Abstract
Purpose
The abundance and effects of structural variation at BRCA1/2 in tumours are not well understood. In particular, the impact of these events on homologous recombination repair deficiency (HRD) has yet to be demonstrated.
Experimental Design
Exploiting a large collection of whole genome sequencing data from high grade serous ovarian carcinoma (N=205) together with matched RNA-seq for the majority of tumours (N=150), we have comprehensively characterised mutation and expression at BRCA1/2.
Results
In addition to the known spectrum of short somatic mutations (SSMs), we discover that multi-megabase structural variants (SVs) are a frequent, unappreciated source of BRCA1/2 disruption in these tumours, and we find a genome wide enrichment for large deletions at the BRCA1/2 loci across the cohort. These SVs independently affect a substantial proportion of patients (16%) in addition to those affected by SSMs (24%), conferring homologous recombination repair deficiency (HRD) and impacting patient survival. We also detail compound deficiencies involving SSMs and SVs at both loci, demonstrating that the strongest risk of HRD emerges from combined SVs at both BRCA1 and BRCA2 in the absence of SSMs. Further, these SVs are abundant and disruptive in other cancer types.
Conclusions
These results extend our understanding of the mutational landscape underlying HRD, increase the number of patients predicted to benefit from therapies exploiting HRD, and suggest there is currently untapped potential in SV detection for patient stratification.
Keywords: high grade serous ovarian cancer, BRCA1, BRCA2, homologous recombination repair deficiency, structural variation, whole genome sequencing
Introduction
Homologous recombination deficiency (HRD) is identifiable in many cancers and is particularly prominent in high grade serous ovarian cancer (HGSOC)1, affecting around 50% of tumours2 and leaving detectable mutational spectra across the tumour genome3. The mutational landscape of HGSOC is dominated by extensive genomic copy number changes and structural rearrangement driven by chromosome instability and defective DNA repair, rather than the patterns of recurrent point mutation in tumour suppressor and oncogenes often observed in other solid tumours4,5.
Germline short mutations (GSM) disrupting the coding sequence of BRCA1 and BRCA2 are the most common types of HRD-associated defect, occurring in 8% and 6% of HGSOC patients respectively, while disruptive somatic short mutations (SSM) in these genes are present in an additional 4% and 3% of HGSOC patients respectively6,7. These GSMs and SSMs include single nucleotide variants (SNVs) as well as short indels, with frameshifts being the predominant mechanism of inactivation. These BRCA-deficient tumours represent approximately 20% of patients with HGSOC. An additional 11% of tumours are thought to be BRCA-deficient through epigenetic silencing of BRCA1 2,8. Mutational or epigenetic inactivation of other genes involved in the HR pathway are also thought to confer HRD in a smaller proportion of HGSOC patients7,9–12. Genome-wide patterns of SNVs, indels and structural variation have been identified as strong predictors of BRCA1/2 deficiency3,13. These mutational signatures of BRCA1/2 deficiency are also found in additional tumours that lack short variants at BRCA1/2, suggesting that other unknown aberrations may also be involved in HRD3. The demonstration of BRCA1/2 loss and detection of HRD is crucial in the management of HGSOC14–19, breast20,21, pancreatic22 and prostate23 cancer to identify patients whose outcome is markedly improved by the administration of PARP inhibitors14–16. PARP inhibitors selectively kill HRD cells since these cells are deficient in HRR (homologous recombination repair) and can neither resolve stalled replication forks nor accurately repair the increased number of double strand breaks that result from the use of these agents24.
The clinical importance of GSMs and SSMs at BRCA1/2 is well established in cancer17,20,25–28. In contrast, the abundance and effects of structural variants (SVs) at BRCA1/2 are not well understood, particularly for large SVs (>1Mb) encompassing multi-megabase regions. Similarly, the compound effects of SVs and short mutations occurring simultaneously at BRCA1 and BRCA2 are poorly studied. Matched tumour-normal whole genome sequencing (WGS) of freshly-frozen tissue is accepted as the best resource to accurately detect SVs in tumours but in the past such data have been scarce for HGSOC29,30. Here we comprehensively characterise the mutational landscape of BRCA1/2 in HGSOC using the largest collection to date of uniformly processed WGS data (N=205), comprising two previously published cohorts5,6, as well as a large novel cohort described here for the first time. We document the prevalence of HRD across these three cohorts to reveal the complexity of the mutations associated with HRD, their impact on gene expression and associations with clinical outcome.
Materials and methods
WGS data from matched tumour and normal blood samples were uniformly remapped and analysed to generate a range of somatic mutation calls (Supplementary Figure 1) for three HGSOC cohorts: chemoresistant or relapsed tumours from the Australian Ovarian Cancer Study5 (AOCS) (N=80), pre-treatment WGS primary tumours from The Cancer Genome Atlas (TCGA)6 (N=44) and the previously unpublished primary tumours from Scottish High Grade Serous Ovarian Cancer (SHGSOC) study (N=81) of which 16 (20%) have had neoadjuvant chemotherapy. The combined cohort (N=205) presented here was uniformly analysed as follows.
Scottish sample collection and preparation for WGS
Scottish HGSOC samples were collected via local Bioresource facilities at Edinburgh, Glasgow, Dundee and Aberdeen and stored in liquid Nitrogen until required. HGSOC patients were determined from pathology records and were included in the study where there was matched tumour and whole blood samples. Tumour samples were divided into two for DNA and RNA extraction and slivers of tissue were taken, fixed in formalin and embedded in paraffin wax (FFPE). Samples were only included if they were confirmed as HGSOC and there was greater than 40% tumour cellularity throughout the tumour, determined using H&E staining of the FFPE sections and pathology review. Somatic DNA was extracted from the tumour and germline DNA was extracted from whole blood. Quality control was then carried out on the resultant DNA to ensure sufficient purity and quality (Supplementary Methods). Only when all quality control requirements were satisfied was the DNA sequenced at the Glasgow Precision Oncology Laboratories.
This study was carried out in accordance with the principles of the Declaration of Helsinki. Ethical approval for the use of human tissue specimens for research was obtained from the Lothian NHS Research Scotland Human Annotated Bioresource (ethics committee reference 15/ES/0094-SR494). Correlation of molecular data to clinical outcome and clinicopathological variables in ovarian cancer was approved by South East Scotland Research Ethics Committee 2 (reference 2007/W/ON/29). All relevant ethical regulations were complied with, including the need for written informed consent where required.
Sequence acquisition and availability
WGS and RNA-seq reads were downloaded in compressed FASTQ format from the sequencing facility (SHGSOC) or in aligned BAM format (including unaligned reads) from the European Genome/Phenome Archive (AOCS) and the Bionimbus Protected Data Cloud (TCGA_US_OV). The reads obtained in BAM format were query-sorted and converted to FASTQ. All whole genome sequence and RNAseq data for the SHGSOC cohort will be made available on publication via EGA.
Primary processing of WGS
Reads were aligned to the GRCh38 reference genome and somatic and germline variant calling was run using a bcbio31 1.0.7 pipeline (see Supplementary Information for full pipeline configuration, program, resource versions and references). Germline SNPs and indels were called with GATK 4.0.0.0 HaplotypeCaller. Somatic SNVs and indels were called as a majority vote between Mutect2, Strelka2 and VarDict. Small variants were annotated with Ensembl Variant Effect Predictor v91 and filtered for oxidation artefacts by GATK 4.0.0.0 FilterByOrientationBias. Somatic structural variants were called with Manta 1.2.1 and somatic copy number variants with CNVkit 0.9.2a0. Loss of heterozygosity and somatic copy number variants were also identified with CLImAT. Sample quality control was performed with Qsignature 0.1 to identify sample mix-ups and VerifyBamId 1.1.3 to identify sample contamination. Tumour cellularity was estimated using both CLImAT’s estimates and p53 variant allele frequency. These measures were compared to the qPure estimates for the AOCS cohort5 and histopathological estimates for the SHGSOC cohort with very good concordance (Supplementary Figure 2). The CLImAT estimates were used as the final estimates of cellularity.
