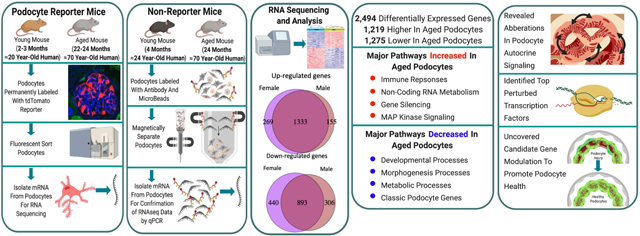
Abstract
Glomerular podocytes undergo structural and functional changes with advanced age, that increase susceptibility of aging kidneys to worse outcomes following superimposed glomerular diseases. To delineate transcriptional changes in podocytes in aged mice, RNA-seq was performed on isolated populations of reporter-labeled (tdTomato) podocytes from multiple young (two to three months) and advanced aged mice (22 to 24 months, equivalent to 70 plus year old humans). Of the 2,494 differentially expressed genes, 1,219 were higher and 1,275 were lower in aged podocytes. Pathway enrichment showed that major biological processes increased in aged podocytes included immune responses, non-coding RNA metabolism, gene silencing and MAP kinase signaling. Conversely, aged podocytes showed downregulation of developmental, morphogenesis and metabolic processes. Canonical podocyte marker gene expression decreased in aged podocytes, with increases in apoptotic and senescence genes providing a mechanism for the progressive loss of podocytes seen with aging. In addition, we revealed aberrations in the podocyte autocrine signaling network, identified the top transcription factors perturbed in aged podocytes, and uncovered candidate gene modulations that might promote healthy aging in podocytes. The transcriptional signature of aging is distinct from other kidney diseases. Thus, our study provides insights into biomarker discovery and molecular targeting of the aging process itself within podocytes.
Keywords: aging, glomerulus, podocyte, RNA sequencing, gene ontology, differentially expressed genes
Graphical abstract
INTRODUCTION
With an aging population, attention has increasingly focused on age-associated changes in many different organs. Several predictable glomerular changes occur in the aged human,1, 2 canine,3 rat4–6 and mouse7–10 kidneys, including fibrosis and hypertrophy, and changes to parietal epithelial cells,11–14 and podocytes.4, 5, 15–17 What is increasingly clear is that the decline in kidney function with aging cannot be solely explained by nephrosclerosis per se.18, 19 Podocyte number and density decrease,20, 21 and because those remaining are unable to replace themselves,22 they undergo maladaptive hypertrophy in aged kidneys.4, 5 Aged podocytes undergo senescence,23 stress,4 and decreased slit diaphragm proteins.4 Understanding the normal aging process of podocytes will allow us to understand their susceptibility to injury and identify targetable protective mechanisms.
The pathways underlying healthy aged-related changes to podocytes are not fully understood.16, 24 Candidate pathways include oxidative stress,25, 26 epigenetic changes,27, 28 senescence,29 sirtuins,23 reduced autophagy30 and increased apoptosis.31, 32 While detailed gene expression analysis has been undertaken on whole portions of the aging kidney,33–35 podocytes are underrepresented. Additionally, transcriptomic analysis of whole tissues cannot distinguish changes in cell number vs. cell type-specific transcriptional changes.36 We therefore sought to directly measure the transcriptomic signature of isolated reporter-labeled aged podocytes and compare them to the expression signature of young podocytes.
RESULTS
Isolation and validation of young and aged podocyte cohorts
The inducible podocyte specific reporter mouse (NPHS2-rtTA|tetO-cre|tdTomato) was used for RNA-seq studies, where a cohort of podocytes permanently labeled in juvenile animals resulted in an irreversible and persistent lifetime labeling of a population of podocytes (Figure S1). Microalbuminuria (Figure S2A) and glomerulosclerosis (Figure S2B) were higher, accompanied by ultrastructural changes in podocytes using super-resolution fluorescence microscopy37, 38(Figure 2C).
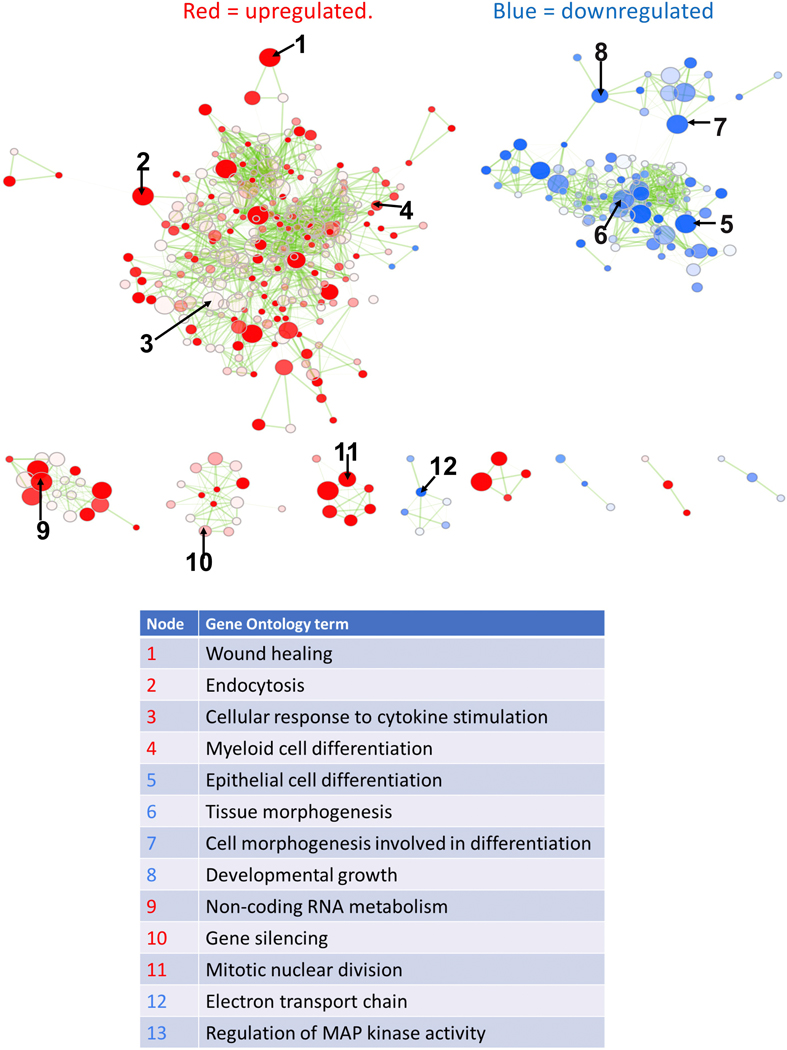
Figure 2. Perturbed biological processes in aged podocytes.
Overall, immune processes are up-regulated, and developmental, differentiation and metabolic processes down-regulated. Each node represents a gene ontology term significantly enriched in genes up-regulated in aged podocytes (red nodes) or down-regulated in aged podocytes (blue nodes). The transparency of the node is proportional to enrichment p-value: more solid-colored nodes represent more significantly enriched GO terms. Node size represent gene set size: GO terms with more genes are larger. Two nodes are connected by an edge if they have overlapping genes. Edge transparency is proportional to the number of genes shared by two GO terms. Select GO terms from each cluster are labeled for each module. Select nodes are numbered and their Gene Ontology terms are listed below.
Live labeled podocytes from young (2–3m) and advanced aged (22–24m) mice were FACS sorted and RNA-seq was used to measure transcriptional changes (Figure S1A). The RNA-seq results reported are from podocyte samples from sex grouped pools of 8 young mice (4 female, 4 male), and 8 individual samples from aged mice (5 female, 3 male). RNA integrity numbers and yields are shown in Figure S3A. The raw and processed data are deposited into the GEO database (GSE136138), and available at https://yuliangwang.shinyapps.io/podo_aging/.
Canonical genes show reduced expression in aged podocytes
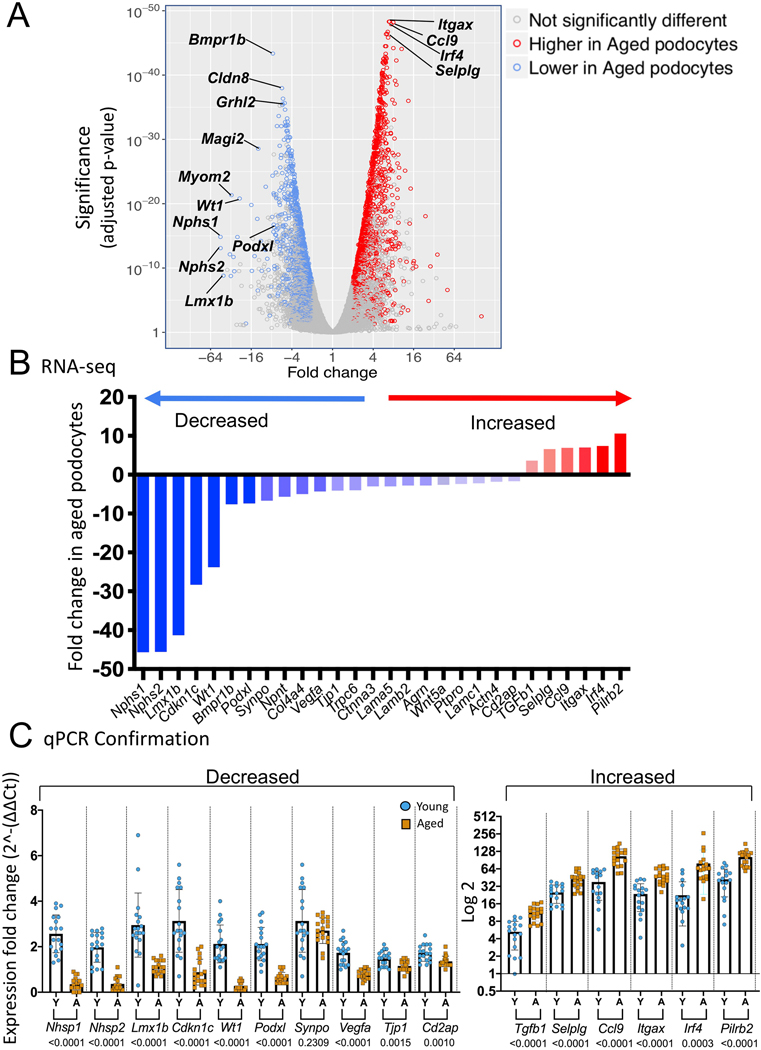
The volcano plot shows changes to podocyte genes (Figure 1A). Examples of well-known canonical podocyte genes were significantly reduced in aged podocytes (Figure 1B)(Table S1). Several genes of interest were highly statistically significantly increased in aged podocytes (Figure 1B).
