Abstract
Sepsis is the major cause of acute kidney injury (AKI) associated with high mortality rates. Mitochondrial dysfunction contributes to the pathophysiology of septic AKI. Mitophagy is an important mitochondrial quality control mechanism that selectively eliminates damaged mitochondria, but its role and regulation in septic AKI remain largely unknown. Here, we demonstrate the induction of mitophagy in mouse models of septic AKI induced by lipopolysaccharide (LPS) treatment or by cecal ligation and puncture. Mitophagy was also induced in cultured proximal tubular epithelial cells exposed to LPS. Induction of mitophagy under these experimental setting was suppressed by pink1 or park2 knockout, indicating the role of the PINK1/PARK2 pathway of mitophagy in septic AKI. In addition, sepsis induced more severe kidney injury and cell apoptosis in pink1 or park2 knockout mice than in wild-type mice, suggesting a beneficial role of mitophagy in septic AKI. Furthermore, in cultured renal tubular cells treated with LPS, knockdown of pink1 or park2 inhibited mitochondrial accumulation of the autophagy adaptor optineurin (OPTN) and silencing Optn inhibited LPS-induced mitophagy. Taken together, these findings suggest that the PINK1/PARK2 pathway of mitophagy plays an important role in mitochondrial quality control, tubular cell survival, and renal function in septic AKI.
Keywords: Acute kidney injury, Mitophgay, Optineurin, PINK1, PARK2, Sepsis
1. Introduction
Acute kidney injury (AKI) is a major kidney disease featured by rapid decline and even loss of renal function [1,2]. The main causes of AKI include sepsis, nephrotoxins, and renal ischemia-reperfusion (IR) [3,4]. Sepsis, which is characterized by systemic inflammation, protracted host immunosuppression and multiple organ dysfunction, contributes to nearly half of AKI in hospitalized patients [5,6]. Unfortunately, septic AKI is associated unacceptably high morbidity and mortality rates [7]. Accumulating evidence suggests that pathogenesis of septic AKI is multifactorial and complex, involving the interplay among inflammation, tubular epithelial cell injury, and endothelial cell injury [8]. Notably, mitochondrial dysfunction has emerged as a converging point of multiple pathological pathways for AKI [[8], [9], [10], [11], [12], [13]]. Specific interventions that preserve and restore mitochondrial function, or timely eliminate damaged mitochondria may provide effective therapeutic strategies for AKI [8,[12], [13], [14], [15]].
Mitophagy is a selective form of autophagy that specifically eliminates unwanted or damaged mitochondria. Under stressful conditions that cause mitochondrial damage, mitophagy represents an important quality control mechanism that maintains mitochondrial homeostasis and thereby preserves cell viability [16,17]. In mammalian cells, mitophagy requires efficient recognition of targeted mitochondria and subsequent engulfment of mitochondria within autophagosome for final lysosomal degradation [18]. Currently, two major pathways of mitophagy have been proposed, which are respectively mediated by the PTEN-induced kinase 1 (PINK1)-parkin RBR E3 ubiquitin protein ligase (PARK2) pathway [19,20] and mitophagy receptors such as BCL2 and adenovirus E1B 19-kDa-interacting protein 3 (BNIP3), BNIP3-like (BNIP3L/NIX), and FUN14 Domain Containing 1 (FUNDC1) [21]. In healthy mitochondria, PINK1 is constitutively imported into mitochondria, where it is cleaved by the intramembrane serine protease PARL and ultimately proteolytically degraded [22]. When mitochondria are depolarized in stressful condition, the import of PINK1 is impeded, leading to PINK1 accumulation at the mitochondrial outer membrane (MOM). PINK1 in MOM recruits PARK2 from cytosol and activates PARK2 E3 ligase activity though direct phosphorylation of PARK2 and ubiquitin. Activated PARK2 builds poly-ubiquitin chains on MOM proteins, which in turn recruits autophagy adaptor proteins, such as NDP52 (calcium binding and coiled-coil domain 2) and optineurin. These adaptor proteins simultaneously bind to poly-ubiquitinated MOM proteins on mitochondria and LC3B (microtubule-associated protein 1 light chain 3 beta) on autophagosome membranes, ultimately resulting in autophagosomal engulfment of poly-ubiquitinated mitochondria for degradation [23,24].
Recent studies have demonstrated that mitophagy in renal tubular cells represents a beneficial response to AKI. For instances, PINK1-PARK2-mediated mitophagy has been shown to play an important role in mitochondrial quality control, tubular cell survival, and renal function during AKI that is induced by ischemia-reperfusion or cisplatin nephrotoxicity [14,[25], [26], [27]]. Mitophagy receptor BNIP3-mediated mitophagy has also been implicated in AKI [14,[25], [26], [27]]. Emerging evidence also suggests a role of mitophagy in the pathogenesis of septic AKI [28]. Despite these studies, the role and regulation of mitophagy in septic AKI remain largely unclear. In the present study, we have shown the activation of the PINK1-PARK2 pathway of mitophagy in septic AKI, demonstrated the protective role of this pathway by using gene knockout models, and suggested the involvement of optineurin as the adaptor protein for mitophagy.
2. Materials and methods
2.1. Antibodies and special reagents
The antibodies used in this study were as follows: anti-Parkin (4211S), anti-TOM20 (42406S), anti-LC3 (12741S), anti-Cleaved caspase-3 (9664S), anti-GAPDH (5174S) and anti-COX IV (4850P) from Cell Signaling Technologies; anti-TIM23 (11123-1-AP), anti-OPTN (10837-1-AP), and anti-p62 (18420-1-AP) from Proteintech; and anti-PINK1 (P0076) from Sigma-Aldrich. All secondary antibodies were purchased from Thermo Fisher Scientific. Lipopolysaccharide (LPS) was purchased from Sigma Aldrich.
2.2. Cell culture and LPS treatment of RPTCs
Immortalized RPTC line was initially from Dr. Ulrich Hopfer (Case Western Reserve University, Cleveland, OH) [29]. The cells were cultured in DMEM/F12 medium with 10% fetal bovine serum (FBS). For LPS treatment, RPTCs at a confluence of 50–60% were incubated in DMEM/F12 medium with 0.2% FBS for 24 h. The cells were then treated with DMEM/F12 medium plus 100 μg/ml LPS for 0, 12, 24, 36 h.
2.3. Plasmids, short interfering RNA, and transfection
The COX8-EGFP-mCherry plasmid was obtained from Addgene (78520; deposited by Dr. David Chan). The short interfering RNA (siRNA) oligonucleotides were synthesized from GenePharma (China). The sequences of Pink1 siRNA, Park2 siRNA and Optn siRNA oligonucleotides were as follows:
Pink1 siRNA, 5′-GCCTTGGGTTCAGCAAACA;
Park2 siRNA, 5′-GGAACAACAGAGTATCGTT;
Optn siRNA, 5′-GCATGGCCTCGGAAATCAA;
Transient transfections of RPTCs with plasmid DNA or siRNA were conducted with Lipofectamine 2000 reagents (Thermo Fisher Scientific, 11668019).
2.4. Animals
8–10-week old male C57/BL6 mice were purchased from SJA Laboratory Animal Corporation (Changsha, Hunan, China). Pink1 KO mice and Park2 KO mice were previously described [30,31]. All mice had free access to water and food in a pathogen-free setting under 12 h light and 12 h dark daily. All animal experiments were performed according to a protocol approved by the Institutional Animal Ethnics Committee of the Second Xiangya Hospital of Central South University.
2.5. Mouse model of septic AKI
Septic AKI was induced by lipopolysaccharides (LPS) treatment or by cecal ligation and puncture (CLP). LPS was administrated by intraperitoneal injection at the dosage of 10 mg/kg body weight, and control mice were injected with 0.9% saline. CLP-induced AKI in mouse was generated as previously described [32]. Briefly, laparotomy was conducted under anesthesia with pentobarbital (60 mg/kg, intraperitoneally) in mice. The cecum was ligated at 1 cm and colon with a 4-0 silk ligature and punctured in two sites with a needle. After removing the needle, a small number of feces was squeezed from the mesenteric and antimesenteric penetration holes to insure patency. Control mice performed the same operation without ligation and perforation. All mice were resuscitated with 40 ml/kg of saline after surgery. The mice were sacrificed at 24 h after LPS injection or CLP, and kidney tissues and blood were harvested for further analysis.
2.6. Renal function analysis
Renal function was indicated by serum creatinine and BUN using commercial kits purchased from BioAssay Systems as previously described [33]. In brief, blood samples were coagulated and then centrifugation at room temperature to gather serum. For serum creatinine measurement, serum samples were added to a pre-warmed reaction solution. The absorbance of 510 nm was monitored at 0 and 5 min of reaction. For BUN measurement, samples were added to reaction mixture at RT for 20 min, and the absorbance at 520 nm was recorded. Serum creatinine and BUN levels were then calculated based on standard curves, and shown as mg/dl.
2.7. Renal histology
For routine analysis, kidney tissues were fixed with 4% paraformaldehyde for paraffin embedding, and kidney section of 4 μM for hematoxylin-eosin (HE) staining. Tissue damage was scored in a blinded manner by the percentage of injured renal tubules and histological injury that was indicated by brush border lost, tubular dilation/flattening, tubular degeneration, tubular cast formation, and vacuolization.
2.8. Apoptosis analysis
Apoptosis were examined in cultured proximal tubular cells by TUNEL assay by using an In Situ Cell Death Detection Kit from Roche Applied Science (Indianapolis, IN) as described previously [34,35]. For quantification, 4 fields with 200 cells per field were determined for TUNEL-positive cells in each experimental condition to evaluate the percentage of apoptosis. Besides, apoptosis was evaluated using flow cytometry. Cell preparation was conducted according to FITC Annexin V Apoptosis Detection Kit I (BD, No.556547). Briefly, cells were washed twice with cold PBS and then resuspended in 1X Binding Buffer. 5 μl of FITC Annexin V and 5 μl PI were added to a 100 μl cell suspension. The cells were then subjected to gentle vortexing and then incubation for 15 min at RT in the dark. Finally, 400 μl of 1X Binding Buffer were added and the cells were analyze by flow cytometry within 1 h. In addition, apoptosis was also assessed by immunoblot analysis of cleaved caspase-3 (C-CAS).
In kidney tissues, apoptosis was also examined by TUNEL assay as described previously. In short, kidney tissue sections were deparaffinized. Sections were then pretreated with 0.1 M sodium citrate, pH 6.0 at 65 °C for 30 min, followed by incubation with a TUNEL reaction solution for 1 h at 37 °C in a humidified-dark chamber. Positive staining was detected by fluorescence microscopy. Ten representative fields were randomly chosen for each section, and the TUNEL-positive cells per mm2 was counted. In addition, apoptosis in kidney tissues was also evaluated by immunoblot analysis of C-CAS.
