Summary
The mitotic kinase AURORA-A is essential for cell cycle progression and is considered a priority cancer target. While the catalytic activity of AURORA-A is essential for its mitotic function, recent reports indicate an additional non-catalytic function, which is difficult to target by conventional small molecules. We therefore developed a series of chemical degraders (PROTACs) by connecting a clinical kinase inhibitor of AURORA-A to E3 ligase-binding molecules (e.g. thalidomide). One degrader induced rapid, durable and highly specific degradation of AURORA-A. In addition ,we found that the degrader complex was stabilized by cooperative binding between AURORA-A and CEREBLON. Degrader-mediated AURORA-A depletion caused an S-phase defect, which is not the cell cycle effect observed upon kinase inhibition, supporting an important non-catalytic function of AURORA-A during DNA replication. AURORA-A degradation induced rampant apoptosis in cancer cell lines, and thus represents a versatile starting point for developing new therapeutics to counter AURORA-A function in cancer.
Keywords: Alisertib, CEREBLON, MLN8237, MLN8054, thalidomide, PROTAC, Degronimid, AURORA-A, cancer, targeted protein degradation, S-phase
Introduction
Multicellular organisms depend on mechanisms that ensure tight control of cell cycle progression during all stages of their life. These control mechanisms are disrupted during malignant transformation, and the resulting deregulated cell cycle is a hallmark of cancer1. As a consequence, drugs that target enzymes driving cell cycle progression, such as cyclin-dependent kinases (CDKs), are being explored for cancer therapy, and some have now entered clinical practice2.
The eukaryotic cell cycle is controlled not only by CDKs but also by mitotic kinases, so named since their activities peak in mitosis3. One of these is AURORA-A kinase, which is part of a small gene family comprising AURORA-A, -B and -C, and is homologous to the yeast kinase Ipl1 (increase-in-ploidy 1). While AURORA-C is mainly active in the testis, AURORA-A and -B are essential for cell cycle progression in most cell types4. AURORA-A and -B phosphorylate multiple proteins during mitosis5 and the catalytic activity of both kinases is necessary for progression throughout mitosis6.
AURORA-A is a priority cancer target. Its gene, AURKA, is a bona fide oncogene that maps to a chromosomal region frequently amplified in epithelial malignancies7,8. Overexpression of AURORA-A transforms immortalized fibroblasts in culture and enables their engraftment in nude mice, directly documenting the enzyme’s oncogenic potency8,9. Ectopic expression of AURORA-A in mammary epithelial cells forms tumors in genetic mouse models, albeit with long latencies10,11. Importantly, inhibition or depletion of AURORA-A can be synthetically lethal with oncogene activation or loss of tumor suppressor genes12–15. Thus, there is a compelling rationale for targeting AURORA-A in cancer.
Potent kinase inhibitors of AURORA-A have been developed,16 and several are being tested clinically. One inhibitor that has progressed to phase III testing is alisertib (MLN8237)17,18. One trial demonstrated significant benefits of AURORA-A inhibition19, but others reported low response rates,18,20,21 raising the question of which mechanism(s) determine therapeutic efficacy.
Besides its well-characterized catalytic activities, AURORA-A is supposed to have also non-catalytic functions. AURORA-A binds to proto-oncoproteins of the MYC family and protects N-MYC15,22 and c-MYC23 from proteasomal degradation; this effect is independent of AURORA-A’s kinase activity. In C. elegans, AURORA-A-mediated stabilization of spindle microtubules is also independent of catalytic activity24. Finally, AURORA-A facilitates a stem cell-like phenotype in breast cancer cells (again independent of kinase activity25), indicating that kinase inhibitors may not abrogate all oncogenic activities of AURORA-A. While many AURORA-A inhibitors bind to the active state of the kinase (type-1)12, others including alisertib26 and CD53227 alter the conformation of the kinase domain and therefore may also disrupt non-catalytic functions. Whether combined targeting of catalytic and non-catalytic functions of AURORA-A is more effective than solely inhibiting its catalytic functions remains to be tested.
An elegant way of simultaneously targeting catalytic and non-catalytic function is possible using bifunctional small molecules such as PROTACs (proteolysis targeting chimeras)28 or Degronimids29. These degraders comprise two chemical moieties: one binds a target of interest while the other binds a cellular E3-ubiquitin ligase such as CEREBLON30 or von Hippel–Lindau tumor suppressor (VHL)31. Depending on the connecting linker, degraders induce productive proximity between the target and the E3 ligase, resulting in target clearance by the ubiquitin/proteasome system32.
Here, we developed a potent AURORA-A degrader by linking alisertib, to the CEREBLON-binding molecule thalidomide. We found that degrader-mediated depletion is highly specific for AURORA-A, as complex formation was supported by cooperative binding between CEREBLON and AURORA-A. Strikingly, degrader-mediated chemical knockdown caused a strong S-phase arrest, which was not observed with ATP-competitive AURORA-A kinase inhibitors, demonstrating that target degradation can cause markedly different phenotypes from that obtained with conventional small molecule inhibitors. Exposure of cancer cell lines to degrader resulted in the strong induction of apoptosis. Our AURORA-A degrader is therefore a powerful new tool for studying scaffolding as well as catalytic functions of AURORA-A and for developing a new class of drugs to counter AURORA-A functions in cancer.
Results
Design of bifunctional AURORA-A degrader molecules
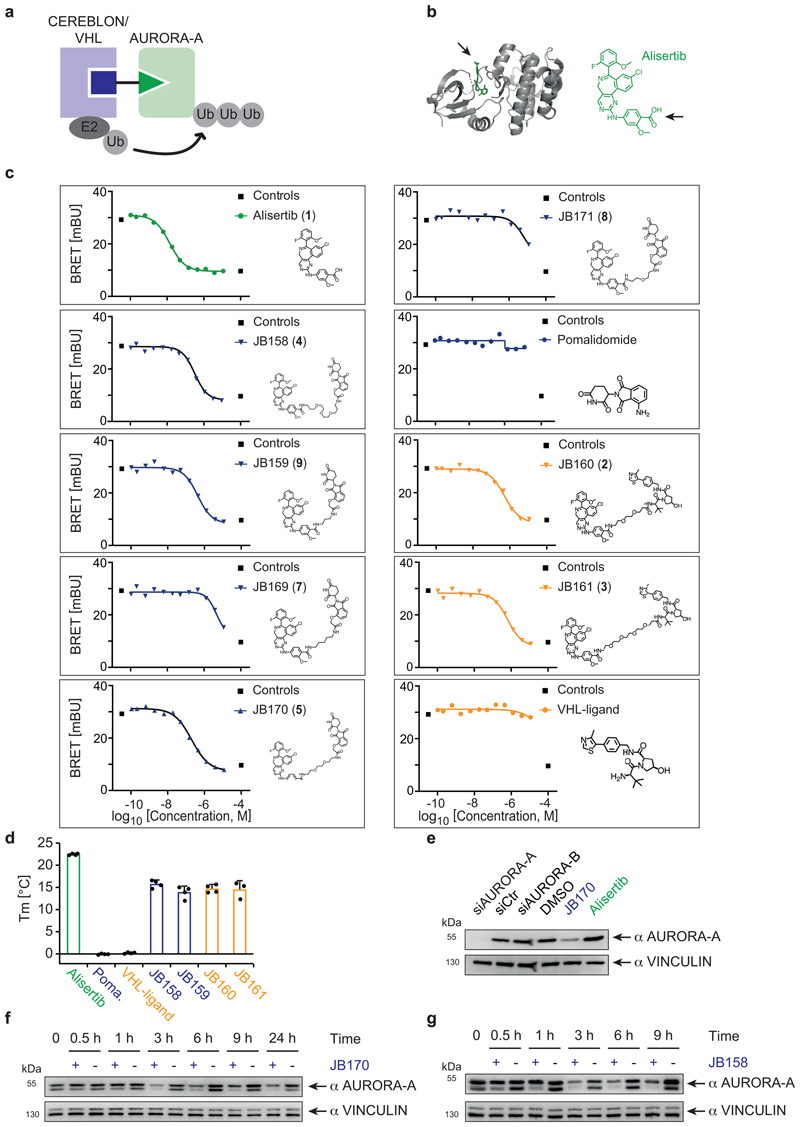
For the development of AURORA-A degraders (Extended Data Fig. 1a), we chose alisertib (MLN8237, 1), a canonical type-1 ATP mimetic kinase inhibitor with known binding mode, high potency for AURORA-A, and ease of derivatization. Crystal structures of AURORA-A in complex with alisertib´s derivative, MLN8054 (PDB 2X81, 2WTV)33,34, revealed a solvent-exposed carboxyl group, to which we attached linkers to connect it to E3 ligase-binding molecules (Extended Data Fig. 1b). We designed AURORA-A-targeting chimeric degraders by linking alisertib to CEREBLON- and VHL-binding moieties through various (poly)ethylene glycol (PEG) and aliphatic linkers (Extended Data Fig. 1c). We synthesized five candidate degraders containing thalidomide as a CEREBLON-recruiting chemical scaffold and two with HIF1-derived peptidomimetics that bind VHL (Supplementary Note 1). The structure and purity of the compounds were confirmed by HPLC, mass spectrometry and NMR (Supplementary Note 1).
JB170 and JB158 induce AURORA-A proteolysis
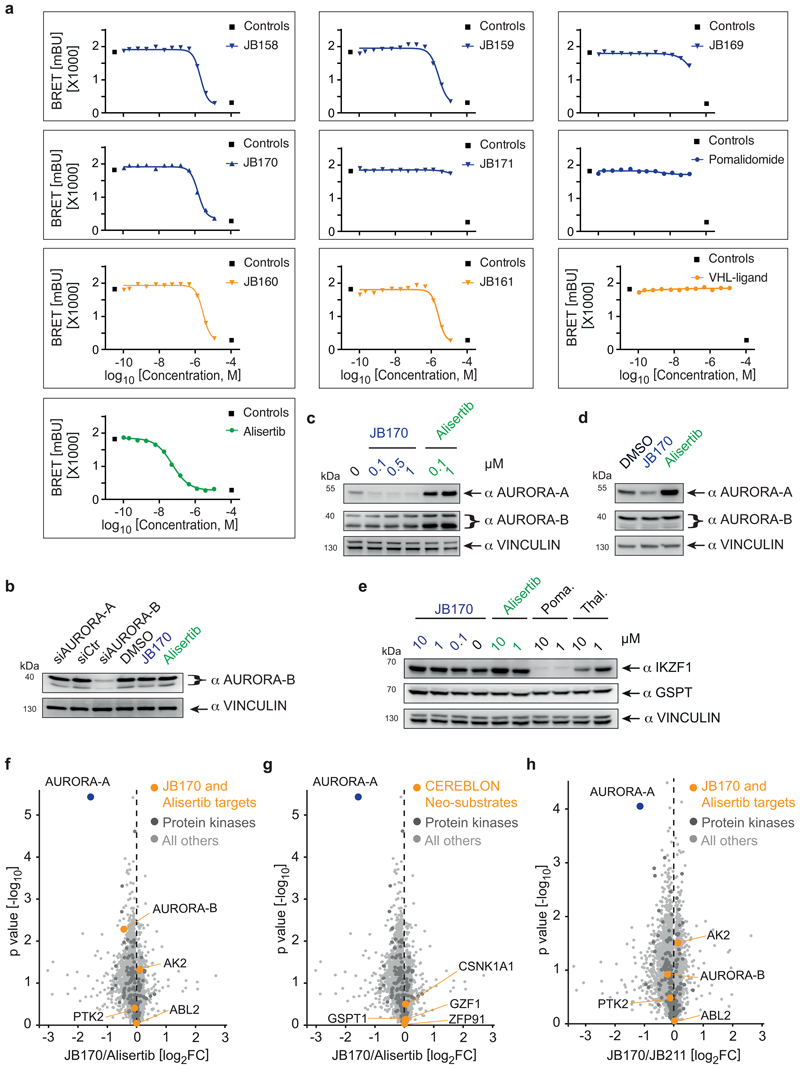
First, we tested whether attachment of an E3 ligase-binding moiety affected the binding of alisertib to AURORA-A in cells. We expressed AURORA-A fused to a minimal luciferase (NanoLuc) and measured bioluminescence resonance energy transfer (BRET) to a fluorescent tracer that bound the AURORA-A active site (Figure 1a, Extended Data Fig. 1c). Five tested molecules replaced the tracer with EC50 values of 193-471 nM, but two degraders with relatively short aliphatic linkers were unable to replace the tracer at nanomolar concentrations (Supplementary Table 1), indicating that either they could not enter cells or their binding to AURORA-A was impaired. We confirmed binding of the compounds to AURORA-A by thermal shift assays with purified AURORA-A protein (Extended Data Fig. 1d).
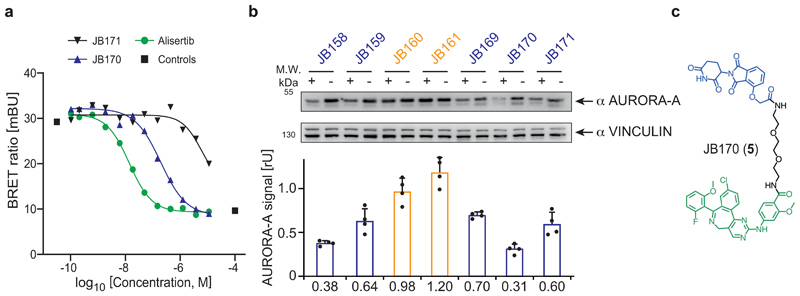
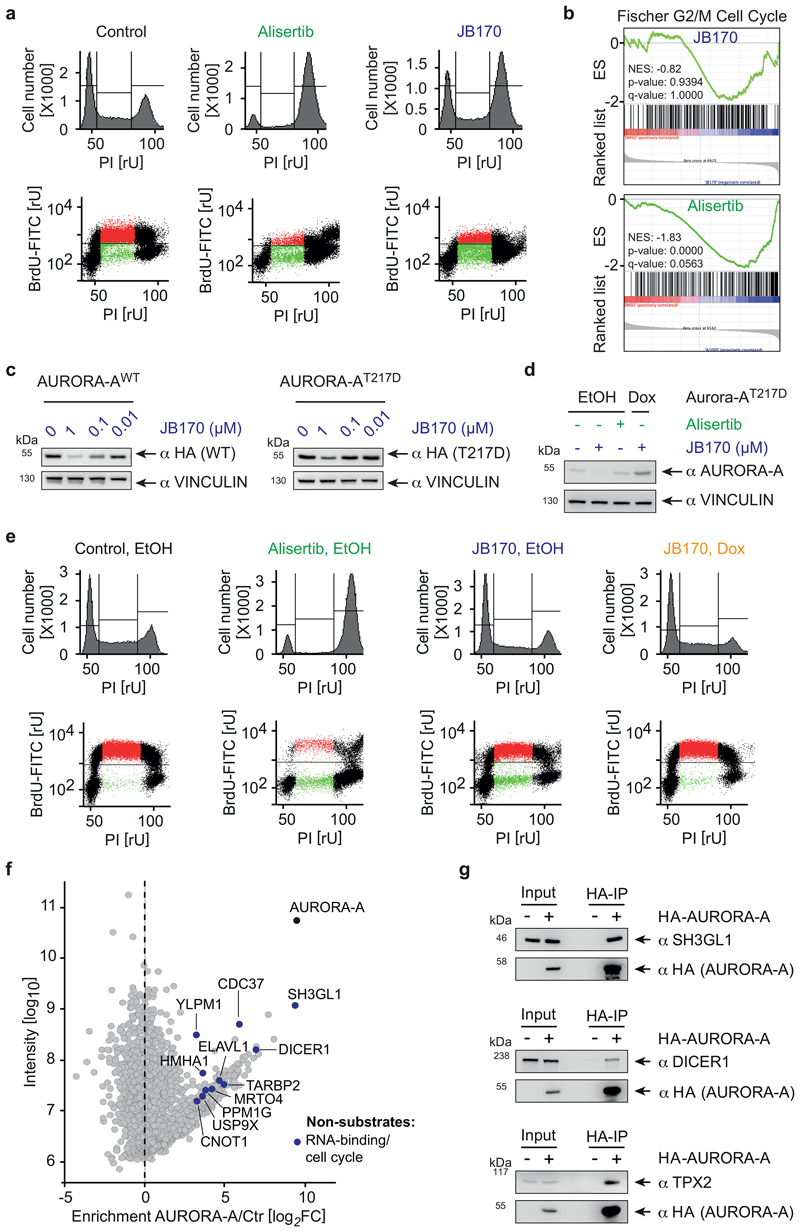
Figure 1. Bifunctional degrader molecules induce depletion of AURORA-A in cells.
