Abstract
Microglia, the tissue resident macrophages of the CNS, play critical roles in immune defence, development and homeostasis. However, isolating microglia from humans in large numbers is challenging. Here, we profiled gene expression variation in primary human microglia isolated from 141 patients undergoing neurosurgery. Using single cell and bulk RNA sequencing, we identify how age, sex and clinical pathology influence microglia gene expression and which genetic variants have microglia-specific functions using expression quantitative trait loci (eQTL) mapping. We follow up one of our findings using an hIPSC-based macrophage model to fine-map a candidate causal variant for Alzheimer’s disease at the BIN1 locus. Our study provides the first population-scale transcriptional map of a critically important cell for human CNS development and disease.
Introduction
Microglia are tissue resident macrophages of the central nervous system (CNS) and play critical roles in synaptic pruning, neuronal plasticity as well as maintaining local immune surveillance within the brain1–3. Disease studies have implicated microglial dysfunction in a number of neurological disorders4–7, but these highly plastic cells have not been studied at a population level. To date, studies of microglial gene expression have been restricted to relatively small samples of either frozen post-mortem tissue from existing brain banks or fresh surgical samples from restricted patient groups, typically temporal lobe resections for epilepsy or peri-tumoral tissue. Single cell transcriptomic studies of similar samples have suggested that microglial function may vary across age, sex and brain region8–13. However, these conclusions are often not replicated in studies of equivalent size.
Here, we performed the first population-scale study of human microglia to understand how age, sex, pathology, cortical anatomy and common germline genetic variation influences the microglia transcriptome. We used a unique cohort of patients who had been sampled within 8 hours of an acute haemorrhage or traumatic brain injury to identify two novel signatures of acute activation in human microglia. Finally, we examined how our results replicated in a scalable cell model system of microglia, using induced pluripotent stem cell derived macrophages (IPSDMac) derived from 133 human IPS lines created by the Human Induced Pluripotent Stem Cell Initiative14.
Characterisation of microglial cell populations
We undertook analysis of human microglia isolated from 141 patients undergoing a range of neurosurgical procedures (Figure 1a). These included a “control” group who had cortical microglia sampled at the beginning of a surgical corridor when the distance to the clinical pathology exceeded 4cm. We also sampled cortical microglia from patients with hydrocephalus, brain tumours, and patients with acute brain injury (spontaneous haemorrhage and trauma) who sustained substantial parenchymal injury, enabling us to capture in vivo microglial activation.
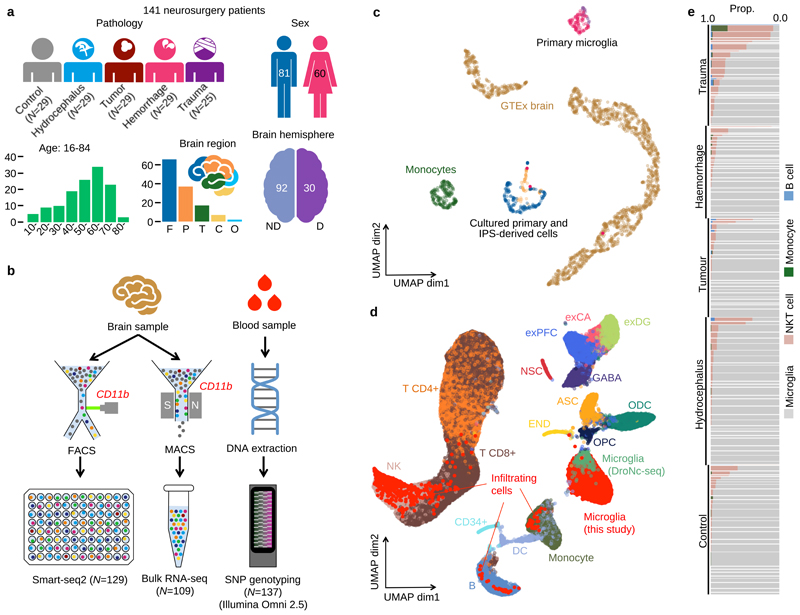
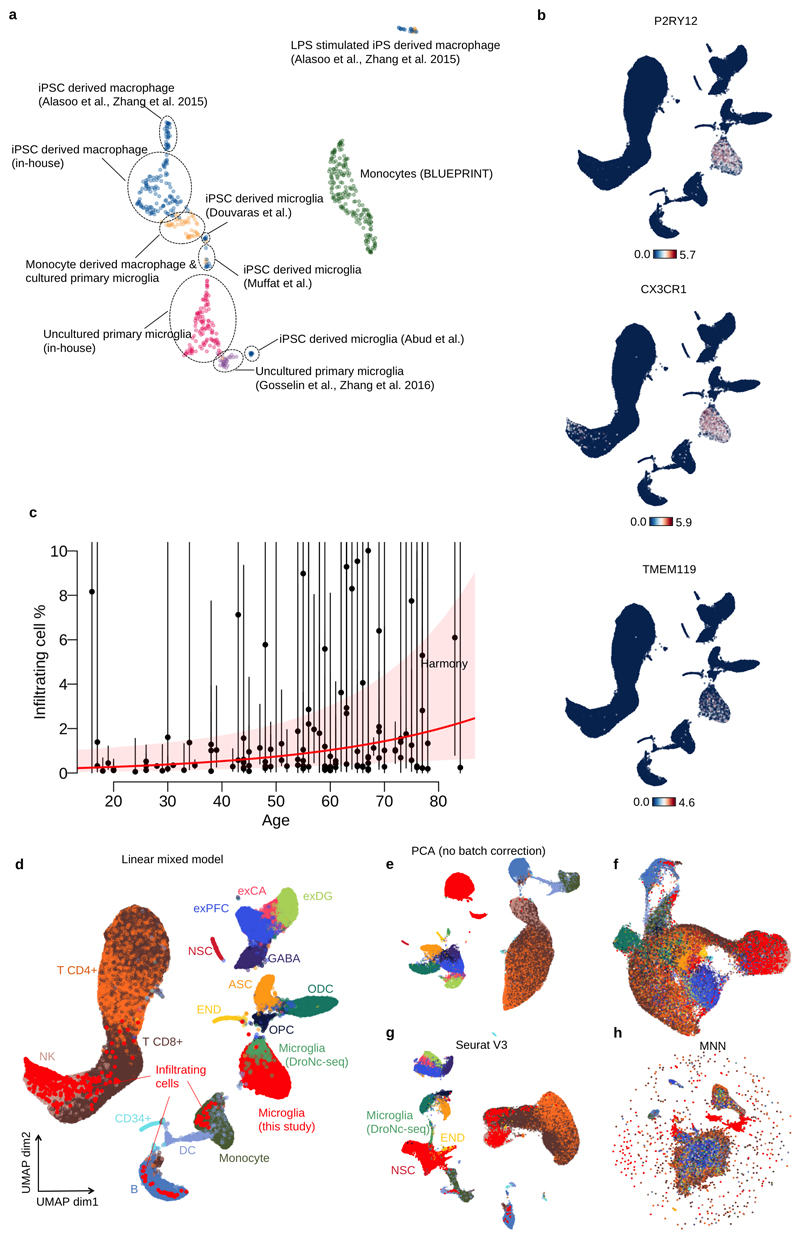
Figure 1. Study design and overview of the data.
a. Metadata from 141 neurosurgery patients enrolled in this study. Brain region annotation: Cerebellum (C); Frontal (F); Occipital (O); Parietal (P); Temporal (T); non-dominant (ND); dominant (D). b. Experimental design using Smart-seq2 and bulk RNA-seq with SNP genotyping. c. UMAP of bulk RNA-seq from myeloid cells and brain tissue. The “Primary microglia” cluster contains samples collected in this study (pink dots) and previous studies (purple dots) (information on the source of previous study data can be found in Supplementary Table 7). “Cultured primary and IPS-derived cells”, includes IPS-derived macrophages and microglia (blue dots), cultured primary microglia and monocyte derived macrophages (orange dots). “Monocytes” (green dots) denotes primary monocytes obtained from the BLUEPRINT project, and “GTEx brain” denotes all brain tissues from GTEx v7. The left cluster of GTEx brain corresponds to cerebellum or cerebellar hemisphere samples and the right cluster contains samples from all other brain regions. d. UMAP of single-cell RNA-seq data combined with 68K PBMC scRNA-seq18 and whole brain DroNc-seq19. Bright red dots represent cells collected in this study. Cell type annotations were obtained from: glutamatergic neurons from the PFC (exPFC); pyramidal neurons from the hip CA region (exCA); GABAergic interneurons (GABA); granule neurons from the hip dentate gyrus region (exDG); astrocytes (ASC); oligodendrocytes (ODC); oligodendrocyte precursor cells (OPC); neuronal stem cells (NSC); endothelial cells (END); dendritic cell (DC); B cell (B); hematopoietic progenitor cell (CD34+); NK T cell (NK). e. Proportions of non microglia for each patient in our data. Each horizontal bar corresponds to one patient. The thickness of each bar is proportional to the number of cells observed for the patient. Patients are stratified by pathology.
For each individual, we isolated CD11b-positive cells and performed both single cell (SmartSeq2)15 and bulk RNA-seq. After QC, we retained 112 bulk RNA-seq samples, and 9,538 single cells from 129 patients (Figure 1b). Our bulk RNA-seq samples clustered closely with microglia from two previous studies16,17, and were distinct from both GTEx brain and BLUEPRINT monocytes (Figure 1c). Additional clustering only with myeloid cell types revealed that iPSC derived microglia were transcriptionally more similar to uncultured primary microglia than iPSC derived macrophages, although substantial between-laboratory effects on both primary and cultured cells were apparent from this analysis (Extended Data Figure 1a).
