Abstract
Targeted gene editing in hematopoietic stem cells (HSCs) is a promising treatment for several diseases. However, the limited efficiency of homology-directed repair (HDR) in HSCs and the unknown impact of the procedure on clonal composition and dynamics upon transplantation have hampered clinical translation. Here, we apply a barcoding strategy to clonal tracking of edited cells (BAR-Seq) and show that editing activates p53, which significantly shrinks the HSC clonal repertoire in hematochimeric mice, although engrafted edited clones preserved multilineage and self-renewing capacity. Transient p53 inhibition restored polyclonal graft composition. We increased HDR efficiency by forcing cell cycle progression and upregulating components of the HDR machinery through transient expression of the Adenovirus 5 E4orf6/7 protein, which recruits the cell cycle controller E2F on its target genes. Combined E4orf6/7 expression and p53 inhibition resulted in HDR editing efficiencies of up to 50% in the long-term human graft, without perturbing repopulation and self-renewal of edited HSCs. This enhanced protocol should broaden applicability of HSC gene editing and pave its way to clinical translation.
The therapeutic potential of HSC gene therapy has been shown in several clinical trials for inherited diseases and may be advanced by targeted genome editing, which allows in situ correction of mutant alleles, restoring function and physiological expression control1. Programmable nucleases, such as CRISPR/Cas, enable gene editing by introducing site-specific DNA double strand breaks (DSBs) into the genome2. DSB repair may occur by non-homologous end-joining (NHEJ), which often introduces small insertion/deletion (indels) at the repaired site, or by the high-fidelity homology-directed repair (HDR), which is exploited for gene correction or targeted integration using an exogenous DNA template. HDR predominantly occurs in S/G2 phases. Unfortunately, HSCs are poorly permissive to HDR, likely due to quiescence and limited template uptake3. Despite recent improvements achieved by culturing hematopoietic stem/progenitor cells (HSPC) with StemRegenin 1 (SR1) and UM1713–6, and using adeno-associated vector serotype 6 (AAV6) for template delivery4,7–11, HDR remains constrained in long-term repopulating HSCs (LT-HSCs) and thus limits applicability of gene correction. Whereas a limited proportion of edited HSCs may successfully treat diseases characterized by selective advantage of the functional progeny4, this might be insufficient in most other conditions, where a large proportion of non-corrected cells competes with the edited ones for engraftment and limits functional hematopoietic reconstitution. Several strategies have been attempted to enhance HDR12 but the efficacy of these approaches in HSPCs is limited. Some helper adenoviral (Ad) proteins which, during AAV infection, promote viral genome processing and modulate host cell responses, have been shown to increase AAV template expression13,14 but not HDR in LT-HSCs10.
We recently showed that transient activation of the p53 pathway occurs in HSPCs even after a single DSB, leading to reversible proliferation delay15. Concomitant exposure to AAV6 led to cumulative and robust p53 activation, causing proliferation arrest and strongly impacting hematopoietic reconstitution upon xenotransplantation in immunodeficient mice. Inhibition of this p53 response by transient expression of a dominant-negative p53 mutant protein (GSE56) during editing increased hematopoietic repopulation by treated cells. It remains unknown whether such outcome was due to altered growth properties or improved preservation of LT-HSCs during editing.
Little information is available on the clonogenic output and multilineage repopulation capacity of individual HSPCs after editing. A low yield of edited HSCs may delay hematopoietic recovery, exposing patients to high risk of infection, and result in oligoclonal hematopoiesis, which may impair graft resilience and potentially increase the risk of leukemia and myelodysplastic syndrome16.
Here, we developed an enhanced gene editing protocol yielding high proportions of edited LT-HSCs by overcoming two major biological barriers, robust p53 response and constrained HDR. Clonal tracking of edited HSPCs proved polyclonal reconstitution and preserved self-renewal and multi-potency of individual edited HSCs, giving confidence to future clinical translation.
Results
Barcoded template enables clonal tracking of edited HSPCs and shows reconstitution by few dominant clones with preserved multilineage potential
We selected the Adeno-Associated Virus Site 1 (AAVS1) as paradigmatic safe harbor for targeted transgene insertion17. We embedded a 22-bp degenerated heritable “barcode” sequence (BAR) in the repair template downstream of a GFP reporter cassette (Fig. 1a) and generated a plasmid library and an AAV6 pool of high and comparable complexity (7.5x105 and 5.9x105 unique BARs, respectively) and nearly homogeneous representation of degenerated consensus sequences (Fig. 1b).
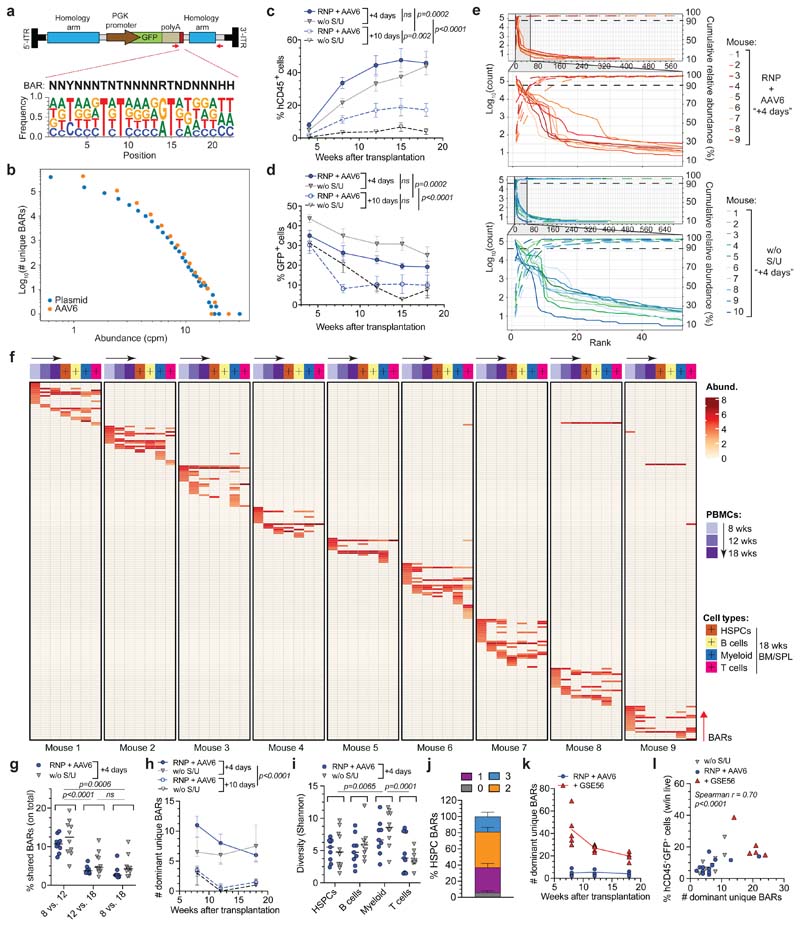
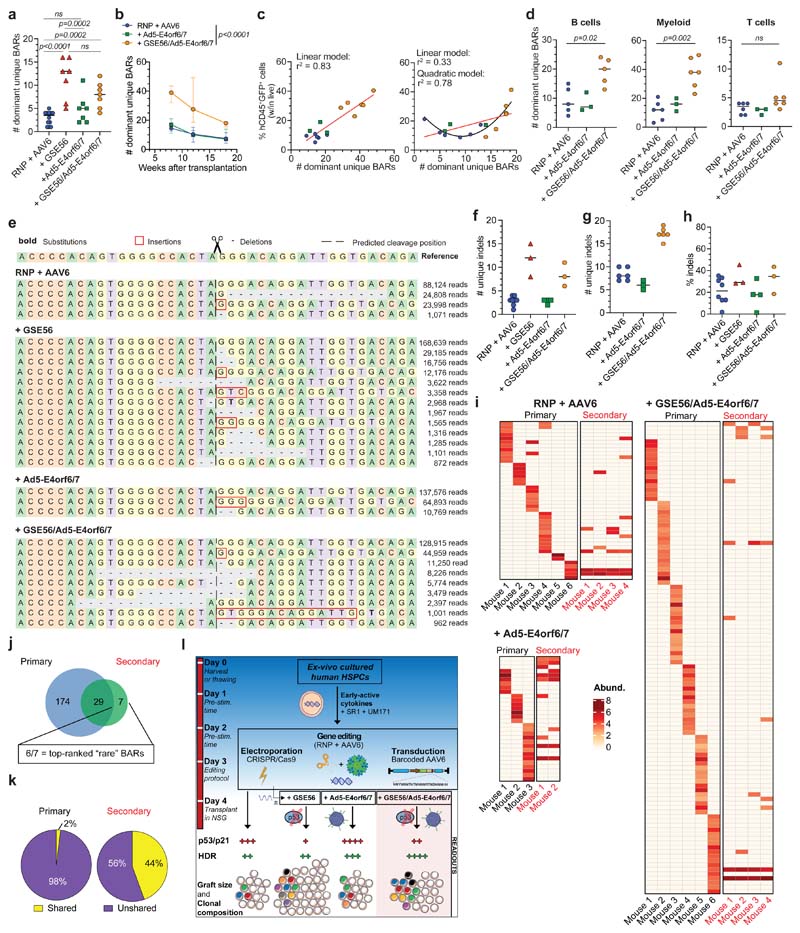
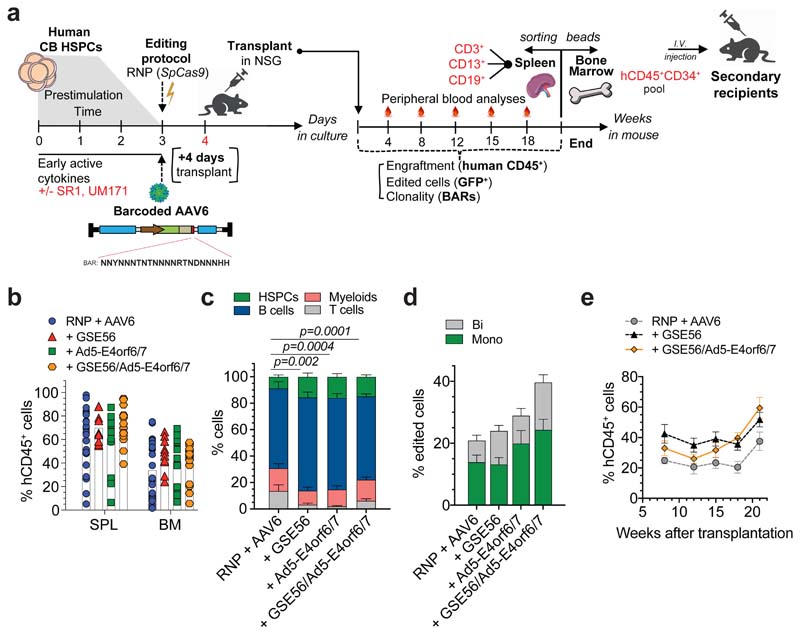
Figure 1. BAR-Seq enables clonal tracking of human HDR-edited HSPCs.
a, Top: schematic of the barcoded AAV6 library for AAVS1 editing and BAR consensus sequence downstream of the GFP reporter. Arrows indicate primer binding sites for plasmid/AAV sequencing. Bottom: logo plot showing the nucleotide frequency in the BAR sequence. b, Number of unique BARs and their abundances (counts per million, cpm) in plasmid and AAV6 libraries. c-d, Percentage of circulating human CD45+ (hCD45+) cells (c) and GFP+ cells within human graft (d) in mice transplanted one day (“+4 days”) or one week (“+10 days”) after editing of HSPCs cultured in presence (“RNP + AAV6”) or absence (“w/o S/U”) of SR1/UM171 (n = 9, 10, 10, 10). Mean ( SEM. Linear Mixed Effects models (LME) followed by post-hoc analysis. Statistics are shown for the last timepoint. e, Abundance of ranked BARs in 18-weeks PBMCs from “+4 days” mice. Solid and dashed lines show absolute and cumulative relative abundance (saturation curves) of ranked BARs, respectively. Magnification of most abundant BARs is shown. f, Heatmap showing the abundance (red-scaled palette) of dominant unique BARs (rows) in “RNP + AAV6 (+4 days)” mice (separated columns) in PBMCs at indicated times after transplant and sorted hCD45+ cell types. g, Percentage of BARs shared between PBMCs harvested at indicated timepoints (“+4 days” mice; n = 9, 10). Median. Friedman test with Dunn’s multiple comparisons. h, Longitudinal PBMC analysis showing the number of dominant unique BARs in analyzed mice from Fig. 1c (n = 9, 10, 6, 3). Median with interquartile range (IQR). Generalized linear Mixed Effects models (GLMER) for count data. i, Clonal diversity within sorted hCD45+ cell types (“+4 days” mice; n = 9, 10). Median. Friedman test with Dunn’s multiple comparisons. j, Percentage of dominant unique HSPC BARs shared with none, 1, 2 or 3 sorted hCD45+ cell lineages (“+4 days” mice; n = 19). Mean M SEM. k, Longitudinal PBMC analysis showing the number of dominant unique BARs in mice transplanted with HSPCs edited in absence or presence of GSE56 and retrieved when including in the analysis >90% of total BAR reads (n = 4, 5). Median. l, Correlation between the percentage of hCD45+GFP+ cells and the number of dominant unique BARs in 18-weeks PBMCs of mice from Fig. 1h, k (n = 28). Spearman correlation coefficient was calculated. For Fig. 1g, h, i, j experimental groups were unified for statistical analysis. All statistical tests are two-tailed. n indicate independent animals.
