Abstract
Colon cancer (CC) is one of the leading causes of cancer-related mortality in China and western countries. Several studies have demonstrated that long non-coding RNAs (lncRNAs) play critical roles in cancer development. However, the function of lncRNA RP11-619L19.2 in colon cancer remains unclear. The aim of the present study was to investigate the expression pattern, function and underlying mechanism of action of RP11-619L19.2 in CC development and metastasis. RP11-619L19.2 was found to be highly expressed in CC tissues and cell lines, and it was associated with advanced TNM stage and lymph node metastasis. Furthermore, knockdown of RP11-619L19.2 inhibited CC cell proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT). It was also observed that RP11-619L19.2 was reciprocally repressed by miR-1271-5p. Of note, miR-1271-5p negatively regulated CD164 expression by directly targeting the 3′-untranslated region of CD164. Overexpression of CD164 reversed the antimetastatic activity of RP11-619L19.2 knockdown in CC cells. Mechanistically, it was demonstrated that lncRNA RP11-619L19.2 played an oncogenic role and promoted CC development and metastasis by regulating the miR-1271-5p/CD164 axis and EMT. In conclusion, the findings of the present study indicated that RP11-619L19.2 regulates CD164 expression and EMT by sponging miR-1271-5p, which may provide novel targets for lncRNA-directed diagnosis and therapy for patients with CC.
Keywords: colon cancer, epithelial-to-mesenchymal transition, RP11-619L19.2, CD164
Introduction
Over 1 million individuals are diagnosed with colon cancer (CC) annually worldwide, resulting in a high rate of morbidity and mortality throughout China and certain western countries (1,2). Despite major advances in diagnostic and therapeutic strategies in recent decades, the prognosis of patients with CC remains poor, which is mainly due to the development of distant metastasis leading to low survival rates (3,4). Moreover, the mechanisms underlying CC tumorigenesis and metastasis remain elusive. Thus, it is crucial to elucidate the molecular mechanism of CC development and progression in order to develop novel effective therapeutic strategies for patients with CC.
Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are two common types of non-coding RNAs with different lengths of nucleotide chains, which play key regulatory roles in human diseases, including cancer (5,6). Mounting evidence has demonstrated that lncRNAs are involved in cancer development, progression and metastasis, and may act as tumor suppressor genes or oncogenes (7,8). miRNAs may suppress or promote tumorigenesis through specifically binding to the 3′-untranslated region (3-UTR) of the target mRNA, leading to mRNA degradation (9). Corroborating evidence revealed that lncRNAs and microRNAs may serve as predictive or therapeutic biomarkers in human cancers (10,11). Interestingly, various lncRNAs share the same targeting sequences with certain miRNAs, while functioning as competing endogenous RNAs (ceRNAs) or miRNA sponges (12–14). Thus, it is crucial to clearly determine the lncRNA-miRNA interaction and their regulatory network in cancer. Multiple ceRNA networks have been investigated in CC (15–17). For example, lncRNA ZNFX1-AS1 was demonstrated to promote colorectal cancer progression and metastasis via interacting with miR-144 to regulate EZH2 expression (18). Yu et al reported that lncRNA CCAT2 regulated miR-145 in CC cells (19). However, the complicated lncRNA-miRNA-mRNA regulatory network in CC requires additional extensive investigation.
In the present study, the altered expression of lncRNAs in CC tissues was screened in comparison with that in normal tissues in order to identify potential lncRNA/miRNA predictive biomarkers in CC. While multiple lncRNAs functioned as potential tumor suppressors or promoters in CC, this investigation was focused on the function of lncRNA RP11-619L19.2, which is highly expressed in CC tissues and cell lines. The effects of RP11-619L19.2 knockdown on CC cell proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT) were investigated, and the association between lncRNA PR11-619L19.2 and miRNA-1271-5p was examined. Additionally, it was investigated whether CD164 is the downstream target of miR-1271-5p, and whether overexpression of CD164 would be able to reverse the effects of RP11-619L19.2 knockdown. In summary, the aim of the present study was to elucidate the detailed function of the RP11-619L19.2/miR-1271-5p/CD164 axis in CC, in the hope of identifying effective and predictive biomarkers as well as therapeutic targets for patients with CC.
Materials and methods
Human samples
A total of 30 pairs of primary CC tissues with matching adjacent normal tissues and 20 CC tissues with distant metastasis were obtained from the Second Affiliated Hospital of Xi'an Jiaotong University. The present study was approved by the Ethics Committee of the Second Affiliated Hospital of Xi'an Jiaotong University, and all patients enrolled provided their signed informed consent. All tissues were frozen in liquid nitrogen and stored at −80°C until total RNA and protein were extracted.
Cell lines
Three human CC cell lines (HCT-116, SW620 and DLD-1) and the normal colonic epithelial cell line FHC were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in DMEM supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 U/ml streptomycin, and incubated at 37°C in a 5% CO2 atmosphere.
Cell transfection
The lentiviral vectors containing short hairpin RNAs (shRNAs) targeting RP11-619L19.2 were designed and constructed by GenePharma. The expression vector for CD164 overexpression was obtained from GeneChem, Inc. The miR-1271-5p mimic and negative control (NC) oligonucleotides were synthesized by Guangzhou RiboBio Co., Ltd. CC cells were transfected with the abovementioned vectors or oligonucleotides by using the Lipofectamine® 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the transfection efficiency was verified by RT-qPCR (Fig. S1).