Filtering of small variants (SNVs and indels) at BRCA1/2
Germline short variants at BRCA1/2 were filtered to include only damaging pathogenic variants for the purposes of establishing BRCA1/2 mutational status. Included variants were all of moderate or high impact according to VEP. Variants with a pathogenic or risk factor annotation according to ClinVar32 were included (n=145). Remaining variants with a ClinVar benign or likely benign status were excluded (n=1147). Remaining frameshift or nonsense (stop gained) or splice donor/acceptor variants were included (n=125). Remaining missense variants with damaging SIFT and PolyPhen predictions were included (n=36). Remaining missense variants called as damaging by only one of SIFT and PolyPhen were considered borderline and were excluded if their CADD score < 2033. Missense or inframe variants with no Clinvar, SIFT or PolyPhen evidence were excluded.
Somatic short variants at BRCA1/2 were also filtered for pathogenicity to include variants that: were annotated by VEP as being of high or moderate impact, were pathogenic according to at least one of SIFT or PolyPhen and had a high CADD score. In addition, we excluded somatic variants with an allele frequency less than 0.4.
Curating a high-confidence list of structural variants at BRCA1/2
We identified structural variants in HGSOC patients using Illumina’s paired and split read based structural variant detection tool, Manta34. However, we observed that Manta was failing to detect a large number of very large deletions (>1Mb) that had been identified using depth of coverage-based approaches in PCAWG35. Therefore, we chose to supplement these calls with copy number variants greater than 1 Mb in size that were called by one caller (CNVkit) using evidence from read depth and were also confirmed by an allele-specific copy number caller (CLImAT) which provides an additional layer of evidence in addition to read depth as it also incorporates the shift in allele frequency of heterozygous SNPs within the potential copy number variant into its variant calling algorithm which also accounts for aneuploidy and sample cellularity. Deletions were assumed to be heterozygous if the copy number as estimated by both CNVkit and CLImAT was 1 although this may be a conservative estimate of allelic loss in the presence of subclones. We visually inspected all the identified structural variants in the Integrative Genomics Viewer (IGV) (v2.4.10). The magnitude of coverage and log fold change were inspected to confirm either duplication or deletion. For structural variants in the range 300bp-30kb, the paired end sequencing reads were manually reviewed including looking at split reads, paired end insert size, read coverage and pair-orientation. We compared our filtered set of large CNVs in the samples included in PCAWG to PCAWG’s copy number calls and found that 40/41 of our variants in these samples were also identified by PCAWG.
Enrichment of structural variation at BRCA1/2
Permutation analyses were carried out using the R package RegioneR36 to investigate whether large deletions overlap more often with BRCA1 and BRCA2 than they do elsewhere in the genome. We carried out 100,000 permutations to simulate the null hypothesis throughout the genome for each gene and judged significance at alpha = 0.05. (Supplementary Figure 3).
We investigated whether large deletions, large duplications and inversions are enriched at BRCA1 and BRCA2 within their respective chromosomes using 100,000 circularised permutations to generate the null distribution of overlaps in each case (Supplementary Figure 4). The observed number of overlaps with BRCA1/2 was well within the range of this null distribution and therefore showed no evidence of within chromosome enrichment which is perhaps unsurprising given the large sizes of these events relative to the length of their chromosomes.
Implementation of HRDetect
To predict the level of HR deficiency in each tumour sample we implemented the HRDetect algorithm as published by Davies and Glodzik et al3. We based our implementation on a Snakemake pipeline made publicly available by Zhao et al37, with some modifications to ensure accurate recapitulation of the original method (Supplementary Methods). As some of the AOCS cohort included here were also used in the validation of HRDetect in the original publication we were able to compare our implementation for the same patients with that of the authors. Our implementation of HRDetect was very highly correlated with the original HRDetect implementation on the same samples (Spearman’s rho = 0.92).
We used the weights for the independent variables that were defined by the original model rather than retraining the model on our data as the original weights trained on a breast cancer dataset have been shown to perform well on ovarian cancer datasets and our total sample size is substantially smaller than that used to train the model originally. As input we used the somatic SNV and indel calls identified by the ensemble calling approach described above; the structural variant calls made by Manta and the copy number segments defined by CNVkit.
Scottish RNA sample preparation and sequencing
HGSOC samples were collected and underwent quality control as described for the DNA samples used for WGS. Somatic RNA was extracted from the resulting RNA sample, underwent quality control and was quantified. RNAseq was carried out by the Edinburgh Clinical Research Facility on an Illumina NExtSeq500 (Further details in Supplementary Methods).
Primary processing of RNA-seq
RNA-seq data was analysed using the Illumina RNA-seq best practice template. Briefly, reads were aligned to the GRCh38 reference genome and quality control was carried out. Salmon quant38 was used to quantify the expression of transcripts against the GRCh38 RefSeq transcript database indexed using the salmon index (k-mers of length 31). Transcript-level abundance estimates were imported into R and summarised for further gene-level analyses. For differential expression analyses, raw expression counts were used by the DESeq2 package39. For visualization of gene expression, counts were normalized using the variance stabilizing transformation. Previously published RNA-seq data available for the AOCS5 (N=80) and TCGA6 (N=30) cohorts, together with novel RNA-seq data for the SHGSOC (N=40) cohort, generated for the present study as detailed above, were processed in this way from FASTQ.
Curation and acquisition of the patient’s clinical information
Scottish High Grade Serous Ovarian Cancer (SHGSOC)
Clinical data for the SHGSOC cohort was retrieved from the Edinburgh Ovarian Cancer Database40, the CRUK Clinical Trials Unit Glasgow and available electronic health records (ethics reference 15/ES/0094-SR751).
Australian Ovarian Cancer Study (AOCS) & The Cancer Genome Atlas (TCGA)
The clinical information including survival end-points, age and stage at diagnosis is available for these patients as part of the PCAWG project35.
Statistical analyses
All downstream statistical analyses were carried out in R (v3.6.0) using Jupyter notebook (v4.3.1).
Differential expression analyses of BRCA1/2 in tumours with and without deletions at BRCA1/2
In order to compare the gene expression levels at BRCA1/2 between tumours with and without BRCA1/2 deletions, we used the package DESeq2 to test for differential expression between the raw gene expression counts at each gene between samples with and without a deletion at that gene. At BRCA1, samples that also had a short variant had significantly lower expression than those that had a deletion alone. As a result we only considered the samples with a deletion in the absence of a short variant. At BRCA2, this was not the case and the samples with short variants in addition to a deletion had comparable levels of expression to those with deletions alone so were included in the analysis. Cohort and tumour sample cellularity were included as covariates in the model formula.
Univariable analyses of genomic features and risk of HR deficiency
The risk of HR deficiency in tumours with BRCA1/2 mutations, grouped by type, relative to those tumours without BRCA1/2 mutations were determined using Fisher’s exact tests. The effect of mutations at BRCA1 and BRCA2 were determined together and, where sample size permitted, separately for mutational categories including: germline short mutations only, somatic short mutations only, single deletion which overlaps at least one exon in the absence of a short variant or other SVs, deletion of at least one exon at both BRCA1 and BRCA2 in the absence of a short variant, inversion spanning BRCA1 in the absence of short variants or deletion and duplication spanning BRCA2 in the absence of short variants or deletion. We also considered the impact of the presence of a short variant accompanied by a deletion at either of the genes. These categories are further described together with their labels and colour coding in the Supplementary Information. The relative risk conferred by each mutational category was calculated in comparison to the group of patients without BRCA1/2 mutations or SVs. Samples where BRCA1/2 promoter methylation had been detected were excluded except for where the effect of BRCA1 promoter methylation was itself being examined. All samples with BRCA1 promoter methylation are predicted to be HR deficient so pseudo counts of 1 are used to estimate the effect size which is therefore a likely underestimate. P-values were adjusted for the impact of multiple testing using Benjamini-Hochberg correction and were considered together with the effect sizes in the reporting of results.