We then edited AAVS1 in human cord blood (CB) HSPCs by electroporating CRISPR/SpCas9 ribonucleoprotein (RNP) with a highly specific chemically modified guide RNA (gRNA)15 and found similar editing efficiency of the barcoded library compared to non-barcoded AAV6, as assessed by GFP+ cells percentage in the treated cells outgrowth (Extended Data Fig. 1a). Deep sequencing of on-target BARs in HSPCs revealed highly diverse repertoire of similar magnitude as the edited cells (54,865 and 27,477 unique BARs retrieved from r200,000 cells edited to 65% efficiency), with only one or two slightly overrepresented (< 0.25%) BAR/sample (Extended Data Fig. 1b).
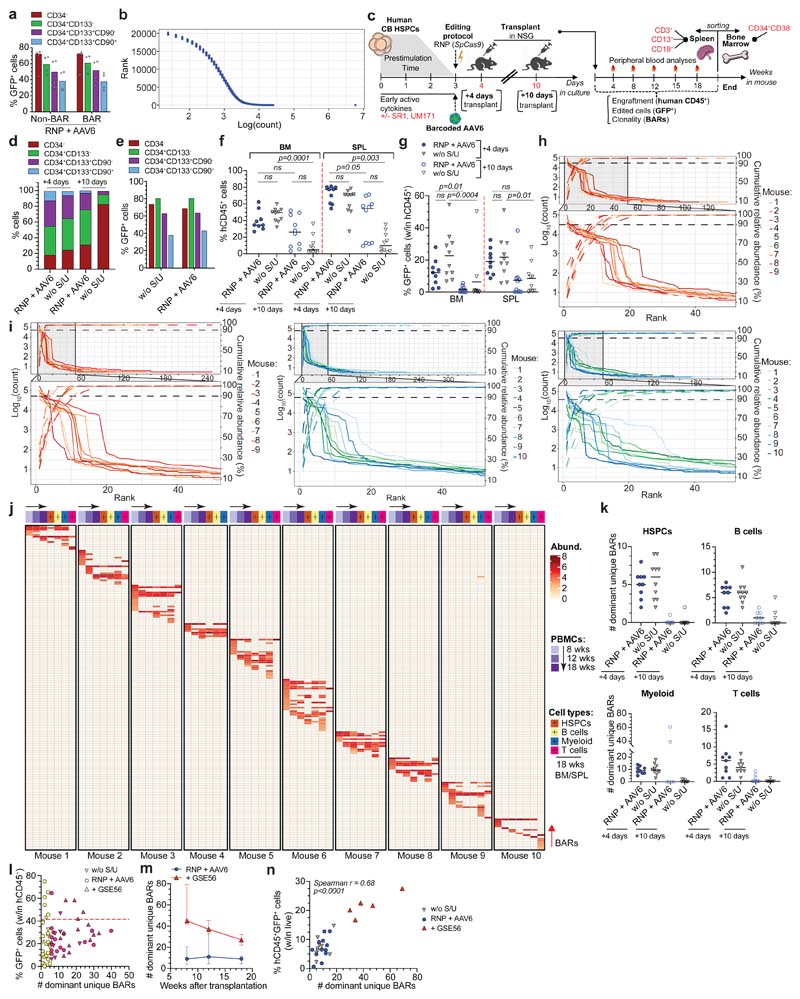
The impact of prolonging culture time on graft clonality and the stem-preserving activity of SR1 and UM171 in the context of gene editing have never been evaluated. We treated the same starting number of HSPCs for AAVS1 editing in presence or absence of SR1/UM171 and transplanted the total outgrowth in NSG mice one day (corresponding to 4th day of culture; “+4 days”) or one week after editing (“+10 days”) (Extended Data Fig. 1c). Analyses of treated cells showed more phenotypically primitive progenitors (CD34+CD133+CD90+, hereafter named “CD90+”) in presence of SR1/UM171 at both times, with comparable editing efficiencies between treatments. CD90+ cells decreased with time in all cultures and became nearly absent without SR1/UM171 (Extended Data Fig. 1d, e). Concordantly, “+4 days”-transplanted mice showed higher human cell engraftment in peripheral blood (PB) and hematopoietic organs compared to the “+10 days” groups. SR1/UM171 increased human engraftment in both comparisons (Fig. 1c and Extended Data Fig. 1f). Despite similar and high percentage of GFP+ cells among all groups in PB at 4 weeks after transplant, “+10 days”-transplanted mice showed decreased GFP marking at later times (Fig. 1d and Extended Data Fig. 1g).
Mice showing detectable (>0.1%) engraftment of human GFP+ cells in PB at 18 weeks were selected for clonal analyses, which included all mice of the “+4 days” and only 6/10 and 3/10 mice of the “+10 days” groups with or without SR1/UM171, respectively. Sequencing of on-target BARs (“BAR-Seq”) from PB mononuclear cells (PBMCs) of “+4 days”-transplanted mice at different times after transplant revealed from f60 to 700 unique BARs/mouse, which segregated in two populations with log-difference in abundance. Upon ranking from the most to the least abundant BAR within each sample, we applied a saturation-based approach and defined “dominant” the small set of BARs accounting for >90% of total abundance and “rare” the remaining ones (Fig. 1e and Extended Data Fig. 1h, i). We then focused our clonal dynamics analyses on dominant BARs, which robustly contribute to hematopoiesis.
Longitudinal analysis within PB of “+4 days”-transplanted mice showed progressive shrinking, up to disappearance, of some dominant clones between 8 and 12-18 weeks, as well as emergence of new dominant BARs at 12 and 18 weeks (Fig. 1f). The fraction of BARs shared between different time points in each mouse was higher in the 8 vs. 12 weeks comparison respect to the 8 or 12 vs. 18 weeks ones (Fig. 1g), independently from ex vivo culture conditions (Extended Data Fig. 1j) and suggested distinct early and steady-state reconstitution phases driven by different clones. The number of dominant BARs was significantly lower in the “+10 days” transplanted groups, suggesting loss of engrafting clones and/or expansion of a limited subset in prolonged culture. Despite a trend for higher number of dominant BARs short term after transplant with HSPCs cultured in presence of SR1/UM171, the edited long-term graft was mostly composed of 6-7 dominant clones per mouse (Fig. 1h).
Contribution to different lineages was similar among treatments, with myeloid and T cells showing highest and lowest clonality, respectively (Fig. 1f, i and Extended Data Fig. 1j, k). Most BARs retrieved from CD34+CD38- HSPCs sorted from bone marrow (BM) of engrafted mice were shared with ≥2 differentiated hematopoietic lineages, confirming at clonal level the multipotent long-term repopulation capacity of individual HDR-edited HSPCs in vivo (Fig. 1f, j and Extended Data Fig. 1j).
Inter-mice BAR sharing was rare but detectable within the same experimental group. The high complexity of the library makes it unlikely that the same BAR integrates in different LT-HSC and collisions during sequencing can be ruled out because we focused our analysis only on abundant clones. Thus, detection of the same BAR among dominant clones of two mice suggests duplication during ex vivo culture of HDR-edited HSPCs which maintain repopulation potential in xenografts (Fig. 1f and Extended Data Fig. 1j). There was no linear correlation between the number of HDR-edited dominant clones and the percentage of GFP+ cells within the human graft. The fewer were the engrafting clones (and, consequently, the smaller the human graft) the higher the variability in GFP marking, as expected from limited sampling of the input population; the more were the engrafting clones (and the larger the size of the human graft) the less the variability in GFP marking and the closer its level to that of the input population (Extended Data Fig. 1l).
Overall, these results show oligoclonal composition of the human graft reconstituted by edited HSPCs which have maintained multi-lineage output and short or long-term self-renewing potential. Addition of SR1/UM171 improved the early phase of reconstitution by increasing the number of short-term contributing clones and the overall extent of repopulation after prolonged ex vivo culture.
P53 activation constrains the number but not output of repopulating edited HSPCs
Co-electroporation of mRNA for the dominant-negative p53 mutant GSE56 with the editing reagents substantially increased (>5 fold) the number of dominant BARs contributing to xenograft hematopoiesis (Fig. 1k), providing a mechanistic explanation for the reported increase in the human graft15. This finding held true also when we expanded the analysis to encompass a larger proportion of BAR reads (Extended Data Fig. 1m). The size of the human edited graft (measured as percentage of human hCD45+GFP+ cells within the total live PBMCs) in mice transplanted with a non-saturating dose of edited cells significantly correlated at early and late times of reconstitution with the number of unique dominant BARs identified, indicating that neither p53 activation induced by gene editing nor its alleviation by GSE56 altered the average clonal output of individual repopulating human HSPCs. Moreover, the average clonal output of individual progenitors was lower at early than late times post-transplant, in line with progressive exhaustion of short-term progenitors (Fig. 1l and Extended Data Fig. 1n).
Adenoviral protein E4orf6/7 improves editing efficiency of human HSPCs
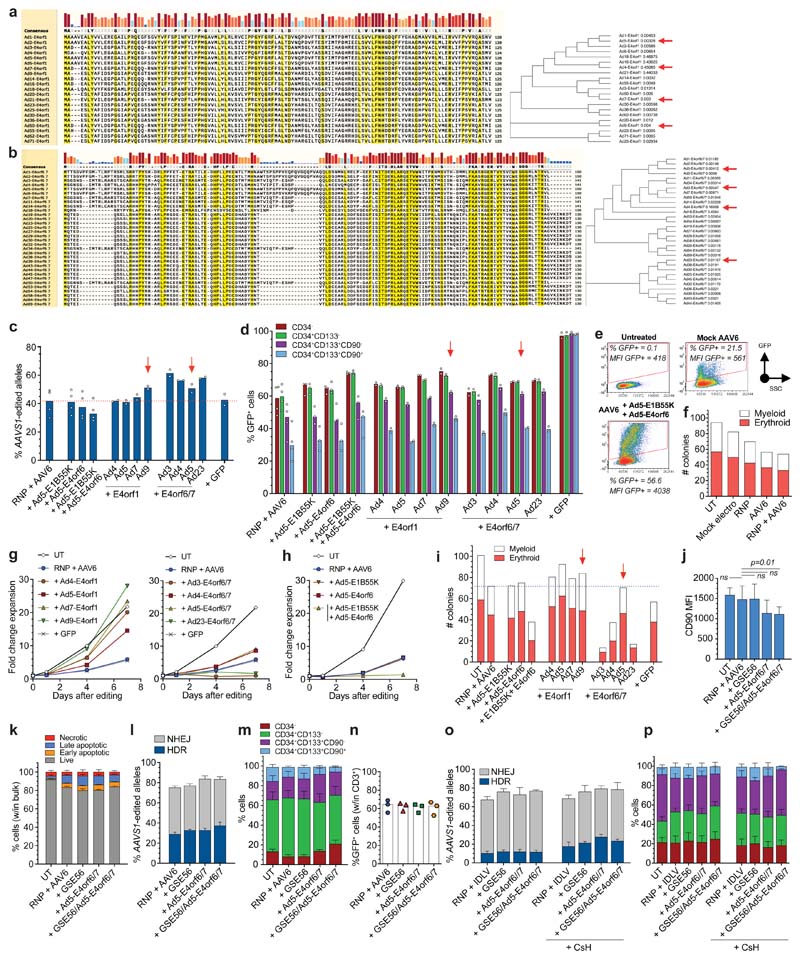
We screened a panel of Ad proteins known to function as helpers in Ad-AAV co-infection18,19. We focused on E4orf1 and E4orf6/7, which interact with cellular components involved in survival20–22 and cell cycle23. Since some viral gene properties differ among Ad serotypes, we screened four serotype variants (Extended Data Fig. 2a, b). We also tested serotype 5 E1B55K and E4orf6 proteins, previously described to increase AAV DNA second strand synthesis10,13,14. Ad proteins were expressed by HPLC-purified mRNAs24 co-delivered with AAVS1-editing reagents (Fig. 2a). All E4orf6/7 variants and Ad9-E4orf1 increased HDR in HSPCs, including the CD90+ fraction (Extended Data Fig. 2c, d). As previously described in primary T cells14, the combination of E1B55K and E4orf6 increased the percentage of GFP+ HSPCs but molecular analysis did not show any HDR improvement, suggesting that increased GFP expression was mainly derived from the AAV6 donor whose double-stranded DNA synthesis was promoted (Extended Data Fig. 2e). As expected from previous reports15,25,26, HSPC editing decreased the proportion of colony-forming cells from the cumulative impact of several steps of the treatment (Extended Data Fig. 2f). E4orf1 variants increased cell proliferation counteracting the editing induced delay, while Ad4- and Ad5-E4orf6/7 did not influence cell growth. Ad3- and Ad23-E4orf6/7 as well as the combination of Ad5-E1B55K and Ad5-E4orf6 strongly decreased cell growth (Extended Data Fig. 2g, h). In agreement, the in vitro colony-forming potential of treated HSPCs was increased by E4orf1 proteins, while it was strongly reduced by the combination of E1B55K and E4orf6 or E4orf6/7 proteins, except for the Ad5-E4orf6/7 variant, which resulted similar to the standard condition (Extended Data Fig. 2i). Overall, this screening prompted further investigation of Ad5-E4orf6/7 and Ad9-E4orf1 (Supplementary Fig. 1) as enhancers of HDR editing.
