Abstract
Single-molecule long-read DNA sequencing with biological nanopores is fast and high-throughput but suffers reduced accuracy in homonucleotide stretches. We now combine the CsgG nanopore with the 35-residue N-terminal region of its extracellular interaction partner CsgF to produce a dual-constriction pore with improved signal and basecalling accuracy for homopolymer regions. The electron cryo-microscopy structure of CsgG in complex with full-length CsgF shows that the 33 N-terminal residues of CsgF bind inside the β-barrel of the pore, forming a defined second constriction. In complexes of CsgG bound to a 35-residue CsgF constriction peptide, the second constriction is separated from the primary constriction by ~25 Å. We find that both constrictions contribute to electrical signal modulation upon ssDNA translocation. DNA sequencing using a prototype CsgG:CsgF protein pore with two constrictions improved single-read accuracy by 25 to 70 % in homopolymers up to 9 nucleotides long.
Introduction
In nanopore sensing applications, the interaction of analyte molecules with a nanometer-scale pore that is inserted in an insulating membrane within an electrical field results in altered ion conductance levels. Together with the frequency and life-time of the interaction, the modulated conductance levels can give information on the size, concentration, and chemical and conformational nature of the analyte1. A practical application of this principle is nanopore sequencing, where a single-stranded polynucleotide is electrophoretically threaded through a protein or solid state nanopore. The differential interaction of nucleotides with the pore’s narrow constriction during translocation contributes to changes in the channel’s ion current and can in principle be used to gain sequence information on the polynucleotide2–4. Because the speed of unperturbed electrophoretic polynucleotide passage precludes the resolution of individual nucleotides, a DNA or RNA-binding motor enzyme is added to the system to gain a processive and slowed down passage (millisecond scale / nucleotide) of the polynucleotide through the nanopore5,6.
Biological nanopore systems offer the advantage over synthetic or solid state nanopores of having a uniform pore size and conformation, and the possibility to make pore alterations through protein engineering and/or alternative combinations with processivity enzymes7. The use of protein nanopores for DNA sequencing was first introduced for α-hemolysin8 and later for MspA9. Both pores were heavily optimised the years after, notably by combining them with phi29 DNA polymerase to slow down DNA strand translocation10–15. Other pores that have been shown to thread ssDNA with some level of nucleotide discrimination include aerolysin, FhuA, Omp F/G, ClyA, PA63, SP1 and viral connectors phi29, SPP1, T3 and T47. Together, the nature of the enzyme, the structure and chemical composition of the nanopore, and the configuration of the nanopore-enzyme complex contribute to the signal-to-noise ratio and therefore sequencing accuracy of the system. In 2016, Oxford Nanopore Technologies (ONT) shifted from R7.4 to R9 chemistry16, implementing the E. coli curli transport channel CsgG17 as the sequencing nanopore. CsgG is a 36-stranded β-barrel responsible for the secretion of the bacterial amyloid curli across the outer membrane of Gram-negative bacteria17–19. The CsgG pore has a nine-fold circular symmetry (C9) and a well-defined ~1 nm wide constriction centrally located in a 4 nm wide channel17,20. During curli secretion, CsgG works in conjunction with the accessory proteins CsgE and CsgF21, which are located in the periplasm and on the cell surface, respectively, and give substrate selection to the channel and ensure the assembly of surface-associated curli fibers22,23. Whilst CsgE was found to bind and gate the CsgG channel17, if and how CsgF alters the CsgG channel properties is currently unknown. Here, we report characterisation of the interaction between CsgG and CsgF, including a single-particle cryo-EM structure and functioning of this complex in sequencing DNA.
Biochemical characterisation of the CsgG:CsgF complex
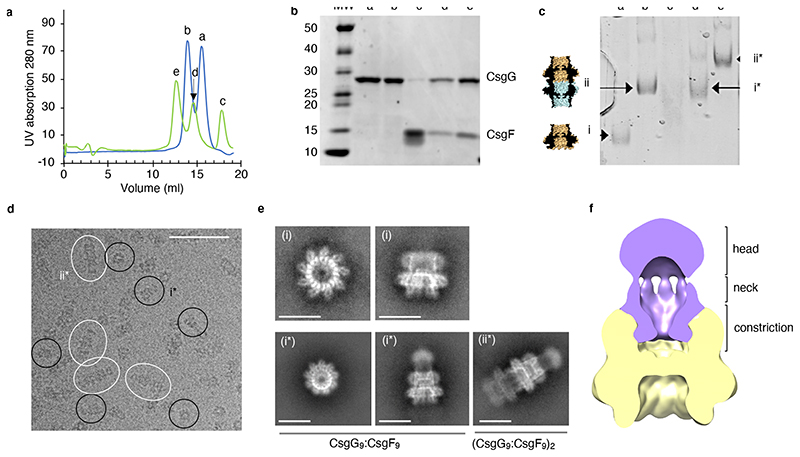
Addition of CsgF to purified, detergent-solubilised CsgG produced a qualitative shift in CsgG retention time on size exclusion chromatography, suggesting an interaction of both proteins (Figure 1a). Wild type E. coli CsgG elutes as two populations corresponding to D9 octadecamers (18-mer) and C9 nonamers (9-mer), the former a known in vitro artefact resulting from tail-to-tail dimerisation of concentrated, membrane-extracted CsgG pores17. Adding fivefold molar excess of CsgF shifted both CsgG populations and showed the presence of CsgF in both CsgG elution fractions as visualised on SDS-PAGE (Figure 1b). Excess CsgF eluted at the apparent molecular weight expected for the monomeric protein. Furthermore, tris-borate native PAGE showed a quasi-quantitative shift in D9 and C9 CsgG retention times, indicating stable and complete complex formation (Figure 1c). Similarly, a tandem affinity pull-down of detergent solubilised proteins from cultures co-expressing StrepII-tagged CsgG and His-tagged CsgF resulted in the isolation of pure CsgG:CsgF complex, indicating that also in vivo the two proteins form a complex in the bacterial outer membrane (Extended Data Figure 1a).
Figure 1. CsgG forms a stable complex with CsgF.
(a) Comparison of size exclusion profiles of CsgG with (green) and without (blue) excess CsgF. (b) 4-20% TGX stain-free SDS-PAGE and (c) tris-borate native PAGE of the elution fractions labelled a-e in panel a, corresponding to, respectively, C9 CsgG single channels (i), D9 CsgG dimeric channels (ii), excess CsgF, and the CsgF complexes of C9 CsgG (i*) and D9 CsgG (ii*). Experiment was repeated 3 times. (d) Cryo-electron micrograph. Single particles of single (i*) and dimeric (ii*) CsgG:CsgF pores are circled black and white, respectively, scale bar is 50 nm. (e) Representative 2D class averages highlighting views along the C9 symmetry axis (left) and side views of single (middle) and double (right) CsgG (upper row) and CsgG:CsgF (lower row) pores. (f) Slice-through 3D volume of CsgG:CsgF complex filtered down to 15 Å using EMAN229 and segmented and displayed at contour of 0.0073 in UCSF Chimera30. Density corresponding to CsgG and CsgF is coloured gold and purple, respectively.
