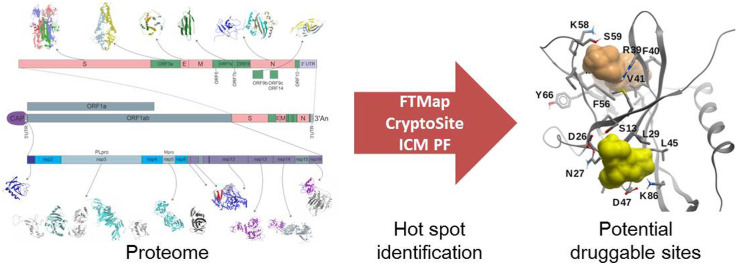
Abstract
The infectious coronavirus disease (COVID-19) pandemic, caused by the coronavirus SARS-CoV-2, appeared in December 2019 in Wuhan, China, and has spread worldwide. As of today, more than 46 million people have been infected and over 1.2 million fatalities. With the purpose of contributing to the development of effective therapeutics, we performed an in silico determination of binding hot-spots and an assessment of their druggability within the complete SARS-CoV-2 proteome. All structural, non-structural, and accessory proteins have been studied, and whenever experimental structural data of SARS-CoV-2 proteins were not available, homology models were built based on solved SARS-CoV structures. Several potential allosteric or protein-protein interaction druggable sites on different viral targets were identified, knowledge that could be used to expand current drug discovery endeavors beyond the currently explored cysteine proteases and the polymerase complex. It is our hope that this study will support the efforts of the scientific community both in understanding the molecular determinants of this disease and in widening the repertoire of viral targets in the quest for repurposed or novel drugs against COVID-19.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, Druggability, Drug discovery, Binding hot-spots
Graphical abstract
1. Introduction
In the past decades, two highly pathogenic coronaviruses (CoVs) known as the severe acute respiratory syndrome coronavirus (SARS-CoV), and the Middle East respiratory syndrome coronavirus (MERS-CoV), triggered global epidemics in 2003 and 2012, respectively (Paules et al., 2020). A new CoV infectious disease (COVID-19), caused by the new pathogenic SARS-CoV-2 appeared in December 2019 in Wuhan, China, spread rapidly worldwide, and was declared a pandemic on March 11th, 2020 by the World Health Organization (WHO). As this work is being written, there have been over 46 million infected cases around the world, with more than 1.2 million fatalities.
Although SARS-CoV-2, SARS-CoV, and MERS-CoV belong to the same genus, SARS-CoV-2 seems to be associated with milder infections. Their fatality rates were 2.3%, 9.5%, and 34.4%, and their basic reproductive numbers (R0) 2.0–2.5, 1.7–1.9, and <1, respectively (Petrosillo et al., 2020). Like SARS and MERS, the typical clinical presentation of severe cases of COVID-19 is pneumonia with fever, cough, and dyspnea. The vast majority of COVID-19 patients have mild disease (around 80%), a lesser percentage (~15%) have moderate disease (with dyspnea, hypoxia or pneumonia with over 50% lung parenchyma involvement), and even a smaller proportion (~5%) of patients develop severe disease with respiratory failure, shock or multiorgan failure (Wu et al., 2020a).
Although the optimal strategy to control this disease would be through vaccination capable of generating a long-lasting and protective immune response, there is an urgent need for the development of a treatment, dictated by: i) the rapid spread of the virus; ii) the increasing number of infected patients with a moderate to severe symptoms that must be treated worldwide; iii) the associated risk of death and depletion of the health systems, particularly in low-income countries.
In terms of antiviral strategies, while no specific therapeutics are available, approved or experimental drugs developed for other diseases are being tested in different clinical trials, in an effort to rapidly find accessible treatments with already established safety profiles, through drug repurposing strategies (Barrantes, 2020; Cavasotto and Di Filippo, 2020; Villoutreix et al., 2020; Zhou et al., 2020). Most of these strategies have been focused in targeting the two viral proteases, and the RNA-dependent RNA polymerase (RdRp) complex. However, protease inhibitors might bind to host proteases, thus resulting in cell toxicity (Kang et al., 2020), and the effectivity of nucleoside inhibitors targeting RdRp is decreased by the highly efficient SARS-CoV-2 proofreading machine. Therefore, alternative therapeutic options to fight COVID-19 are urgently needed.
In this context, the characterization of the druggability of the SARS-CoV-2 proteome is of the utmost importance. In this work, we surveyed the functional role of each SARS-CoV-2 protein, analyzed its key structural features, and using either experimental structural data or homology models, we performed a thorough in silico druggability assessment of the viral proteome. Current drug discovery efforts on the proteases and the RdRp complex with in vitro or in vivo experimental validation are also briefly commented, mainly as a complement to our description of targetability. In addition to the catalytic sites of several proteins, we found potential druggable allosteric and protein-protein interaction (PPI) sites throughout the whole proteome.
We do hope our contribution will help in the development of fast and effective SARS-CoV-2-centered therapeutic options, which considering the high similarity among CoVs, might also be effective against related viruses. Moreover, since it is unlikely that SARS-CoV-2 will be the last CoV to threaten global health, those therapeutics might be instrumental in fighting future epidemics.
2. Computational Methods
2.1. Molecular system setup
All structures were downloaded from the Protein Data Bank (PDB) and prepared using the ICM software (MolSoft LLC, San Diago CA, 2019) in a similar fashion as in earlier works (Cavasotto and Aucar, 2020). Succinctly, hydrogen atoms were added, followed by a short local energy minimization in the torsional space; the positions of polar and water hydrogens were determined by optimizing the hydrogen-bonding network, and then all water molecules were deleted. All Asp and Glu residues were assigned a −1 charge, and all Arg and Lys residues were assigned a +1 charge. Histidine tautomers were chosen according to their corresponding hydrogen-bonding pattern.
2.2. Homology modeling
A crude model was built using the backbone structure of the template, and then refined through local energy minimization using ICM (Domenech et al., 2013). To avoid pocket collapse, and taking into account the complete binding site conservation, whenever available, ligands were kept within the binding site during the refinement process, in a ligand-steered modeling fashion (Cavasotto and Palomba, 2015; Phatak et al., 2010).
2.3. Hot-spots and cryptic sites determination
Identification of binding energy hot-spots was performed using FTMap (https://ftmap.bu.edu/) (Kozakov et al., 2015a). The FTMap algorithm samples a library of 16 small organic probe molecules of different sizes, shapes and polarities on the protein using rigid docking. For each probe, all the poses generated are clustered using a 4 Å clustering radius, and then clusters are ranked on the basis of their average energy, keeping the six lowest-energy clusters for each probe. After the probe clusters of the 16 molecules have been generated, they are then re-clustered into hot-spots or consensus sites (CSs) based on vicinity. These CSs are ranked on the basis of the number of their probe clusters. The program offers a protein-protein interaction (PPI) mode, where the aim is to identify hot-spots on protein-protein interfaces. To identify binding sites, the top ranking CS is considered the kernel of the site, and is expanded by adding neighboring CSs with a center-to-center distance (CD) of less than 8 Å to any existing CS, until no further expansion is possible. The binding site is defined as those residues within 4 Å of any probe of the CSs used. The top ranking CS is removed and the procedure repeated starting from the second ranking CS, and so forth. Considering the druggability criteria (see below), only CSs with at least 13 probe clusters were considered to be expanded.
Cryptic sites on proteins were determined using CryptoSite (Cimermancic et al., 2016) (https://modbase.compbio.ucsf.edu/cryptosite/); cryptic sites are those formed only in ligand-bound structures, but usually “hidden” in unbound structures. Taking into account the analysis of the druggability of cryptic sites, only those cryptic sites with at least 16 probe clusters (determined with FTMap) within 5 Å were considered as potentially druggable (Vajda et al., 2018).
The ICM Pocket Finder method (implemented in the ICM software) predicts the position and shapes of cavities and clefts using a transformation of the Lennard-Jones potential by convolution with a Gaussian kernel of a certain size, and construction of equipotential surfaces along the maps of a binding potential(Abagyan and Kufareva, 2009).
2.4. Druggability criteria
The druggability of a site was characterized based on the CSs generated by FTMap in terms of: i) the number of probe clusters in the primary hot-spot (S), ii) if there are one or more secondary spots with a CD < 8 Å from the primary spot, and iii) the maximum dimension (MaxD) of the connected ensemble (measured as the distance between the two most distant probe atoms within the probe clusters) (Kozakov et al., 2015b). In general, a site was considered druggable if S ≥ 16, CD < 8 Å, and MaxD ≥10 Å; non-druggable if S < 13 or MaxD <7 Å; borderline druggable if 13 ≤ S < 16 and CD < 8 Å, or 13 ≤ S < 16, CD ≥ 8 Å, and MaxD ≥10 Å.
3. The SARS-CoV-2 viral machinery
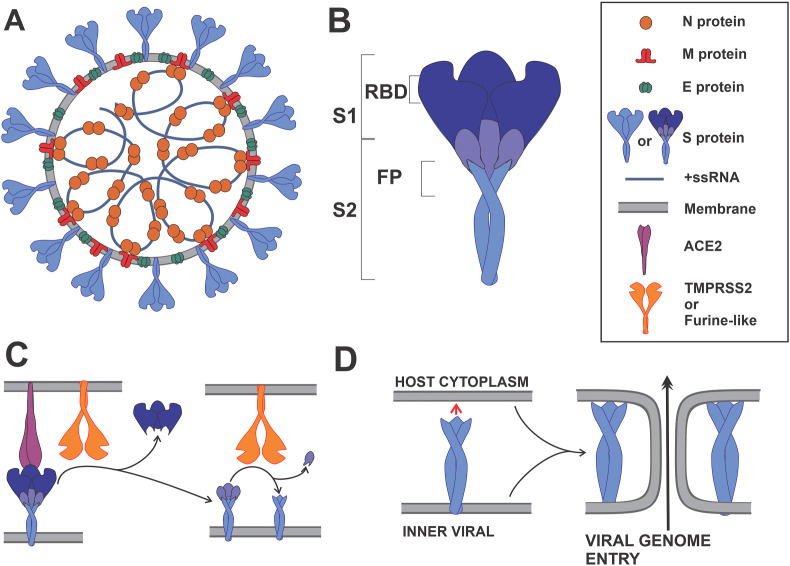
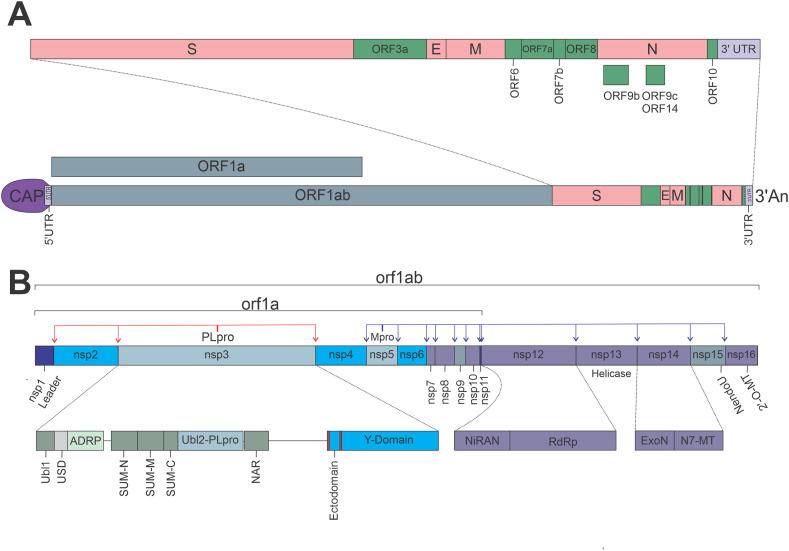
The SARS-CoV-2 genetically clusters with the β-coronavirus genus, and is phylogenetically related to SARS-CoV. It is a positive single-strain RNA (+ssRNA), of ~30 kbs (Wu et al., 2020), and it is enclosed in a spherical lipidic bilayer membrane. About the first two-thirds of the viral RNA genome contain the open reading frames (ORFs) 1a and 1ab, which are translated into the polyproteins (pp) 1a and 1ab, which contain the non-structural proteins (nsp's). The remaining viral genome encode accessory proteins and four essential structural proteins (Cui et al., 2019): the spike (S) receptor binding glycoprotein (ORF2); the nucleocapsid (N) protein (ORF9a); the membrane (M) protein (ORF5), which is a transmembrane (TM) protein involved in the interaction with N; and a small envelope (E) protein (ORF4), which participates in membrane stability and virus assembly (Siu et al., 2008) (Fig. 1 ). The accessory proteins act co-opting host factors, shutting down host functions to redirect resources to viral replication, avoiding immune responses, and inducing pathogenicity.
Fig. 1.
Schematic representation of a SARS-CoV-2 viral particle and key steps in virus entry. (A) The N, S, E and M proteins are represented in their oligomeric state. N protein dimers bind +ssRNA, forming the nucleocapsid. The nucleocapsid is surrounded by the viral membrane that contains S, E and M proteins. The M protein is shown interacting with the S, E and N proteins. (B) Domain localization of the S protein, showing the S1 and S2 fragments; S1 contains the receptor binding domain (RBD), and S2 the fusion peptide (FP). (C) Angiotensin I converting enzyme 2 (ACE2) recognition by RBD, and the subsequent S proteolytic activation by a Furine-like protease or TMPRSS2. (D) Viral and host membrane fusion induction by the exposed FP.
Once in a new host, the viral life-cycle consists basically in four stages: virus entry into a host cell, RNA translation and protein processing, RNA replication, and viral particle assembly and release.
During virus entry, the multidomain S protein binds to the host receptor ACE2, it is proteolyzed by host proteases and activated, and thus triggers a series of events that result in the fusion of the viral membrane with the host membrane, and the subsequent release of the viral genome into the host cytoplasm (Cai et al., 2020) (Fig. 1).
The second step in the virus cycle is the translation of viral structural, non-structural, and accessory proteins. Since CoVs particles do not contain any replicase protein, the translation of viral proteins is the critical step in producing all the necessary machinery for virus RNA replication and assembly. The nsp's are translated as the large pp pp1a and pp1b, and are cleaved by viral proteases to produce fully active proteins. CoVs possess the papain-like protease (PLpro), and the 3-chymotrypsin-like protease (3CLpro) or main protease (Mpro). The PLpro (a domain of nsp3) cleaves nsp1, nsp2 and itself; while Mpro (nsp5) cleaves the remaining nsp's, including itself, resulting in total 16 nsp's (Fig. 2 B).
Fig. 2.
SARS-CoV-2 genome organization. (A) Open reading frames (ORF) distribution in SARS-CoV-2 genome. (B) Non-structural proteins (nsp's) distribution in orf1a and orf1ab, detailing multidomain organization of nsp3, nsp12, and nsp14; red and blue arrows indicate PLpro and Mpro cleaving sites, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In CoVs the RNA replication takes place in double membrane vesicles (DMVs), colloquially known as viral factories (Snijder et al., 2020). DMVs are membrane structures derived from the Endoplasmic Reticulum (ER), formed through the action of proteins nsp3, nsp4 and nsp6 (Angelini et al., 2013). Due to the ability of some nsp3 domains to bind RNA and to the N protein, the viral RNA and the entire replication machinery are located in the DMVs (Snijder et al., 2020). Furthermore, DMVs constitute a barrier that prevents viral RNA from being detected by the cell's antiviral response machinery (den Boon and Ahlquist, 2010).
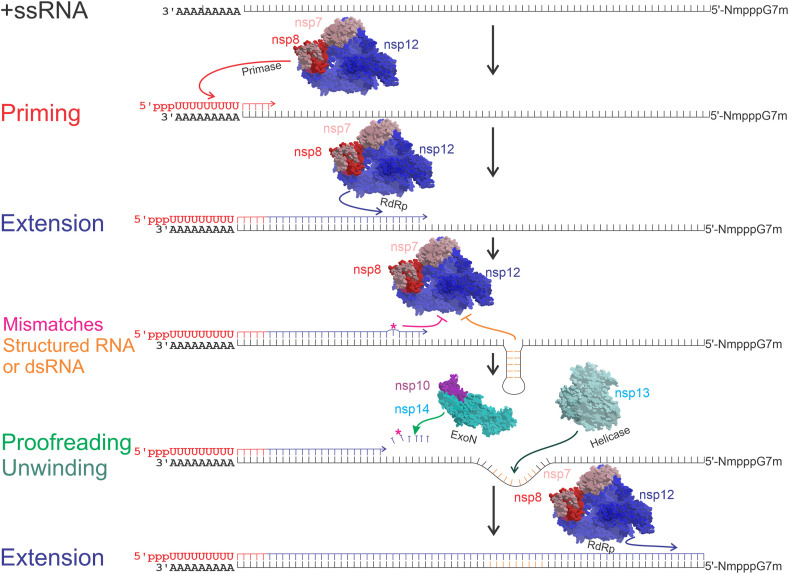
The third step in the viral cycle is the replication of the viral RNA. As all +ssRNA viruses, the genome is copied to a reverse and complementary intermediate -ssRNA, which is copied back to a complete +ssRNA genome and several shorter sub-genomic +ssRNA, that are used as additional templates to translate high amounts of certain structural and accessory proteins, necessary during the viral assembly (Wu et al., 2020b). CoVs require the RdRp for replication, represented by the multi-protein complex nsp12-nsp8-nsp7. The nsp12 has the RdRp activity, nsp8 is the putative primase, while nsp7 acts as a co-factor (Peng et al., 2020). CoVs possess a helicase (nsp13) that unwinds structured or double-strand RNA (dsRNA) to allow the RdRp to proceed unhindered, thus achieving high efficiency and accuracy in RNA replication; they also have a 3′-5′ exoribonuclease (ExoN, nsp14) that is capable of proofreading activity during RNA synthesis, thus lowering the rate of nucleoside misincorporation, while also enhancing resistance to nucleoside analogs (Bouvet et al., 2012; Jia et al., 2019) (Fig. 3 ).
