Abstract
Despite the huge success of tyrosine kinase inhibitors as anticancer agents, severe side effects are a major problem. In order to overcome this drawback, the first hypoxia-activatable 2-nitroimidazole-based prodrugs of the clinically approved ALK and c-MET inhibitor crizotinib were developed. The 2-aminopyridine functionality of crizotinib (essential for target kinase binding) was considered as ideal position for prodrug derivatization. Consequently, two different prodrugs were synthesized with the nitroimidazole unit attached to crizotinib either via carbamoylation (A) or alkylation (B) of the 2-aminopyridine moiety. The successful prodrug design could be proven by docking studies and a dramatically reduced ALK and c-MET kinase-inhibitory potential. Furthermore, the prodrugs showed high stability in serum and release of crizotinib in an enzymatic nitroreductase-based cleavage assay. The in vitro activity of both prodrugs was investigated against ALK- and c-MET-dependent or -overexpressing cells, revealing a distinct hypoxia-dependent activation for prodrug A. Finally, inhibition of c-MET phosphorylation and cell proliferation could also be proven in vivo. In summary of the theoretical, chemical and biological studies, prodrug derivatization of the 2-aminopyridine position can be considered as a promising strategy to reduce the side effects and improve the anticancer activity of crizotinib.
Introduction
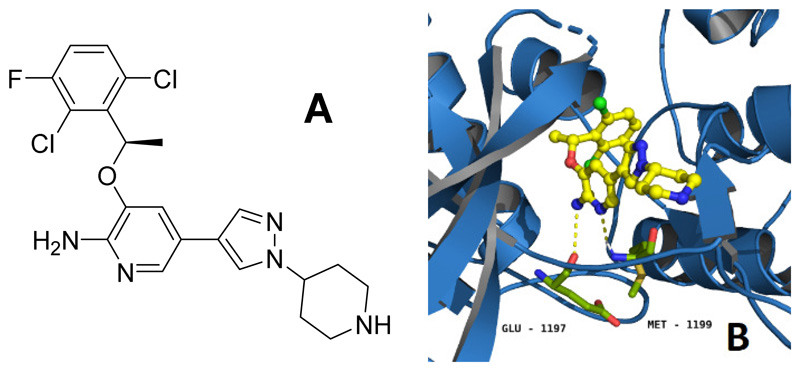
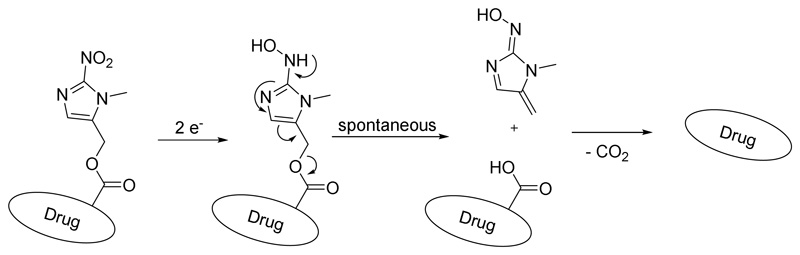
Despite outstanding success in clinical applications1, tyrosine kinase inhibitors (TKIs) evoke strong adverse effects that originate from two different mechanisms: on-target effects on the desired enzymes in healthy tissues and/or off-target effects e.g. inhibition of other enzymes due to insufficient target selectivity2. Notably, combination therapies for simultaneous inhibition of different tyrosine kinases can generate exciting tumor regressions in animal models3 as well as prolonged overall survival in cancer patients4. However, clinical studies also revealed that such combinations often result in accumulated toxicities5. Consequently, the reduction of severe side effects is of central interest to provide improved TKI (pro)drugs. Crizotinib (Figure 1A) is a potent, reversible ATP antagonist mainly targeting anablastic lymphoma kinase (ALK), mesenchymal-epithelial transition factor (MET) and c-ros oncogene 1 (ROS1)6. Currently, crizotinib is approved for treatment of ALK-positive or ROS1-positive non-small cell lung cancer7. The main severe side effects include myelosuppression, pneumonitis and hepatotoxicity as well as pulmonary embolism and the risk of edema formation. Herein, we designed and investigated a rational prodrug approach to reduce the adverse effects of crizotinib. In general, the development of a prodrug first requires suppression of drug activity8, which in case of inhibitors can be achieved by reduction/elimination of the strong interactions with its target(s). Figure 1B depicts the interaction between crizotinib and ALK, showing the 2-aminopyridine residue to form hydrogen bonds with Glu-1197 and Met-11999. In case of c-MET, the main interactions emerge with Pro-1158 and Met-1160 and a crucial π-π stacking of the 2,6-dichloro-3-fluorophenyl ring with Tyr-1230 (Figure S1)10. Therefore, the 2-aminopyridinyl functional group is a reasonable site for prodrug derivatization. Many different trigger moieties for prodrug activation have been described in literature over the past decades, with hypoxia-activated prodrugs as the most prominent strategies, as hypoxia is usually not observed in healthy tissue11, 12. Therefore, the hypoxia-activatable 2-nitroimidazyl trigger-linker unit, a frequently used moiety in research13 and clinical trials (e.g. TH-30214 and TH-400015), was chosen. The activation via an overall two-step one-electron reduction is catalyzed by intracellular oxidoreductases, such as the nicotinamide adenine dinucleotide phosphate (NADPH) cytochrome P450 reductase16. Hereby, a hydroxylamine species is formed which undergoes a spontaneous electron cascade and releases the active drug through an 1,6-elimination process (Scheme 1). Under normoxic conditions, the presence of oxygen quenches the reduction and the nitro group remains stable. In this study, the trigger moiety was introduced either via carbamoylation (A) or alkylation (B) of the 2-aminopyridine moiety of crizotinib (Scheme 2).
Figure 1.
(A) Chemical structure of crizotinib. (B) Interaction of crizotinib (yellow) with ALK (marine blue), hydrogen bonds to the ATP-binding pocket are visualized as dashed yellow lines (PDB ID: 2xp2). The bond forming amino acids Glu-1197 and Met-1199 are coloured in green.
Scheme 1. Schematic illustration of the nitroimidazole trigger activation process after reductive activation into a hydroxylamine and the spontaneous release of the free drug.
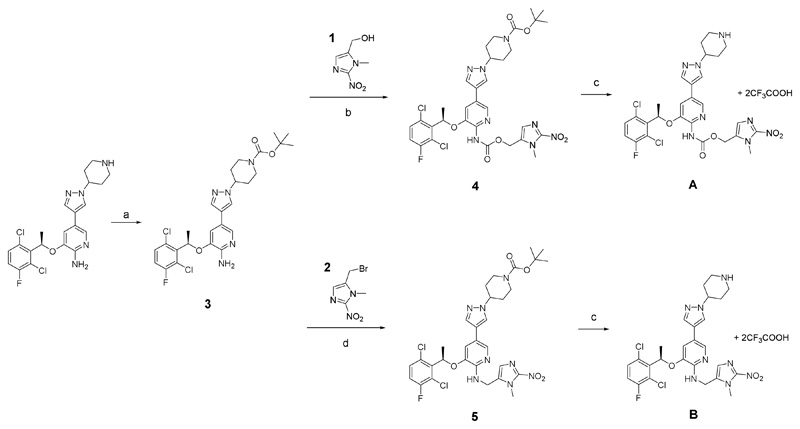
Scheme 2.
Synthesis of the crizotinib prodrugs A and B. a) Boc anhydride, THF, 2 h; b) 1, carbon monoxide, Pd(OAc)2, Cu(OAc)2 x H2O, KI, DMSO, ACN; c) TFA, DCM; d) 2, KI, DMF.
