Abstract
Background
Helicobacter pylori (H. pylori) is ubiquitous among humans and one of the best-studied examples of an intimate association between bacteria and humans. Phylogeny and Phylogeography of H. pylori strains are known to mirror human migration patterns and reflect significant demographic events in human prehistory. In this study, we analyzed the molecular evolution of H. pylori strains detected from different tribes and regions of Sudan using 16S rRNA gene and the phylogenetic approach. Materials and methods. A total of 75 gastric biopsies were taken from patients who had been referred for endoscopy from different regions of Sudan. The DNA extraction was performed by using the guanidine chloride method. Two sets of primers (universal and specific for H. pylori) were used to amplify the 16S ribosomal gene. Sanger sequencing was applied, and the resulted sequences were matched with the sequences of the National Center for Biotechnology Information (NCBI) nucleotide database. The evolutionary aspects were analyzed using MEGA7 software.
Results
Molecular detection of H. pylori has shown that 28 (37.33%) of the patients were positive for H. pylori and no significant differences were found in sociodemographic characteristics, endoscopy series, and H. pylori infection. Nucleotide variations were observed at five nucleotide positions (positions 219, 305, 578, 741, and 763–764), and one insertion mutation (750_InsC_751) was present in sixty-seven percent (7/12) of our strains. These six mutations were detected in regions of the 16S rRNA not closely associated with either tetracycline or tRNA binding sites; 66.67% of them were located in the central domain of 16S rRNA. The phylogenetic analysis of 16S rRNA sequences identified two lineages of H. pylori strains detected from different regions in Sudan. The presence of Sudanese H. pylori strains resembling Hungarian H. pylori strains could reflect the migration of Hungarian people to Sudan or vice versa.
Conclusion
This finding emphasizes the significance of studying the phylogeny of H. pylori strains as a discriminatory tool to mirror human migration patterns. In addition, the 16S rRNA gene amplification method was found useful for bacterial identification and phylogeny.
1. Introduction
Helicobacter pylori (H. pylori) is ubiquitous among humans [1] and one of the best-studied examples of an intimate association between bacteria and humans [2]. It has infected human around 100,000 years ago (range: 88,000–116,000) [1, 3] and has coevolved with humans ancestors for approximately 58000 ± 3500 years, during their first migrations from east Africa [1, 4, 5]. Also, it largely escaped notice until it was cultured by Marshall and Warren [6, 7]. In fact, H. pylori possesses several properties that help this bacterium to persist for several decades, transmit from generation to generation, and make an intimate association with its human host [2, 8]. H. pylori infection is predominantly transmitted vertically from the parent to child and between individuals in close contact such as in a family [2, 9]. This close transmission pattern has resulted in a clear phylogeographic differentiation within these bacteria because of the local dispersion of single nucleotide polymorphisms by high rates of homologous recombination [10, 11]. However, under poverty and inappropriate conditions, especially in the developing countries, H. pylori can be transmitted horizontally which makes infection with multiple strains probably more common than in the developed world [12, 13].
In Sudan, the population is culturally, linguisticall,y and ethnically diverse with more than 597 tribes who speak more than 400 dialects and languages [14, 15]. The majority of the Sudanese population is rural with an urban population of just 33.2%; most of them are in Khartoum [15, 16]. Sudan has severely suffered war, famine, and flood in recent decades and has a large population of internally displaced persons (IDPs) [17, 18]. Although Sudan is rich in terms of natural and human resources, the effects of the civil conflict on health, nutrition, population, and economic and social development have undoubtedly been significant. These several diverse sociodemographic factors lead to a continued increase in the prevalence rate of H. pylori infection ranging from 48% to 65.8% which represents a major public health challenge [19, 20].
However, studying the genetics of the H. pylori population has been of great interest due to its clinical and phylogeographic significance [4]. A number of studies have discovered the importance of evolutionary history to the clinical outcome of H. pylori infection and indicated that the scramble of the relationship between bacterial and human ancestries at the individual level due to a consequence of migrations, invasions, or racial admixture may moderate adverse outcomes for the host and disrupting the selection for a reduced virulence; and this may give some explanation to the continental enigmas of H. pylori [13, 21–24]. Also, phylogeny and phylogeography of H. pylori strains are known to mirror human migration patterns [25–28] and reflect significant demographic events in human prehistory [26, 29]. Therefore, the phylogeographic pattern of H. pylori is a discriminatory tool to investigate human evolution and migration in addition to the traditional human genetic tools, e.g., mitochondrial DNA (mtDNA) and languages [1].
16S ribosomal RNA (16S rRNA) gene amplification and sequencing have been extensively used for bacterial phylogeny and taxonomy and, eventually, the establishment of large public-domain databases [30–33]. Several properties of the 16S rRNA gene make it the “ultimate molecular chronometer” [34], the most common housekeeping genetic marker, and hence, a useful target for clinical identification and phylogeny [35, 36]. These properties include the following: First, it is present in all bacteria, often existing as a multigene family or operons; thus, it is a universal target for bacterial identification [33, 36, 37]. Second, the function of 16S rRNA has not changed over a long period, so random sequence changes are more likely to reflect the microbial evolutionary change (phylogeny) than selected changes which may alter the molecule's function [34]. Finally, the 16S rRNA gene is large enough, approximately 1,500 bp, for informatics purposes [35, 36, 38]. Most importantly, the 16S rRNA gene consists of approximately 50 functional domains and any introduction of selected changes in one domain does not greatly affect sequences in other domains, i.e., less impact selected changes have on phylogenetic relationships [36].
Here, in this study, two sets of primers (universal and specific for H. pylori) were used to amplify the 16S ribosomal gene directly from gastric endoscopic biopsy samples collected from dyspeptic patients who had been referred for endoscopy. Sanger sequencing was performed, and by matching these sequences with those available in the National Center for Biotechnology Information (NCBI) nucleotide database, we analyzed the evolutionary aspects of Sudanese H. pylori strains using a phylogenetic approach. The novelty of our study resides in being the first study to characterize H. pylori Sudanese strains detected from different regions and tribes of Sudan using the 16S rRNA gene.
2. Materials and Methods
2.1. Study Design and Study Settings
A cross-sectional hospital-based study was conducted in public and private hospitals in Khartoum state from June 2018 to September 2019. These hospitals receive patients from all over the regions of Sudan. Currently, there are 16 states in Sudan which are divided between four geographic regions: Eastern, Western, Northern, and Central Sudan which includes the largest metropolitan area, Khartoum (that includes Khartoum, Khartoum North, and Omdurman) [39]. The capital Khartoum is quickly growing and populated with 6 to 7 million, which includes approximately 2 million IDPs from the southern war zone and the drought-affected areas in the west and east [15, 18].
The hospitals include Ibin Sina specialized hospital, Soba teaching hospital, Modern Medical Center, and Al Faisal Specialized Hospital. All laboratory processes were performed in the molecular biological laboratory at the Faculty of Medical Laboratory Sciences at the University of Khartoum.