2.9. Quantification of mitochondrial DNA content
Total cellular DNA was extracted from renal cortex tissue by using the DNeasy Blood and Tissue kit (Qiagen, 69506). The relative content of mtDNA was measured as previously described [36]. In briefly, both mtDNA (evaluated by mitochondrial 16S rRNA gene) and nDNA (evaluated by β2-microglobulin gene) in the same experimental samples were measured by quantitative PCR using a CFX96 Real-Time PCR Detection System from BiO-RAD with the SyBR GreenER qPCR SuperMix (Thermo Fisher Scientific). The primers were as follows:
mt16S forward, 5′-ATTCCAATTCTCCAGGCATACG-3’;
mt16S reverse, 5′-GGGGTTCTTGTTTGCCGAGTT-3’;
β2-microglobulin forward, 5′-AGGGTGTGCAGAATGGGATG-3’;
β2-microglobulin reverse, 5′-GCTTCCCCCAAAGTCTACCC-3’;
GAPDH forward, 5′-AGGTCGGTGAACGGATTG-3’;
GAPDH reverse, 5′-TGTAGACCATGTAGTTGAGGTCA-3’.
2.10. Mitochondrial isolation
Mitochondria were extracted from cultured proximal tubular cells using the mitochondrial extraction kit (Solarbio). In briefly, cells were rinsed in pre-cold PBS and homogenized in 1 ml ice-cold lysis buffer with a pre-cold Dounce-type glass homogenizer. Then the homogenate was centrifuged for 3 times (1000×g for 10 min, 4 °C) to pellet cell debris and nuclei, and collect the supernatant with mitochondria. Finally, mitochondria from the supernatant were pelleted by centrifugation at 12,000×g for 10 min at 4 °C.
2.11. Co-localization of OPTN with mitochondria
Cells were washed with phosphate-buffered saline (PBS). Mitochondria in these cells were labeled with MitoTracker Red FM (Molecular Probes) according to the manufacturer's instructions. The cells were then fixed in pre-cold methanol for 10 min at −20 °C. After additional washing with PBS, the fixed cells were blocked in 10% normal goat serum for 1 h. Cells were then incubated with anti-OPTN antibody overnight at 4 °C, followed by incubation with Alexa Fluor 488-labeled secondary antibodies for 1 h at 37 °C. Then, the cells were stained with DAPI to indicate nuclei. Finally, the cells were examined with an Olympus FV 1000 lasers canning confocal microscope (Olympus Corporation, Tokyo, Japan).
2.12. Transmission electron microscopy
The kidney cortex tissues were fixed with 2.5% glutaraldehyde, and then treated with standard procedures, including dehydration, osmosis, embedding, sectioning and staining. The ultrastructure of renal cells was observed using a Hitachi H7700 electron microscope.
2.13. Immunoblot analysis
Renal cortex tissue and cell lysates were lysed with 2% sodium dodecyl sulfate (SDS) solution with protease inhibitor cocktail (Sigma-Aldrich). Protein concentration was measured with the bicinchoninic acid (BCA) reagent (Thermo Fisher Scientific). 100 μg proteins were loaded per lane and separated on SDS-polyacrylamide electrophoresis gel, and the proteins were then transferred onto polyvinylidene difluoride membrane. After blocking with 5% fat-free milk for 1 h, blots were incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Antigens were visualized with a chemiluminescence kit (Thermo Fisher Scientific). The protein bands were analyzed by Image J software (NIH) for densitometry.
2.14 Statistical analysisIn this study, qualitative data represents at least 3 separate experiments, and quantitative data is expressed as means ± SD. T-test was conducted to examine the statistical significance between 2 groups. ANOVA followed by Tukey's post-tests was used to determine the statistical differences among multiple groups. Statistical analysis was determined with the GraphPad Prism software. P < 0.05 was regarded as significant differences.
3. Results
3.1. Mitophagy is induced in RPTC cells following LPS treatment
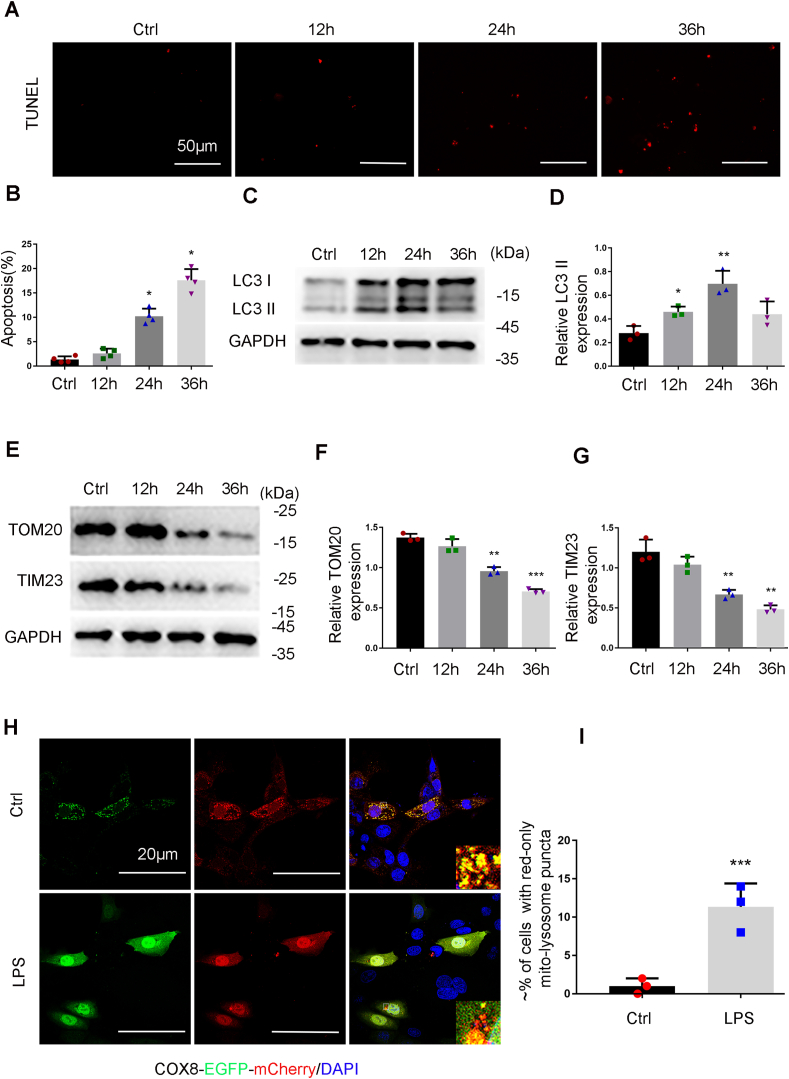
We firstly examined the occurrence of mitophagy in renal proximal tubular cells (RPTCs) exposed to LPS, the main endotoxin in sepsis. To this end, RPTCs were cultured in medium containing 0.2% fetal bovine serum for 24 h before the treatment with 100 μg/ml LPS. TUNEL staining showed an increase of TUNEL-positive cells following LPS treatment, suggesting the occurrence of apoptosis (Fig. 1A and B). Immunoblot analysis showed that following LPS treatment, the levels of LC3 II was increased and peaked at 24 h (Fig. 1C and D), whereas the levels of mitochondrial outer membrane protein TOM20 (translocase of outer mitochondrial membrane 20 homolog) and TIM23 (translocase of inner mitochondrial membrane 23) (Fig. 1E–G) were reduced, suggesting that mitophagy was induced. To further verify the induction of mitophagy, COX8-EGFP-mCherry (a tandem fluorescent-tagged mitochondrial targeting sequence of inner membrane protein COX8) were expressed in RPTCs to monitor the delivery of mitochondria to lysosome based on different pH stability of EGFP and mCherry fluorescent proteins [37]. As shown in Fig. 1H, control cells displayed a yellow staining of tubular or filamentous mitochondria with the merge of green (EGFP) and red (mCherry) signals. In sharp contrast, mitochondria became fragmented with distinct red puncta in LPS-treated cells (Fig. 1H and I), indicating increased delivery of mitochondria into lysosomes. Notably, the distinct red puncta were rarely observed in the cells with typical apoptotic nucleus (Fig.S1). Together, these findings suggest that mitophagy is induced in renal tubular cells in response to LPS treatment.
Fig. 1.
Mitophagy is induced in RPTC cells during LPS treatment. (A-E). RPTC cells were treated with 100 μg/ml LPS for 0 (Ctrl-control) to 36 h. (A) Representative images of TUNEL staining. (B) Apoptosis. The percentage of cells with typical apoptotic morphology was evaluated. (C) Immunoblots of LC3II/I and GAPDH (internal loading control). (D) Densitometry analysis of LC3II. (E) Immunoblots of TIM23 and TOM20. (F, G) Densitometry analysis of TOM20 and TIM23. (H) Mitophagy in RPTC cells transfected with COX8-EGFP-mCherry. After transfection, the cells were subjected to LPS treatment for 24 h. The red puncta in the bottom panel represent mitochondria in lysosomes with acidic pH. (I) Quantification of the cells with red-only puncta. Data in B, F, G and I are expressed as mean ± SD. n = 3. *P < 0.05, **P < 0.01, ***P < 0.001, significantly different from the control group without LPS exposure. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Knockdown of Pink1 or Park2 inhibits LPS-induced mitophagy in RPTC cells
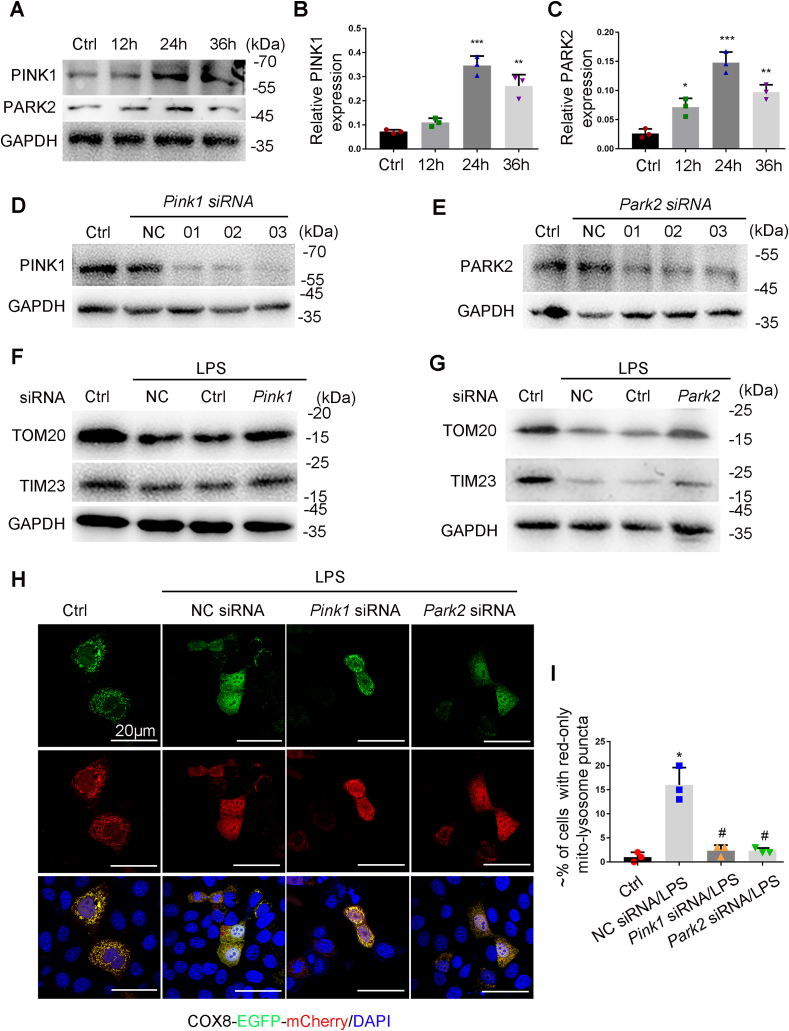
During LPS treatment of RPTCs, PINK1 and PARK2 were increased in a time-dependent manner (Fig. 2A–C). To examine the PINK1-PARK2 pathway in LPS-induced mitophagy, we determined the effect of knock-down of Pink1 or Park2 with specific siRNAs (Fig. 2D and E). As shown in Fig. 2F and G, knockdown of Pink1 or Park2 partially restored TOM20 and TIM23 in RPTCs during LPS treatment (Fig. 2F and G), supporting an important role of PINK1 and PARK2 in mitochondrial degradation in this setting. We further examined the effect of knockdown Pink1 or Park2 on the delivery of mitochondria to lysosomes in COX8-EGFP-mCherry expressing cells. Compared with RPTCs transfected with scrambled siRNA, the cells transfected with either Pink1 siRNA or Park2 siRNA showed a reduction in the levels of mitochondria in lysosomes (Fig. 2H and I). In addition, in response to LPS treatment, mitochondrial translocation of Park2 was dramatically increased (Fig. S2). Together, these results indicate that the PINK1-PARK2 pathway mediates mitophagy in renal tubular cells during LPS treatment.
