Abstract
Parkinson's disease (PD) is the second most common neurodegenerative disease. In recent years, it has been shown that leucine-rich repeat kinase 2 (LRRK2) has a crucial function in both familial and sporadic forms of PD. LRRK2 pathogenic mutations are thought to result in an increase in LRRK2 kinase activity. Thus, inhibiting LRRK2 kinase activity has become a main therapeutic target. Many compounds capable of inhibiting LRRK2 kinase activity with high selectivity and brain availability have been described. However, the safety of long-term use of these ATP-competitive LRRK2 kinase inhibitors has been challenged by several studies. Therefore, alternative ways of targeting LRRK2 activity will have a great benefit. In this review, we discuss the recent progress in the development of allosteric inhibitors of LRRK2, mainly via interfering with GTPase activity, and propose potential new intra and interprotein interactions targets that can lead to open doors toward new therapeutics.
Keywords: dimerization, GTPase, kinase inhibitors, leucine rich repeat kinase, Parkinson's disease
Introduction
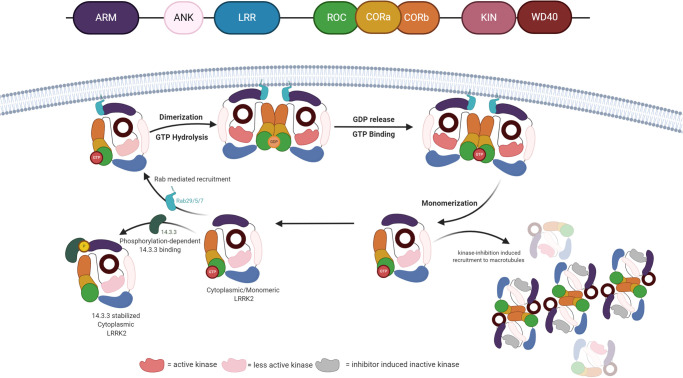
Parkinson's disease (PD) is the second most common neurodegenerative disease [1]. Despite the fact that the etiology of PD is mostly unknown, genome-wide association studies have identified many genetic mutations associated with increased risk of PD. Among all identified risk-genes, missense mutations in the gene encoding leucine-rich repeat kinase 2 (LRRK2) stand as the most common single genetic cause of PD. LRRK2 is an atypical large protein, consisted of four protein–protein interaction (PPI) domains, as well as two domains conferring distinct enzymatic activities, GTPase mediated by the Roc (Ras of complex proteins) domain and a serine–threonine kinase (Figure 1).
Figure 1. LRRK2 oscillate between different conformation for full kinase activation.
LRRK2 is mainly present in a monomeric GTP-bound form in the cytosol. This state is less active and can form a stable cytoplasmic complex with 14-3-3 proteins. Binding of activated Rab proteins induces membrane localization of LRRK2. At the membrane, GTP is hydrolyzed and the protein dimerizes. During the GTPase cycle, the LRRK2 kinase domain gets fully activated and phosphorylates its substrates. The low affinity of LRRK2 for GDP facilitates fast GDP release, rebinding of GTP and, subsequently monomerization of LRRK2 and return to the cytosol. Conventional pharmacological inhibition of LRRK2 induces microtubule recruitment and formation of filamentous LRRK2, dimeric units of LRRK2 extended via WD40 : WD40 or LRR : LRR interaction.
From numerous studies, it is clear that pathogenic mutations of LRRK2 result in a gain-of-function and increased kinase activity [2–4]. Moreover, wild type LRRK2 kinase activity was shown to be increased in patients with idiopathic PD [5], and both LRRK2-related PD and sporadic PD are clinically similar. As a result, considerable efforts were made toward the development of pharmacological inhibitors of LRRK2 kinase activity as a therapeutic intervention. Many compounds capable of inhibiting LRRK2 kinase activity have been described [6,7], and until now, there have been four generations of advancement [8]. The most recently developed (third-generation) ATP-competitive LRRK2 kinase inhibitors, including MLi-2 and PF-06685360, have an optimized potency, selectivity and brain availability [9,10]. Although the first kinase compounds are in clinical trials, several studies have reported that inhibition of LRRK2 with several of these ATP-competitive inhibitors can cause kidney abnormalities in rodents and an accumulation of lamellar bodies in type II pneumocytes in the non-human primate lung [11–13]. While these histological alterations were reversible after drug withdrawal and had no functional consequences [14], it is not completely clear whether the long-term use of such inhibitor will result in different sequels. Furthermore, on a cellular level, it has been shown that these ATP-competitive kinase inhibitors induce cellular recruitment of LRRK2 to microtubules [15–18] which in turn reduces the kinesin — and dynamin-mediated transport along microtubules [19]. It is, therefore, not unlikely that the side effect associated with the use of ATP-competitive LRRK2 kinase inhibitors is a result of this altered localization of LRRK2. Thus, developing other approaches that fine-tune LRRK2 kinase activity might be of an additional benefit.
LRRK2 activation mechanism
Although the exact molecular mechanism is not completely understood, recent data suggest that LRRK2 goes through a complex activation cycle, including membrane-recruitment via binding of upstream effectors to the N-terminus and conformational activation via intramolecular interactions and dimerization (Figure 1). Previous studies showed that LRRK2 exists both in the cytoplasm and as associated with membranous organelles [20–22]. In the cytosol, GTP-bound LRRK2 is mainly monomeric and has a low-kinase activity; while it is predominantly dimeric and active when localized at membranes [20]. The N-terminal segment of LRRK2 interacts with the ubiquitous regulatory protein 14-3-3 in a phosphorylation-dependent manner and thereby stabilizes the cytosolic localization of LRRK2 [23,24]. GTP-bound Rab proteins, including Rab29 and Rab32, bind also to the N-terminus of LRRK2, but thereby recruit GTP-bound LRRK2 to the membrane. At the membrane LRRK2 goes through a multi-step hydrolysis cycle, resulting in a dimeric, GDP-bound conformation of LRRK2 [20]. It is unclear during which step dimerization occurs, however, it is mediated via the Roc and COR domains and is crucial for GTP hydrolysis and to achieve maximum kinase activity. In the active conformation, LRRK2 phosphorylates its substrates, GTP-bound Rab proteins. This shows that the GTPase and kinase activities are clearly linked. However it remains to be determined what exactly is the active conformation of LRRK2 at the membrane. Importantly, all different levels of regulation are essential for proper LRRK2 functioning and therefore are all potential targets to allosterically inhibit LRRK2 activity. In this review, we will highlight the recent advances in developing allosteric LRRK2 inhibitors and discuss our perspective for potential inhibitors for LRRK2 aside from the ATP-binding pocket (Figure 2).