Results and discussion
Docking studies
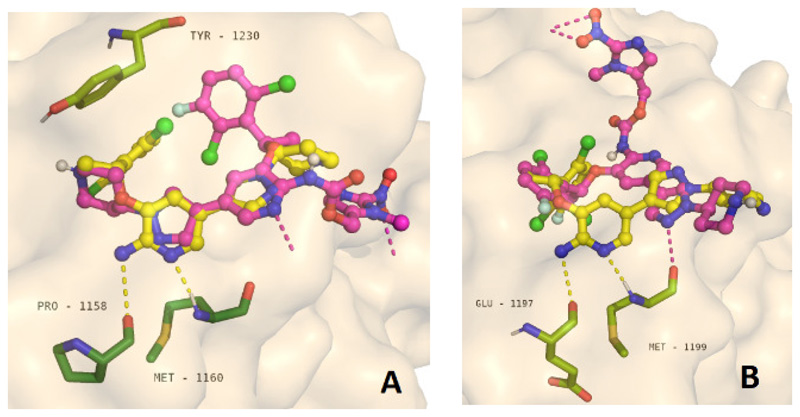
Initially, the spatial interactions between crizotinib and its prodrugs with the tyrosine kinase domains of ALK (PDB ID: 2PX2) and c-MET (PDB ID: 2WGJ) were analyzed by molecular docking studies using AutoDock Vina. Interestingly, the ATP-binding site of c-MET is located in a pocket, while in case of ALK the active center forms a cleft structure. As desired, docking of the prodrugs revealed the prohibition of crucial interactions between the 2-aminopyridine moiety of crizotinib and both tyrosine kinases10 (Figure 2, Figures S1–S4). In case of c-MET, not only the hydrogen bonds with Pro-1158 and Met-1160 residues are suppressed, but also the crucial π-π stacking interaction with the Tyr-1230 moiety (Figure 2A). Consequently, the derivatization of the 2-aminopyridine position of crizotinib can be considered as promising prodrug strategy.
Figure 2.
Visualization of the docking results: Protein - ligand interaction of crizotinib and prodrug A in complex with c-MET (A) and ALK (B). In both structures, the main hydrogen bonds of the 2-aminopyridine moiety of crizotinib that are essential for the tyrosine kinase inhibition are suppressed. In case of c-MET, additionally the crucial π-π interaction between the 2,6-dichloro-3-fluorophenyl moiety and Tyr-1230 is disturbed.
Synthesis
For the synthesis of the crizotinib prodrugs, first the 2-nitroimidazole trigger moiety 1 was synthesized from sarcosine hydrochloride according to a 5-step literature procedure17 (Scheme S1). The respective bromine species 2 (for synthesis of prodrug B) was obtained by stirring the alcohol 1 in thionyl bromide. The formation of the nitroimidazole prodrug A (via carbamoylation) started from commercially available crizotinib, which was tert-butyl carboxyl (Boc)-protected (3) at the piperidine moiety to selectively modify the 2-aminopyridine motif (Scheme 2). To avoid hazardous phosgene-containing coupling methods, a strategy using an activated alcohol based on para-nitrophenyl carbonate13 or an imidazoyl carbamate18 was employed, however, without formation of the desired product. Likewise, the transformation of the amine functionality of crizotinib into an isocyanate19,20 and subsequent addition of 1 did not result in the desired carbamate. Therefore, a catalytic carbonylation method from Wang et al. using carbon monoxide, Pd(OAc)2, Cu(OAc)2 and KI was applied21. The procedure has been adapted by addition of acetonitrile to the DMSO solution (due to the low solubility of carbon monoxide in DMSO), a decrease in the alcohol/amine ratio from a 10-fold excess to equimolar amounts and reduction of the reaction temperature from very harsh 120°C to ~40°C. Especially the need of just one equivalent alcohol was of high importance, considering the five step synthesis of 1. Finally, deprotection was performed using trifluoroacetic acid (TFA) yielding the TFA salt of A. Prodrug B could be synthesized using again the protected crizotinib and 2, which after deprotection also resulted in the TFA salt. Both compounds were fully characterized using NMR spectroscopy, high-resolution mass spectrometry and the purity was confirmed via elemental analysis.
Physicochemical characterization
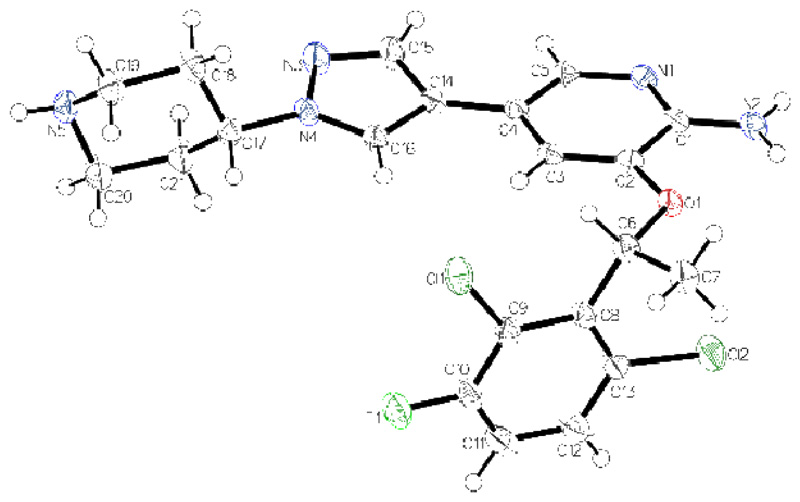
Although crizotinib is an approved anticancer drug, the x-ray crystal structure of this compound has not been reported so far. As depicted in Figure 3, the structure of crizotinib is quite distorted with a planar-conjugated system containing the 2-aminopyridine and pyrazole ring system and a dihedral angle of 83.8° to the 2,6-dichloro-3-fluorophenyl ring. Two hydrogen bonds connect two molecules of crizotinib via the 2-aminopyridine moieties and a third molecule connects the piperidine group with the second hydrogen of the amino functionality (Figure S5).
Figure 3.
ORTEP drawing of the x-ray single crystal structure of crizotinib with thermal ellipsoids depicted at 50 % probability level. The chiral position on C6 in R-conformation is proofed by flack and hooft parameter (0.04(3), 0.03(4)).
The solubility of prodrug A was >20 mg/mL in phosphate-buffered saline (PBS) at room temperature, whereas crizotinib was nearly insoluble under these conditions (<<0.5 mg/mL). Sufficient stability under physiological conditions is one main criterion for the biological integrity of a prodrug system. Therefore, crizotinib and both prodrugs were incubated in phosphate buffer at pH 7.4 and RPMI-1640 medium with 10 % serum at 37 °C. Both prodrugs were stable without significant degradation under these conditions for up to 72 h (Figure S6–S8).
Biochemical investigations
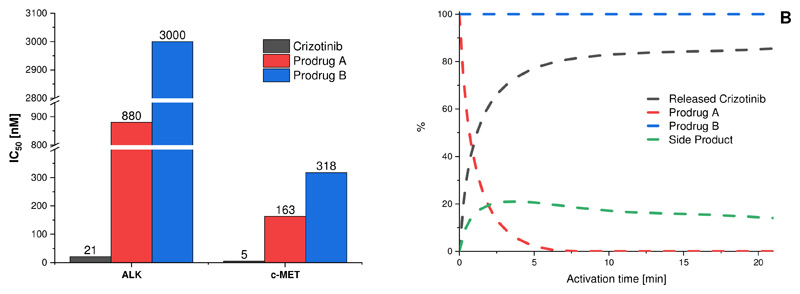
Next, we investigated the kinase-inhibitory potential of the two prodrugs compared to crizotinib in a cell-free kinase assay. The carbamate prodrug A showed a 42-fold and 33-fold lower inhibitory potential compared to crizotinib for ALK and c-MET, respectively. The alkylated prodrug B even revealed an inhibitory reduction of >143-fold for ALK and 64-fold for c-MET (Figure 4A, Figure S9, Table S4). These results are in perfect agreement with the docking studies and indicate promising prodrug properties.
Figure 4.
A. Kinase inhibitory potential of crizotinib and its prodrugs in a cell-free kinase assay. IC50 values of the prodrugs (red and blue) compared to crizotinib (black). B. Activation assay of prodrugs A and B in presence of nitroreductase enzyme (E. coli) and NADH at pH 7 and 37 °C for 20 min (black: released crizotinib; red: prodrug A; green: side product; blue: prodrug B).
