Abstract
The study of ever more complex biomolecular assemblies implicated in human health and disease is facilitated by a suite of complementary biophysical methods. Pulse Dipolar electron paramagnetic resonance Spectroscopy (PDS) is a powerful tool that provides highly precise geometric constraints in frozen solution, however the drive towards PDS at physiologically relevant sub-μM concentrations is limited by the currently achievable concentration sensitivity. Recently, PDS using a combination of nitroxide and CuII based spin labels allowed measuring 500 nM concentration of a model protein. Using commercial instrumentation and spin labels we demonstrate CuII-CuII and nitroxide-nitroxide PDS measurements at protein concentrations below previous examples reaching 500 and 100 nM, respectively. These results demonstrate the general feasibility of sub-μM PDS measurements at short to intermediate distances (~1.5 - 3.5 nm), and are of particular relevance for applications where the achievable concentration is limiting.
Keywords: EPR spectroscopy, Structural biology, PELDOR/DEER, RIDME, double-histidine motif
Introduction
The study of increasingly complex biomolecular assemblies and their interactions with the cellular environment has driven interest towards holistic structural characterization under conditions with high biological validity. Pulse dipolar EPR spectroscopy (PDS) is a powerful tool for such characterization, and complements X-ray crystallography, NMR, Förster resonance energy transfer (FRET), and cryo-EM data by providing solution-state distance constraints in systems of virtually unlimited size and complexity.1–9 Due to these characteristics, PDS is also an emerging technique for conformational studies of protein and nucleic acid complexes in cellulo.10–16 However, physiological concentrations are often in the sub-μM regime. In combination with low numbers of cells within samples, the challenge is to achieve sufficient absolute sensitivity. Analyzing a representative sample of 61 recent applications of nitroxide- nitroxide pulsed electron-electron double resonance (PELDOR)17–18 measurements using the 4- pulse double electron-electron resonance (DEER)19–20 sequence reveals the use of spin concentrations between 5 and 400 μM (median 100 μM, mean 116 ± 90 μM, PubMed search on 30 November 2020 for “PELDOR” or “DEER” and “EPR” covering 2017 to 2020, see Supporting Information (SI) for more details) demonstrating the current state of the art. While measurements down to 1 μM should be feasible we could not identify a single published example. Recently, CuII-nitroxide 5-pulse relaxation induced dipolar modulation enhancement (RIDME)21–22 measurements at 500 nM concentration in a protein in vitro allowed not only precise distance measurements but also determination of the binding affinity.23 Thereby, demonstrating the high-affinity of genetically encoded double-histidine motifs to CuII ions,24–25 and their suitability as labelling sites for low concentration studies.26
Herein, we approach practical concentration limits associated with PDS experiments and found CuII-CuII RIDME measurements and nitroxide-nitroxide PELDOR measurements feasible at 500 nM and 100 nM protein concentration, respectively (corresponding to spin concentrations of 1.6 μM and 200 nM, respectively). Importantly, these measurements were performed in a commercial non-broadband Q-band spectrometer, using well-established spin labels, methanethiosulfonate (MTSL)27–28 and CuII-nitrilotriacetic acid (CuII-NTA)25 (figure 1). PDS measurements in a biological system down to 100 nM protein concentration for nitroxide-nitroxide DEER and 500 nM for CuII-CuII RIDME are unprecedented.
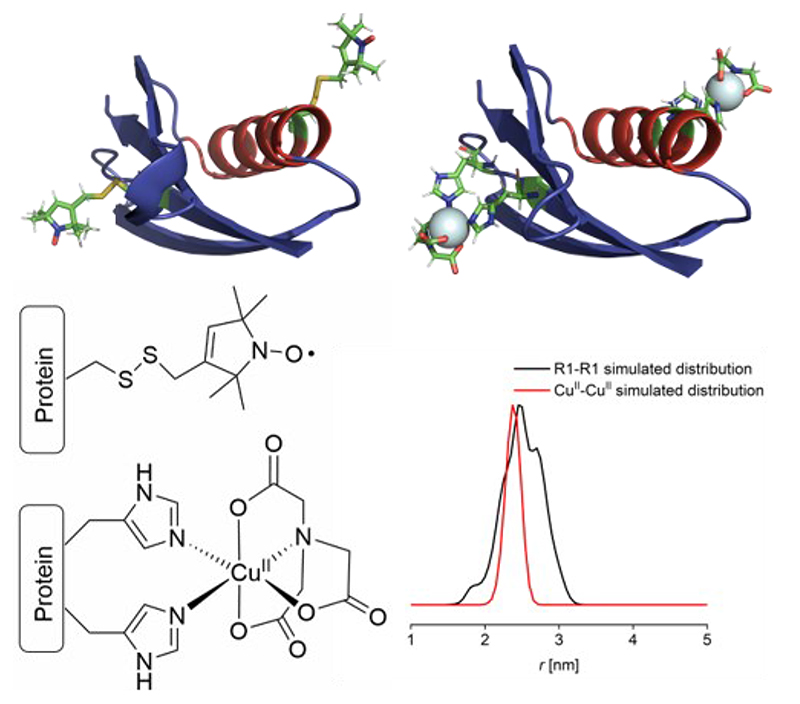
Figure 1.
GB1 constructs, spin label structures and simulated distance distributions. Top: Cartoon representations of GB1 constructs I6R1/K28R1 (left) and I6H/N8H/K28H/Q32H (right), with spin labels shown in stick representation. Bottom: Chemical structures of R1 nitroxide and double histidine CuII-NTA spin labels (left). Corresponding simulated distance distribution (right) for each construct, shown in black and red, respectively.
Commercial instruments have been used successfully for PELDOR measurements at low μM concentration.29 Concentration sensitivity has been demonstrated to further improve in homebuilt high-power resonator-free spectrometers30–31 or by implementation of arbitrary waveform generators (AWGs) and shaped pulses that yield higher spin inversion efficiencies.32–36 Additionally, novel pulse sequences have shown to enhance measurement sensitivity.37–40 Trityl- based radicals41–43 with exquisitely narrow spectral linewidths have been measured at 45 nM protein (90 nM spin) concentration41 employing the single-frequency double quantum coherence (DQC)44 experiment. This is a remarkable achievement owed to bespoke narrow line spin labels allowing use of a single-frequency technique. It is not currently established where the limits are for the most common 4-pulse DEER method applied to the most popular nitroxide labels, nor for the emerging use of RIDME on CuII-labels.
