Abstract
Machine learning methods have been employed to make predictions in psychiatry from genotypes, with the potential to bring improved prediction of outcomes in psychiatric genetics; however, their current performance is unclear. We aim to systematically review machine learning methods for predicting psychiatric disorders from genetics alone and evaluate their discrimination, bias and implementation. Medline, PsychInfo, Web of Science and Scopus were searched for terms relating to genetics, psychiatric disorders and machine learning, including neural networks, random forests, support vector machines and boosting, on 10 September 2019. Following PRISMA guidelines, articles were screened for inclusion independently by two authors, extracted, and assessed for risk of bias. 63 full texts were assessed from a pool of 652 abstracts. Data were extracted for 77 models of schizophrenia, bipolar, autism or anorexia across 13 studies. Performance of machine learning methods was highly varied (0.48-0.95 AUC) and differed between schizophrenia (0.54-0.95 AUC), bipolar (0.48-0.65 AUC), autism (0.52-0.81 AUC) and anorexia (0.62-0.69 AUC). This is likely due to the high risk of bias identified in the study designs and analysis for reported results. Choices for predictor selection, hyperparameter search and validation methodology, and viewing of the test set during training were common causes of high risk of bias in analysis. Key steps in model development and validation were frequently not performed or unreported. Comparison of discrimination across studies was constrained by heterogeneity of predictors, outcome and measurement, in addition to sample overlap within and across studies. Given widespread high risk of bias and the small number of studies identified, it is important to ensure established analysis methods are adopted. We emphasise best practices in methodology and reporting for improving future studies.
Keywords: Machine learning, systematic review, SNPs, polygenic risk score, AUC, psychiatric disorder
Introduction
Machine learning represents a contrasting approach to traditional methods for genetic prediction. It has increased in popularity in recent years following breakthroughs in deep learning [1–4], and the scaling-up of datasets and computing power. The ability to function in high dimensions and detect interactions between loci [5] without assuming additivity makes such methods an attractive option in statistical genetics, where the effects of myriad factors on an outcome is difficult to pre-specify. Calls to address the complexity of disorders like schizophrenia with machine learning have also become more frequent [6–8]. However, the predictive performance of machine learning methods in psychiatric genetics is unclear, and a recent review of clinical prediction models across various outcomes and predictors found them to be no more accurate than logistic regression [9]; it is therefore timely to review their predictive performance in psychiatry.
Genome-wide association studies, genetic prediction and psychiatry have each been reviewed with respect to machine learning [10–16]. Recently, single nucleotide polymorphism (SNP)-based prediction has been reviewed across diseases [17]. However, psychiatry presents a distinct problem from somatic and neurological diseases as a result of genetic correlation between disorders [18] and the risk of class mislabelling due to biological heterogeneity that may underlie symptom-based diagnoses [19].
We systematically reviewed literature related to the question: what is the ability of machine learning (ML) methods to predict psychiatric disorders using only genetic data? We report discrimination, methodology and potential bias for diagnostic or prognostic models and compare to logistic regression (LR) and polygenic risk scores (PRS) where available.
Materials and methods
Search Strategy
Medline via Ovid, PsychInfo, Web of Science and Scopus were searched for journal articles matching terms for machine learning, psychiatric disorders and genetics on 10th September 2019. Searches were broad, with terms for psychiatric disorders including schizophrenia, bipolar, depression, anxiety, anorexia and bulimia, attention-deficit hyperactivity disorder, obsessive compulsive disorder, Tourette’s syndrome or autism. Terms for machine learning were also wide-ranging, including naïve Bayes, k-nearest neighbours (k-NN), penalised regression, decision trees, random forests, boosting, Bayesian networks, Gaussian processes, support vector machines and neural networks, but excluding regression methods without penalty terms, such as logistic regression. Searches were developed and conducted by MBS and were restricted to English language journal articles on humans, with no limits on search dates. Two authors (MBS, KC) independently reviewed all abstracts for inclusion. Full texts were assessed if either author had chosen to access them and independently screened against inclusion criteria. Where conflicts occurred a third author (VEP) was consulted as an arbiter. An example search for Medline (Ovid) is given in the supplementary (Table S1).
Inclusion and Exclusion Criteria
Studies were restricted to cohort, cross-sectional or case-control designs of individuals for binary classification of a single DSM or ICD-recognised psychiatric disorder compared to unaffected individuals, where only genotyping array, exome or whole-genome sequencing data were used as predictors. Studies based solely on gene expression were excluded, but designs which made use of gene expression or functional annotations to inform models of genetic data were accepted. No further restriction was made on participants. Studies were excluded if they only predicted medication response, sub-groups within a psychiatric disorder or a psychiatric phenotype secondary to another disease. Studies were also considered ineligible if they had a clear primary aim of drawing inference at the expense of prediction, if they developed a novel statistical method or only made use of unsupervised or semi-supervised methods. The review was registered to PROSPERO in advance (registration number CRD42019128820).
Extraction and Analysis
A data extraction form was developed through discussion between all authors; items from the critical appraisal and data extraction for systematic reviews of prediction modelling studies (CHARMS) checklist [20] were included as-is or modified, and additional items were included based on expert knowledge and relevance to the review topic, with reference to the genetic risk prediction studies (GRIPS) statement [21] for items pertaining to genetic prediction studies (Table S2). The form was piloted with five publications, containing 40 extracted ML models between them, and updated before being applied to all texts.