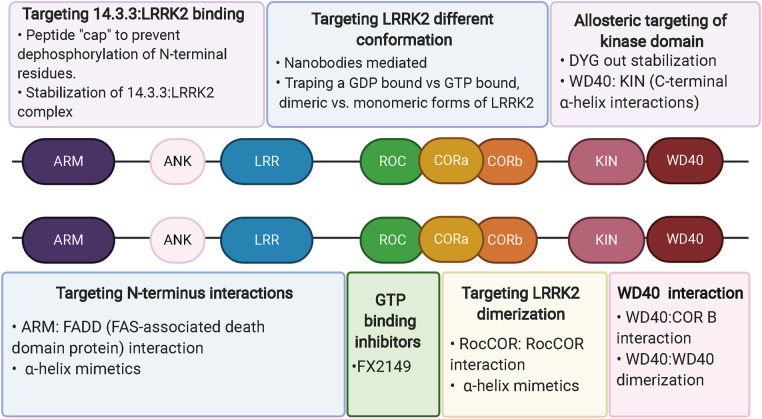
Figure 2. Potential strategies for allosteric inhibition of LRRK2.
Peptidomimetics can be used to disrupt LRRK2 dimerization (RocCOR : RocCOR, WD40 : WD40, WD40 : CORB,) and to interfere with LRRK2 PPIs (ARM domain: FADD). GTP-binding inhibition (FX2149) rescues LRRK2-mediated cellular toxicity. Peptide capping can prevent dephosphorylation of constitutively phosphorylated residues needed for interaction with 14.3.3. Nanobodies can be used to target and stabilize a low-kinase active conformation of LRRK2.
Targeting the LRRK2 kinase domain
In general, classical kinase inhibitors that directly target the ATP-binding pocket (Type I inhibitors) usually have broad selectivity profiles and most have significant off-target effects, unlike allosteric inhibitors which are generally more selective. Whereas the development of classical inhibitors has benefited greatly from the existence of the natural small-molecule substrate, ATP, there are virtually no natural small molecule starting points from which allosteric kinase modulators can be built. This renders allosteric kinase inhibitors more challenging to develop.
Nevertheless, more rational approaches for the development of allosteric inhibitors and efficient high-throughput screening are being utilized more recently and there are already successful examples of allosteric kinase inhibitors in the clinic. This allowed Schaffner et al. [25] to identify a physiological form of vitamin B12, 5′-deoxyadenosylcobalamin (AdoCbl), as the first kinase inhibitor for LRRK2 that is not ATP-competitive. The addition of AdoCbl markedly increased the susceptibility of LRRK2 to proteolysis, whereas conventional LRRK2 kinase inhibitors, protected LRRK2 from proteolytic digestion. This suggests a conformational change in LRRK2 upon binding to AdoCbl that is different from ATP-competitive inhibitors. Keeping in mind the structural similarity between ATP and AdoCbl, both can be considered as adenosine derivatives and it could be possible that vitamin B12 binds the ATP pocket at such a high concentration needed for its kinase inhibitory effect (15 µg/ml vs. 500 pg/ml, the physiological serum concentration). Therefore, it has to be seen how specific AdoCbl is for LRRK2 compared with other kinases.
Another possibility for allosteric targeting of the LRRK2 kinase domain arises from the fact that repositioning of the activation loop of all kinases in the tyrosine kinase-like family is a common conformational change for which LRRK2 is no exception. The conserved DFG motif (DYG motif in LRRK2) in the activation loop tends to create a flexible conformation as a result of the presence of a small glycine residue in the enzyme, which in turn allows the kinase to switch easily between active (DYG-in) and inactive (DYG-out) forms. Using a combination of structure-based analysis and site-directed mutagenesis accompanied by cell-based assays, Schmidt et al. [26] characterized the mechanistic role of the DYG motif in regulating LRRK2 kinase activity. Their data suggest that by stabilizing the DYG-out confirmation (type II inhibitors), it should be possible to inhibit LRRK2 activity. Ray and Liu [27] identified four potential inhibitors of LRRK2 through docking of known DFG-out inhibitors into a LRRK2 model [28]. The next step will be the development of more LRRK2 specific DFG-out inhibitors, which might be challenging especially for the G2019S mutation that lies within the DYG motif and induces a conformational bias towards the active (DYG-in) conformation [28].
Targeting the G-protein cycle
Both the GTPase and the kinase function of LRRK2 are crucial and depend on each other. Furthermore, LRRK2 GTPase activity might be crucial for LRRK2 pathogenicity. In fact, several of the tested pathogenic RocCOR mutations were shown to either increase the affinity for GTP [15,29,30] and/or decrease the rate of GTP hydrolysis [31–36], which is expected to result in enrichment of LRRK2 in the GTP-bound state [34]. In support to this notion, the R1398H protective Roc domain variant is associated with an increased rate of GTP hydrolysis and a weakened binding to GTP [37]. Through a combination of computer-aided drug design and biofunctional screens, Li et al. [38] discovered two compounds that reduce GTP-binding and subsequently inhibit LRRK2 kinase activity. The same group later designed an analog of their parent compound to increase its brain penetrance and in vivo efficacy [39]. Furthermore, reducing GTP-binding activity with these compounds attenuated the impairment of mitochondrial and lysosomal transport in cells expressing R1441C [40] and increased polyubiquitylation of G2019S [41]. Their results highlight the potentiality of targeting LRRK2 kinase activity by targeting the Roc domain.
