Abstract
T cell memory relies on the generation of antigen-specific progenitors with stem-like properties. However, the identity of these progenitors has remained unclear, precluding a full understanding of the differentiation trajectories that underpin the heterogeneity of antigen-experienced T cells. We used a systematic approach guided by single-cell RNA sequencing data to map the organizational structure of the human CD8+ memory T cell pool under physiological conditions. We identified two previously unrecognized subsets of clonally, epigenetically, functionally, phenotypically, and transcriptionally distinct stem-like CD8+ memory T cells. Progenitors lacking the inhibitory receptors programmed death-1 (PD-1) and T cell immunoreceptor with Ig and ITIM domains (TIGIT) were committed to a functional lineage, whereas progenitors expressing PD-1 and TIGIT were committed to a dysfunctional, exhausted-like lineage. Collectively, these data revealed the existence of parallel differentiation programs in the human CD8+ memory T cell pool, with potentially broad implications for the development of immunotherapies and vaccines.
Antigen recognition by CD8+ naive T cells initiates a program of clonal expansion and effector differentiation that leads to the clearance of infected or malignant cells and the subsequent formation of heterogeneous memory populations that confer durable immunity1. These memory populations are thought to be organized in a developmental hierarchy, according to which stem cell memory T (TSCM) cells self-renew and generate long-lived central memory T (TCM) cells and short-lived effector memory T (TEM) cells2–6. However, the mechanisms that underlie the enhanced multipotency of TSCM cells relative to TCM cells have not been clearly defined in molecular terms7.
Memory T cell differentiation can become corrupted under conditions of persistent antigenic stimulation, as observed during chronic viral infections and progressive malignancies, which promote a state of T cell exhaustion, characterized by an orderly loss of effector functions, impaired proliferation, and the upregulation of inhibitory receptors8. This dynamic process occurs over a period of weeks after the initial priming event9,10 and involves the genome-wide accumulation of epigenetic modifications11,12. Recent studies have shown that exhausted T (TEX) cell populations are developmentally and functionally heterogeneous, incorporating stem-like progenitors that express T cell factor 1 (TCF1) which give rise to highly differentiated TEX cells that are constitutively dysfunctional and lack TCF113–16. Importantly, the therapeutic benefits of immune checkpoint blockade in the context of chronic viral infections and various cancers are thought to operate via these TCF1+ progenitors, which appear susceptible to interventions that specifically target the inhibitory receptor programmed death-1 (PD-1)13,15,17–20.
Current evidence therefore suggests that exhausted and functional memory T cells arise from separate populations of stem-like progenitors committed to distinct fates. However, the precise nature of these stem-like progenitors, which shape the adaptive immune response and influence the outcome of many globally relevant diseases, has remained obscure. In this study, we used a comprehensive and unbiased approach to map the origins of dysfunctional and functional human CD8+ memory T cells. Our data identified two distinct subsets of CCR7+ progenitors in healthy individuals, distinguished by the expression of PD-1 and T cell immunoreceptor with Ig and ITIM domains (TIGIT). Progenitors committed to the generation of dysfunctional, exhausted-like progeny expressed both of these inhibitory receptors, whereas progenitors committed to the generation of a more functional progeny lacked both of these inhibitory receptors. Differential inclusion of the transcriptionally distinct PD-1+ TIGIT+ subset also explained most of the differences between TSCM and TCM cells, providing a clearer view of human CD8+ memory T cell differentiation.
Results
Two subsets of stem-like CD8+ memory T cell progenitors exist in humans
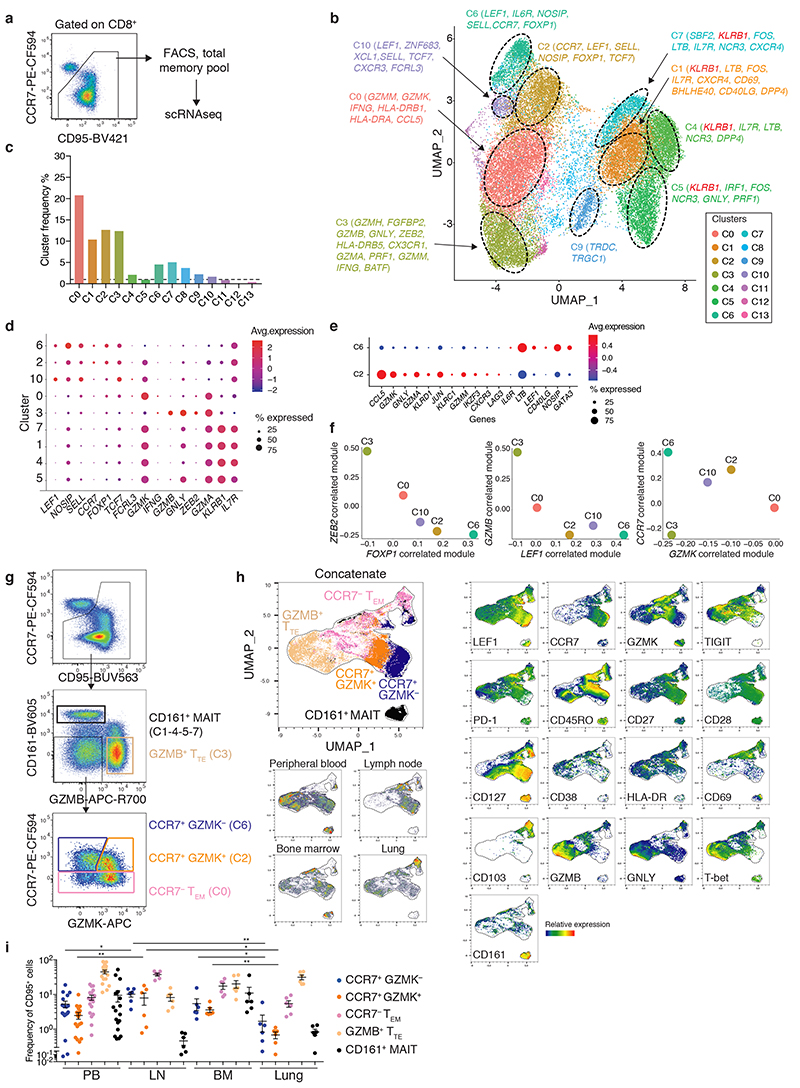
We initially used single-cell RNA sequencing (scRNA-seq; 10X Genomics Platform) to characterize the full spectrum of human CD8+ memory T cells in peripheral blood (PB) samples obtained from healthy donors (n = 4) (Supplementary Table 1). A total of 31,640 cells were isolated for this purpose via fluorescence-activated cell sorting (FACS) based on the expression of CD95 (Fig. 1a and Supplementary Fig. 1a), which identifies a vast majority of all memory T cells in humans4. Bioinformatic analysis of gene expression mapped in two dimensions via Uniform Manifold Approximation and Projection (UMAP)21 identified 14 distinct clusters (denoted individually as C) (Fig. 1b). C1, C4, C5, and C7 were uniformly distant from the other cell populations and expressed high levels of KLRB1, which encodes CD161, and IL7R, which encodes the interleukin (IL)-7 receptor (IL-7R), also known as CD127. These clusters were therefore derived from mucosal associated invariant T (MAIT) cells22. An intermediate cluster, C9, which comprised less than 2% of all cells (Fig. 1c), overexpressed TRDC and TRGC1, which encode the constant regions of the T cell receptor (TCR) δ and γ chains, respectively, suggesting the expression of γδ rather than αβ TCRs.
Fig. 1. Heterogeneity of the human CD8+ memory T cell pool.
a, Strategy for the isolation of CD8+ memory T cells from PB via FACS. b, UMAP plot showing the distribution of 31,640 cells (n = 4 donors). Cluster labels indicate selected DEGs. c, Histogram plot showing the median frequency of each cluster obtained in b. The dashed line is set at 1%. d, Balloon plot showing the average expression levels and expression frequencies of selected genes in each cluster obtained in b. e, Balloon plot showing the average expression levels and expression frequencies of selected genes in C2 versus C6. f, Bivariate plots depicting transcriptional module scores correlated with specific genes for selected clusters obtained in b. g, Flow cytometric gating strategy for the identification of CD8+ memory T cell subsets. Representative data are shown from PB. h, UMAP plot showing the expression of selected markers among CD8+ memory T cells isolated from different tissues (n = 6 donors per tissue with matched peripheral blood samples). Top left: overlays of the cell populations identified in g. i, Dot plot showing the tissue-specific frequencies of each subset identified in g. PB, peripheral blood; LN, lymph node; BM, bone marrow. Each dot represents one donor (n = 6 per tissue with matched peripheral blood samples). Bars indicate mean ± SEM. Statistics were calculated only for the GZMK− and GZMK+ populations. *P < 0.05, **P < 0.01 (two-tailed unpaired t-test for GZMK+ in PB versus LN and lung versus LN, two-tailed Mann-Whitney U test for all other comparisons).
scRNA-seq further identified seven different clusters related to conventional memory T cells (Fig. 1b), the most abundant of which were C0, C2, C3, C6, and C10 (Fig. 1c). C2, C6, and C10 expressed genes associated with early differentiated memory T cells, including CCR7, LEF1, SELL, which encodes L-selectin (CD62L), and FOXP1 (Fig. 1b,d and Supplementary Table 2). In contrast, the highly abundant C0 overexpressed multiple effector transcripts, including GZMK and GZMM, which encode serine proteases termed granzymes, IFNG, which encodes interferon (IFN)-γ, the chemokine (C-C motif) ligands CCL4 and CCL5, and genes encoding human leukocyte antigen (HLA) class II molecules, consistent with the identification of effector memory T (TEM) cells (Fig. 1b,d and Supplementary Table 2). C3 displayed a gene expression profile reminiscent of terminal effector T (TTE) cells, featuring high levels of GZMB, GNLY, NKG7, ZEB2, and GZMA (Fig. 1b,d and Supplementary Table 2). A lack of signature transcripts precluded the identification of C8 based on current knowledge of the T cell differentiation pathway (Supplementary Table 2).