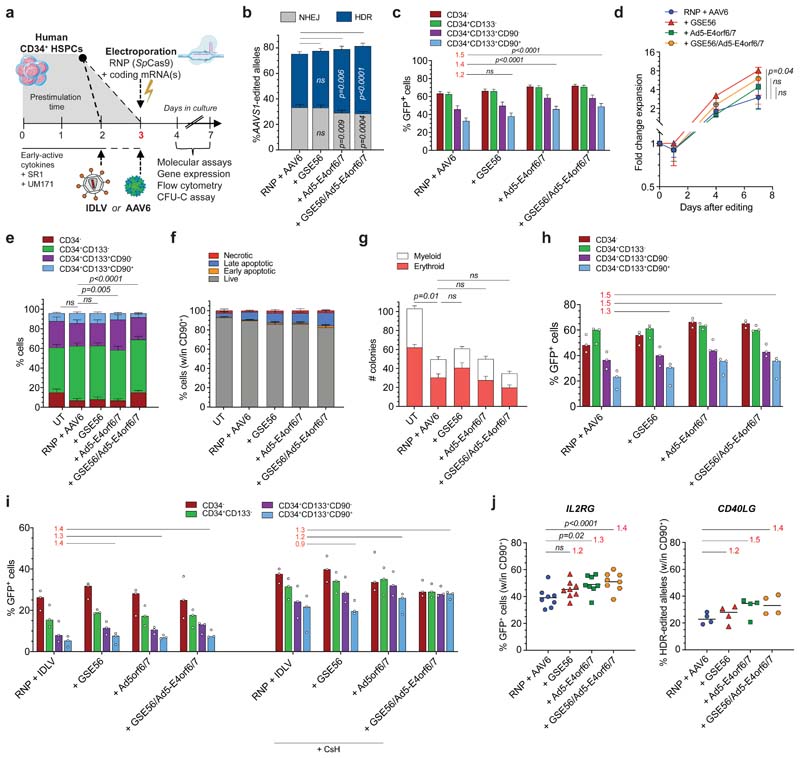
Figure 2. Combined transient expression of Ad5-E4orf6/7 and GSE56 improves editing efficiency in human HSPCs.
a, Experimental workflow. b-c, Percentage of HDR/NHEJ-edited alleles in bulk CB HSPCs (b) and GFP+ cells within subpopulations (c) 96 h after AAVS1 editing with standard protocol (“RNP + AAV6”), in presence of GSE56, Ad5-E4orf6/7 or their combination (35 HSPC donors; n = 15). Mean 3 SEM. d, Fold change expansion of live HSPCs after indicated treatments (9 HSPC donors; n = 5). Median F IQR. Statistical analysis performed at the last timepoint. e, Culture composition 96 h after editing in experiments from Fig. 2b (n = 15). Mean SEM. f, Percentage of live, early/late apoptotic and necrotic CD90+ cells 24 h after editing in the indicated conditions (7 HSPC donors; n = 3). Mean SEM. g, Number of colonies in the indicated conditions (19 HSPC donors; n = 10). Mean N SEM. h, Percentage of GFP+ cells within subpopulations 96 h after AAVS1 editing of mPB HSPCs with the indicated treatments (5 HSPC donors; n = 3). Median. i, Percentage of GFP+ cells within subpopulations 96 h after IDLV-based AAVS1 editing of CB HSPCs with indicated treatments (3 HSPC donors; n = 3). Median. j, Percentage of GFP+ cells (left) and HDR-edited alleles (right) in CD90+ cells 96 h after IL2RG or CD40LG editing, respectively (IL2RG: 12 HSPC donors; n = 8. CD40LG: 4 HSPC donors; n = 4). Median. For all panels with statistical analyses: Friedman test with two-tailed Dunn’s multiple comparisons against “RNP + AAV6”. For panel 2c, h-j: red numbers represent the fold increases of the center values for the percentage of GFP+ cells over “RNP + AAV6” within CD90+ compartment. n indicate independent experiments.
We then evaluated the effect of Ad5-E4orf6/7 together with GSE56, transiently co-expressed by separate mRNAs or single RNA encoding a fusion protein with P2A self-cleaving peptide. Across multiple independent experiments, these combinations increased HDR by an average 50% in CD90+ HSPCs as compared to the standard protocol, and elected the fusion construct for further studies to lower overall mRNA input (Fig. 2b, c). Cell growth was higher when adding GSE56, as previously reported15, although less markedly when combined with Ad5-E4orf6/7 (Fig. 2d). Ad5-E4orf6/7 treatment, with or without GSE56, decreased the fraction of CD90+ cells measured in culture (Fig. 2e), an effect apparently due to lower CD90 expression on cell surface (Extended Data Fig. 2j). Toxicity was mild for all treatments, doubling the fraction of apoptotic/necrotic cells detected in untreated samples up to an average 15% in bulk and CD90+ cells (Extended Data Fig. 2k and Fig. 2f). Colony-forming potential was similarly reduced for all editing treatments as compared to untreated cells, with a trend towards more colonies for GSE56 treated cells and less for the GSE56/Ad5-E4orf6/7 combination, without detectable difference in erythroid to myeloid ratio (Fig. 2g). We also reproduced the increase in HDR editing by Ad5-E4orf6/7 in human mobilized peripheral blood (mPB) HSPCs, reaching up to 1.5-fold within phenotypically primitive cells (Fig. 2h and Extended Data Fig. 2l, m). On the contrary, we did not observe higher efficiency of targeted integration in T cells (Extended Data Fig. 2n).
We then asked whether the effect of Ad5-E4orf6/7 on HDR editing was specific for the AAV donor template. Integrase defective lentiviral vectors (IDLV), although generally less effective than AAV6 as donor template for HSPCs, might be useful to increase cargo capacity and lower predicted immunogenicity. We thus edited AAVS1 with a suitably matching IDLV donor in presence or absence of GSE56, Ad5-E4orf6/7 and cyclosporin H, which increases IDLV transduction27. Ad5-E4orf6/7 boosted HDR up to 1.5 fold in primitive CB HSPCs, reaching up to 35% HDR, with similar effects on culture composition as reported above for the AAV template (Figure 2i and Extended Data Fig. 2o, p).
We then assessed whether our AAV-based protocol was portable to other genomic sites by measuring HDR editing in IL2RG and CD40LG, whose defective mutation cause severe primary immunodeficiencies amenable to HSPC gene therapy. We found a 1.4/1.5-fold increase in HDR by Ad5-E4orf6/7 with or without GSE56 in CD90+ cells as compared to standard condition, reaching up to an average of 50% and 35% GFP+ cells upon IL2RG or CD40LG editing, respectively (Fig. 2j).
Ad5-E4orf6/7 activates the E2F transcriptional pathway upregulating HDR machinery and forcing progression to S/G2 cell cycle phases
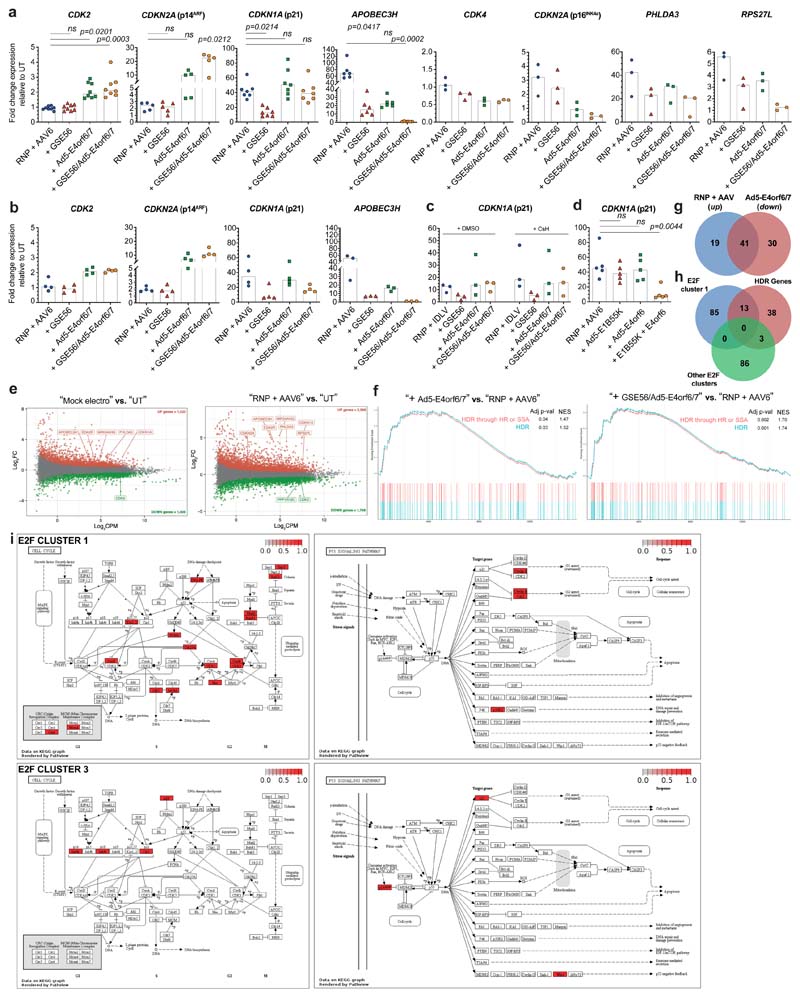
We investigated the cellular response triggered by Ad5-E4orf6/7, which has been reported to directly interact with the master cell cycle regulator E2F, leading to its binding and transcriptional activation of the Ad E2 promoter23,28. We measured expression of cell cycle-related genes upon AAVS1 editing in presence or absence of GSE56 and/or Ad5-E4orf6/7, both in CB and mPB HSPCs. As reported15, GSE56 dampened the editing-induced activation of DNA damage response (DDR) through p53 target genes, such as CDKN1A (p21), RPS27L, PHLDA3 and APOBEC3H 29. Ad5-E4orf6/7 transiently activated CDK2, which promotes S/G2progression30, but also upregulated the E2F target genes CDKN1A (p21) and CDKN2A (p14ARF), which foster cell cycle arrest31,32. Ad5-E4orf6/7 downregulated APOBEC3H, RPS27L, PHLDA3 and CDKN2A (p16INK4a), an effect further increased by combination with GSE56. No differences were found in these transcriptional responses across HSPC sources (Fig. 3a and Extended Data Fig. 3a, b). p21 responses to the different treatments showed similar patterns when using IDLV instead of AAV template (Extended Data Fig. 3c). Whereas editing-induced p53 activation was also dampened by the combination of E1B55K and E4orf6 (Extended Data Fig. 3d) as expected from its p53 degradation activity33, this treatment decreased cell growth and clonogenicity (Extended Data Fig. 2g, i above), showing that pleiotropic proteins interfering with DDR may have drastically different outcomes.
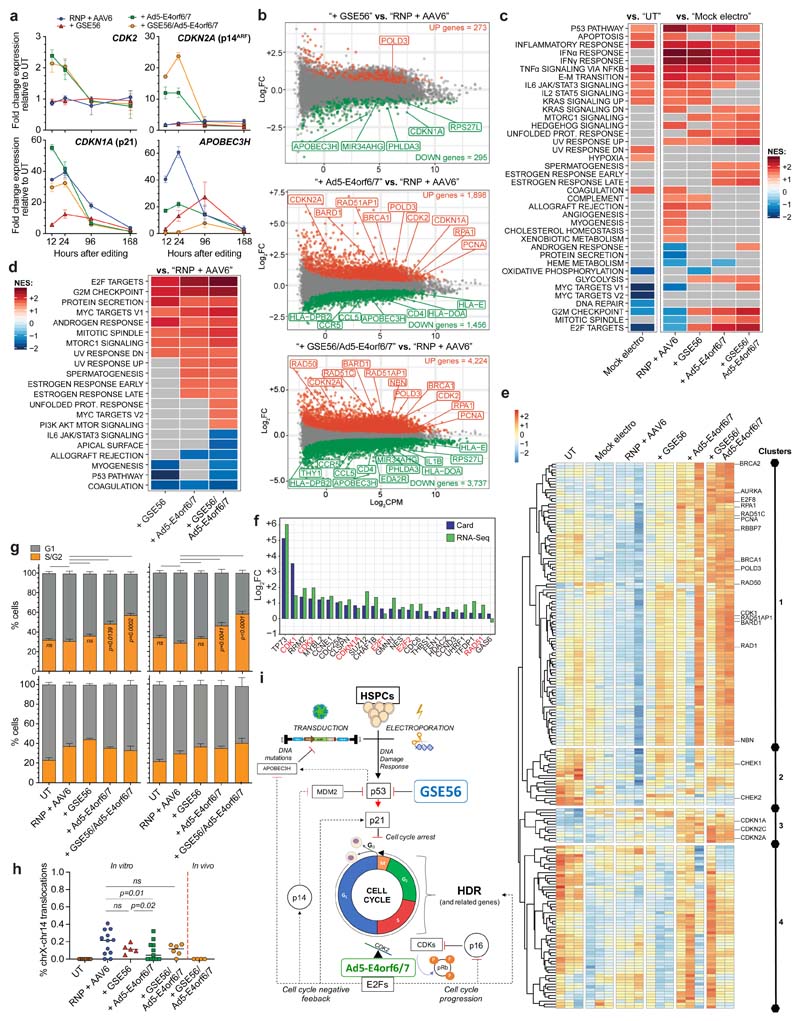
Figure 3. Ad5-E4orf6/7 forces cell cycle progression and upregulates HDR machinery via E2F pathway.
a, Fold change expression over time of CDK2, CDKN2A (p14arf), CDKN1A (p21), APOBEC3H relative to UT (7 HSPC donors; n = 3). Median. b, MA plots showing significant down- (green) and up- (red) regulated genes after AAVS1 editing in presence of GSE56 (top), Ad5-E4orf6/7 (middle) and their combination (bottom) against standard protocol (n = 3). c-d, Heatmaps showing NES from GSEA of indicated comparisons against the Hallmark gene set (Molecular Signatures Database). DEGs were ranked by log2FC expression. e, Heatmap showing normalized read counts for E2F target genes (Hallmark gene set) across samples. Full gene list is available in Supplementary Table 1. f, Log2FC expression values for top 25 up-regulated cell cycle related genes in sorted CD90+ HSPCs edited in presence or absence of Ad5-E4orf6/7 (blue) (n = 1). Comparison with RNA-Seq log2FC expression values (green) is shown. g, Percentage of bulk (left) and CD90+ (right) HSPCs in G1 or S/G2 phases 12-24 h (top; n = 6) and 96 h (bottom; n = 3) after indicated treatments. Mean) SEM. Friedman test with Dunn’s multiple comparisons against “RNP + AAV6” for 12-24 h. h) Percentage of IL2RG alleles harboring chromosome X-14 translocation 3 days after indicated treatments from Fig. 2j and in splenocytes of mice from Figure 4j (n = 8, 12, 5, 11, 6, 4). Median. LME followed by post-hoc analysis. i) Schematic summarizing the molecular mechanisms engaged upon enhanced editing. All statistical tests are two-tailed. n indicate independent experiments, except for Fig. 3b where n indicates independent samples.