The discrimination of machine learning methods was extracted independently by two authors (MBS, KC) as area under the receiver operating characteristic curve (AUC), or c-statistic. Model performance measures for classification by accuracy, sensitivity and specificity were also extracted. 95% confidence intervals for validation were estimated for AUC using Newcombe’s method [22]. Results were not meta-analysed due to sample overlap, present in at least half of studies (see Table S3), which cannot easily be accounted for in the meta-analysis. Information on participants, predictors and model development and validation were also obtained. LR or PRS models were also extracted when present. Though LR can be considered a machine learning approach, for the purpose of this review we regard it as a contrasting method due to its widespread use in classic statistical analysis. The presence of LR and PRS as comparators was not made a requirement due to their sparsity in the literature.
Risk of bias (ROB) and applicability were assessed using the prediction model risk of bias assessment tool (PROBAST) [23]. PROBAST consists of 20 questions designed to signal where ROB may be present in either the development or validation of a model across 4 categories: participants, predictors, outcome and analysis. These include, for instance, questions on how missingness or complexities in study design were handled. Information on handling of population structure, a common confound in genetic association studies, was also extracted to aid ROB assessment. Reporting of the systematic review follows the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines [24]. Extraction and ROB are detailed further in the supplementary.
Results
Selection
1,241 publications were identified through searches in Ovid Medline, PsychInfo, Scopus and Web of Science which included restrictions to English language journal articles (Figure S1). After merging and removing duplicates, 652 studies were assessed for inclusion. Of these, 63 full texts were assessed to determine eligibility. 14 publications were selected, with two merged as publications included the same models on the same dataset. A final total of 13 studies were selected for inclusion, containing 77 distinct machine learning models.
Studies
A wide range of machine learning methods were applied to schizophrenia (7 studies, 47% of models), bipolar disorder (5 studies, 39% of models), autism (3 studies, 10% of models) and anorexia (1 study, 4% of models) (Table 1), with no studies identified for the 6 remaining disorders. Single nucleotide polymorphisms (SNPs) were the most common source of genetic data. Copy number variants (CNVs) and PRSs were each incorporated in models from a single study, and exome-sequencing data formed the basis of two studies. Datasets typically consisted of publicly-available genome-wide association studies (GWAS); potential sample overlap was established for at least 7 studies (Table S3). Briefly, 3 studies [25–27] included controls for the 1958 Birth Cohort [28] or the UK Blood Service [29], 4 studies included controls from Knowledge Networks [25, 30–32], 2 studies used a Swedish population-based sample [32, 33], and 3 studies used the same dataset, or provided a common reference for part of the dataset [25, 30, 31]. The remaining 6 studies [34–40] either gave unclear information, reported no previous reference for the dataset, or used datasets which appear to be separate from other studies. Where samples overlap, all models included in the review are distinct, using different predictors or modelling approaches. Additional overlap or cryptic relatedness may be present between studies.
Table 1. overview of studies.
| First Author (Year) | Disorder | Machine Learning Methods | Data | Models | Comparators |
|---|---|---|---|---|---|
| Aguiar-Pulido et al. (2010; 2013)1 | Schizophrenia | AdaBoost, BFTree, DNTB, decision tables, SVM (kernel not reported), naïve Bayes, Bayesian networks, MDR, neural network (RBF, linear, perceptron), evolutionary computation | SNPs | 12 | |
| Yang et al. (2010) | Schizophrenia | AdaBoost (of SVM (RBF)), SVM (RBF) | SNPs | 2 | |
| Pirooznia et al. (2012) | Bipolar Disorder | Bayesian networks, random forest, neural network (RBF), SVM (kernel not reported) | SNPs | 16 | PRS, LR |
| Li et al. (2014) | Bipolar Disorder, Schizophrenia | LASSO, Ridge, SVM (linear) | SNPs | 6 | |
| Engchuan et al. (2015) | Autism | Neural network (perceptron), SVM (Linear), random forest, CIF | CNVs | 4 | |
| Acikel et al. (2016) | Bipolar Disorder | MDR, random forest, k-NN, naïve Bayes | SNPs | 5 | |
| Guo et al. (2016) | Anorexia nervosa | LASSO, SVM (RBF), GBM | SNPs | 3 | |
| Laksshman et al. (2017) | Bipolar Disorder | Decision tree, random forest, neural network (CNN) | Exomes | 3 | |
| Chen et al. (2018) | Schizophrenia | Neural network (perceptron) | PRS | 4 | PRS, LR |
| Wang et al. (2018) | Schizophrenia, Bipolar Disorder, Autism | Neural networks (cRBM) | SNPs, gene expression | 9 | LR |
| Ghafouri-Fard et al. (2019) | Autism | Neural network (with embedding layer) | SNPs | 1 | |
| Trakadis et al. (2019) | Schizophrenia | LASSO, random forest, SVM (kernel not reported), GBM (XGBoost) | Exomes | 4 | |
| Vivian-Griffiths et al. (2019) | Schizophrenia | SVM (linear, RBF) | SNPs | 8 | PRS |
Merged in extraction [34, 36]. BFTree: best-first decision tree, CIF: conditional inference forest, cRBM: conditional restricted Boltzmann machine, CNN: convolutional neural network, DNTB: Decision table naïve Bayes, GBM: gradient boosting machine, ?-NN: ^-nearest neighbours, LASSO: least absolute shrinkage and selection operator, LR: logistic regression, MDR: multifactor dimensionality reduction, PRS: polygenic risk score, RBF: radial basis function, SVM: support vector machine
Missingness was reported clearly in about half of all studies and models. When reported, it was most commonly handled by imputation after excluding genotypes with high missingness. Studies also reported complete-case analysis and inclusion of missing values in coding of predictors (Table S4).