Luciferase reporter assay
Luciferase reporter plasmids containing RP11-619L19.2 or CD164 with the wild-type (Wt RP11-619L19.2 or Wt CD164) or mutant (Mut RP11-619L19.2 or Mut CD164) miR-1271-5p-binding sites were obtained from GenePharma. HCT-116 or SW620 cells were transfected with the miR-1271-5p mimic or miR-NC, together with the reporter plasmids. Subsequently, 48 h after transfection the luciferase activity was measured by a Dual Luciferase Reporter Gene Assay Kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNAs were purified from cultured cells, CC tissues or adjacent normal tissues by using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse-transcribed into cDNA using the PrimeScript™ kit (Takara Biotechnology Co., Ltd.) at 42°C for 1 h followed by 95°C for 5 min. qPCR was conducted on a 7500 Real-Time PCR System™ (Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR™ Green Master Mix (Takara Biotechnology Co., Ltd.). The thermocycling conditions used were as follows: Initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 sec and annealing/elongation at 60°C for 30 sec. The primers used were as follows: For RP11-619L19.2, 5′-ACTGGGAATGGAGGAAGA-3′ (forward) and 5′-TGAGAAAGGATTGAGGGAAAAG-3′ (reverse); for E-cadherin, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); for N-cadherin, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); for vimentin, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); for β-catenin, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); for Snail, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); for miR-1271-5p, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); and for GAPDH, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse).
Cell migration assay
In vitro wound healing assay was conducted to assess the mobility of CC cells. After different treatments, HCT-116 or SW620 cells were seeded into 6-well plates and cultured until ~100% confluent. Then, each well was scratched with a 200-µl sterile pipette tip, washed with PBS, and incubated with serum-free DMEM at 37°C. The scratch area was photographed at 0 and 48 h to evaluate wound closure rate.
Cell invasion assay
HCT-116 or SW620 cells were suspended in serum-free medium at a density of 4×105 cells/ml at 48 h after transfection. A 100-µl cell suspension was added to the upper Matrigel-coated chambers (8.0 µm pore size, Corning, Inc.) and 600 µl of complete medium was added to the bottom chamber. After a 24 h incubation, the invading cells were fixed in 4% paraformaldehyde for 10 min, and stained with 0.1% crystal violet solution for 30 min at room temperature. The number of cells was calculated and images were captured under a light microscope (Olympus Corporation) at a magnification of ×200.
Cell proliferation assay
HCT-116 or SW620 cells transfected with negative control, sh-RP11-619L19.2, miR-NC, miR-1271-5p mimics, or sh-RP11-619L19.2 + pcDNA3.1-CD164 were seeded into 96-well plates at 20,000 cells/well. After culture for the indicated time, 10 µl CCK-8 reagent (Dojindo Molecular Technologies, Inc.) was added to the cell culture medium and cell viability was assessed following the manufacturer's instructions.
Western blotting
Once different treatments were completed, HCT-116 or SW620 cell lysates were collected by using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) and protein concentration was measured using a BCA assay kit (Pierce; Thermo Fisher Scientific, Inc.). All samples were subjected to 10% SDS-PAGE electrophoresis and transferred to a PVDF membrane (Roche Diagnostics). After blocking in 5% non-fat milk in TBST (containing 0.05% Tween-20) for 1 h at room temperature, the membranes were incubated with the following primary antibodies (all from Abcam): Snail (1:1,000, ab216347), E-cadherin (1:10,000, ab40772), N-cadherin (1:500, ab98952), vimentin (1:1,000, ab20346), β-catenin (1:5,000, ab32572), or β-actin (1:2,000, ab8226) at 4°C overnight. After washing three times with TBST (5 min per wash), the membranes were then incubated with HRP-conjugated goat anti-rabbit IgG (1:20,000, ab205718, Abcam) at room temperature for 1 h. Band detection was performed using an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.).
Immunohistochemical staining
The immunohistochemical staining for CD164 was performed as previously described (20). Briefly, tissue sections were deparaffinized and incubated in an antigen retrieval solution (Target Retrieval; Dako; Agilent Technologies, Inc.) at 95°C for 15 min. The sections were then incubated overnight at 4°C with a monoclonal antibody against CD164 (1:100; HPA010636; Sigma-Aldrich; Merck KGaA). On the following day, the sections were incubated with an HRP-conjugated secondary antibody (1:2,000; 7074, Cell Signaling Technology, Inc.) at room temperature for 30 min; they were then developed with diaminobenzidine at room temperature for 10 min and counterstained with hematoxylin at room temperature for 1 min.
Statistical analysis
All data analyses were conducted using GraphPad Prism 5.0 software (GraphPad Software, Inc.). The data are shown as the mean ± SE. Student's t-test was used when two groups were compared. When more than two groups were compared, one-way ANOVA followed by Tukey's post hoc test was performed. P<0.05 was considered to indicate statistically significant differences.