We compared our single cell data to public datasets of 68K PBMCs isolated from a healthy donor18 and 15K brain cells from 5 GTEx donors19. A total of 8,662 cells in our SmartSeq2 data formed a cluster with the microglia population found in GTEx samples and distinct from PBMCs (Figure 1d) and included a range of known microglial marker genes, including P2RY12, CX3CR1 and TMEM119 at high levels (Extended Data Figure 1b). We defined this population of cells (excluding GTEx samples) as microglia for the remainder of our analysis. We identified three less common, putatively infiltrating populations of cells that closely resembled other blood cell types, including NKT cells, monocytes or B-cells that comprised 8.4%, 0.5% and 0.3% of our single cell dataset, respectively. These cell types may reflect either infiltration of immune cells as a result of blood-brain barrier breakdown or intravascular contamination within the tissue. Abundance of infiltrating cells was strongly correlated with patient pathology, with trauma patients especially enriched (OR=7.6, Fisher exact test P=1.2x10-155) (Figure 1e). We also found a significant effect of age on the abundance of infiltrating cells (3.4% increase per year, Wald test P=0.014) after adjusting for all known confounding factors, which could reflect increasing blood brain barrier permeability over the lifespan (Extended Data Figure 1c). Batch correction using our linear mixed model showed equivalent performance to Seurat V3 (Extended Data Figure 1d-h), but was readily scalable to the 129 donors as batches used in the subsequent analysis (Online Methods).
Within microglia, we found a high level of consistency across all patients. We identified four populations of cells (Figure 2a). By examining their relative abundance in different patient groups, we identified 1 naive and 3 distinct microglial activation states. Population A was most common in control and hydrocephalus patients while Population B, which we identify as sub-acute activation, was enriched in tumour patients (OR=4.9, P=7.6x10-169). Two acute activation populations, C and D, were common in patients with spontaneous haemorrhage and traumatic brain injury (comprising 25-76% of cells), but rare in other pathologies (<5% of cells) (Figure 2b, c).
Figure 2. Transcriptional heterogeneity in human microglia.
a. UMAP of 8,662 microglia cells after removing putative infiltrating cells. Colors show 4 clusters defined using Louvain clustering (Materials and Methods). b. Microglial population variation between patient pathologies. The four different colours in Figure 2a illustrate population compositions for each pathology. Points coloured gray are all other cells. c. Heatmap shows the enrichment (log odds ratio) of microglial populations between pathologies d. Heatmap of averaged, normalised expression level (defined as the posterior mean of pathology random effect term, see Materials and Methods) of differentially expressed genes at local true sign rate (ltsr) greater than 0.9 (see Materials and Methods for details). Heatmap is divided into groups based on all possible pairwise groupings of the four cell populations, with the most transcriptionally distinct population at the top. e. Pathway enrichment analysis of differentially expressed genes between different microglial populations. The x-axis shows the P-value obtained by gProfiler2 with multiple testing corrections (Materials and Methods). Bars are coloured according to the combinations of clusters in which genes are upregulated. The upregulated cluster IDs are also shown besides the bars.
To characterise these populations further, we performed differential expression analysis between cell populations. We observed higher expression of microglial markers including P2RY12 and CX3CR1 in populations A and B. This cohort additionally showed strong upregulation of genes involved in catabolic processes (e.g. GPX1) and phagocytosis (TREM2). Cells within populations B, C and D also had high levels of general immune response and cell activation genes (IL1B, CD83 & CCL3) (Figure 2d, e; Supplementary Table 1). Cells from populations C and D exhibited upregulation of acute immune response pathway genes, including NF-kappa B, STAT3, RUNX1 as well as MHC-I expression. Population C also showed differential expression of genes associated with stress induced senescence and DNA damage (HIST1H2BG), whereas population D expressed genes associated with cell proliferation (FLT1) and chemotaxis (CCL4, CXCL8, CXCL16), the latter also shared with population B.
Populations C and D are of particular interest as they are unlikely to have been previously observed (Figure 2b, c). To test this hypothesis, we compared our population-specific markers with differentially expressed genes from Alzheimer’s disease-associated microglia20 and marker genes from glioma-associated microglia21 (Extended Data Figure 2a). This analysis confirmed that populations of microglia identified in previous studies most closely resembled Populations A or B in our dataset, and that the activated populations C and D were transcriptionally distinct.
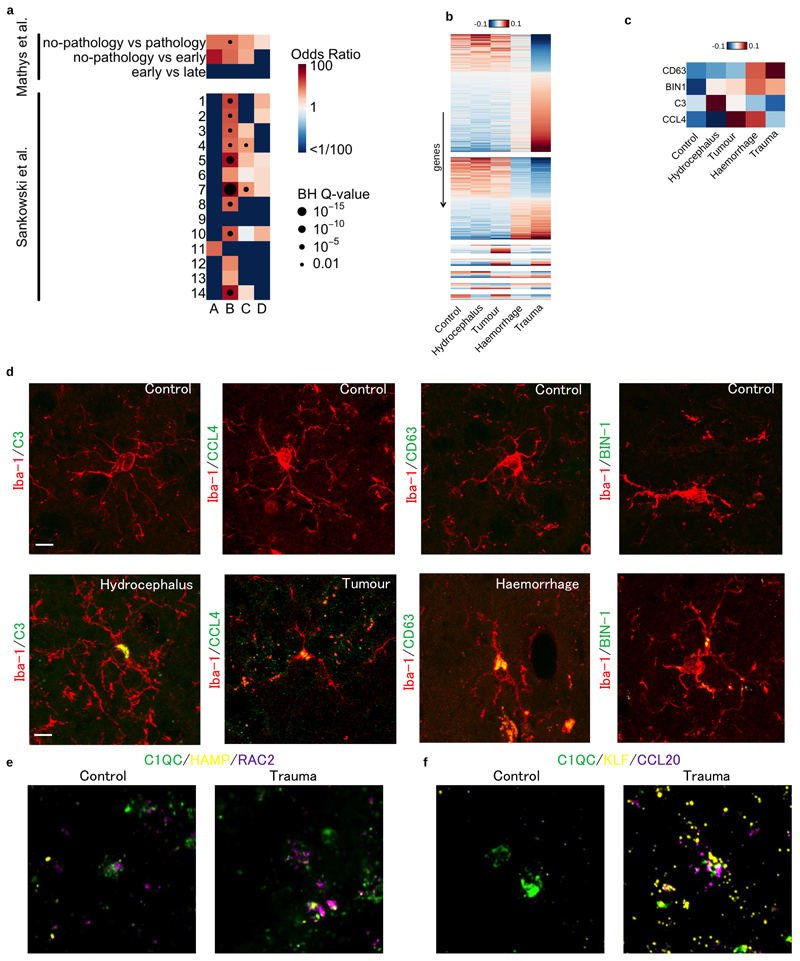
To further validate our findings, we performed differential expression between microglia from different between patient pathologies and controls (Extended Data Figure 2b). We selected 4 candidate marker genes CD63, BIN1, C3 and CCL4 that were upregulated relative to controls in haemorrhage, trauma, hydrocephalus and tumour patient groups, respectively (Extended Data Figure 2c). Immunohistochemistry for each marker in a fixed tissue section confirmed differences in expression between patient groups at the protein level (Extended Data Figure 2d). Finally, we confirmed that expression of activation markers in populations C and D was not driven by sample processing, using RNAScope in fresh frozen tissue sections (Extended Data Figure 2e-f).
Biological determinants of microglial expression
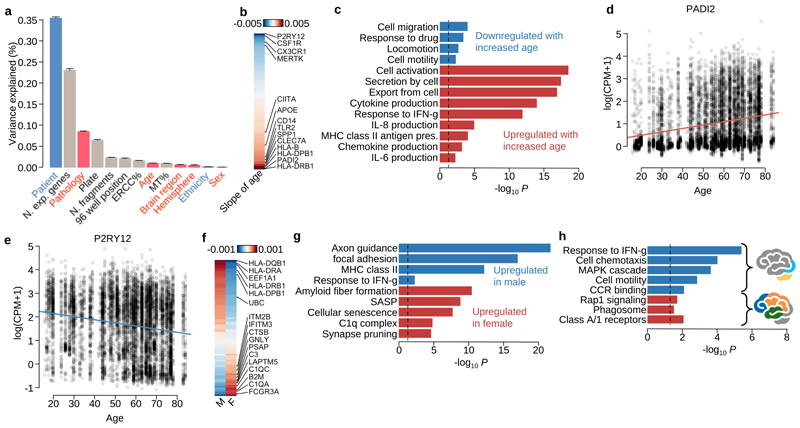
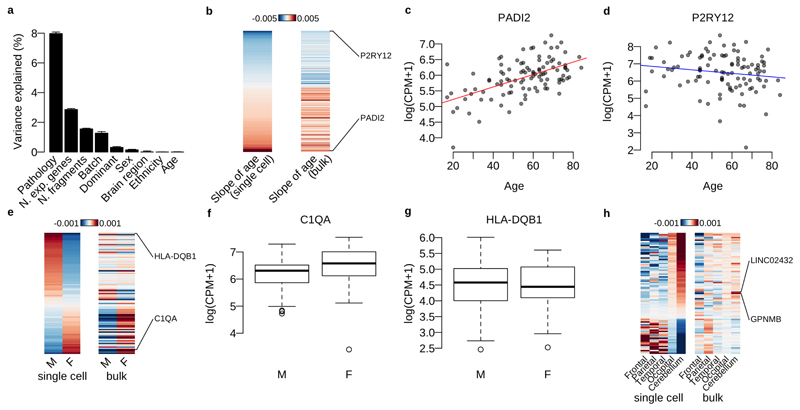
Our sampling design enabled us to explore the relative importance of a wide range of biological factors in determining microglial gene expression, while controlling for important technical confounders, using variance components analysis (Figure 3a). Of the biological factors we examined, clinical pathology explained more variation than all other factors combined, although all factors except sex, including age, brain region, dominant hemisphere and ethnicity, explained some of the variation that was significantly different from zero (LR test FWER 0.05). Individual patient explained the most variability of any single factor in the model. However, although this factor captures the contribution of genetic background, it is also likely to be dominated by unmeasured technical batch effects, such as variability in cell dissociation and surgical sampling. We found 260 genes where age explained a fraction of variation that was significantly different from zero, showing upregulation of genes related to inflammation (CLEC7A, CIITA and TLR2) and downregulation of cell identity (P2RY12, CX3CR1), motility and proliferation genes (CSF1R) with increasing age (Figure 3b-e; Supplementary Table 2). Although sex accounted for little of the overall variation, we found 97 genes that were differentially expressed between males and females (Figure 3f). These included multiple genes in the complement pathway and synaptic pruning mechanisms (C1QA, C1QC and C3) that were more highly expressed in females than males (Figure 3g; Supplementary Table 3). Anatomical region of sampling also had a subtle effect on transcriptional variation, with cerebellar microglia, which are known to exhibit a distinct, less ramified morphology, having increased expression of several recruitment chemokines (CCL4, CCL3, CCL4L2, CCL3L3) (Figure 3h; Supplementary Table 4). We repeated our differential expression analysis using bulk RNA-seq and found a reasonable correlation in effect sizes between bulk and single cell data sets for both the age and sex comparisons (r=0.61 and 0.39 with P=4.1×10-20 and 4.5×10-8 for age and sex respectively) (Supplementary Figure 3).
