Summary
Integral membrane proteins represent approximately 25% of protein-coding genes1. In eukaryotes, the vast majority of membrane proteins are inserted, modified, and folded at the endoplasmic reticulum (ER)2. Extensive work over the past several decades has determined how membrane proteins are targeted to the ER and how individual transmembrane domains (TMDs) are inserted into the lipid bilayer3. By contrast, very little is known about how proteins with multiple TMDs are assembled within the membrane. Assembly of TMDs typically involves polar or charged amino acids whose interactions with each other stabilise the final folded configuration4–8. TMDs with hydrophilic amino acids are likely to be chaperoned during co-translational membrane protein biogenesis, but ER-resident intramembrane chaperones are poorly defined. Here, we identify the PAT complex, an abundant obligate heterodimer of CCDC47 and Asterix, two widely conserved ER resident membrane proteins. The PAT complex engages nascent TMDs containing unshielded hydrophilic side chains within the lipid bilayer and disengages concomitant with substrate folding. Cells lacking either subunit of the PAT complex show reduced biogenesis of numerous multi-spanning membrane proteins. Thus, the PAT complex is an intramembrane chaperone that protects TMDs during their assembly to minimise misfolding of multi- spanning membrane proteins and maintain cellular protein homeostasis.
Multi-spanning membrane proteins have wide-ranging functions including transmembrane transport, cell surface receptors, and intercellular interactions2. Many of these functions require TMDs to serve as more than simple membrane anchors. For example, receptors or transporters need specific hydrophilic surfaces in the plane of the membrane to execute their functions2. For this reason, the TMDs of multi-spanning membrane proteins are highly diverse in length, sequence, and biophysical properties.
When folded, the TMDs of multi-spanning membrane proteins typically pack with their hydrophilic regions facing each other, presenting a mostly uninterrupted hydrophobic surface to the surrounding lipid bilayer4–8. How semi-hydrophilic TMDs are temporarily stabilised within the membrane until their assembly with distal TMDs is not well understood. ER- resident intramembrane chaperones have been postulated but are poorly defined3,9,10. Chaperones that act to facilitate membrane protein biogenesis are likely to be important because impaired folding can cause loss- and gain-of-function phenotypes in many experimental systems and various diseases in humans11,12.
Detection of a potential TMD chaperone
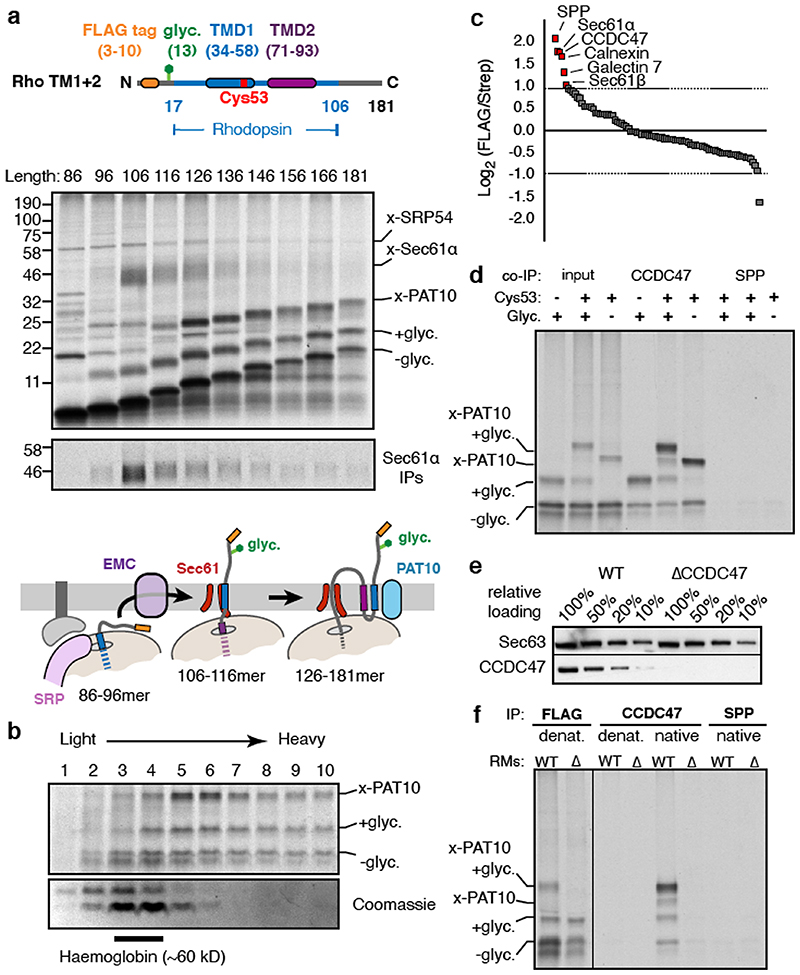
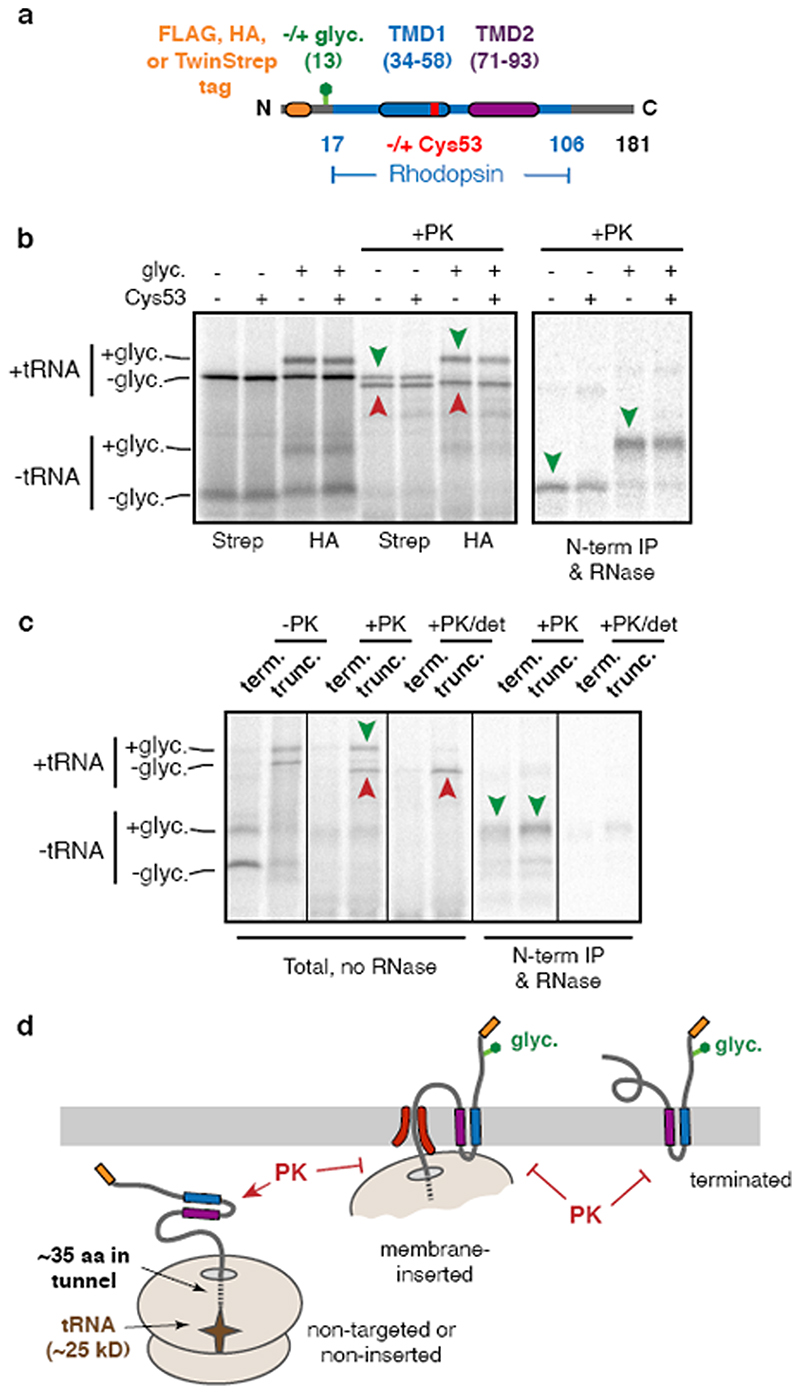
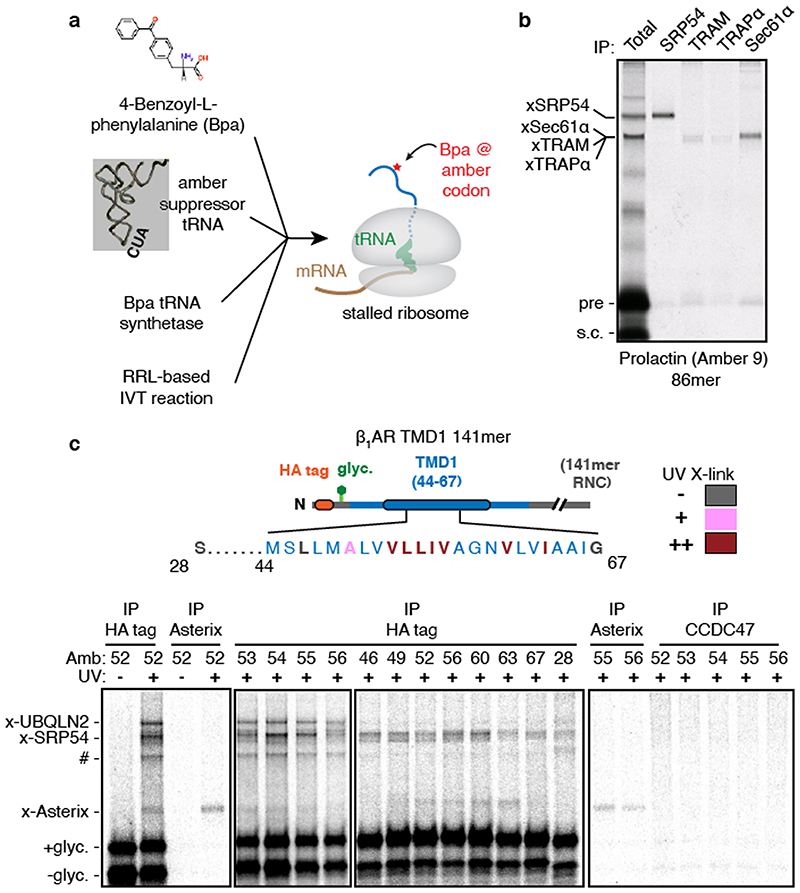
We sought to identify interacting partners of early stage intermediates during biogenesis of G-protein coupled receptors (GPCRs). Tagged constructs coding for the first two TMDs of the GPCR bovine rhodopsin (Rho TM1+2) were inserted in the appropriate orientation in ER-derived rough microsomes (RMs) isolated from HEK293 cells (Extended Data Fig. 1). Stalled translation complexes containing different lengths of nascent polypeptide were crosslinked to adjacent proteins via a single cysteine introduced into TMD1 (Fig. 1a, Extended Data Fig. 2a,b). Early intermediates (lanes 1-3) crosslinked to well-characterised ribosomal and ribosome-associated proteins including the SRP54 subunit of the targeting factor SRP13 and the Sec61α subunit of the translocation channel14. Although EMC (ER membrane protein complex) facilitates TMD1 insertion15, this interaction was not captured by cysteine crosslinking because it probably acts transiently16.
Fig. 1. A protein complex containing CCDC47 engages nascent membrane proteins.