Once the prodrugs reach the tumor site, irreversible enzymatic activation should release crizotinib under hypoxic conditions. The general ability of both prodrugs to be activated was simulated using a nitroreductase enzyme (extracted from E. coli), which is known to reduce the 2-nitroimidazole moiety in presence of NADH at pH 7.413. Formation of crizotinib was monitored via HPLC measurements. For prodrug A, the data clearly showed a complete conversion already after 10 min with release of crizotinib as main product (about 80%; Figure 4B, Figure S10). In contrast, prodrug B, where the nitroimidazole unit was attached via alkylation, could not be activated and, thus, no formation of crizotinib was observed.
Biological investigations
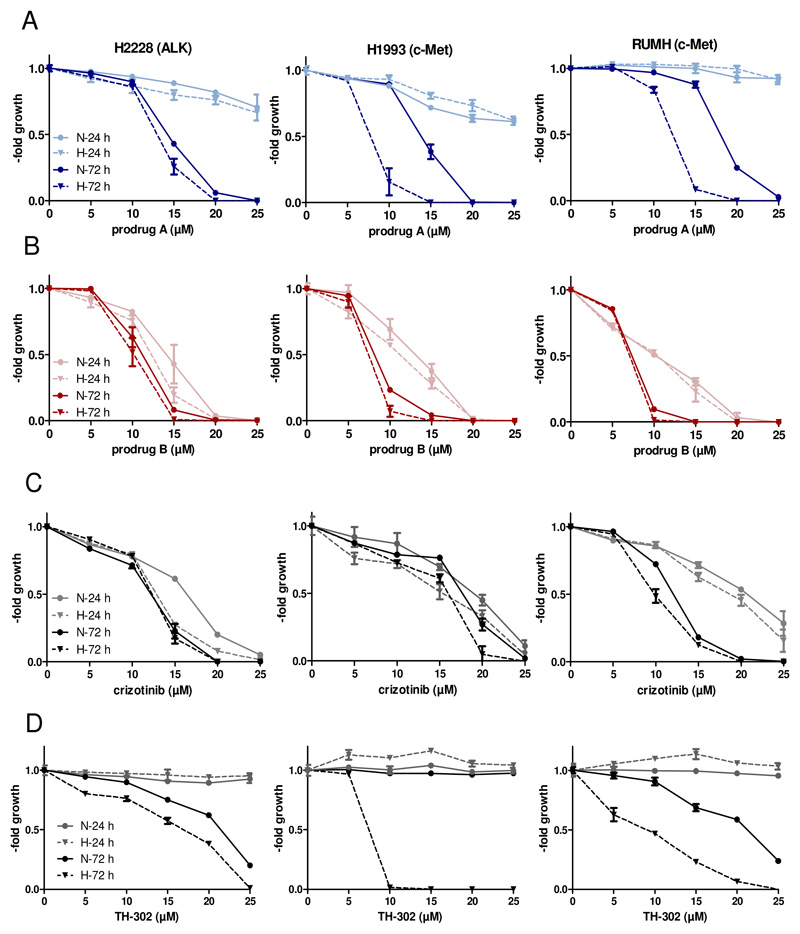
In order to investigate their biological activity, both prodrugs were subsequently tested in cell culture experiments, whereby based on the data of the activation assay, prodrug B was considered as negative control to prodrug A. As a first approach, the prodrug nature as well as the impact of oxygen levels on the anticancer activity in short- and long-term cytotoxicity assays (24 h and 72 h) against three human cancer cell models H2228 (non-small cell lung cancer, ALK-dependent)22, H1993 (non-small cell lung cancer: c-MET-dependent)23 and RUMH (renal cell carcinoma, c-MET-overexpressing) were analyzed. As positive controls for kinase inhibition unmodified crizotinib and for activation under hypoxia the phosphoramidate mustard prodrug TH-302 (Evofosfamide®; which also contains a 2-nitroimidazole-5-yl unit as hypoxic trigger) were used. As shown in Table S5 and Figure 5, after 24 h treatment prodrug A and TH-302 were rather inactive with IC50 values >>25 μM. As expected, crizotinib was active in all three models with IC50 values of ~20 μM. Unexpectedly, also prodrug B displayed distinct anticancer activity in a range similar to crizotinib. The oxygen conditions (1 % O2 vs. 21 % O2) had no relevant impact on the activity of the two prodrugs. In contrast, after 72 h prodrug A showed promising hypoxia-mediated activity in both c-MET-dependent cell lines (H1993 p < 0.05 and RUMH p < 0.01; unpaired t-test with Welsch’s correction), while it was similarly active in case of the ALK-dependent cells. Prodrug B and crizotinib again displayed similar activity patterns under both normoxia and hypoxia. In line with the literature, TH-302 showed a significant anticancer activity under low oxygen levels24. Further, decreasing the O2 content from 1 % to 0.1 % resulted in similar activation profiles (Figure S11). In addition, the activity of prodrug A and crizotinib was evaluated under normoxic conditions in non-malignant, c-MET expressing HACAT (keratinocytes from human skin) and HLF (fibroblasts from human lung) cells for 72 h. As shown in Figure S12, prodrug A exhibits distinctly reduced activity against these cells in comparison to free crizotinib. This strongly indicates reduced toxicity and, thus, enhanced tolerability of prodrug A in healthy tissues compared to crizotinib.
Figure 5.
Cytotoxic activity of prodrug A, prodrug B and crizotinib against the indicated human cancer cells. Incubation time of the compounds on the cells was 24 h or 72 h under normoxic (21% O2) or hypoxic condition (1% O2). Dose-response curves (24 h and 72 h) of prodrug A (A), prodrug B (B), crizotinib (C) and TH-302 (D) in H2228, H1993 and RUMH cells are shown. Values are given as means ± SD of one representative experiment performed in triplicates. N= normoxia, H=hypoxia
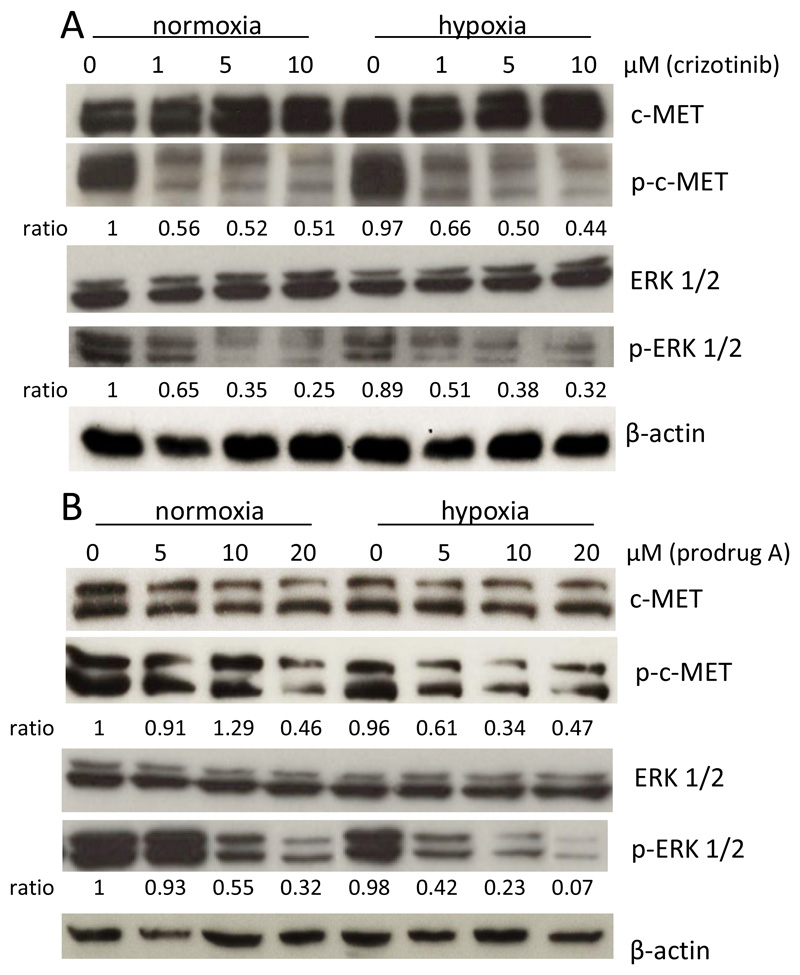
As a next step, the impact of prodrug A on c-MET and the downstream target ERK 1/2 phosphorylation was compared to crizotinib under normoxic and hypoxic conditions by Western blot analysis in H1993 cells (Figure 6). With regard to c-MET, in case of crizotinib no difference in phosphorylation between both oxygen levels was observed, with about 50 % inhibition of the signal at all concentrations tested. In contrast, and in agreement with the cell viability assays, prodrug A was distinctly less effective under normoxia with inhibition of c-MET phosphorylation <10 % for up to 10 μM. Under hypoxic conditions, the inhibitory effect was strongly increased with 40 % and 70 % inhibition at 5 μM and 10 μM, respectively. With regard to pERK 1/2 phosphorylation, crizotinib treatment again showed a similar activity profile in both oxygen conditions with a dose-dependent increase of inhibition. In accordance with the detected p-c-MET levels, the inhibitory potential of prodrug A on ERK 1/2 phosphorylation was markedly reduced under normoxia with less than 10 % inhibition of the signal at 5 μM. In contrast, under hypoxia ~60 % (5 μM) and ~80 % (10 μM) inhibition, respectively, were observed. These results indicate that the derivatization of crizotinib into prodrug A impedes receptor binding and c-MET inhibition under normoxic conditions, while hypoxia leads to the release of the active c-MET inhibitor. To test, whether prodrug A is also sufficiently stable for in vivo application, the compound stability was investigated in pure serum at 37 °C. Indeed, no significant degradation could be observed for up to 72 h (Figure S7C).