Cryo-EM structure of the CsgG:CsgF curli assembly complex
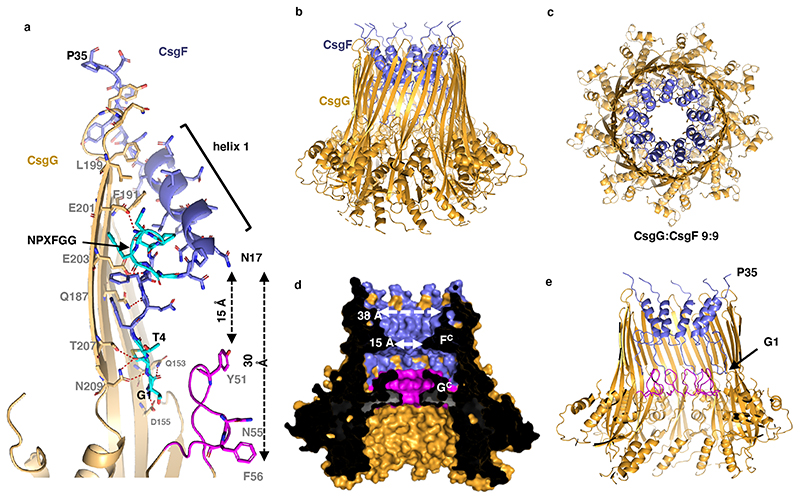
To gain structural insights into the CsgG:CsgF channel, the complex was visualised by single particle electron cryo-microscopy (cryo-EM). Cryo-EM micrographs revealed discrete particles that split into two main populations corresponding to C9 and D9 CsgG channels. Top views show the expected 9-fold symmetry corresponding to the C9/D9 axis in the crystal structure17 (Figure 1e). In the CsgG:CsgF sample, both populations showed the presence of an additional protruding density along the 9-fold symmetry axis, not seen in micrographs of CsgG channels only (Figure 1d, e). Class averages show additional high resolution features inside the main body of the channel as well as a diffuse protrusion from the extracellular side of the channel corresponding to CsgF (Figure 1e). Based on class averages and a C9-symmetrised low-resolution 3D reconstruction, the CsgF density can be separated into a globular extracellular head domain that is linked by a thin neck region to a well-defined constriction region lining the inside of the CsgG barrel (Figure 1f). Following computational enrichment for C9 particles through multiple rounds of 2D and 3D classification in Relion, (cfr class averages in Extended Data Figure 1b) a 3D cryo-EM density map was calculated to a resolution of 3.4 Å (Extended Data Figure 1c, d). Whilst the reconstruction showed unambiguous density for the first 35 residues of mature CsgF (Figure 2, Extended Data Figure 1 c, d), the head and neck density had poor definition and did not allow model building of the corresponding domains. The CsgF N-terminus reaches deep inside the CsgG β-barrel, where the first 4 residues (G1 to T4) make an intricate H-bond network with residues Q153, D155, T207 and N209 in the β-barrel lumen, then followed by a stretch of more hydrophobic residues (Figure 2a). A strictly conserved NPXFGG motif at residues 9-14 makes the CsgF N-terminus kink into the lumen of the CsgG β-barrel, giving rise to a sharp, 15 Å wide constriction of the channel, lined by residue N17 at the constriction apex (Figure 2a, d, 3a). From there the CsgF peptide runs back to the rim of the CsgG β-barrel via a 13-residue helix (helix 1), opening into a 38 Å wide exit of the channel. Notably, the N-terminus was found fully disordered in the solution structure of CsgF24, indicating it adopts a stable conformation only upon interaction with its partner CsgG. In the CsgG:CsgF complex, the CsgF protomers form an interaction interface with CsgG corresponding to 1030 Å2 solvent accessible surface area (corresponding to a calculated solvation free energy of -8.1 kcal/M) and incorporating ten and six putative H-bond and salt bridge interactions, respectively, and have a 330 Å2 solvent accessible surface interface with an adjacent CsgF protomer, the latter lacking polar interactions. Together, these interactions result in a highly stable non-covalent pore complex that withstands heating up to 70 °C, the melting temperature of the CsgG channel (Extended Data Figure 2a). Interestingly, the first 35 residues forming the CsgF constriction peptide (FCP) show a 48 % average pairwise sequence identity amongst CsgF homologs, versus a mere 28 % identity in the neck and head regions, suggesting that the CsgG contact and the conformation of the FCP are the most conserved features in the CsgF accessory protein (Extended Data Figure 3).
Figure 2. CsgG:CsgF cryo-EM structure reveals a dual constriction pore.
(a) Close-up ribbon and sticks (CsgF) representation of a single CsgG (gold) and CsgF (purple) protomer of the CsgG:CsgF cryo-EM structure. The CsgG constriction formed by Y51, N55 and F56 is highlighted magenta, and the N-terminal four residues and the conserved NPXFGG motif in CsgF are highlighted in cyan. Oxygens are red, nitrogens blue. H-bonds anchoring the CsgG:CsgF interaction are depicted as dashed red lines. (b, c, d, e) The CsgG:CsgF pore shown in side (b), top (c) and cross-sectional views (d, e), depicted in ribbon (b, c and e) and solvent-accessible surface (d) representations, coloured as in panel a. GC: CsgG-constriction, FC: CsgF-constriction. Only the CsgF N-terminus (residue G1 to P35) forming the CsgF-constriction peptide (FCP) could be resolved in the cryo-EM density (Extended Data Figure 2).
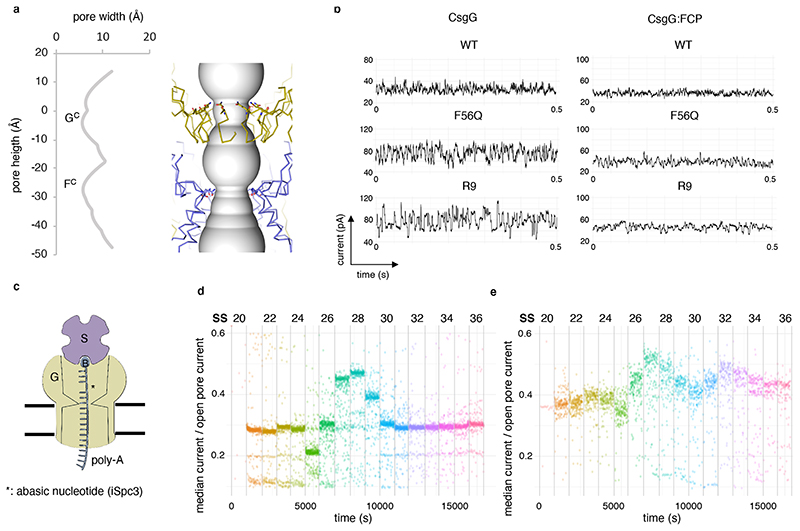
Figure 3. The CsgF constriction peptide creates a second constriction in the CsgG pore.
(a) Channel radii plotted against channel height (left) and its corresponding position in the CsgGF56Q:FCP complex (right). Distances are in angström. CsgG and FCP are depicted in gold and purple, respectively. Q56 in the CsgG constriction and N17 CsgF are shown in stick representation. (b) Representative current signatures during passage of single DNA strands through the WT CsgG, CsgGF56Q, CsgGR9 nanopores and their respective CsgG:FCP complexes. Data measured at -180 mV and representative of >100 capture events and >10 single channels. (Shown experiment representative of at least 3 repeat experiments) (c) Schematic diagram of pore read point detection assay. Pores are probed with oligonucleotides (SS20-SS36; Extended Data Figure 6a) with an abasic nucleotide (asterisk) at a defined distance from a biotin (B) – streptavidin (S) blockage. When the abasic residue resides at the pore constriction, this results in increased conductance levels. (d, e) Current levels for different static oligos (SS20-SS30) bound in CsgGF56Q (d) or CsgGF56Q:FCP (e). Each dot represents a single data point (at least 255 (d) and 76 (e) data points per oligo, measured from at least n=24 pores).
Capture of single-strand DNA by CsgG channels
CsgG channels show stable constitutively open conductance levels in single channel recordings corresponding to the 9 Å solvent-accessible diameter of the CsgG constriction loop17 (Extended Data Figure 4). During nanopore sequencing the CsgG periplasmic vestibule resides on the cis side of the membrane, accepts the translocating polynucleotide and forms the contact face with the processivity enzyme (Extended Data Figure 4a). Despite the narrow constriction defined by Y51, N55 and F56, wild type (WT) CsgG pores capture single stranded DNA, producing a squiggling signal centered around 30 pA with a range of ±10 pA as the DNA strand is translocated through the pore at -180 mV (Figure 3b). WT CsgG pores have been modified significantly to improve their properties in DNA sequencing. One such modified CsgG pore which contains the F56Q mutation (CsgGF56Q) produces a larger open pore current level compared to the WT CsgG pore (Extended Data Figure 4b) and produces a signal with a higher range when a DNA strand is translocated through the pore (Figure 3b). Another modified CsgG channel referred to as R9, proprietary to ONT, with optimised signal-to-noise ratio and improved discrimination of passing nucleotides has formed the baseline for DNA and RNA sequencing nanopores used in their sequencing platforms since 201616 (Figure 3b).
