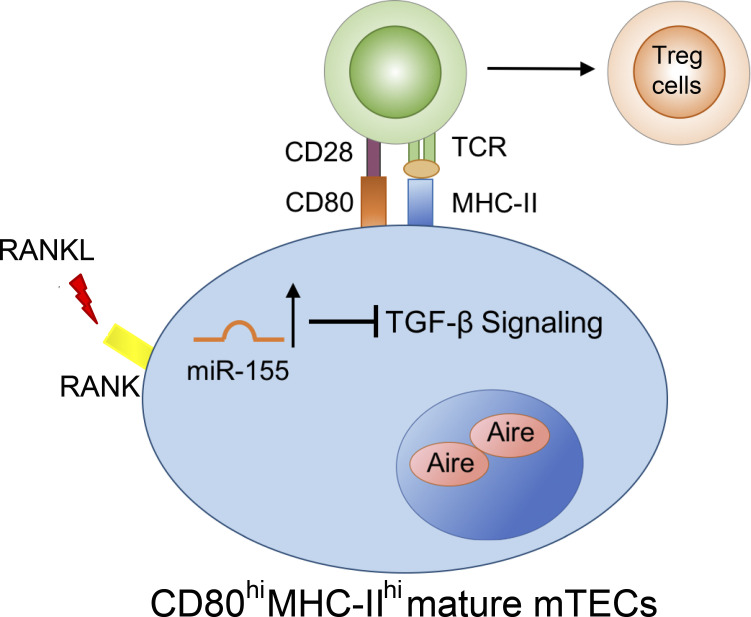
T reg cells are essential for establishing immunological tolerance, and their development in the thymus is tightly regulated. This study demonstrates that miR-155, a miRNA known for conferring T reg cell competitive fitness, can also promote T reg cell differentiation by targeting the TGFβ pathway in the thymic medulla.
Abstract
During thymocyte development, medullary thymic epithelial cells (mTECs) provide appropriate instructive cues in the thymic microenvironment for not only negative selection but also the generation of regulatory T (T reg) cells. Here, we identify that miR-155, a microRNA whose expression in T reg cells has previously been shown to be crucial for their development and homeostasis, also contributes to thymic T reg (tT reg) cell differentiation by promoting mTEC maturation. Mechanistically, we show that RANKL stimulation induces expression of miR-155 to safeguard the thymic medulla through targeting multiple known and previously uncharacterized molecules within the TGFβ signaling pathway, which is recognized for its role in restricting the maturation and expansion of mTECs. Our work uncovers a miR-155–TGFβ axis in the thymic medulla to determine mTEC maturity and, consequently, the quantity of tT reg cells and suggests that miR-155 ensures proper tT reg cell development in both cell-intrinsic and -extrinsic manners.
Graphical Abstract
Introduction
Regulatory T (T reg) cells constitute a specialized tolerogenic subset of cells recognized for maintaining immune homeostasis and preventing inappropriate reactivity to self-antigens and innocuous foreign antigens (Josefowicz et al., 2012a). While T reg cells can be generated in the periphery and play a nonredundant role in restraining allergic-type inflammation at mucosal interfaces, those generated in the thymus (tT reg cells) are absolutely critical for controlling systemic and tissue-specific autoimmunity (Josefowicz et al., 2012b). To this end, the thymic medulla represents a specific site for establishing self-tolerance via the generation of tT reg cells in addition to its known role in mediating negative selection (Hinterberger et al., 2010). In the thymic medulla, medullary thymic epithelial cells (mTECs) express high levels of MHCII molecules, tissue-restricted antigens, and costimulatory ligands CD80/CD86 in order to foster an instructive cross-talk between these specialized thymic stromal cells and developing thymocytes (Lucas et al., 2016). Disruption of the aforementioned interactions results in failed de novo generation of tT reg cells (Aschenbrenner et al., 2007; Malchow et al., 2016; Salomon et al., 2000; Tai et al., 2005). The fact that loss of mTECs leads to an explicit defect in the tT reg cell compartment while leaving conventional CD4 single positive thymocytes unaffected further substantiates an indispensable role for the thymic medulla in tT reg cell differentiation (Cowan et al., 2013).
To support the generation of tT reg cells, mTECs themselves need to differentiate properly. Activation of the RelB-dependent noncanonical NF-κB pathway driven by tumor necrosis factor superfamily cytokines such as receptor activator of NF-κB ligand (RANKL), CD40 ligand, and lymphotoxin β (LTβ) have been shown to be essential for mTEC progenitors to undergo a stepwise differentiation process to generate immature MHCIIloCD80lo mTECs before mature MHCIIhiCD80hi mTECs (Akiyama et al., 2008; Hikosaka et al., 2008; Irla et al., 2008). Among them, RANKL stimulation is particularly important for the induction of autoimmune regulator (AIRE), a transcription factor that plays a major role in driving the expression of tissue-restricted antigens in mature mTECs (Anderson et al., 2002; Rossi et al., 2007; Zuklys et al., 2000). On the other hand, TGFβ has been shown to play a negative role in restraining mTEC maturation by interfering with the noncanonical NF-κB pathway (Hauri-Hohl et al., 2014). However, because TGFβ can be detected in the thymus shortly after birth, and predominantly in the thymic medulla (Konkel et al., 2014), exactly how mTECs shield themselves from persistent TGFβ exposure remains unclear (Hauri-Hohl et al., 2014).
MicroRNAs (miRNAs) comprise a class of small noncoding RNAs that regulate gene expression at the posttranscriptional level and whose roles in controlling the development and function of T cells, including T reg cells, are well established (Chong et al., 2008; Cobb et al., 2006; Cobb et al., 2005; Liston et al., 2008; Zhou et al., 2008). It is also now appreciated that miRNAs can regulate thymic T cell differentiation by maintaining a proper thymic microenvironment, where deletion of the miRNA network within TECs severely compromises thymic infrastructure and largely impacts mTECs (Khan et al., 2014; Papadopoulou et al., 2012; Zuklys et al., 2012). Nevertheless, the current understanding of individual miRNAs crucial for controlling different aspects of TEC biology and, more importantly, their impact on thymic T cell development, remains limited (Khan et al., 2015). Here, we show that miR-155, a prominent miRNA known for its diverse functions in various immune cell populations (Vigorito et al., 2013), plays an equally important role in mTECs. Previously, we and others have shown that elevated expression of miR-155 driven by Foxp3 ensures proper T reg cell homeostasis by maintaining their competitive fitness (Lu et al., 2009). Our current work further demonstrates that miR-155 promotes T reg cell development in the thymus by safeguarding mTEC maturation. Mechanistically, RANK signaling induces miR-155 expression in the thymic medulla to alleviate the negative effects that ensue from the continuous presence of intrathymic TGFβ via targeting multiple known and previously uncharacterized molecules within this cytokine-signaling pathway. As such, the miR-155–TGFβ axis maintains the mature mTEC population and, thus, establishes a thymic microenvironment favorable for tT reg cell development.
