Abstract
The capacity for some pathogens to jump into different host-species populations is a major threat to public health and food security. Staphylococcus aureus is a multi-host bacterial pathogen responsible for important human and livestock diseases. Here, using a population genomic approach we identify humans as a major hub for ancient and recent S. aureus host-switch events linked to the emergence of endemic livestock strains, and cows as the main animal reservoir for the emergence of human epidemic clones. Such host-species transitions are associated with horizontal acquisition of genetic elements from host-specific gene pools conferring traits required for survival in the new host-niche. Importantly, genes associated with antimicrobial resistance are unevenly distributed among human and animal hosts reflecting distinct antibiotic usage practices in medicine and agriculture. In addition to gene acquisition, genetic diversification has occurred in pathways associated with nutrient acquisition, implying metabolic remodeling after a host-switch in response to distinct nutrient availability. For example, S. aureus from dairy cattle exhibit enhanced utilization of lactose, a major source of carbohydrate in bovine milk. Overall, our findings highlight the influence of human activities on the multi-host ecology of a major bacterial pathogen, underpinned by horizontal gene transfer and core genome diversification.
Keywords: Evolution, Staphylococcus aureus, host-species, adaptation, genomics
Introduction
Many bacterial pathogens are host specialists that co-evolve with a single host-species. However, the capacity to switch host species can provide opportunities for expansion into new host populations. The domestication of animals in the Neolithic period (10,000-2,000 BC approximately) and the more recent intensification of livestock farming provided increased opportunities for the movement of bacterial pathogens between humans and animals1. Of note, the majority of emerging human infectious diseases have been traced to an animal origin2. Staphylococcus aureus is associated with a wide spectrum of diseases in humans and strains of both methicillin-sensitive (MSSA) and methicillin-resistant S. aureus (MRSA) are common causes of nosocomial and community-acquired infection3,4. In addition, S. aureus causes an array of infections of livestock that are a major burden on the agricultural industry, including mastitis in cows, sheep and goats 5,6, septicemia and skeletal infections in commercial broiler chickens7, exudative epidermitis in pigs8 and skin abscesses and mastitis in rabbits9.
S. aureus has a clonal population structure defined by a relatively low level of recombination, comprised of lineages that have single or multiple host-tropisms10–12. Inter-host species transmission can be of critical public health importance, as exemplified by the livestock-associated methicillin-resistant clonal complex (CC) 398, which is associated with pigs and other livestock, but can cause zoonotic infections of pig-farmers and their contacts13,14. Previous work employed multi-locus sequence typing (MLST) to provide evidence for the occurrence of host-jump events from humans leading to the emergence of S. aureus clones in livestock populations11,12. More recently, whole genome sequencing has been employed to investigate the evolution of individual clones, providing insights into the emergence, transmission and acquisition of antibiotic resistance in hospital, community, and agricultural settings13,15–17. In addition, a role for specific mobile genetic elements (MGEs) and core gene mutations in the host-adaptation of S. aureus has been identified 9,18,19. For example, the major porcine and avian clones of S. aureus likely originated in humans and the host-jumps were associated with acquisition of MGE not found among human isolates 13,18. Similarly, the major S. aureus clone associated with sheep and goats evolved through a combination of gene acquisition, and allelic diversification including loss of gene function20. Furthermore, several studies have reported the host-specific functional activity of S. aureus effectors such as leucocidins, superantigens, and the von Willebrand factor-binding protein 21–26. In addition, it was demonstrated that for S. aureus strains associated with natural infections of rabbits, a single mutation was responsible for conferring infectivity to the progenitor strain found in human populations9. Taken together, these studies highlight the capacity for bacteria to undergo host-switching events and adapt to different species by multiple evolutionary genetic and functional mechanisms. However, a large-scale, genome-based analysis of the evolutionary history of S. aureus in the context of its host ecology is lacking, and the scale and molecular basis of host-switching events remains poorly understood.
Here, we carry out a population genomic analysis of over 800 S. aureus isolates selected to represent the known breadth of host-species diversity in order to provide a high-resolution picture of the dynamics of S. aureus in the context of its host. The data reveal the impact of human activities such as domestication and the use of antibiotics in medicine and agriculture on the recent evolution of S. aureus, and identify the key evolutionary processes underpinning its multi-host species ecology.
Results
Extensive host-switching events define the evolutionary history of S. aureus
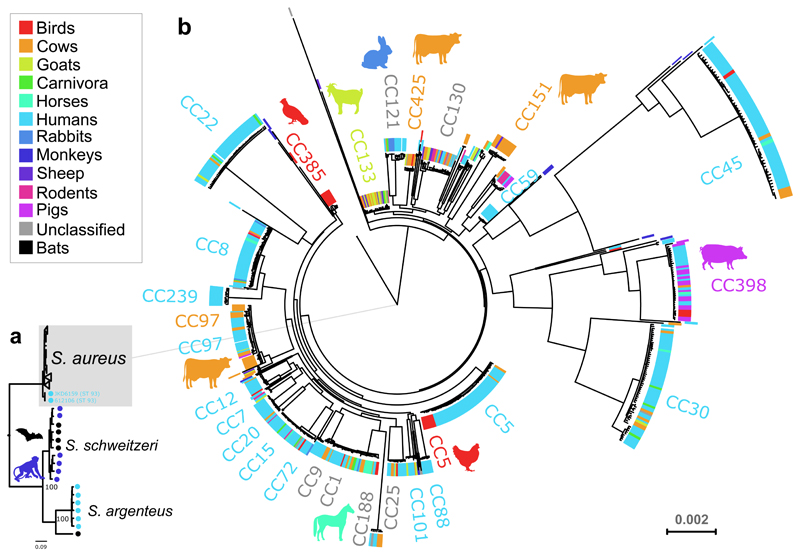
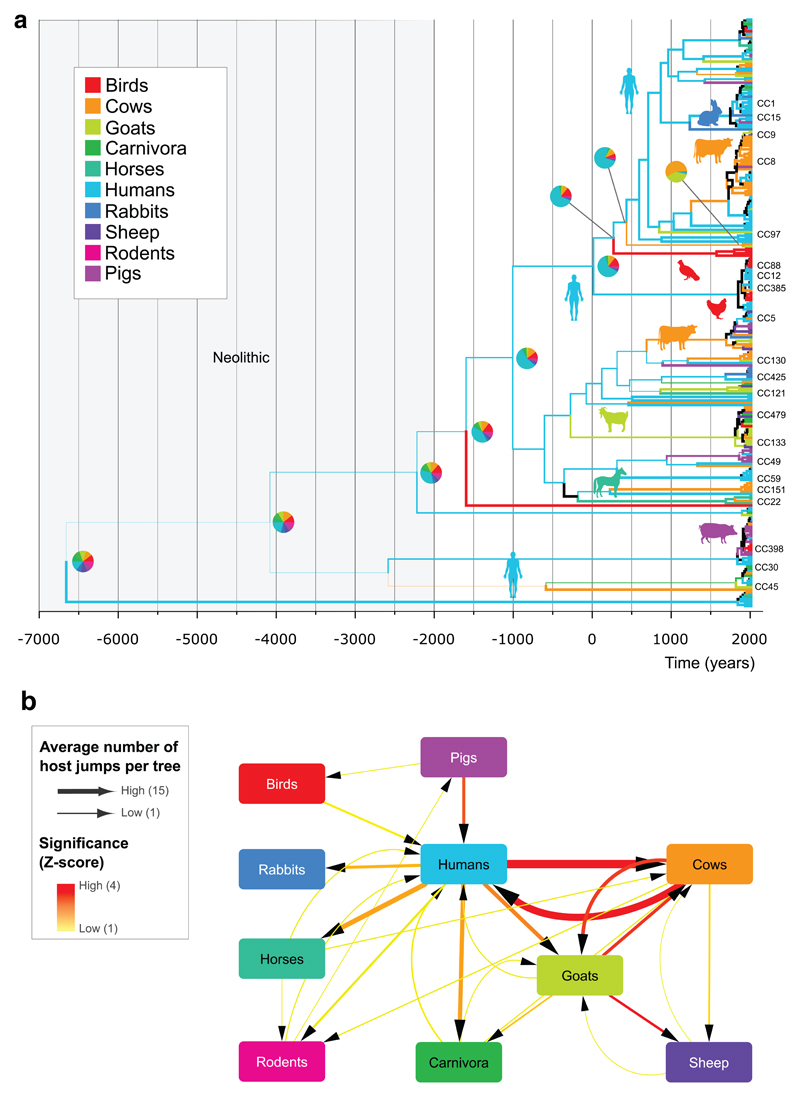
