Abstract
Tissue-like structures from human pluripotent stem cells containing multiple cell types are transforming our ability to model and understand human development and disease. Here we describe a protocol to generate cardiomyocytes (CMs), cardiac fibroblasts and cardiac endothelial cells, the three principal cell types in the heart, from human induced pluripotent stem cells (hiPSCs) and combine these in three-dimensional cardiac microtissues (MTs). We include details of how to differentiate, isolate, cryopreserve and thaw the component cells and how to construct and analyze the MTs. The protocol supports hiPSC-CM maturation and allows replacement of one or more of the three heart cell types in the MTs with isogenic variants bearing disease mutations. Differentiation of each cell type takes approximately 30 days, while MT formation and maturation requires another 20 days. No specialist equipment is needed and the method is low cost, requiring just 5,000 cells per MT.
Keywords: human induced pluripotent stem cell-derived cardiomyocytes, human induced pluripotent stem cell-derived cardiac endothelial cells, human induced pluripotent stem cell-derived cardiac fibroblasts, cardiac microtissue, cardiomyocyte maturation, multicellular cell diseases and drug efficacy platform
Introduction
The dialogue between cells in developing or damaged tissues critically determines the functional phenotype and (patho)physiological properties of the tissue. Most major organs (for example the intestine, liver and pancreas) can now be mimicked in vitro as organoid-like structures1–3. These organoid-like structures have been established from either adult stem cells or hiPSCs and contain various cell types.
The heart contains three major cell types: cardiomyocytes (CMs), cardiac endothelial cells (ECs) and cardiac fibroblasts (CFs). CMs make up approximately 80% of the adult human heart by volume but only 30% of the total cell number4–7. The developing heart, by contrast, contains 60-70% CMs8. The majority of the other cells are ECs, CFs and immune cells, although the estimation of their exact proportion varies between studies5–7. All the cells in the heart communicate via physical interactions and paracrine factors4, thus regulating heart development and function9,10.
Here we describe a protocol to construct three-dimensional (3D) structures here termed “cardiac microtissues” (MTs) composed of 70% CMs, 15% ECs, and 15% CFs derived from human induced pluripotent stem cells (hiPSCs). We used this protocol to generate cardiac MTs from multiple hiPSC lines11. We also describe how to evaluate some standard functional parameters on whole MTs or single CMs dissociated from them. These procedures allow assessment of electrical, structural, mechanical and metabolic maturation of hiPSC-CMs.
Development of the Protocol
During organogenesis, CMs, ECs and CFs differentiate from common precursors in the cardiac mesoderm12–14. We previously described how to co-differentiate CMs and ECs simultaneously from hiPSC-derived cardiac mesoderm progenitors by combining Wnt-inhibition with vascular endothelial growh factor (VEGF) addition15. From the combined culture, ECs and CMs were isolated using the surface proteins CD34 and VCAM1, respectively. In a more recent study11, we further optimized the previous procedures for EC and CM differentiation by driving the CM and EC differentiation pathways independently after cardiac mesoderm induction. Independent culture resulted in production of monotypic cultures, circumventing the need to isolate ECs and CMs. This avoided the need to select CMs with VCAM1, which is also expressed by CFs16. We also described previously how to differentiate hiPSCs to epicardial cells (EPI) that normally cover the surface of the heart17, and which are the major source of CFs developmentally18. We recently extended this differentiation procedure to generate CFs from hiPSC-EPI11. Specifically, we adapted a monolayer differentiation protocol described previously19, based on low insulin BPEL (LI-BPEL) medium for cardiac mesoderm induction and for the initial induction of CFs, then used a commercially available medium (FGM3) for subsequent stages, as this supported CF expansion and increased reproducibility between batches.
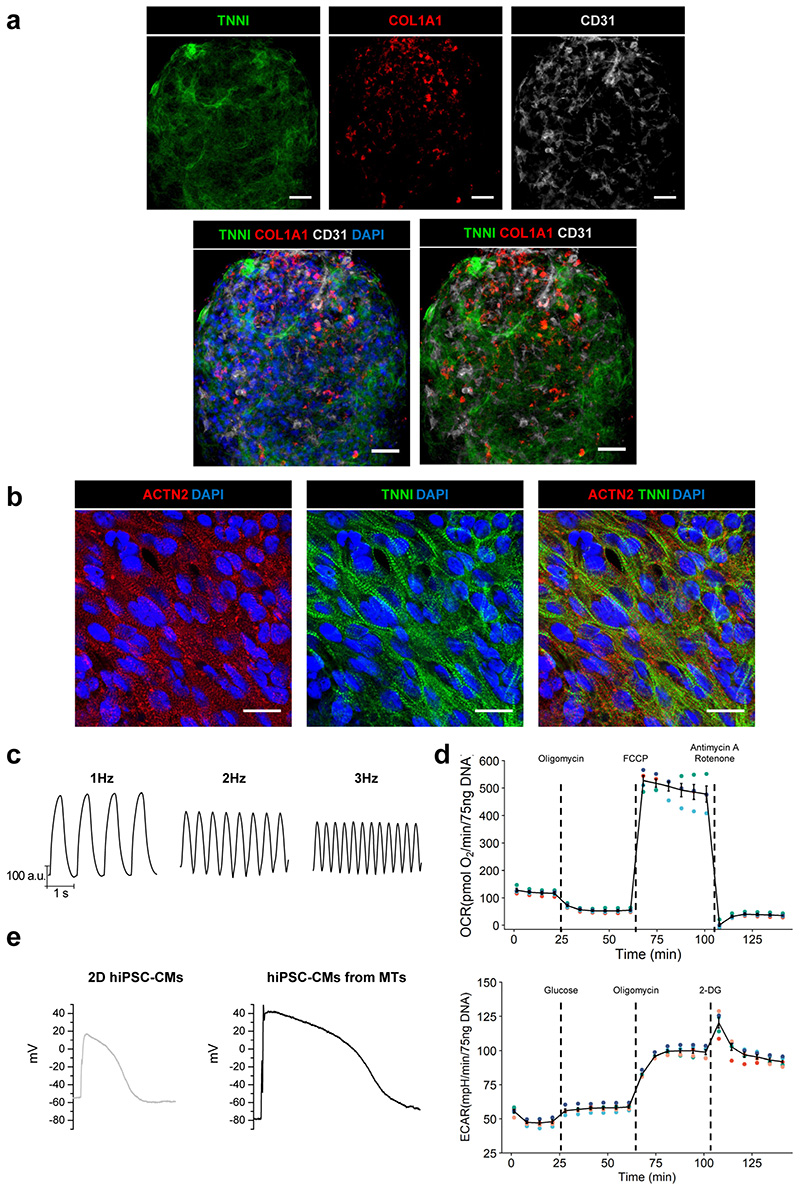
Although adding hiPSC-ECs to 3D aggregates enhanced some aspects of maturation in hiPSC-CMs15, we recently improved this further by also including CFs11. The triple cell type combination supported self-assembly of the MTs in vitro. The features exhibited by the hiPSC-CMs in the three-cell type cardiac MTs that indicated enhanced maturation were: sarcomere length and organization, contraction duration and amplitude, T-tubule-like structures (albeit sparse), action potential profiles characterized by more hyperpolarized resting membrane potential, the presence of an action potential “notch” and expression of postnatal sarcomere isoforms11. We also demonstrated that the features indicating enhanced electrophysiological maturity of the hiPSC-CMs were retained after their dissociation from the MTs11. The dialogue among all three cell types is essential to ensure full maturation of the CMs. Coupling between CMs and CFs through connexin 43 (CX43) gap junctions and cAMP-pathway activation were found to be two key mechanisms contributing to hiPSC-CM maturation11. .
Comparison with other methods
Table 1 compares the cellular composition and substrate/scaffold of the cardiac MTs that we describe in this protocol with those used in various other systems using human pluripotent stem cells (hiPSCs or embryonic stem cells, hESCs, collectively called hPSCs).
Table 1. Cellular composition and scaffold/substrate of published engineered heart tissue models.
| Study | Number of cells per cardiac tissue | CM source | EC source | Stromal cell source | Substrate/scaffold |
|---|---|---|---|---|---|
| (Stevens et al. 2009)52 | 3 x 106 cells | hESCs | hESCs or HUVEC | Neonatal human dermal fibroblasts (NHDFs) | Scaffold-free |
| (Tulloch et al. 2011)31 | 2 x 106 CMs or 2 x 106 CMs + 1 x 106 HUVECor 2 x 106 CMs + 1 x 106 HUVEC + 1 x 106 MSCs or MEFs | hESCs or hiPSCs | HUVEC | Human marrow stromal cells (MSCs) | Type I Collagen; 11% Geltrex |
| (Schaaf et al.2011)32 | 0.5 x 106 cells | hESCs | Fibrinogen/Matrigel plus thrombin; rectangular casting molds with two flexible silicone posts | ||
| (Nunes et al.2013)23 | 0.5 x 106 cells | hESCs and hiPSCs | NA | NA | Type I Collagen; two layers masks made with SU-8 photolithography, propylene glycol monomethyl ether acetate and PDMS |
| (Wang et al. 2014)27 | 1.5 x 106 cells/cm2 | hiPSCs | NA | NA | Elastomers micropatterned with fibronectin lines |
| (Burridge et al. 2014)33 | 2.5 × 105 CMs + 5 × 104 ECs + 5 × 104 MSCs | hESCs | hiPSCs | Human amniotic mesenchymal stem cells (hAMSCs) | 3D Hydrogel platform (Matrigel-based) |
| (Hinson et al. 2015)34 | 1.1 x 106 cells per mold (~1 × 103 cells per tissue) | hiPSCS | NA | Human mesenchymal stem cells(hMSCs) | PDMS, type I collagen and human fibrinogen |
| (Ravenscroft et al. 2016)49 | 5 x 103 cells at ratio CMs:ECs:CFs (or DFs) of 4:1:2 | hESCs or hiPSCs | Human cardiac microvascular ECs (hCMECs) or human coronary artery ECs (hCAECs) or human dermal microvascular ECs (hDMECs) | Primary human cardiac fibroblasts (hCFs) or normal human dermal fibroblasts (NhDFs) | Scaffold-free |
| (Masumoto et al. 2016)46 | 3 x 106 cells | hiPSCs* | hiPSCs* | Mural cells (MCs) from hiPSCs* | Acid-soluble rat-tail type I collagen neutralized with alkalin buffer; Matrigel |
| (Mannhardt et al. 2016)35 | 1 x 106 cells | hiPSCs | NA | NA | Agarose casting molds with solid silicon racks; Fibrin matrix (Matrigel, fibrinogen, thrombin) |
| (Huebsch et al. 2016)47 | 2 × 103 CMs + 2 × 103 stromal cells | hiPSCs* | NA | Fibroblast-like stromal cells from hiPSCs* | PDMS stencils coated with fibronectin |
| (Keung et al. 2016)36 | 1 x 106 cells per mold (~1 × 103 cells per tissue) | hESCs | NA | NA | PDMS, type I collagen and human fibrinogen |
| (Tiburcy et al. 2017)37 | 1.5 x 106 cells | hESCs or hiPSCs | NA | Human fibroblasts | Circular casting molds; Matrigel/rat collagen matrix; bovine collagene (Matrigel-free method) |
| (Gao et al. 2017)24 | 25 × 103 CMs + 12.5 × 103 ECs + 12.5 × 103 SMCs | hiPSCs* | hiPSCs* | Smooth muscle cells from hiPSCs* | 3D-MPE printed scaffold; methacrylated gelatin |
| (Lee et al. 2017)38 | 1 × 105 CMs + 1 × 105 FBs | hESCs | NA | Human foreskin fibroblasts | PDMS elastomer molds; Matrigel/ type I collagen matrix |
| (Giacomelli et al. 2017)15 | 5 x 103 cells at ratio CMs:ECs of 85:15 | hESCs or hiPSCs* | hESCs or hiPSCs* | NA | Scaffold-free |
| (Polonchuk et al. 2017)53 | 12 × 103 cells at ratio CMs:ECs:CFs of 2:1:1 | iCMs from Axiogenesis | hCAECs | iCFs from Axiogenesis | Scaffold-free |
| (Mills et al. 2017)22 | 5 x 104 cells CMs:stromal cells of 70:30 | hESCs and hiPSCs* | NA | CD90+ stromal cells from hESCs and hiPSCs* | SU-8 photolithography and PDMS molds; Matrigel/ type I collagen matrix |
| (Pointon et al. 2017)50 | 5 x 103 cells at ratio CMs:ECs:CFs of 4:1:2 | hiPSCs | hCMECs | hCFs | Scaffold-free |
| (Ong et al. 2017)38 | 33 x 103 cells at ratio CMs:ECs:CFs of 70:15:15 | hiPSCs | HUVEC | Human adult ventricular cardiac fibroblasts | Biomaterial free, 3D bioprinted |
| (Shadrin et al. 2017)39 | 0.5-1 x 106 cells | hESCs and hiPSCs | NA | NA | PDMS square molds; Hydrogel matrix (fibrinogen, Matrigel, thrombin) |
| (Ronaldson-Bouchard et al. 2018)40 | 2 × 106 cells at ratio CMs:FBs of 75:15 | hiPSCs | NA | Skin fibroblasts | PDMS elastomeric pillars with polycarbonate support structures; fibrin hydrogel (fibrinogen, thrombin) |
| (Ma et al. 2018)28 | 3 × 106 cells | hiPSCs | NA | NA | Cell-encapsulation gel-free filamentous matrix (OrmoClear®polymer) |
| (Forsythe et al. 2018)51 | 1.5 x 103 cells at ratio CMs:CFs of 90:10 | iCMs from Axiogenesis | NA | hCFs | Scaffold-free |
| (Tsuruyama et al. 2019)41 | 6 × 106 CMs/sheet + 6 × 106 FBs/sheet | hiPSCs | NA | hDFBs | Cell sheets wrapped around a hollow octagonal tubular column; fibrin and collagen gels to seal the extremities |
| (Zhao et al.2019)30 | 7.47 × 104 CMs + CFs 3.5 × 104 | hESCs and hiPSCs | NA | hCFs | Flexible poly (octamethylene maleate (anhydride) citrate) (POMaC) wires; hydrogel matrix (collagen, Matrigel) |
| (Mills et al. 2019)42 | 5 × 104 cells at ratio CMs:stromal cells of 70:30 | hESCs* | CD90+ stromal cells from hESCs* | PDMS molds; Matrigel/ type I collagen matrix | |
| (Goldfracht et al. 2019)26 | 2 × 106 CMs | hiPSCs | NA | NA | Chitosan-enhanced ECM from decellularized pig heart |
| (Wanjare et al. 2020)29 | 1 × 106 CMs + 4 × 104 ECs | hiPSCs* | hiPSCs* | NA | Circular microfibrous polycaprolactone sheets; Geltrex |
| (Goldfracht et al. 2020)43 | 2 × 106 CMs | hESC-(atrial and ventricular differentiation) | NA | NA | Ring-shaped casting molds; collagen-based hydrogel matrix |
| (Jang et al. 2020)44 | 1 × 106 CMs + 2 × 105 CFs | hESCs and hiPSCs | NA | Human adult ventricular cardiac fibroblasts | PDMS support; collagen I mixture (layer-by-layer deposition) |
| (Sebastiao et al. 2020)54 | 1.5 x 103 CMs | hiPSCs | NA | NA | Scaffold-free |
| (Richards et al. 2020)55 | 1.5 × 105 cells 50% CMs and 50% non-myocyte (at a 4:2:1 ratio of FBs:HUVECs:hADSCs) |
hiPSCs | HUVEC | hCFs Human adipose stem cells from hiPSCs (hADSCs) | Scaffold-free |
| (Dostanic et al. 2020)45 | 4.7 x 104 cells,3.1 x 104 cells or 1.6 x 104 cells (at 4:1 ratio of CMs:CFs) | hiPSCs* | NA | hiPSCs* | PDMS, acid solubilized collagen I and Matrigel |
| (Giacomelli et al. 2020)11 | 5 x 103 cells at ratio CMs:ECs:CFs of 70:15:15 | hiPSCs* | Cardiac specific ECs from hiPSCs* | Cardiac Fibroblasts from hiPSC-epicardium* | Scaffold free |
Indicates hiPSC from the same donor were used to derive the different cell types.
Cell-cell communication in 3D spatial organization is essential for organ formation and to physiologically mimick their multicellularity20,21.
Most of the designs are based on specific tissue-engineered formats using methods such as SU-8 photolithography22,23, bioprinting24,25, decellularization of whole hearts26, surface micropatterning27–29, biowires23,30, or casting molds as hydrogel (collagen, Matrigel or fibrinogens) scaffolds22,31–47. Engineered heart tissues (EHTs) are grown on mechanical supports or pillars providing mechanical load and have been described to induce hiPSC-CM maturation35,37. One system uses forced supraphysiological pacing up to 6 Hz to facilitate this40. The initial designs48 were low-throughput, time-consuming and costly for both academic laboratories and pharma but miniaturized versions have now been made22,30,34,36,42,45,47.
However, EHTs still require specialized technical skills and often costly equipment, thus fewer independent constructs can be made from a single hPSC-CM batch. Moreover, the thickness of the resulting tissue might limit diffusion of, for example, drugs and nutrients. The cell distribution within the EHTs is not often examined and so that it may be unclear whether this is homogenous (as in vivo) or heterogeneous. Other groups have addressed the shortcomings of engineered systems by including hiPSC-CMs in 3D self-assembled (scaffold-free) structures generally using fewer cells and straightforward techniques15,49–55. For both engineered and scaffold-free systems, some use (only) nonpurified CM populations which contain non-CM cells in undefined numbers22,23,27,28,32,35,39,43,54. Others have developed multi-cell type systems that included purified CM population with specific non-CM cardiac cells but most required primary human cardiac microvascular ECs and/or primary human fibroblasts25,30,31,33,34,37,38,40,41,44,49–53,55. Such cells are by definition limited in supply and can be particularly difficult to obtain from patients. Furthermore we have found that dermal fibroblasts express little or no CX43, which can limit their ability to couple with hiPSC-CMs and hence induce CM maturation11. Thus, whilst skin fibroblasts are readily obtainable and expandable they are not able to support hiPSC-CM maturation in the same way as CFs.
Advantages and applications of our protocol
The self-organizing cardiac MT platform we describe here is scaffold-free, hence is easy to fabricate and does not require advanced tissue engineering or specialized apparatus. It requires only 5,000 input cells per MT. A single operator can generate and process up to 1,500-2,000 MTs in a single run, and the 96-well format is compatible with multiple semi-automatic analysis systems for two-dimensional (2D) screening of CMs (e.g. fluorescent plate readers, FLIPR calcium assays, the Hamamatsu FDSS system). Additionally, cells can be cryopreserved at intermediate differentiation stages, so that they are readily available for MT construction from banked stocks. All the cell types necessary for building MTs can be derived from a single hiPSC source, allowing isogenic disease modelling. By systematically interchanging each of the cell types in the cardiac MTs with an isogenic or mutant variant, it is possible to identify the cell type(s) responsible for a particular disease phenotype. We have also demonstrated that the procedure is highly reproducible across multiple hiPSC lines and differentiated cell batches11 (Table 3). All three cell types survive equally well in basal medium and no not require complex media formulations. Because the system does not require specialized equipment and the differentiated cellular components of the cardiac MTs can be cryopreserved, both the procedure and the cells can be easily transferred and distributed to other laboratories in both academia and pharma, enabling multidisciplinary collaborations. Using a 96-well format, production of thousands of MTs is low cost (± 0.22€ per MT; Table 2). In addition, cardiac MTs as decribed here are entirely hiPSC-derived allowing generation of models for investigating patient-specific multicellular responses in diseases and to pharmacological compounds. For this reason, they may have greater utility than unicellular monolayer cultures of hiPSC-CMs in screening drugs that affect the heart56. Moreover, thanks to the enhanced CM maturation that takes place in MTs, they represent a facile solution to an important caveat of modelling cardiac diseases that manifest in adults (or postnally) with hiPSC-CMs, otherwise typically immature57.
Table 3. Variability table of morphological, contraction and single-cell electrophysiological parameters.
The table reports quantitative parameters showing the expected sample-sample, batchbatch and line-line reproducibility. Size: diameter of MTs mounted on coverslips; IBI: inter-beat interval; RMP: resting membrane potential; APA: action potential amplitude; Vmax: maximal upstroke velocity; APD90: action potential duration at 90% repolarization; SD: standard deviation; CV: coefficient of variation (calculated as SD/mean*100). These data have been previously shown in 11. Raw data are reported in Supplementary Table 1.
| Morphology | Contraction | Single-cell electrophysiology | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Size (μm) | Sarcomere length (μm) | Sarcomere alignment index | IBI (ms) | Contraction duration (normalized) | RMP (mV) | APA (mV) | Vmax (dV/dT) | APD90 (ms) | ||
|
Sample to
sample variability |
mean | 482.4 | 1.78 | 0.027 | 3631.4 | 393.9 | -71.0 | 102.4 | 52.8 | 254.9 |
| SD | 38.3 | 0.06 | 0.004 | 819.3 | 118.1 | 6.7 | 10.7 | 32.3 | 89.2 | |
| CV | 7.8 | 3.1 | 13.4 | 22.1 | 31.8 | 9.4 | 10.5 | 61.5 | 33.8 | |
| Batch to batch variability | mean | 471.4 | 1.78 | 0.027 | 3334.2 | 386.6 | -71.2 | 102.9 | 49.4 | 277.1 |
| SD | 31.0 | 0.02 | 0.002 | 1347.9 | 140.7 | 2.3 | 4.2 | 9.3 | 31.8 | |
| CV | 6.8 | 1.3 | 6.6 | 35.9 | 35.4 | 3.2 | 41 | 16.3 | 10.6 | |
| Line to line variability | mean | 471.4 | 1.78 | 0.026 | 3331.2 | 393.5 | -71.5 | 102.9 | 49.9 | 272.0 |
| SD | 46.7 | 0.004 | 0.000 | 584.2 | 24.4 | 1.1 | 0.8 | 11.8 | 68.6 | |
| CV | 9.9 | 0.2 | 0.02 | 17.5 | 6.2 | 1.5 | 0.8 | 23.7 | 25.2 | |
Table 2.