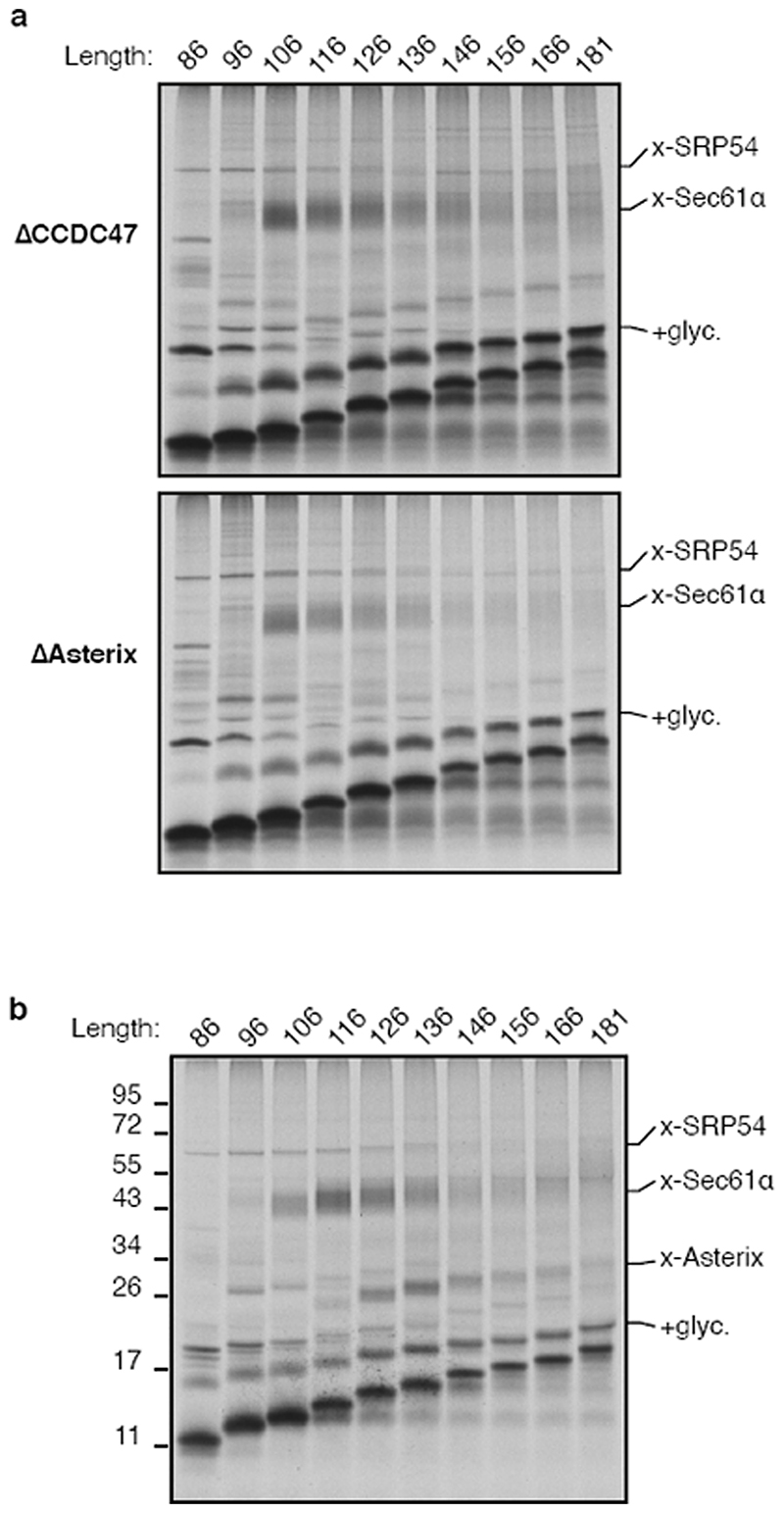
(a) Cysteine-based crosslinking of35S-labelled ribosome nascent chain complexes (RNCs) of the indicated length representing intermediates during targeting and insertion of mammalian rhodopsin. The construct is shown in the upper diagram and a schematic of the results in the lower diagram. RNCs were produced by in vitro translation containing ER-derived rough microsomes (RMs) from HEK293 cells. The upper gel shows the translation products and all of their crosslinks as visualised by autoradiography of immunoprecipitations (IPs) via the nascent chain’s N-terminal FLAG tag. The lower autoradiograph shows the IP products using antibodies against Sec61α. The non-glycosylated (-glyc.) and glycosylated (+glyc) translation products and the crosslinks to PAT10, Sec61α, and SRP54 are indicated. (b) Sucrose gradient separation of the35S-labelled membrane-targeted 146mer RNC after BMH crosslinking, native solubilisation, and release from the ribosome by RNase digestion. Endogenous haemoglobin (∼60 kD) from the translation extract was visualised by Coomassie staining of the same gel. (c) BMH-crosslinked 146mer RNCs containing a FLAG or Strep tag were released from the ribosome by RNase digestion, subjected to native FLAG IPs, and analysed by quantitative mass spectrometry. Proteins enriched 2-fold or more in the FLAG-tagged RNCs are indicated. (d) 146mer RNCs containing or lacking either a glycosylation site (glyc.) or cysteine at position 53 (Cys53) were crosslinked with BMH and analysed directly (input) or after native IP using antibodies against CCDC47 or signal peptide peptidase (SPP). Nascent chains were released from the ribosome with RNase A before IP. (e) RMs prepared from wild type (WT) or CCDC47 knock-out (ΔCCDC47) HEK293 cells were immunoblotted for CCDC47 and Sec63. (f) 146mer RNCs targeted to RMs from WT or ΔCCDC47 (A) cells were treated with BMH, released from the ribosome with RNase, and immunoprecipitated under denaturing (denat.) or native conditions with the antibodies indicated. For gel source data, see Supplementary Figure 1.
Intermediates where TMD2 was emerging from the ribosome (lanes 4-6) showed diminished TMD1-Sec61α crosslinks and the appearance of crosslinks to a ∼10 kD protein which we presume is identical to a previously observed crosslinking partner dubbed PAT10 for “protein associated with the ER translocon of 10 kD”10. At all subsequent lengths, the PAT10 crosslink persisted at approximately the same efficiency while the Sec61α crosslink remained low. Thus, PAT10 engages TMD1 as it departs the vicinity of the Sec61 complex17.
Substrate crosslinking to PAT10 did not depend on glycosylation and crosslinks were observed from other cysteine positions within TMD1 (Extended Data Fig. 2c-e), consistent with earlier observations10. Natively solubilised ribosome-released TMD1-PAT10 complex migrated at an apparent molecular weight of >100 kD by sucrose gradient sedimentation (Fig. 1b). PAT10 is therefore part of a larger assembly (the PAT complex) which is adjacent to a membrane-inserted TMD. Because efficient maleimide-mediated crosslinking requires an aqueous environment, the PAT complex seems to provide a membrane-embedded partially hydrophilic environment where TMD1 binds.
CCDC47 and Asterix form the PAT complex
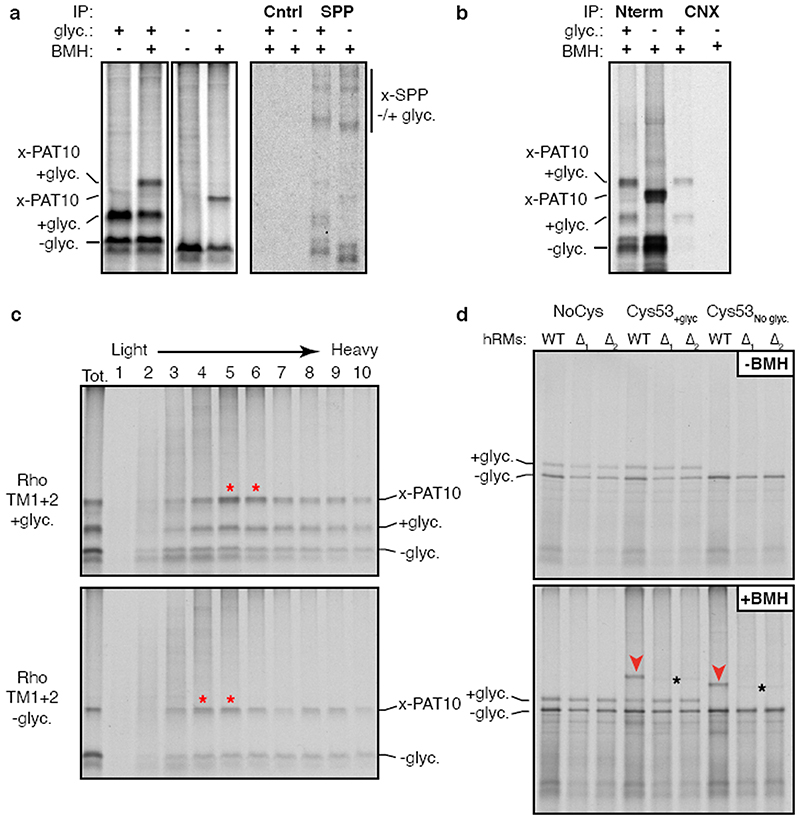
To identify components of the PAT complex, we solubilised the crosslinking reactions under non-denaturing conditions, released the nascent chain from the ribosome, affinity purified via the FLAG tag on the nascent chain, and identified the co-purifying proteins by mass spectrometry (Fig. 1c). Six proteins were enriched more than 2-fold relative to a control substrate containing a Strep tag: Sec61α and Sec61β, the lectins Calnexin and Galectin-7, signal peptide peptidase (SPP), and CCDC47.
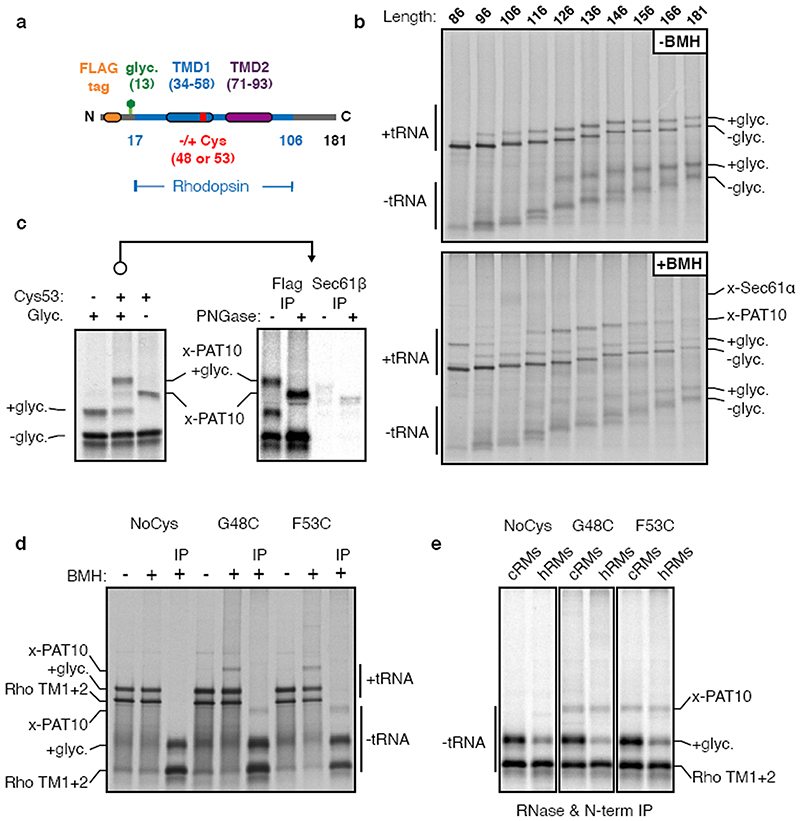
Sec61α, Sec61β, and SPP were apparently recovered due to their direct crosslinking to the nascent chain (Fig. 1a; Extended Data Fig. 3a). Of the remaining candidates, we focused on CCDC47 because unlike the others, the PAT10 crosslink was strongly enriched in native IPs using anti-CCDC47 antibodies (Fig. 1d, Extended Data Fig. 3b,c). Even without crosslinking, the inserted population of nascent chains was enriched by CCDC47 native IPs (but not denaturing IPs) from wild type microsomes, but not from CCDC47 knockout microsomes (Fig. 1e,f). TMD1 crosslinking to PAT10 was lost in microsomes lacking CCDC47 (Fig. 1f, Extended Data Fig. 3d). Thus, CCDC47 is a previously unknown interactor of nascent membrane proteins and a stable subunit of the PAT complex.
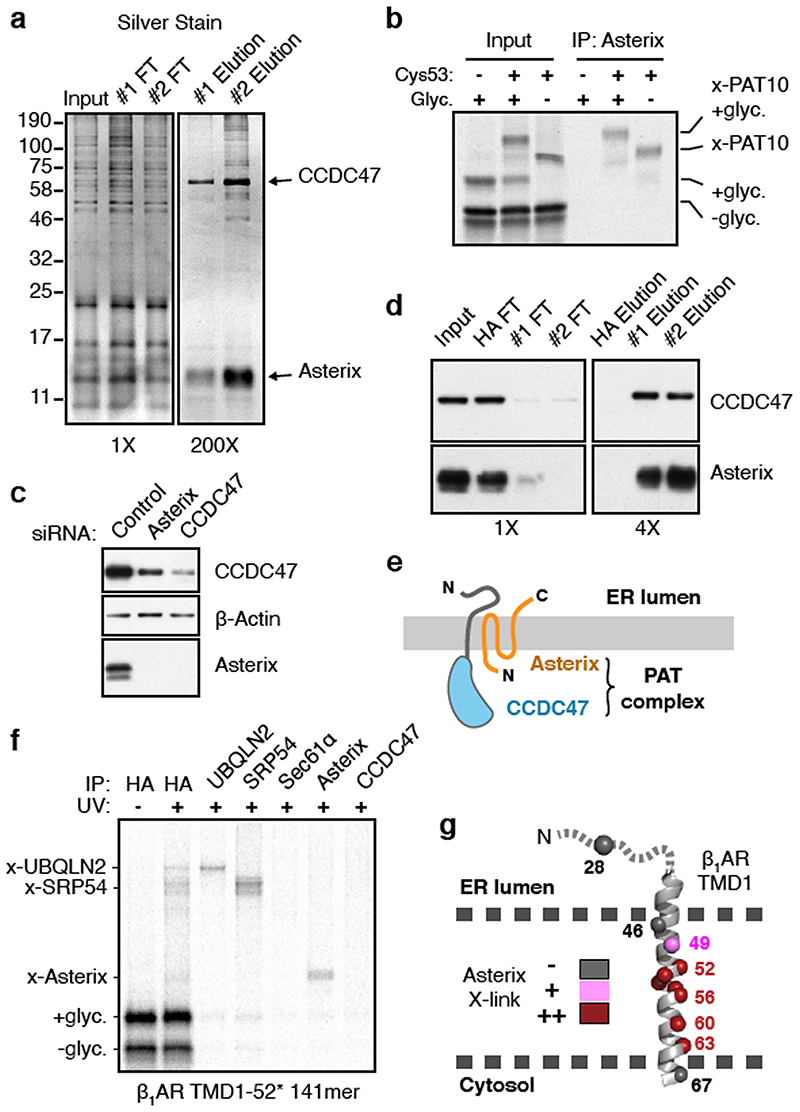
Affinity columns made with two different anti-CCDC47 antibodies each recovered CCDC47 and a prominent ∼10 kD protein (Fig. 2a). While mass spectrometry of tryptic digests failed to identify this protein (explaining why it was not identified in the initial mass spectrometry experiment above), digestion with other proteases produced peptides that matched Asterix (gene name WDR83OS). No other stoichiometric and specific interaction partners were observed in the purifications or identified by mass spectrometry. Antibodies against Asterix specifically recovered the PAT10 crosslink by denaturing IP (Fig. 2b). CCDC47 and Asterix form an obligate complex because knockdown or knockout of either protein results in substantial loss of the other (Fig. 2c), and immunodepletion of CCDC47 from ER extracts efficiently depletes Asterix (Fig. 2d). Thus, the PAT complex contains CCDC47 and Asterix, two poorly studied proteins that are widely expressed in all tissues and widely conserved across eukaryotes.
Fig. 2. Asterix is the substrate-binding subunit of the PAT complex.