Results
RP11-619L19.2 is highly expressed in CC tissues and cell lines
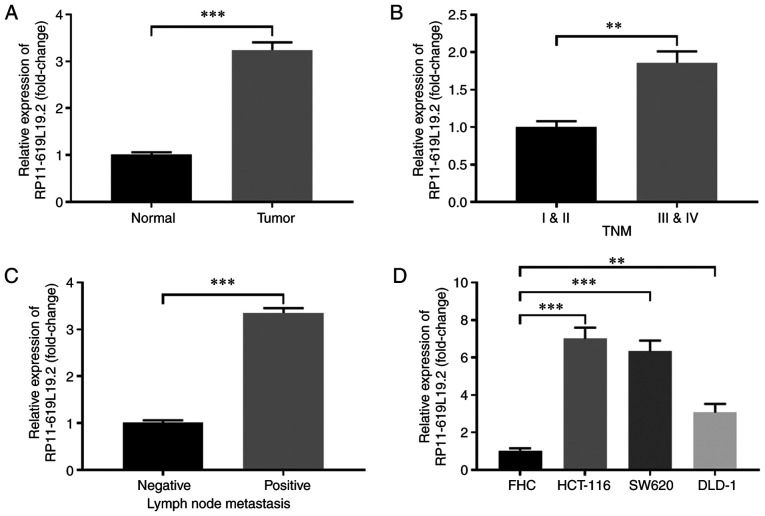
The expression profile of lncRNA RP11-619L19.2 in CC tissues and paired normal tissues was first examined. As shown in Fig. 1A, the expression of RP11-619L19.2 was significantly higher in CC tissues compared with that in normal tissues. Furthermore, CC tissues with advanced TNM stage (III and V) exhibited markedly higher RP11-619L19.2 levels compared with those with TNM stage I and II (Fig. 1B). In addition, CCs with lymph node metastasis exhibited higher expression of RP11-619L19.2 compared with tumors without lymph node metastasis (Fig. 1C). The expression of RP11-619L19.2 in CC cell lines was also analyzed. Compared with the control cell line FHC, the CC cell lines HCT-116, SW620 and DLD-1 had notably higher expression of RP11-619L19.2 (Fig. 1D). Thus, lncRNA RP11-619L19.2 was highly expressed in CC tissues and cell lines.
Figure 1.
RP11-619L19.2 is highly expressed in CC tissues and cell lines. (A) The relative expression of RP11-619L19.2 in CC tissues (tumor) or paired normal tissues (normal) was examined by RT-qPCR analysis. (B) The relative expression of RP11-619L19.2 in CC tissues with different TNM stage was examined by RT-qPCR analysis. (C) The relative expression of RP11-619L19.2 in CC tissues with or without lymph node metastasis was examined by RT-qPCR analysis. (D) The relative expression of RP11-619L19.2 in CC cell lines (HCT-116, SW620 and DLD-1) or control cells (FHC) was examined by RT-qPCR analysis. **P<0.01, ***P<0.001. CC, colon cancer; RT-qPCR, reverse transcription-quantitative PCR.
Knockdown of RP11-619L19.2 inhibits CC cell proliferation, migration, invasion and EMT
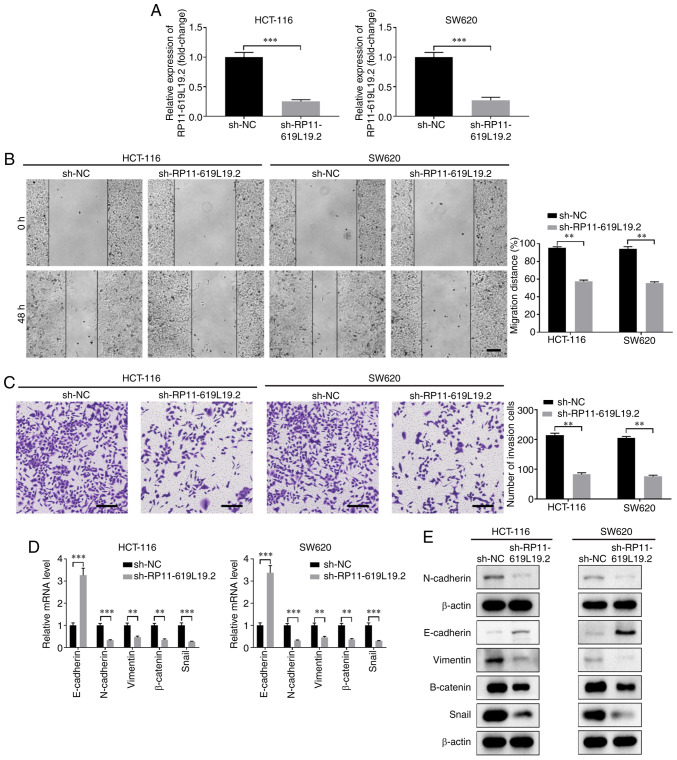
To investigate the function of RP11-619L19.2 in CC, multiple shRNAs were screened to knock down the expression of RP11-619L19.2 (data not shown). The knockdown efficiency of shRNA targeting RP11-619L19.2 was confirmed in HCT-116 and SW620 cells (Fig. 2A). Functionally, knockdown of RP11-619L19.2 suppressed the proliferation of HCT-116 and SW620 cells (Fig. S2A and B). In addition, knockdown of RP11-619L19.2 markedly suppressed the migration and invasion of HCT-116 or SW620 cells, as demonstrated by wound healing and Transwell assays (Fig. 2B and C). EMT is a critical process for tumor invasion and metastasis (21). The expression of EMT-related genes after RP11-619L19.2 knockdown was further analyzed. As shown in Fig. 2D and E, knockdown of RP11-619L19.2 significantly enhanced the expression of E-cadherin, while decreasing the expression of N-cadherin, vimentin, β-catenin and Snail at both the mRNA and protein levels. In conclusion, RP11-619L19.2 regulated EMT-related gene expression and promoted tumor cell proliferation, migration, invasion and metastasis in CC.