2.2. Study Population
The study population composed of 75 patients who had been referred for endoscopy, and most of them were because of dyspepsia. The structured questionnaire, modified from [20], was provided for participants to obtain information about their sociodemographic and clinical characteristics. Patients were selected from those who were not taking antibiotics or nonsteroidal anti-inflammatory drugs (NSAIDs). The patients gave written informed consent before they enrolled in the study. The diagnosis of gastroduodenal diseases was based on the investigation of an experienced gastroenterologist during the upper gastrointestinal (GI) endoscopy procedure, while gastric cancer was diagnosed based on, in addition to gastroscopy, histology.
2.3. Sample Collection
For DNA extraction purposes, gastric biopsies were collected in 1.5 µl Eppendorf tubes with 400 µl phosphate buffer saline (PBS), while for histological examination, the biopsies were transported in 10% formalin. Then, the biopsies were labeled and transported immediately to the laboratory for further processes.
2.4. DNA Extraction
The DNA extraction was performed by using the manual guanidine chloride method [40, 41]. Briefly, biopsies were washed with 400 µl PBS and centrifuged for 5 min at 1000 rpm after each wash. Then, the samples were subjected to digestion by adding 400 µl of WBCs lysis buffer, 200 µl of 6 M guanidine chloride, 50 µl of 7.5 M ammonium acetate, and 5 µl of 20 mg/µl proteinase K and incubated at 37°C overnight. On the following day, samples were cooled down to room temperature; then, 400 µl of cooled prechilled chloroform was added and centrifuged at 1000 rpm for 5 min. Then, three layers were separated, and the supernatant was collected to a new labeled Eppendorf. One ml of cooled prechilled absolute ethanol was added and mixed gently back and forth quickly. Samples were put in −20°C freezer overnight. After that, samples were subjected to quick vortex for one minute and, then, centrifuged for 5 min at 1000 rpm; and the supernatants were discarded. Then, washing with 70% ethanol was performed, and the supernatant was drained with much care to avoid losing the DNA pellet at the bottom of the Eppendorf. The Eppendorf was inverted upside down of a tissue paper leaving the pellet to dry from alcohol for, at least, 2 hours. Finally, the DNA pellet was resuspended in 35 µl of deionized water and was put into −20°C until use. All the used chemical reagents were obtained from iNtRON Biotechnology Inc., Korea.
2.5. 16S rRNA Gene Amplification
Extracted DNA was used to amplify the universal 16S rRNA gene using the following primers: (primers: F:5′-AGAGTTTGATCCTGGCTCAG-3′) (R:5′-CTACGGCTACCTTGTTACGA-3′). PCR amplification was carried out with the Maxime PCR PreMix Kit (i-Taq) (iNtRON Biotechnology, Seongnam, Korea) and a PCR thermocycler (SensoQuest, Germany). The PCR reaction mixtures contained 2.5 U of i-Taq TM DNA polymerase (5 U/µl), 2.5 mM of each deoxynucleoside triphosphates (dNTPs), 1X of PCR reaction buffer (10X), 1X of gel loading buffer, and 1 µl of DNA template. The temperature cycle for the PCR was carried out using a method described previously [42].
To detect the DNA, 3 µl of each PCR products was loaded onto 2% agarose gels stained with 3 µl ethidium bromide (10 mg/ml) and subjected to electrophoresis in 1x Tris EDTA Buffer (TEB buffer) (89 mM of Tris base, 89 mM Boric acid, and 2 mM EDTA dissolved in 1 Litter H2O) for 30 min at 120 V and 50 mA. The gel was visualized under UV light illumination. A 100 bp DNA ladder (iNtRON Biotechnology, Seongnam, Korea) was used in each gel as a molecular size standard. The amplified product for the rrs gene is 1500 bp.
The DNA amplification for the specific 16S rRNA genes was performed to confirm the infection of H. pylori (primers: F:5′-GCGCAATCAGCGTCAGGTAATG-3′) (R:5′-GCTAAGAGAGCAGCCTATGTCC-3′) [43]. The PCR reaction used was an initial step of 3 min at 94°C, followed by denaturation for 30 sec at 94°C, annealing for 30 sec at 53°C, and primer extension for 45 sec at 72°C. After the 40th cycle, the final extension step was prolonged for 5 min to complete the synthesis of strands. The amplified product for the specific rrs gene is 522 bp.
2.6. Sequencing of H. pylori 16S rRNA Gene
The amplified 16S rRNA gene (for the universal and specific) was purified and sequenced, using the Sanger dideoxy sequencing method, commercially by Macrogen Inc, Korea.
2.7. Bioinformatics Analysis
2.7.1. Sequence Analysis
The nucleotide sequence was visualized and analyzed by using the Finch TV program version 1.4.0 [44]. The nucleotide Basic Local Alignment Search Tool (BLASTn; https://blast.ncbi.nlm.nih.gov/) was used for searching about the similarity with other sequences deposited in GenBank [45]. The 16S rRNA genes sequences were submitted in the GenBank nucleotide database under the following accession numbers: from MN845181 to MN845190 and from MN845952 to MN845954.
2.7.2. Molecular Phylogenetic Analysis
Highly similar sequences were retrieved from the NCBI GenBank and subjected to multiple sequence alignment (MSA) using Clustal W2 [46]-BioEdit software [47]. Gblocks was used to eliminate poorly aligned positions and divergent regions of aligned sequences so the alignment becomes more suitable for phylogenetic analysis [48, 49]. The Neighbor-Joining phylogenetic tree [50] of our 16S rRNA sequences with those obtained from the database was constructed using the Jukes–Cantor (JC) model [51] from the substitution (ML) model. [52] The tree was replicated 1000 replicates in which the association with taxa clustered together in the bootstrap test [53]. Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7) was used to conduct evolutionary analyses [54].
2.8. Statistical Analysis
Data were analyzed using the GraphPad Prism 5. Regarding the prevalence of H. pylori infection, differences in frequency distribution by age were examined by the Mann–Whitney test. While bivariate analysis with a categorical variable was assessed by the χ2 test or Fisher's test. The statistical significance level was determined at P < 0.05.
3. Results
3.1. Characteristics of the Study Population
A total of seventy-five patients were included. Forty-one patients (54.67%) were males, and thirty-four (45.33%) were females. Forty-two patients (56%) were urban, and thirty-three (44%) were rural residents. The patients' age ranged from 15 to 85 years, with a mean age of 45.11 ± 17.45 years (Table 1). Most of the participants came from Northern and Central Sudan. Also, their ethnicities were distributed as follows: Shagia (10, 13.33%), Jalyeen (9, 12%), Mahas (8, 10.67%), Rezaigat (4, 5.33%), Zaghawa (4, 5.33%), Kawahla (4, 5.33%), Masalamyia (3, 4%), and others (33, 44%); see Table 1. Regarding clinical symptoms of gastrointestinal disturbances of the participants, abdominal pain was the major symptom (22, 29.33%), followed by nausea (16, 21.33%), and the incidence of H. pylori is more common in patients with gastritis (28.57%) (Table 2). Molecular detection of H. pylori has shown that 28 (37.33%) of patients were positive for H. pylori (Figure 1). Patients from Western Sudan were more prone to H. pylori infection 50% (6/12). Bivariate analysis has found that no significant differences were exhibited across sociodemographic, endoscopy series, and H. pylori infection, as illustrated in Tables 1 and 2.