Selected CsgG:FCP pores capture single-stranded DNA
Our structural studies show that binding of CsgF results in the formation of a second constriction in the CsgG channel, spaced 15 to 30 Å above the exit and entrance of the CsgG constriction (Figure 2d, 3a). With a solvent excluded diameter of 15 Å, the constriction formed by the N-terminal peptide of CsgF is slightly larger than that of CsgG. This is in the range commonly regarded as being useful for nanopore sequencing, with described nanopores having dimensions ranging from 10 Å to 36 Å7. Contrary to the 3-layered native CsgG constriction, CsgF forms a sharp, single layered orifice made by N17 (Figure 2, 3a). The novel structural insights from the CsgG:CsgF complex were used to introduce a second constriction in the CsgG pore, made by the N-terminal peptide of CsgF, dubbed FCP. For production of the CsgG:FCP complex, TEV protease recognition sites were introduced in the CsgF neck region at positions 30, 35 or 40 of the mature protein (Extended Data Figure 2b). Upon in vitro reconstitution of the CsgG:CsgF complex and proteolytic cleavage using TEV-protease, the CsgF neck and head region were removed, obtaining a stable CsgG:FCP channel complex (Extended Data Figure 2c). Using this approach, we formed CsgG:FCP channels with WT CsgG as well as with the basic sequencing pore CsgGF56Q and the different proprietary CsgGR9 pores developed by ONT. The resulting complexes could be reconstituted into artificial membranes of the MinION flow cells developed by ONT and showed stable single channel conductance in agreement with the expected channel dimensions (Extended Data Figure 4b, c). Next, addition of DNA-enzyme complexes to flow cells containing CsgG:FCP, CsgGF56Q:FCP or CsgGR9:FCP resulted in squiggling conductance profiles in agreement with the capture and passage of single stranded DNA (Figure 3b). Remarkably, the conductivity profiles showed that the CsgG:FCP and CsgGR9:FCP pores remain as stable complexes both in solution and in the polymeric membrane. Threading of DNA during the sequencing run had no apparent effect on CsgGR9:FCP single channel life time, where over 90% of the pore complex remained stable over the 24 hours tested (Extended Data Figure 5).
Static DNA strand signal discrimination of hybrid CsgG:FCP pores
To evaluate whether both the CsgG and CsgF constrictions can modulate the global ion conductance during DNA strand passage, we measured single channel conductance of CsgGF56Q:FCP channels exposed to a series of oligonucleotides (dubbed SS20 to SS38) that were modified to lack a single base (iSpc3-positions) at a discrete distance from a biotin – streptavidin complex (Extended Data Figure 6a). The latter results in static pore – oligo complexes with single base aberrations at variable height from the channel entrance that results in an increase in conductance when residing at a channel constriction (Figure 3c). Probing CsgGF56Q channels with the different oligos showed a discrete rise in conductance with SS28 and extending into SS27 and SS29, demonstrating that a significant modulation of channel conductance occurred at 9 ±1 nucleotides from the channel entrance (Figure 3d). When CsgGF56Q:FCP channels were probed, an additional rise in conductance with SS31-33 was observed at 13 ±1 nucleotides from the channel entrance (Figure 3e). Assuming an extended ssDNA with a maximum length per base of 6.3 (±0.8) Å25 the nucleotide distance between these two regions causing a significant channel conductance modulation is in agreement with the ~25 Å spacing of the centers of the CsgG and CsgF constrictions measured from the cryo-EM structure (Figure 3a).
Improved homopolymer resolution by a dual constriction pore
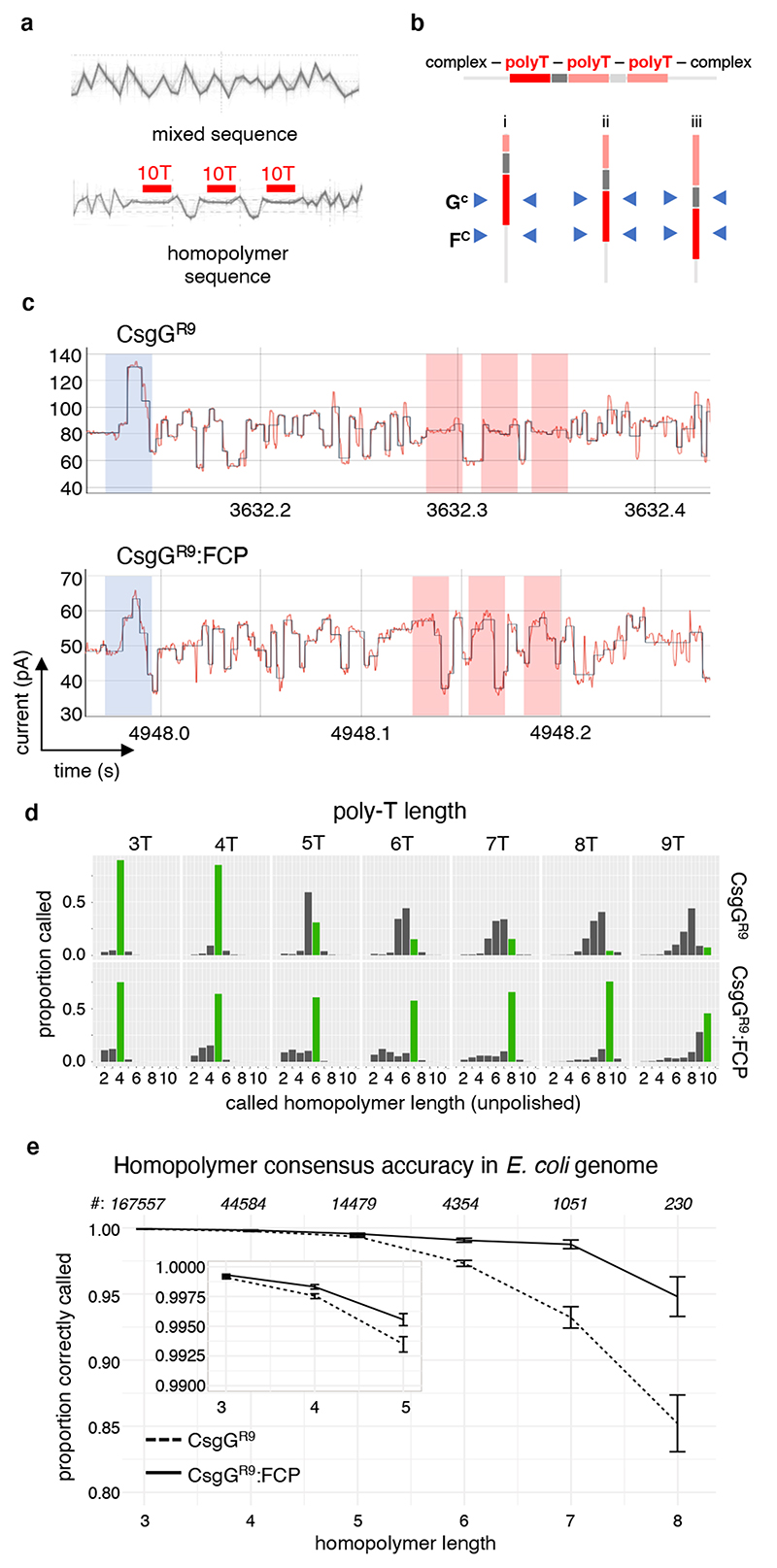
By using immobilised synthetic oligonucleotides, it has been shown that additional sequence information can be gained when two constrictions, rather than one, are employed within a single nanopore11. Therefore, we analysed whether the observed dual constriction nanopores offer opportunities to resolve homopolymer regions during polynucleotide sequencing, a known cause of inaccuracies in nanopore sequencing26,27. Homopolymer DNA translocating CsgG pores shows little to no modulation in conductance, leading to miscalling of the length of the homopolymer and may result in indel errors in basecalling during nanopore sequencing (Figure 4a, b, d), particularly for long homopolymers. To test the effect of a dual constriction, nanopore sequencing reads from oligos containing three regularly spaced stretches of 10 deoxythymidines (T) were compared between the sequencing nanopore CsgGR9 and its complex with FCP (CsgGR9:FCP) (Figure 4a, b). Passage of the poly-T section through CsgGR9:FCP showed increased modulation in pore conductance demonstrating that the conductance signal during single strand passage is likely a function of base interactions with both constrictions (Figure 4b, c). Sequencing of a series of plasmids modified to contain three regularly spaced T homopolymers ranging in length from 3 to 9 showed that the increased signal complexity in the polynucleotide spanning regions observed for CsgGR9:FCP squiggles results in a higher accuracy in homopolymer length calling compared to CsgGR9 (Figure 4d). Whereas for CsgGR9 the unpolished homopolymer length calling started to go down in accuracy from 5-mer poly-T stretches onwards, CsgGR9:FCP showed a 20 to 70% improved length calling up to at least 8- or 9-mer poly-T stretches. To evaluate if the increased homopolymer accuracy extends to the four bases, E. coli genomic DNA was sequenced using both CsgGR9 and CsgGR9:FCP pores. We found no significant effects on average read length in CsgGR9:FCP compared to CsgGR9, a property that largely depends on library preparation rather than pore characteristics. When the single reads of the two pores were aligned and compared, more deletions can be seen at the edge of homopolymer regions read by CsgGR9 compared to the CsgGR9:FCP pore (Extended Data Figure 6b). As a result, the polished consensus accuracy of the homopolymer region drops in CsgGR9 pores when the length of the homopolymer is longer than 5 bases (Figure 4e). Comparatively, CsgGR9:FCP was able to read the homopolymeric region more accurately over a longer length of bases (Figure 4e). Thus, the addition of a second constriction by means of the FCP increased accuracy in calling longer homopolymer regions. Further development of the neural network models used to basecall the electrical signals28 along with further modifications to the pore design may further improve basecalling accuracies of CsgG:FCP dual constriction pores.