To explore the heterogeneity of the CCR7 + memory T cell pool, we focused on C2 and C6, because C10 comprised only ~1% of all sorted cells (Fig. 1c). We identified 160 differentially expressed genes (DEGs) between C2 and C6 (Supplementary Table 3). C2 expressed higher levels of effector molecules, including CCL5, GZMK, GNLY, GZMA, JUN, GZMM, HOPX, IKZF3, RUNX3, and PRF1, which encodes perforin, whereas C6 expressed higher levels of IL6R, LTB, LEF1, NOSIP, GATA3, and SELL (Fig. 1e and Supplementary Table 3). We then used anchor genes selected from the most prominent DEGs to compute transcriptional modules associated with memory differentiation (correlated with CCR7 and LEF1), quiescence (correlated with FOXP1), or cytotoxicity and terminal effector differentiation (correlated with GZMK/GZMB and ZEB2, respectively) among the five conventional memory T cell clusters (C0, C2, C3, C6, and C10). Using this approach, we found that C2 and C10 were similar, exhibiting intermediate memory and effector scores, whereas C6 was skewed toward a high memory score and C0 and C3 were skewed toward high effector scores (Fig. 1f). scRNA-seq analysis therefore identified four major subsets of conventional CD8+ memory T cells, namely early differentiated CCR7 + GZMK − (C6) and CCR7 + GZMK + (C2), TEM-like CCR7 − GZMK hi (C0), and TTE-like GZMB + (C3).
To confirm these findings at the protein level, we designed a high-dimensional flow cytometry panel based on the cluster signature markers CCR7, LEF1, CD161, GZMB, and GZMK (Fig. 1g). This panel was also equipped to detect memory and effector differentiation markers (CD27, CD28, CD45RO, CD127, and T-bet), activation markers (CD38 and HLA-DR), inhibitory receptors (PD-1 and TIGIT), and markers of tissue residency (CD69 and CD103) in CD8+ T cells isolated from the peripheral blood and tissues (Supplementary Table 1). In line with the scRNA-seq data, UMAP analysis revealed that CD161hi MAIT cells were largely distinct from other CD95+ memory T cells (Fig. 1h). Among the effector subsets, CCR7− GZMKhi cells expressed cytolytic molecules and generally lacked the memory markers LEF1, CD27, CD28, and CD127, whereas GZMB+ cells also expressed granulysin (GNLY) and relatively high levels of T-bet (Fig. 1h). In contrast, the CCR7+ GMZK− subset expressed relatively high levels of LEF1, CD27, CD28, and CD127 and lacked effector molecules, activation markers, and inhibitory receptors, whereas the CCR7+ GMZK+ subset expressed intermediate levels of LEF1 alongside PD-1 and TIGIT, which were not detected in the scRNA-seq analysis, together with relatively high levels of CD27, CD28, and CD127 (Fig. 1h). These subsets displayed variable expression of CD45RO (Fig. 1h). As expected, CD69+ CD103+ cells were only detected in tissues (Fig. 1h). A survey of different tissue sites revealed that CCR7+ GZMK− PD-1− TIGIT− and CCR7+ GZMK+ PD-1+ TIGIT+ cells were relatively abundant in PB, lymph nodes (LNs), and bone marrow (BM), whereas CCR7− GZMKhi cells were ubiquitous, and GZMB+ cells predominated in PB and lung tissue (Fig. 1i).
Collectively, these data identified CCR7+ GZMK− and CCR7+ GMZK+ cells as distinct entities in the early differentiated CD8+ memory T cell pool and further showed that these subsets could be distinguished by the expression of PD-1 and TIGIT.
Exhausted-like CD8+ memory T cell progenitors express GZMK, PD-1, and TIGIT
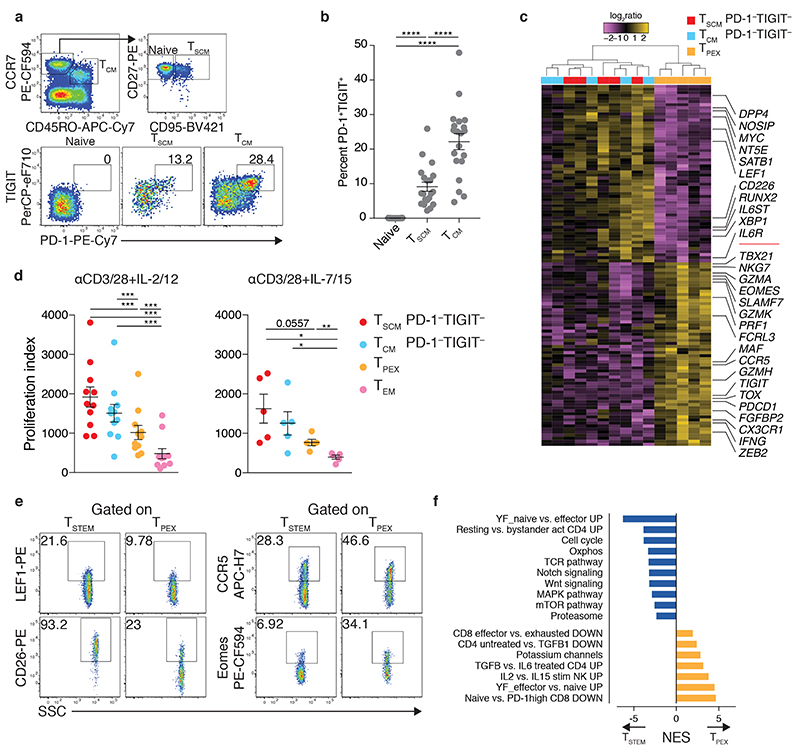
Heterogeneity in the early differentiated memory T cell pool became apparent with the identification of multipotent TSCM cells4,23. These cells exhibit a CCR7+ CD45RO− CD95+ phenotype, in contrast to TCM cells, which exhibit a CCR7+ CD45RO+ CD95+ phenotype. However, our scRNA-seq-guided flow cytometric analyses demonstrated that the CCR7+ GZMK− PD-1− TIGIT− and CCR7+ GZMK+ PD-1+ TIGIT+ subsets could not be distinguished via the expression of CD45RO (Fig. 1h). To place these findings in context, we investigated the expression of PD-1 and TIGIT among classically defined TSCM and TCM cells. We found that 9.1±1.3% of TSCM cells and 22.1±2.3% of TCM cells (mean±SEM) expressed both PD-1 and TIGIT (Fig. 2a,b). Manual gating of the flow cytometry data confirmed that PD-1 and TIGIT were preferentially expressed by CCR7+ GZMK+ cells (Supplementary Fig. 1b).
Fig. 2. Identification of stem-like CD8+ memory T cell progenitors with differential expression of GZMK, PD-1, and TIGIT.
a, Flow cytometric gating strategy for the identification of PD-1+ TIGIT+ cells in the CD8+ naive (CCR7+ CD45RO− CD95−), TSCM (CCR7+ CD45RO− CD95+), and TCM compartments (CCR7+ CD45RO+ CD95+). Numbers indicate percentages in the drawn gates. b, Dot plot summarizing the data obtained as in a. Each dot represents one donor (n = 20 from two independent experiments). Bars indicate mean ± SEM. ****P < 0.0001 (one-way repeated measures ANOVA). c, Heatmap showing DEGs (adjusted P value < 0.01) for the indicated CD8+ memory T cell subsets (n = 5 donors). Labels highlight genes associated with memory or effector differentiation or exhaustion. Significance was evaluated using edgeR analysis with glmQLFTest and Benjamini-Hochberg correction. d, Proliferation of the indicated CD8+ memory T cell subsets in response to stimulation with anti-CD3 plus CD28 for 4 d in the presence of IL-2 and IL-12 (n = 11 donors from six independent experiments) or IL-7 and IL-15 (n = 5 donors from three independent experiments). Index calculations were based on the dilution of carboxyfluorescein succinimidyl ester (CFSE). Each dot represents one donor. Bars indicate mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Wilcoxon signed rank test for each population versus TEM in the presence of IL-2 and IL-12, two-tailed paired t-test for all other comparisons). e, Representative flow cytometric analysis of TSTEM (CCR7+ PD-1– TIGIT–) and TPEX cells (CCR7+ PD-1+ TIGIT+) showing the expression of markers selected from the DEGs identified in c. Numbers indicate percentages in the drawn gates. Similar data were obtained from other donors (n = 5 for LEF1 and CCR5, n = 4 for CD26 and Eomes). f, GSEA based on 1,000 permutations showing manually curated signatures that differed significantly (adjusted P value < 0.05) between TSTEM and TPEX cells. NES, normalized enrichment score.
On the basis of these results, we hypothesized that differential inclusion of the transcriptionally distinct CCR7+ GZMK+ PD-1+ TIGIT+ subset could explain some of the previously reported differences between TSCM and TCM cells. To test this possibility, we analyzed the transcriptomes of TSCM and TCM cells after depletion of the CCR7+ PD-1+ TIGIT+ (GZMK+) population, hereafter termed T progenitor exhausted-like (TPEX) (Supplementary Fig. 1a and Supplementary Fig. 2). In line with our hypothesis, TSCM and TCM cells depleted of TPEX cells were very similar at the transcriptional level and could only be distinguished on the basis of eight DEGs (adjusted P value < 0.01) (Supplementary Fig. 2 and Supplementary Table 4). One of these DEGs was HNRNPLL, which encodes heterogeneous nuclear ribonucleoprotein L-like, a master regulator of alternative splicing responsible for the expression of CD45RO24, which is commonly used as a phenotypic marker to differentiate between TSCM and TCM cells25. In contrast, TPEX cells were largely distinct, featuring lower expression levels of SATB1, which encodes a negative regulator of PD-1 expression26, MYC, DPP4, which encodes CD26, IL6ST, LEF1, IL6R, and NT5E and higher expression levels of transcription factor (TF) genes recently associated with T cell exhaustion, including TOX 27–32, EOMES 33, and MAF 34, and other genes associated with effector differentiation and cytolytic activity, including ZEB2, GZMK, GZMA, TBX21, PRF1, IFNG, and NKG7 (Fig. 2c and Supplementary Table 4). As expected, PDCD1, which encodes PD-1, and TIGIT were also expressed at high levels, validating the integrity of cell isolation via FACS. Several other genes previously found to distinguish TSCM from TCM cells were also identified among these DEGs4 (Fig. 2c and Supplementary Table 4). In line with the transcriptional data, TPEX cells stimulated with anti-CD3 plus CD28 and a combination of effector (IL-2 plus IL-12) or homeostatic cytokines (IL-7 plus IL-15) proliferated less vigorously than PD-1− TIGIT− TSCM and TCM cells under identical conditions (Fig. 2d). However, all three subsets proliferated similarly and remained phenotypically stable in response to IL-15, suggesting equivalent self-renewal capabilities (Supplementary Fig. 3a,b). Accordingly, TSCM and TCM cells were better defined by the CCR7+ PD-1− TIGIT− phenotype, hereafter termed stem-like T (TSTEM), whereas early differentiated memory cells with dysfunctional, exhausted-like traits were characterized by the CCR7+ PD-1+ TIGIT+ phenotype (TPEX). Of note, the gene expression profiles of TSTEM and TPEX cells overlapped significantly with those of C6 and C2, respectively (P < 0.01 for each comparison using a hypergeometric test; not shown), confirming the shared identity of subsets analyzed via scRNA-seq and flow cytometry (Fig. 1e).
