Abstract
The serine hydrolase (SH) superfamily is perhaps, one of the largest functional enzyme classes in all forms of life, and consists of proteases, peptidases, lipases, and carboxylesterases as representative members. Consistent with the name of this superfamily, all members, without any exception to date, use a nucleophilic serine residue in the enzyme active site to perform hydrolytic-type reactions via a two-step ping-pong mechanism involving a covalent enzyme intermediate. Given the highly conserved catalytic mechanism, this superfamily has served as a classical prototype in the development of several platforms of the chemical proteomics technique, activity-based protein profiling (ABPP), to globally interrogate the functions of its different members in various native, yet complex, biological settings. While ABPP-based proteome-wide activity atlas’ for SH activities are available in numerous organisms, including humans, to the best of our knowledge, such an analysis for this superfamily is lacking in any insect model. To address this, here, we initially report a bioinformatics analysis towards the identification and categorization of non-redundant SHs in Drosophila melanogaster. Following up on this in silico analysis, leveraging discovery chemoproteomics, we identify and globally map the full complement of SH activities during various developmental stages and in different adult tissues of Drosophila. Finally, as proof of concept of the utility of this activity atlas, we highlight sexual dimorphism in SH activities across different tissues in adult Drosophila melanogaster, and together, we prospect new research directions, resources and tools that this study can provide to the fly community.
Introduction
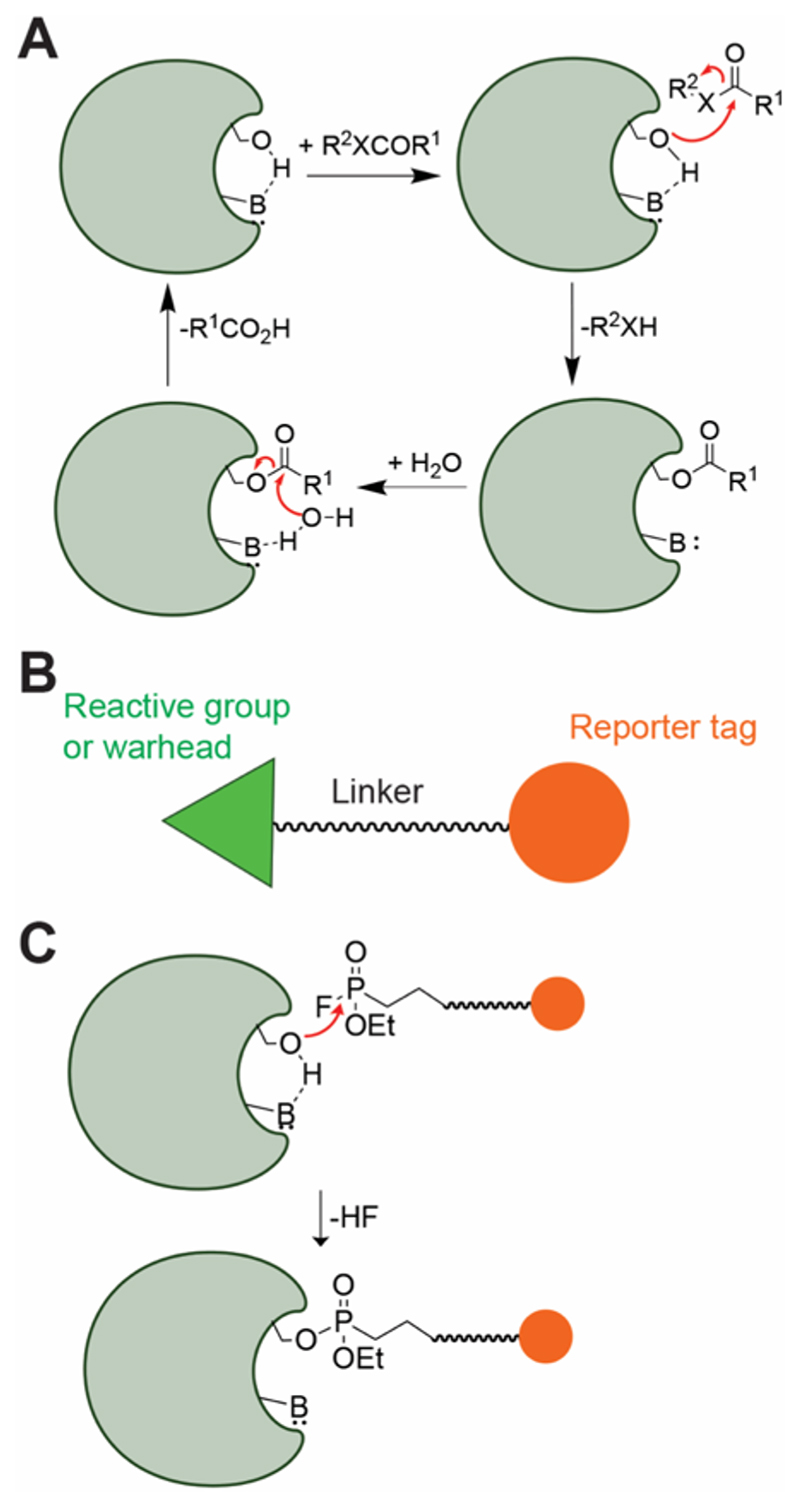
The serine hydrolase (SH) superfamily consists of enzymes that catalyze physiologically important hydrolytic reactions at electron deficient carbon (or phosphorus) centers at amide, ester or thioester functionalities, and consists of proteases, peptidases, esterases, lipases, amidases, and transacylases as prototypical members1–4. This enzyme superfamily is known to be present in both prokaryotes and eukaryotes, and constitutes 1 – 3 % of the total cellular proteome, thus making them, one of the largest functional enzyme classes across all forms of life. All members of this superfamily, without any exceptions known to date, contain a catalytically competent, conserved nucleophilic serine residue in the active site to perform the aforementioned hydrolytic-type reactions via a consensus two-step ping pong mechanism involving a covalent enzyme intermediate (Figure 1A), thus, leading to the “serine hydrolase” moniker for this superfamily1–4. This invariant catalytic serine residue is part of the famous catalytic triad3, 4 (which is the hallmark of this superfamily), and is activated by an active site base (generally a histidine residue) to initiate the enzyme catalytic cycle (Figure 1A). Typically, this catalytic serine residue is part of a canonical GxSxG motif (x = any amino acid), although, exceptions to this motif have been found in all organisms1, 2. From a biological standpoint, deregulation in the activities of different SH enzymes has been linked to detrimental pathophysiological consequences in several organisms, and is often associated with various metabolic and/or autoimmune diseases in humans1.
Figure 1. The SHs and ABPP.
(A) The consensus catalytic mechanism of the enzymes from the SH superfamily. (B) Generic structure of an activity-based probe, having three components, i.e. the reactive group or warhead, a linker and a reporter tag. (C) Reaction of a SH enzyme with a fluorophosphonate (FP) activity probe, leading to the formation of an irreversible covalent adduct, thus showing the utility of FP-probes towards ABPP applications.
Given the conservation in their catalytic ping-pong mechanism, particularly the formation of the covalent enzyme intermediate, the SH enzyme superfamily has served as a classical prototype for the development and advancement of the chemical proteomics (or chemoproteomics) technique termed “activity-based protein profiling” (ABPP), pioneered by Cravatt and co-workers5–9. Briefly, ABPP is a functional proteomics technique that utilizes a biorthogonal “activity” probe with a reactive chemical group (often referred to as a “reactive warhead”) (Figure 1B), which reacts with enzymes (or even proteins) that are mechanistically (or functionally) related, typically from the same enzyme superfamily, and allows for their detection, enrichment and quantitative identification via a reporter tag at a proteome wide scale from complex biological samples like cells or tissues5–9. The fluorophosphonate (FP) moiety in particular, has served as an excellent reactive chemical group (reactive warhead) for the SHs, and has been an integral part of most ABPP probes used for profiling enzymes of this superfamily5–9 (Figure 1C).
ABPP has two popular platforms for assessing superfamily wide enzyme activities: (i) an ingel fluorescence readout using SDS-PAGE analysis, also known as the “gel-based ABPP” platform, which is a medium throughout qualitative (or semi-quantitative) method, used for studying previously known enzymes and their activities in more physiological settings5–9; and (ii) a liquid chromatography coupled to an advanced mass spectrometry (LC-MS/MS) readout, popularly known as “ABPP-MudPIT” (MudPIT = Multidimensional Protein Identification Technology), which is relatively low throughput, but an exhaustive discovery based and/or quantitative method used towards identification and quantification of previously unknown enzyme activities from complex biological settings6, 7, 10. Traditionally, the activity probes used for gel-based ABPP experiments have various fluorophores (like fluorescein, rhodamines, BODIPYs, and their variants) as reporter tags, while biotin (leveraging avidinbased enrichment) has almost exclusively been used as a reporter tag for the activity probes used in the corresponding LC-MS/MS ABPP experiments.
