| LncRNA | Expression | Function | References |
|---|---|---|---|
| TINCR | Downregulated in cSCC | Promotes human epidermal differentiation by stabilization of mRNAs coding for differentiation specific genes | [68] |
| SMRT-2 | Downregulated in cSCC | Induced during keratinocyte differentiation. Knockdown in human organotypic skin downregulates several differentiation specific genes, including ZNF750 and KLF4 | [74] |
| LINC00520 | Downregulated in cSCC | Inhibits cSCC progression by downregulating expression of EGFR and its downstream targets, e.g. PI3K, AKT, and VEGF | [123] |
| PICSAR | Upregulated in cSCC | Promotes cSCC progression by activating ERK1/2 by downregulating DUSP6. Decreases cSCC cell adhesion and increases cSCC cell migration by downregulating integrin expression | [110, 118] |
| PRECSIT | Upregulated in cSCC | Promotes cSCC cell invasion through STAT3-mediated upregulation of production of MMP-13, MMP-3, MMP-1, and MMP-10 | [129] |
| LINC00319 | Upregulated in cSCC | Increases cSCC cell growth, migration, and invasion. Suppresses apoptosis by upregulating cyclin-dependent kinase 3 via miR‐1207‐5p decoy | [130] |
| LINC01048 | Upregulated in cSCC | Interacts with TAF15 transcription factor to induce YAP1 transcription and tumorigenic function via Hippo signaling pathway | [126] |
| MALAT1 | Upregulated in cSCC | Positively regulates EGFR protein expression via c-MYC and KTN1 | [119] |
| lincRNA-p21 | Induced in mouse and human keratinocytes by UVB | Tumor suppressive role by triggering UVB-induced apoptotic death | [105] |
| AK144841 | Induced in mouse DMBA/TPA-induced cSCC | Downregulates several anticancer and cell differentiation genes in mouse | [79] |
| H19,Hottip, Nespas, mHOTAIR, MALAT1, SRA | Upregulated in vitamin D receptor (VDR) deleted mouse keratinocytes and epidermis | Potential oncogenes in skin cancer progression | [108] |
| Kcnq1ot1, lincRNA‐p21, Foxn2‐as, Gtl2‐as, H19‐as | Inhibited in VDR-deleted mouse keratinocytes and epidermis | Potential tumor suppressors in skin cancer formation | [108] |
| H19, CASC15, SPRY4-IT | Upregulated in BCC | Potential oncogenes in BCC | [131] |
Aberrant activation of the ERK1/2 MAPK pathway is one of the central drivers in the molecular pathogenesis of cSCC [113–115]. ERK1/2 pathway is activated by UVA radiation [116]. Moreover, mutational activation of epidermal growth factor receptor (EGFR) results in sustained activation of the RAS-RAF-MEK-ERK signaling pathway and promotes cutaneous carcinogenesis [117].
PICSAR plays a tumorigenic role in cSCC
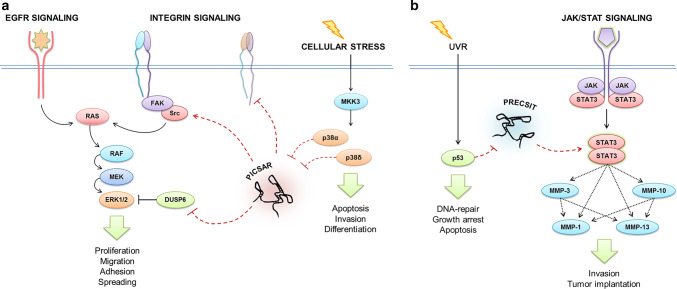
p38-inhibited cutaneous squamous cell carcinoma-associated lincRNA (PICSAR) represents the earliest evidence of a functionally characterized lncRNA in cSCC [110]. The expression of PICSAR is upregulated in cSCC tumor cells in culture and in vivo compared to normal human epidermal keratinocytes (NHEKs) and normal skin [110]. Elevated expression of PICSAR was also noted in vivo in actinic keratosis and cSCC in situ, suggesting a role for PICSAR at the early stage of epidermal carcinogenesis [110]. Silencing of PICSAR expression potently suppresses growth of human cSCC xenografts [110]. Interestingly, PICSAR serves as a regulatory link between p38 and ERK1/2 mitogen-activated protein kinase (MAPK) pathways (Fig. 3a). Inhibition of p38 activity induces PICSAR expression and PICSAR promotes cSCC cell proliferation by promoting ERK1/2 activity via downregulation of dual specificity phosphatase DUSP6 [110]. In addition, PICSAR potently regulates cell adhesion and migration by regulating integrin expression [118], and may this way contribute to cSCC progression and invasion (Fig. 3a).
Fig. 3.
