Abstract
Aim:
Michigan residents were exposed to polybrominated biphenyls (PBBs) when it was accidentally added to the food supply. Highly exposed individuals report sex-specific health problems, but the underlying biological mechanism behind these different health risks is not known.
Materials and methods:
DNA methylation in blood from 381 women and 277 men with PBB exposure was analyzed with the MethylationEPIC BeadChip.
Results:
675 CpGs were associated with PBBs levels in males, while only 17 CpGs were associated in females (false discovery rate <0.05). No CpGs were associated in both sexes. These CpGs were enriched in different functional regions and transcription factor binding sites in each sex.
Conclusion:
Exposure to PBBs may have sex-specific effects on the epigenome that may underlie sex-specific adverse health outcomes.
Keywords: : DNA methylation, EDC, endocrine-disrupting compound, environmental health, epigenetics, epigenome-wide association study, EWAS, polybrominated biphenyl, sex differences, sex specific
Exposure to endocrine-disrupting compounds (EDCs), a diverse class of chemicals defined by their ability to disrupt the function or synthesis of hormones, is common in the modern world because they are present in many products that people come in contact with, such as pesticides, plastics, electronics and household dust [1–6]. Despite their prevalence, increased exposure to EDCs has been linked with several adverse health outcomes. Studies in animal models and human cohorts have demonstrated that increased EDC exposure is associated with altered thyroid hormone levels, lower birth weight and cancer [7–12]. However, there have also been sex-specific effects associated with EDC exposure [13]. For example, in males, EDC exposure has been associated with decreases in sperm count, infertility and prostate cancer [14–18], while in females, EDC exposure has been associated with altered menstrual cycles, infertility and breast cancer [19–24]. Additionally, EDCs have sex-specific effects on endocrine tissues and developmental milestones [25] and can even affect nonendocrine tissues, like neurons, and behaviors in a sex-specific manner [26–29].
How sex-specific health effects occur following exposure to EDCs is not well understood. One potential mechanism that might underlie sex-specific health effects is the activation of distinct biological pathways by EDCs in males and females. These distinct biological pathways would then lead to distinct epigenetic marks and distinct gene expression patterns and thus lead to different health outcomes. In animal models, exposure to other estrogenic EDCs has been linked to reduced gene expression in male gonads and liver [30,31]. Additionally, while female rats exposed to EDCs had more genes with altered expression, the genes altered in male brains were more likely to be genes involved in autism spectrum disorder [32,33]. EDCs also downregulated miRNAs in the hypothalamus of male rats, but upregulated in the hypothalamus of female rats [34]. In human cohorts, environmental exposures like arsenic are associated with global increases in H3K36me2 in men, but are negatively associated with H3K36me2 in women [35]. Polychlorinated biphenyl, an EDC, had a stronger effect in males in one study [36] but in another study, the effect was stronger in females [37]. Additionally, other persistent EDCs, like p,p′-dichlorodiphenyldichloroethylene, have been associated with a much stronger epigenetic effect in males, with many of the associated CpGs being located on the X-chromosome, indicating that there are sex-specific responses to some environmental contaminants [37].
To further study the sex-specific effects of EDCs, we utilized the Michigan Polybrominated Biphenyl (PBB) Registry. This registry was developed after 6–8 million people in Michigan were exposed to PBB, an EDC, that was accidentally introduced into the food supply in 1973 [38,39]. Because PBBs are highly lipophilic and biologically stable, members of this registry still have high levels of exposure to PBBs. Because PBBs can cross the placental barrier and are present in breast milk, offspring of women who were exposed are also exposed to PBBs during development [40–45]. Previous epidemiological research using this registry has reported many associations between PBB exposure and sex-specific health outcomes. For example, women born to exposed mothers have an earlier age of menarche and an increased risk of spontaneous abortion [46,47], whereas men born to exposed mothers have slower pubertal development and an increased risk for genitourinary conditions [48,49]. Additionally, women who were exposed during the incident have an increased risk of breast cancer and thyroid disease [8,50,51]. Previous epigenetic research conducted in both sexes found that PBBs were linked to methylation of 1890 CpGs; the study also reported that PBB-associated CpGs were correlated with estradiol-associated CpGs and were enriched for estrogen-related transcription factor binding sites [52–54]. This supports further epidemiological research which found that higher PBB exposure was associated with lower levels of estrogen and progesterone metabolites [55] and with in vitro work that suggested PBB-153 had some ability to activate the estrogen receptor [56].
While PBBs have been associated with DNA methylation [52], that study would only detect CpGs associated in both sexes, unless the CpG had a large effect in one of the sexes. Additionally, estrogenic EDCs are expected to affect males and females differently and no studies have investigated whether there are sex-specific differences in the association between PBBs and DNA methylation. This study uses Illumina’s MethylationEPIC BeadChip (CA, USA) to test for associations between PBBs and DNA methylation in each sex separately. We then use existing databases to test whether the CpGs significant in each sex differ in the biological pathways or functional positions in a way that may give insight into how PBBs are acting in each sex.
Methods
Participant selection
As previously described [8,52,57], participants were recruited from the Michigan PBB Registry, which was started by the Michigan state health department after the contamination incident. Participants were included in this study if they had existing PBB levels, lipid levels and DNA methylation data from the same blood sample [52,57]. A total of 666 participants (386 women and 280 men) were included. Informed consent was obtained from each participant. The Institutional Review Board at Emory University (Atlanta, GA, USA) approved all study protocols.
Exposure assessment
Exposure to PBBs was assessed as previously described [8,41,52,57]. Briefly, exposure to four congeners of PBB (PBB-153, PBB-101, PBB-77 and PBB-180) was assessed using gas chromatography-tandem mass spectrometry [41]. The value of congeners below the limit of detection in a sample was imputed as the limit of detection divided by the square root of 2 [58]. All congeners were positively correlated with the other congeners (r = 0.23–0.99) and also summed to give a total PBB level and transformed with a natural log for all analyses.
Epigenome-wide methylation assessment
DNA methylation was assessed on the MethylationEPIC BeadChip (Illumina) [59] as previously described [52] and it is available on the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GSE116339). Quality control (QC) and cell type estimation were conducted, as previously described [52] and it was repeated for male and female samples separately in order to protect sex-specific epigenetic patterns. 8106 probes were removed for missing data in the male dataset and 4468 probes were removed for missing data in the female dataset. This resulted in a final dataset of 277 male participants with 814,920 probes and 381 female participants with 818,441 probes. With this number of samples and tests, there is 80% power to detect an association with the interaction between the level of PBBs and sex that explains at least 8% of the variance in methylation levels (in the combined dataset) and 16% of the variance in methylation levels in males and 12% of the variation in methylation levels in females using within group analyses of PBB levels [60].
Statistical analysis & software
R 3.6.1 was used for all analyses. Linear regression models were implemented in CpGassoc and were used to test the interaction between sex and total level of PBBs (sex × total PBBs) and methylation proportion at each CpG that passed QC in the combined sex dataset [61]. For this analysis, total PBB level, sex, age, lipid level and cell type proportions were covariates because of their reported association with PBB and DNA methylation [52–54,62–64]. Because this sample size was underpowered to detect anything but large interactions between sex and PBBs, the association between total PBB level and methylation proportion was next assessed at every probe that passed QC in males and females separately, with age, lipid level and cell type proportions as covariates, using linear models in CpGassoc. A Benjamini–Hochberg false discovery rate (FDR) of less than 0.05 was used to adjust for multiple testing [65]. MissMethyl was used to determine whether any of the gene ontology (GO) pathways were enriched in the results from each sex (FDR <0.05) in order to correct for the number of probes in each gene [66]. Pearson’s correlation coefficients were used to correlate effect sizes from each of the sex-specific epigenetic analyses to the existing PBB epigenetic analysis that was conducted in both sexes combined [52]. We also subset the women who were postmenopausal (either by self-report or over age 55) and analyzed the association between PBBs and DNA methylation. The results in these women were compared with the results from men using a Pearson’s correlation coefficient. We also stratified both male and female groups by age and PBB exposure level (by median split) and compared the results from each subtype using a Pearson’s correlation coefficients.