Figure 3. Single-cell RNA-seq reveals how microglial transcriptional heterogeneity is driven by clinical factors.
a. Barplot shows the variance explained by each factor. Bars coloured gray are technical factors (N.exp.genes: the number of expressed genes in each cell, which is expected to reflect cell health or quality; Plate: cells undergoing library preparation and sequencing together on the same 384 well plate; ERCC%: ERCC spike-in percentage among all mapped fragments for each cell; N.fragments: the number of fragments for each cell mapped on autosomes; 96 well position: the position of a cell on the 96 well plate processed in the SmartSeq2 protocol; MT%: percentage of mitochondrial RNA fragments among all mapped fragments for each cell) and coloured pink are clinical factors (Pathology; Age of patient; Brain region; Brain hemisphere; Sex of patient). Blue bars are partly related to patients’ genetic background (Patient and Ethnicity). b. Heatmap showing the strength and direction of the age effect for differentially expressed (DE) genes at local true sign rate (ltsr) greater than 0.9 (Materials and Methods). The effect size is the posterior mean estimate weighted by the empirical prior distribution, where hyperparameters were estimated from the data using a linear mixed model (Materials and Methods). c. Pathway enrichment for differentially expressed (DE) genes by age. Pathways coloured red are enriched only for DE genes upregulated by age, and the pathways coloured blue are enriched for DE genes downregulated by age. d-e. Example genes upregulated or downregulated by age. f. Heatmap showing the average normalised expression levels for DE genes by sex at ltsr greater than 0.9 (Materials and Methods). The effect size is the posterior mean estimate weighted by the prior distribution calibrated in the linear mixed model (Materials and Methods). g. Pathway enrichment by sex. h. Pathway enrichment for combinations of brain regions. The blue bars show pathways upregulated in cerebellum and occipital lobe. The red bars show pathways upregulated in frontal, parietal and temporal lobes.
eQTL mapping in human microglia
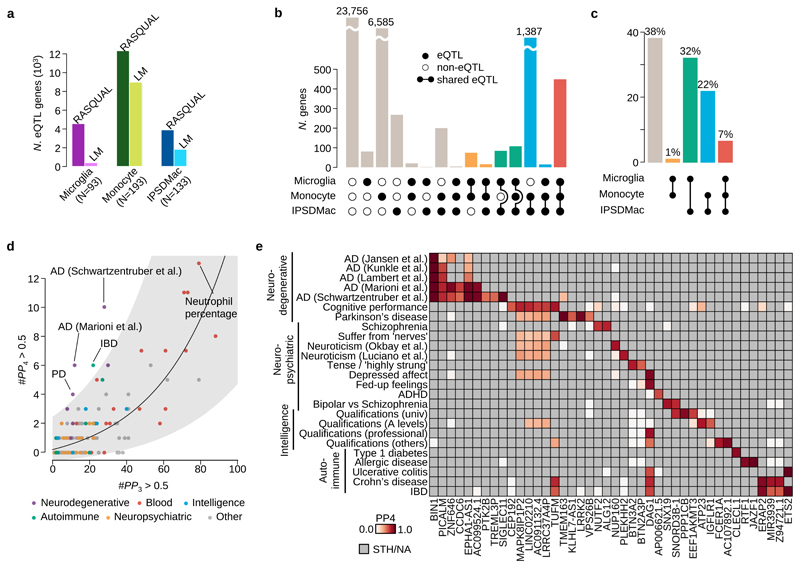
Next, we constructed a map of expression quantitative trait loci (eQTLs). After excluding samples with low genotyping quality or substantial non-European ancestry, we retained bulk RNA-seq data from 93 individuals, and detected 585 eQTLs at FDR 5% using simple linear regression model. For comparison, we mapped eQTLs using the same pipeline and significance thresholds in both the BLUEPRINT monocyte (N=193) and our own IPSDMac data (N=133). This analysis suggested that the number of eQTLs we detected in primary microglia was unexpectedly low. In part, this is likely to be because of the higher between-sample variability in primary microglia, compared with other cell types (Figure 4a). To confirm the eQTLs we detected with linear regression we performed eQTL mapping only using allele-specific expression22. For eQTLs detected at FDR 5%, the effect sizes estimated from linear regression and from allele-specific analysis were highly correlated (r=0.71) suggesting the majority are real (Extended Data Figure 4a).
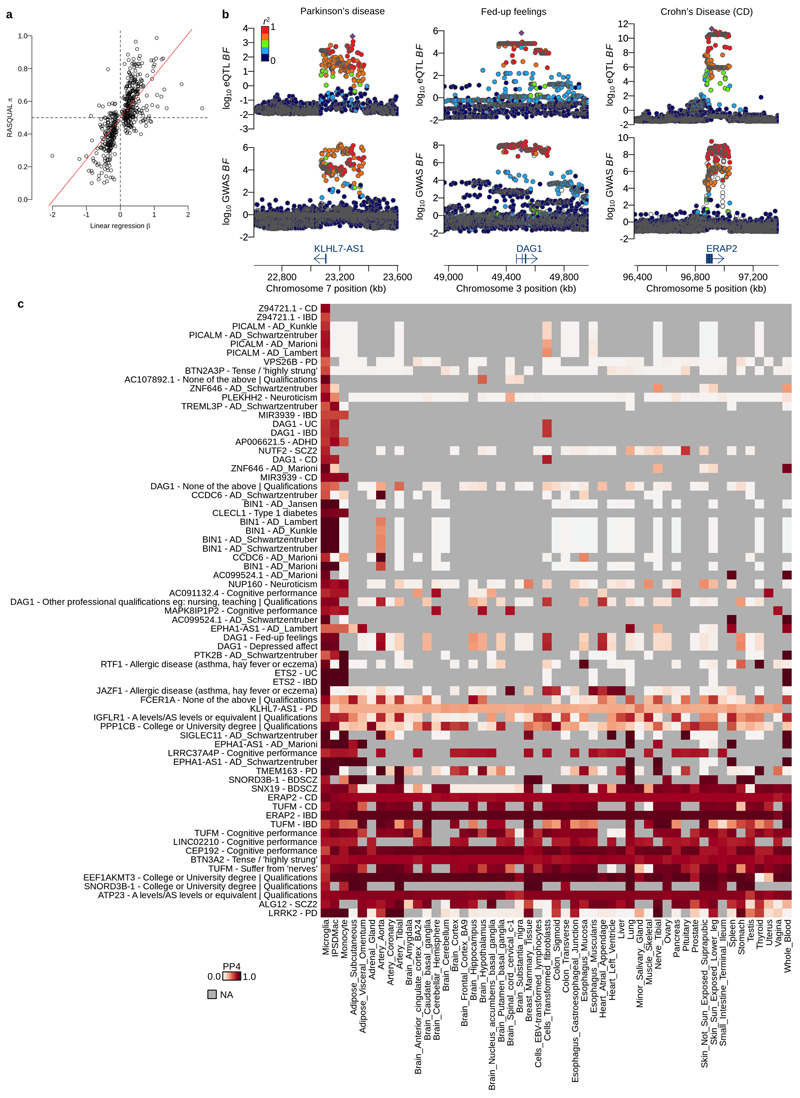
Figure 4. Mapping and colocalisation of microglia eQTLs with various GWAS traits.
a. The numbers of eQTL genes discovered by two different methods, RASQUAL (left bar) or simple linear regression (right bar, LM) in three myeloid cell types at FDR 5% (see Online Methods). b. The number of shared eQTLs across three myeloid cell types obtained by the three-way Bayesian hierarchical model (Online Methods). The combination of genes that are eQTLs (closed dots) or non-eQTLs (open dots) across three different myeloid cell types are shown below each bar. A line connecting two dots indicates a shared eQTL between different cell types. c. Empirical prior probability of eQTL sharing among three different myeloid cell types obtained by the three-way Bayesian hierarchical model (Online Methods). The Y-axis shows the proportion of genes genome-wide and the dots connected by segment illustrate the shared genetic association. d. Colocalisation of microglia eQTLs with 146 GWAS traits. The x-axis shows the number of genes where PP3, the posterior probability of the microglia eQTL and GWAS association being driven by two independent causal variants, was greater than 0.5. The y-axis is the number of colocalised genes where the posterior probability of a single shared causal variant between a microglia eQTL and a GWAS locus (PP4) was greater than 0.5. We subdivided and colored GWAS traits as follows: purple: neurodegenerative diseases; red: blood cell trait; blue: traits related with intelligence; green: autoimmune diseases; yellow: neuropsychiatric diseases; gray: others. The line shows a log-normal linear regression fit with gray shaded area indicating the 95% prediction interval of the fit. e. Heatmap of PP4 for neuro-degenerative/psychiatric diseases, intelligence related traits and autoimmune diseases showing all genes and GWAS trait with a combined PP4 greater than 0.5. Gray cells indicate that the gene-trait combination was not tested because the GWAS locus was not significant (lead SNP P>10-6), or there were no GWAS summary statistics available for secondary hits (PTK2B and TREML3P).