Given their physiological importance, the enzymes from the SH family have been extensively investigated across all forms of life, and leveraging complementary ABPP platforms, superfamily-wide activity maps for this superfamily are available for numerous biological systems (e.g. cell lines, disease tissues, cancers)11–14 and/or model organisms (e.g. rodents, worms, pathogens)15–18. Surprisingly, to the best of our knowledge, such an organismal level superfamily-wide activity map for the SHs (or any enzyme family for that matter) has not been described in literature for Drosophila melanogaster (fruit fly), a popular model organism used extensively for mechanistic discovery using its powerful genetic toolkit19–23. Not surprisingly, our current understanding of SH functions in Drosophila is mostly based on genetic experiments that have uncovered critical roles of SHs, particularly the serine proteases24, to be important in physiological processes related to immune responses25 and embryonic development26 and patterning27, 28.
Therefore, in this paper, we report the first chemoproteomics aided superfamily-wide activity atlas for the SH family in the well-studied fly model, Drosophila melanogaster. Towards achieving this, we initially performed an exhaustive bioinformatics analysis to survey, identify, collate and categorize all known non-redundant SHs from the Drosophila melanogaster genome. Next, leveraging established complementary gel-based and LC-MS/MS-based ABPP platforms, we assessed the superfamily-wide activity status of SHs across the various stages during its developmental life cycle (embryo, larva, pupa, and adult) and also in different tissues of adult flies. Our studies provide interesting new insights into the differential activities of SHs in various developmental life forms in flies, and also suggest sexual dimorphism in their activities. Given the immense value of chemoproteomics, and its poor utilization in fly models, we believe that the data and tools reported here, will be an invaluable resource to the fly community, and will provide new research directions towards connecting enzyme activities (particularly the SHs), to genetic phenotypes in different physiological processes and settings.
Materials and Methods
Materials
Unless mentioned otherwise all reagents and chemicals were purchased from Sigma-Aldrich. FP-rhodamine was a kind gift from Prof. Benjamin F. Cravatt (Scripps Research), and FP-biotin was synthesized in house as per reported protocol5.
Generation and Phylogenetic Characterization of a SH database for Drosophila melanogaster
Genes encoding proteins predicted to be SHs were filtered and collated from four publicly available databases namely: (i) FLYBASE29–35, (ii) NCBI database (Release 6 plus ISO1 MT, GCF_000001215.4, last modified: 2020-04-24) (SH_motif search), (iii) MEROPS36–39, and (iv) HMMER40–42. Within the HMMER database, the filtered fasta sequence of each SH was further analyzed for a Pfam motif using HmmerWeb version 2.41.1. The command for this analysis was ‘hmmscan --cut_ga --hmmdb pfam’ against the Pfam version 32.0 database. The Pfam protein domain, catalytic residue order and Pfam family identity number were retrieved, and the SHs having the same protein motif and catalytic order were grouped together. The SH genes whose catalytic residue were not predicted by the aforementioned HMMER searches, were retrieved using multiple sequence alignments from sequences collated from the other three databases. Proteins with an active SH motif i.e. GxSxG or GxSxxG (where x is any amino acid) were filtered using a Python code on a local computer. The final enzyme list from all the published databases was manually curated to ensure they fit with the SH superfamily, and split into two categories: serine proteases or metabolic SHs, based on sequence alignments to known enzyme activities within this superfamily. Towards generating a phylogenetic tree for the serine proteases and metabolic SHs, multiple sequence alignments for enzymes of each category were performed using the MUSCLE program43, 44 (MEGA-X) with default software defined parameters. The phylogenetic tree was generated by the MEGA-X software using the “Neighbor-Joining” method45, 46, and visualized using the FigTree (version 1.4.4) software.
Fly Experimentation
The Drosophila melanogaster line, w1118, was grown on standard cornmeal agar medium at 25 °C. The embryos (0 – 20 hours) were collected on standard sucrose-agar plate (3% w/v sucrose, 2.5% w/v agar) supplemented with fresh yeast paste. All life stages i.e. the embryo (~ 500 μL per biological replicate), third-instar larva (50 per biological replicate), pupa (50 per biological replicate), 5 – 10 days old adult flies (50 per biological replicate) were eventually lysed in cold 1x Phosphate buffered-saline (PBS), pH 7.4. For head, gut, carcass and hemolymph, adult flies, both males and females were used separately. Flies were beheaded by shaking adults in liquid nitrogen, and heads were subsequently collected using a 600-μ sieve. Gut, carcass, ovary and testis were dissected in ice-cold 1X PBS, pH 7.4, in a batch of 50 flies and then flash-frozen in the liquid nitrogen. For hemolymph extraction, 40 flies were punctured in the thorax using an insulin needle, and collected in a 0.5 mL microcentrifuge tube with a hole at the bottom. Thereafter, the 0.5 mL tube containing the pricked flies was placed in a 1.5 mL microcentrifuge tube, and centrifuged at 4 °C, 5000g for 5 minutes to cause the release of hemolymph into the 1.5 mL microcentrifuge tube. After harvesting the necessary biological samples, they were flash-frozen and stored at -80 °C until further use.
Gel-based ABPP Sample Preparation and Analysis
All the gel-based ABPP experiments were done using established protocols previously reported by us47–49. Briefly, each sample [fly life stages (embryo, larva, pupa or adult), or tissue (head, gut) that was fractionated] was suspended in 500 μL of cold, sterile 1x PBS (pH 7.4) and homogenized using a tissue homogenizer (Bullet Blender24, Next Advance) with one scoop of glass beads (0.5-mm diameter; Next Advance) at a speed setting of 10 for 5 min at 4 °C. Thereafter, an additional 500 μL of cold sterile 1x PBS was added, mixed by pipetting, and centrifuged at 1000g for 5 min at 4 °C to separate the tissue debris. The resulting supernatant (700 μL) was collected and centrifuged at 21,000g for 90 min at 4 °C. Following this higher speed centrifugation step, the resulting supernatant (termed “supernatant proteome”) and the pellet (termed “pellet proteome”) were collected in different tubes. The pellet was washed 3x with cold sterile 1x PBS and re-suspended in 500 μL of cold sterile 1x PBS by pipetting. For tissues not needing fractionation (carcass, hemolymph, and reproductive organs) in supernatant or pellet proteomes, all steps remained the same, except that the higher speed centrifugation step was excluded, and the resulting lysate was used as whole tissue lysate. The protein concentration of each sample lysate was estimated using BCA protein assay kit (Pierce). For the gel-based ABPP assays, 50 μg of each lysate (50 μL of 1 mg/mL concentration) was treated with FP-rhodamine (2 μM, 60 min, 37 °C, with shaking). The reaction was quenched with 12.5 μL of 5X loading buffer and was followed by boiling the samples (95 °C, 10 min). The fluorescently labeled proteomes were resolved on 12% SDS–PAGE gel, and visualized using a Syngene G-Box Chemi-XRQ gel documentation system.
LC-MS/MS based ABPP Sample Preparation and Analysis
For the proteomic preparations for the LC-MS/MS-based ABPP assays, 1 mg of each lysate (1 mg/mL in 1x PBS (pH = 7.4)) were labeled with FP-biotin (200 μM, 60 min, 37 °C, with shaking). After labeling, the proteomes were denatured, reductively alkylated using iodoacetamide, and subsequently digested with trypsin as described earlier using a protocol previously reported by us48, 50. The tryptic peptides were desalted and cleaned using the StageTip protocol51. All LC–MS/MS analysis was performed on a Sciex TripleTOF6600 mass spectrometer interfaced with an Eksigent nano-LC 425. Tryptic peptides (~ 1 μg) were loaded onto an Eksigent C18 trap column (5 μg capacity) and subsequently eluted on an Eksigent C18 analytical column (15 cm × 75-μm internal diameter) with a linear acetonitrile gradient. A typical LC run consisted of 360 min post-loading onto the trap at a constant flow rate of 300 nL/min with solvent A consisting of water + 0.1% formic acid and solvent B consisting of acetonitrile. The gradient schedule for the LC run was 5% (v/v) B for 1 min, a linear gradient of B from 0% to 30% (v/v) over 330 min, 90% (v/v) B for 20 min and equilibration with 5% (v/v) B for 10 min. For all proteomics samples, data were acquired in information-dependent acquisition (IDA) mode over a mass range of 200–2,000 m/z. Each full MS survey scan was followed by MS/MS of the 15 most intense peptides. Dynamic exclusion was enabled for all experiments (repeat count 2; exclusion duration 6 s). Peptide identification52, 53 were carried out using the Protein Pilot (version 2.0.1, Sciex) using Pro-Group™ and Paragon™ algorithms against the RefSeq protein database of the Drosophila melanogastar (Release 6 plus ISO1 MT, GCF_000001215.4, last modified: 2020-04-24). While searching the peptides, iodoacetamide alkylation of cysteine was defined as a static modification, while the oxidation of methionine and N-terminal acetylation were defined as variable modifications. During the peptide searches, the precursor ion and MS/MS mass tolerance were set at 20 and 50 ppm respectively. The Protein Pilot software52–54 defined “Peptide Count” parameter was used for quantifying the extent of activity of a SH enzyme, and only those SHs that were identified in at least two biological replicates were considered for further analysis with Peptide Counts ≥ 1. Precursor ions and MS/MS fragmentations were viewed using the PeakView software (Sciex).