Experimental Methods
Protein expression and purification
Constructs
For this study, two constructs of the immunoglobulin-binding B1 domain of group G streptococcal protein G (GB1) were used: a tetra-histidine mutant with two double-histidine CuII- binding motifs (I6H/N8H/K28H/Q32H) and a double-cysteine mutant (I6C/K28C), both have been described previously.24 The first construct was obtained as a kind gift from Prof. Saxena, the second construct was bought from GenScript, and the pET-11a vector was used for both constructs.
Expression and purification
Both GB1 constructs were expressed and purified according to the procedure described in Wort et al.23 MTSL labelling of the double-cysteine construct was performed as described.23 Spin labelling was confirmed via MALDI-TOF using the in-house mass spectrometry facility, and labelling efficiencies obtained from continuous wave (CW) EPR spectra were ≥ 89 % compared to MTSL as a standard.23
Electron paramagnetic resonance (EPR) spectroscopy
Samples for pulse EPR measurements were prepared with varying protein and CuII-NTA concentrations in deuterated gel filtration buffer and 50% (v/v) deuterated ethylene glycol (Deutero) was used for cryoprotection as previously described.23 The samples with a final volume of 65 μL were transferred to 3 mm quartz EPR tubes which were immediately frozen in liquid nitrogen.
Pulse EPR experiments were performed at Q-band frequency (34 GHz) operating on a Bruker ELEXSYS E580 spectrometer with a 3 mm cylindrical resonator (ER 5106QT-2w in TE012 mode). Pulses were amplified by a pulse travelling wave tube (TWT) amplifier (Applied Systems Engineering) with nominal output of 150 W. Temperature was controlled via a cryogen-free variable temperature cryostat (Cryogenic Ltd) operating in the 3.5 to 300 K temperature range.
Temperature optimization for pulse EPR measurements were performed between 10 K and 50 K. Longitudinal relaxation times (T 1) were estimated from 3-pulse inversion recovery measurements under the mono- or bi-exponential approximation. Transverse dephasing times (T m) were estimated from 2-pulse electron-spin echo decay measurements, data was fitted to a stretched exponential function, with the stretching exponent varying from 1-2. Subsequently, relaxation times were used in downstream processing to determine relative sensitivities per temperature as described previously.23
Instantaneous diffusion (ID, also referred to as dipolar dephasing) was qualitatively assessed by measuring Hahn echo decays with varying flip angles of the second pulse as described.45
Pulse dipolar EPR
PELDOR and RIDME measurements were obtained at Q-band frequency (34 GHz) using a second frequency option (E580-400U).
PELDOR experiments were performed with the 4-pulse DEER18, 20, 46 pulse sequence (π/2(vA) – τ1 – π(vA) – (τ1 + t) – π(vB) – (τ2 - t) – π(vA) – τ2 – echo) at 50 K as described previously,47 with a frequency offset (pump – detection frequency) of +80 MHz (~3 mT). Shot repetition times (SRT) were set to 4 or 5 ms; τ1 was set to 380 ns, and τ2 was set to 1500 ns for the 100 and 500 nM samples (recorded for 47 and 14.5 hours, respectively) and to 3000 ns for the 25 μM sample (recorded for ~70 minutes) of the double-cysteine construct spin-labelled with MTSL (I6R1/K28R1). Pulse lengths were 16 and 32 ns for π/2 and π detection, and 12 or 14 ns for the ELDOR π pump pulse for the micromolar or nanomolar samples, respectively. The pump pulse was placed on the resonance frequency of the resonator and applied to the maximum of the nitroxide field-swept spectrum.
5-pulse RIDME experiments22 were recorded with the pulse sequence (π/2 – τ1 – π – (τ1 + t) – π/2 – Tmix – π/2 – (τ2 - t) –π – τ2 – echo) at 30 K at the field position corresponding to the maximum of the CuII field-swept spectrum with 8-step phase cycling, a τ1 of 400 ns, SRT of 407 μs, and a critically coupled resonator (high Q). The 500 nM sample of the tetra-histidine construct (I6H/N8H/K28H/Q32H) was recorded for 63.5 hours with a mixing time of 34 μs, while the 500 μM sample was recorded for ~15 minutes at three different mixing times (5, 35, and 65 μs, 1 scan) to facilitate the removal of artefacts or residual ESEEM by deconvolution.23
“Dummy” PELDOR and RIDME measurements (1 scan each) were recorded at 500 nM and 25 μM protein concentration for sensitivity estimates as described.23 Briefly, in these measurements, all delays are set to be constant, shifting the entire sequence by the dipolar increment, thus yielding a trace that is only varied by thermal noise on the acquired echo. RIDME experiments were recorded both, with a critically coupled and an over-coupled resonator to determine the effect of high versus low Q on sensitivity.
PDS experiments were analyzed using DeerAnalysis2015.48 PELDOR data were first background-corrected using a 3-dimensional homogeneous background function before Tikhonov regularization followed by statistical analysis using the validation tool in DeerAnalysis2015, varying background start from 5 to 80% of the trace length in 16 trials. Resulting background start time for the best fit was then used as starting point for a second round of Tikhonov regularization followed by a second round of statistical analysis, this time also including the addition of 50% random noise in 50 trials. For the 100 nM and 500 nM I6R1/K28R1 sample the first validation rounds resulted in a best fit where the background fit had a positive and thus unphysiological slope; therefore, data were cut iteratively by 10% and then by 20% of the initial trace length and Tikhonov regularization and validation were repeated. Since cutting of the data did not afford a falling background function while leaving the resulting distributions essentially unchanged, the full-length data were used for further processing.