(a) AURORA-A-NanoLuc target engagement assay. HEK293 cells transfected with an AURORA-A-NanoLuc fusion construct were incubated with various concentrations of alisertib, JB170 or JB171 together with an energy transfer probe for 2 hours and luminescence was measured. The graph shows one of the two biological replicates. (b) Immunoblot and quantification of AURORA-A. MV4-11 cells were treated with VHL- (orange) and thalidomide-based (blue) degraders, and AURORA-A levels were compared to control cells by immunoblotting (top). JB158, JB160, JB161, JB169 and JB170 were used at 0.1 µM whereas JB159 and JB171 at 1 µM. Vinculin was used as a loading control (as in all other immunoblotting experiments). Bar diagram (bottom) shows cellular AURORA-A levels upon degrader treatment. Bars represent mean ± s.d. of n = 4 biological replicates. (c) Structure of the most potent degrader, JB170.
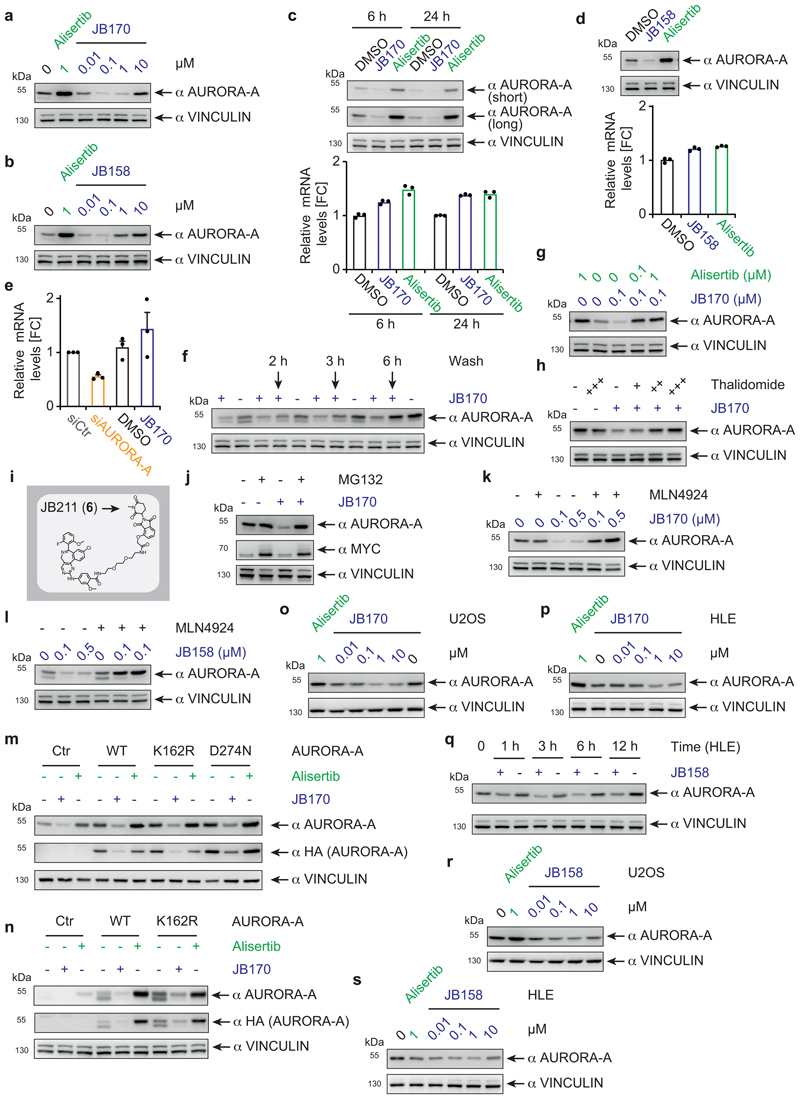
Subsequently, we tested the ability of the seven bifunctional molecules to mediate formation of a productive AURORA-A–E3 ligase complex, by immunoblotting AURORA-A in MV4-11 leukemia cells treated with a single dose of each compound (Figure 1b). Specificity of the anti-AURORA-A antibody was confirmed by siRNA (Extended Data Fig. 1e). While neither of the VHL-based degraders (JB160, 2 and JB161, 3) reduced steady-state AURORA-A levels (Fig. 1b), two CEREBLON-based degraders strongly reduced AURORA-A levels (JB158, 4 62% reduction; JB170, 5 69% reduction) and were selected for further study. These two degraders consisted of alisertib and thalidomide linked via amides by two (JB170; Fig. 1c) or three (JB158) ethylene glycol molecules. Both compounds led to a rapid decrease in cellular AURORA-A levels that was clearly seen after 3 hours (Extended Data Fig. 1f,g).
To further characterize these two compounds, we first determined the optimal concentration for AURORA-A depletion in MV4-11 cells by immunoblotting. JB170 at 10 nM reduced AURORA-A levels modestly, while substantial degradation was observed at 100 nM and 1 µM (Extended Data Fig. 2a). Non-conjugated alisertib did not decrease AURORA-A levels, but instead substantially increased it. We next quantified JB170 potency using a luciferase detection system. We stably expressed AURORA-A fused to a luciferase fragment in MV4-11 cells, treated the cells with various concentrations of JB170, and analyzed luciferase activity as a measure for AURORA-A levels (Figure 2a). Maximal depletion of AURORA-A (DCmax) was achieved at 300 nM JB170 and the half-maximal degradation concentration (DC50) was 28 nM (+/-4 nM). JB170 concentrations ≥10 µM were ineffective in depleting AURORA-A, in both immunoblot and luminescence assays. This observation has already been made for other degraders/PROTACs ("Hook effect"), and is expected because depletion depends on ternary complex formation35. Similar results were obtained with JB158 (Extended Data Fig. 2b).
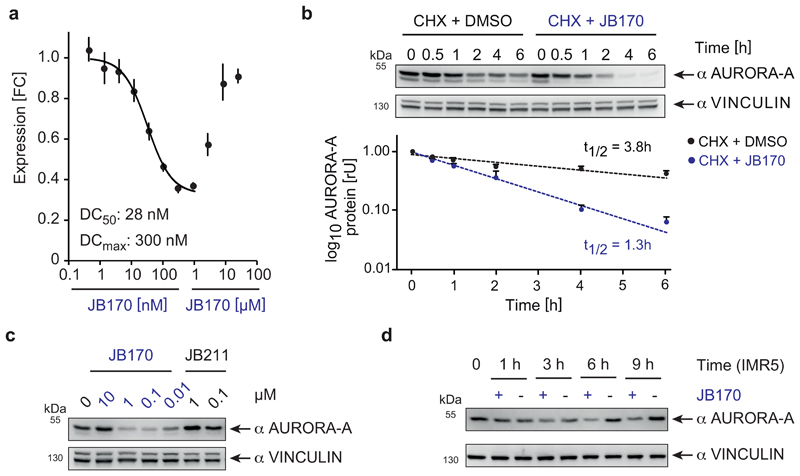
Figure 2. JB170 reduces AURORA-A levels by inducing proteolysis.
(a) AURORA-A levels based on luciferase measurements. AURORA-A was fused to luciferase fragment (HiBiT) and expressed in MV4-11 cells. Cells were treated with different concentrations of JB170 for 6 hours, lysed, complemented with the second luciferase fragment (largeBiT), and measured for luciferase activity (DC50: half maximal degradation concentration, DCmax: maximal degradation concentration). DC50 is calculated with the sigmoidal dose-response (four parameters) equation using only the lower eight concentrations. Data represent mean ± s.d. of n = 3 replicates. (b) Immunoblot and quantification of AURORA-A levels. Protein stability of AURORA-A was analyzed by incubating JB170-treated IMR5 cells and control cells for 0.5, 1, 2, 4, and 6 hours with cycloheximide (CHX). AURORA-A levels were quantified from three biological replicates, and AURORA-A half-life (t1/2) in the presence and absence of JB170 was estimated by linear regression. Data represent mean ± s.e.m. of n = 3 biological replicate experiments. (c) Immunoblot of AURORA-A. MV4-11 cells were treated for 6 hours with different concentrations of JB170 and the related compound JB211 (chemical formula is shown in Extended Data Fig. 2i), which unlike JB170 does not bind CEREBLON. (d) Immunoblot of AURORA-A. IMR5 cells were treated with JB170 (0.1 µM) for the indicated time periods.
To rule out that the compounds reduced AURORA-A levels by lowering AURORA-A mRNA levels, we harvested MV4-11 cells after degrader treatment and quantified mRNA by quantitative PCR. While both JB170 (Extended Data Fig. 2c) and JB158 (Extended Data Fig. 2d) induced depletion of cellular AURORA-A protein, AURORA-A mRNA levels were not reduced. We confirmed the specificity of the PCR using an siRNA targeting AURORA-A (Extended Data Fig. 2e). We then analyzed whether JB170 reduced AURORA-A protein stability, by incubating cells with cycloheximide and JB170 (or vehicle) and estimating AURORA-A levels by immunoblotting at several time points (Fig. 2b). JB170 decreased the half-life of AURORA-A from 3.8 to 1.3 hours. We concluded that JB170 reduces cellular AURORA-A levels by inducing its degradation.
A PROTAC/degrader-like mechanism for JB170 and JB158 was supported by additional observations. First, AURORA-A depletion was reversible in wash-out experiments (Extended Data Fig. 2f). Second, JB170-mediated depletion depended on binding of JB170 to AURORA-A and CEREBLON, as co-incubation of MV4-11 cells with alisertib or thalidomide (Extended Data Fig. 2g,h) abolished its degrader activity. We further tested the specificity of JB170 by synthesizing a compound (JB211, 6) that resembles JB170 but cannot bind to CEREBLON due to a methyl residue at the glutarimide ring of thalidomide (Extended Data Fig. 2i). As expected, JB211 did not deplete AURORA-A, but enhanced AURORA-A protein levels (Fig. 2c), similar to that observed for alisertib. Importantly, both proteasomal inhibition by MG132 and inhibition of cullin neddylation by MLN4924 completely blocked JB170- and JB158-mediated depletion of AURORA-A (Extended Data Fig. 2j,k,l). We concluded that JB170 and JB158 are efficient degraders that induce AURORA-A ubiquitylation by CEREBLON followed by proteolysis via the proteasome.
We wondered if degrader-mediated depletion required AURORA-A catalytic activity, as the phosphorylation status and therefore the activity of AURORA-A varies during the cell cycle. We therefore expressed enzymatically inactive versions of HA-tagged AURORA-A (AURORA-AD274N, AURORA-AK162R)36,37 and found that they were depleted by JB170 to a similar degree as the unmutated protein (Extended Data Fig. 2m,n), indicating that catalytic activity was not required for degrader-mediated depletion of AURORA-A.
We then tested whether degrader-induced AURORA-A depletion was restricted to leukemia cells. We treated human osteosarcoma (U2OS), hepatocellular carcinoma (HLE) and neuroblastoma (IMR5) cell lines with JB170 or JB158, and observed rapid AURORA-A depletion in all cases (Fig. 2d, Extended Data Fig. 2o-s). However, the degrees of depletion, their DC50 values and the concentrations at which the Hook effect became visible varied significantly among the cell lines, potentially due to different cellular concentrations of CEREBLON and AURORA-A. We concluded that JB158 and JB170 induce strong and rapid AURORA-A degradation by a degrader/PROTAC-like mode of action in various cancer cell lines.
JB170 is highly specific for AURORA-A
To investigate the specificity of JB170, we first analyzed if alisertib and JB170 bind proteins other than AURORA-A, by Kinobead selectivity profiling38 in MV4-11 cell lysates. This assay uses immobilized broad-spectrum kinase inhibitors as affinity matrix to enrich kinases and other ATP-binding proteins from samples; titration with competitive inhibitors prevents enrichment, allowing the determination of apparent binding affinities for different proteins (Fig. 3a, Supplementary Dataset 1). Expectedly, alisertib bound AURORA-A with highest affinity (Kd app=7 nM), but also interacted with AURORA-B (Kd app =90 nM), ACAD10 (Kd app =19 nM), ABL2 (Kd app =214 nM), AK2 (Kd app =215 nM) and other kinases. JB170 bound AURORA-A with the lowest Kd (Kd app =99 nM), followed by ACAD10 (Kd app =253 nM) and AURORA-B (Kd app =5.1 µM).
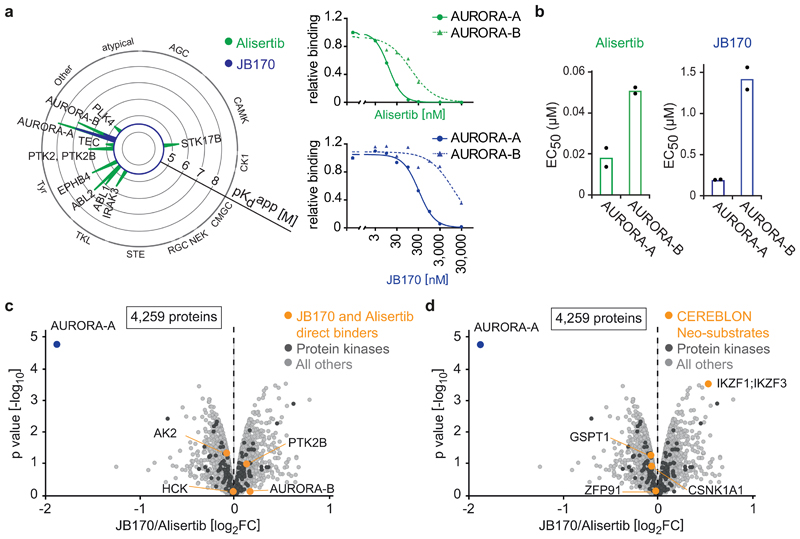
Figure 3. JB170 is highly specific for AURORA-A.
(a) Radar plot and dose-response profile for alisertib and JB170. Radar plot shows the Kinobead selectivity profile of alisertib and JB170 in a lysate of MV4-11 cells. Each spike represents a protein target; binding affinities are depicted as pKd app (negative decadic logarithms of the apparent Kd app value). The MS-based dose-response profiles (right) show binding curves after non-linear regression and the derived apparent Kd for AURORA-A and AURORA-B for alisertib (top) and JB170 (bottom)38. (b) Bar diagram showing EC50 values of AURORA-B-NanoLuc target engagement assay for alisertib (left) and JB170 (right). Values are presented as individual points from n = 2 biological replicates of which one is shown in Fig. S3a. (c,d) Volcano plot showing changes in protein abundance. MV4-11 cells were treated with JB170 (0.1 µM) or alisertib (0.1 µM) for 6 hours, and proteins were analyzed by triple-SILAC mass spectrometry. The X-axis displays the relative abundance of all identified proteins (4,259) in JB170-treated vs. alisertib-treated cells (log2FC). The Y-axis displays the p-value (-log10) from triplicate experiments (p-values were calculated from biological triplicates by the limma package). AURORA-A and other alisertib-binding proteins (c) or neosubstrates of CEREBLON (d) are labeled (orange).
AURORA-B is structurally and functionally related to AURORA-A, so we compared alisertib binding to these proteins in living cells. We analyzed affinities of alisertib and the degrader molecules by measuring BRET to a fluorescent tracer (Extended Data Fig. 3a, Supplementary Table 1). Both alisertib (AURORA-A EC50=18 nM, AURORA-B EC50=51 nM) and JB170 (AURORA-A EC50=193 nM, AURORA-B EC50=1.4 µM) preferentially bound AURORA-A over AURORA-B (Fig. 3b). We concluded that, at high concentrations, alisertib and JB170 bind to more proteins than just AURORA-A, and that their affinities for AURORA-A and AURORA-B differ 3- to 50-fold depending on experimental conditions.
Next, we determined whether JB170-mediated degradation was specific to AURORA-A or if other alisertib-binding proteins were also degraded. MV4-11 cells were treated with JB170 or alisertib, and changes in protein content were determined by SILAC mass spectrometry. JB170 decreased AURORA-A levels by 73% (p=1.68*10-05) compared to alisertib-treated cells (Fig. 3c) and by 57% (p=4.15*10-05) compared to untreated cells. Strikingly, of the 4,259 proteins reliably detected in this experiment, no additional protein was depleted (p<0.05, log2FC<-1) (Fig. 3c, Supplementary Dataset 2), including AURORA-B. We confirmed this observation by treating cells with various concentrations of JB170 and analyzing AURORA-B levels by immunoblotting after having confirmed the specific detection of AURORA-B by siRNA (Extended Data Fig. 3b,c,d).