Using a bacterial homolog of LRRK2, we have shown that the isolated RocCOR domain is primarily dimeric when bound to GDP or when no nucleotides are present, but monomeric when GTP bound [42]. Furthermore, we have revealed that the COR domain is crucial for LRRK2 dimerization [43] and constructs with a truncated COR protein are associated with an impaired dimerization and a significantly lowered (700 times) GTPase activity. Expectedly, impairing dimerization also alters autophosphorylation levels, indicating that both enzymatic activities are dependent on dimerization [34,43–45]. Furthermore, LRRK2 dimerization was shown to be stabilized by multiple domain–domain interactions. Greggio et al. [46] revealed an interaction between Roc and LRR domains. Later, intramolecular interaction was detected between ROC : ROC [45] and ROC : COR [33]. Besides COR domain, the WD40 domain is also involved in LRRK2 dimerization and pathogenicity [47]. Recently, the crystal structure of the WD40 domain of human LRRK2 was reveled at 2.6-Å resolution and showed a dimeric assembly, that was further confirmed by measurements in solution [48]. The deletion of the WD40 repeats led to impaired dimer formation accompanied by diminished kinase activity and altered LRRK2 cellular localization [47]. Although it is not clear how mutations affect LRRK2 dimerization or PPIs, the crystal structure of dimeric LRRK2 WD40 identified surface patches that provide a structure-based template for potential therapeutic interventions. Recently, an atomic model of microtubule-associated LRRK2 was built using cryo-electron tomography [19,49], which further supported the vital need of ROC, COR, and WD40 for LRRK2 dimerization. Thus, any biologic that impairs LRRK2 dimerization is expected to finetune LRRK2 kinase activity.
Interestingly, the most recent model of LRRK2 revealed by Deniston et al. [19] provides a unique feature of LRRK2 represented in a 28-amino acid α-helix located terminally following the WD40 domain and extends along the entire length of the kinase domain. Intriguingly, this α-helix showed extensive interactions with the kinase domain. Furthermore, this helix is juxta-positioned to the COR-B domain. This might suggest a regulatory function of this C-terminal helix in which it acts as an anchoring element that connects COR-B, the kinase, and the WD40 domain [19]. Thus, targeting this particular α-helix (as discussed below) can impair its regulatory function and affect the stability of the kinase domain.
Another successful approach to specifically bind a certain conformation of a protein was achieved via the use of Nanobodies [50,51]. Nanobodies are robust small (15 kDa) single-domain antigen-binding fragments derived from heavy chain-only antibodies, naturally occurring in Camelids, which can be easily produced by a wide range of different hosts. Unlike conventional antibodies, antigen-recognition sites of nanobodies typically form a convex surface with a large elongated complementarity-determining region loop 3 (CDR3) capable of reaching hidden pockets that are less antigenic for other types of binders [52]. This unique feature allowed for a conformation-specific binding between nanobodies and their corresponding antigen [53,54]. Actually, it was shown that nanobodies could be used to trap a kinase in a specific conformation and subsequently affect kinase activity [50,55,56]. Furthermore, Leemans et al. [57] recently used a bacterial LRRK2 homolog and identified two conformation-specific nanobodies that trapped the protein in a GTP-bound state and subsequently increased the GTP turnover rate and reverted the decrease in GTPase activity caused by a PD-analogous mutation. In this regard, the generation of LRRK2 conformation-specific nanobodies has the potential to also trap LRRK2 in both a kinase active or a kinase-inactive conformation. Besides being a valuable tool to understand LRRK2 dynamics and domains plasticity, LRRK2/nanobody structures could form the basis for the future rational design of LRRK2 allosteric inhibitors. Furthermore, nanobodies were shown to be distributed widely and rapidly through the human body once administered [58]. The fusion of nanobodies to cell-penetrating peptides generated nanobodies that are able to get access into cells (transbodies) [59–61], and different approaches were successfully utilized to allow nanobodies to cross the blood-brain barrier (BBB) [62,63]. Nevertheless, the use of nanobodies to target intracellular proteins is still challenged by the short half-life of the nanobodies in the bloodstream and the relatively large amount of nanobodies needed to obtain the desired effect.
Targeting LRRK2 protein–protein interactions
Most kinases exert their biochemical activity as being a partner in a complex network of PPIs. Similarly, LRRK2 was also shown to be functioning as a scaffolding protein [64,65], that pre-assembles signaling complexes at specific cellular localizations [66]. In turn, targeting LRRK2 scaffolding interactions is another attractive strategy to reverse its associated pathogenicity. In this regard, LRRK2 was shown to mediate neuronal cell death through interaction as a scaffold to sequester the ASK1 signaling kinases (ASK1, MKK6, and p38 MAPK) [65]. Also, LRRK2 was shown to bind to synaptic vesicles via an interaction of the WD40 domain and vesicle-associated proteins. Importantly, the toxicity induced by overexpression of full-length LRRK2 in primary neuronal cultures was mimicked by ectopic expression of a construct containing only the C-terminal WD40 domain of LRRK2 [67]. In a similar context, it was shown that LRRK2 neuronal cell death takes place in a FADD-dependent manner [68] and prevention of the physical interaction between FADD and LRRK2 is neuroprotective in primary neurons expressing mutant LRRK2 [69]. The group could also identify the region of LRRK2 involved in the interaction and generated a docking model that uncovered a strong hydrophobic core mediated via α-helices interactions between the LRRK2 ARM domain and FADD protein. This model can provide the basis for a structure-based design of small molecule inhibitors with a potential therapeutic value.