(a) Affinity purification of CCDC47 from natively solubilised RMs using two unrelated CCDC47 antibodies (#1 and #2). The elution samples represent ∼200-fold more than the input and flow-through (FT) samples. (b) 35S-radiolabelled 146mer RNCs of Rho TM1+2 (see Fig. 1a) containing or lacking Cys53 or a glycosylation site were targeted to RMs, treated with BMH, digested with RNase, and analysed directly (input) or after denaturing IP with anti-Asterix antibodies. (c) Either Asterix or CCDC47 was depleted from HEK293 cells using siRNAs and compared to scrambled siRNA treatment (control). The indicated antigens were detected by immunoblotting. (d) Affinity purification via CCDC47 as in panel a, but with a negative control using anti-HA antibodies. Asterix and CCDC47 were detected by immunoblot. The elution samples represent 4-fold more than the input and FT samples. (e) Cartoon depicting topology of the PAT complex as deduced from predictions and direct analysis (see Extended Data Fig. 4). (f) Site-specific photo-crosslinking of a 141mer RNC containing the UV- activated photo-crosslinking amino acid benzoyl-phenylalanine (Bpa) at position 52 within TMD1. Total translation products recovered by IP via an HA tag on the nascent chain are shown adjacent to parallel IPs using the indicated antibodies. RNCs that failed to engage SRP crosslink to UBQLN2, a quality control factor that binds exposed TMDs. A subset of RNCs fail to release from SRP and crosslink to SRP54. Of the membrane-inserted RNCs, the main crosslink is to Asterix. At this length, the TMD has moved away from Sec61α, so crosslinks to this factor are minimal. (g) Summary of Asterix crosslinks observed (or not) from different positions in or near TMD1 of β1AR (see Extended Data Fig. 5). No CCDC47 crosslinks were seen from any of these positions. For gel source data, see Supplementary Figure 1.
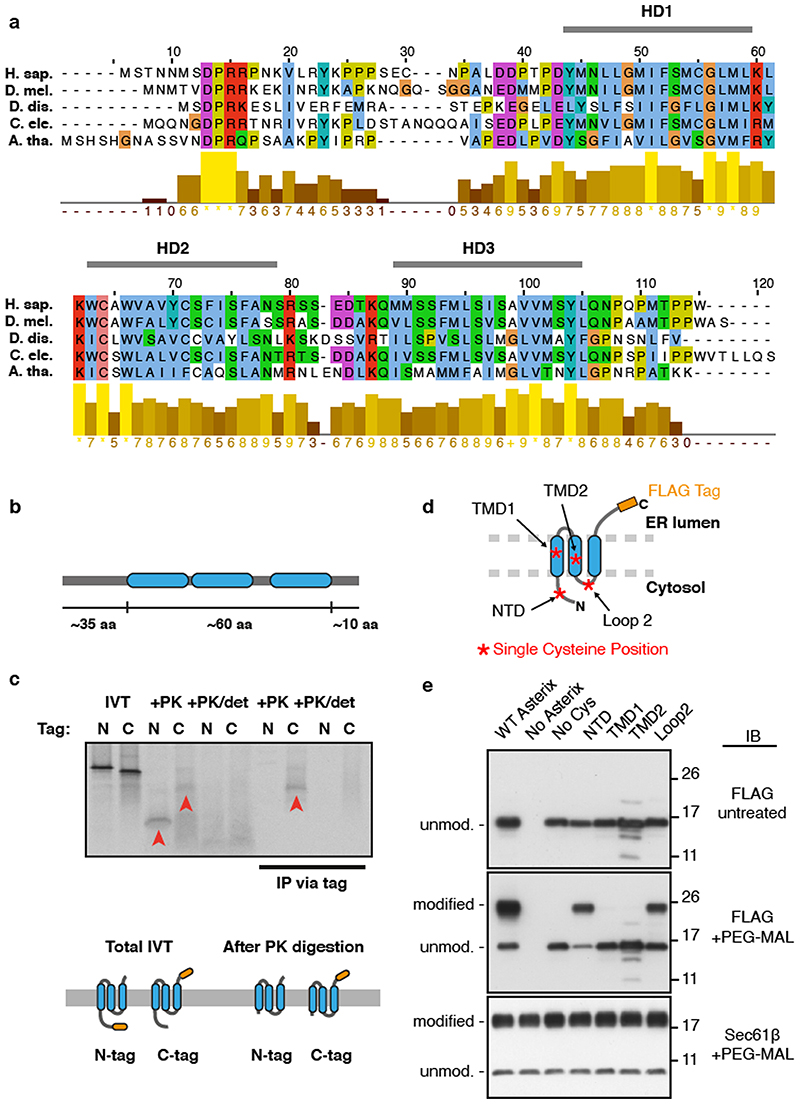
As depicted in Fig. 2e, CCDC47 is an ER-resident single-pass membrane protein with a well-conserved cytosolic domain1. Sequence and topology analysis of Asterix suggests that it is a three-TMD membrane protein whose N-terminus faces the cytosol and C-terminus faces the ER lumen (Extended Data Fig. 4). The three putative TMDs are only ∼15 amino acids each, populated by numerous hydrophilic amino acids, and contain multiple cysteines that explain its crosslinking to nascent TMDs via BMH.
Using a recently optimised site-specific photo-crosslinking approach18 (Extended Data Fig. 5a,b), we introduced the UV-activated crosslinking amino acid benzoyl-phenylalanine (Bpa) at eleven positions within or near TMD1 of β1-adrenergic receptor (β1AR), a class A GPCR like rhodopsin. At a nascent chain length 75 amino acids downstream of TMD1 (when it has been inserted and can diffuse away from Sec61α), several UV-dependent crosslinking products were observed (Fig. 2f). The most prominent of these proved to be SRP54 (bound to non-translocated nascent chains) or UBQLN2, a cytosolic quality control factor for TMDs that fail targeting19. The most prominent remaining crosslink was Asterix as verified by immunoprecipitation. Crosslinking to Asterix was observed from most, but not all positions within the TMD, and was not seen from control positions at the boundaries or outside the TMD (Fig. 2g and Extended Data Fig. 5c). Crosslinks to CCDC47 were never observed. Thus, consistent with the chemical crosslinking experiments, Asterix is the substrate- interacting subunit of the PAT complex, while CCDC47 is needed for Asterix stability.
PAT complex impacts membrane proteins
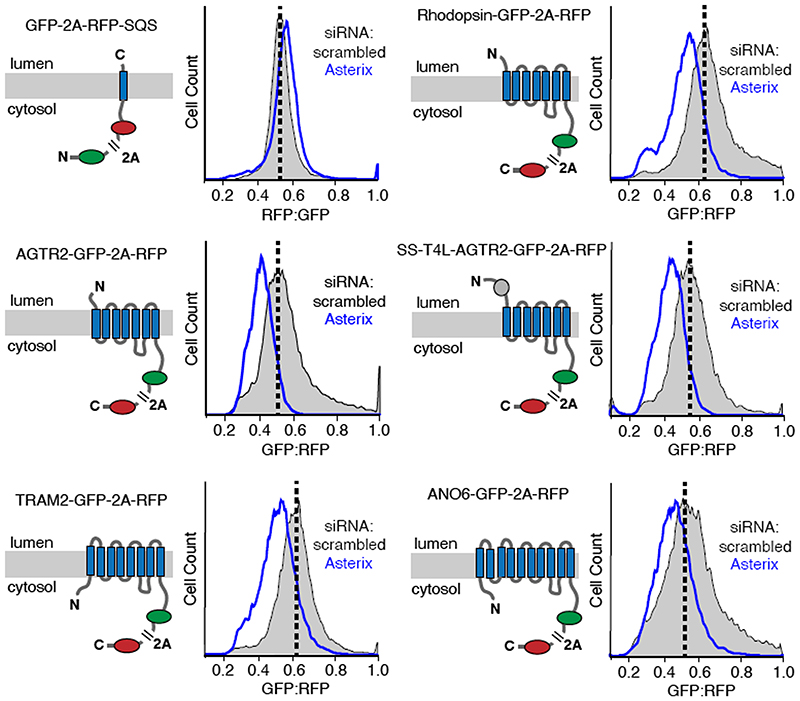
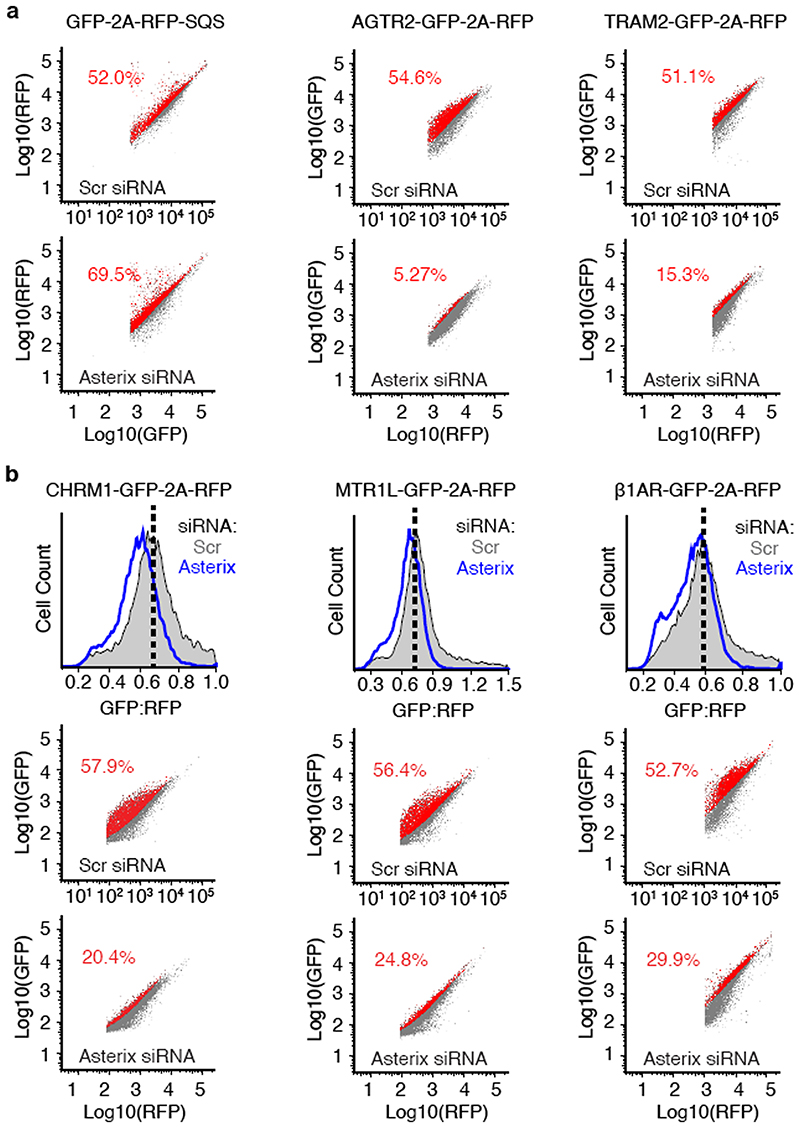
Using a dual-colour ratiometric assay for protein stability15, we found that acute PAT complex depletion using siRNAs against Asterix (which co-depletes CCDC47) had minimal effects on the tail-anchored membrane protein squalene synthase (SQS), but showed reduced biogenesis of five GPCRs, the ER resident protein TRAM2, and the cation channel ANO6 (Fig. 3 and Extended Data Fig. 6). Similar phenotypes were seen with CCDC47 knockdown and with CRISPR-mediated disruption of CCDC47 or Asterix (Extended Data Fig. 7). Thus, the PAT complex is needed for optimal biogenesis of various multi-spanning membrane proteins. This result explains why both genes were hits in a recent screen for factors that impair surface expression of a mutant TRP6 channel20.
Fig. 3. The PAT complex facilitates biogenesis of multi-spanning membrane proteins.
Stable cell lines containing the indicated inducible reporters were treated with scrambled or Asterix-targeting siRNAs, reporter expression was induced for ∼6 hours, and the cells were analysed by flow cytometry. A cartoon depicting the topology, number of TMDs, and fluorescent proteins for each of the membrane protein reporters is show to the left of its respective flow cytometry data. The viral P2A peptide sequence results in two proteins from a single translation reaction as indicated. The plots show histograms of fluorescent protein ratios in control cells (grey) and Asterix-knockdown cells (blue). The dashed black line indicates the mode for the control population.
