Abstract
The mouth presents a multiplicity of local environments in communication with one another via saliva. The spatial organization of microbes within the mouth is shaped by opposing forces in dynamic equilibrium—salivary flow and adhesion, shedding and colonization—and by interactions among and between microbes and the host. Here we review recent evidence confirming that oral microbes are specialized for individual habitats within the mouth and that microbial habitats and niches are defined by micron-scale gradients in combination with short- and long-range interactions. Micron-scale structure illuminates the roles of individual taxa and provides insight into their community ecology and potential pathogenicity.
INTRODUCTION
The human mouth is a natural laboratory for microbial ecology. The mouth presents a range of substrates—such as teeth, tongue, cheeks, and gums—whose differing chemistry, topography, and stability provide differing habitats for microbial communities. Each habitat within the mouth supports a complex, distinctive community, and this distinctiveness presents an opportunity to use the oral microbiome to develop an understanding of fundamental principles of microbial community ecology. Oral microbiome communities are also of immediate practical importance, because they influence health and disease not only in the mouth but also throughout the body, and they play under-appreciated roles in systemic human physiology.
A major goal of microbiome research, from a clinical perspective, is to be able to modulate the microbiome to improve health and treat disease. Microbes colonizing the mouth create spatially organized biofilms (Kolenbrander et al., 2006; Zijnge et al., 2010; Mark Welch et al., 2016; Wilbert et al., 2020; Kim et al., 2020). Understanding the forces governing this spatial organization will be key to success in modulating the microbiome. The concept that spatial order is important in microbial ecosystems is not new; it was introduced implicitly by Winogradsky, the founder of microbial ecology, almost 100 years ago and was explicitly addressed 40 years ago by Wimpenny (1981). Wimpenny discussed the concepts of habitat and niche as they apply to microorganisms. Habitat refers to externalities—the physical space and chemical environment that allow an organism to exist, including contributions from other members of the microbial community. Niche refers to the activity of an organism and the functional role that each member plays in the community. Interactions of the members both with one another and with the habitat drive the emergent organization of the community as a whole.
Recent reviews have highlighted the importance of spatial organization in various aspects of human microbial ecology including extracellular matrix and polymicrobial infections (Stacy et al., 2016; Bowen et al., 2018), the gut microbiome (Tropini et al., 2017), landscape ecology of the upper respiratory tract and mouth (Proctor and Relman, 2017), and implications for caries and periodontal disease (Valm, 2019; Diaz and Valm, 2020; Proctor et al., 2020). In this Minireview, we will focus on how oral microbes are specialized for individual habitats within the mouth and how microbial habitats and niches are defined by micron-scale gradients in combination with short- and long-range interactions. We will discuss how micron-scale structure illuminates the roles of individual taxa and provides insight into their community ecology and potential pathogenicity.
MAJOR HABITATS WITHIN THE MOUTH
The mouth is an open system. Microbes are inhaled with every breath, ingested with every meal or drink, and introduced by close contact with other humans, animals, or our physical surroundings. As a warm, moist, and nutrient-rich environment, the mouth presents a welcoming home for microbes. Yet among the millions of bacterial species on the planet, only approximately 760 are primary residents, rather than transients, in the mouth (Dewhirst et al., 2010; https://www.homd.org, v15.2). These species are not evenly represented; several dozen abundant and prevalent species make up most of the biomass at each oral site, whereas many species have low prevalence as well as low abundance in the healthy human mouth (Eren et al., 2014; Mark Welch et al., 2019). Microbes from other body sites, which are adapted to living on or in the human body and to which there must be fairly continual exposure, are not abundant in the mouth. What accounts for these unique microbial demographics? A major determinant is the interplay between the main habitats in the mouth and the selective forces operating on them.
Nine of the distinctive habitats in the mouth were sampled in the Human Microbiome Project (HMP) (Segata et al., 2012). Although any surface within the mouth is potentially available for colonization, the HMP selected sites that reflected the range of habitat diversity (Figure 1). Surfaces sampled included the tooth surface, both above the gum line (supragingival plaque) and below the gum line (subgingival plaque) (Figure 1A); flexible, non-keratinized epithelia (buccal mucosa and throat); keratinized epithelia (attached gingiva and the hard palate); and specialized epithelia (tonsils and the tongue dorsum) (Figures 1B and 1C). Saliva, although not a site in itself, was also sampled. Each of these sites is not monolithic, because differing environmental conditions even within a single oral site can generate differing micro-habitats. Different sides or aspects of teeth are sheltered or exposed to different degrees. Oxygen is abundant on the crowns of teeth, but the tooth surface in the gingival crevice is in an anoxic environment bathed in gingival crevicular fluid, a protein-rich exudate from the gingival tissues (Jakubovics, 2015a). The film of saliva is thinnest at the roof of the mouth while saliva pools at the floor of the mouth; precise location relative to salivary glands influences the composition and rate of flow of saliva at different sites in the mouth (Proctor and Relman, 2017; Proctor et al., 2020). The dorsum of the tongue is populated by abundant filiform papillae and fungiform papillae, and a row of large circumvallate papillae, foliate papillae, and lingual tonsils add topographic complexity to the back of the tongue (Figure 1C). Additional diversity of oral habitats may exist that is not fully reflected in these nine site categories, as evidenced by the high abundance in saliva of microbes that were not abundant at any of the other sampled sites, suggesting additional unique micro-habitats elsewhere in the mouth (Segata et al., 2012; Eren et al., 2014). Thus, the mouth provides a structurally complex environment with a range of differentiated and, as yet, incompletely explored habitats.
Figure 1. Major Habitats within the Mouth.
