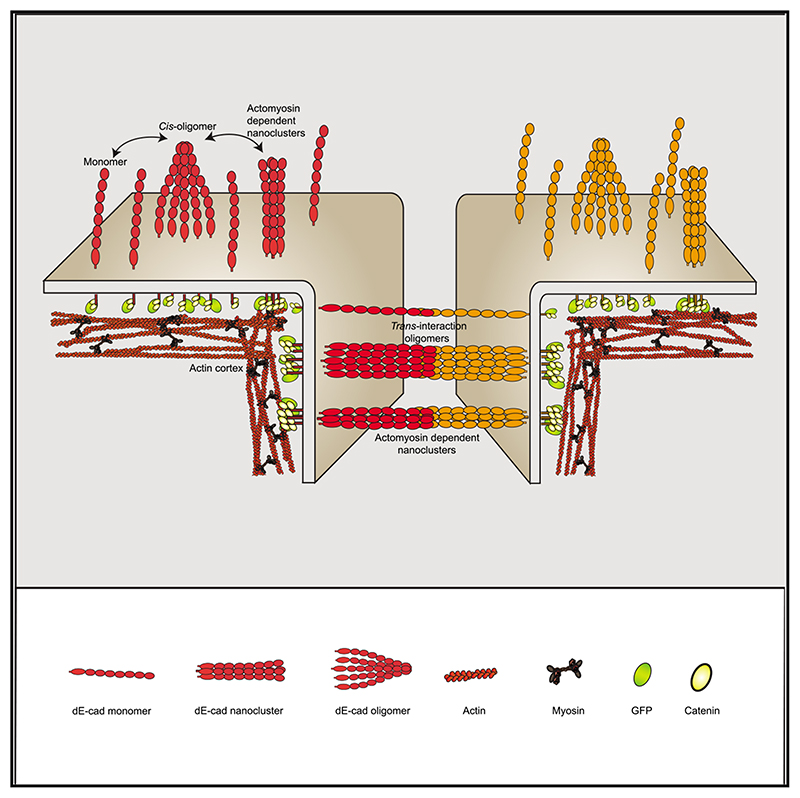
Summary
Cadherins are transmembrane adhesion proteins required for the formation of cohesive tissues.1–4 Intracellular interactions of E-cadherin with the Catenin family proteins, α- and β-catenin, facilitate connections with the cortical actomyosin network. This is necessary for maintaining the integrity of cell-cell adhesion in epithelial tissues.5–11 The supra-molecular architecture of E-cadherin is an important feature of its adhesion function; cis and trans interactions of E-cadherin are deployed12–15 to form clusters, both in cis and trans.11,16–21 Studies in Drosophila embryo have also shown that Drosophila E-cadherin (dE-cad) is organized as finitesized dynamic clusters that localize with actin patches at cell-cell junctions, in continuous exchange with the extra-junctional pool of dE-cad surrounding the clusters.11,19 Here, we use the ectopic expression of dE-cad in larval hemocytes, which lack endogenous dE-cad to recapitulate functional cell-cell junctions in a convenient model system. We find that, while dE-cad at cell-cell junctions in hemocytes exhibits a clustered trans-paired organization similar to that reported previously in embryonic epithelial tissue, extra-junctional dE-cad is also organized as relatively immobile nanoclusters as well as more loosely packed diffusive oligomers. Oligomers are promoted by cis interactions of the ectodomain, and their growth is counteracted by the activity of cortical actomyosin. Oligomers in turn promote assembly of dense nanoclusters that require cortical actomyosin activity. Thus, cortical actin activity remodels oligomers and generates nanoclusters. The requirement for dynamic actin in the organization of dE-cad at the nanoscale may provide a mechanism to dynamically tune junctional strength.
Graphical abstract
Results
Using hemocytes to explore functional organization of dE-cad junctions
The expression of dE-cad::GFP at the endogenous locus (endo-dE-cad::GFP)22 has allowed visualization of dE-cad in the apical regions of embryonic epithelial cells (Figure 1A, top panel).11 The topology of the embryonic tissue, low expression of the protein, and tissue collapse upon perturbation of the adhesion complex limits the exploration of the mechanism behind the organization and function of the dE-cad adhesion complex in a physiological context.2,4,23 To study the organization of dE-cad and its regulation by actomyosin machinery, we ectopically expressed dE-cad in hemocytes derived from wandering third-instar larvae, using upstream activating sequence (UAS)-GAL4 system (see STAR methods). Immunostaining of dE-cad-expressing cells revealed that dE-cad accumulates at the cell-cell junction, mimicking observations in the embryonic epithelium (Figures 1A, 1B, bottom panel, S1A, and S1B). Ectopic expression of dE-cad in hemocytes produces cell aggregates consistent with the presence of functional dE-cadherin on the membrane (Figure 1C). Similar to dE-cad in epithelial cells, dE-cad localization at hemocyte cell surface and its ability to create stable trans homophilic interactions is dependent on Ca+2 (Figures S1C and S1D).24,25
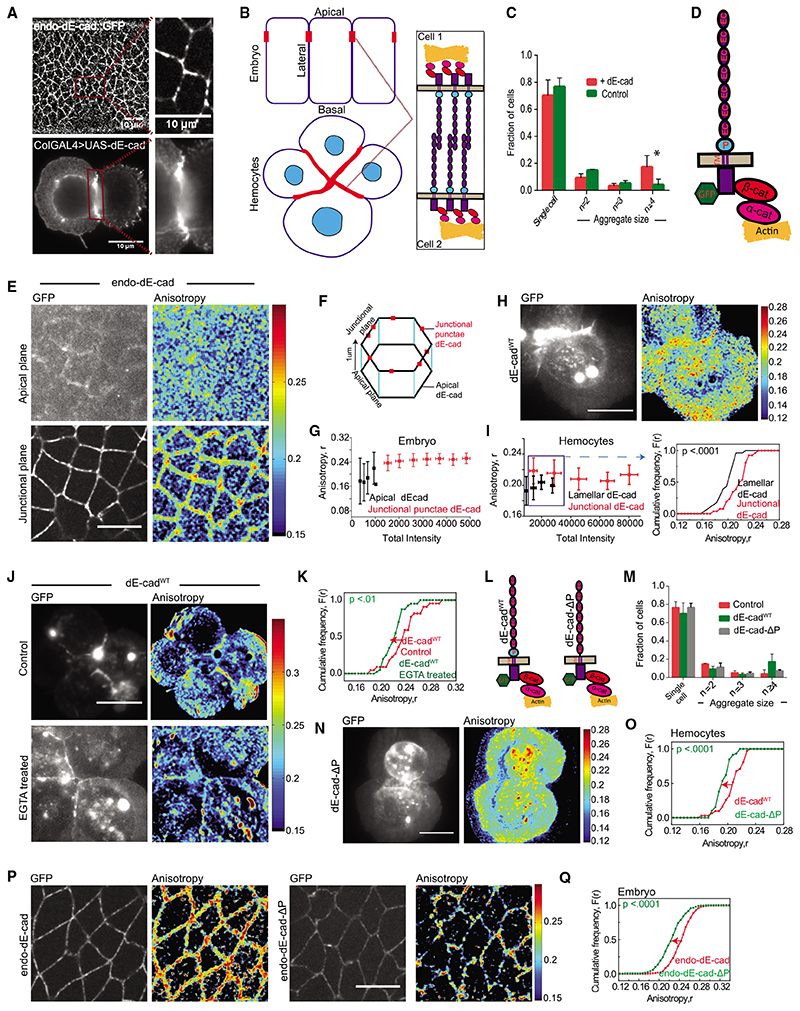
Figure 1. Nanoscale organization of dE-cad in embryo and larval hemocytes probed by homo-FRET microscopy.
(A) Confocal images of dE-cad::GFP-expressing epithelial cells in an early developing embryo (left top) and dE-cad labeled using monoclonal DCAD2 in hemocytes from wandering third-instar larvae from dE-cadWT-expressing flies (left bottom); insets show magnified view of dE-cad in embryos (right top) and hemocytes (right bottom).
(B) Schematic showing Drosophila embryo epithelial cells and larval hemocytes forming cell-cell junctions with a zoomed view of the trans interactions of dE-cad at the junctions.
(C) Histogram showing the percentage of cells as single cells and cell aggregates in control and dE-cadWT::GFP-expressing hemocytes. Dataset contains normalized cell number from 5 control larvae and 6 dE-cadWT larvae, and the bar represents the mean and the standard deviation in each condition.
(D) Schematic showing Drosophila E-cadherin and its interaction machinery.
(E) Total intensity and EA images of endo-dE-cadWT::GFP in the apical and junctional plane in the early gastrulation stage of the embryo.
(F) Representation of epithelial cells of the embryo showing the confocal planes at which apical and junctional EA measurements are taken.
(G) Intensity versus EA plots of endo-dE-cad in the apical and junctional plane from similar stage embryos (n = 5).
(H and I) Total intensity and anisotropy images (H) of dE-cadWT::GFP-expressing hemocytes from wandering third-instar Drosophila larvae, scatterplot (I; left graph) of intensity versus EA values and cumulative frequency distribution of EA values (I; right graph) of lamellar, and junctional dE-cadWT of similar intensity range in the scatterplot (boxed area). Smooth lines indicate data obtained from the lamellar localized proteins, whereas line with square symbols indicates junctional localization.
(J and K)Total intensity and EA images(J) and cumulative frequency distributions of EA values (K) of junctional dE-cadWT::GFP of similar intensity ranges in control and EGTA-treated conditions.