Figure 2.
Knockdown of RP11-619L19.2 inhibits CC cell migration, invasion and epithelial-to-mesenchymal transition. HCT-116 or SW620 cells were transfected with RP11-619L19.2 knockdown vector sh-RP11-619L19.2 or negative control (sh-NC). (A) The knockdown efficiency was evaluated by RT-qPCR analysis. (B) Wound healing assay was performed to evaluate the effect of RP11-619L19.2 knockdown on CC cell migration. Scale bar: 200 µm. (C) Transwell assay was performed to evaluate the effect of RP11-619L19.2 knockdown on CC cell invasion. Scale bar: 200 µm. The (D) mRNA and (E) protein levels of epithelial markers (E-cadherin), mesenchymal markers (N-cadherin, vimentin and Snail) and β-catenin in HCT-116 or SW620 cells transfected with sh-RP11-619L19.2 or sh-NC were assessed by RT-qPCR and western blot analyses. **P<0.01, ***P<0.001. CC, colon cancer; RT-qPCR, reverse transcription-quantitative PCR.
Reciprocal repression is observed between RP11-619L19.2 and miR-1271-5p
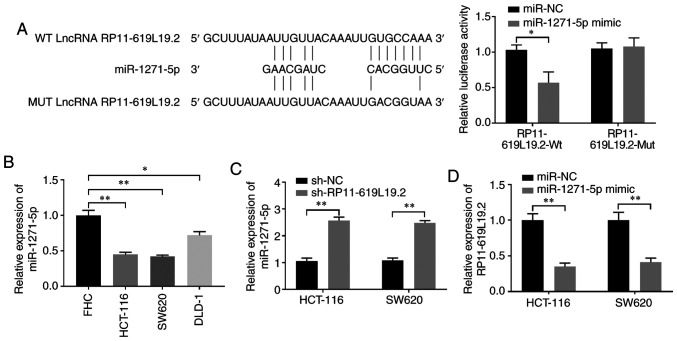
Given the crucial interaction between lncRNA and miRNA, it was further investigated whether miRNA was involved in the regulation of RP11-619L19.2 expression. DIANA tools were employed to search for miRNAs that may interact with RP11-619L19.2 (22). miR-1271-5p was identified to have putative binding sites for RP11-619L19.2 (Fig. 3A). Dual luciferase reporter assay demonstrated that miR-1271-5p mimics inhibited luciferase activity in HCT-116 cells transfected with the reporter vector containing Wt lncRNA RP11-619L19.2 sequences, while no significant suppression was found in HCT-116 cells transfected with the reporter vector containing mutated RP11-619L19.2 sequences (Fig. 3A). In addition, the expression of miR-1271-5p was significantly lower in CC cell lines (HCT-116, SW620 and DLD-1) compared with that in control FHC cells (Fig. 3B). Moreover, it was demonstrated that RP11-619L19.2 knockdown could enhance the expression of miR-1271-5p in HCT-116 and SW620 cells, which further confirmed the reciprocal repression between RP11-619L19.2 and miR-1271-5p (Fig. 3C). By contrast, HCT-116 or SW620 cells transfected with miR-1271-5p mimics significantly downregulated the expression of RP11-619L19.2 compared with cells transfected with the miR-NC control (Fig. 3D).
Figure 3.
Reciprocal repression between RP11-619L19.2 and miR-1271-5p. (A) Schematic representation of the predicted target sequences for miR-1271-5p in lncRNA RP11-619L19.2 (left panel). HCT-116 cells were co-transfected with luciferase reporter plasmid containing RP11-619L19.2-Wt or RP11-619L19.2-Mut, together with miR-NC or miR-1271-5p mimics. The relative luciferase activity was measured 48 h after transfection (right panel). (B) The relative expression of miR-1271-5p in CC cell lines (HCT-116, SW620 and DLD-1) or the control cell line FHC was evaluated by RT-qPCR analysis. (C) The relative expression of miR-1271-5p in HCT-116 or SW620 cells transfected with sh-NC or sh-RP11-619L19.2 was evaluated by RT-qPCR analysis. (D) The relative expression of RP11-619L19.2 in HCT-116 or SW620 cells transfected with miR-NC or miR-1271-5p mimics was evaluated by RT-qPCR analysis. *P<0.05, **P<0.01. RT-qPCR, reverse transcription-quantitative PCR.