Estimated costs to generate hiPSC cardiac MTs based on 2019 quotations including discounts for bulk orders of the suppliers indicated in the Materials. The costs can vary per supplier, applied discounts or change with inflation.
| Material/Reagent/Procedure | Cost (€) |
|---|---|
| Cost to generate ~ 4,000,000 hiPSC-CMs (starting from to one well of a 6-well plate of undifferentiated hiPSCs) | |
| Matrigel | 0,4 |
| Growth factors | 1,13 |
| Culture medium | 0,9 |
| Dissociation | 4,32 |
| Extra medium for dissociation and resuspension | 0,6 |
| Extra material: tips, tubes, plates | 2,17 |
| Total | 9,52 |
| Total for 180 microtissues | 1,5 |
| Total for 1 microtissue | < 0,01 |
| Cost to generate ~ 800,000 hiPSC-ECs (starting from to one well of a 6-well plate of undifferentiated hiPSCs) | |
| Matrigel | 0,4 |
| Growth factors | 1,53 |
| Culture medium | 0,36 |
| Kit for enrichment | 26,87 |
| Extra medium for isolation, dissociation, re-suspension, freezing and thawing | 2 |
| Fibronectin for coating | 1,2 |
| CryoStor CS10 medium for freezing | 2,08 |
| TrypLE 1X for dissociation | 0,85 |
| Extra material: tips, tubes, plates | 3,29 |
| Total | 38,58 |
| Total for 180 microtissues | 6,5 |
| Total for 1 microtissue | < 0,04 |
| Cost to generate ~1,200,000 hiPSC-CFs (starting from to one well of a 6-well plate of undifferentiated hiPSCs) | |
| Matrigel | 0,4 |
| Growth factors | 3,95 |
| Culture medium | 1,54 |
| Fibronectin for coating | 2,7 |
| TrypLE 1X for dissociation | 1,7 |
| FGM3 medium | 11,1 |
| CryoStor CS10 medium for freezing | 2,08 |
| Extra FGM3 medium for dissociation, freezing and thawing | 5,55 |
| Extra material: tips, tubes, plates | 4,96 |
| Total | 33,98 |
| Total for 180 microtissues | 3,82 |
| Total for 1 microtissue | < 0,02 |
| Costs to generate and maintain 180 microtissues (corresponding to 3 V-shaped 96-well plates) | |
| Cells | 11,82 |
| Growth factors | 9,44 |
| Culture medium | 2,16 |
| Material: reservoirs, tips, tubes, plates | 15.4 |
| Total (excluding material) | 23,42 |
| Total (including material) | 38,82 |
| Total (excluding material) for 1 microtissue | ~ 0,13 |
| Total (including material) for 1 microtissue | ~ 0,22 |
Immediate applications for the cardiac MT protocol described include modelling multicellular cardiac diseases, drug screening and studies to understand signalling pathways controlling human heart physiology. The multi-cell type model of the heart we describe allows in-depth mechanistic insight into cardiac diseases which may be poorly characterized because the non-myocyte cells in the heart play active roles in the pathogenesis but cannot be distinguished in end-stage outcomes in patients such as heart failure. Contributions of the non-myocyte cell population to (both genetic and environmental) cardiac diseases is becoming increasingly evident, with fibrosis, arrhythmias and electrical conduction defects all thought to have a non-myocyte contribution. To model these kind of diseases, the versatility of the cardiac MT model described allows modification of non-cardiomyocyte cell ratios during generation, that can be defined depending on the specific application. Moreover, MTs with pre-defined ratios of non-cardiomyocytes enable cellular “culprits” and “victims” in cardiac disease to be distinguished. For example, we demonstrated that modelling Arrhythmogenic Cardiomyopathy (ACM) caused by a mutation in plakophilin 2, a protein of the desmosome, only required mutated CFs for arrhythmias to appear in paced cardiac MTs, indicating CFs and not just the CMs contribute to the disease11. Individual CMs and CFs are known to couple and communicate through homo- and heterocellular gap junctions to control electrical activity58 and conduction in the heart59. This is also evident in the cardiac MT model. In addition, CFs appear to play a specific role in CM maturation during development. This is independent of or additional to the maturation that results from the metabolic switch in energy source, from glucose prenatally to fatty-acids postnatally. Together, these cardiac cell combinations and variations in energy sources can provide opportunities to understand heart failure occurring just after birth in patients with inborn errors of metabolism or mitochondrial disease60. Automated handling systems could further increase throughput and thus facilitate testing of thousands of compounds in parallel in cardiac drug discovery and in assessment of cardiac toxicity. We recently demonstrated the utility of MTs to assess cardiotoxic drug effects in a study designed to identify doxorubicin analogues that do not induce late heart failure, in contrast to doxorubicin itself and other anthracycline cancer chemotherapeutics. Human cardiac MTs were able to distinguish the “safe” variants and gave similar outcomes to 3-month studies in mice56. Cardiac MTs enable elucidation of cellcell interactions and could thus show how individual as well as combinations of genetic variants identified from large-scale genetic studies contribute to the pathogenicity of cardiac diseases in which multicellularity plays a role.
Limitations of the protocol
Mechanical load is a physiological stress that helps heart development and maturation. In our system we are able to achieve a certain degree of cardiomyocyte maturation even without mechanical loading; however, adding load to our system could potentially increase maturation even further. Many key functional parameters, such as electrophysiology, calcium handling, contraction amplitude and dynamics, mitochondrial respiration and glycolytic activity can be measured in cardiac MTs. However, the contraction force determined in EHTs cannot be measured in MTs as it requires contraction against a load. Nevertheless, comparison of contraction amplitude measurements using MUSCLEMOTION software61 with the absolute force measurements assessed by pole deflection in EHTs35 previously showed highly significant correlation (R2=0.879) between force of contraction and cell displacement during contraction61. However, this correlation might be influenced by the viscoelasticity of the MTs, which may change substantially with the state of the cardiomyocytes and microenvironment. Measurements of conduction velocity and propagation via optical mapping are also challenging in MTs, due to their small size. However, appropriate camera setups with high sensitivity or light sheet microscopy62 might overcome this issue.
We demonstrated that in cardiac MTs, key ultrastructural features of hiPSC-CMs improve (mitochondria with well-defined cristae, M-bands and T-tubule-like structures) and that maturity was retained upon isolation of the CMs from the MTs with most CMs displaying typical adult action potential “notches”, hyperpolarized resting membrane potential, and increased upstroke velocity11. Nevertheless, the maturation level achieved was not identical in all cells and did not reach that of CMs in the adult heart. Whether contraction against resistance, fluid flow in organ-on-chip formats allowing perfusion and delivery of nutrients, micropatterned surfaces to induce sarcomere alignment or forced and prolonged pacing would promote further maturation or even aging remains to be investigated. As for all hPSC-derivatives described to date, hiPSC-ECs and hiPSC-CFs are also probably immature: RNA-seq data obtained before incorporation in the 3D MTs showed that hiPSC-CFs clustered closely to both human primary fetal- and adult CFs, and that hiPSC-ECs clustered closely to human primary fetal ECs11. We have not investigated whether the MT environment promotes their maturation, and this warrants further analysis. For this reason, we cannot exclude that possible immaturity of hiPSC-ECs and hiPSC-CFs might affect some aspects of disease modelling, for example cardiac fibrosis, which is not driven only by increased number of fibroblasts but also by a their activation into myofibroblasts.
In the heart other cell types contribute to tissue homeostasis and play a role in disease pathology. Further development of this tricelltype microtissue system will undoubtedly benefit from the addition of other cardiac cells, such as mural cells to promote formation and maintenance of microcapillary structures and macrophages, dendritic cells and neurons, which are relevant for disease modelling.
Experimental design
The procedure describes the steps required to produce scaffold-free 3D cardiac MTs composed of hiPSC-CMs, hiPSC-ECs and hiPSC-CFs and how to analyse their structural, functional (electrical and contraction) and metabolic properties.
hiPSC expansion and initial differentiation
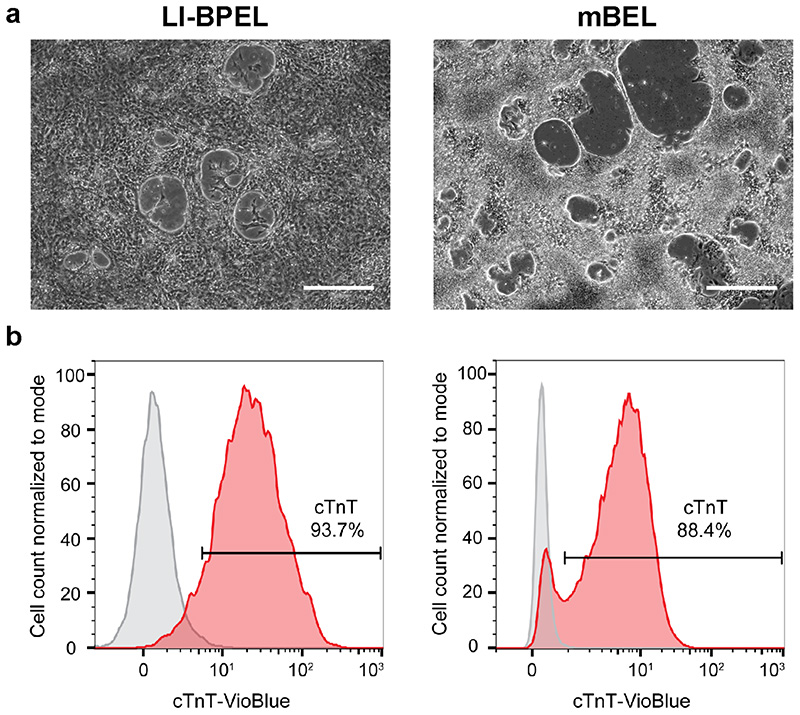
The first part of the procedure describes hiPSC expansion and differentiation into the three hiPSC-derived cardiac cell types. Provided pluripotency of the starting population is high, the hiPSCs can be cultured using various maintenance protocols without affecting differentiation efficiency outcome. The expected results described here were obtained using two independent control hiPSCs lines but we have also successfully applied the protocol to various hiPSC lines for modeling of cardiac diseases (e.g. ACM, long QT syndrome 1)11. The differentiation of CMs, ECs, and CFs starts with induction of cardiac mesoderm, as outlined schematically in Fig. 1. Some optimization of the differentiation protocol may be required for different hiPSC lines, most specifically the starting seeding density. If the optimal seeding density is not pre-determined, variable differentiation efficiencies can result, particularly for CMs. The initial steps of differentiation for the three cell types is carried out in low insulin, BSA-based medium containing polyvinyl alcohol and essential lipids (LI-BPEL medium, see MATERIALS), which was initially optimized for CM differentiation63,64 and subsequently proved efficient for CM and EC co-differentiation15,65. The cytokines, Activin-A and BMP4, together with the Wnt-signalling pathway activator CHIR99021 are added to LI-BPEL for 72 h to direct the differentiation to cardiac mesoderm. Alternatively, CM differentiation can be induced using a modified medium that lacks several of the components present in LI-BPEL, but includes various trace elements66 (mBEL, see Box 1). This circumvents the need to use cytokines as the first induction step requires only CHIR99021 for 48 h.
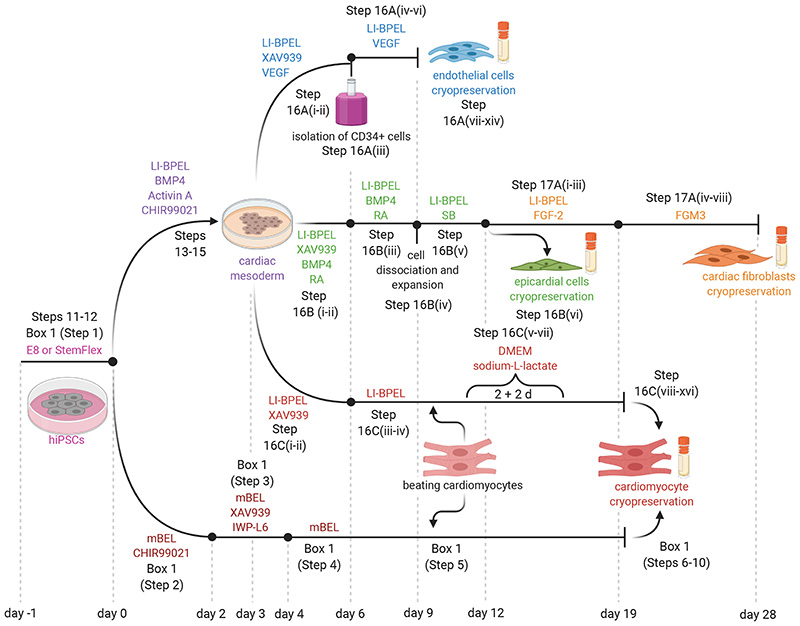
Fig. 1. Outline of in vitro differentiation protocol from hiPSCs to ECs, CFs and CMs.
Schematic of the protocol for differentiation of the three cell types used for MT production with indication of the media and supplements used.
Box 1. Alternative protocol for cardiomyocyte differentiation using small molecules and cryopreservation TIMING 21 d.
This procedure differentiates cardiomyocytes from hiPSCs without requiring growth factors (BMP4 and Activin-A) or a lactate purification step, thereby reducing the cost of generating the hiPSC-CMs. It can thus be used in place of steps 11-15 and 16C, however note that steps 11-15 are still required as part of the process to differentiate cells to CFs and ECs.
Specific differentiation to EC, CF and CMs
Subsequent steps of the procedure are specific for EC, CF and CM differentiation.
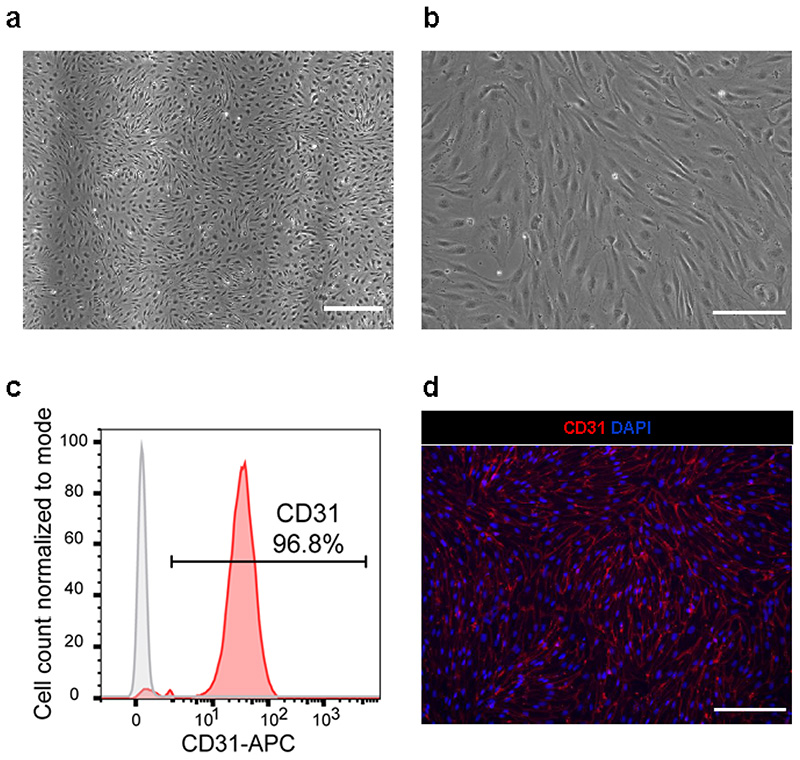
EC differentiation is based on the addition of VEGF and inhibition of the Wnt-pathway with XAV 939 for 72 h. ECs can be cryopreserved once they reach confluence in the plate without affecting their properties. Although ECs do not undergo significant cell death in the freezing/thawing process, they are sensitive to cell contact and will undergo cell cycle withdrawal and death if they become over-confluent in culture. Therefore the density of cell seeding after thawing should take into consideration the time in culture and proliferative capacity of the cells.
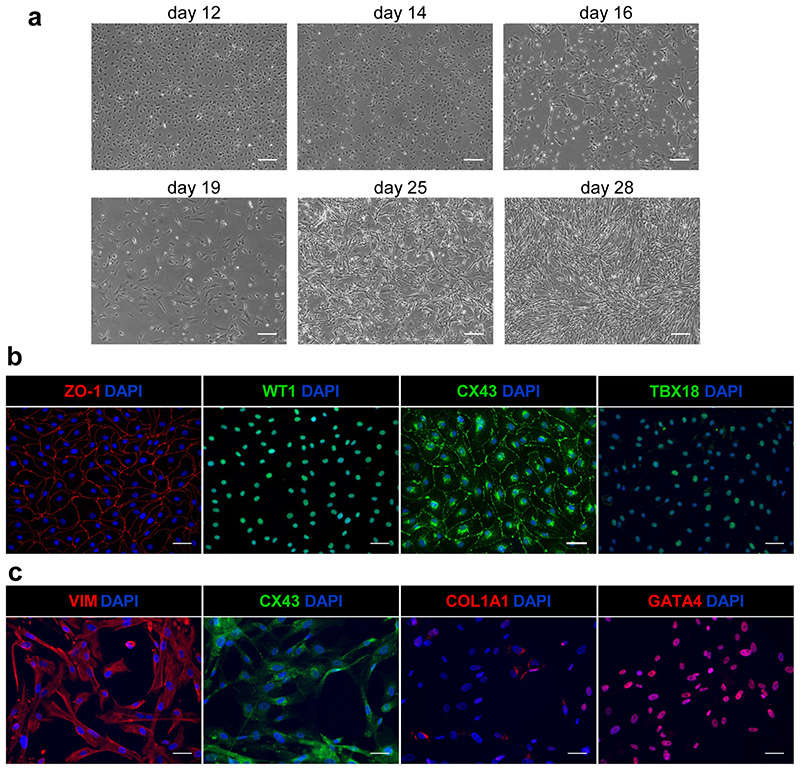
CFs are differentiated from EPI cells that are generated using a protocol we published previously with some modifications17. EPI specification is based on Wnt-inhibition with XAV 939 and addition of BMP4 and retinoic acid (RA) to the basal medium. The differentiation to EPI cells takes 9 d, after which the cells can be expanded and cryopreserved. This creates stocks from which CF differentiation can be initiated and allows large numbers of CFs to be derived from the same EPI batch. CF differentiation is induced by adding basic fibroblast growth factor (FGF-2) to LI-BPEL medium for 5 d. A commercially available medium optimized for fibroblast culture (FGM3 medium, see MATERIALS) is then used. During this step, the proliferation of CFs is slower than, for example, dermal fibroblasts. CFs can be expanded by passaging the culture twice before adapting the cells to grow on uncoated tissue culture plates. CFs can be cryopreserved between passages 2-6. After 7 or more passages, they show reduced proliferation and signs of senescence and should not be used to form MTs.
CMs can be differentiated from cytokine-induced cardiac mesoderm with addition of XAV 93963. Alternatively, the CMs can be differentiated using mBEL medium supplemented with small molecules66 (Box 1), similar to protocols described previously67,68. For the specification to CMs by Wnt-signalling inhibition, the differentiated cells are treated with both XAV 939 and IWP-L6 for 48 h. Small molecules can be useful in improving standardization since there is likely less lot-to-lot variability than with cytokines. Moreover, the use of small molecules is cost-effective compared to cytokines. The small molecule protocol has however only been optimized for CM differentiation and the cytokine protocol should be used if a common cardiac mesoderm for the differentiation of other cell types is required. In general, both CM differentiation protocols are highly efficient and we have seen no differences in CM behaviour in MTs using either method. For hiPSC lines where differentiation efficiencies are poor (< 70% cTnT+ cells), purification based on metabolic selection with lactate can be used to enrich for CMs (> 85-90% cTnT+ cells)67,69. Basic DMEM medium with no glucose or pyruvate but supplemented with lactate supports CM survival only and not that of other glucose-dependent cells69. The CMs can also be cryopreserved without affecting their functional properties66, thereby allowing large cell numbers from the same differentiation batch to be available to form MTs. However, the freeze/thawing procedure does reduce CM recovery to ~30-50%, so this needs to be taken into account for MT formation.
Quality checks of the hiPSC-derivatives are recommended before use in MT formation, as described in the protocol. The methods we describe are based on flow cytometry and immunofluorescence analyses.
MT generation
MTs are generated as embryoid body-like structures by self-aggregation of in total 5,000 of the three cell types in controlled ratios that remain constant over time in culture11. The process is outlined schematically in Fig. 2. MTs can be generated from freshly differentiated cells or cells derived from frozen stocks. Differences in the differentiation protocol for each of the cell types dictates the overall timing of the procedure. Using frozen stocks avoids the need to synchronize differentiation of the three cell types, thereby reducing simultaneous workload and enabling the production of more MTs from the same batches of cell types.
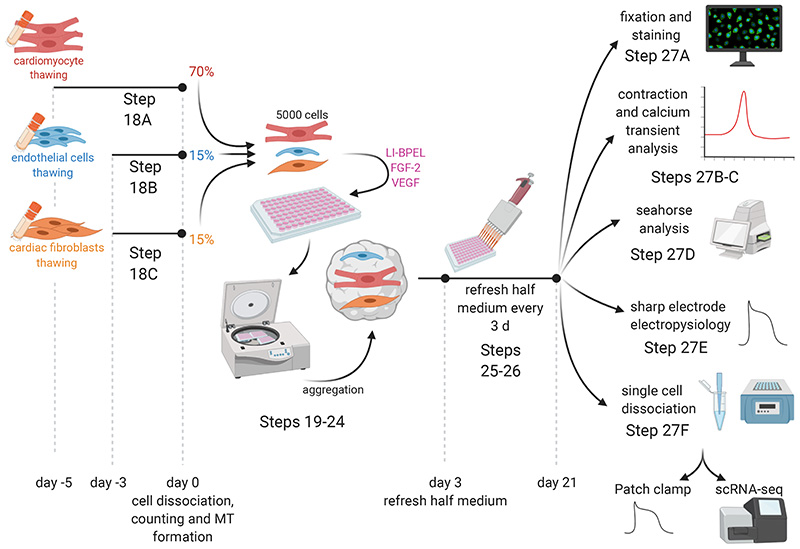
Fig. 2. Outline of in vitro 3D cardiac microtissues (MTs) formation and analysis.
Graphical illustration describing MT generation from frozen stocks of ECs, CFs and CMs and characterization of structural and functional properties using: immunofluorescence, contraction and calcium transient analysis, Seahorse analysis, sharp electrode electrophysiology and single-cell dissociation.
To form MTs, the different hiPSC-cardiac cell types are dissociated, counted and combined in fixed ratios (70:15:15, CMs:ECs:CFs) in homogeneous cell suspension, and a total of 5,000 cells are seeded in each well of V-shaped 96-well plates. Centrifugation is recommended to improve cell self-aggregation and tissue compaction, which is usually completed within 24-48 h. Cells are aggregated in 50 μL LI-BPEL containing VEGF and FGF-2 to support ECs and CFs survival. The MTs start beating spontaneously within 3-4 d of seeding and this is maintained during the entire culture period of 21 d. To refresh the medium, half of the volume is replaced with fresh LI-BPEL containing VEGF and FGF-2 every 3 days.