Proposed molecular functions for lncRNAs PICSAR and PRECSIT in cSCC. a Expression of PICSAR is suppressed by the p38 signaling pathway. PICSAR promotes activity of ERK1/2 and cell proliferation by inhibiting expression of dual-specificity phosphatase DUSP6 in cSCC cells. In addition, PICSAR modulates cSCC cell adhesion and migration by regulating integrin expression on the cell surface. b Expression of PRECSIT is suppressed by functional p53 signaling, and elevated PRECSIT expression in response to p53 inactivation contributes to STAT3 activation, which in turn upregulates matrix metalloproteinases MMP-13, MMP-3, MMP-1, and MMP-10 in the MMP cluster in 11q22.3 and this way promotes proteolytic remodeling of extracellular matrix and basement membrane, and cSCC cell invasion
MALAT1 and LINC00520 play opposite roles in cSCC
MALAT1 is a lncRNA, which has been reported to be deregulated in different types of cancer [85]. Elevated expression of MALAT1 was recently reported in cSCC tumors and it was shown that the expression in cSCC cells is induced by UVB [119]. MALAT1 promotes proliferation, migration, and invasion of cSCC cells and growth of cSCC tumors in vivo and suppresses apoptosis of cSCC cells [119]. Mechanistically, MALAT1 interacts with c-Myc to activate transcription of kinectin 1 (KTN1) gene, which is one of the top downregulated genes after MALAT1 depletion. Knockdown of MALAT1 also results in decreased level of EGFR protein, but not EGFR mRNA [119]. These results suggest that MALAT1 contributes to cSCC pathogenesis by upregulating EGFR protein levels via c-Myc and KTN1 [119].
Marked expression of lncRNA AK144841 has been noted in chemically (DMBA/TPA) induced mouse cSCCs compared to healthy skin [79]. The histology and the genomic background of these tumors are very similar to human cSCCs [120]. Sustained activation of HRAS, which is caused by highly carcinogenic DMBA, results in marked induction of EGFR and its ligands in cSCC mouse model [121, 122]. In this regard, induction of AK144841 in murine cSCC may be related to EGFR activation. A potential human ortholog with homology to AK144841 has been shown to be expressed at high level in cSCC cell lines compared to NHEKs suggesting, that it may be involved in human cSCC progression [79].
Downregulation of LINC00520 has been noted in A431 cSCC cell line, compared to NHEKs, and overexpression of LINC00520 in A431 cells results in suppression of tumor growth and lymph node metastasis [123]. A431 cells express high levels of EGFR [124]. Reduced expression of EGFR and its downstream targets, PI3K, AKT, VEGF, MMP-2, and MMP-9 was noted in A431 cells overexpressing LINC00520, whereas an opposite effect was noted after LINC00520 depletion [123]. Altogether, these results suggest that LINC00520 plays a tumor suppressive role in cSCC by targeting EGFR [123].
TINCR and SMRT-2 are potential tumor suppressors in cSCC
Poor differentiation of cSCC is associated with risk for metastasis and poor prognosis [77, 78]. TINCR and SMRT-2 both promote differentiation of keratinocytes and may this way serve in a protective role in keratinocyte carcinogenesis [68, 74]. Accordingly, decreased expression of TINCR and SMRT-2 has been noted in human cSCCs [68, 74], and a notable decrease in TINCR expression has been reported in DMBA/TPA-induced murine cSCC tumors compared to normal skin [79]. In addition, marked suppression of SMRT-2 expression has been noted in Ras-driven human organotypic epidermal neoplasia [74]. Together, these two lncRNAs may function as potential tumor suppressors in cSCC. In this context, it is interesting that ZNF750 which upregulates the expression of TINCR in keratinocytes, was recently shown to exert a tumor-suppressive role in SCCs of head and neck, lung, cervix, and skin [76].
LINC01048 and Hippo pathway in keratinocyte carcinoma
Hippo pathway is a well-conserved signaling pathway, which is important in skin development, cutaneous homeostasis and tissue regeneration, and aberrant Hippo signaling has been noted in non-melanoma skin cancers [125]. Recently, upregulation of a previously unknown lncRNA, LINC01048, was reported in cSCC associated with lower overall survival of cSCC patients [126]. LINC01048 promotes cSCC cell growth via the Hippo pathway [126]. Depletion of LINC01048 regulates the levels of the downstream effectors of the Hippo signaling, including yes-associated protein 1 (YAP1) and transcriptional co-activator with PDZ-binding motif (TAZ). Mechanistically, LINC01048 interacts with transcription factor TAF15 to promote transcription of YAP1 gene [126]. Accordingly, YAP1 and TAZ function as oncogenes in many cancers, including BCC and cSCC [127, 128]. Together these results provide interesting new evidence for the role of LINC01048/TAF15/YAP1-axis in cSCC progression.
PRECSIT and LINC00319 regulate invasion of cSCC
p53-regulated carcinoma-associated STAT3-activating long intergenic non-protein coding transcript (PRECSIT) is a recently identified lncRNA with elevated expression in cSCC [129]. PRECSIT is a nuclear-enriched lncRNA downregulated by p53 signaling, and a high level of PRECSIT expression is associated with the absence of functional p53 in cSCC tumor cells in vivo [129]. Depletion of PRECSIT inhibits cSCC cell invasion by downregulating STAT3 expression and activation, and production of matrix metalloproteinases (MMPs), MMP-13, MMP-3, MMP-1, and MMP-10 [129], suggesting a tumor-promoting function for PRECSIT (Fig. 3b). These results provide interesting new evidence that p53/PRECSIT/STAT3 axis regulates the expression of invasion proteinases in the MMP gene cluster in 11q22.3: MMP-13/MMP-3/MMP-1/MMP-10.
LINC00319 is a recently identified lncRNA with elevated expression in cSCC shown to correlate with larger tumor size and lymphovascular invasion of cSCC [130]. LINC00319 promotes cSCC cell migration and invasion, and upregulates expression of MMP-2, MMP-9, and markers for epithelial–mesenchymal transition, E-cadherin, and vimentin [130]. PRECSIT regulates the invasion of cSCC cells specifically without affecting cell growth [129], whereas LINC00319 has an anti-apoptotic function and promotes cSCC cell proliferation via miRNA-mediated mechanism [130].