Flow cytometric analyses demonstrated that TSTEM cells expressed CD26 and LEF1 more commonly and CCR5 and Eomes less commonly than TPEX cells (Fig. 2e). Gene set enrichment analysis (GSEA) further revealed that TSTEM cells were characterized by transcripts associated with the naive state, quiescence, oxidative phosphorylation, the Wnt35 and Notch signaling pathways36, and proteasome activity37, whereas TPEX cells were characterized by transcripts associated with the TGF-β signaling pathway38, potassium regulation39, and other mechanistic correlates of exhaustion, including the PD-1hi state (Fig. 2f). Transcripts associated with the cell cycle and the TCR and mTOR signaling pathways, collectively suggesting a predisposition to antigen-driven proliferation and effector differentiation, were also upregulated in TSTEM versus TPEX cells (Fig. 2f). Previous analyses have shown that progenitor exhausted CD8+ T cells from tumors express stem-like genes along with PDCD1, TIGIT and GZMK 16, thereby suggesting a shared identity with TPEX cells. Indeed, the transcriptional features of TPEX cells aligned closely with those reported previously for progenitor exhausted-like (CCR7 hi GZMK hi) but not memory-like CD8+ T cells (CCR7 hi GZMK lo) isolated from melanomas18, whereas the opposite result was obtained in a parallel analysis of TSTEM cells (P < 0.05 for each comparison using a hypergeometric test) (Supplementary Table 4).
Collectively, these data revealed that TSCM and TCM cells were largely homogenous after depletion of the TPEX subset, indicating a need to refine current models of CD8+ memory T cell differentiation.
TSTEM cells are functionally superior to TPEX cells
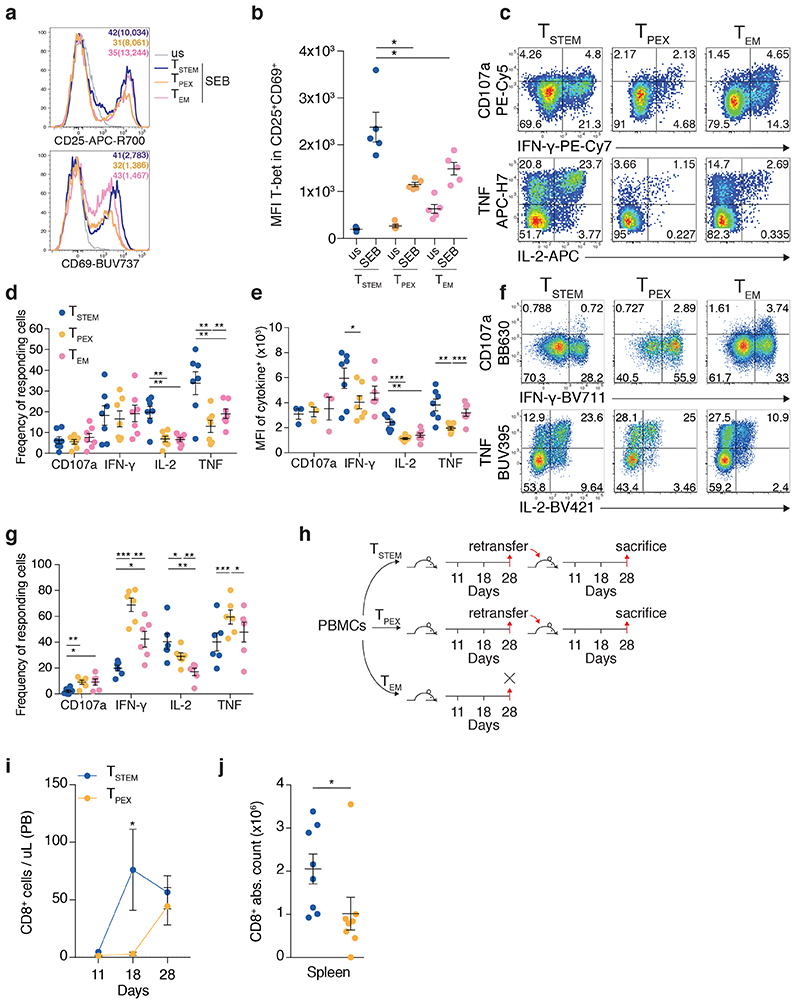
To validate our transcriptional and phenotypic data, we compared the functional properties of FACS-purified TSTEM and TPEX cells. In response to TCR-dependent stimulation with Staphylococcal enterotoxin B (SEB), TSTEM cells upregulated CD25 and CD69 to a greater extent than TPEX cells (Fig. 3a), and activated CD25+ CD69+ TSTEM cells expressed higher levels of T-bet than activated CD25+ CD69+ TPEX cells (Fig. 3b). Likewise, TSTEM cells produced cytokines at higher frequencies (IL-2 and TNF) and at higher levels on a per cell basis (IFN-γ, IL-2, and TNF) than TPEX cells in response to stimulation with anti-CD3 plus CD28 (Fig. 3c–e). No clear differences were observed between TSTEM and TPEX cells with respect to degranulation, measured via the surface mobilization of CD107a (Fig. 3c–e). In response to TCR-independent stimulation with phorbol myristate acetate (PMA) and ionomycin, however, TPEX cells produced IFN-γ and TNF and mobilized CD107a at much higher frequencies than TSTEM cells, the functional superiority of which was therefore limited to conditions that mimicked antigen recognition events (Fig. 3f,g).
Fig. 3. Functional properties of TSTEM and TPEX cells.
a, Representative flow cytometric analysis of FACS-purified TSTEM, TPEX, and TEM cells showing the expression of CD25 and CD69 before (us, unstimulated) and after stimulation with SEB for 24 h. Similar data were obtained from other donors (n = 4). Numbers colored to match each subset indicate percent marker+ cells with the corresponding median fluorescence intensity (MFI) in brackets. b, Dot plot showing the expression of T-bet among CD25+ CD69+ TSTEM, TPEX, and TEM cells before (us) and after stimulation as in a. Data are shown in terms of MFI. Each dot represents one donor (n = 5 from two independent experiments). Bars indicate mean ± SEM. *P < 0.05 (two-tailed paired t-test). c, Representative flow cytometric analysis showing the expression of CD107a, IFN-γ, IL-2, and TNF among TSTEM, TPEX, and TEM cells stimulated with anti-CD3 plus CD28 for 12 h. Numbers indicate percentages in the drawn gates. d, Dot plot summarizing the data obtained as in c. Each dot represents one donor (n = 7 from four independent experiments). Bars indicate mean ± SEM. **P < 0.01 (two-tailed paired t-test). e, Dot plot showing the function+ populations identified in d in terms of MFI. Each dot represents one donor (n = 3 from four independent experiments for CD107a, n = 7 from four independent experiments for IFN-γ, IL-2, and TNF). Bars indicate mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed paired t-test). f, Representative flow cytometric analysis showing the expression of CD107a, IFN-γ, IL-2, and TNF among TSTEM, TPEX, and TEM cells after stimulation of magnetically-enriched CD8+ T cells with PMA and ionomycin for 3 h. Numbers indicate percentages in the drawn gates. Subsets were gated as CCR7+ GZMK− (TSTEM), CCR7+ GZMK+ (TPEX), or CCR7− CD45RO+ CD95+ (TEM). g, Dot plot summarizing the data obtained as in f. Each dot represents one donor (n = 6 from three independent experiments). Bars indicate mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed paired t-test). h, Schematic layout of the serial transfer experiments. i, Line chart showing the absolute numbers of CD8+ T cells in PB on days 11, 18, and 28 after transfer of TSTEM or TPEX cells into secondary NSG recipients. Data were pooled from two independent experiments (total n = 8 mice). Bars indicate mean ± SEM. *P = 0.0117 (two-way ANOVA). j, Dot plot showing the absolute numbers of CD8+ T cells in spleen on day 28 after transfer of TSTEM or TPEX cells into secondary NSG recipients. Each dot represents one mouse (n = 8 from two independent experiments). Bars indicate mean ± SEM. *P = 0.0148 (two-tailed Mann-Whitney U test).
To determine the in vivo relevance of these observations, we performed serial adoptive cell transfers (ACTs) in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) humanized mice (Fig. 3h). TEM cells failed to repopulate these mice efficiently after the first ACT (not shown), as reported previously4. Although the early memory subsets both expanded to similar numbers in primary hosts (not shown), TSTEM cells proliferated more rapidly in PB (Fig. 3i) and repopulated the spleen more efficiently in secondary hosts compared with TPEX cells (Fig. 3j). Of note, the suboptimal proliferative capabilities of TPEX cells observed in vitro and in vivo were not associated with differences in telomere length relative to TSTEM cells, whereas TEM cells harbored shorter telomeres ex vivo compared with either TSTEM or TPEX cells (Supplementary Fig. 3c).
Collectively, these data showed that TSTEM cells were functionally superior to TPEX cells, both under homeostatic conditions and in response to stimulation via the TCR.