Results and discussion
miR-155 promotes tT reg cell development in both T cell–intrinsic and –extrinsic manners
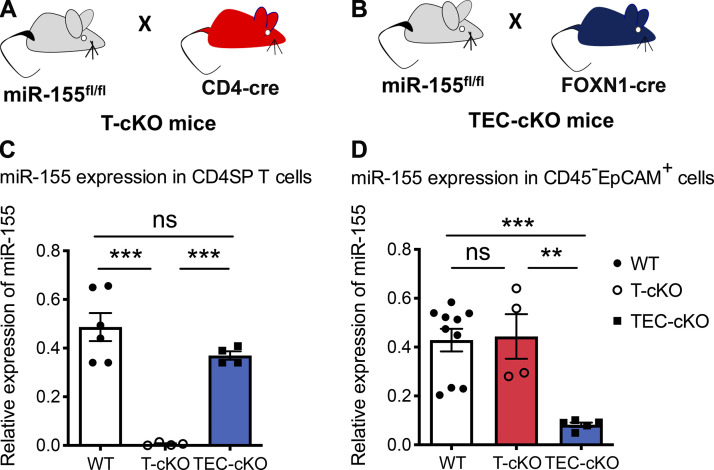
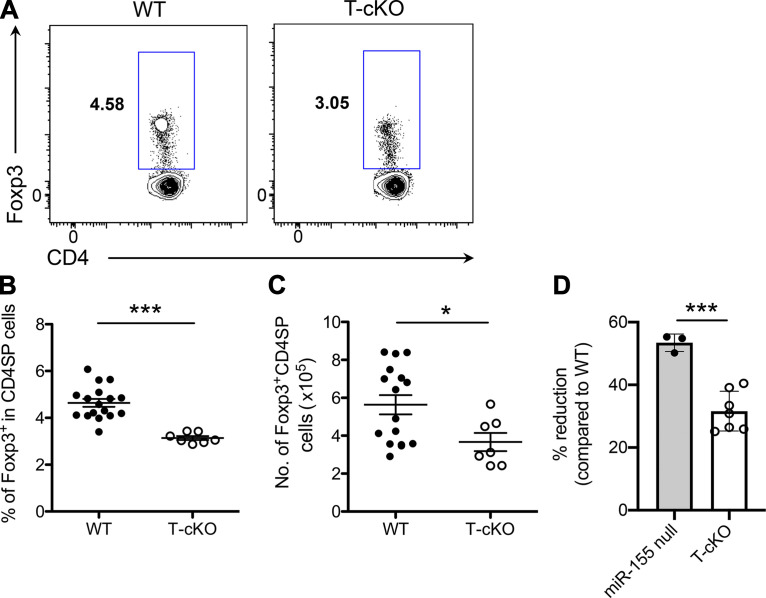
miRNAs play a pivotal role in controlling multiple aspects of T reg cell biology. Among the many miRNAs predominantly expressed in T reg cells, miR-155 is crucial for promoting optimal T reg cell development and homeostasis partly through ensuring responsiveness to IL-2, a cytokine required for thymic and peripheral T reg cell maintenance (Kohlhaas et al., 2009; Lu et al., 2009). Mice harboring a germline deficiency of miR-155 present with diminished T reg cell numbers and frequencies, plus additional studies using mixed bone marrow chimeras have identified a cell autonomous role of miR-155 in controlling T reg cell biology (Kohlhaas et al., 2009; Lu et al., 2009). Consistent with this notion, upon T cell–specific miR-155 ablation (Fig. S1 A), reduced frequencies as well as total numbers of T reg cells in the thymus are also detected (Fig. 1, A–C). Interestingly though, the degree of reduction in T cell–specific miR-155 conditional KO (T-cKO) mice does not fully recapitulate that observed in mice completely devoid of miR-155 (∼30% in T-cKO versus ∼60% in miR-155 null; Fig. 1 D; Kohlhaas et al., 2009; Lu et al., 2009; Sánchez-Díaz et al., 2017). These results suggest that loss of miR-155 expression in other non–T cell populations may also contribute to the impaired tT reg cell phenotype observed in mice containing miR-155 germline deficiency.
Figure S1.
Generation of T-cKO and TEC-cKO mice. (A and B) Schematic of the generation of T-cKO (A) and TEC-cKO (B) mice. (C and D) qPCR analyses of the expression of miR-155 in CD4SP T cells (CD4+CD8−; C) and TECs (CD45−EpCAM+; D) isolated from the thymus of 5–6-wk-old WT, T-cKO, and TEC-cKO mice. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: **, P < 0.01; ***, P < 0.001; ns, not significant.
Figure 1.
miR-155 promotes tT reg cell development in both T cell–intrinsic and –extrinsic manners. (A–C) FACS analysis (A), frequencies (B), and absolute numbers (C) of total Foxp3+ T reg cells in the thymus of 5–6-wk-old WT and T-cKO mice. (D) Percentages of the reduction of tT reg cell frequencies (on the basis of corresponding WT controls) in miR-155 null and T-cKO mice. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: *, P < 0.05; ***, P < 0.001.
miR-155 is preferentially expressed in mature mTECs and is induced by RANKL stimulation
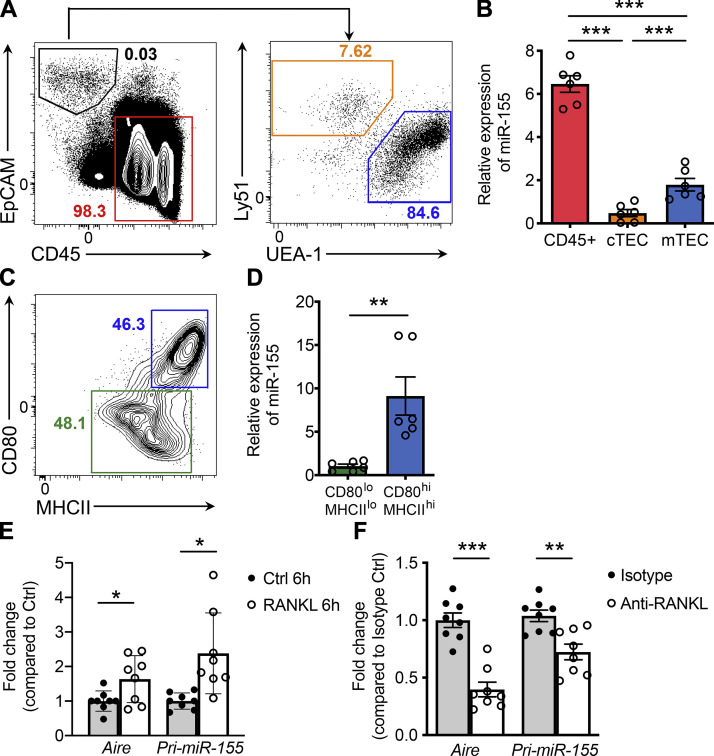
mTECs function as a key stromal cell population crucial for the generation of T reg cells in the thymus, and so it is possible that miR-155 promotes tT reg cell development by regulating mTEC biology. In support of this notion, a previous miRNA profiling study has demonstrated that miR-155 is expressed at elevated levels in mTECs compared with cortical thymic epithelial cells (cTECs), another population of thymic stromal cells critical for early thymocyte development (Khan et al., 2015). In line with this work, our analyses of different thymic resident cell subsets also revealed higher expression levels of miR-155 in mTECs relative to cTECs, albeit lower than that in CD45+ immune cells (Fig. 2, A and B). Moreover, in scrutiny of two major subsets within the mTEC population, CD80loMHCIIlo and CD80hiMHCIIhi (Fig. 2 C), with the latter comprising the more mature subset essential for tT reg cell generation, we found that miR-155 expression is restricted to the CD80hiMHCIIhi mTEC compartment. Together, these data point to a potential role of miR-155–mediated regulation of mTEC maturation.
Figure 2.