Multivariable elastic-net regularised regression model
Given the relative sparsity of the data and the correlation between features we used a multivariable elastic-net regularised regression model for the binary outcome of HRD defined by a probability of HRD greater than 0.7 from HRDetect. The data were partitioned into train and test sets (80:20) and the tuning parameters were optimised, in order to maximise the AUC, using 10-fold cross validation of the training set. The model was then fitted to the training set and the model performance assessed using the test set. The input variables available for selection were: BRCA1 germline short variant status; BRCA1 somatic short variant status; the presence of a large somatic deletion at BRCA1; the presence of a large somatic deletion at BRCA1 and a BRCA1 short variant; all the corresponding variables for BRCA2; the presence of an inversion at BRCA1; the presence of a duplication at BRCA2; the presence of a large somatic deletion at BRCA1 and at BRCA2 (double deletion); BRCA1 promoter hypermethylation; whole genome doubling; genome-wide load of SNVs, large CNVs and SVs in addition to cohort and tumour cellularity. The data was partitioned and the model optimised and fitted to 100 train-test splits of the data in order to assess the robustness of the feature selection.
Survival-time analyses of the impact of HRD on overall survival
Follow up information including overall survival time was available for 190 out of 205 patients, of which 144 were deceased by the time of last follow up. The association between genome-wide patterns of HRD and progression-free survival was also assessed. Progression-free interval time was available for 151 of the patients from the AOCS and SHGSOC cohorts of which 129 relapsed by the time of last follow up. The effect of the HRDetect score, as a measure of the probability of HRD in the tumour, on the length of time that patients survived after diagnosis (overall survival-time) and the time between diagnosis and first radiologically defined progression (progression-free survival-time) was assessed using Cox proportional hazards models stratified by cohort. Multivariable models were also fitted adjusting for age and stage at diagnosis and the Schoenfeld residuals were examined. In all cases hazard ratios reported correspond to a 1 standard deviation increase in HRDetect score. Survival probability through time was compared between HR deficient (HRDetect score > 0.7) and HR proficient (HRDetect score <=0.7) patients in Kaplan-Meier plots. This was repeated excluding the patients with BRCA1/2 short variants to assess the impact of HRD driven by other events on survival.
BRCA1/2 SVs in other cancer types and their impact on expression
We used the consensus SV and CNV calls generated and included in the 2017-01-19 release of PCAWG to investigate the abundance of SVs at BRCA1/2 in other cancer types. Samples (N=2,567) were from all cancer subtypes included in PCAWG that are recommended for use in subtype-specific analyses. Disruptive SVs were identified by intersecting the variant calls with the exons of BRCA1/2. Abundance of SVs, deletions and duplications were tabulated separately for each of the cancer types. Cancer subtypes are ordered from left to right by overall SV burden as determined by PCAWG.
Using gene expression quantifications from matched RNA-seq for N=735 of these samples, we compared BRCA1 expression in samples with and without BRCA1 deletions from each cancer type separately in differential expression analyses carried out using DESeq239. Cancer types were included if there were greater than 15 samples in total and if at least 3 of these samples harboured deletions. This effect was also measured across all samples pan-cancer adjusting for differences in expression between cancer types in the model. These analyses were repeated for BRCA2 expression. Effects on expression and the data behind them were displayed after variance stabilising transformation of the expression counts.
Results
Large structural variants are a frequent source of BRCA1/2 disruption in HGSOC
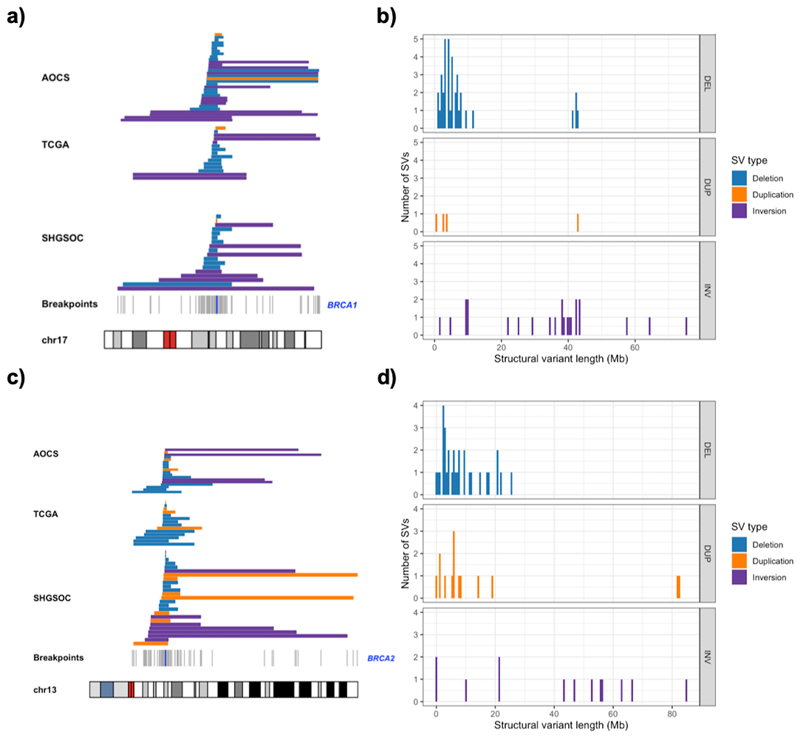
A variety of SVs were detected at the BRCA1/2 loci but large multi-megabase deletions spanning the entirety of BRCA1 or BRCA2 dominated. In all three cohorts, some deletions encompass more than 10% of chromosomes 17 or 13 though the majority are more focal (median BRCA1 deletion = 4.9Mb, median BRCA2 deletion = 6.2Mb) (Figure 1). Heterozygous deletions occur at similar rates at BRCA1 (16%) and BRCA2 (14%) overall, and at comparable rates between the similarly sized AOCS and SHGSOC cohorts (Supplementary Table 1). In 6/205 (3%) samples in the combined cohort, we observe large deletions at both BRCA1 and BRCA2 in the absence of short mutations at both genes. Permutation analyses reveal that BRCA1 and BRCA2 are deleted more than expected given the observed distribution of large deletions throughout the HGSOC genome (Supplementary Figure 3). This is consistent with selection for these events as has been reported for SNVs at BRCA1/2 41 but we can not exclude the role of mutational bias in producing these enrichments of deletions. Inversions occur less often than deletions, but do occur in isolation in 6% of samples, and within groups of large overlapping inversions in 5% of samples. In addition, we observe large duplications that span the entire length of either BRCA1 or BRCA2 in all cohorts (2% and 7% respectively) (Supplementary Table 2). Loss of heterozygosity was near ubiquitous at BRCA1 (202/205, 99%), of which 166 events were copy number neutral, and was present at BRCA2 in more than half of samples (118/205, 58%) of which 91 were copy number neutral. We observe similar genome-wide SV mutational spectra in each cohort despite the clinical differences among them: in particular AOCS represents chemoresistant/relapsed cases, while SHGSOC is composed mainly of samples taken before treatment. This suggests that large SVs predicted to impair BRCA1/2 function are a general feature of HGSOC evolution.
Figure 1. Abundance, location and size of structural variants overlapping BRCA1/2 in three HGSOC cohorts.
a) Alignment of structural variants overlapping BRCA1 across the AOCS, TCGA and SHGSOC cohorts with breakpoints marked in grey according to their position on chromosome 17. Location of BRCA1 marked by a blue line with deletions (blue), duplications (orange) and inversions (purple). b) The distribution of sizes of structural variants (Mb), overlapping BRCA1 across all cohorts. c) Alignment of structural variants overlapping BRCA2 across the three cohorts with breakpoints marked in grey according to their position on chromosome 13. Location of BRCA2 marked by a blue line. d) The distribution of sizes of structural variants (Mb) overlapping BRCA2 across all cohorts with deletions (blue), duplications (orange) and inversions (purple).