TPEX cells are committed to a terminally dysfunctional state
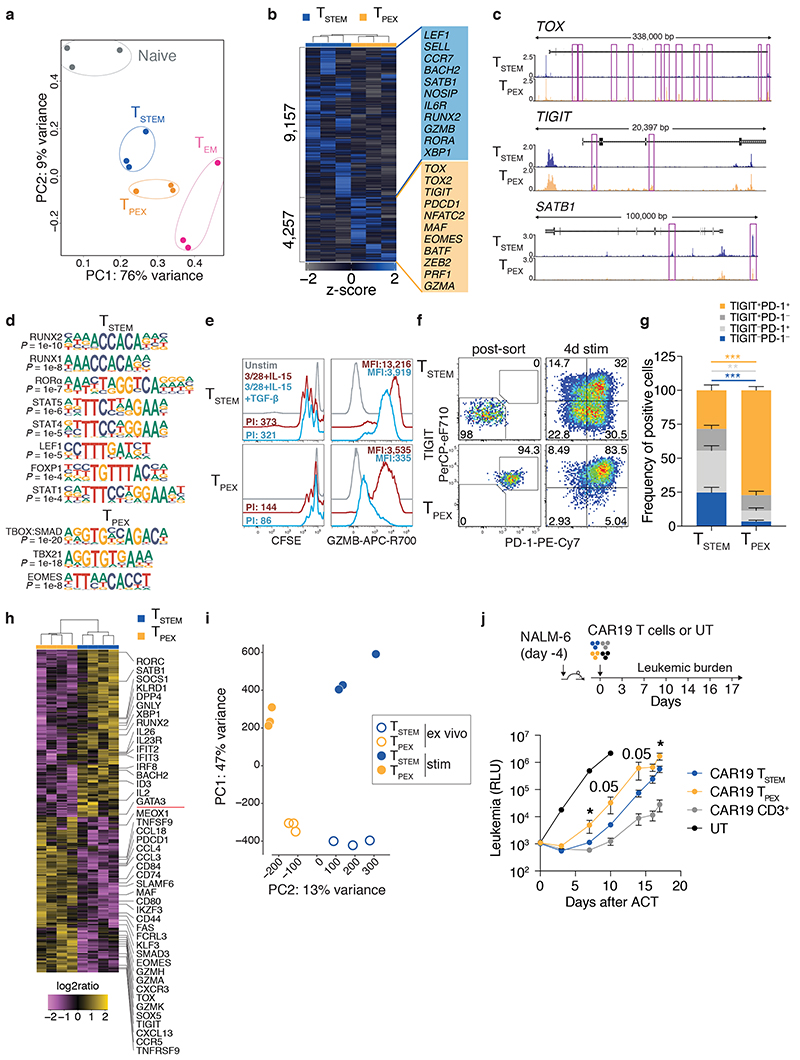
Epigenetic regulation plays a key role in T cell fate decisions40. We therefore employed the Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) to compare the open chromatin landscapes of TSTEM and TPEX cells in terms of differentially accessible regions (DARs). Naive and TEM cells were analyzed in parallel as lineage controls. Principal component analysis (PCA) revealed that TSTEM and TPEX cells were globally similar, although TSTEM cells mapped toward the naive subset, whereas TPEX cells mapped toward the TEM subset (Fig. 4a). However, we also identified a total of 13,414 DARs between TSTEM and TPEX cells (Fig. 4b). Genes associated with T cell dysfunction (e.g., TOX, TOX2, TIGIT, PDCD1, NFATC2, and MAF), terminal differentiation (e.g., ZEB2 and BATF), and other immune-related processes previously identified at the mRNA level (e.g., EOMES and GZMA) were more accessible in TPEX versus TSTEM cells (Fig. 4b,c). In contrast, genes associated with T cell memory (e.g., LEF1, SELL, CCR7, BACH2, and SATB1) and effector functions (e.g., GZMB and RORA 41) were more accessible in TSTEM versus TPEX cells (Fig. 4b,c). Computational analysis of these DARs further identified differentially accessible TF binding motifs (TFBMs). Motifs linked to TFs associated with thymocytes and naive and early memory cells (RUNX2, RUNX1, LEF1, and FOXP1), effector differentiation (RORA)41, and cytokine signaling (STAT5, STAT4, and STAT1) were enriched in TSTEM versus TPEX cells, whereas the TBX21 (T-bet), EOMES, and combined TBOX:SMAD motifs were enriched in TPEX versus TSTEM cells (Fig. 4d).
Fig. 4. Fate commitments of TSTEM and TPEX cells.
a, PCA plot showing the top 1,000 hypervariable peaks obtained from ex vivo ATAC-seq analysis (adjusted P value < 0.01) of TSTEM, TPEX, and TEM cells. Each dot represents one donor (n = 3). b, Heatmap showing DARs. Labels highlight accessible genes associated with memory or effector differentiation or exhaustion. c, Representative genomic regions showing the ATAC-seq profiles of TOX, TIGIT, and SATB1 in TSTEM and TPEX cells. DARs are highlighted in purple. d, TFBMs enriched among the DARs shown in b. Enrichment was assessed using a one-sided hypergeometric test in HOMER with correction for FDR. e, Representative overlay histograms showing CFSE dilution (left) and GZMB expression profiles (right) for TSTEM and TPEX cells stimulated with anti-CD3 plus CD28 and IL-15 for 3 d in the absence or presence of TGF-β. Unstimulated controls are shown for comparison. Similar data were obtained from other donors in the absence (n = 8 from four independent experiments) or presence of TGF-β (n = 6 from four independent experiments). PI, proliferation index. f, Representative flow cytometric analysis of TSTEM and TPEX cells showing the expression of PD-1 and TIGIT after stimulation with anti-CD3 plus CD28 for 4 d in the presence of IL-2 and IL-12. Numbers indicate percentages in the drawn gates. g, Bar graph summarizing the data obtained as in f (n = 5 donors from three independent experiments). Bars indicate mean ± SEM. **P < 0.01, ***P < 0.001 (two-tailed Mann-Whitney U test). h, Heatmap showing selected DEGs (adjusted P value < 0.05) for TSTEM and TPEX cells stimulated as in f (n = 4 donors). Significance was evaluated using edgeR analysis with glmQLFTest and Benjamini-Hochberg correction. i, PCA plot as in a comparing TSTEM and TPEX cells before and after stimulation as in f. j, Top: schematic layout of the adoptive transfer experiment. Bottom: time series plot showing the growth of NALM6 cells in NSG mice (n = 5/group) adoptively transferred with TSTEM or TPEX cells expressing a CAR specific for CD19 (CAR19). RLU, relative light unit; UT, untransduced CD3+ cells. Follow-up was stopped when RLU values ≥ 106 were observed in more than 75% of mice in one of the treated groups. Bars indicate mean ± SEM. *P < 0.05 (two-tailed unpaired t-test for CAR19 TSTEM versus CAR19 TPEX on day 17, two-tailed Mann-Whitney U test for all other comparisons).
The chromatin accessibility data suggested that TPEX cells were predisposed to the generation of dysfunctional progeny and susceptible to the inhibitory effects of TGF-β signaling via SMADs. Accordingly, TPEX cells proliferated to a lesser extent and produced less GZMB than TSTEM cells in response to stimulation with anti-CD3 plus CD28 and IL-15 (Fig. 4e). The addition of TGF-β further inhibited these responses, especially the production of GZMB, in parallel cultures of TPEX cells, whereas minimal effects were observed in parallel cultures of TSTEM cells (Fig. 4e). Importantly, most TPEX cells retained a PD-1+ TIGIT+ phenotype after stimulation with anti-CD3 plus CD28 in the presence of the effector cytokines IL-2 and IL-12 (Fig. 4f,g) or the homeostatic cytokines IL-7 and IL-15 (Supplementary Fig. 3d). In contrast, TSTEM cells generated all possible combinations of phenotypes defined by PD-1 and TIGIT (Fig. 4f,g). We then used RNA-seq to profile the transcriptomes of TSTEM and TPEX cells after stimulation with anti-CD3 plus CD28 in the presence of IL-2 and IL-12 (Fig. 4h). Activated TSTEM cells overexpressed the memory-related genes BACH2, ID3, IL2, and SATB1 alongside the effector-related genes IRF8, RORC, GNLY, XBP1, IL26, and IL23R, whereas activated TPEX cells overexpressed the dysfunction/exhaustion-related molecules TOX, PDCD1, TIGIT, MAF, and CXCL13 42, together with various chemokine genes, IKZF3, which encodes an inhibitor of IL-2 production, SMAD3, and genes associated with cytolytic activity, including GZMK, GZMH, and GZMA (Fig. 4h and Supplementary Table 5). Some of these genes were also differentially expressed between the corresponding subsets in ex vivo analyses (Fig. 2c). GSEA further demonstrated that activated TSTEM cells were preferentially enriched for gene sets associated with early differentiation and proliferation, whereas activated TPEX cells were preferentially enriched for gene sets associated with the TGF-β and PD-1 signaling pathways and exhaustion in the tumor microenvironment18 (Supplementary Fig. 4a). PCA of ATAC-seq data from paired ex vivo and activated samples revealed that stimulation profoundly altered the chromatin accessibility landscape in TSTEM and TPEX cells (Fig. 4i). However, the major epigenetic differences between these subsets in the ex vivo state were maintained after stimulation (Fig. 4i), both at the level of specific genes (Supplementary Fig. 4b) and in terms of enrichment for particular TFBMs (Supplementary Fig. 4c).
To assess the in vivo relevance of these findings, we employed a stringent ACT protocol in which TSTEM and TPEX cells were redirected using a chimeric antigen receptor (CAR) targeting CD19 and transferred in the absence of autologous CD4+ T cells or exogenous cytokines into NSG mice previously injected with the acute lymphoblastic leukemia cell line NALM6. In line with the in vitro data, TSTEM cells displayed enhanced control of leukemic burden compared with TPEX cells at multiple time points after ACT (Fig. 4j).
Collectively, these data indicated that TSTEM cells were relatively resistant to exhaustion, facilitating more efficient control of tumor growth in vivo compared with TPEX cells, which were hardwired to a dysfunctional signature.