Figure 6.
Impact of drug treatment on the c-MET and p-ERK 1/2 phosphorylation under hypoxic (1 % O2) and normoxic (21 % O2) conditions. H1993 cells treated with crizotinib (A) or prodrug A (B). After 24 h cell lysates were prepared and protein expression and phosphorylation levels of p-c-MET and p-ERK 1/2 analyzed by Western blotting. The ratio p-c-MET to c-MET and p-ERK 1/2 to ERK 1/2 (after normalization to the loading control ß-actin) is given below the respective bands.
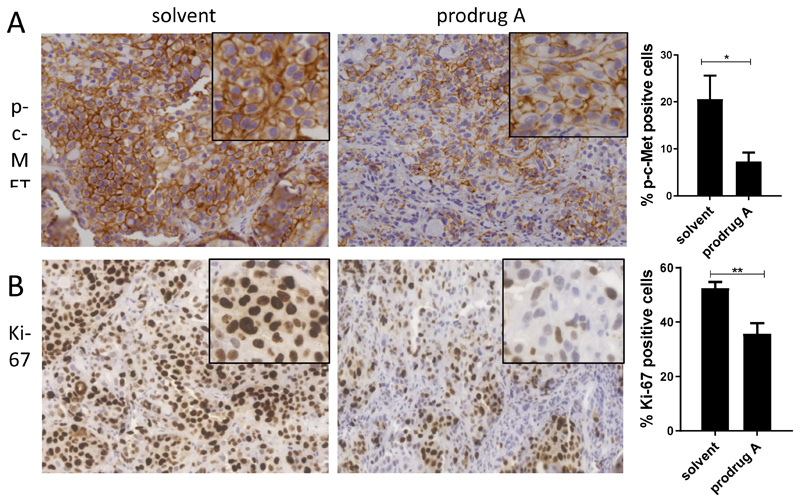
Consequently, the c-MET-inhibitory potential of prodrug A was investigated in vivo. To this end, H1993 cells were implanted s.c. into the right flank of SCID mice and treated once either with 0.9% NaCl or prodrug A (38.5 mg/kg i.v.). After 24 h, animals were sacrificed and tumors harvested for histological evaluation. As shown in Figure 7A, treatment with prodrug A resulted in significant inhibition of the c-MET phosphorylation in the whole tumor tissue. This was accompanied by inhibition of cell proliferation (indicated by the proliferation marker Ki-67, Figure 7B). Interestingly, in addition also a general change in the tumor histology could be observed with a reduction of the tumor:stroma ratio (data not shown).
Figure 7.
Immune-histological analysis of p-c-MET (A) and human Ki-67 (B) expression after prodrug A treatment in vivo. H1993 cells were injected s.c. into SCID mice. After the tumor reached a length of ~10 mm, 38.5 mg/kg prodrug A or solvent were administered i.v.. After 24 h, tumors were paraffin-embedded and stained as described in the M&M section. Evaluation of the stain was performed by Definiens software. Significance was calculated compared to solvent controls with unpaired t-test (**p<0.01, *p<0.05)
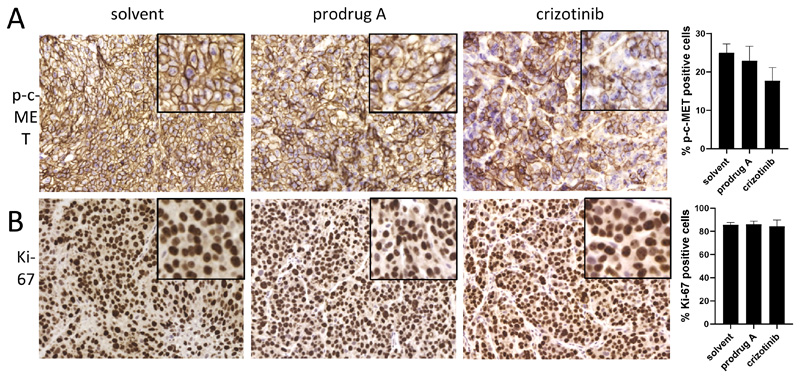
As crizotinib is usually applied orally, an additional in vivo study was performed, in which prodrug A and crizotinib were administered orally to H1993-bearing SCID mice at equimolar concentrations (we first confirmed the stability of prodrug A down to pH 1 for 24 h at 37 °C; data not shown). Next, again phosphorylation levels of c-MET and expression of Ki-67 were stained in the collected tumor tissues 24 h after drug application (Figure 8). The data revealed a slight impact of crizotinib (50 mg/kg p.o.) on reduction of the two visualized markers. In contrast, orally applied prodrug A (89.2 mg/kg p.o.) did not significantly change the level of p-c-MET or Ki-67 expression after 24 h treatment, indicating that prodrug A is not suitable for oral application. However, the usually administered crizotinib scheme of 50 mg/kg (p.o.) turned out to be much less efficient than prodrug A administered i.v. at less as half of the dose (38.5 mg/kg i.v. being equivalent to 21 mg/kg crizotinib). Therefore, further development of our hypoxia-sensitive crizotinib prodrug A could be based on continuing as intravenously applied drug or by designing a derivative of prodrug A with advanced oral bioavailability.
Figure 8.
Immune-histological analysis of p-c-MET (A) and human Ki-67 (B) expression after oral treatment with prodrug A or crizotinib. H1993 cells were injected s.c. into SCID mice. After the tumor reached a length of ~10 mm, 89.2 mg/kg prodrug A, 50 mg/kg crizotinib or solvent were administered p.o.. After 24 h, tumors were paraffin-embedded and stained as described in the M&M section. Evaluation of the stain was performed by Definiens software. Significance was calculated compared to solvent controls with unpaired t-test (no significance was reached).
Conclusion
Tyrosine kinase inhibitors have revolutionized cancer therapy over the past 15 years. Despite their success, clinical application is limited by severe side effects. In this study, we developed the first crizotinib prodrugs with the aim to reduce severe adverse effects and improve the anticancer activity. The prodrug design consists of a hypoxia-activatable, self-immolative trigger moiety at a crucial tyrosine kinase binding position of crizotinib, which should result in strongly reduced affinity to the catalytic pockets of the target kinases c-MET and ALK. Two different prodrug derivatives were synthesized, either with the trigger moiety coupled via carbamoylation (A) or alkylation (B) of the 2-aminopyridine moiety of crizotinib. For both prodrugs, molecular docking studies and a cell-free kinase assay confirmed the inactivation of crizotinib. The hypoxic activation in case of prodrug A could be proven by a nitroreductase-based cleavage assay and comparing normoxic versus hypoxic conditions in cell culture. Furthermore, prodrug A showed high stability in serum, an essential prerequisite for successful prodrug development. Finally, we showed that prodrug A is able to distinctly inhibit c-MET phosphorylation and cell proliferation in tumor tissues in vivo after i.v. application.
