Introductory paragraph
Bile acids (BAs) are signaling molecules that mediate various cellular responses in both physiological and pathological processes. Several studies reported that BAs can be detected in the brain1, yet their physiological role in the central nervous system (CNS) is still largely unknown. Here we show that postprandial BAs can reach the brain and activate a negative feedback loop controlling satiety in response to physiological feeding via TGR5, a G protein-coupled receptor (GPCR) activated by multiple conjugated and unconjugated BAs2 and an established regulator of peripheral metabolism3–8. Notably, peripheral or central administration of a BA mix or a TGR5-specific BA mimetic (INT-777) exerted an anorexigenic effect in wild-type mice while whole-body, neuron-specific or Agouti-Related Peptide (AgRP) neuronal TGR5 deletion caused a significant increase in food intake. Accordingly, orexigenic peptide expression and secretion were reduced after short-term TGR5 activation. In vitro studies demonstrated that activation of the Rho/ROCK/Actin-remodeling pathway decreases orexigenic AgRP/Neuropeptide Y (NPY) release in a TGR5-dependent manner. Taken together, these data identify a signaling cascade by which BAs exert acute effects at the transition between the fasting-feeding state and prime the switch toward satiety, unveiling a previously unrecognized physiological feedback mediated by BAs in the CNS.
Primary BAs are synthesized in the liver from cholesterol and chemically transformed to secondary BAs by the gut microbiota8. Upon feeding, circulating BA levels can reach micromolar concentrations through enterohepatic recirculation and spill over in the systemic circulation9. The presence of BAs in the brain has been reported under both physiological and pathological conditions in rodents and humans1,10–12. Although some of the cholesterol and BA biosynthetic enzymes are expressed in different regions of the brain13, these proteins seem to be involved in cholesterol clearance rather than in BA production14. Brain BAs strongly correlate with circulating levels and are believed to reach the CNS through passive diffusion10. Despite this evidence, a physiological role of BAs in the CNS has not been reported. The recent finding that the main non-genomic BA signaling mediator TGR5 (gene name Gpbar1), is expressed in the CNS15,16, led us to hypothesize that BAs may exert dedicated CNS functions.
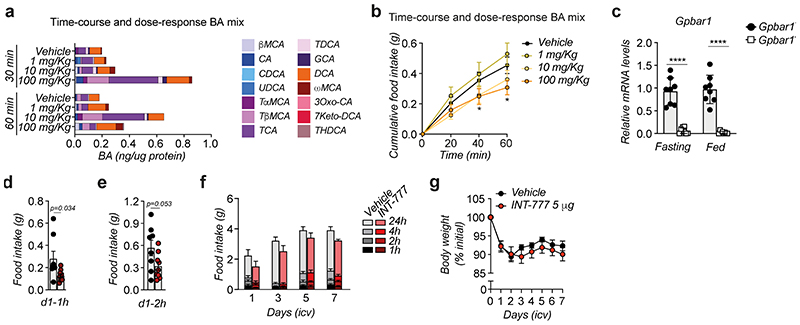
Interestingly, we observed that BAs can reach the hypothalamus, the main central regulator of energy homeostasis, after physiological feeding. The hypothalamic concentration of several endogenous BAs, mainly the tauro-conjugated species, was transiently increased upon food intake at the beginning of the dark phase (Fig. 1a). Oral administration of a BA mix effectively reduced 24-hour food intake (Fig. 1b) suggesting that the feeding-mediated increase in BAs in the CNS may trigger satiety during the fasting-feeding transition through the activation of a BA receptor. Time-course and dose-response studies confirmed that orally administered BAs can rapidly reach the hypothalamus (Extended Data Fig. 1a) and suppress food intake in wild-type mice (Extended Data Fig. 1b). Based on these rapid responses, we reasoned that the anorexigenic phenotype could be mediated by TGR5. Brain expression profiling with RNAscope imaging revealed that endogenous Gpbar1 mRNA is expressed in different brain regions, including the arcuate nucleus (ARC) (Fig. 1c). ARC-enriched hypothalamic punches coupled with qRT-PCR analysis confirmed that Gpbar1 is expressed in this brain region critical for the control of food consumption (Extended Data Fig. 1c).
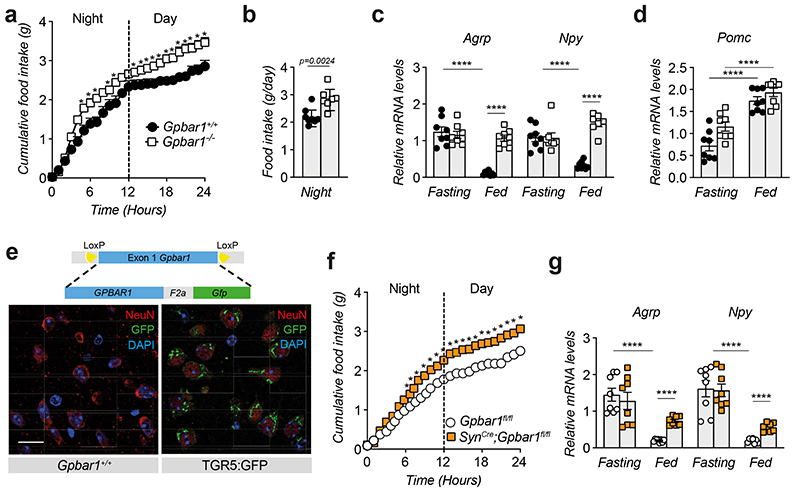
Figure 1. BAs reach the hypothalamus during physiological feeding and suppress food intake through TGR5.
(a) Conjugated and unconjugated BA species measured in the hypothalamus after 30- and 60-min refeeding in wild-type mice. Bars represent the mean from 8 replicates. n=8 animals. (b) 24-hour food intake after oral administration of BA mix (n=5 animals) or vehicle (n=4 animals). (c) Representative images showing Gpbar1 mRNA (in white) using RNAscope technique in the hypothalamus of TGR5 wild-type (Gpbar1+/+) and germline TGR5 knock-out (Gpbar1−/−) mice. ARC = arcuate nucleus, 3V = third ventricle. Scale bar = 50μm and digital zoom. n=4 animals. (d) INT-777 measurement in the hypothalamus after 30 and 60 min of oral administration of the TGR5 specific BA agonist INT-777 at 3 different doses or vehicle in 8-week-old wild-type mice. n=8 (INT-777 10 and 100 mg/Kg 30 min) and n=7 (all the other groups) animals. (e) One-hour cumulative food intake of wild-type mice after oral administration of the TGR5 specific BA agonist INT-777 at three different doses or vehicle. n=6 (INT-777 100 mg/Kg) and n=8 (all other treatments) animals. (f) mRNA levels of orexigenic genes Agrp and Npy 1 hour after oral (gavage) and intracerebroventricular (icv) administration of INT-777 or vehicle in arcuate nucleus (ARC)-enriched hypothalamic punches of wild-type mice. n=6 animals. (g) Pomc mRNA levels in arcuate nucleus (ARC)-enriched hypothalamic punches of mice described in f. n=6 animals. Results represent mean (a) or mean ± SEM (b, d-g). n represents biologically independent replicates. Two-tailed Student’s t-test (b and f), one-way (d) or two-way ANOVA followed by Bonferroni post-hoc correction (e) vs. vehicle group were used for statistical analysis. P values (exact value, * P ≤ 0.05 or **** P ≤ 0.0001) are indicated in the figure.
We next gavaged the TGR5-specific semi-synthetic BA, INT-7774,17, to investigate whether the BA-mediated anorexigenic effect is TGR5-dependent. Time-course and dose-response experiments confirmed that, like endogenous BAs, INT-777 rapidly accumulates in the hypothalamus (Fig. 1d) and reduces food intake at doses ranging between 10-100 mg/Kg (Fig. 1e). At the highest dose, INT-777 reached the hypothalamus at concentrations comparable to those of the endogenous postprandial BAs (Fig. 1a), recapitulating the central physiological concentrations required for BA-mediated feedback regulation of feeding behaviour. Of note, intracerebroventricular (icv) administration of INT-777 to wild-type mice also caused a rapid and significant anorexigenic response after the first injection (Extended Data Fig. 1d–e) but receded over time, and only a trend toward reduced food intake remained (Extended Data Fig. 1f). These data indicate that an acute preprandial stimulation of TGR5 is sufficient to reduce food consumption during the physiological feeding state but that repetitive TGR5 activation in CD-fed mice only marginally affects food intake, without impacting body weight (Extended Data Fig. 1g).
Among the different neuronal populations involved in the regulation of feeding, the orexigenic AgRP/NPY neurons, promoting hunger signaling, and the anorexigenic Proopiomelanocortin (POMC)/Cocaine and Amphetamine-Regulated Transcript (CART)-expressing neurons, coordinating satiety, are known to play a key role18. These neurons are located in the ARC in proximity to a fenestrated blood brain barrier that allows a facilitated exchange with the blood. Analysis of the neuropeptide expression in the ARC showed that Agrp and Npy mRNA was reduced in wild-type mice after 1 hour of either oral or icv administration of the TGR5 agonist (Fig. 1f) while Pomc levels remained unaffected (Fig. 1g).