TPEX cells are abundant in persistent infections and clonally distinct from TSTEM cells
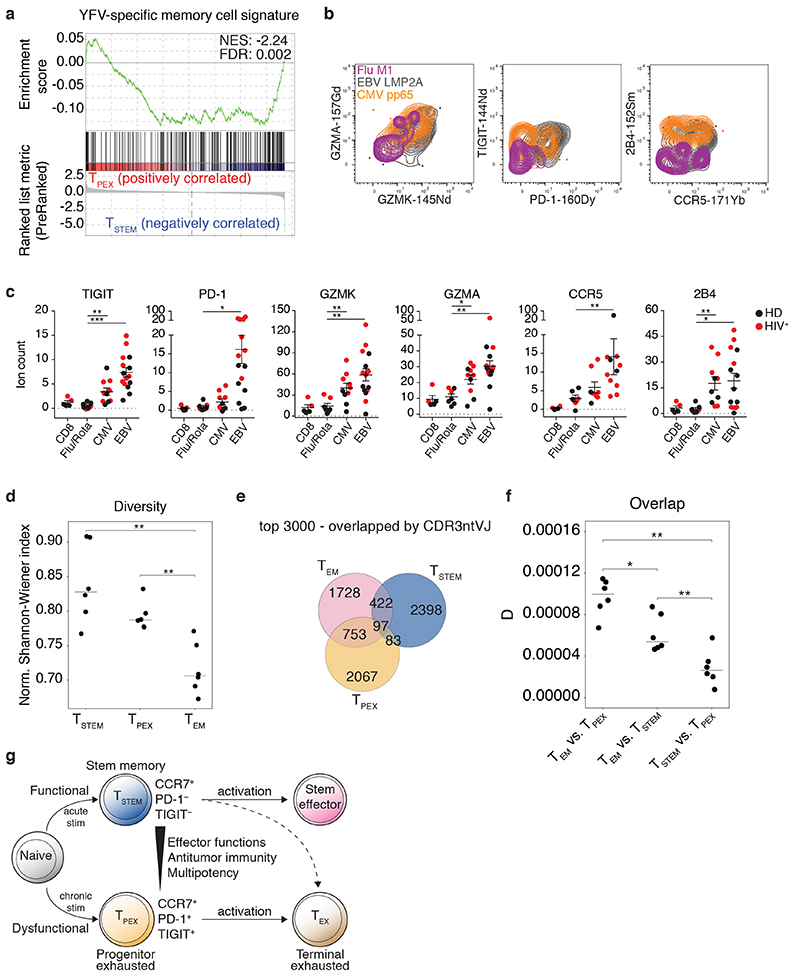
CD8+ T cell dysfunction and exhaustion develop in response to persistent antigenic stimulation via cognate TCRs43. We therefore reasoned that acute viral infections would preferentially generate antigen-specific TSTEM cells, whereas chronic viral infections would preferentially generate antigen-specific TPEX cells. A single round of yellow fever virus (YFV) vaccination is known to induce long-lived memory cells with an early differentiated TSCM-like CCR7+ CD45RA+ (or CD45RO−) CD95+ phenotype44,45. To determine the composition of these TSCM-like populations in terms of CCR7+ PD-1− TIGIT− TSTEM and CCR7+ PD-1+ TIGIT+ TPEX cells, we compared publicly available gene expression data from vaccinated individuals45 with our RNA-seq data (Fig. 2c). In line with our hypothesis, we found that YFV-specific CCR7+ CD45RA+ CD95+ cells analyzed years after vaccination were transcriptionally related to TSTEM but not TPEX cells (Fig. 5a).
Fig. 5. Antigen specificity and clonal identity of TSTEM and TPEX cells.
a, GSEA of the YFV-specific CD8+ memory T cell signature45 in TSTEM versus TPEX cells. b, Representative CyTOF analysis showing the expression of GZMK, GZMA, PD-1, TIGIT, CCR5, and 2B4 among CCR7+ virus-specific CD8+ T cell populations from healthy (n = 3) and HIV+ donors (n = 2). c, Dot plots summarizing the data obtained as in b. Epitopes derived from influenza virus (n = 6) and rotavirus (n = 1) were pooled for simplicity. Comparative data are shown for the corresponding total CD8+ T cell populations. Each dot represents one specificity in one donor (n = 3 healthy donors, n = 2 HIV+ donors). Bars indicate mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Mann-Whitney U test for CCR5 comparisons, two-tailed unpaired t-test for all other marker comparisons). HD, healthy donor. d, Dot plot showing the normalized Shannon-Wiener index for TCRβ repertoires obtained from the TSTEM, TPEX, and TEM subsets. Each dot represents one donor (n = 6). Bars indicate median values. **P < 0.01 (two-tailed paired t-test with Bonferroni correction). e, Venn diagram showing the numbers of shared and unique clonotypes among TSTEM, TPEX, and TEM cells from a representative donor. Similar data were obtained from other donors (n = 5). Analysis was restricted to the top 3,000 clonotypes. f, Dot plot summarizing the pairwise comparisons among subsets illustrated in e. D metric in VDJtools. Each dot represents one donor (n = 6). Bars indicate median values. *P < 0.05, **P < 0.01 (two-tailed paired t-test with Bonferroni correction). g, Proposed model showing the origins and differentiation trajectories of TSTEM and TPEX cells.
To extend these findings, we used peptide-HLA class I tetramers in conjunction with mass cytometry (CyTOF) to investigate the phenotypic characteristics of CCR7+ CD8+ memory T cells specific for acute (influenza virus or rotavirus) or chronic viruses (cytomegalovirus [CMV] or Epstein-Barr virus [EBV]) in healthy donors (n = 3) and HIV+ patients (n = 2). Signature markers of TPEX cells, namely PD-1, TIGIT, GZMK, GZMA, and CCR5, were expressed at higher levels among EBV-specific and, to a lesser extent, CMV-specific versus influenza virus-specific and rotavirus-specific CD8+ T cells (Fig. 5b,c and Supplementary Fig. 5a,b). Chronic virus-specific CD8+ T cells also overexpressed 2B4. Moreover, high-avidity CMV-specific CD8+ T cell populations, selectively identified using a double point-mutated peptide-HLA class I tetramer46, incorporated TPEX cells at frequencies equivalent to those detected among the corresponding total CMV-specific CD8+ T cell populations, suggesting that persistent antigenic drive rather than signal strength determined the acquisition and maintenance of dysfunctional, exhausted-like traits43 (Supplementary Fig. 5c).
In further experiments, we used a high-throughput approach (TCR-seq) to profile the clonotypic repertoires of TSTEM and TPEX cells. As expected, these early differentiated subsets exhibited similarly diverse repertoires, measured via the normalized Shannon-Weiner index, whereas the corresponding TEM subsets exhibited comparatively less diverse repertoires (Fig. 5d). An additional estimator based on abundance, the Chao1 index, which accounts for the distribution of infrequent clonotypes, revealed similar trends and further identified greater levels of diversity among the TSTEM subsets compared with the TPEX subsets, potentially reflecting distinct broadness of specificities (Supplementary Fig. 5d). Although the stem-like subsets both shared clonotypes with the corresponding TEM subsets, minimal repertoire overlap was detected between TSTEM and TPEX cells, quantified in terms of the absolute numbers (Fig. 5e) or normalized counts of shared clonotypes (Fig. 5f), suggesting distinct spectra of recognized antigens. Comparable data were obtained using two additional metrics, F2, which accounts for the size of each clonotype (Supplementary Fig. 5e), and R, which estimates correlations of clonotype frequencies (Supplementary Fig. 5f).
Collectively, these data revealed that TSTEM and TPEX cells were clonally distinct and committed to parallel differentiation programs, the relative prevalence of which was determined by the dynamics of antigen exposure within any given specificity (Fig. 5g).
Discussion
In this study, we used an unbiased approach guided by scRNA-seq to capture the extensive heterogeneity that exists in the human CD8+ memory T cell pool under physiological conditions. We identified two previously unrecognized subsets of stem-like CD8+ memory T cells, neither of which corresponded with previous descriptions of early differentiated progenitors based on the expression of CCR7, CD45RA/RO, and CD95. These subsets were defined by core transcriptional signatures that could be distilled phenotypically into simple profiles, namely CCR7+ PD-1− TIGIT− (TSTEM cells) and CCR7+ PD-1+ TIGIT+ (TPEX cells). Moreover, the distinct gene expression profiles of TSCM and TCM cells were mostly attributable to the differential inclusion of TPEX cells, indicating a need for refined models to understand the process of human CD8+ memory T cell differentiation.
TSTEM cells proliferated vigorously in response to activation and generated a diverse array of memory and effector progeny, collectively enabling functionally superior immunity in vivo. Of note, the ex vivo frequencies of TSTEM cells were sufficient to overcome current limitations associated with the relative paucity of TSCM cells, potentially facilitating immunotherapies that rely on specificity redirection by providing an alternative source of progenitors with self-renewal capabilities and a propensity for effector differentiation. In contrast, TPEX cells were committed to the generation of progeny with reduced functionality and proliferated less efficiently in response to activation, at least via the TCR. Importantly, TSTEM and TPEX cells were also clonally, epigenetically, and transcriptionally distinct, suggesting a branching point in the early memory compartment associated with the initial antigen recognition event(s). This interpretation was supported by the observation that persistent antigenic stimulation was preferentially associated with the development of TPEX cells. On the basis of these findings, we propose a revised model of T cell differentiation, according to which TPEX cells become hardwired to a dysfunctional-like signature after immune activation and effector differentiation, compatible with the generation of a parallel lineage47 defined by genome-wide epigenetic modifications48,49, whereas TSTEM cells remain multipotent and relatively resistant to exhaustion, resulting in enhanced functionality and protective immunity in vivo. This model will likely need further refinement to accommodate a degree of plasticity within the TSTEM subset, given that initially functional CD8+ memory T cells can become exhausted as a result of continuous exposure to high-dose antigen in mice chronically infected with LCMV50.
The acquisition of dysfunctional traits associated with exhaustion was not accompanied by a substantial loss of memory-like features in the TPEX subset. Instead, these characteristics were found to coexist in individual cells, potentially indicating functional adaptation to persistent antigenic stimulation3,16. Such adaptations may be necessary in this context to maintain a diverse repertoire and simultaneously minimize the risk of immunopathology28,30,47. Further studies will nonetheless be required to address these issues in patients with progressive malignancies or uncontrolled viral infections. In contrast, activated TSTEM cells coexpressed stem-like and effector genes, consistent with the notion of a functionally sustainable hybrid state3. Accordingly, we propose that TSTEM cells represent a naturally occurring lineage with optimal features for the induction of potent long-term immunity.