Proteomics Data
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium55 via the PRIDE56 partner repository with the dataset identifier PXD024321.
Data Analysis
All heat map plots presented in this study (Figure 3 & 5) represent mean of three biological replicates, and were made using the Prism 9 (version 9.0.2) for macOS (GraphPad) software.
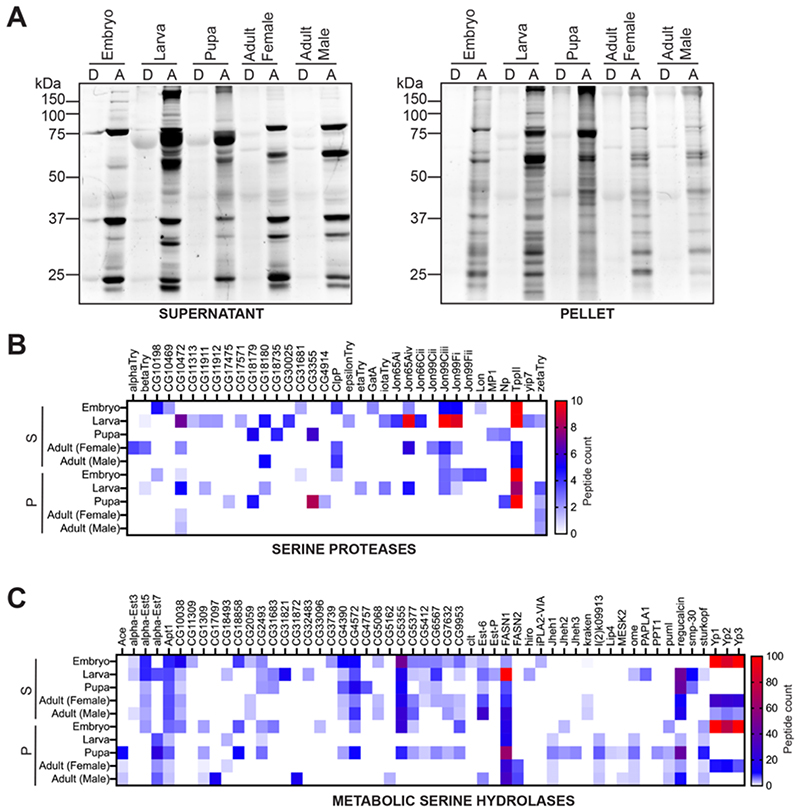
Figure 3. ABPP analysis on lysates from different stages of fly development.
(A) Gelbased ABPP analysis on supernatant and pellet proteomes from different developmental stages of Drosophila labeled with the SH-directed FP-rhodamine probe, and analyzed by SDS-PAGE analysis and in-gel fluorescence scanning. This gel-based ABPP experiment clearly shows that active (A), but not denatured (D) proteomes, have distinct SH activity profiles during different stages of Drosophila development. This gel-based ABPP experiment was done thrice with reproducible results each time. (B, C) Heat map plots for (B) Serine Proteases, and (C) Metabolic Serine Hydrolases, showing SH activity signals identified from a LC-MS/MS based ABPP experiment using the SH-directed FP-biotin probe during various stages of Drosophila development. Data in the heat maps are represented as average peptide counts from three independent biological replicates, and peptide counts above 10 & 100 for the serine proteases and metabolic SHs respectively are colored red. For (B) and (C), S = supernatant proteomic fraction, P = pellet proteomic fraction.
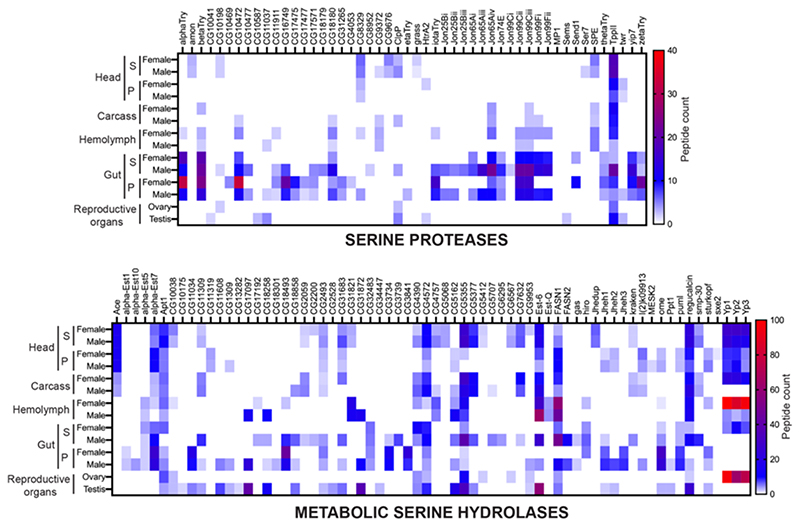
Figure 5. Determining the full complement of active SHs in adult flies.
Heat map plots for the ‘Serine Proteases’, and ‘Metabolic Serine Hydrolases’, showing SH activity signals identified from a LC-MS/MS based ABPP experiment using the SH-directed FP-biotin probe from various tissues and/or anatomical components from adult male and female flies. Data in the heat maps are represented as average peptide counts from three independent biological replicates, and peptide counts above 40 & 100 for the serine proteases and metabolic SHs respectively are colored red. For the head and gut analysis, S = supernatant proteomic fraction, P = pellet proteomic fraction.
Results
Bioinformatics analysis towards identification of SHs in Drosophila melanogaster
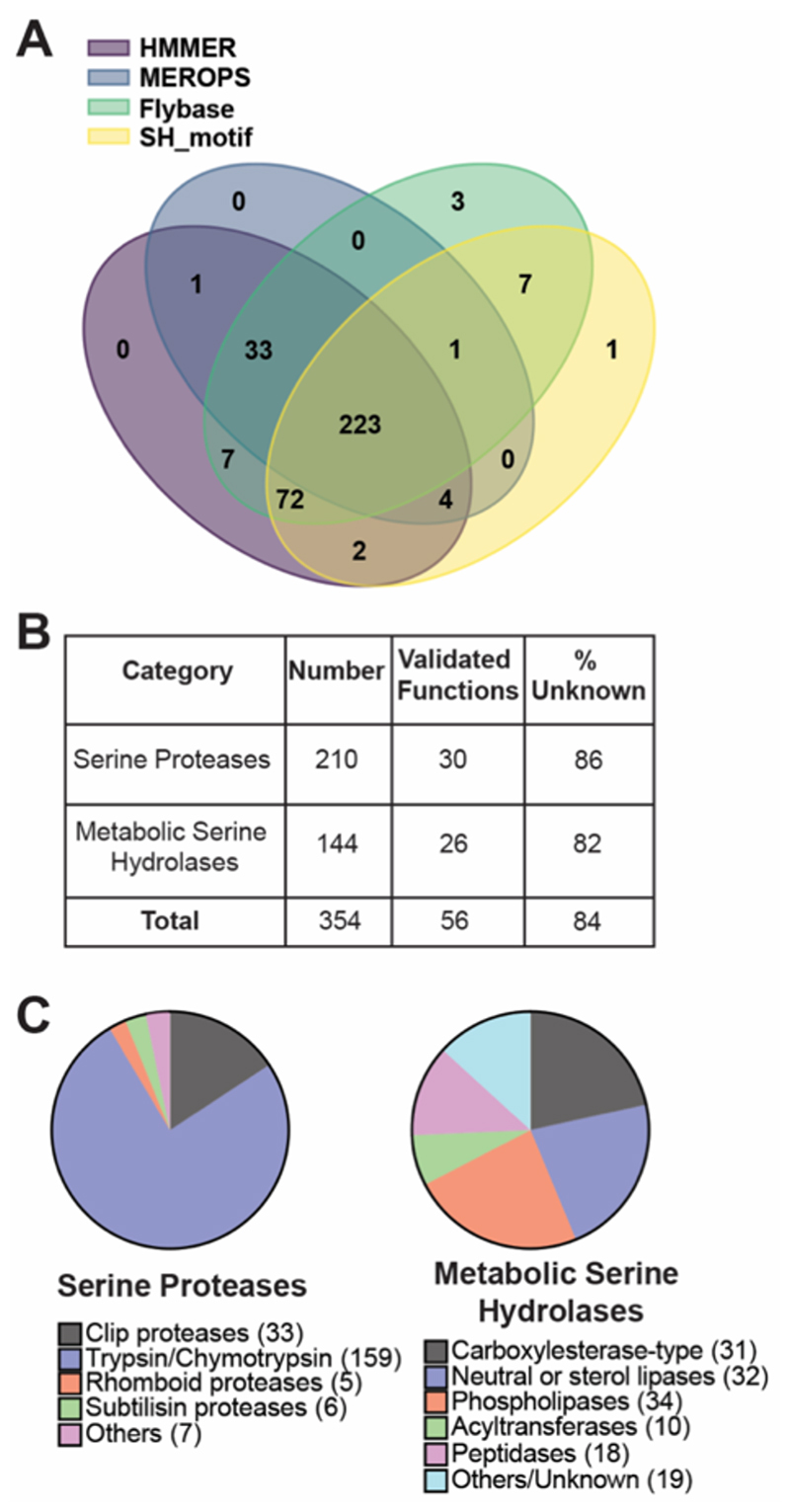
Prior to embarking on the development of a chemoproteomics guided superfamily-wide activity atlas, we first decided to perform a bioinformatics analysis to identify and collate all the protein sequences of SH enzymes in Drosophila melanogaster from publicly available databases, as such an exhaustive analysis for this superfamily, has to the best of our knowledge, not been performed to date. Towards collating this list, we chose 4 publicly available databases: (i) FLYBASE (database for Drosophila genes and genomes)29–32, (ii) NCBI database (Release 6 plus ISO1 MT, GCF_000001215.4, last modified: 2020-04-24) (with a SH_motif search), (iii) MEROPS (peptidase/protease database)36–38, and (iv) HMMER (biosequence search and analysis tool)40–42 (Table S1). We found that the FLYBASE and HMMER databases yielded the most non-redundant SH protein sequences, with 346 and 342 respectively, with a significant overlap (339 common sequences between both databases), while the NCBI and MEROPS databases together provided additional non-redundant SH protein sequences that were not found (or missed while searching) in the FLYBASE or HMMER databases (Figure 2A, Table S1). Together, we identified and collated 354 non-redundant SHs in Drosophila melanogaster, and manually curated them to ensure that they possessed the conserved catalytic serine residue and/or the catalytic triad (Table S1). Our bioinformatics analysis thus shows that the SH enzyme family in Drosophila melanogaster comprises of ~ 2.5 % of the total proteome (354 gene coded serine hydrolase enzymes from a total of 13968 gene coded proteins in Drosophila melanogaster).