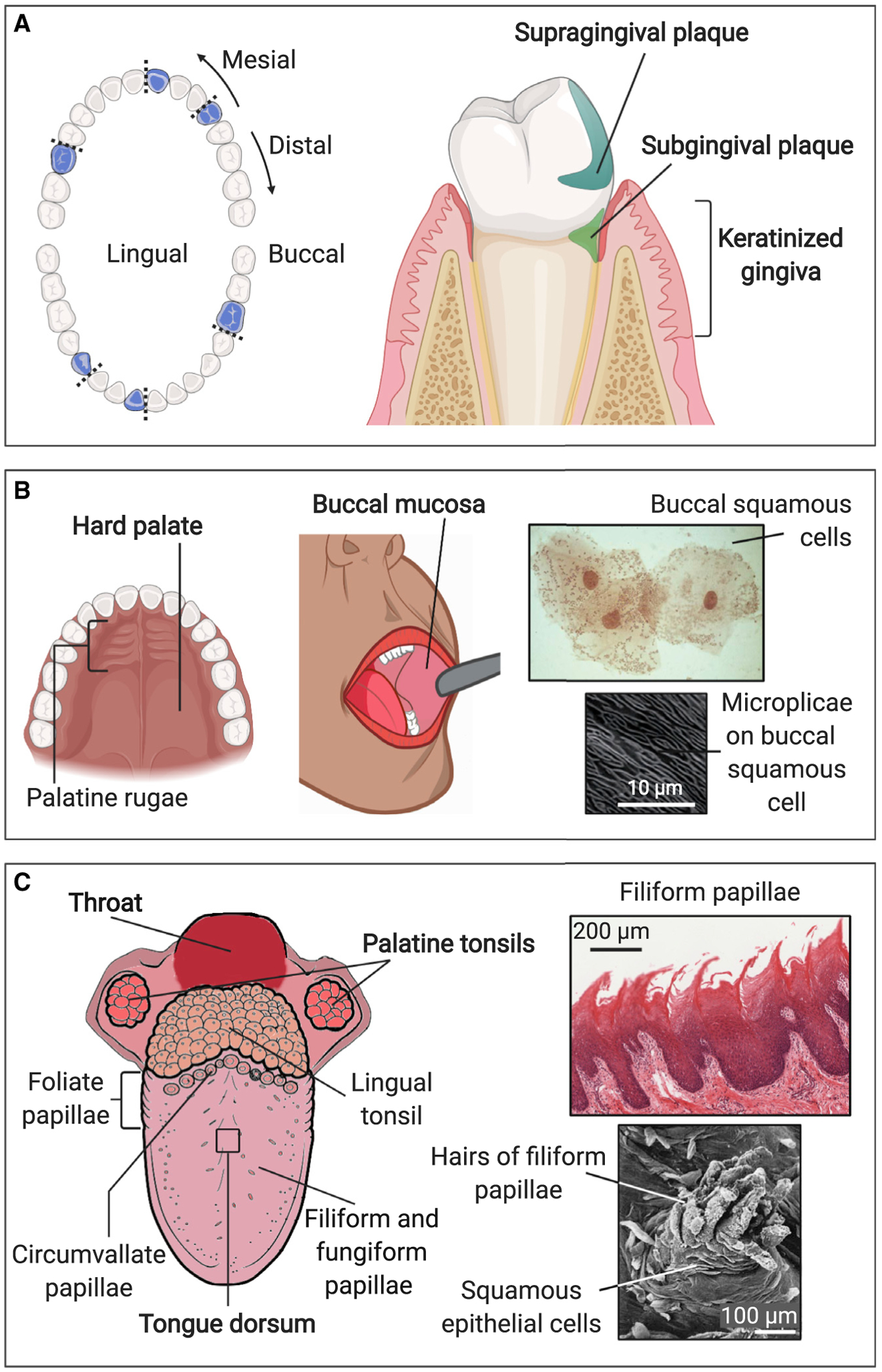
The mouth contains a range of topographies and substrates that present varied habitats for microbes. Samples for DNA sequencing are usually collected by swabbing or scraping an area of 1 cm2 or more, but features at millimeter to micron scales could influence microbial colonization and create specialized micro-habitats supporting distinctive microbiotas. Sites sampled by the HMP are labeled in bold; some additional sites (not bold) are also shown.
(A) Supragingival plaque, subgingival plaque, keratinized gingiva; blue indicates the position of teeth sampled in the HMP.
(B) Hard palate, buccal mucosa; higher-magnification images show micro-habitats on buccal mucosa. Buccal mucosa smear modified from Datar et al. (2013), copyright 2013 Karger Publishers, Basel, Switzerland. Microplicae modified from Kullaa et al. (2014), reprinted by permission of the publisher, Taylor & Francis Ltd, http://www.tandfonline.com.
(C) Palatine tonsils, throat, tongue dorsum; higher-magnification images show micro-habitats on tongue dorsum. Tongue diagram modified from Rice University’s OpenStax (https://openstax.org/books/anatomy-and-physiology/pages/23-3-the-mouth-pharynx-and-esophagus) under a Creative Commons International license (https://creativecommons.org/licenses/by/4.0); filiform papillae modified from https://histology.medicine.umich.edu/ - slide 116 under a Creative Commons Noncommercial-Share Alike license (https://creativecommons.org/licenses/by-nc-sa/3.0); hairs of filiform papillae modified from Kullaa-Mikkonen and Sorvari, 1985, copyright 1985 Karger Publishers, Basel, Switzerland. Illustrations created with BioRender.com.
SELECTIVE FORCES WITHIN THE MOUTH
The mouth’s unique microbial demographics indicate clearly that there are selective forces to which a microbe must be adapted in order to thrive in the mouth and to which the host must be adapted in order to interact effectively with the colonizing microbes. The interaction might be understood as three pairs of opposing factors (Figure 2) that are in dynamic equilibrium in the healthy state.
Figure 2. Selective Forces within the Mouth.