As discussed earlier, LRRK2 kinase activation is regulated by 14-3-3 protein binding, and pathogenic mutations of LRRK2 were reported to impair the protein ability to interact with 14-3-3 protein [70] which in turn is associated with neurite shortening and cell death [71]. Taking into account that 14-3-3 is binding to LRRK2 in a phosphorylation-dependent manner, it has been suggested to protect the phosphorylation residues from being dephosphorylated when unbound to 14-3-3, with a peptide ‘cap’ [72]. Recently, the exact 14-3-3 binding sites in LRRK2 were identified [73,74]. Since both natural and synthetic molecules are already available in the literature that achieve their physiological activities by stabilizing 14-3-3 binding with their target protein (reviewed in [75]), it sounds plausible to utilize available approaches [75,76] to generate a more specific LRRK2-14-3-3 complex stabilizer.
While small molecules have had success as therapeutic kinase inhibitors, they generally failed to address the flat and large nature of PPI surfaces. However, with the recent fast-tracked advance in drug discovery approaches, new PPI-based drugs were delivered leading to an expansive possibility to target what was previously considered ‘undruggable’. Indeed, targeting Kinases PPI interfaces has been shown as a valid strategy to allosterically inhibit kinases. Different approaches have been utilized and can be reflected in LRRK2 targeting. A fluorescence polarization (FP) competitive binding assay can be developed to screen for compounds that disrupt LRRK2 dimerization similar to the approach of Rettenmaier et al. [77] which allowed for the identification of potent disruptors of the interaction between PDK1 and PIFtide. It is convincible to utilize this technique to screen for RocCOR : RocCOR or WD40 : WD40 interactions, provided that both domains are well expressed and stably produced. In this regard, the dimerization interface can be mimicked by fluorescently labeled peptides as FP probes for a competitive binding assay that should be suitable for a high-throughput screen.
Furthermore, a side-directed approach, disulfide trapping (or tethering), was developed previously to allow for a more straightforward characterization and interrogation of allosteric sites on protein kinases with small molecules. Disulfide trapping involves screening disulfide-containing compounds for their ability to form a mixed disulfide with a natural or engineered cysteine residue near a site of interest on a protein target [78,79]. This technique allowed for the identification of potent inhibitors of many disease-related tyrosine kinases [80,81]. This approach can be of great help to discover inhibitors for the WD40 : FADD interaction, or to disrupt C-helix mediated kinase stability; provided that residues involved in each interaction are well identified.
Another potentiality arises from the fact that α-helices are one of the most abundant secondary structural elements involved in PPI interfaces, which confers the ability to bind large and relatively flat surfaces both efficiently and specifically. Accordingly, the use of a peptide to mimic such an α-helix can lead to specific disruption of PPIs. Interestingly, although peptidomimetics are small compared with their parent protein, they have similar specificity toward their targets as biologics. Moreover, peptides are more compelling binders to PPIs interfaces, and exert less toxicity when compared with small molecule drugs, which often produce toxic metabolites. In contrast with small molecules, peptides are degraded into amino acids, which are in turn not toxic or harmful for cells. Different approaches have shown success in producing biologics that therapeutically inhibit PPIs as peptidomimetics. Furthermore, to overcome the rapid degradation with proteases and to stabilize the helical foldability, non-natural amino acids can be used as building blocks of peptides, which in turn create an opportunity to produce diverse scaffolds with modified chemical and functional properties. A step further toward better bioactive peptides was achieved by engineering a non-native carbon–carbon bond constraint with olefin tethers into the peptide sequence [82]. This new hydrocarbon-stapled backbone approach has provided a platform for many successful α-helical peptidomimetics [83–85].
Utilizing any of these approaches in combination with crystal structures or protein models of LRRK2 (or certain domains of interest) is expected to lead to new biologics that can alter LRRK2 kinase and cellular pathogenicity in a completely new mechanism different from conventional kinase inhibitors.
Conclusions
Although potent ATP-competitive kinase inhibitors are available for LRRK2, they induce alteration in LRRK2 conformations and are associated with the recruitment of LRRK2 to microtubules, which in turn results in impaired vesicular transportation within the cell. Allosteric inhibition of LRRK2 is expected to provide an alternative way, potentially without objectionable alteration of cellular localization of the protein. Furthermore, stabilization of different conformation of LRRK2 would be of great importance from a structural point of view. So far, GTP-binding has been successfully targeted and at least two compounds are available with the ability to prevent LRRK2-associated neuronal damage. Although a crystal structure of LRRK2 is still not available yet, recent data revealed a high-resolution structure of LRRK2-bound to microtubules [19,49]. Such advancement should allow for structure-based targeting and design of novel allosteric inhibitors. Furthermore, selective nanobodies have already been identified against conformation-specific forms of a bacterial homolog of LRRK2. Thus, identification of conformation-specific nanobodies against LRRK2 would provide a crucial tool for further characterization of LRRK2 activation steps and biochemical kinetics. The use of peptide-mimetics has been substantially increasing as a successful way of targeting PPIs. In particular, stapled peptides mimicking α-helical structure showed potency to stand as an efficient therapeutic molecule. Accordingly, utilizing such an approach for targeting LRRK2 dimerization interface and/or LRRK2 PPIs is expected to render new useful molecules that can finetune LRRK2 kinase activity. It is worth mentioning that LRRK2 is not only targeted on a protein level, and targeting LRRK2 mRNA with antisense oligonucleotides has shown to be effective in rescuing LRRK2-dependent cellular dysfunction, with a potentiality to correct PD-toxicity [86].
In summary, we have discussed the most promising approaches for allosteric targeting of LRRK2 according to the current knowledge of LRRK2 and its pathogenicity. Besides its potential therapeutic use, such inhibitors would help understanding more about the unique activation mechanism of LRRK2 and its cellular interactions.
Perspectives
Allosteric inhibition of LRRK2 is expected to provide an alternative way potentially without objectionable alteration of cellular localization of the protein. Furthermore, stabilization of different conformation of LRRK2 would be of great importance from a structural point of view.
Although potent ATP-competitive kinase inhibitors are available for LRRK2, they induce alteration in LRRK2 conformations and are associated with the recruitment of LRRK2 to microtubules, which in turn results in impaired vesicular transportation within the cell. So far, GTP-binding has been successfully targeted and at least two compounds are available with the ability to prevent LRRK2-associated neuronal damage.