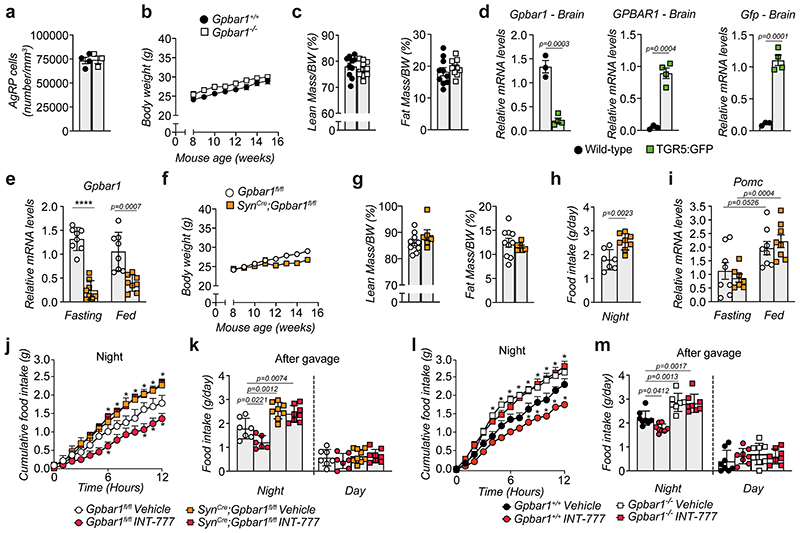
Phenotyping of 8-week-old TGR5 wild-type (Gpbar1 +/+) and knock-out (Gpbar1 −/−) mice revealed that whole-body TGR5 deletion triggers a significant increase in cumulative food intake (Fig. 2a), with an increment in food consumption starting during the night period (Fig. 2b), which coincides with the active and physiological feeding cycle. While wild-type mice showed blunted Agrp and Npy mRNA levels in response to feeding, Gpbar1 −/− mice failed to suppress the expression of these orexigenic neuropeptides (Fig. 2c). On the contrary, the expression of the anorexigenic peptide Pomc was unaffected (Fig. 2d). These data suggest that TGR5 may coordinate satiety in the fed state through the inhibition of AgRP/NPY rather than stimulation of POMC neurons, without affecting the number of AgRP/NPY neurons (Extended Data Fig. 2a). Of note, the increase in food intake did not alter body weight nor fat and lean mass in Gpbar1 −/− mice compared to their controls (Extended Data Fig. 2b–c).
Figure 2. TGR5 is expressed in neurons and its deletion increases food intake.
(a) 24-hour cumulative food intake of 8-week-old TGR5 wild-type (Gpbar1+/+, n=6 animals) and germline TGR5 knock-out (Gpbar1−/−, n=7 animals) mice. (b) 12-hours food intake of Gpbar1+/+ (n=8 animals) and Gpbar1−/− (n=7 animals) mice. (c) mRNA levels of orexigenic genes Agrp and Npy after overnight fasting and fed conditions in arcuate nucleus (ARC)-enriched hypothalamic punches of Gpbar1 +/+ and Gpbar1 −/− mice. n=8 animals. (d) Pomc mRNA levels in arcuate nucleus (ARC)-enriched hypothalamic punches of mice described in c. n=8 animals. (e) Generation of transgenic mice expressing TGR5 and Green Fluorescent Protein (GFP) under the control of the mouse Gpbar1 promoter (TGR5:GFP), and hypothalamic immunodetection of the neuronal marker NeuN in red, TGR5 (GFP) in green and nuclei staining (DAPI) in blue, in TGR5 wild-type (Wild-type) and TGR5:GFP mice. Scale bar = 20 μm. n=4 animals. (f) 24-hour cumulative food intake of Gpbar1fl/fl and SynCre;Gpbar1fl/fl mice. n=8 animals. (g) mRNA levels of orexigenic genes Agrp and Npy after overnight fasting and normal fed conditions in arcuate nucleus (ARC)-enriched hypothalamic punches of Gpbar1fl/fl and SynCre;Gpbar1fl/fl mice. n=8 animals. Results represent mean ± SEM. n represents biologically independent replicates. Two-tailed Student’s t-test (b, c, d and g), or two-way ANOVA followed by Bonferroni post-hoc correction (a and f) vs. Gpbar1+/+ (a-c), fasting (d) or Gpbar1fl/fl (f and g) groups were used for statistical analysis. P values (exact value, * P ≤ 0.05 or **** P ≤ 0.0001) are indicated in the figure.
To analyze the distribution of TGR5 in different cell types in the brain, we used a reporter mouse model (TGR5:GFP), that co-expresses TGR5 and GFP under the control of the mouse Gpbar1 regulatory gene locus (Fig. 2e and Extended Data Fig. 2d). Immunofluorescence analysis using antibodies against GFP and the neuronal marker NeuN confirmed that TGR5 is present in hypothalamic neurons (Fig. 2e), as well as in other brain cells, as previously reported16, including glia and astrocytes. To verify the importance of neuronal TGR5 in controlling food intake, we generated mice with a targeted deletion of Gpbar1 in neurons (SynCre;Gpbar1fl/fl). In line with the expression of Gpbar1 in multiple cell types of the ARC, a substantial, but not complete, reduction of Gpbar1 mRNA levels was observed in the ARC of SynCre;Gpbar1fl/fl mice (Extended Data Fig. 2e). Consistent with the findings observed in the whole-body Gpbar1−/− mice, SynCre;Gpbar1fl/fl mice displayed no apparent changes in body weight and composition (Extended Data Fig. 2f–g)) but a marked increase in cumulative food intake (Fig. 2f and Extended Data Fig. 2h) and in the expression of orexigenic (Fig. 2g), but not anorexigenic (Extended Data Fig. 2i) genes. Moreover, preprandial oral administration of INT-777 decreased cumulative food intake in Gpbar1fl/fl and Gpbar1 +/+ mice (Extended Data Fig. 2j–m)), but not in whole-body Gpbar1 −/− (Extended Data Fig. 2l–m)) and neuron-restricted SynCre;Gpbar1fl/fl mice (Extended Data Fig. 2j–k)), confirming that the modulation of AgRP/NPY expression involved in controlling eating behavior is driven by neuron-specific TGR5.
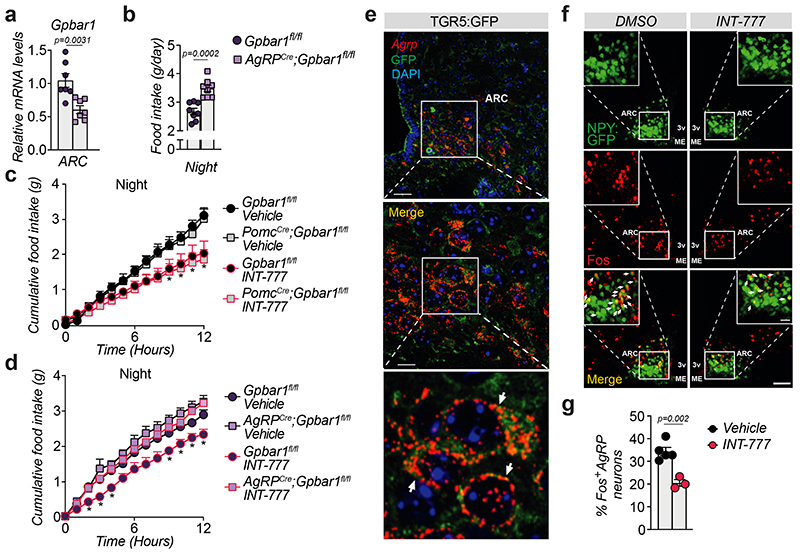
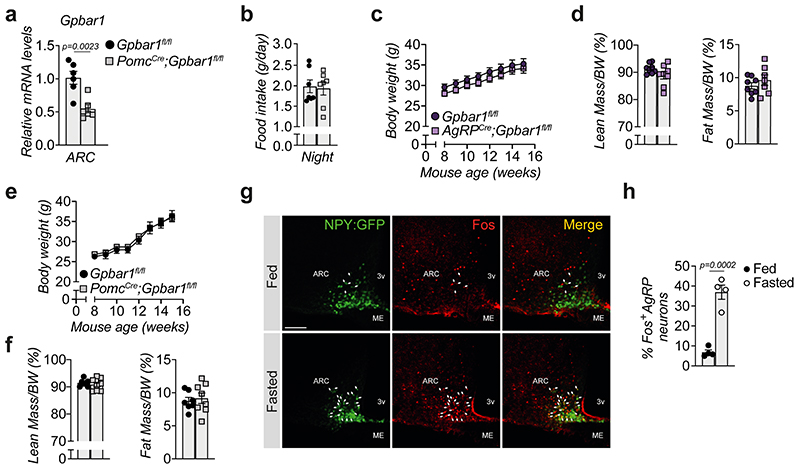
In order to determine which neuronal population in the ARC is responsible for conferring the food intake phenotype, we generated AgRP (AgRPCre;Gpbar1fl/fl) and POMC (PomcCre;Gpbar1fl/fl) neuron-specific Gpbar1 mutant mouse models and confirmed that Gpbar1 expression was reduced by half in the ARC of both strains (Fig. 3a and Extended Data Fig. 3a). AgRPCre;Gpbar1fl/fl but not PomcCre;Gpbar1fl/fl mice displayed a significant increase in food intake during the night phase compared to their controls (Gpbar1fl/fl mice) (Fig. 3b and Extended Data Fig. 3b). Moreover, oral administration of INT-777 exerted an anorexigenic action in the PomcCre;Gpbar1fl/fl mice (Fig. 3c), as well as in Gpbar1fl/fl controls (Fig. 3c–d), while this effect was significantly blunted in AgRPCre;Gpbar1fl/fl mice (Fig. 3d), indicating that TGR5 signaling in AgRP/NPY, but not in POMC neurons is important for the control of food intake. Similar to our previous findings, deletion of TGR5 in both AgRP/NPY and POMC neurons did not affect body weight and composition (Extended Data Fig. 3c–f). Co-staining experiments using Agrp RNAscope probes and GFP immunofluorescence in TGR5:GFP mice confirmed that Gpbar1 is expressed in AgRP/NPY neurons (Fig. 3e). Immunofluorescence experiments showed that INT-777-mediated TGR5 activation in fasted animals significantly decreased the percentage of Fos-positive AgRP/NPY neurons (Fig. 3f–g), mimicking the fed state (Extended Data Fig. 3g–h). Altogether, these data suggest that TGR5 suppresses food intake through inhibition of hypothalamic AgRP/NPY neurons.
