Abstract
Polyether ionophores are complex natural products capable of transporting cations across biological membranes. Many polyether ionophores possess potent antimicrobial activity and a few selected compounds have ability to target aggressive cancer cells. Nevertheless, ionophore function is believed to be associated with idiosyncratic cellu-lar toxicity and, consequently, human clinical development has not been pursued. Here, we demonstrate that structurally novel polyether ionophores can be efficiently constructed by recycling components of highly abundant polyethers to afford analogues with enhanced anti-bacterial selectivity compared to a panel of natural polyether ionophores. We used classic degradation reactions of the natural polyethers lasalocid and monensin and combined the resulting fragments with building blocks provided by total synthesis, including halogen-functionalized tetronic acids as cation-binding groups. Our results suggest that structural optimization of polyether ionophores is possible and that this area represents a potential opportunity for future methodological innovation.
Introduction
Strategic integration of complex molecule synthesis with studies of biological mechanisms1 can catalyze discoveries within the life sciences by providing access to novel chemical space. Diversity-oriented synthesis2 biology-oriented synthesis,3 analogue-oriented synthesis,4 diverted total synthesis,5 complexity-to-diversity-strategies,6 pharmacophore-directed retrosynthesis,7 and pseudo natural products8 are different flavors of a broader objective to construct innovative complex structures with potential pharmacological utility.9,10
The polyether ionophores have been a constant fix-point for organic chemistry since the 1960s. The daunting complexity of compounds such as monensin, salinomycin, and X-206 challenged the abilities of chemists to identify their precise molecular structures11,12 and has served as inspiration for the development of novel synthetic methods and retrosynthetic analysis13,14. Elegant total syntheses of polyether ionophores have been reported15,16 and, more recently, remarkable efficiencies have been achieved for some compounds17,18,19,20.
The main feature of polyether ionophores is their antibiotic activity, including against drug-resistant strains21. However, despite their safe use in the agricultural industry22,23, toxicity is observed in some animals11, which, along with their ‘fuzzy’ mechanisms of perturbing ion-gradients, has earned polyether ionophores a reputation of being unpredictable or pleiotropic. Many analogs of polyether ionophores have been prepared,11,24 but truly systematic efforts to diversify their complex structures and understand the biological consequences have so far been absent, and the prospect of conducting lengthy de novo syntheses within this structural class, without a guidance towards a particular target structure, is not appealing. The question remains whether optimization of polyether ionophores with respect to their anti-bacterial properties is possible. Here, we provide proof-of-principle of an approach for polyether diversification that involves deconstruction of industrial polyether ionophores and subsequent reconstruction to access a new structural domain related to the natural polyether ionophores (Fig. 1a). By subjecting the resulting ‘hybrid polyethers’ to broad biological evaluations, we have identified compounds with improved antibacterial selectivity.
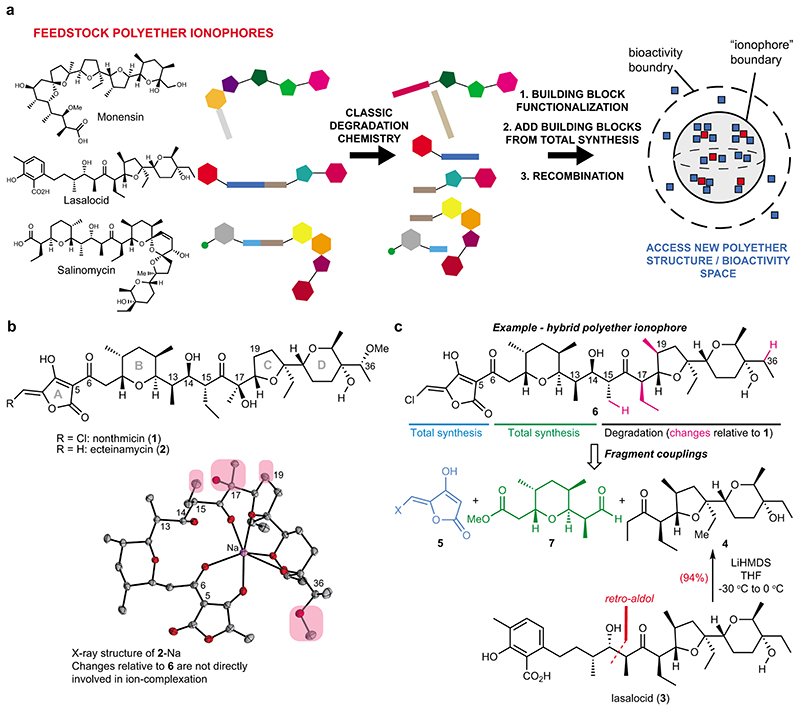
Fig. 1. Accessing structural diversity within the polyether ionophores.
(a) Flowchart depicting the overall concept of reconstructing new polyether scaffolds by recycling elements from abundant feedstock polyether ionophores. The resulting “hybrid” molecules (blue squares) are plotted in a hypothetical structure and bioactivity space to illustrate the relation of these compounds to the natural polyethers (red squares). The compounds that possess ionophore activity constitute a sub-space of a larger bioactivity-space that can be explored using hybrid polyethers. (b) Chemical structures of polyether ionophores nonthmicin and ecteinamycin. Both are active against gram-positive bacteria, with especially strong potency against C. difficile reported for ecteinamycin. The compounds bear resemblance to lysocellin/ferensimycin but the chlorinated methylidene tetronic acid group of nonthmicin is unprecedented. The X-ray structure25 depicts ecteinamycin bound to a single sodium-ion and the chemical groups on the hydrophobic periphery that have been altered in the target hybrid polyether 6 have been circled in pink. No crystal structure of nonthmicin is available. (c) Chemical structure of the hybrid polyether 6 with indication of the required fragments and the origin of these fragments. The main fragment, ketone 4, can be obtained in a single synthetic step from lasalocid.
Results
Degradation of abundant polyether ionophores as the foundation of diversity synthesis of complex polyethers
To test the diversification strategy, we took inspiration from a novel polyether ionophore, nonthmicin (1) (Fig. 1b).25 Nonthmicin is of particular interest for structural reasons due to the unprecedented 5-chloromethylidene tetronic acid that serves as the cation-binding group. Another compound, ecteinamycin (2), which lacks the chlorine-atom, was reported almost simultaneously by the Bugni lab to be an anti-clostridial antibiotic (Fig. 1b).26 Inspection of the structure of 1 and 2 reveals that the eastern portion (the C-D rings) displays significant similarity to lasalocid (3), which is available on multi-kg scale. Based on the classic studies of 3,27,28 we postulated that we could first liberate ketone 4 through a retro-aldol reaction and then develop a concise route to advanced analogs of 1 with compound 6 as one representative target (Fig. 1c). Interestingly, inspection of the published X-ray crystal structure of 2-Na25 suggest that none of the resulting structural changes (pink in Fig. 1b–c) would directly perturb coordination of the metal ion, at least from the solid-state structure.
We started our studies by optimizing the retro-aldol reaction of 3 and found that exposure to LiHMDS could result in the formation of ketone 4 in excellent yield (94%, >10 gram scale). We next focused our attention on the two remaining building blocks, tetrahydropyran-derivative 7 and the halomethylidene tetronic acid moiety 5 found in both 6 and 1.
Construction of tetrahydropyran and halomethylidene tetronic acid building blocks
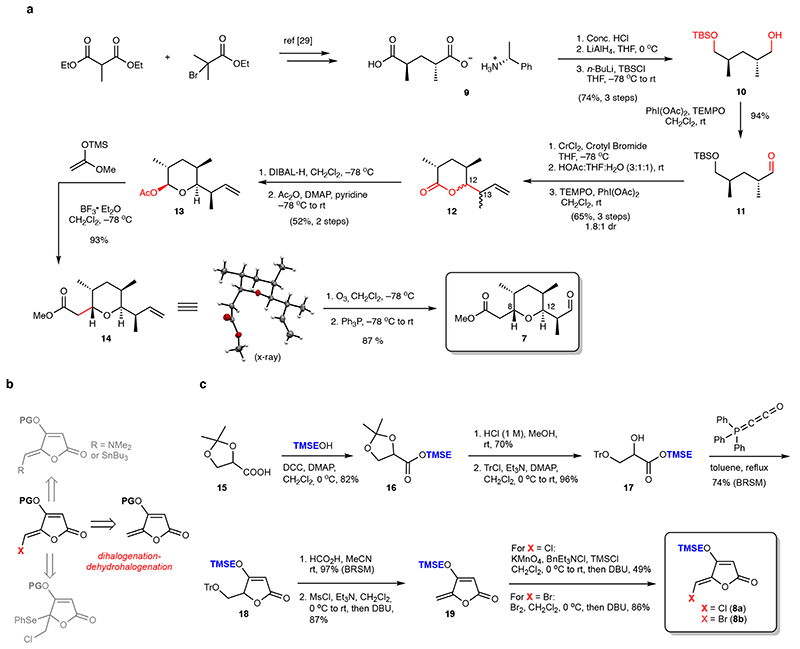
The preparation of the targeted tetrahydropyran-derivative 7 was started from the known compound 9 29, which was readily processed to aldehyde 11. CrCl2-mediated Nozaki-Hiyama-Kishi crotylation30 was conducted and followed by a two-step conversion31 to δ-lactone 12 (dr 1.8:1, 65% isolated yield) which was reduced by DIBAL-H and trapped in situ with Ac2O to afford the corresponding acetate (Fig. 2a). At this stage, the major diastereomer 13 – ultimately found to have the desired configuration at C12 and C13 – could be isolated in 52% yield. The formation of 13 as the major diastereomer is expected as a combination of Felkin-control and the cyclic 6-membered transition state in the crotylation step.30 Using BF3•OEt2, 13 was then exposed to ((1-methoxyvinyl)oxy)trimethylsilane32 at low temperature which provided methyl ester 14 as a single diastereomer (confirmed by X-ray diffraction) in excellent yield. Finally, ozonolysis of 14 followed by reductive work-up generated aldehyde 7 (Fig. 2a).