In JB170, the E3 ligase-recruiting moiety is thalidomide, an immunomodulatory imide that induces the binding of “neosubstrates” to CEREBLON, leading to their ubiquitylation and degradation. We therefore wondered if JB170 depleted any proteins known to be neosubstrates of CEREBLON. While mass spectrometry identified several such proteins (GSPT1, CSNK1A1, ZFP91 and IZKF1/3), none was destabilized by JB170 (Fig. 3d). This result was confirmed by immunoblotting for GSPT and IZKF1 (Extended Data Fig. 3e).
We analyzed the potency and specificity of JB170 by further quantitative mass spectrometry (Supplementary Dataset 3). These experiments confirmed, in another cell line (IMR5), that JB170 induces the specific, efficient degradation of AURORA-A compared to alisertib, and gave similar results when the effects of JB170 were compared to JB211 which resembles JB170 but cannot bind CEREBLON (Extended Data Fig. 3f,g,h). We concluded that JB170-mediated depletion is highly specific to AURORA-A.
Ternary complex formation is supported by cooperativity
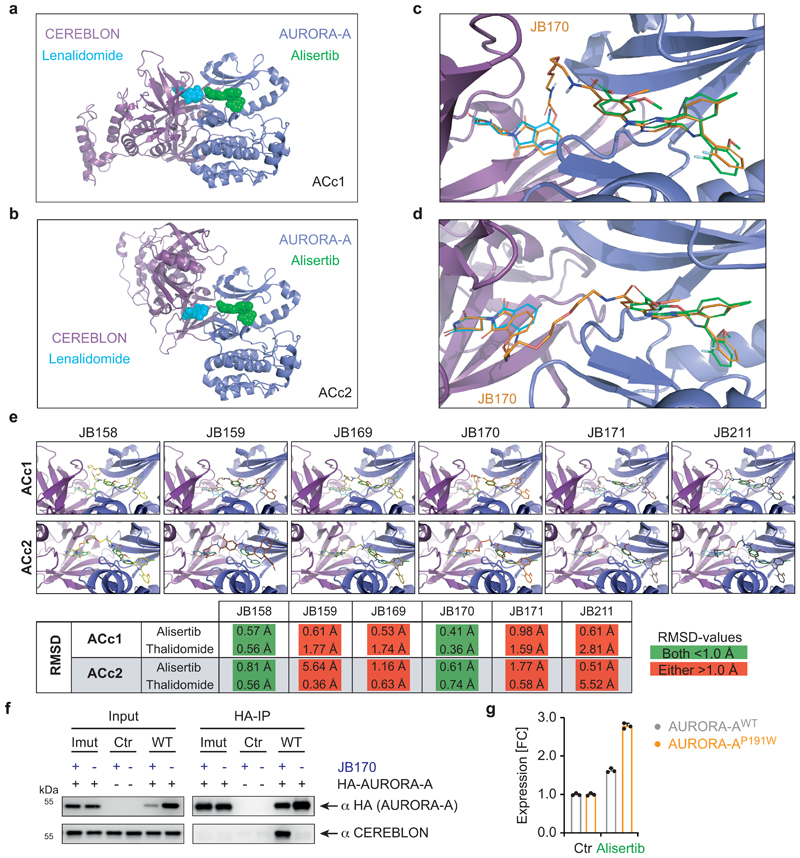
To understand if interactions between AURORA-A and CEREBLON contribute to the high specificity of JB170 for AURORA-A, we used in silico modeling. Based on crystal structures of AURORA-A in complex with MLN8054 (PDB 2X81)33 and of CEREBLON in complex with lenalidomide (PDB 4TZ4)39, we did a protein-protein docking study of AURORA-A and CEREBLON in the absence of JB170. Two complexes were compatible with JB170 binding to its designated binding sites, namely the complex that scored best, AURORA-A-CEREBLON complex 1 (ACc1), and the complex that ranked fifteenth (ACc2). Both complexes had an extensive interface with good shape complementarity, and it was possible to dock the two moieties of JB170 to their known binding sites without major rearrangements (Extended Data Fig. 4a,b,c,d). A subsequent docking study with all thalidomide-based compounds revealed that, besides JB170, only JB158 could be placed in complexes ACc1 and ACc2 without major repositioning of thalidomide and alisertib (Extended Data Fig. 4e). All other compounds required large deviations (>1 Å) of their top-ranking docking poses. We concluded that structural conditions in the ternary complex could explain the distinct degrader efficacies observed.
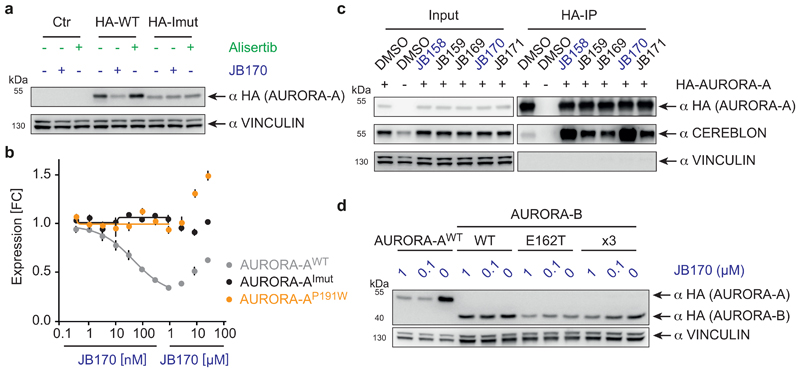
Figure 4. Protein-protein interactions between AURORA-A and CEREBLON support ternary complex formation.
(a) Immunoblot of HA-tagged AURORA-A. MV4-11 cells expressing HA-tagged wildtype AURORA-A (WT), cells expressing a mutated version of AURORA-A (AURORA-AImut) with 12 amino acid substitutions (R137E, K153E, K156E, F157E, I158E, R189E, P191W, K224E, E239R, S266W, A267W and R375E), or cells transduced with an empty vector (control) were treated with 1 µM JB170 or alisertib for 6 hours. (b) AURORA-A levels based on luciferase measurements. Wildtype (WT), AURORA-A with one amino acid substitution (AURORA-AP191W) or AURORA-A with 12 amino acid substitutions (R137E, K153E, K156E, F157E, I158E, R189E, P191W, K224E, E239R, S266W, A267W and R375E, AURORA-AImut) were fused to luciferase fragment (HiBiT) and expressed in MV4-11 cells. Cells were treated with different concentrations of JB170 for 6 hours, lysed, complemented with the second luciferase fragment (largeBiT), and measured for luciferase activity. Data represent mean ± s.d. of n = 3 replicates. (c) Immunoblots of immunoprecipitation experiments. AURORA-A was precipitated with an anti-HA tag antibody from MV4-11 cells stably expressing HA-tagged AURORA-A or control cells transduced with an empty vector after cell lysates were incubated with 0.5 µM degrader. The amount of co-precipitated CEREBLON was analyzed by immunoblotting. (d) Immunoblot of HA-tagged AURORA-A and -B. MV4-11 cells expressing HA-tagged wildtype AURORA-A, wildtype AURORA-B (WT), or mutated versions of AURORA-B mimicking the active site of AURORA-A (AURORA-BE162T; AURORA-BR160L,E162T,K165R, x3) were treated with different concentrations of JB170 for 6 hours.
To investigate how protein-protein interactions support ternary complex formation of AURORA-A, CEREBLON and JB170, molecular dynamics were simulated for both models. Side chain energy contributions to the interactions between AURORA-A and CEREBLON were analyzed to identify residues critical for ternary complex formation. To experimentally validate our in silico binding modes, we constructed an HA- and luciferase-tagged version of AURORA-A with 12 mutated amino acids (R137E, K153E, K156E, F157E, I158E, R189E, P191W, K224E, E239R, S266W, A267W and R375E) in the presumed interface with CEREBLON (AURORA-AImut). When this protein was stably expressed in MV4-11 cells, it was not depleted by JB170, as shown by immunoblotting and luciferase assays (Figure 4a,b), and it did not co-immunoprecipitate with CEREBLON (Extended Data Fig. 4f). To confine the number of amino acids important for ternary complex formation, we expressed additional AURORA-A mutants with fewer changed amino acids. Strikingly, a single change in the predicted interface with CEREBLON (AURORA-AP191W) abolished JB170-mediated depletion (Fig. 4b), but retained stabilization by alisertib, indicating that the protein maintained binding to the kinase ligand of JB170 (Extended Data Fig. 4g). These mutagenesis experiments support the in silico predicted protein-protein interactions and their importance for JB170-mediated AURORA-A degradation.
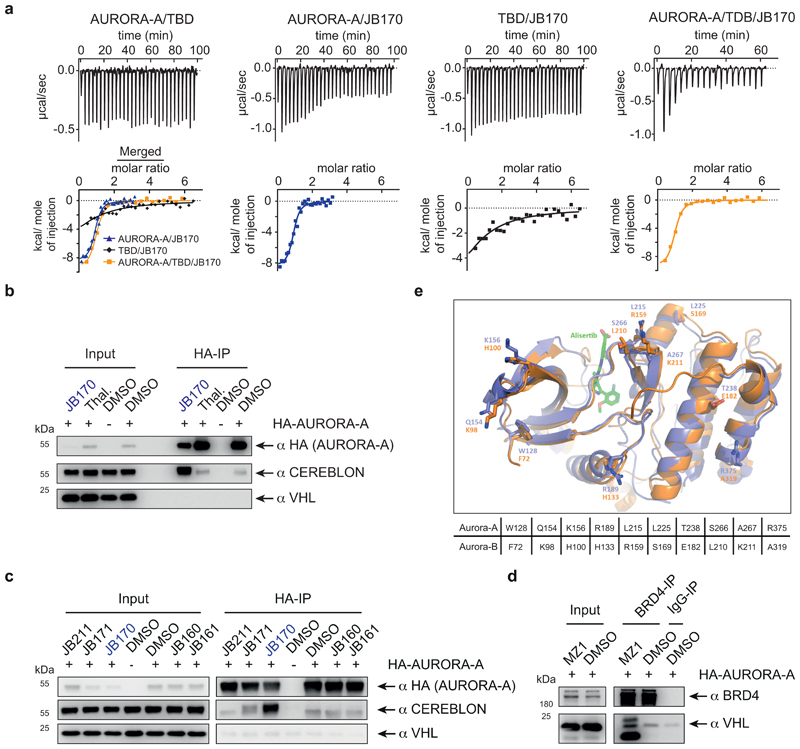
For the BRD4 PROTAC MZ1, ternary complex formation is rate limiting for degrader efficiency and depends on the cooperative binding of proteins in the complex40,41. We therefore measured the affinity of JB170 to CEREBLON and AURORA-A by isothermal titration calorimetry (ITC). For these experiments, we purified the kinase domain of AURORA-A and the thalidomide-binding domain (TBD) of CEREBLON. ITC revealed that JB170 had significantly higher affinity to the complex of two proteins (183 nM +/- 10 nM, n = 2) than to AURORA-A (375 nM +/- 22 nM, n = 4) or CEREBLON (6.88 µM +/- 0.5 µM, n = 3) alone (Extended Data Fig. 5a).
To determine which degraders induce ternary complex formation similar to JB170, we incubated them with MV4-11 cell extracts and then immunoprecipitated HA-tagged AURORA-A. While efficient CEREBLON co-precipitation was observed with JB170 or JB158, lower amounts co-precipitated with the less efficient degraders JB169 (7), JB171 (8), and JB159 (9) (Fig. 4c). This correlation between degrader efficacy and ability to induce ternary complex formation indicates cooperativity as a central feature of potent degraders.
Unexpectedly, we noted that small but significant amounts of CEREBLON co-precipitated with AURORA-A from lysates not incubated with any degrader (Fig. 4c, Extended Data Fig. 5b), indicating binding of AURORA-A to CEREBLON under unperturbed conditions. Differently, VHL did not co-precipitate with AURORA-A, but it did with BRD4 in the presence of the VHL-based PROTAC MZ142 (Extended Data Fig. 5c,d).
Intriguingly, several residues in the interface between AURORA-A and CEREBLON are not conserved in AURORA-B43 (Extended Data Fig. 5e), and protein-protein docking suggested a less stable complex of CEREBLON with AURORA-B (GBVI-score -8.80 kcal/mol) than AURORA-A (GBVI-scores: ACc1, -10.72 kcal/mol; ACc2, -9.21 kcal/mol). We hypothesized that different residues in AURORA-A and AURORA-B at the interface with CEREBLON contributed to the specificity of JB170 in addition to their divergent affinities to alisertib. To test this hypothesis, we constructed two AURORA-B swap mutants, which mimic the active site of AURORA-A (AURORA-BE162T, AURORA-BR160L,E162T,K165R)34,44, and were expected to have affinities to alisertib similar to those of AURORA-A. In line with a cooperative binding model, JB170-mediated degradation was strongly attenuated in mutants with respect to AURORA-A (Fig. 4d). Therefore, interactions between CEREBLON and AURORA-A support degrader-induced complex formation and JB170 specificity.
Degradation of AURORA-A perturbs S-phase progression
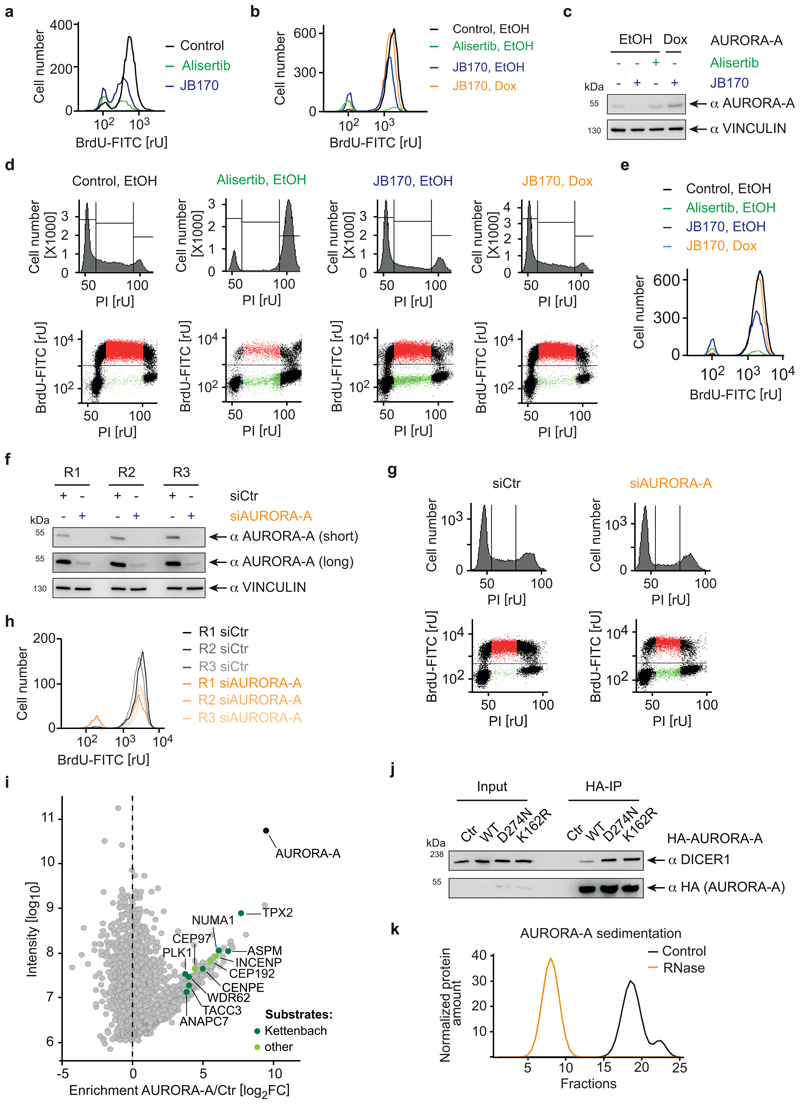
AURORA-A is mainly described as a mitotic kinase4, but reported functions in S-phase may be independent of its catalytic activity45. We therefore analyzed the cell cycle distribution of MV4-11 cells incubated with JB170 or alisertib by BrdU/PI flow cytometry. Consistent with published data17, almost all cells arrested in G2/M after 12 hours of alisertib treatment (Fig. 5a, Extended Data Fig. 6a). In contrast, JB170 treatment induced little accumulation of cells in G2/M. Instead, a major population of cells in S-phase stopped incorporating BrdU, indicating that JB170 delayed or arrested S-phase progression. Consistently, RNA sequencing showed that alisertib induced the expression of genes indicative of G2/M-arrested cells, while the same genes were not activated by JB170 (Figure 5b).