Table 1.
Sociodemographic characteristic of patients.
| Variables | Total n = 75 | H. pylori (+ve) n = 28 | H. pylori (−ve) n = 47 | P value | |
|---|---|---|---|---|---|
| Mean age (years) ± std. deviation (range) | 45.11 ± 17.45 (15–85) | 41.39 ± 17.57 (15–85) | 47.32 ± 17.18 (21–80) | 0.1170 | |
| Gender | Male | 41 (54.67%) | 15 (53.57%) | 26 (55.32%) | 1.0000 |
| Female | 34 (45.33%) | 13 (46.43%) | 21 (44.68%) | ||
| Residence | Urban | 42 (56%) | 15 (53.57%) | 27 (57.45%) | 0.8122 |
| Rural | 33 (44%) | 13 (46.43%) | 20 (42.55%) | ||
| Hospital | Public | 51 (68%) | 20 (71.43%) | 31 (65.96%) | 0.7986 |
| Private | 24 (32%) | 8 (28.57%) | 16 (34.04%) | ||
| History of previous infection | Yes | 42 (56%) | 15 (53.57%) | 27 (57.45%) | 0.8122 |
| No | 33 (44%) | 13 (46.43%) | 20 (42.55%) | ||
| The frequency of recurrence (if yes n = 42) | One time | 32 (76.19%) | 10 (66.67%) | 22 (81.49%) | 0.4508 |
| More than one time | 10 (23.81%) | 5 (33.3%) | 5 (18.52%) | ||
| Family history of infection | Yes | 36 (48%) | 15 (53.58%) | 21 (44.68%) | 0.4833 |
| No | 39 (52%) | 13 (46.43%) | 26 (55.32%) | ||
| Smoking | Yes | 16 (21.33%) | 5 (17.86%) | 11 (23.4%) | 0.7718 |
| No | 59 (78.67%) | 23 (82.14%) | 36 (76.6%) | ||
| Tribe | Shagia | 10 (13.33%) | 4 (14.29%) | 6 (12.77%) | 0.5733 |
| Jalyeen | 9 (12%) | 4 (14.29%) | 5 (10.64%) | ||
| Mahas | 8 (10.67%) | 3 (10.71%) | 5 (10.64%) | ||
| Rezaigat | 4 (5.33%) | 1 (3.57%) | 3 (6.38%) | ||
| Zaghawa | 4 (5.33%) | 3 (10.71%) | 1 (2.13%) | ||
| Kawahla | 4 (5.33%) | 2 (7.14%) | 2 (4.26%) | ||
| Masalamyia | 3 (4%) | 2 (7.14%) | 1 (2.13%) | ||
| Other | 33 (44%) | 9 (32.14%) | 24 (51.06%) | ||
| Region | Northern Sudan | 15 (20%) | 6 (21.43%) | 9 (19.15%) | 0.2697 |
| Central Sudan | 31 (41.33) | 12 (42.86%) | 19 (40.43%) | ||
| Eastern Sudan | 3 (4%) | 2 (7.14%) | 1 (2.13%) | ||
| Western Sudan | 12 (16%) | 6 (21.43%) | 6 (12.77%) | ||
| Southern Sudan | 14 (18.67%) | 2 (7.14%) | 12 (25.53%) | ||
Table 2.
Distribution of participants according to endoscopy series and status of H. pylori infection.
| Endoscopy results | No. (%) | H. pylori (+ve) | H. pylori (−ve) | P value |
|---|---|---|---|---|
| Normal gastric findings | 9 (12%) | 4 (14.29%) | 5 (10.64%) | 0.5259 |
| Esophagitis | 4 (5.33%) | 3 (10.71%) | 1 (2.13%) | |
| Esophageal varices | 6 (8%) | 1 (3.57%) | 5 (10.64%) | |
| Gastritis | 26 (34.67%) | 8 (28.57%) | 18 (38.3%) | |
| Duodenitis | 6 (8%) | 3 (10.71%) | 3 (6.38%) | |
| Peptic ulcer | 6 (8%) | 3 (10.71%) | 3 (6.38%) | |
| Cancer | 18 (24%) | 6 (21.43%) | 12 (25.53%) | |
| Total | 75 (100%) | 28 (37.33%) | 47 (62.67%) |
Figure 1.
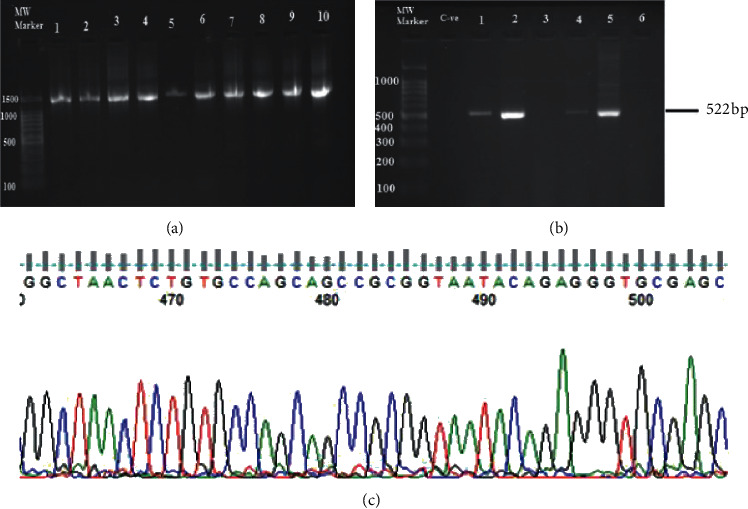
(a) and (b) PCR amplification results of universal and specific H. pylori 16S rRNA gene, respectively, examined on 2% agarose gel electrophoresis. (c) Sequencing result of Acinetobacter radioresistens chromatogram using Finch TV software. The nucleotide sequence was deposited in the GenBank database under the accession number MN845952.
3.2. Analysis of 16S rRNA Sequences
Twelve sequences of 16S rRNA of H. pylori from Sudanese patients were analyzed for mutations and their conservative nature; and findings revealed diversity with few differences, see Table 3 and Figure 2. The description of Sudanese H. pylori strains is shown in Table 3. The amplicon of universal 16S rRNA for patients 22 resulted in Acinetobacter radioresistens (Figure 1). The patient 22 was male with age in the range of [16–36] years, he was Mahasi from Northern Sudan, and he was diagnosed with antral gastritis.
Table 3.