Figure 1. Differentially expressed genes (DEGs) between aged vs. young podocytes.
A) Volcano plot of differentially expressed genes (DEGs) between aged vs. young podocytes. The Y axis shows the statistical significance measured by p-value, and the X axis shows the fold change of aged vs. young podocytes (> 1 reflects higher in aged, < 1 reflects lower in aged podocytes; 4 means 4-fold higher in aged, −4 means 4-fold lower in aged). Each blue circle represents an individual gene that is lower in aged podocytes, and each red circle represents a specific gene higher in aged podocytes.
B.) Graph of critical podocyte genes significantly lower in aged podocytes (blue) and genes that were significantly higher in aged podocytes (red) as measured in the RNA-seq dataset.
C.) Graphs of qPCR of critical podocyte genes on a second cohort of podocytes isolated and analyzed from individual non-reporter mice confirm the significant decrease (left) and increase (right) in aged podocytes (all genes except Synpo had a p value of 0.0015 or lower)
Independent validation of transcriptional changes in a separate mouse podocyte cohort
To avoid potential artifacts from reporter expression or FACS, magnetic bead assisted cell sorting (MACS) isolated podocytes from a second cohort of non-pooled young (n=18) and aged (n=18) non-reporter mice were generated (Figure S1B). While non-podocyte cell fractions did not stain (Figure S3B), Alexa Fluor 488-positive cells indicated the presence of nephrin in the podocyte cell fraction (Figure S3B), validating the separation of podocytes from other kidney cells. qPCR confirmed the canonical podocyte and increased genes (Figure 1C). Staining for nephrin (Nphs1), podocin (Nphs2), synaptopodin (Synpo) and zona occludens 1 (Tjp1) confirmed decreases at the protein level (Figure S4A-D), similar to aged rats,4 and paired immunoglobin-like type 2 receptor beta (Pilrb2) staining confirmed protein increases (Figure S4E). These reproducible results confirmed the RNA-seq dataset.
Global transcriptomic changes in aged podocytes determined using GSEA, gene ontology and hallmark pathway analyses
Table S1 shows 2,494 differentially expressed genes (DEGs) identified between young and aged podocytes using a false discovery rate <0.05, fold change >2, and absolute expression level >4 RPKM normalized for sequencing depth and gene length (Table S2). Of these, 1,219 and 1,275 genes were expressed significantly higher and lower respectively in aged podocytes. Because many perturbed biological processes overlap, we visualized the enrichment results using Enrichment Map (Figure 2, full results in Table S3),39 where similar Gene Ontology (GO) biological processes were clustered together within the network. Major biological processes increased in aged podocytes included immune responses (wound healing, leukocyte activation, cytokine-mediated signaling), non-coding RNA metabolism, gene silencing and MAP kinase signaling (Figure 2). In contrast, developmental and morphogenesis processes, electron transport chain and cellular respiration were down-regulated in aged podocytes (Figure 2).
DAVID enrichment analysis (Tables S4-S5) showed similar findings. Alternative enrichment analysis based on Fisher’s exact test yielded additional insights.40 Fisher’s exact test determines whether being a DEG increases the chances of belonging to a gene set (Figure S5). The 50 Hallmark gene set collection41 identified high-level biological pathways perturbed in podocyte aging, with increases in genes related to immune (allograft rejection), inflammatory and signaling responses (TNF, interferon, IL6, IL2, complement pathways), genes limiting cell cycle, apoptosis, PI3K-mTOR and p53 pathways (Figure S5A). Hallmark analysis showed that genes involved in xenobiotic metabolism, apical junction, and fatty acid metabolism were down-regulated in aged podocytes (Figure S5B).
Further analysis of cellular compartment GO terms showed that genes up-regulated in aged podocytes were located on the external side of the plasma membrane, the immunoglobulin complex and the cytosolic ribosome (Figure S5C). Cellular compartment GO terms showed down-regulated genes for cell-cell junction and apical junction complexes and mitochondria inner membrane (Figure S5D).
Transcriptomic changes in aged podocytes related to sex differences
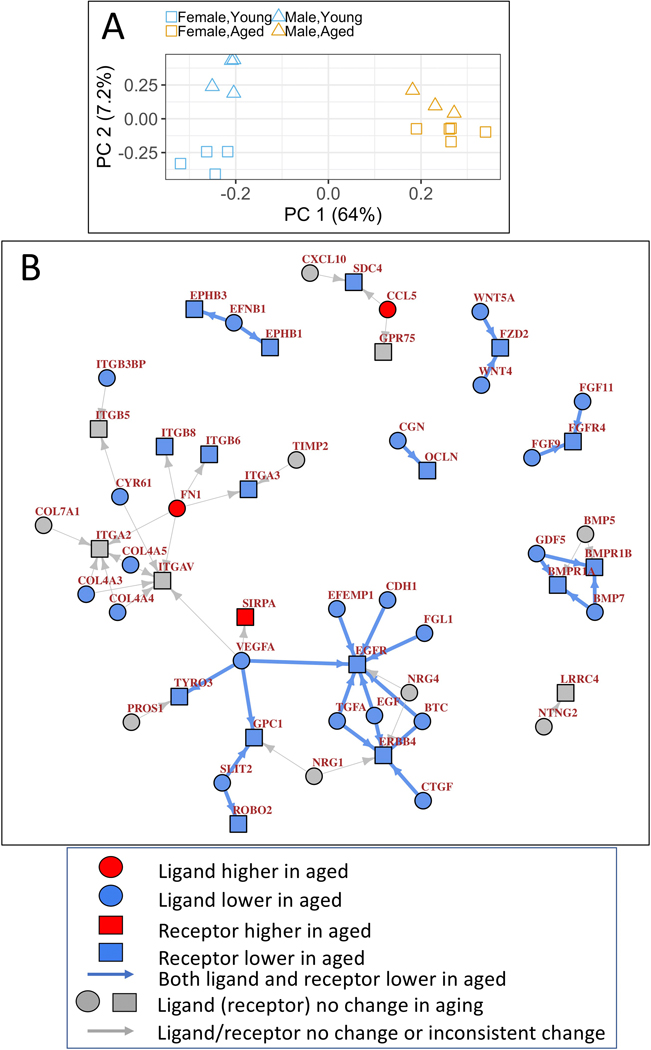
Principal component analysis (Figure 3A) showed that age differences explain most of the expression variance (64%, PC1, x-axis), and overwhelm the small expression variance associated with sex (female vs. male, 7.2%, y-axis). Thus, we focused on the aged vs. young differences in our subsequent analysis. 279 genes significantly contributed to the expression variance along PC2 representing female-male differences: 30 enriched in female, 249 enriched in male podocytes (PC2 loadings 3 standard deviation beyond the mean)(Table S6). As expected, the top female-enriched gene is Xist, while the top male-enriched gene is a Y-linked gene. Male-enriched genes mainly belong to xenobiotic metabolism (Table S7). While female enriched genes did not show any specific pathway enrichment, a few top genes are known to encode immunoglobulin kappa chain and P450 enzymes (Table S6). By contrast, age-related changes dominated the expression landscape in our cohort. Expression changes in aged vs. young mice separated by sex, the up- and down-regulated genes, and pathways, largely overlapped (Figure S6). These trends were similar when female and male samples when combined within each age group (Fig. 2 and Fig. S4), supporting the validity of pooled analysis.
Figure 3. PCA and Podocyte-specific ligand-receptor signaling network based on single cell RNA-seq data from mouse glomeruli.
A.) Principal component analysis (PCA) of all RNA-seq samples using all expressed genes. Age differences explain most of the expression variance (64%, PC1, x-axis), and overwhelm the small expression variance associated with sex (female vs. male, 7.2%, y-axis).
B.) 26 ligand-receptor pairs, defined as podocyte-specific when both the ligand and receptor are two-fold higher in podocytes than in endothelial or mesangial cells in the glomerulus based on single cell RNA-seq, were identified to change in aged podocytes. Circles represent ligands, squares represent receptors, genes with significantly lower expression in aged podocytes are filled with blue color, genes with significantly higher expression in aged podocytes are filled with red color, genes with no significant change are labeled with grey color and gene names are labeled in brown. Arrows represents ligand-receptor interactions, in the direction from ligand to its receptor. Arrows are colored blue if both ligand and receptor are significantly lower in aged podocytes, and grey if ligand/receptor did not show consistent change.
A supervised approach shows podocyte-specific autocrine signaling is decreased in aged podocytes
To connect gene expression changes in disparate genes, we constructed a podocyte-specific autocrine signaling network (autocrine here refers to signaling between the same podocyte but our analysis logic could also apply to paracrine signaling between different podocytes). To achieve this, a manually curated signaling network consisting of 2,557 ligand-receptor pairs42 was used as the input network for a single RNA-seq dataset from healthy mouse glomeruli developed by Karaiskos.43 Next, a ligand-receptor pair was defined as podocyte-specific if the mean expression levels of both ligand and the receptor were at least two-fold higher in podocytes compared to both endothelial cells and mesangial cells based on single cell RNA-seq dataset. Marker genes from the original publication43 were used to define each cell type.