Next, we explored the level of cell-type specificity of the eQTLs we detected by comparing microglia, monocytes and IPSDMac using a three-way empirical Bayesian hierarchical model (Online Methods). Here, numbers of eQTLs were computed by summing over model posterior probabilities and are therefore not expected to be identical to those from our linear regression analysis. We discovered 855 microglia eQTLs of which 108 were microglia-specific, 449 were shared across all three cell types, 192 were shared with IPSDMs but not monocytes, and 106 were shared between microglia and monocytes (Figure 4b). Our model also estimated prior probabilities of shared eQTLs for all possible comparisons. Notably, despite a much larger sample size in monocytes, our model estimated a low probability (1%) of shared eQTLs between microglia and monocytes, with a much higher prior (32%) for shared eQTLs between microglia and IPSDMac, and 7% of eQTLs shared between all three cell types (Figure 4c).
We then tested for colocalization of microglia eQTLs with risk loci from 146 genome wide association studies (GWAS), of which 25 were broadly neurological, including cognitive developmental measures such as intelligence, neuropsychiatric disorders with adolescent/young adult onset, and neurodegenerative diseases (Online Methods). We discovered 245 unique gene-trait combinations with the posterior probability of a single shared causal variant between a microglia eQTL and a GWAS locus (PP4) greater than 0.5 for 84 different traits and 129 unique genes (excluding HLA genes). The number of colocalised genes for each trait most likely reflects the statistical power of the study. For example, we detected 13 colocalisations with neutrophil percentage, which also has a sample size of 349,861. We did, however, observe an excess of colocalised microglial eQTLs for certain traits, including Alzheimer’s disease (AD), Parkinson’s disease (PD) and inflammatory bowel disease (IBD), likely reflecting the known involvement of microglia or macrophages in each of these disease’s pathology (Figure 4d). We also discovered eQTLs that were absent from other tissues, that colocaised with a wide range of GWAS traits. For example, we discovered an eQTL for DAG1, which produces a protein that is involved in the dystrophin-glycoprotein complex with associations with fed-up feelings, intelligence related traits and autoimmune diseases (Figure 4e; Extended Data Figure 4b). Interestingly, this eQTL is detected in both microglia and IPSDMac from this study, but absent from all other cell types and tissues (Extended Data Figure 4c). We also detected 22 gene-trait combinations that colocalised in 10 or more cell types and tissues, an example of which is ERAP2 eQTL colocalised with Crohn’s disease in all 51 cell types and tissues (Figure 4e; Extended Data Figure 4b-c).
Fine-mapping primary microglia eQTLs using an in-vitro model
Given the involvement of microglia in neurodegenerative disease, we next selected Alzheimer’s disease (AD) to undertake a detailed analysis of colocalisation of microglia eQTLs with GWAS loci. Using different AD GWAS23–27, we found between 2-11 AD risk loci with PP4 greater than 0.5 with an eQTL in primary microglia (Figure 4e). These included well-known AD loci, such as BIN1, and less well-studied AD associations, for example EPHA1-AS1. We repeated our analysis using microglia eQTLs mapped by RASQUAL, a method that boosts power to detect eQTLs using allele specific expression (Supplementary Table 5). This analysis detected additional colocalisations at other well-known AD GWAS loci, such as CD33 (Extended Data Figure 5a). Here, analysis of splicing patterns revealed a splice QTL at exon 2 (Extended Data Figure 5b), consistent with previous studies28. One explanation for this result is that the allele-specific signal captured by RASQUAL is more sensitive to the changes in splice pattern. However, we discovered that the test statistics produced by RASQUAL may be inflated by additional overdispersion in our microglia data set (Figure 4a).
The challenges of studying primary microglia make the use of IPSDMac an attractive alternative. We therefore next asked whether any of the 11 primary microglia eQTLs that colocalised with an AD risk association could also be detected as an IPSDMac eQTL. We identified three AD association signals (BIN1, the EPHA1/EPHA1-AS locus and PTK2B) that colocalised with an eQTL both in primary microglia and in IPSDMac eQTLs (Extended Data Figure 4c). At the EPHA1/EPHA1-AS locus, we found an eQTL for the EPHA1-AS1 noncoding RNA that colocalised with the AD risk association, but no equivalent signal for the EPHA1 protein coding gene in most tissues (Extended Data Figure 5c-d). We have previously reported that an eQTL for the gene PTK2B colocalised with an AD risk association on chromosome 829. When we compared this with primary microglia we found a difference in the direction of effect between primary microglia and IPSDMac (Extended Data Figure 5e).
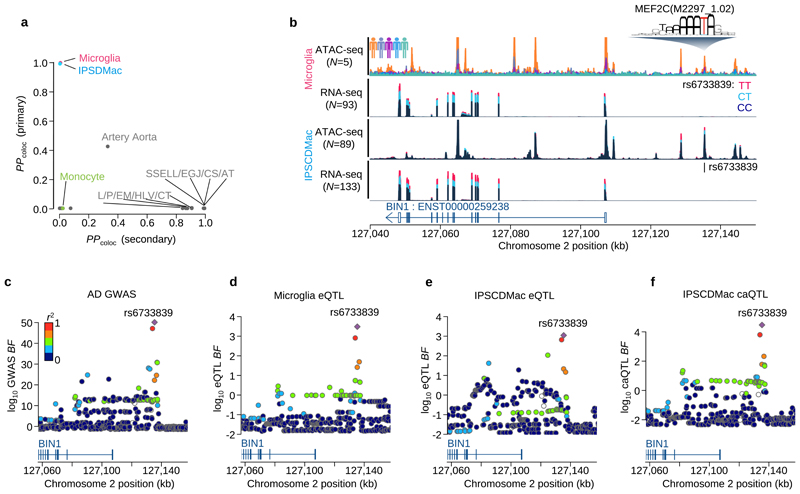
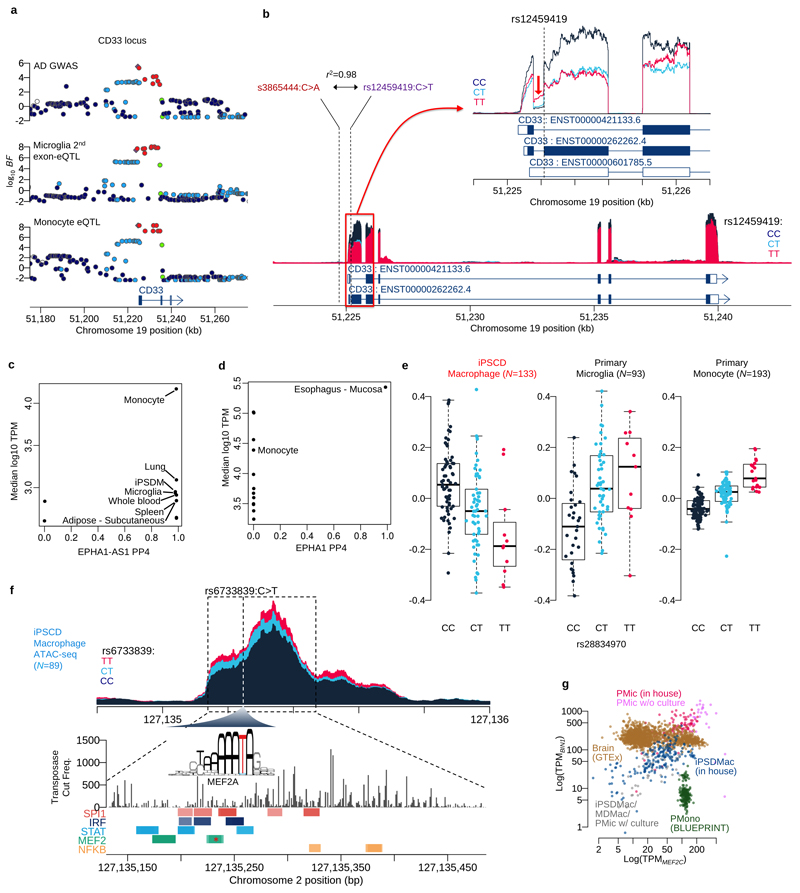
Finally, our analysis revealed that the AD association signal at BIN1 was highly cell type specific, found in primary microglia and IPSDMac, but no other cell types or tissues (Figure 5a). To fine-map causal variants at BIN1, we generated ATAC-seq data from 5 primary microglia and 89 IPSDMac. We found that the lead SNP of this association signal, rs6733839C>T, was located in a region of open chromatin in both microglia and IPSDMac. rs6733839C>T was also associated with a significant change in chromatin accessibility in IPSDMac (a chromatin accessibility QTL, caQTL) (Figure 5b, P<6.1x10-10). This caQTL also colocalised (PP4=0.996) with the AD association signal (Figure 5c-f), strongly suggesting that the causal variant driving the AD risk association directly or indirectly alters chromatin openness in this region. Analysis of the sequence context of this variant revealed that the AD risk allele at rs6733839C>T created a predicted high-affinity binding site for the MEF2C, a transcription factor with established roles in hippocampal learning and memory (PMID: 18599438) (Extended Data Figure 5f). A recent study has examined chromatin interactions between the BIN1 promoter and nearby AD risk variants30. Our results suggest that rs6733839C>T increases AD risk by increasing the binding of MEF2C, in turn, increasing the expression of BIN1. Although BIN1 and MEF2C are broadly expressed in many tissues, co-expression of both genes was found only in primary microglia and IPSDMac (Extended Data Figure 5g). Taken together, our results show that one of the largest common variant associations with AD outside of APOE can be studied using a scalable and relatively straightforward IPS based macrophage model.