Figure 5. Degrader-mediated depletion and kinase inhibition of AURORA-A induce distinct cellular phenotypes.
(a) Cell cycle distribution analyzed by flow cytometry. MV4-11 cells were treated with alisertib (1 µM) or JB170 (0.5 µM) for 12 hours. Cells were labeled with BrdU, stained with propidium iodide (PI) and analyzed by flow cytometry. The amount of intercalating PI (top) and the correlation of BrdU to PI (bottom) are shown. (b) GSEA enrichment plot. MV4-11 cells were treated with JB170 (0.1 µM) or alisertib (1 µM) for 18 hours. cDNA was prepared and subjected to Illumina sequencing (RNA-sequencing). Gene expression of JB170- and alisertib-treated cells was compared to untreated cells by GSEA (gene set enrichment analysis) and enrichment of the gene-set “Fischer G2/M Cell Cycle” is shown. Vertical black bars indicate the position of genes in the ranked gene list; the enrichment score is shown as a green line (NES, normalized enrichment score). In this analysis, the nominal P-value was calculated using an empirical phenotype-based permutation test procedure. The permutation-based false-discovery rate (FDR) Q value was generated by correcting for gene set size and multiple hypothesis testing. (c) Immunoblot of HA-tagged AURORA-A. IMR5 cells expressing HA-tagged mutant AURORA-AT217D or wildtype AURORA-A (WT) were treated with different concentrations of JB170 for 24 hours. (d) Immunoblot of AURORA-A. IMR5 cells expressing AURORA-AT217D upon incubation with doxycycline (Dox) or control cells (EtOH) were treated with JB170 (1 µM) or alisertib (1 µM) for 18 hours. (e) Cell cycle distribution analyzed by flow cytometry. IMR5 cells expressing AURORA-AT217D upon incubation with doxycycline (Dox) or control cells (EtOH, as shown in Fig. 4d) were treated with JB170 (1 µM) or alisertib (1 µM) for 18 hours. Cells were labeled with BrdU, stained with PI, and analyzed by flow cytometry. The amount of intercalating PI (top) and the correlation of BrdU to PI (bottom) are shown. (f) Scatter plot of the AURORA-A interactome with novel binding proteins. The X-axis displays the enrichment (log2FC) of proteins in HA-AURORA-A-expressing cells compared to control cells (Ctr). The Y-axis displays the protein intensities (log10). All AURORA-A-associated proteins are listed in Supplementary Dataset 4. (g) Immunoblots of HA-tagged AURORA-A and interacting proteins. HA precipitations were performed from HEK293 cells transfected to express HA-tagged AURORA-A or control cells. Exogenous AURORA-A was detected with an anti-HA tag antibody, and levels of SH3GL1, DICER1 and TPX2 were analyzed in the input and immunoprecipitant by antibodies recognizing endogenous proteins.
The observation that AURORA-A kinase inhibition and depletion caused distinct cellular phenotypes was surprising and raised the question if the phenotypes reflected an on-target activity of JB170. We therefore performed a genetic rescue experiment using an AURORA-A functional mutant (T217D) with reduced affinity for the alisertib analogue MLN805433. We expressed doxycycline-inducible HA-tagged versions of AURORA-AT217D and the wildtype protein in IMR5 cells. As expected, JB170-mediated depletion of AURORA-AT217D was largely abrogated (Figure 5c). Using this system, we first analyzed the effects of kinase inhibition and depletion without ectopic expression of AURORA-A in IMR5 cells. JB170 induced an accumulation of BrdU-negative cells in S-phase, but did not induce G2/M arrest, while alisertib led to G2/M arrest (Fig. 5d,e, Extended Data Fig. 6b). Strikingly, expression of AURORA-AT217D abolished the JB170-mediated S-phase arrest, indicating that it is exclusively caused by depletion of AURORA-A. JB170 specificity was further confirmed by similar experiments in which we rescued the JB170-mediated S-phase arrest by overexpressing wildtype AURORA-A (Extended Data Fig. 6c,d,e). Furthermore, we depleted AURORA-A by siRNAs and observed an accumulation of BrdU-negative cells in S-phase, similar to JB170-treated cells (Extended Data Fig. 6f,g,h). We concluded that AURORA-A kinase inhibition and depletion induced distinct cellular phenotypes and that JB170-mediated S-phase arrest is caused specifically by AURORA-A depletion.
Different cellular consequences of AURORA-A inhibition and degradation indicated kinase-independent functions. So far, AURORA-A kinase substrates have been characterized5, but AURORA-A-interacting proteins that mediate kinase-independent functions have not been systematically studied. We therefore stably expressed HA-tagged AURORA-A in MV4-11 cells, isolated AURORA-A-associated proteins by HA immunoprecipitation, and analyzed them by quantitative label-free mass spectrometry. Relative to control cells not expressing HA-tagged proteins, this analysis identified 248 proteins (log2FC >3, Supplementary Dataset 4) that were strongly enriched in the AURORA-A precipitate, including a multitude of known AURORA-A substrates and the cofactor TPX2 (Extended Data Fig. 6i). Intriguingly, however, several of the AURORA-A-interacting proteins had never been depicted as substrates (Fig. 5f). Two of them, DICER1 and SH3GL1, were as strongly enriched as TPX2; we confirmed their binding to AURORA-A by co-immunoprecipitation (Fig. 5g). We concluded that AURORA-A interacts with non-substrate proteins, and speculate that these interactions mediate a kinase-independent function of AURORA-A. Consistent with this hypothesis, DICER1 co-precipitated with kinase-dead versions of AURORA-A to an extent similar to or even greater than it did with wildtype AURORA-A (Extended Data Fig. 5j). DICER1 is part of the microprocessor complex. Strikingly, other members of this complex, namely TARBP2, YLPM1 and, to a lesser degree, AGO2 and DROSHA, were also detected in the AURORA-A interactome, indicating that AURORA-A forms large protein complexes. Indeed, it was observed earlier that AURORA-A is part of large RNase-sensitive complexes (Extended Data Fig. 6k)46.
Depletion of AURORA-A induces apoptosis in cancer cells
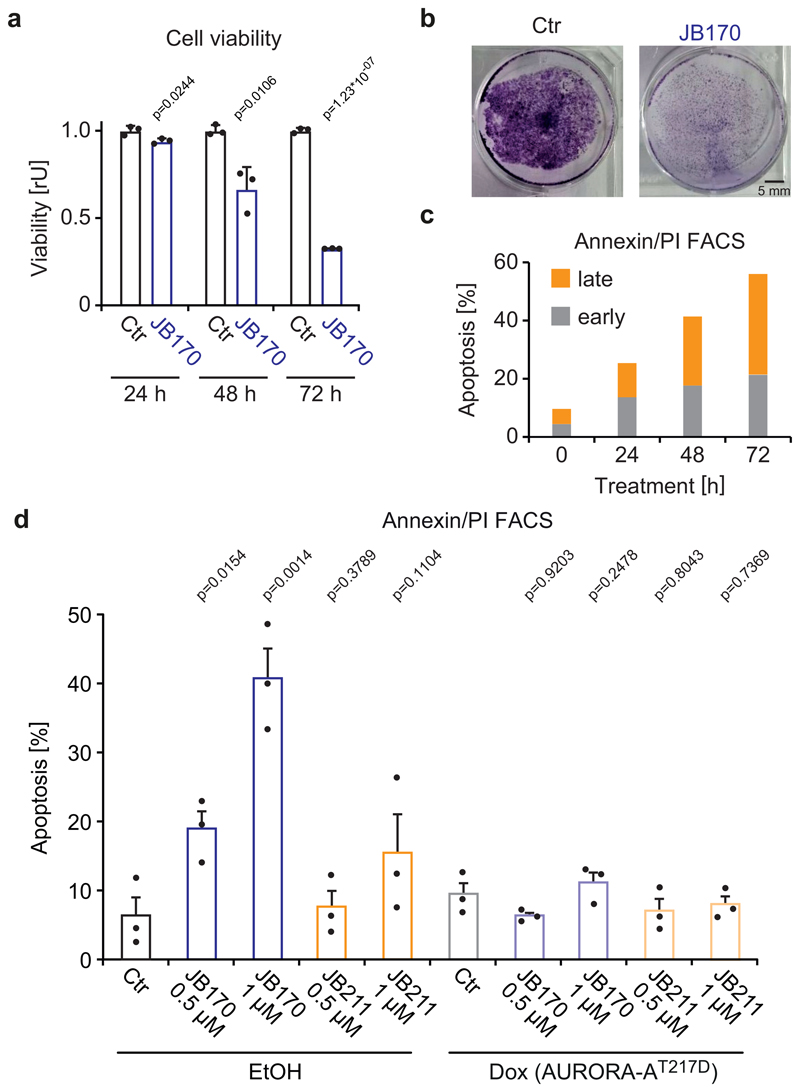
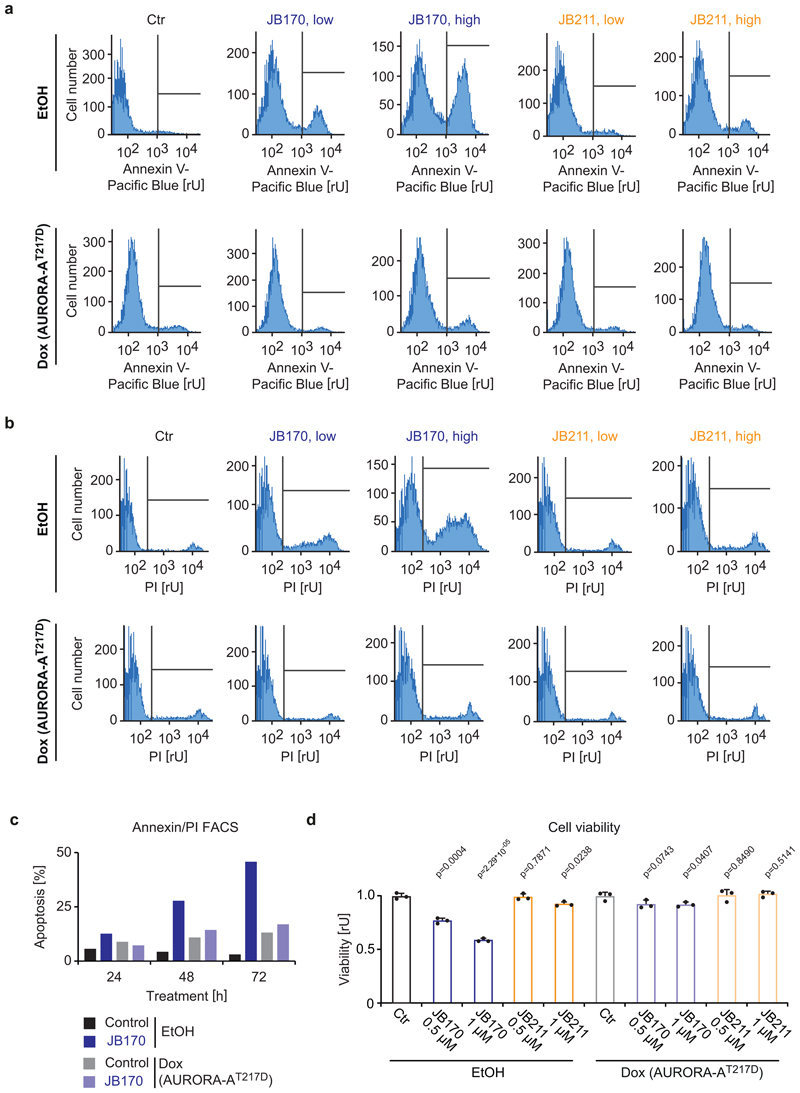
To study the effect of JB170-mediated AURORA-A depletion on cancer cell survival, MV4-11 cells were treated with JB170 and cell viability was measured by the alamarBlue assay. After 72 hours, the number of viable cells was 32% of control levels (Figure 6a). Similar results were observed in a colony formation assay using IMR5 cells (Figure 6b). To study the mechanism of cell death in AURORA-A-depleted cells, we determined the number of apoptotic and necrotic cells by annexin/PI flow cytometry. JB170 increased the fraction of apoptotic cells in culture over time, culminating in 56% annexin-positive MV4-11 cells after 72 hours (Figure 6c). Expression of AURORA-AT217D in IMR5 cells reverted the induction of apoptosis, indicating that JB170-induced apoptosis is exclusively caused by targeting AURORA-A (Figure 6d, Extended Data Fig. 7a,b,c). Importantly, JB211, which binds AURORA-A but does not mediate its degradation, did not significantly induce apoptosis (Figure 6d) or reduce viability (Extended Data Fig. 7d). We concluded that JB170 induces apoptosis in leukemia and neuroblastoma cell lines by depleting AURORA-A.
Figure 6. Degrader-mediated depletion of AURORA-A induces apoptosis in cancer cells.
(a) Bar diagram showing cellular viability. MV4-11 cells were treated with JB170 (1 µM) and cellular viability was measured by the alamarBlue assay at the indicated time points. Bars represent mean ± s.d. for n = 3 replicates. P-values were calculated with two-tailed unpaired t-test assuming equal variance. (b) Colony formation assay. IMR5 cells were treated with JB170 (1 µM) for 4 days and stained with crystal violet. Scale bar represents 5 mm. (c) Cell death analyzed by flow cytometry. MV4-11 cells were treated with JB170 (0.5 µM). Cells were stained with annexin and PI, and early (annexin+, PI-) and late (annexin+, PI+) apoptotic cells were counted by flow cytometry (50,000 sorted events). (d) Quantification of Extended Data Fig. 7a,b. Bars represent mean ± s.e.m. of n = 3 biological replicates. P-values were calculated with one-tailed unpaired t-test assuming equal variance. Homoscedasticity was assessed with Bartlett’s test, and normality was assessed with the Shapiro-Wilk test.
Discussion
We developed AURORA-A degrader molecules to study and ultimately inhibit its kinase-independent functions in cells. We linked the AURORA-A inhibitor alisertib to E3 ligase-binding moieties using various PEG and aliphatic linkers, and evaluated synthesized adducts in cellular assays. Two thalidomide-based drugs with short PEG-based linkers remarkably reduced cellular AURORA-A levels by a mechanism consistent with a PROTAC/degrader-like mode of function: Degrader-induced depletion of AURORA-A was rapid, reversible, caused by proteasomal degradation, and dependent on the E3 ligase CEREBLON. The most efficient degrader, JB170, induced maximal depletion (DCmax) at 300 nM. Despite JB170’s relatively high molecular weight and potentially low cellular permeability, the half-maximal degradation concentration (DC50) was low (28 nM), suggesting a catalytic mode of action as typically observed for degraders/PROTACs28.
Degrader development is challenging since no clear design rules exist, resulting often in inactivity when the linker geometry is changed32. To study E3 ligase recruitment to the target, we used molecular modeling to understand structural details of the AURORA-A–CEREBLON complex. Docking studies predicted complex formation of the two proteins in the absence of degraders via an extensive interaction interface. Because mutation of residues predicted to be critical for the interaction abolished degrader-induced depletion of AURORA-A, our work indicates that ternary complex formation not only depends on degrader binding but also is influenced by protein-protein interactions between AURORA-A and CEREBLON. This observation is important for several reasons.
First, this observation demonstrates that structural modeling is a powerful tool that provide insights into productive target-ligase pair formation and suitable linkers in the degrader. Our docking study correctly indicated that AURORA-A can form a stable complex with CEREBLON, and distinguished between potent and non-productive degraders with incompatible linkers. It therefore made a compelling case for the use of in silico methods for optimizing linkers and reducing the synthetic effort. Structure-guided degrader optimization has been successful for developing improved PROTACs for SMARCA240, indicating ternary complex formation as a rate-limiting step in degrader/PROTAC efficacy. Similarly, cooperative binding between BRD4 and the E3 ligase VHL was shown, by a compelling set of structural–biophysical methods, to be crucial for the efficacy of the BRD4-PROTAC MZ141.