Fig. 2. Building block synthesis.
(a) Route to aldehyde 7 proceeding in ten steps from optically active salt 9. (b) Different synthesis strategies attempted to construct the protected 5-halomethylidene tetronic acid building block. A dihalogenation-dehydrohalogenation of the corresponding 5-methylidene tetronic acid was successful (see also Extended Data Fig. 1). (c) Construction of the required TMSE-protected precursor 19 was carried out in 6 steps from the commercial racemic acetonide-protected glycerate 15. This sequence allowed for preparation of both the chlorine and bromine-variants (8a and 8b). THF = tetrahydrofuran, TBSCl = tert-butyldimethylsilyl chloride, THF = tetrahydrofuran, rt = room temperature, TEMPO = (2,2,6,6-tetramethylpiperidin-1-yl)oxyl, DMAP = N,N-dimethylpyridine-4-amine, DIBAL-H = diisobutylaluminium hydride. Tr = trityl, TMSE = 2-(trimethylsilyl)ethyl, DCC = N,N’-dicyclohexylcarbodiimide, BRSM = based on recovered starting material, Ms = methanesulfonyl, DBU = 1,8-Diazabicyclo[5.4.0]undec-7-ene, TMS = trimethylsilyl.
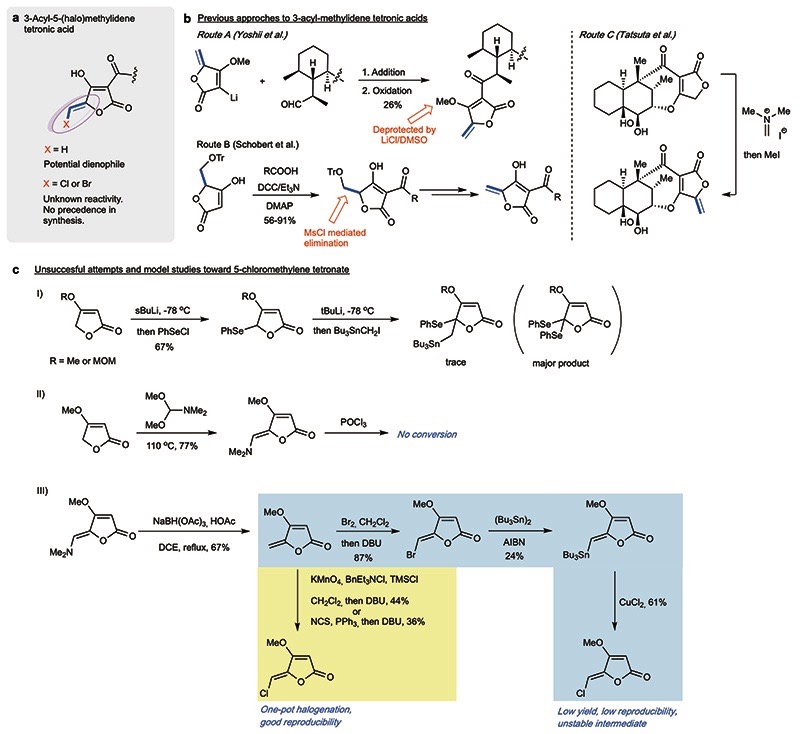
As the most intriguing structural subunit of nonthmicin and 6, the 3-acyl-5-chloromethylidene tetronic acid became the next focus. The ideal approach to this building block should also allow access to other variants such as the methylidene and bromomethylidene species (Fig. 2b). Although the construction of this densely functionalized unit has not previously been realized, syntheses of natural products containing the nonhalogenated version are instructive (Extended Data Fig. 1).33 Mindful of the presumed lability of the halomethylidene species, and the multiple functional groups present in the ultimate coupling partner, we selected a DCC-mediated late-stage coupling as the most appealing strategy and we there-fore decided to target the tetronate building blocks bearing an easily-removable protecting group (TMSE). After a series of unsuccessful attempts (Fig. 2b and Extended Data Fig. 1), we realized the synthesis of the 5-halomethylidene tetronate via a dihalogenation-dehydrohalogenation approach (Fig. 2b). The O-TMSE protected tetronate 19 was prepared via intramolecular Wittig cyclization from TMSE-ester 17 and the Bestmann reagent, and the methylidene group was installed via MsCl-mediated elimination (Fig. 2c). Halogenation was then facilitated by dichlorination (KMnO4-TMSCl)34/dibromination (Br2)35 of 19 and subsequent elimination (DBU) to furnish the target (Z)-halomethylidene tetronates 8a and 8b.
Aldol Fragment coupling
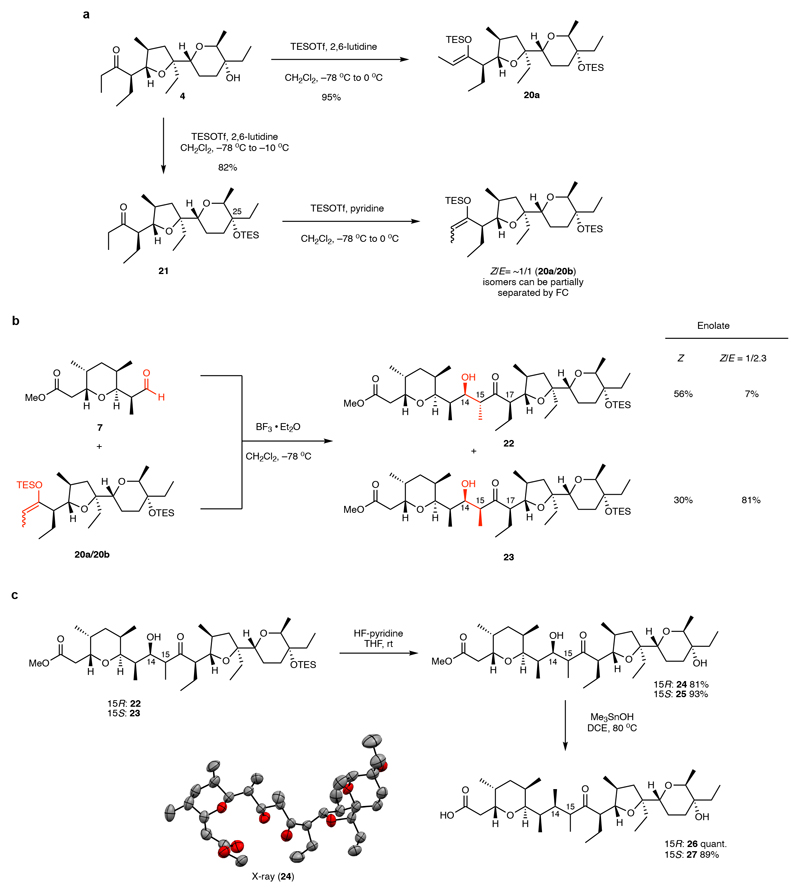
Next, we initiated the fragment coupling sequence. Analysis of the stereotriad at C13-C15 in 6 suggests that the desired configuration could be achieved via an anti-aldol reaction with Felkin-control of the secondary alcohol at C14. This type of stereocontrol is usually reinforcing36 although in the present case, due to the stereogenic center at C17, double stereodifferentiation in the aldol reaction is required.37,38 Following initial tests and further analyses of the specific reaction (Supplementary Discussion 1), we opted for the Mukaiyama aldol reaction to facilitate the fragment coupling. Our first attempts at enolizing ketone 4 afforded some surprising results: While the (Z)-TES-enolate 20a was formed with excess TESOTf and 2,6-lutidine with concomitant protection of the tertiary alcohol at C25, formation of the corresponding (E)-enolate could not be accessed cleanly (Fig. 3a). After extensive experimentation, we found that substitution of 2,6-lutidine with pyridine and inverting the sequence of reagent addition could afford a nearly 1:1 mixture of the (Z)/(E)-TES-enolates (20a/20b) which could be partially separated by flash chromatography. Gratifyingly, upon exposure of the enolates to aldehyde 7 at low temperature in the presence of BF3•OEt2, aldol coupling proceeded in excellent yield (Fig. 3b). We could detect formation of all four putative aldol diastereomers by thin layer chromatography (tentatively designated P1-P4 based on silica gel mobility, see the Supplementary Information), but the reaction using an enriched (E)-TES-enolate was selective for the P3-isomer (P2: 7%, P3: 81%) and the pure (Z)-TES-enolate afforded a mixture of P2 and P3 (P2: 56%, P3: 30%). Subsequent stereochemical assignment by X-ray crystallography revealed P3 to be the (14R,15S)-configured product (23), in accord with our analysis, and P2 to be the desired isomer (14R,15R) (22).
Fig. 3. Fragment-coupling via boron trifluoride-mediated Mukaiyama-aldol reaction.