Description of Sudanese H. pylori strains.
| Strain | Age of the patient (in range) | Clinical origin | Ethnic origin | Province | Region | Accession no. |
|---|---|---|---|---|---|---|
| Strain 12 | [16–36] | Antral gastritis | Rezaigat | Eastern Darfur | West of Sudan | MN845953 |
| Strain 17 | [16–36] | Distal esophagitis and antral gastritis | Halawyeen | Khartoum | Middle of Sudan | MN845187 |
| Strain 21 | [16–36] | Antral gastritis | Kawahla | Sinnar | Southeast of Sudan | MN845181 |
| Strain 23 | [16–36] | GRED | Manaseer | Kartoum | Middle of Sudan | MN845188 |
| Strain 39 | [37–61] | Antral gastritis | Masalamyia | Eastern Darfur | West of Sudan | MN845189 |
| Strain 40 | >60 | Duodenitis | — | Khartoum | Middle of Sudan | MN845182 |
| Strain 44 | [16–36] | Normal gastric finding | Jalyeen | Khartoum | Middle of Sudan | MN845183 |
| Strain 46 | [37–61] | Duodenitis | Shagia | Northern State | North of Sudan | MN845184 |
| Strain 47 | [16–36] | GRED | Shagia | Northern State | North of Sudan | MN845954 |
| Strain 59 | [37–61] | Normal gastric finding | Shagia | Northern State | North of Sudan | MN845190 |
| Strain 101 | [37–61] | Gastric cancer | Zaghawa | North Kordofan | South of Sudan | MN845185 |
| Strain 102 | [37–61] | Gastric cancer | Zaghawa | Northern Darfur | East of Sudan | MN845186 |
Figure 2.
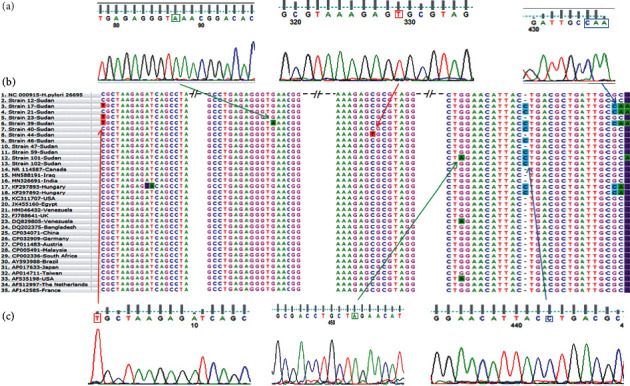
(a) and (c) Sequencing results of chromatograms using Finch TV software show nucleotide variations in the 16S rRNA gene of H. pylori illustrated by squares. (b) Multiple Sequence Alignment (MSA) of 16S rRNA sequences of 12 Sudanese H. pylori strains compared with the rrnA gene of H. pylori strain 26695 (NC_000915) and other selected strains obtained from GenBank databases using Clustal W2.
Regarding mutations, nucleotide variations were found at five nucleotide positions (positions 219, 305, 578, 741, and 763-764), and one insertion mutation (750_InsC_751) was present in sixty-seven percent (7/12) of our strains; the numbering was according to the E.coli sequence [55]. Three strains (17, 21, and 39) revealed a novel transition C⟶T at position 219. A triple base pair substitution (GCC763-764CAA) was detected in strain 17, 23, and 39. However, a cancerous strain 101 had two G⟶A transitions at positions 741 and 765. Strain 44, which was detected in a patient with normal gastric finding and suspected celiac disease and lives in Khartoum, also had a novel C⟶T transversion at nucleotide position 578. Also, strain 39, which was found in a patient from the West of Sudan (Eastern Darfur) and was diagnosed with antral gastritis, revealed a novel G⟶A transition at positions 305. See Figure 2 for more illustration.
3.3. Phylogenetic Analysis
The phylogenetic tree diverged into two lineages (Figure 3). In the lineage one, all the Sudanese H. pylori strains clustered with strains from different countries. The 16S rRNA sequence of strain 44, although clustered with other global strains in one clade, had a novel C⟶T transversion at nucleotide position 578. However, cancerous strain 101 shared a common ancestor with strains from the USA (AF535198) and Venezuela (DQ829805) and represented with them a separated clade with a bootstrap value of 57%. A novel single nucleotide variation (G⟶A) at position 305 of the 16S rRNA of strain 23 made it outgroup as a kind of strain evolution with a bootstrap value of 61%.
Figure 3.
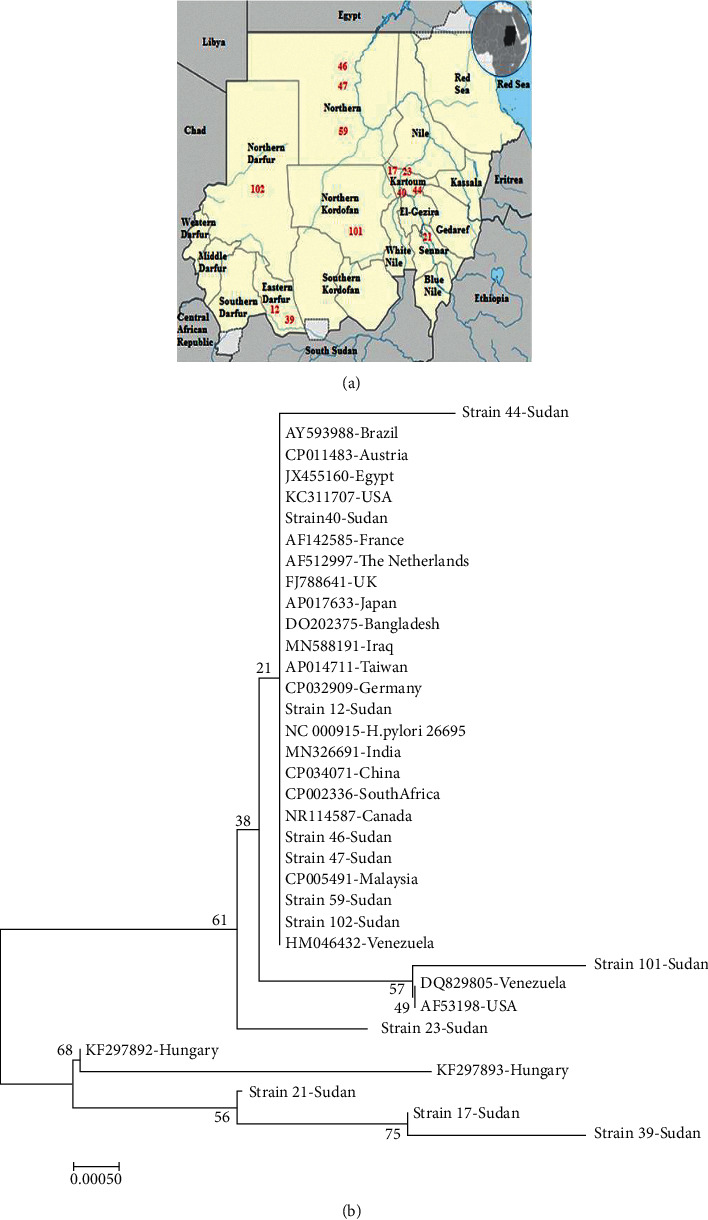
Distribution and evolutionary relationships of Sudanese H. pylori strains. (a) Map of the regional origin of strains in Sudan. (b) The Neighbor-Joining Phylogenetic tree. The percentage of replicate trees (1000 replicates) is shown next to the branches. The evolutionary distance was computed using the JC method and is in the units of the number of base substitution per site. Evolutionary analyses were conducted using MEGA7.