Figure 2. Bioinformatics analysis in building a SH database for Drosophila melanogaster.
(A) Venn diagram representing the number of unique SHs identified and the extent of their redundancies from 4 publicly available databases. (B) Categorization of the SHs into ‘Serine Proteases’ and ‘Metabolic Serine Hydrolases’ along with an analysis of known functions within these categories. (C) List of known functions and/or clans with the ‘Serine Proteases’ and ‘Metabolic Serine Hydrolases’ categories.
Consistent with previous classifications of SHs1, we also decided to broadly categorize members of this superfamily as: (i) serine proteases, and (ii) metabolic SHs. Using the inbuilt multiple protein alignment algorithms in HMMER database, and correlating them to known enzyme activities reported in the FLYBASE and Uniprot databases, we found that 210 protein sequences could be categorized as serine proteases, while 144 protein sequences were classified as metabolic SHs (Figure 2B, Table S1). Upon manually searching these protein sequences for known functions in Uniprot, we found that only small number of the serine proteases (30 members), and metabolic SHs (26 members) had been experimentally validated, with functions and/or associations to established biological activities (Figure 2B, Table S1). Our analysis further showed that approximately 84% members of the SH superfamily (86% of serine proteases, and 82% of metabolic SHs) remain uncharacterized in flies (Figure 2B).
Finally, in an attempt to broadly understand the extent of putative enzymatic activities within the two categories of this superfamily in Drosophila melanogaster, we surveyed these 354 protein sequences across the FLYBASE and MEROPS databases in search of possible clans or sub-families. Based on predicted molecular functions for the protein sequences from all the databases, we further sub-classified the 210 serine proteases as putative clip proteases (33 members), trypsin/chymotrypsin like proteases (159 members), rhomboid proteases (5 members), and subtilisin-like proteases (6 members) (Figure 2C, Table S1). We found that 7 serine proteases could not be classified as any of the above, and hence were classified as “Others” within this category (Figure 2C, Table S1). A similar analysis for the metabolic SH category revealed a greater diversity in putative activities and a more even spread across its members (Figure 2C, Table S1). Here, we sub-classified the 144 metabolic SH enzymes as carboxylesterase-like (31 members), neutral or sterol lipase (32 members)57, phospholipases (34 members)57, acyltransferases (10 members) and peptidases (18 members) (Figure 2C, Table S1). Like the serine proteases, we found that 19 members of the metabolic SH category did not have enough sequence homology to the sub-category members listed above, and were therefore grouped together as “Others/Unknown” members (Figure 2C, Table S1).
Identification of SH activities during different developmental stages in Drosophila melanogaster
Having finalized an updated database of SHs in Drosophila melanogaster, we next assessed the superfamily wide activity status of its different members, and chose to profile these in the four major stages of the Drosophila life cycle, i.e. embryo, larva, pupa and adult. For the adult life form, we decided to separately profile male and female flies, to determine whether SHs displayed any sexual dimorphism in their enzymatic activities (described in a later section). Using established protocols47, 48, we fractionated the different fly life forms into supernatant and pellet proteomes, and performed a gel-based ABPP analysis on them to confirm activity-based labeling of SHs using the FP-rhodamine probe (2 μM, 50 μg proteome in 50 μl, 45 mins, 37 °C, with constant shaking). For this gel-based ABPP experiment, we heat denatured the respective proteomes, to inactivate SH activities, and used them as controls for this assay. Consistent with similar studies using FP probes from other systems, we found that both the supernatant and pellet proteomes from the different stages (embryo, larva, pupa and adult) displayed activity-dependent labeling of SHs, as evident in the active lysates, but not in the denatured lysates (Figure 3A). We confirmed the equal loading of respective lysates by Coomassie Blue staining of the gel (Figure S1). Not surprisingly, we found distinct activity patterns for the supernatant and pellet proteomes, suggesting that different SHs are active during different stages of development in Drosophila (Figure 3A).
Having shown activity dependent labeling by the gel-based ABPP, next, we wanted to enrich and identify the full complement of SHs that were active during the various stages of fly development. Towards this, we performed LC-MS/MS based ABPP experiments on the supernatant and pellet proteomes from the different developmental stages using the FP-biotin probe. Here, using an established protocol48, active SHs from proteomes (supernatant or pellet) were labeled using the FP-biotin probe (200 μM, 1 mg proteome in 1 mL, 60 mins, 37 °C, with constant shaking), enriched by avidin-bead based chromatography and digested on bead using trypsin to yield peptides, that were desalted and subjected to LC-MS/MS to identify active SHs. From this LC-MS/MS based ABPP experiment, we identified a total of 89 SH enzymes, of which 35 were serine proteases (Figure 3B, Table S2), while 54 were metabolic SHs (Figure 3C, Table S2). Consistent with previous reports from mammalian systems showing FP-probes being better suited at enriching metabolic SHs1, 2, we found that, of the total SHs enriched and identified (25.1%, 89 out of 354) using the FP-biotin probe in Drosophila melanogaster, there were substantially more metabolic SHs (37.5%, 54 out of 144) (Figure 3C) than serine proteases (16.7%, 35 out of 210) (Figure 3B). Further consistent with similar previous studies in other systems, we found that of the total identified proteins identified in these LC-MS/MS based ABPP experiments, ~ 20 – 25% of them were SHs. Concomitant with the gel-based ABPP experiments, the LC-MS/MS based ABPP experiments also identified differential SH activity profiles, and showed that certain SHs were exclusively present in only one developmental stage, pointing to a critical and possibly indispensable role for that enzyme in that development stage in flies (Figure 3B, 3C).