(L) Schematic of dE-cadWT::GFP and dE-cad-ΔP::GFP proteins.
(M) Histogram showing the percentage of cells as single cells and cell aggregates in control and dE-cadWT::GFP- and dE-cad-ΔP::GFP-expressing hemocytes. Dataset contains normalized cell number from 5 control larvae, 6 dE-cadWT larvae, and 4 dE-cad-DP larvae, and the bar represents the mean and the standard deviation in each condition.
(N and O) Total intensity and EA images (N) and cumulative frequency distributions (O) of junctional EA values of dE-cadWT- and dE-cad-DP-expressing hemocytes.
(P and Q) Total intensity and EA images (P) and cumulative frequency distributions of EA values (Q) of junctional dE-cad obtained from endo-dE-cad::GFP- and endo-dE-cad-ΔP::GFP-expressing embryos as indicated where 5 embryos each were analyzed for endo-dE-cad endo-dE-cad-DP-expressing animals. Error bars depict standard deviation.
p values are calculated using Mann-Whitney U test. ns (not significant), p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars, 10 μm. Sample size is provided in Table S1. See also Figure S1.
At cell-cell junctions in epithelial tissue, E-cadherin recruits endogenous adaptor proteins, α- and β-catenin (cartoon in Figure 1D).7,11 These proteins accumulate at cell-cell junctions in dE-cad-expressing hemocytes as well (Figure S1E). Immunostaining of actin (Figure S1F) and myosin heavy chain (Figure S1G) also showed an enrichment of actomyosin at cell-cell junctions of dE-cad-expressing cells compared to the control cells. Thus, analogous to its role in the embryonic epithelium, dE-cad interacts with and recruits adaptor proteins, α- and β-catenin, as well as associates with the cortical actomyosin network while supporting cell-cell aggregates. These observations are consistent with functionally active dE-cad expressed in hemocytes.
Fluorescence emission anisotropy tracks trans and cis interactions of dE-cad
We next studied the organization of endo-dE-cad::GFP at cell adherens junctions and in the free apical surface of embryonic epithelial cells. We imaged endo-dE-cad::GFP (Figure 1E) during gastrulation movements of early embryonic development using a spinning disc microscope equipped with the capacity to measure fluorescence intensity and emission anisotropy (EA) (see STAR methods).26 When a fluorophore is excited with plane-polarized light, fluorescence emission may be collected in parallel (I II) and perpendicular (I ⊥) orientations to the excitation polarization. EA is determined as an arithmetic operation [I II - I ⊥)/(I II + 2/⊥)] at every pixel. EA of a fluorophore measures the ability of a fluorophore to preserve its polarized excitation orientation. Therefore, higher (lower) EA indicates decreased (increased) rotational diffusion during the lifetime of a fluorophore that has been excited with plane-polarized excitation.
EA values of the endo-dE-cad::GFP at junctional punctae are higher compared to free apical membrane (Figures 1E-1G). To rule out any artifact of the influence of the location of the membrane protein dynamics, we ascertained that the EA of ectopically expressed GFP-tagged transmembrane protein vesicular stomatitis virus G (VSV-G) (used as control) is similar at the apical surface and at junctional plane of the epithelial cells (Figure S1H). We next imaged GFP-tagged dE-cad in hemocytes using EAbased total internal reflection fluorescence (TIRF) microscopy (see STAR methods).26,27 EA of dE-cadWT::GFP across a similar fluorescence intensity range at the junctions is also higher compared to that measured in lamellar regions corresponding to the extra-junctional apical surface in the embryo (Figures 1H and 1I). We also noted that EA for lamellar dE-cadWT is similar in isolated cells compared to clustered cells (Figure S11), indicating that single cells and cell clusters have similar lamellar EA, although the EA of dE-cadWT::GFP at the junction is distinct.
The higher EA of the ensemble of fluorescent molecules at the junction could be a consequence of extracellular trans interactions of the junctional protein in opposing membranes, resulting in restricting the rotational movement of the membrane protein. To directly test whether the EA of dE-cad::GFP is sensitive to changes in rotational mass, we added secondary antibodies to primary antibodies against the extracellular domain of dE-cad, in cell culture medium with cells expressing dE-cadWT::GFP. The expected increase in the rotational mass of dE-cad::GFP resulted in an increase in EAof dE-cadWT::GFPat both lamella and junction (Figures S1J and S1K). To test whether the higher EA values of dE-cad at the junctions is due to trans pairing of dEcad in the apposed membrane, we depleted extracellular calcium by inclusion of EGTA in the extracellular buffer to abrogate dE-cad-dE-cad trans interactions.24,25 EGTA treatment indeed decreases the EA of the junctional dE-cadWT compared to controls (Figures 1J and 1K). This provides evidence that increase in rotational mass of dE-cadWT::GFP, such as those engaged in trans pairing at the junction, is responsible for the observed increase in EA.
We next utilized a proximal domain deleted mutant of dE-cad (dE-cad-ΔP::GFP; schematic in Figure 1L) that is expected to have defects in homophilic interactions.28 Although dE-cad and dE-cad-DP are expressed at similar levels, dE-cad-DP-expressing hemocytes exhibited fewer cell aggregates as compared to those formed by dE-cadWT (Figure 1M), consistent with its defective capacity to support homophilic interactions. EA values of dE-cad-ΔP at the junction were also not as high compared to junctional dE-cadWT (Figures 1N and 1O). EA of junctional endo-dE-cad-ΔP was also much reduced compared to the endo-dE-cad::GFP expressed under the same conditions in the embryonic epithelium (Figures 1P and 1Q). These observations suggest that strong trans pairing results in restriction in rotational mobility of dE-cad consequently increasing the EA of dE-cad::GFP.
To assess whether dE-cad exhibits tight clustering at the junction at the nanoscale as suggested,19,21 we utilized the propensity of EA to be reduced by fluorescence resonance energy transfer (FRET) between like species (homo-FRET). The extent of reduction in EA is a measure of FRET efficiency between an ensemble of fluorescent molecules29 and hence of their physical proximity.30 This may be conveniently decoupled from effects on rotational motion of the sample, because the extent of homo-FRET may be assessed by photobleaching, which decreases the effective density of fluorescent protein and thereby results in an increase in EA values up to the value of the monomeric species (see schematic in Figure 2A).30 However, if proteins are dispersed as monomers on the membrane and/or are only rotationally constrained (e.g., by trans pairing), they will not exhibit a change in EA values upon photobleaching.
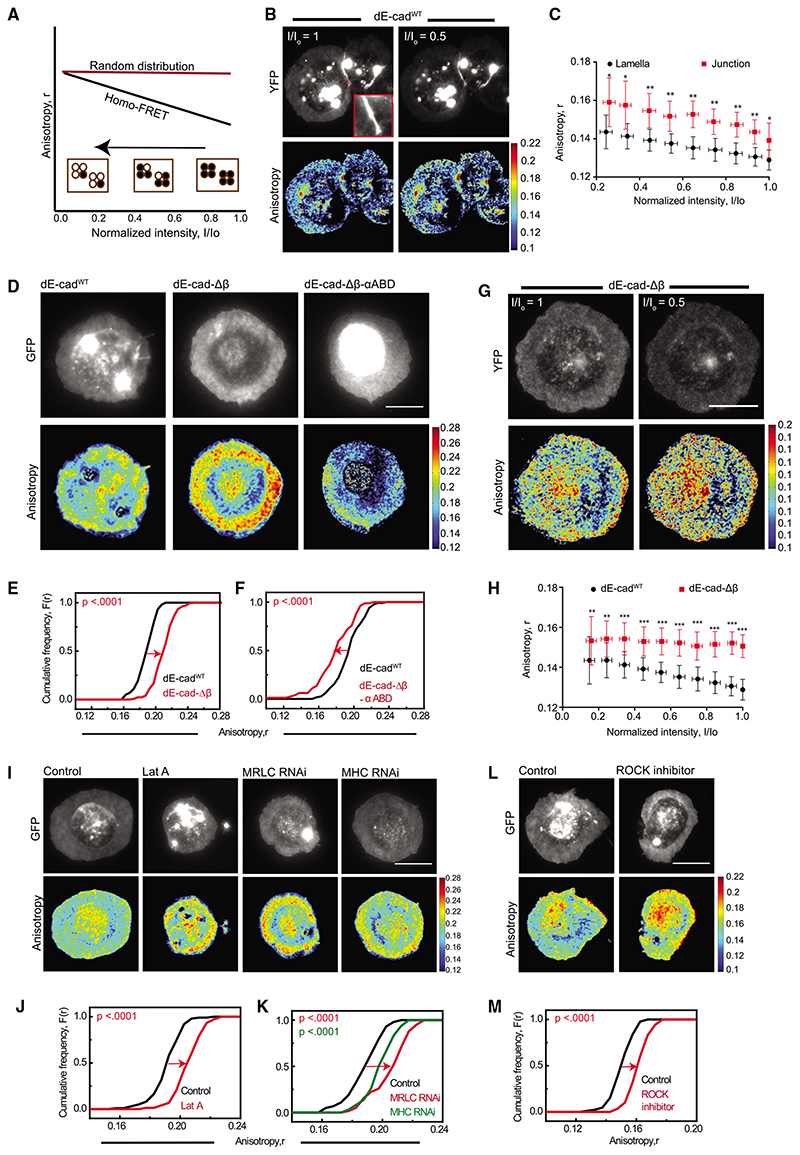
Figure 2. Adaptor complex and actomyosin activity organizes lamellar dE-cad.