Fig. 3.
Representative steps during the synthesis of a complementary RNA strain. Priming of the complementary strain, in this case, a -ssRNA, is catalyzed by nsp8 (red letters and arrows). Primer dependent RNA extension, catalyzed by nsp7-nsp8-nsp12 complex (blue lines and arrows). Magenta asterisk (*) and arrow represent a mis-incorporated nucleotide or nucleotide analog, and nonobligate RNA chain termination, respectively. Orange lines and arrows represent nucleotides forming dsRNA or structured RNA, and extension inhibition, respectively. Light green arrow represents nucleotide excision by nsp14 3′-5′ exonuclease (ExoN). Dark green arrow represents the unwinding activity of nsp13 helicase. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
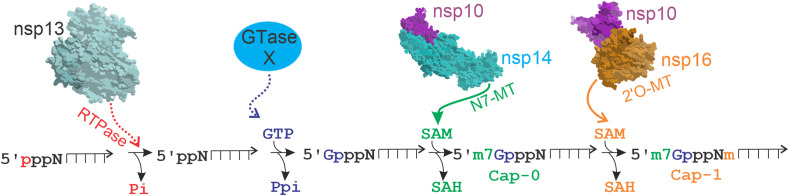
Additionally, for the viral RNA to be translated efficiently and to avoid quick degradation, CoVs have poly-adenylation [poly(A)] and capping modifications, much like host mRNAs. The poly(A) modification is apparently catalyzed by nsp8, which has TAT activity (Tvarogová et al., 2019). The methyl capping consists of a sequence of reactions: 1) the terminal γ-phosphate is removed from the 5′-triphosphate end of RNA by a RNA 5′-triphosphatase (RTPase), probably nsp13; 2) an uncharacterized RNA guanylyltransferase (GTase) adds a GMP molecule to the 5′RNA, resulting in the formation of GpppN-RNA; 3) the GpppN-RNA is methylated at the N7 position of the guanosine by an N7-methyltransferase (N7-MTase, nsp14-nsp10 dimer), yielding m7GpppN (cap-0); 4) finally, a 2′-O-MTase (nsp16-nsp10 dimer) methylates the 2′-O position of the first nucleotide's ribose of m7GpppN, yielding m7GpppNm (cap-1) (Snijder et al., 2016) (Fig. 4 ).
Fig. 4.
Viral RNA CAP synthesis. The capping of viral RNA has 4 enzymatic steps. i) Removal of the first phosphate of the 5′-triphosphate end of RNA by a RNA 5′-triphosphatase (RTPase), probably nsp13 (red arrow); ii) Addition of a GMP molecule to the 5′RNA by an unconfirmed guanylyltransferase (GTase) (blue); iii) Methylation of GpppN-RNA at the N7 position by nsp14 N7-methyltransferase (N7-MTase), forming the cap-0 (green arrow); iv) Methylation at the 2′-O position of the first nucleotide's ribose by the 2′-O-methyltransferase (2′-O-Mtase) (nsp16), yielding the cap-1 (orange arrow). Dotted or arrow lines indicate unconfirmed enzymes. Pi: phosphate, Ppi: pyrophosphate, SAM: S-Adenosyl methionine, SAH: S-Adenosyl homocysteine, m: methyl group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The last step in the viral cycle is the assembly of the viral particles and the release of the viral progeny. During this step, several N proteins bind and compact each copy of the genomic + ssRNA, forming the nucleocapsid. Proteins S, E, and M, accumulated in the ER-Golgi intermediate compartment (ERGIC) (Knoops et al., 2008), bind the nucleocapsid by direct M-N protein interactions (He et al., 2004), and then through the action of E and M proteins on the ERGIC membrane a viral particle is formed in a secretory compartment (Knoops et al., 2008). Finally, the viral progeny particle or virion is released by the exocytosis pathway (Fehr and Perlman, 2015), and the viral cycle is completed.
4. Druggability characterization of SARS-CoV-2 proteins
For each SARS-CoV-2 protein, we highlighted its relevant functional roles and main structural features, and then searched for druggable sites within the protein's structure, analyzing the possible functional consequences of small-molecules binding to these sites. To find and assess the targetability of potential binding sites (including allosteric ones), we used FTMap to identify binding hot-spots on all available (or homology modeled) SARS-CoV-2 structures. FTMap samples a library of small organic probes of different molecular properties through docking, thus identifying consensus sites (CSs) or hot-spots (see Computational Methods for details). Hot-spots are small regions within a binding site considered to strongly contribute to the ligand-binding free energy. This analysis was complemented by the use of ICM Pocket Finder, and by the detection of cryptic sites, which are those usually “hidden” sites in unligated structures, but present only in ligand-bound structures. The strength of a CS is related to the number of fragment-like probe clusters it contains. A strong CS and nearby hot-spots define a potential binding site (Kozakov et al., 2015a), which may be considered druggable if conditions on the number of probe clusters within the primary CS and the presence of other hot-spots in the vicinity are satisfied (see Computational Methods); these conditions mean that a drug-like molecule could bind the receptor with at least micromolar affinity (Kozakov et al., 2015a). It is clear that this assessment of druggability does not necessarily mean that a ligand binding at that site will actually exert an observable biological effect. We also summarized current drug repurposing efforts on the two cysteine proteases and the polymerase, but only including compounds with some kind of experimental validation.
4.1. Non-structural proteins
4.1.1. Non-structural protein 3 (nsp3)
Nsp3 is the largest SARS-CoV-2 protein, with 1945 amino acids. It has at least seven tandem macrodomains involved in different functions. These nsp3 domains are listed in Table S1, and each one is analyzed in the subsequent sections.
4.1.2. The papain-like protease (PLpro, nsp3 domain)
The inhibition of viral proteases might not only be important to block viral polyproteins, but it would be a way to interfere with viral-induced immunosuppression processing, since these proteases also act on host proteins, and are partly responsible for the host cell shutdown. The PLpro has a ubiquitin-like subdomain (Ubl2) located within its N-terminal portion, which is not necessary for PLpro activity in SARS (Clasman et al., 2017), but it is probably involved in Nuclear Factor kappa-light-chain-enhancer of activated B cells (NFκB) signaling, affecting sensibility to interferon (IFN) (Frieman et al., 2009). Through recognition of the consensus cleavage LXGG motif, PLpro hydrolyzes the peptide bond on the carboxyl side of the Gly at the P1 position, thus releasing nsp1, nsp2, and nsp3 proteins. In the case of SARS-CoV, in vitro studies have also shown that this protease has two other proteolytic activities, hydrolyzing ubiquitin (Ub) and ubiquitin-like protein Interferon-Stimulated Gene product 15 (ISG15), from cellular proteins such as the Interferon Regulatory Factor 3 (IRF3), thus suppressing the host's innate immune response (Matthews et al., 2014).
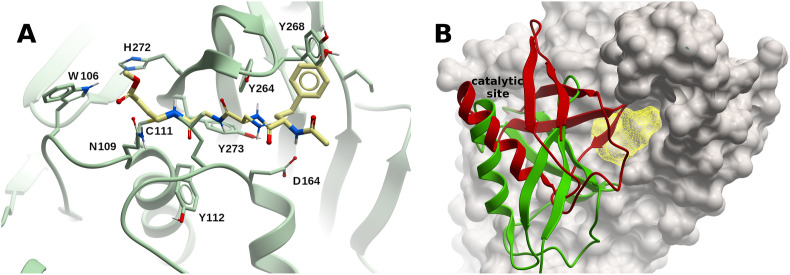
The crystal structures of SARS-CoV-2 PLpro in complex with inhibitors VIR250 and VIR251 covalently attached to the catalytic C111 were solved at 2.8 Å (VIR250, PDB 6WUU) and 1.65 Å (VIR251, PDB 6WX4), respectively (Figs. 5A and S1). These inhibitors displayed similar activities towards SARS-CoV and SARS-CoV-2 PLpros, but a weaker activity towards the MERS-CoV PLpro. Two wild type apo structures (PDBs 6W9C and 6WZU, at 2.7 Å and 1.8 Å, respectively), and the C111S apo structure (PDB 6WRH, at 1.6 Å) have overall main chain RMSD values of ~0.6 Å compared to the lowest resolution PLpro-VIR251 structure, displaying a partial collapse of the catalytic binding site. SARS-CoV-2 PLpro was crystallized also in complex with Ub at 2.7 Å (PDB 6XAA), and with the C-terminal part of ISG15 at 2.9 Å (PDB 6XA9) (Fig. S1). Both structures are very similar to 6XW4, with main chain RMSD values of ~0.5 Å (all available PLpro structures are listed in Table S1).
Fig. 5.
Structure and binding sites of SARS-CoV-2 PLpro. (A) Complex of PLpro with inhibitor VIR251 covalently bound to C111 within the catalytic binding site (PDB 6XW4). (B) Potential binding site (yellow mesh) on SARS-CoV-2 PLpro (ivory surface). Ubiquitin (red) and ISG15 (green) are displayed in ribbon representation. The N-terminus of these two proteins are inserted within the catalytic site. A small-molecule binding to the predicted site would interfere with ubiquitin binding, but not with ISG15. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In agreement with another work (Freitas et al., 2020), it was shown that while SARS-CoV-2 PLpro retained de-ISGylating activity similar to its SARS-CoV counterpart, its hydrolyzing ability of diUbK48 was comparatively decreased (Rut et al., 2020). Interestingly, SARS-CoV-2 PLpro displays a preference for ISG15s from certain species including humans (Freitas et al., 2020). It has yet to be determined whether this functional difference has any implication in the ability of SARS-CoV-2 to evade the human innate immune system.
The catalytic site (Fig. 5 ) would clearly be the first choice for drug discovery (see below an account of successful cases targeting this site). Using FTMap, a borderline druggable binding site delimited by residues R166, L185, L199, V202, E203, M206-M208, and K232, was identified; a small-molecule binding to this site might interfere with Ub binding, but not with ISG15 binding (Fig. 5B). A small hot-spot could also be found limited by amino acids P59, R65, and T74-F79. This site is close to the Ub S2 binding site, and from the structural model of the SARS-CoV-2 PLpro-diUbK48 [generated using the corresponding SARS-CoV structure (PDB 5E6J)], a small-molecule binding to it might interfere with Ub binding. Two other alternative potential binding sites were identified in PLpro, delimited by residues: i) S213-E215, K218, Y252, L254, K255, T258-T260, V304, Y306, and E308; ii) L121-I124, L126-F128, L133, Q134, Y137, Y138, and R141 (Fig. S1). These sites are far from the catalytic, Ub1-, and Ub2-binding sites, so that, even in the case a small-molecule could bind to them, further evidence would be needed to determine whether those amino acids have a functional role, and to assess whether those sites could display allosteric modulation potential.
Considering that the residues lining the catalytic sites of SARS-CoV and SARS-CoV-2 PLpros are identical, Freitas et al. evaluated naphtalene-based SARS-CoV non-covalent inhibitors on SARS-CoV-2, and found that two compounds, GRL-0617 and 6, exhibited IC50 values of 2.4 μM and 5.0 μM, respectively (their corresponding values in SARS-CoV PLpro were 600 nM and 2.6 μM), displaying EC50 values of 27.6 and 21.0 μM, respectively, in antiviral activity assessment, with no cytotoxicity in cell cultures (Freitas et al., 2020). The characterization of the non-covalent inhibition between these ligands and SARS-CoV-2 PLpro was performed using a homology model based on SARS-CoV PLpro bound to GRL-0617 (PDB 3E9S). It should be stressed that a similar approach using MERS-CoV inhibitors (Lee et al., 2019) would not be so straightforward, considering the lower conservation of the catalytic binding site at the sequence level. It was also shown that the synthetic organoselenium drug molecule ebselen, which displays anti-inflammatory, anti-oxidant and cytoprotective activity in mammalian cells, covalently inhibits the enzymatic activity of SARS-CoV-2 PLpro, with an IC50 ~ 2 μM, exhibiting a weaker activity against its SARS-CoV counterpart (Welgarz Tomczak et al., 2020b). In a follow-up contribution, ebselen-derivatives were identified, displaying lower inhibition constants, in the range of 250 nM (Welgarz Tomczak et al., 2020a). Klemm et al. showed that benzodioxolane analogs 3j, 3k, and 5c, which inhibit SARS-CoV PLpro in the sub-micromolar range (Báez-Santos et al., 2014) displayed similar inhibitory activity against SARS-CoV-2 PLpro (Klemm et al., 2020). Two other compounds have been shown to inhibit SARS-CoV-2 PLpro in vitro, inhibiting viral production in cell culture: the approved chemotherapy agent 6-Thioguanine (Swaim et al., 2020), and the anti-dengue protease inhibitor mycophenolic acid (Kato et al., 2020).
4.1.3. Main protease (Mpro, nsp5)
The SARS-CoV-2 Mpro is a cysteine protease involved in most cleavage events within the precursor polyproteins, beginning with the autolytic cleavage of itself from pp1a and pp1ab (Fig. 2). The vital functional role of Mpro in the viral life cycle makes it an attractive antiviral target.
The active form of Mpro is a homodimer containing two protomers composed of three domains each (Jin et al., 2020a): domain I (residues F8–Y101), domain II (residues K102–P184), and domain III (residues T201–V303); domains II and III are connected by a long loop region (residues F185–I200). The Mpro has a Cys-His catalytic dyad (C145–H41), and the substrate binding site is located in the cleft between domains I and II (Fig. S2). The superposition of 12 crystal structures of Mpro from different species (Jin et al., 2020a) showed that the helical domain III and surface loops display the largest conformational variations, while the substrate-binding pocket is highly conserved among CoVs.
Crystal structures of SARS-CoV-2 Mpro were determined in complex with the Michael acceptor covalent inhibitor N3 (Fig. 6 A) at 2.1 Å resolution (PDB 6LU7), and 1.7 Å (PDB 7BQY), respectively (an up-to-date detailed list of Mpro solved structures is shown in Table S2). N3 inhibits the Mpro from multiple CoVs, and has been co-crystalized with Mpro in SARS-CoV (PDB 2AMQ), Infectious Bronchitis Virus (IBV) (PDB 2Q6F), Human coronavirus HKU1 (HCoV-HKU1) (PDB 3D23), Feline Infectious Peritonitis Virus (FIPV) (PDB 5EU8), Human coronavirus NL63 (HCoV-NL63) (PDB 5GWY), Porcine Epidemic Diarrhea Virus (PEDV) (PDB 5GWZ), and Mouse Hepatitis Virus (MHV) (PDB 6JIJ). In the SARS-CoV-2 Mpro-N3 complexes, the Sγ atom of C145 forms a covalent bond with the Cβ atom of the vinyl group of N3 (see Table S2 for available structures solved in the apo form, and with covalent and non-covalent inhibitors; more than a hundred crystal structures with bound fragment-like molecules are not included).
Fig. 6.
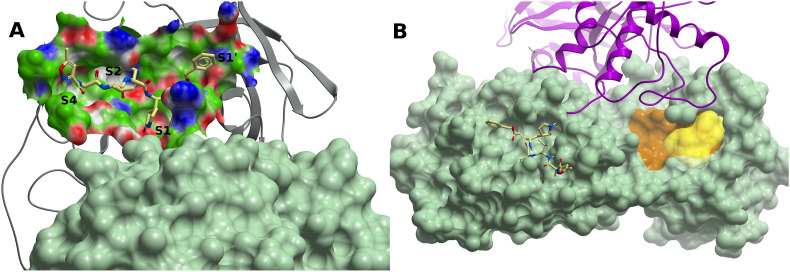
SARS-CoV-2 main protease Mpro binding sites. (A) Mpro (grey ribbon) in complex with peptide inhibitor N3 (PDB 6LU7). The protein subsites S1, S2, S4 and S1′ are labeled. The molecular surface of the other protomer is shown in light green. (B) A cryptic site (brown) on Mpro with a CS site nearby lined up by residues T199, Y237, Y239, L271, L272, G275, M276, and A285-L287 (yellow surface). Mpro is represented by a green molecular surface. N3 is also displayed within the catalytic site (light yellow carbons). The other protomer is represented in magenta ribbon. A small-molecule binding to this potential site might interfere with homodimerization. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The primary choice for drug discovery would be the catalytic site. Zhang et al. crystallized SARS-CoV-2 Mpro in its apo form (PDB 6Y2E) to guide the design of a novel α-ketoamide inhibitor displaying an IC50 = 0.67 μM; crystal structures of SARS-CoV-2 in complex with this inhibitor were determined in the monoclinic and orthorombic form (PDBs 6Y2F and 6Y2G, respectively). The structure of Mpro in complex with antineoplastic drug carmofur, which inhibits viral replication in cells with EC50 = 24.3 μM, was determined at 1.6 Å resolution (Jin et al., 2020b); the carbonyl group of carmofur attaches covalently to catalytic C145 and the fatty acid tail occupies the S2 subsite. Anti-inflammatory ebselen has also shown a promising inhibitory effect in Mpro and reduction of the virus titer in cell culture (Jin et al., 2020a). Starting with the substrate-binding site of the SARS-CoV Mpro, Dai et al. designed and synthesized two novel covalent inhibitors of the SARS-CoV-2 Mpro, displaying anti-SARS-CoV-2-infection activity in cell culture with EC50 values of 0.53 μM and 0.72 μM, respectively, with no significant cytotoxicity (Dai et al., 2020). Some known protease inhibitors have been identified in silico and tested in vitro on Mpro , such as the HIV protease inhibitors ritonavir, nelfinavir, saquinavir, and atazanavir (Fintelman-Rodrigues et al., 2020); the hepatitis C virus (HCV) protease inhibitor boceprevir, the broad-spectrum protease inhibitor GC-376, and calpain inhibitors II and XII were also shown to inhibit viral replication by targeting the Mpro (Ma et al., 2020).