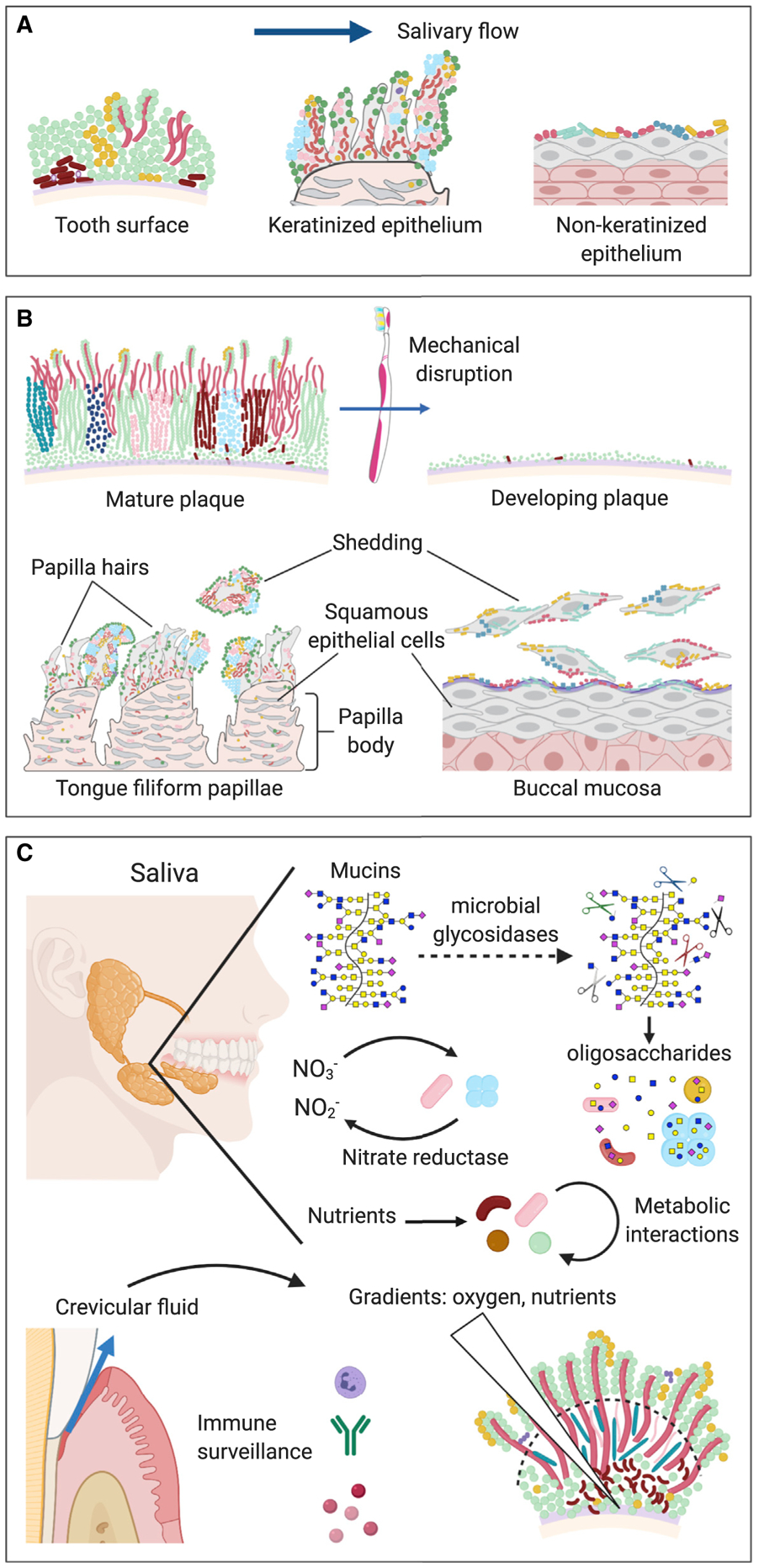
The oral microbiome is shaped by a dynamic equilibrium of opposing factors: flow and adhesion, shedding and colonization, and host and microbe.
(A) Flow and adhesion. Salivary flow exerts a selective requirement for adhesion to oral substrates, both for retention in the mouth and for localization to a favorable metabolic environment. Distinctive oral substrates include the tooth surface, keratinized epithelia, and non-keratinized epithelia.
(B) Shedding and colonization. Mechanical disruption of the biofilm or shedding of the underlying substrate is balanced by colonization of newly available substrate. The development of distinctive communities at the different oral sites occurs by differential binding of microbes to the different oral substrates, regrowth of the residual microbial biofilm, and colonization of the growing biofilm by additional taxa. Long-lived surfaces such as tooth surface and tongue dorsum develop thick biofilms, whereas the biofilm on rapidly shedding surfaces such as buccal mucosa is thin.
(C) Host and microbe. The host and the microbial community exert mutual reciprocal influences on each other through binding interactions, immune surveillance, and gradients of nutrients and solutes. The host secretes salivary mucins, which are complex glycoproteins that support the growth of mixed syntrophic communities of microbes that possess glycosidases capable of releasing oligosaccharides from mucins. Secretion of nitrate and other nutrients into saliva by the host, and release of crevicular fluid from the gingival crevice into the mouth, could also serve to foster the growth of particular microbes, while immune surveillance limits the growth of others. Microbial metabolism, in turn, can generate strong localized gradients of oxygen and nutrients. The positioning of microbes at favorable locations within these gradients can lead to metabolic interaction and spatial structure within the microbial community. Illustrations created with BioRender.com.
Flow and adhesion.
Retention in the oral environment is shaped by the interplay between the opposing forces of flow and adhesion (Figure 2A). Salivary flow imposes a selective requirement for adherence in the oral cavity: microbes can persist in exposed locations in the mouth only if they are adhered to an underlying substrate or to other microbes that are adhered to the substrate (Gibbons and Van Houte, 1975). Flow also prevents the buildup of microbial metabolites in the mouth and therefore imposes a requirement that microbes be in close proximity in order to interact with one another (Egland et al., 2004). In response to these selective pressures, oral microbes developed highly specific adhesin-receptor interactions that form the basis of a phenomenon referred to as coadhesion or coaggregation, in which binding occurs between microbes of different genera (Kolenbrander et al., 2006). The binding interactions are strain specific and, in some cases, also site specific. For example, veillonellae and streptococci are common inhabitants of the oral cavity, yet veillonellae from the tongue bind to streptococci from the tongue while veillonellae from dental plaque bind to streptococci from dental plaque. Thus, flow applies selective pressure for adherence, both for retention in general and for localization to a precise metabolic environment. The microbial response of adhesion to specific targets and partners located in well-defined regions of the mouth suggests that precise spatial positioning is critical for microbial survival.
Shedding and colonization.
The microbial community at each site within the mouth is further shaped by the relative dynamics of shedding of (or removal from) the underlying substrate and re-colonization back to the substrate (Figure 2B). On the permanently exposed enamel surface of teeth, the dynamics of removal are set by oral hygiene or by abrasion caused by chewing of food. In areas sheltered from abrasion and oral hygiene, the plaque biofilm can persist for long periods. This long residence time permits the development of complex communities via succession and diversification (Listgarten et al., 1975; Kolenbrander et al., 2006; Valm, 2019). On mucosal surfaces, by contrast, epithelial cells are regularly shed, along with the attached microbiota, and removed from the mouth by swallowing. The rate of shedding, and the rate and nature of colonization on newly presenting epithelial surfaces, influences the density of the mucosal community as well as its membership and spatial organization (Gibbons and Van Houte, 1975). Based on the surface area of the mucosal surfaces in the mouth and the number of epithelial cells in saliva, Dawes (2003) estimated that the surface layer of oral epithelial cells was replaced every 2.7 h—a rate that precludes the formation of thick biofilms, and indeed Dawes reported only monolayers of bacteria on the rapidly shedding epithelial cells of the palate, gums, and tongue dorsum.