Fig. 2.
Knockdown of Pink1 or Park2 suppresses LPS-induced mitophagy in RPTC cells. (A-C) RPTC cells were subjected to 100 μg/ml LPS treatment for indicated time to collect whole cell lysate for immunoblot analysis of PINK1, PARK2, and GAPDH. (A) Representative blots. (B, C) Densitometry analysis of PINK1 and PARK2. (D, E) Silencing of Pink1 or Park2 expression by siRNA in RPTC cells. RPTC cells were transfected with negative control siRNA (NC siRNA), Pink1 siRNA and Park2 siRNA for 48 h to collect whole cell lysate for immunoblot analysis of PINK1, PARK2 and GAPDH. (F, G) Preservation of TOM20 and TIM23 by silencing of Pink1 and Park2 during LPS treatment of RPTC cells. RPTC cells were transfected with Pink1 siRNA and Park2 siRNA alone or NC siRNA. At 48 h after transfection, the cells were subjected to LPS treatment to collect whole cell lysate for immunoblot analysis of TOM2O, TIM23, and GAPDH. (H, I) Inhibition of LPS-induced mitophagosome formation by silencing Pink1 or Park2. RPTC cells were first transfected NC siRNA, Pink1 siRNA and Park2 siRNA, and 24 h later these cells were transfected with COX8-EGFP-mCherry. After 8 h, the cells were subjected to LPS treatment. Finally, the cells were stained with DAPI (blue) and fixed for confocal microscopy. (H) Representative images. (I) Quantification of the cells with red-only puncta. Data in I are expressed as mean ± SD. n = 3. *P < 0.001 vs. the control group, #P < 0.05 vs. NC siRNA-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Knockdown of Pink1 or Park2 sensitizes RPTC cells to LPS-induced apoptosis
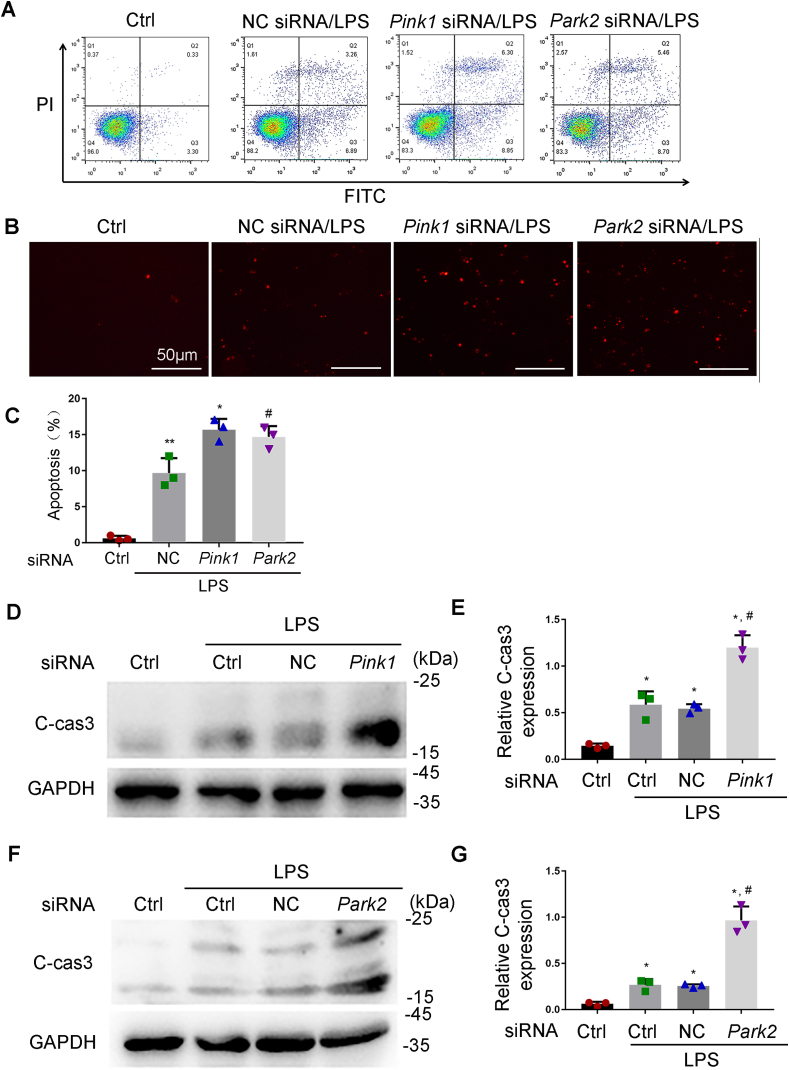
To determine the role of mitophagy, we evaluated the effect of knockdown of Pink1 and Park2 on LPS-induced apoptosis in RPTCs. Flow cytometry analysis of RPTCs stained with FITC Annexin V and PI showed that knockdown of Pink1 or Park2 resulted in an increase of apoptosis during LPS treatment (Fig. 3A). The flow cytometry results were confirmed by TUNEL staining and immunoblot analysis of cleaved caspase3 (C-CAS3). TUNEL assay demonstrated that following LPS treatment, Pink1 siRNA and Park2 siRNA markedly increased the number of TUNEL-positive cells compared to negative control siRNA (NC siRNA) group (Fig. 3B). Quantification by counting TUNEL-positive cells revealed that LPS induced apoptosis in around 10% cells transfected with NC siRNA, but in about 16% and 14.5% apoptosis in Pink1 siRNA or Park2 siRNA-transfected cells, respectively. Consistently, immunoblot analysis showed that Pink1 siRNA or Park2 siRNA-transfected cells had remarkably high levels of C-CAS3 than NC siRNA-transfected cells after LPS treatment (Fig. 3D–G). Collectively, these results suggest that PINK1/PARK2-mediated mitophagy has a cytoprotective role in LPS-treated renal tubular cells.
Fig. 3.