Second, our modeling data indicate that the selectivity of alisertib as a recognition unit for AURORA-A is enhanced by protein-protein interactions between AURORA-A and CEREBLON. This increase in specificity might explain why AURORA-A was the only protein depleted by JB170. Indeed, also other degraders have greater target specificity than the original target-binding moiety47,48; protein-protein interactions in the target-E3 ligase complex likely explain this property. However, the binding preference of our degrader for AURORA-A most likely contributes to its excellent degradation selectivity. It is also possible that rate-limiting steps in the degradation cascade, other than ternary complex formation, contribute to target specificity.
Third, the predicted interface between AURORA-A and CEREBLON suggests a functional interaction between these proteins. Indeed, co-immunoprecipitation experiments demonstrated an interaction of AURORA-A and endogenous CEREBLON in cells, in the absence of any degrader. The biological impact of this interaction is unknown.
Intriguingly, degrader-mediated depletion of AURORA-A induced S-phase arrest, while kinase inhibition induced G2/M arrest in leukemia and neuroblastoma cell lines. The cell cycle defect induced by JB170 was clearly caused by decreased AURORA-A levels, as the S-phase arrest was completely rescued by expressing a JB170/alisertib-insensitive AURORA-A mutant33. Two mechanisms may account for the distinct phenotypes of inhibition and degradation of AURORA-A. First, it is possible that JB170 is more specific than alisertib (see above) and that the observed JB170-induced S-phase arrest results from exclusive targeting of AURORA-A. However, alisertib38 has a relatively narrow selectivity for AURORA-A. Therefore, it seems more likely that the discrepancy between inhibition and degradation results from a scaffolding function of AURORA-A in S-phase. While this role of AURORA-A is less explored than its mitotic functions4, it was also indicated by studies using conformational inhibitors, which induced an S-phase arrest of neuroblastoma cells27,45. Moreover, AURORA-A inhibition in cancer cells has recently been described to be synthetic lethal with mutations in the retinoblastoma protein12, a major regulator of early cell cycle phases. Interestingly, we identified numerous novel AURORA-A interaction partners, not known to be substrates. Several of them, including DICER1, TARBP2, YLPM1, AGO2 and DROSHA, form large cellular complexes and participate in RNA metabolism. In line with our observation, an analysis of RNA-mediated multiprotein complexes46 identified AURORA-A as a component of RNase-sensitive large complexes. We speculate that the interaction of AURORA-A with RNA-binding proteins mediates its non-catalytic functions in S-phase.
Importantly, our observation that degrader-mediated depletion of AURORA-A induced rampant apoptosis in cancer cell lines raises the exciting possibility that targeting this S-phase function, rather than the mitotic function, opens a larger window for tumor therapy. The excellent target specificity of JB170 and its ability to inhibit non-catalytic functions create the basis for developing drug-like degraders for testing in preclinical cancer models.
Methods
Synthesis of compounds and their intermediates is described in Supplementary Note 1. Modeling studies, including docking calculations and molecular dynamics simulations, were performed as described in Supplementary Note 2. Information about antibodies, reagents, commercial kits, cell lines, primers, vectors and software is given in Supplementary Table 2. All biological systems (e.g. cell lines and plasmids) and chemical compounds are available from the corresponding authors on request.
Cell culture
MV4-11 (male), IMR5 (male) and HLE (male) cells were grown in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS (Capricorn Scientific) and 1% penicillin/streptomycin solution (Sigma). HEK293 (female) and U2OS (female) cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin/streptomycin. For stable isotope labeling of MV4-11 cells, cells were cultured in RPMI 1640 medium for SILAC (Thermo Fisher Scientific) with L-lysine and L-arginine (light) or [2H4]-L-lysine and [13C6]-L-arginine (medium labeled) or [13C6, 15N2]-L-lysine and [13C6, 15N4]-L-arginine (heavy labeled) for at least five generations. All light, medium and heavy SILAC media were supplemented with 10% dialyzed FBS and 1% penicillin/streptomycin solution. Moreover, medium and heavy SILAC media were also enriched with proline to prevent arginine-to-proline conversion. Cells were analyzed for labeling efficiency before SILAC proteomics.
Cells were cultured at 37 °C in 5% CO2. Cells were routinely screened for mycoplasma contamination in a PCR-based assay and found negative.
General cloning
All primers along with the template vector used for PCR, gBlock and the vector backbone are listed in Supplementary Table 2. HiBiT-tagged AURORA-A and mutants were cloned by PCR amplification of the template vector and inserted into pRRL vector using AgeI/MluI restriction sites. HA-tagged AURORA-A, AURORA-B (wild type: isoform-5) and corresponding mutants were cloned by PCR amplification and inserted into pRRL using AgeI/SpeI sites. AURORA-A P191W, AURORA-B E162T and AURORA-B x3 were amplified via overlapping PCR using the corresponding vector containing wildtype sequence as template. CRBN-TBD encoding sequence was amplified from the full-length CRBN and inserted into pETM-30 vector using NcoI/NotI sites. HA-tagged AURORA-A Interface mutant (AURORA-AImut) was ordered as a gBlock (IDT) and inserted into pRRL vector using AgeI/SpeI sites. HA-tagged AURORA-A K162R was ordered as two fragments (gBlocks) which were digested by AgeI/BbsI, BbsI/SpeI and inserted into AgeI/SpeI-digested pRRL vector.
Cell line generation and manipulation
Stable cell lines were generated using lentiviral infection. Lentivirus was produced in HEK293 cells using the lentiviral packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259). The virus-containing supernatant was filtered (0.45 µm) before being used for infection. MV4-11 and IMR5 cells were infected twice every 24 h and selected 72 h after infection.
For transfection, polyethylenimine (PEI; Sigma) was used. For immunoprecipitation experiments, transiently transfected HEK293 cells were harvested 24 h after transfection.
For siRNA transfections, RNAiMAX reagent (Thermo Fisher Scientific) was used with siRNA pools against AURORA-A, AURORA-B and negative control siRNA.
Immunoblotting
Cells were treated with compounds for various times and then lysed in RIPA lysis buffer (50 mM HEPES pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate) containing protease and phosphatase inhibitors (Sigma) for 20 min at 4 °C in head-over-tail. Lysates were cleared by centrifugation. Protein quantification was done using BCA assay and equal amounts of protein were separated by BisTris-PAGE and transferred to PVDF membranes (Millipore). The membranes were blocked with 5% (w/v) nonfat dry milk in TBS-T (20 mM Tris HCl, pH 7.5, 150 mM NaCl, and 0.1% (v/v) Tween 20) at room temperature for 1 h and then incubated with the primary antibody overnight at 4 °C. Visualization was done with HRP-labeled secondary antibodies and detected using Chemiluminescent HRP substrate (Millipore) in LAS3000 or LAS4000 Mini (Fuji). The signal was quantified using ImageJ (version 1.52q) or Image Studio Lite (LI-COR Biosciences, version 5.2.5). Vinculin was used as a loading control, if not stated differently.
HA immunoprecipitation
HA immunoprecipitation was done as previously described49. Cells were washed once with ice-cold PBS and harvested in immunoprecipitation buffer (20 mM HEPES pH 7.9, 200 mM NaCl, 0.5 mM EDTA, 10% glycerol, 0.2% NP-40) supplemented with phosphatase and protease inhibitors (Sigma), lysed for 30 min at 4 °C and cleared by centrifugation. For immunoprecipitation, HA-coupled magnetic beads (Pierce Thermo Fisher Scientific) were washed with IP buffer, added to the lysate and incubated for 3 h at 4 °C. Elution was performed in 40 µl 1x LDS buffer (NuPAGE Thermo Fisher Scientific) by incubating at 37 °C with shaking for 30 min.
Cycloheximide chase assay
IMR5 cells were seeded one day before cycloheximide (CHX) treatment. The cells were treated either with 10 µg/ml CHX or with 10 µg/ml CHX and 1 µM JB170 for varying times. They were harvested in RIPA buffer and used in immunoblotting. The band intensity for AURORA-A at time zero was set as 1. The mean ratio ± standard error of mean from three biological replicate experiments were plotted as log10 values and t1/2 was calculated using nonlinear regression (curve fit) with semiolog line (X is linear and Y is log) in GraphPad Prism.
HiBiT assay
MV4-11 cells stably expressing HiBiT-AURORA-A were seeded in 96-well plates and treated with serial dilutions of compounds for 6 h. The HiBiT assay (Promega) was done using the Nano-Glo HiBiT Lytic Detection System. Luminescence was measured on a GloMax 96 Microplate Luminometer (Promega). DC50 was calculated using the eight-fold lower concentrations (which showed sigmoidal behavior) with the sigmoidal dose-response (four parameters) equation in GraphPad Prism.
Flow cytometry
For BrdU-PI flow cytometry, cells were labeled with 10 μM BrdU (Sigma-Aldrich) for 1 h. Cells were collected, washed with ice-cold PBS and fixed in 80% ethanol overnight at -20 °C. Cells were washed once with cold PBS and resuspended in 2 M HCl, 0.5% Triton X-100 for 30 min at room temperature. The cell suspension was neutralized in 0.1 M Na2B4O7 (pH 8.5), and cell pellets were resuspended in 100 μl 1% BSA in PBS-T (0.5% Tween-20 in PBS) containing anti-BrdU-FITC antibody (BioLegend) for 30 min at room temperature in the dark. Cells were washed once with 1% BSA in PBS-T. Cell pellets were resuspended in PBS with RNase A (24 μg/ml) and propidium iodide (54 μM), and incubated for 30 min at 37 °C.
For annexin-PI flow cytometry, the medium in which the cells were cultured was combined with the cells. The cells were washed once with ice-cold PBS, resuspended in 100 µl Annexin V Binding buffer (10 mM HEPES pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) with 2 µl Annexin V/Pacific Blue dye, and incubated for 15 min in the dark at room temperature. 400 µl Annexin V Binding buffer with propidium iodide (18.5 µM) was added. Samples were stored cold in the dark until analysis.
Flow cytometry was done on a BD FACSCanto II flow cytometer. Data were analyzed using BD FACSDIVA software and FlowJo (version 8.8.6).
Cell growth assay
AlamarBlue assay and crystal violet staining were done to analyze cell growth over time. For the alamarBlue assay, 6 x103 MV4-11 cells were seeded in 96-well plates and treated with compounds for various times (refreshed every 24 h). The alamarBlue assay (Thermo Fisher Scientific) was done using HS Cell Viability Reagent. Fluorescence was measured on a Tecan Infinite-200 plate reader using an excitation wavelength of 550 and an emission wavelength of 600 nm.
For crystal violet staining, 1.5 x105 IMR5 cells were plated and treated from the next day onwards with compounds for 4 days (refreshed every 24 h). Cells were stained with crystal violet solution (0.5% crystal violet, 20% ethanol in H2O) for 30 min at room temperature. Cells were washed with H2O and dried for 3-4 h at room temperature.
Quantitative SILAC mass spectrometry
3x106 MV4-11 cells (3x105 cells/ml; light, medium and heavy labeled) were seeded in triplicate one day before use. Cells were treated with DMSO (light), 100 nM JB170 (medium) and 100 nM alisertib (heavy) for 6 h. 3.5x106 cells per treatment (per replicate) were combined and washed twice with ice-cold PBS with protease and phosphatase inhibitors. Cells were lysed in 500 µl 1.5x Laemmli sample buffer, and 25 U benzonase (Novagen) was added to digest DNA for 20 min at room temperature. Samples were then heated at 95 °C for 5 min.
Protein was precipitated overnight at -20 °C with four volumes of acetone. Pellets were washed three times with acetone at -20 °C. Precipitated protein was dissolved in NuPAGE LDS sample buffer (Life Technologies), reduced with 50 mM 1,4-dithiothreitol (DTT) at 70 °C for 10 min, and alkylated with 120 mM iodoacetamide at room temperature for 20 min. Separation was performed on NuPAGE Novex 4-12% Bis-Tris gels (Life Technologies) with MOPS buffer. Gels were washed three times for 5 min with water and stained for 45 min with Simply Blue Safe Stain (Life Technologies). After washing with water for 2 h, each gel lane was cut into 15 pieces. The excised gel bands were destained with 30% acetonitrile in 0.1 M NH4HCO3 (pH 8), shrunk with 100% acetonitrile, and dried under vacuum (Concentrator 5301, Eppendorf, Germany). Digests were performed with 0.1 µg trypsin per gel band overnight at 37 °C in 0.1 M NH4HCO3 (pH 8). After removing the supernatant, peptides were extracted from gel slices with 5% formic acid. Extracted peptides were pooled with the supernatant.
NanoLC-MS/MS was performed on an Orbitrap Fusion (Thermo Scientific) equipped with a PicoView Ion Source (New Objective) and coupled to an EASY-nLC 1000 (Thermo Scientific). Peptides were loaded on capillary columns (PicoFrit, 30 cm x 150 µm ID, New Objective) self-packed with ReproSil-Pur 120 C18-AQ, 1.9 µm (Dr. Maisch) and separated with a 45-min linear gradient from 3% to 30% acetonitrile and 0.1% formic acid, at a flow rate of 500 nl/min.
MS and MS/MS scans were acquired in the Orbitrap analyzer with a resolution of 60,000 and 15,000, respectively. HCD fragmentation with 35% normalized collision energy was applied. A Top Speed data-dependent MS/MS method with a fixed cycle time of 3 s was used. Dynamic exclusion was applied with a repeat count of 1 and an exclusion duration of 30 seconds; singly charged precursors were excluded from selection. Minimum signal threshold for precursor selection was set to 50,000. Predictive AGC was used with AGC a target value of 2e5 for MS scans and 5e4 for MS/MS scans. EASY-IC was used for internal calibration.
Raw MS data files were analyzed with MaxQuant version 1.6.2.250. Andromeda, within MaxQuant, was used to search the UniProt Human database. Additionally, a database of common contaminants was searched. The search was performed with tryptic cleavage specificity with three allowed miscleavages. Protein identification was under control of the false-discovery rate (<1% FDR on protein and peptide levels). In addition to MaxQuant default settings, the search was performed against the following variable modifications: protein N-terminal acetylation, Gln to pyro-Glu formation (N-terminal Gln) and oxidation (Met). Carbamidomethyl (Cys) was set as fixed modification. Further data analysis was performed using R scripts developed in-house. For quantification of pSILAC-labeled proteins, medians were calculated for the log2-transformed heavy to light (H/L), medium to light (M/L), and heavy to medium (H/L) peptide ratios for each protein. Two ratio counts were required for protein quantification. Protein ratios were normalized for each experiment in intensity bins (at least 300 proteins per bin). Outliers were identified by boxplot statistics as those with values outside a 1.5× interquartile range (or 3× for extreme outliers).
Immunoprecipitation mass spectrometry
MV4-11 cells stably expressing HA-tagged AURORA-A or empty vector were seeded one day before harvest. Cells were washed twice with ice-cold PBS supplemented with protease and phosphatase inhibitors. Cells were lysed in 4 ml lysis buffer (20 mM HEPES pH 7.9, 180 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.2% NP-40) supplemented with protease and phosphatase inhibitors, by homogenizing 15 times and shearing by sonication four times for 10 s with 45 s pauses (20% output). Proteins were solubilized from chromatin by incubating with benzonase (100 units/ml; Novagen) for 40 min at 4 °C with rotation. The soluble protein fraction was used for immunoprecipitation with 80 µl HA-coupled magnetic beads (Pierce Thermo Fisher Scientific) for 3 h at 4 °C with rotation. Beads were washed three times with lysis buffer containing 0.1% Triton X-100, incubated 5 min with rotation, and then washed twice in lysis buffer without Triton X-100. Protein complexes were eluted in 100 μl 1x LDS Sample Buffer (NuPAGE Thermo Fisher Scientific) by incubating for 30 min at 37 °C. Then proteins were reduced by adding DTT to a final concentration of 50 mM, and samples were heated at 95 °C for 5 min.
Eluted proteins were alkylated with 120 mM iodoacetamide at room temperature for 20 min. Protein was precipitated overnight at -20 °C with four volumes of acetone. Precipitated proteins were pelleted by centrifugation, washed thrice with acetone and digested by LysC protease in 0.5% sodium deoxycholate for 2 h at 30 °C followed by the addition of trypsin at 37 °C overnight (enzyme/protein, 1/200). Sodium deoxycholate extraction was done with ethyl acetate and 0.5% trifluoroacetic acid. Peptides were dried under vacuum to remove residual ethyl acetate, and desalted with three discs of C18 Empore SPE Disks (3M) in a 200 μl pipet tip. Elution was done twice with 60% acetonitrile and 0.1% formic acid, and the eluates were dried under vacuum and stored at -20 °C. Dried peptides were dissolved in 2% acetonitrile, 0.1% formic acid before nanoLC-MS/MS.