Analysis of MTs after 21 d in culture showed that maturation of hiPSC-CMs was significantly improved at the structural (immunostaining to analyse sarcomere length and organization) and functional level (video analysis of contraction and calcium transients, metabolic function using Seahorse assays, electrophysiology using sharp electrode recordings). For some assays requiring single-cell analysis, we developed a procedure to dissociate MTs, based on cardiac embryoid body dissociation using collagenase II70. This is also described here. Dissociated cells can be used for example, for single-cell RNA-sequencing (scRNA-seq) or plated for single-cell electrophysiology or immunostaining. These assays allow examination of CM maturation and cell type-specific features like gene expression, structure, functional activity. The process is optimized for the recovery of hiPSC-CMs but CFs are also obtained although in a smaller numbers. The survival of ECs after the dissociation and replating is poor, thus alternative procedures are necessary to recover ECs in sufficient numbers for the majority of downstream analyses. Nonetheless, sufficient numbers of each cell type can be recovered after dissociation from MTs for scRNA-seq analysis11.
Materials
Biological Materials
hiPSC. The results we show here used hiPSC line LUMC0020iCTRL-06 (https://hpscreg.eu/cell-line/LUMCi028-A; RRID:CVCL_ZA25: https://scicrunch.org/resolver/RRID:CVCL_ZA25)11,15,71. This protocol has been used successfully on six hiPSC lines to date. These are: LUMC0020iCTRL06 (female), LUMC0099iCTRL04 (https://hpscreg.eu/cell-line/LUMCi004-A; RRID:CVCL_UK77; https://scicrunch.org/resolver/RRID:CVCL_UK77)11 (female), LUMC0059iCTRL0311 (female), LUMC0021iKCNQ-3011 (female), LUMC0153iPKP0311,72 (female), which were reprogrammed from primary skin fibroblasts using Sendai virus by the LUMC hiPSC core facility, and NCRM111 (RRID:CVCL_1E71; https://scicrunch.org/resolver/ RRID:CVCL_1E71) (male), which was obtained from RUDCR Infinite Biologics at Rutgers University.
! CAUTION The stem cell research protocols described here were approved by the medical ethical committee at Leiden University Medical Center.
! CAUTION All hiPSC lines should be routinely tested to ensure pluripotency and genomic integrity by karyotyping (approximately after 15-20 passages)73,74, and to exclude mycoplasma infection.
! CAUTION Sex, defined of being XX or XY, is an important biologic variable in preclinical research75. As such, the sex of the donors of the hPSC lines used should be considered in research areas where sex differences play a role.
! CAUTION hiPSC lines need to be handled following minimum biosafety 1 level precautions, always wearing personal protection equipment and working in a class 1-2 biosafety cabinet.
Reagents
General reagents
! CAUTION Each reagent should be handled based on information reported in reagent specific material safety data sheets and always using proper personal protective equipment.
! CAUTION Room temperature is considered as 20-25 °C.
-
Essential 8™ (E8™) Medium (Thermo Fisher Scientific, cat. no. A1517001) or StemFlex™ Medium (Thermo Fisher Scientific, cat. no. A3349401)
CRITICAL The medium used to maintain the hiPSC lines is dependent on how the user prefers to passage the cells. If the user prefers to passage the hiPSCs as cell clusters using EDTA, then E8 medium should be used for maintenance. If the user prefers to enzymatically passage as single cells (e.g. with 1x TrypLE Select), then the hiPSCs should be cultured with StemFlex Medium.
Matrigel™ hESC-Qualified Matrix (Corning, cat.no. 354277)
Vitronectin, truncated recombinant human (VTN-N) (Thermo Fisher Scientific, cat.no. A14700
Fibronectin bovine plasma (Sigma-Aldrich, cat.no. F1141)
Laminin (BioLamina, cat. no. LN521)
RevitaCell™ Supplement (100X) (Thermo Fisher Scientific, cat.no. A2644501)
UltraPure™ 0.5 M EDTA, pH 8.0 (Thermo Fisher Scientific, cat.no. 15575020)
TrypLE Select, 10X (Thermo Fisher Scientific, cat.no. A1217701)
Phosphate-buffered Saline with calcium, magnesium (PBS+/+) (Thermo Fisher Scientific, cat. no. 14040-091)
Phosphate-Buffered Saline without calcium and magnesium(PBS-/-) (Thermo Fisher Scientific, cat. no. 14190-169)
Hanks’ Balanced Salt Solution (HBSS) no calcium, no magnesium, no phenol red (Thermo Fisher Scientific, cat. no. 14175-053)
DMEM/F-12, Glutamax supplement (Thermo Fisher Scientific, cat. no. 31331-028)
DMEM, no glucose, no pyruvate (Thermo Fisher Scientific, cat. no. 11966-025)
DMEM/F-12, HEPES (Thermo Fisher Scientific, cat. no. 31330-038)
Distilled water (Thermo Fisher Scientific, cat. no. 15230-089)
Fibroblast Growth Medium 3 (FGM3) (PromoCell, cat. no. C-23025)
CryoStor CS10 medium (Stem Cell Technologies, cat. no. 07930)
Fluo-4, AM, cell permeant (Thermo Fisher Scientific, cat. no. F14201)
-
Powdered DMEM, without glucose, L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate (Sigma-Aldrich, cat. no. D5030)
CRITICAL Phenol red has steroid activity and can affect oestrogen receptors; its inclusion could modify results in a sex-specific way.
D-Glucose (Sigma-Aldrich, cat. No. G8270)
Sodium Pyruvate 100mM (Thermo Fisher Scientific, cat. no. 11360070)
L-Glutamine 200mM (Thermo Fisher Scientific, cat. no. 25030024)
Oligomycin (Sigma-Aldrich, cat. no. O4876)
Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma-Aldrich, cat. no. C2920)
Antimycin A (Sigma-Aldrich, cat. no. A8674)
Rotenone (Sigma-Aldrich, cat. no. R8875)
2-Deoxy-D-glucose (2-DG) (Sigma-Aldrich, cat. no. D8375)
EasySep™ Human CD34 Positive Selection Kit II (Stem Cell Technologies, cat. no. 17856)
-
Multi Tissue Dissociation Kit 3 (Miltenyi Biotec, cat. no. 130-110-204)
CRITICAL The dissociation of hiPSC-CMs using Miltenyi Biotec Multi Tissue Dissociation Kit 3 is crucial for MT generation protocol to ensure reproducible single cell suspensions of CMs without affecting their viability.
DNAse Vial (D2) (Worthington-Biochem, cat. no. LK003170)
Quant-iT™ PicoGreen™ dsDNA Assay Kit (Thermo Fisher Scientific, cat. no. P7589)
-
Paraformaldehyde (PFA) (Merck, cat. no. 104005)
! CAUTION Paraformaldehyde is a flammable solid, carcinogen, irritant (skin, eye, and respiratory tract), toxic (by skin contact and inhalation). It can be stored in a flammable storage cabinet. During work, avoid contact and inhalation. Wear protective gloves/protective clothing/eye protection/face protection.
-
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
! CAUTION Triton X-100 can cause acute oral toxicity and serious eye damage/eye irritation. For preventation, wash face, hands and any exposed skin thoroughly after handling. Do not eat, drink or smoke when using this product. Wear protective gloves/protective clothing/eye protection/face protection.
Bovine Serum Albumin BSA (Sigma-Aldrich, cat. no. A3311)
Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, cat. no. 10270-106)
Fixation Medium (Reagent A) (Thermo Fisher Scientific, cat. no. GAS001S5)
Permeabilization Medium (Reagent B) (Thermo Fisher Scientific, cat. no. GAS002S5)
Sodium-L-Lactate (Sigma-Aldrich cat. no. 71718)
-
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, cat. no. D2650)
! CAUTION DMSO is flammable. Keep away from heat/sparks/open flames/hot surfaces. During work, wear protective gloves/protective clothes/eye protection/face protection.
KnockOut™ Serum Replacement (KOSR) (Thermo Fisher Scientific, cat. no. 10828028)
Trypan Blue Stain solution (0.4%) (Thermo Fisher Scientific, cat. no. T10282)
4’,6-Diamidino-2-Phenylindole, Dilactate (DAPI nucleic acid stain) (Thermo Fisher Scientific, cat. no. D3571)
ProLong™ Gold Antifade Mountant (Thermo Fisher Scientific, cat. no. P36930)
Iscove’s Modified Dulbecco’s Medium (IMDM) (Thermo Fisher Scientific, cat. no. 21056-023)
F12 nutrient mixture (Ham) (Thermo Fisher Scientific, cat. no. 31765-027)
Glutamax (Thermo Fisher Scientific, cat. no. 35050-038)
Penicillin/Streptomycin (Thermo Fisher Scientific, cat. no. 15070-063)
Phenol Red (Sigma-Aldrich, cat. no. P3532)
Protein-free Hybridoma Medium-II (PFHMII) (Thermo Fisher Scientific, cat. no. 12040-077)
Bovine Serum Albumin (BSA) (Bovogen Biologicals Australia, cat. no. BSAS05)
Poly(Vinyl Alcohol) (PVA) (Sigma-Aldrich, cat. no. P8136)
Chemically-defined Lipid Concentrate (CDLC) (Thermo Fisher Scientific, cat. no. 11905-031)
Insulin-Transferrin-Selenium Ethanolamine (ITS-X) 100X (Thermo Fisher Scientific, cat. no. 51599-056)
L-ascorbic acid 2-phosphate (Sigma-Aldrich, cat. no. A8960)
α-Monothioglycerol (Sigma-Aldrich, cat. no. M6145)
Recombinant proteins and small molecules
Collagenase type II (Worthington, cat. no. LS004176)
Recombinant Human BMP-4 Protein (R&D Systems, cat. no. 314-BP)
Human Activin A, premium grade (Miltenyi Biotec, cat. no. 130-115-010)
CHIR 99021 (Axon Medchem, cat. no. Axon1386)
XAV 939 (Tocris, cat. no. 3748/10)
Human VEGF, premium grade (Miltenyi Biotec, cat. no. 130-109-386)
Retinoic Acid (Sigma-Aldrich, cat. no. R2625)
TGFβ inhibitor, SB431542 (Tocris, cat. no. 1614/10)
Human FGF-2, premium grade (Miltenyi Biotec, cat. no. 130-093-842)
Primary antibodies
Anti-Troponin I antibody (H-170) (TNNI) (1:500 dilution; Santa Cruz Biotechnology, cat. no. sc-15368; https://scicrunch.org/resolver/RRID:AB_793465) used for immunofluorescence
Anti-alpha-Actinin (Sarcomeric) antibody (ACTN2) (1:800 dilution; Sigma-Aldrich, cat. no. A7811; https://scicrunch.org/resolver/RRID:AB_476766) used for immunofluorescence
Anti-CD31/PECAM-1 antibody (1:200 dilution; R&D Systems, cat. no. AF806; https://scicrunch.org/resolver/RRID:AB_355617) used for immunofluorescence
Anti-Collagen Type I, clone 5D8-G9 antibody (COL1A1) (1:200 dilution; Millipore, cat. no. MAB3391; https://scicrunch.org/resolver/RRID:AB_94839) used for immunofluorescence
Anti-Wilm’s Tumor Protein antibody (WT1) (1:200 dilution; Millipore, cat. no. CA1026; https://scicrunch.org/resolver/RRID:AB_437848) used for immunofluorescence
Anti-TBX18 antibody (TBX18) (1:200 dilution; Sigma-Aldrich, cat. no. HPA029014; https://scicrunch.org/resolver/RRID:AB_10601597) used for immunofluorescence
Anti-ZO-1 antibody (ZO-1) (1:200 dilution; Thermo Fisher Scientific, cat. no. 33-9100; https://scicrunch.org/resolver/RRID:AB_87181) used for immunofluorescence
Anti-Connexin 43/GJA1 antibody (CX43) (1:200 dilution; Abcam, cat. no. ab11370; https://scicrunch.org/resolver/RRID:AB_297976) used for immunofluorescence
Anti-Vimentin, clone V9 antibody (VIM) (1:100 dilution, Sigma-Aldrich, cat. no. V6630; https://scicrunch.org/resolver/RRID:AB_477627) used for immunofluorescence
Anti-GATA4 (1:100 dilution, Santa Cruz Biotechnology, cat. no. sc-25310; https://scicrunch.org/resolver/RRID:AB_627667)
Anti-cardiac Troponin T (cTnT) antibody (clone REA400)-VioBlue(1:50 dilution; Miltenyi Biotec, cat. no. 130-120-402; https://scicrunch.org/resolver/RRID:AB_2783891) used for flow cytometry
REA Control Antibody (I), human IgG1 (clone REA293)-VioBlue (1:50 dilution; Miltenyi Biotec, 130-108-346, https://scicrunch.org/resolver/RRID:AB_2661680) used for flow cytometry
CD31 (PECAM-1) antibody (clone WM59)-APC (1:100 dilution, eBioscience, cat. no. 17-0319-42, https://scicrunch.org/resolver/RRID:AB_10852842) used for flow citometry
IgG1 kappa Isotype Control (clone P3.6.2.8.1)-APC (1:100 dilution, eBioscience, cat. no. 17-4714-42, https://scicrunch.org/resolver/RRID:AB_1603315) used for flow citometry
Secondary antibodies
Cy3-AffiniPure Donkey Anti-Mouse IgG (H+L) antibody (1:100 dilution; Jackson ImmunoResearch Labs, cat. no. 715-165-150; https://scicrunch.org/resolver/RRID:AB_2340813) used for immunofluorescence
-
Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:200 dilution; Thermo Fisher Scientific, cat. no. A-21206;
https://scicrunch.org/resolver/RRID:AB_2535792) used for immunofluorescence
Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (1:200 dilution; Thermo Fisher Scientific, cat. no. A-21203; https://scicrunch.org/resolver/RRID:AB_2535789) used for immunofluorescence
Donkey Anti-Sheep IgG (H+L) Antibody, Alexa Fluor 647 (1:200 dilution; Thermo Fisher Scientific, cat. no. A21448; https://scicrunch.org/resolver/RRID:AB_10374882) used for immunofluorescence
Equipment
6-well cell culture plates (Greiner Bio-one, cat. no. 657 160)
12-well cell culture plates (Greiner Bio-one, cat. no. 665 180)
24-well cell culture plates (Greiner Bio-one, cat. no. 662 160)
96-well black microplates (Corning Falcon 96-Well, cat. no. 353219)
96-well V-bottom microplates (Greiner Bio-one, cat. no. 651161)
5-, 10-, and 25-mL sterile plastic serological pipettes (Greiner Bio-One, cat. no. 606180, 607180, and 760180, respectively)
15- and 50-mL polystyrene conical tubes (Corning Falcon, cat. no. 352097 and 352098, respectively)
1.5-mL microcentrifuge tubes (Eppendorf, cat. no. 0030 120.086)
Flow cytometry tubes with cell strainer snap cap (Corning Falcon, cat. no. 352235)
10-, 200-, and 1000-μL filter tips (Corning, cat. no. 4807, 4810, and 4809, respectively)
12-mm, round, glass coverslips (Menzel-Glaser, cat. no. CB00140RA1)
Vacuum filter/storage bottle system (0.22 μm pore size) (Millipore, cat. no. S2GVU05RE)
Multichannel pipetting reservoirs (VWR, cat. no. 613-1175)
Cryovials (Greiner Bio-One, cat. no. 123263)
Fuchs-Rosenthal counting chamber (VWR, cat. no. 15170-230)
Parafilm (Sigma-Aldrich, cat. no. P7793)
Handheld pipetman (P-10 (10 μL), P-200 (200 μL), P-1000 (1,000 μL) (Gilson international, cat. no. 4807, 4810, and 4809, respectively)
Multichannel, 12-channel 5-50 ul (Finnpipette, cat. no. 4610130)
Nalgene® Mr. Frosty freezing container (Sigma-Aldrich, cat. no. C1562-1EA)
Water bath (Julabo TW20; MCO-18AIC, cat. no. 08010017)
Milli-Q® Water Purification System (Millipore, cat. no. C79625)
Biological safety cabinet/laminar flow-hood (CleanAir)
CO2 cell-culture incubator (Sanyo, cat. no. MCO-15A)
Centrifuge (Eppendorf, cat. no. 5810R)
Seahorse XFe96 Analyzer (Agilent)
Seahorse XFe96 FluxPak (Agilent cat. no. 102601-100)
WTW Lab pH meter inoLab pH 7110 (Xylem analytics cat. No. 1AA11O)
Leica MZ6 stereomicroscope (Leica Microsystems)
Glass pasteur capillary pipettes 150mm (Wilhelm Ulbrich cat. No. 10150)
250 μL pipette tips ultra-low attachment (Mettler Toledo cat. no. 17007958)
MACSQuant®VYB flow cytometry (Miltenyi Biotech, cat. no. 130-096-116)
Nikon Eclipse Ti inverted microscope (or equivalent commercially available device, preferably for phase contrast microscopy) connected to a Nikon DS- 2MBW camera (Nikon) or connected to ThorLabsDCC3240M camera
EVOS M7000 Imaging System (Thermo Fisher Scientific)
LAS AF6000 fluorescent microscope (Leica Microsystems)
SP8WLL fluorescent confocal laser-scanning microscope equipped with a violet (405-nm), blue (488-nm), and orange (532-nm) laser (Leica Mycrosystems)
Thin walled, glass capillaries with filament (World Precision Instruments, cat. no. TW150F-3)
Micropipette puller P-97 (Sutter Instrument)
Software
Fiji-ImageJ (Schindelin, J. et al., 2012 PMID: 22743772) (ImageJ: https://imagej.net/Fiji/Downloads)
FlowJo (FlowJo: https://www.flowjo.com/)
GraphPad Prism 8.2.0 (GraphPad: https://www.graphpad.com/scientific-software/prism/)
-
MUSCLEMOTION (Sala, L. et al., 2018 PMID: 29282212)
(https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.117.312067). MUSCLEMOTION is freely accessible on the web at https://gitlab.com/bjvanmeer/MUSCLEMOTION or https://github.com/l-sala/MUSCLEMOTION/. After downloaging the macro, users can access the code through a text editor or directly through FIJI. For more details, see CODE AVAILABILITY STATEMENT.
Rstudio (RStudio: https://rstudio.com/products/rstudio)
Biorender (https://biorender.com/)
Reagent Setup
Undifferentiated hiPSC culture media
Either E8™ or StemFlex™ medium can be used.
E8™ medium reconstitution: mix E8™ basal medium together with 50X E8™ supplement and 0.5% penicillin-streptomycin.
StemFlex™ reconstitution: mix StemFlex™ basal medium together with 10X StemFlex™ supplement and 0.5% penicillin-streptomycin.
Filter-sterilize the media by passing it through a 0.22-μm filter. Store the media at 4 °C for up to 2 weeks. Following reconstitution, 40-mL aliquots can be prepared and stored at −20 °C for longer period (up to 6 months).
CRITICAL Thaw E8™ and StemFlex™ supplements for 1-2 h at room temperature or overnight at 4 °C. Do not thaw both frozen supplements at 37° C.
CRITICAL Before use, warm the media at room temperature. Do not warm the medium in the water bath at 37° C.
HiPSC dilution medium preparation
HiPSC dilution medium consists of DMEM-F-12, HEPES containing 0.5% BSA. The hiPSC dilution medium can be stored at 4 °C for up to 1 month.
Freezing medium for hiPSC-derived cardiomyocytes (hiPSC-CMs)
1X freezing medium for hiPSC-CMs is composed of 90% (vol/vol) KOSR and 10% (vol/vol) DMSO. The freezing medium can be stored at 4 °C for up to 1 week.
0.5 mM EDTA solution
Dilute 0.5 M EDTA solution in PBS-/- to make 0.5 mM solution (dilution 1:1,000). Store the diluted solution at room temperature for up to 6 months.
1X TrypLE Select solution
Dilute TrypLE Select 10X solution in PBS-/- to make 1X (dilution 1:10) diluted TrypLE Select solution. Store the diluted solution at room temperature for up to 6 months.
Vitronectin (VTN) stock
Thaw the vial of VTN at room temperature. Prepare 60-μL aliquots of VTN and store them at −80 °C.
Laminin stock
Thaw laminin overnight at 4° C. Prepare 1-mL aliquots of undiluted Laminin (100 μg/mL) and store them at at 4° C for up to 3 months under aseptic conditions. Laminin aliquots can be stored at −80 °C for longer period.
CRITICAL Repeated freeze-thaw cycles should be avoided.
Matrigel™ hESC-Qualified Matrix stock
Thaw the bottle of Matrigel overnight at 4° C. Prepare 50- and 100-μL aliquots (containing 0.5 mg and 1 mg of protein, respectively) and store them at −20 °C.
CRITICAL During the entire procedure, it is necessary to always work on ice and use cold tips since Matrigel quickly polymerizes at room temperature. Repeated freeze-thaw cycles should be avoided.
Cardiac fibroblast culture medium (FGM3)
Reconstitute the Fibroblast Growth Medium 3 by mixing the FGM3 basal medium together with the 10X Supplement Mix and 0.5% penicillin-streptomycin. Store the complete medium at 4 °C for up to 6 weeks or prepare 40-mL aliquots and store them at −20 °C for longer period.
CRITICAL Thaw Supplement mix for 1-2 h at room temperature or overnight at 4 °C. Do not thaw supplements at 37 °C.
Seahorse metabolic flux stock medium
Reconstitute 1 bottle of DMEM no glucose powder (sufficient for 1 litre DMEM) in 800 mL Milli-Q water and rinse the stock bottle. Place a large stirrer and mix until all the powder is dissolved. Measure the pH of the reconstituted medium and titrate with 1 M NaOH until a pH of 7.4 is reached. Add Milli-Q water up to 1 L and sterile filtrate using a 0.22 μm filter. This stock medium can be stored at 4 °C for 1 year.
Seahorse metabolic flux assay medium
Two different media are used during the assay, one including glucose to measure the mitochondrial oxidative potential, called SH+ medium, and one without glucose to measure the glycolytic capacity of the MTs, called SH- medium. For SH- medium, take 49 mL of the Seahorse stock medium and add 500 μL of both 200mM L-Glutamine and 100mM sodium pyruvate. For SH+, medium take 48.25 mL of seahorse stock medium and add 750 μL of 1 M D-glucose and 500 μL of 200 mM L-Glutamine and 500 μL 100 mM sodium pyruvate. This will result in media containing 1 mM pyruvate, 2 mM L-glutamine and in the SH+ medium also 15 mM D-Glucose. Warm the medium to 37 °C in a water bath and subsequently adjust the pH to 7.4 with 1 M NaOH. Filter-sterilize the medium through a 0.22 μm filter. Take 35 mL of both media and add to their own tube. To this add 875 μL 8% (wt/vol) BSA, this will yield the media used for washing the MTs prior to plating them, called Wash+ and Wash-.