To further investigate these transcriptional changes, we performed whole transcriptomic analysis on CB HSPCs 12 hours after AAVS1 editing, when the transient response peaked. We tested editing in presence or absence of GSE56, Ad5-E4orf6/7 or their combination. We identified a large subset of significant differentially expressed genes (DEGs) modulated by electroporation per se which was further expanded when performing editing, with p53 targets (APOBEC3H, EDA2R, CDKN1A and MIR34AHG) mostly upregulated (Extended Data Fig. 3e). The number of DEGs upon GSE56 addition compared to standard editing protocol was relatively limited, while addition of Ad5-E4orf6/7 modulated expression of a higher number of genes (including CDK2, CDKN1A and CDKN2A). Combination of GSE56 and Ad5-E4orf6/7 broadened the number of DEGs (Fig. 3b).
Gene Set Enrichment Analysis (GSEA) highlighted significant positive normalized enrichment scores (NES) for p53 pathway and inflammatory/TNFα dependent responses, and negative NES for cell cycle related categories (E2F pathway, G2M checkpoint, c-myc targets) when comparing mock electroporated with untreated cells, suggesting proliferation slowdown. These responses were further exacerbated in cells undergoing standard editing (Fig. 3c). GSEA between cells edited in presence or absence of GSE56 identified negative NES for the p53 pathway and positive for cell cycle-related categories, indicating dampening but not full DDR abrogation. Ad5-E4orf6/7 addition scored the E2F pathway and G2M checkpoints as top ranking positive categories (Fig. 3c, d). Several genes encoding for HDR machinery12,34 (EXO1, DNA2, RBBP8, RPA4, RAD50, NBN, BRCA1/2, RPA1, RAD51C, RAD51AP1, BARD1, POLD3, PCNA) were upregulated and the HDR pathway emerged from a more granular GSEA (Extended Data Fig. 3f). Surprisingly, the allograft rejection category scored with a negative NES in presence of Ad5-E4orf6/7, indicating downregulation of immune response related genes, such as HLAs, CCL5/CCR5, IL1B, IRF7, CD28, CD4 and THY1 (in agreement with the decrease of CD90 surface protein expression in Ad5-E4orf6/7 treated cells) (Extended Data Fig. 3g and Fig. 3d). The combination of GSE56 and Ad5-E4orf6/7 further enriched cell cycle-related categories (Fig. 3d).
Unsupervised clustering of E2F targets highlighted four subsets of genes showing similar expression dynamics across treatments (Fig. 3e and Supplementary Table 1). While editing downregulated genes within all subsets compared to controls, GSE56 partially rescued expression for genes of the first and second cluster, which enriched for genes promoting HDR (Extended Data Fig. 3h) and master regulators of cell cycle (CHEK1, CHEK2), respectively. Expression of the genes in the first cluster was upregulated by Ad5-E4orf6/7 and even more by its combination with GSE56 (Fig. 3e and Extended Data Fig. 3i, top). The third cluster enriched for CDK inhibitor genes (CDKN1A, CDKN2A, CDKN2C), which are involved in cell cycle arrest and were selectively upregulated by Ad5-E4orf6/7, except for CDKN1A, which was already increased by standard editing and further upregulated by Ad5-E4orf6/7, suggesting induction of a feedback loop to limit E2F-driven cell cycle progression (Fig. 3e and Extended Data Fig. 3i, bottom).
Expression analysis on a panel of cell cycle related genes (Supplementary Table 2) in sorted CD90+ cells edited in presence or absence of Ad5-E4orf6/7 showed concordance with RNA-Seq data, suggesting that the transcriptional response described in bulk cultures similarly occurs in primitive HSPCs (Fig. 3f).
To investigate the impact of editing enhancers on cell cycle progression, we performed cell cycle analysis of edited HSPCs. Ad5-E4orf6/7 addition almost doubled the fraction of cells in S/G2 at 12-24 hours after editing, both in bulk and CD90+ cells (Fig. 3g, top). Combination of GSE56 and Ad5-E4orf6/7 showed even more pronounced S/G2 phases transit. As expected from transient expression of the enhancers, their effects on cell cycle extinguished 96 hours after editing (Fig. 3g, bottom).
We then considered the potential genotoxic risk of forcing S/G2 transition and DNA replication in newly activated HSPCs. By exploiting an IL2RG-targeting nuclease which also detectably cleaves one off-target site15, we measured the frequency of chromosomal translocations between on- and off-targets in presence of Ad5-E4orf6/7 and observed similar low levels in the in vitro outgrowth of all treated cells and none in their in vivo outgrowth upon transplantation in NSG mice (Fig. 3h).
Overall, these data suggest that transient overexpression of GSE56 and Ad5-E4orf6/7 triggers E2F-dependent cell cycle progression and upregulation of the HDR machinery, while dampening the editing-induced p53 response (Fig. 3i).
Ad5-E4orf6/7 and GSE56 enhance gene editing in human LT-HSCs
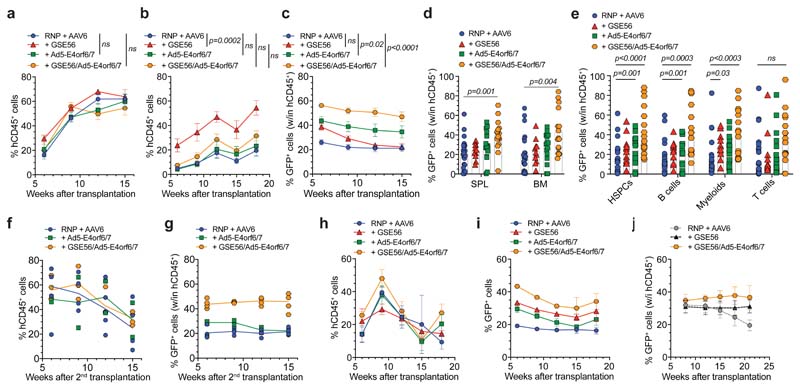
To investigate the repopulation potential of edited HSPCs, we transplanted matched saturating or limiting cell doses into NSG mice (Extended Data Fig. 4a). At saturating cell doses, we observed similar human engraftment across treatments, which reached a plateau of 60% circulating cells (Fig. 4a). At limiting cell doses, we confirmed that GSE56 addition allowed 3-fold higher engraftment than the standard protocol, while its combination with Ad5-E4orf6/7 reduced this increase. Instead, addition of the Ad protein alone showed engraftment comparable to standard treatment (Fig. 4b). Similar patterns of engraftment were found long term after reconstitution in the hematopoietic organs (Extended Data Fig. 4b). Combination of GSE56 and Ad5-E4orf6/7 enabled higher and stable percentages of GFP+ cells across four independent experiments compared to standard protocol, reaching a mean of 50% of the total graft (Fig. 4c). BM analysis showed multi-lineage long-term reconstitution with all treatments with higher proportion of progenitors when using editing enhancers (Extended Data Fig. 4c). The percentage of GFP+ cells within the human graft, sorted progenitors and individual lineages were consistent with the levels observed in the blood, with the combination of GSE56/Ad5-E4orf6/7 outperforming other treatments (Fig. 4d, e). By comparing the percentage of GFP+ cells to the fraction of HDR-edited alleles, we found that GSE56 tended to increase the fraction of biallelic HDR-editing (Extended Data Fig. 4d). To further investigate long-term repopulation capacity of edited HSPCs, we performed secondary transplant (from one primary transplantation experiment) by purifying and pooling human CD34+ cells from BM of primary recipients (Extended Data Fig. 4a). Results confirmed higher and stable fraction of HDR-edited cells when using the GSE56/Ad5-E4orf6/7 combination (Fig. 4f, g). This combination outperformed other treatments even when editing mPB-derived HSPCs, with an average 35% GFP+ cells in long-term human PBMCs (Fig. 4h, i), or when targeting IL2RG in CB HSPCs (Figure 4j and Extended Data Fig. 4e).
Figure 4. Editing enhancers enable high proportion of HDR-edited HSPCs and stable reconstitution in xenograft model.
a-b, Percentage of circulating hCD45+ cells in mice transplanted with the outgrown progeny of starting-matched saturating (a) (n = 11, 4, 7, 9) or limiting (b) (n = 12, 7, 8, 7) doses of CB HSPCs edited in AAVS1 with indicated treatments. Each panel is a pool of two independent experiments. Mean M SEM. Statistics are shown for the last timepoint. c, Percentage of GFP+ cells within human graft in mice from Fig. 4a, b (n = 23, 11, 15, 16). Mean M SEM. Statistics are shown for the last timepoint. d-e, Percentage of GFP+ cells within human graft in hematopoietic organs (d) and lineages (e) of mice from Fig. 4a, b (n = 23, 11, 15, 16). Mean) SEM. f-g, Percentage of circulating hCD45+ cells (f) and GFP+ cells within human graft (g) in secondary recipients transplanted with human BM-derived CD34+ cells harvested from mice of one experiment in Fig. 4a (n = 4, 2, 4). Median. h-i, Percentage of circulating hCD45+ (h) and GFP+ cells within human graft (i) in mice transplanted with the outgrown progeny of starting-matched saturating doses of mPB HSPCs edited in AAVS1 with indicated treatments (n = 3, 5, 5, 5). Mean SEM. j, Percentage of GFP+ cells within human graft in mice transplanted with CB HSPCs edited in IL2RG with editing enhancers (n = 4). Comparison with previously published results for “RNP + AAV6” and “+ GSE56”22 is shown (n = 5, 6). Mean i SEM. For all panels with statistical analyses: LME followed by post-hoc analysis. All statistical tests are two-tailed. n indicate independent animals.
Enhanced gene editing supports polyclonal human graft without perturbing clonal behavior
We then assessed clonal composition and dynamics of host repopulation by enhancer-edited HSPCs. Addition of Ad5-E4orf6/7 when editing cells with standard or GSE56-comprising protocols, while increasing the proportion of HDR-edited cells (see Fig. 4c), did not significantly increase the number of dominant BARs compared to respective controls in limiting dose experiments (Fig. 5a). This finding is consistent with the lower human engraftment obtained with the GSE56/Ad5-E4orf6/7 combination as compared to GSE56 alone (see Fig. 4b). Polyclonal reconstitution after GSE56/Ad5-E4orf6/7 treatment was also confirmed by analyzing PBMCs in the saturating-dose experiment (Fig. 5b). The clonality of HDR-edited cells decreased over time independently from the treatment, as also noted in previous experiments (see Fig. 1h, k), possibly reflecting exhaustion of short-term progenitors. The number of dominant BARs correlated with the percentage of edited cells in PB of recipient mice at early and late timepoints (Fig. 5c), confirming that our treatments did not markedly alter the average clonal output of repopulating HSPCs. Enhanced polyclonal composition was confirmed in B and myeloid cell compartments of GSE56/Ad5-E4orf6/7 mice, while T cells remained oligoclonal (Fig. 5d and Extended Data Fig. 5a, b), suggesting constraints of the model rather than the editing treatment. At the end of the experiment, the majority of dominant repopulating clones showed multilineage output. Few dominant BARs were shared across repopulated mice also upon enhanced editing, indicating the likely occurrence of ex vivo duplication of HDR-edited HSPCs (Extended Data Fig. 5a). Of note, comparable number of BARs were retrieved in mice transplanted with saturating doses of HSPCs edited in presence or absence of GSE56, further indicating that GSE56 treatment does not alter the growth properties of repopulating HSPC (Extended Data Fig. 5c).