Taken together, the data clearly indicates prodrug A as an interesting candidate for further (pre)clinical development and as novel TKI with improved tumor-specific properties.
Experimental section
All solvents and reagents were obtained from commercial suppliers and, unless specified, used without further purification. Anhydrous DMF was bought from Sigma-Aldrich over molecular sieves. Crizotinib was purchased from TripleBond Corporation (Guelph, ON, Canada). Compound 1 was synthesized according to literature-known procedures17. Preparative RP-HPLC was performed on an Agilent 1200 Series system controlled by ChemStation software. The experimental conditions were as follows: stationary phase: ethylene bridged hybrid C18; column: XBridge BEH C18 OBD Prep Column, 130 Å, 10 μm, 19 mm x 250 mm (Waters Corp., Massachusetts, USA); flow rate: 17.06 mL/min, injection volume: 2-4 mL; column temperature: 25 °C. Elemental analyses were performed by the Microanalytical Laboratory of the University of Vienna. ESI-MS spectrometry was carried out with a Bruker Esquire3000 ion trap spectrometer (Bruker Daltonic, Bremen, Germany). Expected and experimental isotope distributions were compared. 1H and 13C NMR spectra were recorded in d6-DMSO with a Bruker FT-NMR AV NEO 500 MHz spectrometer at 500.10 (1H) and 125.75 (13C) MHz at 298 K. Final products were characterized via 2D-NMR in d6-DMSO with a Bruker FT-NMR AVIII 600 MHz spectrometer at 600.25 MHz (1H) and 150.93 MHz (13C). Chemical shifts (ppm) were referenced internal to the solvent residual peaks. For the description of the spin multiplicities the following abbreviations were used: s = singlet, d = duplet, t = triplet, q = quartet, m = multiplet. For biological investigations, crizotinib (LC Laboratory), Evofosfamide® (TH-302, Abmole Bioscience Co. Limited) and all other investigated compounds were dissolved in DMSO. These stock solutions were further diluted into culture media containing 10 % FCS at the indicated concentrations. The final DMSO concentrations were always less than 1 %. All other substances were purchased from Sigma-Aldrich (St. Louis, USA).
Synthesis
5-(bromomethyl)-1-methyl-2-nitro-1H-imidazole (2)
75 mg of 1 17 (1 eq., 0.48 mmol) was dissolved in 702 μL (19 eq., 9.12 mmol) of thionyl bromide and stirred under argon at room temperature for 15 min. The reaction mixture was quenched with 5 mL ice-cold water and extracted with ethyl acetate three times. The combined organic phases were washed with brine, dried over magnesium sulfate and the solvent was removed under reduced pressure. The product was dried in vacuo to give a yellow powder. Yield: 82 mg (78 %). 1H NMR (500 Hz, DMSO-d6) δ 7.12 (s, 1H), 4.55 (s, 2H), 3.92 (s, 3H) ppm. ESI-HRMS (m/z): calc: 219.9716, found: 219.9708.
tert-butyl (R)-4-(4-(6-amino-5-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)pyridin-3-yl)-1H-pyrazol-1-yl)piperidine-1-carboxylate (3)
1 g of crizotinib (1 eq., 2.22 mmol) was dissolved in 20 mL of tetrahydrofuran and 484 mg of di-tert-butyl dicarbonate (1 eq., 2.22 mmol) in 10 mL of tetrahydrofuran was added. The reaction was stirred at room temperature until no educts was indicated by TLC anymore. The reaction mixture was taken up in EtOAc and washed with water and brine. The organic layers were combined, dried over MgSO4 and the solvent was removed under reduced pressure yielding a colorless oil. The oil was taken up in diethyl ether, which was subsequently removed under reduced pressure giving a colorless powder. Yield: 1.16 g (96 %). 1H NMR (500 MHz, DMSO-d6) δ 7.98 (s, 1H), 7.76 (d, J = 1.7 Hz, 1H), 7.58 (dd, J = 9.0, 4.9 Hz, 1H), 7.54 (s, 1H), 7.45 (t, J = 9 Hz, 1H), 6.91 (d, J = 1.7 Hz, 1H), 6.10 (q, J = 6.6 Hz, 1H), 5.66 (s, 2H), 4.38 – 4.27 (m, 1H), 4.04 (bs, 2H), 2.92 (s, 2H), 2.01 (d, 2H), 1.81 (d, J = 6.6 Hz, 3H), 1.79 – 1.70 (m, 2H), 1.43 (s, 9H) ppm. ESI-HRMS (m/z): calc: 550.1782, found: 550.1770.
tert-butyl (R)-4-(4-(5-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-6-((((1-methyl-2-nitro-1H-imidazol-5-yl)methoxy)carbonyl)amino)pyridin-3-yl)-1H-pyrazol-1-yl)piperidine-1-carboxylate (4)
300 mg of 3 (1 eq., 0.55 mmol), 86 mg of 1 (1 eq., 0.55 mmol), 326 mg of Cu(OAc)2 x H2O, 18 mg of potassium iodide (0.2 eq., 0.11 mmol) and 6 mg of Pd(OAc)2 were dissolved in 3 mL of dimethyl sulfoxide and 12 mL acetonitrile. The reaction vessel was set under an atmospheric pressure of carbon monoxide (CAUTION: highly toxic) and shortly warmed with a hot water bath (40 °C). After 1 h the reaction mixture was taken up in EtOAc and washed with sat. NaHCO3 and water. The combined aqueous solutions were washed with EtOAc and the combined organic solutions were washed with brine, dried over MgSO4 and the solvent was removed under reduced pressure. The crude product was dried in vacuo overnight. Purification was performed using column chromatography (EtOAc/petrol ether 9:1). Yield: 160 mg (40 %). 1H NMR (500 MHz, DMSO-d6) δ 9.35 (s, 1H), 8.65 (s, 1H), 8.41 (s, 1H), 8.23 (s, 2H), 7.84 (s, 1H), 7.54 (dd, J = 8.9, 5.0 Hz, 1H), 7.45 (t, J = 8.7 Hz, 1H), 7.38 (s, 1H), 7.28 (s, 1H), 6.15 (d, J = 6.6 Hz, 1H), 5.30 – 5.19 (m, 2H), 4.52 (t, J = 10.9 Hz, 1H), 3.96 (s, 3H), 3.10 (dd, J = 23.1, 11.6 Hz, 2H), 2.23 (d, J = 11.4 Hz, 2H), 2.15 – 2.06 (m, 2H), 1.73 (d, J = 6.6 Hz, 3H), 1.43 (s, 9H) ppm. ESI-MS (M+Na): calc: 755.19 found: 755.39.