Figure 5. Fine-mapping of the BIN1 eQTL / Alzheimer’s disease association.
a. Posterior probability of colocalisation between Alzheimer’s disease27 and the three myeloid cells and GTEx eQTLs for the BIN1 gene. The y-axis is based on the AD GWAS primary signal of the BIN1 locus and the x-axis is based on the secondary signal at BIN1 found by the conditional analysis27. b. Sequencing coverage depth of ATAC-seq and RNA-seq stratified by individuals (top ATAC-seq panel) or the three genotype groups at BIN1 lead eQTL SNP (rs6733839C>T) (bottom three panels). The top two panels show data from the primary microglia (Materials and Methods) and the bottom two panels were obtained from iPS cell derived macrophage (Materials and Methods). The MEF2CA motif overlaps with the lead SNP and the alternative allele (T) increases predicted binding affinity. c. Regional Manhattan plot around the BIN1 gene. The y-axis shows the statistical significance of AD GWAS27 in log10 Bayes factor. d. Regional plot shows the statistical significance of microglia eQTL for BIN1 gene in log10 Bayes factor. e. Regional plot shows the statistical significance of IPSDMac eQTL for BIN1 gene in log10 Bayes factor. f. Regional plot shows the statistical significance of IPSDMac chromatin accessibility QTL (log10 Bayes factor) at the chromatin accessibility peak involving the putative causal variant rs6733839C>T. Tissue type annotation: Artery Tibial (AT), Esophagus Gastroesophageal Junction (EGJ), Colon Sigmoid (CS), Skin Sun Exposed Lower leg (SSELL), Heart Left Ventricle (HLV), Colon Transverse (CT), Esophagus Mucosa (EM), Pituitary (PI).
Discussion
Here we present the first population-level study of human primary microglia. By sampling cells from living donors, we defined transcriptional signatures of in vivo microglial activation, avoiding artefacts from postmortem index and in vitro cell culture. We identified multiple microglial populations and showed how these populations are shaped by pathology and other life history factors. In particular, we identify two populations of microglia that reflect different in vivo acute activation states. We also created the first eQTL map in primary human microglia, identified high confidence causal genes and variants underlying risk loci for a range of neurological traits and identified a subset that replicated in a scalable IPS model system. Among other findings, our study revealed that the well-known AD risk locus near the BIN1 gene on chromosome 2 is likely to be driven by a microglia-specific eQTL and suggested that antagonism of BIN1 in microglia would be therapeutically beneficial in AD.
Our results underscore the variability between microglia from different individuals and clinical pathologies. One implication of the variation we observed between different patient pathologies is that the full spectrum of microglial function, in particular following trauma, is not well captured by small studies of a single patient population. The most obvious example of this are the populations of activated microglia we identified that account for less than 5% of cells in non-trauma patients.
Our analysis also provides a picture of the function of microglia following severe injury, producing cell populations that exhibit a mixture of a proinflammatory and chemotactic phenotypes. Notably, although animal models of acute brain injury suggest rapid expansion of microglia following trauma31, we only observed one population we identified had a proliferative phenotype, and both showed downregulation of CSF1R.
In contrast to previous reports8, we found relatively subtle effects of age on the expression of individual genes in microglia, with the modest changes we did detect consistent with increased inflammatory senescence in microglia over lifespan. However, our single cell data also revealed an increase in the influx of putatively infiltrating cells into the brain with increasing age. One explanation is that this phenomenon reflects decreasing blood-brain barrier integrity with age. Differences in microglia expression between males and females were relatively small, although we did observe increased complement activity in females, perhaps suggesting a role for complement pathways in the higher incidence of AD in women.
Our eQTL analysis revealed a number of candidate risk genes for a range of traits, with function in microglia. This was most obvious for neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease and included well-known risk genes, such as BIN1. At BIN1, we demonstrated how our microglia eQTL map can be used as a reference to establish the validity of different model systems to study the subtle effects of common disease risk variants. At this locus, our study provides evidence that IPSDMac are a suitable model system to explore the role of a putative causal variant, rs6733839C>T, its effects on BIN1 expression, and role in AD risk. More generally, although more complicated protocols for IPS-microglia differentiation exist32, our results highlight that IPSDMac may be sufficient in specific cases. Equally importantly, our results highlight where IPSDMac may not be suitable, for example at the PTK2B locus. Finally, we note that we observe some variability in colocalisation results between different AD GWAS studies. This is likely to reflect differences in power, but also some variation in methodology, for example the use of GWAS by proxy approaches versus direct phenotyping.
An obvious extension of our approach will be to map microglia population-specific genetic effects, for example to detect eQTLs that manifest only in activated microglia. This analysis was not possible here, due to the low number of individuals with 1 or more cells in different populations. In particular, the activated populations C and D are composed of 1,209 and 210 cells from 62 and 23 patients respectively. We anticipate that future studies with larger sample sizes will be sufficiently powered to detect such effects.
In summary, we have generated a population-scale map of gene expression in primary human microglia, across a diverse set of clinical pathologies. We demonstrate the human microglial response to an acute insult of the brain parenchyma. Our study provides a systematic exploration of microglia diversity, defines a reference data set of microglial expression and provides a foundation for robust future functional studies of neurodegenerative disease mechanisms using IPSC-based models.
Methods
Tissue sampling
Human brain tissue was obtained with informed consent under protocol REC 16/LO/2168 approved by the NHS Health Research Authority. Adult brain tissue biopsies were taken from the site of neurosurgery resection for the original clinical indication. Samples were collected into five main categories: a “control” group who had cortical microglia sampled at the beginning of a surgical corridor when the distance to the clinical pathology exceeded 4 cm. This group was utilised to identify any iatrogenic factors influencing the transcriptomics of human microglia. Additionally, samples were obtained from the cortex of patients with hydrocephalus, brain tumours, patients who had sustained a spontaneous haemorrhage and a traumatic brain injury. Paired venous blood was sampled. Tissue was transferred to Hibernate A low fluorescence (HALF) supplemented with 1x SOS (Cell Guidance Systems), 2% Glutamax (Life Technologies), 1% P/S (Sigma), 0.1% BSA (Sigma), insulin (4 g/ml, Sigma), pyruvate (220 g/ml, Gibco) and DNase 1 Type IV (40 g/ml, Sigma) on ice and transported to a CL2 laboratory.
Dissociation of brain tissue
Brain tissue was mechanically digested in fresh ice-cold HALF supplemented with 1x SOS (Cell Guidance Systems), 2% Glutamax (Life Technologies), 1% P/S (Sigma), 0.1% BSA (Sigma), insulin (4 g/ml, Sigma), pyruvate (220 g/ml, Gibco) and DNase 1 Type IV (40 g/ml, Sigma). The prepared mix was spun in HBSS+ (Life Technologies) at 300 g for 5 mins and supernatant discarded. The digested tissue was rigorously triturated at 4°C and filtered through a 70 m nylon cell strainer (Falcon) to remove large cell debris and undigested tissue. Filtrate was spun in a 22% Percoll (Sigma) gradient with DMEM F12 (Sigma) and spun at 800 g for 20 mins. Supernatant was discarded and the pellet was re-suspended in ice cold supplemented HALF.
Fluorescence-activated cell sorting
For single cell smart sequencing, human microglia were using fluorescence-activated cell sorting. The isolated cell suspension was incubated with conjugated PE anti-human CD11b antibody (BioLegend) for 20 mins at 4°C. Cells were washed twice in ice cold supplemented HALF and stained with Helix NP viability marker. Cell sorting was performed on BD AriaIII cell sorter (Becton, Dickinson and Company, Franklin Lakes, New Jersey, US) at the University of Cambridge Cell Phenotyping Hub at Cambridge University Hospital, Cambridge, UK.
Magnetic-activated cell sorting
To avoid sustained stress on microglia as a result of prolonged sorting times for bulk sequencing magnetic-activated cell sorting was performed on these cells. An isolated cell suspension of cells were incubated with anti-CD11b conjugated magnetic beads (1:50, Miltenyi, 130-049-601) for 15 mins at 4°C. Cells were washed twice with supplemented HALF and passed through an MS column (Miltenyi, 130-042-201). Each sample was washed three times in the column and then extracted. Samples were added to a 1.5 ml Eppendorf to which 350 μl of RNAlater (Qiagen) was added, samples were stored at −80°C prior to sequencing.
Immunohistochemistry
Tissue was fixed with 4% PFA at 4°C for 48 hours overnight and subsequently submerged in 30% sucrose w/v in PBS for cryoprotection at 4°C until it settled down to bottom (~48-hours). Cryoprotected brain was then embedded in cryomold filled with OCT. Brain is then frozen in isopentane and stored at −80°C. 12 μm sections were obtained using a cryostat. Tissue sections were air-dried and stored at −80°C. For antigen-retrieval the slides were submerged in preheated citrate buffer pH 6.0 (Sigma) in a water bath at 95°C for 15 min. The slides were washed three times with PBS (5min, RT) and blocked in 0.3% PBST with 10% NDS for 1h at RT. Primary antibodies; Iba-1 (1:1000, Wako, 019-19741), Iba-1(1:300, abcam, ab5076), C3 (1:200 abcam, ab97462), CCL4 (1:50, r&d systems, MAB271), CD63(1:300, abcam, ab59479) and BIN-1 (1:500, abcam, ab182562) were diluted in 0.1% PBST with 5%NDS and incubated overnight at 4°C. The slides were washed 3 times for 10min with PBS. Next, secondary antibodies in blocking solution were applied at a concentration of 1:500 for 2h at RT. Slides were washed 3 times with PBS for 10 min each, whereby the first wash contained Hoechst 33342 nuclear stain (2 μg/ml,). The slides were mounted with coverslips using FluoSave (CalBiochem). Image acquisition was performed using a Leica-SP5 microscope (Leica) and LAS software (Leica). Further image processing and analysis was performed using the ImageJ software package.