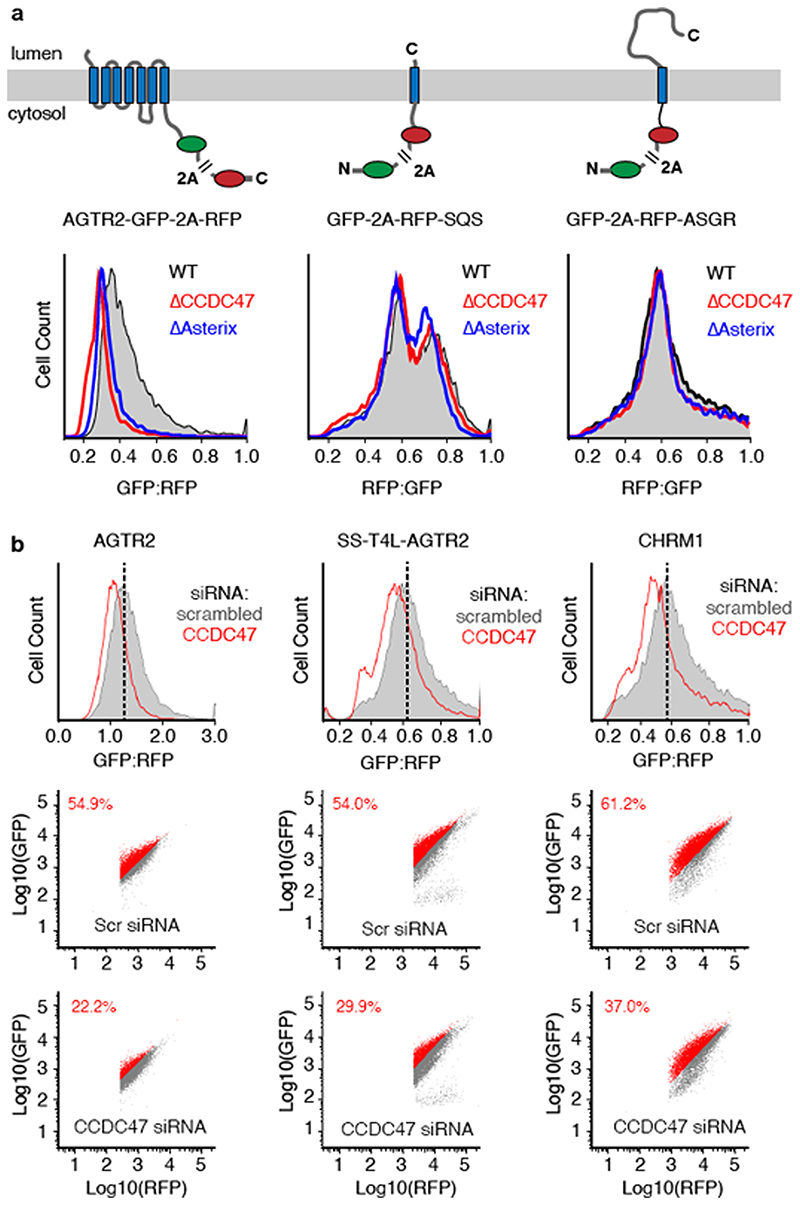
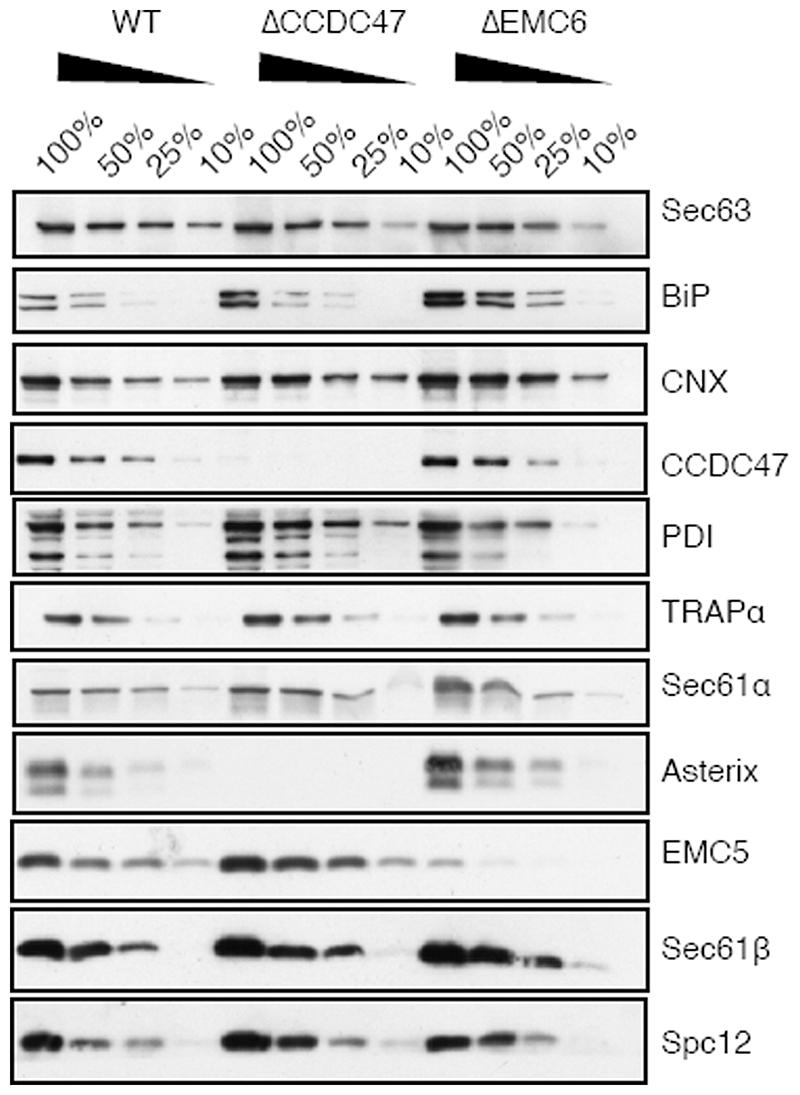
Neither SQS nor the type II single-pass membrane protein asialyloglycoprotein receptor (ASGR) were affected in CCDC47 or Asterix knockout cells using the same assay (Extended Data Fig. 7) under the same conditions. We therefore conclude that the TMD insertase EMC (used by SQS21 and the first TMD of many GPCRs15) and the Sec61 complex (used by ASGR22) are functioning correctly in cells lacking the PAT complex. Neither these nor other biogenesis factors are diminished in cells lacking the PAT complex (Extended Data Fig. 8).
Appending a signal sequence and T4 lysozyme to the N-terminus of AGTR2, which bypasses its requirement for EMC15, does not affect its dependence on the PAT complex (Fig. 3 and Extended Data Fig. 7). Similarly, the EMC-independent substrate TRAM2 is dependent on the PAT complex, suggesting that EMC and the PAT complex facilitate membrane protein biogenesis in different ways. As expected for central ER factors, loss of either EMC or the PAT complex results in an activated unfolded protein response as judged by elevated levels of the ER chaperones BiP and PDI (Extended Data Fig. 8).
Hydrophilic TMDs engage the PAT complex
Analysis of substrate insertion and crosslinking in microsomes lacking the PAT complex showed that the timing of TMD1 insertion and interaction with and release from Sec61α was not appreciably different from wild type microsomes (Extended Data Fig. 9a). This contrasts with the failed TMD1 insertion phenotype seen when microsomes lack the EMC insertase15. Thus, the PAT complex does not participate in targeting and is neither an insertase nor a facilitator of TMD release from Sec61α, explaining why single-pass membrane proteins like SQS and ASGR are unaffected by PAT complex depletion. Instead, the PAT complex acts after a TMD accesses the lipid bilayer.
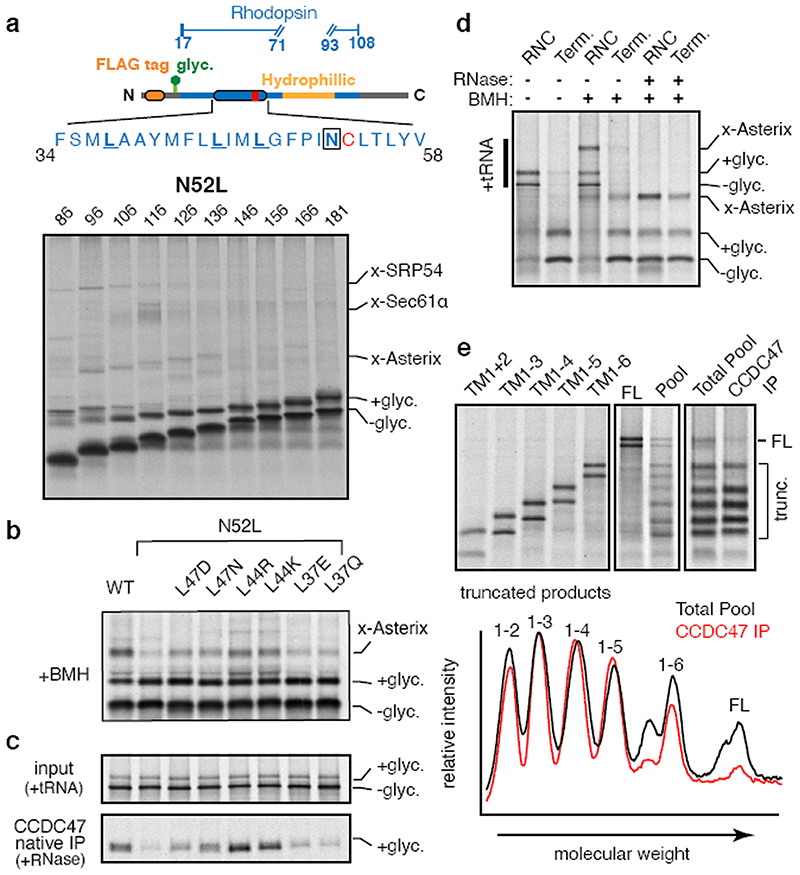
Replacement of the sole polar amino acid (asparagine) in TMD1 with leucine markedly reduced the Asterix-TMD1 interaction at all nascent chain lengths that were examined (Fig. 4a and Extended Data Fig. 9b). Reintroduction of polar or charged residues at other positions in the TMD partially restored crosslinking to Asterix (Fig. 4b). Analysis of substrate interaction with the PAT complex via CCDC47 native IPs led to the same conclusion (Fig. 4c). These data indicate Asterix engages TMDs that expose hydrophilic residues within the lipid bilayer in a mostly position-independent manner. This explains observations that PAT10 can crosslink with TMDs of either orientation and of different unrelated sequences10.
Fig. 4. The PAT complex engages TMDs via exposed polar residues.
(a) The Rho TM1 construct (diagram) containing the N52L mutation was analysed for crosslinking as in Fig. 1a. Compared to the identical construct without the N52L mutation (Extended Data Fig. 9b), crosslinks to Asterix are markedly diminished and strong crosslinks to Sec61α are only seen for the 116mer. (b) Crosslinking reactions of 146mer RNCs of Rho TM1 containing the indicated mutations. (c) Translation reactions as in panel B (but without crosslinking) were either analysed directly (input) or subjected to native IP using anti-CCDC47 antibodies. Nascent chains were released from the ribosome with RNase A before IP. The glycosylated substrate recovered with CCDC47 was visualised by autoradiography. Note that the efficiencies of crosslinking to Asterix in panel b corelate to the efficiencies of recovery with CCDC47. (d) Terminated (Term.) or truncated (RNC) Rho TM1+2 was inserted into RMs and treated with BMH where indicated. Before SDS-PAGE, some of the samples were digested with RNase A as indicated to remove the tRNA. (e) Full length β1AR (FL) or constructs truncated after each TMD were translated in the presence of RMs and analysed either individually (left panel) or pooled before analysis. All constructs contained a stop codon and are terminated. The membrane-targeted population from the pooled reaction was isolated by sedimentation and divided in two aliquots. One aliquot was set aside (Total Pool) and the other was used for a native IP with anti-CCDC47 antibody. Both samples were then subjected to denaturing pulldown via the C-terminal His tag to ensure that only completed translation products were visualised. The graph below the gel represents scanning densitometry of the last two lanes. Note that substantially less full length β1AR is recovered with CCDC47 relative to each of the truncation products. For gel source data, see Supplementary Figure 1.
PAT complex releases folded substrates
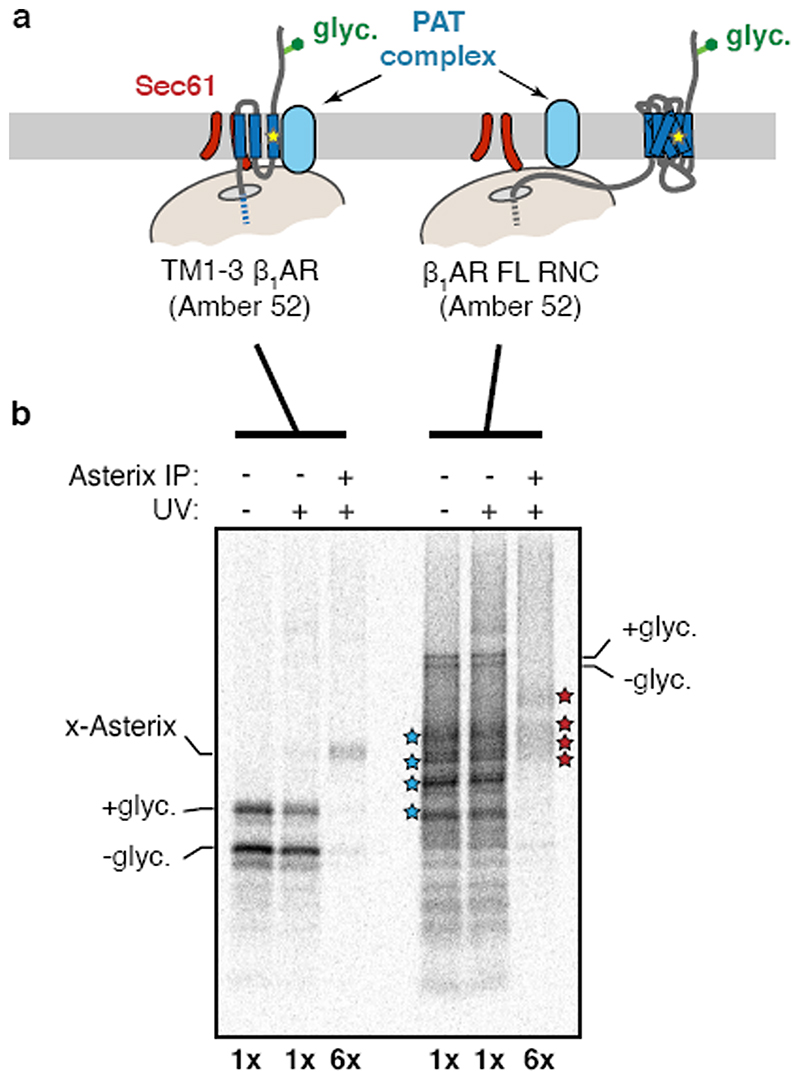
Although the PAT complex initially engages TMDs co-translationally as a peptidyl-tRNA, the complex remains associated even after termination releases the nascent chain from the ribosome-Sec61 complex (Fig. 4d). This observation, together with the finding that exposed hydrophilic residues within a TMD influence PAT complex interaction, suggested that PAT complex dissociation might be triggered when TMDs pack correctly to shield exposed hydrophilicity. To test this, we determined whether PAT complex interaction is selective to immature but not a folded membrane protein. Exploiting the fact that an engineered β1AR folds efficiently after in vitro translation15, we compared its interaction with the PAT complex relative to truncation products intended to mimic biogenesis intermediates. A pooled mixture of intermediates truncated after each TMD produced by in vitro translation was subjected to native IP using anti-CCDC47 antibodies. Full length β1AR was recovered substantially less efficiency than each of the intermediates, suggesting that the PAT complex dissociates upon correct folding of β 1AR (Fig. 4e). A similar conclusion was reached using a site-specific photo-crosslinker installed within TMD1 of β1AR (Extended Data Fig. 10).