Machine Learning Methods
Support vector machines (SVMs) and neural networks were the most popular, followed by random forests and boosting. SVMs were split roughly equally between using a linear kernel (3 studies, 7 models), a radial basis function (RBF) kernel (3 studies, 6 models), or an unreported kernel (3 studies, 6 models). Authors applying neural networks most commonly used multilayer perceptrons (3 studies, 6 models), an RBF network (2 studies, 5 models) or restricted Boltzmann machines (RBMs; 1 study, 9 models), with linear networks, convolutional neural networks (CNNs) and embedding layers each used once. Weak learners in boosted models were mainly decision trees, with the exception of a method which combined feature selection with the boosting of RBF-SVMs in AdaBoost [35]. Penalised regression was employed alongside linear and non-linear methods as least absolute shrinkage and selection operator (LASSO; 3 studies, 4 models) or ridge regression (1 study, 2 models). 51% of all models were implemented in R or WEKA; Matlab and Python were preferred for neural networks (Table S5).
Risk of Bias
Risk of bias was assessed for each model within each study (Figure S2). All models displayed risk of bias, mostly in relation to participants (study design and inclusion/exclusion criteria), outcome (standardised definition and assessment of outcomes) and analysis. Within-study ROB for participants was due to the use of case-control studies. Predictors were mostly rated to have unclear or low ROB; instances of high ROB were limited to predictors which are unavailable at the point of model use. Outcome definitions or measurements often differed between cases and controls.
Models displayed high ROB during analysis. This was often traced to inappropriate or unjustified handling of missingness and removal of enrolled participants prior to analysis, predictor selection using univariable methods and failure to account for overfitting. No studies reported calibration measures. In addition to PROBAST, information on population structure within studies was extracted (Table S6). Most studies did not illustrate genetic ancestry across all observations in the current publication using dimensionality reduction, and none reported any evaluation of the final trained model for bias due to population structure. However, 2 studies (18% of models) visualised principal components for a subsample or showed a table of reported ancestry for participants [31, 39]. Where ancestry was not addressed in a study, it was most often visualised in a referenced publication (55% of all models). 2 studies (13% of models) had no details or references which addressed genetic ancestry.
Across-study ROB was not formally assessed. For schizophrenia, bipolar and autism, studies with smaller numbers of cases in the development set report AUC less often, instead preferring classification metrics such as accuracy, sensitivity and specificity.
PROBAST encourages assessment of studies for applicability to the review question as this is often narrower than inclusion criteria [23]. Concern was identified for models in three studies [30, 39, 41]. All others demonstrated either low concern or unclear applicability. Reasons for concern were attributable to outcomes which combined closely-related disorders, or the use of post-mortem gene expression data, whereas the review question focussed on models of single disorders with potential use in diagnosis or prognosis.
Model Performance
Over half of all models assessed discrimination using AUC (58% models). A wide range of classification metrics and measures of model fit were also reported (Table S7), with less than a quarter of models clearly reporting choosing a decision threshold a priori (Table S8).
Around 79% of models, from 12 studies, reported some form of internal validation (Table S9). The majority of these were k-fold cross-validation (57% of all models; 8 studies), a resampling approach which involves testing a model on each of k independent partitions of a dataset, every time training on the remaining k-1 folds. 10-fold cross-validation (CV) was most commonly used, with just below half of all cross-validated models invoking repeats with different random splits. The remainder of studies using internal validation created a random split between training and testing sets (21% of all models; 5 studies), or applied apparent validation, where training and testing are both done on the whole sample [31]. A minority reported external validation (26% of models; 2 studies). Use of internal validation was not reported for 16 models from a single study [25], but for which geographic and temporal external validation was given. External validation was reported for one other study, but with partly overlapping participants between development and validation sets [32].