Knockdown of Pink1 and Park2 aggravates LPS-induced apoptosis in RPTC cells. RPTC cells were transfected with NC siRNA, Pink1 siRNA or Park2 siRNA. At 48 h after transfection, cells were subjected to 24 h of 100 μg/ml LPS treatment. Apoptosis was assessed by flow cytometry following Annexin V-FITC and PI staining, TUNEL assay, and immunoblot analysis of caspase 3 activation. (A) Flow cytometry following Annexin V-FITC and PI staining. (B) Representative images of TUNEL staining. (C) Percentage of Apoptosis. Greater than 200 cells in each group were evaluated to determine the percentage of TUNEL positive cells. (D, F) Whole cell lysate was collected for immunoblot analysis of active/cleaved caspase-3 (C-cas3) and GAPDH. (E, G) Densitometry of active/cleaved caspase-3. Mean ± SD. n = 3. *P < 0.001, **P < 0.01 vs. the control group, #P < 0.01 vs. NC siRNA-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
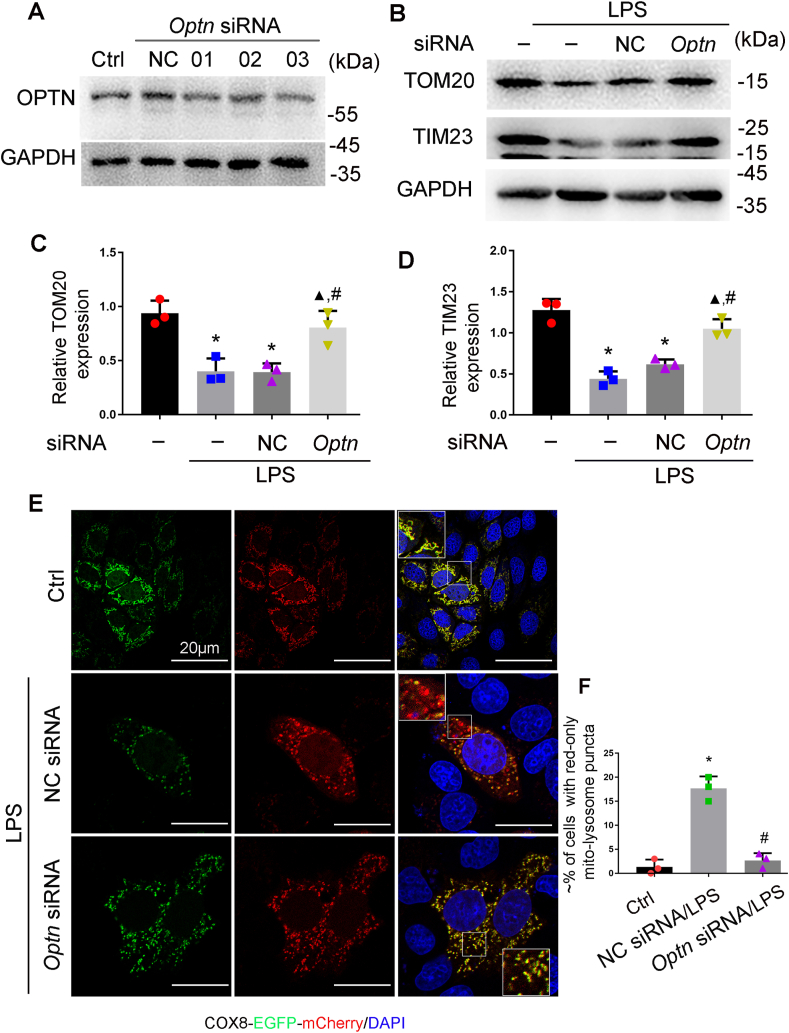
3.4. OPTN participates PINK1-PARK2-mediated mitophagy in RPTCs
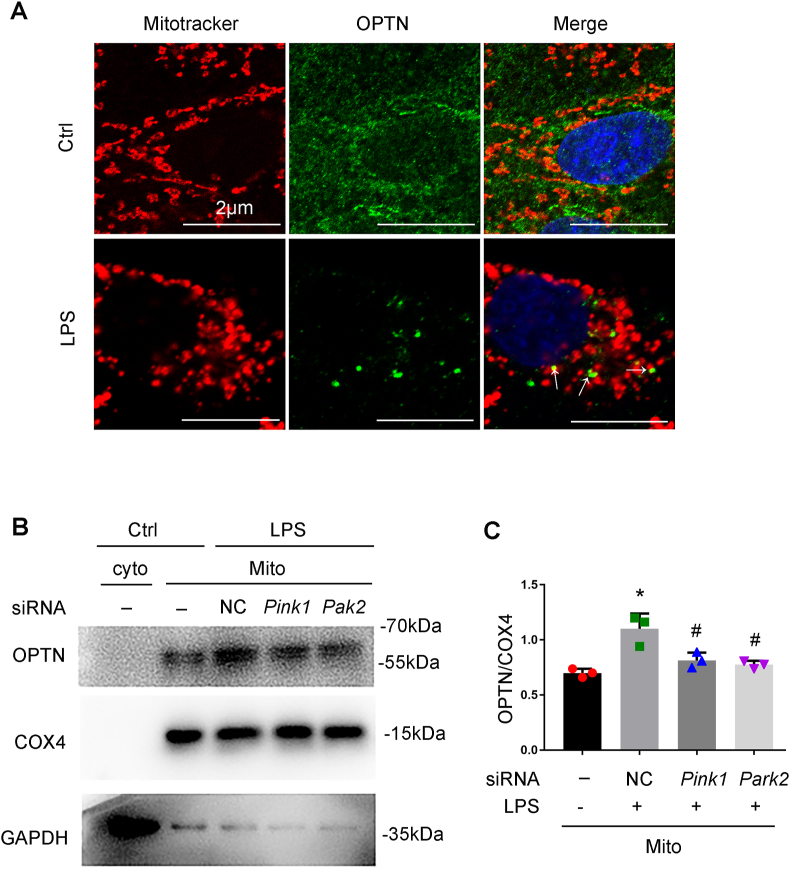
In the PINK1/PARK2 pathway of mitophagy, autophagy adaptor proteins simultaneously bind to poly-ubiquitinated proteins on mitochondria and LC3B on autophagosome membranes, resulting in autophagosomal engulfment of poly-ubiquitinated mitochondria for degradation. Optineurin (OPTN) is one of the selective autophagy adaptors in mitophagy that recognizes the ubiquitin chains on mitochondrial proteins [24]. We firstly determined the subcellular localization of OPTN and its changes during LPS treatment following the expression of GFP-OPTN. In control RPTCs, GFP-OPTN was evenly distributed in cytosol and rarely localized within mitochondria. In LPS-treated cells, GFP-OPTN puncta appeared on fragmented mitochondria, suggesting the translocation of OPTN from cytoplasm to damaged mitochondria under this condition (Fig. 4A). We further analyzed the effect of knockdown of Pink1 and Park2 on the mitochondrial translocation of OPTN. As shown in Fig. 4B and C, LPS increased the levels of OPTN in mitochondrial fraction of RPTCs, and the increase was significantly attenuated in cells transfected with Pink 1 siRNA or Park2 siRNA. In addition, knockdown Pink1 or Park2 dramatically reduced the levels of mitochondrial protein ubiquitination (Fig. S3). To further investigate the role of OPTN in mitophagy in RPTCs, we determined the effect of knockdown of Optn on LPS-induced mitophagy (Fig. 5A). As shown in Fig. 5B–D, LPS-induced reduction of TOM20 and TIM23 was partially prevented in the cells transfected with Optn siRNA. Consistently, suppression of Optn expression significantly inhibited LPS-induced delivery of mitochondria to lysosomes in RPTCs (Fig. 5E and F). Together, these findings support a pivotal role of OPTN in PINK1-PARK2-mediated mitophagy in RPTC cells during LPS treatment.
Fig. 4.
Knockdown of Pink1 or Park2 inhibits OPTN recruitment to mitochondria during LPS treatment. (A) Co-localization of OPTN with mitochondria upon LPS treatment. RPTC cells were subjected to 24 h of 100 μg/ml LPS treatment. Mitochondria in these cells were then labeled with MitoTracker Red FM, while OPTN was stained by immunofluorescence. These cells were examined by confocal microscopy to show the co-localiztion of OPTN (green) and mitochondria (red) in LPS-treated cells (arrows). (B, C) Immunoblot analysis of OPTN accumulation in mitochondria and the effects of Pink1 or Park2 knockdown. RPTC cells transfected with siRNAs for Pink1 or Park2 or negative control siRNA (NC siRNA) were subjected to LPS treatment. These cells were harvested and fractionated to cytosolic (cyto) and mitochondrial (mito) fractions for immunoblot analysis of OPTN, COX4(mitochondrial marker), and GAPDH (cytosolic marker). (B) Representative blots. (C) Densitometry of OPTN. Mean ± SD. n = 3. *P < 0.001 vs. control group. #P < 0.01 vs. siRNA-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
OPTN knockdown inhibits mitophagy in LPS-treated RPTC cells. (A) Immunoblot analysis of Optn knockdown by siRNA. RPTC cells were transfected with Optn siRNA or negative control siRNA (NC) for 48 h to collect lysate for immunoblot analysis. (B–D) RPTC cells transfected with Optn siRNA or negative control siRNA (NC) were subjected to LPS treatment. (B) Representative blots of TOM20 and TIM23. (C–D) Densitometry of TOM20 and TIM23. (E–F) RPTC cells were first transfected Optn siRNA or negative control siRNA (NC), and 24 h later these cells were transfected with COX8-EGFP-mCherry. After another 8 h, the cells were subjected to LPS treatment. Finally, the cells were stained with DAPI and fixed for confocal microscopy. (E) Representative images. (F) Quantification of the cells with red-only puncta-mitolysosomes. Mean ± SD. n = 3. *P < 0.001 vs. control group, ▲P < 0.01 VS. LPS group, #P < 0.01 vs. NC siRNA-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
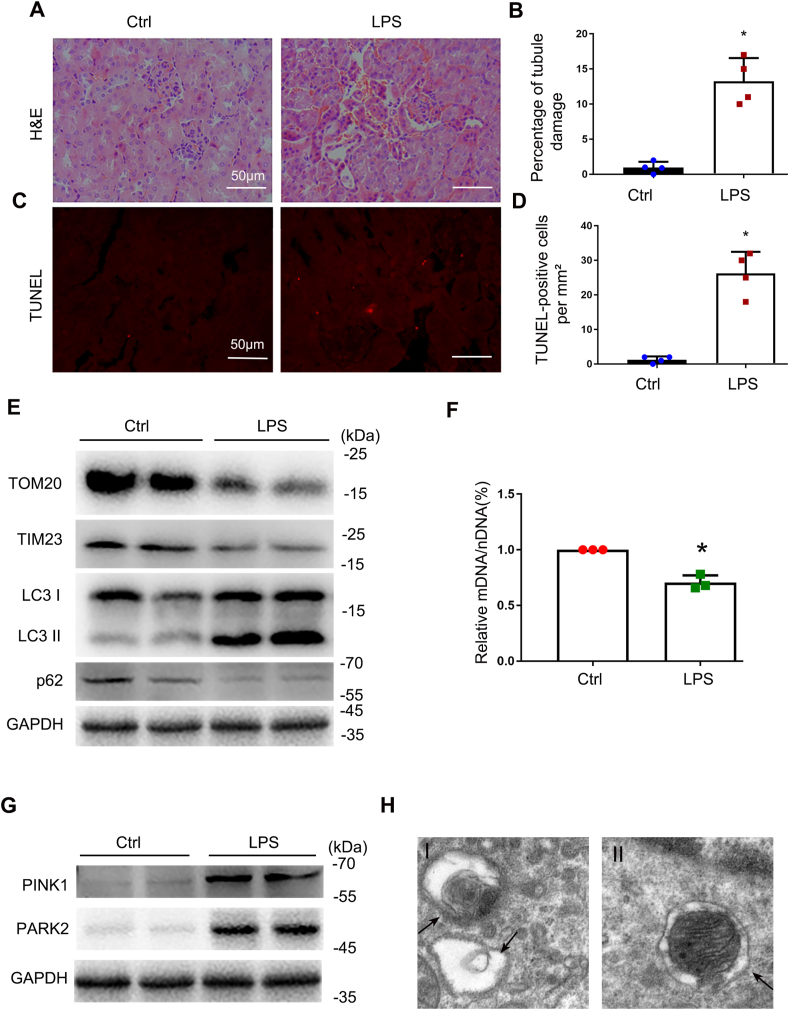
3.5. Induction of mitophagy in renal proximal tubular cells in the mouse model of LPS-induced AKI
To study mitophagy in vivo in septic AKI, we first evaluated the mouse model induced by LPS. To this end, C57/B6 mice were intraperitoneally injected with 10 mg/kg LPS or saline as previously describe [5]. Histological analysis by H&E staining showed a moderate tubule injury in around 12.5% renal tubules in LPS-treated mice (Fig. 6A and B). TUNEL assay showed renal tubular cell apoptosis in kidney tissues of these mice (Fig. 6C and D). Immunoblot analysis demonstrated an increase in LC3II expression and a reduction of p62, along with decreases in TOM20 and TIM23 in kidney tissues of LPS-treated mice compared with untreated control mice (Fig. 6E), suggesting induction of mitophagy in LPS-induced AKI. The ratio of mitochondrial DNA to nuclear DNA (mtDNA: nDNA) decreased in renal cortex tissues of LPS-injected mice compared to control mice (Fig. 6F), further indicating the degradation of mitochondria during LPS treatment. PINK1 and PARK2 were increased in renal tissues of LPS-treated mice (Fig. 6G). Transmission electron microscopy (TEM) showed the formation of autophagosome and mitophagosome in renal tubular cells in mice after LPS treatment (Fig. 6H), which was rarely seen in renal tissues of saline-injected mice. Together, these findings indicate that mitophagy is activated in renal tissues during LPS-induced septic AKI in vivo.