RIDME data were first background-corrected using a homogeneous 6-dimensional background function before Tikhonov regularization followed by statistical analysis varying background start from 5 to 30% of the trace length in 8 trials and varying the background dimension from 3 to 6 in 7 trials. Resulting background start time and dimension for the best fit were then used as starting points for a second round of Tikhonov regularization followed by a second round of statistical analysis, this time also including the addition of 50% random noise in 16 trials. Validation trials from the second validation round for PELDOR and RIDME data were pruned with a prune level of 1.15, where trials exceeding the root mean square deviation of the best fit by at least 15% are discarded. In all cases the regularization parameter α was chosen according to the L-curve criterion49 and the goodness-of-fit. RIDME data obtained at 500 μM protein concentration were cut at a trace length of 2200 ns to improve estimation of the background function and fitting.
Simulations and Modelling
Distance distributions were modelled based on the I6H/N8H/K28H/Q32H construct (PDB ID: 4WH4);24 all modelling was performed using MMM 2018.50–51 For the I6R1/K28R1 construct, histidine residues at positions 6 and 28 of the I6H/N8H/K28H/Q32H construct were mutated to cysteine residues, while histidine residues at positions 8 and 32 were mutated to asparagine and glutamine residues, and R1 moieties were introduced at residues 6 and 28 in MMM 2018. Labelling with either MTSL or CuII-NTA was modelled under ambient temperature (298 K), and the corresponding predicted distance distributions are shown in the main text. For the CuII-NTA labelling, a correction factor of 1.13 was applied to the distance axis to correct for the g-value change from a free electron to bound CuII, assuming a giso of 2.13 for bound CuII-NTA.52
Cartoon structural representations of I6H/N8H/K28H/Q32H and I6R1/28H/32H GB1 constructs were generated using Pymol53 and MTSL wizard,54 respectively.
Results and Discussion
In the current study, Streptococcus sp. Group G protein G, B1 domain (GB1) constructs (I6R1/K28R1 and I6H/N8H/K28H/Q32H) were used as biological model systems (figure 1). GB1 has been used extensively in previous EPR methodology studies.23–26, 55–58 We have shown previously that nitroxide-detected CuII-nitroxide and CuII-CuII RIDME are similar in sensitivity and roughly two orders of magnitude more sensitive than CuII-CuII PELDOR when limited to rectangular pulses.23 Here, we endeavored to test the sensitivity of the most widespread pulse dipolar EPR methodology, nitroxide-nitroxide PELDOR.59 Therefore CuII-CuII RIDME and nitroxide-nitroxide PELDOR were measured at 500 nM concentration for a direct comparison of experiment sensitivity (figure 2).
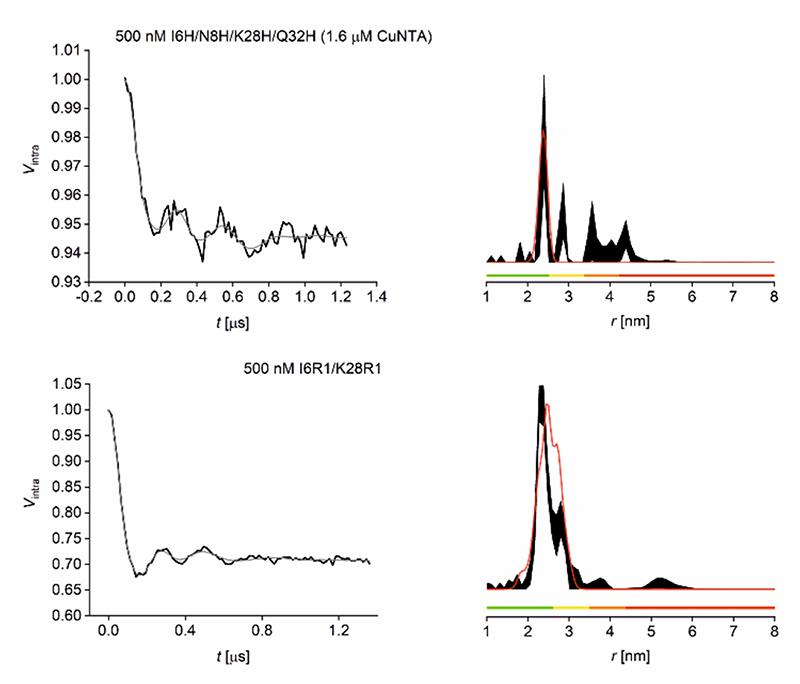
Figure 2.
500 nM GB1 CuII-CuII RIDME and nitroxide-nitroxide PELDOR Q-band data at 30 and 50 K, respectively. Top: RIDME data for 500 nM GB1 tetra-histidine with 1.6 μM CuII-NTA added. Bottom: PELDOR data for 500 nM GB1 I6R1/K28R1. Left: Background-corrected data (black) and fit (grey). Right: Corresponding distance distributions given as 95% confidence intervals (± 2σ) with 50% noise added for error estimation during statistical analysis; simulated distance distributions are shown in red. Color bars represent reliability ranges (green: shape reliable; yellow: mean and width reliable; orange: mean reliable; red: no quantification possible).
The optimum temperatures with respect to sensitivity were found to be 30 K and 50 K, respectively (see SI). As RIDME is a single frequency technique, it can be performed with all pulses coinciding with the resonance frequency of the resonator and thus benefits in sensitivity compared to double frequency techniques, such as the 4-pulse DEER sequence where detection is generally performed off-resonance. This sensitivity gain in dependence of the cavity quality factor being adjusted to meet the required bandwidth could be quantified as approximately a factor 2 (see SI). Furthermore, the influence of instantaneous diffusion (that occurs when dephasing is induced by dipolarly coupled spins being inverted by detection pulses reducing the detected echo) was shown to be negligible in the I6R1/K28R1 construct at both 500 nM and 25 μM concentrations (see SI).