(A) Schematic of the photobleaching assay shows the effect of photobleaching on EA where there are a fraction of nanoclusters and monomers initially present (black line) versus when there are no nanoclusters present (red line). A typical photobleaching profile of the EA versus normalized intensity of fluorescent molecules is shown where EA values of regions of interest (ROIs) are plotted against the intensity values, normalized to the starting total intensity value of ROI. Filled circles depict molecules that fluoresce and open circles bleached fluorophores.
(B) Total intensity and EA images of dE-cadWT::YFP-expressing cells at the beginning of photobleaching and at 50% photobleaching. Inset showed the zoomedin view of the junction marked in the cell.
(C) Intensity versus EA plots of lamellar and junctional dE-cad in dE-cadWT-expressing cells show changes in EA values upon photo bleaching. Each data point represents average EA values taken from multiple regions from cells(n = 9 [lamella] and 5 [junction])for the corresponding intensity bins normalized to the starting intensity value. Error bars represent standard deviations.
(D–F) Total intensity and EA images (D) of dE-cadWT::GFP-(left), dE-cad-Δβ::GFP-(middle), and dE-cad-Δβ-αABD::GFP (right)-expressing hemocytes and cumulative frequency distributions of lamellar EA values of dE-cadWT-compared to dE-cad-Δβ- (E) and to dE-cad-Δβ-αABD (F)-expressing hemocytes.
(G and H) Total intensity and EA images (G) of dE-cad-Δβ::YFP-expressing cells at the beginning of photobleaching and at 50% photobleaching and intensity versus EA plots (H) of dE-cadWT::YFP- (as shown in C, lamella above) and dE-cad-Δβ::YFP-expressing cells depicting changes in EA values upon photobleaching. Error bars represent standard deviations.
(I–K) Total intensity and EA images (I) of dE-cadWT::GFP in control, Lat-A-treated, and MRLC RNAi- and MHC RNAi-expressing cells and corresponding cumulative frequency distributions (J and K) of EA values of lamellar dE-cadWT::GFP.
(L and M) Total intensity and EA images (L) of dE-cadWT::GFP in control and ROCK-inhibitor-treated hemocytes and corresponding cumulative frequency distributions of EA values (M).
p values are calculated using Mann-Whitney U test. ns, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars, 10 μm. Sample size is provided in Table S1. See also Figure S2.
Photobleaching of dE-cadWT::yellow fluorescent protein (YFP) results in a linear increase in junctional EA (Figures 2B and 2C), consistent with a fraction of the dE-cadWT::YFP species engaging in homo-FRET at the junction. The linear nature of the increase in EA upon photobleaching for dE-cadWT::YFP at the junction is characteristic of the presence of a fraction of dispersed nanoclusters of dE-cadWT::YFP and not a uniform large-scale patch of densely packed molecules.30 These data are entirely consistent with the observation of a collection of nanoscale clusters as evidenced from stochastic optical reconstruction microscopy (STORM) studies in E-cad expressed in mammalian epithelia21 and from photoactivated localization microscopy (PALM) studies of the junctional dE-cad protein in Drosophila embryos.19
Surprisingly, we also observed a linear increase in EA of dE-cadWT::YFP localized to the lamella, indicating the presence of very close packed dE-cad (within FRET distances <6 nm for YFP) at the lamellar region in a fraction of molecules. Because the YFP moiety is located in the cytoplasmic domain of the proteins and are close enough to undergo homo-FRET,30 this implies that dE-cad engages in measurable cis interactions at the junctions and on the free surface. The organization of cadherin clusters has been proposed in Drosophila embryonic epithelium,16,19 and the adhesion-independent cadherin clusters were also investigated in cell lines and C. elegans embryo.31–33
We also conducted experiments that are similar in principle to photobleaching, i.e., by chemical destruction of a fluorophore (chemical quenching of GFP fluorophore by dithionite, a reducing agent that destroys the GFP chromophore), to observe a change in EA of GFP-tagged proteins. Chemical quenching also resulted in an increase in EAof dE-cadWT and dE-cad-ΔP, consistent with the presence of nanoclusters of these proteins at the junction and lamella (Figures S1M and S1N). After chemical quenching, the EA of dE-cadWT::GFP remains significantly higher than the EA of dE-cad-ΔP::GFP at the junction, consistent with dE-cad-ΔP::GFP having weaker trans interactions than dE-cadWT::GFP (Figure S1O). Together, these results provide strong confirmation of the presence of dense nanoclusters of dE-cad both at the junction and in extra-junctional sites as proposed in Drosophila embryonic epithelium.19
Molecular machinery regulating dE-cad cis clustering
The hemocyte system developed here allows an exploration of the molecular mechanism of the nanoclustering of extra-junctional lamellar dE-cad. cis clustering of mammalian E-cad at extra-junctional sites has been reported to be mediated mainly by interactions of the cytoplasmic domain, with some contribution from homophilic interactions of the extracellular domain in increasing the packing density of the protein.21 We first addressed whether this type of clustering requires the recruitment of the adhesion complex machinery by utilizing dE-cad mutants defective in binding to adaptor proteins β-catenin34 and a-catenin34,35 (Figure S2A). Immunostaining of the dE-cad-Δβ-expressing hemocytes showed that they do not recruit β-catenin to the cell-cell junctions, compared to the junctional recruitment of β-catenin in the presence of wild-type protein, confirming the loss of interaction of the mutant protein with β-catenin (Figure S2B). Lamellar dE-cad-Δβ::GFP has significantly higher EA compared to dE-cadWT protein (Figures 2D, middle panel, and 2E), and importantly, lamellar dE-cad-Δβ::YFP did not show a detectable change in EA values upon photobleaching (Figures 2G and 2H), consistent with a lack of nanoclustering of this mutant protein on the lamella membrane.
Next, we expressed chimeric proteins of dE-cad-Δβ with fulllength or truncated α-catenin domains that carries only the actin binding domain of α-catenin in hemocytes (domain structure of Drosophila α-catenin schematized in Figure S2C).34,35 Only dE-cad-Δβ with α-catenin actin binding domain (dE-cad-Δβ-αABD) is trafficked to the cell surface in hemocytes, whereas the others are sequestered in intracellular compartments (Figure S2D). EA values of lamellar dE-cad-Δβ-αABD::GFP were in fact marginally lower compared to dE-cadWT (Figures 2D, right panel, and 2F), suggesting that the stable association with actin binding domain of α-catenin restores36 clustering of dE-cad-Δβ on the lamella membrane. Together, these results suggest that at least a fraction of lamellar dE-cad exists as nanoclusters and its ability to interact with the adaptor proteins, α- and α-catenin, drives the nanoclustering of dE-cad on the lamellar membrane.
dE-cad is known to associate with actomyosin network via its interaction with α-catenin21,35–38; therefore, we next probed whether perturbations of the actomyosin could alter the nanoclustering characteristics of lamellar dE-cadWT. Disruption of actin filaments by actin depolymerizing drug, Lat A, generated higher EA values compared to control cells (Figures 2I and 2J). Genetic perturbations of non-muscle myosin 2, RNAi against myosin regulatory light chain (MRLC), and myosin heavy chain (MHC) (Figures 2I and 2K) or inhibiting MRLC kinase Rho-associated protein kinase (ROCK), using the drug, H1152,39 also resulted in an increase in the EA values (Figures 2L and 2M). These results are consistent with a role for actin and myosin in building nanoclusters of lamellar dE-cad, as previously observed for transmembrane proteins with actin-binding motifs.40
We next investigated whether the machinery regulating nanoclustering of lamellar dE-cad also mediates the organization of junctional protein by investigating homo-FRET of dE-cad-Δβ. EA of junctional dE-cad-Δβ is higher than dE-cadWT at junctions (Figures 3A and 3B). dE-cad-Δβ is also more polarized at the junction compared to its counterpart in the lamella. However, unlike lamellar dE-cad-Δβ (Figures 2G and 2H), we see a small but significant increase in EA of junctional dE-cad-Δβ::YFP upon photobleaching, indicative of the presence of packed dE-cad-Δβ species capable of homo-FRET in this location (Figure 3E, red plot). By contrast, the profile of photobleaching of dE-cad-Δβ::YFP is distinct from that obtained with junctional dE-cadWT::YFP (Figure 3E, black plot). The lack of a change in EA until significant fraction of the fluorophore is bleached (30%), followed by a transition to a rise in EA, is entirely consistent with that expected from patches or aggregates much larger than the range of the homo-FRET process (Forster’s radius ~6 nm).30 Thus, although nanoclustering is altered at the junctions when the cytoplasmic connection is lost, our observations suggest that trans interactions are sufficient to promote a dense association of the dE-cad proteins, albeit of different local structure of dE-cadWT. This structure is consistent with observations of crystalline packing of trans-paired E-cadherin, nucleated on E-cad-Fc-coated surfaces.15
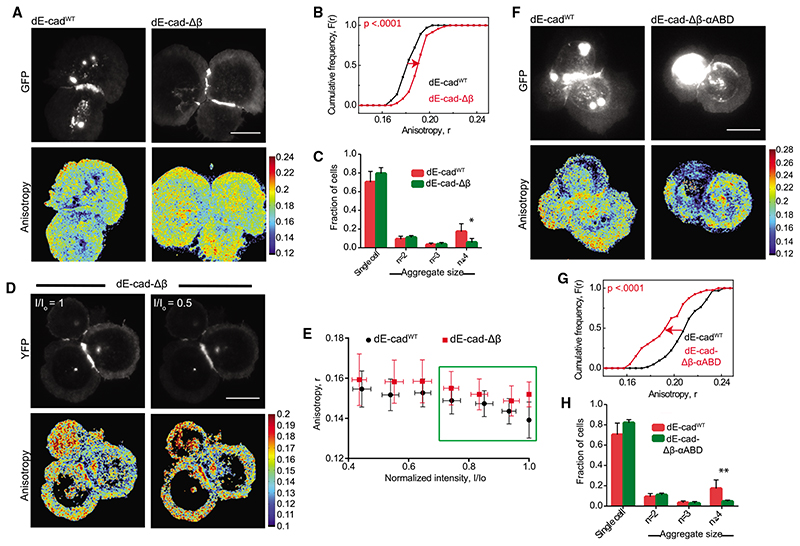
Figure 3. Adaptor protein-dependent clustering of junctional dE-cad.