Chi-square tests were used to determine whether any CpG sites most associated with PBBs (FDR <0.2) were enriched based on their functional role. This relaxed statistical cut-off was selected to make the groups of CpGs between males and females more comparable. As previously described, CpG positions and potential functional role by the annotation provided by the manufacturer and the publicly available ChromHMM tracks [52,67–69]. An α level of 0.05 was used to determine statistical significance for the positional and functional enrichment. Enrichment of transcription factor binding sites in the PBB-associated CpGs (FDR <0.05) was determined by oPOSSUM-3 as previously described [52,70]. A Benjamini–Hochberg FDR of less than 0.05 was used to adjust for multiple testing.
Results
Study population characteristics
The study population is composed of 381 women and 277 men who are primarily White/non-Hispanic. The population’s racial composition is indicative of the population structure of rural Michigan in the 1970s when the exposure incident occurred. The male participants had a higher PBB exposure on an average than the female participants (range in males: 0.01–236.73 ng/ml, range in females: 0.01–129.05 ng/ml, Supplementary Figure 1, Table 1). The male participants were also older than the female participants (range in males: 33.1–88.4 years, median: 57.8 years; range in females: 23.0–85.3 years, median: 50.0 years; Supplementary Figure 2). Age at sample collection and current PBB level were correlated in male participants (r = 0.39, p = 1.64e-11), but not in female participants (r = 0.06, p = 0.21). Because a majority of the study population was exposed during the year-long contamination incident, age when exposed to PBBs is highly correlated with current age in both males (r = 0.99, p < 2.2e-16) and females (r = 0.96, p < 2.2e-16). Current lipid levels and current PBB levels were not correlated in either sex (males: r = -0.07, p = 0.18; females: r = -0.02, p = 0.63). Estimated cell proportions also differed by sex with women having higher proportions of CD8T and CD4T cells and men having higher proportions of granulocytes, monocytes and natural killer cells (Supplementary Table 1).
Table 1. . Demographics table. Age, race and exposure information for the members of Michigan PBB Registry by sex.
| Males (n = 277) | Females (n = 381) | p-value | |
|---|---|---|---|
| Age when sample collected (years)† | 58.58 ± 11.96 | 51.16 ± 12.40 | 4.44e-14 |
| Age when exposed (years)† | 18.54 ± 11.79 | 12.73 ± 10.73 | 2.05e-10 |
| Total PBB level (ng/ml)‡ | 0.69 (5.17) | 0.37 (4.14) | 4.54e-7 |
| Total PBB level (ng/g lipid)‡ | 109.99 (5.43) | 54.29 (4.29) | 3.53e-8 |
| Race/ethnicity§ | 0.45 | ||
| White/non-Hispanic | 266 (96.03%) | 372 (97.64%) | |
| White/Hispanic | 11 (3.97%) | 9 (2.36%) |
Mean ± standard deviation.
Geometric mean (standard deviation).
Number (percentage).
PBB: Polybrominated biphenyl.
Interaction between PBB & sex
The interaction between PBBs and sex was tested at each CpG that passed QC in the dataset that contained both sexes. No CpGs were genome-wide significant, which is likely due to the sample size of this study and the multiple comparisons threshold needed for statistical significance. Six probes were associated with a p < 1e-5 (Supplementary Table 2).
Differences in DNA methylation by sex
Separating the sexes prior to filtering and normalizing the DNA methylation allowed for differences in DNA methylation in each sex to be maintained. For probes on autosomes, slightly fewer were retained in males than in females (Supplementary Table 3). However, for the sex chromosome probes, males had substantially fewer X chromosome probes pass QC (odds ratio [OR] = 0.74, p = 1.64e-15) and substantially more Y chromosome probes pass QC (OR = 14.68, p < 2.2e-16). Additionally, for the 814,635 probes that were retained in each sex, the mean methylation across all probes was highly correlated between the sexes (r = 0.99, p < 2.2e-16), but there were several probes that had large differences between mean methylation in females and males (range: -0.63–0.64; Supplementary Figure 3A). Likewise, the distribution of methylation was similar in each sex (r = 0.97, p < 2.2e-16), but there were several probes that had large differences between the interquartile range in females and the interquartile range in males (range: -0.43–0.40; Supplementary Figure 3B).
Association between PBB & DNA methylation differs by sex
To test for sex-specific associations between PBB levels and DNA methylation, the association between each probe and level of PBBs was assessed in each sex separately, controlling for age, lipid levels and cell type composition. In males, 675 of the 814,920 CpGs were associated with PBB levels (Figure 1A, Supplementary Table 4), while in females 17 of the 818,441 CpGs were associated with PBB levels (Figure 1B, Supplementary Table 5). Six CpGs on the X chromosome associated with PBBs in males, while none of the results in females were on the sex chromosomes.
Figure 1. . Total polybrominated biphenyl level is associated with different CpGs in each sex.
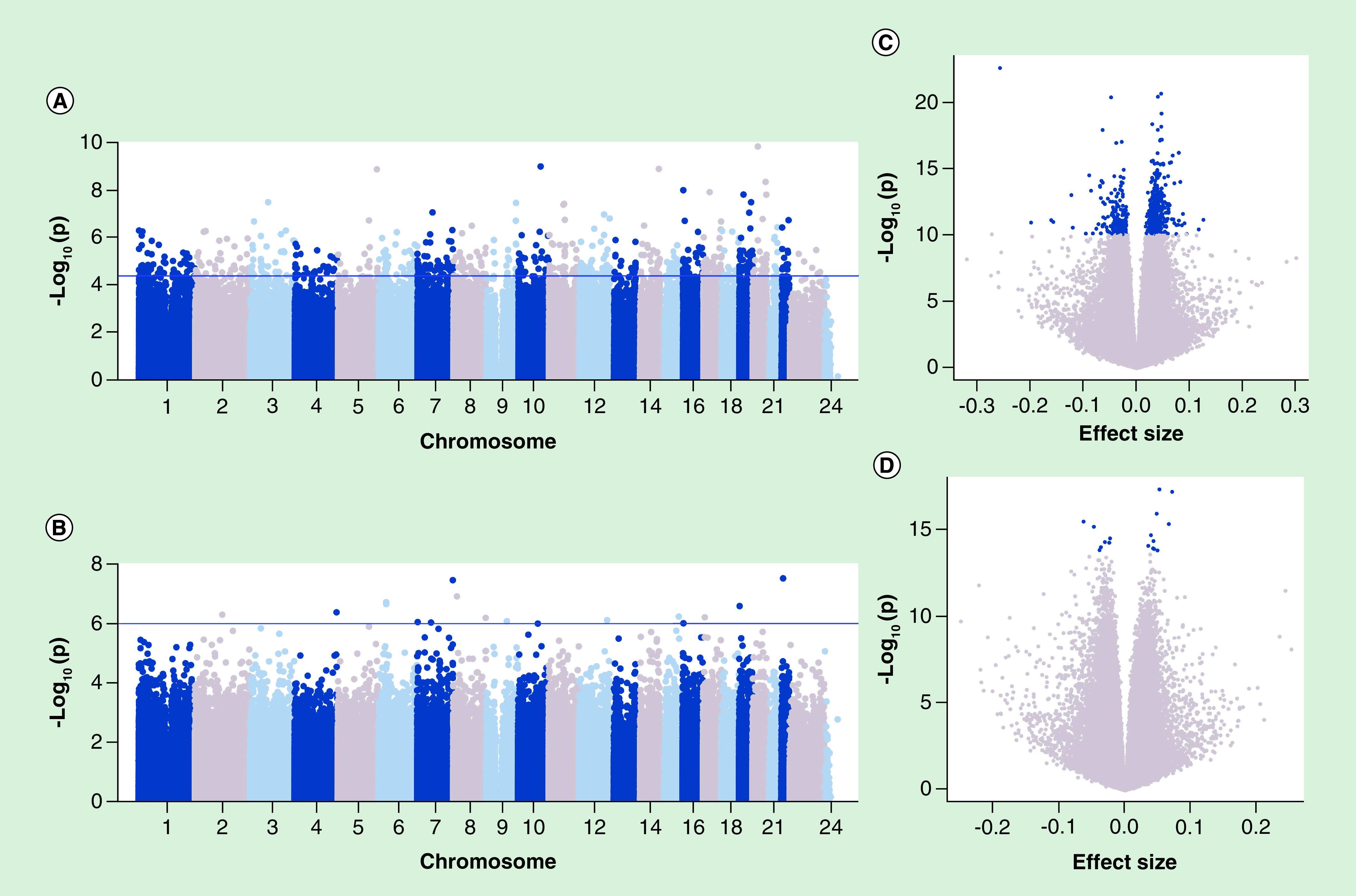
A Manhattan plot of the association of total PBB level with DNA methylation proportion at 814,920 sites in males (A) or 818,441 sites in females (B). The x-axis is the location of each site across the genome. The y-axis is the –log10 of the p-value for the association with PBBs. The blue line indicates statistical significance (FDR <0.05). In males, 675 CpGs had a statistically significant association with total PBB level, while in females, 17 CpGs were associated. In males, 71.85% of these significant CpGs (blue) had increasing methylation levels with higher PBB levels (C), while in females, 58.82% of these significant CpGs (blue) had increasing methylation levels with higher PBB levels (D). The effect size is the beta coefficient from the regression of DNA methylation proportion on total PBB level, controlling for age, lipids and estimates cell type proportions.