Large deletions spanning BRCA1/2 are associated with lower gene expression
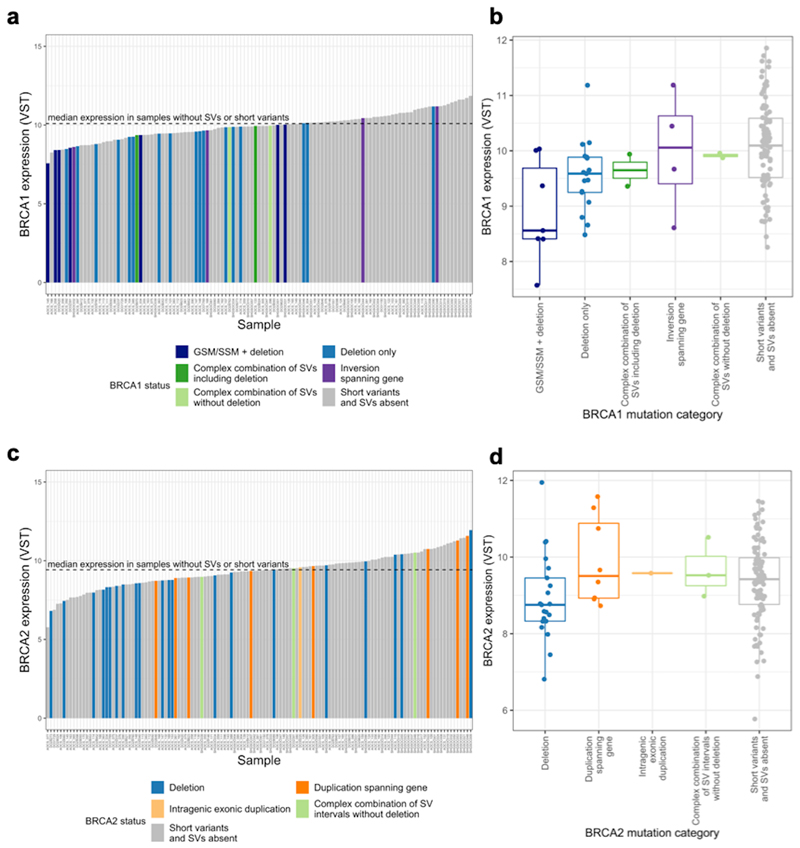
We found that patients with a large deletion spanning BRCA1, in the absence of a GSM or SSM, had lower BRCA1 expression than those patients with no BRCA1 GSM, SSM or SVs (log2 fold change of no GSM/SSM/SVs versus a deletion only = 0.45, p-value=0.0093) (Figure 2). We also found that tumours with a large deletion spanning BRCA2 had lower BRCA2 expression than those without GSM/SSM/SVs at BRCA2 (log2 fold change in expression between no GSM/SSM/SV versus a deletion = 0.43, p-value = 0.037). In the analysis of BRCA2 expression, we included tumours with large deletions that also had GSM or SSM in the remaining copy, having shown that in our data BRCA2 expression is not significantly different in these tumours (Supplementary Figure 5). In spite of the unavoidable heterogeneity in tumour expression data among samples although variation in tumour purity was accounted for, the trends observed here are consistent with large BRCA1/2 deletions, reducing BRCA1/2 expression, though indirect mechanisms cannot be excluded.
Figure 2. Structural variation and expression of BRCA1/2 in the combined HGSOC cohort.
a)c) Expression of BRCA1/2 (variance stabilising transformed RNA-seq counts) across samples ordered from lowest to highest expression. Median BRCA1/2 expression is indicated by a black dashed line. Sample bars are coloured by BRCA1/2 mutational category. b)d) Boxplot of BRCA1/2 expression for each category of BRCA1/2 mutation. The BRCA1 deletion category is split into those samples with SNVs and deletions and those with only deletions as their expression is significantly different (Supplementary Figure 5a). This is not the case for BRCA2 so all samples with deletions are considered together to maximise the available sample size (Supplementary Figure 5b).
Large deletions spanning BRCA1/2 contribute to HRD independently of pathogenic SNVs and indels
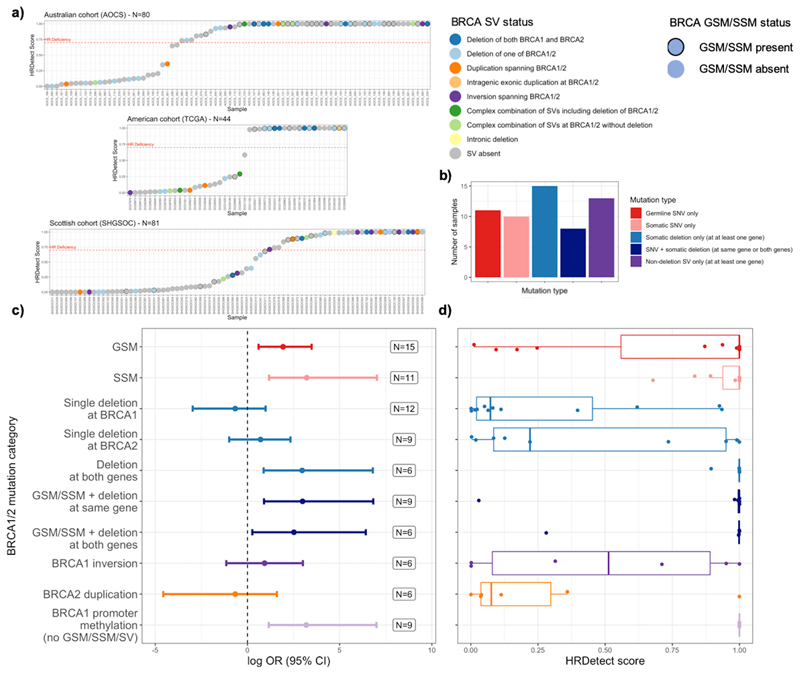
We examined the functional impact of all BRCA1/2 mutations detected across all cohorts using an established method, HRDetect3, which predicts HRD based upon genome-wide mutational spectra, and therefore provides a functional readout for the HRR pathway in tumours (Figure 3).
Figure 3. BRCA1/2 mutation classes and repair deficiency in three HGSOC cohorts.
a) Predictions of HRD for three large cohorts of HGSOC coloured by BRCA1/2 structural variant status and outlined by BRCA1/2 mutation status. Categories of mutation include GSMs and SSMs. Categories of structural variation include deletions, duplications, inversions and complex overlapping combinations thereof as formally described in the methods and absence of structural variants at BRCA1/2. The HRDetect scores range from 0, least likely to be HR deficient to 1, most likely to be HR deficient. The red dashed line represents the threshold of 0.7 representing HRD<sup>3</sup><sup>3</sup><sup>3</sup><sup>3</sup><sup>3</sup><sup>3</sup>. b) The number of HRD tumours with different categories of BRCA1/2 short variants, deletions or non-deleting SVs. c),d) The increase in log odds ratio of HRD (HRDetect score > 0.7) associated with different categories of mutation and structural variation at BRCA1/2 in comparison to the frequency of the reference category where samples lack evidence of BRCA1/2 inactivation (GSV, SSV, SV). All categories apart from the BRCA1 promoter methylation category itself also exclude tumours with BRCA1 promoter methylation where this is known (TCGA and AOCS). ORs are defined using Fisher’s Exact tests for enrichment. Error bars represent 95% confidence intervals. Mutually exclusive categories of mutation examined include GSV only, SSV only, the presence of a deletion at one or both genes without a GSV or SSV, the presence of a short variant together with deletion of one or both genes, non-deleting SVs in samples without short variants or deletions, samples with BRCA1 promoter methylation and no mutational BRCA1/2 deficiencies.
As expected, tumours with GSMs or SSMs in BRCA1/2, are more likely to have HR deficient tumours (HRDetect scores >0.7) than those tumours without GSMs, SSMs or SVs at these genes (GSM OR 6.9, 95% CI 1.8 – 33, p-value=2.3x10-3, adj. p-value= 3x10-2; SSM OR 25, 95% CI 3.2-1121, p-value = 1.7x10-4, adj. p-value = 2.3x10-3) (Figure 3). Four samples with GSMs at BRCA2 demonstrated low HRDetect scores and accordingly showed no evidence for subsequent loss of the wild-type allele in the tumour which suggests that certain GSMs that are predicted to be disruptive are insufficient to generate HRD (Supplementary Table 3a).