We selected S. aureus strains to represent the breadth of the known clonal, geographic, and host-species diversity (Isolate selection details in Methods section). Overall, we included 800 isolates representative of 43 different host species, 77 clonal complexes (CCs), isolated in 50 different countries across 5 continents (Supplementary Figure 1-3; Supplementary Table 1). Among the 800 isolates, a total of 115,149 SNPs were identified in a core genome of 711,562 bp and used for reconstruction of the maximum-likelihood (ML) phylogeny for the S. aureus species (Fig. 1). The S. aureus species tree indicates the existence of highly divergent clades representative of the recently-described Staphylococcus argenteus and Staphylococcus schweitzeri species which belong to the extended S. aureus-related complex (Fig. 1a)27. S. argenteus, an emerging cause of human clinical infection28, is more closely related to bat and monkey isolates than to other human S. aureus sequence types (STs), consistent with a possible non-human evolutionary origin for S. argenteus. Removal of isolates from the divergent clades resulted in a phylogeny of 783 isolates that segregated according to clonal complexes defined by MLST (Fig. 1b). The phylogeny indicates the broad diversity of isolates of human origin with expansion of several successful epidemic hospital and community-associated clones including CC22, CC30, and ST45, as previously described29 (Fig. 1). Animal isolates are typically found in discrete host-specific clades interspersed among human lineages, consistent with ancient and recent host-switching events across the phylogenetic tree (Fig. 1). In order to examine the frequency and timing of host-switching events during the evolution of S. aureus, we employed Bayesian evolutionary analysis by sampling trees (BEAST) using substitution rates from published datasets (Fig. 2; Supplementary Table 2). We estimated the number of cross-species transmissions for 10 major host categories (Supplementary Table 3, Supplementary Figures 2-5) using BEAST with Markov Jumps30. In order to reduce bias caused by the larger numbers of sequences from human and cow hosts compared to the other host types we used 10 stratified subsamples containing 252 sequences each, designed to maintain geographic, host-type and temporal diversity while reducing over-representation. To assess the robustness of the main analysis, we performed additional analyses as outlined in Supplementary Material (Supplementary Notes; Supplementary Figures 4-11; Supplementary Tables 4-5), that included ‘severe balanced’ subsamples of 97 taxa each containing 18-20 taxa of 5 host-types, and ancestral state and host-jumps using the BASTA approximation to the structured coalescent31. However, we had difficulty in getting BASTA to run and converge possibly due to its assumptions about the structure of the data and numerical instability. Each subsampled sequence set was analyzed separately within BEAST and resulted in a collection of posterior trees per dataset (Supplementary Figures 6-10). In each case, the analysis revealed extensive host-switching events that occurred over a time-frame spanning several thousand years up to the present decade (Fig. 2a).
Figure 1. Staphylococcus aureus phylogeny according to host-species origin.
(a) Phylogenetic tree of 800 isolates constructed using the maximum liklihood (ML) method demonstrating the relationship between S. aureus and other members of the S. aureus complex; S. schweitzeri and S. argenteus; (b) Phylogenetic (ML) tree of 783 S. aureus isolates, with host species indicated in colour or animal symbols indicating major domesticated animal clones that are largely host-specific. The evolutionary history of S. aureus was calibrated using well-established substitution rates from published datasets (see methods).
Figure 2. S. aureus has undergone extensive ancient and recent host-switching events with humans acting as a major hub.
(a) Time-scaled phylogeny of a subsample of the S. aureus sequences with clonal complexes (CCs) labeled, branches colored according to host-species group. Pie charts indicate relative probability of host origin at the ancestral nodes, and line thickness corresponds to probability of the majority host (see Supplementary Figures 6-10 for all subsamples). Major clonal complexes (CC)s are indicated. (b) Quantification of the number of host-switch events: Host transition count network from BEAST Markov Jumps models averaged over all subsamples of the data. Line-width represents the average Markov Jump count per tree, averaged over all subsamples (Supplementary Figures 4-5) and line color represents the significance compared to permuted label analysis (Z-score). Only transitions with higher counts compared to models with permuted host-labels are shown (Z-score >= 0.5).
Our analysis identifies humans as a major donor with host-jumps identified from humans into all other host-species groups examined (Fig. 2b, Supplementary Figure 4). The most common recipient for S. aureus jumps from humans was cows with a median of 14 jumps (HPD 3-22) between the years -2000 and 2012. Cows also represented a major donor for host-switching events back into humans (n=10; HPD 2-26). In addition, there were numerous S. aureus host-switches among ruminants, particularly between cattle and goats in both directions and into sheep. However, host jumps from sheep into other species are rare and not strongly supported by our analyses suggesting that although a common host for S. aureus 5, sheep do not represent a major reservoir for the spread of S. aureus to other animals.
Host-specific accessory gene pools promote adaptive evolution after host-switching events
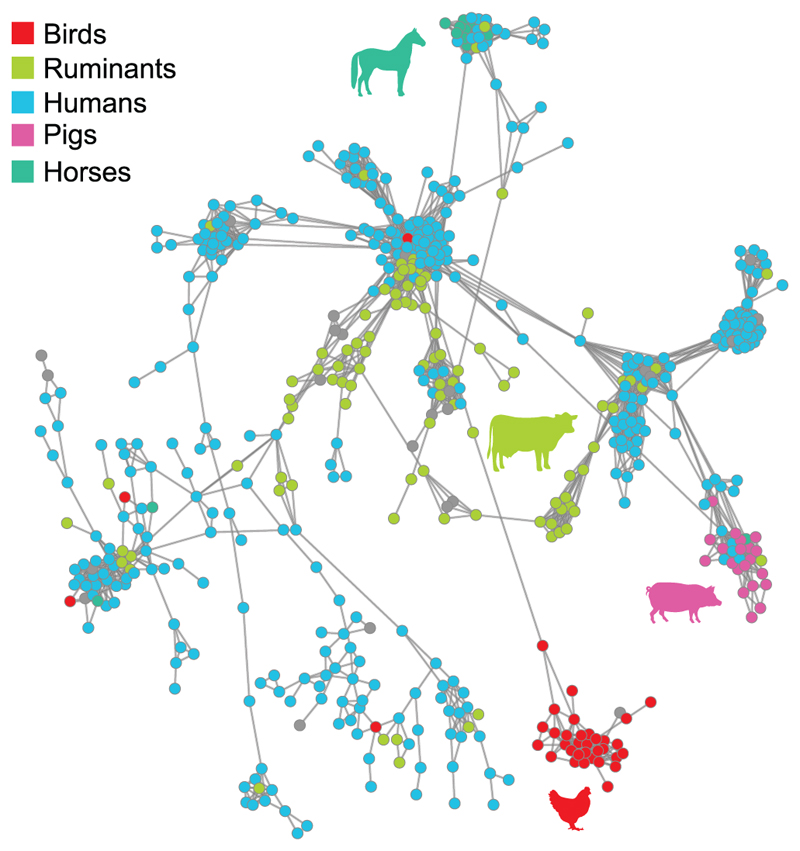
In order to investigate the distribution of MGEs on a population level across human and animal isolates, we employed a pangenome-wide association analysis approach to identify genes that were enriched among isolates from specific host-species. First, to account for phylogeny we removed genes identified among all strains within clonal complexes associated with multiple host-species (lineage-dependent genes). Network analysis indicated a remarkable correlation between accessory genome and host-species revealing that diverse clonal complexes can share highly similar accessory genomes that are specific for birds, pigs or horses, respectively. This strongly points to the existence of a host-specific gene pool required for S. aureus host-adaptation. Although accessory genomes of S. aureus obtained from humans, and from cows, sheep and goats also tended to cluster together in a host-specific manner, there was greater diversity in gene content (Fig. 3). This may reflect the existence of multiple cryptic niches that exist within a single host-species such as those proposed previously for Campylobacter jejuni 32. We note the existence of a small number of clusters made of isolates from multiple host-species. The existence of these clusters suggests that some accessory gene combinations may confer a more generalist host tropism with the capacity to infect multiple host-species. Alternatively, insufficient time may have passed since the host-transition event for loss of dispensable MGE to occur. Of note, antibiotic resistance gene determinants influenced the clustering of equine and pig isolates suggesting a role for acquisition of resistance in host-adaptation (Supplementary Figure 12).