Overexpression of miR-1271-5p suppresses CC cell proliferation, migration, invasion and EMT
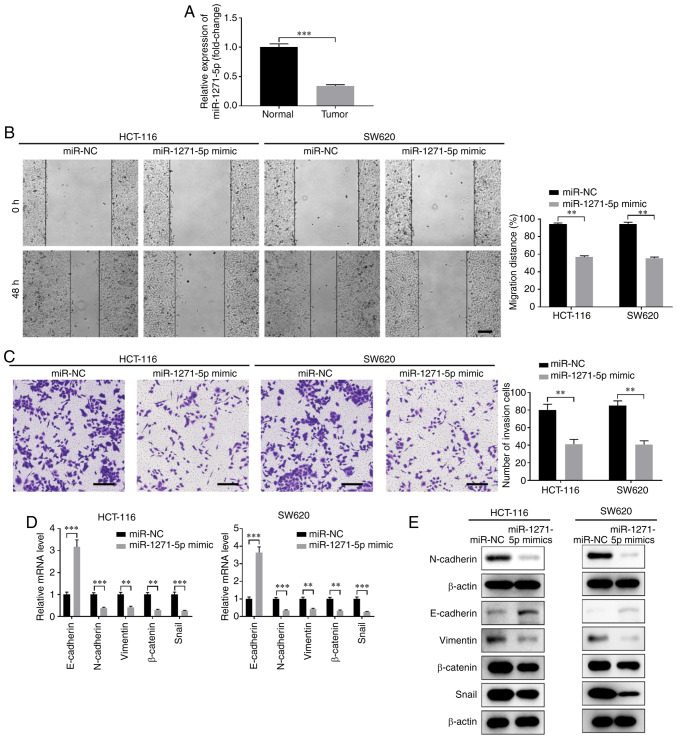
Next, the function of miR-1271-5p in CC development was evaluated. The expression of miR-1271-5p was markedly lower in CC tissues compared with that in normal tissues (Fig. 4A). Overexpression of miR-1271-5p using miR-1271-5p mimics significantly inhibited the proliferation, migration and invasion of HCT-116 and SW620 cells (Fig. 4B and C, Fig. S2C and D). Furthermore, overexpression of miR-1271-5p suppressed EMT, with enhanced expression of E-cadherin and decreased expression of N-cadherin, vimentin, β-catenin and Snail at both the mRNA and protein levels (Fig. 4D and E).
Figure 4.
Overexpression of miR-1271-5p suppresses CC cell migration, invasion and epithelial-to-mesenchymal transition. (A) The relative expression of miR-1271-5p in CC tissues (tumor) and paired normal tissues (normal) was measured by RT-qPCR analysis. HCT-116 or SW620 cells were transfected with miR-1271-5p mimics or negative control miR-NC. (B) Wound healing assay was performed to evaluate the effect of miR-1271-5p overexpression on CC cell migration. Scale bar: 200 µm. (C) Transwell assay was performed to evaluate the effect of miR-1271-5p overexpression on CC cell invasion. Scale bar: 200 µm. The (D) mRNA and (E) protein levels of epithelial markers (E-cadherin, N-cadherin and vimentin) and mesenchymal markers (β-catenin and Snail) in HCT-116 or SW620 cells transfected with miR-1271-5p mimics or miR-NC were assessed by RT-qPCR and western blot analyses, respectively. **P<0.01, ***P<0.001. CC, colon cancer; RT-qPCR, reverse transcription-quantitative PCR.
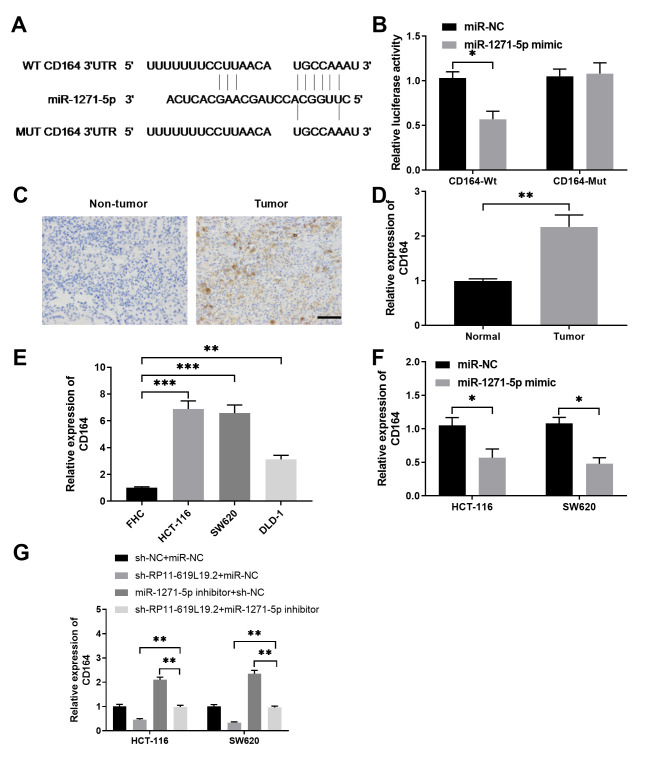
CD164 is a direct target of miR-1271-5p
To explore how miR-1271-5p regulates CC metastasis, bioinformatics analysis was performed and the targets of miR-1271-5p were predicted by TargetScan. As shown in Fig. 5A, miR-1271-5p had the complementary binding sequences targeting the 3′-UTR of CD164. Luciferase reporter assay demonstrated that miR-1271-5p specifically inhibited the luciferase activity in HCT-116 cells transfected with reporter vector containing the Wt 3′-UTR of CD164, but not in HCT-116 cells transfected with reporter vector containing the Mut 3′-UTR of CD164 (Fig. 5B). Immunohistochemical staining of CD164 was performed, and the results revealed that CD164 was highly expressed in CC tissues compared with that in non-tumor control tissues (Fig. 5C and D). It was also confirmed that CD164 expression was significantly higher in CC cell lines compared with that in FHC control cells, indicating that the expression of CD164 was negatively correlated with miR-1271-5p expression (Fig. 5E). Furthermore, overexpression of miR-1271-5p was able to suppress the expression of CD164 in HCT-116 or SW620 cells (Fig. 5F), validating that CD164 was a direct target of miR-1271-5p. Moreover, while knockdown of RP11-619L19.2 inhibited CD164 expression and inhibition of miR-1271-5p enhanced CD164 expression in HCT-116 or SW620 cells, inhibition of miR-1271-5p antagonized the inhibitory effect of RP11-619L19.2 knockdown (Fig. 5G).