Although a crystal structure of LRRK2 is still not available yet, recent data revealed a 14-A° structure of LRRK2-bound to microtubules. Such advancement should allow for structure-based targeting and design of novel allosteric inhibitors. Furthermore, selective nanobodies have already been identified against conformation-specific forms of a bacterial homolog of LRRK2. Thus, identification of conformation-specific nanobodies against LRRK2 would provide a crucial tool for further characterization of LRRK2 activation steps and biochemical kinetics.
Abbreviations
- FP
fluorescence polarization
- LRRK2
leucine-rich repeat kinase 2
- PD
Parkinson's disease
- PPI
protein–protein interaction
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
Work in the author's laboratories is supported by The Michael J. Fox Foundation for Parkinson's Research (A.K.) and a Tubitak 2232 grant (A.K.).
Open Access
Open access for this article was enabled by the participation of University of Groningen in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society.
Author Contribution
A.S., F.N.C. and A.K. wrote and edited the manuscript.
References
- 1.Poewe W., Seppi K., Tanner C.M., Halliday G.M., Brundin P., Volkmann J. et al. (2017) Parkinson disease. Nat. Rev. Dis. Prim. 3, 17013 10.1038/nrdp.2017.1 [DOI] [PubMed] [Google Scholar]
- 2.West A.B., Moore D.J., Biskup S., Bugayenko A., Smith W.W., Ross C.A. et al. (2005) Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U.S.A. 102, 16842–16847 10.1073/pnas.0507360102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A. et al. (2006) Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 23, 329–341 10.1016/j.nbd.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 4.Berwick D.C., Javaheri B., Wetzel A., Hopkinson M., Nixon-Abell J., Grannò S. et al. (2017) Pathogenic LRRK2 variants are gain-of-function mutations that enhance LRRK2-mediated repression of β-catenin signaling. Mol. Neurodegener. 12, 9 10.1186/s13024-017-0153-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Maio R., Hoffman E.K., Rocha E.M., Keeney M.T., Sanders L.H., De Miranda B.R. et al. (2018) LRRK2 activation in idiopathic Parkinson's disease. Sci. Transl. Med. 10, eaar5429 10.1126/scitranslmed.aar5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domingos S., Duarte T., Saraiva L., Guedes R.C. and Moreira R. (2019) Targeting leucine-rich repeat kinase 2 (LRRK2) for the treatment of Parkinson's disease. Future Med. Chem. 11, 1953–1977 10.4155/fmc-2018-0484 [DOI] [PubMed] [Google Scholar]
- 7.Vancraenenbroeck R., Lobbestael E., De Maeyer M., Baekelandt V. and Taymans J.-M. (2012) Kinases as targets for Parkinson's disease: from genetics to therapy. CNS Neurol. Disord. - Drug Targets 10, 724–740 10.2174/187152711797247858 [DOI] [PubMed] [Google Scholar]
- 8.West A.B. (2017) Achieving neuroprotection with LRRK2 kinase inhibitors in Parkinson disease. Exp. Neurol. 298, 236–245 10.1016/j.expneurol.2017.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott J.D., DeMong D.E., Greshock T.J., Basu K., Dai X., Harris J. et al. (2017) Discovery of a 3-(4-Pyrimidinyl) indazole (MLi-2), an orally available and selective leucine-rich repeat kinase 2 (LRRK2) inhibitor that reduces brain kinase activity. J. Med. Chem. 60, 2983–2992 10.1021/acs.jmedchem.7b00045 [DOI] [PubMed] [Google Scholar]
- 10.Fell M.J., Mirescu C., Basu K., Cheewatrakoolpong B., Demong D.E., Ellis J.M. et al. (2015) MLi-2, a potent, selective, and centrally active compound for exploring the therapeutic potential and safety of LRRK2 kinase inhibition. J. Pharmacol. Exp. Ther. 355, 397–409 10.1124/jpet.115.227587 [DOI] [PubMed] [Google Scholar]
- 11.Andersen M.A., Wegener K.M., Larsen S., Badolo L., Smith G.P., Jeggo R. et al. (2018) PFE-360-induced LRRK2 inhibition induces reversible, non-adverse renal changes in rats. Toxicology 395, 15–22 10.1016/j.tox.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 12.Fuji R.N., Flagella M., Baca M., Baptista MA S., Brodbeck J., Chan B.K. et al. (2015) Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci. Transl. Med. 7, 273ra15 10.1126/scitranslmed.aaa3634 [DOI] [PubMed] [Google Scholar]
- 13.Herzig M.C., Kolly C., Persohn E., Theil D., Schweizer T., Hafner T. et al. (2011) LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum. Mol. Genet. 20, 4209–4223 10.1093/hmg/ddr348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baptista M.A.S., Merchant K., Barrett T., Bhargava S., Bryce D.K., Michael Ellis J. et al. (2020) LRRK2 inhibitors induce reversible changes in nonhuman primate lungs without measurable pulmonary deficits. Sci. Transl. Med. 12, eaav0820 10.1126/scitranslmed.aav0820 [DOI] [PubMed] [Google Scholar]
- 15.Ramírez M.B., Ordóñez A.J.L., Fdez E., Madero-Pérez J., Gonnelli A., Drouyer M. et al. (2017) GTP binding regulates cellular localization of Parkinson's disease-associated LRRK2. Hum. Mol. Genet. 26, 2747–2767 10.1093/hmg/ddx161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X., Dzamko N., Prescott A., Davies P., Liu Q., Yang Q. et al. (2011) Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat. Chem. Biol. 7, 203–205 10.1038/nchembio.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dzamko N., Deak M., Hentati F., Reith A.D., Prescott A.R., Alessi D.R. et al. (2010) Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser 910/Ser935, disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem. J. 430, 405–413 10.1042/BJ20100784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kett L.R., Boassa D., Ho C.C.Y., Rideout H.J., Hu J., Terada M. et al. (2012) LRRK2 Parkinson disease mutations enhance its microtubule association. Hum. Mol. Genet. 