Figure 3. Network analysis of S. aureus accessory genome indicates clustering according to host species group.
Network graph of pairwise distances of accessory genome gene content between isolates. Each node represents an isolate, colour-coded to indicate host species origin, and each edge indicates greater than 50% of shared accessory genome content with the length of the edges weighted by distance (proportion of shared accessory genes; shorter edges have more genes in common). All edges with <50% shared accessory genome content were removed.
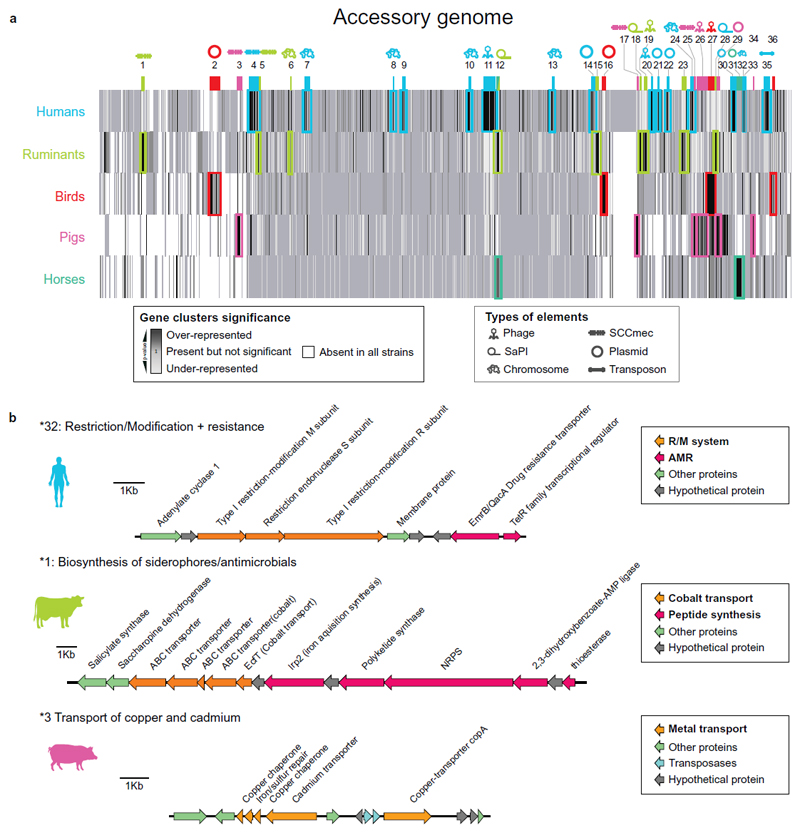
Further examination of the impact of the accessory genome on successful host-switching events was carried out by identifying gene acquisition or loss events that correlated with host-switching events identified on the phylogeny of S. aureus. A total of 36 distinct MGEs including predicted plasmids, transposons, S. aureus Pathogenicity Islands (SaPIs) and prophages were identified to be associated with host-switch events (p< 0.0001) (Fig. 4a, Supplementary Table 6). Several of the MGEs have previously been identified and demonstrated to encode proteins with host-specific activity. For example, the β-converting phage ϕSa3 encodes modulators of the human innate immune response, and pathogenicity islands encode superantigens or von Willebrand factor-binding proteins with ruminant-specific activity 19,33. In addition, equine isolates contain a phage encoding a novel equine allele of the staphylococcal inhibitor of complement (scn) which also encodes the LukP/Q toxin, recently characterized to have equine-specific activity22,25. However, numerous uncharacterized MGEs have been identified in the current study to be linked to successful host-switch events providing many novel avenues for characterizing the molecular basis of S. aureus host-adaptation (Fig 4b). For example, in isolates from pigs, a putative novel plasmid linked to SCCmec encoding resistance to heavy metal ions, a common supplement in pig-feed, was linked to host-switching events from humans into pigs (Fig. 4b). Finally, several gene clusters encoding bacterocins were enriched in isolates from specific host-species (p< 0.0001) or were linked to host-switching events (p<0.0001), consistent with the need to compete with resident bacteria for survival (Supplementary Table 6). Taken together, these data suggest that successful host-switch events are associated with acquisition of MGEs from an accessory gene pool that exists in the recipient host-species, and/or loss of MGEs linked to the source species.
Figure 4. Identification of horizontally-acquired genetic elements correlated with host-adaptation.
(a) Schematic representation of the S. aureus pan-genome with gene clusters linked to host-species indicated by shading. Coloured symbols indicate the nature of the mobile genetic element and the associated host-species (b) Annotated gene maps of selected novel genetic elements linked to specific host species. associated with acquisition of MGEs from an accessory gene pool that exists in the recipient host-species, and/or loss of MGEs linked to the source species.
In order to investigate the potential origin of MGEs horizontally acquired after a host-switch event, we examined the codon usage bias of host-specific MGE, and found that MGEs enriched in pig isolates had significantly elevated %GC content and reduced codon adaptive index (CAI) indicative of a distinct genealogical origin (Supplementary Figures 13-15). Of note, an MGE found in pig isolates had highest BLASTn similarity to a putative pathogenicity island previously identified in the pig-associated zoonotic pathogen Streptococcus suis (GC content of ~41%) (Supplementary Table 6).
Both gain and loss of gene function are associated with S. aureus host-adaptation
Determination of the number of predicted functional genes in each S. aureus genome identified a significantly higher number of genes in bird strains compared to S. aureus from any other host-species (Supplementary Figures 16-17). In contrast, the number of pseudogenes per genome is significantly higher (p<0.0001-0.02) in ruminant strains compared to those from other host-species suggesting that the niche occupied by S. aureus in cows may provide stronger selection for loss of gene function compared to the niches for S. aureus in birds and pigs. Numerous pseudogenes associated with transport of nutrients in S. aureus including carbohydrates, are over-represented in ruminant isolates implying metabolic remodeling in response to distinct nutrient availabilities in the bovine niche (Supplementary Table 8).
Refinement of host adaptation involves modification of biological pathways in response to nutrient availability
In addition to accessory genes, adaptive mutations in the core genome may be selected for in response to environmental changes such as antibiotic exposure or a switch in host-species 9,34. In order to examine the impact of host-species on diversification of the S. aureus core genome, we identified groups of related isolates (e.g. within CCs or STs) associated with a specific host-species for genome-wide analysis of positive selection (Supplementary Table 9; Supplementary Figure 18). Positive selection was identified across all host-associated groups examined, with an average of 68 genes (33 to 129) representing approximately 2.7% (1.3% to 5.1%) of a clade-specific core genome (Supplementary Table 10). A limited number of genes were under diversifying selection across multiple host species, including several that encode membrane proteins, lipoproteins and a protein involved in biofilm formation. Some genes were identified as undergoing positive selection in distinct lineages that were associated with the same host-species (mostly human), suggesting strong selective pressure leading to convergent evolution. However, for the most part, our analysis detected distinct sets of genes under positive selection in different lineages, suggesting that signatures of host-adaptation are dependent on the genetic background of the strain, and that host-adaptation can occur via multiple trajectories involving modification of distinct pathways.
We predicted functional categories of genes under positive selection and the biological pathways affected revealing several functional groups that were enriched for positively selected genes independently of the host species including genes linked to pathogenesis, immune evasion and maintenance of MGEs ((Supplementary Table 11; Supplementary Figure 19). However, the majority of the functional categories were host-species dependent, consistent with distinct mechanisms underpinning adaptation to different host-species (Supplementary Table 11; summarized Fig. 5). In particular, biological pathways associated with amino acid metabolism and iron acquisition were under positive selection in several host-species suggesting diversification in response to distinct nutrient availability in different host niches. In addition, genes associated with transport and metabolism of carbohydrates demonstrated signatures of positive selection in S. aureus clones from humans and cows (Fig. 5).