FDR: False discovery rate; PBB: Polybrominated biphenyl.
Results from both analyses were moderately correlated with the results from the analysis in the combined dataset (males: r = 0.69, p < 2.2e-16; females: r = 0.70, p < 2.2e-16; Supplementary Figures 5 & 6) [52]. In males, methylation was positively associated with PBB levels in 71.85% of the significant CpGs (Figure 1C), while in females, a smaller proportion of CpGs (58.82%) had a positive association between methylation and PBB levels (Figure 1D). Additionally, none of the CpGs that were significant in the male analysis were significant in the female analysis. In general, there was little overlap among the significant CpGs identified in either sex (Figure 3, Table 2). Furthermore, while the direction of effect was the same 64% of the time, which is more than one would expect due to chance (p = 1.87e-14), 36% of the time the effects were in opposite directions. For the 692 CpGs significant in either sex, the effect sizes in males and females were only moderately correlated (r = 0.37, p < 2.2e-16; Figure 2). The correlation in effect sizes between males and females was similar if the association was compared in just people with higher exposure (r = 0.32, p < 2.2e-16), in older people (r = 0.29, p = 1.38e-14), in younger people (r = 0.36, p = 2.2e-16) and when postmenopausal women were compared with men (r = 0.37, p = 2.2e-16). The correlation in effect sizes between males and females was lower in people with lower exposure (r = 0.11, p = 0.002).
Figure 3. . Scatterplots of the CpGs most highly associated with total polybrominated biphenyl levels.
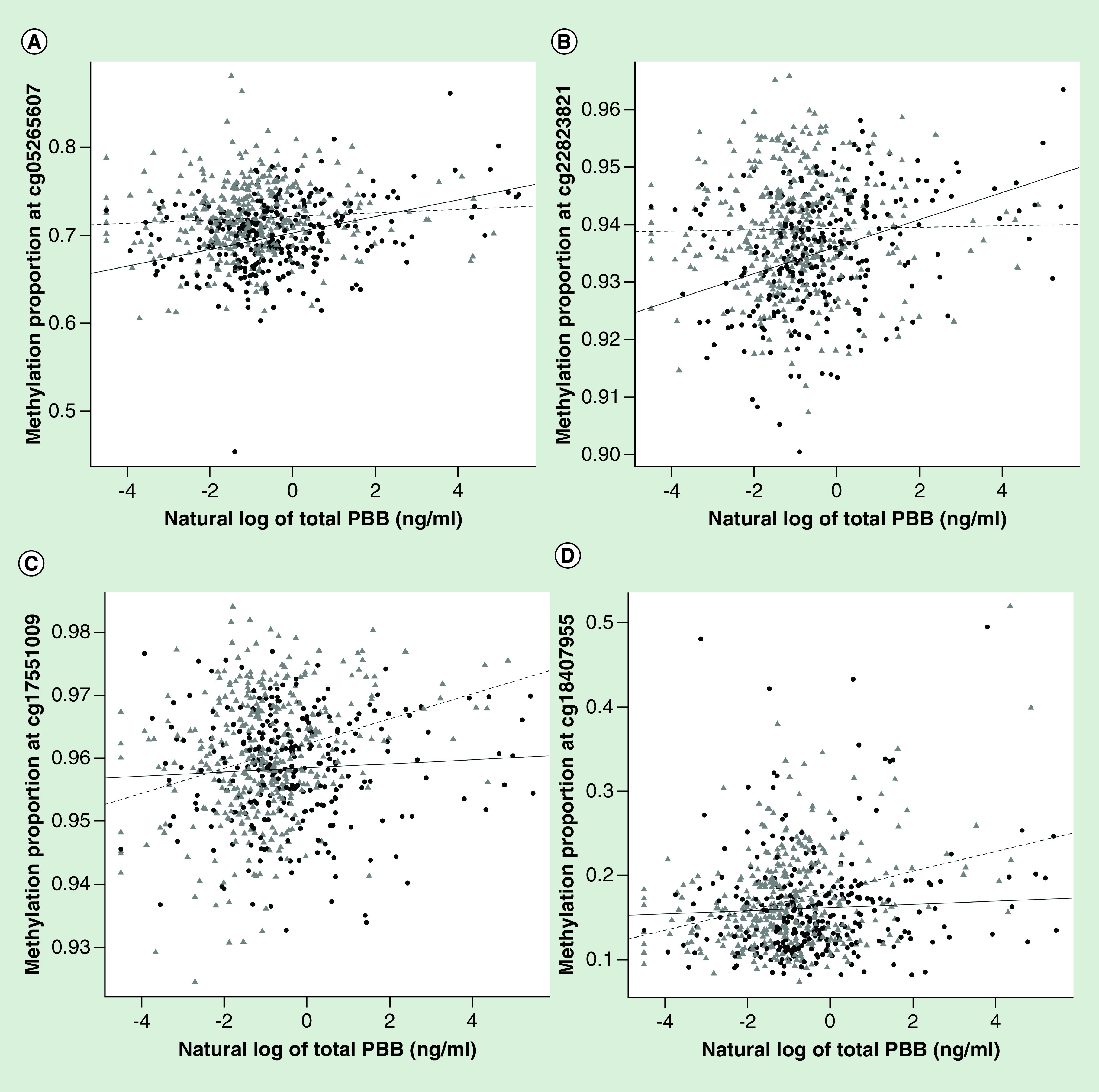
The top CpGs most associated with total PBB levels were only associated in either males (black circles) or females (gray triangles): cg05265607 (SLIT1, males: p = 1.02e-9; females: p = 0.17) (A) cg22823821 (MIR377, males: p = 1.28e-9; females: p = 0.72) (B) cg17551009 (MIF-AS1, males: p = 0.34; females: p = 2.97e-8) (C) cg18407955 (PTPRN2, males: p = 0.32; females: p = 3.41e-8) (D). The solid line is the regression of DNA methylation proportion at that CpG on total PBB level in males (controlling for age, lipids and estimates cell type proportions) and the dashed line is the regression of DNA methylation proportion at that CpG on total PBB levels in females (controlling for age, lipids and estimates cell type proportions).
PBB: Polybrominated biphenyl.