For the CuII-CuII RIDME data shown in figure 2, only the distance peak at ~2.5 nm was shown to be stable upon data validation. Additional measurements at 500 μM protein concentration suggested the distribution peaks above 2.5 nm were artefacts, insignificant in the 95% confidence interval (see SI). This indicated that measurements at 500 nM tetra-histidine protein concentration likely approached the lower concentration limit for CuII-CuII RIDME in our hands. It should be noted that the poor modulation depth (5.5%) is a result of the limiting affinity of CuII-NTA for the β-sheet double histidine motif.23 Pulse dipolar EPR methods allow precise determination of binding affinities from PELDOR60–62 and RIDME63 data. The observed modulation depth is consistent with predictions using binding affinities previously derived from CuII-nitroxide RIDME pseudo-titration23 and extrapolated ITC data (see SI). Conversely, for the nitroxide-nitroxide PELDOR data the bimodal distribution shown in figure 2 was recapitulated in additional measurements at 25 μM I6R1/K28R1 protein concentration (see SI). This suggested that measurements at 500 nM protein concentration were not yet testing the lower concentration limit for nitroxide-nitroxide PELDOR. To test this hypothesis, nitroxide-nitroxide PELDOR was also measured at 100 nM protein (200 nM spin) concentration (figure 3).
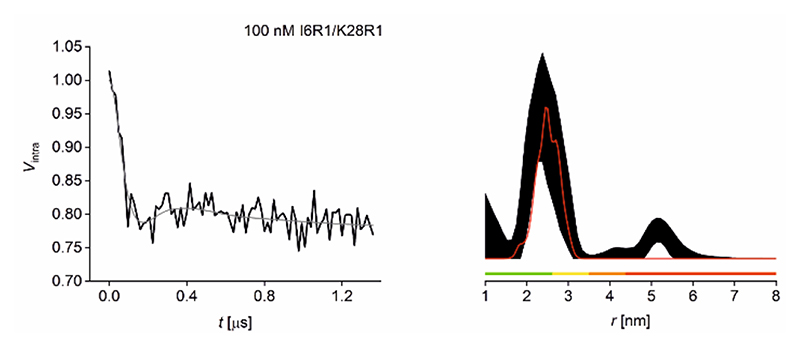
Figure 3.
100 nM GB1 Q-band PELDOR data at 50K. Left: Background-corrected PELDOR data (black) and fit (grey) for 100 nM I6R1/K28R1 GB1. Right: Corresponding distance distribution given as 95% confidence intervals (± 2σ) with 50% noise added for error estimation during statistical analysis; simulated distance distributions are shown in red. Color bars represent reliability ranges (green: shape reliable; yellow: mean and width reliable; orange: mean reliable; red: no quantification possible).
The nitroxide-nitroxide PELDOR data shown in figure 3, measured with the same dipolar evolution time for comparison, indicate that at 100 nM the retrieved experimental distribution is no longer bimodal, however the mean distance is still retrieved as the only significant peak following data validation. Nevertheless, the relatively poor signal-to-noise ratio mandates a regularization parameter that does not allow resolving both distance populations (SI). This loss in resolution has been confirmed using other processing approaches (see SI for details). This experiment thus highlights the dependence of distance resolution on achievable signal-to-noise and thus, on spin concentration. Together, this suggests that 100 nM approaches the minimum concentration achievable for reliable determination of the mean distance from nitroxide-nitroxide PELDOR under our conditions; it is already beyond a reliable determination of the distance distribution shape. Sensitivity analysis shows that measurement of I6R1/K28R1 at 100 nM is a factor ~15 noisier than measurement at 500 nM, rather than the factor 5 expected from the concentration difference. The additional factor 3 can be considered a penalty for the challenging measurement optimization at these very low concentrations (see SI).
Comparing the relative sensitivities of nitroxide-nitroxide PELDOR and CuII-CuII RIDME reveals the former to be approximately 10-fold more sensitive (see SI). The 3 main factors contributing to the relative sensitivity are echo amplitude (i.e., signal-to-noise), modulation depth, and averaging rate. For CuII-CuII RIDME the smaller signal is largely compensated by the faster averaging. However, the modulation depth is a limiting factor for sensitivity. This is to be expected for a non-covalent spin-label with dissociation constants in the high nM (α-helix) and low μM (β-sheet) regime. Overcoming the low modulation depth will make sensitivity of CuII-CuII RIDME competitive. Simulation of a tetra-histidine construct containing a pair of α-helical binding sites reveals modulation depths > 12% for CuII-CuII RIDME under otherwise identical conditions (see SI). Possible strategies to further improve modulation depths include insertion of an artificial amino-acid bearing a covalent CuII centre to overcome the limiting equilibrium constants,64 or measuring nitroxide detected CuII-nitroxide RIDME in excess CuII chelate spin label. This will shift the binding equilibrium into saturation of the binding site and achieve modulation depths approaching 50%, independent of protein concentration.
Comparing nitroxide-nitroxide PELDOR with available CuII-nitroxide RIDME data23 suggests the latter to be an additional factor ~1.5 more sensitive potentially allowing measurements even below 100 nM protein concentration (see SI). Additionally, RIDME measurements may be less prone to the optimization penalty found for 4-pulse DEER at 100 nM as the single-frequency method will need fewer parameters to be set. These findings showcase that in favorable circumstances superb concentration sensitivities are achievable using commercial instrumentation and spin labels. In the case of the widely applied nitroxide-nitroxide 4-pulse DEER experiment, concentration sensitivities orders of magnitude greater than routinely applied (≥10 μM) are possible using rectangular pulses at Q-band frequencies. Additionally, CuII-CuII RIDME measurements showcase that systems not amenable to conventional thiol-based covalent spin labelling are also accessible in the sub-μM concentration regime, when used in conjunction with double-histidine motifs. Concentrations realized here for CuII-CuII RIDME are more than an order of magnitude below published applications of the 5-pulse RIDME experiment on metal-metal spin systems (≥ 25 μM; see SI).
Nevertheless, at these low concentrations long distances or more complex distance distributions will be a challenge for both, nitroxide-nitroxide DEER and CuII-CuII RIDME experiments. The presence of conformational flexibility or two (or more) conformational states will result in broad or multimodal distance distributions, respectively. The superposition of frequencies will lead to less pronounced oscillations and a larger ambiguity in the distance analysis.65 While the distance distribution mean and width were recoverable from the 100 nM GB1 sample the bimodal distribution was only resolved in the 500 nM sample. Long distances and highly resolved distance distributions require longer dipolar evolution times. In these cases, sensitivity enhancement is achievable by full deuteration of protein and matrix due to decreased echo dephasing (increased phase memory time T m).66–67 The benefit that can be realised from deuteration will strongly depend on the required trace lengths.