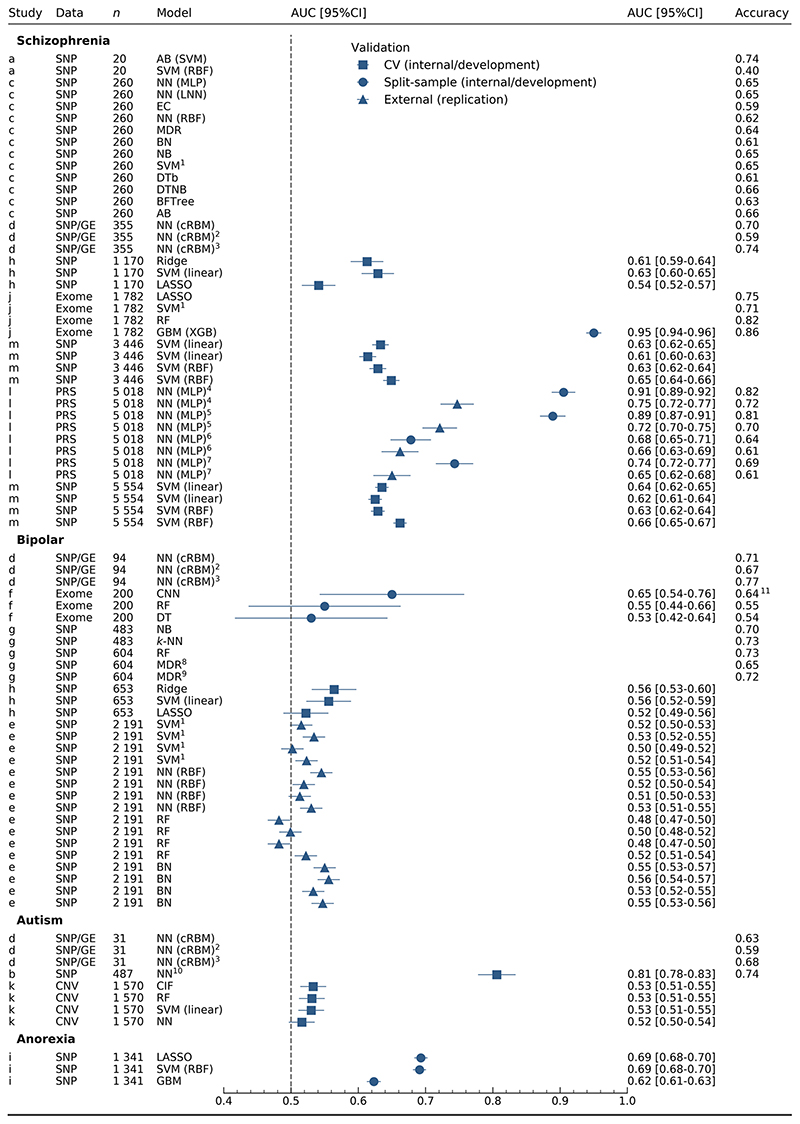
Model performance varied by choice of statistical method, sample size and number of predictors within studies (Table S10). Discrimination for models of schizophrenia (Figure 1) was extremely varied (0.541-0.95 AUC), with the highest AUC from exome data using XGBoost (0.95 AUC) [33]. In this study, Trakadis et al. (2019) used counts of variants in each gene, after annotation and predictor selection, on participants with part-Finnish or Swedish ancestry [42]. Similarly high AUC (0.905 AUC) made use of multiple schizophrenia-associated PRS [32]. However, the authors identify the presence of both the development and validation samples in the psychiatric genomics consortium (PGC) GWAS used to generate the schizophrenia PRS [43], in addition to having overlapping controls between internal validation (model development) and external validation (replication) samples. All other schizophrenia models involved learning from SNPs [27, 30, 34–36], with the exception of Wang et al. (2018) [39] where gene expression data from post-mortem samples informed the weights in a conditional RBM trained on genotypes.
Figure 1. discrimination for all models.
n: number of cases in training set. Studies: a [35], b [40], c [34, 36], d [39], e [25], f [38], g [31], h [30], i [26], j [33], k [37], l [32], m [27]. 1SVM kernel not reported. 2Modified architecture with intermediate phenotypes in training set only. 3Modified architecture with intermediate phenotypes for training and test sets. 4,5,6,7Internal and external validation are shown for study l, where validations for the same model are denoted with the same number. 8Two-way MDR. 9Three-way MDR. 10Neural network embedding layer. 11Accuracy calculated from confusion matrix. AB: AdaBoost, BN: Bayesian networks, BFTree: best-first tree, CIF: conditional inference forest, cRBM: conditional restricted Boltzmann machine, CI: confidence interval, CNN: convolutional neural network, CNV: copy number variation, DTb: decision tables, DTNB: decision table naïve Bayes, DT: decision tree, EC: evolutionary computation, GE: gene expression, GBM: gradient boosting machine, k-NN: k-nearest neighbours, LASSO: least absolute shrinkage and selection operator, LNN: linear neural network, MDR: multifactor dimensionality reduction, MLP: multi-layer perceptron, NB: naïve Bayes, NN: neural network, PRS: polygenic risk scores, RBF: radial basis function, RF: random forests, SNP: single nucleotide polymorphisms, SVM: support vector machine, XGB: extreme gradient boosting.
Predictive ability for bipolar disorder (Figure 1) was consistently lower than for schizophrenia, frequently overlapping with chance (0.482-0.65 AUC). Models were trained on genotypes, excepting a study [38] using exome data to train a CNN as part of the Critical Assessment of Genome Interpretation (CAGI) competition [44], for which moderate discrimination was achieved (0.65 AUC).
Significantly fewer models were reported for autism (8 models, 3 studies) and anorexia (3 models, 1 study) (Figure 1). Varying predictive performance was illustrated in autism (0.516-0.806 AUC). High AUC (0.806 AUC) was shown for a single prediction model [40], while models developed with a greater sample size by Engchuan et al. (2015) using CNVs were closer to or overlapping with chance (0.516-0.533 AUC) [37]. The only models predicting anorexia nervosa had moderate discriminative ability between cases and controls (0.623-0.693 AUC) [26].
Logistic regression and polygenic risk scores
Three studies reported AUC for either logistic regression (5 models) or polygenic risk scores (12 models) alongside machine learning methods. PRS were weighted by summary statistics from a GWAS on the same disorder as the outcome and used as the sole predictor in a logistic regression model. Though discrimination shows some difference between model types, the number of studies for comparison is low and results are clustered by study and type of validation (Figure S3).
Predictors
Coding of predictors was mostly unclear or unreported (7 studies, 55% of models). Coding was unclear if it was implied through the description of the type of classifier or software but not clearly articulated for the reported study. PRS were continuous [32] while counts of variants-per-gene or genes-per-gene-set were used for exomes and CNVs respectively [33, 37]. SNPs were coded under an additive model, a z-transformation of additive coding, or one-hot encoded (one predictor per genotype at a locus) (Table S11). GWAS summary statistics from external datasets were also used in the selection, weighting or combining of predictors (9 studies, 64% models; Table S12).