Figure 3. Selective deletion of TGR5 in AgRP/NPY but not in POMC neurons abrogates satiety induced by BA derivatives.
(a) Gpbar1 mRNA levels in arcuate nucleus (ARC)-enriched hypothalamic punches of control mice (Gpbar1fl/fl) and AgRP neuron-specific TGR5 knock-out (AgRPCre;Gpbar1fl/fl) mice. n=7 animals. (b) 12-hours food intake of Gpbar1fl/fl and AgRPCre;Gpbar1fl/fl mice. n=8 animals. (c) 12-hour cumulative food intake of control (Gpbar1fl/fl) and POMC neuron-specific TGR5 knock-out (PomcCre;Gpbar1fl/fl) mice after oral administration of TGR5 agonist INT-777 or vehicle. n=6 (Gpbar1fl/fl INT-777), n=7 (Vehicle groups) and n=8 (PomcCre;Gpbar1fl/fl INT-777) animals. (d) 12-hour cumulative food intake of control (Gpbar1fl/fl) and AgRP neuron-specific TGR5 knock-out (AgRPCre;Gpbar1fl/fl) mice after oral administration of TGR5 agonist INT-777 or vehicle. n=7 (Gpbar1fl/fl INT-777) n=8 (all other groups) animals. (e) Representative images showing Agrp mRNA (in red) using RNAscope technique together with TGR5 immunodetection (GFP) in green and nuclei staining (DAPI) in blue in the acuate nucleus (ARC) of TGR5:GFP mice (n=4 animals). Arrows in the lower panel (digital zoom image) indicate cells that co-express Agrp mRNA (in red) and GFP protein (in green). Colocalization between Agrp and GFP is shown in orange (merge). Scale bars = 50 μm and 10μm plus digital zoom in the image below. (f-g) Representative images (f) and quantification (g) of Fos immunoreactivity (in red) in AgRP/NPY neurons (in green) and merge (in orange) in the hypothalamus of AgRP/NPY-GFP expressing mice after oral administration of INT-777 (n=3 animals) or vehicle (n=5 animals). Arrows in images indicate colocalization of Fos and GFP. Scale bar = 100 μm. Results represent mean ± SEM. n represents biologically independent replicates. Two-tailed Student’s t-test (a, b and g) or two-way ANOVA followed by Bonferroni post-hoc correction (c and d) vs. Gpbar1fl/fl (a and b), Gpbar1fl/fl vehicle for PomcCre;Gpbar1fl/fl (c), Gpbar1fl/fl vehicle for AgRPCre;Gpbar1fl/fl (d), or vehicle (g) groups were used for statistical analysis. P values (exact value or * P ≤ 0.05) are indicated in the figure.
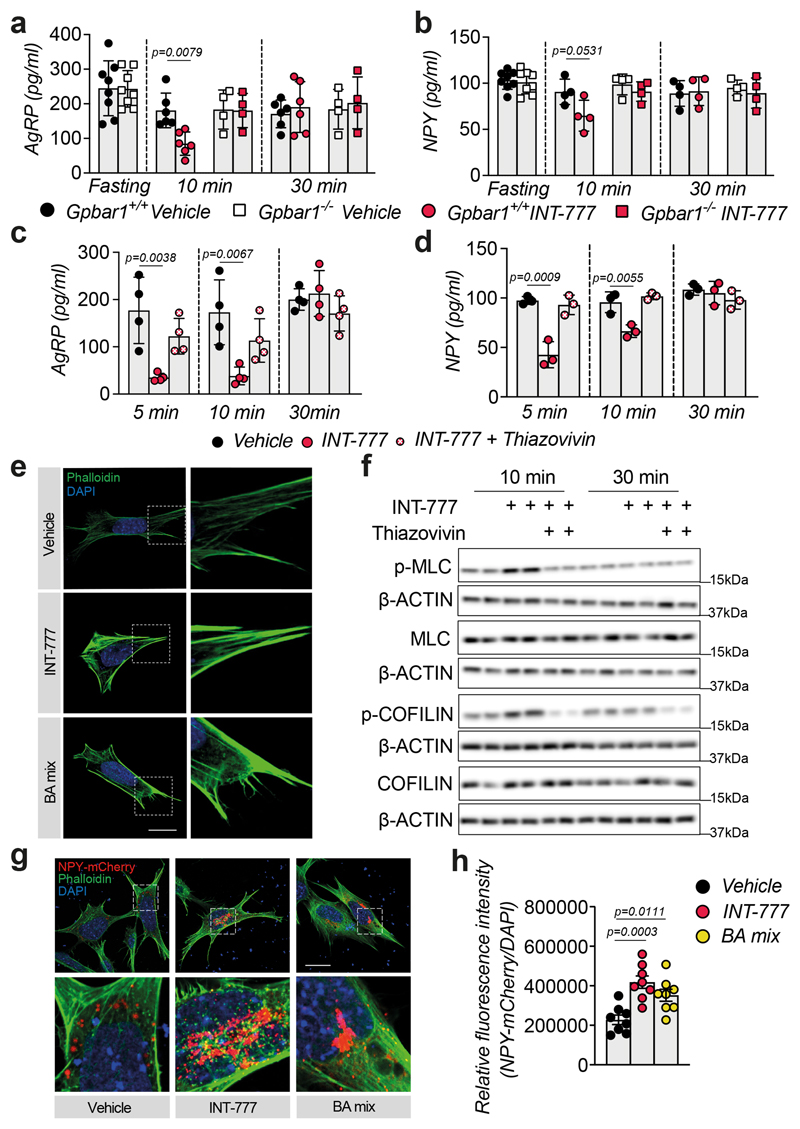
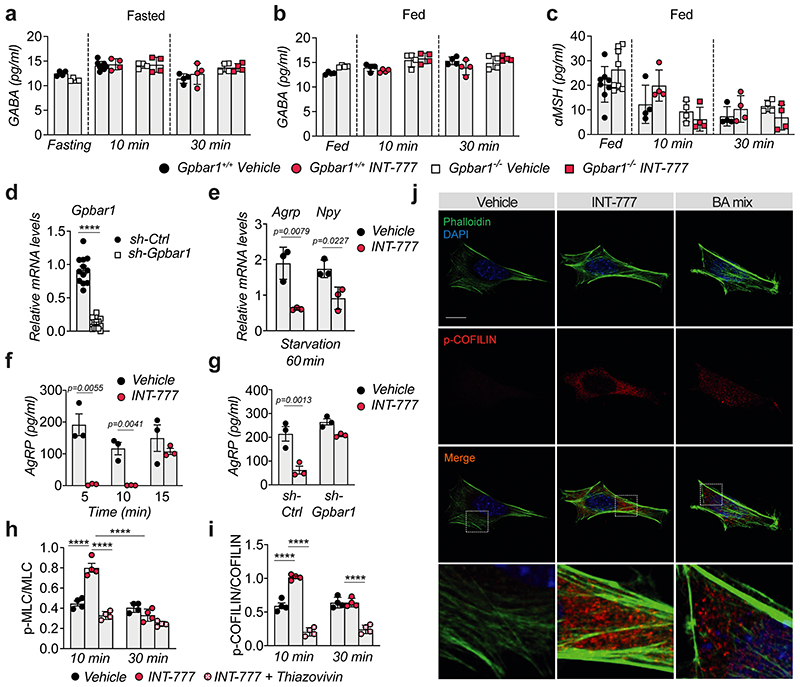
Since TGR5 mediates acute signaling responses, we reasoned that, in addition to its effect on orexigenic gene expression, this receptor could also be involved in blocking neuropeptide secretion. To test this hypothesis, we conducted ex vivo experiments on hypothalamic explants. Upon fasting, TGR5 activation with INT-777 acutely blunted the release of the orexigenic neuropeptides AgRP and NPY in Gpbar1 +/+, but not in Gpbar1 −/− explants (Fig. 4a–b). Importantly, TGR5 stimulation did not affect the secretion of gamma-aminobutyric acid (GABA), a fast-acting small molecule transmitter that is also released from AgRP/NPY neurons18 (Extended Data Fig. 4a–b). In contrast to AgRP and NPY neuropeptides, which are stored in large dense-core vesicles (DCVs), GABA is quickly released from small clear synaptic vesicles, which are docked closely to the plasma membrane in axon terminals19. The distinct processing and secretion mode could account for the observed difference in neuropeptide release. In addition, TGR5 stimulation did not affect POMC-derived alpha-melanocyte-stimulating hormone (αMSH) secretion (Extended Data Fig. 4c). These results confirm that the physiological anorexigenic effect of BA-TGR5 signaling during the postprandial state is achieved through the selective reduction of AgRP/NPY secretion.