Figure 5.
miR-1271-5p negatively regulates CD164 expression by binding to 3′-UTR of CD164. (A) Schematic representation of the potential miR-1271-5p target sequences in CD164 mRNA 3′-UTR predicted by TargetScan. (B) HCT-116 cells were transfected with luciferase reporter plasmid containing wild-type CD164-3′-UTR (CD164-Wt) or mutant CD164-3′-UTR (CD164-Mut), together with miR-1271-5p mimics or negative control miR-NC. The relative luciferase activity was measured 48 h after transfection. (C) The expression of CD164 in the CC tissues and non-tumor control tissues was examined by immunohistochemical staining. Scale bar: 200 µm. (D) The relative expression of CD164 in CC tissues or normal control tissues was evaluated by qPCR analysis. (E) The relative expression of CD164 in CC cell lines (HCT-116, SW620 and DLD-1) or the control cell line FHC was evaluated by RT-qPCR analysis. (F) The relative expression of CD164 in HCT-116 or SW620 cells transfected with miR-1271-5p mimics or miR-NC was evaluated by RT-qPCR analysis. (G) HCT-116 or SW620 cells were transfected with control, sh-RP11-619L19.2, miR-1271-5p inhibitor or sh-RP11-619L19.2 + miR-1271-5p inhibitor. The relative expression of CD164 was evaluated by RT-qPCR analysis 48 h later. *P<0.05, **P<0.01, ***P<0.001. UTR, untranslated region; RT-qPCR, reverse transcription-quantitative PCR.
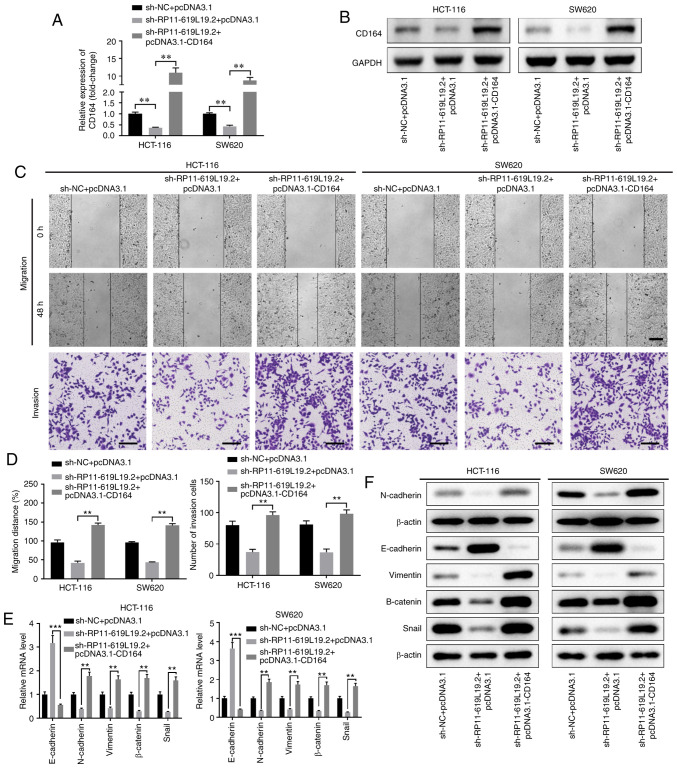
Overexpression of CD164 reverses the antimetastatic activity of RP11-619L19.2 knockdown in CC cells
To determine whether RP11-619L19.2 exerted its effects through the miR-1271-5p/CD164 axis in CC cells, rescue experiments were performed. CD164 overexpression reversed the inhibitory effect of RP11-619L19.2 knockdown on CD164 expression (Fig. 6A and B). Functionally, while sh-RP11-619L19.2 inhibited CC cell prolifer-ation, migration and invasion, overexpression of CD164 together with RP11-619L19.2 antagonized the inhibition mediated by RP11-619L19.2 knockdown (Fig. 6C and D, Fig. S2E and F).
Figure 6.
The antimetastatic activity of RP11-619L19.2 knockdown in CC cells is mediated by CD164. HCT-116 or SW620 cells were transfected with negative control sh-NC, sh-RP11-619L19.2, or sh-RP11-619L19.2 + CD164 overexpression vector. The relative expression levels of CD164 (A) mRNA or (B) protein were evaluated by RT-qPCR or western blot analyses, respectively. (C and D) Wound healing and Transwell assays were performed to evaluate the HCT-116 or SW620 cell migration and invasion. Scale bar: 200 µm. The mRNA or protein expression levels of epithelial markers (E-cadherin, N-cadherin and vimentin) and mesenchymal markers (β-catenin and Snail) in HCT-116 or SW620 cells transfected with sh-NC, sh-RP11-619L19.2, or sh-RP11-619L19.2 + CD164 overexpression vector were analyzed by (E) RT-qPCR analysis and (F) western blotting. **P<0.01, ***P<0.001. CC, colon cancer; RT-qPCR, reverse transcription-quantitative PCR.
Moreover, overexpression of CD164 promoted EMT, with decreased expression of E-cadherin and enhanced expression of N-cadherin, vimentin, β-catenin and Snail at both the mRNA and protein levels (Fig. 6E and F). Taken together, these results indicated that the antimetastatic activity of RP11-619L19.2 knockdown in CC cells was mediated by CD164.