Conclusions
Benchmarking the nanomolar sensitivity is truly promising as a pathway to novel applications and may facilitate study of systems previously thought to be beyond the scope of pulse EPR spectroscopy. Perhaps most importantly, our results emphasize that commercial instrumentation and standard labelling protocols already yield sufficient concentration sensitivity for applications in the sub-μM regime. Indeed, a very recent study published after our initial report68 has demonstrated DEER distance measurements in cell for GdIII at ~350 nM protein concentrations and in vitro for nitroxides down to 120 nM protein / 200 nM spin concentration.69 The imminent adoption of cryogenically cooled preamplifiers for EPR spectroscopy70 heralds a further order of magnitude of sensitivity improvement. Embracing the opportunity to measure at concentrations two to three orders of magnitude below past practice will bring new science into reach that is currently sample limited in either concentration or absolute amount.
Open data statement
The research data supporting this publication can be accessed at https://doi.org/10.17630/6ca0de75-904d-4768-8a21-2c235a717412.71
Supplementary Material
Acknowledgements
We thank Dr Angeliki Giannoulis for insightful discussions. This research was funded, in whole or in part, by the Wellcome Trust (099149/Z/12/Z and 204821/Z/16/Z). A CC BY or equivalent license is applied to the Author Accepted Manuscript arising from this submission, in accordance with the grant’s open access conditions. BEB and KA acknowledge support by the Leverhulme Trust (RPG-2018–397). JLW is supported by the BBSRC DTP Eastbio. BEB acknowledges equipment funding by BBSRC (BB/R013780/1).
Footnotes
Notes
The authors declare no competing financial interests.
References
- (1).Constantinescu-Aruxandei D, Petrovic-Stojanovska B, Schiemann O, Naismith JH, White MF. Taking a Molecular Motor for a Spin: Helicase Mechanism Studied by Spin Labeling and PELDOR. Nucleic Acids Res. 2016;44:954–968. doi: 10.1093/nar/gkv1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FHT. Structural Basis of the Non-Coding RNA RsmZ Acting as a Protein Sponge. Nature. 2014;509:588–592. doi: 10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- (3).Ghosh S, Lawless MJ, Brubaker HJ, Singewald K, Kurpiewski MR, Jen-Jacobson L, Saxena S. Cu2+-Based Distance Measurements by Pulsed EPR Provide Distance Constraints for DNA Backbone Conformations in Solution. Nucleic Acids Res. 2020;48:e49-e49. doi: 10.1093/nar/gkaa133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hagelueken G, Ingledew WJ, Huang H, Petrovic-Stojanovska B, Whitfield C, ElMkami H, Schiemann O, Naismith JH. PELDOR Spectroscopy Distance Fingerprinting of the Octameric Outer-Membrane Protein Wza from Escherichia coli . Angew Chem Int Ed. 2009;48:2904–2906. doi: 10.1002/anie.200805758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Joseph B, Sikora A, Bordignon E, Jeschke G, Cafiso DS, Prisner TF. Distance Measurement on an Endogenous Membrane Transporter in E. coli Cells and Native Membranes Using EPR Spectroscopy. Angew Chem Int Ed. 2015;54:6196–6199. doi: 10.1002/anie.201501086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kapsalis C, Wang B, El Mkami H, Pitt SJ, Schnell JR, Smith TK, Lippiat JD, Bode BE, Pliotas C. Allosteric Activation of an Ion Channel Triggered by Modification of Mechanosensitive Nano-Pockets. Nat Commun. 2019;10:4619–4619. doi: 10.1038/s41467-019-12591-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sameach H, Ghosh S, Gevorkyan-Airapetov L, Saxena S, Ruthstein S. EPR Spectroscopy Detects Various Active State Conformations of the Transcriptional Regulator CueR. Angew Chem Int Ed. 2019;58:3053–3056. doi: 10.1002/anie.201810656. [DOI] [PubMed] [Google Scholar]
- (8).Schmidt T, Wälti MA, Baber JL, Hustedt EJ, Clore GM. Long Distance Measurements up to 160 Å in the GroEL Tetradecamer Using Q-Band DEER EPR Spectroscopy. Angew Chem Int Ed. 2016;55:15905–15909. doi: 10.1002/anie.201609617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Verhalen B, Dastvan R, Thangapandian S, Peskova Y, Koteiche HA, Nakamoto RK, Tajkhorshid E, McHaourab HS. Energy Transduction and Alternating Access of the Mammalian ABC Transporter P-Glycoprotein. Nature. 2017;543:738–741. doi: 10.1038/nature21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Azarkh M, Bieber A, Qi M, Fischer JWA, Yulikov M, Godt A, Drescher M. Gd(III)-Gd(III) Relaxation-Induced Dipolar Modulation Enhancement for In-Cell Electron Paramagnetic Resonance Distance Determination. J Phys Chem Lett. 2019;10:1477–1481. doi: 10.1021/acs.jpclett.9b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Igarashi R, Sakai T, Hara H, Tenno T, Tanaka T, Tochio H, Shirakawa M. Distance Determination in Proteins inside Xenopus laevis Oocytes by Double Electron-Electron Resonance Experiments. J Am Chem Soc. 2010;132:8228–8229. doi: 10.1021/ja906104e. [DOI] [PubMed] [Google Scholar]