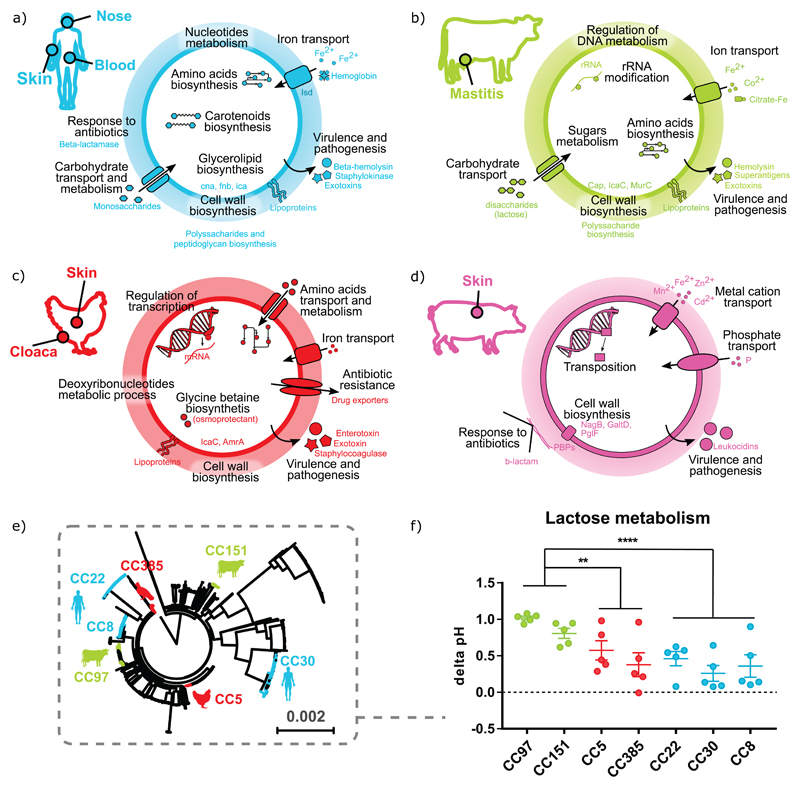
Figure 5. Summary of biological pathways under positive selection in different host-species and evidence for phenotypic adaptation.
The main anatomical isolation sites on each host group are indicated by filled circles. Functional categories virulence and pathogenesis, resistance to antibiotics, transport of ions and cell wall biosynthesis were under positive selection in all 4 host-species groups. In humans (a) and ruminants (b) the categories amino acids biosynthesis and transport/metabolism of carbohydrates were positively selected. The categories amino-acid transport/metabolism and biosynthesis of osmoprotectants were under positive selection in birds (c) and transposable elements in pigs (d). (e) Phylogenetic tree indicating the distinct lineages selected for comparative analysis of lactose fermentation,(f) Fermentation of the disaccharide lactose is enhanced in bovine lineages. Acidification of S. aureus culture supernatant in presence of 100 mM lactose as indicated by the delta pH. Experiments were performed in triplicate with 5 strains per clonal lineage. Each dot represents the average delta pH per strain and bars indicate the SEM per clonal lineage (n = 5). Asterisks indicate significant differences between bovine (CC97 and CC151, n = 10), avian (CC5 and CC385, n = 10) and human lineages (CC22, CC30, and CC8, n = 15) with **P<0.005, ***P<0.001 and ****P<0.0001 using One-Way ANOVA followed by Tukey’s multiple comparison test.
Bovine S. aureus strains utilize lactose with higher efficiency compared to human or avian strains
Considering the signatures of positive selection identified among pathways associated with carbohydrate and amino acid metabolism, we investigated differences in growth phenotype of selected host-specific S. aureus strains using a metabolic phenotype microarray (Biolog), and observed preliminary strain-dependent differences in growth that were influenced by the availability of specific amino acids or carbohydrates. For example, S. aureus strains from cows had higher relative growth in the presence of lactose, the primary disaccharide available in bovine milk. The genome-wide positive selection analysis indicated that in bovine strains, genes associated with the functional category of transport of disaccharides and oligosaccharides were impacted by positive selection. To further investigate this, we carried out phenotypic analysis of S. aureus strains from bovine, human and avian host- species of different clonal complexes when grown in the presence of lactose (Fig. 5e). As lactic acid is produced by S. aureus as a by-product of fermentation, we measured pH levels in culture media containing lactose and identified a decrease in pH levels for bovine S. aureus clones in comparison to human or avian clones, consistent with increased efficiency of fermentation of lactose (Fig. 5f). These data support the concept that S. aureus undergoes genetic diversification in response to the nutrients that differ in availability in different niches.
Resistance to antimicrobials differs among human and pig S. aureus
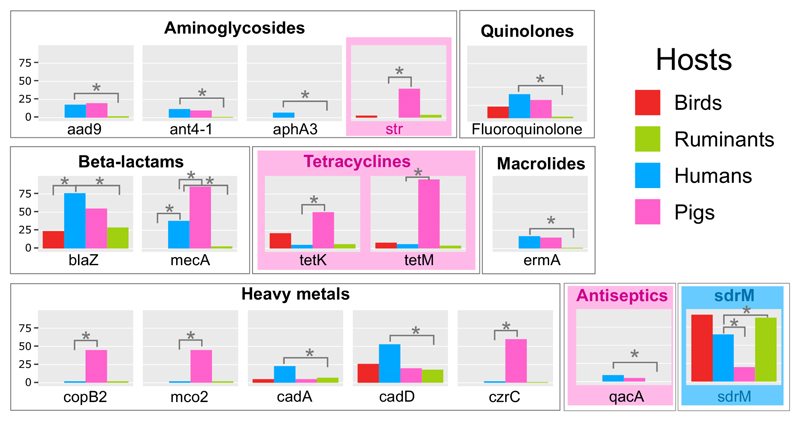
Our understanding of the relative contribution of the use of antibiotics in human medicine and agriculture to the emergence of antibiotic resistance is very limited. To address this question for the model human and animal pathogen S. aureus, we examined the distribution of antibiotic, antiseptic and heavy metal-ion resistance determinants among human and livestock isolates, and then accounted for phylogenetic relatedness for resistance to different classes of antibiotic (Supplementary Table 12). An array of resistance determinants were significantly enriched in human, ruminant, and pig isolates, respectively, but not among avian isolates, consistent with a limited role for the poultry industry in the emergence of antibiotic resistance in S. aureus.(Fig. 6; Table S12). When testing for phylogenetic independence, we aimed to maximize statistical power by including all gene determinants into groups specific for each class of antimicrobial, and also examined selected individual determinants str and sdrM. The analysis indicated that resistance to streptomycin, antiseptics, and tetracyclines were all significantly associated with pig isolates, whereas sdrM was enriched in human isolates. However, fluoroquinolone and heavy metal ion resistance did not correlate with hosts after correction for phylogeny implying that expansion of specific clones has contributed to the high frequency of those resistance determinants among human and pig hosts, respectively. Taken together, these data demonstrate that resistance to specific classes of antimicrobial in S. aureus is host species-dependent providing evidence for distinct antibiotic selective pressures in humans and livestock. Of note tetracyclines, and aminoglycosides (such as streptomycin) are used in much higher amounts in farmed animals compared to human medicine 35. Zoonotic transmission of S. aureus is a relatively common occurrence for some clones, particularly between pigs and humans in the case of CC398, providing a route for the transmission of resistant strains and associated resistance determinants to humans36.
Figure 6. Resistance to antimicrobials is non-randomly associated with host species.
Proportion (%) of isolates examined which contain the specified resistance determinant (Supplementary Table 12). Asterisks indicate significant association of resistance determinants with host-species (Fisher exact test), and colored borders indicate antibiotic class or single determinants (sdrM and str) that are associated with host-species group after testing for phylogenetic independence.