RANKL stimulation results in elevated miR-155 expression in mature mTECs. (A) Representative FACS profiles with gating strategy for different thymic subsets are shown. Hematopoietic cells were defined as CD45+EpCAM−. TECs were defined as CD45−EpCAM+ and further divided into cTEC (Ly51+UEA-1−) and mTEC (Ly51−UEA-1+) populations. (B) qPCR analyses of miR-155 expression in hematopoietic cells, cTECs, and mTECs. (C) Immature and mature mTECs within the mTEC population were further discriminated based on the levels of expression of CD80 and MHCII molecules. (D) qPCR analyses of miR-155 expression in CD80loMHCIIlo immature mTECs and CD80hiMHCIIhi mature mTECs. (E and F) qPCR analyses for Aire and pri-miR-155 expressions in sorted primary immature mTECs stimulated with 500 ng/ml RANKL for 6 h in vitro (E) or in total mTECs isolated from mice administrated with anti-RANKL or isotype control antibodies (F). The n-fold changes on the basis of corresponding untreated control groups were shown. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Among different tumor necrosis factor superfamily members that have been characterized in mTEC development, RANKL–RANK is the foremost determinant of mTEC development (Akiyama et al., 2008). To examine whether elevated miR-155 expression in mature mTECs is induced by RANK signaling, primary CD80loMHCIIlo immature mTECs were isolated and stimulated with RANKL. Along with prior work (Rossi et al., 2007), we noted an induction of AIRE mRNA in these cells as early as 6 h following RANKL stimulation (Fig. 2 E). Moreover, along with up-regulation of AIRE, an increase in the primary transcript of miR-155 (pri-miR-155) was also readily detectable (Fig. 2 E). Contrarily, a reduction in pri-miR-155 was observed concomitantly with diminished expression of Aire in mTECs isolated from mice treated with RANKL-blocking antibodies (Fig. 2 F). Collectively, these results locate RANK signaling in driving miR-155 expression in mTECs and imply functional relevance for miR-155 during mTEC development.
Expression of miR-155 in TECs is required to maintain optimum mTEC maturation and tT reg cell development
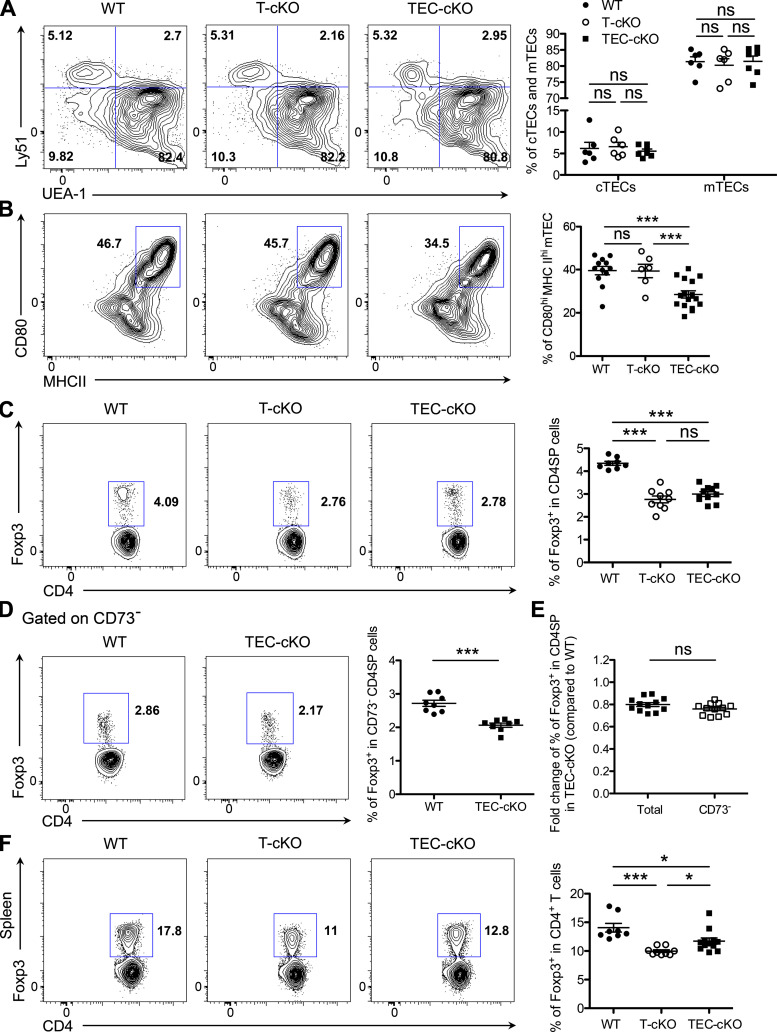
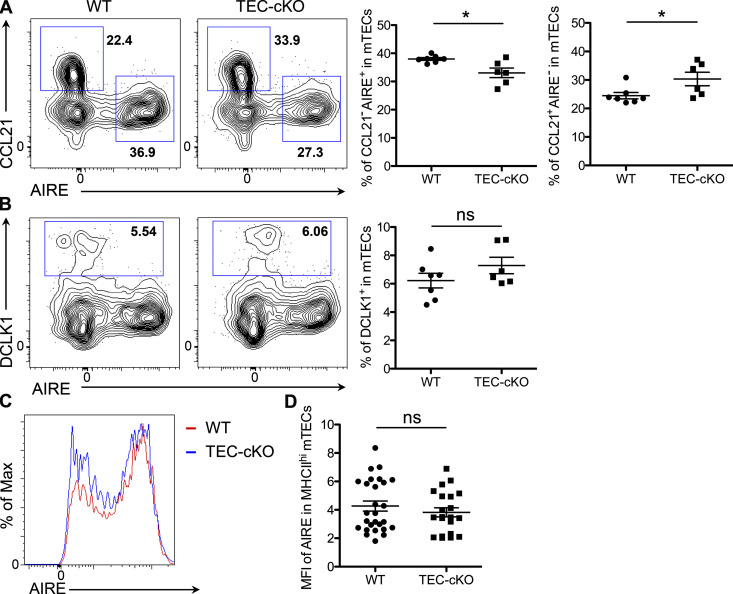
To investigate the potential role of miR-155 in mTEC maturation and its subsequent impact on tT reg cell development, we generated mice with TEC-specific ablation of miR-155 by crossing miR-155 floxed mice (miR-155fl) to FOXN1-Cre mice (Fig. S1, B–D). Considering that deletion of miR-155 is not restricted to mTECs but rather encompasses the entire thymic epithelium, we first sought to determine whether the cellularity and phenotype of the thymic epithelia would be impacted by the loss of miR-155. As depicted in Fig. 3 A (and data not shown), we did not detect any alterations in total thymic cellularity, including proportions of cTECs and mTECs, upon deletion of miR-155 in TECs relative to control WT or T-cKO mice. In contrast, we found that the frequency of CD80hiMHCIIhi mature mTECs was significantly reduced in TEC-specific miR-155 conditional knockout mice (TEC-cKO) mice in comparison to both WT and T-cKO controls, supporting our proposed function of miR-155 during mTEC maturation (Fig. 3 B). Accompanied by a decrease in mature mTECs that also express AIRE, we detected an increase in the frequency of a subset of AIRE−CCL21+ mTECs in TEC-cKO mice (Fig. S2 A). Unlike AIRE-expressing mTECs, the development of this specific mTEC subset relies on LTβ receptor signaling and has been implicated in recruiting (and/or retaining) positively selected CCR7+ thymocytes to the medulla (Lkhagvasuren et al., 2013; Zhang and Bhandoola, 2014). On the other hand, no significant change was observed in DCLK1-expressing thymic tuft cells (Fig. S2 B), another mTEC subset that is transcriptionally distinct from the AIRE+ mature mTECs (Bornstein et al., 2018). Interestingly, despite the reduction of mature mTECs in TEC-cKO mice, on a per-cell basis, levels of AIRE in mature mTECs were comparable between TEC-cKO and control mice (Fig. S2, C and D), implying that miR-155 does not directly regulate AIRE expression despite its role in mTEC maturation.
Figure 3.
miR-155 deficiency in TECs leads to a diminished mature mTEC population and impaired tT reg cell development. (A) FACS analysis and frequencies of Ly51+UEA-1− cTEC and Ly51−UEA-1+ mTEC populations from 5–6-wk-old WT, T-cKO, and TEC-cKO mice. (B) FACS analysis and frequencies of CD80hiMHCIIhi mTECs from 5–6-wk-old WT, T-cKO, and TEC-cKO mice. (C and D) FACS analysis and frequencies of total Foxp3+ T reg cells (C) or CD73− Foxp3+ T reg cells (D) in the thymus of 5–6-wk-old WT, T-cKO, and TEC-cKO mice. (E) Fold change of percentage of total or CD73− tT reg cells in TEC-cKO mice compared with WT littermates. (F) FACS analysis and frequencies of Foxp3+ T reg cells in the spleen of 5–6-wk-old WT and TEC-cKO mice. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: *, P < 0.05; ***, P < 0.001; ns, not significant.
