Abstract
14-3-3 proteins are mostly expressed in the brain and are closely involved in numerous brain functions and various brain disorders. Among the isotypes of the 14-3-3 proteins, 14-3-3γ is mainly expressed in neurons and is highly produced during brain development, which could indicate that it has a significance in neural development. Furthermore, the distinctive levels of temporally and locally regulated 14-3-3γ expression in various brain disorders suggest that it could play a substantial role in brain plasticity of the diseased states. In this review, we introduce the various brain disorders reported to be involved with 14-3-3γ, and summarize the changes of 14-3-3γ expression in each brain disease. We also discuss the potential of 14-3-3γ for treatment and the importance of research on specific 14-3-3 isotypes for an effective therapeutic approach.
Keywords: 14-3-3 gamma, 14-3-3 protein, 14-3-3γ, Brain disorders, Glioma, Inflammatory disease, Neurodegenerative disease, Neuro-developmental disease, YWHAG
INTRODUCTION
The 14-3-3 proteins were first extracted from the bovine brain in the 1960s (1) and were identified as activators of tyrosine and tryptophan hydroxylases involved in the biosynthesis of neurotransmitters (2). Subsequently, intensive follow-up studies revealed that 14-3-3 proteins are involved in diverse cellular functions, including cell survival, growth, differentiation, migra-tion, and signaling. The naming the protein ‘14-3-3’ was based on the initial discovery that the proteins were extracted from the 14th fraction in a diethylaminoethyl-cellulose chromato-graphy and were located at the 3.3 position on a starch gel electrophoresis (1). The 14-3-3 proteins are highly conserved, approximately 30-kDa acidic molecules that form dimers (3) and bind to various intracellular proteins. These partners are transmembrane receptors, cytoskeletal proteins, and signal-transducing proteins, such as kinases and phosphatases (4). They participate in the regulation of transcription, cell-cycle control, protein trafficking, metabolism, signal transduction, stress res-ponse, and apoptosis (5).
Isotypes and general functions of 14-3-3 proteins
The 14-3-3 proteins are a family of highly homologous mole-cules expressed in all eukaryotic cells. About fifteen isotypes have been reported in plants, two in yeast, and two in C. elegans and Drosophila (6-8). In mammals, seven isotypes (β, ε, γ, η, τ/θ, ξ, σ) have been identified, and each of them is encoded by different genes (YWHAB, YWHAE, YWHAG, YWHAH, YWHAQ, YWHAZ and SFN, arranged in the order listed above). The previously named α and δ are phosphoㄴrylated β and ξ, respectively (Table 1) (9).
Table 1.
The isotypes of the 14-3-3 proteins
| Gene | Chromo-some | Isotypes | ||
|---|---|---|---|---|
| YWHAB | 20q13.1 | Beta/Alpha | β/α | α is a phosphorylated form of β |
| YWHAE | 17p13.3 | Epsilon | ε | |
| YWHAG | 7q11.23 | Gamma | γ | |
| YWHAH | 22q12.3 | Eta | η | |
| YWHAQ | 2p25.1 | Tau/Theta | τ/θ | θ is an alternative name for τ |
| YWHAZ | 8q23.1 | Zeta/Delta | ξ/δ | δ is a phosphorylated form of ξ |
| SFN | 1p35.3 | Sigma | σ | |
Highly conserved seven isoforms of the 14-3-3 proteins have been identified. The α and δ are phosphorylated forms of the β and ξ, respectively and theta is an alternative name for tau. Genes encoding these isotypes are located at different positions of the human chromo-somes.
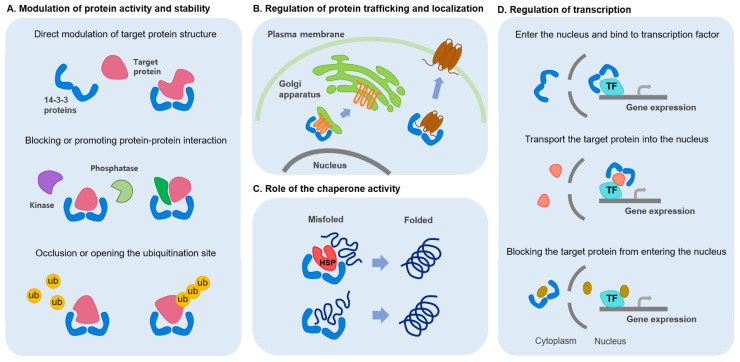
The 14-3-3 proteins regulate the activity of binding proteins by blocking or promoting protein-protein interactions with other proteins (10-12). In addition, the 14-3-3 proteins can regulate stability of binding proteins by occluding or opening the ubiquitination site and also control their trafficking and localization (13). In addition, 14-3-3 proteins interact with the chaperone proteins, such as the heat shock protein (HSP), and has the chaperone-like activity itself (13, 14). Moreover, the 14-3-3 proteins have a nuclear localization sequence (NLS), which can regulate the gene expression by transporting target proteins to the nucleus (Fig. 1) (15, 16).
Fig. 1.
Functional characteristics of the 14-3-3 proteins. The 14-3-3 proteins form dimers and bind to various proteins in the cell to regulate the function of the target proteins. (A) The target proteins can be structurally modified by binding to the 14-3-3 proteins. The 14-3-3 proteins bind to the target proteins and modulate the activity of the binding protein by blocking or promoting protein-protein interaction with other proteins. In addition, by blocking or opening sites where ubiquitin (ub) is attached to the target protein, 14-3-3 proteins can be involved in protein degradation to regulate stability. (B) The 14-3-3 proteins can regulate the trafficking and localization of binding proteins. (C) The 14-3-3 proteins interact with chaperone proteins such as heat shock proteins (HSP) and have chaperone-like activity themselves. (D) The 14-3-3 proteins have a nuclear localization sequence (NLS), can enter the nucleus, and can bind to transcription factors (TF). Moreover, the 14-3-3 proteins can regulate gene expression by transporting the target protein to the nucleus, or by blocking the target protein from entering the nucleus.
Structural features of 14-3-3 proteins for dimerization and binding motif
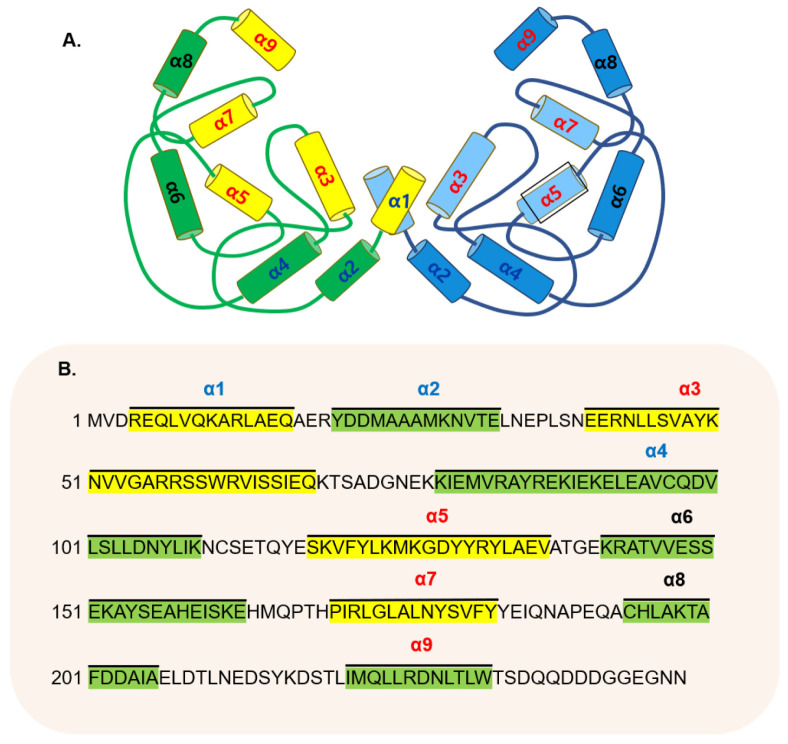
The 14-3-3 proteins form dimers, which make a U-shaped binding groove (17, 18). One monomer has a bundle of nine α-helices; especially the first four helices are essential for dimer formation. First two helices at the N-terminal of one monomer are connected to the fourth helix of the other monomer by the salt bridge formation (Fig. 2). The dimer interface buries several hydrophobic and polar residues (Ll3, Al7, S58, V62, I66, and Y85), with the salt bridge including Arg-Glu (R19 and E92) and the side chain containing Asp-Lys (D21 and K88). All residues are based on human 14-3-3γ.
Fig. 2.