In the second lineage, strains 17, 23, and 39 were closely related to strains from Hungary (KF297893 and KF297892). Strain 17 and Strain 39, from Khartoum and Eastern Darfur, respectively, were sisters with a bootstrap value of 75%, as presented in Figure 3.
4. Discussion
In this study, the phylogenetic analysis of 16S rRNA sequences identified two lineages of H. pylori strains detected from different regions in Sudan which suggested differential evolution. This finding is in agreement with a number of studies that found an unusually high degree of genetic diversity in genomic sequence analyses of H. pylori strains in connection with their housekeeping and virulence-associated genes [26, 56, 57]. Strain 17, Strain 21, and Strain 39 were derived from one lineage along with two strains from Hungary. They shared a double base pair substitution (GC763-764CA) and one insertion mutation (751_InsC_752). Both mutations were located in the central domain of the 16S rRNA gene. The presence of Sudanese H. pylori strains resembling Hungarian H. pylori strains could reflect the migration of Hungarian people to Sudan or vice versa. In 1935, Almásy and his colleague von der Esch became the first Europeans to re-establish contact with the Magyarab tribe [58]. Magyarab name is a concatenation of “Magyar” (Hungarian) and “ab” which means in Nubian simply “tribe.” So the name of the tribe combined translates to “Tribe of the Magyars” [59]. However, Hungarian ancestries came to Sudan in the late 16th century as a part of the Ottoman Empire army. Some of the Hungarian troops were sent to Wadi Halfa, a place located in the Northern state of Sudan. A portion or the entirety of them remained there and intermarried with local Nubian women, generating Magyarab tribe who have lived there ever since [60, 61]. But, because of the construction of the Aswan Dam and the flooding of Lake Nasser, most of Magyarab's villages and the ancient city of Faras were submerged [62, 63]. Therefore, some of them were resettled to New Halfa in the Butana region, the area between Al-Khartoum and Blue Nile, and others spread in many directions throughout Sudan [63, 64]. This could explain, to some extent, the variation in the regional origin of the strains (Strain 17, Strain 21, and Strain 39) that resemble Hungarian strains (Table 3). On the other hand, a significant number of Sudanese, especially from Western State, has left to Europe seeking safety and better life [65, 66].
Laboratory diagnosis of H. pylori by the conventional cultural methodology and phenotypic identification tests is still less specific, time-consuming, incurs increased capital costs, needs highly skilled personnel, and also is difficult to diagnose reinfections [67]. However, molecular tests and PCR technology have been raised as a useful alternative means of bacterial identification, which may circumvent some of these difficulties [67, 68] but has limitations mainly contamination and inadequate sensitivity issues [69]. In this study, molecular detection of H. pylori has shown that 37.33% of patients were positive for H. pylori in comparison to other studies conducted in different regions of Sudan, 22.2%, 59%, 65.8%, 40.1%, 48%, and 21.8% by Abdalsadeg et al., respectively, using different detection methods [19, 20, 70–73]. The present study revealed that there was no statistically significant association between the incidence of H. pylori infection and age, gender, and residence (Table 1) which are in agreement with previous studies [20, 74–77]. However, some other studies have shown different results [78–82]. These contradictory findings may be attributable to the difference in sample size and studied population. [83] Moreover, no significant association (P=0.7718) between the rate of infection and history of smoking was found in this study, which is similar to previous studies [20, 80, 84]. Cigarette smoking stimulates the excretion of hydrochloric acid which is regarded as a natural protective barrier against infective microorganism such as H. pylori [85]. Although no significant differences were found in frequency distribution of H. pylori infection by tribes or regions of participants, patients from Western Sudan were more prone to H. pylori infection 6/12 (50%). This could be explained by the low socioeconomic status, sanitary conditions, and level of educational background caused by drought, war, and civil conflict in Western Sudan compared to other regions [18]. Also, the study showed no significant difference in endoscopy series and status of H. pylori infection; however, the incidence of H. pylori is more common in patients with gastritis (28.57%) which is not totally unexpected since the first presentation of the infection with H. pylori is a symptomatic gastritis [86].
Tetracycline is one of the 30S-targeting antibiotics that inhibit translation elongation by sterically interfering with the binding of aminoacyl-tRNA to the A-site of the ribosome [87]. Therefore, mutations located in two domains (III and IV) in 16S rRNA: helix 34 and the loop next to helix 31 [88] can affect the conformation of the tetracycline-binding site, leading to high-level resistance [87]. In this study, six mutations were detected in two domains (I and II) which are in regions of the 16S rRNA not closely associated with either tetracycline or tRNA binding sites. This finding is partially in agreement with a study conducted by Catharine et al., who found six nucleotide changes in two tetracycline-resistance strains. Two of these changes were located in domain I and domain II, G360 A and deletion of G771, respectively [89]. Thus, further studies for in vitro phenotypic resistance are encouraged to investigate the association between these mutations and phenotypic tetracycline-resistance and examine the reliability of these mutations as molecular markers for tetracycline-resistance. Furthermore, we observed that 66.67% of nucleotide variations were located in the central domain of 16S rRNA (nucleotides 567–915) which is associated with five ribosomal proteins (S15, S6, and S18) folds into the platform of the small subunit [90–93]. The deleterious mutations (C18G, A55G, A161G, A373G, G521A, C614A, A622G, and deletion of one A in a triple-A cluster 607–609) in domains I and II which were suggested by Aymen et al., in purposes of revealing covered putative functional regions of the ribosome and aiding in the development of new antibiotics [94], were found conserved in our strains, as illustrated in Figure 2(b).
Interestingly, the insertion mutation (InsC) between two nucleotides (750 and 751), which was detected in sixty-seven percent (7/12) of our strains, is located at the lower portion of helix 22 (H22), which is one of the lower three-helix junctions (3HJ) of 16S rRNA that bind with S15 [95, 96]. Also, the mutation in cancerous strain 101 (G ⟶ A transition at positions 741) was located in the upper portion of helix 22 (H22). The molecular dynamic (MD) simulations conducted by Wen et al. showed that 16S rRNA and S15 bind across the major groove of H22 via electrostatic interactions, i.e., the negatively charged phosphate groups of G658, U740, G741, and G742 bind to the positively charged S15 residues Lys7, Arg34, and Arg37 [95]. However, studying the effect of these mutations on the functions of 16S rRNA molecules in protein synthesis and antibiotic resistance is of great importance especially for essential regions such as the central domain. [97] For example, Prescott and Dahlberg in 1990 found that the presence of substitution from a C to G at position 726 induces the synthesis of heat shock protein and affects the expression levels of various proteins [98]. Also, mutations in the sequence 911–915 were conferred streptomycin-resistance by impaired the binding of this antibiotic [99–101].
We acknowledge that limitations of the study are the small sample size, and the phylogenetic tree was built based on the 16S rRNA gene only which is unable to cover more complex evolutionary events and distinguish between closely related strains or species [4]. Therefore, further national studies with a large sample size are recommended; also, building a phylogeny by increasing the number of genes analyzed like multilocus sequence analysis (MLST).