Fig. 6.
Mitophagy is activated in LPS-induced AKI in C57/BL6 mice. C57BL/6 mice (male, 8–10 weeks old) were injected with 10 mg/kg LPS or saline for 24 h. (A) Representative histology of kidney cortex shown by hematoxylin and eosin (H&E) staining. (B) Tubular injury evaluated by counting the renal tubules with signs of injury. (C) Representative images of TUNEL staining of kidney cortex. (D) Quantification of TUNEL positive cells. (E) Immunoblot analysis of TOM20, TIM23, LC3 II/I, p62 and GAPDH (loading control) in kidney tissues. (F) Relative mitochondrial DNA content (mtDNA/nDNA). (G) Immunoblot analysis of PINK1 and PARK2. (H) Representative electron micrographs showing autophagosomes (arrowhead in the left panel) and mitophagosomes (arrowhead in the right panel) in renal proximal tubule cells after LPS treatment. Mean ± SD. n ≥ 3. *P < 0.001 and #P < 0.01 vs. the control group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
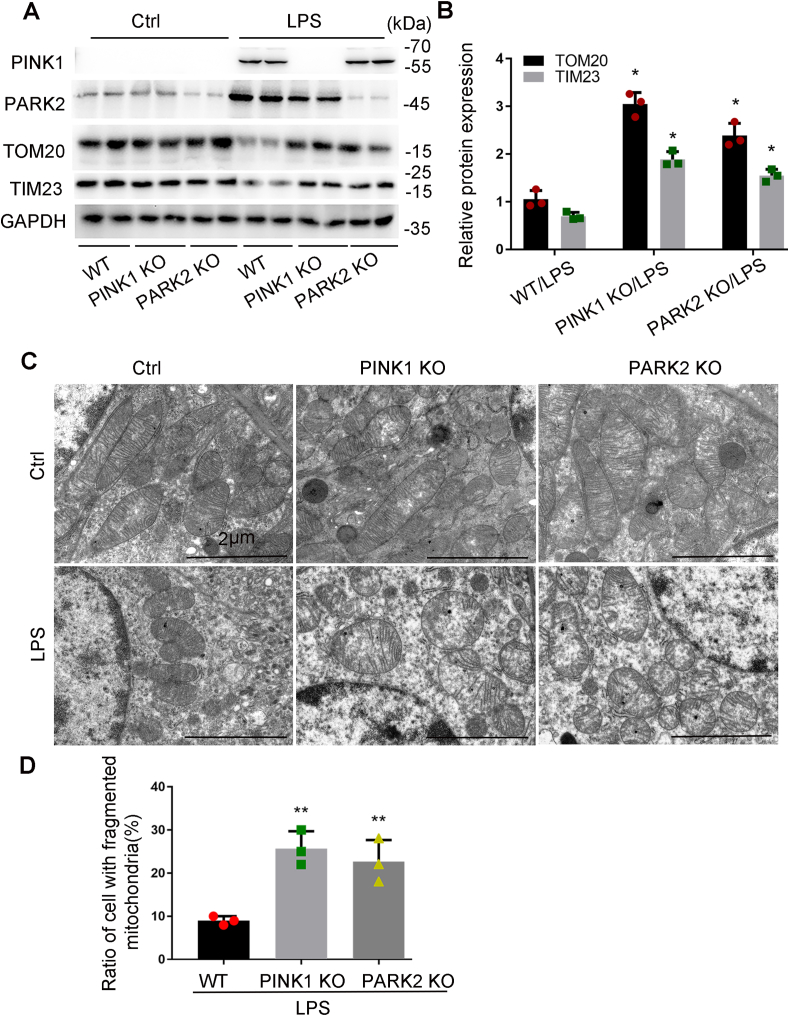
3.6. Pink1 or Park2 deficiency reduces mitophagy and aggravates LPS-induced mitochondrial injury in mice
To determine the role of PINK1/PARK2 pathway-mediated mitophagy in the pathogenesis of septic AKI in vivo, we tested pink1 knockout (KO) or park2 KO mouse models. As shown in Fig. 7A and B, LPS treatment reduced the mitochondrial proteins TOM20 and TIM23 in kidney tissues in wild-type (WT) mice, and this reduction was partially inhibited in pink1-or park2-KO mice, suggesting the involvement of PINK1 and PARK2 in mitophagy in septic AKI. We also examined mitochondrial morphology in kidney tubular cells in these mice by electron microscopy (Fig. 7C). Without treatment, KO and WT mice showed similar mitochondrial size and shape in kidney tubular cells with most mitochondria showing a tubular morphology of greater than 2 μm in length. After LPS treatment, kidney tubular cells in WT mice showed many small and round mitochondria, indicating mitochondrial fragmentation. In pink1-or park2-KO mice, mitochondria in kidney tubular cells were more severely injured showing loss of cristae, fragmentation, swelling, and vacuoles in the mitochondria matrix. Quantitative analysis verified that KO mice had significantly more kidney tubular cells with fragmented mitochondria than WT mice (Fig. 7D). Together, these results support a critical role of PINK1-PARK2-mediated mitophagy in preserving mitochondrial integrity during septic AKI.
Fig. 7.
Pink1 or Park2 deficiency inhibits mitophagy and aggravates LPS-induced mitochondrial injury in mice. Pink1 KO, park2 KO, and WT mice were injected with 10 mg/kg LPS or saline. (A) Kidney cortex tissues were collected from pink1-or park2- KO mice and WT littermates for immunoblot analysis of PINK1, PARK2 TOM20, TIM23, and GAPDH. (B) Densitometry of TOM20 and TIM23 signals. (C–D) Renal cortex was fixed and processed for transmission electron microscopy. (C) Representative electron micrographs. (D) Quantification of mitochondrial fragmentation. The length of mitochondria was evaluated in each tubular cell to calculate the percentage of cells in which mitochondrial length >2 μm was less than 1%. A total of 30 cells in control and 30 cells in LPS-treated kidneys from three animals in each condition were assessed. Mean ± SD. n = 3. *P < 0.01 and #P < 0.05 vs. WT-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
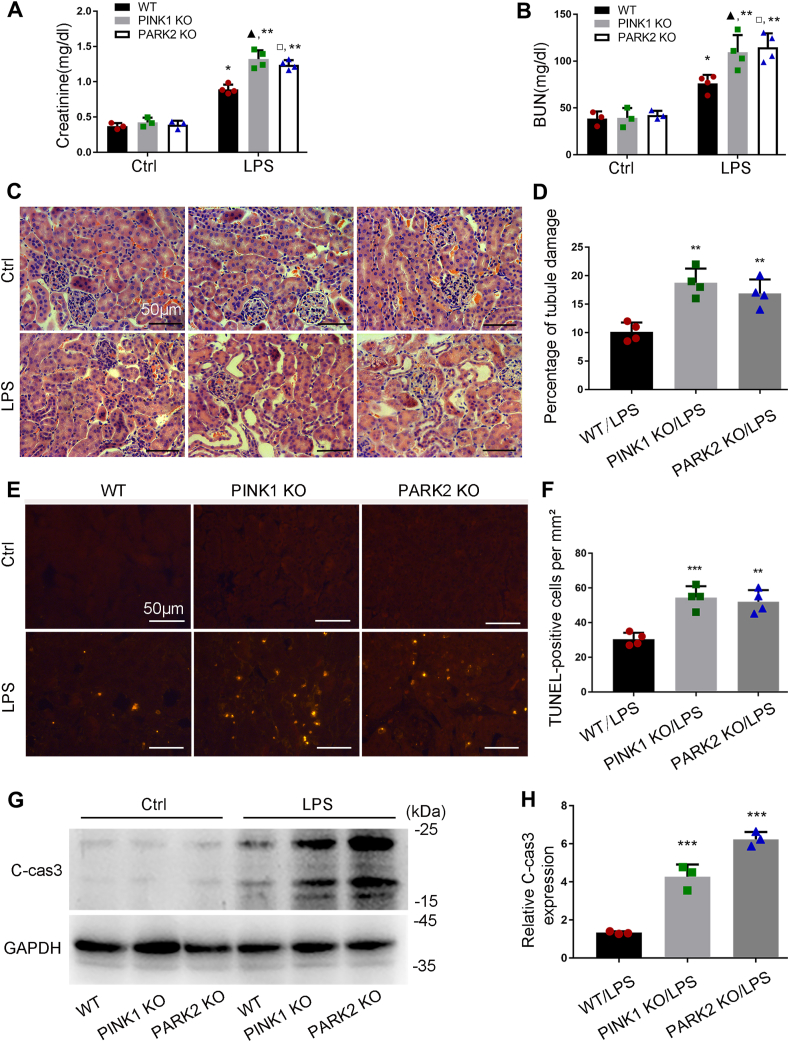
3.7. Pink1 or Park2 knockout aggravates kidney damage and tubular cell apoptosis during LPS treatment
Control Pink1 KO or Park2 KO mice and their WT littermates had similarly low levels of serum creatinine and blood urea nitrogen (BUN), indicating normal renal function in these mice (Fig. 8A and B). At 24 h after LPS treatment, both WT and KO mice showed increases in serum creatinine, but the increase in KO mice was significantly higher (Fig. 8A). Consistently, KO mice had higher levels of BUN than WT mice following LPS treatment (Fig. 8B). In HE staining of histology, pink1-and park2-KO mice showed more severe renal tubular injury than WT mice, as indicated by tubular swelling, vacuolization, dilatation and occasional detachment (Fig. 8C and D). We further examined renal tubular apoptosis by TUNEL staining and by immunoblot analysis of caspase-3 cleavage. TUNEL assay showed more apoptotic cells in kidney tissues of KO mice compared to WT mice after LPS injection (Fig. 8E and F). Consistently, LPS induced higher levels of cleaved caspase-3 in KO mouse kidneys than in WT (Fig. 8G and H). Together, these in vivo results support that the PINK1/PARK2 pathway of mitophagy plays a protective role in septic AKI.
Fig. 8.