Structural features of the 14-3-3γ. (A) The 14-3-3 proteins con-sist of nine α--helical bundles to form a dimer for making a U-shaped groove. The first and second helices of the N-terminus of one monomer are connected to the fourth helix of the other monomer by the salt bridge. The first, third, fifth and seventh helices of one monomer are located inside the cup, and the third, fifth, seventh and ninth helices form a conserved amphiphilic groove that binds to the target protein. (B) The 14-3-3γ is a highly conserved protein that has the same protein sequence in humans, rat, and mice. Like most isotypes, the 14-3-3γ consists of nine helical structures.
Most isotypes show a similar preference for either homodi-mer or heterodimer formation, but σ prefers homodimer (19), while ε prefers heterodimer formation (20). Different preferences for homo- or heterodimer formation of each 14-3-3 isotype appears to result from their detailed structural differences (21). For example, the side chains of Asp-Lys (D21 and K88) are preserved in most isotypes except for ε, which has Glu-Met (E22 and M88) instead. Given this difference, ε can easily form a heterodimer instead of a homodimer (22). Meanwhile, most 14-3-3 proteins generally act as a dimer, but sometimes they function as a monomer. For example, 14-3-3ξ can regulate the activity of Raf kinase when they are dimers (23), but the activity of the Drosophila calcium-dependent potassium channel can be controlled by a monomeric form (24).
For binding with the target proteins, the first, third, fifth, and seventh helices are located inside the cup, and the third, fifth, seventh, and ninth helices form a conserved amphipathic binding groove, which has a hydrophobic patch on one side and a cluster of charged and polar residues on the other side (Fig. 2) (25). On one side of the groove, the seventh and ninth helices form a hydrophobic surface which includes four leucine side chains (L173, L211, L221, and L226). On the other side, the third helix has three basic side chains (K49, R56, and R61) and the fifth helix has polar and charged groups (K120, D129, Y130, and Rl32) (26). The phosphate group of the interacting protein binds to the binding site formed by the conserved Lys-Arg-Arg-Tyr (K50, R57, R132, and Y133) in the third and fifth helices inside the cup (Fig. 2) (17).
The 14-3-3 proteins recognize the high-affinity binding motifs containing phosphoserine or phosphothreonine (pSer/pThr) (18, 27), including RSX[pS/pT]XP and RXXX[pS/pT] XP, or the C-terminal phosphorylation motif (28, 29), such as pS/TX-COOH of target proteins and bind them in a phosphoryla-tion-dependent or -independent manner. The X represents any residue and the p indicates phosphorylation. Furthermore, other nonphosphorylated binding motifs have also been found (30-32). The basic cluster of binding grooves consisting of K49, R56, and R127 in 14-3-3 proteins mediates the interaction with the phosphoamino acid of target proteins, and R19 is involved in the binding of the non-phosphorylated proteins (33, 34).
Binding specificity of 14-3-3 proteins
Since most residues involved in dimer formation and protein-binding sites are conserved in most isotypes of 14-3-3 proteins, it could be speculated that the 14-3-3 proteins can randomly form dimers and interact with numerous binding proteins. However, 14-3-3 isotypes can specifically select their target proteins by forming a complex tertiary structure via dimeriza-tion (18, 21, 22). Homo- or heterodimers of 14-3-3 isotypes can create various structural differences in position, angle, and specificity of binding groove (21, 22, 35). In addition to the conserved amphipathic binding groove, the C-terminal region of 14-3-3 proteins can be involved in regulation of binding specificity with target proteins (36, 37). Since C-terminal region of 14-3-3 proteins is the most flexible region, relatively freely rotating C-terminus of 14-3-3 proteins can help to permit isotype-specific interactions with target proteins. It has been reported that various factors such as stress, drugs, and disease can regulate gene expression of 14-3-3 isotypes (16, 38). Altered gene expression of specific 14-3-3 isotype could contribute to the changes in the ratio of 14-3-3 isotypes and the contents of 14-3-3 dimers. Therefore, regulated gene expression of specific 14-3-3 isotype also can affect binding specifi-city of other 14-3-3 proteins.
14-3-3γ in brain function
The 14-3-3 proteins are ubiquitously expressed in almost all organs (39, 40), and are most expressed in the brain, except for σ, also known as stratifin, which is predominantly expres-sed in epithelial cells (41). It is not surprising that 14-3-3 proteins are involved in numerous brain disorders, considering that 14-3-3 proteins bind to several critical proteins that have cellular and molecular functions in the brain. Among the 14-3-3 proteins, the 14-3-3γ is encoded by the gene YWHAG, located on the 7th chromosome (7q11.23) (Table 1) and is most frequently expressed in neurons at the mRNA level (42), unlike other isotypes that were relatively evenly expressed in various brain cells. Thus, the 14-3-3γ is expected to be importantly involved in neuronal functions by binding to several signaling proteins, such as β-catenin (43), RGS14 (44) and LRRK2 (45). Moreover, the 14-3-3γ participates in neural processes such as ion channel regulation and receptor trafficking (46-48). Although 14-3-3γ studies on brain and nerve function are still lacking, the possibility of 14-3-3γ playing a significant role is emerging.
14-3-3γ IN BRAIN DISORDERS
The 14-3-3γ protein has been implicated in neurodegenerative diseases as well as neurodevelopmental and neuropsychiatric disorders (Table 2). Moreover, the 14-3-3γ has also been re-ported in several neurological disorders, such as neuroinflam-matory disease and glioma (Table 2).
Table 2.
The 14-3-3γ in brain disorders
| Category of disease | Name of disease | Expression pattern or potential role in disease | References |
|---|---|---|---|
| Neurodevelopmental disorders | Williams syndrome | Associated with epilepsy phenotypes | 54-57 |
| Down syndrome (DS) | Decreased in the cortex of fetal DS patients | 63 | |
| Increased in the cortex of aged DS patients | 62 | ||
| Neurodegenerative disorders | Parkinson’s disease (PD) | Colocalized with the Lewy body of PD | 70-72 |
| Decreased in the transgenic a-syn mouse | 73 | ||
| Neuroprotection against the rotenone or MPTP induced cytotoxicity | 74 | ||
| Promote the a-syn aggregation | 75, 76 | ||
| Alzheimer’s disease (AD) | Colocalized with the NFT in hippocampus of AD patients | 83 | |
| Increased in overall cortical regions of aged AD patients | 84 | ||
| Decreased in the frontal cortex of postmortem AD patients | 85 | ||
| Creutzfeldt-Jakob disease (CJD) | Useful marker for CJD diagnosis using CSF | 88-95 | |
| Neuroinflammatory disease | Inflammatory joint disease (IJD) | Highly observed in SF and serum of patients with IJD | 96 |
| Cruciate ligament rupture (CCLR) | A marker for osteoarthritis caused by CCLR | 97 | |
| Multiple sclerosis (MS) | Increased in the spinal cord of chronic MOG-EAE | 98 | |
| A protective factor for OL against autoimmune dehydration | 98 | ||
| Ischemic damaged brain | Increased in infarct lesions of ischemic brain | 102, 103 | |
| Detected in CSF of MELAS patients | 104 | ||
| A survival factor for ischemic-induced cell death in neurons | 106-108 | ||
| A protectant against ischemic cortical astrocytes | 109-115 | ||
| CNS cancer | Glioma | Promote glioblastoma progression | 126-128 |
The 14-3-3γ is implicated in neurodegenerative and neurodevelopmental disorders, and is also associated with neuroinflammatory diseases and CNS cancer such as glioma. The 14-3-3γ is likely to be involved in the development of various brain diseases, and is useful as a biomarker for diagnosis and a potential therapeutic target for treatment. (α-syn: α-synuclein; NFT: neurofibrillary tangles; SF: synovial fluid; CSF: cerebrospinal fluid; CCLR: cranial cruciate ligament rupture; MOG-EAE: murine myelin oligodendrocyte-induced experimental autoimmune encephalomyelitis; OL: oligodendrocyte; MELAS: mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes).
14-3-3γ in neurodevelopmental disorders
It has been reported that 14-3-3 proteins are a key regulator in neurodevelopmental processes, including neuronal differentia-tion, migration, and morphogenesis including synaptogenesis (49). Among the studies of each 14-3-3 isotype associated with neuronal development, those on 14-3-3ε and ξ are relatively advanced, but studies on other isotypes are still lacking.