Predictor selection was adopted by most (12, 73% of models) and limited to filter-based selection, used prior to modelling, and embedded selection, an integral part of the prediction model (Table S13). The latter involved LASSO regression, or ensembles and hybrids of decision trees and decision tables, in addition to a modified AdaBoost [35]. Filters were based on internal or external univariable association tests (GWAS). Embedded and wrapper-based methods, which typically ‘wrap’ a model in forward or backward-selection, were both also used prior to any predictive modelling. Modification of predictors using information from the test set was the most common cause of information ‘leaking’ from the test set to the training set, a source of inflation in performance measures (Table S14).
Sample size
Total sample size was generally low where a single sample had been used, but higher if genotypes from publicly-available amalgamated datasets used in a GWAS had been downloaded (median 3486, range 40-11853) (Table S10). Number of events in development followed a similar pattern (median 1341, range 20-5554) as class imbalance was minimal (median 1, range 0.65-2.93, calculated as non-events over events). Around half of studies gave sufficient information to calculate events per variable (EPV) (median 0.69, range 0.00063-74.6). It could not be calculated where the number of candidate predictors where not reported for models in 2 studies [25, 39]; approximations are given in the supplementary where reporting was unclear in a further 5 studies [26, 32–34, 36, 38] (Table S10).
Hyperparameter Search
Hyperparameter search was mostly unreported or unclear (41 models, 9 studies), with some models reported as having been used with default settings. Ambiguous reporting resulted from description of search and tuning for a specific model, with no clarity as to whether these conditions applied to other models in the study. Only 19% of models clearly reported attempting different hyperparameters for the extracted models (Table S15). Studies also report non-standard final hyperparameters, such as uneven batch size in neural networks, or showed good accuracy for a model which is highly sensitive to tuning of crucial hyperparameters, yet few reported tuning (Table S16). It is therefore likely that most studies evaluated several hyperparameter choices but did not report this.
Discussion
All studies displayed high risk of bias in model development and validation with infrequent reporting of standard modelling steps. Performance measures consequently demonstrated a wide range of abilities to discriminate between cases and controls (0.482-0.95 AUC). These are likely optimistic owing to the high risk of bias identified through PROBAST and unaddressed sample overlap and population structure, as two studies showing the highest AUCs left these issues unresolved [32, 33]. Though potential bias and effective sample size limit overall interpretation of discrimination, low standards of model development, validation and reporting are a clear and consistent theme throughout all studies. Broad discrimination has also been observed for machine learning studies in cancer genomics [45]; more established fields with clearer predictor-response relationships, such as medical imaging, are much more consistent [46].
Issues relating to ROB often rest on distinctions in methodology between clinical prediction modelling, machine learning and genetic association studies. For instance, genetic studies most commonly employ a case-control design. Such studies are extremely useful for identifying genetic risk factors for rare outcomes, but are considered inadequate for prediction modelling as absolute risks cannot be estimated; instead, case-cohort, nested case-control, or prospective cohort designs are preferred [47]. Case-cohort and nested casecontrol designs involve sampling from an existing cohort and can be used for prediction models if the sampling fraction in controls is accounted for in analysis [48]. To project the prediction to the whole population in case-control studies, positive and negative predictive values should be corrected in accordance with the disease prevalence in the population and ratio of cases and controls in the sample [49]. Similarly, univariable tests of association are applied routinely in GWAS, and are often used in selection of predictors for genetic prediction models. Their application in prediction modelling though is usually discouraged, as predictors may differ in their importance when evaluated in isolation as compared to when considered concurrently with other variables [50].
Lack of adherence to appropriate procedures for machine learning are also a common cause of a model being assessed as at high risk of bias. Standard model validation procedures were followed by some researchers; however, many ‘leaked’ information between training and testing sets through not applying predictor manipulations or selection in only the training set/fold, or using the testing set/fold to adjust model hyperparameters, which can impose significant bias on estimates of prediction performance [51].
Most studies provided a measure of classification or discrimination for each model; none reported a measure of calibration. Model calibration compares observed and predicted probabilities of the outcome occurring, and is a crucial part of model development [52] which has been noted for its absence in genetic prediction literature [53]. Authors reporting only classification measures, such as accuracy, sensitivity or specificity, should also note that measures of discrimination are preferred as they use all the information over predicted probabilities and delay any thresholding of risks to a more appropriate time. Of discrimination measures, the AUC is the most widely used in both machine learning and genetics [54, 55].
Hyperparameter optimisation is an essential part of developing machine learning models as it determines how they navigate the bias-variance trade-off and learn from data [56]. It is therefore surprising that it was so often unreported or subject to a small number of manual experiments. Hyperparameters should be systematically searched to ensure a model is not over or under-fit. Randomised search has been shown to be more effective than grid search where two or more such parameters require tuning [57], though grid search is also recommended by practitioners for SVMs, often with an initial ‘coarse’ search followed by a more thorough exploration of a finer grid of values [58]. The importance of search is particularly relevant in domains where there are a small number of events per candidate predictor [59], such as genomics, as appropriate hyperparameter choices can reduce overfitting.
Split-sample approaches were used by several studies, but should be avoided in favour of resampling methods such as bootstrapping or k-fold cross-validation [60]. The latter is an appropriate form of internal validation for traditional statistical methods; however, estimated prediction accuracies become overly-optimistic if done repeatedly, as when used for hyperparameter tuning through repeated rounds of CV. Nested cross-validation, where hyperparameters are optimised in an inner-fold and evaluated in the outer-fold, has been shown to give more realistic estimates [51, 61] but was not used in any studies. A single study presented both internal and external validation of models [32], for which a large drop in performance is seen upon replication. Though partly due to sample overlap between the development set and the summary statistics used for generating a PRS, difficulty with replication is a wider issue in polygenic risk prediction. Risk scores for psychiatric disorders typically explain a small proportion of variance in a trait [62], with generalisation issues compounded by variants with small effect sizes and different allele frequencies between populations. Risk scores generated through machine learning methods have the potential to be more affected by these issues if appropriate modelling procedures are not followed.