Discussion
Our findings indicate that the PAT complex is a widely conserved ER resident complex that directly interacts with nascent TMDs, releases its substrates upon correct folding, and is needed for optimal membrane protein biogenesis. These features define the PAT complex as an intramembrane chaperone. Its preference for TMDs with exposed hydrophilic amino acids within the lipid bilayer is analogous to soluble chaperones that prefer hydrophobic patches exposed to the aqueous environment23,24. The more favourable TMD-TMD interactions that accompany correct folding likely displaces the PAT complex, whose interaction must necessarily be more generic and hence weaker. Just as the binding cleft of chaperones like Hsp70 are hydrophobic25, the substrate-interaction domain of Asterix may be hydrophilic, consistent with a large number of conserved hydroxylated amino acids in its TMDs.
Analogous intramembrane interactions with hydrophilic TMDs have been proposed to facilitate recognition of misfolded membrane proteins during quality control26–28. By binding to and shielding similar motifs in a nascent protein, the highly abundant PAT complex29 probably prevents promiscuous degradation. This would explain why its depletion leads to post-translational reduction of various multi-spanning membrane proteins. Loss of CCDC47 in mice is embryonic lethal with a wide range of developmental problems30 and mutations in humans causes numerous developmental phenotypes with early death31. Cells lacking either subunit of the PAT complex show reduced fitness across multiple mammalian cell lines32 but are viable, show activated ER stress responses, and have several otherwise unrelated membrane-associated phenotypes30. These observations are consistent with loss of an intramembrane chaperone radiating to numerous cellular and organismal functions.
Despite such pleiotropic phenotypes, individual chaperone disruptions often have only modest consequences at a cellular level due to their considerable redundancy32. Thus, there may be many other intramembrane chaperones that can partially compensate for the PAT complex in its absence. While several substrate-specific chaperones have been proposed for particular membrane proteins33–35, our findings suggest that the PAT complex is an abundant generalist for semi-hydrophilic TMDs that are found in nearly all of the over 2500 multi- spanning human membrane proteins. Furthermore, the PAT complex, or factors like it, might play a similar role in chaperoning integral membrane subunits of multi-protein complexes.
Methods
Constructs
Sequences encoding 1xFLAG-Rho TM1+2 and 1xFLAG-Rho TM1+2 (F53C) were ordered as gene blocks (IDT) and inserted into a parent vector containing an SP6 promoter and the flexible N-terminus of Sec61β with a 6His tag appended to the C-terminus. A glycosylation acceptor site (NGT) was introduced at amino acids 13-16. Variants of these parent constructs with a TwinStrep tag, HA tag, or lacking a glycosylation site were generated by standard subcloning methods. Constructs encoding Rho TM1, where TM2 is replaced by a flexible hydrophilic linker (SGSGSGSGSGGSSGGMGGSGS), were ordered as gene blocks containing a 5’ SP6 promoter and transcribed directly for in vitro translation. Similar methods were used for all Rho TM1+linker polar residue point mutants. The in vitro expression plasmid encoding HA- β1AR containing a C-terminal His tag has been described previously15. Truncated versions and variants containing amber codons at specific sites were created by PCR-based cloning methods and site-directed mutagenesis, respectively, and verified by sequencing. Constructs encoding the mutant E.coli tyrosyl-tRNA synthetase in the pET21 vector and B. stearothermophilus suppressor tRNATyr sequence in the pRSET-A vector have been described18. Templates for in vitro translation of tagged Asterix constructs were ordered as gene blocks containing a 5’ SP6 promoter and used directly for transcription and translation. Mammalian expression constructs for C-terminally FLAG-tagged human Asterix and the various cysteine variants were produced as gene blocks, sub-cloned into pcDNA3.1-based vectors, and verified by sequencing. For the creation of stable cells lines expressing various membrane protein reporters, the coding sequences for CHRM1 (NP_000729.2), bovine rhodopsin (NP_001014890.1), Anoctamin-6 (NP_001020527.2), and MTR1L (NP_004215.2) were PCR amplified and placed into a parent pcDNA5/FRT/TO vector backbone with a C-terminal GFP-P2A-RFP using Gibson Assembly (NEB). Analogous constructs encoding ASGR, SQS, TRAM2, β1AR, AGTR2, and SS-T4L-AGTR2 have been described previously15. CRISPR-Cas9 mediated gene disruptions of CCDC47 and Asterix were performed using the pSpCas9(BB)-2A-Puro (PX459) plasmid (Addgene) containing the guide RNAs 5’-GTATGGACTGCCGGACTCTT-3’ (CCDC47) and 5’- AAGGCCGGGTTACATTCGCT-3’ (Asterix).
Antibodies
CCDC47 antibody #1 (A305-100A) and CCDC47 antibody #2 (A305-101A) were obtained from Bethyl-laboratories. FLAG immunoprecipitations were performed using FLAG-M2 affinity gel (Sigma). Anti-HA antibody was generated in house19. Signal Peptide Peptidase (SPP) antibody was purchased from Bethyl Laboratories (A304-404A). Anti-Asterix (WDR83OS) antibody was purchased from Sigma/Human Protein Atlas (HPA065685). Anti- UBQLN2 antibody was clone 5F5 obtained from Sigma (WH0029978M3). Anti-SRP54 was from BD Biosciences (610940). Custom antibodies against Sec61α, Sec61β, TRAPα, and TRAM have been described and characterised previously36. All antibodies for western blotting in Extended Data Fig. 8 have been described previously15.
Cell Lines
Flp-In TRex 293 cells (Invitrogen) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal calf serum (FCS). The authentication of this cell line was ensured by the antibiotic resistance markers within its genome and by its unique FRT site downstream of a doxycycline-inducible promoter. Cell lines were tested monthly for mycoplasma contamination and verified to be negative. For cell lines containing a stably expressed doxycycline-induced reporter, tetracycline-free FCS was used in conjunction with 15μg/ml blasticidin and 100μg/ml hygromycin. Stable cells lines were generated using the FRT Flp-In system according to manufacturer’s protocol (Invitrogen). Stably expressed cell lines included GFP-2A-RFP-SQS, β1AR-GFP-2A-RFP, AGTR2-GFP-2A-RFP, SS-T4L- AGTR2-GFP-2A-RFP, TRAM2-GFP-2A-RFP, Ano6-GFP-2A-RFP, CHRM1-GFP-2A-RFP, MTR1L-GFP-2A-RFP, and Rhodosin-GFP-2A-RFP. CCDC47 and Asterix knockout cells were generated by transient transfection with the px459 plasmids containing the appropriate guide RNAs. After 24 h, transfected cells were selected for 48hrs with 1μg/ml puromycin then trypsinised and diluted into 96-well plates at a concentration of 0.5cells/well to select for single cell colonies. After ∼2weeks, single colonies were expanded and clones displaying undetectable CCDC47 or Asterix were chosen for further analysis.
Flow-cytometry analysis
For experiments using knockout cell lines, ΔCCDC47 and ΔAsterix cells were transiently transfected with 1 μg/ml of the appropriate pcDNA5 expression constructs ∼24 h before induction with 100 ng/ml doxycycline. All transfections were performed using Mirus-Transit 293 according to manufacturer’s instructions. For experiments using stably expressed reporter cell lines, siRNA depletion was performed over a ∼96 h period using the Lipofectamine RNAiMAX reagent according to manufactures instructions (Thermo). Briefly, a first round of siRNA treatment was performed in the presence of DMEM and 10% tetracycline-free FCS. Cells were incubated for 48 h before a second round of siRNA treatment was performed under the exact same conditions. Following a second ∼48 h incubation, expression of fluorescent reporter constructs was induced with 100 ng/ml doxycycline for 6 h before analysis by flow cytometry. Acute expression of reporters was essential to accurately monitor degradation of reporter constructs and avoid saturation of degradation pathways. In all experiments the cells were collected by trypsinisation, washed once in ice-cold PBS, then resuspended in the equivalent culture volume of PBS and 1μ/ml DAPI stain (Thermo). Cells were passed through a 70μm filter before flow cytometry analysis using a Beckton Dickinson LSRII instrument. 20,000 GFP positive (or RFP positive for SQS and ASGR) were collected. Further gating for live cells (negative for DAPI stain) and relatively high soluble fluorescent protein was used to focus on the population of cells with productive translation of reporter constructs.
Preparation of ER-derived rough microsomes
Rough microsomes (RM) derived from adherent wild type, ΔCCDC47, or AAsterix HEK293 cells were prepared as described previously15. Wild type RM were also obtained from HEK293 cells grown in suspension (Expi293F cells) and prepared similarly to RM from adherent cells with a few minor modifications to adjust for the larger scale. In brief, ∼2 L of cells were grown to a concentration of 5x106 cells/mL then collected by centrifugation. Cell pellets were washed twice with ice-cold PBS and pooled as necessary. A ∼30 mL pellet was resuspended in 60 mL of sucrose buffer (50 mM HEPES, pH 7.4, 50 mM KOAc, 6 mM Mg(OAc)2, 1 mM EDTA, 250 mM sucrose, 1 mM DTT) and lysis was carried out in a glass dounce homogeniser. Lysate was cleared twice by centrifugation at 3,500 x g for 30 min at 4°C. Supernatant was recovered and underlaid with one-third the volume of sucrose cushion (1.3 M sucrose, 50 mM HEPES, pH 7.4, 50 mM KOAc, 6 mM Mg(OAc)2, 1mM EDTA, 1 mM DTT) and centrifuged for 1 h at 371,000 x g (60,000 rpm) and 4°C for 1 h in the Type 70Ti rotor (Beckman). The supernatant was removed by aspiration, and the pellets were resuspended and pooled by manual homogenisation in a dounce using 6 mL resuspension buffer (250 mM sucrose, 50 mM HEPES, pH 7.4, 1 mM DTT). The final preparation was adjusted to an absorbance of 75 when measured at 280 nm in 1% SDS. Canine pancreas- derived rough microsomes (cRM) were prepared as described previously37, and were used in very few experiments where explicitly stated in the figure legends. All microsome preparations were flash frozen in liquid nitrogen and stored at −80°C.
In vitro transcription and translation
In vitro transcription was performed with SP6 polymerase using PCR products as the template38,39. The transcription reactions were conducted with 5-20 ng/ml PCR product at 37°C. In vitro translation reactions were in rabbit reticulocyte lysate (RRL) as described previously in detail38,39. All translation reactions were carried out at 32°C for 15-30 min. For translation reactions in the presence of RM, 0.25-1.0μl (at an absorbance of 75) of RMs were added to a 10μl translation reaction. All batches of microsomes were individually titrated to achieve optimal translation and insertion as monitored by glycosylation efficiency. For pooled translation reactions in Figure 4E, initial experiments provided the relative translation efficiency of each intermediate. The levels of each transcript were titrated accordingly to achieve roughly equal translation of each individual intermediate and the full-length product.
Generation of ribosome-nascent chain complexes (RNCs)
Truncated mRNA templates were generated via PCR using 5’ primer that annealed upstream of the SP6 promoter and a 3’primer that annealed at the site of truncation to generate the desired nascent chain length. The 3’ primer also encoded the residues “MLKV” at the very C-terminus to improve radiolabelling (via the added methionine) and minimise hydrolysis of the tRNA-peptidyl bond during electrophoresis40. For all experiments, RM were included during translation which was carried out at 32°C for 15-20 min, then returned to ice for subsequent crosslinking and protease-protection analysis.
Site-specific photo-crosslinking
Incorporation of benzoyl-phenylalanine (Bpa) at specific positions during in vitro translation was accomplished by amber suppression as described18. In short, an amber codon containing template was translated in RRL as above but with 5 μM B. Stearothermophilus tRNATyr, 0.25 μM Bpa tyrosyl-tRNA synthetase, and 0.1 mM Bpa. UV irradiation was for 12 min with a hand-held UV lamp (ThermoFisher Scientific, cat. #95034) at 254 nm positioned ∼2.5 cm above the sample sitting on ice. As described before18, E.coli Bpa tyrosyl-tRNA synthetase was purified via the C-terminal His tag on a 5 mL HiTrap Ni-NTA column (GE), desalted by a gel filtration column on FPLC and concentrated by Amicon Ultra centrifugal filter (Millipore, Z717185-8EA). B. stearothermophilus tRNATyr, was synthesised by in vitro transcription as before18. The pRSET-based construct encoding the tRNA was digested with BstN1, yielding a DNA fragment containing the exact tRNATyr sequence under a T7 promoter. 5 mL transcription reaction was carried out with 1.2 mg DNA template, 1 mM spermidine, 5 mM DTT, 0.1% Triton, 5 mM NTPs, 25 μM MgCl2, 20 μg/mL E. coli pyrophosphatase, 20 p,g/mL T7 polymerase and 125 U Recombinant RNasin (Promega) for 4 hours at 37 °C. The reaction product was digested with Turbo DNase (Ambion) and extracted by acid phenol chloroform extraction to yield purified tRNA.