(a) The (Z)-TES-enolate 20a could be readily obtained, but special procedures had to be developed to access mixtures of (E)- and (Z)-TES-enolates. Purification could be used to further enrich the (E)-TES-enolate 20b. (b) Aldol reaction affords two major products (22 and 23) depending on the configuration of the silyl-enolate derived from ketone 4. Compound 22 was confirmed by X-ray analysis of derivative 24 to be the initially targeted aldol-product (c) Both aldol products 22 and 23 could be processed towards the final fragment coupling in two high-yielding steps. TESOTf = triethylsilyltrifluoromethanesulfonate, THF = tetrahydrofuran, rt = room temperature, DCE = 1,2-dichloroethane.
End game coupling and purification
Both aldol products (22 and 23) underwent a two-step deprotection sequence, involving first Olah’s reagent to remove the C25-OTES group and then trimethyltin hydroxide to cleave the methyl ester (Fig. 3c)39. The latter conditions were found to be critical to avoid retro-aldol cleavage.
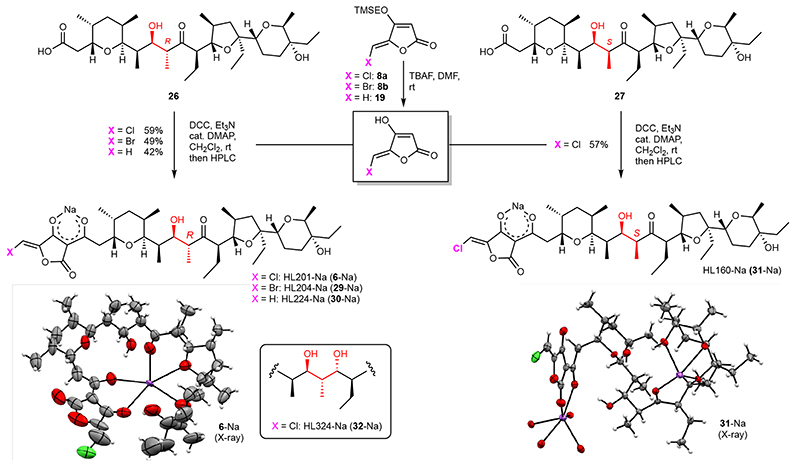
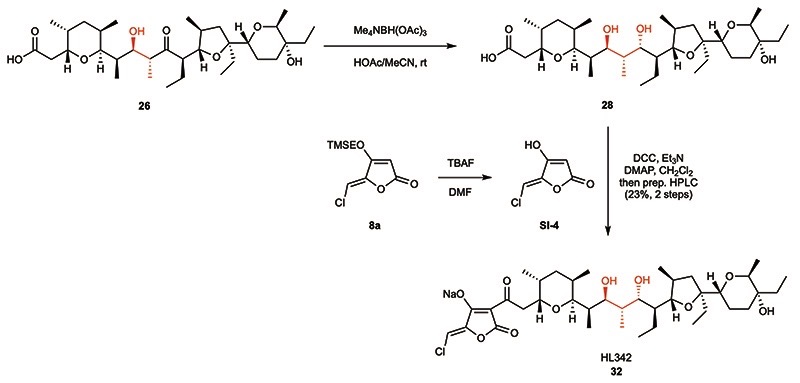
With the desired acid fragments (26 and 27) and O-TMSE protected tetronates (19 and 8a-b) in hand, we started to investigate the final fragment coupling reaction. After TBAF-mediated deprotection of the O-TMSE group on 8a and simple extraction, the crude tetronic acid was submitted to DCC coupling with carboxylic acid 26 (Fig. 4). Fortunately, the desired product HL201 (6) was smoothly formed in 24 hours with full conversion of 26, and 57% yield (as the sodium salt, 6-Na) was obtained by preparative HPLC. It is worth to note that the sodium salt (formed by subsequent NaHCO3 treatment and extraction) show much better solubility in organic solvents than the acid form, which indicates the formation of a lipophilic complex, a featured property of polyether ionophores.40 Encouraged by this result, the bromine-analog HL204 (29) and hydrogen-analog HL224 (30), as well as the chlorine-analog HL160 (31) bearing 15-(S) configuration, were synthesized following the same procedure in 42-57% yields (Fig. 4). We managed to prepare crystals for X-ray diffraction from both 6, 29, and 31 which revealed the formation of a cage-like structure by the “naturally-configured” 15-(R) analogs 6 and 29 while a dimeric complex was formed by the corresponding 15-(S)-configured compound 31. Anticipating that small structural changes could potentially have a large impact on the biological activities, we also generated derivative HL324 (32, Fig. 4) by performing an anti-selective Evans-Saksena41 reduction of the carbonyl group in 26 followed by coupling with the tetronic acid derived from 8a (Extended Data Fig. 2).
Fig. 4. Final coupling of tetronic acid derivatives.
Mild conditions were developed for effecting the coupling of unprotected 5-(halo)methylidene tetronates to carboxylic acids 26 or 27. The structures of HL201 (6), HL204 (29, see Supplementary Information) and HL160 (31), all as the sodium salts, were solved by X-ray diffraction. TMSE = 2-(trimethylsilyl)ethyl, TBAF = tetrabutylammonium fluoride, DMF = N,N-dimethylformamide, DCC = N,N’-dicyclohexylcarbodiimide, DMAP = 4-dimethylaminopyridine, rt = room temperature, HPLC = high pressure liquid chromatography.
Hybrid tetronate polyethers derived from monensin
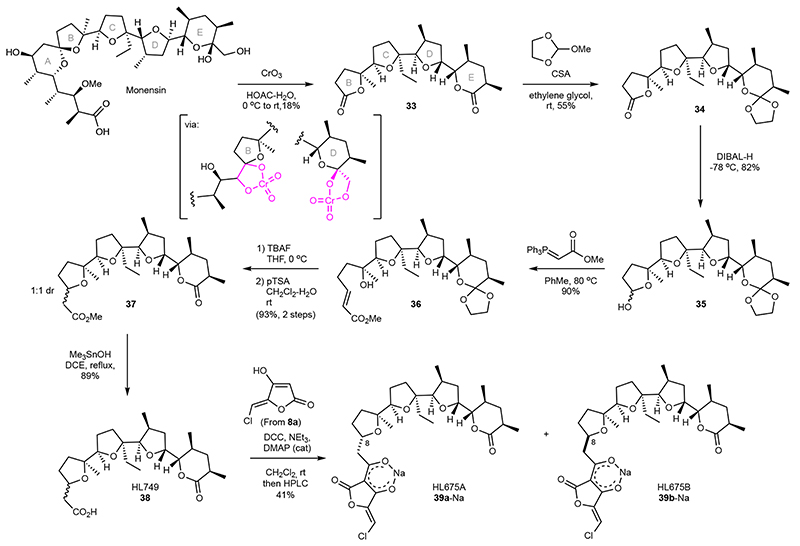
Finally, we aimed to demonstrate that degradation of another industrial polyether ionophore, monensin, could also be employed for diversification. We studied the classic conditions employing chromic acid to oxidize both the B and E rings to the corresponding γ- and δ-lactones (Fig. 5).42 This transformation, which likely proceeds via the intermediacy of the cyclic chromate esters43 affords 33 in 18% yield. While the yield is somewhat modest, the reaction is scalable and the formation of several other products indicates that this could be an interesting starting point for future exploration of new deconstruction methods of this natural product. To re-expand one end of the compound, we next selectively installed an orthoester at the E-ring δ-lactone.42 A high-yielding, five-step sequence was developed to transform the γ-lactone to the corresponding tetrahydrofuran ring with an added carboxymethylene unit. This derivative, 38, formed as a 1:1 mixture of inseparable diastereomers generated in the preceding oxa-Michael cyclization, was finally coupled with the tetronic acid derived from 8a to afford the 5-chloromethylidine tetronate derivatives HL675A (39a) and HL675B (39b) after preparative HPLC.
Fig. 5. Oxidative deconstruction of monensin for hybrid polyether synthesis.
Single-step Cr-mediated conversion of monensin to complex bis-lactone 33 can be used to generate novel derivatives. Selective protection of the δ-lactone in 33 allows reconstruction of a THF-unit through an oxa-Michael cyclization and subsequent coupling of the 5-chloromethylidene tetronate. Assignment of stereochemistry at C8 in 39a/39b could not be unambiguously done, see Supplementary Information. rt = room temperature, CSA = camphorsulfonic acid, DIBAL-H = Diisobutylaluminium hydride, TBAF = tetrabutylammonium fluoride, THF = tetrahydrofuran, pTSA = p-toluenesulfonic acid, DCE = 1,2-dichloroethane, DCC = N,N’-dicyclohexylcarbodiimide, DMAP = 4-dimethylaminopyridine, HPLC = high pressure liquid chromatography
Characterization of the antibacterial selectivity of hybrid tetronate polyethers
To characterize the biological properties of the hybrid polyethers we conducted a multipronged testing campaign. While natural polyether ionophores have undergone extensive assessments of their antibiotic and antiproliferative effects over the years, comparative data sets that describe their antibacterial selectivity is to our knowledge not available. We there-fore assembled a panel of nine natural polyether ionophores to benchmark the synthetic compounds against. For antibiotic activities, we used an agar-based droplet screen of all compounds, including synthetic intermediates, against 8 different bacterial strains as an initial gating experiment (Extended Data Fig. 3). This was followed by MIC measurements for some compounds in selected strains. (Supplementary Fig. 1). Activity in mammalian cells were characterized by viability experiments in five different cell lines (cancer: MCF-7, U-2OS, HepG2; non-cancer: BJ, Vero, Supplementary Fig. 2) and by morphological profiling through cell painting (Extended data Fig. 4 and Supplementary Fig. 3–9)44,45 conducted in both U-2OS and Vero cells. The complete dataset is shown in Table 1 (see also Supplementary Table 1).