Concluding remarks
The role of lncRNAs in epidermal biology is slowly emerging. The recent findings summarized here elucidate the functional role of lncRNAs in physiological conditions and keratinocyte cancer development, specifically in cSCC (Fig. 2, Table 1). It is noteworthy, that none of the BCC-associated lncRNAs have been functionally characterized yet. Moreover, considering UVR as a common nominator for the development of cSCC and BCC, it remains unclear whether they share the same UV-regulated lncRNAs. These cancers have distinct mutational background and different oncogenic signaling pathways. Therefore, it is likely that there are also specific lncRNAs which, by function, are associated with either cSCC or BCC development by co-operating with various signaling molecules to mediate the expression of tumor promoting or tumor suppressing genes. LncRNAs present great potential in developing new diagnostic and therapeutic approaches. Along with conventional molecular markers, distinct lncRNA expression signature may provide better diagnostic accuracy of the disease. Moreover, therapeutic targeting of tumorigenic lncRNAs may enhance the efficacy of cancer therapy.
Acknowledgements
Open access funding provided by University of Turku (UTU) including Turku University Central Hospital. The original work of authors has been supported by grants from the Finnish Cancer Research Foundation, The Cancer Society of South-West Finland, Jane and Aatos Erkko Foundation, Sigrid Jusélius Foundation, The state research funding of the Turku University Hospital (project 13336), The Kymenlaakso Regional Fund of the Finnish Cultural Foundation, Ida Montin Foundation, Instrumentarium Science Foundation, The Paulo Foundation, The Maud Kuistila Memorial Foundation, University of Turku Graduate School Funding and Turku University Foundation.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Asgari MM, Moffet HH, Ray GT, Quesenberry CP. Trends in basal cell carcinoma incidence and identification of high-risk subgroups, 1998–2012. JAMA Dermatol. 2015;151:976–981. doi: 10.1001/jamadermatol.2015.1188. [DOI] [PubMed] [Google Scholar]
- 2.Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US Population, 2012. JAMA Dermatol. 2015;151:1081–1086. doi: 10.1001/jamadermatol.2015.1187. [DOI] [PubMed] [Google Scholar]
- 3.Liu-Smith F, Jia J, Zheng Y. UV-induced molecular signaling differences in melanoma and non-melanoma skin cancer. Adv Exp Med Biol. 2017;996:27–40. doi: 10.1007/978-3-319-56017-5_3. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 5.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García-Sancha N, Corchado-Cobos R, Pérez-Losada J, Cañueto J. MicroRNA dysregulation in cutaneous squamous cell carcinoma. Int J Mol Sci. 2019;20:E2181. doi: 10.3390/ijms20092181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syed DN, Lall RK, Mukhtar H. MicroRNAs and photocarcinogenesis. Photochem Photobiol. 2015;91:173–187. doi: 10.1111/php.12346. [DOI] [PubMed] [Google Scholar]
- 8.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, Tsai KY, Curry JL, Tetzlaff MT, Lai SY, Yu J, Muzny DM, Doddapaneni H, Shinbrot E, Covington KR, Zhang J, Seth S, Caulin C, Clayman GL, El-Naggar AK, Gibbs RA, Weber RS, Myers JN, Wheeler DA, Frederick MJ. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20:6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inman GJ, Wang J, Nagano A, Alexandrov LB, Purdie KJ, Taylor RG, Sherwood V, Thomson J, Hogan S, Spender LC, South AP, Stratton M, Chelala C, Harwood CA, Proby CM, Leigh IM. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat Commun. 2018;9:3667. doi: 10.1038/s41467-018-06027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.South AP, Purdie KJ, Watt SA, Haldenby S, den Breems N, Dimon M, Arron ST, Kluk MJ, Aster JC, McHugh A, Xue DJ, Dayal JH, Robinson KS, Rizvi SH, Proby CM, Harwood CA, Leigh IM. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J Invest Dermatol. 2014;134:2630–2638. doi: 10.1038/jid.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res. 2015;21:1447–1456. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Rohil RN, Tarasen AJ, Carlson JA, Wang K, Johnson A, Yelensky R, Lipson D, Elvin JA, Vergilio JA, Ali SM, Suh J, Miller VA, Stephens PJ, Ganesan P, Janku F, Karp DD, Subbiah V, Mihm MC, Ross JS. Evaluation of 122 advanced-stage cutaneous squamous cell carcinomas by comprehensive genomic profiling opens the door for new routes to targeted therapies. Cancer. 2016;122:249–257. doi: 10.1002/cncr.29738. [DOI] [PubMed] [Google Scholar]