21, 890–899 10.1093/hmg/ddr526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deniston C.K., Salogiannis J., Mathea S., Snead D.M., Lahiri I., Matyszewski M. et al. (2020) Structure of LRRK2 in Parkinson's disease and model for microtubule interaction. Nature 10.1038/s41586-020-2673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger Z., Smith K.A. and Lavoie M.J. (2010) Membrane localization of LRRK2 is associated with increased formation of the highly active lrrk2 dimer and changes in its phosphorylation. Biochemistry 49, 5511–5523 10.1021/bi100157u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James N.G., Digman M.A., Gratton E., Barylko B., Ding X., Albanesi J.P. et al. (2012) Number and brightness analysis of LRRK2 oligomerization in live cells. Biophys. J. 102, 41–43 10.1016/j.bpj.2012.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schapansky J., Nardozzi J.D., Felizia F. and LaVoie M.J. (2014) Membrane recruitment of endogenous LRRK2 precedes its potent regulation of autophagy. Hum. Mol. Genet. 23, 4201–4214 10.1093/hmg/ddu138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichols R.J., Dzamko N., Morrice N.A., Campbell D.G., Deak M., Ordureau A. et al. (2010) 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem. J. 430, 393–404 10.1042/BJ20100483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynolds A., Doggett E.A., Riddle S.M., Lebakken C.S. and Nichols R.J. (2014) LRRK2 kinase activity and biology are not uniformly predicted by its autophosphorylation and cellular phosphorylation site status. Front. Mol. Neurosci. 7, 1–14 10.3389/fnmol.2014.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffner A., Li X., Gomez-Llorente Y., Leandrou E., Memou A., Clemente N. et al. (2019) Vitamin B12 modulates Parkinson's disease LRRK2 kinase activity through allosteric regulation and confers neuroprotection. Cell Res. 29, 313–329 10.1038/s41422-019-0153-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt S.H., Knape M.J., Boassa D., Mumdey N., Kornev A.P., Ellisman M.H. et al. (2019) The dynamic switch mechanism that leads to activation of LRRK2 is embedded in the DFGψ motif in the kinase domain. Proc. Natl. Acad. Sci. U.S.A. 116, 14979-14988 10.1073/pnas.1900289116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray S. and Liu M. (2012) Current understanding of LRRK2 in Parkinsons disease: biochemical and structural features and inhibitor design. Future Med. Chem. 4, 1701–1713 10.4155/fmc.12.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M., Bender S.A., Cuny G.D., Sherman W., Glicksman M. and Ray S.S. (2013) Type II kinase inhibitors show an unexpected inhibition mode against Parkinson's disease-linked LRRK2 mutant G2019S. Biochemistry 52, 1725–1736 10.1021/bi3012077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West A.B., Moore D.J., Choi C., Andrabi S.A., Li X., Dikeman D. et al. (2007) Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 16, 223–232 10.1093/hmg/ddl471 [DOI] [PubMed] [Google Scholar]
- 30.Aasly J.O., Vilariño-Güell C., Dachsel J.C., Webber P.J., West A.B., Haugarvoll K. et al. (2010) Novel pathogenic LRRK2 p.Asn1437His substitution in familial Parkinson's disease. Mov. Disord. 25, 2156–2163 10.1002/mds.23265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis P.A., Greggio E., Beilina A., Jain S., Baker A. and Cookson M.R. (2007) The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 357, 668–671 10.1016/j.bbrc.2007.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Tan Y.-C., Poulose S., Olanow C.W., Huang X.-Y. and Yue Z. (2007) Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J. Neurochem. 103, 238–247 10.1111/j.1471-4159.2007.04743.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danie˜ ls V., Vancraenenbroeck R., Law B.M.H., Greggio E., Lobbestael E., Gao F. et al. (2011) Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. J. Neurochem. 116, 304–315 10.1111/j.1471-4159.2010.07105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao J., Wu C.-X., Burlak C., Zhang S., Sahm H., Wang M. et al. (2014) Parkinson disease-associated mutation R1441H in LRRK2 prolongs the “active state” of its GTPase domain. Proc. Natl. Acad. Sci. U.S.A. 111, 4055–4060 10.1073/pnas.1323285111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong Y., Coombes C.E., Kilaru A., Li X., Gitler A.D., Bowers W.J. et al. (2010) GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet. 6, e1000902 10.1371/journal.pgen.1000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C.X., Liao J., Park Y., Reed X., Engel V.A., Hoang N.C. et al. (2019) Parkinson's disease-associated mutations in the GTPase domain of LRRK2 impair its nucleotide-dependent conformational dynamics. J. Biol. Chem. 294, 5907–5913 10.1074/jbc.RA119.007631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nixon-Abell J., Berwick D.C., Grannó S., Spain V.A., Blackstone C. and Harvey K. (2016) Protective LRRK2 R1398H variant enhances GTPase and Wnt signaling activity. Front. Mol. Neurosci. 9, 18 10.3389/fnmol.2016.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T., Yang D., Zhong S., Thomas J.M., Xue F., Liu J. et al. (2014) Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson's disease cell and mouse models. Hum. Mol. Genet. 23, 6212–6222 10.1093/hmg/ddu341 [DOI] [PubMed] [Google Scholar]