The 14-3-3γ is abundantly expressed in the cerebral cortex of developing mice. The mRNA of 14-3-3γ is highly expressed in the embryonic state and decreases from 30 days after birth. (50). Therefore, 14-3-3γ is expected to play an important role in cortical development. Indeed, 14-3-3γ directly binds to Copine1, known as a calcium-dependent membrane-binding protein, and overexpression of 14-3-3γ and Copine1 causes an increase in neuronal differentiation via protein kinase B (AKT) phosphorylation in the hippocampal progenitor cell line (51). But to date, research into the molecular mechanisms of 14-3-3γ in neuronal development and differentiation during cortical development is still lacking. Interestingly, there was a report of 14-3-3γ playing an important role in neuronal migration (50). In the cerebral cortex of postnatal P3 mice in which 14-3-3γ knockdown was induced at fetal-stage embryo-nic day (E)14.5 or E16.5 in utero, 14-3-3γ deficient neurons generated from the ventricular zone and subventricular zone did not properly migrate to the cortical plate but rather stayed in the ventricular zone or the intermediate zone (50). There-fore, it has been suggested that 14-3-3γ knockdown leads to the migration delay of pyramidal neurons in the cerebral cortex and plays an important role in the late developmental stage. Interestingly, the delay of neuronal migration was also observed in the 14-3-3γ overexpression model using the same strategy (52), which suggests the importance of the balance of 14-3-3γ. However, the detailed molecular mechanisms how 14-3-3γ regulates neuronal migration are still unknown.
Williams Syndrome (WS) is a neurodevelopmental disorder caused by a deletion on chromosome 7q11.23 and is accom-panied by symptoms of delayed developmental and intellec-tual disabilities (53). As previously reported, typical WS patients showed deletions of 1.5-1.8 Mb in size, whereas atypical patients showed deletions greater than 1.8 Mb; the deletions of the atypical patients include the YWHAG gene that encoded the 14-3-3γ (54-56). Interestingly, atypical patients have epile-psy symptoms that do not occur in typical patients, and epile-psy has also been reported in WS patients with a reciprocal duplication syndrome of the 7q11.23 gene (57). These reports suggest that changes in the level of 14-3-3γ may be associated with epilepsy phenotypes in WS patients.
Also, a more direct link between 14-3-3γ and seizures was observed in temporal lobe epilepsy (58-60). The 14-3-3γ and ξ were detected in mitochondrial and microsome-enriched frac-tions, whereas other isotypes are expressed only in the cyto-plasmic compartment of the hippocampus. Especially, the ex-pression of 14-3-3γ was significantly reduced in the microsome-enriched compartment of the rat hippocampus, when experi-enced the kainic acid (KA)-induced acute seizure (58). How-ever, in the human hippocampus of chronic temporal lobe epilepsy, there was no difference in 14-3-3γ expression, but the levels of 14-3-3ε and ξ were increased in the micro-somal-rich fraction. Thus, this report identified microsomal-rich fractions in the hippocampus of epileptic rat as major sites for altered 14-3-3 protein levels, which suggests changes in isotype distribution in acute and chronic seizures (58). Re-cently in a KA-induced rat model, it has been shown that 14-3-3γ, ε, η, σ and τ/θ were abundantly detected in the frontal cortex, whereas levels of β and ξ isoforms were rela-tively low, and the down-regulation of total 14-3-3 proteins was also observed (59). Both BAX and phosphorylated ERK bind to 14-3-3 proteins, showing increased levels after KA intraperitoneal injection (59). Recently, high enrichment of de novo variants in the YWHAG gene of the developmental and epilep-tic encephalopathies have been identified by in silico model-ing, suggesting that 14-3-3γ de novo mutations are implicated in early-onset epilepsy (60). Taken together, 14-3-3γ appears to be involved in early events related to acute epileptic seizures and excitatory toxicity.
Down syndrome (DS), also known as trisomy 21, is another genetic neurodevelopmental disorder characterized by a third copy on chromosome 21, which has symptoms of delayed physical growth and intellectual disability, along with charac-teristic facial features (61). Notably, compared to healthy con-trols, the aged DS patients displayed an increase in expression of 14-3-3γ and ε in the cortical regions (62). In contrast, by subsequent matrix-assisted laser/desorption ionization (MALDI) identification, the decreased 14-3-3γ expression was detected in the fetal brain cortex of DS (63). These results suggest that 14-3-3γ might be involved in the pathology of DS, but that the expression of 14-3-3γ may differ depending on the age and brain regions involved.
14-3-3γ in neurodegenerative disorders
Parkinson’s disease (PD) is an age-related neurodegenerative disease with symptoms of motor dysfunction, such as progres-sive rigidity, bradykinesia, tremor, and postural instability (64). Some PD patients have symptoms of dementia and mood problems such as depression and anxiety (65, 66). PD is characterized by a loss of dopaminergic neurons and Lewy body formation in the substantia nigra pars compacta as well as the cortical regions and brainstem (67). The Lewy body is mainly composed of α-synuclein (α-syn), which contributes to PD pathogenesis (68, 69). The interaction of the 14-3-3 pro-teins and α-syn was confirmed by co-immunoprecipitation in the rat brain (70), and the 14-3-3 proteins were also observed by immunohistochemistry in Lewy bodies of PD and the Diffuse Lewy body disease (LBD) (71). Among the 14-3-3 family, 14-3-3γ, ε, ξ, and τ/θ were observed to be colocalized with the Lewy body of PD (72). In addition, a decrease in 14-3-3γ, ε and τ/θ was detected in transgenic α-syn mouse, an animal model of PD (73). It was also observed that over-expression of 14-3-3τ/θ reduced cytotoxicity of dopaminergic neurons in MPTP mouse model of PD (74). Mechanistically, it has been suggested that 14-3-3γ, ε, η, and ξ promote the formation of α-syn aggregates and participate in oligomer degradation and aggregation (75, 76). Furthermore, it was proposed that 14-3-3η affects α-syn aggregation by binding with the α-syn oligomer, and that overexpression of 14-3-3η reduces α-syn toxicity and binds with the parkin, which is ano-ther factor of PD pathogenesis (77). According to other studies, 14-3-3ξ reportedly inhibits the ubiquitin-ligase activity of Parkin (78) and activates the tyrosine hydroxylase (TH), a rate-limiting enzyme in catecholamine biosynthesis (79). Thus, the 14-3-3 proteins generally appear to influence aggregation and neurotoxicity of α-syn and may be involved in the dopamine synthesis by interacting with α-syn. Since 14-3-3γ, along with η and ξ, is responsible for the α-syn aggregates (75), it will likely play a critical role and be the potential target for neuro-protective effects on dopaminergic neurons (77-79).
Alzheimer’s disease (AD) is also a neurodegenerative dis-ease with the symptoms of dementia accompanied by memory loss and cognitive impairment (80). AD is characterized by the accumulation of amyloid plaques caused by neurotoxic extracellular β-amyloid (Aβ) aggregation (81) and intracellular neurofibrillary tangles (NFT) composed of the abnormally hyper-phosphorylated microtubule-associated protein tau (82). Although the 14-3-3 proteins were not observed in amyloid plaques, there is growing evidence of the link between NTF and the 14-3-3 proteins. The expression of the 14-3-3 proteins in hippocampal NFT of AD have been shown by immuno-histochemistry (83), and the increased 14-3-3γ and ε in overall cortical regions of aged AD patients were observed (84). However, in a recent report, the levels of 14-3-3γ and η shown to be significantly decreased in the frontal cortex of postmor-tem AD patients (85). Therefore, it is possible that 14-3-3γ ex-pression can be differently regulated in the different stages of AD progression or the different cortical regions of AD patients. Research into the association of NFT with specific isotypes of the 14-3-3 protein is still missing.
Creutzfeldt-Jakob Disease (CJD) is a fatal degenerative brain disorder caused by a misfolded protein, PrPSc prion (scrapie isoform of the prion protein), with the symptoms of confusion, depression, abnormal body sensations, autonomic nervous sys-tem disorders, and dementia (86, 87). The importance of the 14-3-3 proteins as a biomarker for the diagnosis of CJD has been frequently discussed over the past decade. Since the 14-3-3 proteins were first proposed as a potential CJD marker for the rapid detection in cerebrospinal fluid (CSF) (88), many studies have confirmed that the 14-3-3 proteins are a useful diagnostic marker for CJD (89, 90). Interestingly, the 14-3-3 proteins were not observed in the CSF of patients with some brain destructions, such as Rasmussen’s encephalitis, Schilder’s disease, or diffuse large B-cell lymphoma (91). These results may indicate that the increased level of 14-3-3 proteins in CSF is not caused by protein leakage due to cell death, it is more likely that the 14-3-3 proteins are specifically involved in the pathological mechanism of CJD. Early-stage CJD diagnostic antibodies detected all 14-3-3 isotypes, but a recent study of the diagnostic accuracy of CJD reported that 14-3-3γ and β were more reliable (92, 93). More recently, it has been sug-gested that 14-3-3γ could be used as a specific marker for neuronal damage caused by CJD (94, 95). Therefore, this sug-gests that 14-3-3γ is closely related to the pathogenesis of CJD.