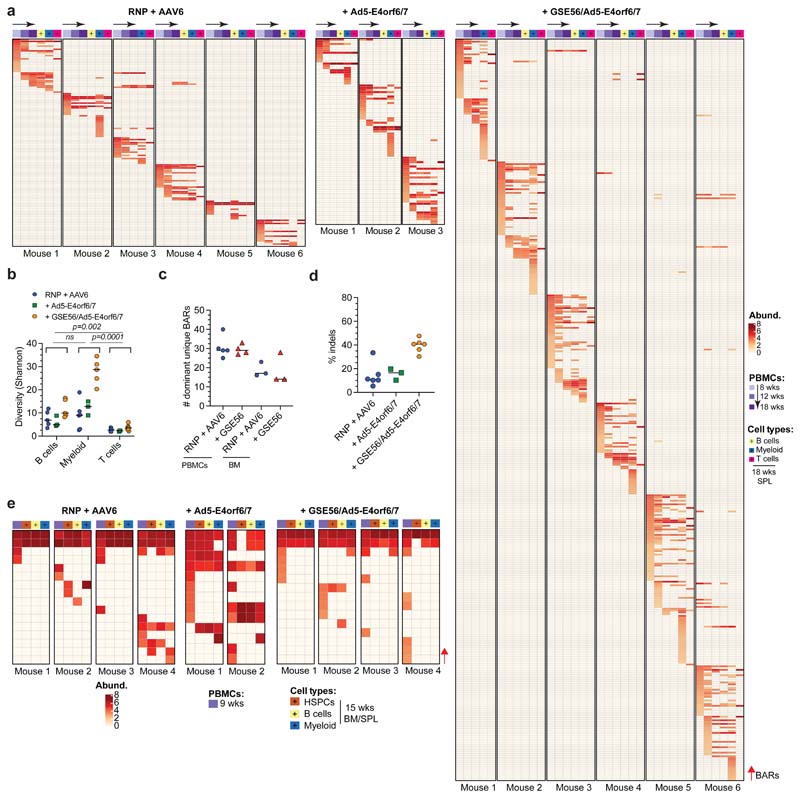
Figure 5. Editing enhancers allow polyclonal composition of the human edited graft without perturbing clonal dynamics.
a, Number of dominant unique BARs in human splenocytes of mice in Fig. 4b (n = 12, 7, 7, 7). Median. GLMER for count data. b, Longitudinal PBMC analysis showing the number of dominant unique BARs in mice from one experiment in Fig. 4a (n = 6, 3, 6). Median with IQR. GLMER for count data. c, Linear/quadratic regression showing the relationship between the number of dominant unique BARs from Fig. 5b and the percentage of hCD45+GFP+ cells at the 8 weeks (left) and 18 weeks (right) (n = 6, 3, 6). d, Number of dominant unique BARs in sorted hCD45+ cell lineages of mice from one experiment in Fig. 4a (n = 6, 3, 6). Median. Mann-Whitney test. e, Deep sequencing analysis of AAVS1 in human edited splenocytes from one experiment in Fig. 4b. Dashed line indicates Cas9 cleavage site. The reference wild type allele and representative plots for one mouse/group are shown. f-g, Number of unique indels in human splenocytes of mice from one experiment at starting-matched limiting (f) (n = 8, 3, 4, 3) and saturating (g) (n = 6, 3, 6) HSPC doses. Median. h, Percentage of NHEJ-edited alleles within the non-HDR edited fraction from Fig. 5f (n = 8, 3, 4, 3). Median. i, Heatmaps showing dominant unique BARs (rows) and relative abundances in PBMCs of primary (18 weeks) and secondary (9 weeks) transplant from Fig. 4a, f. j, Venn diagram showing the number of dominant unique BARs shared between PBMCs of primary and secondary recipients. k, Pie charts showing the percentage of shared/unshared BARs in primary and secondary recipients. Fisher's exact test (p<0.0001). l) Schematic summary of the editing strategies and their outcomes in HSPCs. All statistical tests are two-tailed. n indicates independent animals.
To assess the impact of the editing procedure also on HSPCs undergoing NHEJ-mediated repair of DNA DSBs, we deep sequenced the AAVS1 locus in the long-term human graft and measured indel diversity35. GSE56 increased the number of unique indels, while Ad5-E4orf6/7 protein per se did not affect indel diversity (Fig. 5e-g). Moreover, the fraction of NHEJ-edited alleles within the non-HDR edited subset was tendentially higher in presence of GSE56 (Fig. 5h and Extended Data Fig. 5d). These findings support the contention that editing-induced DDR shrinks clonal repertoire of the edited human graft independently of the pathway engaged for DNA DSB repair and that our enhanced editing protocol rescues its polyclonal composition.
HDR-edited LT-HSCs perform symmetrical and asymmetrical divisions in xenotransplantation settings
To assess self-renewal and clonal dynamics of HDR-edited HSPCs, we performed clonal tracking on secondary transplanted mice from Fig. 4f, g. We observed a strong contraction in the total number of dominant clones in PB of secondary recipients, uncovering a “bottleneck” effect during engraftment of human HSPCs. About 80% of dominant BARs were recaptured from those retrieved in PBMC at long term in primary recipients, while remaining BARs were either identified as dominant within sorted cell lineages or within the “rare” BAR populations (Fig. 5i, j). These results confirmed that individual HDR-edited LT-HSCs retain self-renewing capacity in serial transplantation. Remarkably, r44% of the BARs identified in secondary recipients where shared among different mice (Fig. 5k), suggesting that some HDR-edited HSPCs underwent symmetric self-renewing divisions in primary recipients. These clones robustly contributed to hematopoietic lineages and were present within CD34+ progenitors in the BM (Extended Data Fig. 5e). Overall, these data provide stringent evidence at single cell level that human HDR-edited HSPCs are able to perform symmetric and asymmetric divisions long term after transplantation.
Discussion
Our findings elucidate and overcome two major biological barriers to efficient HDR-mediated gene editing in HSPC and show by clonal tracking that our enhanced editing protocol preserves their multilineage and self-renewal capacity long term after serial transplant (see schematic in Fig. 5l).
The substantially lower number of repopulating HSPC clones well explains the lower human engraftment reported after transplanting edited vs. untreated cells15. The mechanism underlying this loss remains to be fully understood, although the robust activation of p53 pathway and its downstream effectors, such as p21, p14 and p16 suggests induction of detrimental processes like permanent growth arrest, senescence and apoptosis36. Although we measured some increase in apoptosis among treated HSPCs, its extent was limited and could not explain the several-fold loss in engrafting clones. Because clonal dynamics was not different among all treatments, there might be a threshold of p53 activation37 leading to all or none outcome when LT-HSCs are treated for editing, i.e. full preservation or irreversible loss of repopulation potential. In support of this hypothesis are the increased indels frequency, diversity and biallelic HDR targeting when GSE56 is added to the treatment, suggesting preferential rescue of cells undergoing higher DDR burden from multiple DNA DSBs and/or increased template uptake. It should be mentioned that our clonal dynamics analysis could not investigate quiescence and short-lived progenitors providing limited output and was limited to dominant clones within the edited cell graft. However, if we consider that dominant clones accounted for 1 every 2x103-2x104 edited CD34+ cells throughout our study, such frequency is consistent with previous estimates of SCID-repopulating cells in cultured CB CD34+ cells assayed by limiting dilution transplantation38–41, suggesting preservation of the normal repopulation capacity by individual HDR/NHEJ-edited HSPC.
Our data clearly show that cell cycle regulation represents a fundamental rate-limiting step for HDR-editing in HSPCs. However, despite the fraction of cells in S/G2 was similar at the time of editing between bulk and CD90+ HSPCs, HDR efficiency was always lower in the latter cells, as previously reported15,42. This observation suggests that, beside the requirement for progression to S/G2, other factors account for lower HDR efficiency in CD90+ cells, such as low expression and activity of HDR machinery15,43 and delayed transit through the G1/S checkpoint44, which receives multiple inputs to adjust metabolic regulation of growth rate to cell size and cell cycle progression. Ad5-E4orf6/7 is known to bind and stably recruit active E2F transcription factors to the adenoviral E2 and cellular E2F-1 promoters and activate downstream gene expression45,46. Transient expression of Ad5-E4orf6/7 in HSPCs triggered an E2F-driven pleiotropic response coupling promotion of G1/S transition47 and enhanced expression of HDR machinery, which increased HDR efficiency preferentially in the most primitive cells. Such pervasive modulation of highly integrated cellular networks by a viral protein naturally evolved to capture the benefits of cell proliferation for viral infection might be difficult to replicate with small drugs or other strategies targeting individual genes engaged in the process.
Notably, the HDR increase by Ad5-E4orf6/7 was further enhanced by combination with p53 inhibition, which can be explained by counteracting the p21 and p14 mediated negative feedback triggered by E2F activation, a previously reported finding31,32 also shown by our data. This feedback might also explain why Ad5-E4orf6/7 did not increase engraftment of standard edited cells and lowered the GSE56-dependent graft increase. Of note, the number of clones upon Ad5-E4orf6/7 addition might be underestimated if upregulation of the HDR machinery increased the proportion of cells undergoing template integration before replication of the targeted locus, thus producing two clones with the same BAR. Intriguingly, granular inspection of Fig. 5c shows that Ad5-E4orf6/7-treated long-term engrafting clones tend to have higher output than their experimental counterparts, as shown by better fit of data to a quadratic regression model. The decreased percentage and MFI of CD90+ cells upon Ad5-E4orf6/7 treatment is likely due to the observed transcriptional downregulation of the CD90 gene rather than differentiation. This proposition is further supported by the observation that other LT-HSC markers, such as CD133 (PROM1)48, CD49f (ITGA6)49 and CD201 (EPCR)50, were not downregulated by Ad5-E4orf6/7 treatments.
The detrimental effects of p53 activation might confer selective advantage to rare p53-/- cells51,52. Limited and transient inhibition of the editing-induced p53 response would reduce the risk of selecting for p53 mutant clones and mono/oligo-clonal expansion. Robust p53-dependent transcriptional activation of the DNA cytidine deaminase APOBEC3H upon editing15 might also raise concerns for mutagenesis targeting single-stranded genomic DNA intermediates during repair, replication and transcription53. GSE56 and Ad5-E4orf6/7 together nearly abolished APOBEC3H induction, thus potentially protecting edited cells from a further source of genotoxicity. Importantly, the use of mRNA for transient expression of the p53 inhibitor and Ad5-E4orf6/7 rules out the risk of genomic integration of these potentially transforming factors. As first readout of genomic alterations, we did not detect increased occurrence by single or combined addition of GSE56 and Ad5-E4orf6/7 of a chromosomal translocation specifically traceable to the activity of an editing nuclease.
An unexpected benefit of Ad5-E4orf6/7 treatment is the downregulation of some immune response/chemokine genes, which may contribute to the immune evasive strategy of the parental virus. This response might also decrease the risk of antigen presentation and immune effector recruitment by the administered HSPCs, which shortly after editing still contain immunogenic proteins of bacterial and viral origin, such as Cas nuclease and AAV capsid proteins.
Overall, the gains in clonal repertoire and percentage of edited HSPCs obtained by our enhanced protocol are relevant for clinical translation. Indeed, oligoclonal composition might delay hematopoietic recovery after conditioning and limit the size, long-term stability and safety of the engineered cell graft. Moreover, the higher the proportion of HDR-edited cells in the cell product the less is the competition with unedited and residual HSPCs in the host to achieve sufficient chimerism for therapeutic benefit. These benefits may well balance the inherent risk of first-in-human clinical testing in suitable disease contexts, such as primary immunodeficiencies, where HSPC gene editing may eventually provide effective treatment.
Online Methods
Vectors and nucleases
AAV6 donor templates were generated from a construct containing AAV2 inverted terminal repeats (ITRs), produced at the TIGEM Vector Core (Pozzuoli (NA), Italy) by triple-transfection method and purified by ultracentrifugation on a cesium chloride gradient. Design of the non-barcoded AAV6 donor templates carrying homologies for AAVS1 or IL2RG (both encompassing a PGK.GFP reporter cassette) were previously reported15. Design of the AAV6 donor template with homologies for CD40LG will be reported elsewhere. The barcoded vector was obtained by subcloning a degenerated BAR sequence downstream of the GFP reporter cassette in the reference AAV backbone for AAVS1 editing. For molecular cloning of the barcoded AAV, a single stranded ODN embedding the 22-bp BAR sequence flanked by unique restriction sites (Bsu36I and SphI, New England Biolabs) was purchased from Sigma Aldrich. Theoretical complexity of the ssODN was estimated in 2.9x1010. BAR consensus sequence was designed to contain some invariant positions (7, 9, 15) and others limited to few bases (3, 14, 17, 21, 22) to avoid generating Bsu36I and SphI restriction sites. To generate the complementary strand, 50 pmol of the ssODN underwent 10 PCR cycle with Easy-A High-Fidelity enzyme (Agilent Technologies) using the appropriate primers (see Supplementary Table 3) and according to manufacturer instruction. The amplified product was purified with MinElute PCR Purification kit (QIAGEN), digested with the restriction enzymes and verified by capillary electrophoresis. 2 µg of this purified product were ligated with the digested reference backbone (molar ratio 7:1) using T4 DNA Ligase (New England Biolabs) by scaling up the manufacturer protocol. XL-10 Gold Ultracompetent Cells (Agilent Technologies) were transformed with the ligation product, plated and incubated for 12 h at 30°C to minimize the occurrence of recombination events. Colonies were scraped, mixed, grown in LB medium for additional 6 h and processed with NucleoBond Xtra MaxiPrep (Machery Nagel) according to manufacturer instruction. The plasmid prep was screened with MscI and XmaI restriction enzymes (New England Biolabs) for ITRs and plasmid integrity.
IDLV donor was generated using HIV-derived, third-generation self-inactivating transfer construct and the IDLV stock was prepared by transient transfection of HEK293T, as previously described27. At 30 hours post-transfection, vector-containing supernatant was collected, filtered, clarified, DNAse treated and loaded on a DEAE-packed column for Anion Exchange Chromatography. The vector-containing peak was collected, subjected to a second round of DNAse treatment, concentration by Tangential Flow Filtration and a final Size Exclusion Chromatography separation followed by sterilizing filtration and titration of the purified stock as previously described27.