- 15.Pellegrini C, Maturo MG, Di Nardo L, Ciciarelli V, Gutiérrez García-Rodrigo C, Fargnoli MC. Understanding the molecular genetics of basal cell carcinoma. Int J Mol Sci. 2017;18:E2485. doi: 10.3390/ijms18112485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, Seplyarskiy VB, Sharpe HJ, McKee T, Letourneau A, Ribaux PG, Popadin K, Basset-Seguin N, Ben Chaabene R, Santoni FA, Andrianova MA, Guipponi M, Garieri M, Verdan C, Grosdemange K, Sumara O, Eilers M, Aifantis I, Michielin O, de Sauvage FJ, Antonarakis SE, Nikolaev SI. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 17.Jayaraman SS, Rayhan DJ, Hazany S, Kolodney MS. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Invest Dermatol. 2014;134:213–220. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- 18.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissinen L, Farshchian M, Riihilä P, Kähäri VM. New perspectives on role of tumor microenvironment in progression of cutaneous squamous cell carcinoma. Cell Tissue Res. 2016;365:691–702. doi: 10.1007/s00441-016-2457-z. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Adli M. Mapping and making sense of noncoding mutations in the genome. Cancer Res. 2019;79:4309–4314. doi: 10.1158/0008-5472.CAN-19-0905. [DOI] [PubMed] [Google Scholar]
- 21.Gao P, Wei GH. Genomic insight into the role of lncRNA in cancer susceptibility. Int J Mol Sci. 2017;18:E1239. doi: 10.3390/ijms18061239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nehal KS, Bichakjian CK. Update on keratinocyte carcinomas. N Engl J Med. 2018;379:363–374. doi: 10.1056/NEJMra1708701. [DOI] [PubMed] [Google Scholar]
- 23.Nagarajan P, Asgari MM, Green AC, Guhan SM, Arron ST, Proby CM, Rollison DE, Harwood CA, Toland AE. Keratinocyte carcinomas: current concepts and future research priorities. Clin Cancer Res. 2019;25:2379–2391. doi: 10.1158/1078-0432.CCR-18-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riihilä P, Nissinen L, Knuutila J, Rahmati Nezhad P, Viiklepp K, Kähäri VM. Complement system in cutaneous squamous cell carcinoma. Int J Mol Sci. 2019;20:E3550. doi: 10.3390/ijms20143550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomas A, Leonardi-Bee J, Bath-Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol. 2012;166:1069–1080. doi: 10.1111/j.1365-2133.2012.10830.x. [DOI] [PubMed] [Google Scholar]
- 26.Burton KA, Ashack KA, Khachemoune A. Cutaneous squamous cell carcinoma: a review of high-risk and metastatic disease. Am J Clin Dermatol. 2016;17:491–508. doi: 10.1007/s40257-016-0207-3. [DOI] [PubMed] [Google Scholar]
- 27.Small J, Barton V, Peterson B, Alberg AJ. Keratinocyte carcinoma as a marker of a high cancer-risk phenotype. Adv Cancer Res. 2016;130:257–291. doi: 10.1016/bs.acr.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Gaulin C, Sebaratnam DF, Fernández-Peñas P. Quality of life in non-melanoma skin cancer. Australas J Dermatol. 2015;56:70–76. doi: 10.1111/ajd.12205. [DOI] [PubMed] [Google Scholar]
- 29.Rognoni E, Watt FM. Skin cell heterogeneity in development, wound healing, and cancer. Trends Cell Biol. 2018;28:709–722. doi: 10.1016/j.tcb.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Paredes M, Bormann F, Raddatz G, Gutekunst J, Lucena-Porcel C, Köhler F, Wurzer E, Schmidt K, Gallinat S, Wenck H, Röwert-Huber J, Denisova E, Feuerbach L, Park J, Brors B, Herpel E, Nindl I, Hofmann TG, Winnefeld M, Lyko F. Methylation profiling identifies two subclasses of squamous cell carcinoma related to distinct cells of origin. Nat Commun. 2018;9:577. doi: 10.1038/s41467-018-03025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandiver AR, Irizarry RA, Hansen KD, Garza LA, Runarsson A, Li X, Chien AL, Wang TS, Leung SG, Kang S, Feinberg AP. Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin. Genome Biol. 2015;16:80. doi: 10.1186/s13059-015-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Wu R, Sargsyan D, Yin R, Kuo HC, Yang I, Wang L, Cheng D, Wang C, Li S, Hudlikar R, Lu Y, Kong AN. UVB drives different stages of epigenome alterations during progression of skin cancer. Cancer Lett. 2019;449:20–30. doi: 10.1016/j.canlet.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brinkhuizen T, van den Hurk K, Winnepenninckx VJ, de Hoon JP, van Marion AM, Veeck J, van Engeland M, van Steensel MA. Epigenetic changes in basal cell carcinoma affect SHH and WNT signaling components. PLoS ONE. 2012;7:e51710. doi: 10.1371/journal.pone.0051710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao RC, Chan MP, Andrews CA, Kahana A. Epigenetic markers in basal cell carcinoma: universal themes in oncogenesis and tumor stratification?—a short report. Cell Oncol (Dordr) 2018;41:693–698. doi: 10.1007/s13402-018-0402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cockerell CJ. Histopathology of incipient intraepidermal squamous cell carcinoma ("actinic keratosis") J Am Acad Dermatol. 2000;42:11–17. doi: 10.1067/mjd.2000.103344. [DOI] [PubMed] [Google Scholar]
- 36.Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: management of advanced and high-stage tumors. J Am Acad Dermatol. 2018;78:249–261. doi: 10.1016/j.jaad.2017.08.058. [DOI] [PubMed] [Google Scholar]
- 37.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, Marmur E, Rudin CM, Chang AL, Low JA, Mackey HM, Yauch RL, Graham RA, Reddy JC, Hauschild A. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Migden MR, Rischin D, Schmults CD, Guminski A, Hauschild A, Lewis KD, Chung CH, Hernandez-Aya L, Lim AM, Chang ALS, Rabinowits G, Thai AA, Dunn LA, Hughes BGM, Khushalani NI, Modi B, Schadendorf D, Gao B, Seebach F, Li S, Li J, Mathias M, Booth J, Mohan K, Stankevich E, Babiker HM, Brana I, Gil-Martin M, Homsi J, Johnson ML, Moreno V, Niu J, Owonikoko TK, Papadopoulos KP, Yancopoulos GD, Lowy I, Fury MG. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med. 2018;379:341–351. doi: 10.1056/NEJMoa1805131. [DOI] [PubMed] [Google Scholar]