14-3-3γ in neuroinflammatory disease
In several studies, there is evidence that 14-3-3γ is closely related to the inflammatory conditions (96, 97). High levels of 14-3-3γ and η were observed in synovial fluid (SF) and serum obtained from patients with inflammatory joint disease, and their expression levels correlated strongly with the levels of matrix metalloprotease (MMP)-1 and MMP-3, two biomarkers for rheumatoid arthritis (96). A subsequent study confirmed that 14-3-3γ and η were abundantly detected in the SF from dogs with cranial cruciate ligament rupture (CCLR). Thus, these results suggest that 14-3-3γ and η are potent stimulators of MMPs and could be a marker for osteoarthritis caused by CCLR (97).
The 14-3-3γ is also shown to be involved in inflammatory diseases in the central nervous system (CNS) (98). Multiple sclerosis (MS) is a demyelinating disease caused by damage to the insulating sheath of neurons in the brain and spinal cord. MS is the most common immune-mediated disorder with neurodegenerative symptoms, such as sensory nerve damage and movement disorder (99). Experimental autoimmune encephalomyelitis (EAE) is a model that mimics the pathological features of MS, showing oligodendrocyte (OL) damage, inflam-matory demyelination, and astrocyte activation (100, 101). The mRNA expression of 14-3-3γ was increased in the spinal cord of chronic murine myelin oligodendrocyte-induced EAE (98). Thus, the deficiency of 14-3-3γ promotes demyelination and increases the sensitivity of OL to inflammatory attacks. There-fore, 14-3-3γ is likely to act as a protective factor for OL against autoimmune demyelination by participating in the apoptotic mechanism (98).
The importance of 14-3-3γ in the inflammatory conditions can be further explored in ischemic stroke (102-104). The intensity of 14-3-3 staining was increased in cortical neurons of rat brains with ischemic damage (102), and 14-3-3γ, β, ε, σ, and τ/θ were detected in the CSF of ischemic patients (103). An increase of these isotypes was also observed in infarct lesions of the human brain obtained from an autopsy. This increase was proportional to the severity of the ischemia pathology, particularly abundant in the chronic phase (103). Furthermore, 14-3-3γ and τ/θ were also detected in the CSF of patients with mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), such as severe mental re-gression and limb paralysis (104). The detection of these pro-teins was correlated with the degree of damage to brain ob-served by magnetic resonance imaging (104). Therefore, it has been suggested that some 14-3-3 isoforms may be potential biomarkers for ischemic CNS damage.
Moreover, several studies have reported that 14-3-3γ acts as a survival factor for cortical neurons in ischemia (43, 105). The nuclear translocation of Akt-phosphorylated SRPK2 inactivates p53 in the ischemic-attacked brain and induces an increase in cyclin D1 to promote cell-cycle progression and apoptosis. The 14-3-3 proteins have been proposed to inhibit cell-cycle and neuronal cell death by interacting with and inhibiting SRPK2, a protein kinase for serine/arginine (SR)-rich splicing factor (106). In another study, an increase of 14-3-3γ was ob-served in primary cortical neurons under oxygen-glucose depri-vation (105). Moreover, 14-3-3γ overexpression reduced oxygen glucose deprivation (OGD)-induced cell death in cultured neurons, whereas 14-3-3γ knockdown increased the expres-sion of BAX to promote neuronal cell death (105). It was con-firmed that OGD promotes entry into nuclei of 14-3-3γ, induces binding with increased nuclear phosphorylated-β-catenin, and reduces BAX expression, thereby suppressing neuronal apoptosis (105).
In inflammatory conditions, the 14-3-3 proteins have been reported to be involved in the activation of the NFκB-signaling pathway for the regulation of immune responses and apoptosis (107). After ischemia-reperfusion (I/R) in transient middle cere-bral artery occlusion, an animal model that mimics ischemic stroke, upregulation of 14-3-3γ and increased translocation from the cytoplasm to the nucleus were observed in cortical neurons. In this process, 14-3-3γ interacts with p65 to regulate its expression (107). Therefore, 14-3-3γ may be a potential therapeutic target for stroke.
Recently, the role of 14-3-3γ in starvation-activated neuronal autophagic influx signaling has been suggested (43). By immuno-staining, the increased expression levels of Beclin-1 and 14-3-3γ, and the colocalization of 14-3-3γ, Beclin-1, and LC3, were detected in starvation-treated neuroblastoma (N2a) cells and cultured cortical neurons of ischemic brains. Inhibition of 14-3-3γ reduced hunger-induced activation of Beclin-1 and LC3, and overexpression of 14-3-3γ increase Beclin-1/LC3 signal transduction. Under starvation, 14-3-3γ preferred binding to phosphorylated-β-catenin rather than Beclin-1. Thus, it is suggested that 14-3-3γ overexpression reactivates Beclin-1-LC3 signaling because of the inhibition of β-catenin in starving neurons (43). It is also reported that pre-ischemic exercise increases the expression of 14-3-3γ, thereby reducing ischemia-induced neuronal cell death through anti-apoptotic pathways including phosphorylated β-catenin, BAX, and caspase 3 (108). Therefore, 14-3-3γ is proposed to act as an initial survival factor for ischemic-induced cell death in neurons.
In addition, a potential protective role of 14-3-3γ has been proposed in astrocytes, which play an important regulatory role in the inflammatory and apoptotic conditions of stroke-induced ischemic attacks. The 14-3-3γ is mainly expressed in neurons, but was also observed in primary cultures of mouse cortical astrocytes (109, 110). Moreover, the increased level of 14-3-3γ was detected in ischemic cortical astrocytes (111) and only 14-3-3γ was observed in the astrocytic nuclei in the ischemic human brain (112). Another study has confirmed that overexpression of 14-3-3γ promotes the survival of astrocytes under ischemia, whereas 14-3-3γ knockdown increases apoptosis of astrocytes, and suggested as a protective mechanism of 14-3-3γ that 14-3-3γ binds to a phosphorylated BAD, thereby preventing BAD from entering the mitochondria and conse-quently inhibiting the initiation of apoptosis (113). Moreover, 14-3-3γ upregulation by ischemia in astrocytes was observed to be blocked by the inhibitions of c-Jun N-terminal kinase (JNK) and activator protein 1 (AP-1) (114). A subsequent study confirmed that an increase of 14-3-3γ during ischemic precon-ditioning reduced astrocytic death caused by ischemic damage via the JNK pathway (115). Thus, 14-3-3γ was selectively increased during ischemia and has a protective role against astrocytes, which seem to be implicated in the JNK signaling pathway.
Taken together, 14-3-3γ could be a biomarker for ischemic stroke and can act as a cell protectant in inflammatory con-ditions against ischemic damage. The protective mechanism of 14-3-3γ appears to involve the pathways associated with apop-tosis and immune responses. Therefore, it is worth considering 14-3-3γ as a new strategic target to reduce cell damage in astrocytes as well as neurons and to improve the recovery of stroke.
14-3-3γ in glioma
Research into the role of 14-3-3γ in astrocytes is ongoing in glioma, a CNS tumor associated with glial cells. These studies have been linked to the function of the 14-3-3 proteins on the regulation of cell cycle and apoptosis, which contribute to cell growth and cancer progression (116-118). The increase of 14-3-3γ expression has been observed in several cancers, and it has been suggested that the mechanism may promote cancer development via pathways associated with P53 and BAD (105, 119, 120). However, it is still unclear whether the role of 14-3-3γ is pro-apoptotic or anti-apoptotic in glioma (40).
Analysis of the 14-3-3 isotype-specific expression in early glioma studies showed contradictory results. An increase of all 14-3-3 isotypes was observed in most primary nervous-system tumors (121), but 14-3-3γ and σ were not detected in astro-cytomas (122, 123). Moreover, in human glioma tissues and para-cancerous brain tissues, 14-3-3γ expression was detected but no increase was reported (124). However, using Isotope-Coded Protein Label technology, an increase of 14-3-3γ was observed in the tumor zone of glioblastoma patients and the overexpression of 14-3-3γ was confirmed using immunoblot-ting and immunohistochemistry (125). Thus, the expression of 14-3-3γ might be regulated under certain glioma cases.
Furthermore, 14-3-3 has been proposed as a potential mole-cular target for the development of anti-cancer drugs. It has been reported that inhibition of 14-3-3 by difopein or siRNA suppresses glioma growth and improves sensitivity of glioma cells to apoptosis (126). It has been also observed that 14-3-3γ knockdown reduces the migration and invasion of glioblastoma cells, and a deficiency of 14-3-3γ results in a decrease in sur-face expression and channel activity of ANO1, a calcium-acti-vated chloride channel, which binds directly to 14-3-3γ. There-fore, 14-3-3γ may mediate the progression of glioblastoma by interacting with ANO1 (46), which has been shown to in-crease its expression in various cancer cells, including gliobla-stoma (127). More recently, it has been observed that Comp 5, a potential activator of sirtuin-1, which is a highly conserved NAD+-dependent protein deacetylase and an emerging tumor suppressor, down-regulates 14-3-3γ to inhibit autophagy/mito-phagy in glioblastomas (128). Therefore, the regulation of 14-3-3γ may provide new opportunities for glioma treatment.