4.1. Conclusions
In this study, the 16S rRNA gene amplification method was found useful for bacterial identification and phylogeny. The positive H. pylori infection rate among participants was 37.33%, and no significant differences were found in sociodemographic, endoscopy series, and H. pylori infection. Regarding mutations, six nucleotide variations were detected in regions of the 16S rRNA not closely associated with either tetracycline or tRNA binding sites; 66.67% of them were located in the central domain of 16S rRNA. The phylogenetic analysis of 16S rRNA sequences identified two lineages of H. pylori strains detected from different regions in Sudan. The presence of Sudanese H. pylori strains resembling Hungarian H. pylori strains could reflect the migration of Hungarian people to Sudan or vice versa. This finding emphasizes the significance of studying the phylogeny of H. pylori strains as a discriminatory tool to mirror human migration patterns.
Acknowledgments
The authors would like to thank patients who participated in the study and the staff of the gastroscopic unit in Ibin Sina Specialized Hospital, Soba Teaching Hospital, Modern Medical Centre, and Al Faisal Specialized Hospital.
Data Availability
All data generated or analyzed during this study are included in this published article.
Ethical Approval
Approval to conduct this study was obtained from the Khartoum Ministry of the Health Research Department, University of Khartoum, Faculty of Medical Laboratory Sciences ethical review board, and Research Ethics Committees of hospitals.
Consent
The patients gave written informed consent before they enrolled in the study.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors' Contributions
MAH and EMI supervised the methodology and revised the manuscript. ABI, HGH, LBI, AMA, and MMAI collected the samples. ABI, MASA, and SME extracted the DNA. ABI, MASA, and HNA amplified the 16S rRNA gene. ABI analyzed the data and wrote the manuscript. MAH edited and revised the final manuscript. Hadeel Gassim Hassan, Maryam Atif Salaheldin Ali, and Sulafa Mohammed Eltaher contributed equally to this work.
References
- 1.Linz B., Balloux F., Moodley Y., et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falush K., Yahara K. Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS Genetics. 2017;13(2) doi: 10.1371/journal.pgen.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moodley Y., Linz B., Bond R. P., et al. Age of the association between Helicobacter pylori and man. PLoS Pathogens. 2012;8(5) doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Ramirez Z. Y., Mendez-Tenorio A., Kato I., et al. Whole genome sequence and phylogenetic analysis show Helicobacter pylori strains from Latin America have followed a unique evolution pathway. Frontiers in Cellular and Infection Microbiology. 2017;7 doi: 10.3389/fcimb.2017.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falush D., Stephens M., Pritchard J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren J. R., Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. The Lancet. 1983;321(8336):1273–1275. doi: 10.1016/s0140-6736(83)92719-8. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki R., Shiota S., Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infection, Genetics and Evolution. 2012;12(2):203–213. doi: 10.1016/j.meegid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Testerman T. L., Morris J. Beyond the stomach: an updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment. World Journal of Gastroenterology. 2014;20(36):12781–12808. doi: 10.3748/wjg.v20.i36.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz S., Morelli G., Kusecek B., et al. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathogens. 2008;4(10) doi: 10.1371/journal.ppat.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morelli G., Didelot X., Kusecek B., et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genetics. 2010;6(7) doi: 10.1371/journal.pgen.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Achtman M., Azuma T., Berg D. E., et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Molecular Microbiology. 1999;32(3):459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 12.Suerbaum C., Perez-Perez G. I., van Doorn L. J., Domínguez-Bello M. G., Blaser M. J. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. Journal of Clinical Microbiology. 2005;43(6):2635–2641. doi: 10.1128/jcm.43.6.2635-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodaman N., Pazos A., Schneider B. G., et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proceedings of the National Academy of Sciences. 2014;111(4):1455–1460. doi: 10.1073/pnas.1318093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul Lewis M. Ethnologue: Languages of the World. Dallas, TX, USA: SIL International; 2009. [Google Scholar]
- 15. Sudan Population (2019-05-11), http://worldpopulationreview.com/countries/sudan/
- 16. T. W. Sicinschi, Sudan 2019, https://www.cia.gov/library/publications/the-world-factbook/geos/su.html.
- 17.Sharaf Eldin G. S., Fadl-Elmula I., Ali M. S., et al. Tuberculosis in Sudan: a study of Mycobacterium tuberculosis strain genotype and susceptibility to anti-tuberculosis drugs. BMC Infectious Diseases. 2011;11:219. doi: 10.1186/1471-2334-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Sony A. I., Khami A. H., Enarson D. A., Baraka O., Mustafa S. A., Bjune G. Treatment results of DOTS in 1797 Sudanese tuberculosis patients with or without HIV co-infection. International Journal of Tuberculosis and Lung Disease. 2002;6(12):1058–1066. [PubMed] [Google Scholar]
- 19.Abdalsadeg N., Adam A., Abdul-Aziz H., Omer W., Osman H., Bolad A. Comparison of different diagnostic methodsof Helicobacter pylori infection in Sudanese patients. Al Neelain Medicine Journal. 2012;2(4):27–34. [Google Scholar]
- 20.Abdallah T. M., Mohammed H. B., Mohammed M. H., Ali A. A. A. Sero-prevalence and factors associated with Helicobacter pylori infection in Eastern Sudan. Asian Pacific Journal of Tropical Disease. 2014;4(2):115–119. doi: 10.1016/s2222-1808(14)60326-1. [DOI] [Google Scholar]
- 21.Sitaraman R. Allergies, Helicobacter pylori and the continental enigmas. Frontiers in Microbiology. 2015;6(578) doi: 10.3389/fmicb.2015.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galvani A. P. Epidemiology meets evolutionary ecology. Trends in Ecology & Evolution. 2003;18(3):132–139. doi: 10.1016/s0169-5347(02)00050-2. [DOI] [Google Scholar]
- 23.Herre E. A. Population structure and the evolution of virulence in nematode parasites of fig wasps. Science. 1993;259(5100):1442–1445. doi: 10.1126/science.259.5100.1442. [DOI] [PubMed] [Google Scholar]
- 24.Stewart A. D., Logsdon J. M., Jr., Kelley S. E. An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution. 2005;59(4):730–739. doi: 10.1554/03-330. [DOI] [PubMed] [Google Scholar]
- 25.Latifi-Navid S., Ghorashi S. A., Siavoshi F., et al. Ethnic and geographic differentiation of Helicobacter pylori within Iran. PLoS one. 2010;5(3) doi: 10.1371/journal.pone.0009645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salmanian D., Wirth T., Linz B., et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 27.Wirth T., Wang X., Linz B., et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proceedings of the National Academy of Sciences. 2004;101(14):4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morelli Y., Linz B., Yamaoka Y., et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323(5913):527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maady Y., Linz B. Helicobacter pylori sequences reflect past human migrations. Microbial Pathogenomics. 2009;6:62–74. doi: 10.1159/000235763. [DOI] [PubMed] [Google Scholar]