In addition to the catalytic site, we identified a cryptic site with a CS within 5 Å defined by residues T199, Y237, Y239, L271, L272, G275, M276, and A285-L287; this site is not too distant from the partner protomer, and as such should be explored whether a molecule binding to it might preclude dimer formation (Fig. 6B). A second borderline druggable site delimited by residues Q107-Q110, V202, N203, H246, I249, and T292-F294 was also identified. This site lies on the opposite side of the dimerization interface, and the functional consequences of a molecule binding to it have yet to be determined.
4.1.4. The RNA polymerase complex (nsp12-nsp7-nsp8)
In +ssRNA viruses, the synthesis of RNA is catalyzed by the RdRp, in a primer-dependent manner. In SARS-CoV-2 and other CoVs, the RdRp complex consists of a catalytic subunit (nsp12), and co-factors nsp7 and nsp8 (Gao et al., 2020; Kirchdoerfer and Ward, 2019).
The SARS-CoV-2 RdRp experimental structures contain two nsp8 molecules: one forming a heterodimer with nsp7 (nsp8-2), and a second one bound at a different site (nsp8-1), in a similar fashion as in SARS-CoV (Kirchdoerfer and Ward, 2019) (cf. Table S3 for a list of the available RdRp complex experimental structures). Interaction with nsp7 and nsp8 provides stability to nsp12 (Peng et al., 2020), consistent with the observation that nsp12 in isolation displays little activity, while the presence of nsp7 and nsp8 enhance template binding and processivity (Kirchdoerfer and Ward, 2019; Yin et al., 2020). The overall structure of the SARS-CoV-2 RdRp complex is very similar to that of SARS-CoV, with a Cα RMSD of ~0.8 Å, consistent with the high degree of sequence similarity (nsp7, 99%; nsp8, 97%; nsp12, 97%). Although the amino acid substitutions are not located in the catalytic site, the SARS-CoV-2 RdRp complex displays a 35% lower efficiency for RNA synthesis than its SARS-CoV counterpart; this lower efficiency is due to changes restricted to nsp8 and nsp12 only (Peng et al., 2020).
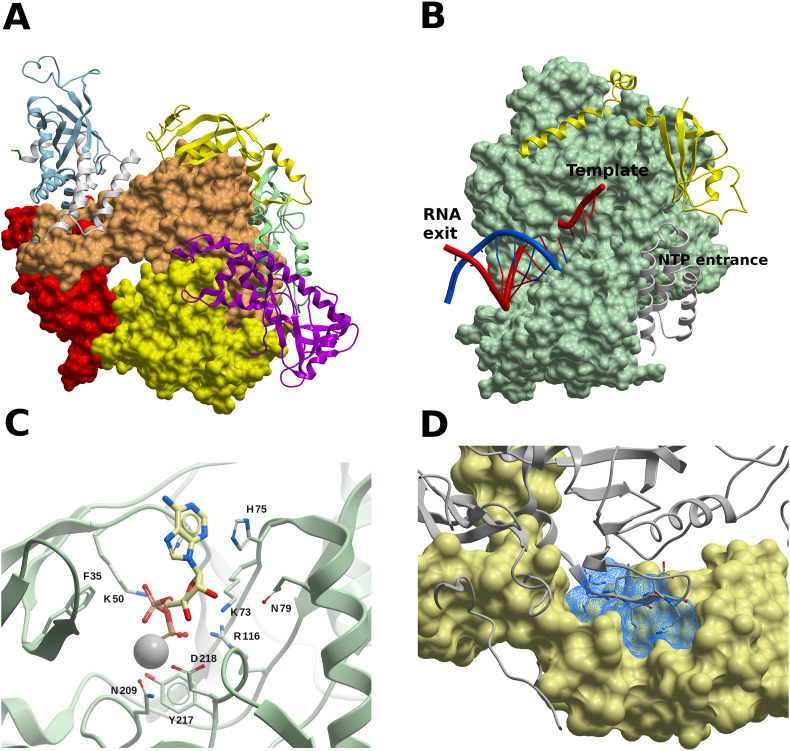
The nsp12 contains a right-hand RdRp domain (residues L366-F932) −a conserved architecture in viral RdRps− and an N-terminal nido-virus RdRp-associated nucleotidyl-transferase (NiRAN) domain (residues D51-R249); these two domains are linked by an interface domain (residues A250 to R365). The polymerase domain is formed by three subdomains: a fingers subdomain (residues L366-A581, and K621-G679), a palm subdomain (residues T582-P620 and T680-Q815), and a thumb subdomain (residues H816-E932) (Fig. 7 A). An N-terminal β-hairpin (residues D29-K50) establishes close contacts with the NiRAN domain and the palm subdomain, and contributes to stabilize the structure. As in the corresponding SARS-CoV nsp12 (Kirchdoerfer and Ward, 2019), the integrity of the overall structure is maintained by two zinc ions that are present within metal-binding sites composed by residues H295–C301–C306–C310 and C487–H642–C645–C646, respectively.
Fig. 7.
The SARS-CoV-2 RdRp complex (nsp12-nsp8-nsp7). (A) Structure of the nsp12-nsp7-nsp8 complex. The channel in the middle corresponds to the catalytic active site. Color code: nsp7, white ribbon; nsp8-2, light blue ribbon; nsp8-1, yellow ribbon. The nsp12 domains are colored as follows: palm, yellow; fingers, tans; palm, red; interface, pale green ribbon; NiRAN, magenta ribbon. (B) RdRp complex bound to RNA. Nsp12 is displayed as a green surface. Color code: Primer RNA, blue; template RNA, red; nsp7, yellow ribbon; nsp8-1, grey ribbon. (C) Molecule of ADP-Mg2+ within the NiRAN domain of nsp12. Interacting residues are shown, in what may constitute a druggable binding site. (D) Target site in nsp8 (light yellow surface). The predicted binding site is represented using a blue mesh representation, and nsp12 is shown as grey ribbon. Nsp12 residues N386–K391 are displayed (though not labeled) to highlight that a small molecule binding to these potential sites might interfere with PPIs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The RdRp-RNA structures show that RNA mainly interacts with nsp12 through its phosphate-ribose backbone, especially involving the 2′-OH groups. Except for three specific and localized structural changes (Yin et al., 2020), the overall structures of the apo and RNA-bound complexes are very similar, displaying a main chain RMSD value of ~0.5 Å. The absence of relevant conformational rearrangements from the apo state implies that the enzyme can start to function as replicase upon RNA binding, which also correlates with its high processivity. The RNA binding residues are highly conserved across viral polymerases (teVelthuis, 2014). It should be noted that in RdRp structures with longer RNA products (PDBs 6YYT, 26 bases, and 6XEZ, 34 bases), the long α-helical extensions of both nsp8 molecules form positively charged ‘sliding poles’ along the exiting RNA, in what could be a supporting scaffold for processing the large CoV genome. Since these nsp8 extensions are mobile in free RdRp (Gao et al., 2020; Kirchdoerfer and Ward, 2019), it is possible that they adopt an ordered conformation upon dsRNA exit from nsp12.
Remdesivir, a drug designed for Ebola virus, has received a great deal of attention as a RdRp inhibitor and potential treatment for COVID-19 (Wang et al., 2020a). Remdesivir is a prodrug that is converted into its active triphosphate form [remdesivir triphosphate (RTP)] by host enzymes. Two cryo–electron microscopy structures of the SARS-CoV-2 RdRp complex have been solved, one in the apo form, and the other in complex with a template-primer RNA and with remdesivir covalently bound to the primer strand (Yin et al., 2020) (PDBs 7BV1 and 7BV2, respectively) (Fig. 7B). At high RTP concentration, the mono-phosphate form of remdesivir (RMP) is added (at position i) into the primer-strand (Yin et al., 2020), thus causing termination of RNA synthesis at position i+3, in SARS-CoV-2, SARS-CoV, and MERS-CoV (Gordon et al., 2020a). Favipiravir (also called avifavir), used in Hepatitis C Virus (HCV) treatment, is also a promising drug (Wang et al., 2020a). Other antivirals, such as sofosbuvir, alovudine, tenofovir alafenamide, AZT, abacavir, lamivudine, emtricitabine, carbovir, ganciclovir, stavudine, and entecavir, are also incorporated by SARS-CoV-2 RdRp and block replication in vitro (Chien et al., 2020). Another type of broad spectrum ribonucleoside analog, β-D-N4-hydroxycytidine (EIDD-1931), has been shown to inhibit SARS-CoV-2, SARS-CoV and MERS-CoV in cell culture by increasing the mutation transition rate, probably exceeding the proofreading ability granted by the ExoN (Sheahan et al., 2020).
In the RdRp-nsp13 complex (PDB 6XEZ), the NiRAN domain is occupied by an ADP-Mg2+ molecule (Fig. 7C). While the target of the NiRAN nucleotidyltransferase activity is unknown, the activity itself is essential for viral propagation (Lehmann et al., 2015). Thus, the ADP-binding site might constitute an interesting druggable site in nsp12, and it should be further assessed whether a small-molecule binding to it would interfere with the viral cycle.
The design of non-nucleoside inhibitors calls for the analysis of alternative disruptive sites within the RdRp complex, considering that the assembly of the nsp12-nsp7-nsp8 complex is needed for RNA synthesis. We used FTMap to identify druggable sites on PPI interfaces in nsp12, nsp7, and nsp8. Several hot-spots were identified on nsp8, three of which formed a potential druggable site, outlined by residues P121, A125, K127–P133, T137, T141, F147, and W154 (Fig. 7D). This site lies within the nsp12-nsp8 interface, and a molecule binding to this site could interfere with the interaction of the β-strand N386–K391 of nsp12 with nsp8 (Fig. 7D). A borderline druggable target site was identified on nsp12, lined up by residues L270, L271, P323, T324, F326, P328–V330, R349, F396, V398, M666, and V675 (Fig. S3), where a molecule binding to it might interfere with nsp8 binding by clashing with its segment V115–I119. On nsp7, a borderline druggable site was identified within the PPI interface with nsp8-2, defined by residues K2, D5, V6, T9, S10, V12, F49, and M52-S54.
In addition to its functionality within the RdRp complex, the nsp7-nsp8 hexadecamer has de novo initiation of RNA synthesis capability, also known as primase activity, while nsp8 has also displayed TATase activity. Analysis of the SARS-CoV nsp7-nsp8 hexadecameric structure (PDB 2AHM) shows several sites which could be targeted to interfere with its primase activity, making impossible for nsp12 to extend the complementary strand due to the lack of primer. Moreover, it is also possible that small-molecules could interfere with the conformational dynamics needed to interact within the RdRp complex. However, further structural and computational studies (such as molecular dynamics, MD) would be needed to confirm these hypotheses.
4.1.5. Helicase (nsp13)
The SARS-CoV-2 nsp13 possesses helicase activity, thus playing a key role in catalyzing the unwinding in the 5′ to 3′ direction of dsRNA or structured RNA into single strands. It has been demonstrated that in SARS-CoV this happens in an NTP-dependent manner (Jia et al., 2019). Additionally, it has been shown that nsp13 has RNA 5′-triphosphata (RTPase) activity (which may be the first step for the formation of the 5′ cap structure of viral RNAs). The helicase-associated NTPase and RTPase activities share a common active site, both in SARS-CoV (Ivanov et al., 2004) and the human CoV 229E (HCoV-229E) (Ivanov and Ziebuhr, 2004).
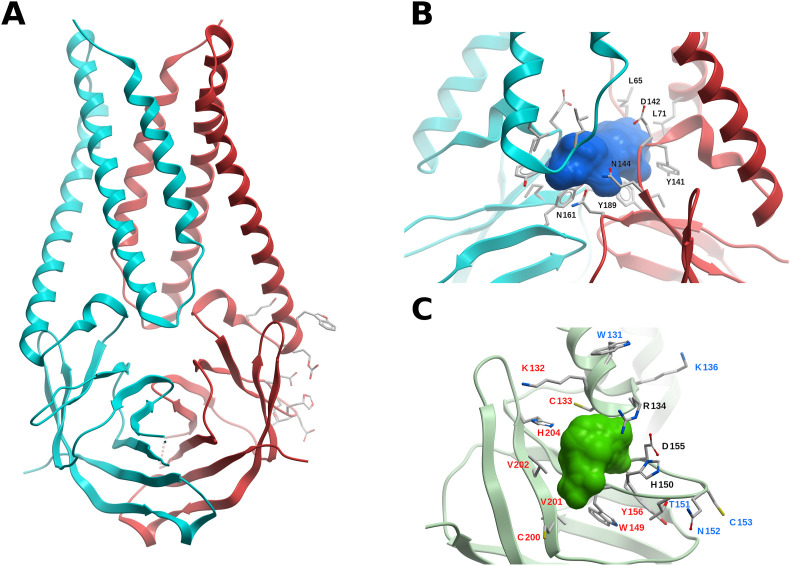
The crystal structure of the SARS-CoV-2 nsp13 has been solved at 1.9 Å (PDB 6ZSL). A structure solved by cryo-EM of nsp13 in complex with RdRp (nsp12-nsp8-nsp7) is also available (PDB 6XEZ). Nsp13 has the form of a triangular pyramid with five domains. At the top, the N-terminal zinc binding domain with three zinc fingers (A1-S100) is connected with the stalk domain (D101-G150); then, at the base of the pyramid, three domains (1B, I151-E261; 1A, F262-R442; 2A, R443-N596) form the triangular base (Fig. S4). Compared to its SARS-CoV and MERS-CoV counterparts, the SARS-CoV-2 nsp13 shares a 99.8% (100%), and 71% (82%) sequence identity (similarity), respectively. The nsp13 structures are also very similar, with backbone RMSD values of ~1.9 Å between SARS-CoV-2 nsp13 and the corresponding SARS-CoV (PDB 6JYT) and MERS-CoV (PDB 5WWP).
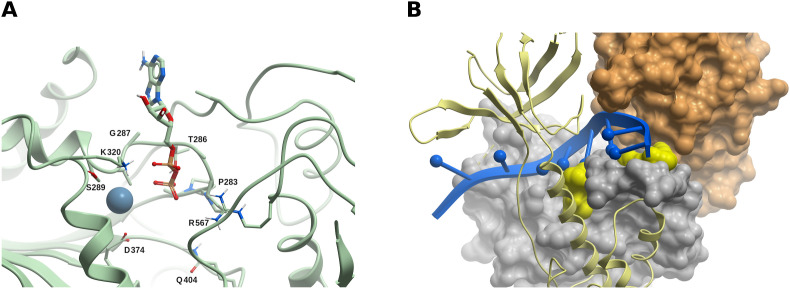
In the SARS-CoV nsp13, six residues were identified as being involved in NTP hydrolysis (K288, S289, D374, E375, Q404, and R567), and mutations of any of these residues to alanine resulted in high unwinding deficiency and decreased ATPase activity (Jia et al., 2019). Moreover, it was also shown in the same study that, for all six mutants, changes in helicase activity are consistent with changes in their ATPase activity, thus demonstrating that nsp13 performs its helicase activity in an NTP-dependent manner. As stated above, this site would also correspond to the RTPase activity. The SARS-CoV-2 nsp13 structure PDB 6XEZ features an ADP-Mg2+ molecule in the vicinity of those residues (Fig. 8 A), suggesting that targeting this site may interfere with both NTPase or RTPase functions.
Fig. 8.
The SARS-CoV-2 helicase/NTPase/RTPase (nsp13). (A) The ADP-Mg2+ bound within nsp13. This site has been identified as being involved in NTP hydrolysis in SARS-CoV, and could constitute a druggable site. (B) Two potential borderline druggable binding sites identified in nsp13. The structure of RNA (blue) was modeled based on the yeast Upf1-RNA complex structure (PDB 2XZL). Nsp13 is represented as a green ribbon, but nsp13 domains 1A and 2A are displayed as grey and tan molecular surfaces, respectively. The potential sites are shown in yellow. Based on our model, molecules binding to these sites might interfere with RNA binding. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The zinc binding and stalk domains are important for the helicase activity in SARS-CoV (Jia et al., 2019). Mutations to alanine of the zinc binding domain residue N102 (which interacts with the stalk domain) and of the stalk residue K131 (which interacts with the 1A domain) resulted in decreased helicase activity. This appears to be a top-to-bottom signaling system, and it is difficult to figure out a way to interfere in this process with small molecules, with the information available up to today. The zinc binding domain might be targeted with metal chelators such as bismuth complexes.
Based on an interaction model of dsRNA with SARS-CoV nsp13, it was hypothesized that residues 176–186 (1B domain), 209–214 (1B domain), 330–350 (1A domain) and 516–541 (2A domain) constitute a probable nucleic acid binding region (Jia et al., 2019). While there is no CoV nsp13 structure with a nucleic acid substrate bound, we performed a crude model of SARS-CoV-2 nsp13 in complex with RNA based on a Yeast-Upf1-RNA structure (PDB 2XZL), in a similar fashion as in an earlier work (Jia et al., 2019), and in agreement with a recent model (Chen et al., 2020) (Fig. 8B). Using FTMap, two potential borderline druggable sites were identified delimited by amino acids P406–409, L412, T413, G415, L417, F422, N557, and R560, and K139, E142, C309, M378-D383, P408, and T410, which could interfere with RNA binding, according to our artificial model (Fig. 8B).