Fixative solution
Dilute 8% (wt/vol) PFA in 0.2 M phosphate buffer, pH 7.4 in order to make 4% (vol/vol) PFA solution. Store the solution at 4 °C up to 1 month.
! CAUTION PFA is carcinogen and it causes skin, eye and respiratory tract irritation. Always wear the appropriate personal protection equipment and work under a fume hood.
Permeabilization solution for immunostaining on 2D cells
Add 50 μL of Triton X-100 to 50 mL of PBS+/+ in order to make 0.1% (vol/vol) Triton X-100 solution. Store the solution up to 3 months at 4 °C.
Permeabilization solution for immunostaining on 3D cardiac MTs
Add 100 μL of Triton X-100 to 50 mL of PBS+/+ in order to make 0.2% (vol/vol) Triton X-100 solution. Store the solution up to 3 months at 4 °C.
Blocking solution for immunostaining on 2D cells and 3D cardiac MTs
Prepare blocking solution for 3D MTs with PBS+/+ containing 10% (vol/vol) FBS. Store the solution up to 1 week at 4 °C.
CM dissociation enzyme mix (Multi Tissue Dissociation Kit 3)
To prepare CM dissociation enzyme mix, add Enzyme T to Buffer X of the Multi Tissue Dissociation Kit 3 in a ratio of 1:10. Prepare fresh each time and use immediately.
CRITICAL Upon receipt of the kit, immediately prepare 500-μL and 1-mL aliquots and store them at −20 °C up to 6 months. Avoid repeated freeze-thaw cycles of the aliquots.
DNAse solution
Dissolve in PBS to a final concentration of 1000 units/mL. Add to CM dissociation enzyme mix to a final concentration of 100 units/mL.
Collagenase type II enzymatic solution for cardiac microtissue dissociation
Dissolve Collagenase type II in HBSS solution in order to make 290U/mg solution. Filter-sterilize the medium by passing through a 0.22-μm filter. Prepare fresh each time, pre-warm the enzymatic solution at 37 °C and use it immediately.
BMP4 stock solution
Reconstitute BMP4 powder at 100 μg/mL in sterile 4 mM HCl 0.1% BSA solution and then dilute in sterile 0.1% BSA in PBS-/- to make BMP4 stock at 25 μg/mL. Prepare 30-μL aliquots of BMP4 and store them at −80 °C.
CRITICAL Thaw BMP4 aliquot on ice and spin down before using. Store the residual aliquot up to 7 d at 4 °C. Avoid freeze-thaw cycles.
Activin A stock solution
Reconstitute Activin A powder at 100 μg/mL in distilled sterile-filtered water and then proceed with further a dilution at 25 μg/mL in sterile 0.1% BSA in PBS-/-. Prepare 30-μL aliquots of Activin A and store them at −80 °C.
CRITICAL Thaw Activin A aliquot on ice and spin down before using. Store the residual aliquot up to 7 d at 4 °C. Avoid freeze-thaw cycles.
CHIR99021 stock solution
Dissolve CHIR99021 at 4mM stock solution in DMSO. Prepare 50- and 100- μL aliquots and store them at −20 °C.
CRITICAL Thaw CHIR99021 aliquot at room temperature, protecting the aliquot from light. Before using, spin it down. After thawing, store the aliquot up to 10 d at 4 °C.
XAV 939 stock solution
Dissolve XAV 939 at 5mM stock solution in DMSO. Prepare 100-μL aliquots and store them at −20 °C. CRITICAL Thaw XAV 939 aliquot at room temperature and spin it down, before using. After thawing, store the aliquot up to 10 d at 4 °C.
VEGF stock solution
Reconstitute VEGF powder at 100 μg/mL in distilled sterile-filtered water and then proceed with further a dilution at 50 μg/mL sterile in 0.1% BSA in PBS-/-. Prepare 50-μL and 100- μL aliquots of VEGF and store them at −80 °C.
CRITICAL Thaw VEGF aliquot on ice and spin down before using. Store the residual aliquot up to 7 d at 4 °C. Avoid freeze-thaw cycles.
FGF-2 stock solution
Reconstitute FGF-2 powder at 100 μg/mL in distilled sterile-filtered water and then proceed with further a dilution at 10 μg/mL sterile in 0.1% BSA in PBS-/-. Prepare 50-μL aliquots of FGF-2 and store them at −80 °C.
CRITICAL Thaw FGF-2 aliquot on ice and spin down before using. Store the residual aliquot up to 7 d at 4 °C. Avoid freeze-thaw cycles.
SB431542 (TGFβ inhibitor) stock solution
Dissolve SB431542 at 20 mM stock solution in DMSO. Prepare 50-μL aliquots and store them at −20 °C. CRITICAL Thaw SB431542 at room temperature and spin it down, before using. After thawing, store the aliquot up to 10 d at 4 °C.
RA stock solution
Dissolve RA at 100 μM stock solution in DMSO. Prepare 50-μL and 100-μL aliquots and store them at −20 °C.
CRITICAL Thaw RA at room temperature and spin it down, before using. After thawing, discard the leftover.
CRITICAL RA is sensitive to air and light. Always protect it from light, switching off the light of the safety cabinet.
Sodium-L-Lactate stock solution
Dissolve Sodium-L-Lactate at 100 mM in DMEM no glucose, no pyruvate. Prepare 500-μL and 1-mL aliquots and store them at −20 °C.
Phenol Red solution for LI-BPEL medium
Dissolve 1 mg of Phenol Red in 25 mL of IMDM medium, vortex to properly mix and incubate at 37 °C in a water bath for 10 min. Then, use it immediately for LI-BPEL medium preparation.
Bovine Serum Albumin (BSA) stock solution for LI-BPEL medium
Dissolve 10% (wt/vol) BSA (Bovogen Biologicals Australia, cat. no. BSAS05) in IMDM medium. Place the tube(s) on a rollerbank at 4 °C for 2 h to properly ensure BSA is totally dissolved. Filter-sterilize BSA stock solution by passing it through a 0.22-μm filter and store it at 4 °C up to 1 month.
Polyvinyl Alcohol (PVA) stock solution for LI-BPEL medium
Dissolve 5% (wt/vol) PVA in distilled sterile-filtered water and place the tube(s) on a rollerbank at least overnight (up to 2 d) at 4 °C. Incubate PVA solution at 75 °C for 10-15 min, vortexing in between. Once PVA solution is completely dissolved, store it at 4 °C up to 1 month.
CRITICAL PVA is hard to dissolve. Following these instructions is essential to ensure proper PVA dissolution.
L-ascorbic acid 2-phosphate (AA2P) stock solution for LI-BPEL medium
Dissolve AA2P at 5 mg/mL stock solution in distilled sterile-filtered water. Prepare 1-mL aliquots and store them at −20 °C.
Low insulin BSA-based, Polyvinyl Alcohol, Essential Lipids (LI-BPEL) medium preparation
To prepare 100 mL of LI-BPEL medium, combine all the components/supplements as listed in the table below. Filter the medium using a 0.22-μm filter system and store the medium at 4 °C for up to 2 weeks.
| Component | Quantity for 100 mL |
|---|---|
| IMDM | 43 mL |
| F12 | 43 mL |
| Protein Free Hybridoma Medium-II (PFHMII) | 5 mL |
| BSA (10% wt/vol in IMDM) | 2.5 mL |
| PVA (5% wt/vol in distilled water) | 2.5 mL |
| Chemically Defined Lipid Concentrate (CDLC) 100X | 1 mL |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) 1000X | 0.1 mL |
| α-Monothioglycerol (13 μL in 1 mL IMDM) | 0.3 mL |
| L-ascorbic acid 2-phosphate (5 mg/mL in distilled water) | 1 mL |
| Glutamax | 1 mL |
| Penicillin-streptomycin | 0.5 mL |
| Phenol red | 1mg |
FACS buffer
FACS buffer consists of PBS-/- containing 2% (vol/vol) FBS and 2 mM EDTA. Store at 4 °C for up to 3 months.
Bovine Serum Albumin (BSA) stock solution for Seahorse medium
Dissolve 8% (wt/vol) BSA (Bovogen Biologicals Australia, cat. no. BSAS05) in Seahorse stock medium. Place the tube(s) on a roller bank at 4 °C for 2 hours to properly ensure BSA is dissolved. Filter-sterilize BSA stock solution by passing it through a 0.22-μm filter and store it at 4 °C up to 1 month or in smaller stocks at −20 °C for up to a year.
Oligomycin stock solution
Dissolve the entire 5 mg content of the Oligomycin bottle in 2.52 mL of DMSO. This will result in a 2.5 mM stock which can be frozen in 50 μL stocks at −20 °C.
FCCP stock solution
Add 1.3 mL DMSO to 10 mg of FCCP to achieve a concentrated superstock solution of 30 mM. This can be stored at −20 °C. Regular stocks are made by further dilution of the superstock by 10X. These stocks can be frozen in 40 μL aliquots at −20 °C.
Antimycin-A stock solution
Weigh 25 mg of Antimycin-A and add 4.5 mL of Ethanol (DMSO can also be used if preferred) to make a 10 mM superstock which can be stored at −20 °C. Regular stocks are made by further dilution of the superstock by 10X. These stocks can be frozen in 60 μL aliquots at −20 °C.
Rotenone stock solution
Weigh 10 mg of Rotenone and add 2.5 mL of DMSO to make a 10 mM superstock which can be stored at −20 °C. Regular stocks are made by further dilution of the superstock by 10X. These stocks can be frozen in 60 μL aliquots at −20 °C.
2-DG stock solution
Dissolve 5 g of 2-DG powder in 15.15 mL of seahorse stock medium. This will make a 2 M solution at close to max solubility. If small clumps remain a short period of heating at 37 °C is sufficient to dissolve all the powder. Subsequently, freeze in small aliquots of 1 mL at −20 °C.
D-Glucose stock solution
Weigh 1.8 g of D-glucose powder and add it to 10 mL of seahorse stock medium. Freeze in aliquots of 1 mL at −20 °C.
Fluo-4 AM stock solution
Reconstitute 50 μg of Fluo-4 AM in 10 μL of DMSO to make a stock solution of 5 mM, mixing thoroughly to ensure it is completely dissolved. Store Fluo-4 AM stock solution up to 1 week at 4 °C.
CRITICAL Protect the solution from light to avoid bleaching of the fluorescent dye.
Fluo-4 AM loading solution
Dilute 5 mM Fluo-4 AM stock solution in LI-BPEL medium to make a 5 μM Fluo-4 AM loading solution (dilution 1:1,000). Vortex briefly the diluted solution to ensure homogenous distribution of the fluorescent dye. Prepare fresh each time and use immediately.
CRITICAL Protect the solution from light to avoid bleaching of the fluorescent dye.
Equipment Setup
VTN-coated plates
Remove an aliquot of VTN from −80 °C and thaw it at room temperature. Dilute VTN in sterile PBS-/- (dilution 1:100) at room temperature, gently pipetting the VTN dilution up and down. Coat the growth surface of the plates, using 1 mL per 10-cm2, in order to obtain a final concentration of 0.5 μg/cm2. Incubate the plates at room temperature for 1 h and use them immediately or store at 4 °C for up to 2 weeks. Before using the coated plates, pre-warm them at room temperature for at least 30 min.
CRITICAL Do not allow the coated surface to dehydrate as this will not support cell growth. Prior to storing at 4 °C, top up the plates with additional PBS-/- and seal with parafilm (to prevent evaporation and contamination).
Laminin-coated plates
Remove an aliquot of laminin from −80 °C and thaw slowly at 4 °C. Dilute the laminin stock (100 μg/mL) to a final concentration of 5μg/mL in sterile PBS+/+ (dilution 1:20) at room temperature. Coat the growth surface of the plates (1 mL per 10-cm2) and incubate the coated plates for a minimum of 2 h at 37 °C or overnight at 4 °C for a more reliable coating. Store the plates for up to 4 weeks at 4 °C, adding extra PBS+/+ and sealing with parafilm to prevent the plates from drying out.
CRITICAL The use of PBS+/+ is crucial for proper coating since divalent cations are important for the protein structure and function.
Matrigel-coated plates
Thaw Matrigel aliquot on ice and dilute it in ice-cold DMEM/F12 (dilution 1:120). Coat the growth surface of the plates (1 mL per 10-cm2) and incubate them at room temperature for 1 h. Use them immediately or top up with extra DMEM/F12 and store them at 4 °C up to 2 weeks.
CRITICAL During the entire procedure, keep Matrigel and DMEM/F12 on ice and use prechilled conical tubes and pipette tips, in order to prevent Matrigel polymerization.
Fibronectin-coated plates
Dilute fibronectin in sterile PBS-/- at final concentration of 5μg/mL (dilution 1:200) at room temperature and coat the growth surface of the plates (1 mL per 10-cm2). Incubate the plates at room temperature for 1 h, then use them immediately or add extra PBS without calcium and magnesium, seal with parafilm and store at 4 °C up to 2 weeks.
Cell culture incubators for Seahorse experiments
Set cell culture incubators to 37 °C and 0% CO2 for incubation of the cartridge and cardiac microtissues prior Seahorse experiments.
Seahorse assay Matrigel-coated plates
Thaw Matrigel aliquot on ice and dilute it in ice-cold DMEM/F12 (dilution 1:60). Add 80 μL to each well of the seahorse tissue culture plate and incubate at room temperature for 1 h. The plate can be used immediately or stored at 4 °C for up to 2 weeks.
CRITICAL During the entire procedure, keep Matrigel and DMEM/F12 on ice and use prechilled conical tubes and pipette tips, to prevent Matrigel polymerization.
Procedure
Thawing and expansion of undifferentiated hiPSCs TIMING 10 min plus 3-4 d for expansion before passaging hiPSCs
Prepare a 15-mL conical tube filled with 5 mL of hiPSC medium (E8 or StemFlex), previously equilibrated at room temperature.
Remove the cryovial of hiPSCs from liquid nitrogen storage tank and transfer it on dry ice or in a portable liquid nitrogen container to the cell culture room.
-
Immerse the cryovial in a 37 °C water bath or incubate in a 37 °C, 5% CO2 cell culture incubator and swirl it gently, until only an ~2 mm3 ice crystal remains.
CRITICAL STEP To prevent contamination, do not submerge the cap of the cryovial.
Remove the cryovial from the water bath or from the 37 °C, 5% CO2 cell culture incubator, spray with 70% ethanol and place it into the biological safety cabinet.
-
Gently transfer the content of the cryovial into the 15-mL conical tube containing the hiPSC medium (E8 or StemFlex). Rinse the vial with additional 1 mL of hiPSC medium to recover any residual cells and add it into the same 15-mL conical tube.
CRITICAL STEP Transfer the content of the cryovial drop-wise to minimize osmotic shock
Centrifuge the cells at 300g for 3 min at room temperature, aspirate and discard the supernatant. Resuspend the cell pellet in the desired volume of hiPSC medium supplemented with RevitaCell (1:200), by gently pipetting the cells up and down a few times.
Dispense the cell suspension onto VTN or laminin-coated plate (for cells to be incubated with E8 or StemFlex, respectively) and place it gently into a 37 °C, 5% CO2 cell culture incubator, moving the plate side-to-side and back-and-forth to ensure a homogenous distribution of the cells.
Refresh the medium (2 mL per 10-cm2 growth surface) after 24 h with fresh hiPSC medium (without RevitaCell).
-
Refresh the hiPSC medium daily or every-other-day (for E8 or StemFlex, respectively), until the cells reach approximately 70% or 80% (for E8 or StemFlex respectively) of confluency. This generally takes 3-4 days.
CRITICAL STEP From initial thawing, hiPSCs require 1 week (2 passages) to adapt to culture conditions before starting any differentiation protocol.
? TROUBLESHOOTING
Passaging of undifferentiated hiPSCs
-
10.
Passage hiPSCs cultured using E8 medium/VTN-coated plate as described in option A and hiPSCs using StemFlex medium/laminin-coated plate as described in option B.
(A) Passaging undifferentiated hiPSCs cultured using E8 medium/VTN-coated plate TIMING 10 min
-
(i)
When the cells are 70% confluent, aspirate the culture medium and wash with room temperature PBS-/-.
-
(ii)
Incubate hiPSCs at room temperature with 0.5 mM EDTA for 3 min in 37 °C, 5% CO2 cell culture incubator (1 mL of EDTA per 10-cm2 growth surface).
CRITICAL STEP Monitor the cells under a microscope. Different cell lines may require different incubation times in EDTA. EDTA should be removed when cells start to round up and the colonies appear to have holes in them.
-
(iii)
Aspirate and discard EDTA solution and add E8 medium (1 mL of E8 per 10-cm2 growth surface).
-
(iv)
Detach the cells from the surface of the well(s) by gently pipetting the medium up and down until all cells are in suspension. Collect the cells in a 15-mL conical tube.
CRITICAL STEP Do not pipette the cells too vigorously, cell clusters are required rather than a single cell suspension.
CRITICAL STEP Some hiPSC lines re-adhere very rapidly after the addition of hiPSC medium to neutralize EDTA. For this reason, it is advisable to work with no more than 1 to 3 wells at a time.
CRITICAL STEP To avoid cross contamination between cell lines, it is advisable to work with 1 cell line at a time.
-
(v)
Rinse the well surface with E8 medium (1 mL per 10 cm2 growth surface) and put this medium in the 15-mL conical tube containing the cell suspension.
-
(vi)
Add an appropriate volume of hiPSC medium supplemented with RevitaCell, 1:200 to each well of a VTN-coated 6-well plate so that each well contains a total volume of 2 mL after addition of the cell suspension.
-
(vii)
Plate the cells at a density such that they reach 80% confluency and are ready to be passaged after 3 or 4 days, respectively.
CRITICAL STEP The appropriate plating ratio must be determined empirically and may vary among different cell lines.
-
(viii)
Incubate the plate in the 37 °C, 5% CO2 incubator and change the medium (without RevitaCell) every 24 h.
? TROUBLESHOOTING
(B) Passaging undifferentiated hiPSCs cultured using StemFlex medium/laminin-coated plate TIMING 10 min
-
(i)
When the cells are 80% confluent, aspirate the culture medium and wash with room temperature PBS-/-.
-
(ii)
Incubate hiPSCs with room temperature 1X TrypLE Select for 5 min in a 37 °C, 5% CO2 cell culture incubator (1 mL of 1X TrypLE Select per 10-cm2 growth surface).
CRITICAL STEP Monitor the cells under a microscope. The cells need to be harvested, when they round up and start to detach.
-
(iii)
Detach the cells from the surface of the well(s) by gently pipetting the dissociation reagent up and down using a P-1000 pipette.
-
(iv)
Collect the cells in a 15-mL conical tube containing 3x volume of hiPSC dilution medium to dilute the dissociation reagent.
-
(v)
Wash the well with an additional 1 mL of hiPSC dilution medium to collect any remaining cells and add to the conical tube.
-
(vi)
Centrifuge the cells at 300g for 3 min.
-
(vii)
Aspirate the supernatant and resuspend the cells in 1 mL of StemFlex medium.
(viii) Aspirate the laminin solution from the coated surface of a new plate and add an appropriate volume of StemFlex medium so that each well will contain a total of 2 mL of medium per 10-cm2 after addition of the cell suspension.
-
(ix)
Plate the cells at a density such that they reach 80% confluency and are ready to be passaged after 3 or 4 days.
CRITICAL STEP The appropriate plating ratio must be determined empirically and may vary among different cell lines.
-
(x)
Incubate the plate in a humidified incubator at 37 °C and 5% CO2. Change the medium 24 h later, followed by every 48 h thereafter until the cells are confluent.
Alternatively, 24 h after passaging the cells can be fed with double the volume of StemFlex medium, which means that they do not require feeding for a further 72 h.
? TROUBLESHOOTING
Cardiac mesoderm induction with cytokines TIMING 5 d
CRITICAL STEP To ensure optimal differentiation success rate, use high quality hiPSC cultures at passages lower than 40-50.
CRITICAL STEP The cardiac mesoderm induction with cytokines has been optimized for hiPSCs cultured in E8 medium.
-
11.
Repeat step 10 option A i-v. Take a 20-μL aliquot of cell suspension and count the cell number using a hemocytometer and microscope.
-
12.
Seed 1.5 x 104 (for differentiation of cardiac endothelial cells) and 2.5 x 104 cells per cm2 (for differentiation of epicardial cells and cardiomyocytes) on Matrigel-coated plate in E8 medium supplemented with RevitaCell (1:200). Incubate the plate overnight into the 37 °C, 5% CO2 incubator. This is day −1 of differentiation.
CRITICAL STEP The differentiation efficiency is profoundly affected by the initial seeding density of undifferentiated hiPSCs (Fig.3) . When starting differentiation of new hiPSC lines, it is crucial to identify the optimal seeding density to get highly efficient differentiation, thus we recommend seeding plates at 2 different densities.
-
13.
After 24 h, prepare the appropriate volume of cardiac mesoderm induction medium composed of LI-BPEL medium supplemented with 20 ng/mL BMP4, 20 ng/mL Activin A, and 1.5 μM CHIR 99021.
CRITICAL STEP Warm LI-BPEL medium at room temperature. Add the cytokines fresh each time and use the medium immediately.
-
14.
Aspirate E8 medium and wash the cells once with PBS-/-, followed by an additional washing step with LI-BPEL medium.
-
15.
Add cardiac mesoderm induction medium to the well(s) (3 mL per 10-cm2 growth growth surface). This is day 0 of differentiation. Incubate cells. Do not change the medium until day 3, to avoid disturbing cardiac mesoderm differentiation.
CRITICAL STEP To improve differentiation efficiency and reproducibility, cardiac mesoderm induction should be started exactly 24 h after the seeding of hiPSCs.
CRITICAL STEP To guarantee the success of subsequent differentiation, the next step should be started exactly 96 h after the seeding of hiPSCs.
? TROUBLESHOOTING
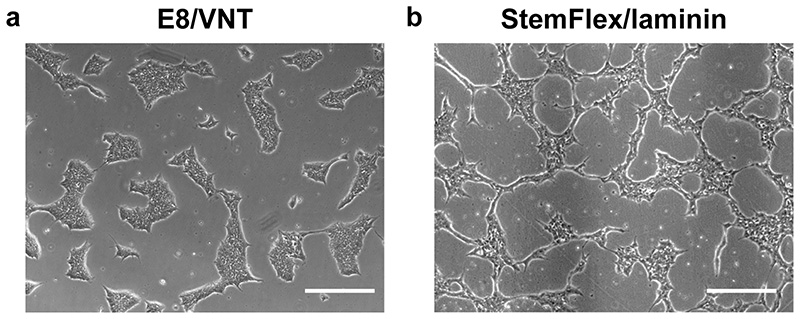
Fig. 3. Morphology of undifferentiated hiPSCs.