Using these filtering criteria, 55 podocyte-specific ligand-receptor interactions were observed out of 2,557 possible ligand-receptor interactions. Next, the podocyte-specific autocrine signaling network was analyzed using the igraph R package. Mapping gene expression changes in our podocyte aging RNA-seq onto the podocyte-specific autocrine signaling network identified 26 ligand-receptor pairs, where both the ligand and the receptor showed consistent downregulated changes (Figure 3B). Surprisingly, no ligand-receptor pairs showed consistent up-regulation of gene expression. The 26 ligand-receptor pairs include many pathways required for normal podocyte function, including FGF, VEGFA, WNT, BMP, and SLIT2/ROBO2.
The same analysis on bulk RNA-seq of FACS-purified podocytes, mesangial cells and podocytes from healthy mouse glomeruli44 showed a similar podocyte autocrine signaling network, with age-associated declines in VEGFA, WNT, BMP pathways, and laminin-integrin interactions (Figure S7).
Apoptosis and senescence-associated genes were significantly up-regulated in aged podocytes, and tight junction proteins were down-regulated
Differential expression and pathway enrichment analysis provided a list of perturbed genes and pathways in aged podocytes, which included the apoptosis pathway. In order to better visualize and understand the interactions between differentially expressed genes in a larger context within perturbed pathways, the RNA-seq data was mapped onto the KEGG pathway database45 using the PathView tool.46 Figure S8 shows down- and up-regulated genes within the apoptosis pathway in aged podocytes. Most were significantly up-regulated, including tumor necrosis factor (4.3 fold), Fas-L (7.4 fold), Bim (3.1 fold), Bcl-2 (1.8 fold) and Perforin (3.8 fold). Many p53 pathway genes were up-regulated (Figure S8).
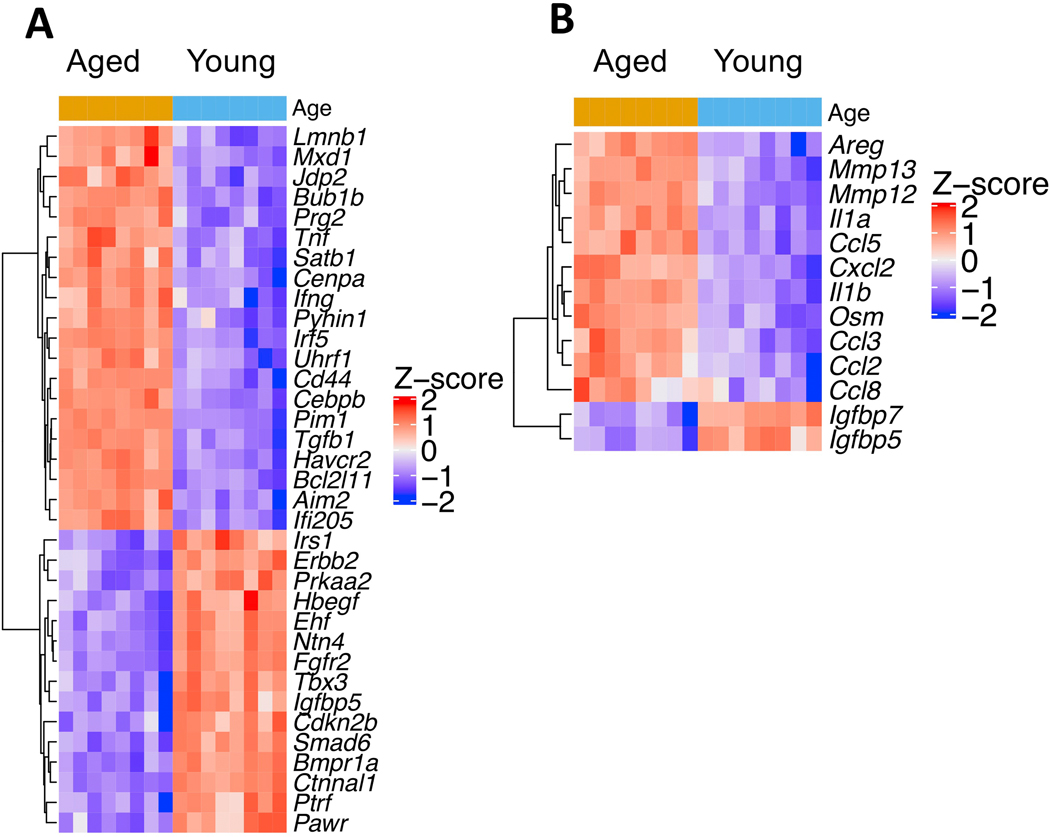
Since cellular senescence is a hallmark of aging,47 we compiled a list of senescence related genes by combining genes from CSGene,48 a literature-based database for senescence genes, with genes from the Gene Ontology term “cellular senescence” (GO:0090398). This yielded 504 senescence genes in total, which were then intersected with DEGs from aged podocytes. The results showed that up-regulated genes in aged podocytes were significantly enriched for senescence (2-fold more senescence genes among up-regulated genes than expected by chance, hypergeometric test p-value 2.9×10−12), while down-regulated genes did not show significant enrichment for senescence genes (p-value 0.72). Figure 4A shows the top differentially expressed senescence genes using the criteria: fold change >3, false discovery rate <0.01, and absolute expression>4 RPKM in either aged or young samples. Figure 4B shows 13 differentially expressed SASP (Senescence-Associated Secretory Phenotype) genes, 11 of which are up-regulated such as Ccl2, 3, 5, and 8, and MMPs 12 and 13.
Figure 4. Increased senescent genes in aged podocytes.
A.) Heatmap of top differentially expressed senescence genes in aged vs. young podocytes (adjusted p-value <0.01, >3 fold change in either direction, expressed above 4 RPKM in either condition).
B.) Heatmap of all differentially Senescence-Associated Secretory Phenotype (SASP) genes (adjusted p-value <0.05, fold change >2, expressed above 4 RPKM in either condition) showed that chemokines and matrix metalloproteases are enriched in aged podocytes.
The KEGG pathway database45 and PathView tool46 visualized interactions between DEGs involved in the tight junctions, essential for normal podocyte function and shape.49 Aged podocytes had reduced expression for occludins (−4 fold), ZO-150 (−4 fold) Cingulin51 (−4.6 fold), MAGI-152 (−2.9), claudins53 (claudin 8, −5.6 fold; claudin 4, −4.4 fold) and JAM (junctional adhesion molecule)49 (Figure S9). Many regulators of these genes were decreased, including PAR3 (−2.9 fold), PAR6 (−4.4 fold),54 aPKC,55, 56 and Nedd4–2 (−2.1 fold).57 Actin increased in aged podocytes (Figure S9).
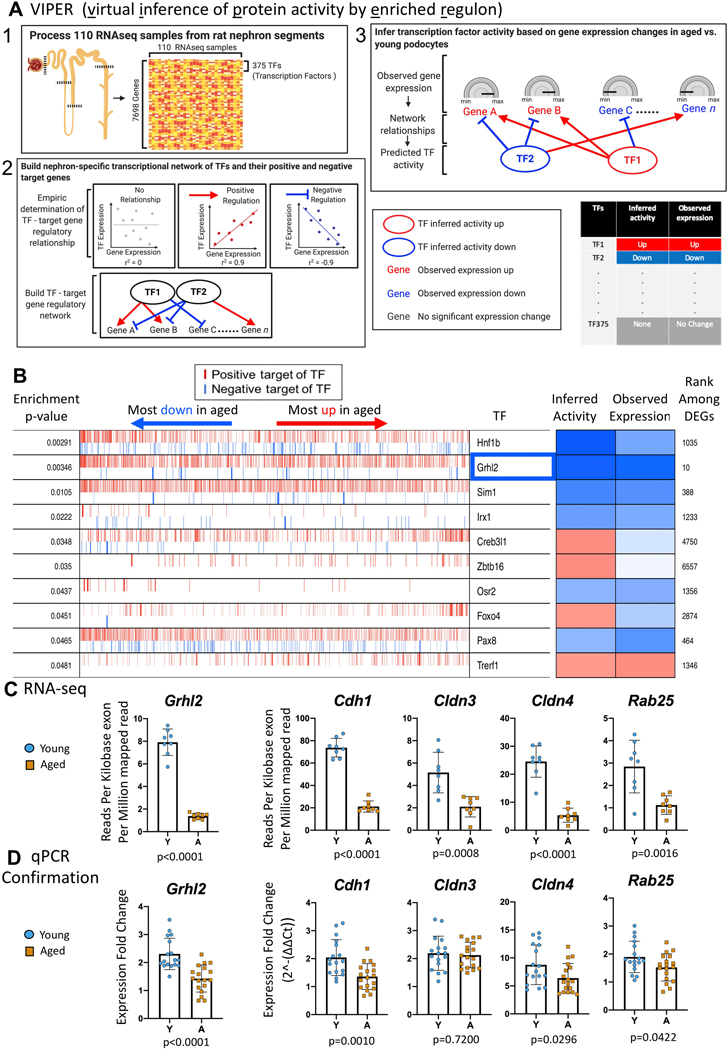
VIPER analysis identified master transcription factor gene expression in aged podocytes
To identify transcriptional regulators of the changes observed, we used VIPER (“virtual inference of protein activity by enriched regulon analysis”) software.58 The rationale is that because of post-transcriptional regulation or low abundance, the expression level of a transcription factor (TF) itself may not be a good proxy for its functional activity. Instead, by using the expression changes of the TF’s predicted target genes, VIPER provides a buffer against weak or inconsistent TF gene expression. By way of example, VIPER infers that if the majority of genes positively regulated by a particular TF are down-regulated in aged podocytes and genes negatively regulated by the TF are up-regulated, then that TF is likely less active in aged podocytes.