- 39.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayupe AC, Tahira AC, Camargo L, Beckedorff FC, Verjovski-Almeida S, Reis EM. Global analysis of biogenesis, stability and sub-cellular localization of lncRNAs mapping to intragenic regions of the human genome. RNA Biol. 2015;12:877–892. doi: 10.1080/15476286.2015.1062960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez Y, Segura V, Marín-Béjar O, Athie A, Marchese FP, González J, Bujanda L, Guo S, Matheu A, Huarte M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, HenryBezy M, Lawrence JB, O'Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fico A, Fiorenzano A, Pascale E, Patriarca EJ, Minchiotti G. Long non-coding RNA in stem cell pluripotency and lineage commitment: functions and evolutionary conservation. Cell Mol Life Sci. 2019;76:1459–1471. doi: 10.1007/s00018-018-3000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. TheGENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kopp F. Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blythe AJ, Fox AH, Bond CS. The ins and outs of lncRNA structure: How, why and what comes next? Biochim Biophys Acta. 2016;1859:46–58. doi: 10.1016/j.bbagrm.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Amin V, Harris RA, Onuchic V, Jackson AR, Charnecki T, Paithankar S, Lakshmi Subramanian S, Riehle K, Coarfa C, Milosavljevic A. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nat Commun. 2015;6:6370. doi: 10.1038/ncomms7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, Tsai S, Kohli M, Boardman L, Patel T, Wang L. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. doi: 10.1038/srep19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umu SU, Langseth H, Bucher-Johannessen C, Fromm B, Keller A, Meese E, Lauritzen M, Leithaug M, Lyle R, Rounge TB. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2018;15:242–250. doi: 10.1080/15476286.2017.1403003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9:e1471. doi: 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belokhvostova D, Berzanskyte I, Cujba AM, Jowett G, Marshall L, Prueller J, Watt FM. Homeostasis, regeneration and tumour formation in the mammalian epidermis. Int J Dev Biol. 2018;62:571–582. doi: 10.1387/ijdb.170341fw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuchs E. Skin stem cells: rising to the surface. J Cell Biol. 2008;180:273–284. doi: 10.1083/jcb.200708185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miroshnikova YA, Le HQ, Schneider D, Thalheim T, Rübsam M, Bremicker N, Polleux J, Kamprad N, Tarantola M, Wang I, Balland M, Niessen CM, Galle J, Wickström SA. Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat Cell Biol. 2018;20:69–80. doi: 10.1038/s41556-017-0005-z. [DOI] [PubMed] [Google Scholar]
- 61.Kypriotou M, Huber M, Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the 'fused genes' family. Exp Dermatol. 2012;21:643–649. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 62.Cavazza A, Miccio A, Romano O, Petiti L, Malagoli Tagliazucchi G, Peano C, Severgnini M, Rizzi E, De Bellis G, Bicciato S, Mavilio F. Dynamic transcriptional and epigenetic regulation of human epidermal keratinocyte differentiation. Stem Cell Reports. 2016;6:618–632. doi: 10.1016/j.stemcr.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubin AJ, Barajas BC, Furlan-Magaril M, Lopez-Pajares V, Mumbach MR, Howard I, Kim DS, Boxer LD, Cairns J, Spivakov M, Wingett SW, Shi M, Zhao Z, Greenleaf WJ, Kundaje A, Snyder M, Chang HY, Fraser P, Khavari PA. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat Genet. 2017;49:1522–1528. doi: 10.1038/ng.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazar J, Sinha S, Dinger ME, Mattick JS, Perera RJ. Protein-coding and non-coding gene expression analysis in differentiating human keratinocytes using a three-dimensional epidermal equivalent. Mol Genet Genomics. 2010;284:1–9. doi: 10.1007/s00438-010-0543-6. [DOI] [PubMed] [Google Scholar]
- 65.Ørom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harb Symp Quant Biol. 2010;75:325–331. doi: 10.1101/sqb.2010.75.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, Rinn JL, Chang HY, Siprashvili Z, Khavari PA. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao S, Kretz M, Khavari PA. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. 2015;32:693–706. doi: 10.1016/j.devcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziegler C, Graf J, Faderl S, Schedlbauer J, Strieder N, Förstl B, Spang R, Bruckmann A, Merkl R, Hombach S, Kretz M. The long non-coding RNA LINC00941 and SPRR5 are novel regulators of human epidermal homeostasis. EMBO Rep. 2019;20:e46612. doi: 10.15252/embr.201846612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-K. [DOI] [PubMed] [Google Scholar]