- 39.Li T., He X., Thomas J.M., Yang D., Zhong S., Xue F. et al. (2015) A novel GTP-binding inhibitor, FX2149, attenuates LRRK2 toxicity in Parkinson's disease models. PLoS One 10, e0122461 10.1371/journal.pone.0122461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas J.M., Li T., Yang W., Xue F., Fishman P.S. and Smith W.W. (2017) 68 and FX2149 attenuate mutant LRRK2-R1441C-induced neural transport impairment. Front. Aging Neurosci. 8, 337 10.3389/fnagi.2016.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas J.M., Wang X., Guo G., Li T., Dai B., Nucifora L.G. et al. (2020) GTP-binding inhibitors increase LRRK2-linked ubiquitination and Lewy body-like inclusions. J. Cell. Physiol. 235, 7309–7320 10.1002/jcp.29632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deyaert E., Wauters L., Guaitoli G., Konijnenberg A., Leemans M., Terheyden S. et al. (2017) A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover. Nat. Commun. 8, 1008 10.1038/s41467-017-01103-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gotthardt K., Weyand M., Kortholt A., Van Haastert P.J.M. and Wittinghofer A. (2008) Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 27, 2239–2249 10.1038/emboj.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greggio E., Taymans J.M., Zhen E.Y., Ryder J., Vancraenenbroeck R., Beilina A. et al. (2009) The Parkinson's disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites. Biochem. Biophys. Res. Commun. 389, 449–454 10.1016/j.bbrc.2009.08.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng J., Lewis P.A., Greggio E., Sluch E., Beilina A. and Cookson M.R. (2008) Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc. Natl. Acad. Sci. U.S.A. 105, 1499–1504 10.1073/pnas.0709098105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J.M., Daniëls V. et al. (2008) The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 283, 16906–16914 10.1074/jbc.M708718200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen N.D., Peng Y., Ho C.C.Y., Rideout H.J., Petrey D., Liu P. et al. (2009) The WD40 domain is required for LRRK2 neurotoxicity. PLoS One 4, e8463 10.1371/journal.pone.0008463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P., Fan Y., Ru H., Wang L., Magupalli V.G., Taylor S.S. et al. (2019) Crystal structure of the WD40 domain dimer of LRRK2. Proc. Natl. Acad. Sci. U.S.A. 116, 1579–1584 10.1073/pnas.1817889116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe R., Buschauer R., Böhning J., Audagnotto M., Lasker K., Lu T.-W. et al. (2020) The in situ structure of Parkinson's disease-linked LRRK2. Cell. 182, 1508–1518.e16 10.1016/j.cell.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaikuad A., Keates T., Vincke C., Kaufholz M., Zenn M., Zimmermann B. et al. (2014) Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies. Biochem. J. 459, 59–69 10.1042/BJ20131399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bayliss R., Burgess S.G. and McIntyre P.J. (2017) Switching aurora-A kinase on and off at an allosteric site. FEBS J. 284, 2947–2954 10.1111/febs.14069 [DOI] [PubMed] [Google Scholar]
- 52.Muyldermans S. (2013) Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- 53.Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A. et al. (2010) Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–139 10.1038/nsmb.1727 [DOI] [PubMed] [Google Scholar]
- 54.Kaur H., Hartmann J.B., Jakob R.P., Zahn M., Zimmermann I., Maier T. et al. (2019) Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J. Biomol. NMR. 73, 375–384 10.1007/s10858-019-00250-8 [DOI] [PubMed] [Google Scholar]
- 55.Zorba A., Nguyen V., Koide A., Hoemberger M., Zheng Y., Kutter S. et al. (2019) Allosteric modulation of a human protein kinase with monobodies. Proc. Natl. Acad. Sci. U.S.A. 116, 13937–13942 10.1073/pnas.1906024116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingram J.R., Knockenhauer K.E., Markus B.M., Mandelbaum J., Ramek A., Shan Y. et al. (2015) Allosteric activation of apicomplexan calciumdependent protein kinases. Proc. Natl. Acad. Sci. U.S.A. 112, E4975–E4984 10.1073/pnas.1505914112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leemans M., Galicia C., Deyaert E., Daems E., Krause L., Paesmans J. et al. (2020) Allosteric modulation of the GTPase activity of a bacterial LRRK2 homolog by conformation-specific nanobodies. Biochem. J. 477, 1203–1218 10.1042/BCJ20190843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Roy M., Ververken C., Beirnaert E., Hoefman S., Kolkman J., Vierboom M. et al. (2015) The preclinical pharmacology of the high affinity anti-IL-6R Nanobody® ALX-0061 supports its clinical development in rheumatoid arthritis. Arthritis Res. Ther. 17, 135 10.1186/s13075-015-0651-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jittavisutthikul S., Thanongsaksrikul J., Thueng-In K., Chulanetra M., Srimanote P., Seesuay W. et al. (2015) Humanized-VHH transbodies that inhibit HCV protease and replication. Viruses 7, 2030–2056 10.3390/v7042030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tabtimmai L., Suphakun P., Srisook P., Kiriwan D., Phanthong S., Kiatwuthinon P. et al. (2019) Cell-penetrable nanobodies (transbodies) that inhibit the tyrosine kinase activity of EGFR leading to the impediment of human lung adenocarcinoma cell motility and survival. J. Cell. Biochem. 120, 18077–18087 10.1002/jcb.29111 [DOI] [PubMed] [Google Scholar]
- 61.Thueng-in K., Thanongsaksrikul J., Srimanote P., Bangphoomi K., Poungpair O., Maneewatch S. et al. (2012) Cell penetrable humanized-VH/VHH that inhibit RNA dependent RNA polymerase (NS5B) of HCV. PLoS One 7, e49254 10.1371/journal.pone.0049254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li T., Bourgeois J.P., Celli S., Glacial F., Le Sourd A.M., Mecheri S. et al. (2012) Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: Application to brain imaging. FASEB J. 26, 3969–3979 10.1096/fj.11-201384 [DOI] [PubMed] [Google Scholar]