- 30.Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proceedings of the National Academy of Sciences. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van de Peer Y., Jansen J., De Rijk P., De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Research. 1997;25(1):111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maidak B., Olsen G. J., Larsen N., Overbeek R., McCaughey M. J., Woese C. R. The ribosomal database project (RDP) Nucleic Acids Research. 1996;24(1):82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.-P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. Journal of Clinical Microbiology. 2000;38(10):3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese C. R. Bacterial evolution. Microbiological Reviews. 1987;51(2):221–271. doi: 10.1128/mmbr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janda J. M., Abbott S. L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. Journal of Clinical Microbiology. 2007;45(9):2761–2764. doi: 10.1128/jcm.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel J. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Molecular Diagnosis. 2001;6(4):313–321. doi: 10.2165/00066982-200106040-00012. [DOI] [PubMed] [Google Scholar]
- 37.Tang Y.-W., Ellis N. M., Hopkins M. K., Smith D. H., Dodge D. E., Persing D. H. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. Journal of Clinical Microbiology. 1998;36(12):3674–3679. doi: 10.1128/jcm.36.12.3674-3679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.EMRO. Health Systems Profile-Sudan. Cairo, Egypt: World Health Organization; 2006. [Google Scholar]
- 40.Strohman R. C., Moss P. S., Micou-Eastwood J., Spector D., Przybyla A., Paterson B. Messenger RNA for myosin polypeptides: isolation from single myogenic cell cultures. Cell. 1977;10(2):265–273. doi: 10.1016/0092-8674(77)90220-3. [DOI] [PubMed] [Google Scholar]
- 41.Gassoum A., Arbab M. A. R., Aldeaf S. A. H., Elhassan L. A., Elshibli E., Elhassan A. M. Allele frequency of P53 gene Arg72Pro in Sudanese meningioma patients and controls. International Journal of Scientific & Technology Research. 2014;3(6):243–248. [Google Scholar]
- 42.Liu W., Bao Q., Jirimutu, et al. Isolation and identification of lactic acid bacteria from Tarag in Eastern Inner Mongolia of China by 16S rRNA sequences and DGGE analysis. Microbiological Research. 2012;167(2):110–115. doi: 10.1016/j.micres.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 43.SunChen M., Maugeri T. L., Gugliandolo C., La Camera E., Biondo C., Fera M. T. Occurrence of Helicobacter pylori DNA in the coastal environment of southern Italy (Straits of Messina) Journal of Applied Microbiology. 2005;98(3):768–774. doi: 10.1111/j.1365-2672.2004.02517.x. [DOI] [PubMed] [Google Scholar]
- 44.Geospiza Inc. FinchTV. 1.4.0. Seattle, WA, USA: Geospiza, Inc.; 2012. [Google Scholar]
- 45.Altschul S., Madden T. L., Schäffer A. A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McWilliam H., Li W., Uludag M., et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Research. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowley T. A. BioEdit: a user-friendly biologicalsequence alignment editor and analysis programfor Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 48.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 49.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology. 2007;56(4):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 50.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Jukes T. H., Cantor C. R. Chapter 24-evolution of protein molecules. In: Munro H. N., editor. Mammalian Protein Metabolism. Cambridge, MA, USA: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 52.Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- 53.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution; International Journal of Organic Evolution. 1985;39(4):783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 54.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proceedings of the National Academy of Sciences. 1978;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laith, Hassan A., Al-Obaidi L., Jabbar A. M. Sequencing and phylogenetic analysis of Helicobacter pylori through 16S rRNA gene isolated from gastritis biopsies. International Journal of Applied Biology and Forensics. 2017;1:9–5. [Google Scholar]
- 57.Taylor D. E., Eaton M., Chang N., Salama S. M. Construction of a Helicobacter pylori genome map and demonstration of diversity at the genome level. Journal of Bacteriology. 1992;174(21):6800–6806. doi: 10.1128/jb.174.21.6800-6806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almásy L. (Air... On Sand...) Budapest, Hungary: Franklin-Társulat; 1937. Levegőben... homokon... [Google Scholar]
- 59.Cartledge B. The Will to Survive: A History of Hungary. New York, NY, USA: Columbia University Press; 2011. [Google Scholar]
- 60.Spiers E. M. Sudan: The Reconquest Reapprasied. London, UK: Frank Cass & Co. Ltd; 1998. [Google Scholar]
- 61.Boldizsár I. The New Hungarian Quarterly. Budapest, Hungary: Lapkiadó Publishing House; 1966. [Google Scholar]
- 62. Faras University of Warsaw: Polish Centre of Mediterranean Archaeology, 2019, https://pcma.uw.edu.pl/en/2019/03/20/faras-2/
- 63.Cumming D. The Nile. New York, NY, USA: World Almanac Library; 2003. [Google Scholar]
- 64.Scudder T. Aswan High Dam Resettlement of Egyptian Nubians. Singapore: Springer; 2016. [Google Scholar]
- 65.Jaspars S., Buchanan-Smith M. The Research and Evidence Facility. London, UK: Overseas Development Institute; 2018. Darfuri migration from Sudan to Europe: from displacement to despair. [Google Scholar]
- 66.Migration IOM. Migrationa Crisis Operational Framwork: MCOF Sudan. Khartoum, Sudan: IOM Sudan Country Office; 2017. [Google Scholar]
- 67.Banerjee H. N., Gramby M., Hawkins Z. Molecular diagnosis of Helicobacter pylori strain by 16S rDNA PCR amplification and direct sequencing. Journal of Bioprocessing & Biotechniques. 2011;1 doi: 10.4172/2155-9821.1000105e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proceedings of the National Academy of Sciences. 1985;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petti C. A. Detection and identification of microorganisms by gene amplification and sequencing. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2007;44(8):1108–1114. doi: 10.1086/512818. [DOI] [PubMed] [Google Scholar]
- 70.Salih K. M. A., Elfaki O. A., Hamid Y. H. M., Eldouch W. M. A., Diab M., Abdelgadir S. O. Prevalence of Helicobacter Pylori among Sudanese children admitted to a specialized children hospital. Sudanese Journal of Paediatrics. 2017;17(1):14–18. [PMC free article] [PubMed] [Google Scholar]
- 71.Abbas M., Sharif F. A., Osman S. M., et al. Prevalence and associated symptoms of Helicobacter pylori infection among school children in Kassala state, east of Sudan. Interdisciplinary Perspectives on Infectious Diseases. 2018;2018:p. 5. doi: 10.1155/2018/4325752.4325752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mirghani Y. A. A., Ahmed S., Ahmed M., et al. Detection ofHelicobacter pyloriin endoscopic biopsies in Sudan. Tropical Doctor. 1994;24(4):161–163. doi: 10.1177/004947559402400407. [DOI] [PubMed] [Google Scholar]