(A and B) Total intensity and EA images (A) of dE-cadWT::GFP- and dE-cad-Db::GFP-expressing hemocytes and corresponding cumulative frequency distributions (B) of EA values of junctional dE-cad.
(C) Histogram showing percentage of cell aggregates in dE-cadWT::GFP- and dE-cad-Db::GFP-expressing cells. Data points show average normalized cell number from 6 dE-cadWT larvae and 7 dE-cad-Db larvae. Error bar represents the standard deviation in each condition.
(D and E) Total intensity and EA images (D) of dE-cadWT::YFP- and dE-cad-Δβ::YFP-expressing cells at the beginning of photo bleaching and at 50% photobleaching and corresponding normalized intensity versus EA plots (E) of junctional dE-cad obtained upon photobleaching. Each data point represents average EA values taken from multiple regions of different junctions (n = 5 [dE-cadWT and 4 [dE-cad-Δβ]). Error bars show standard deviation for the corresponding intensity bins normalized to the starting intensity value.
(F and G) Total intensity and EA images (F) of dE-cadWT::GFP- and dE-cad-Δβ-αABD::GFP-expressing hemocytes and corresponding cumulative frequency distributions (G) of EA values.
(H) Histogram showing percentage of cell aggregates in dE-cadWT::GFP- and dE-cad-Δβ-αABD::GFP-expressing cells. Dataset contains average normalized cell number from 6 dE-cadWT larvae and 5 dE-cad-Δβ-αABD larvae. Error bar represents the mean and the standard deviation in each condition.
p values are calculated using Mann-Whitney U test. Scale bars, 10 μm. Sample size is provided in Table S1. See also Figure S3.
Expression of dE-cad-Δβ-αABD::GFP resulted in a lowering of EA for the junctional protein compared to dE-cadWT, indicating that the nanocluster organization of dE-cad at the junctions is restored by linking it up to actin network, confirming the role of a dynamic actomyosin network in the nanoscale organization of dE-cad at the junction (Figures 3F and 3G). However, both dE-cad-Δβ or dE-cad-Δβ-αABD-expressing cells showed reduced cell aggregation compared to dE-cadWT-expressing cells (Figures 3C and 3H), suggesting that a dynamic association with cortical actin is important for creating adhesion complexes capable of supporting stable cell-cell interactions.
Next, we addressed the role of junctional actomyosin in nanoclustering using chemical inhibitors and genetic perturbations. Treatment with Lat A, H1152, and RNAi against MRLC and MHC all showed increased EA at the hemocyte junctions (Figures S3A–S3F). We also addressed whether the same molecular machinery, which regulates the nanoscale organization of dE-cad in hemocytes, also operates in embryos. The role of myosin-II in the remodeling of epithelial cell junctions in Drosophila embryo has been studied extensively.16,39,41 We also observed an increase in EA, indicative of loss of clustering of the junctional dE-cad in ROCK inhibitor H1152-injected embryos compared to controls (Figures S3G and S3I). Similarly, an increase in EA in Lat-A-injected embryos compared to controls confirmed the role of actomyosin network in regulating the organization of junctional dE-cad (Figures S3H and S3J).
Monitoring membrane diffusion of dE-cad at the lamellar membrane identifies multiple higher order species
Membrane diffusion offers a different method for exploring molecular organization, and here, we utilized both fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy (FCS) to monitor these organizational states.42 Actomyosin-sensitive nanoclusters of dE-cad as described above are expected to exhibit turnover rates on the order of 0.1–1 s–1, as noted from previous experiments characterizing the dynamics of such types of nanoclustering.43 Coupled with the expectation that these nanoclusters are relatively immobile, this slow turnover rate is likely to result in a pool of effectively immobilized dE-cad on the seconds timescales. FRAP using an ~3-μm-diameter spot probes these timescales, and indeed, in the hemocyte lamella, these experiments detected a small but significant immobile pool for dE-cadWT. This was significantly larger than that observed for dE-cad-Δβ (Figures S4A and S4B), consistent with the role for interactions with actomyosin resulting in immobile clusters.
To characterize the mobile fraction (~70%) of dE-cad species, we used FCS44,45 (see also STAR methods), which determines diffusion coefficient, number, and the average brightness of the diffusing entities in the confocal volume. Ascertaining the brightness of single cytoplasmic GFP molecules (Figure S4C) would also allow us to estimate the oligomeric state of the GFP-tagged diffusing species, composed of dE-cadWT::GFP or its variants. We first compared the brightness of cytoplasmic GFP to VSV-G::GFP, a known trimeric membrane protein.46 The measured brightness indicated that, although VSV-G::GFP is brighter than cytoplasmic GFP (~10 cpm versus 6.2), its “effective brightness per molecule in the trimer” is about 54% of cytoplasmic GFP (Figure S4D; Data S1). This is lower than the expected brightness of the diffusing VSV-G::GFP trimer, which should be ~3× cytoplasmic GFP. Assuming a similar “effective brightness per molecule” for individual dE-cad::GFP molecules, we can conclude that diffusing species of the dE-cadWT::GFP resemble an oligomeric species that is tetrameric, on an average, because they are approximately 1.25-fold brighter than VSV-G::GFP.
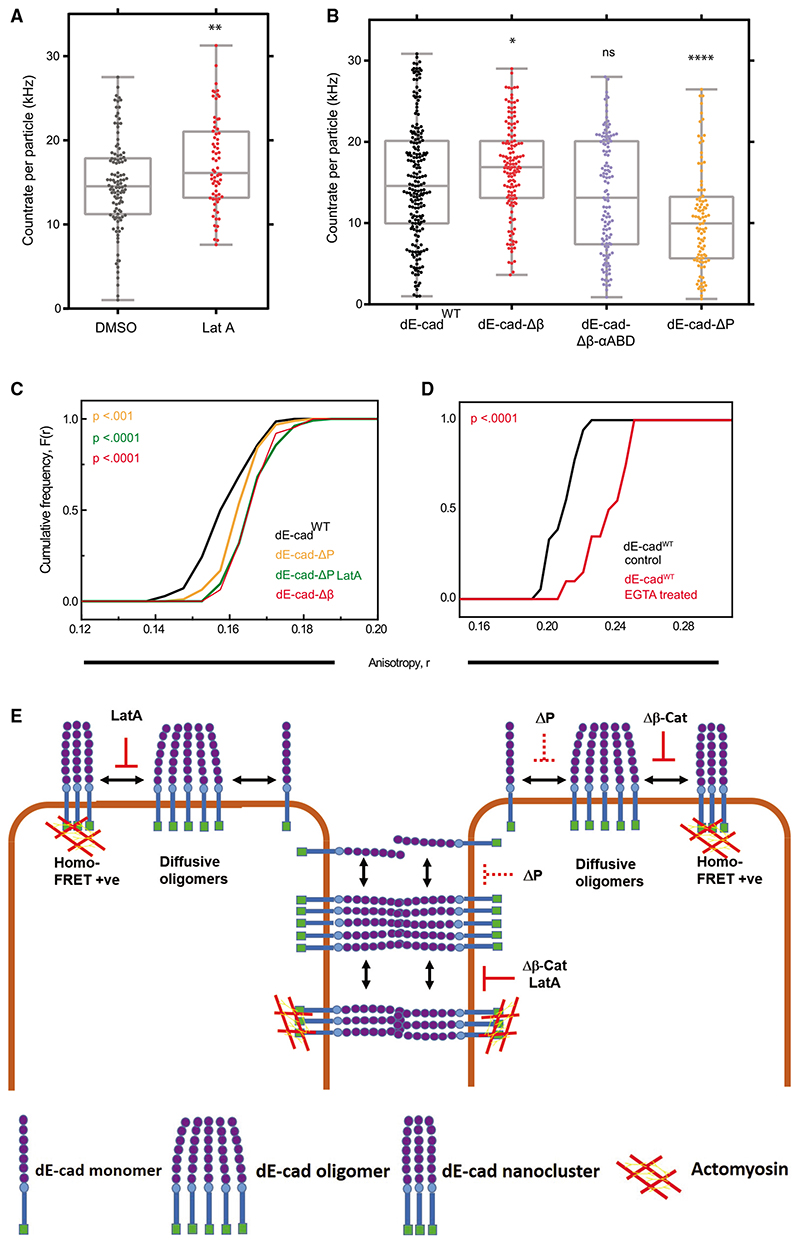
We then depolymerized F-actin, using Lat A, and found that dE-cadWT makes brighter diffusing species on lamella, as compared to control (Figure 4A; Data S2). Furthermore, dE-cad-Δβ diffusive species are brighter than the dE-cadWT species, whereas the brightness of dE-cad-Δβ-αABD was similar to that of dE-cadWT (Figure 4B; Data S3). This indicated that the cytoplasmic interactions with F-actin counteracts the tendency of dE-cad to make larger diffusing species.