Single-molecule fluorescent in situ hybridization
Human tissue smFISH was performed using the RNAScope LS Multiplex Assay (Advanced Cell Diagnostics (ACD)). Before staining, slides were directly transferred from −80°C into pre-chilled 4% PFA (methanol-free) in PBS for 45-min and then submerged in boiling ER1 buffer (Advanced Cell Diagnostics, Bio-Techne) for 15-min. After the antigen-retrieval, slides were serially dehydrated through 50%, 70%, 100%, and 100% ethanol for 5 minutes each. Tissue sections were then processed using a Leica BOND RX to automate staining with the RNAScope Multiplex Fluorescent Reagent Kit v2 Assay and RNAScope 4-plex Ancillary Kit for Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics, Bio-Techne) following the manufacturers’ instructions. Automated processing included heat-induced epitope retrieval at 95°C for 10 minutes in ER2 buffer and digestion with Protease III for 10 minutes. Tyramide signal amplification with 1:300 Opal 520, Opal 570, and Opal 650 (Akoya Biosciences) was used to develop three probe channels. Nuclei staining was performed with 1:50,000 DAPI (Life Technologies Ltd). Stained sections were imaged with a Perkin Elmer Opera Phenix High-Content Screening System, in confocal mode with 1 μm z-step size, using a 20x water-immersion objective (NA 0.16, 298.99 nm/pixel). Channels: DAPI (Excitation 375 nm; Emission 435-480 nm), Opal 520 (Ex. 488 nm; Em. 500-550 nm), Opal 570 (Ex. 561 nm; Em. 570-630 nm), Opal 650 (Ex. 640 nm; Em. 650-760 nm).
Blood preparation
DNA extraction was performed from the venous blood. 10 ml of whole blood was washed with 1% phosphate buffered saline (PBS) and layered on pancoll human (PAN biotech) and spun at 500 g for 25 mins. The white cell component was extracted and transferred to a 1.5ml Eppendorf and stored as a frozen pellet at −80C prior to sequencing.
SNP genotyping
Genomic DNA was extracted from blood using the QIAamp DNA mini and blood mini kit (Qiagen, 51104). 200 ng of gDNA was used for input for the SNP array (Infinium Omni2.5-8 v1.4 Kit) and genotyping was performed according to the manufacturer’s instructions. We discarded 3 samples that showed the genotyping call rate below 95% (see Supplementary Table 6 for details). We used the 1000 Genomes Phase III integrated variant set (Data availability) as the reference haplotype data and performed whole genome imputation by using the Beagle software (version 4.0; https://faculty.washington.edu/browning/beagle/b4_0.html). We converted the genome coordinate from GRCh37 to GRCh38 using CrossMap (version 0.5.2; http://crossmap.sourceforge.net/).
iPS cell culture and macrophage differentiation
We cultured 133 iPS cell lines from HipSci14. iPS cell culture and macrophage differentiation was carried as previously described29 with some minor modifications (see Supplementary Note for details).
Single cell RNA-seq of primary microglia
Single primary microglia cells were processed as previously described15, but with some minor modifications to the Nextera library making process: 0.5 ng of cDNA was used as input for the tagmentation process with all Nextera (Illumina, FC-121-1030) reagent volumes scaled down 100-fold. Tagmentation was quenched with 0.2 % sodium dodecyl sulphate. Libraries were amplified with KAPA HiFi (Kapa Biosystems, KK2601) with indexing primers ordered from Integrated DNA Technologies.
Low-input bulk RNA-seq and ATAC-seq library preparation for primary microglia and iPS-derived macrophages
For RNA-seq samples, between 0.3 ng and 10 ng of bulk total RNA from primary microglia cells or iPS-derived macrophage cells was used as input for a modified Smart-seq2 library preparation15 (see Supplementary Note for detailed protocol). ATAC-seq library preparation was performed as previously described29. Pools of 96 libraries were sequenced over 8 lanes or 24 lanes of a HiSeq SBS v4 for RNA-seq and ATAC-seq preparations, respectively, collecting 75 bp paired-end reads.
Bulk RNA-seq data of other myeloid cells and brain tissues
We downloaded fastq files of the bulk RNA-seq of 6 primary microglias (pMICs) and 9 iPS cell derived microglia (iMICs)32, 10 monocyte derived macrophages (MDMs) and iPS cell derived macrophages (IPSDMac)33, 10 iMICs, 8 MDMs and 4 pMICs34, 45 pMICs17, 9 iMICs and 3 pMICs35, 18 IPSDMac and 9 MDMs36, and 3 pMICs16. See Supplementary Table 7 for details of cell types and sample sizes. For brain tissues, we downloaded the count table of RNA-seq data for all tissues from GTEx (V7; Data availability) and extracted 1,671 brain samples. We also downloaded fastq files of the BLUEPRINT monocyte37 RNA-seq data from EGA (Data availability) and processed the same as our sample.
Sequencing data preprocessing
All sequence data sets were aligned to human genome assembly GRCh38. We performed adapter trimming of Tn5 transposon and PCR primer sequences for our RNA-seq (both single-cell and bulk) and ATAC-seq data using skewer38 (version 0.1.127; https://github.com/relipmoc/skewer) before alignment. Both Smart-seq2 and bulk RNA-seq data were aligned using STAR39 (version 2.5.3a; https://github.com/alexdobin/STAR/releases) using ENSEMBL human gene assembly 90 as the reference transcriptome. All other RNA-seq data were also aligned as same as our RNA-seq data without adapter trimming. Following alignment, we used featureCounts40 (version 1.5.3; http://subread.sourceforge.net/) to count fragments for each annotated gene. The ATAC-seq data were aligned using bwa41 (version 0.7.4; https://sourceforge.net/projects/bio-bwa/files/). We performed peak calling as described in42 by pooling all five samples.
Smart-seq2 scRNA-seq quality control with other public data sets
In total we sequenced 26,496 cells, of which 9,538 cells passed the quality control criteria: the minimum number of sequenced fragments (>10,000 autosomal fragments), the minimum number of expressed genes (>500 autosomal genes), mitochondrial fragment percentage (<20%) and the library complexity (percentage of autosomal fragment counts for the top 100 highly expressed genes <30%). We also performed demuxlet43 to remove doublets from two different patients with different genetic background. We then performed cell type clustering with other primary single cell RNA-seq of 68k PBMCs18 and GTEx brain tissues characterised by DroNc-seq19 (Data availability). The count data from two studies were joined by gene IDs and converted into CPM (count per million) along with our primary microglia read count data. We fit the latent factor linear mixed model in which the three different studies were treated as a random effect (see Supplementary Note Section 1 for details). We obtained the 12 latent factors which were subsequently used for UMAP clustering. We extracted 8,662 microglia cells in the UMAP plot (Figure 1d) for downstream analysis, which were distinct from other circulating blood cell types (such as NK T cells, Monocytes and B cells). To ensure our batch correction approach was valid, we also compared with the three established batch correction methods: Harmony45, Seurat V346 and MNN correct47. Our model returned a reasonable clustering of cells that was comparable to that from Seurat V3 (Extended Data Figure 1d-h). It was not possible to compare the performance of our method when fitting donor and plate as batch effects, because these existing methods do not scale to large numbers of batches (129 donors and 67 plates).
Characterisation of infiltrating cells
We used the lme4 package implemented in R to fit the generalised linear mixed model for infiltrating cell status (microglia/non-microglia) as a binary outcome. We used all possible clinical confounders (patient, pathology, brain region, brain hemisphere, ethnicity and sex) and technical confounders (the number of expressed genes for each cell, 384 plate on which each cell undergone library preparation and sequencing, the number of mapped fragments, 96 well plate position where each cell was sorted, ERCC% among all mapped fragments and mitochondrial RNA fragment percentage among all mapped fragments) as random effects and the patients’ age as the fixed effect to investigate the statistical significance of age effect.
Variance component analysis
A linear mixed model of log(CPM+1) values across genome-wide genes (whose CPM>0 for 10% of total cells) was used to estimate the transcriptional variation. The 13 different factors (Patient, the number of expressed genes per cell, pathology, plate ID, ERCC percentage, the number of expressed genes in each cell, 96 well plate position, age of patient, mitochondria RNA percentage, brain region, brain hemisphere, ethnicity and sex) were fitted as random effects with independent variance parameters ϕk 2. The variance explained by the factor k was measured by the intraclass correlation ϕk 2/(1+ϕk 2, where the other 12 factors were fixed constant. The standard error of the intraclass correlation was computed by the delta method with the standard error of the variance parameter estimator. See Supplementary Note Section 1.1 for details.
Detection of microglia populations
We used the linear mixed model to estimate the latent factors with the 13 known confounding factors (see Supplementary Note Section 1.2 for details). We used 15 first latent factors to cluster cells into populations. We utilised the Shared Nearest Neighbour Clustering implemented in Seurat (version 3.0.2) with resolution parameter of 0.2 to identify the four microglia populations.