Table 1. Biological activities of natural polyether ionophores and hybrid polyethers a .
| Strain MIC (μg/mL) |
Cell line IC50 (μM) |
Bioactivi-ty threshold (μM) |
Antibacterial selectivity |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus DSM 20231 | S. aureus MRSA USA 300 je2 | S. aureus ATCC 29213 | B. cereus DSM 31 | Cancer | Non-cancer | U-2OS | Vero | ||||||
| MCF7 | HepG2 | U-2OS | BJ | Vero | |||||||||
| Natural products | Lasalocid | 1 | 3 | 2 | 1 | 6.0 | 5.2 | 10 | 1.5 | 9.8 | 1.4 | 4.2 | 2.3 |
| Salinomycin | 0.75 | 2 | 2 | 0.5 | 4.6 | 3.9 | 5.9 | 0.31 | 5.2 | 0.46 | 0.46 | 2.3 | |
| Monensin | 6 | 8 | 4 | 2 | >5 | 0.84 | >5 | 0.035 | 2.8 | >0.02 | 1.4 | >0.4 | |
| X-206 | 0.25 | 1 | 0.5 | 0.25 | 0.52 | 1.6 | 0.94 | 0.059 | 0.50 | >0.05 | >0.05 | 1.3 | |
| Calci-my-cin | 0.094 | 0.13 | 0.13 | 0.13 | 0.41 | 0.40 | 0.37 | 0.22 | 0.62 | 0.46 | 0.15 | 1.8 | |
| Ion-omycin | 4 | 8 | 6 | 4 | 2.1 | 2.1 | 3.6 | 1.4 | 4.2 | 1.4 | 4.2 | 0.4 | |
| Narasin | 0.38 | 0.5 | 0.5 | 0.38 | 0.95 | 1.7 | 1.5 | 0.15 | 1.3 | 0.46 | 0.15 | 2.0 | |
| Ma-duramy-cin | 2 | 4 | 4 | 1 | 3.8 | 2.8 | 2.3 | 0.34 | 2.5 | 0.15 | 0.46 | 0.8 | |
| Nan-changmycin | 2 | 2 | 2 | 1 | 2.1 | 1.2 | 2.0 | 0.084 | 0.97 | 0.05 | 0.15 | 0.6 | |
| Synthetic | HL201 (6) | 2 | 2 | 2 | 1 | 17 | 6.6 | 7.9 | 17 | 19 | 13 | >13 | 5.7 |
| HL204 (29) | 2 | 2 | 2 | 1 | 13 | 6.1 | 6.8 | 12 | 14 | 13 | >13 | 4.6 | |
| HL160 (31) | >32 | >32 | >32 | >32 | >35 | 29 | 34 | >35 | >35 | >13 | >13 | ND | |
| HL224 (30) | 4 | 8 | 8 | 2 | 35 | 14 | 22 | 38 | 37 | 13 | >13 | 3.7 | |
| HL342 (32) | 4 | 8 | 8 | 2 | 7.1 | 3.7 | 5.9 | 7.8 | 8.5 | 4.2 | 13 | 0.9 | |
MIC-data are the averages of two biological repeats, each performed in technical triplicates (distinct wells). Cell viability data are the averages of two (HepG2, BJ, Vero) or three (MCF7 U-2OS) biological repeats, each performed in technical triplicates (distinct wells). The morphological profiling assays were conducted once in technical quadruplicates. In U-2OS cells (light grey columns, blue font) and Vero cells (dark gray columns, yellow font), the bioactivity threshold designates the first concentration where significant perturbation of the cells is observed using morphological profiling (see Extended data Fig. 4). The bioactivity threshold values should be compared to the respective IC50-values. Data for HL675A (39a) and HL675B (39b), which were inactive in all assays at the highest tested concentrations, has not been included in the table. See Supplementary Table 1 for all MIC and cell viability data including standard deviations.
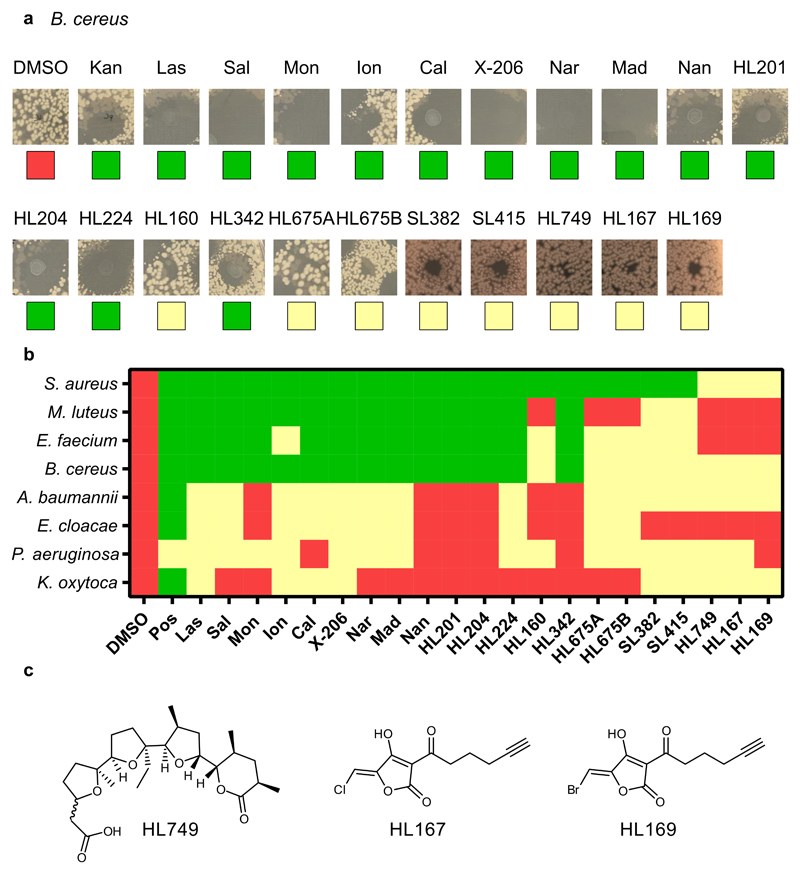
Hybrid polyether ionophores HL201 (6) and HL204 (29) did indeed maintain antibacterial activity with a potency comparable to most of the natural polyether ionophores against the tested gram-positive strains (wild-type B. cereus and both wild-type and drug-resistant variants of S. aureus). Analogs HL224 (30) and HL324 (32) were 2 to 4-fold less potent and HL160 (31), epimeric to 6 at C15, was completely devoid of activity (MIC >32 μg/mL). The monensin-derivatives HL675A/B (39a/39b) did not show appreciable anti-bacterial activity (MIC > 32 μg/mL, Extended Data Fig. 3, Supplementary Fig. 1, Supplementary Table 1). As expected, all of the natural polyether ionophores were active in the gram-positive strains and some, e.g. calcimycin, with very potent activity (Table 1). We also tested all compounds in E. coli and re-confirmed the general lack of activity of these compounds against gram negative strains as seen in the droplet screen although an E. Coli strain with increased permeability was sensitive to some ionophores at high concentrations (16-32 μg/mL, Supplementary Table 1).
The effects on mammalian cell viability, however, differentiated the synthetic compounds from the natural polyether ionophores. These trends can be seen in various ways, including a significantly increased sensitivity of BJ fibroblasts (8 - 27 fold, relative to HepG2) to a subset of the natural polyether ionophores salinomycin, monensin, X-206, narasin, maduramycin, and nanchangmycin (Table 1). Lasalocid, ionomycin, and calcimycin still displayed increased activity in BJ fibroblasts, but only marginally (1.5 – 3.5 fold). In comparison, HL201 (6) and HL204 (29) were quite inactive in all of the cell lines, with BJ fibroblasts among the least sensitive cell lines. This indicates different mechanistic behavior. Calculation of the antibacterial selectivity (ratio of average molar MICs in bacteria vs. IC50’s mammalian cells) revealed that HL201 (6) and HL204 (29) outperformed all of the natural polyether ionophores (selectivity ratios 5.7 and 4.6, respectively). Indeed, several of the natural products had a selectivity ratio below 1, which underscores their significant toxicity to mammalian cells. Finally, we used morphological profiling to provide a more sensitive assessment of compound-induced perturbation of mammalian cells. We used the mp-value46 to determine the first bioactive concentration for each compound in each cell line (Supplementary Discussion 2, Extended Data Fig. 4, Supplementary Fig. 3–9). HL201 (6) and HL204 (29) were inactive at all concentrations in Vero cells but active at the highest concentration (13 μM) in U-2OS cells, the ketone-reduced analog HL324 (32) had active profiles at 4 μM (U-2OS) and 13 μM (Vero), and the C15-epimer HL160 (31) as well as monensin derivatives 39a and 39b were inactive at all tested concentrations in both cell lines (Table 1, Extended Data Fig. 4, Supplementary Fig. 3–9). In contrast, the natural polyether ionophores afforded active profiles at concentrations significantly below their IC50 values in the two cell lines, typically by 5-10 fold but for some compounds, such as X-206, active profiles were detected even at the lowest concentration used in both cell lines. For several compounds active profiles could be observed that were not accompanied by any measurable loss in viability (Supplementary Discussion 2). Monensin displayed an interesting pattern of having a very wide span between the first bioactive concentration and the IC50-value in U-2OS cells (>250 fold) but not in Vero cells (2 fold). Collectively, our data suggests that HL201 (6) and HL204 (29) have strongly diminished effects on mammalian cells while maintaining antibacterial activity on par with many of the natural polyether ionophores.