Figure 4. Homopolymer basecalling by a prototype CsgG:FCP nanopore.
(a) Overlaid single molecule conductance profiles of a mixed ssDNA sequence (top) and a trial ssDNA sequence (bottom) containing three consecutive homopolymers of ten deoxythymidines (10T) spaced by GGAA intervals as read using the CsgGR9 nanopore (>500 individual traces coming from at least 50 pores). (b) Schematic representation of three interaction scenarios of the trial ssDNA sequence by the CsgG and FCP constrictions, labelled GC and FC respectively. The dual constriction is expected to increase the inclusion of sequence outside the homonucleotide stretch during passage through the pore (scenario i and iii).
(c) Single molecule conductance signals of the 10T homopolymer containing trial sequence (shown in panel a) analysed by CsgGR9 (upper) or CsgGR9:FCP (lower) pores. Shaded zones correspond to the adaptor (blue) and the 10T (red) regions. Traces are representative for >1000 capture events and >50 pores. (d) Histograms of single read homopolymer length calling of ssDNA containing poly-T stretches ranging 3 to 9 nucleotides in length, sequenced by CsgGR9 or CsgGR9:FCP. Correctly called homopolymer lengths are shown in green (plots contain at least 166,000 single reads per oligo). (e) Comparison of the proportion (± SD) of correctly called homopolymers versus homopolymer length for CsgGR9 and CsgGR9:FCP pores. The plot shows consensus accuracies across the four bases, using data polished with the medaka software package developed at ONT31, and is based on an E. coli assembly of depth 100x. Occurrences of the respective homopolymer lengths in the E. coli genome are indicated on top. Nonameric (n=22) and longer (n=2) homopolymers become too rare to provide statistically relevant numbers.
Discussion
Our data provide proof-of-principle for the construction of a dual constriction nanopore using the curli secretion channel CsgG or its nanopore derivatives and the N-terminal 35 residues of its secretion partner CsgF, and demonstrate the combined contribution of the two constriction points to conductance signals during passage of a polymer through the nanopore. Using this concept, we built a prototype dual constriction sequencing nanopore made of FCP-modified CsgGR9, and show its principle advance in homopolymer calling. Thus, FCP peptides form a convenient tool to modify the channel properties of CsgG-based nanopores. Since FCP peptides are binding the channel trans side, modifying CsgG-based channels with FCP does not affect cis side properties such as contact with nucleotide binding enzymes and analyte delivery to the pore. Besides applications in polynucleotide sequencing as shown in this study, we anticipate that dual constriction pores may have advantages in small molecule analyte sensing due to the consecutive passage of analytes through two chemically distinct points of interaction.
Online Methods
Strains and protein expression constructs
E. coli Top10 was used for all cloning procedures. E. coli C43(DE3) and BL21(DE3) were used for protein production (Supplementary Table 1). Expression of C-terminally StrepII-tagged E. coli CsgG as outer membrane localised pore made use of plasmid pPG117. For the expression of C-terminally 6x-His tagged CsgF in the E. coli cytoplasm, a PCR product encompassing the coding sequence for mature E. coli CsgF (i.e. CsgF without its signal sequence, Supplementary Table 1, sequence 1; primers 1 and 2) was cloned into pET22b via the NdeI and EcoRI sites, resulting in the CsgF-His expression plasmid pNA101. CsgG-step and CsgF-His were co-expressed using plasmid pNA62. pNA62 is a pTrc99a based vector encoding csgF-His and csgG-strep, and was created based on pNA152, a modified pTrc99a with the pDEST14 Gateway® cassette and the ampR resistance gene replaced by the streptomycin/spectinomycin resistance gene aadA. A PCR fragment encompassing part of the E. coli MC4100 csgDEFG operon corresponding to the coding sequences of csgE, csgF and csgG was generated with primers 3 and 4 (Supplementary Table 1; primer 4 adds the Strep-II tag sequence SAWSHPQFEK to the CsgG C-terminus) and was inserted into pDONR221 (ThermoFisher Scientific) via BP Gateway® recombination to generate pNA41. The recombinant csgEFG operon was subsequently inserted in pNA152 via LR Gateway® recombination to yield pNA43. The coding sequence for a 6×His-tag was added to the CsgF C-terminus in pNA43 by Quick-change mutagenesis PCR using primers 5 and 6 (Supplementary Table 1), yielding pNA60b, followed by removal of csgE by outward PCR using primers 7 and 8 (Supplementary Table 1) to obtain pNA62. Coding sequences corresponding to the TEV recognition side (ELNYFQS) were introduced after positions 30, 35 and 45 of mature CsgF by Quick-change mutagenesis PCR.
Recombinant protein expression
CsgG-StrepII was expressed in E. coli BL21 (DE3) cells transformed with plasmid pPG117. The cells were grown at 37°C to an OD 600 nm of 0.6 in Terrific Broth medium. Recombinant protein production was induced with 0.0002% anhydrotetracyclin and the cells were grown at 25°C for a further 16 h before being harvested by centrifugation at 5500 g. CsgF-His was expressed in the cytoplasm of E. coli BL21(DE3) cells transformed with plasmid pNA101. Cells were grown at 37°C to an OD of 600 nm followed by induction by 1mM IPTG and left to express protein for 15h at 37°C before being harvested by centrifugation at 5500 g. Co-expression of CsgG-strep and CsgF-His was performed using E. coli C43(DE3) cells transformed with plasmid pNA62 and grown at 37°C in Terrific Broth medium. When the cell culture reached an optical density (OD) at 600 nm of 0.7, recombinant protein expression was induced with 0.5 mM IPTG and cells were left to grow for 15 hours at 28°C, before being harvested by centrifugation at 5500 g.