Figure 4. Analysis of E-cadherin oligomerization.
(A) Brightness distribution of dE-cadWT oligomers in the lamella of dE-cadWT::GFP-expressing cells treated with DMSO or Lat A. Parameters, such as the number of diffusing species and diffusion timescales estimated through related FCS analysis, are detailed in Figure S4 and Data S2.
(B) Brightness distribution of oligomers of various GFP-tagged constructs of dE-cad in the lamella of cells expressing these constructs. Parameters, such as the number of diffusing species and diffusion timescales estimated via related FCS analysis, are detailed in Figure S4 and Data S3.
(C) Cumulative frequency distributions of EA values of lamellar dE-cad in dE-cadWT-, dE-cad-ΔP-, and dE-cad-Δβ-expressing hemocytes.
(D) Cumulative frequency distributions of EA values of lamellar dE-cadWT::GFP of similar intensity range in control and EGTA-treated conditions.
(E) Schematic model for the formation of cis oligomers and actomyosin-sensitive nanoscale clusters of dE-cad at the lamellar membrane or trans oligomers and nanoclusters at cell-cell junctions under different conditions. Solid red line indicates complete loss, and dashed red line indicates partial loss of nanoclustering or oligomerization.
p values are calculated using Mann-Whitney U test. ns, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Sample size is provided in Table S1. See also Figure S4 and Data S1, S2, and S3.
The increased brightness of the large diffusible species (as detected by FCS) does not result in a homo-FRET readout in the lamellar membrane, neither for the dE-cad-Δβ species (Figure 2H) nor for the Lat-A-treated dE-cadWT protein (Figure 2J). Thus, in the absence of actomyosin-driven clustering at the cytoplasmic leaflet, dE-cad forms a distinct type of large diffusible oligomeric species that is not sufficiently packed to elicit homo-FRET between the cytoplasmic-tail-attached GFP moieties. We thus conclude that extra-junctional dE-cad exists in at least two different homomeric states, namely, loosely packed diffusive oligomers and relatively immobile densely packed actomyosin-dependent nanoclusters.
This prompted us to ask whether this organization is hierarchical. We considered two possibilities: (1) oligomers form spontaneously due to protein-protein interactions in cis, and these in turn promote nanoscale clusters mediated by interaction with dynamic actin, or (2) actively assembled nanoclusters promote the formation of oligomers. The second alternative is unlikely, as the oligomers exist in the absence of nanoclusters. To test the first alternative, we ascertained the nanoclustering propensity of dE-cad constructs. dE-cad-ΔP, which forms oligomers very poorly when compared to dE-cadWT (Figure 4B), also exhibits reduced actin-dependent nanoclustering (Figure 4C), in support of the first alternative. The first alternative also implies that a complete abrogation of extracellular cis interactions will prevent nanoclustering. EGTA treatment of dE-cadWT reduces nanoclustering of the protein at the lamella, consistent with a role of Ca+2-dependent ectodomain cis interactions in promoting nanoscale clusters (Figure 4D). We do not find any condition where nanoclusters are formed in the absence of oligomers of dE-cad. Together, these data indicate that cis interactions of dE-cad are necessary both for forming diffusive oligomers and for efficient actomyosin-dependent dE-cad nanoclustering.
Discussion
Previous studies in cultured mammalian cells have characterized a higher level organization of E-cadherin in the context of cis and trans interactions of the E-cadherin protein.15,21,32 Indeed, Wu et al.21 showed that cis interactions of the cytoplasmic tail-deleted molecules resulted in lower density clusters and ectodomain-deleted molecules continue to exhibit nanoscale clustering. However, a mechanistic understanding of the construction of oligomers and nanoclusters and the relationship between these species was yet unexplored.
Here, we identify different types of higher level organization of dE-cad at the free membrane as well as at cell-cell junctions. We fnd that there are (1) diffusive cis oligomers in the lamella, (2) actomyosin-based cis nanoclusters both at cell-cell junctions and in the contact-free (lamellar or apical) membrane, and (3) trans-paired dE-cad at cell-cell junctions. A consistent model that reconciles all our observations (see model in Figure 4E) suggests a hierarchical assembly mechanism for the generation of the actomyosin-based nanoclusters, wherein spontaneous cis interactions of the dE-cad protein generate actin-limited oligomers, in extra-junctional regions, and these facilitate the actomyosin-based nanoclustering step. It is conceivable that the dense cis oligomerization may be seeded by trans-engaged dE-cad molecules, similar to previous observations,15 which in turn could be remodeled by the actomyosin activity to form trans nanoclusters. We believe such trans-engaged nanoclusters to be the building blocks of strong, stable, and yet tunable cell-cell adhesion. It is possible a threshold size or number of oligomers is required for assembling the activity-driven nanoclusters, providing a potential for non-linearity in the formation of nanoclusters and stable adhesions. Further studies elucidating the exchange between the oligomeric, nanoclustered, and trans-engaged dE-cad species can shed more light on the nature of the non-linearity.
The study of the organization of the cytoplasmic tail-deleted dE-cad-Δβ and dE-cad-Δβ-αABD isoform as well as the proximal ectodomain deleted dE-cad-ΔP variant has provided insights into how cell-cell junctions may be established. Available evidence suggests that strong cell-cell interactions not only require trans pairing of the cis oligomers but also require tunable interaction with cytoplasmic actin machinery necessary for junctional strengthening. This is consistent with the view that energy dissipation and rheology at adhesive interfaces are critical elements of adhesion.47 Although dE-cad-Δβ exhibits trans interactions, it lacks association with F-actin. It is known that there is a force-dependent strengthening of dE-cad-dE-cad trans interactions via α-catenin coupling to F-actin.6,8,48,49 Furthermore, trans interactions of dE-cad are likely to be strengthened by transmitting tensile forces generated by association with the actomyosin cortex in adjacent cells.39 In this context, it is likely that the medial apical contractile actomyosin network in Drosophila epithelial cells and in other organisms strengthens trans interactions acting to exert tensile stresses perpendicular to the junction.39 At the same time, the actomyosin network that acts parallel to the junction may assist in the disassembly of the trans-paired dE-cad, for instance by exerting shear stress.39
Actin coupling to loosely packed oligomers effectively reduces their size but also endows them with remodeling capacity. This feature, we propose, is a prerequisite for their evolution to more stable actin-dependent nanoclusters and eventually their functional trans-paired association. Equilibrium models based on surface tension have been successful at explaining some features of adhesion-based cell organization in vivo.50 However, our results further document that adhesion should rather be considered as an out-of-equilibrium, active, and dissipative process that can be dynamically tuned by actin interactions with E-cad oligomers/nanoclusters. This may endow cells with the capacity to actively tune trans pair assembly by adjusting to the dynamically changing mechanical/contractile environment of a cell.
Star* Methods
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
-
RESOURCE AVAILABILITY
Lead contact
Materials availability
Data and code availability
-
EXPERIMENTAL MODELAND SUBJECT DETAILS
Hemocyte isolation and culturing
Embryo Preparation
-
METHOD DETAILS
Transgenic lines and genetics
Drug injection
Acto-myosin perturbations in hemocytes
Immunofluorescence imaging/microscopy
Fluorescence Emission Anisotropy (EA) measurements and image analysis
Fluorescence Recovery After Photobleaching (FRAP)
Fluorescence correlation spectroscopy (FCS)
-
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
Graph Plots
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-E-cadherin | DSHB | Cat#DCAD2, RRID:AB_528120 |
| Mouse monoclonal anti-β-catenin | DSHB | Cat#N27A1, RRID:AB_528089 |
| Rat monoclonal anti-α-catenin | DSHB | Cat#DCAT-1, RRID:AB_532377 |
| Alexa Fluor 568 Phalloidin | Thermo Fisher Scientific | Cat#A12380 |
| Rabbit monoclonal anti-zipper | Lecuit Laboratory | 16 |
| Chemicals, peptides, and recombinant proteins | ||
| Ethyleneglycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) | Sigma Aldrich | Cat#E0396 |
| ROCK Inhibitor, H 1152 dihydrochloride | Tocris | Cat#2414 |
| Actin inhibitor, Lat A | Thermo Fisher Scientific | Cat#L12370 |
| Sodium Dithionite | Sigma Aldrich | Cat#157953 |
| Experimental models: organisms/strains | ||
| D.melanogaster: endo dE-cad::GFP,sqh Sqh::mCherry | Sqh-mCherry gift from Adam C Martin | Recombinant used in51 |
| D.melanogaster: endo dE-cadΔP::GFP/Cy | This manuscript, Lecuit Laboratory | 28 |
| D.melanogaster: Ubi-VSV-G::GFP | Lecuit Laboratory | 44 |
| D.melanogaster: UAS-dE-cad::GFP | Gift from H.Oda | 52 |
| D.melanogaster: Col-GAL4 | Gift from C. Dearolf | 53 |
| D.melanogaster: UAS-dE-cad-Δβcat::GFP | This manuscript, Lecuit laboratory | 34 |
| D.melanogaster: UAS-dE-cad-Δβcat-αcatABD::GFP | This manuscript, Lecuit laboratory | 34 |
| D.melanogaster: UAS-dE-cad::YFP | This manuscript, Lecuit laboratory | 52 |
| D.melanogaster: UAS-dE-cad-Δβcat::YFP | This manuscript, Lecuit laboratory | 34 |
| D.melanogaster: w1118; P{GD1695}v7916 | Vienna Drosophila Resource Center | VDRC:v7916 |
| D.melanogaster: w1118; P{GD1566}v7819 | Vienna Drosophila Resource Center | VDRC:v7819 |
| D.melanogaster: UAS-dEcad-ΔP | Gift from H.Oda | 28 |
| D.melanogaster: Hml-GAL4,UAS-2xEGFP | Bloomington Drosophila Stock Center | BL-30140 |
| Software and algorithms | ||
| MATLAB, Image processing toolbox | Mathworks Inc | https://www.mathworks.com/products/matlab.html |
| ImageJ | N/A | https://imagej.nih.gov/ij/index.html |
| Metamorph 7.0 software | Molecular Devices Corporation, CA, USA | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientifc-software/prism/ |
| OriginPro 8 | Origin Lab | https://www.originlab.com/ |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Satyajit Mayor (mayor@ncbs.res.in).