Sequences of the gRNAs were designed using an online tool54 and selected for predicted specificity score and on-target activity. Genomic sequences recognized by the gRNAs were previously reported (AAVS1, IL2RG)15 or will be reported elsewhere (CD40LG). RNP complexes were assembled by incubating at 1:1.5 molar ratio Streptococcus pyogenes (Sp)Cas9 protein (Aldevron) with pre-annealed synthetic Alt-R® crRNA:tracrRNA (Integrated DNA Technologies) for 10’ at 25°C. together with 0.1 nmol of Alt-R® Cas9 Electroporation Enhancer (Integrated DNA Technologies) was added prior to electroporation according to manufacturer’s instructions.
Vector maps were designed with SnapGene software v5.0.7 (from GSL Biotech; available at snapgene.com) or Vector NTI® Express v1.6.2 (from Thermo Fisher Scientific, available at thermofisher.com).
Multiple sequences alignment
Multiple sequences alignments were performed with E4orf1 and E4orf6/7 variants derived from different Ad serotypes using the T-Coffee algorithm55.
mRNA in vitro transcription
The GSE56 construct was previously described15. For other constructs, DNA coding sequences were synthetized (GeneArt™, Thermo Fisher) using Homo sapiens codon-usage optimized Ad-E4orf1 and E4orf6/7 sequences. Coding sequences were subcloned in “pVax” plasmids under the control of the T7 promoter and followed by WHP posttranscriptional regulatory element (WPRE) and a 64-bp polyA sequence. For GSE56 and Ad5-E4orf6/7 co-expression, we used separate mRNA in initial setup experiments and a fusion construct, in which the coding sequences were part of the same ORF and separated by a nucleotide sequence encoding for the P2A self-cleaving peptide in most of the follow-up experiments (Supplementary Table 4). For mRNA in vitro transcription, pVax plasmids were linearized with SpeI (New England Biolabs) restriction enzyme and purified by phenol-chloroform extraction. mRNA was in vitro transcribed using the commercial 5X MEGAscript T7 kit (Thermo Fisher) and capped with the Anti-Reverse Cap Analog (ARCA) 3′-O-Me-mG(5′) ppp(5′)G (New England Biolabs). mRNA was purified using RNeasy Plus Mini Kit (Qiagen) followed by HPLC purification (ADS BIOTEC WAVE® System) and Amicon Ultra-15 (30K) tube (Millipore) concentration. mRNA productions were aliquoted and stored at -80°C.
Cell lines and primary cell culture
HEK293T cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Euroclone), 100 IU/ml penicillin, 100 µg/ml streptomycin and 2% glutamine.
Primary T cells were isolated from healthy male donors’ peripheral blood mononuclear cells (PBMCs) purified from buffy coats by sequential centrifugations in a Ficoll gradient according to a protocol approved by the Ospedale San Raffaele Scientific Institute bioethical committee (TIGET-HPCT). CD3+ T cells were stimulated using magnetic beads (1:3 cell:beads ratio) conjugated with anti-CD3/anti-CD28 antibodies (Dynabeads human T-activator CD3/CD28, Thermo Fisher). Cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM; Corning) supplemented with 10% heat-inactivated FBS, 100 IU/ml penicillin, 100 µg/ml streptomycin, 2% glutamine, 5 ng/ml hIL-7 (PreproTech) and 5 ng/ml hIL-15 (PreproTech)56. Dynabeads were removed after 6 days of culture.
CB CD34+ HSPCs were purchased frozen from Lonza upon approval by the Ospedale San Raffaele Bioethical Committee (TIGET-HPCT) and were seeded at the concentration of 5x105 cells/ml in serum-free StemSpan medium (StemCell Technologies) supplemented with 100 IU/ml penicillin, 100 µg/ml streptomycin, 2% glutamine, 100 ng/ml hSCF (PeproTech), 100 ng/ml hFlt3-L (PeproTech), 20 ng/ml hTPO (PeproTech), and 20 ng/ml hIL-6 (PeproTech) and 10 μM PGE2 (at the beginning of the culture; Cayman). Culture medium was also supplemented with 1 μM SR1 (Biovision) and 50 nM UM171 (STEMCell Technologies), unless otherwise specified.
G-CSF mPB CD34+ HSPCs were purified with the CliniMACS CD34 Reagent System (Miltenyi Biotec) from Mobilized Leukopak (AllCells) upon approval by the Ospedale San Raffaele Bioethical Committee (TIGET-HPCT) according to manufacturer’s instructions. HSPCs were seeded at the concentration of 5x105 cells/ml in serum-free StemSpan medium (StemCell Technologies) supplemented with 100 IU/ml penicillin, 100 µg/ml streptomycin, 2% glutamine, 300 ng/ml hSCF, 300 ng/ml hFlt3-L, 100 ng/ml hTPO and 10 μM PGE2 (at the beginning of the culture). Culture medium was also supplemented with 1 μM SR1 and 35 nM UM171.
All cells were cultured in a 5% CO2 humidified atmosphere at 37°C.
Gene editing of human T cells and analyses
After three days of stimulation, 5-10x105 T cells were electroporated using P3 Primary Cell 4D-Nucleofector X Kit and program DS-130 (Lonza). Cells were electroporated with RNP at final concentration 1.25 μM (Integrated DNA Technologies) together with 0.1 nmol of Alt-R® Cas9 Electroporation Enhancer(Integrated DNA Technologies) and transduced with 5x104 vg/cell of AAV6 15’ after electroporation. Where specified, mRNAs were added to the electroporation mixture at the final concentrations reported in Supplementary Table 4. T cells were expanded for 14 days to perform flow cytometry.
Gene editing of human HSPCs and analyses
For AAV6-based gene editing, after 3 days of stimulation 1-5x105 cells were washed with Dulbecco’s Phosphate-Buffered Saline without Ca2+ and Mg2+ (DPBS, Corning) and electroporated using P3 Primary Cell 4D-Nucleofector X Kit and program EO-100 (Lonza). Cells were electroporated with RNPs at final concentration 1.25-2.5 μM together with 0.1 nmol of Alt-R® Cas9 Electroporation Enhancer (Integrated DNA Technologies), according to manufacturer’s instructions. AAV6 transduction was performed at a dose of 1-2x104 vg/cell 15’ after electroporation. For IDLV-based gene editing, after 2 days of stimulation 1-5x105 cells were treated with 8 μM cyclosporin H (CsH, Sigma) or DMSO vehicle and transduced with purified IDLV at multiplicity of infection of 100-200 (vector concentration = 1.1x1010 Transducing Units293T/mL). After 24 h, cells were washed with DPBS and electroporated using P3 Primary Cell 4D-Nucleofector X Kit and program EO-100 (Lonza), as described above.
Additional mRNAs were added to the electroporation mixture at the final concentrations reported in Supplementary Table 4. Three/four days after editing procedure cells were harvested to analyze by flow cytometry the percentage of cells expressing the GFP marker within HSPC subpopulations and to extract genomic DNA for molecular analyses, unless otherwise indicated.
CFU-C assay was performed 24 h after editing procedure by plating 600 cells in methylcellulose-based medium (MethoCult H4434, StemCell Technologies) supplemented with 100 IU/ml penicillin and 100 µg/ml streptomycin. Three technical replicates were performed for each condition. Two weeks after plating, colonies were counted and identified according to morphological criteria.
Mice
All experiments and procedures involving animals were performed with the approval of the Animal Care and Use Committee of the San Raffaele Hospital (IACUC: #749) and authorized by the Italian Ministry of Health and local authorities accordingly to Italian law. NOD-SCID-IL2Rg-/- (NSG) female mice (The Jackson Laboratory) were held in specific pathogen-free (SPF) conditions.
CD34+ HSPC xenotransplantation experiments in NSG mice
For transplantation of CB and G-CSF mPB CD34+ HSPCs, the outgrowth of 1-3x105 and 1x106 HSPCs, respectively, at the start of the culture were injected intravenously 24 h after editing into sub-lethally irradiated NSG mice (150-180 cGy). Sample size for each experiment was determined by the total number of available treated cells. Mice were randomly distributed to each experimental group. Human CD45+ cell engraftment and the presence of edited cells were monitored by serial collection of blood from the mouse tail and, at the end of the experiment (>18 weeks after transplantation), BM and SPL were harvested for end-point analyses.
Secondary transplantation was performed upon injection of 2x106 beads-purified human CD34+ cells (Miltenyi Biotec) harvested from the BM of primary engrafted NSG mice and pooled for each experimental group.
Flow cytometry
Immunophenotypic analyses were performed on FACS Canto II (BD Pharmingen) or CytoFLEX LX Flow Cytometer (Beckman Coulter). From 0.5 to 2x105 cells (either from culture or mouse samples) were analyzed by flow cytometry. Cells were stained for 15’ at 4°C with antibodies listed in the Reporting Summary in a final volume of 100 μl and then washed with DPBS + 2% heat-inactivated FBS. Single stained and FMO stained cells were used as controls. Live/Dead Fixable Dead Cell Stain Kit (Thermo Fisher) or 7-Aminoactinomycin D (Sigma Aldrich) were included during sample preparation according to the manufacturer’s instructions to identify dead cells. Apoptosis analysis was performed as previously described3. Cell sorting was performed on a BD FACSAria Fusion (BD Biosciences) using BDFACS Diva software and equipped with four lasers: blue (488 nm), yellow/green (561 nm), red (640 nm) and violet (405 nm). Cells were sorted with an 85 mm nozzle. Sheath fluid pressure was set at 45 psi. A highly pure sorting modality (4-way purity sorting) was chosen. Sorted cells were collected in 1.5 ml Eppendorf tubes containing 500 μl of DPBS. Gating strategies for flow cytometry analyses are provided in Supplementary Figure 2. Data were analyzed with FCS Express 6 Flow.
Molecular analyses
For molecular analyses, genomic DNA was isolated with QIAamp DNA Micro Kit (QIAGEN) according to manufacturer’s instructions. Nuclease activity was measured by mismatch-sensitive endonuclease T7 assay (New England Biolabs) or Surveyor> mutation detection kit (IDT) on PCR-based amplification products of the targeted locus, as previously described4. Digested DNA fragments were resolved and quantified by capillary electrophoresis on LabChip® GX Touch HT (Perkin Elmer) or 4200 TapeStation System (Agilent) according to the manufacturer’s instructions.
For HDR digital droplet PCR (ddPCR) analysis, 5-50 ng of genomic DNA were analyzed using the QX200 Droplet Digital PCR System (Bio-Rad) according to the manufacturer’s instructions. HDR ddPCR primers and probes were designed on the junction between the vector sequence and the targeted locus. Human TTC5 (Bio-Rad) was used for normalization. The percentage of cells harboring biallelic integration was calculated with the following formula: (# of AAVS1 + droplets / # of TTC5 + droplets x 200) - % GFP+ cells. The percentage of monoallelic integration was then calculated with the following formula: % GFP+ cells - % cells with biallelic integration. For chromosomes X-14 translocation, ddPCR was performed as previously reported15.
For gene expression analyses, total RNA was extracted using RNeasy Plus Micro Kit (QIAGEN), according to the manufacturer’s instructions and DNase treatment was performed using RNase-free DNase Set (QIAGEN). cDNA was synthetized with SuperScript VILO IV cDNA Synthesis Kit (Thermo Fisher) with EzDNase treatment. cDNA was then used for qPCR in a Viia7 Real-time PCR thermal cycler using TaqMan Gene Expression Assays (Applied Biosystems) mapping to genes listed in Supplementary Table 3. Data were analyzed with QuantStudio™ Real-Time PCR software v1.1 (Applied Biosystem). Relative expression of each target gene was first normalized to HPRT and then represented as fold changes (2-ΔΔCt) relative to the untreated cells.
For ddPCR array card, CD34+CD133+CD90+ cells were sorted 12 h after HSPC editing in presence or absence of Ad5-E4orf6/7. After RNA isolation and reverse transcription as described above, gene expression was performed with “Cell Cycle Generic H384” predesigned 384-well panel (PrimePCR Arrays, Bio-Rad) with SYBR Green system. Data were analyzed with QuantaSoft™ Software v1.7.4 (Bio-Rad).
BAR-Seq library preparation
PCR amplicons for individual samples were generated by nested PCR using primers listed in Supplementary Table 3 and starting from >50-100 ng of purified gDNA. The first PCR step was performed with GoTaq G2 DNA Polymerase (Promega) according to manufacturer instruction using the following amplification protocol: 95°C x 5’, (95°C x 0.5’, 60°C x 0.5’, 72°C x 0.5’) x 20 cycles, 72°C x 5’. Forward primer was designed to bind donor template upstream the BAR sequence, while the reverse primer annealed outside the homology arm, thus amplifying 328 bp of the on-target integrated cassette. For targeted deep sequencing of the plasmid and AAV libraries, the reverse primer annealed to the homology arm. The second PCR step was performed with GoTaq G2 DNA Polymerase (Promega) according to manufacturer instruction using 5 µl of the first-step PCR product and the following amplification protocol: 95°C x 5’, (95°C x 0.5’, 60°C x 0.5’, 72°C x 0.5’) x 20 cycles, 72°C x 5’. Second-step PCR primers were endowed with tails containing P5/P7 sequences, i5/i7 Illumina tags to allow multiplexed sequencing and R1/R2 primer binding sites (Supplementary Table 3). PCR amplicons were separately purified using MinElute PCR Purification kit (QIAGEN) and AmpPure XP beads (Beckman Coulter). Library quality was assessed by Agilent Tapestation (Agilent Technologies). Amplicons were multiplexed and run on MiSeq 2x75bp or 2x150bp paired end (Illumina).