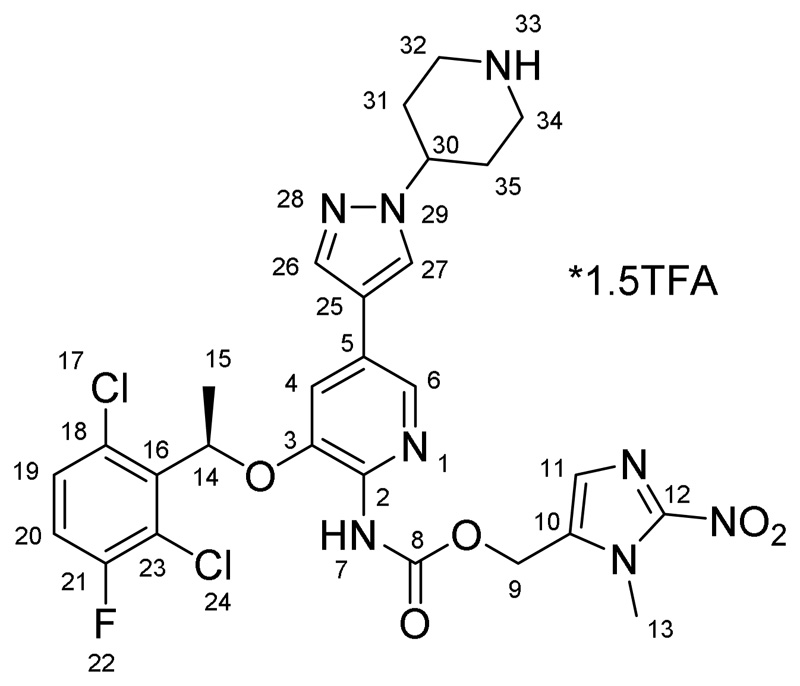
(1-methyl-2-nitro-1H-imidazol-5-yl)methyl (R)-(3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-yl)carbamate*1.5TFA (A)
160 mg (0.22 mmol) of 4 was suspended in 2 mL of dichloromethane and 1 mL of trifluoroacetic acid was added. The reaction mixture was stirred for 1 h and the volatiles were removed under reduced pressure. The precipitate was suspended in diethyl ether, filtered and dried in vacuo yielding a grey powder. Yield: 171 mg (96 %). 1H NMR (600 MHz, DMSO-d6) δ 9.31 (bs, 1H, H7), 8.69 (dd, J = 168.7, 6.2 Hz, 2H, H33), 8.22 (s, 2H, H26/H27 + H6), 7.83 (s, 1H, H26/H27), 7.53 (dd, J = 8.0, 4.8 Hz, 1H, H19), 7.44 (t, J = 8.5 Hz, 1H, H20), 7.37 (s, 1H, H4), 7.27 (s, 1H, H11), 6.14 (q, J = 6.5 Hz, 1H, H14), 5.24 (m, 2H, H9), 4.52 (m, 1H, H30), 3.96 (s, 3H, H13), 3.42 (m, J = 10.9 Hz, 2Haeq, H32/H34), 3.12 (m, J = 10.5 Hz, 2Hax, H34/H32), 2.22 (m, 2Haeq, H31/H35), 2.11 (m, 2Hax, H31/H35), 1.72 (d, J = 6.2 Hz, 3H, H15) ppm. 13C NMR (150.95 MHz, DMSO-d6) δ 156.82 (d, J = 247 Hz, C21), 152.87 (C8), 146.50 (C3), 146.01 (C12), 139.21 (C2), 136.54 (C6), 136.46 (C16), 136.25 (C26/C27), 133.49 (C10), 130.52 (C19), 128.66 (C18), 128.58 (C11), 127.01 (C5/C25), 125.95 (C26/C27), 121.04 (d, J = 19 Hz, C23), 117.94 (C4), 117.78 (C5/C25), 117.52 (d, J = 23 Hz,C20), 73.14 (C14), 55.33 (C9), 55.27 (C30), 42.17 (C32/C34), 34.19 (C13), 28.66 (C31/C35), 18.43 (C15) ppm. ESI-HRMS (m/z): calc: 633.1538, found: 633.1537. Anal. calcd for C27H27Cl2FN8O5*1.5CF3COOH (MR = 803.14 g/mol): C, 44.79; H, 3.57; N, 13.93; O, 15.91. Found: C, 44.92; H, 3.60s
tert-butyl (R)-4-(4-(5-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-6-(((1-methyl-2-nitro-1H-imidazol-5-yl)methyl)amino)pyridin-3-yl)-1H-pyrazol-1-yl)piperidine-1-carboxylate (5)
100 mg of 3 (1 eq., 0.18 mmol) was dissolved in 2 mL of DMF abs. under an argon atmosphere and 15 mg of KI (0.5 eq., 0.09 mmol), 31 μL of DIPEA (1 eq., 0.18 mmol) and 40 mg of 2 (1 eq., 0.18 mmol) were added. The reaction mixture was stirred at room temperature overnight. The reaction progress was monitored via HPLC. The solvent was removed under reduced pressure and purification was performed via column chromatography (chloroform/methanol 9:1). Yield: 37 mg (32 %). 1H NMR (500 MHz, DMSO-d6) δ 8.01 (s, 1H), 7.87 (d, J = 1.7 Hz, 1H), 7.63 (dd, J = 9.0, 4.9 Hz, 1H), 7.58 (s, 1H), 7.50 (t, J = 9.0 Hz, 1H), 7.06 (s, 1H), 6.91 (d, J = 1.7 Hz, 1H), 6.65 (t, J = 5.9 Hz, 1H), 6.18 (q, J = 6.7 Hz, 1H), 4.69 (ddd, J = 46.2, 15.9, 5.9 Hz, 2H), 4.38 (tt, J = 11.4, 3.9 Hz, 1H), 4.15 – 4.05 (m, 2H), 4.04 (s, 3H), 2.97 (s, 2H), 2.04 (d, 2H), 1.89 (d, J = 6.6 Hz, 3H), 1.80 (qd, J = 12.4, 4.4 Hz, 2H), 1.48 (s, 9H) ppm. ESI-MS (m/z): calc: 689.57 found: 689.24.
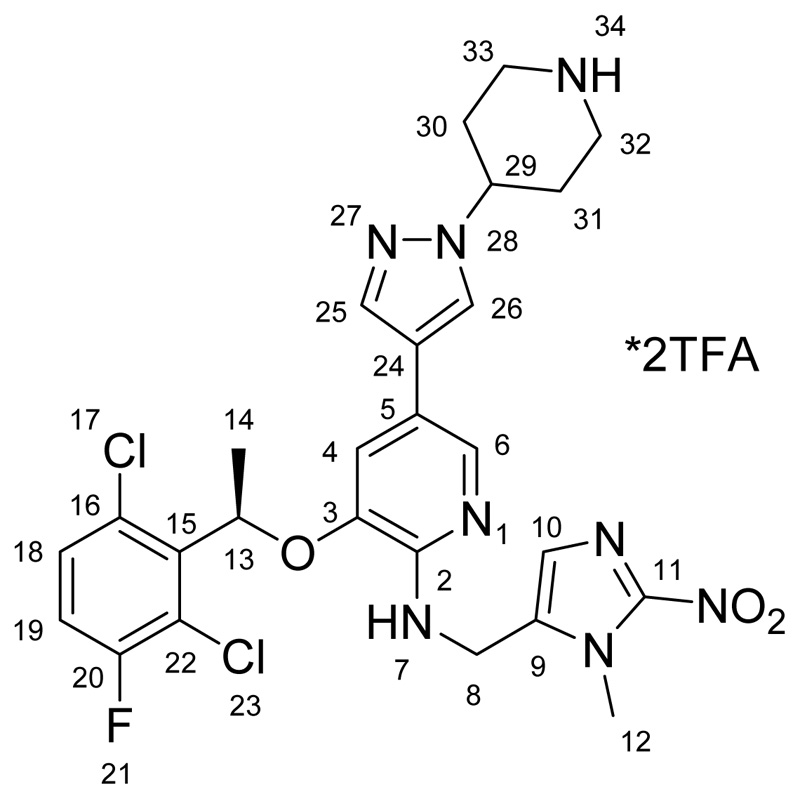
(R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-N-((1-methyl-2-nitro-1H-imidazol-5-yl)methyl)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine *2TFA (B)
37 mg (0.05 mmol) of 5 was dissolved in 2 mL of dichloromethane and 1 mL of trifluoroacetic acid was added. The reaction mixture was stirred at room temperature overnight and subsequently all volatiles were removed under reduced pressure. Purification was performed via preparative HPLC using a gradient from 30-40 % acetonitrile (+0.1 % TFA) in Millipore water (+0.1 % TFA) yielding a yellow powder. Yield: 22.4 mg (50 %). 1H NMR (600 MHz, DMSO-d6) δ 8.76 (s, 1H, N34), 8.61 (s, 2H, N7), 8.53 (s, 1H, N34), 8.00 (s, 1H, H25/H26), 7.92 (s, 1H, H6), 7.67 (s, 1H, H26/H27), 7.63 (dd, 1H, H18), 7.51 (t, 1H, H19), 7.15 (t, 1H, H4), 6.84 (s, 1H, H10), 6.32 (q, 1H, H13), 5.56 (m, 2H, H8), 4.52 (tt, J = 10.9, 4.0 Hz, 1H, H29), 3.41 (d, J = 12.7 Hz, 2Haeq, H33/H32), 3.08 (dd, J = 22.9, 11.1 Hz, 2Hax, H33/H32), 2.18 (d, J = 12.5 Hz, 2Haeq, H30/H31), 2.07 (dd, J = 24.3, 11.7 Hz, 2Hax, H30/H31), 1.85 (d, J = 6.6 Hz, 3H, H14) ppm. 13C NMR (150.95 MHz, DMSO-d6) δ 156.98 (d, J = 257 Hz, C20), 146.85 (C2), 146.33 (C11), 142.13 (C3), 135.74 (C25/C26), 135.04 (C15), 130.89 (C9), 130.85 (C18), 128.77 (C16), 126.12 (C10), 125.62 (C25/C26), 125.50 (C6), 121.23 (d, J = 20 Hz, C22), 118.15 (d, J = 23 Hz, C19), 117.50 (C5), 116.53 (C4), 116.00 (C24), 74.20 (C13), 55.41 (C29), 48.45 (C8), 42.14 (C32/C33), 34.55 (C12), 28.71 (C31/C30), 18.45 (C14) ppm. ESI-HRMS (m/z): calc: 589.1640, found: 589.1638. Anal. Calcd. For C26H27Cl2FN8O3*2CF3COOH*0.5H2O (MR = 826.51 g/mol): C, 43.60; H, 3.66; N, 13.41. Found: C, 43.28; H, 3.43; N, 13.41.