Discussion
The cross-membrane ion transport activity of polyether ionophores depends on several factors. The association-dissociation dynamics for binding to different cations is influenced both by the molecular structure of the ionophore and by the specific lipid microenvironment. Furthermore, the equilibrium of membrane adsorption/desorption including the detailed molecular process of membrane insertion47 will determine the overall distribution – and thus potential ion-transport activity – of a polyether ionophore in any cellular system. Even in the relative simplicity of a bacterial cell this is a complex situation especially when considering the multiple, interrelated, ion-gradients that may be differentially affected by the ionophore. In mammalian cells, with their multiple endomembrane systems, the complexity increases significantly. Additionally, polyether ionophores could also have effects, that do not involve ion-transport, as has been recently demonstrated for salinomycin.48,49,50
Given the fact that the canonical ion-transport activity of polyether ionophores must be strongly dependent on the specific lipid composition, it should be possible to achieve selectivity for bacterial cells and thus increase the relative therapeutic window of the compounds. We are, however, not aware of prior attempts to systematically address this question. Here, we demonstrate, for the first time to our knowledge, that it is possible to make synthetic polyether ionophores that exceed the activities of a series of the naturally occurring compounds by head-to-head comparison of their relative inhibitory activities in several bacterial and mammalian cells. By additionally including a very sensitive assay technique such as morphological profiling, we have shown that these differences may be even more significant as the natural products cause disturbances in mammalian cells at very low (non-toxic) concentrations. In conclusion, our study suggests the exciting prospect of optimizing polyether ionophores for use as antibiotics.
Our data, however, has also illuminated how minute structural differences can result in e.g. loss of antibacterial selectivity (HL342, reduction of carbonyl group relative to HL201, selectivity ratio 0.9 vs. 5.7) or loss of antibacterial activity altogether (HL160, epimer relative to HL201) within this class of compounds. The loss of antibacterial selectivity displayed by HL324 is especially intriguing to us and by studying the mechanistic underpinnings we may learn how to prepare even more selective compounds in the future. Likewise, the lack of cellular activity of LH675A/B, despite preserving major structural elements of monensin and approximate distance to the cation-binding group, also need further studies and could be an opportunity to explore how to back-engineer ionophore activity into this scaffold. Collectively, our findings underscore the value of adopting a diversity-focused synthetic approach to these compounds which we suggest should be significantly expanded in the future. By starting from the industrially produced agricultural antibiotics or – alternatively – family members which have previously been fermented on large scale, it should, at least at the origin, be possible to scale the production of any derived synthetic polyether ionophore which would be required for further pre-clinical development. There are important challenges within this area ranging from finding new creative ways to recycle the components of the available polyether ionophores to studying the molecular details of the ion-transport process/membrane interactions using e.g. computational or spectroscopic tools.
Methods
Organic Synthesis
All reactions were conducted in flame-dried glassware under an atmosphere of argon unless otherwise stated. CH2Cl2, MeCN, THF and PhMe were dried over aluminium oxide via an MBraun SPS-800 solvent purification system. DCE, DMF, MeOH and pyridine were purchased as anhydrous. The dryness of solvents was controlled via Karl Fischer titration. Reagents were used as received from commercial suppliers unless otherwise stated (Sigma Aldrich, Merck, AK Scientific, Fluorochem, and TCI). Et3N and DIPEA were dried by stirring for at least 30 minutes over CaH2 followed by distillation onto preactivated molecular sieves (4 Å). Concentration in vacuo was performed using a rotary evaporator with the water bath temperature at 30 °C, or 40 °C, followed by further concentration using a high vacuum pump. TLC analysis was carried out on silica coated aluminum foil plates (Merck Kieselgel 60 F254). The TLC plates were visualized by UV irradiation and/or by staining with either CAM stain ((NH4)6Mo7O24·4H2O (10 g), Cerric ammonium sulfate (4 g), 10% H2SO4 (aq., 400 mL)), ninhydrin stain (ninhydrin (12 g) and AcOH (12 mL) in n-butanol (400 mL)) or KMnO4 stain (KMnO4 (5.0 g), 5 % NaOH (aq., 8.3 mL) and K2CO3 (33.3 g) in H2O (500 mL)). Molecular sieves were activated by drying in the oven at 120 °C for at least 24 hours, before they were heated in a microwave at maximum power for 2 minutes, followed by evaporation of the formed vapour on the high vacuum line. This was repeated 3-4 times, and finished by gently flame-drying the flask containing the molecular sieves. Flash column chromatography (FCC) was carried out using silica gel (230-400 mesh particle size, 60 Å pore size) as stationary phase. Infrared spectra (IR) were acquired on a PerkinElmer Spectrum TwoTM UATR. Mass spectra (HRMS) were recorded on a Bruker Daltonics MicrOTOF time-of-flight spectrometer with positive electrospray ionization, or negative ionization when stated. Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker BioSpin Co. AVANCE III 500 MHz spectrometer, a Varian Mercury 400 MHz spectrometer or a Bruker BioSpin GmbH 400 MHz spectrometer, running at 500/400 and 125/101 MHz for 1H and 13C, respectively. Chemical shifts (δ) are reported in ppm relative to the residual solvent signals (CDCl3: 7.26 ppm 1H NMR, 77.16 ppm 13C NMR, CD3OD: 3.31 ppm 1H NMR, 49.00 ppm 13C NMR, d6-DMSO: 2.50 ppm 1H NMR, 39.52 ppm 13C NMR. Multiplicities are indicated using the following abbreviations: s = singlet, d = doublet, t = triplet, q = quartet, h = heptet, m = multiplet, br = broad. Preparative HPLC purification were performed using a Gilson HPLC system.
All synthesis protocols and characterization data is available in the supplementary information.
Supplementary Discussion 1 – Aldol fragment coupling
With the aim of conducting the aldol coupling between ketone 4 and aldehyde 7, we first attempted formation of the (E)-boron enolate from 4 or the C25-OTES protected derivative 21 using (cHex)2BCl-NEt3 but useful conversion to the enolate could not be achieved. In fact, the only known aldol reactions using 4 were carried out in the classic syntheses of lasalocid 3 (ref. 13). and utilized in situ formation of the zinc-enolate from 4 which generates preferentially the configuration found in 3 – also anti-aldol-Felkin – and which differs from the configuration needed to make 6 at all of the three stereocenters. Although this aldol coupling utilized a different aldehyde compared to 7, we evaluated the possibilities for favoring the desired aldol product as being low. Consequently, we decided to instead rely on the Lewis acid catalyzed Mukaiyama aldol reaction which is known to maintain Felkin-selectivity.51 Importantly, double stereodifferentiation in Mukaiyama aldol reactions is established from studies by Evans38 thus providing a theoretical framework for the analysis of the key fragment coupling reaction. Despite the lack of very closely related reactions in the literature, we expected that formation of the desired anti-aldol isomer (14R,15R; anti 15,17-Me↔Et) would still be challenging due to the intrinsic preference for syn-aldol products in the Mukaiyama-aldol reaction and the combined diastereofacial bias from the stereocenters already present in aldehyde 7 and ketone 4. Formation of the anti-aldol products would demand use of the (Z)-silylenolate, whereas the corresponding (E)-enolate could be expected to reinforce the formation of a syn-aldol product (14R,15S; syn 15,17-Me↔Et).
Supplementary Discussion 2 – Cell Painting activity
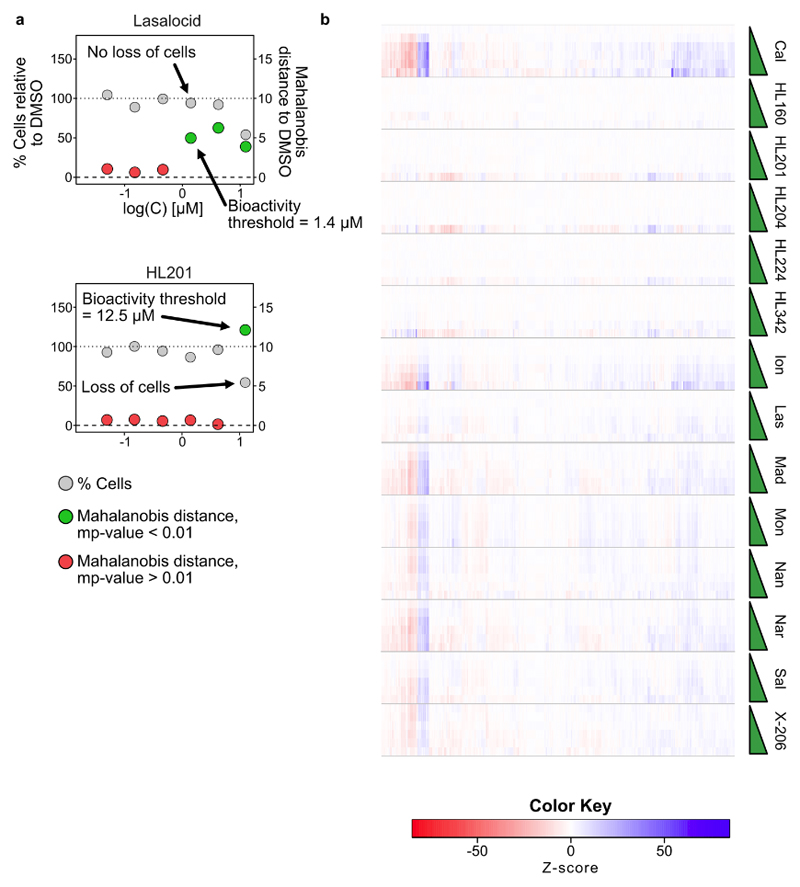
To determine bioactivity, we used the Mahalanobis distance and multidimensional perturbation value (mp-value) as described by Hutz et al.46 using non-normalized per-well profiles. For each batch of four 96-well plates was included 48 DMSO control wells and four replicates of treatment (one on each plate). We note that often only two principal components are kept by the mp-value workflow to describe > 90% variation of the 48 DMSO controls and one com-pound at one concentration. We also included quadruplicates of DMSO treated wells (i.e. we treated these additional DMSO wells as a compound by itself, with four replicates distributed over four plates) in U-2OS cells. We chose a stricter threshold for activity of mp-value < 0.01 instead of mp-value < 0.05. We based this on the fact that HL160 at two concentrations (0.46 μM and 4.2 μM) gave mp-values < 0.05 (0.042 and 0.022 respectively) but drops to inactive mp-values at higher concentrations (in U2OS cells). We also occasionally observe quadruplicates of DMSO wells with mp-values < 0.05 but > 0.01.