Anatomical profiling of SH activities in adult Drosophila melanogaster
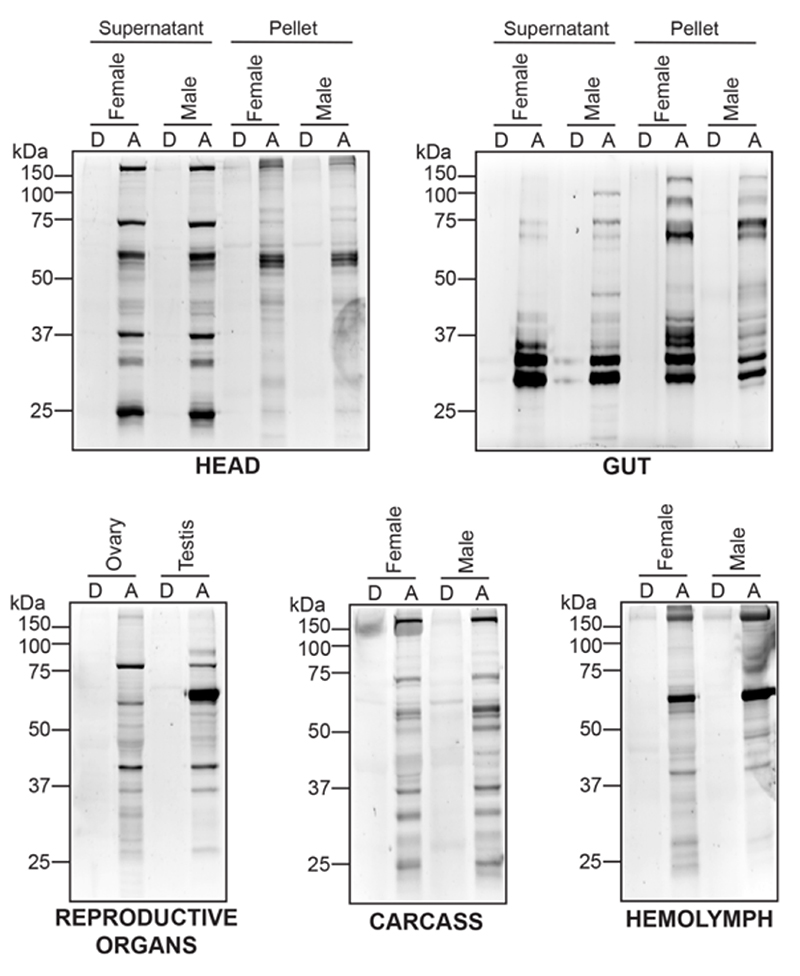
Having identified unique active SHs during the different developmental stages in Drosophila melanogaster, next, we wanted to increase their superfamily wide coverage. Toward this, we decided to isolate different tissues and/or anatomical components in an adult fly (male and female done separately), and perform tandem ABPP experiments to enrich and identify additional active SHs. Here, we isolated the head, gut, reproductive organs (testis for males, and ovaries for females), hemolymph, and the remaining carcass from adult male and female flies separately for this chemoproteomics analysis. Since some tissues (reproductive organs, hemolymph, and the carcass) were difficult to get in sufficient amounts from adult flies, we decided to profile them as whole tissue lysates, while, more abundantly available tissues (head and gut) were fractionated into supernatant and pellet proteomes. First, leveraging established gel-based ABPP protocols47, 48, using the FP-rhodamine probe (2 μM, 50 μg proteome in 50 μl, 45 mins, 37 °C, with constant shaking), we found that all the tissues showed activity-dependent labeling of SHs in the active lysates, but not in the corresponding denatured controls (Figure 4). We confirmed equal loading of respective corresponding active and denatured lysates by Coomassie Blue staining of the gel after in-gel fluorescence imaging for SH activity (Figure S2). Analogous to results from the developmental stages, not surprisingly again, we found distinct activity profiles for SHs in the different tissues and/or anatomical components, suggesting that specific SHs have distinct spatiotemporal expression and are functionally active in different tissues of flies (Figure 4).
Figure 4. Gel-based ABPP analysis on fly tissue lysates.
A panel of Drosophila melanogaster tissue and/or anatomical component proteomes labeled with the SH-directed FP-rhodamine probe and analyzed by SDS-PAGE and in-gel fluorescence scanning. This gel-based ABPP experiment clearly shows that active (A), but not denatured (D) lysates, have distinct SH activity profiles in different Drosophila tissues and/or anatomical components. This gel-based ABPP experiment was done thrice with reproducible results each time.
Having confirmed efficient activity-dependent labeling of SHs by a FP-probe in adult fly tissue lysates by gel-based ABPP experiments, next, we wanted to identify the SHs that were active in the aforementioned tissues using LC-MS/MS based ABPP. Here, using an established protocol48, with the FP-biotin probe (200 μM, 1 mg proteome in 1 mL, 60 mins, 37 °C, with constant shaking), we enriched SHs, and identified them by LC-MS/MS. From this LC-MS/MS based ABPP experiment, we identified 118 unique SHs, of which 50 and 68 belonged to serine proteases and metabolic SHs categories respectively (Figure 5, Table S2). Here too, consistent with similar ABPP studies done in other systems previously, we found that of the total identified proteins identified in these LC-MS/MS based ABPP experiments, ~ 20 – 30% of them were SHs. Consistent with the gel-based ABPP experiments (Figure 4), we found several SHs were uniquely enriched in certain tissues in adult flies, and possibly suggests an important role for these SHs in regulating this tissue’s physiology. Not surprisingly, we found that a large fraction of active SHs were present in the adult fly gut, consistent with the role of this organ in digestion of food, and its metabolism.
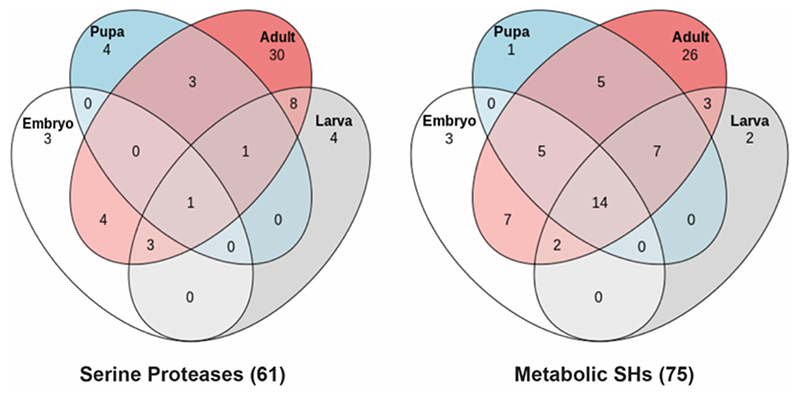
Of note, this LC-MS/MS based ABPP experiment led to a significant increase in the overall number of non-redundant active SHs identified in adult flies. Specifically, we identified an additional 40 and 34 serine proteases and metabolic SHs respectively (Figure 5), from this anatomical profiling in adult flies, relative to previous whole adult fly profiling (Figure 3), which had identified only 10 and 35 serine proteases and metabolic SHs respectively. Next, upon collating all the data from LC-MS/MS based ABPP experiments in total, we have now identified 136 non-redundant active SHs in flies, thus reaching a coverage of 38.4% (136 out of 354) for the SH superfamily with a single FP-biotin probe (Figure 6). Further, we find that amongst the total non-redundant active SHs identified, 61 are serine proteases (29%, 61 out of 210 total serine proteases), while 75 are metabolic SHs (52%, 75 out of 144 total metabolic SHs) (Figure 6, Table S2). Interestingly, our chemoproteomics profiling data further shows that only a small portion of SHs (11 out of 61 serine proteases, and 6 out of 75 metabolic SHs) are actually exclusively present in early developmental stages (embryo or larva or pupa) in flies, relative to those found exclusively found in adults (30 out of 61 serine proteases, and 26 out of 75 metabolic SHs) (Figure 6, Table S2). Not surprisingly, several of the identified SHs were present in at least 1 early developmental stage (embryo or larva or pupa) and also in adult flies (20 out of 61 serine proteases, and 43 out of 75 metabolic SHs) (Figure 6, Table S2).
Figure 6. Mapping SH activities during fly development.
A Venn diagram analysis showing the complete distribution of the full complement of active SHs identified by us using a LC-MS/MS based ABPP approach, during various life stages of Drosophila development.
Sexual dimorphism of SH activities in adult Drosophila melanogaster
Having mapped the entire complement of active SHs in flies, as a proof of concept of the utility of such a superfamily-wide activity atlas, next, we assessed whether there exists any sexual dimorphism in SH activities in the different tissues and/or anatomical components in adult flies. To collate a list of such SHs, we looked at tissues and/or anatomical components (head, gut, carcass and hemolymph) other than the reproductive organs, and filtered those SHs that were present only in adult males or females, and had average peptide counts ≥ 2 from 3 biological replicates. Additionally, we also looked for SHs in the same tissues that had average peptide counts ≥ 2 from 3 biological replicates, and were enriched ≥ 5-fold in one gender over the other. Based on this criteria for selection, we found 16 serine proteases, and 19 metabolic SHs, in adult flies that displayed sexual dimorphism in their activities (Table 1). Not surprisingly, a large fraction of the differentially active SHs identified, were found in the gut, given this organ’s important role in metabolism/digestion, and in modulating sexual dimorphism in flies58 (Table 1). Interestingly, a significant majority of the identified serine proteases (11 out of 16), and metabolic SHs (16 out of 19) were present in adult male flies (Table 1). Amongst the metabolic SHs, the enrichment of Yp1, Yp2 and Yp3 in females was expected, as these SHs are needed for necessary for egg formation59, 60. However, the enrichment of other metabolic SHs displaying sexual dimorphism (Table 1) raises intriguing new possibilities of sex-specific regulation of enzymatic activities in flies, as most of these metabolic SHs lack proper functional annotation experimentally, despite having predicted molecular functions. To gain confidence in the assignment of the aforementioned active SHs as sexually dimorphic, we turned to published literature and databases that relates to the differential expression of genes between male and female flies. The refinement of microarray technology and the development of next-generation sequencing (NGS) methods has led to a thorough and quantitative characterization of gene expression patterns during Drosophila development61, 62. Several studies are now available that report the differences in sex-specific gene expression in various somatic tissues and reproductive organs63–67. In each case where literature is available for a particular SH, we found that published literature correlated directly with our assignment, i.e. SHs that we have listed as ‘Male-specific’ have higher mRNA levels in males and vice-versa (Table 1). Our study also appears to have uncovered as-of-yet unknown sexually dimorphic SHs, which have not been classified in the aforementioned transcriptome studies, and would need further validation. We would also like to make a note that the given the low peptide counts for some SHs that display sexual dimorphism in their activities (Table 1), our chemoproteomics data in such cases only, may be interpreted as “relative activity” for such candidates, rather than the complete absence of such SHs from one gender.