Representative brightfield images of hiPSCs cultured in E8/VTN (a) or StemFlex/Laminin (b). Scale bars, 200 μm.
Differentiation into cardiac endothelial cells, epicardial cells and cardiomyocytes
-
16.
On day 3 of differentiation, start the process of differentiating the common mesodermal precursors into cardiac endothelial cells and epicardial cells by following A and B. Also, either differentiate common mesodermal precursors to cardiomyocytes by following C or differentiate from hiPSCs without requiring growth factors (BMP4 and Activin-A) as described in Box 1. All three cell types can be cryopreserved once generated so differentiations can be carried out in parallel or sequentially.
(A) Cardiac endothelial cell differentiation, isolation, culture and cryopreservation TIMING 6 d
-
(i)
On day 3 of differentiation, prepare endothelial cell specification medium composed of LI-BPEL medium supplemented with 5 μM XAV 939 and 50 ng/mL VEGF.
CRITICAL STEP Warm LI-BPEL medium at room temperature. Add the cytokines fresh each time and use the medium immediately.
-
(ii)
Aspirate and discard the cardiac mesoderm induction medium from the well(s) seeded with 1.5 x 104 cells per cm2 and add endothelial cell specification medium (3 mL per 10-cm2 growth surface).
CRITICAL STEP To guarantee the success of the differentiation, cardiac mesoderm induction should be started exactly 96 h after the seeding of hiPSCs. Different cell lines may require different seeding density thus this might require optimization.
-
(iii)
On day 6 of differentiation, isolate endothelial cells (ECs) by immunomagnetic selection of CD34+ cells by using EasySep Human CD34 Positive Selection Kit II as described in the manufacturer’s instructions and outlined previously15. After immunomagnetic isolation, collect a 20-μL aliquot of freshly isolated CD34+ ECs for cell count.
-
(iv)
Dilute ECs in endothelial cell medium composed of LI-BPEL medium supplemented with 50 ng/mL VEGF and plate the cells on fibronectin (FBN)-coated plate at 2.0 x 104 per cm2 seeding density.
CRITICAL STEP Different cell lines may require different seeding density thus seeding density might require optimization.
-
(v)
Incubate the plate overnight into the 37°C, 5% CO2 incubator and then replace medium with LI-BPEL medium supplemented with 50 ng/ml VEGF after 24 h.
-
(vi)
Check cells regularly. On day 9-10 of differentiation, or when cells reach 90-95% confluency (Fig. 4a, b), move to the next step to dissociate EC cells.
-
(vii)
Aspirate cell culture medium and wash with room temperature PBS-/-.
-
(viii)
Incubate the cells with 1X TrypLE Select for 5 min in 37°C, 5% CO2 cell culture incubator (1 ml of 1X TrypLE Select per 10-cm2 growth surface).
-
(ix)
Neutralize 1X TrypLE Select by diluting with the same amount of LI-BPEL medium per well.
-
(x)
Detach the cells from the surface of the well(s) by pipetting the medium up and down and collect the cells in a 15-mL conical tube.
-
(xi)
Wash each well with 1 mL of LI-BPEL per 10-cm2 growth surface, in order to collect residual cells and transfer them into the same 15-mL tube.
CRITICAL STEP Sample an aliquot of cell suspension (approximately 250,000-300,000 cells) to run flow cytometry for endothelial marker CD31 as quality control to evaluate the purity of EC population (CD31+ cells > 90%) (see Box 2 and Fig. 4c).
-
(xii)
Centrifuge the cells at 300g for 3 min at room temperature, aspirate and discard the supernatant. Proceed to next step to cryopreserve cells or start to expand cells for MT production as described in step 18Biii.
(xiii) Resuspend the cell pellet in appropriate volumes of CryoStor CS10 freezing medium to transfer 3.5, 7.0, or 10 x 104 cells/500 μL into a cryovial. Transfer appropriate volumes of cells to cryovials.
CRITICAL STEP Cryovials are prepared to contain different numbers of frozen cells so that only the right amounts (without waste) are available for further experiments.
-
(xiv)
Transfer the cryovial(s) into a Mr. Frosty freezing container and place it at −80 °C. Approximately 24-48 h later, transfer the cryovials in liquid nitrogen tank for longer storage.
? TROUBLESHOOTING
PAUSEPOINT Cells can be stored in liquid nitrogen indefinitely.
Fig. 4. hiPSC-EC characterization.
(a) Representative brightfield image of CD34+ hiPSC-ECs 6 d postisolation and before freezing. Scale bar, 200 μm. (b) Representative brightfield image of CD34+ hiPSC-ECs after thawing at higher magnification. Scale bar, 200 μm. (c) Flow cytometry analysis of CD34+ hiPSC-ECs showing a representative histogram for the expression of the EC marker CD31. Isotype negative control (grey) and hiPSC-ECs are stained with CD31 (red). (d) Representative immunofluorescence images of the endothelial marker CD31 in hiPSC-ECs (day 12). Nuclei are stained with DAPI (blue). Scale bar, 50 μm.
Box 2. Characterization of ECs by FACS analysis TIMING 1.5 h for sample preparation plus 15 min for sample acquisition.
Dissociate ECs as described in ‘Cardiac endothelial cell differentiation, isolation, culture and cryopreservation’ section (Step 16 Avii-xi)
Take an aliquot of cell suspension (approximately 250,000-300,000 cells) and transfer it into a FACS tube.
Centrifuge the cells at 300g for 3 min at room temperature, aspirate and discard the supernatant.
Resuspend the pellet in 1 mL of FACS buffer and split 500 μL of cell suspension into two FACS filter-cap tubes, passing the cells through the filter-caps.
-
Centrifuge the cells at 300g for 3 min at room temperature, and discard the supernatant.
CRITICAL STEP This additional step of centrifugation in FACS buffer is required to remove the residual culture medium, before proceeding with the staining.
Resuspend the cells in one tube with 100 μL of FACS buffer containing 1 μL of directly labelled antibody (Anti-CD31-APC, dilution 1:100). Resuspend the cells in the other tube with 100 μL of FACS buffer containing 1 μL of the correspondent isotype control (IgG1-APC, dilution 1:100).
Incubate the cells for 1 h at 4 °C in the dark. After incubation, wash the cells with 500 μL of FACS buffer.
Centrifuge the cells at 300g for 3 min at room temperature, aspirate and discard the supernatant.
-
Resuspend the cells in 200 μL of FACS buffer and proceed with flow cytometry analysis, adjusting the gates according to the isotype control, and analyze the data in FlowJo software (Supplementary Figure 1).
END BOX
(B) Epicardial cell differentiation, culture and cryopreservation TIMING 9 d
-
(i)
On day 3 of differentiation, prepare epicardial cell induction medium I composed of LI-BPEL medium supplemented with 5 μM XAV 939, 30 ng/mL BMP4, and 1 μM RA. CRITICAL STEP Warm LI-BPEL medium at room temperature. Add the cytokines fresh each time and use the medium immediately. RA is light sensive, thus perform such procedure in the dark.
-
(ii)
Aspirate the cardiac mesoderm induction medium from the well(s) seeded with 2.5 x 104 cells per cm2 and add epicardial cell induction medium (3 mL per 10-cm2 growth surface). Incubate cells.
CRITICAL STEP To guarantee the success of the differentiation, cardiac mesoderm induction should be started exactly 72 h after the seeding of hiPSCs. Different cell lines may require different seeding density thus seeding density might requireoptimization.
-
(iii)
On day 6 of differentiation, prepare the appropriate volume of epicardial cell specification medium II composed of LI-BPEL medium supplemented with 30 ng/mL BMP4, 1 μM RA. Replace medium on the cells with this medium (3 mL per 10-cm2 growth surface) and incubate cells.
CRITICAL STEP Warm LI-BPEL medium at room temperature. Add the cytokines fresh each time and use the medium immediately. RA is light sensive, thus carry out this step in the dark.
-
(iv)
On day 9 of differentiation, follow step 16 A vii-Xii to dissociate the newly formed epicardial cells (EPI).
-
(v)
Resuspend EPI cells in epicardial cell maintenance medium composed of LI-BPEL medium supplemented with 10 μM SB431542 and plate the cells on FBN-coated plate at 1.5 x 104 per cm2 seeding density.
CRITICAL STEP Different cell lines may require different seeding densities, thus seeding density might require further optimization.
-
(vi)
Checks cells regularly. On day 12 of differentiation, EPI cells should be 90-95% confluent. Either cryopreserve (as described in step 16A vii-Xiv) or proceed with cardiac fibroblast differentiation as described in the next step (Fig. 5a).
CRITICAL STEP To evaluate efficiency of epicardial differentiation, perform immunofluorescence for WT1, CX43, TBX18, and ZO-1 epicardial markers (see Box 3 and Fig. 5b).
? TROUBLESHOOTING
PAUSEPOINT Cells can be stored in liquid nitrogen indefinitely.
Fig. 5. hiPSC-EPI and hiPSC-CF characterization.
(a) Brightfield images at different time points showing the epithelial mesenchymal transition of hiPSC-EPI (day 12) toward hiPSC-CFs (day 28) during cardiac fibroblast differentiation. Scale bar, 200 μm. (b) Representative immunofluorescence images of the epicardial markers WT1, TBX18, CX43 (green) and ZO-1 (red) in hiPSC-EPI (day 12). Nuclei are stained with DAPI (blue). Scale bar, 50 μm. (c) Representative immunofluorescence images of the fibroblast markers VIM, COL1A1, GATA4 (red) and CX43 (green) in hiPSC-CFs (day 28). Nuclei are stained with DAPI (blue). Scale bar, 50 μm.
Box 3. Characterization of ECs, EPI and CFs by immunofluorescence TIMING 2 d.
-
Dissociate ECs, EPI and CFs as described in Step 16 Avii-xii and re-plate the cells on FBN-(for ECs and EPI: 1.0 x 104 cells) or VTN-(for CFs: 1.0 x 104 cells) coated black-walled 96-well plates with clear bottom in LI-BPEL medium supplemented with 50 ng/ml VEGF (for ECs), 10 μM SB431542 (for EPI) and FGM3 medium (for CFs).
CRITICAL STEP The use of opaque black-walled 96-well plates with clear bottom is crucial for immunofluorescence assays to reduce fluorescent signal crosstalk and background among different wells.
After 2-3 d in culture, aspirate the medium and wash twice with 50 μL of PBS+/+ to completely remove residual culture medium.
Fix the cells with 50 μL of 4% (vol/vol) PFA solution for 15 min at room temperature.
-
Aspirate the fixative solution and wash three times (5 min each) with PBS+/+ to completely remove residual fixative solution.
PAUSEPOINT Fixed cells can be stored in 100 μL of PBS+/+ at 4 °C up to 2 -3 weeks, sealing the plate with parafilm, before proceeding with immunostaining.
Permeabilize the cells with 50 μL of permeabilization solution 0.1% (vol/vol) Triton X-100 for 10 min at room temperature.
Block to minimize unspecific binding of the primary antibody by adding 50 μL of blocking solution for 1-2 h at room temperature.
Stain with 30 μL of blocking solution containing primary antibodies: CD31 for ECs; WT1, TBX18, ZO-1 and CX43 for EPI cells; CX43, COL1A1, VIM, GATA4 for CFs (see recommended dilution in ‘Materials’ section).
Incubate the plate overnight at 4 °C on a rocking platform, with gentle shaking.
The following day, wash three times with 50 μL of PBS+/+, each time for 10 min on a rocking platform, with gentle shaking.
Stain with 30 μL of blocking solution containing secondary antibodies (see dilution in ‘Materials’ section). Incubate the plate for 1 h at room temperature in the dark on a rocking platform, with gentle shaking.
Wash three times with 50 μL of PBS+/+ in the dark, each time for 10 min on a rocking platform, with gentle shaking.
Stain with 30 μl of DAPI at 1:500 dilution in PBS+/+ for 10 min at room temperature in the dark on a rocking platform, with gentle shaking.
Wash once with 50 μl of PBS+/+ to remove residual DAPI solution.
-
After completion of immunostaining, proceed immediately with image acquisition or store the plates at 4 °C in the dark, sealing the plate with parafilm.
PAUSEPOINT Stained cells can be stored in 100 μL of PBS+/+ at 4 °C for up to 1-2 months, sealing the plate with parafilm, before proceeding with image acquisition.
END BOX
(C) Cardiomyocyte differentiation, lactate purification, culture and cryopreservation TIMING 15-17 d
-
(i)
On day 3 of differentiation, prepare cardiomyocyte specification medium composed of LI-BPEL medium supplemented with 5 μM XAV 939.
CRITICAL STEP At this stage the cells should have proliferated, formed a confluent monolayer and medium should be yellow. If the monolayer is not confluent, this will negatively affect the efficiency of cardiomyocyte differentiation, resulting in a low percentage of cTnT positive cells in the populations and the formation of non homogenous beating monolayer.
CRITICAL STEP Warm LI-BPEL medium at room temperature. Add the cytokines fresh each time and use the medium immediately.
-
(ii)
Aspirate the cardiac mesoderm induction medium from the well(s) seeded with 2.5 x 104 cells per cm2 and add cardiomyocyte specification medium (3 mL per 10-cm2 growth surface). Incubate cells.
CRITICAL STEP To improve differentiation efficiency and reproducibility, perform this step 72 h after cardiac mesoderm induction. Different cell lines may require different seeding density thus seeding density might require optimization.
-
(iii)
On day 6 of differentiation, replace the medium with LI-BPEL without supplements (3 mL per 10-cm2 growth surface).
-
(iv)
On day 9 of differentiation, replace the medium with fresh LI-BPEL without supplements. Beating CMs should start to appear at day 9-10. Replace medium with fresh LI-BPEL without supplements every 2-3 d until the beating cells appear.
-
(v)
When the wells show a homogenous beating monolayer (usually at day 10-12 of differentiation), aspirate LI-BPEL medium and replace with lactate purification medium composed of DMEM (no glucose, no pyruvate) supplemented with 4 mM Sodium-L-Lactate. Further incubate cells.
-
(vi)
Replace medium on beating CMs with fresh lactate purification medium after 48 h (day 12-14 of differentiation).
CRITICAL STEP Metabolic selection of CMs with lactate treatment maximizes CM enrichment (> 90% cTnT positive cells)
-
(vii)
After an additional 48 h of lactate purification treatment (day 14-16 of differentiation), replace medium with LI-BPEL medium.
CRITICAL STEP Before proceeding to CM dissociation for cryopreservation, it is crucial to let CMs recover from metabolic selection by culturing and re-adapting the CMs to LI-BPEL culture condition for at least 24 h.
-
(viii)
Check cells. On day 18-20 of differentiation, beating CMs should be ready to be dissociated as detailed in the following steps (Fig. 6a and Supplementary Video 1).
-
(ix)
Aspirate and discard cell culture medium and wash cells with room temperature PBS -/-.
-
(x)
Incubate the cells with CM dissociation enzyme mix for 10 min in 37 °C, 5% CO2 cell culture incubator (1 mL of enzyme mix per 10-cm2 growth surface).
-
(xi)
Neutralize the enzyme mix by diluting with double the amount of LI-BPEL medium per well (2 mL of LI-BPEL per 10-cm2 growth surface).
-
(xii)
Dislodge the cells from the surface of the well(s) and dissociate the CMs by gently pipetting the medium up and down several times using a P-1000 pipette (orifice 1.5 mm). Collect the cell suspension in a 15-mL conical tube.
CRITICAL STEP To avoid too much mechanical stress and damage of the CM cell membranes, stop pipetting as soon as big clusters are no longer visible.
? TROUBLESHOOTING
-
(xiii)
Rinse each well with an additional 1 mL of LI-BPEL per 10-cm2 growth surface to collect residual cells and transfer into the 15-mL tube used in the previous step. Collect a 20-μL aliquot of cell suspension and count cell number.
CRITICAL STEP To check for the extent of the differentiation, also remove an aliquot of cell suspension (approximately 250,000-300,000 cells) and run flow cytometry, checking for the presence of the cardiomyocyte marker cTnT (see Box 4 and Fig. 6b).
-
(xiv)
Centrifge the cells at 300g for 3 min at room temperature, aspirate and discard the supernatant. Proceed to the next step for cryopreservation or use freshly dissociated CMs to proceed directly to MT generation (step 20).
-
(xv)
Resuspend the cell pellet in appropriate volume of CM freezing medium (90% KOSR with 10% DMSO) and transfer 2.5-3.0 x 106 cells/300 μL into a cryovial.
-
(xvi)
Transfer the cryovial(s) into a Mr. Frosty freezing container and place it at −80 °C. Approximately 24-48 h later, transfer the cryovials in liquid nitrogen tank for longer storage.
? TROUBLESHOOTING
PAUSEPOINT Cells can be stored in liquid nitrogen indefinitely.
Fig. 6. hiPSC-CM characterization.
(a) Brightfield images of hiPSC-CMs after 21 d of differentiation using LI-BPEL protocol with lactate purification (left panel) or mBEL protocol (right panel). Scale bars, 200 μm. (b) Expression of cardiac Troponin T (cTnT) by hiPSC-CMs. Representative flow cytometry analysis of hiPSC-CMs generated using either the LI-BPEL with lactate purification (left panel) or mBEL (right panel). Histograms show hiPSC-CMs stained with isotype control (LI-BPEL with lactate purification) or unstained (mBEL) (grey), and stained with cTnT (red).
Box 4. Characterization of cardiomyocytes (CMs) by flow cytometric analysis for the cardiomyocyte marker, cardiac troponin T (cTnT).
Dissociate CMs as described in Box 1 steps 6-9 or Step 16 Cix-xiii, take an aliquot of cell suspension (approximately 250,000-300,000 cells) and transfer it into a FACS tube.
Centrifuge the cells at 300g for 3 min at room temperature. Discard the supernatant.
Resuspend the cell pellet in 500 μL FACS buffer. Filter the cell suspension using a 5 mL tube with a 35 μm cell strainer cap.
-
Pellet the cells at 300g for 3 min at room temperature and discard the supernatant.
CRITICAL STEP Resuspending and pelleting the cells in FACS buffer is important to remove residual culture medium.
Resuspend the cells in 50 μL Fixation medium. Briefly vortex to ensure the cells are completely resuspended.
Fix the cells for 15 min at room temperature. Flick the tube after ~7.5 min to ensure the cells remain in suspension.
-
Dilute the Fixation medium with 1 mL FACS buffer. Pellet the cells at 300g for 3 min at room temperature. Discard the supernatant.
PAUSEPOINT Fixed cells can be stored in 1 mL of FACS buffer at 4 °C in a sealed tube for up to 7 d. When ready to continue, pellet the cells and proceed with next step.
Resuspend the cells in 1 mL of FACS buffer. Divide the cell suspension evenly into 2 tubes, filtering the cells through a 35 μm cell strainer cap.
Pellet the cells at 300g for 3 min at room temperature and discard the supernatant.
Resuspend the cells in one tube with 50 μL of Permeabilization medium containing 5 μL of directly labelled antibody (Anti-cTnT-VioBlue, dilution 1:50. Resuspend the cells in the other tube with 50 μL of Permeabilization medium containing 5 μL of the correspondent isotype control (REA Control (I)-VioBlue, dilution 1:50).
Briefly vortex to ensure the pellet is properly resuspended and incubate the cells for 20 min at room temperature in the dark.
After incubation, wash the cells with 1 mL of FACS buffer.
Pellet the cells at 300g for 3 min at room temperature, aspirate and discard the supernatant.
Resuspend the cells in 200-500 μL of FACS buffer and run the samples on a flow cytometer. Analyze the data using FlowJo software, adjusting the gates according to the isotype control (Supplementary Figure 2).
Cardiac fibroblasts differentiation, culture and cryopreservation
-
17.
If differentiating cardiac fibroblasts (CFs) from freshly dissociated EPI cells, follow option A. To differentiate CFs from cryopreserved EPI cells, follow option B.
(A) Cardiac fibroblast differentiation from freshly dissociated EPI cells TIMING 16 d
-
(i)
On day 12 of differentiation, dissociate the EPI cells as described in step 16 A vii-Xii . Then resuspend EPI cells in CF induction medium composed of LI-BPEL medium supplemented with 10 ng/mL FGF-2 and plate the cells on a VTN-coated plate at 2.5 x 104 per cm2 seeding density.
CRITICAL STEP Different cell lines may require different seeding density thus seeding density might require optimization.
-
(ii)
Incubate the plate overnight in the 37 °C, 5% CO2 incubator and replace the medium with fresh LI-BPEL supplemented with 10 ng/mL FGF-2 after 24 h.
-
(iii)
Replace CF induction medium every 2 d for the next 6 d (i.e. until day 19 of differentiation).
-
(iv)
On day 19 of differentiation, start culturing the newly formed CFs by replacing the medium with Fibroblast Growth Medium 3 (FGM3), to support the expansion of CFs.
-
(v)
Replace the medium with FGM3 every 2 d for approximately 9-10 d.
-
(vi)
On day 28-29 of differentiation, CFs should be 90-95% confluent (Fig. 5a). Repeat step 16 A vii-Xii to dissociate CFs.
-
(vii)
Resuspend CFs in FGM3 medium and plate the cells on VTN-coated plate at a split ratio of 1:2.
CRITICAL STEP The first amplification step should be carried out on a VTN-coated plate. Note that different cell lines may require different seeding ratios.
-
(viii)
After 3-4 d, CFs should be confluent and can be amplified and adapted to grow on (uncoated) tissue culture plates by repeating step 16 A vii-Xii and plating in FGM3 medium at a split ratio of 1:2.
CRITICAL STEP After this passage (passage 2), CFs can be further amplified, or cryopreserved (as described in step 16A vii-Xiv) or immediately expanded for MT production (by proceeding with step 18Cii).
CRITICAL STEP To evaluate efficiency of cardiac fibroblast differentiation, perform IF for COL1A1, Vimentin, GATA4, and CX43 (see Box 3 and Fig. 5c).
? TROUBLESHOOTING
(B) Cardiac fibroblast differentiation from cryopreserved EPI cells TIMING 10 min for thawing plus 3-4 d for expansion of EPI cells, and 16 d for CF differentiation
-
(i)
To thaw EPI cells, prepare a 15-mL conical tube filled with 5 mL of LI-BPEL medium, previously equilibrated at room temperature.
-
(ii)
Remove the cryovial of frozen EPI cells from liquid nitrogen storage tank and transfer it on dry ice or in a portable liquid nitrogen container to the cell culture room.