Cysteine based crosslinking
Unless explicitly stated, the crosslinking reactions were performed on isolated RMs. Here, the translation reactions were placed over a 20% sucrose cushion in physiological salt buffer [PSB - 100mM KOAc, 50mM HEPES pH 7.4, 2.5mM Mg(OAc)2], centrifuged in the TLA- 55 rotor for 20 min at 4°C, and the pelleted RMs resuspend in one-half the original translation reaction volume of PSB. Bismaleimidohexane (BMH) was added to a final concentration of 250 μM and the reaction was incubated on ice for 1hr to allow the crosslinking reaction to occur. BMH was quenched with 5 mM 2-mercaptoethanol. Aliquots of the reaction were removed at different stage of the process for analysis, as indicated in the individual figure legends. The samples were either used directly for downstream applications (primarily immunoprecipitations as described below) or flash frozen in liquid nitrogen and stored at −80°C for later analysis.
Large scale affinity purification of crosslinked substrate complexes
1 mL translation reactions in the presence of 0.8 mM cold methionine were carried out to produce 1xFLAG tagged or TwinStrep tagged (as a control) Rho TM1+2 146mer RNCs. Membranes were isolated and cysteine-based crosslinking was carried out as described above. Isolated membranes resuspended in PSB were solubilised on ice by addition of an equal volume of 2x solubilisation buffer (300 mM KOAc, 2% deoxyBigChap, 20 mM EDTA). RNase A was added to a final concentration of 10 ng/ml and RNA digestion was carried out on ice for 20 min before the solubilised extracts were cleared by centrifugation at 100,000 rpm in the TLA120.2 (Beckman). Cleared extracts were directly immunoprecipitated with FLAG M2 affinity resin (Sigma) in batch at 4°C with end-over-end rotation for 2.5 h. Unbound fraction was removed, and beads were washed 4 times in 1x NSB [200mM KOAc, 1%DBC, 10mM EDTA, 25mM HEPES, 1mM Mg(OAc)2], then twice in 1x NSB without detergent. Beads were resuspended in 1.5x the bead volume of 200 mM KOAc and 25mM HEPES pH 7.4 for direct trypsinisation and analysis by quantitative mass spectrometry.
Quantitative mass spectrometry using TMT labelling
Proteins samples on beads were reduced with 5 mM DTT at 56°C and alkylated with 10 mM iodoacetamide (IAA) in the dark at 22°C. The alkylation reaction was quenched by the addition of DTT and the samples were digested overnight with trypsin (Promega, 0.1 μg) at 37°C. After digestion, each supernatant was transferred to a fresh Eppendorf tube, the bead samples were extracted once with 50% acetonitrile/ 0.1% TFA and combined with the corresponding supernatant. The peptide mixtures were then partially dried in Speed Vac and desalted using home-made C18 (3M Empore) stage tip filled with 2 μl of poros R3 (Applied Biosystems) resin. Bound peptides were eluted sequentially with 30%, 50% and 80% acetonitrile in 0.1%TFA and lyophilised. Dried peptide mixtures from each condition were re-suspended in 20 ul of 7% MeCN and 1 M triethyl ammonium bicarbonate was added to a final concentration of 200 mM. 0.8 mg of TMT10plex reagents (Thermo Fisher Scientific) was re-constituted in 41 μl anhydrous MeCN. 10 μl of TMT (130C or 131) reagent was added to each peptide mixture and incubated for 1 hr at 20°C. The labelling reactions were terminated by incubation with 2.5 μl of 5% hydroxylamine for 15min. The labelled samples were pooled into one Eppendorf tube and the speed Vac was used to evaporate acetonitrile. Peptides were separated on an Ultimate 3000 RSLC nano System (Thermo Scientific), using an acetonitrile gradient, consisting of buffer A (2% MeCN, 0.1% formic acid) and buffer B (80% MeCN, 0.1% formic acid). Eluted peptides were introduced directly via a nanospray ion source into a Q Exactive Plus hybrid quardrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in standard data dependent mode, performed survey full-scan (MS, m/z 380-1600) with a resolution of 70000, followed by MS2 acquisitions of the 15 most intense ions with a resolution of 35000 and NCE of 33%. MS target values of 3e6 and MS2 target values of 1e5 were used. The isolation window was set as 0.7 m/z and dynamic exclusion was enabled for 40s. The acquired MS/MS raw files were processed using Proteome Discoverer (version 2.1, Thermo Scientific). MS/MS spectra were searched against a Human Reviewed, UniProt Fasta database (download in 2016), using Mascot (version 2.4, Matrix Science) search engine. Carbamidomethylation of cysteines, TMT6plex (N-term) and TMT6plex (K) were set as fixed modifications, while methionine oxidation and N-terminal acetylation (protein) were selected as variable modifications. For reporter ion quantification, the co-isolation threshold is 30 and average reporter S/N threshold is 10. The output file from Proteome Discoverer, the proteins table was filtered for proteins FDR of 1% and exported as excel files used to produce the plot in Fig. 1c.
Preparation of anti-CCDC47 affinity columns
A 50μl bead volume of protein A agarose was diluted in 750μl PBS, placed in 1 mL Pierce Spin columns, then pre-equilibrated with two 800 μl PBS washes. 100 μg of CCDC47 antibody #1 (A305-100A), CCDC47 antibody #2 (A305-101A), or an anti-HA antibody were diluted in 1 mL of PBS then placed over the prepared protein A columns and allowed to pass through the resin by gravity flow. Flow through was collected and passed over the column a second time. Columns were equilibrated with 800 μl of 0.1 M Na-Borate pH 9.0, then antibodies were conjugated to resin by passing through a 1 mL solution by gravity flow of 0.1 M Na-Borate pH 9.0 containing 5 mg/mL dimethyl pimelimidate (DMP). DMP was quenched with 1 mL of 0.2M ethanolamine pH 8.0 and columns were re-equilibrated in 1x PBS and 0.02% NaN3 for storage at 4°C until use.
Native affinity purification of CCDC47
The purification was performed at the bench on ice. CCDC47 and HA affinity columns were pre-washed with 300 μl of 0.1 M glycine-HCl pH 2.3, neutralised with 800 μl of 1x PBS, then equilibrated in 800 μl of 1x native solubilisation buffer (NSB): 200mM KOAc, 10mM EDTA, 1% DBC, 25mM HEPES pH 7.4. 3 mL of RMs, prepared as described above (at an A280 value of 75), were solubilised by addition of an equal volume of 2x NSB. RNase A was added to a final concentration of 10 ng/mL and the samples incubated on ice for 30 min during column preparation. The solubilised extract was cleared by centrifugation at 100,000 rpm in the TLA100.3 (Beckman) for 1 h at 4°C. Supernatant was removed, divided into 3 equal parts, and passed twice over each column (anti-CCDC47 Ab #1, #2, or an anti-HA control). Each column was washed once with 1 mL of 1x NSB. Recovered proteins were eluted with 300 μl of 0.1 M glycine pH 2.3 and elutions were immediately neutralised with 1 M Tris-Cl pH 8.8. For initial small scale experiments using only CCDC47 antibodies (Fig 2A), the protocol was the exact same except 100μl of starting RMs was used and final elutions were TCA precipitated using standard procedures before loading on a gel.
Identification of Asterix by mass spectrometry
Protein samples eluted from the CCDC47 affinity resin were reduced with DTT and alkylated with iodoacetamide. Because initial efforts to identify the co-purifying 10 kD protein via analysis of tryptic digests failed, we re-did the analysis using other proteases. The samples were digested overnight either with trypsin, Glu-C, chymotrypsin or elastase (Promega). Digest mixtures were acidified with formic acid (FA) and a portion of each of these samples was analysed by nano-scale capillary LC-MS/MS (Ultimate U3000 HPLC, Thermo Scientific Dionex) at a flow of 300 nL/min. A C18 Acclaim PepMap100 5 μm, 100 μm x 20 mm nanoViper (Thermo Scientific Dionex), trapped the peptides prior to separation on an EASY- Spray columns with an acetonitrile gradient. The Eluted peptides were introduced directly via a EASY-Spray ion source into a Q Exactive mass spectrometer (Thermo Scientific). Data dependent analysis was performed using a resolution of 35,000 for the full MS spectrum, followed by ten MS/MS spectra in the orbitrap. MS spectra were collected over a m/z range of 350-1600 m/z. LC-MS/MS data were searched against the UniProt KB database using Mascot (Matrix Science), with a precursor tolerance of 10 ppm and a fragment ion mass tolerance of 0.1 Da. Two missed enzyme cleavages (or no enzyme for elastase) and variable modifications for oxidised methionine, carbamidomethyl cysteine and pyroglutamic acid, were included. MS/MS data were validated using Scaffold (Proteome Software Inc).
Immunoprecipitations
Unless otherwise indicated, all CCDC47 IPs were performed with CCDC47 antibody #1. For immunoprecipitations under denaturing conditions, samples of interest were first denatured in 1% SDS and 100 mM Tris pH 8.0 and boiled for 2-5 min. Samples were diluted 10-fold in IP buffer (100 mM NaCl, 50 mM HEPES pH 7.4, 1% TritonX-100) then immunoprecipitated in batch with desired antibodies at 4°C rotating end-over-end for 2-4 h. Unbound supernatant was removed by aspiration and beads were washed 3x in IP buffer before elution in 2x SDS- PAGE sample buffer. Native immunoprecipitation followed similar protocols but were performed in the presence of NSB (200 mM KOAc, 25 mM HEPES pH 7.4, 1% DBC). EDTA and RNase A was added prior to solubilisation where RNase treatment is indicated in the figure legends. Pulldowns of His-tagged β1AR intermediates and full length β1AR were performed using Ni-NTA affinity resin (Invitrogen) in 1xPBS +250 mM NaCl, 0.5% TritonX-100, 10 mM Imidazole.
Sucrose gradient separation
The products of the quenched crosslinking reaction was solubilised in NSB (200 mM KOAc, 25 mM HEPES pH 7.4, 1% DBC). EDTA and RNase A were added to release the nascent chain from the ribosome and digest the attached tRNA. The sample (typically 10 or 20 μL volume) was loaded onto 200 μL micro sucrose gradient (5-25% sucrose in NSB), centrifuged at 55,000 rpm in the TLS-55 rotor (with suitable adaptors) for 2 h 20 min, and fractionated manually into 11 fractions from the top. The final fraction, which contains aggregates and non-solubilised material, was not analysed. Fractions 1-10 were analysed by SDS-PAGE and autoradiography (or Coomassie staining to detect endogenous proteins).
Protease protection assays
Immediately following the translation reaction, the samples were placed on ice and 10% of the reactions were set aside for analysis by SDS-PAGE and autoradiography of total products. The remainder was subjected to protease digestion by the addition of proteinase K (PK) to a final concentration of 0.5 mg/ml and incubated on ice for 50 min. To stop the digestion reaction, PMSF was added to 5 mM, incubated on ice for 2-5 min, and the entire reaction transferred to 10 volumes of boiling 1% SDS, 100 mM Tris-Cl, pH 8.0. 10% of the reaction was set aside for analysis by SDS-PAGE and autoradiography of total digestion. The remainder of the sample was prepared for immunoprecipitation as described above.
Topology mapping by cysteine accessibility
Asterix-knockout HEK293 T-Rex cells were transfected with constructs encoding human Asterix tagged at the C-terminus with a FLAG tag, including variants lacking all cysteines or containing single cysteines as described in the figure. After 48 h, the cells from each well of a 6-well plate were washed once and collected in ice-cold PBS, pelleted by centrifugation, resuspended on ice in 500 μl permeabilisation buffer (100 mM KOAc, 10 mM Tris, pH 7.4, 10 mM MgCl2), and adjusted to 0.01% digitonin. After 5 min on ice, the cells were re- sedimented by centrifugation, washed once in permeabilisation buffer lacking digitonin, sedimented again, and resuspended in 30 μl of PSB on ice. An aliquot was removed for the untreated sample, and the remainder was adjusted to 1 mM final concentration of 5,000 Dalton PEG-maleimide prepared in DMSO. After 1 h on ice, the reaction was quenched with 10 mM 2-mercaptoethanol, the cells recovered by centrifugation, washed once with 600 μl PSB, and finally prepared for analysis by SDS-PAGE and immunoblotting.
Statistics and Reproducibility
No statistical analyses were applied to any of the data. The following figure panels show representative data from at least two independent biological replicates that showed similar results: Figure 1b, 1f, 2a, 2d, 4a, 4b, 4c, 4d, Extended Data Figures 1b, 1c, 2b, 2d, 2e, 3c, 3d, 4c, 6b, 8, 9b, 10b. The following figure panels show representative data from at least three independent biological replicates that showed similar results: Figures 1a, 1d, 1e, 2b, 2c, 2f, 4e, Extended Data Figures 2c, 3a, 3b, 5b, 5c, 7a, 7b, 9a. The following figure panels show representative data from at least four independent biological replicates that showed similar results: Figure 3, Extended Data Figure 6a. No attempts at replication failed. The mass spectrometry experiment in Fig. 1c was performed once but all identified proteins indicated in the figure were directly validated in follow up experiments that are shown within the paper. The experiment in Extended Data Fig. 4e was performed once, but is internally controlled for both positive and negative results.
Extended Data
Extended Data Fig. 1. Characterisation of Rho TM1+2 insertion.