Table 1. List of SHs showing sexual dimorphism in activities in adult fly.
| Gene ID | Predicted Molecular Function | Gender | Tissue(s) | Literature Reference |
|---|---|---|---|---|
| SERINE PROTEASES | ||||
| CG10469 | Serine-type endopeptidase | Female | Gut | Parisi et. al. 2004; Hudry et. al., 2016 |
| Send1 | Serine-type endopeptidase | Female | Gut | Arbeitman et. al., 2004 |
| CG4053 | Serine-type endopeptidase | Female | Gut | Hudry et al,2016; Ahmed et. al., 2020 |
| Jon65Aiii | Serine-type endopeptidase | Female | Gut | Hudry et. al., 2016 |
| HtrA2 | Serine-type endopeptidase | Female | Head | Hudry et. al., 2016 |
| CG17477 | Serine-type endopeptidase | Male | Gut | Hudry et. al., 2016 |
| CG8952 | Serine-type endopeptidase | Male | Gut | Ahmed et. al., 2020; Hudry et. al., 2016 |
| Jon99Ci | Serine-type endopeptidase | Male | Gut | Hudry et. al., 2016 |
| etaTry | Serine-type endopeptidase | Male | Gut | Ahmed et. al., 2020; Hudry et. al., 2016 |
| CG18179 | Serine-type endopeptidase | Male | Gut | Ahmed et. al., 2020; Hudry et. al., 2016 |
| Jon25Biii | Serine-type endopeptidase | Male | Gut | Lebo et. al. 2009; Hudry et. al., 2016 |
| Jon25Bi | Serine-type endopeptidase | Male | Gut | Ahmed et. al., 2020; Hudry et. al., 2016 |
| CG11037 | Serine-type endopeptidase | Male | Hemolymph | Parisi et. al. 2004 |
| CG10477 | Serine-type endopeptidase | Male | Gut | |
| Jon25Bii | Serine-type endopeptidase | Male | Gut | |
| Ser7 | Serine-type endopeptidase | Male | Head | |
| METABOLIC SHs | ||||
| Yp1 | Carboxylic ester hydrolase | Female | Hemolymph | Lebo et. al. 2009; Parisi et. al. 2004; Hudry et. al., 2016 |
| Yp2 | Carboxylic ester hydrolase | Female | Hemolymph | Lebo et. al. 2009; Parisi et. al. 2004 |
| Yp3 | Carboxylic ester hydrolase | Female | Hemolymph | Hudry et. al., 2016 |
| CG31872 | Sterol esterase | Male | Hemolymph, Gut | Parisi et. al. 2004 |
| CG17097 | Sterol esterase | Male | Hemolymph, Gut | Parisi et. al. 2004 |
| CG18258 | Carboxylic ester hydrolase | Male | Hemolymph, Gut | Parisi et. al. 2004 |
| CG3739 | Serine-type peptidase | Male | Gut | Parisi et. al. 2004; Ahmed et. al., 2020 |
| Alpha-Est10 | Carboxylic ester hydrolase | Male | Gut | Parisi et. al. 2004 |
| CG6295 | Phospholipase | Male | Gut | Ahmed et. al., 2020 |
| MESK2 | Unknown hydrolase | Male | Head | Ahmed et. al., 2020 |
| CG1309 | Phospholipase | Male | Head | |
| Est-Q | Unknown hydrolase | Male | Hemolymph | |
| FASN2 | Fatty acid synthesis | Male | Gut | |
| Ppt1 | Palmitoyl-(protein) hydrolase | Male | Gut | |
| CG18858 | O-acyltransferase | Male | Gut, Carcass | |
| CG5707 | Unknown hydrolase | Male | Gut | |
| Gas | Unknown hydrolase | Male | Gut | |
| CG11608 | Triglyceride lipase | Male | Gut | |
| CG18301 | Triglyceride lipase | Male | Gut | |
Discussion
From its early beginnings as a model organism for genetics in the Columbia ‘Fly room’ in 1910, Drosophila continues to be a powerful experimental tool in the fields of genetics, cell biology, developmental biology, neurobiology, and evolutionary biology19–23. Many fundamental discoveries have been based on work done in Drosophila, and this has led to six Nobel prizes, in diverse fields of biology. It’s continued popularity is a consequence of the sustained upgradation of its genetic toolkit, with quantum improvements seen every decade. Increasingly, the fly model is seen as a translational tool towards improving human health, with a focus on understanding human genetic diseases19–23. Chemoproteomics has proved to be an invaluable and powerful tool in assigning functions to proteins68, 69, particularly enzymes, in the post-genomic era, and yet, to the best of our knowledge, there are no reports describing its utility in Drosophila melanogaster, despite fly being such a popular biological model. The functional proteomics strategy, also popularly called ABPP, developed initially by Cravatt and co-workers, has been extensively used to study the SH superfamily5–9. SHs are known to function as metabolic lynchpins, regulate several physiologically important processes, and their deregulation is often associated with detrimental effects in in all forms of life1. However, despite their biological importance, the SH superfamily remains poorly explored in flies (Figure 7).
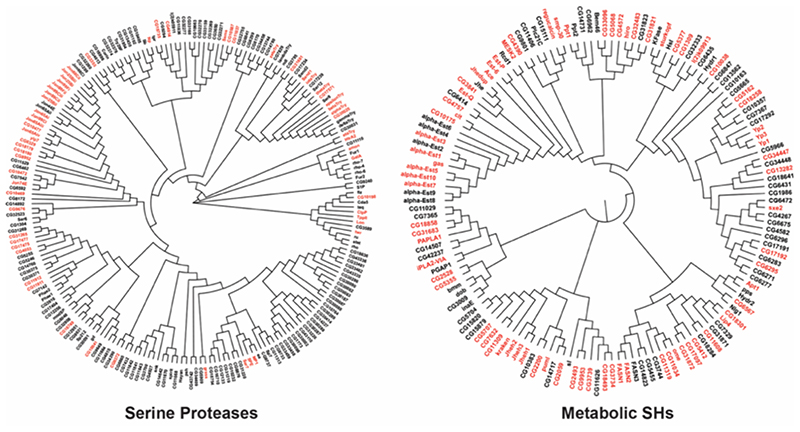
Figure 7. A chemoproteomic atlas of SH activities in Drosophila melanogaster.
A dendrogram analysis of all the predicted serine proteases and metabolic SHs in Drosophila melanogaster, in which, the red and black coloring represents enzymes that were enriched and not enriched respectively in the LC-MS/MS based ABPP experiments using the SH-directed FP-biotin probe. All protein sequences of the respective category (serine proteases and metabolic SHs) were aligned using the MUSCLE program of the MEGA-X software by the neighbor joining method, where the branch length denotes relatedness between protein sequences.
To address this problem, using bioinformatics, we began collating a list all the known non-redundant SHs in Drosophila melanogaster from various publicly available databases, and found that flies genetically encodes 354 of them (Figure 2, Table S1). Consistent with previous categorizations, we classified these 354 non-redundant SHs based on sequence homology to known molecular functions into serine proteases and metabolic SHs, and found that these two categories contained 210 and 144 members respectively, of which a fair majority lacked any known physiological function Figure 2, Table S1). Next, leveraging gelbased and LC-MS/MS based ABPP approaches, we profiled and identified active SHs in various developmental stages in flies (Figure 3, Table S2) and in different tissues and/or anatomical components in adult male and female flies (Figure 4-5, Table S1) in an attempt to develop an activity atlas for SHs in Drosophila melanogaster. From this ABPP study, we found a total of 136 non-redundant active SHs with a superfamily wide coverage of 38.4%, of which 61 were serine proteases (29% coverage for this category), while 75 belonged to the metabolic SH category (52% coverage for this category) (Figure 7). Further, we found that, several of identified SHs were present exclusively in the adult life form, and suggest an important (and perhaps conserved) role for these enzymes adult metabolism (Figure 6, Table S2). During the course of our ABPP studies of the adult life form, we purposefully decided to profile males and females separately. Interestingly, we found consistent with published comprehensive transcriptomic studies in adult flies, that several SHs display sexual dimorphism in their enzymatic activities, and most of them have unknown physiological and/or molecular functions (Table 1). Overall, our studies reported here, to the best of knowledge, describe the first ABPP enabled superfamily wide activity atlas for any enzyme family in Drosophila melanogaster.