VIPER analysis requires 3 steps (Figure 5A). Step 1: We used the ARACNE method59 to build a TF–target gene regulatory network based on the co-expression patterns between TF and non-TF genes across 110 RNA-seq samples of rat kidney glomeruli and nephron tubule segments.60 Step 2: VIPER then determined whether a target gene was positively or negatively regulated by the putative TF. Step 3: VIPER used the aged vs. young podocyte RNA-seq samples to infer the activity of TFs in podocyte aging by examining whether the positively-regulated TF target genes were up-regulated in aged podocytes compared to young podocytes, and if the negatively-regulated TF target genes were down-regulated, thus inferring the TF active in aged podocytes. The opposite pattern inferred a less active TF.
Figure 5. Transcription factor (TF) changes in aged podocytes.
A.) Overview of the VIPER (virtual inference of protein activity by enriched regulon analysis) method. There are three steps in VIPER analysis. First, we processed a publicly available RNA-seq dataset 49 and obtained an expression matrix that includes 7698 detected genes (375 of them are TFs) across 110 rat kidney nephron samples. Second, we used the ARACNE method,50 one of the most popular method used to infer TF-target interactions in mammalian cells, to infer a nephron-specific TF-target gene regulatory network. VIPER takes inferred TF-target interactions from ARACNE and assigns the targets of each TF into positive or negative categories based on the direction of expression correlation in the rat nephron RNA-seq dataset. In the final step, we inferred activity changes of TFs in podocyte aging based on aging-associated expression changes of TF target genes in aged vs. young podocyte RNA-seq data from the current study. If a significant fraction of a TF’s positive targets are up-regulated, and its negative targets down-regulated in aged podocytes, this TF is inferred to be activated in aged podocytes (inactivated in young podocytes); if a significant fraction of a TF’s positive targets are down-regulated and its negative targets are up-regulated in aged podocytes, the TF is inferred to be inactivated in aged podocytes (activated in young podocytes).
B.) VIPER analysis results. The columns from left to right are as follows: the first column is the enrichment p-value of TF target genes in the up- and down-regulated genes in aged vs. young podocytes. The second column represents all genes detected in our podocyte RNA-seq data, sorted from the most down-regulated to the most up-regulated in aged podocytes. A red vertical bar represents the gene at that sorted position is a positive target gene of a TF; a blue vertical bar represents the gene at that sorted position is a negative target of a TF. The total number of target genes vary across TFs. For example, Grhl2 has many target genes, whereas Irx1 have much fewer. The third “TF” column indicates the name of the individual TF. The “Inferred activity” column denotes the activity of the TF based the distribution of its target genes in the sorted list of differentially expressed genes in aged vs. young podocytes, blue represents the TF is inferred to be inactivated; red represents the TF is activated. The “observed expression” column denotes the observed expression change of the TF itself in aged vs. young podocytes regardless of the expression pattern of its targets. The furthermost right column of integer numbers (e.g. “1035, 10, 388 etc.) denotes the rank of the TF among all differentially expressed genes (DEGs). Smaller numbers indicate that the TF is highly differentially expressed between aged and young podocytes. TFs that are inferred to be inactivated based on the expression of their targets (e.g., positive targets down-regulated, negative targets up-regulated) and also have decreased expression themselves (i.e. blue squares in both “inferred activity” and “observed expression” columns) are most interesting because they potentially mediate the down-regulation of kidney developmental processes, metabolic activities and cell-cell junction genes observed in aged podocytes. Grhl2 (highlighted in blue rectangle) is the most down-regulated TF based on its own expression change, and its positive targets are significantly down-regulated in aged podocytes.
C.) The transcription factor Grhl2, a master regulator known to be involved in cell junction regulation, and its key target genes known to mediate the effects of Grhl2, were significantly down-regulated in aged podocytes based on our RNA-seq data, and can potentially explain the global down-regulation of cell-cell junction pathway identified in Figure S5. Y axis denotes absolute expression level of Grhl2 and its targets.
D.) Graphs of qPCR analysis of Grhl2 and its key target genes on a second cohort of podocytes isolated from non-reporter mice confirmed the significant decreases in aged reporter podocytes (all genes except Cldn3 had a p value of 0.04 or lower)
Figure 5B lists the top ten transcription factors in the RNA-seq dataset identified by VIPER in aged podocytes: Hnf1b, Grhl2, Sim1, Lrx1, Creb3l1, Zbtb16, Osr2, Foxo4, Pax8, and Trerf1. For example, the expression level of grainyhead-like transcription factor 2 (Grhl2) itself and its inferred activity based on target gene expression changes was significantly lower in aged podocytes, including E-cadherin (Cdh1), claudin 3 (Cldn3) and 4 (Cldn4), and Ras-Related Protein Rab-25 (Rab25), which were all significantly lower in aged podocytes (Figure 5C).
qPCR performed on the second cohort of isolated podocytes confirmed that of the 5 selected genes for validation, all were also significantly decreased in aged podocytes (Figure 5D, p value ≤0.04).
Bulk RNA-seq from isolated podocytes compared to published single cell RNA-seq
We compared our podocyte aging bulk RNA-seq with the published single cell (sc)RNA-seq of kidney aging from the Tabula Muris Senis project (https://tabula-muris-senis.ds.czbiohub.org). A limitation of scRNA-seq was the number of podocytes available for study: only 58 and 171 podocytes in the 3 and 30-month mice respectively. By comparison, each of our bulk RNA-seq samples included between 104,615 and 401,350 cells (Figure S9). In 3m mice, an average 2,308 genes were detected per cell in the scRNA-seq; by comparison, 8,043 genes were expressed above 4 RPKM in our bulk RNA-seq data. We detected 2,494 DEGs with stringent cutoff (FDR<0.05, fold change >2, absolute expression level >4 RPKM in either aged or young mice; 1219 up, 1,275 down). Even using a more lenient cutoff (FDR<0.1, fold change >1.5, in order to get sufficient number of genes for enrichment analysis) resulted in only 323 DEGs in the scRNA-seq data (97 up, 226 down, Figure S10A). Therefore, our RNA-seq data has more statistical power and detected a larger number of DEGs than the scRNA-seq experiment. Notably, none of the up- and down-regulated genes detected in our RNA-seq data and validated by qPCR in another batch of biological samples (Fig. 1C) were detected as DEGs by scRNA-seq (Figure S10B & C).
At the pathway level, only 9 GO terms were significantly enriched in the 226 genes down-regulated in the podocyte aging scRNA-seq data (FDR<0.05), compared to 500 GO terms significantly enriched in the down-regulated genes of our data. 4 of the 9 GO terms enriched in the scRNA-seq data are kidney and nephron developmental processes, all of which were also in our RNA-seq data, with higher statistical confidence (smaller p-value) in our data (Figure S10D). For the remaining 5 GO terms that did not achieve significance (circadian rhythm, EGF signaling), our data still captured many DEGs in those functional processes (Figure S10D). 496 of the highly significant functional processes we discovered (FDR<0.02) were missed by the scRNA-seq data, and few genes in these processes were found as DEGs by scRNA-seq (Figure S10E shows 10 such examples).
Comparison of differentially expressed genes in aging with diabetic kidney disease (DKD) and focal segmental glomerulosclerosis (FSGS)
Lastly, we used published data to intersect with our mouse aged podocyte data to determine any gene overlaps with two common glomerular diseases characterized by podocyte injury. FDR<0.05 and fold change >1.5 was used to match both studies. Using Diabetes Affymetrix microarray data,61 we matched 1,379 of the 1,700 differentially expressed probes with 1,174 unique genes in our RNA-seq data (Figure S11A). Only 102 of the 345 genes up-regulated in human DKD were also up-regulated in our podocyte aging RNA seq dataset; only 155 of the 829 down-regulated genes in DKD are also down-regulated in podocyte aging (Figure S11A). Immune regulatory genes commonly up-regulated (Ccl5, Runx3, C1qa, C1qb, Tlr7); genes important for podocyte function (Nphs1, Nphs2, Wt1), cell-cell junction (Tjp1, Synpo), and Wnt (Tcf7, Tcf21) pathways were commonly down-regulated in podocyte aging and DKD. Genes uniquely up-regulated in DKD were enriched for extracellular matrix (ECM) organization, endopeptidase activity, and vitamin D signaling; genes uniquely down-regulated in DKD were enriched for actin filament assembly and RNA polymerase II transcriptional initiation (Table S8).
We also compared our podocyte-aging RNA-seq data with Affymetrix microarray from human FSGS patients.62 Of the 784 genes up-regulated in FSGS, 148 of were also up-regulated in podocyte aging (Figure S11B). Of the 181 genes down-regulated in FSGS, 63 were down-regulated in podocyte aging. Commonly up-regulated genes include caspases 1 and 4, immune response genes (e.g, Cx3cr1, Tyrobp, C1qa, C1qb, Tlr1), and commonly down-regulated genes include growth factors (e.g., Egf, Ghr) and transporters (e.g., Oat3, Npt2, NaDC-3). Genes uniquely up-regulated in FSGS were enriched for angiogenesis, ribosomal RNA processing, and protein folding; genes uniquely down-regulated in FSGS were enriched for cAMP signaling, steroid hormone signaling, and would healing.
Taken together, pathways up- and down-regulated in podocyte aging were largely distinct from those perturbed in DKD and FSGS, represent a distinct biological signature.