Figure 4. TGR5 regulates orexigenic neuropeptide secretion though transient activation of the Rho/ROCK signaling pathway.
(a-b) AgRP (a) or NPY (b) release in hypothalamic explants from TGR5 wild-type (Gpbar1+/+) and germline TGR5 knock-out (Gpbar1−/−) mice after starvation or 10- and 30-min stimulation with TGR5 agonist INT-777 or vehicle. n= 4 (Gpbar1−/− 10 and 30 min), n= 6 (Gpbar1+/+ 10 and 30 min) and n=8 (fasting) (a); n= 4 (Gpbar1+/+ and Gpbar1−/− 10 and 30 min) and n=8 (fasting) (b) animals. (c-d) AgRP (c) or NPY (d) release after starvation followed by short-term (5, 10 and 30 min) stimulation with TGR5 agonist INT-777, vehicle or preincubation (30 min) with specific ROCK inhibitor (thiazovivin – 10μM) followed by INT-777 stimulation in mouse embryonic hypothalamic N41 cell line (mHypoE-N41). n=4 (c) and n=3 (d) samples. (e) Representative images of phalloidin staining to detect actin fibers (in green) and DAPI staining to detect nuclei (in blue) after 10 minutes stimulation with INT-777, bile acid (BA) mix or vehicle in mHypoE-N41 cells. Scale bar = 10μm and digital zoom. n=4 samples. (f) Representative Western blot of phosphorylated or total ROCK signaling targets (MLC and COFILIN) from the cells described in c, β-ACTIN was used as loading control. n=4 samples. (g-h) Representative images (g) or quantification of fluorescence intensity (h) of NPY vesicles (in red), phalloidin staining to detect actin fibers (in green) and DAPI staining to detect nuclei (in blue) after 10 minutes stimulation with INT-777, bile acid (BA) mix or vehicle in mHypoE-N41 transfected cells. Scale bars = 10μm and digital zoom. n=8 samples. Results represent mean ± SEM. n represents biologically independent replicates. One-way ANOVA followed by Bonferroni post-hoc correction vs. Gpbar1+/+ vehicle (a and b) or vehicle (c, d and h) groups was used for statistical analysis. P values (exact value) are indicated in the figure.
To further explore the molecular mechanism of the observed phenotype, we conducted a series of experiments in the mouse embryonic hypothalamic cell line N41 (mHypoE-N41), an AgRP/NPY positive cell line that also expresses Gpbar1 (Extended Data Fig. 4d). In line with the results obtained in vivo and ex vivo, INT-777 treatment rapidly decreased Agrp and Npy expression in mHypoE-N41 cells (Extended Data Fig. 4e) and transiently blocked AgRP release (Extended Data Fig. 4f). This effect was TGR5-dependent since it was lost upon silencing of the receptor (Extended Data Fig. 4g). Previous evidence has shown that at the early stages of DCV mobilization, the cortical actin filaments represent a primary physical barrier between the secretory vesicles and the plasma membrane and that rearrangements of the actin network are established mechanisms to restrict peptide secretion in different cell types, including neurons20. Actin depolymerization and stabilization can be modulated by the Rho/ROCK signaling pathway21,22. Specifically,activation of ROCK promotes actin fiber stabilization and thus blocks DCV exocytosis23,24. To test the hypothesis that TGR5 could transiently modulate neuropeptide secretion through the Rho/ROCK/actin axis, we evaluated AgRP and NPY secretion upon INT-777 treatment and concomitant ROCK inhibition with thiazovivin. Inhibition of ROCK abrogated the TGR5-dependent block of AgRP and NPY secretion (Fig. 4c–d). Moreover, short-term TGR5 stimulation with INT-777 or BA mix, modulated the cell cytoskeleton, favoring the stabilization and polymerization of cortical actin fibers (Fig. 4e). In line with these findings, TGR5 activation triggered transient phosphorylation of the two canonical ROCK targets, myosin light chain (MLC) and COFILIN, while concomitant thiazovivin treatment counteracted this process (Fig. 4f and Extended Data Fig. 4h–i). In addition to INT-777, short stimulation with BA mix induced COFILIN phosphorylation in vitro (Extended Data Fig. 4j) indicating that also BAs can induce ROCK signaling. In line with our previous observations, the release of NPY-containing granules in mHypoE-N41 cells was markedly reduced upon acute TGR5 activation with INT-777 or BA mix (Fig. 4g), as reflected by the increase in intracellular accumulation of fluorescent NPY-mCherry vesicles (Fig. 4h). Altogether, these results show that TGR5 activation decreases the mRNA abundance and transiently blocks the release of orexigenic neuropeptides in AgRP/NPY expressing neurons, contributing to the inhibition of food intake after physiological feeding.
BAs have been established as versatile signaling molecules mediating the communication between the enterohepatic and peripheral metabolic organs, where circulating BAs coordinate adaptation to nutritional and physiological cues8. While growing evidence is supporting the presence of BAs and their transporters in the brain, very little is known about their central actions. Our current studies reveal that a multitude of BA species can reach the hypothalamus shortly after feeding and trigger a molecular response in the hypothalamic AgRP/NPY neurons, leading to a TGR5-mediated repression of food intake during the physiological fasting-feeding transition. The anorexigenic action of BAs is furthermore supported by oral or icv administration of INT-777, a BA derivative with selective TGR5 agonistic properties, indicating that neuronal TGR5 activation is essential in this process. In line with the rapid non-genomic actions of GPCR signaling, our findings support a model in which TGR5 activation by BAs represses appetite via a dual mechanism, involving an acute inhibition of AgRP/NPY vesicle release, followed by a repression of Agrp/Npy transcript levels within the first hour after BA exposure. While the latter mechanism likely engages similar signaling transduction pathways as those by which leptin and insulin inhibit Agrp/Npy expression25, the early repressive actions of BAs on Rho/ROCK/actin-dependent AgRP/NPY exocytosis represent an unanticipated pathway by which BAs initiate repression of appetite.
While our findings demonstrate that BAs transiently accumulate in the hypothalamus to coordinate satiety during the homeostatic fasting-feeding transition, they also consolidate that the BA-TGR5 pathway is not essentially required to maintain normal energy balance during normal chow feeding, as was reported earlier4. This is not surprising as homeostasis pertains to a self-regulating process in which systems tend to maintain stability. Homeostatic circuits however can be compromised or damaged by disease or chronic HFD feeding, and it is possible that distinct TGR5-triggered circuits may prevail in these pathophysiological settings. Further studies will be needed to explore this possibility.
Unlike its function in the CNS, the role of TGR5 in peripheral metabolism has been widely studied. Activation of this receptor exerts beneficial metabolic effects such as the induction of the thermogenic program in brown3 and white7 adipose tissue to confer protection against diet-induced obesity3,4,7 and drives anti-inflammatory processes preventing the development of atherosclerosis5 and insulin resistance6. In addition, TGR5 signaling regulates glucose homeostasis4,26 by controlling the secretion of gut hormones, including glucagon-like peptide-1 (GLP-1). Besides its incretin effect, GLP-1 also signals to neurons via the systemic circulation27,28 or through activation of vagal nerves afferents to the intestine and reduces food intake29. While our findings support a role for the CNS in mediating BA-induced satiety, it cannot be excluded that TGR5 expressed in peripheral neurons could also contribute to the anorexigenic actions of BAs. Indeed, we observed that synapsin-directed recombination of the Gpbar1 locus in both peripheral and central neurons leads to a significant rise in spontaneous feeding. Consistently, a recent study showed that BAs synergistically act with the gut peptide cholecystokinin (CCK) to reduce appetite through activation of TGR5 and CCK-A receptors in the nodose ganglia neurons, an effect abolished by vagotomy30. These data, together with ours, indicate that TGR5 signaling primes the satiety signal upon feeding via both direct and indirect actions in the gut-brain axis of mice. Whether these findings will translate to human subjects remains to be investigated.