- (12).Jassoy JJ, Berndhäuser A, Duthie F, Kühn SP, Hagelueken G, Schiemann O. Versatile Trityl Spin Labels for Nanometer Distance Measurements on Biomolecules In Vitro and within Cells. Angew Chem Int Ed. 2017;56:177–181. doi: 10.1002/anie.201609085. [DOI] [PubMed] [Google Scholar]
- (13).Joseph B, Sikora A, Cafiso DS. Ligand Induced Conformational Changes of a Membrane Transporter in E. coli Cells Observed with DEER/PELDOR. J Am Chem Soc. 2016;138:1844–1847. doi: 10.1021/jacs.5b13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Krstić I, Hänsel R, Romainczyk O, Engels JW, Dötsch V, Prisner TF. Long-Range Distance Measurements on Nucleic Acids in Cells by Pulsed EPR Spectroscopy. Angew Chem Int Ed. 2011;50:5070–5074. doi: 10.1002/anie.201100886. [DOI] [PubMed] [Google Scholar]
- (15).Theillet F-X, Binolfi A, Bekei B, Martorana A, Rose HM, Stuiver M, Verzini S, Lorenz D, van Rossum M, Goldfarb D, et al. Structural Disorder of Monomeric α-Synuclein Persists in Mammalian Cells. Nature. 2016;530:45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- (16).Yang Y, Chen S-N, Yang F, Li X-Y, Feintuch A, Su X-C, Goldfarb D. In-Cell Destabilization of a Homodimeric Protein Complex Detected by DEER Spectroscopy. Proc Natl Acad Sci U S A. 2020;117:20566–20575. doi: 10.1073/pnas.2005779117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Milov AD, Ponomarev AB, Tsvetkov YD. Electron-Electron Double Resonance in Electron Spin Echo: Model Biradical Systems and the Sensitized Photolysis of Decalin. Chem Phys Lett. 1984;110:67–72. doi: 10.1016/0009-2614(84)80148-7. [DOI] [Google Scholar]
- (18).Milov AD, Salikov KM, Shirov MD. Application of ELDOR in Electron-Spin Echo for Paramagnetic Center Space Distribution in Solids. Fiz Tverd Tela. 1981;23:975–982. [Google Scholar]
- (19).Martin RE, Pannier M, Diederich F, Gramlich V, Hubrich M, Spiess HW. Determination of End-to-End Distances in a Series of TEMPO Diradicals of up to 2.8 nm Length with a New Four-Pulse Double Electron Electron Resonance Experiment. Angew Chem Int Ed. 1998;37:2833–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20. <2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- (20).Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-Time Free Measurement of Dipole-Dipole Interactions between Spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- (21).Kulik LV, Dzuba SA, Grigoryev IA, Tsvetkov YD. Electron Dipole–Dipole Interaction in ESEEM of Nitroxide Biradicals. Chem Phys Lett. 2001;343:315–324. doi: 10.1016/S0009-2614(01)00721-7. [DOI] [Google Scholar]
- (22).Milikisyants S, Scarpelli F, Finiguerra MG, Ubbink M, Huber M. A Pulsed EPR Method to Determine Distances Between Paramagnetic Centers with Strong Spectral Anisotropy and Radicals: The Dead-time Free RIDME Sequence. J Magn Reson. 2009;201:48–56. doi: 10.1016/i.imr.2009.08.008. [DOI] [PubMed] [Google Scholar]
- (23).Wort JL, Ackermann K, Giannoulis A, Stewart AJ, Norman DG, Bode BE. Sub-Micromolar Pulse Dipolar EPR Spectroscopy Reveals Increasing CuII-labelling of Double-Histidine Motifs with Lower Temperature. Angew Chem Int Ed. 2019;58:11681–11685. doi: 10.1002/anie.201904848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Cunningham TF, Putterman MR, Desai A, Horne WS, Saxena S. The Double-Histidine Cu2+-Binding Motif: a Highly Rigid, Site-Specific Spin Probe for Electron Spin Resonance Distance Measurements. Angew Chem Int Ed. 2015;54:6330–6334. doi: 10.1002/anie.201501968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ghosh S, Lawless MJ, Rule GS, Saxena S. The Cu2+-Nitrilotriacetic Acid Complex Improves Loading of α-Helical Double Histidine Site for Precise Distance Measurements by Pulsed ESR. J Magn Reson. 2018;286:163–171. doi: 10.1016/Jjmr.2017.12.005. [DOI] [PubMed] [Google Scholar]
- (26).Wort JL, Arya S, Ackermann K, Stewart AJ, Bode BE. Pulse Dipolar EPR Reveals Double-Histidine Motif CuII–NTA Spin-Labeling Robustness against Competitor Ions. J Phys Chem Lett. 2021;12:2815–2819. doi: 10.1021/acs.ipclett.1c00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Berliner LJ, Grunwald J, Hankovszky HO, Hideg K. A Novel Reversible Thiol Specific Spin Label: Papain Active Site Labeling and Inhibition. Anal Biochem. 1982;119:450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- (28).Hubbell WL, Altenbach C. Investigation of Structure and Dynamics in Membrane Proteins Using Site-Directed Spin Labeling. Current Opinion in Structural Biology. 1994;4:566–573. doi: 10.1016/S0959-440-X(94)90219-4. [DOI] [Google Scholar]
- (29).Babaylova ES, Malygin AA, Lomzov AA, Pyshnyi DV, Yulikov M, Jeschke G, Krumkacheva OA, Fedin MV, Karpova GG, Bagryanskaya EG. Complementary-Addressed Site-Directed Spin Labeling of Long Natural RNAs. Nucleic Acids Res. 2016;44:7935–7943. doi: 10.1093/nar/gkw516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cruickshank PAS, Bolton DR, Robertson DA, Hunter RI, Wylde RJ, Smith GM. A Kilowatt Pulsed 94 GHz Electron Paramagnetic Resonance Spectrometer with High Concentration Sensitivity, High Instantaneous Bandwidth, and Low Dead Time. Rev Sci Instrum. 2009;80:103102. doi: 10.1063/1.3239402. [DOI] [PubMed] [Google Scholar]