(a) Diagram of Rho TM1+2 constructs used throughout this study. Variations on this construct include different N-terminal epitope tags, the presence or absence of a glycosylation site near the N-terminus, the presence, absence, or position of a cysteine within TMD1, various mutations within TMD1, and the presence or absence of TMD2. All of the constructs were tested either by protease protection or glycosylation to verify that no appreciable differences were observed in their insertion efficiencies. Note that although the exact amino acid numbering varies depending on the N-terminal tag, the numbering system corresponding to the FLAG-tagged version is used throughout. Thus, the 146mer refers to a truncation at the 146th codon in the numbering scheme indicated in Fig. 1a and in this diagram, even in constructs containing a different tag. (b) Representative example of insertion assays on two different tagged versions of Rho TM1+2. The TwinStrep tagged version (Strep) lacking a glycosylation site was compared to an HA tagged version containing a glycosylation site. Identical constructs containing either the wild-type Rho TM1 sequence or a point mutant (F53C) were tested in parallel to confirm no insertion defects result from insertion of a cysteine in TM1 (used for BMH-mediated crosslinking in later experiments). In this experiment,35S-methionine labelled ribosome nascent chain complexes (RNCs) of 181 amino acids were produced by in vitro translation using rabbit reticulocyte lysate (RRL) in the presence of ER-derived rough microsomes (RMs) after which the microsomes were isolated and resuspended. Aliquots of the reactions were left untreated or digested with proteinase K (PK) and analysed directly by SDS-PAGE and visualised by autoradiography (left panel). Green arrow heads represent the fully inserted and PK-protected population and red arrowheads denote the non-inserted and proteolytically cleaved products. The cleaved product contains the region of polypeptide protected by the ribosomal tunnel and the attached tRNA. Aliquots of the PK-digested sample were treated with EDTA and RNase to release the polypeptide from the ribosome and tRNA and immunoprecipitated (IP) via the N-terminal tag. Only the fully inserted products are recovered by IP (green arrowheads). (c) Comparison of the topology of truncated RNCs and terminated Rho TM1+2. In this experiment, the FLAG-tagged Rho TM1+2 containing a glycosylation site with (term.) or without (trunc.) a stop codon was translated in the presence of RM after which the RMs were isolated by centrifugation. Aliquots of the isolated RMs were analysed directly (-PK), after PK digestion (+PK), or after PK digestion in the presence of detergent (+PK/det). Where indicated, the +PK and +PK/det samples were released from the attached tRNA and immunoprecipitated via the N-terminal tag. Note comparable glycosylation near the N-terminus and complete protection from PK for both the truncated and terminated products. (d) Diagram representing the interpretation of the experiments in panels b and c. The relatively short cytosolic loop between TMD1 and TMD2 is not accessible to PK digestion either as an RNC or a terminated product. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 2. Additional characterisation of crosslinking to PAT10.
(a) Diagram of constructs either lacking or containing a cysteine in place of Gly48 or Phe53 within TMD1 of a Rho TM1+2 cassette. (b) 35S-labelled FLAG Rho TM1+2 ribosome nascent chains (RNCs) of varying lengths (as described in Figure 1a) containing a Cys at position 53 were generated by in vitro translation in the presence of RMs. Membranes were isolated by centrifugation through a sucrose cushion and resuspended in physiological salt buffer (PSB). An equivalent amount of each translation reaction was taken before (-BMH) and after (+BMH) the addition BMH for analysis by SDS-PAGE. The tRNA-linked nascent chains and free nascent chains are indicated. The free nascent chains arise from partial hydrolysis of the tRNA during electrophoresis under moderately basic conditions. Glycosylation is first observed at the 96mer length, which is 38 amino acids downstream of the end of TMD1. This matches the length of the ribosome tunnel, and indicates that membrane insertion and glycosylation occurs only after TMD1 is fully exposed outside the ribosome. Crosslinks to Sec61α are most prominent for the 106mer. Crosslinks to PAT10 are most prominent from the 126mer onwards, after the Sec61α crosslinks diminish. Note that all of the crosslinked adducts are seen to the tRNA-attached nascent chain, verifying that they are co-translational. The Sec61α crosslink and others are not as visible when total translation products are analysed, which is why we typically immunoprecipitate the sample via the nascent chain (e.g., in Fig. 1a). This reduces the background, allowing otherwise obscured crosslinks to be visualised clearly. Furthermore, we usually digest the samples with RNase after the experiment but before SDS-PAGE to remove the tRNA, thereby avoiding the heterogeneity that results from partial tRNA hydrolysis during sample handling and SDS-PAGE. All of the indicated crosslinking adducts observed were completely dependent on the presence of BMH. (c) The indicated “Input” crosslinking sample from Fig. 2b (reproduced here on the left) was subjected to immunoprecipitation (IP) using anti-FLAG or anti-Sec61p antibodies under denaturing conditions. The IP samples were either left untreated or digested with PNGase F to remove N-linked glycans. Equivalent amounts were loaded in each lane. Note that Sec61p is not an appreciable crosslinking partner of these RNCs, and to the extent a crosslink is observed, it migrates slightly faster than the PAT10 crosslink. (d) The indicated Rho TM1+2 variants were translated in vitro in the presence of RMs and treated with BMH as indicated. In this experiment, the crosslinking was done directly on total translation reactions, not after isolation of the microsome fraction. Instead, translation reactions were diluted 5-fold with buffer to dilute the reduced glutathione and minimise quenching of BMH. An aliquot of each reaction was analysed directly by SDS-PAGE. Cross-linking efficiency is reduced compared to other experiments because membranes were not isolated by centrifugation through a sucrose cushion to remove reduced glutathione from translation extract. One aliquot of the BMH-treated translation reactions were treated with RNase A and EDTA, denatured, and IPed via the N-terminal FLAG tag (IP). (e) As in panel d except Rho TM1+2 variants were generated in the presence of RMs derived from either canine pancreas (cRM) or HEK293 cells (hRM). In this experiment, the microsomes were isolated by centrifugation through a sucrose cushion prior to BMH crosslinking (note higher cross linking efficiency). While the PAT10 crosslink is seen in both cRM and hRM, cross-linking efficiency of the inserted (glycosylated) product is significantly lower in cRMs, which is one reason we used HEK293- derived RMs for most of the experiments in this study. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 3. Analysis of PAT complex candidates.
(a) Radiolabelled 146mer RNCs of Rho TM1+2 constructs containing or lacking a glycosylation site were generated by in vitro translation in the presence of RMs. Membranes were isolated by centrifugation through a sucrose cushion, subjected to BMH crosslinking where indicated, treated with RNase A, then analysed by SDS-PAGE. One aliquot of the BMH treated reactions were solubilised under native conditions and IPed with either an antibody raised against the HA epitope tag (Cntrl) for a specificity control, or an antibody against signal peptide peptidase (SPP). Direct crosslinks to SPP are observed as two distinct adducts seen on very long exposures (compare to Fig. 1d, which is a shorter exposure), but native IPs do not enriched for a PAT10 engaged substrate. (b) Radiolabelled 146mer RNCs of FLAG tagged Rho TM1+2 containing (+) or lacking (-) a glycosylation site (glyc.) were generated by in vitro translation with RM, crosslinked with BMH, then immunoprecipitated under native conditions via the N-terminal FLAG tag on the substrate (Nterm) or with an antibody recognising the C-terminus of calnexin (CNX). Only the glycosylated substrate is recovered with CNX, consistent with its binding via the glycan. (c) Aliquots of the crosslinked and natively solubilised samples from panel b were run on a 5-25% sucrose gradient before analysis by SDS-PAGE and autoradiography. Red asterisks denote peak fractions containing the PAT complex as detected by the PAT10 crosslinking product. The PAT complex crosslinked to unglycosylated Rho TM1+2 migrates slightly smaller on the gradient than glycosylated Rho TM1+2, likely the result of CNX (∼90 kD) no longer associated with the nascent chain. (d) The insertion and BMH mediated cross-linking for 146mer RNCs of the parent Rho TM1+2 construct or versions lacking a cysteine (NoCys) or lacking a glycosylation site (Cys53No glyc). The radiolabelled RNCs were produced by in vitro translation in the presence of RMs isolated from wild type (WT) or two different CCDC47 KO cell lines (A1 and A2) generated from two different guide RNAs. Aliquots of the reaction before (-BMH) and after (+BMH) addition BMH were analysed by SDS-PAGE. No appreciable difference in insertion efficiency was observed in KO microsomes for Rho TM1+2 as monitored by glycosylation efficiency. Red arrowheads indicate the PAT10 crosslink which is lost upon CCDC47 KO. The faint crosslinked adduct observed in the KO samples (black asterisks) migrates slightly faster on the gel and likely represents weak Rho TM1+2 crosslinks to Sec61p (see Extended Data Fig. 2c). For gel source data, see Supplementary Figure 1.
Extended Data Fig. 4. Conservation and topology of Asterix.
(a) Alignment of Asterix homologs for five divergent species with a bar chart representing conservation scores of each amino acid. Indicated are three hydrophobic domains (HD1 to HD3), each of ∼15 amino acids, that are candidate TMDs. (b) Representation of human Asterix amino acid sequence and the relative lengths of hydrophilic (grey bar) and hydrophobic (blue) regions. (c) Two matched human Asterix constructs containing either an N- or C-terminal FLAG tag were generated by in vitro translation in the presence of RMs. One aliquot of the reaction was set aside for analysis by SDS-PAGE of total translation products (IVT). The remainder of the reaction was treated with proteinase K (+PK) without or with detergent (det). These protease- digested samples were either analysed directly or after immunoprecipitation (IP) via the FLAG tag. Red arrowheads indicate fragments protected from PK in the absence, but not presence of detergent. The PK-protected fragment from the C-terminally tagged Asterix was recovered by IP, suggesting that the C-terminus is located within the ER lumen and the N- terminus is located in the cytosol. The relative size difference between the N-and C- terminally tagged constructs observed after PK digestion can be attributed to digestion or protection of the FLAG tag. Below the gel is a cartoon depiction of one possible topology based on the results and the protease protected fragments that remain after digestion with PK. The other possible topology is a single-spanning orientation with HD2 and HD3 in the lumen. (d) Schematic of human Asterix with a C-terminal FLAG tag in its predicted 3-TMD topology based on the protease digestion results in panel c. To test this prediction, a cysteine- free version of Asterix (No Cys) was modified with single cysteines at the position indicated by the red-asterisks. If the topology prediction is correct, only the N-terminal domain (NTD) cysteine and the Loop 2 cysteine should be accessible to sulfhydryl modifying reagents added to the cytosolic side of them membrane. If the protein only spans the membrane once with the N-terminus facing the cytosol, then the Loop 2 cysteine should not be modified. As shown in panel a, wild type Asterix naturally has four cysteines, only one of which should be exposed to the cytosol because it is in the NTD. (e) Asterix KO HEK-293 cells were transiently transfected with the indicated Asterix-FLAG constructs, semi-permeabilised in 0.01% digitonin, washed to remove digitonin, and treated with PEG-Maleimide (average molecular weight 5 kDa) in order to modify any cytosolically exposed cysteine residues. WT Asterix contains 4 native cysteine residues, one in the N-terminus preceding TMD1 and 3 others within the putative TMD regions. Modification was only observed for the NTD cysteine and the cysteine in Loop 2, supporting a 3-TMD topology as depicted in panel d. The single cysteine present in the cytosolic domain of Sec61p was used as a positive control demonstrating equal modification efficiency in all samples, and the No Cys construct verifies sulfhydryl-dependent modification. Protection of the cysteines in TMD1 and TMD2 from modification verifies that membrane integrity was maintained in the experiment. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 5. Characterisation of site-specific photo-crosslinking.
(a) Schematic of the strategy for site-specific incorporation of the photo-crosslinking amino acid Benzoyl- phenylalanine (Bpa) during in vitro translation (IVT). Bpa, a synthetic amber-suppressor tRNA, and recombinant Bpa tRNA synthetase are added to an IVT reaction. The nascent protein that is produced incorporates Bpa at an amber codon. UV irradiation results in Bpa activation and crosslinking to adjacent proteins. (b) The photo-crosslinking strategy was tested using a well-validated translocation intermediate: the 86mer of the secretory protein prolactin. The amber codon was installed at position 9, within the hydrophobic core of the signal sequence. At this length, the majority of the nascent chain is precursor (pre), with a small amount that is signal-cleaved (s.c.). The primary crosslinks to SRP54 and components of the translocation site (Sec61α, TRAM, and TRAPa) were verified by immunoprecipitation. (c) Site-specific photo-crosslinking of a 141mer RNC containing the UV-activated photo-crosslinking amino acid benzoyl-phenylalanine (Bpa) at the indicated amber positions (Amb). The diagram above the autoradiographs shows a schematic of the construct with the appropriate amino acid numbering. Amino acids in red show the strongest crosslinks to Asterix, pink show reduced crosslinks, and grey no detectable crosslinks. Total translation products recovered by IP via an HA tag on the nascent chain are shown adjacent to parallel IPs of selected samples using the indicated antibodies. Although not all IPs are shown, each position was tested for crosslinking to Asterix and CCDC47. RNCs that failed to engage SRP crosslink to UBQLN2, a quality control factor that binds exposed TMDs. Note that this crosslink diminishes markedly when increased RMs are used in the reaction (lanes 916, compared to lanes 5-8), presumably because the RMs contribute SRP, which is otherwise limiting in the reaction. A subset of RNCs fail to release from SRP and crosslink to SRP54. The crosslink indicated by the hashtag (#) is likely to be a mixture of similarly migrating crosslinks. Because this crosslink diminishes substantially with increased RMs (similar to the UBQLN2 crosslink), it is likely to be SGTA, another TMD-binding factor in the cytosol of this size. A small proportion of this crosslink could be the similarly sized Sec61α or TRAM. Of the membrane-inserted RNCs, the main crosslink is to Asterix, seen prominently for residues 52 to 63. At this length, the TMD has moved away from Sec61α, so crosslinks to this factor are minimal. No crosslinking to CCDC47 were ever observed. Note that by testing five sequential positions in the centre of the TMD, all sides of the helix have been sampled.
Extended Data Fig. 6. Effect of Asterix depletion on multi-spanning membrane proteins.