4.1.6. Exoribonuclease/guanine-N7 methyltransferase (nsp14-nsp10 complex)
While the RdRp complex allows the synthesis of large viral RNA due to an increased processivity conferred by the heteromeric complex, to achieve high accuracy, CoVs possess 3′-5′ exoribonuclease (ExoN) activity located in the N-terminal domain of nsp14, to proofread the viral genome synthesis. The ExoN excises mutagenic nucleotides misincorporated by the RdRp, thus conferring potential drug resistance to nucleoside analog inhibitors. It has been reported that the ExoN activity protects SARS-CoV from the effect of base analog 5-fluorouracil (Smith et al., 2013), and that guanosine analog ribavirin (Rbv) 5′-monophosphate is incorporated at the 3′-end of RNA by the SARS-CoV RdRp, but excised from RNA by the nsp14-nsp10 ExoN (to a lesser degree by the nsp14-ExoN alone), which could account for the poor effect of Rbv in treating CoV-infected patients (Al-Tawfiq and Memish, 2017). Since it was shown that SARS-CoV displays a significantly lower nucleotide insertion fidelity in vitro than that of the Dengue RdRp (Ferron et al., 2018), it could be concluded that the low mutation rate of SARS-CoV is related to the ExoN activity. This shows that targeting the ExoN function could be an excellent strategy for the development of pan-CoV therapeutics, complementing existing RdRp inhibitors, like Remdesivir. In fact, MHV lacking the ExoN activity was shown to be more susceptible to Remdesivir (Agostini et al., 2018).
The C-terminal domain of nsp14 functions as a guanine-N7 methyltransferase (N7-MTase), where S-adenosyl-L-methionine (SAM) is demethylated to produce S-adenosyl-L-homocysteine (SAH), transferring the methyl group to the N7 position of the guanine in 5′GpppN in viral RNAs, and forming m7GpppN (cap-0). This capping is followed by a second methylation by the 2′-O-MTase (nsp16), forming m7GpppNm (cap-1) (Fig. 4). The capping structure is a protective and pro-transductional modification of the viral RNA, and blocking MTase activity may increase viral RNA decay and suppress viral RNA translation.
While there is no experimental structure available of SARS-Cov-2 nsp14, crystal structures of the SARS-CoV nsp10-nsp14 dimer are available, with nsp14 in complex with SAM at 3.2 Å (PDB 5C8T), with SAH and guanosine-P3-adenosine-5′,5′-triphosphate (GpppA) at 3.3 Å (PDB 5C8S), and in its unbound form (PDBs 5C8U and 5NFY, both at 3.4 Å). The sequence identity between SARS-Cov-2 and SARS-CoV nsp14 is 95% (98% similarity), with no gaps in the alignment and full conservation in all functionally relevant sites. SARS-CoV nsp10 and its SARS-CoV-2 counterpart share 98% sequence identity (similarity 100%). We thus built a homology model of the SARS-CoV-2 nsp10-nsp14 dimer using the corresponding SARS structure 5C8T as a template (see Methods); the missing S454-D464 segment within the template was included in the model, and optimized. Since there are no gaps, the numbering scheme of template and model coincides.
In nsp14, amino acids A1-C285 fold into the ExoN domain, and the N7-MTase function lies within amino acids D301-Q527; both domains are connected by a loop (amino acids F286-G300), and its abolition was shown to suppress the N7-MTase function in SARS-CoV (Chen et al., 2013) (Fig. 9 A). The architecture of the catalytic core and active sites of the ExoN domain are similar to those of the DEDD superfamily exonucleases, though exhibiting some differences (Ferron et al., 2018; Ma et al., 2015). The catalytic residues D90, E92, E191, D273 and H268 (DEEDh motif) display similar structural arrangements to other proofreading homologs, such as the DNA polymerase I (1KLN) and the ε subunit of DNA polymerase III of E. coli (1J53), but with only a single Mg2+ ion at its active center. Mutating any of these residues to alanine either impaired the ExoN activity or severely reduced the ability to degrade RNA in SARS-CoV (Ma et al., 2015). The ExoN domain also contains two zinc fingers, the first one comprising residues C207, C210, C226 and H229, and the second one in proximity to the catalytic site, comprising residues H257, C261, H264 and C279. In SARS-CoV, none of the mutants of zinc finger 1 could be expressed as soluble proteins, thus revealing its importance in protein stability, and mutants of zinc finger 2 had their enzymatic activity abolished (Ma et al., 2015). The ExoN domain interacts with nsp10 (Fig. 9A), exhibiting an interaction surface of ~7800 Å2, more than twice that of the nsp10-nsp16 complex. It has been reported that SARS-CoV nsp10 is necessary for the correct positioning of the residues of the ExoN catalytic site, which partially collapses in the absence of nsp10, what explains the reduced ExoN activity of isolated nsp14 (Bouvet et al., 2012).
Fig. 9.
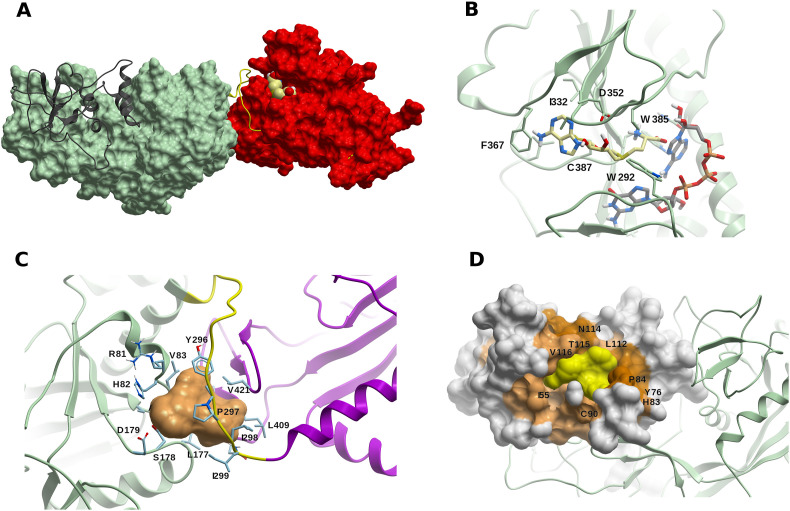
The ExoN/MTase complex (nsp14-nsp10) (A) The ExoN nsp14 domain (in green) and the MTase domain (in red) are connected by the hinge loop F286-G300 (yellow). Nsp10 is shown in dark grey ribbon, and a S-adenosyl-L-methionine (SAM) molecule (light yellow carbons) is displayed within the catalytic site. (B) A model of SAM (light yellow carbons) within the catalytic site of the MTase domain of SARS-CoV-2 nsp14. A molecule of guanosine-P3-adenosine-5′,5′-triphosphate (GpppA) (green carbons) is added as reference. (C) Potential druggable (allosteric) binding site in the vicinity of the hinge region F286-G300 (in yellow), including Y296 and P297. The linked nsp14 domains ExoN and MTase are displayed in green and magenta, respectively. (D) Druggable site (yellow surface) within a cryptic site on nsp10 (colored in lighter or darker brown according to the cryptic score). The ExoN domain of nsp14 is shown as green ribbon. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As it has been pointed out in SARS-CoV (Ferron et al., 2018; Ma et al., 2015), nsp14 has an atypical MTase fold, with an additional α-helix in the last 12 amino acids which stabilizes the neighboring environment; deletion of this helix has been shown to decrease or abolish the MTase activity of nsp14. A third zinc finger is also present in this domain formed by C452, C477, C484 and H487, but it is distant from the active site, and mutations of amino acids from the zinc finger 3 have a very marginal effect on MTase activity (Ma et al., 2015). But it has been hypothesized that it might be important in binding with the nsp16-nsp10 dimer to accomplish the second methylation for completion of the capped structure.
The SARS-CoV-2 N7-MTase domain in complex with methyl donor SAM is shown in Fig. 9B; the binding site residues are fully conserved, thus the contacts are similar to the SARS-CoV nsp14-SAM complex. Methyl acceptor GpppA binds close to methyl donor SAM (cf. PDB 5C8S) to facilitate methyl transfer. Comparing the N7-MTase catalytic sites of SARS-CoV nsp14 structures bound to SAM (5C8T), and to SAH-GpppA (5C8S), no significant structural changes are found, thus supporting the hypothesis that ligand binding sites are pre-formed.
The hinge region (amino acids F286-G300, Fig. 9A) is conserved across CoVs, thus suggesting it might have a functional role. In SARS-CoV, lateral and rotational movements of the C-terminal domain relative to the N-terminal domain of up to 13 Å have been observed (Ferron et al., 2018), and crystallographic and SAXS data have shown that nsp14 undergoes important conformational changes, where the hinge might act as a molecular switch; moreover, hinge residues Y296 and P297 are essential for ExoN activity in SARS-CoV.
In fact, nsp14 is involved in two processes that use RNA substrates in different ways: a newly synthesized RNA strand with a mismatch should be translocated by the polymerase complex to the ExoN catalytic site of nsp14, whereas during replication, the 5′-mRNA should go into the catalytic tunnel of the N7-MTase for methyl capping. These results could be rationalized in terms of nsp14 flexibility. Moreover, the mutual dependence of the ExoN domain on N7-MTase function (Chen et al., 2013), and of the N7-MTase domain on ExoN activity (Ferron et al., 2018) have been established. Ferron et al. further showed that both the ExoN domain (excluding the first 71 residues) and the N7-MTase domain interact with nsp12-RdRp (Ferron et al., 2018), a biologically relevant interaction due to the possibility of tandem associated activities that makes RNA replication more efficient.
Considering the structural and functional features of the SARS-CoV-2 nsp10-nsp14 dimer just discussed, the following small-molecule targeting strategies could be suggested:
-
•
The SAM binding site is an attractive target to develop CoVs inhibitors using small-molecules that could preclude SAM or GpppA binding, thus suppressing the N7-MTase activity of nsp14. In fact, the N7-MTase catalytic tunnel lies close to the SAM binding site, and blocking it would also preclude mRNA binding.
-
•
A potential druggable (allosteric) site was identified using FTMap and ICM Pocket Finder defined by residues R81-A85, L177-D179, Y296–I299, N408, L409, L411, and V421. This site lies in the vicinity of hinge residues Y296 and P297 (Fig. 9C). A small-molecule binding within this site could interfere with the dynamic behavior of nsp14 and its associated conformational changes.
-
•
There is a small hot-spot in nsp14 (lined up by residues F60, M62, L192, M195, and K196) in the vicinity of a cryptic site, in which a molecule binding in this region would clash with nsp10 binding.
-
•
Considering the high sequence identity between the SARS-CoV and SARS-CoV-2 nsp10-nsp14 dimers, and that the contact residues are fully conserved, blocking the nsp10-nsp14 interaction to decrease or abolish full ExoN activity could be a valid strategy against CoV diseases. A potential borderline druggable site on the PPI surface of nsp10 with nsp14 and lined up by residues T5, E6, N40, A71, S72, C77, R78, H80, L92, K95, and Y96 was identified using FTMap; this site is defined by two hot-spots separated by ~9 Å, which could impose a limit on the expected affinity of a potential ligand (Kozakov et al., 2015b). However, it should also be mentioned that in the absence of nsp10, the nsp14 ExoN catalytic site partially collapses and its activity decreases (Bouvet et al., 2012); thus, it would be interesting to further explore the behavior of this site using MD simulations.
-
•
A druggable site was identified within a region of a high cryptic site score in nsp10 (Fig. 9D). This site does not overlap with the nsp14-nsp16 PPI interface, and while its functional role is uncertain, it may have allosteric modulation potential.
4.1.7. RNA nucleotide-2′O-methyltransferase complex (nsp16-nsp10)
The nsp 16 possesses a SAM-dependent RNA 2′O-MTase activity that is capable of cap-1 formation. It adds a methyl group to the m7GpppN previously capped by nsp14 (cap-0) to form m7GpppNm (cap-1), by methylating the ribose of the first nucleotide at position 2′-O (Fig. 4). Like nsp14, nsp16 uses SAM as methyl donor (Decroly et al., 2008). In SARS-CoV, nsp16 requires nsp10 to execute its activity, since nsp10 is necessary for nsp16 to bind both m7GpppA-RNA and SAM (Chen et al., 2011); moreover, nsp16 was found to be an unstable protein in isolation, and most of the disruptions in the interface of nsp16-nsp10 eliminate the methylation activity (Rosas-Lemus et al., 2020). In humans, most mRNAs include the cap-1 modification; while cap-0 appears to be sufficient to recruit the entire translational machinery, cap-1 modification is necessary to evade recognition by host RNA sensors, such as the Retinoic acid-Inducible Gene I (RIG-I), MDA-5, and IFN induced proteins with tetratricopeptide repeats (IFIT), and to resist the IFN-mediated antiviral response (Devarkar et al., 2016).
Several crystal structures of the nsp16-nsp10 heterodimer have been recently solved (cf. Table S4) (Fig. 10 A). SARS-CoV-2 nsp10 shares 99% and 59% sequence identity with SARS-CoV and MERS-CoV, respectively. Similarly, nsp16 is highly similar to SARS-CoV (95% sequence identity), but only 66% identical to MERS-CoV. Both proteins interact through a large network of hydrogen-bonds, water-mediated interactions, and hydrophobic contacts (Krafcikova et al., 2020). The high conservation of nsp10 and nsp16 sequences, and the complete conservation of catalytic and substrate-binding residues strongly support the idea that nsp16-mediated 2′-O-MTase mechanism and functionality are highly conserved in CoVs. Nsp10 exhibits two zinc fingers: the first one is coordinated by C74, C77, H83 and C90, and the second one is coordinated by C117, C120, C128, and C130. The zinc fingers residues are 100% conserved across β-CoVs, highlighting the relevance of this motif in the replication process.
Fig. 10.
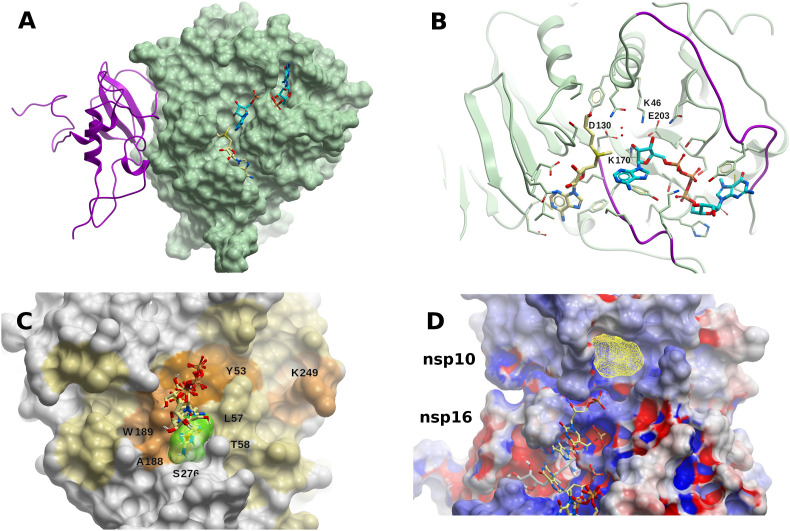
The SARS-CoV-2 RNA nucleoside-2′O-methyltransferase complex (nsp16-nsp10). (A) Structure of the 2′O-MTase (nsp16, green molecular surface) heterodimer with nsp10 (magenta ribbon). Molecules of SAM (light yellow carbons) and m7GpppA (cyan carbons) are shown within the catalytic site. (B) Catalytic site of nsp16 with methyl donor SAM (light yellow carbons) and methyl acceptor m7GpppA (cyan carbons). The nucleotide binding site flexible loops (D26-K38, M131-N138) are colored in blue. The highly conserved KDKE motif (K46, D130, K170, E203) for methyl-transfer, found in many 2′O-MTases, is highlighted in magenta, and oxygen water molecules are displayed in red. (C) Binding hot-spot (transparent yellow) identified with FTMap in the vicinity of residues L57, T58, A188, C209, N210, and S276, on the surface of nsp16, within an extended cryptic site identified in the same region using CryptoSite (brown colored surface). Small-molecules bound within that site (taken from crystal structures) are also displayed (light yellow carbon atoms): [adenosine, 2-(n-morpholino)-ethanesulfonic acid, β-D-fructopyranose, 7-methyl-guanosine-5′-triphosphate, and 7-methyl-guanosine-5′-diphosphate]. This site lies on the opposite side of the catalytic site, ~25 Å away from it, in what thus could be an allosteric site. (D) Extension of the RNA groove in nsp16 towards nsp10. Five RNA nucleotides are shown (light yellow carbon atoms), which correspond to those of the human mRNA 2′O-MTase (PDB 4N48), after structural superposition of the binding site residues. SAM is displayed in grey carbon atoms. The consensus site identified with FTMap is shown in yellow mesh. Nsp16 and nsp10 are colored according to their electrostatic potential (blue, positively charged; red, negatively charged; white, neutral). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Most nsp16-nsp10 structures feature SAM (or the close analog pan-MTase inhibitor sinefungin) in their substrate binding site, being coordinated by residues N43, Y47, G71, A72, S74, G81, D99, N101, L100, D114, and M131, together with few water molecules (Fig. 10B). All these residues are also 100% conserved in SARS-CoV. Some structures also include m7GpppA within the catalytic site (PDBs 6WQ3 and 6WRZ in presence of SAH, and 6WKS and 6WVN in presence of SAM). The nucleotide binding site is surrounded by flexible loops comprised of amino acids D26-K38, M131-N138, and the highly conserved KDKE motif (K46, D130, K170, E203) for methyl-transfer, which is found in many 2′-O-MTases (including the phylogenetically more distant MERS-CoV), is also present (Fig. 10B). The MTase active site is clearly an attractive target for antiviral drug discovery, but the structural conservation among CoVs and host cellular MTases might pose a challenge for the development of a specific compound.