The majority of samples (65%) with BRCA1/2 deletions were HRD (HRDetect score >0.7), though some deletions occur in tumours that also harbour short mutations (19/49). Since many of the samples with BRCA1/2 deletions lack short mutations (30/49, 61%), analysis of the effects of deletions independent of short variants was undertaken. The compound effect of deletions at both the BRCA1 and BRCA2 loci in the absence of short variants is particularly pronounced, demonstrating a significantly increased risk of HRD (OR 19, CI 2.4-896, p=1.3x10-3, adj p =1.7x10-2) and consistently high HRDetect scores. Furthermore, compound deletions at both loci generate an OR that is comparable with other classes of HRD mutations known to have clinical importance, including the well-studied disruptive BRCA1/2 short variants (Figure 3c),d)). Single deletions at either BRCA1 or BRCA2 do not consistently confer an increased risk of HRD in the absence of a BRCA1/2 short variant (Figure 3) though the analysis may be underpowered to detect small effects given the current sample size. The HRDetect scores for samples with these single deletions form a bimodal distribution for which we have been unable to find a defining characteristic for the difference, such as the length of the deletion, resultant level of gene expression or a background of whole genome doubling. The estimated effect that we observe of a single deletion at BRCA2 merits further investigation although it does not achieve statistical significance in our data (OR 2, 95% CI 0.37-10.3). Previous studies have identified rearrangement signatures associated with HR deficiency42. We observe an elevation in these signatures, particularly in the case of rearrangement signature 5, in samples with deletion at BRCA1/2, which is broadly consistent with the levels of elevation observed in the presence of short mutations at BRCA1/2 (Supplementary Figure 6). However, HRDetect incorporates additional genome-wide sources of information including the more powerful presence of indel microhomology leading to more accurate HRD prediction.
Beyond deletions the functional impact of other classes of SV, such as inversions or duplications, is less well studied. Half of the samples with only BRCA1 inversions bear an HR deficient signature which suggests that although in isolation their presence is not associated with HR deficiency these samples can show evidence of HRD perhaps via another unappreciated route (Figure 3). In contrast, only one of the samples with only BRCA2 duplications is HR deficient, which suggests potential for enrichment in HR proficient samples but this would need to be further explored in greater sample sizes.
Deletions are a frequent source of BRCA1/2 inactivation in repair deficiency
Samples across the combined cohort never had more than one short mutation across BRCA1 and BRCA2. This suggests that SSMs are not a mechanism for biallelic inactivation of a gene affected by a GSM. This is consistent with reports from a previous study of HGSOC41 (Supplementary Tables 3a,b). In contrast, of the HGSOC tumours with a GSM at BRCA1/2 predicted to cause HRD, we find that 11/32 (34%) show evidence for an SV at the same gene which, may contribute to HRD if the SV occurs on the other allele (Supplementary Tables 2 and 3). We find that most of these somatic events (8/11 = 73%) are large deletions, while two tumours possess more than one SV spanning the same gene as the GSM and one more shows evidence of somatic duplication. The importance of ‘second hit’43 mutations in tumours is well established44 but these data suggest that multi-megabase deletions have an under-appreciated role in this phenomenon in HGSOC. Across the three cohorts, 24% (50/205) of patients have a disruptive short variant at either BRCA1 or BRCA2, and 30% (15/50) of these patients also carry a BRCA1/2 deletion at the same locus. Also, it appears that SVs, including deletions, can occur at both BRCA1 and BRCA2 in the same sample. Large somatic deletions occur at both BRCA1 and BRCA2 in 13 samples and in 7 of these samples there is no short variant at either gene, although 1 sample had a hypermethylated BRCA1 promoter. These data suggest that large deletions and other SVs disproportionately contribute to biallelic inactivation in HGSOC, driven by the unusually high rates of structural variation seen in this cancer45.
Integrative modelling reveals complex mechanisms underlying repair deficiency
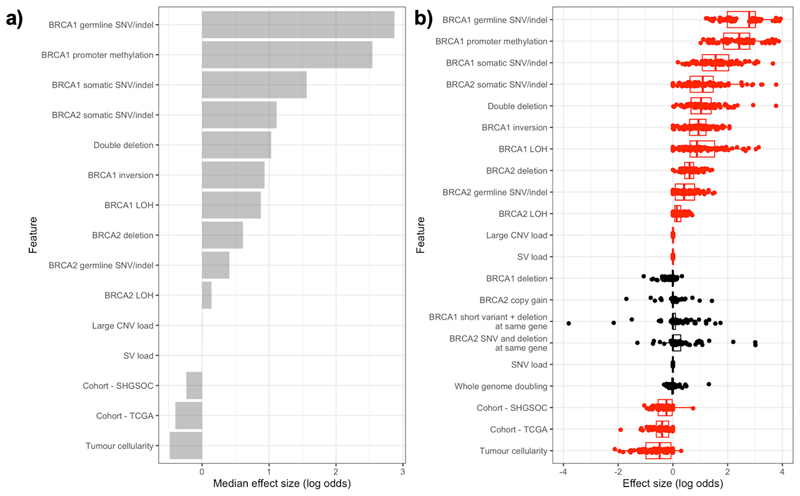
We comprehensively modelled the effects of a range of genomic alterations at the BRCA1/2 loci on HRD, to investigate the relative importance of these features in explaining the patterns of HRD observed. Given the relative sparsity of the data and the correlation between features we used a multivariable elastic net regularised regression model.
In addition to the previously reported impact of short variants at BRCA1/2 and BRCA1 promoter hypermethylation on HRD, large deletions at BRCA2 confer an increased risk of HRD (Figure 4). Furthermore, samples with double deletions, where deletions are found at both BRCA1 and BRCA2, are more likely to be HR deficient. Importantly, the influence of these double deletions on HRD exceeds that of genome-wide large CNV loads and genome-wide SV loads. Also, large inversions at BRCA1 are independently associated with an increased risk of HRD. The functional impact of these events on the gene is currently unknown but this suggests that these events may either be markers for processes that impact the gene’s function or may even directly impact the function of the gene themselves. The model’s ability to predict HRD was good with a mean ROC curve AUC of 0.75, which although promising suggests that there are additional unknown sources of HRD.
Figure 4. Integrative modelling of repair deficiency in HGSOC.
a) Median effect sizes of genomic features selected to predict HRD, using elastic net regularised regression on 100 training/test set splits. Model performance was measured for each split and average AUC = 0.75. Binary mutational status variables (e.g. presence/absence of BRCA1 somatic SNV) were included as factors and continuous variables were standardised to allow comparisons between variables. b) Distributions of effect size for each variable on HRD (log odds) in each training/test set split. Variables in red are selected for inclusion by the model in more than half of the training sets.
We can explain the observed pattern of HRD by the presence of mutational or epigenetic defects at the BRCA1/2 genes in 81 out of 106 samples with predicted HRD (72 GSM/SSM/SV at BRCA1/2, 9 with BRCA1 promoter methylation) but a further 25 samples with HRD remain unexplained. On further examination, we found that all of these samples harboured damaging GSMs and/or SSMs at other HR genes (defined by KEGG pathway annotation; Supplementary Tables 5, 6 and 7), motivating analysis of the potential roles of mutations at loci other than BRCA1/2 and their inclusion in an expanded model. However, we found no convincing evidence for a strong influence of mutations at other HR genes or the combined expression of genes dysregulated in the presence of HRD (Supplementary Figure 7).