DISCUSSION
The purpose of this study was to use a global and unbiased approach to define the transcriptomic differences between podocytes derived from aged mice and young mice. Our study leverages two methodologic strengths. First, we utilized inducible podocyte labeling in which podocytes were permanently labeled at a juvenile age (3–6 weeks), and allowed each mouse to age to 2–3 months or 22–24 months (equivalent to a 70+ year old human), thereby capturing truly aged podocytes. Second, direct isolation of reporter-labeled podocytes and unbiased gene expression quantification by bulk RNA-seq overcomes the problem of change in relative cellular composition of glomeruli in aged mice due to progressive loss of podocytes. Still, since fewer podocytes were isolated from aged mice, we normalized gene expression levels by the total sequenced read depth, which resulted in roughly equal numbers of significantly up- (1,219) and down- (1,275) regulated genes in aged vs. young podocytes. These steps ensured the differential expression changes observed were directly attributable to the age of the podocytes and not confounded by cellular composition (in the case of whole glomerular RNA-seq analyses) or the absolute number of isolated podocytes at each timepoint.
Expression of canonical podocyte genes was decreased in aged podocytes, such as slit diaphragm (Nphs1, Nphs2, Cd2ap,Trpc6), actin cytoskeleton (Synpo, Actn4), apical charge (Podxl), transcription factor (Wt1, Lmx1B) and the podocyte marker Cdkn1c (p57) genes. Podocyte number and density decrease with advancing age.20, 21 Pathways predicted to cause podocyte loss such as apoptosis, p53 and senescence were all significantly up-regulated in aged podocytes. Additionally, tight junction related genes such as ZO-1 (Tjp1),50, 63 MAGI-1,52, 57, 64 claudins53 and JAM 49 were decreased (Figure S7). With the exception of TGFß1, the upregulation of immune response genes (allograft rejection, TNF, interleukin and interferon signaling, complement pathway) was unexpected and needs further investigation. Interestingly, inflammatory genes increase in neuronal cells.65
We also analyzed sex differences. In podocyte aging, over 83% of up-regulated genes, and 67% of down-regulated genes, were shared between male and female mice. PCA analysis showed that sex differences account for a much smaller proportion of expression variance than age (7.2% vs. 64%). Genes preferentially expressed in female podocytes include the classical X-chromosome gene Xist, P450 enzymes (Cyp4a14, Cyp4a10), immunoglobulin kappa chain genes, and glutathione S-transferases. Genes preferentially expressed in male podocytes include Y chromosome genes Ddx37, Eif2s3y, Uty, Kdm5d and genes in xenobiotic detoxification. This contrasts with the kidney aging of cells of renin lineage, where most aging-associated genes were sex-specific.7
Taken together, our findings indicate that the decreased expression of canonical podocyte marker genes, junctional and adhesion proteins and pro-survival pathways synergize with the increase of inflammatory response pathways to provide a comprehensive mechanistic foundation for the well-established decrease in podocyte number and function with aging.
A second major finding was that many key podocyte-specific signaling activities were down in aged podocytes, likely leading to reduced function. By enriching for podocyte expressed genes in publicly available single cell RNA-seq data,43 we built a podocyte-specific ligand-receptor interaction network.42 Mapping our RNA-seq data onto this network revealed a set of podocyte-specific ligand-receptor pairs that show consistent and significant decline in aged podocytes. The receptors EGFR (−2.9 fold), ErBB4 (−2.7 fold) and many of their ligands that promote kidney injury66, 67 significantly declined. Other critical signaling pathways that declined included BMP, WNT, VEGF, FGF and SLIT2/ROBO2. Augmenting these declining signaling pathways may be protective in the aging kidney.
A third major finding of our study was that analysis of transcription factor gene expression and regulatory networks identified a candidate regulator of the aging response in podocytes. Grainyhead-like transcription factor 2 (Grhl2) and its key targets were significantly down-regulated in aged podocytes. Grhl2 null mice are embryonically lethal at approximately E11.5.68 Grhl2 is important for the formation of functional tight junctions and the apical junction complex by regulating the expression of junction genes E-cadherin, Claudin 4, 3, and Rab25.69–72 Grhl2 regulates renal epithelial barrier function,69 but neither its expression, nor function, have been previously reported in podocytes, in health or disease. Since down-regulation of cell-cell junctions was a major transcriptomic signature of aged podocytes, we speculate that Grhl2 may be a regulator of podocyte functional decline in aging. Our data included long non-coding RNAs, which represent a diverse set of RNA species that have been implicated in a wide variety of processes including kidney disease.73, 74 Some of these may reflect epigenetic activation of enhancers75 and can mediate sensitivity to drugs76 or contribute to pathways associated with sterile inflammation in aging.77
We compared our aging data to several published datasets. First, we asked if the transcriptomic signals of aging were distinct from other glomerular injury. While we found some common pathways shared with DKD and FSGS, 83% of DEGs were not differentially regulated in either disease. By contrast, the top upregulated pathway implicated in DKD is ECM organization while actin filament assembly is the most significantly downregulated pathways. FSGS is characterized by upregulation of angiogenesis pathways and downregulation of cAMP signaling. However, this conclusion must be balanced by noting that the two studies used for comparison used whole glomeruli, while we have reported data from isolated podocytes.
Important scRNA-seq studies have provided invaluable information about the podocyte transcriptome, but do not specifically address changes seen with aging. Kimmel included scRNA-seq of multiple kidney cell types, but unfortunately did not capture podocytes.78 Park79 and Han80 included podocytes at a single time point, but not in aging. The most comparable dataset is scRNA-seq of 10 kidney cell types in the Tabula Muris Senis project. Reassuringly, we found genes down-regulated in our dataset overlapped significantly with genes down-regulated in their scRNA-seq in aged podocytes. Kidney developmental GO terms were down-regulated in our bulk and their scRNA-seq of podocytes. No other kidney cell type in their scRNA-seq data displayed down-regulation of kidney developmental GO terms (data not shown), suggesting that this aging associated change is unique to podocytes. Therefore, our bulk RNA-seq data and analyses represents a richer exploration of the range of pathways affected during aging, many of which were missed in scRNA-seq due to the reduced sensitivity of the method.
Long non-coding RNAs have been implicated in a variety of processes including kidney disease,73, 74 may reflect epigenetic enhancer activation,75 drug sensitivity76 or contribute to pathways associated with sterile inflammation of aging.77 Reflecting the complexity of their regulation and function, many have been implicated in diverse pathways including development, metabolism, inflammation and epigenetic regulation.
We recognize our study limitations, including one aged time point, that changes might be species specific, and the use of warm digestion in Liberase versus cold digestion protocols.81Accordingly, we performed bulk RNAseq to overcome artifacts due to the relatively small cell number and lower quantity of RNA utilized for scRNAseq.
In conclusion, we generated a rich reference data set of transcriptional changes associated with aging in murine podocytes. Our findings reveal a transcriptional program that alters cell junctional complexes, increases senescence pathways and apoptosis that synergizes to promote podocyte loss and detachment over time. We have identified autocrine/paracrine signaling perturbations that provide an opportunity for pharmacologic modulation of podocyte aging. Our analysis of gene regulatory networks identified Grhl2 as a potential master regulatory of podocyte aging. While some pathways are shared with DKD and FSGS, the majority of the transcriptional changes are unique to aging. In summary, our data provides unique insight into podocyte aging and identifies novel targetable pathways with potential to reverse the decline of podocyte number and function with age.
METHODS
See Supplementary Methods for detailed descriptions.
Animals:
Podocyte reporter mice (inducible): Inducible NPHS2-rtTA|tetO-cre|tdTomato mice were generated as previously reported.82–84
Non-reporter mice: NIA aged C57BL/6 colony were analyzed.
Podocyte reporter mice (constitutive): Nphs1-FLPo/FRT-EGFP mice were utilized.
Animal protocols were approved (2968–04) by the University of Washington Institutional Animal Care and Use Committee.
Isolation of podocytes for RNA-seq
From podocyte reporter mice:
Kidney cortex was digested and podocytes isolated using fluorescence-activated cell sorting (FACS) (Figure S1A).
From non-reporter mice:
Kidney cortex was digested and podocytes isolated using magnetic-activated cell sorting (MACS) (Figure S1B).
RNA isolation
RNA purification was performed using commercial kits. A summary of the RNA integrity numbers and yields are shown in Figure S2A.
RNA-seq library preparation and sequencing
A detailed description of the RNA-seq library preparation and sequencing is provided in the Supplemental Methods.
RNA-seq Data Analysis
A detailed description of the RNA-seq data analysis is provided in the Supplemental Methods.
Quantitative real-time PCR Analysis
A detailed description of quantitative real-time PCR analysis is provided in the Supplemental Methods.
Immunohistochemistry
Immunofluorescent38, 85–87 and immunoperoxidase8, 11, 88 staining were performed as previously described.
Assessment of glomerular scarring and extracellular matrix
Jones’ staining was performed as previously described.11
Podocyte ultrastructure analysis using kidney expansion and microscopy
Expansion microscopy was performed using a previously described protocol.37, 38
Supplementary Material
Figure S1 Overview of Methodology.
A.) Chronological schema of overall experimental approach for fluorescence-activated cell sorting and RNA-Seq of reporter labeled young and aged podocytes.
B.) Chronological schema of overall experimental approach for magnetic activated cell sorting of non-reporter labeled young and aged podocytes for qPCR confirmation.
Figure S2. Functional, histological and morphological changes in aged kidneys.
A.) Graph showing albumin to creatinine ratio measurements in young mice (3months) and aged mice (22 months). Microalbuminuria was higher in aged mice.