Materials availability
Fly lines generated in this study are available in Mayor Laboratory.
Experimental Model and Subject Details
Hemocyte isolation and culturing
Hemocytes were cultured in Schneider’s complete medium (SCM) containing Schneider’s insect medium (GIBCO-BRL, Gaithersberg, MD) supplemented with 10% non-heat inactivated serum (GIBCO-BRL) and 1μg/ml bovine pancreatic insulin (Sigma-Aldrich). SCM was aged overnight at 4°C prior to use. Hemocytes were dissected from the third instar larvae as previously described54 in 150μl of SCM and were incubated in a BOD incubator at 23°C. Hemocytes were imaged 2.5-hour post-dissection.
Embryo Preparation
Drosophila melanogaster flies were maintained at 22°C in glass vials containing standard fly media. For embryo collection flies were maintained in cages with apple juice agar plates, supplemented with yeast paste. Embryos collected on these plates were washed with water and then treated with bleach to remove chorions. Early gastrulation embryos were aligned on the coverslips and covered with Halocarbon oil 200 for fluorescence imaging.
Method Details
Transgenic lines and genetics
yellow white (y w) flies were used as a control for immunostaining. UAS-dE-cad::GFP and UAS-SHGDP#10 were gift from H. Oda. UAS-dE-cad-Δβ::GFP, UAS-dE-cad-Δβ-a-catABD::GFP, UAS-dE-cad::YFP and UAS-dE-cad-Δβ::YFP lines were generated in Lecuit lab. Collagen-GAL4 (gift from C. Dearolf) was used to drive expression of UAS constructs in hemocytes. endo-dE-cad::GFP is a knock in line of E-cadherin at the locus22. Myo-RLC::mCherry (gift from A. Martin) is also imaged along with dE-cad::GFP in embryo for drug perturbations. endo-dE-cad-DP/CyO flies was generated in Lecuit lab. Myosin-II RNAi lines, MRLC RNAi and MHC RNAi, were obtained from VDRC. Ubi-VSV-G::GFP line (Thomas Lecuit) was used as control for EA measurements in embryo. Hml-GAL4, UAS-2xEGFP (Bloomington) line was obtained from Dr. Tina Mukherjee lab.
Drug injection
Embryos were prepared as described before. For drug injections, embryos were kept in box containing Drierite and injected with either drugs or similar volumes of their solvents (RNase free water or DMSO), at the end of cellularisation or early gastrulation. For ROCK inhibition, embryos were injected with 20mM H1152 (Tocris) and imaged immediately. For actin perturbations, embryos were injected with 5mM Lat A (Invitrogen) and imaged immediately. For anisotropy measurements, 15-20 Z sections separated by 0.25μm were acquired from the most apical membrane of the embryo.
Acto-myosin perturbations in hemocytes
Actin perturbations were carried out using Lat A (Invitrogen); at 20μM for 15 minutes in case of anisotropy measurements, and at 1 μM for 1 hour in case of FCS. ROCK perturbations were carried out using 10μM H1152 (Sigma) for 1 hour. Imaging was carried out in the presence of the drug. All drug treatments were carried out in M1 buffer (150mM NaCl, 5mM KCl, 1mM CaCl2, 1mM MgCl2 and 20mM HEPES - pH 6.9) containing 1mg/ml BSA and 2mg/ml glucose at 23°C BOD incubator for the indicated time periods.
Immunofluorescence imaging/microscopy
Hemocytes were fixed with 2.5% paraformaldehyde for 20 minutes at room temperature and permeabilized with 0.37% Igepal for 13 minutes at room temperature for whole cell protein labeling. Cells were blocked using M1 containing 2mg/ml BSA and 2mg/ml glucose (blocking buffer) for 30 minutes and labeled with rat anti-E-cadherin (1:10 DCAD2, DSHB), mouse anti-beta catenin (1:10 N27A1, DSHB), rabbit anti-alpha catenin (1:10 DCAT2, DSHB) or rabbit anti-zipper (1:150, previously used in16) for 1 hour and then labeled with corresponding fluorescent secondary antibody. For actin labeling cells were labeled with Alexa 568 labeled phalloidin (Molecular probes). Immunostained samples were imaged using a wide field microscope (Nikon) using a Plan Apochromat 60X/ 1.4 NA oil immersion objective except MHC antibody labeled cells in Figure S1G, which were imaged using 100X objective.
Fluorescence Emission Anisotropy (EA) measurements and image analysis
Anisotropy measurements of hemocytes were carried out in TIRF microscope setup on Nikon TE2000 body, having polarized 488 laser, using a 100X objective with NA of 1.4940. Embryo measurements were carried out in confocal Spinning disk microscope equipped with a Yokogawa CSU-22 unit using 100X objective with NA of 1.4, custom fitted with excitation anisotropy imaging capabilities26. Andor laser combiners emitting 488 and 561 nm were used to image embryos and the intensities were collected using Andor ixon+897 EMCCD cameras40.
In this method polarized excitation was used to illuminate the sample and the resultant fluorescence emission is collected in parallel and perpendicular orientations (with respect to the axis of excitation polarization) to determine emission anisotropy (EA). The emission was split into parallel (Ipa) and perpendicular (Ipe) components with the help of a polarizer fitted in the path and collected simultaneously by two EMCCD cameras. Fluorescence anisotropy was calculated from the intensities collected in the parallel and perpendicular cameras using the formula
Rotational diffusion of the fluorophore will reduce emission anisotropy (EA), a situation also referred to below as depolarization of EA. Conversely, restriction of this motion (for example by increasing the rotational mass of the protein complex) will result in an increase of emission anisotropy, also referred to as polarization of EA. The emission anisotropy is dependent on the intermolecular distance between fluorophores. The random organization of proteins results in concentration dependent anisotropy distribution whereas the organization of protein into clusters results in concentration independent anisotropy distribution (Figure S2E). Construction of EA maps of the fluorescent protein therefore provide a spatial distribution of regions of the cell membrane where the protein is organized at different scales. Correspondingly, polarized (high) EA values (depicted as red pixels in the EA maps) reflect less cis-clustered or a more rotationally constrained fluorophore (e.g., by trans-interactions) and depolarized (low) EA values (depicted as blue pixels in the EA maps), also indicate the location of cis-clustered species.
The parallel and perpendicular fluorescent images were background subtracted using M1 buffer image taken at the same imaging conditions. Both images were then aligned using MATLAB code in MATLAB (Mathworks, USA) and G-factor corrected. Hemocyte lamellar EA measurements were taken from multiple 20 × 20-pixel (1.7*1.7 μm) regions selected on the flat lamellopodium of the hemocyte (excluding the central body of the hemocyte having punctae and large intracellular structures) using Metamorph™ software. Hemocyte junctional measurements were taken by dividing the junctions into multiple small regions. The intensities of parallel and perpendicular images were extracted and EA values were calculated using the formula described above. EA values from similar total intensity range were plotted as cumulative frequency distributions in Origin software. EA maps of the cells were generated from the aligned images using a code written in MATLAB.
Embryo images were processed through the same steps before EA measurements. Apical EA measurements were taken from 5 × 5-pixel (0.68 × 0.68 μm) regions from the apical most membrane of the embryo. Junctional punctae measurements were taken from junctional membrane plane, which is approximately 1 μm from the apical most membrane. Punctae measurements were taken from around 5-6 Z-planes, separated by 0.25μm, of the junctional membrane. For junctional punctae analysis, images were subtracted using a 50-pixel radius median filter. Embryos depending on the stage or its generation (F0 versus F2) were having different medial intensities and affecting the anisotropy values. Since different embryos were having different medial intensities, this step was introduced to reduce the effect of medial pool on the anisotropy measurements.
Photobleaching experiments were done in TIRF microscope using stream acquisition with an interval of 50-100ms between frames. For photobleaching analysis on hemocyte lamella and junction, 20 × 20-pixel (1.7*1.7 μm) regions were selected on the flat lamellopodium for lamellar measurements and on the cell-cell junctions for junctional measurements. Intensities and EA values of all frames were calculated using the formula described above. The intensities in the other frames were normalized to the intensity in the first frame of the ROI. Photobleaching graph was plotted using average EA values of different ROIs from different cells under similar experimental conditions.
Fluorescence Recovery After Photobleaching (FRAP)
We used Zeiss LSM 780 Confocor 3 System for the FRAP experiments using a 40X 1.2 NA UV-VIS-IR C Achromat water-immersion objective. The laser power at the back-focal plane of the objective was kept constant at 6μW for the 488nm line from Argon-laser. A circular ROI (diameter 3μm) was photobleached by switching the laser power to 100%. Another reference ROI of the same dimensions was drawn in the same cell to estimate imaging related photobleaching. A third ROI outside the cell was used to account for background. Images were acquired every 500ms, with 5 frames prior to photobleaching, and 120 frames after. The intensity traces in the bleach region were corrected for background and photobleaching, and were normalized using the averaged intensity in the prebleach region. Individual FRAP trace profiles were fitted to:
where, I is intensity at time-point t post-photobleaching. The bleaching is never 100% and the residual initial intensity is estimated through I0. The intensity recovers till the mobile fraction (F) is exchanged, at characteristic half-life ln2/tau. Here we are more interested in the immobile fraction which is estimated as 1-(F+I0).
