Abstract
The transcription factor Pax5 controls B cell development, but its role in mature B cells is largely enigmatic. Here, we demonstrated loss of Pax5 by conditional mutagenesis in peripheral B lymphocytes led to the significant reduction of B-1a, marginal zone (MZ) and germinal center (GC) B cells as well as plasma cells. Follicular (FO) B cells tolerated the loss of Pax5 but had a shortened half-life. The Pax5-deficient FO B cells failed to proliferate upon B cell receptor or toll-like receptor stimulation due to impaired PI3K-AKT signaling, which was caused by increased expression of PTEN, a negative regulator of the PI3K pathway. Pax5 restrained PTEN protein expression at the posttranscriptional level, likely involving Pten-targeting microRNAs. Additional PTEN loss in Pten,Pax5 double-mutant mice rescued FO B cell numbers and the development of MZ B cells, but did not restore GC B cell formation. Hence, the posttranscriptional downregulation of PTEN expression is an important function of Pax5 that facilitates the differentiation and survival of mature B cells, thereby promoting humoral immunity.
Keywords: Pax5, mature B cell types, class switch recombination, B cell receptor signaling, Toll-like receptor signaling, PI3K-AKT signaling, microRNA-mediated control of PTEN expression
Introduction
B cell immunity provides humoral protection against infections through the generation and secretion of high-affinity antibodies that recognize an almost unlimited diversity of pathogens. This enormous adaptive potential of B cells is generated through V(D)J recombination of the immunoglobulin heavy-chain (Igh) and light-chain (Igk and Igl) genes during early B cell development (1), which results in the emergence of immature B cells in the bone marrow. Upon migration to the spleen, these immature B cells differentiate into distinct mature B cell types present in peripheral lymphoid organs. The innate-like B-1a cells in the peritoneal and pleural cavities and the marginal zone (MZ) B cells in the spleen provide the first line of defense against pathogens by rapidly developing to antibody-secreting plasma cells in a T cell-independent manner (2, 3). In response to antigen stimulation and T cell help, follicular (FO) B cells in the spleen and lymph nodes differentiate into germinal center (GC) B cells that undergo class switch recombination (CSR) (4) and somatic hypermutation (SHM) (5) at immunoglobulin genes.
While CSR exchanges Igh constant (CH) exon regions to generate IgH isotypes with distinct effector functions (4), SHM alters the antigen-binding variable (V) sequences of immunoglobulin heavy- and light-chains (5). Affinity-based selection in the GC subsequently leads to the clonal expansion of B cells expressing high-affinity B cell receptors, which then differentiate into proliferating, antibody-secreting plasmablasts (5). Upon migration to specialized bone marrow niches, plasmablasts differentiate into long-lived non-proliferating plasma cells secreting high amounts of antibodies (6). While these B cell responses are orchestrated by many transcription factors, we describe here the role of Pax5 in controlling these processes.
The transcription factor Pax5 (7) is an essential regulator of B cell commitment (8) and development (9), which is exclusively expressed in the B-lymphoid lineage within the hematopoietic system (10). At the molecular level, Pax5 fulfills a dual role in B lymphopoiesis as it acts as a transcriptional repressor to suppress B-lineage-inappropriate genes (11, 12) and as an activator to induce gene expression required for B cell development and function (12, 13). Pax5 regulates its gene expression program in part by inducing active chromatin at activated target loci and eliminating active chromatin at repressed target loci through recruitment of chromatin-remodeling and histone-modifying complexes (12, 14).
Pax5 is expressed throughout B cell development from pro-B cells in the bone marrow to mature B cells in peripheral lymphoid organs (10), where it is required for the generation of mature B cells (9). Pax5 maintains the B cell gene expression program also in mature B cells, as conditional inactivation of Pax5 leads to the downregulation of B cell-specific genes and reactivation of lineage-inappropriate genes in these B cells (9, 11–13). Importantly, the conditional loss of Pax5 results in the conversion of mature B cells into functional T cells by dedifferentiation to uncommitted progenitors in the bone marrow and, with progressing age of the mice, gives rise to the development of an aggressive progenitor cell leukemia (15). Hence, loss of the B cell phenotype upon Pax5 inactivation highlights an important role of Pax5 in maintaining B cell identity throughout B lymphopoiesis (15, 16) and in suppressing B cell leukemia (15) in the mouse, which is consistent with its function as a haploinsufficient tumor suppressor in B cell acute lymphoblastic leukemia in humans (17).
In contrast to early B cell development, little is known about the role of Pax5 in controlling late B lymphopoiesis. Here, we used conditional Pax5 inactivation in mature B cell types to demonstrate that B-1a, MZ B, GC B and plasma cells were not generated upon Pax5 loss. Pax5-deficient FO B cells had a shortened half-life, which was caused by impaired phosphoinositide 3-kinase (PI3K) signaling due to posttranscriptionally increased protein expression of PTEN, a negative regulator of the pathways. Our study therefore identified Pax5 as an essential regulator of different aspects of B cell immunity.
Results
Reduced numbers and shortened lifespan of follicular B cells lacking Pax5
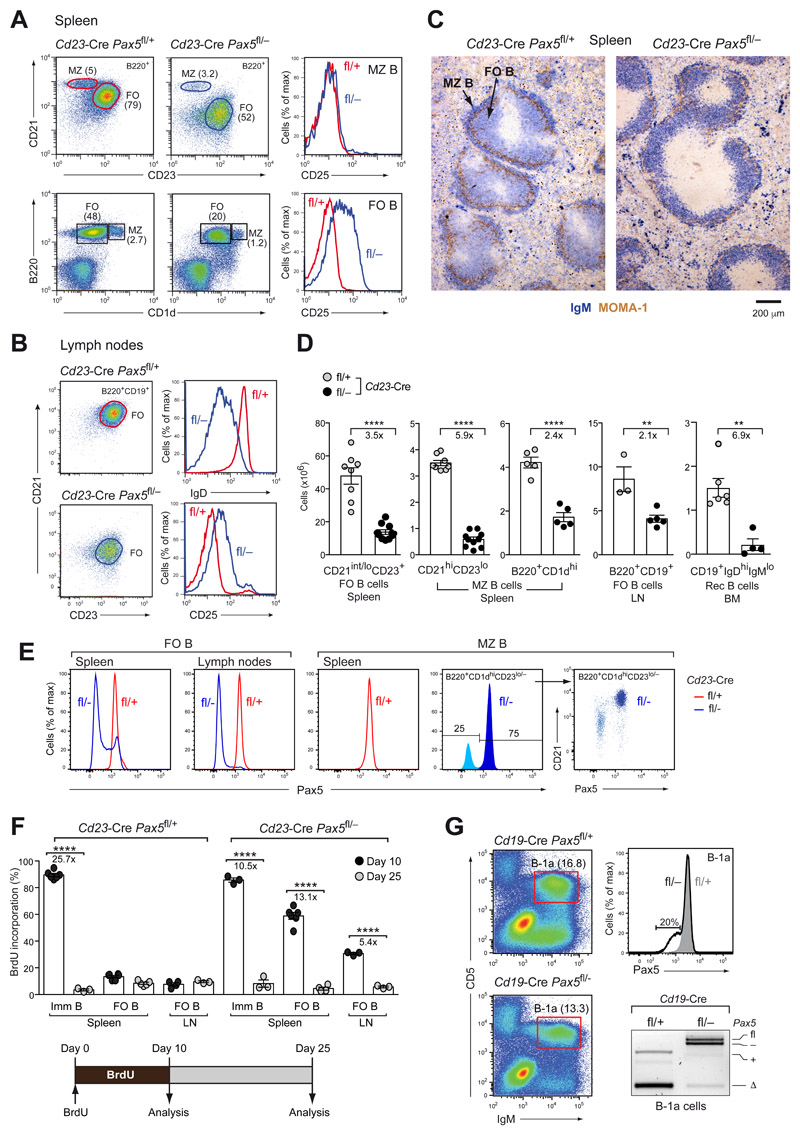
To study the role of Pax5 in mature B cell types, we used the Cd23-Cre line, which initiates Cre-mediated recombination in immature B cells of the spleen (18), to delete the floxed (fl) exon 2 of Pax5 (9) in control Cd23-Cre Pax5 fl/+ mice and Cd23-Cre Pax5 fl/− littermates, which additionally contained the Pax5 null (–) allele (19). Flow-cytometric analysis revealed a prominent CD21lo B cell population (CD21loCD23hiB220+) in the spleen of Cd23-Cre Pax5 fl/− mice instead of the CD21int FO B cells (CD21intCD23hiB220+) detected in control Cd23-Cre Pax5fl/+ mice (Fig. 1A), consistent with the fact that Pax5 directly activates the Cr2 (CD21) gene in mature B cells (9, 12). The Pax5 mutant CD21lo B cell population could reflect a differentiation arrest at an aberrant transitional B cell stage or correspond to Pax5-deficient FO B cells. As lymph nodes that lack transitional B cells (CD21−CD23−) contained only CD21loCD23hi B cells in Cd23-Cre Pax5 fl/− mice (Fig. 1B), we will herein refer to the CD21loCD23hi B cell population as Pax5 mutant FO B cells. These mutant FO B cells were reduced 3.5-fold in the spleen and 2.1-fold in the lymph nodes and mutants recirculating B cells were reduced 6.9-fold in the bone marrow of Cd23-Cre Pax5 fl/− mice compared with Cd23-Cre Pax5 fl/+ mice (Fig. 1D). The upregulated expression of CD25, encoded by the repressed Pax5 target gene Il2ra (12), and the downregulated expression of IgD and CD21 in FO B cells of Cd23-Cre Pax5 fl/− mice (Fig. 1A,B) suggested that Pax5 was lost in Pax5 mutant FO B cells. Intracellular Pax5 staining confirmed the loss of Pax5 protein in all Pax5 mutant FO B cells of the lymph node and in most of the Pax5 mutant FO B cells of the spleen (Fig. 1E). This was further confirmed by the full deletion of the floxed Pax5 exon 2 and an almost complete absence of the Pax5 protein in sorted Pax5 mutant FO B cells, as revealed by PCR and immunoblot analyses, respectively (Fig. S1A,B). Hence, FO B cells were severely reduced in the absence of Pax5.
Figure 1. Loss of mature B cell types upon Pax5 inactivation.