BAR-Seq analyses
BAR-Seq data were processed with TagDust57 (v2.33) to identify and extract the BAR from each sample by taking advantage of the structural composition of the reads. Each putative BAR was then examined to filter out those having an incorrect nucleotide at the fixed positions or BAR length different from the expected one (22 bp). BAR abundance was quantified by summing the number of identical sequences. Since amplification and sequencing errors may produce highly similar barcodes, a graph-based procedure was employed. For each sample a graph structure was created in which BARs represent nodes and two nodes are linked with an edge if the corresponding sequences have edit distance < 3. Ego subnetworks, i.e. subgraphs focalized on highly abundant BARs, were iteratively identified and collapsed into a single node and, consequently, into a single BAR sequence. More precisely, nodes were ranked based on their counts, and at each iteration the ego network composed of the most abundant BAR and its neighbors were merged into a single BAR (the focal node) and its nodes were removed from the graph. The rationale behind this approach was that, although sequencing errors could produce different sequences, the parental BAR, which constitutes the focal node of the network, would have the highest count. BARs with read count lower than 3 were discarded and the remaining set of BARs were identified as the valid BARs of this sample. To verify that all the samples used in the analysis were informative after the filtering process, we employed a previously described approach to estimate the richness of each sample58, verifying that such value was above the threshold of 95% in all the samples. After BAR ranking from the most to the least abundant, a saturation-based approach was implemented. The dominant set of BARs for each sample was defined as the pool of BARs representing >90% of the total abundance of valid BARs, while the remaining <10% were comprised of rare BARs.
Total RNA-Seq library preparation and analysis
Whole transcriptomic analysis was performed on a pool of HSPCs derived from 5 CB donors. All conditions were performed in triplicate. Total RNA was isolated at 12 h after editing using miRNeasy Micro Kit (QIAGEN), and DNase treatment was performed using RNase-free DNase Set (QIAGEN), according to the manufacturer’s instructions. RNA was quantified with The Qubit 2.0 Fluorometer (ThermoFisher) and its quality was assessed by a 2100 Agilent Bioanalyser (Agilent Technologies). Minimum quality was defined as RNA integrity number (RIN)>8. 300 ng of total RNA were used for library preparation with TruSeq Stranded mRNA (Illumina) and sequenced on a NextSeq 500 High 75 (Illumina). Read quality was determined using FastQC and low-quality sequences were trimmed using trimmomatic. Reads were then aligned to the human reference genome (GRCh38/hg38) using STAR, with standard input parameters, and gene counts were produced using Subread featureCounts and Genecode v31 as gene annotation. Transcript counts were processed by R/Bioconductor package edgeR, normalizing for library size using trimmed mean of M-values, and correcting p-values using FDR.
Cell cycle analysis
Cell cycle was analyzed by flow cytometry 12-24 or 96 h after editing by collecting 1-2x105 bulk cultured HSPCs. Cells were stained for >1 h at room temperature with 5 μl of solution 1 μg/μl Hoechst 33342 (Sigma-Aldrich) in a final volume of 100 μl and then washed with DPBS + 2% heat-inactivated FBS. Gating strategy for flow cytometry analysis is provided in Supplementary Figure 2. Data were analyzed with FCS Express 6 Flow.
Indels-based clonal tracking library preparation
PCR amplicons for individual samples were generated by nested PCR using primers listed in Supplementary Table 3 and starting from >50-100 ng of purified gDNA. The first PCR step was performed with GoTaq G2 DNA Polymerase (Promega) according to manufacturer instruction using the following amplification protocol: 95°C x 5’, (95°C x 0.5’, 60°C x 0.5’, 72°C x 0.25’) x 20 cycles, 72°C x 5’. The second PCR step was performed with GoTaq G2 DNA Polymerase (Promega) according to manufacturer instruction using 5 µl of the first-step PCR product and the following amplification protocol: 95°C x 5’, (95°C x 0.5’, 60°C x 0.5’, 72°C x 0.3’) x 20 cycles, 72°C x 5’. Second-step PCR primers were endowed with tails containing P5/P7 sequences, i5/i7 Illumina tags to allow multiplexed sequencing and R1/R2 primer binding sites (Supplementary Table 3). PCR amplicons were separately purified performing double-side selection with AmpPure XP beads (Beckman Coulter). Library quality was assessed by LabChip® GX Touch HT (Perkin Elmer). Amplicons were multiplexed and sequenced by GeneWiz on MiSeq 2x300bp paired end sequencing (Illumina).
Indels-based clonal tracking analyses
Samples for Indels-based clonal tracking were analyzed with CRISPResso235, a suite of software developed to detect and quantify insertions, mutations and deletions in reads from gene editing experiments. In details, the CRISPRessoBatch pipeline was used to filter NGS reads relying on the phred33 score, getting rid of low-quality sequences, and to remove Illumina TruSeq3-PE adapters using Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic). Then, each couple of paired-end reads was merged using FLASh to produce a single sequence, which was mapped to the input amplicon reference sequence using a global alignment method. The gRNA sequence was uploaded in CRISPResso2 to focus the analysis on the target region. Quantification window was set to 10 nts. As suggested in CRISPResso2 guidelines, the sgRNA was provided without including the PAM sequence. For each sample, identified alleles were quantified by measuring the number of reads and their relative abundance based on total read counts. Alleles showing a relative abundance lower than the false positive threshold (set at 0.3%, based on untreated control) were filtered out.
Quantifications and statistical analyses
“n” indicates biologically independent samples/animals/experiments. For some experiments, different HSPC donors were pooled. Data were summarized as median (interquartile range) or mean ii SEM depending on data distribution. Inferential techniques were applied in presence of adequate sample sizes (n ≥ 5), otherwise only descriptive statistics are reported. Two-sided tests were performed. Association between categorical variables was evaluated by means of Fisher’s Exact test. Spearman’s correlation coefficient was calculated to evaluate the presence of a monotonic relationship between variables. Linear and quadratic regression models were fitted to test for the presence of linear/nonlinear relationships. Mann-Whitney test was performed to compare two independent groups, while in presence of more than two independent groups Kruskal-Wallis test followed by post-hoc analysis using Dunn’s test was used. In presence of dependent observations, Friedman test with Dunn’s multiple comparisons or Linear/Generalized Mixed-Effects models (LME/GLMER)59,60 were performed. The latter procedures were applied to properly account for the dependence structure among observations, by including additional (nested) random-effect terms, thus considering in the model unobservable sources of heterogeneity among experimental units. When analyzing time courses, treatment group indicator and time variables, along with their interaction, were included as covariates in the model to identify potential differences in growth dynamics of treatment groups. A random intercept model was estimated and, when necessary, nested random effects were considered (e.g., to account for repeated measures of cells/mouse within experiments). GLMER models were applied to properly analyze count data: in particular Poisson mixed models were estimated. Logarithmic and square root transformations of the outcome were also considered to satisfy underlying model assumptions. Post-hoc analysis after LME was performed, considering all the pairwise comparisons of treatment groups at a fixed time point. p-values were adjusted using Bonferroni’s correction. In all the analyses, the significance threshold was set at 0.05, while “ns” means non significance. Analyses were performed using GraphPad Prism v8.4.2 (GraphPad) and R statistical software. Detailed results of statistical analyses are shown in Supplementary Table 5.
Life Sciences Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. BAR-Seq dissects clonal dynamics of HDR-edited cells.
a, Percentage of GFP+ cells within subpopulations 96 h after AAVS1 editing with the barcoded or non-barcoded AAV6 (3 HSPC donors; n = 4). Median. b, Number of unique BARs and relative abundances in bulk cultured HSPCs 72 h after editing. One representative sample out of two is shown. c, Experimental scheme. d-e, Culture composition (d) and percentage of GFP+ cells within subpopulations (e) of AAVS1 edited HSPCs with the indicated treatments at the time of transplant and 96 h after editing, respectively (10 HSPC donors; n = 1). f-g, Percentage of hCD45+ cells (f) and GFP+ cells within human graft (g) in BM or spleen (SPL) of mice from Fig. 1c (n = 9, 10, 6, 3). Median. Kruskal-Wallis test. h-i, Abundance of ranked BARs from PBMCs collected at 8 (h) and 12 (i) weeks after transplant, as in Fig. 1e. j, Heatmap as in Fig. 1f for “w/o S/U (+4 days)”-transplanted mice. k, Number of dominant unique BARs in sorted hCD45+ cell lineages and HSPCs of mice from Fig. 1c. Mice with % of circulating hCD45+GFP+ cells at 18 weeks timepoints < 0.1 were plotted with BAR count = 0 (n = 9, 10, 10, 10). Median. l, Correlation between the percentage of GFP+ cells (within hCD45+) and the number of dominant unique BARs in “w/o S/U”, “RNP + AAV6” and “+ GSE56” mice of this study (n = 71). Each dot represents one mouse. Mice with number of dominant unique BARs ≥6 (arbitrary threshold) are shown in magenta (coefficient of variation (CV) = 0.51); mice with number of dominant unique BARs <5 are shown in yellow (CV = 0.87). Dashed line indicates the median percentage of GFP+ cells within CD90+ HSPCs in the in vitro outgrown of transplanted edited cells. m, Longitudinal PBMC analysis as in Fig. 1k but including in the analysis >95% of total BAR reads (n = 4, 5). Median. n, Correlation as in Fig. 1l at 8 weeks after transplant (n = 28). Spearman correlation coefficient was calculated. All statistical tests are two-tailed. n indicate independent animals.
Extended Data Fig. 2. Identification of Ad protein variants improving HDR efficiency.
a-b, Multiple sequences alignment of E4orf1 (a) and E4orf6/7 (b) Ad variants. Sequences were collected from online RCSB Protein Data Bank and UniProt. c-d, Percentage of HDR-edited alleles (c) and GFP+ cells within subpopulations (d) 96 h after AAVS1 editing in bulk CB HSPCs with indicated treatments (n = 4, 4, 4, 4; other treatments: n = 2). Median. e, FACS plots of untreated (UT) and AAV6-transduced HSPCs in absence (“Mock AAV6”) or presence of Ad5-E1B55K+Ad5-E4orf6 measured 24 h after treatments. The results of one representative experiment out of three is shown. f, Number of colonies from bulk edited HSPCs in the indicated treatments (n = 2). Mean. g-h, Fold change expansion of live HSPCs after indicated treatments from Extended Data Fig. 2c (n = 2). Median. i, Number of colonies from bulk edited HSPCs with the indicated treatments (n = 2). Mean. j, CD90 MFI in edited HSPCs measured 96 h after editing with indicated treatments (n = 6). Median with IQR. Friedman test with two-tailed Dunn’s multiple comparisons. k, Percentage of live, early/late apoptotic and necrotic bulk HSPCs 24 h after editing with the indicated treatments (7 HSPC donors; n = 3). Mean (SEM. l-m, Percentage of HDR/NHEJ-edited alleles (l) and culture composition (m) 96 h after editing of bulk mPB HSPCs from Fig. 2h (n = 3). Mean (SEM. n) Percentage of GFP+ T cells 14 days after AAVS1 editing with indicated treatments (n = 3). Median. o-p, Percentage of HDR/NHEJ-edited alleles (o) and culture composition (p) 96 h after IDLV-based editing of bulk CB HSPCs from Fig. 2i (n = 3). Mean (SEM. Red arrows indicate Ad protein variants selected for further investigation. n indicate independent experiments.
Extended Data Fig. 3. Investigating the transcriptional response upon enhanced editing.
a-b, Fold change expression of cell cycle related genes relative to UT 24 h after AAV-based editing with the indicated treatments in CB (a) or mPB (b) HSPCs (CB: n = 8, 5, 7, 6, 3, 3, 3, 3; mPB: n = 4, 4, 4, 3). Median. c, Fold change expression of CDKN1A relative to UT 24 h after IDLV-based editing with indicated treatments in CB HSPCs (n = 3). Median. d, Fold change expression of CDKN1A relative to UT at 24 h after AAV-based editing with indicated treatments in CB HSPCs (n = 5). Median. e) MA plots showing significant down- (green) and up- (red) regulated genes after AAVS1 editing in mock electroporated (left) and standard edited (right) compared to UT (n = 3). PPP1R12C, the AAVS1 hosting gene appears among the downregulated genes, concordantly with previous reports showing transient transcriptional repression at the site of DNA DSB15. f) Random walk plots for the indicated Reactome categories. Relative adjusted p-values and NES are shown. g) Venn diagram showing the number of genes related to the “Allograft rejection” category upregulated upon standard editing and downregulated in presence of “+ Ad5-E4orf6/7” treatment. h) Venn diagram showing the number of HDR genes (“Homology directed repair” category from Reactome database) shared with E2F pathway target genes (Hallmark gene set) from cluster 1 or other clusters from Fig. 3e. i) Schematic of “cell cycle” and “p53 pathway” KEGG gene ontologies highlighting genes (red) belonging to clusters 1 (top) and 3 (bottom) of Fig. 3e. For all panels with statistical analysis: Friedman test with two-tailed Dunn’s multiple comparisons. n indicate independent experiments, except for Extended Data Fig. 3e where n indicates independent samples.