DISCUSSION
Alteration in the expression levels of the 14-3-3 proteins detected under disease are related to their function. Accumulating evidence on the signaling pathways involved in the 14-3-3 binding proteins reveals their contribution to the diseases and narrows the scope of the targets for the therapeutic approach. However, there are still pieces to be put together. The differ-ence in 14-3-3 isotypes in various brain disorders makes it clear that they have different roles. Also, the expression of the same isotype is regulated temporally and regionally in different ways in the same disease, even with opposite expression patterns. For example, the level of 14-3-3γ was reduced in the rat hippocampus of KA-induced acute seizure but there was no difference in the human hippocampus of chronic epilepsy (58). Similarly, the decreased 14-3-3γ in the cortex of fetal DS (63) was observed to increase in aged DS patients (62). In addition, 14-3-3γ was increased in the overall cortex of AD patients (84) but decreased in the frontal cortex (85). There-fore, further research into the multifaceted function of 14-3-3γ is needed to overcome their structural similarity, reduce side effects, and enable accurate drug treatment for the diseases involving 14-3-3γ.
As described above, based on evidence that 14-3-3γ is involved in brain pathogenesis and is a useful biomarker of brain diseases, generating knockout mice of 14-3-3γ is worthy. However, unexpectedly, previously generated 14-3-3γ knockout mice showed normal behavior and no significant defects (129). In contrast, the recently generated 14-3-3γ-deficient mice were lethal in the prenatal period, and the haploin-sufficient heterozygous mice were smaller in body size and weight compared to the wild-type mice, which means that 14-3-3γ may play an important role in the development (130). Moreover, these mice showed hyperactive locomotion and stress-sensitive behaviors, demonstrating that 14-3-3γ can be implicated in various brain diseases (130). In fact, in the case of the ξ and ε, their deficient mice show abnormal behavioral patterns that show symptoms of disease. The 14-3-3ξ-deficient mice exhibit neuropsychiatric behaviors, cognitive defects, and hyperactivity locomotion (131-133). The 14-3-3ε-deficient mice showed schizophrenia-like behavior, hyperactive loco-motion, and working memory defect (134, 135). Considering the behavioral patterns of mice lacking ξ or ε, and 14-3-3γ-related brain diseases as described in this review, the abnor-mal phenotypes in recently produced 14-3-3γ-deficient mice is very reliable. The 14-3-3γ-deficient mice will be used as a powerful tool for identifying the functions of 14-3-3γ and developing new therapeutic methods in studies including vari-ous brain disorders summarized in this review.
In conclusion, this review is focused on the involvement of 14-3-3γ in various brain disorders. This review also can pro-vide a helpful guidance for future studies about crucial roles of 14-3-3γ in brain disorders.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation (NRF) of Korea (NRF-2016M3C7A1904149, NRF2017M3C7A1079694 and NRF-2017R1A2B3012502).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Moore BE, Perez VJ. Specific acidic proteins of the nervous system. Physiological and Biochemical aspects of Nervous integration. A symposium; Prentice Hall, Englewood Cliffs, NJ. 1967. pp. 343–359. [Google Scholar]
- 2.Ichimura T, Isobe T, Okuyama T, et al. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxy-genase in the presence of Ca2+,calmodulindependent protein kinase II. FEBS Letts. 1987;219:79–82. doi: 10.1016/0014-5793(87)81194-8. [DOI] [PubMed] [Google Scholar]
- 3.Jones DH, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368:55–58. doi: 10.1016/0014-5793(95)00598-4. [DOI] [PubMed] [Google Scholar]
- 4.Jin J, Smith FD, Stark C, et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 5.Darling DL, Yingling J, Wynshaw-Boris A. Role of 14-3-3 Proteins in Eukaryotic Signaling and Development. Curr Top Dev Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Shakes DC. Molecular evolution of the 14-3-3 protein family. J Mol Evol. 1996;43:384–398. doi: 10.1007/BF02339012. [DOI] [PubMed] [Google Scholar]
- 7.Rosenquist M, Alsterfjord M, Larsson C, Sommarin M. Data mining the arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001;127:142–149. doi: 10.1104/pp.127.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su TT, Parry DH, Donahoe B, Chien CT, O'Farrell PH, Purdy A. Cell cycle roles for two 14-3-3 proteins during drosophila development. J Cell Sci. 2001;114:3445–3454. doi: 10.1242/jcs.114.19.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aitken A, Howell S, Jones D, Madrazo J, Patel Y. 14-3-3 alpha and delta are the phosphorylated forms of Raf-activating 14-3-3 beta and zeta. J Biol Chem. 1995;270:5706–5709. doi: 10.1074/jbc.270.11.5706. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 11.van Heusden GP. 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life. 2005;57:623–629. doi: 10.1080/15216540500252666. [DOI] [PubMed] [Google Scholar]
- 12.Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2004;296:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- 13.Sluchanko NN, Gusev NB. Moonlighting chaperone-like activity of the universal regulatory 14-3-3 proteins. FEBS J. 2017;284:1279–1295. doi: 10.1111/febs.13986. [DOI] [PubMed] [Google Scholar]
- 14.Fan X, Cui L, Zeng Y, et al. 14-3-3 proteins are on the crossroads of cancer, aging, and age-related neurodegenerative disease. Int J Mol Sci. 2019;20:3518. doi: 10.3390/ijms20143518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/S0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 16.Obsil T, Obsilova V. Structural basis of 14-3-3 protein functions. Semin Cell Dev Biol. 2011;22:663–672. doi: 10.1016/j.semcdb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 18.Yaffe MB, Volinia S, Caron PR, et al. The Structural basis for 14-3-3: phosphopeptide Binding Specificity. Cell. 1997;91:961–971. doi: 10.1016/S0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 19.Wilker EW, Grant RA, Artim SC, Yaffe MB. 14-3-3 proteins: a family of versatile molecular regu-lators. J Biol Chem. 2005;280:18891–18898. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhri M, Scarabel M, Aitken A. Mam-malian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem Biophys Res Commun. 2003;300:679–685. doi: 10.1016/S0006-291X(02)02902-9. [DOI] [PubMed] [Google Scholar]
- 21.Gardino AK, Smerdon SJ, Yaffe MB. Structural determinants of 14-3-3 binding specificities and regu-lation of subcellular localization of 14-3-3-ligand com-plexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Lee WH, Sobott F, et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci U S A. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzivion G, Luo Z, Avruch J. A Dimeric 14-3-3 protein is an essential cofactor for Raf Kinase Activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Reddy S, Murrey H, Fei H, Levitan IB. Monomeric 14-3-3 protein is sufficient to modulate the activity of the drosophila slowpoke calcium-dependent potassium channel. J Biol Chem. 2003;278:10073–10080. doi: 10.1074/jbc.M211907200. [DOI] [PubMed] [Google Scholar]
- 25.Xiao B, Smerdon SJ, Jones DH, et al. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 26.Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 27.Rittinger K, Budman J, Xu J, et al. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/S1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 28.Coblitz B, Shikano S, Wu M, et al. C-terminal recognition by 14-3-3 proteins for surface expression of membrane receptors. J Biol Chem. 2005;280:36263–36272. doi: 10.1074/jbc.M507559200. [DOI] [PubMed] [Google Scholar]
- 29.Coblitz B, Wu M, Shikano S, Li M. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 2006;580:1531–1535. doi: 10.1016/j.febslet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Petosa C, Masters SC, Bankston LA, et al. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- 31.Masters SC, Pederson KJ, Zhang L, Barbieri JT, Fu H. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry. 1999;38:5216–5221. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- 32.Zhai J, Lin H, Shamim M, Schlaepfer WW, Canete-Soler R. Identification of a novel interaction of 14-3-3 with p190RhoGEF. J Biol Chem. 2001;276:41318–41324. doi: 10.1074/jbc.M107709200. [DOI] [PubMed] [Google Scholar]