B-C.) Representative images of Jones’ staining for basement membrane from the same young (2–3 months) and aged (22–24 months) male and female mice from which the RNAseq dataset was obtained. Lower magnification (upper panels) and higher magnification (lower panels show the yellow dashed boxes in the upper panels) are shown. Aged mice have thickening of the glomerular basement membrane and Bowman’s capsule, mesangial matrix expansion and focal areas of scarring.
D-G.) Representative images of glomeruli from young (3 months) and aged (24 months) Nphs1-FLPo/FRT-EGFP podocyte reporter mice using a super-resolution fluorescence microscopy method, called expansion microscopy (ExM), for volumetric nanoscale interrogation of mouse kidney tissue. Glomeruli from aged mice show focal loss of podocyte foot process structures compared to glomeruli from young mice.
Figure S3. Quality Control.
A.) Table lists the podocyte yield, RNA Integrity Number, RNA concentration and RNA yield for each sample used to generate the RNA-seq dataset.
B.) Images of non-podocyte cell fractions (no staining) and Alexa Fluor® 488- positive cells in the podocyte cell fraction, indicated the presence of nephrin. qPCR for a panel of known podocyte genes showed significantly increased expression in the podocyte cell fractions compared to the non-podocyte cell fractions. Both findings confirm the isolation of podocytes from other kidney cells by magnetic-activated cell sorting.
Figure S4. Reduced protein expression mirrors transcriptomic changes in aged podocytes.
Kidney sections from young and aged mice used for FACS sorting was used for immunofluorescent staining for A.) nephrin (Nhsp1), B.) podocin (Nhsp2), C.) synaptopodin (Synpo) and D.) zona occludens 1 (Tjp1). Similar to the transcripts, staining intensity was reduced in aged glomeruli. Similar to the transcript, immunoperoxidase staining for E.) paired immunoglobin-like type 2 receptor beta (Pilrb2) was increased in aged glomeruli.
Figure S5. Perturbed pathways in podocyte aging.
(A, B) Hallmark Pathways. (A): Based on Hallmark gene sets, aged podocytes showed up-regulation of inflammatory and immune responses, apoptosis, PI3K-AKT-mTOR and p53 pathways. (B) Multiple metabolic pathways (xenobiotic metabolism fatty acid metabolism, glycolysis etc.) are down-regulated in aged podocytes
(C, D) Gene Ontology Cellular Compartments. (C): Aged podocytes showed up-regulation of MHC complexes and nucleus compartments; (D): Genes involved cell-cell junctions are down-regulated in aged podocytes.
Figure S6. A) Aged vs. young analysis for female and male mice separately showed that most up and down-regulated genes in podocyte aging are shared between the sexes.
B) and C) the functional processes perturbed in female and male podocyte aging were also shared and overlapped with both sexes as follows: a significant up-regulation of immune response, TNF signaling, apoptosis, p53 pathways and down-regulation of multiple kidney developmental processes and cell-cell junction.
Figure S7. Podocyte-specific ligand-receptor signaling network based on bulk RNA-seq of FACS-purified podocytes, mesangial cells and endothelial cells from healthy mouse glomeruli.
19 ligand-receptor pairs, defined as podocyte-specific when both the ligand and receptor are two-fold higher in podocytes than in endothelial or mesangial cells in the glomerulus based on FACS-purified RNA-seq data, were identified to change in aged podocytes. Circles represent ligands, squares represent receptors, genes with significantly lower expression in aged podocytes are filled with blue color, genes with significantly higher expression in aged podocytes are filled with red color, genes with no significant change are labeled with grey color. gene names are labeled in brown. Arrows represents ligand-receptor interactions, in the direction from ligand to its receptor. Arrows are colored blue if both ligand and receptor are significantly lower in aged podocytes, and grey if ligand/receptor did not show consistent change.
Figure S8. Genes in the apoptosis pathway were up-regulated in aged podocytes.
Gene expression data was mapped to the Apoptosis pathway in KEGG pathway database. Red rectangles denote up-regulated genes (aged/young expression ratio >1), blue rectangles denote down-regulated genes (aged/young expression ratio <1); grey rectangles denote genes with no significant change. Edges represent interactions between genes/proteins. “+p” denotes phosphorylation. Sharp arrows indicate positive, while dashed arrows indicate negative regulation. The results showed an overall significant increase in genes regulating apoptosis in aged podocytes.
Figure S9. Gene expression data mapped to the Tight Junction pathway in KEGG pathway database.
Red rectangles denote up-regulated genes (aged/young expression ratio >1), blue rectangles denote down-regulated genes (aged/young expression ratio <1); grey rectangles denote genes with no significant change. Edges represent interactions between genes/proteins. “+p” denotes phosphorylation. Sharp arrows indicate positive, while dashed arrows indicate negative regulation. Most components of the tight junction pathway, including several critical for podocyte function (e.g. ZO-1, MAGI-1, JAM, occludins) are down-regulated in aged podocytes.
Figure S10. Comparison with kidney aging single cell RNA-seq data
A). The bulk podocyte RNA-seq from this study captured 14 times more up-regulated genes and 8 fold more down-regulated genes than single cell RNA-seq of podocyte aging from the Tabula Muris Senis project.
B) and C). Most significantly up- and down-regulated genes, including those known to be critical for podocyte function (e.g., Wt1, Nphs1, Nphs2) are captured by our bulk RNA-seq and independently validated in qPCR, but none are identified as differentially expressed by single cell RNA-seq
D). Down-regulated biological processes identified by podocyte single cell RNA-seq are mostly shared with our bulk RNA-seq.
E). Bulk RNA-seq of podocyte aging uniquely captured many kidney-relevant biological processes missed by single cell RNA-seq.
Figure S11. Most podocyte-aging associated genes are not shared with DKD or FSGS.
A). Venn diagram of unique and overlapping genes between podocyte aging and DKD. Genes uniquely up- and down-regulated in DKD are enriched for ECM organization and actin filament assembly, respectively (Table S8).
B). Venn diagram of unique and overlapping genes between podocyte aging and FSGS. Genes uniquely up- and down-regulated in FSGS are enriched for angiogenesis/protein folding, and cAMP signaling, respectively (Table S9).
Table S1: 2494 differentially expressed genes.
Table S2: RPKM values.
Table S3: GSEA results.
Table S4: DAVID GO BP DIRECT PODO UP.
Table S5: DAVID GO BP DIRECT PODO DOWN.
Table S6: Top genes contributing to PC2 sex differences.
Table S7: GO analysis for PC2 male enriched genes.
Table S8: GO analysis for genes uniquely up or down in DKD.
Table S9: GO analysis for genes uniquely up or down in FSGS.
Acknowledgements:
Grants Supporting Work: 5 R01 DK 056799–10, 5 R01 DK 056799–12, 1 R01 DK097598–01A1 (All Funded to S.J.S)
Footnotes
DISCLOSURES
None of the authors have any financial or other conflicts of interest. The results presented in this paper have not been published previously, in whole or part.
SUPPLEMENTAL MATERIAL
Supplementary information is available on Kidney International’s website.
Figure S1: Overview of Methodology.
Figure S2: Functional, histological and ultrastructural changes in aged kidneys.
Figure S3: Quality Control.
Figure S4: Reduced podocyte protein expression mirrors transcriptomic changes in aged podocytes.
Figure S5: Perturbed pathways in podocyte aging.
Figure S6: Up- and down-regulated genes and pathways in female and male podocyte aging are mostly shared.
Figure S7: Podocyte-specific ligand-receptor signaling network based on bulk RNA-seq of FACS-sorted podocytes, mesangial cells and endothelial cells data from mouse glomeruli.
Figure S8: Genes in the apoptosis pathway were up-regulated in aged podocytes.
Figure S9: Gene expression data mapped to the Tight Junction pathway in KEGG pathway database.
Figure S10: Comparison with kidney aging single cell RNA-seq data.
Figure S11: Comparing genes perturbed in aging, DKD and FSGS.