Table 2. . CpGs most associated with polybrominated biphenyl levels in each sex.
| CpGs most associated with PBB in males | ||||||
|---|---|---|---|---|---|---|
| CpG | Gene | Position | T-statistic in males | p-value in males | T-statistic in females | p-value in females |
| cg01471923 | SSTR4 | chr20:23015091 | -6.67 | 1.47E-10 | -0.84 | 0.39 |
| cg05265607 | SLIT1 | chr10:98900817 | 6.33 | 1.02E-09 | 1.35 | 0.17 |
| cg22823821 | MIR377 | chr14:101528332 | 6.29 | 1.28E-09 | 0.35 | 0.72 |
| cg07463740 | chr5:172737556 | -6.28 | 1.35E-09 | -1.81 | 0.07 | |
| cg12560768 | STX16 | chr20:57248316 | 6.06 | 4.54E-09 | 2.49 | 0.01 |
| cg08131081 | ABCA3 | chr16:2347567 | 5.91 | 1.01E-08 | 0.01 | 0.98 |
| cg14692377 | SLC6A4 | chr17:28562685 | 5.87 | 1.23E-08 | 1.52 | 0.12 |
| cg09934938 | CERS1 | chr19:19000923 | 5.83 | 1.55E-08 | 1.06 | 0.28 |
| cg16491680 | SSTR4 | chr20:60528285 | -5.83 | 1.57E-08 | -0.45 | 0.65 |
| cg16558906 | SLIT1 | chr3:78546329 | 5.69 | 3.26E-08 | 1.33 | 0.18 |
| CpGs most associated with PBB in females | ||||||
| CpG | Gene | Position | T-statistic in males | p-value in males | T-statistic in females | p-value in females |
| cg17551009 | MIF-AS1 | chr22:24242020 | 0.93 | 0.34 | 5.66 | 2.97E-08 |
| cg18407955 | PTPRN2 | chr7:158110685 | 0.99 | 0.32 | 5.63 | 3.41E-08 |
| cg20108327 | MTUS1 | chr8:17534512 | 0.64 | 0.52 | 5.39 | 1.20E-07 |
| cg13703714 | COL11A2 | chr6:33159593 | 1.14 | 0.25 | -5.31 | 1.88E-07 |
| cg15808668 | IP6K3 | chr6:33710837 | 2.31 | 0.02 | 5.28 | 2.17E-07 |
| cg06488678 | TMPRSS9 | chr19:2390040 | 1.40 | 0.16 | -5.25 | 2.54E-07 |
| cg16402161 | ACSL1 | chr4:185677585 | 1.22 | 0.22 | 5.15 | 4.13E-07 |
| cg27234239 | RALB | chr2:121010047 | -1.17 | 0.24 | -5.11 | 4.97E-07 |
| cg17095257 | PDE8A | chr15:85609662 | -0.10 | 0.91 | 5.08 | 5.79E-07 |
| cg01078620 | chr17:7548313 | -1.22 | 0.22 | -5.07 | 6.13E-07 | |
The ten CpGs most associated with PBBs in each sex were not associated with PBBs in the other sex.
PBB: Polybrominated biphenyl.
ƒ. . Effect sizes of polybrominated biphenyl-associated CpGs is only moderately correlated between males and females.
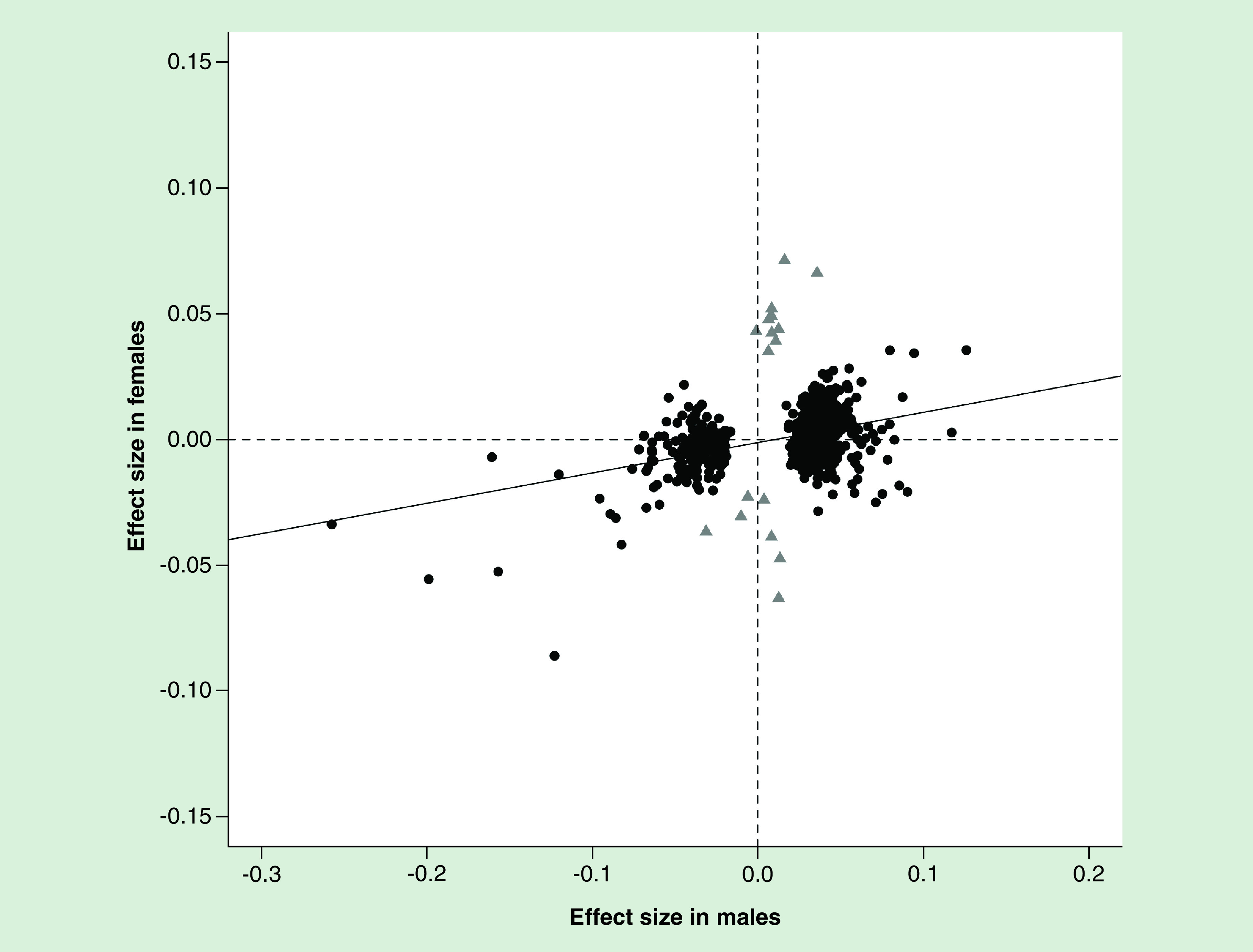
Out of the 692 CpGs associated with PBBs in either sex (FDR <0.05), the effect size for the analysis in males was only moderately correlated with the effect size in females (r = 0.37, p < 2.2e-16). Generally, the CpGs associated with PBBs in females (gray triangles) had a very small effect in males and the CpGs associated with PBBs in males (black circles) had a very small effect in females. The effect size is the β coefficient from the regression model. The solid line is the regression line between the two effect sizes. The dashed lines are an effect size of zero.
FDR: False discovery rate; PBB: Polybrominated biphenyl.
In males, 484 of the 675 CpGs (71.7%) associated with PBBs were annotated to genes. Many of the results were in genes that are members of the cytochrome P450 family (CYP26C1, CYP2S1 and CYP3A7), are endocrine-related (SSTR4 and STX16) or are involved in gene regulation (HDAC4, MIR1268A, MIR1975, MIR377, ZNF211, ZNF418, ZNF507, ZNF 509 and ZNF71, Supplementary Table 3). In females, 14 of the 17 CpGs (82.4%) associated with PBBs were annotated to genes. Some of these results are in genes that are involved in endocrine-related disease (PTPRN2) or immune function (SEMA4D, Supplementary Table 4).
To test whether there were distinct biological pathways enriched in males and females, enrichment for GO terms was investigated in the results from each sex. Two terms were significantly enriched in the results from males (GO:0007156: homophilic cell adhesion via plasma membrane adhesion molecules and GO:0098742: cell-to-cell adhesion via plasma-membrane adhesion molecules), but no GO terms were enriched in the results from females. Importantly, the top terms enriched in males were not enriched in the top terms in females and the top terms enriched in or the top terms in females were not enriched in males (Table 3, Supplementary Table 6).