More recently, by gentle scraping of the tongue dorsum, we detected not only thinly colonized epithelial cells but also much thicker biofilms with complex highly structured organization (Wilbert et al., 2020), suggesting that the epithelia of the tongue dorsum are a mixture of both rapidly shedding, thinly colonized cells and longer-lived structures on which a more substantial biofilm can develop. Not only the overall thickness but also the spatial structure of the biofilm are influenced by the rate of shedding. Oral biofilms can be analyzed as a mosaic of patches of distinctive microbial composition (Proctor and Relman, 2017). One could infer that the rate of shedding would influence the size of such patches. Thin biofilms on oral surfaces are likely to be well oxygenated throughout, but as the biofilm increases in thickness, anoxic habitats can develop, and the abundance of anaerobes could increase in these micro-habitats (Mark Welch et al., 2016; Wilbert et al., 2020). Thus, the host property of epithelial shedding contributes to structuring the composition and spatial organization of microbial biofilms in the mouth.
The microbial response to shedding, and to the loss of microbes from non-shedding surfaces by abrasion, is colonization of fresh substrates. A clean enamel surface in the mouth immediately acquires a salivary pellicle or protein-based covering, to which pioneer colonizing microbes bind with high specificity. For example, cell surface adhesins on pioneer colonizing streptococci bind to cysteine repeat domains within glycoproteins or sialic acid of mucin in the enamel pellicle (Jakubovics, 2015b; Diaz and Valm, 2020). Similarly, newly exposed epithelial cells rapidly acquire a salivary mucosal pellicle (Carpenter, 2020). A set of pioneer colonizers, different from those that colonize enamel, adheres more readily to these mucosal surfaces (Gibbons and Van Houte, 1975). Adherence of specific bacteria to the mucosa could be mediated in part by secretory immunoglobulin A, which attaches both to the mucosal pellicle and to a set of oral commensal bacteria (Carpenter, 2020). Thus, colonization is a property of microbes but is dependent on molecules from both host and microbial sources.
Host and microbe.
Host and microbe act as selective forces on one another, each shaping the other through sustained interaction and adaptation (Figure 2C). Salivary flow and immune surveillance are properties of the host that reduce the microbial load. However, saliva is also a vehicle for positive selection of microbes in the mouth, because mucins and nutrients such as lactate, bicarbonate, nitrate, and vitamins are actively secreted into saliva (Carpenter, 2020). Salivary mucins support the growth of microbes that possess the glycosidases capable of releasing oligosaccharides from these complex host substrates, as well as mixed syntrophic communities that can benefit from their digestion (Jakubovics, 2015a; Carpenter, 2020). The complexity and refractory nature of mucins could therefore encourage the growth of structured microbial communities and discourage overgrowth of single taxa. Synergistic interactions among oral bacteria involve food webs in which one bacterium produces a waste product, such as lactate, that serves as an important nutrient for other lactate-consuming bacteria (Jakubovics 2015b). Such metabolic cooperation occurs when cells are in close proximity and thus selects for coaggregation and spatial mixing rather than segregation of the interacting taxa (Jakubovics 2015b; Stacy et al., 2016). The secretion of lactate, and possibly also bicarbonate, by the host could selectively promote growth of the bacteria participating in these food webs (Carpenter, 2020) and be a mechanism by which the host influences the spatial organization of the microbiota.
Host secretion of nitrate into saliva could promote the growth of facultatively anaerobic oral bacteria that use nitrate as an electron acceptor for respiration; the microbial conversion of nitrate to nitrite in turn permits the efficient generation of nitric oxide from dietary nitrate which helps maintain nitric oxide homeostasis (Hezel and Weitzberg, 2015). Nitric oxide provides important benefits to the host including lowering of blood pressure, improved endothelial function, reversal of metabolic syndrome, and reduction of oxidative stress (Lundberg et al., 2018). Other potentially beneficial functions of the oral microbiota include restricting access of non-commensal microbes to the host epithelium by physically blocking the epithelium and by secreting antimicrobials (Valm, 2019). Thus, the interaction between host and microbiota is a dynamic equilibrium in which both sides benefit, and each modulates the metabolism of the other.
SITE-SPECIALIST COMMUNITIES IN THE MAJOR ORAL HABITATS
The concept that different sites within the mouth support distinctive microbiotas was first introduced by pioneering studies almost 50 years ago (Socransky and Manganiello, 1971; Gibbons and Van Houte, 1975) and is well supported by an array of recent studies. High-throughput identification of microbes with DNA sequencing allows characterization of communities at a systems level and identifies a major distinction between the microbial community making up dental plaque and the community on mucosal surfaces (Mager et al., 2003; Segata et al., 2012; Simón-Soro et al., 2013; Eren et al., 2014; Lloyd-Price et al., 2017). In healthy people, supra- and subgingival plaque resembled one another. The microbiota of the tongue dorsum was similar to that sampled from the throat and the palatine tonsils; the microbiota of the cheeks, gums, and the roof of the mouth (buccal mucosa, keratinized gingiva, and hard palate) were similar to one another. Thus, sites within the mouth were distinguishable by their resident microbiota and divided roughly into three categories of microbial composition: dental plaque; tongue and allied sites; and the gums, cheeks, and hard palate.
A rich and deeply rooted literature, however, suggests a stronger conclusion: that each microbe in the mouth is specialized for one habitat or another, so that the microbiota at one oral site is different from the microbiota at other oral sites not only in overall composition and proportions of common taxa but also in specific membership. Early studies investigating cultivable members of the oral microbiota showed that most are not cosmopolitan but grow in selected habitats within the mouth (Socransky and Manganiello, 1971; Gibbons and Van Houte, 1975). Each taxon had a “primary ecologic niche”—teeth, tongue, or buccal mucosa for example (Socransky and Manganiello, 1971). High-throughput studies of 16S ribosomal RNA sequences, analyzed with single-nucleotide resolution, buttressed this conclusion, showing that closely related taxa differing by as little as a single nucleotide in the sequenced region of the 16S rRNA gene had dramatically different distributions among the oral sites (Eren et al., 2014). Analysis of whole-genome sequences provided yet higher resolution, distinguishing among strains of a species and likewise finding high site specificity among closely related strains as well as species (Lloyd-Price et al., 2017). Based on a review of this collective literature and data, we proposed the site-specialist hypothesis (Mark Welch et al., 2019) for oral microbiota.
The site-specialist hypothesis has important implications for the structure and function of oral microbial communities. Namely, it suggests that the most significant factor that determines the niche for a microbe is its local habitat, which includes its immediate neighbors. It implies that the co-evolution of microbes within the mouth has led to highly specific taxon-taxon interactions that result in most microbes being restricted to the habitat type in the mouth that is occupied by those neighbors. Microbial communities at different sites in the mouth are organized similarly, in that they are composed predominantly of several dozen abundant and prevalent taxa from core genera that are represented in each of the site microbiomes. However, at the species level, the sites are distinct (Eren et al., 2014; Mark Welch et al., 2019). The genus Actinomyces, for example, has species strongly specialized for plaque and other species strongly specialized for the tongue dorsum. The same is true of genus Fusobacterium, genus Leptotrichia, genus Neisseria, genus Rothia, genus Streptococcus, and genus Veillonella. This pattern—of a common set of genera but different species from site to site—suggests that the different members of the microbial community are adapted to one another and co-evolve as the community adapts from one oral site to the next. Such co-evolution suggests that the microbes themselves are an important part of the environment for one another and that taxon-taxon relationships are highly specific, and one taxon cannot simply swap with its relative from a different oral site.