Moving ahead, we anticipate that this study will lead to chemoproteomics being more extensively used in fly models, and more ABPP enabled superfamily-wide activity atlas’ for different well studied enzyme families in Drosophila melanogaster will emerge, towards understanding endogenous function of unannotated enzymes, and uncovering of new biological pathways. Here, we show that a large fraction of the SH superfamily, particularly metabolic SHs, lack defined and/or experimentally validated physiological functions, but are amenable to labeling/enrichment using FP-probes. Therefore, following up on this study, assigning the endogenous function to such SHs, and mapping the biological pathways they regulate in flies, should now be more tractable using the rich array of established pharmacological tools and chemical genetic screening methods used for members of this superfamily in other organisms15, 70. Next, we also report several candidate SHs that show sexual dimorphism in their activity in adult flies, and characterizing the basis of this differential activity using established genetic and pharmacological tools, will certainly advance our understanding of hormonal regulation of enzymatic activities. Further, we acknowledge that a large portion of the SHs (> 60%) are still not covered using the FP-biotin probe and the ABPP approaches we describe here. We attribute this to two major factors: (i) We profiled adult male and female flies between 5 – 10 days of age, under normal conditions and environment, and we posit that several SHs display age and/or context (e.g. exposure to infection, inflammatory stimuli, stress etc.) dependent expression and in turn activity; and (ii) Previous reports from mammalian systems have shown that several SHs don’t react with FP-biotin15, 70, and therefore the use of established tailored activity probes (e.g. HT01, WHP01, MB064)71–74 and/or the development of new activity based probes will be needed to increase the chemoproteomic coverage of members of this family in flies. As more ABPP profiling studies in flies with FP-probes emerge, it will be interesting to also determine whether non-SH enzymes (e.g. threonine hydrolases like AIG1 and ADTRP)75 are also active and FP reactive, and if such enzyme classes also exist in flies. Lastly, we would like to note that while reporting “active” enzymes is the major advantage of ABPP, this chemoproteomics technique provides very little information on the spatiotemporal expression (and in turn activity) of particular enzymes in comparison to some other biological techniques (e.g. immunofluorescence, in situ hybridization). Therefore, in the context of functionally annotating SHs, using ABPP in conjunction with complementary genetic tools and other biological techniques, will great facilitate the characterization of this enzyme superfamily, and will provide new biological insights into various physiological processes in flies.
Conclusions
The rapid progress in genome sequencing technologies have made a large number of protein sequences available for numerous organisms, and assigning function to these proteins represents a major challenge for researchers in the post-genomic era76. Over the past decade, the chemoproteomics technique ABPP has emerged as a powerful and popular tool in bridging this knowledge gap, and has greatly facilitated the functional annotation of enzymes from diverse enzyme families6, 7. The SHs catalyze a large number of physiologically important hydrolysis reactions, and this superfamily remains the most investigated enzyme class using ABPP5–9. While ABPP aided superfamily-wide activity atlas’ are available for SHs in various organisms, both ABPP and the SH superfamily remain largely unexplored in the fly model, Drosophila melanogaster. Here, using gel-based and LC-MS/MS based ABPP, we build an activity atlas map of SHs in flies, and report the activity profiles of enzymes from this superfamily during various developmental stages and in different adult tissues. Our studies also suggest sexual dimorphism in SH activities, and the resource we provide here, will greatly facilitate the use of ABPP, and aid functional annotation of hitherto unknown SHs in various fly models.
Supplementary Material
All the proteomics data are available via ProteomeXchange with identifier PXD024321.
Acknowledgements
Members of the S.S.K., and G.S.R. labs are thanked for providing critical comments and helpful discussions on this study. Saddam Shekh (IISER Pune) is thanked for technical assistance.
Funding
This work has been supported by: DBT/Wellcome Trust India Alliance Fellowship (grant number IA/I/15/2/502058) and an EMBO Young Investigator Award to S.S.K.; Department of Biotechnology – Genome Engineering technology grant (BT/PR26095/GET/119/199/2017) and Ministry of Human Resource Development – Scheme for Translational and Advanced Research in the Sciences (MHRD-STARS) to G.S.R.; Department of Science and Technology Fund for Improvement of S&T Infrastructure (DST-FIST) (grant number SR/FST/LSII-043/2016) to the IISER Pune Biology Department for building a proteomics facility. Part of the work was carried at the IISER Drosophila media and stock-keeping facility supported by the National Facility for Gene Function in Health and Disease (NFGFHD) at IISER Pune, supported by a grant from the Department of Biotechnology, Govt. of India (BT/INF/22/SP17358/2016). K.K. is supported by a graduate student fellowship from the Council of Scientific and Industrial Research (CSIR).
Footnotes
Accession Codes
The NCBI accession IDs for all the SH enzymes in Drosophila melanogaster can be found in Table S1.
Notes
The authors declare no competing financial interests.
Author Contributions
K.K. performed all the experiments. A.M. synthesized all compounds. K.K., G.S.R., and S.S.K. analysed all the data. G.S.R. and S.S.K. conceived and supervised the project, and acquired funding. S.S.K. wrote the paper with inputs from all authors.
References
- [1].Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- [4].Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- [5].Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- [7].Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu Rev Biochem. 2014;83:341–377. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- [8].Jessani N, Young JA, Diaz SL, Patricelli MP, Varki A, Cravatt BF. Class assignment of sequence-unrelated members of enzyme superfamilies by activity-based protein profiling. Angew Chem Int Ed Engl. 2005;44:2400–2403. doi: 10.1002/anie.200463098. [DOI] [PubMed] [Google Scholar]
- [9].Jessani N, Cravatt BF. The development and application of methods for activity-based protein profiling. Curr Opin Chem Biol. 2004;8:54–59. doi: 10.1016/j.cbpa.2003.11.004. [DOI] [PubMed] [Google Scholar]
- [10].Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- [11].Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat Rev Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, 3rd, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Keller LJ, Lentz CS, Chen YE, Metivier RJ, Weerapana E, Fischbach MA, Bogyo M. Characterization of Serine Hydrolases Across Clinical Isolates of Commensal Skin Bacteria Staphylococcus epidermidis Using Activity-Based Protein Profiling. ACS Infect Dis. 2020;6:930–938. doi: 10.1021/acsinfecdis.0c00095. [DOI] [PubMed] [Google Scholar]
- [15].Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen AL, Lum KM, Lara-Gonzalez P, Ogasawara D, Cognetta AB, 3rd, To A, Parsons WH, Simon GM, Desai A, Petrascheck M, Bar-Peled L, et al. Pharmacological convergence reveals a lipid pathway that regulates C. elegans lifespan. Nat Chem Biol. 2019;15:453–462. doi: 10.1038/s41589-019-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yoo E, Schulze CJ, Stokes BH, Onguka O, Yeo T, Mok S, Gnadig NF, Zhou Y, Kurita K, Foe IT, Terrell SM, et al. The Antimalarial Natural Product Salinipostin A Identifies Essential alpha/beta Serine Hydrolases Involved in Lipid Metabolism in P. falciparum Parasites. Cell Chem Biol. 2020;27:143–157.:e145. doi: 10.1016/j.chembiol.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lentz CS, Sheldon JR, Crawford LA, Cooper R, Garland M, Amieva MR, Weerapana E, Skaar EP, Bogyo M. Identification of a S. aureus virulence factor by activity-based protein profiling (ABPP) Nat Chem Biol. 2018;14:609–617. doi: 10.1038/s41589-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hales KG, Korey CA, Larracuente AM, Roberts DM. Genetics on the Fly: A Primer on the Drosophila Model System. Genetics. 2015;201:815–842. doi: 10.1534/genetics.115.183392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rajan A, Perrimon N. Of flies and men: insights on organismal metabolism from fruit flies. Bmc Biol. 2013;11(38):1–6. doi: 10.1186/1741-7007-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jennings BH. Drosophila - a versatile model in biology & medicine. Mater Today. 2011;14:190–195. [Google Scholar]