Docking studies
Prodrug structures were drawn using the ChemBioDraw® Office Suite and energy was minimized using the ChemBio3D® program. This structure was further optimized via DFT calculations implemented in the Gaussian09 program package25, using the B3LYP functional and the 6-31G(d) basis set. Addition and merging of non-polar hydrogens of the receptors was performed using the AutoDockTools 1.5.6 software26. The grid box size was set as follow: ALK (size_x = 22, size_y = 22, size_z = 26, center_x = 33, center_y = 49, center_z = 13, Spacing = 1 Angström), c-MET (size_x = 30, size_y = 30, size_z = 30, center_x = 15.235, center_y = 85.36, center_z = 6.139, Spacing = 1 Angström). Docking studies were performed using AutoDock Vina27 using an exhaustiveness factor of 50 in all experiments. Crystal structures of crizotinib in complex with ALK (PDB ID: 2PX2) and c-MET (PDB ID: 2WGJ) were manipulated by removing and redocking the ligands to evaluate the docking process. Subsequently, the ligand-free protein shell was used to investigate both prodrug compounds. All 3D models were visualized using PyMol28 and 2D interactions using the LigPlot+ software package29. Docking procedures were performed in quintuplicates.
Kinase inhibition assay
The ALK and c-MET kinase-inhibitory potentials of prodrug A, prodrug B and crizotinib were evaluated using the Select Screen® Biochemical Kinase Profiling Service at Life Technologies (ThermoFisher Scientific, Madison, USA). The test compounds were screened in a final concentration of 1 % DMSO using the Z′-LYTE® Assay. Results are shown in Table S4 and Fig. S9.
Activation assay
E. coli nitroreductase (NTR) and 1,4-dihydronicotinamide adenine dinucleotide (NADH) were purchased from Sigma Aldrich Inc.. Drug stock solutions (1 mM) were prepared in DMSO, NADH (1 mM) and the enzyme (50 μg/mL) were prepared in 50 mM phosphate buffer at pH 7.4. As a first step, the stability of all compounds under the assay conditions was tested in absence of NTR. Therefore, 7.5 μL of the drug stock solution was added to 37.5 μL of the NADH (5 eq.) stock solution, diluted to 750 μL using buffer solution and investigated via HPLC. The NTR activation assay was performed via addition of the drug (7.5 μL) to NADH solution (37.5 μL, 37 °C), diluted with buffer (205 μL, 37 °C) and addition of the enzyme solution (500 μL, 37 °C). All samples were incubated at 37 °C and equivalents were removed and injected into the HPLC every 8 minutes. All measurements were performed on a ThermoFisher Ultimate 3000RS system using a Water’s Acquity BEH C18 column (3x50 mm, 1.7 μm) and a 5-95 % gradient of ACN (+0.1 % TFA) in water (+0.1 % TFA). Results are shown in Figure 4B and Figure S10.
Stability assay
Stock solutions of all drugs were prepared in concentrations of 10 mM in DMSO. These solutions were diluted 1:10 by addition of phosphate buffer (PB) at pH 7.4. 1 μL of the diluted samples were added to 19 μL of PB at pH 7.4, RPMI-1640 cell culture medium + 10 % fetal calf serum (FCS) or FCS alone and incubated for different time intervals at 37 °C. After incubation, drug extraction was performed as followed (also for the PB7.4 sample): 40 μL of acetonitrile was added to precipitate macromolecules. The samples were vortexed, centrifuged for 10 min at 6000 U/min and the supernatant was analyzed via HPLC-MS. For the HPLC-MS experiments an Agilent 1260 Infinity system with a Waters Atlantis T3 column 150 mm x 4.6 mm coupled to a Bruker AmaZon SL ESI-IT mass spectrometer was used. Milli-Q water, containing 0.1 % formic acid, and acetonitrile containing 0.1 % formic acid were used as eluents. A gradient of 1-99 % acetonitrile in 20 min was used. Results are shown in Fig. S6–S8.
Cell culture
The human cancer cell lines H2228 (non-small cell lung cancer), H1993 (non-small cell lung cancer), HACAT (keratinocytes from human skin) and HLF cells (fibroblasts from human lung) were purchased from American Type Culture Collection (ATCC) (Rockville, MD, USA), whereas the RUMH (renal cell carcinomas) was established at the Institute of Cancer Research (Medical University of Vienna). All cell lines were grown unless otherwise indicated in humidified incubators (37 °C, 21 % O2, 5 % CO2) in RPMI 1640 medium containing 10 % FCS (PAA, Linz, Austria). In case of hypoxic conditions, plated and treated cells in 96- or 60 mm petri-dishes were incubated in an humidified incubators with 1 % or 0.1 % O2/ 5 % CO2 level for indicated time points (24-72 h for MTT assays and 18 h for protein isolation) before analysis/harvesting.
Cytotoxicity assays
Cells were seeded (3-6 x 103 cells/well, depending on cell line) in 96-well plates and allowed to recover for 24 h. Subsequently, the dissolved drugs were added in increasing concentrations. After 24 h or 72 h drug exposure, the proportion of viable cells was determined by MTT assay following the manufacturer’s recommendations (EZ4U, Biomedica, Vienna, Austria). Cytotoxicity was expressed as IC50 values calculated from full dose-response curves using Graph Pad Prism software.
Western blot analysis
To assess the impact of the new drugs on the signaling pathway of c-MET, Western blot analysis were performed. Therefore, H1993 cells were plated (8x105 cells/dish) and after 24 h recovery, treated with the drugs in different concentrations for 24 h. Then, cells were harvested, proteins were isolated, resolved by SDS/PAGE, and transferred onto a polyvinylidene difluoride membrane for Western blotting as previously described30. The following antibodies were used: c-MET (#8198), p-c-MET (Tyr1234/1235) (#3077), ERK 1/2 (#9102) and p-ERK 1/2 (Thr202/Tyr204) (#9101) (all four monoclonal rabbit, Cell Signaling Technology, Beverly, MA, USA) and β-actin (monoclonal mouse, Sigma). All primary antibodies were used in 1:1000 dilutions. Additionally, horseradish peroxidase-labeled secondary antibodies anti-mouse (#7076) and anti-rabbit (#7074) were used at working dilutions of 1:10000.