A source of bias not explicitly covered in PROBAST is population structure. Genetic ancestry has the potential to bias both associations [63, 64] and predictions [65, 66] from genetic data. Supervised machine learning methods have proved particularly sensitive in detecting ancestry [67–69]. Few researchers discussed visualising ancestry or reported exclusions, and none reported modelling adjustments, even when previous association studies on the same datasets had demonstrated stratification and included principal components as covariates. The extent of the bias introduced in these studies is not clear: evidence mostly relates to deliberately predicting populations in humans using ML or looking at bias in complex trait prediction from PRS. While the potential for population stratification to impact predictions is apparent, the method for dealing with it when using machine learning methods is not. Several techniques have been proposed, including modifications to random forests [70]; exclusions by, or inclusion of, principal components; and regressing-off the linear effects of principal components on SNPs before modelling (for example [71, 72]). Whether any combination of these is sufficient to reduce the effects of population stratification in nonlinear machine learning predictions has not been demonstrated.
General reporting guidelines for machine learning prediction models are yet to be developed [73], though recommendations for undertaking [74, 75] evaluating [76] or reporting [77] exist for machine learning in omics data, psychiatry and medicine respectively, in addition to reporting guidelines outside of machine learning [21, 78]. We encourage authors to report on implementation, samples, predictors, missingness, hyperparameters and handling of potential information leakage, and consult guidelines where needed. Finally, we advocate for machine learning methods to be reported alongside polygenic risk scores as a standard baseline model for comparison. The potential for machine learning methods to provide improved prediction has received heightened attention in recent years. Any such outcome cannot occur without adherence to standards for the development, validation and reporting of models.
Supplementary Material
Acknowledgements
The authors wish to thank the Dementia Research Institute (UKDRI-3003) and MRC Centre for Neuropsychiatric Genetics and Genomics Centre (MR/L010305/1) and Program Grants (MR/P005748/1).
Footnotes
Conflict of Interest
All authors report no potential conflicts of interest.
References
- 1.Glorot X, Bordes A, Bengio Y. Deep Sparse Rectifier Neural Networks. Proc fourteenth Int Conf Artif Intell Stat. 2011:315–323. [Google Scholar]
- 2.Hinton G, Deng L, Yu D, Dahl G, Mohamed AR, Jaitly N, et al. Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal Process Mag. 2012;29:82–97. [Google Scholar]
- 3.Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. Adv Neural Inf Process Syst. 2012:1097–1105. [Google Scholar]
- 4.Sutskever I, Vinyals O, Le QV. Sequence to Sequence Learning with Neural Networks. Adv Neural Inf Process Syst. 2014:3104–3112. [Google Scholar]
- 5.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krystal JH, Murray JD, Chekroud AM, Corlett PR, Yang G, Wang X-J, et al. Computational Psychiatry and the Challenge of Schizophrenia. Schizophr Bull. 2017;43:473–475. doi: 10.1093/schbul/sbx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnack HG. Improving individual predictions: Machine learning approaches for detecting and attacking heterogeneity in schizophrenia (and other psychiatric diseases) Schizophr Res. 2019;214:34–42. doi: 10.1016/j.schres.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Tandon N, Tandon R. Will Machine Learning Enable Us to Finally Cut the Gordian Knot of Schizophrenia. Schizophr Bull. 2018;44:939–941. doi: 10.1093/schbul/sby101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22. doi: 10.1016/j.jclinepi.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99:323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okser S, Pahikkala T, Aittokallio T. Genetic variants and their interactions in disease risk prediction - machine learning and network perspectives. BioData Min. 2013;6:5. doi: 10.1186/1756-0381-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iniesta R, Stahl D, McGuffin P. Machine learning, statistical learning and the future of biological research in psychiatry. Psychol Med. 2016;46:2455–2465. doi: 10.1017/S0033291716001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Librenza-Garcia D, Kotzian BJ, Yang J, Mwangi B, Cao B, Pereira Lima LN, et al. The impact of machine learning techniques in the study of bipolar disorder: A systematic review. Neurosci Biobehav Rev. 2017;80:538–554. doi: 10.1016/j.neubiorev.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, Ragguett R-M, Mansur RB, Boutilier JJ, Rosenblat JD, Trevizol A, et al. Applications of machine learning algorithms to predict therapeutic outcomes in depression: A meta-analysis and systematic review. J Affect Disord. 2018;241:519–532. doi: 10.1016/j.jad.2018.08.073. [DOI] [PubMed] [Google Scholar]
- 16.Durstewitz D, Koppe G, Meyer-Lindenberg A. Deep neural networks in psychiatry. Mol Psychiatry. 2019;24:1583–1598. doi: 10.1038/s41380-019-0365-9. [DOI] [PubMed] [Google Scholar]
- 17.Ho DSW, Schierding W, Wake M, Saffery R, O’Sullivan J. Machine Learning SNP Based Prediction for Precision Medicine. Front Genet. 2019;10:267. doi: 10.3389/fgene.2019.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapur S, Phillips A, Insel T. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol Psychiatry. 2012;17:1174–1179. doi: 10.1038/mp.2012.105. [DOI] [PubMed] [Google Scholar]