Discussion
Many new pathogens emerge following zoonotic or anthroponotic events providing the opportunity for spread within a new host population2. S. aureus is considered a generalist bacterial species, capable of colonizing a wide range of hosts5. However, the species is composed of distinct sub-lineages that are commonly associated with particular hosts or host groups10,14. Accordingly, S. aureus represents an excellent model to explore the dynamics of a bacterial pathogen at the human-animal interface. Here, we demonstrate that the segregated host-specialism of S. aureus arose via multiple cross-species transmission events that occurred over the last 5,000-6,000 years, leading to the emergence of successful endemic and epidemic clones circulating in distinct host-species populations. We identify humans as a major reservoir for the spread of S. aureus to livestock, reflecting the role of humans in domestication of animals, and subsequent opportunities for cross-species transmission events consistent with analysis using MLST12. Importantly, we also identify cows as the main animal source for the emergence of S. aureus clones that are epidemic in human populations consistent with a previous study that identified a bovine origin for emergent CC97 clones causing human infections across multiple continents17.
The identification of combinations of MGEs that are associated with specific host-species and linked to host-switching events provides compelling evidence for the key role of horizontal gene acquisition in the adaptation of S. aureus to their hosts. While several MGEs have been identified to be associated with host-specific clones18,19,22,24, our species-wide analysis reveals combinations of MGEs linked to specific host species providing many new avenues for investigating mechanisms of bacterial host-adaptation. Overall, the data suggest that host-specific accessory gene pools presumably present in the microbiota of the new host-species promote the host-adaptive evolution of S. aureus.
In addition to gene acquisition associated with host-switch events, we identified evidence of adaptive evolution in the core genome consistent with host-specific selective pressure driving the diversification of biological pathways that are involved in survival or transmission. Furthermore, in some cases, distinct pathways were under positive selective pressure in different clones associated with the same host-species, implying that multiple distinct pathways may mediate host-adaptation depending on the genetic background of the strain. In particular, pathways linked to carbohydrate transport exhibited signatures of host-adaptation and phenotypic analysis revealed enhanced utilization by bovine S. aureus clones of the disaccharide lactose, the major carbohydrate available in bovine milk.
These findings inform a model of S. aureus host-adaptation in which acquisition of a specific set of MGEs occur rapidly after a host-switch event (although we can’t rule out this could occur prior to the jump in some cases), conferring the capacity for survival in the new host, largely through targeting of the innate immune response via bacterial effectors such as leukocidins, superantigens and other immune-modulators. Other MGEs confer resistance to antibiotics and heavy metal ions allowing survival under strong anti-microbial selective pressures. Subsequently, positive selection acts on the core genome via point mutation and/or recombination 37 causing allelic variation and loss of gene function that results in modification of metabolism in response to distinct nutrient availability.
Our findings suggest that since human-driven domestication, interactions with livestock have provided opportunities for numerous successful host-switch events between humans and livestock hosts. Further, industrialization of agriculture including use of antibiotics and feed supplements in intensive farming have directly influenced the evolution of S. aureus clones resulting in the emergence of resistance in response to distinct antibiotic selective pressures in human medicine and agriculture18,38. These data support the idea that surveillance could play a critical role in the early identification of emerging clones that have jumped host.
Taken together, our data provide a high-resolution view of the capacity for a model multi-host pathogen to undergo radical changes in host ecology by genetic adaptation. Investigation into the functional basis of these genetic changes will reveal key host-pathogen interactions that could be targeted for novel therapies. Further, the identification of the common routes for S. aureus livestock-human host-species switches and distinct types of antimicrobial resistance in humans and livestock species could inform the design of more effective farm security and antibiotic treatment practices to limit the emergence of new resistant clones. These findings will be relevant to other major bacterial pathogens with the capacity to spread between livestock and humans.
Methods
Isolate selection
For selection of isolates, the literature was reviewed (date: November 2013) and all available S. aureus strains associated with animals and humans for which genomes had been determined were identified. We aimed to include isolates to represent the breadth of clonal complexes, host-species diversity, geographical locations and as wide a temporal scale as possible (Supplementary Tables 1-3). Publicly available sequences were selected as follows; 74 reference genomes, 302 from the EARSS project29, and 252 from other published studies of the authors (Supplementary Table 1). Furthermore, to be as representative of the known S. aureus host, clonal, and geographic diversity as possible we selected an additional 172 isolates for whole genome sequencing (Supplementary Table 1). Our dataset is biased towards human isolates which represent approximately 60% of the total with 40% approximately from animal sources. This reflects that fact that much of the known diversity of the S. aureus species is of human origin12, and also that fewer number of isolates that have been obtained from animals. Given the predominant European origin of the animal isolates (due to the contemporary interest in animal S. aureus in Europe), we chose to enrich the number of human isolates with the EARSS collection representative of the diversity of invasive S. aureus circulating among humans in Europe in 200629. Accordingly, there is a European bias to the sample dataset and we can’t rule out that we have under-sampled the S. aureus diversity that exists in other parts of the world. Nonetheless, our dataset contained isolates from 50 different countries across 5 continents and many sequence types are widely distributed on an intercontinental scale. In addition, our dataset includes isolates from the years 1930 to 2014, although the majority have been isolated since 2005 reflecting the greater availability of recent clinical isolates (particularly from animals). It should therefore also be considered that the dataset is biased towards contemporary S. aureus and that older lineages that are now less abundant or extinct may not be represented in our dataset. In order to partially address the uneven distributions of isolates by host, space and time, where appropriate, we have carried out experimental replicates based on severe subsampling of the dataset that provide more evenly distributed groups. In addition, we have drawn conclusions that are consistent across subsampled data and, when appropriate, multiple different analytic approaches. Overall, we included 800 isolates representative of 43 different host species and 77 clonal complexes (CCs), isolated in 50 different countries across 5 continents (Supplementary Table 1). All sequences and associated metadata have been uploaded to Microreact a publicly accessible database that allows visualization and analysis of the data https://microreact.org/project/shacdata 39.
Sequencing, genome assemblies, variant calling and phylogenetic reconstruction
For the current study, bacterial DNA was extracted and sequenced using Illumina HiSeq2000 with 100-cycle paired-end runs at the Wellcome Trust Sanger Institute or Illumina HiSeq2000 at Edinburgh Genomics. The nucleotide sequence data were submitted to the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena) with the accession numbers listed in Supplementary Table 1. Completed genomes downloaded from the NCBI database were converted into pseudo-fastq files using Wgsim (https://github.com/lh3/wgsim). For each isolate the sequence reads were used to create multiple assemblies using VelvetOptimiser v2.2.540 and Velvet v1.241. The assemblies were improved by scaffolding the best N50 and contigs using SSPACE42 and sequence gaps filled using GapFiller43. Isolates were excluded from the analysis for the following reasons that are indicative of contamination or poor quality sequence data; a large number of contigs and a large number of ‘N’s in the assemblies or large genome size (>2.9 Mb). Sequence types were determined from the assemblies using MLST check (https://github.com/sanger-pathogens/mlst_check), which was used to compare the assembled genomes against the MLST database for S. aureus (http://pubmlst.org/saureus/). Sequence reads were mapped to a relevant reference genome (European Nucleotide Archive (ENA) ST425 (strain LGA251, accession number FR821779), using SMALT (http://www.sanger.ac.uk/science/tools/smalt-0) following the default settings to identify single nucleotide polymorphisms (SNPs). Consensus sequences were obtained using samtools and concatenated into core genome alignments44. SNPs located in mobile genetic elements were removed from the alignments and a maximum likelihood tree was constructed using RAxML following default settings and 1000 bootstrap replicates45.
Time scaled trees and estimation of the number of host jumps
Time scaled trees were generated using BEAST 1.8.246. All isolates with unknown date, unknown host species or unknown geographical location were removed in addition to the diverse BAPs groups 12 and 14 leaving a total of 696 isolates. Sites determined to be affected by recombination from the BNG analysis of the individual BAPs groups, were coded as missing data. Since missing data can effect phylogenetic inference and contribute to heavy likelihood calculations, sites that had more than one missing state in the alignment (either from missing mapped reads or recombination) were excluded from further analyses, leaving a total of 55,778 sites (4306 segregating sites).