HRD is associated with longer survival in the absence of disruptive short variants at BRCA1/2
In HGSOC, while HRD as a result of GSMs and SSMs at BRCA1/2 is associated with response to platinum and PARP inhibition and improved survival, the relationship between HRD resulting from disruption of BRCA1/2 via other mechanisms is less clear1,6,8,11,46–49
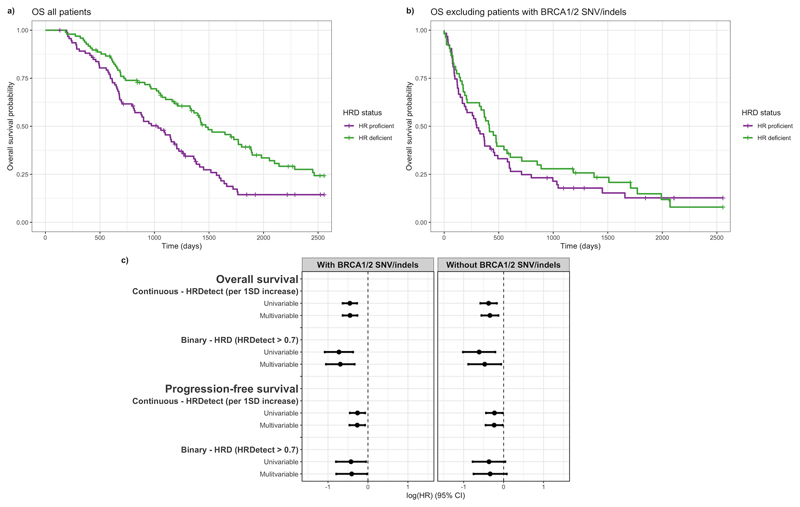
A higher probability of HRD is significantly associated with longer overall survival in our combined cohort (HR = 0.64, 95% CI 0.53–0.76 (per 1 standard deviation increase), p-value = 8.4x10-7) and this effect is only slightly attenuated by adjustment for patient age and tumour stage at diagnosis (Figure 5a). Notably, this effect persists (HR=0.67, 95% CI 0.56-0.84 (per 1 standard deviation increase), p-value=3.6x10-4) (Figure 5b) when we exclude the patients with BRCA1/2 GSM/SSM, who are already known to have longer survival. We see a similar effect when we examine the effect of HRD on progression-free survival (PFS) (HR=0.77, 95% CI 0.64 – 0.93 (per 1 standard deviation increase), p-value = 0.007) which also is robust to the exclusion of patients with BRCA1/2 GSM/SSM (HR=0.79, 95% CI 0.64-0.98 (per 1 standard deviation increase), p-value=0.03). Correspondingly, in all of these instances patients with HRD tumours (HRDetect >0.7) have longer survival times than patients with non-HRD tumours. The effects observed are consistent with the presence of HRD through mechanisms other than BRCA1/2 GSMs and SSMs affecting overall survival.
Figure 5. Predicted HRD is associated with patient survival in the absence of short variants at BRCA1/2.
a) The effect of HRD on overall survival time after diagnosis (in days) in HGSOC. (N (events) =190 (144)). b) The effect of HRD on overall survival time after diagnosis (in days) in HGSOC patients without BRCA1/2 GSV/SSV. (N (events) =145 (113)). c) Forest plot showing the effects of HRD on overall and progression-free survival, unadjusted and adjusted for age and stage at diagnosis in a multivariable model. Estimates are also shown with tumours with SNVs/indels at BRCA1/2 excluded. Kaplan-Meier plots compare survival times between HR deficient and HR proficient patients as defined by HRDetect score above and below 0.7. Hazard ratio estimates (on a log scale) are taken from Cox proportional hazards models and correspond to a 1 standard deviation increase in HRDetect score, stratified by cohort and adjusted for age and stage at diagnosis.
Large deletions at BRCA1/2 are abundant in other cancer types and disrupt expression
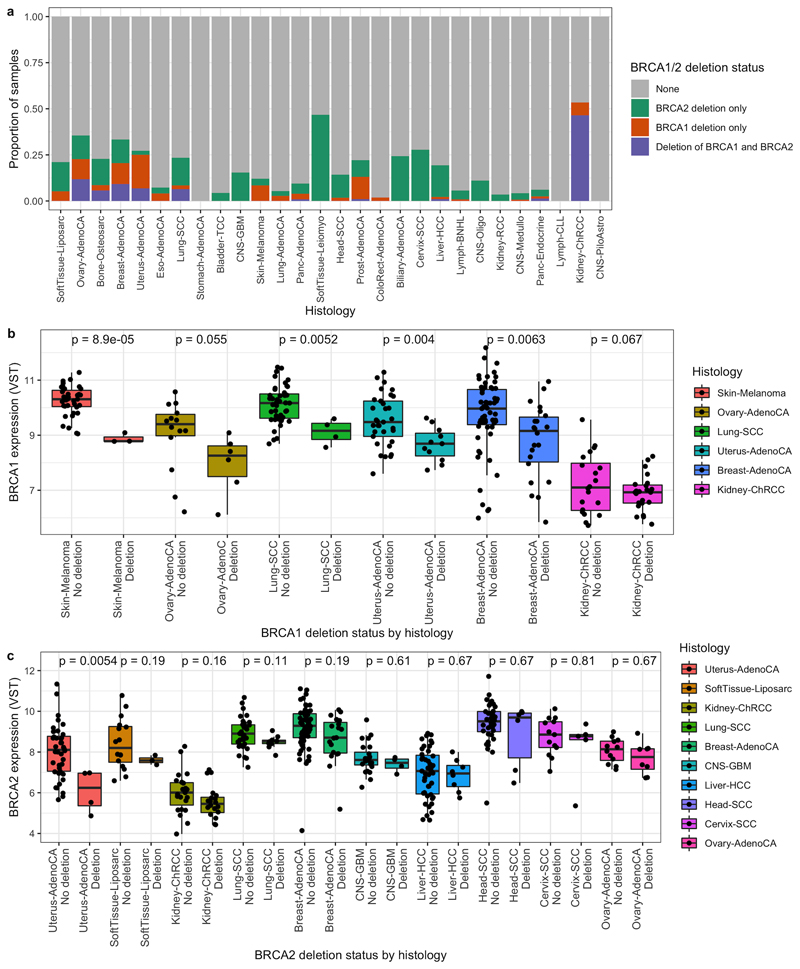
Cancers with other sites of origin vary in their level of structural variation. It is possible that large deletions may compromise BRCA1/2 function in these cancers also. The Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium recently generated conservative consensus SV calls based upon WGS data across many tumour types but did not report specifically on SV patterns at BRCA1/250. These data illustrate that large deletions (Figure 6a) (and other SVs (Supplementary Figure 8)) at BRCA1/2 are common across a variety of non-HGSOC cancer types, including in cancers also known to show evidence of HRD such as breast and prostate cancer. In addition, two rare tumour types show notably higher frequencies of BRCA1/2 deletions: chromophobe renal cell carcinoma (CHRCC) and leiomyosarcoma. Exploiting matched RNA-seq for N=735 of the PCAWG samples we found that BRCA1 and BRCA2 have significantly lower expression in those samples with deletions at BRCA1 and BRCA2 respectively, in comparison to in those samples without a deletion (log2 fold change of no BRCA1 deletion versus a BRCA1 deletion= 0.94, p-value=5.9x10-12; log2 fold change of no BRCA2 deletion versus a BRCA2 deletion= 0.6, p-value=2.9x10-6) after adjusting for primary site. Furthermore, BRCA1 expression was consistently compromised within all cancer types with sufficient samples to test individually (Figure 6b),c)).
Figure 6. Deletions at BRCA1/2 in other cancer types and their impact on expression.
a) The proportion of samples with deletions at BRCA1, BRCA2 or both, by primary site in PCAWG. b) BRCA1 expression in tumours with and without deletions at BRCA1 coloured by primary site. c) BRCA2 expression in tumours with and without deletions at BRCA2 coloured by primary site. Gene expression visualised in variance stabilising transformed (VST) counts. P-values are from differential expression analyses conducted using DESeq2 for each primary site separately and are adjusted for multiple testing using the Benjamini-Hochberg approach. Only sites with at least 15 samples in total and at least 3 samples with a deletion at the gene in question were included.
Discussion
DNA repair deficiency in general and HRD in particular represent cancer cell vulnerabilities that have recently been exploited to exceptional patient benefit14,15,17,19–24,46. However, the understanding of the genomic mechanisms that give rise to HRD is incomplete and the identification of patients whose tumours are homologous recombination deficient remains inaccurate.