Using FTMap, a small hot-spot was found in the vicinity of residues L57, T58, A188, C209, N210, and S276, within an extended cryptic site on the surface of nsp16 (Fig. 10D). Interestingly, recent crystal structures of the nsp16-nsp10 dimer feature small-molecules bound within that site, such as adenosine (PDB 6WKS), 2-(n-morpholino)-ethanesulfonic acid (PDB 6YZ1), β-D-fructopyranose (PDB 6W4H), 7-methyl-guanosine-5′-triphosphate (PDBs 6WVN and 6WRZ), and 7-methyl-guanosine-5′-diphosphate (PDB 6WQ3) (Fig. 10C). This site lies on the opposite side of the catalytic site, 25 Å away from it, in what could thus be an allosteric site (Viswanathan et al., 2020). Further studies would be necessary to clarify the function of this binding site, and its impact and feasibility in terms of druggability.
Considering that m7GpppAC5 acts as an effective substrate of SARS-CoV nsp16-nsp10 (Züst et al., 2011), Chen et al. performed docking of m7GpppGAAAAA and m7GpppAAAAAA within the SARS-CoV nsp16-nsp10 dimer (Chen et al., 2011), and found that while the first three nucleotides contact nsp16, the following ones were in contact with nsp10, which extends the positively charged area of the RNA-binding groove. Interestingly, all the residues defining this groove extension are also conserved in SARS-CoV-2. The extension to the RNA-binding site provided by nsp10 may serve to accommodate the RNA chain and stabilize the interaction between m7GpppA-RNA within the catalytic site, which could also be observed by superposing the nsp16 SAM binding site with that of the human mRNA 2′O-MTase (PDB 4N48) (Krafcikova et al., 2020) (Fig. 10D). Using FTMap, we identified a small borderline druggable site within nsp10, in the area where RNA would extend (Fig. 10D). This pocket lies within the RNA binding groove extension described above, and is lined up by the side chains of amino acids P37, T39, C41, K43, F68, A104, and P107 of nsp10. A molecule binding to it might interfere with extended RNA binding. It should be stressed that, due to its zinc fingers, nsp10 has the ability to bind poly-nucleotides non-specifically (Matthes et al., 2006).
It was also shown that 12-mer and 29-mer peptides extracted from the dimerization surface of SARS-CoV nsp10 (F68–H80 and F68–Y96, respectively) were found to inhibit the activity of nsp16 at IC50 ~ 160 μM (Ke et al., 2012); this suggests that using short peptides might be a possible strategy, considering there is 100% sequence conservation within the nsp10/nsp16 interface for SARS-CoV and SARS-CoV-2. Interfering with the nsp16-nsp10 dimer formation using small molecules might still be challenging, however, due to the large area of contact and the absence of buried pockets at the nsp10-nsp16 interaction surfaces. In spite of this, a cryptic site was identified on nsp16 in the region of the PPI interface with nsp10 (Fig. S5), close to where the 12-mer and 29-mer peptides would bind. However, no CSs were found near this cryptic site, so further studies are needed to confirm this site as a potential target to inhibit oligomerization.
4.1.8. RNA uridylate-specific endoribonuclease (nsp15)
The nsp15 harbors a nidoviral RNA uridylate-specific endoribonuclease (NendoU) that belongs to the EndoU family, whose activity is to cleave downstream of uridylate, releasing 2′-3′ cyclic phosphodiester and 5′-hydroxyl termini. The NendoU cleaves polyuridines produced during the priming of the poly(A) ssRNA in replication (Fig. 3), which helps to dampen dsRNA melanoma differentiation-associated protein 5 (MDA5)-dependent antiviral IFN responses (Hackbart et al., 2020). Clearly, small molecules blocking RNA catalytic sites, or interfering with nsp15 oligomerization, would inhibit the enzyme activity and would help trigger cellular antiviral mechanisms.
The SARS-CoV-2 nsp15 structure displays an N-term oligomerization domain, a middle domain, and a C-term NendoU catalytic domain (Kim et al., 2020) (Fig. S6A). Nsp15 has been crystallized in its apo form (PDB 6VWW at 2.2 Å resolution), and in complex with citric acid (PDBs 6XDH at 2.35 Å, and 6W01 at 1.9 Å), uridine-3′-monophosphate (PDB 6X4I at 1.85 Å), uridine-5′-monophosphate (PDB 6WLC at 1.85 Å), drug Tipiracil (PDB 6WXC at 1.85 Å), and product di-nucleotide GpU (PDB 6X1B at 1.97 Å). All structures are very similar, with main chain RMSD values of less than 0.5 Å between any pair of them. Taking structure 6W01 as a reference, SARS-CoV-2 nsp15 exhibits ~0.5 Å RMSD with respect to its SARS-CoV counterpart (88% sequence identity, 95% similarity), and 1.1 Å with respect to MERS-CoV nsp15 (51% sequence identity, 65% similarity) [cf. Ref (Kim et al., 2020). for a detailed comparison of SARS-CoV-2 nsp15 with SARS-CoV and MERS-CoV structures].
The catalytic active site within the NendoU domain is formed by residues H235, H250, K290, V292, S294, T341, and Y343 (Fig. S6B) (also conserved in SARS-CoV and MERS-CoV), with H235, H250, and K290 as the proposed catalytic triad (Kim et al., 2020). The catalytic residues also form a druggable binding site, as can be confirmed by its shape and the crystal structure of drug Tipiracil in complex with nsp15 (PDB 6WXC). The catalytic activity has been observed to be metal-dependent in most NendoU proteins, although there could be exceptions in other nidoviruses (Nedialkova et al., 2009). Although no crystal structure has been found with a metal ion (nor complexed with RNA), a metal binding site required for maintaining the conformation of the active site and substrate during catalysis has been proposed, coordinated by the carboxylate group of D283, the hydroxyl group of S262, and the carbonyl oxygen of P263.
SARS-CoV-2 nsp15 folds into a hexamer (a dimer of trimers) (Kim et al., 2020), in agreement with an earlier work showing that SARS-CoV nsp15 conformationally exists as a hexamer (PDB 2RHB), and also suggesting oligomerization-dependent endoribonuclease activity (Guarino et al., 2005). The channel of the hexamer is ~10 Å wide, open from top to bottom, and the hexamer is stabilized by extensive contacts between monomers. As such, it might be disrupted or destabilized by mutations or small molecules (Kim et al., 2020). Using FTMap, we identified two potential druggable sites; the first one is delimited by residues K71, N74, N75, M272, S274–N278, I328, L346, and Q347 (Fig. S6C), and close to the binding area of another protomer within the hexamer (Fig. S6C); a small-molecule binding to this site might disrupt PPI; the second one, delimited by residues E69, K71, K90, T196, S198, L252, D273, K277, Y279, V295, and D297, is deep and rather buried, within an area with a higher-than-average cryptic site score and an opening towards the hexamer channel.
4.1.9. Non-structural protein 9 (nsp9)
Nsp9 acts as a hub that interacts with several viral components; it binds ssRNA, nsp8, and the N protein (Gordon et al., 2020b; Sutton et al., 2004). Proteomic experiments showed that SARS-CoV-2 nsp9 also binds to several nuclear pore proteins (Gordon et al., 2020b), a process well characterized for other viruses that affect nuclear shuttling as part of their replicative and host shutdown mechanisms. SARS-CoV nsp9 exists as a homodimer in solution formed through the interaction between the GXXXG motifs in parallel α-helices of the opposite protomers (Sutton et al., 2004); mutations in any of the glycines inducing dimer disruption have been associated with impeded viral replication and reduced RNA binding in SARS-CoV (Miknis et al., 2009), and in porcine delta coronavirus (PDCoV) (Zeng et al., 2018). Thus, as it has been already suggested, disruption of the homodimeric interface could be an appealing strategy against CoV-associated diseases (Hu et al., 2017). Homologs of nsp9 have been found in other β-coronaviruses such as SARS-CoV, MERS-CoV, HCoV-229E, and the avian IBV, with different degrees of sequence similarity compared to SARS-CoV-2. The mechanism of RNA binding to nsp9 and how it enhances viral replication, are not yet fully understood, but apparently depend on several positively charged amino acids on the surface (Zeng et al., 2018). While it is not known whether SARS-CoV-2 nsp9 plays an analog role like its SARS-CoV counterpart, their high sequence identity (97%), and their strong structural similarity (see below), might suggest a highly conserved functional role.
The structure of the SARS-CoV-2 nsp9 homodimer has been solved at 2.0 Å (PDB 6WXD) and at 3.0 Å (PDB 6W4B); both structures share a backbone RMSD of 0.5 Å and 0.9 Å for the monomer and dimer, respectively. The monomer has a backbone RMSD of 0.94 Å compared to that of SARS-CoV (PDB 1QZ8); the RMSD value decreases to 0.44 Å when considering α-helices and β-sheets only. The protomers interact mainly through van der Waals contacts of the backbone from the conserved GXXXG motifs within the opposed α-helices, near the C-terminus of the protein (Fig. S7A), as in other CoVs (Egloff et al., 2004; Hu et al., 2017). Another nsp9 structure was determined featuring a rhinoviral 3C protease sequence LEVLFQGP in the N-terminal tag (PDB 6W9Q). The inserted peptide folds back and forms a β-sheet with the N-terminal of the protein. Residues LEVLF of the peptide make hydrophobic contacts with residues P6, V7, A8, L9, Y31, M101, S105, and L106 of nsp9, and is hydrogen-bonded with residues P6, V7, L9 and S105. These nsp9 residues are conserved among other nsp9 homologs, including full conservation in SARS-CoV. This provides evidence that this binding site could be targeted by a peptidomimetic small molecule to disrupt dimer formation and thus reduce RNA binding and viral replication.
Using FTMap, two potential druggable sites were identified in nsp9. One site is defined by residues R39-V41, F56–S59, I65-E68, I91, and K92, the other one by residues S13, C14, D26-L29, L45-L48, and K86 (Fig. S7B). These two sites lie in regions of intermediate-to-high cryptic site score, and do not overlap with the dimerization interface, but are close to positively charged residues that might be involved in RNA binding, as was postulated for IBV (Chen et al., 2009a); it should also be considered that they might interfere in nsp9-nsp8 PPI, though this is subject to validation. A hot-spot within a region of moderate-high cryptic site score was identified in the C-terminus of nsp9, delimited by residues C73, F75, L88, L103, L106, A107, and L112 (Fig. S7C); a molecule binding in this region might clash with the N-terminal part of the other protomer.
4.1.10. ADP-ribose-phosphatase (nsp3 domain)
The first nsp3 macrodomain (Mac 1) is conserved throughout CoVs and has an ADP-ribose phosphatase (ADRP) activity, by which ADRP binds to and hydrolyzes mono-ADP-ribose (Alhammad et al., 2020). This appears to be related to the removal of ADP-ribose from ADP-ribosylated proteins or nucleic acids (RNA and DNA); it should be pointed out that ADRP is able to remove mono-ADP-ribose, but not poly-ADP-ribose (Alhammad et al., 2020). Anti-viral ADP-ribosylation is a host post-translational modification in response to viral infections, since many of the IFN and cytokine signaling components, such as NF-κB essential modulator (NEMO), TANK-binding kinase 1 (TBK1), and NFκB need to be ribosylated to be fully active (Fehr et al., 2020). Although ADRP is not an essential protein for viral replication, it has been shown to be an essential pathogenesis factor in animal models for CoV infection; for example, mutations of ADRP in SARS-CoV enhanced IFN response and reduced viral loads in vivo in mice models (Fehr et al., 2016). Thus, its role against host-induced anti-viral activity makes it an attractive target for drug design.
Recently, five crystal structures of SARS-CoV-2 ADRP were solved (see Table S1), including the apo form (PDBs 6VXS at resolution 2.0 Å, 6WEN at 1.35 Å), and in complex with 2-(N-morpholino)ethanesulfonic acid (MES) (PDB 6WCF at 1.09 Å), AMP (PDB 6W6Y at 1.45 Å), and ADP-ribose (PDB 6W02 at 1.50 Å) (Fig. S8). Another structure of ADRP complexed with ADP-ribose (PDB 6WOJ) at 2.2 Å is available. These structures exhibit low main chain atom RMSD between any pair of them, with values in the range 0.25–0.65 Å. The SARS-CoV-2 ADP-bound ADRP structures share structural similarity to related homologs from SARS-CoV (71% sequence identity, 82% similarity) and MERS-CoV (40% sequence identity, 61% similarity), with main chain RMSD values of 0.6 Å (PDB 2FAV) and 1.4 Å (PDB 5HOL), respectively.
The binding site of SARS-CoV-2 ADRP (Fig. S8) also bears high similarity with those of SARS-CoV and MERS-CoV. The ADP-ribose is stabilized within the binding site through hydrophobic interactions, and direct and solvent-mediated hydrogen-bond interactions; it should be highlighted that most of the hydrogen-bonds involve main chain atoms. The binding mode of ADP-ribose is conserved in those three CoV (RMSD values of 0.3 Å and 1.2 Å, respectively, with respect to SARS-CoV-2 ADRP), with ADP-ribose exhibiting similar affinities towards the three CoVs (Alhammad et al., 2020). Within the overall conserved binding site conformation of the SARS-CoV-2 ADRP structures, some shifts are observed when comparing the apo structure, and those in complex with AMP, ADP-ribose, and MES, mainly around the proximal ribose. The rotameric states of several side chains (F132, I131, F156) adopt a ligand-dependent conformation, and a flip in the A129-G130 peptide bond could be observed, dependent on the presence of the phosphate group in ADP-ribose (phosphate 2), or MES.
Considering its highly conserved structural features in CoVs, especially SARS-CoV and MERS-CoV, and its role in countering host-induced antiviral responses, ADRP appears as an attractive therapeutic target. It should be noted, however, that no other druggable binding site could be identified other than the ADP-ribose pocket using FTMap or ICM Pocket Finder, nor could cryptic sites be found on the surface.
4.1.11. Ubiquitin-like 1 domain (nsp3)
The first ~110 residues of nsp3 have an ubiquitin-like fold, and are thus named the Ubl1 domain. The function of Ubl1 in CoVs is related to ssRNA binding, while probably interacting with the N protein too. In SARS-CoV Ubl1 has been shown to bind ssRNA with AUA patterns, and since the SARS-CoV 5′-UTR (un-translated region) is rich in AUA repeats, it probably binds to it (Serrano et al., 2007); in fact, SARS-CoV-2 has 439 AUA within its genome. In MHV, this Ubl1 domain binds the structural N protein (Hurst et al., 2013). It was also shown that MHV-Ubl1 is essential for viral replication, since viable virae could not be recovered from the Ubl1 full deletion mutant (Hurst et al., 2013).
There is yet no available experimentally solved structure of SARS-CoV-2 Ubl1. Considering that the sequence identity and similarity with its SARS-CoV counterpart is 79% and 93%, respectively (with only one deletion in the alignment, close to the N-terminal), a structural model was built using SARS-CoV Ubl1 (PDB 2GRI) as template (Fig. S9A). There are several positive residues on the protein surface, compatible with ssRNA binding. These residues are conserved in SARS-CoV, with the exception of R23N, R102H, and N98K (SARS-CoV numbering). Two small potentially druggable sites were identified with FTMap and ICM Pocket Finder, which lie in areas of above-average cryptic site score. One site is defined by the side chains of Y42, T43, T48, E52, F53, C55 and V56 (Fig. S9B), and the second one by F25-D28, T86, Y87, W82, and C104–F106 (Fig. S9C). These sites are near positively charged residues, but do not overlap with them. While a molecule binding to them might interfere with ssRNA binding, it is also possible that it might disrupt PPIs with partner proteins, such as the N protein. Further biochemical and functional characterization of Ubl1 is needed to shed light on the actual value of these sites.
4.1.12. SARS-unique domain (SUD) (nsp3 domain)
The SUDs is part of nsp3 and it is implicated in tracking the viral RNA to DMVs. In SARS-CoV-2 and SARS-CoV there are three SUD domains connected by short peptide linkers: SUD-N, SUD-M, and SUD-C, indicating the N-terminal, the middle, and the C-terminal regions of SUD, respectively. SUD-N (macromolecular domain 2, Mac 2) binds G-quadruplexes, an unusual nucleic-acid structures formed by guanidine-rich nucleotides in ssRNAs; it shares 71% of identity and 85% of similarity with its SARS-CoV counterpart. SUD-M/Mac3 (macromolecular domain 3, Mac3) has a single-stranded purine rich (G and/or A) RNA binding activity (Johnson et al., 2010) which includes poly(A) and G-quadruplexes; this allows SUD-M to act as a poly(A)-binding-protein and thus protect the poly(A) tail at the 3' end of viral RNA from host 3’ exonucleases. SUD-C/DPUP (Domain Preceding Ubl2 and PLpro) also binds to ssRNA, and recognizes purine bases more strongly than pyrimidine bases (Johnson et al., 2010). This RNA binding activity is apparently stabilized by the presence of SUD-M, while SUD-C seems to modulate the sequence specificity of SUD-M (Johnson et al., 2010). Mutagenesis analysis in SARS-CoV showed that SUD-M is indispensable for virus replication, while the absence of SUD-N or SUD-C barely reduce the virus titer (Kusov et al., 2015).