(A,B) Flow-cytometric analysis of MZ B (B220+CD21hiCD23lo/−) and FO B (B220+CD21int/loCD23hi) cells from the spleen (A) and FO B cells from lymph nodes (B) of Cd23-Cre Pax5 fl/− (fl/–) and Cd23-Cre Pax5 fl/+ (fl/+) mice. The percentages of cells in each gate are indicated. (C) Staining of spleen sections for MOMA-1 (brown) and IgM (blue) expression. (D) Absolute numbers of the indicated cell types in the spleen, lymph nodes (LN) and bone marrow (BM) of Cd23-Cre Pax5 fl/+ mice (gray dots) and Cd23-Cre Pax5 fl/− mice (black dots). (E) Intracellular Pax5 staining of FO B and MZ B (CD19+B220+CD 1dhiCD23lo/−) cells. (F) BrdU labeling of splenic immature and FO B cells and lymph node FO B cells of 2-month-old Cd23-Cre Pax5 fl/+ and Cd23-Cre Pax5 fl/− mice. BrdU+ B cells were identified by flow cytometry after 10 days of BrdU labeling (black dots) or after a subsequent 15-day chase period (gray dots) without BrdU in the drinking water (Fig. S1C), as shown by the diagram below. (G) Flow-cytometric analysis of B-1a cells (IgMhiCD5+) from the peritoneum of Cd19-Cre Pax5 fl/− (fl/–; black) and Cd19-Cre Pax5 fl/+ (fl/+; gray) mice. Upper right: Intracellular Pax5 staining of B-1a cells. The percentage of Cd19-Cre Pax5 fl/− B-1a cells with reduced Pax5 expression is shown. Lower right: PCR analysis of the deletion of the floxed Pax5 allele in B-1a cells (IgMhiCD5+). PCR fragments corresponding to the deleted (Δ) or intact (fl) floxed Pax5 allele and the wild-type (+) or null (–) Pax5 allele are indicated. Statistical data (D, F) are shown as mean value with SEM and were analyzed by two-tailed unpaired Student’s t-test (D) or two-way ANOVA with Šídák’s multiple comparison test (F); **P < 0.01, ****P < 0.0001. Each dot represents one mouse.
To investigate the lifespan of Pax5-deficient FO B cells, we continuously labeled Cd23-Cre Pax5 fl/− and Cd23-Cre Pax5 fl/+ mice with the thymidine analogue bromodeoxyuridine (BrdU) for 10 days prior to flow-cytometric analysis of BrdU incorporation in FO B cells (Fig. 1F). BrdU was incorporated in 13.4% of all splenic FO B cells and 89.5% of all immature B cells (CD21−CD23−B220+CD19+) in control Cd23-Cre Pax5 fl/+ mice (Figs. 1F and S1C), confirming that only a few immature B cells are integrated into the quiescent FO B cell pool during the 10-day labeling period (20). In contrast, 59% of the splenic FO B cells and 31% of the lymph node FO B cells in Cd23-Cre Pax5 fl/− mice incorporated BrdU during the first 10 days (Fig. 1F), but were then efficiently replaced by unlabeled Pax5 mutant FO B cells during the subsequent 15-day chase period in a manner similar to immature B cells (Figs. 1F and S1C). Moreover, short-term labeling for 2 hours demonstrated that the Pax5 mutant FO B cells did not proliferate similar to control FO B cells (Fig. S1D). Together, these data revealed a shortened lifespan and rapid turnover of Pax5-deficient FO B cells in the spleen and lymph nodes. Expression of the pro-survival protein Bcl2 from the Vav-Bcl2 transgene (21) could, however, not rescue the FO B cell numbers in the spleen and lymph nodes of Vav-Bcl2 Cd23-Cre Pax5 fl/− mice (Fig. S1E).
Loss of B-1a and marginal zone B cells upon conditional Pax5 inactivation
MZ B cells, which were defined as B220+CD21hiCD23lo/− or B220+CD1dhi cells, were reduced 5.9- and 2.4-fold, respectively, in the spleen of Cd23-Cre Pax5 fl/− mice compared with Cd23-Cre Pax5 fl/+ littermates (Fig. 1A,D). Consequently, few IgM+ B cells were detected on histological spleen sections in the marginal zone outside of the MOMA-1+ macrophage ring in B cell follicles of Cd23-Cre Pax5 fl/− mice in contrast to Cd23-Cre Pax5 fl/+ mice (Fig. 1C). As expected, the MZ B cells of Cd23-Cre Pax5 fl/− mice, which were defined by the Pax5-regulated marker CD21 (B220+CD21hiCD23lo/−), did not upregulate CD25 expression (Fig. 1A) and retained the intact floxed Pax5 allele (Fig. S1A), indicating that a sizeable fraction of MZ B cells did not delete Pax5 (Fig. 1D). Intracellular Pax5 staining of the B220+CD1dhiCD23lo/− MZ B cells confirmed that 75% of the MZ B cells in the Cd23-Cre Pax5 fl/− mice expressed the Pax5 protein at the same level as the MZ B cells of control littermates (Fig. 1E). Notably, 25% of MZ B cells lost Pax5 and downregulated CD21, but failed to accumulate (Fig. 1E), demonstrating that MZ B cells stringently depend on Pax5 function.
To study B-1a cells, we used the Cd19-Cre line, which initiates Cre-mediated recombination in early B cell development (22). Most B-1a cells (IgMhiCD5+) in the peritoneum of Cd19-Cre Pax5 fl/− mice also expressed Pax5 at a normal level, consistent with efficient retention of the floxed Pax5 allele, as opposed to the efficient Pax5 deletion observed in B-1a cells of Cd19-Cre Pax5 fl/+ mice (Fig. 1G). Although 20% of the B-1a cells in Cd19-Cre Pax5 fl/− mice were losing Pax5 expression, these cells did not accumulate, indicating that B-1a cells were lost upon Pax5 inactivation. These data demonstrated that B-1a and MZ B cells did not tolerate the loss of Pax5.
Pax5-dependent initiation and maintenance of germinal center B cell differentiation
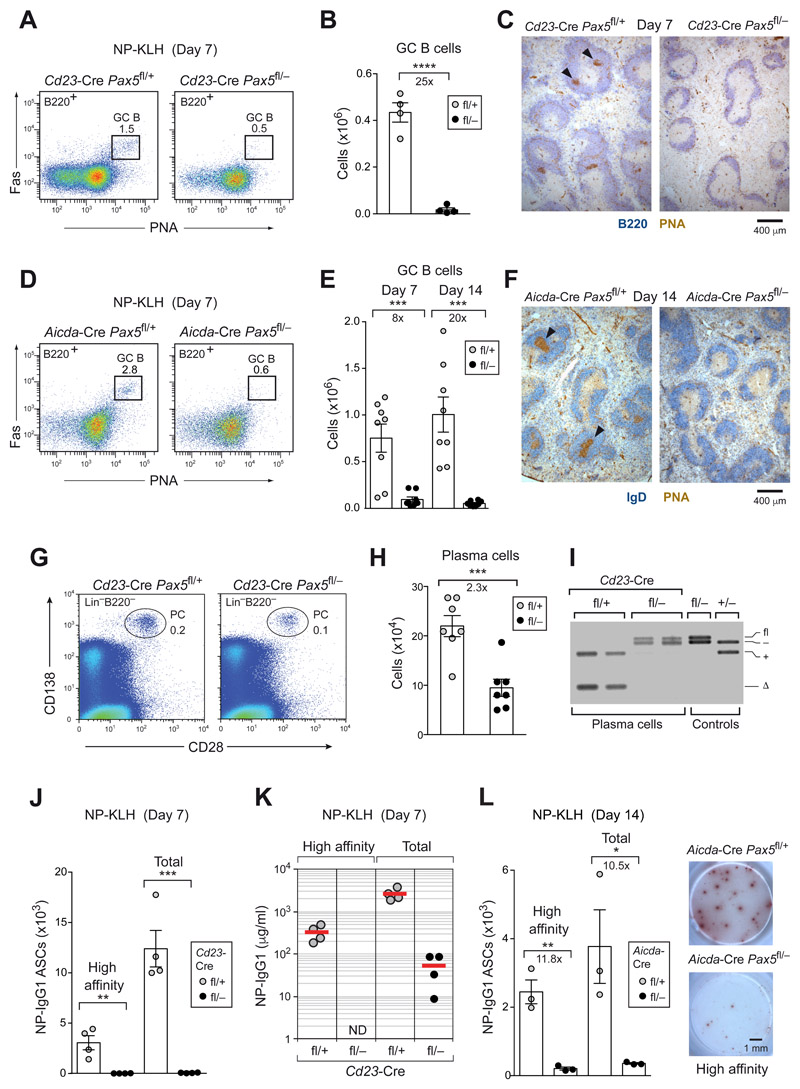
As Pax5 is already lost in Cd23-Cre Pax5 fl/− FO B cells prior to germinal center (GC) B cell formation (Fig. 1E), we immunized Cd23-Cre Pax5 fl/− and control Cd23-Cre Pax5 fl/+ mice with the T cell-dependent antigen NP-KLH (in alum) to study the role of Pax5 at the start of the GC B cell response. At day 7 after immunization, no GC B cells could be detected in the spleen of Cd23-Cre Pax5 fl/− mice both by flow cytometry (Fig. 2A,B) and histological analysis (Fig. 2C).
Figure 2. The initiation and maintenance of GC B cell differentiation depended on Pax5.
(A-C) GC B cell differentiation in the spleen of Cd23-Cre Pax5 fl/+ (fl/+; gray dots) and Cd23-Cre Pax5 fl/− (fl/–; black dots) mice at day 7 after immunization with NP-KLH (in alum). Absolute numbers of GC B cells (B220+Fas+PNA+) were determined by flow cytometry (A,B), and PNA+ GC B cells were visualized by staining of spleen sections for PNA (brown) and B220 (blue) expression (C). Arrowheads indicate GCs. (D-F) GC B cell differentiation in the spleen of Aicda-Cre Pax5 fl/+ and Aicda-Cre Pax5 fl/− mice at day 7 and 14 after immunization with NP-KLH (in alum) was analyzed, as described above. (G-I) Flow-cytometric analysis of plasma cells (CD28+CD138+Lin−) from the bone marrow (femur and tibia of hind legs) of non-immunized Cd23-Cre Pax5 fl/+ and Cd23-Cre Pax5 fl/− mice (G,H). PCR determination of Pax5 exon 2 deletion in sorted plasma cells (I), as described in Fig. 1G. (J-L) T cell-dependent immune responses. Pax5 fl/+ and Pax5 fl/− mice carrying Cd23-Cre (J) or Aicda-Cre (L) were immunized with NP-KLH (in alum) and analyzed at the indicated days after immunization by ELISPOT assay to determine NP-specific IgG1 antibody-secreting cells (ASCs) in the spleen. NP4-BSA- or NP23-BSA-coated plates were used for detecting ASCs secreting high-affinity or total anti-NP-IgG1 antibodies, respectively. Representative ELISPOT images are shown. (K) ELISA analysis of serum titers of NP-specific IgG1 antibodies using NP7-BSA- or NP30-BSA-coated plates. ND, not detected. Statistical data (B,E,H,J,L) are shown as mean value with SEM and were analyzed by the two-tailed unpaired Student’s t-test; *P < 0.05; ***P < 0.001 and ****P < 0.0001. Each dot represents one mouse.
Consistent with this result, no Pax5-deleted GC B cells were present in Cd19-Cre Pax5 fl/− mice that were immunized for 10 days with sheep red blood cells (SRBCs; Fig. S2A-C). As Aicda (AID) and the Aicda-Cre transgene (18) are activated at an early phase of GC B cell differentiation (23), we assessed the function of Pax5 at subsequent stages of the GC B cell response by NP-KLH immunization of Aicda-Cre Pax5 fl/− and control Aicda-Cre Pax5 fl/+ mice.
GC B cells in Aicda-Cre Pax5 fl/− mice were significantly reduced at day 7 and 14 after immunization (Fig. 2D,E) and could not be detected on histological sections at day 14 (Fig. 2F). Importantly, GC B cells with complete deletion of the floxed Pax5 allele were initially observed in the spleen of Aicda-Cre Pax5 fl/− mice at day 5 after SRBC immunization, but were subsequently lost at day 8, as shown by flow-cytometric and histological analyses (Fig. S2D,E). Moreover, the differentiation of GC B cells in Peyer’s patches, which are exposed to antigens of gastrointestinal microbiota, was also strongly impaired in Aicda-Cre Pax5 fl/− mice (Fig. S2F).