We used the mp-value measurement to determine the lowest concentration at which a compound displays a significantly active effect on the cells. Interestingly, for the natural polyether ionophores this was generally seen before any loss of cell viability was detected (Extended Data Fig. 4, Supplementary Fig. 5 and Supplementary Fig. 8) while active profiles for the hybrid compounds were always correlated to a loss of cell viability, indicating a general “toxic profile”.
Agar diffusion test
An agar diffusion test served as a primary screen for antimicrobial activity against Staphylo-coccus aureus DSM 20231, Micrococcus luteus DSM 20030, Enterococcus faecium DSM 20477, Bacillus cereus DSM 31, Acinetobacter baumannii DSM 300007, Enterobacter cloacae DSM 30054, Pseudomonas aeruginosa DSM 19880, and Klebsiella oxytoca DSM 5175. All test strains were grown aerated at 37 °C until an OD600 of 0.4-0.6 was reached. The bacterial cultures were diluted in 0.9% saline solution and inoculated on Müller-Hinton agar (MHA). The test compounds were dissolved (10 mM) in dimethyl sulfoxide (DMSO) and ten-microliter volumes were loaded on the inoculated MHA plates. DMSO served as negative control and kanamycin or ampicillin was used as positive control. The inoculated plates were incubated for 12-24 h at 37 °C and inspected for clearing zones. Antimicrobial activity was evaluated by the size of the clearing zone and test compounds were categorized as either inhibiting, slightly inhibiting or not inhibiting.
MIC-determination
MIC of all compounds was determined in Staphylococcus aureus DSM 20231, Staphylococcus aureus MRSA USA 300 je2, Staphylococcus aureus ATCC 29213 (weak β-lactamase producer), Bacillus cereus DSM 31, Escherichia coli BW25113 (wt) (from the Keio Collection, CGSC at Yale University), and Escherichia coli BW25113 ΔBamB ΔTolC (a gift from the laboratory of Prof. Gerry Wright at McMaster University). The bacterial cultures were grown aerated at 37 °C for 16-18 hours and diluted to a final density of 5x105 CFU/mL. The inoculation density was validated in parallel using agar plate counts. Stocks of the compounds were prepared in serial dilutions in DMSO (final conc. <5%) in the concentration range 32 μg/mL to 0.03 μg/mL (for monensin 16 μg/mL to 0.016 μg/mL). The MIC values were tested in 96-well microtiter plates using a microbroth dilution method in Luria Broth (Miller). The microtiter plates were incubated at 37 °C for 20-24 hours. Growth was measured as absorbance at 600 nm using a Tecan Nanoquant infinite M200Pro plate reader. The absorbance value correlating to a difference seen by the unaided eye was determined to 0.07. Measurements above this value is deemed as growth when determining the MIC values. Replicate measurements were performed in distinct wells.
Mammalian cell culture
U-2OS cells (ATCC HTB-96) were cultured in McCoy’s 5A (Sigma, Cat#: M9309) supplemented with fetal bovine serum (FBS, 10%, Gibco, Cat#: A3160802) and penicillin/streptomycin (1%, Sigma, Cat#: P4333). MCF-7 cells (ATCC HTB-22) were cultured in DMEM (Gibco, Cat#: 21969035) supple-mented with Ala-Gln (Sigma, Cat#: G8541), 10 μg/mL human recombinant insulin (Sigma, Cat#: I9278), FBS (10%) and penicillin/streptomycin (1%). HepG2 (ATCC HB-8065) and BJ cells (ATCC CRL-2522) were cultured in MEM (Gibco, Cat#: 41090-028) supplemented with FBS (10%), non-essential amino acids (Gibco, Cat#: 11140-035), sodium pyruvate (Gibco, Cat#: 11360-039) and penicillin/streptomycin (1%). Vero E6 (ATCC CRL-1586) were cultured in DMEM supplemented with Ala-Gln, FBS (10%) and penicillin/streptomycin (1%).
Cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. Every third-fourth day, the cells were washed in PBS (2×5 mL, Sigma, Cat#: D8537) and dissociated from the culture flask by trypsin-EDTA (Sigma, Cat#: T4049) and one fifth (BJ: one third) of the cells were reseeded in fresh complete medium in a T75 flask (Thermo Scientific, Cat#: 130190)
Cell Viability assay
MCF-7, Vero, U-2OS and HepG2 cells were seeded in black 96-well plates (Thermo Scientific, Cat#: 137101) at a density of 2000 cells/well in complete medium (75 μL/well). BJ cells were seeded as the other cell lines but at a density of 1500 cells/well. After seeding the cells were left to adhere to the substratum overnight. Compounds were dosed in the designated culture plates in triplicates as 4X solutions in 25 μL medium with a normalized DMSO concentration (0.5%) in all treatments, and the plates were placed at 37 °C in a humidified atmosphere containing 5% CO2. After 46.5 h, plates were added CellTiter-Blue (Promega G8081) (20 μL/well) and incubated for 90 min after which the plates were analyzed in a Tecan Spark 10 M multimode plate reader for fluorescence (552 ± 10 nm excitation; 598 ± 10 nm emission). Replicate measurements were performed in distinct wells. Average growth of treated cells was calculated by correcting fluorescence values for background fluorescence and subsequently normalized to the average of the two lowest treatments of the compound in question. Data was plotted and fitted to a four-parameter dose−response curve using Prism 8.4.2 for Windows (GraphPad Software, La Jolla, CA, www.graphpad.com).
Morphological profiling
The cell painting protocol is adapted from Bray et al 201644 to a 96-well plate format. In brief: Cells (U-2OS: 4000 cells/well; Vero E6: 5000 cells/well) were seeded into the inner 60 wells of a 96-well plate with optical bottom (Corning Cat#: 3603) in complete medium (75 μL) and incubated (37 °C, 5% CO2, humid) for 24 h. Compounds or DMSO were dosed in the designated culture plates in quadruplicates, distributed over 4 plates, as 4X solutions in 25 μL medium with a normalized DMSO concentration (0.5%). A total of 12 DMSO control wells were included on each plate for normalization. After 24 h, 75 μL medium was removed and re-placed with 75 μL complete medium containing 500 nM MitoTracker Deep Red (final C = 325 nM) and plates were incubated in the dark for 30 min. Wells were then aspirated and 75 μL medium were added, before adding 25 μL 16% paraformaldehyde (Electron Microscopy Sciences 15710-S) (final PFA = 4%) and incubating in the dark for 20 min. Plates were washed once with 1X HBSS (Invitrogen Cat#: 14065-056) and 75 μL 0.1% (vol/vol) Triton X-100 (BDH Cat#: 306324N) in 1X HBSS was added and incubated for 15 min in the dark. Plates were washed twice with 1X HBSS before addition of 75 μL multiplex staining solution (Hoechst 33342 (U-2OS: 5 μg/mL; Vero: 5 μg/mL), Concanavalin-Alexa Fluor 488 conjugate (U-2OS: 35 μg/mL; Vero: 8.75 μg/mL), SYTO 14 Green Fluorescence Nuclei Acid Stain (U-2OS: 3 μM; Vero: 0.75 μM), Phalloidin-Alexa Fluor 568 conjugate (U-2OS: 5 μL/mL; Vero: 2.5 μL/mL), Wheat-Germ agglutinin-Alexa Fluor 555 conjugate (U-2OS: 1.5 μg/mL, Vero: 0.75 μg/mL)) in HBSS containing 1% BSA (Sigma-Aldrich Cat# A9647) and incubation for 30 min in the dark. Plates were then washed three times with 1X HBSS with no final aspiration and imaged immediately in a Zeiss Celldiscoverer 7 automated microscope.