- 20.Moons KGM, de Groot JAH, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, et al. Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies: The CHARMS Checklist. PLoS Med. 2014;11:e1001744. doi: 10.1371/journal.pmed.1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janssens ACJ, Ioannidis JP, van Duijn CM, Little J, Khoury MJ. Strengthening the reporting of genetic risk prediction studies: the GRIPS statement. Genome Med. 2011;3:16. doi: 10.1186/gm230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debray TPA, Damen JAAG, Snell KIE, Ensor J, Hooft L, Reitsma JB, et al. A guide to systematic review and meta-analysis of prediction model performance. BMJ. 2017;356:i6460. doi: 10.1136/bmj.i6460. [DOI] [PubMed] [Google Scholar]
- 23.Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann Intern Med. 2019;170:51. doi: 10.7326/M18-1376. [DOI] [PubMed] [Google Scholar]
- 24.Moher D, Liberati A, Tetzlaff J, Altman DG, Group TP. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirooznia M, Seifuddin F, Judy J, Mahon PB, Potash JB, Zandi PP, et al. Data mining approaches for genome-wide association of mood disorders. Psychiatr Genet. 2012;22:55–61. doi: 10.1097/YPG.0b013e32834dc40d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y, Wei Z, Keating BJ, Hakonarson H, Nervos GCA, Consor WTCC, et al. Machine learning derived risk prediction of anorexia nervosa. BMC Med Genomics. 2016;9:4. doi: 10.1186/s12920-016-0165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivian-Griffiths T, Baker E, Schmidt KM, Bracher-Smith M, Walters J, Artemiou A, et al. Predictive modeling of schizophrenia from genomic data: Comparison of polygenic risk score with kernel support vector machines approach. Am J Med Genet Part B Neuropsychiatr Genet. 2019;180:80–85. doi: 10.1002/ajmg.b.32705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 29.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Yang C, Gelernter J, Zhao H. Improving genetic risk prediction by leveraging pleiotropy. Hum Genet. 2014;133:639–650. doi: 10.1007/s00439-013-1401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acikel C, Son YA, Celik C, Gul H. Evaluation of potential novel variations and their interactions related to bipolar disorders: Analysis of genome-wide association study data. Neuropsychiatr Dis Treat. 2016;12:2997–3004. doi: 10.2147/NDT.S112558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Wu J, Mize T, Shui D, Chen X. Prediction of Schizophrenia Diagnosis by Integration of Genetically Correlated Conditions and Traits. J Neuroimmune Pharmacol. 2018;13:532–540. doi: 10.1007/s11481-018-9811-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trakadis YJ, Sardaar S, Chen A, Fulginiti V, Krishnan A. Machine learning in schizophrenia genomics, a case-control study using 5,090 exomes. Am J Med Genet Part B Neuropsychiatr Genet. 2019;180:103–112. doi: 10.1002/ajmg.b.32638. [DOI] [PubMed] [Google Scholar]
- 34.Aguiar-Pulido V, Seoane JA, Rabuñal JR, Dorado J, Pazos A, Munteanu CR. Machine learning techniques for single nucleotide polymorphism - disease classification models in schizophrenia. Molecules. 2010;15:4875–4889. doi: 10.3390/molecules15074875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Liu J, Sui J, Pearlson G, Calhoun VD. A Hybrid Machine Learning Method for Fusing fMRI and Genetic Data: Combining both Improves Classification of Schizophrenia. Front Hum Neurosci. 2010;4:192. doi: 10.3389/fnhum.2010.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguiar-Pulido V, Gestal M, Fernandez-Lozano C, Rivero D, Munteanu CR. Applied Computational Techniques on Schizophrenia Using Genetic Mutations. Curr Top Med Chem. 2013;13:675–684. doi: 10.2174/1568026611313050010. [DOI] [PubMed] [Google Scholar]
- 37.Engchuan W, Dhindsa K, Lionel AC, Scherer SW, Chan JH, Merico D. Performance of case-control rare copy number variation annotation in classification of autism. BMC Med Genomics. 2015;8:S7. doi: 10.1186/1755-8794-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laksshman S, Bhat RR, Viswanath V, Li X, Sundaram L, Bhat RR, et al. DeepBipolar: Identifying genomic mutations for bipolar disorder via deep learning. Hum Mutat. 2017;38:1217–1224. doi: 10.1002/humu.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science (80-) 2018;362:eaat8464. doi: 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghafouri-Fard S, Taheri M, Omrani MD, Daaee A, Mohammad-Rahimi H, Kazazi H. Application of Single-Nucleotide Polymorphisms in the Diagnosis of Autism Spectrum Disorders: A Preliminary Study with Artificial Neural Networks. J Mol Neurosci. 2019;68:515–521. doi: 10.1007/s12031-019-01311-1. [DOI] [PubMed] [Google Scholar]
- 41.Pirooznia SK, Chiu K, Chan MT, Zimmerman JE, Elefant F. Epigenetic Regulation of Axonal Growth of Drosophila Pacemaker Cells by Histone Acetyltransferase Tip60 Controls Sleep. Genetics. 2012;192:1327. doi: 10.1534/genetics.112.144667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daneshjou R, Wang Y, Bromberg Y, Bovo S, Martelli PL, Babbi G, et al. Working toward precision medicine: Predicting phenotypes from exomes in the Critical Assessment of Genome Interpretation (CAGI) challenges. Hum Mutat. 2017;38:1182–1192. doi: 10.1002/humu.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patil S, Habib Awan K, Arakeri G, Jayampath Seneviratne C, Muddur N, Malik S, et al. Machine learning and its potential applications to the genomic study of head and neck cancer-A systematic review. J Oral Pathol Med. 2019;48:773–779. doi: 10.1111/jop.12854. [DOI] [PubMed] [Google Scholar]