Together, these data demonstrated an essential role of Pax5 in the initiation and maintenance of GC B cell differentiation. Moreover, NP-IgG1-specific memory B cells (NP+IgG1+CD38hiCD19+B220+Lin−) were absent in the spleen of Aicda-Cre Pax5 fl/− mice at day 14 and 28 after NP-KLH immunization (Fig. S2G,H), indicating that IgG1+ memory B cells of GC-dependent or GC-independent origin were not generated upon conditional Pax5 loss in activated B cells.
Pax5 was required for the generation of plasma cells in vivo
Long-lived plasma cells (CD28+CD138+Lin−) (11) in the bone marrow of non-immunized Cd23-Cre Pax5 fl/− mice were 2.3-fold reduced compared with control Cd23-Cre Pax5 fl/+ mice (Fig. 2G,H) and retained the intact floxed Pax5 allele (Fig. 2I), suggesting that they could be derived from MZ and B-1a cells, which failed to undergo Pax5 deletion (Fig. 1E,G). Moreover, plasma cells secreting high-affinity or total NP-specific IgG1 antibodies were not detected by ELISPOT assay at day 7 and 14 after NP-KLH immunization in the spleen of Cd23-Cre Pax5 fl/− and Aicda-Cre Pax5 fl/− mice (Fig. 2J,L), respectively, consistent with the absence of high-affinity NP-specific IgG1 antibodies in the serum of Cd23-Cre Pax5 fl/− mice (Fig. 2K). Hence, T cell-dependent B cell immune responses were effectively lost upon Pax5 inactivation in mature B cells.
Pax5 regulated CSR, but not proliferation, upon anti-CD40 and IL-4 stimulation
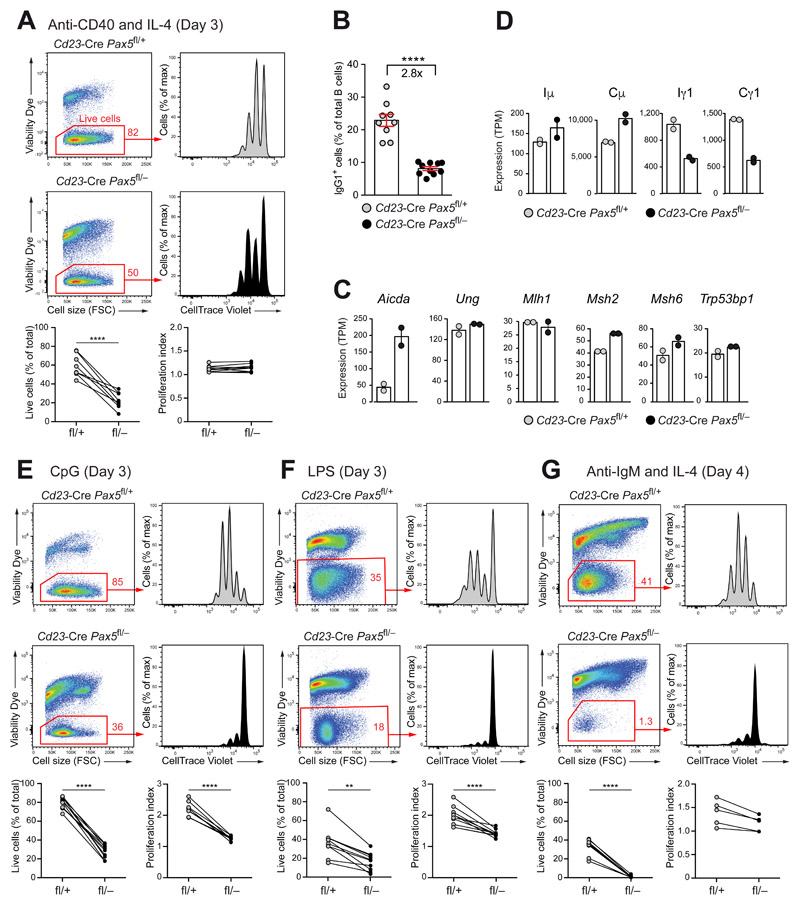
To investigate whether Pax5 controls cell proliferation and class switch recombination (CSR), we stimulated CellTrace Violet-labeled FO B cells from lymph nodes of Cd23-Cre Pax5 fl/− and Cd23-Cre Pax5 fl/+ mice with anti-CD40 and IL-4, which mimics T cell help. Consistent with the observed shortened lifespan (Fig. 1F), the viability of the Pax5-deficient B cells was greatly decreased relative to that of control B cells after 3 days of stimulation (Fig. 3A). The viable Pax5-deficient and control B cells diluted CellTrace Violet at a comparable frequency, resulting in a similar proliferation index for both cell types (Fig. 3A). Notably, the percentage of IgG1+ B cells at day 4 of stimulation was reduced 2.8-fold upon Pax5 inactivation (Fig. 3B), demonstrating that Pax5 was required for efficient CSR to IgG1.
Figure 3. Pax5 controlled B cell proliferation in response to BCR and TLR signaling.
(A,B) Proliferation and IgG1 CSR response to anti-CD40 plus IL-4 stimulation. CellTrace Violet-labeled FO B cells from lymph nodes of Cd23-Cre Pax5 fl/− (fl/–; black) and Cd23-Cre Pax5 fl/+ (fl/+; gray) mice were stimulated with anti-CD40 and IL-4 for 3 (A) or 4 (B) days and then stained with the Viability Dye eFluor™ 780. The cell viability and proliferation index of the stimulated cells (A) and the percentage of IgG1+ B cells (B) were determined by flow-cytometric analysis. Lines connect the results obtained with Pax5-deficient and control B cells in the same stimulation experiment. (C,D) Gene expression in lymph node B cells stimulated with anti-CD40 and IL-4 for 2 days, as determined by RNA-seq (Fig. S3E). The expression of selected genes involved in CSR (C) and the abundance of germline transcripts at the Iμ and Ig1 exons and transcripts at the Cμ and Cg1 exons (D) are shown as mean expression value (TPM, transcripts per million) with SEM based on two independent RNA-seq experiments per genotype. (E-G) Stimulation of lymph node FO B cells of the indicated genotypes with CpG oligodeoxynucleotides (E) and LPS (F) for 3 days or with anti-IgM and IL-4 (G) for 4 days, as described (A). The data (A,E,F,G) were statistically analyzed by the two-tailed unpaired Student’s test: **P < 0.01, ****P < 0.0001. Each dot represents one mouse.
To study the molecular mechanism by which Pax5 controls CSR, we determined the genome-wide Pax5 binding (by ChIP-seq), DNase I hypersensitive (DHS) sites (by DHS-seq) and gene expression (by RNA-seq) in Pax5-deficient and control FO B cells after 2 days of anti-CD40 plus IL-4 stimulation (Fig. S3A-E and Table S1). Genes with important functions in CSR, base-excision repair, mismatch repair and non-homologous end joining were similarly expressed in stimulated Pax5-deficient and control B cells except for Aicda, encoding the central regulator AID of CSR and SHM (24), whose expression was 4-fold upregulated in the absence of Pax5 (Figs. 3C and S3G). Hence, the CSR defect in Pax5-deficient B cells was not caused by decreased expression of genes implicated in CSR or repair pathways.
The switch region located 5’ of a constant gene exon is made accessible for CSR by germline transcription from an upstream I promoter that is activated by cytokine signaling (4, 25). The Iγ1 germline transcript (GLT) was strongly induced by anti-CD40 plus IL-4 stimulation in control B cells, whereas its expression was 2-fold reduced in Pax5-deficient B cells (Fig. 3D). The Cγ1 gene region contained Pax5-binding sites at the Iγ1 promoter and at a downstream enhancer known as Cγ1-2b DHS site 1 (26) (Fig. S3H). The DNase I hypersensitivity at the Iγ1 promoter and downstream enhancer was strongly reduced in stimulated Pax5-deficient B cells compared with control B cells (Fig. S3H) in contrast to the DHS sites at the control Tbp locus (Fig. S3I). These data therefore indicated that Pax5 promoted CSR to IgG1 by inducing the formation of open chromatin at the Iγ1 promoter and downstream enhancer.
Pax5 controlled B cell proliferation in response to BCR and TLR signaling
We next stimulated Pax5-deficient and control FO B cell by activating the Toll-like receptor 9 (TLR9) with CpG oligodeoxynucleotides and TLR4 with lipopolysaccharide (LPS) for 3 days, or the BCR with anti-IgM antibody and IL-4 for 4 days (Fig. 3E-G). The viability of Pax5-deficient B cells was greatly reduced relative to that of control B cells under all stimulation conditions (Fig. 3E-G). The viable Pax5-deficient B cells showed little dilution of CellTrace Violet, thus resulting in a strongly decreased proliferation index (Fig. 3E-G). As Pax5-deficient FO B cells undergo normal cell divisions upon anti-CD40 plus IL-4 treatment (Fig. 3A), we concluded that the loss of Pax5 did not affect IL-4 signaling and that the proliferation defect of Pax5-deficient FO B cells upon stimulation with anti-IgM and IL-4 must be caused by impaired BCR signaling. Together, these data revealed an essential role of Pax5 in the control of BCR and TLR signaling.