To account for different evolutionary processes acting at synonymous and non-synonymous sites, RNA and noncoding sites, the evolutionary model was partitioned in to 1+2nd sites, 3rd sites, non-coding sites, and RNAs according to the reference strain LGA251. Pseudogenes were partitioned in to 1+2nd and 3rd sites with the rationale that they may be functional in other isolates. For overlapping reading frames, sites were assigned to the region of highest constraint (e.g. when coding and RNA, sites assigned as RNA, when 1st and 3rd, sites assigned as 1+2nd etc.). For all partitions, we used an HKY + Γ substitution model.
Dating
We treated all sequences as contemporaneous but assigned a median prior of 1.61 [0.604, 2.9] substitutions per site per million years on to 3rd positions, which are less likely to be subject to strong purifying selection (known to affect rates over different timescales). The prior comes from previous studies of S. aureus using tip dates on different strain types (Supplementary Table 3). An uncorrelated lognormal model of changes in substitution rate across different branches was employed. An initial MCMC run with this model was performed in BEAST v1.8.246 using Beagle47 with two independent chains, removing the appropriate burnin and run for approximately 100,000,000 generations.
In addition, a 10 further subsampled datasets were produced that included only sequences from the 10 major host types. These were stratified subsamples containing 252 samples each, designed to maintain the host species, geographic and temporal diversity. The major hosts types are birds, cows, goats, carnivores, horses, humans, rabbits, sheep, rodents and pigs. From these analyses, we subsampled an empirical distribution of 1000 trees post burnin which were used for all further BEAST analyses.
Markov Jump analysis
In order to reconstruct host transition events, we used an asymmetric discrete state phylogeographic analysis with Markov Jumps (REF: 51) applied to the 10 major host types with default priors. We employed the Markov jump analysis to estimate the posterior expectation of the number of host change events across the branches of the phylogeny30, using posterior sets of 1000 time scaled trees from the initial BEAST analyses on the subsampled data sets. The trait models were used in an MCMC chain of 1,100,000 steps, sampling every 100 steps and discarding the first 10% as burn-in, leaving 10,000 trees annotated with the host information (i.e. approximately 10 model instances per tree of the original posterior set). Since biased sampling can lead to biased results when using these trait models, in addition to using the 10 stratified subsamples of 252 sequences each we performed additional analyses with host state randomization and using 100 bootstrapped maximum likelihood trees (RAxML) in place of the 1000 original BEAST trees (for the 10 stratified subsamples) To balance the numbers of isolates per host category further, we also created 10 ‘severe’ stratified subsamples containing 97 sequences each with 20 from Humans, Cows and Sheep+Goats combined and 19 and 18 from Birds and Pigs respectively. Necessarily in these severe subsamples it was not possible to maintain the full human and cattle diversity, although sequences from different geographic locations and years were chosen. We applied BEAST with Markov Jumps on these 5 host categories using: full joint inference of trees using sequences and traits together; trees using the sequences only followed by the trait mapping as before; and the BASTA structured coalescent approximation 31.
Pseudogene analysis
Pseudogenes were predicted during the PROKKA annotation process48. Specifically, each protein in a genome was searched against UniProtKB (Swiss-Prot) using BLASTp49 or UniProtKB (TrEMBL). If no significant hits were identified, proteins were examined for conserved motifs. Any proteins exhibited less than 95% coverage of their top hit were listed as potential pseudogenes. The region of the top hit that was not present in the protein sequence was then interrogated against all contigs using BLASTn49. Hits that were in the correct orientation and on the same contigs were accepted as pseudogenes and labelled according to their type (frameshift, stop codon, insertion). Proteins that were less 95% coverage of their top hit and on the edge of a contig with their counterpart on another contigs were not labelled as pseudogenes, rather CDS that have split due to the assembly breaking at this point.
The UniProt ID Mapping tool was used to assign Gene ontology (GO) terms to all pseudogenes by transferring the GO terms assigned to the closest reference (identified during the annotation process described above). GO was assigned to all non-pseudogenes (CDS features) using the same method and InterProScan50. The R package topGO51 with Fisher’s exact test was used to identify enriched GO terms whilst taking into account the GO hierarchy (the p-value was adjusted using Bonferroni correction).
Pan-genome association analysis
All genomes in this study were organised into a list of reference genomes followed by assembled contigs. The second genome in the list was aligned to the first genome using Nucmer52 and any regions larger than 100 bp that did not map to the first genome were appended to the end of it to produce a pangenome representing the unique regions in the first two genomes. Each subsequent genome aligned to the combination of all unique regions from the previously aligned genomes in the list producing a pangenome that represents all of the nucleotide sequences of all genomes. All genes were organised into groups of orthologues using the bi-directional best hits algorithm in Get_homologues with a minimum coverage setting of 50% and minimum sequence identity setting of 80%53. The pangenome was used as the reference and the coding sequences (CDS) predicted in the annotations described previously were compared to all CDS within the pangenome. Features annotated as pseudogenes were excluded from this analysis. The Get_homologues compare_clusters perl script was used to create a pangenome matrix of all identified gene clusters against all genomes. All core gene clusters (clusters that contain genes from every genome) were removed from the pangenome matrix. Further to this all clusters that only contained genes from one genome or all genomes except one were removed. Furthermore, gene clusters that were found in all members of any STs associated with multiple host species were removed on the basis that they are not specific to a single host species. This has the effect of removing lineage-associated genes resulting in a set of gene clusters that are strain-dependent and largely independent of phylogeny. Hypergeometric testing was used to find over- and under- represented gene clusters for each host (the p-value was adjusted using Bonferroni correction). All gene clusters were searched against the NCBI non-redundant nucleotide database using blastn to provide the most up to date annotation and to examine the likely bacterial species origin of each MGE.
A pairwise distance matrix was calculated from the pangenome matrix using the distmat function in EMBOSS54. The matrix was converted into a bidirectional graph with distance as the edge weight parameter. The graph was processed in BioLayout with an edge weight threshold of 0.555.
Identification of gene acquisitions or losses associated with host-switching events
The R package APE (Analysis of Phylogenetics and Evolution)56 was used to fit a single discrete trait model and get the ancestral state of each node for each gene cluster against the phylogenetic tree. From this, a vector for every gene cluster was created with gene acquisition/loss events by comparing every child node in the tree to its parent node to determine if there was no change, a gene acquisition or a gene loss event. This was performed separately for each host type (i.e. human, ruminant, bird, horse and pig) to identify which nodes are associated with a host-switching event. All gene state vectors were compared to all host state vectors using a Fisher’s exact test to show whether a gene loss/acquisition even is related to a host switch event. The p-values were adjusted using Bonferroni correction.
Codon usage bias analysis
The codon adaptation index is used to calculate codon usage bias by comparing the CAI of a gene against the codon usage table of a reference set of genes. The codon usage table was calculated using the EMBOSS tool cusp54. For this study, the codon usage table was comprised of all genes that were not significantly over represented in a host or significantly associated with a host switch. The codon adaptation index (CAI) for all genes significantly over represented in a host and significantly associated with a host jump was calculated using the EMBOSS tool cai54. The codon adaptation index was also calculated using five random subsets of 50 genes as controls. A one-way ANOVA test was used to test whether there was a significant effect of host upon CAI. A Tukey HSD test was then applied to compare the CAIs between host species.