NanoLC-MS/MS was performed on an Orbitrap Fusion (Thermo Scientific) equipped with a PicoView Ion Source (New Objective) and coupled to an EASY-nLC 1000 (Thermo Scientific). Peptides were loaded on capillary columns (PicoFrit, 30 cm x 150 µm ID, New Objective) self-packed with ReproSil-Pur 120 C18-AQ, 1.9 µm (Dr. Maisch) and separated with a 45-min linear gradient from 3% to 30% acetonitrile and 0.1% formic acid, at a flow rate of 500 nl/min.
MS and MS/MS scans were acquired in the Orbitrap analyzer with resolutions of 60,000 and 15,000, respectively. HCD fragmentation with 35% normalized collision energy was applied. A Top Speed data-dependent MS/MS method with a fixed cycle time of 3 s was used. Dynamic exclusion was applied with a repeat count of 1 and an exclusion duration of 30 s; singly charged precursors were excluded from selection. Minimum signal threshold for precursor selection was set to 50,000. Predictive AGC was used with AGC a target value of 5e5 for MS scans and 5e4 for MS/MS scans. EASY-IC was used for internal calibration.
Raw MS data files were analyzed with MaxQuant version 1.6.2.2. Andromeda, within MaxQuant, was used to search the UniProt Human database. Additionally, a database of common contaminants was searched. The searches were performed with tryptic cleavage specificity with three allowed miscleavages. Protein identification was under control of the false-discovery rate (<1% FDR on protein and peptide levels). In addition to MaxQuant default settings, searches were performed against the following variable modifications: protein N-terminal acetylation, Gln to pyro-Glu formation (N-terminal Gln) and oxidation (Met). For protein quantitation, LFQ intensities were used. Proteins with less than two identified razor/unique peptides were dismissed. Missing LFQ intensities in control samples were imputed with values close to baseline if intensities in the corresponding IP samples were present. Data imputation was performed with values from a standard normal distribution with a mean of the 5% quantile of the combined LFQ intensities and a standard deviation of 0.1. Missing logFC values for individual samples were imputed via MICE package in R, and p values were recalculated using a linear method in limma package in R.
Quantitative TMT mass spectrometry
IMR5 cells (4x106) were seeded in quadruplicate one day before treatment with 1 µM JB170, JB211 or alisertib for 6 h. Cells were washed twice with ice-cold PBS with protease and phosphatase inhibitors, and then lysed in SDS lysis buffer (2% SDS, 40 mM Tris-HCl pH 7.6). To reduce viscosity, samples were sonicated using a Diagenode Bioruptor (15 cycles, 30 s sonication, 30 s pause, 4 °C). Samples were boiled at 95 °C for 10 min, and trifluoroacetic acid was added to a final concentration of 1%. Then the pH was neutralized (final pH, 7.6-8.0) by adding 300 mM N-methylmorpholine to a final concentration of 2%. Protein concentrations of cell lysates were determined using the BCA Protein Assay Kit (Thermo Scientific).
The bead suspension for sp3 sample workup was prepared by mixing magnetic SeraMag-A and SeraMag-B beads (10 µl per sample of each type; Cytiva) in a 1:1 ratio, washing three times with ultrapure H2O and resuspending in 10 µl ultrapure H2O per sample. A total of 50 µg per sample was mixed with 10 µl bead suspension. Acetonitrile was added to a final concentration of 70%, and samples were incubated at room temperature for 18 min with rotation at 800 rpm. The supernatant was discarded, and beads were washed twice with 200 µl 80% ethanol. For reduction and alkylation, beads were resuspended in 50 μl of 8 M urea in 40 mM Tris pH 7.6. Proteins were reduced with 10 mM DTT for 45 min at 37 °C with rotation at 800 rpm, and alkylated with 55 mM chloroacetamide at room temperature in the dark for 30 min. Acetonitrile was added to a final concentration of 70%, and samples were incubated at room temperature for 18 min with rotation at 800 rpm. The supernatant was discarded, and beads were washed twice with 200 µl 80% ethanol and once with 200 µl acetonitrile. The supernatant was discarded, and the beads were air-dried for 30 s before 100 µl of 25 mM HEPES pH 8.5 was added. Protein was digested (1:100 trypsin/substrate weight) for 3 h, and after a second addition of trypsin, incubated overnight at 37 °C with rotation at 1200 rpm. Samples were centrifuged (5 min at 1200 x g), sonicated 3 times for 30 s, and the supernatant was collected. Beads were washed once with 20 µl ultrapure H2O, sonicated 3 times for 30 s, and supernatants were combined with the previous supernatants. Peptide concentration was determined using a NanoDrop spectrophotometer. Samples were frozen at -80 °C and dried in a SpeedVac. The desalted peptides (20 μg per condition) were labeled with tandem mass tags 11 (TMT11)-plex (Thermo Fisher Scientific) as previously described51. One TMT channel was used for each treatment condition and replicate (126=JB170_repl1, 127N=JB170_repl2, 127C=JB170_repl3, 128N=JB170_repl4, 128C=JB211_repl1, 129N=JB211_repl2, 129C=JB211_repl3, 130N=Alisertib_repl1, 130C=Alisertib_repl2, 131N=Alisertib_repl3, 131C=Alisertib_repl4). After labeling, peptides were pooled and dried in a SpeedVac. Samples were reconstituted in 0.1% formic acid, and desalted using tC18 RP solid-phase extraction cartridges (Waters; wash solvent, 0.1% formic acid; elution solvent, 0.1% formic acid in 50% acetonitrile). A Dionex Ultra 3000 HPLC system operating a Waters XBridge BEH130 C18 3.5 µm 2.1 × 150 mm column was used to fractionate the pooled peptides (reconstituted in solvent A). Solvent A was 25 mM ammonium bicarbonate pH 8.0, solvent B was 100% ultrapure water (Elga), and solvent C was 100% acetonitrile. The proportion of solvent A was kept at 10% during separation. Pooled peptides (220 µg) were separated by a linear gradient from 7% to 45% solvent C over 45 min at a flow rate of 200 µl/min, followed by a linear gradient from 45% to 85% C in 6 min. Fractions were collected every minute into a 96-well plate and subsequently pooled into 48 fractions by adding fraction 49 to fraction 1, fraction 50 to fraction 2, and so forth. Peptide fractions were frozen at -80 °C, dried in a SpeedVac without prior desalting, and stored at -20 °C until LC-MS3 analysis.
For microflow LC-MS/MS, a Dionex UltiMate 3000 RSLCnano System was coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) as previously described52. Peptides were dissolved in 0.1% formic acid, and one half was directly injected onto the microflow LC system. Online chromatography was performed using an Acclaim PepMap 100 C18 LC column (2 µm particle size, 1 mm ID × 150 mm; Thermo Fisher cat. no. 164711). Column temperature was maintained at 55 °C using the integrated column oven. Peptides were delivered at a flow rate of 50 µl/min and separated using a 27-min linear gradient from 4% to 32% LC solvent B (0.1% formic acid, 3% DMSO in acetonitrile) in LC solvent A (0.1% formic acid, 3% DMSO). The Orbitrap Fusion Lumos was operated as follows: positive polarity; spray voltage, 3.5 kV, capillary temperature, 325 °C; vaporizer temperature, 125 °C. The flow rates of sheath gas, aux gas and sweep gas were set to 32, 5, and 0, respectively. Cycle time was set to 1.2 s. Full MS was readout in the Orbitrap, resolution was set to 60,000, and the mass range was set to 360–1560. Full MS AGC target value was 4E5 with a maximum IT of 50 ms and RF lens value was set to 50. The MIPS properties were set to peptide. Default charges were set to state 2–6. The dynamic exclusion duration was set to 50 s, exclude after one time. For readout of MS2 spectra, the ion trap was used applying the rapid scan function. The isolation width was set to 0.6 m/z, the first mass was fixed at 100 m/z, activation type was HCD, HCD collision energy [%] was 32. The AGC target value was set to 1.2E4 at a maximum IT of 40 ms. The precursor selection range was set to 400–2000, exclusion mass widths were set to 20 m/z for low and 5 m/z for high. For MS3 spectra readout, the Orbitrap was used at 50,000 resolution and over a scan range of 100–1000. Synchronous precursor selection (SPS) was enabled, and the number of SPS precursors was set to 8. MS2 isolation window was 3 m/z, activation type was HCD, and HCD collision energy was 55%. The AGC target was 1E5 with a maximum IT of 86 ms.
Proteins and peptides were identified and quantified using MaxQuant50 (version 1.6.2.10) by searching the MS3 data against all canonical protein sequences as annotated in the UniProt reference database (human proteins only, 20,205 SwissProt entries, downloaded 3 December 2015, internally annotated with PFAM domains) using the search engine Andromeda53. Carbamidomethylated cysteine (due to low CAA labeling efficiency), oxidation of methionine and N-terminal protein acetylation were set as variable modification. Trypsin/P was specified as the proteolytic enzyme and up to two missed cleavage sites were allowed. The minimum peptide length was set to seven and all data were adjusted to 1% peptide-spectrum match (PSM) and 1% protein FDR. MS3-based TMT quantification was enabled, taking TMT correction factors as supplied by the manufacturer into account.
Data analysis was performed using the Perseus software suite54 (version 1.5.8.5) and Microsoft Excel on identified and quantified protein groups as provided in the proteinGroups.txt file. proteinGroups.txt was filtered for contaminants and reverse hits, and total sum normalization and log2 transformation were performed. The JB170 replicate 4 showed large differences from the other replicates and was not considered for further analysis. Entries were filtered for at least three valid values in one condition, and remaining missing values were replaced from a normal distribution (width, 0.3; down shift, 1.8). A two-sample t-test was performed between JB170 and alisertib or JB211 (S0:0; permutation-based FDR, 5%; number of randomizations, 250).
The experimental design involved TMT11plex reagents enabling the analysis of JB170 treatment in quadruplicates, alisertib treatment in quadruplicates, and JB211 in triplicates within one TMT batch. Data quality control detected replicate 4 of JB170 treatment to be a technical outlier showing low TMT reporter intensities (about 10% of intensity of other channels) and distinct composition in a principal component analysis, despite consistent input amounts for protein digestion (by BCA assay) and subsequent peptide labeling (by NanoDrop quantification). Therefore, the outlier is most probably of technical nature, e.g. due to inefficient TMT labeling, and was excluded from further analysis.
Kinobead selectivity profiling
Kinobead pulldown experiments were performed as previously described38. Briefly, cells were lysed in 0.8% NP-40, 50 mM Tris-HCl pH 7.5, 5% glycerol, 1.5 mM MgCl2, 150 mM NaCl, 1 mM Na3VO4, 25 mM NaF, 1 mM DTT, protease inhibitors (SigmaFast, Sigma) and phosphatase inhibitors (prepared in-house according to phosphatase inhibitor cocktail 1, 2 and 3 from Sigma-Aldrich). The MV4-11 cell lysate was ultracentrifuged and diluted with 50 mM Tris-HCl pH 7.5, 5% glycerol, 1.5 mM MgCl2, 150 mM NaCl, 1 mM Na3VO4, 25 mM NaF, 1 mM DTT, protease inhibitors and phosphatase inhibitors to a final concentration of 5 mg/ml as determined by Bradford assay. For selectivity profiling of small molecule inhibitors, the cell lysate (2.5 mg total protein per pulldown) was pre-incubated with increasing compound concentrations (DMSO vehicle, 3 nM, 10 nM, 30 nM, 100 nM, 300 nM, 1000 nM, 3 µM and 30 µM) for 45 min at 4 °C in an end-over-end shaker. Subsequently, lysates were incubated with Kinobeads epsilon (17 µL settled beads) for 30 min at 4 °C. Beads were washed, and bound proteins were eluted with 40 µL 2x NuPAGE LDS sample buffer (Invitrogen) containing 50 mM DTT and incubated for 30 min at 50 °C. To assess the degree of protein depletion from the lysates by Kinobeads, a second Kinobead pulldown was performed with fresh beads and the flow through of the DMSO control55. For LC-MS/MS analysis, Kinobead eluates were alkylated with 55 mM chloroacetamide (4 µL of 550 mM), and protein was desalted and concentrated by a short electrophoresis on a 4-12% NuPAGE gel (Invitrogen). Tryptic in-gel digestion was performed according to standard procedures.
Peptides were measured on a Dionex Ultimate3000 nano HPLC coupled online to an Orbitrap Fusion Lumos (Thermo Fisher Scientific) mass spectrometer. Peptides were dissolved in 0.1% formic acid and delivered to a trap column (ReproSil-pur C18-AQ, 5 μm, Dr. Maisch, 20 mm × 75 μm, self-packed) at a flow rate of 5 μl/min in LC solvent A (0.1% formic acid, 3% DMSO). After 10 min of loading, peptides were transferred to an analytical column (ReproSil Gold C18-AQ, 3 μm, Dr. Maisch, 400 mm × 75 μm, self-packed) and separated using a 70-min linear gradient from 4% to 32% LC solvent B (0.1% formic acid, 3% DMSO in acetonitrile) in LC solvent A.
The Orbitrap Fusion Lumos was operated as follows: positive polarity; spray voltage, 2.0 kV, capillary temperature, 275 °C. Full MS was readout in the Orbitrap, resolution was set to 60,000, and the mass range was set to 360–1300. Full MS AGC target value was 4E5 with a maximum IT of 50 ms and RF lens value was set to 30. The MIPS properties were set to peptide. Default charges were set to state 2–6. The dynamic exclusion duration was set to 20 s, exclude after one time. For readout of MS2 spectra, the Orbitrap was used at a resolution of 15,000. Up to 10 peptide precursors were isolated, isolation window was set to 1.3, the first mass was fixed at 100 m/z, activation type was HCD, HCD collision energy [%] was 30. The AGC target value was set to 5E4 at a maximum IT of 22 ms.
Peptides and proteins were identified and quantified using MaxQuant50 (version 1.6.2.10) by searching the MS2 data against all canonical protein sequences as annotated in the UniProt reference database (human proteins only, 20,205 SwissProt entries, downloaded 3 December 2015, internally annotated with PFAM domains) using the search engine Andromeda53. Carbamidomethylated cysteine was set as fixed modification, and phosphorylation of serine, threonine and tyrosine, oxidation of methionine and N-terminal protein acetylation were set as variable modifications. Trypsin/P was specified as the proteolytic enzyme, and up to two missed cleavage sites were allowed. Precursor tolerance was set to 10 ppm and fragment ion tolerance to 20 ppm. The minimum length of amino acids was set to seven and all data were adjusted to 1% PSM and 1% protein FDR. Label-free quantification56 and match between runs were enabled within MaxQuant.
For Kinobead competition binding assays, relative binding was calculated based on the LFQ intensity ratio to the DMSO control for every single inhibitor concentration. EC50 values were derived from a four-parameter log-logistic regression using an internal pipeline that uses the ‘drc’ package in R. Multiplying the EC50 values by a correction factor generates an apparent binding constant (Kd app). The correction factor for a protein is defined as the ratio of the amount of protein captured from two consecutive pulldowns of the same DMSO control lysate55. Targets of the inhibitors were annotated manually. A protein was considered a target if the resulting binding curve had a sigmoidal shape with a dose-dependent decrease of binding to the beads. Additionally, the number of unique peptides and MS/MS counts per condition were taken into account. Protein intensity in the DMSO control sample was also included in the target annotation process. A protein was considered a direct binder if annotated in Uniprot.org as a protein kinase, lipid kinase, nucleotide binder, helicase, ATPase, GTPase, or FAD- or heme-containing protein. Most other proteins that were annotated as targets are interaction partners and were termed indirect binders.