Impaired intracellular BCR and TLR signaling in the absence of Pax5
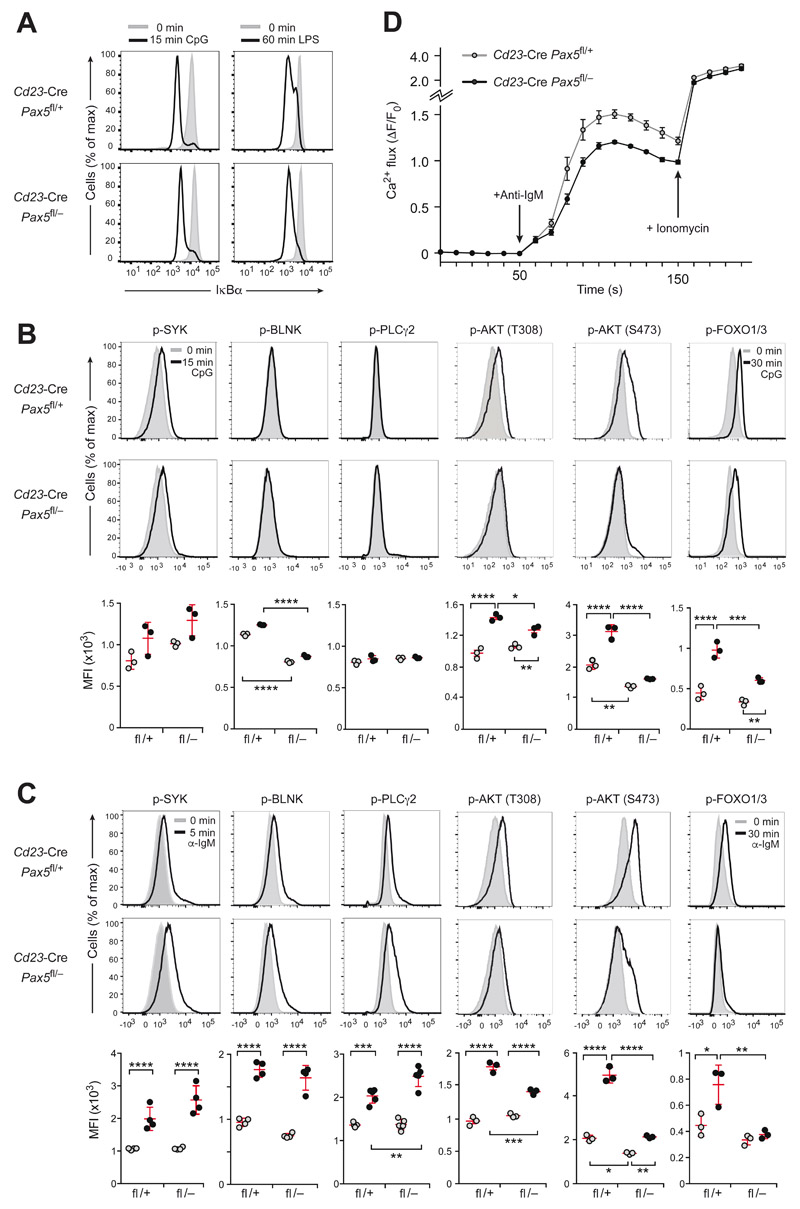
As TLR signaling results in MyD88-dependent activation of the transcription factor NF-κB (27), we investigated whether intracellular signaling leading to the degradation of the NF-κB inhibitor IκBα was impaired in Pax5-deficient FO B cells. As revealed by intracellular staining, IκBα was efficiently degraded in Pax5-deficient and control FO B cells within 15 min of CpG oligodeoxynucleotide addition or 60 min of LPS stimulation, indicating normal NF-κB activation in the absence of Pax5 (Fig. 4A). TLR signaling also engages the PI3K pathway (27), which depends on a crosstalk with BCR signaling in B cells (28, 29). While CpG-mediated TLR9 stimulation for 15 min did not induce phosphorylation of the signal transducers BLNK and PLCγ2, it resulted in moderately increased SYK phosphorylation in Pax5-deficient and control FO B cells (Fig. 4B). In contrast, TLR9 signaling induced phosphorylation of AKT (at Thr308 and Ser473) and FOXO1,3 strongly in control FO B cells, but weakly in Pax5-deficient FO B cells (Figs. 4B and S4A). Hence, the loss of Pax5 did not affect NF-κB activation, but instead impaired PI3K signaling in response to TLR9 signaling.
Figure 4. Intracellular signaling upon TLR9 and BCR activation in Pax5-deficient FO B cells.
(A) IκBα degradation upon TLR signaling. The IkBa protein amount was determined by intracellular staining of CD43− FO B cells from lymph nodes of Cd23-Cre Pax5 fl/− or control Cd23-Cre Pax5 fl/+ mice before (gray surface) and after (black line) stimulation for 15 min with CpG oligodeoxynucleotides or for 60 min with LPS. (B,C) Intracellular TLR9 and BCR signaling. The phosphorylation (p-) status of signal transducers of the calcium and PI3K signaling pathways (Fig. S4A) was determined in lymph node FO B cells of the indicated genotypes before or after stimulation with CpG oligodeoxynucleotides for 15 min (B) or stimulation with anti-IgM for 5 min (C) except for a 30-min stimulation with either stimuli for analyzing p-FOXO1,3. (B,C). Flow cytometry (top and middle) was performed with antibodies specific for p-AKT (p-Thr308) p-AKT (p-Ser473), p-BLNK (p-Tyr84), p-PLCγ2 (p-Tyr759), p-SYK (p-Tyr525/526) and p-FOXO1 (p-Thr24)/p-FOXO3 (p-Thr32). Bottom, the median fluorescence intensity (MFI) for untreated (gray dots) and stimulated (black dots) FO B cells of the indicated genotypes is shown. Statistical data are indicated as mean value with SEM and were analyzed by two-way ANOVA with Tukey’s multiple comparison test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Each dot represents one mouse. One of at least three experiments is shown. (D) Calcium mobilization in response to BCR signaling. Intracellular Ca2+ fluxes in CD43− FO B cells from lymph nodes of the indicated genotypes were recorded as an increase of the fluorescent emission of a Ca2+ sensor dye after addition of anti-IgM (arrow, after 50 s) or ionomycin (arrow, after 150 s) and are presented as ΔF/F0 (F0, average fluorescence before antibody addition; ΔF, fluorescence at time ‘t’ - F0). Mean values with SEM are shown for 3 independent experiments.
Phosphorylation of AKT (at Thr308 and Ser473) and FOXO1,3 in response to anti-IgM stimulation of the BCR was also strongly impaired in Pax5-deficient FO B cells relative to control B cells (Figs. 4C and S4A). Moreover, phosphorylation of the 4E-BP1 and S6 proteins downstream of mTORC1 signaling was also affected (Fig. S4B), in agreement with a critical role of the PI3K-AKT pathway in controlling mTORC1 activity (30) (Fig. S4A). In contrast, the phosphorylation of SYK, BLNK and PLCγ2 was similarly increased in Pax5-deficient and control FO B cells upon anti-IgM treatment (Fig. 4C). Consistent with normal phosphorylation of these three signaling molecules, intracellular calcium mobilization (31) was efficiently induced in Pax5-deficient FO B cells in response to anti-IgM stimulation (Figs. 4D and S4A). The moderately attenuated calcium fluxes of the Pax5-deficient FO B cells could be explained by the loss of PI3K signaling, which minimally affects calcium signaling in anti-IgM-treated B cells (32). In summary, PI3K-AKT signaling, which is essential for cell proliferation and survival (30), was impaired in response to both TLR9 and BCR activation in Pax5-deficient FO B cells, while BCR-induced calcium signaling and TLR9-mediated NF-κB activation were largely normal in the absence of Pax5.
The genes coding for different components of the PI3K-AKT pathway were similarly expressed in Pax5-deficient and control FO B cells, except for Cd19 coding for an essential upstream activator of the PI3K (33) (Fig. S4C). Pax5-deficient FO B cells in the lymph node and spleen expressed the CD19 protein at a 2.6- and 1.9-fold lower level relative to control B cells, respectively (Fig. S4D). Heterozygous Cd19 +/− FO B cells with their 1.9-fold lower CD19 expression (Fig. S4D) could efficiently induce phosphorylation of AKT (S473) upon anti-IgM stimulation in contrast to the strongly impaired AKT phosphorylation observed in Pax5-deficient FO B cells (Fig. S4E). We therefore concluded that the lower expression of CD19 in Pax5-deficient FO B cells dud not explain the strong AKT signaling defect of these cells.
Pax5-dependent activation of immediate-early genes in response to BCR signaling
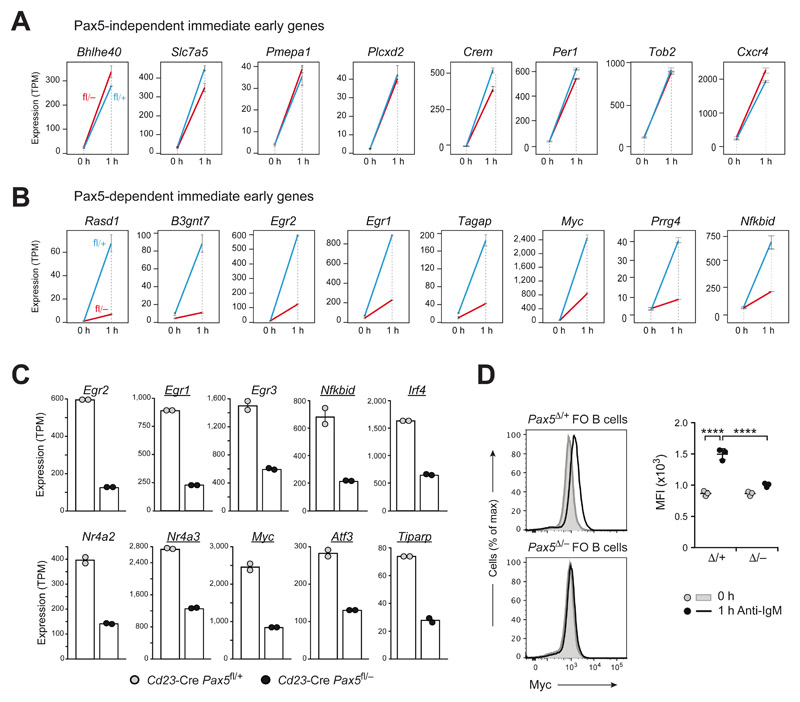
To investigate a role of Pax5 in controlling immediate-early gene activation, we performed RNA-seq analysis of FO B cells from lymph nodes of Cd23-Cre Pax5 fl/− and Cd23-Cre Pax5 fl/+ mice before and after anti-IgM stimulation for 1 h (Fig. S5A-C and Table S2). The bioinformatic analysis of the RNA-seq data obtained at two time points of BCR stimulation and in B cells of two different genotypes (see Materials and Methods and Fig. S5D, E) resulted in the identification of 11 Pax5-independent and 39 Pax5-dependent immediate-early genes that were induced > 9-fold in control FO B cells (Fig. S5F and Table S3). The Pax5-independent and Pax5-dependent expression of immediate-early genes (Fig. 5A, B) was consistent with our finding that the intracellular signaling pathways of the BCR were either largely unaffected (SYK-BLNK-PLCγ2) or impaired (PI3K-AKT) in the absence of Pax5 (Fig. 4C). Notably, 11 of the 39 Pax5-dependent immediate-early genes coded for known transcriptional regulators (Fig. 5C, D and Table S3). In summary, these data demonstrated that Pax5 directly or indirectly activated the expression of several transcription factors in response to BCR signaling.
Figure 5. Activation of immediate-early genes in response to BCR signaling.