Marker gene enrichment analysis for microglia in different studies
We downloaded the differentially expressed genes from20 found in the three different comparisons (1) no-pathology vs Alzheimer’s disease pathology (2) no-pathology and early stage of Alzheimer’s disease pathology (3) early vs late Alzheimer’s disease pathology. We also downloaded the marker genes for the 14 different clusters found in the comparison between healthy and glioma-associated microglia21. We performed the Fisher exact test on the 2-by-2 table of the marker genes for one of our microglia populations (A-D) and one of the three comparisons for Alzheimer’s disease microglia or one of 14 different clusters for the glioma associated microglia data. Fisher exact P-values were adjusted by Bejamini-Hochberg method.
Differential expression analysis of clinical factors
We utilised the same linear mixed model we employed for the variance component analysis to adjust for 13 known confounding effects and the effect of four cell population (see above) as a random effect in differential expression analysis. We fit the model on gene-by-gene basis using the estimated variance parameters {} to test each factor k explaining a significant amount of transcription variation. If the focal factor k is numerical (e.g., age of patients), the Bayes factor of effect size was computed by comparing the full model and the reduced model without the factor k. If the focal factor k is a categorical variable with l levels (e.g., pathology with 5 levels), we partitioned the levels into any of two groups. There are 2l-1 contrasts which were tested against the null model (removing the focal factor k in the model) to compute Bayes factors. Then, those Bayes factors were used for fitting a finite mixture model to compute the posterior probability as well as the local true sign rate (ltsr) (see Supplementary Note Section 1.3 for more details). We used g:Profiler 2 implemented in R (version 2.0.1.5) to perform which pathways are enriched for differentially expressed genes for each contrast. We used genes whose ltsr is greater than 0.5 to perform the analysis (both upregulated and downregulated genes separately).
We also repeated the same analysis using the bulk RNA-seq data. We only used a part of bulk samples from patients with genotype data to estimate ethnicity (N=102). We normalised the raw count data into CPM (counts per million) and then log transformed. The linear mixed model with 9 technical and biological factors (pathology, the number of genes expressed, the number of total fragments, sequencing batch, dominant hemisphere, sex, brain region, ethnicity and age) was used to adjust confounding effects between factors. The effect size and its significance (ltsr value) of differential expression for each gene were computed as described above (see also Supplementary Note for more details).
Expression QTL mapping using linear regression and RASQUAL
We used simple linear regression to map eQTLs. The fragment counts were GC corrected as described before22, normalised into TPM (transcripts per million) and then log transformed (log of TPM+1). 25 principal components (PCs) were calculated and regressed out from the normalised expression levels. The 25 PCs were determined by comparing the eigenvalues between original and permuted count tables. If a real eigenvalue in descending order was greater than that from permuted data at the same rank, we used the corresponding PC as the covariates. We note that patient pathology was well captured by PCs of gene expression: including patient pathology as an additional covariate in our model did not improve power to detect eQTLs (575 eQTLs detected at FDR 5% with pathology in the model, vs 585 without). For each gene, we applied Benjamini-Hochberg (BH) FDR correction across all variants tested in the cis-regulatory region to obtain the minimum Q-value. Then, the minimum Q-values across all genes are adjusted again by BH FDR method to compute the genome-wide FDR. We also mapped eQTLs using RASQUAL (version 0.1; https://github.com/natsuhiko/rasqual) with the raw count data and the same 25 PCs used in linear regression as the covariates. We used --no-posterior-update option to keep the posterior genotype dosage identical to the prior genotype dosage, that allowed us to stabilise the convergence of model fitting. We picked up the minimum BH Q-value for each gene to perform the multiple testing correction genome-wide. We performed a permutation test once for each gene and constructed the empirical null distribution to which the real Q-values were compared to calibrate the FDR threshold22. Colocalisation analysis with GWAS traits was performed using the COLOC47 implemented on R.
Bayesian hierarchical model
We extended a standard Bayesian hierarchical model48 to jointly map eQTLs in three different cell types. We employed the association Bayes factor at each variant for each gene to compute the regional Bayes factors (RBFs) in a cis region of 1Mb centred at transcription start site (TSS) under 15 different hypotheses. Those RBFs were used in a hierarchical model to estimate prior probabilities that eQTLs are colocalised between any two of the three cell types as well as shared among three cell types. It can provide posterior probability that a gene is an eQTL for each cell type. See Supplementary Note Section 2 for more details.
Extended Data
Extended Data Fig. 1. Overview of bulk and single cell RNA-seq data.
UMAP of bulk RNA-seq for myeloid cells. The “Primary microglia” cluster contains samples collected in this study (pink dots) and previous studies (purple dots) (information on the source of previous study data can be found in Supplementary Table 7). “Cultured primary and IPS-derived cells”, includes IPS-derived macrophages and microglia (blue dots), cultured primary microglia and monocyte derived macrophages (orange dots). “Monocytes” (green dots) denotes primary monocytes obtained from the BLUEPRINT project. b. Feature plots of three microglia marker genes (P2RY12, CX3CR1 and TMEM119) using the same UMAP coordinates as Figure. 1d. c. Age versus percentage of infiltrating cells. Red line shows the logistic regression line, the red transparent band shows the 95% confidence interval estimated using a generalised linear mixed model for the binary outcome (Materials and Methods). d. UMAP plot identical to Figure 1d. e. UMAP plot from the first 12 principal components computed from the same input data for the linear mixed model without any batch correction. f. UMAP of the same 12 PCs where the batch effect was corrected by using Harmony45. g. UMAP of batch corrected data using the canonical correlation analysis method implemented in Seurat V3 46 with a default setting. We computed the 12 PCs from the integrated data for UMAP plot. h. UMAP of batch corrected data using MNN correct47. Note that points were coloured according to the cell types (same as Figure 1d): glutamatergic neurons from the PFC (exPFC); pyramidal neurons from the hip CA region (exCA); GABAergic interneurons (GABA); granule neurons from the hip dentate gyrus region (exDG); astrocytes (ASC); oligodendrocytes (ODC); oligodendrocyte precursor cells (OPC); neuronal stem cells (NSC); endothelial cells (END); dendritic cell (DC); B cell (B); hematopoietic progenitor cell (CD34+); NK T cell (NK).
Extended Data Fig. 2. Microglia marker gene comparisons and validations.
a. Marker gene enrichment analysis with Alzheimer’s disease associated microglia20 and glioma associated microglia21. There are three different comparisons for Alzheimer’s disease associated microglia and 14 different populations for glioma associated microglia. Heatmap shows odds ratios and Benjamini-Hochberg (BH) Q-values of the Fisher exact tests between our marker genes and differentially expressed genes in other studies. b. Differentially expressed genes between microglia from different patient pathologies using single cell RNA-seq data. Heatmap shows averaged, normalised expression level (defined as the posterior mean of pathology random effect term, see Materials and Methods) of differentially expressed genes at local true sign rate (ltsr) greater than 0.9 ((Urbut et al. 2019); see Materials and Methods for details). Heatmap is divided into groups based on all possible pairwise groupings of the four cell populations, ordered by most transcriptionally distinct, such that the most different grouping, trauma versus all non-trauma, appears at the top. c. Differential expression of candidate marker genes for immunohistochemistry in fresh frozen patient tissue samples. d. Immunohistochemistry panel of each pathology to validate expression of a differentially expressed gene at the protein level; hydrocephalus (C3), tumour (CCL4), haemorrhage (CD63) and trauma (BIN-1) compared to control. Iba-1 (red) and protein of interest (green). e. RNAScope image of differentially expressed gene panel for cluster C; HAMP (yellow) and RAC2 (purple) with C1QC (green) used to identify microglia. f. RNAScope image of differentially expressed gene panel for cluster D; KLF (yellow) and CCL20 (purple). Scale bar 10μM.
Extended Data Fig. 3. Differential expression analysis with bulk RNA-seq data.
a. Variance components analysis of log CPM values for the bulk RNA-seq data (N=102) with biological and technical factors using the linear mixed model (Online methods). b. Heatmap shows the effect size of age for each gene (each row) estimated by the linear mixed model (Online methods). The genes with LTSR>0.9 in single-cell data are shown. c. PADI2 normalised expression in bulk RNA-seq data against patients’ age. d. P2RY12 expression in bulk RNA-seq data against patients’ age. e. Heatmap shows the average expression of males and females for each gene (each row) estimated by the linear mixed model (Online methods). The genes with LTSR>0.9 in single-cell data are shown. f. C1QA normalised expression in bulk RNA-seq data for males (M) and females (F). g. HLA-DQB1 normalised expression in bulk RNA-seq data for males (M) and females (F). h. Heatmap shows the average expression for 5 different brain regions estimated by the linear mixed model (Online methods). The genes differentially expressed between a combination of Occipital and Cerebellum and the 3 other regions (LTSR>0.9) in single-cell data are shown.
Extended Data Fig. 4. Colocalisation of eQTLs with various GWAS traits.
a. eQTL effect size comparison for 502 eQTL genes at FDR 5% (linear regression) whose gene body contains at least one feature SNP with sufficient coverage (greater than 5% of average coverage across coding regions). The x-axis shows the eQTL effect size (beta) estimated from linear regression and the y-axis shows the eQTL effect size (pi value) from RASQUAL using only allele-specific count data. The red line shows the least square line crossing (0, 0.5). Note that, x=0 is the null hypothesis for linear regression and y=0.5 is the null hypothesis for RASQUAL. b. Examples of colocalised eQTLs in microglia. Colocalisation with Parkinson’s disease at KLHL7-AS1 eQTL (left column), colocalisation with Fed-up feelings at DAG1 eQTL (middle column) and colocalisation with Crohn’s disease at ERAP2 eQTL (right column). The y-axis of each panel shows log10 association Bayes factor for the eQTL or the GWAS trait. The colour of each point indicates LD index (r2 value) to the lead eQTL variant shown by the purple diamond. c. Heatmap of the posterior probability for colocalisation (PP4) between various GWAS traits and cell types/tissues. Each row corresponds to a specific combination of gene and a GWAS trait. Each column corresponds to eQTLs discovered in different cell types and tissues. The first column of the heatmap corresponds to microglia eQTLs, the second column corresponds to eQTLs in IPS cell derived macrophage (IPSDMac) from this study (Materials and Methods), the third column shows eQTLs in primary monocytes from the BLUEPRINT project (Materials and Methods) and the remaining 48 tissues are eQTLs from GTEx V7 (Materials and Methods). The colour of each grid shows the strength of PP4 (white: PP4=0.0 and red: PP4=1.0). Gray indicates that the gene was very weakly or not expressed, and therefore no eQTL summary statistics were available.