Methods
Animal studies
Generation of mouse models and tissue collection
The whole body Gpbar1 −/− and the NPY:GFP mouse models have been described previously4,31. Mice carrying floxed alleles for the Gpbar1 gene were crossed with different Cre lines (purchased from Jackson Laboratory) to achieve selective deletion of Gpbar1 in all neurons (Synapsin1-Cre, Tg(Syn1-cre)671Jxm, JAX stock #003966,32), in AgRP neurons (Agrp-IRES-Cre (Agrptm1(cre)Lowl/J, JAX stock #012899,33) or in POMC neurons (Pomc1-Cre (Tg(Pomc1-cre)16Lowl/J, JAX stock #005965,34). 8-to 15-week-old male mice were used in the study and the different lines were validated by analysis of Gpbar1 mRNA expression in the brain regions of interest. Mice were housed with ad libitum access to water and food (chow diet DS-SAFE150) and kept under a 12-hour dark/12-hour light cycle with a temperature of 22°C±1°C and a humidity of 60%±20%. Body weight was monitored weekly over an 8-week period. Body composition was analyzed by noninvasive monitoring of fat and lean mass using EchoMRI (Echo Medical Systems), as previously described35. Mice were sacrificed in the morning after an overnight fasting (12-16 hours), in the fed state (9-10 am) or after physiological feeding at the onset of the dark phase for the indicated time points after a 12-hour light-phase fasting. For biochemical analysis, tissues were collected and flash-frozen in isopentane and dry ice and stored at −80°C. For histological analysis, mice were anesthetized and transcardially perfused with 0.9% saline with heparin (10 mg/L, Sigma-Aldrich, Cat#:H3393), followed by fixative solution (4% paraformaldehyde in 0.1 M phosphate buffer). Brains were collected, post-fixed overnight, then cryo-protected with a 30% sucrose solution in PBS at 4°C and frozen.
To evaluate the expression and localization of Gpbar1 in the brain we used a transgenic mouse reporter model (TGR5:GFP), that co-expresses human GPBAR1 and the enhanced Green Fluorescent Protein (Gfp) linked by the foot-and-mouth disease virus (F2a) cleaving peptide sequence under the control of the endogenous Gpbar1 mouse promoter.
Food intake recording and oral administration of TGR5 agonist
Food intake was recorded at regular intervals by means of a TSE PhenoMaster system (TSE Systems GmbH, Germany). Each mouse was placed individually in a cage with normal bedding and free access to food and water during the acclimatation (48h) and the test (30 minutes, 1 hour or 24 hours). The system was set to have 12/12 hours light/dark cycle. The oral administration of INT-777 (TES Pharma, 1 mg/Kg, 10 mg/Kg or 100 mg/Kg in 2% CMC), BA mix (a crude bile extract containing the sodium salt of taurocholic, glycocholic, deoxycholic and cholic acids - Sigma, Cat#:S9875, 1 mg/Kg, 10 mg/Kg or 100 mg/Kg in 2% CMC) or vehicle (CMC 2% final solution) was performed through gavage 30 minutes before the start of the dark phase.
Oral administration of TGR5 agonists
Mice of the indicated genotypes were fasted overnight (12-15 hours) and gavaged with vehicle, the selective TGR5 agonist INT-777 (TES Pharma, 100 mg/Kg in 2% CMC) or BA mix (Sigma, Cat#:S9875, 10μg/ml in DMSO).
Stereotaxic administration
TGR5 agonist INT-777 (30μM) or Vehicle (DMSO) were administered by intracerebroventricular injection (icv) in the third ventricle. For the stereotaxic injections, mice were anesthetized by ip injection of Xylazine (5-10 mg/kg) plus Ketamine (80-100 mg/kg) followed by the placement on stereotaxic frame on a heating pad and with eye gel cover (Viscotears). Before the intracerebral injection the region was disinfected with Betadine. After scalp shaving, a local anesthesia was performed with 20 μl of a lidocaine (6 mg/kg) and bupivacaine (2.5 mg/kg) mix. The skin incision was of 1 cm, piercing the skull with a suitable drilling machine (Dremel), mounted on the arm of the stereotaxic frame. Stereotaxic coordinates were determined using the mouse brain atlas (K. Franklin and G. Paxinos), according to bregma: anterio-posterior (AP) −1.46 mm; dorso-ventral (DV) −5.6 mm; lateral (ML) 0 mm. The speed of injection was 0.2 μl/min via an automatic injector and the injected volume was 2 μl. After injection, suture of the skin was performed with 6-0 bioresorbable thread and the skin was disinfected. The mice were sacrificed 1 hour after icv injection and the brain was harvested and flash-frozen in isopentane and dry ice for further analysis.
Chronic icv administration of TGR5 agonists
Male C57BL/6J mice (Janvier, France) were anesthetized using isofluorane. Subcutaneous buprenorphine (0.1 mg/kg) and lidocaine at the level of the skull (0.1mL at 0.5%) were injected prior to surgery to reduce discomfort. Unconscious mice were placed on a stereotactic frame (David Kopf Instruments, USA) and an incision was made to the skin to expose the skull after fur removal and asepsis with an iodine solution (Betadine®). A burr hole for the stainless steel guide cannula (C313GS−5/SPC, G22; Plastics One®) was drilled on the skull (AP: −0.5; ML: −1.2; DV: −2.1). Following surgery, all mice received saline solution subcutaneously, and meloxicam (5 mg/kg) for 2 days. Animals were housed individually and body weight was monitored daily during one week to assess recovery. Icv treatments were performed with a 28-gauge cannula, which was fitted into the guide cannula and connected by a polyethylene tube to a micro syringe. Correct cannula placement was assessed in vivo by icv infusion of NPY (Phoenix Pharmaceuticals Inc.) in free-fed mice, as described previously36. Mice that ate >0.5g after 2 h of injection were included in the study. All infusions were carried out in free-fed mice and compounds were administered just before the dark phase. INT−777 (5 μg in 1μl 60% DMSO−40% aCSF) injections were done daily during 1 week. Body weight was measured before and 24 h after the icv administrations; food intake was measured daily after 1, 2, 4 and 24 h from the icv administrations.
Ethical approval
These experiments were authorized by the Veterinary Office of the Canton of Vaud, Switzerland (authorizations#:3143 and 3143.1), the French Ministry of Agriculture and Fisheries and the Ethical Committee of the University of Bordeaux for Animal Experimentation (authorization #13394) and by the Yale University Institutional Animal Care and Use Committee (authorization#:10670).
RNAscope
RNAscope Multiplex Fluorescent V2 assay (Bio-techne, Cat#:323110) was performed according to manufacturer’s instruction on 4 μm paraffin sections, hybridized with the probes Mm-Gpbar1-C1(Bio-techne, Cat#:318451), Mm-Agrp-C2 (Bio-techne, Cat#:400711-C2), Mm-Ppib-C1 (Bio-techne, Cat#:313911) as positive control and DapB-C1 (Bio-techne, Cat#:310043) as negative control at 40°C for 2 hours and revealed with TSA Plus Cy3 (Perkin Elmer, Cat#:NEL744E001KT). Tissues were counterstained with DAPI and mounted with FluoromountG (Bioconcept, Cat#:100.01). When indicated, sections were incubated overnight at 4°C with a goat anti-GFP primary antibody (Abcam, Cat#:ab6673 – 1:200 dilution). After incubation with a donkey anti-goat Alexa488 secondary antibody (Thermo Fisher, Cat#:A11055, 1:1000 dilution), tissues were counterstained with DAPI and mounted with FluoromountG (Bioconcept, Cat#:100.01). The imaging was performed with Zeiss LSM 700 confocal microscope (Carl Zeiss Microscopy, Germany).
Fluorescent in situ hybridization
Brain coronal sections (30μm) were cut with a CM1950 cryostat (Leica, Germany), collected, and stored in an antifreeze solution (30% ethylene glycol, 30% glycerol in KPBS) at −20°C until analysis. Fluorescent in situ hybridization with digoxigenin-labeled riboprobe against mouse AgRP (AgRP-DIG, Allen Brain Atlas, #RP_051027_01_B05) was performed as previously described37. Briefly, free-floating sections were treated with 0.2M HCl then acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine, pH=8.0 for 10 min. Between all steps, sections were rinsed in PBS with 0.01% diethylpyrocarbonate. After hybridization with AgRP-DIG (1:1000) overnight at 70°C, sections were washed at 65°C with increased stringency buffers and incubated 30 min in 3% H2O2 PBS, 1h in blocking buffer (Akoya Biosciences, Cat#:FP1012), 2h in antidigoxigenin-POD antibody (Sigma, Cat#:11207733910 ; 1:1000) and 30 min in TSA plus Cy3 system (Akoya Biosciences, Cat#:TS-000202 ; 1:100). Between all steps, sections were rinsed in TNT (100mM Tris-HCl, 150mM NaCl, 0.05% Tween20). Slices were counterstained with DAPI and mounted on gelatinized slides, cover-slipped with Prolong®. Images were acquired on a Leica DM6000 CFS TCS SP8 confocal microscope using a 20X/0.75 objective (Leica HC HPL Apo CS2). An engineer of the Bordeaux Imaging Center designed a toolset within Image J in order to automatize the quantification of AgRP positive cells within the volume of the ARC. Analyses were performed by an experimenter who was blinded to the experimental group on 3-7 ARC from 2-4 sections per animal. The number per mm3 of AgRP positive cells was then calculated.