(A,B) Pax5-independent (A) and Pax5-dependent (B) immediate-early genes that were induced > 9-fold upon anti-IgM stimulation in control FO B cells and were further defined, as described in Fig. S5D-F and the Materials and Methods. The expression of activated genes in Cd23-Cre Pax5 fl/+ (fl/+; blue) and Cd23-Cre Pax5 fl/− (fl/–; red) FO B cells before (0 h) and after (1 h) of anti-IgM stimulation is shown as mean expression value (TPM) with SEM based on two independent RNA-seq experiments for each genotype and treatment condition. (C) mRNA expression of Pax5-dependent immediate-early genes, coding for known transcription factors, is shown for FO B cells of the indicated genotypes after 1 h of anti-IgM stimulation. Genes bound by Pax5 (12) are underlined. (D) Intracellular Myc staining of Cd23-Cre Pax5 fl/+ (Pax5 Δ/+) and Cd23-Cre Pax5 fl/− (Pax5 Δ/−) FO B cells before (gray) and after (black) stimulation for 1 h with anti-IgM (left). Myc expression (right) is shown as mean MFI value with SEM and was analyzed by twoway ANOVA with Tukey’s multiple comparison test; ****P < 0.0001. Each dot represents one mouse.
miRNA-mediated downregulation of PTEN expression by Pax5 in mature B cells
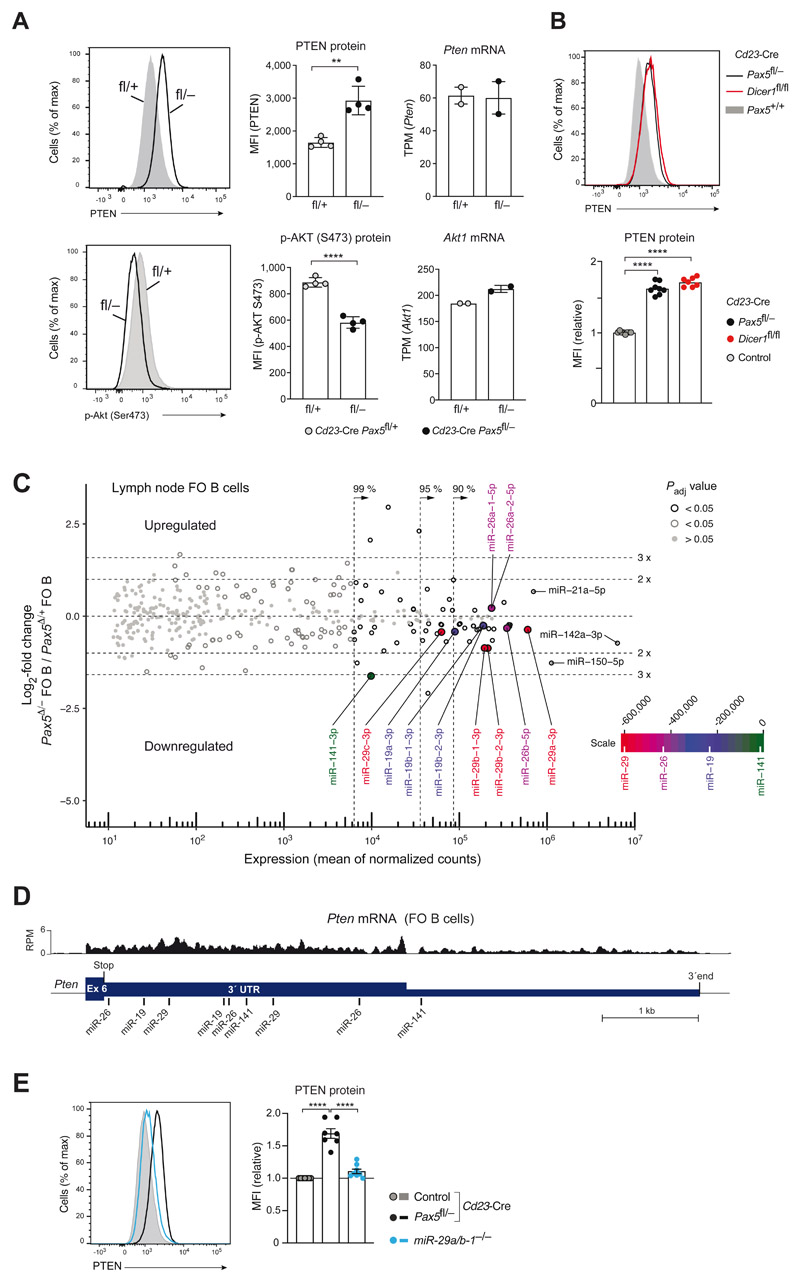
As genes coding for essential components of PI3K-AKT signaling were not significantly regulated by Pax5 (Fig. S4C), we investigated the expression of the lipid phosphatase PTEN, which antagonizes the function of the phosphoinositide 3-kinase (PI3K) by converting phosphatidylinositol-3,4,5-triphosphate (PI(3,4,5)P3) to PI(4,5)P2 (30) (Fig. S4A). As shown by intracellular staining, PTEN protein expression was significantly increased in unstimulated Pax5-deficient FO B cells compared with control FO B cells, while phosphorylation of AKT (Ser473) was concomitantly reduced in the absence of Pax5 (Fig. 6A). Pten mRNA expression was, however, similar in FO B cells of both genotypes (Fig. 6A). PTEN expression is regulated at the posttranscriptional level by different microRNAs (miRNAs) that target the 3’ untranslated region (3’UTR) of the Pten mRNA (34). To investigate whether the loss of miRNAs may cause the increased PTEN expression, we analyzed FO B cells lacking the RNase III enzyme Dicer, which is essential for processing of all pre-miRNAs to mature miRNAs (35). Importantly, Dicer-deficient (Cd23-Cre Dicer1 fl/fl) FO B cells expressed the PTEN protein at a similarly high level as Pax5-deficient FO B cells (Fig. 6B), although the Pten mRNA was not increased compared with control FO B cells (Fig. S6A). These data indicated that Pax5 may be involved in the generation of miRNAs that target the Pten mRNA.
Figure 6. Pax5 downregulated PTEN expression by controlling the abundance of Pten-targeting microRNAs.
(A) PTEN protein expression and phosphorylation of AKT (Ser473) in unstimulated lymph node FO B cells of Cd23-Cre Pax5 fl/− (back) and Cd23-Cre Pax5 fl/+ (gray) mice, as determined by intracellular staining (left) and MFI quantification (middle). Pten and Akt1 mRNA expression in the same FO B cell types (right) is shown as mean expression value (TPM) with SEM, as determined by RNA-seq. (B) PTEN expression in lymph node FO B cells of Cd23-Cre Dicer1 fl/fl (red), Cd23-Cre Pax5 fl/− (black) and Cd23-Cre Pax5 +/+ (gray) mice, as determined by intracellular staining and quantification of the MFI values relative to the control genotypes (Cd23-Cre Pax5 +/+ Dicer1 +/+, Pax5+/+ Dicer1 +/+, Pax5 fl/+ Dicer1 +/+, Pax5 +/+ Dicer1 fl/+ or Pax5+/+ Dicer1 fl/fl). (C) MA plot of miRNA expression differences between Cd23-Cre Pax5 fl/− (Pax5 Δ/−) and Cd23-Cre Pax5 fl/+ (Pax5 Δ/+) FO B cells, which were isolated from lymph nodes as CD23+ cells by immunomagnetic sorting. Two small-RNA-seq experiments per genotype were performed. The abundance of individual miRNAs in the two B cell types is plotted as mean value of the normalized counts versus the log2-fold change in abundance between Pax5 Δ/− and Pax5 Δ/+ FO B cells (Table S4 and S6). Dotted lines indicate the area containing 99%, 95% or 90% of the total normalized counts. The statistical significance of the observed differences is indicated by gray and black circles (P value < 0.05) or gray dots (P value > 0.05). Adjusted P values were determined by DESeq2. Each symbol represents one miRNA species. Pten-targeting miRNAs are highlighted by the color corresponding to their position on the scale bar, which was generated by multiplying the sum of the normalized read counts of all members of a miRNA family with the total context+/+ score of the miRNA family (Table S5). (D) Location of the target sites of the indicated miRNAs in the 3’UTR of the mouse Pten mRNA, as predicted by the TargetScanMouse algorithm (v7.2; targetscan.org; (37)). The mRNA-seq profile of Pten exon 6 in control FO B cells is shown. (E) PTEN expression in unstimulated lymph node FO B cells of miR-29a/b-1 −/− (blue), Cd23-Cre Pax5 fl/− (back) and control Cd23-Cre Pax5 fl/+ (gray) mice, as determined by intracellular staining (left) and quantification of the MFI values (right) relative to control FO B cells (Cd23-Cre Pax5 +/+, Cd23-Cre Pax5 fl/+, Pax5 +/+ or Pax5 fl/fl). Statistical data (A,B,E) are shown as mean value with SEM and were statistically analyzed by the two-tailed unpaired Student’s t-test (A) or by one-way ANOVA with Tukey’s multiple comparison test (B,E): **P < 0.01, ****P < 0.0001.
As Pax5 did not regulate genes involved in the synthesis (Drosha, DGCR8 or Dicer) or function (Argonaute proteins) of miRNAs, we used small-RNA-sequencing (36) to determine the differential abundance of mature miRNAs in Cd23-Cre Pax5 fl/− versus Cd23-Cre Pax5 fl/+ FO B cells from lymph nodes (Fig. 6C and Table S4) or spleen (Fig. S6B and Table S4). We next analyzed the miRNA dataset of lymph node FO B cells for differentially expressed Pten-targeting miRNAs according to the following criteria; a Pten-targeting miRNA should have a predicted ‘total context++ score’ (37) of < −0.47 (Fig. S6C) for targeting Pten 3’UTR sequences, should be part of the miRNAs accounting for 99% of the normalized read counts and should be differentially expressed with a Padj value of < 0.05 (see Materials and methods). This analysis identified members of the miR-29, miR-26, miR-19 and miR-141 families as differentially expressed miRNAs, with strongly predicted Pten targeting, in lymph node FO B cells (Fig. 6C,D and Table S5). miRNAs of these four families were also differentially expressed in splenic FO B cells (Fig. S6B). Moreover, target sites in the Pten 3’UTR (Fig. 6D) have been experimentally validated for all four miRNA families; miR-19 (38), miR-26 (39), miR-29 (40–42) and miR-141 (43–45).
Of the Pten-targeting miRNAs, the four members of the miR-29 family were together most highly expressed (Figs. 6C and S6B) and were downregulated between 2.9- and 1.3-fold in Pax5-deficient FO B cells from lymph nodes and spleen (Fig. S6D). As the four miR-29 isoforms are encoded by the two loci miR-29a/b-1 and miR-29b-2/c, we next analyzed the effect of deleting miR-29a and miR-29b-1 on PTEN expression in FO B cells from lymph nodes of miR-29a/b-1 –/− mice (46, 47). Intracellular staining revealed a small increase of PTEN expression in miR-29a/b-1 −/− FO B cells compared with control FO B cells (Fig. 6E), while phosphorylation of AKT (S473) was reduced in miR-29a/b-1 −/− FO B cells relative to control FO B cells upon anti-IgM stimulation (Fig. S6E). Given the small effect on PTEN expression in miR-29a/b-1 −/− FO B cells, it is conceivable that the cumulative loss of other Pax5-deregulated Pten-targeting miRNAs (Table S5) may have contributed to the strongly increased PTEN expression observed in Pax5-deficient FO B cells. Together, these data indicated that Pax5 restrained PTEN expression in mature B cells, likely by controlling the abundance of Pten-targeting miRNAs.