Discussion
Genomic and transcriptomic analysis by next-generation sequencing has led to a well-characterized molecular profiling of CC. Moreover, numerous lncRNAs have been found to be dysregulated in CC (23,24). We also screened for lncRNAs with altered expression in CC tissues in the present study (data not shown). To the best of our knowledge, this is the first study to report that lncRNA RP11-619L19.2 is highly expressed in CC tissues and cell lines. Furthermore, the expression level of RP11-619L19.2 was found to be positively correlated with advanced TNM stage and lymph node metastasis of CC. Functionally, knockdown of RP11-619L19.2 inhibited cell proliferation, migration, invasion and EMT, which indicated that RP11-619L19.2 may act as an oncogene promoting CC development.
As there are only few studies investigating the function and mechanism of action of RP11-619L19.2 in CC, we performed bioinformatics analysis using DIANA tools to identify the potential miRNAs involved in the regulation of RP11-619L19.2 expression. RP11-619L19.2 was shown to act as a miR-1271-5p sponge, and there was a reciprocal repression between the expression of RP11-619L19.2 and that of miR-1271-5p. miR-1271-5p has been reported as a tumor suppressor in several different types of tumors. Chen et al reported that miR-1271-5p suppressed cell proliferation and induced cell apoptosis via regulating ZIC2 in acute myeloid leukemia (25). In another study, miR-1271-5p negatively regulated FOXK2 and inhibited cell growth and hepatocellular carcinoma development (26). lncRNA UCA was demonstrated to regulate the miR-1271-5p/HGF axis in multiple myeloma by controlling cell apoptosis and proliferation (27). Our findings revealed that miR-1271-5p also acted as a tumor suppressor in CC, while overexpression of miR-1271-5p suppressed cell proliferation, migration, invasion and EMT.
To further address the mechanism underlying the role of miR-1271-5p in CC, CD164 was identified as a direct target of miR-1271-5p by bioinformatics analysis. CD164 belongs to the sialomucin family, which regulates cell proliferation, adhesion and migration of hematopoietic progenitor cells (28). In addition, CD164 has been reported to regulate tumor development and progression (29). Tang et al reported that inhibition of CD164 resulted in suppressed tumor cell proliferation, mobility and metastasis in CC cell lines (30). Furthermore, CD164 has been identified as a new biomarker in acute lymphoblastic leukemia (31). The present study also demonstrated that CD164 had a function similar to that of RP11-619L19.2, promoting cell proliferation, migration, invasion, and EMT in CC cell lines.
Although there was sufficient evidence to support the presence of an interaction network involving RP11-619L19.2, miR-1271-5p and CD164 in CC, whether other miRNAs may also interact with RP11-619L19.2 and whether miR-1271-5p controls multiple targets besides CD164 were not fully addressed. Furthermore, it was demonstrated that RP11-169L192 and CD164 acted as oncogenes, while miR-1271-5p acted as a tumor suppressor in regulating CC cell migration, invasion and EMT; however, in vivo CC models must be employed to further investigate the function of the RP11-619L19.2/miR-1271-5p/CD164 axis.
In conclusion, the results of the present study revealed that lncRNA RP11-619L19.2 functions as a ceRNA in regulating miR-1271-5p/CD164 and controlling CC cell migration and invasion. These findings provide new insight into the lncRNA/miRNA/mRNA network in CC, which may represent a novel diagnostic and predictive biomarker, as well as a target for the treatment of patients with CC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the Key Research and Development Project of Shaanxi Province (grant no. S2018YBSF0274).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XZ, XS and HZ conceived and designed the experiments; XZ, SL and DZ performed the experiments; XZ and DZ analyzed and interpreted the data; XZ wrote the manuscript; XS and HZ revised the manuscript. All the authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients and the study protocol was approved by the Ethics Committee of The Second Affiliated Hospital of Xi'an Jiaotong University.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Favoriti P, Carbone G, Greco M, Pirozzi F, Pirozzi RE, Corcione F. Worldwide burden of colorectal cancer: A review. Updates Surg. 2016;68:7–11. doi: 10.1007/s13304-016-0359-y. [DOI] [PubMed] [Google Scholar]
- 3.Zhang ML, Peng P, Wu CX, Gong YM, Zhang SW, Chen WQ, Bao PP. Report of breast cancer incidence and mortality in China registry regions, 2008–2012. Zhonghua Zhong Liu Za Zhi. 2019;41:315–320. doi: 10.3760/cma.j.issn.0253-3766.2019.04.013. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 4.Telang NT, Li G, Katdare M. Prevention of early-onset familial/hereditary colon cancer: New models and mechanistic biomarkers (R-eview) Int J Oncol. 2006;28:1523–1529. [PubMed] [Google Scholar]
- 5.Yang S, Sun Z, Zhou Q, Wang W, Wang G, Song J, Li Z, Zhang Z, Chang Y, Xia K, et al. MicroRNAs, long noncoding RNAs, and circular RNAs: Potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res. 2018;10:2249–2257. doi: 10.2147/CMAR.S166308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue M, Zhuo Y, Shan B. MicroRNAs, long noncoding RNAs, and their functions in human disease. Methods Mol Biol. 2017;1617:1–25. doi: 10.1007/978-1-4939-7046-9_1. [DOI] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 8.Renganathan A, Felley-Bosco E. Long Noncoding RNAs in Cancer and Therapeutic Potential. Adv Exp Med Biol. 2017;1008:199–222. doi: 10.1007/978-981-10-5203-3_7. [DOI] [PubMed] [Google Scholar]