Protein purification of the CsgG:CsgF complex, CsgG, and CsgF
For CsgG:CsgF tandem affinity purification E. coli cells transformed with pNA62 were resuspended in 50 mM Tris–HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 0.4 mM AEBSF, 1 μg/mL Leupeptin, 0.5 mg/mL DNase I and 0.1 mg/mL lysozyme. The cells were disrupted at 20 kPsi using a TS series cell disruptor (Constant Systems Ltd) and the lysed cell suspension was incubated 30’ with 1% n-dodecyl-β-d-maltopyranoside (DDM; Inalco) for extraction of outer membrane components. Next, remaining cell debris and membranes were spun down by ultracentrifugation at 100,000 g for 40’. Supernatant was loaded onto a 5 mL HisTrap column (GE Healthcare) equilibrated in buffer A (25 mM Tris pH8, 200 mM NaCl, 10 mM imidazole, 10% sucrose and 0.06% DDM). Column was washed with >10 column volumes (CVs) of 5% buffer B (25 mM Tris pH8, 200 mM NaCl, 500 mM imidazole, 10% sucrose and 0.06% DDM) in buffer A, and eluted with a gradient of 5-100% buffer B over 60 mL. Eluens was diluted 2-fold before loading overnight on a 5 mL Strep-tactin column (IBA GmbH) equilibrated with buffer C (25 mM Tris pH8, 200 mM NaCl, 10% sucrose and 0.06% DDM). The column was washed with >10 CVs buffer C and the bound protein was eluted in buffer C complemented with 2.5 mM desthiobiotin. CsgG-strep purification for in vitro reconstitution follows the protocol for CsgG:CsgF, except omitting sucrose in the buffers and bypassing the IMAC step. CsgF-His purification for in vitro reconstitution was performed by resuspension of the cell mass in 50 mM Tris–HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 0.4 mM AEBSF, 1 μg/mL Leupeptin, 0.5 mg/mL DNase I and 0.1 mg / mL lysozyme. The cells were disrupted at 20 kPsi using a TS series cell disruptor (Constant Systems Ltd) and the lysed cell suspension was centrifuged at 10,000 g for 30 minutes to remove intact cells and cell debris. Supernatant was added to 5 mL Ni-IMAC-beads (Workbeads 40 IDA, Bio-Works Technologies AB) equilibrated with buffer D (25 mM Tris pH8, 200 mM NaCl, 10 mM imidazole) and left incubating for 1 hour at 4°C. Ni-NTA beads were pooled in a gravity flow column and washed with 100 mL of 5% buffer E (25 mM Tris pH8, 200 mM NaCl, 500 mM imidazole diluted in buffer D. Bound protein was eluted by stepwise increase of Buffer E (10% steps of each 5mL). For electron microscopy and electrophysiology, CsgG:CsgF and CsgG:FCP protein complexes were obtained by either in vitro reconstitution or co-expression. For reconstitution, purified CsgG and CsgF were mixed at a molar ratio of 1 CsgG: 5 CsgF to saturate the CsgG barrel with CsgF. Next, the reconstituted or co-expressed complex was injected on a Superose 6 10/30 column (GE Healthcare) equilibrated with buffer F (25 mM Tris pH8, 200 mM NaCl and 0.03% DDM), and run at 0.5 mL/min. For CsgG:FCP, the CsgF forms correspond to CsgF35TEV and reconstituted CsgG:CsgF complexes were digested at room temperature overnight with TEV protease in buffer F. The mixture was then run back through a 5 mL HisTrap (GE Healthcare) column and the flow through was collected, heated at 60°C for 15 minutes and centrifuged at 21,000 g for 10 minutes prior to use in electrophysiology. Protein concentrations were determined based on calculated absorbance at 280 nm and assuming 1/1 stoichiometry.
SDS and native PAGE
Fractions from size exclusion were boiled for 5 minutes in loading buffer (final concentrations of 60 mM Tris-HCl pH 6.8, 2% SDS, 10% v/v glycerol and 0.01% bromophenol blue) and run on Mini-Protean 4-15% TGX-SDS-PAGE in llaemli running buffer (25 mM Tris, 192 mM Glycine, 0.1% SDS pH 8.3) at 300 V for 20 minutes followed by staining with InstantBlue (Expedeon). Molecular weight ladder used is PageRuler Unstained (ThermoFisher). Samples for native PAGE were mixed 3:1 with sample buffer (1x TBE, 20% v/v glycerol, 0.04% Bromophenol blue) and run at 90 V for 2h on a 4.5% native PAGE in 0.5x TBE buffer (1x TBE is 90 mM Tris-borate pH 8.3; 1mM EDTA).
Structural analysis using cryogenic electron microscopy
Sample behaviour of the size exclusion fractions was probed using negative stain electron microscopy. Samples are stained with 1% uranyl formate and imaged using an in-house 120 kV JEM 1400 (JEOL) microscope equipped with a LaB6 filament. For high resolution cryo-EM analysis, CsgG:CsgF samples were prepared by spotting 3 μl sample on R2/1 Holey grids (Quantifoil) coated with graphene oxide (Sigma Aldrich), manually blotted and plunged in liquid ethane using a CP3 plunger (Gatan). Sample quality was screened on the in-house JEOL JEM 1400 before collecting a dataset on a 300 kV TITAN KRIOS (FEI, Thermo-Scientific) microscope equipped K2 Summit direct electron detector (Gatan) at the Astbury Centre in Leeds, UK (CsgG:CsgF) or at Diamond – eBIC, Harwell Science and Innovation Campus, UK (CsgG). The detector was used in counting mode with a cumulative electron dose of 56 electrons per Å2 spread over 50 frames. For CsgG:CsgF, 2045 images were collected with a pixel size of 1.07Å. Images were motion-corrected with MotionCor2.136 and defocus values were determined using ctffind437. No dose-weighting scheme was applied and the first 2 frames were discarded as the result of excessive motion. Data was further analysed using a combination of EMAN229, RELION2.038 and SIMPLE39. 50 micrographs were manually picked using e2boxer.py from the EMAN2 package resulting in 4000 particles for which 2D templates were generated using Relion 2D classification. Next, particles were picked automatically using Gautomatch (Dr. Kai Zhang, https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/), yielding a total of 750,000 particles. Subsequent 2D stack cleaning, discarding graphene oxide lines and the dimeric barrels (Figure 1e), yielded a stack of 135,000 particles for 3D refinement. C9 symmetry was imposed during 3D model generation and refinement. Initial model was computed using e2initialmodel.py from the EMAN2 package. 3D classification in Relion was used to enrich for head-containing particles, indicative for presence of CsgF. This yielded a stack of 62,000 particles used to calculate the final map at 3.4 Å resolution (Supplementary Table 2). De novo model building of CsgF was done with COOT40 and iterative cycles of model building and refinement of the full complex comprising nine CsgG copies ranging from residues 10 to 260 (the loop spanning residues 102-111 was disordered and is absent from the model) and nine copies of CsgF ranging from residue 1 to 35 (mature protein numbering) was done with PHENIX real-space refinement41 in combination with COOT (data and model statistics are found in Table S2). Surface and cartoon representations were made with The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. Pore plots in Figure 3a were made with HOLE42, using the endrad 10 parameter, plotted with MS Excel 2013 and dumbbell representation was visualised in PyMol (https://pymol.org/2/).
Testing CsgG and CsgG:FCP pores on MinIONs
All CsgG or CsgG:FCP samples were incubated with Brij58 (final concentration of 0.1%) for 10 minutes at room temperature before diluting CsgG (1 mg/mL) samples 1:100,000 and CsgG:FCP samples (1 mg/mL) in 1:5000 in MinION flow cell buffer (25 mM potassium phosphate, 150 mM potassium ferrocyanide, 150 mM potassium ferricyanide, pH 8.0) for pore insertion. All pore experiments were done on MinION flow cells (lacking any pre-inserted nanopores) using MinION devices developed by ONT. MinKNOW core 1.11.5 version software developed and provided by ONT was used to control scripts during all experiments. For insertion of CsgG or CsgG:FCP pores, 300 μL of diluted pore samples were loaded into the priming port of the flow cell. The pore insertion script of MinKNOW was used to apply voltage starting from -150 mV, increasing 10 mV every 15 seconds up until -450 mV. 1 mL of flow cell buffer was perfused through the priming port to remove any excess pores. Groups and positions with single pores were evaluated using the standard flow cell check protocol using MinKNOW. Open pore currents of CsgG or CsgG:FCP pores were recorded at -180 mV before adding any DNA (Extended Data Figure 4b).