-
(iii)
Immerse the cryovial in a 37 °C water bath and swirl it gently, until only an ice crystal remains.
CRITICAL STEP To prevent contamination, do not submerge the cap of the cryovial.
-
(iv)
Remove the cryovial from the water bath, spray with 70% ethanol and place it into the biological safety cabinet.
-
(v)
Gently transfer the content of the cryovial into the 15-mL conical tube containing the LI-BPEL medium. Rinse the vial with additional 1 mL of LI-BPEL medium to recover any residual cells and add it into the 15-mL conical tube.
-
(vi)
Centrifuge the cells at 300g for 3 min at RT, aspirate and discard the supernatant.
-
(vii)
Resuspend the cell pellet in EPI medium composed of LI-BPEL medium supplemented with 10 μM SB431542 and re-plate 2.5-3.0 x 104 cells per cm2 on a FBN-coated plate.
-
(viii)
Place the cells gently into a 37 °C, 5% CO2 cell culture incubator and replace the medium (2 mL per 10-cm2 growth surface) after 24 h with fresh LI-BPEL medium supplemented with 10 μM SB431542. After 3 d, the cells should have reached approximately 85-90% of confluency. Proceed with step 17Ai-viii to dissociate and differentiate to CF.
? TROUBLESHOOTING
Additional Materials Required
Trace elements B (Corning, cat. no. 25022CI)
Trace elements C (Corning, cat. no. 25023CI)
IWP-L6 (AbMole, cat. no. M2781)
Additional Reagent Setup
IWP-L6 stock solution
Dissolve IWP-L6 at 25 mM stock solution in DMSO. From this stock solution, dilute futher to 0.25 mM in DMSO. Prepare 10-μL aliquots and store at −20 °C.
CRITICAL Thaw IWP-L6 aliquots at room temperature. Before use, briefly centrifuge to collect solution at the bottom of tube . After thawing, store the aliquot up to 10 d at 4 °C.
3-step Modified LI-BPEL (mBEL) medium preparation
To prepare 100 mL of each of the 3-step basal mBEL medium, combine all the components/supplements listed in the tables below. Filter the medium using a 0.22-μm filter system and store the medium at 4 °C for up to 1 month.
Cardiomyocyte Induction Medium (Day 0-4) - mBEL minus insulin and essential lipids:
| Component | Quantity for 100 mL |
|---|---|
| IMDM | 47.25 mL |
| F12 | 47.25 mL |
| BSA (10% wt/vol in IMDM) | 2.5 mL |
| α-Monothioglycerol (13 μL in 1 mL IMDM) | 0.3 mL |
| L-ascorbic acid 2-phosphate (5 mg/mL in distilled water) | 1 mL |
| Glutamax | 1 mL |
| Trace elements B | 0.01 mL |
| Trace elements C | 0.1 mL |
| Penicillin-streptomycin | 0.5 mL |
Cardiomyocyte Specification Medium (Day 4-onwards) - mBEL minus essential lipids:
| Component | Quantity for 100 mL |
|---|---|
| IMDM | 47.25 mL |
| F12 | 47.25 mL |
| BSA (10% wt/vol in IMDM) | 2.5 mL |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) 100X | 0.1 mL |
| α-Monothioglycerol (13 μL in 1 mL IMDM) | 0.3mL |
| L-ascorbic acid 2-phosphate (5 mg/mL in distilled water) | 1 mL |
| Glutamax | 1 mL |
| Trace elements B | 0.01 mL |
| Trace elements C | 0.1 mL |
| Penicillin-streptomycin | 0.5 mL |
Cardiomyocyte Maintenance Medium (after thawing and replating) - mBEL:
| Component | Quantity for 100 mL |
|---|---|
| IMDM | 47.25 mL |
| F12 | 47.25 mL |
| BSA (10% wt/vol in IMDM) | 2.5 mL |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) 100X | 0.1 mL |
| Chemically Defined Lipid Concentrate (CDLC) | 1 mL |
| α-Monothioglycerol (13 μL in 1 mL IMDM) | 0.3mL |
| L-ascorbic acid 2-phosphate (5 mg/mL in distilled water) | 1 mL |
| Glutamax | 1 mL |
| Trace elements B | 0.01 mL |
| Trace elements C | 0.1 mL |
| Penicillin-streptomycin | 0.5 mL |
Procedure
-
Seed 4.3-5.7 x 104 hiPS cells per cm2 on Matrigel-coated plates (i.e. 150,000-200,000 cells per well of a 12-well plate) in StemFlex or E8 medium supplemented with RevitaCell (1:200). Incubate the plate overnight in a humidified incubator at 37 °C and 5% CO2. This is day −1 of differentiation
CRITICAL STEP The differentiation efficiency is affected by the confluency of the undifferentiated hiPSCs when the differentiation is initiated. It is crucial to identify the optimal seeding density for individual hiPSC lines to maximise the purity and total number of CMs acquired.
-
After 24 h (day 0 of differentiation), remove the StemFlex or E8 medium and add cardiomyocyte induction medium supplemented with 5 μM CHIR 99021 (1 mL per well of a 12-well plate).
CRITICAL STEP To improve differentiation efficiency and reproducibility, the differentiation should be started exactly 24 h after seeding the hiPSCs. Do not change the medium until day 2 of differentiation.
On day 2 of differentiation, replace the medium with cardiomyocyte induction medium supplemented with 5 μM XAV 939 and 0.25 μM IWP-L6 (1 mL per well of a 12-well plate). CRITICAL STEP To improve differentiation efficiency and reproducibility, perform this step exactly 48 h after the last medium replacement.
On day 4 of differentiation, replace the medium with cardiomyocyte specification medium (1 mL per well of a 12-well plate).
Completely replace the medium 2 d later, and then every 2-3 d thereafter. Spontaneously beating CMs should be apparent between day 8-10 of differentiation.
On day 21 of differentiation, the cells can be dissociated (Fig. 6a and Supplementary Video 2). First, aspirate the culture medium and wash the cells with room temperature PBS-/-.
Incubate the cells with 5X TrypLE Select for 10 min in a humidified incubator at 37°C and 5% CO2 (0.5 mL of 5X TrypLE Select per well of a 12-well plate).
-
Detach the cells from the tissue culture plate by gently pipetting the medium up and down using a P-1000 pipette (orifice 1.5 mm). Collect the cells in a 15-mL conical tube containing 3X volume of cardiomyocyte specification medium.
CRITICAL STEP Generate a single cell suspension of CMs by very gentle pipetting to avoid damaging the CMs.
-
Wash the tissue culture plate with cardiomyocyte specification medium (1 mL per well of a 12 well plate) to collect residual cells and transfer to the same 15-mL tube. Collect an aliquot to count the cells.
CRITICAL STEP Remove an aliquot of the cell suspension (approximately 200,000-300,000 cells) to evaluate by flow cytometry the purity of CM population based on cTnT expression (see Box 4 and Fig. 6b). The differentiated cells should be >80% cTnT+.
-
If you wish to cryopreserve cells, follow steps described in step 16C Xiii-Xv. Otherwise cells are ready to be used in step 20.
? TROUBLESHOOTING
END BOX
Thawing and re-plating the three cardiac cell types (ECs, CFs and CMs) for cardiac microtissue generation
-
18.
Thaw (if necessary) CMs (cytokine/growth factor and small molecule protocols), thaw (if necessary) and expand ECs and CFs as described in options A, B and C, respectively. Option A needs to be started 5 days before step 19, while options B and C need to be started 3 days before step 19 . If cells are already thawed, start at steps Avii, Biii and Cii respectively.
(A) Thawing and re-plating of cardiomyocytes (CMs) TIMING 10 min plus 5 d for cell recovery before cardiac microtissue generation
-
(i)
5 d before MT production, remove a cryovial of CMs from the liquid nitrogen storage tank and transfer to a biological safety cabinet on dry ice or in a portable liquid nitrogen container.
-
(ii)
Immerse the cryovial in a 37 °C water bath or into a 37°C, 5% CO2 cell culture incubator, holding the tube stationary, without swirling it, until only a 2 mm3 ice crystal remains.
CRITICAL STEP To prevent contamination, do not submerge the cap of the cryovial. It is recommended to use a floating microcentrifuge tube rack.
-
(iii)
Immediately remove the cryovial from the water bath, spray with 70% ethanol and place it into the biological safety cabinet.
-
(iv)
Gently transfer the contents of the cryovial (2.5-3.0 x 106 cells in 300 μL of CM freezing medium) to a 15-mL conical tube using a P1000 pipette (orifice 1.5 mm). Avoid pipetting the cell suspension up and down.
-
(v)
Rinse the empty cryovial with 1 mL of LI-BPEL medium or cardiomyocyte maintenance mBEL medium, previously equilibrated at RT. Using a P1000 pipette, add the medium drop-wise to the 15-ml tube containing the cells while gently swirling the tube for 1-5 seconds after the addition of each drop.
CRITICAL STEP The drop-wise addition of the medium facilitates suitable mixing of the cells thus minimizing osmotic shock and increasing CM recovery and viability.
-
(vi)
Slowly add an additional 4.7 mL of medium (LI-BPEL or mBEL medium) to the 15-ml tube containing the cell suspension using a 5-ml pipette. Collect an aliquot of cell suspension for cell count.
-
(vii)
Centrifuge the cells at 300g for 3 min at room temperature and discard the supernatant.
-
(viii)
Gently resuspend the cell pellet from the previous step, Box 1 or step 16Cxiv in the appropriate volume of the original culture medium (LI-BPEL or mBEL medium) and plate 1.0-4.0 x 105 cells per cm2 seeding density on Matrigel-coated plate.
-
(ix)
Place the cells gently into a humidified incubator at 37°C, and 5% CO2. Refresh the medium after 24 h. The CMs should start spontaneously beating 24-48 h post thawing.
-
(x)
Refresh with the appropriate medium every 2-3 d. After 5 d, CMs should be ready for use in step 19.
CRITICAL STEP Expected survival after thawing is approximately 30-50% of the initial number of cells cryopreserved. Before MT production, it is crucial to wait at least 5 d to ensure complete recovery of the CMs from the thawing procedure.
? TROUBLESHOOTING
(B) Thawing and expansion of endothelial cells (ECs) TIMING 10 min plus 3 d for cell expansion before cardiac microtissue generation
-
(i)
3 d before you plan to produce cardiac microtissues (MTs), prepare a 15-mL conical tube filled with 5 mL of LI-BPEL medium, previously equilibrated at room temperature.
-
(ii)
Thaw ECs (from step 16Axvi) as described in step 17B(ii-vi).
-
(iii)
Resuspend the cell pellet (from previous step or step 16Axiv) in EC medium composed of LI-BPEL medium supplemented with 50 ng/ml VEGF and re-plate 2.5-3.0 x 104 cells per cm2 on FBN-coated plate.
-
(iv)
Place the cells gently into a 37 °C, 5% CO2 cell culture incubator and replace the medium (2 mL per 10-cm2 growth surface) after 24 h with fresh LI-BPEL medium supplemented with 50 ng/mL VEGF.
-
(v)
After 3 d, the cells should reach approximately 85-90% of confluency and are ready for use in step 19.
CRITICAL STEP Expected survival after thawing should be > 80%. For MT production, it is crucial to let ECs recover from thawing for at least 3 d, being careful that cells do not become over confluent and detach.
(C) Thawing and expansion of cardiac fibroblasts (CFs) TIMING 10 min plus 3 d for cell expansion before cardiac microtissue generation
-
(i)
Thaw CFs by following steps 17B(ii-vi)..
-
(ii)
Resuspend the cell pellet from previous step or step 17Aviii in FGM3 medium and re-plate 25-30x 103 cells per cm2 on (uncoated) plastic plates tissue culture treated.
-
(iii)
Place the cells gently into a 37°C, 5% CO2 cell culture incubator and replace the medium (2 ml per 10-cm2 growth surface) after 24 h with fresh FGM3. After 3 d, the cells should reach approximately 85-90% of confluency and are ready for use in step 19.
CRITICAL STEP Expected survival after thawing should be > 80%. Before MT production, it is crucial to let CFs recover from thawing for at least 2 days, being careful that cells do not become over confluent and detach.
Generation and culture of 3D cardiac microtissues (MTs) composed of hiPSC-CMs, -ECs and -CFs TIMING 2h for cardiac microtissue generation plus 21 d for cardiac microtissue culture
-
19.
Dissociate ECs and CFs as described in step 16Avii-xii and dissociate CMs as described in step 16Cix-xiv.
CRITICAL STEP Monitor the results of each cell type dissociation under the microscope to ensure single-cell detachment. Single-cell suspension and absence of cell clumps are crucial for precise cell counting and proper MT production.
-
20.
Resuspend all three cell types in 1 mL of LI-BPEL medium and collect a 20-μL aliquot of each cell type suspension. Count cells using a hemocytometer and microscope.
-
21.
Prepare a 15-mL conical tube and dilute ECs, CFs and CMs in LI-BPEL medium supplemented with 50 ng/mL VEGF and 5ng/mL FGF-2 combining 5,000 cells in the defined ratio of 70% CMs, 15% ECs and 15% CFs (3,500 CMs, 750 ECs and 750 CFs for each MT) per 50 μL of medium.
CRITICAL STEP 50 μL cell suspension will be required per well (cells are later plated in a 96-well V-bottom microplate).
-
22.
Gently resuspend the cell suspension using a 5- or 10-mL sterile plastic serological pipette and transfer to a sterile plastic reservoir.
-
23.
Fill each external well with 100 μL of PBS. Seed 50 μL of cell suspension per internal well (for each MT) of a 96-well V-bottom microplate using a multichannel pipette.
CRITICAL STEP Do not seed MTs in the outermost wells of the 96-well V-bottom microplate to avoid possible medium evaporation. Each 96-well microplate will contain a maximum of 60 MTs.
CRITICAL STEP For initial optimization of the protocol it is recommended to prepare 120 MTs corresponding to two 96-well plates.
CRITICAL STEP Move the plastic reservoir side-to-side after plating each 3 rows of the 96-well microplate to assure maintenance of a homogeneous cell suspension. This is critical to obtain MTs which are similar in size and cellular composition.
-
24.
Centrifuge the microplate(s) at 300g for 10 min at room temperature to facilitate the aggregation of the cell suspension on the bottom of the well(s). This is day 0 of MT formation.
-
25.
Place the microplates gently into a 37°C, 5% CO2 cell culture incubator. To allow cellular aggregation and proper MT formation, do not move the plates for 3 d.
CRITICAL STEP Proper cell aggregation and subsequent tissue formation is evident as compact and round-shaped MTs observed after 24h (Fig. 7).
-
26.
Every 3 d, refeed MTs by removing 25 μL of old medium and replacing it with 25 μL of fresh LI-BPEL supplemented with 50 ng/ml VEGF and 5ng/ml FGF-2. After 21 d, MTs should be ready for the next step (further characterization and analyses).
? TROUBLESHOOTING
CRITICAL STEP Note that for subsequent video analysis of calcium transients (step 27 option C) cells need to be transferred to a black plate 2 days prior to analysis, i.e. on day 19.
Fig. 7. 4: Examples of optimal and sub-optimal cardiac MTs after 24h after formation.
Representative images of compact and round-shaped MT (a) and not properly formed MT (b). Scale bar 200 μm.
Assays that can be undertaken on 3D cardiac microtissues
-
27.
After 21 d in culture from initial aggregation, 3D whole MTs can used for various assays. Option A describes how to undertake structural analysis by immunofluorescence staining. Option B describes video analysis to assess contraction. Option C describes video analysis of intracellular calcium transients. Option D describes how to use the Seahorse XFe96 assay to assess metabolic functionality. Option E describes how to assess electrical properties by sharp electrode electrophysiology. Option F describes how to dissociate MTs into single cells and then undertake scRNAseq or electrophysiology or immunostaining of single cells.
(A) Immunofluorescence staining TIMING 3 d
-
(i)
Collect at least 30 MTs and transfer them to a 15-mL conical tube using a P-1000 pipet. Wait for 3-5 min until all MTs sediment at the bottom of the tube.
-
(ii)
Carefully aspirate the supernatant using a 1-mL pipet and wash MTs with 1 mL of PBS+/+ to completely remove residual culture medium. Transfer PBS solution containing MTs to a 1.5-mL tube and wait for 3-5 min until all MTs sediment at the bottom of the tube.
CRITICAL STEP Avoid pipetting MTs up and down in order to preserve the integrity of microtissues.
-
(iii)
Carefully aspirate the supernatant using a 1-mL pipet and fix MTs with 100 μL of 4% (vol/vol) PFA solution for 1 h at 4° C.
-
(iv)
Aspirate and discard the fixative solution without disturbing MTs on the bottom of the tube and wash three times (5 min each) with 100 μL of PBS+/+ to completely remove residual fixative solution.
CRITICAL STEP Avoid pipetting MTs up and down in order to preserve the integrity of microtissues.
PAUSEPOINT Fixed MTs can be stored in 1 mL of PBS+/+ at 4 °C up to 2-3 weeks before proceeding with immunostaining protocol.
-
(v)
Permeabilize and immunostain with primary and secondary antibodies, as previously described15,65.
-
(vi)
Carefully aspirate the supernatant using a 1-mL pipet and permeabilize MTs with 100 μL of permeabilization solution 0.2% (vol/vol) Triton X-100 for 30 min at room temperature.
-
(vii)
Aspirate and discard the permeabilization solution without disturbing MTs on the bottom of the tube and wash three times (5 min each) with 100 μL of PBS+/+ to completely remove residual permeabilization solution.
-
(viii)
Block to minimize non-specific binding of the primary antibodies by adding 100 μL of blocking solution composed of PBS+/+ containing 10% (vol/vol) FBS for 2-3 h at room temperature.
-
(ix)
Stain MTs with 60 μL of blocking solution containing primary antibodies: α-Sarcomeric Actinin (ACTN2), Troponin I (TNNI) for CMs; CD31 for ECs; Collagen Type I (COL1A1) for CFs (see recommended dilution in ‘Materials’ section) and incubate them overnight at 4 °C.
-
(x)
The following day, wash three times with 100 μl of PBS+/+, each time for 20 min, waiting for microtissues to sediment on the bottom of the tube.
-
(xi)
Stain MTs with 60 μL of blocking solution containing secondary antibodies (see dilution in ‘Materials’ section) and incubate them overnight at 4 °C in the dark.
-
(xii)
The following day, wash three times with 100 μl of PBS+/+ in the dark, each time for 20 min, waiting for microtissues to sediment on the bottom of the tube.
(xiii) Stain with 60 μl of DAPI at 1:500 dilution in PBS+/+ for 30 min at room temperature in the dark.
-
(xiv)
At the end of staining procedure, carefully aspirate the supernatant using a 200-μl pipet to completely remove residual DAPI solution.
-
(xv)
Resuspend no more than 25-30 MTs in 10 μL of liquid mountant, transfer MTs on microscope slides and mount them on top of 12-mm round, glass coverslips, avoiding bubbles and making sure MTs are well distributed throughout the coverslip to avoid MT overlap. After completion of the immunostaining protocol, proceed with the image acquisition of whole 3D MTs mounted on microscope slides on the top of glass coverslips using a confocal laser-scanning microscope.
CRITICAL STEP Before starting with image acquisition, allow the slides to dry at least 24 h at room temperature in the dark.
-
(xvi)
In order to evaluate the architecture and cell composition of whole 3D MTs, acquire images using either a 20× or 25× objective magnification and Z-stack acquisition. MTs should stain positive for cardiac marker TNNI, endothelial marker CD31 and fibroblast marker COL1A1.
(xvii) In order to perform structural analysis of sarcomeres (length and organization), acquire images of 3D whole 3D MTs using 100× objective magnification and Z-stack acquisition. MTs stain positive for cardiac markers TNNI and ACTN2.
(xviii) Determine sarcomere length by measuring the average distance between Z-bands (stained with ACTN2) using standard analysis plugin in open-source Fiji Software, ImageJ.
(xix) Evaluate sarcomere organization as sarcomere alignment index based on the regularity and periodicity of ACTN2 positive Z-bands (stained with ACTN2) using a standard fast Fourier transform (FFT) analysis (Fiji Software, ImageJ) as previously described 76,77 .
? TROUBLESHOOTING
(B) Video analysis of contraction TIMING 2-3 min per cardiac microtissue
CRITICAL It is advisable to avoid re-plating of MTs to reduce possible variation of contraction parameters due to potential attachment of MTs on flat bottom plates, thus analyse MTs in the V-shaped culture microplate in which they were generated.
-
(i)
Record movies of MTs spontaneously beating or paced at 1, 2 or 3 Hz (20 V, 3 ms) using custom-made electrodes fitting the V-shaped 96-wells (Supplementary Figure 3) for at least 20 s at video acquisition speed of 100 frames per s (f.p.s.), using 10x objective phase containing the entire tissue within the field of view.
CRITICAL STEP To adequately perform contraction video analysis, use a high-speed recording camera for the movie acquisition. Avoid variations of the lighting conditions during measurements.
CRITICAL STEP Record videos for at least 20 s in order to capture multiple contraction cycles within each video and possibly identify beating abnormalities.
-
(ii)
Perform contraction video analysis using the MUSCLEMOTION ImageJ macro as described previously61 , including in a detailed protocol61’78.
(C) Video analysis of calcium transients TIMING 1 h for dye incubation and 2-3 min for analysis of single cardiac microtissue
CRITICAL For analysis on day 21, MTs need to be picked 18-19d following initial MT formation (by aspirating the content of each well with a P1000 pipette) and seeded on Matrigel-coated 96-well black plates. It is important to seed no more than 3-4 MTs per well to ensure homogenous distribution within the surface of the well, avoiding direct contact between different MTs.
-
(i)
On day 21 (48-72 h after re-plating) aspirate LI-BPEL medium from the well(s) and add to each well containing MTs 50 μL of 5 μM Fluo-4 AM loading solution for 30 min into a 37 °C, 5% CO2 cell culture incubator.
-
(ii)
Remove Fluo-4 AM loading solution using P200 pipet and replace with LI-BPEL medium. Incubate loaded MTs for an additional 30 min into a 37 °C, 5% CO2 cell culture incubator, before proceeding to the next step for recordings.
CRITICAL STEP Carefully aspirate Fluo-4 AM loading solution to remove any dye nonspecifically associated with the surface of MTs. A further 30 min incubation before recordings is crucial to allow complete de-esterification of intracellular AM esters.
-
(iii)
Transfer the plate with loaded MTs under a fluorescence microscope (with proper filters for excitation at 494 nm and emission at 516 nm) equipped with an environmental chamber to perform measurements at 37 °C and 5% CO2.