- 9.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Pian C, Chen Z, Zhang J, Xu M, Zhang L, Chen Y. Identification of cancer-related miRNA-lncRNA biomarkers using a basic miRNA-lncRNA network. PLoS One. 2018;13:e0196681. doi: 10.1371/journal.pone.0196681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hon KW, Abu N, Ab Mutalib NS, Jamal R. miRNAs and lncRNAs as predictive biomarkers of response to FOLFOX therapy in colorectal cancer. Front Pharmacol. 2018;9:846. doi: 10.3389/fphar.2018.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao M, Li W. Transcriptional factor regulation network and competitive endogenous RNA (ceRNA) network determining response of esophageal squamous cell carcinomas to neoadjuvant chemoradiotherapy. PeerJ. 2019;7:e6668. doi: 10.7717/peerj.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Li H, Zheng B, Sun L, Yuan Y, Xing C. Competitive endogenous RNA (ceRNA) regulation network of lncRNA-miRNA-mRNA in colorectal carcinogenesis. Dig Dis Sci. 2019;64:1868–1877. doi: 10.1007/s10620-019-05506-9. [DOI] [PubMed] [Google Scholar]
- 14.Zhou RS, Zhang EX, Sun QF, Ye ZJ, Liu JW, Zhou DH, Tang Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19:779. doi: 10.1186/s12885-019-5983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y, Geng L, Wang K, Sun J, Xu W, Gong S, Zhu Y. Long noncoding RNA expression signatures of colon cancer based on the ceRNA network and their prognostic value. Dis Markers. 2019;2019:7636757. doi: 10.1155/2019/7636757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Z, Fu P, Yu Z, Zhen F, Gu Y. Comprehensive Analysis of lncRNA-miRNA- mRNA network ascertains prognostic factors in patients with colon cancer. Technol Cancer Res Treat. 2019;18:1533033819853237. doi: 10.1177/1533033819853237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang XJ, Wang W, Hann SS. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie. 2019;163:58–72. doi: 10.1016/j.biochi.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Hong X, Ba L, He X, Xiong Y, Ding Q, Yang S, Peng G. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10:150. doi: 10.1038/s41419-019-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Nangia-Makker P, Farhana L, Majumdar APN. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16:155. doi: 10.1186/s12943-017-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang AF, Chen MW, Huang SM, Kao CL, Lai HC, Chan JY. CD164 regulates the tumorigenesis of ovarian surface epithelial cells through the SDF-1α/CXCR4 axis. Mol Cancer. 2013;12:115. doi: 10.1186/1476-4598-12-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K, Willbanks A, Sarkar S. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG. DIANA-LncBase: Experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Xu J, Hong J, Tang R, Zhang X, Fang JY. Long noncoding RNA profiles identify five distinct molecular subtypes of colorectal cancer with clinical relevance. Mol Oncol. 2014;8:1393–1403. doi: 10.1016/j.molonc.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Yang S, Zeng J, Chen M. miR12715p inhibits cell proliferation and induces apoptosis in acute myeloid leukemia by targeting ZIC2. Mol Med Rep. 2019;19:508–514. doi: 10.3892/mmr.2018.9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin MF, Yang YF, Peng ZP, Zhang MF, Liang JY, Chen W, Liu XH, Zheng YL. FOXK2, regulted by miR-1271-5p, promotes cell growth and indicates unfavorable prognosis in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017;88:155–161. doi: 10.1016/j.biocel.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Chen L. Downregulation of lncRNA UCA1 facilitates apoptosis and reduces proliferation in multiple myeloma via regulation of the miR-1271-5p/HGF axis. J Chin Med Assoc. 2019;82:699–709. doi: 10.1097/JCMA.0000000000000145. [DOI] [PubMed] [Google Scholar]
- 28.Doyonnas R, Yi-Hsin Chan J, Butler LH, Rappold I, Lee-Prudhoe JE, Zannettino AC, Simmons PJ, Bühring HJ, Levesque JP, Watt SM. CD164 monoclonal antibodies that block hemopoietic progenitor cell adhesion and proliferation interact with the first mucin domain of the CD164 receptor. J Immunol. 2000;165:840–851. doi: 10.4049/jimmunol.165.2.840. [DOI] [PubMed] [Google Scholar]
- 29.Lin J, Xu K, Wei J, Heimberger AB, Roth JA, Ji L. MicroRNA-124 suppresses tumor cell proliferation and invasion by targeting CD164 signaling pathway in non-small cell lung cancer. J Gene Ther. 2016;2:6. doi: 10.13188/2381-3326.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang J, Zhang L, She X, Zhou G, Yu F, Xiang J, Li G. Inhibiting CD164 expression in colon cancer cell line HCT116 leads to reduced cancer cell proliferation, mobility, and metastasis in vitro and in vivo. Cancer Invest. 2012;30:380–389. doi: 10.3109/07357907.2012.666692. [DOI] [PubMed] [Google Scholar]
- 31.Coustan-Smith E, Song G, Clark C, Key L, Liu P, Mehrpooya M, Stow P, Su X, Shurtleff S, Pui CH, et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2011;117:6267–6276. doi: 10.1182/blood-2010-12-324004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.