Generation of IV curves on MinION
To obtain current-voltage (IV) curves of CsgG or CsgG:FCP pores in Extended Data Figure 4c, flow cells with different pores were prepared as shown above. Different electric potentials were applied using MinKNOW, ranging from -200 mV to +200 mV, where every 30 s the potential was increased 25 mV. For both CsgG and CsgG:FCP pores, data was analysed over multiple pores (as indicated) within a flow cell by measuring the mean open pore current at each potential. The mean open pore current and the 95% confidence level was calculated for each potential and plotted using R43.
DNA sequencing experiments on MinION
To evaluate the performance of CsgG and CsgG:FCP pores, DNA experiments were performed using a 3.6kb ssDNA section from the 3’ end of the lambda genome, linearised pTrc99a vectors with synthetic poly-T inserts (see below) or genomic DNA from E. coli. For nanopore sequencing, DNA strands need to be ligated to the ONT adapter mix (AMX) in order to optimise the capture and threading of the DNA strands into the pore. Unless stated otherwise, all components used for ligation and sequencing are provided in the ONT-SQK-LSK109 kit developed by ONT. Ligation was performed as per instructions provided with the kit. Briefly, 1 μg of dA tailed 3.6kb lambda DNA was mixed with 40 nM of adapter mix in the presence of 50 μL of NEB Blunt/TA Ligase Master Mix (NEB). The reaction was incubated for 10 minutes at room temperature. The ligation mixture was purified from un-ligated/free adapter using Agencourt AMPure XP Spri beads (Beckmann coulter). 0.4x (v/v) of spri beads were added to the ligation mixture and incubated for a further 10 minutes. The beads were then washed twice using 250 μL of short fragment buffer (SFB) for 10 minutes each. The DNA was then eluted from the beads using 25 μL of elution buffer. Final sequencing library mixture for each flow cell was prepared by adding 37.5 μL of sequencing buffer, 25.5 μL library loading beads and 12 μL of eluted DNA. Before loading the sequencing mix, the flow cell was initially flushed with 800 μL of priming buffer through the priming port. The SpotON port cover was opened and an additional 200 μL of priming buffer was flushed through the priming port. Finally, 75 μL of sequencing library mix was added to the flow cell using the SpotON port. All recordings were conducted at -180 mV and data acquisition and analysis was performed using MinKNOW.
The DNA squiggles shown in Figure 3b were generated using E. coli genomic DNA and data was plotted using ONT in-house software. Figure 4a and 4b were generated with a 3.6 kb ssDNA section from the 3’ end of the lambda genome. This strand contains 3 stretches of 10 deoxythymidine nucleotides, spaced by GGAA and flanked by mixed sequence preceding and following the homopolymer regions. To test homopolymer calling accuracy (Figure 4d), a series of oligonucleotides with three stretches of deoxythymidine nucleotides, ranging from 3 to 9 in length and spaced by GGAA was cloned into the pTrc99a vector using XmaI and HindIII restriction (called 3x_T3 to 3x_T9, Table S1). Prior to sequencing, the modified plasmids were linearised by PCR using primers 9 and 10 (Supplementary Table 1). The different constructs were confirmed by Sanger sequencing and run in different parallel MinION sequencing runs. Data acquisition and analysis was performed using MinKNOW and basecalling was performed locally using Guppy v2.3.5 software. Per construct, the length of the poly-T insert as called in the unpolished single reads was plotted in histograms, using at least 166,000 single reads. E. coli genomic DNA was used as a reference for comparing the homopolymer basecalling of CsgGR9 and CsgGR9:FCP across the four bases and in the context of random flanking sequence (Figure 4e, Extended Data Figure 4b). Data acquisition and analysis as described above. The pile up of basecalls in Figure 4d was plotted using the Integrative Genomics Viewer software33.
Preparation and testing of DNA-biotin-streptavidin static strand complexes on MinION
A set of polyA DNA strands (SS20 to SS38; Figure 3c) in which one base is missing from the DNA backbone (iSpc3) was obtained by Integrated DNA Technologies (IDT). The 3’ end of each of these strands comprises a biotin modification. The DNA strands were incubated with monovalent streptavidin in a 1:1 ratio (9.9 μM final concentration) at room temperature for 20 minutes, resulting in the DNA-biotin-streptavidin static strand complexes for each polyA DNA strand. The complexes were diluted to 2 μM with priming buffer. MinION flow cells with CsgG or CsgG:FCP pores were prepared as described above. 800 μL of priming buffer was flushed into the flow cell via the priming port in preparation for the static strands. The script used for the static-strands experiment was run at -200 mV for CsgG:FCP pores and -180 mV for CsgG pores with a reverse potential flick every minute (0 mV for 2 seconds, 100 mV for 2 seconds and then 0 mV for 2 seconds). 75 μL of each DNA-biotin-streptavidin complex was added to the flow cell sequentially via the SpotOn port and data was recorded for 15 minutes for each complex. 800 μL of priming buffer was flushed via the priming port in between each addition of the complex to make sure the first DNA-biotin-streptavidin complex is removed before the addition of the next. This process was repeated for all static strands. Once the final static strand complex had been incubated on the flow cell, 800 μL of priming buffer was flushed via the priming port and 10 minutes of open pore recording was generated before finishing the experiment.
The median block current level for each static strand complex, SS20-SS38, was measured by filtering the data to show only ‘block events’ over time. Block events are defined as being longer than 5 seconds, less than 60 seconds in duration and less than 150 pA median current for the CsgGF56Q or less than 80 pA CsgGF56Q:FCP. A scatter plot was generated for both CsgGF56Q and CsgGF56Q:FCP by plotting each data point over time (s), where each data point represents a single block event normalised against the open pore current of that event.
Extended Data
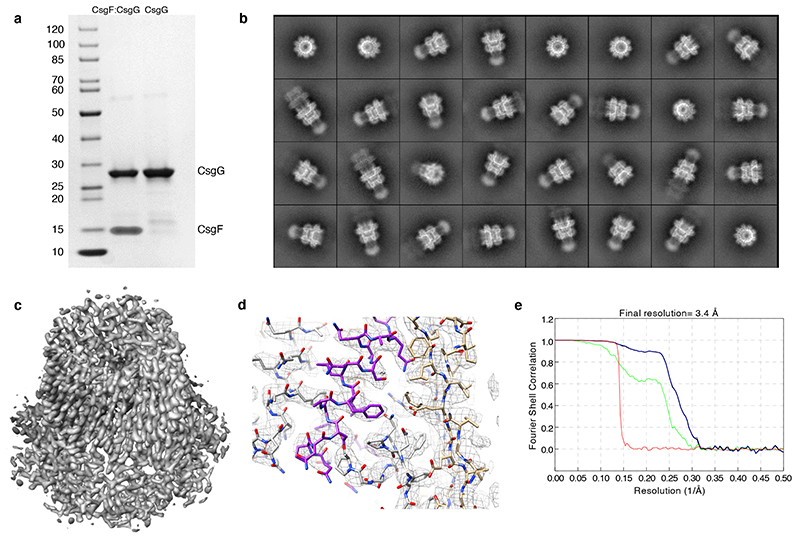
Extended Data Fig. 1. Electron cryo-microscopy of the CsgG:CsgF complex.
(a) SDS PAGE of CsgG or CsgG:CsgF complex obtained by tandem affinity purification of the outer membrane proteins extracted from cells expressing CsgG-strep (pPG1) or CsgG-strep and CsgF-His (pNA62), respectively. Gel representative for n>10 experiments. (b) Representative 2D class averages for the CsgG:CsgF dataset enriched for single pores (i.e. C9 CsgG:CsgF complexes), generated using SIMPLE and used for 3D reconstruction using Relion-2.0. (c) Off-axis top view and cross-sectional side view of the CsgG:CsgF cryo-EM 3D density reconstructed to 3.4 Å. (d) Representative region of electron density of the CsgG:CsgF complex. Region of focus is the constriction helix of FCP, stacking against the lumen of the CsgG β-barrel. One CsgF protomer is highlighted in purple, the others in grey; CsgG is depicted in gold. Heteroatoms are in blue (nitrogens) and red (oxygens). The density map is cut-off at a contour of 0.5, shown in stick and mesh representation, rendered using UCSF Chimera 1.10.2. (e) Fourier Shell Correlation (FSC) curves of the final 3D reconstruction (black: FSC corrected map, green: FSC unmasked map, blue: FSC masked map, red: FSC phase randomized unmasked map).