-
(iv)
Acquire calcium transients of MT paced at 1.5 Hz, 20 V, 3 ms by recording videos for at least 20 s at video acquisition speed of at least 56 f.p.s., using 10x objective phase containing the entire tissue within the field of view.
CRITICAL STEP Record videos for at least 20 s in order to capture multiple calcium transient cycles within each video and possibly identify calcium abnormalities.
CRITICAL STEP The use of opaque black-walled 96-well plates with clear bottom is crucial for assays using fluorescent dye as Fluo-4 AM to reduce fluorescent signal crosstalk and background.
? TROUBLESHOOTING
(D) Metabolic functionality of whole MTs using Seahorse XFe96 TIMING 6 h for plating MTs plus 3 h for measurements
CRITICAL The Seahorse XFe96 sensor cartridge must be hydrated one day prior to the assay . To do this, open the Seahorse Flux assay kit in a sterile biosafety cabinet. Take the sensor cartridge off the utility plate and place it upside down, sensors facing up. Add 200 μL of ddH2O to all the wells of the utility plate and place the sensor cartridge back into the utility plate. Additionally, take 25 mL of seahorse calibration medium and place in a 50 mL falcon tube. Place both the plate and the tube in a 0% CO2 incubator overnight.
CRITICAL STEP Make sure the incubator is set to 0% CO2 and that it is properly humidified.
CRITICAL STEP Make sure the Probes of the sensor cartridge do not touch any surface as they are sensitive to damage
-
(i)
On the day of the assay, take both the Seahorse calibration medium and the Seahorse XFe96 sensor cartridge out of the incubator. Again, take off the sensor cartridge, place it upside down, and aspirate the ddH2O from all wells. Subsequently, add 200 μL of Seahorse Calibration medium to all the wells. Leave for at least one hour.
CRITICAL STEP Ensure that all the wells are filled, including those that are not used during the assay.
CRITICAL STEP Allow at least 1 h of incubation in the calibration fluid. The cartridge should be used on the day it is placed in the calibration fluid.
-
(ii)
Prepare the Seahorse assay media. (This can also be prepared the day beforeassay).
-
(iii)
Using a P1000 carefully take 20 MTs for the mitochondrial stress test plus 20 additional MTs for the glycolytic capacity test and place them in separate 15 mL Falcon tubes.
CRITICAL STEP Each MT measurement condition should consist of 4 technical repeats each containing a total of 4 MTs. Therefore, a total of 16 MTs per measurement condition is required. However, MTs can be lost or damaged during transfer and plating. We therefore recommend taking 20 MTs per condition.
-
(iv)
Let the MTs sediment for 30 s and aspirate the supernatant without disturbing the MT pellet
-
(v)
Wash the pellets intended for the mitochondrial stress test with Wash+ medium and the glycolytic capacity pellets with Wash- medium 3 times, allowing at least 30 s after medium addition for the MTs to settle.
CRITICAL STEP BSA must be included in the wash medium, in its absence, the MTs will stick to both the surface of the Falcon tube and the pipette tips.
-
(vi)
Prepare a 6-well ultra-low attachment plate and for each condition put a total of 2 mL of the Wash media in a well.
-
(vii)
After the last wash, resuspend the MTs in 750 μL of their wash medium and take up all the MTs with a P1000 and transfer them to a pre-filled well in the ultra-low attachment plate.
CRITICAL STEP Ensure that MTs are added underneath the surface of the liquid as they tend to float on top of the medium.
CRITICAL STEP The medium in the wells must contain BSA otherwise the MTs will stick to the surface of the well and pipette tips.
-
(viii)
Take the coated Seahorse tissue culture plate and aspirate the Matrigel. Add
160 μL of preheated SH+ or SH-medium to the wells for which assay will be run and keep the plate in the biosafety cabinet.
CRITICAL STEP At least 4 wells must be filled with the medium but without MTs. These wells serve as blank measurements for correction of the data.
CRITICAL STEP Although all wells can be used in the assay, it is recommended to use the centre 60 wells as these provide the most reliable data due to less evaporation compared to wells at the edge.
-
(ix)
Place the plate in a stereomicroscope and using a P20 pipette pick 4 MTs in a single 20 μL volume and place them in the centre of a Seahorse tissue culture well. Repeat until all desired wells are filled.
-
(x)
Take a Glass Pasteur pipette and pull the tip by holding it in a gas flame to create a sealed blunt tip. Subsequently, bend the thin part of the pipette in a 90° angle in the gas flame (Fig. 8a).
-
(xi)
Under a stereomicroscope, use the blunt Pasteur pipette to position the four MTs carefully in the centre of the well without the MTs touching each other (Fig. 8b, c).
-
(xii)
Carefully place the Seahorse tissue culture plate in a 0% CO2 incubator for 4 h to allow the MTs to lightly attach to the plate
CRITICAL STEP Do not allow the MTs to attach overnight. This will cause the CFs to migrate out of the MT and attach to the bottom of the plate changing the MT composition.
-
(xiii)
Thaw all the prepared drug stocks (oligomycin, antimycin A, rotenone) at room temperature. Fill all ports that will not be used for drug injection in the next steps with equal amounts of liquid to ensure proper ejection of the compounds during the assay.
CRITICAL STEP Drug concentrations might require optimisation for use with different cell lines. Due to the 3D nature of the tissues, high concentrations are usually required compared to those used for cells growing in 2D culture.
-
(xiv)
For the mitochondrial stress test, load the desired cartridge drug injection ports using a multichannel pipette and Mettler Toledo pipette tips as follows: add 41 μL of the oligomycin stock to 3 mL of SH+ medium yielding a final concentration of 35 μM and load 20 μL into port A. Put 20 μL FCCP in 1.5 mL of SH+ medium at a final concentration of 40 μM and load 22 μL in port B.
Finally add 30 μL of both antimycin A and rotenone to 1.5 mL of SH+ medium, final concentration 2 μM, and load 25 μL into port C.
-
(xv)
To test glycolytic capacity, load the cartridge drug injection ports using a multichannel pipette and Mettler Toledo pipette tips as follows:, dilute 225 μL of the glucose stock in 1275 μL of SH-medium and load 20 μL in port A. Port B will be loaded with 22 μL of oligomycin, which was already prepared in the previous step. Finally, dilute 750 μL of 2-DG stock in 750 μL of SH-medium and put 25 μL in port C.
-
(xvi)
Run the Seahorse according to Seahorse manual and using a 3-injection scheme. Set 4 measurements in the base measurements and 6 measurements for all subsequent injections.
CRITICAL STEP Longer measurement periods compared to 2D cell experiments are necessary after each injection to allow for proper diffusion of the drugs into the 3D tissues. The above recommendations are based on oligomycin addition. It is recommended measuring until the curve has flattened and the OCR no longer decreases compared to the previous measuring point.
-
(xvii)
Normalize the data per well based on either protein measurement, using Bradford or BCA, or DNA content, using Picogreen, or any other preferred normalisation method.
PAUSEPOINT After the addition of the lysis buffer the plate can be kept at −20°C for several weeks before performing the normalisation assay.
? TROUBLESHOOTING
Fig. 8. Overview of correct Seahorse plating conditions.
(a) Bent and stubbed Pasteur pipette used for MT positioning. Scale in cm. (b) MTs directly after plating at an incorrect position in the plate. Scale bar, 400 μm. (c) Correct positioning of MTs in the middle of the Seahorse well. Scale bar, 400 μm.
(E) Sharp electrode recordings TIMING 10-15 min for recording of single cardiac microtissue
-
(i)
On day 21 after the initial MT formation, collect MTs as in option A (i) and transfer them to Matrigel-coated plastic coverslips.
-
(ii)
48-72 h after re-plating MTs, check they are attached to the plate and ready to be recorded.
-
(iii)
Immediately before recording, cut MTs into 2-3 smaller pieces using a syringe needle.
-
(iv)
Place the coverslip onto the patch clamp setup and perfuse with Tyrode solution at a controlled temperature of 37 °C.
-
(v)
Prepare sharp electrodes with a final resistance of 80-100 MOhM from glass capillaries using a micropipette puller.
-
(vi)
Gently impale a single MT piece using sharp electrode and penetrate it using the zap function of the amplifier.
-
(vii)
Start recording action potentials of MT spontaneous activity.
(F) Dissociation of cardiac microtissues into single-cells TIMING 2-3 h
CRITICAL The efficient dissociation of MTs at single-cell level is crucial for several downstream characterizations such as single-cell RNA sequencing, single-cell electrophysiology analysis, or single-cell immunostaining.
-
(i)
Collect MTs and transfer them to a 15-mL conical tube using a P-1000 pipet. Wait for 3-5 min until all MTs sediment at the bottom of the tube.
CRITICAL STEP The number of MTs to use for dissociation is strongly dependent on downstream application and needs to be optimized.
-
(ii)
Carefully aspirate and discard the supernatant using a P-1000 pipet (orifice 1.5 mm) and wash MTs with 0.5 mL of HBSS buffer to completely remove residual culture medium.
-
(iii)
Transfer HBSS containing MTs to a 1.5-mL tube. Wash the original 15-mL tube with 0.5 mL HBSS and add it to the same 1.5-mL tube. Wait until all MTs sediment at the bottom of the tube.
-
(iv)
Carefully aspirate and discard the HBSS solution using a P-1000 pipet and add 300 μL of collagenase II enzymatic solution (290U/mg) pre-warmed at 37 °C.
-
(v)
Incubate the 1.5-mL tube for 20 min at 37 °C, under constant shaking at 450 rpm in a heating block.
-
(vi)
Let MTs sediment on the bottom of the tube, transfer the supernatant to a 15-mL tube containing 4 mL of LI-BPEL medium supplemented with 10% FBS.
-
(vii)
Add 300 μL fresh collagenase II enzymatic solution pre-warmed at 37 °C to MTs in the 1.5-mL tube and incubate for 10 min at 37 °C, under constant shaking at 450 rpm in a heating block.
-
(viii)
Gently pipette the dissociated cells 3 times up and down using P-1000 pipet and let
undissociated MTs sediment, before transferring the supernatant into the same 15-mL tube as that used in step ii.
-
(ix)
Repeat steps vii and vii an additional 3-8 times, until MTs are completely dissociated.
CRITICAL STEP The number of dissociation cycles required for single cell dissociation may vary for each experiment. Check if aggregates are still visible before starting a new dissociation cycle. Avoid over-digestion of MTs to recover as many single cells as possible.
-
(x)
Collect a 20-μL aliquot of cell suspension and count. Centrifuge the remaining cells at 300g for 3 min at room temperature, aspirate and discard the supernatant.
-
(xi)
Depending on the downstream application, proceed as follows:
>For single-cell RNA-sequencing:
Prepare a single-cell suspension in LI-BPEL medium following the manufacturer’s instructions. For 10x Genomics®, for example, if the total number of input cells required is 1700, then the optimal concentration is in the range of ~200 to 700 cells/μL. >For single-cell electrophysiology using patch clamp:
Resuspend the cell pellet in the appropriate volume of LI-BPEL medium supplemented with 50 ng/mL VEGF and 5ng/mL FGF-2 and plate 20-100 x 103cells on Matrigel-coated glass coverslips placed into 24-well plate in a final volume of 700 μL.
Refresh half of the volume of the medium (350 μL) 24 h after dissociation with fresh LI-BPEL medium and keep cells in culture for at least 5 d until patch clamp experiment. This delay is needed as cells require time to recover from dissociation. Although cells may appear recovered after 1-2 d, plasma membranes result too fragile for patch clamp recordings. Keeping cell in culture for longer than 10 d will lead however to some cell death.
>For single-cell immunostaining: resuspend the cell pellet in the appropriate volume of LI-BPEL medium and plate 25-50 x 103 cells per cm2 seeding density on Matrigel-coated glass coverslips. Refresh the cells 24 h after dissociation with fresh LI-BPEL medium. Cells need time to recover from dissociation so keep the cells in culture for at least 3-5 d, before proceeding with immunostaining protocols.
? TROUBLESHOOTING
Timing
Steps 1-9, Thawing and expansion of hiPSCs: 10 min plus 3-4 d for cell expansion
Step 10A, Passaging hiPSCs in E8 medium/VTN-coated plate: 10 min
Step 10B, Passaging hiPSCs in hiPSCs in StemFlex medium/laminin-coated plate: 10 min
Steps 11-15, Cardiac mesoderm induction with growth factor: 5 d
Step 16A, Cardiac endothelial cell differentiation, isolation, culture and cryopreservation: 6 d
Step 16B, Epicardial cell differentiation, culture and cryopreservation: 9 d
Step 16C, Cardiomyocyte differentiation, lactate purification, and cryopreservation: 15-17 d Step Step 17A, Cardiac fibroblast differentiation from freshly dissociated epicardial cells: 16 d
Step 17B, Cardiac fibroblast differentiation from cryopreserved epicardial cells: 10 min for thawing plus 3-4 d for expansion of EPI cells, and 16 d for CF differentiation
Step 18A, Thawing and re-plating of cardiomyocytes (CMs): 10 min plus 5 d for cell recovery
Step 18B, Thawing and expansion of endothelial cells (ECs): 10 min plus 3 d for cell expansion
Step 18C, Thawing and expansion of cardiac fibroblasts (CFs): 10 min plus 3 d for cell expansion Steps 19-26, Generation and culture of 3D cardiac microtissues: 2h for cardiac microtissue generation plus 21 d for cardiac microtissue culture
Step 27A, Immunofluorescence statining: 3 d
Step 27B, Video analysis of contraction: 2-3 min per cardiac microtissue
Step 27C, Video analysis of calcium transients: 1 h for dye incubation and 2-3 min for analysis of single cardiac microtissue
Step 27D, Metabolic functionality of whole MTs using Seahorse XFe96: 6 h for plating plus 3 h for measurements
Step 27E, Sharp electrode recordings: 10-15 min for recording of single cardiac microtissue
Step 27F, Dissociation of cardiac microtissues into single-cells: 2-3 h
Anticipated Results
For succesful cardiac MT generation, highly efficient differentiation of all three input cell types is essential. For this reason, high-quality undifferentiated hiPSC cultures are required to start the differentiation protocols. This can be achieved in various ways with different hiPSC culture media and coatings (E8/VTN and StemFlex/laminin) that provide optimal growth and expansion of undifferentiated hiPSCs, retaining typical hiPSC morphology (Fig. 3). Quality control of the differentiated cell types is necessary to determine the efficiency of differentiation and purity of the final cell populations, before proceeding with cryopreservation and further incorporation of the cells into MTs.
High quality EC populations (Box 2, 3) should display typical endothelial (cobblestone) morphology (Fig. 4a,b) and be positive for endothelial markers, which can be assessed by flow cytometry and immunofluorescence (as VE-cadherin, CD31+ cells > 90%) (Fig. 4c,d). The quality of the EPI population is also crucial since it represents a key intermediate stage required for successful CF differentiation. During differentiation, EPI cells undergo epithelial-to-mesenchymal transition to form CFs and brightfield images at different time points (Fig. 5a) show the change of cell morphology from epithelial to elongated fibroblast-like shape. Epicardial differentiation should give rise to cells exhibiting characteristic cobblestone-like morphology (Fig. 5a) that are positive for epicardial cell markers E-cadherin, ZO-1, and CX43 (junctional localization), as well as nuclear WT1 and TBX18, as seen by immunofluorescence (Fig. 5b). Efficiency of cardiac fibroblast differentiation is assessed by immunofluorescence for markers such as Vimentin, CX43, COL1A1, and GATA4 (Fig. 5c) (Box 3).
In this protocol, we describe two alternative and equally efficient methods for CM generation, one based on cytokines starting from the same common mesoderm to obtain the other cell types11 and the other using a modified protocol based on small molecules66 (Step 16C and Box 1, respectively). Notably, the differentiation method does not influence MT characteristics, although high CM differentiation efficiency is crucial and should be at least 80%. As shown in Box 4, the presence of a homogenous beating monolayer of cells starting from day 8-10 of differentiation is evidence of optimal CM differentiation (Fig. 6a and Supplementary Video 1, 2). Differentiated CM also show positive staining for the sarcomeric marker Troponin T (cTnT+ cells > 80-90%) when analysed by flow citometry (Fig. 6b).
Cellular components of MTs can be produced, characterized and stored frozen in large batches, for rapid use post-thaw in MTs. After MT generation, time-lapse video shows the rapid cell aggregation of the three cell types in compact round-shaped spheroids reaching a cell density of approximately 600-900 cells/mm3 within 24-48 h (Fig. 7 and Supplementary Video 3) and with the MTs starting to beat normally within 1-4 d after initial formation. We characterized the structural and functional parameters of MTs 21 d after their generation and the results obtained using this protocol provide evidence of extensive CM maturation, with significant enhancement of structure and contractile, metabolic, and electrophysiological function11. Representative immunostaining of 3D whole MT at low magnification shows the overall morphology and homogeneous distribution of the three different cell types which stain for the cardiac marker Troponin I (TNNI), endothelial marker CD31 and fibroblast marker Collagen Type I (COL1A1) (Fig. 9a). Notably, CD31 positive cells display a vascular-like network organization within the MT architecture (Supplementary Video 4). Immunofluorescence images of MTs for cardiac sarcomeric proteins TNNI and alpha-2 Actinin (ACTN2) at high magnification shows highly organized sarcomere structures (Fig. 9b). Expected values and variability of sarcomere length and alignment are shown in Table 3, with sarcomere length being the most reproducible parameter among different batches and lines.
Fig. 9. Characterization of 3D MTs after 21 d in culture.
(a) Immunofluorescence analysis of cardiac-(TNNI, green), fibroblast-(COL1A1, red) and endothelial-(CD31, grey) markers in d21-MTs. Nuclei are stained in blue with DAPI. Scale bar, 50 μm. (b) Representative immunofluorescence images for cardiac sarcomeric proteins ACTN2 (red) and TNNI (green), showing the organization of sarcomere structures in 3D MTs. Nuclei are stained in blue with DAPI. Scale bar, 20 μm. (c) Representative contraction traces from MTs stimulated at 1Hz, 2Hz and 3Hz, as indicated. (d) Representative traces of mitochondrial stress test (oxygen consumption rate, OCR, upper panel) and glycolytic stress test (extracellular acidification rate, ECAR, lower panel) from Seahorse measurements of MTs. N = 16 MTs/4 wells. Data are shown as mean ± SEM of 4 wells for the line and mean for each well of 4 MTs for the scatter. (e) Representative action potential (AP) traces recorded from hiPSC-CMs from standard 2D culture (left panel) and dissociated from MTs (right panel). The cells were not artificially hyperpolarized and APs were recorded using the perforated patch clamp technique in current clamp mode.
To evaluate mechanical properties of MTs, contraction video analysis can be performed on spontaneously beating MTs or stimulated at increasing pacing frequencies (Fig. 9c and Supplementary Video 5) and different parameters can be quantified by MUSCLEMOTION software61. Virtually all MTs formed with wild type cells can be expected to follow pacing at 1-2 Hz, whereas only around 50% follow at 3 Hz11. Higher pacing frequency was not tested. To characterize Ca2+ handling in MTs, Ca2+ transients can be assessed by video analysis of MTs loaded with Ca2+-sensitive dye Fluo-4 AM and paced at 1.5 Hz (Supplementary Video 6). Of note, we developed a method to perform metabolic analysis using Seahorse XFe96 assay to measure the metabolic activity on whole 3D MTs11. Usually, MTs are dissociated and re-plated as single-cells which is a less than ideal reflection of the metabolic environment. In this protocol, OXPHOS and glycolytic activity of the cells can be characterized under the same conditions used during regular tissue culture, allowing much more accurate probing of cellular metabolism (Fig. 9d). Electrical maturity of MTs can be determind in whole MTs in 3D using sharp electrode analysis or after single-cell dissociation and single-cell electrophysiology. The action potential (AP) parameters are typically characterized by more hyperpolarized resting membrane potential compared to 2D cultured hiPSC-CMs and by the presence of an AP notch in the majority (> 50%) of the cells/clusters (Fig. 9e). The expected AP parameters from single CMs dissociated from MTs and their expected batch-to-batch and line-to-line variability is shown in Table 3.
In summary, by following this protocol contracting 3D MTs should be obtained in which CMs are characterized by an enhanced maturation. Because differentiated cellular components of MTs can be frozen in large batches, rapid availability for large-scale production of MTs with high reproducibility across different lines and batches, as shown in Table 3, is expected.
Supplementary Material
Editorial Summary.
Human induced pluripotent stem cells are differentiated to the three main cell types found in the heart (cardiomyocytes, cardiac fibroblasts and cardiac endothelial cells) and then combined to form three-dimensional cardiac microtissues.
TWEET Generation of isogenic three-dimensional self-aggregating #CardiacMicrotissue from #hiPSC.
COVER TEASER hiPSC-derived cardiac microtissues
Key Papers.
Giacomelli, E., Meraviglia V., Campostrini G. et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 26, 862-879 e811, doi:10.1016/j.stem.2020.05.004 (2020).
Giacomelli E. et al. Co-Differentiation of Human Pluripotent Stem Cells-Derived Cardiomyocytes and Endothelial Cells from Cardiac Mesoderm Provides a Three-Dimensional Model of Cardiac Microtissue. Curr Protoc Hum Genet. 2017 Oct 18, ’95:21.9.1-21.9.22. doi: 10.1002/cphg.46. PMID: 29044469.
Giacomelli, E. et al. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008-1017, doi:10.1242/dev.143438 (2017).