9 images are acquired in each well with 2x2 binning using the AxioCam 702 CMOS 12-bit camera with 4x analog gain in Zen 2.6 and 3.0 software for Celldiscoverer 7 using the imaging settings described in the Supplementary Information
To generate the bioactivity profiles the workflow outlined in Svenningsen & Poulsen45 was followed. In short, CellProfiler 2.1.152 was used to correct images for uneven illumination followed by image segmentation and extraction of 1476 features across nuclei, cytoplasm and the whole cell on a per-cell basis. Features were then averaged to per-well profiles after which the data was normalized on a per-plate basis followed by per-treatment aggregation which affords the final profiles using the cytominer 0.1.0 package53 in R 3.6.0 (ref 54).
The heatmap of morphological profiles is visualized with heatmap.2 in the gplots 3.0.3 package55. The Pearson correlation matrix is calculated using the stats package in R 3.6.0 and visualized using the corrplot 0.84 package56.
Hierarchical clustering of the correlation matrix is performed using the stats package using Pearson correlation coefficients as distance metric and average linkage method.
To determine activity scores and thresholds the workflow described by Hutz et al.46 was fol-lowed. We chose more stringent threshold of mp-value < 0.01 instead of mp-value < 0.05 (see Supplementary Discussion 2)
Extended Data
Extended Data Fig. 1. Strategies for the syn-thesis of protected 5-(halo)methylidene tetronic acid building blocks and subsequent coupling.
(a) Chemical structure of the targeted 3-acyltetronic acid-derivatives found in nonthmicin/ecteinamycin and 6. No previous syntheses of the halogenated variants have been reported. (b) Examples of known methods used to prepare non-halogenated variants. (c) Failed attempts and model studies toward 5-chloromethylidene tetronate. I) Unsuccessful synthesis of 5-tributylstannylmethylidene tetronate. II) Unsuccessful chlorination of 5-dimethylaminomethylidene tetronate. III) Direct and indirect chlorination of 5-methylidene tetronate methyl ether. In practice, the removal of the methyl group from the chlorinated tetronate product using LiCl/DMSO or BBr3 was difficult. Thus, we moved to synthesize the tetronate derivatives bearing a TMSE group, which can be removed under much milder condition (TBAF/DMF). DMSO = dimethylsulfoxide DCC = N,N’-dicyclohexylcarbodiimide, DMAP = 4-dimethylaminopyridine, Ms = methanesulfonyl, MOM = methoxymethyl, DCE = 1,2-dichloroethane, DBU = 1,8-diazabicyclo[5.4.0]undec-7-ene, TMS = trimethylsilyl, NCS = N-chlorosuccinimide, AIBN = azobisisobutyronitrile
Extended Data Fig. 2. Synthesis of hybrid polyether HL324 (32).
Preparation of analog HL324 (32) was performed through anti-selective (Evans-Saksena) reduction of the ketone-functionality of 26 followed by DCC-mediated coupling with 5-chloromethylidene tetronate. rt = room temperature, TBAF = tetrabutylammonium fluoride, DMF = N,N-dimethylformamide, DCC = N,N’-dicyclohexylcarbodiimide, DMAP = 4-dimethylaminopyridine, HPLC = high pressure liquid chromatography.
Extended Data Fig. 3. Droplet screen for anti-bacterial activity in B. cereus .
a) Representative images of inhibition zones of all com-pounds from B. cereus. b) Heatmap representing no (red), slight (yellow) or large (green) inhibition zones. All compounds were tested at 10 mM in DMSO except monensin (1 mM) and HL160 (7 mM). c) Chemical struc-tures of acid-containing synthetic fragments. SL382 = 27; SL415 = 26 – see Fig. 3.
Extended Data Fig. 4. Use of morphological profiling via cell painting to determine bioactivity threshold in U-2OS cells.
a) Plots of cell count (grey) and Mahalanobis distance (‘Bioactivity’; red and green) against concentration of compound. Green points indicate significant activity (mp-value < 0.01) while red points indicate no significant activity (mp-value > 0.01). Many ionophores induce an active profile without loss of cell viability while bioactivity of the hybrid ionophores is correlated with a loss of cells, indicating a profile representing toxicity. The bioactivity threshold is determined as the first concentration to reach significance (mp-value < 0.01) b) Dose-dependent morphological profiles from which the bioactivity threshold is determined. Note that some ionophores change profile at higher concentrations, e.g. calcimycin (Cal) and nan-changmycin (Nan), typically correlated with a loss of cells. See Supplementary Fig. 3–9 for representative images, all dose-response plots, profiles and correlation matrices in both U-2OS and Vero cells.
Supplementary Material
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 865738). Financial support from the Carlsberg foundation (grant CF17-0800), Novo Nordisk Foundation (grant NNF19OC0054782) and Independent Research Fund Denmark (grant 9040-00117B and grant 6110-00600B) is acknowledged. We are grateful to Erik Jung and Anja Johnbeck for technical assistance with organic synthesis, Anders Bodholt Nielsen for technical assistance with NMR spectroscopy, and Iben Charlotte Stensgaard Jensen for technical assistance with microbiology. Correspondence and requests for materials should be addressed to Thomas B. Poulsen (thpou@chem.au.dk).
Footnotes
Author contributions
TBP conceived and supervised the study. TBP, SL, HL, EBS designed experiments. SL, HL, JMV, and CNP performed organic synthesis. EBS and KMJ conducted cell biological experiments and analyzed data. MW and FDA conducted microbiology experiments. TT supervised microbiology experiments and analyzed data. PN carried out X-ray crystallographic analyses. TBP, SL, HL, and EBS wrote the manuscript with input from all authors. SL and HL contributed equally to this work.
Competing interests
The authors declare no competing interests.
Data availability
MIC and IC50-values for cell viability for each biological replicate is available in Supplementary Table 1. The morphological profiling data (aggregated profiles and correlation matrix), included in Supplementary Fig. 3–9, has been deposited at Mendeley Data (DOI 10.17632/jv3sjk8wy4.2). The images from morphological profiling are too large to be deposited at Mendeley Data (which has a limit of 10 GB per data set), but can be obtained upon request. The X-ray crystallography data have been deposited in the Cambridge Crystallographic Data Centre (CCDC) using the following identifiers (www.ccdc.cam.ac.uk/structures/): 1920656 (compound 6-Na), 1920657 (compound 14), 1920658 (compound 24), 1920659 (compound 29-Na) and 1920660 (compound 31-Na).
Code availability
Code/scripts for analysis of morphological profiling data and Cell Profiler pipelines has been deposited at Mendeley Data (DOI 10.17632/jv3sjk8wy4.2).
References
- 1.Huffman BJ, Shenvi RA. Natural products in the ‘marketplace’: Interfacing synthesis and biology. J Am Chem Soc. 2019;141:3332–3346. doi: 10.1021/jacs.8b11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 3.Wetzel S, Bon RS, Kumar K, Waldmann H. Biology-oriented synthesis. Angew Chem Int Ed. 2011;50:10800–10826. doi: 10.1002/anie.201007004. [DOI] [PubMed] [Google Scholar]
- 4.Könst ZA, et al. Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides. Nat Chem. 2017;9:1140–1149. doi: 10.1038/nchem.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson RM, Danishefsky SJ. Small molecule natural products in the discovery of therapeutic agents: the synthesis connection. J Org Chem. 2006;71:8329–8351. doi: 10.1021/jo0610053. [DOI] [PubMed] [Google Scholar]
- 6.Huigens RW, et al. A ring-distortion strategy to construct stereochemically complex and structurally diverse compounds from natural products. Nat Chem. 2013;5:195–202. doi: 10.1038/nchem.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbasov ME, et al. Simplified immunosuppressive and neuroprotective agents based on gracilin A. Nat Chem. 2019;11:342–350. doi: 10.1038/s41557-019-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karageorgis G, Foley DJ, Laraia L, Waldmann H. Principle and design of pseudo-natural products. Nat Chem. 2020;12:227–235. doi: 10.1038/s41557-019-0411-x. [DOI] [PubMed] [Google Scholar]
- 9.Seiple IB, et al. A platform for the discovery of new macrolide antibiotics. Nature. 2016;533:338–345. doi: 10.1038/nature17967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter MF, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westley J, editor. Polyether antibiotics - naturally occurring acid ionophores. 1st. Marcel Dekker; New York: 1982. [Google Scholar]
- 12.Dutton CJ, Banks BJ, Cooper CB. Polyether ionophores. Nat Prod Rep. 1995;12:165–181. doi: 10.1039/np9951200165. [DOI] [PubMed] [Google Scholar]
- 13.Nakata T, et al. A Total synthesis of lasalocid A. J Am Chem Soc. 1978;100:2933–2935. [Google Scholar]
- 14.Fukuyama T, et al. Total synthesis of monensin. 3. Stereocontrolled total synthesis of monensin. J Am Chem Soc. 1979;101:262–263. [Google Scholar]
- 15.Faul MM, Huff BE. Strategy and methodology development for the total synthesis of polyether ionophore antibiotics. Chem Rev. 2000;100:2407–2473. doi: 10.1021/cr940210s. [DOI] [PubMed] [Google Scholar]