In the largest collection of HGSOC WGS data examined to date, with matched expression data for most of the 205 tumours included, we have revealed new insights into the genesis of HRD in HGSOC based upon genome-wide mutational spectra. We show that structural variation at BRCA1/2 in HGSOC is frequent and dominated by multi-megabase deletions which are significantly enriched at BRCA1 and BRCA2 relative to the rest of the HGSOC genome. Examining transcriptomic data for the same samples we have shown that large deletions overlapping BRCA1/2 are associated with lower BRCA1/2 expression, suggesting a direct impact of these deletions on gene function in many cases. Large deletions spanning BRCA1/2 contribute to HRD independently of short variants, and samples with compound deletions affecting both BRCA1 and BRCA2 generate the highest risk of HRD. The frequent inactivation of BRCA1/2 by large deletions in HGSOC is novel to our knowledge, and the original analysis of the AOCS cohort reported only one BRCA1/2 large deletion (>1Mb)5 (Patch et al (2015), Supplementary Table 4.2). The original TCGA cohort analysis did report frequent losses of the chr13q and chr17q chromosome arms (including the BRCA1/2 loci) based upon SNP microarray data6 (Bell et al (2011), Supplementary Table S5.1), but such data are known to generate high levels of false positives and negatives47,48 and these losses were not postulated to affect BRCA1/2 function. Thus previous assessments of SVs impacting the BRCA1/2 loci have been characterised by under-reporting, likely to be a result of the use of less sensitive algorithms tuned to detect smaller focal deletions5 as well as CNA estimates derived from SNP microarray data5,6 and exome-restricted sequencing data6,51. Other types of structural variation are less frequent but still evident, such as large inversions at BRCA1 and duplications at BRCA2. The impact of these categories of mutation on the function of the gene is less well studied but our data suggest that when BRCA1 inversions in particular are considered together with the other mutational events in the tumour, their presence may aid prediction of HRD.
Our data also suggest a significant frequency of BRCA1 and BRCA2 loss by structural variation in a series of other cancers including both cancers known to suffer BRCA1 and BRCA2 loss due to SNVs or indels such as breast, pancreatic and prostate cancer as well as a number of cancers outside of this group including squamous cell lung cancer and cervical cancer. Perhaps most notably soft tissue leiomyosarcoma had a high incidence of large deletions at BRCA2 (47%) and chromophobe renal cancers (CHRCC) had a high incidence of large deletions of both BRCA1 (53%) and BRCA2 (47%). CHRCC often undergoes concurrent deletion of both BRCA1 and BRCA2 as a result of whole chromosome arm losses as has been previously reported52 and trials of PARP inhibition as a therapy for renal cell carcinoma are ongoing53. The fact that across these cancer types, large BRCA1 and BRCA2 deletions were associated with loss of gene expression suggests functionality is lost and that strategies to detect these genomic events and trials of PARP inhibition should also be considered in these patients.
Finally, we have constructed an integrated model of HRD in HGSOC, including a large variety of mutation and expression-based variables across the combined cohort. This model supports an independent role for structural variation at BRCA1/2 in HRD and highlights the diversity of routes that tumours may follow to reach HRD. Given this diversity, and the substantial fraction of samples where HRD is detected in the absence of any detectable BRCA1/2 mutations, we conclude that the direct detection of HRD in HGSOC using genome-wide sequencing data is a valuable addition to the search for inactivating mutations in HR pathway genes. This is likely to be the case for other cancers showing evidence for HRD, such as uterine, lung squamous, oesophageal, sarcoma, bladder, lung adenocarcinoma, head and neck, and gastric carcinomas1. The variety of events sufficient for a tumour to develop HRD is not well understood, but recent studies suggest that there is selective pressure for biallelic inactivation leading to HRD in cancer types with predisposing germline variants in the HR pathway, such as breast, ovarian, pancreatic and prostate cancers54.
One of the key challenges in studies of this type is deciding upon a ‘gold standard’ test of HRD. Current functional, clinical and molecular tests all have advantages and disadvantages. The limitations of HRDetect that we use here include that it was developed and trained using breast tumour data and is predicated on BRCA1/2 deficiency arising from SSV disruption and promoter methylation, rather than any form of disruption to any HR gene. Although in the context of the current study, of BRCA1/2 disruption by SV, the latter is of less importance. All current genomic HRD tests are further limited to demonstrating that HRD once existed in the evolution of a tumour, and are blind to the restoration of HR by events such as secondary mutations, hypomorphic HRD variants and epigenomic changes.
There is an urgent clinical need to better understand the processes that give rise to both BRCA1/2 loss and more broadly contribute to HRD. Our study demonstrates that BRCA1/2 loss by structural variation may have a comparable impact on HRD and patient survival to short variants at BRCA1/2. Furthermore, these events are not specific to HGSOC, they are abundant, they compromise gene expression and are likely to be functionally important in other cancer types including in those types where HRD has been previously identified. However, these variants are unlikely to be detected by sequencing methods currently employed in the clinic. A change in sequencing approach to identify all BRCA1/2 loss may be required to maximise the potential of PARP inhibition.
Supplementary Material
Statement of significance.
We demonstrate that BRCA1/2 loss by structural variation (predominantly large deletion) are novel and unappreciated events, leading to loss of gene expression in ovarian and other cancers. Structural variants contribute to homologous recombination deficiency and their detection could identify more patients suitable for PARP inhibition, a therapy with proven benefit.
Acknowledgements
A. Ewing is supported by a UKRI Innovation Fellowship (MR/RO26017/1). C.A. Semple, A. Meynert and G.R. Grimes are supported by MRC core funding to the MRC Human Genetics Unit (MRC grant MC_UU_00007/16). R.L. Hollis is supported by an MRC-funded Research Fellowship. S. Dowson received funding from AstraZeneca and the Beatson Cancer Charity. I.A.McNeish acknowledges funding from Ovarian Cancer Action and the NIHR Imperial Biomedical Research Centre. A.V. Biankin acknowledges funding from Cancer Research UK (C29717/A17263, C29717/A18484, C596/A18076, C596/A20921, A23526), Wellcome Trust Senior Investigator Award (103721/Z/14/Z), Pancreatic Cancer UK Future Research Leaders Fund (FLF2015_04_Glasgow), MRC/EPSRC Glasgow Molecular Pathology Node, The Howat Foundation.
Sequencing of the SHGSOC cohort was supported by AstraZeneca, the Medical Research Council and the Scottish Chief Scientist through a Precision Medicine Scotland Innovation Centre/Scottish Genome Partnership (SEHHD-CSO 1175759/2158447) collaboration. This Scottish Genomes Partnership is funded by the Chief Scientist Office of the Scottish Government Health Directorates [SGP/1] and The Medical Research Council Whole Genome Sequencing for Health and Wealth Initiative (MC/PC/15080). This study would not be possible without the families, patients, clinicians, nurses, research scientists, laboratory staff, informaticians and the wider Scottish Genomes Partnership team to whom we give grateful thanks. Members of the Scottish Genome Partnership (SGP) include Timothy J. Aitman, Andrew V. Biankin, Susanna L. Cooke, Wendy Inglis Humphrey, Sancha Martin, Lynne Mennie, Alison Meynert, Zosia Miedzybrodzka, Fiona Murphy, Craig Nourse, Javier Santoyo-Lopez, Colin A. Semple, and Nicola Williams. More information about SGP can be found at www.scottishgenomespartnership.org. The authors would also like to acknowledge the Edinburgh Clinical Research Facility for the sequencing of RNA samples from the SHGSOC cohort.
The authors would also like to extend our thanks to the Nicola Murray Foundation, and the Edinburgh Ovarian Cancer Database, from which the clinical data for much of the Scottish cohort were retrieved. We also thank the NRS Lothian Human Annotated Bioresource, NHS Lothian Department of Pathology, the Edinburgh Experimental Cancer Medicine Centre, the Biorepository at the Glasgow Queen Elizabeth University Hospital and the Tayside Biorepository for their support.