The structure of the SARS-CoV construct SUD-N/SUD-M (SUD-NM) has been solved by crystallography at 2.2 Å resolution (PDB 2W2G). The solution structures of SARS-CoV SUD-M and SUD-C within a SUD-MC construct were obtained using NMR (PDBs 2KQV and 2KQW, respectively), together with the isolated SUD-C (PDB 2KAF). The isolated SARS-CoV SUD-M has also been solved by NMR (PDBs 2JZD and 2JZE), together with the structure of an N-terminal extended SUD-M (PDBs 2RNK and 2JZF). Main chain RMSD values among corresponding solved structures are within 0.8 Å. It has been shown that SUD-NM is monomeric in solution (Johnson et al., 2010), and the absence of evidence suggesting a tight transient or static contacts in solution showed that SUD could be modeled as three flexible linked globular domains (Johnson et al., 2010).
The SARS-CoV-2 SUD domains are very similar to their SARS-CoV counterpart, with sequence identity (similarity) of 71% (91%), 79% (94%), and 72% (92%) for SUD-N, SUD-M, and SUD-C, respectively. Considering that only SUD-M appears to be essential to viral replication, we focused our druggability analysis on it. We thus built a model by homology using PDB 2AKF as template. For SARS-CoV, it was shown that upon poly(A) binding to SUD-MC, the molecular surface area of SUD-M affected by NMR chemical shift perturbation experiments was mapped to a positively charged surface cavity (Johnson et al., 2010), defined by residues N532, L533, I556, M557, A558, T559, Q561, and V611 (Chatterjee et al., 2009). This area is wholly conserved in SARS-CoV-2, and a potential druggable binding site within it was identified using FTMap and ICM Pocket Finder (Fig. S10). It could be hypothesized that a small-molecule binding to this site might preclude RNA binding.
4.1.13. Nucleic-acid binding region (NAB, nsp3 domain)
NAB is a small domain of ~120 amino acids that binds ssRNA, strongly preferring sequences containing repeats of three consecutive guanosines (Serrano et al., 2009). In SARS-CoV, NAB binds ssRNA through a positively charged surface patch defined by the residues K75, K76, K99, and R106, while the neighboring residues N17, A18, S19, D66, H69, T97 are also affected by RNA binding (Serrano et al., 2009). Interestingly, this RNA binding site bears similarity to that of the sterile alpha motif of the Saccharomyces cerevisiae Vts1p protein (Serrano et al., 2009). Experiments have shown that N-terminal and C-terminal extensions, corresponding to links with the PLpro and the TM-Lumen/Ectodomain, respectively, behave as flexibly disordered segments (Serrano et al., 2009).
While the SARS-CoV-2 NAB structure is not yet available, the SARS-CoV counterpart has been solved by NMR (PDB 2K87). We used the latter to build a structural model of SARS-CoV-2 NAB by homology (sequence identity 81%, similarity 94%, no gaps in the alignment). All the positive residues are conserved, with two additions, T43K and S51K (Fig. S11A).
No druggable binding sites were identified within the positive charged patch on the surface where ssRNA binds in SARS-CoV; however, two cryptic sites with nearby CSs were predicted on the sides of that patch (Fig. S11B). It should be further explored whether these sites are involved in PPI, and whether small-molecules binding to them might allosterically modulate ssRNA binding.
4.1.14. Other nsp3 domains
The DUF3655/HVR/Acidic-domain, TMs-Lumen/Ectodomain, and the C-Terminal domain/Y-Domain (Table S1) are structurally uncharacterized nsp3 domains, which bear little similarity to any other experimentally solved structure, thus ruling out the possibility of homology modeling.
DUF3655-HVR is a hypervariable, Glu and Asp rich domain, probably implicated in protein-protein interactions with N, as in MHV (Keane and Giedroc, 2013). Since it is not essential to viral replication in MHV (Hurst et al., 2013), it would be a second priority for drug targeting.
The TMs Lumen/Ectodomain is flanked by two transmembrane domains (TMs), exposed to the ER lumen, and works by binding to the lumen domain of nsp4, necessary to form the DMVs where CoVs replicate, anchoring the whole nsp3 protein to membranes through the TMs. While DMVs formation is essential, the lack of structural information poses an insurmountable hurdle for drug discovery (Hagemeijer et al., 2014).
The C-Terminal domain/Y-Domain is conserved in CoVs, and it seems to be involved in the DMVs formation, probably interacting with nsp6, and improving nsp3-nsp4 interaction (Hagemeijer et al., 2014). Although breaking this interaction would seriously impact viral replication, the lack of structural and biochemical information precludes any targeting attempt.
4.1.15. Other non-structural proteins: nsp1, nsp2, nsp4, and nsp6
The nsp1 is the leader protein and the first translated and PLpro-processed protein. It has the capacity to bind to the 40S ribosome subunit to inactivate the translation of host mRNAs (Thoms et al., 2020), also selectively promoting host mRNAs degradation, which makes it the main actor in host cell shutdown (Thoms et al., 2020), and indirectly in the immune response evasion by delaying IFN responses (Lei et al., 2020). Additionally, based on recent proteomics analyses, SARS-CoV-2 nsp1 seems to interfere with the host DNA duplication by interacting with the DNAPolA complex, which is the host primase complex, and the first step in DNA synthesis (Gordon et al., 2020b). Although nsp1 is not essential for viral replication, its absence makes the virus susceptible to IFN (Lei et al., 2020), which would make it an important pathogenic factor and a good target for drug design. However, the structural information available of this 180-amino acids protein is limited. An NMR structure from SARS-CoV (PDB 2GDT) of 116 amino acids (corresponding to H13 to G127 in SARS-CoV-2) does not contain the 40S binding region (which includes residues K164 and H165), and only partially represents the active site for RNA binding (Lokugamage et al., 2012). There is a recent cryo-EM structure of the nsp1 C-terminus (E148-G180) in complex with the 40S ribosomal subunit and RNA (PDB 6ZLW). While amino-acids K164 and H165 are present, the C-terminal portion of only 33 residues cannot be used for structure-based drug design.
The nsp2 is a membrane protein not essential for the viral production in SARS-CoV and MHV homologs. However, in MHV, the presence of nsp2 positively affects the viral titer (Gadlage et al., 2008). It is a delocalized membrane protein, but is recruited to replication sites by other viral components (Graham et al., 2005), probably by the M protein (Li et al., 2020a). In SARS-CoV-2, proteomic data showed that nsp2 interacts with cellular components associated with vesicles formation and translational regulators (Gordon et al., 2020b), which might hint that its function is associated with the formation of membrane structures and co-opting host components. However, up to the present date there is no structural data of nsp2 from SARS-CoV-2 or related proteins.
The nsp4 is essential for viral replication in MHC (Sparks et al., 2007). The main function of nsp4 is the formation of DMVs in SARS-CoV and MERS-CoV (Angelini et al., 2013). Nsp4 has four TM domains, a ER-Lumen exposed domain, and a cytoplasmatic exposed C-Terminal domain (Clementz et al., 2008). In SARS and MHV, the lumen domain of nsp4 interacts strongly with the nsp3 lumen domain/ectodomain to induce DMVs formation (Hagemeijer et al., 2014). Thus, an nsp3-nsp4 structure might help to design a PPI inhibitor that could prevent DMVs formation. The cytoplasmatic C-Terminal domain (~90 amino acids) of MHV and Feline CoV nsp4 structures are available (3VC8, 3GZF, respectively), but SARS-CoV-2 nsp4 shares with them only 59% and 39% sequence identity, respectively.
The nsp6 is a membrane protein with a medium conservation degree between different CoVs. In SARS-CoV, nsp6 collaborates with nsp4 and nsp3 in DMVs formation, also inducing the formation of membrane vesicles (Angelini et al., 2013). Additionally, in several CoVs, nsp6 activates omegasome and autophagosome formation independently of starvation, to degrade cellular components and so increase the availability of resources for viral replication (Cottam et al., 2011). Nevertheless, there is no structural information on any related protein which would allow homology modeling and structure-based drug discovery.
4.2. Structural proteins
4.2.1. S protein
The SARS-CoV-2 spike-protein (S protein, ORF2) is a large homotrimeric multidomain glycoprotein. The small C-terminal transmembrane attaching domain is followed by the S2 and S1 subunits. During infection, the receptor binding domain (RBD) in S1 is exposed and recognized by ACE2, the junction S1/S2 is cleaved by a furin-like protease releasing the S1 domain, while S2 is cleaved again by the metalloprotease TMPRSS2, to expose the FP, which is responsible in inducing the membrane fusion mechanisms (Wrapp et al., 2020) (Fig. 1).
Several strategies are focused on inhibiting the viral entry, either by interfering with the binding of S to ACE2, or with the membrane fusion induction (Zhou et al., 2020). The first approach was the development of inhibitory antibodies or recombinant ACE2 proteins that block the S protein (Zhou et al., 2020); as the outer surface component of CoVs, the S protein is a major target of antibodies, and the main focus of vaccine development. Using a different approach, based on structural information and the knowledge of the membrane fusion mechanisms, several peptides that mimic neutralizing antibodies have been developed, such as a series of lipo-peptides EK1C1-EK1C7 and IPB02, that target the subdomain HR1 in the S2 fragment to inhibit membrane fusion by interfering with the FP, and which showed a decrease of SARS-CoV-2 virus titer in cell cultures (Zhu et al., 2020).
As the mechanism of the virus entry depends on host factors, targeting ACE2 and TMPRSS2 is being explored. There are no commercial ACE2-inhibitors. The main candidates for inhibiting the ACE2-Spike interaction appear to be specific antibodies, which are currently raising great expectations, and are completing their clinical phases. The inhibition of TMPRSS2 is also being explored, by blocking the proteolytic priming of the S2 fragment, necessary to induce membrane fusion; the inhibitors Nafamostat, Gabexate, and Calmostat (Shrimp et al., 2020) have been shown to inhibit viral production in cell cultures, and will be tested in humans; however, their low half-life suggests a need for developing more efficient new drugs [cf. Ref (Singh et al., 2020). for new options targeting TMPRSS2].
4.2.2. E protein
It is a small homopentameric membrane protein (75 amino-acids per protomer) (Pervushin et al., 2009), which has been shown to be essential for viral particle assembly in SARS-CoV (Siu et al., 2008). The N-terminal region of the protein spans the lipid bilayer twice, while the C-terminal is exposed to the interior of the virus (Shen et al., 2003). In IBV and SARS-CoV, the E protein interacts with the viral M protein through an undefined region (Chen et al., 2009b); and also through its C-terminus with the host Protein Associated with Lin Seven 1 (PALS1), a factor associated with pathogenesis (Teoh et al., 2010). In many CoVs, the E protein works as an ion-channelling viroporin (Pervushin et al., 2009), which affects the production of cytokines, and consequently the inflammatory response (Nieto-Torres et al., 2014).
The NMR structures of the SARS-CoV-2 and SARS-CoV E protein homopentamers (PDB 7K3G and 5X29, respectively) are shown in Fig. S12. Only the TM regions were solved, showing 31 and 58 residues per helix, respectively. While in SARS-CoV-2 the helices are parallel, and in SARS-CoV they exhibit a tilt angle of ~15°, the size of the channel is roughly conserved. In fact, based on structural and functional considerations, the E protein channel would be the primary target site for drug development. Moreover, hexamethylene amiloride (HMA) has been reported to bind to the SARS-CoV E protein homopentamer, but not to an isolated protomer (Surya et al., 2018).
4.2.3. N protein
β-coronavirus nucleocapsid (N) proteins are involved in the packing of viral +ssRNA to form a ribonucleoprotein (RNP) complex, which interacts with the M protein (He et al., 2004). They share an overall conserved domain structure, with an RNA-binding N-terminal domain (NTD, amino acids 49–175), and a dimerization C-terminal domain (CTD, amino acids 247–365); these two domains are connected by a disordered region, and the C-terminal tail at the CTD (366–419) is termed the B/N3 domain. The CTD forms a homodimer in solution, while the addition of the B/N3 spacer results in homotetramer formation (Ye et al., 2020). It has been suggested that the assembly of β-coronavirus N protein filaments may consist of at least three steps, namely, dimerization through the CTD, tetramerization mediated by the B/N3 region, and further filament assembly through both viral RNA binding and association of N protein homotetramers (Ye et al., 2020). Since the formation of the RNP complex is essential for viral replication, identification of small-molecule modulators of the nucleocapsid assembly, interfering with NTD RNA binding, or precluding CTD dimerization or oligomerization, would be valid therapeutic strategies against SARS-CoV-2.
Several structures of the N protein NTD and CTD have recently become available (see Table S5). Structures of the NTD (and CTD) superimpose closely among themselves. With respect to SARS-CoV, the NTD and CTD share 88% (96%), and 96% (98%) sequence identity (similarity), respectively, and the corresponding structures overlay with RMSD values within 0.8 Å.
The structure of the NTD solved by NMR (PDB 6YI3) showing that amino-acids A50, T57, H59, R92, I94, S105, R107, R149, and Y172 participate in RNA binding (Dinesh et al., 2020) is shown in Fig. S13A. A druggable site identified near those residues partially overlaps with the AMP binding site in the NTD of the homolog protein human CoV OC43 (HCoV-OC43) (PDB 4LI4), in agreement with an earlier hypothesis (Kang et al., 2020). Two small borderline druggable sites have also been identified with FTMap (site 1: L159-P162, T165, L167, A173, S176; site 2: Q70-N75, Q83, T135, P162). These sites would not overlap with RNA binding, and further experiments are needed to confirm whether they could be allosteric sites, or near PPI interfaces.
The CTD homodimer structure is shown in Fig. S13B (PDB 6WZQ), also displaying two potential druggable sites. A molecule binding to any of them might interfere with dimer formation. Two other distinct sites were predicted on the surface of the homodimer, one over the central four-stranded β-sheet, and the other opposite to it, near the C-terminal α-helices (Fig. S13C). Since it is not clear yet how the homotetramer is formed (Ye et al., 2020), it should be further explored whether any of these sites might overlap with PPI interfaces.
4.2.4. M protein
It is a membrane homodimeric glycoprotein that forms part of the virion. In SARS-CoV, the M protein interacts with the N protein, being a nexus between virus membrane formation and RNA association in the virion (Nal et al., 2005), also inhibiting IFN production in SARS and MERS-related diseases (Lui et al., 2016; Siu et al., 2014). While it is established that dimerization and N-M PPI motifs reside in the C-terminus of the protein (Kuo et al., 2016), the lack of structural data for SARS-CoV-2 and other CoV homologs precludes further drug discovery targeting protein-M.
4.3. Accessory proteins
4.3.1. Orf3a/X1/U274
The orf3a was characterized as a potassium ion channel in SARS-CoV, involved in inducing caspase-dependent apoptosis under different pathological conditions (Chan et al., 2009). It is included in the virion (Shen et al., 2005), and interacts specifically with the M, E, and S structural proteins, as well as with orf7a/U122 (Tan et al., 2004). In SARS-CoV, orf3a expression increases the mRNA levels of all three subunits of fibrinogen, thus promoting fibrosis, one of the serious pathogenic aspects of SARS (Tan et al., 2005), and the expression of NFκB, IL8, and JNK, all involved in inflammatory responses (Kanzawa et al., 2006). In this scenario, design therapeutics that suppress its function could be very important; moreover, the presence of similar proteins to orf3a in other β-CoVs (SARS-CoV-2 orf3a shares ~73% identity and ~85% similarity with its SARS-CoV counterpart), and in α-CoVs, suggests that drugs targeting orf3a might be a therapeutic option against a broad range of CoV-related diseases (Kern et al., 2020).
Recently the SARS-CoV-2 orf3a structure was solved by cryo-EM (PDB 6XDC). The N-terminus (amino acids 1–39), the C-terminus (239–275), and a short loop (175–180) were not observed, probably due to molecular disorder. The orf3a was solved as a homodimer, although the authors were able to reconstruct the tetramer at a lower final resolution of ~6.5 Å, also showing a model of the neighboring dimers, and inferring from this model that residues W131, R134, K136, H150, T151, N152, C153, and D155 were involved in a network of interactions that would mediate tetramerization (Kern et al., 2020) (Fig. 11 A). Considering that SARS-CoV orf3a has been identified as an emodin-sensitive potassium-permeable cation channel, the narrow size of the pore in the SARS-CoV-2 orf3a structure strongly suggests that it is in the closed or inactive conformation (Kern et al., 2020).
Fig. 11.
Structure and potential binding sites of the SARS-CoV-2 accessory protein orf3a. (A) Ribbon representation of the orf3a homodimer (cyan and red). Residues W131, R134, K136, H150, T151, N152,C153, and D155, which might be involved in homo-tetramerization, are displayed (not labeled for the sake of clarity). (B) Potential druggable site within the orf3a homodimer interface (blue surface). The neighboring residues are displayed, and labeled for one protomer. (C) Tetramerization interface and a partially overlapping potentially druggable site (green molecular surface). Residue labels have been colored red (binding site), blue (tetramerization interface), black (common residues). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Using FTMap and ICM Pocket Finder, a druggable site was found within the dimer, lined up by the side chains L65, L71, Y141, D142, N144, P159, N161, and Y189 of both protomers (Fig. 11B). This cavity has also been identified in Ref. (Kern et al., 2020). Since conformational changes of the TMs are needed for channel opening, we could hypothesize that a small-molecule binding within this site would interfere with this rearrangement, or directly block the channel. Another potentially druggable site delimited by residues K132-R134, C148–H150, D155, Y156, C200–V202, and H204, was identified, which partially overlaps with the proposed tetramerization interface, and thus it could be explored for a possible PPI inhibitor (Fig. 11C).