Fluorescence correlation spectroscopy (FCS)
We used a point scanning confocal; Zeiss LSM 780 Confocor 3 System for the FCS measurements using a 40X 1.2 NA UV-VIS-IR C Achromat water-immersion objective. The back-focal plane of the objective was overfilled using 488nm line from an Argon-laser (at 6 μW, corresponding to about 5-10 kW/cm2) in order to create a diffraction-limited confocal volume that was calibrated on each day before the experiment by maximizing the count-rate per particle of Rhodamine 6G55,56. Hemocytes were cultured and pretreated as describe above, depending on the experiment. Confocal spot was focused on the ventral membrane of the hemocytes in the lamellar region of the cell. The confocal spot was parked in the center of the field and region of interest in individual cell was moved there by moving the stage. The correct focal distance was determined each time as the z-distance where the initial estimate of counts per molecule were highest. Next, the emission photon stream was recorded with the same objective, descanned, through an aligned pinhole (32 μm), wavelength selected between 491-571 and detected on a gallium arsenide detector array.
Two intensity in time traces (I(t)) of 60 s each were recorded for each cell as 6 iterations of 10 s. Each 10 s trace was autocorrelated into an autocorrelation curve G(τ) using the Zeiss onboard autocorrelator which calculates the self-similarity through:
Here CD denotes the time-average, δI(t) = I(t) - 〈I(t)〉 and τ is called the timelag. The 10 s measurement is long enough to contain sufficient events and short enough to avoid each trace to be contaminated by events that do not arise from Ecad::GFP diffusing in-plane of the plasma membrane. 1ms-binned intensity traces were manually assessed for data quality. Traces that contained monotonous changes in intensity (most likely arising due to photobleaching and/or z-drift during the measurement) or contained short high-intense bursts (most likely arising from endosomes passage) were discarded from further analysis. If the first set of traces were useful upon visual inspection, another set of traces were recorded the same point in the cell. This way, each cell yielded either 12 or 24 traces of 10 s each, along with corresponding autocorrelations generated by the Confocor.
A potential distribution of fluctuation timescales was previously determined44 for membranous proteins. This article indicated existence of 3 timescales as follows. First, a 20-120 us timescale corresponding to the triplet state of EGFP45; then second, a 0.5-5ms timescale corresponding to the intracellular/ luminal GFP that has also been observed previously in cells expressing EGFP tagged membrane protein57,58; and third, a timescale of ranging 10-100ms corresponds to the lateral diffusion of GFP-tagged membrane protein through the confocal spot.
We manually went through all 1ms binned intensity traces to discard the ones that show artifacts as mentioned above. For the traces that remain, we fitted their corresponding raw 10 s autocorrelation. The autocorrelation G(τ) was fitted to:
N reflects the number of moving particles in the confocal volume (including both 2D and 3D diffusing species) and GT(τ) is correlation function associated to blinking/triplet kinetics:
Where T is the fraction of molecules in the dark state and τt corresponds to the lifetime of the dark state. GD(τ) is correlation function associated to diffusion, which in this case contains two diffusional timescales corresponding to either 2D or 3D diffusion as follows:
The fraction f corresponds to the intracellular 3D diffusing pool of EGFP that has a timescale of τD1. S is the structure factor that accounts for timescales arising from the fact that the intracellular EGFP diffuses in a volume rather that a plane. Free fitting this parameter converges the value to about 0.2, consistent with earlier reports on cultured cells. In order to contain the number of free parameters we decided to fix this value to 0.2. The diffusion time associated with laterally diffusing dE-cad::GFP is finally calculated with τD2. When fitting the autocorrelation traces for GFP in solution or cytoplasm, the 2D component was excluded from the fit equation.
For estimating the brightness of 2D diffusing species of dE-cadWT or its variants, we first used the fraction f to estimate the number of diffusing entities in 2D and 3D as:
Then, solving following equation gives the brightness of the 2D diffusing component Q2D:
when, we plug in the brightness of cytosolic monomeric EGFP in place of Q3D, which was estimated in an independent experiment (see Figure S4C). The fit values for the free parameters in this analysis are presented in Data S1.
Quantification and Statistical Analysis
Statistical Analysis
Each hemocyte EA experiment reported here has dataset from one experiment which was performed at least twice with similar results. For lamellar data quantification, a 20 × 20-pixel (1.7 × 1.7 μm) region of interest (ROI) based analysis was performed on 10-20 cells. For junctional data quantification, different area regions were drawn over 10-20 junctions in each experiment. Apical and junctional data quantification was done on 5 × 5-pixel (0.68 × 0.68 μm) region of interest drawn on embryos with same experimental conditions. Embryo experiments has been conducted over different days and the data was generated by pooling from different days embryos. N refers to the number of embryos taken for measurements. Non-parametric Mann-Whitney test was performed to check the statistical significance between different experimental conditions and to calculate p values. p values are indicated in the figure panel itself. ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Graph Plots
All the graphs were plotted either in OriginPro 8 or in Prism Graph-Pad software.
Supplementary Material
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.01.059.
In Brief.
Chandran et al. demonstrate that the Drosophila E-cadherin forms two kinds of nanoscale assemblies: loosely packed oligomers and dense nanoclusters. Extracellular cis interactions produce the oligomers, and the cytoplasmic interactions with actomyosin remodel the oligomers into nanoclusters, necessary for regulating strong stable cell adhesions.
Highlights.
Drosophila primary hemocytes serve as a model for dE-cadherin-based cell adhesion
dE-cad forms adhesion-independent dense nanoclusters and loosely packed oligomers
cis interactions of dE-cad seed the oligomers necessary for nanoclusters assembly
Actomyosin activity restricts oligomer growth and remodels them into nanoclusters
Acknowledgments
We thank H. Oda and P. Rørth for the gift of flies and plasmid constructs, Bloomington fly facility and Vienna Drosophila Resource Center for providing transgenic flies, Thomas Van Zanten for the help on FCS, Lokavya Kurup and Screening facility for cell aggregate imaging, D. Trivedi and NCBS fly facility for microinjection system, and the Central Imaging and Flow Cytometry Facility at NCBS. We thank Mayor lab members for their valuable comments on the manuscript. R.C. acknowledges CSIR-UGC JRF fellowship from CSIR (Government of India). G.K. acknowledges bridging post-doctoral fellowship from NCBS. This project benefited from support from the CNRS (Laboratoire International Associé, [SysTiM] between IBDM and NCBS) and Aix-Marseille Université, A*MIDEX, ANR-11-IDEX-0001–02, funded by the Investissements d’Avenir French government program (BioTIM) and JC Bose Fellowship from DST (Government of India), Margadarshi fellowship (IA/M/15/1/502018) to S.M., and support from the Department of Atomic Energy (Government of India) under project no. RTI 4006 to NCBS.
Footnotes
Author Contributions
R.C., T.L., and S.M. conceived the project. R.C. did EA and homo-FRET experiments and related data analysis. G.K. did FRAP and FCS experiments and related data analysis. J.-M.P. did all the constructs and generated transgenic flies for this study. S.M. and T.L. provided the resources for the project. R.C., G.K., and S.M. wrote the original draft with comments from all the authors.
Declaration of Interests
The authors declare no competing interests.
Data and code availability
This study did not generate datasets or code.