- 71.Tanis SEJ, Köksal ES, van Buggenum JAGL, Mulder KW. BLNCR is a long non-coding RNA adjacent to integrin beta-1 that is rapidly lost during epidermal progenitor cell differentiation. Sci Rep. 2019;9:31. doi: 10.1038/s41598-018-37251-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai P, Otten ABC, Cheng B, Ishii MA, Zhang W, Huang B, Qu K, Sun BK. A genome-wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res. 2020;30:22–34. doi: 10.1101/gr.251561.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li CX, Li HG, Huang LT, Kong YW, Chen FY, Liang JY, Yu H, Yao ZR. H19 lncRNA regulates keratinocyte differentiation by targeting miR-130b-3p. Cell Death Dis. 2019;8:e3174. doi: 10.1038/cddis.2017.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee CS, Mah A, Aros CJ, Lopez-Pajares V, Bhaduri A, Webster DE, Kretz M, Khavari PA. Cancer-associated long noncoding RNA SMRT-2 controls epidermal differentiation. J Invest Dermatol. 2018;138:1445–1449. doi: 10.1016/j.jid.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hazawa M, Lin DC, Handral H, Xu L, Chen Y, Jiang YY, Mayakonda A, Ding LW, Meng X, Sharma A, Samuel S, Movahednia MM, Wong RW, Yang H, Tong C, Koeffler HP. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36:2243–2254. doi: 10.1038/onc.2016.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thieu K, Ruiz ME, Owens DM. Cells of origin and tumor-initiating cells for nonmelanoma skin cancers. Cancer Lett. 2013;338:82–88. doi: 10.1016/j.canlet.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuri P, Rompolas P. Differentiation keeps skin cancer at bay. Nat Cell Biol. 2018;20:1237–1239. doi: 10.1038/s41556-018-0222-0. [DOI] [PubMed] [Google Scholar]
- 79.Ponzio G, Rezzonico R, Bourget I, Allan R, Nottet N, Popa A, Magnone V, Rios G, Mari B, Barbry P. A new long noncoding RNA (lncRNA) is induced in cutaneous squamous cell carcinoma and down-regulates several anticancer and cell differentiation genes in mouse. J Biol Chem. 2017;292:12483–12495. doi: 10.1074/jbc.M117.776260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toriseva M, Kähäri VM. Proteinases in cutaneous wound healing. Cell Mol Life Sci. 2009;66:203–224. doi: 10.1007/s00018-008-8388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trent JT, Kirsner RS. Wounds and malignancy. Adv Skin Wound Care. 2003;16:31–34. doi: 10.1097/00129334-200301000-00014. [DOI] [PubMed] [Google Scholar]
- 82.Liu Z, Ren L, Tian J, Liu N, Hu Y, Zhang P. Comprehensive analysis of long noncoding RNAs and messenger RNAs expression profiles in patients with Marjolin ulcer. Med Sci Monit. 2018;24:7828–7840. doi: 10.12659/MSM.911177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goustin AS, Thepsuwan P, Kosir MA, Lipovich L. The growth-arrest-specific (GAS)-5 long non-coding RNA: a fascinating lncRNA widely expressed in cancers. Noncoding RNA. 2019;5:E46. doi: 10.3390/ncrna5030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sawaya AP, Pastar I, Stojadinovic O, Lazovic S, Davis SC, Gil J, Kirsner RS, Tomic-Canic M. Topical mevastatin promotes wound healing by inhibiting the transcription factor c-Myc via the glucocorticoid receptor and the long non-coding RNA Gas5. J Biol Chem. 2018;293:1439–1449. doi: 10.1074/jbc.M117.811240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat 1: its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–1714. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cooper DR, Wang C, Patel R, Trujillo A, Patel NA, Prather J, Gould LJ, Wu MH. Human adipose-derived stem cell conditioned media and exosomes containing MALAT1 promote human dermal fibroblast migration and ischemic wound healing. Adv Wound Care (New Rochelle) 2018;7:299–308. doi: 10.1089/wound.2017.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu XQ, Duan LS, Chen YQ, Jin XJ, Zhu NN, Zhou X, Wei HW, Yin L, Guo JR. lncRNA MALAT1 accelerates wound healing of diabetic mice transfused with modified autologous blood via the HIF-1α signaling pathway. Mol Ther Nucleic Acids. 2019;17:504–515. doi: 10.1016/j.omtn.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Tao SC, Rui BY, Wang QY, Zhou D, Zhang Y, Guo SC. Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 2018;25:241–255. doi: 10.1080/10717544.2018.1425774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo JR, Yin L, Chen YQ, Jin XJ, Zhou X, Zhu NN, Liu XQ, Wei HW, Duan LS. Autologous blood transfusion augments impaired wound healing in diabetic mice by enhancing lncRNA H19 expression via the HIF-1α signaling pathway. Cell Commun Signal. 2018;16:84. doi: 10.1186/s12964-018-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li D, Kular L, Vij M, Herter EK, Li X, Wang A, Chu T, Toma MA, Zhang L, Liapi E, Mota A, Blomqvist L, Gallais Sérézal I, Rollman O, Wikstrom JD, Bienko M, Berglund D, Ståhle M, Sommar P, Jagodic M, Landén NX. Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration. Proc Natl Acad Sci USA. 2019;116:9443–9452. doi: 10.1073/pnas.1814097116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herter EK, Li D, Toma MA, Vij M, Li X, Visscher D, Wang A, Chu T, Sommar P, Blomqvist L, Berglund D, Ståhle M, Wikstrom JD, Xu Landén N. WAKMAR2, a long noncoding RNA downregulated in human chronic wounds, modulates keratinocyte motility and production of inflammatory chemokines. J Invest Dermatol. 2019;139:1373–1384. doi: 10.1016/j.jid.2018.11.033. [DOI] [PubMed] [Google Scholar]