Loss of PTEN rescued PI3K signaling and the survival of Pax5 mutant FO B cells
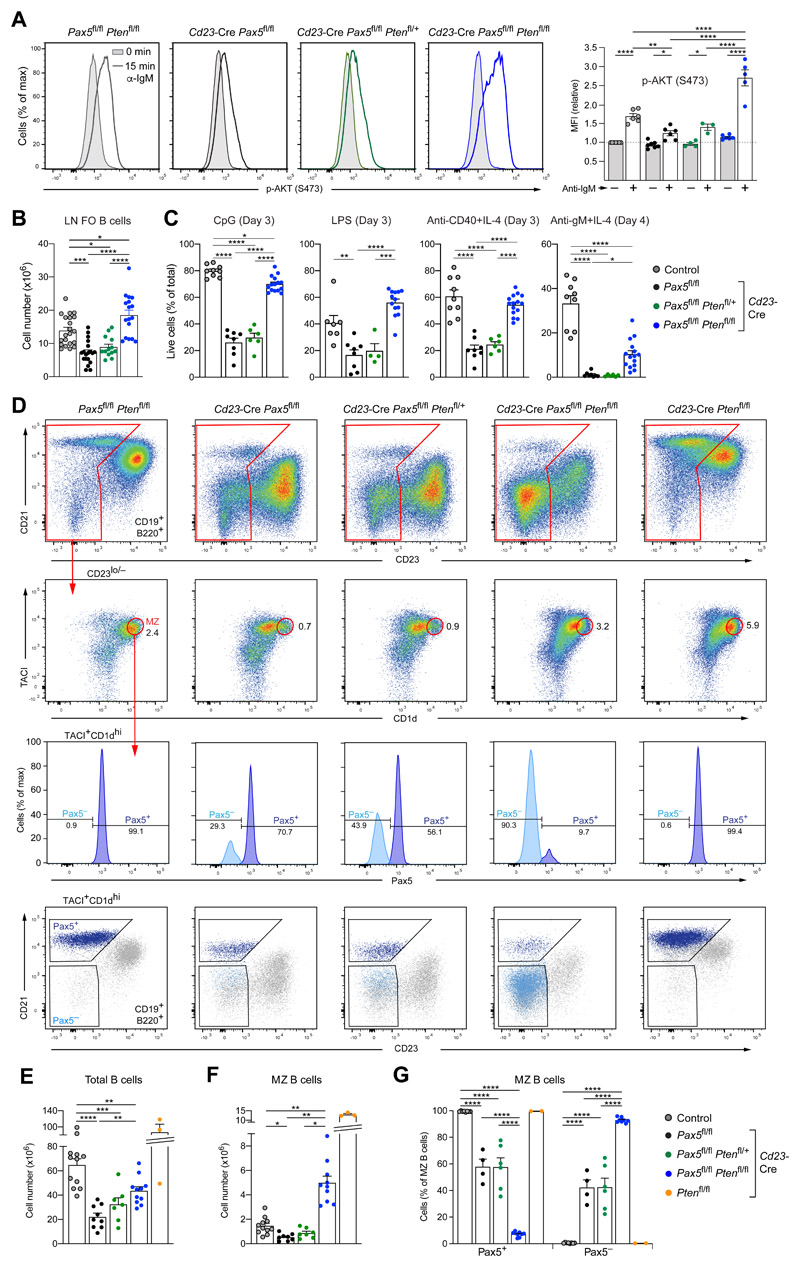
To investigate whether the loss of PTEN may restore PI3K signaling in Pax5 mutant FO B cells, we deleted the Pten gene in Cd23-Cre Pax5 fl/fl Pten fl/+ and Cd23-Cre Pax5 fl/fl Pten fl/fl mice. Intracellular staining and immunoblot analyses revealed that the PTEN protein was lost in Cd23-Cre Pax5 fl/fl Pten fl/fl FO B cells, while it was still expressed at elevated levels in Cd23-Cre Pax5 fl/fl Pten fl/+ FO B cells compared with control FO B cells (Fig. S7A,B). Anti-IgM stimulation for 15 min resulted in significantly increased phosphorylation of AKT (Ser473) in Cd23-Cre Pax5 fl/fl Pten fl/fl FO B cells in contrast to Cd23-Cre Pax5 fl/fl Pten fl/+ FO B cells that exhibited the same low AKT phosphorylation levels as Pax5 mutant FO B cells (Fig. 7A). Notably, FO B cell numbers were restored in lymph nodes of Cd23-Cre Pax5 fl/fl Pten fl/fl mice in contrast to Cd23-Cre Pax5 fl/fl Pten fl/+ mice (Fig. 7B). Stimulation with CpG oligodeoxynucleotides, LPS or anti-CD40 and IL-4 for 3 days revealed that the survival of Cd23-Cre Pax5 fl/fl Pten fl/fl FO B cells was rescued compared with Cd23-Cre Pax5 fl/fl Pten fl/+ FO B cells, while a partial rescue was observed upon anti-IgM plus IL-4 treatment for 4 days (Figs. 7C and S7C). The proliferation of Cd23-Cre Pax5 fl/fl Pten fl/fl FO B cells was, however, not restored in response to stimulation with CpG, LPS or anti-IgM and IL-4 (Fig. S7C,D). Together, these data indicated that the combined loss of Pax5 and PTEN rescued PI3K signaling and cell survival, but not the proliferation of FO B cells.
Figure 7. Loss of PTEN rescued PI3K signaling, FO B and MZ B cell numbers in Pax5 mutant mice.
(A) Rescue of PI3K signaling. CD43− FO B cells from lymph nodes of Cd23-Cre Pax5 fl/fl (black), Cd23-Cre Pax5 fl/fl Pten fl/+ (green), Cd23-Cre Pax5 fl/fl Pten fl/fl (blue) and control Pax5 fl/fl Pten fl/fl (gray) mice were either left untreated (gray surface) or stimulated (colored line) for 15 min with anti-IgM prior to intracellular staining with an anti p-AKT (Ser473) antibody (left). Quantification of the MFI values relative to the unstimulated FO B cells of the control genotype is shown to the right. (B,C) Rescue of the FO B cell survival before and after stimulation. FO B cells from lymph nodes (LN) of the indicated genotypes were analyzed directly ex vivo (B) or after stimulation (C) with CpG oligodeoxynucleotides, LPS, anti-CD40 and IL-4 or anti-IgM and IL-4 for the indicated days prior to staining with the Viability Dye eFluor™ 780. The frequency of viable B cells is plotted. (D) Flow-cytometric analysis of MZ B cells in the spleen of the indicated genotypes. MZ B cells were identified as CD19+B220+CD23lo/−TACI+CD1dhi cells and analyzed for Pax5 protein expression by intracellular staining. As shown by backgating, the Pax5+ MZ B cells expressed CD21, whereas the Pax5− MZ B cells lost CD21 expression. (E-G) Statistical analysis indicating the number of total B cells (E) and MZ B cells (F) as well as the relative frequency of Pax5+ and Pax5− MZ B cells (G) in the spleen of mice of the five indicated genotypes. Statistical data are shown as mean value with SEM and were analyzed by two-way ANOVA with Šídák’s multiple comparison test (A) or one-way (B,C,E,F) or two-way (G) ANOVA with Tukey’s multiple comparison test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Each dot represents one mouse. The control genotypes (A,B,E,F,G) were Pax5 fl/fl Pten fl/fl, Pax5 fl/fl Pten fl/+, Pax5 fl/fl Pten +/+ or Cd23-Cre Pax5 fl/+ Pten +/+.
Rescue of MZ B cell development in Pten,Pax5 double-mutant mice
Splenic B cell numbers were partially restored in Cd23-Cre Pax5 fl/fl Pten fl/fl mice (Fig. 7D,E). In contrast, GC B cells were present at very low numbers in the spleen of non-immunized Cd23-Cre Pax5 fl/fl Pten fl/fl mice, while the residual GC B cells expressed normal Pax5 levels due to strong counterselection against Pax5 deletion (Fig. S7E). Similarly, the numbers of GC B cells in Peyer’s patches were low in Cd23-Cre Pax5 fl/fl mice and increased in Cd23-Cre Pax5 fl/fl Pten fl/fl mice, although Pax5 displayed normal expression in GC B cells of both genotypes (Fig. S7F). Hence, these data demonstrated that the loss of PTEN could not rescue the development of Pax5-deficient GC B cells.
To study MZ B cell differentiation, we took advantage of the fact that the higher expression of CD1d and TACI on MZ B cells compared with FO B cells could be used to distinguish these two cell types (Fig. S7G). MZ B cells (CD19+B220+CD23lo/−TACI+CD1dhi) were present at similarly low abundance in Cd23-Cre Pax5 fl/fl Pten fl/+ mice as in Cd23-Cre Pax5 fl/fl mice (Fig. 7D,F), and a major fraction of these residual MZ B cells failed to delete Pax5 (Fig. 7D,G), thus indicating that the loss of one Pten allele did not restore MZ B cell numbers. In marked contrast, the number of MZ B cells was 3.4-fold higher in Cd23-Cre Pax5 fl/fl Pten fl/fl mice relative to Pten-expressing control mice, while MZ B cells were even more abundant in Cd23-Cre Pten fl/fl mice (Fig. 7D,F), confirming previous published work (48, 49). Notably, most MZ B cells (93%) in Cd23-Cre Pax5 fl/fl Pten fl/fl mice did not express Pax5 and CD21, indicating that counterselection against Pax5 deletion was no longer observed upon further PTEN loss (Fig. 7D,G). We therefore concluded that the additional loss of PTEN allowed Pax5-deficient B cells to differentiate to MZ B cells.
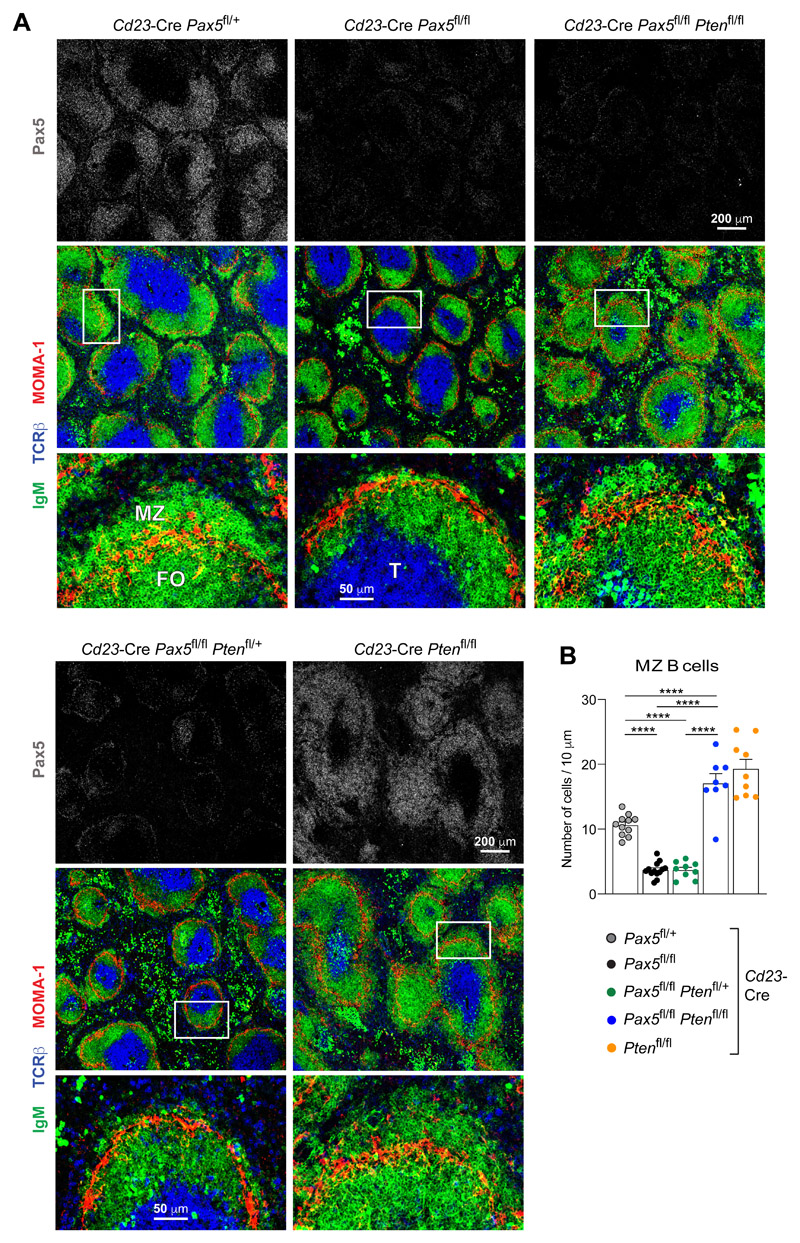
To confirm these results, we analyzed spleen sections from mice of all five genotypes by immunofluorescence analysis with antibodies detecting Pax5, IgM (B cells), MOMA-1 (metallophilic macrophages) or TCRβ (T cells; Fig. 8A). Staining with the anti-Pax5 antibody confirmed the loss of Pax5 in most B lymphocytes of Cd23-Cre Pax5 fl/fl, Cd23-Cre Pax5 fl/fl/ Pten fl/+ and Cd23-Cre Pax5 fl/fl Pten fl/fl mice (Fig. 8A). Moreover, MZ B cells located outside of the MOMA-1+ macrophage ring were largely absent in Cd23-Cre Pax5 fl/fl and Cd23-Cre Pax5 fl/fl Pten fl/+ mice, but present in Cd23-Cre Pax5 fl/+, Cd23-Cre Pten fl/fl and Cd23-Cre Pax5 fl/fl Pten fl/fl mice (Fig. 8A). We next measured the cellularity of the MZ B cell layer outside of the macrophage ring with a custom-made program (see Methods). This statistical evaluation corroborated that MZ B cell numbers were rescued only upon complete loss of PTEN in Cd23-Cre Pax5 fl/fl Pten fl/fl mice (Figs. 8B and S7H). Together, these data demonstrated that the rescue of PI3K signaling in Cd23-Cre Pax5 fl/fl Pten fl/fl mice restored MZ B cell development.