Protein expression and purification
The AURORA-A (122-403) kinase domain was recombinantly expressed as a fusion protein incorporating an N-terminal His6-TEV (tobacco etch virus protease) tag. The CEREBLON thalidomide-binding domain (S320-D428) was expressed as an N-terminal-His6-GST-TEV fusion protein. Briefly, E. coli, with and without additional expression vector for lambda-phosphatase cultured in TB media, was initially grown at 37 °C until reaching an OD600 of 1.5. Cells were subsequently cooled to 18 °C until the culture reached an OD600 of 2.8. Then, protein expression was induced with 0.5 mM IPTG overnight. Harvested cells were lysed by sonication, and tagged proteins were purified using immobilized metal affinity chromatography with a resin charged with Ni2+ ions. The eluted proteins were buffer-exchanged into 50 mM HEPES, pH 7.5, 500 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP) and 5% glycerol, and their expression tags were cleaved using His6-labeled TEV. The cleaved proteins were passed through Ni2+ beads again, and further purified by size exclusion chromatography. Protein purity and phosphorylation status was assessed by intact mass analysis on an HPLC-ESI-TOF mass spectrometer. The pure proteins were stored in 50 mM HEPES, pH 7.5, 500 mM NaCl, 0.5 mM TCEP supplemented with 5% glycerol at -80 °C. For isothermal titration calorimetry, the buffer for both AURORA-A and CEREBLON-TBD was changed via size exclusion chromatography to the same buffer containing 25 mM HEPES, pH 7.5, 200 mM NaCl, 0.5 mM TCEP and 5% glycerol.
Thermal stability shift assay
Recombinant AURORA-A (2 μM in 10 mM HEPES, pH 7.5, 500 mM NaCl) was mixed with 10 μM each inhibitor and incubated at room temperature for ~10 min. SyPRO orange dye (Invitrogen) was subsequently added, and the fluorescence signals corresponding to temperature-dependent protein unfolding were measured using a Real-Time PCR Mx3005p instrument (Stratagene). Data were evaluated as described57.
Quantitative real-time PCR
Total RNA was extracted using peqGOLD TriFast reagent (Peqlab). cDNA synthesis was done with 1 µg total RNA using MLV reverse transcriptase, random primers, Ribolock RNAse inhibitor, and dNTPs in MLV buffer. cDNA was diluted five times in ultrapure H2O and 10 µl was used as template for quantitative real-time PCR (qRT-PCR) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). For analysis, expression was normalized to β2-microglobulin expression. Primers for qRT-PCR are listed in Supplementary Table 2.
RNA sequencing
RNA was isolated from MV4-11 cells using the miRNeasy Mini kit (Qiagen). RNA quality was determined by the Fragment Analyzer (Agilent). Total RNA (1 µg) was used for poly-A+ RNA isolation using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB). NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) was used for library preparation. cDNA libraries were amplified with 8 PCR cycles. Library size and amount was determined by Fragment Analyzer High Sense DNA kit (Agilent). Next generation sequencing was performed on a NextSeq500 Illumina platform for 75 cycles.
Cell transfections and BRET measurements
C-terminal NanoLuc-AURORA-A/B kinase fusions, encoded in pFC32K expression vectors (Promega), were used. For cellular BRET target engagement experiments, HEK293T cells were transfected with NanoLuc/target fusion constructs using FuGENE HD (Promega) according to the manufacturer’s protocol.
For all experiments, energy transfer probe 5 (Promega) was used in duplicate at a concentration of 10 nM. Compound and energy transfer probe handling were performed by an ECHO acoustic dispenser. Cells were incubated with compound dilutions and energy probes for 2 h at 37 °C in 5% CO2 prior to substrate addition.
Luminescence was measured on a PheraStar FSX microplate reader (BMG Labtech), and milli-BRET units (mBU) were calculated as a ratio of BRET signal to the overall luciferase signal. Inhibitory constants were calculated using the sigmoidal dose-response (four parameters) equation in GraphPad Prism. Reported values are the average of two biological replicates.
Isothermal titration calorimetry
All titrations were performed on a Nano ITC calorimeter (TA Instruments) at 25 °C. The titrations of the binary complexes (AURORA-A into JB170 and CEREBLON-TBD into JB170) were performed as reverse titrations. Protein concentrations were determined spectroscopically at 280 nm using calculated extinction coefficients and a Thermo Scientific NanoDrop spectrophotometer and a buffer of 25 mM HEPES pH 7.5, 200 mM NaCl, 0.5 mM TCEP, 5% glycerol was used. For AURORA-A, concentrations in the injector (between 57 and 110 µM) had to be optimized due to protein stability issues matching JB170 concentrations between 1.0 and 10.0 µM. Values were calculated from four titrations. Best conditions were achieved at 110 µM AURORA-A and 10 µM JB170. For the CEREBLON (TBD) titration concentrations between 88 and 100 µM were used for the protein and 2.0 and 3.5 µM for JB170. Dissociation constants were calculated from three independent titrations.
Titrations for the ternary complexes were determined as previously described41. Briefly, CEREBLON(TBD) at 0.1 µM was titrated as described above. The binary complex remained in the calorimeter and the excess of solution after the titration was removed using a syringe. AURORA-A (110 µM) was titrated into the binary complex which had a JB170 concentration of 3.2 µM and 2.8 µM in two independent titration experiments. All data were fitted using a single binding site model in NanoAnalyse software (TA Healthcare) to obtain Kd values and thermodynamic binding parameters.
Bioinformatics analysis and statistics
Quantitative data are shown in bar graphs as mean and standard deviation from replicate experiments, if not stated differently. Mean values and number of experiment replicates are given in figure legends. Significance between two conditions (p-value) was calculated with two-tailed unpaired t-test assuming equal variances, if not stated differently. Figure 1a, 1b, 2a, 2b, 2c, 2d, 3b, 4a, 4b, 4c, 5a, 5c, 5d, 5e, 5g, 6a, 6b, 6d, Extended Data Figure 1c, 1d, 1e, 1f, 2a, 2b, 2c, 2d, 2e, 2f, 2g, 2h, 2j, 2k, 2m, 2o, 2p, 2s, 3a, 3b, 3c, 3d, 3e, 4f, 4g, 5a, 6c, 6d, 6f, 6g, 6j, 7a, 7b and 7d were repeated at least two times biologically. Figure 4d, Extended Data Figure 2l, 2n, 5b, 5c and 5d were repeated at least two times technically. Figure 6c, Extended Data Figure 1g, 2q, 2r, and 7c were only performed once. For RNA sequencing, FASTQ files were checked for quality using FASTQC (Babraham Bioinformatics) followed by alignment to hg19 human genome build using Bowtie2 (v2.3.4.1) using -N 1 option. All aligned reads-containing files were normalized to same reads depth using Samtools (v1.3). These read-normalized bam files were then used for differential expression regulation analysis using edgeR protocol58 implemented in R v3.5.2. Briefly, the reads from bam files were read into R using readGAlignments function, followed by extraction of read counts for every gene with summarizeOverlaps function. Non-expressed genes were removed, and dispersion was calculated followed by p- and q- value calculation using Benjamini Hochberg correction. All further graphs were generated using ggplot2.
Extended Data
Extended Data Figure 1.
(a) Model of bifunctional degrader molecules (PROTACs) of AURORA-A. (b) Ribbon model of AURORA-A and structural formula of alisertib. The structure of AURORA-A bound to MLN8054 (PDB: 2X81)33 identified a solvent-exposed carboxyl group on the compound to which linkers could be attached by amide bonds. (c) Degrader and ligand structures and their effects on AURORA-A-NanoLuc target engagement. HEK293 cells transfected with an AURORA-A-NanoLuc fusion construct were incubated with various concentrations of the AURORA-A degrader molecules or their components together with an energy transfer probe for 2 hours and luminescence was measured. EC50 values were calculated by assuming a sigmoidal dose-response relationship (four parameters). The graph shows one of the two biological replicates. The graphs display the profile of each compound for the assay as shown in Fig. 1a. (d) Bar diagram showing the binding of degrader molecules to AURORA-A. AURORA-A was incubated with degrader molecules and binding was analyzed by thermal shift assays and compared to alisertib. Pomalidomide and VHL-binding moieties were used as controls. Bars represent mean ± s.d. for n = 4 replicates (n = 3 for JB161) (e) Immunoblot of AURORA-A. AURORA-A and AURORA B were depleted in IMR5 cells by siRNA and AURORA-A levels were compared to cells treated with JB170 (1 µM) or alisertib (1 µM) and to control cells (Ctr). (f) Immunoblot of AURORA-A. MV4-11 cells were treated with 0.1 µM JB170 for the indicated times and AURORA-A levels were compared to control cells. (g) Immunoblot of AURORA-A. MV4-11 cells were treated with 0.1 µM JB158 for the indicated times, and AURORA-A levels were compared to control cells.
Extended Data Figure 2.
(a) Immunoblot of AURORA-A. MV4-11 cells were treated with various concentrations of JB170 and alisertib for 6 hours, and AURORA-A levels were compared to control cells. (b) Immunoblot of AURORA-A. MV4-11 cells were treated with various concentrations of JB158 and alisertib for 6 hours and AURORA-A levels were compared to control cells. (c) Immunoblot of AURORA-A and bar diagram of RT-qPCR analysis. RNA and protein were isolated from MV4-11 cells treated with JB170 (0.1 μM) and alisertib (0.1 μM) for 6 and 24 hours. AURORA-A protein (top) and RNA levels (bottom) were analyzed. Short and long exposures are shown for AURORA-A. AURORA-A expression levels were normalized to control cells (DMSO). Bars represent mean of technical replicates. (d) Immunoblot of AURORA-A and bar diagram of RT-qPCR analysis. RNA and protein were isolated from MV4-11 cells treated with JB158 (0.1 µM) or alisertib (0.1 µM) for 6 h. AURORA-A protein (left) and RNA levels (right) were analyzed. Vinculin was used as a loading control. The AURORA-A expression levels are normalized to control cells (DMSO). Bars represent mean of technical replicates. (e) Bar diagram of RT-qPCR analysis. RNA was isolated from IMR5 cells treated with an siRNA against AURORA-A, a non-targeting control (siCtr), JB170 and control cells (DMSO). AURORA-A RNA levels were analyzed by RT-qPCR in comparison to beta-2 microglobulin and normalized to control cells (siCtr). Bars represent mean ± s.e.m. of relative expression for n = 3 biological replicates. (f) Immunoblot of AURORA-A upon washout of JB170. MV4-11 cells were treated with JB170 (0.1 µM) for 3 hours before they were either cultured in JB170-containing or JB170-free medium for up to 6 hours. (g) Immunoblot of AURORA-A. MV4-11 cells were treated with different concentrations JB170 and alisertib for 6 hours and compared to cells treated with one compound or untreated cells. (h) Immunoblot of AURORA-A. MV4-11 cells were treated for 6 hours with JB170 (0.1 µM) and unconjugated thalidomide (+: 1 µM, ++: 10 µM, +++: 20 µM). (i) Structure of JB211 that is an inactive analogue of JB170. (j) Immunoblot of AURORA-A. MV4-11 cells were treated with JB170 (0.1 µM) and proteasomal inhibitor MG132 (10 µM) for 6 hours. (k) Immunoblot of AURORA-A. MV4-11 cells were treated with different concentrations JB170 and the NEDD8 activating enzyme (NAE) inhibitor MLN4924 (3 µM) for 6 hours. (l) Immunoblot of AURORA-A. MV4-11 cells were treated with different concentrations JB158 and the NEDD8-activating enzyme inhibitor MLN4924 (3 µM) for 6 hours. (m) Immunoblot of HA-tagged AURORA-A. Kinase-dead versions of HA-tagged AURORA-A (AURORA-AK162R, AURORA-AD274N) and the HA-tagged wildtype protein (WT) were expressed in IMR5 cells. Cells were then treated with alisertib (1 µM) and JB170 (1 µM) for 18 hours, and AURORA-A levels were analyzed using an anti-HA tag antibody and an anti-AURORA-A antibody. (n) Immunoblot of HA-tagged AURORA-A. Kinase-dead version of HA-tagged AURORA-A, AURORA-AK162R and the HA-tagged wildtype protein (WT) were expressed in MV4-11 cells. Cells were then treated with alisertib (1 µM) or JB170 (1 µM) for 18 hours, and AURORA-A levels were analyzed with an anti-HA tag antibody and an anti-AURORA-A antibody (o) Immunoblot of AURORA-A. U2OS cells were treated with different concentrations of JB170 for 6 hours. (p) Immunoblot of AURORA-A. HLE cells were treated with different concentrations of JB170 for 6 hours. (q) Immunoblot of AURORA-A. HLE cells were treated with JB158 (1 µM) for the indicated time periods. (r) Immunoblot of AURORA-A. U2OS cells were treated with different concentrations of JB158 for 6 hours. (s) Immunoblot of AURORA-A. HLE cells were treated with different concentrations of JB158 for 6 hours.
Extended Data Figure 3.
(a) Effects of ligands and degraders on AURORA-B-NanoLuc target engagement. HEK293 cells transfected with an AURORA-B-NanoLuc fusion construct were incubated with various concentrations of the AURORA-A degraders or their components together with an energy transfer probe for 2 hours and luminescence was measured. EC50 values were calculated by assuming a sigmoidal dose-response relationship (four parameters). The graph shows one of the two biological replicates. (b) Immunoblot of AURORA-B. AURORA-A and AURORA B were depleted in IMR5 cells by siRNA and AURORA-B levels were compared to cells treated with JB170 (1 µM) or alisertib (1 µM) and to control cells (Ctr). (c) Immunoblot of AURORA-A and AURORA-B. MV4-11 cells were treated with various concentrations of JB170 and alisertib for 24 hours, and protein levels were compared to control cells. (d) Immunoblot of AURORA-A and AURORA-B. IMR5 cells were treated with DMSO, JB170 or alisertib for 6 hours and protein levels were compared. (e) Immunoblot of AURORA-A, GSPT and IKZF1. MV4-11 cells were treated with JB170, alisertib, pomalidomide (Pom.) and thalidomide (Thal.) for 18 hours. (f, g) Volcano plot showing changes in protein abundance. IMR5 cells were treated with JB170 (1 μM) or alisertib (1 μM) for 6 hours and proteins were analyzed by isobaric labeling (TMT: tandem mass tag) followed by mass spectrometry. The X-axis displays the relative abundance of all identified proteins (6485) in JB170-treated vs. alisertib-treated cells (log2FC). The Y-axis displays the p-value (-log10) from triplicate experiments (Student's t-test, two-sided, permutation-based FDR correction, FDR: 5%). AURORA-A and other alisertib-binding proteins (f) or neosubstrates of CEREBLON (g) are labeled (orange). (h) Volcano plot showing changes in protein abundance. IMR5 cells were treated with JB170 (1 µM) or JB211 (1 µM) for 6 hours, and proteins were analyzed by isobaric labeling (TMT: tandem mass tag) followed by mass spectrometry. The X-axis displays the relative abundance of all identified proteins (6485) in JB170-treated vs. JB211-treated cells (log2FC). The Y-axis displays the p-value (-log10) from triplicate experiments (Student's t-test, two-sided, permutation-based FDR correction, FDR: 5%). AURORA-A, other alisertib-binding proteins and protein kinases are labeled.
Extended Data Figure 4.
(a) Model of the AURORA-A (blue) / CEREBLON (purple) complex. Structures of AURORA-A with alisertib and of CEREBLON with lenalidomide39 were used for protein-protein docking. The top-ranking solution is displayed (ACc1: AURORA-A-CEREBLON complex 1). Alisertib and lenalidomide are shown in green and aqua, respectively. (b) Model of the second-best AURORA-A (blue)-CEREBLON (purple) complex compatible with JB170 binding (ACc2). Structures of AURORA-A with alisertib and of CEREBLON with lenalidomide were used for protein-protein docking. Alisertib and lenalidomide are shown in green and aqua, respectively. (c) Modeled ternary complex structure of JB170 (orange) bound to the ACc1 shown in Extended Data Fig. 4a. Alisertib (green) and lenalidomide (aqua) were modified, connected and minimized to give JB170. (d) Modeled ternary complex of JB170 (orange) bound to the ACc2 shown in Extended Data Fig. 4b. Alisertib (green) and lenalidomide (aqua) were modified, connected and minimized to give JB170. (e) Docking solutions of the active degraders JB170 and JB158 as well as the less functional degraders JB159, JB169, JB171 and negative control JB211 in the AURORA-A (blue)-CEREBLON (purple) complexes ACc1 and ACc2. Scaffolds of thalidomide and alisertib used as light constraints during docking and as reference structures for the substructure root-mean-square deviation of atomic positions (RMSD) measurements are shown. Active degraders (labelled in green) achieve RMSD values below 1 Å with respect to both alisertib and the thalidomide scaffold. (f) Immunoblots of immunoprecipitation experiments. Cells expressing HA-tagged wildtype AURORA-A (WT), a version of AURORA-A (AURORA-AImut) with 12 amino acid substitutions (R137E, K153E, K156E, F157E, I158E, R189E, P191W, K224E, E239R, S266W, A267W and R375E) or an empty vector (Ctr) were treated with 0.5 µM JB170 for 6 hours. AURORA-A was precipitated with an anti-HA tag antibody, and the amount of co-precipitated CEREBLON was tested by immunoblotting. (g) AURORA-A levels based on luciferase measurements. Wildtype (WT) or AURORA-A with one amino acid substitution (AURORA-AP191W) were fused to luciferase fragment (HiBiT) and expressed in MV4-11 cells. Cells were treated with 1 µM alisertib for 6 hours, lysed, complemented with the second luciferase fragment (largeBiT), and measured for luciferase activity. Bars represent mean ± s.d. of n = 3 replicates.