- 92.Bernard JJ, Gallo RL, Krutmann J. Photoimmunology: how ultraviolet radiation affects the immune system. Nat Rev Immunol. 2019;19:688–701. doi: 10.1038/s41577-019-0185-9. [DOI] [PubMed] [Google Scholar]
- 93.Chen H, Weng QY, Fisher DE. UV signaling pathways within the skin. J Invest Dermatol. 2014;134:2080–2085. doi: 10.1038/jid.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kripke ML, Cox PA, Alas LG, Yarosh DB. Pyrimidine dimers in DNA initiate systemic immunosuppression in UV-irradiated mice. Proc Natl Acad Sci USA. 1992;89:7516–7520. doi: 10.1073/pnas.89.16.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vink AA, Moodycliffe AM, Shreedhar V, Ullrich SE, Roza L, Yarosh DB, Kripke ML. The inhibition of antigen-presenting activity of dendritic cells resulting from UV irradiation of murine skin is restored by in vitro photorepair of cyclobutane pyrimidine dimers. Proc Natl Acad Sci USA. 1997;94:5255–5260. doi: 10.1073/pnas.94.10.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garssen J, van Steeg H, de Gruijl F, de Boer J, van der Horst GT, van Kranen H, van Loveren H, van Dijk M, Fluitman A, Weeda G, Hoeijmakers JH. Transcription-coupled and global genome repair differentially influence UV-B-induced acute skin effects and systemic immunosuppression. J Immunol. 2000;164:6199–6205. doi: 10.4049/jimmunol.164.12.6199. [DOI] [PubMed] [Google Scholar]
- 97.Vieyra-Garcia PA, Wolf P. From early immunomodulatory triggers to immunosuppressive outcome: therapeutic implications of the complex interplay between the wavebands of sunlight and the skin. Front Med (Lausanne) 2018;5:232. doi: 10.3389/fmed.2018.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim KH, Kim HJ, Lee TR. Epidermal long non-coding RNAs are regulated by ultraviolet irradiation. Gene. 2017;637:196–202. doi: 10.1016/j.gene.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 99.Zeng Q, Wang Q, Chen X, Xia K, Tang J, Zhou X, Cheng Y, Chen Y, Huang L, Xiang H, Cao K, Zhou J. Analysis of lncRNAs expression in UVB-induced stress responses of melanocytes. J Dermatol Sci. 2016;81:53–60. doi: 10.1016/j.jdermsci.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 100.Zheng Y, Xu Q, Peng Y, Gong Z, Chen H, Lai W, Maibach HI. Expression profiles of long noncoding RNA in UVA-induced human skin fibroblasts. Skin Pharmacol Physiol. 2017;30:315–323. doi: 10.1159/000477972. [DOI] [PubMed] [Google Scholar]
- 101.Yo K, Rünger TM. UVA and UVB induce different sets of long noncoding RNAs. J Invest Dermatol. 2017;137:769–772. doi: 10.1016/j.jid.2016.10.041. [DOI] [PubMed] [Google Scholar]
- 102.Peng Y, Song X, Zheng Y, Wang X, Lai W. Circular RNA profiling reveals that circCOL3A1-859267 regulate type I collagen expression in photoaged human dermal fibroblasts. Biochem Biophys Res Commun. 2017;486:277–284. doi: 10.1016/j.bbrc.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 103.Lei L, Zeng Q, Lu J, Ding S, Xia F, Kang J, Tan L, Gao L, Kang L, Cao K, Zhou J, Xiao R, Chen J, Huang J. MALAT1 participates in ultraviolet B-induced photo-aging via regulation of the ERK/MAPK signaling pathway. Mol Med Rep. 2017;15:3977–3982. doi: 10.3892/mmr.2017.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y, Dobozy A, Kemeny L, Szell M. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 105.Hall JR, Messenger ZJ, Tam HW, Phillips SL, Recio L, Smart RC. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015;6:e1700. doi: 10.1038/cddis.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin S, Yang X, Li J, Yang W, Ma H, Zhang Z. p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol Cancer. 2019;18:38. doi: 10.1186/s12943-019-0993-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reichrath J, Saternus R, Vogt T. Endocrine actions of vitamin D in skin: Relevance for photocarcinogenesis of non-melanoma skin cancer, and beyond. Mol Cell Endocrinol. 2017;453:96–102. doi: 10.1016/j.mce.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 108.Jiang YJ, Bikle DD. LncRNA: a new player in 1α, 25(OH)(2) vitamin D(3) /VDR protection against skin cancer formation. Exp Dermatol. 2014;23:147–150. doi: 10.1111/exd.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Expression profiles of long noncoding RNAs in cutaneous squamous cell carcinoma. Epigenomics. 2016;8:501–518. doi: 10.2217/epi-2015-0012. [DOI] [PubMed] [Google Scholar]
- 110.Piipponen M, Nissinen L, Farshchian M, Riihilä P, Kivisaari A, Kallajoki M, Peltonen J, Peltonen S, Kähäri VM. Long noncoding RNA PICSAR promotes growth of cutaneous squamous cell carcinoma by regulating ERK1/2 activity. J Invest Dermatol. 2016;136:1701–1710. doi: 10.1016/j.jid.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 111.Volders PJ, Lefever S, Baute S, Nuytens J, Vanderheyden K, Menten B, Mestdagh P, Vandesompele J. Targeted genomic screen reveals focal long non-coding RNA copy number alterations in cancer cell lines. Noncoding RNA. 2018;4:E21. doi: 10.3390/ncrna4030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, Almeida R, Zhernakova A, Reinmaa E, Võsa U, Hofker MH, Fehrmann RS, Fu J, Withoff S, Metspalu A, Franke L, Wijmenga C. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lambert SR, Mladkova N, Gulati A, Hamoudi R, Purdie K, Cerio R, Leigh I, Proby C, Harwood CA. Key differences identified between actinic keratosis and cutaneous squamous cell carcinoma by transcriptome profiling. Br J Cancer. 2014;110:520–529. doi: 10.1038/bjc.2013.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, Chapman PB, Kim MJ, Hayward R, Martin M, Yang H, Wang Q, Hilton H, Hang JS, Noe J, Lambros M, Geyer F, Dhomen N, Niculescu-Duvaz I, Zambon A, Niculescu-Duvaz D, Preece N, Robert L, Otte NJ, Mok S, Kee D, Ma Y, Zhang C, Habets G, Burton EA, Wong B, Nguyen H, Kockx M, Andries L, Lestini B, Nolop KB, Lee RJ, Joe AK, Troy JL, Gonzalez R, Hutson TE, Puzanov I, Chmielowski B, Springer CJ, McArthur GA, Sosman JA, Lo RS, Ribas A, Marais R. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Einspahr JG, Calvert V, Alberts DS, Curiel-Lewandrowski C, Warneke J, Krouse R, Stratton SP, Liotta L, Longo C, Pellacani G, Prasad A, Sagerman P, Bermudez Y, Deng J, Bowden GT, Petricoin EF., 3rd Functional protein pathway activation mapping of the progression of normal skin to squamous cell carcinoma. Cancer Prev Res (Phila) 2012;5:403–413. doi: 10.1158/1940-6207.CAPR-11-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bachelor MA, Bowden GT. UVA-mediated activation of signaling pathways involved in skin tumor promotion and progression. Semin Cancer Biol. 2004;14:131–138. doi: 10.1016/j.semcancer.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 117.Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9:E52. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Piipponen M, Heino J, Kähäri VM, Nissinen L. Long non-coding RNA PICSAR decreases adhesion and promotes migration of squamous carcinoma cells by downregulating α2β1 and α5β1 integrin expression. Biol Open. 2018;7:bio037044. doi: 10.1242/bio.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Y, Gao L, Ma S, Ma J, Wang Y, Li S, Hu X, Han S, Zhou M, Zhou L, Ding Z. MALAT1-KTN1-EGFR regulatory axis promotes the development of cutaneous squamous cell carcinoma. Cell Death Differ. 2019;26:2061–2073. doi: 10.1038/s41418-019-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nassar D, Latil M, Boeckx B, Lambrechts D, Blanpain C. Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat Med. 2015;21:946–954. doi: 10.1038/nm.3878. [DOI] [PubMed] [Google Scholar]
- 121.Casanova ML, Larcher F, Casanova B, Murillas R, Fernández-Aceñero MJ, Villanueva C, Martínez-Palacio J, Ullrich A, Conti CJ, Jorcano JL. A critical role for ras-mediated, epidermal growth factor receptor-dependent angiogenesis in mouse skin carcinogenesis. Cancer Res. 2002;62:3402–3407. [PubMed] [Google Scholar]
- 122.Dlugosz AA, Cheng C, Williams EK, Darwiche N, Dempsey PJ, Mann B, Dunn AR, Coffey RJ, Jr, Yuspa SH. Autocrine transforming growth factor alpha is dispensible for v-rasHa-induced epidermal neoplasia: potential involvement of alternate epidermal growth factor receptor ligands. Cancer Res. 1995;55:1883–1893. [PubMed] [Google Scholar]
- 123.Mei XL, Zhong S. Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin Med J (Engl) 2019;132:454–465. doi: 10.1097/CM9.0000000000000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Merlino GT, Xu YH, Ishii S, Clark AJ, Semba K, Toyoshima K, Yamamoto T, Pastan I. Amplification and enhanced expression of the epidermal growth factor receptor gene in A431 human carcinoma cells. Science. 1984;224:417–419. doi: 10.1126/science.6200934. [DOI] [PubMed] [Google Scholar]
- 125.Rognoni E, Walko G. The roles of YAP/TAZ and the Hippo pathway in healthy and diseased skin. Cells. 2019;8:E411. doi: 10.3390/cells8050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen L, Chen Q, Kuang S, Zhao C, Yang L, Zhang Y, Zhu H, Yang R. USF1-induced upregulation of LINC01048 promotes cell proliferation and apoptosis in cutaneous squamous cell carcinoma by binding to TAF15 to transcriptionally activate YAP1. Cell Death Dis. 2019;10:296. doi: 10.1038/s41419-019-1516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Debaugnies M, Sánchez-Danés A, Rorive S, Raphaël M, Liagre M, Parent MA, Brisebarre A, Salmon I, Blanpain C. YAP and TAZ are essential for basal and squamous cell carcinoma initiation. EMBO Rep. 2018;19:e45809. doi: 10.15252/embr.201845809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Piipponen M, Nissinen L, Riihilä P, Farshchian M, Kallajoki M, Peltonen J, Peltonen S, Kähäri VM. p53-regulated long non-coding RNA PRECSIT promotes progression of cutaneous squamous cell carcinoma via STAT3 signaling. Am J Pathol. 2019;190:503–517. doi: 10.1016/j.ajpath.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 130.Li F, Liao J, Duan X, He Y, Liao Y. Upregulation of LINC00319 indicates a poor prognosis and promotes cell proliferation and invasion in cutaneous squamous cell carcinoma. J Cell Biochem. 2018;119:10393–10405. doi: 10.1002/jcb.27388. [DOI] [PubMed] [Google Scholar]
- 131.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Long-noncoding RNAs in basal cell carcinoma. Tumor Biol. 2016;37(8):10595–10608. doi: 10.1007/s13277-016-4927-z. [DOI] [PubMed] [Google Scholar]