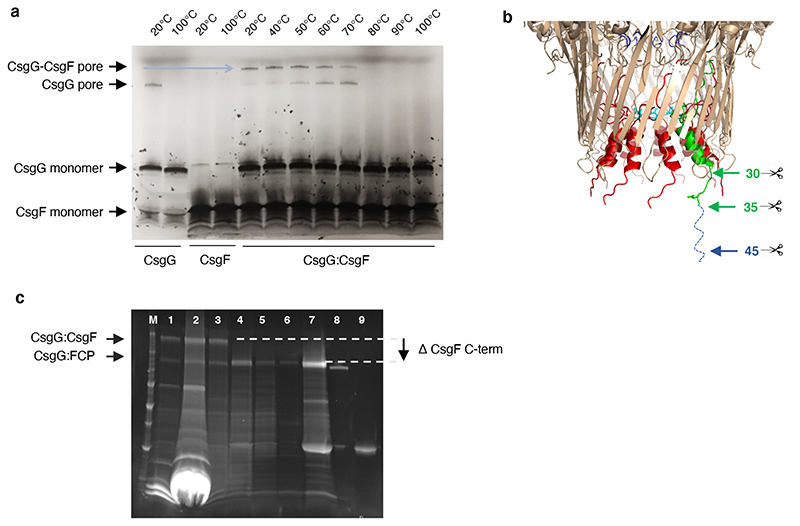
Extended Data Fig. 2. Production and thermal stability of CsgG:CsgF and CsgG:FCP pores.
(a) Production and temperature stability assessment of the CsgG:CsgF pore complex. Incubation of purified CsgG and CsgF in a 1:1 ratio results in the formation of a SDS stable CsgG:CsgF pore complex that is heat stable up to 70 °C. (b) The N-terminal residues of CsgF insert into the CsgG channel and form a second region of constriction, whilst the remaining ~100 residues form a cap like head structure (Figure 1e, f). For nanopore sensing purposes, we sought to produce a complex of CsgG with the CsgF constriction peptide (FCP), lacking the neck and head domains. To do so, CsgG was complexed with CsgF mutants modified to insert a TEV cleavage site at position 30, 35 or 45. The reconstituted CsgG:CsgF pore complexes were digested with TEV protease and analysed by SDS-PAGE (c). M: molecular mass marker, Lane 1, 2: Strep-tag affinity purified CsgG:CsgF complex and excess CsgG, Lane 3: isolation of CsgG:CsgF complex by size exclusion chromatography, Lane 4: CsgG:CsgF35-TEV cleaved with TEV protease to generate CsgG:FCP complex, Lane 5: flow through of CsgG:FCP after strep purification, Lane 6: CsgG:FCP heated to 60°C for 10 minutes. Lane 7: Eluted CsgG:FCP complex from strep column, Lane 8: CsgG pore as the control, Lane 9: TEV protease as the control.
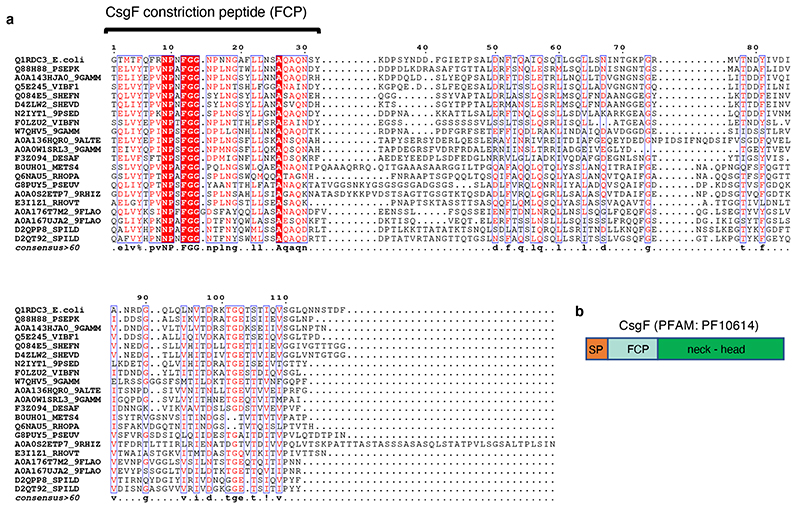
Extended Data Fig. 3. Multiple sequence alignment of CsgF-homologues.
(a) Multiple sequence alignment (Multalin32) of 22 representative CsgF sequences. Aligned sequences are shown as mature proteins (i.e. lacking their N-terminal signal peptide). The N-terminal 33 residues of the mature protein form a continuous stretch of high sequence conservation (48% average pairwise sequence identity) encompassing the region interacting with CsgG and forming the CsgF constriction peptide. CsgF homologues included in the multiple sequence alignment are UniProt entries Q88H88; A0A143HJA0; Q5E245; Q084E5; F0LZU2; A0A136HQR0; A0A0W1SRL3; B0UH01; Q6NAU5; G8PUY5; A0A0S2ETP7; E3I1Z1; F3Z094; A0A176T7M2; D2QPP8; N2IYT1; W7QHV5; D4ZLW2; D2QT92; A0A167UJA2. (b) Schematic diagram of CsgF protein architecture. (SP) signal peptide, cleaved upon secretion; (FCP) CsgF constriction peptide, CsgF neck and head region are coloured green.
Extended Data Fig. 4. Sequencing setup and channel characteristics of CsgG and CsgG:FCP nanopores.
(a) Schematic representation of the electrophysiology setup of CsgG-based nanopores as used for polynucleotide sequencing. CsgG-based channels (G) are reconstituted into artificial membranes with the periplasmic vestibule and β-barrel exposed to the cis and trans sides, respectively. Polynucleotide – enzyme (E) complexes are added to the cis side and current reads are recorded under an electric potential (Δψ) of 100 to 300 mV. (b, c) Representative single channel traces (b) and current - voltage (IV) curves (c) for wildtype CsgG, CsgGF56Q and CsgGR9 and their FCP complexes: CsgG:FCP, CsgGF56Q:FCP and CsgGR9:FCP. IV curves show mean ± 95% confidence interval of at least 60 single channels per pore, with the exception of wildtype CsgG (36 single channels) and CsgG:FCP (14 single channels).
Extended Data Fig. 5. Single channel stability of CsgGR9:FCP complexes.
(a, b) Single channel conductance trace of two representative CsgGR9:FCP nanopores during a 24 hour sequencing run, recorded at -180 mV. The data show both CsgGR9 and CsgGR9:FCP are predominantly in a sequencing, DNA-occupied state, with apo pores capturing new DNA strands within seconds. The two traces show a CsgGR9:FCP pore complex that stays intact of the 24h sequencing run (a), as well as a pore complex that shows dissociation of the FCP peptides during the sequencing run (at ~ 19h; b). Upon FCP dissociation, the channel continues sequencing now as a CsgGR9 apo pore (labelled CsgGR9). Arrows indicate the average conductance levels of the open pore and the DNA-occupied pore during sequencing intervals. The zoomed in panels show two representative 30s time windows of the sequencing run of the intact CsgGR9:FCP channel (left) and the CsgGR9 channel following dissociation of FCP (right). The full and zoomed in sequencing runs show high DNA capture rates for CsgGR9:FCP channels throughout the 24h sequencing run. (c) Scatter plot of the open pore current of 25 CsgGR9:FCP channels during 24h sequencing runs, recorded at -180 mV. Open pore plots for CsgGR9:FCP pores that stay intact throughout the 24h run (n=22), and pores that lose FCP (n=3) are coloured blue and red, respectively.