- (31).Hofbauer W, Earle KA, Dunnam CR, Moscicki JK, Freed JH. High-Power 95 GHz Pulsed Electron Spin Resonance Spectrometer. Rev Sci Instrum. 2004;75:1194–1208. doi: 10.1063/E1710700. [DOI] [Google Scholar]
- (32).Breitgoff FD, Keller K, Qi M, Klose D, Yulikov M, Godt A, Jeschke G. UWB DEER and RIDME Distance Measurements in Cu(II)-Cu(II) Spin Pairs. J Magn Reson. 2019;308:106560. doi: 10.1016/i.jmr.2019.07.047. [DOI] [PubMed] [Google Scholar]
- (33).Doll A, Pribitzer S, Tschaggelar R, Jeschke G. Adiabatic and Fast Passage Ultra-Wideband Inversion in Pulsed EPR. J Magn Reson. 2013;230:27–39. doi: 10.1016/jjmr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- (34).Motion CL, Cassidy SL, Cruickshank PAS, Hunter RI, Bolton DR, El Mkami H, Van Doorslaer S, Lovett JE, Smith GM. The use of Composite Pulses for Improving DEER Signal at 94GHz. J Magn Reson. 2017;278:122–133. doi: 10.1016/jjmr.2017.03.018. [DOI] [PubMed] [Google Scholar]
- (35).Spindler PE, Glaser SJ, Skinner TE, Prisner TF. Broadband Inversion PELDOR Spectroscopy with Partially Adiabatic Shaped Pulses. Angew Chem Int Ed. 2013;52:3425–3429. doi: 10.1002/anie.201207777. [DOI] [PubMed] [Google Scholar]
- (36).Spindler PE, Zhang Y, Endeward B, Gershernzon N, Skinner TE, Glaser SJ, Prisner TF. Shaped Optimal Control Pulses for Increased Excitation Bandwidth in EPR. J Magn Reson. 2012;218:49–58. doi: 10.1016/jjmr.2012.02.013. [DOI] [PubMed] [Google Scholar]
- (37).Bahrenberg T, Yang Y, Goldfarb D, Feintuch A. rDEER: A Modified DEER Sequence for Distance Measurements Using Shaped Pulses. Magnetochemistry. 2019;5 doi: 10.3390/magnetochemistry5010020. [DOI] [Google Scholar]
- (38).Borbat PP, Georgieva ER, Freed JH. Improved Sensitivity for Long-Distance Measurements in Biomolecules: Five-Pulse Double Electron–Electron Resonance. J Phys Chem Lett. 2013;4:170–175. doi: 10.1021/jz301788n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Doll A, Qi M, Pribitzer S, Wili N, Yulikov M, Godt A, Jeschke G. Sensitivity Enhancement by Population Transfer in Gd(III) Spin Labels. Phys Chem Chem Phys. 2015;17:7334–7344. doi: 10.1039/C4CP05893C. [DOI] [PubMed] [Google Scholar]
- (40).Spindler PE, Waclawska I, Endeward B, Plackmeyer J, Ziegler C, Prisner TF. Carr–Purcell Pulsed Electron Double Resonance with Shaped Inversion Pulses. J Phys Chem Lett. 2015;6:4331–4335. doi: 10.1021/acs.ipclett.5b01933. [DOI] [PubMed] [Google Scholar]
- (41).Fleck N, Heubach C, Hett T, Spicher S, Grimme S, Schiemann O. Ox-SLIM: Synthesis of and Site-Specific Labelling with a Highly Hydrophilic Trityl Spin Label. Chem Eur J. 2021;27:5292–5297. doi: 10.1002/Chem202100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Reginsson GW, Kunjir NC, Sigurdsson ST, Schiemann O. Trityl Radicals: Spin Labels for Nanometer-Distance Measurements. Chem Eur J. 2012;18:13580–13584. doi: 10.1002/Chem201203014. [DOI] [PubMed] [Google Scholar]
- (43).Yang Z, Liu Y, Borbat P, Zweier JL, Freed JH, Hubbell WL. Pulsed ESR Dipolar Spectroscopy for Distance Measurements in Immobilized Spin Labeled Proteins in Liquid Solution. J Am Chem Soc. 2012;134:9950–9952. doi: 10.1021/ia303791p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Borbat PP, Freed JH. Multiple-Quantum ESR and Distance Measurements. Chem Phys Lett. 1999;313:145–154. doi: 10.1016/S0009-2614(99)00972-0. [DOI] [Google Scholar]
- (45).Ackermann K, Pliotas C, Valera S, Naismith JH, Bode BE. Sparse Labeling PELDOR Spectroscopy on Multimeric Mechanosensitive Membrane Channels. BioPhys J. 2017;113:1968–1978. doi: 10.1016/JbpJ2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Larsen RG, Singel DJ. Double Electron-Electron Resonance Spin-Echo Modulation - Spectroscopic Measurement of Electron-Spin Pair Separations in Orientationally Disordered Solids. J Chem Phys. 1993;98:5134–5146. doi: 10.1063/1.464916. [DOI] [Google Scholar]
- (47).Kerry PS, Turkington HL, Ackermann K, Jameison SA, Bode BE. Analysis of Influenza A Virus NS1 Dimer Interfaces in Solution by Pulse EPR Distance Measurements. J Phys Chem B. 2014;118:10882–8. doi: 10.1021/jp508386r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006 - a Comprehensive Software Package for Analyzing Pulsed ELDOR Data. Appl Magn Reson. 2006;30:473–498. doi: 10.1007/BF03166213. [DOI] [Google Scholar]
- (49).Chiang YW, Borbat PP, Freed JH. The Determination of Pair Distance Distributions by Pulsed ESR using Tikhonov Regularization. J Magn Reson. 2005;172:279–95. doi: 10.1016/Jjmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- (50).Ghosh S, Saxena S, Jeschke G. Rotamer modelling of Cu(II) spin labels based on the double-histidine motif. Appl Magn Reson. 2018;49:1281–1298. doi: 10.1007/s00723-0181052-8. [DOI] [Google Scholar]
- (51).Polyhach Y, Bordignon E, Jeschke G. Rotamer Libraries of Spin Labelled Cysteines for Protein Studies. Phys Chem Chem Phys. 2011;13:2356–2366. doi: 10.1039/C0CP01865A. [DOI] [PubMed] [Google Scholar]
- (52).Singewald K, Bogetti X, Sinha K, Rule GS, Saxena S. Double Histidine Based EPR Measurements at Physiological Temperatures Permit Site-Specific Elucidation of Hidden Dynamics in Enzymes. Angew Chem Int Ed. 2020;59:23040–23044. doi: 10.1002/anie.202009982. [DOI] [PubMed] [Google Scholar]