Immunofluorescence
Brains from NPY:GFP mice31 were collected, post-fixed overnight and sectioned (50 μm) using a vibratome. Brain sections were processed for immunofluorescence staining using anti-GFP antibody (Abcam, Cat#:ab13970, 1:5000 dilution) and/or anti-Fos antibody (Santacruz, Cat#:sc−52, 1:2000 dilution). After overnight incubation and washes, sections were incubated with a secondary antibody (Alexa Fluor 488–coupled goat anti-chicken, 1:500 dilution; Life Technologies, Cat#:A11039) or with biotinylated goat anti-rabbit immunoglobulin G (Vector Laboratories, Cat#:BA-1000, 1:200 dilution) and then further incubated with streptavidin-conjugated Alexa Fluor 594 (Life technologies, Cat#:S32356, 1:2000 dilution) for 2 hours at room temperature. Sections were then cover slipped with VectaShield antifade (Vector Laboratories, Cat#:H-1000) and fluorescent images were captured with a fluorescence microscope (KEYENCE, Model BZ-X710).
Hypothalamic explants
Whole brain from overnight fasted (fasting condition) or fed (fed condition) mice was dissected in the morning and placed into a cutting solution (2.5 mM KCl, 6 mM MgCl2, 1 mM CaCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3, 10 mM Glucose, and 250 mM Sucrose). A 2 mm thick slice (at bregma levels from approx. −0.4 mm to −2.65 mm) of mediobasal forebrain including the PVN and ARC was prepared using a vibratome (Leica Microsystems, Leica VT100S), and the hypothalamus was cut from the rest of the brain in the cooled cutting solution and immediately transferred to room temperature in a 96 well plate. For fed conditions, hypothalami were incubated for 1 hour at 37 °C with 95% O2 and 5% CO2 in aCSF (124 mM NaCl, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 1.23 mM NaH2PO4, 26 mM NaHCO3, and 10 mM Glucose). After equilibration, hypothalami were incubated for 45 min in aCSF (15mM glucose-basal) and then for 10 and 30 min in a high glucose (15 mM glucose) aCSF with either INT-777 (30μM) or vehicle (DMSO). For the fasting conditions, hypothalami were incubated at 37 °C with low-glucose (6 mM glucose) aCSF. After 1 h equilibration with 95% O2 and 5% CO2, hypothalami were incubated for 45 min in low-glucose aCSF (basal) and then for 10 and 30 min in low-glucose aCSF with either INT-777 (30μM) or vehicle. Tissue viability was verified by exposure to 56 mM KCl for 45 min. At the end of each stimulation, supernatants were collected and frozen immediately in liquid nitrogen.
Cell culture, transfection and stimulation
The mouse embryonic hypothalamic mHypoE-N41 (Tebu-bio, Cat#:CLU121-A) cells were cultured in complete medium (Dulbecco’s modified Eagle’s medium (DMEM) containing glucose (4.5 g/l), 10% fetal calf serum (FBS) and penicillin/streptomycin (PS-10 mg/ml). Cells were tested for mycoplasma using MycoProbe (R&D Systems, Cat#:CUL001B), following the manufacturer’s instructions.
Transfection of the cells was performed with the jetPEI reagent (Polyplus transfection, Cat#:101-10N) according to manufacturer’s instructions. For the silencing of TGR5, cells were transfected with 2.5 μg of DNA (sh-Gpbar1 or control (sh-Ctrl)) plasmids. For the vesicle tracking experiment, cells were transfected with 2.5 μg DNA of a NPY-mCherry plasmid (Addgene, Cat#:67156).
For acute stimulation of TGR5 signaling pathway, mHypoE-N41 cells were starved for 4h with low-glucose DMEM (1g/l) FBS free medium with 0.1% free fatty acid-free bovine serum albumin (BSA), followed by treatment with INT-777 (30 μM), BA mix (10 μg/ml), or vehicle (DMSO or Ethanol) for 5, 10, 15, 30 or 60 minutes.
Neuropeptide release measurements
Detection of AgRP, NPY and GABA neuropeptides from mHypoE-N41 cell or hypothalamic explant supernatants, was performed using ready-to-use enzyme-linked immunosorbent assay (ELISA) kits (LSBio, Cat#:LS-F32623-1, LS-F5409 and LS-F4121-1, respectively) according to manufacturer’s instructions. αMSH detection was performed by radioimmunoassay (RIA) (Phoenix Pharmaceuticals, Cat#:RK-043-01) according to manufacturer’s instructions.
Immunocytochemistry
mHypoE-N41 cells were rinsed with 1X ice-cold PBS and fixed with 4% paraformaldehyde for 15 min at room temperature (RT). After three washes, the cells were permeabilized with 0.1% Triton X-100 for 15 min at RT. Blocking and antibody incubation were performed in 3% BSA. Blocking was for 1h at RT, and phospho-COFILIN Ser3 (Cell Signaling, Cat#:3311, 1:200 dilution) primary antibody was incubated overnight at 4°C. Three washes were performed before incubation with secondary anti-rabbit Alexa Fluor®647 (Jackson Immunoresearch, Cat#:711-605-152, 1:1000 dilution) antibody and phalloidin-conjugated Alexa Fluor™ 488 Phalloidin (ThermoFisher Scientific, Cat#:A12379, 1:200 dilution) for 1h at RT. Cells were washed, incubated with DAPI for 10 min and mounted. The imaging was performed with Zeiss LSM 700 confocal microscope (Carl Zeiss Microscopy, Germany).
Quantitative real-time qPCR
RNA was extracted from ARC brain punches, tissues or mHypoE-N41 cells using the RNAqueous™ Kit (ThermoFisher Scientific, Cat#:AM1912) following the manufacturer’s instructions and was transcribed to complementary DNA using QuantiTect Reverse Transcription Kit (Qiagen, Cat#:205311) following manufacturer’s instructions. Expression of selected genes was analyzed using the LightCycler 480 System (Roche) and SYBR Green chemistry. The oligonucleotide sequences used are listed in the Supplementary Table 2. All quantitative polymerase chain reaction (PCR) results were presented relative to the mean of 36b4 and B2m housekeeping genes (DDCt method). The average of at least three technical repeats was used for each biological data point.
Western blotting
Samples were lysed in RIPA buffer [50mM Tris-HCl (pH 7.4), 150mM KCl, 1mM EDTA, 1% Triton X-100, 0.5% Sodium deoxycholate, protease and phosphatase inhibitors]. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes. Blocking and antibody incubations were performed in 5% BSA. Myosin light chain 2 (MLC2) (Cat#:3672, 1:1000 dilution), phospho-MLC2 (Ser19) (Cat#:3671, 1:1000 dilution), COFILIN (Cat#:5175, 1:1000 dilution) and phospho-COFILIN (Cat#:3311, 1:1000 dilution) antibodies were purchased from Cell Signaling; beta-ACTIN (Sigma-Aldrich, Cat#:A5441, 1:1000 dilution) antibody detection was used as loading control. Antibody detection reactions were developed by enhanced chemiluminescence (Advansta) and imaged using the c300 imaging system (Azure Biosystems). Quantification was done using ImageJ software.
BA quantification
BAs and INT-777 were profiled by stable isotope dilution mass spectrometry assay at the Metabolomics Platform of University of Lausanne. Standard solutions for calibration and samples were prepared as described previously38. Briefly, calibrators were prepared by serial dilutions with stripped serum (C0-C7). Internal standard (IS) mixture contained 13 deuterium-labeled BAs. Sample preparation. The frozen hypothalamus was homogenized in tissue homogenizer (Precellys) by the addition of MeOH:H2O (2:1) with 0.1% FA (300 μL) and ceramic beads (for 2 × 20 seconds at 10,000 rpm). Homogenized extracts were centrifuged for 15 minutes at 14,000g at 4°C, and the supernatants were further processed using solid phase extraction (SPE) as described previously38. LC-HRMS analysis: BA quantification was performed on a Vanquish Horizon (ThermoFisher Scientific) ultra-high performance liquid chromatography (UHPLC) system coupled to Q-Exactive Focus mass spectrometer interfaced with a HESI source operating in negative ionization mode. Chromatographic separation was carried out using an Acquity UPLC® HSS T3 1.8 μm 2.1 x 100 mm column. The mobile phase was composed of A = 5 mM Ammonium Acetate and 0.1% formic acid in H2O and B = 0.1% formic acid in ACN. Gradient elution was set as described in Supplementary Table 1. The flow rate was 350 μL/min, column temperature 30°C and the injection volume 20 μL. Mass spectrometry parameters were set to: full scan in narrow mass range m/z 370-522, mass resolving power = 70,000 FWHM and AGC target = 5e5. Data were processed as described previously38. BA concentrations were reported to the protein concentration.
Statistics
Differences between two groups were assessed using two-tailed t tests. Differences between more than two groups were assessed using one-way analysis of variance (ANOVA). To compare the interaction between two factors, two-way ANOVA tests were performed. ANOVA, assessed by Bonferroni’s post-test, was used when comparing more than two groups. GraphPad Prism 9 was used for all statistical analyses. The studies were either replicated or the results were confirmed by using different approaches (RT-qPCR, Western blot, immunohistochemistry) yielding similar results. All P values ≤0.05 were considered significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
Extended Data
Extended Data Fig. 1. BA mix and INT-777 reach the hypothalamus after oral or icv administration, respectively, and reduce food intake by activating TGR5.