4.3.2. Orf7a/X4/U122
The SARS-CoV-2 orf7a is a transmembrane protein of 121 amino acids (106 if only the mature protein is considered, excluding the signaling peptide), with 86% identity and 94% similarity with respect to its SARS-CoV counterpart. Interestingly, it exhibits structural similarity to Igs (Nelson et al., 2005; Tan et al., 2020), but shares poor sequence identity with proteins of the Ig superfamily (within the 2–16% range). Like orf3a, orf7a is also included in the virion, and both proteins have been shown to interact with each other in SARS-CoV (Tan et al., 2004) and in SARS-CoV-2 (Li et al., 2020a). In SARS-CoV, orf7a expression increases the expression of NFκB, IL8 and JNK (Kanzawa et al., 2006), while the deletion of orf7a reduces the virus titer 30-fold. Orf7a appears to be unique to SARS CoVs, showing no significant similarity to any other protein, either viral or non-viral (Nelson et al., 2005). In SARS-CoV-2, orf7a interacts with midasin AAA ATPase 1 (MDN1) and HEAT repeat containing 3 (HEATR3) (Gordon et al., 2020b). MDN1 is a protein involved in the maturation of ribosomes in eucaryotes (Garbarino and Gibbons, 2002), while HEATR3 has a positive role in Nucleotide-binding oligomerization domain-containing protein 2 (NOD2)-mediated NF-κB signaling (Zhang et al., 2013), and is also involved in the assembly of the 5S ribosomal subunit (Calvino et al., 2015). From this information it can be expected that orf7a works by regulating inflammation like its SARS-CoV homolog, and also that it is involved in protein translation regulation. The N-terminal ectodomain structure of orf7a (amino acids 1–66, using the numbering of the mature protein) is available for SARS-CoV, either from crystallography (PDB 1XAK at 1.8 Å) or NMR (PDB 1OY4), and recently for SARS-CoV-2 (PDB 6W37). The structure displays a seven-stranded β-sandwich fold with two disulfide bonds (Fig. S14), and exhibits main chain RMSD values of 0.4 Å and 0.9 Å with the corresponding SARS-CoV orf7a structures, respectively. The SARS-CoV NMR structure also features a disordered part corresponding to the stalk region, between the ectodomain and the membrane (residues 68–82). The transmembrane region comprises approximately amino acids 83–101.
While the functionality and interaction partners of orf7a are not clearly known, we identified two hot-spots on the surface of orf7a. An extended cryptic site was predicted lined up by residues H4, Q6, L16, P19, Y60, L62, with a CS nearby which includes a deep hydrophobic pocket already identified in SARS-CoV as a potential PPI site (Nelson et al., 2005) (Fig. S14). A hot-spot defined by residues E18, T24, Y25, F31, P33, A35, N37, and F50 was also identified (Fig. S14). Both sites might have functional roles, and further studies are needed to confirm this hypothesis, and whether small-molecules binding to any of them would interfere with the viral mechanism.
4.3.3. Orf8
Orf8 is one of the fastest evolving genes in SARS-CoV-2, as can be inferred by its high variability (Tan et al., 2020). The orf8 expression is not essential for SARS-CoV and SARS-CoV-2 replication; it was observed, however, that a 29 nucleotide deletion in SARS-CoV may correlate with milder disease (Muth et al., 2018). SARS-CoV-2 orf8 is involved in inhibiting IFN signaling (Li et al., 2020b) and also downregulates signaling MHC-I in most cells (Zhang et al., 2020).
The SARS-CoV-2 orf8 shares less than 20% sequence identity with its SARS-CoV counterpart, which might be taken into consideration to understand the lethal characteristics of SARS-CoV-2. The crystal structure of SARS-CoV-2 orf8 solved at 2.04 Å exhibits an Ig-like fold, and revealed two novel dimerization interfaces unique to SARS-CoV-2: i) a covalent dimer-interface formed through a sequence-specific region, and linked by an intermolecular disulfide-bond mediated by the unique C20 of each monomer, and ii) a non-covalent interface stabilized by the SARS-CoV-2-specific motif Y73–I74-D75-I76 (Pancer et al., 2020). The presence of these interfaces may account for oligomerization and explain how SARS-CoV-2 orf8 forms unique large-scale assemblies not possible in SARS-CoV. Interestingly, we identified a druggable site in the vicinity of the Y73–I76 motif defined by residues I47-L60, V62, D63, Y73–I76, Y79, T80, Q91, K94, and L95, together with a borderline druggable site in the neighborhood of the covalent-dimerization interface defined by residues E19, R48, A51, K53, S97, V99, and D113-R115.
4.3.4. Orf9b
In SARS-CoV, orf9b is a non-essential dimeric membrane protein, produced from an alternative start codon to N-orf9a, with a lipid-binding-like structure (Meier et al., 2006), and with the ability to bind several other viral proteins (von Brunn et al., 2007), including structural proteins, which allows orf9b to be incorporated into the virion (Xu et al., 2009). In SARS-CoV, its action interfers with mitochondrial factors that limit IFN responses and with immune response-related apoptosis (Han et al., 2020). In SARS-CoV-2, it suppresses IFN-I and IFN-III responses, induced by host dsRNA sensing components (Han et al., 2020). Additionally, in SARS-CoV-2, orf9b has been reported to interfere with microtubule organization and IRES dependent translation factors (Gordon et al., 2020b).
The homodimer structure of SARS-CoV-2 orf9b has been solved by x-ray crystallography at 2.0 Å (PDB 6Z4U); the SARS-CoV structure at 2.8 Å is also available (PDB 2CME). The M26-G38 loop is not present in the structure, likely due to its high flexibility. The sequence identity and similarity of orf9b between SARS-CoV and SARS-CoV-2 is 73% and 83%, respectively, and the main chain RMSD values of the orf9b dimers corresponding to both species (measured using residues with defined secondary structure) is 0.65 Å (a higher value of ~2 Å is obtained using the full length, due to the high B-factor loops). The structure features a 2-fold symmetric dimer, where both protomers are in a highly interlocked architecture, as in a handshake (Fig. S15A).
The dimer exhibits a central hydrophobic cavity (Fig. S15A) lined up by residues V15, I19, L21, I44, L46, L52-L54, I74, V76, M78, and V94 of both protomers; in the crystal structure PDB 6Z4U this central cavity is filled with polyethylene glycol (in the corresponding SARS-CoV structure, a decane molecule is present). In SARS-CoV, it was hypothesized that orf9b immerses its positively charged surface into the negatively charged lipid head groups of the membrane, while becoming anchored by lipid tails that could bind to this hydrophobic cavity (Meier et al., 2006). In this context, a small-molecule which could bind to the central cavity would impede membrane attachment by competing with the lipid tails.
Using FTMap, two potentially druggable sites were identified, where a molecule binding to them might interfere with homodimerization (Fig. S15). These sites are defined by residues: i) D2-I5, M8, L12, I45, R47, L87, D89, F91, and V93; ii) V15, P17-L21, V41, I44, L46, S53, L54, V76, and V94; the latter site also lies in a region with above-average cryptic site score, and overlaps with the central cavity mentioned above.
4.3.5. Other accessory proteins
SARS-CoV-2 possesses at least another five accessory proteins: orf3b, orf6, orf7b, orf9c/orf14 and orf 10. Some of them are produced by alternative start codons of the same orf. These proteins are more divergent among different CoVs than the previously described; not being essential for replication, they have a lower natural selection pressure, and therefore a higher mutation rate. In fact, the deletion of orf3a, orf3b, orf6, orf7a, and orf7b in SARS-CoV do not eliminate viral production in vitro (Yount et al., 2005), nevertheless, it does affect pathogenicity and virulence.
The following proteins lack experimentally solved structure, or they cannot be modeled due to the absence of similar experimentally solved proteins which could serve as templates:
Orf3b is a short protein observed in some β- and γ-coronaviruses, and produced by an alternative start-codon. In SARS-CoV it was shown to inhibit the expression of IFN-β during synthesis and signaling (Kopecky-Bromberg et al., 2007). It differs considerably among different CoVs, but maintains its pathogenic function (Zhou et al., 2012).
The SARS-CoV orf6 is a small membrane protein (Tangudu et al., 2007) sharing 69% of identity with its SARS-CoV-2 counterpart. It acts as a pathogenicity factor, due to its ability to convert a sublethal MHV infection into a lethal one (Pewe et al., 2005); this property only depends on the N-terminal transmembrane segment (Netland et al., 2007). The SARS-CoV orf6 enhances viral replication (Zhao et al., 2009) through interaction with components of the viral replication machinery, such as nsp8 (Kumar et al., 2007). In addition, it shows pathogenic activity due to its ability to induce apoptosis, similar to orf3a and orf7a. Additionally, SARS-CoV-2 orf6 is involved in the inhibition of IFN signaling, as its SARS-CoV paralog (Li et al., 2020b).
In SARS-CoV, orf7b is a small membrane protein that could be included in the virion (Schaecher et al., 2007), and is probably involved in attenuating viral production (Pfefferle et al., 2009).
Orf9c/orf14 is the third putative protein translated from the orf 9 (Shukla and Hilgenfeld, 2015), and the least characterized. It enables immune evasion and coordinates cellular changes, impairing IFN signaling, antigen presentation, and complement signaling, while inducing interleukin-6 (IL-6) signaling (Dominguez Andres et al., 2020).
Orf10 is a putative membrane protein present only in SARS related CoVs (Cagliani et al., 2020). No functional activity has been determined experimentally, but it is inferred from proteomic data that it is involved in targeting host proteins for proteasome degradation (Gordon et al., 2020b).
5. SARS-CoV-2 polymorphisms
Understanding the evolution of pathogens is important for the effective development of new antiviral strategies to circumvent drug resistance.
Since the outbreak in December 2019, several genetic variants have appeared compared to the first SARS-CoV-2 genome sequenced in Wuhan (Mercatelli and Giorgi, 2020). These polymorphic amino acids are found in structural, non-structural, and accessory proteins, and the most frequent ones are listed in Table S6. In structural proteins, the most notorious case is the D614G mutation in the RBD domain of the S protein, which became predominant during the first months of the outbreak (Plante et al., 2020), along with other changes such as the D936Y in the FP domain (Cavallo and Oliva, 2020). In non-structural proteins, the P504L and Y541C are the predominant mutations in the helicase nsp13 (Table S6), and they were reported to affect the RNA binding (Begum et al., 2020). Among accessory proteins, the mutation Q57H in orf3a is becoming dominant and appears to be associated with disease severity; orf3a also possesses other relevant polymorphism such as S171L, G196V and G251V (Issa et al., 2020) (Table S6).
Knowledge of polymorphic amino-acids could be taken into consideration when selecting molecular targets for drug development to avoid or reduce potential drug resistance.
6. Discussion and perspective
The appearance of COVID-19 caused by the SARS-CoV-2, its fast spread throughout the world, and the mounting number of infected persons have triggered a prompt and resolute quest for therapeutic options to treat this serious infectious disease.
In a recent work, the SARS-CoV-2-human interactome was characterized, and +300 PPIs were identified. It was found that 66 of the interacting host factors could be modulated with 69 compounds, including some approved drugs (Gordon et al., 2020b). This study will certainly enhance the possibilities of drug discovery efforts targeting host cells receptors. As of today, candidates include the antimalarial chloroquine and its derivatives (Zou et al., 2020), anti-inflammatory drugs and immunomodulators, and even anticoagulants and anti-fibrinolytic drugs (Hoffmann et al., 2020).
Although the factors that condition the progression of the disease in severe cases are not fully understood, evidence seems to show that the symptoms in patients with moderate to severe disease due to SARS-CoV-2 are related not only to viral proliferation, but also to two factors related to its pathogenesis: i) an exacerbated inflammatory response associated with increased concentrations of proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and ILs, including IL-1 and IL-6 (Hadjadj et al., 2020; Mehta et al., 2020), and ii) abnormalities in coagulation and thrombosis, similar to a combination of mild disseminated intravascular coagulopathy and a localized pulmonary thrombotic microangiopathy, which could have a substantial impact on organ dysfunction in the more severely affected patients (Levi et al., 2020).
It is clear that a thorough characterization of the druggability of the SARS-CoV-2 proteome would provide a rich array of alternative targets for drug discovery. In this work we present an in-depth functional, structural and druggability assessment of all non-structural, structural, and accessory proteins of SARS-CoV-2, identifying potential druggable allosteric and PPI sites throughout the whole proteome, thus broadening the repertoire of current targetable proteins. It should be stressed that druggability characterization of a site does not necessarily imply that a compound binding at that site will modulate that target and exhibit an observable biological effect.
We would like to highlight three interesting drug discovery strategies:
-
•
The helicase (nsp13), and methyltransferases N7-Mtase (nsp14) and 2′-O-MTase (nsp16) contribute to genome stability through their involvement in the capping process; interfering with the functioning of one or more of these proteins could be an excellent antiviral approach.
-
•
The ExoN (nsp14) is responsible for proofreading, and thus for the extremely low mutation rate and nucleoside analogs resistance of SARS-CoV-2; a therapeutic combo of Remdesivir (or other RdRp nucleoside compound) in combination with an ExoN inhibitor could be a valid approach to boost the efficacy of nucleoside-based therapies.
-
•
In CoVs, a delayed IFN response is a redundant pattern that allows robust viral replication, and also induces the accumulation of cytokine-producing macrophages, thus increasing the severity of the disease (Chen and Subbarao, 2007). Targeting one (or a combination) of NendoU (nsp15), ADRP, PLpro, orf6, and orf9b, would certainly obstruct viral replication.
We are convinced that our work will contribute to the quick development of an effective SARS-CoV-2 antiviral strategy which, in view of the high similarity among CoVs, might be useful to fight related viruses. Moreover, these therapeutic options might be instrumental in fighting different CoV-associated diseases that could threaten global health in the future.
Competing interests
The Authors declare that no competing interests exist.
CRediT authorship contribution statement
Claudio N. Cavasotto: conceived the original idea of the work, designed the research process, performed the functional, structural and druggability, Formal analysis, interpreted the results, wrote the manuscript. Maximiliano Sánchez Lamas: contributed to the, Formal analysis, interpretation of results, and to the manuscript. Julián Maggini: Formal analysis, interpretation of results, and to the manuscript, All authors have reviewed the manuscript and approved the submission.