(a) The raw data for three of the histograms of the GFP:RFP ratio (or RFP:GFP ratio in the case of GFP-2A-RFP-SQS) shown in Fig. 3. The mode of the control histogram (dotted black line in Fig. 3) was used to determine the statistical mode of GFP:RFP (or RFP:GFP) ratio. This mode was used as a gate to colour the dot plots shown below the histograms such that all cells above the gate were coloured red and those below the gate were coloured grey. The percent of cells above the gate for each plot is indicated. (b) Flow- cytometry analysis of the indicated GPCR reporters using the dual-colour assay system exactly as in Fig. 3. Cell lines containing the reporter stably integrated at a single FRT site located downstream of a doxycycline-inducible reporter were used for these assays. This allows assay of cells using a single transfection (which proved to be less toxic than sequential transfections with both the siRNA and the reporter), and provided control of the length of time of reporter expression. Each reporter cell line was treated with scrambled (Scr) or Asterix-targeting siRNAs, then at the time of effective knockdown (verified in separate experiments using immunoblotting), the reporter was induced for ∼6-8 hours. Induction only after knockdown allows us to monitor the reporter that was produced in the absence of Asterix rather than a heterogeneous mixture of reporter expressed during the knockdown. The histograms of the GFP:RFP ratio in the scrambled- versus Asterix-siRNA cells are shown in grey and blue, respectively, in the upper plot for each construct. The two dot plots below the histogram are the corresponding raw data plotted as described in panel a. Each reporter shows a distribution of lower GFP:RFP ratio, with some reporters being more impacted than others. This is not seen with the tail-anchored protein SQS using the same assay format.
Extended Data Fig. 7. Effect of CCDC47 depletion on membrane protein biogenesis.
(a) The diagrams depict dual-colour fluorescent reporters for protein stability as an indirect measure of successful biogenesis. The membrane protein of interest is tagged with one fluorescent protein (FP), which is separated from a second FP by the viral 2A peptide sequence. When the 2A sequence is translated, peptide bond formation is skipped without perturbing elongation by the ribosome. Thus, translation results in two separate proteins made in a 1:1 stoichiometry that are separated at the 2A sequence. If biogenesis of the membrane protein is impaired, it will be degraded along with its tagged FP, resulting in an altered ratio of the two FPs. Thus, treatment conditions that impair biogenesis of the membrane protein will be reflected as a relative change in the ratio of FPs. The three reporters encoding angiotensin type-2 receptor II (AGTR2), squalene synthase (SQS) and Asialglycoprotein receptor (ASGR) were transiently transfected into wild-type (WT), CCDC47 KO (ΔCCDC47) or Asterix KO (AAsterix) HEK293 cells and analysed by dual- colour flow cytometry. Histograms represent the distribution of FP ratio in WT (grey), ΔCCDC47 (red) and AAsterix (blue) cells. A biogenesis defect is only seen for the multi- spanning membrane protein AGTR2, but not for the tail-anchored protein SQS or the signal- anchored single pass protein ASGR. (b) Assays similar to those in Fig. 3, but for cell lines treated with scrambled versus CCDC47 siRNAs as indicated. We find that the phenotypes for Asterix and CCDC47 knockdowns are very similar for all reporters (three are shown here), with CCDC47 consistently being somewhat more modest. The reason for this seems to be that CCDC47 knockdown is slower and less efficient than Asterix knockdown. Note that similar phenotypes are seen for AGTR2 and SS-T4L-AGTR2, a version that contains an N- terminal signal sequence and T4 lysozyme preceding TMD1. In earlier studies, we found that initiating translocation with a signal sequence completely bypasses the requirement for EMC- mediated TMD1 insertion. The fact that SS-T4L-AGTR2 remains sensitive to PAT complex depletion (as judged by either Asterix or CCDC47 knockdowns) argues that the PAT complex acts independently of EMC.
Extended Data Fig. 8. Expression of ER biogenesis factors in ΔCCDC47 and AEMC6 cells.
ER rough microsomes were isolated from WT, ΔCCDC47 and AEMC6 HEK293 cells and normalised to an absorbance of 75 at 280nm. Serial dilutions of each sample were analysed by SDS-PAGE and immunoblotting for the indicated antigens. Note that BiP levels are elevated in both knockout cell lines consistent with an activated UPR caused by altered ER homeostasis. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 9. TMD1 insertion does not require the PAT complex or TMD2.
(a) Rho TM1+2 RNCs of varying nascent chain lengths (indicated at top of gels) were translated in vitro in the presence of RM prepared from ΔCCDC47 or AAsterix HEK293 cells as indicated. Membranes were isolated by centrifugation through a sucrose cushion and treated with the chemical cross linking reagent BMH. The samples were denatured in 1% SDS and immunoprecipitated via the substrates’s N-terminal FLAG tag. Notice that the glycosylation of substrate is very similar in efficiency and timing as that seen in RM prepared from wild type HEK293 cells (see Fig. 1a for comparison). Furthermore, the appearance and disappearance of the SRP54 and Sec61α cross linking adducts are not appreciably altered from the results seen in RM prepared from wild type cells. Thus, the early steps of Rhodopsin biogenesis are not impaired appreciably in the absence of Asterix or CCDC47. As expected, the crosslink to Asterix/PAT10 is not seen (verified by anti-Asterix immunoprecipitation; not shown). Crosslinking products seen at the approximate size of the Asterix crosslink are therefore other protein(s). (b) The Rho TM1 construct in which TMD2 is replaced with a hydrophilic linker sequence (see diagram in Fig. 4a) was analysed for crosslinking as in Fig. 1a. Note that the absence of TMD2 does not affect the crosslinking between TMD1 and Asterix. By contrast, mutation of the most polar residue in TMD1 (N52) markedly reduces Asterix crosslinking and reduces TMD1 proximity to Sec61α (see Fig. 4a).
Extended Data Fig. 10. Analysis of Asterix interaction with TMD1 by photo- crosslinking.
(a) Experimental strategy for comparing Asterix interaction with a membrane protein intermediate versus full length product. In this experiment, the photo-crosslinking amino acid Bpa (yellow star) is incorporated into position 52 within TMD1 of β1AR by in vitro translation. The intermediate is represented by the TM1-3 product containing the first three TMDs of β1AR. The full length (FL) β1AR contains all seven TMDs followed by a long flexible linker. TM1-3 is stalled 35 amino acids downstream of TMD3 (with TMD4 inside the ribosomal tunnel), allowing TMD3 to be outside the ribosome. β1AR FL is stalled 152 amino acids downstream of TMD7, providing a sufficiently long tether for all seven TMDs to have emerged, inserted into the membrane, and assembled together. The translation products are then irradiated with UV light to activate the Bpa and any crosslinking to Asterix is subsequently detected by denaturing IP via Asterix. (b) Results from a photo-crosslinking experiment as depicted in panel a. The microsomes from the IVT reaction were isolated, resuspended, irradiated with UV light (or left untreated), and denatured. The samples were then divided in two and immunoprecipitated via the nascent chain or via Asterix. Six-fold more of the Asterix IPs were loaded on the gel relative to the nascent chain IPs. As expected, the Bpa in TMD1 crosslinks to Asterix in the TM1-3 intermediate. Note that the crosslinked band in the 6x Asterix IP is the same intensity as the glycosylated band in the 1x nascent chain IP. Although elongation to the full length product was somewhat inefficient, clear glycosylated and non-glycosylated products are observed in the nascent chain IPs. No band is seen in the 6x Asterix IP sample that is of comparable intensity to the glycosylated band in the 1x nascent chain IP. This argues that the proximity of TMD1 to Asterix has diminished substantially in the full length nascent chain relative to the TM1-3 intermediate. Of note, a heterogeneous set of crosslinks (marked by red stars) are seen at a lower part of the gel in the 6x Asterix IP. These correspond to the sizes expected for Asterix crosslinks (i.e., shifted by ∼10 kD) to the major incomplete translation products (marked by blue stars). These crosslinks provide an internal control and further supports the conclusion that incomplete products engage Asterix, while a complete 7-TMD product does not.
Supplementary Material
Acknowledgements
We thank S.-Y. Peak-Chew, F. Begum, and M. Skehel for mass spectrometry analysis; S. Juszkiewicz, J. O’Donnell, and E. Zavodszky for productive discussions and advice; N. Peters and H. Damstra for initial characterisation of photo- crosslinking methods. This work was supported by the UK Medical Research Council (MC_UP_A022_1007 to R.S.H.) and a studentship (to PJC) from the MRC International PhD Programme.
Footnotes
Reporting summary:
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Author contributions: P.J.C. and R.S.H. conceived the project, analysed the results, and wrote the manuscript. P.J.C. did all of the experimental work with advice from R.S.H.
Competing interests: The authors declare no competing interests.
Additional Information: Supplementary Information is available for this paper. Correspondence and requests for materials should be addressed to R.S.H. Reprints and permissions information is available at www.nature.com/reprints.
Data availability
All data supporting the findings of this study are available within the paper, its figures, its extended data figures, and its supplementary figure. Uncropped images of all gels and autoradiographs in the figures are provided in Supplementary Figure 1.
References
- 1.UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699–2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Heijne G. The membrane protein universe: what’s out there and why bother? J. Intern. Med. 2007;261:543–57. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 3.Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu P, et al. Accurate computational design of multipass transmembrane proteins. Science. 2018;359:1042–1046. doi: 10.1126/science.aaq1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrington SE, Ben-Tal N. Structural determinants of transmembrane helical proteins. Structure. 2009;17:1092–103. doi: 10.1016/j.str.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 2000;7:154–60. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 7.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–94. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 8.Lear JD, DeGrado WF, Choma C, Gratkowski H. Asparagine-mediated self- association of a model transmembrane helix. Nat. Struct. Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 9.Shurtleff MJ, et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife. 2018 doi: 10.7554/eLife.37018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meacock SL, Lecomte FJL, Crawshaw SG, High S. Different Transmembrane Domains Associate with Distinct Endoplasmic Reticulum Components during Membrane Integration of a Polytopic Protein. Mol. Biol. Cell. 2002;13:4114–4129. doi: 10.1091/mbc.E02-04-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park PS-H. Rhodopsin Oligomerization and Aggregation. J. Membr. Biol. 2019;252:413–423. doi: 10.1007/s00232-019-00078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estabrooks S, Brodsky JL. Regulation of CFTR Biogenesis by the Proteostatic Network and Pharmacological Modulators. Int. J. Mol. Sci. 2020;21:452. doi: 10.3390/ijms21020452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keenan RJ, Freymann DM, Stroud RM, Walter P. The Signal Recognition Particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 14.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–9. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 15.Chitwood PJ, Juszkiewicz S, Guna A, Shao S, Hegde RS. EMC Is Required to Initiate Accurate Membrane Protein Topogenesis. Cell. 2018;175:1507–1519.e16. doi: 10.1016/j.cell.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chitwood PJ, Hegde RS. The Role of EMC during Membrane Protein Biogenesis. Trends Cell Biol. 2019;29:371–384. doi: 10.1016/j.tcb.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Ismail N, Crawshaw SG, High S. Active and passive displacement of transmembrane domains both occur during opsin biogenesis at the Sec61 translocon. J. Cell Sci. 2006;119:2826–36. doi: 10.1242/jcs.03018. [DOI] [PubMed] [Google Scholar]
- 18.Lin Z, et al. TTC5 mediates autoregulation of tubulin via mRNA degradation. Science. 2020;367:100–104. doi: 10.1126/science.aaz4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itakura E, et al. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol. Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talbot BE, Vandorpe DH, Stotter BR, Alper SL, Schlondorff JS. Transmembrane insertases and N-glycosylation critically determine synthesis, trafficking, and activity of the nonselective cation channel TRPC6. J. Biol. Chem. 2019;294:12655–12669. doi: 10.1074/jbc.RA119.008299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guna A, Volkmar N, Christianson JC, Hegde RS. The ER membrane protein complex is a transmembrane domain insertase. Science. 2018;359:470–473. doi: 10.1126/science.aao3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–30. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 23.Doring K, et al. Profiling Ssb-Nascent Chain Interactions Reveals Principles of Hsp70-Assisted Folding. Cell. 2017;170:298–311.e20. doi: 10.1016/j.cell.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein KC, Kriel A, Frydman J. Nascent Polypeptide Domain Topology and Elongation Rate Direct the Cotranslational Hierarchy of Hsp70 and TRiC/CCT. Mol. Cell. 2019;75:1117–1130.e5. doi: 10.1016/j.molcel.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato BK, Schulz D, Do PH, Hampton RY. Misfolded Membrane Proteins Are Specifically Recognized by the Transmembrane Domain of the Hrd1p Ubiquitin Ligase. Mol. Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato K, Sato M, Nakano A. Rer1p, a Retrieval Receptor for ER Membrane Proteins, Recognizes Transmembrane Domains in Multiple Modes. Mol. Biol. Cell. 2003;14:3605–3616. doi: 10.1091/mbc.E02-12-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan N, Foresti O, Wendrich K, Stein A, Carvalho P. Quality Control of Protein Complex Assembly by a Transmembrane Recognition Factor. Mol. Cell. 2020;77:108–119.e9. doi: 10.1016/j.molcel.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itzhak DN, Tyanova S, Cox J, Borner GH. Global quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5 doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto S, et al. Contribution of calumin to embryogenesis through participation in the endoplasmic reticulum-associated degradation activity. Dev. Biol. 2014;393:33–43. doi: 10.1016/j.ydbio.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 31.Morimoto M, et al. Bi-allelic CCDC47 Variants Cause a Disorder Characterized by Woolly Hair, Liver Dysfunction, Dysmorphic Features, and Global Developmental Delay. Am. J. Hum. Genet. 2018;103:794–807. doi: 10.1016/j.ajhg.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, et al. Identification and characterization of essential genes in the human genome. Science (80-.) 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kota J, Ljungdahl PO. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 2005;168:79–88. doi: 10.1083/jcb.200408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu S, et al. Brain a7 Nicotinic Acetylcholine Receptor Assembly Requires NACHO. Neuron. 2016;89:948–955. doi: 10.1016/j.neuron.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Brechet A, et al. AMPA-receptor specific biogenesis complexes control synaptic transmission and intellectual ability. Nat. Commun. 2017;8:15910. doi: 10.1038/ncomms15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–39. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- 38.Sharma A, Mariappan M, Appathurai S, Hegde RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol. Biol. 2010;619:339–63. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Q, Shao S. In vitro reconstitution of translational arrest pathways. Methods. 2018;137:20–36. doi: 10.1016/j.ymeth.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Shao S, Von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper, its figures, its extended data figures, and its supplementary figure. Uncropped images of all gels and autoradiographs in the figures are provided in Supplementary Figure 1.