Extended Data Figure 5.
(a) Isothermal titration calorimetry experiments of AURORA-A into TBD (left panel, no measurable binding was observed), the binary complexes of AURORA-A/JB170, CEREBLON(TBD)/JB170 and ternary complex of AURORA-A/CEREBLON(TBD)/JB170. Shown are the raw heat rates on the top panel with integrated, baseline-corrected heats per injection and the corresponding fits below. An overlay of all curves (merged) is shown on the bottom left. (b) Immunoblots of immunoprecipitation experiments. AURORA-A was precipitated with an anti-HA tag antibody from MV4-11 cells stably expressing HA-tagged AURORA-A or control cells transduced with an empty vector after treatment with 0.5 µM JB170 or 5 µM thalidomide (Thal.). The amount of co-precipitated CEREBLON and VHL was tested by immunoblotting. (c) Immunoblots of immunoprecipitation experiments. AURORA-A was precipitated with an anti-HA tag antibody from MV4-11 cells stably expressing HA-tagged AURORA-A (+) or from control cells (-) after treatment with CEREBLON-based degraders (JB170, JB171, JB211), VHL-based degraders (JB160, JB161) or DMSO. The amount of co-precipitated CEREBLON and VHL was tested by immunoblotting. (d) Immunoblots of immunoprecipitation experiments. BRD4 was precipitated with a specific antibody from MV4-11 cells after treatment with the VHL-based PROTAC MZ1 or DMSO in the presence of 5 mM MG132 for 1 hour. The amount of co-precipitated VHL was tested by immunoblotting. (e) Structural superposition of AURORA-A (blue, with bound alisertib in green) and AURORA-B. Residues with a maximum distance of 5 Å from CEREBLON in the AURORA-A-CEREBLON complex models are shown in cyan. Non-conserved amino acids near the proposed interaction interfaces of AURORA-A with CEREBLON are highlighted as labeled sticks and provided as a tabular list below the figure.
Extended Data Figure 6.
(a) Histogram of BrdU incorporation. The amount of incorporated BrdU is shown for cells in the S-phase in Fig. 5a. (b) Histogram of BrdU incorporation. The amount of incorporated BrdU is shown for cells in the S-phase in Fig 5e. (c) Immunoblot of AURORA-A. IMR5 cells overexpressing HA-tagged AURORA-A upon incubation with doxycycline (Dox) or control cells (EtOH) were treated with JB170 (1 µM) or alisertib (1 µM) for 18 hours. (d) Flow cytometry plots showing cell cycle distribution. IMR5 cells overexpressing AURORA-A upon incubation with doxycycline (Dox) or control cells (EtOH, as shown in Extended Data Fig. 6c) were treated with JB170 (1 µM) or alisertib (1 µM) for 18 hours. Cells were labeled with BrdU, stained with PI, and analyzed by flow cytometry. The amount of intercalating PI (top) and the correlation of BrdU to PI (bottom) are shown. (e) Histogram showing BrdU incorporation for cells in the S-phase in Extended Data Fig. 6d. (f) Immunoblot of AURORA-A. AURORA-A was depleted in IMR5 cells by an siRNA and AURORA-A levels were analyzed by immunoblotting for biological triplicates. Cells were analyzed by flow cytometry as shown in Extended Data Fig. 6g,h. (g) Cell cycle distribution analyzed by flow cytometry. IMR5 cells depleted for AURORA-A by siRNA were labeled with BrdU, stained with PI, and analyzed by flow cytometry. The amount of intercalating PI (top) and the correlation of BrdU to PI (bottom) are shown. (h) Histogram showing BrdU incorporation for cells in the S-phase in Extended Data Fig. 6g and two additional biological replicates. (i) Scatter plot of the AURORA-A interactome. The X-axis displays the enrichment (log2FC) of proteins in HA-AURORA-A-expressing cells compared to control cells (Ctr). The Y-axis displays the protein intensities (log10). Substrates of AURORA-A5 are labeled. (j) Immunoblots of HA-tagged AURORA-A and DICER1. HA-precipitations were performed from HEK293 cells transfected to express versions of HA-tagged catalytically inactive AURORA-A (AURORA-AD274N, AURORA-AK162R) or the wildtype protein. Exogenous AURORA-A was detected with an anti-HA tag antibody, and levels of DICER1 were analyzed in the input and immunoprecipitant by antibodies recognizing the endogenous protein. Control cells (Ctr) did not express HA-tagged protein. (k) AURORA-A sedimentation profile in the presence and absence of RNases. The graph is a re-analysis of published data46 retrieved from http://r-deep.dkfz.de/.
Extended Data Figure 7.
(a, b) Cell death assay. IMR5 cells expressing AURORA-AT217D upon incubation with doxycycline (Dox) or control cells (EtOH) were treated with JB170 (0.5 μM, low; 1 μM, high) or JB211 (0.5 μM, low; 1 μM, high) for 72 hours. Cells were stained with annexin (a) and PI (b), and apoptotic cells were counted by flow cytometry. (c) Fractions of apoptotic cells analyzed by flow cytometry. IMR5 cells expressing AURORA-AT217D upon incubation with doxycycline (Dox) or control cells (EtOH) were treated with JB170 (1 µM) for indicated times. Cells were stained with annexin and PI, and apoptotic cells were counted by flow cytometry (50,000 sorted events) (d) Bar diagram showing cellular viability. IMR5 cells expressing AURORA-AT217D upon incubation with doxycycline (Dox) or control cells (EtOH) were treated with 0.5 µM and 1 µM JB170 or JB211 for 72 hours, and cellular viability was measured by the alamarBlue assay. Bars represent mean ± s.d. of n = 3 replicate experiments. P-values were calculated with two-tailed unpaired t-test assuming equal variance.
Extended Data Figure 8. Visualization of the flow cytometry gating.
(a) FACS gating strategy for cell cycle distribution (BrdU-PI flow cytometry). The gating strategy is shown for one exemplary experiment. (b) FACS gating strategy for the analysis of annexin-positive cells.
Supplementary Material
Editorial Summary.
A bifunctional AURORA-A degrader induces the fast, specific degradation of this kinase in cancer cell lines, which then arrest in S-phase and undergo rampant apoptosis. This degrader is a prototype molecule for targeting non-catalytic, oncogenic functions of AURORA-A in cancer cells.
Acknowledgements
We thank André Kutschke for excellent technical assistance. Manuscript editing was provided by Valerie Matarese. We appreciate the scientific discussions with Martin Eilers and Kaspar Burger about the biology of AURORA-A and DICER1 and thank the Eilers lab for cell lines and plasmids. This work was supported by grants from the German Research Foundation (WO 2108/1-1 to EW, GRK 2243 to CS), the European Research Council (TarMYC to EW) and the Federal Ministry of Education and Research (CANCER to EW and SK). SM and MS are grateful for support by the SGC, a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [EUbOPEN, agreement No 875510], Janssen, Merck KGaA, MSD, , Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. MW is grateful for support by the Else Kröner-Fresenius PhD program (TRIP) and JB by the German Cancer Research Center DKTK.
Footnotes
Author Contributions
JB synthesized all degrader molecules and BA performed most of the cell-based experiments. MS performed BRET and calorimetry assays, MW synthesized E3-ligands and linkers. MD performed all computational modeling. JDS, JH, AN, MV, NDS, LE, AB, PB contributed experimental data. LS expressed recombinant proteins. AS, SH and BK performed and analyzed mass spectrometry experiments together with BA and JH. SH and BK analyzed alisertib and JB170 protein binding. CS supervised computational modeling studies. SK supervised the synthesis and biophysical experiments, and EW supervised the cell-based assays. SK and EW designed the study and wrote the manuscript with the help of all authors.
Declaration of Interests
BK is cofounder and shareholder of msAId GmbH and OmicScouts GmbH. BK has no operational role in either company. The University of Würzburg filed a patent for degraders described in the study, and EW is listed as an inventor.
Data Availability Statement
Sequence data have been deposited at GEO with accession number GSE141911. SILAC mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017342. TMT mass spectrometry proteomics data have been deposited with the data set identifier PXD019585 (PRIDE). AURORA-A interactome data have been deposited with the data set identifier PXD019684 (PRIDE). AURORA-A sedimentation profile in the presence and absence of RNases was obtained from http://r-deep.dkfz.de (AURKA_HUMAN.pdf).
Code Availability Statement
Data analysis was mostly performed with published analysis tools (as described above). Custom code was only used for standard operations and will be made available by the corresponding authors on request.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 4.Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 5.Kettenbach AN, et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochegger H, Hegarat N, Pereira-Leal JB. Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol. 2013;3:120185. doi: 10.1098/rsob.120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner MM, et al. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–9. [PubMed] [Google Scholar]
- 8.Bischoff JR, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–65. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–93. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–58. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 11.Treekitkarnmongkol W, et al. Aurora kinase-A overexpression in mouse mammary epithelium induces mammary adenocarcinomas harboring genetic alterations shared with human breast cancer. Carcinogenesis. 2016;37:1180–1189. doi: 10.1093/carcin/bgw097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong X, et al. Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of the RB1 Tumor Suppressor Gene. Cancer Discov. 2019;9:248–263. doi: 10.1158/2159-8290.CD-18-0469. [DOI] [PubMed] [Google Scholar]
- 13.Wu C, et al. Targeting AURKA-CDC25C axis to induce synthetic lethality in ARID1A-deficient colorectal cancer cells. Nat Commun. 2018;9:3212. doi: 10.1038/s41467-018-05694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollaoglu G, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell. 2017;31:270–285. doi: 10.1016/j.ccell.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Borisa AC, Bhatt HG. A comprehensive review on Aurora kinase: Small molecule inhibitors and clinical trial studies. Eur J Med Chem. 2017;140:1–19. doi: 10.1016/j.ejmech.2017.08.045. [DOI] [PubMed] [Google Scholar]
- 17.Gorgun G, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115:5202–13. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connor OA, et al. Randomized Phase III Study of Alisertib or Investigator's Choice (Selected Single Agent) in Patients With Relapsed or Refractory Peripheral T-Cell Lymphoma. J Clin Oncol. 2019;37:613–623. doi: 10.1200/JCO.18.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DuBois SG, et al. Phase II Trial of Alisertib in Combination with Irinotecan and Temozolomide for Patients with Relapsed or Refractory Neuroblastoma. Clin Cancer Res. 2018;24:6142–6149. doi: 10.1158/1078-0432.CCR-18-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosse YP, et al. A Phase 2 Study of Alisertib in Children with Recurrent/Refractory Solid Tumors or Leukemia: Children's Oncology Group Phase 1 and Pilot Consortium ADVL0921) Clin Cancer Res. 2019 doi: 10.1158/1078-0432.CCR-18-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beltran H, et al. A Phase II Trial of the Aurora Kinase A Inhibitor Alisertib for Patients with Castration-resistant and Neuroendocrine Prostate Cancer: Efficacy and Biomarkers. Clin Cancer Res. 2019;25:43–51. doi: 10.1158/1078-0432.CCR-18-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brockmann M, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013;24:75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dauch D, et al. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med. 2016;22:744–53. doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
- 24.Toya M, Terasawa M, Nagata K, Iida Y, Sugimoto A. A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat Cell Biol. 2011;13:708–14. doi: 10.1038/ncb2242. [DOI] [PubMed] [Google Scholar]
- 25.Zheng F, et al. Nuclear AURKA acquires kinase-independent transactivating function to enhance breast cancer stem cell phenotype. Nat Commun. 2016;7:10180. doi: 10.1038/ncomms10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards MW, et al. Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc Natl Acad Sci U S A. 2016;113:13726–13731. doi: 10.1073/pnas.1610626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustafson WC, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014;26:414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burslem GM, Crews CM. Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell. 2020 doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzl A, Winter GE. Targeted protein degradation: current and future challenges. Curr Opin Chem Biol. 2020;56:35–41. doi: 10.1016/j.cbpa.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter GE, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science (New York, N Y ) 2015;348:1376–81. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bondeson DP, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–7. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniaci C, Ciulli A. Bifunctional chemical probes inducing protein-protein interactions. Curr Opin Chem Biol. 2019;52:145–156. doi: 10.1016/j.cbpa.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Sloane DA, et al. Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem Biol. 2010;5:563–76. doi: 10.1021/cb100053q. [DOI] [PubMed] [Google Scholar]
- 34.Dodson CA, et al. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem J. 2010;427:19–28. doi: 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]
- 35.Douglass EF, Jr, Miller CJ, Sparer G, Shapiro H, Spiegel DA. A comprehensive mathematical model for three-body binding equilibria. J Am Chem Soc. 2013;135:6092–9. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crosio C, et al. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22:874–85. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114:3749–57. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 38.Klaeger S, et al. The target landscape of clinical kinase drugs. Science. 2017;358 doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamberlain PP, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21:803–9. doi: 10.1038/nsmb.2874. [DOI] [PubMed] [Google Scholar]
- 40.Farnaby W, et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat Chem Biol. 2019;15:672–680. doi: 10.1038/s41589-019-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gadd MS, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zengerle M, Chan KH, Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem Biol. 2015;10:1770–7. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elkins JM, Santaguida S, Musacchio A, Knapp S. Crystal structure of human aurora B in complex with INCENP and VX-680. J Med Chem. 2012;55:7841–8. doi: 10.1021/jm3008954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuchiya Y, et al. Covalent Aurora A regulation by the metabolic integrator coenzyme A. Redox Biol. 2020;28:101318. doi: 10.1016/j.redox.2019.101318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buchel G, et al. Association with Aurora-A Controls N-MYC-Dependent Promoter Escape and Pause Release of RNA Polymerase II during the Cell Cycle. Cell Rep. 2017;21:3483–3497. doi: 10.1016/j.celrep.2017.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caudron-Herger M, et al. R-DeeP: Proteome-wide and Quantitative Identification of RNA-Dependent Proteins by Density Gradient Ultracentrifugation. Mol Cell. 2019;75:184–199 e10. doi: 10.1016/j.molcel.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bondeson DP, et al. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem Biol. 2018;25:78–87 e5. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remillard D, et al. Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands. Angew Chem Int Ed Engl. 2017;56:5738–5743. doi: 10.1002/anie.201611281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baluapuri A, et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol Cell. 2019;74:674–687 e11. doi: 10.1016/j.molcel.2019.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 51.Zecha J, et al. TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach. Mol Cell Proteomics. 2019;18:1468–1478. doi: 10.1074/mcp.TIR119.001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bian Y, et al. Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC-MS/MS. Nat Commun. 2020;11:157. doi: 10.1038/s41467-019-13973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 54.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–40. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 55.Medard G, et al. Optimized chemical proteomics assay for kinase inhibitor profiling. J Proteome Res. 2015;14:1574–86. doi: 10.1021/pr5012608. [DOI] [PubMed] [Google Scholar]
- 56.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–26. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fedorov O, Niesen FH, Knapp S. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol Biol. 2012;795:109–18. doi: 10.1007/978-1-61779-337-0_7. [DOI] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited at GEO with accession number GSE141911. SILAC mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017342. TMT mass spectrometry proteomics data have been deposited with the data set identifier PXD019585 (PRIDE). AURORA-A interactome data have been deposited with the data set identifier PXD019684 (PRIDE). AURORA-A sedimentation profile in the presence and absence of RNases was obtained from http://r-deep.dkfz.de (AURKA_HUMAN.pdf).
Data analysis was mostly performed with published analysis tools (as described above). Custom code was only used for standard operations and will be made available by the corresponding authors on request.