Extended Data Fig. 4. Transplantation of enhancer-edited HSPCs in NSG mice.
a, Experimental workflow. b, Percentage of hCD45+ cells in SPL and BM of mice from Fig. 4a and 4b (n = 23, 11, 15, 16). LME followed by post-hoc analysis. Mean (SEM. c, BM cell composition in mice from Fig. 4a and 4b. LME followed by post-hoc analysis for HSPCs (n = 23, 11, 15, 16). Mean (SEM. d, Percentage of cells harboring monoallelic or biallelic integration(s) in SPL of mice from Fig. 4a and 4b (n = 23, 11, 15, 16). Mean (SEM. e, Percentage of circulating hCD45+ cells in mice transplanted with CB HSPCs IL2RG-edited in presence of GSE56 and Ad5 E4orf6/7 (n = 4). Comparison with the previously published results for “RNP + AAV6” and “+ GSE56” groups22 is shown (n = 5, 6). All statistical tests are two-tailed. n indicate independent animals.
Extended Data Fig. 5. Enhanced editing preserves multilineage repopulation capacity and self-renewing potential of individual edited HSPC clones.
a, Heatmap showing the abundance (red-scaled palette) of dominant unique BARs (rows) retrieved in PBMCs at indicated times after transplant and sorted hCD45+ cell lineages of mice from one experiment of Fig. 4a (separated columns). b, Clonal diversity within sorted hCD45+ cell lineages in mice from Extended Data Fig. 5a (B cells: n = 5, 3, 5; Myeloid and T cells: n= 6, 3, 3). Median. Two-tailed Friedman test with Dunn’s multiple comparisons. Experimental groups were unified for statistical analysis. c, Number of dominant unique BARs in PBMCs or BM of mice from one experiment in Fig. 4a (PBMCs: n = 5, 4; BM: n = 3, 3). Median. d, Percentage of NHEJ-edited alleles within the non-HDR edited fraction from Fig. 5g (n = 6, 3, 6). Median. e, Heatmaps as in Extended Data Fig. 5a showing the dominant unique BARs in 9-weeks PBMCs and in sorted hCD45+ cell lineages (15 weeks) of secondary recipients. n indicate independent animals.
Supplementary Material
Acknowledgments
We thank all members of LN’s laboratory for discussion, the IRCCS San Raffaele Hospital Flow Cytometry facility (FRACTAL), the IRCCS San Raffaele Center for Omics Sciences (COSR), A. Auricchio and M. Doria (Telethon Institute of GEnetics and Medicine; TIGEM Vector Core, Pozzuoli (NA), Italy) for providing AAV6 vectors, E. Ayuso (INSERM UMR1089, Nantes, France) for providing comments on AAV biology, L. Periè (Institute Curie, Paris, France) and J. Urbanus (Netherlands Cancer Institute; NKI, Amsterdam, Netherlands) for advices on barcoding cloning strategy, G. Schiroli for initial help with the design of the BAR-Seq strategy, R. Di Micco and B. Gentner for critical reading of the manuscript. We thank T. Plati for technical support in ddPCR analyses, T. Di Tomaso and G. Desantis for purifying mPB HSPCs, F. Benedicenti for helping in library preparation for NHEJ clonal tracking, L. Sergi Sergi, I. Cuccovillo, M. Biffi and M. Soldi for IDLV production and purification (SR-Tiget, IRCCS San Raffaele Scientific Institute), C. Di Serio for coordinating CUSSB support (Vita-Salute San Raffaele University). This work was supported by grants to: LN from Telethon (TIGET grant E4), the Italian Ministry of Health (PE-2016-02363691; E-Rare-3 JTC 2017), the Italian Ministry of University and Research (PRIN 2017 Prot. 20175XHBPN), the EU Horizon 2020 Program (UPGRADE), and from the Louis-Jeantet Foundation through the 2019 Jeantet-Collen Prize for Translational Medicine; PG from Telethon (TIGET grant E3) and the Italian Ministry of Health (GR-2013-02358956); AKR from the ERC (ImmunoStem – 819815). SF, VV and GU conducted this study as partial fulfillment of their Ph.D. in Molecular Medicine, International Ph.D. School, Vita-Salute San Raffaele University (Milan, Italy). AJ conducted this study as partial fulfillment of his Ph.D. in Translational and Molecular Medicine - DIMET, Milano-Bicocca University (Monza, Italy) with M. Serafini acting as University tutor.
Footnotes
Author Contributions
SF and AJ performed research, interpreted data and wrote the manuscript; GU performed experiments with IDLV based repair template supervised by AKR; LA provided technical help with mouse experiments; VV performed CD40LG and T cell experiments; SB, DC, DL and IM performed bioinformatic analysis; CB and FC performed statistical analyses; PG and LN designed the study, interpreted data, supervised research and wrote the manuscript; LN coordinated the work.
Competing Interests
LN, PG, AKR are inventors of patents on applications of gene editing in HSPCs owned and managed by the San Raffaele Scientific Institute and the Telethon Foundation, including a patent application on improved gene editing filed by SF, AJ, PG and LN. LN is founder and quota holder and PG is quota holders of GeneSpire, a startup company aiming to develop ex vivo gene editing in genetic diseases. All other authors declare no conflict of interests.
Data and Software Availability
All relevant data are included in the manuscript. BAR-Seq and RNA-Seq data are deposited in GEO with the following access codes: GSE143995 (for RNA-Seq) and GSE144340 (BAR-Seq). The reagents described in this manuscript are available under a material transfer agreement with IRCCS Ospedale San Raffaele and Fondazione Telethon; requests for materials should be addressed to LN.
Code Availability
BAR-Seq computer code are available from the corresponding author on reasonable request.
References
- 1.Naldini L. Genetic engineering of hematopoiesis: current stage of clinical translation and future perspectives. EMBO Mol Med. 2019 doi: 10.15252/emmm.201809958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll D. Genome Engineering with Targetable Nucleases. Annu Rev Biochem. 2014 doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 3.Genovese P, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014 doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiroli G, et al. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- 5.Boitano AE, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science (80.) 2010 doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fares I, et al. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science (80.) 2014 doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015 doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dever DP, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016 doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Ravin SS, et al. Targeted gene addition in human CD34(>+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat Biotechnol. 2016;34:1–8. doi: 10.1038/nbt.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo CY, et al. Site-Specific Gene Editing of Human Hematopoietic Stem Cells for X-Linked Hyper-IgM Syndrome. Cell Rep. 2018 doi: 10.1016/j.celrep.2018.04.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavel-Dinu M, et al. Gene correction for SCID-X1 in long-term hematopoietic stem cells. Nat Commun. 2019 doi: 10.1038/s41467-019-09614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeh CD, Richardson CD, Corn JE. Advances in genome editing through control of DNA repair pathways. Nat Cell Biol. 2019;21:1468–1478. doi: 10.1038/s41556-019-0425-z. [DOI] [PubMed] [Google Scholar]
- 13.Chu VT, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015 doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 14.Gwiazda KS, et al. High Efficiency CRISPR/Cas9-mediated Gene Editing in Primary Human T-cells Using Mutant Adenoviral E4orf6/E1b55k ‘Helper’ Proteins. Mol Ther. 2016;24:1–11. doi: 10.1038/mt.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiroli G, et al. Precise Gene Editing Preserves Hematopoietic Stem Cell Function following Transient p53-Mediated DNA Damage Response. Cell Stem Cell. 2019 doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steensma DP. Myelodysplastic syndromes current treatment algorithm 2018. Blood Cancer J. 2018 doi: 10.1038/s41408-018-0085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lombardo A, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 18.Zhao H, Dahlö M, Isaksson A, Syvänen AC, Pettersson U. The transcriptome of the adenovirus infected cell. Virology. 2012 doi: 10.1016/j.virol.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Täuber B, Dobner T. Adenovirus early E4 genes in viral oncogenesis. Oncogene. 2001;20:7847–7854. doi: 10.1038/sj.onc.1204914. [DOI] [PubMed] [Google Scholar]
- 20.Seandel M, et al. Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proc Natl Acad Sci. 2008;105:19288–19293. doi: 10.1073/pnas.0805980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frese KK, et al. Selective PDZ protein-dependent stimulation of phosphatidylinositol 3-kinase by the adenovirus E4-ORF1 oncoprotein. Oncogene. 2003 doi: 10.1038/sj.onc.1206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javier RT, Rice AP. Emerging Theme: Cellular PDZ Proteins as Common Targets of Pathogenic Viruses. J Virol. 2011 doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang MM, Hearing P. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 1989;3:1699–1710. doi: 10.1101/gad.3.11.1699. [DOI] [PubMed] [Google Scholar]
- 24.Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattabhi S, et al. In Vivo Outcome of Homology-Directed Repair at the HBB Gene in HSC Using Alternative Donor Template Delivery Methods. Mol Ther - Nucleic Acids. 2019 doi: 10.1016/j.omtn.2019.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero Z, et al. Editing the Sickle Cell Disease Mutation in Human Hematopoietic Stem Cells: Comparison of Endonucleases and Homologous Donor Templates. Mol Ther. 2019 doi: 10.1016/j.ymthe.2019.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrillo C, et al. Cyclosporine H Overcomes Innate Immune Restrictions to Improve Lentiviral Transduction and Gene Editing In Human Hematopoietic Stem Cells. Cell Stem Cell. 2018 doi: 10.1016/j.stem.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obert S, O’Connor RJ, Schmid S, Hearing P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol Cell Biol. 1994;14:1333–46. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menendez D, Nguyen TA, Snipe J, Resnick MA. The cytidine deaminase APOBEC3 family is subject to transcriptional regulation by p53. Mol Cancer Res. 2017 doi: 10.1158/1541-7786.MCR-17-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005 doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 31.Radhakrishnan SK, et al. Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle. Oncogene. 2004 doi: 10.1038/sj.onc.1207571. [DOI] [PubMed] [Google Scholar]
- 32.Komori H, Enomoto M, Nakamura M, Iwanaga R, Ohtani K. Distinct E2F-mediated transcriptional program regulates p14ARF gene expression. EMBO J. 2005 doi: 10.1038/sj.emboj.7600836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Querido E, et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mjelle R, et al. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst) 2015 doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Clement K, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nature Biotechnology. 2019 doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milyavsky M, et al. A Distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 37.van den Berg J, et al. A limited number of double-strand DNA breaks is sufficient to delay cell cycle progression. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang JCY, Doedens M, Dick JE. Primitive human hematopoietic cells are enriched in cord blood compared with adult bone marrow or mobilized peripheral blood as measured by the quantitative in vivo SCID-repopulating cell assay. Blood. 1997 doi: 10.1182/blood.v89.11.3919. [DOI] [PubMed] [Google Scholar]
- 39.Zonari E, et al. Efficient Ex Vivo Engineering and Expansion of Highly Purified Human Hematopoietic Stem and Progenitor Cell Populations for Gene Therapy. Stem Cell Reports. 2017;8:977–990. doi: 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagenblast E, et al. Functional profiling of single CRISPR/Cas9-edited human long-term hematopoietic stem cells. Nat Commun. 2019 doi: 10.1038/s41467-019-12726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai T, et al. Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat Med. 2019 doi: 10.1038/s41591-019-0601-5. [DOI] [PubMed] [Google Scholar]
- 42.Hoban MD, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015 doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurenti E, et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaley J, O’Connor RJ, Taylor LJ, Bar-Sagi D, Hearing P. Induction of the Cellular E2F-1 Promoter by the Adenovirus E4-6/7 Protein. J Virol. 2000 doi: 10.1128/jvi.74.5.2084-2093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaley JE, Polonskaia M, Hearing P. The Adenovirus E4-6/7 Protein Directs Nuclear Localization of E2F-4 via an Arginine-Rich Motif. J Virol. 2005 doi: 10.1128/jvi.79.4.2301-2308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanelle J, Stiewe T, Theseling CC, Peter M, Pützer BM. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 2002 doi: 10.1093/nar/30.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin AH, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997 doi: 10.1182/blood.v90.12.5002.5002_5002_5012. [DOI] [PubMed] [Google Scholar]
- 49.Notta F, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science (80.) 2011 doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 50.Fares I, et al. EPCR expression marks UM171-expanded CD34 + cord blood stem cells. Blood. 2017 doi: 10.1182/blood-2016-11-750729. blood-2016-11-750729. [DOI] [PubMed] [Google Scholar]
- 51.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018 doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 52.Ihry RJ, et al. P53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018 doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 53.Sakofsky CJ, et al. Repair of multiple simultaneous double-strand breaks causes bursts of genome-wide clustered hypermutation. PLoS Biol. 2019 doi: 10.1371/journal.pbio.3000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013 doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Notredame C, Higgins DG, Heringa J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000 doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 56.Provasi E, et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat Med. 2012 doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lassmann T. TagDust2: A generic method to extract reads from sequencing data. BMC Bioinformatics. 2015 doi: 10.1186/s12859-015-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Core Luca, Montini Eugenio, Di Serio Clelia, C A. Dealing with Data Evolution and Data Integration: An approach using Rarefaction. 49th Scientific meeting of the Italian Statistical Society; 2018. [Google Scholar]
- 59.Pinheiro J, Bates D, DebRoy S, Sarkar D, Team, the R. C The nlme Package: Linear and Nonlinear Mixed Effects Models. R-project. 2007 [Google Scholar]
- 60.Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015 doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the manuscript. BAR-Seq and RNA-Seq data are deposited in GEO with the following access codes: GSE143995 (for RNA-Seq) and GSE144340 (BAR-Seq). The reagents described in this manuscript are available under a material transfer agreement with IRCCS Ospedale San Raffaele and Fondazione Telethon; requests for materials should be addressed to LN.
BAR-Seq computer code are available from the corresponding author on reasonable request.