Figure 8. Rescue of MZ B cell development in Pten,Pax5 double-mutant mice.
(A) Immunohistological analysis of spleen sections from 12-week-old mice of the indicated genotypes. The sections were stained with antibodies detecting IgM (green), MOMA-1 (red), TCRβ (blue) and Pax5 (gray). Selected areas (boxed) of B cell follicles are shown at higher magnification. T, FO B and MZ B cell zones are indicated. One of three experiments is shown. (B) Quantification of the MZ B cells on histological sections. The average number of IgM+ B cells outside of the MOMA-1+ macrophage ring was determined per 10 μm length of the perimeter of the MOMA-1+ ring (see Methods). Each dot represents the measurement of one follicle. The mean values determined for the indicated genotypes is shown with SEM and were analyzed by one-way ANOVA with Tukey’s multiple comparison test; ****P < 0.0001.
Discussion
Pax5 is a key regulator of B lymphocytes in health and disease, as it controls B cell lineage commitment (8), development (9) and identity (15), and functions as a prominent tumor suppressor (15, 17) or oncoprotein (50) in B cell leukemia. Here, we have studied the function of Pax5 in late B lymphopoiesis and demonstrated that the innate-like B-1 and MZ B cells stringently depended on this transcription factor, whereas FO B cells tolerated the loss of Pax5 but had a considerably shortened half-life. Immunization with T cell-dependent antigens revealed an essential role for Pax5 in the initiation and maintenance of GC B cell development and the subsequent generation of memory B cells and plasma cells. At the molecular level, Pax5 controlled PI3K signaling, which promoted the survival and proliferation of B cells upon BCR or TLR stimulation. The severe impairment of BCR and TLR signaling in response to adaptive and innate signals likely explains the loss of all mature B cell types in the absence of Pax5, which identifieed Pax5 as a central regulator of B cell immunity.
Here, we showed Pax5 was essential for efficient CSR to IgG1 by activating Iγ1 germline transcription by binding to and inducing open chromatin at the Iγ1 promoter and an enhancer located downstream of the Cγ1 gene. Pax5 was previously implicated in controlling CSR by binding to and activating the Aicda gene (51, 52), which encodes the essential CSR regulator AID (24). Unexpectedly, our genetic analysis revealed that Aicda expression in response to anti-CD40 plus IL-4 stimulation was strongly increased in Pax5-deficient FO B cells. While the transcription factor FOXO1 promotes CSR by activating Aicda expression (53, 54), its activity is negatively regulated through phosphorylation by the AKT kinase (30) (Fig. S4A). Hence, Pax5 has two opposing effects on the regulation of CSR to IgG1. The loss of Pax5 directly interfered with Iγ1 promoter activity and indirectly led to increased Aicda expression by inhibiting PI3K signaling leading to enhanced FOXO1 activity. Notably, IgG1+ B cells were decreased rather than increased in the absence of Pax5, suggesting that the direct transcriptional effect on the Iγ1 promoter is dominant over the indirect posttranslational control leading to enhanced Aicda expression.
In contrast to the activation of CD40 and IL-4 receptor pathways, Pax5-deficient FO B cells failed to proliferate upon anti-IgM plus IL-4, LPS or CpG stimulation, thus revealing a severe impairment of BCR and TLR signaling in the absence of Pax5. Phosphorylation of signal transducers and analysis of intracellular calcium mobilization in response to BCR engagement demonstrated that Pax5-deficient FO B cells could efficiently activate the SYK-BLNK-PLCγ2-dependent calcium signaling pathway, which is essential for controlling B cell differentiation and cell fate decisions (31). Likewise, TLR signaling in Pax5-deficient FO B cells efficiently induced NF-κB activation via the MyD88 pathway, which stimulates pro-inflammatory cytokine gene expression (55). In contrast, signaling along the PI3K-AKT pathway, which is essential for cell proliferation and survival (30, 56), was severely impaired in Pax5-deficient FO B cells upon BCR or TLR stimulation. This common defect may explain the failure of Pax5-deficient FO B cells to proliferate in response to BCR or TLR signals. Consistent with this conclusion, B cells lacking the regulatory PI3K subunit p85a (Pik3r1) fail to proliferate in response to anti-IgM plus IL-4 treatment, but undergo normal proliferation upon anti-CD40 and IL-4 stimulation (57, 58), similar to our finding with Pax5-deficient FO B cells.
B-1a and MZ B cells stringently depend on PI3K signaling as they are lost in mice lacking the following positive regulators of this pathway: CD19 (59–61), the regulatory PI3K subunit p85a (Pik3r1) (57, 58), the catalytic PI3K subunit p110δ (Pik3cd) (32, 62) and both AKT1 and AKT2 (63). Moreover, Akt1 −/− Akt2 −/− mice have reduced numbers of splenic FO B cells and lose B-1a and MZ B cells (63), which resembles the Pax5 mutant phenotype. In contrast, conditional loss of PTEN, a negative regulator of PI3K signaling, leads to hyperactivation of the pathway and thus to increased B-1a and MZ B cell development at the expense of FO B cell generation (48, 49, 62). Notably, the PI3K-AKT-FOXO1 signaling axis is essential for controlling the survival of mature B cells in response to ‘tonic’ BCR signaling (64). Consequently, the impaired PI3K signaling in the absence of Pax5 likely explains the loss of B-1 and MZ B cells as well as the shortened half-life of FO B cells.
While Pax5 did not transcriptionally regulate genes implicated in PI3K signaling including Pten, Pax5-deficient FO B cells exhibited significantly elevated expression of the PTEN protein, which antagonizes PI3K signaling by converting PI(3,4,5)P3 to PI(4,5)P2 (30). Expression of the PTEN protein is under posttranscriptional control by different miRNAs that target the Pten 3’UTR (34). Our observation that the PTEN protein levels were equally high in Dicer- and Pax5-deficient FO B cells strongly suggests a role for Pax5 in the control of miRNA expression, although genes implicated in miRNA processing or function are not deregulated by Pax5. By small-RNA-seq, we identified four distinct Pten-targeting miRNA families (miR-29, miR-26, miR-19 and miR-141), whose abundance was deregulated in Pax5-deficient versus Pax5-expressing FO B cells. Interestingly, the four miRNAs of the miR-29 family were not only abundantly expressed in FO B cells, but were also present at a 2.9- to 1.3-fold lower abundance in Pax5-deficient FO B cells relative to control B cells in the spleen and lymph nodes. However, loss of miR-29a and miR-29b-1 led only to a small increase of PTEN expression and concomitant decrease of AKT phosphorylation in FO B cells of miR-29a/b-1 −/− mice. A significant loss of mature B cells and plasma cells was recently reported upon deletion of both the miR-29a/b-1 and miR-29c/b-2 loci, although deletion of all four miR-29 genes still resulted in a modest increase of PTEN expression (65). It is thus conceivable that the cumulative loss of other Pax5-regulated Pten-targeting miRNAs may have contributed to the high PTEN expression observed in Pax5-deficient FO B cells. While we detected robust Pax5 binding at the miR-29 loci, future experiments will be required to address whether Pax5 directly controls expression of the primary miRNA transcripts or indirectly determines the abundance of the mature miRNAs.
Additional loss of PTEN restored AKT phosphorylation and cell survival in Pten,Pax5 double-mutant FO B cells in response to BCR and TLR stimulation. However, in vitro B cell proliferation was not rescued, suggesting that Pax5 may, in addition to its effect on PI3K signaling, activate genes implicated in cell cycle entry or progression, as exemplified by the immediate-early genes Myc, Egr2 and Egr3, which are essential for antigen-induced B cell proliferation (66, 67). Nevertheless, FO B cell numbers were restored in the lymph nodes, and MZ B cells were rescued and properly located in the marginal zone of lymphoid follicles in the spleen of Pten,Pax5 double-mutant mice. As the B cell-specific loss of PTEN alone already increases MZ B cell development (48, 49, 62), two different scenarios might explain the MZ B cell rescue in Pten,Pax5 double-mutant mice. First, in addition to controlling PI3K signaling, Pax5 may fulfill another essential function to promote MZ B cell development. In this case, PTEN loss should only rescue the majority of the residual MZ B cells that fail to delete Pax5 in Cd23-Cre Pax5 fl/fl mice. Second, the main function of Pax5 in MZ B cell generation may be to restrain PTEN protein expression to promote PI3K signaling. In this scenario, the Pax5-deleted MZ B cells, which are prone to die, should be rescued by PTEN loss, thus giving rise to increased MZ B cell development. Our observation that the majority of rescued MZ B cells in Pten,Pax5 double-mutant mice have lost Pax5 strongly indicates that the loss of PTEN can rescue the Pax5 mutant phenotype, as Pax5 and PTEN fulfill opposing roles in controlling PI3K signaling and MZ B cell development.
Our study furthermore demonstrated that Pax5 was essential for the initiation and maintenance of GC B cell differentiation. At the start of this developmental process, quiescent FO B cells are activated by antigen-mediated stimulation of the BCR and by interaction with T helper cells, which triggers a cascade of events leading to the formation of GC B cells (68). As BCR activation starts this process, it is conceivable that quiescent FO B cells lacking Pax5 may fail to initiate efficient B cell activation due to impaired PI3K signaling. However, GC B cells are generated upon B cell-specific loss of PTEN (48, 53) or the catalytic PI3K subunit p110δ (69). Hence, PI3K signaling does not play an essential role in the generation of GC B cells, which likely explains why GC B cells cannot be rescued upon additional loss of PTEN in Pten,Pax5 double-mutant mice. Pax5 must therefore regulate GC B cell differentiation through another important, yet unknown, function possibly involving Pax5-mediated control of cell proliferation.