MICROBIAL HABITATS AND NICHES AT THE MICRON SCALE
Recognition of the importance of gradients in spatially structured microbial communities goes back to Winogradsky in the 1880s and 1890s. The concepts were elaborated by Wimpenny, who pointed out the importance of diffusion in the transport of solutes in a spatially organized ecosystem and concluded that an organism using a solute should be as close to its source as possible (Wimpenny, 1981). These proximity considerations suggest that close cell-to-cell associations and steep gradients are important in forming the habitat for each microbe. Close cell-to-cell associations are evident in dental plaque, as seen by both electron microscopy and confocal microscopy (Listgarten et al., 1975; Zijnge et al., 2010; Dige et al., 2014). These methods showed that dental plaque is a densely packed microbial biofilm that is stratified into layers of differing microbial composition. The stratified morphology itself is suggestive of the presence of steep gradients. However, subsequent microprobe analysis (Koley et al., 2011) and fluorescence imaging (Kim et al., 2020) of oral biofilms provided direct evidence for micron-scale gradients of redox potential and pH respectively.
Identifying microbes by using combinatorial labeling and spectral imaging FISH (CLASI-FISH) allowed us to go beyond the characterization of dental plaque as a layered structure. We were able to identify a distinctive community that we called a hedgehog, in which aerobes and facultative anaerobes occupied an outer shell approximately 20 to 30 μm wide (Mark Welch et al., 2016). Inside this outer aerobic shell lay a middle layer occupied by taxa that grow well in micro-aerobic conditions; filaments of Corynebacterium spp. were densely packed at its base and extended through the middle layer to the outer shell. Similarly, in the tongue dorsum biofilm described by Wilbert et al. (2020), aerobes or facultative anaerobes tended to occupy the outer cortex, and the deep core of the structure was rich in taxa that grow anaerobically. The distribution of taxa within oral biofilms, as well as the known growth requirements of these taxa, suggests that the observed stratification is evidence of habitat zones with differing conditions of oxygen and other small molecules, presumably generated by microbial activity itself, and that the gradients so generated are sharp enough to produce distinctive microbial composition in bands a few microns to tens of microns wide.
In addition to habitat zones and gradients, tight cell-to-cell associations between disparate taxa are characteristic of oral biofilms and are the micron-scale manifestation of the molecular-level coadhesion interactions among taxa. The mixing of different taxa can be seen by morphology in electron micrographs (Listgarten et al., 1975; Kullaa-Mikkonen and Sorvari, 1985; Jakubovics, 2015b). Discrimination and identification of taxa can be achieved by FISH with probes targeting species-or genus-level taxa. FISH experiments examining the early stages of formation of dental plaque showed intimate contact between members of different taxa, suggesting that coadhesion is important for forming multi-generic microcolonies in which metabolic interactions between disparate taxa can take place (Diaz et al., 2006). A recent study made the connection between coadhesion and spatial organization explicit, demonstrating extensive coaggregation interactions in strains isolated from single individuals and showing that strains that participate in large numbers of coaggregations were present in small clusters or dispersed throughout the undisturbed biofilm (Palmer et al., 2017). Thus, both spatial organization and molecular interactions indicate the importance of cell-cell associations in localizing microbes to their fine-scale habitat in oral biofilms.
Distinct differences in microbial composition occur in different regions of disease-associated biofilms. In teeth with advanced generalized periodontitis, subgingival biofilms were characterized by facultatively or obligately anaerobic taxa such as Actinomyces, Fusobacterium, and Treponema, as well as periodontal pathogens that appeared to be colonizing pre-existing biofilms and forming microcolonies within them (Zijnge et al., 2010). Supragingival biofilms, by contrast, had a different and more heterogeneous architecture and composition (Zijnge et al., 2010). Similarly, in teeth affected by caries, different habitats were characterized by different microbial composition; disease-associated taxa such as Lactobacillus and Bifidobacterium were present in carious lesions as well as in outer layers of biofilm adjacent to the lesions, whereas Actinomyces spp. were present along the entrance of fissures in the enamel (Dige et al., 2014). A direct connection between spatial organization and biofilm pathogenicity was demonstrated by Kim et al. (2020) studying Streptococcus mutans in caries. They showed that early biofilms were thin, flat, and characterized by intermixing between S. mutans and the non-pathogenic S. oralis, whereas later biofilms formed a domed structure in which the two taxa segregated from one another. Production of glucans by S. mutans was required for the segregation, as well as for the formation of the domed structure which was associated with demineralization of enamel.
The patchy, heterogeneous nature of biofilms in the mouth shows that to understand the dynamics and function of oral biofilms, it will be important to understand the range of partners with which a taxon can interact. The site-specialist hypothesis holds that each taxon is restricted to a single category of site within the mouth and therefore is restricted to a limited set of partners, yet both spatial nearest-neighbor arrangements of taxa in oral biofilms, and the molecular interactions that underlie them, indicate that many taxa have a range of potential partners within a site. In coaggregation interactions in vitro, the same receptor can be recognized by adhesins on multiple different partners, which then compete for binding; some cells can participate in interactions with a wide range of potential partners and some only with a few (Kolenbrander et al., 2006; Palmer et al., 2017).
A range of specificity of interaction is also observed in images of oral biofilms ex vivo. The structures in dental plaque called corncobs have a highly stereotypical arrangement, with a central filament surrounded by a single or double layer of cocci (Figure 3A). The participating cocci are not non-specific; they are drawn from species in the Streptococcus, Porphyromonas, and Haemophilus/Aggregatibacter genera. In natural healthy plaque, these corncobs form selectively around Corynebacterium spp. even when other filament-forming cells such as Fusobacterium spp., Capnocytophaga spp., and Leptotrichia spp. are present in the immediate environs (Mark Welch et al., 2016). A different but superficially similar corncob-like arrangement results when blooms of Candida spp. appear (Zijnge et al., 2010; Kim et al., 2020). Thus, corncobs are an example of fairly specific taxon-taxon interactions but with some flexibility in membership. The characteristic tongue dorsum consortia have not only a more variable shape and size but also a more flexible arrangement. Tongue consortia are made up of a well-defined set of taxa arranged in patches and stripes. There is a modest but measurable tendency for certain of these taxa—S. mitis, S. salivarius, and Veillonella spp.—to be adjacent, but a patch of each of the taxa can be found next to any other (Wilbert et al., 2020; Figures 3B and 3C). Similarly in dental plaque biofilms, each of the visualized taxa can be found in a variety of nearest-neighbor relationships (Mark Welch et al., 2016). These complex and variable structures indicate that most oral microbes are capable of growth next to a variety of partners drawn from the limited set of site-specialists present in a given habitat.