Figure S2.
Effects of miR-155 deficiency on different mTEC subsets. (A and B) FACS analysis and frequencies of CCL21−AIRE+ and CCL21+AIRE− mTECs (A) as well as DCLK1+ tTuft cells (B) from 5–6-wk-old WT and TEC-cKO mice. (C and D) FACS analyses (C) and mean fluorescence intensity (MFI; D) of AIRE in MHCIIhi mature mTECs of TEC-cKO mice compared with WT controls. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: *, P < 0.05; ns, not significant.
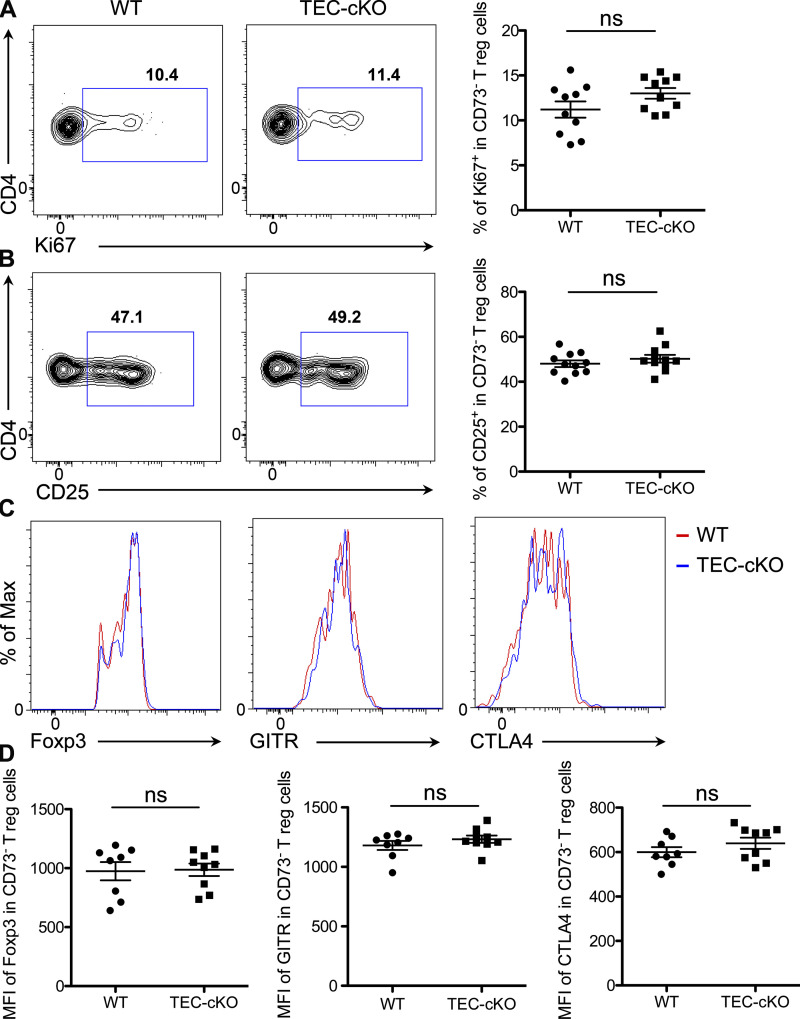
Due to the aforementioned role of mature mTECs in tT reg cell development and the impairment in mature mTECs from mice bearing TEC-specific ablation of miR-155, we next asked whether the generation of tT reg cells is similarly affected in TEC-cKO mice. Analogous to what has been discerned in T-cKO mice, a comparable reduction in tT reg cell frequency was detected in TEC-cKO mice (Fig. 3 C), highlighting that miR-155 as expressed in T cells and TECs contributes equally to T reg cell development in the thymus. Further supporting this notion, a similar reduction in the frequency of nascent tT reg cells in TEC-cKO mice was also seen when mature recirculating CD73+ T reg cells were excluded from the total tT reg cell population (Fig. 3, D and E). It should be noted that while miR-155 deletion in mTECs results in a reduction in tT reg cell frequency, the proliferative capacity as well as expression levels of Foxp3 and other T reg cell–associated molecules were unaffected in these T reg cells, relative to WT controls (Fig. S3). In the spleen, even though the frequency of T reg cells in TEC-cKO mice remained lower than WT controls, significantly more T reg cells in the spleen of TEC-cKO mice were seen when compared with T-cKO mice (Fig. 3 F). While the difference between the results obtained from the thymus and the spleen is intriguing, it is not surprising, seeing as miR-155 is a known crucial regulator conferring competitive fitness to T reg cells both in the thymus and in the periphery in a cell autonomous manner (Lu et al., 2009). Taken together, these data clearly demonstrate an indispensable role of TEC-derived miR-155 in controlling mTEC maturation and, subsequently, the generation of T reg cells in the thymus.
Figure S3.
Despite reduced frequencies, tT reg cells developed in TEC-cKO mice exhibited normal phenotype. (A and B) FACS analysis and frequencies of Ki67+ (A) and CD25+ (B) in thymic CD73− Foxp3+ T reg cells from 5–6-wk-old WT and TEC-cKO mice. (C and D) FACS analyses (C) and mean fluorescence intensity (MFI; D) of Foxp3, GITR, and CLTA4 in thymic CD73− Foxp3+ T reg cells from 5–6-wk-old WT and TEC-cKO mice. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: ns, not significant.
miR-155 limits TGFβ signaling via targeting multiple components of the TGFβ signaling pathway
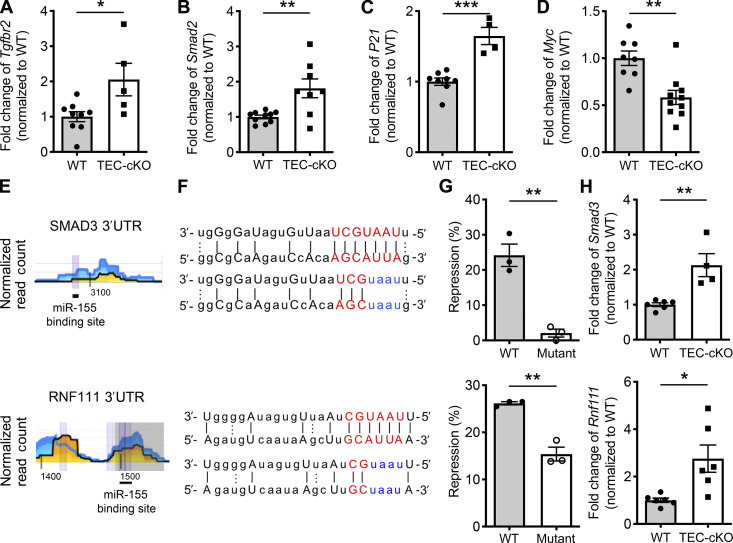
The TGFβ signaling cascade has previously been reported to play a regulatory role in limiting the establishment and function of the thymic medulla, specifically by influencing the differentiation of mTECs (Konkel et al., 2014). In addition, miR-155 has also been implicated in regulating TGFβ signaling by directly targeting Tgfbr2 and Smad2 in human lung fibroblasts and in THP-1 monocyte cell lines, respectively (Chu et al., 2017; Louafi et al., 2010). Therefore, it is possible that miR-155 also promotes mTEC maturation by restraining TGFβ signaling. Indeed, we observed increased levels of both Tgfbr2 and Smad2 in mature mTECs isolated from TEC-cKO mice, indicating that these two genes are also subject to regulation by miR-155 in mature mTECs (Fig. 4, A and B). Consistently, we detected higher levels of P21, an established TGFβ-induced gene (Datto et al., 1995), in mature mTECs devoid of miR-155. The levels of Myc, a gene known to be repressed by TGFβ activity (Frederick et al., 2004), were, alternatively, reduced (Fig. 4, C and D). Overall, these results are suggestive of enhanced TGFβ signaling in the absence of miR-155–mediated gene regulation.