Distribution of antibiotic resistance genes analysis
Antimicrobial resistance genes were identified as described by Holden et al15. Resistance genes were identified by a combination of BlastN and mapping against assemblies and as previously described 57 and resistance SNPs identified by mapping against a pseudomolecule of genes with previously reported resistance-conferring mutations. Isolates were grouped into human isolates and all animals and them human, rabbits, companion animals (horses, dogs, cats), marine, pigs, primates, ruminants (goats, sheep, cows) and small mammals (rats, mice, other small mammals). The proportions of isolates with each resistance gene and ≥1 resistance conferring SNP for each antibiotic was compared to identify enrichment using a two-tailed Fishers Exact test with a Bonferroni correction for multiple testing. Determinates with a P value <9.9 x 10-5 were considered statistically significantly enriched. To examine whether the Fisher Exact tests of independence were robust when accounting for population structure, we tested whether resistance phenotypes and host were correlated across the S. aureus phylogeny. We conducted these for pig/human and ruminant/human since these were the only comparisons where significant differences were observed according to the Fisher’s exact test. In order to maximise statistical power, we grouped all gene determinants into specific classes of antimicrobial (e.g. Tetracycline-resistant if encoding any tet allele) and tested for correlation with host species using the program BayesTraits58 (using the posterior sample of trees from our earlier BEAST analysis). We note that the correlated evolutionary analysis may be overly conservative in cases where horizontal gene transfer is rampant or homoplasies are high. BayesTraits uses a continuous-time Markov model to estimate transition rates between the presence and absence of a gene or SNP and between human and non-human hosts. We allowed the transition rates to evolve in either a correlated fashion (where the rate of change in one trait depends on the state found in the other trait) or independently. Posterior distributions of parameters were estimated from up to 4 million iterations of the MCMC with default priors. After discarding burn-in, the marginal likelihoods of the dependent and independent models were obtained using the Akaike Information Criterion (AICM) estimated using the methods-of-moment estimator in Tracer 1.659
Genome-wide positive selection analysis
To identify genes under positive selection in different host groups, we first identified lineages (STs or CCs) correlated with particular hosts. As the power of the selection analysis is determined by the number of isolates included, only clades with more than 10 isolates associated with a host were considered. Based on these criteria, 15 CCs from four groups of hosts were analyzed: 9 for humans (CC30, CC5, CC59, CC15, CC12, ST239, ST8, CC22 and CC45), 3 for ruminants (CC133, primarily associated with sheep and goats and the cows related CC151 and CC97), 2 for birds (CC5 and CC385) and one for pigs (CC398) (Supplementary Table 7). Although the CC398 clade also contained several human isolates, these mostly represent spill-over events rather than an established association so the CC398-human group was not included for the analysis. Given the variable number of isolates of each CC-host group, in order to standardize the analysis while preventing the underestimation of genes under positive selection, 10 isolates linked with a host were analyzed at a time. Replicates or triplicates of different subsets of genomes using sampling with replacement was carried out if the number of isolates for that lineage was large enough. Next, we identified orthologous genes in each of these groups using the algorithm OrthoMCL integrated in get_homologues (identity >70%, similarity >75%, f50, e-value = 1e-5)53. Genes were considered orthologous if they were present in at least 70% of the genomes. Since alignment of coding DNA sequences may insert gaps in codons and produce frame-shifts, we aligned genes at the protein level using MUSCLE 3.8.3160 and translated these sequences back to DNA using pal2nal v1461. Genes identified as inparalogous that turned out to be duplications were kept for further analyses, otherwise discarded. For every alignment, recombination was detected using the NSS, Max Chi and Phi tests included in PhiPack62 and recombinant genes removed from further analyses. For the gene clusters containing 10 isolates, phylogenetic trees were extracted from the 783 isolates ML tree. For clusters with less than 10 genomes, subtrees were produced from the general tree using the tree prune function in ete263. The DNA alignments and trees were used for PAML analysis64. We employed the site evolution models of Codeml (M1a, M2a, M7, M8 and M8a) to perform codon-by-codon analysis of dN/dS ratios (nonsynonymous to synonymous substitution, ω) of genes and a likelihood ratio test (LRT) was used to determine significant differences between nested models M1a-M2a, M7-M8, M8a-M8, where one accounts for positive selection (alternative hypothesis) and the other specifies a neutral model (null hypothesis). Statistic tests were assessed to a chi-square distribution with 2 and 1 degrees of freedom64. Bayes Empirical Bayes65 was used to calculate the posterior probabilities of amino acid sites under positive selection of proteins that had significant LRTs. As independent replicates from similar CC/Host groups resulted in slightly different genes positively selected, we used get_homologues to merge the core genomes and genes selected for each group using same parameters as above. Genes under positive selection were considered when they were in common for different replicates with a p-value of 0.05 or were identified in different replicates with a stringent p-value (0.05/number of genes per core genome).
To explore functional categories under positive selection we performed classification of Clusters of Orthologous Groups (COGs), annotated Gene Ontology terms (GO) and analysed metabolic pathways (KEGG). To assign COG terms, we performed BLASTp of single representatives of the orthologous clusters against the prot2003-2014 database, retrieving the top 5 hits to include alternative annotations. We mapped the gene IDS obtained to the cog2003-2014.csv database from which the COGs were inferred. Frequencies of COGs for positively selected genes in each CC-host were compared with the average COG frequencies in the respective core genomes. GO annotations were obtained by mapping the genes to the go_20151121-seqdb, uniprot_sprot and uniprot_trembl databases using BLASTp. From these, the UniProtKB were mapped to the gene_association_goa database and filtered by bacteria domain to obtain the GO categories. To visualize and identify overrepresented GO categories of positively selected genes in different hosts, we used BiNGO66. We identified overrepresented categories using the hypergeometric test with the Benjamini & Hochberg False Discovery Rate (FDR) multiple testing correction at a significance level of 5%. We chose the ‘Biological Process’ category and the prokaryotic ontology file (gosubset_prok.obo). However, as most groups did not show significant overrepresentation, we visualized all the GO categories of genes under positive selection and used REVIGO67 with the p-values from BiNGO in order to obtain summaries of non-redundant GO terms classified into functional categories.
Analysis of lactose fermentation
S. aureus was cultured in Tryptic Soy Broth (TSB) in presence or absence of 100 mM lactose at 37°C for 17 h with shaking at 200 rpm. OD600 was measured and culture supernatants were collected by centrifugation. Subsequently, the pH of the supernatants was measured using a pH meter (Sartorius, UK). Delta pH values were calculated by subtracting the pH values of TSB cultures supplemented with 100 mM lactose from the pH values of normal TSB cultures. Statistical analysis was performed in Graphpad Prism 7 using One-Way ANOVA followed by Tukey’s multiple comparison test.
Supplementary Material
Acknowledgments
The study was supported by a project grant (BB/K00638X/1) and institute strategic grant funding ISP2: BB/P013740/1 from the Biotechnology and Biological Sciences Research Council (UK) to J.R.F, Medical Research Council (UK) grant MRNO2995X/1 to J.R.F. and Wellcome Trust collaborative award 201531/Z/16/Z to J.R.F. S.Y.C.T. is an Australian National Health and Medical Research Council Career Development Fellow (#1065736). L.A.W is supported by a Dorothy Hodgkin Fellowship funded by the Royal Society (Grant Number DH140195) and a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 109385/Z/15/Z). S.L. is supported by a Chancellor’s Fellowship from the University of Edinburgh. M.T.G.H was supported by the Scottish Infection Research Network and Chief Scientist Office through the Scottish Healthcare Associated Infection Prevention Institute consortium funding (CSO Reference: SIRN10). E.M.H. and S.J.P were funded by The Health Innovation Challenge Fund (WT098600, HICF-T5-342), a parallel funding partnership between the Department of Health and Wellcome Trust, and the UKCRC Translational Infection Research Initiative, and the Medical Research Council (Grant Number G1000803). S.J.P. is a National Institute for Health Research Senior Investigator. M.T.G.H was supported by the Scottish Infection Research Network and Chief Scientist Office through the Scottish Healthcare Associated Infection Prevention Institute consortium funding (CSO Reference: SIRN10) M.T.G.H was supported by the Scottish Infection Research Network and Chief Scientist Office through the Scottish Healthcare Associated Infection Prevention Institute consortium funding (CSO Reference: SIRN10). P.A.H. is supported by Natural Environment Research Council for Grant NE/M001415/1. We thank Beth Blane, Nick Brown and Estee Torok for their role in the original study that isolated and sequenced S. aureus from patients at the Cambridge University Hospitals NHS Foundation Trust 68, from which 76 genomes were downloaded from the ENA and used in this study. We also thank Edinburgh Genomics for sequencing, and all those who made isolates available for the study including Zoological Society London, G. Foster, H. Hasman, S. Monecke, E. Smith, D. Smyth, and H. Jorgensen.