References
- 1.Oda H, Tsukita S, Takeichi M. Dynamic behavior of the cad-herin-based cell-cell adhesion system during Drosophila gastrulation. Dev Biol. 1998;203:435–450. doi: 10.1006/dbio.1998.9047. [DOI] [PubMed] [Google Scholar]
- 2.Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Török T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- 3.Niewiadomska P, Godt D, Tepass U. DE-cadherin is required for intercellular motility during Drosophila oogenesis. J Cell Biol. 1999;144:533–547. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uemura T, Oda H, Kraut R, Hayashi S, Kotaoka Y, Takeichi M. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes Dev. 1996;10:659–671. doi: 10.1101/gad.10.6.659. [DOI] [PubMed] [Google Scholar]
- 5.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 6.Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17:533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 7.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. α-catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 8.Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. Cell adhesion. The minimal cad-herin-catenin complex binds to actin filaments under force. Science. 2014;346:1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu Y-S, Thomas WA, Eder O, Pincet F, Perez E, Thiery JP, Dufour S. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J Cell Biol. 2004;167:1183–1194. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maître J-L, Berthoumieux H, Krens SFG, Salbreux G, Jülicher F, Paluch E, Heisenberg C-P. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science. 2012;338:253–256. doi: 10.1126/science.1225399. [DOI] [PubMed] [Google Scholar]
- 11.Cavey M, Rauzi M, Lenne P-F, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453:751–756. doi: 10.1038/nature06953. [DOI] [PubMed] [Google Scholar]
- 12.Takeda H, Shimoyama Y, Nagafuchi A, Hirohashi S. E-cadherin functions as a cis-dimer at the cell-cell adhesive interface in vivo. Nat Struct Biol. 1999;6:310–312. doi: 10.1038/7542. [DOI] [PubMed] [Google Scholar]
- 13.Troyanovsky RB, Laur O, Troyanovsky SM. Stable and un-stable cadherin dimers: mechanisms of formation and roles in cell adhesion. Mol Biol Cell. 2007;18:4343–4352. doi: 10.1091/mbc.E07-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci USA. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strale P-O, Duchesne L, Peyret G, Montel L, Nguyen T, Png E, Tampe R, Troyanovsky S, Hénon S, Ladoux B, Mége RM. The formation of ordered nanoclusters controls cadherin anchoring to actin and cell-cell contact fluidity. J Cell Biol. 2015;210:333–346. doi: 10.1083/jcb.201410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levayer R, Pelissier-Monier A, Lecuit T. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocy-tosis during epithelial morphogenesis. Nat Cell Biol. 2011;13:529–540. doi: 10.1038/ncb2224. [DOI] [PubMed] [Google Scholar]
- 17.Adams CL, Chen Y-T, Smith SJ, Nelson WJ. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kametani Y, Takeichi M. Basal-to-apical cadherin flow at cell junctions. Nat Cell Biol. 2007;9:92–98. doi: 10.1038/ncb1520. [DOI] [PubMed] [Google Scholar]
- 19.Truong Quang B-A, Mani M, Markova O, Lecuit T, Lenne P-F. Principles of E-cadherin supramolecular organization in vivo. Curr Biol. 2013;23:2197–2207. doi: 10.1016/j.cub.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Engl W, Arasi B, Yap LL, Thiery JP, Viasnoff V. Actin dy-namics modulate mechanosensitive immobilization of E-cadherin at adherens junctions. Nat Cell Biol. 2014;16:587–594. doi: 10.1038/ncb2973. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Kanchanawong P, Zaidel-Bar R. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev Cell. 2015;32:139–154. doi: 10.1016/j.devcel.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Zhou W, Dong W, Watson AM, Hong Y. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci USA. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223:1019–1026. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim SA, Tai C-Y, Mok L-P, Mosser EA, Schuman EM. Calcium-dependent dynamics of cadherin interactions at cell-cell junctions. Proc Natl Acad Sci USA. 2011;108:9857–9862. doi: 10.1073/pnas.1019003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh S, Saha S, Goswami D, Bilgrami S, Mayor S. Dynamic imaging of homo-FRET in live cells by fluorescence anisotropy microscopy. Methods Enzymol. 2012;505:291–327. doi: 10.1016/B978-0-12-388448-0.00024-3. [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan V, Kalappurakkal JM, Mehta SB, Nordenfelt P, Moore TI, Koga N, Baker DA, Oldenbourg R, Tani T, Mayor S, et al. Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc Natl Acad Sci USA. 2017;114:10648–10653. doi: 10.1073/pnas.1701136114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haruta T, Warrior R, Yonemura S, Oda H. The proximal half of the Drosophila E-cadherin extracellular region is dispensable for many cadherin-dependent events but required for ventral furrow formation. Genes Cells. 2010;15:193–208. doi: 10.1111/j.1365-2443.2010.01389.x. [DOI] [PubMed] [Google Scholar]
- 29.Perrin F. Polarisation de la lumière de fluorescence. Vie moyenne des molécules dans l’etat excité. J Phys Radium. 1926;7:390–401. [Google Scholar]
- 30.Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 31.Padmanabhan A, Ong HT, Zaidel-Bar R. Non-junctional E-cadherin clusters regulate the actomyosin cortex in the C. elegans zygote. Curr Biol. 2017;27:103–112. doi: 10.1016/j.cub.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 32.Indra I, Choi J, Chen C-S, Troyanovsky RB, Shapiro L, Honig B, Troyanovsky SM. Spatial and temporal organization of cad-herin in punctate adherens junctions. Proc Natl Acad Sci USA. 2018;115:E4406–E4415. doi: 10.1073/pnas.1720826115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sako Y, Nagafuchi A, Tsukita S, Takeichi M, Kusumi A. Cytoplasmic regulation of the movement of E-cadherin on thefree cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J Cell Biol. 1998;140:1227–1240. doi: 10.1083/jcb.140.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pacquelet A, Rørth P. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J Cell Biol. 2005;170:803–812. doi: 10.1083/jcb.200506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong S, Troyanovsky RB, Troyanovsky SM. Binding to F-actin guides cadherin cluster assembly, stability, and movement. J Cell Biol. 2013;201:131–143. doi: 10.1083/jcb.201211054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. Monomeric α-catenin links cadherin tothe actin cytoskeleton. Nat Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- 37.Levayer R, Lecuit T. Oscillation and polarity of E-cadherin asymmetries control actomyosin flow patterns during morphogenesis. Dev Cell. 2013;26:162–175. doi: 10.1016/j.devcel.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 38.Chen C-S, Hong S, Indra I, Sergeeva AP, Troyanovsky RB, Shapiro L, Honig B, Troyanovsky SM. α-catenin-mediated cadherin clustering couples cadherin and actin dynamics. J Cell Biol. 2015;210:647–661. doi: 10.1083/jcb.201412064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kale GR, Yang X, Philippe J-M, Mani M, Lenne P-F, Lecuit T. Distinct contributions of tensile and shear stress on E-cadherin levels during morphogenesis. Nat Commun. 2018;9:5021. doi: 10.1038/s41467-018-07448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gowrishankar K, Ghosh S, Saha S, R C, Mayor S, Rao M. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Munjal A, Philippe J-M, Munro E, Lecuit T. A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature. 2015;524:351–355. doi: 10.1038/nature14603. [DOI] [PubMed] [Google Scholar]
- 42.Scarselli M, Annibale P, McCormick PJ, Kolachalam S, Aringhieri S, Radenovic A, Corsini GU, Maggio R. Revealing G-protein-coupled receptor oligomerization at the single-molecule level through a nanoscopic lens: methods, dynamics and biological function. FEBS J. 2016;283:1197–1217. doi: 10.1111/febs.13577. [DOI] [PubMed] [Google Scholar]
- 43.Goswami D, Gowrishankar K, Bilgrami S, Ghosh S, Raghupathy R, Chadda R, Vishwakarma R, Rao M, Mayor S. Nanoclusters ofGPI-anchored proteinsareformed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jha A, van Zanten TS, Philippe J-M, Mayor S, Lecuit T. Quantitative control ofGPCR organization and signaling by endocytosis in epithelial morphogenesis. Curr Biol. 2018;28:1570–1584.:e6. doi: 10.1016/j.cub.2018.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altan-Bonnet N, Altan-Bonnet G. Fluorescence correlation spectroscopy in living cells: a practical approach. Curr Protoc Cell Biol Chapter. 2009;Chapter 4 doi: 10.1002/0471143030.cb0424s45. Unit 4.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doms RW, Keller DS, Helenius A, Balch WE. Role for adenosine triphosphate in regulating the assembly and transport ofvesic-ular stomatitis virus G protein trimers. J Cell Biol. 1987;105:1957–1969. doi: 10.1083/jcb.105.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yap AS, Duszyc K, Viasnoff V. Mechanosensing and me-chanotransduction at cell-cell junctions. Cold Spring Harb Perspect Biol. 2018;10:a028761. doi: 10.1101/cshperspect.a028761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci USA. 2012;109:18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manibog K, Li H, Rakshit S, Sivasankar S. Resolving the molecular mechanism of cadherin catch bond formation. Nat Commun. 2014;5:3941. doi: 10.1038/ncomms4941. [DOI] [PubMed] [Google Scholar]
- 50.Lecuit T, Lenne P-F. Cell surface mechanics and the control ofcell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 51.Kerridge S, Munjal A, Philippe J-M, Jha A, de las Bayonas AG, Saurin AJ, Lecuit T. Modular activation of Rho1 by GPCR signalling imparts polarized myosin II activation during morphogenesis. Nat Cell Biol. 2016;18:261–270. doi: 10.1038/ncb3302. [DOI] [PubMed] [Google Scholar]
- 52.Oda H, Tsukita S. Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins. Dev Biol. 1999;216:406–422. doi: 10.1006/dbio.1999.9494. [DOI] [PubMed] [Google Scholar]
- 53.Asha H, Nagy I, Kovacs G, Stetson D, Ando I, Dearolf CR. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics. 2003;163:203–215. doi: 10.1093/genetics/163.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sriram V, Krishnan KS, Mayor S. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila mel-anogaster. J Cell Biol. 2003;161:593–607. doi: 10.1083/jcb.200210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gendron P-O, Avaltroni F, Wilkinson KJ. Diffusion coeffi-cients of several rhodamine derivatives as determined by pulsed field gradient-nuclear magnetic resonance and fluorescence correlation spectroscopy. J Fluoresc. 2008;18:1093–1101. doi: 10.1007/s10895-008-0357-7. [DOI] [PubMed] [Google Scholar]
- 56.Majer G, Melchior JP. Characterization of the fluorescence correlation spectroscopy (FCS) standard rhodamine 6G and calibration of its diffusion coefficient in aqueous solutions. J Chem Phys. 2014;140:094201. doi: 10.1063/1.4867096. [DOI] [PubMed] [Google Scholar]
- 57.Brock R, Vámosi G, Vereb G, Jovin TM. Rapid character-ization of green fluorescent protein fusion proteins on the molecular and cellular level by fluorescence correlation microscopy. Proc Natl Acad Sci USA. 1999;96:10123–10128. doi: 10.1073/pnas.96.18.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiantia S, Ries J, Schwille P. Fluorescence correlation spectroscopy in membrane structure elucidation. Biochim Biophys Acta. 2009;1788:225–233. doi: 10.1016/j.bbamem.2008.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.01.059.
Data Availability Statement
This study did not generate datasets or code.