Figure 4.
miR-155 regulates TGFβ signaling in mature mTECs via targeting multiple key components. (A–D) qPCR analyses for the expressions of Tgfbr2 (A), Smad2 (B), P21 (C), and Myc (D) in sorted CD80hiMHCIIhi mature mTECs from 5–6-wk-old WT and TEC-cKO mice. The n-fold changes on the basis of each corresponding WT controls were shown. (E and F) High-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation analyses (the underlying numbers represent the nucleotide position related to the start of the 3′UTR (E) and sequence alignments (F) of the putative miR-155 binding sites in 3′UTR of SMAD3 (upper) and RNF111 (lower). Mutations of the corresponding miR-155 target sites are shown in blue. (G) Ratios of repressed luciferase activity of cells with SMAD3 3′UTR (upper) and RNF111 3′UTR (lower) with or without mutations in the seed sequences in the presence of miR-155 compared with cells transfected with empty vector. (H) qPCR analyses of the expressions of Smad3 (upper) and Rnf111 (lower) in sorted CD80hiMHCIIhi mature mTECs from 5–6-wk-old WT and TEC-cKO mice. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Since miRNAs usually exert their regulatory effects by targeting multiple genes in a shared pathway or protein complex to ensure biological impact (Ebert and Sharp, 2012), we sought to define additional targets in the TGFβ signaling pathway that may be controlled by miR-155 in mature mTECs. After analyzing previous results obtained from high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP), a biochemical approach that affords the identification of functional miRNA–mRNA interaction in a given tissue/cell sample (Loeb et al., 2012), we identified SMAD3, a molecule that acts cooperatively with SMAD2 to form major TGFβ signaling transducers, as another potential miR-155 target (Fig. 4, E and F). Supporting this notion, our luciferase reporter studies confirmed that miR-155 directly represses SMAD3 (Fig. 4 G), and mature mTECs isolated from TEC-cKO mice express significantly higher amounts of Smad3 transcript (Fig. 4 H). Moreover, by taking similar approaches, we also identified and confirmed RNF111 as a direct target of miR-155 (Fig. 4, E–H). RNF111 (or “Arkadia”) is an E3 ubiquitin ligase recognized for its role in enhancing TGFβ responses by promoting the degradation of c-SKI, a known negative regulator of TGFβ signaling that blocks TGFβ-driven transcriptional activation and repression by forming an inhibitory complex with SMAD proteins (Sharma et al., 2011; Suzuki et al., 2004). Thus, our data show that through targeting multiple requisite components within the TGFβ signaling pathway, ranging from the receptor to major signal transducers and to the E3 ligase that degrades the TGFβ signaling inhibitor, miR-155 acts as a key molecule involved in attenuating TGFβ signaling in mTECs amid their maturation process.
Diminished mTECs and tT reg cells in TEC-cKO mice are largely rescued by partial TGFβ receptor deletion
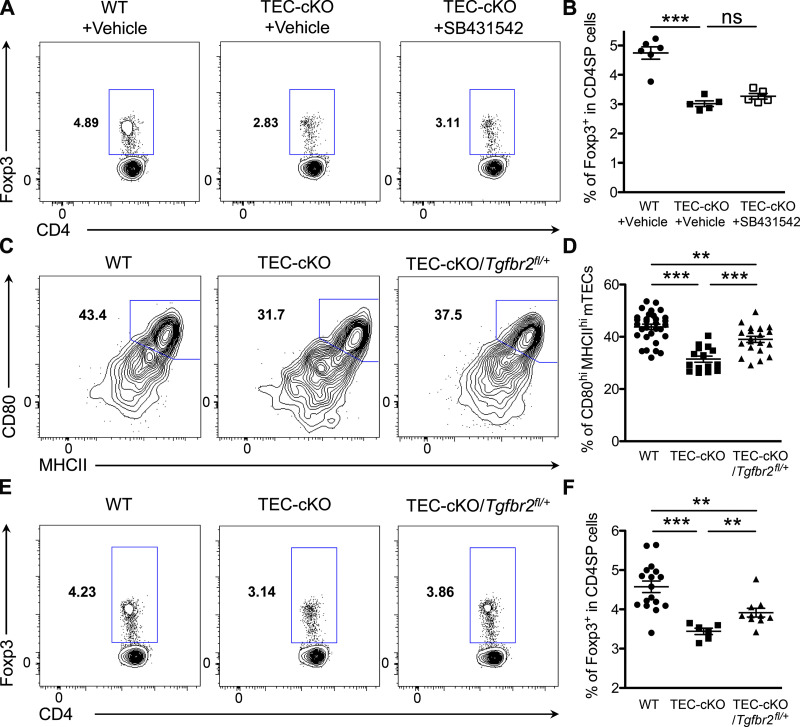
While our results demonstrate that TEC-specific miR-155 ablation leads to enhanced TGFβ signaling in mTECs, it remains obscure as to whether loss of miR-155–dependent regulation of TGFβ signaling is responsible for the mTEC and tT reg cell phenotypes observed in TEC-cKO mice. Previously, it has been shown that short-term systemic pharmacological inhibition of TGFβ signaling with the TGFβ receptor Ι (ΤGFβRI) kinase inhibitor selectively increases the number of mature mTECs (Hauri-Hohl et al., 2014). Interestingly, however, such treatment did not seem to rescue the defective tT reg cell phenotype observed in mice with TEC-specific miR-155 ablation; only a modest increase of tT reg cell frequencies was detected in TEC-cKO mice following treatment of the TGFβRI kinase inhibitor compared with the untreated group (Fig. 5, A and B). It should be noted that TGFβ signaling in thymocytes has previously been reported to be needed for both induction of Foxp3 and the differentiation of tT reg cells (Konkel et al., 2014). It is therefore possible that any positive effect resulting from blockade of TGFβ signaling in mTECs is masked by the loss of TGFβ-driven tT reg cell induction in mice with systemic TGFβRI kinase inhibitor administration.
Figure 5.
Tgfbr2 heterozygosity in TECs restores mature mTEC and tT reg cell phenotypes in TEC-cKO mice. (A and B) FACS analyses (A) and frequencies (B) of Foxp3+ T reg cells in the thymus of 6–8-wk-old WT and TEC-cKO mice treated daily with the TGFβRI kinase inhibitor SB431542 or vehicle only via i.p. injection from day 1 to day 6 and analyzed 12 d after the first injection. (C and D) FACS analysis (C) and frequencies (D) of CD80hiMHCIIhi mature mTECs in the thymus of 5–6-wk-old WT, TEC-cKO, and TEC-cKO/Tgfbr2fl/+mice. (E and F) FACS analysis (E) and frequencies (F) of Foxp3+ T reg cells in the thymus of 5–6-wk-old WT, TEC-cKO, and TEC-cKO/Tgfbr2fl/+mice. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: **, P < 0.01; ***, P < 0.001; ns, not significant.