In summary, although the pleiotropic transcription factor Pax5 controls many genes (12) (this study) and may thus contribute to the regulation of diverse aspects of B cells, we have shown here that the posttranscriptional downregulation of PTEN expression is an important function of Pax5 in mature B cells. As a consequence, Pax5 facilitates PI3K signaling leading to the differentiation and survival of distinct mature B cell types, which jointly cooperate to provide humoral immunity.
Supplementary Material
One Sentence Summary.
Pax5 controls the generation, proliferation and survival of all mature B cells by promoting PI3K signaling via PTEN downregulation.
Acknowledgements
We thank Karin Aumayr’s team for flow-cytometric sorting, Thomas Lendl for image analysis and Andreas Sommer’s team at the Vienna BioCenter Core Facilities for Illumina sequencing.
Funding
This research was funded by Boehringer Ingelheim, the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreements 291740-LymphoControl and 740349-PlasmaCellControl), the Austrian Research Promotion Agency (Early Stage Grant ‘Molecular Control’ FFG-878286) and the Human Frontier Science Program fellowship LT00427/2013 (to L. Calderón).
Footnotes
Author contributions: L.Calderón performed all cell proliferation and survival experiments, the rescue experiments with Pten,Pax5 mutant mice, the respective phosflow and immunofluorescence experiments, the analysis of miR29-a/b-1 mutant mice; K.S. performed the phosflow analysis of Pax5-deficient FO B cells, intracellular PTEN staining, RNA-seq analysis of anti-IgM-stimulated B cells and contributed to small-RNA-sequencing; S.G.M. performed the flow-cytometric analysis of mature B cell types, immunization experiments, H3K4me2 ChIP and RNA-seq analysis of anti-CD40 plus IL-4 activated B cells; A.S. analyzed the Cd19-Cre Pax5 fl/− mice; Q.S. performed the BrdU experiments, ELISPOT assays and histological spleen analysis of immunized mice; T.S. provided advice on flow-cytometric and immunofluorescence analyses; C.A. performed the small-RNA-sequencing; H.T. generated the DHS-seq data; A.E. did the Pax5 Bio-ChIP-seq analysis of activated B cells; M.M. analyzed the bone marrow plasma cells; M.F. and M.J. performed the bioinformatic analysis of mRNA-seq, small-RNA-seq and ChIP-seq data; L.Cochella provided advice on microRNA experiments; A.L. provided the miR29-a/b-1 −/− mouse; M.B. supervised the study and wrote the manuscript with L.Calderón.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability
RNA-seq, ChIP-seq, DHS-seq and small-RNA-seq data, which were generated for this study (Table S6), are available at the Gene Expression Omnibus (GEO) repository under the accession number GSE103260. All data needed to evaluate the conclusions in the paper are present in the paper of the Supplementary Material.
References and Notes
- 1.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 2.Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- 3.Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol. 2013;13:118–132. doi: 10.1038/nri3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 5.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 6.Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 7.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 8.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 9.Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14:779–790. doi: 10.1016/s1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- 10.Fuxa M, Busslinger M. Reporter gene insertions reveal a strictly B lymphoid-specific expression pattern of Pax5 in support of its B cell identity function. J Immunol. 2007;178:3031–3037. doi: 10.4049/jimmunol.178.5.3031. [DOI] [PubMed] [Google Scholar]
- 11.Delogu A, et al. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Revilla-i-Domingo R, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–3146. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schebesta A, et al. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 14.McManus S, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–2404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 16.Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–113. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- 17.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 18.Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Urbánek P, Wang Z-Q, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 20.Rolink AG, Andersson J, Melchers F. Characterization of immature B cells by a novel monoclonal antibody, by turnover and by mitogen reactivity. Eur J Immunol. 1998;28:3738–3748. doi: 10.1002/(SICI)1521-4141(199811)28:11<3738::AID-IMMU3738>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 21.Ogilvy S, et al. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci USA. 1999;96:14943–14948. doi: 10.1073/pnas.96.26.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family, in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 24.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 25.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medvedovic J, et al. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity. 2013;39:229–244. doi: 10.1016/j.immuni.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawlings DJ, Schwartz MA, Jackson SW, Meyer-Bahlburg A. Integration of B cell responses through Toll-like receptors and antigen receptors. Nat Rev Immunol. 2012;12:282–294. doi: 10.1038/nri3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otipoby KL, et al. The B-cell antigen receptor integrates adaptive and innate immune signals. Proc Natl Acad Sci USA. 2015;112:12145–12150. doi: 10.1073/pnas.1516428112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schweighoffer E, Nys J, Vanes L, Smithers N, Tybulewicz VLJ. TLR4 signals in B lymphocytes are transduced via the B cell antigen receptor and SYK. J Exp Med. 2017;214:1269–1280. doi: 10.1084/jem.20161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharenberg AM, Humphries LA, Rawlings DJ. Calcium signalling and cell-fate choice in B cells. Nat Rev Immunol. 2007;7:778–789. doi: 10.1038/nri2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okkenhaug K, et al. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, et al. The physiologic role of CD19 cytoplasmic tyrosines. Immunity. 2002;17:501–514. doi: 10.1016/s1074-7613(02)00426-0. [DOI] [PubMed] [Google Scholar]
- 34.Sellars E, Gabra M, Salmena L. The complex landscape of PTEN mRNA regulation. Cold Spring Harb Perspect Med. 2020;10:a036236. doi: 10.1101/cshperspect.a036236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dexheimer PJ, Cochella L. MicroRNAs: from mechanism to organism. Front Cell Dev Biol. 2020;8:409. doi: 10.3389/fcell.2020.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reimao-Pinto MM, et al. Uridylation of RNA hairpins by Tailor confines the emergence of microRNAs in Drosophila . Mol Cell. 2015;59:203–216. doi: 10.1016/j.molcel.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huse JT, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tumaneng K, et al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou H, et al. MicroRNA-29A/PTEN pathway modulates neurite outgrowth in PC12 cells. Neuroscience. 2015;291:289–300. doi: 10.1016/j.neuroscience.2015.01.055. [DOI] [PubMed] [Google Scholar]
- 42.Lin X, et al. MicroRNA-29 regulates high-glucose-induced apoptosis in human retinal pigment epithelial cells through PTEN. In Vitro Cell Dev Biol Anim. 2016;52:419–426. doi: 10.1007/s11626-015-9990-z. [DOI] [PubMed] [Google Scholar]
- 43.Ji J, et al. Mitochondria-related miR-141-3p contributes to mitochondrial dysfunction in HFD-induced obesity by inhibiting PTEN. Sci Rep. 2015;5:16262. doi: 10.1038/srep16262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin Y-Y, et al. Involvement of microRNA-141-3p in 5-fluorouracil and oxaliplatin chemo-resistance in esophageal cancer cells via regulation of PTEN. Mol Cell Biochem. 2016;422:161–170. doi: 10.1007/s11010-016-2816-9. [DOI] [PubMed] [Google Scholar]
- 45.Li X-Y, et al. Triptolide restores autophagy to alleviate diabetic renal fibrosis through the miR-141-3p/PTEN/Akt/mTOR pathway. Mol Ther Nucleic Acids. 2017;9:48–56. doi: 10.1016/j.omtn.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papadopoulou AS, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-a receptor. Nat Immunol. 2011;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dooley J, et al. The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes. 2016;65:53–61. doi: 10.2337/db15-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anzelon AN, Wu H, Rickert RC. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat Immunol. 2003;4:287–294. doi: 10.1038/ni892. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki A, et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med. 2003;197:657–667. doi: 10.1084/jem.20021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smeenk L, et al. Molecular role of the PAX5-ETV6 oncoprotein in promoting B-cell acute lymphoblastic leukemia. EMBO J. 2017;36:718–735. doi: 10.15252/embj.201695495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonda H, et al. The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran TH, et al. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat Immunol. 2010;11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- 53.Omori SA, et al. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 54.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 56.Deane JA, Fruman DA. Phosphoinositide 3-kinase: diverse roles in immune cell activation. Annu Rev Immunol. 2004;22:563–598. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- 57.Fruman DA, et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85a. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki H, et al. Xid-like immunodeficiency in mice with disruption of the p85a subunit of phosphoinositide 3-kinase. Science. 1999;283:390–392. doi: 10.1126/science.283.5400.390. [DOI] [PubMed] [Google Scholar]
- 59.Rickert RC, Rajewsky K, Roes J. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature. 1995;376:352–355. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
- 60.Engel P, et al. Abnormal B lymphocyte development, activation, and differentiation in mice that lack or overexpress the CD19 signal transduction molecule. Immunity. 1995;3:39–50. doi: 10.1016/1074-7613(95)90157-4. [DOI] [PubMed] [Google Scholar]
- 61.Martin F, Kearney JF. Positive selection from newly formed to marginal zone B cells depends on the rate of clonal production, CD19, and btk . Immunity. 2000;12:39–49. doi: 10.1016/s1074-7613(00)80157-0. [DOI] [PubMed] [Google Scholar]
- 62.Janas ML, et al. The effect of deleting p110d on the phenotype and function of PTEN-deficient B cells. J Immunol. 2008;180:739–746. doi: 10.4049/jimmunol.180.2.739. [DOI] [PubMed] [Google Scholar]
- 63.Calamito M, et al. Akt1 and Akt2 promote peripheral B-cell maturation and survival. Blood. 2010;115:4043–4050. doi: 10.1182/blood-2009-09-241638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–686. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hines MJ, et al. miR-29 sustains B cell survival and controls terminal differentiation via regulation of PI3K signaling. Cell reports. 2020;33:108436. doi: 10.1016/j.celrep.2020.108436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 67.Li S, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37:685–696. doi: 10.1016/j.immuni.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 69.Rolf J, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185:4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki A, et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14:523–534. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- 71.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith KG, Light A, Nossal GJ, Tarlinton DM. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 1997;16:2996–3006. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minnich M, et al. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol. 2016;17:331–343. doi: 10.1038/ni.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alberti C, et al. Cell-type specific sequencing of microRNAs from complex animal tissues. Nat Methods. 2018;15:283–289. doi: 10.1038/nmeth.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adams B, et al. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 81.öhner M, et al. Molecular functions of the transcription factors E2A and E2-2 in controlling germinal center B cell and plasma cell development. J Exp Med. 2016;213:1201–1221. doi: 10.1084/jem.20152002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thijs G, et al. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Computational Biol. 2002;9:447–464. doi: 10.1089/10665270252935566. [DOI] [PubMed] [Google Scholar]
- 84.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends in genetics : TIG. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 85.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luo W, et al. The AKT kinase signaling network is rewired by PTEN to control proximal BCR signaling in germinal center B cells. Nat Immunol. 2019;20:736–746. doi: 10.1038/s41590-019-0376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq, ChIP-seq, DHS-seq and small-RNA-seq data, which were generated for this study (Table S6), are available at the Gene Expression Omnibus (GEO) repository under the accession number GSE103260. All data needed to evaluate the conclusions in the paper are present in the paper of the Supplementary Material.