Supplemental Methods: Detailed descriptions of all methods used.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Denic A, Mathew J, Lerman LO, et al. Single-Nephron Glomerular Filtration Rate in Healthy Adults. N Engl J Med 2017; 376: 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denic A, Lieske JC, Chakkera HA, et al. The Substantial Loss of Nephrons in Healthy Human Kidneys with Aging. J Am Soc Nephrol 2017; 28: 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cianciolo RE, Benali SL, Aresu L. Aging in the Canine Kidney. Vet Pathol 2016; 53: 299–308. [DOI] [PubMed] [Google Scholar]
- 4.Wiggins JE, Goyal M, Sanden SK, et al. Podocyte hypertrophy, “adaptation,” and “decompensation” associated with glomerular enlargement and glomerulosclerosis in the aging rat: prevention by calorie restriction. J Am Soc Nephrol 2005; 16: 2953–2966. [DOI] [PubMed] [Google Scholar]
- 5.Wiggins JE. Aging in the glomerulus. J Gerontol A Biol Sci Med Sci 2012; 67: 1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floege J, Hackmann B, Kliem V, et al. Age-related glomerulosclerosis and interstitial fibrosis in Milan normotensive rats: a podocyte disease. Kidney Int 1997; 51: 230–243. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Eng DG, Pippin JW, et al. Sex differences in transcriptomic profiles in aged kidney cells of renin lineage. Aging (Albany NY) 2018; 10: 606–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamatani H, Eng DG, Kaverina NV, et al. Lineage tracing aged mouse kidneys shows lower number of cells of renin lineage and reduced responsiveness to RAAS inhibition. Am J Physiol Renal Physiol 2018; 315: F97–F109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oxburgh L, Carroll TJ, Cleaver O, et al. (Re)Building a Kidney. J Am Soc Nephrol 2017; 28: 1370–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefanska A, Eng D, Kaverina N, et al. Interstitial pericytes decrease in aged mouse kidneys. Aging (Albany NY) 2015; 7: 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider RR, Eng DG, Kutz JN, et al. Compound effects of aging and experimental FSGS on glomerular epithelial cells. Aging (Albany NY) 2017; 9: 524–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweetwyne MT, Pippin JW, Eng DG, et al. The mitochondrial-targeted peptide, SS-31, improves glomerular architecture in mice of advanced age. Kidney Int 2017; 91: 1126–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNicholas BA, Eng DG, Lichtnekert J, et al. Reducing mTOR augments parietal epithelial cell density in a model of acute podocyte depletion and in aged kidneys. Am J Physiol Renal Physiol 2016; 311: F626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roeder SS, Stefanska A, Eng DG, et al. Changes in glomerular parietal epithelial cells in mouse kidneys with advanced age. Am J Physiol Renal Physiol 2015; 309: F164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt R, Melk A. Molecular mechanisms of renal aging. Kidney Int 2017; 92: 569–579. [DOI] [PubMed] [Google Scholar]
- 16.Camici M, Carpi A, Cini G, et al. Podocyte dysfunction in aging--related glomerulosclerosis. Front Biosci (Schol Ed) 2011; 3: 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiggins J. Podocytes and glomerular function with aging. Semin Nephrol 2009; 29: 587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glassock RJ, Rule AD. The implications of anatomical and functional changes of the aging kidney: with an emphasis on the glomeruli. Kidney Int 2012; 82: 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hommos MS, Glassock RJ, Rule AD. Structural and Functional Changes in Human Kidneys with Healthy Aging. J Am Soc Nephrol 2017; 28: 2838–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puelles VG, Cullen-McEwen LA, Taylor GE, et al. Human podocyte depletion in association with older age and hypertension. Am J Physiol Renal Physiol 2016; 310: F656–F668. [DOI] [PubMed] [Google Scholar]
- 21.Hodgin JB, Bitzer M, Wickman L, et al. Glomerular Aging and Focal Global Glomerulosclerosis: A Podometric Perspective. J Am Soc Nephrol 2015; 26: 3162–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shankland SJ, Freedman BS, Pippin JW. Can podocytes be regenerated in adults? Curr Opin Nephrol Hypertens 2017; 26: 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuang PY, Cai W, Li X, et al. Reduction in podocyte SIRT1 accelerates kidney injury in aging mice. Am J Physiol Renal Physiol 2017; 313: F621–F628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noordmans GA, Hillebrands JL, van Goor H, et al. A roadmap for the genetic analysis of renal aging. Aging Cell 2015; 14: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell 2005; 120: 483–495. [DOI] [PubMed] [Google Scholar]
- 26.Sun N, Youle RJ, Finkel T. The Mitochondrial Basis of Aging. Mol Cell 2016; 61: 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiels PG, McGuinness D, Eriksson M, et al. The role of epigenetics in renal ageing. Nat Rev Nephrol 2017; 13: 471–482. [DOI] [PubMed] [Google Scholar]
- 28.Pal S, Tyler JK. Epigenetics and aging. Sci Adv 2016; 2: e1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valentijn FA, Falke LL, Nguyen TQ, et al. Cellular senescence in the aging and diseased kidney. J Cell Commun Signal 2018; 12: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenoir O, Tharaux PL, Huber TB. Autophagy in kidney disease and aging: lessons from rodent models. Kidney Int 2016; 90: 950–964. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Jung KJ, Kim JW, et al. Suppression of apoptosis by calorie restriction in aged kidney. Exp Gerontol 2004; 39: 1361–1368. [DOI] [PubMed] [Google Scholar]
- 32.Tower J. Programmed cell death in aging. Ageing Res Rev 2015; 23: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wheeler HE, Metter EJ, Tanaka T, et al. Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet 2009; 5: e1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melk A, Mansfield ES, Hsieh SC, et al. Transcriptional analysis of the molecular basis of human kidney aging using cDNA microarray profiling. Kidney Int 2005; 68: 2667–2679. [DOI] [PubMed] [Google Scholar]
- 35.Rodwell GE, Sonu R, Zahn JM, et al. A transcriptional profile of aging in the human kidney. PLoS Biol 2004; 2: e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu J, Wei C, Lee K, et al. Comparison of Glomerular and Podocyte mRNA Profiles in Streptozotocin-Induced Diabetes. J Am Soc Nephrol 2016; 27: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chozinski TJ, Mao C, Halpern AR, et al. Volumetric, Nanoscale Optical Imaging of Mouse and Human Kidney via Expansion Microscopy. Sci Rep 2018; 8: 10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaverina NV, Eng DG, Freedman BS, et al. Dual lineage tracing shows that glomerular parietal epithelial cells can transdifferentiate toward the adult podocyte fate. Kidney Int 2019; 96: 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merico D, Isserlin R, Stueker O, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010; 5: e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006; 22: 1600–1607. [DOI] [PubMed] [Google Scholar]
- 41.Liberzon A, Birger C, Thorvaldsdottir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015; 1: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramilowski JA, Goldberg T, Harshbarger J, et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 2015; 6: 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karaiskos N, Rahmatollahi M, Boltengagen A, et al. A Single-Cell Transcriptome Atlas of the Mouse Glomerulus. J Am Soc Nephrol 2018; 29: 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potter AS, Drake K, Brunskill EW, et al. A bigenic mouse model of FSGS reveals perturbed pathways in podocytes, mesangial cells and endothelial cells. PLoS One 2019; 14: e0216261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013; 29: 1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Otin C, Blasco MA, Partridge L, et al. The hallmarks of aging. Cell 2013; 153: 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao M, Chen L, Qu H. CSGene: a literature-based database for cell senescence genes and its application to identify critical cell aging pathways and associated diseases. Cell Death Dis 2016; 7: e2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukasawa H, Bornheimer S, Kudlicka K, et al. Slit diaphragms contain tight junction proteins. J Am Soc Nephrol 2009; 20: 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnabel E, Anderson JM, Farquhar MG. The tight junction protein ZO-1 is concentrated along slit diaphragms of the glomerular epithelium. J Cell Biol 1990; 111: 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boerries M, Grahammer F, Eiselein S, et al. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int 2013; 83: 1052–1064. [DOI] [PubMed] [Google Scholar]
- 52.Semeniuk Ia P. [Characteristics of the examination and registration of the case histories of military personnel subjected to the action of occupational hazards]. Voen Med Zh 1976: 73–74. [PubMed] [Google Scholar]
- 53.Gong Y, Sunq A, Roth RA, et al. Inducible Expression of Claudin-1 in Glomerular Podocytes Generates Aberrant Tight Junctions and Proteinuria through Slit Diaphragm Destabilization. J Am Soc Nephrol 2017; 28: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartleben B, Schweizer H, Lubben P, et al. Neph-Nephrin proteins bind the Par3-Par6-atypical protein kinase C (aPKC) complex to regulate podocyte cell polarity. J Biol Chem 2008; 283: 23033–23038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satoh D, Hirose T, Harita Y, et al. aPKClambda maintains the integrity of the glomerular slit diaphragm through trafficking of nephrin to the cell surface. J Biochem 2014; 156: 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartleben B, Widmeier E, Suhm M, et al. aPKClambda/iota and aPKCzeta contribute to podocyte differentiation and glomerular maturation. J Am Soc Nephrol 2013; 24: 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shirata N, Ihara KI, Yamamoto-Nonaka K, et al. Glomerulosclerosis Induced by Deficiency of Membrane-Associated Guanylate Kinase Inverted 2 in Kidney Podocytes. J Am Soc Nephrol 2017; 28: 2654–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alvarez MJ, Shen Y, Giorgi FM, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet 2016; 48: 838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 2006; 7 Suppl 1: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JW, Chou CL, Knepper MA. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am Soc Nephrol 2015; 26: 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woroniecka KI, Park AS, Mohtat D, et al. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011; 60: 2354–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shved N, Warsow G, Eichinger F, et al. Transcriptome-based network analysis reveals renal cell type-specific dysregulation of hypoxia-associated transcripts. Sci Rep 2017; 7: 8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Itoh M, Nakadate K, Matsusaka T, et al. Effects of the differential expression of ZO-1 and ZO-2 on podocyte structure and function. Genes Cells 2018; 23: 546–556. [DOI] [PubMed] [Google Scholar]
- 64.Ni J, Bao S, Johnson RI, et al. MAGI-1 Interacts with Nephrin to Maintain Slit Diaphragm Structure through Enhanced Rap1 Activation in Podocytes. J Biol Chem 2016; 291: 24406–24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 2015; 21: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bollee G, Flamant M, Schordan S, et al. Epidermal growth factor receptor promotes glomerular injury and renal failure in rapidly progressive crescentic glomerulonephritis. Nat Med 2011; 17: 1242–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee HW, Khan SQ, Khaliqdina S, et al. Absence of miR-146a in Podocytes Increases Risk of Diabetic Glomerulopathy via Up-regulation of ErbB4 and Notch-1. J Biol Chem 2017; 292: 732–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rifat Y, Parekh V, Wilanowski T, et al. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev Biol 2010; 345: 237–245. [DOI] [PubMed] [Google Scholar]
- 69.Aue A, Hinze C, Walentin K, et al. A Grainyhead-Like 2/Ovo-Like 2 Pathway Regulates Renal Epithelial Barrier Function and Lumen Expansion. J Am Soc Nephrol 2015; 26: 2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Senga K, Mostov KE, Mitaka T, et al. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell 2012; 23: 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Werth M, Walentin K, Aue A, et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 2010; 137: 3835–3845. [DOI] [PubMed] [Google Scholar]
- 72.Tanimizu N, Mitaka T. Role of grainyhead-like 2 in the formation of functional tight junctions. Tissue Barriers 2013; 1: e23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol 2016; 12: 360–373. [DOI] [PubMed] [Google Scholar]
- 74.Ignarski M, Islam R, Muller RU. Long Non-Coding RNAs in Kidney Disease. Int J Mol Sci 2019; 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010; 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bester AC, Lee JD, Chavez A, et al. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell 2018; 173: 649–664 e620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Cecco M, Ito T, Petrashen AP, et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019; 566: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kimmel JC, Penland L, Rubinstein ND, et al. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res 2019; 29: 2088–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park J, Shrestha R, Qiu C, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 2018; 360: 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han X, Wang R, Zhou Y, et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 2018; 172: 1091–1107 e1017. [DOI] [PubMed] [Google Scholar]
- 81.Adam M, Potter AS, Potter SS. Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 2017; 144: 3625–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shigehara T, Zaragoza C, Kitiyakara C, et al. Inducible podocyte-specific gene expression in transgenic mice. J Am Soc Nephrol 2003; 14: 1998–2003. [DOI] [PubMed] [Google Scholar]
- 83.Perl AK, Wert SE, Nagy A, et al. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A 2002; 99: 10482–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010; 13: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eng DG, Kaverina NV, Schneider RRS, et al. Detection of renin lineage cell transdifferentiation to podocytes in the kidney glomerulus with dual lineage tracing. Kidney Int 2018; 93: 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaverina NV, Eng DG, Largent AD, et al. WT1 Is Necessary for the Proliferation and Migration of Cells of Renin Lineage Following Kidney Podocyte Depletion. Stem Cell Reports 2017; 9: 1152–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaverina NV, Kadoya H, Eng DG, et al. Tracking the stochastic fate of cells of the renin lineage after podocyte depletion using multicolor reporters and intravital imaging. PLoS One 2017; 12: e0173891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suzuki T, Eng DG, McClelland AD, et al. Cells of NG2 lineage increase in glomeruli of mice following podocyte depletion. Am J Physiol Renal Physiol 2018; 315: F1449–F1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldberg S, Adair-Kirk TL, Senior RM, et al. Maintenance of glomerular filtration barrier integrity requires laminin alpha5. J Am Soc Nephrol 2010; 21: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marshall CB, Krofft RD, Pippin JW, et al. CDK inhibitor p21 is prosurvival in adriamycin-induced podocyte injury, in vitro and in vivo. Am J Physiol Renal Physiol 2010; 298: F1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heeringa SF, Vlangos CN, Chernin G, et al. Thirteen novel NPHS1 mutations in a large cohort of children with congenital nephrotic syndrome. Nephrol Dial Transplant 2008; 23: 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013; 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao Y, Hsiao TH, Suresh U, et al. A novel significance score for gene selection and ranking. Bioinformatics 2014; 30: 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Overview of Methodology.