- 16.Song Z, Lohse AG, Hsung RP. Challenges in the synthesis of a unique mono-carboxylic acid antibiotic, (+)-zincophorin. Nat Prod Rep. 2009;26:560–571. doi: 10.1039/b821450f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasun ZA, Gao X, Lipinski RM, Krische MJ. Direct generation of triketide stereopolyads via merged redox-construction events: Total synthesis of (+)-zincophorin methyl ester. J Am Chem Soc. 2015;137:8900–8903. doi: 10.1021/jacs.5b05296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Krische MJ. Total synthesis of (+)-SCH 351448: Efficiency via chemoselectivity and redox-economy powered by metal catalysis. J Am Chem Soc. 2016;138:8088–8091. doi: 10.1021/jacs.6b04917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L-A, Ashley MA, Leighton JL. Evolution of an efficient and scalable nine-step (longest linear sequence) synthesis of zincophorin methyl ester. J Am Chem Soc. 2017;139:4568–4573. doi: 10.1021/jacs.7b01590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Lin S, Jacobsen KM, Poulsen TB. Chemical syntheses and chemical biology of carboxyl polyether ionophores: Recent highlights. Angew Chem Int Ed. 2019;58:13630–13642. doi: 10.1002/anie.201812982. [DOI] [PubMed] [Google Scholar]
- 21.Kevin II DA, Meujo DA, Hamann MT. Polyether ionophores: broad-spectrum and promising biologically active molecules for the control of drug-resistant bacteria and parasites. Expert Opin Drug Discov. 2009;4:109–146. doi: 10.1517/17460440802661443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman HD, Jeffers TK, Williams RB. Forty years of monensin for the control of coccidiosis in poultry. Poultry Sci. 2010;89:1788–1801. doi: 10.3382/ps.2010-00931. [DOI] [PubMed] [Google Scholar]
- 23.Goodrich RD, et al. Influence of monensin on the performance of cattle. J Anim Sci. 1984;58:1484–1498. doi: 10.2527/jas1984.5861484x. [DOI] [PubMed] [Google Scholar]
- 24.Antoszczak M, et al. Biological activity of doubly modified salinomycin analogs - evaluation in vitro and ex vivo. Eur J Med Chem. 2018;156:510–523. doi: 10.1016/j.ejmech.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi Y, et al. Nonthmicin, a polyether polyketide bearing a halogen-modified tetronate with neuroprotective and antiinvasive activity from Actinomadura sp. Org Lett. 2017;19:1406–1409. doi: 10.1021/acs.orglett.7b00318. [DOI] [PubMed] [Google Scholar]
- 26.Wyche TP, et al. Chemical genomics, structure elucidation, and in vivo studies of the marine-derived anticlostridial ecteinamycin. ACS Chem Biol. 2017;12:2287–2295. doi: 10.1021/acschembio.7b00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westley JW, Evans RH, Williams T, Stempel A. Structure of antibiotic X-537A. J Chem Soc D. 1970:71–72. [Google Scholar]
- 28.Westley JW, Evans RH, Williams T, Stempel A. Pyrolytic cleavage of antibiotic x-537A and related reactions. J Org Chem. 1973;38:3431–3433. doi: 10.1021/jo00959a050. [DOI] [PubMed] [Google Scholar]
- 29.Gruenfeld N, et al. Angiotensin converting enzyme inhibitors: 1-glutarylindoline-2-carboxylic acids derivatives. J Med Chem. 1983;26:1277–1282. doi: 10.1021/jm00363a012. [DOI] [PubMed] [Google Scholar]
- 30.Hiyama T, Kimura K, Nozaki H. Chromium (II) mediated threo selective synthesis of homoallyl alcohols. Tetrahedron Lett. 1981;22:1037–1040. [Google Scholar]
- 31.Hansen TM, et al. Highly chemoselective oxidation of 1,5-diols to δ-lactones with TEMPO/BAIB. Tetrahedron Lett. 2003;44:57–59. [Google Scholar]
- 32.Brazeau J-F, et al. Stereocentrolled synthesis of C1-C17 fragment of narasin via a free radical-based approach. Org Lett. 2010;12:36–39. doi: 10.1021/ol902414u. [DOI] [PubMed] [Google Scholar]
- 33.Zografos AL, Georgiadis D. Synthetic strategies towards naturally occurring tetronic acids. Synthesis. 2006:3157–3188. [Google Scholar]
- 34.Markó IE, Richardson PR, Bailey M, Maguire AR, Coughlan N. Selective manganese-mediated transformations using the combination: KMnO4/Me3SiCl. Tetrahedron Lett. 1997;38:2339–2342. [Google Scholar]
- 35.Sabbah M, Bernollin M, Doutheau A, Soulère L, Queneau Y. A new route towards fimbrolide analogues: Importance of the exomethylene motif in LuxR dependent quorum sensing inhibition. Med Chem Commun. 2013;4:363–366. [Google Scholar]
- 36.Roush WR. Concerning the diastereofacial selectivity of the aldol reactions of α-methyl chiral aldehydes and lithium and boron propionate enolates. J Org Chem. 1991;56:4151–4157. [Google Scholar]
- 37.Masamune S, Choy W, Petersen JS, Sita LR. Double asymmetric synthesis and a new strategy for stereochemical control in organic synthesis. Angew Chem Int Ed. 1985;24:1–30. [Google Scholar]
- 38.Evans DA, Yang MG, Dart MJ, Duffy JL, Kim AS. Double stereodifferentiating lewis acid-promoted (Mukaiyama) aldol bond constructions. J Am Chem Soc. 1995;117:9598–9599. [Google Scholar]
- 39.Nicolaou KC, Estrada AA, Zak M, Lee SH, Safina BS. A mild and selective method for the hydrolysis of esters with trimethyltin hydroxide. Angew Chem Int Ed. 2005;44:1378–1382. doi: 10.1002/anie.200462207. [DOI] [PubMed] [Google Scholar]
- 40.Vögtle F, Weber E. Multidentate acyclic neutral ligands and their complexation. Angew Chem Int Ed. 1979;18:753–776. [Google Scholar]
- 41.Evans D, Chapman K, Carreira EM. Directed reduction of β-hydroxy ketones employing tetramethylammonium triacetoxylborohydride. J Am Chem Soc. 1988;110:3560–3578. [Google Scholar]
- 42.Cai D, Still WC. Synthesis of monensin. Reconstruction from degradation products. J Org Chem. 1988;53:4641–4643. [Google Scholar]
- 43.Baskaran S, Islam I, Raghaven M, Chandrasekaran S. Pyridinium chlorochromate in organic synthesis. A facile and selective oxidative cleavage of enol ethers. Chem Lett. 1987;16:1175–1178. [Google Scholar]
- 44.Bray M-A, et al. Cell painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc. 2016;11:1757–1774. doi: 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svenningsen EB, Poulsen TB. Establishing cell painting in a smaller chemical biology lab – A report from the frontier. Bioorg Med Chem. 2019;27:2609–2615. doi: 10.1016/j.bmc.2019.03.052. [DOI] [PubMed] [Google Scholar]
- 46.Hutz JE, et al. The multidimensional perturbation value: A single metric to measure similarity and activity of treatments in high-throughput multidimensional screens. J Biomol Screen. 2013;18:367–377. doi: 10.1177/1087057112469257. [DOI] [PubMed] [Google Scholar]
- 47.Golbek TW, Schmüsen L, Rasmussen MH, Poulsen TB, Weidner T. Lasalocid acid antibiotic at a membrane surface probed by sum frequency generation spectroscopy. Langmuir. 2020;36:3184–3192. doi: 10.1021/acs.langmuir.9b03752. [DOI] [PubMed] [Google Scholar]
- 48.Versini A, et al. Chemical biology of salinomycin. Tetrahedron. 2018;74:5585–5614. [Google Scholar]
- 49.Mai TT, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem. 2017;9:1025–1033. doi: 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F, et al. Nucleolin is a functional binding protein for salinomycin in neuroblastoma stem cells. J Am Chem Soc. 2019;141:3613–3622. doi: 10.1021/jacs.8b12872. [DOI] [PubMed] [Google Scholar]
- 51.Heathcock CH, Flippin LA. Acyclic stereoselection. 16. High diastereofacial selectivity in Lewis acid mediated additions of enol silanes to chiral aldehydes. J Am Chem Soc. 1983;105:1667–1668. [Google Scholar]
- 52.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfilerTM: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 53.Becker T, Goodman A, McQuin C, Rohban M, Singh S. cytominer: Methods for Image-Based Cell Profiling. 2017 R package version 0.1.0 Available at: https://cran.r-project.org/package=cytominer. [Google Scholar]
- 54.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. Available at: https://www.R-project.org/ [Google Scholar]
- 55.Warnes GR, et al. gplots: Various R Programming Tools for Plotting Data. 2020 R package version 3.0.3 Available at: https://cran.r-project.org/package=gplots. [Google Scholar]
- 56.Wei T, Simko V. R package ‘corrplot’: Visualization of a Correlation Matrix. 2017 R package versionversion 084 Available at: https://github.com/taiyun/corrplot. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MIC and IC50-values for cell viability for each biological replicate is available in Supplementary Table 1. The morphological profiling data (aggregated profiles and correlation matrix), included in Supplementary Fig. 3–9, has been deposited at Mendeley Data (DOI 10.17632/jv3sjk8wy4.2). The images from morphological profiling are too large to be deposited at Mendeley Data (which has a limit of 10 GB per data set), but can be obtained upon request. The X-ray crystallography data have been deposited in the Cambridge Crystallographic Data Centre (CCDC) using the following identifiers (www.ccdc.cam.ac.uk/structures/): 1920656 (compound 6-Na), 1920657 (compound 14), 1920658 (compound 24), 1920659 (compound 29-Na) and 1920660 (compound 31-Na).
Code/scripts for analysis of morphological profiling data and Cell Profiler pipelines has been deposited at Mendeley Data (DOI 10.17632/jv3sjk8wy4.2).