Figure 3. Diversity of Micron-Scale Associations in the Oral Microbiome.
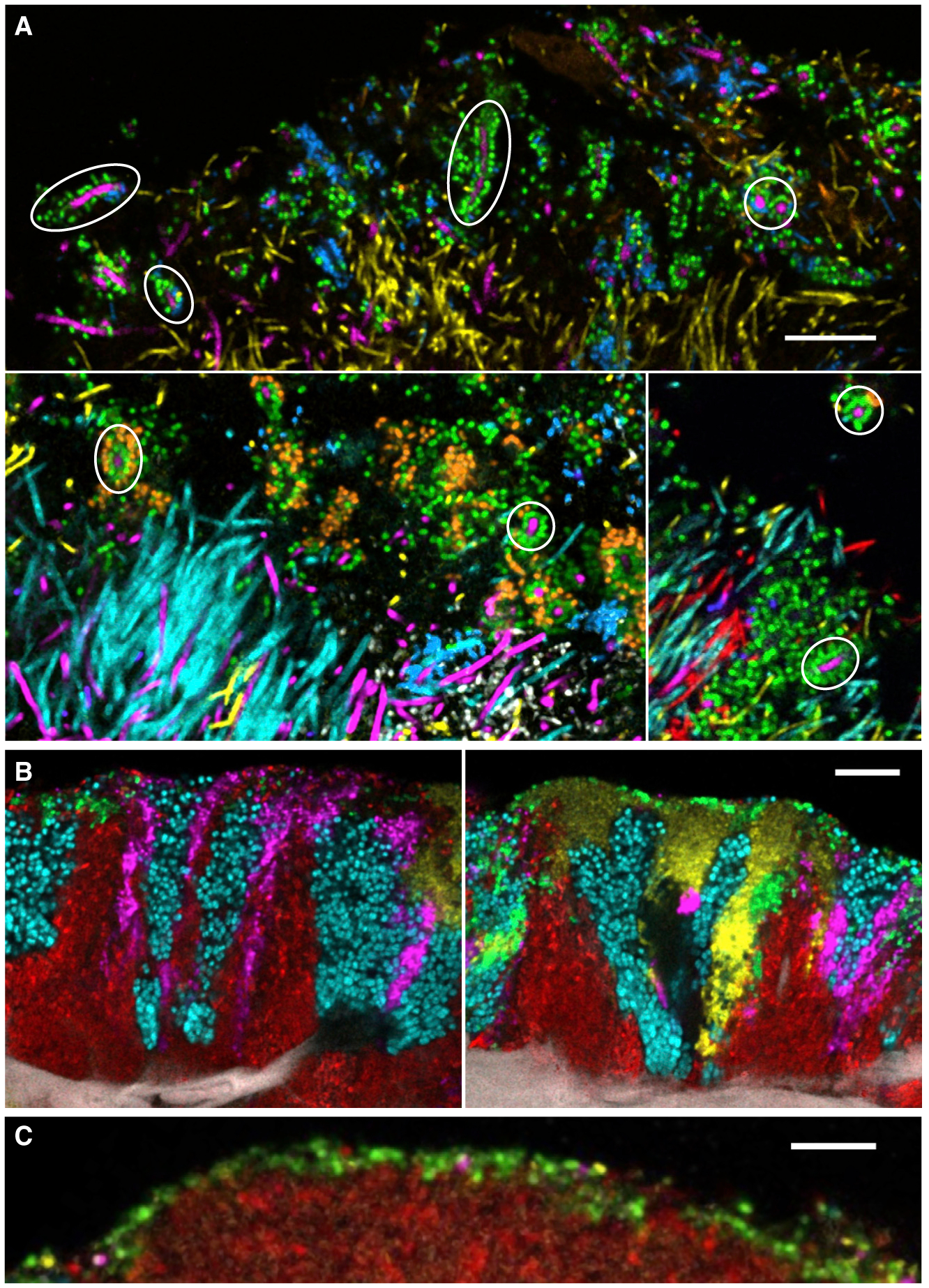
(A) Some microbial taxa have highly specific structural associations. Corncobs in dental plaque are characterized by specific binding of Streptococcus sp. (green) or Porphyromonas sp. (blue) to Corynebacterium sp. (magenta), even when other filamentous taxa such as Fusobacterium (yellow), Leptotrichia (cyan), and Capnocytophaga (red) are present and apparently available for binding in the same area. Circles indicate examples of corncobs formed around Corynebacterium filaments. Scale bar, 10 μm. Modified from Mark Welch et al. (2016).
(B) Some microbial taxa tend to form clonal clusters. The five taxa visualized here on the tongue dorsum occur in patches that have different characteristic shapes and relations to the underlying host epithelial cells but show clonal boundary relationships with each of the other 4 taxa. For example, Rothia (cyan) can be found next to Actinomyces (red), Neisseriaceae (yellow), Veillonella (magenta), and Streptococcus (green). Scale bar, 10 μm. Image credit: Steven Wilbert.
(C) Streptococcus (green) often forms a thin (one-two cell thick) layer at the periphery of a tongue dorsum consortium—in this case, over a cluster of Actinomyces (red). Scale bar, 10 μm. Modified from Wilbert et al. (2020).
Fusobacterium nucleatum is widely thought to occupy a special position in the development and structure of oral biofilms. It is capable of coaggregation with an exceptionally broad range of taxa including both early and late colonizers; it enters the biofilm at a temporally intermediate stage of development, between the early and the late colonizers, and its appearance seems to be associated with the entry into the biofilm of late-colonizing anaerobes (Kolenbrander et al., 2006). Thus, Kolenbrander et al. (2006) proposed “that fusobacteria are central structural components of plaque and essential for plaque maturation and an increase in plaque diversity.”