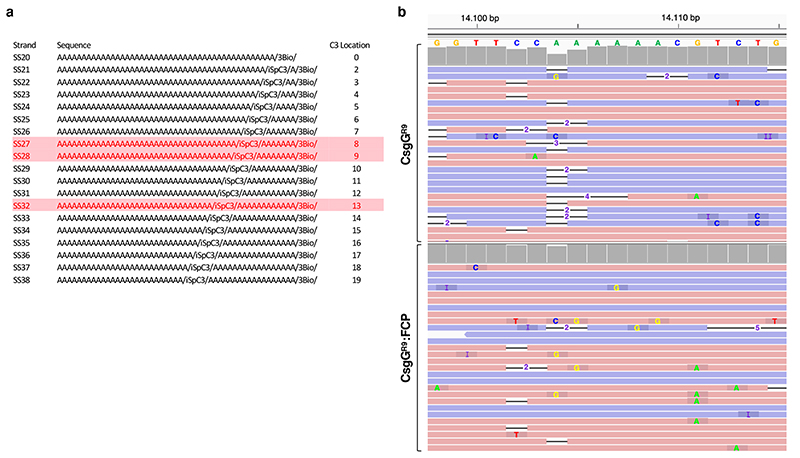
Extended Data Fig. 6. Constriction mapping oligos and single read basecalls for CsgGR9 and CsgGR9:FCP nanopores.
(a) Set of static polyA ssDNA oligonucleotides in which one base is missing from the DNA backbone (iSpc3). These oligos that have differing location of the abasic nucleotide, dubbed SS20 to SS38, were used to map the constriction position in CsgGF56Q or CsgGF56Q:FCP (Figure 3d). Biotin modification at the 3’ end of each strand is complexed with monovalent streptavidin to block translocation of the oligo and give a defined distance marker between the pore entrance (block site) and pore constriction (site of increased conductance when occupied by the abasic nucleotide; Figure 3c). SS27-SS28 and SS32 (highlighted red) have their abasic nucleotide located at the CsgG and FCP constriction, respectively (Figure 3d, e). (b) Comparison of errors in single read (n=26) basecalls from CsgGR9 and CsgGR9:FCP pores that have been aligned to a representative region of the E. coli reference genome sequence. The region displayed corresponds to the locus 14,098 to 14,115. The figure is plotted using the Integrative Genomics Viewer software33. Pink/purple bars correspond to single reads in the forward and reverse directions respectively. Black horizontal bars correspond to deletions in the basecalls, where the number corresponds to the number of deletions at the specific loci. Individual substitutions are labeled with the miscalled nucleotide (C in blue, T in red, G in orange and A in green). Insertions are labelled “I” (purple). Grey bars on top of the list of single reads of the CsgGR9 and CsgGR9:FCP pores correspond to the consensus accuracy per position.
Supplementary Material
Acknowledgements
We are thankful to R. Thompson and J. van Rooyen for assistance during cryo-EM data collection on Titan Krios 1 at the Astbury Biostructure Laboratory, Leeds, and Krios m02 at Diamond - eBIC, Harwell Science and Innovation Campus, UK, respectively. We thank R. Efremov for advice on cryo-EM image processing, and are thankful to S. Young at ONT for helpful discussion and advice on MinION data analysis. This work received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 649082 (BAS-SBBT). SVDV is recipient of a PhD fellowship of the Flanders Research Foundation (FWO).
Footnotes
Author Contributions
SVDV produced and characterised CsgG:CsgF complexes and determined their cryo-EM structure, supervised by HR. NVG and WJ produced and analysed CsgG, CsgF and CsgG:FCP constructs and protein. PS, RH, JK, MJ and JW produced CsgG:FCP pores, recorded and analysed electrophysiology data. LJ and HR supervised the study and analysed data. SVDV, JW, LJ and HR wrote the paper, with contributions of all authors.
Competing interests
VIB and ONT have jointly filed two provisional patent applications on the construction and use of dual constriction pores in nanopore sensing applications (PCT/GB2018/051858 and PCT/GB2018/051191). VIB has a funded research collaboration agreement with VIB related to CsgG-derived nanopores. ONT uses CsgG-derived nanopores in its MinION, GridION and PromethION nanopore sequencing devices. As inventors on VIB IP, SVDV, NVG and HR receive a share in royalty payments. RH, PS, JK, MJ, EJW and LJ are employees of ONT and own company share options.
Data Availability
Coordinates and the electron density maps for the CsgG:CsgF cryo-EM structure have been deposited in the PDB and EMDB under accession codes 6SI7 and EMD-10206, respectively. R9 pores are proprietary mutants of E. coli CsgG developed by ONT and are available as membrane embedded single pores incorporated in Flongle, MinION, GridION and PromethION flow cells.
Code Availability
The ONT software MinKnow and Guppy is made available through https://community.nanoporetech.com/downloads, and Medaka is made available through https://github.com/nanoporetech/medaka.
References
- 1.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 2.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nature biotechnology. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 3.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophysical journal. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benner S, et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nature nanotechnology. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olasagasti F, et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nature nanotechnology. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Zhao Z, Haque F, Guo P. Engineering of protein nanopores for sequencing, chemical or protein sensing and disease diagnosis. Current opinion in biotechnology. 2018;51:80–89. doi: 10.1016/j.copbio.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoddart D, Franceschini L, Heron A, Bayley H, Maglia G. DNA stretching and optimization of nucleobase recognition in enzymatic nanopore sequencing. Nanotechnology. 2015;26 doi: 10.1088/0957-4484/26/8/084002. 084002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoddart D, et al. Nucleobase recognition in ssDNA at the central constriction of the alpha-hemolysin pore. Nano Lett. 2010;10:3633–3637. doi: 10.1021/nl101955a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci U S A. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maglia G, Heron AJ, Stoddart D, Japrung D, Bayley H. Analysis of single nucleic acid molecules with protein nanopores. Methods Enzymol. 2010;475:591–623. doi: 10.1016/S0076-6879(10)75022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nature biotechnology. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nature biotechnology. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown CG. No Thanks, I’ve already got one. Clive G Brown, CTO of Oxford Nanopore Technologies; 2016. www.youtube.com/watch?v=nizGyutn6v4. [Google Scholar]
- 17.Goyal P, et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature. 2014;516:250–253. doi: 10.1038/nature13768. nature13768 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. MMI4997 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Gerven N, Van der Verren SE, Reiter DM, Remaut H. The Role of Functional Amyloids in Bacterial Virulence. Journal of Molecular Biology. 2018:10–16. doi: 10.1016/j.jmb.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao B, et al. Structure of the nonameric bacterial amyloid secretion channel. Proc Natl Acad Sci U S A. 2014;111:E5439–5444. doi: 10.1073/pnas.1411942111. 1411942111 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. 295/5556/851 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A. 2009;106:900–905. doi: 10.1073/pnas.0812143106. 0812143106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nenninger AA, et al. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol. 2011;81:486–499. doi: 10.1111/j.1365-2958.2011.07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubeis T, et al. Structural and functional characterization of the Curli adaptor protein CsgF. FEBS Letters. 2018;592:1020–1029. doi: 10.1002/1873-3468.13002. [DOI] [PubMed] [Google Scholar]
- 25.Chi Q, Wang G, Jian J. The persistence length and length per base of single-stranded DNA obtained from fluorescence correlation spectroscopy measurements using mean field theory. Physica A: Statistical Mechanics and its Applications. 2013;392:1072–1079. [Google Scholar]
- 26.Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome biology. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter JM, Hussain S. Robust long-read native DNA sequencing using the ONT CsgG Nanopore system. Wellcome open research. 2017;2:23. doi: 10.12688/wellcomeopenres.11246.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Oxford Nanopore Technologies, Medaka 0.8.1. 2018 https://nanoporetech.github.io/medaka/
- 32.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 35.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 36.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5 doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reboul CF, Eager M, Elmlund D, Elmlund H. Single-particle cryo-EM-Improved ab initio 3D reconstruction with SIMPLE/PRIME. Protein Sci. 2018;27:51–61. doi: 10.1002/pro.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14(376):354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 43.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. URL https://www.R-project.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and the electron density maps for the CsgG:CsgF cryo-EM structure have been deposited in the PDB and EMDB under accession codes 6SI7 and EMD-10206, respectively. R9 pores are proprietary mutants of E. coli CsgG developed by ONT and are available as membrane embedded single pores incorporated in Flongle, MinION, GridION and PromethION flow cells.
The ONT software MinKnow and Guppy is made available through https://community.nanoporetech.com/downloads, and Medaka is made available through https://github.com/nanoporetech/medaka.