- (53).The PyMOL Molecular Graphics System, Version 1.2r3pre. SchrÖdinger, LLC; [Google Scholar]
- (54).Hagelueken G, Ward R, Naismith JH, Schiemann O. MtsslWizard: In Silico Spin Labeling and Generation of Distance Distributions in PyMOL. Appl Magn Reson. 2012;42:377–391. doi: 10.1007/s00723-012-0314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Gamble Jarvi A, Cunningham TF, Saxena S. Efficient Localization of a Native Metal Ion within a Protein by Cu2+-Based EPR Distance Measurements. Phys Chem Chem Phys. 2019;21:10238–10243. doi: 10.1039/C8CP07143H. [DOI] [PubMed] [Google Scholar]
- (56).Gamble Jarvi A, Ranguelova K, Ghosh S, Weber RT, Saxena S. On the Use of Q-Band Double Electron–Electron Resonance To Resolve the Relative Orientations of Two Double Histidine-Bound Cu2+Ions in a Protein. J Phys Chem B. 2018;122:10669–10677. doi: 10.1021/acs.jpcb.8b07727. [DOI] [PubMed] [Google Scholar]
- (57).Giannoulis A, Yang Y, Gong Y-J, Tan X, Feintuch A, Carmieli R, Bahrenberg T, Liu Y, Su X-C, Goldfarb D. DEER Distance Measurements on Trityl/Trityl and Gd(III)/Trityl Labelled Proteins. Phys Chem Chem Phys. 2019;21:10217–10227. doi: 10.1039/C8CP07249C. [DOI] [PubMed] [Google Scholar]
- (58).Wort JL, Ackermann K, Norman DG, Bode BE. A General Model to Optimise CuIILabelling Efficiency of Double-Histidine Motifs for Pulse Dipolar EPR Applications. Phys Chem Chem Phys. 2021;23:3810–3819. doi: 10.1039/D0CP06196D. [DOI] [PubMed] [Google Scholar]
- (59).Polyhach Y, Bordignon E, Tschaggelar R, Gandra S, Godt A, Jeschke G. High Sensitivity and Versatility of the DEER Experiment on Nitroxide Radical Pairs at Q-Band Frequencies. Phys Chem Chem Phys. 2012;14:10762–10773. doi: 10.1039/C2CP41520H. [DOI] [PubMed] [Google Scholar]
- (60).Ackermann K, Giannoulis A, Cordes DB, Slawin AMZ, Bode BE. Assessing Dimerisation Degree and Cooperativity in a Biomimetic Small-Molecule Model by Pulsed EPR. Chem Commun. 2015;51:5257–5260. doi: 10.1039/C4CC08656B. [DOI] [PubMed] [Google Scholar]
- (61).Bode BE, Margraf D, Plackmeyer J, Dürner G, Prisner TF, Schiemann O. Counting the Monomers in Nanometer-Sized Oligomers by Pulsed Electron-Electron Double Resonance. J Am Chem Soc. 2007;129:6736–6745. doi: 10.1021/ja065787t. [DOI] [PubMed] [Google Scholar]
- (62).Giannoulis A, Ackermann K, Spindler PE, Higgins C, Cordes DB, Slawin AMZ, Prisner TF, Bode BE. Nitroxide–Nitroxide and Nitroxide–Metal Distance Measurements in Transition Metal Complexes with Two or Three Paramagnetic Centres Give Access to Thermodynamic and Kinetic Stabilities. Phys Chem Chem Phys. 2018;20:11196–11205. doi: 10.1039/C8CP01611A. [DOI] [PubMed] [Google Scholar]
- (63).Giannoulis A, Oranges M, Bode BE. Monitoring Complex Formation by Relaxation-Induced Pulse Electron Paramagnetic Resonance Distance Measurements. ChemPhysChem. 2017;18:2318–2321. doi: 10.1002/cphc.201700666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Merz GE, Borbat PP, Muok AR, Srivastava M, Bunck DN, Freed JH, Crane BR. Site-Specific Incorporation of a Cu2+Spin Label into Proteins for Measuring Distances by Pulsed Dipolar Electron Spin Resonance Spectroscopy. J Phys Chem B. 2018;122:9443–9451. doi: 10.1021/acs.ipcb.8b05619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Jeschke G. Interpretation of Dipolar EPR Data in Terms of Protein Structure. In: Timmel CR, Harmer JR, editors. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013. pp. 83–120. [DOI] [Google Scholar]
- (66).Jeschke G. DEER Distance Measurements on Proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- (67).Ward R, Bowman A, Sozudogru E, El-Mkami H, Owen-Hughes T, Norman DG. EPR Distance Measurements in Deuterated Proteins. J Magn Reson. 2010;207:164–167. doi: 10.1016/Jjmr.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Ackermann K, Wort JL, Bode BE. Nanomolar Pulse Dipolar EPR Spectroscopy in Proteins; the Copper(II)-Copper(II) and Nitroxide-Nitroxide Cases. ChemRxiv Preprint. 2020 doi: 10.26434/chemrxiv.13370924.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Kucher S, Elsner C, Safonova M, Maffini S, Bordignon E. In-Cell Double Electron–Electron Resonance at Nanomolar Protein Concentrations. J Phys Chem Lett. 2021;12:3679–3684. doi: 10.1021/acs.jpclett.1c00048. [DOI] [PubMed] [Google Scholar]
- (70).Šimėnas M, O’Sullivan J, Zollitsch CW, Kennedy O, Seif-Eddine M, Ritsch I, Hülsmann M, Qi M, Godt A, Roessler MM, et al. A Sensitivity Leap for X-Band EPR Using a Probehead with a Cryogenic Preamplifier. J Magn Reson. 2021;322:106876. doi: 10.1016/Jjmr.2020.106876. [DOI] [PubMed] [Google Scholar]
- (71).Ackermann K, Wort JL, Bode BE. Nanomolar Pulse Dipolar EPR Spectroscopy in Proteins; the CuII-CuIIand Nitroxide-Nitroxide Cases (Dataset) University of St Andrews Research Portal. 2021 doi: 10.17630/6ca0de75-904d-4768-8a21-2c235a717412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The research data supporting this publication can be accessed at https://doi.org/10.17630/6ca0de75-904d-4768-8a21-2c235a717412.71