To directly examine the role of the miR-155–TGFβ signaling axis in mTECs and its subsequent impact on tT reg cell development, we opted for a genetic approach by removing one allele of Tgfbr2 specifically in the thymic epithelia of TEC-cKO mice (TEC-cKO/Tgfbr2fl/+). As shown in Fig. 5, C and D, a significantly enlarged mature CD80hiMHCIIhi mTEC population was seen in TEC-cKO/Tgfbr2fl/+ mice compared with TEC-cKO mice. Accordant with the previously established role of mature mTECs in driving tT reg cell development, substantially augmented tT reg cell frequencies were also detected in accompany with an increase in the mature mTEC population in TEC-cKO mice containing Tgfbr2 heterozygosity (Fig. 5, E and F). Notably, the frequencies of mature mTECs and tT reg cells in TEC-cKO/Tgfbr2fl/+mice remained lower than those in WT controls (Fig. 5, C–F), indicating that additional miR-155 targets independent of TGFβ signaling might also contribute to the mTEC and tT reg cell phenotypes observed in TEC-cKO mice. Nevertheless, our data effectively demonstrate that miR-155 supports mTEC maturation and the resultant tT reg cell development by restricting TGFβ signaling.
In the past decade, the role of miRNAs in controlling diverse immune responses has become a focal point of intense investigation. Previously, we have shown that the same miRNA can impose its gene regulatory effect in different immune cell subsets, enabling them to play their specialized roles in producing a concerted response to a particular environmental stimulus (Cho et al., 2018). Therefore, while numerically speaking, it seems that combining the loss of miR-155 in T cells and TECs largely accounts for the tT reg cell phenotype in mice with the germline miR-155 deletion, it remains possible that impaired function of other immune cells such as dendritic cells (DCs) could also contribute to the reduction in T reg cell numbers seen in miR-155 null mice. After all, miR-155 deficiency is already known to diminish the antigen-presenting and costimulatory capacities of DCs (Rodriguez et al., 2007), and like mTECs, DCs are also crucial for optimal tT reg cell generation (Perry et al., 2014; Proietto et al., 2008). Still, despite our accumulating knowledge of miRNA-mediated gene regulation in different immune cell populations, relatively limited efforts have been spent on understanding how miRNAs impact the immune system by shaping the environmental cues supplied by nonimmune cells. Here, our results demonstrate that miR-155 can facilitate T reg cell development in the thymus by ensuring the proper maturation of the thymic medulla and, thus, suggest that additional attention be paid to stromal cells residing in the same microenvironments in order to better understand the biological impact of a given miRNA on a selective immunological process.
Materials and methods
Mice
miR-155fl mice (Hu et al., 2014) were bred with CD4-Cre mice (Lee et al., 2001) and FOXN1-Cre mice (Gordon et al., 2007), respectively, to obtain mice with T cell–specific deletion (T-cKO) and TEC-specific deletion (TEC-cKO) of miR-155. Tgfbr2fl mice (Levéen et al., 2002) were bred with TEC-cKO mice to obtain TEC-cKO/Tgfbr2fl/+ mice. Unless otherwise indicated, 5–6-wk-old mice were used. All mice were maintained and handled in accordance with the Institutional Animal Care and Use Guidelines of University of California, San Diego and National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and the Animal Research: Reporting of In Vivo Experiments Guidelines.
Tissue preparation
For T cell analysis, single-cell suspensions of thymus and spleen were prepared by slide mechanical disruption. TECs were isolated as previously described (Seach et al., 2013). In brief, thymus was mechanically disrupted and digested with DNase I (Sigma-Aldrich) and Liberase (Roche). The resulting single-cell suspension was filtered and washed once in magnetic-activated cell sorting buffer. In some experiments, cells were used immediately for FACS analysis. Otherwise, density-gradient centrifugation was performed for TEC enrichment before FACS analysis or sorting. Briefly, cells were resuspended in 2 ml of 1.115 g/ml isotonic Percoll and overlaid with 1.5 ml of 1.065 g/ml isotonic Percoll, followed by a layer of 1 ml of PBS. Samples were then centrifuged at 600 g at 4°C for 30 min with the brakes off. The thymic stroma accumulated between the top and middle layers and was collected and washed for subsequent FACS analysis or cell sorting.
Flow cytometry and antibodies
For FACS analysis, cells were first stained with Ghost Dye Red 780 (Tonbo Biosciences) followed by surface antibody staining for CD4 (RM4-5), CD8a (53–6.7), CD45 (30-F11), EpCAM (G8.8), Ly51 (6C3), BiotinylatedUEA-1 (Vector Labs), CD80 (16-10A1), MHCII (M5/114.15.2), TCRβ (H57-597), Streptavidin (eBioscience), CD44 (IM7), CD62L (MEL-14), CD25 (PC61), GITR (DTA-1). Intracellular staining for Foxp3 (FJK-16s), Ki67 (SolA15), AIRE (5H12), CTLA-4 (UC10-4B9), CCL21 (59106), DCLK1 (Abcam polyclonal ab31704) and AlexaFluor-488–conjugated donkey anti-rabbit IgG Ab (Poly4064) was completed after fixation and permeabilization with Foxp3/Transcription Factor Staining Kit according to manufacturer protocol (Tonbo). Cells were fixed in 2% paraformaldehyde before analysis by flow cytometry. For T reg cell analysis, total tT reg cells were gated on Foxp3+ cells from the CD4+CD8− population. For nascent tT reg cells, CD73− cells were first gated from the CD4+CD8− population, followed by Foxp3+ gating. As for TEC analysis, cells were first gated/sorted from the CD45−EpCAM+ population followed by using Ly51 and UEA-1 to separate cTECs (Ly51+UEA-1−) and mTECs (Ly51−UEA-1+). Within the mTECs, immature (or mTEClo) and mature (or mTEChi) mTECs were separated by the expression levels of CD80 and MHCII. Moreover, AIRE and CCL21 were used to identify the CCL21+AIRE− mTEC subset while DCLK1 was used to label the thymic tuft cells. FACS data were collected by BD LSRFortessa or BD LSRFortessa X-20 (BD Biosciences) and analyzed by FlowJo (Tree Star). Finally, BD BD FACSAria Fusion (BD Biosciences) was used for cell sorting.
Quantitative PCR analysis
Different thymic cell subsets, CD45+(CD45+EpCAM−), cTEC (CD45−EpCAM+Ly51+UEA-1−), mTEC (CD45−EpCAM+Ly51−UEA-1+) cells, immature mTEC (CD45−EpCAM+Ly51−UEA-1+CD80loMHCIIlo), and mature mTEC (CD45−EpCAM+Ly51−UEA-1+CD80hiMHCIIhi) cells were sorted on a FACSAria Fusion (BD Biosciences), and then total mRNA was isolated using a miRNeasy kit (Qiagen) according to the manufacturer’s instructions. To determine expression of miR-155, TaqMan MicroRNA Assay (Thermo Fisher Scientific) was performed. For other gene detection, cDNA was generated using iScript cDNA Synthesis Kit (Bio-Rad), followed by quantitative real-time PCR (qPCR) reactions using SYBR Green PCR Mix (Thermo Fisher Scientific). The primer sequences used were as follows: Tgfbr2: 5′-GAGAAGCCGCATGAAGTCTG-3′ (F), 5′-CATGAAGAAAGTCTCGCCCG-3′ (R); Smad2: 5′-ATATAGGAAGGGGAGTGCGC-3′ (F), 5′-AAACGGCTTCAAAACCCTGG-3′ (R); Smad3: 5′-ACTTGGACCTACAGCCAGTC-3′ (F), 5′-TGCATTCCGGTTGACATTGG-3′ (R); Rnf111: 5′-CAGCCTTCCACAGTGTCAGA-3′ (F), 5′-GGTGTGCTAATGCATGATGG-3′ (R); Myc: 5′-CACTCACCAGCACAACTACG-3′ (F), 5′-GTTCCTCCTCTGACGTTCCA-3′ (R); P21: 5′-GCAGATCCACAGCGATATCC-3′ (F), 5′-CAACTGCTCACTGTCCACGG-3′ (R); Aire: 5′-AGGTCAGCTTCAGAGAAAACCA-3′ (F), 5′-TCATTCCCAGCACTCAGTAGA-3′ (R); pri-miR-155: 5′-ACCCTGCTGGATGAACGTAG-3′ (F), 5′-CATGTGGGCTTGAAGTTGAG-3′ (R); Gapdh: 5′-CGTCCCGTAGACAAAATGGT-3′ (F), 5′-TCAATGAAGGGGTCGTTGAT-3′ (R); pri-miR-29a: 5′-AACCGATTTCAGATGGTGCT-3′ (F), 5′-AAGCCTTCTCTGGAAGTGGAC-3′ (R).