(a) Conjugated and unconjugated BA species measured in the hypothalamus 30- and 60-min after oral administration of a BA mix at three different doses or vehicle in wild-type mice (n=8 animals). Bars represent the mean from 8 replicates. (b) One-hour cumulative food intake of wild-type mice after oral administration of a BA mix at three different doses or vehicle. n=8 (Vehicle and 1 mg/Kg) and n=10 (10 mg/Kg and 100 mg/Kg) animals. (c) Gpbar1 mRNA levels in arcuate nucleus (ARC)-enriched hypothalamic punches of TGR5 wild-type (Gpbar1+/+) and germline TGR5 knock-out (Gpbar1−/−) mice. n=8 animals. (d) One-hour (1h) food intake measured after the first icv injection (d1 – 1h) of vehicle (n=8 animals) or INT-777 (n=9 animals) in wild-type mice. (e) Two-hour (2h) food intake measured after the first icv injection (d1 – 2h) of vehicle (n=8 animals) or INT-777 (n=9 animals) in wild-type mice. (f) Food intake measured after a daily (right before the dark phase) icv injection of vehicle (n=8 animals) or INT-777 (n=9 animals) in wild-type mice at the indicated time points. (g) Body weight change (expressed as a percentage of the initial body weight) of vehicle (n=8 animals) or INT-777 (n=9 animals) in wild-type mice at the indicated time points. Results represent mean (a) or mean ± SEM (b-g). n represents biologically independent replicates. Two-tailed Student t-test (c, d and e) or two-way ANOVA followed by Bonferroni post-hoc correction (b) vs Vehicle (b, d-e) or Gpbar1+/+ (c) were used for statistical analysis. P values (exact value, * P ≤ 0.05 or **** P ≤ 0.0001) are indicated in the figure.
Extended Data Fig. 2. TGR5 activation in neurons reduces food intake.
(a) Quantification of AgRP neurons in the arcuate nucleus of over-night fasted TGR5 wild-type (Gpbar1+/+, 4 animals) and TGR5 knock-out (Gpbar1−/−, 3 animals) mice. (b) Body weight curve of 8-week-old Gpbar1+/+ and Gpbar1−/− mice over an 8-week period. n=8 animals. (c) Body composition (lean and fat mass expressed as a percentage over the body weight (BW)) of Gpbar1+/+ and Gpbar1−/− mice. n=10 animals. (d) Validation of the TGR5:GFP knock-in mouse model (n=4 animals) in which the mouse Gpbar1 gene was substituted with a construct containing the human GPBAR1, the F2a sequence and the Gfp. Brains from wild-type mice (n=3 animals) were used as controls. (e) Gpbar1 mRNA levels in arcuate nucleus (ARC)-enriched hypothalamic punches of control mice (Gpbar1fl/fl) and neuron-specific TGR5 knock-out (SynCre;Gpbar1fl/fl) mice. n=8 animals. (f) Body weight curve of 8-week-old of control mice (Gpbar1fl/fl, 10 animals) and neuron-specific TGR5 knock-out (SynCre;Gpbar1fl/fl, 9 animals) mice over an 8-week period. (g) Body composition (lean and fat mass expressed as percentage over body weight (BW)) of Gpbar1fl/fl (n=10 animals) and SynCre;Gpbar1fl/fl (n=6 animals) mice. (h) 12-hour food intake of Gpbar1fl/fl (n=7 animals) and SynCre;Gpbar1fl/fl (n=8 animals) mice. (i) Pomc mRNA levels in ARC-enriched hypothalamic punches of Gpbar1fl/fl and SynCre;Gpbar1fl/fl mice. n=7 (SynCre;Gpbar1fl/fl fasting) and n=8 (all other groups) animals. (j) 12-hour cumulative food intake of control mice (Gpbar1fl/fl) and neuron-specific TGR5 knock-out (SynCre;Gpbar1fl/fl) mice after oral administration of INT-777 (100 mg/Kg) or vehicle. n=5 (Gpbar1fl/fl), n=8 (SynCre;Gpbar1fl/fl INT-777) and n=9 (SynCre;Gpbar1fl/fl vehicle) animals. (k) Total food intake of Gpbar1fl/fl and SynCre;Gpbar1fl/fl during night and day. n=6 (Gpbar1fl/fl INT-777 night), n=7 (Gpbar1fl/fl other groups), n=8 (SynCre;Gpbar1fl/fl other groups) and n=9 (SynCre;Gpbar1fl/fl vehicle day) animals. (l) 12-hour cumulative food intake of control mice (Gpbar1+/+) and germline TGR5 knock-out (Gpbar1−/−) mice after oral administration of the TGR5 specific BA agonist INT-777 (100 mg/Kg) or vehicle. n=6 (Gpbar1+/+ vehicle), n=7 (Gpbar1−/− vehicle) and n=8 (all other groups) animals. (m) Total food intake of Gpbar1+/+ and Gpbar1−/− mice during night and day. n=7 (Gpbar1−/− vehicle night) and n=8 (all other groups) animals. Results represent mean ± SEM. n represents biologically independent replicates. Two-tailed Student t-test (d, e, h and i), one-way ANOVA (k and m) or two-way ANOVA (j and l) followed by Bonferroni post-hoc corrections vs. Wild-type (d), Gpbar1fl/fl (e, h and i), Gpbar1fl/fl Vehicle (j and k) or Gpbar1+/+ Vehicle (l and m) were used for statistical analysis. P values (exact value, * P ≤ 0.05 or **** P ≤ 0.0001) are indicated in the figure.
Extended Data Fig. 3. Disruption of TGR5 in AgRP or POMC neurons in CD-fed mice does not change body weight and composition.
(a) Gpbar1 mRNA levels in arcuate nucleus (ARC)-enriched hypothalamic punches of control mice (Gpbar1fl/fl) and POMC neuron-specific TGR5 knock-out (PomcCre;Gpbar1fl/fl) mice. n=6 animals. (b) 12-hours food intake of Gpbar1fl/fl and PomcCre;Gpbar1fl/fl mice. n=7 animals. (c) Body weight curve of 8-week-old of control mice (Gpbar1fl/fl, n=7 animals) and AgRP neuron-specific TGR5 knock-out (AgRPCre;Gpbar1fl/fl, n=9 animals) mice over an 8-week period. (d) Body composition (lean and fat mass expressed as percentage over body weight (BW)) of Gpbar1fl/fl and AgRPCre;Gpbar1fl/fl mice. n=8 animals. (e) Body weight curve of 8-week-old of control mice (Gpbar1fl/fl) and POMC neuron-specific TGR5 knock-out (PomcCre;Gpbar1fl/fl) mice over an 8-week period. n=10 animals. (f) Body composition (lean and fat mass expressed as percentage over body weight (BW)) of Gpbar1fl/fl (n=7 animals) and PomcCre;Gpbar1fl/fl (n=8 animals) mice. (g-h) Representative images (g) and quantification (h) of Fos immunoreactivity (in red) in AgRP/NPY neurons (in green) and merge (in orange) in the hypothalamus of fed or fasted AgRP/NPY:GFP reporter mice. Arrows in images indicate colocalization of Fos and GFP. Scale bar = 100 μm. n=4 animals. Results represent mean ± SEM. n represents biologically independent replicates. Two-tailed Student t-test (a and h) vs. Gpbar1fl/fl (a) or fed AgRP/NPY-GFP (h) groups were used for statistical analysis. P values are indicated in the figure.
Extended Data Fig. 4. TGR5 inhibits AgRP secretion through activation of the Rho/ROCK/actin signaling axis.
(a) GABA release in ex vivo hypothalamic explants from TGR5 wild-type (Gpbar1+/+) and germline TGR5 knock-out (Gpbar1−/−) mice after starvation or starvation followed by 10- and 30-minutes stimulation with TGR5 agonist INT-777 or vehicle. n=4 (all other groups) and n=8 (Gpbar1+/+ vehicle 10 min) animals. (b-c) GABA (b) or αMSH (c) release in ex vivo hypothalamic explants from Gpbar1+/+ and Gpbar1−/− mice after high-glucose (15mM) solution to mimic fed conditions, followed by 10- and 30-minutes stimulation with TGR5 agonist INT-777 or vehicle. n=4 (b), n=4 (all other groups in c) and n=8 (Gpbar1+/+ and Gpbar1−/− fed, c) animals. (d) Gpbar1 mRNA levels in mouse embryonic hypothalamic N41 cell line (mHypoE-N41) after Gpbar1 silencing using transient transfection of sh-Gpbar1 or control vector (sh-Ctrl). n=12 samples. (e) Agrp and Npy mRNA levels in mHypoE-N41 cells after 60 min starvation conditions followed by 60 min stimulation with INT-777 or vehicle. n=3 samples. (f) AgRP release after starvation followed by short-term (5, 10 and 15 min) stimulation with TGR5 agonist INT-777 or vehicle in mHypoE-N41 cells. n=3 samples. (g) AgRP release after starvation followed by 10 min stimulation with INT-777 or vehicle in mHypoE-N41 Gpbar1-silenced cells using transient transfection of sh-Gpbar1 or control vector (sh-Ctrl). n=3 samples. (h and i) Quantitative densitometry of phosphorylated vs total ROCK signaling targets (MLC and COFILIN) from the cells described in Fig. 4f. n=4 samples. (j) Representative images of phalloidin staining to detect actin fibers (in green) and DAPI staining to detect nuclei (in blue), phosphorylated COFILIN (p-COFILIN) immunodetection (in red) and merge, after 10 min stimulation with INT-777, BA mix or vehicle in mHypoE-N41 cells. Scale bar = 10μm and digital zoom. n=4 samples. Results represent mean ± SEM. n represents biologically independent replicates. Two-tailed Student t-test (d-f) or one-way ANOVA followed by Bonferroni post-hoc correction (g, h and i) vs. sh-Ctrl (d) or vehicle (e-i) groups were used for statistical analysis. P values (exact value or **** P ≤ 0.0001) are indicated in the figure.