- 33.Wang B, Yang H, Liu Y, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry. 1999;38:12499–12504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- 34.Aitken A, Baxter H, Dubois T, et al. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem Soc Trans. 2002;30:351–360. doi: 10.1042/bst0300351. [DOI] [PubMed] [Google Scholar]
- 35.Aitken A. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin Cell Dev Biol. 2011;22:673–680. doi: 10.1016/j.semcdb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Truong AB, Masters SC, Yang H, Fu H. Role of the 14-3-3 C-terminal loop in ligand interaction. Proteins. 2002;49:321–325. doi: 10.1002/prot.10210. [DOI] [PubMed] [Google Scholar]
- 37.Silhan J, Obsilova V, Vecer J, et al. 14-3-3 protein C-terminal stretch occupies ligand binding groove and is displaced by phosphopeptide binding. J Biol Chem. 2004;279:49113–49119. doi: 10.1074/jbc.M408671200. [DOI] [PubMed] [Google Scholar]
- 38.Pennington KL, Chan TY, Torres MP, Andersen JL. The Dynamic and stress-adaptive signaling hub of 14-3-3: Emerging mechanisms of regulation and context-dependent protein-protein interactions. Oncogene. 2018;37:5587–5604. doi: 10.1038/s41388-018-0348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boston PF, Jackson P, Kynoch PA, Thompson RJ. Human 14-3-3 protein: radioimmunoassay, tissue distribution, and cerebrospinal fluid levels in patients with neurological disorders. J Neurochem. 1982;38:1466–1474. doi: 10.1111/j.1471-4159.1982.tb07928.x. [DOI] [PubMed] [Google Scholar]
- 40.Aghazadeh V, Papadopoulos Y. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov Today. 2016;21:278–287. doi: 10.1016/j.drudis.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Leffers H, Madsen P, Rasmussen HH, et al. Molecular cloning and expression of the transformation sensitive epithelial marker stratifin. A member of a protein family that has been involved in the protein kinase C signalling pathway. J Mol Biol. 1993;231:982–998. doi: 10.1006/jmbi.1993.1346. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe M, Isobe T, Ichimura T, Kuwano R, Takahashi Y, Kondo H. Molecular cloning of rat cDNAs for beta and gamma subtypes of 14-3-3 protein and developmental changes in expression of their mRNAs in the nervous system. Brain Res Mol Brain Res. 1993;17:135–146. doi: 10.1016/0169-328X(93)90082-Z. [DOI] [PubMed] [Google Scholar]
- 43.Xiong XX, Hu DX, Xu L, et al. Selective 14-3-3γ upregulation promotes Beclin-1-LC3-Autophagic Influx via β-catenin interaction in starved neurons in vitro and in vivo. Neurochem Res. 2019;44:849–858. doi: 10.1007/s11064-019-02717-4. [DOI] [PubMed] [Google Scholar]
- 44.Gerber KJ, Squires KE, Hepler JR. 14-3-3γ binds regulator of G protein signaling 14 (RGS14) at distinct sites to inhibit the RGS14:Gαi-AlF4− signaling complex and RGS14 nuclear localization. J Biol Chem. 2018;293:14616–14631. doi: 10.1074/jbc.RA118.002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Civiero L, Cogo S, Kiekens A, et al. PAK6 Phosphorylates 14-3-3γ to Regulate Steady State Phos-phorylation of LRRK2. Front Mol Neurosci. 2017;10:417. doi: 10.3389/fnmol.2017.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee YS, Lee JK, Bae Y, et al. Suppression of 14-3-3γ-mediated surface expression of ANO1 inhibits cancer progression of glioblastoma cells. Sci Rep. 2016;6:26413. doi: 10.1038/srep26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh SJ, Woo J, Lee YS, et al. Direct interaction with 14-3-3γ promotes surface expression of Best1 channel in astrocyte. Mol Brain. 2016;10:51. doi: 10.1186/s13041-017-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho CH, Kim E, Lee YS, et al. Depletion of 14-3-3γ reduces the surface expression of Transient Receptor Potential Melastatin 4b (TRPM4b) channels and attenuates TRPM4b-mediated glutamate-induced neuronal cell death. Mol Brain. 2014;7:52. doi: 10.1186/s13041-014-0052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornell B, Toyo-Oka K. 14-3-3 proteins in brain development: neurogenesis, neuronal migration and neuromorphogenesis. Front Mol Neurosci. 2017;10:318. doi: 10.3389/fnmol.2017.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wachi T, Cornell B, Marshall C, Zhukarev V, Baas PW, Toyo-oka K. Ablation of the 14-3-3gamma protein results in neuronal migration delay and morpho-logical defects in the developing cerebral cortex. Dev Neurobiol. 2016;76:600–614. doi: 10.1002/dneu.22335. [DOI] [PubMed] [Google Scholar]
- 51.Yoo JC, Park N, Lee B, et al. 14-3-3γ regulates Copine1-mediated neuronal differentiation in HiB5 hippocampal progenitor cells. Exp Cell Res. 2017;356:85–92. doi: 10.1016/j.yexcr.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 52.Cornell B, Wachi T, Zhukarev V, Toyo-Oka K. Overexpression of the 14-3-3γ protein in embryonic mice results in neuronal migration delay in the develo-ping cerebral cortex. Neurosci Lett. 2016;628:40–46. doi: 10.1016/j.neulet.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Hillier L, Fulton R, Fulton L, et al. The DNA sequence of chromosome 7. Nature. 2003;424:157–164. doi: 10.1038/nature01782. [DOI] [PubMed] [Google Scholar]
- 54.Ramocki MB, Bartnik M, Szafranski P, et al. Recurrent distal 7q11.23 deletion including HIP1 and YWHAG identified in patients with intellectual disabili-ties, epilepsy, and neurobehavioral problems. Am J Hum Genet. 2010;87:857–865. doi: 10.1016/j.ajhg.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fusco C, Micale L, Augello B, et al. Smaller and larger deletions of the Williams Beuren syndrome region implicate genes involved in mild facial phenotype, epilepsy and autistic traits. Eur J Hum Genet. 2014;22:64–70. doi: 10.1038/ejhg.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicita F, Garone G, Spalice A, et al. Epilepsy is a possible feature in Williams-Beuren syndrome patients harboring typical deletions of the 7q11.23 critical region. Am J Med Genet A. 2016;170A:148–155. doi: 10.1002/ajmg.a.37410. [DOI] [PubMed] [Google Scholar]
- 57.Morris CM, Mervis CB, Paciorkowski AP, et al. Osborne 7q11.23 duplication syndrome: physical characteristics and natural history. Am J Med Genet A. 2015;167A:2916–2935. doi: 10.1002/ajmg.a.37340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler CK, Heverin M, Henshall DC. Isoform- and subcellular fraction-specific differences in hippocampal 14-3-3 levels following experimentally evoked seizures and in human temporal lobe epilepsy. J Neurochem. 2016;99:561–569. doi: 10.1111/j.1471-4159.2006.04153.x. [DOI] [PubMed] [Google Scholar]
- 59.Smani D, Sarkar S, Raymick J, Kanungo J, Paule MG, Gu Q. Downregulation of 14-3-3 proteins in a kainic acid-induced neurotoxicity model. Mol Neurobiol. 2018;55:122–129. doi: 10.1007/s12035-017-0724-y. [DOI] [PubMed] [Google Scholar]
- 60.Guella I, McKenzie MB, Evans DM, et al. De Novo mutations in YWHAG cause early-onset epilepsy. Am J Hum Genet. 2017;101:300–310. doi: 10.1016/j.ajhg.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bull MJ. Down syndrome. N Engl J Med. 2020;382:2344– 2352. doi: 10.1056/NEJMra1706537. [DOI] [PubMed] [Google Scholar]
- 62.Fountoulakis M, Cairns N, Lubec G. Increased levels of 14-3-3γ and ε proteins in brain of patients with Alzheimer's disease and Down syndrome. J Neural Transm Suppl. 1999;57:323–335. doi: 10.1007/978-3-7091-6380-1_23. [DOI] [PubMed] [Google Scholar]
- 63.Peyril A, Weitzdoerfer R, Gulesserian T, Fountoulakis M, Lubec G. Aberrant expression of signaling-related proteins 14-3-3γ and RACK1 in fetal Down syndrome brain (trisomy 21) Electrophoresis. 2002;23:152–157. doi: 10.1002/1522-2683(200201)23:1<152::AID-ELPS152>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 64.Hughes AJ, Ben-Shlomo Y, Daniel SE, Lees AJ. What features improve the accuracy of clinical diagnosis in Parkinson's disease: a clinicopathologic study. Neurology. 1992;42:1142–1146. doi: 10.1212/WNL.42.6.1142. [DOI] [PubMed] [Google Scholar]