Footnotes
Author contributions
C.G, C.A.S, C.S.H, I.A.M, T.M, M.F, N.M, A.M, B.D, R.M, J.C.B, A.V.B conceived the study within the broader remit of the Scottish molecular ovarian cancer collaboration. A.E, C.A.S, C.G conceived and designed the analysis of this cohort. M.C, R.L.H, M.F, M.L, T.M, N.M, N.S, S.D, M.M, F.N, R.G, P.R and C.G acquired patient samples. C.S.H performed histo-pathological review. M.C, I.C, D.E and B.M performed sample processing. B.M and A.V.B performed whole genome sequencing. A.Me, A.E, G.R.G processed the sequencing data. T.R, R.L.H, C.B, T.M, M.F, N.M, M.L, S.D. R.G, M.M, F.N, P.R and C.G provided clinical data. A.E performed statistical analyses. P.R, C.G, C.A.S, L.M, A.M, B.D, R.M, J.C.B and A.V.B provided strategic direction to the project. SGP and AstraZeneca funded the work. A.E, C.A.S and C.G drafted the manuscript. All authors read and commented on the manuscript and approved the final version.
Conflicts of interest
J.C.B, A.M and B.D are employees and stock holders of AstraZeneca. I.A.M is on the advisory boards for Clovis Oncology, Tesaro, AstraZeneca, Carrick Therapeutics, Roche and ScanCell. I.A.M also benefits from institutional funding from AstraZeneca. C.G. has received research funding from AstraZeneca, Aprea, Nucana, Tesaro, GSK and Novartis; honoraria/consultancy fees from Roche, AstraZeneca, Tesaro, GSK, Nucana, MSD, Clovis, Foundation One, Sierra Oncology and Cor2Ed; and is named on issued/pending patents relating to predicting treatment response in ovarian cancer unrelated to this work. R.G is or has been on the advisory boards of AstraZeneca, GSK, Tesaro and Clovis; has received speaker fees and funding to attend medical conferences from GSK and Tesaro and is a UK co-ordinating investigator or site principal investigator for studies sponsored by Astrazeneca, GSK, Pfizer and Clovis. F.N has been or is a site principal investigator for studies sponsored by AstraZeneca and Clovis. P.R has received research funding from AstraZeneca and Tesaro and honoraria/consultancy fees from AstraZeneca and GSK.
Data availability
Previously published WGS and RNA-seq data that were reanalysed here are available via EGA at accession code EGAS00001001692 (ICGC PCAWG). WGS, RNA-seq and clinical data from the Scottish cohort (SHGSOC) will be made available via EGA at accession code EGAS00001004410. Other supporting data have been provided in the Supplementary Tables.
Code availability
All code will be made available at: https://github.com/ailithewing/Structural_variants_BRCA1_2_HRD_inHGSOC.
References
- 1.Knijnenburg TA, et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23:239–254.:e6. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowtell DD, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–79. doi: 10.1038/nrc4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017 doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patch A-M, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 6.Bell D, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollis RL, Gourley C. Genetic and molecular changes in ovarian cancer. Cancer Biol Med. 2016;13:236–47. doi: 10.20892/j.issn.2095-3941.2016.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang JW, Karlan BY, Cass llana, Baldwin RL. BRCA1 promoter methylation predicts adverse ovarian cancer prognosis. Gynecol Oncol. 2006;101:403–410. doi: 10.1016/j.ygyno.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 9.Hughes-Davies L, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–35. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 10.Brown LA, et al. Amplification of EMSY, a novel oncogene on 11q13, in high grade ovarian surface epithelial carcinomas. Gynecol Oncol. 2006;100:264–270. doi: 10.1016/j.ygyno.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Hollis RL, et al. High EMSY expression defines a BRCA-like subgroup of high-grade serous ovarian carcinoma with prolonged survival and hypersensitivity to platinum. Cancer. 2019;125:2772–2781. doi: 10.1002/cncr.32079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: Exploiting the fundamental vulnerability of ovarian cancer. Cancer Discovery. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen LWM, Martens J, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-19406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledermann J, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 15.Mirza MR, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 16.Coleman RL, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore K, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 18.Ray-Coquard I, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 19.González-Martín A, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 20.Robson M, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 21.Litton JK, et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golan T, et al. Maintenance Olaparib for Germline BRCA -Mutated Metastatic Pancreatic Cancer. N Engl J Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bono J, et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 24.Gourley C, et al. Moving From Poly (ADP-Ribose) Polymerase Inhibition to Targeting DNA Repair and DNA Damage Response in Cancer Therapy. J Clin Oncol. 2019;37:2257–2269. doi: 10.1200/JCO.18.02050. [DOI] [PubMed] [Google Scholar]
- 25.Wooster R, Bignell G, Lancaster J, Swift S. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1996 doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 26.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 27.Paluch-Shimon S, et al. Prevention and screening in BRCA mutation carriers and other breast/ovarian hereditary cancer syndromes: ESMO Clinical Practice Guidelines for cancer prevention and screening †. 2016 doi: 10.1093/annonc/mdw327. [DOI] [PubMed] [Google Scholar]
- 28.Lord CJ, Ashworth A. BRCAness revisited. 2016 doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 29.Guan P. Structural variation detection using next-generation sequencing data: A comparative technical review. Methods. 2016;102:36–49. doi: 10.1016/j.ymeth.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Ewing A, Semple C. Breaking point: the genesis and impact of structural variation in tumours. F1000Research. 2018;7:1814. doi: 10.12688/f1000research.16079.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. [Accessed: 11th November 2019];GitHub - bcbio/bcbio-nextgen: Validated, scalable, community developed variant calling, RNA-seq and small RNA analysis. Available at: https://github.com/bcbio/bcbio-nextgen.
- 32.Landrum MJ, Kattman BL. ClinVar at five years: Delivering on the promise. Hum Mutat. 2018;39:1623–1630. doi: 10.1002/humu.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 35.Campbell PJ, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gel B, et al. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. doi: 10.1093/bioinformatics/btv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao EY, et al. Homologous Recombination Deficiency and Platinum-Based Therapy Outcomes in Advanced Breast Cancer. Clin Cancer Res. 2017;23:7521–7530. doi: 10.1158/1078-0432.CCR-17-1941. [DOI] [PubMed] [Google Scholar]
- 38.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irodi A, et al. Patterns of clinicopathological features and outcome in epithelial ovarian cancer patients: 35 years of prospectively collected data. BJOG An Int J Obstet Gynaecol. 2020 doi: 10.1111/1471-0528.16264. [DOI] [PubMed] [Google Scholar]
- 41.Dougherty BA, et al. Biological and clinical evidence for somatic mutations in BRCA1 and BRCA2 as predictive markers for olaparib response in high-grade serous ovarian cancers in the maintenance setting. Oncotarget. 2017;8:43653–43661. doi: 10.18632/oncotarget.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 45.Priestley P, et al. Pan-cancer whole genome analyses of metastatic solid tumors. bioRxiv. 2019:415133. doi: 10.1101/415133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swisher EM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 47.Haraksingh RR, Abyzov A, Urban AE. Comprehensive performance comparison of high-resolution array platforms for genome-wide Copy Number Variation (CNV) analysis in humans. BMC Genomics. 2017;18:321. doi: 10.1186/s12864-017-3658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou B, et al. Whole-genome sequencing analysis of CNV using low-coverage and paired-end strategies is efficient and outperforms array-based CNV analysis. J Med Genet. 2018;55:735–743. doi: 10.1136/jmedgenet-2018-105272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger AC, et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 2018;33:690–705.:e9. doi: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger AC, et al. A comprehensive Pan-Cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018;33:690. doi: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rathmell KW, Chen F, Creighton CJ. Genomics of chromophobe renal cell carcinoma: Implications from a rare tumor for pan-cancer studies. Oncoscience. 2015;2:81–90. doi: 10.18632/oncoscience.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilié PG. Genomic Instability in Kidney Cancer: Etiologies and Treatment Opportunities. Kidney Cancer. 2019;3:143–150. [Google Scholar]
- 54.Jonsson P, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–579. doi: 10.1038/s41586-019-1382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published WGS and RNA-seq data that were reanalysed here are available via EGA at accession code EGAS00001001692 (ICGC PCAWG). WGS, RNA-seq and clinical data from the Scottish cohort (SHGSOC) will be made available via EGA at accession code EGAS00001004410. Other supporting data have been provided in the Supplementary Tables.
All code will be made available at: https://github.com/ailithewing/Structural_variants_BRCA1_2_HRD_inHGSOC.