Supplementary Material
Acknowledgments
We thank Andréane Fouassier, Sabrina Bichet, Thibaud Clerc, Philippe Colin, Aline Aebi, Tony Teav (Metabolomics Platform), the Histology facility and the Phenotyping Unit (UDP) for technical assistance, the Mouse Clinical Institute in Strasbourg for the generation of the TGR5:GFP mouse strain, and the Bordeaux Imaging Center (CNRS-INSERM and Bordeaux University; ANR-10-INBS-04) for confocal microscopy.
Funding
This work was funded by the Swiss National Science Foundation (SNSF N° 310030_189178 and CRSII5_180317/1, the Kristian Gehrard Jebsen Foundation, and the Ecole Polytechnique Fédérale de Lausanne (EPFL) (to K.S.), NIH DK097566 (to S.D.) and INSERM, ANR-17-CE14-0007, ANR-10-EQX-008-1 OPTOPATH (to D.C.). L.A.V.V. and A.P. were supported by a postdoctoral fellowship from the AXA Research Fund.
Footnotes
Author contributions: A.P., L.A.V.V. and K.S. conceived and designed the project. B.S., S.D., and D.C. provided critical expertise. A.G. and R.P. provided chemical compounds. A.P., L.A.V.V., N.B., Y.S., Q.H., V.F., A.C-J., P.Z., G.B., S.J. and A.B. performed experiments. A.P., L.A.V.V., N.B., V.F., A.C-J., P.Z., G.B., S.J. and J.I. analyzed the data. A.P., L.A.V.V., and K.S. wrote the manuscript. All the authors read and approved the manuscript.
Competing interests: A.G. and R.P. are cofounders of TES Pharma. R.P. is the president and CEO of TES Pharma. The other authors declare no conflict of interest.
Data availability
All the datasets generated during the current study are available upon request to the corresponding author..
References
- 1.McMillin M, DeMorrow S. Effects of bile acids on neurological function and disease. FASEB J. 2016;30:3658–3668. doi: 10.1096/fj.201600275R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 4.Thomas C, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pols TWH, et al. TGR5 Activation Inhibits Atherosclerosis by Reducing Macrophage Inflammation and Lipid Loading. Cell Metabolism. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perino A, et al. TGR5 reduces macrophage migration through mTOR-induced C/EBPβ differential translation. J Clin Invest. 2014;124:5424–5436. doi: 10.1172/JCI76289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velazquez-Villegas LA, et al. TGR5 signalling promotes mitochondrial fission and beige remodelling of white adipose tissue. Nat Commun. 2018;9:245. doi: 10.1038/s41467-017-02068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perino A, Demagny H, Velazquez-Villegas LA, Schoonjans K. Molecular Physiology of Bile Acid Signaling in Health, Disease and Aging. Physiol Rev. 2020 doi: 10.1152/physrev.00049.2019. [DOI] [PubMed] [Google Scholar]
- 9.Angelin B, Björkhem I, Einarsson K, Ewerth S. Hepatic uptake of bile acids in man. Fasting and postprandial concentrations of individual bile acids in portal venous and systemic blood serum. J Clin Invest. 1982;70:724–731. doi: 10.1172/JCI110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higashi T, et al. Unconjugated bile acids in rat brain: Analytical method based on LC/ESI-MS/MS with chemical derivatization and estimation of their origin by comparison to serum levels. Steroids. 2017;125:107–113. doi: 10.1016/j.steroids.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Mano N, et al. Presence of protein-bound unconjugated bile acids in the cytoplasmic fraction of rat brain. J Lipid Res. 2004;45:295–300. doi: 10.1194/jlr.M300369-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X, et al. The Brain Metabolome of Male Rats across the Lifespan. Sci Rep. 2016;6:24125. doi: 10.1038/srep24125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiriyama Y, Nochi H. The Biosynthesis, Signaling, and Neurological Functions of Bile Acids. Biomolecules. 2019;9 doi: 10.3390/biom9060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorbek G, Lewinska M, Rozman D. Cytochrome P450s in the synthesis of cholesterol and bile acids--from mouse models to human diseases. FEBS J. 2012;279:1516–1533. doi: 10.1111/j.1742-4658.2011.08432.x. [DOI] [PubMed] [Google Scholar]
- 15.Yanguas-Casás N, Barreda-Manso MA, Nieto-Sampedro M, Romero-Ramírez L. TUDCA: An Agonist of the Bile Acid Receptor GPBAR1/TGR5 With Anti-Inflammatory Effects in Microglial Cells. J Cell Physiol. 2017;232:2231–2245. doi: 10.1002/jcp.25742. [DOI] [PubMed] [Google Scholar]
- 16.Keitel V, et al. The bile acid receptor TGR5 (Gpbar-1) acts as a neurosteroid receptor in brain. Glia. 2010;58:1794–1805. doi: 10.1002/glia.21049. [DOI] [PubMed] [Google Scholar]
- 17.Pellicciari R, et al. Discovery of 6alpha-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. J Med Chem. 2009;52:7958–7961. doi: 10.1021/jm901390p. [DOI] [PubMed] [Google Scholar]
- 18.Waterson MJ, Horvath TL. Neuronal Regulation of Energy Homeostasis: Beyond the Hypothalamus and Feeding. Cell Metab. 2015;22:962–970. doi: 10.1016/j.cmet.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 19.Schöne C, Burdakov D. Glutamate and GABA as rapid effectors of hypothalamic ‘peptidergic’ neurons. Front Behav Neurosci. 2012;6:81. doi: 10.3389/fnbeh.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meunier FA, Gutiérrez LM. Captivating New Roles of F-Actin Cortex in Exocytosis and Bulk Endocytosis in Neurosecretory Cells. Trends Neurosci. 2016;39:605–613. doi: 10.1016/j.tins.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Amano M, Nakayama M, Kaibuchi K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken) 2010;67:545–554. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sit S-T, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124:679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 23.Symons M, Rusk N. Control of vesicular trafficking by Rho GTPases. Curr Biol. 2003;13:R409–418. doi: 10.1016/s0960-9822(03)00324-5. [DOI] [PubMed] [Google Scholar]
- 24.Wolf M, et al. ADF/Cofilin Controls Synaptic Actin Dynamics and Regulates Synaptic Vesicle Mobilization and Exocytosis. Cereb Cortex. 2015;25:2863–2875. doi: 10.1093/cercor/bhu081. [DOI] [PubMed] [Google Scholar]
- 25.Varela L, Horvath TL. Leptin and insulin pathways in POMC and AgRP neurons that modulate energy balance and glucose homeostasis. EMBO Rep. 2012;13:1079–1086. doi: 10.1038/embor.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perino A, Schoonjans K. TGR5 and Immunometabolism: Insights from Physiology and Pharmacology. Trends in Pharmacological Sciences. 2015;36:847–857. doi: 10.1016/j.tips.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Orskov C, Poulsen SS, Møller M, Holst JJ. Glucagon-like peptide I receptors in the subfornical organ and the area postrema are accessible to circulating glucagon-like peptide I. Diabetes. 1996;45:832–835. doi: 10.2337/diab.45.6.832. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto H, et al. Glucagon-like peptide-1-responsive catecholamine neurons in the area postrema link peripheral glucagon-like peptide-1 with central autonomic control sites. J Neurosci. 2003;23:2939–2946. doi: 10.1523/JNEUROSCI.23-07-02939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rüttimann EB, Arnold M, Hillebrand JJ, Geary N, Langhans W. Intrameal hepatic portal and intraperitoneal infusions of glucagon-like peptide-1 reduce spontaneous meal size in the rat via different mechanisms. Endocrinology. 2009;150:1174–1181. doi: 10.1210/en.2008-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, et al. Satiety induced by bile acids is mediated via vagal afferent pathways. JCI Insight. 2020;5 doi: 10.1172/jci.insight.132400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto S, et al. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science. 2004;304:110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong Q, Ye C-P, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci. 2008;11:998–1000. doi: 10.1038/nn.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balthasar N, et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Argmann CA, Champy M-F, Auwerx J. Evaluation of energy homeostasis. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb29b01s73. Chapter 29, Unit 29B.1. [DOI] [PubMed] [Google Scholar]
- 36.Bellocchio L, et al. Activation of the sympathetic nervous system mediates hypophagic and anxiety-like effects of CB1 receptor blockade. Proc Natl Acad Sci U S A. 2013;110:4786–4791. doi: 10.1073/pnas.1218573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saucisse N, et al. POMC neurons functional heterogeneity relies on mTORC1 signaling. 2020 doi: 10.1101/2020.03.25.007765. [DOI] [PubMed] [Google Scholar]
- 38.Sorrentino G, et al. Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the datasets generated during the current study are available upon request to the corresponding author..