In summary, we have identified two subsets of human stem-like CD8+ memory T cell progenitors with distinct fate commitments and lineage relationships. Although further work is required to characterize the molecular mechanisms that underlie the early dichotomy between TSTEM and TPEX cells, we anticipate that such efforts will reveal novel targets for therapeutic interventions designed to inhibit or reverse the process of exhaustion, with obvious implications for the treatment of persistent infections and various cancers.
Methods
Study approval
The use of human samples was approved by the Humanitas Clinical and Research Center Institutional Review Board under the following protocols: buffy coats from healthy donors (28/01/2016), LNs and PB from patients with head and neck cancer (700/2010), adjacent cancer-free lung tissue and PB from patients with non-small cell lung cancer (1501), and BM and PB from healthy donors (1397). Healthy and HIV+ donors from the Fred Hutchinson Cancer Research Center were obtained via the HIV Vaccine Trials Network (HVTN). All donors provided written informed consent in accordance with the Declaration of Helsinki. Mice were housed and bred in a specific pathogen-free animal facility and treated in accordance with the European Union Guideline on Animal Experiments. Mouse protocols were approved by the Italian Ministry of Health, the Humanitas Institutional Animal Care and Use Committee (256/2015-PR), and the San Raffaele Institutional Animal Care and Use Committee (646).
Cells
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats via density gradient separation and either used fresh or cryopreserved in fetal bovine serum (FBS) supplemented with 10% dimethyl sulfoxide (DMSO). Tissue samples were processed as described previously16,51,52. Total CD8+ T cells were enriched via negative magnetic separation using an EasySep Human CD8+ T Cell Isolation Kit (Stem Cell Technologies) or a MojoSort Human CD8+ T Cell Isolation Kit (BioLegend). Total CD3+ T cells were enriched via negative magnetic separation using a MojoSort Human CD3+ T Cell Isolation Kit (BioLegend). CD8-depleted PBMCs were obtained via negative magnetic separation using CD8 MicroBeads (Miltenyi Biotec). The human NALM-6 cell line (DSMZ) was tested for Mycoplasma (Eurofins Genomics) and transduced with a lentiviral vector encoding secreted luciferase (Lucia+/NGFR+/NALM-6)53.
Flow cytometry and cell sorting
High-dimensional flow cytometry was performed as described previously54. Dead cells were excluded from all analyses using Zombie Aqua (BioLegend). Fluorochrome-conjugated monoclonal antibodies were purchased from commercial vendors (Supplementary Table 6). All reagents were titrated prior to use to determine optimal concentrations. Cells were fixed/permeabilized for intracellular analyses using a Cytofix/Cytoperm Kit (BD Biosciences). Transcription factors and intranuclear molecules were measured in conjunction with a FoxP3 Transcription Factor Staining Buffer Set (eBioscience) or a Transcription Factor Buffer Set (BD Biosciences). Cell proliferation was determined by measuring the progressive dilution of CFSE (Thermo Fisher Scientific). The proliferation index (PI) was calculated as follows: (MFI of the non-proliferating fraction / MFI of the proliferating fraction) × (% cells with diluted CFSE)16. Samples were acquired using a FACSymphony A5 or an LSR Fortessa equipped with FACSDiva software version 8.0.1 (all from BD Biosciences). Electronic compensation was performed using single-stained controls prepared with antibody-capture beads (BD Biosciences)54. T cell subsets were sorted to purity using a FACSAria III (BD Biosciences) as shown in Supplementary Fig. 1a.
Flow cytometry standard (FCS) 3.0 files were imported into FlowJo software version 9 (FlowJo LLC). A conventional gating strategy was used to remove aggregates and dead cells, and 5,000 CD95+ bulk CD8+ memory T cells per sample (Supplementary Table 1) were exported into FlowJo software version 10 (FlowJo LLC). Data were then biexponentially transformed and exported for further analysis in Python version 3.7.3 using a custom-written script incorporating PhenoGraph retrieved from the scikit-learn package (https://github.com/luglilab/Cytophenograph). Tissue samples were labeled with a unique computational barcode for further identification, converted into comma separated (CSV) files, and concatenated in a single matrix using the merge function in pandas (https://pandas.pydata.org/). The K value, indicating the number of nearest neighbors identified in the first iteration of the algorithm, was set to 1,000. UMAP was retrieved from Python. Data were visualized using FlowJo version 10 (FlowJo LLC).
Peptide-HLA class I tetramers
Biotinylated wildtype and D227K/T228A (KA) HLA-A*0201 complexes refolded with CMV pp65495–503 NLVPMVATV (NV9) were multimerized with streptavidin-PE (Sigma-Aldrich) as described previously52. Cells were stained with each tetramer at a concentration of 5 μg/ml for 15 min at 37°C.
Mass cytometry by time of flight (CyTOF)
Cryopreserved PBMCs from three healthy donors and two HIV+ donors were obtained from the HVTN. Purified monoclonal antibodies were purchased from commercial vendors (Supplementary Table 7) and labeled according to the Maxpar Antibody Labeling Kit Protocol (Fluidigm). Streptavidin was produced and labeled as described previously55. Myc-tagged peptide-HLA class I monomers were synthesized and biotinylated as described previously (Supplementary Table 7)56. Peptide-HLA class I tetramers were generated via the addition of heavy metal-labeled streptavidins in a triple coding scheme and used to stain cells in a cocktail format as described previously55. All reagents were titrated prior to use to determine optimal concentrations. Antibody staining, live/dead discrimination, and DNA staining were performed as described previously57.
Cell culture and stimulation conditions
Cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin, and 2 mM L-glutamine. To induce cytokine production, magnetically-enriched CD8+ T cells were stimulated for 3 h with phorbol 12-myristate 13-acetate (PMA, 10 ng/ml) and ionomycin (500 ng/ml) (both from Sigma-Aldrich) in the presence of anti-CD107a (clone H4A3, BD Biosciences) and the protein transport inhibitors GolgiPlug (brefeldin A, 1 μl/ml, BD Biosciences) and GolgiStop (monensin, 0.67 μl/ml, BD Biosciences). Subsets were identified among bulk CD8+ memory T cells by gating as specified in the legend to Fig. 3f. Alternatively, FACS-purified CD8+ T cell subsets were stimulated for 12 h with anti-CD3 plus CD28 DynaBeads (bead-to-cell ratio 1:2, Thermo Fisher Scientific). To evaluate the expression of activation markers and T-bet, FACS-purified CD8+ T cell subsets were mixed with CD8-depleted autologous PBMCs (cell-to-cell ratio 1:4) and stimulated for 24 h with SEB (1 μg/ml, Sigma-Aldrich). To evaluate differentiation and proliferation, FACS-purified CD8+ T cell subsets were stimulated for 3 or 4 d with anti-CD3 plus CD28 DynaBeads (bead-to-cell ratio 1:2, Thermo Fisher Scientific) in combination with various cocktails of human cytokines, including TGF-β, IL-2, IL-7, IL-12, and IL-15 (each at 10 ng/ml, Peprotech). To evaluate self-renewal capacity, FACS-purified CD8+ T cell subsets were stimulated for 10 d with IL-15 (25 ng/ml, Peprotech). Unstimulated samples were used as controls in all assays.
scRNA-seq
FACS-purified CD95+ CD8+ T cells were resuspended in 1 ml of PBS−/− containing 0.04% bovine serum albumin, washed twice by centrifugation at 450 rcf for 7 min, resuspended in 100 μl of the same medium, and counted using a Countess II Automatic Cell Counter (Thermo Fisher Scientific). Approximately 20,000 cells from each sample were then loaded into one channel of Single Cell Chip A using a Chromium Single Cell 3’ v2 Reagent Kit (10X Genomics). After capture and lysis, cDNA was synthesized and amplified over 14 cycles according to the manufacturer’s protocol (10X Genomics). Libraries were prepared from 50 ng of amplified cDNA. Sequencing was performed using a NovaSeq 6000 System (Illumina). An average sequencing depth of at least 50,000 reads/cell was obtained for each sample.
scRNA-seq data analysis
Sample demultiplexing, barcode processing, and unique molecular identifier (UMI) counting were performed using Cell Ranger version 2.1.1 (10X Genomics). Briefly, raw base call files were demultiplexed in FASTQ format using the “cellranger mkfastq” pipeline, and the “cellranger count” pipeline was run with “--transcriptome=refdata-cellranger-GRCh38-1.2.0” for each sample. Outputs from “cellranger count” were concatenated in a single matrix. Libraries were then normalized to an identical sequencing depth using the “cellranger aggr” pipeline. Pooled data were imported into R version 3.5.1 using Seurat version 3.0.1 (ref. 58). Genes detected in less than three cells or cells containing less than 200 features were excluded from the analysis. Cells with unique feature counts less than 200 or greater than 3,500 were also filtered out, along with cells containing mitochondrial counts above 10%. The resulting dataset was normalized using a global scaling method converted by a scale factor (10,000) and log-transformed using the “ScaleData” function in Seurat version 3.0.1. Data were then subjected to cluster analysis using standard package procedures and the “FindClusters” function in Seurat version 3.0.1. Parameters were set to the first 20 principal components and a resolution of 0.6. DEGs for each cluster were identified using the Wilcoxon rank sum test with default correction for multiple comparisons in Seurat version 3.0.1.
Anchor gene analysis of memory cell clusters from scRNA-seq data
Six different gene modules were computed from the scRNA-seq dataset using selected genes as anchors. Transcriptional scores were built by calculating the mean expression profiles of the top 100 genes most correlated with the anchors (Pearson’s correlation).
Bulk RNA-seq
RNA was extracted from 50,000 FACS-purified CD8+ T cells per subset using a Direct-Zol RNA Microprep Kit (Zymo Research) and stored at −80°C. Quality control was performed using a High Sensitivity RNA ScreenTape Assay with a 4200 TapeStation System (Agilent). Libraries for mRNA sequencing were prepared from 5 ng of total RNA using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech-Takara). Full-length cDNAs were processed using a Nextera XT DNA Library Preparation Kit (Illumina). Quality control was performed using a High Sensitivity DNA ScreenTape Assay with a 4200 TapeStation System (Agilent). Libraries were then multiplexed in an equimolar pool and sequenced using a NextSeq 500/550 Platform (Illumina). An average of 11 million single-end 75 base pair (bp) reads were generated per sample. Libraries for total RNA sequencing were prepared from 1 ng of total RNA using a SMART-Seq Stranded Kit (Clontech-Takara). Quality control and sequencing were performed as described for the mRNA libraries, generating an average of 103 million paired-end 75 bp reads per sample.