- [22].Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- [23].Bilder D, Irvine KD. Taking Stock of the Drosophila Research Ecosystem. Genetics. 2017;206:1227–1236. doi: 10.1534/genetics.117.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cao XL, Jiang HB. Building a platform for predicting functions of serine protease-related proteins in Drosophila melanogaster and other insects. Insect Biochem Molec. 2018;103:53–69. doi: 10.1016/j.ibmb.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- [26].Cho YS, Stevens LM, Sieverman KJ, Nguyen J, Stein D. A Ventrally Localized Protease in the Drosophila Egg Controls Embryo Dorsoventral Polarity. Current Biology. 2012;22:1013–1018. doi: 10.1016/j.cub.2012.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Misra S, Hecht P, Maeda R, Anderson KV. Positive and negative regulation of Easter, a member of the serine protease family that controls dorsal-ventral patterning in the Drosophila embryo. Development. 1998;125:1261–1267. doi: 10.1242/dev.125.7.1261. [DOI] [PubMed] [Google Scholar]
- [28].Morisato D, Anderson KV. Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu Rev Genet. 1995;29:371–399. doi: 10.1146/annurev.ge.29.120195.002103. [DOI] [PubMed] [Google Scholar]
- [29].Larkin A, Marygold SJ, Antonazzo G, Attrill H, Dos Santos G, Garapati PV, Goodman JL, Gramates LS, Millburn G, Strelets VB, Tabone CJ, et al. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021;49:D899–D907. doi: 10.1093/nar/gkaa1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, et al. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rey AJ, Attrill H, Marygold SJ, FlyBase, C Using FlyBase to Find Functionally Related Drosophila Genes. Methods Mol Biol. 2018;1757:493–512. doi: 10.1007/978-1-4939-7737-6_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marygold SJ, Crosby MA, Goodman JL, FlyBase, C Using FlyBase, a Database of Drosophila Genes and Genomes. Methods Mol Biol. 2016;1478:1–31. doi: 10.1007/978-1-4939-6371-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gelbart WM, Crosby M, Matthews B, Rindone WP, Chillemi J, Russo Twombly S, Emmert D, Ashburner M, Drysdale RA, Whitfield E, Millburn GH, et al. FlyBase: a Drosophila database. The FlyBase consortium. Nucleic Acids Res. 1997;25:63–66. doi: 10.1093/nar/25.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ashburner M, Drysdale R. FlyBase--the Drosophila genetic database. Development. 1994;120:2077–2079. doi: 10.1242/dev.120.7.2077. [DOI] [PubMed] [Google Scholar]
- [36].Rawlings ND, Bateman A. How to use the MEROPS database and website to help understand peptidase specificity. Protein Sci. 2021;30:83–92. doi: 10.1002/pro.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rawlings ND, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2000;28:323–325. doi: 10.1093/nar/28.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rawlings ND, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 1999;27:325–331. doi: 10.1093/nar/27.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Prakash A, Jeffryes M, Bateman A, Finn RD. The HMMER Web Server for Protein Sequence Similarity Search. Curr Protoc Bioinformatics. 2017;60:3 15 11–13 15 23. doi: 10.1002/cpbi.40. [DOI] [PubMed] [Google Scholar]
- [42].Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Saitou N, Nei M. The Neighbor-Joining Method - a New Method for Reconstructing Phylogenetic Trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- [46].Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rajendran A, Vaidya K, Mendoza J, Bridwell-Rabb J, Kamat SS. Functional Annotation of ABHD14B, an Orphan Serine Hydrolase Enzyme. Biochemistry. 2020;59:183–196. doi: 10.1021/acs.biochem.9b00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kelkar DS, Ravikumar G, Mehendale N, Singh S, Joshi A, Sharma AK, Mhetre A, Rajendran A, Chakrapani H, Kamat SS. A chemical-genetic screen identifies ABHD12 as an oxidized-phosphatidylserine lipase. Nat Chem Biol. 2019;15:169–178. doi: 10.1038/s41589-018-0195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Joshi A, Shaikh M, Singh S, Rajendran A, Mhetre A, Kamat SS. Biochemical characterization of the PHARC-associated serine hydrolase ABHD12 reveals its preference for very-long-chain lipids. Journal of Biological Chemistry. 2018;293:16953–16963. doi: 10.1074/jbc.RA118.005640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kulkarni A, Soni I, Kelkar DS, Dharmaraja AT, Sankar RK, Beniwal G, Rajendran A, Tamhankar S, Chopra S, Kamat SS, Chakrapani H. Chemoproteomics of an Indole-Based Quinone Epoxide Identifies Druggable Vulnerabilities in Vancomycin-Resistant Staphylococcus aureus. J Med Chem. 2019;62:6785–6795. doi: 10.1021/acs.jmedchem.9b00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- [52].Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 2008;7:3661–3667. doi: 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- [53].Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- [54].Jagtap P, Bandhakavi S, Higgins L, McGowan T, Sa R, Stone MD, Chilton J, Arriaga EA, Seymour SL, Griffin TJ. Workflow for analysis of high mass accuracy salivary data set using MaxQuant and ProteinPilot search algorithm. Proteomics. 2012;12:1726–1730. doi: 10.1002/pmic.201100097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Deutsch EW, Bandeira N, Sharma V, Perez-Riverol Y, Carver JJ, Kundu DJ, Garcia-Seisdedos D, Jarnuczak AF, Hewapathirana S, Pullman BS, Wertz J, et al. The ProteomeXchange consortium in 2020: enabling ‘big data’ approaches in proteomics. Nucleic Acids Research. 2020;48:D1145–D1152. doi: 10.1093/nar/gkz984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Horne I, Haritos VS, Oakeshott JG. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem Molec. 2009;39:547–567. doi: 10.1016/j.ibmb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- [58].Millington JW, Rideout EJ. Sexual Dimorphism: Ecdysone Modulates Sex Differences in the Gut. Curr Biol. 2020;30:R1327–R1330. doi: 10.1016/j.cub.2020.08.088. [DOI] [PubMed] [Google Scholar]
- [59].Bownes M. The regulation of the yolk protein genes, a family of sex differentiation genes in Drosophila melanogaster. Bioessays. 1994;16:745–752. doi: 10.1002/bies.950161009. [DOI] [PubMed] [Google Scholar]
- [60].Belote JM, Handler AM, Wolfner MF, Livak KJ, Baker BS. Sexspecific regulation of yolk protein gene expression in Drosophila. Cell. 1985;40:339–348. doi: 10.1016/0092-8674(85)90148-5. [DOI] [PubMed] [Google Scholar]
- [61].Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- [62].Johnston R, Wang B, Nuttall R, Doctolero M, Edwards P, Lu J, Vainer M, Yue H, Wang X, Minor J, Chan C, et al. FlyGEM, a full transcriptome array platform for the Drosophila community. Genome Biol. 2004;5:R19. doi: 10.1186/gb-2004-5-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lebo MS, Sanders LE, Sun F, Arbeitman MN. Somatic, germline and sex hierarchy regulated gene expression during Drosophila metamorphosis. BMC Genomics. 2009;10:80. doi: 10.1186/1471-2164-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Arbeitman MN, Fleming AA, Siegal ML, Null BH, Baker BS. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development. 2004;131:2007–2021. doi: 10.1242/dev.01077. [DOI] [PubMed] [Google Scholar]
- [65].Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lu J, Doctolero M, Vainer M, Chan C, Malley J, Eastman S, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hudry B, Khadayate S, Miguel-Aliaga I. The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature. 2016;530:344–348. doi: 10.1038/nature16953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ahmed SMH, Maldera JA, Krunic D, Paiva-Silva GO, Penalva C, Teleman AA, Edgar BA. Fitness trade-offs incurred by ovary-to-gut steroid signalling in Drosophila. Nature. 2020;584:415–419. doi: 10.1038/s41586-020-2462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Saghatelian A, Cravatt BF. Assignment of protein function in the postgenomic era. Nat Chem Biol. 2005;1:130–142. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]
- [69].Saghatelian A, Cravatt BF. Global strategies to integrate the proteome and metabolome. Curr Opin Chem Biol. 2005;9:62–68. doi: 10.1016/j.cbpa.2004.12.004. [DOI] [PubMed] [Google Scholar]
- [70].Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of the serine hydrolases. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kamat SS, Camara K, Parsons WH, Chen DH, Dix MM, Bird TD, Howell AR, Cravatt BF. Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay. Nat Chem Biol. 2015;11:164–171. doi: 10.1038/nchembio.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Baggelaar MP, den Dulk H, Florea BI, Fazio D, Bernabo N, Raspa M, Janssen APA, Scavizzi F, Barboni B, Overkleeft HS, Maccarrone M, et al. ABHD2 Inhibitor Identified by Activity-Based Protein Profiling Reduces Acrosome Reaction. ACS Chem Biol. 2019;14:2295–2304. doi: 10.1021/acschembio.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Janssen APA, van der Vliet D, Bakker AT, Jiang M, Grimm SH, Campiani G, Butini S, van der Stelt M. Development of a Multiplexed Activity-Based Protein Profiling Assay to Evaluate Activity of Endocannabinoid Hydrolase Inhibitors. ACS Chem Biol. 2018;13:2406–2413. doi: 10.1021/acschembio.8b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Parsons WH, Kolar MJ, Kamat SS, Cognetta AB, 3rd, Hulce JJ, Saez E, Kahn BB, Saghatelian A, Cravatt BF. AIG1 and ADTRP are atypical integral membrane hydrolases that degrade bioactive FAHFAs. Nat Chem Biol. 2016;12:367–372. doi: 10.1038/nchembio.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gerlt JA, Allen KN, Almo SC, Armstrong RN, Babbitt PC, Cronan JE, Dunaway-Mariano D, Imker HJ, Jacobson MP, Minor W, Poulter CD, et al. The Enzyme Function Initiative. Biochemistry. 2011;50:9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.