In vitro RANKL stimulation
CD80loMHCIIlo immature mTECs were sorted on a FACSAria Fusion (BD Biosciences) into complete RPMI, followed by stimulation with 500 ng/ml recombinant mouse RANKL (BioLegend) for 6 h at 37°C.
In vivo mouse treatment
For in vivo RANKL blockade, anti-RANKL antibody (IK22/5) or isotype control antibody (RTK2758) was administrated to 4-wk-old mice at a dose of 250 µg in PBS every other day via i.p. injection for a total of three injections. Mouse thymi were collected for further study 1 d after the last injection. For systemic administration of TGFβRI kinase inhibitor SB431542 (Selleckchem; 10 mg per kg body weight, 0.2 mg in 100 µl of 2% DMSO + 30% PEG300 + PBS) or vehicle control was administrated via i.p. injection once a day from day 1 to day 6 and analyzed 12 d after the first injection.
Luciferase reporter assay
The 3′UTR sequences of SMAD3 and RNF111 were amplified from mouse genomic DNA and cloned into pSiCheck2 vector (Promega). Site-direct mutagenesis (Agilent) was performed to obtain mutants of SMAD3 3′UTR and RNF111 3′UTR, respectively. Indicated 3′UTR WT or mutant plasmids were transfected into HEK293T (ATCC CRL-3216) cells along with either a miR-155–expressing plasmid or a control empty vector. Luciferase activity was determined by the Dual luciferase reporter assay system (Promega) according to the manufacturer’s instructions at 24 h after transfection.
Statistical analyses
An unpaired, two-tailed Student’s t test (or one-way ANOVA for studies with more than two groups) was done on all reported data using Prism software (GraphPad; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant). All experiments were performed independently at least three times to ensure the reproducibility of the data.
Online supplemental material
Fig. S1 shows the generation of T-cKO and TEC-cKO mice. Fig. S2 shows the effects of miR-155 deficiency on different mTEC subsets. Fig. S3 shows that tT reg cells developed in TEC-cKO mice exhibited normal phenotypes.
Acknowledgments
We thank all members of our laboratory for discussions.
This work was supported by National Institutes of Health grants AI103646, AI108651, and AI140095 (to L.-F. Lu) and AG047956 (to R.M. O’Connell).
Author contributions: Conceived and designed the experiments: J. Dong, L.M. Warner, and L.-F. Lu. Performed the experiments: J. Dong, L.M. Warner, M.-C. Chen, and L.-L. Lin. Analyzed the data: L.M. Warner, J. Dong, and L.-F. Lu. Contributed reagents/materials/analysis tools: R.M. O’Connell. Wrote the paper: J. Dong, L.M. Warner, and L.-F. Lu.
References
- Akiyama, T., Shimo Y., Yanai H., Qin J., Ohshima D., Maruyama Y., Asaumi Y., Kitazawa J., Takayanagi H., Penninger J.M., et al. . 2008. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 29:423–437. 10.1016/j.immuni.2008.06.015 [DOI] [PubMed] [Google Scholar]
- Anderson, M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J., von Boehmer H., Bronson R., Dierich A., Benoist C., and Mathis D.. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298:1395–1401. 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- Aschenbrenner, K., D’Cruz L.M., Vollmann E.H., Hinterberger M., Emmerich J., Swee L.K., Rolink A., and Klein L.. 2007. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8:351–358. 10.1038/ni1444 [DOI] [PubMed] [Google Scholar]
- Bornstein, C., Nevo S., Giladi A., Kadouri N., Pouzolles M., Gerbe F., David E., Machado A., Chuprin A., Tóth B., et al. . 2018. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature. 559:622–626. 10.1038/s41586-018-0346-1 [DOI] [PubMed] [Google Scholar]
- Cho, S., Lee H.M., Yu I.S., Choi Y.S., Huang H.Y., Hashemifar S.S., Lin L.L., Chen M.C., Afanasiev N.D., Khan A.A., et al. . 2018. Differential cell-intrinsic regulations of germinal center B and T cells by miR-146a and miR-146b. Nat. Commun. 9:2757 10.1038/s41467-018-05196-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, M.M., Rasmussen J.P., Rudensky A.Y., and Littman D.R.. 2008. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205:2005–2017. 10.1084/jem.20081219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, L., Li Y., Zhang X., Fu X., and Zhu Z.. 2017. MicroRNA-155 inhibits the pro-fibrogenic activities of TGF-β1 in human lung fibroblasts. Int. J. Clin. Exp. Med. 10:347–356. [Google Scholar]
- Cobb, B.S., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., and Merkenschlager M.. 2005. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201:1367–1373. 10.1084/jem.20050572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, B.S., Hertweck A., Smith J., O’Connor E., Graf D., Cook T., Smale S.T., Sakaguchi S., Livesey F.J., Fisher A.G., and Merkenschlager M.. 2006. A role for Dicer in immune regulation. J. Exp. Med. 203:2519–2527. 10.1084/jem.20061692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan, J.E., Parnell S.M., Nakamura K., Caamano J.H., Lane P.J., Jenkinson E.J., Jenkinson W.E., and Anderson G.. 2013. The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+ thymocyte development. J. Exp. Med. 210:675–681. 10.1084/jem.20122070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto, M.B., Li Y., Panus J.F., Howe D.J., Xiong Y., and Wang X.F.. 1995. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc. Natl. Acad. Sci. USA. 92:5545–5549. 10.1073/pnas.92.12.5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, M.S., and Sharp P.A.. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell. 149:515–524. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, J.P., Liberati N.T., Waddell D.S., Shi Y., and Wang X.F.. 2004. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 24:2546–2559. 10.1128/MCB.24.6.2546-2559.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, J., Xiao S., Hughes B. III, Su D.M., Navarre S.P., Condie B.G., and Manley N.R.. 2007. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7:69 10.1186/1471-213X-7-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri-Hohl, M., Zuklys S., Holländer G.A., and Ziegler S.F.. 2014. A regulatory role for TGF-β signaling in the establishment and function of the thymic medulla. Nat. Immunol. 15:554–561. 10.1038/ni.2869 [DOI] [PubMed] [Google Scholar]
- Hikosaka, Y., Nitta T., Ohigashi I., Yano K., Ishimaru N., Hayashi Y., Matsumoto M., Matsuo K., Penninger J.M., Takayanagi H., et al. . 2008. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 29:438–450. 10.1016/j.immuni.2008.06.018 [DOI] [PubMed] [Google Scholar]
- Hinterberger, M., Aichinger M., Prazeres da Costa O., Voehringer D., Hoffmann R., and Klein L.. 2010. Autonomous role of medullary thymic epithelial cells in central CD4(+) T cell tolerance. Nat. Immunol. 11:512–519. 10.1038/ni.1874 [DOI] [PubMed] [Google Scholar]