Bulk RNA-seq data analysis
Raw sequence data were quality-controlled using FastQC version 0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Single-end reads were aligned to the human genome (GENCODE Human Release 29, reference genome sequence GRCh38/hg38) using STAR version 2.5.1b (ref. 59). Alignments were performed using default parameters. Reads associated with annotated genes were counted using the STAR aligner option “-quantMode geneCounts”. Differential gene expression was assessed using the edgeR package version 3.20.9. Benjamini-Hochberg correction was applied to estimate the false discovery rate (FDR). Paired-end reads were processed similarly after removing adapter sequences and poor-quality bases with Trimmomatic version 0.36.
Overrepresentation analysis
GSEA was applied to the entire list of genes in the RNA-seq expression matrix. Genes were ranked based on log2 fold changes calculated using the edgeR package version 3.20.9. GSEA was performed in preranked mode using a “classic” enrichment statistic. Gene sets of interest were retrieved from collections C2 and C7 in the Molecular Signatures Database version 6.2 and integrated with those corresponding to exhausted T cell clusters G6 and G9 in Sade-Feldman et al. 18 or with those obtained via a reanalysis of the dataset in Akondy et al. 45.
Enrichment analysis
Normalized scRNA-seq counts were downloaded from the Gene Expression Omnibus (GSE120575). Analysis was restricted to cells belonging to clusters G5 or G10 as defined in Sade-Feldman et al. 18. DEGs with an adjusted P value < 0.01 in the pairwise comparisons of G5 versus G10 were identified using the “FindAllMarkers” function in Seurat version 3.0.1. Hypergeometric tests were used to compare the G5 or G10 signatures with the combined TSCM/TCM or TPEX signatures in “phyper” R.
Microarray data analysis
Normalized data matrices from Akondy et al. 45 were downloaded from GEO: GSE26347. To identify the signatures “YF_naive vs. effector UP” and “YF_effector vs. naive UP”, the expression profiles of effector CD8+ T cells isolated 14 d after vaccination with YF-17D (GSM837587, GSM837588, GSM837589, and GSM837590) were compared with those of naive CD8+ T cells (GSM837584, GSM837585, and GSM837586) using the limma algorithm in R version 3.34.9 (ref. 60). The gene set of interest arbitrarily included the top 200 genes with the highest log fold change among DEGs with an adjusted P value < 0.05. All samples hybridized on the Human Genome U133 Plus 2.0 Array (the second set of samples from the GSE26347 dataset) were concatenated with those hybridized on the HT Human Genome U133A Array (the third set of samples from the GSE26347 dataset). Probe sets were matched on both chips. Batch effects were eliminated by adjusting gene expression values in the combined data matrix with the empirical Bayes method ComBat in SVA version 3.26.0 (ref. 61). ComBat was applied with default parameters, except for the adjustment variables, which were imputed as a vector of platform type labels. To identify genes comprising the “YFV-specific memory cell signature” shown in Fig. 5a, the expression profiles of YF-17D virus NS4B-214 epitope-specific human CD8+ memory T cells (GSM837594, GSM837595, GSM837596, GSM837597, GSM837598, and GSM837599) were compared with those of YFV-tetramer+ effector CD8+ T cells (GSM837587, GSM837588, GSM837589, and GSM837590) using the limma algorithm, arbitrarily selecting the top 200 genes with the highest log fold change among DEGs with an adjusted P value < 0.05.
ATAC-seq
Libraries were prepared using a protocol adapted from Buenrostro et al. 62. Briefly, 50,000 FACS-purified CD8+ T cells per subset were washed in PBS−/− and resuspended in 50 μl of lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM MgCl2, 0.1% IPEGAL CA-630). Nuclei were pelleted by centrifugation for 10 min at 500 g and resuspended in a final reaction volume of 50 μl comprising 1 μl of Tn5 transposase (made in-house), 10 μl of 5x transposase buffer (50 mM Tris-HCl pH 8.4, 25 mM MgCl2), and 39 μl of ultrapure water (Milli-Q). The reaction was incubated with mixing at 300 rpm for 30 min at 37°C, supplemented with 10 μl of clean-up buffer (900 mM NaCl, 30 mM EDTA), 5 μl of 20% SDS, 0.7 μl of ultrapure water (Milli-Q), and 4.3 μl of proteinase K (18.6 μg/μl, Thermo Fisher Scientific), and incubated for a further 30 min at 40°C. Tagmented DNA was isolated using 2x SPRI Beads (Beckman Coulter) and amplified via PCR. Fragments smaller than 600 bp were isolated via negative size selection using 0.65x SPRI Beads (Beckman Coulter) and purified using 1.8x SPRI Beads (Beckman Coulter). Quality control was performed using a 4200 TapeStation System (Agilent) in conjunction with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific). Libraries were then multiplexed in an equimolar pool and sequenced using a NextSeq 500/550 Platform (Illumina). At least 20 million single-end 75 bp reads were generated per sample.
ATAC-seq data analysis
Read quality was assessed using FastQC version 0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Adaptors and poor-quality bases were trimmed using Cutadapt version 1.16 (ref. 63). Samples were aligned to the human reference genome GRCh38 using default parameters in BWA-MEM version 0.7.17. Mitochondrial reads were removed using SAMtools version 1.9 (ref. 64). PCR duplicates were removed using the “MarkDuplicates” function in Picard Tools version 2.19 (http://broadinstitute.github.io/picard/). Open chromatin was detected using MACS2 version 2.1.2 (ref. 65) with an FDR < 0.01. The number of reads in each peak was determined using featureCounts version 1.6.4 (ref. 66). Differentially accessible peaks were identified using an FDR cut-off below 0.05 after normalization in DESeq2 version 1.20 (Bioconductor). Peaks were annotated using the “annotatePeaks.pl” function and scanned for motifs using the “findMotifsGenome.pl” function in HOMER version 4.9.1.
Single telomere length analysis
DNA was extracted from 6,000 FACS-purified CD8+ T cells per subset using a QIAamp DNA Micro Kit (Qiagen). Single telomere length analysis was carried out at the XpYp telomere as described previously67. Briefly, genomic DNA was eluted in 35 μl of Tris (10 mM) containing 0.75 μl of the Telorette-2 linker (10 μM). Multiple PCRs were then performed for each test DNA. Each reaction was set up in a final volume of 10 μl containing 1 μl of DNA and 0.5 μM of the telomere-adjacent and teltail primers in 75 mM Tris-HCl pH 8.8, 20 mM (NH4)2SO4, 0.01% Tween-20, and 1.5 mM MgCl2, with 0.5 U of a 10:1 mixture of Taq (Thermo Fisher Scientific) and Pwo polymerase (Roche Molecular Biochemicals). The reactions were processed in a Tetrad2 Thermal Cycler (Bio-Rad). DNA fragments were resolved via 0.5% Tris-acetate-EDTA agarose gel electrophoresis and identified via Southern hybridization with a random-primed anti-32P-labeled (PerkinElmer) TTAGGG repeat probe, together with probes specific for molecular weight markers at 1 kb (Stratagene) and 2.5 kb (Bio-Rad). Hybridized fragments were detected using a Typhoon FLA 9500 Phosphorimager (GE Healthcare). The molecular weights of the DNA fragments were calculated using a Phoretix 1D Quantifier (Nonlinear Dynamics).
TCR-seq
Total RNA was extracted separately from duplicate vials of 150,000 FACS-purified CD8+ T cells per subset using an RNeasy Mini Kit (Qiagen). UMI-labeled 5′ RACE TCRβ sequencing libraries were prepared using a Human TCR Profiling Kit (MiLaboratory LLC). Libraries were prepared in parallel using the same number of PCR cycles and sequenced using a NextSeq 500/550 High-Output Kit with a NextSeq 500 Platform (Illumina). Approximately 3 × 107 reads were obtained in total and assembled into 2.2 × 106 UMI-labeled cDNA molecules (up to 105 per library). UMI extraction and consensus assembly were performed using MIGEC software version 1.2.9 (ref. 68) with a threshold of at least three reads per UMI. In-frame CDR3β repertoires were extracted using MiXCR software version 3.0.3 (ref. 69). Each library contained from 3,000 to 18,000 functional CDR3β clonotypes. Diversity metrics were calculated using VDJtools software version 1.2.1 (ref. 70) after normalization to 42,000 randomly selected UMIs per sample. D, R, and F2 metrics were calculated for the top 3,000 clones from each pair of samples using VDJtools software version 1.2.1 (ref. 70).
T cell transduction and culture conditions
FACS-purified CD8+ T cell subsets were stimulated with a MACS-GMP T Cell TransAct Cocktail (Miltenyi Biotec). Stimulated cells were transduced the following day with a bidirectional lentiviral vector encoding a CD19-specific CAR with a CD28 costimulus in sense and the LNGFR marker gene in antisense and then cultured for 13 d in TexMACS Medium (Miltenyi Biotec) supplemented with 3% FBS, 1% penicillin/streptomycin, IL-7 (25 U/ml, Miltenyi Biotec), and IL-15 (50 U/ml, Miltenyi Biotec). Magnetically purified CD3+ cells were processed similarly for control purposes.
Mouse studies
TSTEM and TPEX cells were isolated from the PB of healthy donors based on differential expression of PD-1. Eight-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Charles River) were infused retroorbitally with FACS-purified TSTEM, TPEX or TEM cells (1 × 106 per mouse) and autologous CD8− PBMCs (6 × 106 per mouse) and sacrificed on day 28. To maximize recovery, spleen and lung cells were mixed from the same experimental group, normalized in terms of the CD4 to CD8 ratio, and injected as above into secondary NSG recipients (1 × 106 CD8+ T cells per mouse). Spleens were harvested on day 28 and processed to single cell suspensions. Absolute numbers of T cells in blood were determined using CountBright Absolute Counting Beads (Thermo Fisher Scientific). The frequencies of human CD4+ and CD8+ T cell subsets were determined by flow cytometry. For tumor experiments, eight-week-old female NSG mice were injected intravenously with 0.5 × 106 Lucia+/NGFR+/NALM-6 cells. After 4 d, mice were further injected with 3 × 106 CAR19-redirected TSTEM, TPEX, or total CD3+ T cells. Untransduced CD3+ T cells were used as controls. Tumor progression was monitored weekly via bioluminescence detection using QUANTI-Luc (InvivoGen) and expressed as relative light units (RLUs).