- 73.Saidia M., M. E. S., Enan K. A., Abdo A. E., Hassan M. A. Molecular identification 0f 16s ribosomal RNA gene of Helicobacter pylori isolated from gastric biopsies in Sudan. American Journal of Microbiological Research. 2015;3(2):50–54. [Google Scholar]
- 74.Tkachenko M. A., Zhannat N. Z., Erman L. V., et al. Dramatic changes in the prevalence of Helicobacter pylori infection during childhood: a 10-year follow-up study in Russia. Journal of Pediatric Gastroenterology and Nutrition. 2007;45(4):428–432. doi: 10.1097/mpg.0b013e318064589f. [DOI] [PubMed] [Google Scholar]
- 75.Graham S., Adel S., Alireza R. Seroprevalence study of Helicobacter pylori infection among visitors of cardiac patients in Razi hospital in Ahvaz, Iran. Jundishapur Journal of Microbiology. 2010;3 [Google Scholar]
- 76.Alvarado-Esquivel C. Seroepidemiology of Helicobacter pylori infection in tepehuanos aged 15 Years and older in durango, Mexico. Journal of Pathogens. 2013;2013:5. doi: 10.1155/2013/243246.243246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bureš J., Kopacova M., Koupil I., et al. Significant decrease in prevalence of Helicobacter pylori in the Czech Republic. World Journal of Gastroenterology. 2012;18:4412–4418. doi: 10.3748/wjg.v18.i32.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marie M. A. Seroprevalence of Helicobacter pylori infection in large series of patients in an urban area of Saudi arabia. The Korean Journal of Gastroenterology = Taehan Sohwagi Hakhoe Chi. 2008;52(4):226–229. [PubMed] [Google Scholar]
- 79.Valliani A., Khan F., Chagani B., et al. Factors associated with Helicobacter pylori infection, results from a developing country-Pakistan. Asian Pacific Journal of Cancer Prevention. 2013;14(1):53–56. doi: 10.7314/apjcp.2013.14.1.53. [DOI] [PubMed] [Google Scholar]
- 80.Nanji I. S., Boccio J., Santos A. S., et al. Prevalence of Helicobacter pylori infection and associated factors among adults in Southern Brazil: a population-based cross-sectional study. BMC Public Health. 2005;5:118. doi: 10.1186/1471-2458-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoang T. T. H., Bengtsson C., Phung D. C., Sörberg M., Granström M. Seroprevalence of Helicobacter pylori infection in urban and rural Vietnam. Clinical Diagnostic Laboratory Immunology. 2005;12(1):81–85. doi: 10.1128/cdli.12.1.81-85.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim S. H., Kwon J. W., Kim N., et al. Prevalence and risk factors of Helicobacter pylori infection in Korea: nationwide multicenter study over 13 years. BMC Gastroenterology. 2013;13:104. doi: 10.1186/1471-230x-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsongo L., Nakavuma J., Mugasa C., Kamalha E. Helicobacter pylori among patients with symptoms of gastroduodenal ulcer disease in rural Uganda. Infection Ecology & Epidemiology. 2015;5(1) doi: 10.3402/iee.v5.26785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi R., Xu S., Zhang H., et al. Prevalence and risk factors for Helicobacter pylori infection in Chinese populations. Helicobacter. 2008;13(2):157–165. doi: 10.1111/j.1523-5378.2008.00586.x. [DOI] [PubMed] [Google Scholar]
- 85.Chen A., Kikuchi S., Hasegawa A., et al. Relationship between Helicobacter pylori infection and smoking and drinking habits. Journal of Gastroenterology and Hepatology. 2000;15(3):271–276. doi: 10.1046/j.1440-1746.2000.02077.x. [DOI] [PubMed] [Google Scholar]
- 86.Mizukoshi J. R. Spiral bacteria of the gastric antrum. The Medical Journal of Australia. 1984;141(7):477. doi: 10.5694/j.1326-5377.1984.tb132878.x. [DOI] [PubMed] [Google Scholar]
- 87.Wilson D. N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nature Reviews Microbiology. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 88.Dailidiene D., Bertoli M. T., Miciuleviciene J., et al. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrobial Agents and Chemotherapy. 2002;46(12):3940. doi: 10.1128/aac.46.12.3940-3946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kupcinskas C. A., Taylor D. E. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. Journal of Bacteriology. 2002;184(8):2131–2140. doi: 10.1128/jb.184.8.2131-2140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu Z., Culver G. M. Differential assembly of 16S rRNA domains during 30S subunit formation. RNA. 2010;16(10):1990–2001. doi: 10.1261/rna.2246710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Batey R. T., Williamson J. R. Interaction of theBacillus stearothermophilusRibosomal protein S15 with 16 S rRNA: I. Defining the minimal RNA site. Journal of Molecular Biology. 1996;261(4):536–549. doi: 10.1006/jmbi.1996.0481. [DOI] [PubMed] [Google Scholar]
- 92.Agalarov S. C., Williamson J. R. A hierarchy of RNA subdomains in assembly of the central domain of the 30 S ribosomal subunit. RNA. 2000;6(3):402–408. doi: 10.1017/s1355838200991945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agalarov S. C., Zheleznyakova E. N., Selivanova O. M., et al. In vitro assembly of a ribonucleoprotein particle corresponding to the platform domain of the 30S ribosomal subunit. Proceedings of the National Academy of Sciences. 1998;95(3):999–1003. doi: 10.1073/pnas.95.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spirin A., Fredrick K., Mankin A. S. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proceedings of the National Academy of Sciences. 2005;102(46):16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li W., Ma B., Shapiro B. Binding interactions between the core central domain of 16S rRNA and the ribosomal protein S15 determined by molecular dynamics simulations. Nucleic Acids Research. 2003;31(2):629–638. doi: 10.1093/nar/gkg149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agalarov S. C., Sridhar Prasad G., Funke P. M., Stout C. D., Williamson J. R. Structure of the S15, S6, S18-rRNA complex: assembly of the 30S ribosome central domain. Science (New York, NY) 2000;288(5463):107–112. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 97.Brink M. F., Nels R. N. M., Verbeet M. P., de Boer H. A. Specialized ribosomes allow for the study of mutations in functionally important regions in 16 S rRNA, without affecting cell growth. Journal of Molecular Biology. 1994;237(4):368–377. doi: 10.1006/jmbi.1994.1240. [DOI] [PubMed] [Google Scholar]
- 98.Prescott C. D., Dahlberg A. E. A single base change at 726 in 16S rRNA radically alters the pattern of proteins synthesized in vivo. The EMBO Journal. 1990;9(1):289–294. doi: 10.1002/j.1460-2075.1990.tb08107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Montandon P. E., Wagner R., Stutz E. E. coli ribosomes with a C912 to U base change in the 16S rRNA are streptomycin resistant. The EMBO Journal. 1986;5(13):3705–3708. doi: 10.1002/j.1460-2075.1986.tb04703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leclerc D., Melançon P., Brakier-Gingras L. Mutations in the 915 region of Escherichia coli 16S ribosomal RNA reduce the binding of streptomycin to the ribosome. Nucleic Acids Research. 1991;19(14):3973–3977. doi: 10.1093/nar/19.14.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frattali A. L., Flynn M. K., De Stasio E. A., Dahlberg A. E. Effects of mutagenesis of C912 in the streptomycin binding region of Escherichia coli 16S ribosomal RNA. Biochimica et biophysica acta. 1990;1050(1-3):27–33. doi: 10.1016/0167-4781(90)90136-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