Table 3. . Gene ontology pathways most enriched in polybrominated biphenyl-associated CpGs.
| Gene ontology | Term | p-value in males | FDR in males | p-value in females | FDR in females |
|---|---|---|---|---|---|
| GO:0007156 | Homophilic cell adhesion via plasma membrane adhesion molecules | 6.30E-09 | 0.0001 | 1 | 1 |
| GO:0098742 | Cell-cell adhesion via plasma-membrane adhesion molecules | 1.12E-06 | 0.01 | 1 | 1 |
The top GO pathways with PBB-associated CpGs in males were not associated enriched in the females (no pathways were significantly enriched in females).
FDR: False discovery rate; GO: Gene ontology; PBB: Polybrominated biphenyl.
Enrichment in functional regions differs by sex
Next we sought to test whether different functional regions were impacted by PBBs in males and females. The top CpGs associated with PBBs in males were enriched in polycomb regions (OR = 1.08, 95% CI: 1.04–1.12, p = 3.73e-5), regions of low activity near high activity areas (OR = 1.19, 95% CI: 1.14–1.24, p = 1.50e-15), transcription-associated regions (OR = 1.30, 95% CI: 1.25–1.36, p < 2.2e-16) and promoter flanking regions (OR = 1.12, 95% CI: 1.00–1.24, p = 0.03). The same were depleted in heterochromatin regions (OR = 0.93, 95% CI: 0.90–0.97, p = 0.002), insulator regions (OR = 0.87, 95% CI: 0.79–0.95, p = 0.002) and inactive and active promoters (inactive promoter: OR = 0.75, 95% CI: 0.68–0.82, p = 6.30e-10; active promoter: OR = 0.71, 95% CI: 0.68–0.74, p < 2.2e-16). In females, the top CpGs associated with PBBs were enriched with transcription-associated regions (OR = 1.37, 95% CI: 1.16–1.60, p = 0.0001) and strong and weak enhancers (strong enhancers: OR = 1.39, 95% CI: 1.00–1.87, p = 0.04; weak enhancers: OR = 1.32, 95% CI: 1.03–1.67, p = 0.02) and were depleted in active promoters (OR = 0.70, 95% CI: 0.58–0.83, p = 0.0001; Figure 4).
Figure 4. . CpGs most associated with polybrominated biphenyls in each sex are enriched in positional and functional regions.
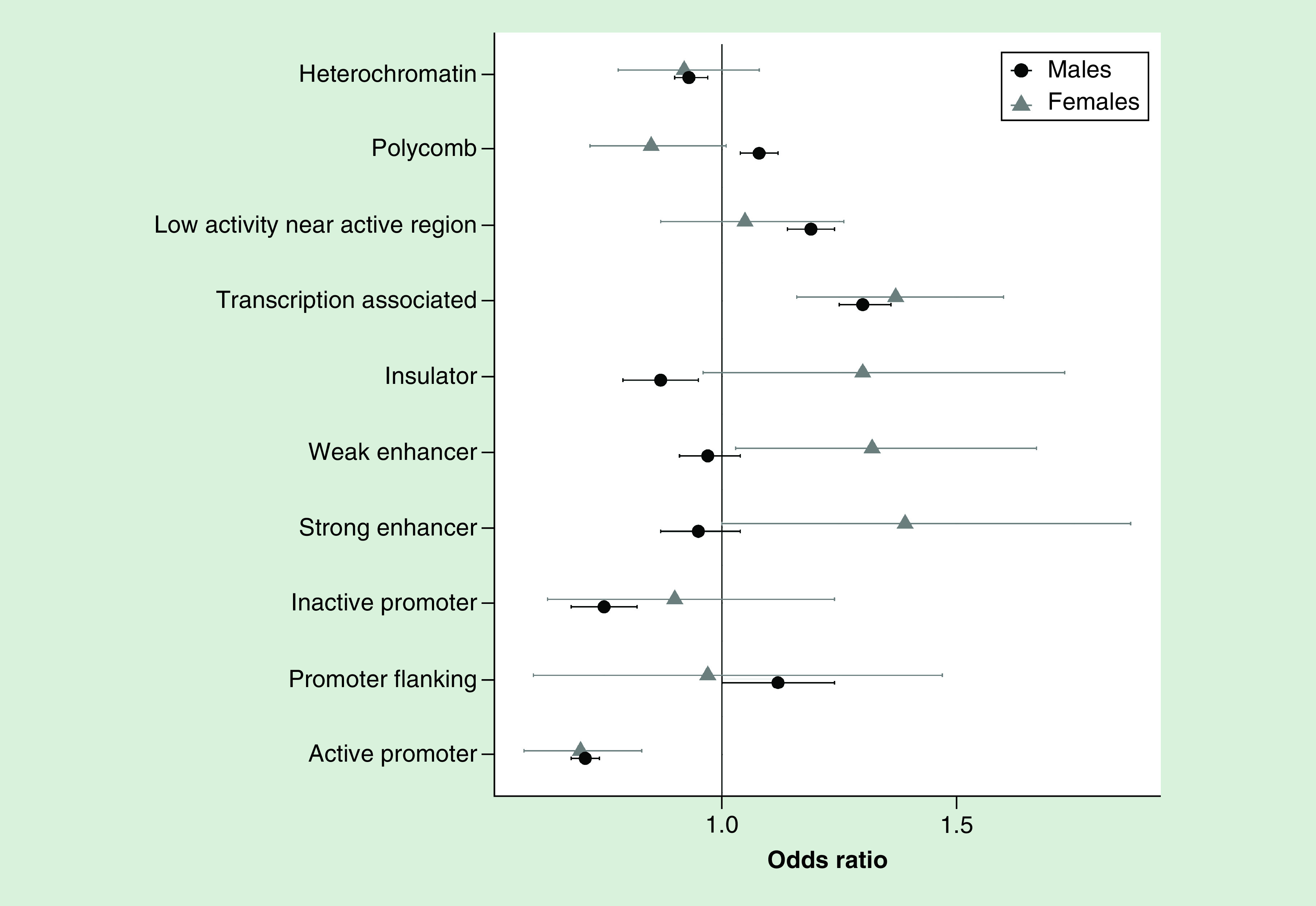
Enrichment tests were conducted to determine if the CpGs most associated with polybrominated biphenyl (PBB) were enriched in certain functional regions. PBB-associated CpGs in males were enriched in polycomb regions, areas of low activity near regions of high activity, transcription-associated regions and promoter flanking regions and depleted in active and inactive promoters, insulator and heterochromatin regions; in females, PBB-associated CpGs were enriched in transcription-associated regions and strong and weak enhancer and depleted in active promoters.
Different transcription factor binding sites enriched in each sex
To better assess whether different biological pathways were affected by PBBs in males and females, enrichment for transcription factor binding sites was tested for the PBB-associated CpGs in each sex separately. In males, the binding sites for 42 transcription factors were significantly enriched or depleted in the PBB-associated CpGs (Supplementary Table 7). In females, the binding sites for 18 transcription factors were significantly enriched or depleted in the PBB-associated CpGs (Supplementary Table 8). Only 10 transcription factors were enriched in both sexes and 20% of the shared transcription factors had their association in the opposite direction. Both males and females had nuclear hormone receptors and transcription factors involved in xenobiotic metabolism enriched in the results (females: Esrrb, NR4A2, Arnt, HIF1A::ARNT; males: PPARG::RXRA, Esrrb; ESR2, NR4A2, RORA_1; RXR::RAR_DR5; Arnt::Ahr; HIF1A::ARNT; Arnt, PLAG1). However, males had more transcription factors enriched in these pathways than females. Males also had enrichment for transcription factors involved in immune function (NFATC2, ARID3A, Tal1::Gata1, EBF1, NFKB1, Foxd3, STAT1, Stat3, TLX1::NFIC), male reproductive function (Spz1, SRY) and females had members of cancer-related pathways, like C-MYC enriched (MAX, Myc).