We suggest that the imaging data now available show that F. nucleatum does not constitute a physical bridge between early- and late-colonizing dental plaque organisms. Instead, we hypothesize that F. nucleatum is an opportunistic colonizer and indicator of the maturation of plaque to the point where anoxic niches become available. In the dental plaque hedgehog structure, filaments of Corynebacterium spp. form the structural bridge, reaching from the base of the structure to the tip where they form the core of the corncob structures that fringe the hedgehog. Fusobacterium spp. are present, but in patches or as sporadic filaments rather than as a consistent physical bridge, and Leptotrichia spp. and Capnocytophaga spp. occupy a similar position to Fusobacterium spp. (Mark Welch et al., 2016). Consistent with this view, Zijnge et al. (2010) described varied and complex supragingival plaque structures in which Fusobacterium spp. were not prominent; rather, they found Fusobacterium sp. as a dominant member of subgingival plaque (Zijnge et al., 2010), although without an obvious structural localization. In supragingival biofilms, Dige et al. (2014) observed Fusobacterium spp. scattered beneath the surface in the outer layer of plaque. Fusobacterium spp. are obligate anaerobes. The most important habitat requirement for them could be low oxygen concentration. The ability of F. nucleatum to coaggregate with a wide variety of partners suggests that it establishes itself by adhering to whichever partners are available in its preferred low oxygen-concentration habitat.
Given the importance of syntrophy to the oral microbiome, the question arises of how syntrophy is maintained during growth of the biofilm. If a microbe grows most efficiently when it can receive resources from disparate microbes a few microns away, its growth will slow or cease when it gets too far from those syntrophic partners—yet the process of cell division itself will tend to produce large clonal clusters. Imaging the spatial organization of oral biofilms suggests two answers: growth in vertical columns and growth in a filamentous morphology. Mature biofilms of both dental plaque and the tongue dorsum are characterized by a columnar growth outward from the substrate—a growth habit that could be a natural result of growth on a crowded substrate, but it also permits syntrophy across micron-scale gradients to continue throughout the biofilm growth cycle. A filamentous growth habit accomplishes the same result, and clusters of filaments are especially prominent in dental plaque (Listgarten et al., 1975; Zijnge et al., 2010; Mark Welch et al., 2016) and also occur in tongue dorsum biofilms (Wilbert et al., 2020). A prominent exception to syntrophy appears to be members of the genus Actinomyces, which form large patches both at the base of dental plaque biofilms (Zijnge et al., 2010; Dige et al., 2014; Mark Welch et al., 2016) and also at the base of tongue dorsum biofilms near host epithelial cells (Wilbert et al., 2020). Comparatively large and apparently single-taxon patches of Rothia mucilaginosa and of Streptococcus salivarius also occur on the tongue dorsum (Wilbert et al., 2020). The meaning of these large clonal clusters cannot be determined from imaging studies alone. Clearly, it will be necessary to employ other approaches to test mechanistic hypotheses.
SHORT-RANGE AND LONG-RANGE FACTORS INTERACT
Although short-range factors such as direct adhesion and strong micron-scale gradients have a powerful influence on microbial habitats at micron scales, longer-range factors interact with these short-range factors and likely influence the composition of oral biofilms in complex ways. Interactions might occur between taxa separated by many centimeters across the mouth, through small molecules diffusing through the air or transported via salivary flow. The tongue microbiota is high in biomass and its secreted products could conceivably influence the biology of dental plaque or the communities at other mucosal sites.
In two recent comprehensive reviews, Proctor and colleagues suggest that localized shifts in community composition could occur as a result of differences in the composition and velocity of flow of saliva from place to place in the mouth (Proctor and Relman, 2017; Proctor et al., 2020). Changes in the velocity of salivary flow result in changes in the clearance rate of substances from the surface of the biofilm, which presumably strengthen or attenuate the micron-scale gradients within the biofilm. Effects of flow velocity could also directly influence sites deeper in the biofilm in cases where there are channels through the biofilm (Wood et al., 2000).
Differentiated habitats can be defined within a site at scales of millimeters by sequencing approaches if the sampling is done with spatial precision. Simón-Soro et al. (2013) sampled 2 individuals at 112 sites each and found Streptococcus in very high abundance at vestibular (buccal) sites and Veillonella highest on the canines. Proctor et al. (2018) sampled buccal and lingual aspects of teeth in 30 individuals and found that the most abundant taxa were consistent regardless of location, but less-abundant taxa showed a gradient in abundance from front to back of the mouth, particularly at lingual sites. Although both studies found evidence for shifts in the proportions of the dental plaque microbiota on different teeth or different aspects of teeth, the details of their findings differed. It is possible that the question of how dental plaque communities shift across the mouth has not yielded a straightforward answer because of a mismatch between the sampling method and the size and spatial organization of the communities under study. Sampling from a single aspect of a single tooth results in the collection of minute samples of plaque, as noted by Simón-Soro et al. (2013). The studies working with these low-biomass samples are a technical tour de force. Complicating the interpretation of results, the community at any such site could be influenced not only by deterministic factors but also by drift, as noted by Proctor et al. (2020). Key factors influencing the details of community composition, including local topography and historical accident, could produce locally significant effects. In addition, sampling methods such as swab, paper point, or scrape of a curette could incidentally collect microbes from saliva or adjacent gingival tissue as well as the intended target plaque, and microbes in saliva or dislodged from adjacent non-plaque sites could become incorporated into the plaque biofilm and might or might not be active while there. In short, DNA sequencing approaches can now tell us with great accuracy and completeness what microbes are present in the samples we collected and homogenized, but the sampling technology for sequencing lacks the requisite spatial resolution to investigate community structure. To address questions of how microbes are organized, how they interact, and what is the functional role of each microbe in the physiology of an oral biofilm at the sampled sites, we need analysis methods with a higher level of spatial resolution and in which spatial organization remains as intact as possible.
CONCLUSION
This Minireview has focused on the micron-scale habitats and niches of the oral microbiome. Although multiplexed imaging has revealed rich ecological interactions, unraveling the role of spatial patterning in oral microbial ecology will require new approaches and the overcoming of technological barriers. Identifying microbes by in situ hybridization requires fixation of the sample and thus provides only snapshots in time. Clearly it will be necessary to devise a way of carrying out taxonomically resolved live cell imaging in order to capture the dynamics of microbial community assembly and turnover. Mechanistic understanding will require in vitro experiments that permit perturbation and assay of individual components. Yet, these in vitro approaches cannot be so simplified that they lose the relevant biology. Bacteria growing on surfaces in the mouth experience varied topography, localized gradients, and interactions with multiple different microbial species. Informative in vitro experiments will require engineered microenvironments that capture these complex properties.