A.) Chronological schema of overall experimental approach for fluorescence-activated cell sorting and RNA-Seq of reporter labeled young and aged podocytes.
B.) Chronological schema of overall experimental approach for magnetic activated cell sorting of non-reporter labeled young and aged podocytes for qPCR confirmation.
Figure S2. Functional, histological and morphological changes in aged kidneys.
A.) Graph showing albumin to creatinine ratio measurements in young mice (3months) and aged mice (22 months). Microalbuminuria was higher in aged mice.
B-C.) Representative images of Jones’ staining for basement membrane from the same young (2–3 months) and aged (22–24 months) male and female mice from which the RNAseq dataset was obtained. Lower magnification (upper panels) and higher magnification (lower panels show the yellow dashed boxes in the upper panels) are shown. Aged mice have thickening of the glomerular basement membrane and Bowman’s capsule, mesangial matrix expansion and focal areas of scarring.
D-G.) Representative images of glomeruli from young (3 months) and aged (24 months) Nphs1-FLPo/FRT-EGFP podocyte reporter mice using a super-resolution fluorescence microscopy method, called expansion microscopy (ExM), for volumetric nanoscale interrogation of mouse kidney tissue. Glomeruli from aged mice show focal loss of podocyte foot process structures compared to glomeruli from young mice.
Figure S3. Quality Control.
A.) Table lists the podocyte yield, RNA Integrity Number, RNA concentration and RNA yield for each sample used to generate the RNA-seq dataset.
B.) Images of non-podocyte cell fractions (no staining) and Alexa Fluor® 488- positive cells in the podocyte cell fraction, indicated the presence of nephrin. qPCR for a panel of known podocyte genes showed significantly increased expression in the podocyte cell fractions compared to the non-podocyte cell fractions. Both findings confirm the isolation of podocytes from other kidney cells by magnetic-activated cell sorting.
Figure S4. Reduced protein expression mirrors transcriptomic changes in aged podocytes.
Kidney sections from young and aged mice used for FACS sorting was used for immunofluorescent staining for A.) nephrin (Nhsp1), B.) podocin (Nhsp2), C.) synaptopodin (Synpo) and D.) zona occludens 1 (Tjp1). Similar to the transcripts, staining intensity was reduced in aged glomeruli. Similar to the transcript, immunoperoxidase staining for E.) paired immunoglobin-like type 2 receptor beta (Pilrb2) was increased in aged glomeruli.
Figure S5. Perturbed pathways in podocyte aging.
(A, B) Hallmark Pathways. (A): Based on Hallmark gene sets, aged podocytes showed up-regulation of inflammatory and immune responses, apoptosis, PI3K-AKT-mTOR and p53 pathways. (B) Multiple metabolic pathways (xenobiotic metabolism fatty acid metabolism, glycolysis etc.) are down-regulated in aged podocytes
(C, D) Gene Ontology Cellular Compartments. (C): Aged podocytes showed up-regulation of MHC complexes and nucleus compartments; (D): Genes involved cell-cell junctions are down-regulated in aged podocytes.
Figure S6. A) Aged vs. young analysis for female and male mice separately showed that most up and down-regulated genes in podocyte aging are shared between the sexes.
B) and C) the functional processes perturbed in female and male podocyte aging were also shared and overlapped with both sexes as follows: a significant up-regulation of immune response, TNF signaling, apoptosis, p53 pathways and down-regulation of multiple kidney developmental processes and cell-cell junction.
Figure S7. Podocyte-specific ligand-receptor signaling network based on bulk RNA-seq of FACS-purified podocytes, mesangial cells and endothelial cells from healthy mouse glomeruli.
19 ligand-receptor pairs, defined as podocyte-specific when both the ligand and receptor are two-fold higher in podocytes than in endothelial or mesangial cells in the glomerulus based on FACS-purified RNA-seq data, were identified to change in aged podocytes. Circles represent ligands, squares represent receptors, genes with significantly lower expression in aged podocytes are filled with blue color, genes with significantly higher expression in aged podocytes are filled with red color, genes with no significant change are labeled with grey color. gene names are labeled in brown. Arrows represents ligand-receptor interactions, in the direction from ligand to its receptor. Arrows are colored blue if both ligand and receptor are significantly lower in aged podocytes, and grey if ligand/receptor did not show consistent change.
Figure S8. Genes in the apoptosis pathway were up-regulated in aged podocytes.
Gene expression data was mapped to the Apoptosis pathway in KEGG pathway database. Red rectangles denote up-regulated genes (aged/young expression ratio >1), blue rectangles denote down-regulated genes (aged/young expression ratio <1); grey rectangles denote genes with no significant change. Edges represent interactions between genes/proteins. “+p” denotes phosphorylation. Sharp arrows indicate positive, while dashed arrows indicate negative regulation. The results showed an overall significant increase in genes regulating apoptosis in aged podocytes.
Figure S9. Gene expression data mapped to the Tight Junction pathway in KEGG pathway database.
Red rectangles denote up-regulated genes (aged/young expression ratio >1), blue rectangles denote down-regulated genes (aged/young expression ratio <1); grey rectangles denote genes with no significant change. Edges represent interactions between genes/proteins. “+p” denotes phosphorylation. Sharp arrows indicate positive, while dashed arrows indicate negative regulation. Most components of the tight junction pathway, including several critical for podocyte function (e.g. ZO-1, MAGI-1, JAM, occludins) are down-regulated in aged podocytes.
Figure S10. Comparison with kidney aging single cell RNA-seq data
A). The bulk podocyte RNA-seq from this study captured 14 times more up-regulated genes and 8 fold more down-regulated genes than single cell RNA-seq of podocyte aging from the Tabula Muris Senis project.
B) and C). Most significantly up- and down-regulated genes, including those known to be critical for podocyte function (e.g., Wt1, Nphs1, Nphs2) are captured by our bulk RNA-seq and independently validated in qPCR, but none are identified as differentially expressed by single cell RNA-seq
D). Down-regulated biological processes identified by podocyte single cell RNA-seq are mostly shared with our bulk RNA-seq.
E). Bulk RNA-seq of podocyte aging uniquely captured many kidney-relevant biological processes missed by single cell RNA-seq.
Figure S11. Most podocyte-aging associated genes are not shared with DKD or FSGS.
A). Venn diagram of unique and overlapping genes between podocyte aging and DKD. Genes uniquely up- and down-regulated in DKD are enriched for ECM organization and actin filament assembly, respectively (Table S8).
B). Venn diagram of unique and overlapping genes between podocyte aging and FSGS. Genes uniquely up- and down-regulated in FSGS are enriched for angiogenesis/protein folding, and cAMP signaling, respectively (Table S9).
Table S1: 2494 differentially expressed genes.
Table S2: RPKM values.
Table S3: GSEA results.
Table S4: DAVID GO BP DIRECT PODO UP.
Table S5: DAVID GO BP DIRECT PODO DOWN.
Table S6: Top genes contributing to PC2 sex differences.
Table S7: GO analysis for PC2 male enriched genes.
Table S8: GO analysis for genes uniquely up or down in DKD.
Table S9: GO analysis for genes uniquely up or down in FSGS.