Discussion
In this study, we investigated sex specific associations between PBBs and DNA methylation by utilizing a cohort that was highly exposed to PBBs. We then tested whether the sex-specific epigenetic marks were correlated between the sexes and whether they were enriched for any biological pathways, transcription factor binding sites or functional regions that could give insight into how PBBs may influence gene regulation in each sex. While no CpGs were associated with the interaction between PBBs and sex, this is most likely due to the sample size being too small to detect an association given the number of tests which needed to be corrected for. However, when the association between PBBs and DNA methylation was tested in each sex separately, we found that men had 675 CpGs that significantly associated with current PBB level, with 71.85% of them having higher methylation with higher PBB concentrations and that women had 17 CpGs that were significantly associated with current PBB levels, with 58.82% of them having higher methylation with higher PBB concentrations. Additionally, finding a stronger effect of PBBs on the male epigenome is consistent with the majority of other studies that found a stronger effect or more CpGs associated with arsenic or EDC exposure in males [35–37]. However, it is not consistent with the study that found more CpGs associated with polychlorinated biphenyls in females [37].
It is possible that the stronger effect seen in men could be due to men having a more sensitive response to estrogen-like chemicals than women who have a wider range in natural estrogen levels. Since PBBs have been associated with estrogen signaling previously [52] and with estrogen-related transcription factors in this analysis, the stronger effect seen in men could be due to PBBs acting as a weak estrogen. It is also possible that the stronger effect seen in men is due to men having higher body burdens of PBBs. However, if the stronger effect was simply due to men having higher PBB levels, then one would expect that the same CpGs would have the largest association in both sexes, but they would only reach statistical significance in men. Contrary to that, men and women had distinct CpGs associated with PBBs and none of the CpGs that were significant in one sex were near significance in the other sex. Additionally, the effect sizes of significant CpGs were only moderately correlated between the sexes, in both the total study population and the strata, with the most significant CpGs in one sex having a small effect size in the other sex. The direction of effect in one sex was also in the opposite direction of the effect in the other sex in 36% of the significant CpGs. These different CpGs associated with PBB levels were also enriched in different biological pathways. The CpGs in males were enriched for two GO terms involved in cell adhesion, which plays a critical role in many different biological processes including immune function. However, the GO terms enriched in males had no evidence of enrichment in females and the top GO terms were not similar between the two analyses. Taken together, this suggests that PBBs could have a sex-specific effect on the epigenome with CpGs in different biological pathways being affected, which could provide a mechanism for sex-specific health effects from environmental exposures. However, it is also possible that the difference in CpGs associated with PBBs in each sex is a consequence of the difference in health outcomes experienced by men and women. Additionally, most of the CpGs identified in this study were also not identified in the epigenome-wide association study (EWAS) that analyzed both sexes together [52], indicating that sex-specific effects of EDCs can be missed in combined analyses. A sex-specific analysis in epigenome wide analyses should be considered in addition to the combined-sex analysis in order to get a more complete picture of how environmental factors are affecting the epigenome.
We also found that the CpGs most associated with PBBs in males had different functional enrichment from the CpGs most associated with PBBs in females, further emphasizing the difference in how PBBs affect the male and female epigenome. In functional categories defined by ChIP-seq data, the most associated CpGs in both males and females were enriched in transcription-associated regions and depleted in active promoters. However, only males had enrichment in polycomb regions, promoter flanking regions and regions of low activity near regions of high activity and depletion in insulator regions and inactive promoters; and only females had enrichment in strong and weak enhancers. Because the relationship between positional methylation and gene expression is unclear [71,72] and it is unclear which genes enhancers might be acting upon, it is difficult to say exactly how these functional enrichments could affect health outcomes. However, finding differences in functional enrichment in the results from men and women indicate that different biological pathways are being involved in each sex following PBB exposure. This could explain some of the sex-specific health outcomes from PBB exposure. While some of these enrichment differences may be due to the differences in the number of CpGs most associated in males and females, the threshold for association was lowered to FDR <0.2 in order to minimize this difference. No studies have investigated functional enrichment differences by sex and so it is not known whether there are sex-specific functional enrichments with other environmental exposures.
Complementary to finding differences in function enrichment, we also found differences in the transcription factor binding sites enriched in men and women. The binding sites for more transcription factors were enriched in the results from men compared with women. While both males and females had CpGs enriched near the binding sites for transcription factors that are nuclear hormone receptors, estrogen-related signaling transcription factors and related to xenobiotic metabolism, males had more of each category enriched in their results (n = 10) than women did (n = 4). Together, this may further support that PBBs have a stronger effect in men. Additionally, transcription factor binding sites related to immune function and male reproductive function were only enriched in the CpGs significant in males and transcription factor binding sites related to cancer development, like C-MYC, were only enriched in CpGs significant in females. Furthermore, out of the 10 transcription factor binding sites enriched in both sexes, there was no correlation in their Z-scores and 20% of them were associated in the opposite direction. This demonstrates the different biological pathways associated with PBBs in each sex and that the effects of PBBs on the male and female epigenome are distinct. These distinct epigenetic effects could underlie sex-specific health effects from PBB exposure like genitourinary conditions in men or increased breast cancer risk in women [48,50]. No other study has tested for sex-specific enrichment of transcription factor binding sites with an environmental contaminant, so it is not known if these results would be seen in sex-specific studies of other EDCs. The differences in transcription factors enriched in the results of each sex and the differences in direction of effect further highlights that there may be sex-specific differences from environmental exposures.
The findings of this study should be considered in light of its limitations. We were only able to test peripheral blood samples and there are most likely sex-specific effects in other tissues. Additionally, our sample size is not large enough to detect interactions between sex and PBB levels that account for less than 8% of the variance in methylation levels so we were unable to determine if there are smaller effects common to both sexes. There is also no other study population that has been assessed for their exposure to PBBs, making it impossible to replicate our results at this time. However, this study population is unique because they were highly exposed to a single EDC during a distinct time frame and they have been followed for the past 40 years, making them a valuable resource for environmental health studies.
Conclusion
This study found the first evidence that PBBs may have sex-specific effects on the epigenome, with 675 CpGs associating with PBBs in men and 17 different CpGs associating with PBBs in women. We also found that the most associated CpGs in men and women were enriched in different functional and positional regions. Finally, men and women had different transcription factor binding sites enriched near their most associated CpGs and even the transcription factors in common had differences in direction of association for 50% of the transcription factors. This suggests that there may be different transcription factors affected by PBB exposure in men and women, and therefore different CpGs associated with PBB exposure in men and women. Sex-specific epigenetic associations have the potential to underlie sex-specific health risks from environmental exposures.
Future perspective
Recent scientific studies have shown an increased appreciation of sex as a biological variable, which has been shown to influence a wide variety of physiological processes, such as cellular metabolism, disease manifestation and progression, and hormonal and reproductive health outcomes [73]. This study further emphasizes the importance of sex in biological studies by indicating that there are sex-specific effects on the epigenome from environmental exposures that are missed by analyses that combine the sexes. These sex-specific associations can increase our understanding of how environmental exposures affect human health and future studies should consider sex-specific biological responses to environmental stimuli.
Summary points.
We analyzed the association between exposure to polybrominated biphenyls (PBBs) and genome-wide DNA methylation levels in 277 men and 381 women in order to test for sex-specific epigenetic effects.
675 CpGs were associated with PBB levels in men and 17 CpGs were associated in women (false discovery rate: <0.05). No CpGs were associated in both sexes, and the effect size for associated CpGs were only moderately correlated.
Most of the CpGs identified in this analysis were distinct from those identified in the analysis with the sexes combined, indicating that a combined-sex analysis may miss sex-specific CpGs.
The CpGs associated with PBB levels in men were enriched in cell adhesion pathways and were not enriched in the results from women.
The CpGs most associated in men were in distinct function regions than the CpGs most associated in women, with men having enrichment in transcription-associated regions and women having enrichment in enhancer regions.