Animals
Eight- to twelve-week-old male CB-17 (SCID) mice were purchased from Envigo Laboratories (San Pietro al Natisone, Italy). The animals were kept in a pathogen-free conditions and controlled environment with 12 h light–dark cycle. All procedures were performed in a laminar flow hood. Tumor growth and effects of treatment were evaluated by daily recording of tumor size by caliper measurement and distress development. The experiments were done according to the regulations of the Ethics Committee for the Care and Use of Laboratory Animals at the Medical University Vienna (proposal number BMWF-66.009/0157-V/3b/2019), The U.S. Public Health Service Policy on Human Care and Use of Laboratory Animals as well as the United Kingdom Coordinating Committee on Cancer Prevention Research’s Guidelines for the Welfare of Animals in Experimental Neoplasia.
Xenograft experiments
H1993 cells (1.7 × 106 cells in 50 μl) were injected subcutaneously into the right flank. Animals were randomly assigned to treatment groups. In treatment scheme 1, prodrug A was intravenously injected with 38.5 mg/kg (equimolar to 40 % bioavailability of 50 mg/kg crizotinib) dissolved in PBS, and in treatment scheme 2, prodrug A (89.2 mg/kg in PBS, equimolar to crizotinib) and crizotinib (50 mg/kg in H2O containing 5% DMSO, 10% cremophore and 10% EtOH) were administered p.o., when tumor nodules reached a length of ~10 mm. The control groups received PBS alone either i.v. or p.o.. After 24 h the animals were sacrificed by cervical dislocation. Tumor were collected and further processed for histological studies.
Immunohistochemistry
Tumors were fixed in 4 % formaldehyde for 24 h (Carl Roth, # P087.3) and paraffin-embedded using a KOS machine (Milestone). For histological evaluation, tumor tissues were sliced in 3 μm thick sections. The c-MET inhibitory potential of prodrug A was observed by staining with p-c-MET (#3077, Cell Signaling Technology). Additionally, a staining of Ki-67 (DAKO, human-specific) was performed to analyze the amount of proliferating tumor cells in the tissue. Therefore, the samples were heated for 10 min in EDTA buffer (pH 8.6) followed by incubation with the primary antibody (overnight at 4 °C). Binding of primary antibodies was detected with the UltraVision LP detection system according to the manufacturer’s instructions (Thermo Fisher Scientific), followed by incubation with 3,3′-diaminobenzidine and counterstaining with haematoxylin. Evaluation and quantification of the staining was done by Definiens software.
Supplementary Material
Acknowledgments
This work was financially supported by the Austrian Science Fund (FWF) [grant number P28853, to C.R.K.]. We thank Ing. Alexander Roller and Natalie Lukic for X-ray crystallographic measurements.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Bhullar KS, Lagarón NO, McGowan EM, Parmar I, Jha A, Hubbard BP, Rupasinghe HPV. Mol Cancer. 2018;17:48. doi: 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moslehi JJ. New Engl J Med. 2016;375:1457–1467. doi: 10.1056/NEJMra1100265. [DOI] [PubMed] [Google Scholar]
- 3.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, Frias RL, Gainor JF, Amzallag A, Greninger P, Lee D, et al. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong CWS, Wu WKK, Loong HHF, Cho WCS, To KKW. Cancer Lett. 2017;405:100–110. doi: 10.1016/j.canlet.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Solomon B. Journal of Thoracic Oncology. 2017;12:9–11. doi: 10.1016/j.jtho.2016.11.2214. [DOI] [PubMed] [Google Scholar]
- 6.Landi L, Cappuzzo F. Expert Opin Pharmacother. 2014;15:2587–2597. doi: 10.1517/14656566.2014.970174. [DOI] [PubMed] [Google Scholar]
- 7.Kazandjian D, Blumenthal GM, Chen H-Y, He K, Patel M, Justice R, Keegan P, Pazdur R. The Oncologist. 2014;19:e5–e11. doi: 10.1634/theoncologist.2014-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rautio J, Kumpulainen H, Heimbach T, Oliyai R, Oh D, Järvinen T, Savolainen J. Nat Rev Drug Discov. 2008;7:255–270. doi: 10.1038/nrd2468. [DOI] [PubMed] [Google Scholar]
- 9.Wang W-C, Shiao H-Y, Lee C-C, Fung K-S, Hsieh H-P. MedChemComm. 2014;5:1266–1279. [Google Scholar]
- 10.Cui JJ, Tran-Dubé M, Shen H, Nambu M, Kung P-P, Pairish M, Jia L, Meng J, Funk L, Botrous I, McTigue M, et al. J Med Chem. 2011;54:6342–6363. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 11.Baran N, Konopleva M. Clin Cancer Res. 2017;23:2382–2390. doi: 10.1158/1078-0432.CCR-16-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A, Arambula JF, Koo S, Kumar R, Singh H, Sessler JL, Kim JS. Chem Soc Rev. 2019;48:771–813. doi: 10.1039/c8cs00304a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karnthaler-Benbakka C, Groza D, Koblmüller B, Terenzi A, Holste K, Haider M, Baier D, Berger W, Heffeter P, Kowol CR, Keppler BK. ChemMedChem. 2016;11:2410–2421. doi: 10.1002/cmdc.201600417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutsem EV, Lenz H-J, Furuse J, Tabernero J, Heinemann V, Ioka T, Bazin I, Ueno M, Csõszi T, Wasan H, Melichar B, et al. J Clin Oncol. 2016;34:4007–4007. [Google Scholar]
- 15.Patterson AV, Silva S, Guise C, Bull M, Abbattista M, Hsu A, Sun JD, Hart CP, Pearce TE, Smaill JB. J Clin Oncol. 2015;33:e13548-e13548 [Google Scholar]
- 16.Wilson WR, Hay MP. Nat Rev Canc. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 17.O’Connor LJ, Cazares-Körner C, Saha J, Evans CNG, Stratford MRL, Hammond EM, Conway SJ. Nat Protoc. 2016;11:781–794. doi: 10.1038/nprot.2016.034. [DOI] [PubMed] [Google Scholar]
- 18.Lanzillotto M, Konnert L, Lamaty F, Martinez J, Colacino E. ACS Sust Chem Engin. 2015;3:2882–2889. [Google Scholar]
- 19.Zhang Y, Anderson M, Weisman JL, Lu M, Choy CJ, Boyd VA, Price J, Sigal M, Clark J, Connelly M, Zhu F, et al. ACS Med Chem Lett. 2010;1:460–465. doi: 10.1021/ml100083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knölker H-J, Braxmeier T, Schlechtingen G. Angew Chem Int Ed. 1995;34:2497–2500. [Google Scholar]
- 21.Guan ZH, Lei H, Chen M, Ren ZH, Bai Y, Wang YY. Adv Synth Catal. 2012;354:489–496. [Google Scholar]
- 22.Wilson FH, Johannessen CM, Piccioni F, Tamayo P, Kim JW, Van Allen EM, Corsello SM, Capelletti M, Calles A, Butaney M, Sharifnia T, et al. Cancer Cell. 2015;27:397–408. doi: 10.1016/j.ccell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Cancer Res. 2010;70:1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun JD, Liu Q, Wang J, Ahluwalia D, Ferraro D, Wang Y, Duan JX, Ammons WS, Curd JG, Matteucci MD, Hart CP. Clin Cancer Res. 2012;18:758–770. doi: 10.1158/1078-0432.CCR-11-1980. [DOI] [PubMed] [Google Scholar]
- 25.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, et al. Gaussian, Rev 09. Gaussian Inc; Wallingford, CT (USA): 2009. [Google Scholar]
- 26.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Comput chemistry. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trott O, Olson AJ. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMol User’s Manual. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- 29.Laskowski RA, Swindells MB. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 30.Heffeter P, Pongratz M, Steiner E, Chiba P, Jakupec MA, Elbling L, Marian B, Körner W, Sevelda F, Micksche M, Keppler BK, Berger W. J Pharmacol Exp Ther. 2005;312:281–289. doi: 10.1124/jpet.104.073395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.