Acknowledgments
The Authors thank Lucía Cavasotto, M. Laura Fernández, and Pilar Cossio for critical reading of the manuscript. This work was supported by the National Agency for the Promotion of Science and Technology (ANPCyT) (PICT-2017-3767). CNC thanks Molsoft LLC (San Diego, CA) for providing an academic license for the ICM program.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejphar.2020.173705.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abagyan R., Kufareva I. The flexible pocketome engine for structural chemogenomics. Methods Mol. Biol. 2009;575:249–279. doi: 10.1007/978-1-60761-274-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J.A., Memish Z.A. Update on therapeutic options for Middle East respiratory syndrome coronavirus (MERS-CoV) Expert Rev. Anti Infect. Ther. 2017;15:269–275. doi: 10.1080/14787210.2017.1271712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad Y.M.O., Kashipathy M.M., Roy A., Johnson D.K., McDonald P., Battaile K.P., Gao P., Lovell S., Fehr A.R. The SARS-CoV-2 conserved macrodomain is a highly efficient ADP-ribosylhydrolase enzyme. bioRxiv. 2020 doi: 10.1101/2020.05.11.089375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y.M., Barraza S.J., Wilson M.W., Agius M.P., Mielech A.M., Davis N.M., Baker S.C., Larsen S.D., Mesecar A.D. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrantes F.J. While we wait for a vaccine against SARS-CoV-2, why not think about available drugs? Front. Physiol. 2020;11:820. doi: 10.3389/fphys.2020.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum F., Banerjee A.K., Tripathi P.P., Ray U. Two mutations P/L and Y/C in SARS-CoV-2 helicase domain exist together and influence helicase RNA binding. bioRxiv. 2020 doi: 10.1101/2020.05.14.095224. [DOI] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R., Forni D., Clerici M., Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83:104353. doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvino F.R., Kharde S., Ori A., Hendricks A., Wild K., Kressler D., Bange G., Hurt E., Beck M., Sinning I. Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA-binding site. Nat. Commun. 2015;6:6510. doi: 10.1038/ncomms7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo L., Oliva R. D936Y and Other Mutations in the Fusion Core of the SARS-Cov-2 Spike Protein Heptad Repeat 1 Undermine the Post-Fusion Assembly. bioRxiv. 2020 doi: 10.1101/2020.06.08.140152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasotto C.N., Aucar M.G. High-Throughput docking using quantum mechanical scoring. Front. Chem. 2020;8:246. doi: 10.3389/fchem.2020.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasotto C.N., Di Filippo J.I. In silico drug repurposing for COVID-19: targeting SARS-CoV-2 proteins through docking and consensus ranking. Mol. Inform. 2020;39:2000115. doi: 10.1002/minf.202000115. [DOI] [PubMed] [Google Scholar]
- Cavasotto C.N., Palomba D. Expanding the horizons of G protein-coupled receptor structure-based ligand discovery and optimization using homology models. Chem. Commun. 2015;51:13576–13594. doi: 10.1039/c5cc05050b. [DOI] [PubMed] [Google Scholar]
- Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K., Chan H.Y. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Johnson M.A., Serrano P., Pedrini B., Joseph J.S., Neuman B.W., Saikatendu K., Buchmeier M.J., Kuhn P., Wüthrich K. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J. Virol. 2009;83:1823–1836. doi: 10.1128/JVI.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Fang S., Tam J.P., Liu D.X. Formation of stable homodimer via the C-terminal α-helical domain of coronavirus nonstructural protein 9 is critical for its function in viral replication. Virology. 2009;383:328–337. doi: 10.1016/j.virol.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Subbarao K. The immunobiology of SARS*. Annu. Rev. Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- Chen S.C., Lo S.Y., Ma H.C., Li H.C. Expression and membrane integration of SARS-CoV E protein and its interaction with M protein. Virus Gene. 2009;38:365–371. doi: 10.1007/s11262-009-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tao J., Sun Y., Wu A., Su C., Gao G., Cai H., Qiu S., Wu Y., Ahola T., Guo D. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J. Virol. 2013;87:6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M., Anderson T.K., Jockusch S., Tao C., Li X., Kumar S., Russo J.J., Kirchdoerfer R.N., Ju J. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J. Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P., Weinkam P., Rettenmaier T.J., Bichmann L., Keedy D.A., Woldeyes R.A., Schneidman-Duhovny D., Demerdash O.N., Mitchell J.C., Wells J.A., Fraser J.S., Sali A. CryptoSite: expanding the druggable proteome by characterization and prediction of cryptic binding sites. J. Mol. Biol. 2016;428:709–719. doi: 10.1016/j.jmb.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasman J.R., Baez-Santos Y.M., Mettelman R.C., O'Brien A., Baker S.C., Mesecar A.D. X-ray structure and enzymatic activity profile of a core papain-like protease of MERS coronavirus with utility for structure-based drug design. Sci. Rep. 2017;7:40292. doi: 10.1038/srep40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz M.A., Kanjanahaluethai A., O'Brien T.E., Baker S.C. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375:118–129. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zhang B., Su H., Li J., Zhao Y., Xie X., Jin Z., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (Nucleoside-2′O)-Methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J. Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh D.C., Chalupska D., Silhan J., Veverka V., Boura E. Structural Basis of RNA Recognition by the SARS-CoV-2 Nucleocapsid Phosphoprotein. bioRxiv. 2020 doi: 10.1101/2020.04.02.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech R., Hernandez-Cifre J.G., Bacarizo J., Diez-Pena A.I., Martinez-Rodriguez S., Cavasotto C.N., de la Torre J.G., Camara-Artigas A., Velazquez-Campoy A., Neira J.L. The histidine-phosphocarrier protein of the phosphoenolpyruvate: sugar phosphotransferase system of Bacillus sphaericus self-associates. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez Andres A., Feng Y., Campos A.R., Yin J., Yang C.-C., James B., Murad R., Kim H., Deshpande A.J., Gordon D.E., Krogan N., Pippa R., Ronai Z.e.A. SARS-CoV-2 ORF9c Is a Membrane-Associated Protein that Suppresses Antiviral Responses in Cells. bioRxiv. 2020 doi: 10.1101/2020.08.18.256776. [DOI] [Google Scholar]
- Egloff M.-P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7 doi: 10.1128/mBio.01721-16. 1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Singh S.A., Kerr C.M., Mukai S., Higashi H., Aikawa M. The impact of PARPs and ADP-ribosylation on inflammation and host–pathogen interactions. Genes Dev. 2020;34:341–359. doi: 10.1101/gad.334425.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fintelman-Rodrigues N., Sacramento C.Q., Lima C.R., da Silva F.S., Ferreira A.C., Mattos M., de Freitas C.S., Soares V.C., Gomes Dias S.d.S., Temerozo J.R., Miranda M., Matos A.R., Bozza F.A., Carels N., Alves C.R., Siqueira M.M., Bozza P.T., Souza T.M.L. Atazanavir Inhibits SARS-CoV-2 Replication and Pro-inflammatory Cytokine Production. Antimicrob Agents Chemother. 2020;64(10):e00825–820. doi: 10.1128/AAC.00825-20. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadlage M.J., Graham R.L., Denison M.R. Murine coronaviruses encoding nsp2 at different genomic loci have altered replication, protein expression, and localization. J. Virol. 2008;82:11964–11969. doi: 10.1128/JVI.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J.E., Gibbons I.R. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genom. 2002;3:18. doi: 10.1186/1471-2164-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Sims A.C., Brockway S.M., Baric R.S., Denison M.R. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U.S.A. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J., Reggiori F., de Haan C.A. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Zhuang M.-W., Zheng Y., Zhang J., Nan M.-L., Wang P.-H., Gao C. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. bioRxiv. 2020 doi: 10.1101/2020.08.16.252973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., Theriault S., Cutts T., Cao J., Booth T.F., Plummer F.A., Tyler S., Li X. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Schroeder S., Kleine-Weber H., Muller M.A., Drosten C., Pohlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64:e00754–20. doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Chen C., Li H., Dou Y., Zhou M., Lu D., Zong Q., Li Y., Yang C., Zhong Z., Singh N., Hu H., Zhang R., Yang H., Su D. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017;26:1037–1048. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5:e00266–20. doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu J., Yang X., Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Yang H., Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- Johnson M.A., Chatterjee A., Neuman B.W., Wuthrich K. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J. Mol. Biol. 2010;400:724–742. doi: 10.1016/j.jmb.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;7:1128–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S., Niiro A., Yasui F., Kohara M., Morita K., Matsushima K., Le M.Q., Masuda T., Kannagi M. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato F., Matsuyama S., Kawase M., Hishiki T., Katoh H., Takeda M. Antiviral activities of mycophenolic acid and IMD-0354 against SARS-CoV-2. Microbiol. Immunol. 2020;64:635–639. doi: 10.1111/1348-0421.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M., Chen Y., Wu A., Sun Y., Su C., Wu H., Jin X., Tao J., Wang Y., Ma X., Pan J.-A., Guo D. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012;167:322–328. doi: 10.1016/j.virusres.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Giedroc D.P. Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J. Virol. 2013;87:3502–3515. doi: 10.1128/JVI.03112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D.M., Sorum B., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Brohawn S.G. Cryo-EM Structure of the SARS-CoV-2 3a Ion Channel in Lipid Nanodiscs. bioRxiv. 2020 doi: 10.1101/2020.06.17.156554. [DOI] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G.C., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E, Blackmore T.R., van der Heden van Noort G.J., Geurink P.P., Ovaa H., Newman J., Riboldi-Tunnicliffe A., Czabotar P.E., Mitchell J.P., Feltham R., Lechtenberg B.C., Lowes K.N., Dewson G., Pellegrini M., Lessene G., Komander D. Mechanism and inhibition of the papain‐like protease, PLpro, of SARS‐CoV‐2. EMBO J. 2020;39:e106275. doi: 10.15252/embj.2020106275. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D., Grove L.E., Hall D.R., Bohnuud T., Mottarella S.E., Luo L., Xia B., Beglov D., Vajda S. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D., Hall D.R., Napoleon R.L., Yueh C., Whitty A., Vajda S. New frontiers in druggability. J. Med. Chem. 2015;58:9063–9088. doi: 10.1021/acs.jmedchem.5b00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Gunalan V., Liu B., Chow V.T., Druce J., Birch C., Catton M., Fielding B.C., Tan Y.J., Lal S.K. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366:293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Hurst-Hess K.R., Koetzner C.A., Masters P.S. Analyses of coronavirus assembly interactions with interspecies membrane and nucleocapsid protein chimeras. J. Virol. 2016;90:4357–4368. doi: 10.1128/JVI.03212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Ren J., Pesavento R.P., Ojeda I., Rice A.J., Lv H., Kwon Y., Johnson M.E. Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via high-throughput screening and molecular modeling. Bioorg. Med. Chem. 2019;27:1981–1989. doi: 10.1016/j.bmc.2019.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi M., Thachil J., Iba T., Levy J.H. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet. Haematol. 2020;7:e438–e440. doi: 10.1016/S2352-3026(20)30145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., Wang C., Zhang Y., Zhu Z., Liu Z., Peng C., Zhu T., Liang Q. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2020 doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., Ge X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Narayanan K., Huang C., Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui P.Y., Wong L.Y., Fung C.L., Siu K.L., Yeung M.L., Yuen K.S., Chan C.P., Woo P.C., Yuen K.Y., Jin D.Y. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg. Microb. Infect. 2016;5:e39. doi: 10.1038/emi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes N., Mesters J.R., Coutard B., Canard B., Snijder E.J., Moll R., Hilgenfeld R. The non-structural protein Nsp10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 2006;580:4143–4149. doi: 10.1016/j.febslet.2006.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K., Schafer A., Pham A., Frieman M. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol. J. 2014;11:209. doi: 10.1186/s12985-014-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J., Grimes J.M., Stuart D.I. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miknis Z.J., Donaldson E.F., Umland T.C., Rimmer R.A., Baric R.S., Schultz L.W. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 2009;83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., Gloza-Rausch F., Balboni A., Battilani M., Rihtarič D., Toplak I., Ameneiros R.S., Pfeifer A., Thiel V., Drexler J.F., Müller M.A., Drosten C. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Nedialkova D.D., Ulferts R., van den Born E., Lauber C., Gorbalenya A.E., Ziebuhr J., Snijder E.J. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J. Virol. 2009;83:5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., Ferraro D., Pewe L., Olivares H., Gallagher T., Perlman S. Enhancement of murine coronavirus replication by severe acute respiratory syndrome coronavirus protein 6 requires the N-terminal hydrophobic region but not C-terminal sorting motifs. J. Virol. 2007;81:11520–11525. doi: 10.1128/JVI.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Verdia-Baguena C., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer K., Milewska A., Owczarek K., Dabrowska A., Branicki W., Sanak M., Pyrc K. The SARS-CoV-2 ORF10 Is Not Essential in Vitro or in Vivo in Humans. bioRxiv. 2020 doi: 10.1101/2020.08.29.257360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections—more than just the common cold. J. Am. Med. Assoc. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., Gao G.F., Shi Y. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Soong T.W., Liu D.X., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L., Zhou H., Netland J., Tangudu C., Olivares H., Shi L., Look D., Gallagher T., Perlman S. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 2005;79:11335–11342. doi: 10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Krahling V., Ditt V., Grywna K., Muhlberger E., Drosten C. Reverse genetic characterization of the natural genomic deletion in SARS-Coronavirus strain Frankfurt-1 open reading frame 7b reveals an attenuating function of the 7b protein in-vitro and in-vivo. Virol. J. 2009;6:131. doi: 10.1186/1743-422X-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatak S.S., Gatica E.A., Cavasotto C.N. Ligand-steered modeling and docking: a benchmarking study in Class A G-Protein-Coupled Receptors. J. Chem. Inf. Model. 2010;50:2119–2128. doi: 10.1021/ci100285f. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.-Y. Spike Mutation D614G Alters SARS-CoV-2 Fitness and Neutralization Susceptibility. bioRxiv. 2020 doi: 10.1101/2020.09.01.278689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Jaroszewski L., Godzik A., Joachimiak A., Satchell K.J.F. The Crystal Structure of Nsp10-Nsp16 Heterodimer from SARS-CoV-2 in Complex with S-Adenosylmethionine. bioRxiv. 2020 doi: 10.1101/2020.04.17.047498. [DOI] [Google Scholar]
- Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity Profiling and Structures of Inhibitor-Bound SARS-CoV-2-PLpro Protease Provides a Framework for Anti-COVID-19 Drug Design. bioRxiv. 2020 doi: 10.1101/2020.04.29.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., Neuman B.W., Subramanian V., Saikatendu K.S., Buchmeier M.J., Stevens R.C., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:12049–12060. doi: 10.1128/JVI.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Chatterjee A., Neuman B.W., Joseph J.S., Buchmeier M.J., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3. J. Virol. 2009;83:12998–13008. doi: 10.1128/JVI.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G., Hong W., Tan Y.J. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Xue J.H., Yu C.Y., Luo H.B., Qin L., Yu X.J., Chen J., Chen L.L., Xiong B., Yue L.D., Cai J.H., Shen J.H., Luo X.M., Chen K.X., Shi T.L., Li Y.X., Hu G.X., Jiang H.L. Small envelope protein E of SARS: cloning, expression, purification, CD determination, and bioinformatics analysis. Acta Pharmacol. Sin. 2003;24:505–511. [PubMed] [Google Scholar]
- Shrimp J.H., Kales S.C., Sanderson P.E., Simeonov A., Shen M., Hall M.D. An enzymatic TMPRSS2 assay for assessment of clinical candidates and discovery of inhibitors as potential treatment of COVID-19. ACS Pharmacol. Trans. Sci. 2020;3:997–1007. doi: 10.1021/acsptsci.0c00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Hilgenfeld R. Acquisition of new protein domains by coronaviruses: analysis of overlapping genes coding for proteins N and 9b in SARS coronavirus. Virus Gene. 2015;50:29–38. doi: 10.1007/s11262-014-1139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Decroly E., Khatib A.M., Villoutreix B.O. Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages. Eur. J. Pharmaceut. Sci. 2020;153:105495. doi: 10.1016/j.ejps.2020.105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Chan C.P., Kok K.H., Chiu-Yat Woo P., Jin D.Y. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 2014;11:141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F., Koster A.J., Barcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks J.S., Lu X., Denison M.R. Genetic analysis of Murine hepatitis virus nsp4 in virus replication. J. Virol. 2007;81:12554–12563. doi: 10.1128/JVI.01257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim. Biophys. Acta Biomembr. 2018;1860:1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L.M., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaim C.D., Perng Y.C., Zhao X., Canadeo L.A., Harastani H.H., Darling T.L., Boon A.C.M., Lenschow D.J., Huibregtse J.M. 6-Thioguanine blocks SARS-CoV-2 replication by inhibition of PLpro protease activities. bioRxiv. 2020 doi: 10.1101/2020.07.01.183020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Schneider T., Leong M., Aravind L., Zhang D. Novel immunoglobulin domain proteins provide insights into evolution and pathogenesis of SARS-CoV-2-related viruses. mBio. 2020;11 doi: 10.1128/mBio.00760-20. e00760-00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Tham P.Y., Chan D.Z., Chou C.F., Shen S., Fielding B.C., Tan T.H., Lim S.G., Hong W. The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J. Virol. 2005;79:10083–10087. doi: 10.1128/JVI.79.15.10083-10087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81:1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- teVelthuis A.J.W. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 2014;71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh K.T., Siu Y.L., Chan W.L., Schluter M.A., Liu C.J., Peiris J.S., Bruzzone R., Margolis B., Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Sturzel C.M., Frohlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvarogová J., Madhugiri R., Bylapudi G., Ferguson L.J., Karl N., Ziebuhr J. Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3′-terminal adenylyltransferase activity. J. Virol. 2019;93 doi: 10.1128/JVI.00291-19. 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda S., Beglov D., Wakefield A.E., Egbert M., Whitty A. Cryptic binding sites on proteins: definition, detection, and druggability. Curr. Opin. Chem. Biol. 2018;44:1–8. doi: 10.1016/j.cbpa.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villoutreix B.O., Beaune P.H., Tamouza R., Krishnamoorthy R., Leboyer M. Prevention of COVID-19 by drug repurposing: rationale from drugs prescribed for mental disorders. Drug Discov. Today. 2020;25:1287–1290. doi: 10.1016/j.drudis.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., Park J.-G., Oladunni F., Kovalskyy D., Hromas R.A., Martinez-Sobrido L., Gupta Y.K. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11:3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Węglarz-Tomczak E., Tomczak J.M., Giurg M., Burda-Grabowska M., Brul S. Discovery of Potent Inhibitors of PLpro CoV2 by Screening a Library of Selenium-Containing Compounds. bioRxiv. 2020 doi: 10.1101/2020.05.20.107052. [DOI] [Google Scholar]
- Węglarz-Tomczak E., Tomczak J.M., Talma M., Brul S. Ebselen as a Highly Active Inhibitor of PLpro CoV2. bioRxiv. 2020 doi: 10.1101/2020.05.17.100768. [DOI] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in wuhan, China. JAMA Int. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Zheng B.J., Zeng R., Lu W., Lin Y.P., Xue L., Li L., Yang L.L., Xu C., Dai J., Wang F., Li Q., Dong Q.X., Yang R.F., Wu J.R., Sun B. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29:1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.-C., Tian G., Jiang H.-W., Tao S.-C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Deng F., Shi K., Ye G., Wang G., Fang L., Xiao S., Fu Z., Peng G. Dimerization of coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. J. Virol. 2018;92 doi: 10.1128/JVI.00692-18. 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Hui K.Y., Gusev A., Warner N., Ng S.M., Ferguson J., Choi M., Burberry A., Abraham C., Mayer L., Desnick R.J., Cardinale C.J., Hakonarson H., Waterman M., Chowers Y., Karban A., Brant S.R., Silverberg M.S., Gregersen P.K., Katz S., Lifton R.P., Zhao H., Nunez G., Pe'er I., Peter I., Cho J.H. Extended haplotype association study in Crohn's disease identifies a novel, Ashkenazi Jewish-specific missense mutation in the NF-kappaB pathway gene. HEATR3. Genes Immun. 2013;14:310–316. doi: 10.1038/gene.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., Yang T., Yu F., Liu J., Liu B., Song Z., Chen J., Pan T., Zhang X., Li Y., Li R., Huang W., Xiao F., Zhang H. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv. 2020 doi: 10.1101/2020.05.24.111823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Falcon A., Zhou H., Netland J., Enjuanes L., Perez Brena P., Perlman S. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 2009;83:2368–2373. doi: 10.1128/JVI.02371-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Li H., Wang H., Wang L.F., Shi Z. Bat severe acute respiratory syndrome-like coronavirus ORF3b homologues display different interferon antagonist activities. J. Gen. Virol. 2012;93:275–281. doi: 10.1099/vir.0.033589-0. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Kato-Weinstein J., Li Y., Deng Y., Granet R., Garner L., Liu C.Y., Polshakov D., Gessner C., Watkins S. Potential therapeutic agents and associated bioassay data for COVID-19 and related human coronavirus infections. ACS Pharmacol. Trans. Sci. 2020;3:813–834. doi: 10.1021/acsptsci.0c00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Yan H., Chong H., He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 2020;94 doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Dai L., Zhang X., Zhang Z., Zhang Z. Hydroxychloroquine and chloroquine: a potential and controversial treatment for COVID-19. Arch Pharm. Res. (Seoul) 2020;43:765–772. doi: 10.1007/s12272-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.