Pink1 or Park2 deficiency increases kidney damage and tubular apoptosis during LPS treatment in mice.Pink1 KO, park2 KO, and WT mice were injected with 10 mg/kg LPS or saline. (A–B) Blood samples were harvested for measurement of serum creatinine (A) and BUN (B) to indicate renal function decline. (C) Representative images of HE staining. (D) Quantification of tubular damage by counting the renal tubules with signs of injury. (E) Representative images of TUNEL staining. (F) Quantification of TUNEL-positive cells in renal tissues. (G) Representative immunoblots of cleaved/active caspase-3 (C-CAS3) in kidney tissues. (H) Densitometry of C-CAS3 signals. Mean ± SD. n ≥ 3. #P < 0.001 vs. WT-Ctrl group, ▲P < 0.001 vs. PINK1 KO-Ctrl group, □P < 0.001 vs. PARK2 KO-Ctrl group, ***P < 0.001 and **P < 0.01 vs. WT-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.8. CLP-induced mitophagy is suppressed in Pink1-or Park2-KO mice
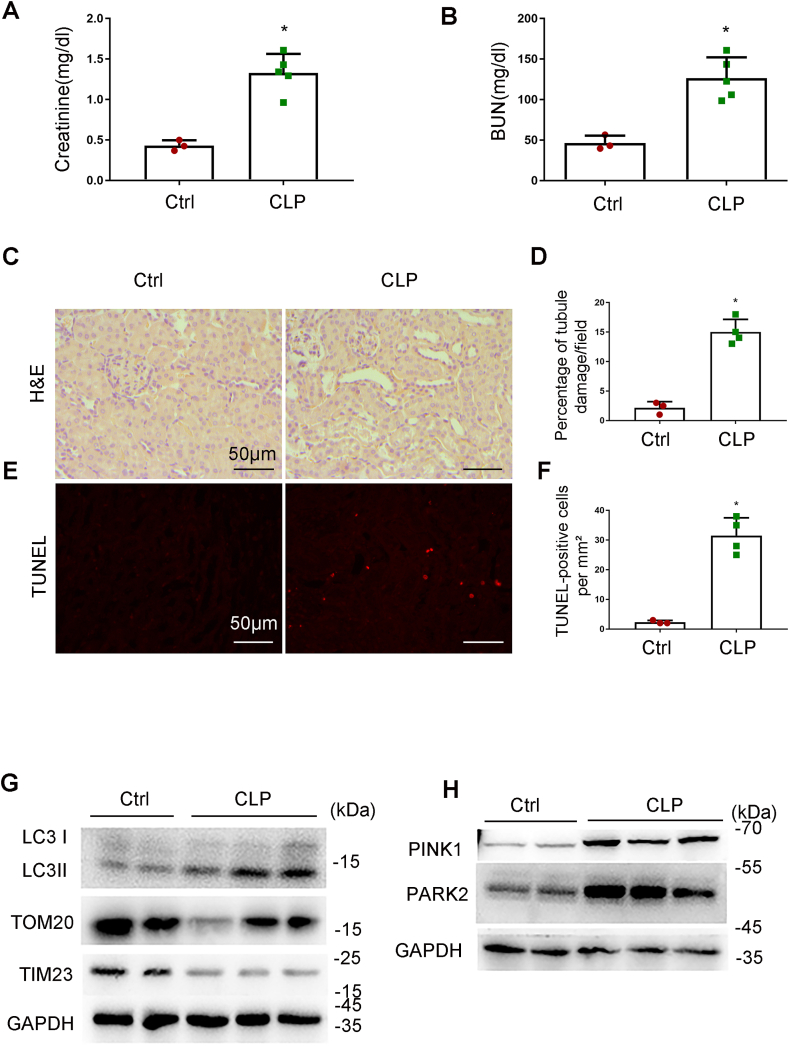
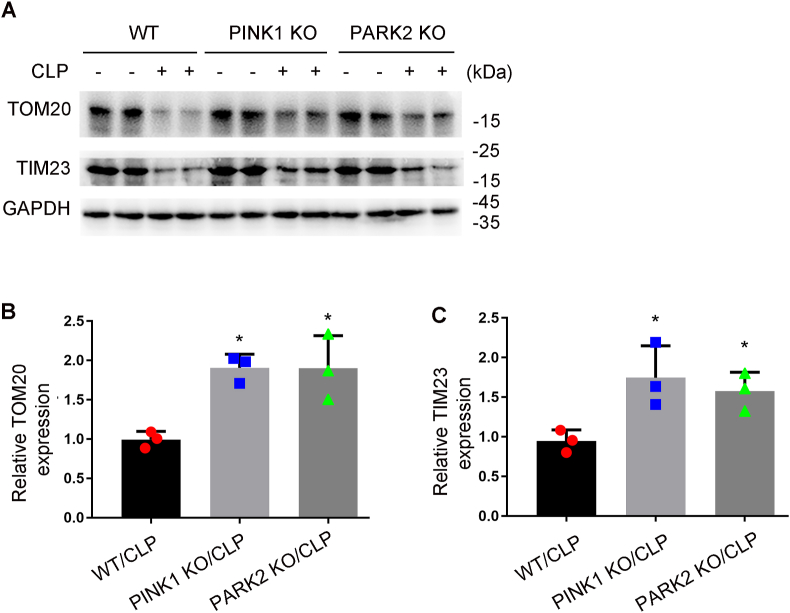
While endotoxic injury by LPS is commonly used to study septic AKI, the model of cecal ligation puncture (CLP) may better mimic the clinical condition [38]. Thus, we further examined mitophagy in a mouse model of CLP. C67/B6 mice were subjected to CLP as describe [32] and samples were collect 24 h later for analysis. Following CLP, the mice showed increases in both serum creatinine and BUN (Fig. 9A and B). Histological analysis and TUNEL staining also revealed renal tubular injury and apoptosis in CLP-treated mice (Fig. 9C–F). In addition, these mice showed increases in LC3II, PINK1 and PARK2 in kidney tissues, which were associated by decreases in TOM20 and TIM23, suggesting an induction of mitophagy (Fig. 9G and H). Notably, CLP-induced reduction of TOM20 and TIM23 in kidney tissues was partially inhibited by pink1-or park2 knockout (Fig. 10A and B). These findings suggest that the PINK1-PARK2 pathway has an important role in mitophagy in CLP-induced AKI.
Fig. 9.
Mitophagy is induced in CLP-induced AKI in mice. C57BL/6 mice (male, 8–10 weeks old) were subjected to CLP or sham-surgery for analysis 24 h later. Blood samples were collected for measurements of serum creatinine (A) and BUN (B). (C) Representative images of HE staining. (D) Percentage of damaged tubules. (E) Representative images of TUNEL staining, (F) Quantification of TUNEL positive cells. (G) Immunoblot analysis of TOM20, TIM23 and LC3 II/I. (H) Immunoblot analysis of PINK1 and PARK2. Mean ± SD. n ≥ 4. *P < 0.001 vs. the control group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 10.
CLP-induced mitophagy is inhibited in Pink1-or Park2-KO mice Pink1 KO, park2 KO, and WT mice were subjected to CLP or sham-surgery for analysis 24 h later. Kidney tissues were collected for immunoblot analysis of TOM20 and TIM23. (A) Representative blots. (B–C) Densitometry analysis. Mean ± SD. n = 3. *P < 0.01 vs. WT-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
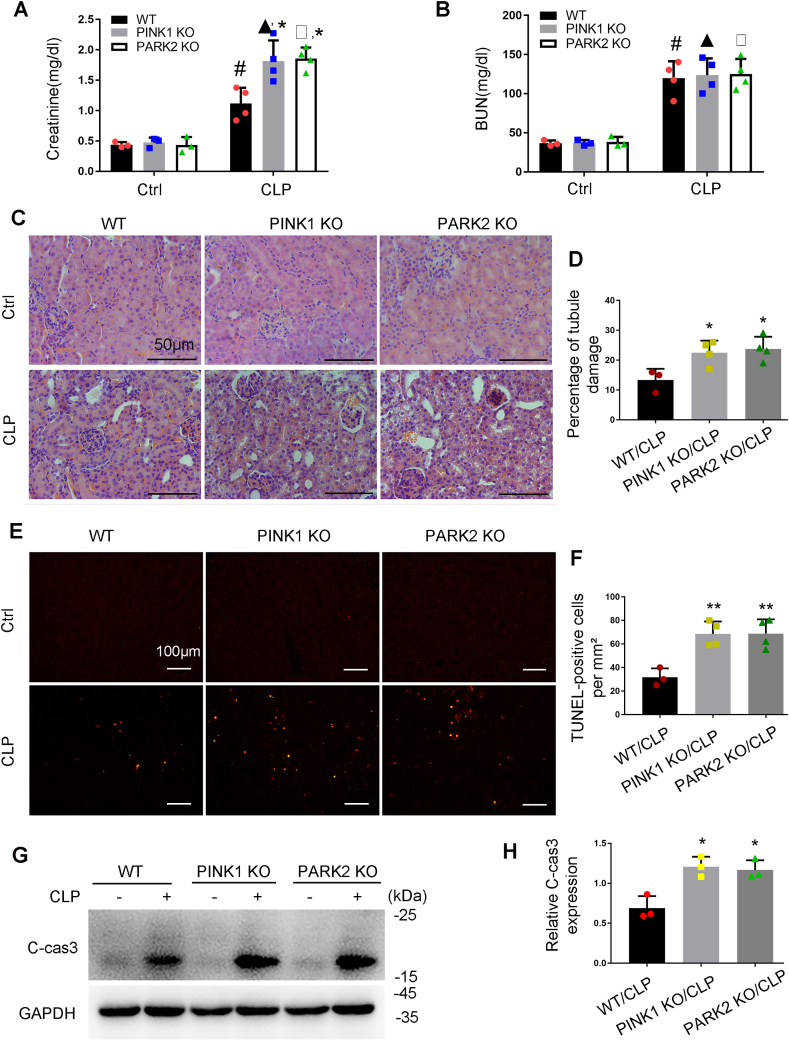
3.9. Pink1 or Park2 knockout aggravates CLP-induced AKI
Following CLP, Pink1 KO or Park2 KO mice showed a significantly higher increase in serum creatinine than WT mice, although they had similar levels of BUN (Fig. 11A and B). HE staining of histology revealed more severe kidney tissue damage in KO mice than in WT mice. The tubular damage score was 13.4% for WT, 22.5% for pink1-KO mice, and 23.5% for park2-KO mice (Fig. 11C and D). In TUNEL assay, CLP induced a higher level of apoptosis in kidney tissues of KO mice than in WT mice (Fig. 11E and F). Consistently, CLP-induced a higher level of cleaved caspase-3 in Pink1 or Park2 KO kidney tissues (Fig. 11G and H). These results support a protective role of PINK1/PARK2-mediated mitophagy in CLP-induced septic AKI.
Fig. 11.