- Hu, R., Kagele D.A., Huffaker T.B., Runtsch M.C., Alexander M., Liu J., Bake E., Su W., Williams M.A., Rao D.S., et al. . 2014. miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation. Immunity. 41:605–619. 10.1016/j.immuni.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irla, M., Hugues S., Gill J., Nitta T., Hikosaka Y., Williams I.R., Hubert F.X., Scott H.S., Takahama Y., Holländer G.A., and Reith W.. 2008. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity. 29:451–463. 10.1016/j.immuni.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Josefowicz, S.Z., Lu L.F., and Rudensky A.Y.. 2012a. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz, S.Z., Niec R.E., Kim H.Y., Treuting P., Chinen T., Zheng Y., Umetsu D.T., and Rudensky A.Y.. 2012b. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 482:395–399. 10.1038/nature10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, I.S., Taniguchi R.T., Fasano K.J., Anderson M.S., and Jeker L.T.. 2014. Canonical microRNAs in thymic epithelial cells promote central tolerance. Eur. J. Immunol. 44:1313–1319. 10.1002/eji.201344079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, I.S., Park C.Y., Mavropoulos A., Shariat N., Pollack J.L., Barczak A.J., Erle D.J., McManus M.T., Anderson M.S., and Jeker L.T.. 2015. Identification of MiR-205 As a MicroRNA That Is Highly Expressed in Medullary Thymic Epithelial Cells. PloS One. 10:e0135440 10.1371/journal.pone.0135440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaas, S., Garden O.A., Scudamore C., Turner M., Okkenhaug K., and Vigorito E.. 2009. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J. Immunol. 182:2578–2582. 10.4049/jimmunol.0803162 [DOI] [PubMed] [Google Scholar]
- Konkel, J.E., Jin W., Abbatiello B., Grainger J.R., and Chen W.. 2014. Thymocyte apoptosis drives the intrathymic generation of regulatory T cells. Proc. Natl. Acad. Sci. USA. 111:E465–E473. 10.1073/pnas.1320319111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. . 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Levéen, P., Larsson J., Ehinger M., Cilio C.M., Sundler M., Sjöstrand L.J., Holmdahl R., and Karlsson S.. 2002. Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood. 100:560–568. 10.1182/blood.V100.2.560 [DOI] [PubMed] [Google Scholar]
- Liston, A., Lu L.F., O’Carroll D., Tarakhovsky A., and Rudensky A.Y.. 2008. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205:1993–2004. 10.1084/jem.20081062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lkhagvasuren, E., Sakata M., Ohigashi I., and Takahama Y.. 2013. Lymphotoxin β receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. J. Immunol. 190:5110–5117. 10.4049/jimmunol.1203203 [DOI] [PubMed] [Google Scholar]
- Loeb, G.B., Khan A.A., Canner D., Hiatt J.B., Shendure J., Darnell R.B., Leslie C.S., and Rudensky A.Y.. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell. 48:760–770. 10.1016/j.molcel.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louafi, F., Martinez-Nunez R.T., and Sanchez-Elsner T.. 2010. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-beta. J. Biol. Chem. 285:41328–41336. 10.1074/jbc.M110.146852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L.F., Thai T.H., Calado D.P., Chaudhry A., Kubo M., Tanaka K., Loeb G.B., Lee H., Yoshimura A., Rajewsky K., and Rudensky A.Y.. 2009. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 30:80–91. 10.1016/j.immuni.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, B., McCarthy N.I., Baik S., Cosway E., James K.D., Parnell S.M., White A.J., Jenkinson W.E., and Anderson G.. 2016. Control of the thymic medulla and its influence on αβT-cell development. Immunol. Rev. 271:23–37. 10.1111/imr.12406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchow, S., Leventhal D.S., Lee V., Nishi S., Socci N.D., and Savage P.A.. 2016. Aire Enforces Immune Tolerance by Directing Autoreactive T Cells into the Regulatory T Cell Lineage. Immunity. 44:1102–1113. 10.1016/j.immuni.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou, A.S., Dooley J., Linterman M.A., Pierson W., Ucar O., Kyewski B., Zuklys S., Hollander G.A., Matthys P., Gray D.H., et al. . 2012. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat. Immunol. 13:181–187. 10.1038/ni.2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J.S.A., Lio C.J., Kau A.L., Nutsch K., Yang Z., Gordon J.I., Murphy K.M., and Hsieh C.S.. 2014. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 41:414–426. 10.1016/j.immuni.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietto, A.I., van Dommelen S., Zhou P., Rizzitelli A., D’Amico A., Steptoe R.J., Naik S.H., Lahoud M.H., Liu Y., Zheng P., et al. . 2008. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc. Natl. Acad. Sci. USA. 105:19869–19874. 10.1073/pnas.0810268105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. . 2007. Requirement of bic/microRNA-155 for normal immune function. Science. 316:608–611. 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, S.W., Kim M.Y., Leibbrandt A., Parnell S.M., Jenkinson W.E., Glanville S.H., McConnell F.M., Scott H.S., Penninger J.M., Jenkinson E.J., et al. . 2007. RANK signals from CD4(+)3(-) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 204:1267–1272. 10.1084/jem.20062497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon, B., Lenschow D.J., Rhee L., Ashourian N., Singh B., Sharpe A., and Bluestone J.A.. 2000. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 12:431–440. 10.1016/S1074-7613(00)80195-8 [DOI] [PubMed] [Google Scholar]
- Sánchez-Díaz, R., Blanco-Dominguez R., Lasarte S., Tsilingiri K., Martín-Gayo E., Linillos-Pradillo B., de la Fuente H., Sánchez-Madrid F., Nakagawa R., Toribio M.L., and Martín P.. 2017. Thymus-Derived Regulatory T Cell Development Is Regulated by C-Type Lectin-Mediated BIC/MicroRNA 155 Expression. Mol. Cell. Biol. 37:e00341-16 10.1128/MCB.00341-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seach, N., Hammett M., and Chidgey A.. 2013. Isolation, characterization, and reaggregate culture of thymic epithelial cells. Methods Mol. Biol. 945:251–272. 10.1007/978-1-62703-125-7_15 [DOI] [PubMed] [Google Scholar]
- Sharma, V., Antonacopoulou A.G., Tanaka S., Panoutsopoulos A.A., Bravou V., Kalofonos H.P., and Episkopou V.. 2011. Enhancement of TGF-β signaling responses by the E3 ubiquitin ligase Arkadia provides tumor suppression in colorectal cancer. Cancer Res. 71:6438–6449. 10.1158/0008-5472.CAN-11-1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Yagi K., Kondo M., Kato M., Miyazono K., and Miyazawa K.. 2004. C-Ski inhibits the TGF-beta signaling pathway through stabilization of inactive Smad complexes on Smad-binding elements. Oncogene. 23:5068–5076. 10.1038/sj.onc.1207690 [DOI] [PubMed] [Google Scholar]
- Tai, X., Cowan M., Feigenbaum L., and Singer A.. 2005. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 6:152–162. 10.1038/ni1160 [DOI] [PubMed] [Google Scholar]
- Vigorito, E., Kohlhaas S., Lu D., and Leyland R.. 2013. miR-155: an ancient regulator of the immune system. Immunol. Rev. 253:146–157. 10.1111/imr.12057 [DOI] [PubMed] [Google Scholar]
- Zhang, S.L., and Bhandoola A.. 2014. Trafficking to the thymus. Curr. Top. Microbiol. Immunol. 373:87–111. [DOI] [PubMed] [Google Scholar]
- Zhou, X., Jeker L.T., Fife B.T., Zhu S., Anderson M.S., McManus M.T., and Bluestone J.A.. 2008. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 205:1983–1991. 10.1084/jem.20080707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuklys, S., Balciunaite G., Agarwal A., Fasler-Kan E., Palmer E., and Holländer G.A.. 2000. Normal thymic architecture and negative selection are associated with Aire expression, the gene defective in the autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). J. Immunol. 165:1976–1983. 10.4049/jimmunol.165.4.1976 [DOI] [PubMed] [Google Scholar]
- Zuklys, S., Mayer C.E., Zhanybekova S., Stefanski H.E., Nusspaumer G., Gill J., Barthlott T., Chappaz S., Nitta T., Dooley J., et al. . 2012. MicroRNAs control the maintenance of thymic epithelia and their competence for T lineage commitment and thymocyte selection. J. Immunol. 189:3894–3904. 10.4049/jimmunol.1200783 [DOI] [PMC free article] [PubMed] [Google Scholar]