PBB-associated CpGs in men were enriched near more and distinct transcription factor binding sites than PBB-associated CpGs in women, including near transcription factor binding sites involved in male reproduction.
Taken together, this could indicate that PBBs may have sex-specific effects on the epigenome and that a stronger effect is seen in men, consistent with PBB acting weakly estrogenic.
This is the first study of the sex-specific epigenetic effects of PBB exposure and finding sex-specific epigenetic marks could suggest a mechanism for sex-specific health problems from PBB exposure.
Supplementary Material
Acknowledgments
We are grateful to the members of the Michigan polybrominated biphenyl (PBB) Registry for their participation and engagement with research studies over the past 40 years, the Michigan Department of Health and Human Services, which had the foresight to create the Registry and to our community partners (PBB Citizen Advisory Board, Pine River Superfund Citizen Task Force and the Mid-Michigan District Health Department) who continue to provide guidance and insight to the Michigan PBB Research.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2019-0179
Financial & competing interests disclosure
This work was supported by the National Institute of Environmental Health Sciences (NIEHS; R01ES024790, R01ES025775, R24ES028528 and P30ES019776) and the National Institute of General Medical Sciences (T32GM008490). This study was supported in part by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under Award Number UL1TR002378. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
Study protocols were approved by the Institutional Review Board at Emory University. Informed consent was obtained from all participants involved.
Data sharing statement
The DNA methylation data can be accessed on NCBI’s Gene Expression Omnibus (GSE116339).
References
Papers of special note have been highlighted as: • of interest
- 1.Porta M, Pumarega J, Gasull M, Lopez T. Contamination from endocrine disrupters of the general population at low and high concentrations. Vitam. Horm. 94, 167–192 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Fromme H, Hilger B, Kopp E, Miserok M, Volkel W. Polybrominated diphenyl ethers (PBDEs), hexabromocyclododecane (HBCD) and “novel” brominated flame retardants in house dust in Germany. Environ. Int. 64, 61–68 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Bergman A, Heindel JJ, Jobling S, Kidd KA, Zoeller RT. State of the science of endocrine disrupting chemicals. World Health Organization. (2012). www.who.int/ceh/publications/endocrine/en/ [Google Scholar]
- 4.Frederiksen M, Vorkamp K, Thomsen M, Knudsen LE. Human internal and external exposure to PBDEs–a review of levels and sources. Int. J. Hyg. Environ. Health 212(2), 109–134 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Cowell WJ, Stapleton HM, Holmes D. et al. Prevalence of historical and replacement brominated flame retardant chemicals in New York City homes. Emerg. Contam. 3(1), 32–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munn S, Goumenou M-P. Thresholds for endocrine disrupters and related uncertainties. (2013) (Epub ahead of print).
- 7.Birks L, Casas M, Garcia AM. et al. Occupational exposure to endocrine-disrupting chemicals and birth weight and length of gestation: a european meta-analysis. Environ. Health Perspect. 124(11), 1785–1793 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobson MH, Darrow LA, Barr DB. et al. Serum polybrominated biphenyls (PBBs) and polychlorinated biphenyls (PCBs) and thyroid function among Michigan adults several decades after the 1973–1974 PBB contamination of livestock feed. Environ. Health Perspect. 125(9), 097020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrne JJ, Carbone JP, Hanson EA. Hypothyroidism and abnormalities in the kinetics of thyroid hormone metabolism in rats treated chronically with polychlorinated biphenyl and polybrominated biphenyl. Endocrinology 121(2), 520–527 (1987). [DOI] [PubMed] [Google Scholar]
- 10.Kitamura S, Kato T, Iida M. et al. Anti-thyroid hormonal activity of tetrabromobisphenol A, a flame retardant, and related compounds: affinity to the mammalian thyroid hormone receptor, and effect on tadpole metamorphosis. Life Sci. 76(14), 1589–1601 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Hoque A, Sigurdson AJ, Burau KD, Humphrey HE, Hess KR, Sweeney AM. Cancer among a Michigan cohort exposed to polybrominated biphenyls in 1973. Epidemiology 9(4), 373–378 (1998). [PubMed] [Google Scholar]
- 12.Aleksandra Buha Djordjevic EA, Marijana Curcic, Vesna Milovanovic, Biljana Antonijevic. Endocrine-disrupting mechanisms of polychlorinated biphenyls. Curr. Opini. Toxicol. 19, 42–49 (2020). [Google Scholar]
- 13.Kundakovic M. Sex-specific epigenetics: implications for environmental studies of brain and behavior. Curr. Environ. Health Rep. 4(4), 385–391 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Axelstad M, Hass U, Scholze M, Christiansen S, Kortenkamp A, Boberg J. EDC IMPACT: reduced sperm counts in rats exposed to human relevant mixtures of endocrine disrupters. Endocr. Connect. 7(1), 139–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giwercman A. Estrogens and phytoestrogens in male infertility. Curr. Opin. Urol. 21(6), 519–526 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Hu WY, Shi GB, Hu DP, Nelles JL, Prins GS. Actions of estrogens and endocrine disrupting chemicals on human prostate stem/progenitor cells and prostate cancer risk. Mol. Cell. Endocrinol. 354(1–2), 63–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steves AN, Bradner JM, Fowler KL. et al. Ubiquitous flame-retardant toxicants impair spermatogenesis in a human stem cell model. iScience 3, 161–176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steves AN, Turry A, Gill B. et al. Per- and polyfluoroalkyl substances impact human spermatogenesis in a stem-cell-derived model. Syst. Biol. Reprod. Med. 64(4), 225–239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Rocca C, Tait S, Guerranti C. et al. Exposure to endocrine disrupters and nuclear receptor gene expression in infertile and fertile women from different Italian areas. Int. J. Environ. Res. Public Health 11(10), 10146–10164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler PA, Bellingham M, Sinclair KD. et al. Impact of endocrine-disrupting compounds (EDCs) on female reproductive health. Mol. Cell. Endocrinol. 355(2), 231–239 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Hernandez-Hernandez KL, Tapia-Orozco N, Gimeno M. et al. Exposure to bisphenol A: current levels from food intake are toxic to human cells. Mol. Biol. Rep. (2019) (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 22.Hafezi SA, Abdel-Rahman WM. The endocrine disruptor bisphenol A (BPA) exerts a wide range of effects in carcinogenesis and response to therapy. Curr. Mol. Pharmacol. (2019) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park C, Lee J, Kong B. et al. The effects of bisphenol A, benzyl butyl phthalate, and di(2-ethylhexyl) phthalate on estrogen receptor alpha in estrogen receptor-positive cells under hypoxia. Environ. Pollut. 248, 774–781 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Lambrecht LK, Barsotti DA, Allen JR. Responses of nonhuman primates to a polybrominated biphenyl mixture. Environ. Health Perspect. 23, 139–145 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker DM, Goetz BM, Gore AC. Dynamic postnatal developmental and sex-specific neuroendocrine effects of prenatal polychlorinated biphenyls in rats. Mol. Endocrinol. 28(1), 99–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates how prenatal exposure to estrogenic exposure to endocrine-disrupting compounds have sex-specific effects on gene expression in certain brain regions in rats.