Pink1 or Park2 deficiency worsens CLP-induced AKI in mice. Pink1 KO, park2 KO, and WT mice were subjected to CLP or sham-surgery for analysis 24 h later. Blood samples were harvested for measurements of serum creatinine (A) and BUN (B). (C) Representative images of HE staining. (D) Percentage of damaged tubules. (E) Representative images of TUNEL staining. (F) Quantification of TUNEL-positive cells. (G) Representative immunoblots of cleaved/active caspase-3 (C-CAS3). (H) Densitometry of cleaved/active caspase-3. Mean ± SD. n ≥ 3. #P < 0.001 vs. WT-Ctrl group, ▲P < 0.001 vs. Pink1 KO-Ctrl group, □P < 0.001 vs. Park2 KO-Ctrl group, *P < 0.01 vs. WT-LPS group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Mitochondrial dysfunction has been implicated in the pathogenesis of septic AKI [8], but the mechanism of mitochondrial quality control in this devastating disease remains largely unknown. In the present study, we have demonstrated that mitophagy was induced in cultured RPTCs treated with LPS, and in renal tubules in mouse models of septic AKI induced by LPS and CLP. Mitophagy induction in these experimental settings was suppressed by Pink1 or Park2 deficiency. Functionally, Pink1 or Park2 deficiency aggravated LPS- or CLP-induced AKI in mice. We further show that knockdown of Pink1 or Park2 reduced the translocation of autophagic adaptor OPTN to damaged mitochondria in cultured tubular cells exposed to LPS, and silencing Optn inhibited mitophagy following LPS treatment. These findings support an important role for the PINK1/PARK2 pathway of mitophagy in mitochondrial quality control, tubular cell survival, and renal function during septic AKI.
Recent studies demonstrate that mitochondrial injury contributes to the pathophysiology of sepsis-induced AKI [[39], [40], [41]]. Especially, specific interventions to increase mitochondrial biogenesis or mitochondrial oxidative capacity ameliorated septic-AKI [40]. Tran and colleagues demonstrated that loss of PGC1α, the master regulator of mitochondrial biogenesis, facilitated sepsis AKI and delayed renal recovery [40], suggesting a critical role of loss of mitochondrial homeostasis in the pathogenesis of septic AKI. Mitochondrial biogenesis and mitophagy are two important mitochondrial quality control mechanisms. Mitochondrial biogenesis is essential for replacing the damaged mitochondria removed by mitophagy and also repopulating regenerating renal tubular cells, while timely removal of damaged mitochondrial by mitophagy may prevent the replication of damaged mitochondria. Mitophagy is an important mitochondrial quality control mechanism that selectively eliminates damaged mitochondria. A very recent study by Liu et al. demonstrated the co-localization of mitochondrial cytochrome c oxidase IV (COX IV) and autophagy marker LC3 in CLP-induced AKI, suggesting mitophagy [41]. Dai et al. further suggested an increase in the formation of mitophagosome in renal tubular cells at 4 h after CLP in mice by electronic microscopy [28]. In the present study, we have provided compelling evidence that mitophagy was induced in renal tubular cells during septic AKI, which includes 1) increases in mitophagosome formation and in mitochondrial protein degradation in RPTCs exposed to LPS, 2) increases in mitophagosome formation and in mitochondrial protein degradation, and reduction in mtDNA mass in the kidney of mouse model of AKI induced by LPS or CLP; and 3) mitophagosome formation and mitochondrial clearance in septic AKI are suppressed by blocking the PINK1-PARK2 pathway. Future studies by using the mitophagy reporter mouse model [14] will provide more information on the spatial and temporal changes of mitophagy in the kidney during septic AKI. Notably, induction of PINK1-PARK2 pathway of mitophagy has also been demonstrated as a protective mechanism in other tissues during sepsis, including heart, lung, and liver [[42], [43], [44]],suggesting that mitophagy may represent a common defense mechanism in different tissues during sepsis [45]. In the present study, since Pink1 or park2 was globally deleted in mice, the potential effects of the loss of PINK1-PARK2 pathway of mitophagy in those tissues beyond kidney on sepsis-induced AKI awaits future investigation.
The PINK1-PARK2 pathway of mitophagy has been implicated in ischemic and nephrotoxic AKI [25,26,46]. Mitophagy receptor BNIP3-mediated mitophagy has also been shown in renal tubular cells of ischemic AKI [47]. Liu et al. showed that PINK1 and PARK2 expression was increased within 4 h after CLP, which was in parallel to the changes in the colocalization of mitochondrial protein and LC3 in renal tubular cell in the AKI models, suggesting a potential role of PINK1-PARK2 pathway in the regulation of mitophagy in this experimental setting. Dai and colleagues further demonstrated that Pink1 knockdown significantly attenuated LC3-II protein expression in during LPS treatment of RPTC cells, whereas PINK1 overexpression enhanced LC3-II protein expression [28]. Despite these initial studies, it remains unclear whether the PINK1-PARK2 pathway is critical to mitophagy, mitochondrial homeostasis, tubular cell survival and kidney function in septic AKI. In the present study, we provide both in vitro and in vivo evidence that the expression of PINK1 and PARK2 was increased in renal tubular cells during septic AKI, and moreover, Pink1 or Park2 deficiency markedly reduced mitophagy and exaggerated tubular damage and septic AKI, proving an important role of the PINK1-PARK2 pathway in mitophagy and tubular cell viability in septic AKI. These observations, the together previous studies, indicate that induction of the PINK1-PARK2 pathway of mitophagy in renal tubular cells represents a common feature in AKI. In this pathway, PINK1 acts upstream of PARK2 by recruiting PARK2 to damaged/depolarized mitochondria and regulating PARK2's ubiquitin E3 ligase activity through direct phosphorylation of PARK2 and ubiquitin. The PARK2-mediated ubiquitination of mitochondrial surface proteins leads to the recruitment of autophagy adaptor proteins such as TAX1BP1, NBR1, p62, NDP52, and OPTN [48], which simultaneously bind poly-ubiquitin chains and LC3, and thus results in the engulfment of target mitochondria within autophagosomes. In the present study, we show that inhibition of PINK1-PARK2 pathway in RPTCs exposed to LPS reduced mitochondrial translocation of OPTN to mitochondria, and moreover, knockdown Optn reduced mitophagy, suggesting that OPTN is an important adaptor in the PINK1-PARK2 pathway of mitophagy in septic AKI. These results provide new insights into the molecular pathway of mitophagy in septic AKI. Of note, PARK2 was induced by LPS in PINK1 KO mice (Fig. 7) albeit at a lower level than in WT mice. This observation suggests that PARK2 expression may not depend on PINK1, but the pathway that regulates PARK2 expression during LPS treatment awaits further investigation.
Induction of mitophagy may act as a protective mechanism in ischemic and nephrotoxic AKI [[25], [26], [27],46]. Our present study has provided further evidence that the PINK1-PARK2 pathway of mitophagy is protective against sepsis-induced AKI. We demonstrated that in vitro, silencing Pink1 or Park2 sensitized RPTCs to LPS-induced apoptosis. In vivo, Pink1 or Park2 knockout aggravated LPS- or CLP-induced tubular cell apoptosis, kidney tissue damage, and kidney dysfunction. It is conceivable that, at the initial stage of kidney injury, induction of mitophagy removes damaged mitochondria, thereby preventing ROS accumulation, proapoptotic factor release and ultimately decreasing tubular cell injury and death. Elimination of damaged mitochondria may also prevent the release of damage-associated molecular patterns from mitochondria, such as mtDNA, thus mitigating the stimulation of immune response or inflammation that has been implicated in AKI pathogenesis [49]. However, as the disease progresses or under the context of severe kidney injury, when the number of damaged mitochondria exceeds the capacity of mitophagy or the mitophagy becomes impaired, cell death will become dominant to induce excessive tissue damage. Thus, pharmacological enhancement of mitophagy may represents a novel therapeutic approach for AKI. It is noteworthy that the PINK1-PARK2 pathway has also been implicated in the regulation of a mitophagy-independent quality control mechanism called mitochondria-derived vesicles (MDVs), which selectively transport cargo from mitochondria to lysosomes [50]. PINK1 and PARK2 may also function as repressors of adaptive immunity by suppress mitochondrial antigen presentation in mitophagy-independent mechanism [50]. The contribution of these mechanisms to septic AKI remains to be investigated. Moreover, Bhatia and colleagues recently demonstrated that the PINK1-PARK2 pathway of mitophagy has a role in the regulation of macrophage reprogramming [51]. Thus, it is important to determine the role of mitophagy in different cell types in the pathogenies of septic AKI by using cell-types specific mitophagy deficiency animal models. Moreover, Park2-or Pink1-independent mitophagy has also been reported in the pathogenesis of AKI [18]. For instance, Tang et al. demonstrated that activation of BNIP3-mediated mitophagy in renal tubular cells was protective against renal ischemia-reperfusion-induced AKI [47]. More recently, Wang et al. showed that Fundc1-dependent mitophagy was critically important to ischemic preconditioning-conferred renoprotection in ischemic AKI [52]. However, the involvement of these pathways of mitophagy in septic AKI awaits future investigation.
In summary, this study has provided substantial evidence that mitophagy is induced in renal tubular cells during septic AKI. Under this condition, mitophagy is mainly mediated by the PINK1-PARK2 pathway where OPTN is the major adaptor protein. Functionally, induction of this pathway of mitophagy in renal tubular cells is a renoprotective response in septic AKI. Therefore, mitophagy may represent a promising therapeutic target for the prevention and treatment of septic AKI, a devastating disease.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study was supported in part by the National Key R&D Program of China (2018YFC1312700) and National Natural Science Foundation of China (81720108008, 81870474).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101767.
Contributor Information
Chengyuan Tang, Email: tangchengyuan@csu.edu.cn.
Yu Liu, Email: rory@csu.edu.cn.
Zheng Dong, Email: zdong@augusta.edu.