Extended Data Fig. 5. Finemapping of microglia eQTLs.
a. Regional association plots at the CD33 locus. b. Coverage plot shows the normalised expression level around the CD33 gene stratified by genotype at the putative splice variant (rs12459419C>T). The zoom-in panel shows a coverage plot of expression level around the second exon (ENST0000262262.4). The coverage shows the first intron expression is negatively correlated with the second exon expression, suggesting the expression of non-coding isoform (ENST00000601785.5) is increased by the alternative allele (T) of the splicing QTL. c. Colocalisation between an association with risk for Alzheimer’s disease on chromosome 2 and an eQTL for the noncoding RNA gene EPHA1-AS1 in microglia, GTEx tissues and myeloid cell types. The x-axis shows the posterior probability of colocalisation (PP4) and y-axis shows the average expression level (log10 TPM) for each tissue or cell type. d. Colocalisation between AD risk and expression of the protein-coding EPHA1 gene. The x-axis shows the posterior probability of colocalisation (PP4) and y-axis shows the average expression level (log10 TPM) for each tissue or cell type. e. Boxplots show the relationship between expression at the PTK2B gene and genotype at the lead eQTL variant (rs28834970C>T) three myeloid cell types. The y-axis shows normalised expression levels (log TPM value). Each dot on the box shows the expression level of a single sample. f. Coverage plot shows chromatin accessibility in iPS cell derived macrophages stratified by three genotype groups of the lead AD GWAS/BIN1 eQTL variant g. Scatter plot of MEF2C (x-axis) and BIN1 (y-axis) expression in GTEx brain tissues and myeloid cell type.
Supplementary Material
Acknowledgements
R.F was supported by funding from the UK Multiple Sclerosis Society (MS50), The Adelson Medical Research Foundation and a core support grant from the Wellcome Trust and MRC to the Wellcome Trust-Medical Research Council Cambridge Stem Cell Institute (203151/Z/16/Z). A.Y is supported by a Wellcome Trust Clinicians PhD Fellowship (RRZD/029). All data for this study was generated under Open targets project OTAR039. N.K and D.J.G. were funded by the Wellcome Trust grant WT206194. We thank the staff in the Cellular Genetics and Phenotyping and Sequencing core facilities at the Wellcome Sanger Institute.
Footnotes
Author contributions
The brain samples were obtained under neurosurgery by A.Y, H.B, J.C, K.P.B, A.J, M.R.G, R.T, R.K, R.M, M.R.G, I.T, I.J, K.H, R.M, R.M, C.W, S.J.P, P.J.K, T.S and P.J.H. Cell isolation protocols were performed by A.Y, M.S, N.M and C.E.M. Microglia isolation strategies were designed by A.Y, T.R.H, B.S, R.F. Single cell and bulk RNA-seq were performed by A.Y, A.K, and F.C. Cell culture experiments iPSCDMacs were performed by A.K and N.P. The main analyses and data preparations were performed by N.K E.M and M.G processed GWAS summary statistics for colocalisation analysis. J.S and J.L provided the summary statistics of Alzheimer’s disease. K.K and N.S provided the imputed BLUEPRINT genotype data. N.P preprocessed the iPSCDMac RNA-seq data. J.S.P and O.A.B performed the RNA-scope and immunohistochemistry assay. A.Y, N.K, R.F and D.J.G wrote the manuscript; T.R.H and B.S assisted in editing the manuscript.
Competing Interests Statement
D.J.G. and E.M. were employees of Genomics PLC at the time the manuscript was submitted.
Data availability
Patients were consented to share both expression and raw genotype data under managed access and all data are available under managed access from the EGA, upon approval by the Wellcome Sanger Institute Data Access Committee. More details on how to access these data can be found at https://ega-archive.org/datasets/EGAD00001005736. Raw data (fastq files and CRAM files) of Smart-Seq2 and bulk RNA-seq for the primary microglia samples as well as the raw genotype data (Illumina Omni 2.5) and imputed genotype data by Beagle software are available from European phenome-Genome Archive (EGA) (Accession ID: EGAD00001005736). Summary statistics of eQTLs mapped by linear regression and RASQUAL for primary microglia are also available from EGA (Accession ID: EGAD00001005736). The 1000 Genomes Phase III integrated variant set can be obtained from the project website (http://www.internationalgenome.org/data). GTEx V7 summary statistics and brain DroNc-seq data with cell type annotation data can be obtained from the GTEx project website (https://www.gtexportal.org/home/datasets). PBMC 68k single cell data is available from the project GitHub page (https://github.com/10XGenomics/single-cell-3prime-paper/tree/master/pbmc68k_analysis). The BLUEPRINT monocyte RNA-seq data is available from EGA (Accession ID: EGAD00001002674). For details on how to access these data, please visit https://ega-archive.org/datasets/EGAD00001002674.
References
- 1.Schafer DP, Stevens B. Microglia Function in Central Nervous System Development and Plasticity. Cold Spring Harb Perspect Biol. 2015;7:a020545. doi: 10.1101/cshperspect.a020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. 2018;18:225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 3.Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med. 2017;23:1018–1027. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- 4.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tansey KE, Cameron D, Hill MJ. Genetic risk for Alzheimer’s disease is concentrated in specific macrophage and microglial transcriptional networks. Genome Med. 2018;10:14. doi: 10.1186/s13073-018-0523-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gjoneska E, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olah M, et al. A transcriptomic atlas of aged human microglia. Nat Commun. 2018;9:539. doi: 10.1038/s41467-018-02926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keren-Shaul H, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169:1276–1290.:e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Hammond TR, et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity. 2019;50:253–271.:e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda T, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–392. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 12.Mrdjen D, et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity. 2018;48:380–395.:e6. doi: 10.1016/j.immuni.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Mathys H, et al. Temporal Tracking of Microglia Activation in Neurodegeneration at Single-Cell Resolution. Cell Rep. 2017;21:366–380. doi: 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilpinen H, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosselin D, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356 doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habib N, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sankowski R, et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat Neurosci. 2019;22:2098–2110. doi: 10.1038/s41593-019-0532-y. [DOI] [PubMed] [Google Scholar]
- 22.Kumasaka N, Knights AJ, Gaffney DJ. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen I, Savage J, Watanabe K, Bryois J, Williams D. Genetic meta-analysis identifies 10 novel loci and functional pathways for Alzheimer’s disease risk. bioRxiv. 2018 [Google Scholar]
- 24.Kunkle BW, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marioni RE, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiatry. 2018;8:99. doi: 10.1038/s41398-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartzentruber J, et al. Genome-wide meta-analysis, fine-mapping, and integrative prioritization identify new Alzheimer’s disease risk genes. medRxiv. 2020:2020.01.22.20018424 [Google Scholar]
- 28.Raj T, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet. 2014;23:2729–2736. doi: 10.1093/hmg/ddt666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alasoo K, et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nott A, et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 2019;366:1134–1139. doi: 10.1126/science.aay0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vela JM, Yáñez A, González B, Castellano B. Time course of proliferation and elimination of microglia/macrophages in different neurodegenerative conditions. J Neurotrauma. 2002;19:1503–1520. doi: 10.1089/089771502320914723. [DOI] [PubMed] [Google Scholar]
- 32.Abud EM, et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017;94:278–293.:e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alasoo K, et al. Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription. Sci Rep. 2015;5:12524. doi: 10.1038/srep12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douvaras P, et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports. 2017;8:1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muffat J, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, et al. Functional analysis and transcriptomic profiling of iPSC-derived macrophages and their application in modeling Mendelian disease. Circ Res. 2015;117:17–28. doi: 10.1161/CIRCRESAHA.117.305860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414.:e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H, Lei R, Ding S-W, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumasaka N, Knights AJ, Gaffney DJ. High-resolution genetic mapping of putative causal interactions between regions of open chromatin. Nat Genet. 2019;51:128–137. doi: 10.1038/s41588-018-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang HM, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36:89–94. doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018 doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veyrieras J-B, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Patients were consented to share both expression and raw genotype data under managed access and all data are available under managed access from the EGA, upon approval by the Wellcome Sanger Institute Data Access Committee. More details on how to access these data can be found at https://ega-archive.org/datasets/EGAD00001005736. Raw data (fastq files and CRAM files) of Smart-Seq2 and bulk RNA-seq for the primary microglia samples as well as the raw genotype data (Illumina Omni 2.5) and imputed genotype data by Beagle software are available from European phenome-Genome Archive (EGA) (Accession ID: EGAD00001005736). Summary statistics of eQTLs mapped by linear regression and RASQUAL for primary microglia are also available from EGA (Accession ID: EGAD00001005736). The 1000 Genomes Phase III integrated variant set can be obtained from the project website (http://www.internationalgenome.org/data). GTEx V7 summary statistics and brain DroNc-seq data with cell type annotation data can be obtained from the GTEx project website (https://www.gtexportal.org/home/datasets). PBMC 68k single cell data is available from the project GitHub page (https://github.com/10XGenomics/single-cell-3prime-paper/tree/master/pbmc68k_analysis). The BLUEPRINT monocyte RNA-seq data is available from EGA (Accession ID: EGAD00001002674). For details on how to access these data, please visit https://ega-archive.org/datasets/EGAD00001002674.