- 65.Chen H, Burton EA, Webster RG, et al. Research on the premotor symptoms of Parkinson's disease: clinical and etiological implications. Environ. Health Perspect. 2013;121:11–12. doi: 10.1289/ehp.121-A342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sveinbjornsdottir S. The clinical symptoms of Parkinson's disease. J Neurochemistry. 2016;139:318–324. doi: 10.1111/jnc.13691. [DOI] [PubMed] [Google Scholar]
- 67.Jellinger KA. Formation and development of Lewy pathology: a critical update. J Neurology. 2009;256:270–279. doi: 10.1007/s00415-009-5243-y. [DOI] [PubMed] [Google Scholar]
- 68.Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9:13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- 69.Wong YC, Krainc D. α-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nat Med. 2017;23:1–13. doi: 10.1038/nm.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostrerova N, Petrucelli L, Farrer M, et al. alpha-Synuclein shares physical and functional homology with 14-3-3 proteins. J Neurosci. 1999;19:5782–5791. doi: 10.1523/JNEUROSCI.19-14-05782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawamoto Y, Akiguchi I, Nakamura S, Honjyo Y, Shibasaki H, Budka H. 14-3-3 proteins in Lewy bodies in Parkinson disease and diffuse Lewy body disease brains. J Neuropathol Exp Neurol. 2002;61:245–253. doi: 10.1093/jnen/61.3.245. [DOI] [PubMed] [Google Scholar]
- 72.Berg D, Riess O, Bornemann A. Specification of 14-3-3 proteins in Lewy bodies. Ann Neurol. 2003;53:135. doi: 10.1002/ana.10621. [DOI] [PubMed] [Google Scholar]
- 73.Yacoubian TA, Slone SR, Harrington AJ, et al. Differential neuroprotective effects of 14-3-3 proteins in models of Parkinson's disease. Death Dis. 2010;1:e2. doi: 10.1038/cddis.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding H, Underwood R, Lavalley N, Yacoubian TA. 14-3-3 inhibition promotes dopaminergic neuron loss and 14-3-3q overexpression promotes recovery in the MPTP mouse model of Parkinson's disease. Neuroscience. 2015;307:73–82. doi: 10.1016/j.neuroscience.2015.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu Z, Graham K, Foote M, et al. 14-3-3 protein targets misfolded chaperone-associated proteins to aggresomes. J Cell Sci. 2013;126:4173–4186. doi: 10.1242/jcs.126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jia B, Wu Y, Zhou Y. 14-3-3 and aggresome formation: implications in neurodegenerative diseases. Prion. 2014;8:173–177. doi: 10.4161/pri.28123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plotegher N, Kumar D, Tessari I, et al. The chaperone-like protein 14-3-3η interacts with human α-synuclein aggregation intermediates rerouting the amyloidogenic pathway and reducing α-synuclein cellu-lar toxicity. Hum Mol Genet. 2014;23:5615–5629. doi: 10.1093/hmg/ddu275. [DOI] [PubMed] [Google Scholar]
- 78.Sato S, Chiba T, Sakata E, et al. 14-3-3eta is a novel regulator of parkin ubiquitin ligase. EMBO J. 2006;25:211–221. doi: 10.1038/sj.emboj.7600774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J, Lou H, Pedersen CJ, Smith AD, Perez RG. 14-3-3zeta contributes to tyrosine hydroxylase activity in MN9D cells: localization of dopamine regulatory proteins to mitochondria. J Biol Chem. 2009;284:14011– 14019. doi: 10.1074/jbc.M901310200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scheltens P, Blennow K, Breteler MM, et al. Alzheimer's disease. Lancet. 2016;388:505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 81.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol Med. 2016;8:595– 608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iqbal K, Liu F, Gong CX, Grundke-Iqbal I. Tau in Alzheimer disease and related tauopathies. Curr Alzheimer Res. 2010;7:656–664. doi: 10.2174/156720510793611592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Layfield R, Fergusson J, Aitken A, Lowe J, Landon M, Mayer RJ. Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci Lett. 1996;209:57–60. doi: 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- 84.Fountoulakis M, Cairns N, Lubec G. Increased levels of 14-3-3γ and ε proteins in brain of patients with Alzheimer's disease and Down syndrome. J Neural Transm Suppl. 1999;57:323–335. doi: 10.1007/978-3-7091-6380-1_23. [DOI] [PubMed] [Google Scholar]
- 85.Gu O, Cuevas E, Raymick J, Kanungo J, Sarkar S. Downregulation of 14-3-3 proteins in Alzheimer's Disease. Mol Neurobiol. 2020;57:32–40. doi: 10.1007/s12035-019-01754-y. [DOI] [PubMed] [Google Scholar]
- 86.National Institute of Neurological Disorders and Stroke, author. Creutzfeldt-Jakob Disease Fact Sheet. NINDS. 2003 https://www.ninds.nih.gov/Disorders/Patient-Caregiver-Education/Fact-Sheets/Creutzfeldt-Jakob-Disease-Fact-Sheet .
- 87.Spero M, Lazibat I. Creutzfeldt-Jakob disease: case report and review of the literature. Acta Clin Croat. 2010;49:181–187. [PubMed] [Google Scholar]
- 88.Hsich G, Kenney K, Gibbs CJ, Lee KH, Harrington MG. The 14-3-3 brain protein in cerebrospinal fluid as a marker for transmissible spongiform encepha-lopathies. N Engl J Med. 1996;335:24–30. doi: 10.1056/NEJM199609263351303. [DOI] [PubMed] [Google Scholar]
- 89.Zerr I, Bodemer M, Gefeller O, et al. Detection of 14-3-3 protein in the cerebrospinal fluid supports the diagnosis of Creutzfeldt-Jakob disease. Ann Neurol. 1998;43:32–40. doi: 10.1002/ana.410430109. [DOI] [PubMed] [Google Scholar]
- 90.Lemstra AW, van Meegen MT, Vreyling JP, et al. 14-3-3 testing in diagnosing Creutzfeldt-Jakob disease: a prospective study in 112 patients. Neurology. 2000;55:514–516. doi: 10.1212/WNL.55.4.514. [DOI] [PubMed] [Google Scholar]
- 91.Burkhard PR, Sanchez JC, Landis T, Hochstrasser DF, et al. CSF detection of the 14-3-3 protein in unselec-ted patients with dementia. Neurology. 2001;56:1528–1533. doi: 10.1212/WNL.56.11.1528. [DOI] [PubMed] [Google Scholar]
- 92.Wiltfang J, Otto M, Baxter HC, et al. Isoform pattern of 14-3-3 proteins in the cerebrospinal fluid of patients with Creutzfeldt-Jakob disease. J Neurochem. 1999;73:2485–2490. doi: 10.1046/j.1471-4159.1999.0732485.x. [DOI] [PubMed] [Google Scholar]
- 93.Shiga Y, Wakabayashi H, Miyazawa K, Kido H, Itoyama Y. 14-3-3 protein levels and isoform patterns in the cerebrospinal fluid of Creutzfeldt-Jakob disease patients in the progressive and terminal stages. J Clin Neurosci. 2006;13:661–665. doi: 10.1016/j.jocn.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 94.Matsui Y, Satoh K, Miyazaki T, et al. High sensitivity of an ELISA kit for detection of the gamma-isoform of 14-3-3 proteins: usefulness in laboratory diagnosis of human prion disease. BMC Neurol. 2011;11:120. doi: 10.1186/1471-2377-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leitão MJ, Baldeiras I, Almeida MR, et al. Sporadic Creutzfeldt-Jakob disease diagnostic accuracy is improved by a new CSF ELISA 14-3-3γ assay. Neuroscience. 2016;322:398–407. doi: 10.1016/j.neuroscience.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 96.Kilani RT, Maksymowych WP, Aitken A, et al. Detection of high levels of 2 specific isoforms of 14-3-3 proteins in synovial fluid from patients with joint inflam-mation. J Rheumatol. 2007;34:1650–1657. [PubMed] [Google Scholar]
- 97.Sardari K, Chavez-Muñoz C, Kilani RT, Schiller T, Ghahary A. Increased levels of the 14-3-3 η and γ proteins in the synovial fluid of dogs with unilateral cranial cruciate ligament rupture. Can J Vet Res. 2011;75:271–277. [PMC free article] [PubMed] [Google Scholar]
- 98.Lee D, Steinacker P, Seubert S, et al. Role of glial 14-3-3 gamma protein in autoimmune demyelination. J Neuroinflammation. 2015;6:187. doi: 10.1186/s12974-015-0381-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dendrou CA, Fugger L, Friese MA. Immuno-pathology of multiple sclerosis. Nat Rev Immunol. 2015;15:545–558. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- 100.Nakahara J, Maeda M, Aiso S, Suzuki N. Current concepts in multiple sclerosis: autoimmunity versus oligodendrogliopathy. Clin Rev Allergy Immunol. 2012;42:26–34. doi: 10.1007/s12016-011-8287-6. [DOI] [PubMed] [Google Scholar]