Author's contribution
ZD, CT and YW designed this study; YW performed the experiments; all authors contributed to data analysis and discussion; ZD, CT and YW contributed to manuscript preparation.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Guo C., Dong G., Liang X., Dong Z. Epigenetic regulation in AKI and kidney repair: mechanisms and therapeutic implications. Nat. Rev. Nephrol. 2019;15(4):220–239. doi: 10.1038/s41581-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He L., Wei Q., Liu J., Yi M., Liu Y., Liu H., Sun L., Peng Y., Liu F., Venkatachalam M.A. AKI on CKD: heightened injury, suppressed repair, and the underlying mechanisms. Kidney Int. 2017;92(5):1071–1083. doi: 10.1016/j.kint.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucaloiu I.D., Kirchner H.L., Norfolk E.R., Hartle J.E., 2nd, Perkins R.M. Increased risk of death and de novo chronic kidney disease following reversible acute kidney injury. Kidney Int. 2012;81(5):477–485. doi: 10.1038/ki.2011.405. [DOI] [PubMed] [Google Scholar]
- 4.Venkatachalam M.A., Weinberg J.M., Kriz W., Bidani A.K. Failed tubule recovery, AKI-CKD transition, and kidney disease progression. J. Am. Soc. Nephrol. 2015;26(8):1765–1776. doi: 10.1681/ASN.2015010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mei S., Livingston M., Hao J., Li L., Mei C., Dong Z. Autophagy is activated to protect against endotoxic acute kidney injury. Sci. Rep. 2016;6:22171. doi: 10.1038/srep22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emlet D.R., Shaw A.D., Kellum J.A. Sepsis-associated AKI: epithelial cell dysfunction. Semin. Nephrol. 2015;35(1):85–95. doi: 10.1016/j.semnephrol.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Gomez H., Kellum J.A. Sepsis-induced acute kidney injury. Curr. Opin. Crit. Care. 2016;22(6):546–553. doi: 10.1097/MCC.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikh S.M., Yang Y., He L., Tang C., Zhan M., Dong Z. Mitochondrial function and disturbances in the septic kidney. Semin. Nephrol. 2015;35(1):108–119. doi: 10.1016/j.semnephrol.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szeto H.H. Pharmacologic approaches to improve mitochondrial function in AKI and CKD. J. Am. Soc. Nephrol. 2017;28(10):2856–2865. doi: 10.1681/ASN.2017030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Cai J., Tang C., Dong Z. Mitophagy in acute kidney injury and kidney repair. Cells. 2020;9(2) doi: 10.3390/cells9020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J., Zhang J., Tian J., Virzì G.M., Digvijay K., Cueto L., Yin Y., Rosner M.H., Ronco C. Mitochondria in sepsis-induced AKI. J. Am. Soc. Nephrol. 2019;30(7):1151–1161. doi: 10.1681/ASN.2018111126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhargava P., Schnellmann R.G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 2017;13(10):629–646. doi: 10.1038/nrneph.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang C., Cai J., Yin X., Weinberg J., Venkatachalam M.A., Dong Z. Mitochondrial quality control in kidney injury and repair. Nat. Rev. Nephrol. 2020 doi: 10.1038/s41581-020-00369-0. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livingston M.J., Wang J., Zhou J. Clearance of damaged mitochondria via mitophagy is important to the protective effect of ischemic preconditioning in kidneys. Autophagy. 2019;15(12):2142–2162. doi: 10.1080/15548627.2019.1615822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan M., Usman I.M., Sun L., Kanwar Y.S. Disruption of renal tubular mitochondrial quality control by Myo-inositol oxygenase in diabetic kidney disease. J. Am. Soc. Nephrol. 2015;26(6):1304–1321. doi: 10.1681/ASN.2014050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H., Li G., Liu L., Feng L., Wang X., Jin H. Regulation and function of mitophagy in development and cancer. Autophagy. 2013;9(11):1720–1736. doi: 10.4161/auto.26550. [DOI] [PubMed] [Google Scholar]
- 17.Zhan M., Brooks C., Liu F., Sun L., Dong Z. Mitochondrial dynamics: regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int. 2013;83(4):568–581. doi: 10.1038/ki.2012.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang C., Livingston M., Liu Z., Dong Z. Autophagy in kidney homeostasis and disease. Nat. Rev. Nephrol. 2020;16:489–508. doi: 10.1038/s41581-020-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang C., He L., Liu J., Dong Z. Mitophagy: basic mechanism and potential role in kidney diseases. Kidney Dis. 2015;1(1):71–79. doi: 10.1159/000381510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheibye-Knudsen M., Fang E.F., Croteau D.L., Wilson D.M., 3rd, Bohr V.A. Protecting the mitochondrial powerhouse. Trends Cell Biol. 2015;25(3):158–170. doi: 10.1016/j.tcb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang M., Linn B.S., Zhang Y., Ren J. Mitophagy and mitochondrial integrity in cardiac ischemia-reperfusion injury. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019;1865(9):2293–2302. doi: 10.1016/j.bbadis.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Riley B.E., Lougheed J.C., Callaway K., Velasquez M., Brecht E., Nguyen L., Shaler T., Walker D., Yang Y., Regnstrom K. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 24.Wong Y.C., Holzbaur E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U. S. A. 2014;111(42):E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang C., Han H., Yan M., Zhu S., Liu J., Liu Z., He L., Tan J., Liu Y., Liu H. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy. 2018;14(5):880–897. doi: 10.1080/15548627.2017.1405880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Tang C., Cai J., Chen G., Zhang D., Zhang Z., Dong Z. PINK1/Parkin-mediated mitophagy is activated in cisplatin nephrotoxicity to protect against kidney injury. Cell Death Dis. 2018;9(11):1113. doi: 10.1038/s41419-018-1152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao C., Chen Z., Xu X., An X., Duan S., Huang Z., Zhang C., Wu L., Zhang B., Zhang A. Pink1/Parkin-mediated mitophagy play a protective role in cisplatin induced renal tubular epithelial cells injury. Exp. Cell Res. 2017;350(2):390–397. doi: 10.1016/j.yexcr.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Dai X.G., Xu W., Li T., Lu J.Y., Yang Y., Li Q., Zeng Z.H., Ai Y.H. Involvement of phosphatase and tensin homolog-induced putative kinase 1-Parkin-mediated mitophagy in septic acute kidney injury. Chin Med J (Engl) 2019;132(19):2340–2347. doi: 10.1097/CM9.0000000000000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woost P.G., Orosz D.E., Jin W., Frisa P.S., Jacobberger J.W., Douglas J.G., Hopfer U. Immortalization and characterization of proximal tubule cells derived from kidneys of spontaneously hypertensive and normotensive rats. Kidney Int. 1996;50(1):125–134. doi: 10.1038/ki.1996.295. [DOI] [PubMed] [Google Scholar]
- 30.Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C., Xia K., Jiang W., Ronai Z., Zhuang X. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Invest. 2009;119(3):650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choo Y.S., Tang C., Zhang Z. Critical role of PINK1 in regulating Parkin protein levels in vivo. Arch. Neurol. 2011;68(5):684–685. doi: 10.1001/archneurol.2011.95. [DOI] [PubMed] [Google Scholar]
- 32.Rittirsch D., Huber-Lang M.S., Flierl M.A., Ward P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009;4(1):31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang M., Wei Q., Dong G., Komatsu M., Su Y., Dong Z. Autophagy in proximal tubules protects against acute kidney injury. Kidney Int. 2012;82(12):1271–1283. doi: 10.1038/ki.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooks C., Wei Q., Cho S.G., Dong Z. Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J. Clin. Invest. 2009;119(5):1275–1285. doi: 10.1172/JCI37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo C., Pei L., Xiao X., Wei Q., Chen J.K., Ding H.F., Huang S., Fan G., Shi H., Dong Z. DNA methylation protects against cisplatin-induced kidney injury by regulating specific genes, including interferon regulatory factor 8. Kidney Int. 2017;92(5):1194–1205. doi: 10.1016/j.kint.2017.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bueno M., Lai Y.C., Romero Y., Brands J., St Croix C.M., Kamga C., Corey C., Herazo-Maya J.D., Sembrat J., Lee J.S. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J. Clin. Invest. 2015;125(2):521–538. doi: 10.1172/JCI74942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rojansky R., Cha M.-Y., Chan D.C. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife. 2016;5 doi: 10.7554/eLife.17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doi K., Leelahavanichkul A., Yuen P.S., Star R.A. Animal models of sepsis and sepsis-induced kidney injury. J. Clin. Invest. 2009;119(10):2868–2878. doi: 10.1172/JCI39421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Q., Ren J. Urinary mitochondrial DNA identifies renal dysfunction and mitochondrial damage in sepsis-induced acute kidney injury. Oxid Med Cell Longev. 2018;2018:8074936. doi: 10.1155/2018/8074936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran M., Tam D., Bardia A., Bhasin M., Rowe G.C., Kher A., Zsengeller Z.K., Akhavan-Sharif M.R., Khankin E.V., Saintgeniez M. PGC-1α promotes recovery after acute kidney injury during systemic inflammation in mice. J. Clin. Invest. 2011;121(10):4003–4014. doi: 10.1172/JCI58662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J.X., Yang C., Zhang W.H., Su H.Y., Liu Z.J., Pan Q., Liu H.F. Disturbance of mitochondrial dynamics and mitophagy in sepsis-induced acute kidney injury. Life Sci. 2019;235:116828. doi: 10.1016/j.lfs.2019.116828. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z., Chen Z., Liu R., Liang Q., Peng Z., Yin S., Tang J., Gong T., Liu Y. Bcl-2 proteins regulate mitophagy in lipopolysaccharide-induced acute lung injury via PINK1/parkin signaling pathway. Oxi Med Cell longev. 2020;2020 doi: 10.1155/2020/6579696. 6579696-6579696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J.S., Choi H.S., Yim S.Y., Lee S.M. Heme oxygenase-1 protects the liver from septic injury by modulating TLR4-mediated mitochondrial quality control in mice. Shock. 2018;50(2):209–218. doi: 10.1097/SHK.0000000000001020. [DOI] [PubMed] [Google Scholar]
- 44.Li J., Shi W., Zhang J., Ren L. To explore the protective mechanism of PTEN-induced kinase 1 (PINK1)/parkin mitophagy-mediated extract of Periplaneta americana on lipopolysaccharide-induced cardiomyocyte injury. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2019;25:1383–1391. doi: 10.12659/MSM.912980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumida M., Doi K., Ogasawara E., Yamashita T., Hamasaki Y., Kariya T., Takimoto E., Yahagi N., Nangaku M., Noiri E. Regulation of mitochondrial dynamics by dynamin-related protein-1 in acute cardiorenal syndrome. J. Am. Soc. Nephrol. 2015;26(10):2378–2387. doi: 10.1681/ASN.2014080750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Q., Li S., Jiang N., Shao X., Zhang M., Jin H., Zhang Z., Shen J., Zhou Y., Zhou W. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019;26:101254. doi: 10.1016/j.redox.2019.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang C., Han H., Liu Z., Liu Y., Yin L., Cai J., He L., Liu Y., Chen G., Zhang Z. Activation of BNIP3-mediated mitophagy protects against renal ischemia-reperfusion injury. Cell Death Dis. 2019;10(9):677. doi: 10.1038/s41419-019-1899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524(7565):309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuji N., Tsuji T., Ohashi N., Kato A., Fujigaki Y., Yasuda H. Role of mitochondrial DNA in septic AKI via toll-like receptor 9. J. Am. Soc. Nephrol. 2016;27(7):2009–2020. doi: 10.1681/ASN.2015040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts R.F., Fon E.A. Presenting mitochondrial antigens: PINK1, Parkin and MDVs steal the show. Cell Res. 2016;26(11):1180–1181. doi: 10.1038/cr.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatia D., Chung K.-P., Nakahira K., Patino E., Rice M.C., Torres L.K., Muthukumar T., Choi A.M., Akchurin O.M., Choi M.E. Mitophagy-dependent macrophage reprogramming protects against kidney fibrosis. JCI insight. 2019;4(23) doi: 10.1172/jci.insight.132826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J., Zhu P., Li R., Ren J., Zhou H. Fundc1-dependent mitophagy is obligatory to ischemic preconditioning-conferred renoprotection in ischemic AKI via suppression of Drp1-mediated mitochondrial fission. Redox Biol. 2020;30:101415. doi: 10.1016/j.redox.2019.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.