Table 4. Troubleshooting advice.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 9: hiPSC thawing | Poor attachment and survival of hiPSCs | hiPSC medium was not supplemented with RevitaCell | Add RevitaCell to hiPSC medium before plating the cells |
| Imperfect coating | Passage hiPSCs to properly coated new plates | ||
| 9: hiPSC expansion | Spontaneous differentiation of hiPSCs | Plating density was not optimal (cells too sparse or too dense) | Adjust passage ratio/plating density |
| hiPSCs are proliferating very slowly | hiPSCs should be passaged several times to completely recover from thawing | Passage hiPSCs more | |
| Mycoplasma contamination | Test for Mycoplasma | ||
| Karyotype rearrangements | Perform karyotyping analysis | ||
| 10A(viii) and 10B(x): Passaging hiPSCs | Many dead cells after replating | EDTA/enzyme incubation time was too long | Monitor the cells under a microscope and stop dissociation earlier |
| 15: Cardiac mesoderm induction | Massive cell death during the first 24 h of cardiac mesoderm induction | Poor quality of hiPSC culture | See above problems and solutions related to issues during hiPSC expansion Do not use hiPSCs passaged more than 50-60 times |
| Timing of mesoderm induction was not optimal | To improve differentiation efficiency, start cardiac mesoderm induction ~ 23-24 h after the seeding of hiPSCs | ||
| Seeding density was not optimal | Adjust and optimize the seeding density at day -1 | ||
| Cytokines/small molecules were not added or added incorrectly in the medium | Carefully add the correct molecules (double-check concentration/dilution) to the medium at day 0 of differentiation Adjust the cytokine concentration based on the specific activity of the lot in use | ||
| Cytokines/small molecules were too old or not properly handled/stored | Thaw new aliquots of cytokine/small molecule stocks at the correct temperature and store them no longer than indicated | ||
| Low efficiency of differentiation and | Seeding density of hiPSCs was not optimal | Adjust and optimize the seeding density at day -1 | |
| Cytokines/small molecules were not added or added incorrectly in the medium during cardiac mesoderm induction | Carefully add the correct molecules (double-check concentration/dilution)to the medium exactly after 72h from mesoderm induction | ||
| 17A(viii) and 17B(viii): CF differentiation | Cytokines/small molecules were too old or not properly handled/stored | See above solution for mesoderm induction problems | |
| 16C(xii): CM dissociation | Incomplete dissociation of CM clusters | Insufficient pipetting during dissociation | Examine a small sample of cell suspension under the microscope to check whether dissociation has taken place properly |
| High cell death | Cell line is particularly sensitive to dissociation | Repeat gentle pipetting if necessary | |
| Use 100 units/mL DNAse (see Reagents and Reagent setup) to reduce viscosity during dissociation | |||
| 16C(xvi) and Box 1: CM differentiation | No monolayer confluency at day 6 of differentiation | Seeding density of hiPSCs was not optimal | Adjust and optimize the seeding density at day -1 |
| Cytokines/small molecules were not added or were incorrectly added in the medium during cardiac mesoderm induction | See above solution for problems in the other cell type differentiations | ||
| No spontaneously contracting cells at day 10-12 of differentiation | Monolayer confluency at day 6 of differentiation was not optimal | See solutions above for monolayer confluency problem | |
| Cells were not properly fed and nutrients in the medium are exhausted | Refresh LI-BPEL medium every 2-3 d after day 6 of differentiation and increase the amount of medium per well if medium turns yellow. If cells are still not beating, discard the differentiation and start a new one. | ||
| 18A(x): CM thawing and re-plating | Many dead cells after thawing and re-plating | Freezing medium was unsuitable | Use 90% (vol/vol) KSR with 10% DMSO (vol/vol) for freezing CMs |
| Freezing process damaged CMs | Pipette CMs carefully and gently to preserve cell membrane | ||
| Always use isopropanol freezing cryoboxes and freeze at −80 °C. Store the cryovials in liquid nitrogen tank after 24-48 h | |||
| Thawing process damaged CMs | Thaw CMs as quickly as possible but add medium slowly to minimize osmotic shock | ||
| Revitacell (dilution 1:100 to 1:200) can be added to the medium to improve recovery. | |||
| 26: Cardiac MT generation | Cardiac MTs are not compact and/or are not beating well | CM differentiation efficiency was too low | Only use differentiated cell populations with > 80% TnT positive cells |
| Cell dissociation was too harsh | Monitor cell dissociation under the microscope | ||
| Carefully and gently pipette the cells and do not centrifuge at more than 300g | |||
| 96-well plates were not centrifuged after plating | Plates need to be centrifuged for 10 min at 300g to promote cell aggregation | ||
| MTs were not properly fed and nutrients in the medium are exhausted | Refresh medium every 3 d Add growth factors to LI-BPEL just before refreshing medium | ||
| Extensive medium evaporation occurred | Check cell incubator settings to maintain proper humidity | ||
| Do not seed MTs in the outermost wells of the plate. Add 50uL of PBS to the outermost wells of the plate to prevent medium evaporation | |||
| 27A(xix): Immunostaining of whole 3D cardiac MTs | Immunostaining of cardiac MTs is not homogenous | Under- or over fixation of cardiac MTs | Fix the cardiac MTs under the correct conditions: 1h at 4° C |
| High fluorescent background in immunostained cardiac MTs | Inadequate permeabilization step | Avoid under- or over permeabilization (30 min-1 h at room temperature is the optimal condition) | |
| Insufficient washing | Wash cardiac MTs no less than three times (20-30 min each) | ||
| 27C(iv): Video analysis of calcium transients | Cardiac MTs are not properly and homogenously loaded with Fluo-4 AM fluorescent calcium dye | Fluo-4 AM loading solution was not properly mixed | Vortex Fluo-4 AM solution before loading cardiac MTs to ensure homogenous distribution of the fluorescent dye |
| Fluo-4 AM loading solution concentration and incubation conditions were incorrect | Carefully dilute the correct amount of Fluo-4 AM stock to LI-BPEL medium. Ensure that incubation time (30 min) and temperature (37 °C, 5% CO2) are correct. | ||
| High fluorescence background of cardiac MTs after Fluo-4 AM | Incubation time was too long or Fluo-4 AM loading solution concentration was too high | Do not exceed 30 min as incubation time. Do not exceed 5 μM as concentration of Fluo-4 AM loading solution | |
| Fluo-4 AM loading solution was not properly washed out after incubation | Carefully wash out and remove any dye not specifically associated with cardiac MTs | ||
| 27D(xvii): Metabolic analysis using Seahorse | Injections do not show expected results | Incubation time too short | Longer exposure to drugs is necessary, rerun assay with more measurement points and wait for curve to flatten out |
| Drug concentrations are not optimal | Depending on cell line, drug concentrations might need additional adjustment. Note that concentrations can be both too low and too high. A broad screen is recommended | ||
| Injection ports are clogged | Make sure to use medium without large proteins, such as albumin. These can cause the small injection ports to clog resulting in the drug not being added to the medium | ||
| MTs detach after assay | Aspiration of medium too disruptive | Use a pipette to carefully remove medium manually | |
| Normalisation has very low signal | Assay not sensitive enough | Total cell count per well is low, decrease lysis buffer used to increase concentration or use a different assay. Picogreen assay has necessary sensitivity | |
| 27F(xi): Dissociation of cardiac MTs at single-cell level | Many dead cells after replating | Enzymatic dissociation was too harsh | Carefully and gently pipette the cells to to preserve cell membrane |
| Reduce or increase the rounds of dissociation, and reduce the mechanical pipetting |
Acknowledgements
We thank D. Ward-van Oostwaard, M.P.H. Mol, L.G.J. Tertoolen, A. Krotenberg Garcia and A. Cochrane for technical assistance.
This project was supported by European Research Council (ERCAdG 323182 STEMCARDIOVASC; ERCStG 91715303 STEMCARDIORISK); European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement No. 602423; European Union’s Horizon 2020 research and innovation Programme under grant agreement No. 668724; Netherlands Organ-on-Chip Initiative, an NWO Gravitation project funded by the Ministry of Education, Culture and Science of the government of the Netherlands (024.003.001); Transnational Research Project on Cardiovascular Diseases (JTC2016_FP-40-021 ACM-HF); The Netherlands Organisation for Health Research and Development ZonMW (MKMD project No. 114022504; VIDI project No. 91715303); Health~Holland TKI-LSH PPP-allowance (LSHM17013-H007); European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska Curie Grant Agreement No. 707404; Individual Fellowship under the Marie Sklodowska Curie Grant Agreement No. 838985.
Footnotes
Author Contributions
G.C. and V.M. designed and performed the experiments, analysed the data, wrote the manuscript. E.G developed and optimized the cardiac microtissue protocol, acquired and analysed the data, wrote the manuscript. R.v.H. and L.Y acquired and analysed the data and wrote the manuscript. R.P.D. contributed in drafting the manuscript and revising it for important intellectual content. M.B., V.O. and C.L.M. conceived, designed and supervised the protocol, acquired the funding, wrote and approved the final manuscript.
Competing Interests
C.L.M. is co-founder of Pluriomics (now Ncardia) bv.
Data Availability
The datasets generated and/or analysed during the current study are available in Giacomelli et al. 202011 and from the corresponding authors upon request.
Code Availability Statement
MUSCLEMOTION is freely accessible on the web at https://gitlab.com/bjvanmeer/MUSCLEMOTION or https://github.com/l-sala/MUSCLEMOTION/. The license is a GNU GENERAL PUBLIC LICENSE version 3 (GPL v3), which can be found here: https://github.com/l-sala/MUSCLEMOTION/blob/master/LICENSE. After downloading the macro, users can access the code through a text editor or directly through FIJI. When using MUSCLEMOTION, users are encouraged to read Sala, L. et al., 2018 PMID: 29282212 (https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.117.312067)61.
References
- 1.Broutier L, et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc. 2016;11:1724–1743. doi: 10.1038/nprot2016.097. [DOI] [PubMed] [Google Scholar]
- 2.Koike H, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature. 2019;574:112–116. doi: 10.1038/s41586-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/Jgastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 4.Tirziu D, Giordano FJ, Simons M. Cell communications in the heart. Circulation. 2010;122:928–937. doi: 10.1161/CIRCULATIONAHA.108.847731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann O, et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell. 2015;161:1566–1575. doi: 10.1016/Jcell2015.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Zhou P, Pu WT. Recounting Cardiac Cellular Composition. Circ Res. 2016;118:368–370. doi: 10.1161/CIRCRESAHA.116.308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinto AR, et al. Revisiting Cardiac Cellular Composition. Circ Res. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293:H1883–1891. doi: 10.1152/ajpheart00514.2007. [DOI] [PubMed] [Google Scholar]
- 9.Furtado MB, Nim HT, Boyd SE, Rosenthal NA. View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development. 2016;143:387–397. doi: 10.1242/dev120576. [DOI] [PubMed] [Google Scholar]
- 10.Brutsaert DL. Cardiac endothelial-myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol Rev. 2003;83:59–115. doi: 10.1152/physrev00017.2002. [DOI] [PubMed] [Google Scholar]
- 11.Giacomelli E, et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell. 2020;26:862–879.:e811. doi: 10.1016/Jstem.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi: 10.1016/Jcell2006.11.031. [DOI] [PubMed] [Google Scholar]
- 13.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/Jcell2006.10.029. [DOI] [PubMed] [Google Scholar]
- 14.van Wijk B, van den Hoff M. Epicardium and myocardium originate from a common cardiogenic precursor pool. Trends Cardiovasc Med. 2010;20:1–7. doi: 10.1016/Jtcm.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Giacomelli E, et al. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development. 2017;144:1008–1017. doi: 10.1242/dev143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwamiya T, Matsuura K, Masuda S, Shimizu T, Okano T. Cardiac fibroblast-derived VCAM-1 enhances cardiomyocyte proliferation for fabrication of bioengineered cardiac tissue. Regen Ther. 2016;4:92–102. doi: 10.1016/Jreth.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guadix JA, et al. Human Pluripotent Stem Cell Differentiation into Functional Epicardial Progenitor Cells. Stem Cell Reports. 2017;9:1754–1764. doi: 10.1016/Jstemcr2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tallquist MD, Molkentin JD. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol. 2017;14:484–491. doi: 10.1038/nrcardio.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, et al. Efficient Differentiation of TBX18(+)/WT1(+) Epicardial-Like Cells from Human Pluripotent Stem Cells Using Small Molecular Compounds. Stem Cells Dev. 2017;26:528–540. doi: 10.1089/scd.2016.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bich L, Pradeu T, Moreau JF. Understanding Multicellularity: The Functional Organization of the Intercellular Space. Front Physiol. 2019;10:1170. doi: 10.3389/fphys.2019.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan ACA. Recapitulating Cell-Cell Interactions for Organoid Construction - Are Biomaterials Dispensable? Trends Biotechnol. 2016;34:711–721. doi: 10.1016/Jtibtech.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 22.Mills RJ, et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunes SS, et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao L, et al. Myocardial Tissue Engineering With Cells Derived From Human-Induced Pluripotent Stem Cells and a Native-Like, High-Resolution, 3-Dimensionally Printed Scaffold. Circ Res. 2017;120:1318–1325. doi: 10.1161/CIRCRESAHA.116.310277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong CS, et al. Biomaterial-Free Three-Dimensional Bioprinting of Cardiac Tissue using Human Induced Pluripotent Stem Cell Derived Cardiomyocytes. Sci Rep. 2017;7:4566. doi: 10.1038/s41598-017-05018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldfracht I, et al. Engineered heart tissue models from hiPSC-derived cardiomyocytes and cardiac ECM for disease modeling and drug testing applications. Acta Biomater. 2019;92:145–159. doi: 10.1016/Jactbio.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Z, et al. Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nat Biomed Eng. 2018;2:955–967. doi: 10.1038/s41551-018-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wanjare M, et al. Vascularization of Engineered Spatially Patterned Myocardial Tissue Derived From Human Pluripotent Stem Cells in vivo. Front Bioeng Biotechnol. 2019;7:208. doi: 10.3389/fbioe.2019.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell. 2019;176:913–927.:e918. doi: 10.1016/Jcell2018.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tulloch NL, et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaaf S, et al. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One. 2011;6:e26397. doi: 10.1371/journalpone.0026397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burridge PW, et al. Multi-cellular interactions sustain long-term contractility of human pluripotent stem cell-derived cardiomyocytes. Am J Transl Res. 2014;6:724–735. [PMC free article] [PubMed] [Google Scholar]
- 34.Hinson JT, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannhardt I, et al. Human Engineered Heart Tissue: Analysis of Contractile Force. Stem Cell Reports. 2016;7:29–42. doi: 10.1016/Jstemcr2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keung W, et al. Non-cell autonomous cues for enhanced functionality of human embryonic stem cell-derived cardiomyocytes via maturation of sarcolemmal and mitochondrial KATP channels. Sci Rep. 2016;6:34154. doi: 10.1038/srep34154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiburcy M, et al. Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair. Circulation. 2017;135:1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee EK, et al. Machine Learning of Human Pluripotent Stem Cell-Derived Engineered Cardiac Tissue Contractility for Automated Drug Classification. Stem Cell Reports. 2017;9:1560–1572. doi: 10.1016/Jstemcr2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shadrin IY, et al. Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat Commun. 2017;8:1825. doi: 10.1038/s41467-017-01946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronaldson-Bouchard K, et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuruyama S, Matsuura K, Sakaguchi K, Shimizu T. Pulsatile tubular cardiac tissues fabricated by wrapping human iPS cells-derived cardiomyocyte sheets. Regen Ther. 2019;11:297–305. doi: 10.1016/Jreth.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills RJ, et al. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell. 2019;24:895–907.:e896. doi: 10.1016/Jstem.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Goldfracht I, et al. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat Commun. 2020;11:75. doi: 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang Y, et al. Modulating cardiomyocyte and fibroblast interaction using layer-by-layer deposition facilitates synchronisation of cardiac macro tissues. Soft Matter. 2020;16:428–434. doi: 10.1039/c9sm01531k. [DOI] [PubMed] [Google Scholar]
- 45.Dostanic M, et al. A Miniaturized EHT Platform for Accurate Measurements of Tissue Contractile Properties. Journal of Microelectromechanical Systems. 2020;29:881–887. doi: 10.1109/jmems.2020.3011196. [DOI] [Google Scholar]
- 46.Masumoto H, et al. The myocardial regenerative potential of three-dimensional engineered cardiac tissues composed of multiple human iPS cell-derived cardiovascular cell lineages. Sci Rep. 2016;6:29933. doi: 10.1038/srep29933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huebsch N, et al. Miniaturized iPS-Cell-Derived Cardiac Muscles for Physiologically Relevant Drug Response Analyses. Sci Rep. 2016;6:24726. doi: 10.1038/srep24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmermann WH, et al. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 2002;90:223–230. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- 49.Ravenscroft SM, Pointon A, Williams AW, Cross MJ, Sidaway JE. Cardiac Nonmyocyte Cells Show Enhanced Pharmacological Function Suggestive of Contractile Maturity in Stem Cell Derived Cardiomyocyte Microtissues. Toxicol Sci. 2016;152:99–112. doi: 10.1093/toxsci/kfw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pointon A, et al. From the Cover: High-Throughput Imaging of Cardiac Microtissues for the Assessment of Cardiac Contraction during Drug Discovery. Toxicol Sci. 2017;155:444–457. doi: 10.1093/toxsci/kfw227. [DOI] [PubMed] [Google Scholar]
- 51.Forsythe SD, et al. Environmental Toxin Screening Using Human-Derived 3D Bioengineered Liver and Cardiac Organoids. Front Public Health. 2018;6:103. doi: 10.3389/fpubh.2018.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens KR, Pabon L, Muskheli V, Murry CE. Scaffold-free human cardiac tissue patch created from embryonic stem cells. Tissue Eng Part A. 2009;15:1211–1222. doi: 10.1089/tentea.2008.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polonchuk L, et al. Cardiac spheroids as promising in vitro models to study the human heart microenvironment. Sci Rep. 2017;7:7005. doi: 10.1038/s41598-017-06385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sebastiao MJ, et al. Bioreactor-based 3D human myocardial ischemia/reperfusion in vitro model: a novel tool to unveil key paracrine factors upon acute myocardial infarction. Transl Res. 2020;215:57–74. doi: 10.1016/Jtrsl2019.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Richards DJ, et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat Biomed Eng. 2020;4:446–462. doi: 10.1038/s41551-020-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiao X, et al. Uncoupling DNA damage from chromatin damage to detoxify doxorubicin. Proc Natl Acad Sci U S A. 2020;117:15182–15192. doi: 10.1073/pnas.1922072117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veerman CC, et al. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev. 2015;24:1035–1052. doi: 10.1089/scd.2014.0533. [DOI] [PubMed] [Google Scholar]
- 58.Gaudesius G, Miragoli M, Thomas SP, Rohr S. Coupling of cardiac electrical activity over extended distances by fibroblasts of cardiac origin. Circ Res. 2003;93:421–428. doi: 10.1161/01.RES.0000089258.40661.0C. [DOI] [PubMed] [Google Scholar]
- 59.De Simone SA, et al. The Role of Membrane Capacitance in Cardiac Impulse Conduction: An Optogenetic Study With Non-excitable Cells Coupled to Cardiomyocytes. Front Physiol. 2020;11:194. doi: 10.3389/fphys.2020.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quaife-Ryan GA, et al. Multicellular Transcriptional Analysis of Mammalian Heart Regeneration. Circulation. 2017;136:1123–1139. doi: 10.1161/CIRCULATIONAHA.117.028252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sala L, et al. MUSCLEMOTION: A Versatile Open Software Tool to Quantify Cardiomyocyte and Cardiac Muscle Contraction In Vitro and In Vivo. Circ Res. 2018;122:e5–e16. doi: 10.1161/CIRCRESAHA.117.312067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mastikhina O, et al. Human cardiac fibrosis-on-a-chip model recapitulates disease hallmarks and can serve as a platform for drug testing. Biomaterials. 2020;233:119741. doi: 10.1016/Jbiomaterials.2019.119741. [DOI] [PubMed] [Google Scholar]
- 63.van den Berg CW, Elliott DA, Braam SR, Mummery CL, Davis RP. Differentiation of Human Pluripotent Stem Cells to Cardiomyocytes Under Defined Conditions. Methods Mol Biol. 2016;1353:163–180. doi: 10.1007/7651_2014_178. [DOI] [PubMed] [Google Scholar]
- 64.Sala L, Ward-van Oostwaard D, Tertoolen LGJ, Mummery CL, Bellin M. Electrophysiological Analysis of human Pluripotent Stem Cell-derived Cardiomyocytes (hPSC-CMs) Using Multi-electrode Arrays (MEAs) J Vis Exp. 2017 doi: 10.3791/55587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giacomelli E, Bellin M, Orlova VV, Mummery CL. Co-Differentiation of Human Pluripotent Stem Cells-Derived Cardiomyocytes and Endothelial Cells from Cardiac Mesoderm Provides a Three-Dimensional Model of Cardiac Microtissue. Curr Protoc Hum Genet. 2017;95:21 29 21–21 29 22. doi: 10.1002/cphg.46. [DOI] [PubMed] [Google Scholar]
- 66.van den Brink L, et al. Cryopreservation of human pluripotent stem cell-derived cardiomyocytes is not detrimental to their molecular and functional properties. Stem Cell Res. 2020;43:101698. doi: 10.1016/Jscr2019.101698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burridge PW, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tohyama S, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/Jstem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 70.Moretti A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 71.Zhang M, et al. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc Natl Acad Sci U S A. 2014;111:E5383–5392. doi: 10.1073/pnas.1419553111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meraviglia V, et al. Generation of human induced pluripotent stem cell line LUMCi027-A and its isogenic gene-corrected line from a patient affected by arrhythmogenic cardiomyopathy and carrying the c.2013delC PKP2 mutation. Stem Cell Res. 2020;46:101835. doi: 10.1016/Jscr2020.101835. [DOI] [PubMed] [Google Scholar]
- 73.Assou S, Bouckenheimer J, De Vos J. Concise Review: Assessing the Genome Integrity of Human Induced Pluripotent Stem Cells: What Quality Control Metrics? Stem Cells. 2018;36:814–821. doi: 10.1002/stem.2797. [DOI] [PubMed] [Google Scholar]
- 74.Stacey GN, et al. Stem cell culture conditions and stability: a joint workshop of the PluriMes Consortium and Pluripotent Stem Cell Platform. Regen Med. 2019;14:243–255. doi: 10.2217/rme-2019-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller LR, et al. Considering sex as a biological variable in preclinical research. FASEB J. 2017;31:29–34. doi: 10.1096/fJ201600781R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pioner JM, et al. Isolation and Mechanical Measurements of Myofibrils from Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Reports. 2016;6:885–896. doi: 10.1016/Jstemcr2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferrantini C, et al. Impact of detubulation on force and kinetics of cardiac muscle contraction. J Gen Physiol. 2014;143:783–797. doi: 10.1085/jgp.201311125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Meer BJ, et al. Quantification of Muscle Contraction In Vitro and In Vivo Using MUSCLEMOTION Software: From Stem Cell-Derived Cardiomyocytes to Zebrafish and Human Hearts. Curr Protoc Hum Genet. 2018;99:e67. doi: 10.1002/cphg.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in Giacomelli et al. 202011 and from the corresponding authors upon request.
MUSCLEMOTION is freely accessible on the web at https://gitlab.com/bjvanmeer/MUSCLEMOTION or https://github.com/l-sala/MUSCLEMOTION/. The license is a GNU GENERAL PUBLIC LICENSE version 3 (GPL v3), which can be found here: https://github.com/l-sala/MUSCLEMOTION/blob/master/LICENSE. After downloading the macro, users can access the code through a text editor or directly through FIJI. When using MUSCLEMOTION, users are encouraged to read Sala, L. et al., 2018 PMID: 29282212 (https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.117.312067)61.