- 101.Constantinescu CS, Farooqi N, O'Brien K, Gran B. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS) Br J Pharmacol. 2011;164:1079–1106. doi: 10.1111/j.1476-5381.2011.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pirim I. Ischemic rat brains contain immuno-reactivity of 14-3-3 proteins. Int J Neurosci. 1998;95:101–106. doi: 10.3109/00207459809000653. [DOI] [PubMed] [Google Scholar]
- 103.Kawamoto Y, Akiguchi I, Tomimoto H, Shirakashi Y, Honjo Y, Budka H. Upregulated expression of 14-3-3 proteins in astrocytes from human cerebrovas-cular ischemic lesions. Stroke. 2006;37:830–835. doi: 10.1161/01.STR.0000202587.63936.37. [DOI] [PubMed] [Google Scholar]
- 104.Fujii K, Tanabe Y, Kobayashi K, Uchikawa H, Kohno Y. Detection of 14-3-3 protein in the cerebro-spinal fluid in mitochondrial encephalopathy with lactic acidosis and stroke-like episodes. J Neurol Sci. 2005;239:115– 118. doi: 10.1016/j.jns.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 105.Lai XJ, Ye SQ, Zheng L, et al. Selective 14-3-3γ induction quenches p-β-catenin Ser37/Bax-enhanced cell death in cerebral cortical neurons during ischemia. Cell Death Dis. 2014;5:e1184. doi: 10.1038/cddis.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jang SW, Liu X, Fu H, et al. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. J Biol Chem. 2009;284:24512–24525. doi: 10.1074/jbc.M109.026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou XY, Hu DX, Chen RQ, et al. 14-3-3 Isoforms differentially regulate NFκB signaling in the brain after ischemia-reperfusion. Neurochem Res. 2017;42:2354–2362. doi: 10.1007/s11064-017-2255-3. [DOI] [PubMed] [Google Scholar]
- 108.Otsuka S, Sakakima H, Terashi T, Takada S, Nakanishi K, Kikuchi K. Preconditioning exercise reduces brain damage and neuronal apoptosis through enhanced endogenous 14-3-3γ after focal brain ischemia in rats. Brain Struct Funct. 2019;224:739–740. doi: 10.1007/s00429-018-1815-x. [DOI] [PubMed] [Google Scholar]
- 109.Dong Y, Liu HD, Zhao R, et al. Ischemia activates JNK/c-Jun/AP-1 pathway to up-regulate 14-3-3gamma in astrocyte. J Neurochem. 2009;109:182–188. doi: 10.1111/j.1471-4159.2009.05974.x. [DOI] [PubMed] [Google Scholar]
- 110.Chen XQ, Yu AC. The association of 14-3-3gamma and actin plays a role in cell division and apoptosis in astrocytes. Biochem Biophys Res Commun. 2002;296:657–663. doi: 10.1016/S0006-291X(02)00895-1. [DOI] [PubMed] [Google Scholar]
- 111.Chen XQ, Chen JG, Zhang Y, Hsiao WW, Yu AC. 14-3-3gamma is upregulated by in vitro ischemia and binds to protein kinase Raf in primary cultures of astrocytes. Glia. 2003;42:315–324. doi: 10.1002/glia.10185. [DOI] [PubMed] [Google Scholar]
- 112.Umahara T, Uchihara T, Tsuchiya K, et al. Intra-nuclear localization and isoform-dependent translocation of 14-3-3 proteins in human brain with infarction. J Neurol Sci. 2007;260:159–166. doi: 10.1016/j.jns.2007.04.053. [DOI] [PubMed] [Google Scholar]
- 113.Chen XQ, Fung YW, Yu AC. Association of 14-3-3gamma and phosphorylated bad attenuates injury in ischemic astrocytes. J Cereb Blood Flow Metab. 2005;25:338–347. doi: 10.1038/sj.jcbfm.9600032. [DOI] [PubMed] [Google Scholar]
- 114.Dong Y, Liu HD, Zhao R, et al. Ischemia activates JNK/c-Jun/AP-1 pathway to up-regulate 14-3-3gamma in astrocyte. J Neurochem. 2009;109:182–188. doi: 10.1111/j.1471-4159.2009.05974.x. [DOI] [PubMed] [Google Scholar]
- 115.Pang Y, Chai CR, Gao K, et al. Ischemia preconditioning protects astrocytes from ischemic injury through 14-3-3γ. J Neurosci Res. 2015;93:1507–1518. doi: 10.1002/jnr.23574. [DOI] [PubMed] [Google Scholar]
- 116.Hermeking H, Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol. 2006;16:183–192. doi: 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 117.Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gardino AK, Yaffe MB. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol. 2011;22:688–695. doi: 10.1016/j.semcdb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jin Y, Dai MS, Lu SZ, et al. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 2006;25:1207–1218. doi: 10.1038/sj.emboj.7601010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee JH, Lu H. 14-3-3gamma inhibition of MDMX-mediated p21 turnover independent of p53. J Biol Chem. 2011;286:5136–5142. doi: 10.1074/jbc.M110.190470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cao WD, Zhang X, Zhang JN, et al. Immuno-cytochemical detection of 14-3-3 in primary nervous system tumors. J Neurooncol. 2006;77:125–130. doi: 10.1007/s11060-005-9027-7. [DOI] [PubMed] [Google Scholar]
- 122.Cao L, Cao W, Zhang W, et al. Identification of 14-3-3 protein isoforms in human astrocytoma by immunohistochemistry. Neurosci Lett. 2008;432:94–99. doi: 10.1016/j.neulet.2007.11.071. [DOI] [PubMed] [Google Scholar]
- 123.Yang X, Cao W, Lin H, et al. Isoform-specific expression of 14-3-3 proteins in human astrocytoma. J Neurol Sci. 2009;276:54–59. doi: 10.1016/j.jns.2008.08.040. [DOI] [PubMed] [Google Scholar]
- 124.Liang S, Xu Y, Shen G, et al. Quantitative protein expression profiling of 14-3-3 isoforms in human renal carcinoma shows 14-3-3 epsilon is involved in limitedly increasing renal cell proliferation. Electrophoresis. 2009;30:4152–4162. doi: 10.1002/elps.200900249. [DOI] [PubMed] [Google Scholar]
- 125.Com E, Clavreul A, Lagarrigue M, Michalak S, Menei P, Pineau C. Quantitative proteomic Isotope-Coded Protein Label (ICPL) analysis reveals alteration of several functional processes in the glioblastoma. J Proteomics. 2012;75:3898–3913. doi: 10.1016/j.jprot.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 126.Cao W, Yang X, Zhou J, et al. Targeting 14-3-3 protein, difopein induces apoptosis of human glioma cells and suppresses tumor growth in mice. Apoptosis. 2010;15:230–241. doi: 10.1007/s10495-009-0437-4. [DOI] [PubMed] [Google Scholar]
- 127.Liu J, Liu Y, Ren Y, Kang L, Zhang L. Transmembrane protein with unknown function 16A overexpression promotes glioma formation through the nuclear factor-kappaB signaling pathway. Mol Med Rep. 2014;9:1068–1074. doi: 10.3892/mmr.2014.1888. [DOI] [PubMed] [Google Scholar]
- 128.Yao ZQ, Zhang X, Zhen Y, et al. A novel small-molecule activator of Sirtuin-1 induces autophagic cell death/mitophagy as a potential therapeutic strategy in glioblastoma. Cell Death Dis. 2018;9:767. doi: 10.1038/s41419-018-0799-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Steinacker P, Schwarz P, Reim K, et al. Unchanged Survival Rates of 14-3-3γ Knockout Mice after Inocula-tion with Pathological Prion Protein. Mol Cell Biol. 2005;25:1339–1346. doi: 10.1128/MCB.25.4.1339-1346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim DE, Cho CH, Sim KM, et al. 14-3-3γ haplo-insufficient mice display hyperactive and stress-sensitive behaviors. Exp Neurobiol. 2019;28:43–45. doi: 10.5607/en.2019.28.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheah PS, Ramshaw HS, Thomas PQ, et al. Neurodevelopmental and neuropsychiatric behavior defects arise from 14-3-3zeta deficiency. Mol Psychiatry. 2012;17:451–466. doi: 10.1038/mp.2011.158. [DOI] [PubMed] [Google Scholar]
- 132.Xu X, Jaehne EJ, Greenberg Z, et al. 14-3-3zeta deficient mice in the BALB/c background display behavioral and anatomical defects associated with neurodevelopmental disorders. Sci Rep. 2015;5:12434. doi: 10.1038/srep12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jaehne EJ, Ramshaw H, Xu X, et al. In-vivo administration of clozapine affects behaviour but does not reverse dendritic spine deficits in the 14-3-3ξ KO mouse model of schizophrenia-like disorders. Pharmacol Biochem Behav. 2015;138:1–8. doi: 10.1016/j.pbb.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 134.Ikeda M, Hikita T, Taya S, et al. Identification of YWHAE, a gene encoding 14-3-3epsilon, as a possible susceptibility gene for schizophrenia. Hum Mol Genet. 2008;17:3212–3222. doi: 10.1093/hmg/ddn217. [DOI] [PubMed] [Google Scholar]
- 135.Wachi T, Cornell B, Toyo-Oka K. Complete ablation of the 14-3-3epsilon protein results in multiple defects in neuropsychiatric behaviors. Behav Brain Res. 2017;319:31–36. doi: 10.1016/j.bbr.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]