Footnotes
Data availability. The sequence datasets generated during the current study are available in the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena) with the accession number PRJEB20741. Accession numbers of previously published sequences analyzed in the current study are listed in Supplementary Table 1. All data analysed during this study are included in this published article (and its supplementary information files).
Author contributions
J.R.F. S.J.P, J.P, M.H., E.M.H, L.A.W., and M.T.G.H., conceived and designed the study. E.J.R., R.B., E.M.H., L.A.W., S.L, M.V. and K.R. carried out experiments. E.J.R., R.B., E.M.H., L.A.W., S.L., G.K.P, D.M.A., M.T.H., E.J.F., J.C., M.V., P.A.H., K.R. and J.R.F analysed data. S.Y.C.T., A.S., and W.vW. provided isolates. E.J.R., R.B., E.M.H., S.L. and J.R.F. wrote the manuscript. All authors contributed to manuscript editing.
Competing Interests
The authors declare no competing interests.
References
- 1.Morand S, McIntyre KM, Baylis M. Domesticated animals and human infectious diseases of zoonotic origins: domestication time matters. Infect Genet Evol. 2014;24:76–81. doi: 10.1016/j.meegid.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Woolhouse ME, Haydon DT, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20:238–44. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowy FD. Staphylococcus aureus Infections. New England Journal of Medicine. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 4.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peton V, Le Loir Y. Staphylococcus aureus in veterinary medicine. Infect Genet Evol. 2014;21:602–15. doi: 10.1016/j.meegid.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Bradley AJ, Leach KA, Breen JE, Green LE, Green MJ. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet Rec. 2007;160:253–7. doi: 10.1136/vr.160.8.253. [DOI] [PubMed] [Google Scholar]
- 7.McNamee PT, Smyth JA. Bacterial chondronecrosis with osteomyelitis ('femoral head necrosis') of broiler chickens: a review. Avian Pathol. 2000;29:477–95. doi: 10.1080/030794500750047243. [DOI] [PubMed] [Google Scholar]
- 8.van Duijkeren E, et al. Methicillin-resistant Staphylococcus aureus in pigs with exudative epidermitis. Emerg Infect Dis. 2007;13:1408–10. doi: 10.3201/eid1309.061268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viana D, et al. A single natural nucleotide mutation alters bacterial pathogen host tropism. Nat Genet. 2015;47:361–6. doi: 10.1038/ng.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil EJ, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shepheard MA, et al. Historical zoonoses and other changes in host tropism of Staphylococcus aureus, identified by phylogenetic analysis of a population dataset. PLoS One. 2013;8:e62369. [Google Scholar]
- 12.Weinert LA, et al. Molecular dating of human-to-bovid host jumps by Staphylococcus aureus reveals an association with the spread of domestication. Biol Lett. 2012;8:829–32. doi: 10.1098/rsbl.2012.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price LB, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio. 2012;3 doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald JR. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 2012;20:192–8. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Holden MT, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23:653–64. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAdam PR, et al. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2012;109:9107–12. doi: 10.1073/pnas.1202869109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spoor LE, et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. MBio. 2013;4 doi: 10.1128/mBio.00356-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowder BV, et al. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:19545–50. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viana D, et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol Microbiol. 2010;77:1583–94. doi: 10.1111/j.1365-2958.2010.07312.x. [DOI] [PubMed] [Google Scholar]
- 20.Guinane CM, et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol Evol. 2010;2:454–66. doi: 10.1093/gbe/evq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koymans KJ, Vrieling M, Gorham RD, Jr, van Strijp JA. Staphylococcal Immune Evasion Proteins: Structure, Function, and Host Adaptation. Curr Top Microbiol Immunol. 2016 doi: 10.1007/82_2015_5017. [DOI] [PubMed] [Google Scholar]
- 22.Koop G, et al. Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci Rep. 2017;7:40660. doi: 10.1038/srep40660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loffler B, et al. Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010;6:e1000715. doi: 10.1371/journal.ppat.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vrieling M, et al. LukMF' is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci Rep. 2016;6:37759. doi: 10.1038/srep37759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jong NWM, et al. Identification of a staphylococcal complement inhibitor with broad host specificity in equid Staphylococcus aureus strains. J Biol Chem. 2018;293:4468–4477. doi: 10.1074/jbc.RA117.000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson GJ, et al. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 2011;7:e1002271. doi: 10.1371/journal.ppat.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong SY, et al. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol. 2015;65:15–22. doi: 10.1099/ijs.0.062752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaipadungpanit J, et al. Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. J Clin Microbiol. 2015;53:1005–8. doi: 10.1128/JCM.03049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aanensen DM, et al. Whole-Genome Sequencing for Routine Pathogen Surveillance in Public Health: a Population Snapshot of Invasive Staphylococcus aureus in Europe. MBio. 2016;7 doi: 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minin VN, Suchard MA. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol. 2008;56:391–412. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- 31.De Maio N, Wu CH, O'Reilly KM, Wilson D. New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation. PLoS Genet. 2015;11:e1005421. doi: 10.1371/journal.pgen.1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheppard SK, et al. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol. 2014;23:2442–51. doi: 10.1111/mec.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deringer JR, Ely RJ, Monday SR, Stauffacher CV, Bohach GA. Vbeta-dependent stimulation of bovine and human T cells by host-specific staphylococcal enterotoxins. Infect Immun. 1997;65:4048–54. doi: 10.1128/iai.65.10.4048-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howden BP, Peleg AY, Stinear TP. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol. 2014;21:575–82. doi: 10.1016/j.meegid.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 35.Directorate, P.H.E.a.V.M. UK One Health report: antibiotics use in humans and animals. 2015 [Google Scholar]
- 36.Ward MJ, et al. Time-scaled evolutionary analysis of the transmission and antibiotic resistance dynamics of Staphylococcus aureus CC398. Appl Environ Microbiol. 2014 doi: 10.1128/AEM.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray S, et al. Recombination-Mediated Host Adaptation by Avian Staphylococcus aureus. Genome Biol Evol. 2017;9:830–842. doi: 10.1093/gbe/evx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward MJ, et al. Identification of source and sink populations for the emergence and global spread of the East-Asia clone of community-associated MRSA. Genome Biol. 2016;17:160. doi: 10.1186/s13059-016-1022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Argimon S, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1105s31. Chapter 11, Unit 11 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–9. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 43.Nadalin F, Vezzi F, Policriti A. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics. 2012;13(Suppl 14):S8. doi: 10.1186/1471-2105-13-S14-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayres DL, et al. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol. 2012;61:170–3. doi: 10.1093/sysbio/syr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aibar S, Fontanillo C, Droste C, De Las Rivas J. Functional Gene Networks: R/Bioc package to generate and analyse gene networks derived from functional enrichment and clustering. Bioinformatics. 2015;31:1686–8. doi: 10.1093/bioinformatics/btu864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–83. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 55.Wright DW, Angus T, Enright AJ, Freeman TC. Visualisation of BioPAX Networks using BioLayout Express (3D) F1000Res. 2014;3:246. doi: 10.12688/f1000research.5499.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paradis E. Analysis of diversification: combining phylogenetic and taxonomic data. Proc Biol Sci. 2003;270:2499–505. doi: 10.1098/rspb.2003.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David S, et al. Evaluation of an Optimal Epidemiological Typing Scheme for Legionella pneumophila with Whole-Genome Sequence Data Using Validation Guidelines. J Clin Microbiol. 2016;54:2135–48. doi: 10.1128/JCM.00432-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barker D, Meade A, Pagel M. Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics. 2007;23:14–20. doi: 10.1093/bioinformatics/btl558. [DOI] [PubMed] [Google Scholar]
- 59.Rambaut ASM, S M, Xie D, Drummond AJ. Tracer v 1.6. 2014 [Google Scholar]
- 60.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–12. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruen T, Bruen T. PhiPack: PHI test and other tests of recombination. McGill University; Montreal, Quebec: 2005. [Google Scholar]
- 63.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol Biol Evol. 2016;33:1635–8. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–9. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 66.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–9. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 67.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reuter S, et al. Building a genomic framework for prospective MRSA surveillance in the United Kingdom and the Republic of Ireland. Genome Res. 2015 doi: 10.1101/gr.196709.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.