- 26.Patisaul HB, Todd KL, Mickens JA, Adewale HB. Impact of neonatal exposure to the ERalpha agonist PPT, bisphenol-A or phytoestrogens on hypothalamic kisspeptin fiber density in male and female rats. Neurotoxicology 30(3), 350–357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobolewski M, Conrad K, Allen JL. et al. Sex-specific enhanced behavioral toxicity induced by maternal exposure to a mixture of low dose endocrine-disrupting chemicals. Neurotoxicology 45, 121–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebuli ME, Gibson P, Rhodes CL, Cushing BS, Patisaul HB. Sex differences in microglial colonization and vulnerabilities to endocrine disruption in the social brain. Gen. Comp. Endocrinol. 238, 39–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebuli ME, Patisaul HB. Assessment of sex specific endocrine disrupting effects in the prenatal and pre-pubertal rodent brain. J. Steroid Biochem. Mol. Biol. 160, 148–159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Xiao Y, Gai Z. et al. Reproductive toxicity of low level bisphenol A exposures in a two-generation zebrafish assay: evidence of male-specific effects. Aquat. Toxicol. 169, 204–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strakovsky RS, Wang H, Engeseth NJ. et al. Developmental bisphenol A exposure leads to sex-specific modification of hepatic gene expression and epigenome at birth that may exacerbate high-fat diet-induced hepatic steatosis. Toxicol. Appl. Pharmacol. 284(2), 101–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arambula SE, Jima D, Patisaul HB. Prenatal bisphenol A (BPA) exposure alters the transcriptome of the neonate rat amygdala in a sex-specific manner: a CLARITY-BPA consortium study. Neurotoxicology 65, 207–220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thongkorn S, Kanlayaprasit S, Jindatip D, Tencomnao T, Hu VW, Sarachana T. Sex differences in the effects of prenatal bisphenol a exposure on genes associated with autism spectrum disorder in the hippocampus. Sci. Rep. 9(1), 3038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reports that bisphenol A (BPA) exposure in developing male rates was associated with differences in gene expression in genes associated with autism spectrum disorder.
- 34.Topper VY, Walker DM, Gore AC. Sexually dimorphic effects of gestational endocrine-disrupting chemicals on microRNA expression in the developing rat hypothalamus. Mol. Cell. Endocrinol. 414, 42–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates how prenatal exposure to an exposure to endocrine-disrupting compounds has sex-specific effects on microRNAs in certain brain regions in rats.
- 35.Howe CG, Liu X, Hall MN. et al. Associations between blood and urine arsenic concentrations and global levels of post-translational histone modifications in Bangladeshi men and women. Environ. Health Perspect. 124(8), 1234–1240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgiadis P, Gavriil M, Rantakokko P. et al. DNA methylation profiling implicates exposure to PCBs in the pathogenesis of B-cell chronic lymphocytic leukemia. Environ. Int. 126, 24–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reports the association between polychlorinated biphenyl exposure and DNA methylation in a human cohort. It also reports that the effect of polychlorinated biphenyl exposure is stronger in men.
- 37.Leung YK, Ouyang B, Niu L. et al. Identification of sex-specific DNA methylation changes driven by specific chemicals in cord blood in a Faroese birth cohort. Epigenetics 13(3), 290–300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fries GF. The PBB episode in Michigan: an overall appraisal. Crit. Rev. Toxicol. 16(2), 105–156 (1985). [DOI] [PubMed] [Google Scholar]
- 39.Kay K. Polybrominated biphenyls (PBB) environmental contamination in Michigan, 1973–1976. Environ. Res. 13(1), 74–93 (1977). [DOI] [PubMed] [Google Scholar]
- 40.Safe S. Polychlorinated biphenyls (PCBs) and polybrominated biphenyls (PBBs): biochemistry, toxicology, and mechanism of action. Crit. Rev. Toxicol. 13(4), 319–395 (1984). [DOI] [PubMed] [Google Scholar]
- 41.Marder ME, Panuwet P, Hunter RE, Ryan PB, Marcus M, Barr DB. Quantification of polybrominated and polychlorinated biphenyls in human matrices by isotope-dilution gas chromatography-tandem mass spectrometry. J. Anal. Toxicol. 40(7), 511–518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terrell ML, Manatunga AK, Small CM. et al. A decay model for assessing polybrominated biphenyl exposure among women in the Michigan long-term PBB Study. J. Expo. Sci. Environ. Epidemiol. 18(4), 410–420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanck HM, Marcus M, Hertzberg V. et al. Determinants of polybrominated biphenyl serum decay among women in the Michigan PBB cohort. Environ. Health Perspect. 108(2), 147–152 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brilliant LB, Wilcox K, Van Amburg G. et al. Breast-milk monitoring to measure Michigan's contamination with polybrominated biphenyls. Lancet 2(8091), 643–646 (1978). [DOI] [PubMed] [Google Scholar]
- 45.Joseph AD, Terrell ML, Small CM, Cameron LL, Marcus M. Assessing inter-generational transfer of a brominated flame retardant. J. Environ. Monit. 11(4), 802–807 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Small CM, Cheslack-Postava K, Terrell M. et al. Risk of spontaneous abortion among women exposed to polybrominated biphenyls. Environ. Res. 105(2), 247–255 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanck HM, Marcus M, Tolbert PE. et al. Age at menarche and tanner stage in girls exposed in utero and postnatally to polybrominated biphenyl. Epidemiology 11(6), 641–647 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Small CM, Decaro JJ, Terrell ML. et al. Maternal exposure to a brominated flame retardant and genitourinary conditions in male offspring. Environ. Health Perspect. 117(7), 1175–1179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Small CM, Terrell ML, Cameron LL, Wirth J, Monteilh CP, Marcus M. In utero exposure to a brominated flame retardant and male growth and development. Int. J. Child and Adolescent Health 2(3), 11 (2009). [PMC free article] [PubMed] [Google Scholar]
- 50.Terrell ML, Rosenblatt KA, Wirth J, Cameron LL, Marcus M. Breast cancer among women in Michigan following exposure to brominated flame retardants. Occup. Environ. Med. 73(8), 564–567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henderson AK, Rosen D, Miller GL. et al. Breast cancer among women exposed to polybrominated biphenyls. Epidemiology 6(5), 544–546 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Curtis SW, Cobb DO, Kilaru V. et al. Exposure to polybrominated biphenyl (PBB) associates with genome-wide DNA methylation differences in peripheral blood. Epigenetics (2019) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Houseman EA, Accomando WP, Koestler DC. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 14(10), R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howards PP, Terrell ML, Jacobson MH. et al. Polybrominated biphenyl exposure and menstrual cycle function. Epidemiology 30(5), 687–694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakari T, Pessala P. In vitro estrogenicity of polybrominated flame retardants. Aquat. Toxicol. 74(3), 272–279 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Curtis SW, Cobb DO, Kilaru V. et al. Exposure to polybrominated biphenyl and stochastic epigenetic mutations: application of a novel epigenetic approach to environmental exposure in the Michigan polybrominated biphenyl registry. Epigenetics (2019) (Epub aheadof print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helsel DR. Less than obvious - statistical treatment of data below the detection limit. Environ. Sci. Technol. 24(12), 1766–1774 (1990). [Google Scholar]
- 59.Pidsley R, Zotenko E, Peters TJ. et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17(1), 208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Champely S. pwr: Basic Functions for Power Analysis. R package version 1.2-2.(2018). https://CRAN.R-project.org/package=pwr [Google Scholar]
- 61.Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics 28(9), 1280–1281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie T, Gorenjak V, GS M. et al. Epigenome-wide association study (EWAS) of blood lipids in healthy population from stanislas family study (SFS). Int. J. Mol. Sci. 20(5), (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braun KVE, Dhana K, De Vries PS. et al. Epigenome-wide association study (EWAS) on lipids: the Rotterdam Study. Clin. Epigenetics 9, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yousefi P, Huen K, Dave V, Barcellos L, Eskenazi B, Holland N. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genomics 16, 911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 57(1), 289–300 (1995). [Google Scholar]
- 66.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics 32(2), 286–288 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9(3), 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol 28(8), 817–825 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Outlines a method for annotating cytosine-guanine dinucleotides (or other genomic regions) to function regions defined by ChIP-seq data, leading to a better biological understanding of the results.
- 69.Hoffman MM, Ernst J, Wilder SP. et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 41(2), 827–841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 2(9), 987–1002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Outlines a method for annotating cytosine-guanine dinucleotides (or other genomic regions) to potential transcription factors binding motifs, leading to a better biological understanding of the results
- 71.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26(4), 577–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teissandier A, Bourc'his D. Gene body DNA methylation conspires with H3K36me3 to preclude aberrant transcription. EMBO J. 36(11), 1471–1473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clayton JA. Applying the new SABV (sex as a biological variable) policy to research and clinical care. Physiol. Behav. 187, 2–5 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