- 46.Islam MM, Yang HC, Poly TN, Jian WS, (Jack)Li YC. Deep learning algorithms for detection of diabetic retinopathy in retinal fundus photographs: A systematic review and meta-analysis. Comput Methods Programs Biomed. 2020;191:105320. doi: 10.1016/j.cmpb.2020.105320. [DOI] [PubMed] [Google Scholar]
- 47.Moons KGM, Kengne AP, Woodward M, Royston P, Vergouwe Y, Altman DG, et al. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 2012;98:683–690. doi: 10.1136/heartjnl-2011-301246. [DOI] [PubMed] [Google Scholar]
- 48.Biesheuvel CJ, Vergouwe Y, Oudega R, Hoes AW, Grobbee DE, Moons KGM. Advantages of the nested case-control design in diagnostic research. BMC Med Res Methodol. 2008;8:1–7. doi: 10.1186/1471-2288-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kallner A. Bayes’ theorem, the roc diagram and reference values: Definition and use in clinical diagnosis. Biochem Medica. 2018;28:16–25. doi: 10.11613/BM.2018.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun G-W, Shook TL, Kay GL. Inappropriate use of bivariable analysis to screen risk factors for use in multivariable analysis. J Clin Epidemiol. 1996;49:907–916. doi: 10.1016/0895-4356(96)00025-x. [DOI] [PubMed] [Google Scholar]
- 51.Vabalas A, Gowen E, Poliakoff E, Casson AJ. Machine learning algorithm validation with a limited sample size. PLoS One. 2019;14:e0224365. doi: 10.1371/journal.pone.0224365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steyerberg EW. Clinical Prediction Models. 2nd. Springer International Publishing; 2019. [Google Scholar]
- 53.Janssens ACJ, Ioannidis JP, Bedrosian S, Boffetta P, Dolan SM, Dowling N, et al. Strengthening the reporting of genetic risk prediction studies (GRIPS): explanation and elaboration. Eur J Hum Genet. 2011;19:615–615. doi: 10.1038/ejhg.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997;30:1145–1159. [Google Scholar]
- 55.Wray NR, Yang J, Goddard ME, Visscher PM. The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning. Springer New York; New York, NY: 2013. [Google Scholar]
- 57.Bergstra J, Bengio Y. Random Search for Hyper-Parameter Optimization. J Mach Learn Res. 2012;13:281–305. [Google Scholar]
- 58.Ben-Hur A, Weston J. A User’s Guide to Support Vector Machines. Data MiN Tech. life Sci. Humana Press; 2010. pp. 223–239. [DOI] [PubMed] [Google Scholar]
- 59.Pavlou M, Ambler G, Seaman SR, Guttmann O, Elliott P, King M, et al. How to develop a more accurate risk prediction model when there are few events. BMJ. 2015;351:h3868. doi: 10.1136/bmj.h3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steyerberg EW, Harrell FE, Borsboom GJJ, Eijkemans MJ, Vergouwe Y, Habbema JDF. Internal validation of predictive models: Efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54:774–781. doi: 10.1016/s0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]
- 61.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 64.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 65.Belgard TG, Jankovic I, Lowe JK, Geschwind DH. Population structure confounds autism genetic classifier. Mol Psychiatry. 2014;19:405–407. doi: 10.1038/mp.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bridges M, Heron EA, O’Dushlaine C, Segurado R, Morris D, Corvin A, et al. Genetic Classification of Populations Using Supervised Learning. PLoS One. 2011;6:e14802. doi: 10.1371/journal.pone.0014802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schrider DR, Kern AD. Supervised Machine Learning for Population Genetics: A New Paradigm. Trends Genet. 2018;34:301–312. doi: 10.1016/j.tig.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flagel L, Brandvain Y, Schrider DR. The Unreasonable Effectiveness of Convolutional Neural Networks in Population Genetic Inference. Mol Biol Evol. 2019;36:220–238. doi: 10.1093/molbev/msy224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stephan J, Stegle O, Beyer A. A random forest approach to capture genetic effects in the presence of population structure. Nat Commun. 2015;6:7432. doi: 10.1038/ncomms8432. [DOI] [PubMed] [Google Scholar]
- 71.Zhao Y, Chen F, Zhai R, Lin X, Wang Z, Su L, et al. Correction for population stratification in random forest analysis. Int J Epidemiol. 2012;41:1798–1806. doi: 10.1093/ije/dys183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheutlin AB, Chekroud AM, Polimanti R, Gelernter J, Sabb FW, Bilder RM, et al. Multivariate Pattern Analysis of Genotype-Phenotype Relationships in Schizophrenia. Schizophr Bull. 2018;44:1045–1052. doi: 10.1093/schbul/sby005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–1579. doi: 10.1016/S0140-6736(19)30037-6. [DOI] [PubMed] [Google Scholar]
- 74.Boulesteix A-L, Wright MN, Hoffmann S, König IR. Statistical learning approaches in the genetic epidemiology of complex diseases. Hum Genet. 2019:1–12. doi: 10.1007/s00439-019-01996-9. [DOI] [PubMed] [Google Scholar]
- 75.Teschendorff AE. Avoiding common pitfalls in machine learning omic data science. Nat Mater. 2019;18:422–427. doi: 10.1038/s41563-018-0241-z. [DOI] [PubMed] [Google Scholar]
- 76.Tandon N, Tandon R. Machine learning in psychiatry-standards and guidelines. Asian J Psychiatr. 2019;44:A1–A4. doi: 10.1016/j.ajp.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 77.Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J Med Internet Res. 2016;18:e323. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Ann Intern Med. 2015;162:55. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.