Statistics
Statistical analyses were performed using Prism version 7.0c (GraphPad) or R software version 3.4.4. Significance was assigned at P < 0.05 unless stated otherwise. Specific tests are indicated in the relevant figure legends.
Supplementary Material
Acknowledgements
The authors thank G. Natoli (European Institute of Oncology, Milan) for assistance with the ATAC-seq protocol, R. Roychoudhuri (University of Cambridge) and M. Iannacone (San Raffaele Scientific Institute, Milan) for critical discussions, and G. Cugini and G. Colombo (Humanitas Clinical and Research Center, Milan) for the provision of lymph node samples. This work was funded by the European Research Council (ERC_StG_2014 PERSYST #640511 to E.L.). Additional support was provided by the Associazione Italiana per la Ricerca sul Cancro (AIRC IG 21567 to D.M.), the Italian Ministry of Health (Bando Ricerca Finalizzata PE-2016-02363915 to D.M.), the Intramural Research Fund of the Humanitas Clinical and Research Center (5 X 1000 2019 Program to D.M.), and Cancer Research UK (C17199/A18246/A29202 to D.M.B.). G.G., G.D.S., S.P., and E.S. were supported by Fellowships from the Fondazione Italiana per la Ricerca sul Cancro-Associazione Italiana per la Ricerca sul Cancro (FIRC-AIRC). A.N.D. and M.M. were supported by the Ministry of Education, Youth, and Sports of the Czech Republic (CEITEC 2020 LQ1601). D.A.P. was supported by a Wellcome Trust Senior Investigator Award (100326/Z/12/Z). D.M.C. was supported by the Ministry of Health of the Russian Federation (0908300057056). The purchase of a FACSSymphony A5 was defrayed in part by a grant from the Italian Ministry of Health (Agreement 82/2015).
Footnotes
Author contributions
G.G. and E.L. conceived the study; G.G., G.D.S., E.M.C.M., S.P., C.M., T.M.B., A.N.D., M.M., E.S., G.A., F.D.P., V.Z., A.S., B.C., F.S.C., A.A., C.P., S.P., L.G., R.E.J., D.M.B., E.G., S.L.L., and K.L. performed experiments; G.G., G.D.S., E.M.C.M., S.P., T.M.B., A.N.D., D.M.B., D.M.C., E.W.N., M.C., and E.L. analyzed data; D.M., S.K.B., B.A.Y., and D.A.P. provided critical expertise and resources; E.L. supervised the study; G.G., D.A.P., and E.L. wrote the manuscript. All authors contributed intellectually and approved the manuscript.
Competing Interests
The Laboratory of Translational Immunology receives reagents in kind as part of a collaborative research agreement with BD Biosciences (Italy). L.G. and E.L. are inventors on a patent describing methods for the generation and isolation of TSCM cells. L.G. has consulting agreements with Lyell Immunopharma and Advaxis Immunotherapies. E.W.N. is a cofounder and advisor for ImmunoScape Pte. Ltd. The other authors have no conflicts of interest to disclose.
Reporting summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Publicly available scRNA-seq data were retrieved from the Gene Expression Omnibus via accession code GSE120575 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120575). Microarray data from YFV-17D-specific CD8+ T cells were retrieved from the Gene Expression Omnibus via accession code GSE26347 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26347). Gene sets of interest were retrieved from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp). The ATAC-seq data reported in this paper are available on request. The bulk RNA-seq and scRNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus under accession code GSE147398. The TCR-seq data reported in this paper have been deposited at the European Bioinformatic Institute under accession code E-MTAB-8892. All other data that support the findings of this study are available on request from the corresponding author.
Code availability
Scripts used to analyze the ATAC-seq data are available at https://github.com/luglilab/SP018_CD8_Galletti_et_al. All other codes are available on request.
References
- 1.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 2.Gattinoni L, Speiser DE, Lichterfeld M, Bonini C. T memory stem cells in health and disease. Nat Med. 2017;23:18–27. doi: 10.1038/nm.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lugli E, Galletti G, Boi SK, Youngblood BA. Stem, effector, and hybrid states of memory CD8+ T cells. Trends Immunol. 2020;41:17–28. doi: 10.1016/j.it.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biasco L, et al. In vivo tracking of T cells in humans unveils decade-long survival and activity of genetically modified T memory stem cells. Sci Transl Med. 2015;7:273ra13. doi: 10.1126/scitranslmed.3010314. [DOI] [PubMed] [Google Scholar]
- 6.Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. The who’s who of T-cell differentiation: human memory T-cell subsets. Eur J Immunol. 2013;43:2797–2809. doi: 10.1002/eji.201343751. [DOI] [PubMed] [Google Scholar]
- 7.Graef P, et al. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8+ central memory T cells. Immunity. 2014;41:116–126. doi: 10.1016/j.immuni.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 9.Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol. 2012;86:8161–8170. doi: 10.1128/JVI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schietinger A, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 2016;45:389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philip M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leong YA, et al. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 2016;17:1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 15.Utzschneider DT, et al. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity. 2016;45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J Exp Med. 2018;215:2520–2535. doi: 10.1084/jem.20180684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He R, et al. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature. 2016;537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 18.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013.:e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddiqui I, et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50:195–211.:e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 20.Miller BC, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 22.Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 23.Lugli E, et al. Superior T memory stem cell persistence supports long-lived T cell memory. J Clin Invest. 2013;123:594–599. doi: 10.1172/JCI66327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberdoerffer S, et al. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lugli E, et al. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat Protoc. 2013;8:33–42. doi: 10.1038/nprot.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephen TL, et al. SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity. 2017;46:51–64. doi: 10.1016/j.immuni.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao C, et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat Immunol. 2019;20:890–901. doi: 10.1038/s41590-019-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alfei F, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571:265–269. doi: 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 29.Scott AC, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571:270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan O, et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 2019;571:211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J Hepatol. 2019;71:731–741. doi: 10.1016/j.jhep.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Seo H, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc Natl Acad Sci. 2019;116:12410–12415. doi: 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, He Y, Hao J, Ni L, Dong C. High levels of Eomes promote exhaustion of anti-tumor CD8+ T cells. Front Immunol. 2018;9:2981. doi: 10.3389/fimmu.2018.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giordano M, et al. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 2015;34:2042–2058. doi: 10.15252/embj.201490786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo T, et al. Notch-mediated conversion of activated T cells into stem cell memory-like T cells for adoptive immunotherapy. Nat Commun. 2017;8:15338. doi: 10.1038/ncomms15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Widjaja CE, et al. Proteasome activity regulates CD8+ T lymphocyte metabolism and fate specification. J Clin Invest. 2017;127:3609–3623. doi: 10.1172/JCI90895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang ZZ, et al. TGF-β upregulates CD70 expression and induces exhaustion of effector memory T cells in B-cell non-Hodgkin’s lymphoma. Leukemia. 2014;28:1872–1884. doi: 10.1038/leu.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vodnala SK, et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science. 2019;363:eaau0135. doi: 10.1126/science.aau0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henning AN, Roychoudhuri R, Restifo NP. Epigenetic control of CD8+ T cell differentiation. Nat Rev Immunol. 2018;18:340–356. doi: 10.1038/nri.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doering TA, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thommen DS, et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utzschneider DT, et al. High antigen levels induce an exhausted phenotype in a chronic infection without impairing T cell expansion and survival. J Exp Med. 2016;213:1819–1834. doi: 10.1084/jem.20150598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuertes Marraco SA, et al. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci Transl Med. 2015;7:282ra48. doi: 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- 45.Akondy RS, et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price DA, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blank CU, et al. Defining ‘T cell exhaustion’. Nat Rev Immunol. 2019;19:665–674. doi: 10.1038/s41577-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghoneim HE, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170:142–157.:e19. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West EE, et al. Tight regulation of memory CD8+ T cells limits their effectiveness during sustained high viral load. Immunity. 2011;35:285–298. doi: 10.1016/j.immuni.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lugli E, et al. IL-15 delays suppression and fails to promote immune reconstitution in virally suppressed chronically SIV-infected macaques. Blood. 2011;118:2520–2529. doi: 10.1182/blood-2011-05-351155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberto A, et al. Role of naive-derived T memory stem cells in T-cell reconstitution following allogeneic transplantation. Blood. 2015;125:2855–2864. doi: 10.1182/blood-2014-11-608406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falcone L, Casucci M. Exploiting secreted luciferases to monitor tumor progression in vivo. Methods Mol Biol. 2016;1393:105–111. doi: 10.1007/978-1-4939-3338-9_10. [DOI] [PubMed] [Google Scholar]
- 54.Brummelman J, et al. Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nat Protoc. 2019;14:1946–1969. doi: 10.1038/s41596-019-0166-2. [DOI] [PubMed] [Google Scholar]
- 55.Newell EW, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakker AH, et al. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc Natl Acad Sci. 2008;105:3825–3830. doi: 10.1073/pnas.0709717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simoni Y, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 58.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- 64.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 67.Capper R, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 69.Bolotin DA, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 70.Shugay M, et al. VDJtools: unifying post-analysis of T cell receptor repertoires. PLoS Comput Biol. 2015;11:e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available scRNA-seq data were retrieved from the Gene Expression Omnibus via accession code GSE120575 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120575). Microarray data from YFV-17D-specific CD8+ T cells were retrieved from the Gene Expression Omnibus via accession code GSE26347 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26347). Gene sets of interest were retrieved from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp). The ATAC-seq data reported in this paper are available on request. The bulk RNA-seq and scRNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus under accession code GSE147398. The TCR-seq data reported in this paper have been deposited at the European Bioinformatic Institute under accession code E-MTAB-8892. All other data that support the findings of this study are available on request from the corresponding author.
Scripts used to analyze the ATAC-seq data are available at https://github.com/luglilab/SP018_CD8_Galletti_et_al. All other codes are available on request.