Historically, geography has provided a roadmap for exploration of the earth and a baseline for re-engineering of it. Similarly, the added value of mapping oral microbiome geography could lie in providing, on the one hand, a roadmap for future spatial studies in oral microbial ecology and, on the other hand, a baseline for how to rationally engineer microbial communities to promote oral and systemic human health.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health NIDCR grant DE022586 (to G.G.B.). We thank Steven Wilbert for images of the tongue dorsum and Floyd Dewhirst for helpful discussions. Because the number of citations in the review is limited by the journal, we unfortunately cannot cite all papers relevant to the topic, and we regret any omissions that might have resulted.
REFERENCES
- Bowen WH, Burne RA, Wu H, and Koo H (2018). Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 26, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter GH (2020). Salivary Factors that Maintain the Normal Oral Commensal Microflora. J. Dent. Res 99, 644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar U, Angadi PV, Hallikerimath S, and Kale AD (2013). Cytological assessment of Barr bodies using aceto-orcein and papanicolaou stains in buccal mucosal smears and their sex estimation efficacy in an Indian sample. Acta Cytol. 57, 516–521. [DOI] [PubMed] [Google Scholar]
- Dawes C (2003). Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch. Oral Biol 48, 329–336. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, Lakshmanan A, and Wade WG (2010). The human oral microbiome. J. Bacteriol 192, 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, and Valm AM (2020). Microbial Interactions in Oral Communities Mediate Emergent Biofilm Properties. J. Dent. Res 99, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ Jr., and Kolenbrander PE (2006). Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol 72, 2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dige I, Grønkjær L, and Nyvad B (2014). Molecular studies of the structural ecology of natural occlusal caries. Caries Res. 48, 451–460. [DOI] [PubMed] [Google Scholar]
- Egland PG, Palmer RJ Jr., and Kolenbrander PE (2004). Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101, 16917–16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, and Mark Welch JL (2014). Oligotyping analysis of the human oral microbiome. Proc. Natl. Acad. Sci. USA 111, E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, and Van Houte J (1975). Bacterial adherence in oral microbial ecology. Annu. Rev. Microbiol 29, 19–44. [DOI] [PubMed] [Google Scholar]
- Hezel MP, and Weitzberg E (2015). The oral microbiome and nitric oxide homoeostasis. Oral Dis. 21, 7–16. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS (2015a). Saliva as the Sole Nutritional Source in the Development of Multispecies Communities in Dental Plaque. Microbiol. Spectr 3, MBP-0013–MBP-2014. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS (2015b). Intermicrobial Interactions as a Driver for Community Composition and Stratification of Oral Biofilms. J. Mol. Biol 427, 3662–3675. [DOI] [PubMed] [Google Scholar]
- Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, Scisci EL, Hajishengallis E, Whiteley M, and Koo H (2020). Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc. Natl. Acad. Sci. USA 117, 12375–12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ Jr., Rickard AH, Jakubovics NS, Chalmers NI, and Diaz PI (2006). Bacterial interactions and successions during plaque development. Periodontol. 2000 42, 47–79. [DOI] [PubMed] [Google Scholar]
- Koley D, Ramsey MM, Bard AJ, and Whiteley M (2011). Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc. Natl. Acad. Sci. USA 108, 19996–20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullaa AM, Asikainen P, Herrala M, Ukkonen H, and Mikkonen JJW (2014). Microstructure of oral epithelial cells as an underlying basis for salivary mucosal pellicle. Ultrastruct. Pathol 38, 382–386. [DOI] [PubMed] [Google Scholar]
- Kullaa-Mikkonen A, and Sorvari TE (1985). A scanning electron microscopic study of the dorsal surface of the human tongue. Acta Anat. (Basel) 123, 114–120. [DOI] [PubMed] [Google Scholar]
- Listgarten MA, Mayo HE, and Tremblay R (1975). Development of dental plaque on epoxy resin crowns in man. A light and electron microscopic study. J. Periodontol 46, 10–26. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, et al. (2017). Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg JO, Carlström M, and Weitzberg E (2018). Metabolic effects of dietary nitrate in health and disease. Cell Metab. 28, 9–22. [DOI] [PubMed] [Google Scholar]
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, and Socransky SS (2003). Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol 30, 644–654. [DOI] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, and Borisy GG (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 113, E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Dewhirst FE, and Borisy GG (2019). Biogeography of the Oral Microbiome: The Site-Specialist Hypothesis. Annu. Rev. Microbiol 73, 335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RJ Jr., Shah N, Valm A, Paster B, Dewhirst F, Inui T, and Cisar JO (2017). Interbacterial Adhesion Networks within Early Oral Biofilms of Single Human Hosts. Appl. Environ. Microbiol 83, e00407–e00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DM, and Relman DA (2017). The Landscape Ecology and Micro-biota of the Human Nose, Mouth, and Throat. Cell Host Microbe 21, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DM, Fukuyama JA, Loomer PM, Armitage GC, Lee SA, Davis NM, Ryder MI, Holmes SP, and Relman DA (2018). A spatial gradient of bacterial diversity in the human oral cavity shaped by salivary flow. Nat. Commun 9, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DM, Shelef KM, Gonzalez A, Davis CL, Dethlefsen L, Burns AR, Loomer PM, Armitage GC, Ryder MI, Millman ME, et al. (2020). Microbial biogeography and ecology of the mouth and implications for periodontal diseases. Periodontol. 2000 82, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, and Izard J (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Soro A, Tomás I, Cabrera-Rubio R, Catalan MD, Nyvad B, and Mira A (2013). Microbial geography of the oral cavity. J. Dent. Res 92, 616–621. [DOI] [PubMed] [Google Scholar]
- Socransky SS, and Manganiello SD (1971). The oral microbiota of man from birth to senility. J. Periodontol 42, 485–496. [DOI] [PubMed] [Google Scholar]
- Stacy A, McNally L, Darch SE, Brown SP, and Whiteley M (2016). The biogeography of polymicrobial infection. Nat. Rev. Microbiol 14, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C, Earle KA, Huang KC, and Sonnenburg JL (2017). The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe 21, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM (2019). The Structure of Dental Plaque Microbial Communities in the Transition from Health to Dental Caries and Periodontal Disease. J. Mol. Biol 431, 2957–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert SA, Mark Welch JL, and Borisy GG (2020). Spatial Ecology of the Human Tongue Dorsum Microbiome. Cell Rep. 30, 4003–4015.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimpenny JWT (1981). Spatial Order in Microbial Ecosystems. Biol. Rev. Camb. Philos. Soc 56, 295–342. [Google Scholar]
- Wood SR, Kirkham J, Marsh PD, Shore RC, Nattress B, and Robinson C (2000). Architecture of intact natural human plaque biofilms studied by confocal laser scanning microscopy. J. Dent. Res 79, 21–27. [DOI] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmur R, and Harmsen HJM (2010). Oral biofilm architecture on natural teeth. PLoS One 5, e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]