- 63.Boado R.J., Hui E.K.W., Lu J.Z. and Pardridge W.M. (2012) Glycemic control and chronic dosing of rhesus monkeys with a fusion protein of iduronidase and a monoclonal antibody against the human insulin receptor. Drug Metab. Dispos. 40, 2021–2025 10.1124/dmd.112.046375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berwick D.C. and Harvey K. (2012) LRRK2 functions as a wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6. Hum. Mol. Genet. 21, 4966–4979 10.1093/hmg/dds342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoon J.H., Mo J.S., Kim M.Y., Ann E.J., Ahn J.S., Jo E.H. et al. (2017) LRRK2 functions as a scaffolding kinase of ASK1-mediated neuronal cell death. Biochim. Biophys. Acta - Mol. Cell Res. 1864, 2356–2368 10.1016/j.bbamcr.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 66.Berwick D.C. and Harvey K. (2011) LRRK2 signaling pathways: The key to unlocking neurodegeneration? Trends Cell Biol. 21, 257–265 10.1016/j.tcb.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 67.Piccoli G., Onofri F., Cirnaru M.D., Kaiser C.J.O., Jagtap P., Kastenmuller A. et al. (2014) Leucinerich repeat kinase 2 binds to neuronal vesicles through protein interactions mediated by Its C-terminal WD40 domain. Mol. Cell. Biol. 34, 2147–2161 10.1128/MCB.00914-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melachroinou K., Leandrou E., Valkimadi P.E., Memou A., Hadjigeorgiou G., Stefanis L. et al. (2016) Activation of FADD-dependent neuronal death pathways as a predictor of pathogenicity for LRRK2 mutations. PLoS One 11, e0166053 10.1371/journal.pone.0166053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antoniou N., Vlachakis D., Memou A., Leandrou E., Valkimadi P.-E., Melachroinou K. et al. (2018) A motif within the armadillo repeat of Parkinson's-linked LRRK2 interacts with FADD to hijack the extrinsic death pathway. Sci. Rep. 8, 3455 10.1038/s41598-018-21931-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X., Wang Q.J., Pan N., Lee S., Zhao Y., Chait B.T. et al. (2011) Phosphorylation-dependent 14-3-3 binding to LRRK2 is impaired by common mutations of familial Parkinson's disease. PLoS One 6, e17153 10.1371/journal.pone.0017153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lavalley N.J., Slone S.R., Ding H., West A.B. and Yacoubian T.A. (2016) 14-3-3 proteins regulate mutant LRRK2 kinase activity and neurite shortening. Hum. Mol. Genet. 25, 109–122 10.1093/hmg/ddv453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rudenko I.N., Chia R. and Cookson M.R. (2012) Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson's disease? BMC Med. 10, 20 10.1186/1741-7015-10-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manschwetus J.T., Wallbott M., Fachinger A., Obergruber C., Pautz S., Bertinetti D. et al. (2020) Binding of the human 14-3-3 isoforms to distinct sites in the leucine-rich repeat kinase 2. Front. Neurosci. 14, 302 10.3389/fnins.2020.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stevers L.M., de Vink P.J., Ottmann C., Huskens J. and Brunsveld L. (2018) A thermodynamic model for multivalency in 14-3-3 protein–protein interactions. J. Am. Chem. Soc. 140, 14498–14510 10.1021/jacs.8b09618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ballone A., Centorrino F. and Ottmann C. (2018) 14-3-3: a case study in ppi modulation. Molecules 23, 1386 10.3390/molecules23061386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao J., Meyerkord C.L., Du Y., Khuri F.R. and Fu H. (2011) 14-3-3 proteins as potential therapeutic targets. Semin. Cell Dev. Biol. 22, 705–712 10.1016/j.semcdb.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rettenmaier T.J., Sadowsky J.D., Thomsen N.D., Chen S.C., Doak A.K., Arkin M.R. et al. (2014) A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1. Proc. Natl. Acad. Sci. U.S.A. 111, 18590–18595 10.1073/pnas.1415365112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Erlanson D.A., Braisted A.C., Raphael D.R., Randal M., Stroud R.M., Gordon E.M. et al. (2000) Site-directed ligand discovery. Proc. Natl. Acad. Sci. U.S.A. 97, 9367–9372 10.1073/pnas.97.17.9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heikal A., Nakatani Y., Jiao W., Wilson C., Rennison D., Weimar M.R. et al. (2018) ‘Tethering’ fragment-based drug discovery to identify inhibitors of the essential respiratory membrane protein type II NADH dehydrogenase. Bioorganic Med. Chem. Lett. 28, 2239–2243 10.1016/j.bmcl.2018.05.048 [DOI] [PubMed] [Google Scholar]
- 80.Hopkins B.T., Bame E., Bell N., Bohnert T., Bowden-Verhoek J.K., Bui M. et al. (2019) Optimization of novel reversible Bruton's tyrosine kinase inhibitors identified using tethering-fragment-based screens. Bioorganic Med. Chem. 27, 2905–2913 10.1016/j.bmc.2019.05.021 [DOI] [PubMed] [Google Scholar]
- 81.Miller R.M., Paavilainen V.O., Krishnan S., Serafimova I.M. and Taunton J. (2013) Electrophilic fragment-based design of reversible covalent kinase inhibitors. J. Am. Chem. Soc. 135, 5298–5301 10.1021/ja401221b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schafmeister C.E., Po J. and Verdine G.L. (2000) An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J. Am. Chem. Soc. 122, 5891–5892 10.1021/ja000563a [DOI] [Google Scholar]
- 83.Cowell J.K., Teng Y., Bendzunas N.G., Ara R., Arbab A.S. and Kennedy E.J. (2017) Suppression of breast cancer metastasis using stapled peptides targeting the WASF regulatory complex. Cancer Growth Metastasis 10, 117906441771319 10.1177/1179064417713197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fulton M.D., Hanold L.E., Ruan Z., Patel S., Beedle A.M., Kannan N. et al. (2018) Conformationally constrained peptides target the allosteric kinase dimer interface and inhibit EGFR activation. Bioorganic Med. Chem. 26, 1167–1173 10.1016/j.bmc.2017.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teng Y., Bahassan A., Dong D., Hanold L.E., Ren X., Kennedy E.J. et al. (2016) Targeting the WASF3-CYFIP1 complex using stapled peptides suppresses cancer cell invasion. Cancer Res. 76, 965–973 10.1158/0008-5472.CAN-15-1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Korecka J.A., Thomas R., Hinrich A.J., Moskites A.M., Macbain Z.K., Hallett P.J. et al. (2020) Splice-switching antisense oligonucleotides reduce LRRK2 kinase activity in human LRRK2 transgenic mice. Mol. Ther. - Nucleic Acids 21, 623–635 10.1016/j.omtn.2020.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]